Deubiquitinase
Deubiquitinases (DUBs) are a large group of proteases that cleave ubiquitin from proteins and other molecules. Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate thecellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions. DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases. DUBs play several roles in the ubiquitin pathway. One of the best characterised functions of DUBs is the removal of monoubiqutin and polyubiquitin chainsfrom proteins.
Targets for Deubiquitinase
Products for Deubiquitinase
- Cat.No. Product Name Information
-
GC39625
6RK73
6RK73 is a covalent irreversible and specific UCHL1 inhibitor with an IC50 of 0.23 ?M. 6RK73 shows almost no inhibition of UCHL3 (IC50=236 ?M). 6RK73 specifically inhibit UCHL1 activity in breast cancer.
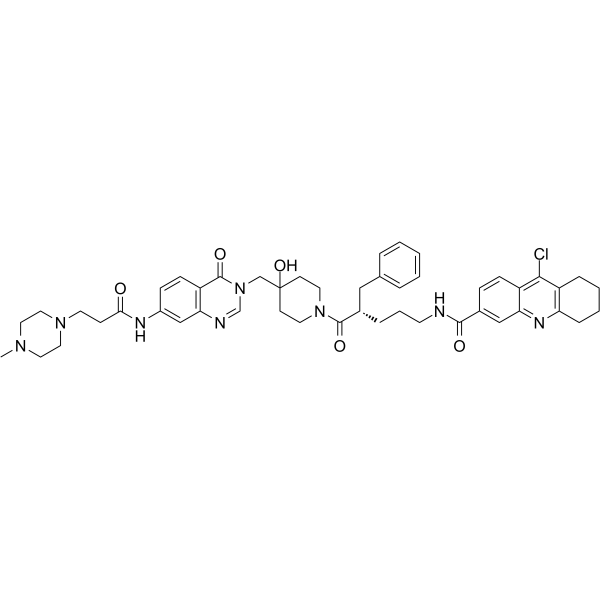
-
GC13035
Bay 11-7821
A selective and irreversible NF-κB inhibitor
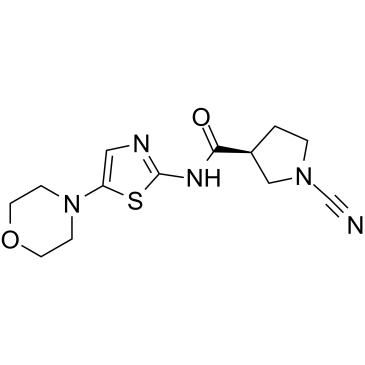
-
GC65259
BC-1471
BC-1471 is a STAM-binding protein (STAMBP) deubiquitinase inhibitor.
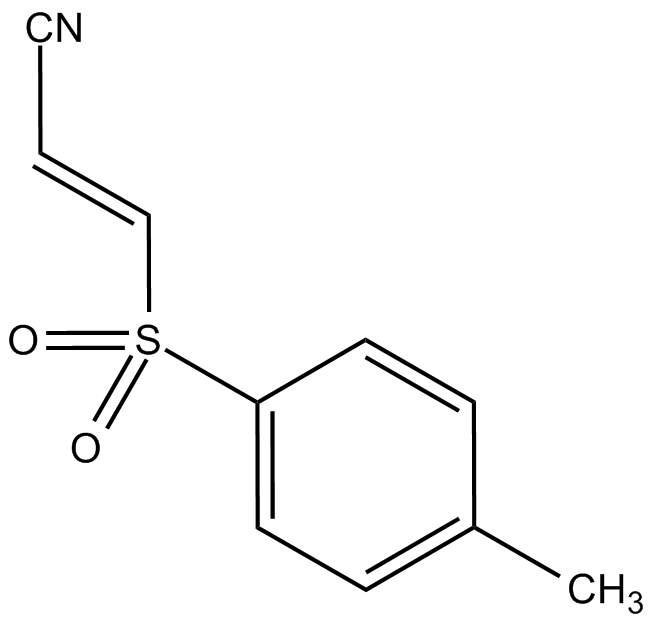
-
GC13892
C527
Inhibitor of USP1/USF1 complex
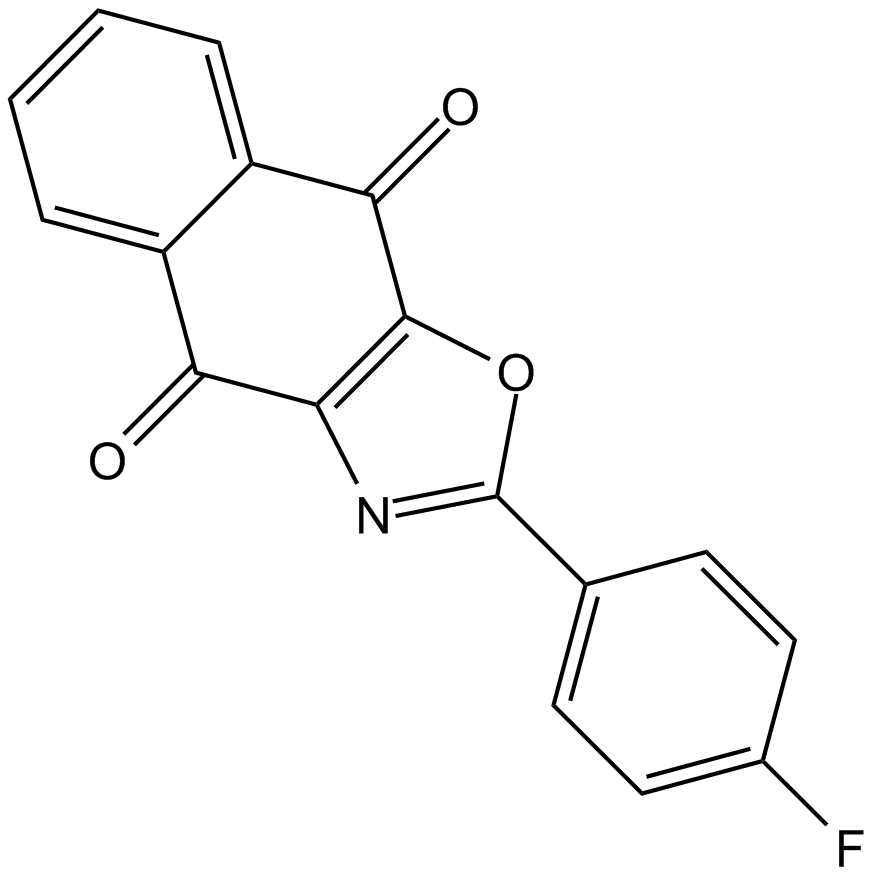
-
GC67886
DC-U4106

-
GC35906
DUBs-IN-1
DUBs-IN-1 is an active inhibitor of ubiquitin-specific proteases (USPs), with an IC50 of 0.85 μM for USP8.
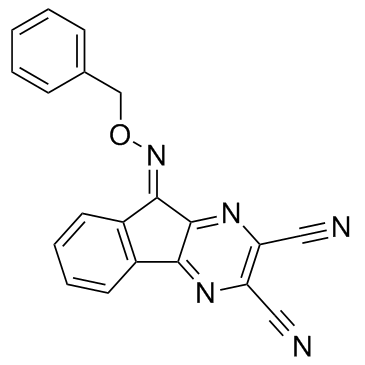
-
GC17125
DUBs-IN-2
DUBs-IN-2 is a potent deubiquitinase inhibitor with an IC50 of 0.28 μM for USP8.
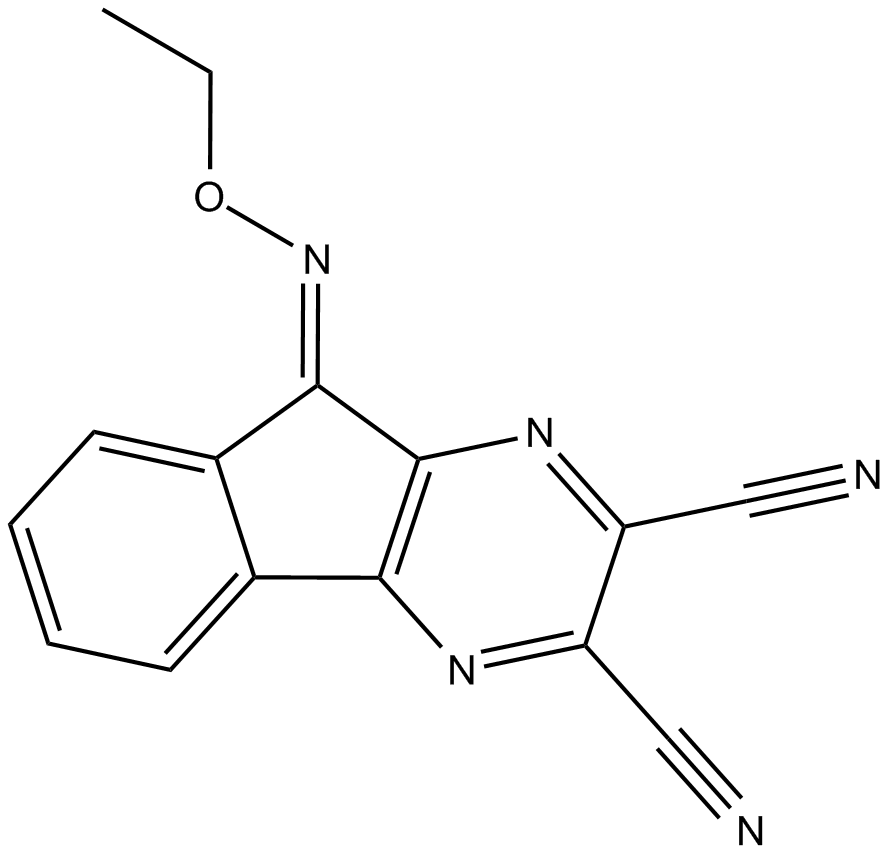
-
GC10356
DUBs-IN-3
DUBs-IN-3 is a potent deubiquitinase (USP) enzyme inhibitor extracted from reference compound 22c with an IC50 of 0.56 μM for USP8.
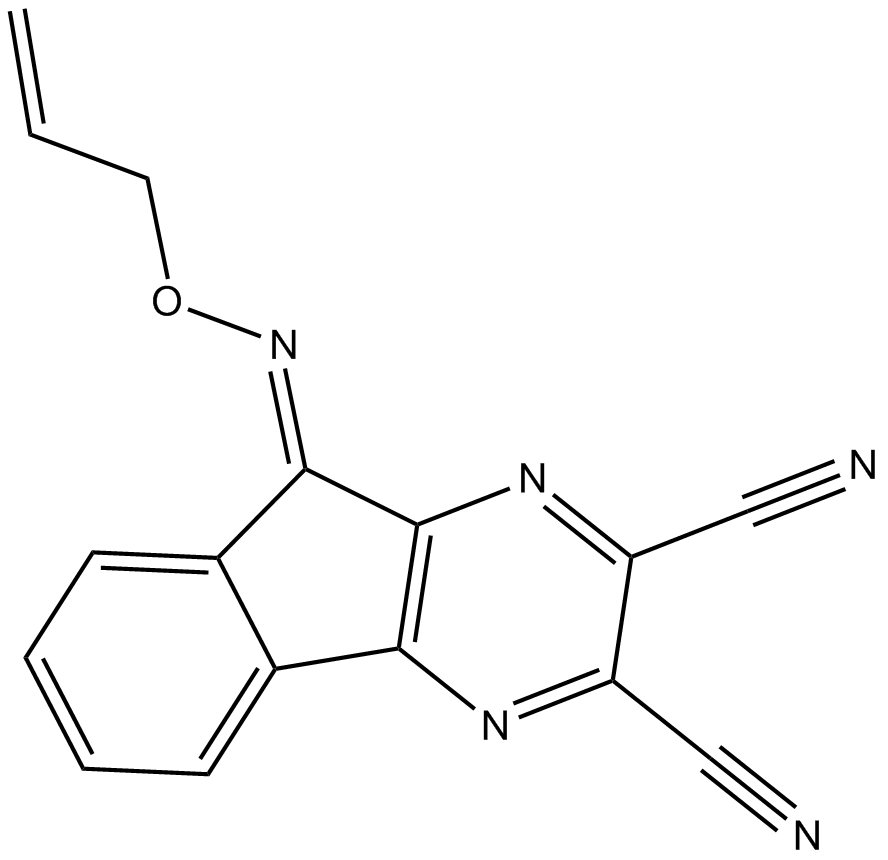
-
GC34083
EOAI3402143
EOAI3402143 is a deubiquitinase (DUB) inhibitor, which inhibits dose-dependently inhibits Usp9x/Usp24 and Usp5.
-
GC62979
FT206
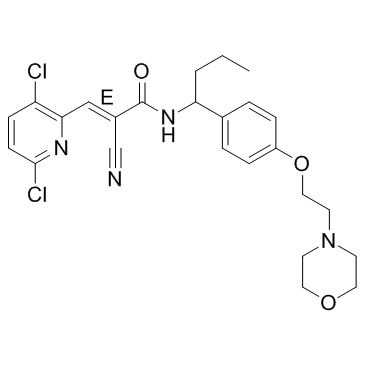
FT206 is an inhibitor of carboxamides as ubiquitin-specific protrase extracted from patent WO 2020033707 A1, example 11-1.
-
GC67915
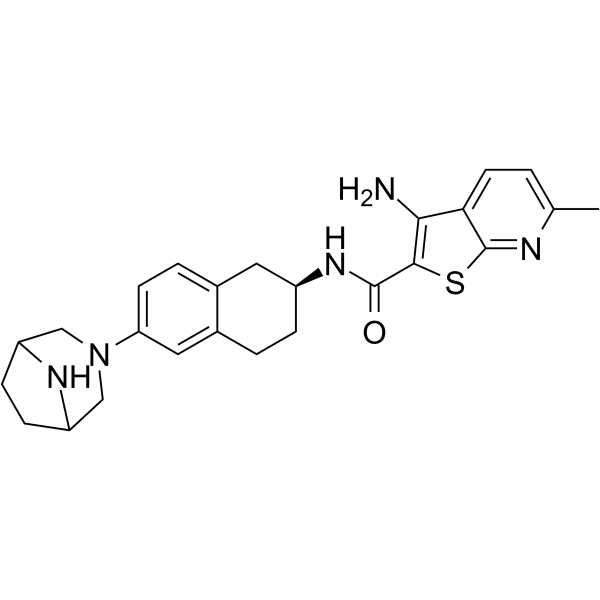
FT3967385

-
GC32786
FT671
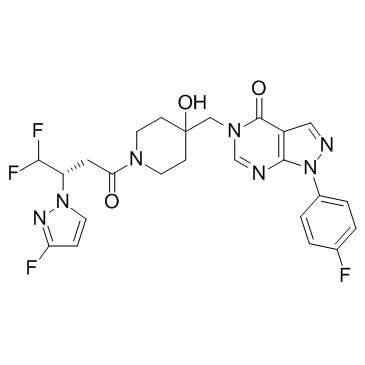
FT671 is a potent, non-covalent and selective USP7 inhibitor with an IC50 of 52 nM and binds to the USP7 catalytic domain with a Kd of 65 nM.
-
GC69134
FT709
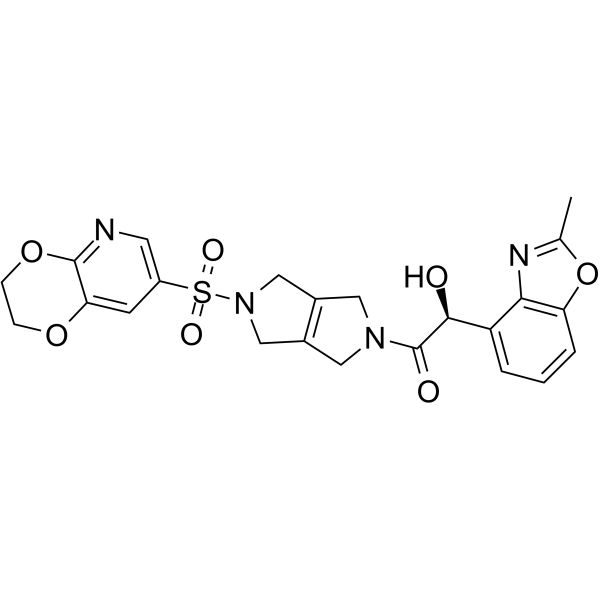
FT709 is an effective selective inhibitor of USP9X with an IC50 value of 82 nM. USP9X is involved in centrosome function, chromosome alignment during mitosis, EGF receptor degradation, chemical sensitization and circadian rhythms.
-
GC32917
FT827
FT827 is a selective and covalent ubiquitin-specific protease 7 (USP7) inhibitor (Ki=4.2 ?M). FT827 binds to the USP7 catalytic domain (USP7CD; residues 208-560) with an apparent Kd value of 7.8 ?M.
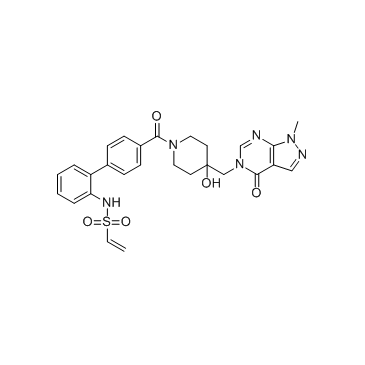
-
GC32856
GNE-6640
An inhibitor of USP7
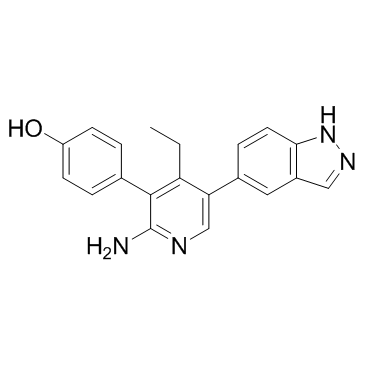
-
GC32727
GNE-6776
GNE-6776 is a selective and orally bioavailable USP7 inhibitor.
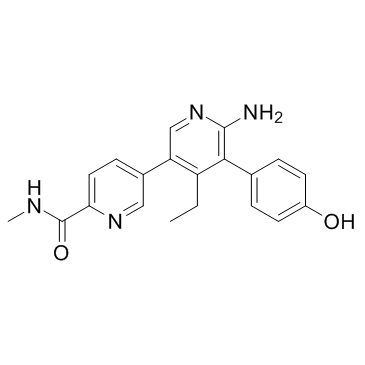
-
GC39172
GRL0617
A SARS-CoV and SARS-CoV-2 PLpro inhibitor
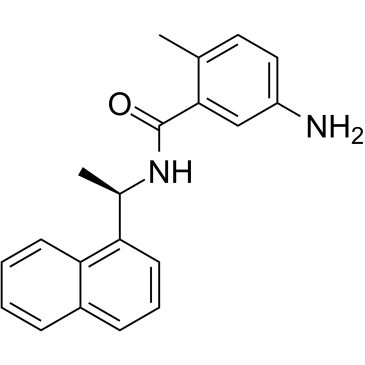
-
GC34354
GSK2643943A
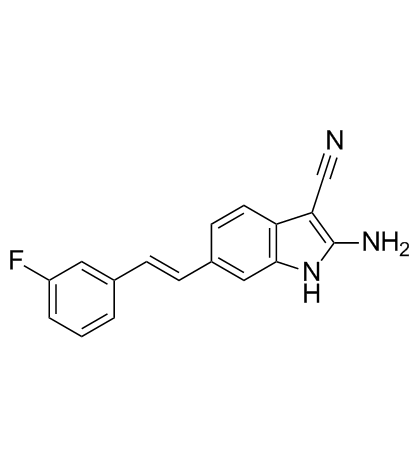
GSK2643943A is a deubiquitinating enzyme (DUB) inhibitor targeting USP20. GSK2643943A has affinity with an IC50 of 160 nM for USP20/Ub-Rho. GSK2643943A has anti-tumor efficacy and can be used for the research of oral squamous cell carcinoma (OSCC) .
-
GC36211
HBX 19818
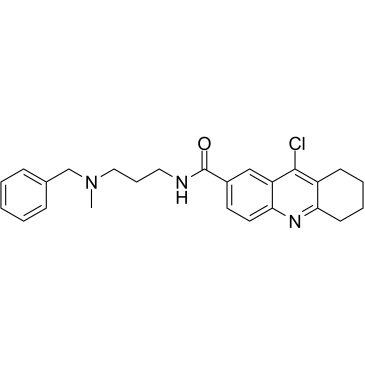
HBX 19818 is a specific inhibitor of ubiquitin-specific protease 7 (USP7), with an IC50 of 28.1 μM.
-
GC14797
IU1
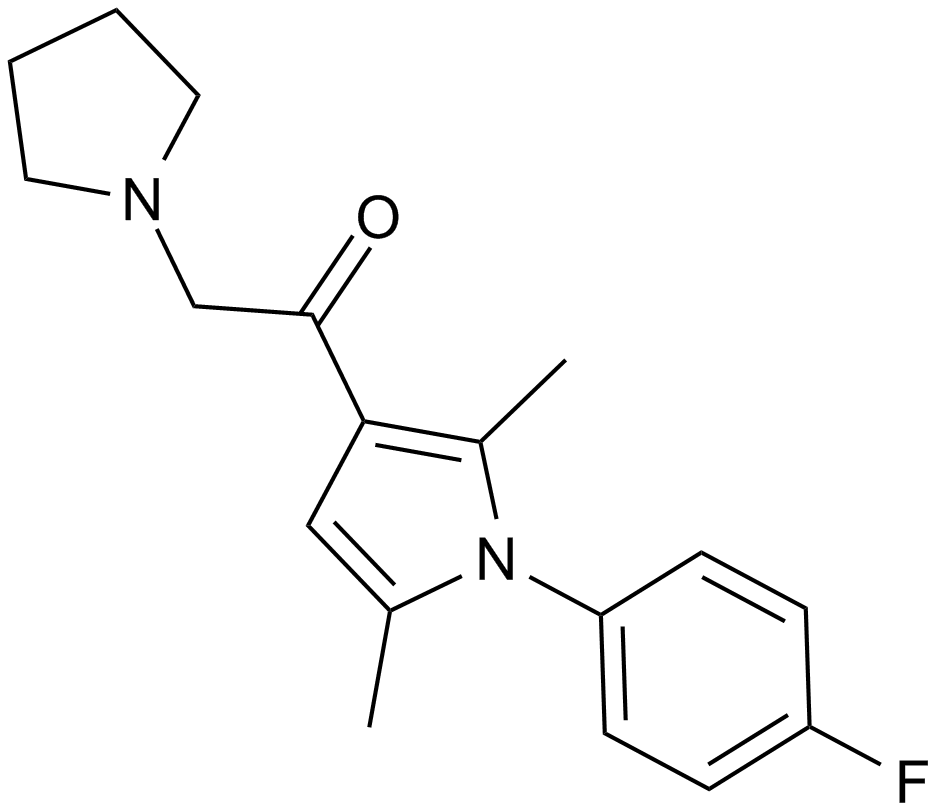
A deubiquitinating enzyme inhibitor
-
GC63027
IU1-248
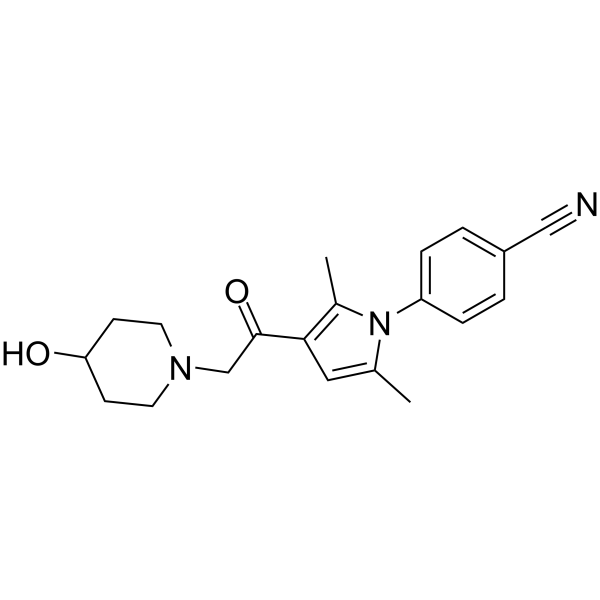
IU1-248, a derivative of IU1, is a potent and selective USP14 inhibitor with an IC50 of 0.83?μM.
-
GC64956
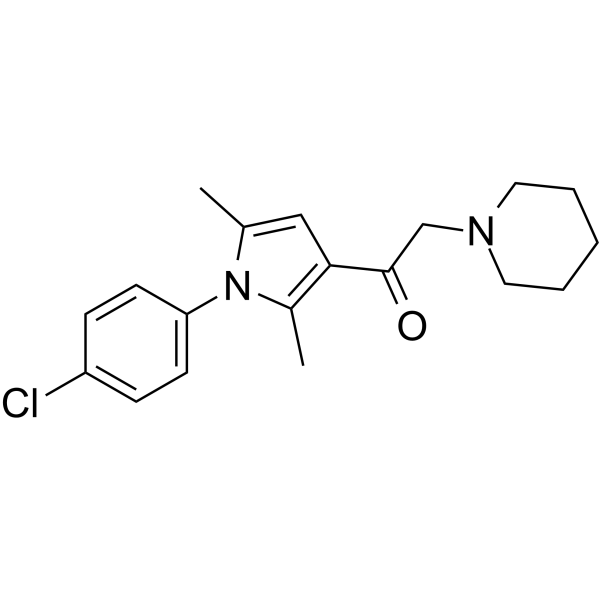
IU1-47
IU1-47 is a potent and specific USP14 inhibitor with an IC50 of 0.6 μM.
-
GC62460
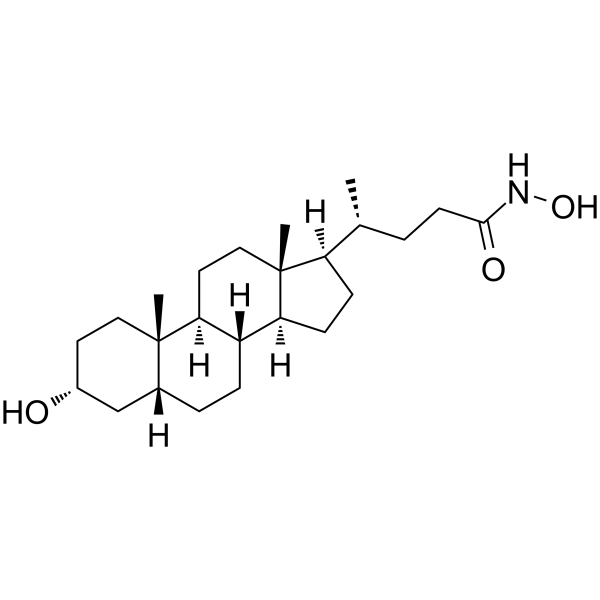
LCAHA
LCAHA (LCA hydroxyamide) is a deubiquitinase USP2a inhibitor with IC50s of 9.7 μM and 3.7μM in Ub-AMC Assay and Di-Ub Assay, respectively. LCAHA destabilizes Cyclin D1 and induces G0/G1 arrest by inhibiting deubiquitinase USP2a.
-
GC10510
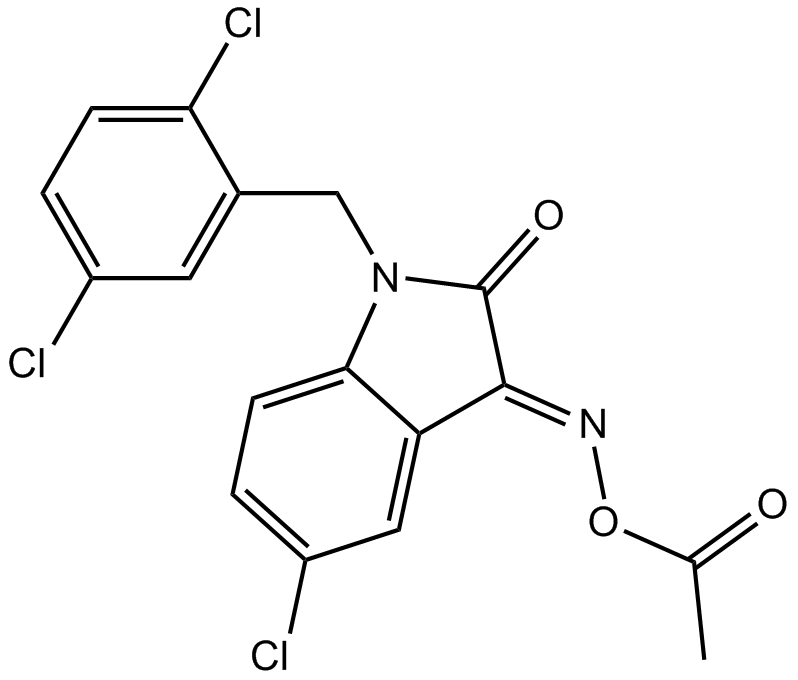
LDN 57444
An inhibitor of UCH-L1
-
GC60981
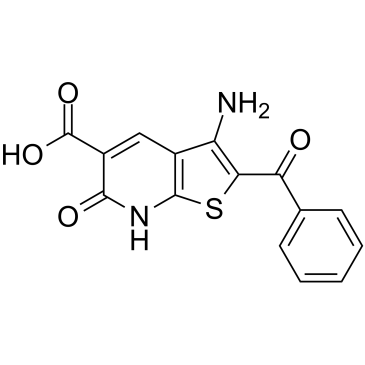
LDN-91946
LDN-91946 is a potent, selective and uncompetitive ubiquitin C-terminal hydrolase-L1 (UCH-L1) inhibitor with a Ki app of 2.8 μM.
-
GC34670
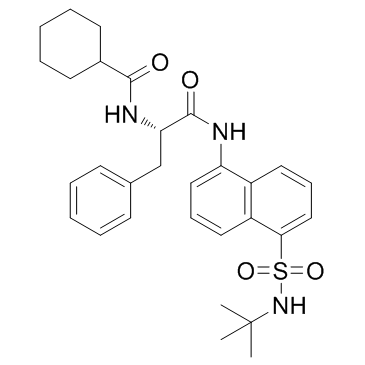
MF-094
MF-094 is a potent and selective USP30 inhibitor with an IC50 of 120 nM. MF-094 increases protein ubiquitination and accelerates mitophagy.
-
GC17635
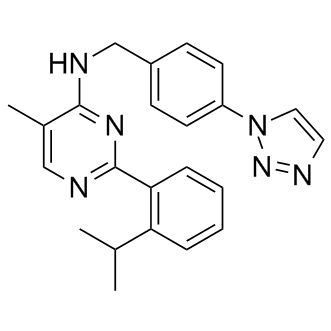
ML-323
USP1-UAF1 inhibitor
-
GC32720
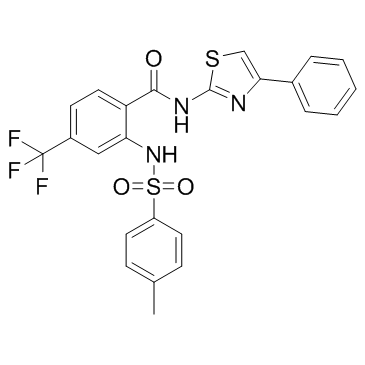
ML364
An inhibitor of USP2
-
GC12475
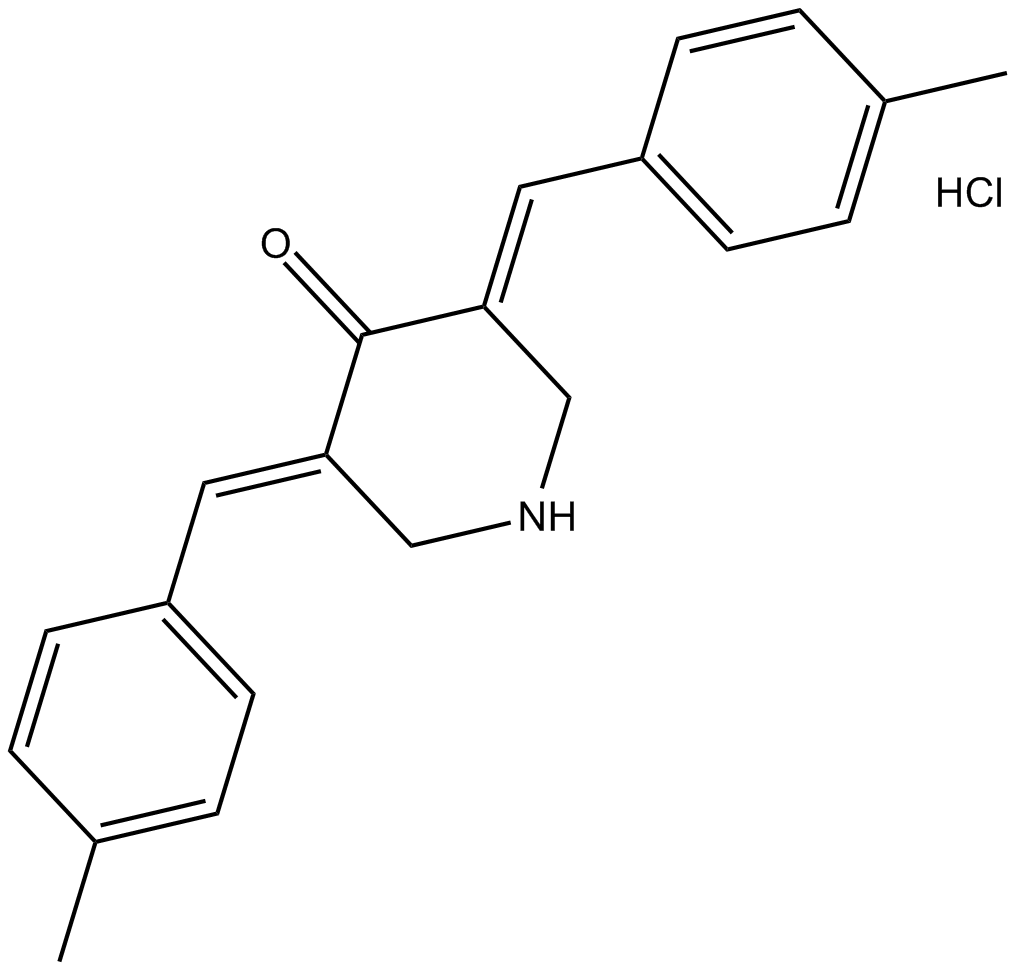
NSC 632839 hydrochloride
A deubiquitylase and deSUMOylase inhibitor
-
GC15503
NSC 687852 (b-AP15)
An inhibitor of the deubiquitinases USP14 and UCHL5
-
GC69637
OTUB1/USP8-IN-1
OTUB1/USP8-IN-1 is an effective dual inhibitor of OTUB1 and USP8, with IC50 values of 0.17 and 0.28 nM, respectively. It can be used for cancer research.
-
GC10379
P 22077
-
GC12067
P005091
A selective inhibitor of ubiquitin-specific protease 7
-
GC13208
PR-619
PR-619 is a broad-spectrum DUB (deubiquitinating enzyme) inhibitor. PR-619 has demonstrated robust DUB inhibitory activity (5-20 µM) and growth inhibitory activity with IC50 of 2 µM in HEK 293T cells..
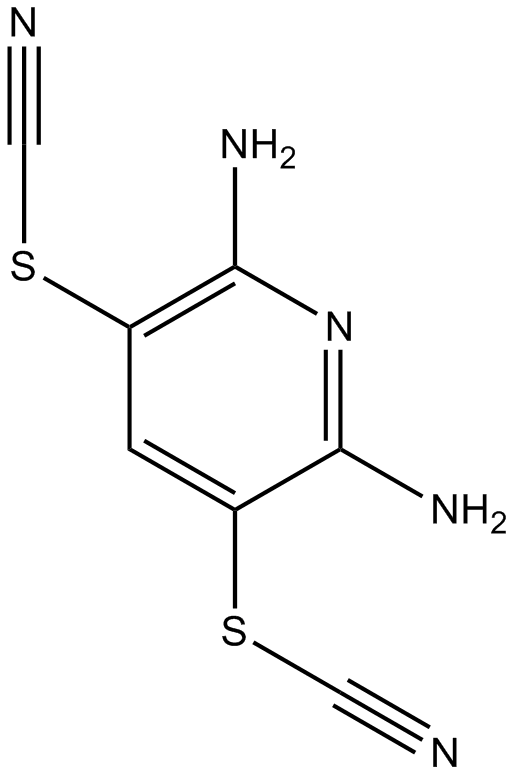
-
GC60317
RA-9
RA-9 is a potent and selective proteasome-associated deubiquitinating enzymes (DUBs) inhibitor with favorable toxicity profile and anticancer activity. RA-9 blocks ubiquitin-dependent protein degradation without impacting 20S proteasome proteolytic activity. RA-9 selectively induces onset of apoptosis in ovarian cancer cell lines and primary cultures derived from donors. RA-9 induces endoplasmic reticulum (ER)-stress responses in ovarian cancer cells.
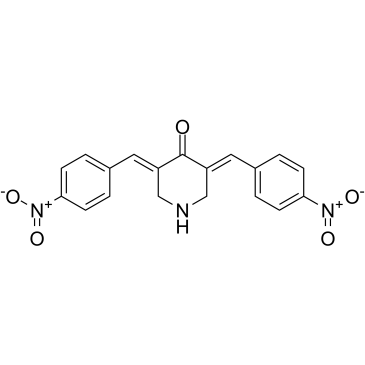
-
GC17698
SJB2-043
An inhibitor of the USP1-UAF1 complex and SARS-CoV-2 PLpro
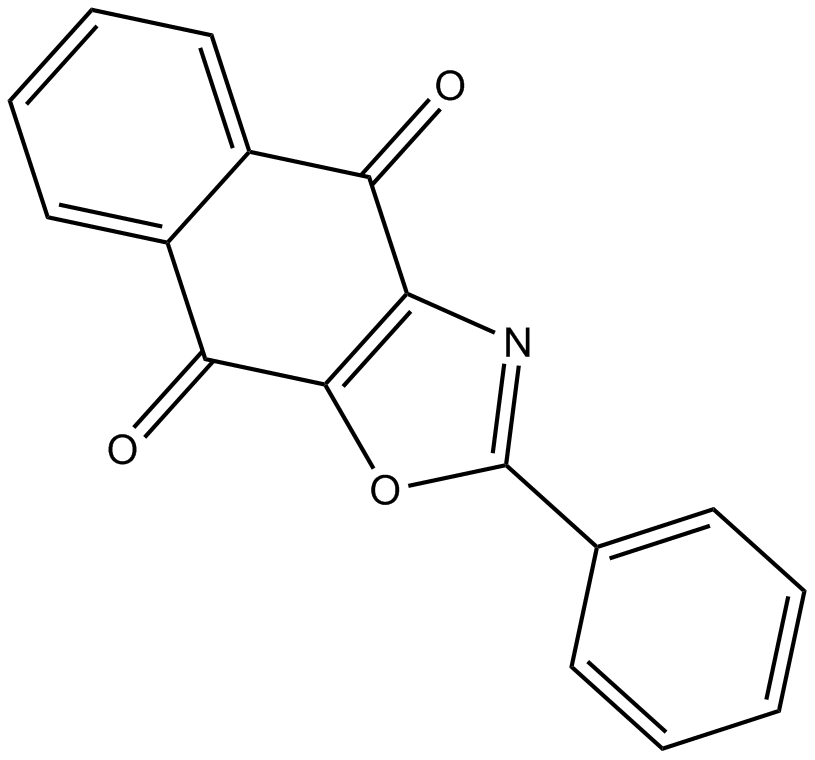
-
GC13401
SJB3-019A
Usp1 inhibitor
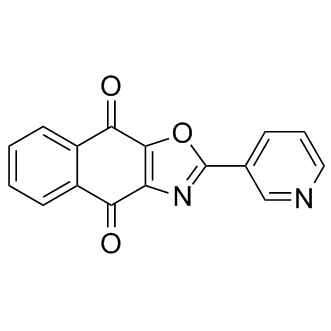
-
GC67712
STAMBP-IN-1
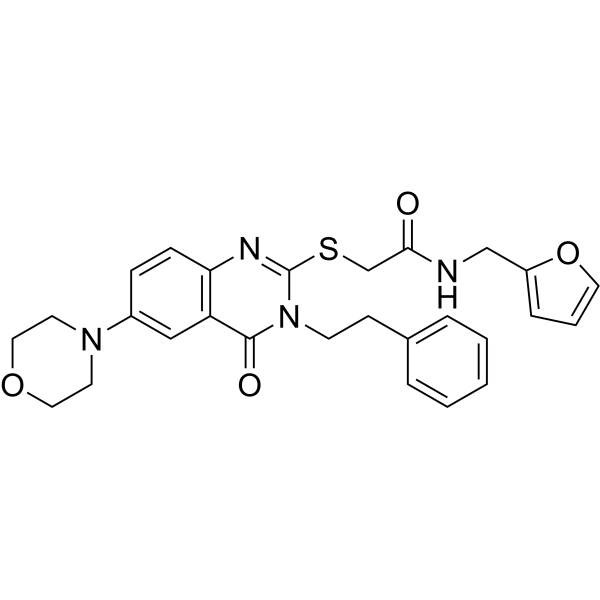
-
GC62509
STD1T
STD1T is a deubiquitinase USP2a inhibitor with an IC50 of 3.3 μM in Ub-AMC Assay.
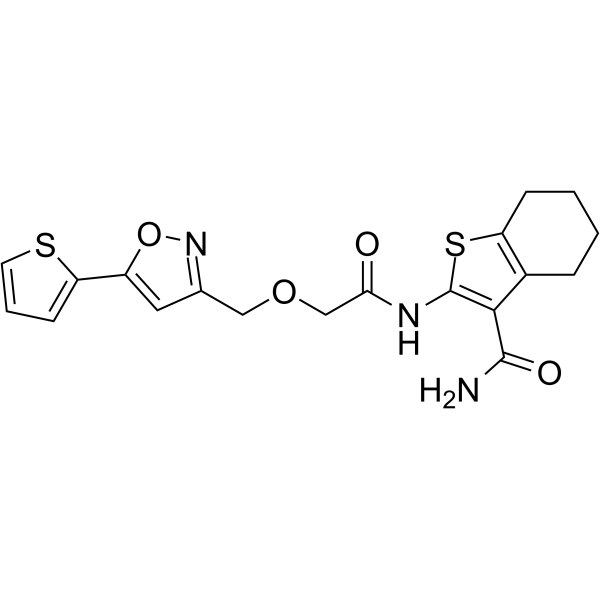
-
GC13013
TCID
UCHL3 inhibitor, cell-permeable
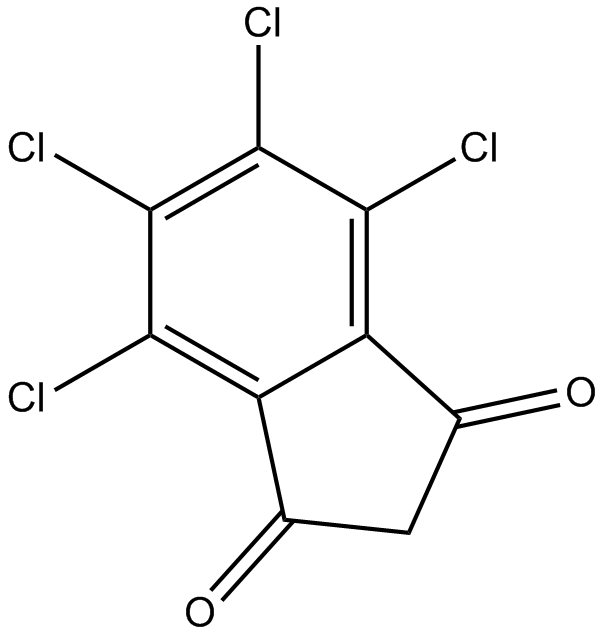
-
GC14366
Thioguanine
A thiopurine analog
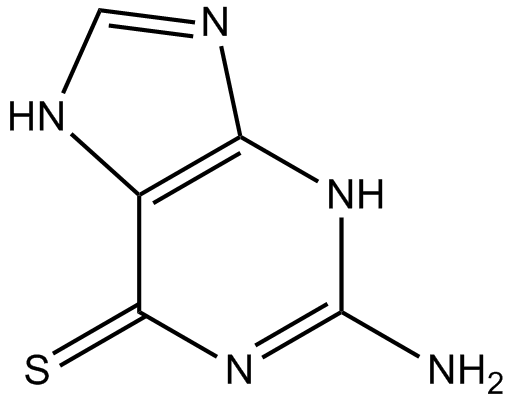
-
GC70098
USP1-IN-2
USP1-IN-2 (Compound I-193) is an effective inhibitor of USP1 with an IC50 less than 50 nM.
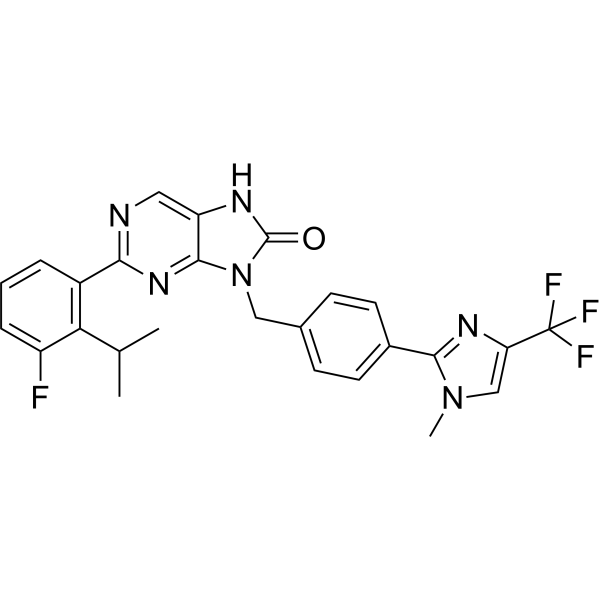
-
GC67963
USP15-IN-1
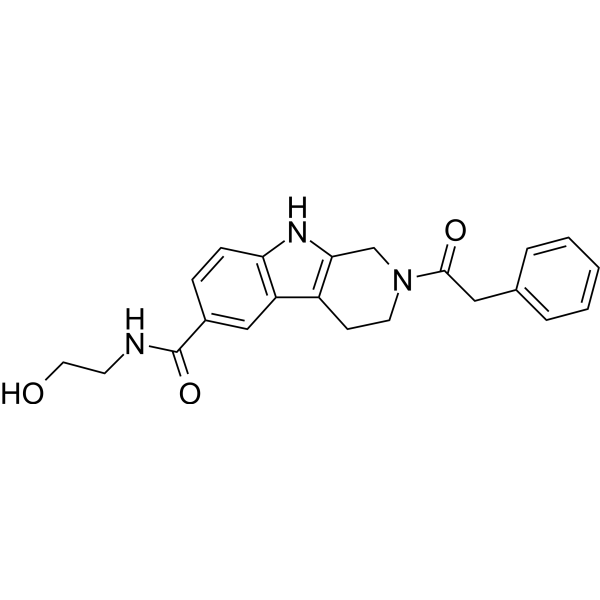
-
GC38043
USP25/28 inhibitor AZ1
USP25/28 inhibitor AZ1 (AZ1) is an orally active, selective, noncompetitive, dual ubiquitin specific protease (USP) 25/28 inhibitor with IC50s of 0.7 μM and 0.6 μM, respectively
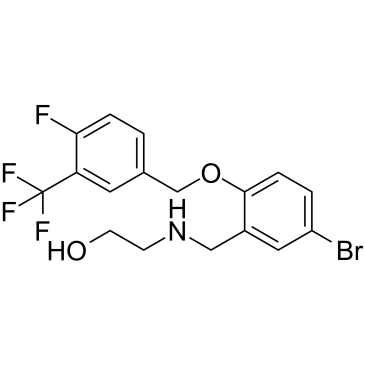
-
GC70099
USP30 inhibitor 11
USP30 inhibitor 11 is a selective and effective inhibitor of USP30, with an IC50 value of 0.01 µμ. It is Example 83 derived from patent document WO2017009650A1, used for research on cancer and diseases involving mitochondrial dysfunction.
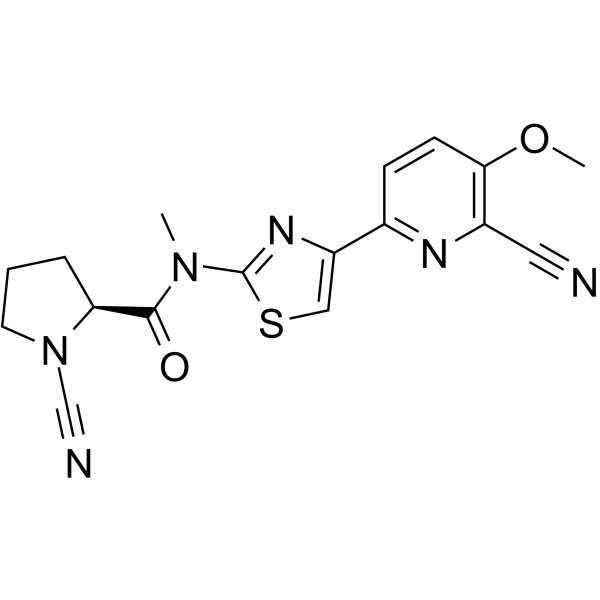
-
GC62410
USP30 inhibitor 18
USP30 inhibitor 18 is a selective USP30 inhibitor with an IC50 of 0.02 μM. USP30 inhibitor 18 increases protein ubiquitination and accelerates mitophagy.
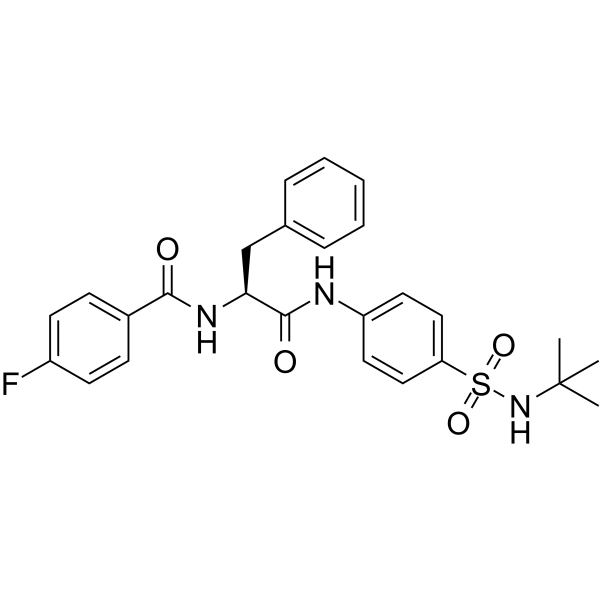
-
GC66438
USP5-IN-1
USP5-IN-1 (compound 64), a potent deubiquitinase USP5 inhibitor, binds to the USP5 ZnF-UBD with a KD of 2.8 μM. USP5-IN-1 is selective over nine proteins containing structurally similar ZnF-UBD domains. USP5-IN-1 inhibits the USP5 catalytic cleavage of a di-ubiquitin substrate.
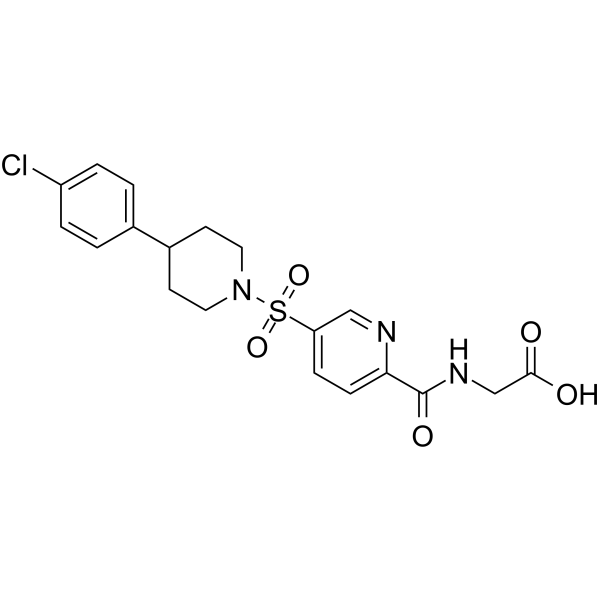
-
GC37875
USP7-IN-1
USP7-IN-1 is a selective and reversible inhibitor of ubiquitin-specific protease 7 (USP7), with an IC50 of 77 μM, and can be used for the research of cancer.
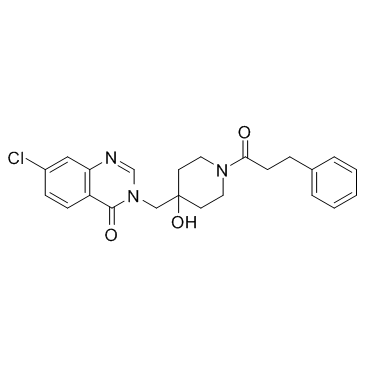
-
GC70100
USP7-IN-11
USP7-IN-11 is an effective inhibitor of ubiquitin-specific protease 7 (USP7) (Deubiquitinase), with an IC50 of 0.37 nM. USP7-IN-11 has anti-cancer properties (WO2022048498A1; Example 187).
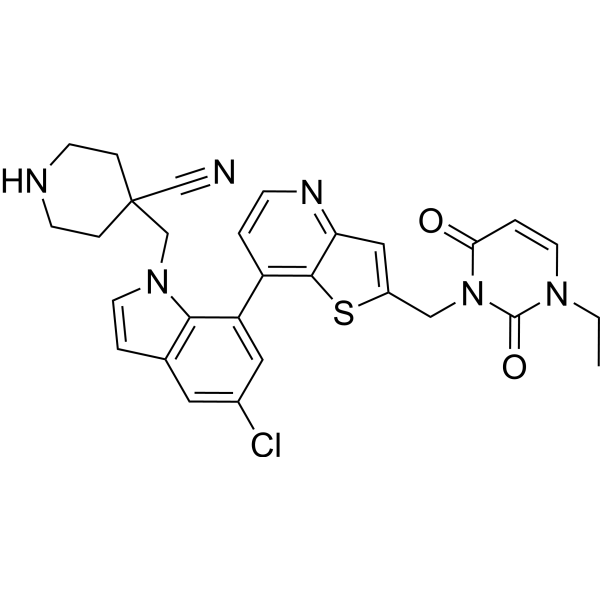
-
GC65275
USP7-IN-3
USP7-IN-3 (Compound 5) is a potent and selective allosteric ubiquitin-specific protease 7 (USP7) inhibitor.
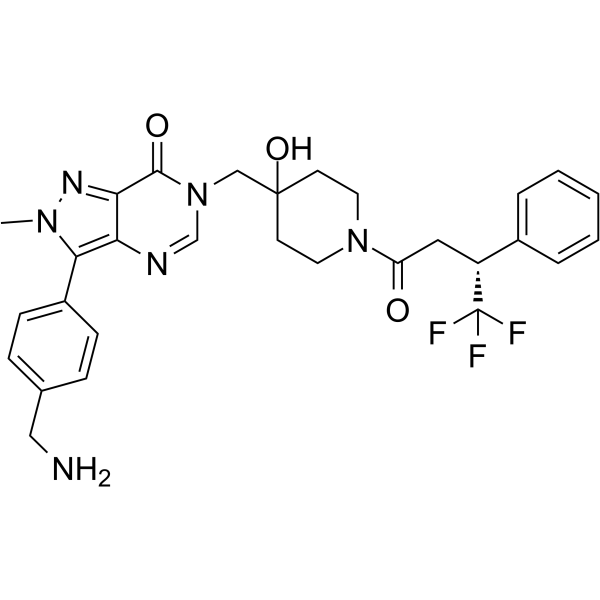
-
GC70101
USP7-IN-7
USP7-IN-7 (compound 124) is a USP7 inhibitor with an IC50 value of <10 nM. It has low nanomolar cell toxicity against p53 mutant cancer cell lines, p53 wild-type blood tumors, and neuroblastoma cell lines. USP7-IN-7 can be used for tumor research.
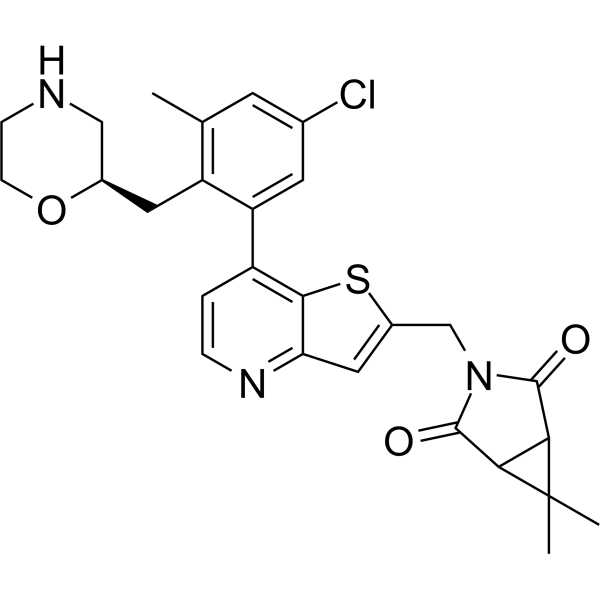
-
GC62377
USP7-IN-8
USP7-IN-8 (example 81) is a selective ubiquitin-specific protease 7 (USP7) inhibitor with an IC50 of 1.4 μM in an Ub-Rho110 assay. USP7-IN-8 shows no activity against USP47 and USP5. USP7-IN-8 has anticancer effects.
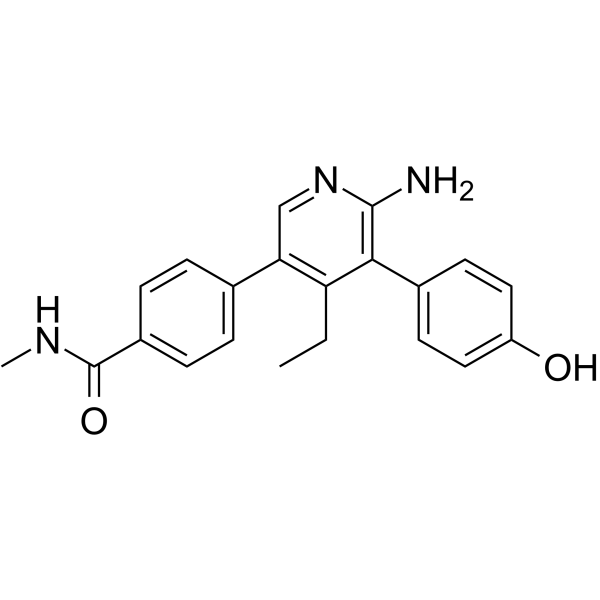
-
GC70102
USP7-IN-9
USP7-IN-9 is an efficient inhibitor of USP7 with an IC50 of 40.8 nM. It can induce apoptosis in RS4;11 cells and block the cell cycle at G0/G1 and S phase. USP7-IN-9 reduces the levels of cancer proteins MDM2 and DNMT1, while increasing the levels of tumor suppressor proteins p53 and p21.
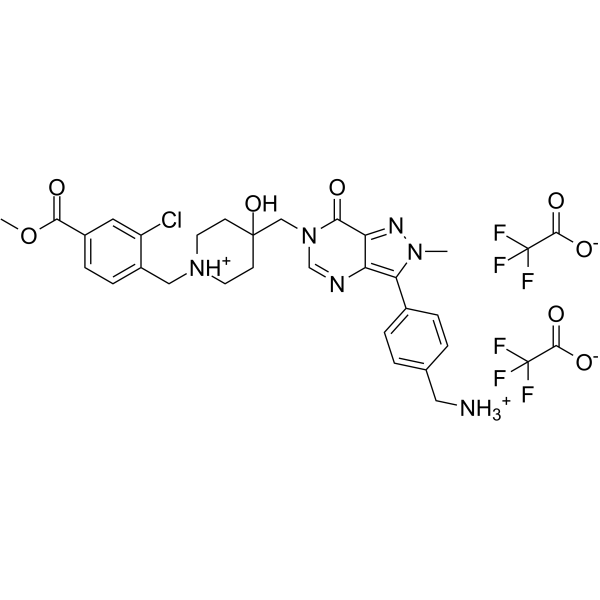
-
GC18160
USP7/USP47 inhibitor
?A selective inhibitor of USP7/USP47
-
GC64537
USP8-IN-1
USP8-IN-1 is a USP8 inhibitor with an IC50 of 1.9 μM. USP8-IN-1 inhibits H1975 cell growth with a GI50 of 82.04 μM (CN111138358A; U10).
-
GC70103
USP8-IN-3
USP8-IN-3 (Compd U51) is a deubiquitinase USP8 inhibitor with an IC50 value of 4.0 μM. USP8-IN-3 also inhibits the proliferation of GH3 and H1957 cells, with GI50 values of 37.03 μM and 6.01 μM, respectively.
-
GC19378
VLX1570
VLX1570, an analogue of b-AP15, is a competitive inhibitor of deubiquitinase activity, with an IC50 of appr 10 uM.
-
GC10970
WP1130
WP1130 (WP1130) is a cell-permeable deubiquitinase (DUB) inhibitor, directly inhibiting DUB activity of USP9x, USP5, USP14, and UCH37. WP1130 has been shown to downregulate the antiapoptotic proteins Bcr-Abl and JAK2.
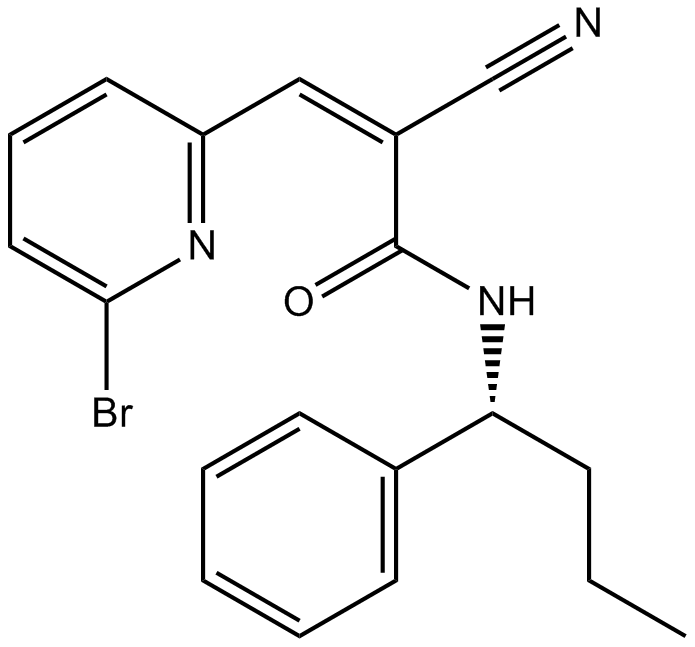
-
GC62517
XL177A
XL177A is a highly potent and selective irreversible USP7 inhibitor with an IC50 of 0.34?nM. XL177A elicits cancer cell killing through a p53-dependent mechanism.