Cell Cycle/Checkpoint
Cell Cycle
Cells undergo a complex cycle of growth and division that is referred to as the cell cycle. The cell cycle consists of four phases, G1 (GAP 1), S (synthesis), G2 (GAP 2) and M (mitosis). DNA replication occurs during S phase. When cells stop dividing temporarily or indefinitely, they enter a quiescent state called G0.
Targets for Cell Cycle/Checkpoint
- ATM/ATR(26)
- Aurora Kinase(47)
- Cdc42(4)
- Cdc7(4)
- Chk(16)
- c-Myc(20)
- CRM1(8)
- Cyclin-Dependent Kinases(91)
- E1 enzyme(1)
- G-quadruplex(14)
- Haspin(7)
- HMTase(1)
- Kinesin(26)
- Ksp(6)
- Microtubule/Tubulin(243)
- Mps1(15)
- Mitotic(11)
- RAD51(18)
- ROCK(71)
- Rho(13)
- PERK(11)
- PLK(37)
- PTEN(8)
- Wee1(7)
- PAK(21)
- Arp2/3 Complex(8)
- Dynamin(12)
- ECM & Adhesion Molecules(40)
- Cholesterol Metabolism(3)
- Endomembrane System & Vesicular Trafficking(26)
- G1(38)
- G2/M(26)
- G2/S(10)
- Genotoxic Stress(18)
- Inositol Phosphates(18)
- Proteolysis(99)
- Cytoskeleton & Motor Proteins(53)
- Cellular Chaperones(8)
Products for Cell Cycle/Checkpoint
- Cat.No. Product Name Information
-
GC11905
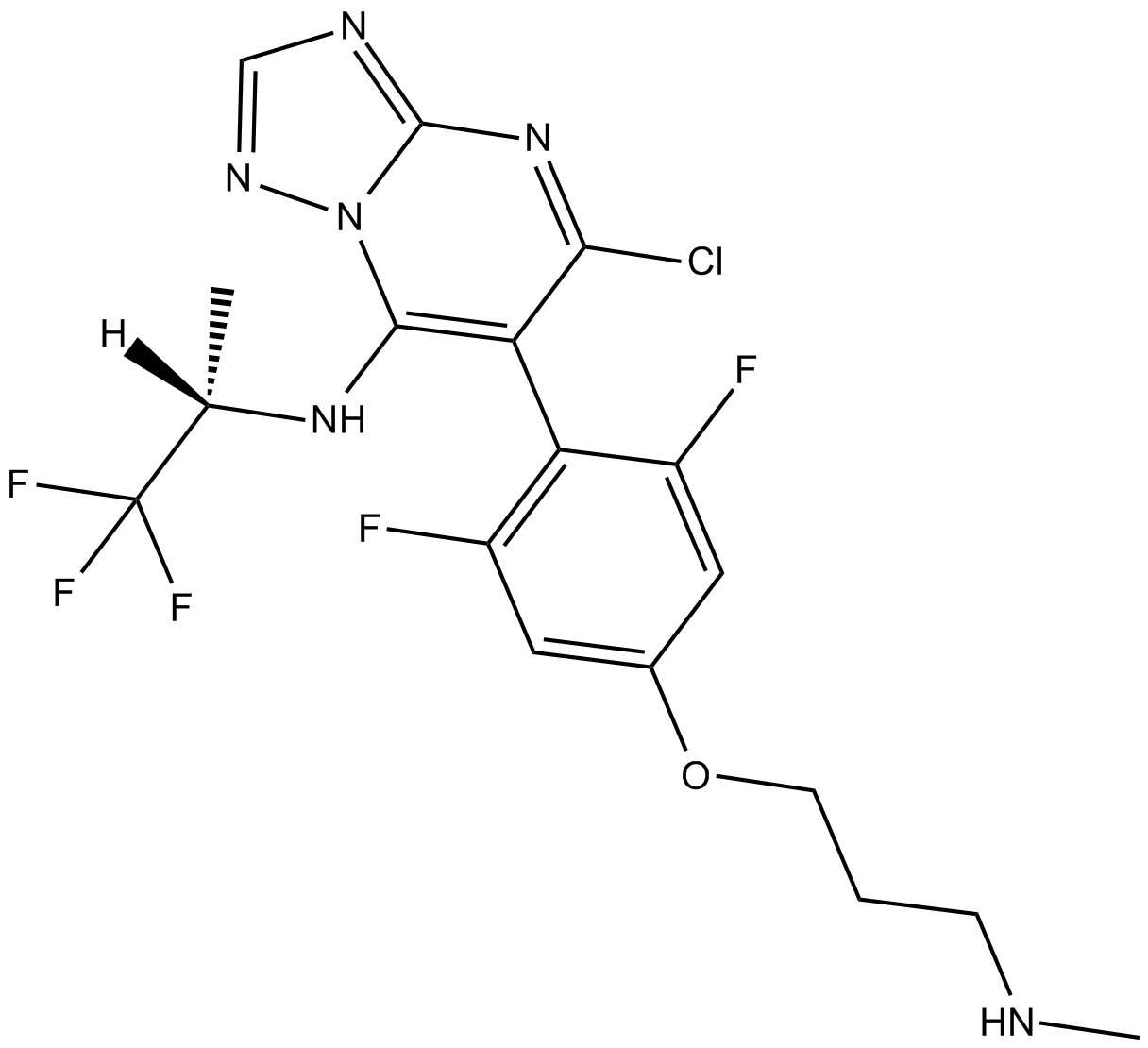
Cevipabulin
Anti-microtubule agent
-
GC35659
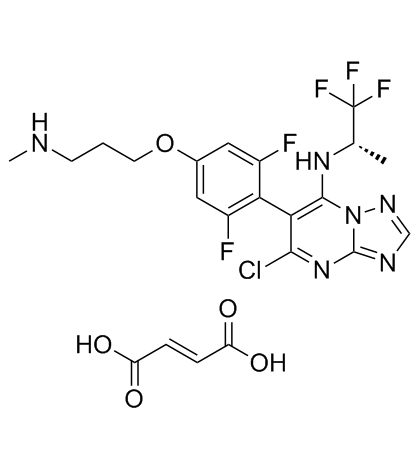
Cevipabulin fumarate
Cevipabulin fumarate (TTI-237 fumarate) is an oral, microtubule-active, antitumor compound and inhibits the binding of [3H]NSC 49842 to tubulin, with an IC50 of 18-40 nM for cytotoxicity in human tumor cell line.
-
GC13606
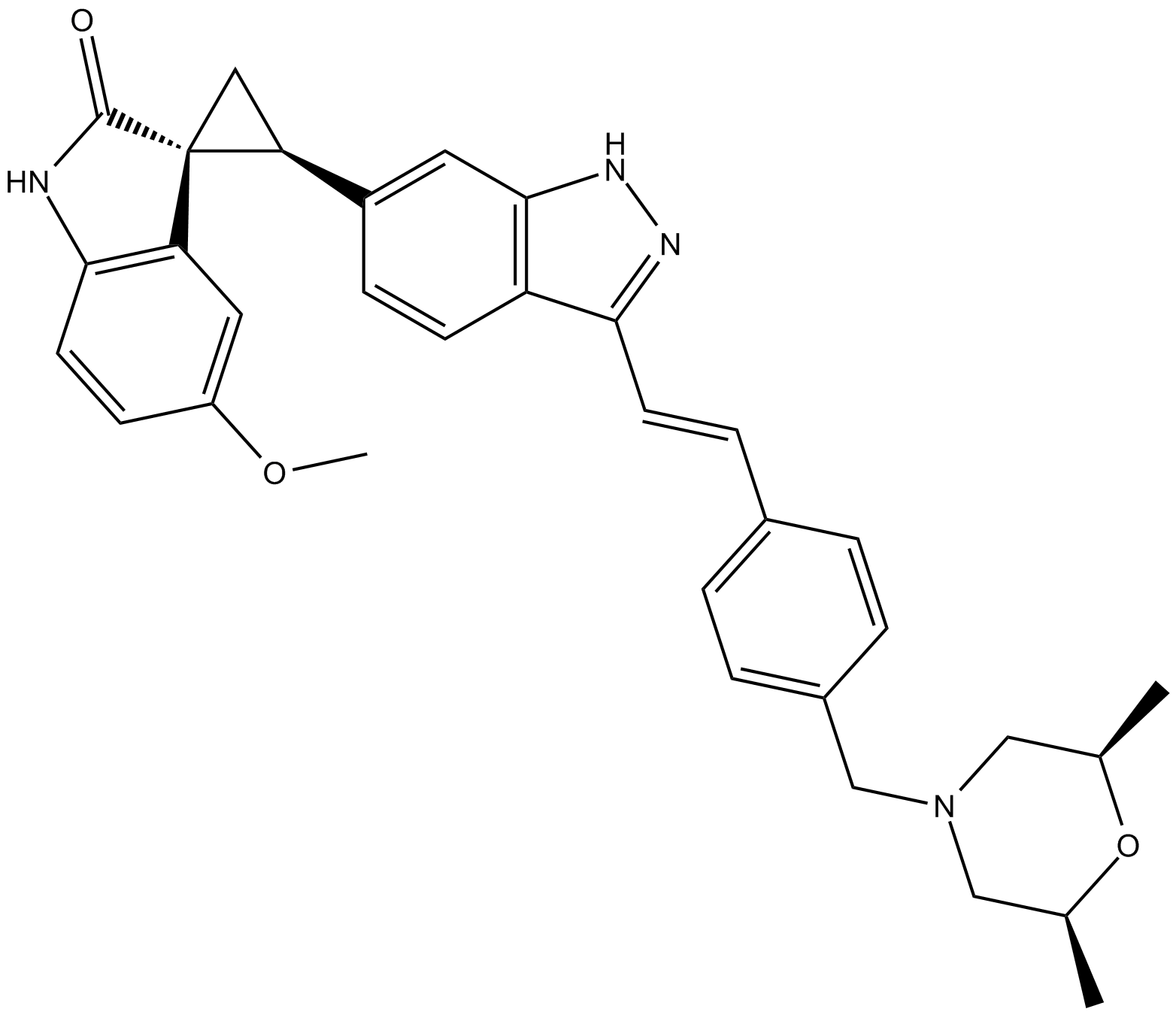
CFI-400945
orally available, selective inhibitor of polo-like kinase 4 (PLK4)
-
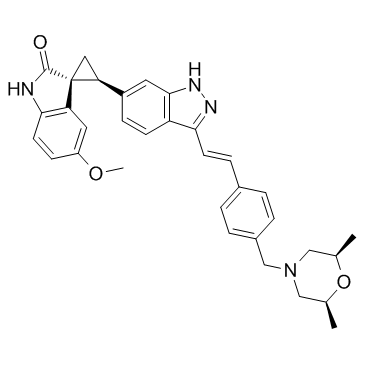
GC35660
CFI-400945 free base
-
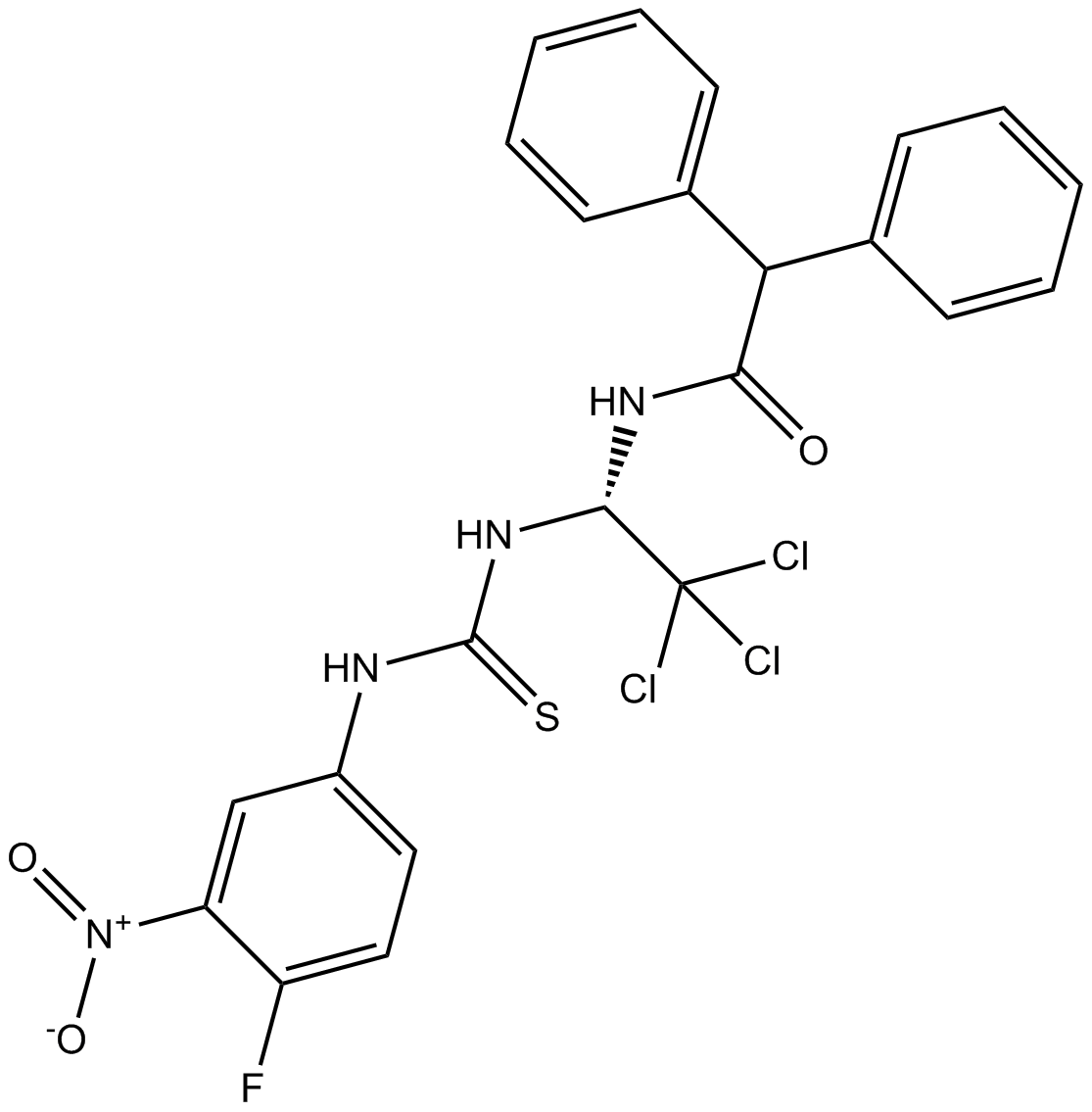
GC14526
CGK733
ATM/ATR inhibitor,potent and selective
-
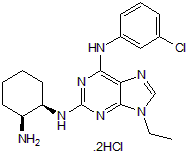
GC50175
CGP 74514 dihydrochloride
Potent cdk1 inhibitor
-
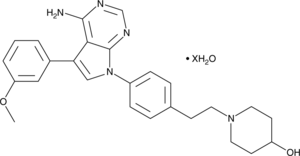
GC48992
CGP 77675 (hydrate)
An inhibitor of Src family kinases
-
GC14650
CGP60474
A CDK inhibitor
-
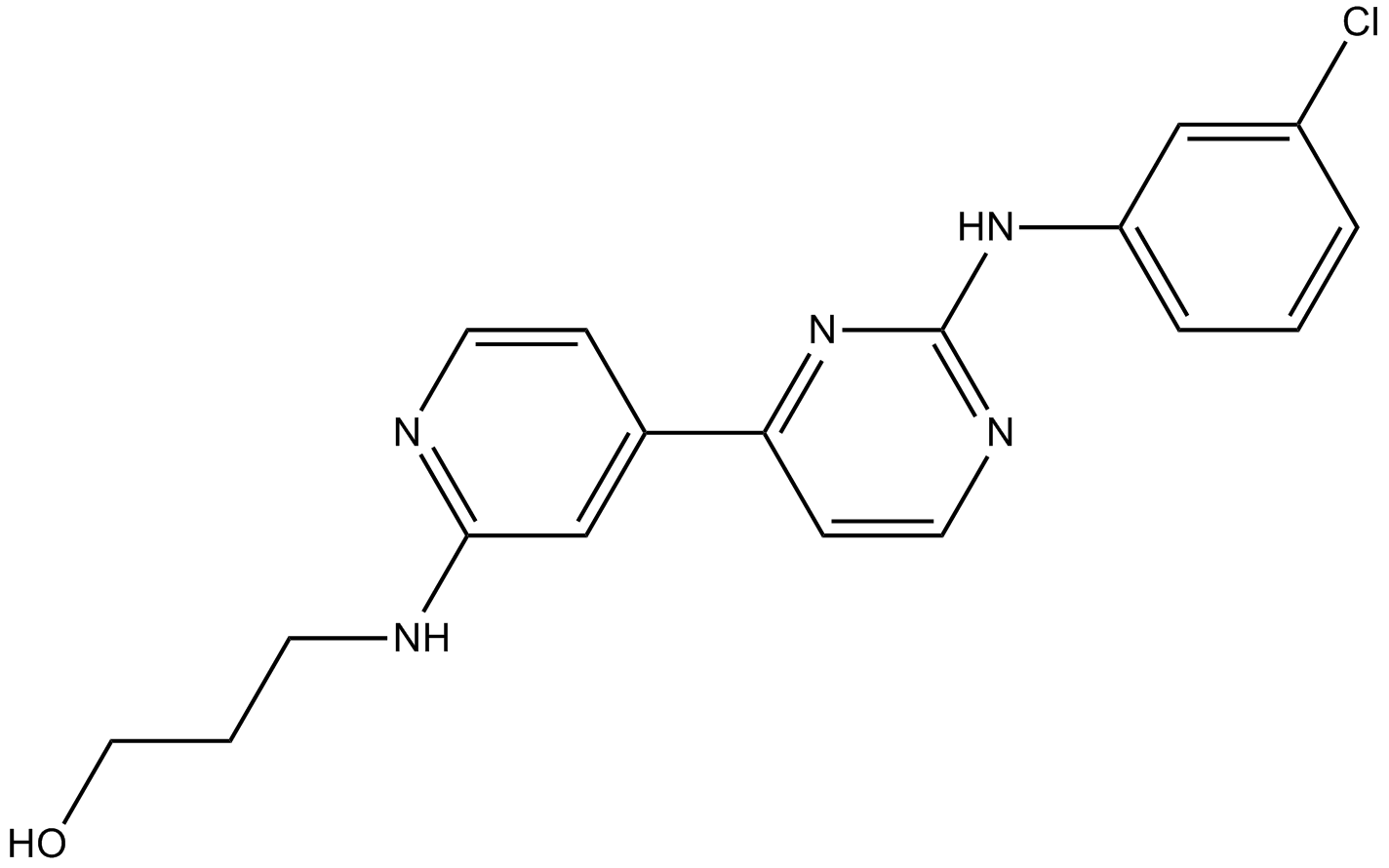
GC15739
CHIR-124
Chk1 inhibitor,novel and potent
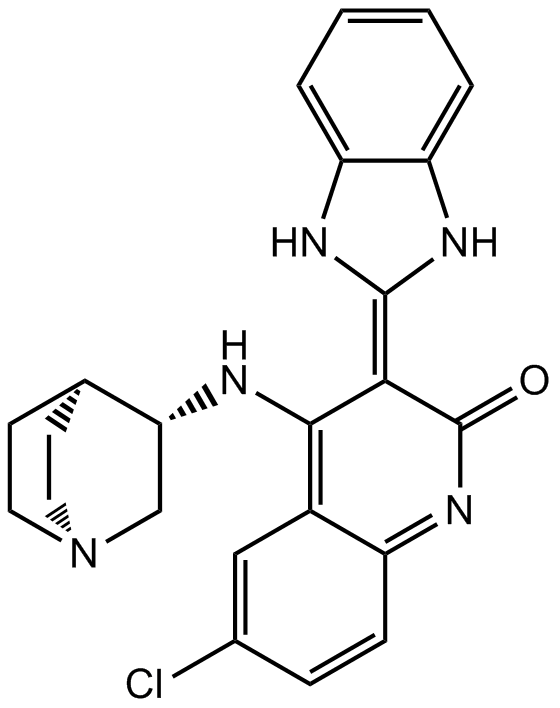
-
GC43239
Chk2 Inhibitor
Chk2 Inhibitor (compound 1) is a potent and selective inhibitor of checkpoint kinase 2 (Chk2), with IC50s of 13.5 nM and 220.4 nM for Chk2 and Chk1, respectively. Chk2 Inhibitor can elicit a strong ataxia telangiectasia mutated (ATM)-dependent Chk2-mediated radioprotection effect.
-
GC47079
Chloramine-T (hydrate)
A common reagent
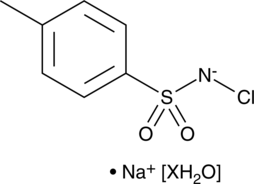
-
GC43250
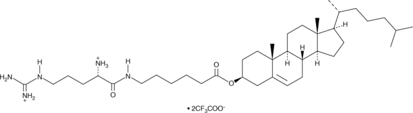
Cho-Arg (trifluoroacetate salt)
Cho-Arg is a steroid-based cationic lipid that contains a cholesterol skeleton coupled to an L-arginine head group.
-
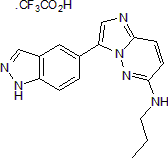
GC50145
CHR 6494 trifluoroacetate
CHR 6494 trifluoroacetate is a potent inhibitor of haspin, with an IC50 of 2 nM. CHR 6494 trifluoroacetate inhibits histone H3T3 phosphorylation. CHR 6494 trifluoroacetate induces the apoptosis of cancer cells, including melanoma and breast cancer. CHR 6494 trifluoroacetate can be used in the research of cancer.
-
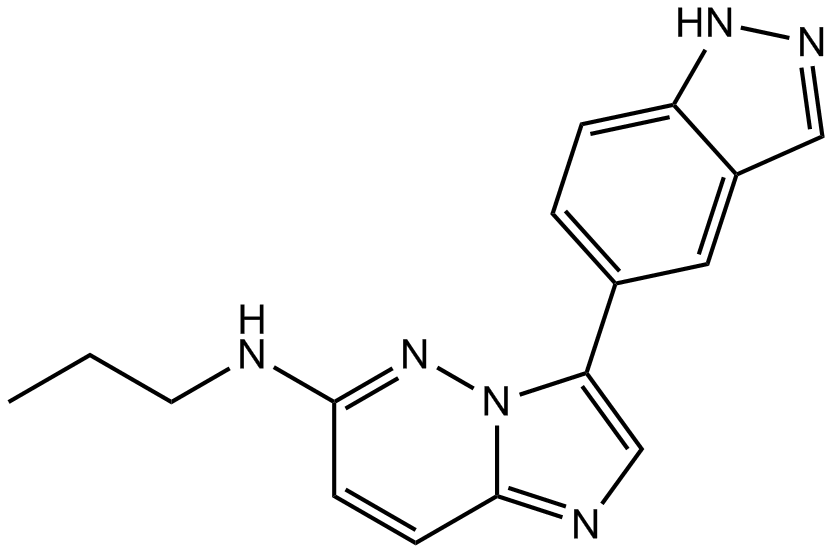
GC17254
CHR-6494
A selective Haspin protein kinase inhibitor
-
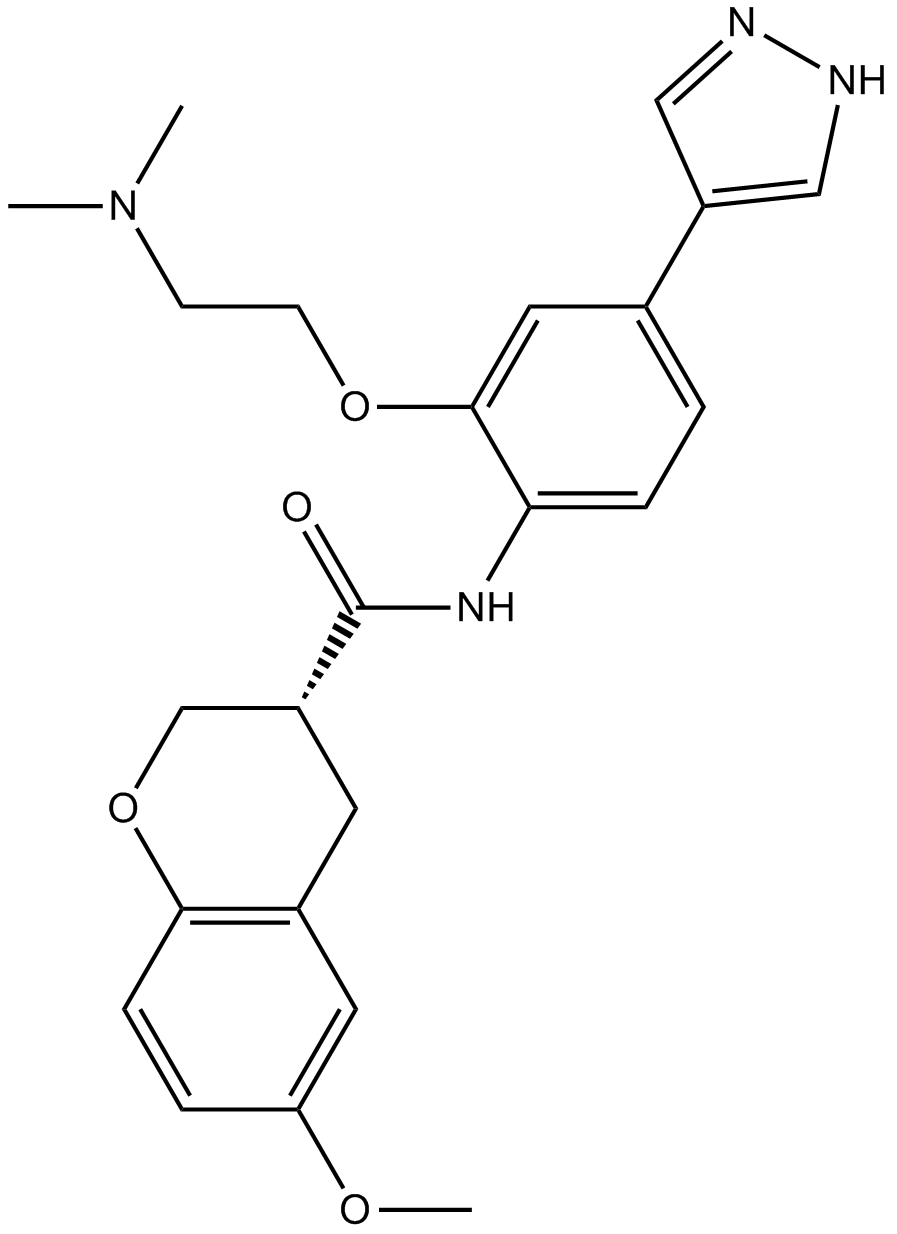
GC16097
Chroman 1
A ROCK2 inhibitor
-
GC49334
Chroman 1 (hydrochloride hydrate)
A ROCK2 inhibitor
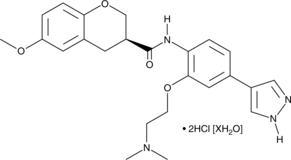
-
GC43265
Chromomycin A2
Chromomycin A2 is an aureolic acid that has been found in several marine actinomycetes and has antibacterial and anticancer activities.
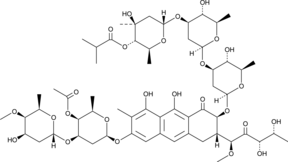
-
GC52360
Chymotrypsin Substrate I, Colorimetric (trifluoroacetate salt)
A colorimetric chymotrypsin substrate
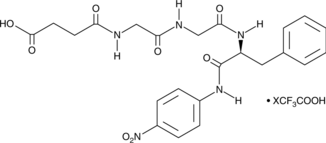
-
GC12352
cis-trismethoxy Resveratrol
Cis-trismethoxy resveratrol is a potent anti-mitotic reagent.Cis-trismethoxy resveratrol inhibits tubulin polymerization with an IC50 value of 4 μM.
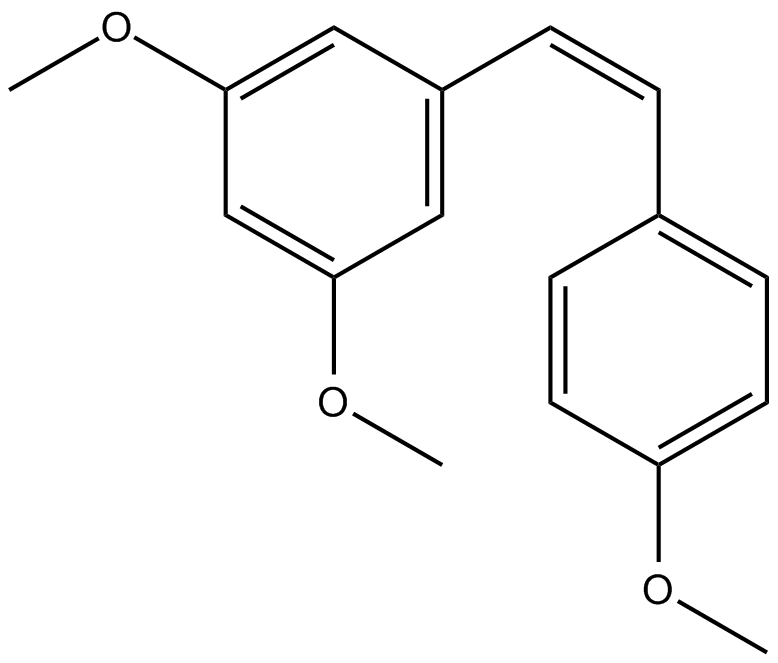
-
GC52367
Citrullinated Vimentin (G146R) (R144 + R146) (139-159)-biotin Peptide
A biotinylated and citrullinated mutant vimentin peptide

-
GC52370
Citrullinated Vimentin (R144) (139-159)-biotin Peptide
A biotinylated and citrullinated vimentin peptide

-
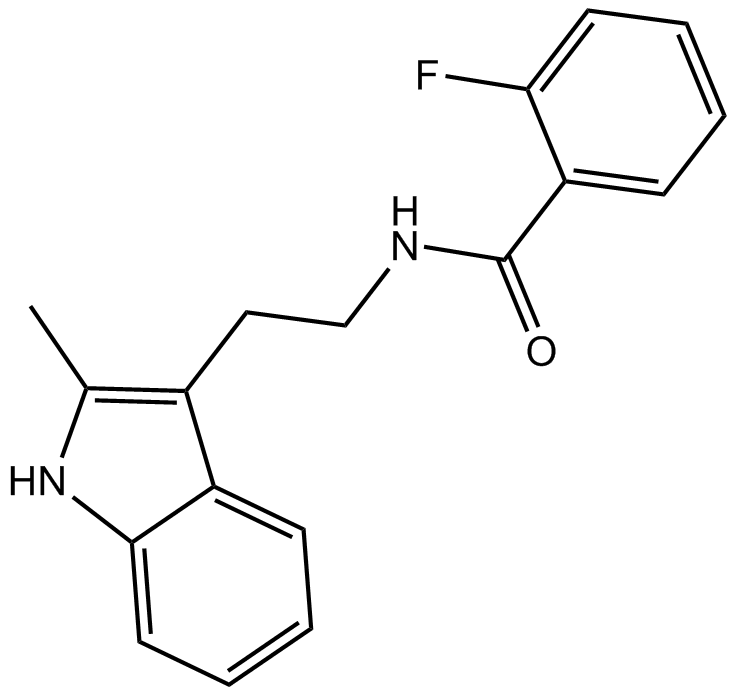
GC14060
CK 666
Arp2/3 complex inhibitor that inhibits actin polymerization
-
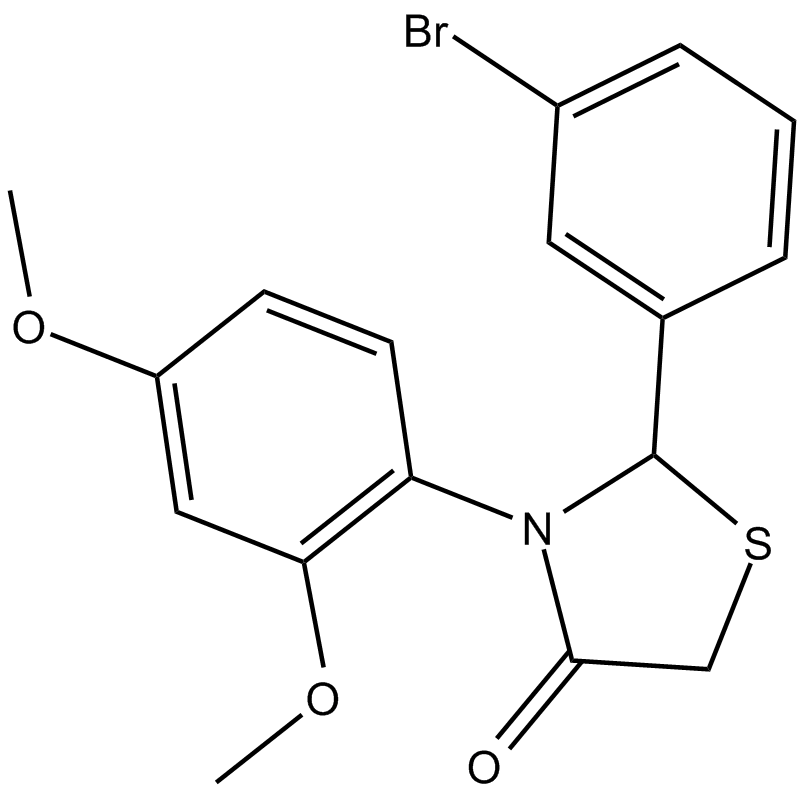
GC15076
CK 869
actin-related protein 2/3 (Arp2/3) complex inhibitor
-
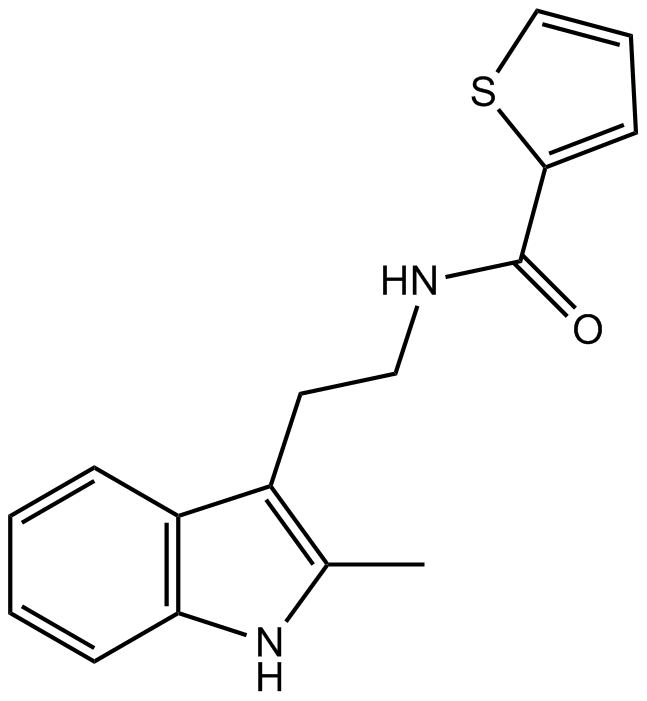
GC15894
CK-636
Arp2/3 complex inhibitor
-
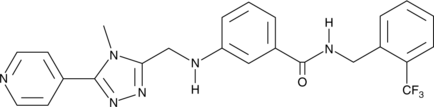
GC43286
CMPD101
A GRK2 and GRK3 inhibitor
-
GC49345
Coelenterazine hcp
A synthetic bioluminescent luciferin
-
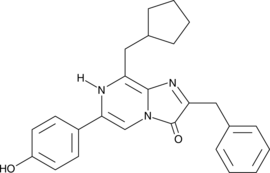
GC40664
Colcemid
Colcemid is a cytoskeletal inhibitor that induces mitotic arrest in the G2/M phase or meiotic arrest in the vesicle rupture (GVBD) phase in mammalian cells or oocytes, respectively.
-
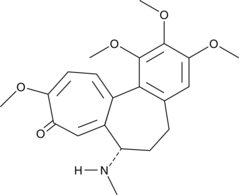
GC13261
Colchicine
An inhibitor of microtubule polymerization
-
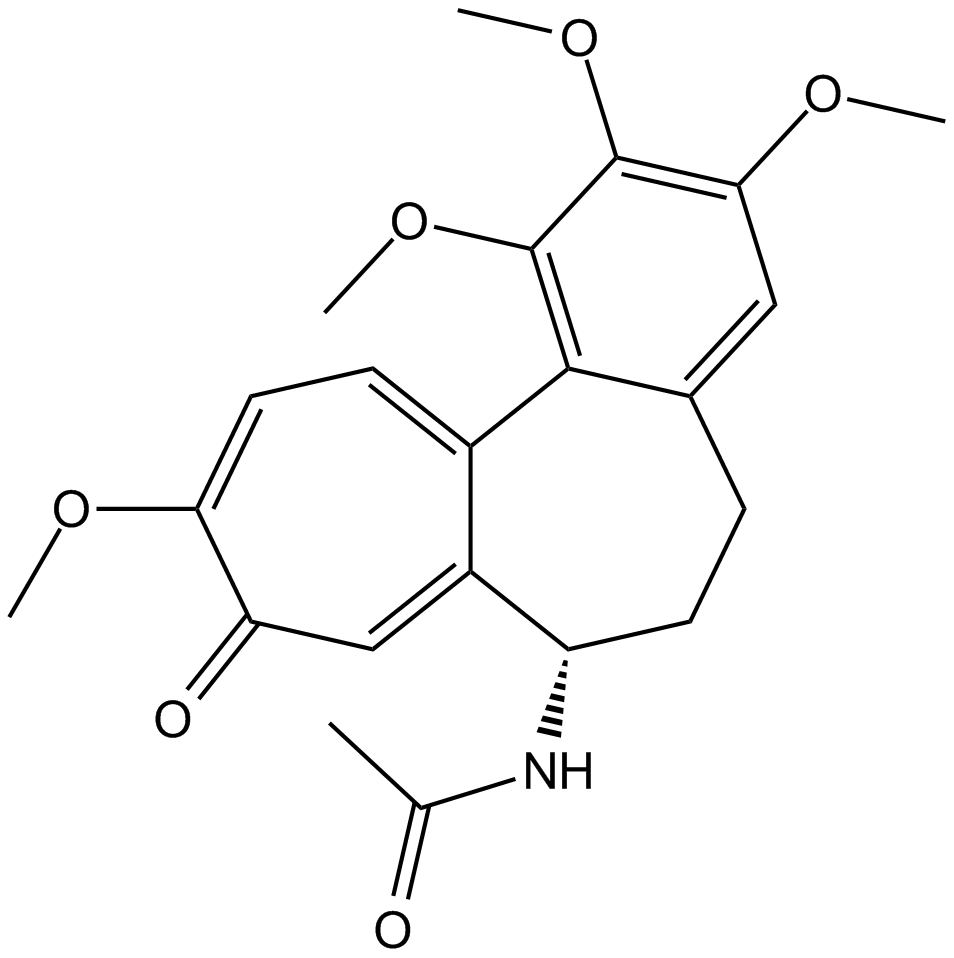
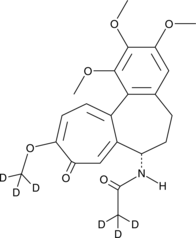
GC47117
Colchicine-d6
An internal standard for the quantification of colchicine
-
GC43300
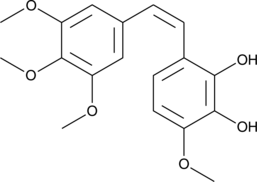
Combretastatin A1
Combretastatin A1 is a cis-stilbene originally isolated from C.
-
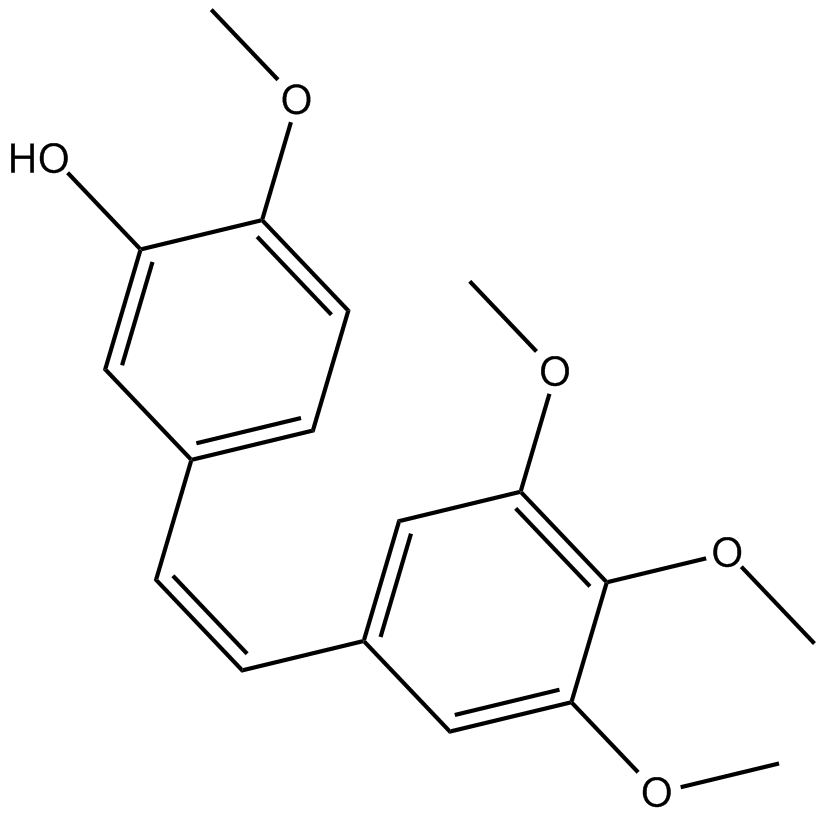
GC17631
Combretastatin A4
tubulin polymerization inhibitor
-
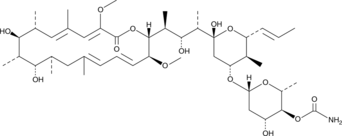
GC43307
Concanamycin B
Concanamycin B is a macrolide antibiotic that selectively inhibits vacuolar type H+-ATPases, also known as V-ATPases (IC50 = 5 nM).
-
GC43315
Corynecin IV
Corynecin IV is a chloramphenicol-like bacterial metabolite originally isolated from Corynebacterium.
-
GC40663
Corynecin V
Corynecin V is a chloramphenicol-like bacterial metabolite originally isolated from Corynebacterium.
-
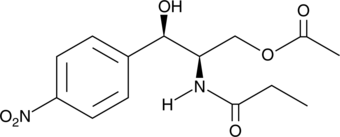
GC63333
Cotosudil
-
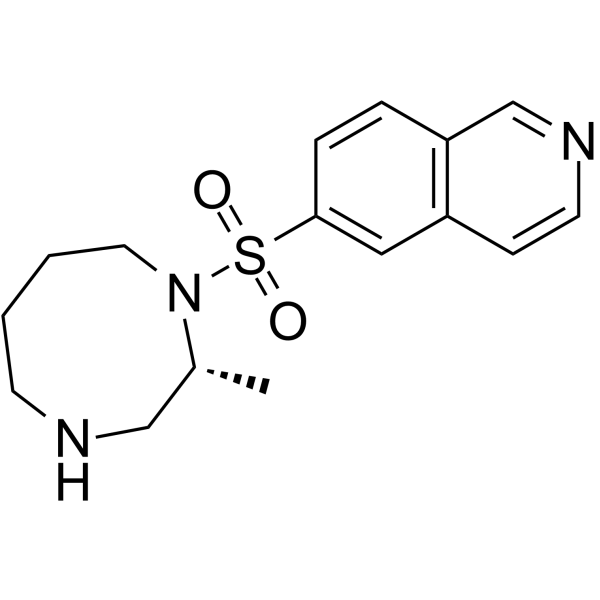
GC16489
CP-466722
ATM inhibitor,potent and reversible
-
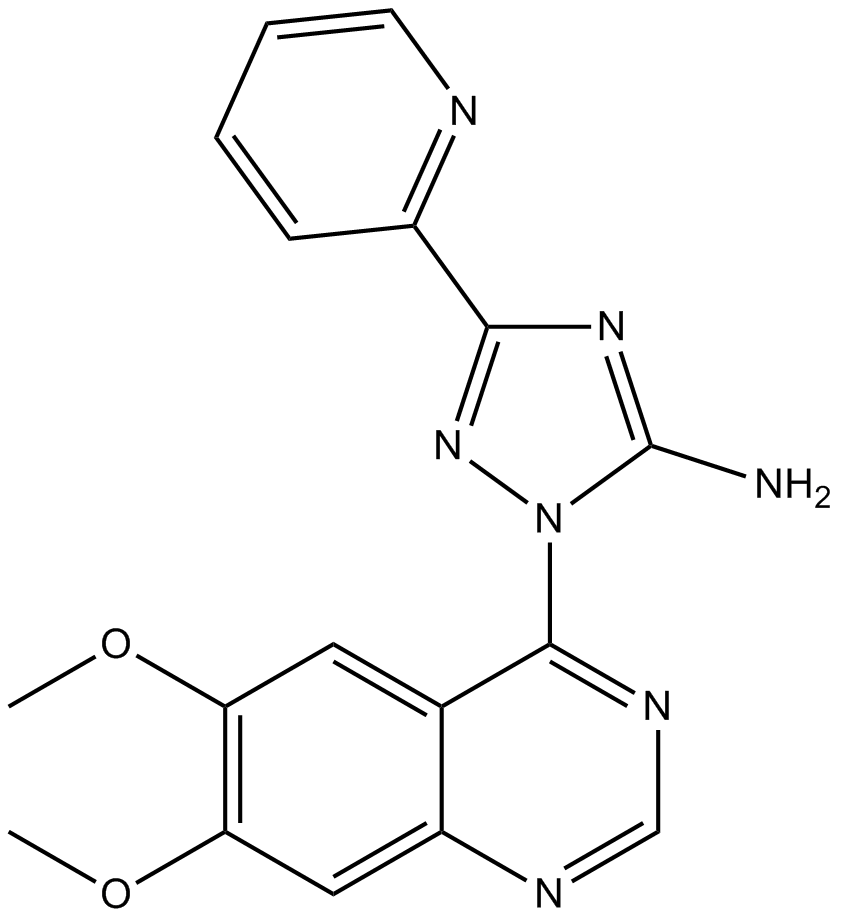
GC19112
Crolibulin
Crolibulin is a small molecule tubulin polymerization inhibitor.
-
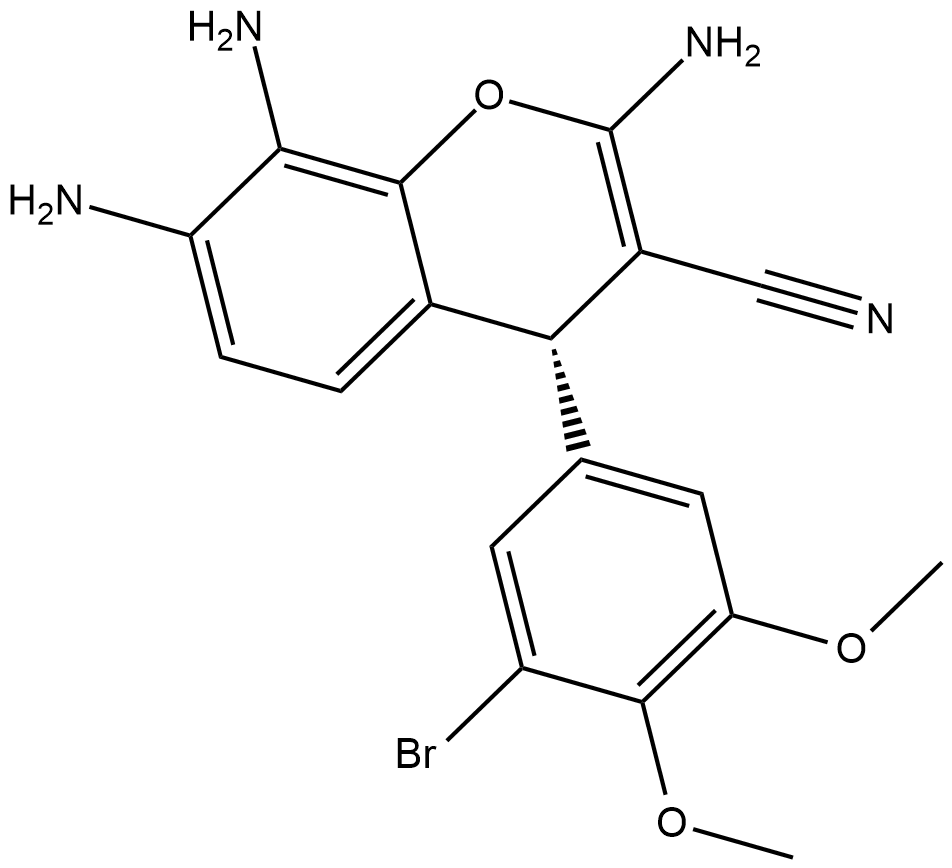
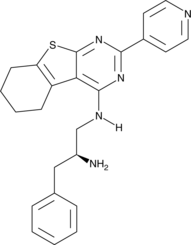
GC45414
CRT0066854
-
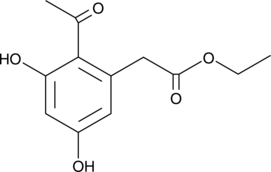
GC40482
Curvulin
Curvulin is a phytotoxin first isolated from several species of the mold Curvularia.
-
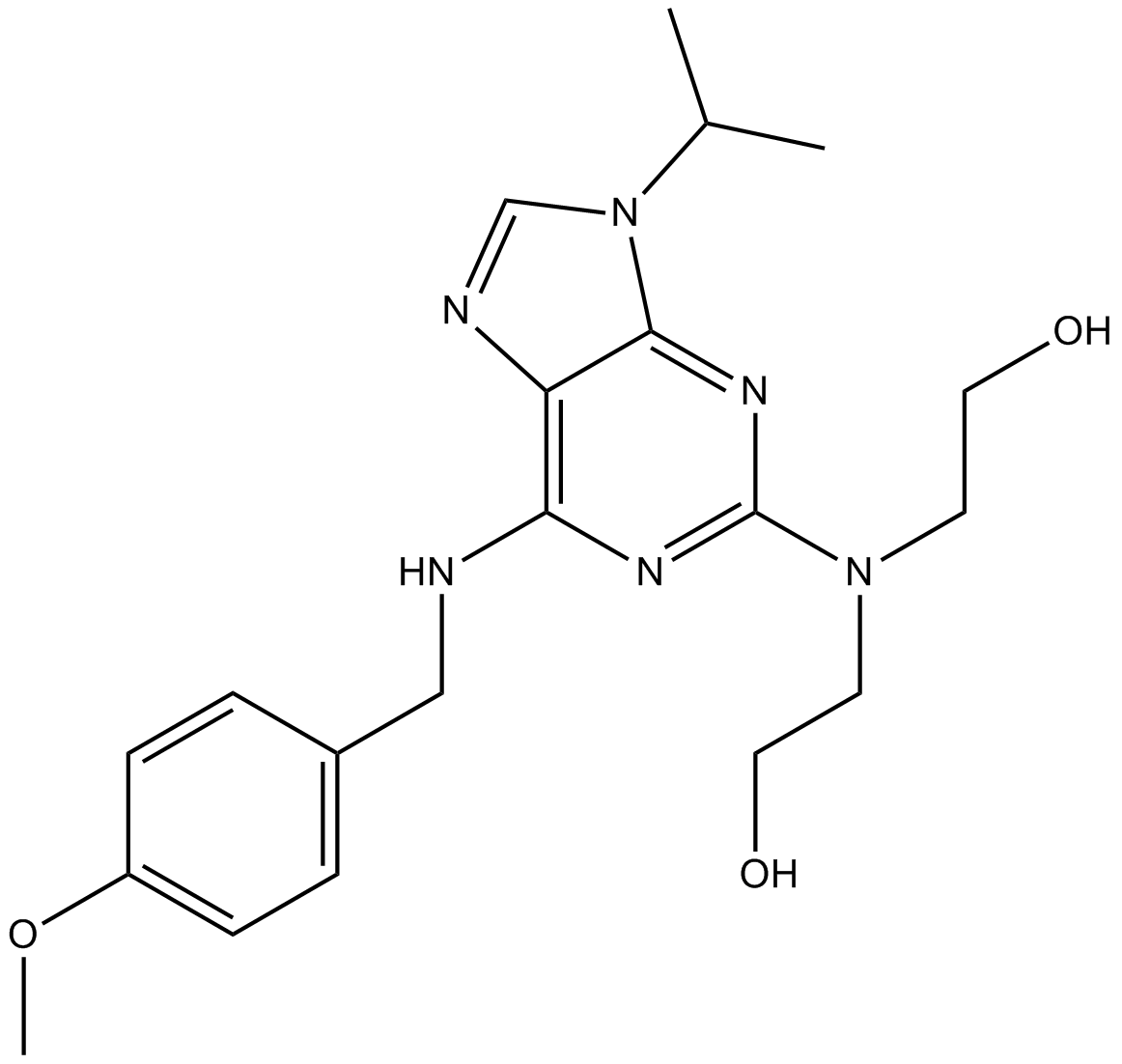
GC18028
CVT-313
A Cdk2 inhibitor
-
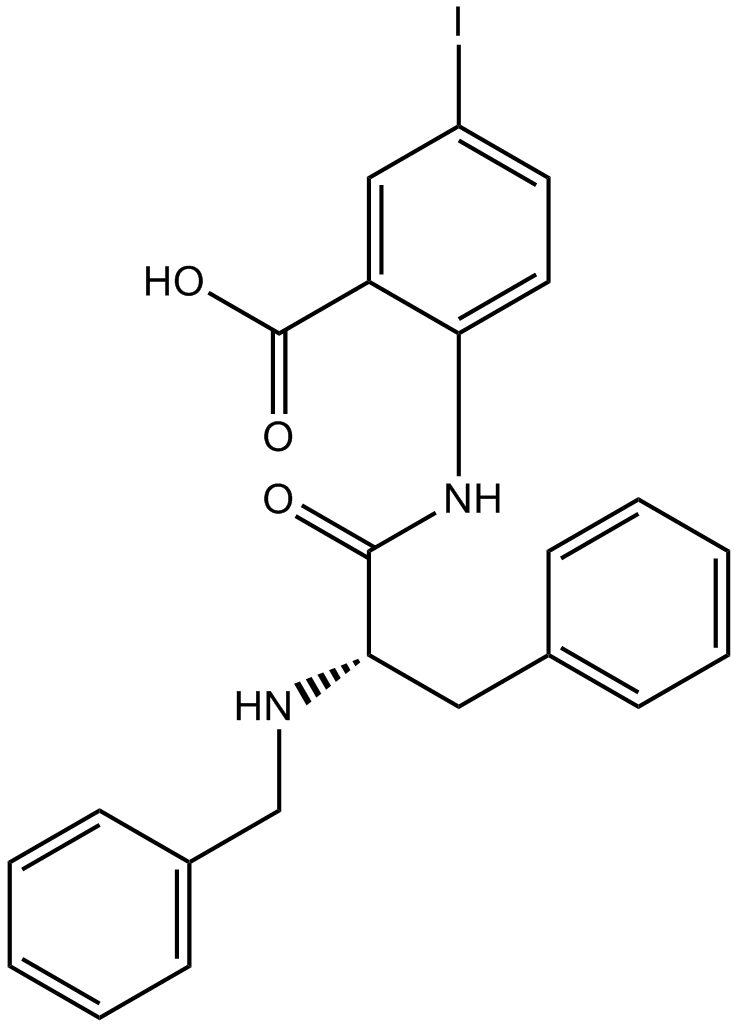
GC11252
CW069
allosteric inhibitor of HSET, selective
-
GC13268
Cyclapolin 9
PLK1 inhibitor
-
GC49709
cyclo(RGDyC) (trifluoroacetate salt)
A cyclic pentapeptide
-
GC49716
Cyclo(RGDyK) (trifluoroacetate salt)
A cyclic peptide ligand of αVβ3 integrin
-
GC43346
Cyclopamine-KAAD
Cyclopamine-KAAD, a hedgehog signaling inhibitor, is a smoothened antagonist.

-
GC43351
Cylindrospermopsin
Cylindrospermopsin, a tricyclic uracil derivative, is a cyanobacterial toxin that was first discovered in an algal bloom contaminating a local drinking supply on Palm Island in Queensland, Australia after an outbreak of a mysterious disease.
-
GC45877
CYM 5478
An S1P2 agonist
-
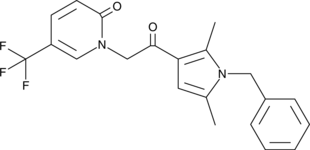
GC35789
Cys-mcMMAD
Cys-mcMMAD is a drug-linker conjugate for ADC. MMAD is a potent tubulin inhibitor.
-
GC43354
Cysmethynil
Post-translational protein prenylation is a 3-step process that occurs at the C-terminus of a number of proteins involved in cell growth control and oncogenesis.

-
GC11383
CYT997 (Lexibulin)
CYT997 (Lexibulin) (CYT-997) is a potent and orally active tubulin polymerisation inhibitor with IC50s of 10-100 nM in cancer cell lines; with potent cytotoxic and vascular disrupting activity in vitro and in vivo. CYT997 (Lexibulin) induces cell apoptosis and induces mitochondrial ROS generation in GC cells.
-
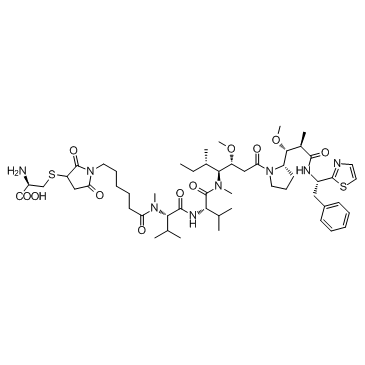
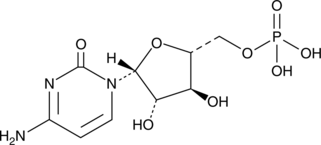
GC49863
Cytarabine 5′-monophosphate
An active metabolite of cytarabine
-
GC32890
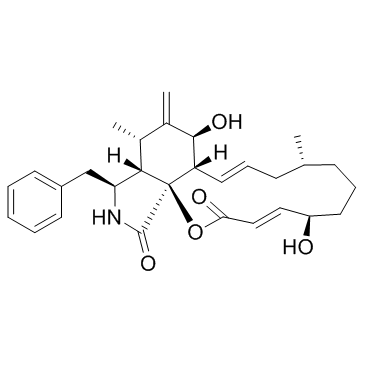
Cytochalasin B (Phomin)
Cytochalasin B is a cyto-permeable mycotoxin, and it is isolated from an ascomycete fungus belonging to the Phoma genus[1-2].
-
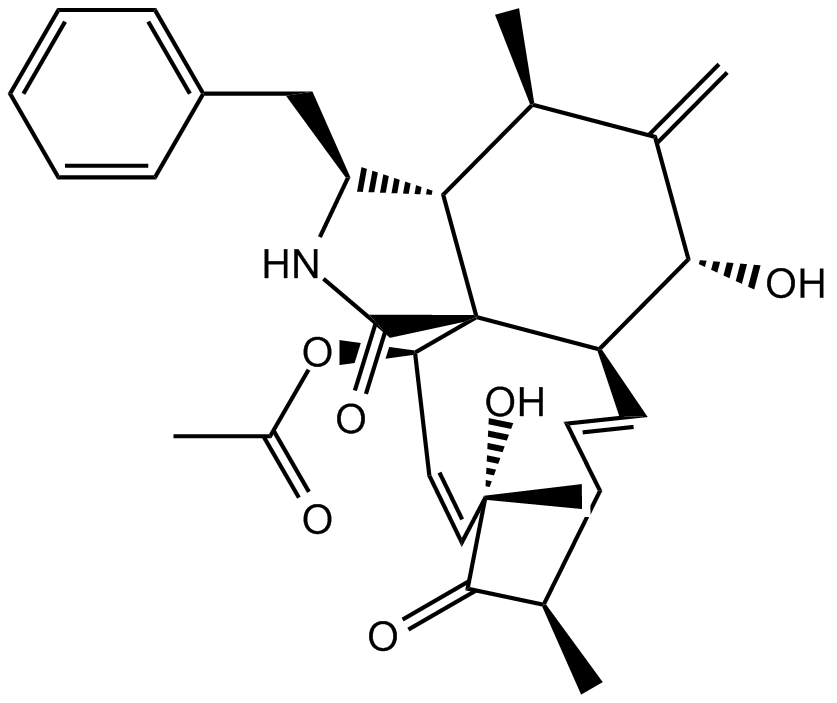
GC13440
Cytochalasin D
An inhibitor of actin polymerization, selective
-
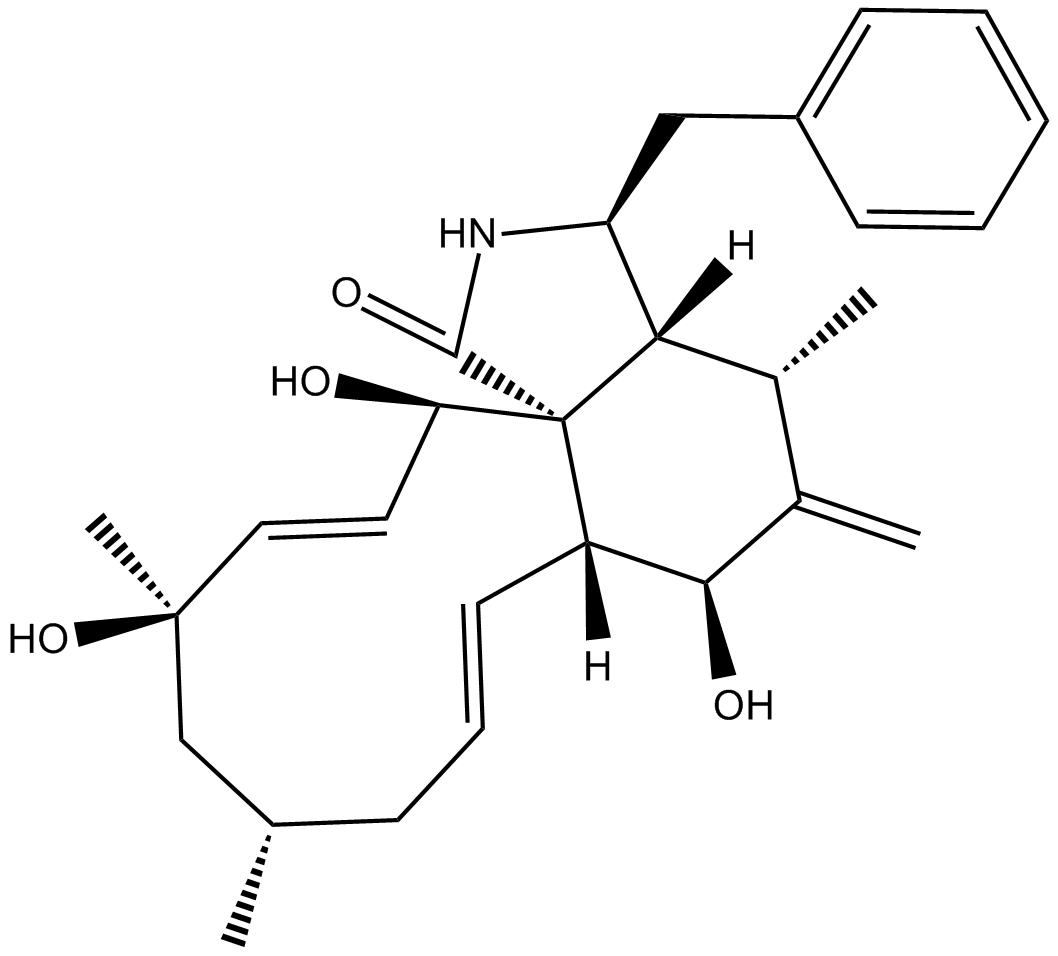
GC10870
Cytochalasin J
alters mitotic spindle microtubule organization and kinetochore structure
-
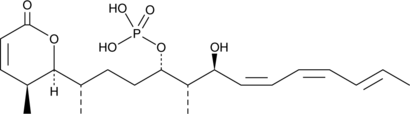
GC43360
Cytostatin
Cytostatin is a natural antitumor inhibitor of cell adhesion to extracellular matrix, blocking adhesion of B16 melanoma cells to laminin and collagen type IV in vitro (IC50s = 1.3 and 1.4 μg/ml, respectively) and B16 cells metastatic activity in mice.
-
GC43361
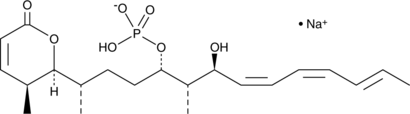
Cytostatin (sodium salt)
Cytostatin is a natural antitumor inhibitor of cell adhesion to extracellular matrix, blocking adhesion of B16 melanoma cells to laminin and collagen type IV in vitro (IC50s = 1.3 and 1.4 μg/ml, respectively) and B16 cells metastatic activity in mice.
-
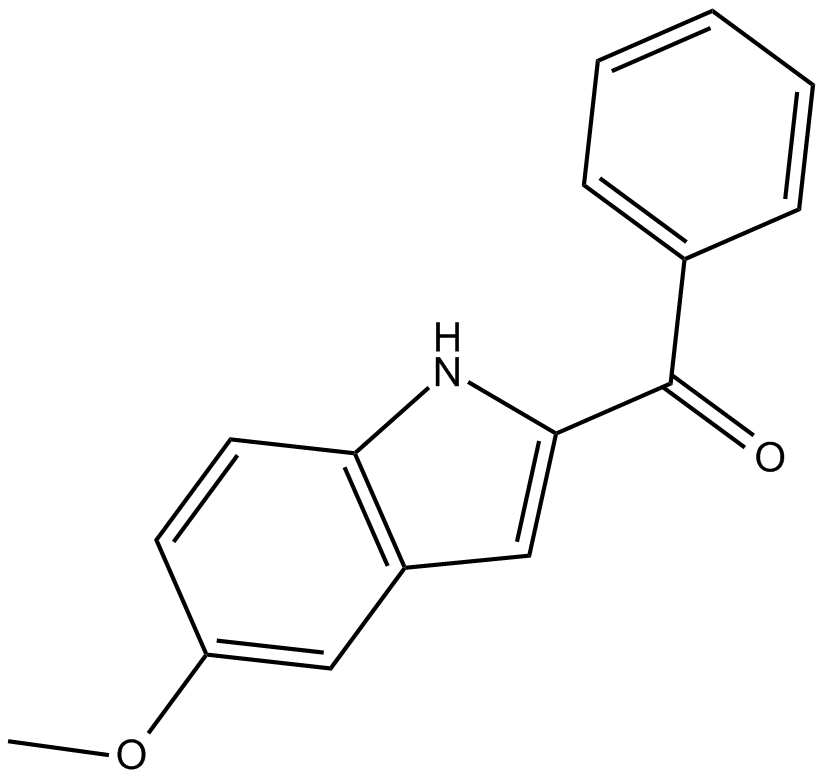
GC12384
D-64131
An inhibitor of tubulin polymerization
-
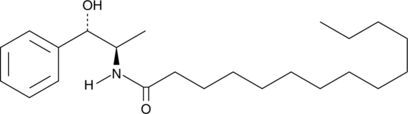
GC40296
D-erythro-MAPP
D-erythro-MAPP is a derivative of ceramide and an inhibitor of alkaline ceramidase (Ki = 2-13 μM; IC50 = 1-5 μM).
-
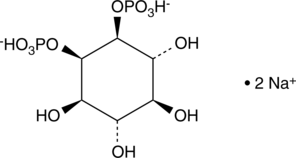
GC43513
D-myo-Inositol-1,2-diphosphate (sodium salt)
Ins(1,2)P2 (sodium salt) is one of the many inositol phosphate (InsP) isomers that could act as small, soluble second messengers in the transmission of cellular signals.
-
GC43516
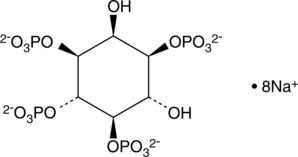
D-myo-Inositol-1,3,4,5-tetraphosphate (sodium salt)
D-myo-Inositol-1,3,4,5-tetraphosphate (Ins(1,3,4,5)-P4) is formed by the phosphorylation of Ins(1,4,5)P3 by inositol 1,4,5-triphosphate 3-kinase.
-
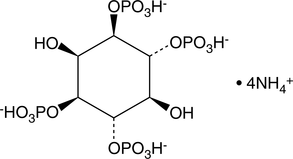
GC43517
D-myo-Inositol-1,3,4,6-tetraphosphate (ammonium salt)
The inositol phosphates (IPs) are a family of molecules produced by altering the phosphorylation status of each of the six carbons on the cyclic inositol structure.
-
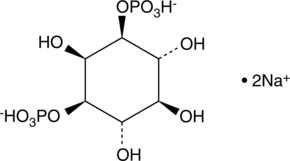
GC43520
D-myo-Inositol-1,3-diphosphate (sodium salt)
D-myo-Inositol-1,3-phosphate (Ins(1,3)P) is a member of the inositol phosphate (InsP) molecular family that play critical roles as small, soluble second messengers in the transmission of cellular signals.
-
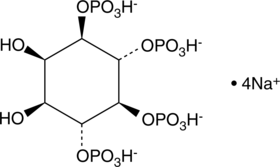
GC43521
D-myo-Inositol-1,4,5,6-tetraphosphate (sodium salt)
D-myo-Inositol-1,4,5,6-tetrahosphate (sodium salt) (Ins(1,4,5,6)-P4) is one of several different inositol oligophosphate isomers implicated in signal transduction.
-
GC43522
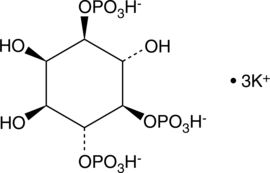
D-myo-Inositol-1,4,5-triphosphate (potassium salt)
D-myo-Inositol-1,4,5-triphosphate (Ins(1,4,5)P3) is a second messenger produced in cells by phospholipase C (PLC) mediated hydrolysis of phosphatidyl inositol-4,5-biphosphate.
-
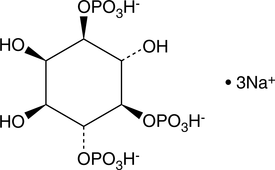
GC43523
D-myo-Inositol-1,4,5-triphosphate (sodium salt)
Primary intracellular IP3 receptor agonist
-
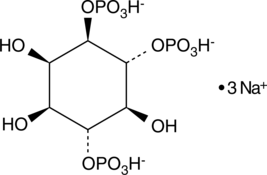
GC43524
D-myo-Inositol-1,4,6-triphosphate (sodium salt)
D-myo-Inositol-1,4,6-phosphate (Ins(1,4,6)-P3) is a member of the inositol phosphate (InsP) family that play critical roles as small, soluble second messengers in the transmission of cellular signals.
-
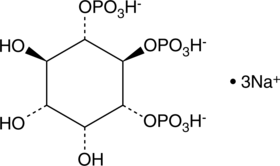
GC43526
D-myo-Inositol-1,5,6-triphosphate (sodium salt)
The inositol phosphates are a family of mono- to poly-phosphorylated compounds that act as messengers, regulating cellular functions including cell cycling, apoptosis, differentiation, andmotility.
-
GC43540
D-myo-Inositol-4-phosphate (ammonium salt)
D-myo-Inositol-4-phosphate (Ins(4)P1) is a member of the inositol phosphate (InsP) molecular family that play critical roles as small, soluble second messengers in the transmission of cellular signals.
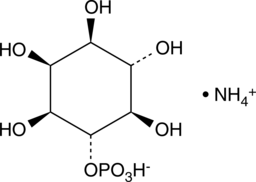
-
GC35797
D8-MMAD
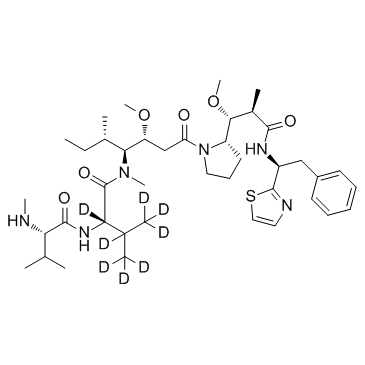
-
GC35798
D8-MMAF
D8-MMAF hydrochloride is a deuterated form of MMAF hydrochloride. MMAF Hydrochloride, a potent tubulin polymerization inhibitor, is used as a antitumor agent and a cytotoxic component of antibody-drug conjugates (ADCs).
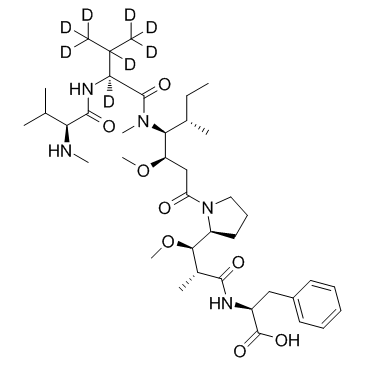
-
GC35799
D8-MMAF hydrochloride
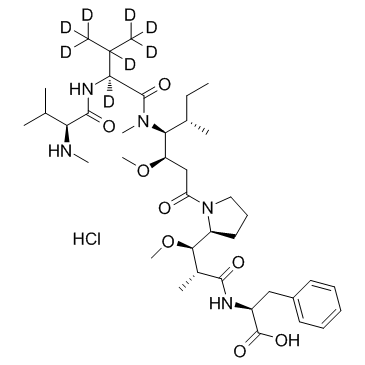
-
GC41305
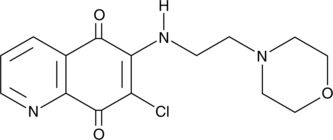
DA-3003-2
Cdc25 dual-specific protein tyrosine phosphatases are important for cell cycle progression and often overexpressed in cancers.
-
GC18421
Dabcyl-YVADAPV-EDANS
Dabcyl-YVADAPV-EDANS is a fluorogenic substrate for caspase-1.
-
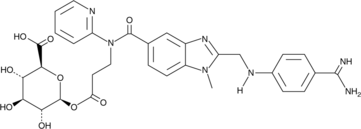
GC43371
Dabigatran Acyl-β-D-Glucuronide
Dabigatran acyl-β-D-glucuronide is a major active metabolite of the thrombin inhibitor dabigatran.
-
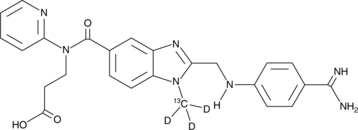
GC49684
Dabigatran-13C-d3
An internal standard for the quantification of dabigatran
-
GC49682
Dabigatran-d3
An internal standard for the quantification of dabigatran
-
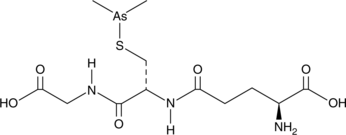
GC43379
Darinaparsin
A dimethylated arsenic linked to glutathione
-
GC68147
dAURK-4 hydrochloride
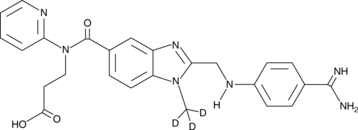
-
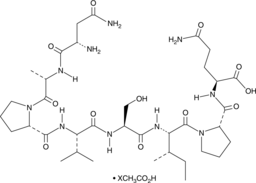
GC49913
Davunetide (acetate)
A neuroprotective ADNP-derived peptide
-
GC43385
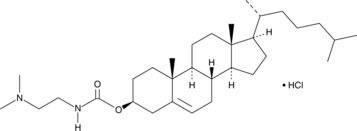
DC-Chol (hydrochloride)
DC-Chol(hydrochloric acid) can inhibit the formation of Aβ40 fibers,DC-Chol(hydrochloric acid) can inhibit the amyloid formation of oxidized hCT[1,2].
-
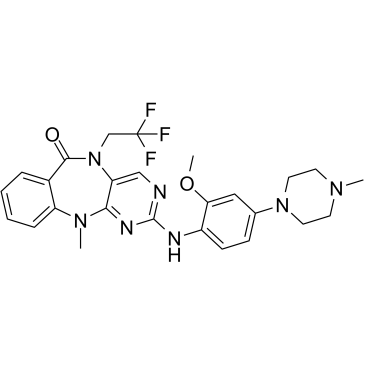
GC39486
DCLK1-IN-1
DCLK1-IN-1 is a selective, oral bioavailability in vivo-compatible chemical probe of the doublecortin like kinase 1 (DCLK1 kinase) domain. DCLK1-IN-1 inhibits DCLK1 and DCLK2 kinases (IC50: DCLK1=9.5/57.2 nM and DCLK2=31/103 nM in binding and kinase assay, respectively). DCLK1-IN-1 shows low toxicity, and can investigate DCLK1 biology and establish its role in cancer, like DCLK1+ pancreatic ductal adenocarcinoma (PDAC).
-
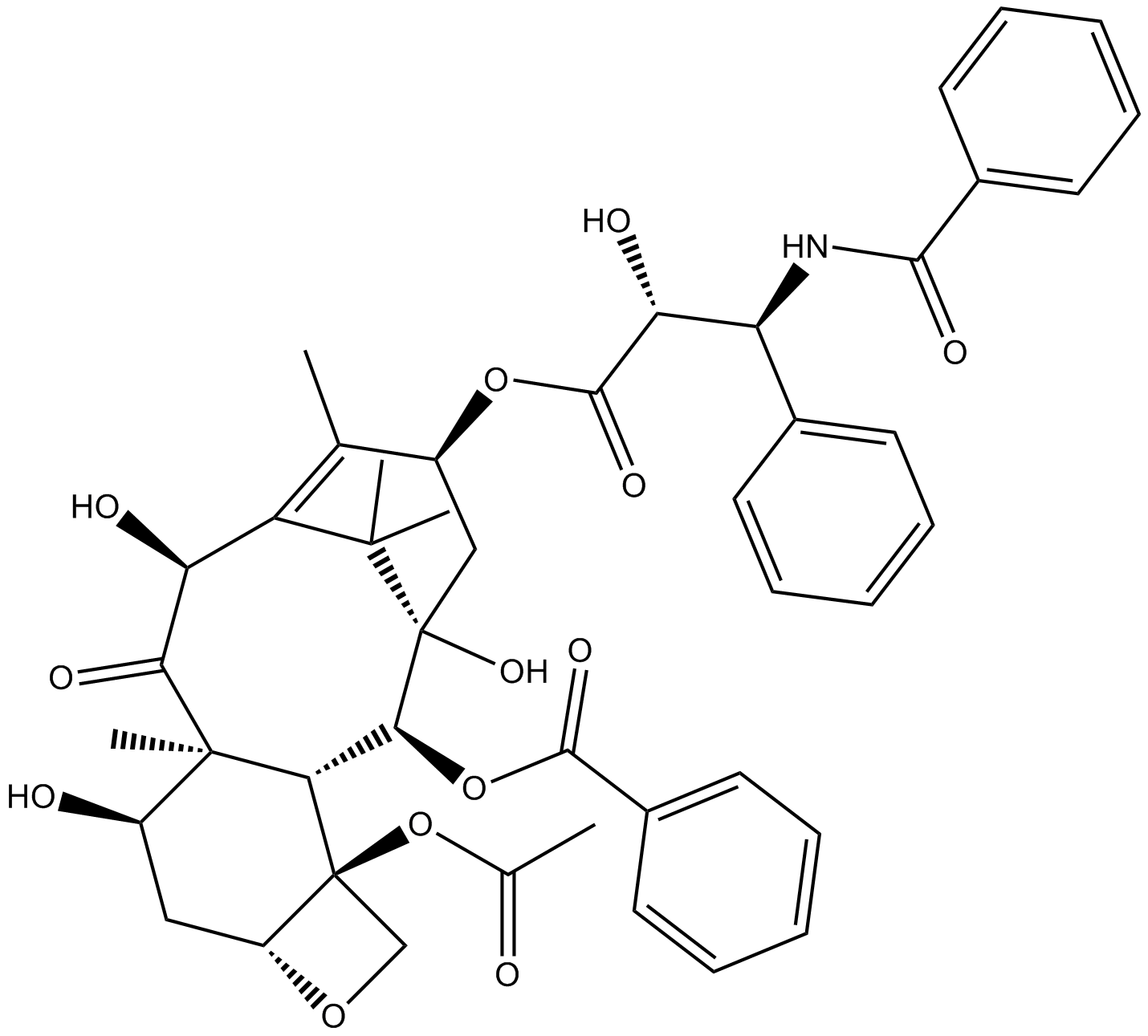
GN10426
Deacetyltaxol
-
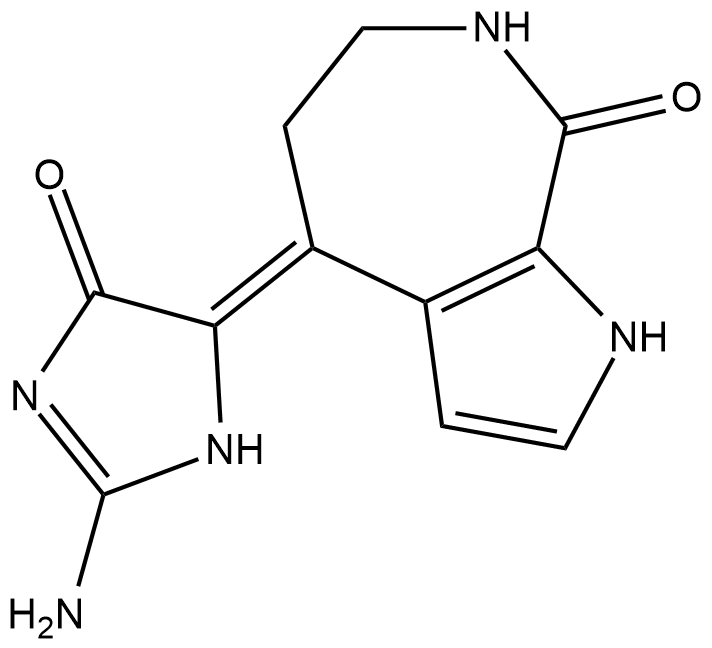
GC18666
Debromohymenialdisine
Damaged DNA in humans is detected by sensor proteins that transmit a signal through checkpoint kinases (Chks) Chk1 and Chk2.
-
GC11835
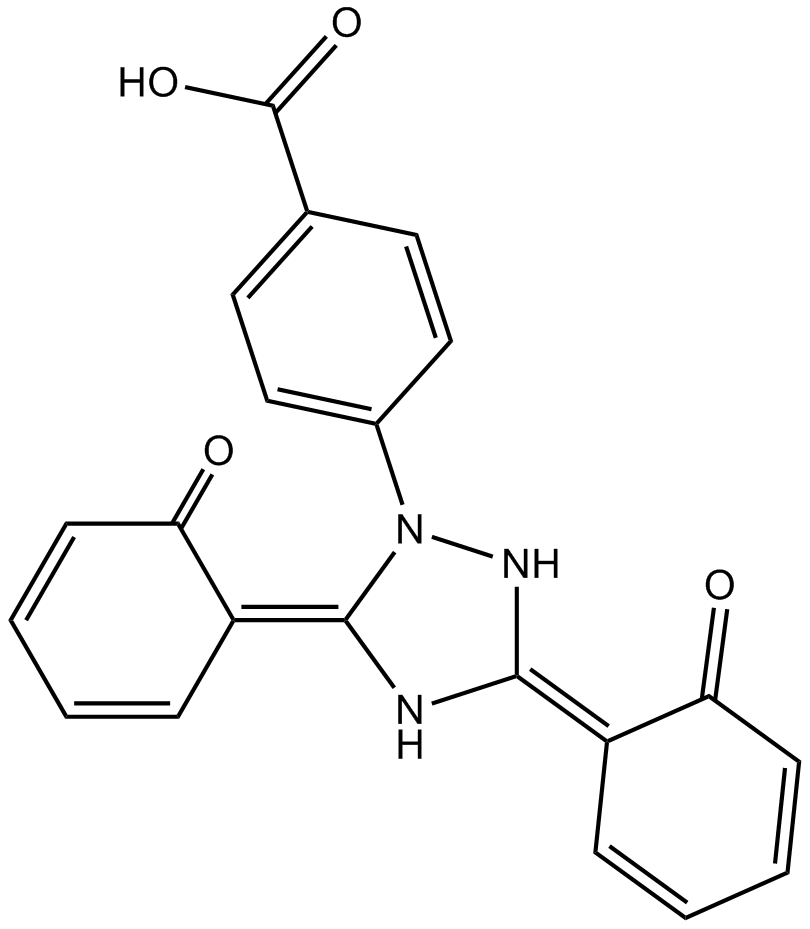
Deferasirox
Oral iron chelator
-
GC41304
Deoxybrevianamide E
Deoxybrevianamide E is an alkaloid fungal metabolite that has been found in Aspergillus.
-
GC38564
Deoxypodophyllotoxin
Deoxypodophyllotoxin (DPT), a derivative of podophyllotoxin, is a lignan with potent antimitotic, anti-inflammatory and antiviral properties isolated from rhizomes of Sinopodophullumhexandrum (Berberidaceae).
-
GC47211
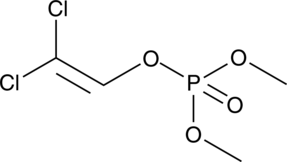
Dichlorvos
An organophosphate insecticide
-
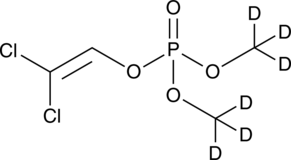
GC49293
Dichlorvos-d6
An internal standard for the quantification of dichlorvos
-
GC43461
Dihydrocytochalasin B
Dihydrocytochalasin B (DCB) is a member of the cytochalasin mycotoxin family that inhibits actin assembly.
-
GC43467
Dimethyldioctadecylammonium (bromide)
Dimethyldioctadecylammonium (DDA) is a cationic amphipathic lipid.
-
GC32969
Dimethylenastron
An inhibitor of EG5
-
GC43469
Diminutol
Diminutol is a cell-permeable purine derivative that inhibits the NADP-dependent oxidoreductase, NQO1 (Ki = 1.72 μM), to destabilize microtubules and disrupt mitosis.
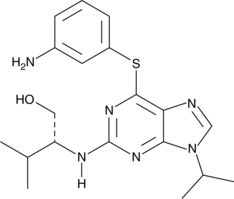
-
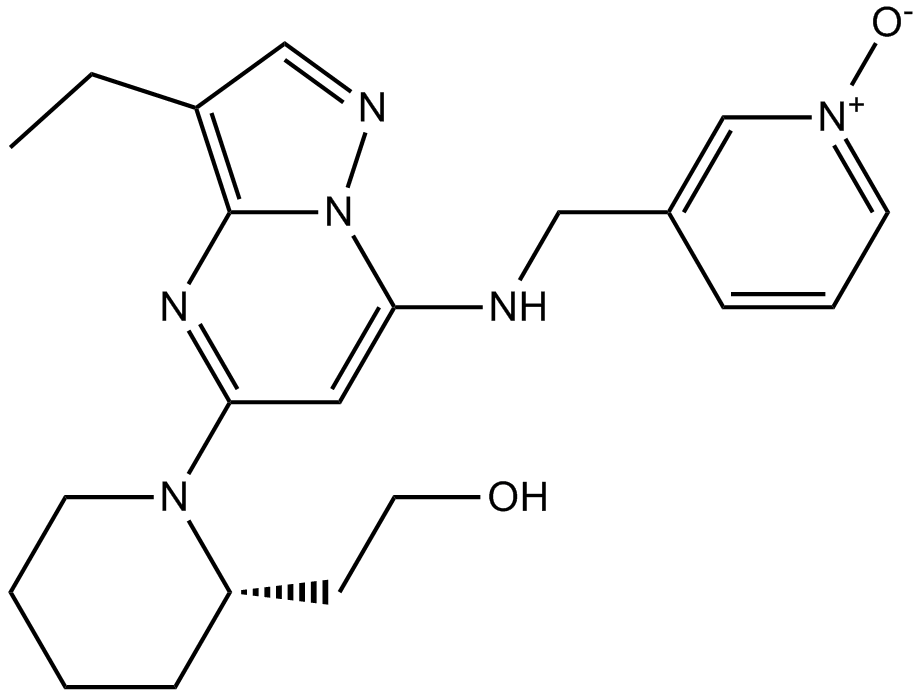
GC17648
Dinaciclib(SCH727965)
Dinaciclib(SCH727965) (SCH 727965) is a potent inhibitor of CDK, with IC50s of 1 nM, 1 nM, 3 nM, and 4 nM for CDK2, CDK5, CDK1, and CDK9, respectively.
-
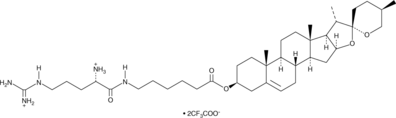
GC43472
Dios-Arg (trifluoroacetate salt)
Dios-Arg is a steroid-based cationic lipid that contains a diosgenin skeleton coupled to an L-arginine head group.
-
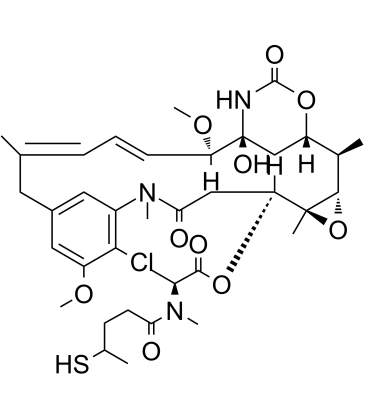
GC60143
DM3
DM3 (Maytansinoid DM3) is a maytansine analog bearing disulfide or thiol groups and a tubulin inhibitor, and is a cytotoxic moiety of antibody-drug conjugates (ADCs).
-
GC38355
DM3-SMe
DM3-SMe is a maytansine derivative and a tubulin inhibitor, and is a cytotoxic moiety of antibody-drug conjugates (ADCs), which can be linked to antibody through disulfide bond or stable thioether bond. DM3-SMe shows highly cytotoxic activity in vitro with an IC50 of 0.0011 nM.
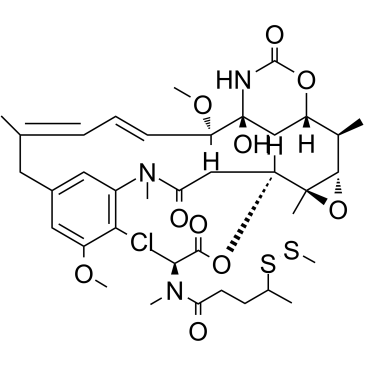
-
GC32810
DM4
DM4 is is an antitubulin agent that inhibit cell division. DM4 can be used in the preparation of antibody drug conjugate.
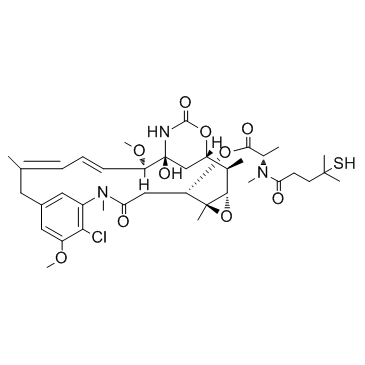
-
GC38468
DM4-SMe
DM4-SMe is a metabolite of antibody-maytansin conjugates (AMCs) and a tubulin inhibitor, and also a cytotoxic moiety of antibody-drug conjugates (ADCs), which can be linked to antibody through disulfide bond or stable thioether bond. DM4-SMe inhibits KB cells with an IC50 of 0.026 nM.
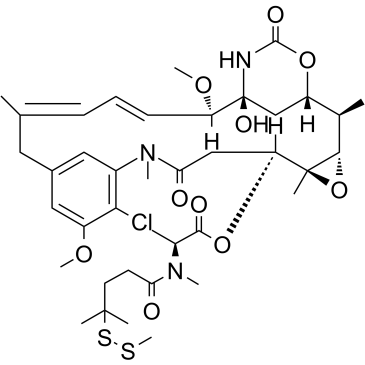
-
GC38460
DM4-SPDP
DM4-SPDP is a drug-linker conjugate composed of a potent antitubulin agent DM4 and a linker SMCC to make antibody drug conjugate. SPDP is a short-chain crosslinker for amine-to-sulfhydryl conjugation via NHS-ester and pyridyldithiol reactive groups that form cleavable (reducible) disulfide bonds with cysteine sulfhydryls.
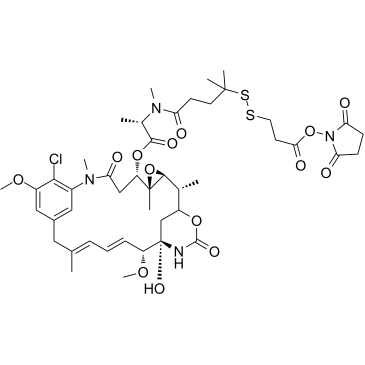
-
GC43545
Dnp-P-Cha-G-Cys(Me)-HA-K(Nma)-NH2
Dnp-P-Cha-G-Cys(Me)-HA-K(Nma)-NH2 is a fluorogenic substrate for matrix metalloproteinase-1 (MMP-1) and MMP-9.



