Li Jin's Team Reveals RNA Editing Underlies Genetic Risk for Common Inflammatory Diseases
The Human Genome Project (HGP) has successfully mapped the human genome, and human beings have also obtained their own "natural book". However, in the following 20 years, the interpretation of this "book from the sky" has been very difficult, and until now, we still lack sufficient understanding of the expression patterns of the human genome.

Today, genome-wide association studies (GWAS) have identified hundreds of thousands of risk variants associated with traits and disease etiology, and quantitative trait locus (QTL) studies have successfully linked GWAS variants to their molecular mechanisms. However, other post-transcriptional processes, such as RNA editing, remain largely unexplored, despite the growing appreciation for their important functions in health and disease.
Recently, the team of Professor Jin Billy Li from the Department of Genetics of Stanford University (Li Qin is the first author) published a paper in Nature entitled: RNA editing underlies genetic risk of common inflammatory diseases. genetic risk) research paper [1].
This study demonstrates that adenosine-inosine (A→I) RNA editing mediated by the RNA editing protein ADAR is an important post-transcriptional event that prevents the cell's own double-stranded RNA (dsRNA) from triggering an innate immune interferon response, caused by genetic factors Decreased levels of double-stranded RNA editing are important factors leading to increased genetic risk of inflammatory diseases.

The innate immune system is the vanguard of the body's fight against viral infections, in which MDA5 is an intracellular dsRNA monitor that can sense viral dsRNA and then produce interferons required for anti-virus. But in fact, MDA5 can also recognize endogenous dsRNA and trigger autoimmune diseases.
Adenosine deaminase (ADAR) is an RNA editing enzyme that catalyzes the deamination of adenosine (A) of dsRNA to yield inosine (I). Normally, ADARs edit the host's own dsRNA to prevent it from being recognized by MDA5, thereby avoiding unnecessary autoimmune responses.
As early as 2015, Professor Li Jin's team published a paper in Science [2], revealing that ADAR gene-deficient mice died at embryonic day 13.5 and exhibited multi-organ inflammatory responses caused by interferon immune responses. If the MDA5 gene was knocked out at the same time, the mice survived.

In this latest study, the research team used data from the GTEx project to study the genome and RNA sequences of 49 human tissues from 838 people, and analyzed how RNA editing patterns varied between individuals to identify cis-parameters associated with RNA editing. These variants are called edited quantitative trait loci (edQTLs).
The research team identified a total of 30,319 edQTLs with cis-regulatory effects on RNA editing levels, of which nearly one-third were tissue-specific. They found that the effect strength of these edQTLs was significantly correlated with the predicted ADAR binding strength, suggesting that genetic variation can alter RNA editing levels by affecting ADAR binding strength. In addition, changes in RNA secondary structure and RNA motifs also alter RNA editing levels.
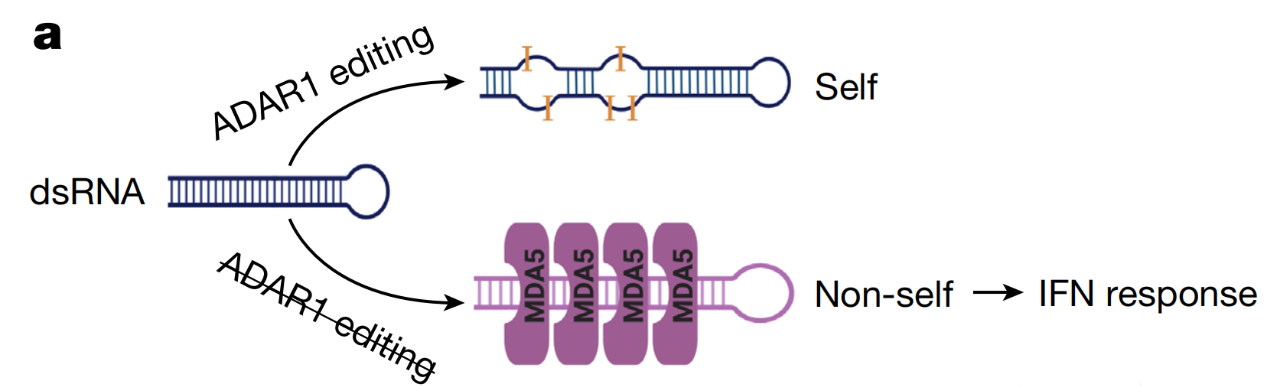
ADAR edits endogenous dsRNA to avoid autoimmune response
To assess the potential role of changes in RNA editing levels caused by edQTLs in common genetic diseases and traits, the research team performed a combined analysis of data from multiple GWAS studies. The findings suggest that these edQTLs are significantly enriched in GWAS signaling in autoimmune diseases such as psoriasis, rheumatoid arthritis, and systemic lupus erythematosus. In addition, significant enrichment of edQTLs was also observed in some non-autoimmune diseases but closely related to immune function, such as coronary heart disease, amyotrophic lateral sclerosis, and Parkinson's disease.
This indicated that edQTLs associated with decreased RNA editing levels were significantly different among different organs and tissues, and were especially highly enriched in immune tissues and immune-disease-associated tissues.
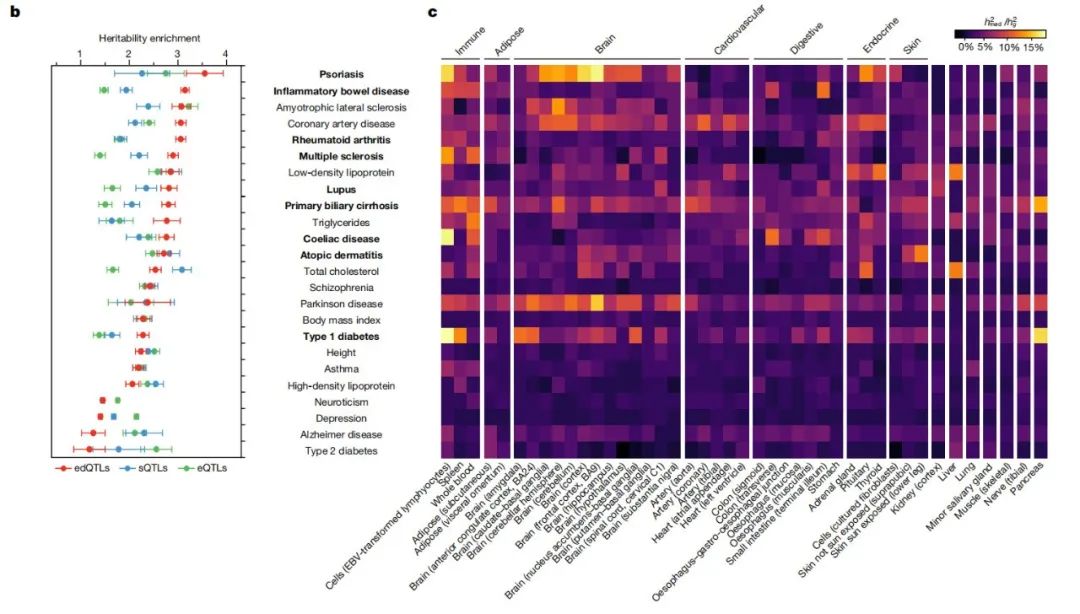
edQTLs associated with decreased levels of RNA editing are highly enriched in immune and immune-disease-associated tissues
Based on this, the research team further conducted co-localization analysis of 17 immune-related diseases and edQTL, and identified 194 immunogenic dsRNAs, of which 178 (92%) were located in exonic regions, especially in UTRs. Among the 194 immunogenic dsRNAs, at least 42 (22%) were shared between at least two diseases, suggesting that the immunogenicity of dsRNAs may be an important factor in causing autoimmune diseases.
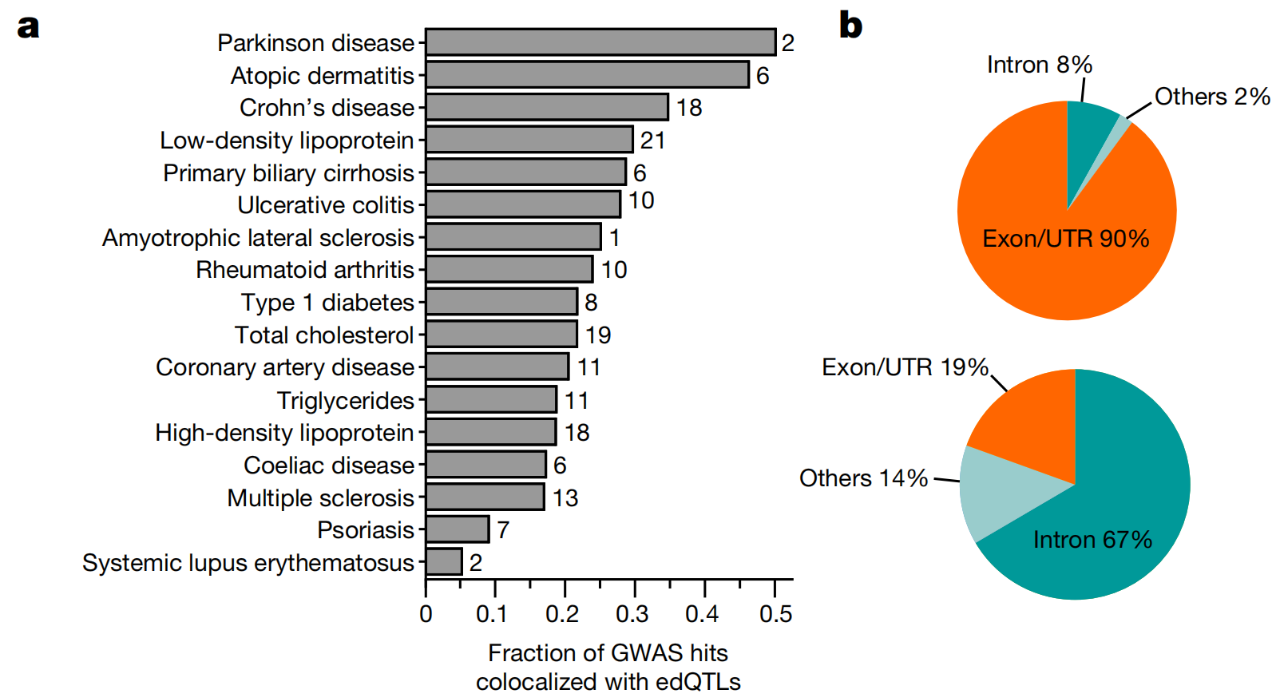
Most (92%) of immunogenic dsRNAs are located in exonic regions
To further search for evidence in disease tissue, the researchers performed analysis of allele-specific RNA editing (ASED) on hundreds of patient samples. They found a significant positive correlation between decreased dsRNA editing levels and increased levels of interferon immune responses.
More interestingly, more than 95% of RNA editing sites in the human transcriptome are located in inverted repeats, but the team was surprised to find that about 33% of the 194 immunogenic dsRNAs were located in protein-coding regions. This also raises a different view than before, that at these sites, the risk of disease is not due to changes in protein function, but the formation of dsRNA leads to the activation of the innate immune system, and the function of the encoded protein may even be related to this. The disease is completely unrelated.
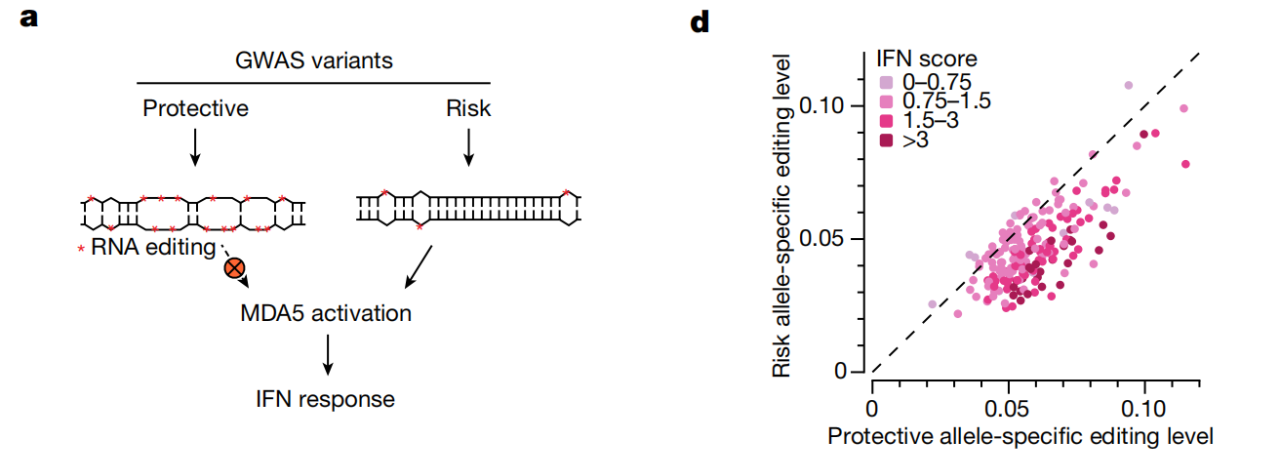
Risk variants in inflammatory diseases are associated with reduced levels of nearby dsRNA editing and induction of interferon responses
Overall, this study demonstrates that decreased levels of dsRNA editing caused by genetic variation are an important mechanism in the initiation of autoimmune and inflammatory diseases. These common genetic variants reduce the editing levels of at least hundreds of immunogenic dsRNAs simultaneously in patients, thereby triggering an MDA5-mediated interferon immune response that ultimately leads to chronic inflammation.
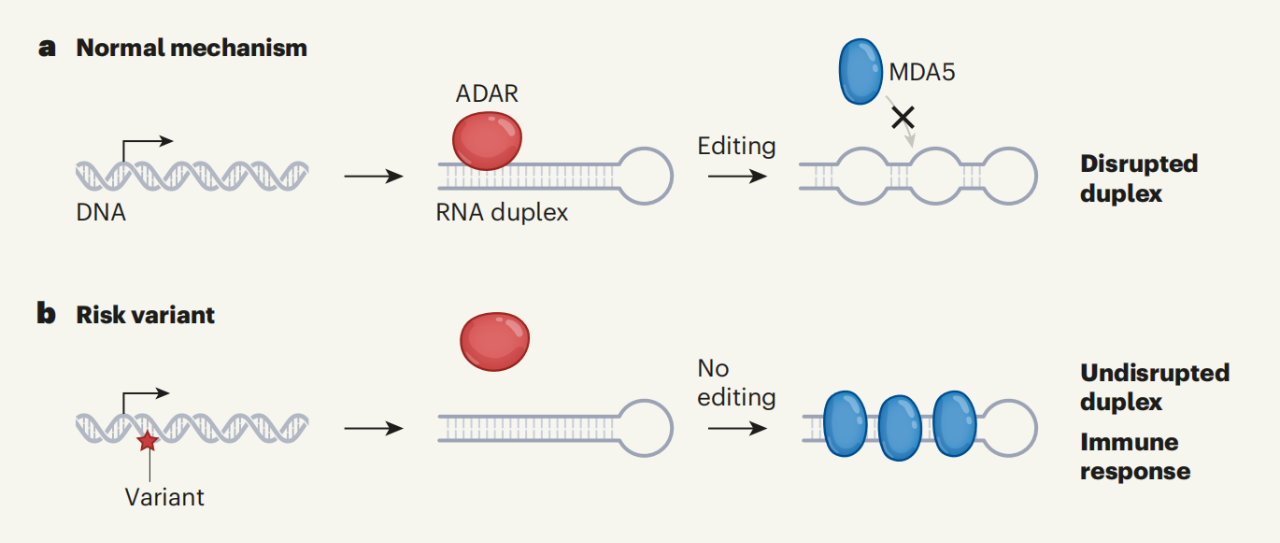
Decreased levels of dsRNA editing due to genetic variation trigger autoimmune disease
Not only that, this discovery also provides new ideas for the screening, diagnosis and treatment of autoimmune and inflammatory diseases, suggesting that inhibiting the abnormal activation of MDA5 in disease-related tissues and cells may be a potential new target.
Paper link:
1.https://www.nature.com/articles/s41586-022-05052-x
2.https://www.science.org/doi/10.1126/science.aac7049











Comments