PAR1
PAR1 (protease-activated receptor 1) is a subfamily of related G protein-coupled receptors that are activated by cleavage of part of their extracellular domain.
Products for PAR1
- Cat.No. Product Name Information
-
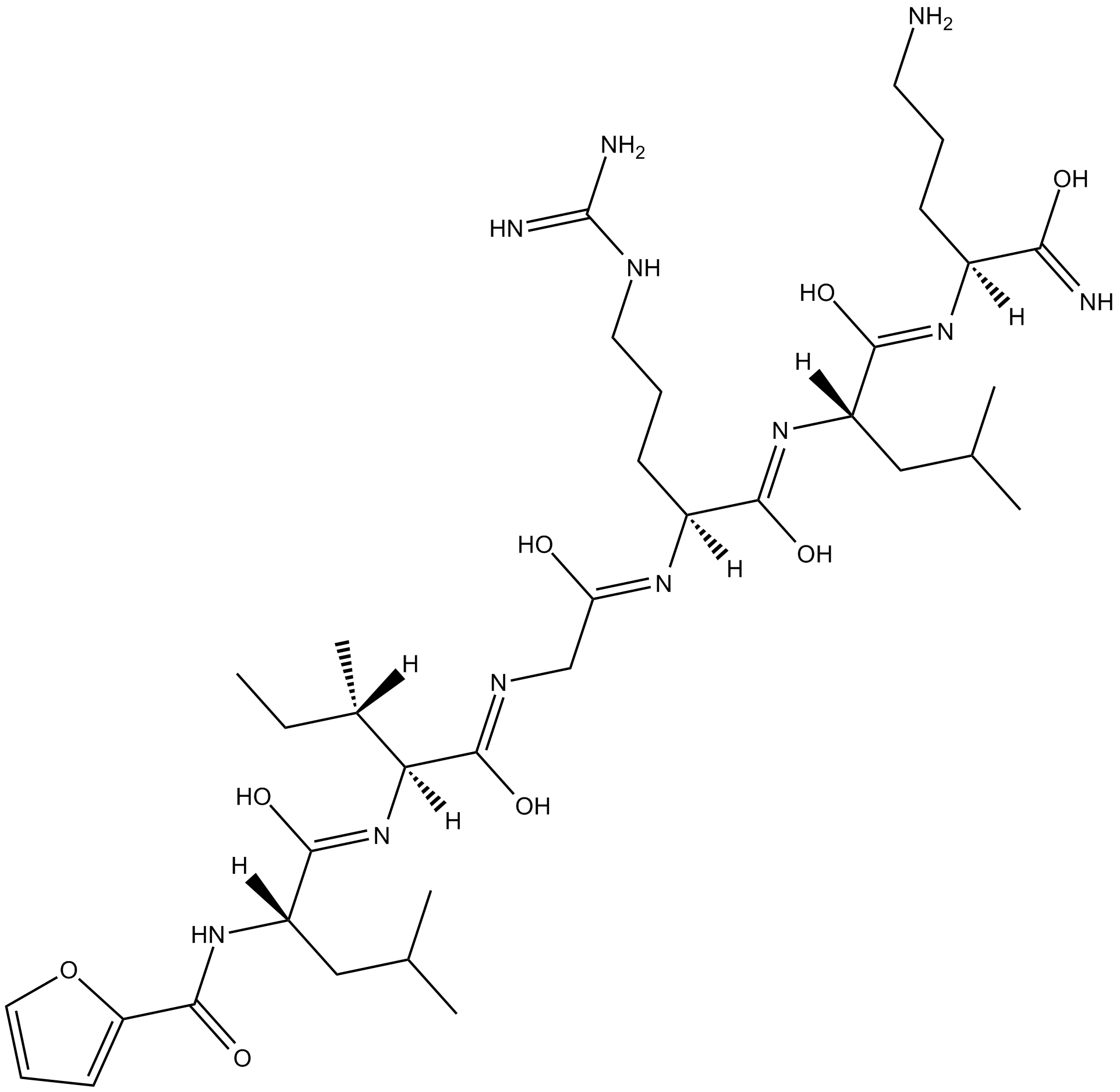
GC17368
2-Furoyl-LIGRLO-amide
Protease-activated receptor agonist
-
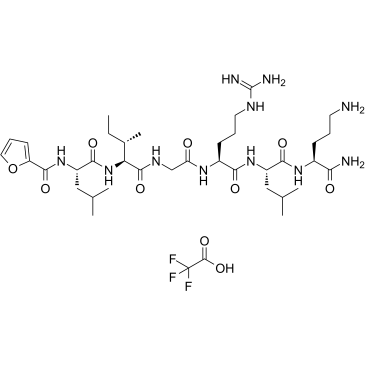
GC38731
2-Furoyl-LIGRLO-amide TFA
2-Furoyl-LIGRLO-amide TFA is a potent and selective proteinase-activated receptor 2 (PAR2) agonist with a pD2 value of 7.0.
-
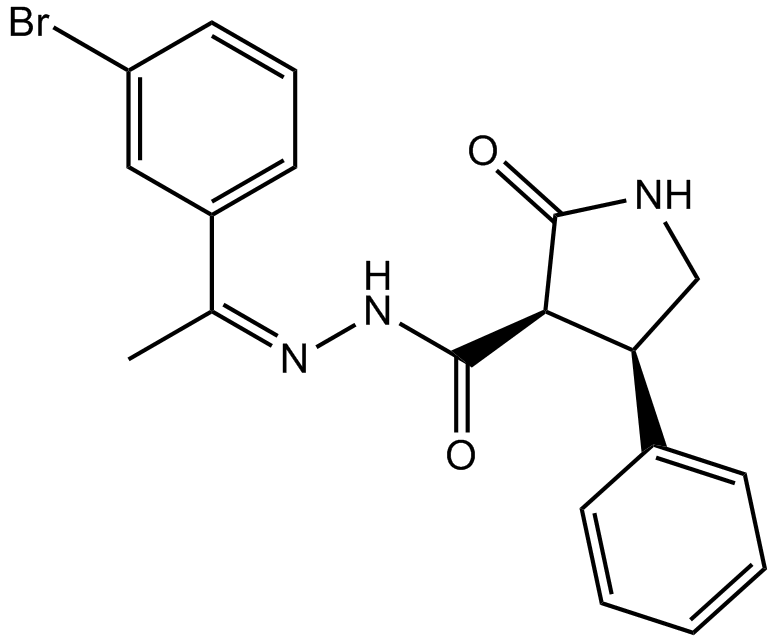
GC14831
AC 264613
PAR2 agonist,potent and selective
-
GC15290
AC 55541
PAR2 agonist,potent and selective
-
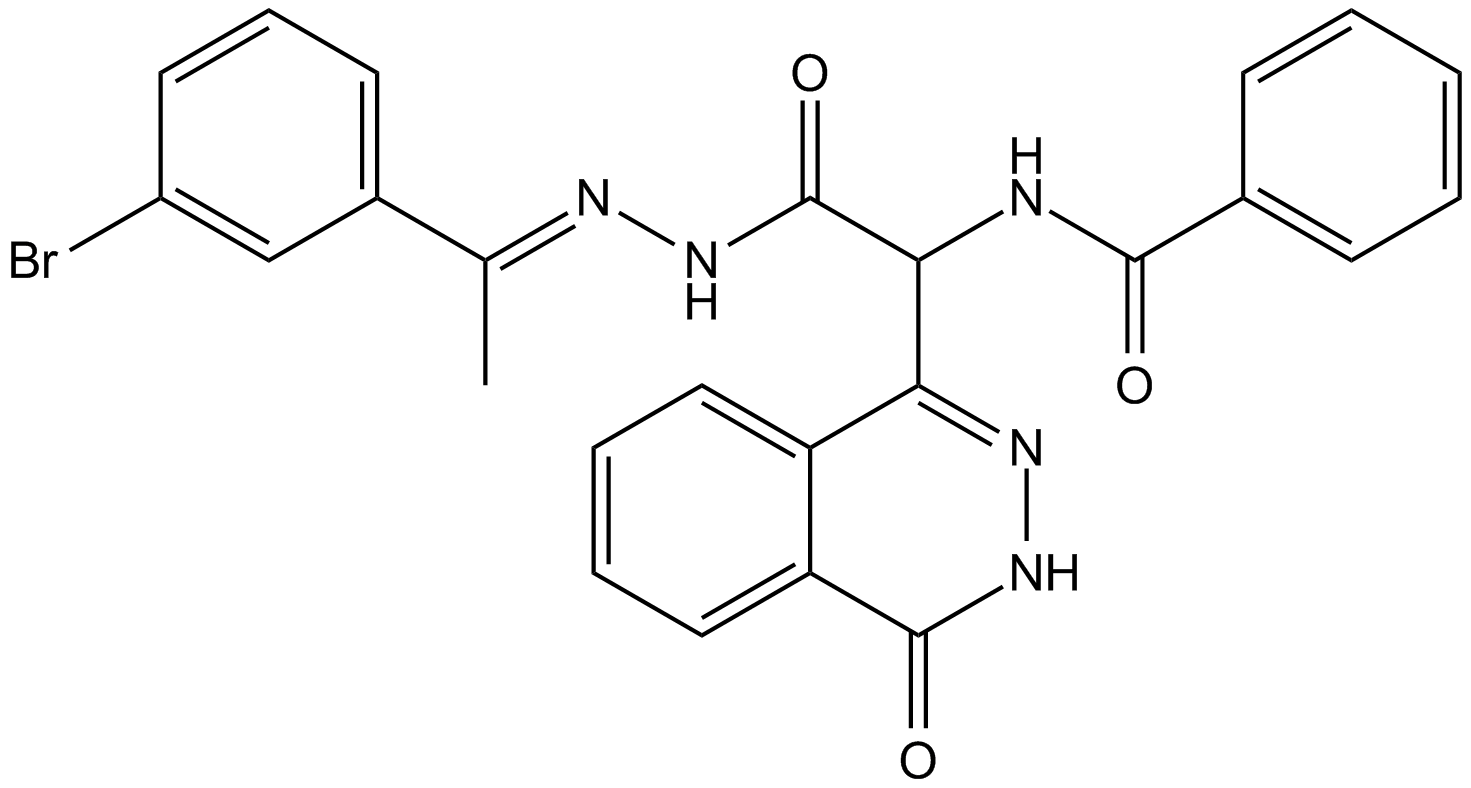
GC63658
Atopaxar
Atopaxar (E5555) is a potent, orally active, selective and reversible thrombin receptor protease-activated receptor-1 (PAR-1) antagonist.
-
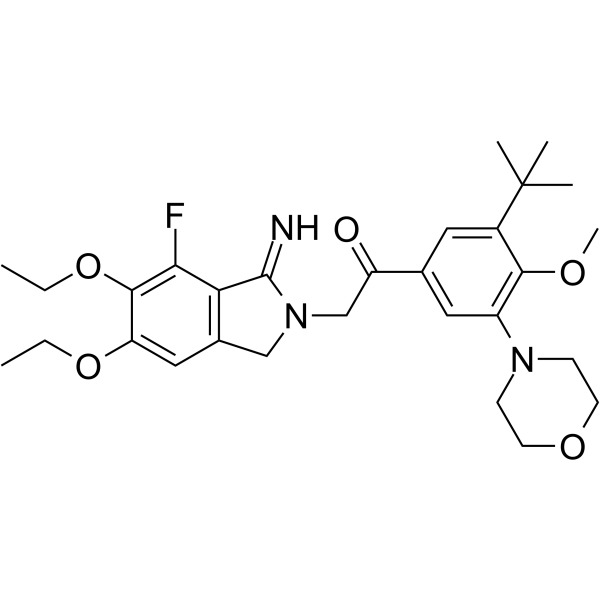
GC18152
AZ-3451
A potent, and selective PAR2 antagonist with Kd of 13.5 nM.
-
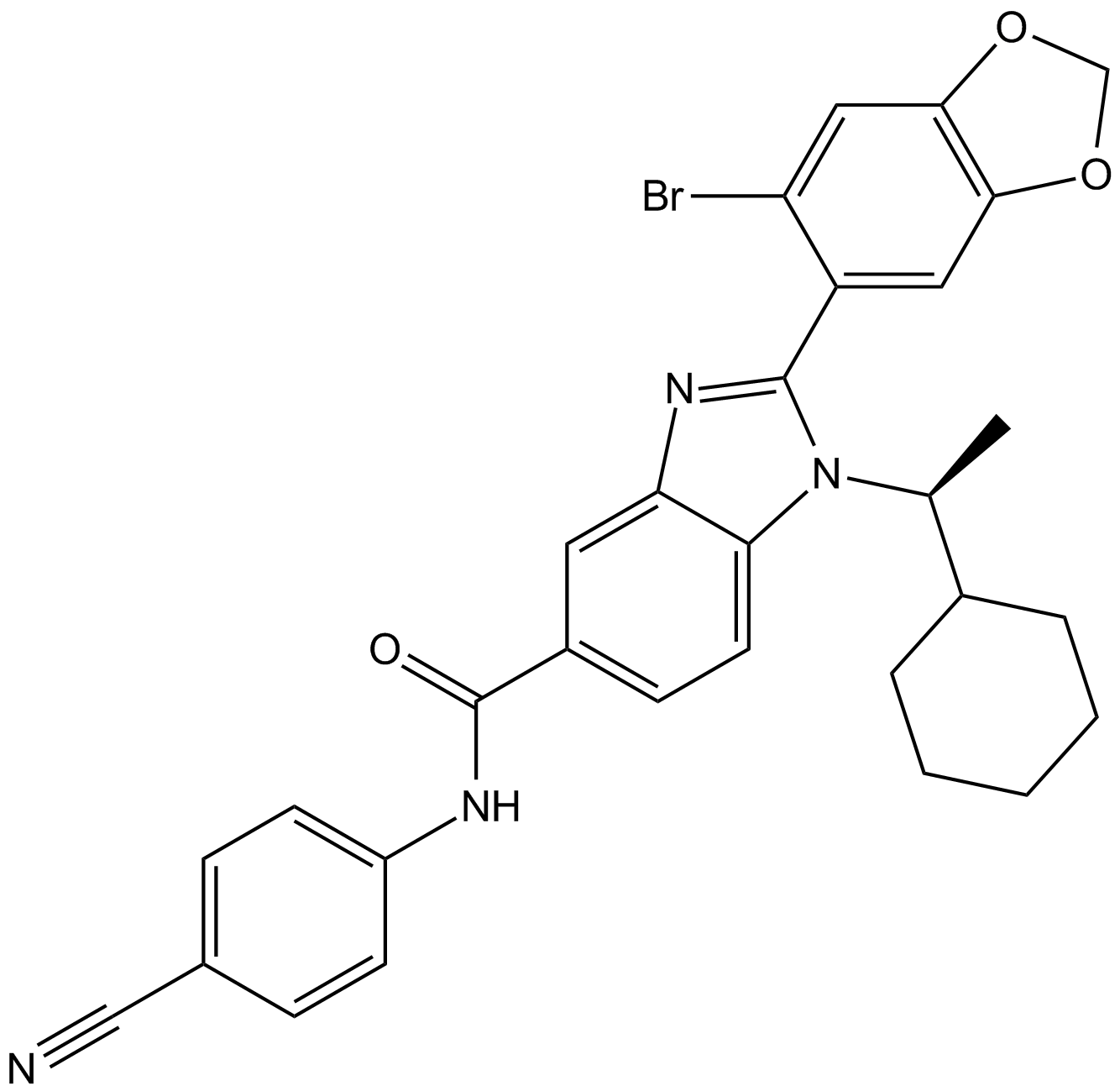
GC65312
AZ8838
AZ8838 is a potent, competitive, allosteric, orally active non-peptide small molecule antagonist of PAR2 with a pKi of 6.4 for hPAR2.
-
GC18717
BMS 986120
A selective PAR4 antagonist
-
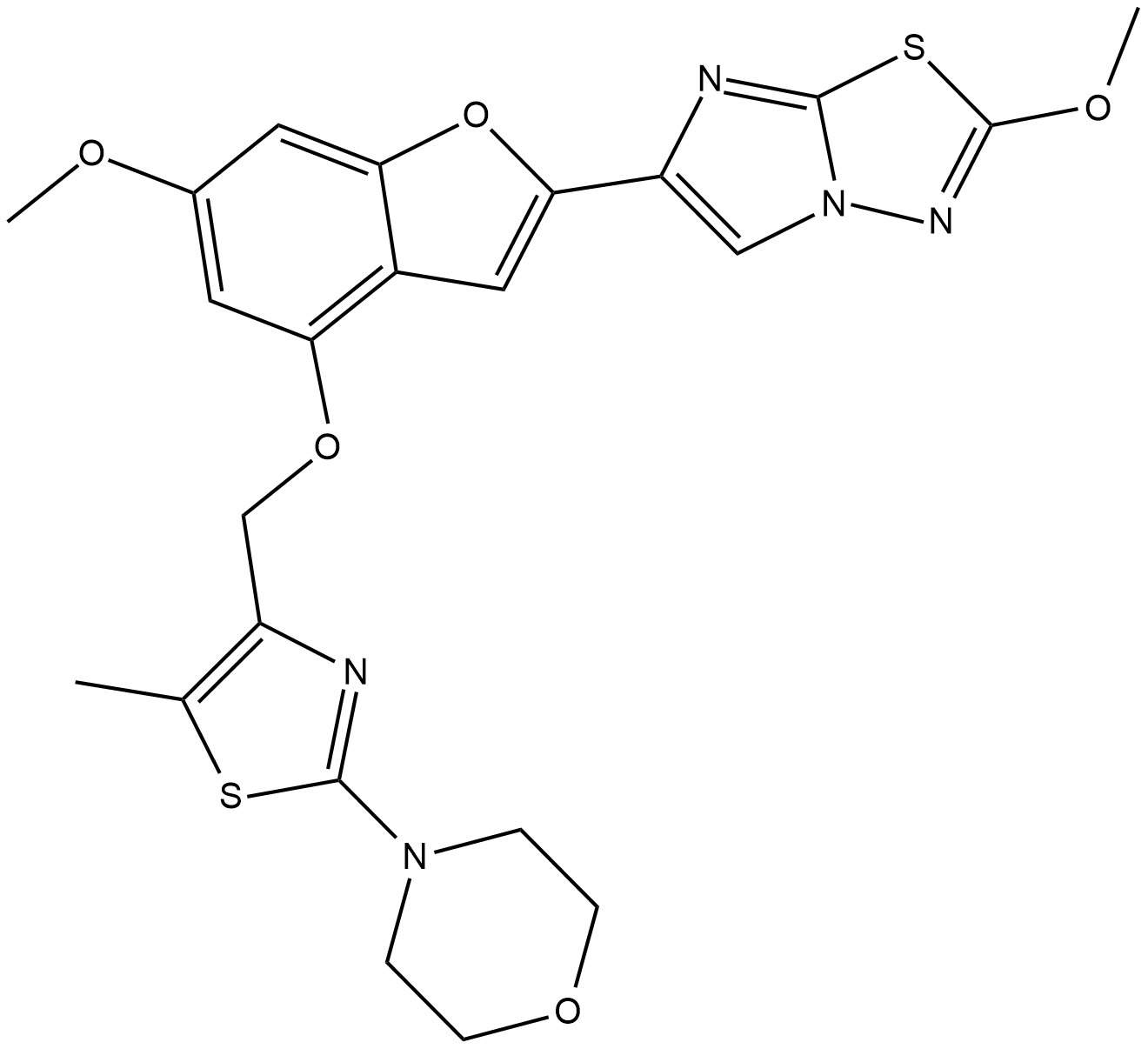
GC60807
ENMD-1068 hydrochloride
Selective protease-activated receptor 2 (PAR2) anatagonist
-
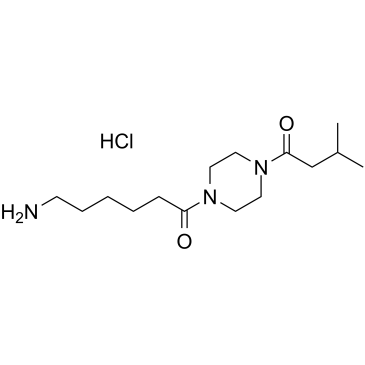
GC61501
FSLLRY-NH2 TFA
FSLLRY-NH2 TFA is a protease-activated receptor 2 (PAR2) inhibitor.
-
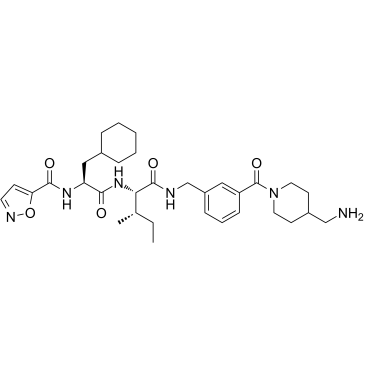
GC36125
GB-110
GB-110 is a potent, orally active, and nonpeptidic protease activated receptor 2 (PAR2) agonist.
-
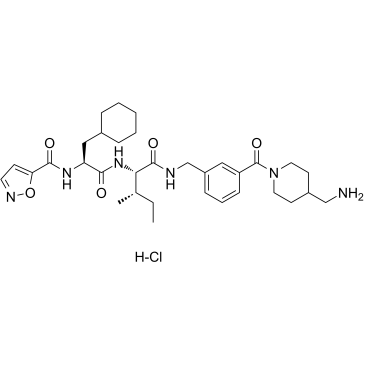
GC38329
GB-110 hydrochloride
GB-110 hydrochloride is a potent, orally active, and nonpeptidic protease activated receptor 2 (PAR2) agonist.
-
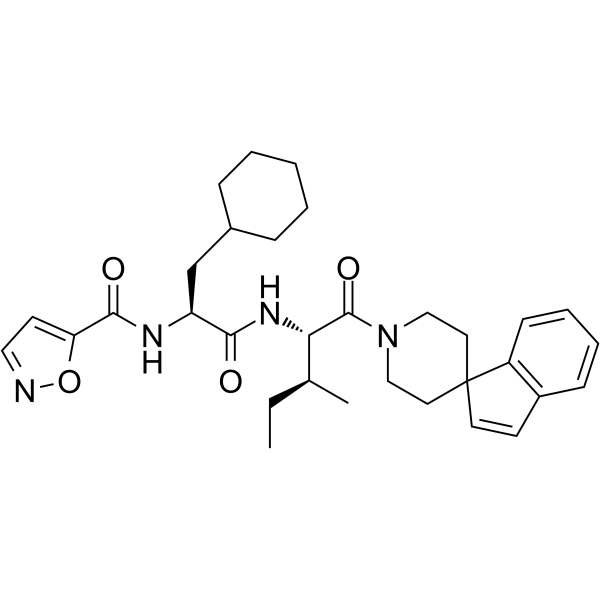
GC65439
GB-88
GB-88 is an oral, selective non-peptide antagonist of PAR2, inhibits PAR2 activated Ca2+ release with an IC50 of 2 ?M.
-
GC64979
I-191
I-191 is a potent, selective protease-activated receptor 2 (PAR2) antagonist.

-
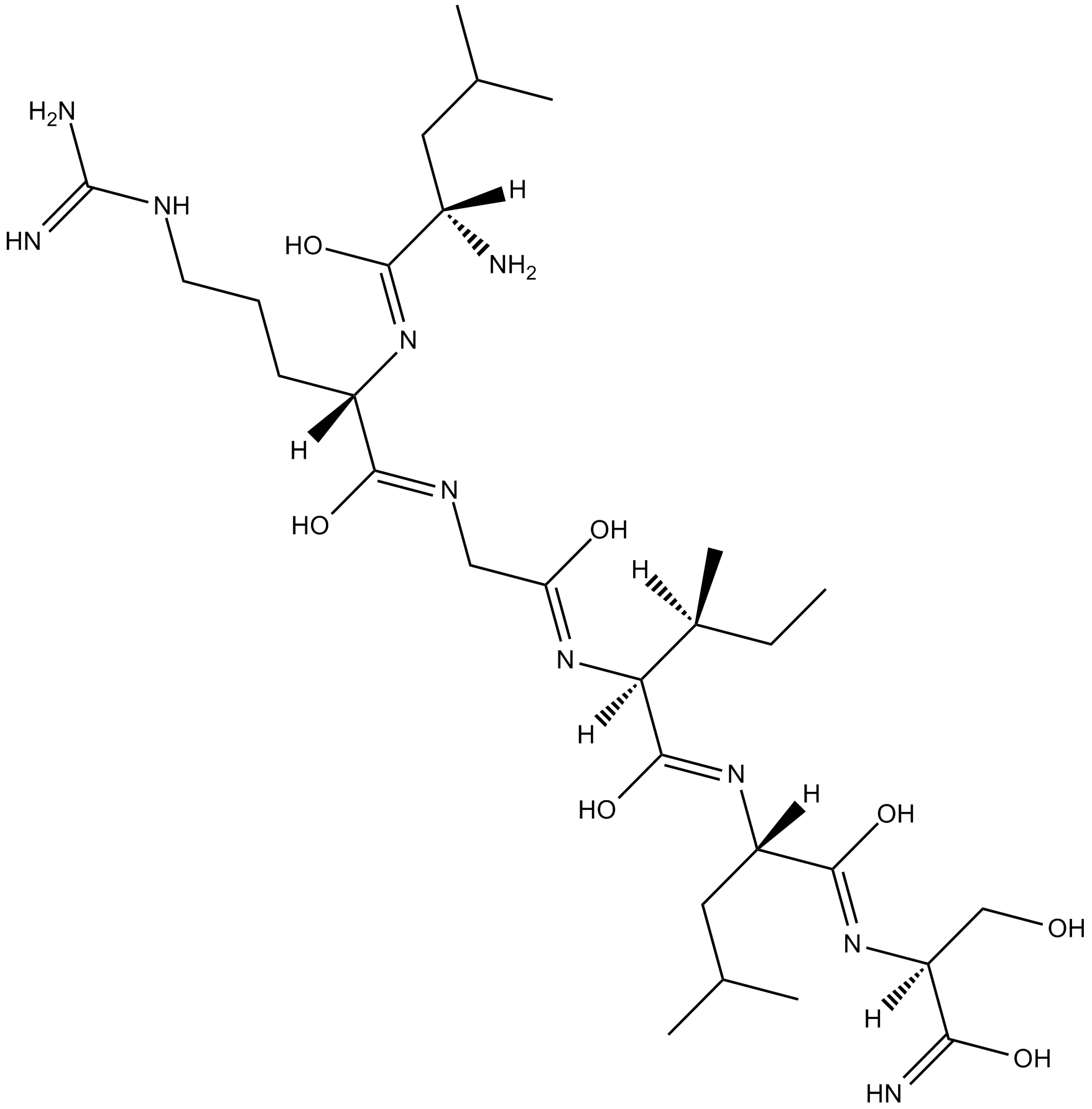
GC13227
LRGILS-NH2
Protease-activated receptor agonist
-
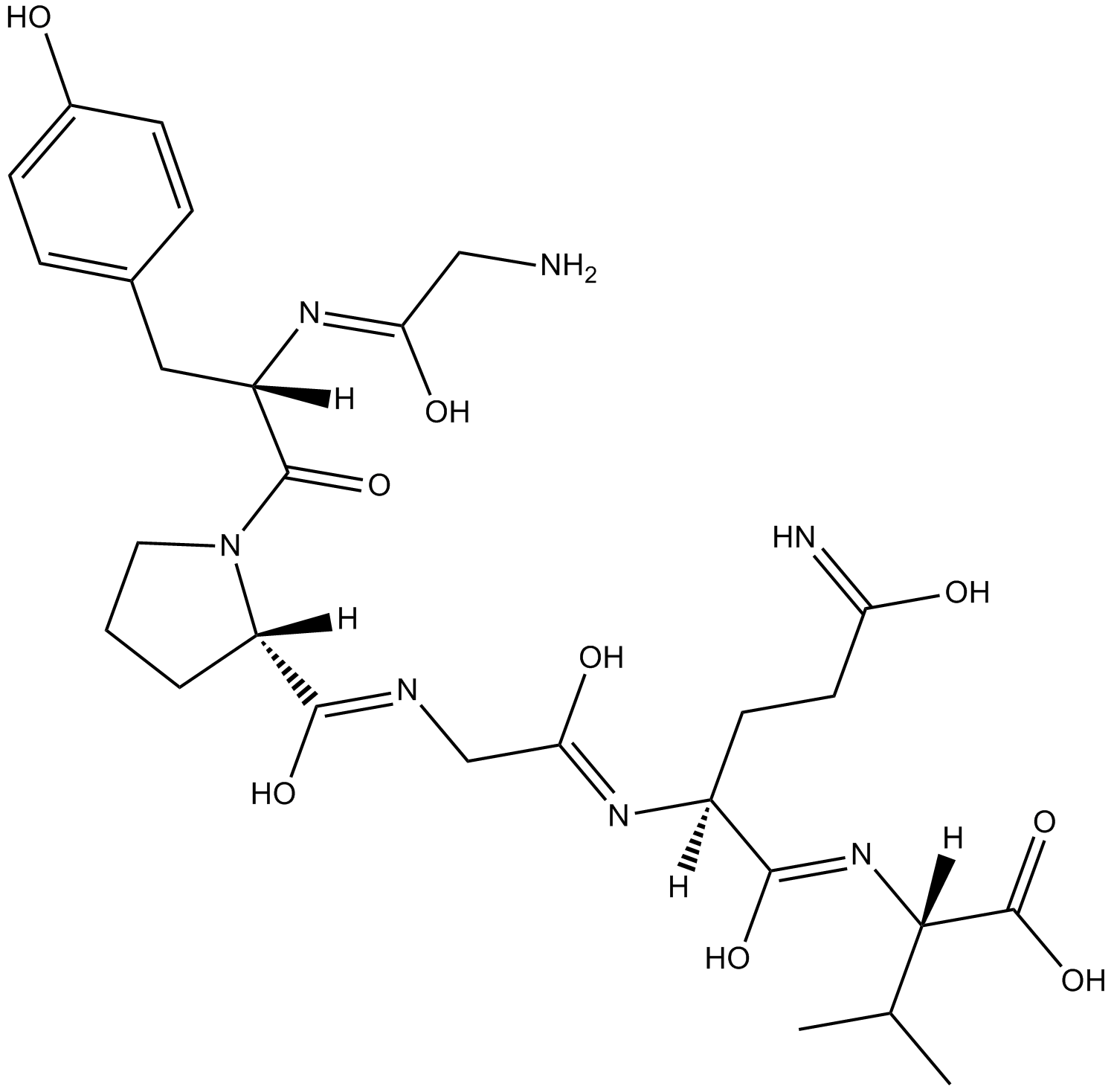
GC11951
PAR 4 (1-6)
PAR4 agonist
-
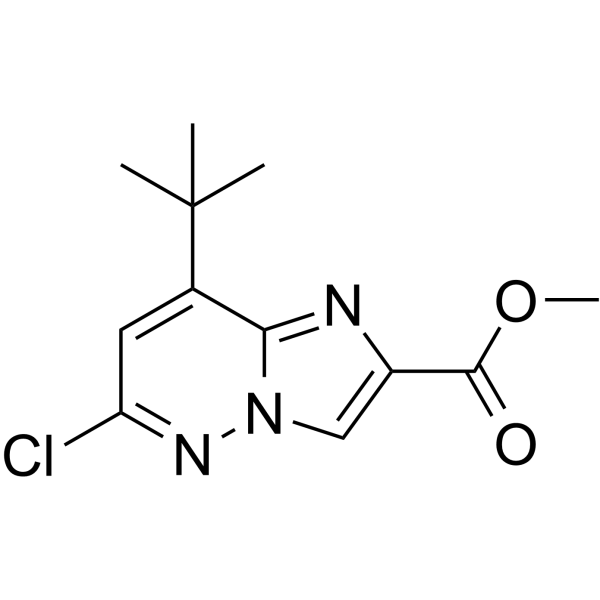
GC62101
PAR-2-IN-1
PAR-2-IN-1 is a protease-activated receptor-2 (PAR2) signaling pathway inhibitor with anti-inflammatory and anticancer effects.
-
GA23350
PAR-3 (1-6) amide (human)
PAR-3 (1-6) amide (human) is a proteinase-activated receptor (PAR-3) agonist peptide.

-
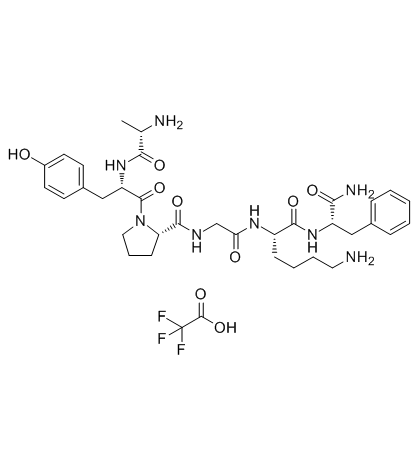
GC33599
PAR-4 Agonist Peptide, amide TFA (PAR-4-AP (TFA))
PAR-4 Agonist Peptide, amide TFA (PAR-4-AP (TFA)) (PAR-4-AP TFA; AY-NH2 TFA) is a proteinase-activated receptor-4 (PAR-4) agonist, which has no effect on either PAR-1 or PAR-2 and whose effects are blocked by a PAR-4 antagonist.
-
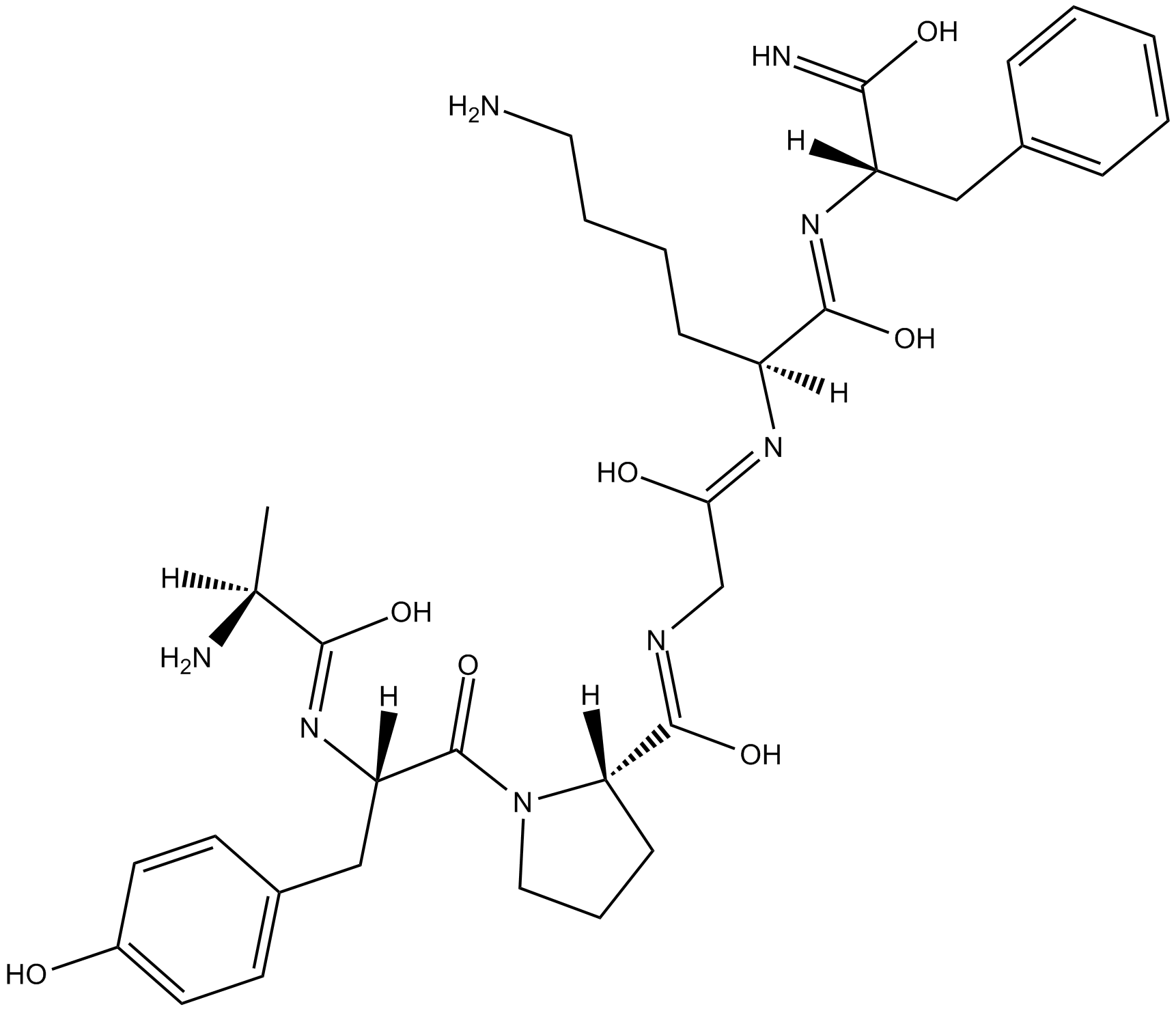
GC15817
PAR-4 Agonist Peptide, amide (AY-NH2)
PAR-4 Agonist Peptide, amide(AY-NH2) (PAR-4-AP; AY-NH2) is a proteinase-activated receptor-4 (PAR-4) agonist, which has no effect on either PAR-1 or PAR-2 and whose effects are blocked by a PAR-4 antagonist.
-
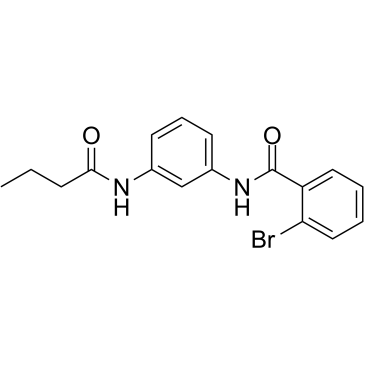
GC38134
Parmodulin 2
Reversible inhibitor of PAR1mediated platelet activation
-
GC69663
Parstatin(mouse) TFA
Parstatin (mouse) TFA is a peptide agonist of the PAR-1 thrombin receptor with cell-penetrating properties and an effective inhibitor of angiogenesis.

-
GC69768
Protease-Activated Receptor-1, PAR-1 Agonist
Protease-Activated Receptor-1, PAR-1 Agonist is a selective peptide that activates Protease-Activated Receptor-1 (PAR-1), which corresponds to the PAR1 ligand and selectively mimics the action of thrombin through this receptor.
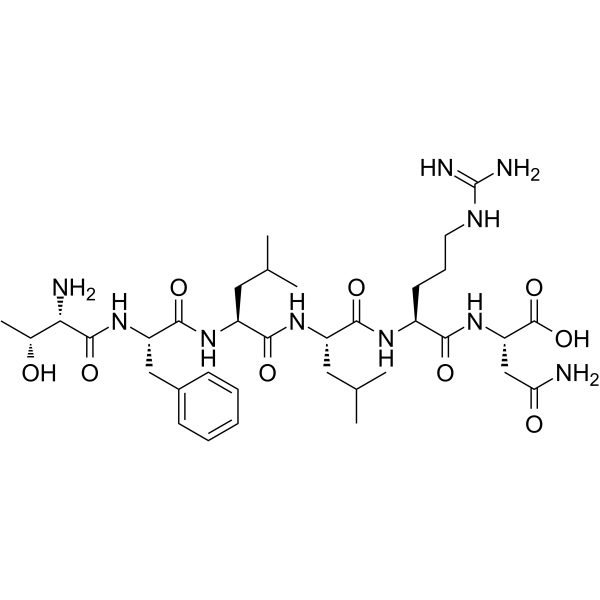
-
GC62469
Protease-Activated Receptor-1, PAR-1 Agonist TFA
Protease-Activated Receptor-1, PAR-1 Agonist TFA is a selective proteinase-activated receptor1 (PAR-1) agonist peptide. Protease-Activated Receptor-1, PAR-1 Agonist TFA corresponds to PAR1 tethered ligand and which can selectively mimic theactions of thrombin via this receptor.
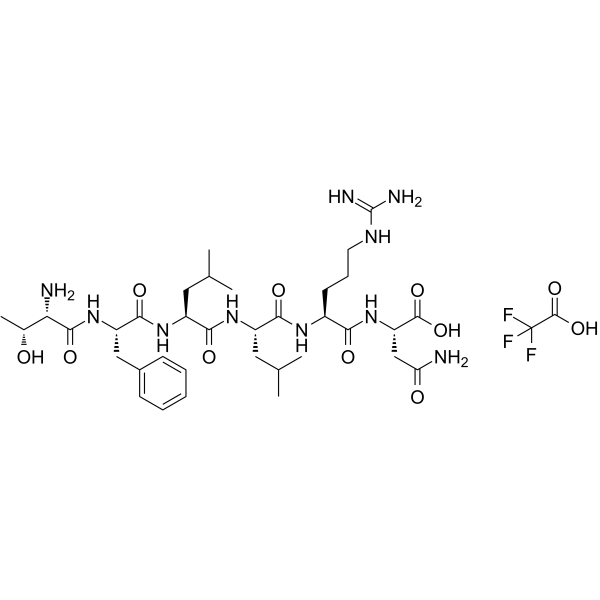
-
GC61456
Protease-Activated Receptor-3 (PAR-3) (1-6), human TFA
Protease-Activated Receptor-3 (PAR-3) (1-6), human TFA is a proteinase-activated receptor (PAR-3) agonist peptide.
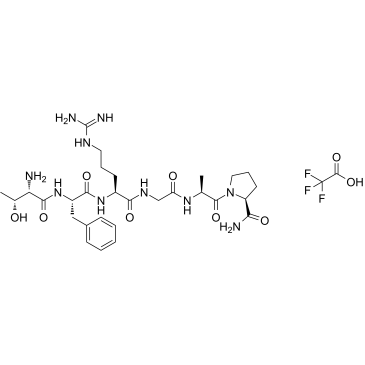
-
GC33429
Protease-Activated Receptor-4
Protease-Activated Receptor-4 is the agonist of proteinase-activated receptor-4 (PAR4).
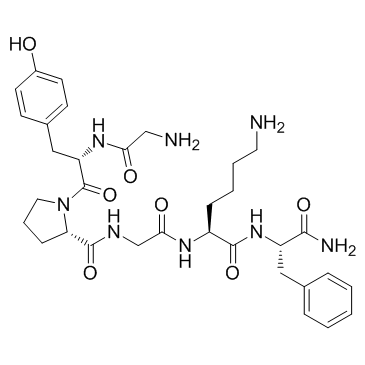
-
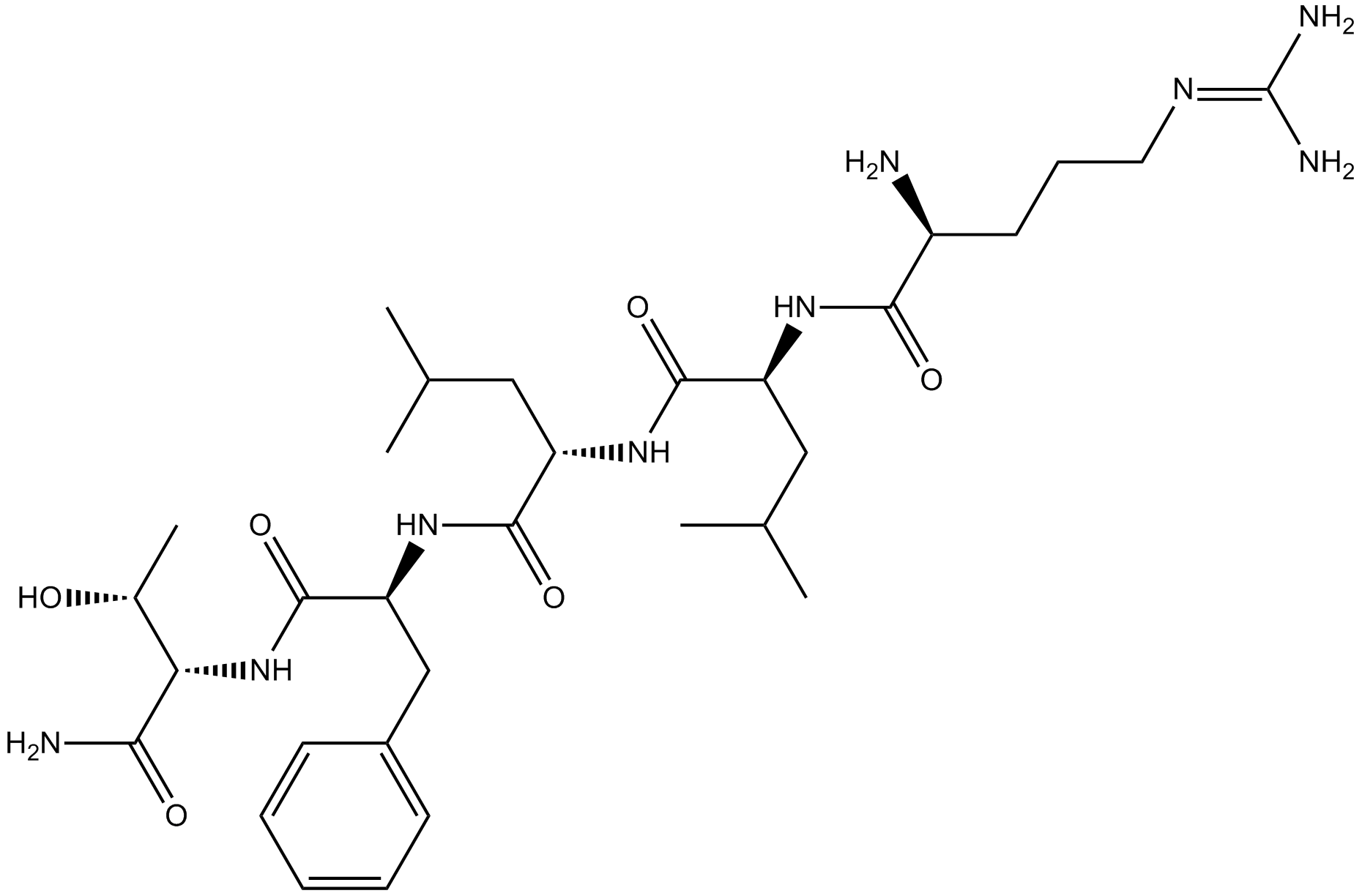
GC17991
RLLFT-NH2
Protease-activated receptor agonist
-
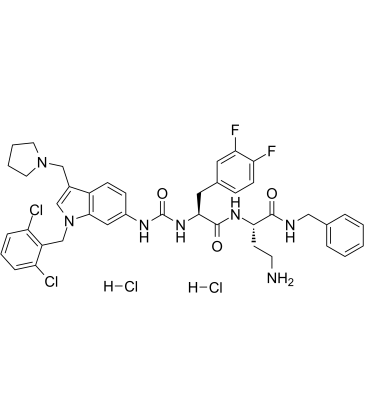
GC61777
RWJ-56110 dihydrochloride
RWJ-56110 dihydrochloride is a potent, selective, peptide-mimetic inhibitor of PAR-1 activation and internalization (binding IC50=0.44 uM) and shows no effect on PAR-2, PAR-3, or PAR-4.
-
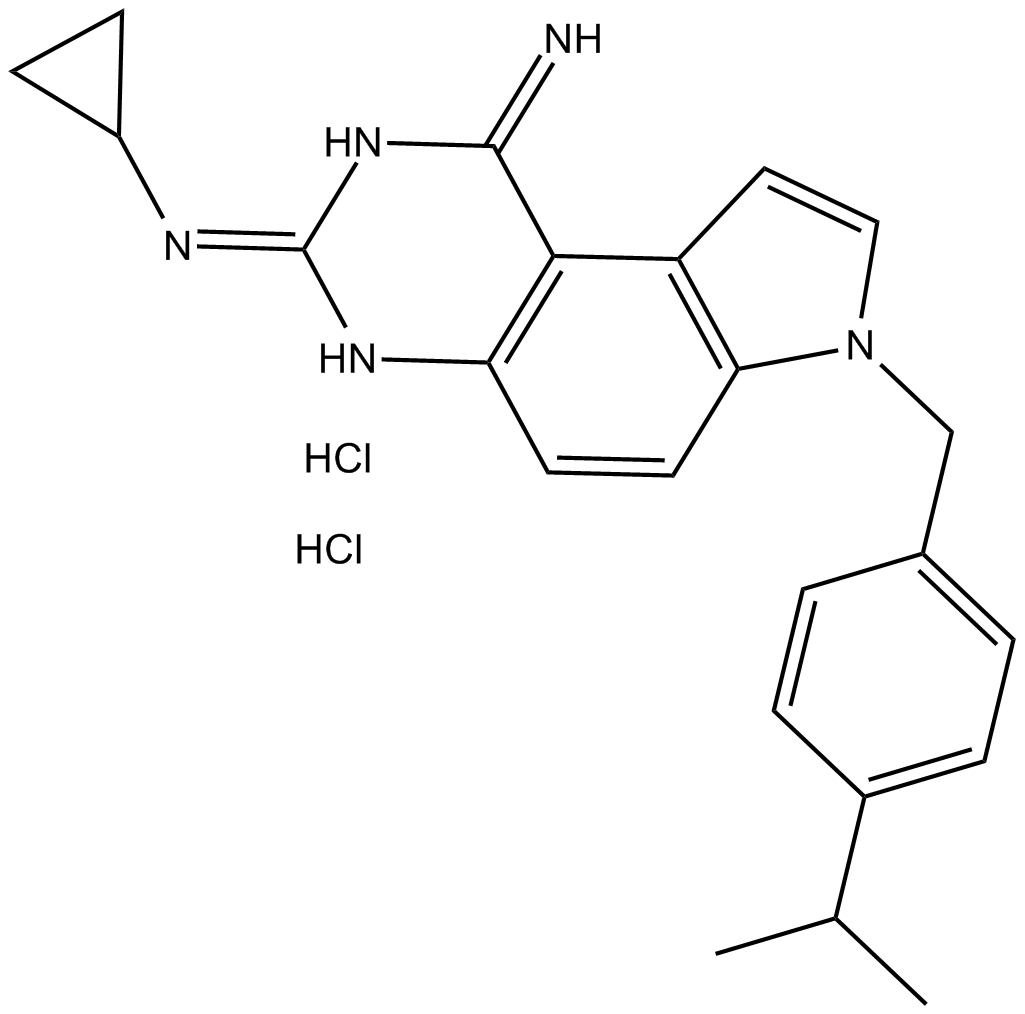
GC10291
SCH 79797 dihydrochloride
PAR1 receptor antagonist
-
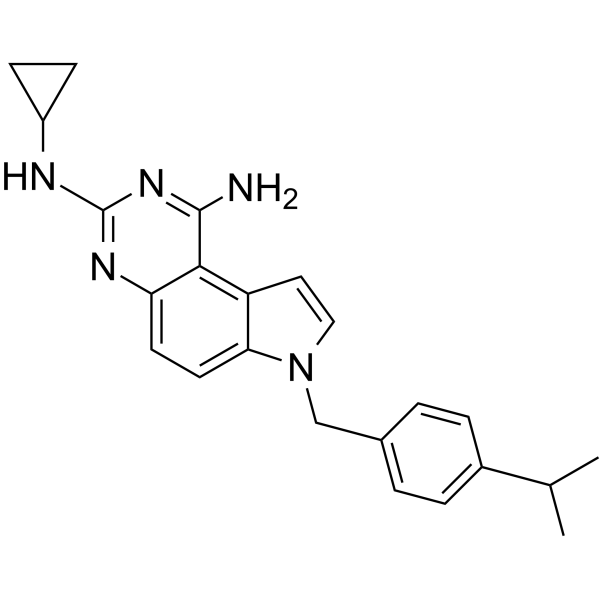
GC63540
SCH79797
SCH79797 is a highly potent, selective nonpeptide protease activated receptor 1 (PAR1) antagonist.
-
GC14540
SLIGKV-NH2
SLIGKV-NH2 (SLIGKV-NH2) is a highly potent protease-activated receptor-2 (PAR2) activating peptide.
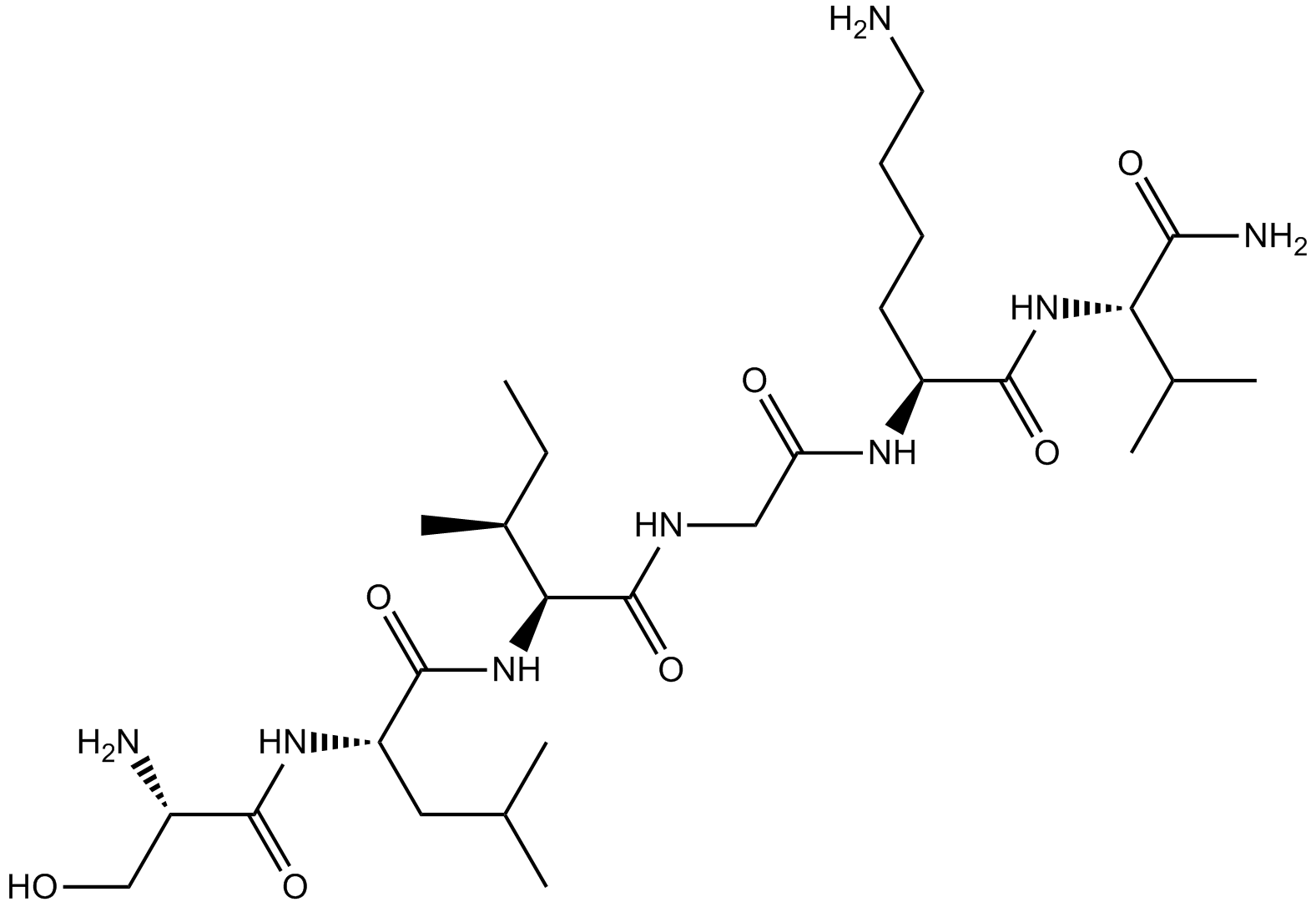
-
GC17514
SLIGRL-NH2
PAR2 activator
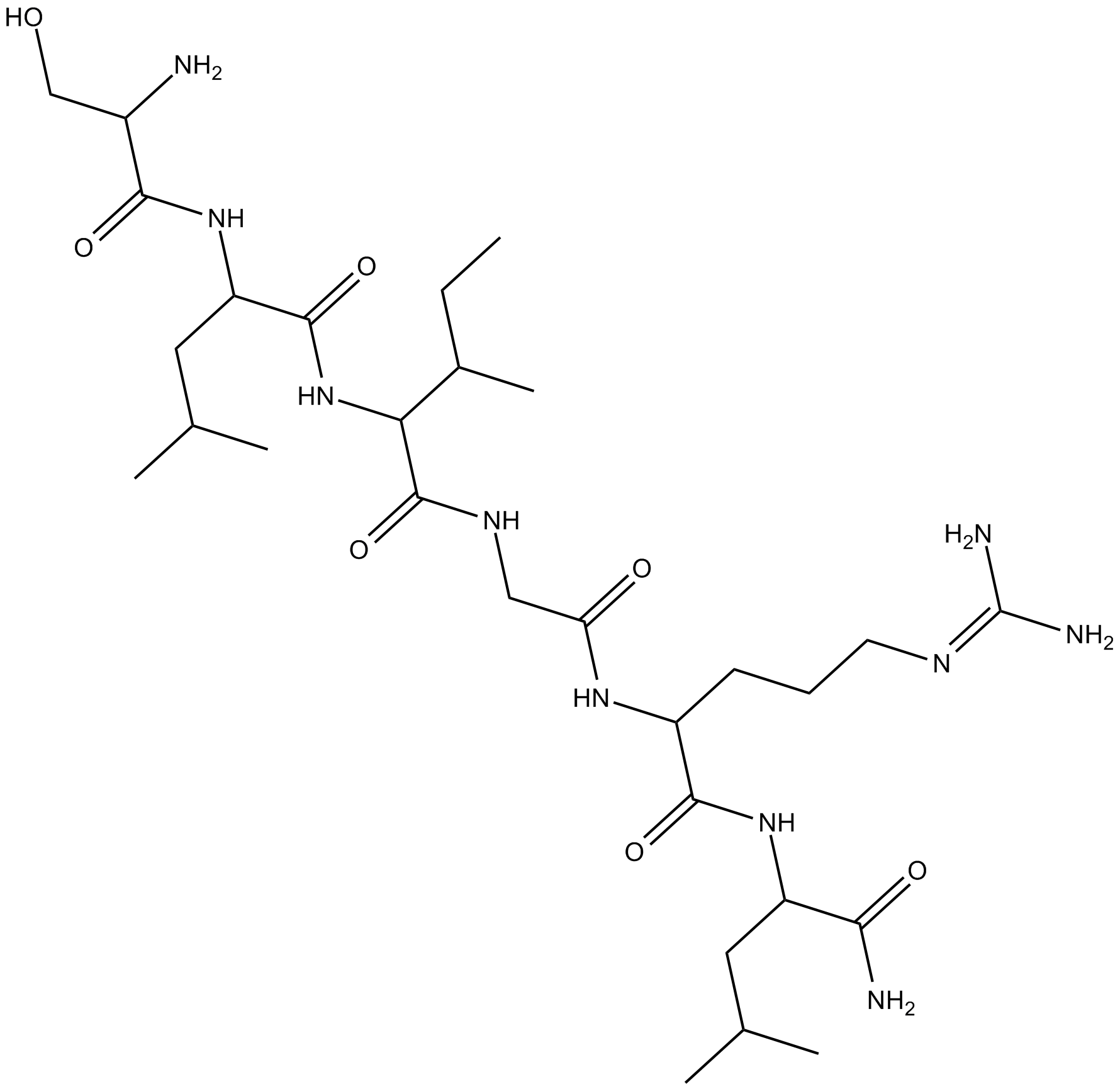
-
GC61316
tcY-NH2 TFA
tcY-NH2 ((trans-Cinnamoyl)-YPGKF-NH2) TFA is a potent selective PAR4 antagonist peptide.
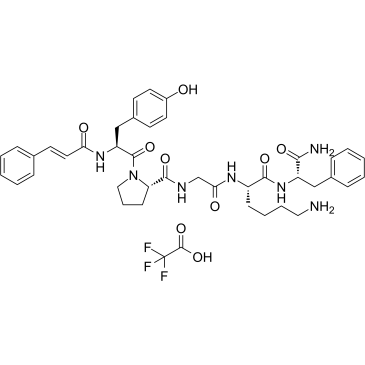
-
GC17444
TFLLR-NH2
PAR1 selective agonist
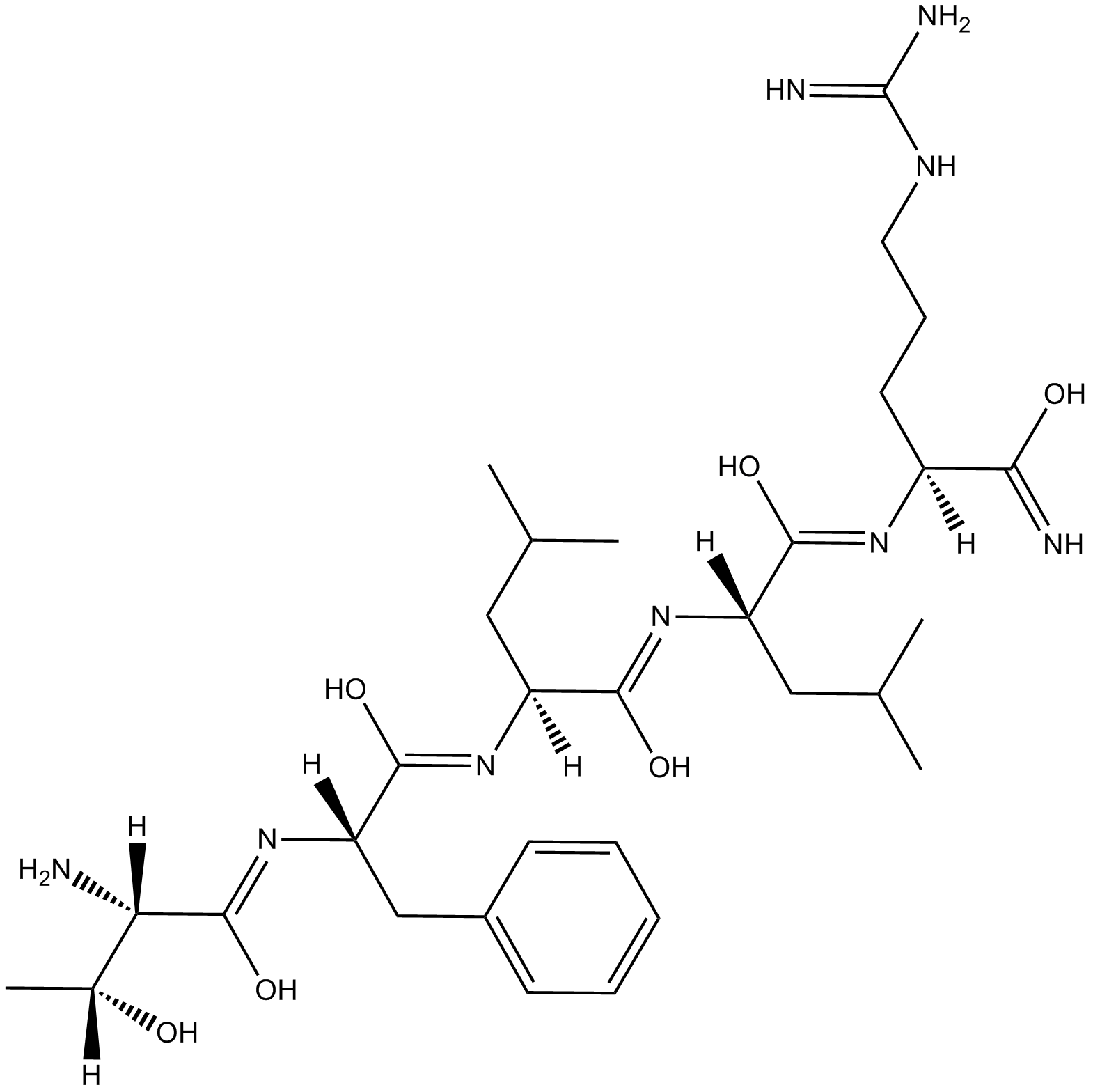
-
GC37770
TFLLR-NH2(TFA)
TFLLR-NH2 (TFA) is a selective PAR1 agonist with an EC50 of 1.9 μM.
-
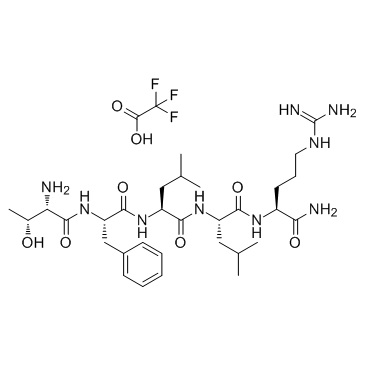
GP10085
Thrombin Receptor Activator for Peptide 5 (TRAP-5)
-
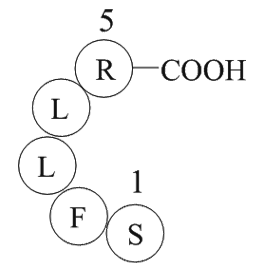
GC10140
Thrombin Receptor Agonist Peptide
Thrombin Receptor Agonist Peptide is a synthetic thrombin receptor agonist peptide.
-
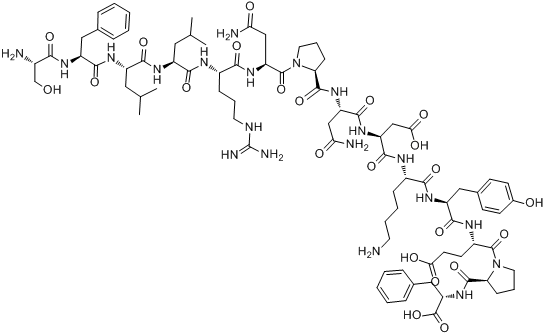
GC11662
TRAP-6
PAR1 agonist
-
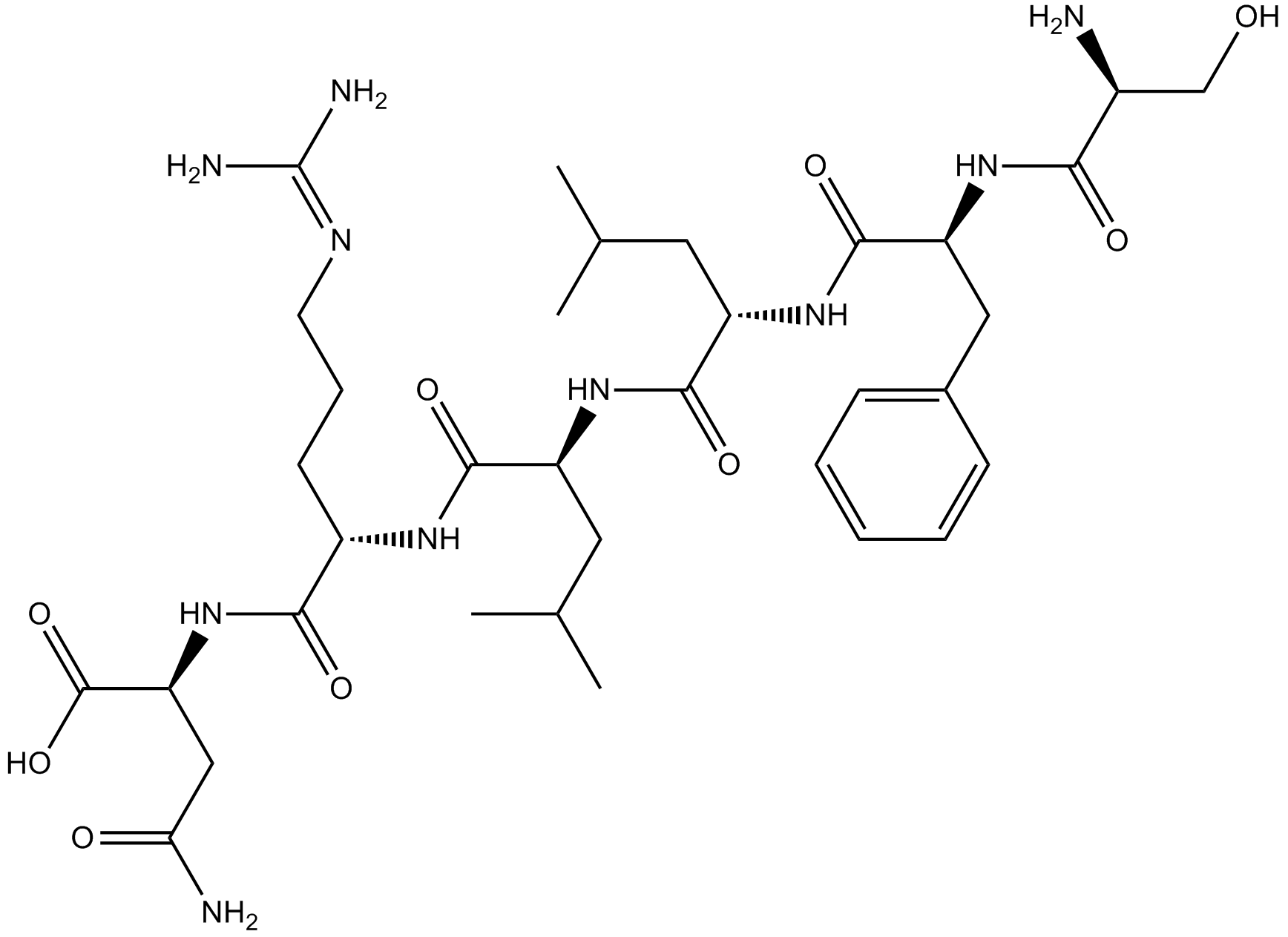
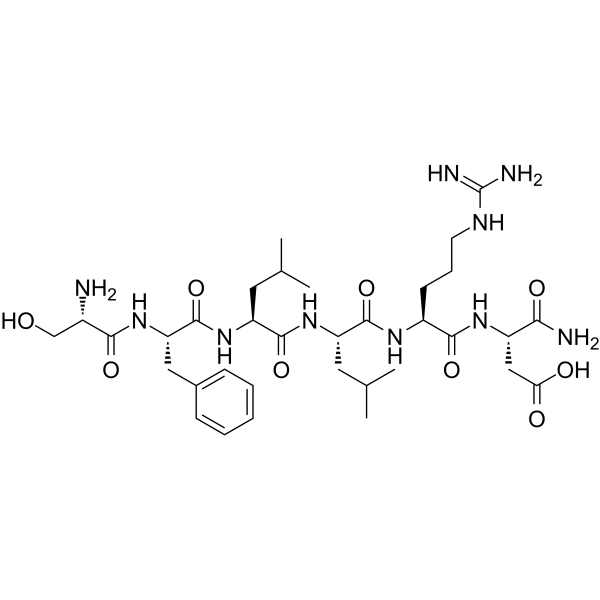
GC65529
TRAP-6 amide
TRAP-6 amide is a PAR-1 thrombin receptor agonist peptide.
-
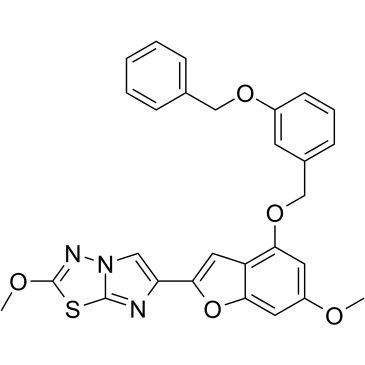
GC37852
UDM-001651
UDM-001651 is a potent, selective, and orally bioavailable protease-activated receptor 4 (PAR4) antagonist (IC50=4 nM; Kd=1.4 nM).
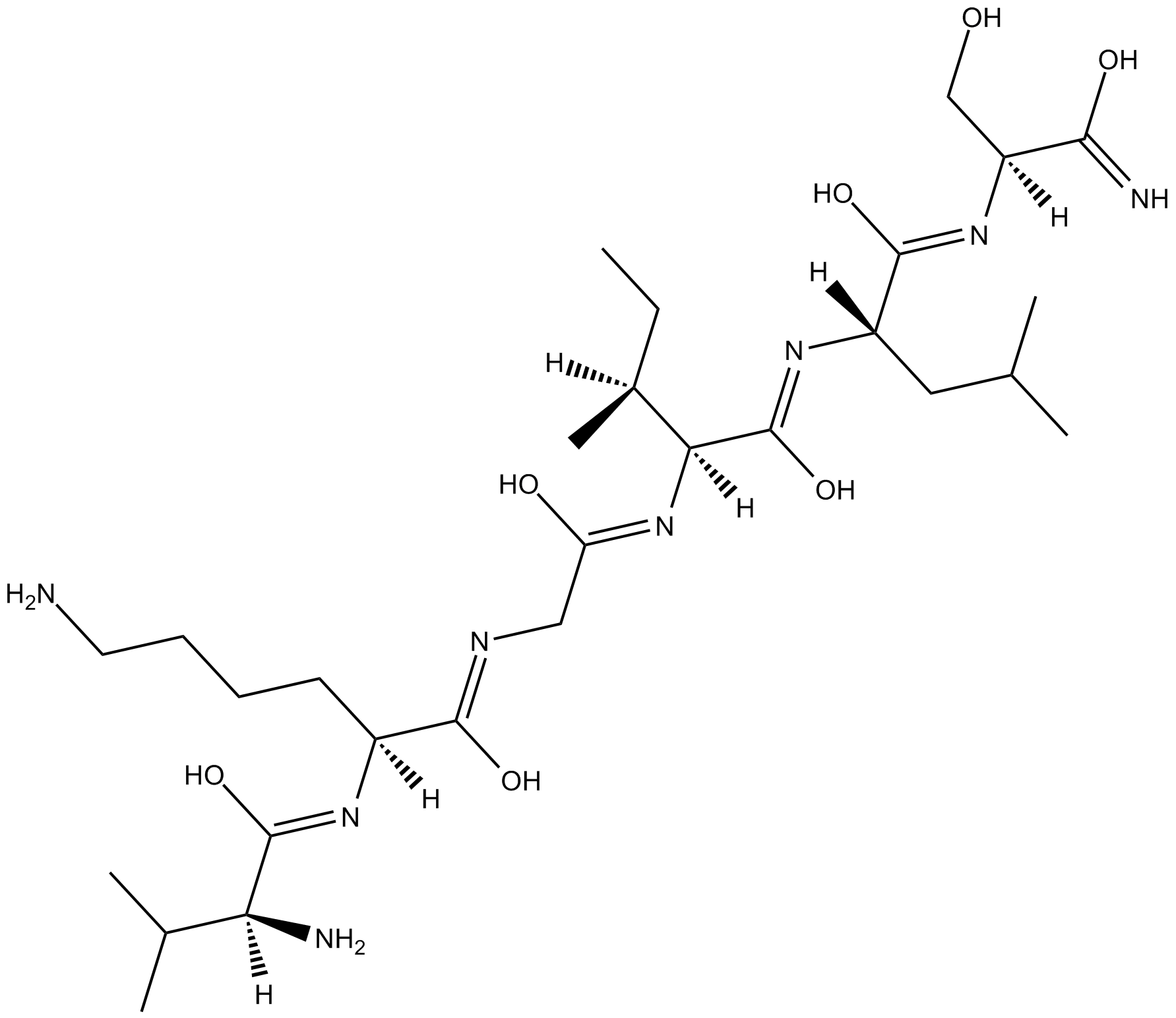
-
GC11727
VKGILS-NH2
control peptide for SLIGKV-NH2, PAR1 agonist
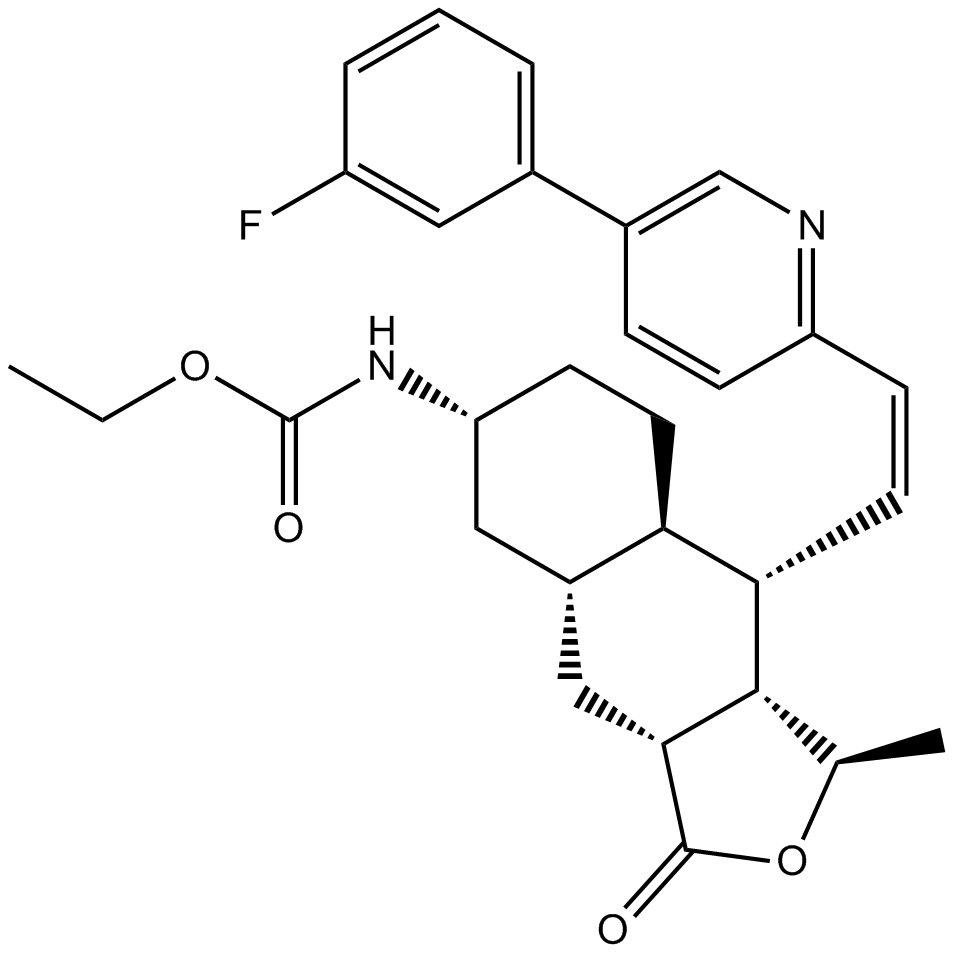
-
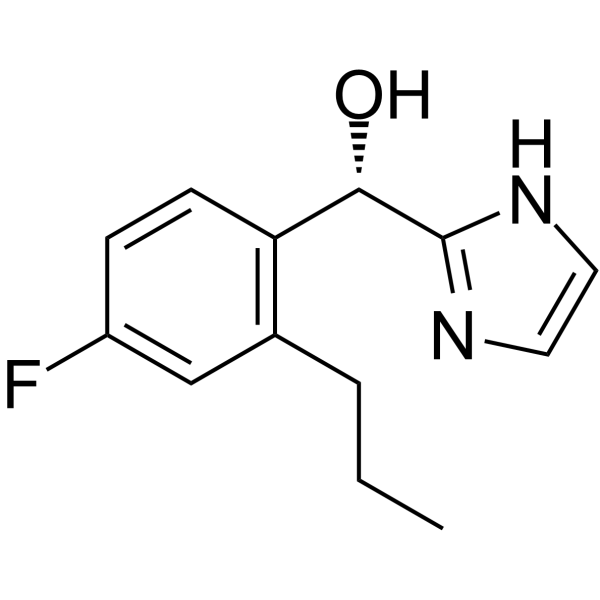
GC17545
Vorapaxar
PAR-1 antagonist,potent and orally active
-
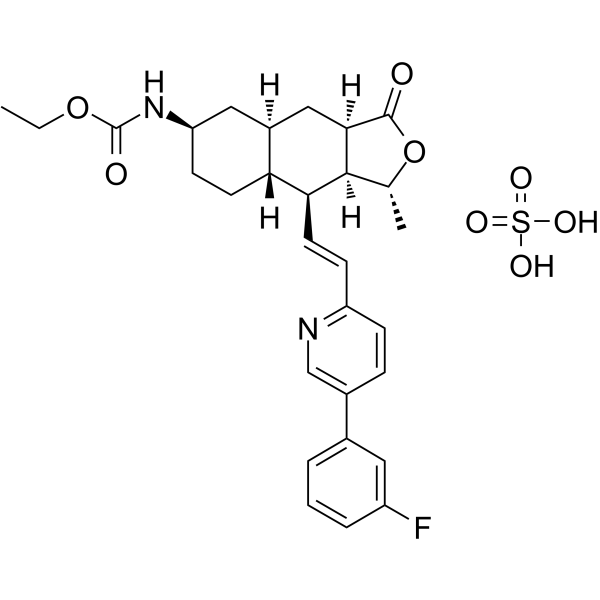
GC64347
Vorapaxar sulfate
Vorapaxar sulfate (SCH 530348 sulfate), an antiplatelet agent, is a selective, orally active, and competitive thrombin receptor protease-activated receptor (PAR-1) antagonist (Ki=8.1 nM).