Proteasome
The proteasome is a large multisubunit complex of approximately 2.5 MDa that mediates a wide range of physiological and pathological cellular processes by selectively degrading unnecessary proteins in eukaryotes. The structure of a proteasome is comprised of the catalytic core particle (CP) and two terminal regulatory particles (RPs). The CP (also known as the 20S proteasome) is a barrel shaped multisubunit complex (approximately 750 kDa) consisting of four axially stacked heptameric rings (two outer α-rings and two inner β-rings) with 7 subunits in each ring, where three β subunits (β1, β2 and β5) contain catalytically active threonine residues and are associated with caspase-like, trypsin-like and chymotrypsin-like activities respectively.
Products for Proteasome
- Cat.No. Product Name Information
-
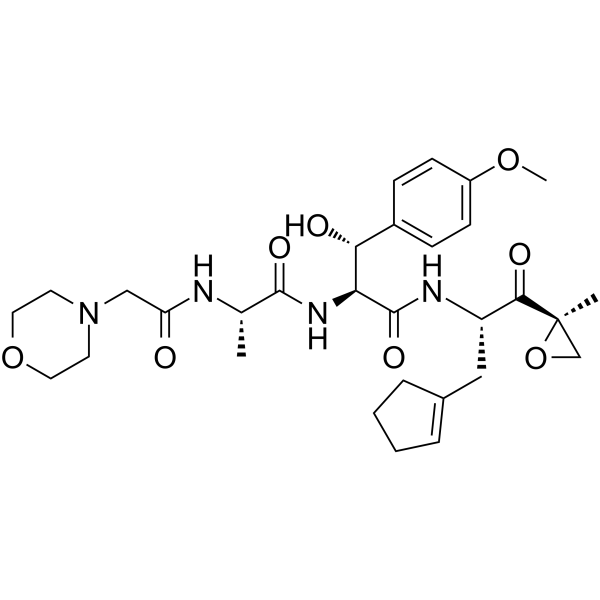
GC65547
(1S,2R)-Alicapistat
(1S,2R)-Alicapistat ((1S,2R)-ABT-957) is an orally active selective inhibitor of human calpains 1 and 2 for the potential application of Alzheimer's disease (AD).
-
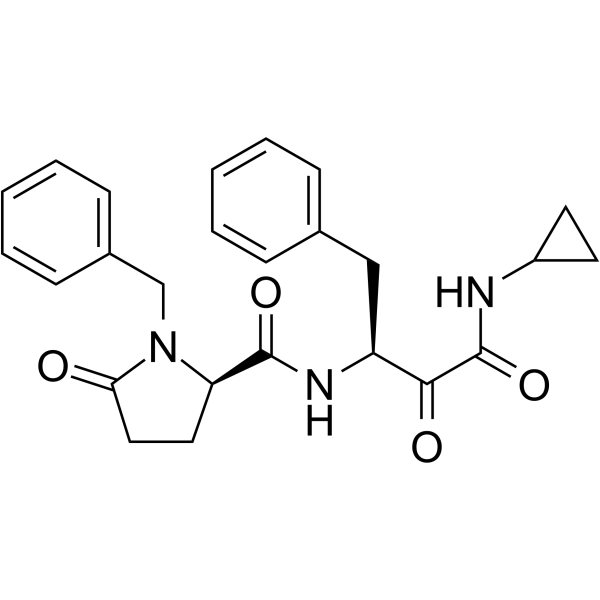
GC62193
(1S,2S)-Bortezomib
(1S,2S)-Bortezomib is an enantiomer of Bortezomib. Bortezomib is a cell-permeable, reversible, and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki of 0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is an anti-cancer agent and the first therapeutic proteasome inhibitor to be used in humans.
-
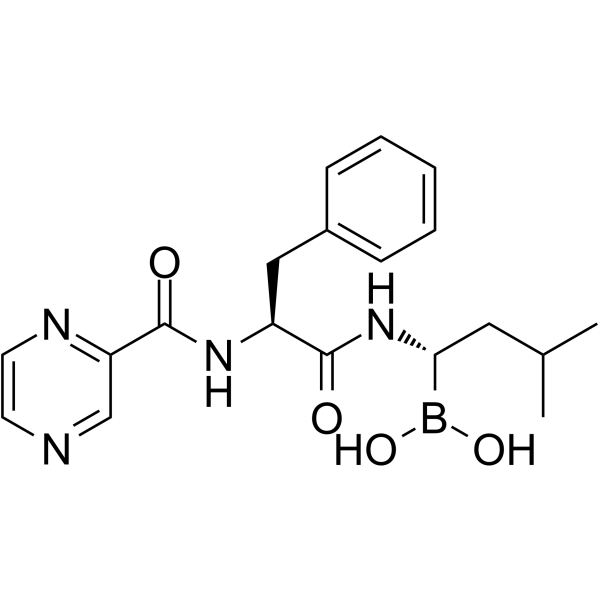
GC41233
(R)-MG132
A potent proteasome inhibitor
-
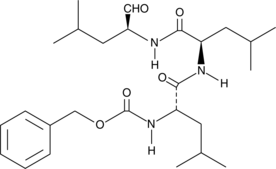
GC68503
20S Proteasome activator 1
20S Proteasome activator 1 is an effective activator of the 20S protease, with IC50 values of 0.3 μM, 0.7 μM and 1.8 μM for trypsin-like sites, chymotrypsin-like sites and caspase-like sites respectively. It can be translated in cellular systems to prevent accumulation of pathogenic A53T mutant alpha-synuclein. 20S Proteasome activator 1 can be used for research on neurodegenerative diseases.
-
GC64472
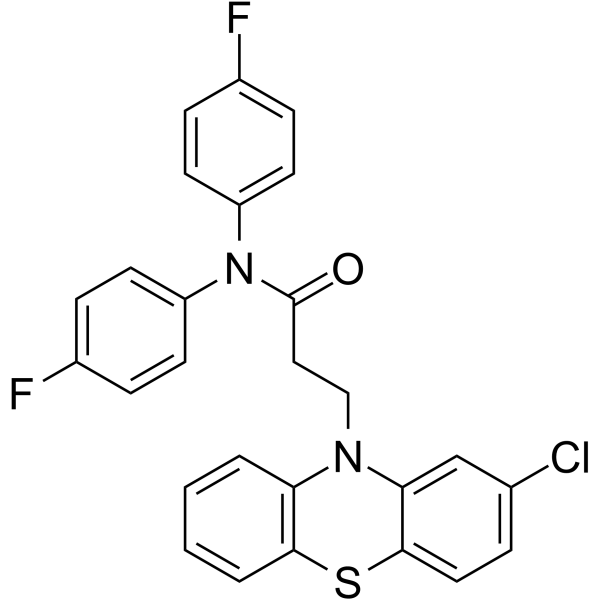
20S Proteasome-IN-1
20S Proteasome-IN-1 is a 26S proteasome inhibitor extracted from patent WO2006128196A2 compound 2.
-
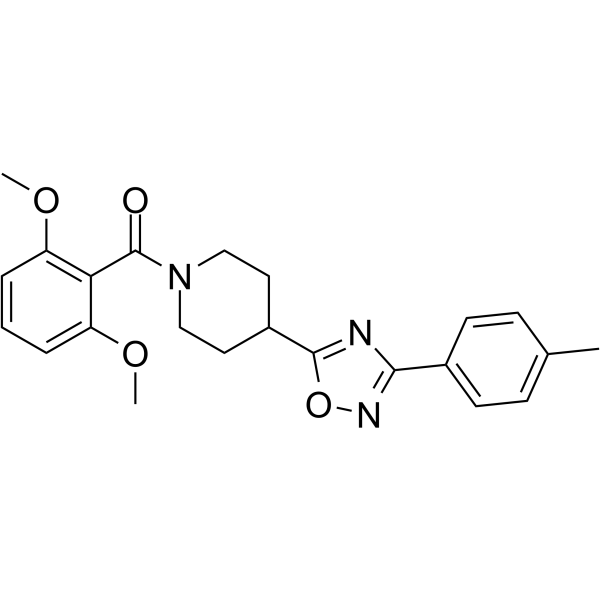
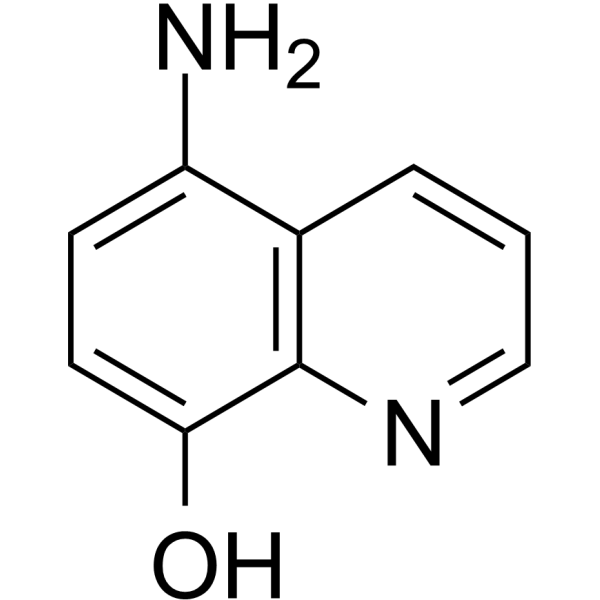
GC65668
5-Amino-8-hydroxyquinoline
5-Amino-8-hydroxyquinoline (5A8HQ), a potential anticancer candidate, has promising proteasome inhibitory activity.
-
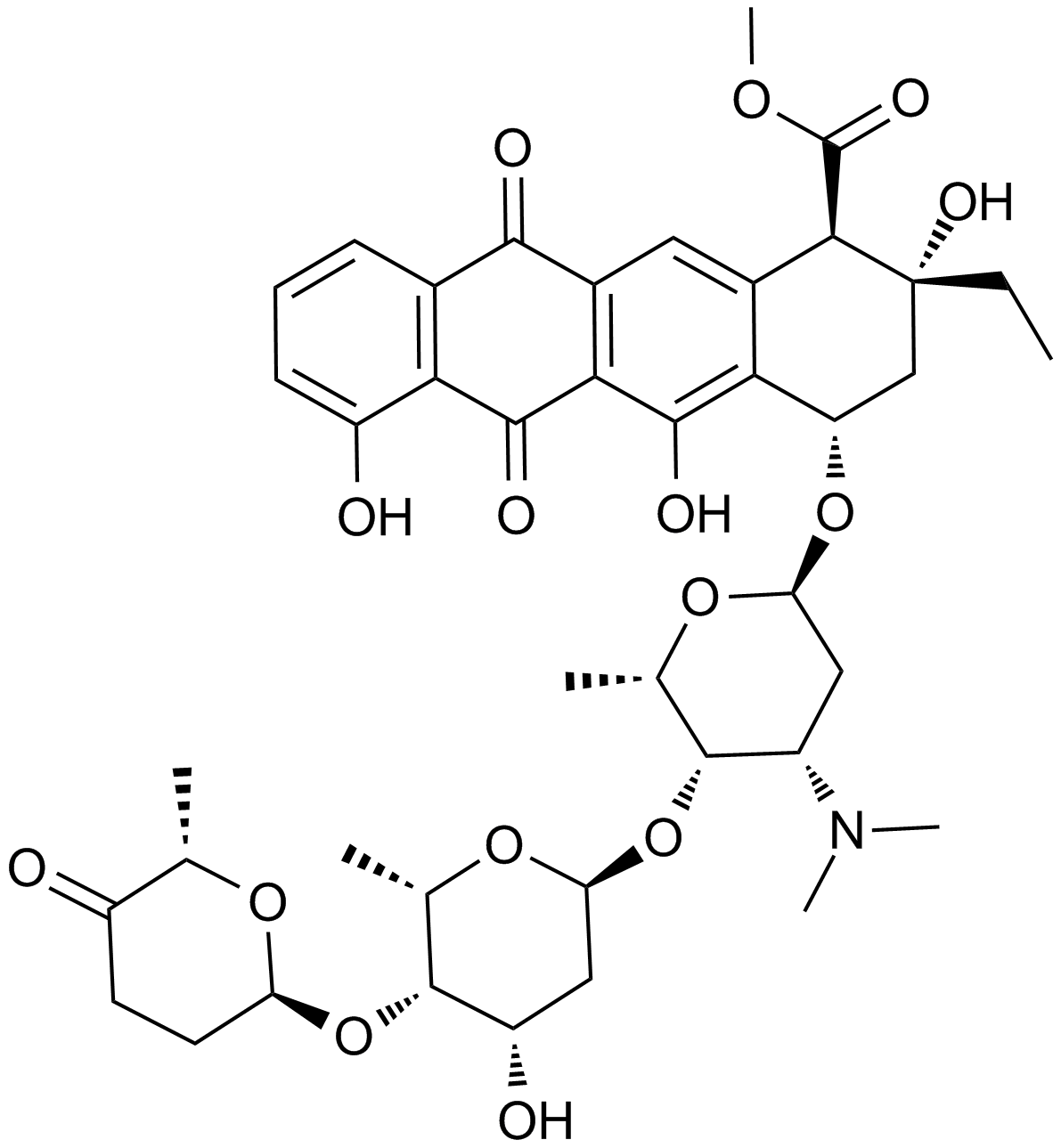
GC10102
Aclacinomycin A
An anthracycline with antibiotic and anticancer activities
-
GC35238
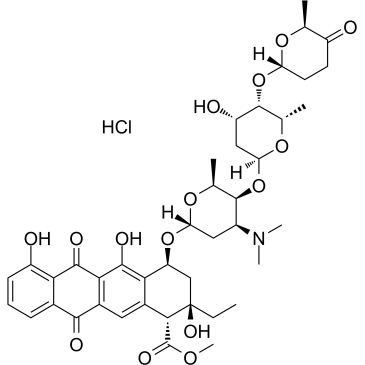
Aclacinomycin A hydrochloride
Aclacinomycin A hydrochloride (Aclarubicin hydrochloride), a fluorescent molecule and the first described non-peptidic inhibitor showing discrete specificity for the CTRL (chymotrypsin-like) activity of the 20S proteasome. Aclacinomycin A hydrochloride is also a dual inhibitor of topoisomerase I and II. An effective anthracycline chemotherapeutic agent for hematologic cancers and solid tumors.
-
GC33743
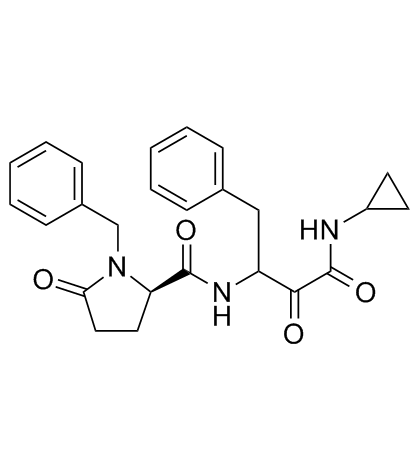
Alicapistat (ABT-957)
Alicapistat (ABT-957) (ABT-957) is an orally active selective inhibitor of human calpains 1 and 2 for the potential application of Alzheimer's disease (AD).
-
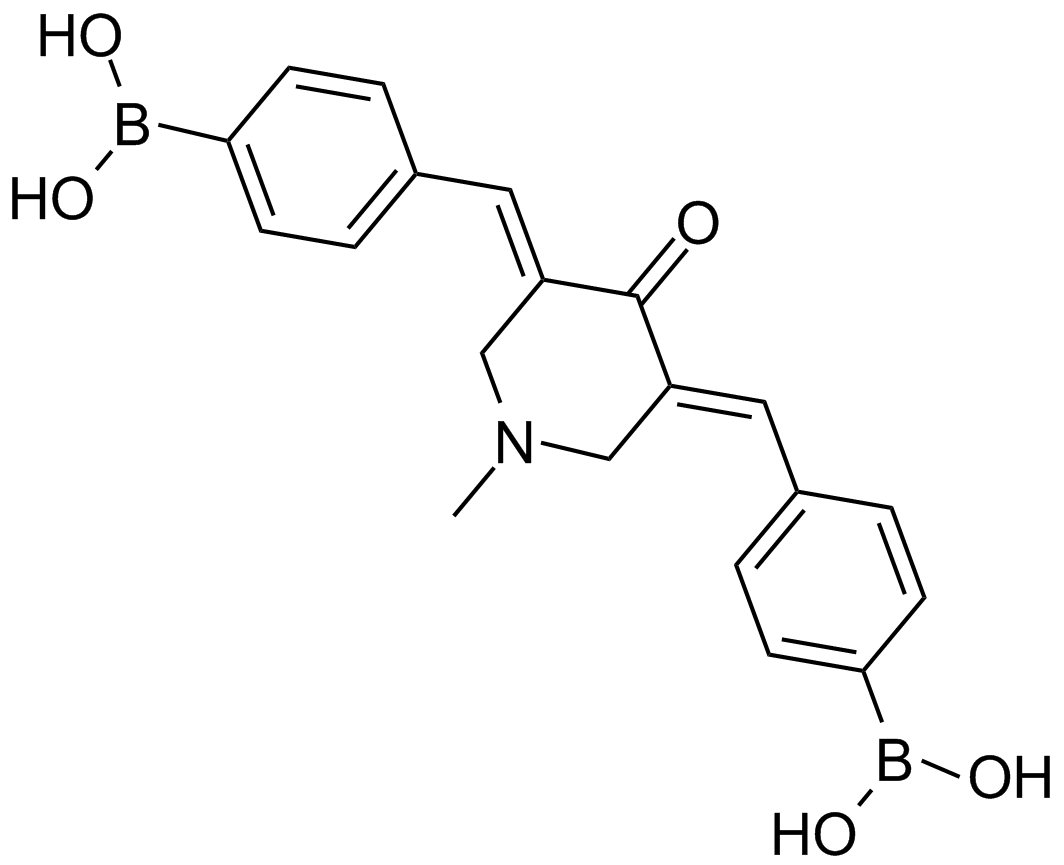
GC13789
AM 114
-
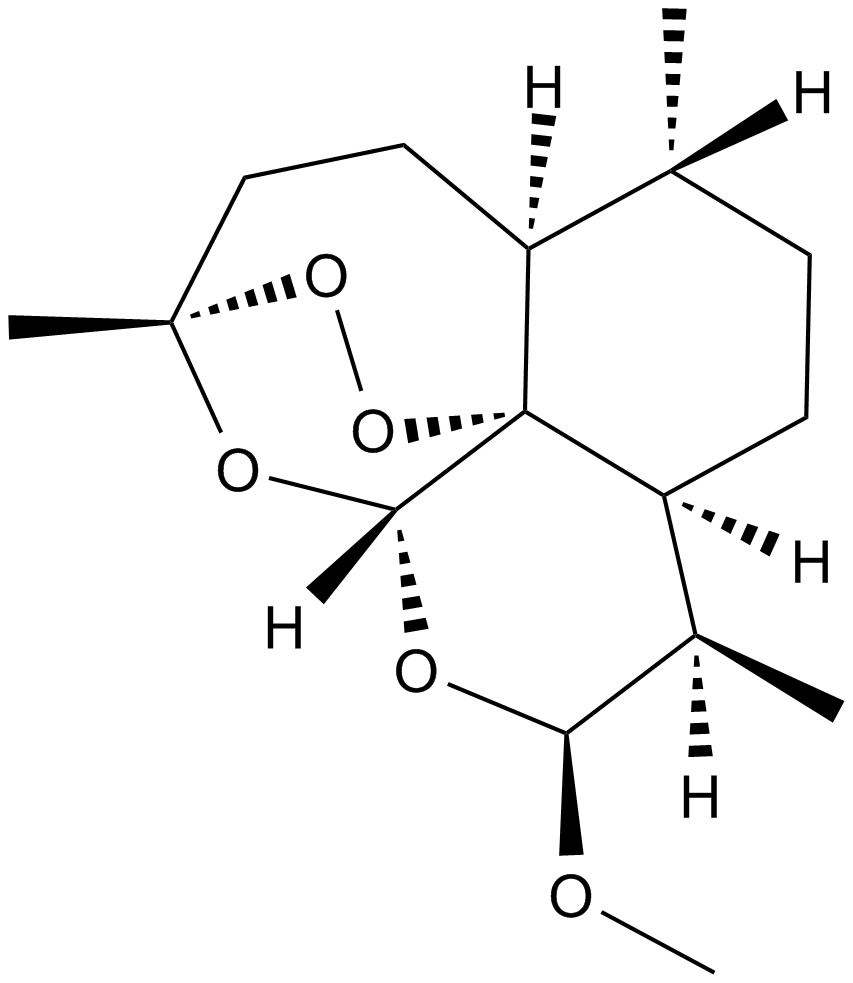
GC12784
Artemether (SM-224)
An antiparasitic compound
-
GC17644
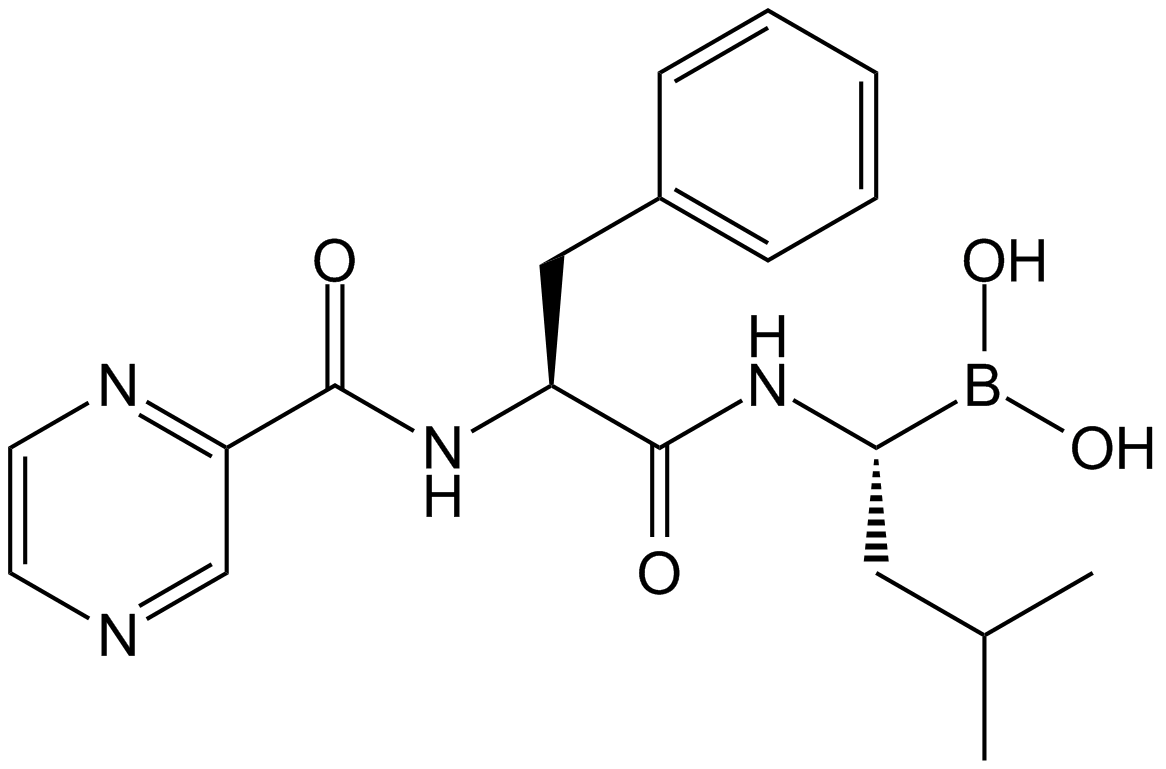
Bortezomib (PS-341)
Bortezomib (PS-341), as a dipeptide boronic acid proteasome inhibitor with antitumor activity, can potently inhibiit 20S proteasome with Ki of 0.6 nM by targeting a threonine residue..
-
GC65010
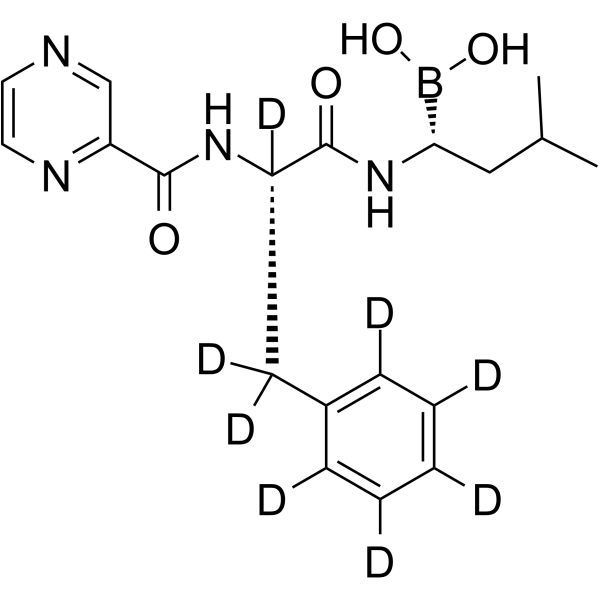
Bortezomib-d8
Bortezomib-d8 (PS-341-d8) is the deuterium labeled Bortezomib. Bortezomib (PS-341) is a reversible and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki=0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is the first proteasome inhibitor anticancer agent. Anti-cancer activity.
-
GC12527
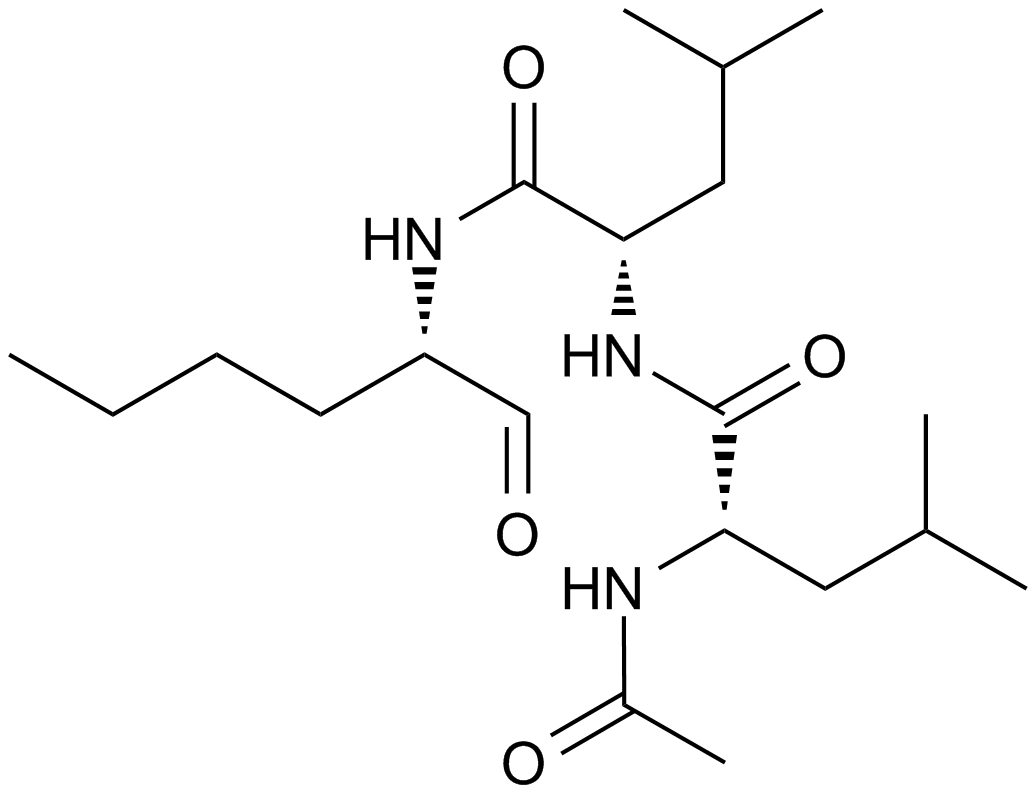
Calpain Inhibitor I, ALLN
A non-selective cysteine protease inhibitor
-
GC40694
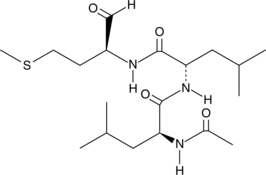
Calpain Inhibitor II
Calpain Inhibitor II (Calpain inhibitor II) is a potent inhibitor of calpain and cathepsin proteases.
-
GC67908
Calpain-2-IN-1
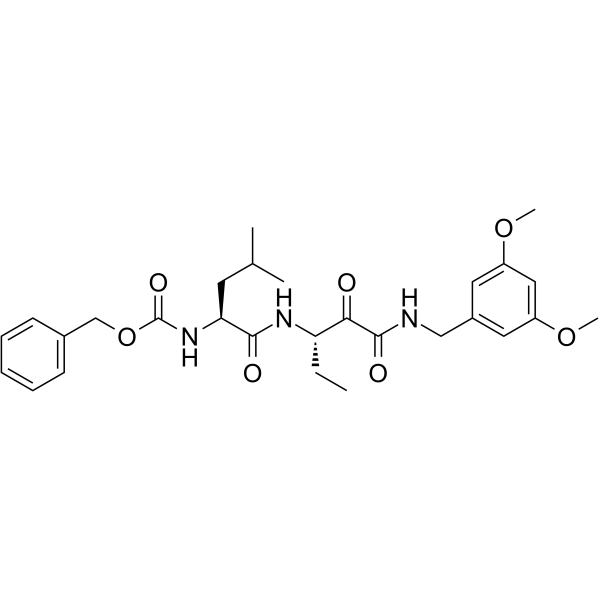
-
GC10342
Calpeptin
A calpain inhibitor
-
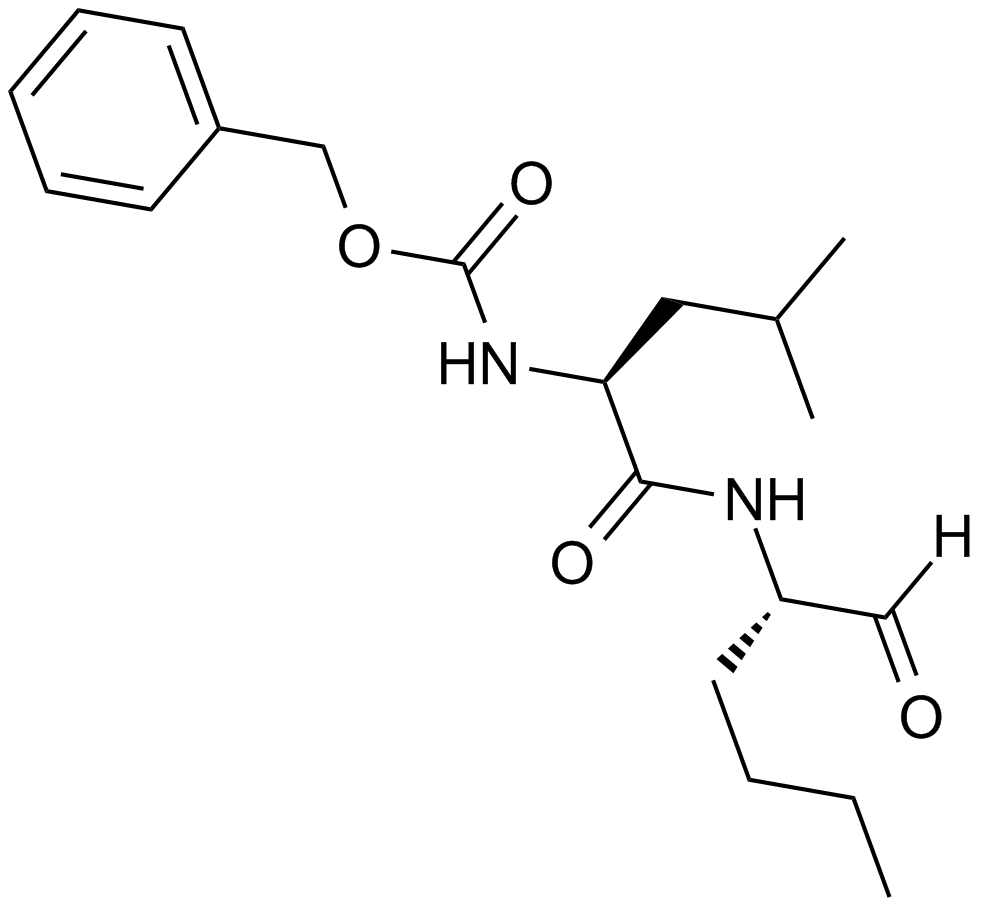
GC32815
Capzimin
Capzimin is a potent and moderately specific proteasome isopeptidase Rpn11 inhibitor.
-
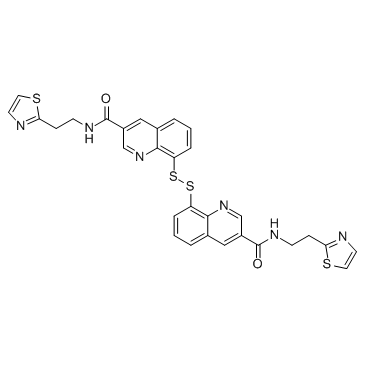
GC15089
Carfilzomib (PR-171)
A proteasome inhibitor
-
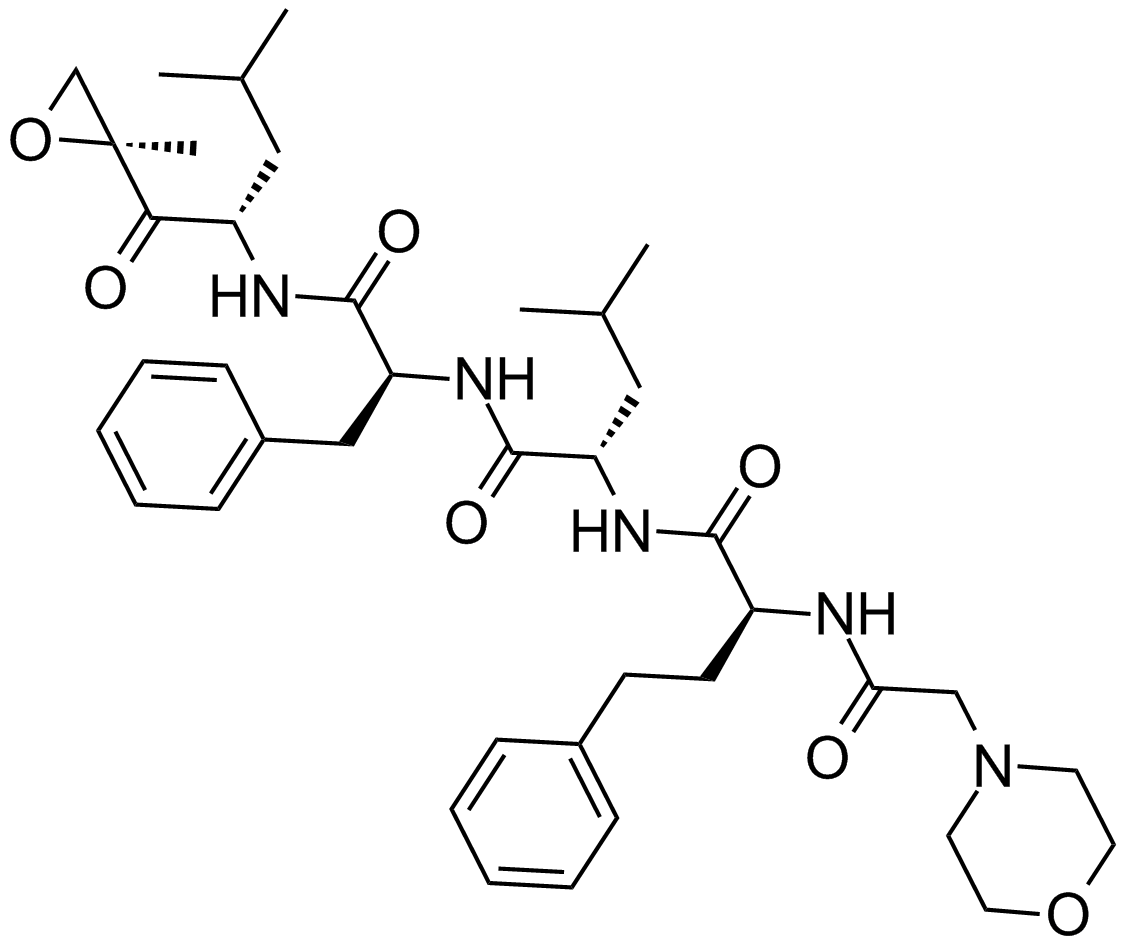
GC15083
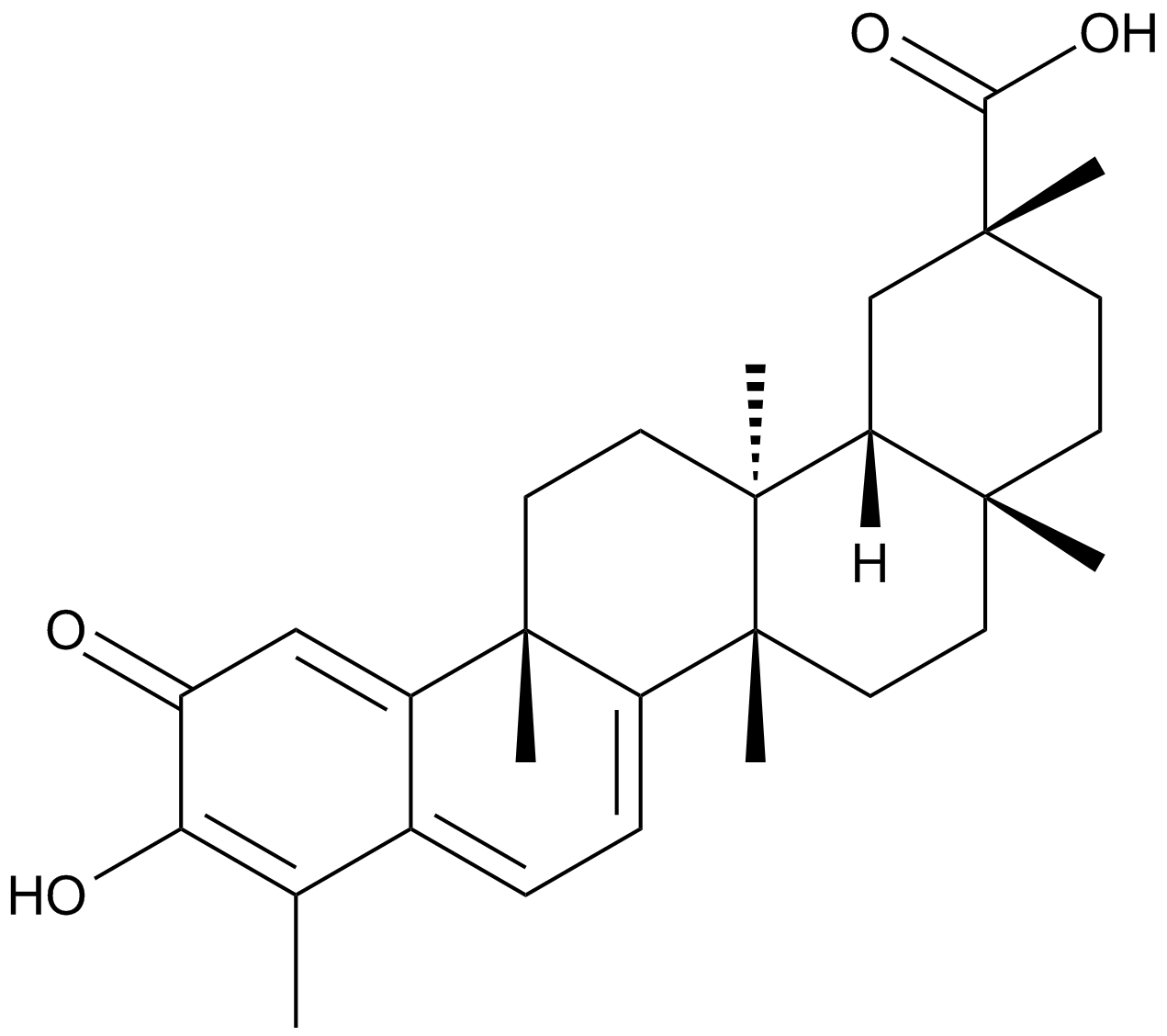
Celastrol
A triterpenoid antioxidant
-
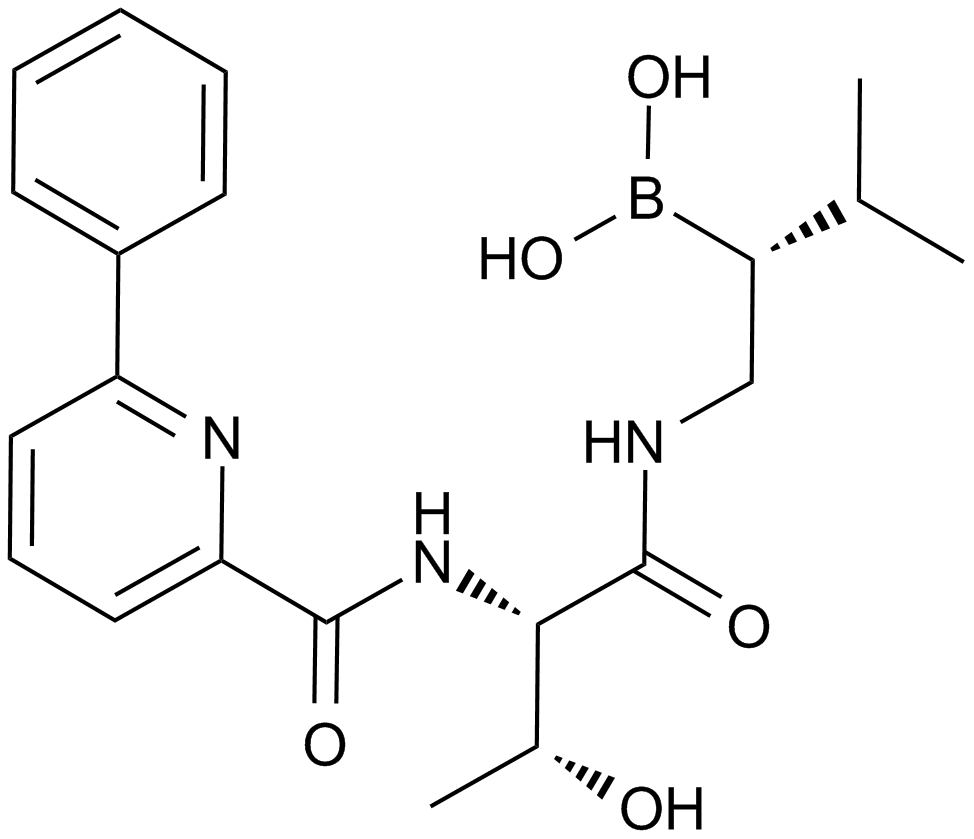
GC12272
CEP-18770
An inhibitor of chymotrypsin-like proteasome activity
-
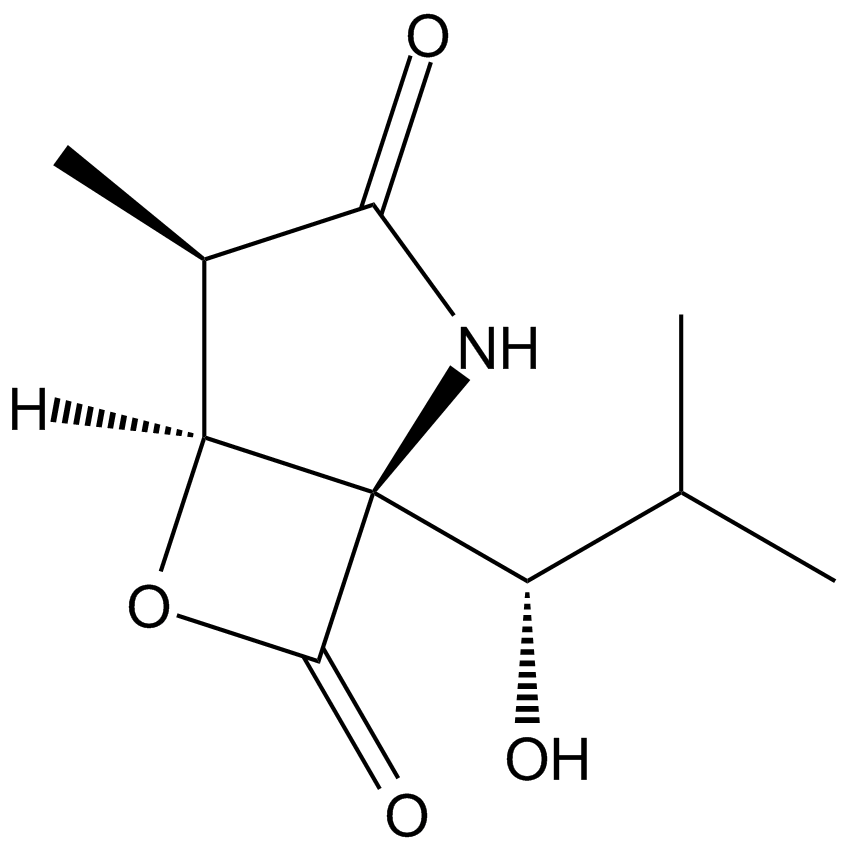
GC16581
Clasto-Lactacystin β-lactone
A selective inhibitor of the 20S proteasome
-
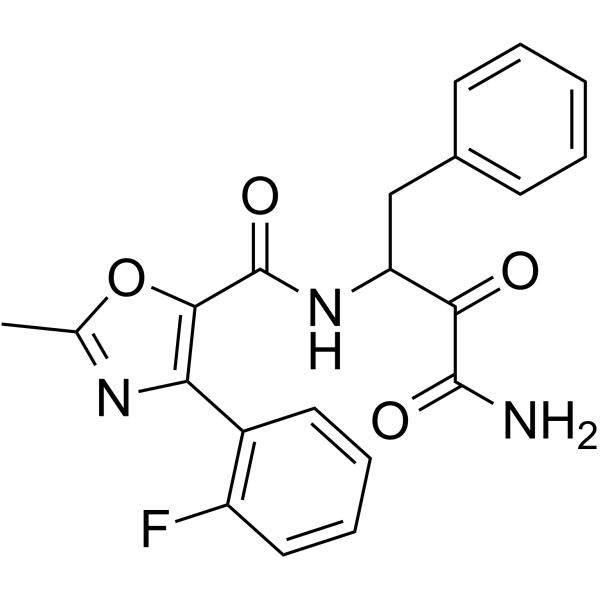
GC64607
Dazcapistat
Dazcapistat is a potent calpain inhibitor, with IC50s of <3 μM for calpain 1, calpain 2 and calpain 9, respectively (patent WO2018064119A1, compound 405).
-
GC16747
Dihydroeponemycin
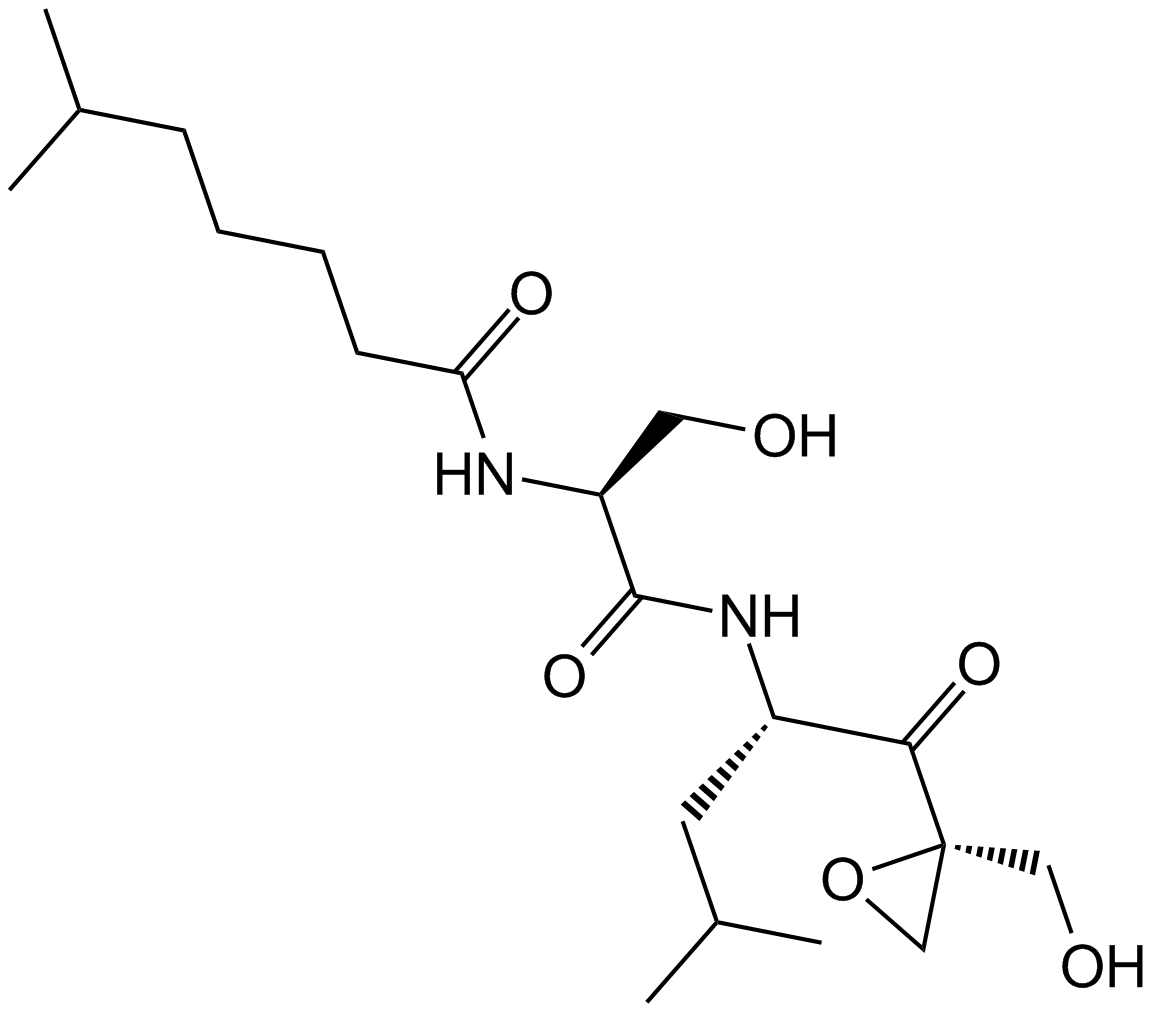
-
GC12494
Epoxomicin
A highly selective proteasome inhibitor
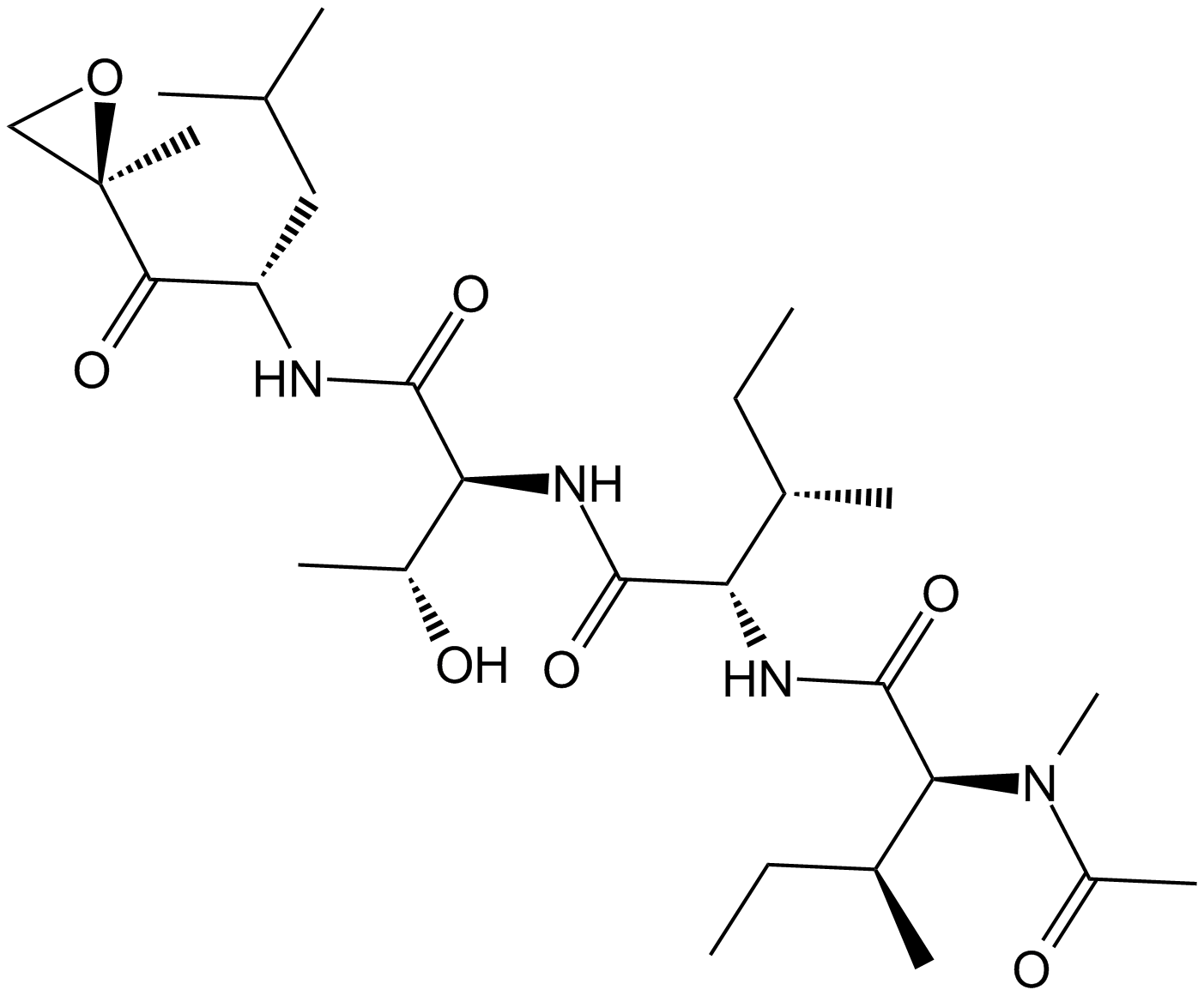
-
GC31563
FK-448 Free base
FK-448 Free base is an effective and specific inhibitor of chymotrypsin, with an IC50 of 720 nM.
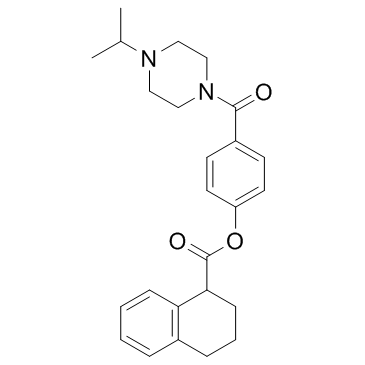
-
GC13711
Gabexate mesylate
A serine protease inhibitor
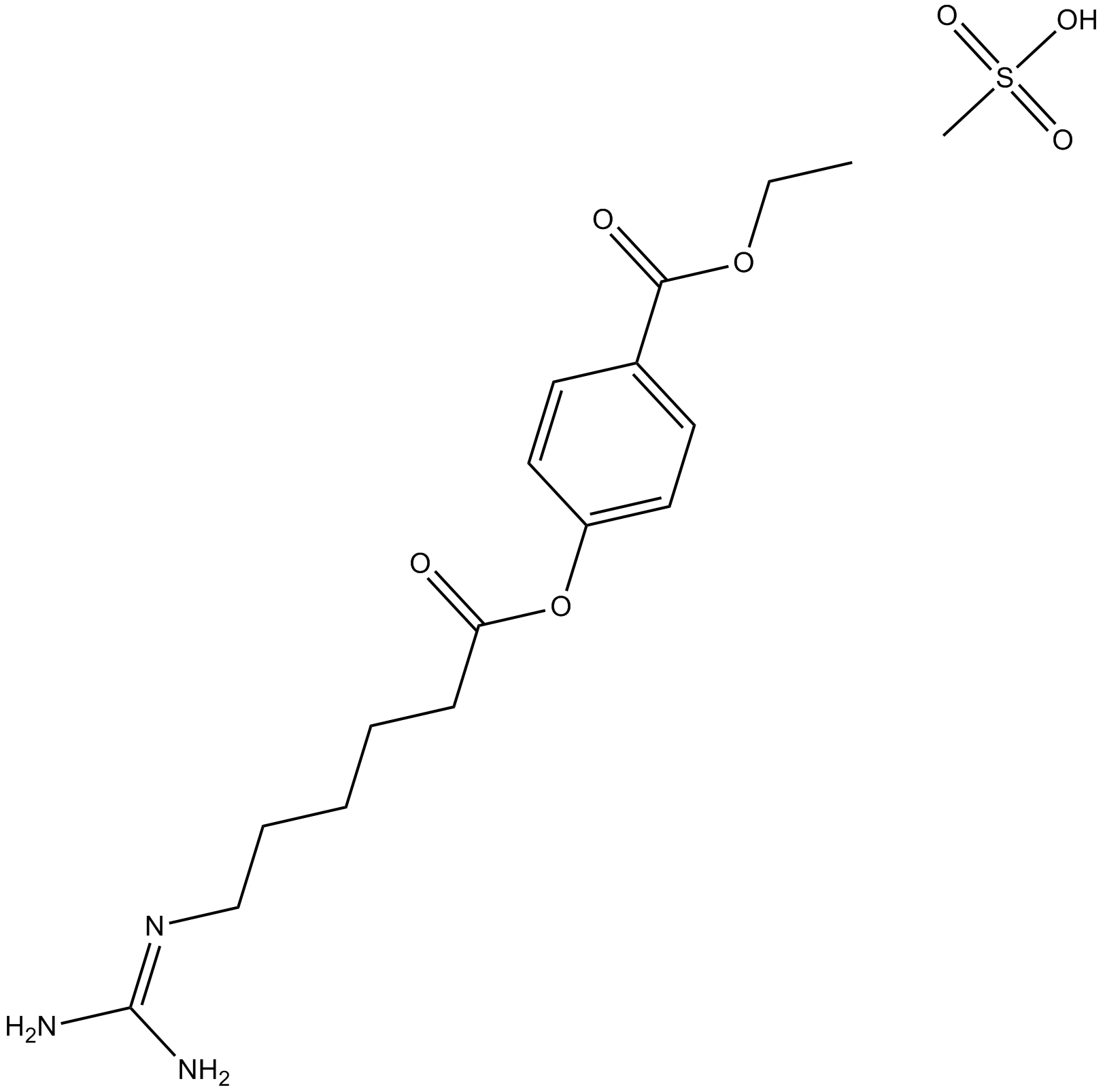
-
GC17255
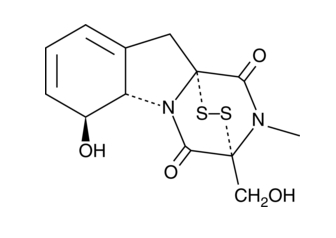
Gliotoxin
An immunosuppressive mycotoxin with diverse biological effects
-
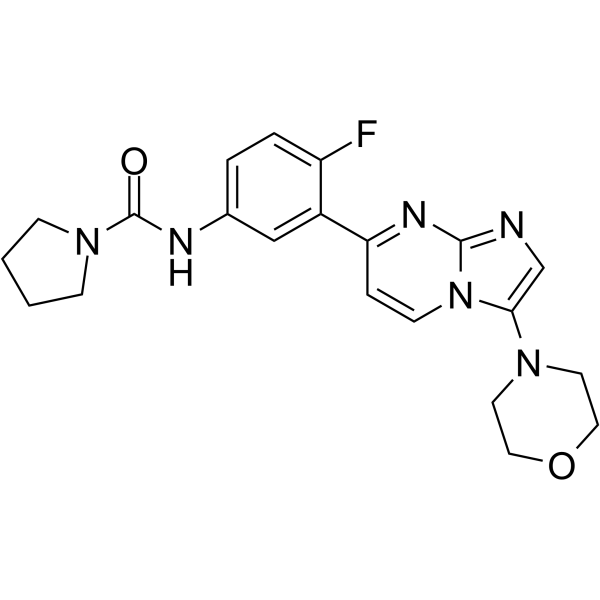
GC64213
GSK3494245
GSK3494245 (DDD01305143) is a potent, orally active, and selective inhibitor of the chymotrypsin-like activity of the parasite proteasome binding in a site sandwiched between the β4 and β5 subunits (IC50=0.16 μM for WT L.
-
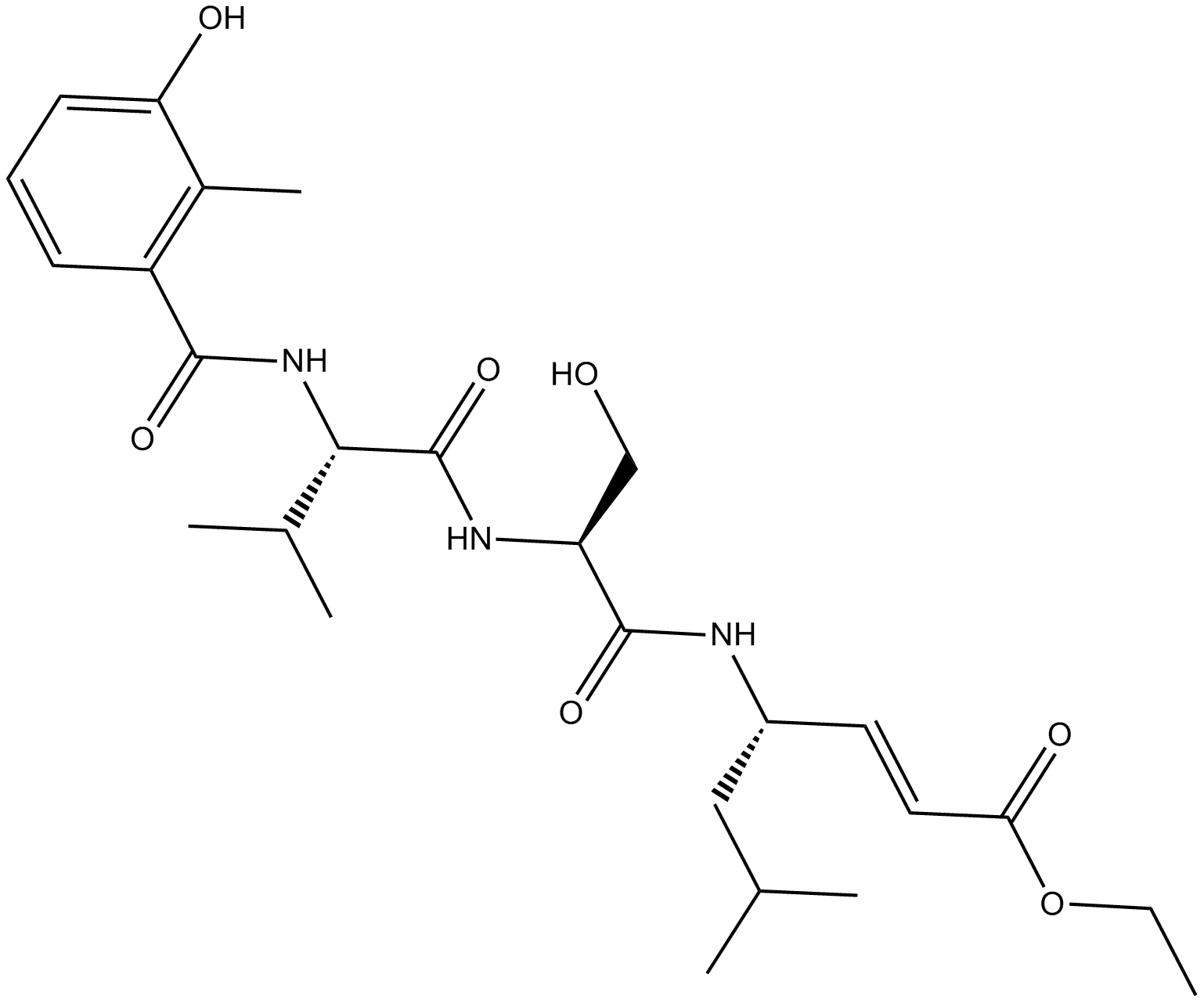
GC16699
HMB-Val-Ser-Leu-VE
inhibitor of the trypsin-like activity of the 20S proteasome
-
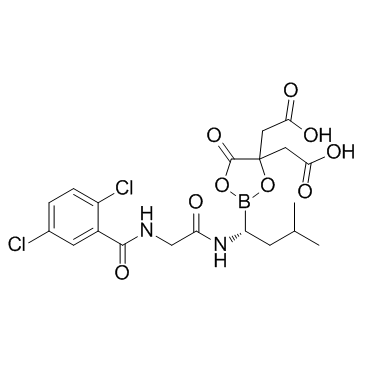
GC36356
Ixazomib citrate
Ixazomib citrate (MLN9708) is a reversible inhibitor of the chymotrypsin-like proteolytic β5 site of the 20S proteasome with an IC50 of 3.4 nM and a Ki of 0.93 nM.
-
GC31899
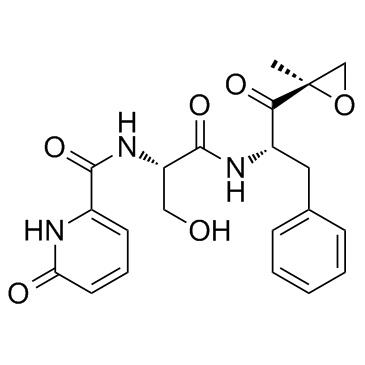
KZR-504
KZR-504 is a highly selective inhibitor of immunoproteasome low molecular mass polypeptide 2 (LMP2), with IC50s of 51 nM, 4.274 μM for LMP2 and LMP7, respectively.
-
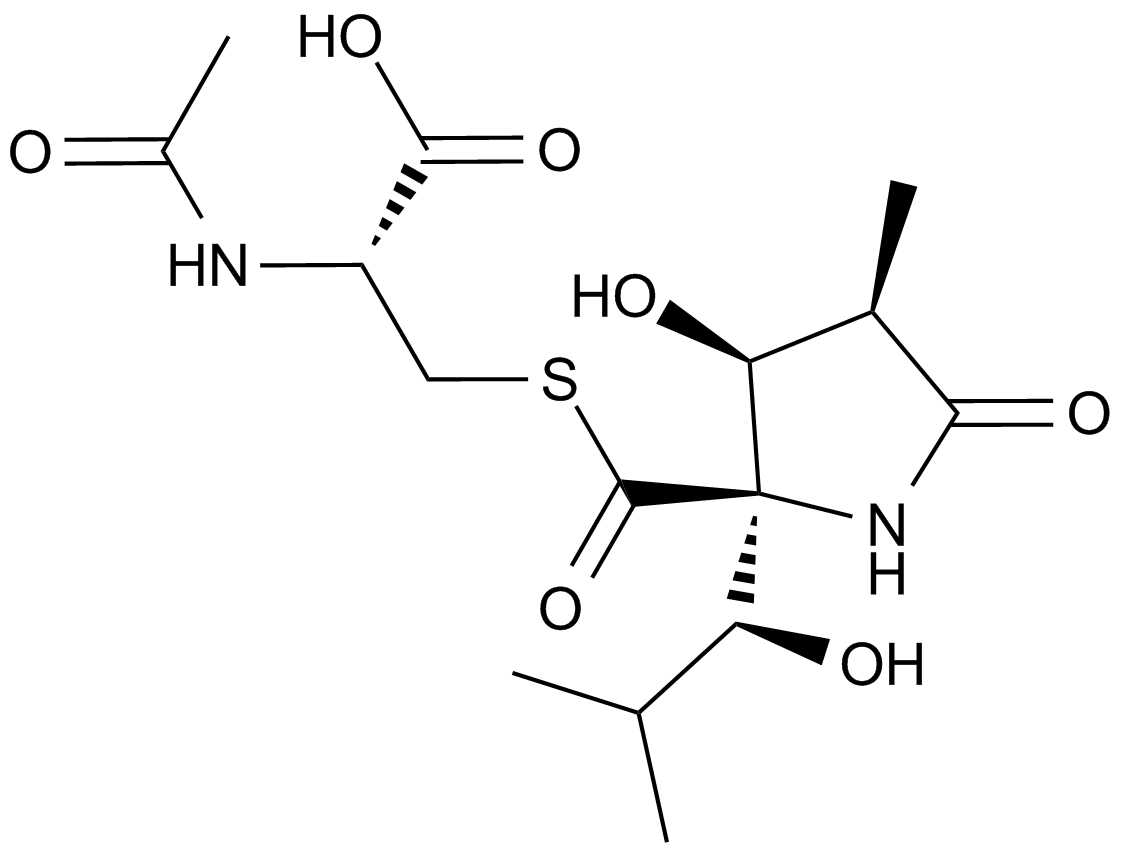
GC13123
Lactacystin (Synthetic)
A selective inhibitor of the 20S proteasome
-
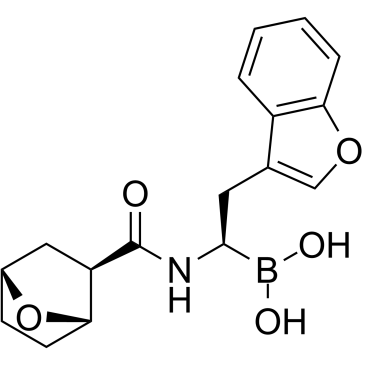
GC34649
LMP7-IN-1
LMP7-IN-1 is an orally bioavailable, potent, reversible and highly selective immunoproteasome subunit LMP7 (β5i) inhibitor. LMP7-IN-1 exerts high biochemical (IC50=3.6 nM) and cellular (IC50=3.4 nM) potency against the LMP7 subunit. LMP7-IN-1 shows strong antitumor efficacy in multiple myeloma xenograft models. LMP7-IN-1 leads to a significant and prolonged suppression of tumor LMP7 activity and ubiquitinated protein turnover and the induction of apoptosis in multiple myeloma cells.
-
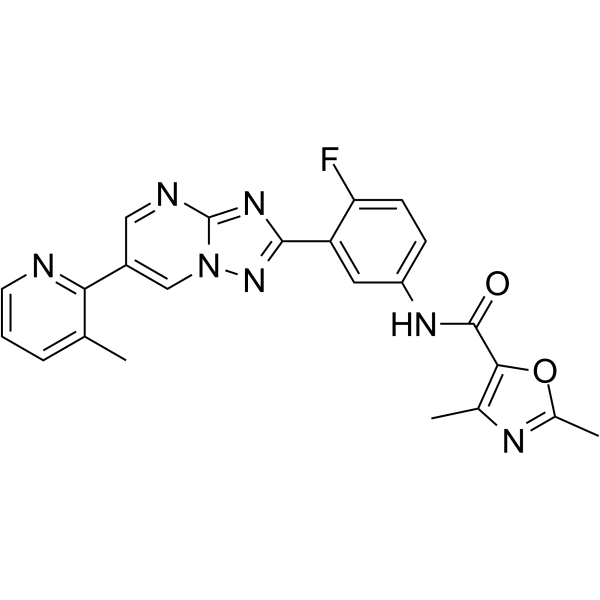
GC64282
LXE408
LXE408 is an orally active, non-competitive and kinetoplastid-selective proteasome inhibitor.
-
GC11640
MDL 28170
A cell permeable, selective calpain inhibitor
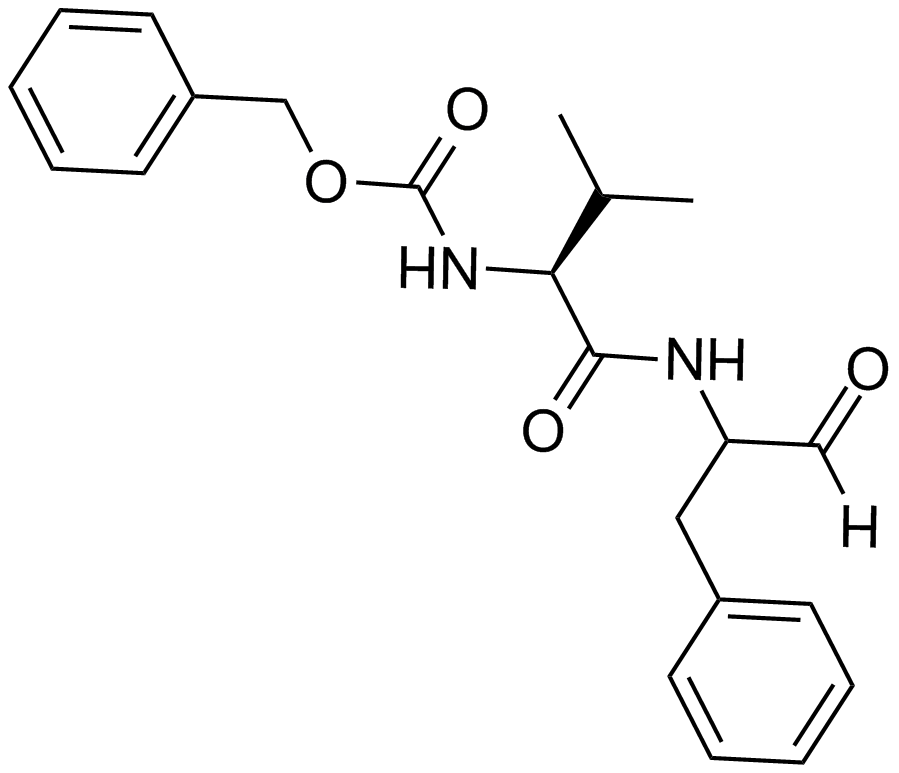
-
GC10178
MG-115
A potent, reversible proteasome inhibitor
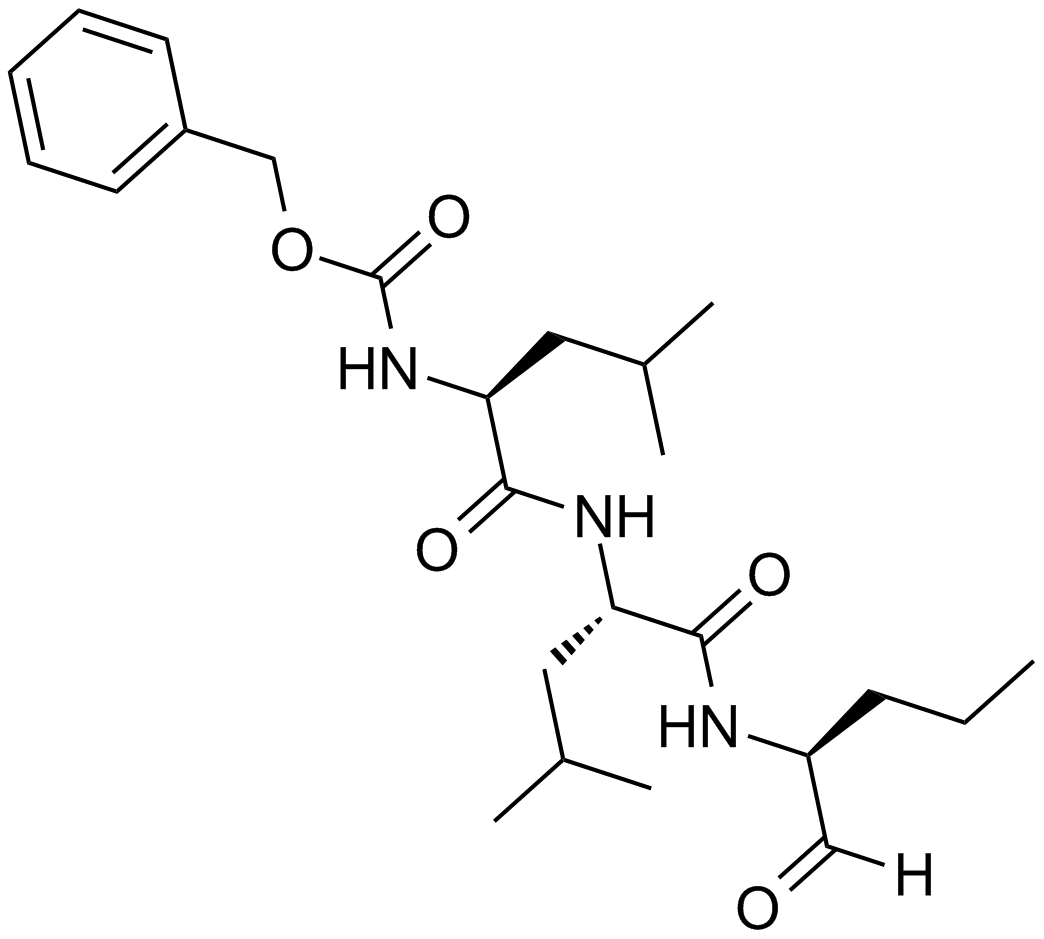
-
GC10383
MG-132
A potent proteasome inhibitor
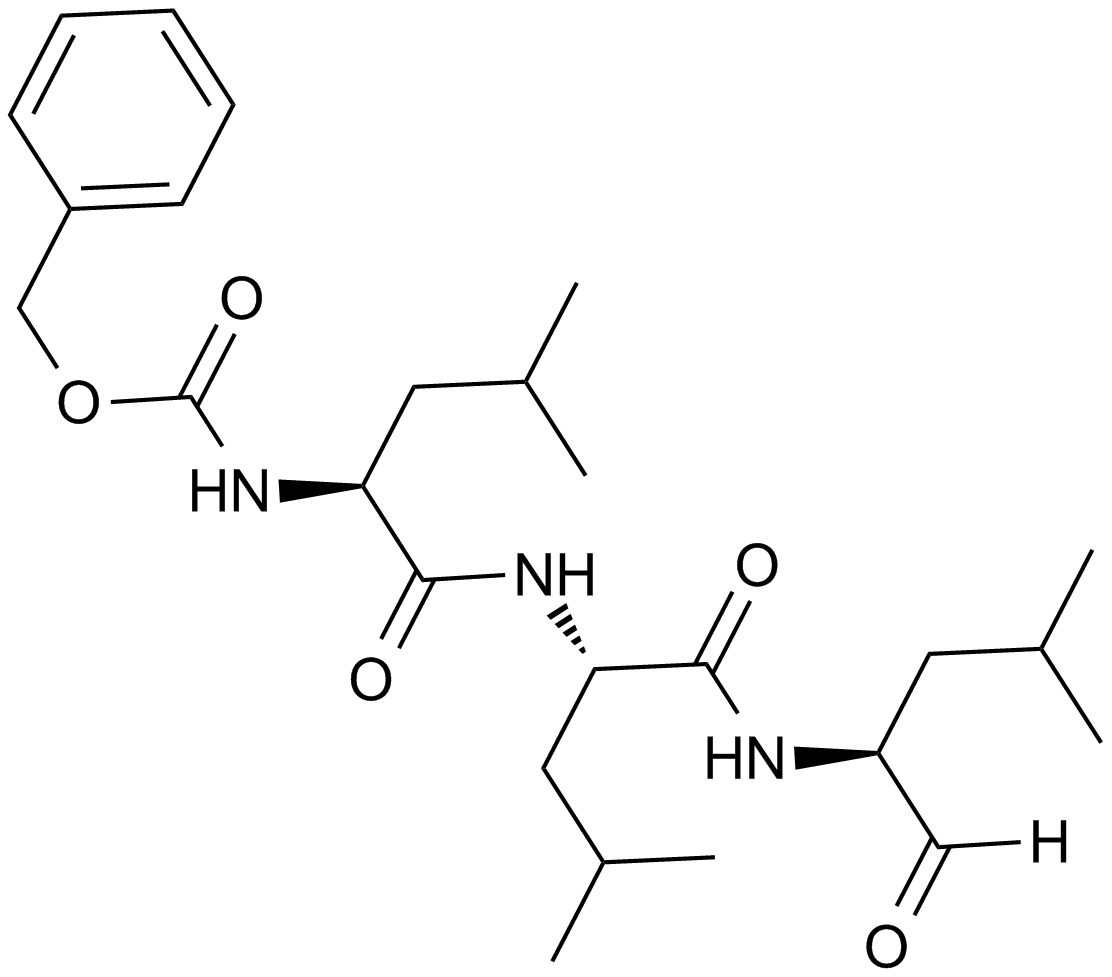
-
GC15517
MG-262
A proteasome inhibitor with diverse biological activities
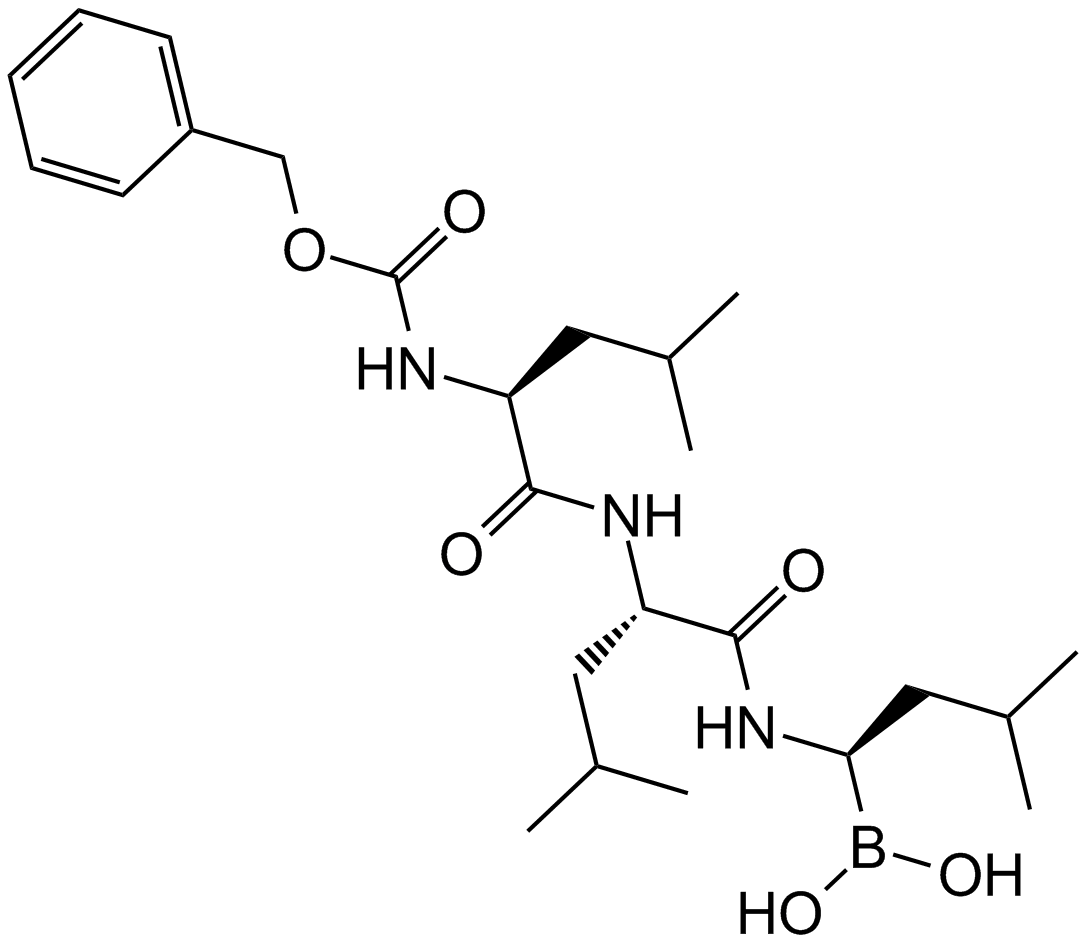
-
GC16146
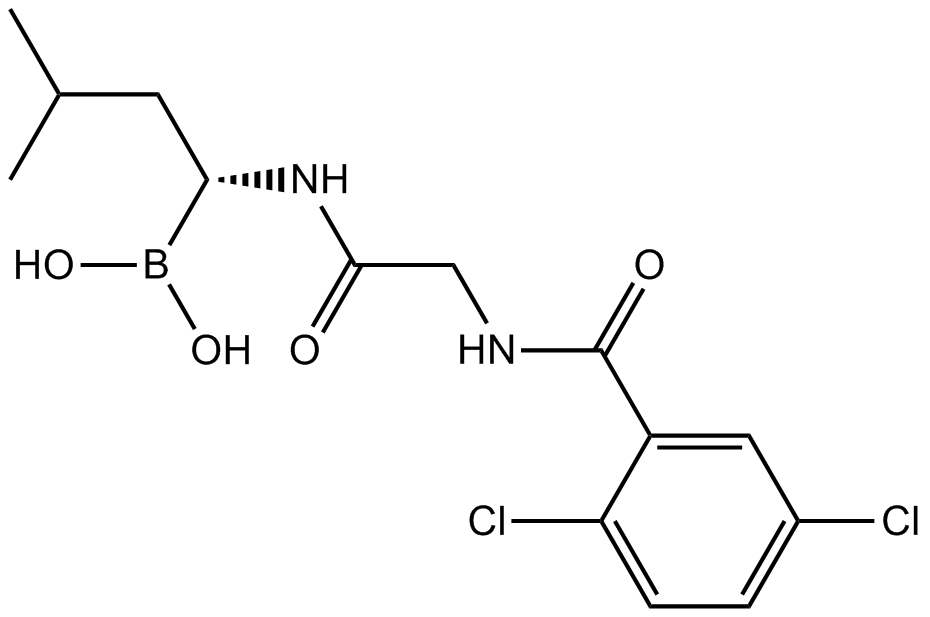
MLN2238
A 20S proteasome inhibitor and an active metabolite of MLN9708
-
GC13718
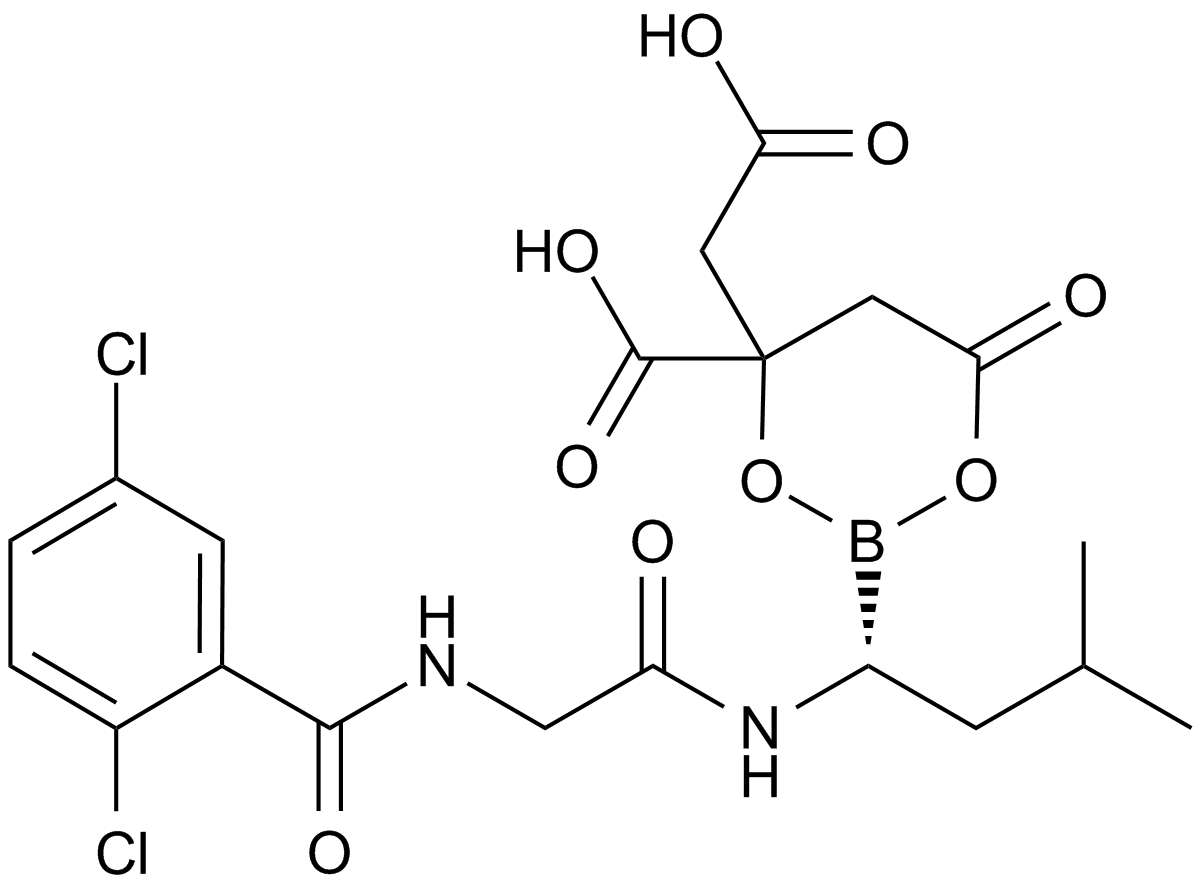
MLN9708
A prodrug form of MLN2238
-
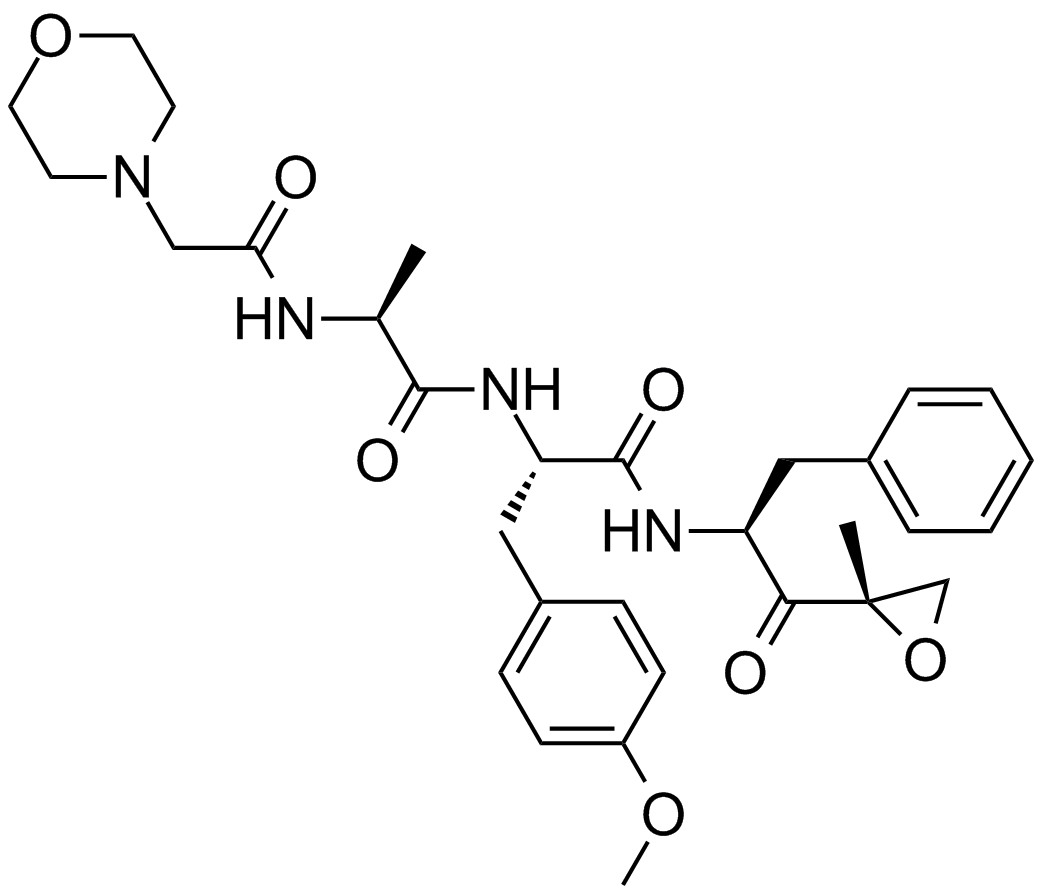
GC15778
ONX-0914 (PR-957)
A selective inhibitor of the β5i (LMP7) subunit of the immunoproteasome
-
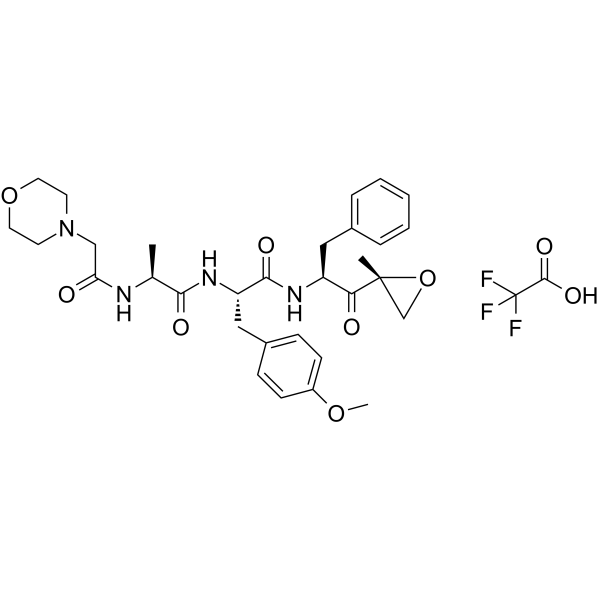
GC62210
ONX-0914 TFA
ONX-0914 (PR-957) TFA is a selective inhibitor of low-molecular mass polypeptide-7 (LMP7), the chymotrypsin-like subunit of the immunoproteasome.
-
GC15447
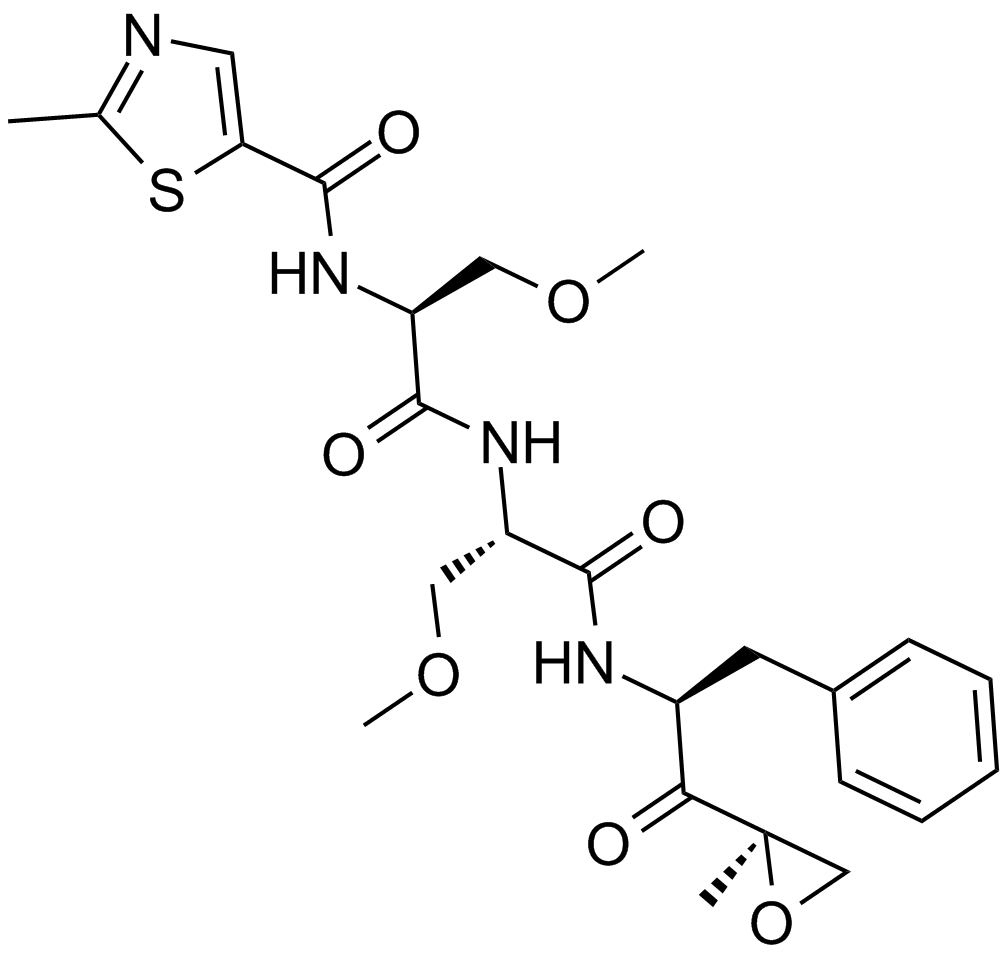
Oprozomib (ONX-0912)
An orally available proteasome inhibitor
-
GC16912
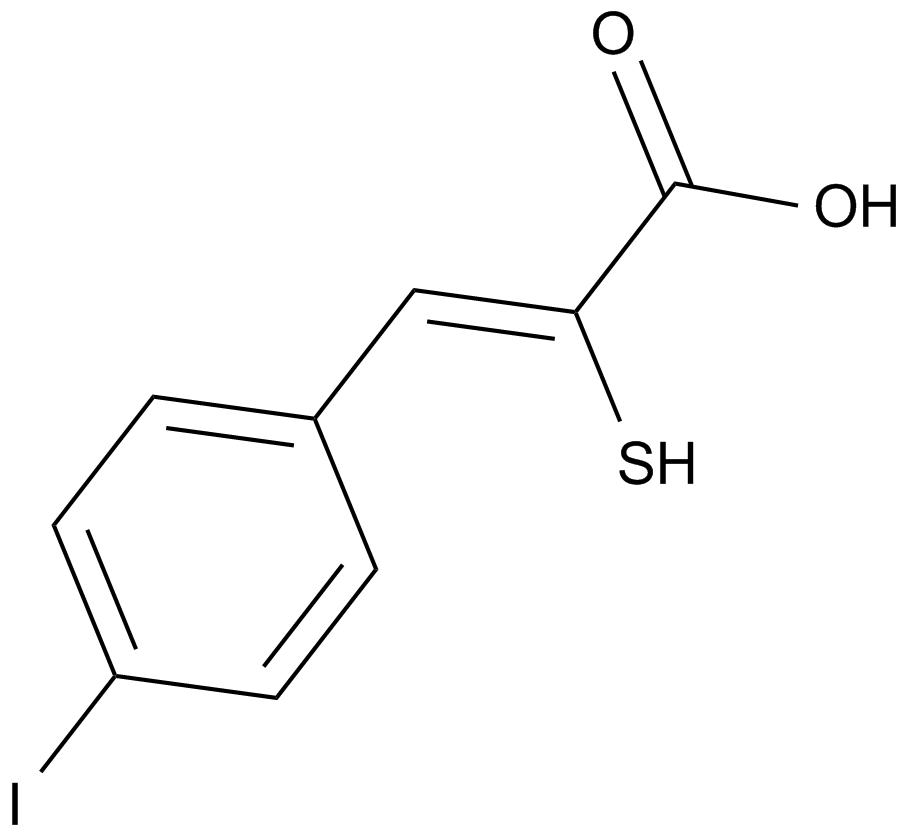
PD 150606
An inhibitor of calpains
-
GC10561
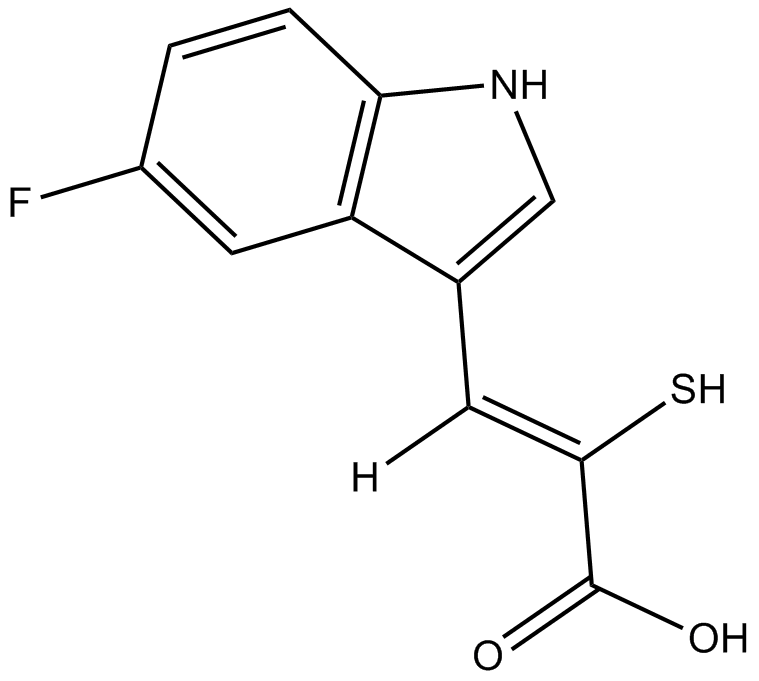
PD 151746
calpain inhibitor, cell-permeable
-
GC11974
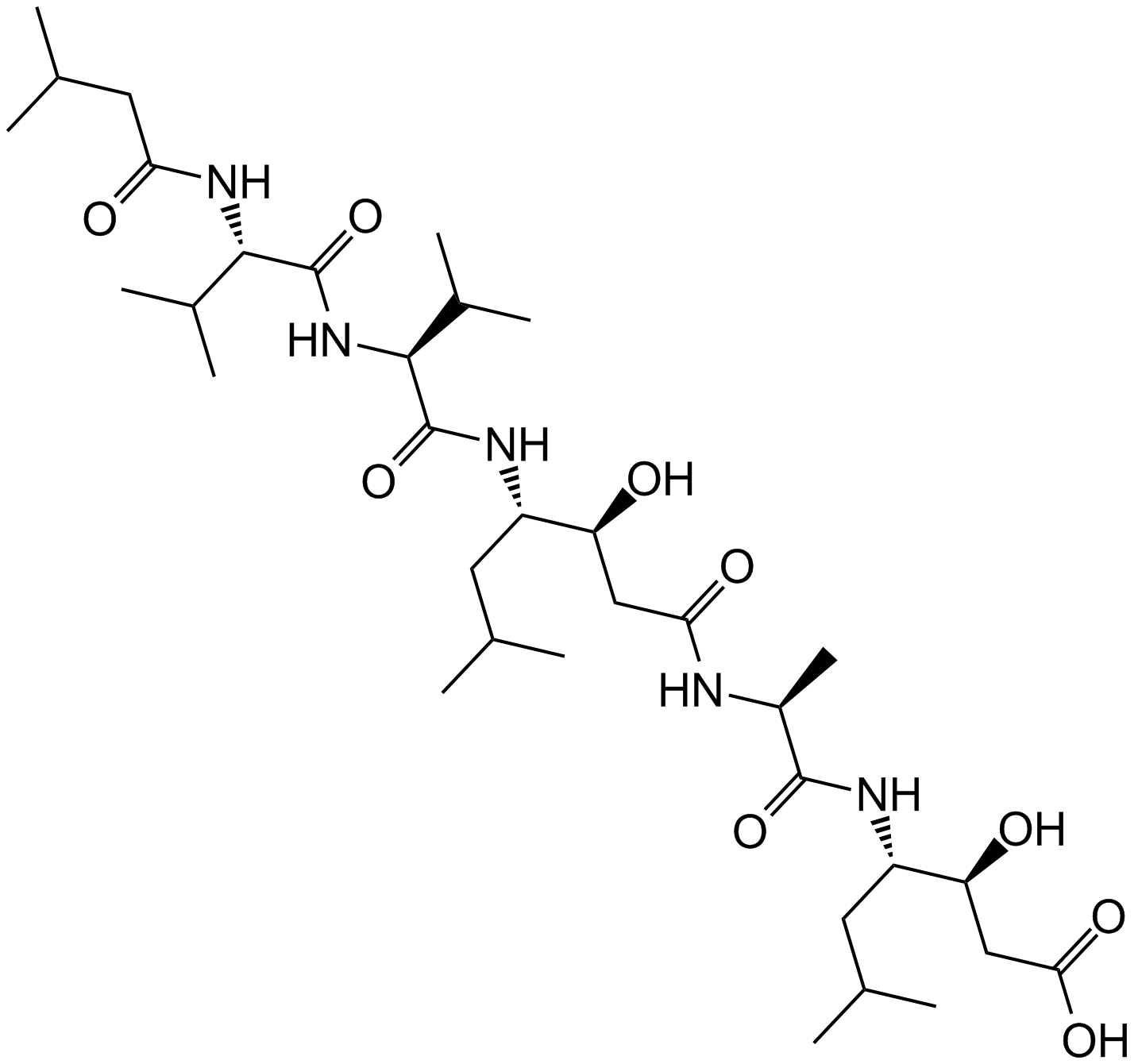
Pepstatin A
Pepstatin A is an orally active inhibitor of aspartic proteases, which is produced by actinomycetes.
-
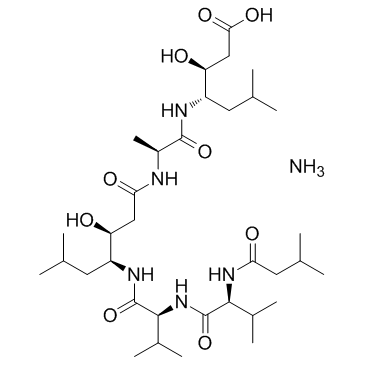
GC36871
Pepstatin Ammonium
Pepstatin Ammonium is a specific aspartic protease inhibitor produced by actinomycetes, with IC50s of 4.5 nM, 6.2 nM, 150 nM, 290 nM, 520 nM and 260 nM for hemoglobin-pepsin, hemoglobin-proctase, casein-pepsin, casein-proctase, casein-acid protease and hemoglobin-acid protease, respectively.
-
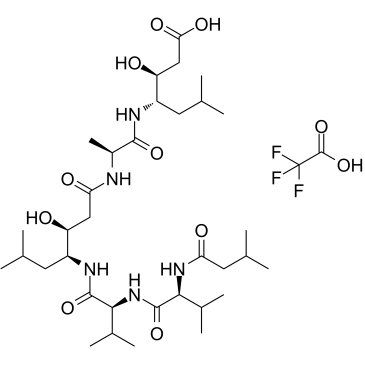
GC36872
Pepstatin Trifluoroacetate
Pepstatin Trifluoroacetate (Pepstatin A Trifluoroacetate) is a specific aspartic protease inhibitor produced by actinomycetes, with IC50s of 4.5 nM, 6.2 nM, 150 nM, 290 nM, 520 nM and 260 nM for hemoglobin-pepsin, hemoglobin-proctase, casein-pepsin, casein-proctase, casein-acid protease and hemoglobin-acid protease, respectively.
-
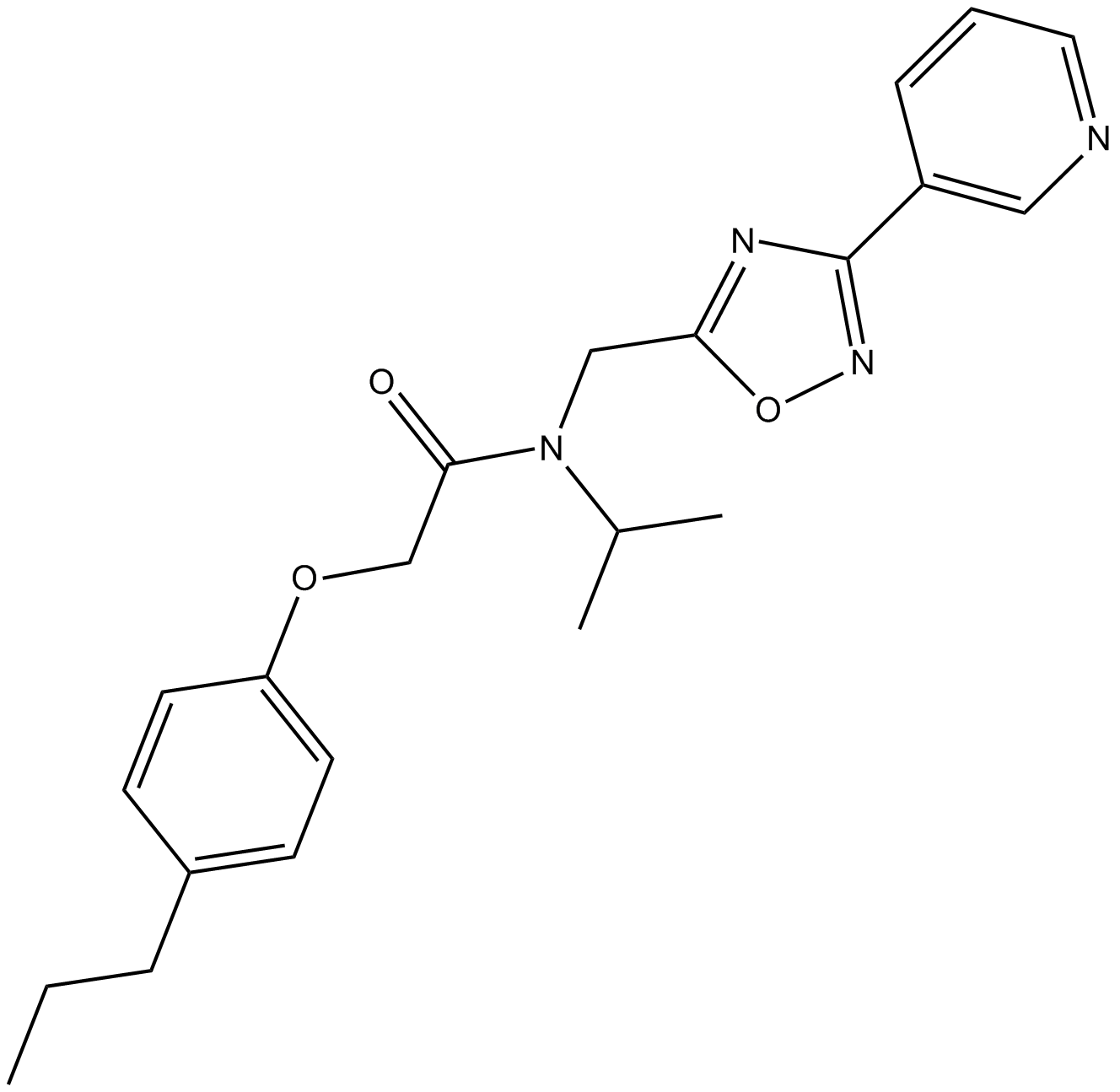
GC15802
PI-1840
Chymotrypsin-like (CT-L) inhibitor
-
GC63964
PR-39 TFA
PR-39 TFA, a natural proline- and arginine-rich antibacterial peptide, is a noncompetitive, reversible and allosteric proteasome inhibitor.
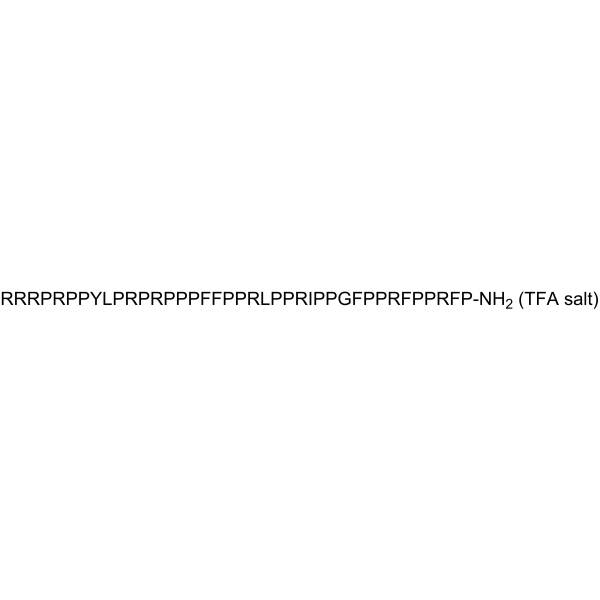
-
GC61945
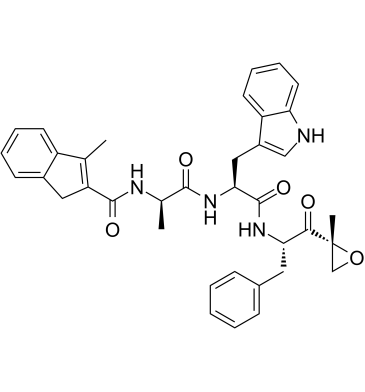
PR-924
PR-924 is a selective tripeptide epoxyketone immunoproteasome subunit LMP-7 inhibitor with an IC50 of 22 nM. PR-924 covalently modifies proteasomal N-terminal threonine active sites. PR-924 inhibits growth and triggers apoptosis in multiple myeloma (MM) cells. PR-924 has antitumor activities.
-
GC33311
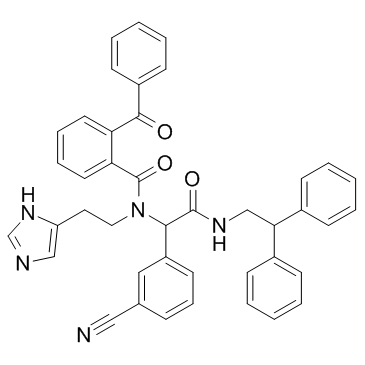
Proteasome-IN-1
Proteasome-IN-1 is a proteasome inhibitor extracted from patent WO 2013142376 A1.
-
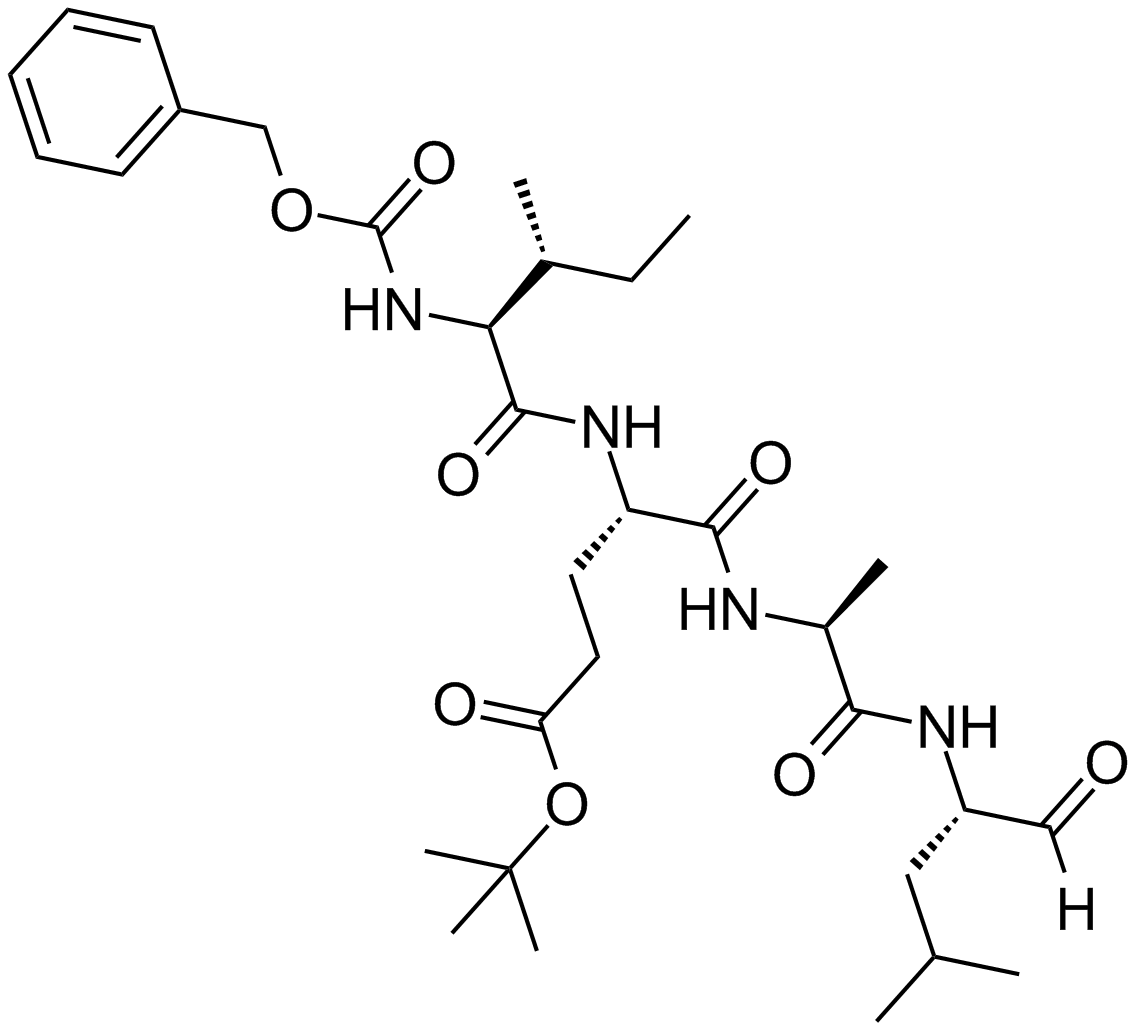
GC13865
PSI
-
GC61221
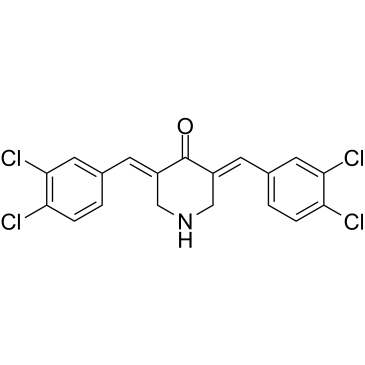
PTP1B-IN-9
PTP1B-IN-9 is a ubiquitin-proteasome system (UPS)-stressor. PTP1B-IN-9 inhibits ubiquitin-mediated protein degradation upstream of the 20S proteasomal catalytic activites. PTP1B-IN-9 triggers a ubiquitin-proteasome-system (UPS)-stress response without affecting 20S proteasome catalytic activities. Anticancer activity.
-
GC32765
RA190
RA190, a bis-benzylidine piperidon, inhibits proteasome function by covalently binding to cysteine 88 of ubiquitin receptor RPN13.
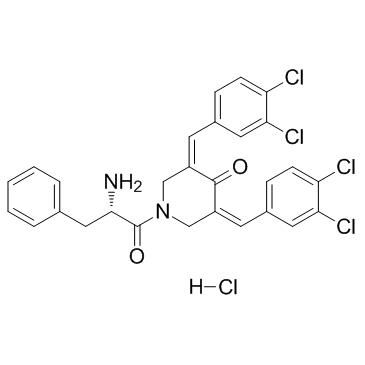
-
GC62399
RA375
RA375 is a RPN13 (26S proteasome regulatory subunit) inhibitor. RA375 activates UPR signaling, ROS production and apoptosis. RA375 exhibits ten-fold greater activity against cancer lines than RA190, reflecting its nitro ring substituents and the addition of a chloroacetamide warhead.
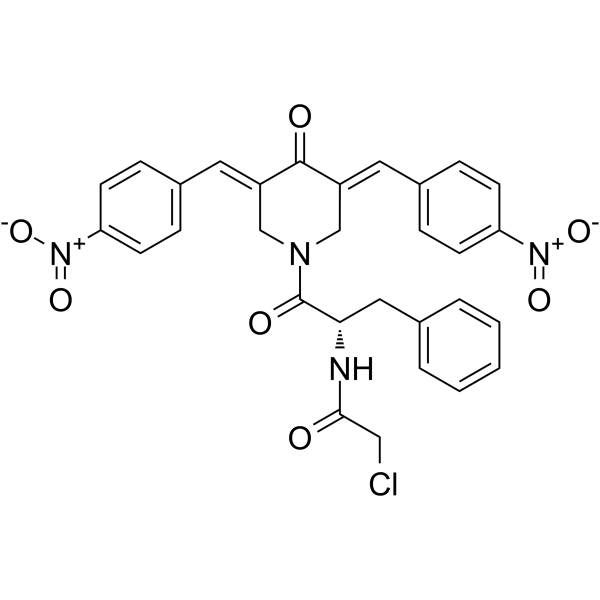
-
GC32407
Rpn11-IN-1
Rpn11-IN-1 (Capzimin intermediate) is a potent and selective inhibitor of proteasome subunit Rpn11 with an IC50 of 390 nM.
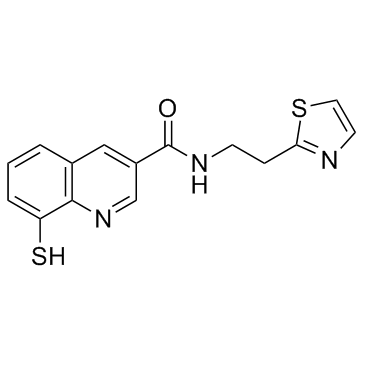
-
GC10486
Salinosporamide A (NPI-0052, Marizomib)
An orally bioactive proteasome inhibitor
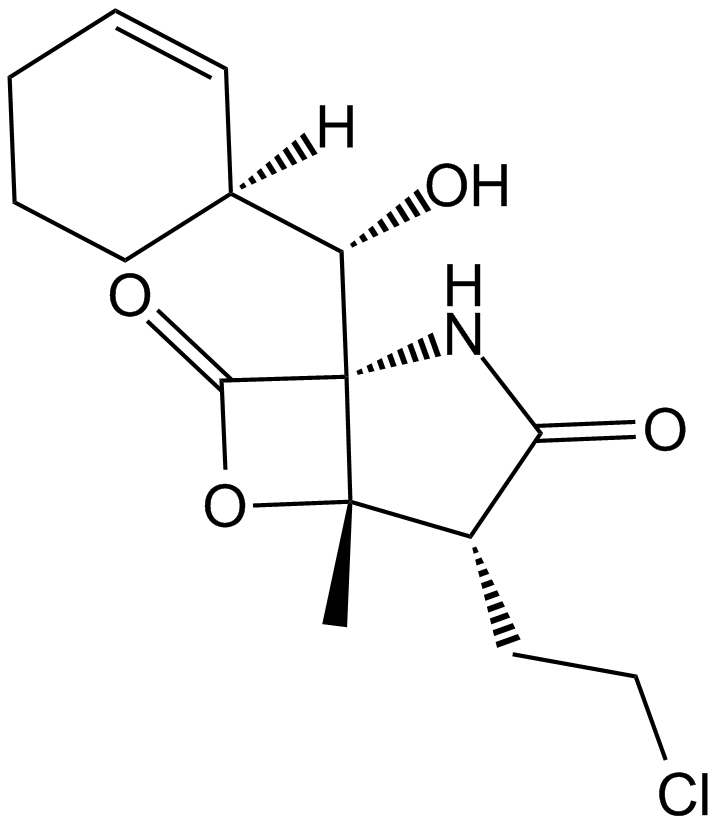
-
GC39166
TCH-165
TCH-165 is a small molecule modulator of proteasome assembly, which increases 20S levels and facilitates 20S-mediated protein degradation.
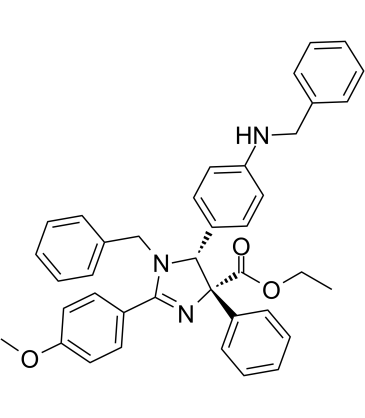
-
GC65938
Thrombin inhibitor 5
Thrombin inhibitor 5 (compound 385) is a thrombin inhibitor, with IC50s ranging from 0.1 μM to 1 μM. Thrombin inhibitor 5 can be used for research of venous thromboembolism.
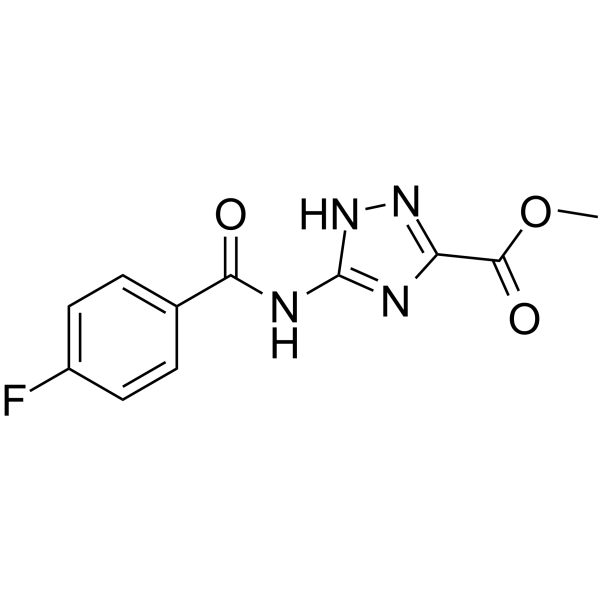
-
GC37814
Tomatine
Tomatine is a glycoalkaloid, found in the tomato plant (Lycopersicon esculentum Mill.
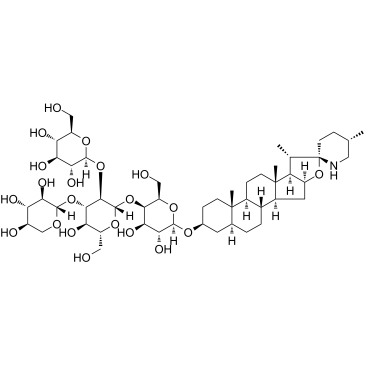
-
GC68437
Vimentin-IN-1

-
GC12003
VR23
proteasome inhibitor
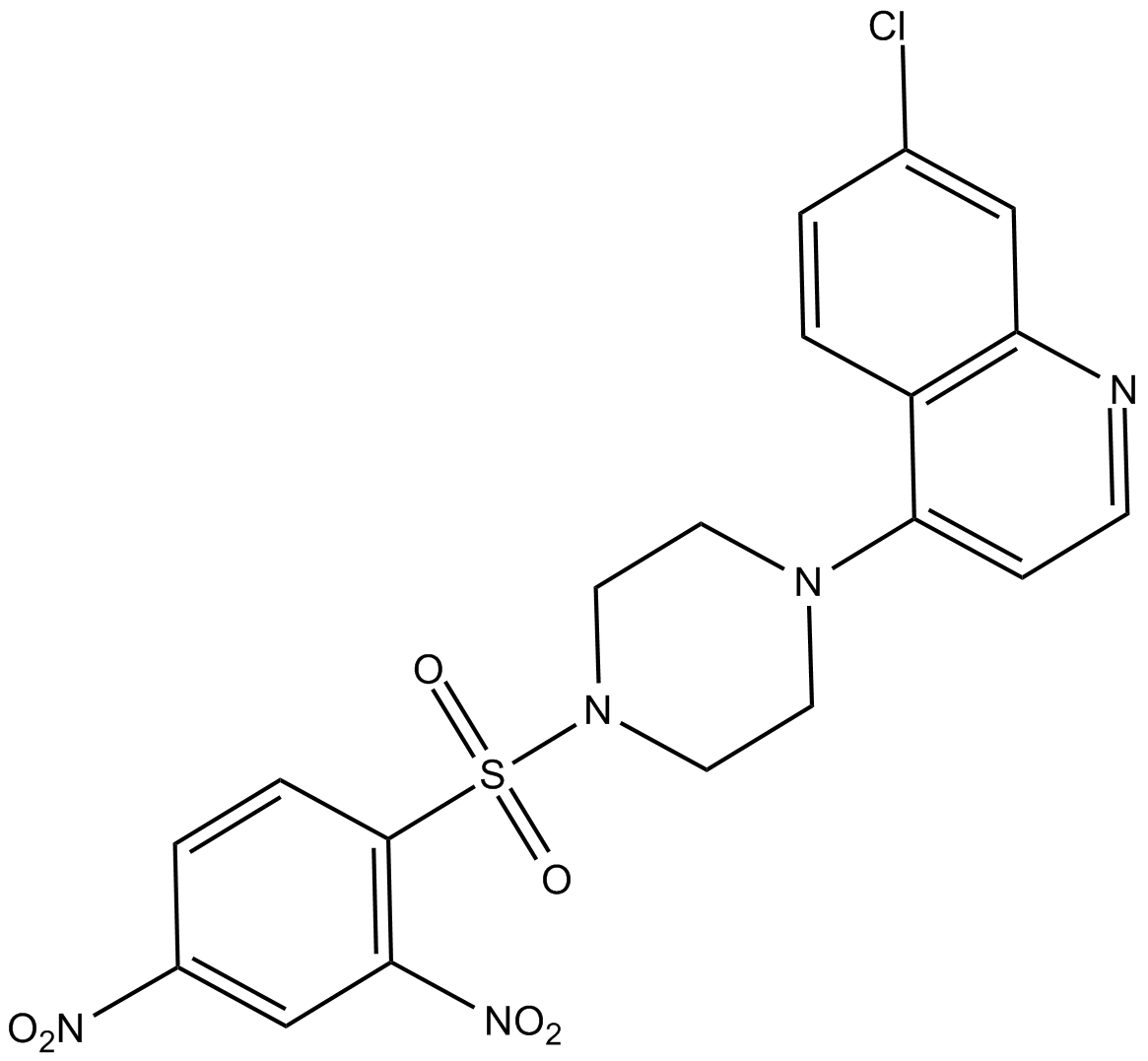
-
GA23841
Z-Ile-Glu(OtBu)-Ala-Leu-aldehyde
Z-IE(OtBu)AL-CHO is an inhibitor of chymotrypsin-like activity of the multicatalytic proteinase complex (MPC; 20S proteasome) in HT4 cells. It is the first inhibitor reported so far which can cause accumulation of ubiquitinylated proteins in neuronal cells. Furthermore, this compound induced massive apoptosis in murine leukaemia L1210 cells. Therefore, proteasome inhibitors may be considered as potential anti-neoplastic agents.

-
GC62239
Zetomipzomib
Zetomipzomib (KZR-616), a first-in-class inhibitor of the immunoproteasome, selectively targets the LMP7 (IC50: 39/57 nM=hLMP7/mLMP7) and LMP2 (IC50: 131/179 nM=hLMP7/mLMP7) subunits of the immunoproteasome.