Adaptive Immunity
Products for Adaptive Immunity
- Cat.No. Product Name Information
-
GC45194
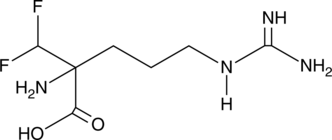
α-(difluoromethyl)-DL-Arginine
Bacteria synthesize the cellular growth factor putrescine through a number of pathways.
-
GC50708
(±)-ML 209
An RORγt antagonist
-
GC41703
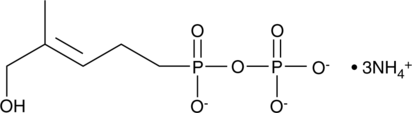
(E)-C-HDMAPP (ammonium salt)
Synthetic and natural alkyl phosphates, also known as phosphoantigens, stimulate the proliferation of γδ-T lymphocytes.
-
GC49028
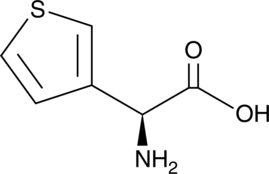
(S)-3-Thienylglycine
A thienyl-containing amino acid
-
GC49768
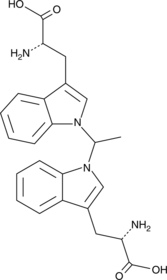
1,1’-Ethylidene-bis-(L-tryptophan)
A potential impurity found in commercial preparations of L-tryptophan
-
GC40910
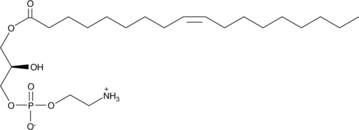
1-Oleoyl-2-hydroxy-sn-glycero-3-PE
1-Oleoyl-2-hydroxy-sn-glycero-3-PE is a naturally-occurring lysophospholipid and an analog of plasmalogen lysophosphatidylethanolamine.
-
GC18376
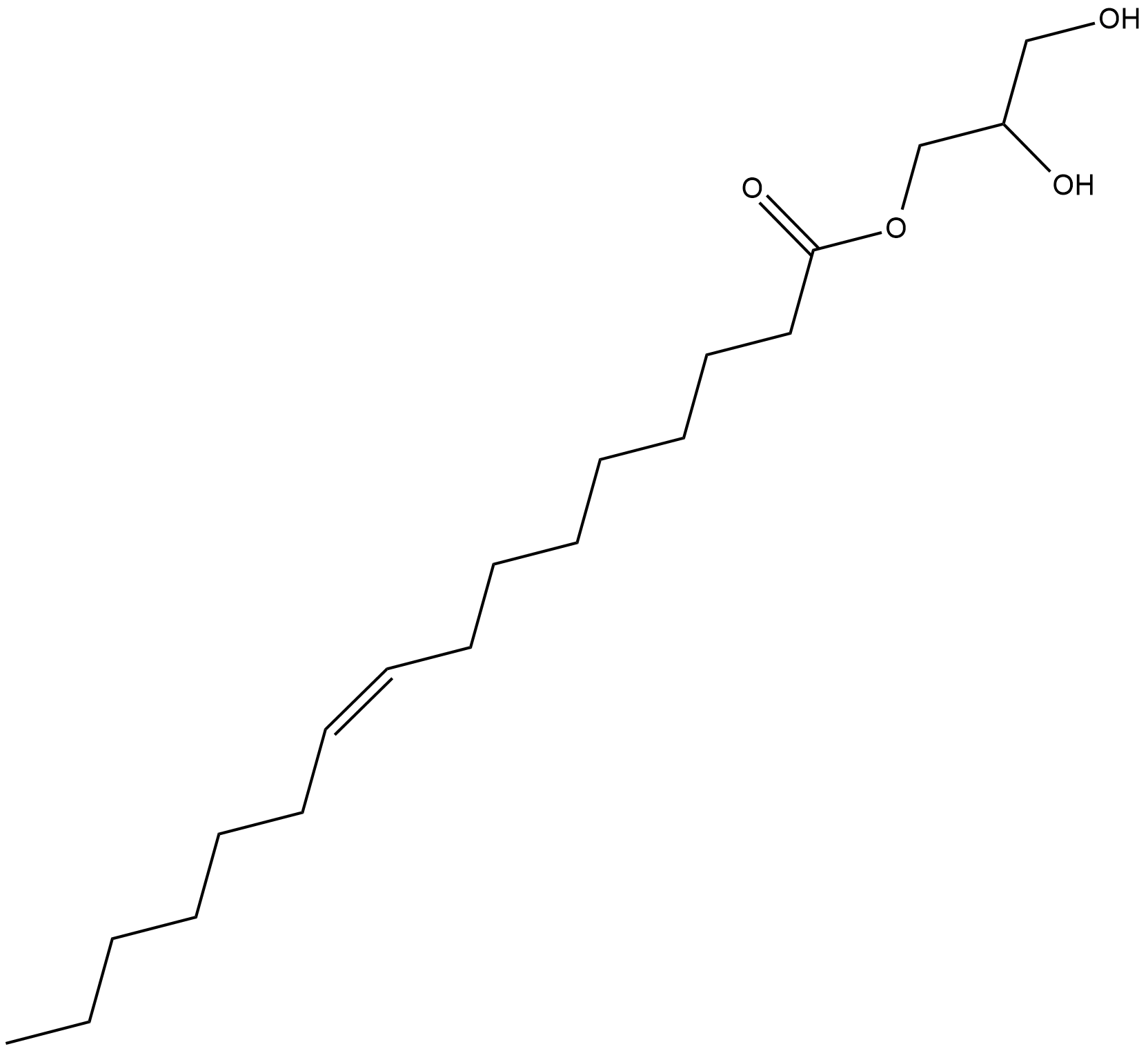
1-Palmitoleoyl glycerol
1-Palmitoleoyl glycerol is a bioactive monoacylglycerol.
-
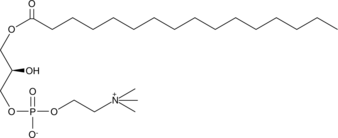
GC42026
1-Palmitoyl-2-hydroxy-sn-glycero-3-PC
1-Palmitoyl-2-hydroxy-sn-glycero-3-PC is an abundant gonadal LPC (lysophosphatidylcholine).
-
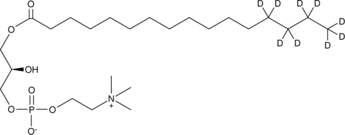
GC45693
1-Palmitoyl-d9-2-hydroxy-sn-glycero-3-PC
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
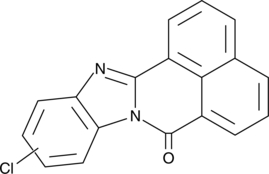
GC46400
10(11)-Cl-BBQ Mixture
A mixture that acts as an AhR agonist
-
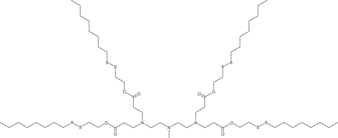
GC52343
113-O12B
An ionizable cationic lipidoid
-
GC46415
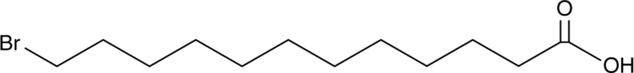
12-Bromododecanoic Acid
A halogenated form of lauric acid
-
GC49759
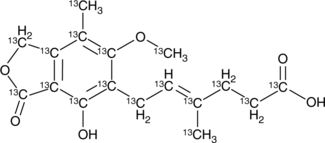
13C17-Mycophenolic Acid
An internal standard for the quantification of mycophenolic acid
-
GC18778
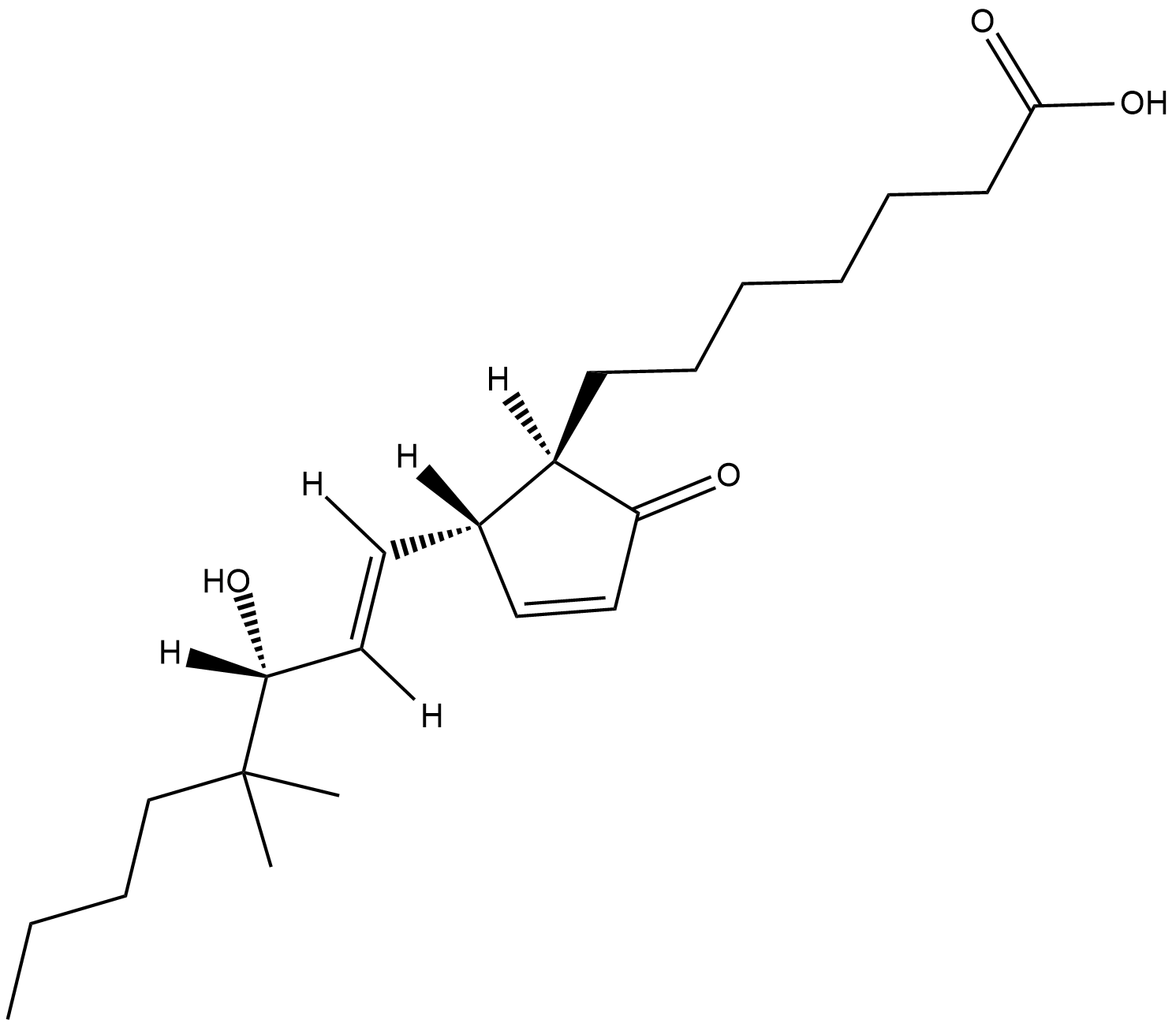
16,16-dimethyl Prostaglandin A1
16,16-dimethyl PGA1 is a metabolism resistant analog of PGA1.
-
GC42112
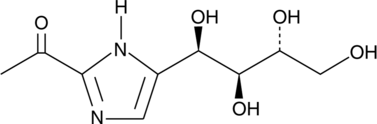
2-Acetyl-5-tetrahydroxybutyl Imidazole
Sphingosine-1-phosphate (S1P) lyase catalyzes the irreversible decomposition of S1P to hexadecanaldehyde and phosphoethanolamine.
-
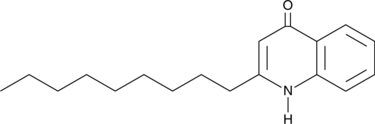
GC46553
2-Nonylquinolin-4(1H)-one
A quinolone alkaloid with diverse biological activities
-
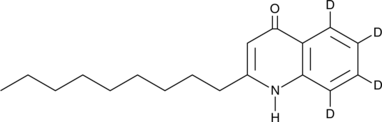
GC52446
2-Nonylquinolin-4(1H)-one-d4
An internal standard for the quantification of 2-nonylquinolin-4(1H)-one
-
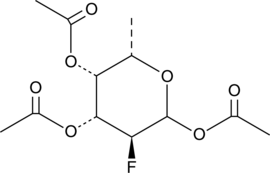
GC46549
2F-Peracetyl-Fucose
An inhibitor of protein fucosylation
-
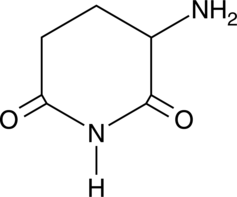
GC46583
3-Amino-2,6-Piperidinedione
An active metabolite of (±)-thalidomide
-
GC42259
3-Deaza-2'-deoxyadenosine
3-Deaza-2'-deoxyadenosine strongly inhibits lymphocyte-mediated cytolysis with low cytotoxicity when applied at 100 μM.
-
GC49244
4-oxo Isotretinoin
An active metabolite of isotretinoin
-
GC49206
7α-hydroxy Dehydroepiandrosterone
An active metabolite of dehydroepiandrosterone
-
GC46740
7β,27-dihydroxy Cholesterol
An oxysterol and agonist of RORγ and RORγt
-
GC46733
7,12-Dimethylbenz[a]anthracene
7,12-Dimethylbenz[a]anthracene has carcinogenic activity as a polycyclic aromatic hydrocarbon (PAH). 7,12-Dimethylbenz[a]anthracene is used to induce tumor formation in various rodent models.
![7,12-Dimethylbenz[a]anthracene Chemical Structure 7,12-Dimethylbenz[a]anthracene Chemical Structure](/media/struct/GC4/GC46733.png)
-
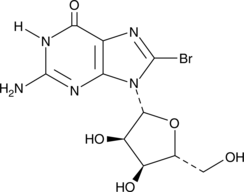
GC42623
8-Bromoguanosine
8-Bromoguanosine is a brominated derivative of guanosine.
-
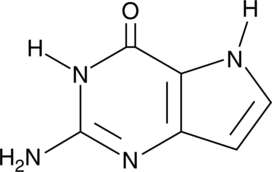
GC42644
9-Deazaguanine
9-Deazaguanine is an analog of guanine that acts as an inhibitor of purine nucleoside phosphorylase (PNP; Kd = 160 nM).
-
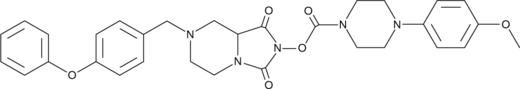
GC42668
ABC34
ABC34 is an inactive control probe for JJH260, the inhibitor of androgen-induced gene 1 (AIG1), an enzyme that hydrolyzes fatty acid esters of hydroxy fatty acids (FAHFAs).
-
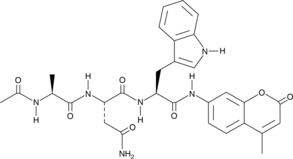
GC42685
Ac-ANW-AMC
Ac-ANW-AMC is a fluorogenic substrate for the β5i/LMP7 subunit of the 20S immunoproteasome.
-
GC18443
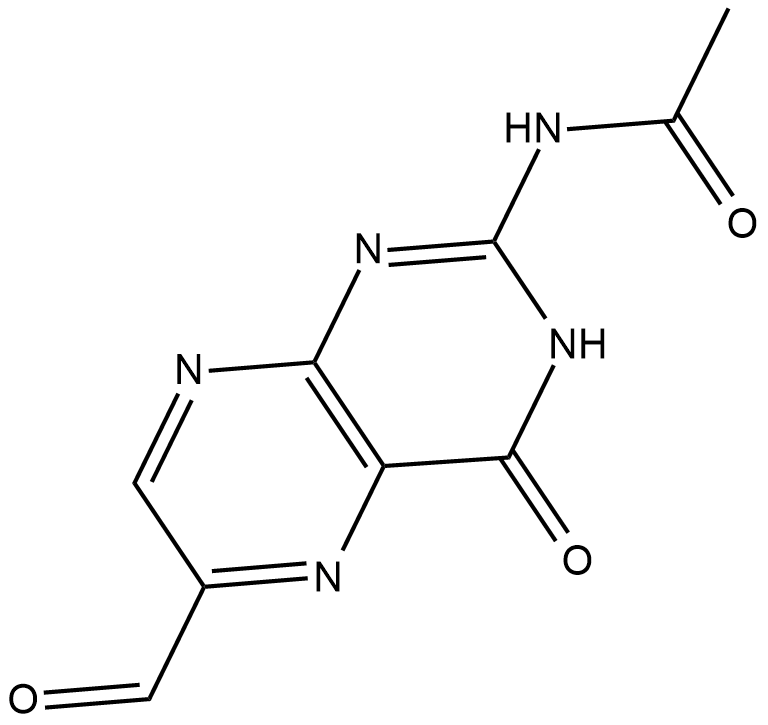
Acetyl-6-formylpterin
Acetyl-6-formylpterin is an inhibitor of mucosal-associated invariant T (MAIT) cell activation.
-
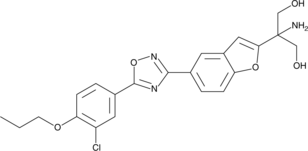
GC45681
AKP-11
An S1P1 receptor agonist
-
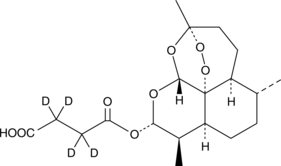
GC45790
Artesunate-d4
An internal standard for the quantification of artesunate
-
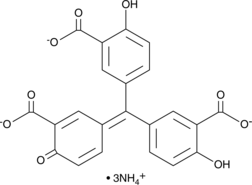
GC46895
Aurintricarboxylic Acid (ammonium salt)
A protein synthesis inhibitor with diverse biological activities
-
GC42877
AUY954
AUY954 is an orally bioavailable and selective agonist of the sphingosine-1-phosphate receptor 1 (S1P1; EC50 = 1.2 nM for stimulating GTPγS binding to S1P1 in CHO cells).
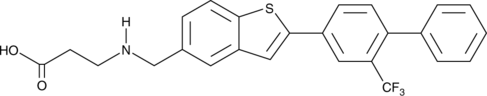
-
GC42891
azido-FTY720
FTY720 is a derivative of ISP-1 (myriocin), a fungal metabolite of the Chinese herb Iscaria sinclarii as well as a structural analog of sphingosine.
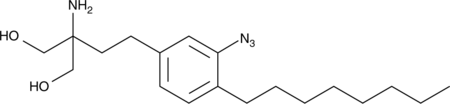
-
GC49044
Benastatin C
A bacterial metabolite with diverse biological activities
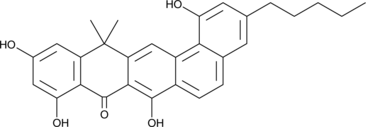
-
GC52326
Biotin-PEG4-LL-37 (human) (trifluoroacetate salt)
A biotinylated and pegylated form of LL-37

-
GC18716
Bisindolylmaleimide XI (hydrochloride)
Bisindolylmaleimide XI (BIM XI) is a selective, cell-permeable protein kinase C (PKC) inhibitor that displays 10-fold greater selectivity for PKCα (IC50 = 9 nM) and 4-fold greater selectivity for PKCβI (IC50 = 28 nM) over Ca2+-independent PKCε (IC50 = 108 nM).
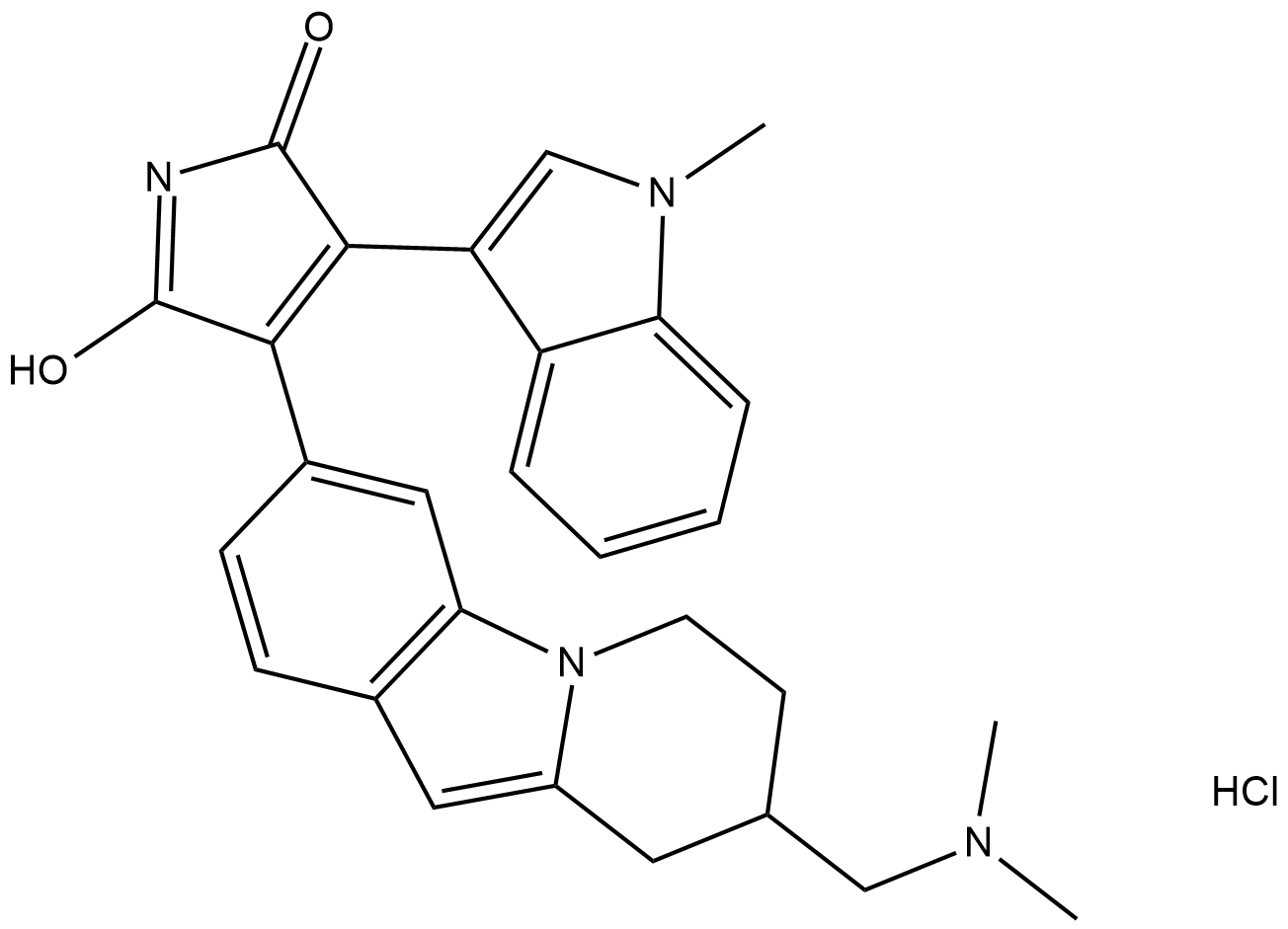
-
GA20972
Boc-D-Leu-OSu
An amino acid-containing building block

-
GC52101
Brazilein
Brazilein is an important immunosuppressive component isolated from Caesalpinia sappan L.
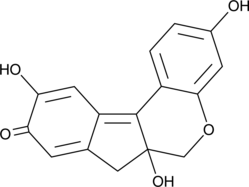
-
GC43007
C12 Galactosylceramide (d18:1/12:0)
C12 Galactosylceramide is a bioactive sphingolipid.
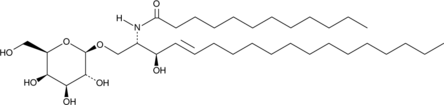
-
GC43032
C16 Globotriaosylceramide (d18:1/16:0)
C16 globotriaosylceramide is an endogenous sphingolipid found in mammalian cell membranes that is synthesized from C16 lactosylceramide.
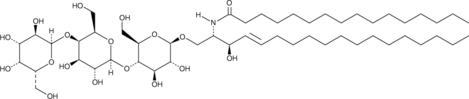
-
GC43049
C18 Globotriaosylceramide (d18:1/18:0)
C18 globotriaosylceramide is an endogenous sphingolipid found in mammalian cell membranes that is synthesized from lactosylceramide.
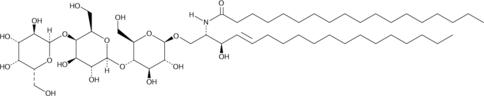
-
GC40709
C2 L-threo Ceramide (d18:1/2:0)
C2 L-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
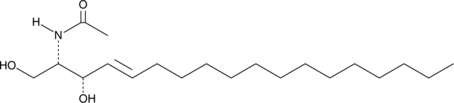
-
GC43084
C4 Ceramide (d18:1/4:0)
C4 Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
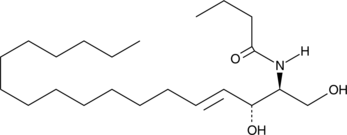
-
GC40688
C6 D-threo Ceramide (d18:1/6:0)
C6 D-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides., C6 D-threo Ceramide is cytotoxic to U937 cells in vitro (IC50 = 18 μM).
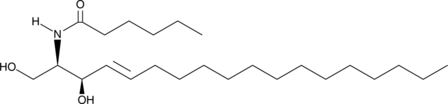
-
GC40690
C6 L-threo Ceramide (d18:1/6:0)
C6 L-threo Ceramide (d18:1/6:0) is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
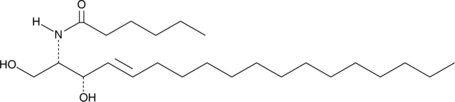
-
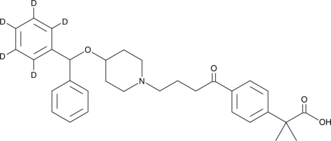
GC47040
Carebastine-d5
An internal standard for the quantification of carebastine
-
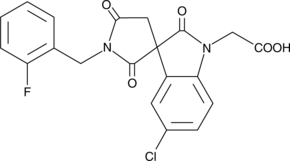
GC43181
CAY10597
The biological effects of prostaglandin D2 (PGD2) are transduced by at least two 7-transmembrane G protein-coupled receptors, designated DP1 and CRTH2/DP2.
-
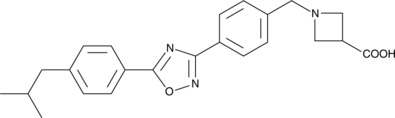
GC40757
CAY10734
An S1P1 receptor agonist
-
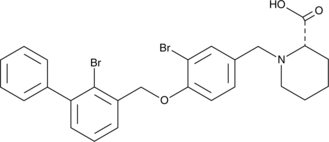
GC48427
CAY10774
A PD-1/PD-L1 interaction inhibitor
-
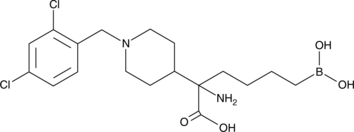
GC49664
CB-1158
An arginase inhibitor
-
GC49584
CD74 Monoclonal Antibody (Clone PIN1)
For immunochemical analysis of CD74

-
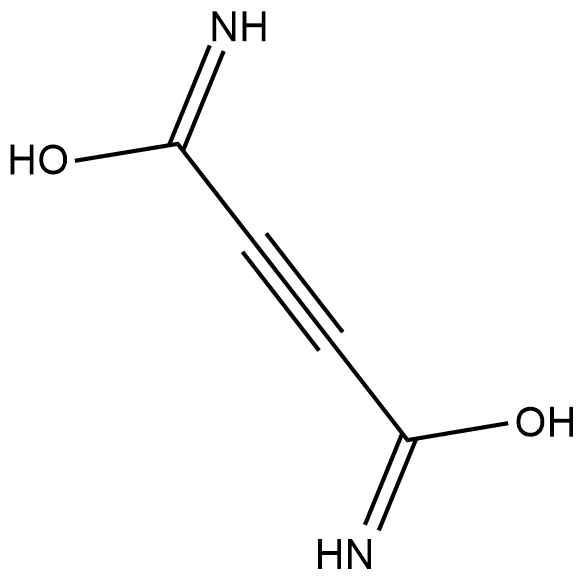
GC18392
Cellocidin
Cellocidin is an antibiotic originally isolated from S.
-
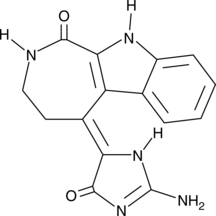
GC43239
Chk2 Inhibitor
Chk2 Inhibitor (compound 1) is a potent and selective inhibitor of checkpoint kinase 2 (Chk2), with IC50s of 13.5 nM and 220.4 nM for Chk2 and Chk1, respectively. Chk2 Inhibitor can elicit a strong ataxia telangiectasia mutated (ATM)-dependent Chk2-mediated radioprotection effect.
-
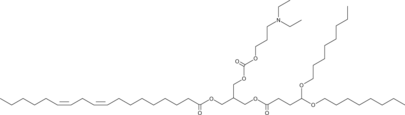
GC52153
CIN-16645
CIN-16645 is a cationic lipid useful in the delivery of biologically active agents to cells and tissues (extracted from patent WO2015095340 A1).
-
GC47109
Clotrimazole-d5
An internal standard for the quantification of clotrimazole
-
GC43288
Cochlioquinone A
Cochlioquinone A, a bioactive compound isolated from D.
-
GC43307
Concanamycin B
Concanamycin B is a macrolide antibiotic that selectively inhibits vacuolar type H+-ATPases, also known as V-ATPases (IC50 = 5 nM).
-
GC18572
Concanavalin A
Concanamycin A belongs to the concanamycins, a family of macrolide antibiotics isolated from Streptomyces diastatochromogenes that are highly active and selective inhibitors of the vacuolar proton-ATPase (v-[H+]ATPase).

-
GC49021
CXCR3 Antagonist 6c
A CXCR3 antagonist
-
GC43340
Cyclic di-IMP (sodium salt)
Cyclic di-IMP (sodium salt) (c-di-IMP) is a synthetic second messenger structurally related to the bacterial second messengers cyclic di-GMP and cyclic di-AMP.
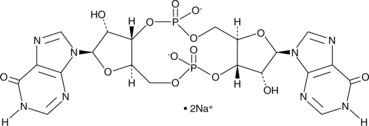
-
GC43350
Cyclosporin B
Cyclosporin B is a minor cyclopeptide metabolite produced by T.
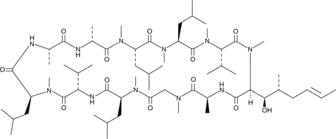
-
GC49526
Cytidine-d2
An internal standard for the quantification of cytidine
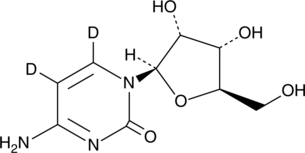
-
GC49153
Didemnin B
Didemnin B is a cyclic depsipeptide produced by marine tunicates that specifically binds the GTP-bound conformation of EEF1A.
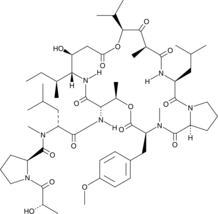
-
GC40629
Dimethoxycurcumin
Naturally occurring phytochemicals such as turmeric (curcumin) have been found to inhibit the growth of tumor cells.
-
GC43467
Dimethyldioctadecylammonium (bromide)
Dimethyldioctadecylammonium (DDA) is a cationic amphipathic lipid.
-
GC48352
Diprovocim-1
An agonist of TLR1/TLR2
-
GC47257
Dodecanoyl D-Sucrose
A nonionic surfactant
-
GC40082
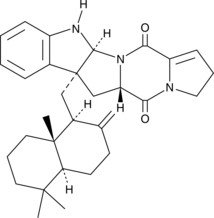
Drimentine B
Drimentine B is a terpenylated diketopiperazine antibiotic originally isolated from Actinomycete bacteria.
-
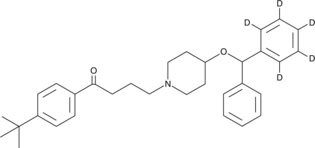
GC47278
Ebastine-d5
An internal standard for the quantification of ebastine
-
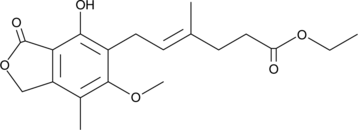
GC52303
Ethyl Mycophenolate
A potential impurity found in commercial preparations of mycophenolate mofetil
-
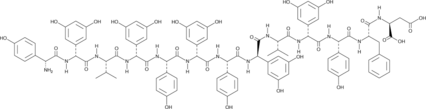
GC43659
Feglymycin
Feglymycin is a 13-amino acid peptide originally isolated from Streptomyces that has antibacterial and antiviral activities.
-
GC18325
Ferrichrome (iron-free)
Ferrichrome is a hydroxamate siderophore produced by various fungi, including U.
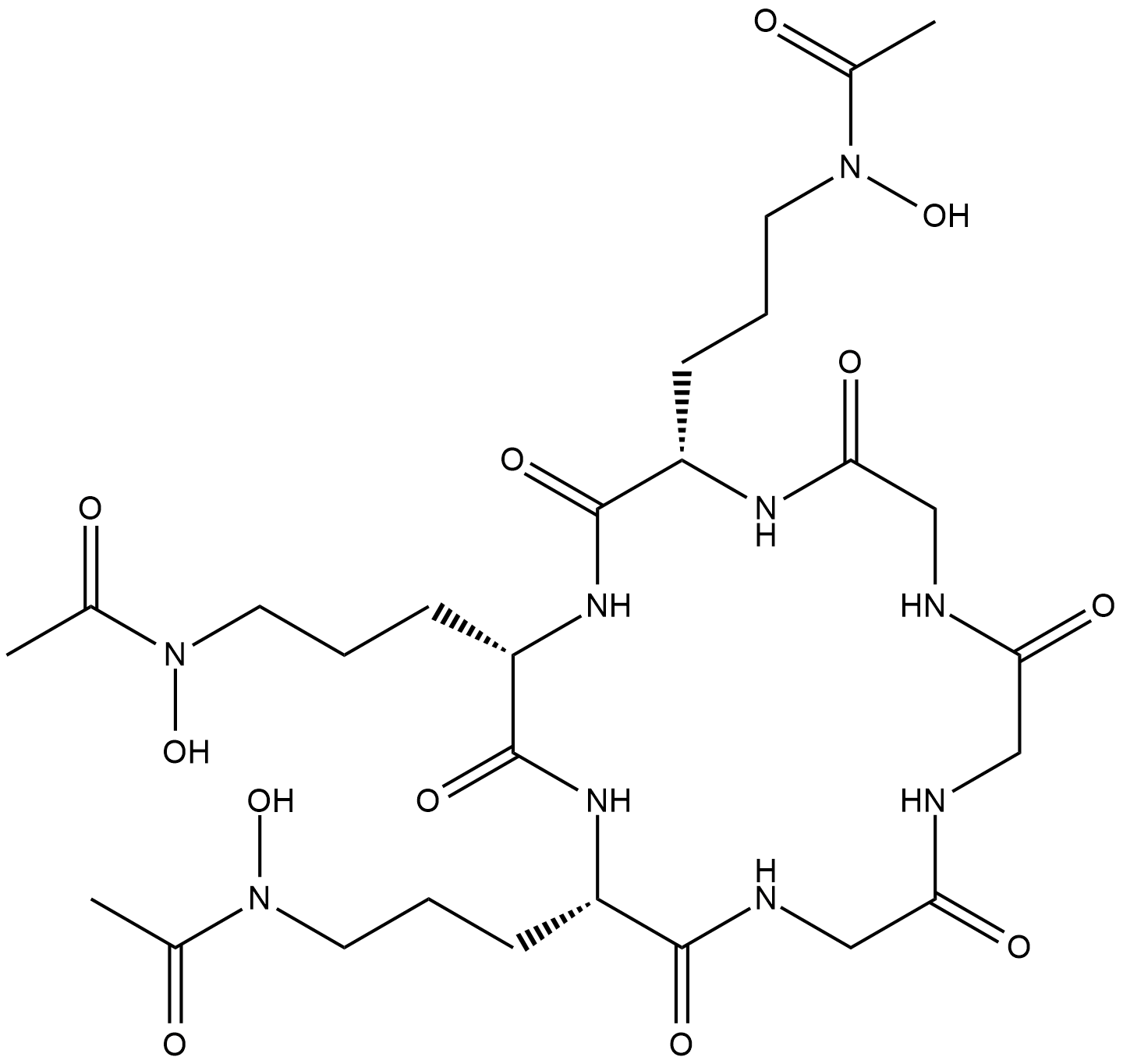
-
GC46148
Filgotinib-d4
An internal standard for the quantification of filgotinib
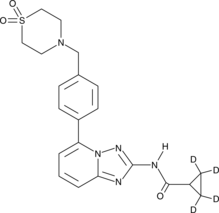
-
GC47351
Fingolimod-d4
An internal standard for the quantification of fingolimod
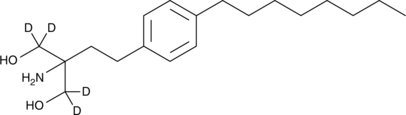
-
GC47352
FK-506-13C-d2
An internal standard for the quantification of FK-506
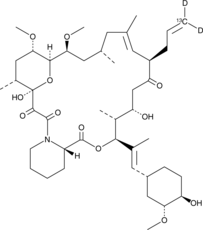
-
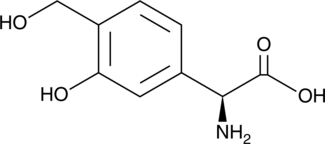
GC52014
Forphenicinol
An immunomodulator and a derivative of forphenicine
-
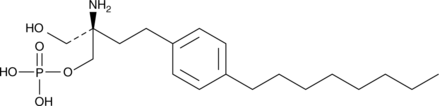
GC40887
FTY720 (R)-Phosphate
FTY720 is a novel immune modulator that prolongs allograft transplant survival in numerous models by inhibiting lymphocyte emigration from lymphoid organs.
-
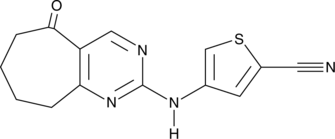
GC47387
G6PDi-1
An inhibitor of G6PDH
-
GC18469
Galactosylcerebrosides (bovine)
Galactosylcerebrosides are glycosphingolipids that contain a galactose attached to a ceramide acylated with hydroxy and non-hydroxy fatty acids.

-
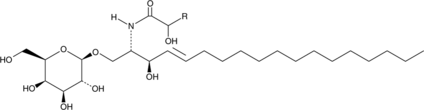
GC43721
Galactosylcerebrosides (hydroxy)
Galactosylcerebrosides are glycosphingolipids that contain a galactose attached to a ceramide acylated with a hydroxy or non-hydroxy fatty acid.
-
GC43722
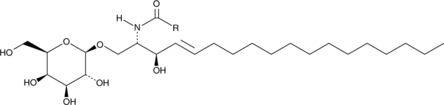
Galactosylcerebrosides (non-hydroxy)
Galactosylcerebrosides are glycosphingolipids that contain a galactose attached to a ceramide acylated with a hydroxy or non-hydroxy fatty acid.
-
GC43730
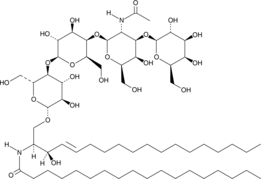
Ganglioside GM1 Asialo Mixture
Ganglioside GM1 asialo is a component of cellular lipid rafts and can be formed by the cleavage of the sialic acid residue from ganglioside GM1 by neuraminidase.
-
GC47392
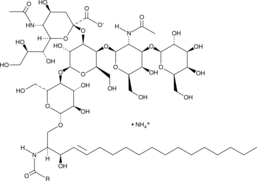
Ganglioside GM1 Mixture (ovine) (ammonium salt)
A mixture of ganglioside GM1
-
GC47398
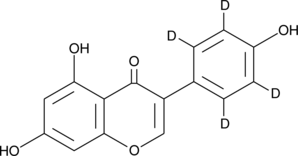
Genistein-d4
An internal standard for the quantification of genistein
-
GC47403
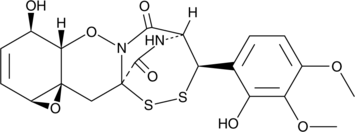
Gliovirin
A fungal metabolite
-
GC49798
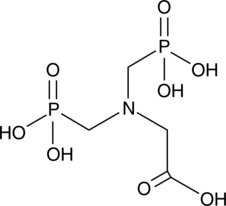
Glyphosine
A plant growth regulator
-
GC43804
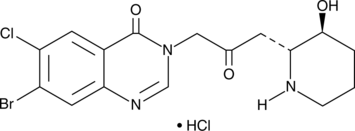
Halofuginone (hydrochloride)
Halofuginone is a halogenated derivative of febrifugine, a natural quinazolinone-containing compound found in the Chinese herb D.
-
GC43807
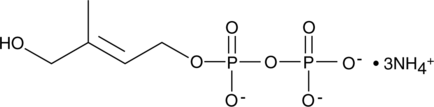
HDMAPP (ammonium salt)
HDMAPP is a metabolite of the microbial dioxyxylulose-phosphate pathway, which is analogous to the isopentenyl pyrophosphate pathway in mammals.
-
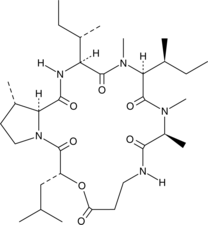
GC47434
Homodestcardin
A fungal metabolite with immunosuppressant activity
-
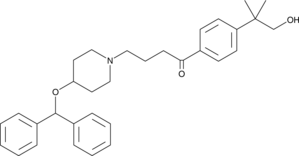
GC43879
Hydroxy Ebastine
Hydroxy Ebastine is an ebastine metabolite.
-
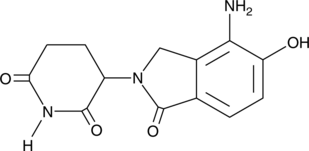
GC49282
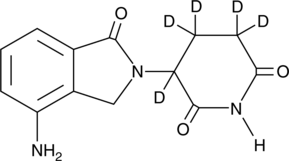
Hydroxy Lenalidomide
A metabolite of lenalidomide
-
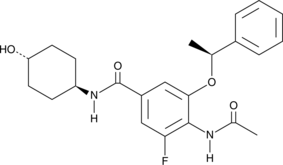
GC48548
iBET-BD2
A BD2 bromodomain inhibitor
-
GC47456
Indole-3-pyruvic Acid
Indole-3-pyruvic Acid, a keto analogue of tryptophan, is an orally active AHR agonist.
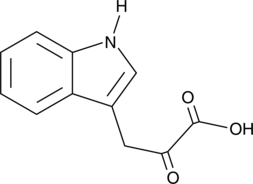
-
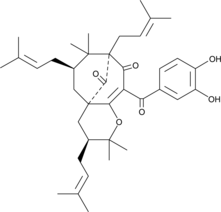
GC40901
Isogarcinol
Isogarcinol is a natural polyisoprenylated benzophenone first isolated from plant species in the genus Garcinia.
-
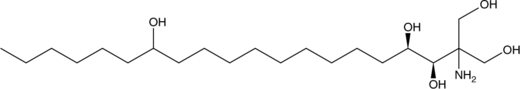
GC52321
ISP-I-28
An immunosuppressant
-
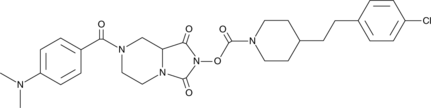
GC43928
JJH260
JJH260 is an N-hydroxy hydantoin carbamate that inhibits androgen-induced gene 1 (AIG1), an enzyme that hydrolyzes fatty acid esters of hydroxy fatty acids (FAHFAs).
-
GC47533
L-778,123
A dual inhibitor of FTase and GGTase I
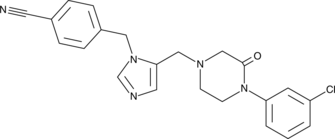
-
GC47552
Leflunomide-d4
An internal standard for the quantification of leflunomide
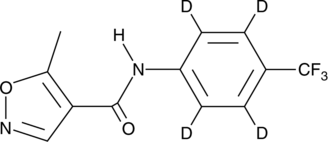
-
GC47553
Lenalidomide-d5
An internal standard for the quantification of lenalidomide