Inositol Phosphates
Products for Inositol Phosphates
- Cat.No. Product Name Information
-
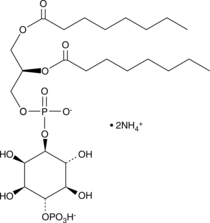
GC43513
D-myo-Inositol-1,2-diphosphate (sodium salt)
Ins(1,2)P2 (sodium salt) is one of the many inositol phosphate (InsP) isomers that could act as small, soluble second messengers in the transmission of cellular signals.
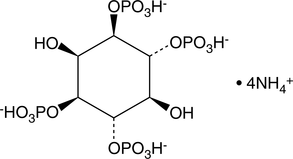
-
GC43516
D-myo-Inositol-1,3,4,5-tetraphosphate (sodium salt)
D-myo-Inositol-1,3,4,5-tetraphosphate (Ins(1,3,4,5)-P4) is formed by the phosphorylation of Ins(1,4,5)P3 by inositol 1,4,5-triphosphate 3-kinase.
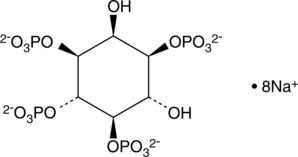
-
GC43517
D-myo-Inositol-1,3,4,6-tetraphosphate (ammonium salt)
The inositol phosphates (IPs) are a family of molecules produced by altering the phosphorylation status of each of the six carbons on the cyclic inositol structure.
-
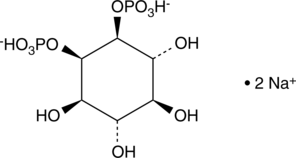
GC43520
D-myo-Inositol-1,3-diphosphate (sodium salt)
D-myo-Inositol-1,3-phosphate (Ins(1,3)P) is a member of the inositol phosphate (InsP) molecular family that play critical roles as small, soluble second messengers in the transmission of cellular signals.
-
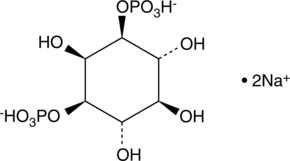
GC43521
D-myo-Inositol-1,4,5,6-tetraphosphate (sodium salt)
D-myo-Inositol-1,4,5,6-tetrahosphate (sodium salt) (Ins(1,4,5,6)-P4) is one of several different inositol oligophosphate isomers implicated in signal transduction.
-
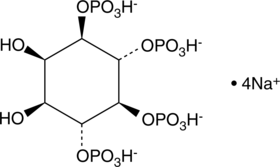
GC43522
D-myo-Inositol-1,4,5-triphosphate (potassium salt)
D-myo-Inositol-1,4,5-triphosphate (Ins(1,4,5)P3) is a second messenger produced in cells by phospholipase C (PLC) mediated hydrolysis of phosphatidyl inositol-4,5-biphosphate.
-
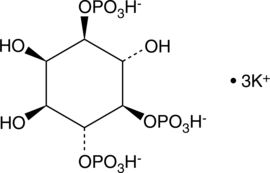
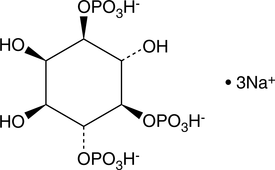
GC43523
D-myo-Inositol-1,4,5-triphosphate (sodium salt)
Primary intracellular IP3 receptor agonist
-
GC43524
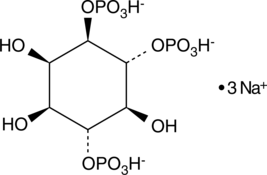
D-myo-Inositol-1,4,6-triphosphate (sodium salt)
D-myo-Inositol-1,4,6-phosphate (Ins(1,4,6)-P3) is a member of the inositol phosphate (InsP) family that play critical roles as small, soluble second messengers in the transmission of cellular signals.
-
GC43526
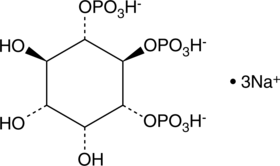
D-myo-Inositol-1,5,6-triphosphate (sodium salt)
The inositol phosphates are a family of mono- to poly-phosphorylated compounds that act as messengers, regulating cellular functions including cell cycling, apoptosis, differentiation, andmotility.
-
GC43540
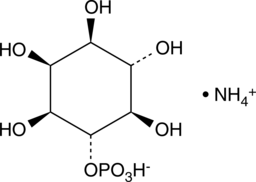
D-myo-Inositol-4-phosphate (ammonium salt)
D-myo-Inositol-4-phosphate (Ins(4)P1) is a member of the inositol phosphate (InsP) molecular family that play critical roles as small, soluble second messengers in the transmission of cellular signals.
-
GC44081
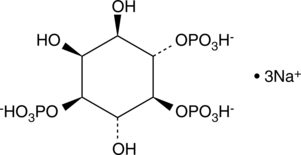
L-myo-Inositol-1,4,5-triphosphate (sodium salt)
Ins(1,4,5)P3 is an isomer of the biologically important D-myo-inositol-1,4,5-triphosphate.
-
GC44260
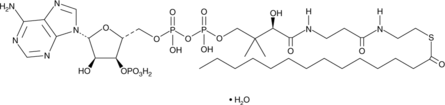
Myristoyl Coenzyme A (hydrate)
Myristoyl coenzyme A (myristoyl-CoA) is a derivative of CoA that contains the long-chain fatty acid myristic acid.
-
GC44740
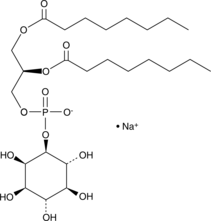
PtdIns-(1,2-dioctanoyl) (sodium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
-
GC44743
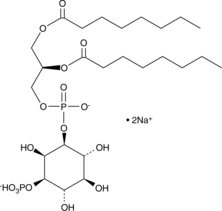
PtdIns-(3)-P1 (1,2-dioctanoyl) (sodium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
-
GC44748
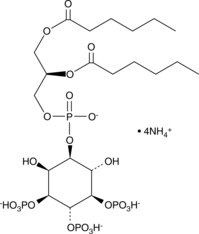
PtdIns-(3,4,5)-P3 (1,2-dihexanoyl) (ammonium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
-
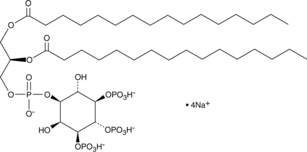
GC44750
PtdIns-(3,4,5)-P3 (1,2-dipalmitoyl) (sodium salt)
The phosphatidylinositol phosphates represent a small percentage of total membrane phospholipids.
-
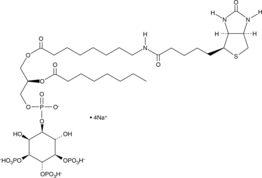
GC44753
PtdIns-(3,4,5)-P3-biotin (sodium salt)
The PtdIn phosphates play an important role in the generation and transduction of intracellular signals.
-
GC44759
PtdIns-(4)-P1 (1,2-dioctanoyl) (ammonium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.