PI3K
PI3K (phosphatidylinositol-4,5-bisphosphate 3-kinase) is a family of enzymes involved in cellular functions such as cell growth, proliferation, differentiation, motility, survival and intracellular trafficking, which in turn are involved in cancer.
Products for PI3K
- Cat.No. Product Name Information
-
GC62450
(S)-PI3Kα-IN-4
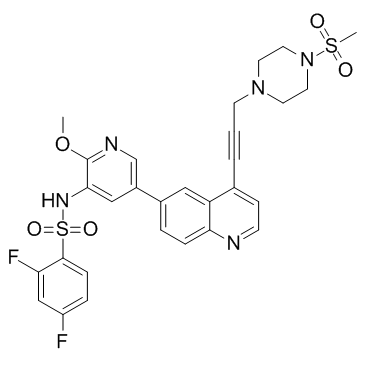
(S)-PI3Kα-IN-4 is a potent inhibitor of PI3Kα, with an IC50 of 2.3 nM. (S)-PI3Kα-IN-4 shows 38.3-, 4.25-, and 4.93-fold selectivity for PI3Kα over PI3Kβ, PI3Kδ, and PI3Kγ, respectively. (S)-PI3Kα-IN-4 can be used for the research of cancer.
-
GC35037
1,3-Dicaffeoylquinic acid
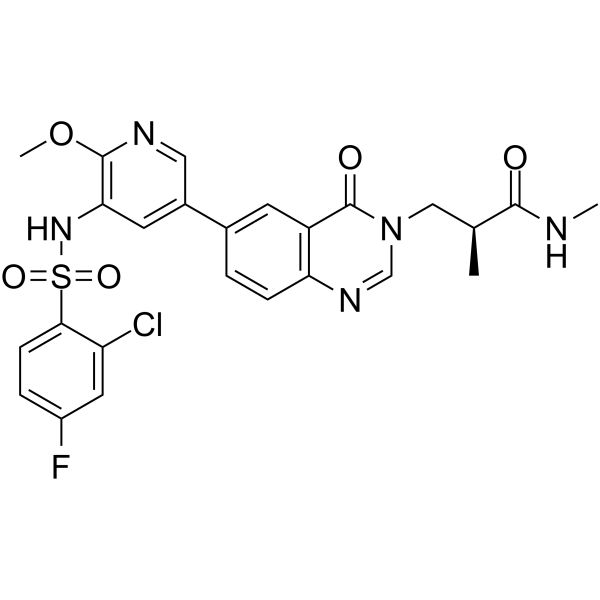
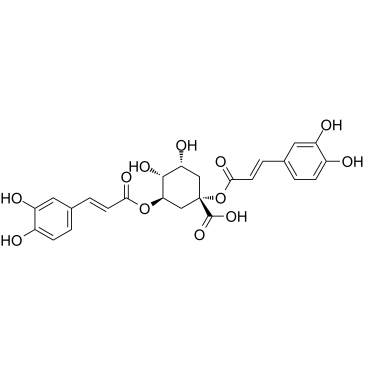
1,3-Dicaffeoylquinic acid is a caffeoylquinic acid derivative, and activates PI3K/Akt.
-
GC41992
1-Deoxynojirimycin (hydrochloride)
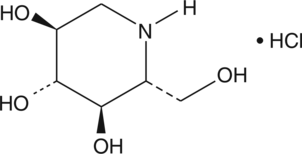
1-Deoxynojirimycin (1-dNM) (hydrochloride), produced by Bacillus species, is a glucose analog that potently inhibits α-glucosidase I and II.
-
GC10710
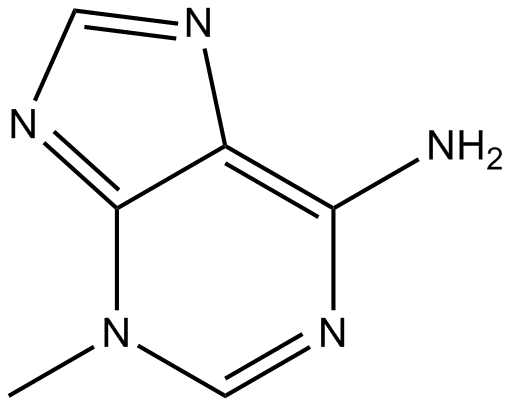
3-Methyladenine
3-Methyladenine is a classic autophagy inhibitor.
-
GC68539
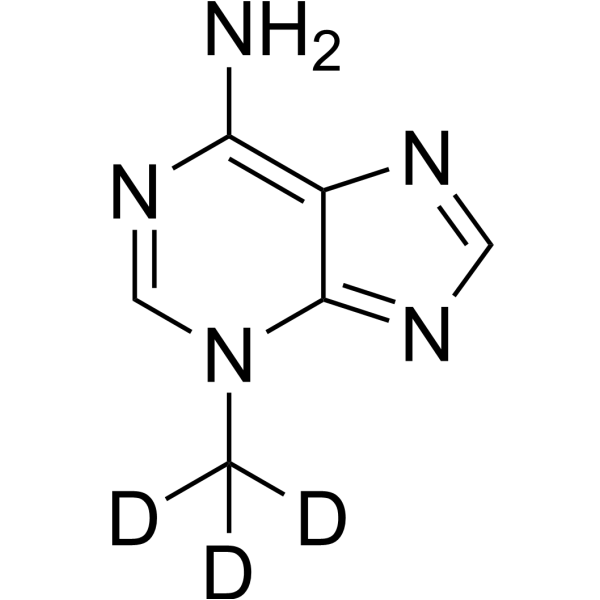
3-Methyladenine-d3
3-Methyladenine-d3 is the deuterated form of 3-Methyladenine. 3-Methyladenine (3-MA) is an inhibitor of PI3K. It is widely used as an autophagy inhibitor by inhibiting class III PI3K.
-
GC16157
740 Y-P
PI 3-kinase activator,cell permeable
-
GC17550
A66
P110α inhibitor
-
GC10860
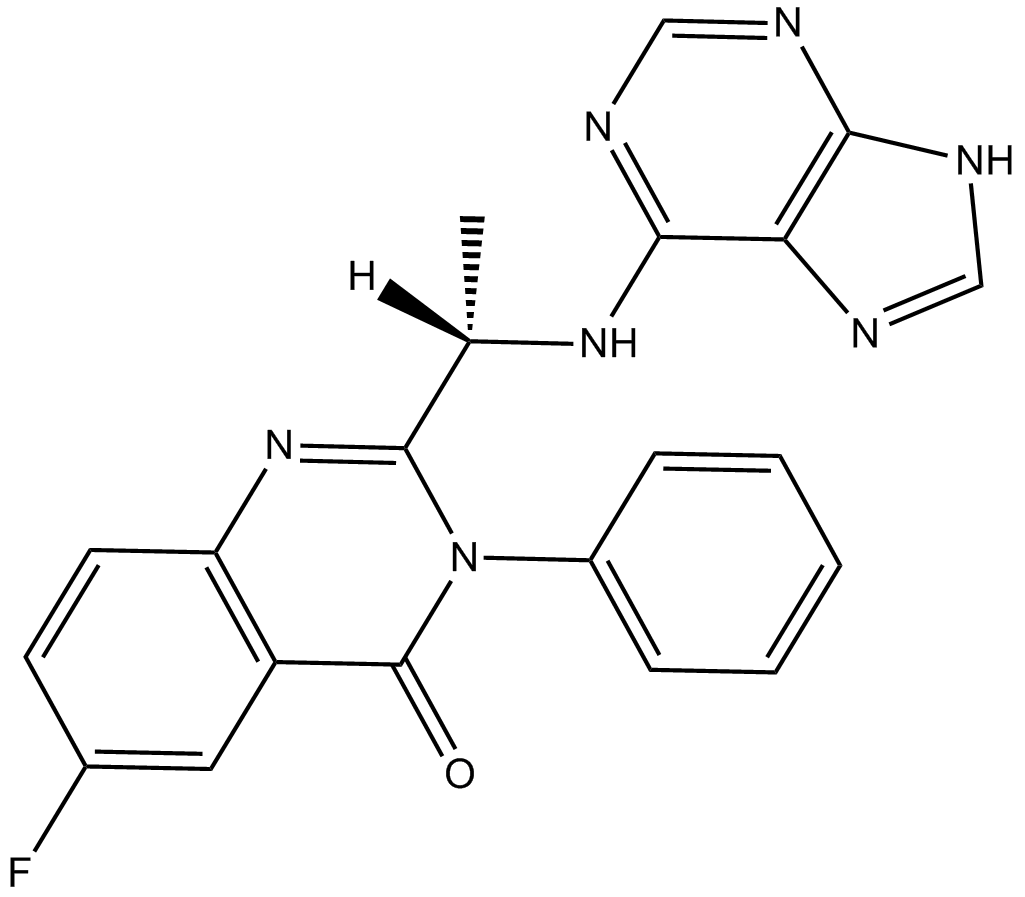
Acalisib (GS-9820)
Acalisib (GS-9820) is a potent and selective PI3Kδ inhibitor with an IC50 of 12.7 nM.
-
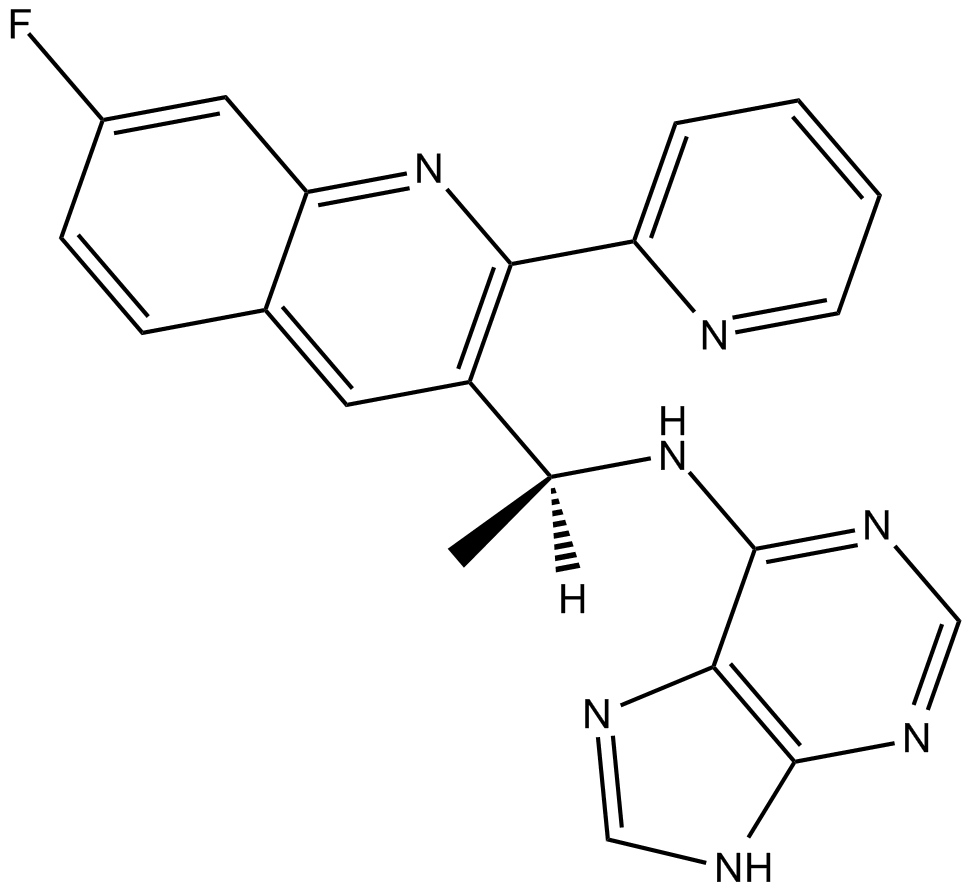
GC13913
AMG319
PI3Kδ inhibitor
-
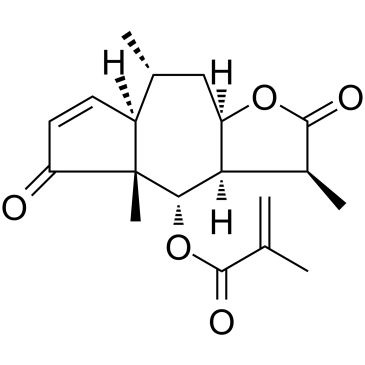
GC35395
Arnicolide D
Arnicolide D is a sesquiterpene lactone isolated from Centipeda minima. Arnicolide D modulates the cell cycle, activates the caspase signaling pathway and inhibits the PI3K/AKT/mTOR and STAT3 signaling pathways. Arnicolide D inhibits Nasopharyngeal carcinoma (NPC) cell viability in a concentration- and time-dependent manner.
-
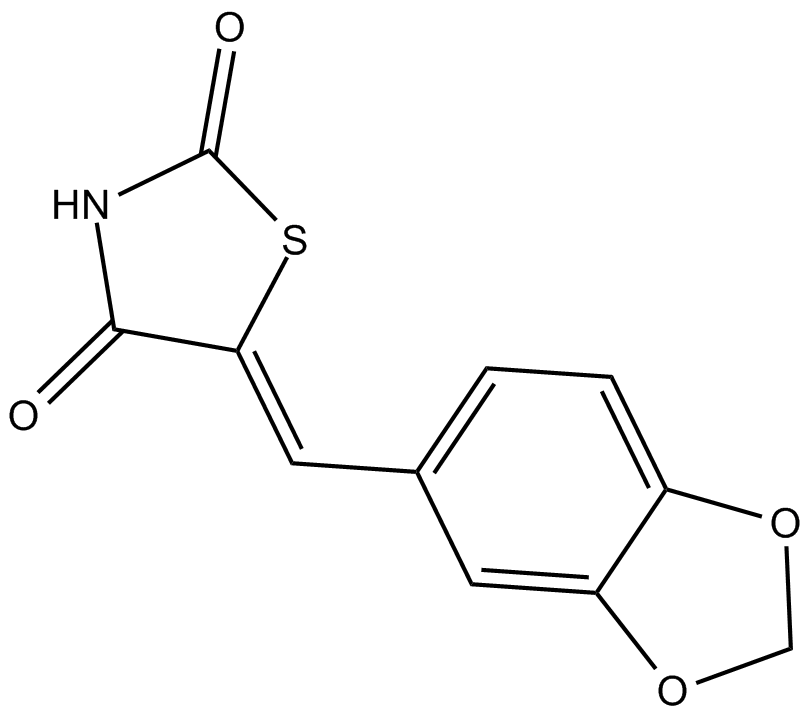
GC11910
AS-041164
PI3Kγ inhibitor
-
GC13901
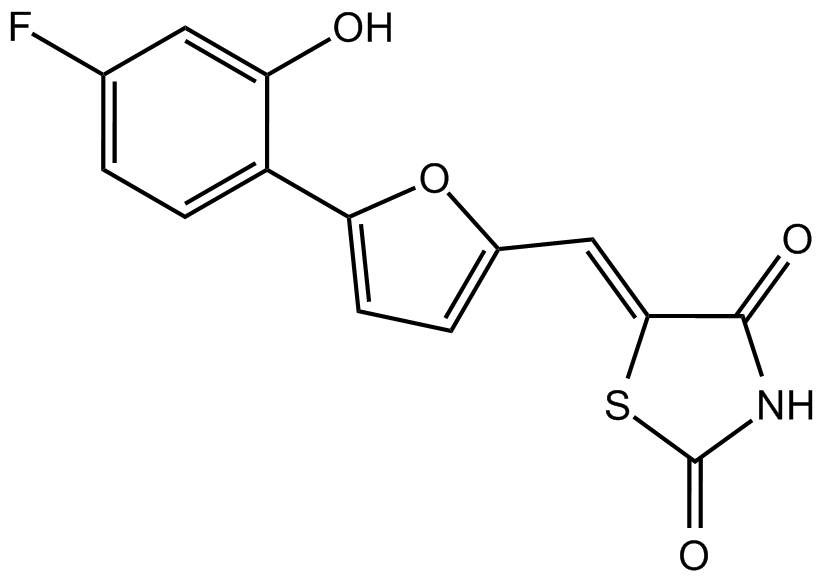
AS-252424
PI3Kγ inhibitor,novel and potent
-
GC38061
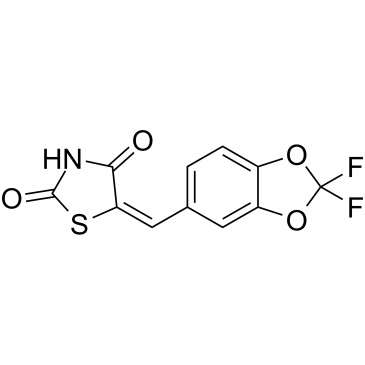
AS-604850
A selective inhibitor of PI3Kγ
-
GC12080
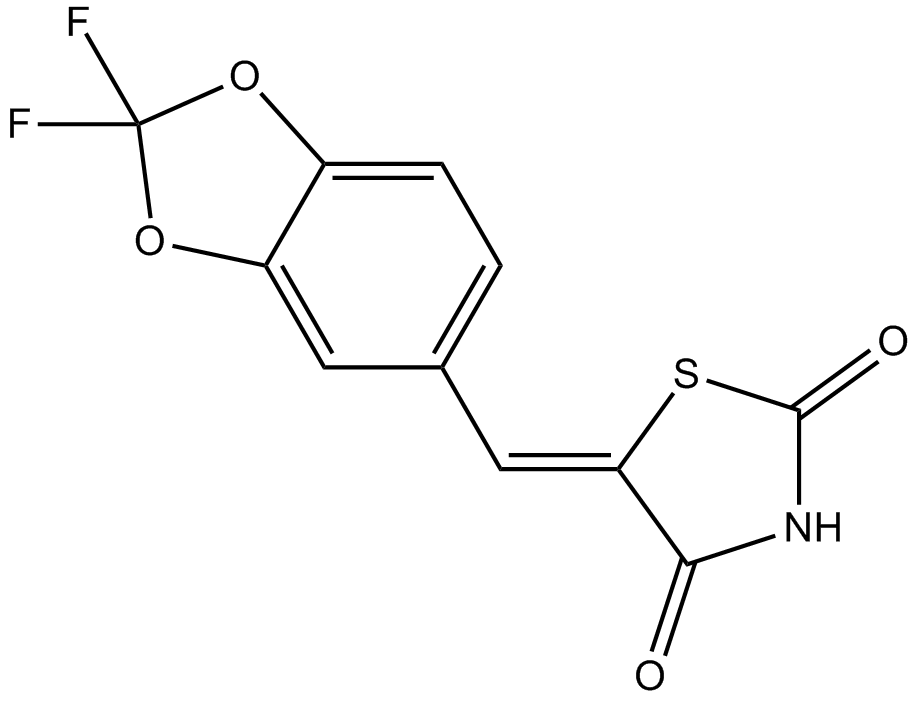
AS-604850
PI3Kγ inhibitor,selective and ATP-competitve
-
GC12474
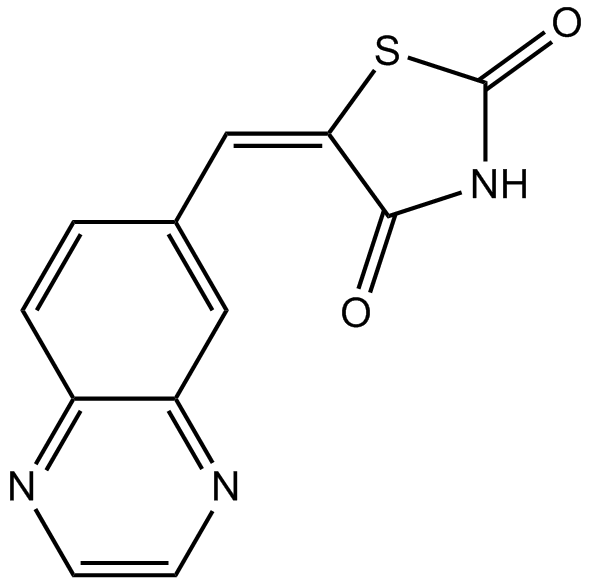
AS-605240
Potent and selective PI 3-Kγ inhibitor
-
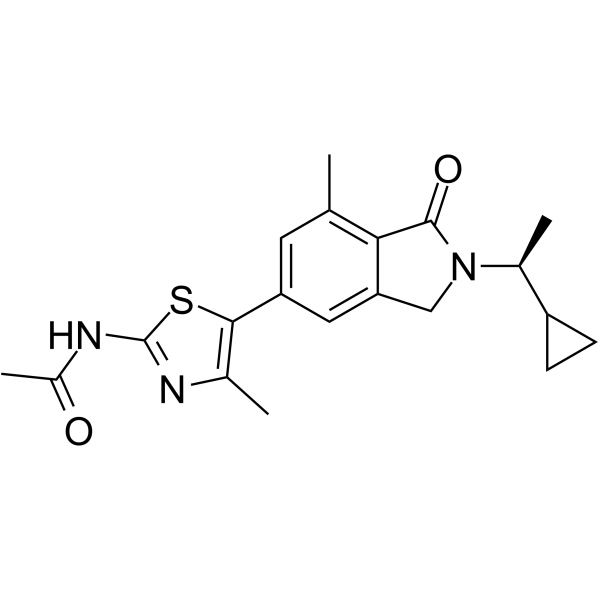
GC62335
AZ2
AZ2 is a highly selective PI3Kγ inhibitor (The pIC50 value for PI3Kγ is 9.3).
-
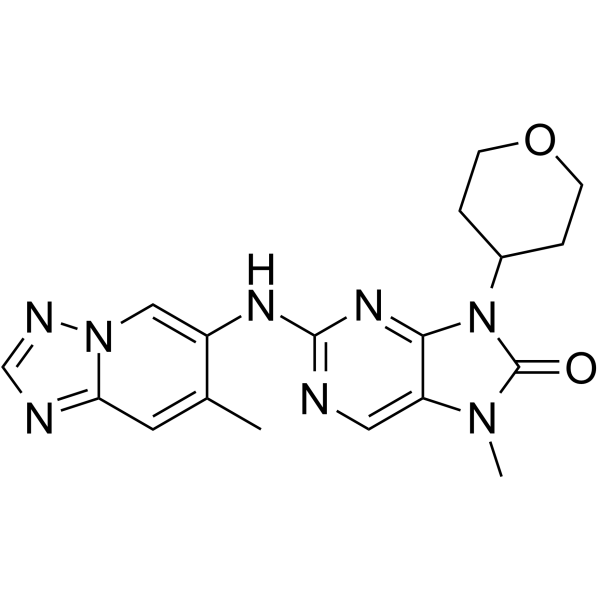
GC64938
AZD-7648
AZD-7648 is a potent, orally active, selective DNA-PK inhibitor with an IC50 of 0.6 nM. AZD-7648 induces apoptosis and shows antitumor activity.
-
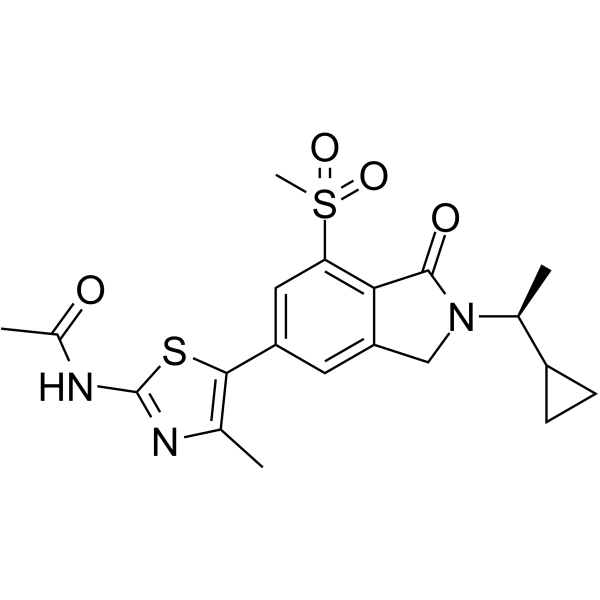
GC65149
AZD3458
AZD3458 is a potent and remarkably selective PI3Kγ inhibitor with pIC50s of 9.1, 5.1, <4.5, and 6.5 for PI3Kγ, PI3Kα, PI3Kβ, and PI3Kδ, respectively.
-
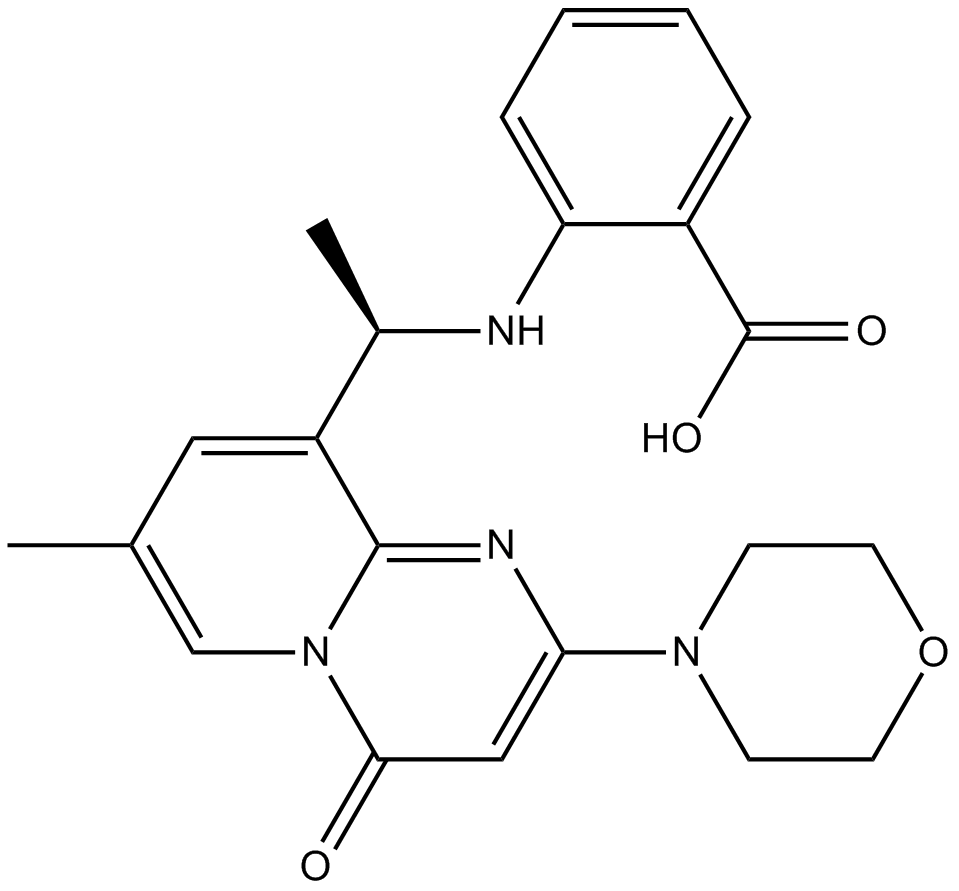
GC16851
AZD6482
PI3Kβ inhibitor,potent and selective
-
GC65507
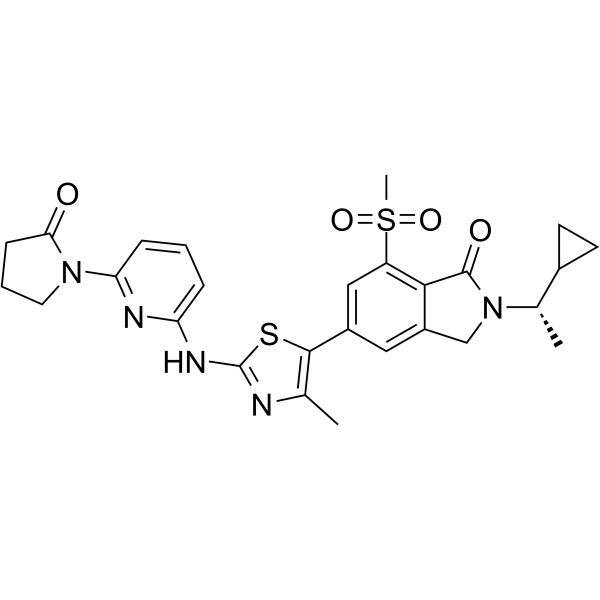
AZD8154
AZD8154 is a novel inhaled selective PI3Kγδ dual inhibitor targeting airway inflammatory disease.
-
GC12576
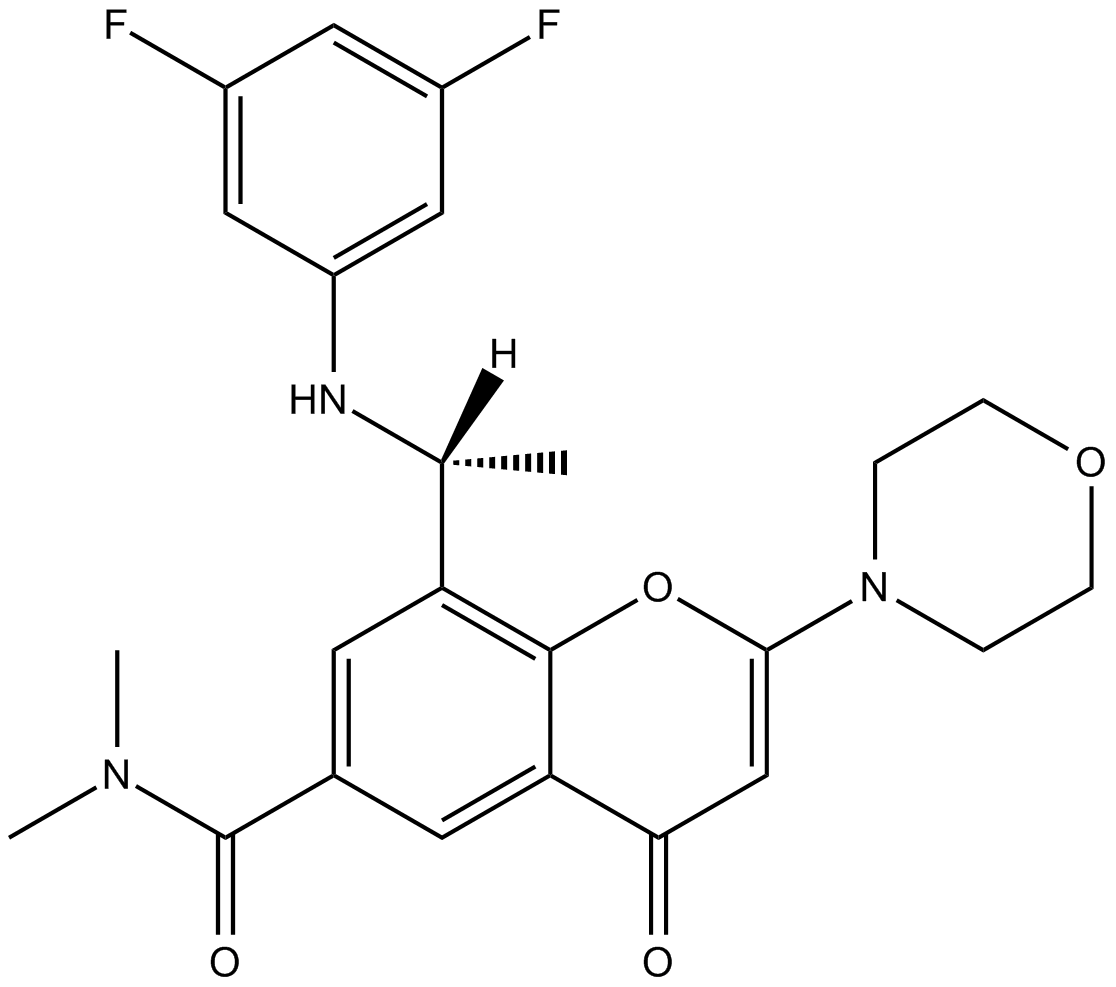
AZD8186
Potent and selective inhibitor of PI3Kβ and PI3Kδ
-
GC11248
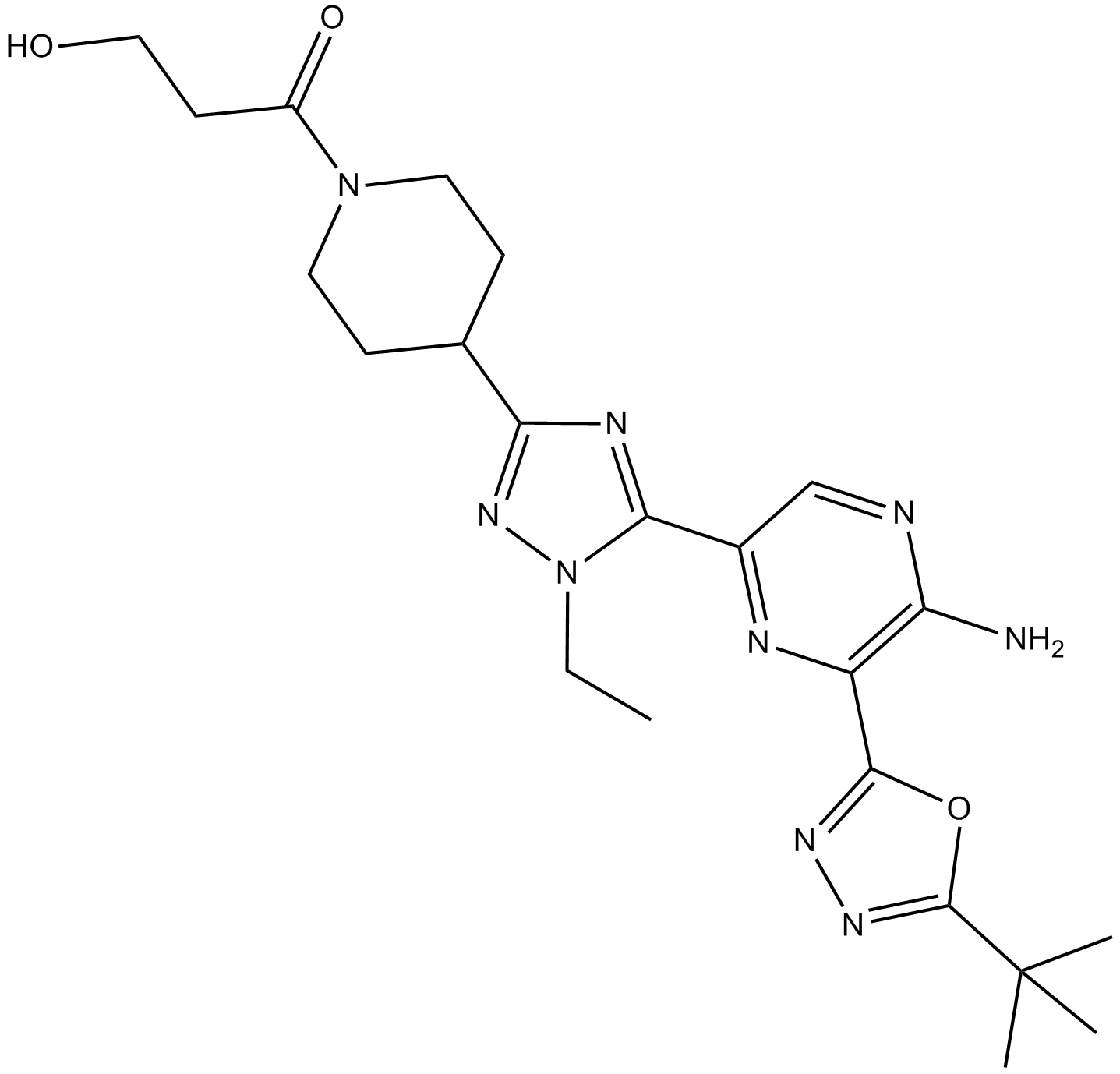
AZD8835
PI3Kα and PI3Kδ inhibitor
-
GC17766
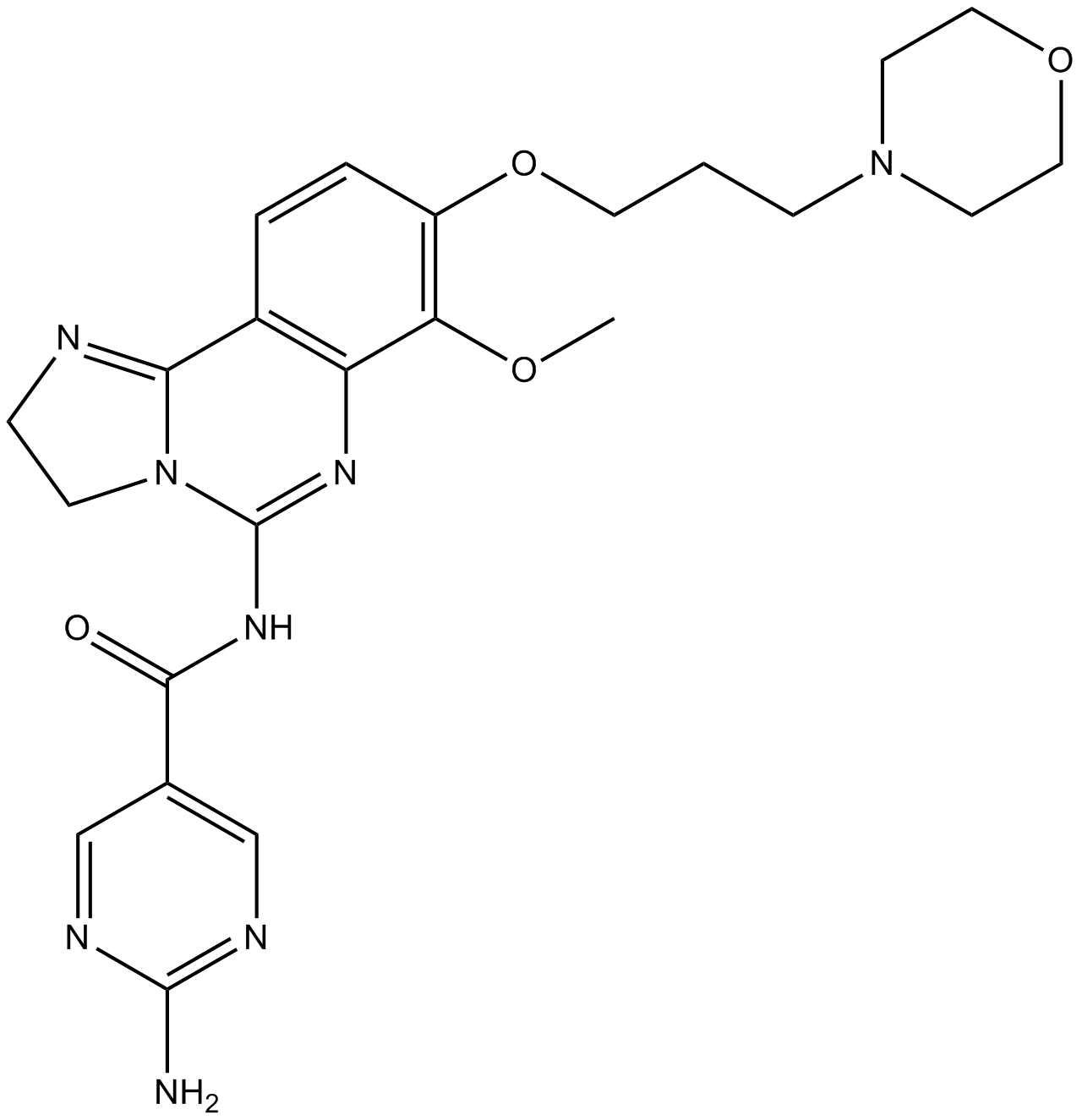
BAY 80-6946 (Copanlisib)
BAY 80-6946 (Copanlisib) (BAY 80-6946) is a potent, selective and ATP-competitive pan-class I PI3K inhibitor, with IC50s of 0.5 nM, 0.7 nM, 3.7 nM and 6.4 nM for PI3Kα, PI3Kδ, PI3Kβ and PI3Kγ, respectively. BAY 80-6946 (Copanlisib) has more than 2,000-fold selectivity against other lipid and protein kinases, except for mTOR. BAY 80-6946 (Copanlisib) has superior antitumor activity.
-
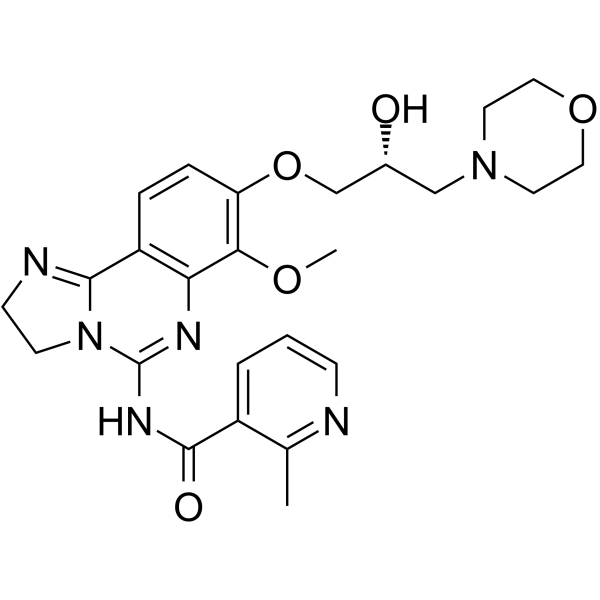
GC62164
BAY1082439
BAY1082439 is an orally bioavailable, selective PI3Kα/β/δ inhibitor. BAY1082439 also inhibits mutated forms of PIK3CA. BAY1082439 is highly effective in inhibiting Pten-null prostate cancer growth.
-
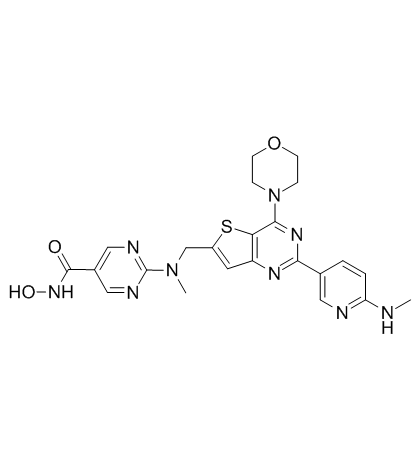
GC33145
BEBT-908 (PI3Kα inhibitor 1)
BEBT-908 (PI3Kα inhibitor 1) (PI3Kα inhibitor 1) is a selective PI3Kα inhibitor extracted from patent US/20120088764A1, Compound 243, has an IC50<0.1 μM, PI3Kα inhibitor 1 also inhibits HDAC (0.1 μM≤IC50≤1 μM) .
-
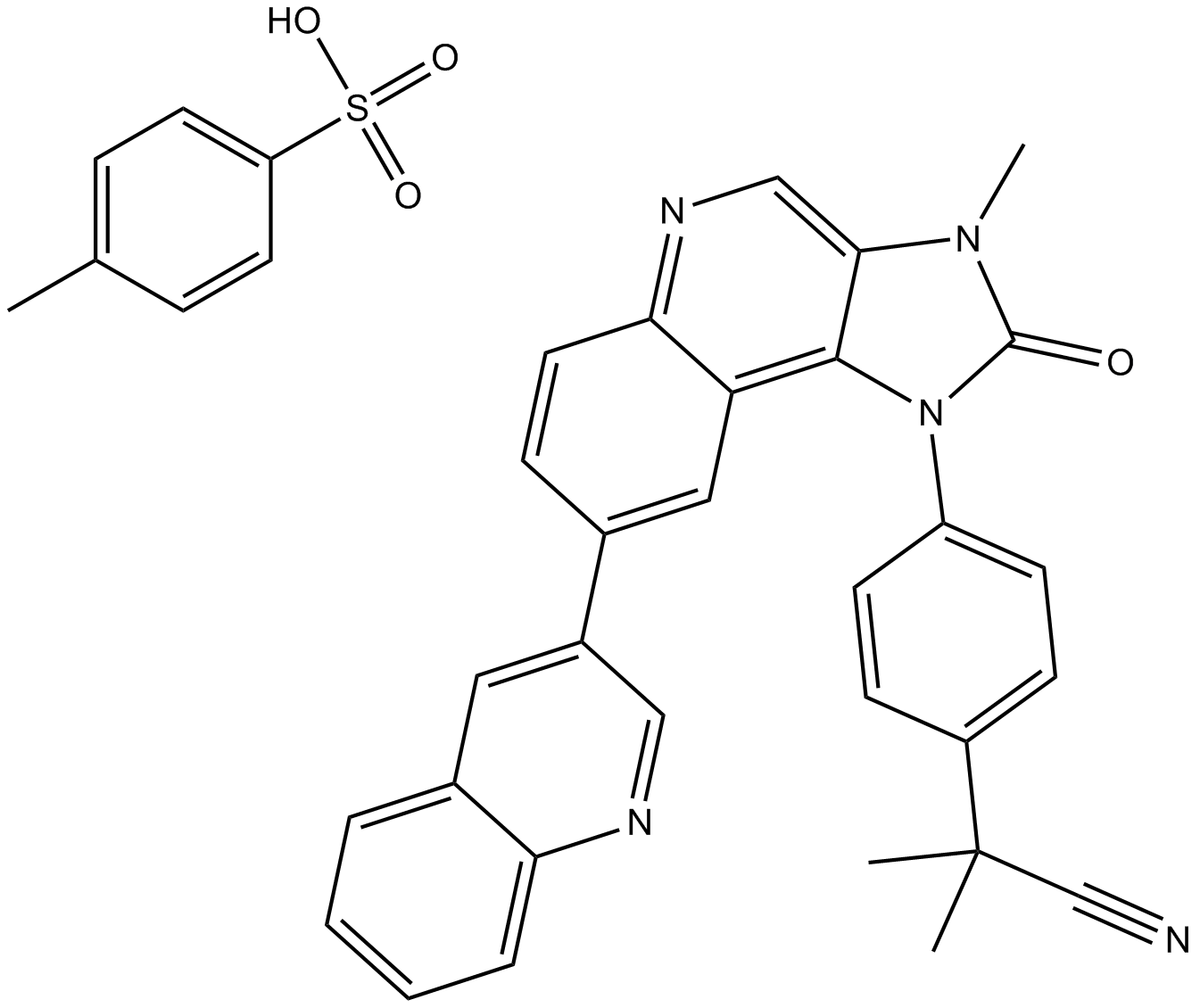
GC13271
BEZ235 Tosylate
-
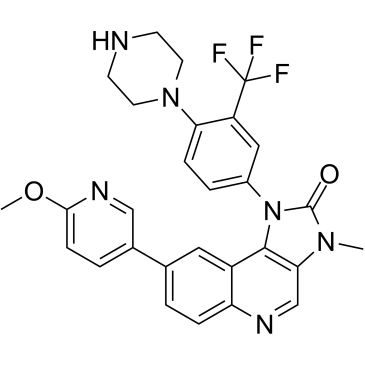
GC35509
BGT226
BGT226 (NVP-BGT226) is a PI3K (with IC50s of 4 nM, 63 nM and 38 nM for PI3Kα, PI3Kβ and PI3Kγ)/mTOR dual inhibitor which displays potent growth-inhibitory activity against human head and neck cancer cells.
-
GC11931
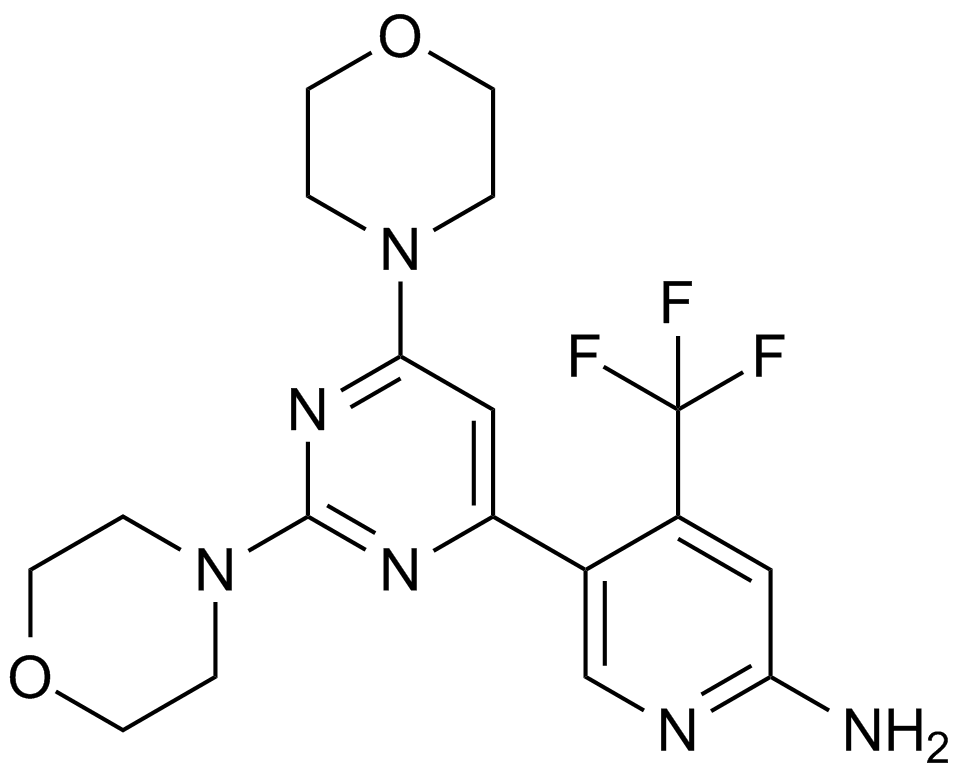
BKM120
An inhibitor of class I PI3K isoforms
-
GC65534
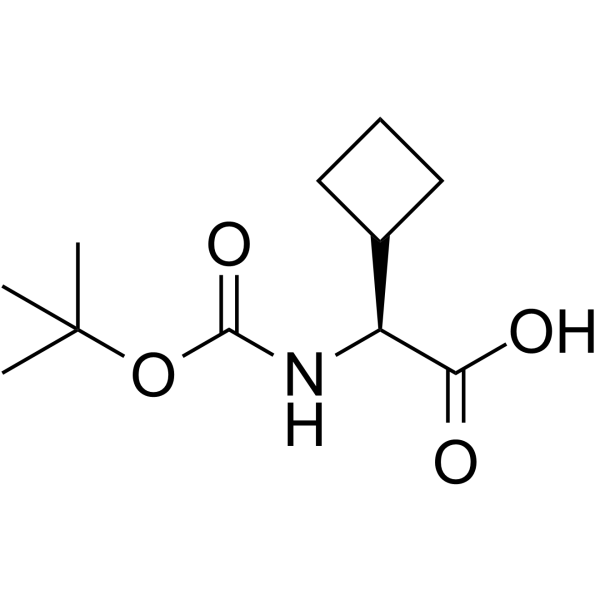
Boc-L-cyclobutylglycine
Boc-L-cyclobutylglycine is a glycine derivative that can be used for PI3K inhibitor synthesis.
-
GC30156
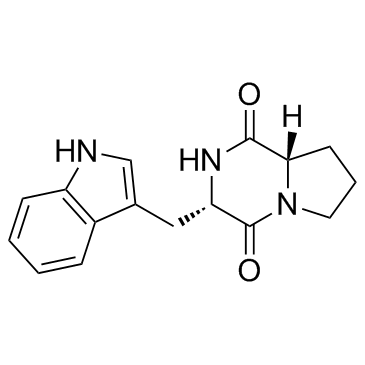
Brevianamide F (Cyclo(L-Pro-L-Trp))
Brevianamide F (Cyclo(L-Pro-L-Trp)) (Cyclo(L-Pro-L-Trp)) is a mycotoxin isolated from Colletotrichum gloeosporioides, with antibacterial activity.
-
GC16462
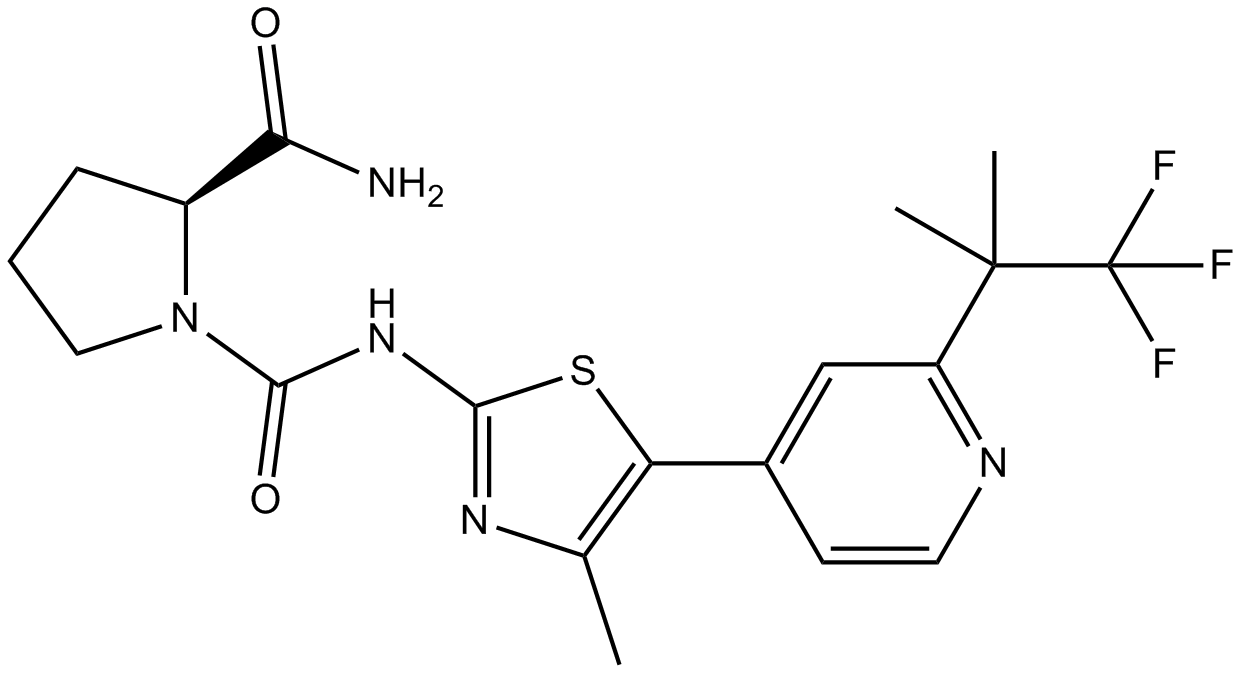
BYL-719
BYL-719 (BYL-719) is a potent, selective, and orally active PI3Kα inhibitor. BYL-719 (BYL-719) shows efficacy in targeting PIK3CA-mutated cancer. BYL-719 (BYL-719) also inhibits p110α/p110γ/p110δ/p110β with IC50s of 5/250/290/1200 nM, respectively. Antineoplastic activity.
-
GC13396
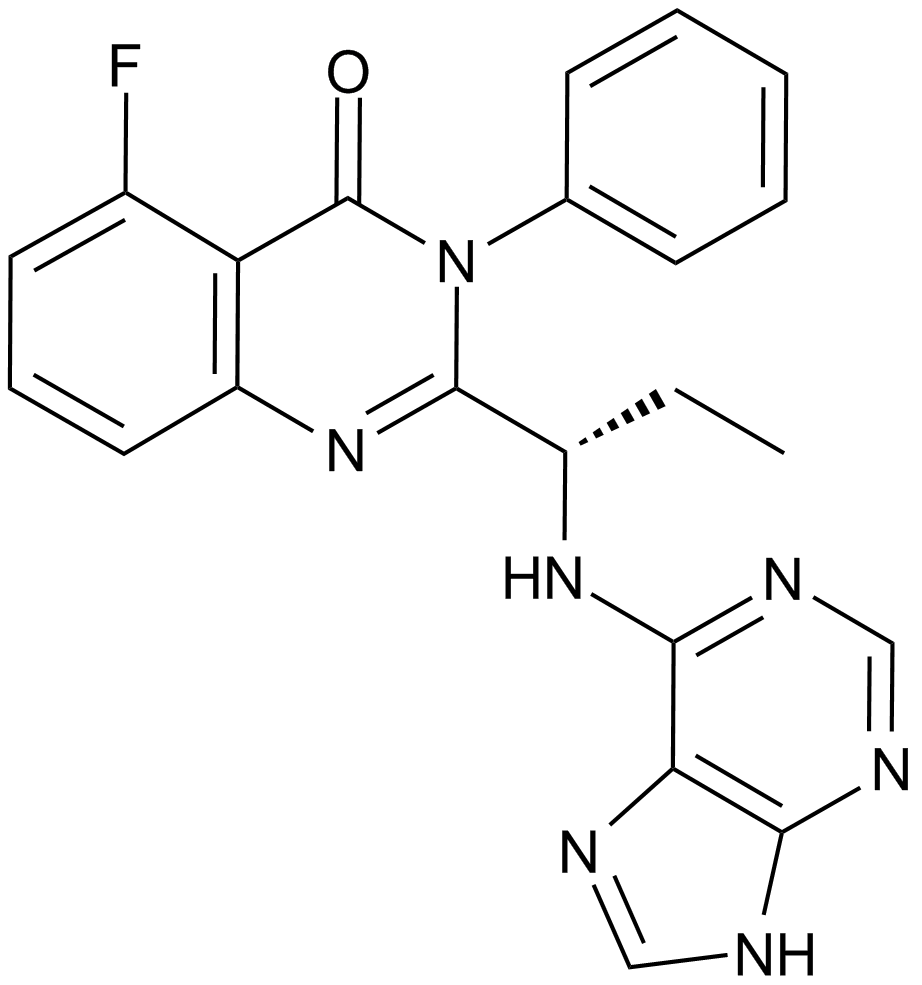
CAL-101 (Idelalisib, GS-1101)
A selective PI3K p110δ inhibitor
-
GC35579
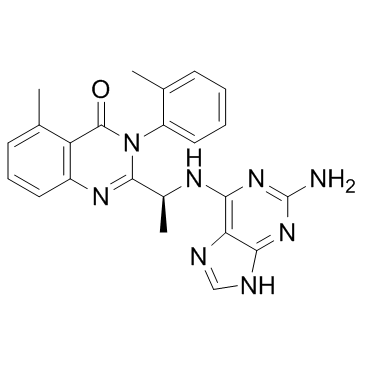
CAL-130
CAL-130 is a PI3Kδ and PI3Kγ inhibitor with IC50s of 1.3 and 6.1 nM, respectively.
-
GC11135
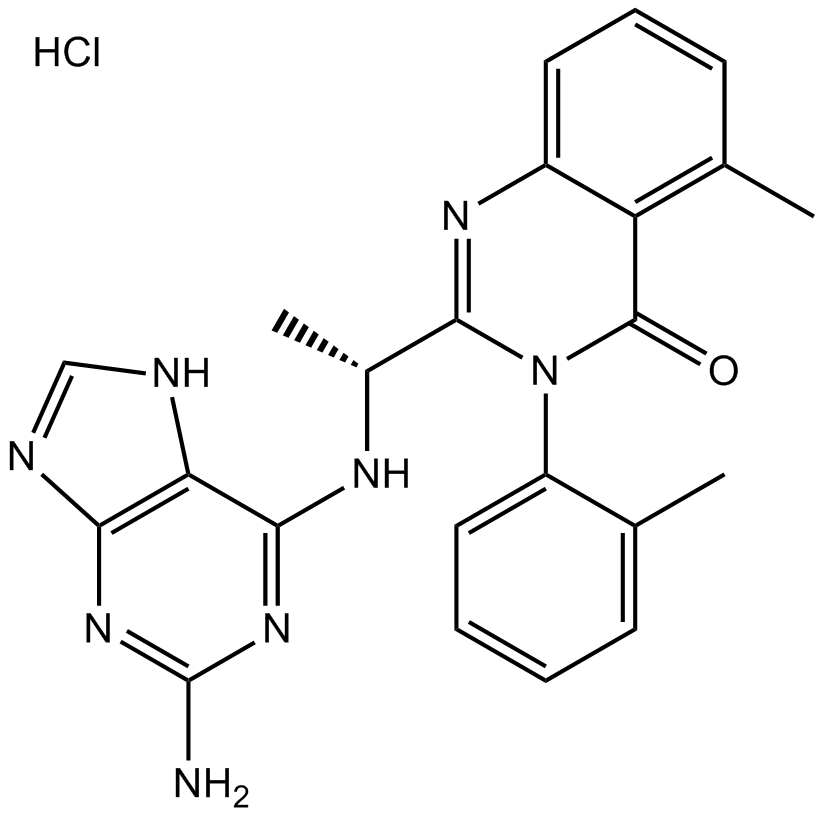
CAL-130 Hydrochloride
-
GC35580
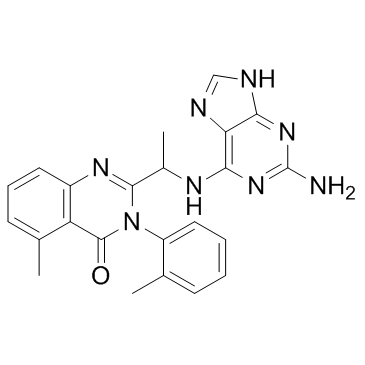
CAL-130 Racemate
CAL-130 Racemate is the racemate of CAL-130. CAL-130 Racemate is a PI3Kδ inhibitor.
-
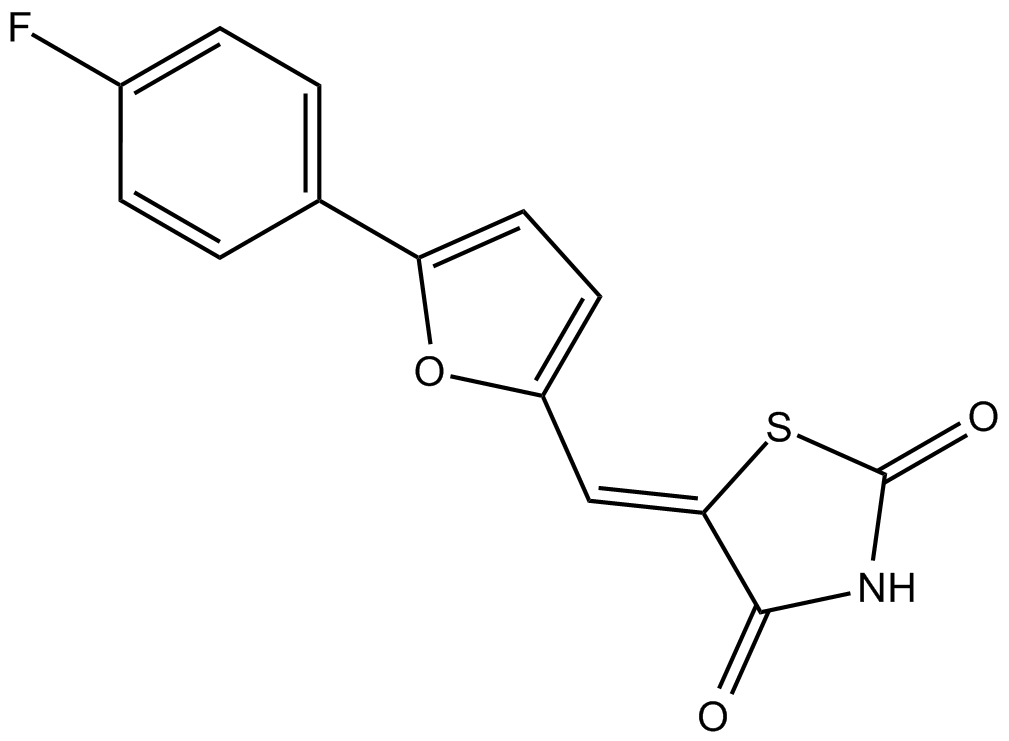
GC14318
CAY10505
Potent PI3Kγ inhibitor
-
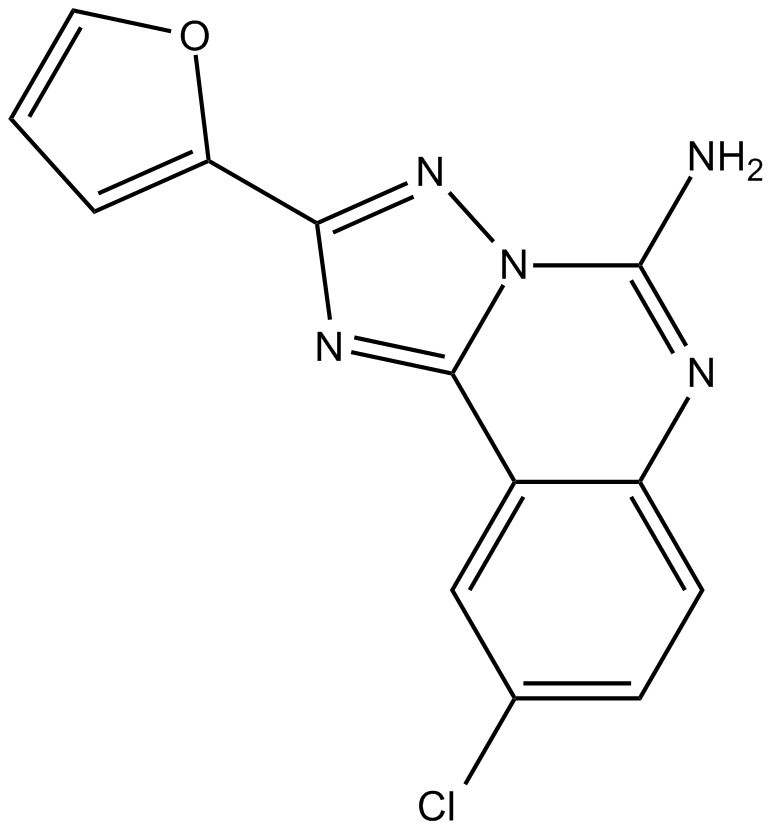
GC10070
CGS 15943
adenosine receptor antagonist
-
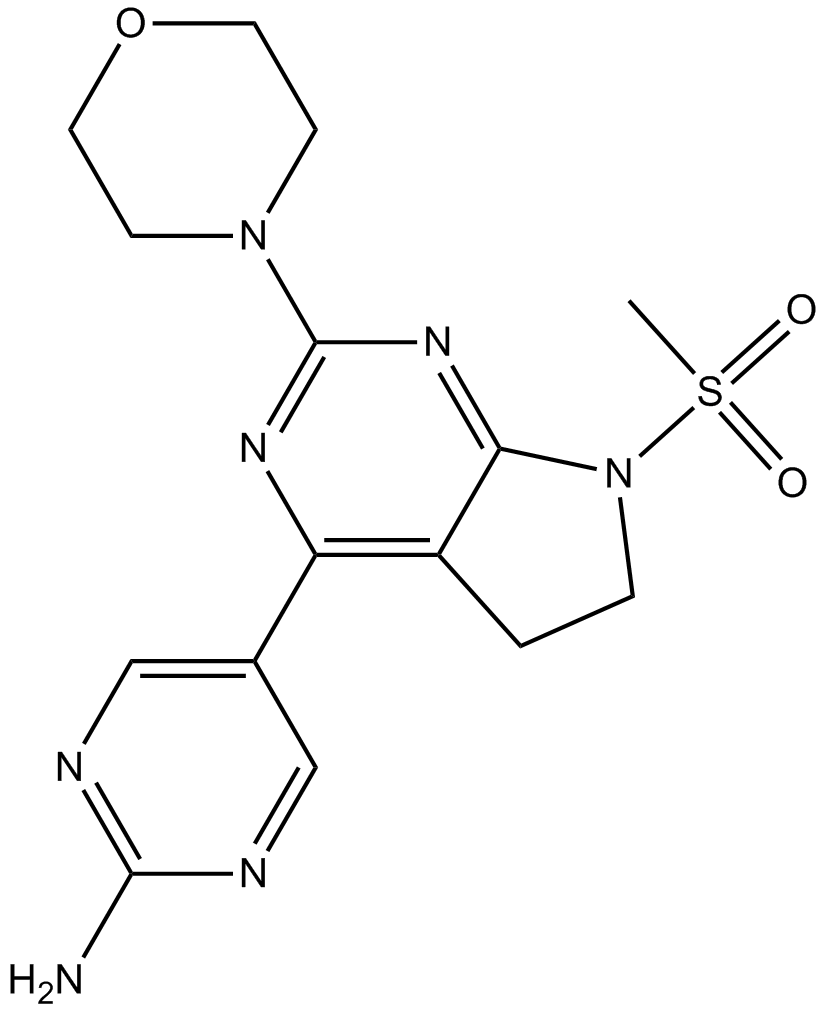
GC11868
CH5132799
Class I PI3K inhibitor
-
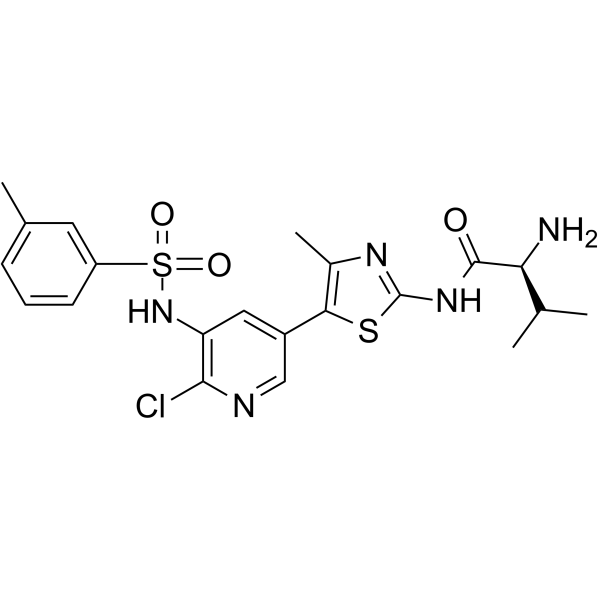
GC65969
CHMFL-PI3KD-317
CHMFL-PI3KD-317 is a highly potent, selective and orally active PI3Kδ inhibitor, with an IC50 of 6 nM, and exhibits over 10-1500 fold selectivity over other class I, II and III PIKK family isoforms, such as PI3Kα (IC50, 62.6 nM), PI3Kβ (IC50, 284 nM), PI3Kγ (IC50, 202.7 nM), PIK3C2A (IC50, >10000 nM), PIK3C2B (IC50, 882.3 nM), VPS34 (IC50, 1801.7 nM), PI4KIIIA (IC50, 574.1 nM) and PI4KIIIB (IC50, 300.2 nM). CHMFL-PI3KD-317 inhibits PI3Kδ-mediated Akt T308 phosphorylation in Raji cells, with an EC50 of 4.3 nM. CHMFL-PI3KD-317 has antiproliferative effects on cancer cells.
-
GC19109
CNX-1351
CNX-1351 is a potent and isoform-selective targeted covalent PI3Kα inhibitor with IC50 of 6.8 nM.
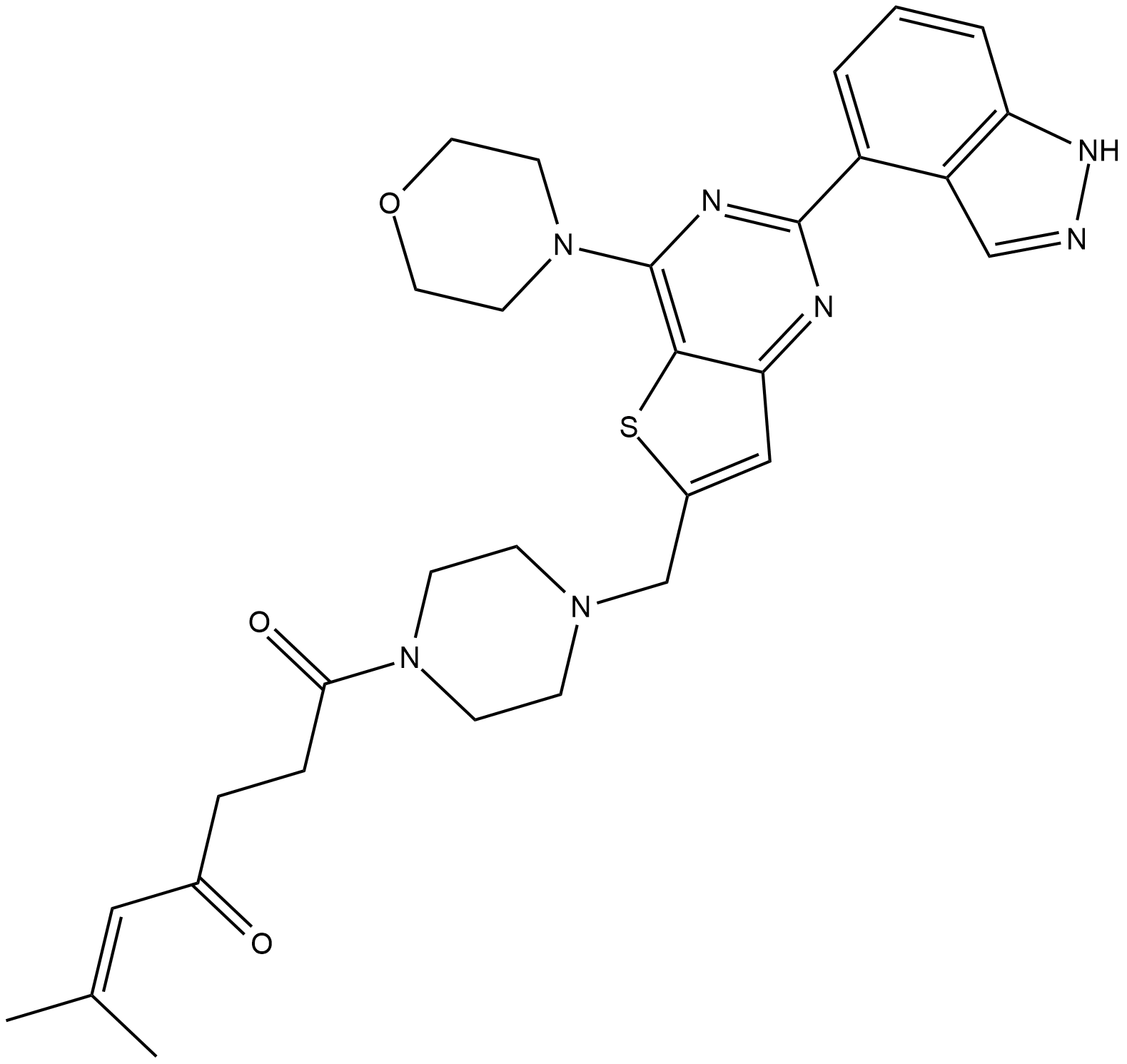
-
GC12115
CUDC-907
A dual inhibitor of HDACs and PI3Ks
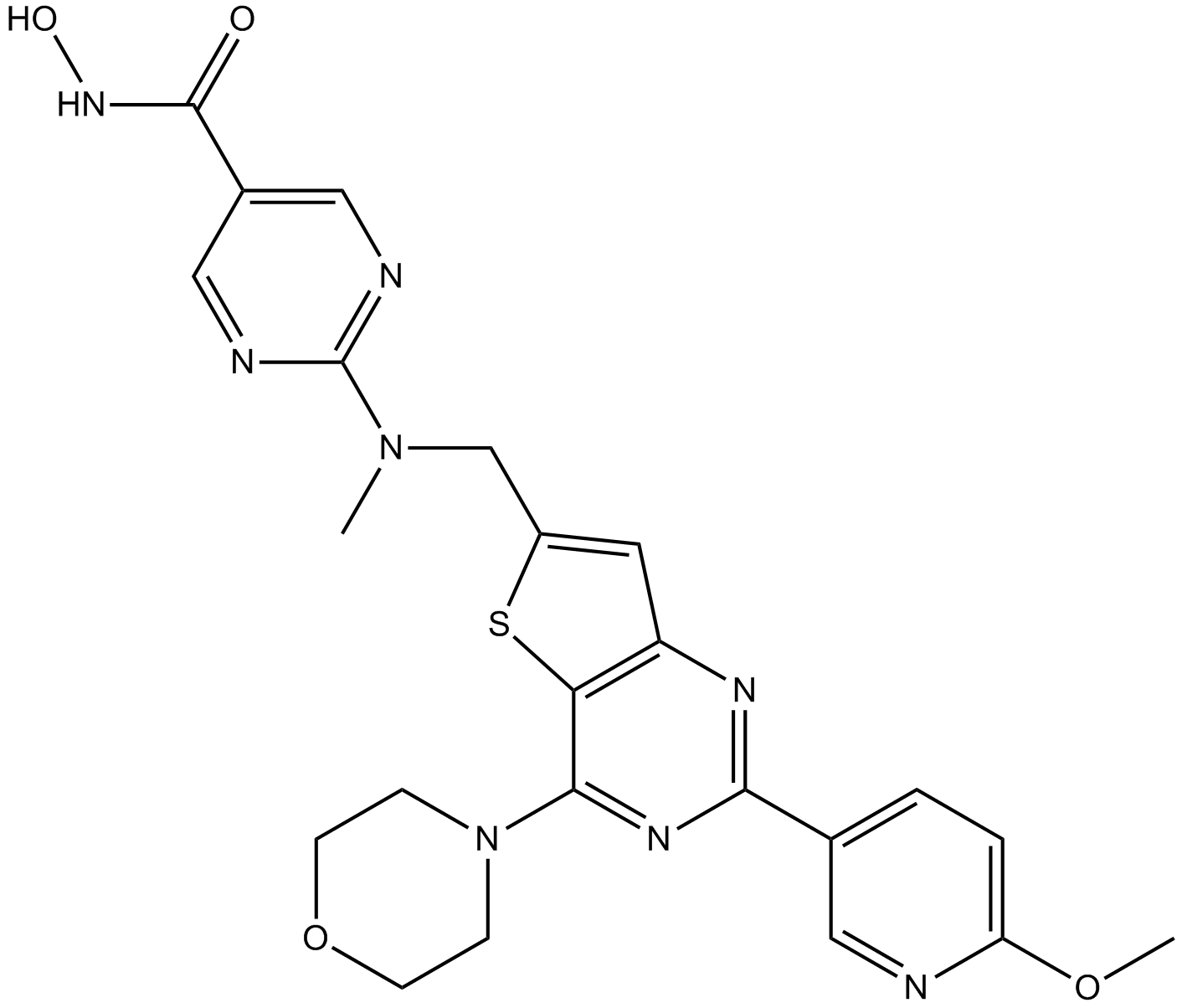
-
GC68413
CYH33
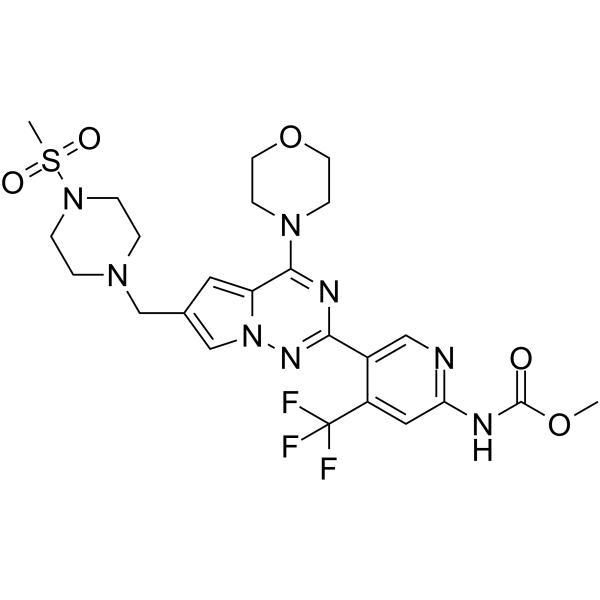
-
GC63391
CYH33 methanesulfonate
CYH33 methanesulfonate is an orally active, highly selective PI3Kα inhibitor with IC50s of 5.9 nM/598 nM/78.7 nM/225 nM against α/β/δ/γ isoform, respectively. CYH33 methanesulfonate inhibits phosphorylation of Akt, ERK and induces significant G1 phase arrest in breast cancer cells and non-small cell lung cancer (NSCLC) cells. CYH33 methanesulfonate has potent activity against solid tumors.
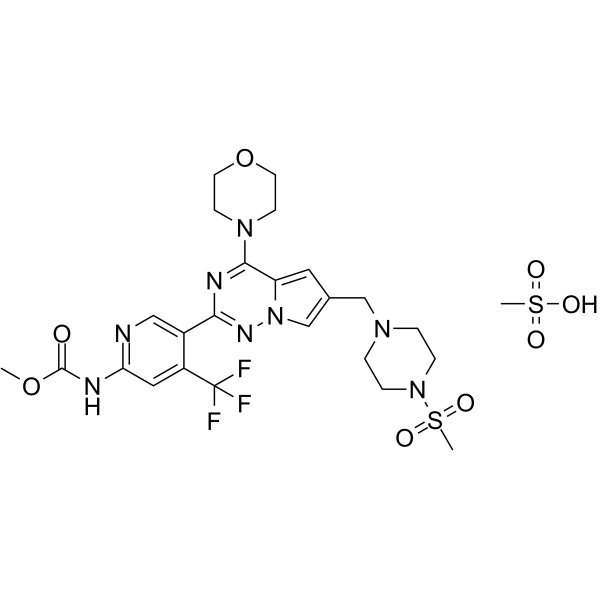
-
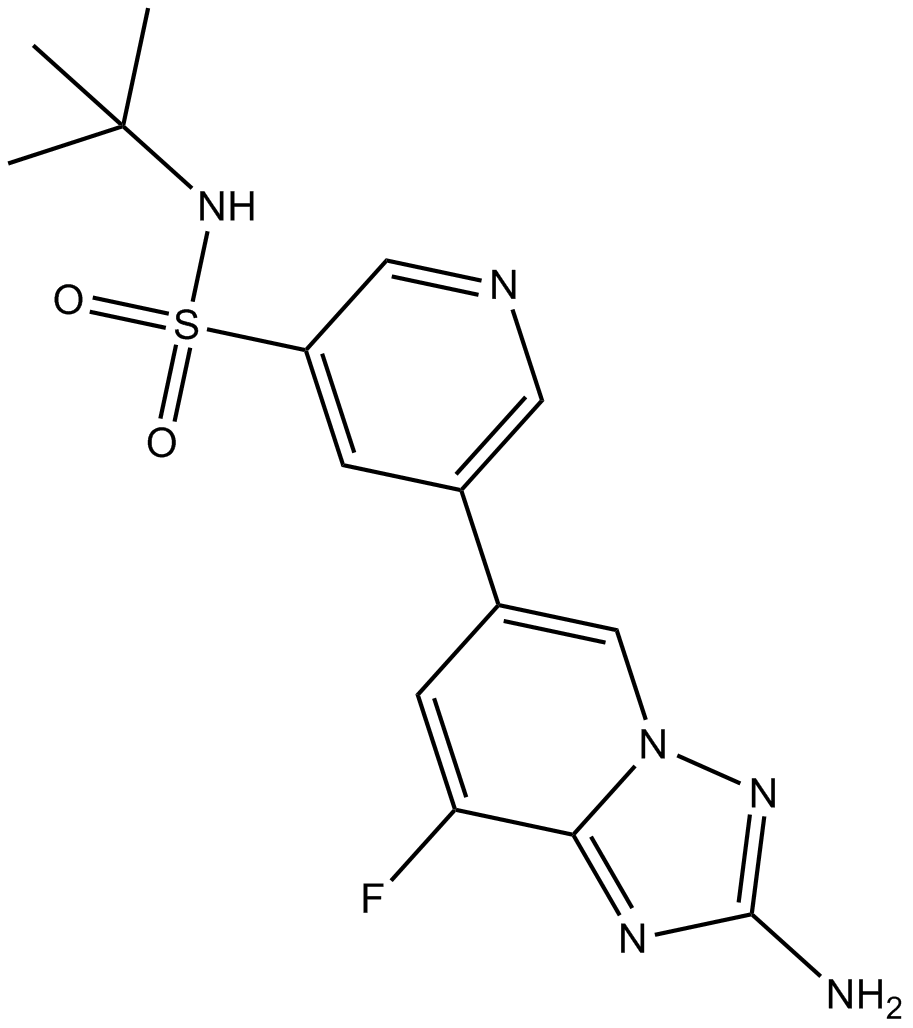
GC14798
CZC24832
A PI3Kγ inhibitor
-
GC38482
Desmethylglycitein
Desmethylglycitein (4',6,7-Trihydroxyisoflavone), a metabolite of daidzein, sourced from Glycine max with antioxidant, and anti-cancer activities.Desmethylglycitein binds directly to CDK1 and CDK2 in vivo, resulting in the suppresses CDK1 and CDK2 activity. Desmethylglycitein is a direct inhibitor of protein kinase C (PKC)α, against solar UV (sUV)-induced matrix matrix metalloproteinase 1 (MMP1). Desmethylglycitein binds to PI3K in an ATP competitive manner in the cytosol, where it inhibits the activity of PI3K and downstream signaling cascades, leading to the suppression of adipogenesis in 3T3-L1 preadipocytes.

-
GC60782
Disitertide TFA
Disitertide (P144) TFA is a peptidic transforming growth factor-beta 1 (TGF-β1) inhibitor specifically designed to block the interaction with its receptor. Disitertide (P144) TFA is also a PI3K inhibitor and an apoptosis inducer.

-
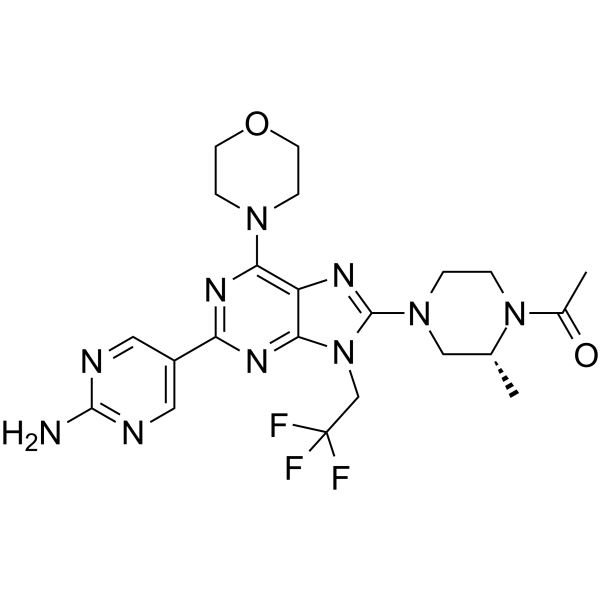
GC62495
DS-7423
DS-7423 is a dual PI3K and mTOR inhibitor, with IC50 values of 15.6 nM, 34.9 nM for PI3Kα and mTOR, respectively. DS-7423 possesses anti-tumor activity.
-
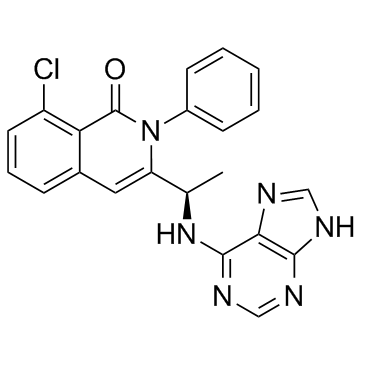
GC33162
Duvelisib R enantiomer (IPI-145 R enantiomer)
Duvelisib R enantiomer is a PI3K inhibitor, which is the less active enantiomer of Duvelisib.
-
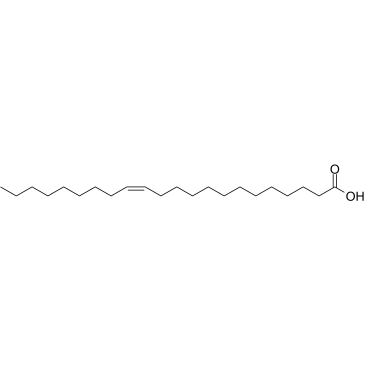
GC38694
Erucic acid
A 22carbon monounsaturated fatty acid
-
GC38236
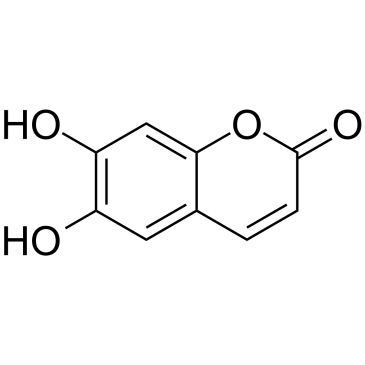
Esculetin
A coumarin with diverse actions
-
GC19145
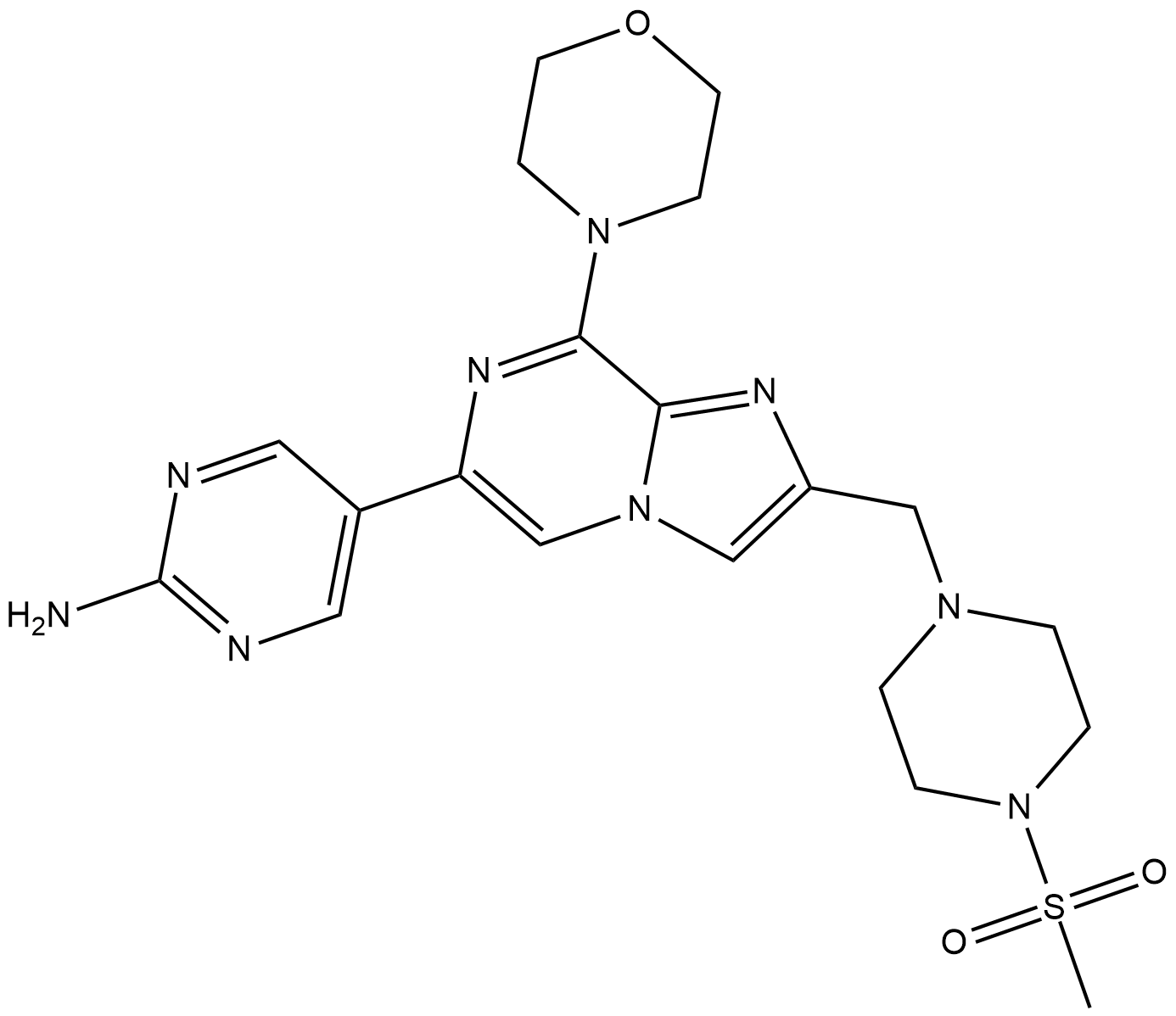
ETP-46321
ETP-46321 is a potent and orally bioavailable PI3Kα and PI3Kδ inhibitor with Kiapps of 2.3 and 14.2 nM, respectively.
-
GC38392
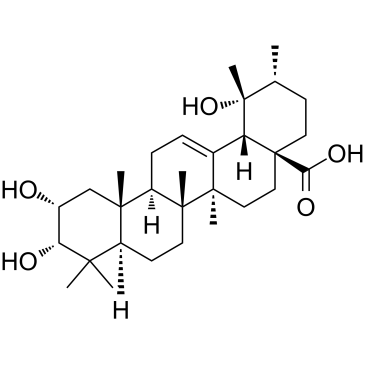
Euscaphic acid
Euscaphic acid, a DNA polymerase inhibitor, is a triterpene from the root of the R. alceaefolius Poir. Euscaphic inhibits calf DNA polymerase α (pol α) and rat DNA polymerase β (pol β) with IC50 values of 61 and 108 μM. Euscaphic acid induces apoptosis.
-
GC62973
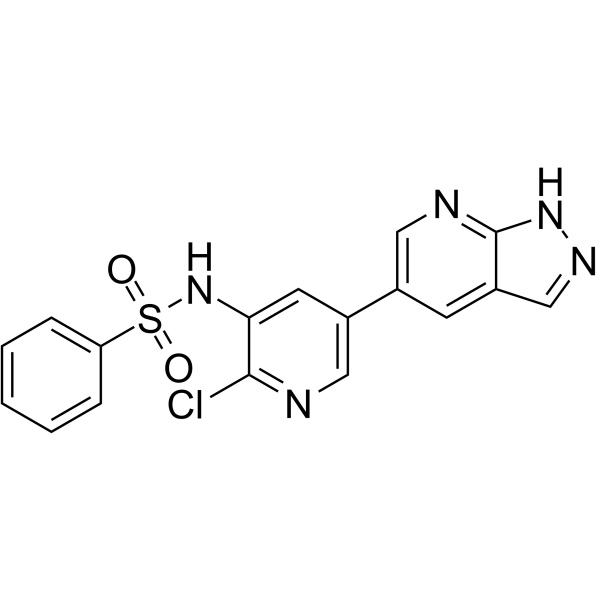
FD223
FD223 is a potent and selective phosphoinositide 3-kinase delta (PI3Kδ) inhibitor. FD223 displays high potency (IC50=1 nM) and good selectivity over other isoforms (IC50s of 51 nM, 29 nM and 37 nM, respectively for α, β and γ). FD223 exhibits efficient inhibition of the proliferation of acute myeloid leukemia (AML) cell lines by suppressing p-AKT Ser473 thus causing G1 phase arrest during the cell cycle. FD223 has potential for the research of leukemia such as AML.
-
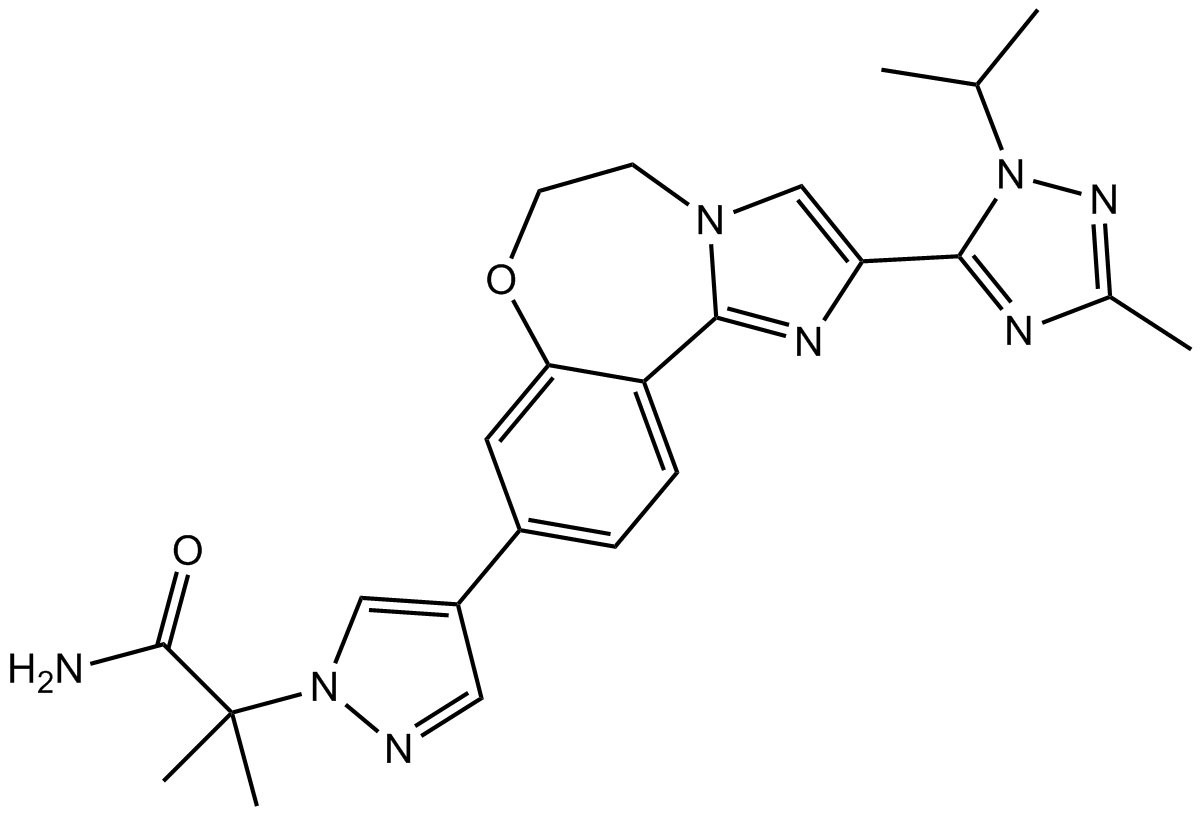
GC17967
GDC-0032
GDC-0032 (GDC-0032) is a potent PI3K inhibitor targets PIK3CA mutations, with Kis of 0.12 nM, 0.29 nM, 0.97 nM, and 9.1 nM for PI3Kδ, PI3Kα, PI3Kγ and PI3Kβ, respectively.
-
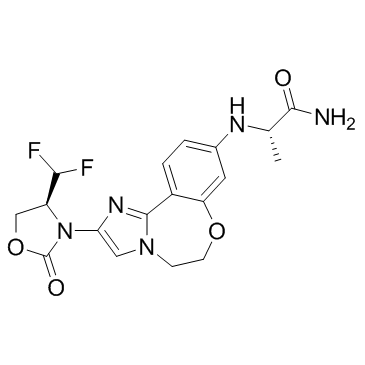
GC32710
GDC-0077 (RG6114)
GDC-0077 (RG6114) is a potent, orally available, and selective PI3Kα inhibitor (IC50=0.038 nM). GDC-0077 (RG6114) exerts its activity by binding to the ATP binding site of PI3K, thereby inhibiting the phosphorylation of PIP2 to PIP3. GDC-0077 (RG6114) is more selective for mutant versus wild-type PI3Kα.
-
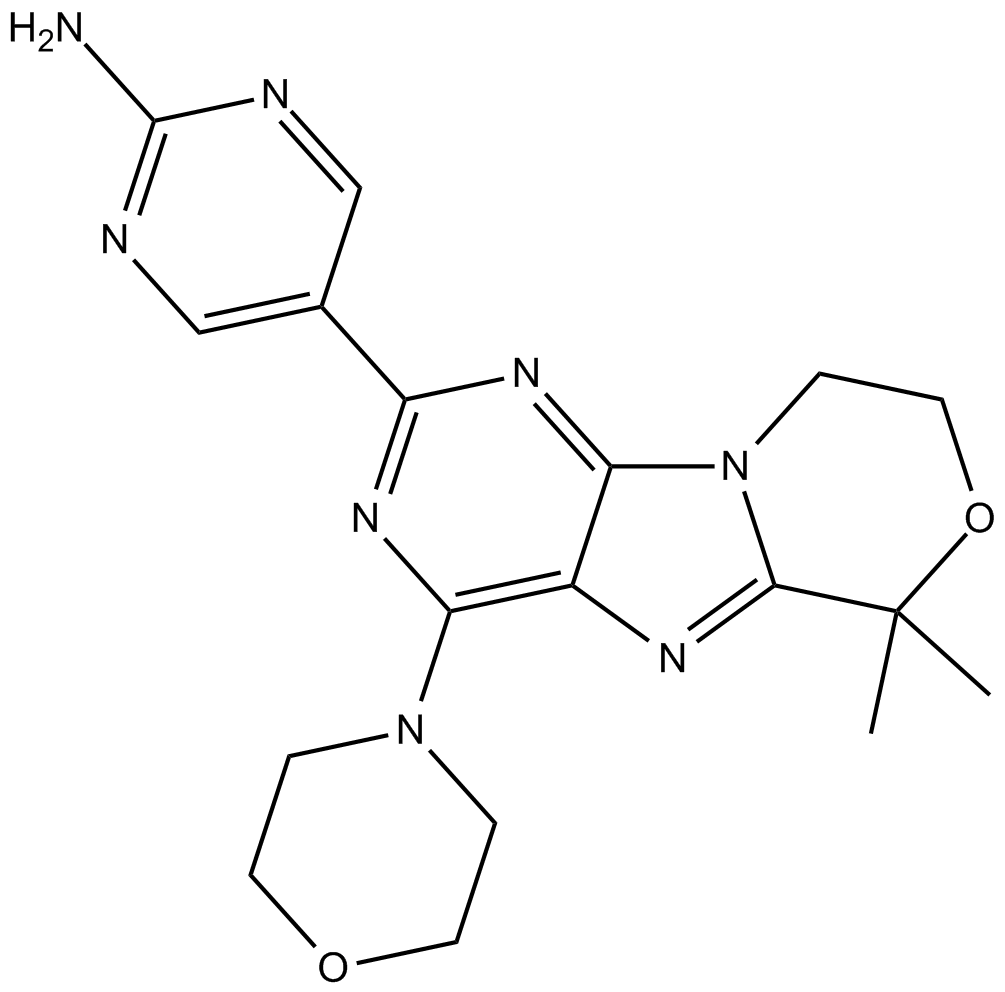
GC10228
GDC-0084
PI3K and mTOR inhibitor, brain-permeable
-
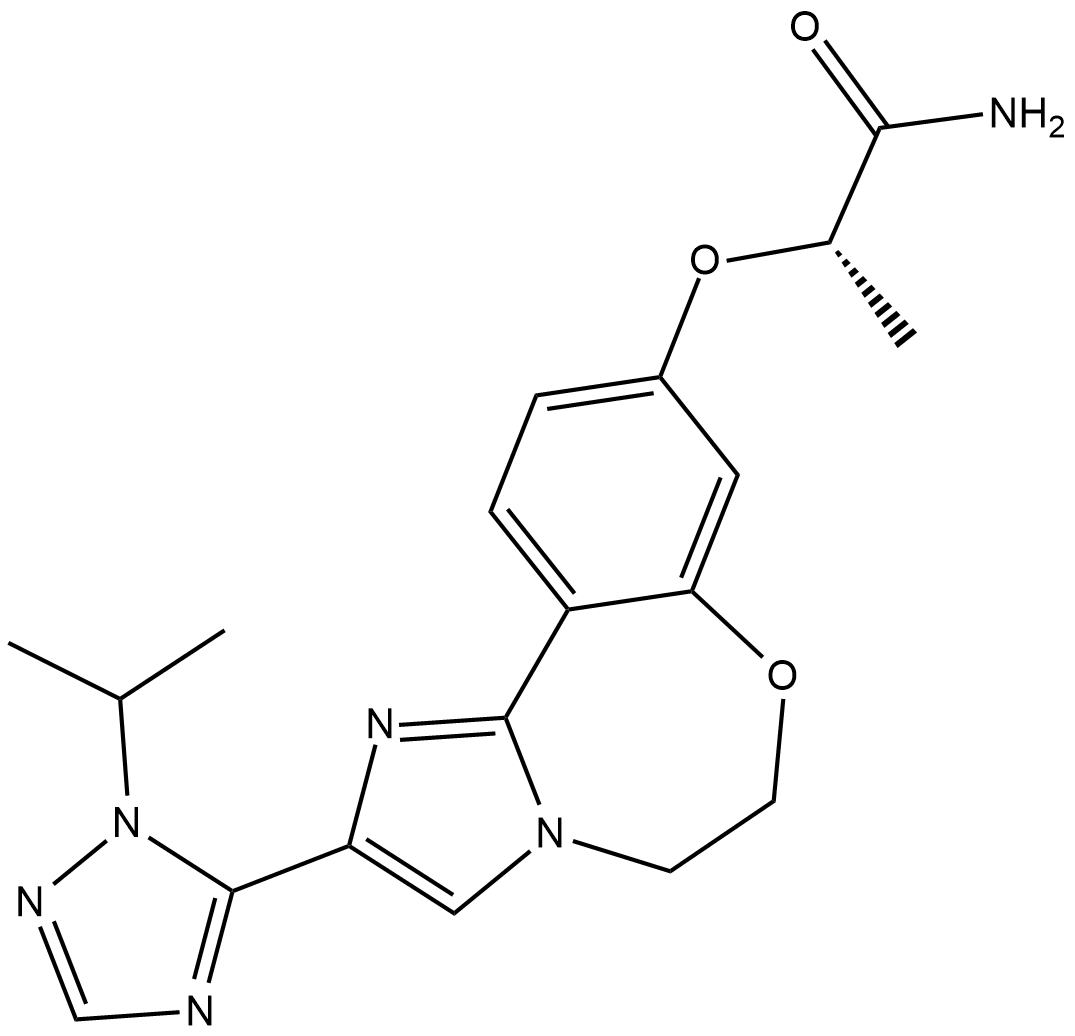
GC19161
GDC-0326
GDC-0326 is a potent and selective PI3Kα inhibitor with a Ki of 0.2 nM.
-
GC16154
GDC-0941 dimethanesulfonate
A pan inhibitor of class I PI3K isoforms
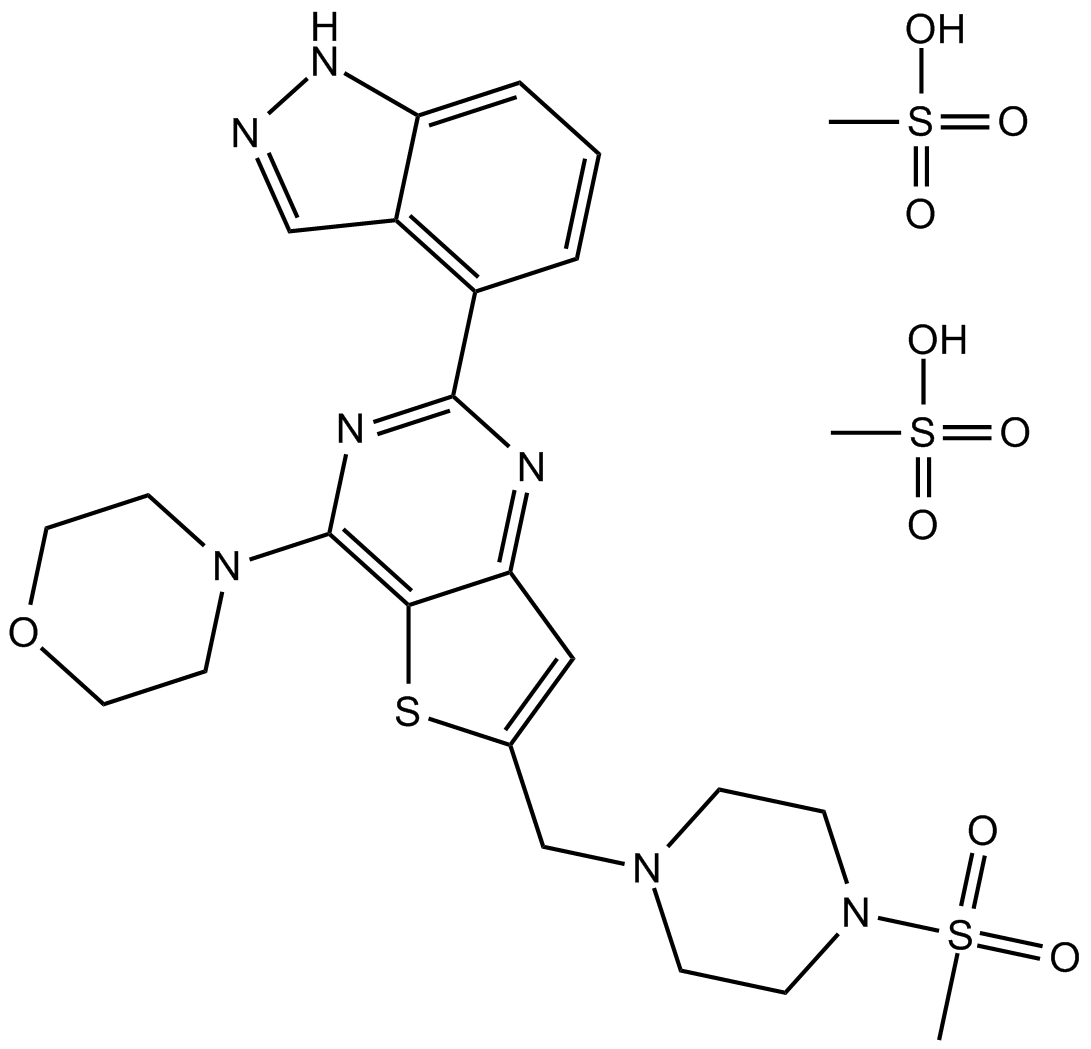
-
GC11177
GDC-0980 (RG7422)
GDC-0980 (RG7422) (GDC-0980; GNE 390; RG 7422) is a selective, potent, orally bioavailable Class I PI3 kinase and mTOR kinase (TORC1/2) inhibitor with IC50s of 5 nM/27 nM/7 nM/14 nM for PI3Kα/PI3Kβ/PI3Kδ/PI3Kγ, and with aKiof 17 nM for mTOR.
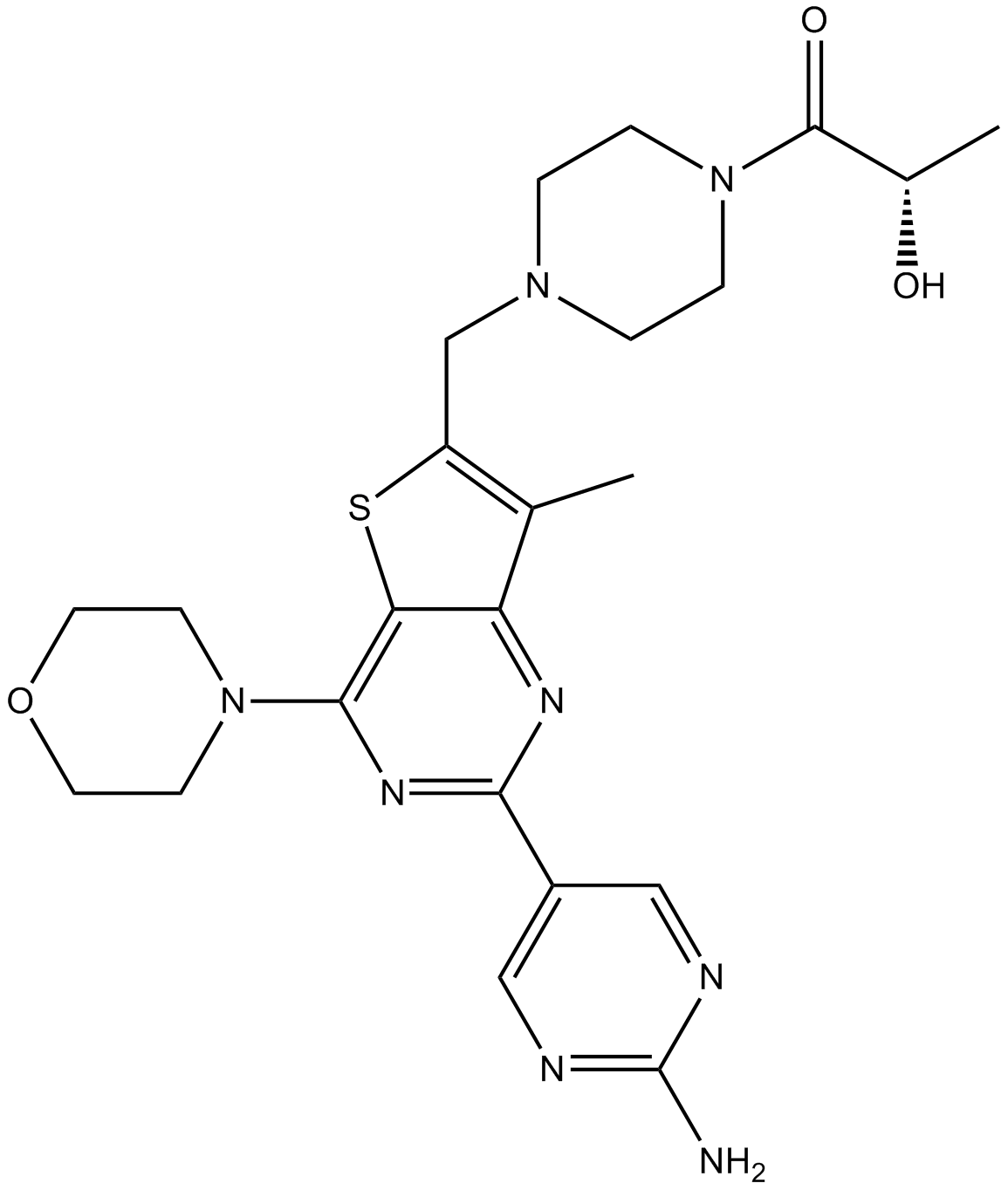
-
GC64778
Gilmelisib
Gilmelisib is an antineoplastic. Gilmelisib is a PI3K inhibitor (IC50 <1 nM for PI3K p110α) extracted from WO2017101847 A1, compound 1.
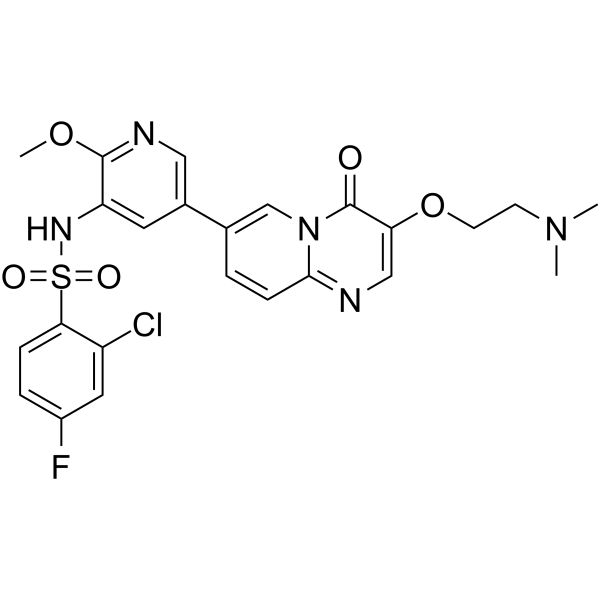
-
GN10584
Ginsenoside Rk1
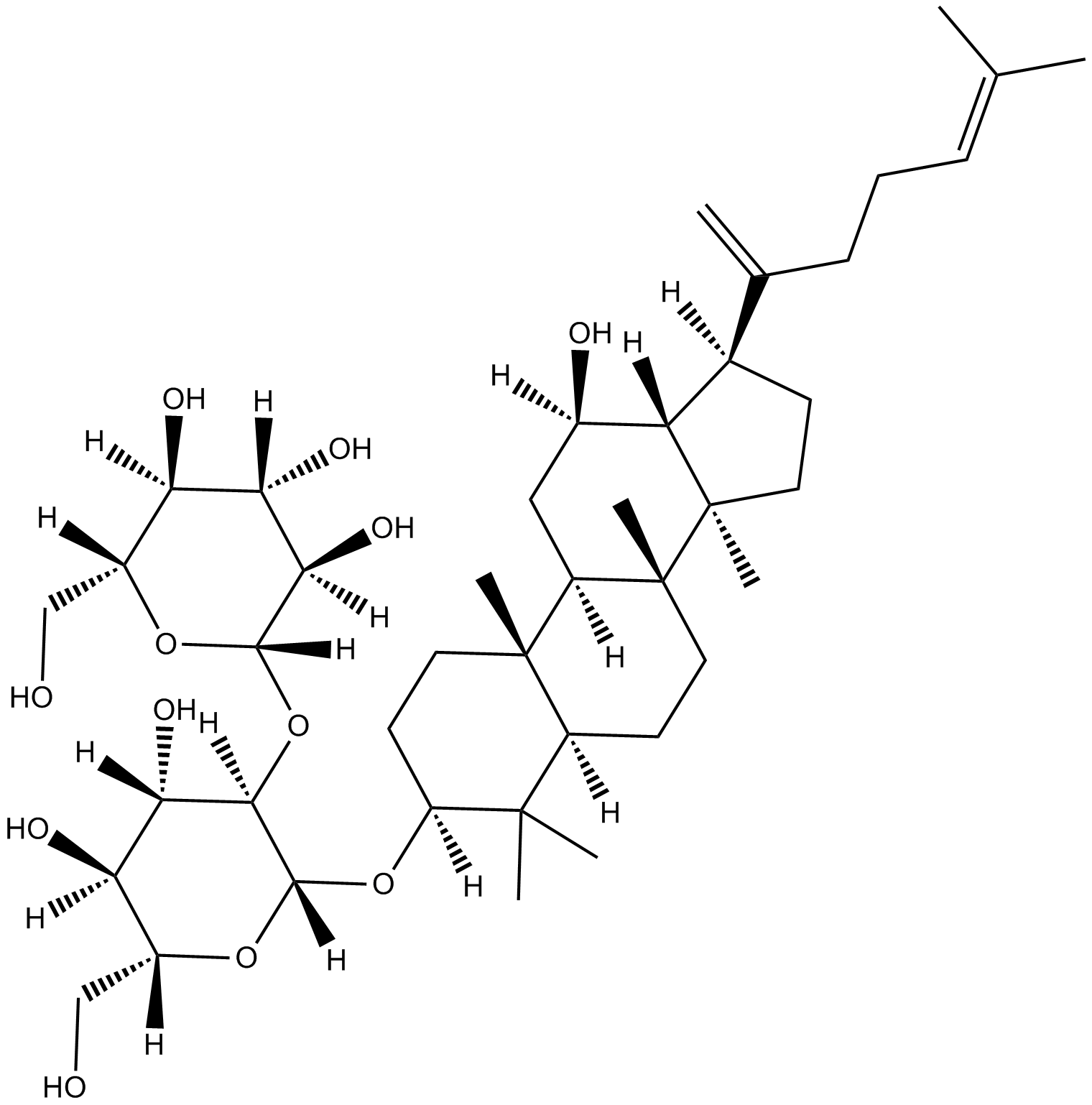
-
GC38606
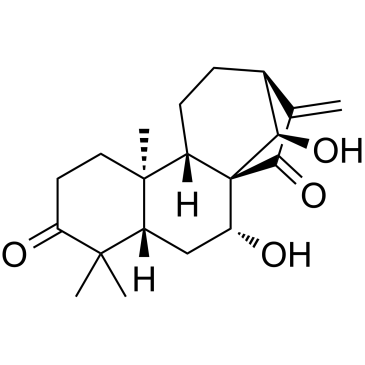
Glaucocalyxin A
Glaucocalyxin A, an ent-kauranoid diterpene from Rabdosia japonica var., induces apoptosis in osteosarcoma by inhibiting nuclear translocation of Five-zinc finger Glis 1 (GLI1) via regulating PI3K/Akt signaling pathway. Glaucocalyxin A has antitumor effect.
-
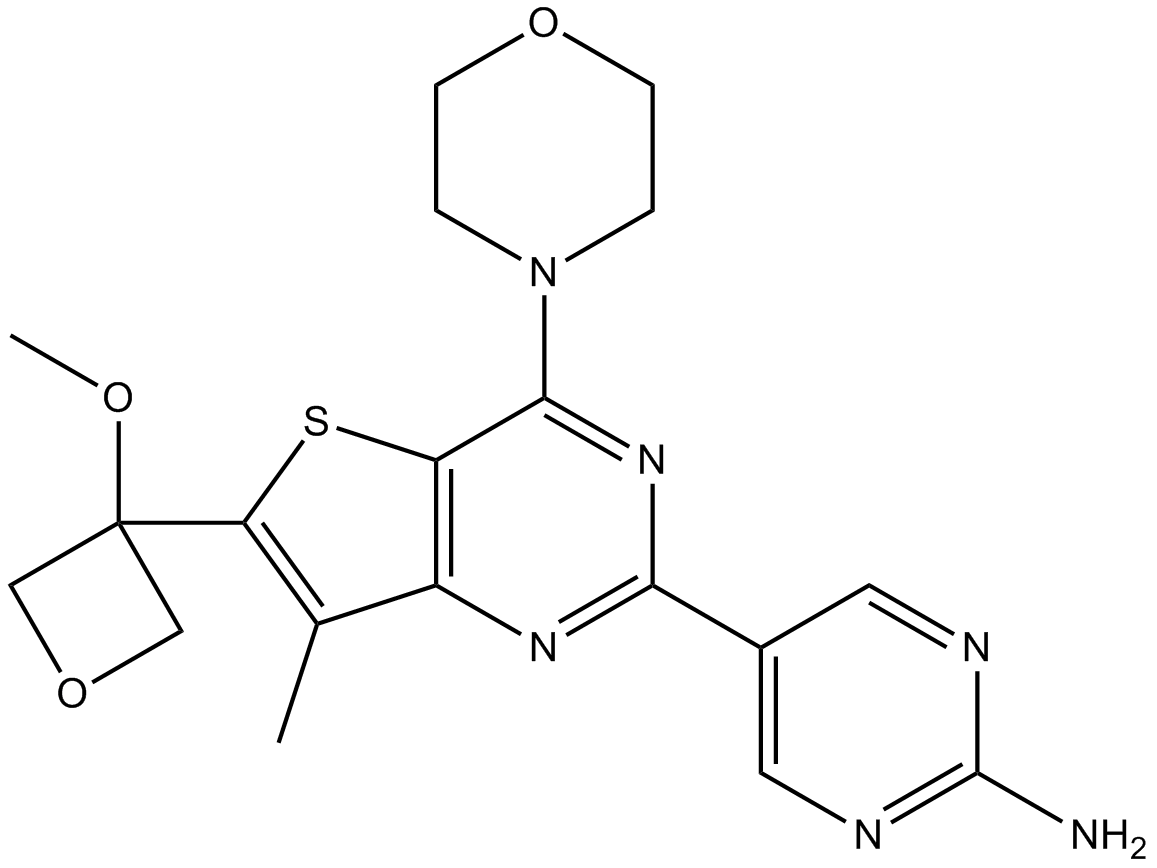
GC17338
GNE-317
potent, brain-penetrant PI3K inhibitor
-
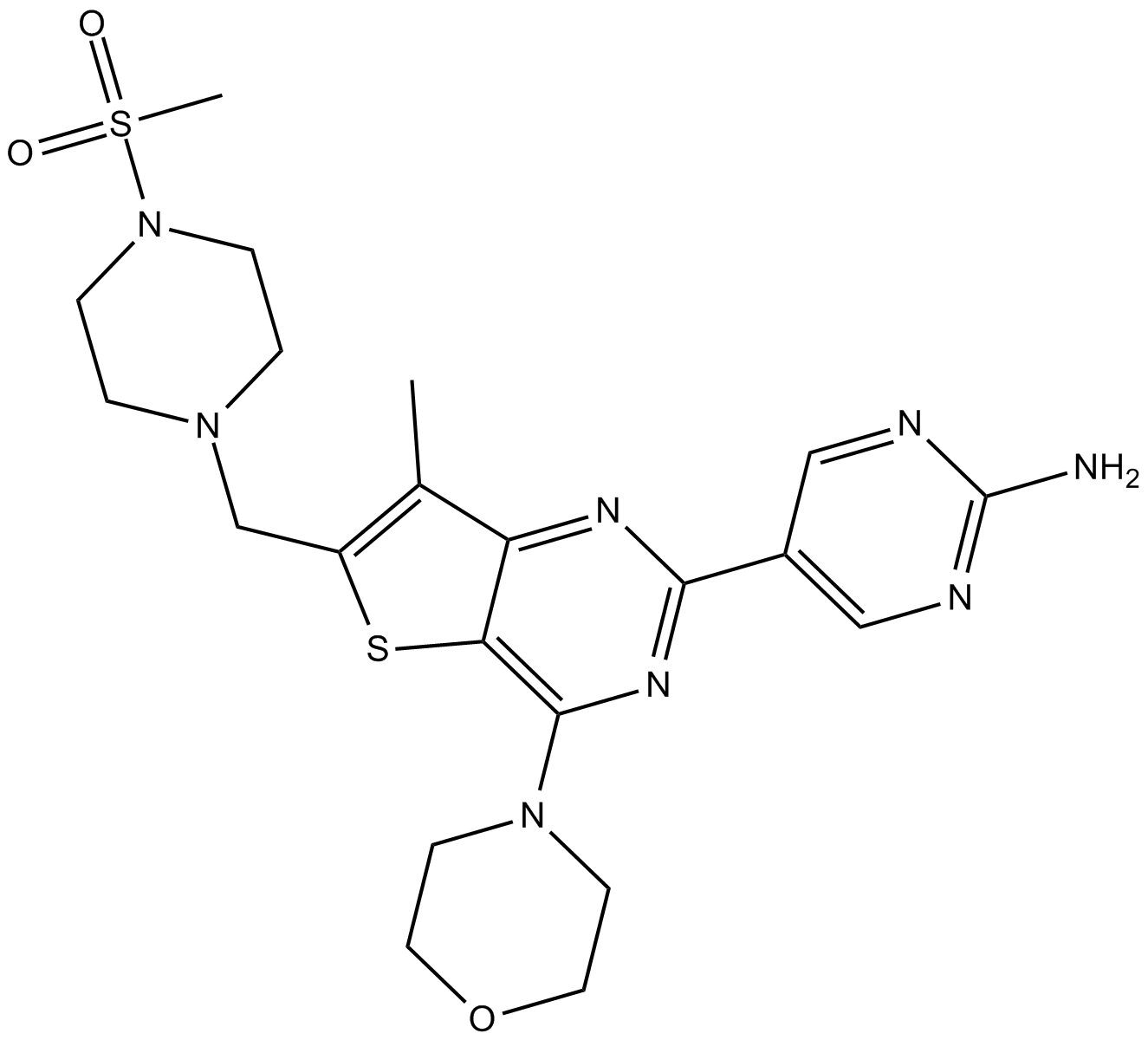
GC10340
GNE-477
dual PI3K/mTOR inhibitor
-
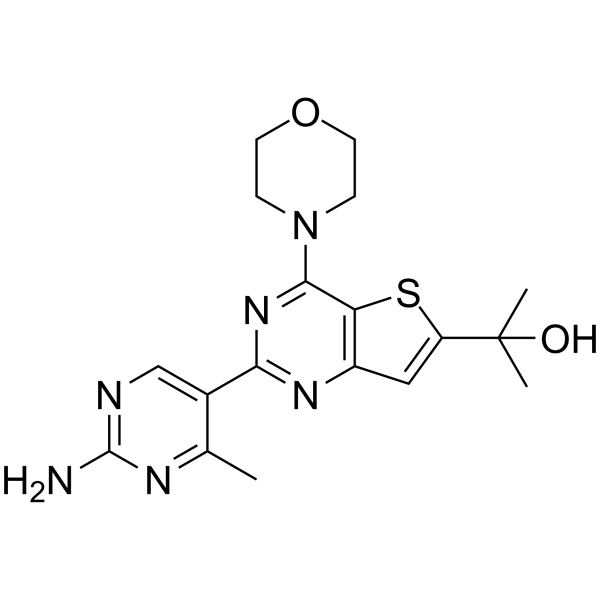
GC68429
GNE-490
-
GC15423
GNE-493
Pan-PI3K/mTOR inhibitor
-
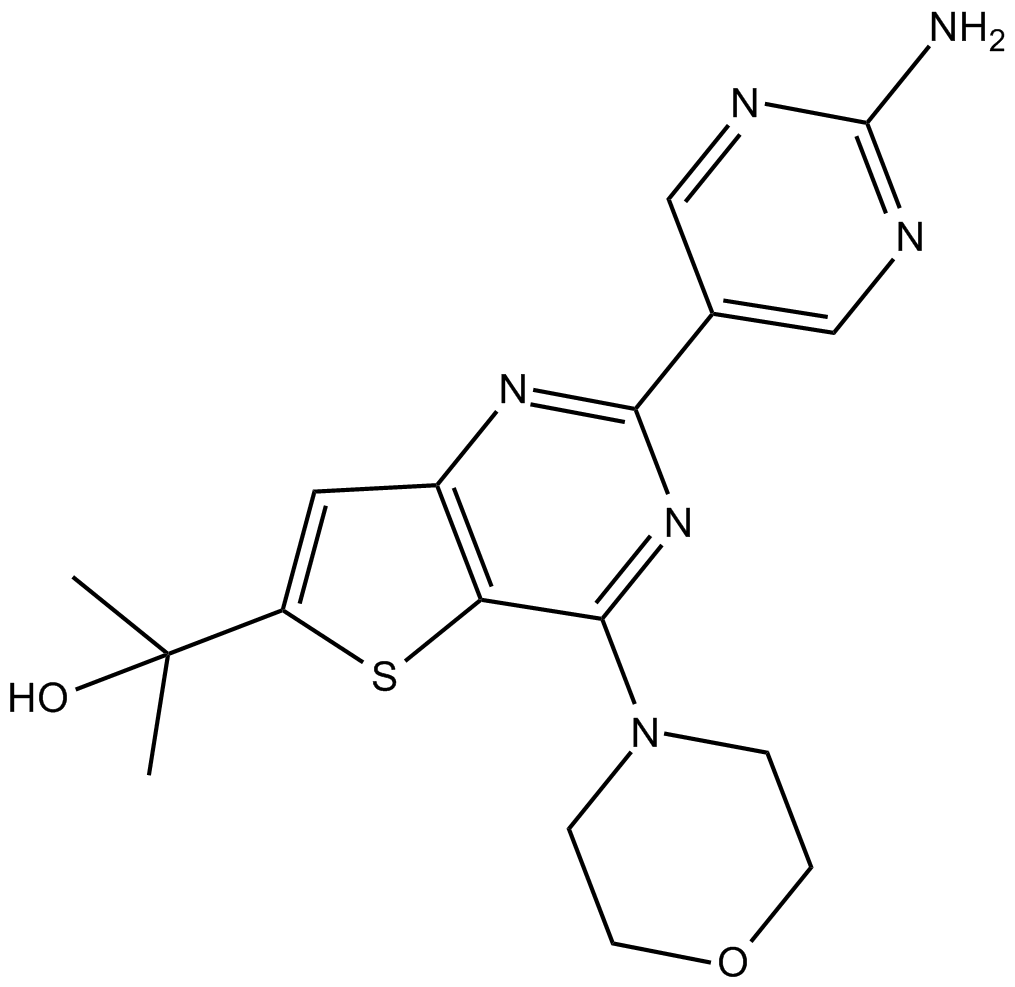
GC36186
GS-9901
GS-9901 is a highly selective and orally active PI3Kδ inhibitor, with an IC50 of 1 nM.
-
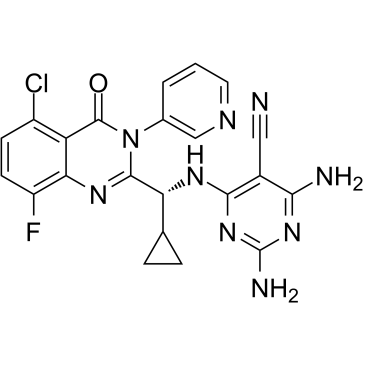
GC65309
GSK-F1
GSK-F1 (Compound F1) is an orally active PI4KA inhibitor with pIC50 values of 8.0, 5.9, 5.8, 5.9, 5.9 and 6.4 against PI4KA, PI4KB, PI3KA, PI3KB, PI3KG and PI3KD, respectively.
-
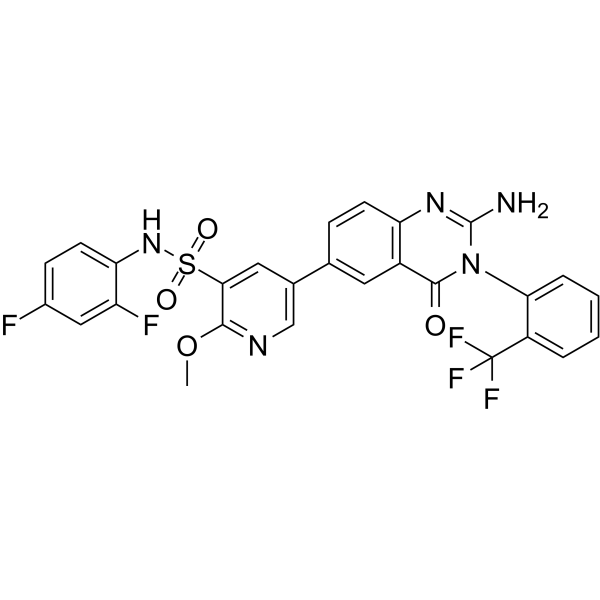
GC11018
GSK1059615
PI3K and mTOR inhibitor,potent and reversible
-
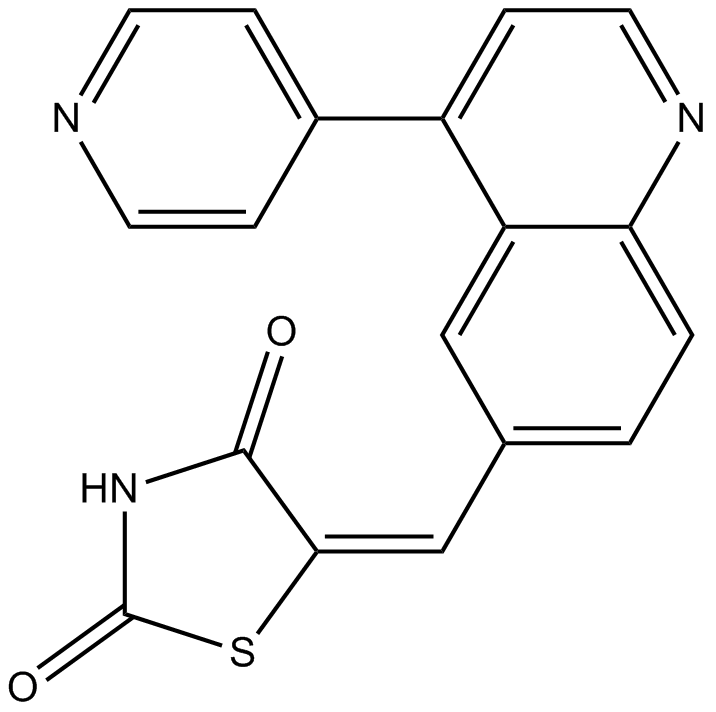
GC15072
GSK2126458
GSK2126458 (GSK2126458) is an orally active and highly selective inhibitor of PI3K with Kis of 0.019 nM/0.13 nM/0.024 nM/0.06 nM and 0.18 nM/0.3 nM for p110α/β/δ/γ, mTORC1/2, respectively. GSK2126458 has anti-cancer activity.
-
GC15595
GSK2269557
inhibitor of PI3Kδ
-
GC15679
GSK2292767
PI3Kδ inhibitor,potent and selective
-
GC64612
GSK251
GSK251 is a highly potent, highly selective, orally bioavailable inhibitor of PI3Kδ with a novel binding mode.
-
GC17109
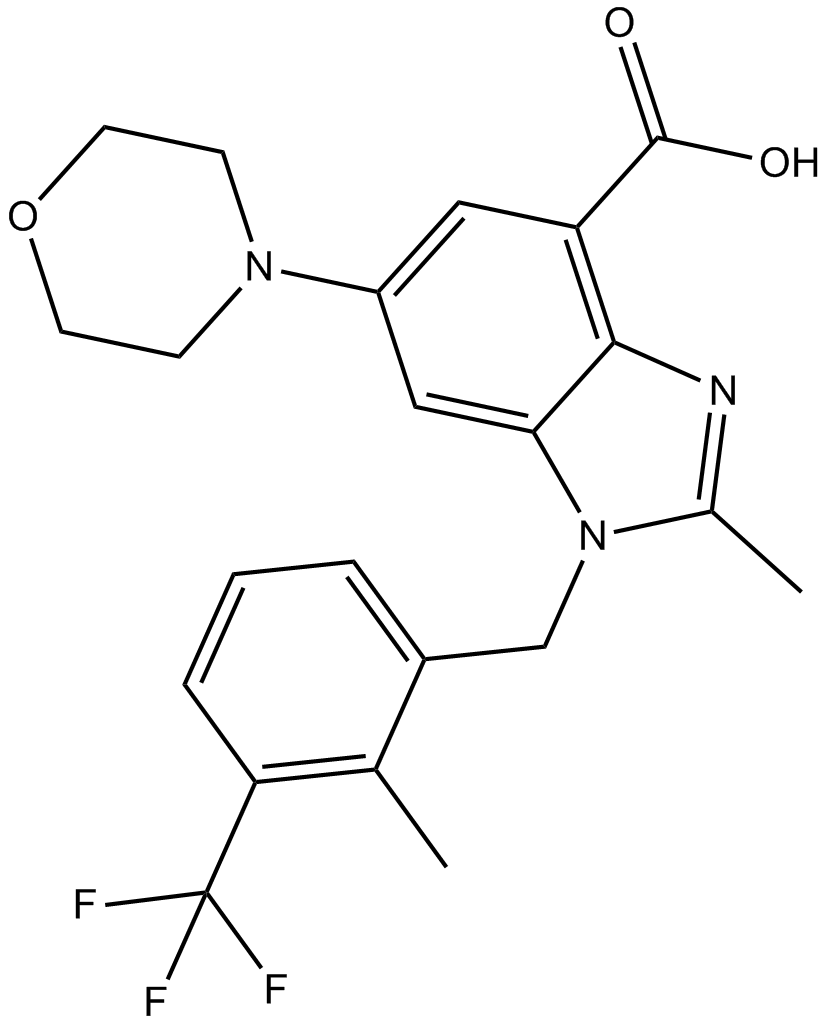
GSK2636771
GSK2636771 is a potent adenosine triphosphate competitive oral inhibitor of PI3Kβ, with an IC50 of 5.
-
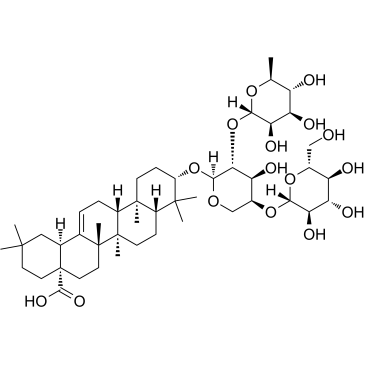
GC38088
Hederacolchiside A1
-
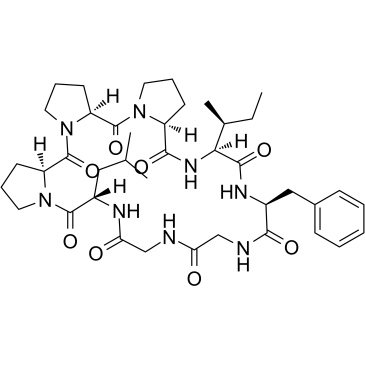
GC38168
Heterophyllin B
Heterophyllin B is an active cyclic peptide isolated from Pseudostellaria heterophylla. Heterophyllin B provides a novel strategy for the treatment of esophageal cancer.
-
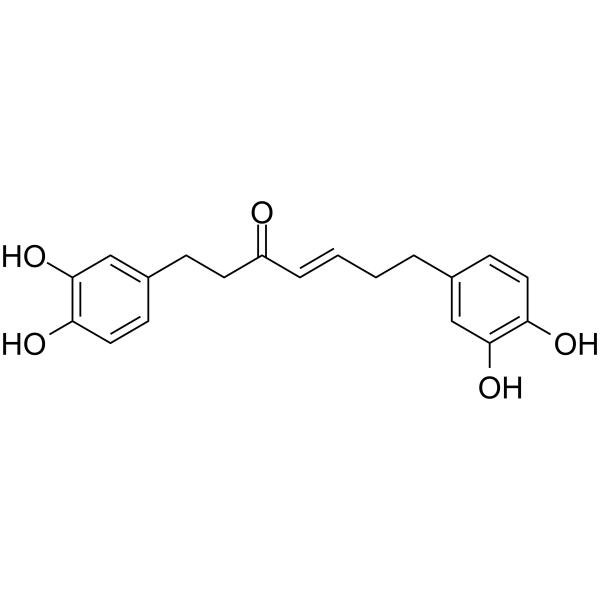
GC64035
Hirsutenone
Hirsutenone is an active botanical diarylheptanoid present in Alnus species and exhibits many biological activities, including anti-inflammatory, anti-tumor promoting and anti-atopic dermatitis effects.
-
GC16713
HS-173
novel PI3K inhibitor
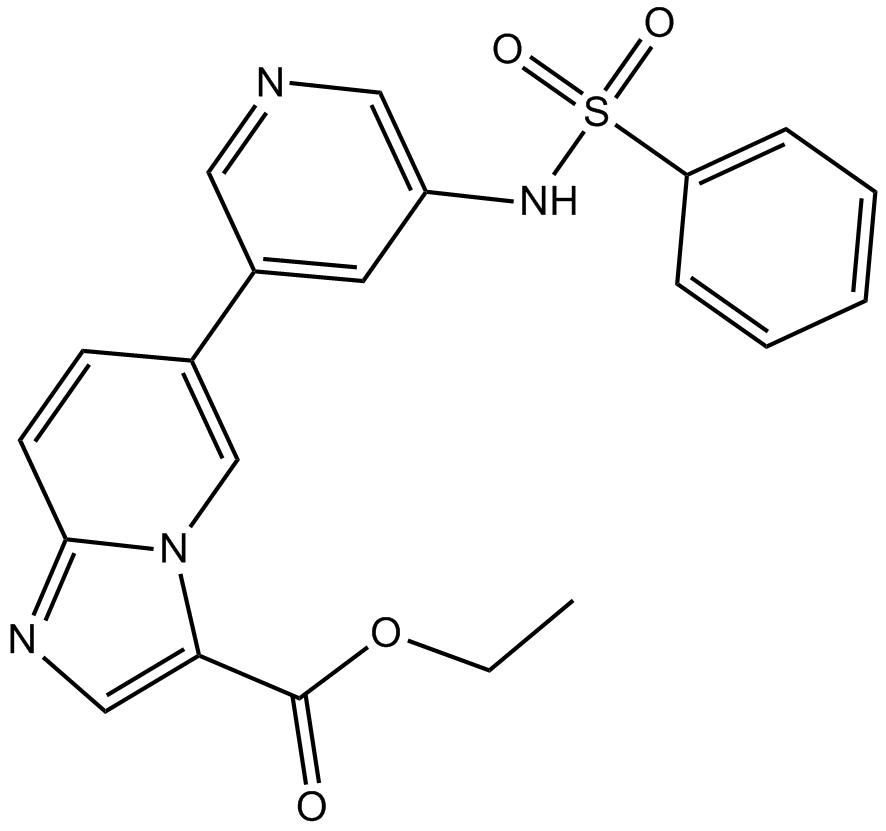
-
GC62572
hSMG-1 inhibitor 11e
hSMG-1 inhibitor 11e is a potent and selective hSMG-1 kinase inhibitor with an IC50 of 900-fold selectivity over mTOR (IC50 of 45 nM), PI3Kα/γ (IC50s of 61 nM and 92 nM) and CDK1/CDK2 (IC50s of 32 μM and 7.1 μM).
-
GC61925
hSMG-1 inhibitor 11j
hSMG-1 inhibitor 11j, a pyrimidine derivative, is a potent and selective inhibitor of hSMG-1, with an IC50 of 0.11 nM. hSMG-1 inhibitor 11j exhibits >455-fold selectivity for hSMG-1 over mTOR (IC50=50 nM), PI3Kα/γ (IC50=92/60 nM) and CDK1/CDK2 (IC50=32/7.1 μM). hSMG-1 inhibitor 11j can be used for the research of cancer.
-
GC15540
IC-87114
PI3Kδ inhibitor
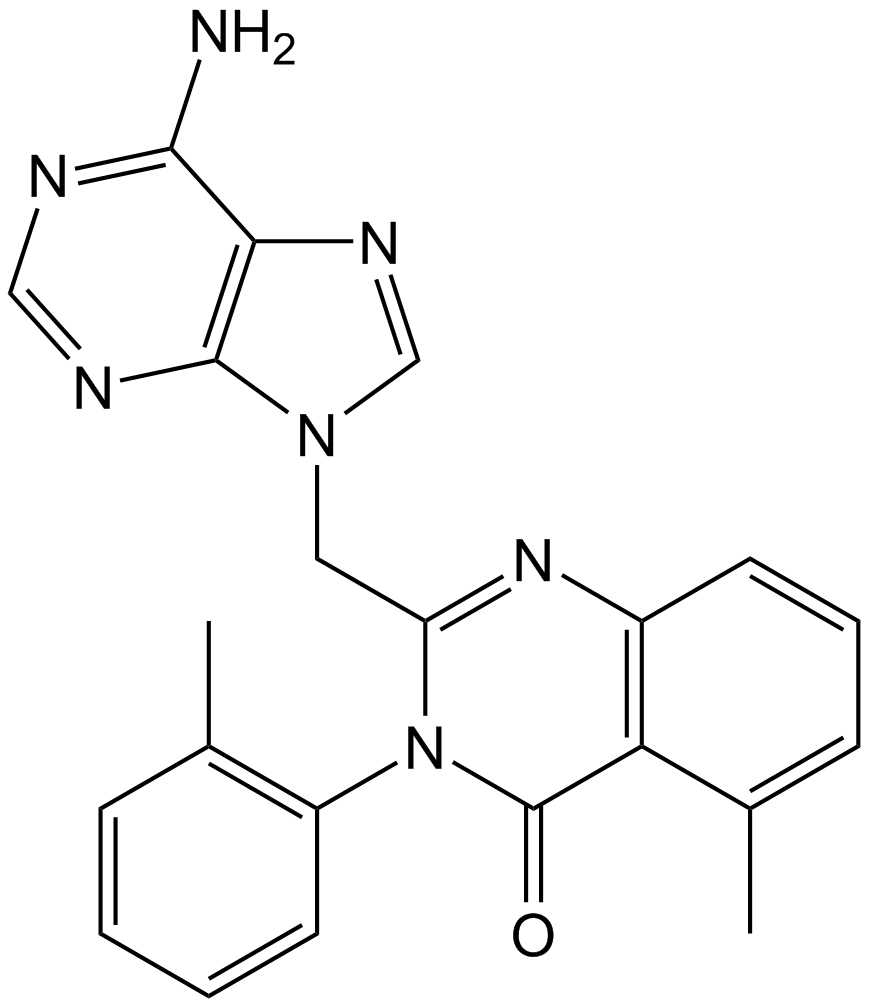
-
GC62465
Idelalisib D5
Idelalisib D5 is a deuterium labeled Idelalisib. Idelalisib is a highly selective and orally bioavailable p110δ inhibitor.
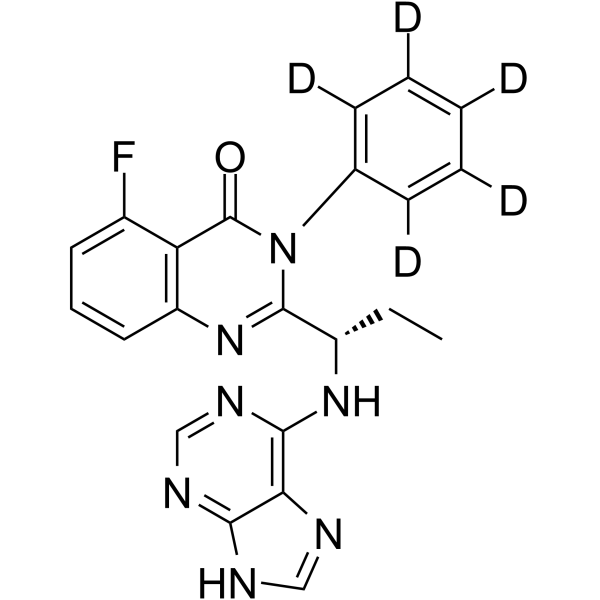
-
GC63017
IHMT-PI3Kδ-372
IHMT-PI3Kδ-372 is a potent and selective PI3Kδ inhibitor with an IC50 of 14 nM.
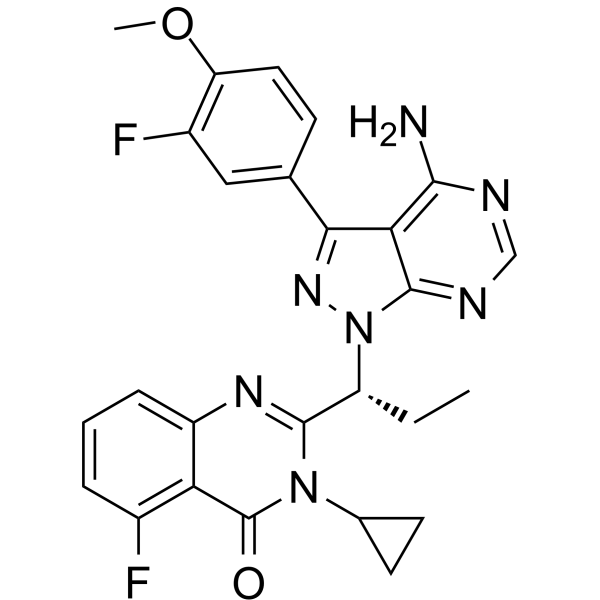
-
GC19411
IITZ-01
IITZ-01 is a potent lysosomotropic autophagy inhibitor with single-agent antitumor activity, with an IC50 of 2.62 μM for PI3Kγ.
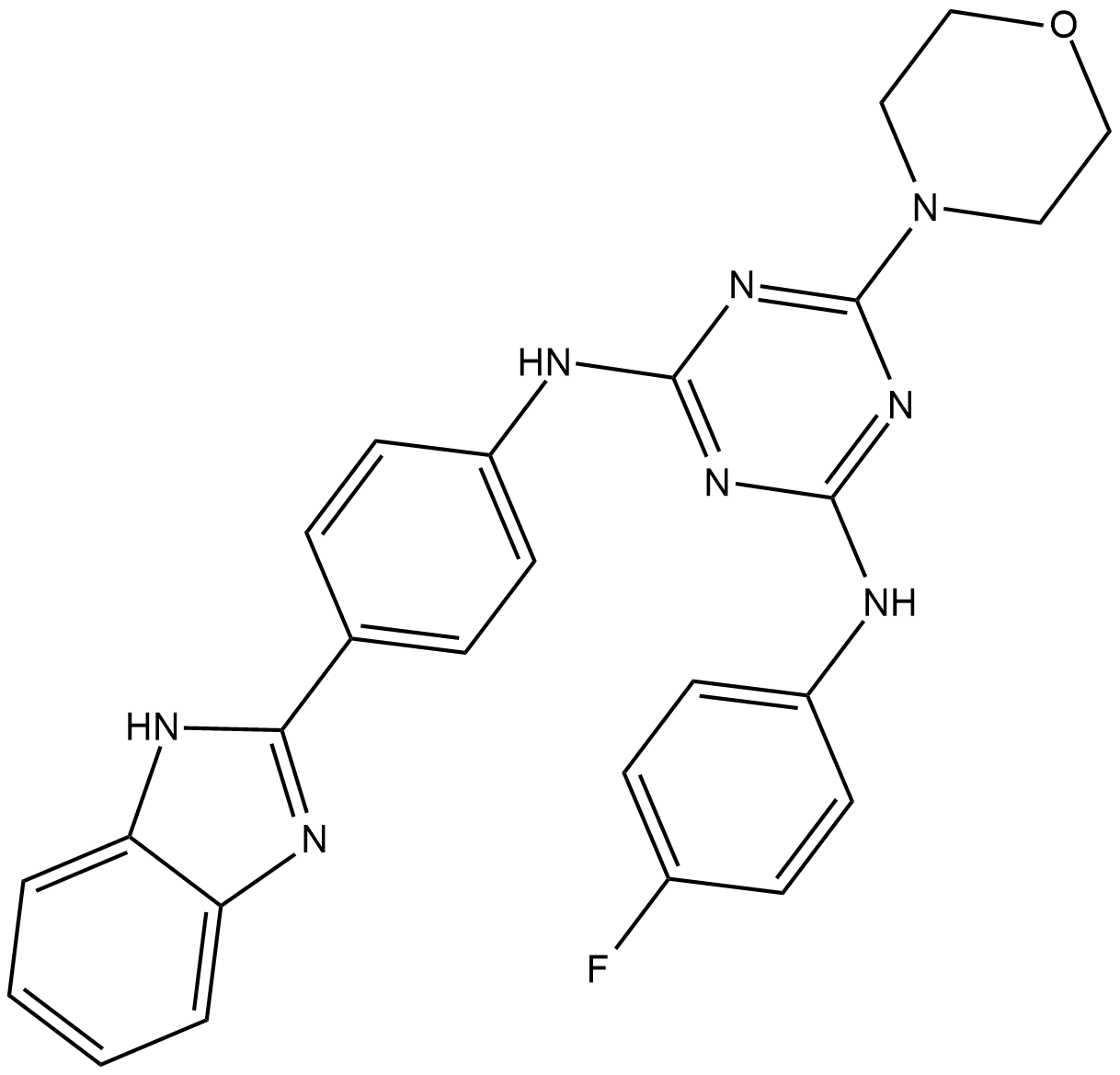
-
GC13503
iMDK
PI3K and endogenous midkine (MDK) expression inhibitor
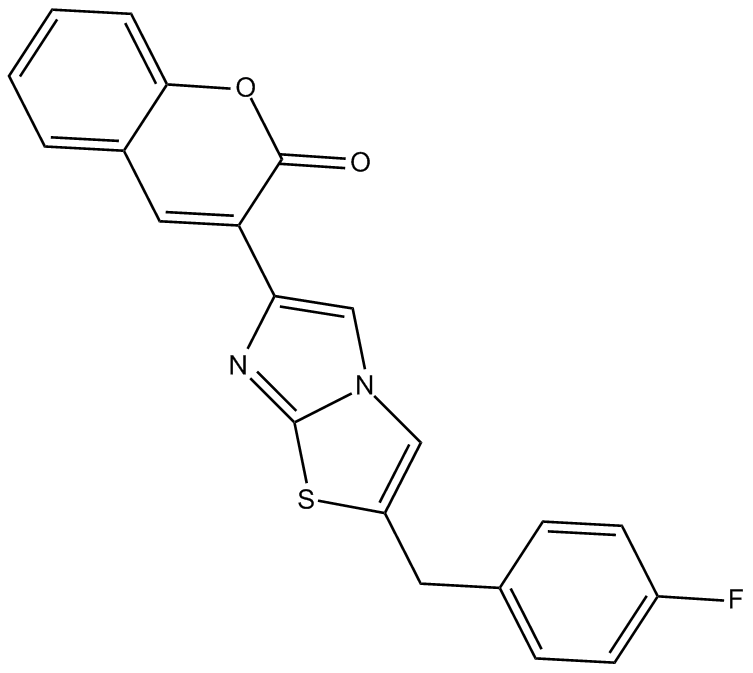
-
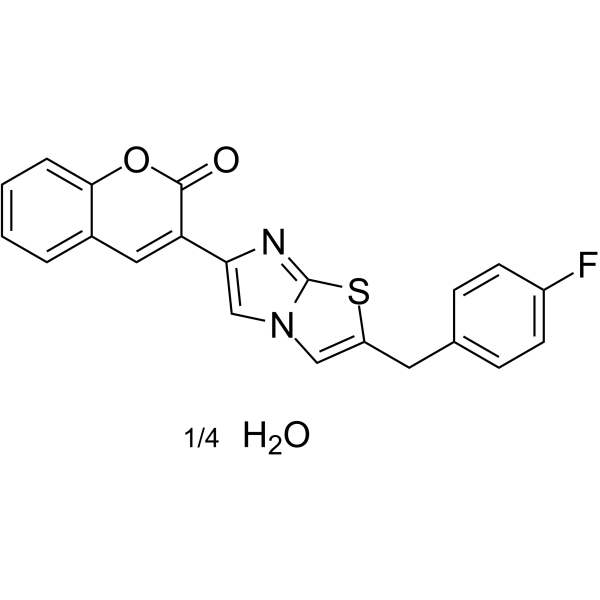
GC62376
iMDK quarterhydrate
iMDK quarterhydrate is a potent PI3K inhibitor and inhibits the growth factor MDK (also known as midkine or MK). iMDK quarterhydrate suppresses non-small cell lung cancer (NSCLC) cooperatively with A MEK inhibitor without harming normal cells and mice.
-
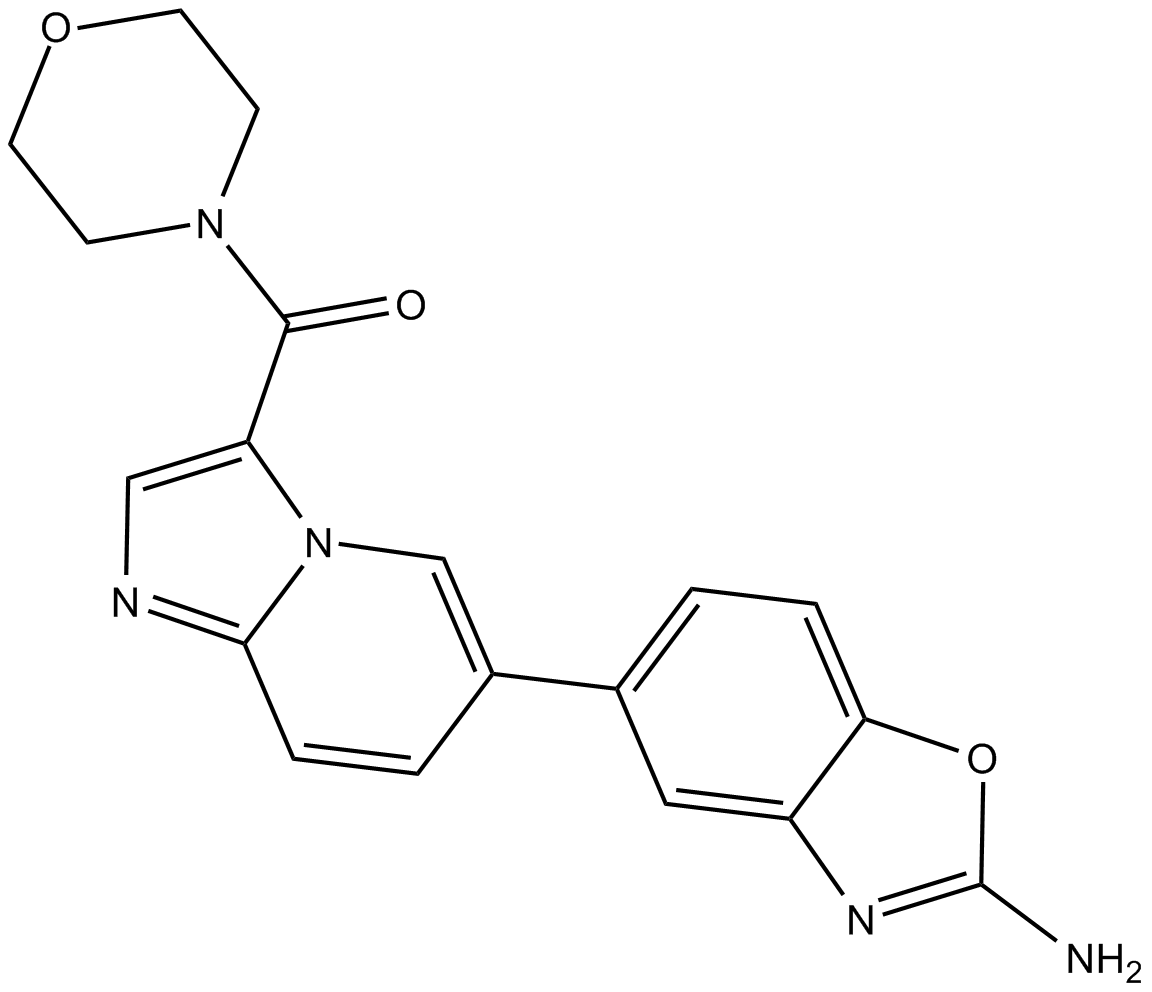
GC12416
INK1117
INK1117 (MLN1117) is a selective p110α inhibitor with an IC50 of 15 nM.
-
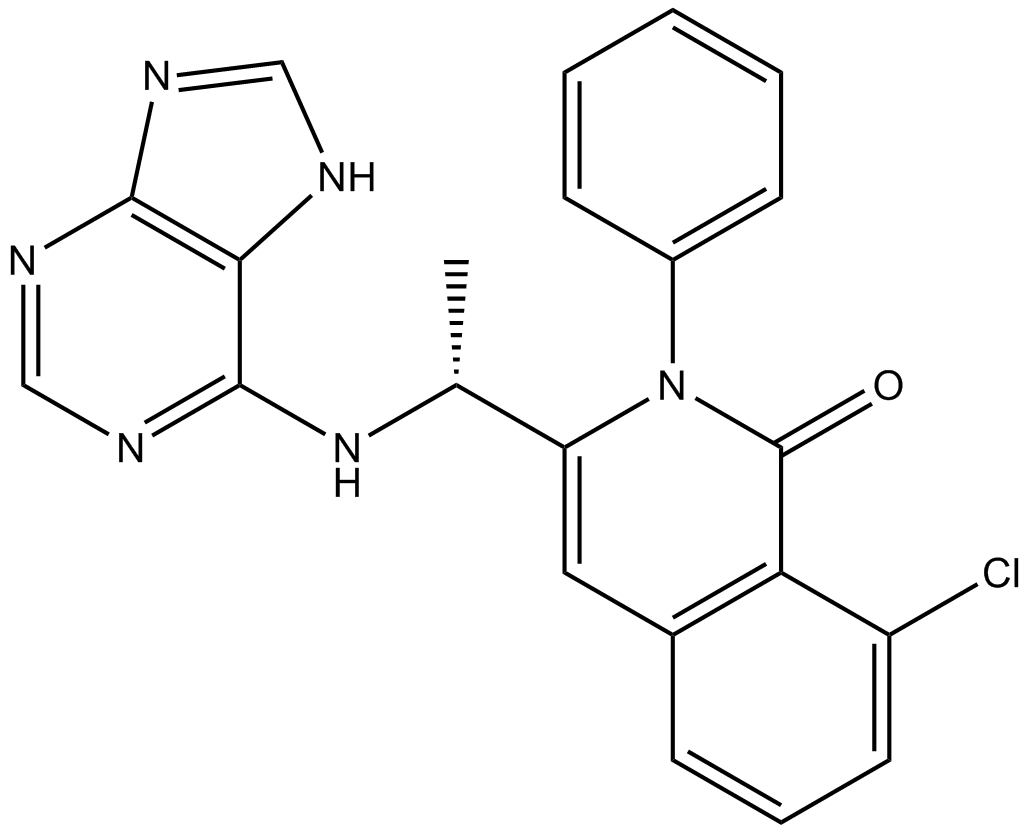
GC10749
IPI-145 (INK1197)
IPI-145 (INK1197) (IPI-145) is a selectivite p100δ inhibitor with IC50 of 2.5 nM, 27.4 nM, 85 nM and 1602 nM for p110δ, P110γ, p110β and p110α, respectively.
-
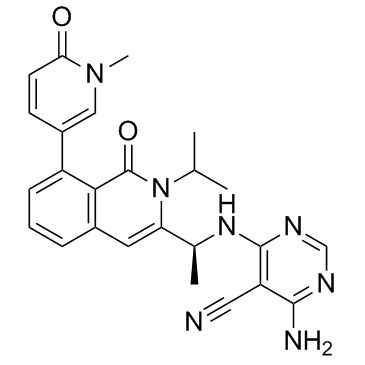
GC31756
IPI-3063
IPI-3063 is a potent and selective PI3K p110δ inhibitor with an IC50 of 2.5?±?1.2 nM.
-
GC19202
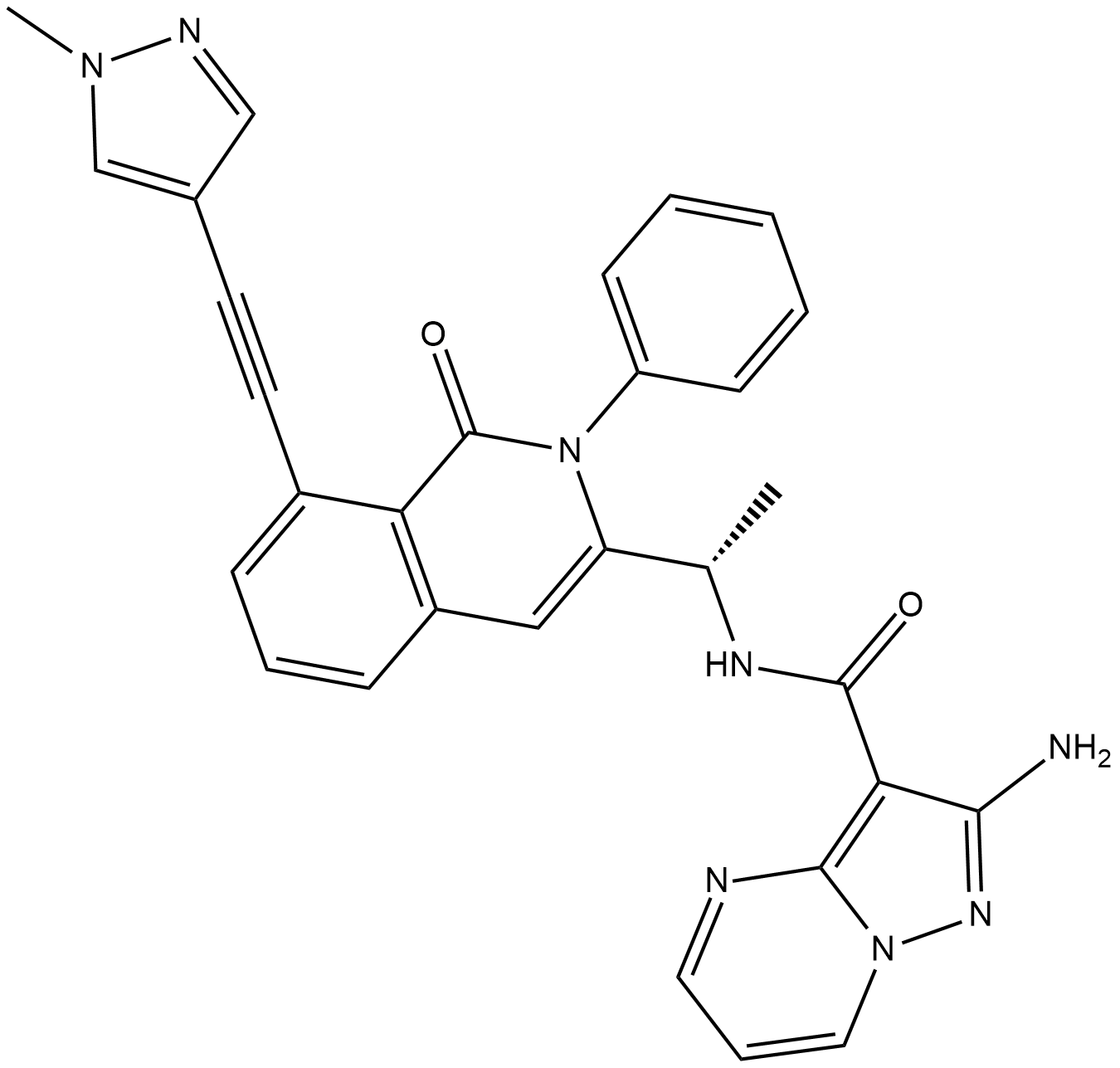
IPI549
IPI549 (IPI549) is a potent and selective PI3Kγ inhibitor with an IC50 of 16 nM. IPI549 shows >100-fold selectivity over other lipid and protein kinases.
-
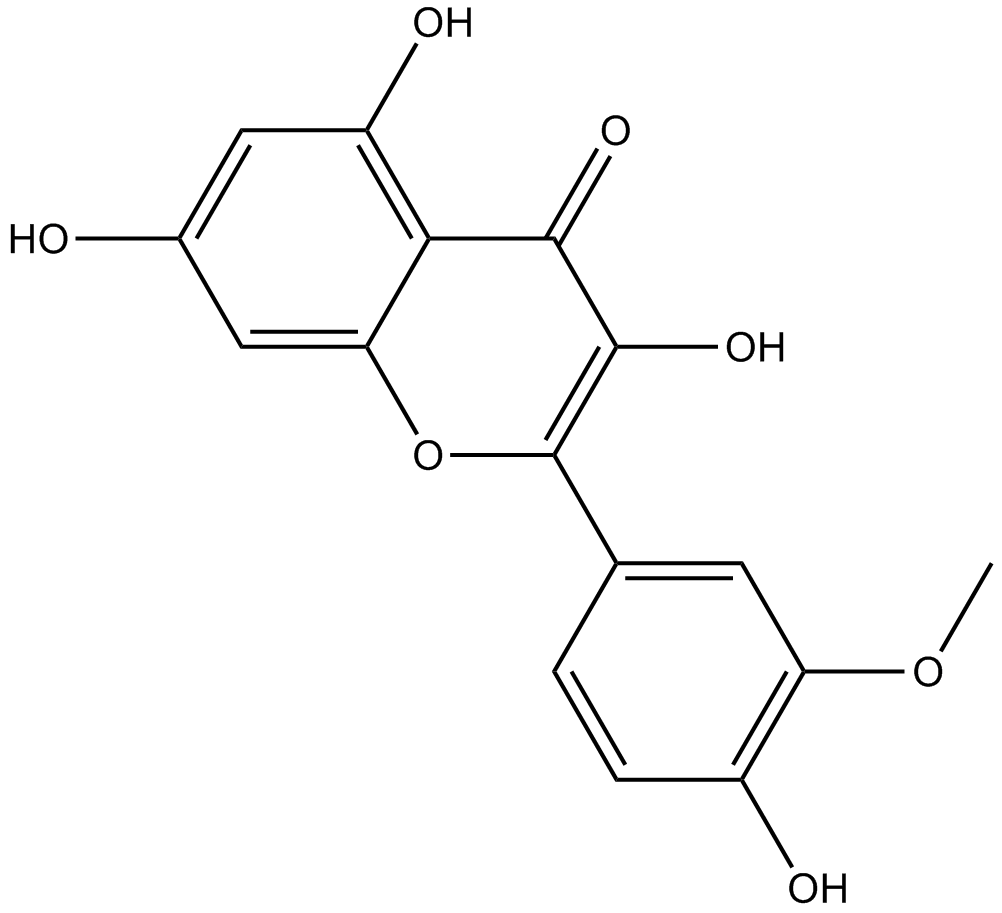
GN10023
Isorhamnetin
-
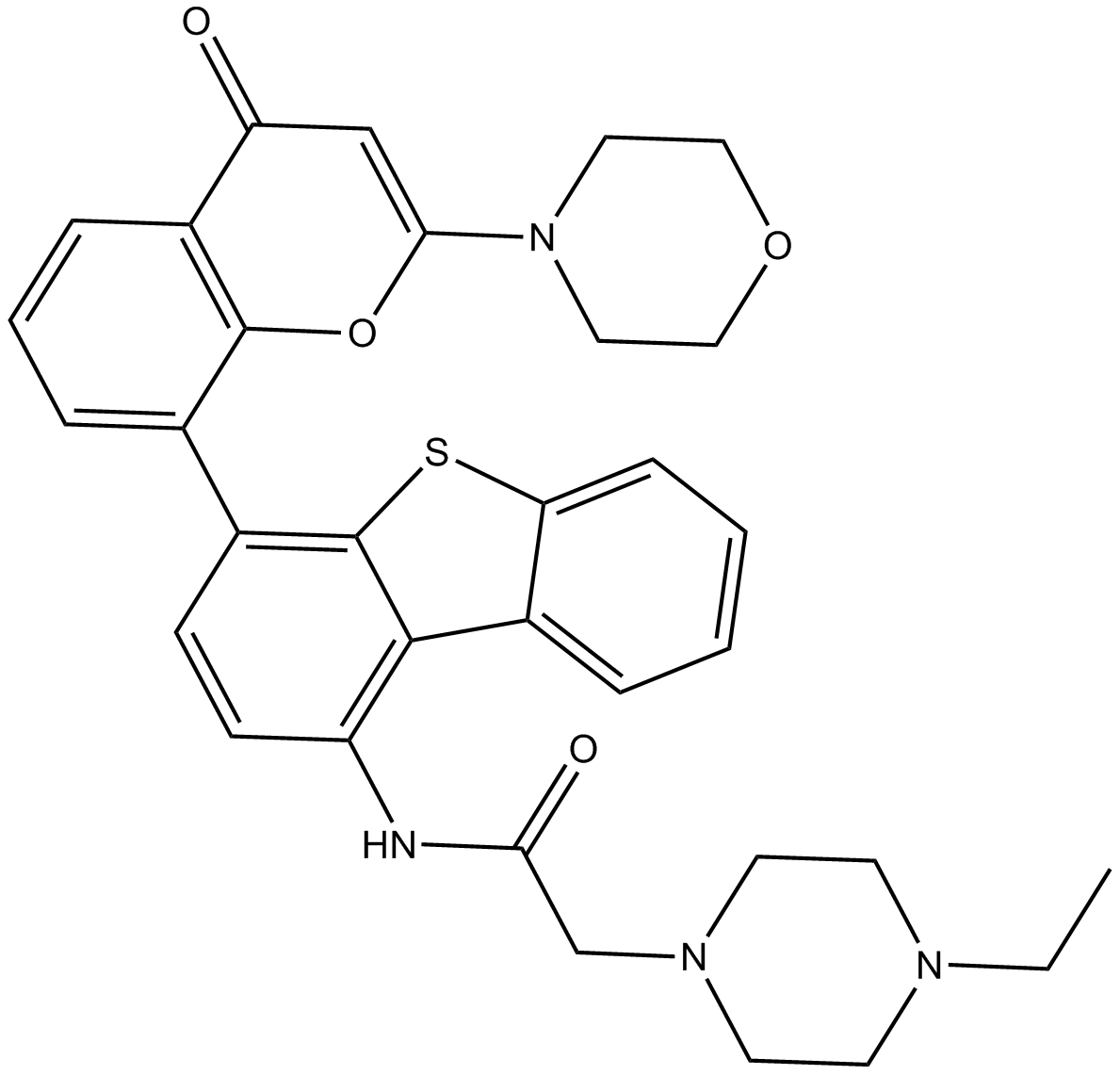
GC14346
KU-0060648
Dual DNA-PK/PI3-K inhibitor, ATP-competitive
-
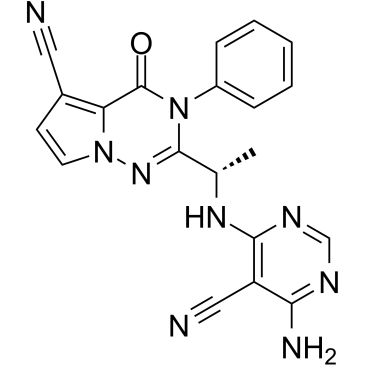
GC36426
LAS191954
LAS191954 is a potent, selective and orally active PI3Kδ inhibitor for inflammatory diseases treatment, with an IC50 of 2.6 nM.
-
GC19471
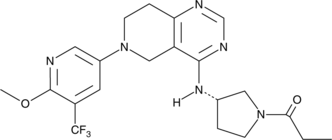
Leniolisib
A potent and selective PI3Kδ inhibitor
-
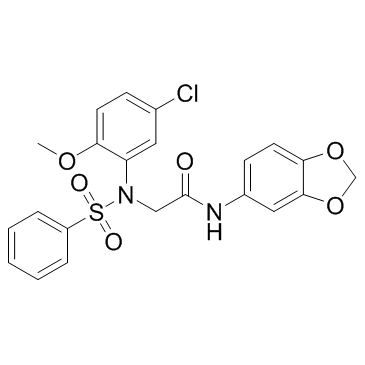
GC30884
LX2343
An inhibitor of BACE1 and PI3K
-
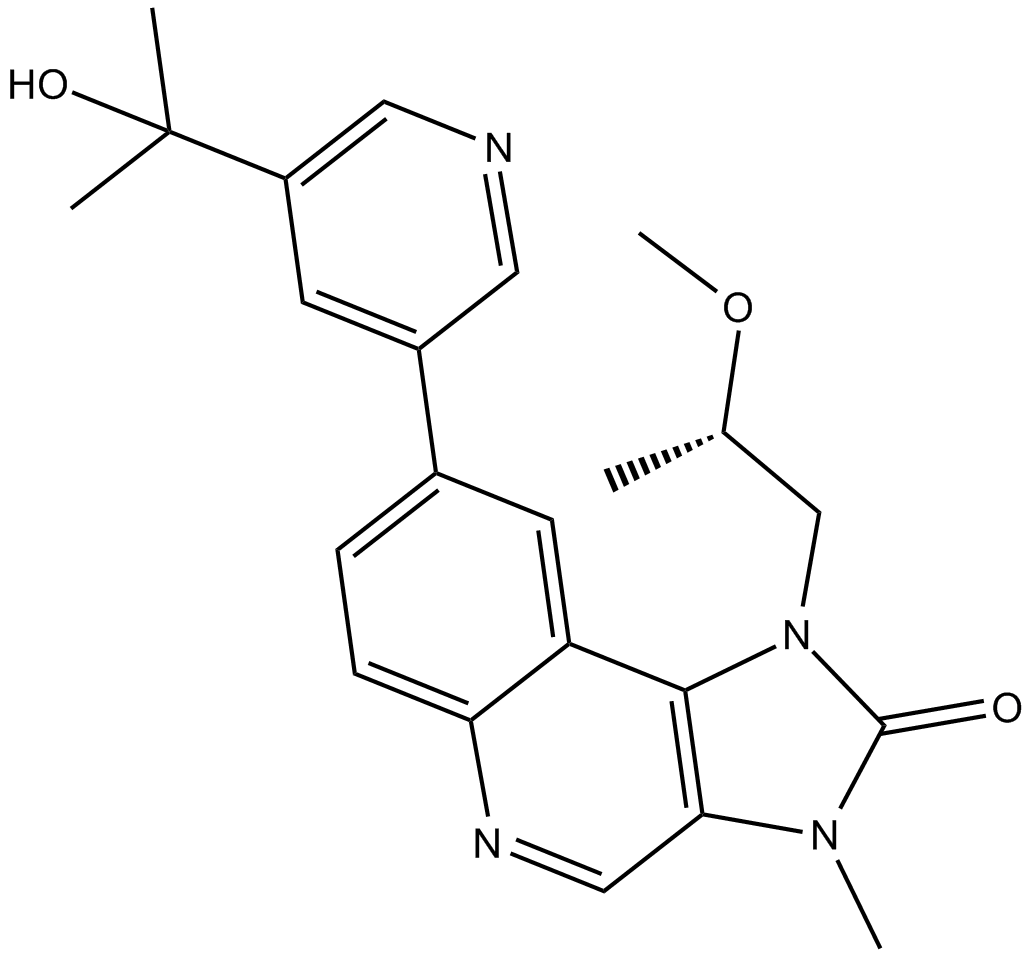
GC14371
LY3023414
LY3023414 (LY3023414) potently and selectively inhibits class I PI3K isoforms, DNA-PK, and mTORC1/2 with IC50s of 6.07 nM, 77.6 nM, 38 nM, 23.8 nM, 4.24 nM and 165 nM for PI3Kα, PI3Kβ, PI3Kδ, PI3Kγ, DNA-PK and mTOR, respectively. LY3023414 potently inhibits mTORC1/2 at low nanomolar concentrations.
-
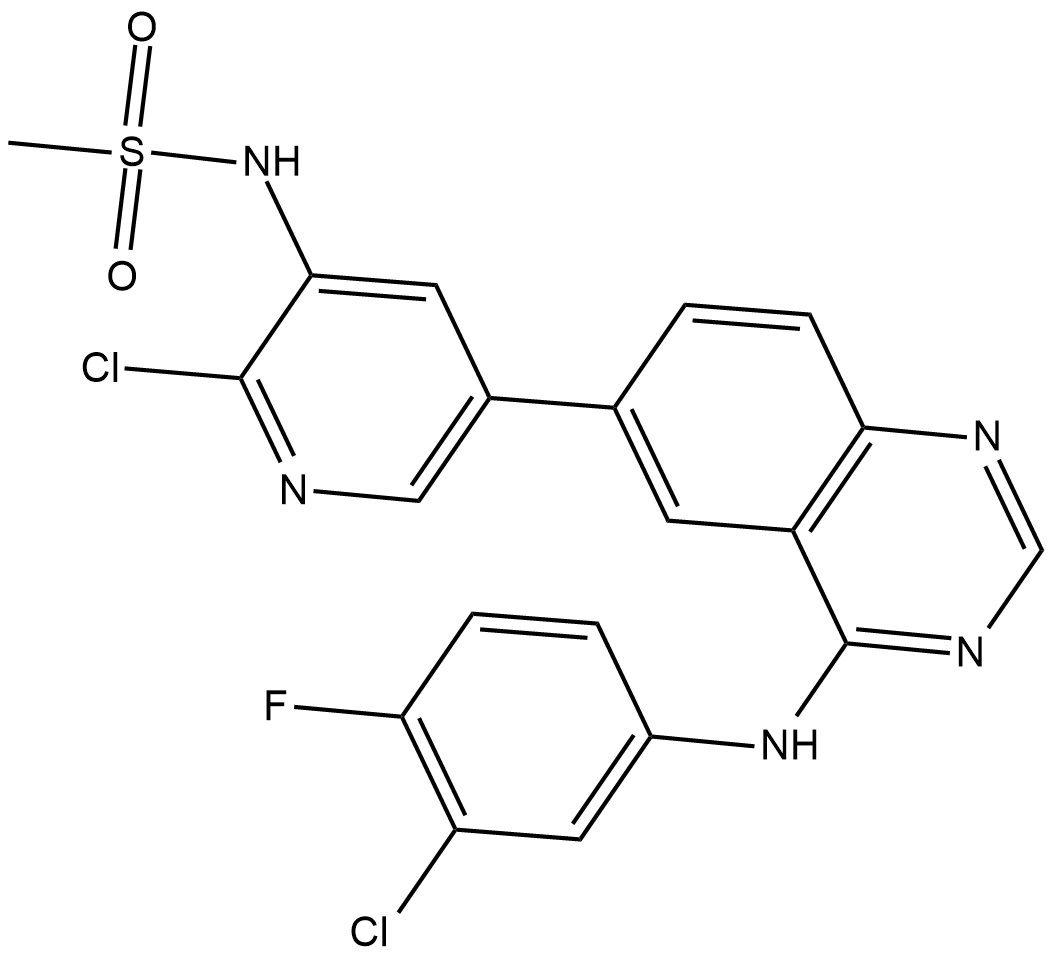
GC19256
MTX-211
MTX-211 is a dual inhibitor of EGFR and PI3K, used for the treatment of cancer and other diseases.
-
GC65442
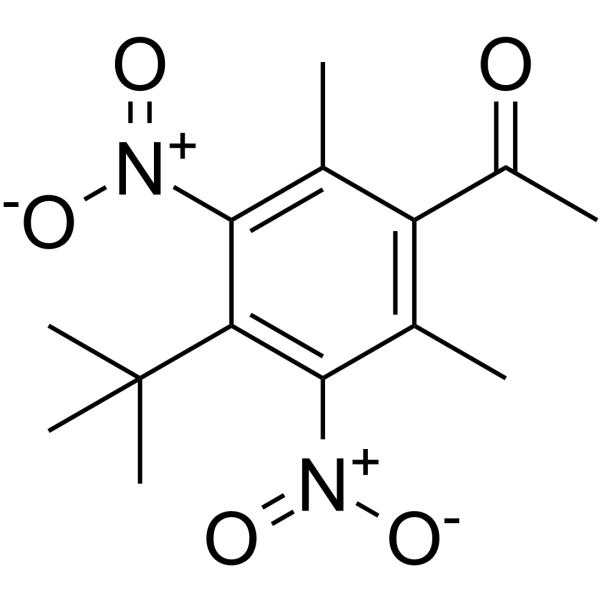
Musk ketone
Musk ketone (MK) is a widely used artificial fragrance.
-
GC36714
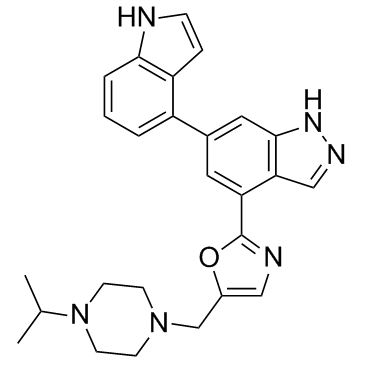
Nemiralisib
Nemiralisib (GSK2269557 free base) is a potent and highly selective PI3Kδ inhibitor with a pKi of 9.9.
-
GC33014
NSC781406
A dual inhibitor of PI3K and mTOR