Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(15)
- Caspase(77)
- 14.3.3 Proteins(3)
- Apoptosis Inducers(71)
- Bax(15)
- Bcl-2 Family(136)
- Bcl-xL(13)
- c-RET(15)
- IAP(32)
- KEAP1-Nrf2(73)
- MDM2(21)
- p53(137)
- PC-PLC(6)
- PKD(8)
- RasGAP (Ras- P21)(2)
- Survivin(8)
- Thymidylate Synthase(12)
- TNF-α(141)
- Other Apoptosis(1145)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(6)
- PD-1/PD-L1 interaction(60)
- ASK1(4)
- PAR4(2)
- RIP kinase(47)
- FKBP(22)
Products for Apoptosis
- Cat.No. Product Name Information
-
GC15355
2-Trifluoromethyl-2'-methoxychalcone
Nrf2 activator
-
GN10800
20(S)-NotoginsenosideR2
-
GC46528
25-hydroxy Cholesterol-d6
An internal standard for the quantification of 25hydroxy cholesterol
-
GC48482
28-Acetylbetulin
A lupane triterpenoid with anti-inflammatory and anticancer activities
-
GC35112
3'-Hydroxypterostilbene
3'-Hydroxypterostilbene is a Pterostilbene analogue. 3'-Hydroxypterostilbene inhibits the growth of COLO 205, HCT-116 and HT-29 cells with IC50s of 9.0, 40.2 and 70.9 ?M, respectively. 3'-Hydroxypterostilbene significantly down-regulates PI3K/Akt and MAPKs signaling pathways and effectively inhibits the growth of human colon cancer cells by inducing apoptosis and autophagy. 3'-Hydroxypterostilbene can be used for the research of cancer.
-
GC12791
3,3'-Diindolylmethane
A phytochemical with antiradiation and chemopreventative effects

-
GC42237
3,5-dimethyl PIT-1
PtdIns-(3,4,5)-P3 (PIP3) serves as an anchor for the binding of signal transduction proteins bearing pleckstrin homology (PH) domains such as phosphatidylinositol 3-kinase (PI3K) or PTEN.
-
GC64762
3,6-Dihydroxyflavone
3,6-Dihydroxyflavone is an anti-cancer agent. 3,6-Dihydroxyflavone dose- and time-dependently decreases cell viability and induces apoptosis by activating caspase cascade, cleaving poly (ADP-ribose) polymerase (PARP). 3,6-Dihydroxyflavone increases intracellular oxidative stress and lipid peroxidation.
-
GC46583
3-Amino-2,6-Piperidinedione
An active metabolite of (±)-thalidomide
-
GC49849
3-Aminosalicylic Acid
A salicylic acid derivative
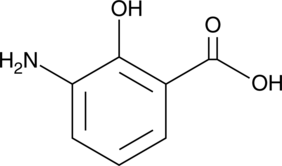
-
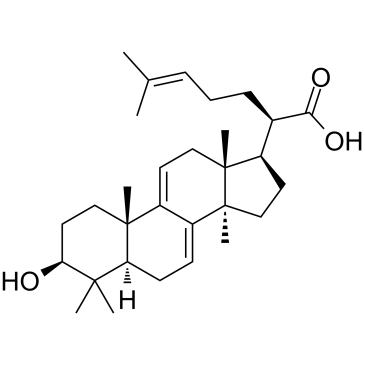
GC35106
3-Dehydrotrametenolic acid
3-?Dehydrotrametenolic acid, isolated from the sclerotium of Poria cocos, is a lactate dehydrogenase (LDH) inhibitor. 3-?Dehydrotrametenolic acid promotes adipocyte differentiation in vitro and acts as an insulin sensitizer in vivo. 3-?Dehydrotrametenolic acid induces apoptosis and has anticancer activity.
-
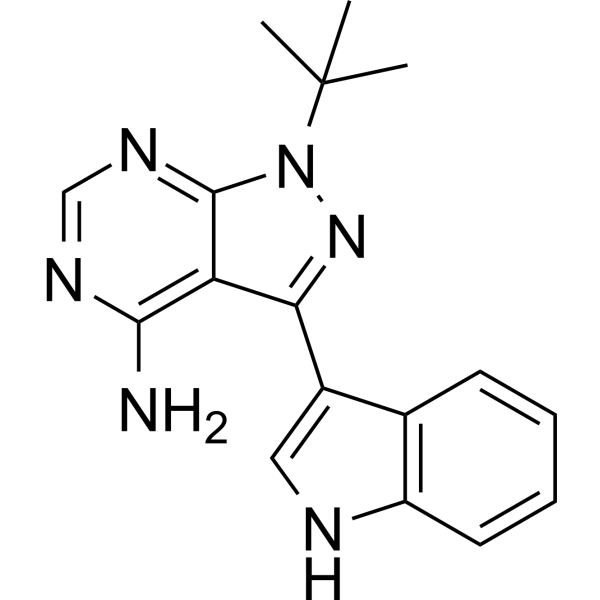
GC68537
3-IN-PP1
3-IN-PP1 is a protein kinase D (PKD) inhibitor. It has effective and broad PKD inhibitory activity against PKD1, PKD2, and PKD3 with IC50 values of 108, 94, and 108 nM respectively. Additionally, 3-IN-PP1 is a broad-spectrum anticancer agent that inhibits the growth of various tumor cells. It can be used in cancer research.
-
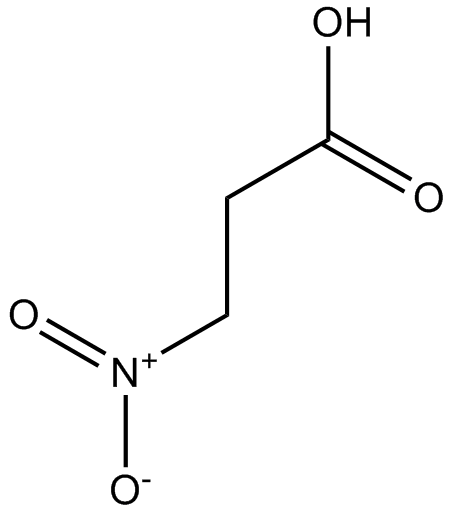
GC17394
3-Nitropropionic acid
3-Nitropropionic acid (β-Nitropropionic acid) is an irreversible inhibitor of succinate dehydrogenase.
-
GC35099
3-O-Acetyloleanolic acid
3-O-Acetyloleanolic acid (3AOA), an oleanolic acid derivative isolated from the seeds of Vigna sinensis K., induces in cancer and also exhibits anti-angiogenesis activity.
-
GC60507
3-O-Methylgallic acid
3-O-Methylgallic acid (3,4-Dihydroxy-5-methoxybenzoic acid) is an anthocyanin metabolite and has potent antioxidant capacity. 3-O-methylgallic acid inhibits Caco-2 cell proliferation with an IC50 value of 24.1 μM. 3-O-methylgallic acid also induces cell apoptosis and has anti-cancer effects.
-
GC32767
3BDO
A butyrolactone derivative and autophagy inhibitor
-
GC45354
4β-Hydroxywithanolide E
A withanolide with anti-inflammatory and anticancer activities
-
GC48437
4'-Acetyl Chrysomycin A
A bacterial metabolite with antibacterial and anticancer activities
-
GC42346
4-bromo A23187
4-bromo A23187 is a halogenated analog of the highly selective calcium ionophore A23187.
-
GC42401
4-hydroperoxy Cyclophosphamide
An activated analog of cyclophosphamide
-
GC30896
4-Hydroxybenzyl alcohol
A phenol with diverse biological activities
-
GC33815
4-Hydroxyphenylacetic acid
A phenolic acid with anti-inflammatory activity
-
GC35138
4-Methyldaphnetin
4-Methyldaphnetin is a precursor in the synthesis of derivatives of 4-methyl coumarin. 4-Methyldaphnetin has potent, selective anti-proliferative and apoptosis-inducing effects on several cancer cell lines. 4-Methyldaphnetin possesses radical scavenging property and strongly inhibits membrane lipid peroxidation.
-
GC68231
4-Methylsalicylic acid

-
GC31648
4-Octyl Itaconate
4-Octyl Itaconate?(4-OI) is a cell-permeable itaconate derivative. Itaconate and 4-Octyl Itaconate?had similar thiol reactivity, making 4-Octyl Itaconate?a suitable itaconate surrogate to study its biological function.
-
GC49127
4-oxo Cyclophosphamide
An inactive metabolite of cyclophosphamide
-
GC45352
4-oxo Withaferin A
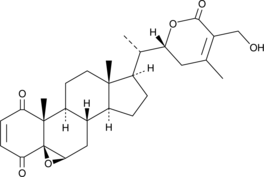
4-oxo Withaferin A is the analogue of withaferin A. Withaferin A is a withanolide isolated from Withania somnifera. 4-oxo Withaferin A has the potential for the research of multiple myeloma.
-
GC45353
4-oxo-27-TBDMS Withaferin A
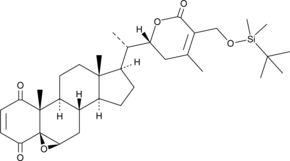
4-oxo-27-TBDMS Withaferin A, a withaferin A derivative, exhibits potent antiproliferative effects on the tumor cells.4-oxo-27-TBDMS Withaferin A induces tumor cells apoptosis. 4-oxo-27-TBDMS Withaferin A is a anticancer agent.
-
GC60525
4-Vinylphenol (10%w/w in propylene glycol)
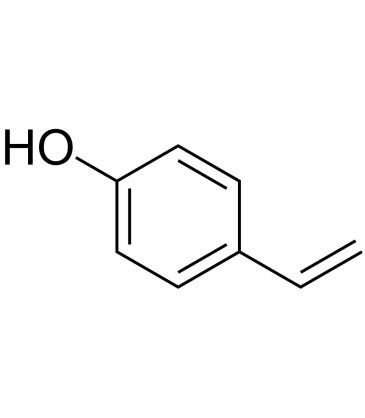
4-Vinylphenol is found in the medicinal herb Hedyotis diffusa Willd, wild rice and is also the metabolite of p-coumaric and ferulic acid by lactic acid bacteria in wine. 4-Vinylphenol induces apoptosis and inhibits blood vessels formation and suppresses invasive breast tumor growth in vivo.
-
GC10468
4EGI-1
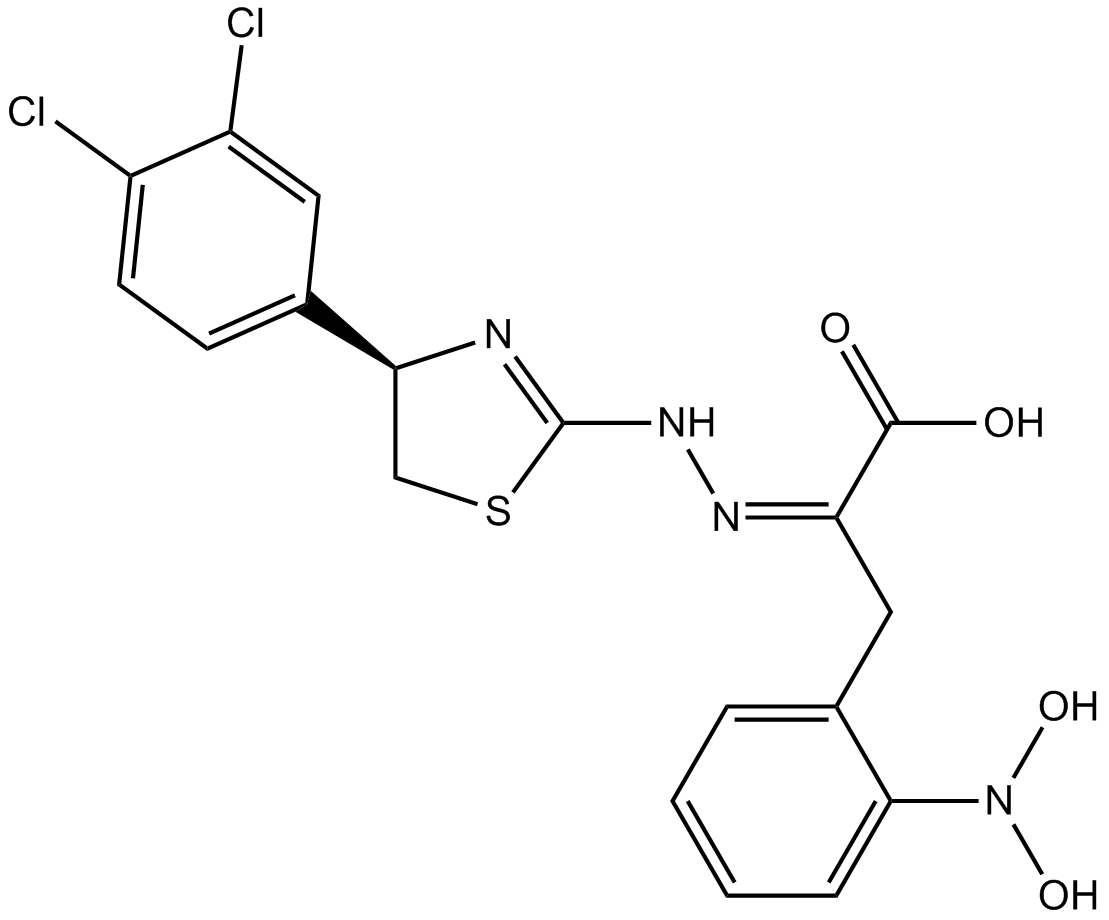
Competitive eIF4E/eIF4G interaction inhibitor
-
GC35150
5,7,4'-Trimethoxyflavone
5,7,4'-Trimethoxyflavone is isolated from Kaempferia parviflora (KP) that is a famous medicinal plant from Thailand. 5,7,4'-Trimethoxyflavone induces apoptosis, as evidenced by increments of sub-G1 phase, DNA fragmentation, annexin-V/PI staining, the Bax/Bcl-xL ratio, proteolytic activation of caspase-3, and degradation of poly (ADP-ribose) polymerase (PARP) protein.5,7,4'-Trimethoxyflavone is significantly effective at inhibiting proliferation of SNU-16 human gastric cancer cells in a concentration dependent manner.
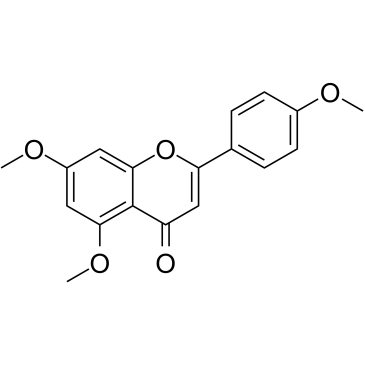
-
GN10629
5,7-dihydroxychromone
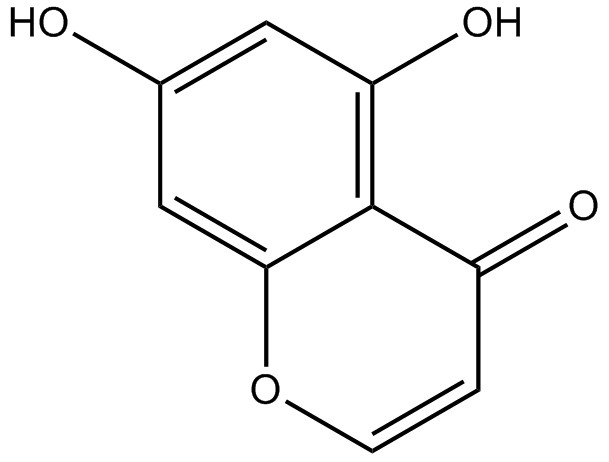
-
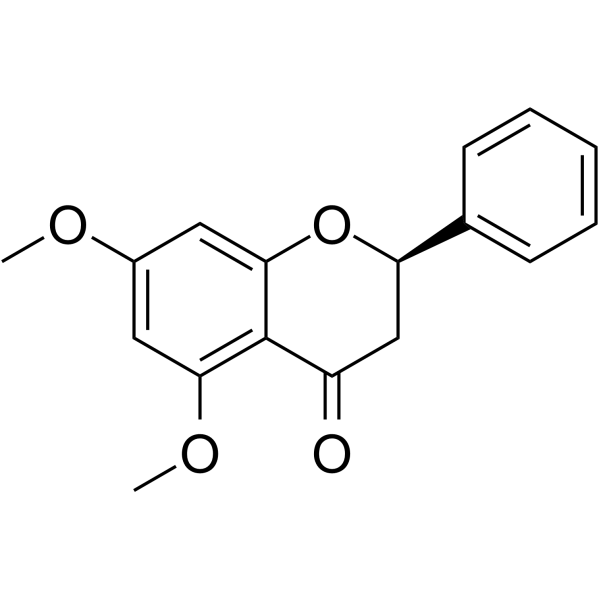
GC63972
5,7-Dimethoxyflavanone
5,7-Dimethoxyflavanone shows potent antimutagenic activity against MeIQ mutagenesis in Ames test using the S.
-
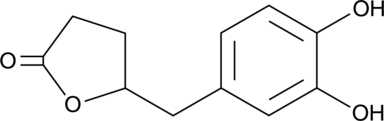
GC52227
5-(3',4'-Dihydroxyphenyl)-γ-Valerolactone
An active metabolite of various polyphenols
-
GC35147
5-(N,N-Hexamethylene)-amiloride
An amiloride derivative with diverse biological activities
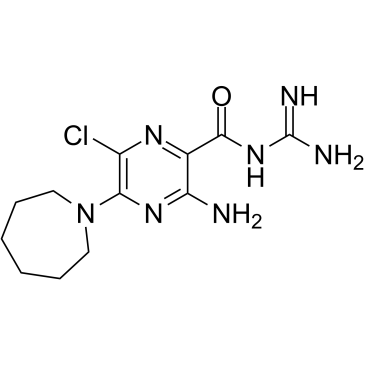
-
GC45356
5-Aminolevulinic Acid (hydrochloride)
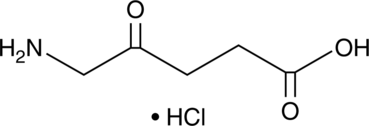
-
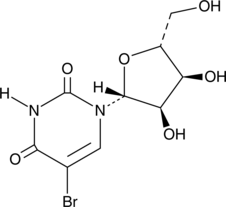
GC46681
5-Bromouridine
A brominated uridine analog
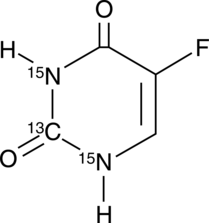
-
GC42545
5-Fluorouracil-13C,15N2
5-Fluorouracil-13C,15N2 is intended for use as an internal standard for the quantification of 5-flurouracil by GC- or LC-MS.
-
GC46705
5-Methoxycanthinone
5-Methoxycanthinone is an orally active inhibitor of Leishmania strains.
-
GC42586
6α-hydroxy Paclitaxel
6α-hydroxy Paclitaxel is a primary metabolite of the anticancer compound paclitaxel, produced by the action of the cytochrome P450 isoform CYP2C8.
-
GC45772
6(5H)-Phenanthridinone
An inhibitor of PARP1 and 2
-
GN10093
6-gingerol
-
GC49429
6-keto Lithocholic Acid
A metabolite of lithocholic acid
-
GC35184
7,3',4'-Tri-O-methylluteolin
7,3',4'-Tri-O-methylluteolin (5-Hydroxy-3',4',7-trimethoxyflavone), a flavonoid compound, possesses potent anti-inflammatory effects in LPS-induced macrophage cell line mediated by inhibition of release of inflammatory mediators, NO, PGE2, and pro-inflammatory cytokines.
-
GC45673
7,8-Dihydroneopterin
An antioxidant
-
GC16853
7,8-Dihydroxyflavone
Tyrosine kinase receptor B (TrkB) agonist
-
GC42616
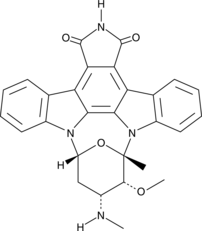
7-oxo Staurosporine
7-oxo Staurosporine is an antibiotic originally isolated from S.
-
GC16037
7BIO
caspase independent nonapoptotic cell death inducer
-
GC46741
8(E),10(E),12(Z)-Octadecatrienoic Acid
A conjugated PUFA

-
GC42622
8-bromo-Cyclic AMP
8-bromo-Cyclic AMP is a brominated derivative of cAMP that remains long-acting due to its resistance to degradation by cAMP phosphodiesterase.
-
GC49275
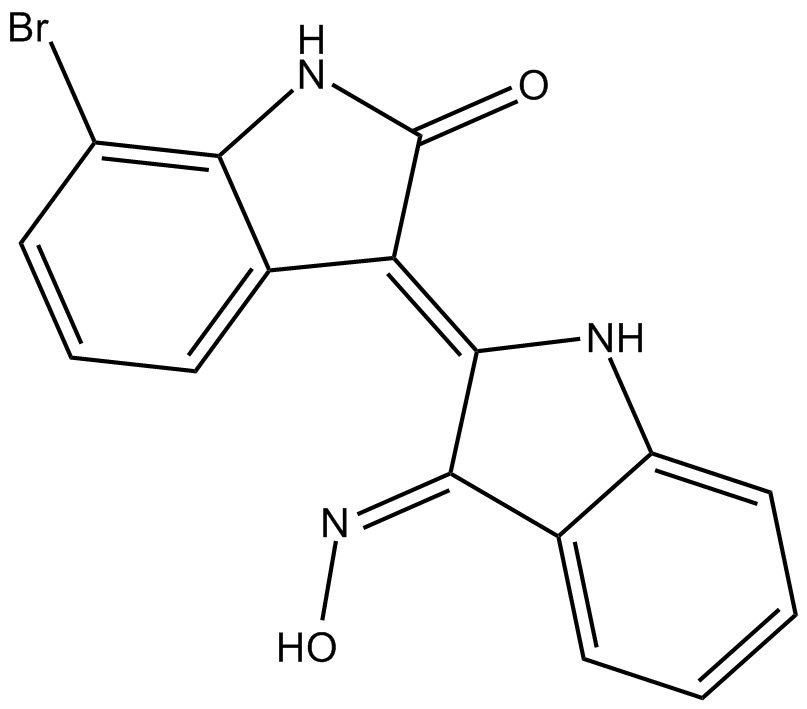
8-Oxycoptisine
8-Oxycoptisine is a natural protoberberine alkaloid with anti-cancer activity.
-
GC17119
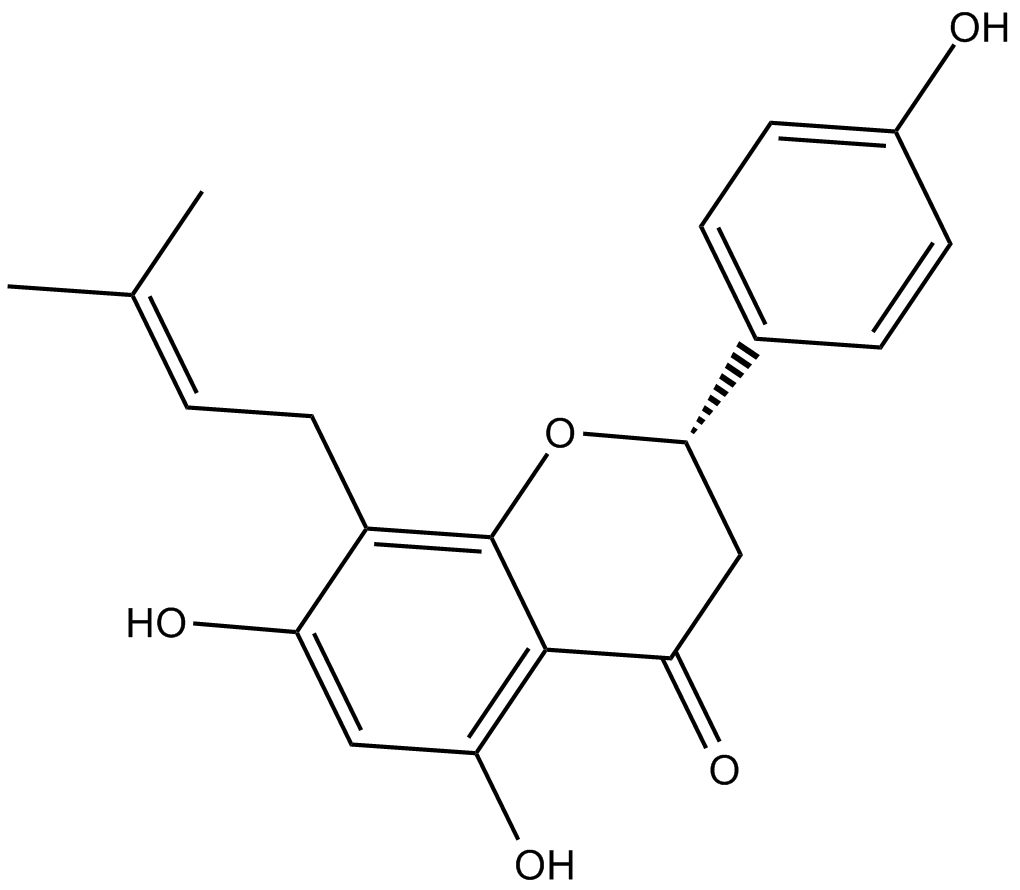
8-Prenylnaringenin
estrogen receptor inhibitor
-
GC41642
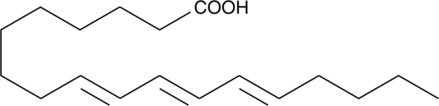
9(E),11(E),13(E)-Octadecatrienoic Acid
9(E),11(E),13(E)-Octadecatrienoic acid (β-ESA) is a conjugated polyunsaturated fatty acid that is found in plant seed oils and in mixtures of conjugated linolenic acids synthesized by the alkaline isomerization of linolenic acid.
-
GC41643
9(Z),11(E),13(E)-Octadecatrienoic Acid
9(Z),11(E),13(E)-Octadecatrienoic Acid (α-ESA) is a conjugated polyunsaturated fatty acid commonly found in plant seed oil.

-
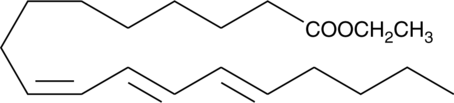
GC40785
9(Z),11(E),13(E)-Octadecatrienoic Acid ethyl ester
9(Z),11(E),13(E)-Octadecatrienoic Acid ethyl ester (α-ESA) is a conjugated polyunsaturated fatty acid commonly found in plant seed oil.
-
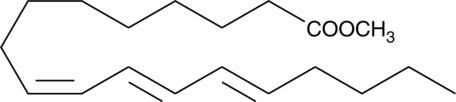
GC40710
9(Z),11(E),13(E)-Octadecatrienoic Acid methyl ester
9Z,11E,13E-octadecatrienoic acid (α-ESA) is a conjugated polyunsaturated fatty acid commonly found in plant seed oil.
-
GC39152
9-ING-41
9-ING-41 is a maleimide-based ATP-competitive and selective glycogen synthase kinase-3β (GSK-3β) inhibitor with an IC50 of 0.71 μM. 9-ING-41 significantly leads to cell cycle arrest, autophagy and apoptosis in cancer cells. 9-ING-41 has anticancer activity and has the potential for enhancing the antitumor effects of chemotherapeutic drugs.
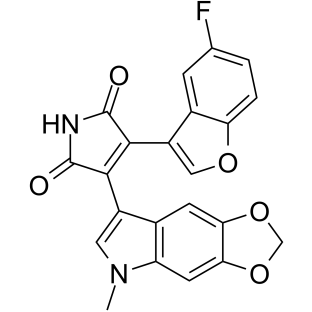
-
GN10035
9-Methoxycamptothecin
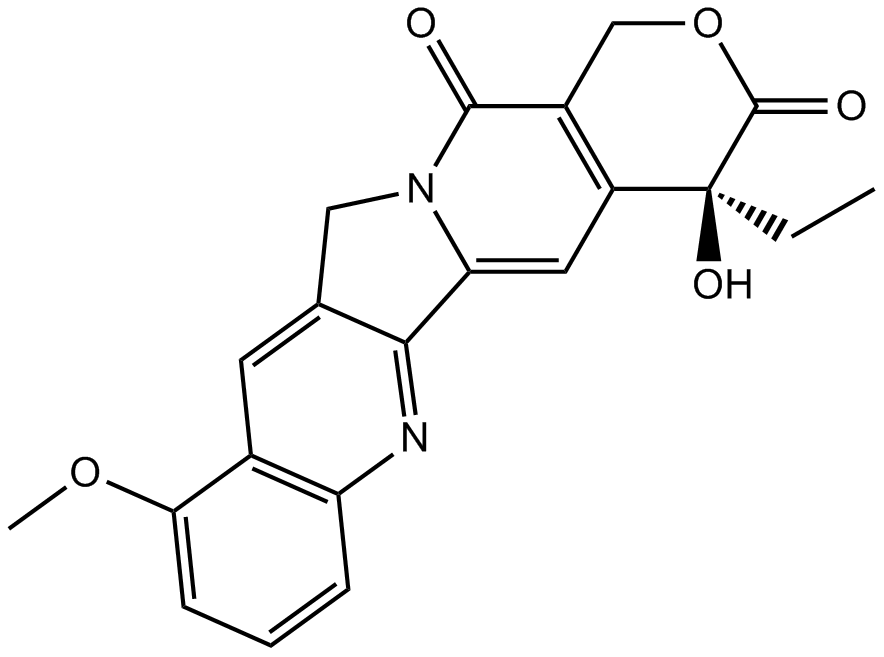
-
GC45960
9c(i472)
9c(i472) is a potent inhibitor of 15-LOX-1 (15-lipoxygenase-1) with an IC50 value of 0.19 μM.
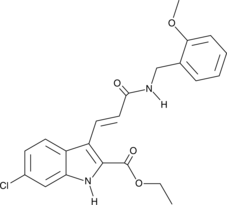
-
GC50465
A 410099.1
High affinity XIAP antagonist; active in vivo
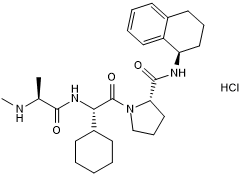
-
GC17512
A-1155463
BCL-XL inhibitor, potent and selective
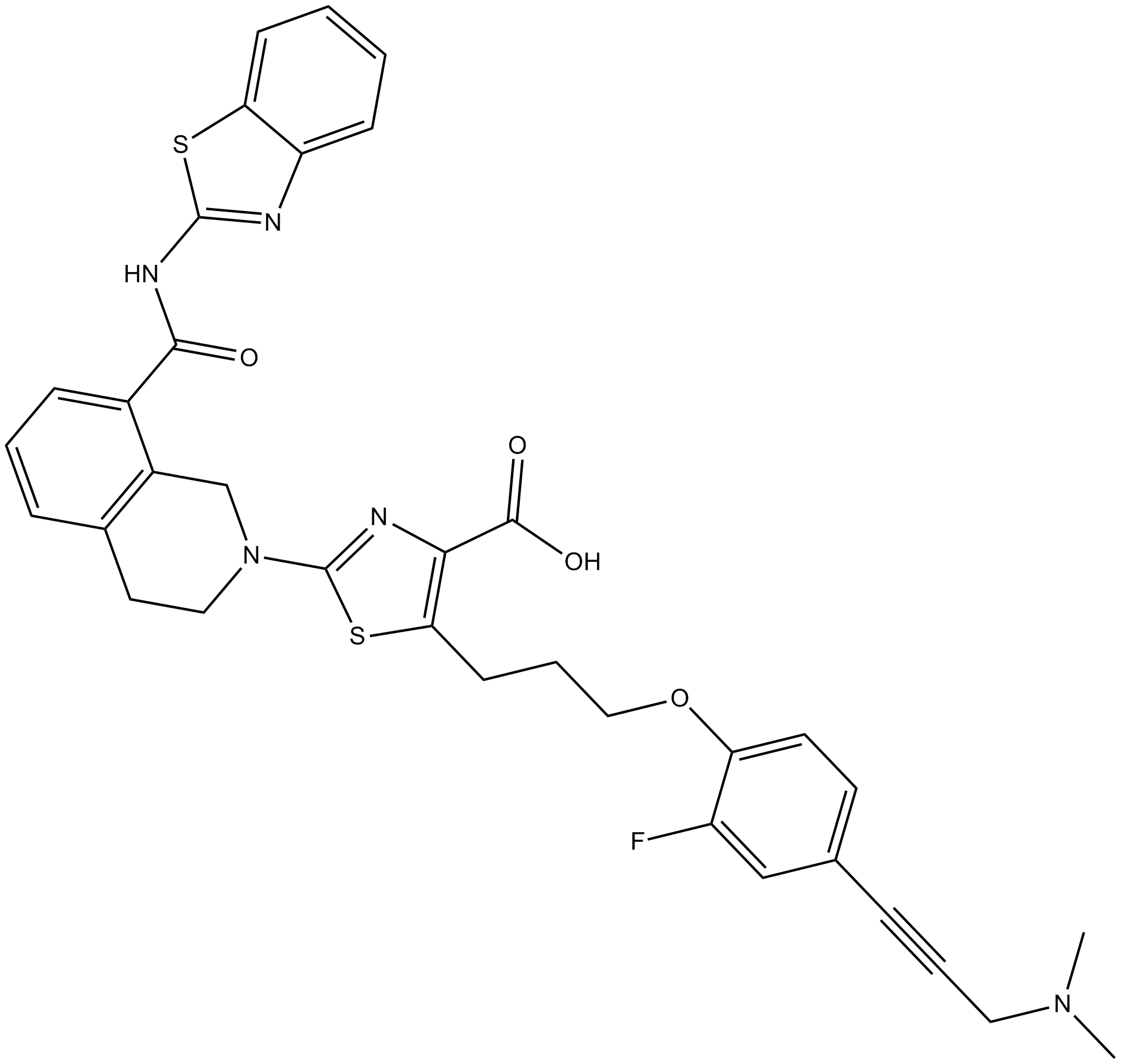
-
GC16278
A-1210477
MCL-1 inhibitor
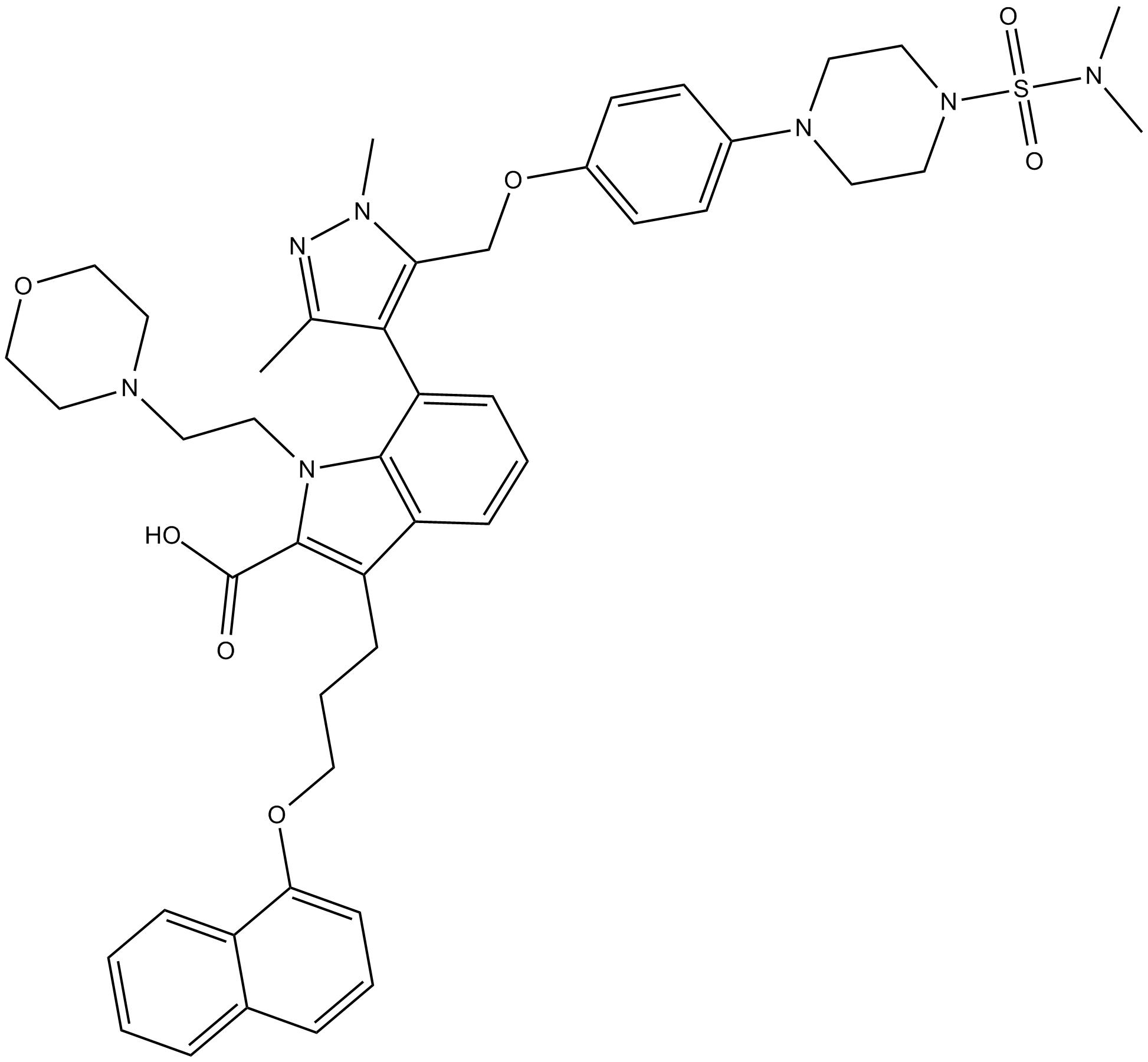
-
GC17513
A-1331852
BCL-XL inhibitor, potent and selective
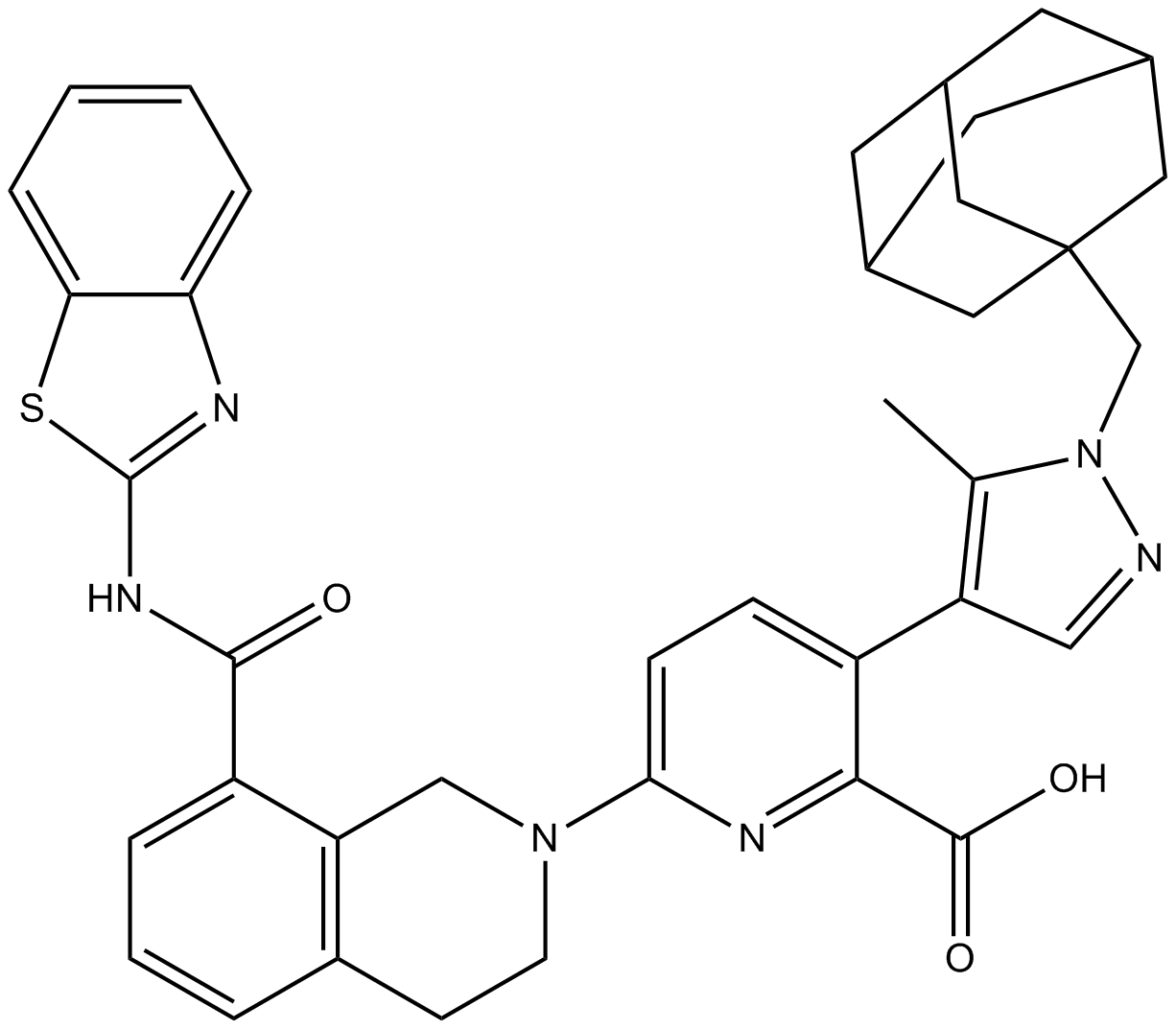
-
GC60544
A-192621
A-192621 is a potent, nonpeptide, orally active and selective endothelin B (ETB) receptor antagonist with an IC50 of 4.5 nM and a Ki of 8.8 nM.
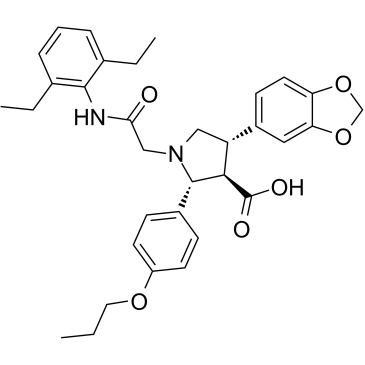
-
GC32981
A-385358
A-385358 is a selective inhibitor of Bcl-XL with Kis of 0.80 and 67 nM for Bcl-XL and Bcl-2, respectively.
-
GC11200
A23187
A23187, free acid is a Ca2+ ionophore
-
GC42659
A23187 (calcium magnesium salt)
A23187 is a divalent cation ionophore.
-
GC35216
AAPK-25
AAPK-25 is a potent and selective Aurora/PLK dual inhibitor with anti-tumor activity, which can cause mitotic delay and arrest cells in a prometaphase, reflecting by the biomarker histone H3Ser10 phosphorylation and followed by a surge in apoptosis. AAPK-25 targets Aurora-A, -B, and -C with Kd values ranging from 23-289 nM, as well as PLK-1, -2, and -3 with Kd values ranging from 55-456 nM.
-
GC13805
Abacavir
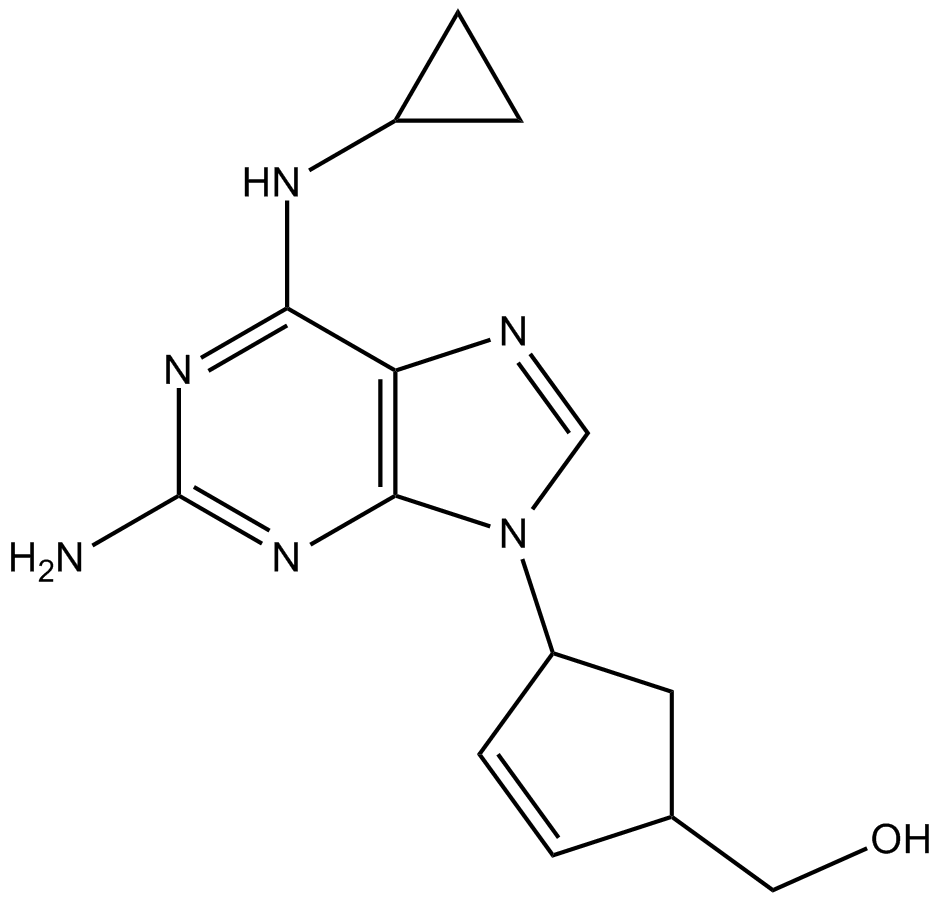
-
GC64674
ABBV-167
ABBV-167 is a phosphate prodrug of the BCL-2 inhibitor venetoclax.
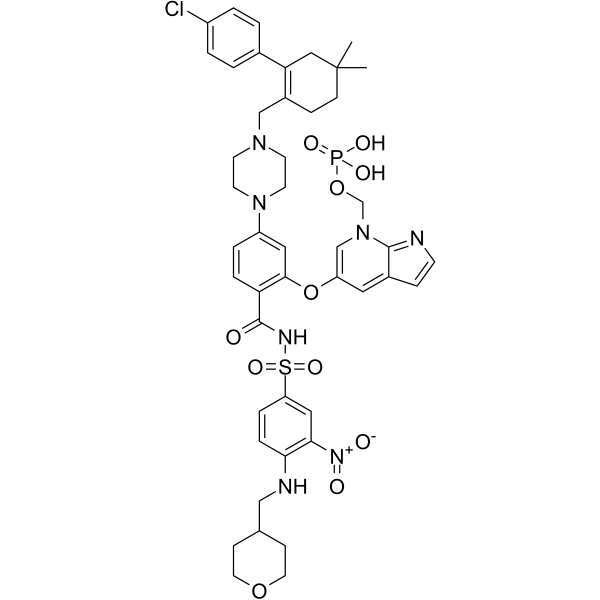
-
GC60548
ABT-100
ABT-100 is a potent, highly selective and orally active farnesyltransferase inhibitor. ABT-100 inhibits cell proliferation (IC50s of 2.2 nM, 3.8 nM, 5.9 nM, 6.9 nM, 9.2 nM, 70 nM and 818 nM for EJ-1, DLD-1, MDA-MB-231, HCT-116, MiaPaCa-2, PC-3, and DU-145 cells, respectively), increases apoptosis and decreases angiogenesis. ABT-100 possesses broad-spectrum antitumor activity.
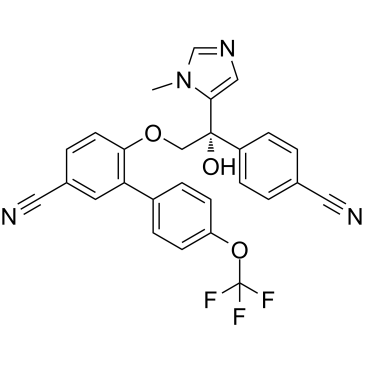
-
GC14069
ABT-199
A Bcl-2 inhibitor
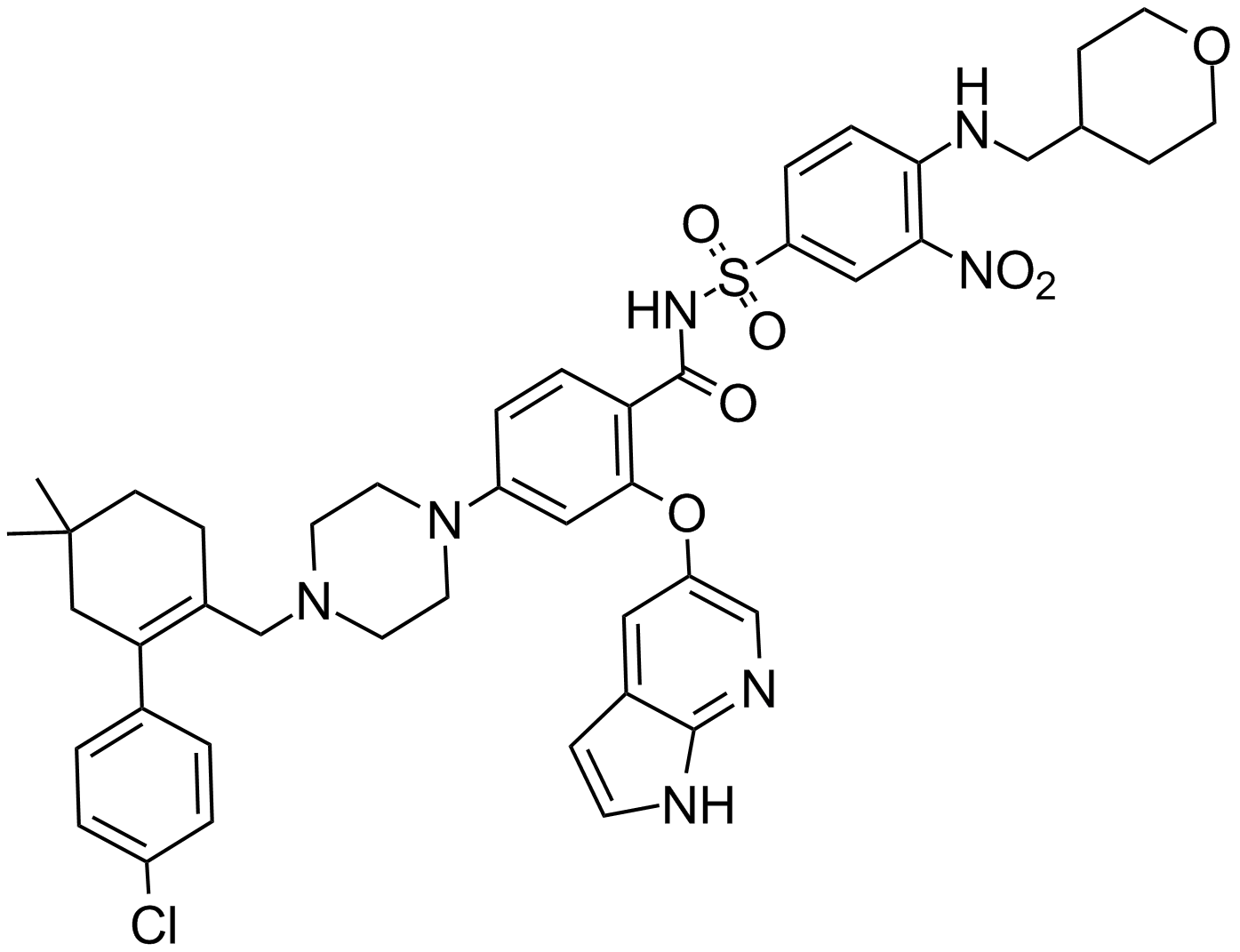
-
GC12405
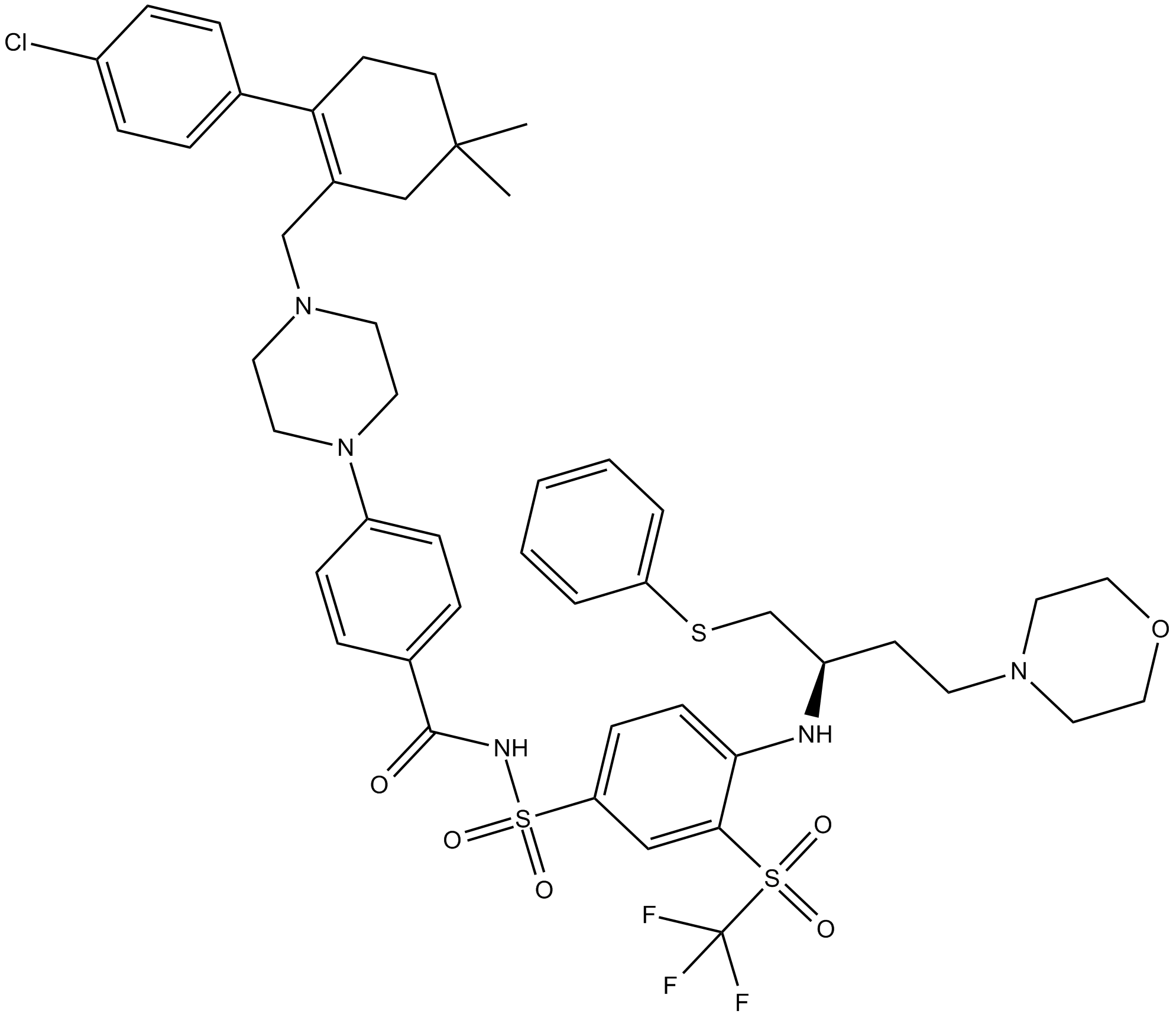
ABT-263 (Navitoclax)
ABT-263 (Navitoclax) is a inhibitor of Bcl-xL, Bcl-2 and Bcl-w, with Ki ≤0.5 nM, ≤1 nM and ≤1 nM respectively.
-
GC49745
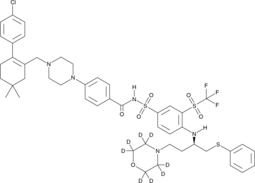
ABT-263-d8
ABT-263-d8 is the deuterium labeled Navitoclax. Navitoclax (ABT-263) is a potent and orally active Bcl-2 family protein inhibitor that binds to multiple anti-apoptotic Bcl-2 family proteins, such as Bcl-xL, Bcl-2 and Bcl-w, with a Ki of less than 1 nM.
-
GC17234
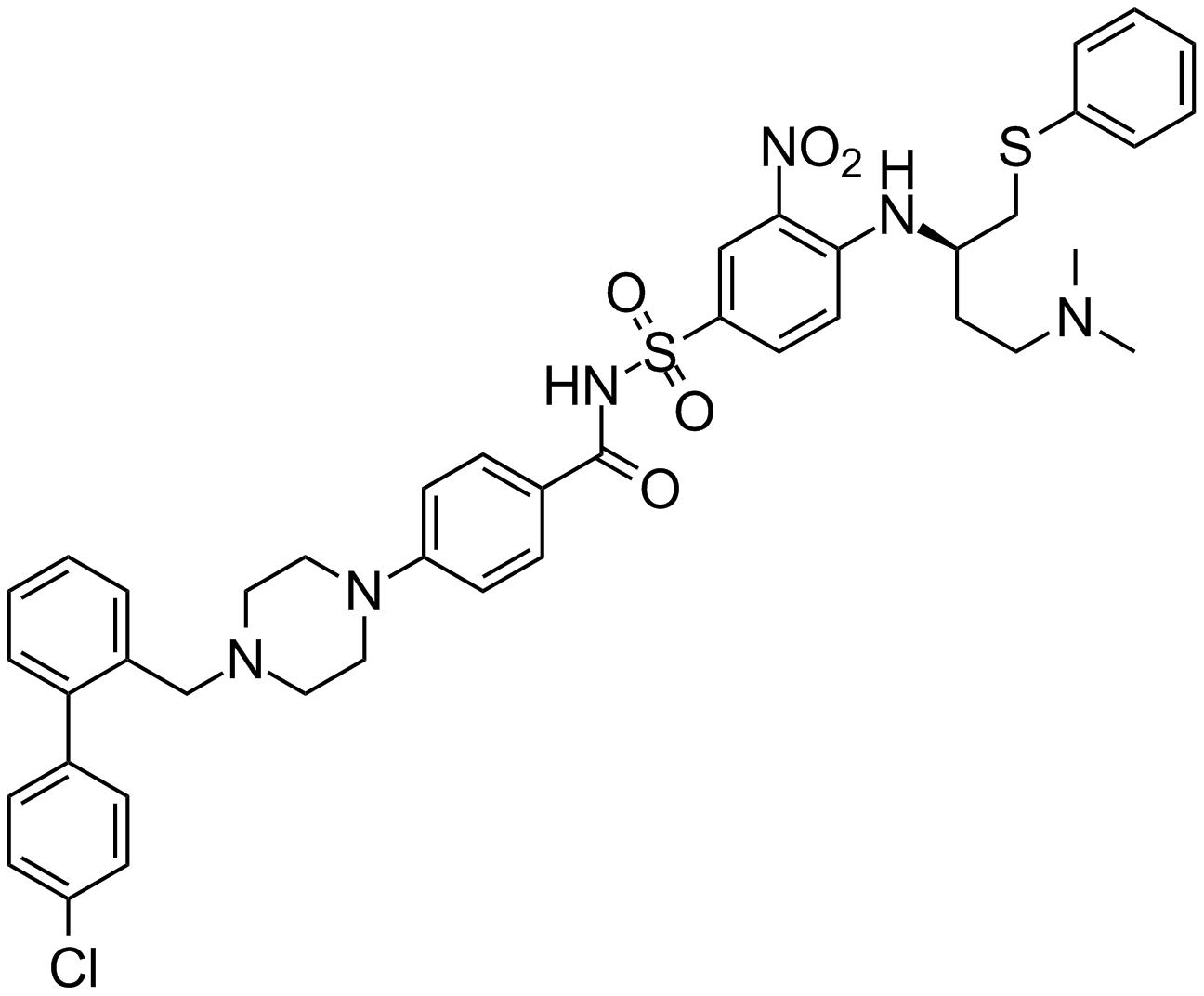
ABT-737
An inhibitor of anti-apoptotic Bcl-2 proteins
-
GA20494
Ac-Asp-Glu-Val-Asp-pNA
The cleavage of the chromogenic caspase-3 substrate Ac-DEVD-pNA can be monitored at 405 nm.

-
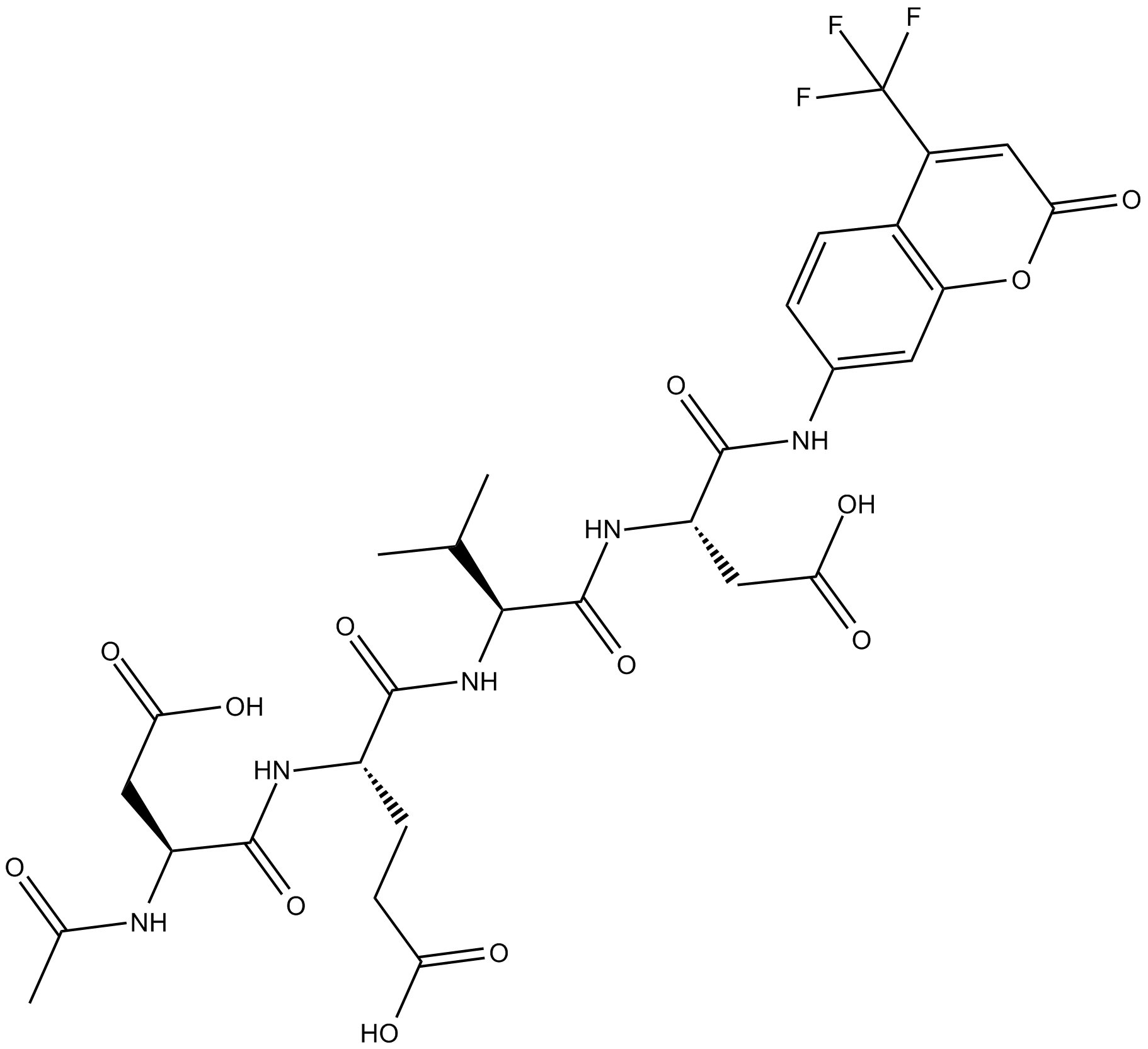
GC17602
Ac-DEVD-AFC
fluorogenic substrate for activated caspase-3
-
GC32695
Ac-DEVD-CHO
Ac-DEVD-CHO is a specific Caspase-3 inhibitor with a Ki value of 230 pM.
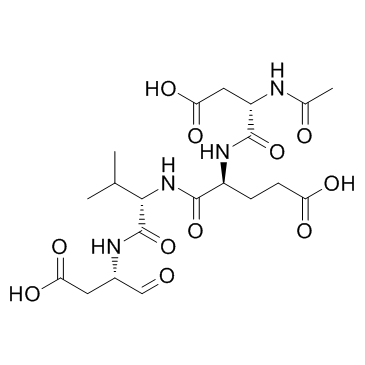
-
GC48470
Ac-DEVD-CHO (trifluoroacetate salt)
A dual caspase3/caspase7 inhibitor
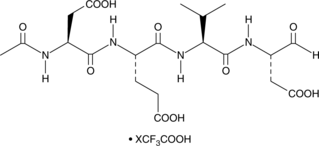
-
GC10951
Ac-DEVD-CMK
cell-permeable, and irreversible inhibitor of caspase
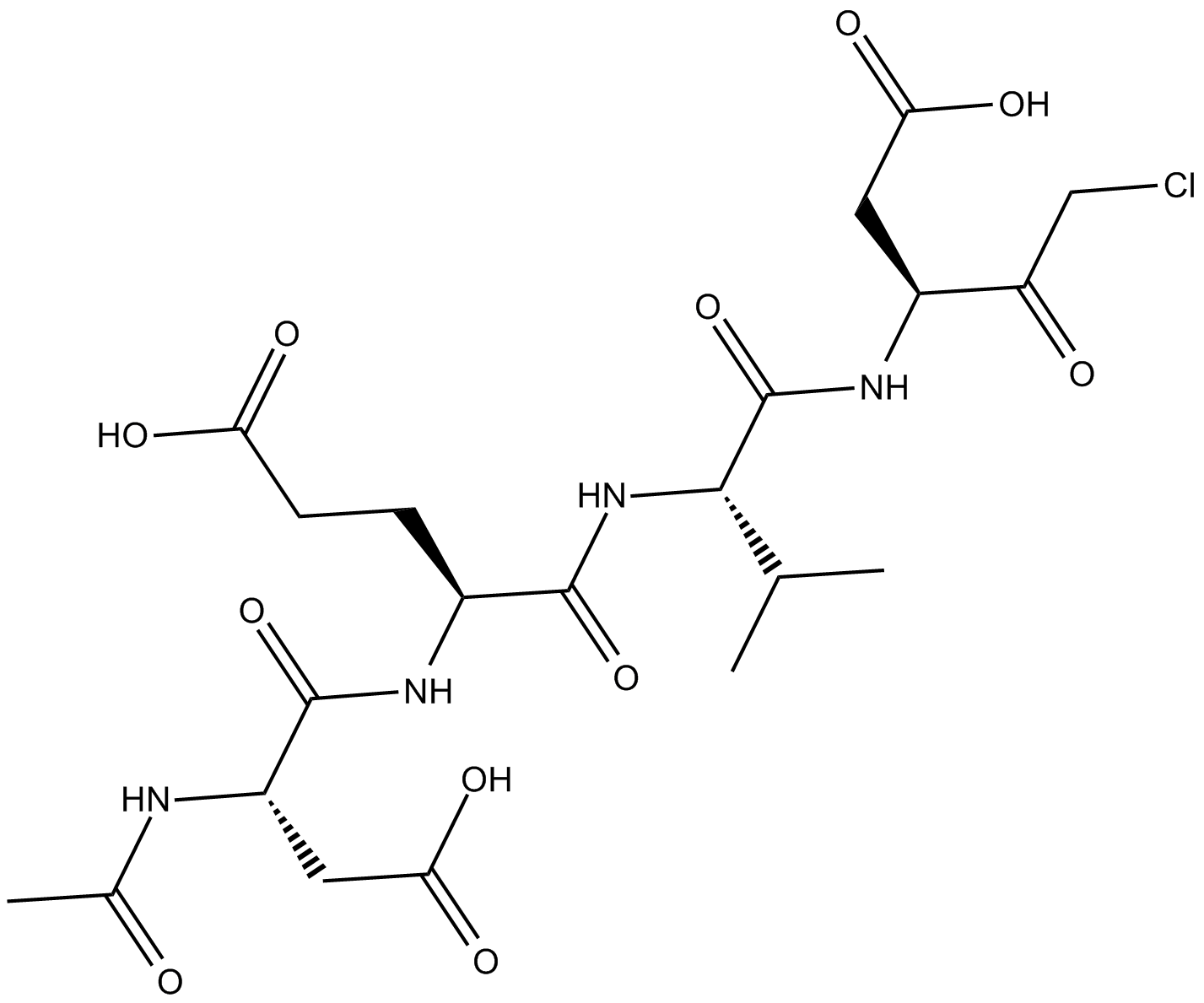
-
GC42689
Ac-DNLD-AMC
Ac-WLA-AMC is a fluorogenic substrate of caspase-3.
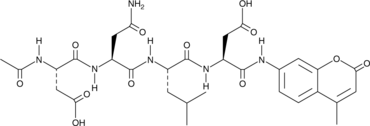
-
GC65107
Ac-FEID-CMK TFA
Ac-FEID-CMK TFA is a potent zebrafish-specific GSDMEb-derived peptide inhibitor.
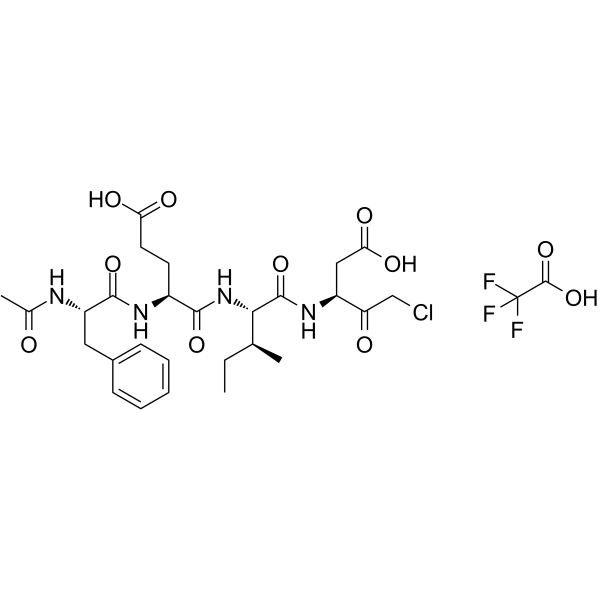
-
GC60558
Ac-FLTD-CMK
Ac-FLTD-CMK, a gasdermin D (GSDMD)-derived inhibitor, is a specific inflammatory caspases inhibitor.
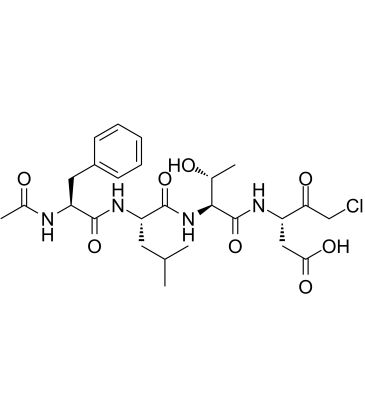
-
GC49704
Ac-FLTD-CMK (trifluoroacetate salt)
An inhibitor of caspase-1, -4, -5, and -11

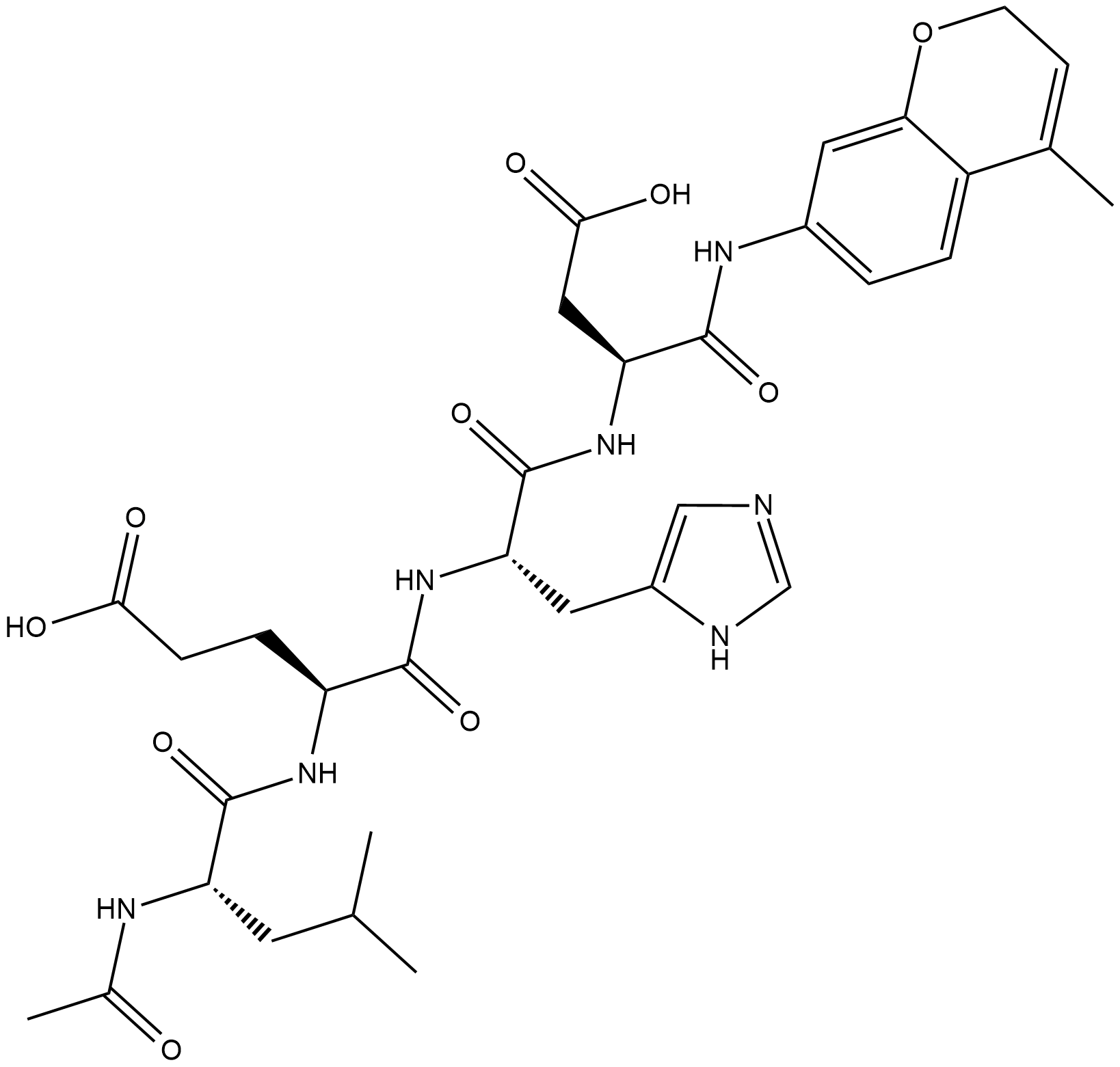
-
GC18226
Ac-LEHD-AMC (trifluoroacetate salt)
Ac-LEHD-AMC (trifluoroacetate salt) is a fluorogenic substrate for caspase-9 (Excitation: 341 nm; Emission: 441 nm).
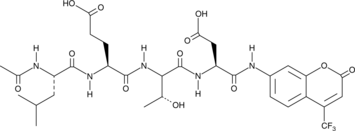
-
GC40556
Ac-LETD-AFC
Ac-LETD-AFC is a fluorogenic substrate that can be cleaved specifically by caspase-8.
-
GC13400
Ac-VDVAD-AFC
Ac-VDVAD-AFC is a caspase-specific fluorescent substrate. Ac-VDVAD-AFC can measure caspase-3-like activity and caspase-2 activity and can be used for the research of tumor and cancer.
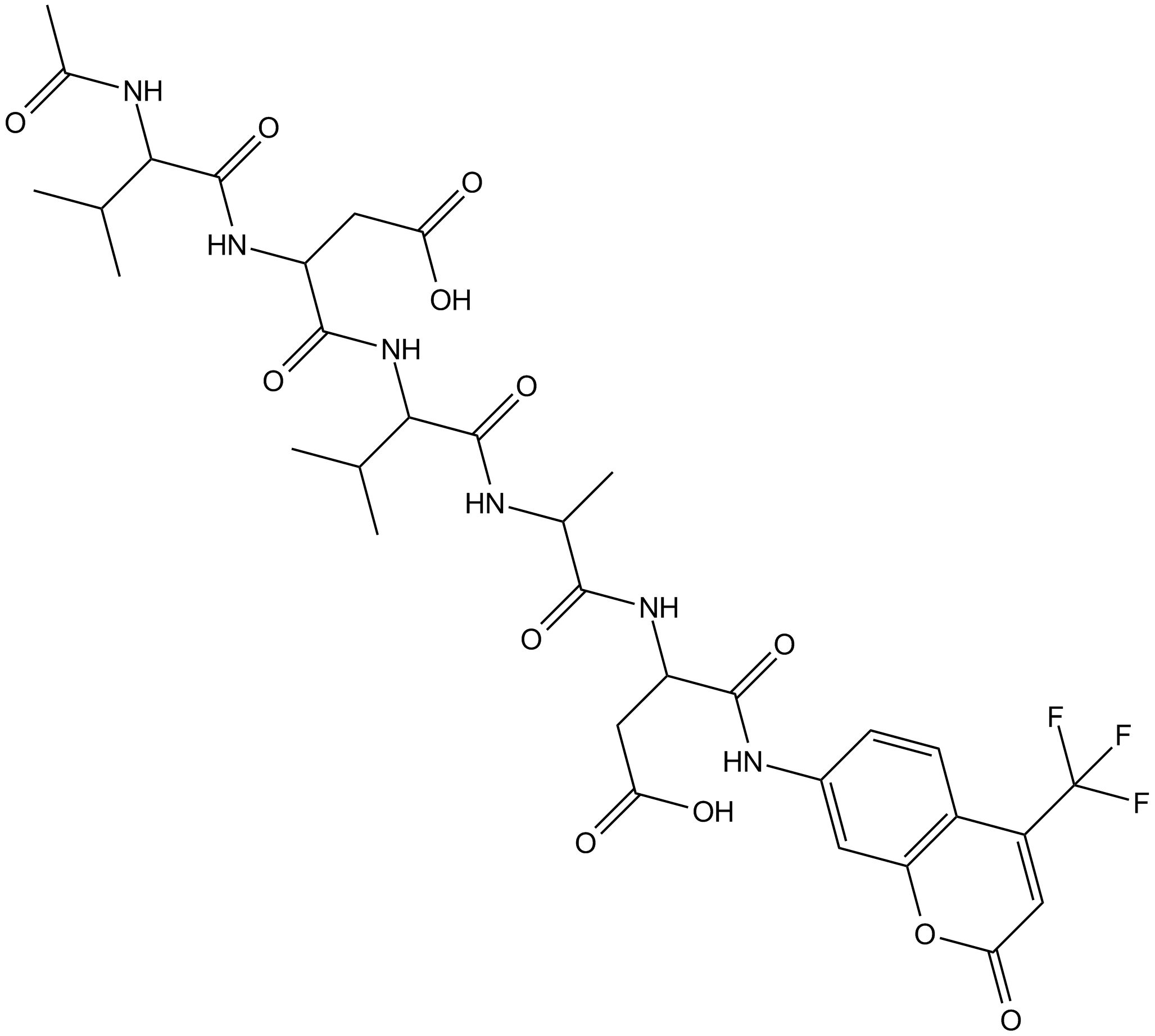
-
GC52372
Ac-VDVAD-AFC (trifluoroacetate salt)
A fluorogenic substrate for caspase-2
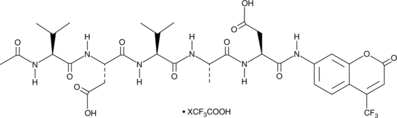
-
GC48974
Ac-VEID-AMC (ammonium acetate salt)
A caspase-6 fluorogenic substrate
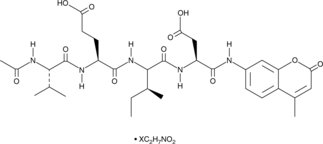
-
GC18021
Ac-YVAD-CHO
Selective inhibitor of interleukin-1β converting enzyme (ICE; Caspase-1)
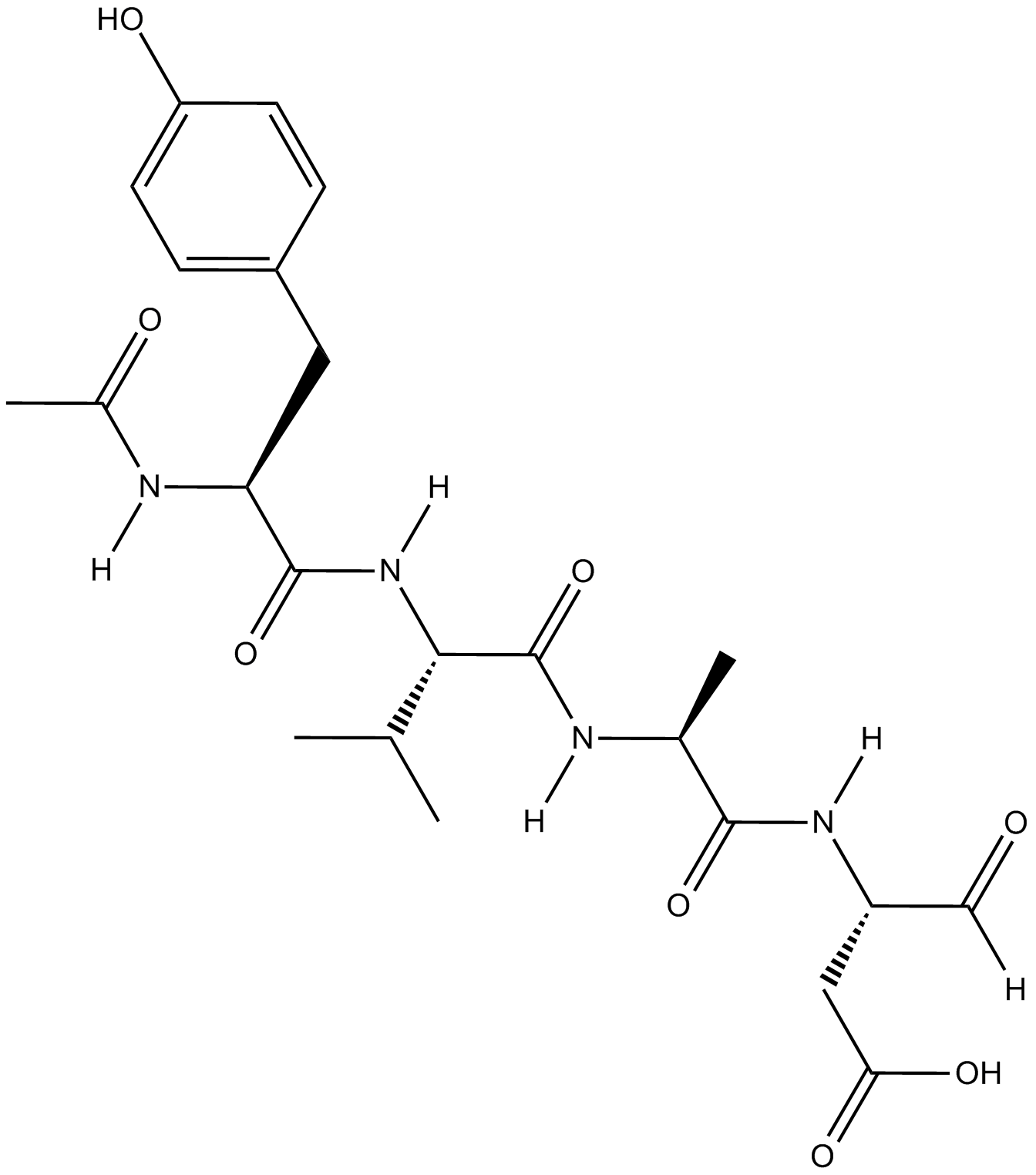
-
GC42721
Ac-YVAD-CMK
Ac-YVAD-CMK is a selective irreversible inhibitor of caspase-1 (Ki=0.8nM), which can prevent the proinflammatory cytokine IL-1β activation. Ac-YVAD-CMK can reduce the inflammatory response and induce a long-lasting neuroprotective effect.
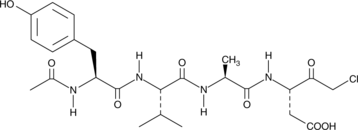
-
GC35227
ACBI1
ACBI1 is a potent and cooperative SMARCA2, SMARCA4 and PBRM1 degrader with DC50s of 6, 11 and 32 nM, respectively. ACBI1 is a PROTAC degrader. ACBI1 shows anti-proliferative activity. ACBI1 induces apoptosis.
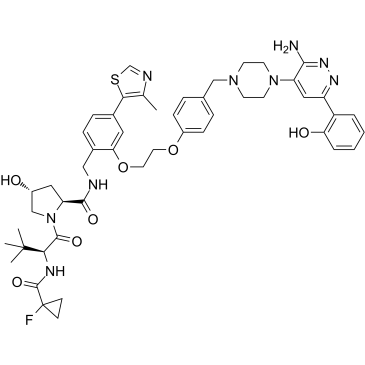
-
GN10341
Acetate gossypol
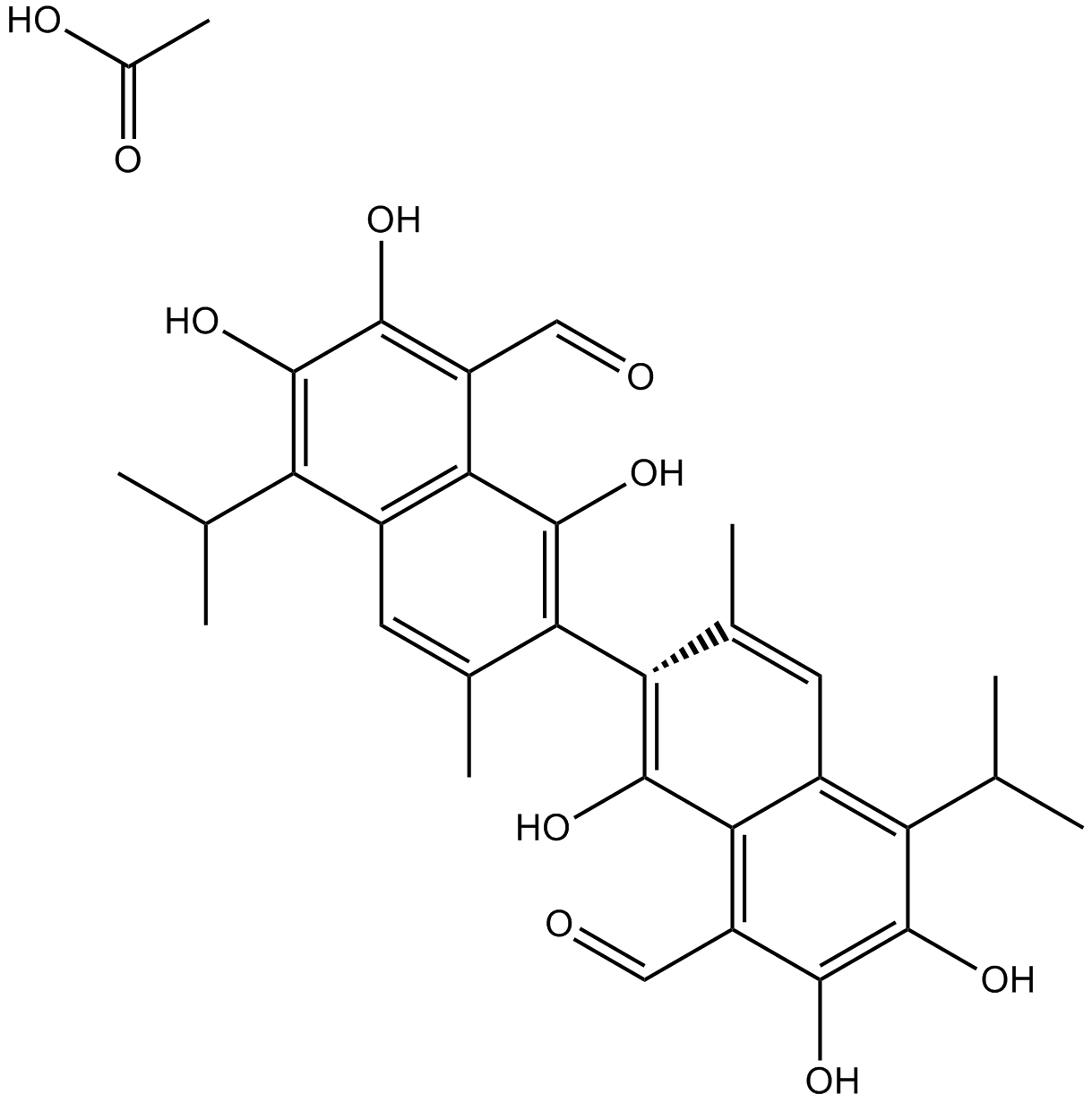
-
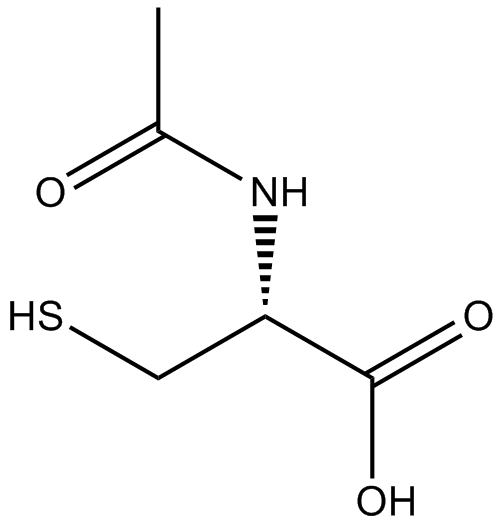
GC11786
Acetylcysteine
Acetylcysteine is the N-acetyl derivative of CYSTEINE.
-
GC17094
Acitretin
Metabolite of etretinate
-
GC35242
Actein
Actein is a triterpene glycoside isolated from the rhizomes of Cimicifuga foetida. Actein suppresses cell proliferation, induces autophagy and apoptosis through promoting ROS/JNK activation, and blunting AKT pathway in human bladder cancer. Actein has little toxicity in vivo.
-
GC16866
Actinomycin D
A DNA-interacting transcription blocker with anti-cancer activity
-
GC16350
Actinonin
Peptidomimetic antibiotic that inhibits aminopeptidases
-
GC16362
AD57 (hydrochloride)
polypharmacological cancer therapeutic that inhibits RET.
-
GC34214
Adalimumab (Anti-Human TNF-alpha, Human Antibody)
Adalimumab (Anti-Human TNF-alpha, Human Antibody) is a human monoclonal IgG1 antibody targeting tumour necrosis factorα (TNF-α).



