Apoptosis
Products for Apoptosis
- Cat.No. Product Name Information
-
GC63941
α-Solanine
α-solanine, a bioactive component and one of the major steroidal glycoalkaloids in potatoes, has been observed to inhibit growth and induce apoptosis in cancer cells.
-
GC67618
α-Tocopherol phosphate disodium
α-Tocopherol phosphate (alpha-Tocopherol phosphate) disodium, a promising antioxidant, can protect against long-wave UVA1 induced cell death and scavenge UVA1 induced ROS in a skin cell model. α-Tocopherol phosphate disodium possesses therapeutic potential in the inhibition of apoptosis and increases the migratory capacity of endothelial progenitor cells under high-glucose/hypoxic conditions and promotes angiogenesis.
-
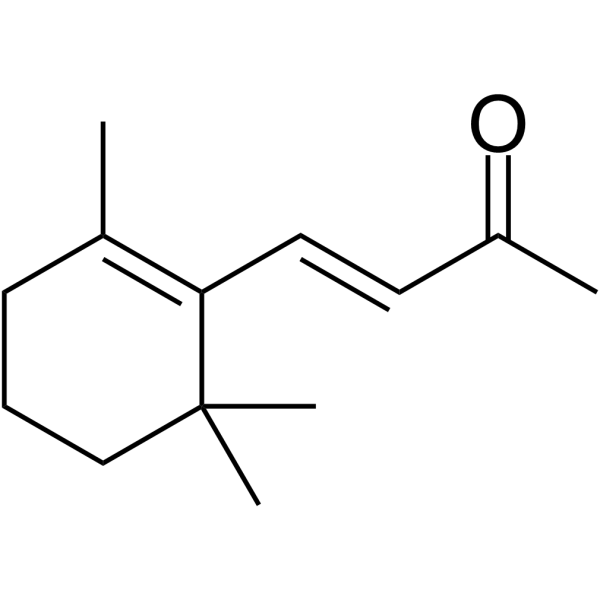
GC64619
β-Ionone
β-Ionone is effective in the induction of apoptosis in gastric adenocarcinoma SGC7901 cells. Anti-cancer activity.
-
GC52192
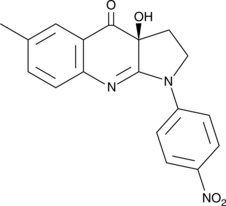
(S)-4'-nitro-Blebbistatin
(S)-4'-nitro-Blebbistatin is a non-cytotoxic, photostable, fluorescent and specific Myosin II inhibitor, usd in the study of the specific role of myosin II in physiological, developmental, and cell biological studies.
-
GC68452
2,4,6-Triiodophenol

-
GC68043
2-tert-Butyl-1,4-benzoquinone

-
GC64762
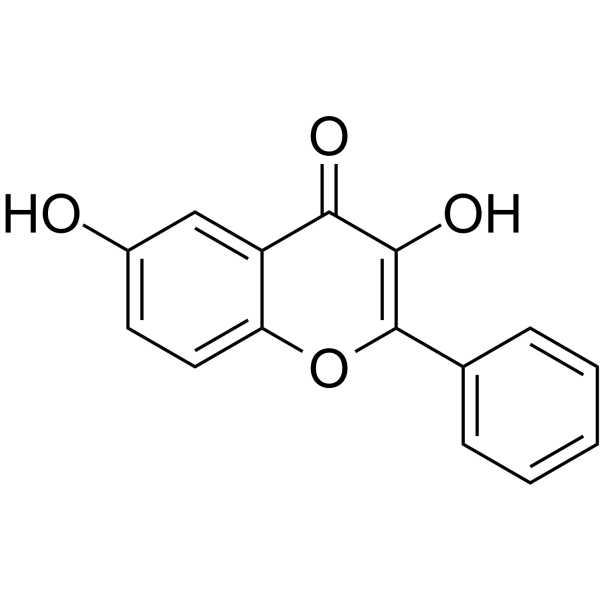
3,6-Dihydroxyflavone
3,6-Dihydroxyflavone is an anti-cancer agent. 3,6-Dihydroxyflavone dose- and time-dependently decreases cell viability and induces apoptosis by activating caspase cascade, cleaving poly (ADP-ribose) polymerase (PARP). 3,6-Dihydroxyflavone increases intracellular oxidative stress and lipid peroxidation.
-
GC52227
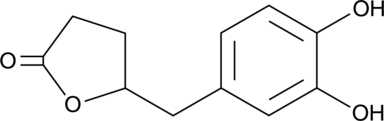
5-(3',4'-Dihydroxyphenyl)-γ-Valerolactone
An active metabolite of various polyphenols
-
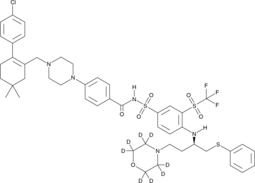
GC49745
ABT-263-d8
ABT-263-d8 is the deuterium labeled Navitoclax. Navitoclax (ABT-263) is a potent and orally active Bcl-2 family protein inhibitor that binds to multiple anti-apoptotic Bcl-2 family proteins, such as Bcl-xL, Bcl-2 and Bcl-w, with a Ki of less than 1 nM.
-
GC52372
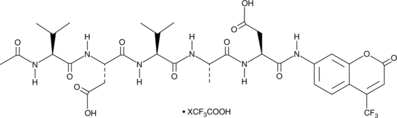
Ac-VDVAD-AFC (trifluoroacetate salt)
A fluorogenic substrate for caspase-2
-
GC63932
Amsilarotene
Amsilarotene (TAC-101; Am 555S), an orally active synthetic retinoid, has selective affinity for retinoic acid receptor α (RAR-α) binding with Ki of 2.4, 400 nM for RAR-α and RAR-β. Amsilarotene induces the apoptotic of human gastric cancer, hepatocellular carcinoma and ovarian carcinoma cells. Amsilarotene can be used for the research of cancer.
-
GC65004
Apostatin-1
Apostatin-1 (Apt-1) is a potent TRADD inhibitor.
-
GC65163
Ardisiacrispin B
Ardisiacrispin B displays cytotoxic effects in multi-factorial drug resistant cancer cells via ferroptotic and apoptotic cell death.
-
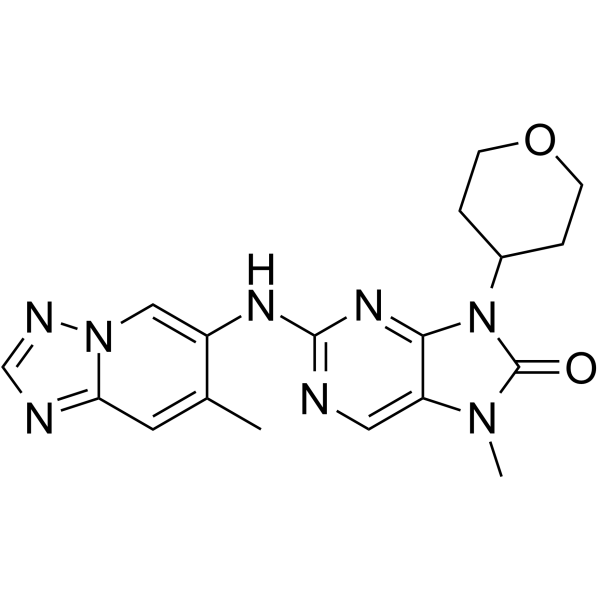
GC64938
AZD-7648
AZD-7648 is a potent, orally active, selective DNA-PK inhibitor with an IC50 of 0.6 nM. AZD-7648 induces apoptosis and shows antitumor activity.
-
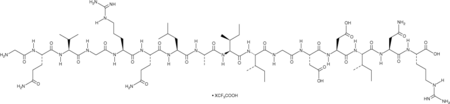
GC52344
Bak BH3 (72-87) (human) (trifluoroacetate salt)
A Bak-derived peptide
-
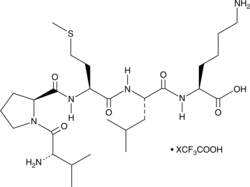
GC52476
Bax Inhibitor Peptide V5 (trifluoroacetate salt)
A Bax inhibitor
-
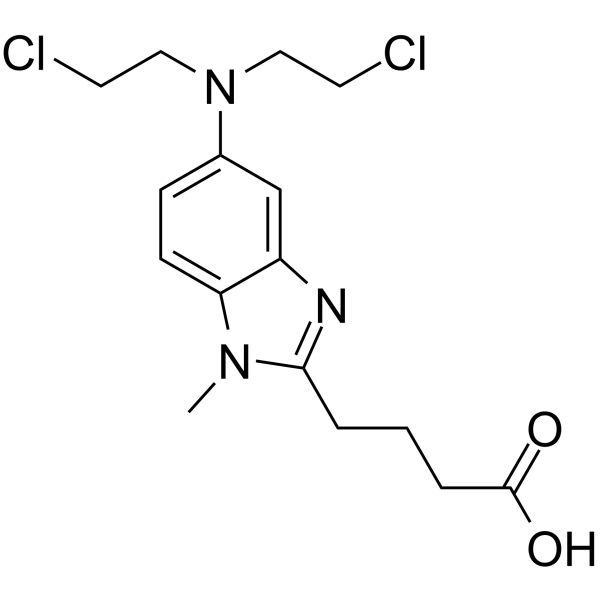
GC64354
Bendamustine
Bendamustine (SDX-105 free base), a purine analogue, is a DNA cross-linking agent. Bendamustine activates DNA-damage stress response and apoptosis. Bendamustine has potent alkylating, anticancer and antimetabolite properties.
-
GC49513
Bim/BOD (IN) Polyclonal Antibody
For immunodetection of Bim-
related proteins 
-
GC52355
BimS BH3 (51-76) (human) (trifluoroacetate salt)
A Bim-derived peptide

-
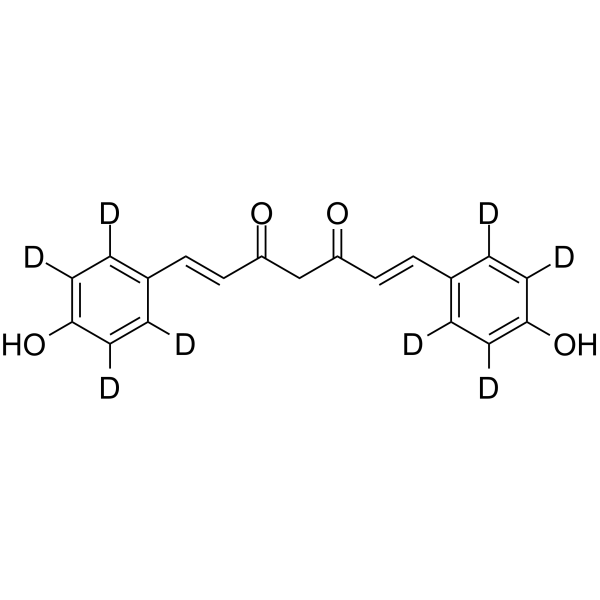
GC68308
Bisdemethoxycurcumin-d8
-
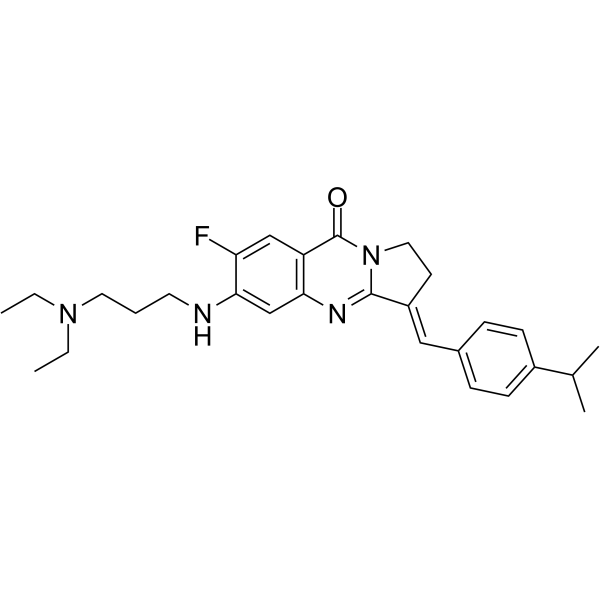
GC65428
BLM-IN-1
BLM-IN-1 (compound 29) is an effective Bloom syndrome protein (BLM) inhibitor, with a strong BLM binding KD of 1.81 μM and an IC50 of 0.95 μM for BLM. Induces DNA damage response, as well as apoptosis and proliferation arrest in cancer cells.
-
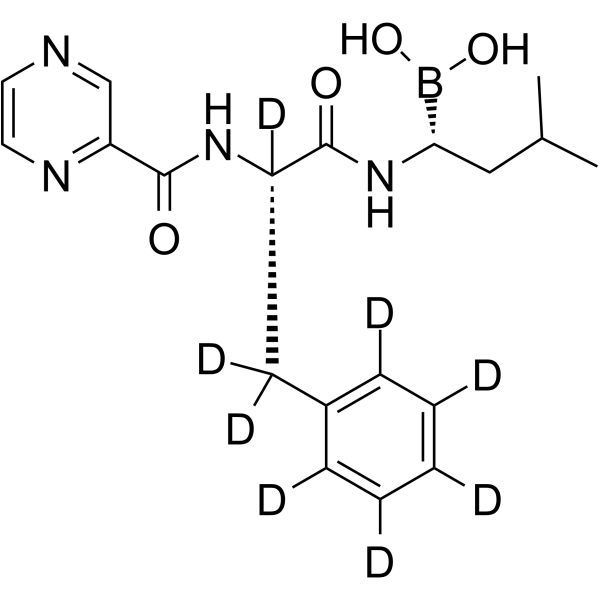
GC65010
Bortezomib-d8
Bortezomib-d8 (PS-341-d8) is the deuterium labeled Bortezomib. Bortezomib (PS-341) is a reversible and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki=0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is the first proteasome inhibitor anticancer agent. Anti-cancer activity.
-
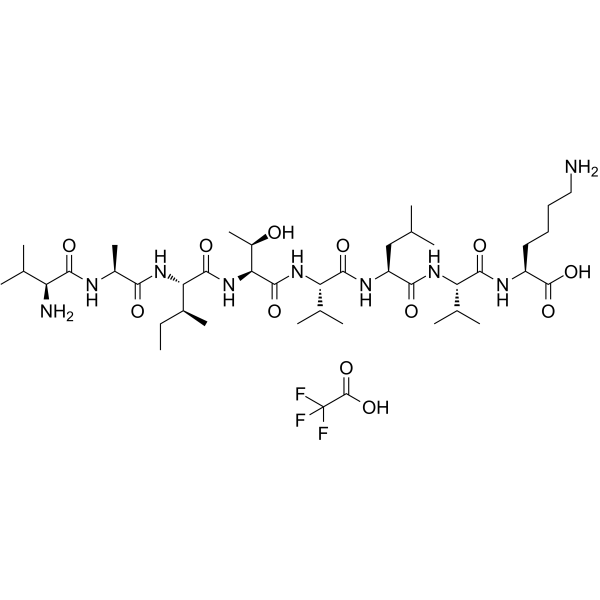
GC65081
CALP1 TFA
CALP1 TFA is a calmodulin (CaM) agonist (Kd of 88 ?M) with binding to the CaM EF-hand/Ca2+-binding site.
-
GC64110
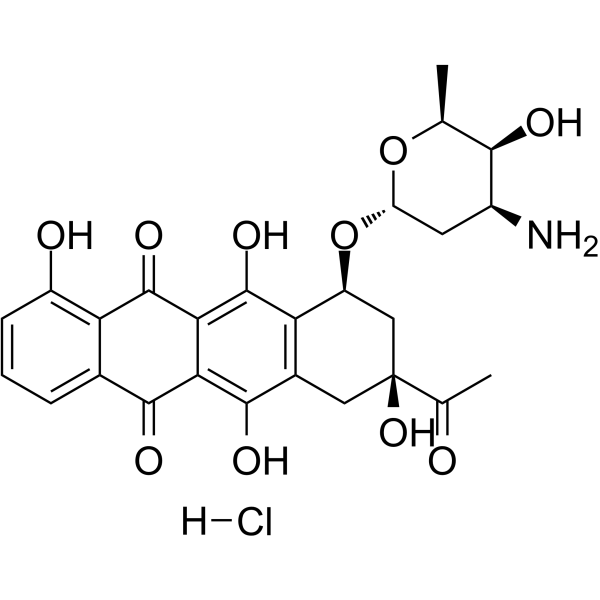
Carubicin hydrochloride
Carubicin hydrochloride is a microbially-derived compound.
-
GC52245
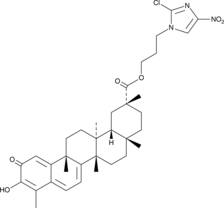
CAY10792
An anticancer agent
-
GC52489
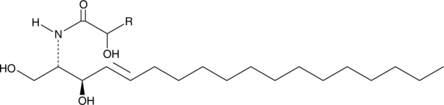
Ceramide (hydroxy) (bovine spinal cord)
A sphingolipid
-
GC52485
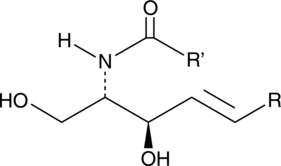
Ceramide (non-hydroxy) (bovine spinal cord)
A sphingolipid
-
GC52486
Ceramide Phosphoethanolamine (bovine)
A sphingolipid
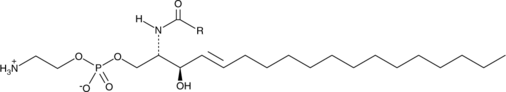
-
GC64993
Chicoric acid
Chicoric acid (Cichoric acid), an orally active dicaffeyltartaric acid, induces reactive oxygen species (ROS) generation.
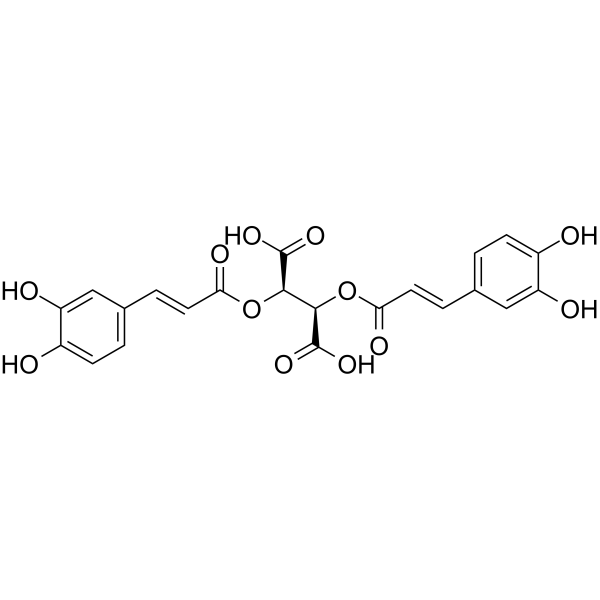
-
GC64028
Chrysosplenol D
Chrysosplenol D is a methoxy flavonoid that induces ERK1/2-mediated apoptosis in triple negative human breast cancer cells.
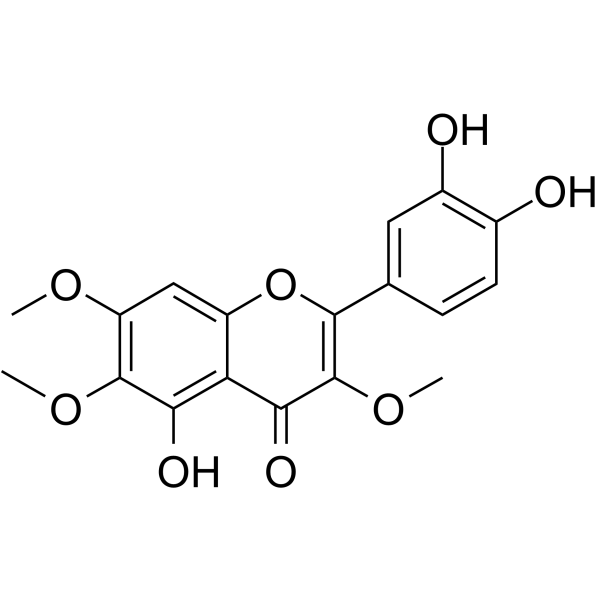
-
GC52269
Cinnabarinic Acid-d4
An internal standard for the quantification of cinnabarinic acid
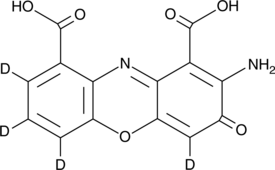
-
GC68051
Citric acid-d4

-
GC52367
Citrullinated Vimentin (G146R) (R144 + R146) (139-159)-biotin Peptide
A biotinylated and citrullinated mutant vimentin peptide

-
GC52370
Citrullinated Vimentin (R144) (139-159)-biotin Peptide
A biotinylated and citrullinated vimentin peptide

-
GC49454
Complex 3
A fluorescent copper complex with anticancer activity
-
GC66356
Cusatuzumab
Cusatuzumab is a human αCD70 monoclonal antibody. Cusatuzumab shows cytotoxicity activity with enhanced antibody-dependent cellular. Cusatuzumab reduces leukemia stem cells (LSCs) and triggers gene signatures related to myeloid differentiation and apoptosis. Cusatuzumab has the potential for the research of Acute myeloid leukemia (AML).

-
GC63967
Cycleanine
Cycleanine is a potent vascular selective Calcium antagonist.
-
GC65565
Cyproheptadine
Cyproheptadine is a potent and orally active 5-HT2A receptor antagonist, with antidepressant and antiserotonergic effects.
-
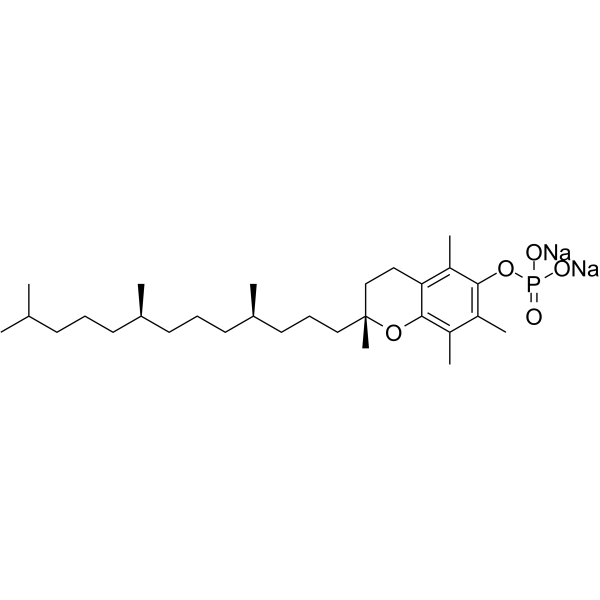
GC66824
D-α-Tocopherol Succinate
D-α-Tocopherol Succinate (Vitamin E succinate) is an antioxidant tocopherol and a salt form of vitamin E. D-α-Tocopherol Succinate inhibits Cisplatin -induced cytotoxicity. D-α-Tocopherol Succinate can be used for the research of cancer.
-
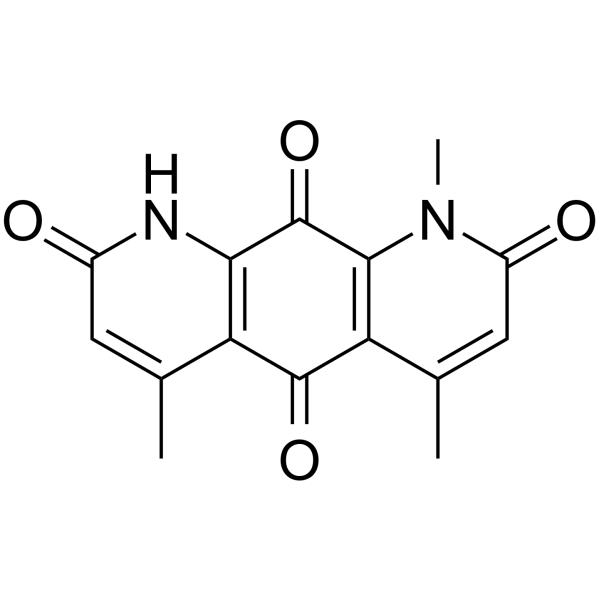
GC68306
Deoxynyboquinone
-
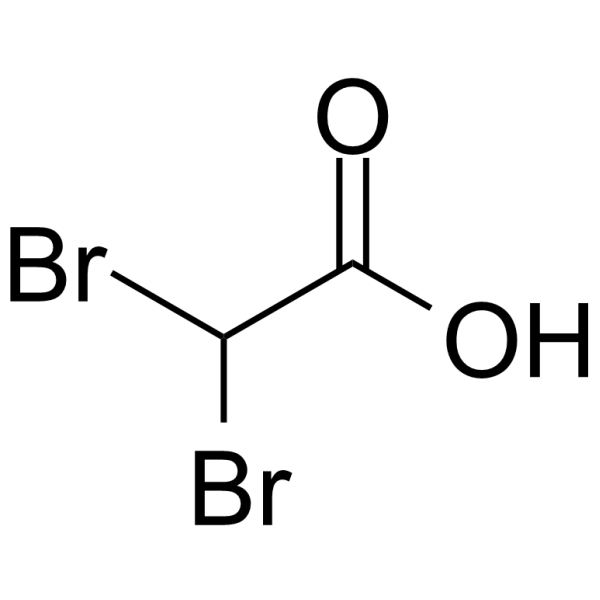
GC64139
Dibromoacetic acid
-
GC64375
Difopein TFA
Difopein (TFA), a specific and competitive inhibitor of 14-3-3 protein (a highly conserved eukaryotic regulatory molecule), blocking the ability of 14-3-3 to bind to target proteins and inhibits 14-3-3/Ligand interactions. Difopein (TFA) leads to induction of apoptosis and enhances the ability of cisplatin to kill cells.

-
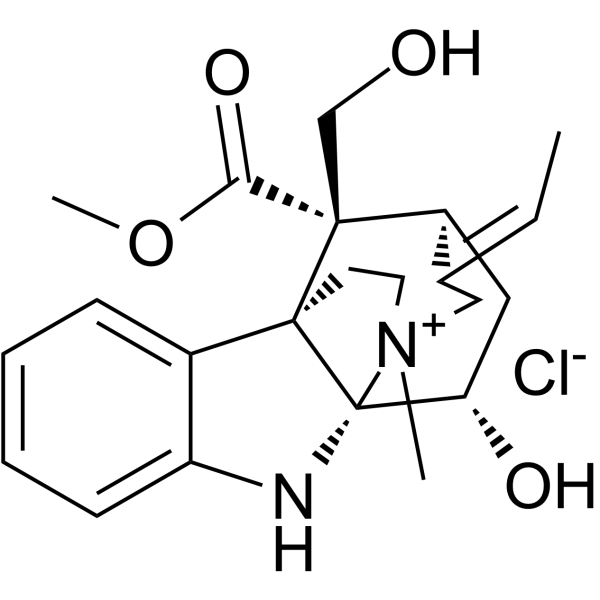
GC63871
Echitamine chloride
Echitamine chloride is the major monoterpene indole alkaloid present in Alstonia with potent anti-tumour activity. Echitamine chloride induces DNA fragmentation and cells apoptosis. Echitamine chloride inhibits pancreatic lipase with an IC50 of 10.92 μM.
-
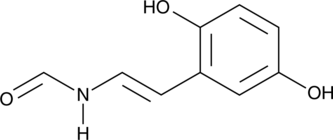
GC52516
Erbstatin
A tyrosine kinase inhibitor
-
GC63845
Eribulin-d3 mesylate
Eribulin-d3 mesylate is a deuterium labeled Eribulin mesylate. Eribulin mesylate is a microtubule targeting agent that is used for the research of cancer.
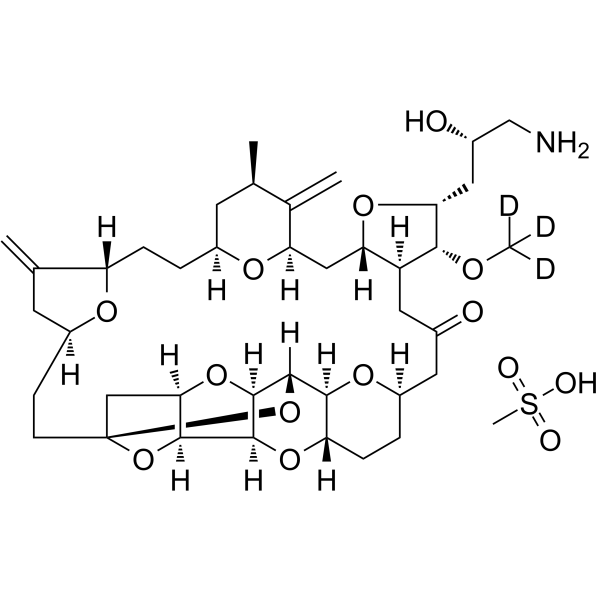
-
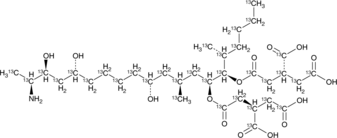
GC52288
Fumonisin B1-13C34
An internal standard for the quantification of fumonisin B1
-
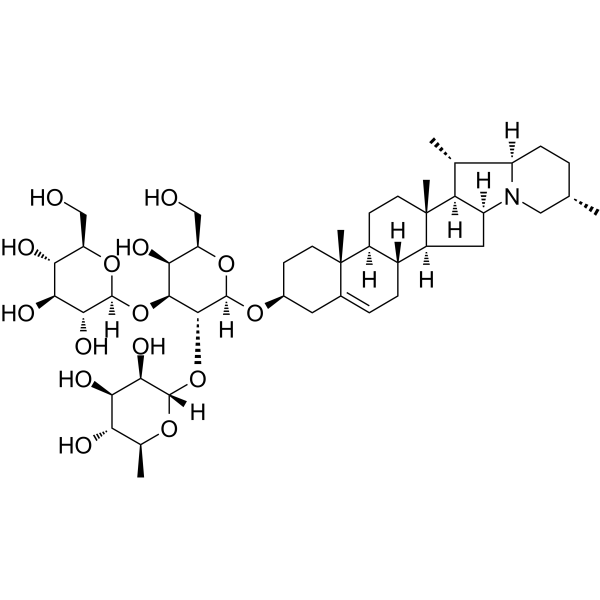
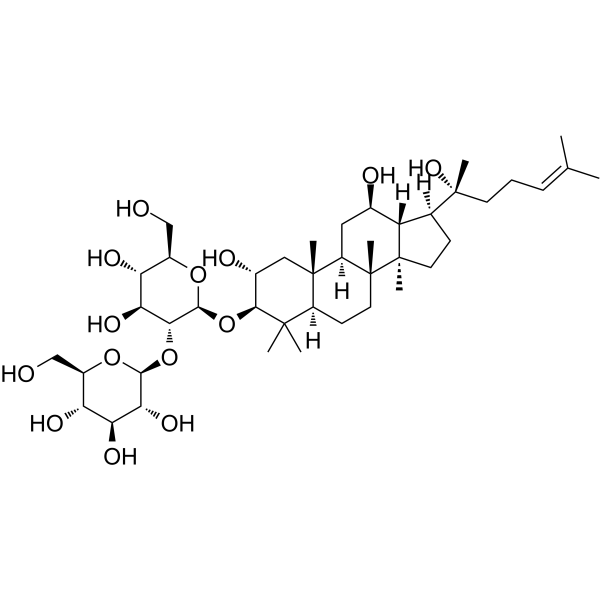
GC64115
Gypenoside LI
Gypenoside LI, a gypenoside monomer, possesses anti-tumor activity. Gypenoside LI induces cell apoptosis, cell cycle and migration.
-
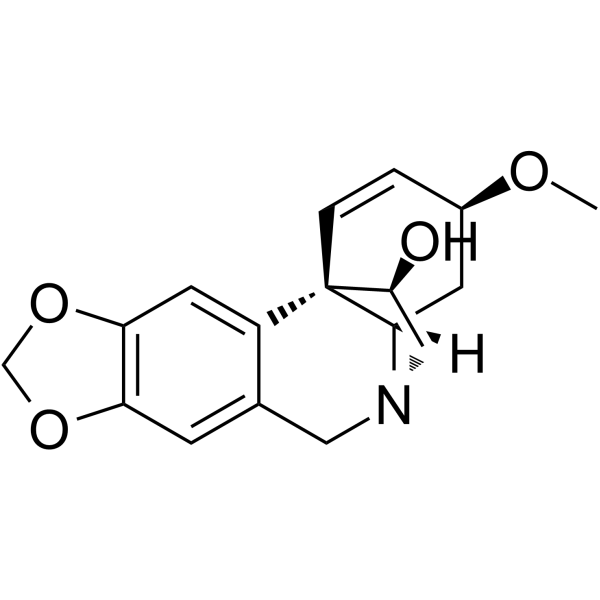
GC65043
Haemanthamine
Haemanthamine is a crinine-type alkaloid isolated from the Amaryllidaceae plants with potent anticancer activity.
-
GC65460
HDACs/mTOR Inhibitor 1
HDACs/mTOR Inhibitor 1 is a dual Histone Deacetylases (HDACs) and mammalian target of Rapamycin (mTOR) target inhibitor for treating hematologic malignancies, with IC50s of 0.19 nM, 1.8 nM, 1.2 nM and >500 nM for HDAC1, HDAC6, mTOR and PI3Kα, respectively. HDACs/mTOR Inhibitor 1 stimulates cell cycle arrest in G0/G1 phase and induce tumor cell apoptosis with low toxicity in vivo.
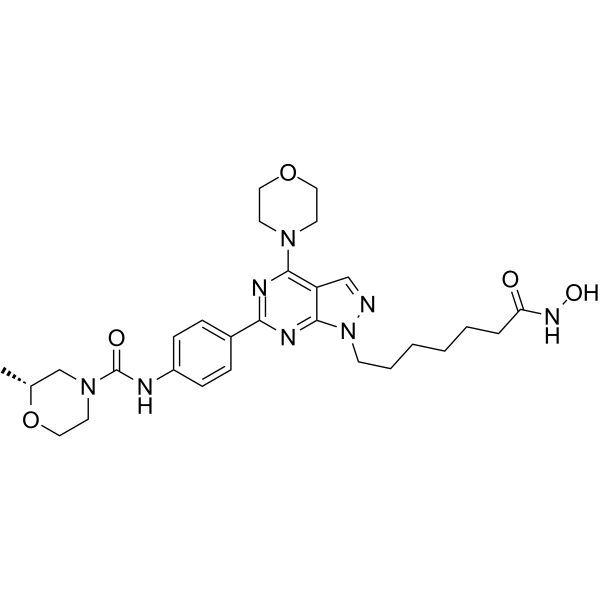
-
GC64662
Helichrysetin
Helichrysetin, isolated from the flowers of Helichrysum odoratissimum, is an ID2 (inhibitor of DNA binding 2) inhibitor, and suppresses DCIS (ductal carcinoma in situ) formation. Helichrysetin possess strong inhibitory effects on cell growth and is capable of inducing apoptosis in A549 cells.
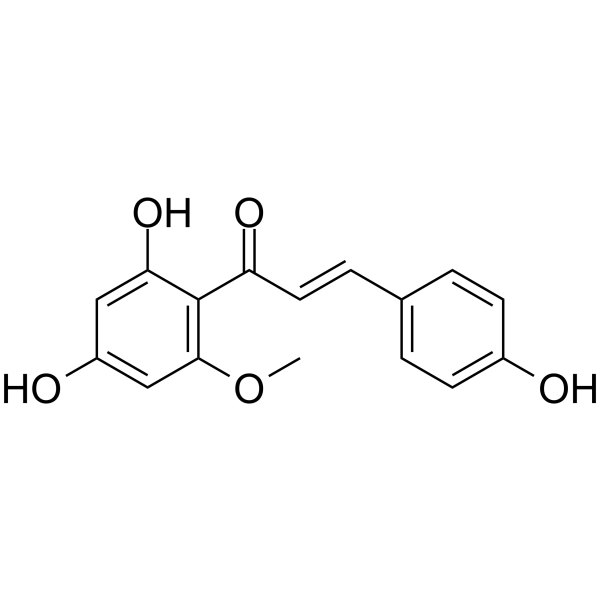
-
GC64786
Hellebrigenin
Hellebrigenin, one of bufadienolides belonging to cardioactive steroids, is isolated from traditional Chinese medicine Venenum Bufonis. Hellebrigenin induces DNA damage and cell cycle G2/M arrest. Hellebrigenin triggers mitochondria-mediated apoptosis.
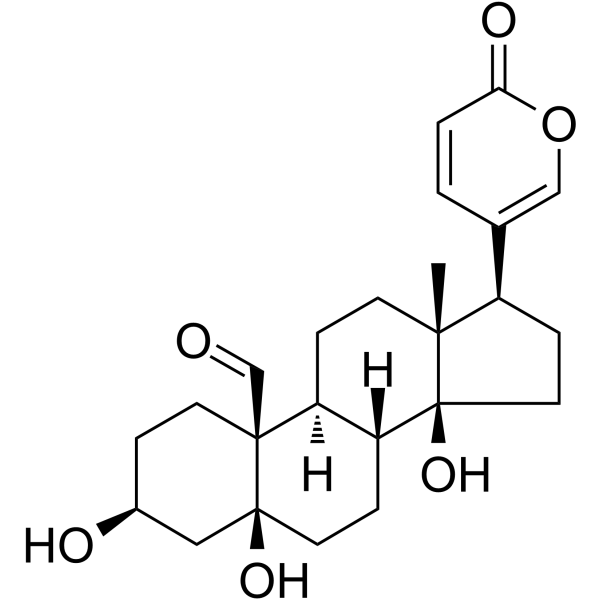
-
GC63902
Hematoporphyrin monomethyl ether
Hematoporphyrin monomethyl ether, second generation of porphyrin-related photosensitizer, is characterized by its single form, high yield of singlet oxygen, high selectivity, and low toxicity, which has been widely used in the diagnosis and treatment of various tumors, including lung cancer, bladder cancer, and nevus flammeus and brain glioma.
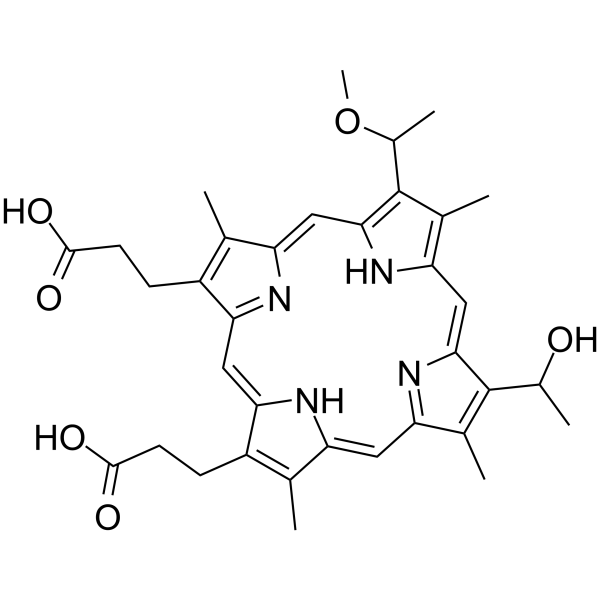
-
GC64485
HXR9 hydrochloride
HXR9 hydrochloride is a cell-permeable peptide and a competitive antagonist of HOX/PBX interaction. HXR9 hydrochloride antagonizes the interaction between HOX and a second transcrip-tion factor (PBX), which binds to HOX proteins in paralogue groups1 to 8. HXR9 hydrochloride selectively decreases cell proliferation and promotes apoptosis in cells with a high level of expression of the HOXA/PBX3 genes, such as MLL-rearranged leukemic cells.
-
GC49479
Hypoxanthine-d4
An internal standard for the quantification of hypoxanthine
-
GC65168
Imifoplatin
Imifoplatin (PT-112) is a platinum-based agent belonging to the phosphaplatin family. Imifoplatin exhibits antineoplastic activity.
-
GC49670
Indium (III) thiosemicarbazone 5b
An anticancer agent
-
GC52472
Inostamycin A (sodium salt)
A bacterial metabolite with anticancer activity
-
GC63934
Karanjin
Karanjin is a major active furanoflavonol constituent of Fordia cauliflora. Karanjin induces GLUT4 translocation in skeletal muscle cells by increasing AMPK activity. Karanjin can induce cancer cell death through cell cycle arrest and enhance apoptosis.
-
GC64370
Kongensin A
Kongensin A is a natural product isolated from Croton kongensis.
-
GC64352
L-Glutamic acid-15N
-
GC65095
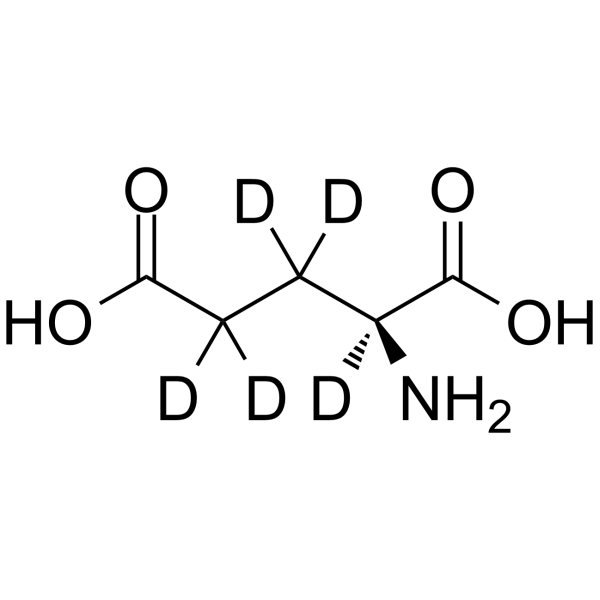
L-Glutamic acid-d5
L-Glutamic acid-d5 is the deuterium labeled L-Glutamic acid.
-
GC64325
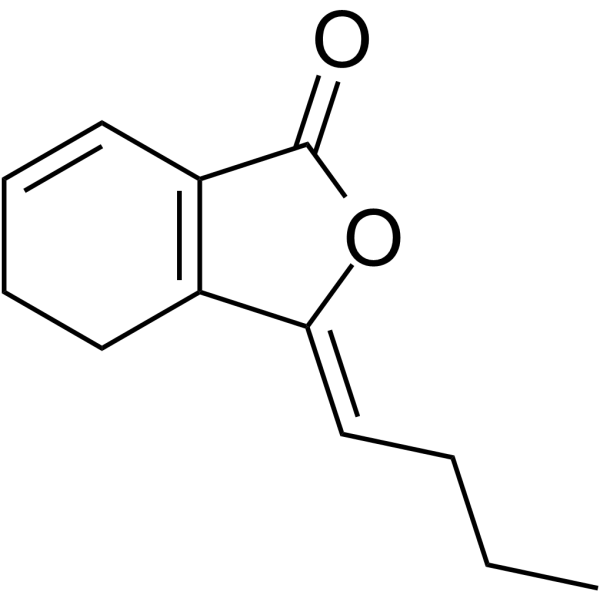
Ligustilide
Ligustilide is is a bioactive phthalide derivative isolated from Angelica sinensis and Chuanxiong.
-
GC67936
Lupiwighteone

-
GC67966
Methylstat
-
GC66462
MGH-CP1
MGH-CP1 is a potent and orally active TEAD2 and TEAD4 auto-palmitoylation inhibitor with IC50s of 710 nM and 672 nM, respectively. MGH-CP1 can decrease the palmitoylation levels of endogenous or ectopically expressed TEAD proteins in cells. MGH-CP1 can suppress Myc expression, inhibit epithelial over-proliferation, and induce apoptosis when together with Lats1/2 deletion.
-
GC68213
MitoBloCK-6
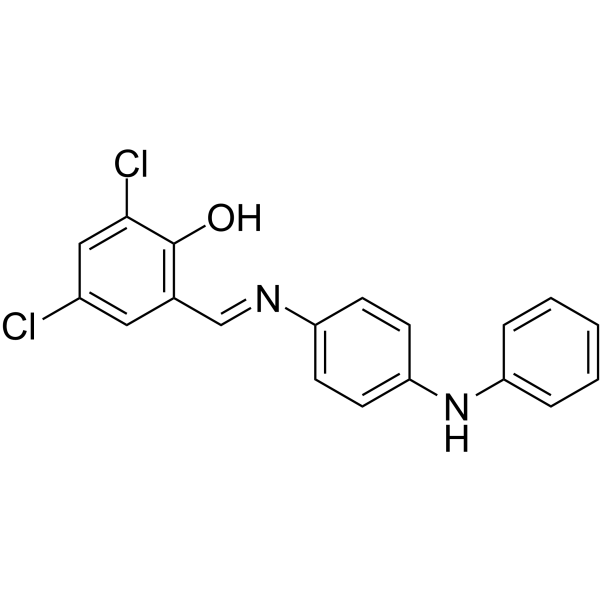
-
GC65143
MKC-1
MKC-1 (Ro-31-7453) is an orally active and potent cell cycle inhibitor with broad antitumor activity. MKC-1 inhibits the Akt/mTOR pathway. MKC-1 arrests cellular mitosis and induces cell apoptosis by binding to a number of different cellular proteins including tubulin and members of the importin β family.
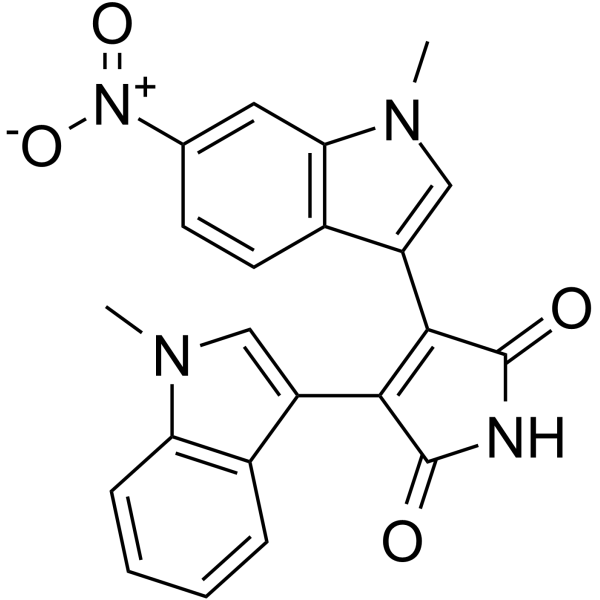
-
GC65442
Musk ketone
Musk ketone (MK) is a widely used artificial fragrance.
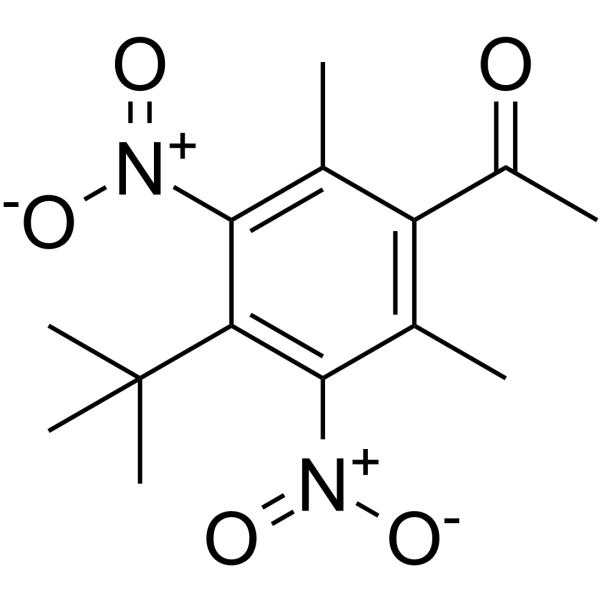
-
GC64980
MV1
MV1 is an antagonist of IAP (inhibitor of apoptosis protein), leads to protein knockdown of HaloTag-fused proteins when combined with HaloTag ligand.
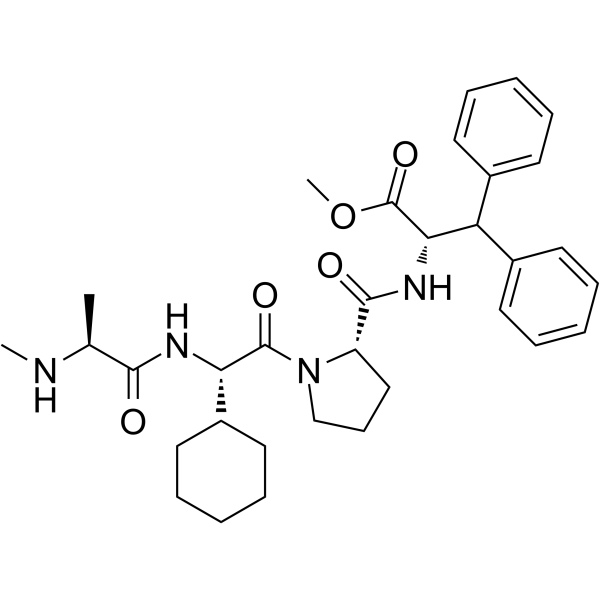
-
GC66343
n-Butyl-β-D-fructofuranoside
n-Butyl-β-D-fructofuranoside could be isolated from kangaisan. n-Butyl-β-D-fructofuranoside induces apoptosis through the mitochondrial pathway. n-Butyl-β-D-fructofuranoside can be used for cancer research.
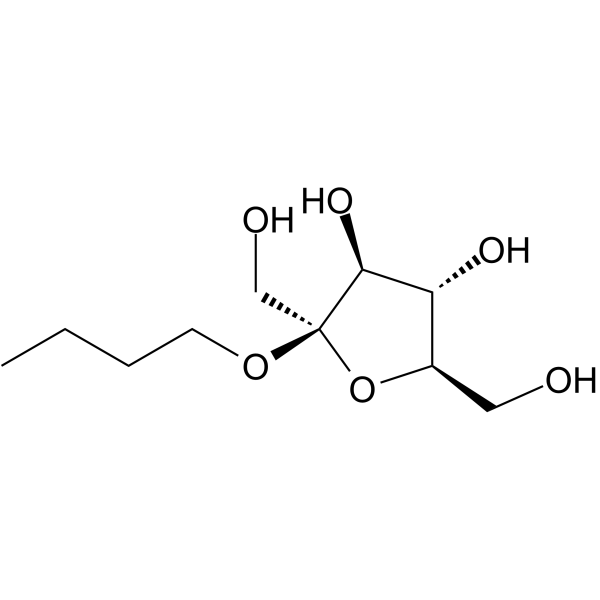
-
GC67272
N6-Benzyladenosine
N6-Benzyladenosine is an adenosine receptor agonist, has a cytoactive activity. N6-Benzyladenosine arrests cell cycle at G0/G1 phase and induces cell apoptosis. N6-Benzyladenosine also exerts inhibitory effect on T. gondii adenosine kinase and glioma-.
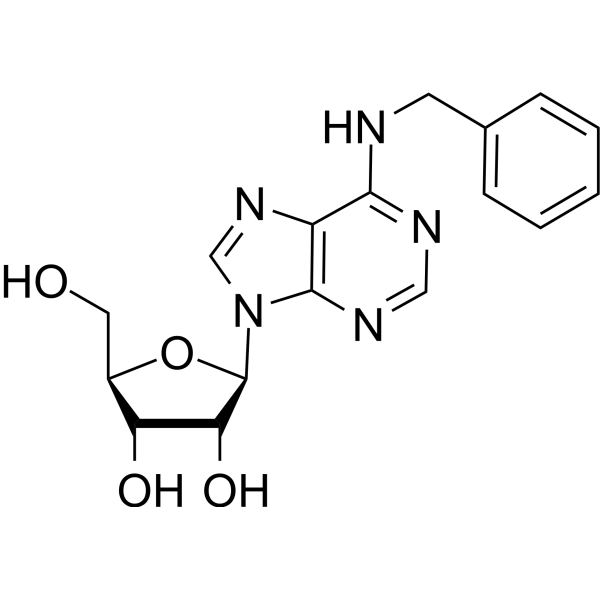
-
GC49681
Necrosulfonamide-d4
Necrosulfonamide-d4 is the deuterium labeled Necrosulfonamide. Necrosulfonamide is a necroptosis inhibitor acting by selectively targeting the mixed lineage kinase domain-like protein (MLKL). Necrosulfonamide prevents MLKL-RIP1-RIP3 necrosome complex from interacting with its downstream effectors. MLKL is a critical substrate of RIP3 during the induction of necrosis.
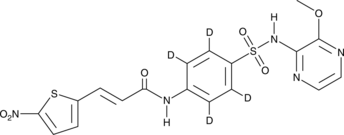
-
GC64126
Neoechinulin A
Neoechinulin A is an isoprenyl indole alkaloid that exhibits scavenging, neurotrophic factor-like, and anti-apoptotic activities.
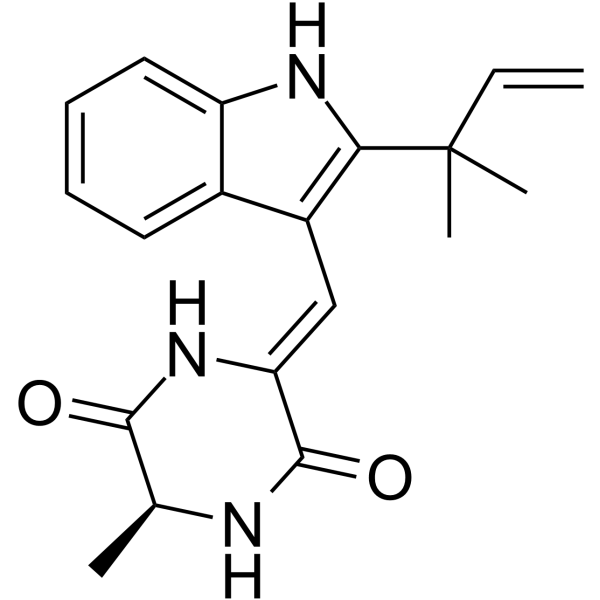
-
GC65200
Nevanimibe hydrochloride
Nevanimibe hydrochloride (PD-132301 hydrochloride) is an orally active and selective acyl-coenzyme A:cholesterol O-acyltransferase 1 (ACAT1) inhibitor with an EC50 of 9 nM. Nevanimibe hydrochloride inhibits ACAT2 with an EC50 of 368 nM. Nevanimibe hydrochloride induces cell apoptosis and has the potential for adrenocortical cancer.
-
GC65542
NSC-87877 disodium
NSC-87877 disodium is a potent inhibitor of Shp2 and Shp1 protein tyrosine phosphatases (SH-PTP2 and SH-PTP1), with IC50 values of 0.318 μM, 0.355 μM shp2 and shp1, respectively. NSC-87877 also inhibits dual-specificity phosphatase 26 (DUSP26).
-
GC67792
NSC49652
-
GC52318
Oleic Acid-13C5
An internal standard for the quantification of oleic acid
-
GC63942
Oxysophoridine
Oxysophoridine (Sophoridine N-oxide) is a bioactive alkaloid extracted from the Sophora alopecuroides Linn.
-
GC65253
PCC0208017
PCC0208017 is a microtubule affinity regulating kinases (MARK3/MARK4) inhibitor with IC50s of 1.8 and 2.01?nM, respectively. PCC0208017 has much lower inhibitory activity against MARK1 and MARK2, with IC50s of 31.4 and 33.7?nM, respectively. PCC0208017 suppresses glioma progression?in?vitro?and?in?vivo. PCC0208017 disrupts microtubule dynamics and induces G2/M phase cell cycle arrest and cell apoptosis. PCC0208017 demonstrates robust antitumor activity?in?vivo?and displays good BBB permeability.
-
GC69705
Physalin A
Physalin A is a withanolide isolated from Physalis alkekengi var franchetii. It induces apoptosis and upregulates the expression of caspase-3 and caspase-8. Physalin A also induces autophagy, which has been found to counteract apoptosis in HT1080 cells. Physalin A has potential for researching cancer diseases.
-
GC52104
Ponatinib (hydrochloride)
An inhibitor of native and mutant Bcr-Abl
-
GC65066
Prodigiosin hydrochloride
Prodigiosin (Prodigiosine) hydrochloride is a red pigment produced by bacteria as a bioactive secondary metabolite.
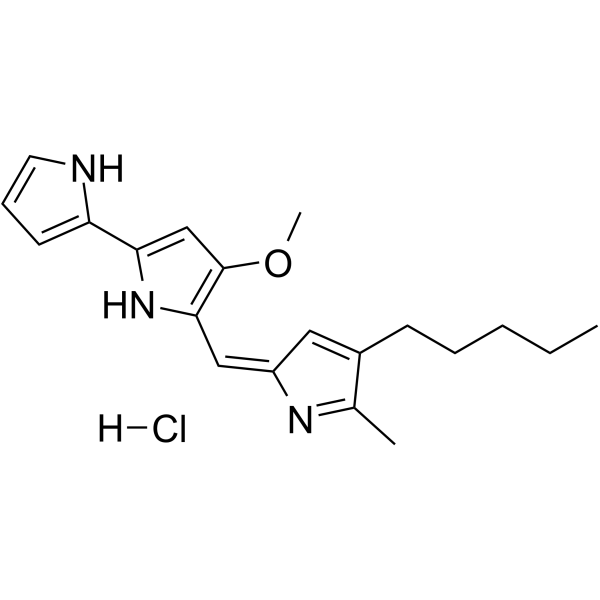
-
GC65555
PROTAC FLT-3 degrader 1
PROTAC FLT-3 degrader 1 is a von Hippel-Lindau-based PROTAC FLT-3 internal tandem duplication (ITD) degrader with an IC50 0.6 nM. Anti-proliferative activity; apoptosis induction.
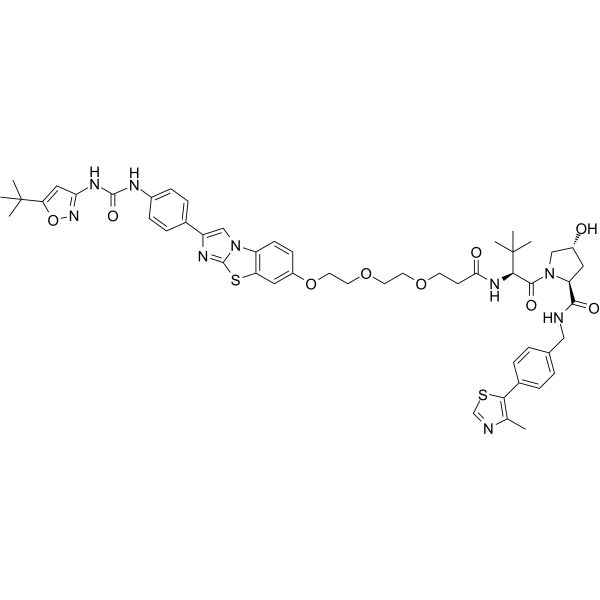
-
GC63916
PROTAC-O4I2
PROTAC-O4I2 is a PROTAC targets splicing factor 3B1 (SF3B1). PROTAC-O4I2 induces FLAG-SF3B1 degradation with an IC50 value of 0.244 μM in K562 cells. PROTAC-O4I2 also induces cellular apoptosis in K562 WT cells.
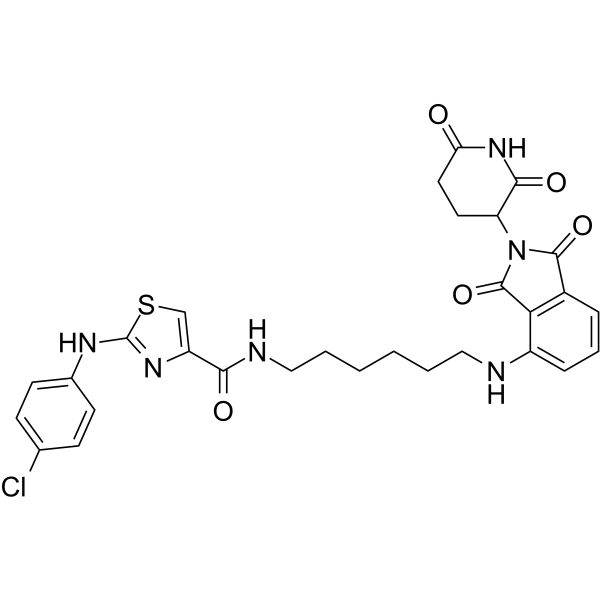
-
GC63860
Rapanone
Rapanone is a natural benzoquinone.
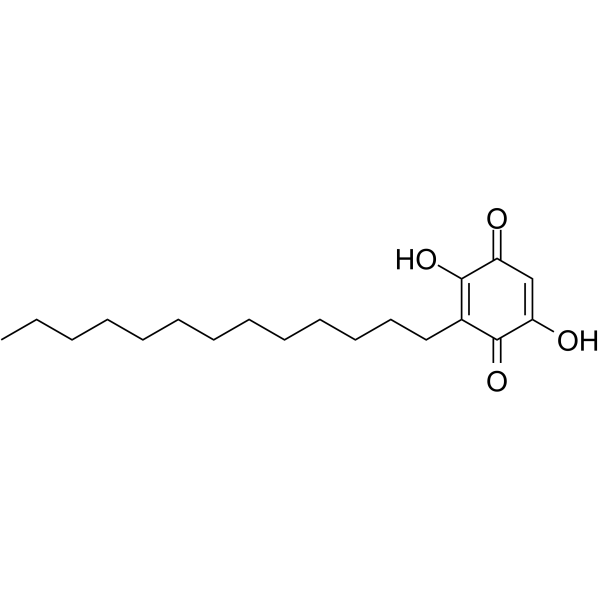
-
GC52196
RGD Peptide
RGD Peptide acts as an inhibitor of integrin-ligand interactions and plays an important role in cell adhesion, migration, growth, and differentiation.
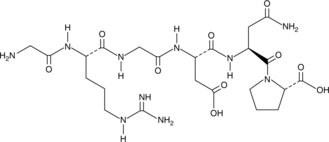
-
GC64995
RIPGBM
RIPGBM is a selective inducer of apoptosis in glioblastoma multiforme (GBM) cancer stem cells (CSCs) with an EC50 of ≤500 nM.
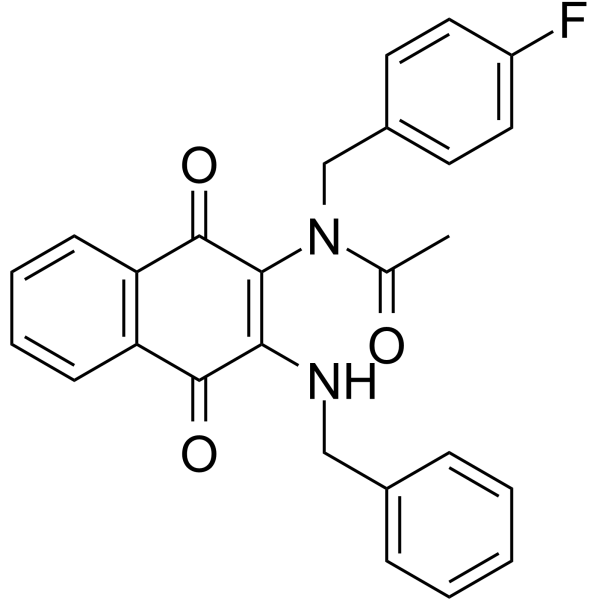
-
GC64331
Ritonavir-13C,d3
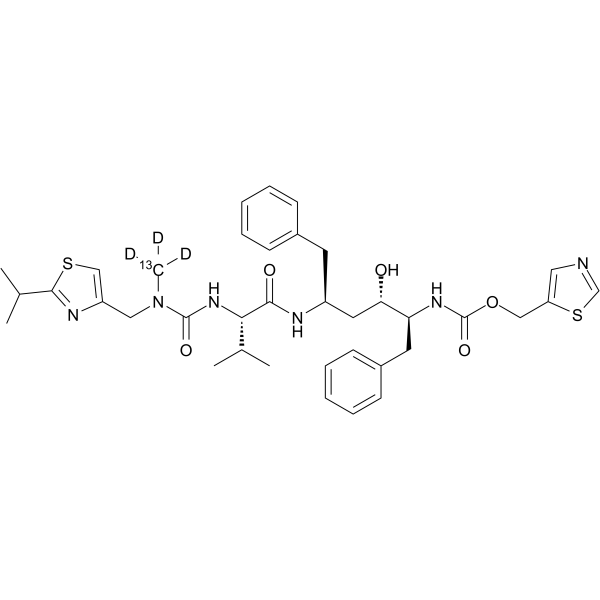
-
GC63979
Ro24-7429
Ro24-7429 is a potent and orally active HIV-1 transactivator protein Tat antagonist.
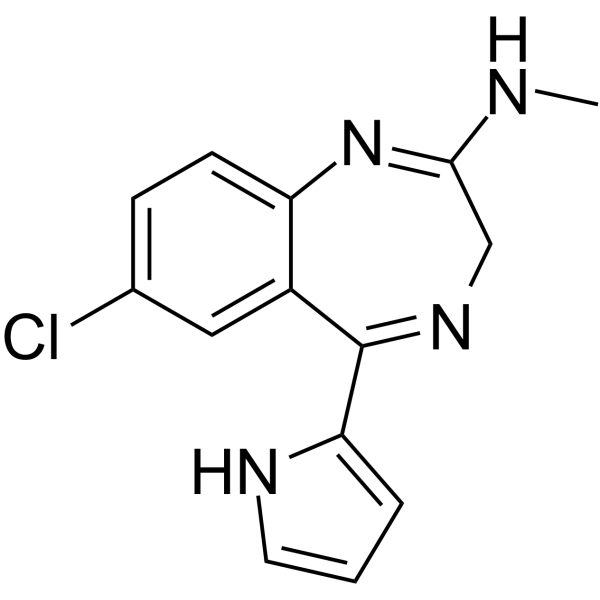
-
GC69841
Rubropunctatin
Rubropunctatin is an orange nitrogen-containing ketone pigment isolated from the extract of red yeast rice (Monascus purpureus). Rubropunctatin has anti-inflammatory, immunosuppressive and antioxidant properties, as well as anti-tumor activity.
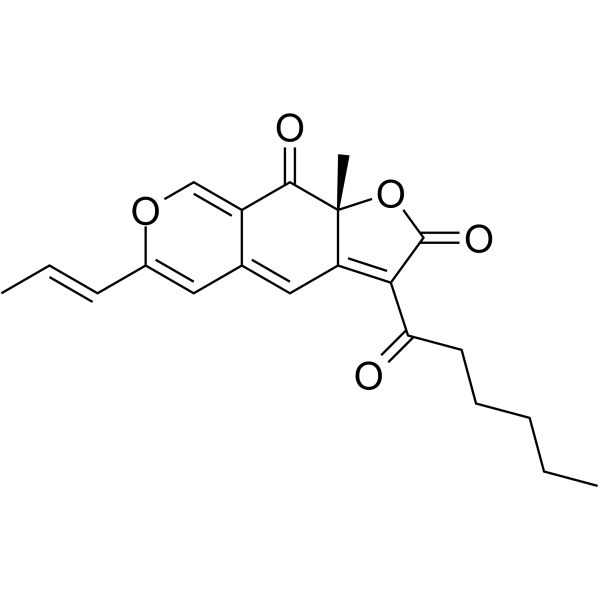
-
GC52115
S-99
An inhibitor of ASK1
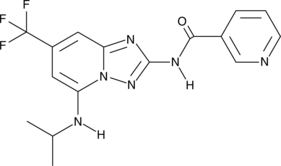
-
GC63933
S-Allylmercaptocysteine
S-allylmercaptocysteine, an organic sulfur compound extracted from garlic, has anti-inflammatory and anti-oxidative effects for various pulmonary diseases.
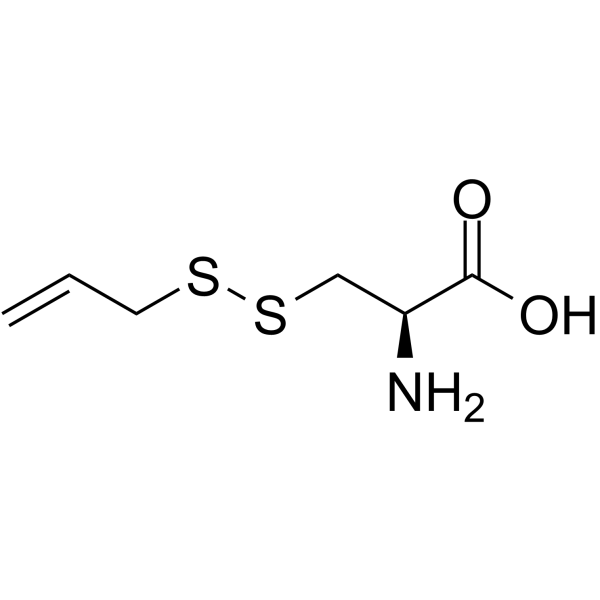
-
GC65307
S130
S130 is a high affinity, selective inhibitor of ATG4B (a major cysteine protease) with an IC50 of 3.24 ?M. S130 suppresses autophagy flux.
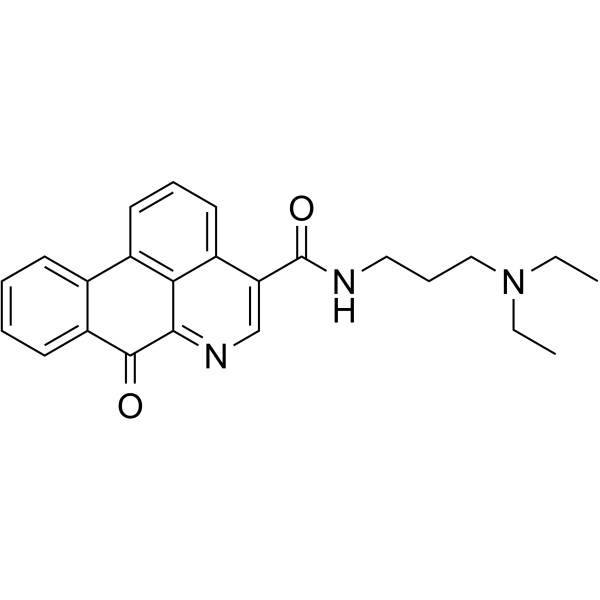
-
GC64303
S2116
S2116, a N-alkylated tranylcypromine (TCP) derivative, is a potent lysine-specific demethylase 1 (LSD1) inhibitor. S2116 increases H3K9 methylation and reciprocal H3K27 deacetylation at super-enhancer regions. S2116 induces apoptosis in TCP-resistant T-cell acute lymphoblastic leukemia (T-ALL) cells by repressing transcription of the NOTCH3 and TAL1 genes. S2116 significantly retardes the growth of T-ALL cells in xenotransplanted mice.
-
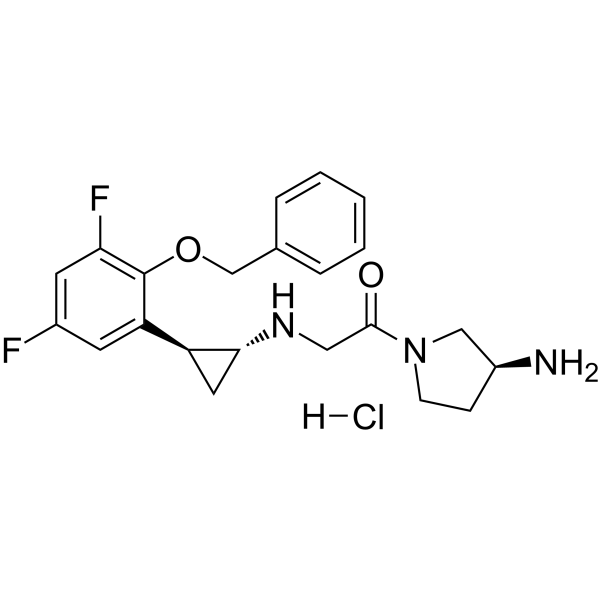
GC49632
SACLAC
An inhibitor of acid ceramidase
-
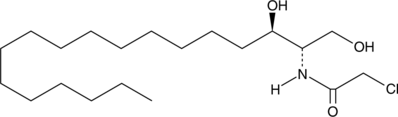
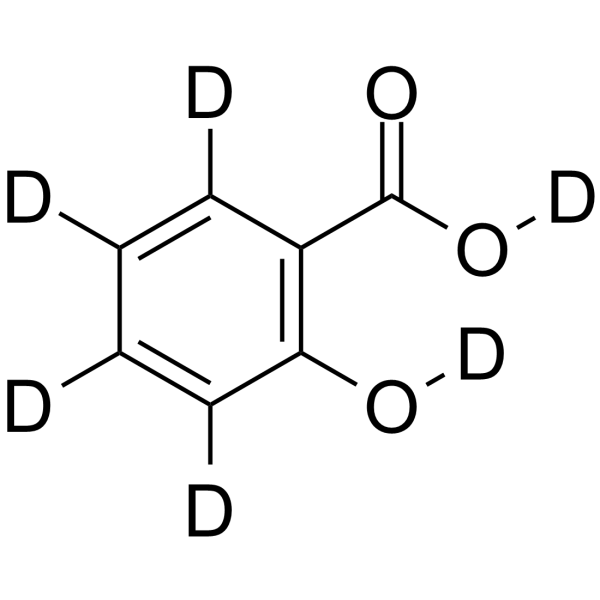
GC64032
Salicylic acid-d6
Salicylic acid-D6 (2-Hydroxybenzoic acid-D6) is a deuterium labeled Salicylic acid.
-
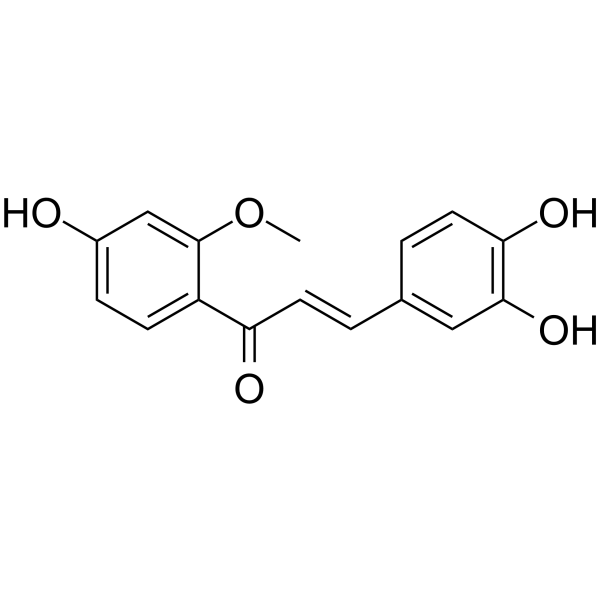
GC64645
Sappanchalcone
Sappanchalcone, a flavonoid isolated from Caesalpinia sappan L., induces caspase-dependent and AIF-dependent apoptosis in human colon cancer cells.
-
GC69899
SIRT6 activator 12q
SIRT6 activator 12q is an effective, selective, and orally active SIRT6 activator with IC50 values of 171.20, >200, >200, >200, and 0.58 μM for SIRT1, SIRT2, SIRT3, SIRT5 and SIRT6 respectively. It inhibits cell growth and migration and induces apoptosis (cell death) and G2 phase cell cycle arrest. Additionally, it exhibits anti-cancer activity.
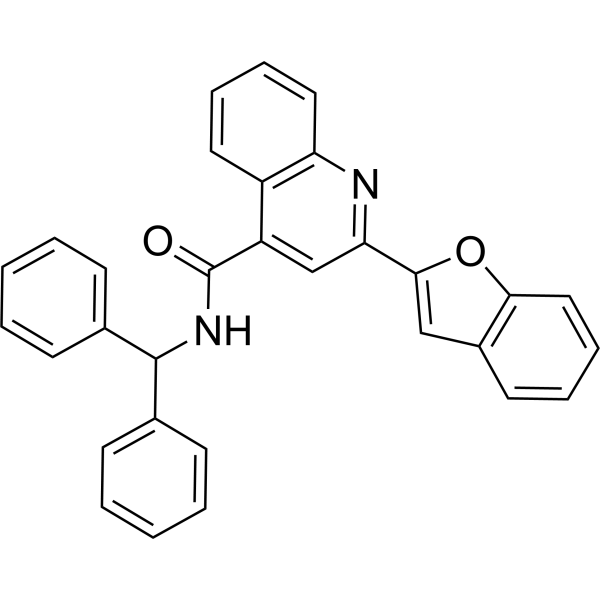
-
GC69912
SLC7A11-IN-1
SLC7A11-IN-1 is an effective inhibitor of SLC7A11. It exhibits anti-proliferative activity and inhibits cell invasion and metastasis. SLC7A11-IN-1 induces apoptosis and cell cycle arrest in the S phase. It has anti-tumor activity.
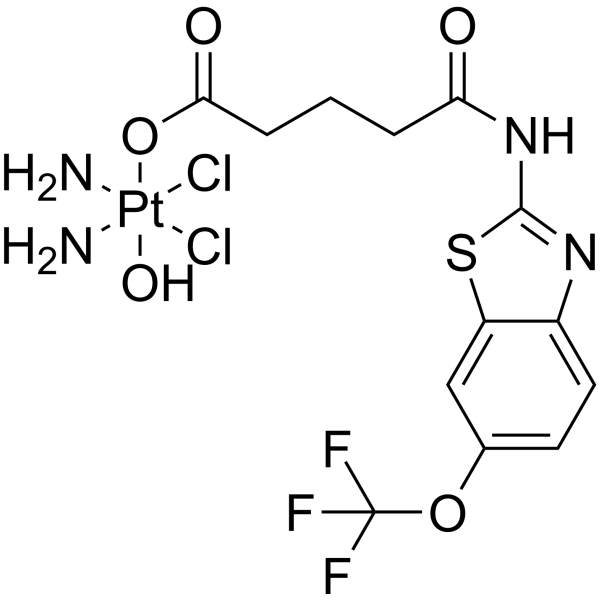
-
GC64223
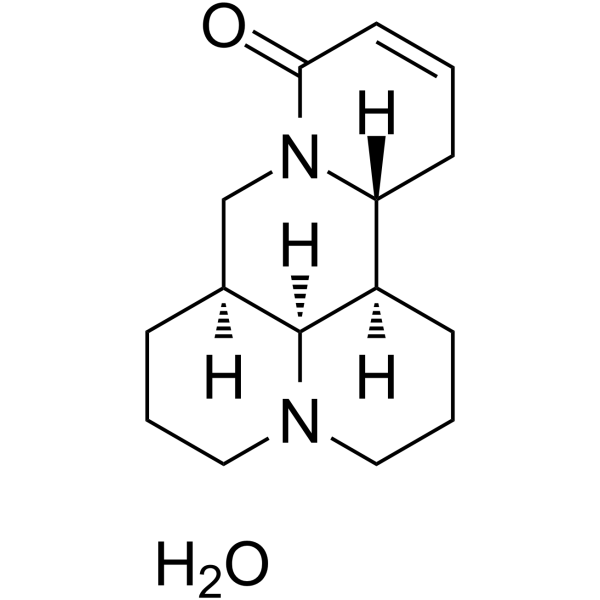
Sophocarpine monohydrate
Sophocarpine (monohydrate) is one of the significant alkaloid extracted from the traditional herb medicine Sophora flavescens which has many pharmacological properties such as anti-virus, anti-tumor, anti-inflammatory.