HCV
HCV (hepatitis C virus ) is a enveloped single-stranded RNA virus of the Flavivridae family. It lead to acute hepatitis, chronic liver disease and liver cancer.
Products for HCV
- Cat.No. Product Name Information
-
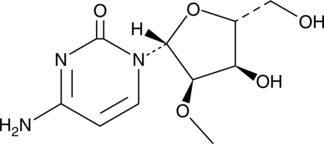
GC48873
2′-O-Methylcytidine
An inhibitor of HCV NS5B
-
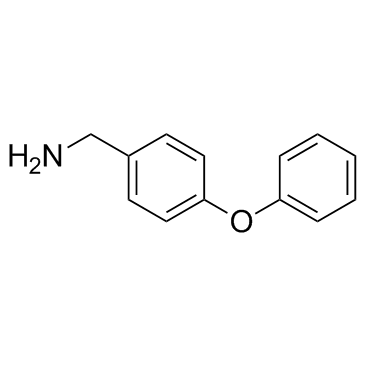
GC32354
4-Phenoxybenzylamine
4-Phenoxybenzylamine inhibits the function of the NS3 protein by stabilizing an inactive conformation with an IC50 of about 500 μM against FL NS3/4a.
-
GC33977
ABT-072
ABT-072 is an orally active and potent non-nucleoside HCV NS5B polymerase inhibitor (HCV GT1a EC50=1 nM; HCV GT1b EC50=0.3 nM).
-
GC63300
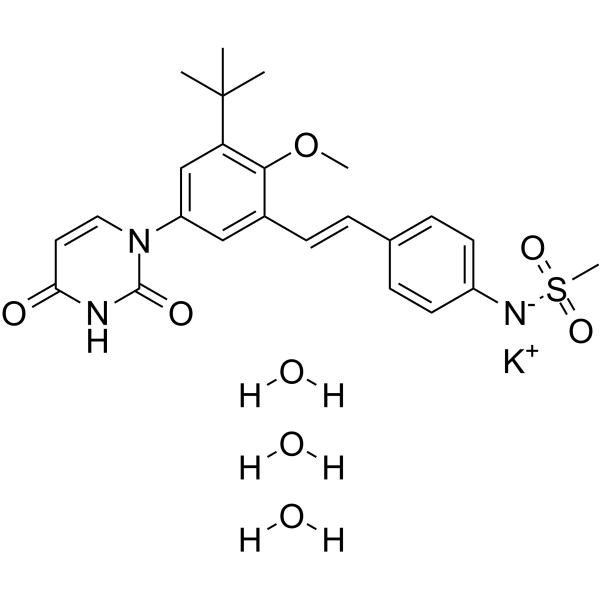
ABT-072 potassium trihydrate
ABT-072 (potassium trihydrate) is an orally active and potent non-nucleoside HCV NS5B polymerase inhibitor (HCV GT1a EC50=1 nM; HCV GT1b EC50=0.3 nM).
-
GC32305
ACH-806 (GS9132)
ACH-806 (GS9132) is an NS4A antagonist which can inhibit Hepatitis C Virus (HCV) replication with an EC50 of 14 nM.
-
GC50013
AG 1478 hydrochloride
Highly potent EGFR-kinase inhibitor
-
GC32072
Alisporivir (DEB-025)
Alisporivir (DEB-025) (Debio-025) is a cyclophilin inhibitor molecule with potent anti-hepatitis C virus (HCV) activity.
-
GC14937
Anguizole
-
GN10647
Artemisinine
-
GC14458
Asunaprevir (BMS-650032)
An HCV NS3/4A protease inhibitor
-
GC62320
AT-511
AT-511 (AT-511) is a potent and orally active HCV viral replication inhibitor.
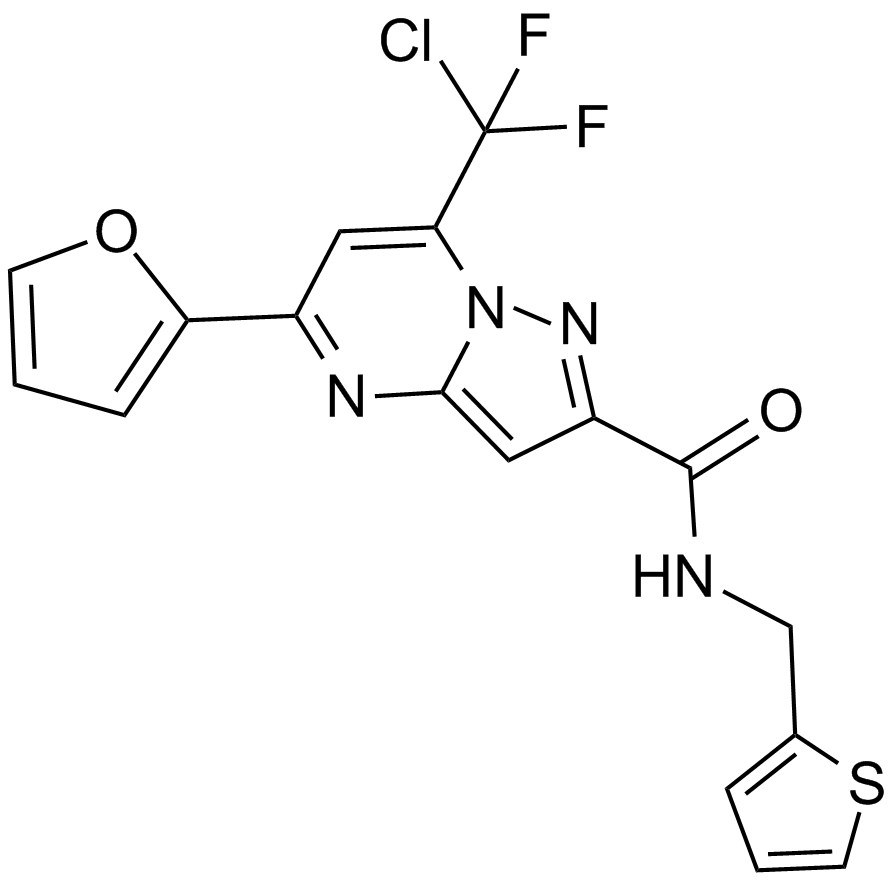
-
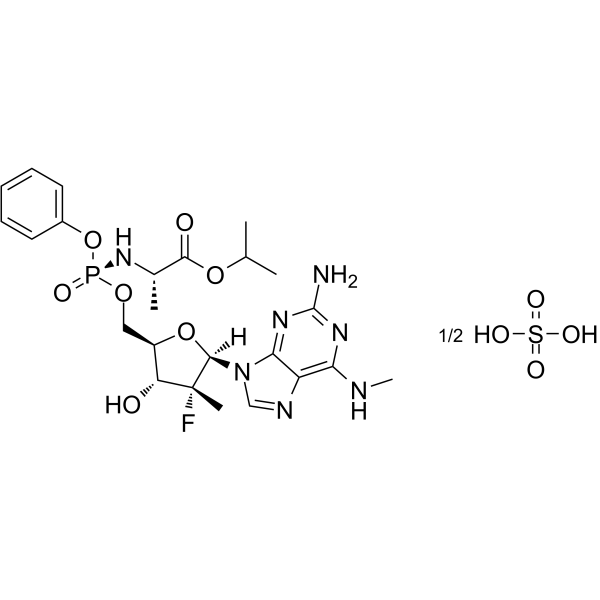
GC62321
AT-527
AT-527(Bemnifosbuvir hemisulfate), a hemisulfate salt of AT-511, a guanosine nucleotide proagent, is a potent and orally active HCV viral replication inhibitor.
-
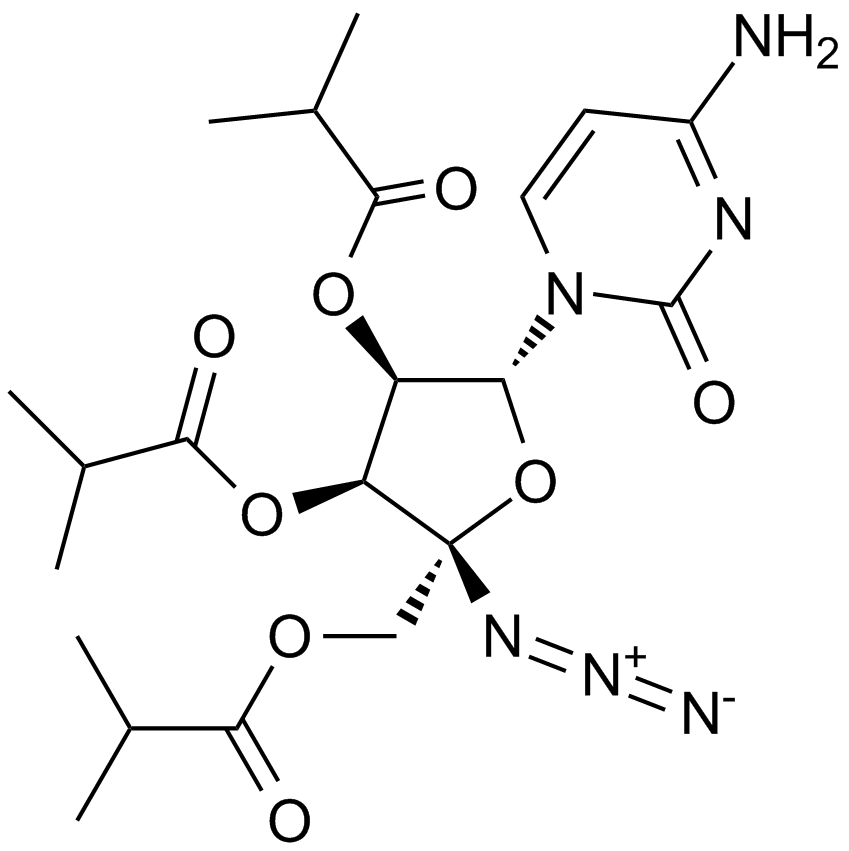
GC10930
Balapiravir
-
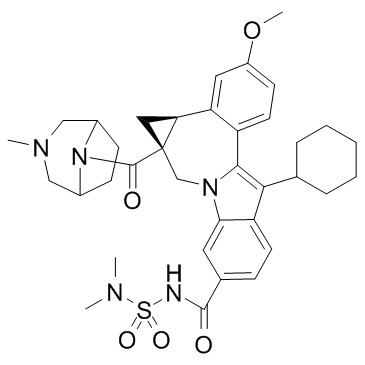
GC33925
Beclabuvir (BMS-791325)
Beclabuvir (BMS-791325) is an allosteric inhibitor that binds to thumb site 1 of the hepatitis C virus (HCV) NS5B RNA-dependent RNA polymerase, and inhibits recombinant NS5B proteins from HCV genotypes 1, 3, 4, and 5 with IC50 of < 28 nM.
-
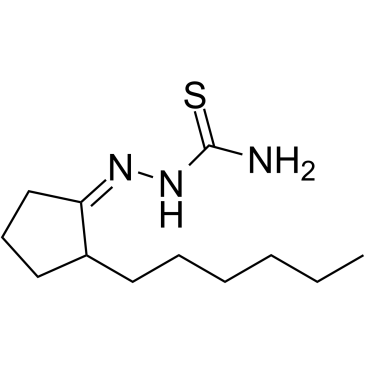
GC60648
BLT-1
BLT-1, a thiosemicarbazone copper chelator, is a selective scavenger receptor B, type 1 (SR-BI) inhibitor.
-
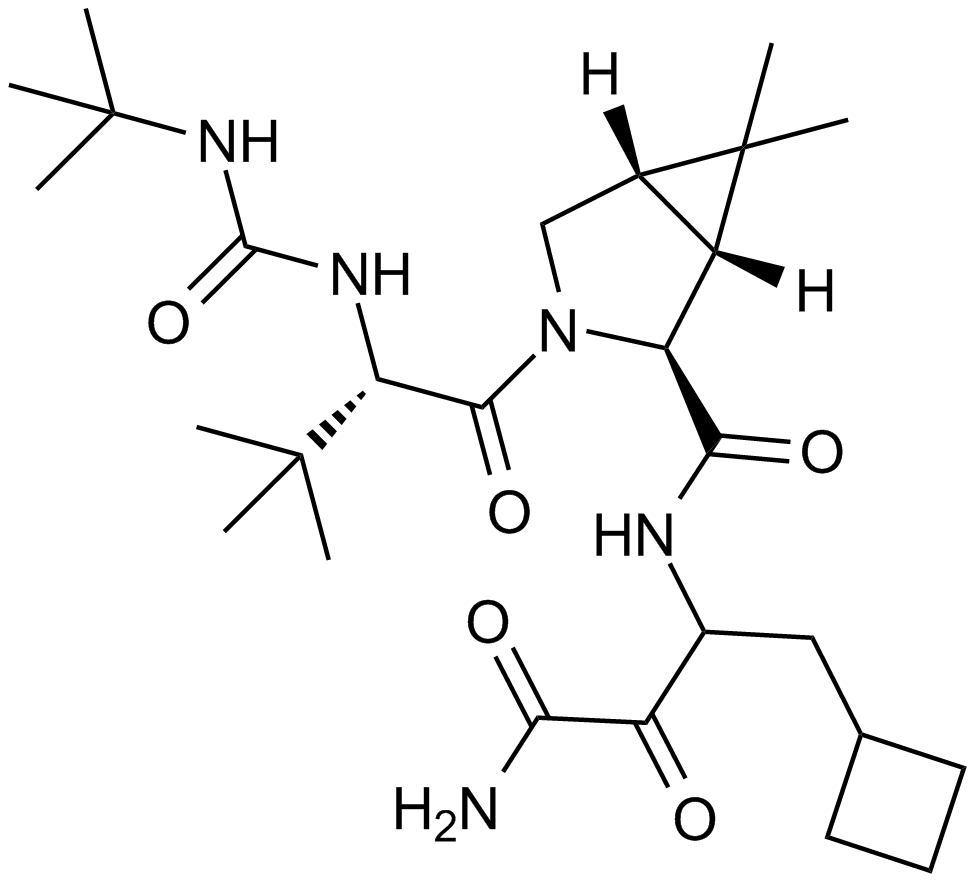
GC10959
Boceprevir
An NS3/4A protease inhibitor
-
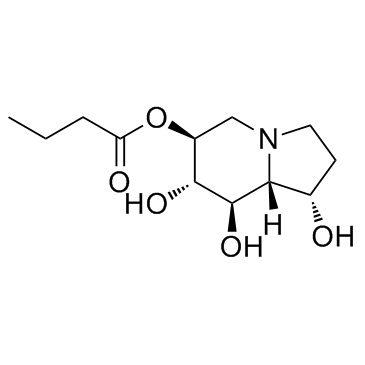
GC32191
Celgosivir (MBI 3253)
Celgosivir (MBI 3253) (MBI 3253; MDL 28574; MX3253) is an α-glucosidase I inhibitor; inhibits bovine viral diarrhoea virus (BVDV) with an IC50 of 1.27 μM in in vitro assay.
-
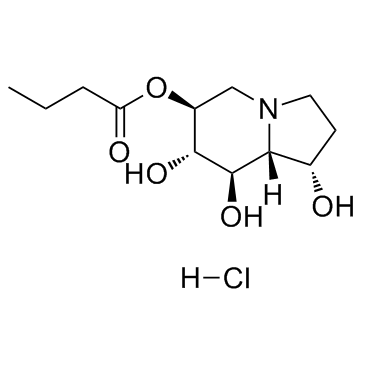
GC32074
Celgosivir hydrochloride (MBI 3253 (hydrochloride))
Celgosivir hydrochloride (MBI 3253 (hydrochloride)) (MBI 3253 hydrochloride; MDL 28574 hydrochloride; MX3253 hydrochloride) is an α-glucosidase I inhibitor; inhibits bovine viral diarrhoea virus (BVDV) with an IC50 of 1.27 μM in in vitro assay.
-
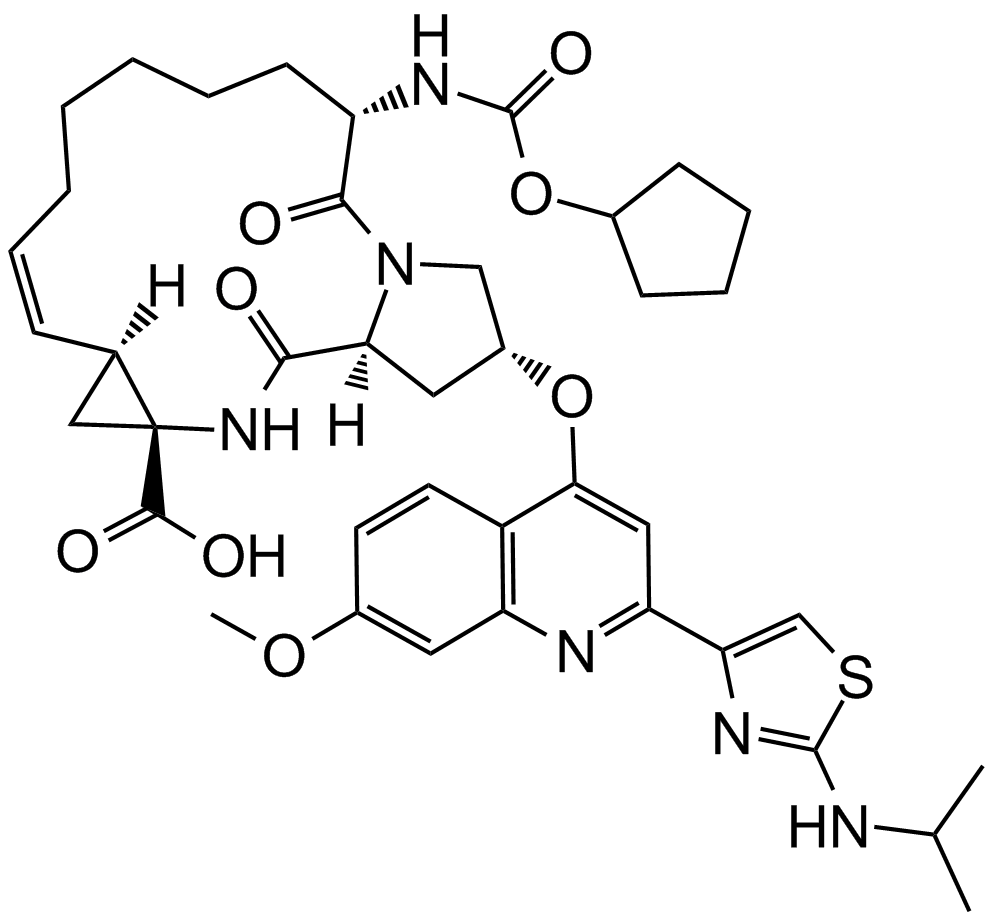
GC11840
Ciluprevir (BILN-2061)
Inhibitor of HCV NS3 protease
-
GC12444
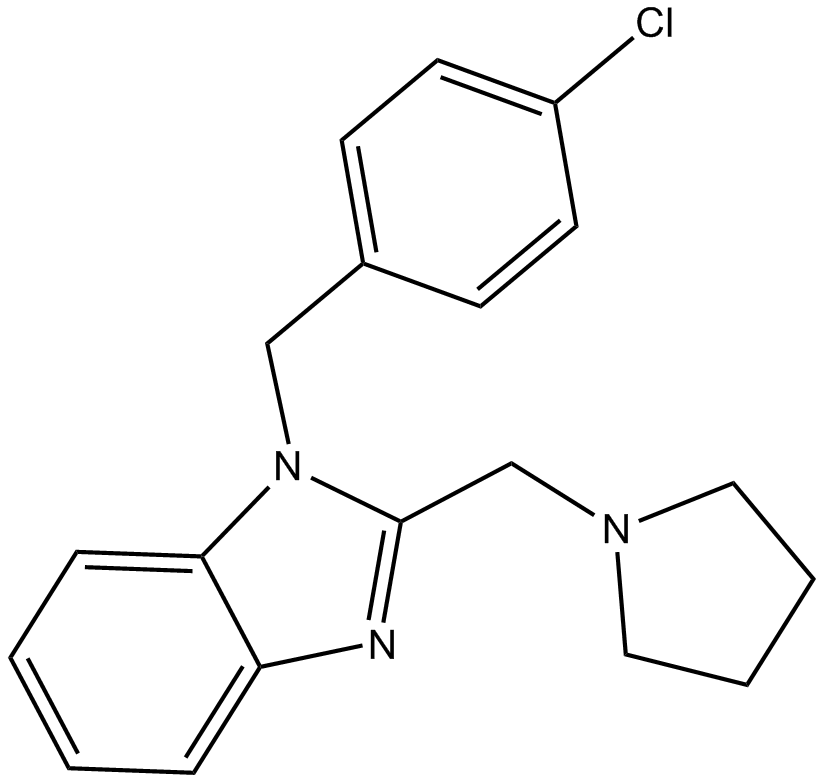
Clemizole
An antihistamine and TRPC5 channel blocker
-
GC17701
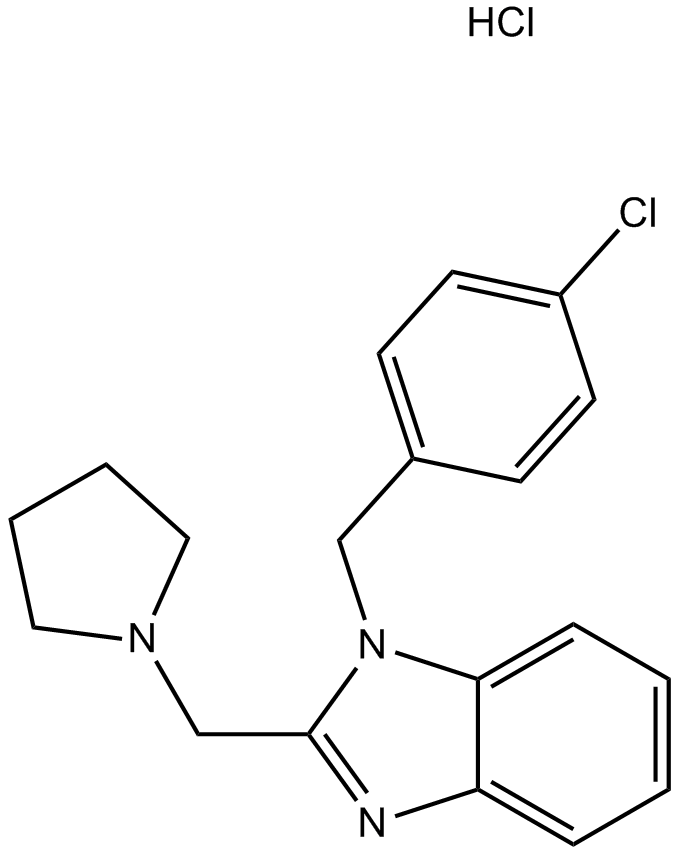
Clemizole hydrochloride
An antihistamine and TRPC5 channel blocker
-
GC64065
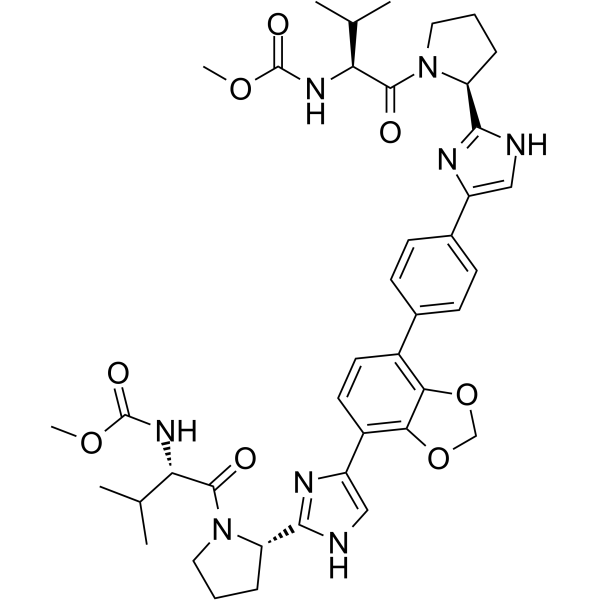
Coblopasvir
Coblopasvir (KW-136) is a pangenotypic non-structural protein 5A (NS5A) inhibitor.
-
GC64066
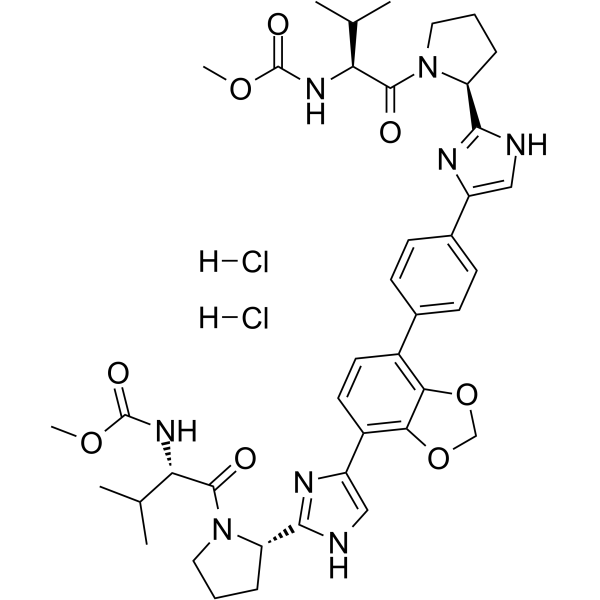
Coblopasvir dihydrochloride
Coblopasvir (KW-136) dihydrochloride is a pangenotypic non-structural protein 5A (NS5A) inhibitor.
-
GC40756
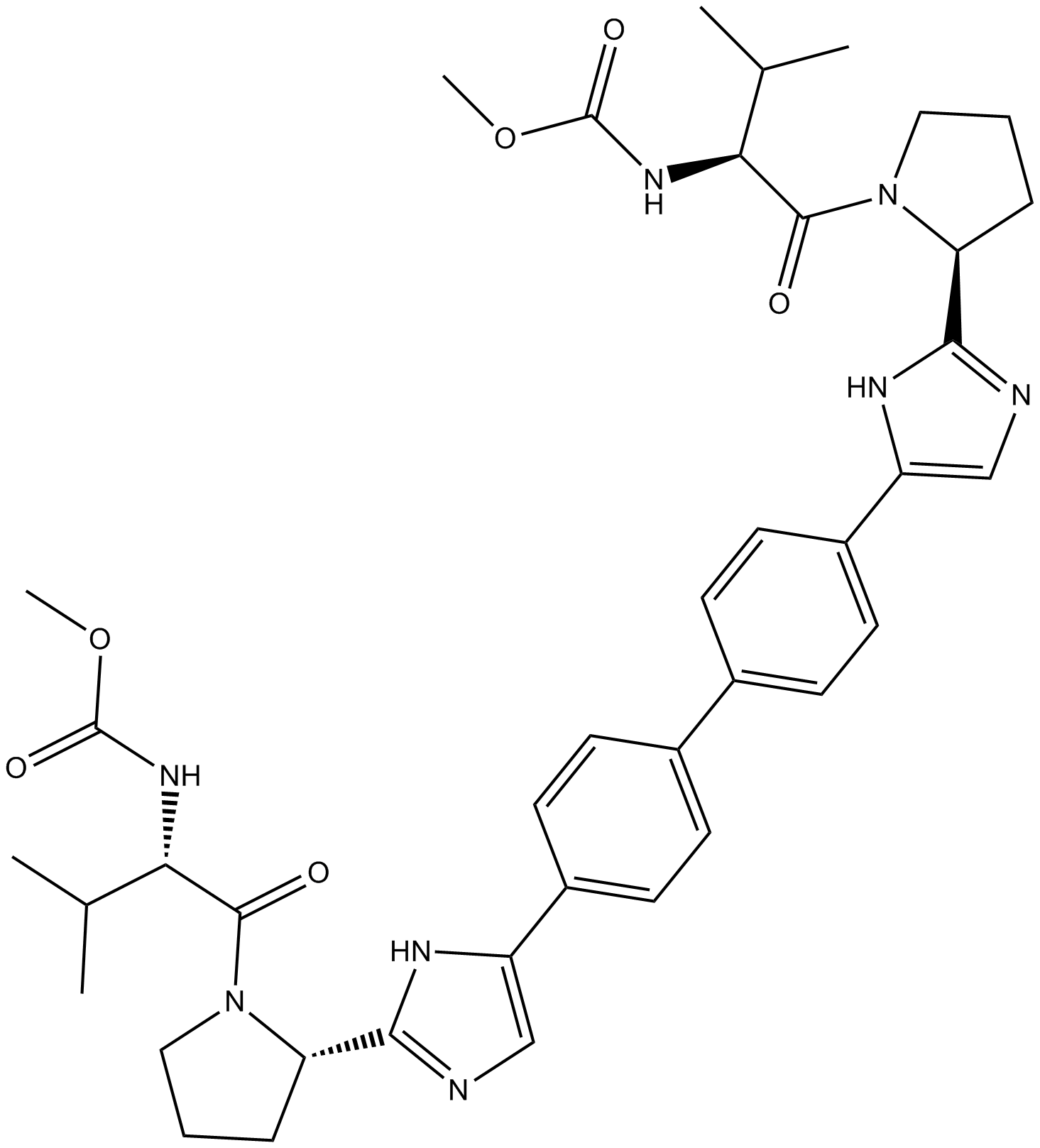
Daclatasvir
Daclatasvir is a first generation direct-acting inhibitor of hepatitis C virus (HCV) non-structural protein 5A (NS5A; Kds = 8 and 210 nM for the NS5A33-202 and NS5A26-202 residues of HCV genotype 1b, respectively).
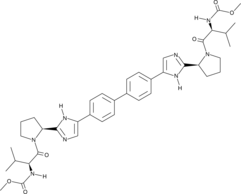
-
GC12057
Daclatasvir (BMS-790052)
HCV NS5A inhibitor
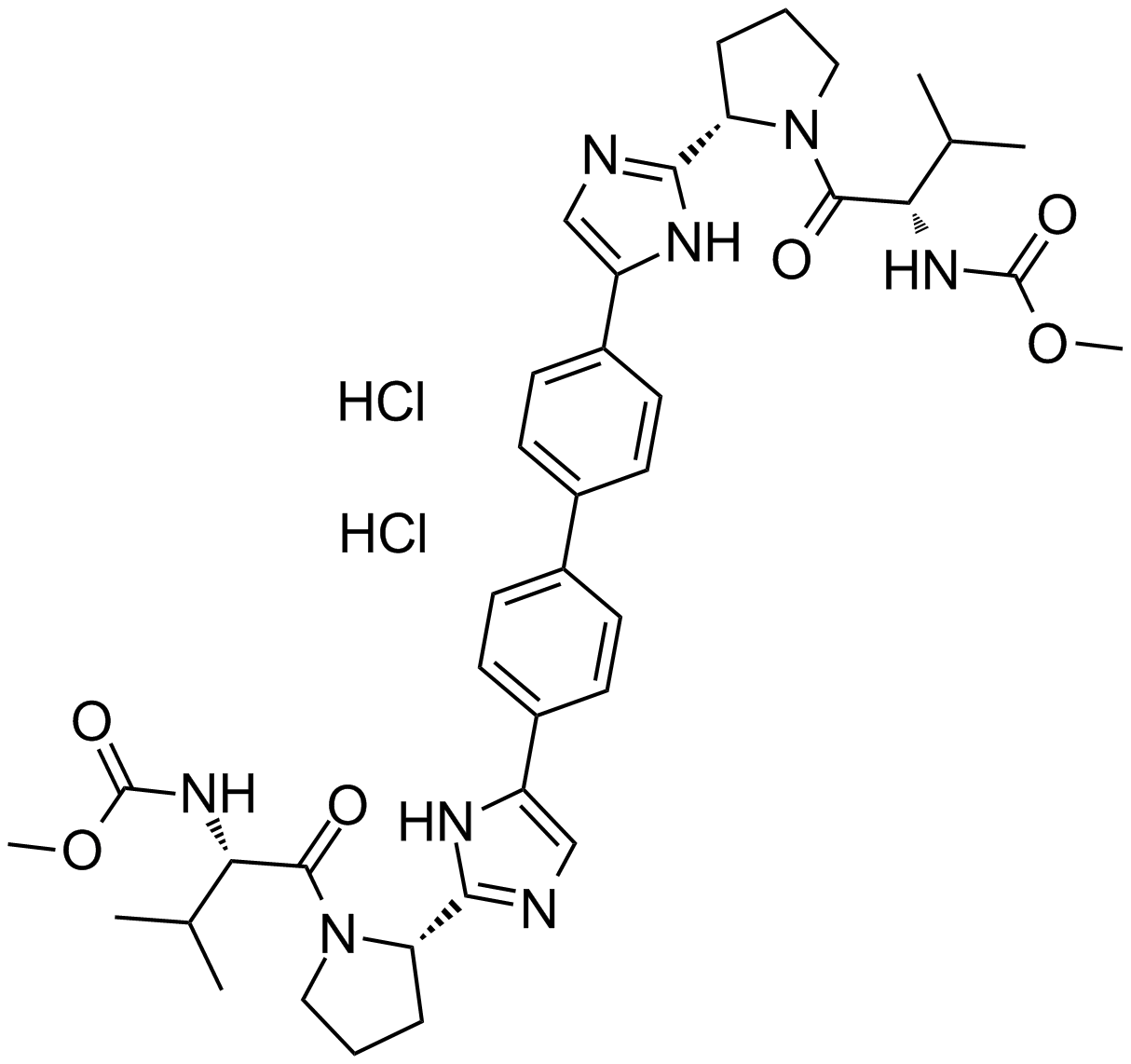
-
GC12236
Daclatasvir dihydrochloride
An NS5A inhibitor
-
GC12879
Danoprevir (RG7227)
An HCV NS3/4A protease inhibitor
-
GC19015
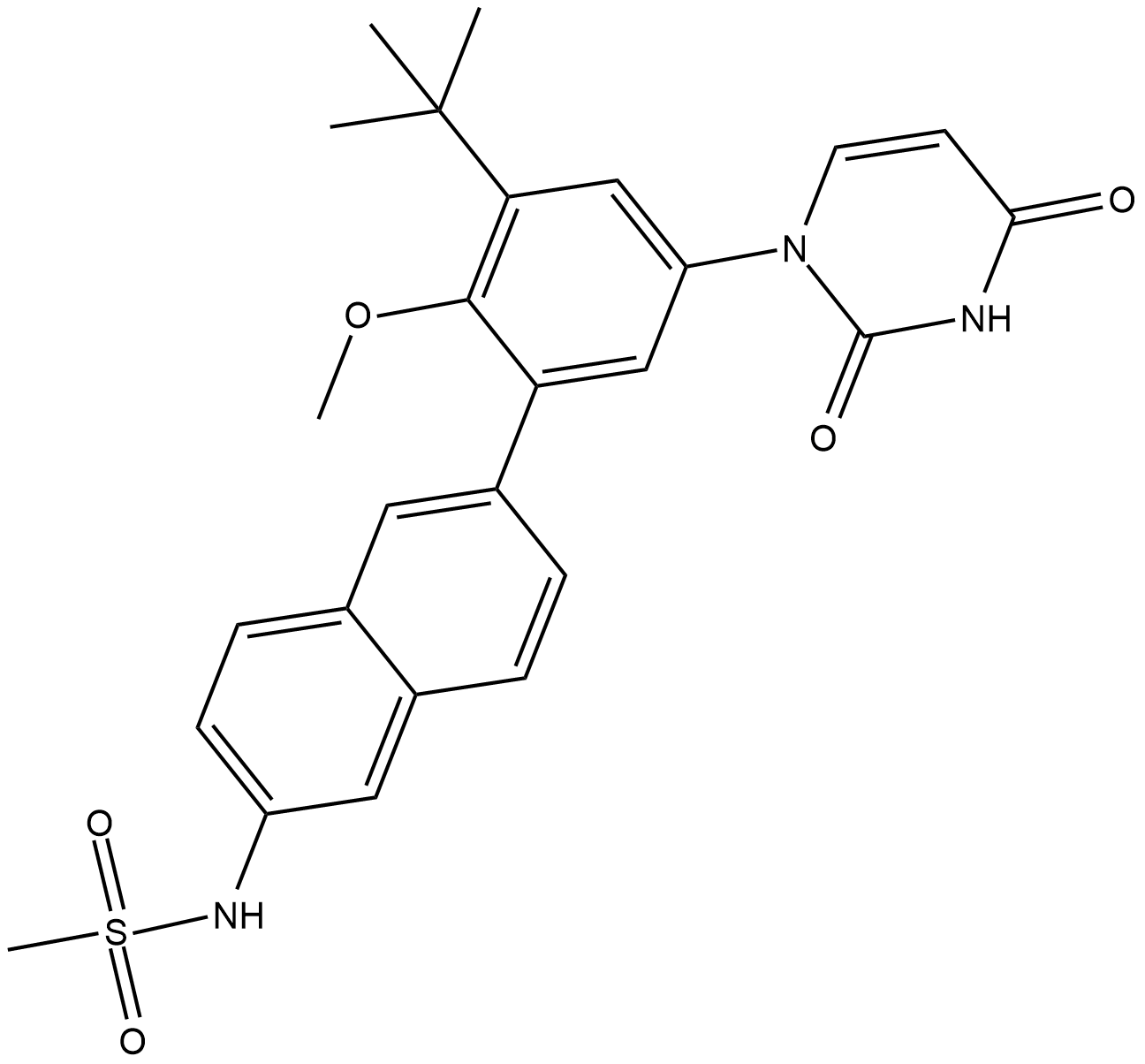
Dasabuvir
Dasabuvir is a nonnucleoside inhibitor of the RNA-dependent RNA polymerase encoded by the HCV NS5B gene, inhibits recombinant NS5B polymerases derived from HCV genotype 1a and 1b clinical isolates, with IC50 between 2.2 and 10.7 nM.
-
GC35823
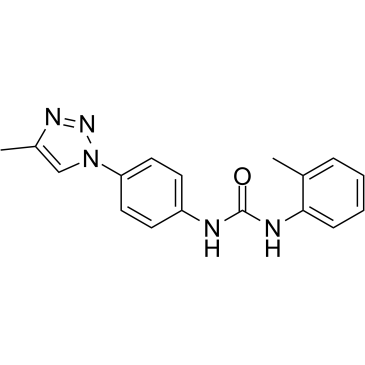
DDX3-IN-1
DDX3-IN-1 (Compound 16f) is a DEAD-box polypeptide 3 (DDX3) inhibitor with CC50s of 50 and 36 μM for HIV and HCV, respectively.
-
GN10458
Deapio platycodin D
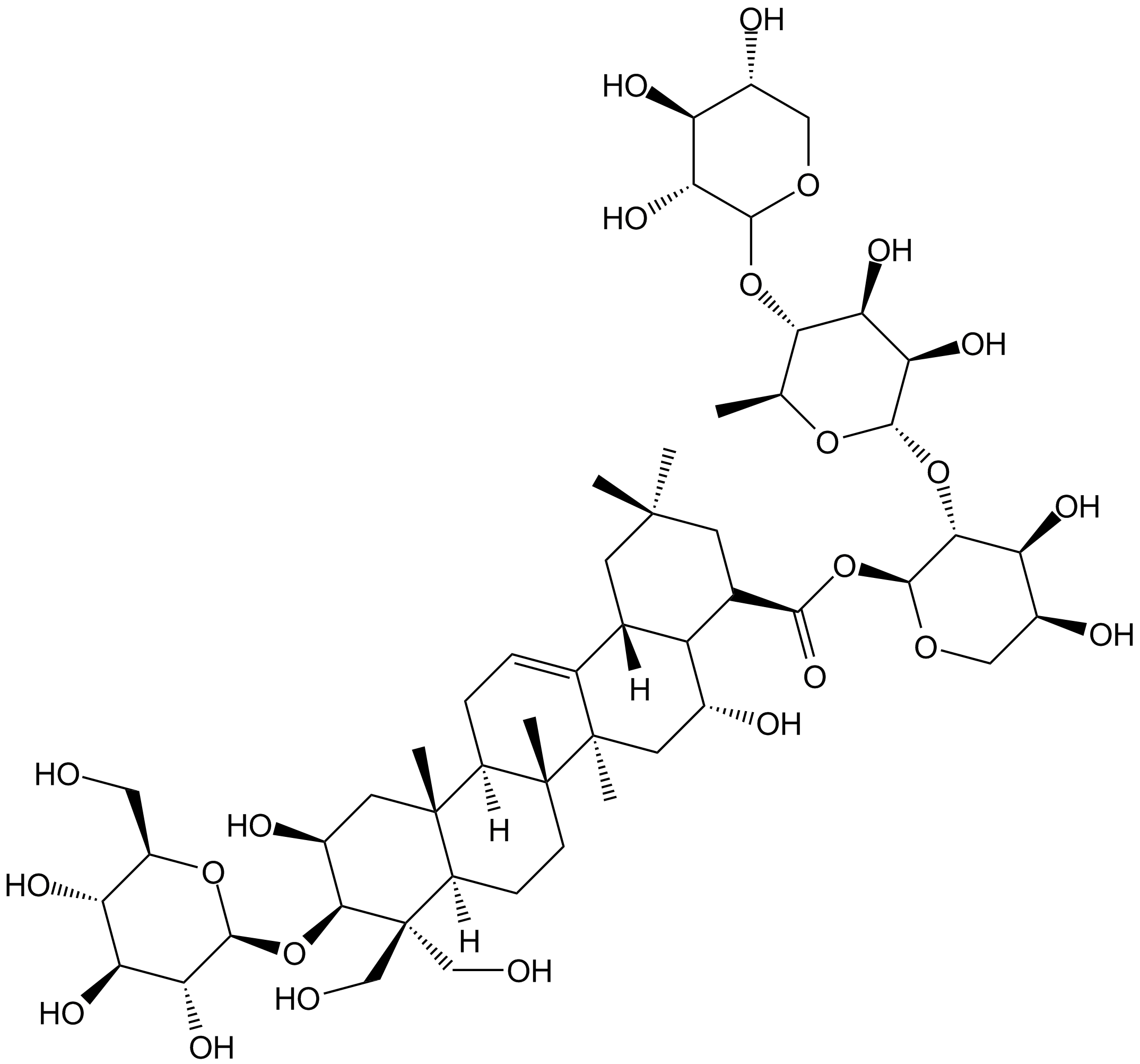
-
GC18101
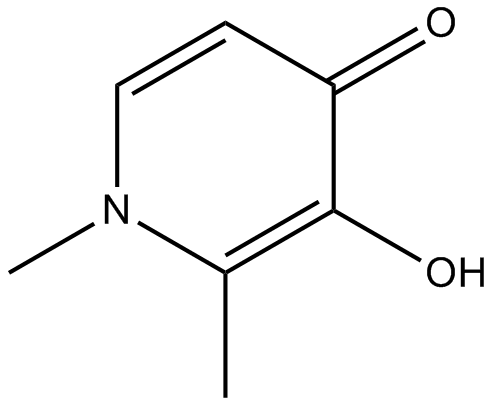
Deferiprone
Chelating agent
-
GC17567
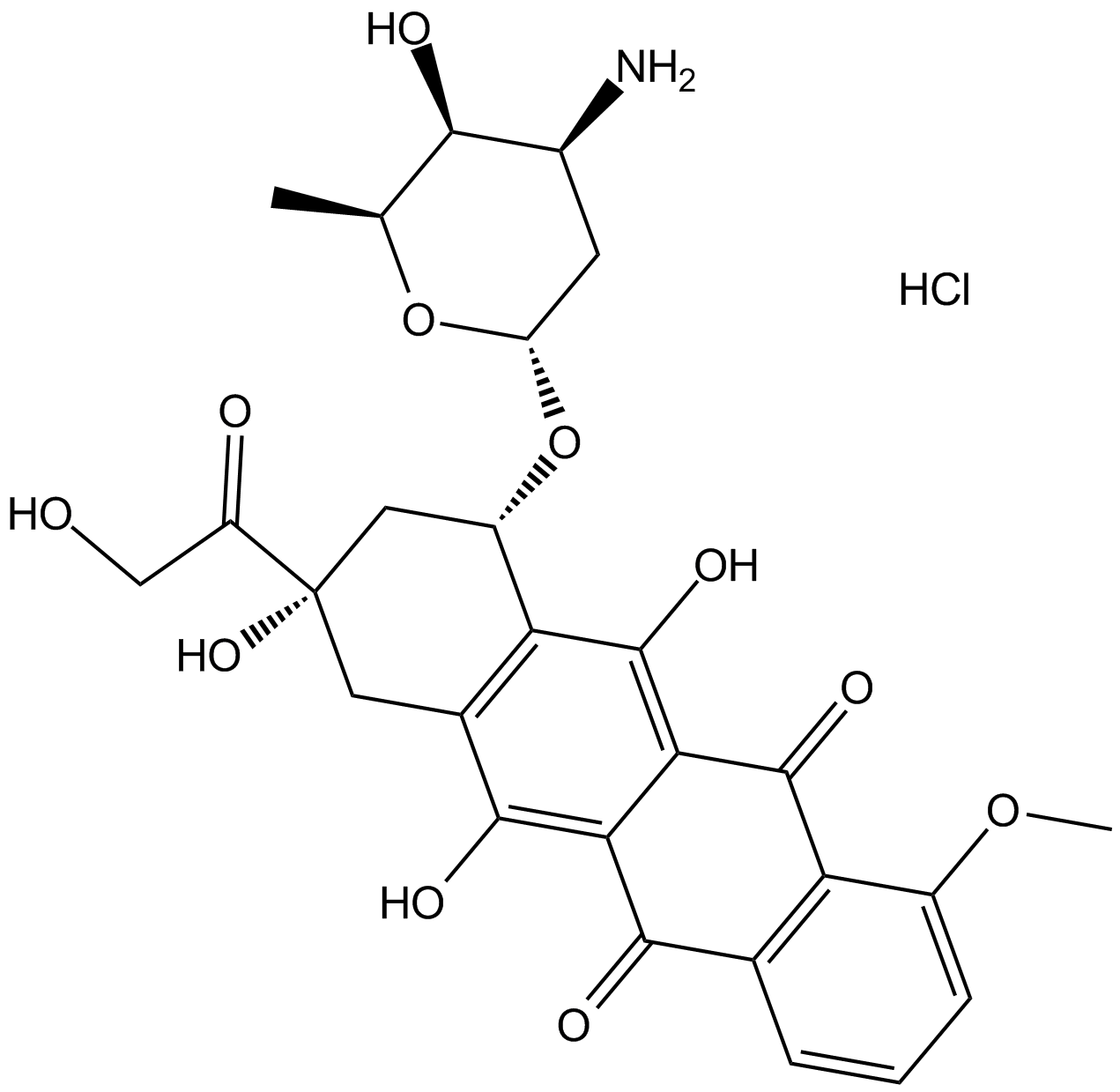
Doxorubicin (Adriamycin) HCl
Doxorubicin (Hydroxydaunorubicin) hydrochloride, a cytotoxic anthracycline antibiotic, is an anti-cancer chemotherapy agent.
-
GC18620
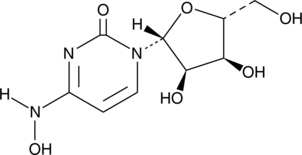
EIDD-1931
A ribonucleoside analog with antiviral activity
-
GC32062
Elbasvir (MK-8742)
Elbasvir (MK-8742) (MK-8742) is a hepatitis C virus nonstructural protein 5A (HCV NS5A) inhibitor with EC50s of 4, 3 and 3 nM against genotype 1a, 1b, and 2a, respectively.
-
GC60840
FGI-106 tetrahydrochloride
FGI-106 tetrahydrochloride is a potent and broad-spectrum inhibitor with inhibitory activity against multiple viruses.
-
GC36045
Filibuvir
Filibuvir is an orally active, selective non-nucleoside inhibitor of the HCV nonstructural 5B protein (NS5B) RNA-dependent RNA polymerase (RdRp).
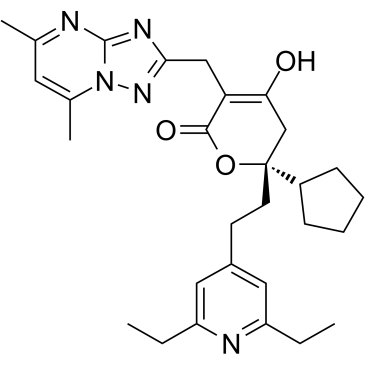
-
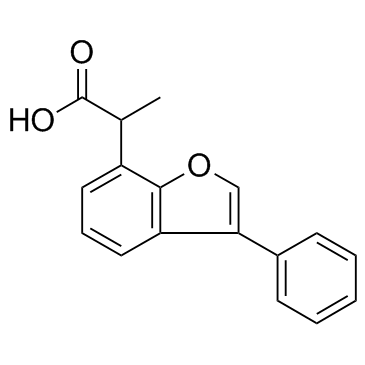
GC32341
Furaprofen (R803)
Furaprofen (R803) (R803) is an effective HCV replication inhibitor.
-
GN10720
Gentiopicrin
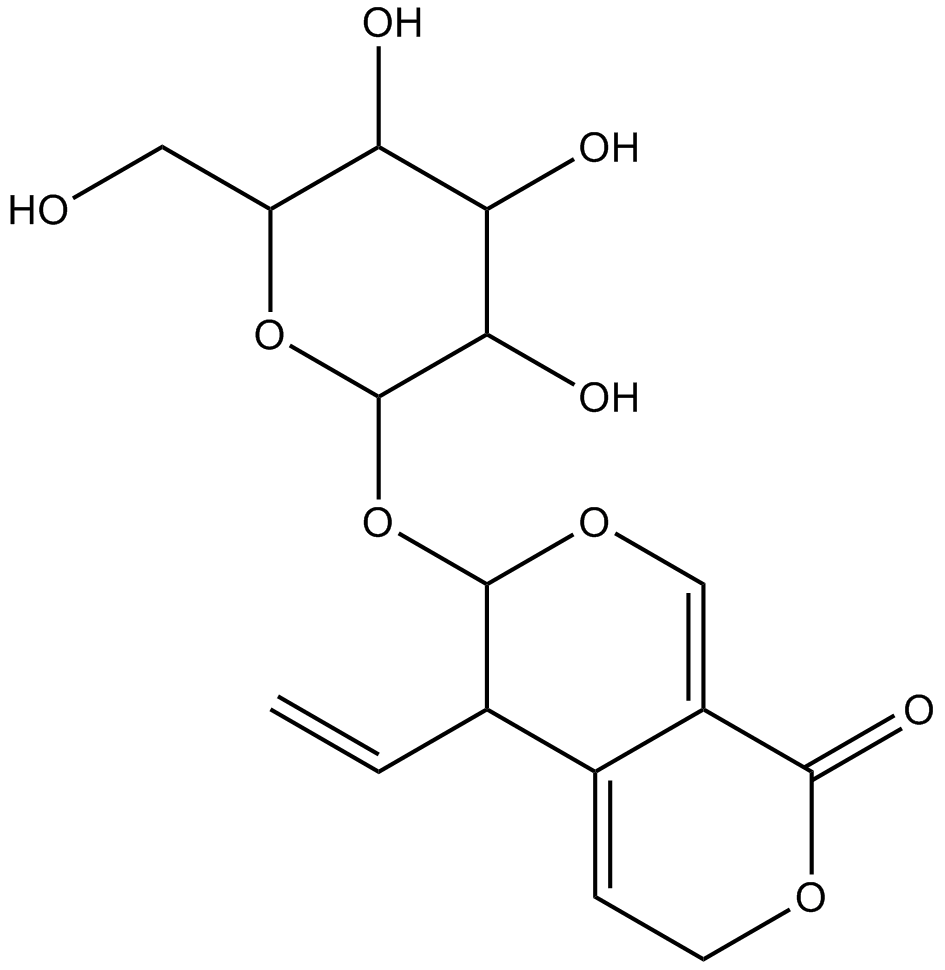
-
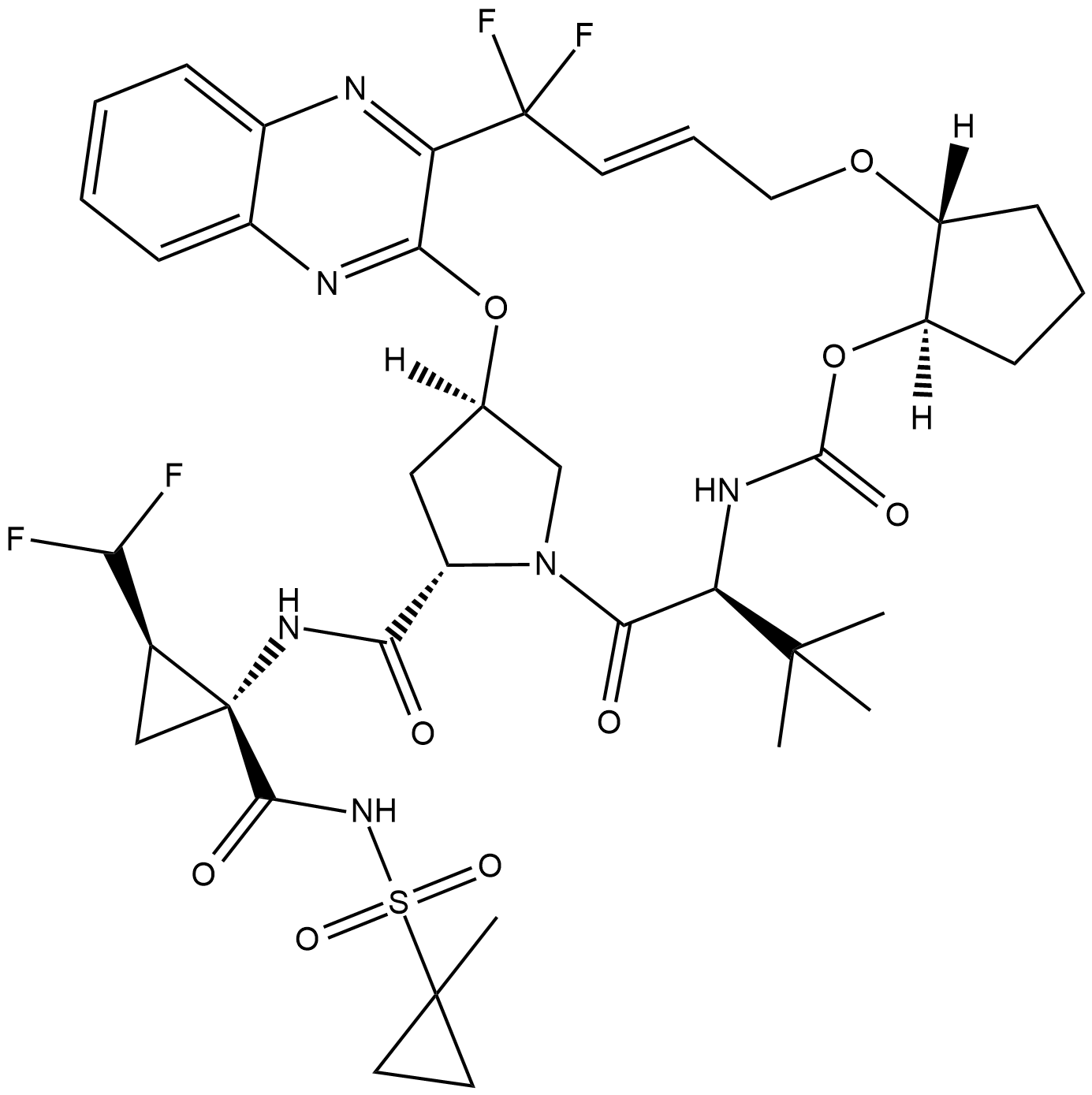
GC19165
Glecaprevir
Glecaprevir is a novel HCV NS3/4A protease inhibitor, with IC50 values ranging from 3.5 to 11.3 nM.
-
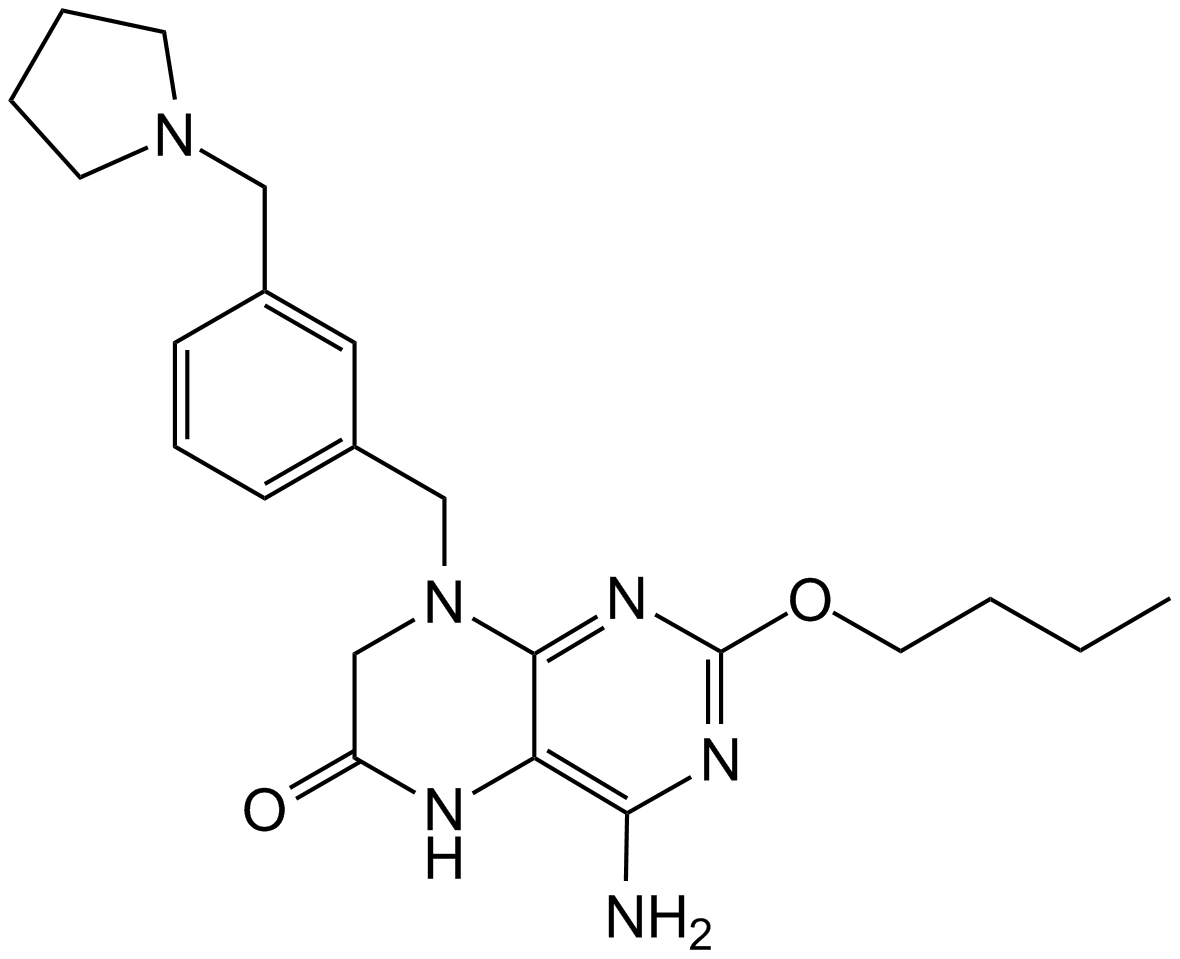
GC12555
GS-9620
A TLR7 agonist
-
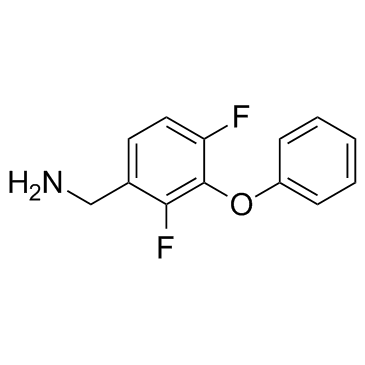
GC30967
HCV-IN-3
HCV-IN-3 is a hepatitis C virus (HCV) NS3/4a protein inhibitor, with an IC50 of 20 μM, a Kd of 29 μM.
-
GC68217
HCV-IN-30
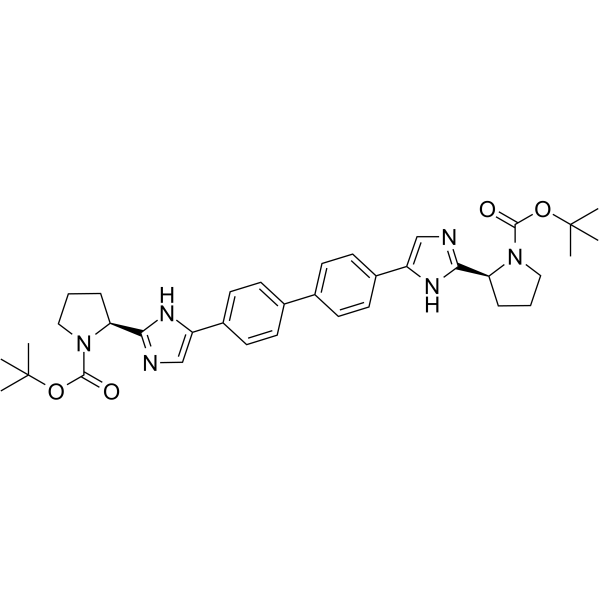
-
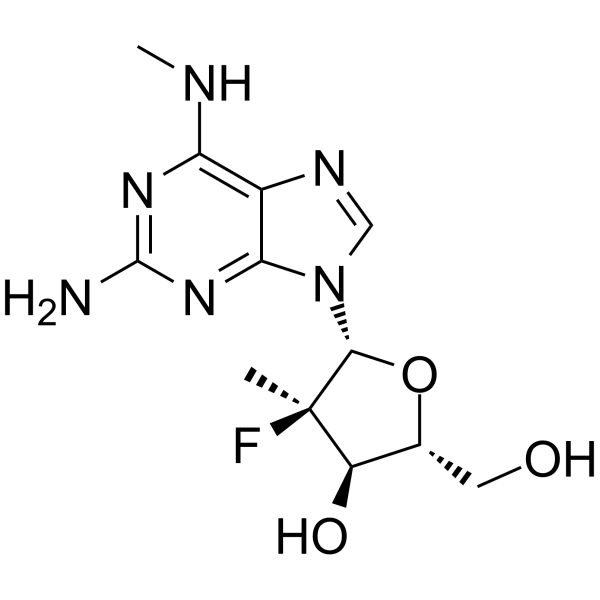
GC62138
HCV-IN-31
HCV-IN-31 (compound 4) is a HCV inhibitor, with an EC50/EC95 of 15.7 μM for HCV replicon.
-
GC36213
HCV-IN-4
HCV-IN-4 is a potent and orally active HCV NS5A inhibitor, shows great potency against GT1a, GT2b, GT3a, GT1a Y93H and GT1a L31V, with EC90s of 3 pM, 0.3 nM, 0.01 nM, 0.5 nM and 0.02 nM, respectively.
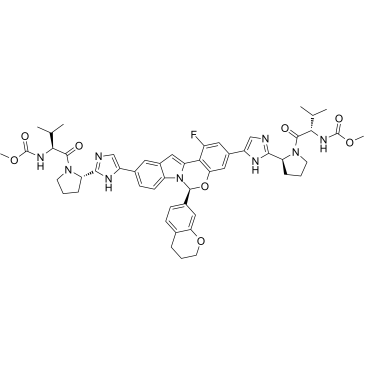
-
GP10026
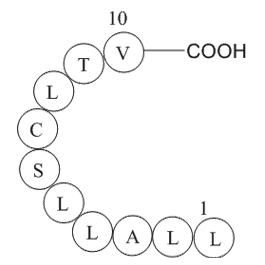
Hepatitis C virus
Hepatitis C virus, (C48H88N10O13S), a peptide with the sequence H2N-Leu-Leu-Ala-Leu-Leu-Ser-Cys-Leu-Thr-Val-OH, MW= 1045.34.
-
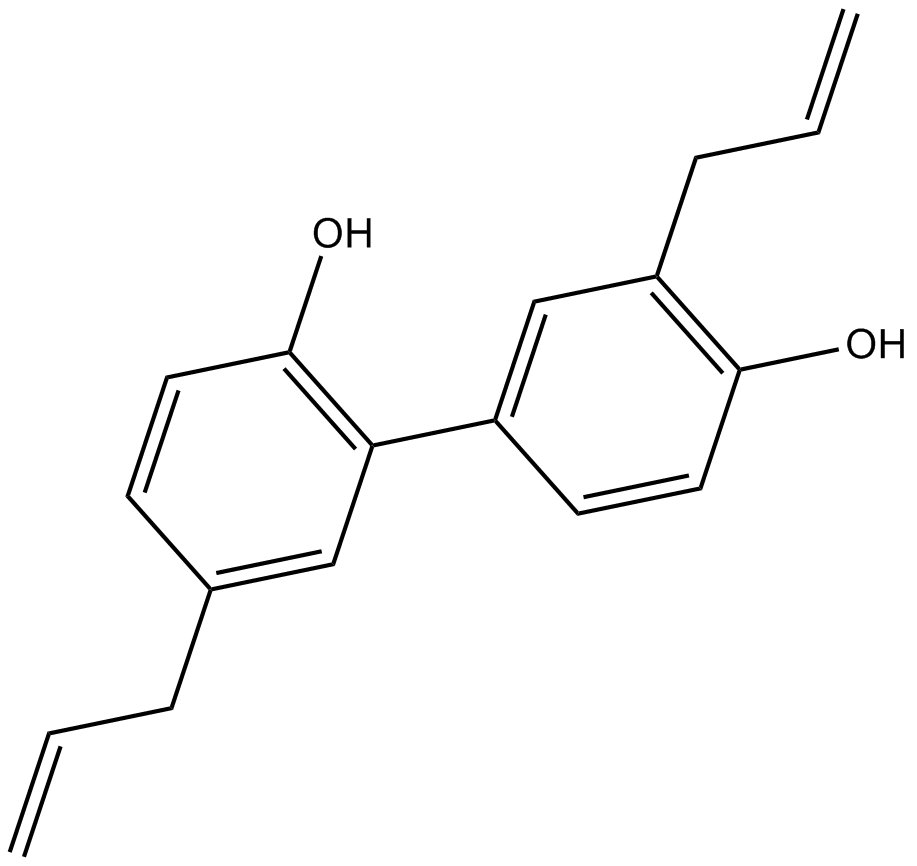
GN10664
Honokiol
-
GC30794
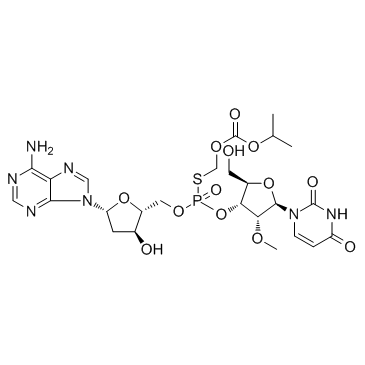
Inarigivir soproxil (SB9200)
Inarigivir soproxil (SB9200) (SB9200) is an agonist of innate immunity and shows potent antiviral activity against resistant HCV variants, with EC50s of 2.2 and 1.0 μM for HCV 1a/1b in cells of genotype 1 HCV replicon systems.
-
GC32209
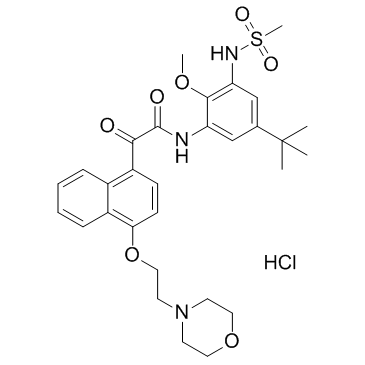
ITX5061
ITX5061 is a type II inhibitor of p38 MAPK and also an antagonist of scavenger receptor B1 (SR-B1).
-
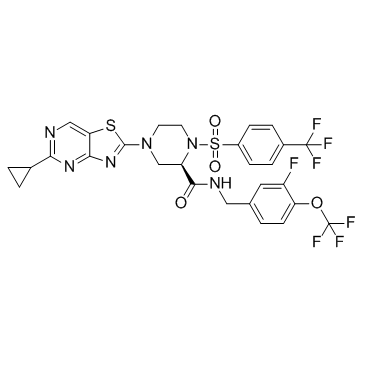
GC32384
JTK-853
JTK-853 is a novel, non-nucleoside Hepatitis C Virus (HCV) polymerase inhibitor which shows effective antiviral activity in HCV replicon cells with EC50s of 0.38 and 0.035 ?M in genotype 1a H77 and 1b Con1 strains, respectively.
-
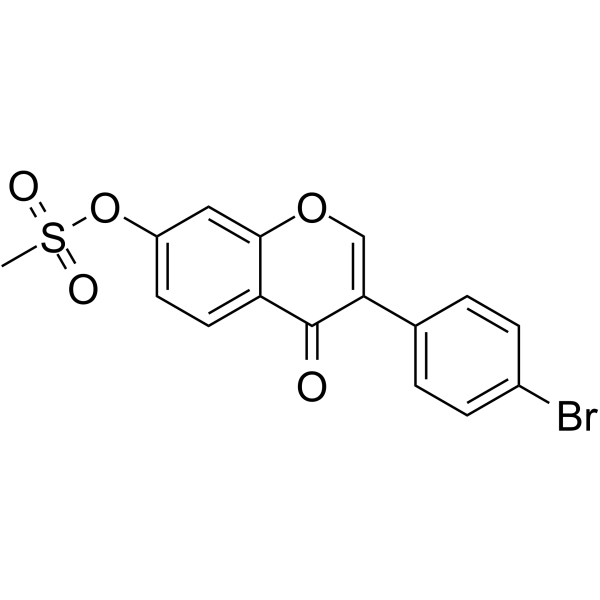
GC64253
KIN101
KIN101 is a potent RNA viral inhibitor with IC50s of 2 μM, >5 μM for influenza virus and Dengue virus (DNV), respectively.
-
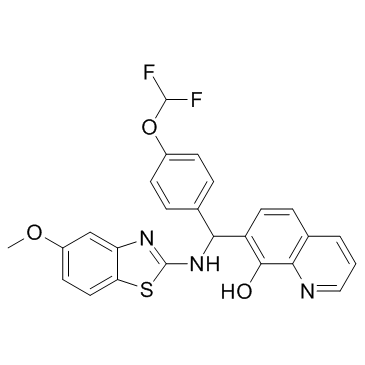
GC32122
KIN1408
An activator of the RLR pathway
-
GC17439
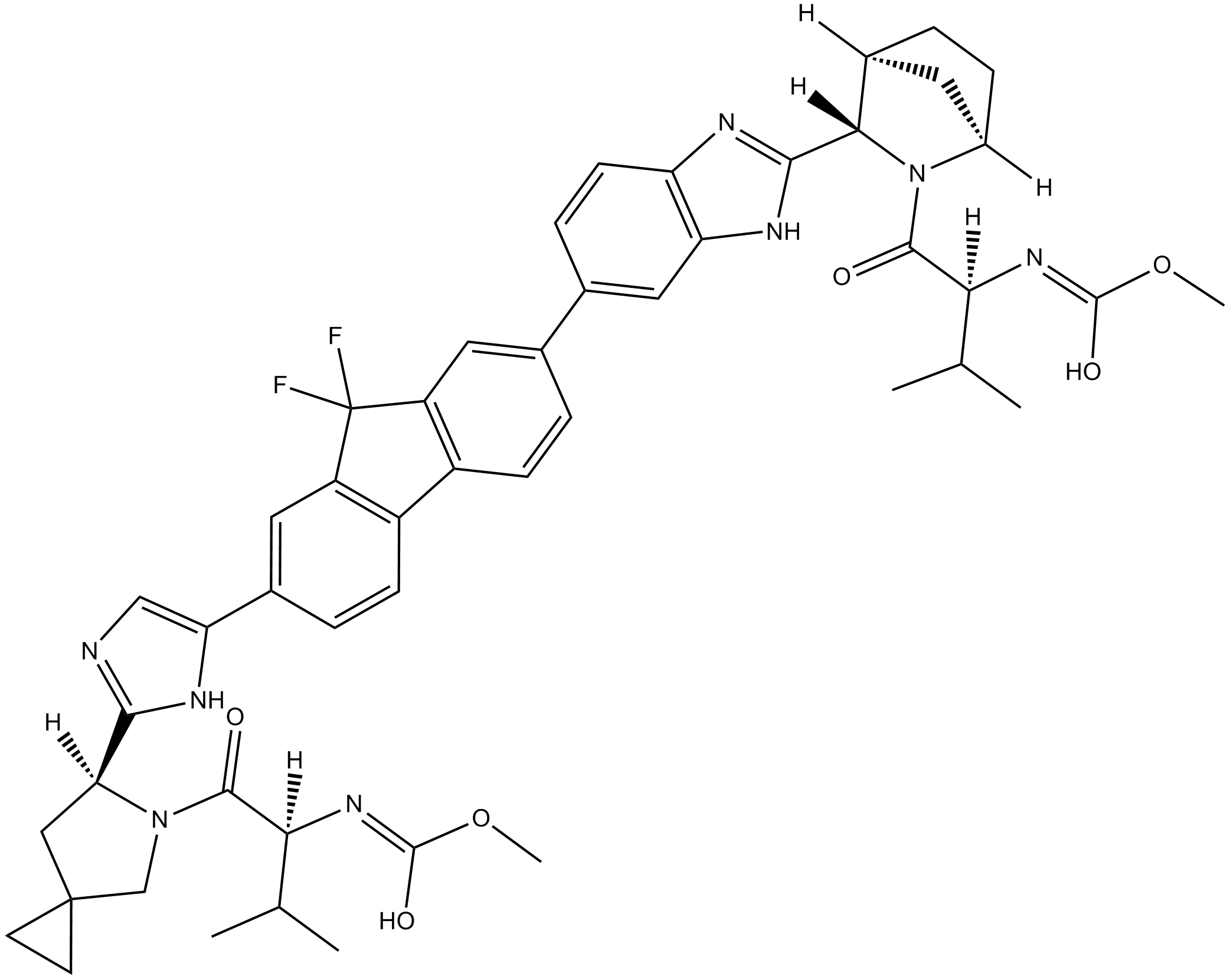
Ledipasvir
An HCV NS5A inhibitor
-
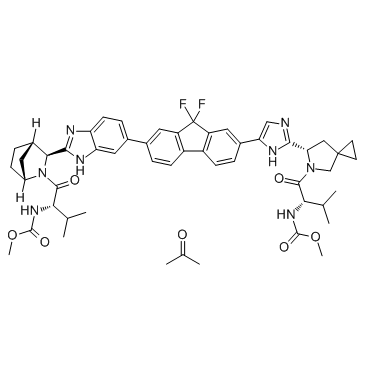
GC36435
Ledipasvir acetone
Ledipasvir acetone (GS-5885 acetone) is the active ingredient of Ledipasvir.
-
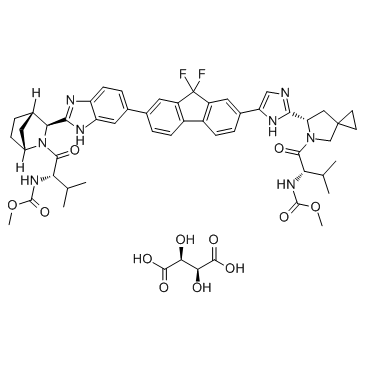
GC36437
Ledipasvir D-tartrate
Ledipasvir D-tartrate is an inhibitor of the hepatitis C virus NS5A, with EC50 values of 34 pM against GT1a and 4 pM against GT1b replicon.
-
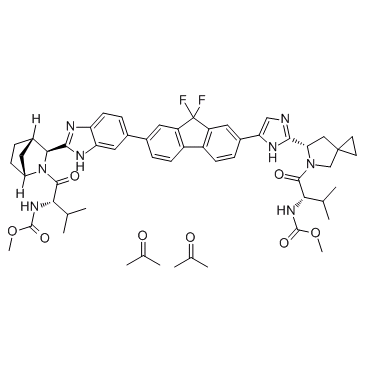
GC36436
Ledipasvir diacetone
Ledipasvir diacetone (GS-5885 diacetone) is the active ingredient of Ledipasvir.
-
GC11047
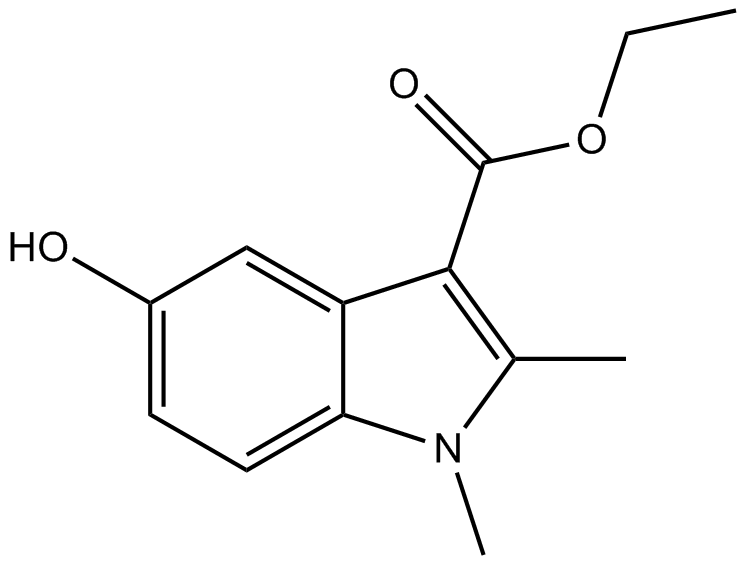
Mecarbinate
chemical intermediate of arbidol hydrochloride
-
GC13522
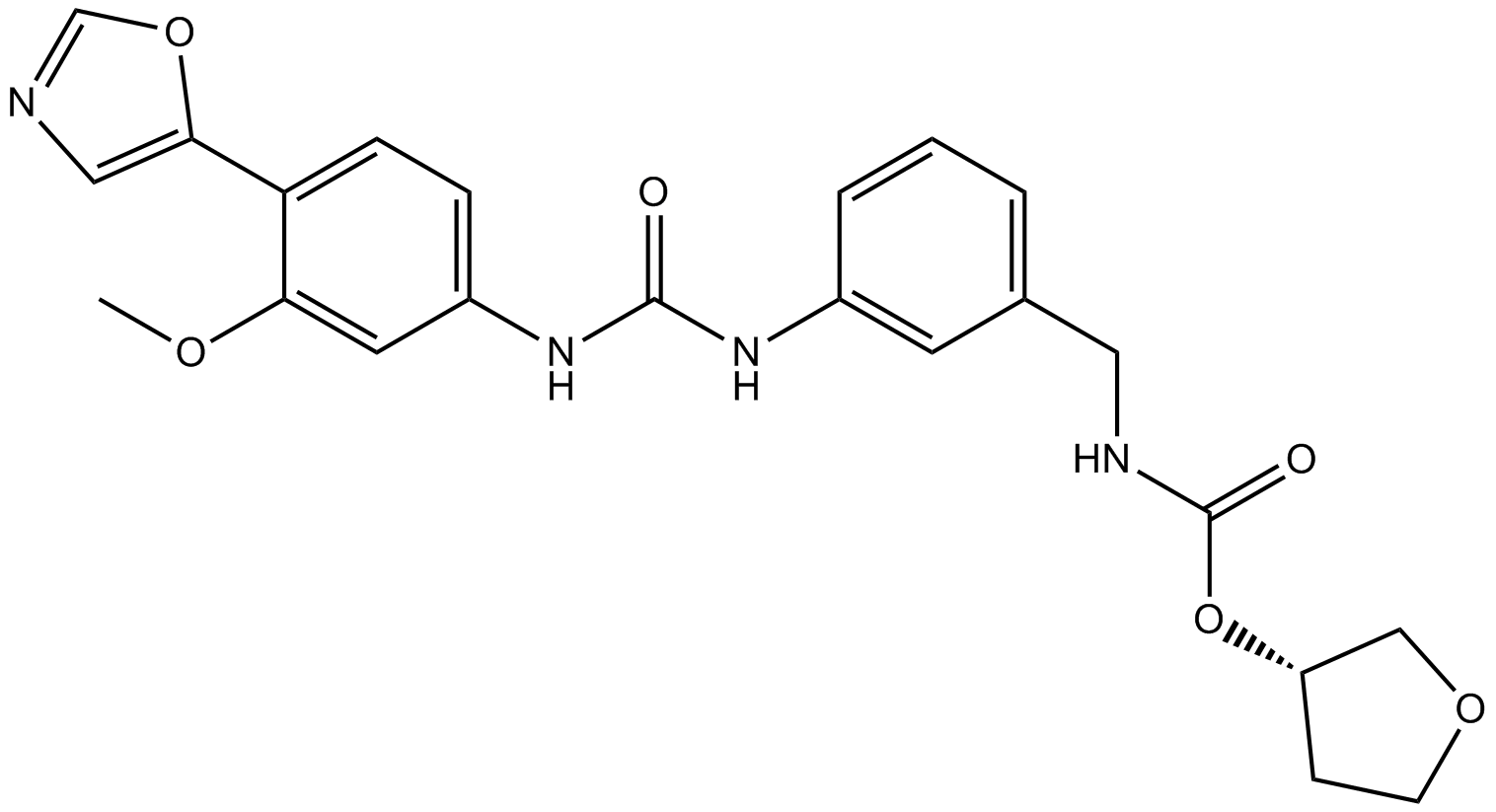
Merimepodib
Novel noncompetitive inhibitor of IMPDH(Inosine monophosphate dehydrogenase).
-
GC44192
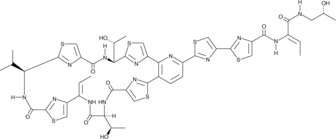
Micrococcin P1
Micrococcin P1 is a macrocyclic peptide antibiotic that functions as an acceptor-site-specific inhibitor of ribosomal protein synthesis, effectively preventing the growth of Gram-positive bacteria without affecting that of Gram-negative bacteria.
-
GC18004
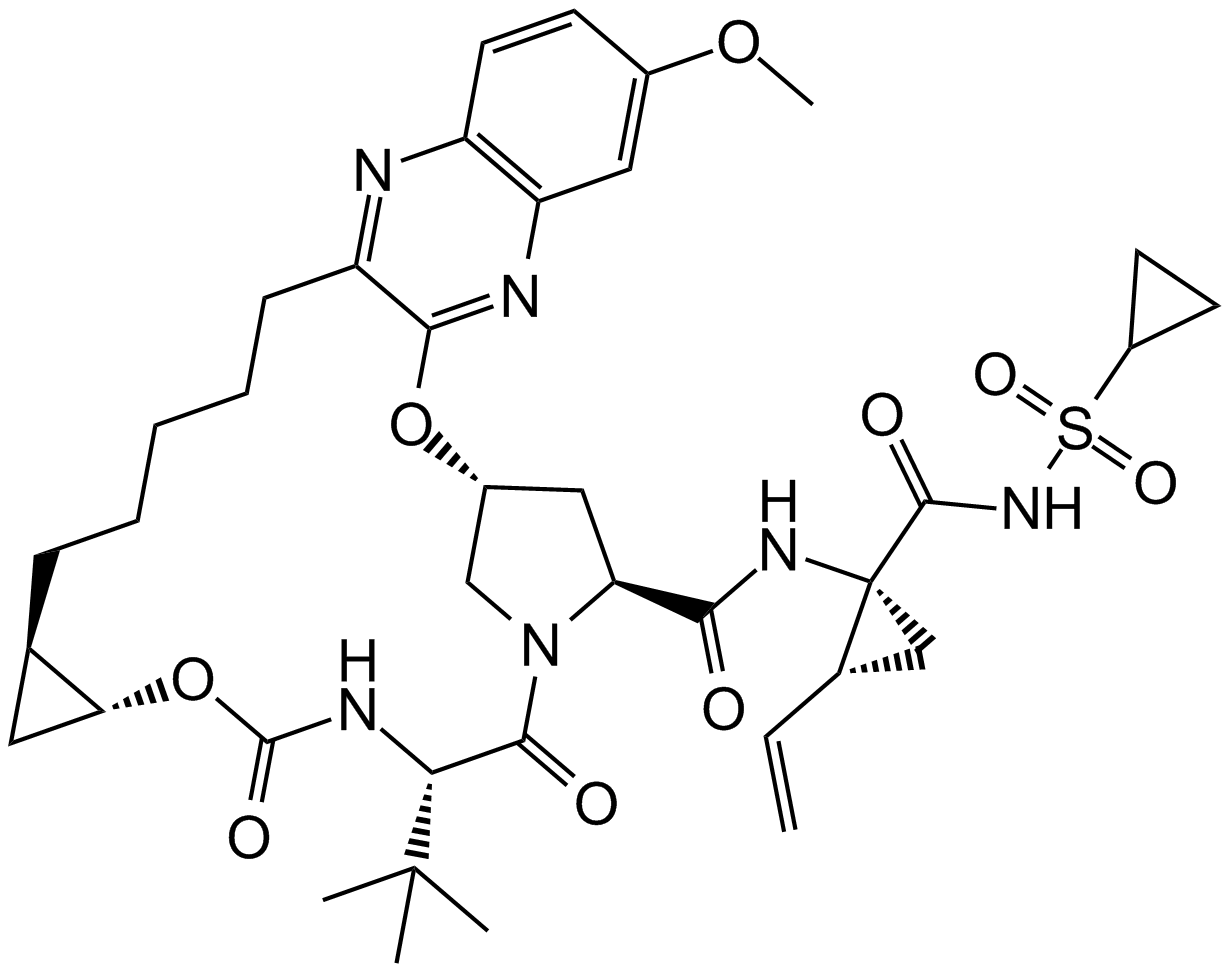
MK-5172
An NS3/4A protease inhibitor
-
GC16098
MK-5172 hydrate
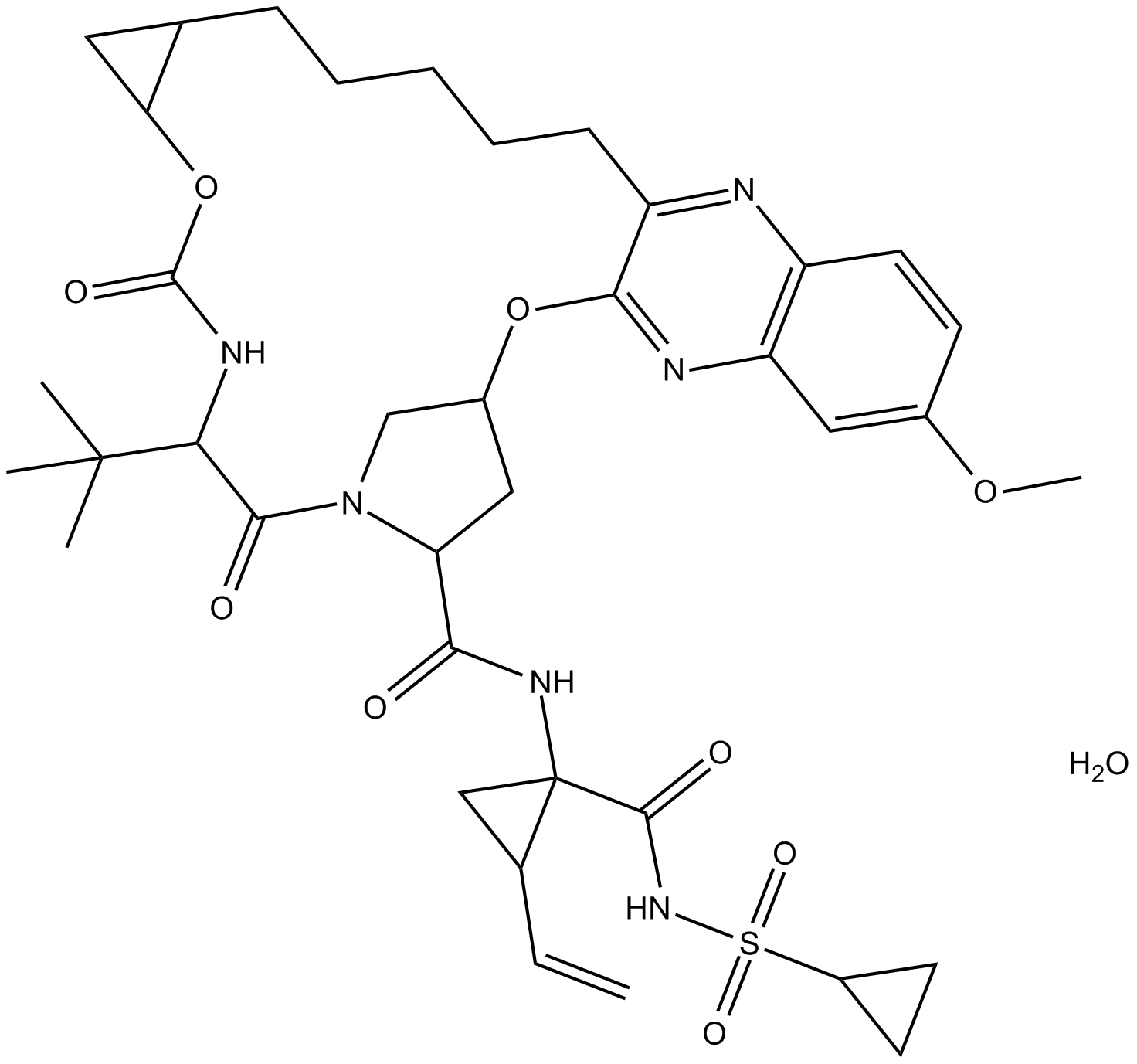
-
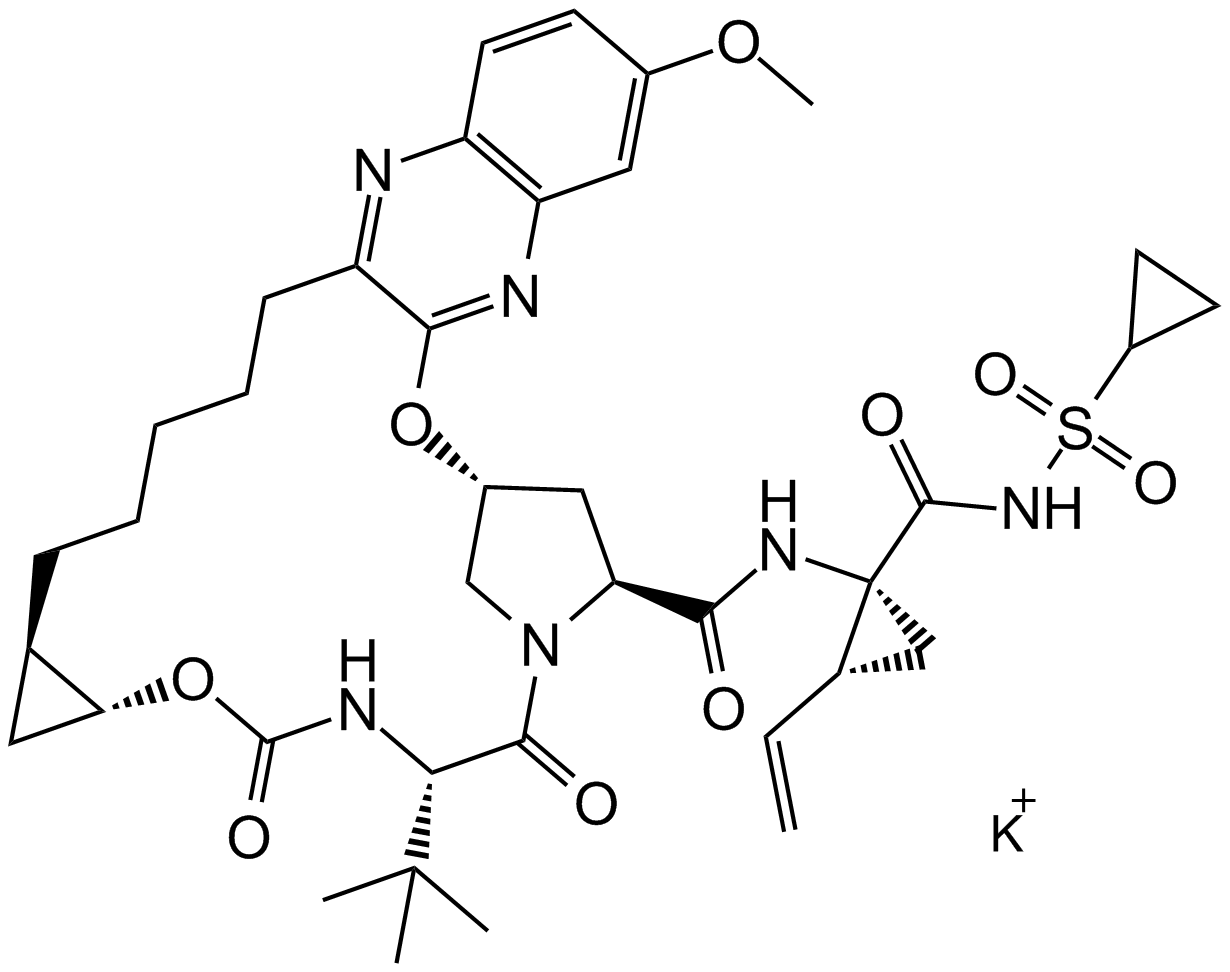
GC17114
MK-5172 potassium salt
-
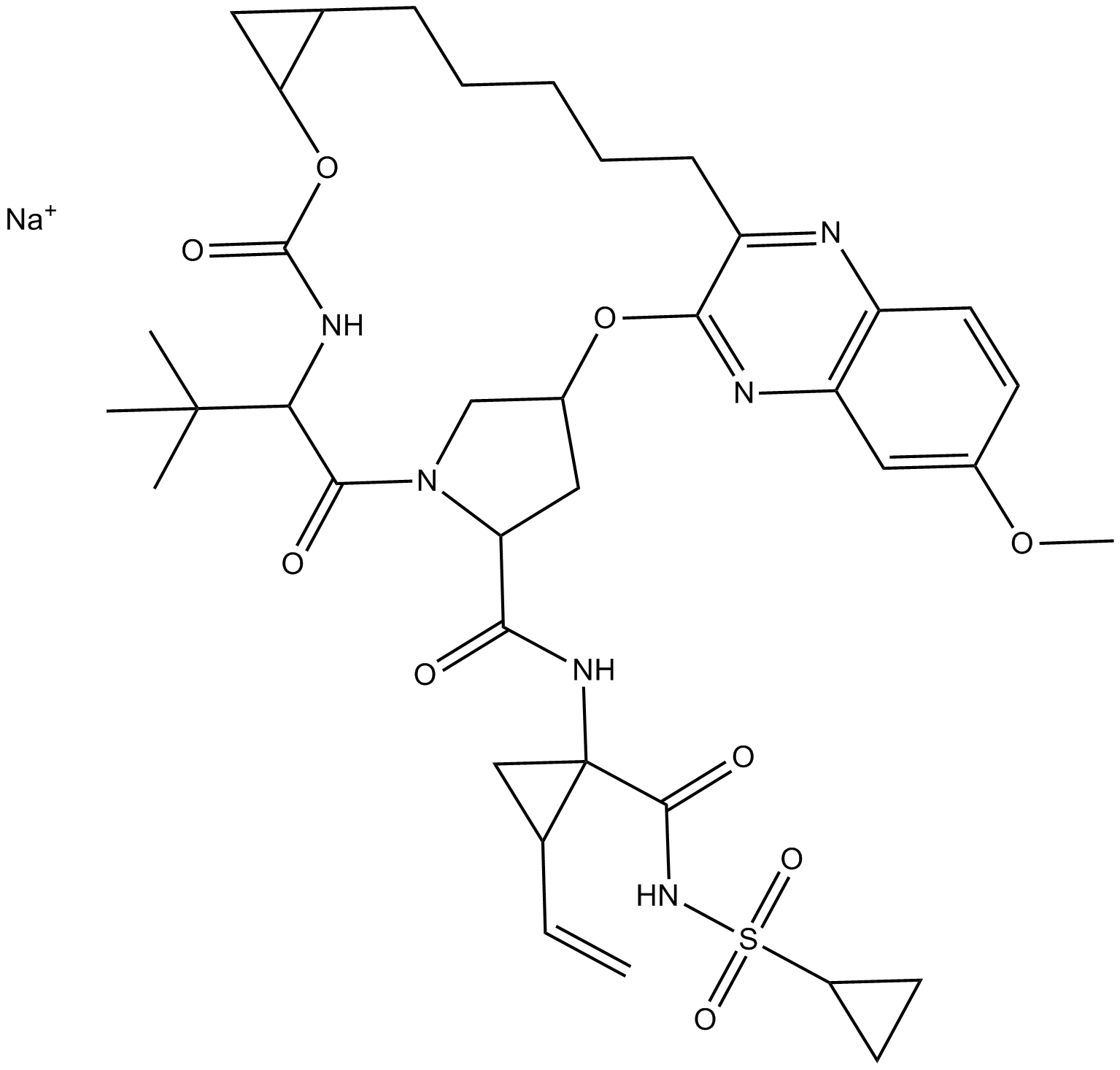
GC11463
MK-5172 sodium salt
-
GC61620
Monodes(N-carboxymethyl)valine Daclatasvir
Monodes(N-carboxymethyl)valine Daclatasvir (Daclatasvir Impurity A) is the main degradation product of Daclatasvir.
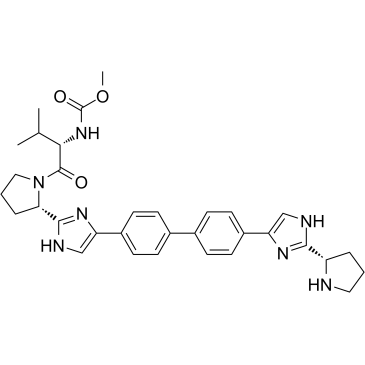
-
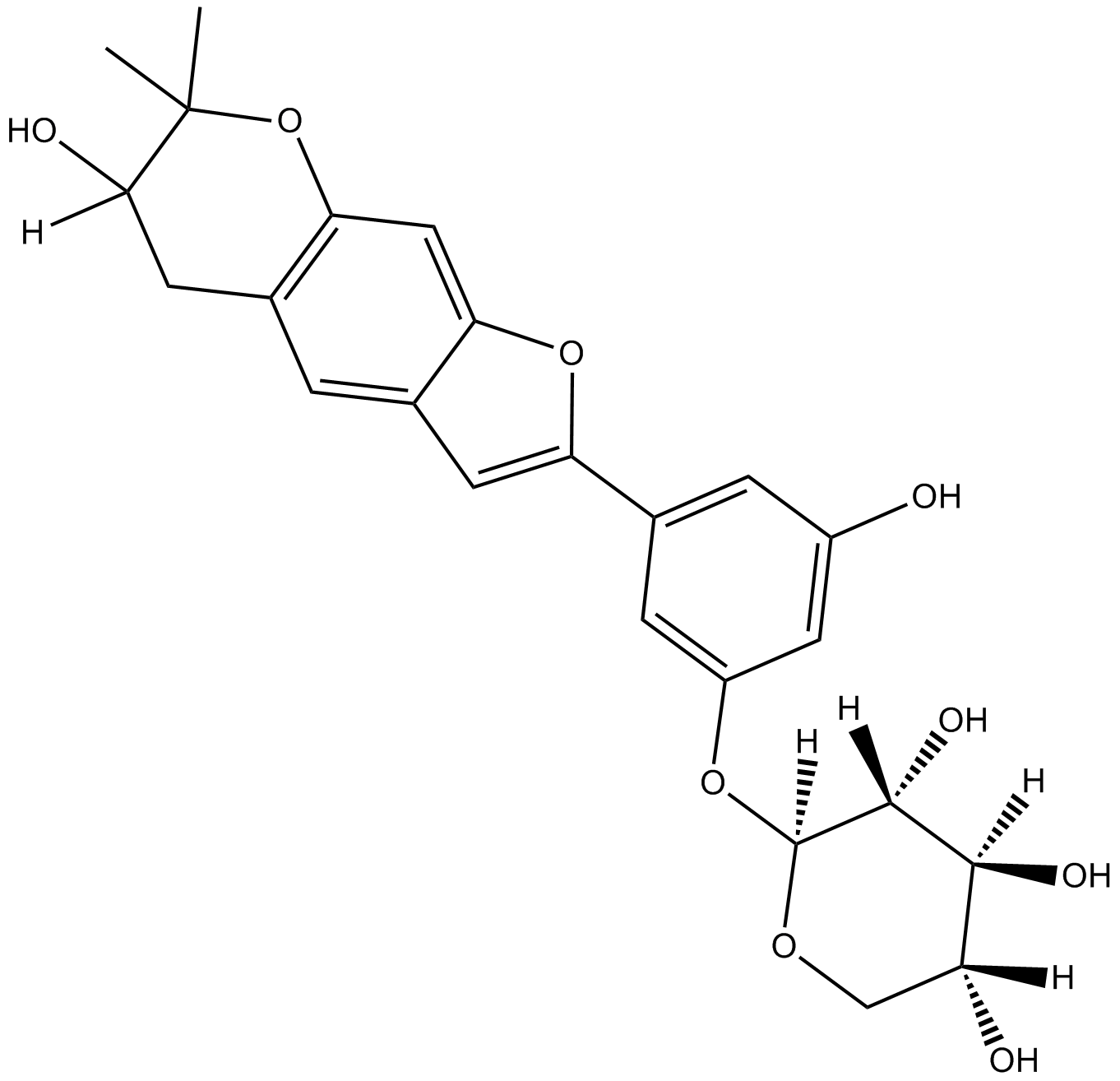
GN10230
Mulberroside C
-
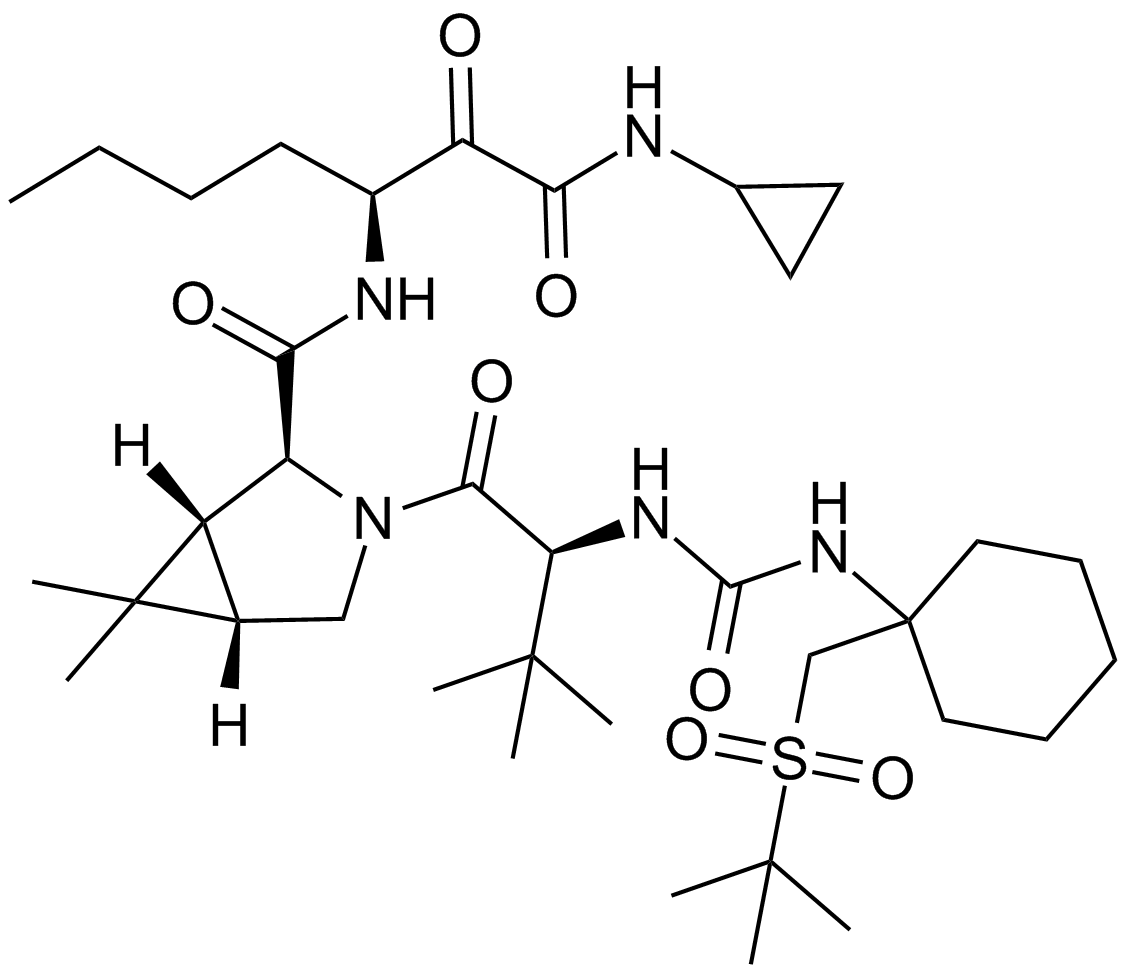
GC14074
Narlaprevir
-
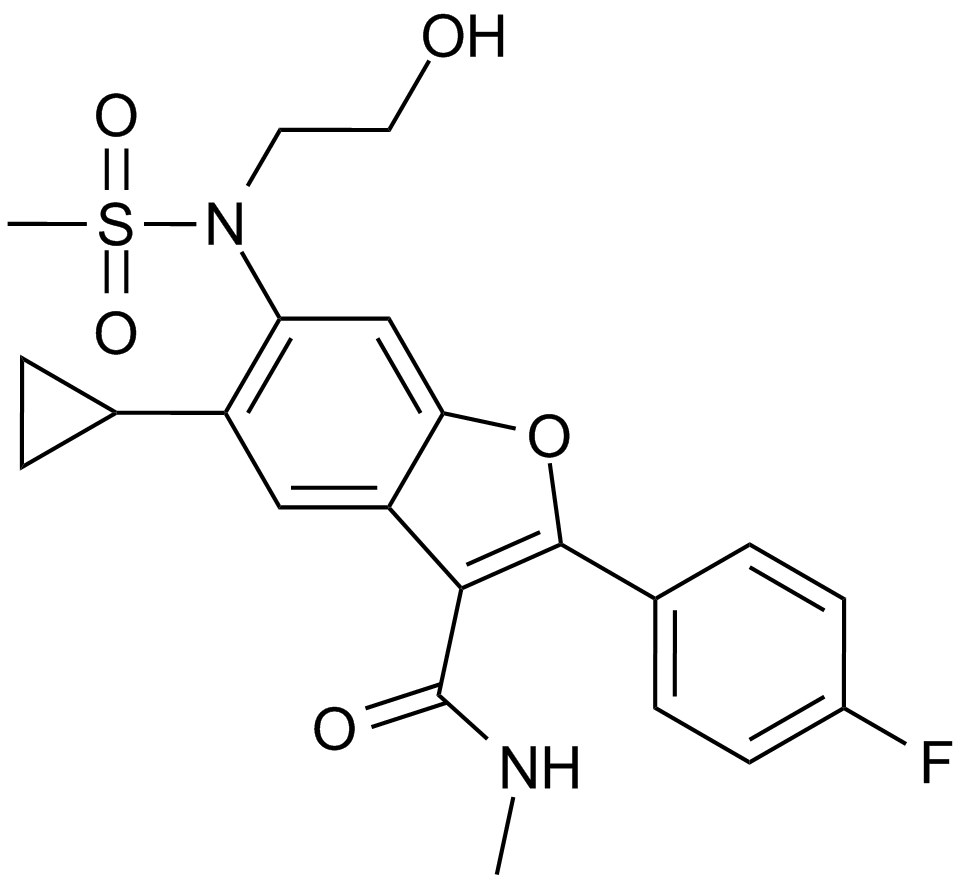
GC15891
Nesbuvir
-
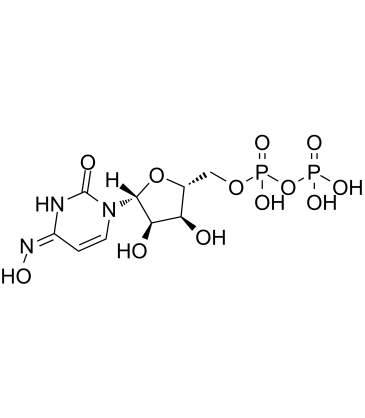
GC61956
NHC-diphosphate
NHC-diphosphate is an active phosphorylated?intracellular metabolite?of β-d-N4-Hydroxycytidine (NHC) as a diphosphate form.
-
GC61130
NHC-triphosphate
NHC-triphosphate is an active phosphorylated intracellular metabolite of β-d-N4-Hydroxycytidine (NHC) as a triphosphate form.
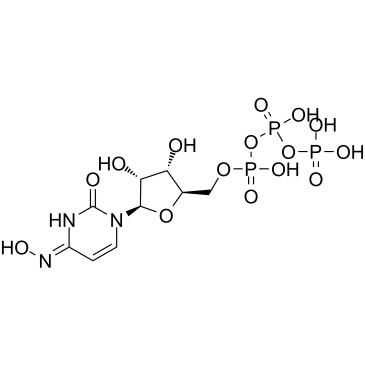
-
GC61961
NHC-triphosphate tetrasodium
NHC-triphosphate tetrasodium is an active phosphorylated intracellular metabolite of β-d-N4-Hydroxycytidine (NHC) as a triphosphate form.
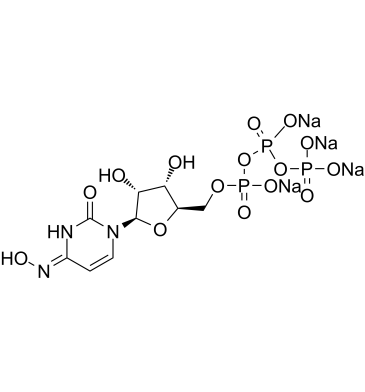
-
GC31646
NIM811 ((Melle-4)cyclosporin)
NIM811 ((Melle-4)cyclosporin) ((Melle-4)cyclosporin; SDZ NIM811 ((Melle-4)cyclosporin)) is an orally bioavailable mitochondrial permeability transition and cyclophilin dual inhibitor, which exhibits potent in vitro activity against hepatitis C virus (HCV) .
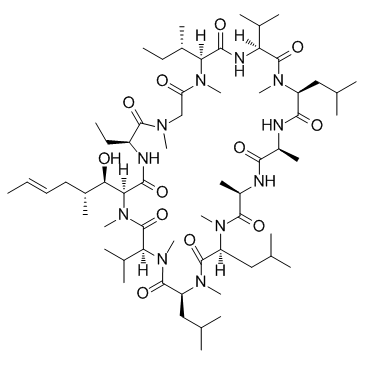
-
GC33936
NM107 (2'-C-Methylcytidine)
NM107 (2'-C-Methylcytidine) (2'-C-Methylcytidine) is an nucleoside inhibitor of the hepatitis C virus (HCV) NS5B polymerase, the EC50 of NM107 (2'-C-Methylcytidine) in the wild-type replicon cells is 1.85 μM.
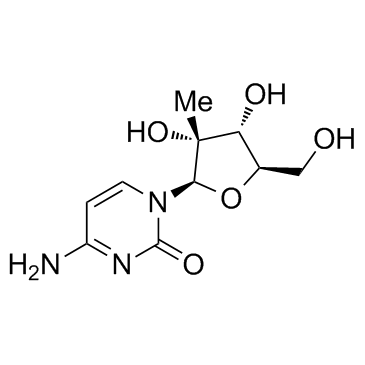
-
GC33956
Nucleoside-Analog-1
Nucleoside-Analog-1 is a 4′-Azidocytidine analogue against Hepatitis C virus replication.
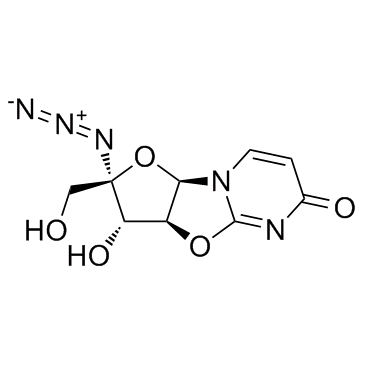
-
GC33959
Nucleoside-Analog-2
Nucleoside-Analog-2 is a 4'-Azidocytidine analogue against Hepatitis C virus (HCV) replication.
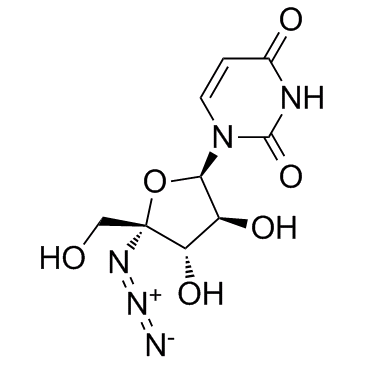
-
GC32502
Oglufanide (H-Glu-Trp-OH)
Oglufanide (H-Glu-Trp-OH) (H-Glu-Trp-OH) is a dipeptide immunomodulator isolated from calf thymus.
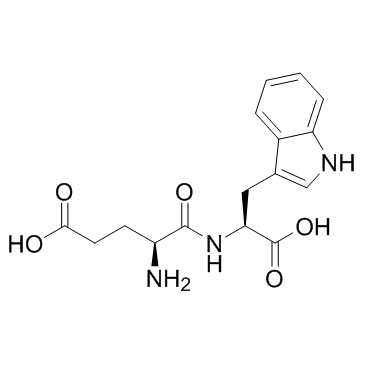
-
GC33914
Ombitasvir (ABT-267)
Ombitasvir (ABT-267) is a potent inhibitor of the hepatitis C virus protein NS5A, with EC50s of 0.82 to 19.3 pM against HCV genotypes 1 to 5 and 366 pM against genotype 6a.
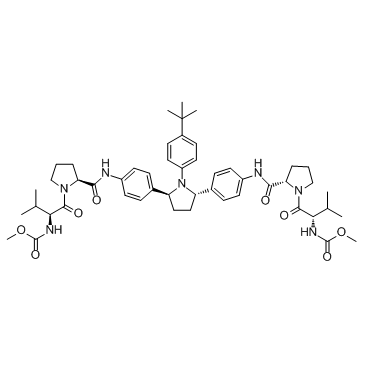
-
GC19275
Paritaprevir
Paritaprevir (ABT-450) is a potent non-structural protein 3/4A (NS3/4A) protease inhibitor with EC50s of 1 and 0.21 nM against HCV 1a and 1b, respectively.
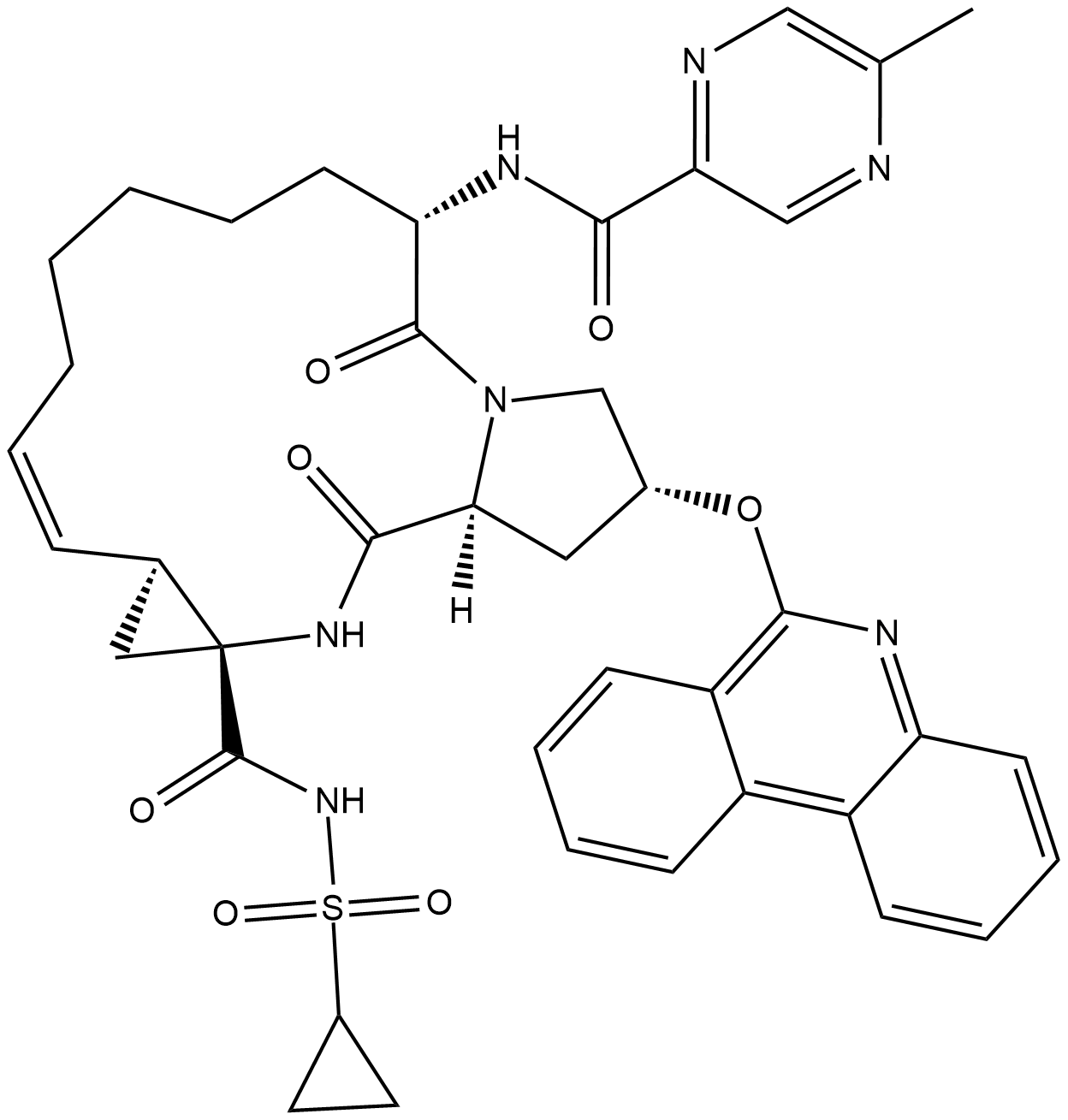
-
GC32107
Peretinoin (NIK333)
Peretinoin (NIK333) is an oral acyclic retinoid with a vitamin A-like structure that targets retinoid nuclear receptors such as retinoid X receptor (RXR) and retinoic acid receptor (RAR).
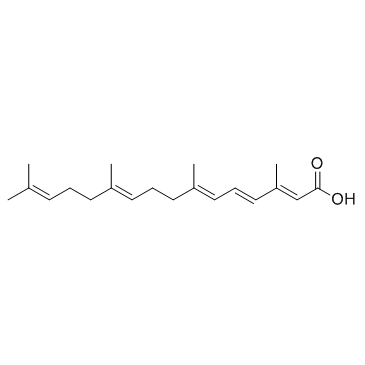
-
GC19454
Pibrentasvir
Pibrentasvir is a novel and pan-genotypic hepatitis C virus (HCV) NS5A inhibitor

-
GC36937
Platycodin D3
Platycodin D3 is a triterpenoid saponin isolated from Platycodon grandiflorum, with anti-HCV activity.
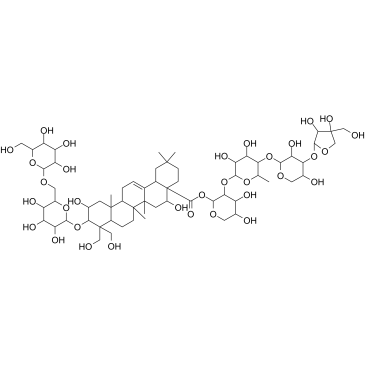
-
GC33971
PSI-352938 (PSI-938)
PSI-352938 (PSI-938) (PSI-938) is a hepatitis C virus (HCV) nucleotide inhibitor.
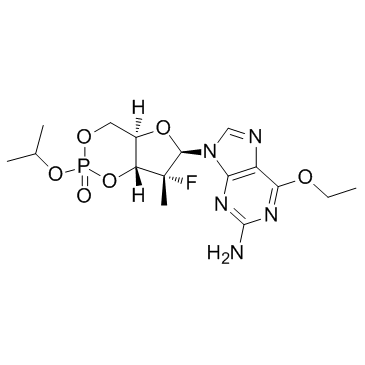
-
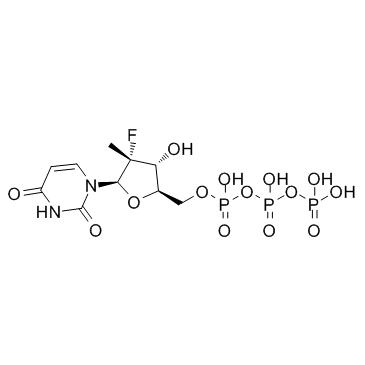
GC37028
PSI-6130
PSI-6130 is a potent and selective inhibitor of HCV NS5B polymerase, and inhibits HCV replication with a mean IC50 of 0.6 μM.
-
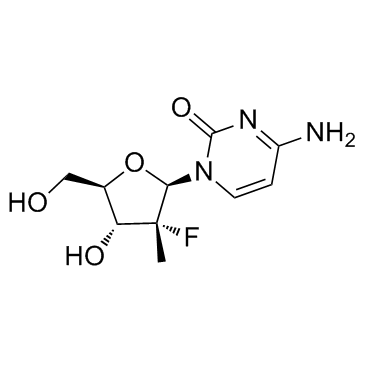
GC16901
PSI-6206
An inactive derivative of a HCV NS5B inhibitor
-
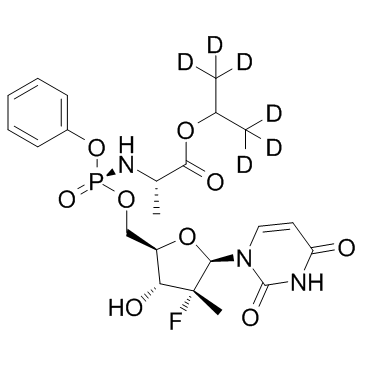
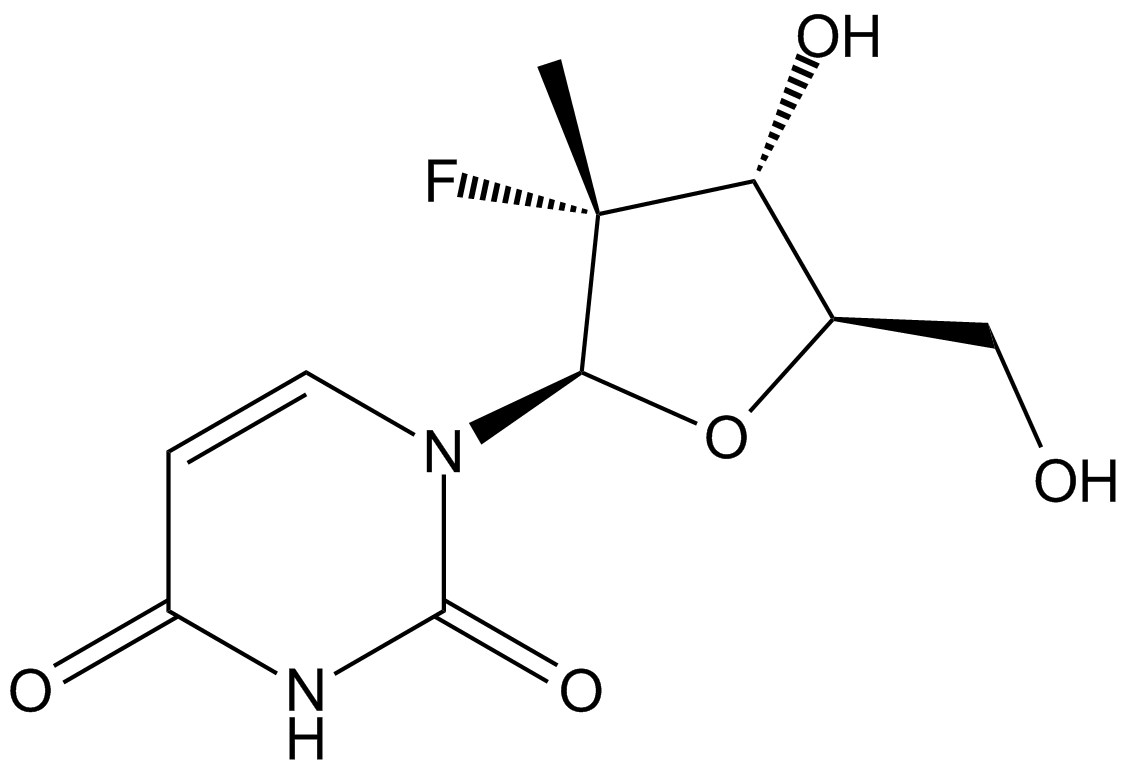
GC37029
PSI-6206 13CD3
PSI-6206 13CD3 is the deuterium labeled PSI-6206.
-
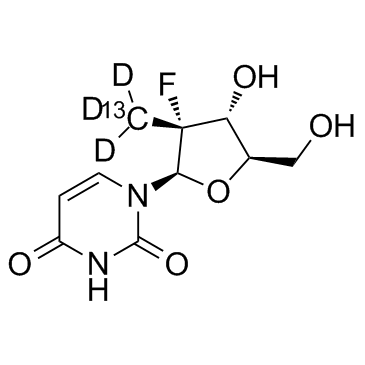
GC37031
PSI-7409
PSI-7409 is the active 5'-triphosphate metabolite of Sofosbuvir (PSI-7977). Sofosbuvir (PSI-7977) is a selective and highly active nucleotide analog inhibitor of HCV.
-
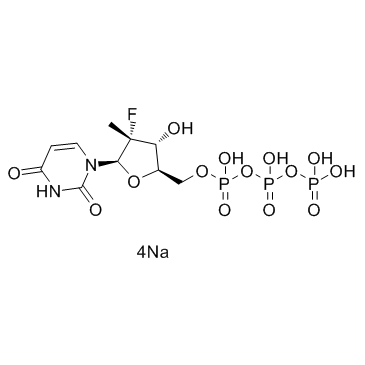
GC33946
PSI-7409 tetrasodium
PSI-7409 tetrasodium is an active 5'-triphosphate metabolite of sofosbuvir (PSI-7977), inhibiting HCV NS5B polymerases, with IC50s of 1.6, 2.8, 0.7 and 2.6 μM for GT 1b_Con1, GT 2a_JFH1, GT 3a, and GT 4a NS5B polymerases, respectively.
-
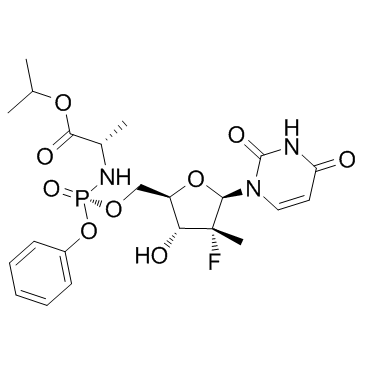
GC37032
PSI-7976
PSI-7976 is the isomer of PSI-7977.
-
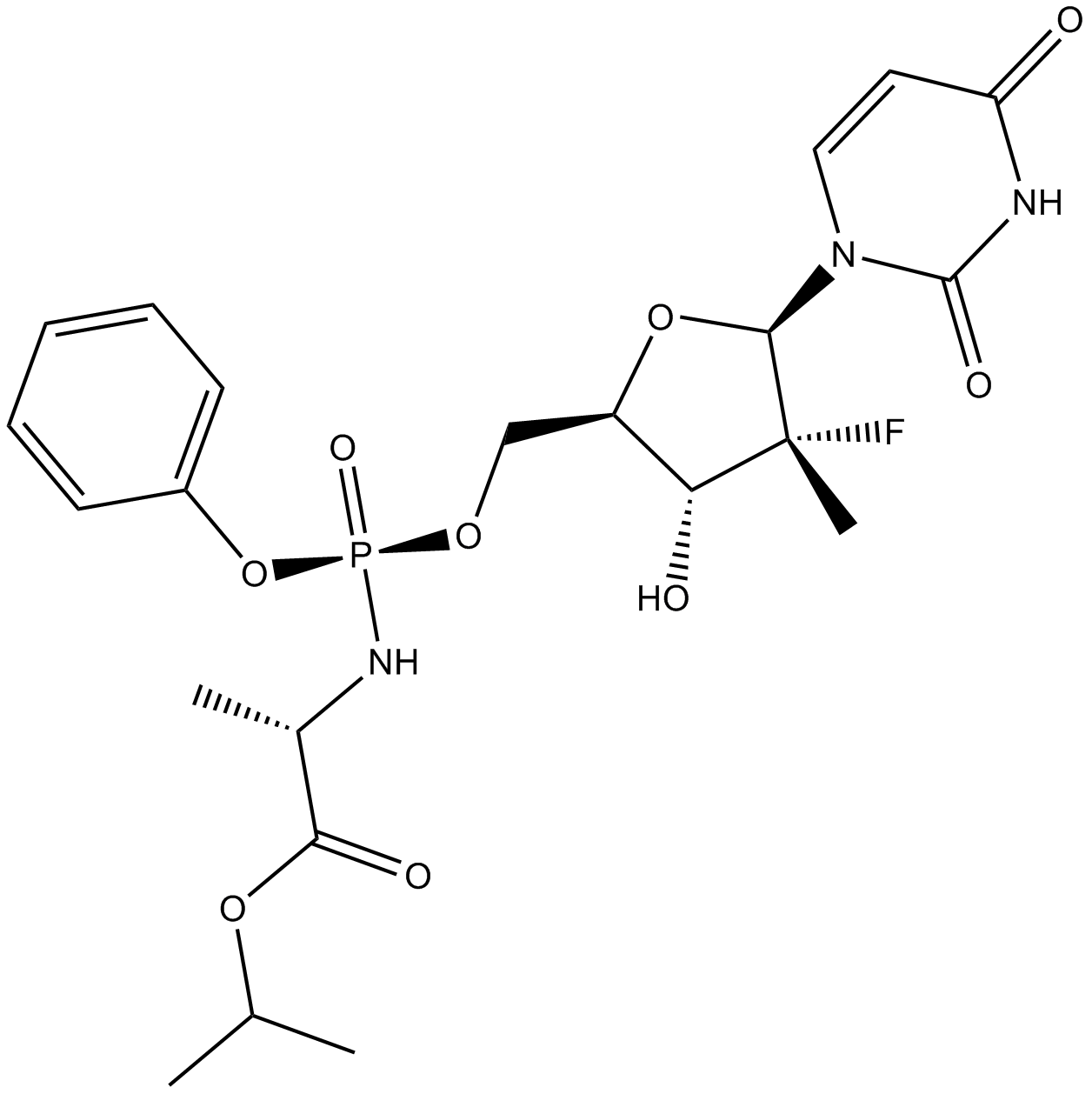
GC16367
PSI-7977
A prodrug form of PSI-7411
-
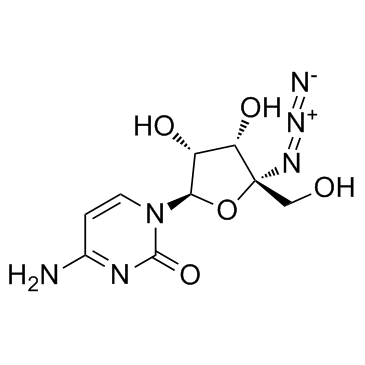
GC37063
R-1479
R-1479 (4'-Azidocytidine), a nucleoside analogue, is a specific inhibitor of RNA-dependent RNA polymerase (RdRp) of HCV.
-
GC10158
R-7128
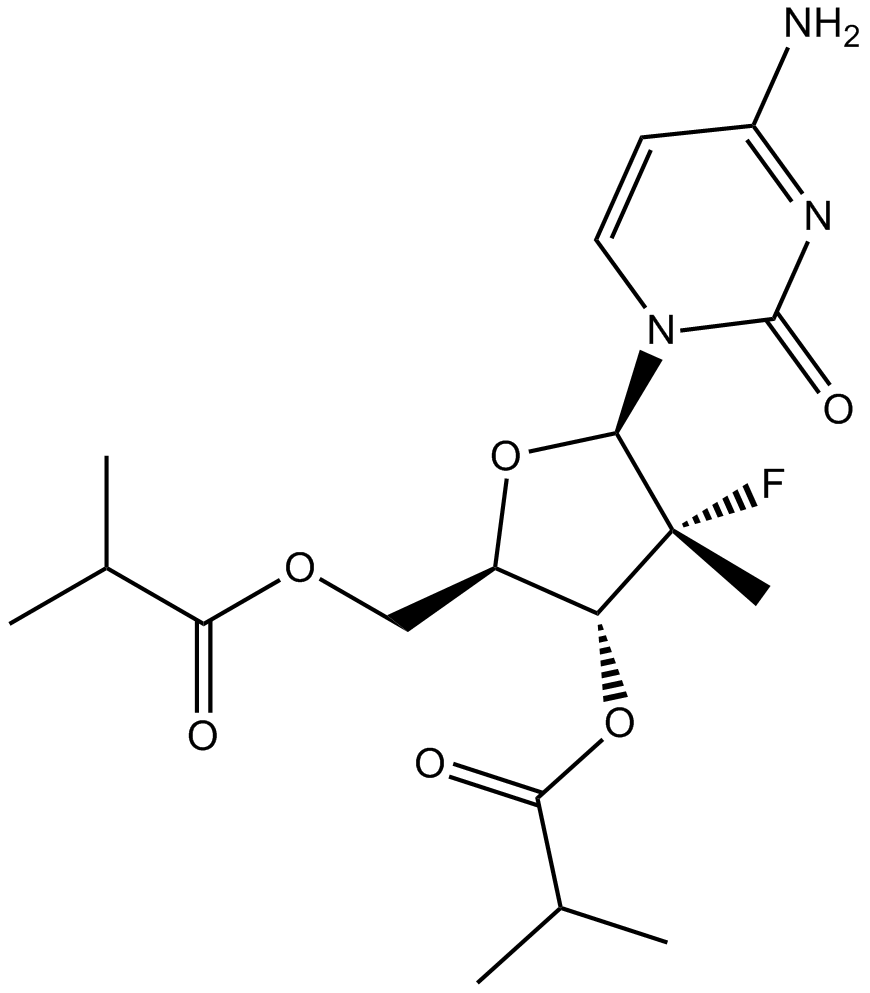
-
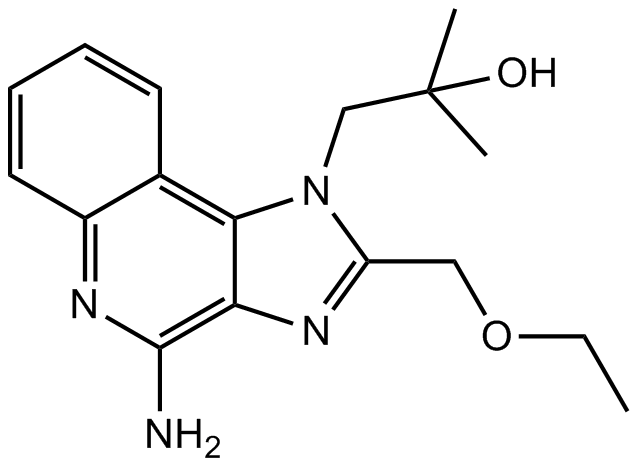
GC13650
Resiquimod (R-848)
Resiquimod (R-848) is a Toll-like receptor 7 and 8 (TLR7/TLR8) agonist that induces the upregulation of cytokines such as TNF-α, IL-6 and IFN-α.
-
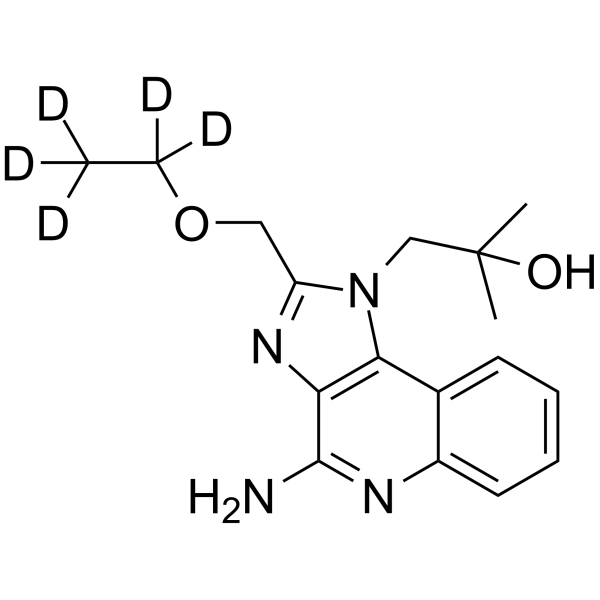
GC62636
Resiquimod-d5
Resiquimod-d5 (R848-d5) is deuterium labeled Resiquimod.
-
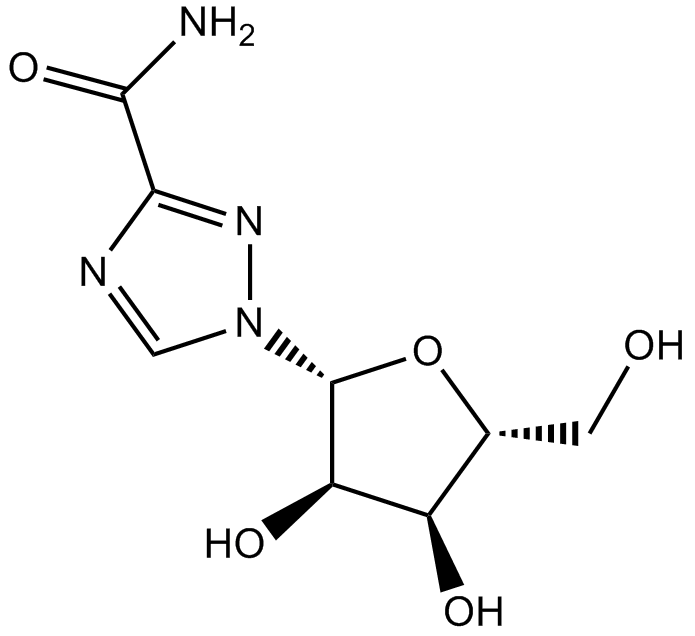
GC11518
Ribavirin
Antiviral guanosine ribonucleoside analog
-
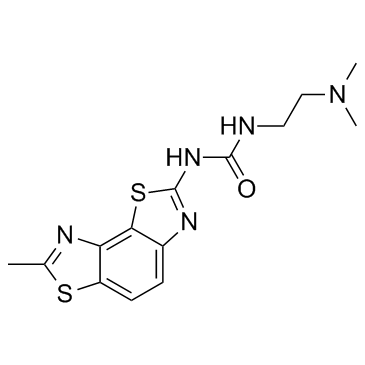
GC33932
RIG-1 modulator 1
RIG-1 modulator 1 is an anti-viral compound which can be useful for the treatment of viral infections including influenza virus, HBV, HCV and HIV extracted from patent WO 2015172099 A1.
-
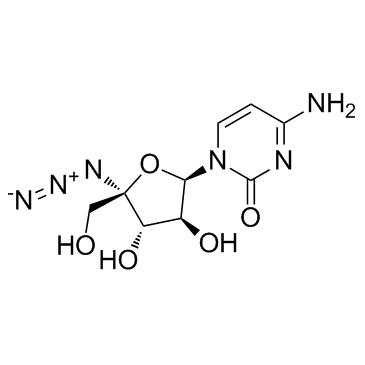
GC37550
RO-9187
RO-9187 is a potent inhibitor of HCV virus replication with an IC50 of 171 nM.
-
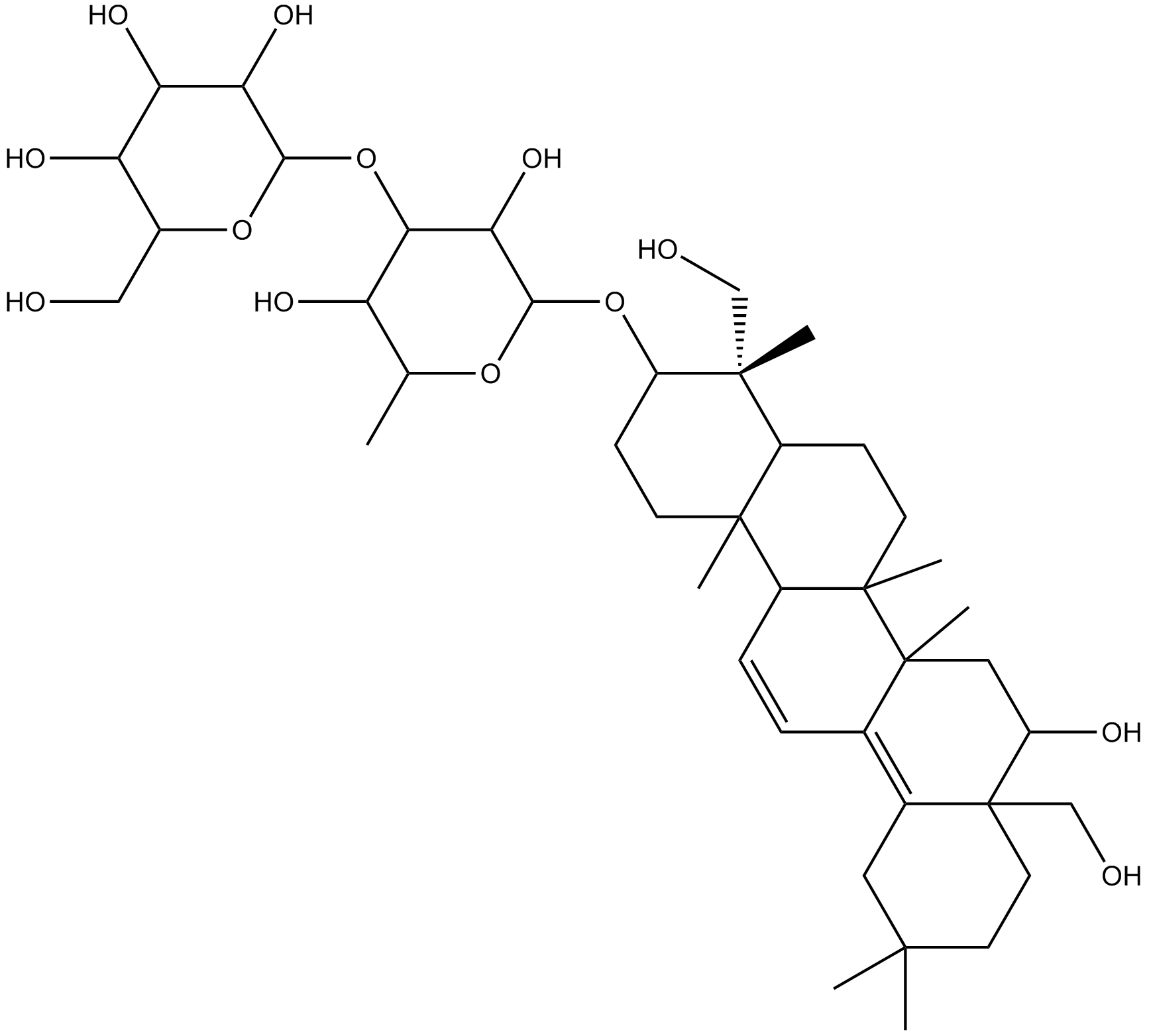
GN10209
Saikosaponin B2
-
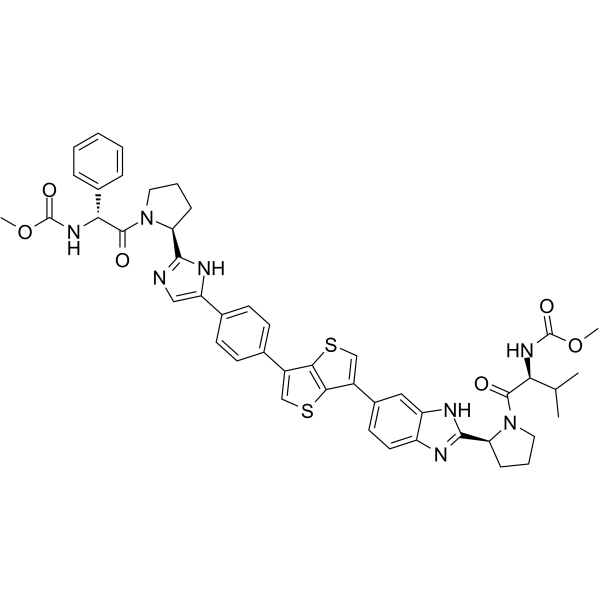
GC64016
Samatasvir
Samatasvir (IDX71) is a potent, orally active NS5A inhibitor of HCV replication.
-
GC17270
Simeprevir
A potent NS3/4A protease inhibitor
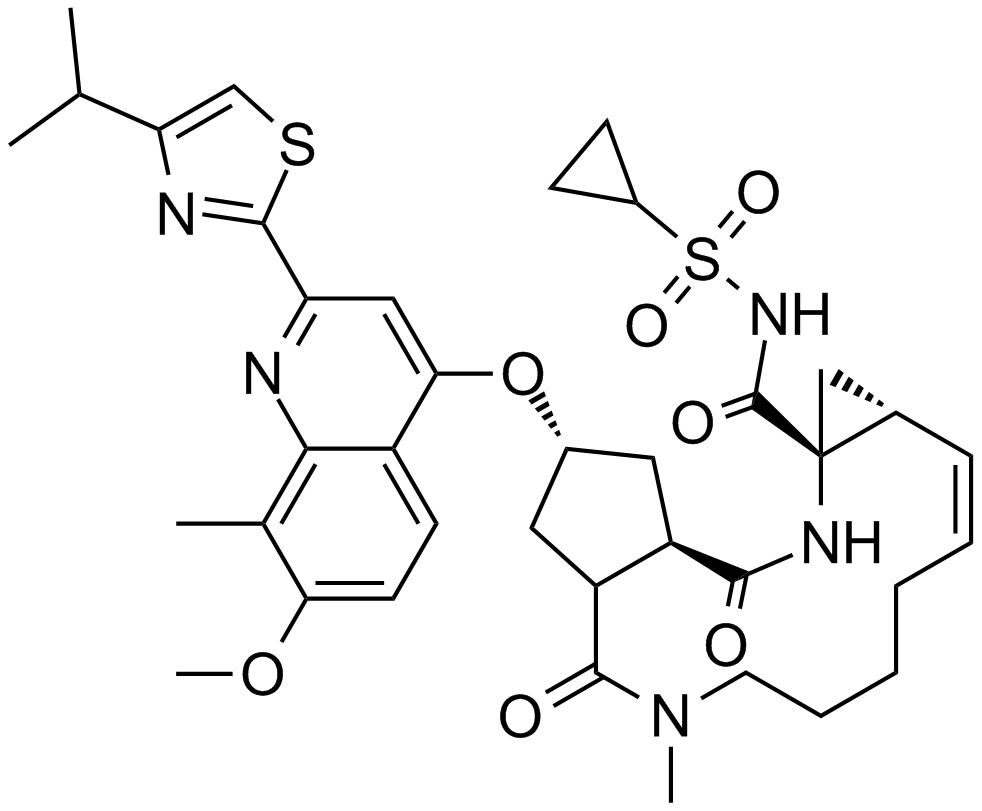
-
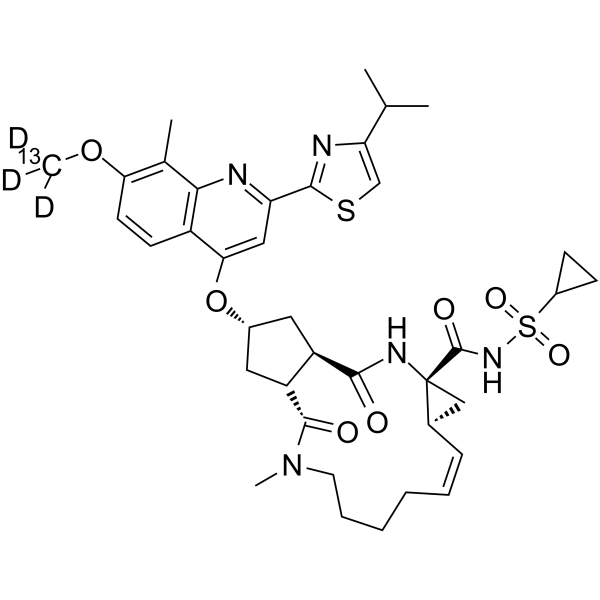
GC64038
Simeprevir-13C,d3
Simeprevir-13C,d3 (TMC435-13C,d3) is the 13C- and deuterium labeled Simeprevir.
-
GC34420
Sofosbuvir 13CD3 (PSI-7977 13CD3)
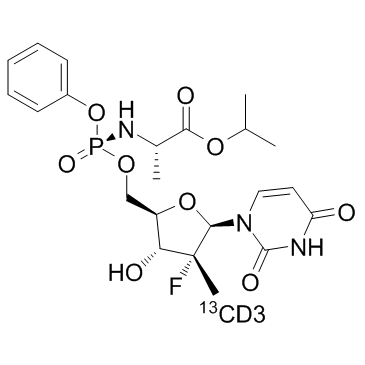
-
GC34419
Sofosbuvir D6 (PSI-7977 D6)