Signaling Pathways
The antibody-drug conjugate (ADC), a humanized or human monoclonal antibody conjugated with highly cytotoxic small molecules (payloads) through chemical linkers, is a novel therapeutic format and has great potential to make a paradigm shift in cancer chemotherapy. The three components of the ADC together give rise to a powerful oncolytic agent capable of delivering normally intolerable cytotoxins directly to cancer cells, which then internalize and release the cell-destroying drugs. At present, two ADCs, Adcetris and Kadcyla, have received regulatory approval with >40 others in clinical development.
ADCs are administered intravenously in order to prevent the mAb from being destroyed by gastric acids and proteolytic enzymes. The mAb component of the ADC enables it to circulate in the bloodstream until it finds and binds to tumor-specific cell surface antigens present on target cancer cells. Linker chemistry is an important determinant of the safety, specificity, potency and activity of ADCs. Linkers are designed to be stable in the blood stream (to conform to the increased circulation time of mAbs) and labile at the cancer site to allow rapid release of the cytotoxic drug. First generation ADCs made use of early cytotoxins such as the anthracycline, doxorubicin or the anti-metabolite/antifolate agent, methotrexate. Current cytotoxins have far greater potency and can be divided into three main groups: auristatins, maytansines and calicheamicins.
The development of site-specific conjugation methodologies for constructing homogeneous ADCs is an especially promising path to improving ADC design, which will open the way for novel cancer therapeutics.
References:
[1] Tsuchikama K, et al. Protein Cell. 2016 Oct 14. DOI:10.1007/s13238-016-0323-0.
[2] Peters C, et al. Biosci Rep. 2015 Jun 12;35(4). pii: e00225. doi: 10.1042/BSR20150089.
Targets for Signaling Pathways
- Proteases(29)
- Apoptosis(824)
- Chromatin/Epigenetics(15)
- Metabolism(336)
- MAPK Signaling(26)
- Tyrosine Kinase(73)
- DNA Damage/DNA Repair(49)
- PI3K/Akt/mTOR Signaling(38)
- Microbiology & Virology(42)
- Cell Cycle/Checkpoint(168)
- Ubiquitination/ Proteasome(24)
- JAK/STAT Signaling(10)
- TGF-β / Smad Signaling(21)
- Angiogenesis(59)
- GPCR/G protein(3)
- Stem Cell(19)
- Membrane Transporter/Ion Channel(161)
- Cancer Biology(366)
- Endocrinology and Hormones(152)
- Neuroscience(367)
- Obesity, Appetite Control & Diabetes(17)
- Peptide Inhibitors and Substrate(2)
- Other Signal Transduction(156)
- Immunology/Inflammation(934)
- Cardiovascular(98)
- Vitamin D Related(0)
- Antibody-drug Conjugate/ADC Related(0)
- PROTAC(212)
- Ox Stress Reagents(24)
- Others(4010)
- Antiparasitics(6)
- Toxins(62)
Products for Signaling Pathways
- Cat.No. Product Name Information
-
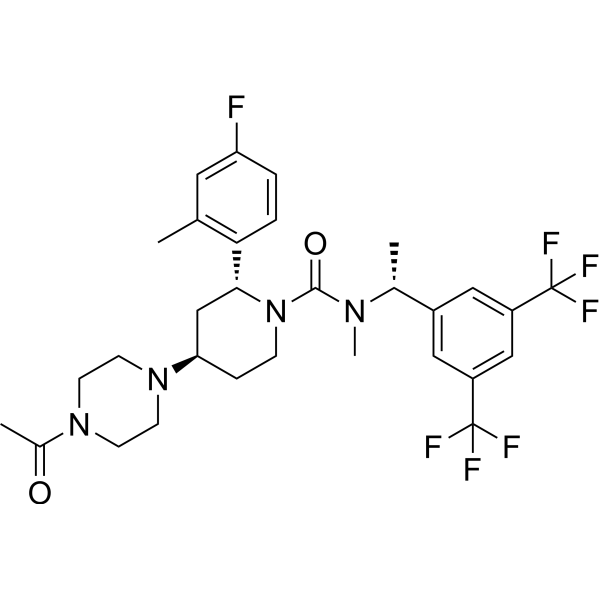
GC66692
(+)-Biotin-SLC
(+)-Biotin-SLC is an alkyl chain-based PROTAC linker that can be used in the synthesis of PROTACs.
-
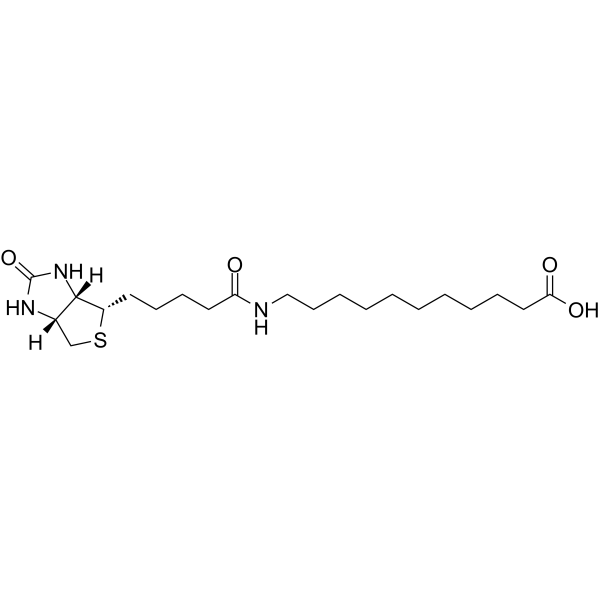
GN10605
(+)-Catechin hydrate
-
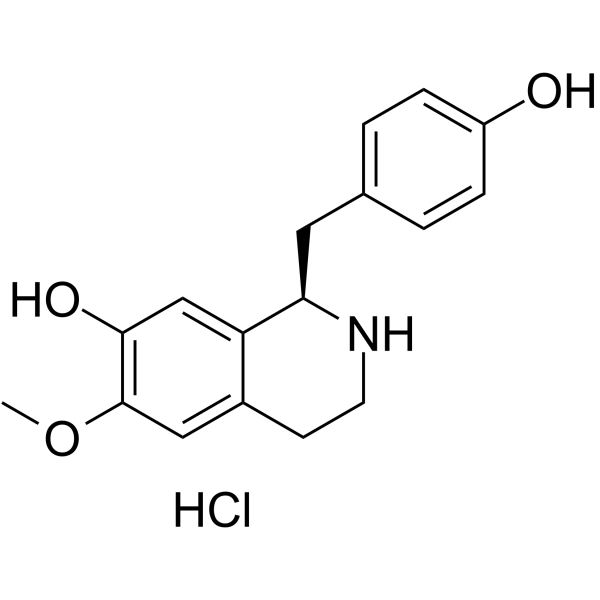
GC63999
(+)-Coclaurine hydrochloride
(+)-Coclaurine ((+)-(R)-Coclaurine) hydrochloride, benzyltetrahydroisoquinoline alkaloid isolated from a variety of plant sources.
-
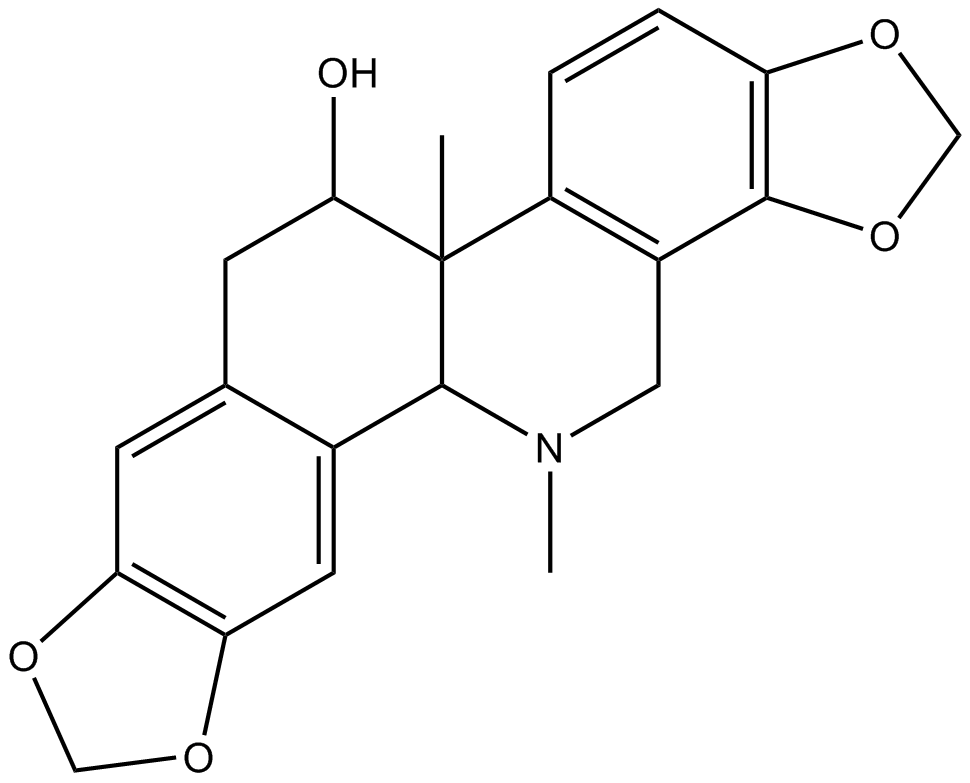
GN10654
(+)-Corynoline
-
GC61765
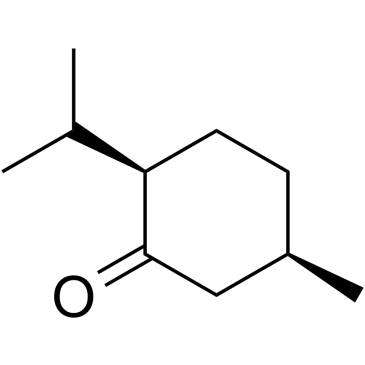
(+)-Isomenthone
(+)-Isomenthone is an isomenthone isolated from Ziziphora clinopodioides Lam.
-
GC64207
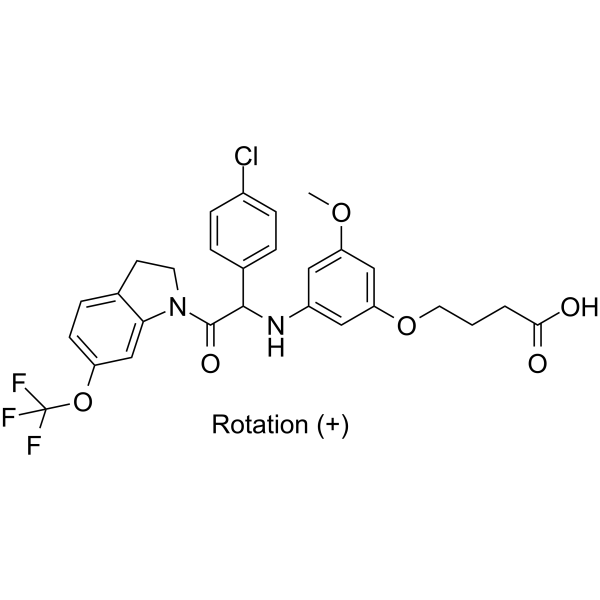
(+)-JNJ-A07
(+)-JNJ-A07 is a highly potent, orally active pan-serotype dengue virus inhibitor targeting the NS3-NS4B interaction.
-
GC61595
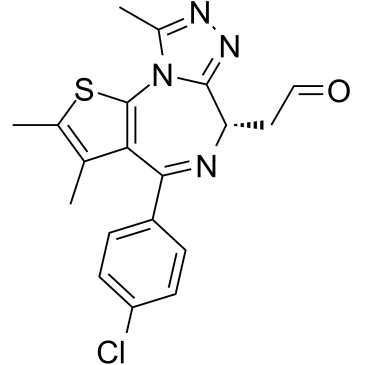
(+)-JQ-1-aldehyde
(+)-JQ-1-aldehyde is the aldehyde form of (+)-JQ1. (+)-JQ-1-aldehyde can be uesd as a precursor to synthesize PROTACs, which targets BET bromodomains.
-
GC61647
(+)-Longifolene
(+)-Longifolene is a sesquiterpenoid and a metabolite in rabbits.
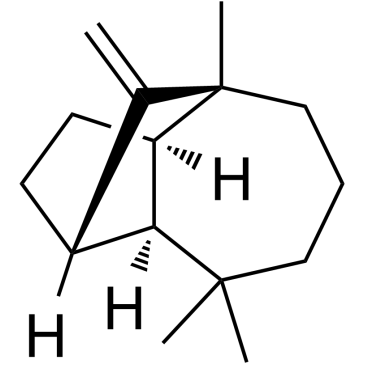
-
GC67931
(+)-Medioresinol
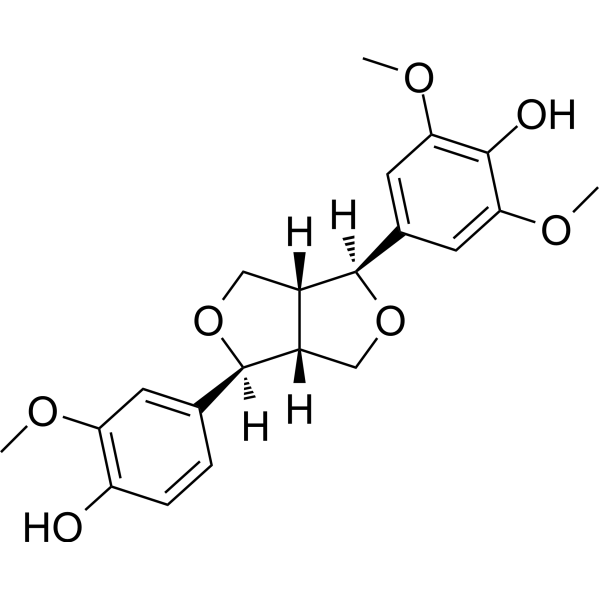
-
GC16616
(+)-MK 801
-
GC68210
(+)-Norfenfluramine

-
GC63969
(+)-Schisandrin B
(+)-Schisandrin B is an enantiomer of Schisandrin B.
-
GC48783
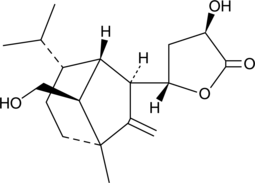
(+)-Sorokinianin
A phytotoxic fungal metabolite
-
GC69720
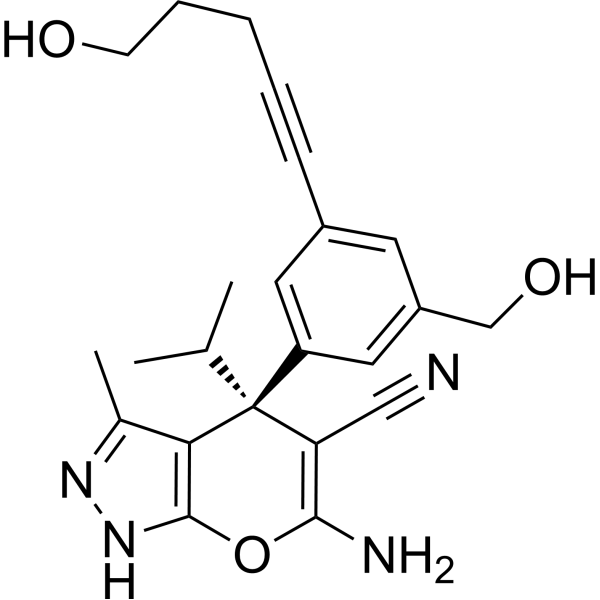
(+)SHIN2
(+)SHIN2 is a serine hydroxymethyltransferase (SHMT) inhibitor, and its in vivo target can be traced using 13C-serine. (+)SHIN2 increases the survival rate of mice with primary acute T-cell lymphoblastic leukemia (T-ALL) driven by Notch1 and has a synergistic effect with Methotrexate.
-
GC49502
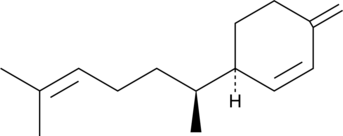
(-)-β-Sesquiphellandrene
A sesquiterpene with antiviral and anticancer activities
-
GC61646
(-)-Camphoric acid
-
GC48635
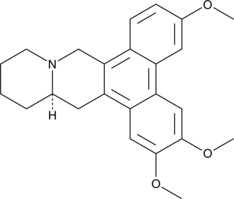
(-)-Cryptopleurine
An alkaloid with diverse biological activities
-
GC63940
(-)-Denudatin B
(-)-Denudatin B is an antiplatelet agent.

-
GC14049
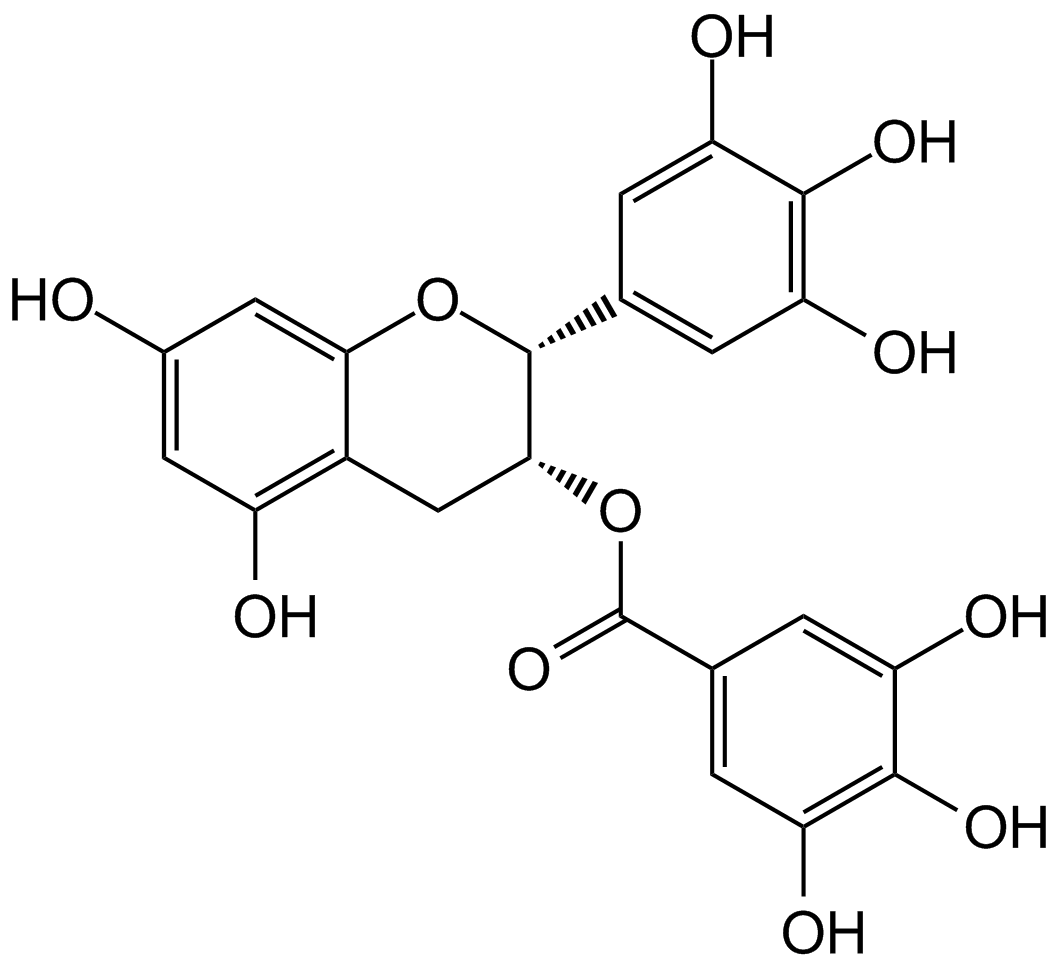
(-)-Epigallocatechin gallate (EGCG)
(-)-Epigallocatechin Gallate sulfate (EGCG) is a major polyphenol in green tea that inhibits cell proliferation and induces apoptosis. In addition, it inhibits the activity of glutamate dehydrogenase 1/2 (GDH1/2, GLUD1/2) ..
-
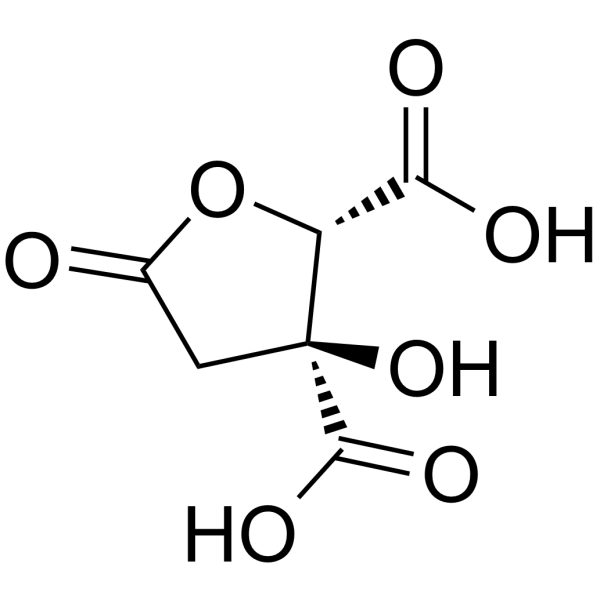
GC63570
(-)-Hydroxycitric acid lactone
(-)-Hydroxycitric acid lactone (Garcinia lactone) is an anti-obesity agent and a popular weight loss food supplement.
-
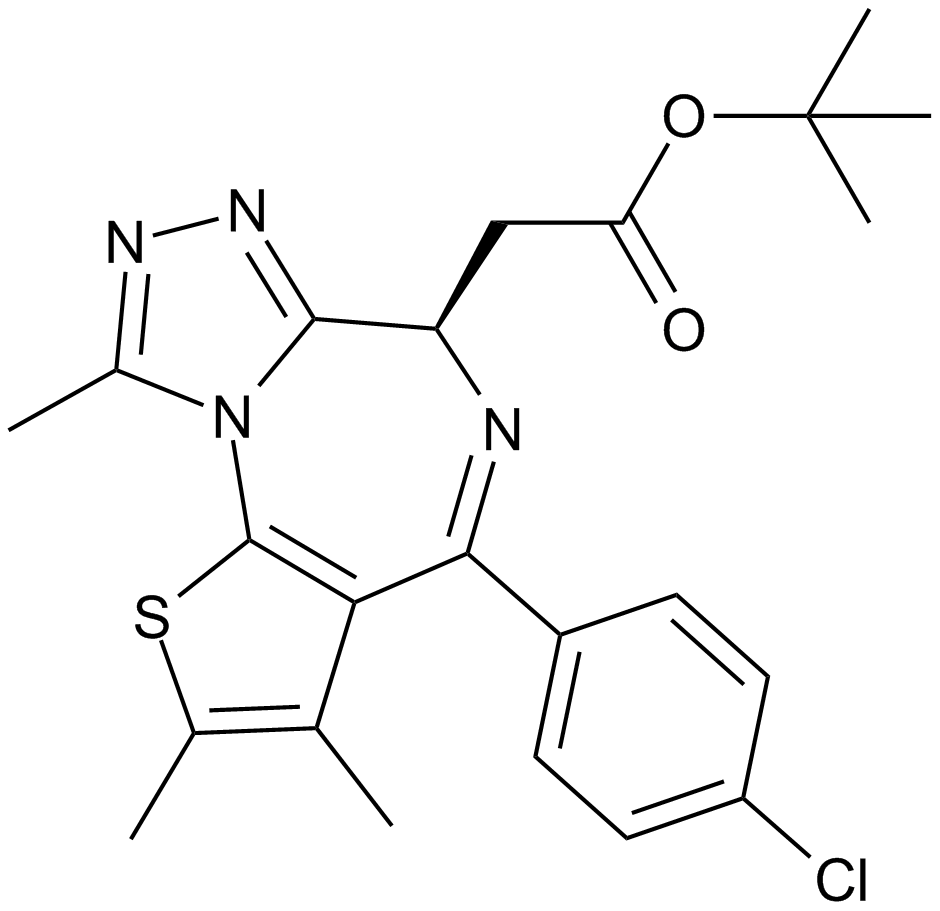
GC13822
(-)-JQ1
A selective inhibitor of BET bromodomains
-
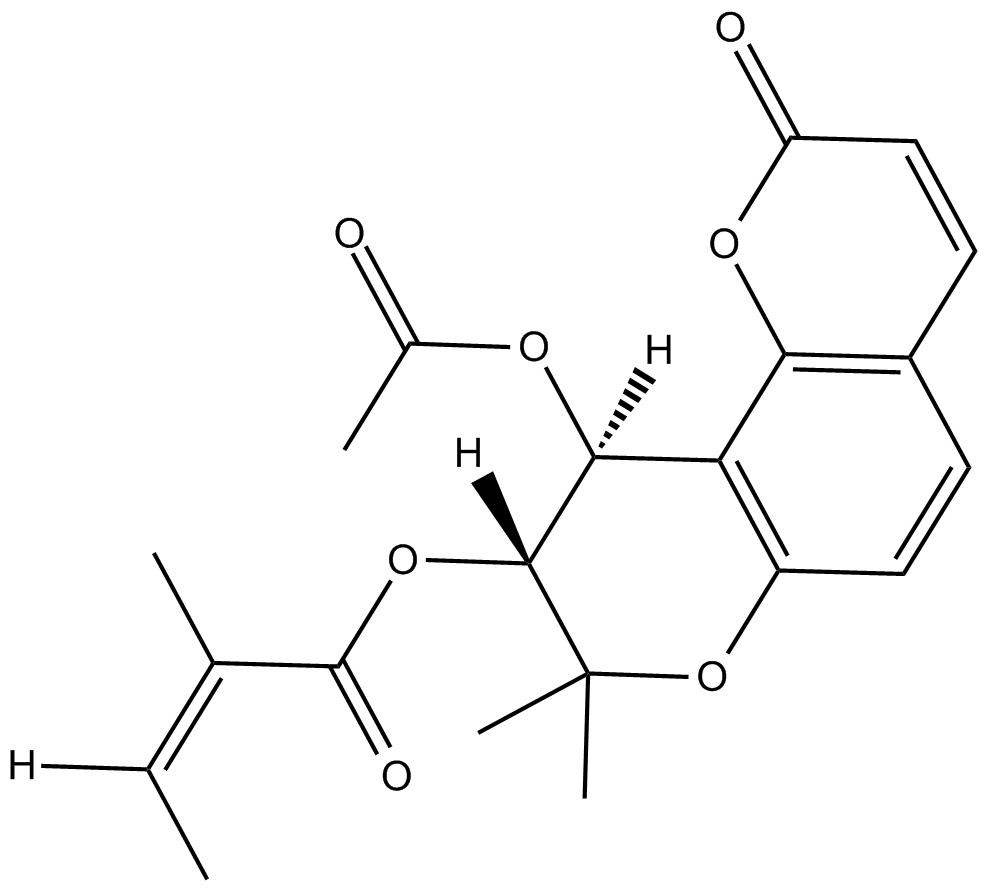
GN10445
(-)-pareruptorin A
-
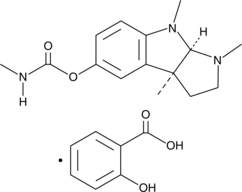
GC48551
(-)-Physostigmine (salicylate)
An alkaloid and cholinergic agent
-
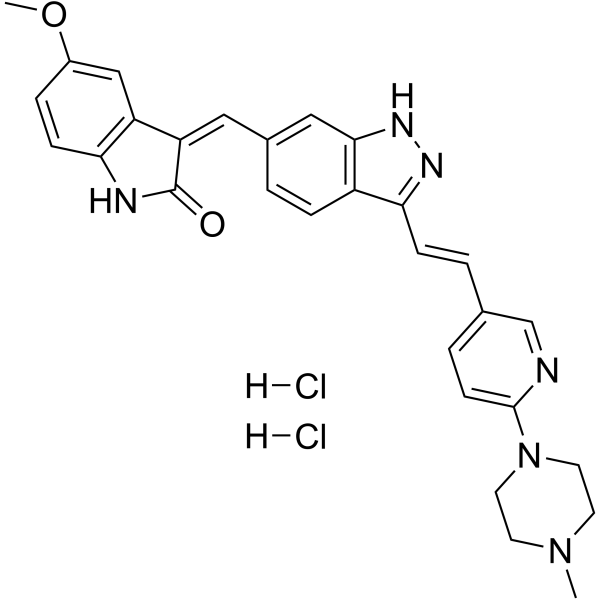
GC62728
(1E)-CFI-400437 dihydrochloride
(1E)-CFI-400437 dihydrochloride is a potent PLK4 (IC50= 0.6 nM) inhibitor and selective against other members of the PLK family (>10 μM). (1E)-CFI-400437 dihydrochloride inhibits Aurora A, Aurora B, KDR and FLT-3 with IC50s of 0.37, 0.21, 0.48, and 0.18 μM, respectively. Antiproliferative activity.
-
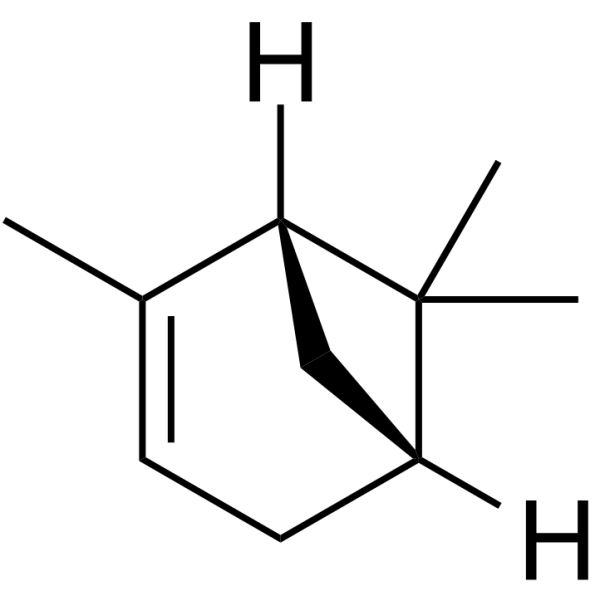
GC62729
(1R)-α-Pinene
(1R)-α-Pinene is a volatile monoterpene with antimicrobial activities.
-
GC65363
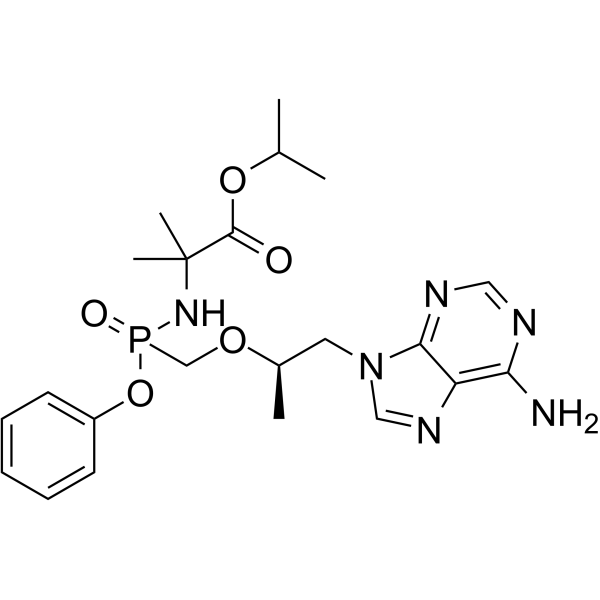
(1R)-Tenofovir amibufenamide
(1R)-Tenofovir amibufenamide ((1R)-HS-10234) is the isomer of Tenofovir amibufenamide, is an orally active antiviral agent.
-
GC64062
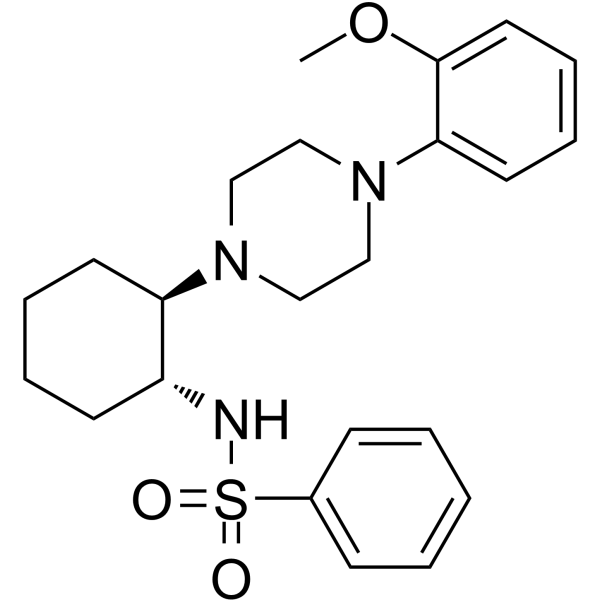
(1R,2R)-ML-SI3
(1R,2R)-ML-SI3 is a potent inhibitor of both TRPML1 and TRPML2 (IC50 values of 1.6 and 2.3 μM) and a weak inhibitor (IC50 12.5 μM) of TRPML3.
-
GC70081
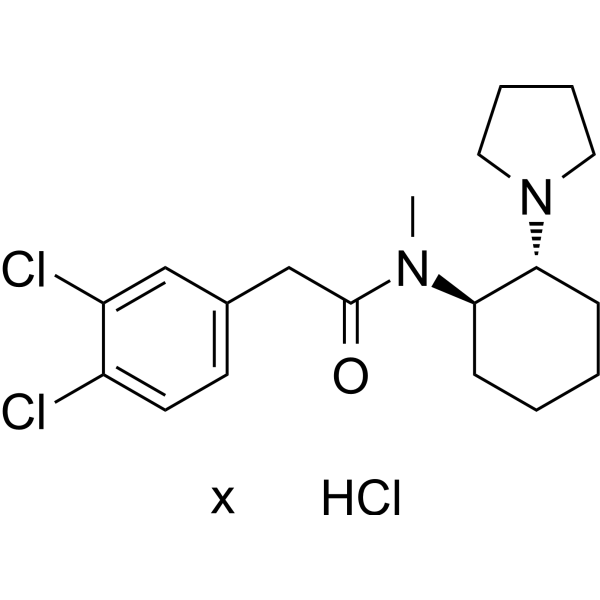
(1R,2R)-U-50488 hydrochloride
(1R,2R)-U-50488 hydrochloride is the absolute configuration of (±)-U-50488 hydrochloride. (±)-U-50488 hydrochloride is a κ opioid receptor agonist.
-
GC67880
(1R,2S)-Xeruborbactam disodium

-
GC62730
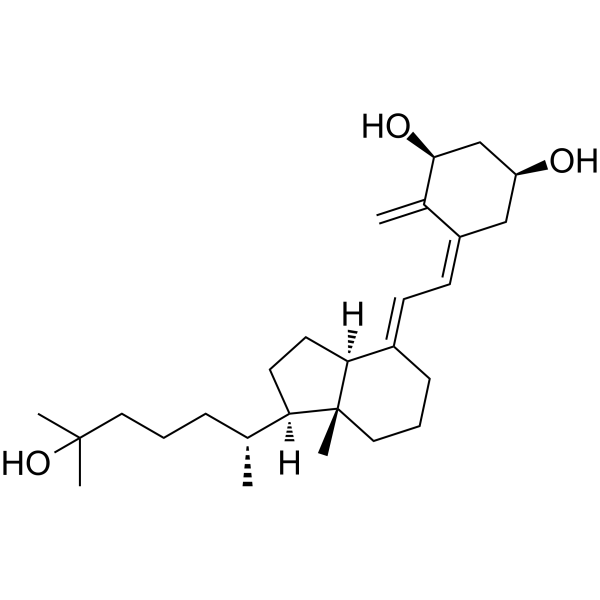
(1S)-Calcitriol
(1S)-Calcitriol (1α,25-Dihydroxy-3-epi-vitamin-D3) is a natural metabolite of 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3).
-
GC65547
(1S,2R)-Alicapistat
(1S,2R)-Alicapistat ((1S,2R)-ABT-957) is an orally active selective inhibitor of human calpains 1 and 2 for the potential application of Alzheimer's disease (AD).
-
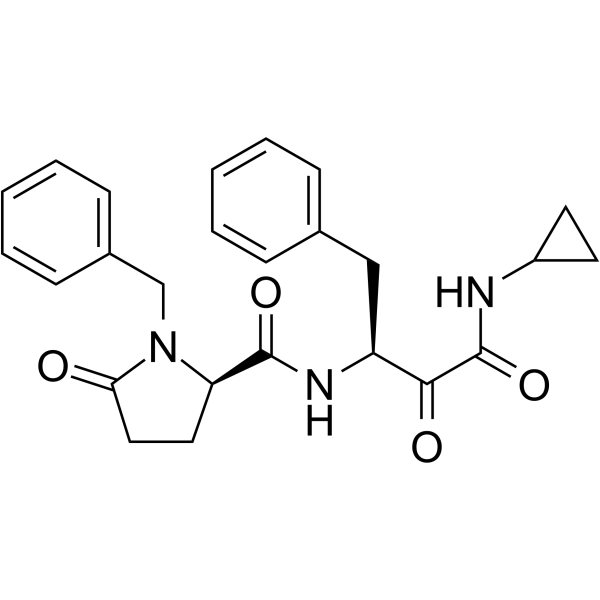
GC62193
(1S,2S)-Bortezomib
(1S,2S)-Bortezomib is an enantiomer of Bortezomib. Bortezomib is a cell-permeable, reversible, and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki of 0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is an anti-cancer agent and the first therapeutic proteasome inhibitor to be used in humans.
-
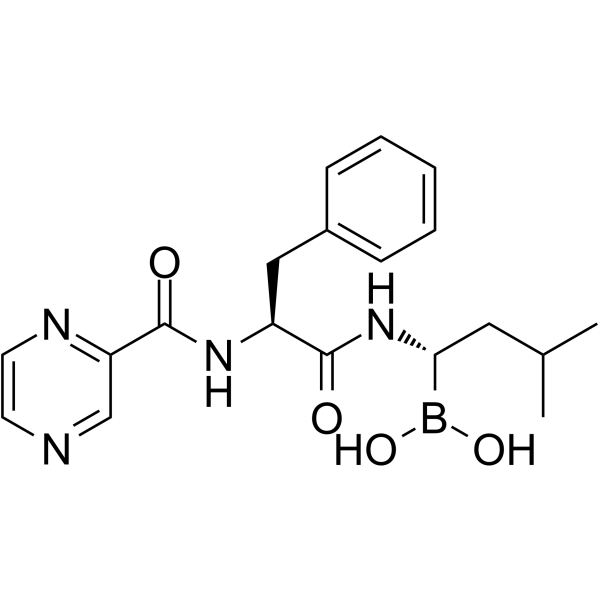
GC65885
(1S,3R)-GNE-502
(1S,3R)-GNE-502 (compound 179) is a potent ERα degrader with an EC50 value of 13 nM against ERα in MCF7 HCS. (1S,3R)-GNE-502 can be used to research cancer related with estrogen receptor.
-
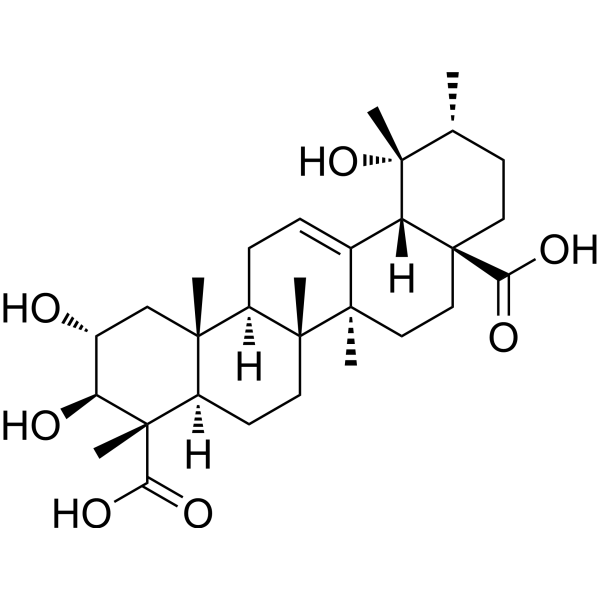
GC68525
(2α,3β,4α)-2,3,19-Trihydroxyurs-12-ene-23,28-dioic acid
(2α,3β,4α)-2,3,19-Trihydroxyurs-12-ene-23,28-dioic acid is a saponin that can be isolated from Rubus ellipticus var. obcordatus. It inhibits α-Glucosidase with an IC50 of 1.68 mM.
-
GC63900
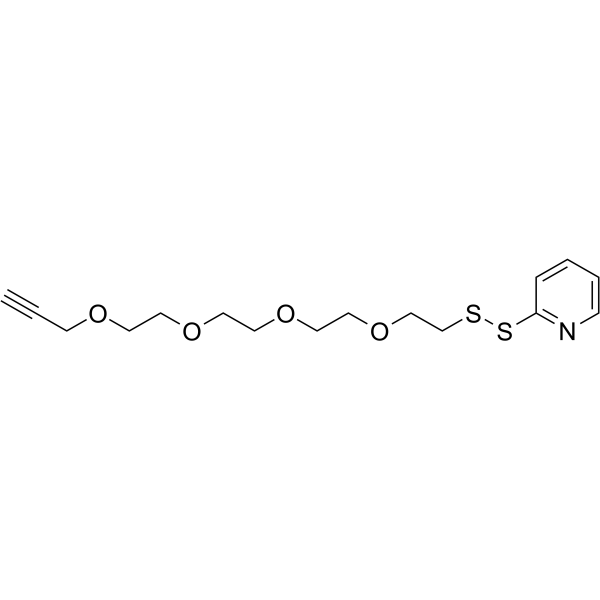
(2-pyridyldithio)-PEG1-hydrazine
(2-pyridyldithio)-PEG1-hydrazine is a cleavable 1 unit PEG ADC linker used in the synthesis of antibody-drug conjugates (ADCs).
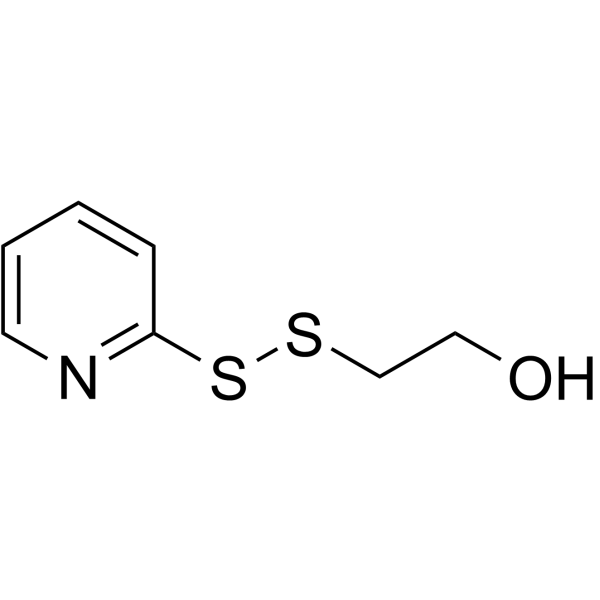
-
GC65777
(2-Pyridyldithio)-PEG2-Boc
(2-Pyridyldithio)-PEG2-Boc is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
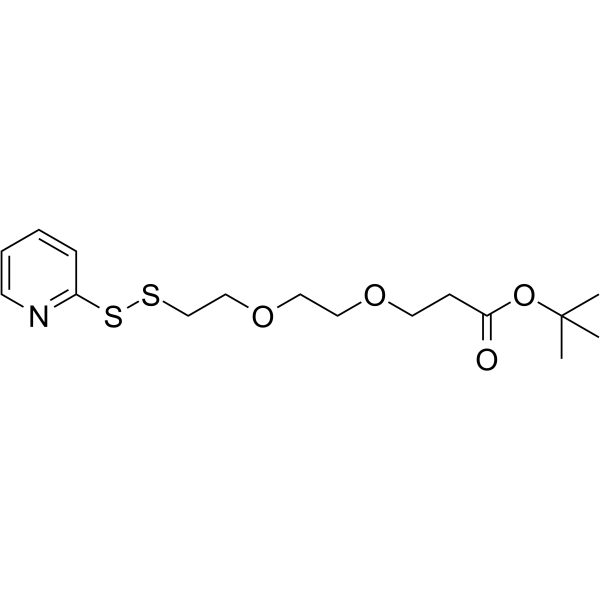
-
GC65623
(2-pyridyldithio)-PEG4 acid
(2-pyridyldithio)-PEG4 acid is a cleavable 4 unit PEG ADC linker used in the synthesis of antibody-drug conjugates (ADCs).
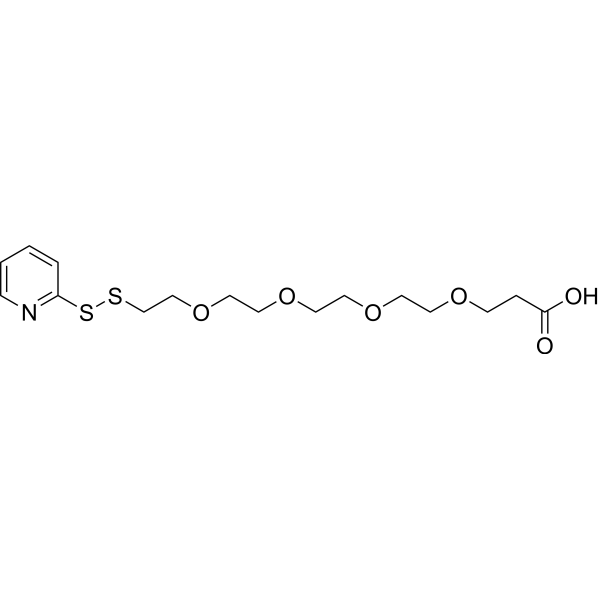
-
GC65703
(2-Pyridyldithio)-PEG4-alcohol
(2-Pyridyldithio)-PEG4-alcohol is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
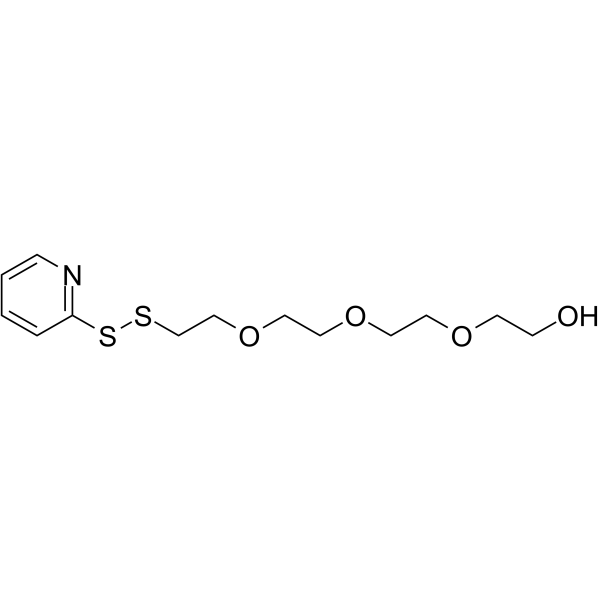
-
GC65337
(2-Pyridyldithio)-PEG4-propargyl
(2-Pyridyldithio)-PEG4-propargyl is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
-
GC11717
(24S)-MC 976
-
GC68509
(25R)-12α-Hydroxyspirost-4-en-3-one
(25R)-12α-Hydroxyspirost-4-en-3-one is a secondary metabolite produced by Nocardia globerula from Hecogenin.
-
GC62731
(2R,3R)-Butane-2,3-diol
(2R,3R)-Butane-2,3-diol is an endogenous metabolite.

-
GC68522
(2R,3S)-Brassinazole
Brassinazole (0.5, 1, 5 μM) significantly caused morphological abnormalities in seedlings similar to BR-deficient mutants. Brassinazole resulted in dwarfism and altered leaf morphology, such as the typical downward curling and dark green appearance seen in Arabidopsis BR-deficient mutants. However, treatment with 10 nM BR reversed the dwarfism.
-
GC62130
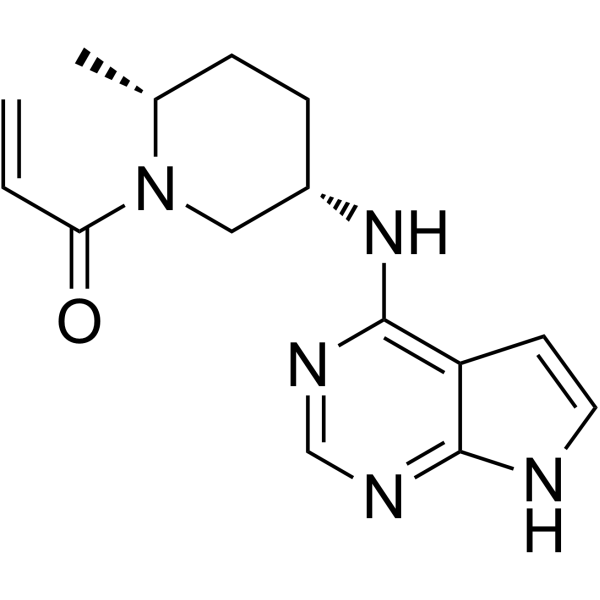
(2R,5S)-Ritlecitinib
(2R,5S)-Ritlecitinib ((2R,5S)-PF-06651600) is a potent and selective JAK3 inhibitor (IC50=144.8 nM) extracted from patent US20150158864A1, example 68.
-
GC68524
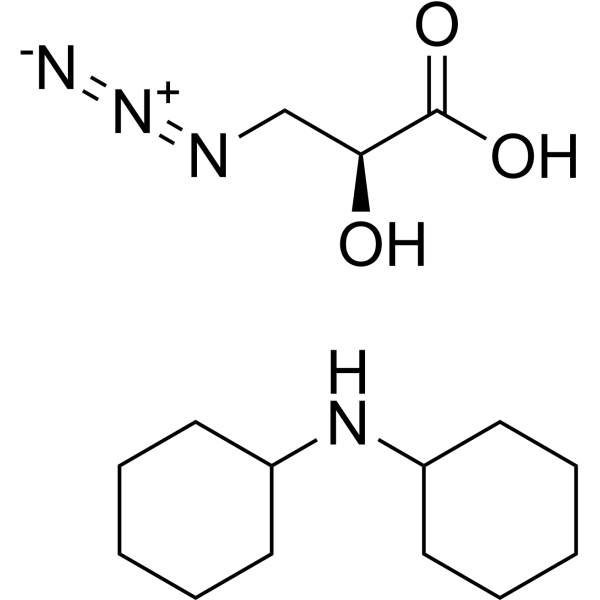
(2S)-N3-IsoSer (DCHA)
"(2S)-N3-IsoSer DCHA" is a click chemistry reagent, which is a chiral α-hydroxy acid containing azide."
-
GC68523
(2S,3R)-Brassinazole
(2S,3R)-Brassinazole is an isomer of brassinazole, which is a compound that inhibits the biosynthesis of brassinosteroids (BR), a class of plant steroids, by acting on the oxidation process from 6-oxo-campestanol to teasterone. (2S,3R)-Brassinazole may be the most active form of brassinazole.

-
GC64260
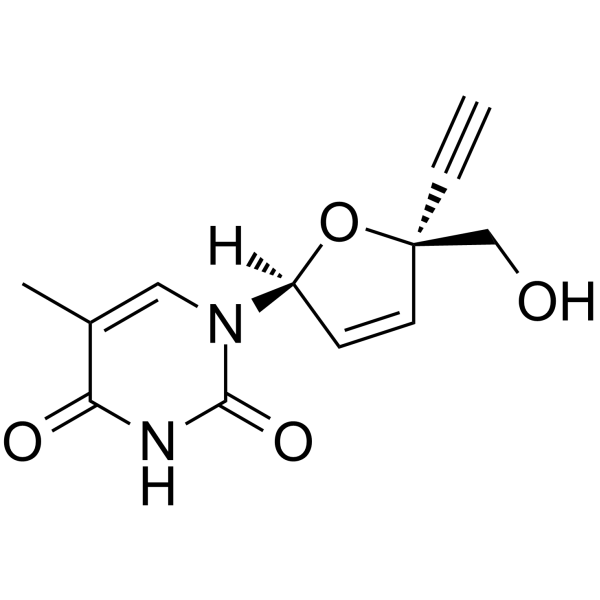
(2S,5S)-Censavudine
(2S,5S)-Censavudine ((2S,5S)-OBP-601) is the (2S,5S)-enantiomer of Censavudine.
-
GC63524
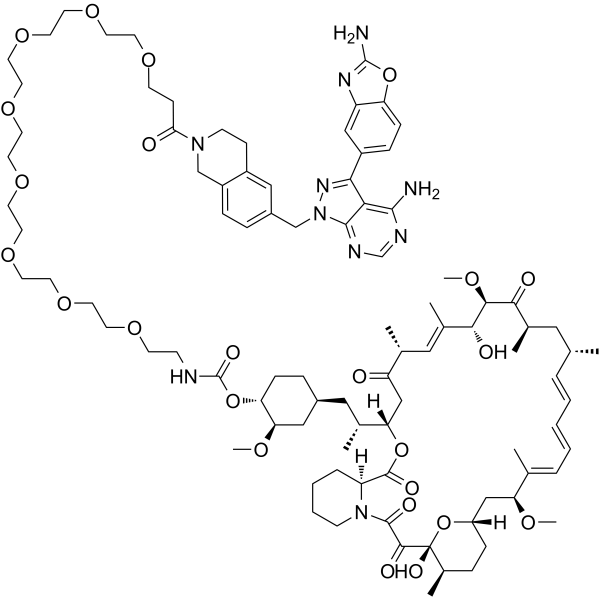
(32-Carbonyl)-RMC-5552
(32-Carbonyl)-RMC-5552 is a potent mTOR inhibitor. (32-Carbonyl)-RMC-5552 inhibits mTORC1 and mTORC2 substrate (p-P70S6K-(T389), p-4E-BP1-(T37/36), AND p-AKT1/2/3-(S473)) phosphorylation with pIC50s of > 9, >9 and between 8 and 9, respectively (patent WO2019212990A1, example 2).
-
GC49690
(3R,5R)-Rosuvastatin (calcium salt)
A potential impurity found in bulk preparations of rosuvastatin
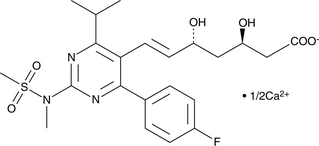
-
GC68317
(3S,5R)-Fluvastatin-d6 sodium
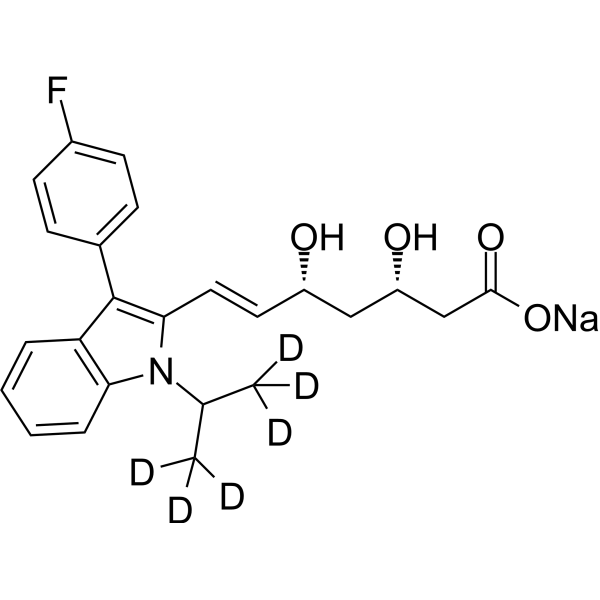
-
GC65358
(4-NH2)-Exatecan
(4-NH2)-Exatecan, a topoisomerase inhibitor derivative extracted from patent US20200306243A1, compound A. (4-NH2)-Exatecan can be used in the synthesis of antibody-drug conjugates (ADCs).

-
GC62710
(5α)-Stigmastane-3,6-dione
(5α)-Stigmastane-3,6-dione is a naturally occurring sterol that could be isolated from fruits of Ailanthus altissima Swingle.

-
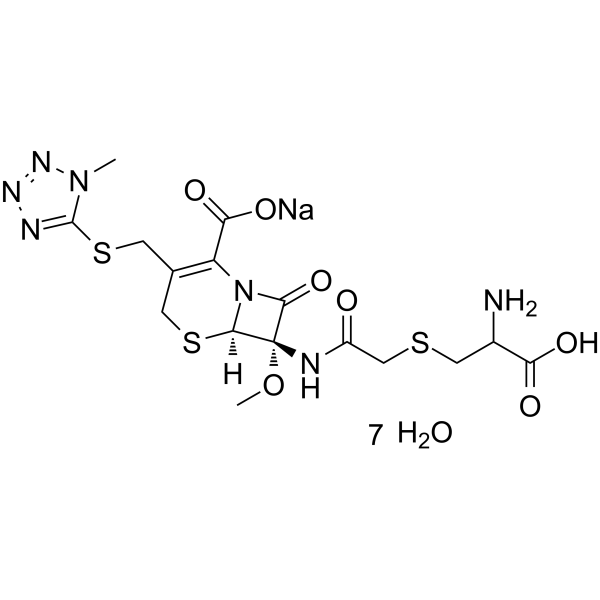
GC63685
(6R,7S)-Cefminox sodium heptahydrate
(6R,7S)-Cefminox sodium heptahydrate is an isomer of Cefminox sodium heptahydrate.
-
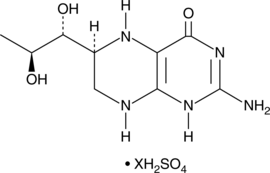
GC49519
(6S)-5,6,7,8-tetrahydro-L-Biopterin (sulfate)
A diastereomer of (6R)-5,6,7,8-tetrahydro-L-biopterin
-
GC52100
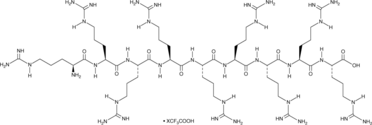
(Arg)9 (trifluoroacetate salt)
A cationic cell-penetrating peptide
-
GC52442
(D)-PPA 1 (trifluoroacetate salt)
An inhibitor of the PD-1-PD-L1 protein-protein interaction
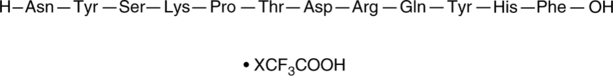
-
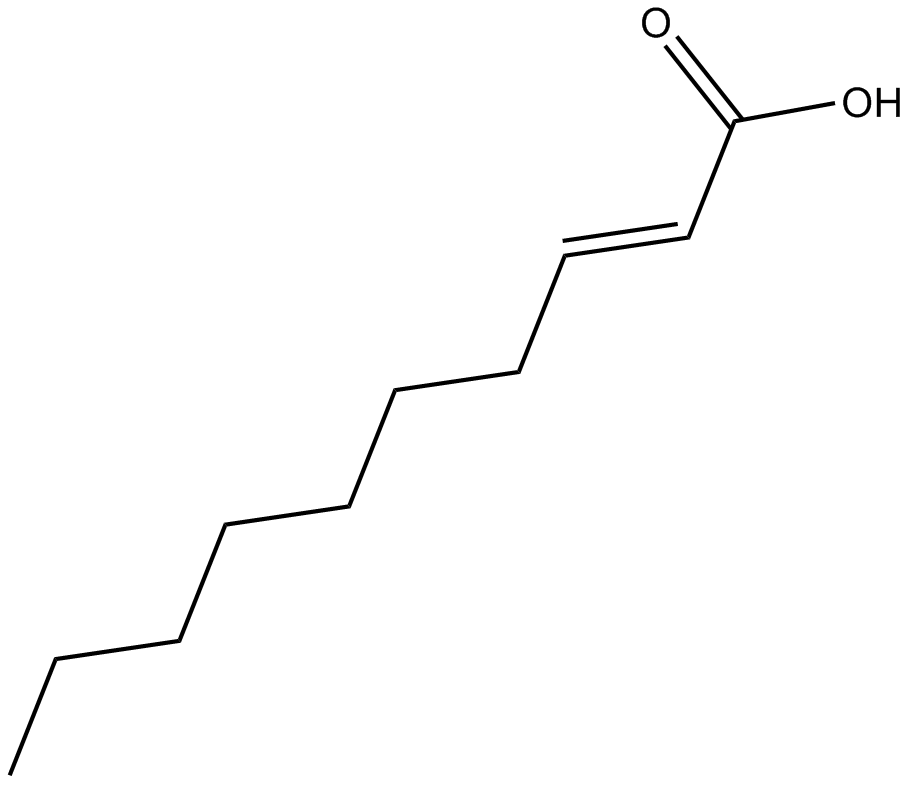
GC15373
(E)-2-Decenoic acid
An unsaturated fatty acid found in royal jelly
-
GC66255
(E)-3,4,5-Trimethoxycinnamic acid
(E)-3,4,5-Trimethoxycinnamic acid (TMCA) is a cinnamic acid substituted by multi-methoxy groups. (E)-3,4,5-Trimethoxycinnamic acid is an orally active and potent GABAA/BZ receptor agonist. (E)-3,4,5-Trimethoxycinnamic exhibits favourable binding affinity to 5-HT2C and 5-HT1A receptor, with IC50 values of 2.5 and 7.6 μM, respectively. (E)-3,4,5-Trimethoxycinnamic acid shows anticonvulsant and sedative activity. (E)-3,4,5-Trimethoxycinnamic acid can be used for the research of insomnia, headache and epilepsy.

-
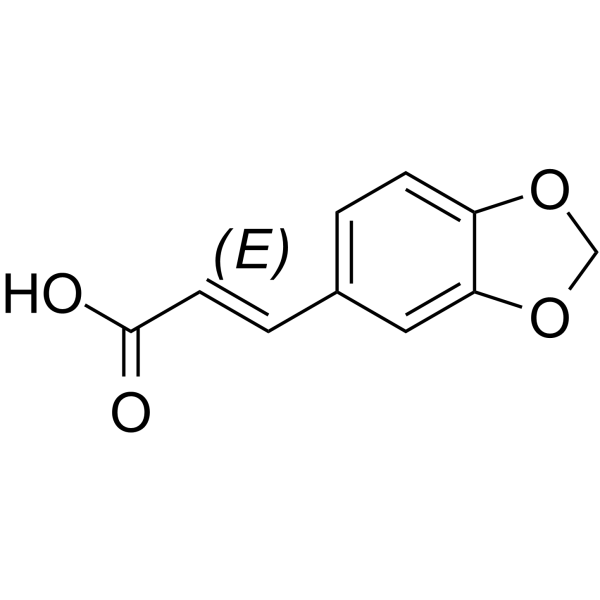
GC65239
(E)-3,4-(Methylenedioxy)cinnamic acid
(E)-3,4-(Methylenedioxy)cinnamic acid is a cinnamic acid derivative obtained from the stem bark of Brombya platynema.
-
GC61668
(E)-3,4-Dimethoxycinnamic acid
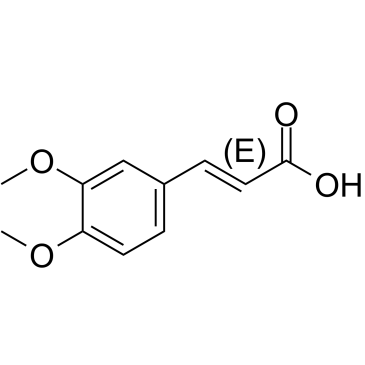
(E)-3,4-Dimethoxycinnamic acid is the less active isomer of 3,4-Dimethoxycinnamic acid.
-
GC49003
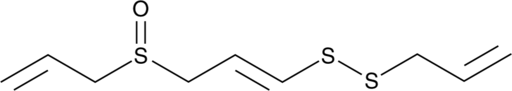
(E)-Ajoene
A disulfide with diverse biological activities
-
GC62122
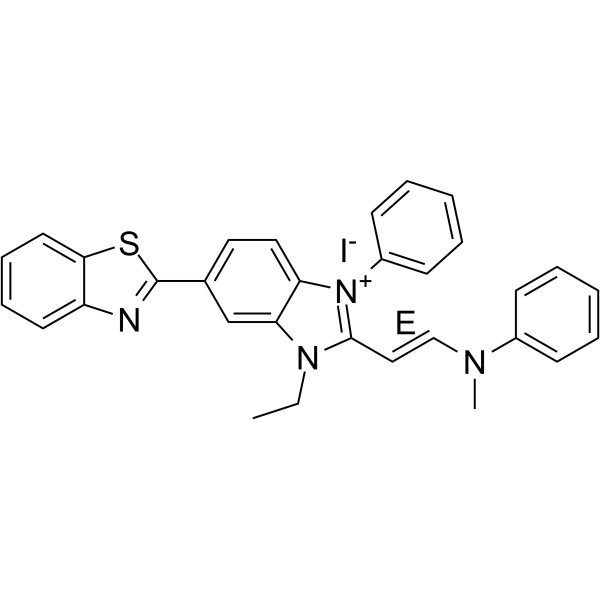
(E)-Akt inhibitor-IV
(E)-Akt inhibitor-IV ((E)-AKTIV) is a PI3K-Akt inhibitor, with potent cytotoxic.
-
GC62733
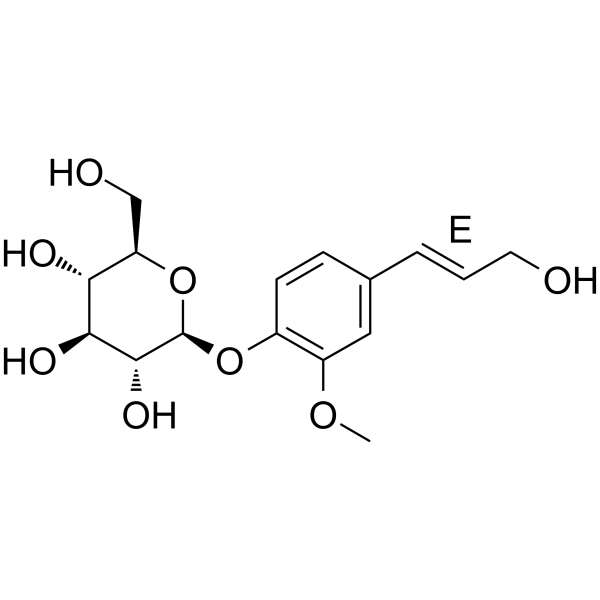
(E)-Coniferin
(E)-Coniferin is the isomer of Coniferin.
-
GC66371
(E)-Ferulic acid-d3
(E)-Ferulic acid-d3 ((E)-Coniferic acid-d3) is the deuterium labeled (E)-Ferulic acid. (E)-Ferulic acid is a isomer of Ferulic acid which is an aromatic compound, abundant in plant cell walls. (E)-Ferulic acid causes the phosphorylation of β-catenin, resulting in proteasomal degradation of β-catenin and increases the expression of pro-apoptotic factor Bax and decreases the expression of pro-survival factor survivin. (E)-Ferulic acid shows a potent ability to remove reactive oxygen species (ROS) and inhibits lipid peroxidation. (E)-Ferulic acid exerts both anti-proliferation and anti-migration effects in the human lung cancer cell line H1299.
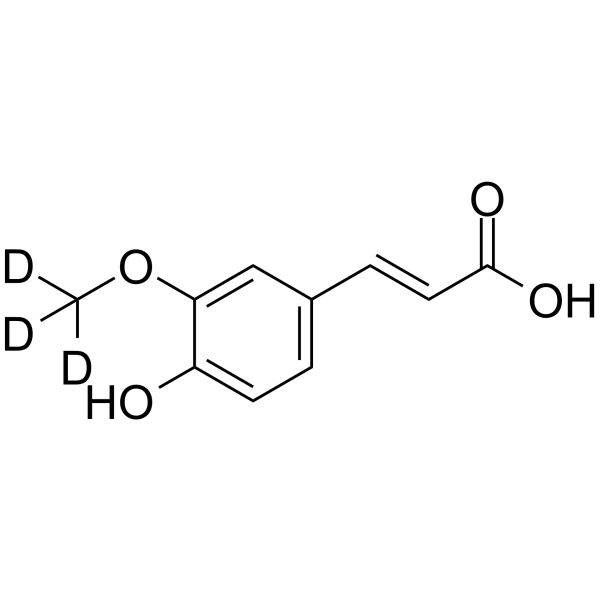
-
GC61437
(E)-Methyl 4-coumarate
(E)-Methyl 4-coumarate (Methyl 4-hydroxycinnamate), found in several plants, such as green onion (Allium cepa) or noni (Morinda citrifolia L.
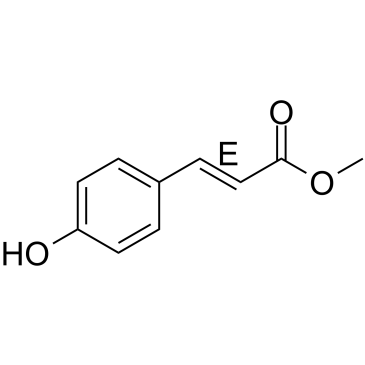
-
GC62734
(E)-Oct-2-enoic acid
(E)-Oct-2-enoic acid is an endogenous metabolite.
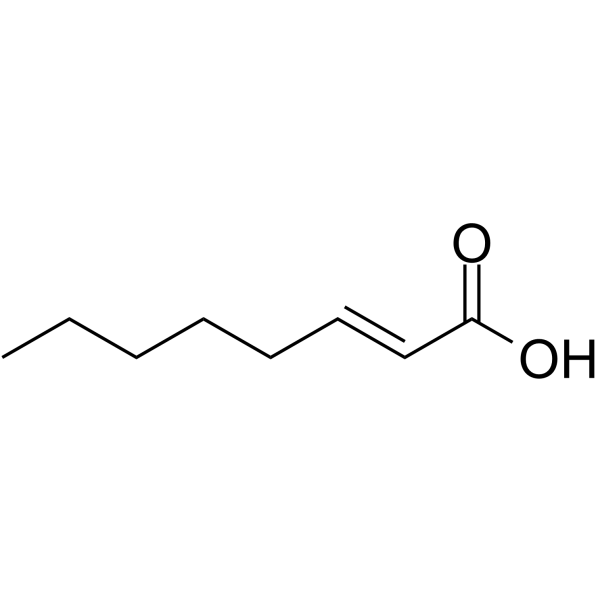
-
GC63903
(E)-Osmundacetone
(E)-Osmundacetone is the isomer of Osmundacetone.
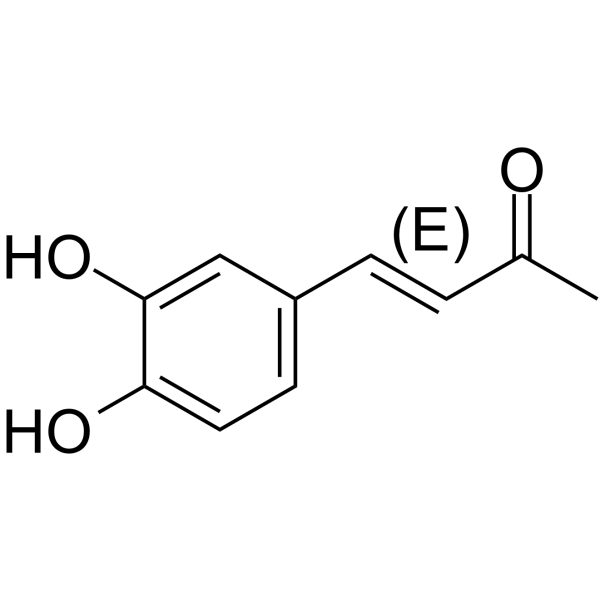
-
GC67663
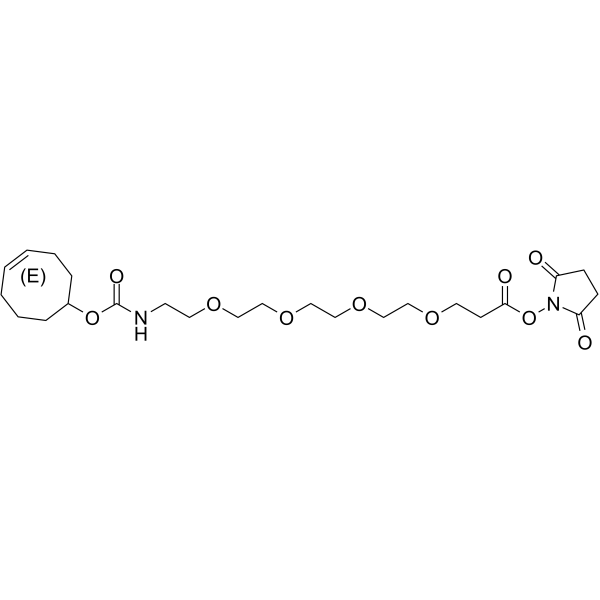
(E)-TCO-PEG4-NHS ester
(E)-TCO-PEG4-NHS ester is a PEG-based PROTAC linker that can be used in the synthesis of PROTACs.
-
GC49189
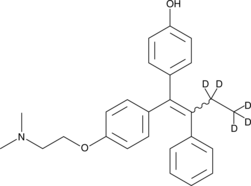
(E/Z)-4-hydroxy Tamoxifen-d5
An internal standard for the quantification of (E/Z)-4-hydroxy tamoxifen
-
GC61807
(E/Z)-AG490
(E/Z)-AG490 ((E/Z)-Tyrphostin AG490) is a racemic compound of (E)-AG490 and (Z)-AG490 isomers. (E)-AG490 is a tyrosine kinase inhibitor that inhibits EGFR, Stat-3 and JAK2/3.

-
GC25000
(E/Z)-BCI
(E/Z)-BCI (BCI, NSC 150117) is an inhibitor of dual specific phosphatase 1/6 (DUSP1/DUSP6) and mitogen-activated protein kinase with EC50 of 13.3 μM and 8.0 μM for DUSP6 and DUSP1 in cells, respectively. (E)-BCI induces apoptosis via generation of reactive oxygen species (ROS) and activation of intrinsic mitochondrial pathway in H1299 lung cancer cells.

-
GC62407
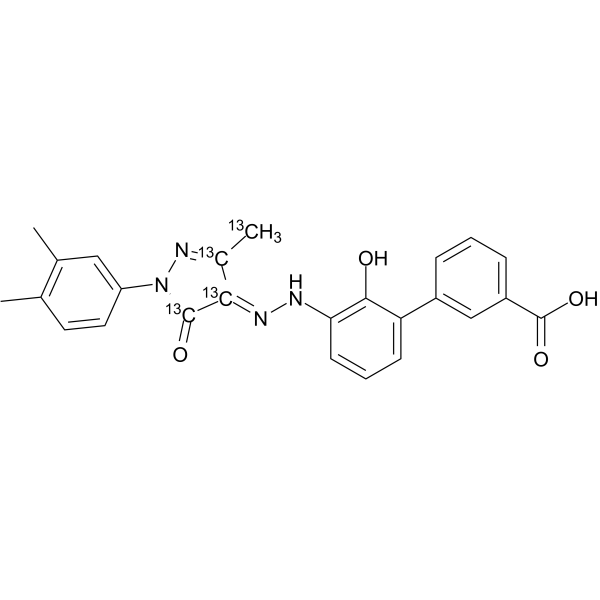
(E/Z)-Eltrombopag 13C
(E/Z)-Eltrombopag 13C ((E/Z)-SB-497115 13C4) is a mixture complex of E-Eltrombopag and Z-Eltrombopag, with 13C labeled.
-
GC62735
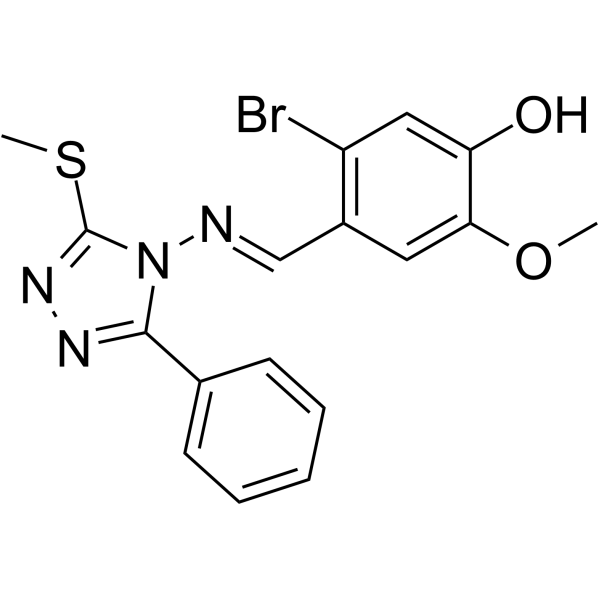
(E/Z)-GO289
(E/Z)-GO289 is a potent and selective casein kinase 2 (CK2) inhibitor (IC50=7 nM). (E/Z)-GO289 strongly lengthens circadian period. (E/Z)-GO289 exhibits cell type–dependent inhibition of cancer cell growth that correlated with cellular clock function.
-
GC62736
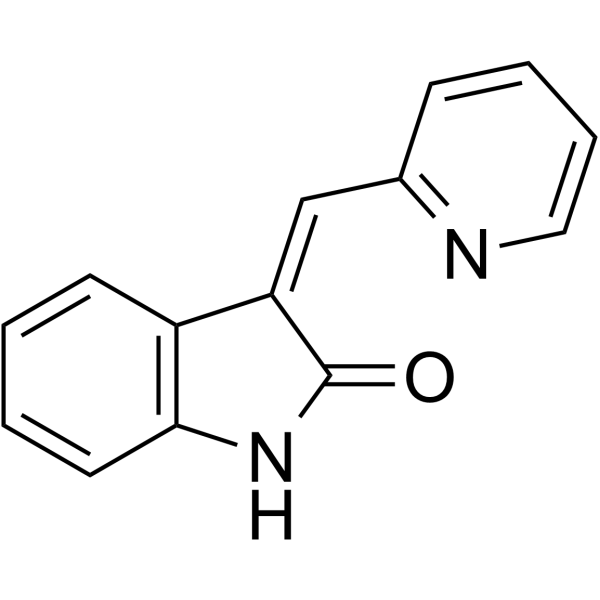
(E/Z)-GSK-3β inhibitor 1
(E/Z)-GSK-3β inhibitor 1 is a racemic compound of (E)-GSK-3β inhibitor 1 and (Z)-GSK-3β inhibitor 1 isomers.
-
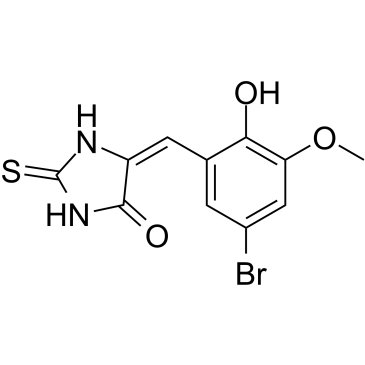
GC61564
(E/Z)-IT-603
(E/Z)-IT-603 is a mixture of E-IT-603 and Z-IT-603 (IT-603).
-
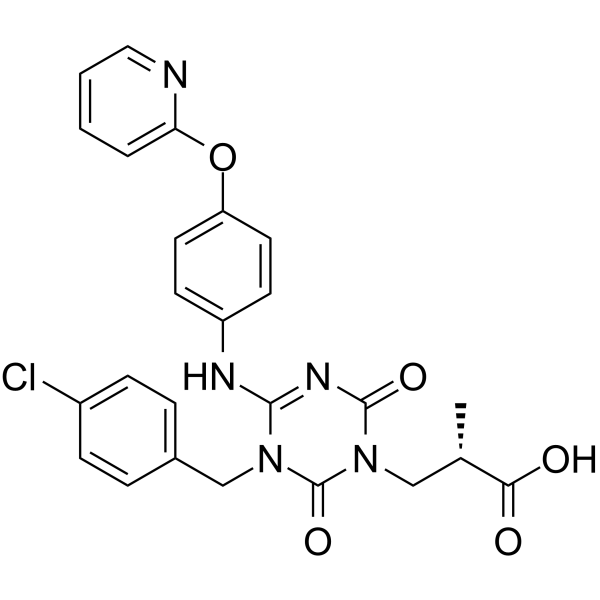
GC62560
(E/Z)-Sivopixant
(E/Z)-Sivopixant ((E/Z)-S-600918) is a potent P2X3 receptor antagonist with an IC50 of 4 nM.
-
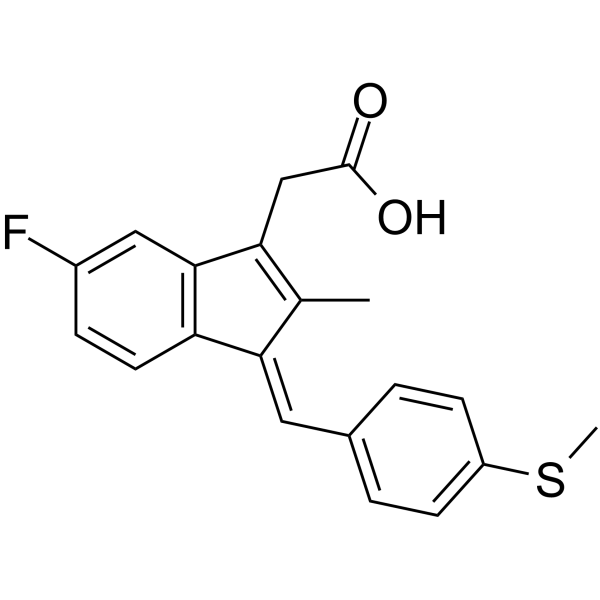
GC67476
(E/Z)-Sulindac sulfide
(E/Z)-Sulindac sulfide is a potent γ-secretase modulator (GSM). (E/Z)-Sulindac sulfide selectively reduces Aβ42 production in favor of shorter Aβ species. (E/Z)-Sulindac sulfide can be used for researching Alzheimer's disease.
-
GC64657
(E/Z)-ZINC09659342
(E/Z)-ZINC09659342 is an inhibitor of Lbc-RhoA interaction.
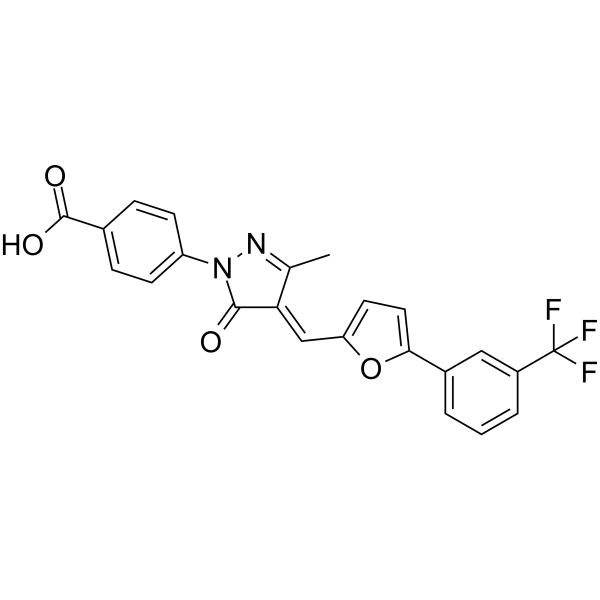
-
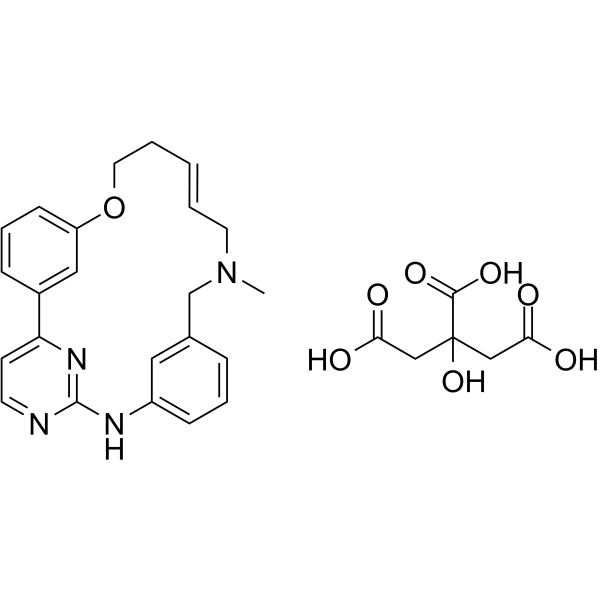
GC64429
(E/Z)-Zotiraciclib citrate
(E/Z)-Zotiraciclib citrate is a potent CDK2, JAK2, and FLT3 inhibitor.
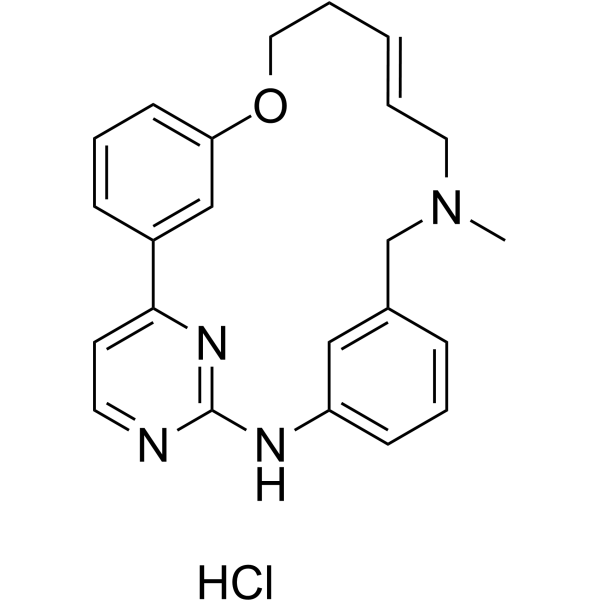
-
GC63864
(E/Z)-Zotiraciclib hydrochloride
(E/Z)-Zotiraciclib ((E/Z)-TG02) hydrochloride is a potent CDK2, JAK2, and FLT3 inhibitor.
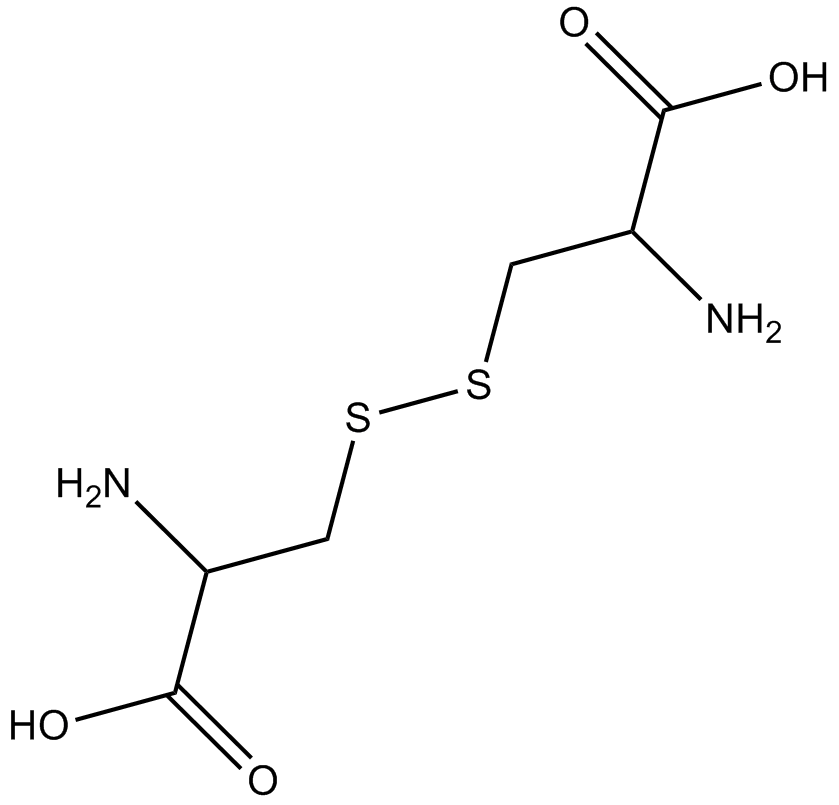
-
GA11210
(H-Cys-OH)2
-
GN10783
(R) Ginsenoside Rh2
-
GC69834
(R)-(+)-Dimethindene maleate
(R)-(+)-Dimethindene maleate is an orally active H1-receptor blocker with anti-histamine properties in pigs.
-
GC49167
(R)-(+)-Trityl glycidyl ether
A synthetic precursor
-
GC61425
(R)-(-)-1,2-Propanediol
(R)-(-)-1,2-Propanediol is a (R)-enantiomer of 1,2-Propanediol that produced from glucose in Escherichia coli expressing NADH-linked glycerol dehydrogenase genes.
-
GC61694
(R)-(-)-2-Butanol
(R)-(-)-2-Butanol is released by the females of the white grub beetle, Dasylepida ishigakiensis, to attract males.
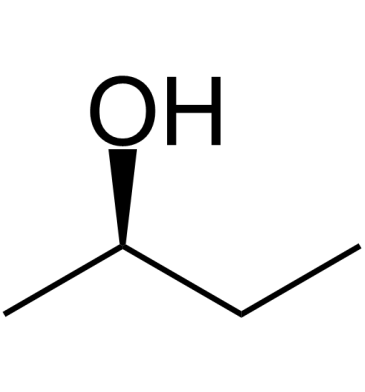
-
GC69823
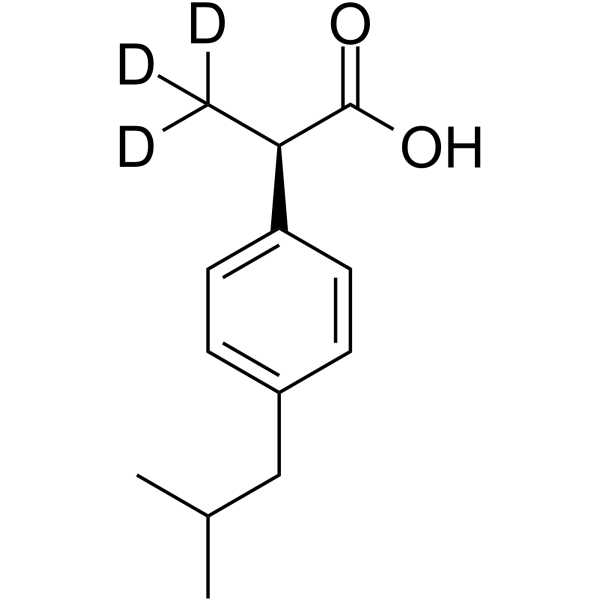
(R)-(-)-Ibuprofen-d3
(R)-(-)-Ibuprofen-d3 is the deuterated form of (R)-(-)-Ibuprofen. (R)-(-)-Ibuprofen is the R enantiomer of Ibuprofen, which has no effect on COX and can inhibit NF-κB activation. (R)-(-)-Ibuprofen has anti-inflammatory properties and can be used in pain relief research.
-
GC63791
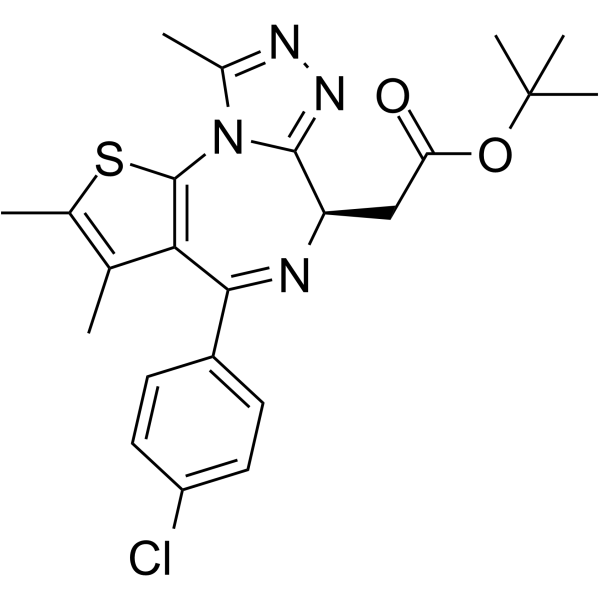
(R)-(-)-JQ1 Enantiomer
(R)-(-)-JQ1 Enantiomer is the stereoisomer of (+)-JQ1. (+)-JQ1 potently decreases expression of both BRD4 target genes, whereas (R)-(-)-JQ1 Enantiomer has no effect.
-
GC62737
(R)-(-)-O-Desmethyl Venlafaxine D6

-
GC61759
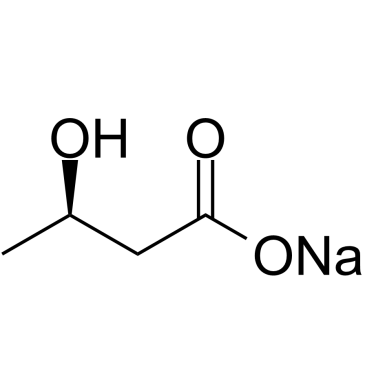
(R)-3-Hydroxybutanoic acid sodium
(R)-3-Hydroxybutanoic acid sodium ((R)-3-Hydroxybutyric acid) is a metabolite converted from acetoacetic acid catalyzed by 3-hydroxybutyrate dehydrogenase.
-
GC65610
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone is a diarylheptanoid that can be found in Alpinia officinarum.

-
GC69793
(R)-5-O-Benzoyl-1,2-di-O-isopropylidene-alpha-D-xylofuranose
(R)-5-O-Benzoyl-1,2-di-O-isopropylidene-alpha-D-xylofuranose is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity and target inert lymphoid malignancies. The anticancer mechanism in this process depends on inhibiting DNA synthesis, inducing apoptosis (cell death), etc.
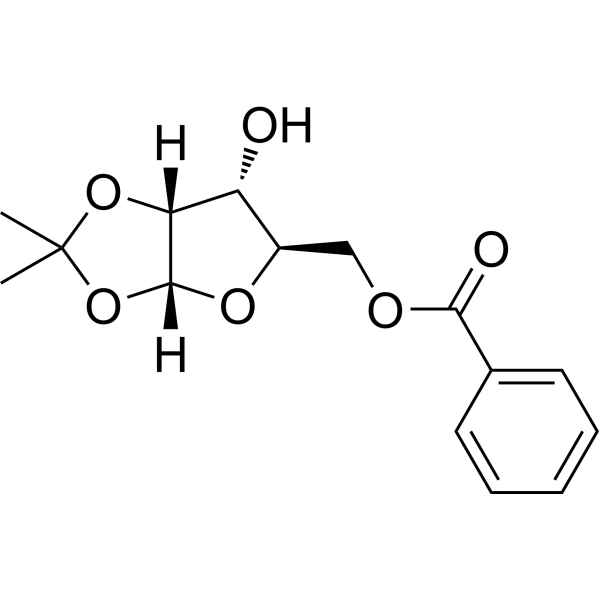
-
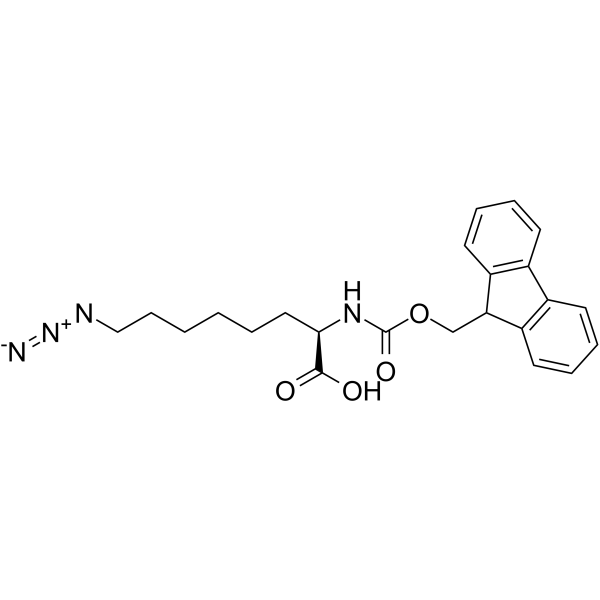
GC66904
(R)-8-Azido-2-(Fmoc-amino)octanoic acid
(R)-8-Azido-2-(Fmoc-amino)octanoic acid is a non-cleavable ADC linker used in the synthesis of antibody-drug conjugates (ADCs).
-
GC67688
(R)-Asundexian
-
GC25001
(R)-Avanafil
R-Avanafil is a strong competitive inhibitor of phosphodiesterase 5 (PDE5) with a demonstrated in vitro IC 50 of 5.2 nM.
-
GC64091
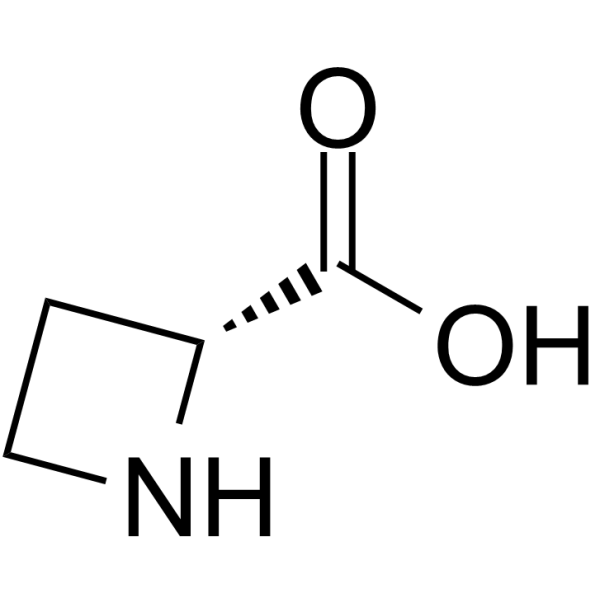
(R)-Azetidine-2-carboxylic acid
(R)-Azetidine-2-carboxylic acid is a non-cleavable?ADC linker?used in the synthesis of antibody-drug conjugates (ADCs). (R)-Azetidine-2-carboxylic acid is also a alkyl chain-based PROTAC linker?that can be
-
GC63781
(R)-BAY-899
(R)-BAY-899 is the R-enantiomer of BAY-899.

-
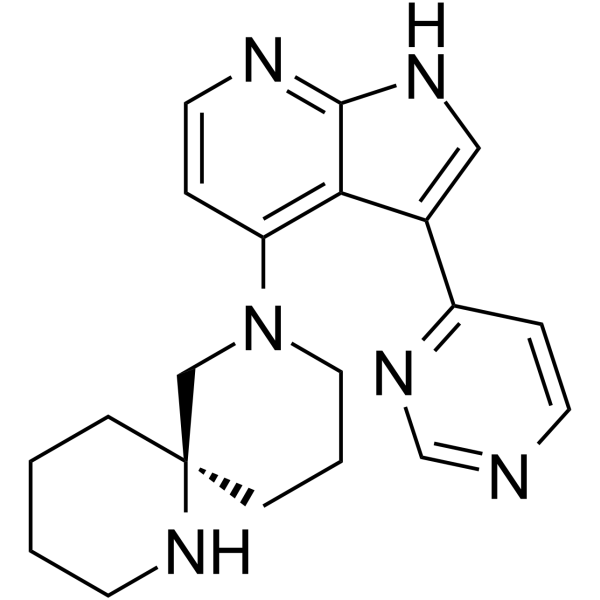
GC63906
(R)-BDP9066
(R)-BDP9066 is a potent inhibitor of myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK). (R)-BDP9066 blocks cancer cell invasion. (R)-BDP9066 has the potential for the research of proliferative diseases, such as cancer.
-
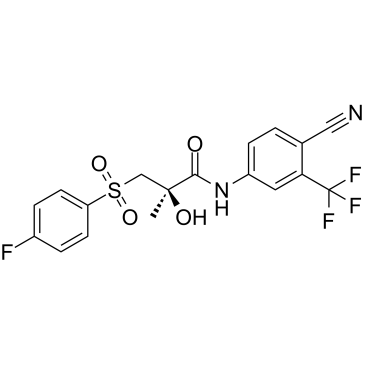
GC61491
(R)-Bicalutamide
(R)-Bicalutamide is the (R)-enantiomer of Bicalutamide. (R)-Bicalutamide is an androgen receptor (AR) antagonist, with antineoplastic activity. (R)-Bicalutamide is widely used for the research of prostate cancer.
-
GC69805
(R)-Casopitant
(R)-Casopitant ((R)-GW679769) is an isomer of Casopitant. Casopitant is an NK(1) receptor antagonist that can be used in the research of chemotherapy-induced nausea and vomiting.