CXCR
CXCR (CXC chemokine receptor) is a group of receptors that belong to GPCRs and specifically binds and responds to cytokines of the CXC chemokine family.
Products for CXCR
- Cat.No. Product Name Information
-
GC62588
(R,R)-CXCR2-IN-2
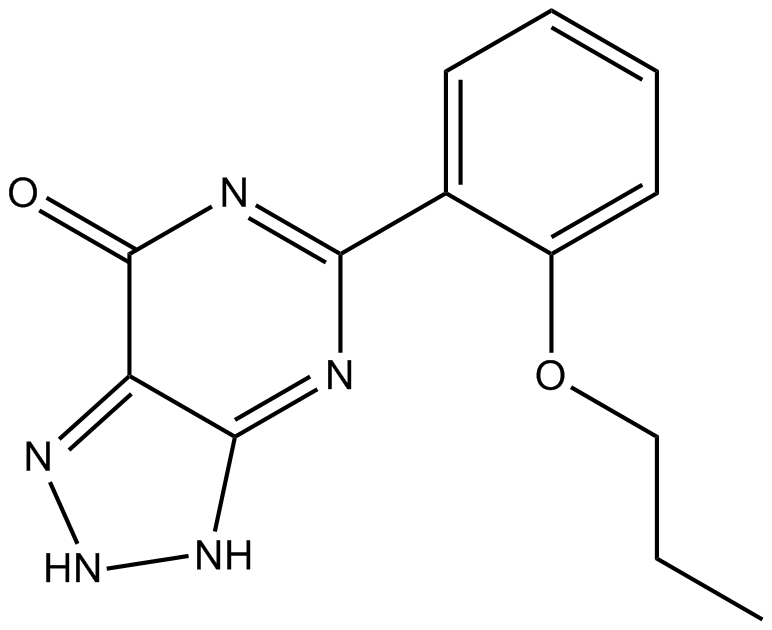
(R,R)-CXCR2-IN-2, diastereoisomer of CXCR2-IN-2 (compound 68), is a brain penetrant CXCR2 antagonist with a pIC50 of 9 and 6.8 in the Tango assay and d in the HWB Gro-α induced CD11b expression assay, respectively.
-
GC32208
ALX 40-4C
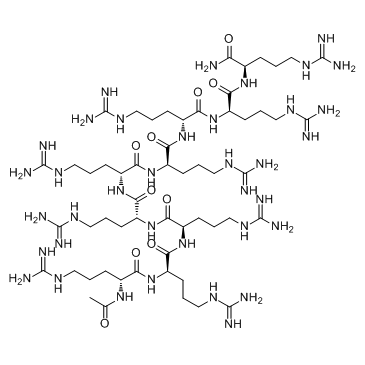
ALX 40-4C is a small peptide inhibitor of the chemokine receptor CXCR4, inhibits SDF-1 from binding CXCR4 with a Ki of 1 μM, and suppresses the replication of X4 strains of HIV-1; ALX 40-4C Trifluoroacetate also acts as an antagonist of the APJ receptor, with an IC50 of 2.9 μM.
-
GC34386
ALX 40-4C Trifluoroacetate
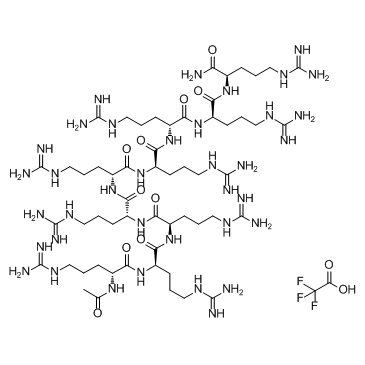
ALX 40-4C Trifluoroacetate is a small peptide inhibitor of the chemokine receptor CXCR4, inhibits SDF-1 from binding CXCR4 with a Ki of 1 μM, and suppresses the replication of X4 strains of HIV-1; ALX 40-4C Trifluoroacetate also acts as an antagonist of the APJ receptor, with an IC50 of 2.9 μM.
-
GC10473
AMD 3465
CXCR4 antagonist,potent and selective
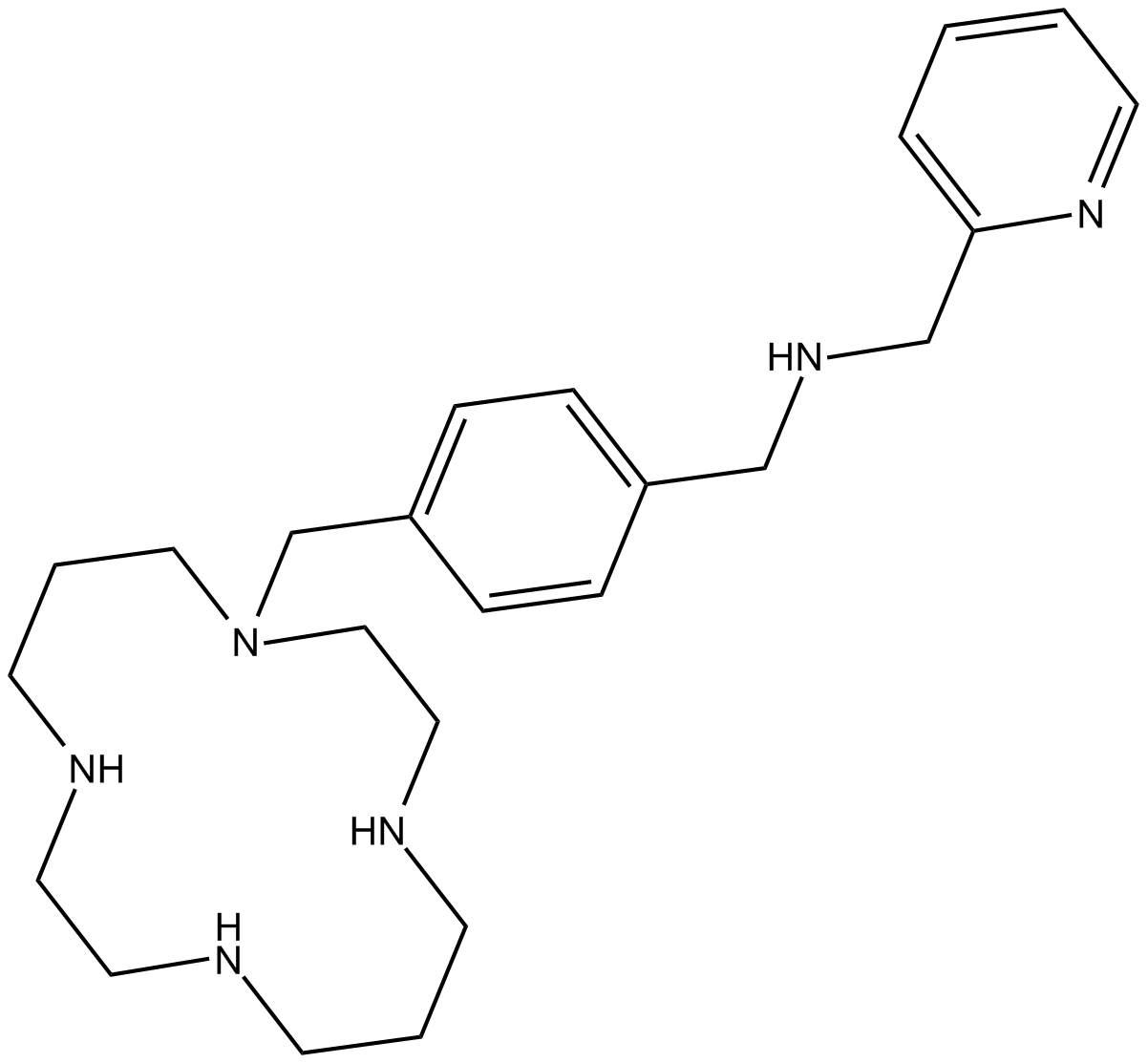
-
GC16706
AMD 3465 hexahydrobromide
potent, selective CXCR4 antagonist
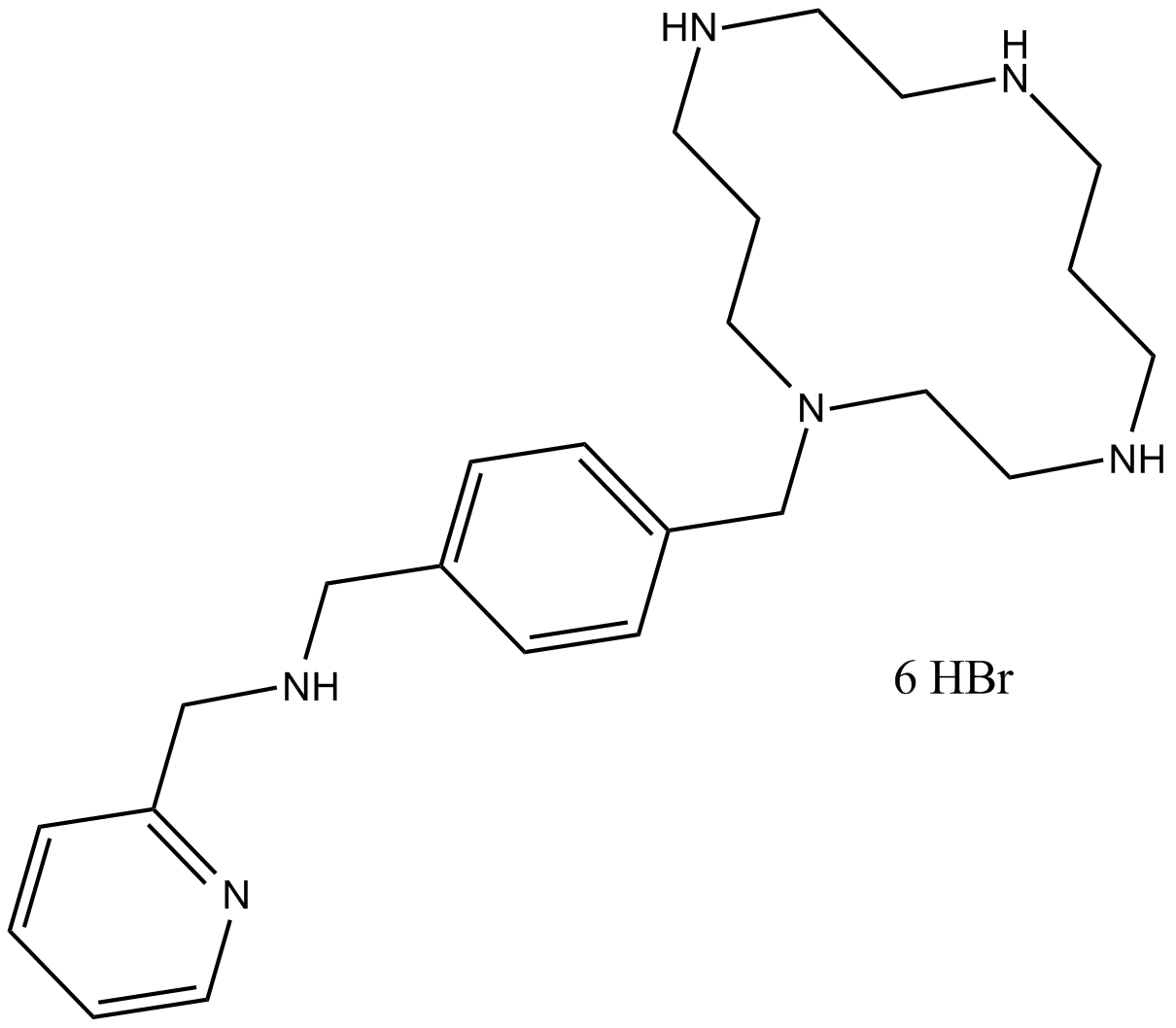
-
GC12196
AMD-070
A CXCR4 antagonist
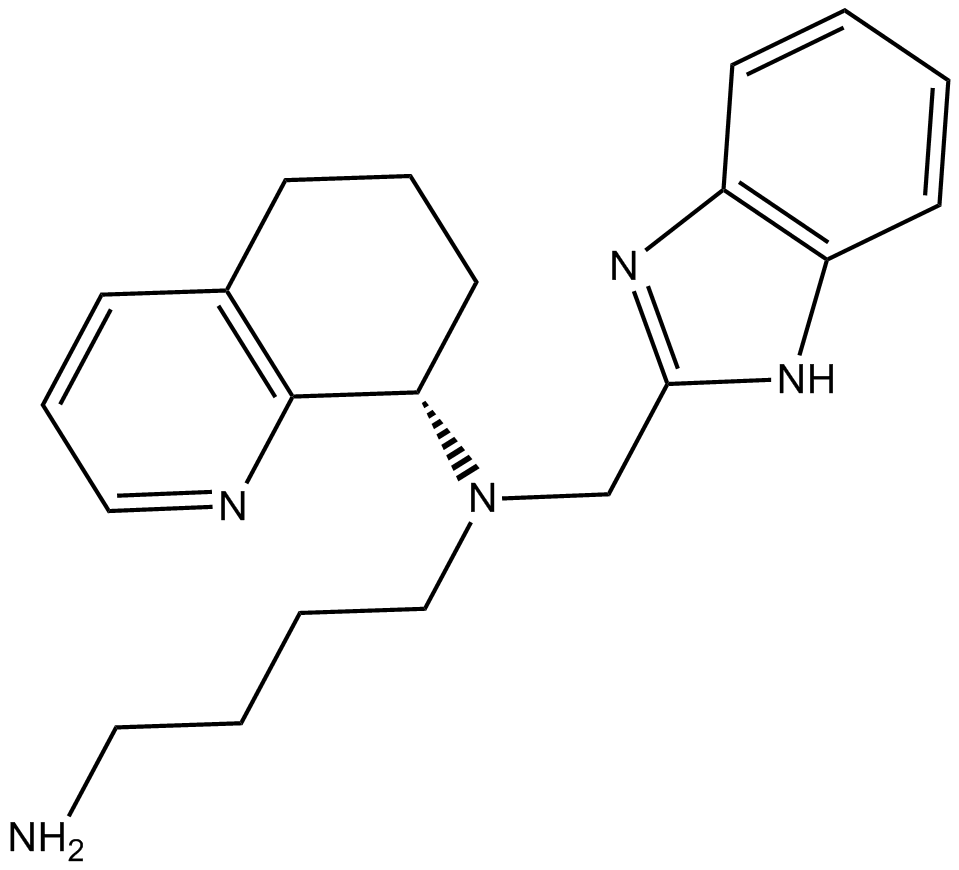
-
GC17018
AMD-070 hydrochloride
CXCR4 antagonist,potent and selective
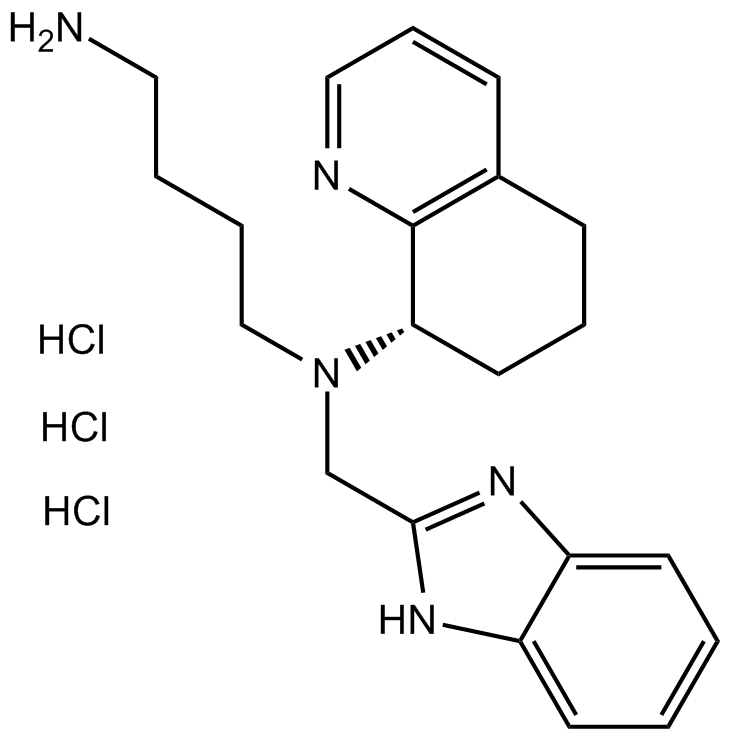
-
GC11939
AMG 487
AMG 487 is an oral, selective chemokine receptor 3 (CXCR3) antagonist that inhibits CXCL10 and CXCL11 binding to CXCR3 with IC50 values of 8.0 and 8.2 nM, respectively.
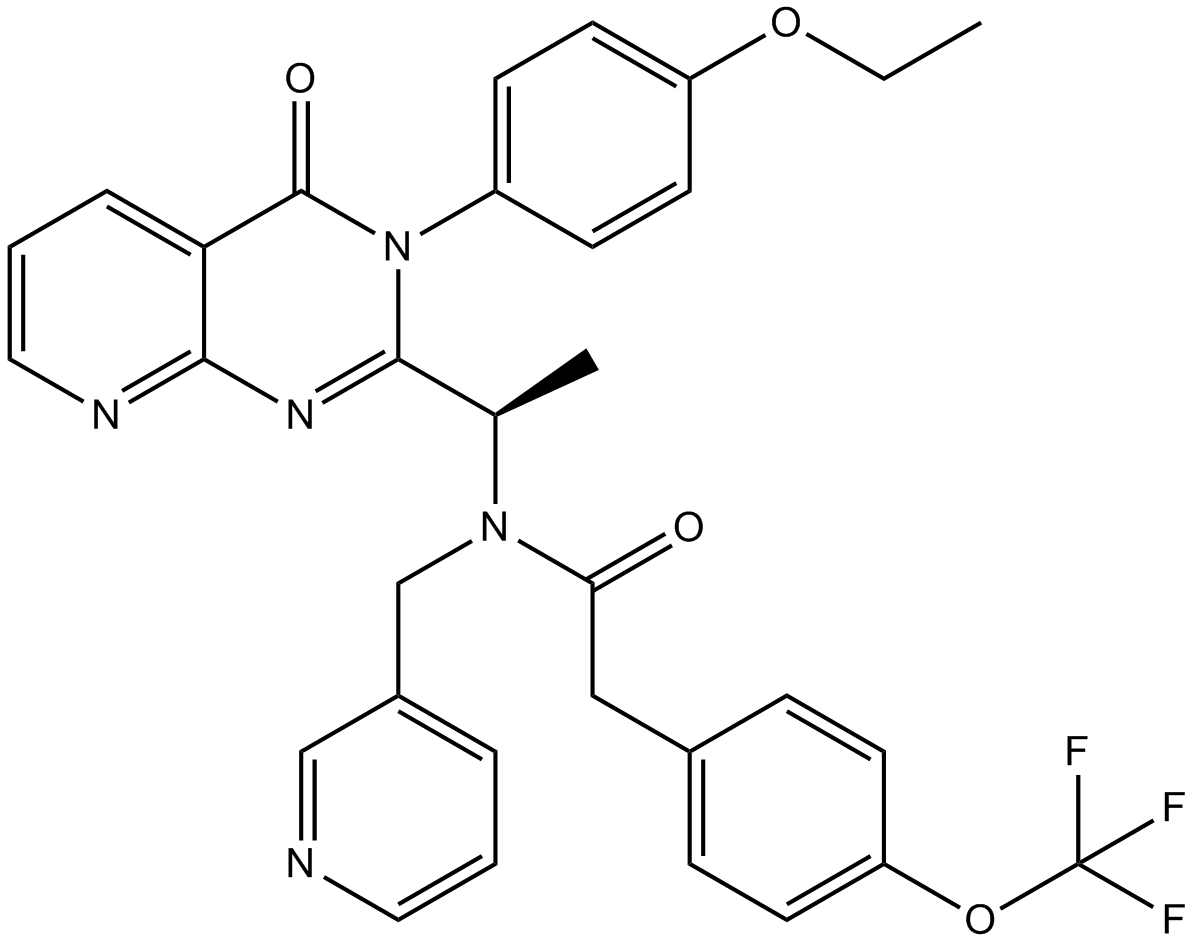
-
GC42817
Antileukinate
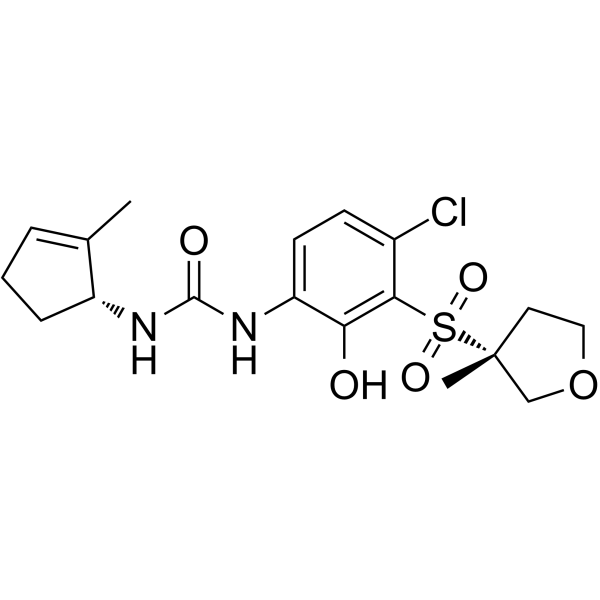
Antileukinate is a synthetic hexapeptide with an acetylated amino terminus and an amidated carboxyl terminus that inhibits the binding of CXC chemokines to the chemokine receptor CXCR2.
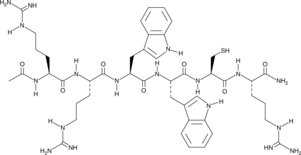
-
GC14711
ATI-2341
CXCR4 allosteric agonist

-
GC38541
ATI-2341 TFA
ATI-2341 is a potent and functionally selective allosteric agonist of C-X-C chemokine receptor type 4 (CXCR4), which functions as a biased ligand, favoring Gαi activation over Gα13.
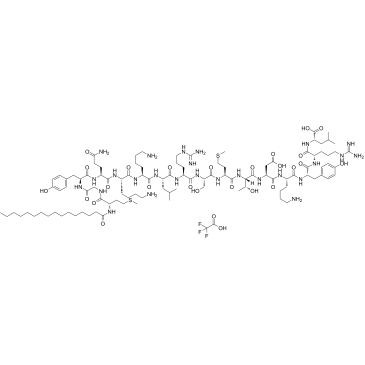
-
GC32707
AZD-5069
A CXCR2 antagonist
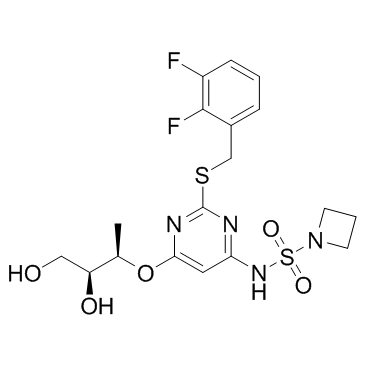
-
GC64312
AZD4721
AZD4721 (RIST4721) is the potent and orally active antagonist of acidic CXC chemokine receptor 2 (CXCR2).
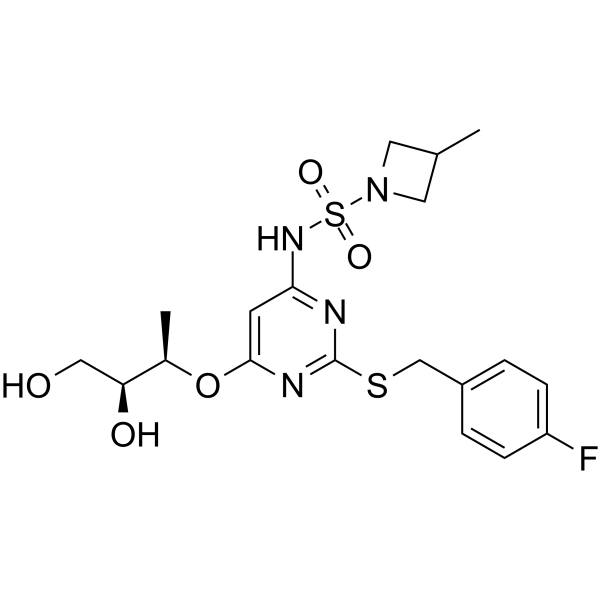
-
GC31653
AZD8797
AZD8797 (KAND567) is an allosteric non-competitive and orally active antagonist of the human CX3CR1 receptor; antagonizes CX3CR1 and CXCR2 with Kis of 3.9 and 2800 nM, respectively.
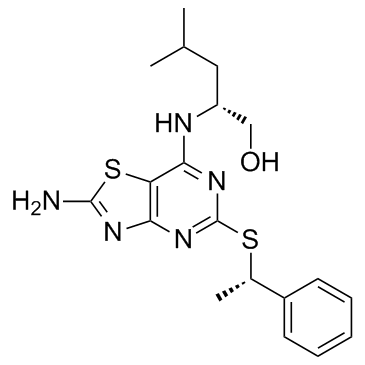
-
GC25120
Balixafortide (POL6326)
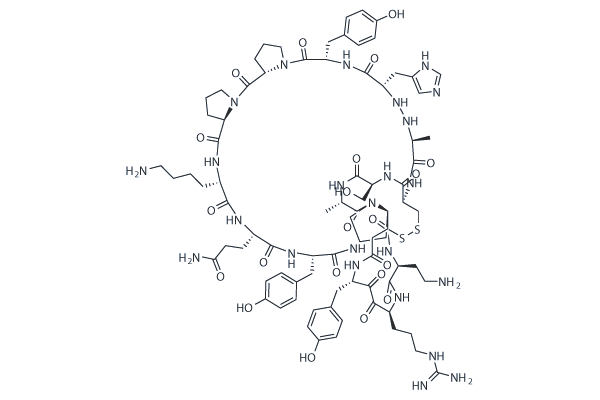
-
GN10507
Baohuoside I
Baohuoside I, a flavonoid isolated from Epimedium koreanum Nakai, acts as an inhibitor of CXCR4, downregulates CXCR4 expression, induces apoptosis and shows anti-tumor activity.
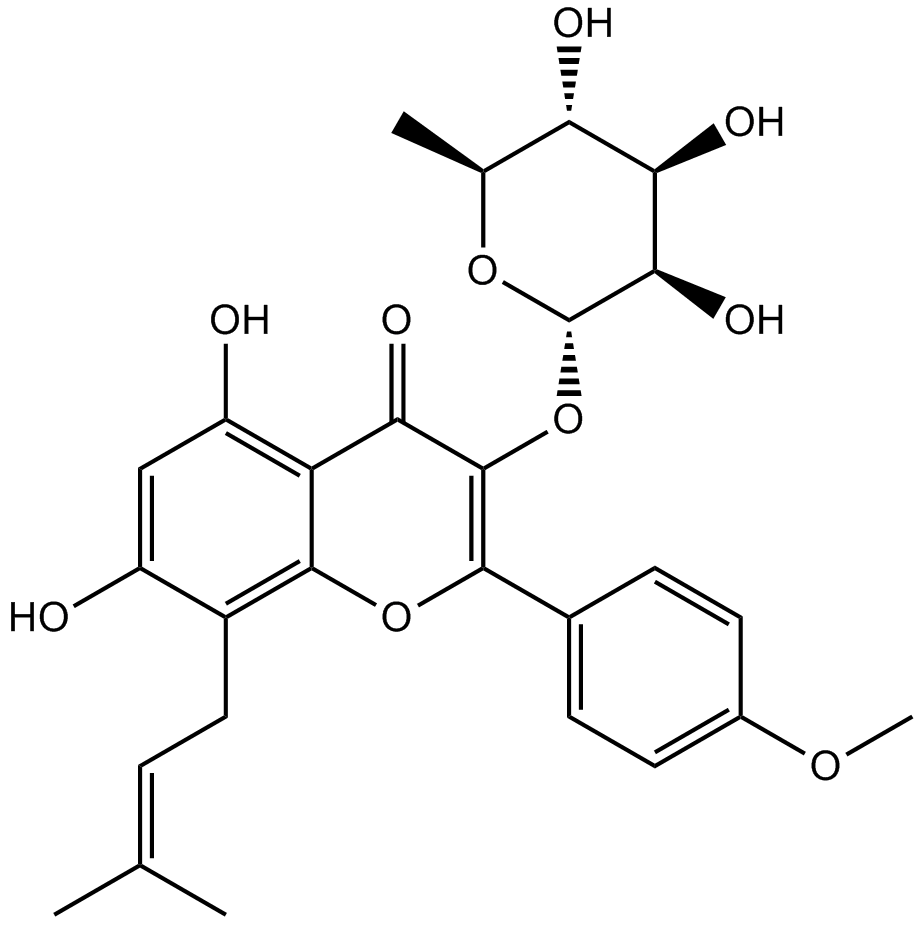
-
GC17526
BKT140
BKT140 (BKT140 4-fluorobenzoyl) is a novel CXCR4 antagonist with an IC50 vakue of ~1 nM.
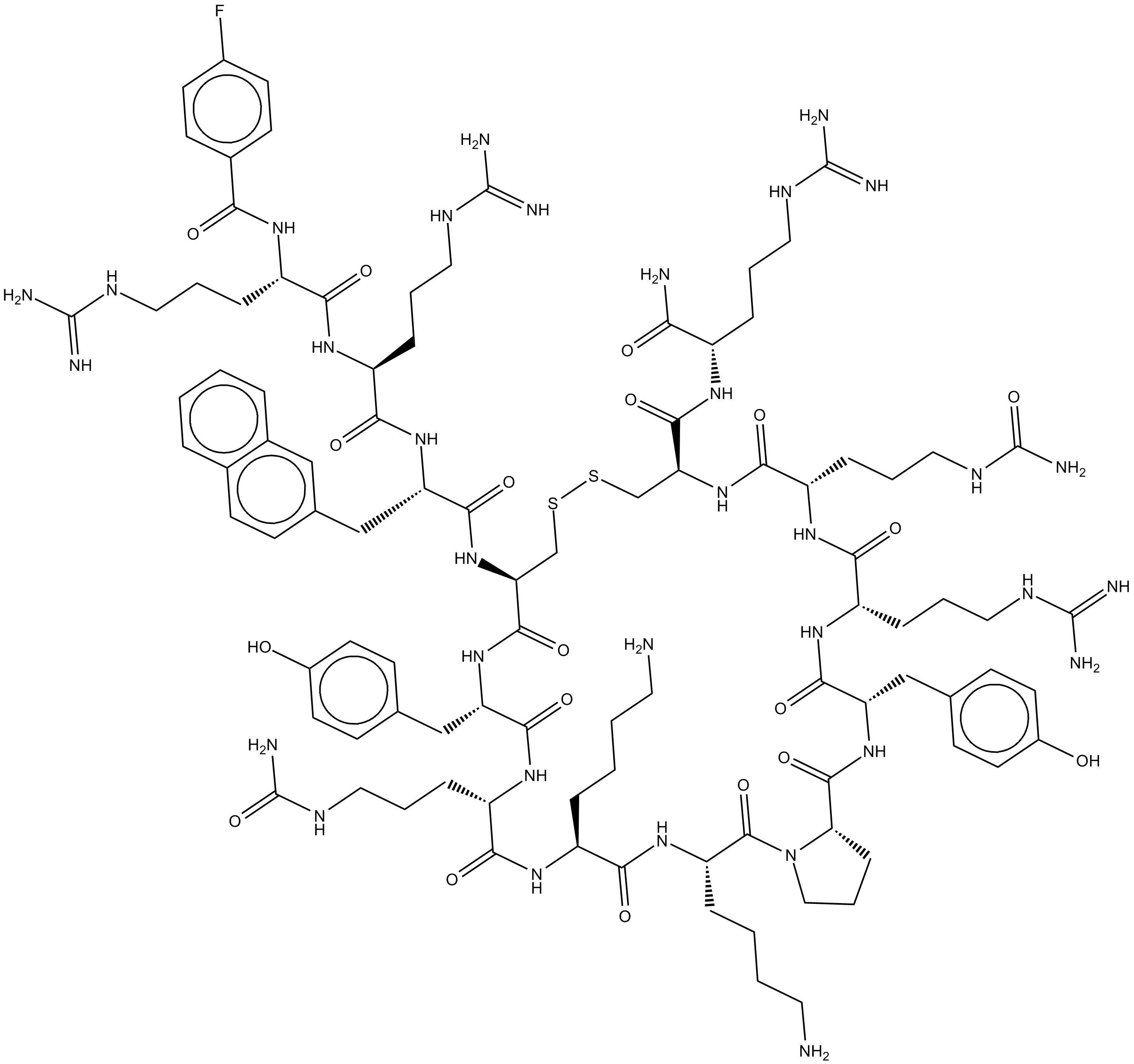
-
GC30625
Burixafor hydrobromide (TG-0054 hydrobromide)
Burixafor hydrobromide (TG-0054 hydrobromide) (TG-0054 hydrobromide) is an orally bioavailable and potent antagonist of CXCR4 and a well anti-angiogenic drug that is of potential value in treating choroid neovascularization.
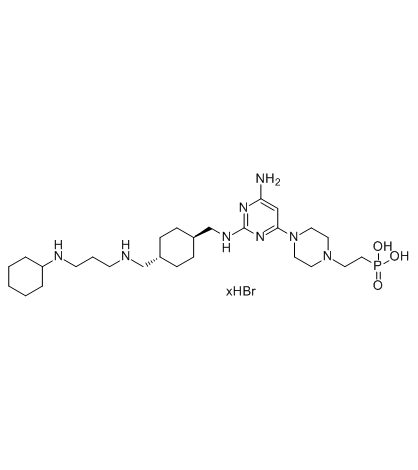
-
GC64090
Corydalmine
Corydalmine (L-Corydalmine) inhibits spore germination of some plant pathogenic as well as saprophytic fungi.
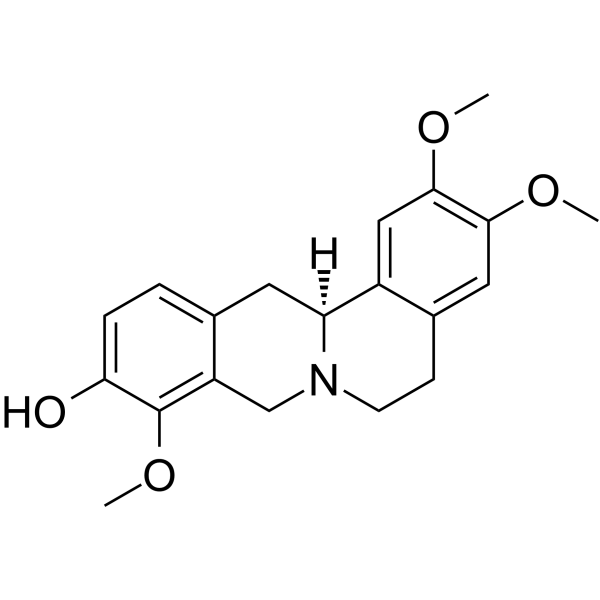
-
GC65908
CXCR2 antagonist 2
CXCR2 antagonist 2 is a potent CXCR2 antagonist for cancer immunotherapy with an IC50 value of 95 nM.
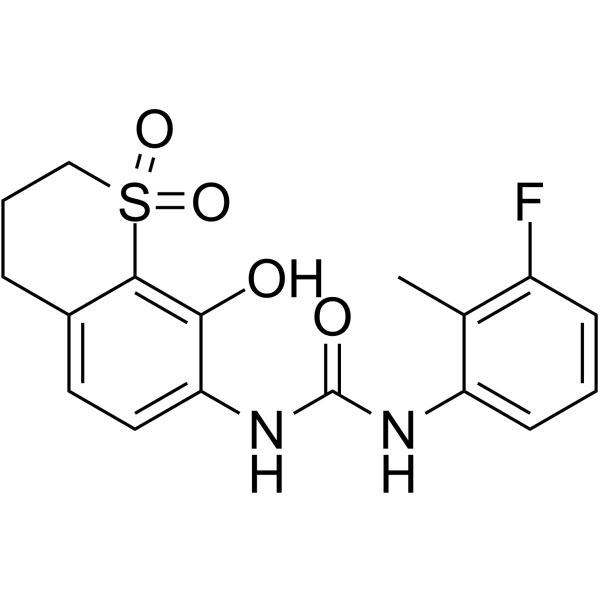
-
GC31754
CXCR2-IN-1
CXCR2-IN-1 is a central nervous system penetrant CXCR2 antagonist with a pIC50 of 9.3.
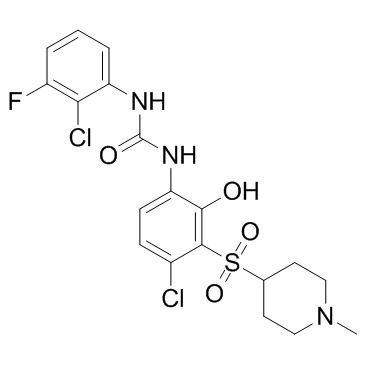
-
GC62475
CXCR2-IN-2
CXCR2-IN-2 is a selective, brain penetrant, and orally bioavailable CXCR2 antagonist (IC50=5.2 nM/1 nM in β-arrestin assay/CXCR2 Tango assay, respectively).
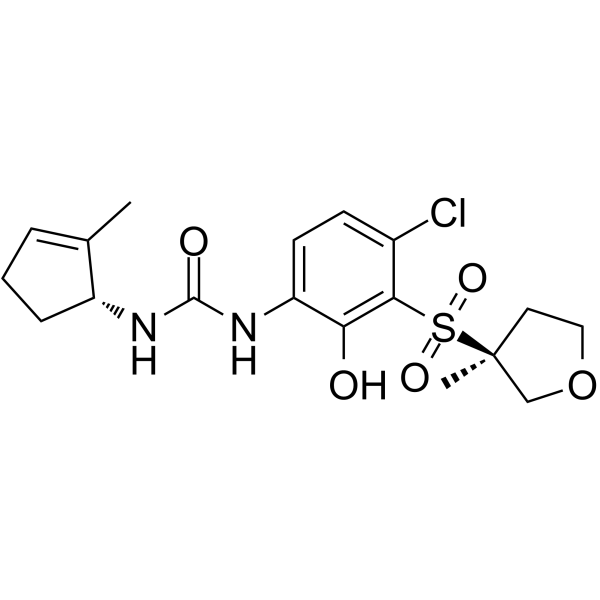
-
GC63411
CXCR7 antagonist-1
CXCR7 antagonist-1 is a CXCR7 antagonist that inhibits the binding of the SDF-1 chemokine (also known as the CXCL12 chemokine) or I-TAC (also known as CXCL11) to the chemokine receptor CXCR7.
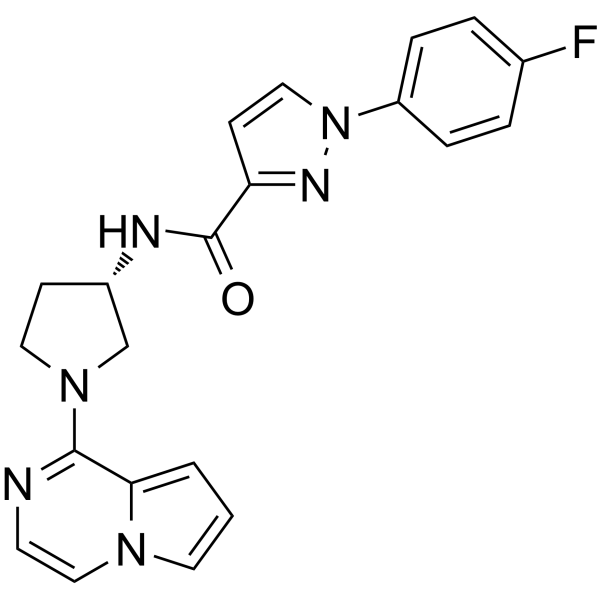
-
GC65476
CXCR7 antagonist-1 hydrochloride
CXCR7 antagonist-1 hydrochloride is a CXCR7 antagonist that inhibits the binding of the SDF-1 chemokine (also known as the CXCL12 chemokine) or I-TAC (also known as CXCL11) to the chemokine receptor CXCR7.
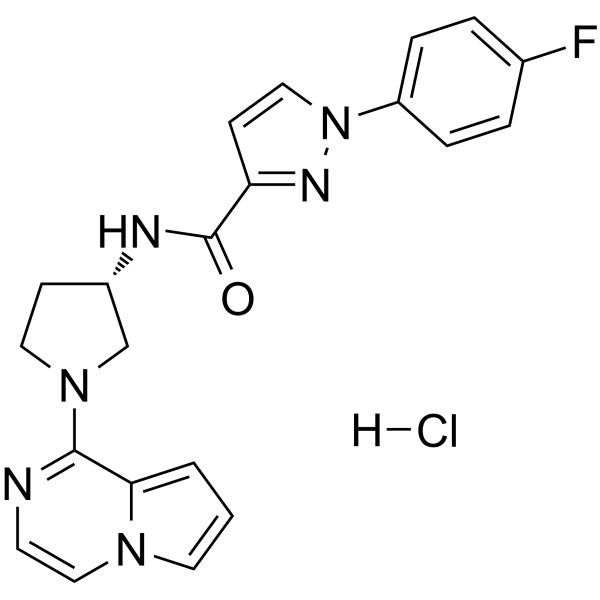
-
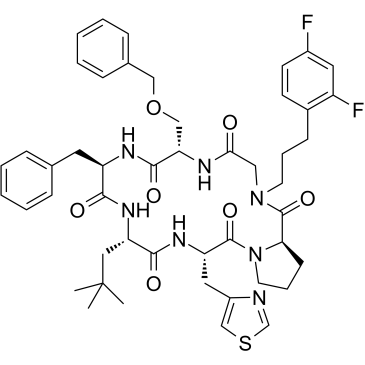
GC35763
CXCR7 modulator 1
CXCR7 modulator 1 (compound 25) is a potent and orally bioavailable peptoid hybrid CXCR7 modulator, with a Ki of 9 nM.
-
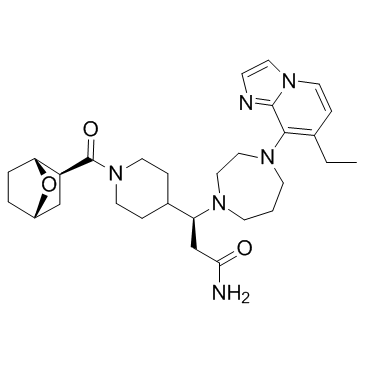
GC34339
CXCR7 modulator 2
CXCR7 modulator 2 is a modulator of C-X-C Chemokine Receptor Type 7 (CXCR7), with a Ki of 13 nM.
-
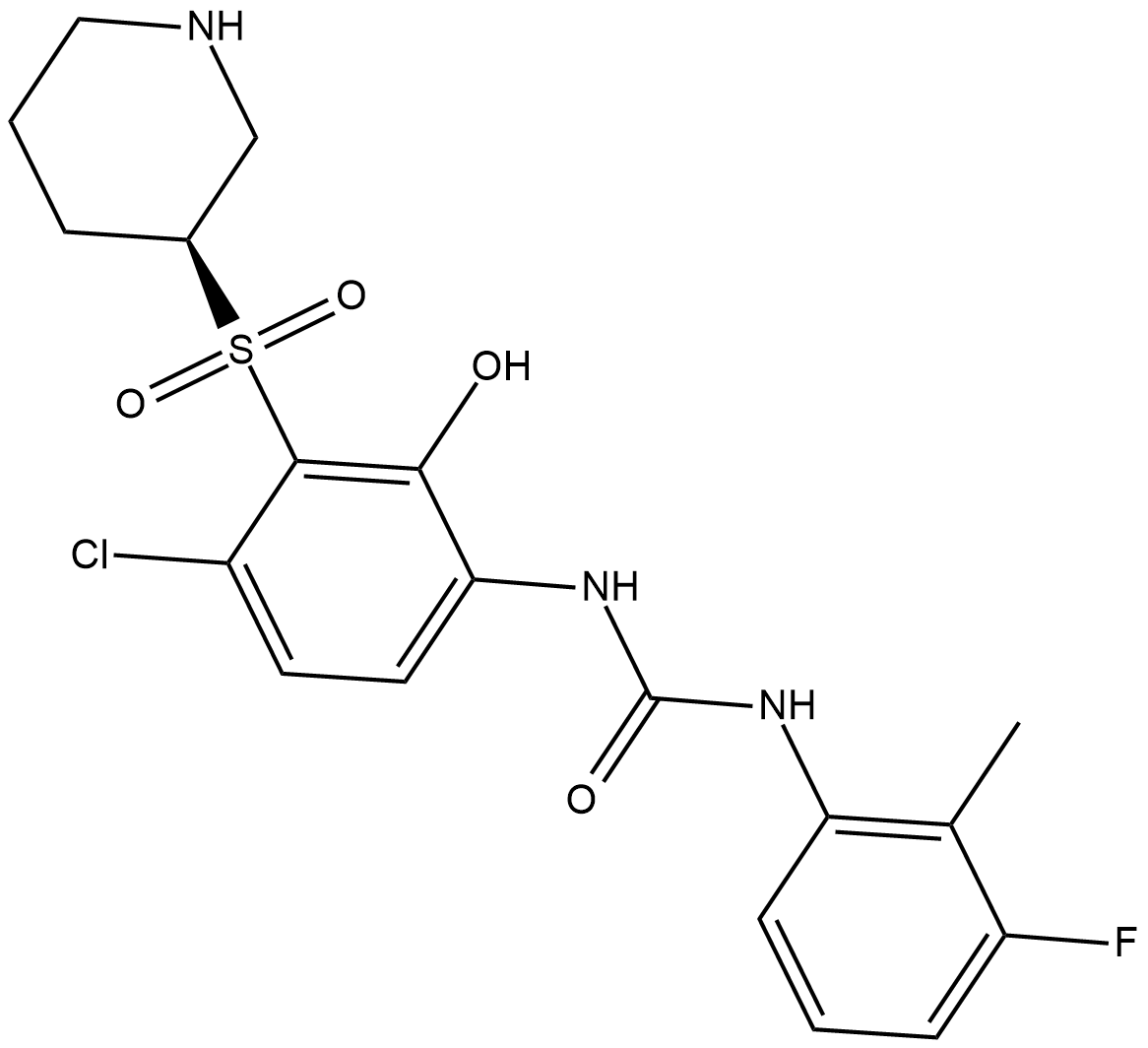
GC19116
Danirixin
Danirixin is a selective, and reversible CXCR2 antagonist, with IC50 of 12.5 nM for CXCL8.
-
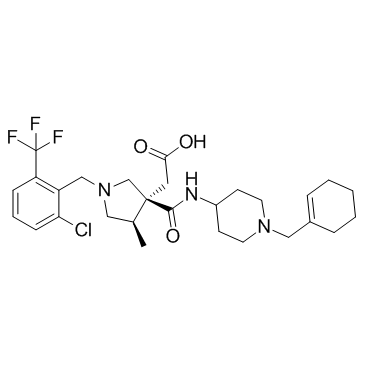
GC31763
E6130
E6130 is an orally active and highly selective CX3CR1 modulator, that may be effective for treatment of inflammatory bowel disease.
-
GC66332
Eldelumab
Eldelumab (BMS-936557) is a humanised anti-CXCL10 (IP-10) monoclonal antibody (IgG1 type). Eldelumab selectively binds to CXCL10 and blocks CXCL10-induced calcium flux and cell migration. Eldelumab can be used in studies of autoimmune and auto-inflammatory diseases such as rheumatoid arthritis, ulcerative colitis and crohn's disease.

-
GC36033
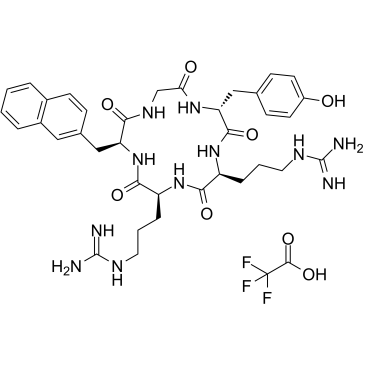
FC131 TFA
FC131 TFA is a CXCR4 antagonist, inhibits [125I]-SDF-1 binding to CXCR4, with an IC50 of 4.5 nM.
-
GC36184
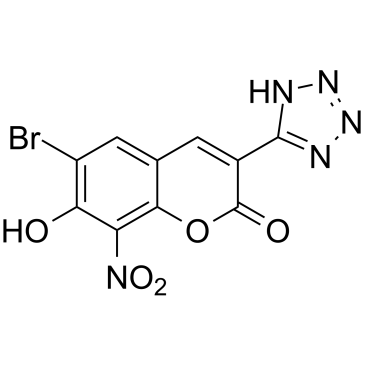
GPR35 agonist 1
GPR35 agonist 1 (compound 50) is a potent and specific G protein-coupled receptor-35 (GPR35)/CXCR8 agonist with an EC50 of 5.8 nM, displays good druggability.
-
GC36350
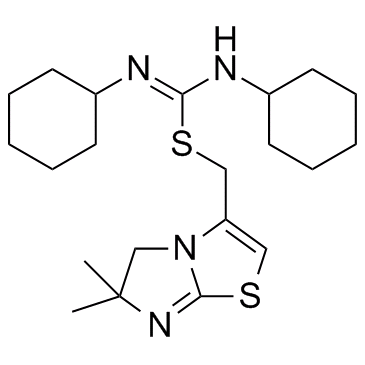
IT1t
IT1t is a potent CXCR4 antagonist; inhibits CXCL12/CXCR4 interaction with an IC50 of 2.1 nM.
-
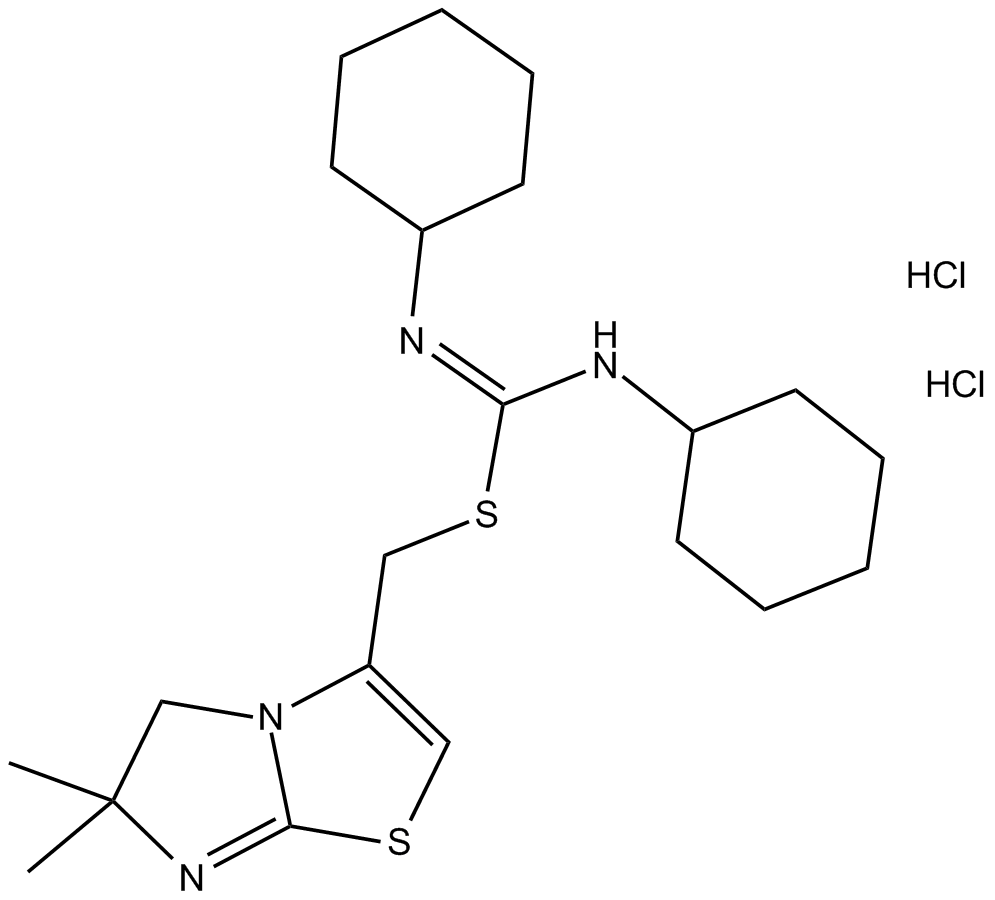
GC10432
IT1t dihydrochloride
CXCR4 antagonist, potent
-
GC36411
Kynurenic acid sodium
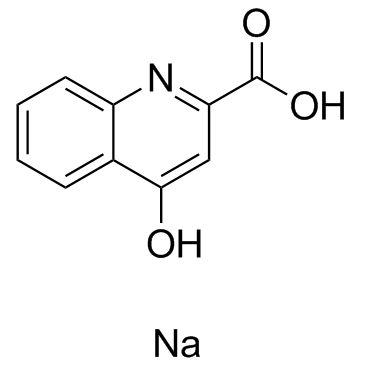
-
GC62256
Ladarixin
Ladarixin (DF 2156A free base) is an orally active, allosteric non-competitive and dual CXCR1 and CXCR2 antagonist.
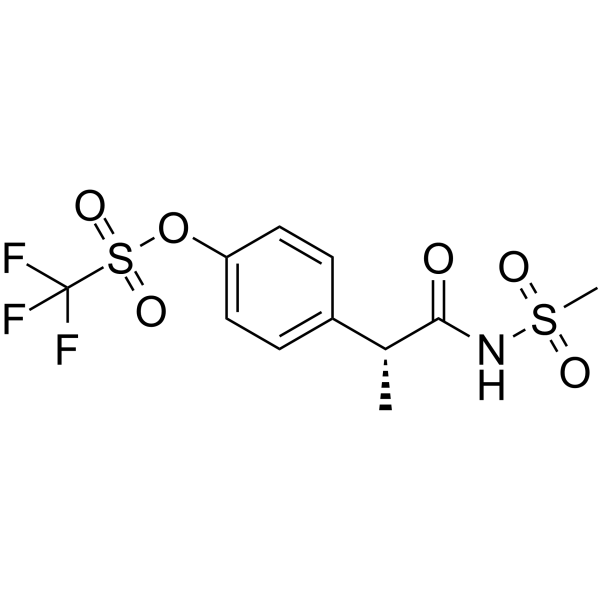
-
GC62296
Ladarixin sodium
Ladarixin sodium (DF 2156A) is an orally active, allosteric non-competitive and dual CXCR1 and CXCR2 antagonist.
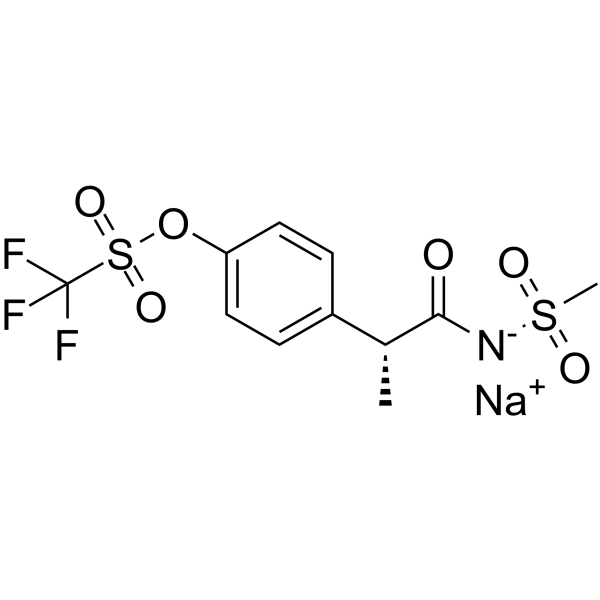
-
GC19486
LIT-927
LIT-927 is a locally and orally active CXCL12 neutraligand

-
GC19230
LY2510924
LY2510924 is a potent and selective CXCR4 antagonist; blocks SDF-1 binding to CXCR4 with an IC50 of 0.079 nM.
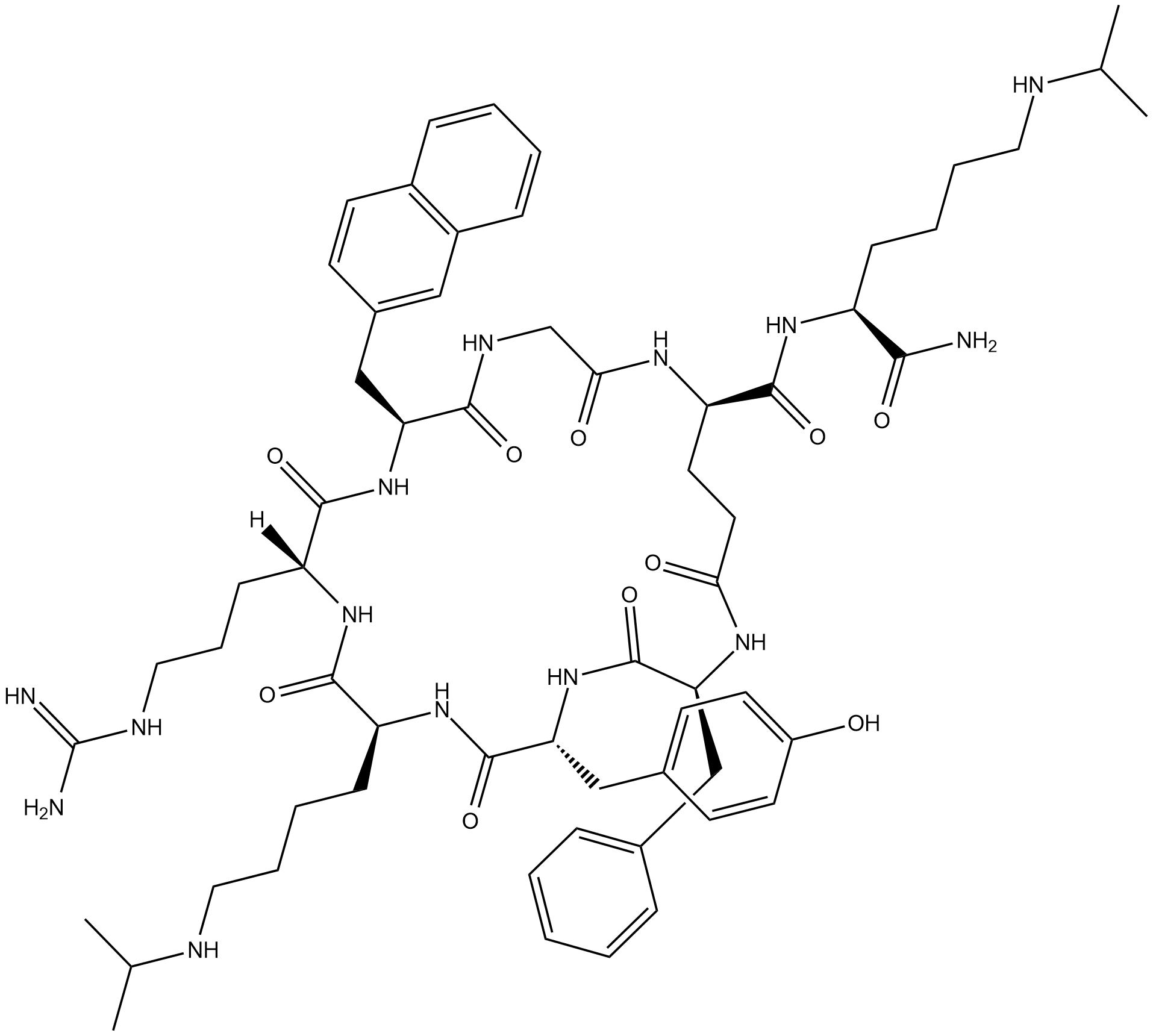
-
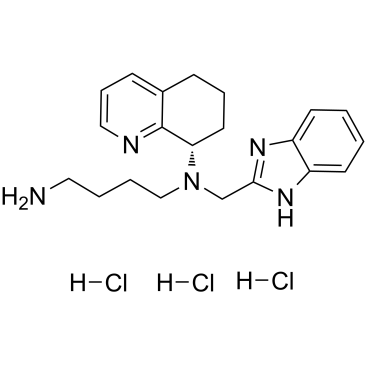
GC36550
Mavorixafor trihydrochloride
Mavorixafor trihydrochloride (AMD-070 trihydrochloride) is a potent, selective and orally available CXCR4 antagonist, with an IC50 value of 13 nM against CXCR4 125I-SDF binding, and also inhibits the replication of T-tropic HIV-1 (NL4.3 strain) in MT-4 cells and PBMCs with an IC50 of 1 and 9 nM, respectively.
-
GC17174
ML 145
GPR35 antagonist
-
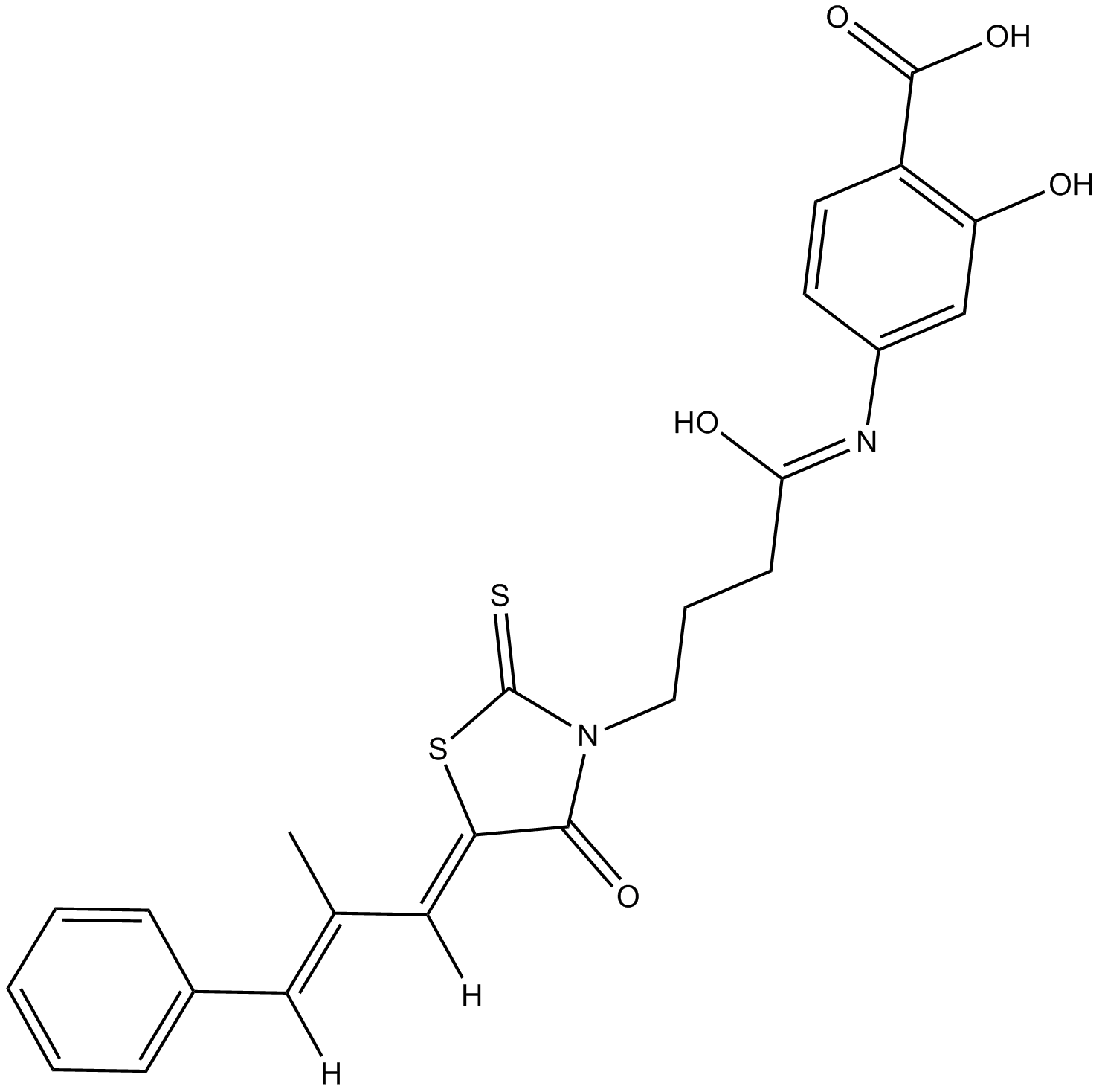
GC19255
MSX-122
MSX-122 is a partial antagonist of CXCR4, inhibiting CXCR4/CXCL12 actions, with an IC50 of ?10 nM; MSX-122 has anti-inflammatory and anti-metastatic activity.
-
GC68040
MSX-127

-
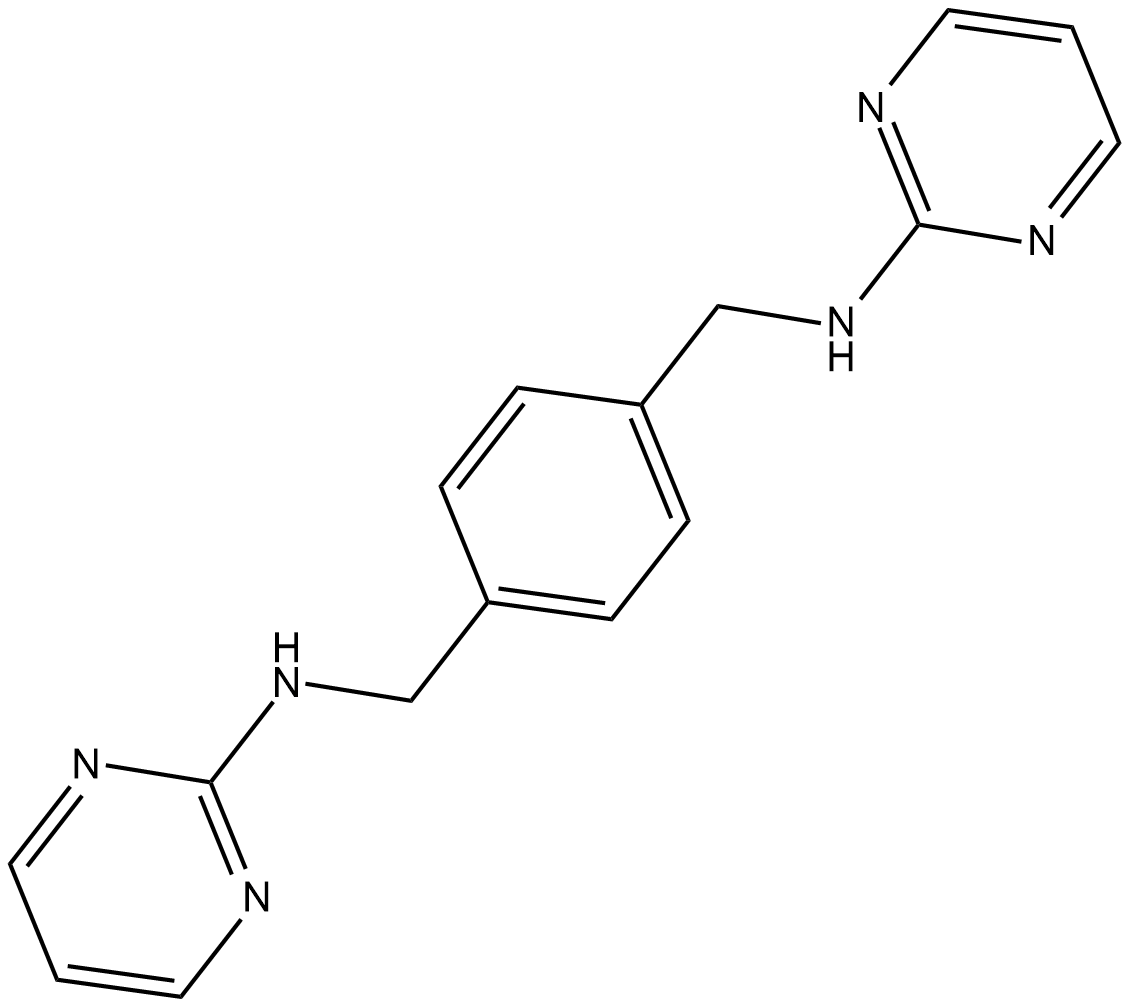
GC36704
NBI-74330
A CXCR3 antagonist
-
GC36737
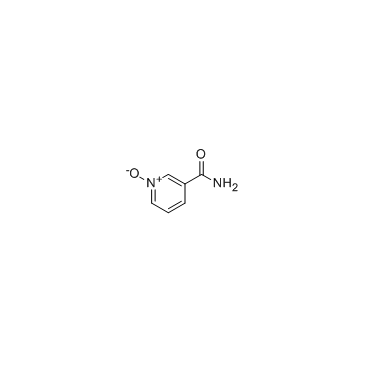
Nicotinamide N-oxide
Nicotinamide N-oxide, an in vivo nicotinamide metabolite, is a potent, and selective antagonist of the CXCR2 receptor.
-
GC14745
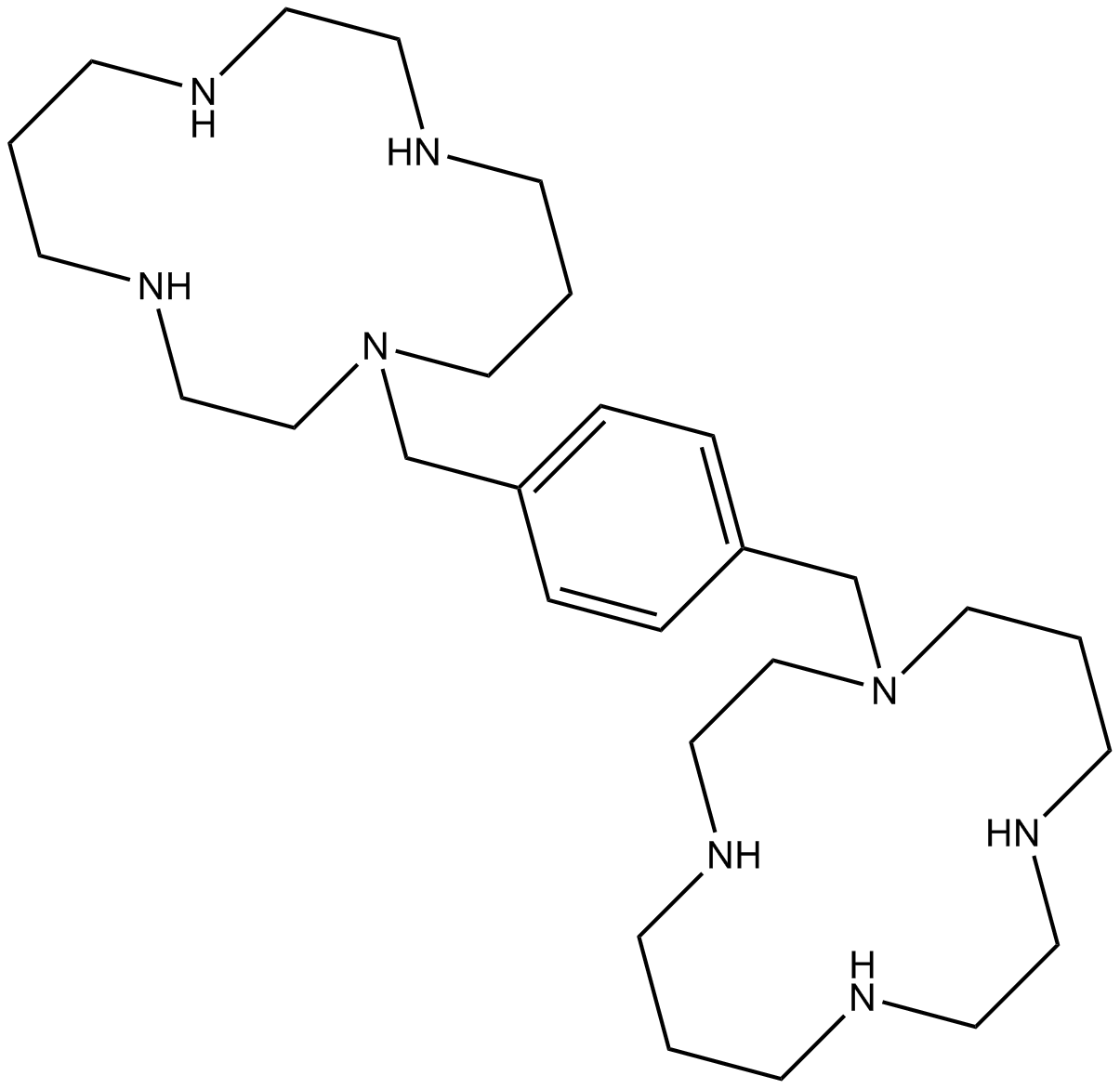
Plerixafor (AMD3100)
Plerixafor (AMD3100) (AMD 3100) is a selective CXCR4 antagonist with an IC50 of 44 nM.
-
GC17949
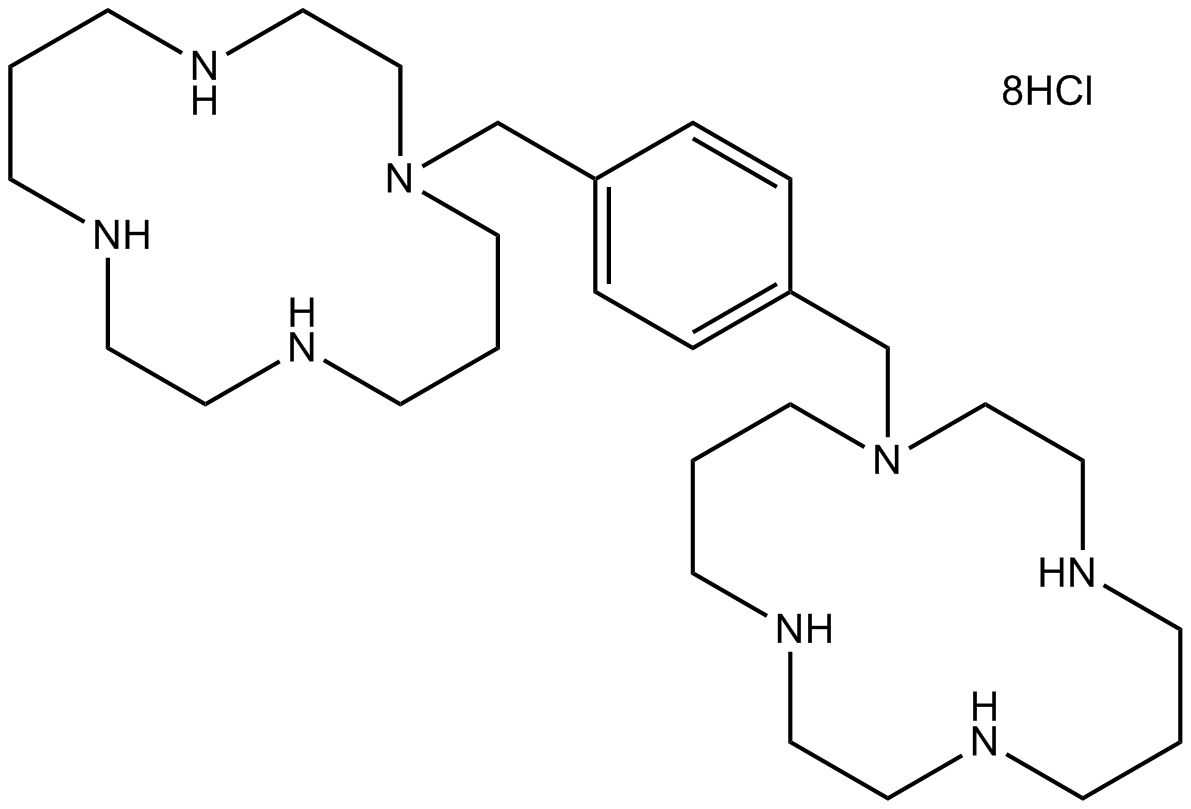
Plerixafor 8HCl (AMD3100 8HCl)
Plerixafor 8HCl (AMD3100 8HCl) (AMD3100 octahydrochloride) is a selective CXCR4 antagonist with an IC50 of 44 nM.
-
GC37022
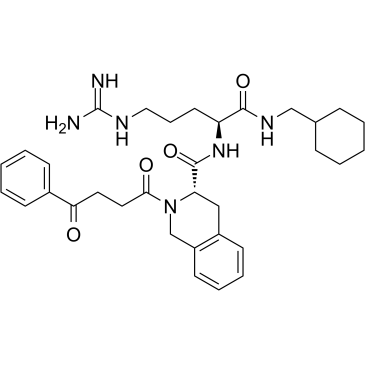
PS372424
PS372424, a three amino-acid fragment of CXCL10, is a specific human CXCR3 agonist with anti-inflammatory activity.
-
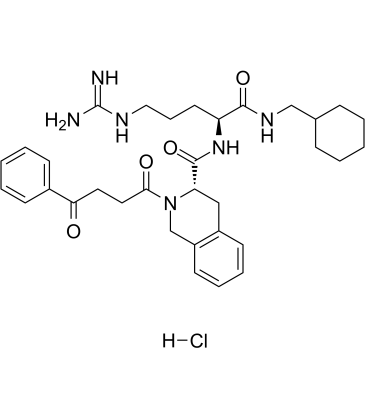
GC61219
PS372424 hydrochloride
PS372424 hydrochloride, a three amino-acid fragment of CXCL10, is a specific human CXCR3 agonist with anti-inflammatory activity.
-
GC69789
Quetmolimab
Quetmolimab is a monoclonal antibody that targets humanized fractalkine (CX3CL1), which is a chemokine that regulates chemotaxis and adhesion.

-
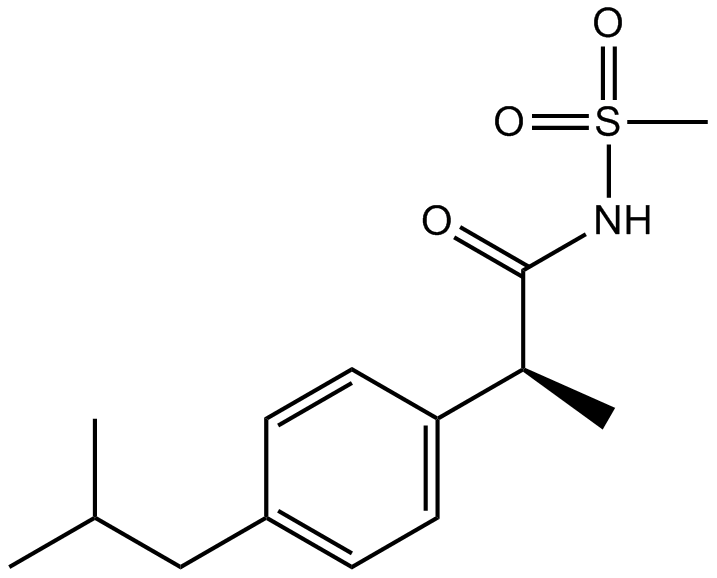
GC12849
Reparixin
An allosteric inhibitor of CXCR1 and CXCR2
-
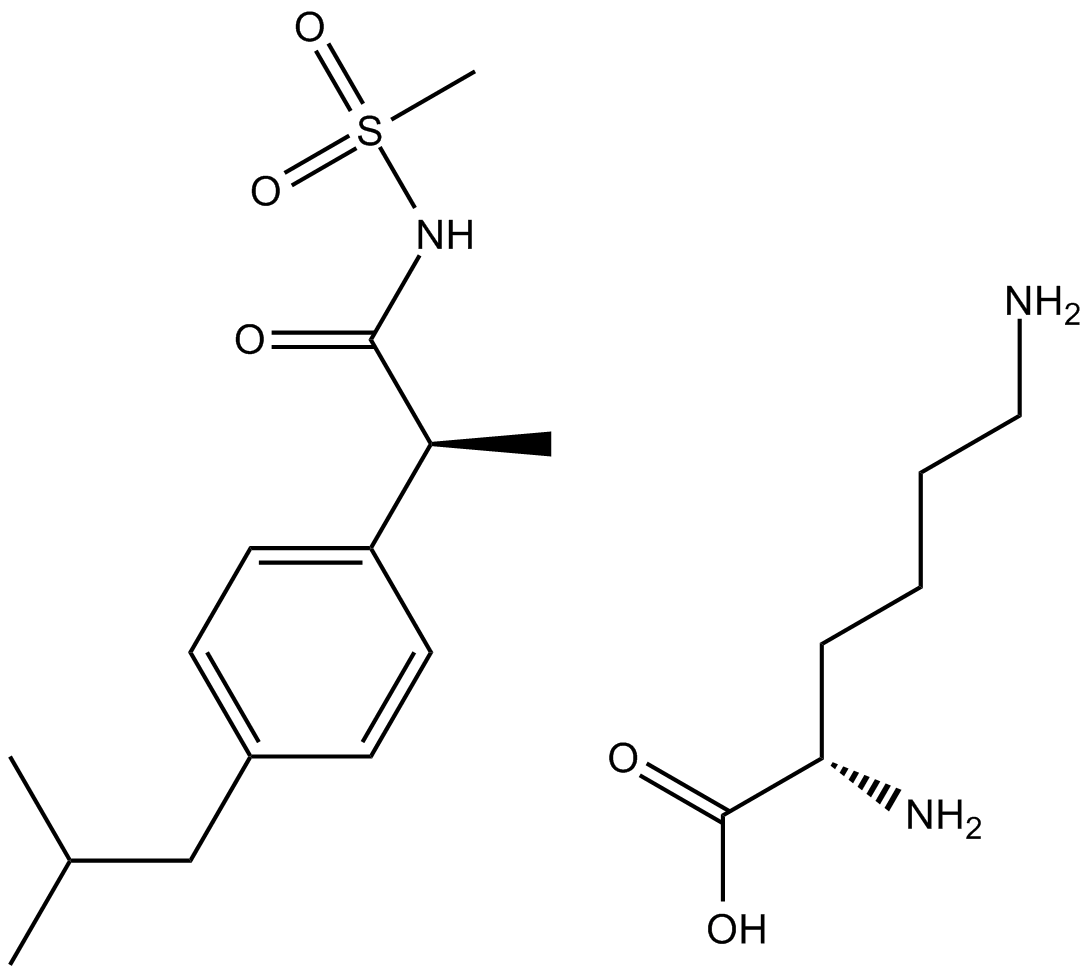
GC13813
Reparixin L-lysine salt
Perifosine is an orally active alkyl phospholipid analogue that has shown antitumor activity in a variety of cancers by interfering .
-
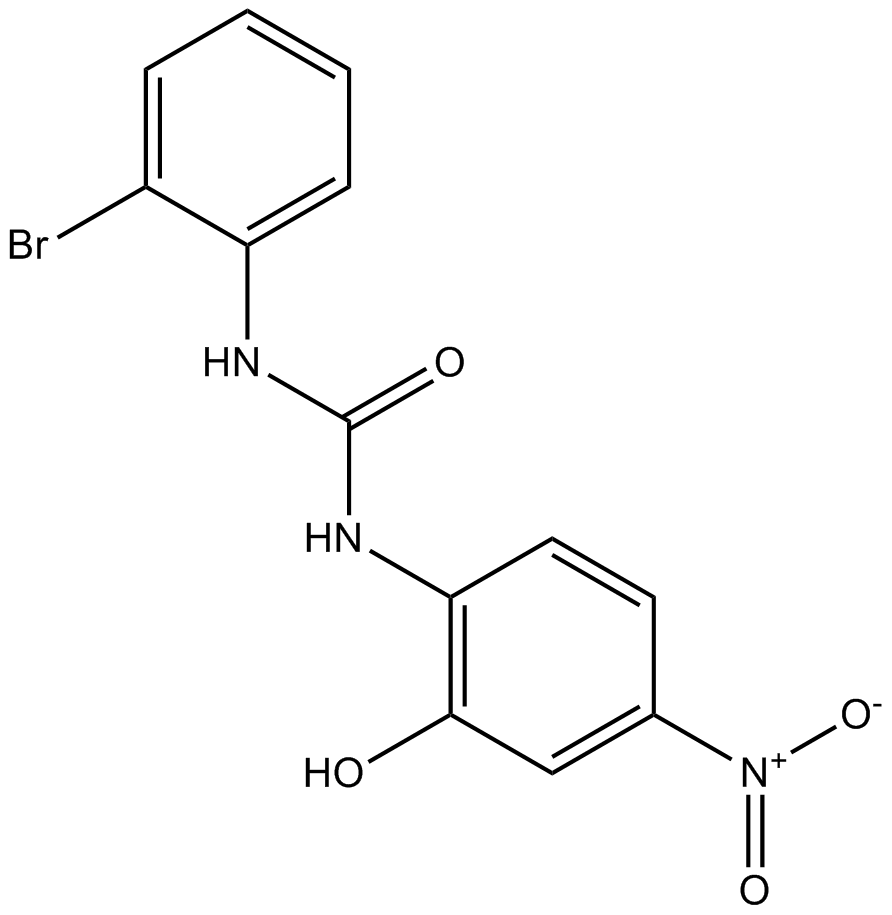
GC16465
SB 225002
CXCR2 antagonist, potent and selective
-
GC13458
SB 265610
CXCR2 antagonist, potent
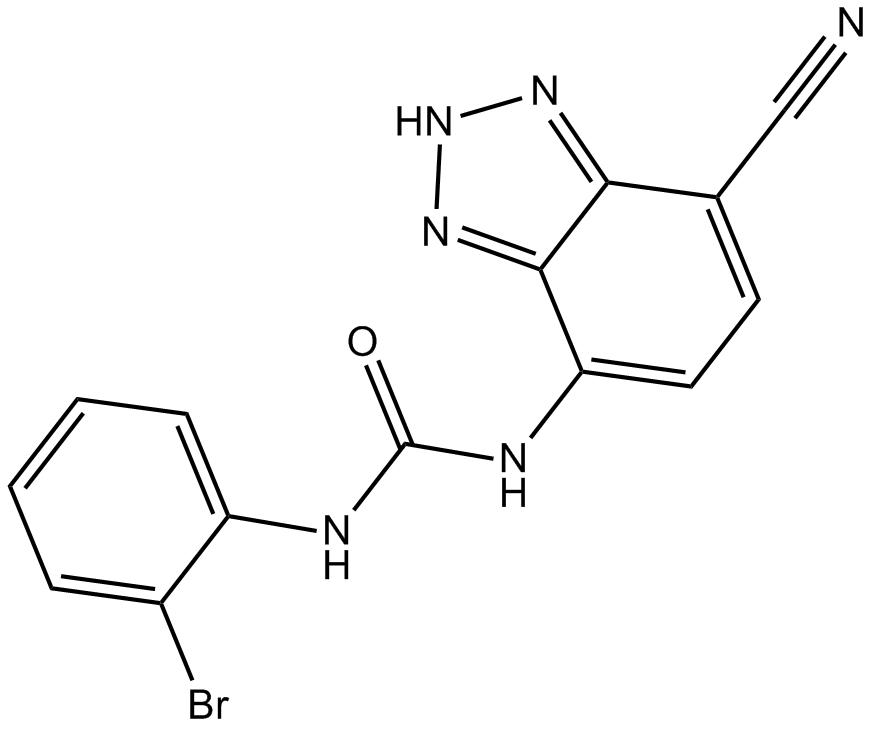
-
GC50230
SB 332235
Potent CXCR2 antagonist
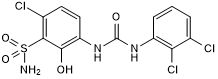
-
GC16100
SCH 527123
An allosteric antagonist of CXCR1 and CXCR2
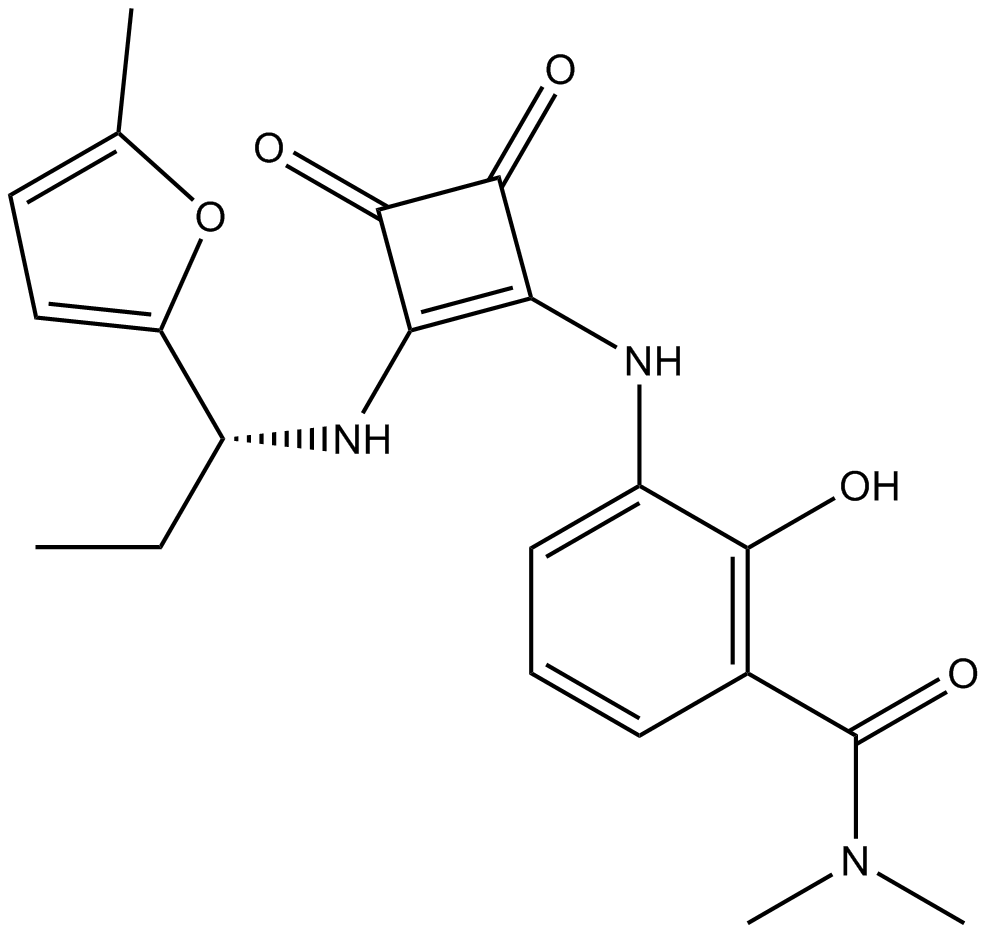
-
GC37605
SCH 546738
SCH 546738 is a potent, orally active and non-competitive CXCR3 antagonist, the affinity constant (Ki) of SCH 546738 binding to human CXCR3 receptor is determined to be 0.4 nM in multiple experiments.
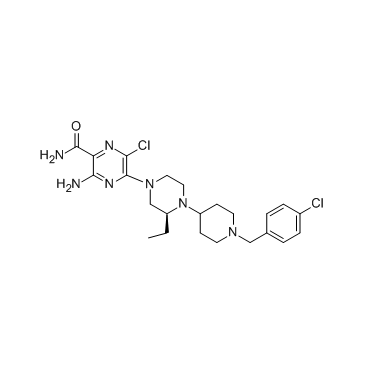
-
GC15401
SCH 563705
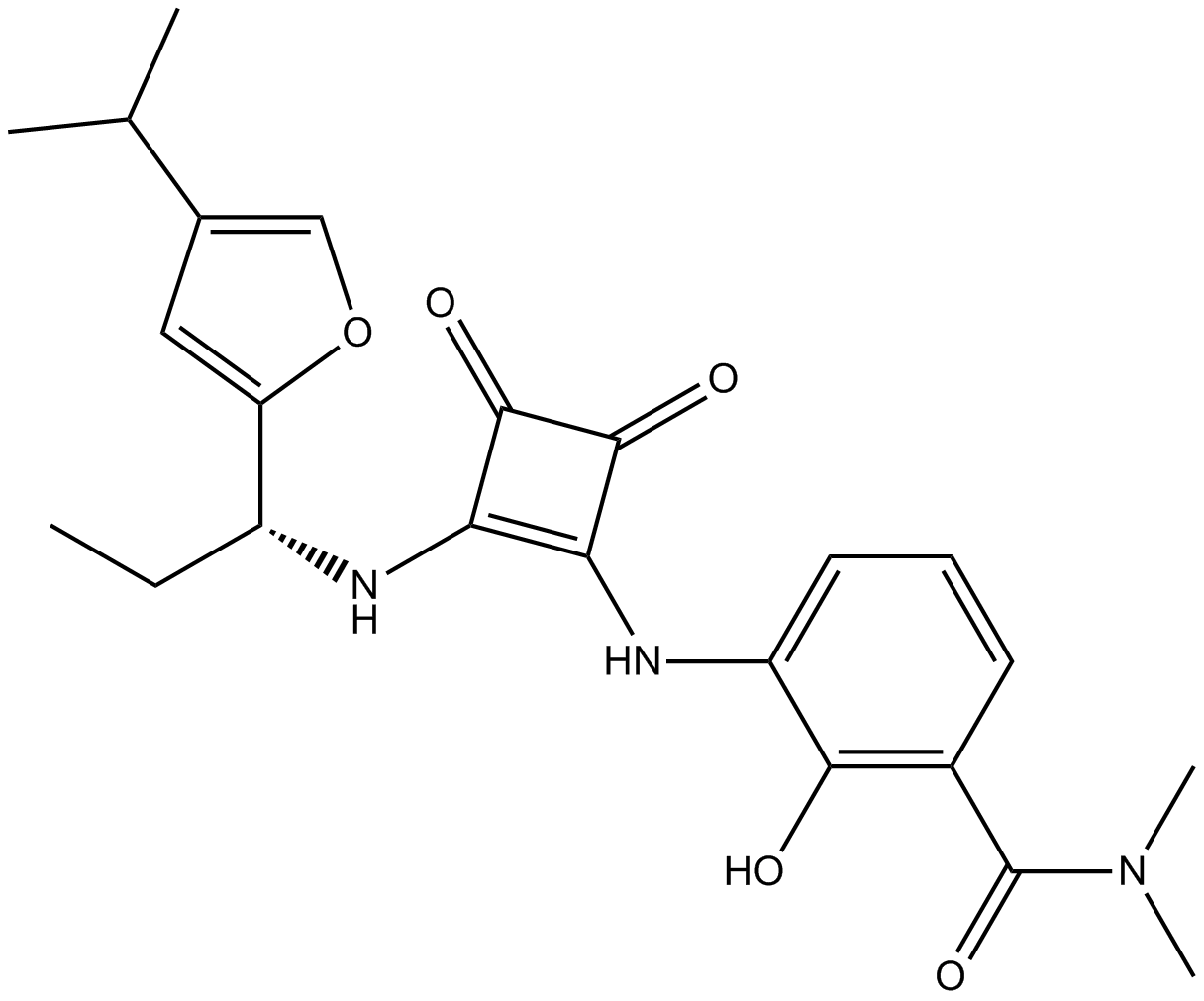
-
GC16710
SRT3109
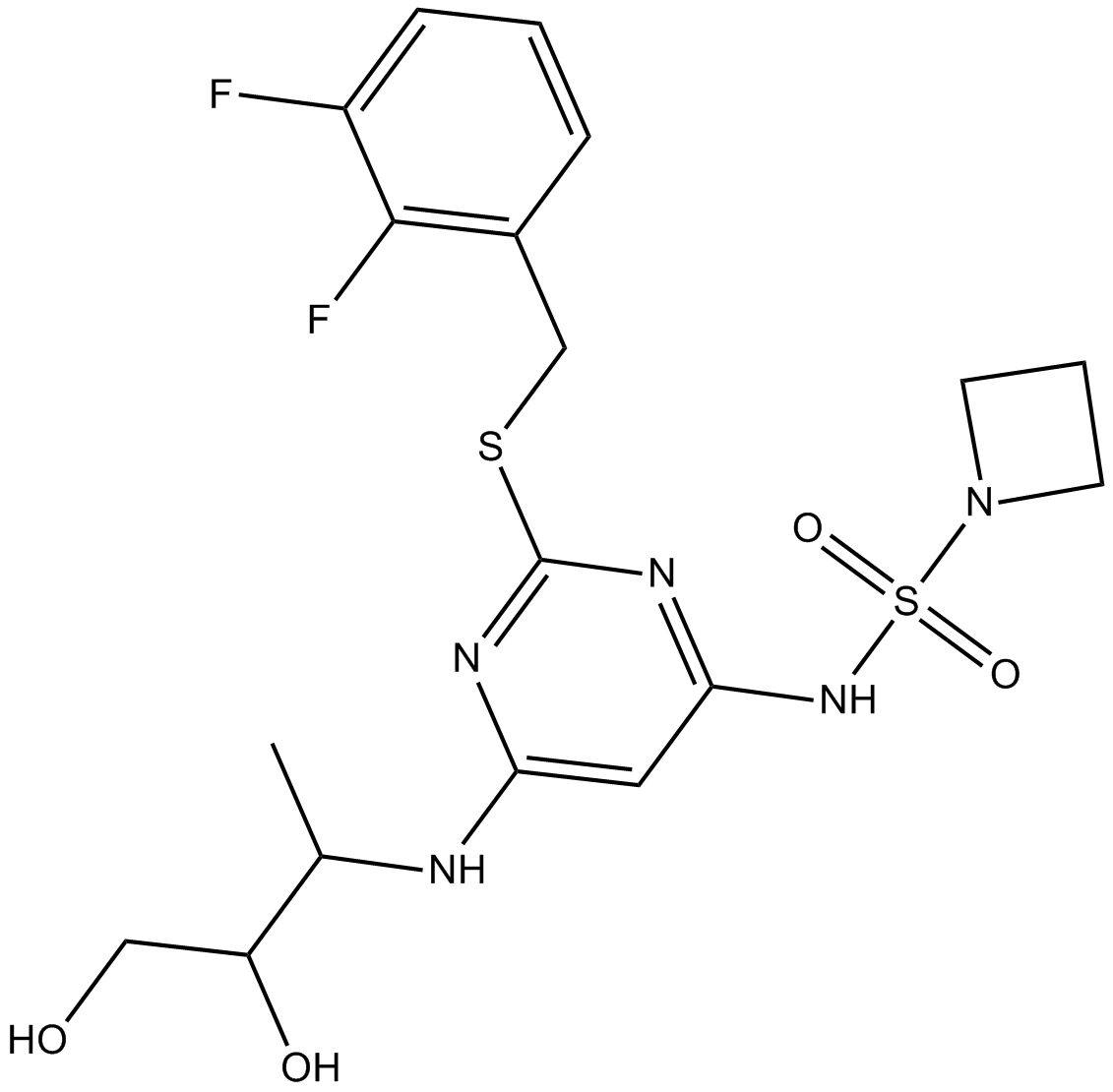
-
GC16377
SRT3190
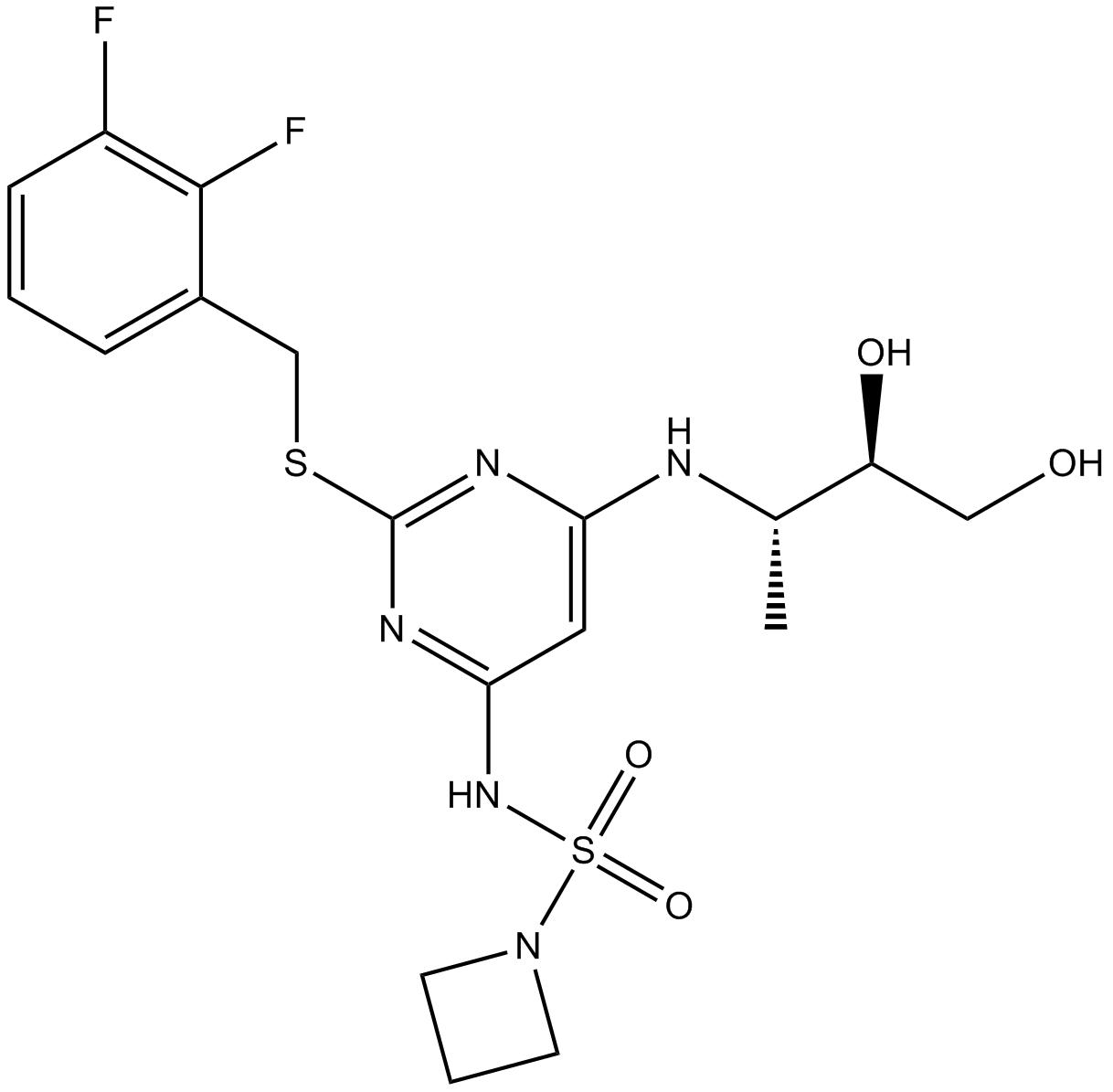
-
GC38204
SX-682
SX-682 is an orally bioavailable, potent allosteric inhibitor of CXCR1 and CXCR2.
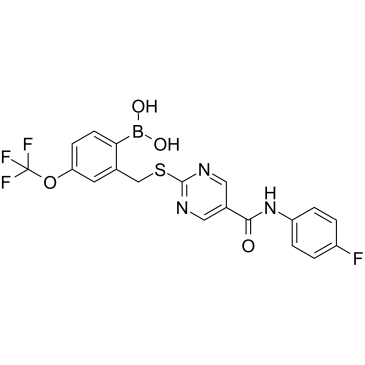
-
GC37724
TAK-779
An antagonist of CCR5, CXCR3, and CCR2b
-
GC17538
UNBS 5162
A pan-antagonist of CXCL chemokines
-
GC33413
USL311
USL311 is a potent and selective CXCR4 antagonist, with anti-tumor activity.
-
GC38217
VUF11207 fumarate
VUF11207 fumarate is a CXCR7 agonist that binds specifically to CXCR7.
-
GC15989
WZ811
Competitive CXCR4 antagonist,highly potent
-
GC12909
Zaprinast
phosphodiesterase inhibitor and GPR35 agonist