Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(15)
- Caspase(77)
- 14.3.3 Proteins(3)
- Apoptosis Inducers(71)
- Bax(15)
- Bcl-2 Family(136)
- Bcl-xL(13)
- c-RET(15)
- IAP(32)
- KEAP1-Nrf2(73)
- MDM2(21)
- p53(137)
- PC-PLC(6)
- PKD(8)
- RasGAP (Ras- P21)(2)
- Survivin(8)
- Thymidylate Synthase(12)
- TNF-α(141)
- Other Apoptosis(1145)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(6)
- PD-1/PD-L1 interaction(60)
- ASK1(4)
- PAR4(2)
- RIP kinase(47)
- FKBP(22)
Products for Apoptosis
- Cat.No. Product Name Information
-
GC10610
Adapalene
RARβ and RARγ agonist

-
GC46798
Adapalene-d3
An internal standard for the quantification of adapalene
-
GC13959
Adarotene
An atypical retinoid
-
GC65880
ADH-6 TFA
ADH-6 TFA is a tripyridylamide compound. ADH-6 abrogates self-assembly of the aggregation-nucleating subdomain of mutant p53 DBD. ADH-6 TFA targets and dissociates mutant p53 aggregates in human cancer cells, which restores p53's transcriptional activity, leading to cell cycle arrest and apoptosis. ADH-6 TFA has the potential for the research of cancer diseases.
-
GC42735
Adipostatin A
Adipostatin A (Adipostatin A) is a glycerol-3-phosphate dehydrogenase (GPDH) inhibitor with an IC50 of 4.1 μM.
-
GC11892
AEE788 (NVP-AEE788)
AEE788 (NVP-AEE788) is an inhibitor of the EGFR and ErbB2 with IC50 values of 2 and 6 nM, respectively.
-
GC42743
AEM1
Cancer cell survival appears partly dependent on antioxidative enzymes, whose expression is regulated by the Keap1-Nrf2 pathway, to quench potentially toxic reactive oxygen species generated by their metastatic transformation.
-
GC13168
AG 825
Selective ErbB2 inhibitor
-
GC13697
AG-1024
Selective IGF-1R inhibitor
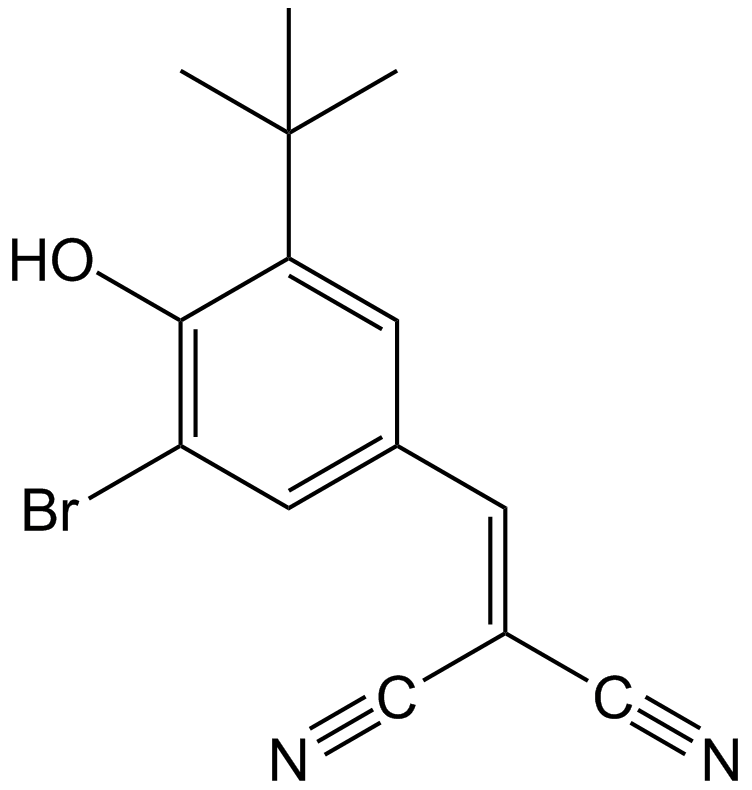
-
GC17881
AGK 2
AGK2 is a selective SIRT2 inhibitor, with an IC50 of 3.
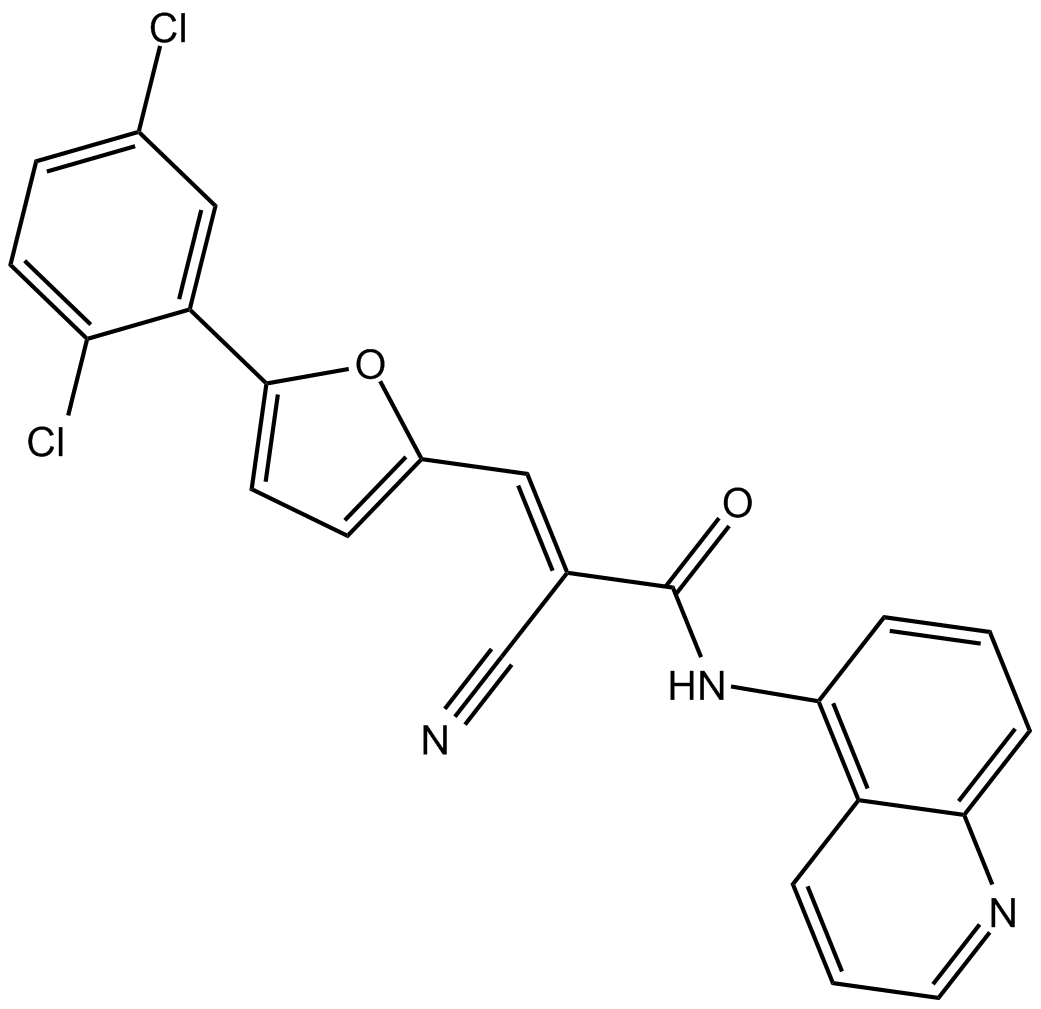
-
GC39584
AGN194204
AGN194204 (IRX4204) is an orally active and selective RXR agonist with Kd values 0.4 nM, 3.6 nM and 3.8 nM and EC50s of 0.2 nM, 0.8 nM and 0.08 nM for RXRα, RXRβ and RXRγ, respectively.
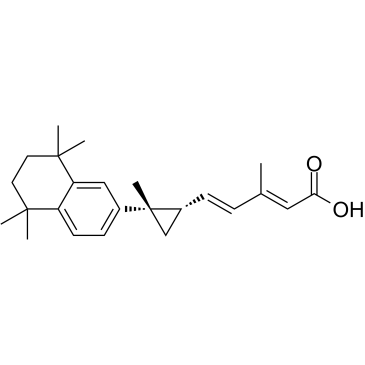
-
GC16120
AI-3
Nrf2/Keap1 and Keap1/Cul3 interaction inhibitor
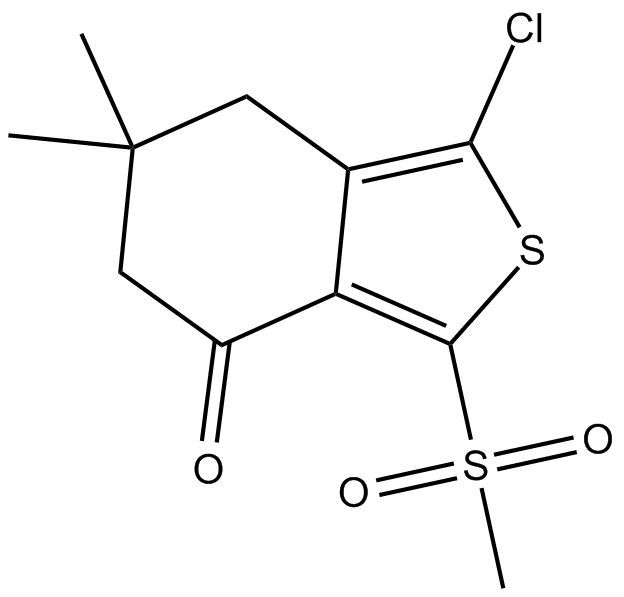
-
GC46821
Ajoene
A disulfide with diverse biological activities
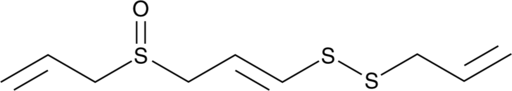
-
GC39620
AKOS-22
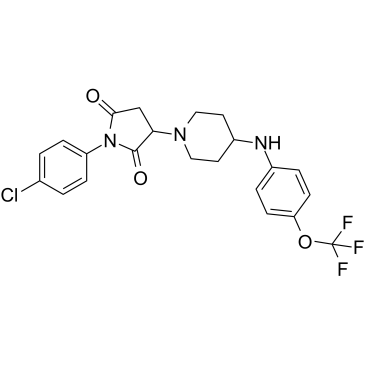
-
GC11589
AKT inhibitor VIII
A potent inhibitor of Akt1 and Akt2

-
GC35275
AKT-IN-3
AKT-IN-3 (compound E22) is a potent, orally active low hERG blocking Akt inhibitor, with 1.4 nM, 1.2 nM and 1.7 nM for Akt1, Akt2 and Akt3, respectively. AKT-IN-3 (compound E22) also exhibits good inhibitory activity against other AGC family kinases, such as PKA, PKC, ROCK1, RSK1, P70S6K, and SGK. AKT-IN-3 (compound E22) induces apoptosis and inhibits metastasis of cancer cells.
-
GC49773
Albendazole sulfone-d3
An internal standard for the quantification of albendazole sulfone
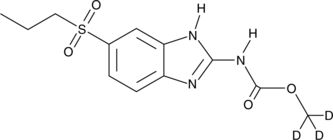
-
GC48848
Albendazole-d7
An internal standard for the quantification of albendazole
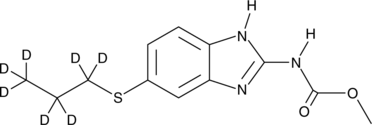
-
GC41080
Albofungin
Albofungin is a xanthone isolated from A.
-
GC16597
Alda 1
ALDH2 activator
-
GC35288
Alkannin
A naphthoquinone with diverse biological activities
-
GC49393
all-trans-13,14-Dihydroretinol
A metabolite of all-trans retinoic acid

-
GC32127
Alofanib (RPT835)
Alofanib (RPT835) (RPT835) is a potent and selective allosteric inhibitor of fibroblast growth factor receptor 2 (FGFR2).

-
GC14314
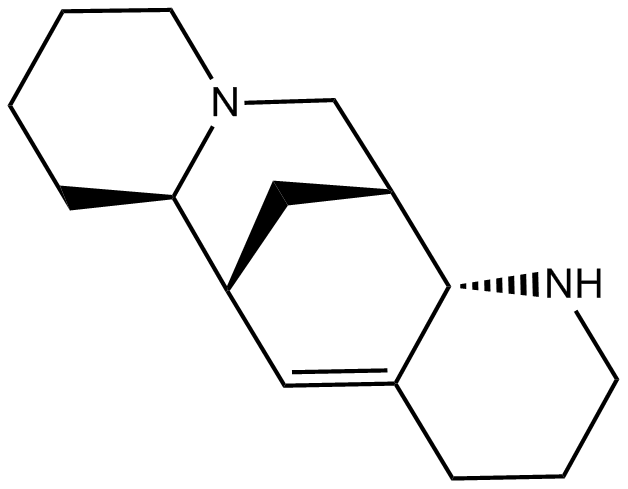
Aloperine
An alkaloid
-
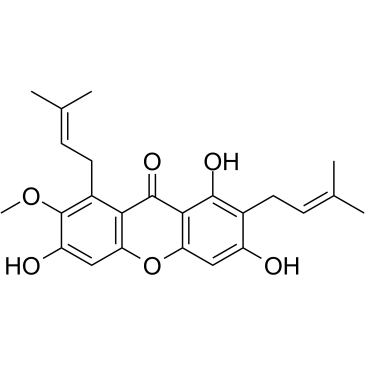
GC35306
alpha-Mangostin
alpha-Mangostin (α-Mangostin) is a dietary xanthone with broad biological activities, such as antioxidant, anti-allergic, antiviral, antibacterial, anti-inflammatory and anticancer effects. It is an inhibitor of mutant IDH1 (IDH1-R132H) with a Ki of 2.85 μM.
-
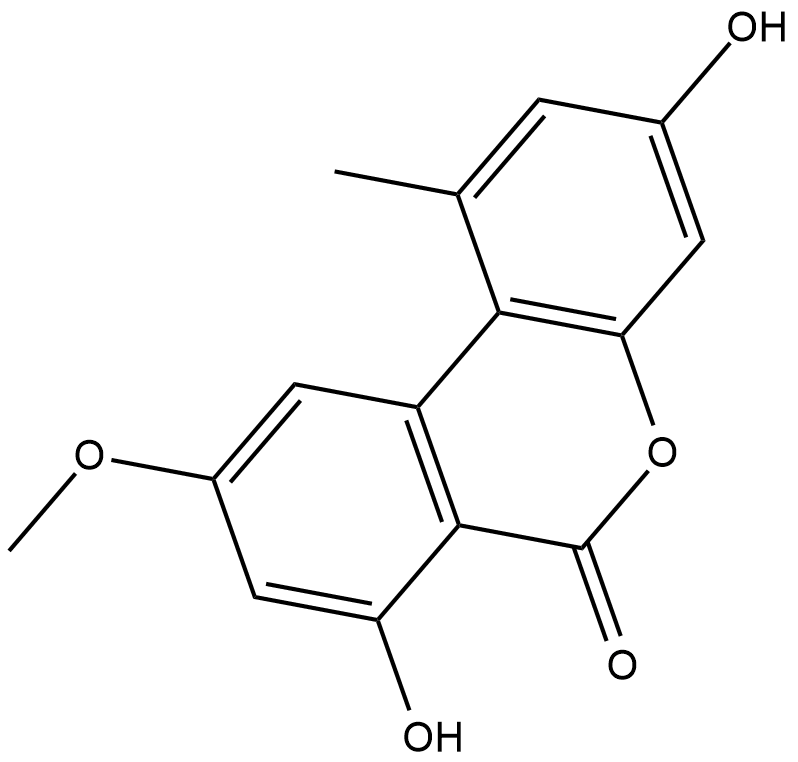
GC18437
Alternariol monomethyl ether
Alternariol monomethyl ether, isolated from the roots of Anthocleista djalonensis (Loganiaceae), is an important taxonomic marker of the plant species.
-
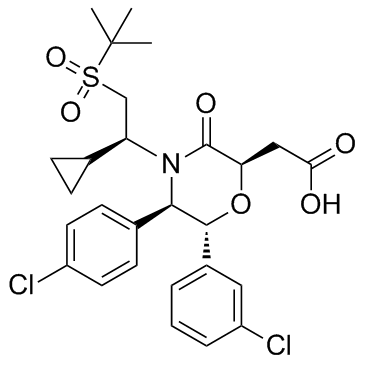
GC33356
AM-8735
AM-8735 is a potent and selective MDM2 inhibitor with an IC50 of 25 nM.
-
GC42776
Amarogentin
A secoiridoid glycoside with diverse biological activities
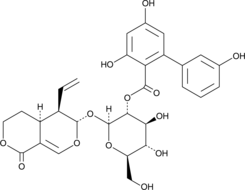
-
GN10484
Amentoflavone
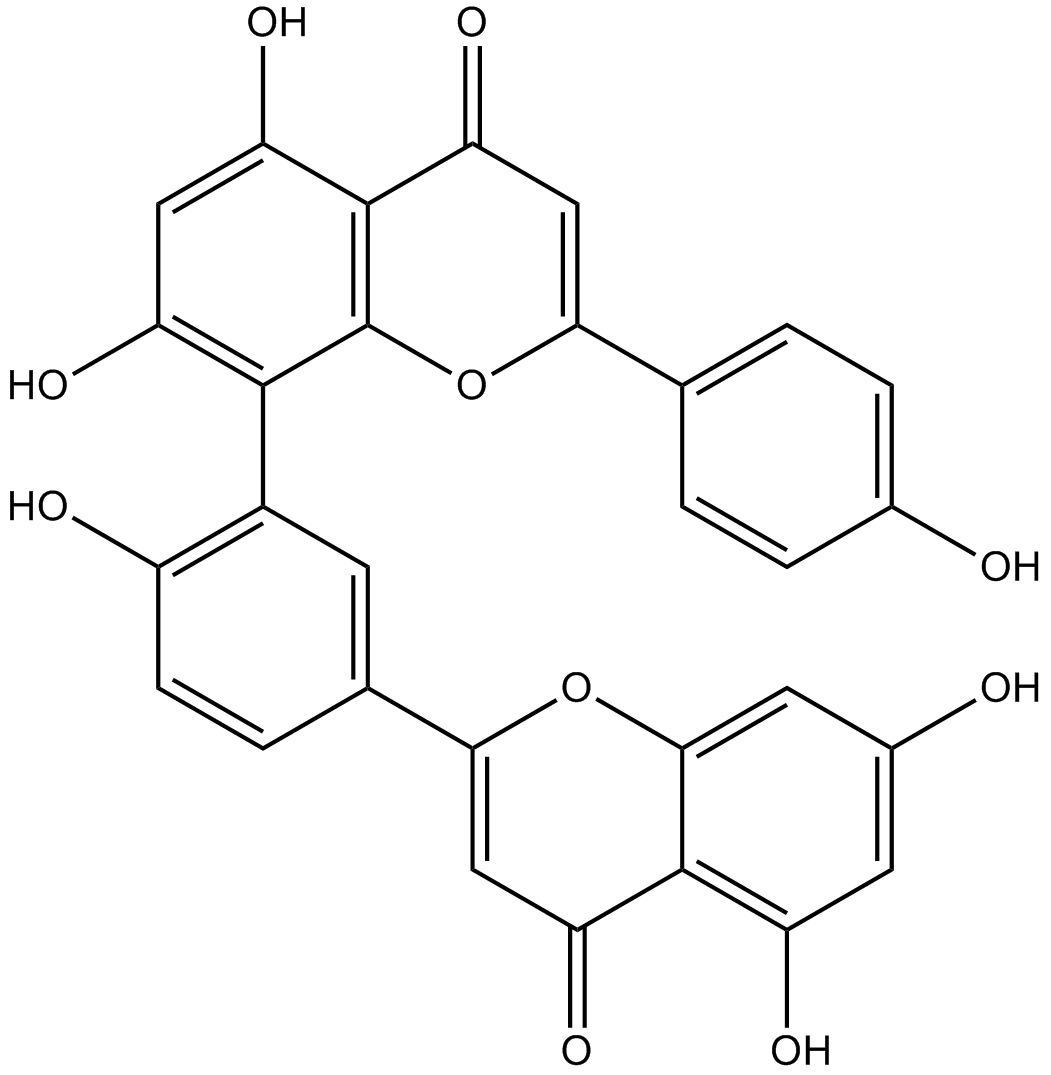
-
GC42783
Ametantrone
Ametantrone (NSC 196473) is an antitumor agent that intercalates into DNA and induces topoisomerase II (TOP2)-mediated DNA break.
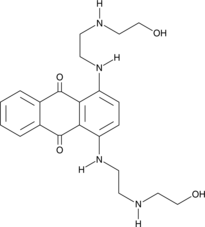
-
GC19452
AMG-176
AMG-176 (AMG-176) is a potent, selective and orally active MCL-1 inhibitor, with a Ki of 0.13 nM.
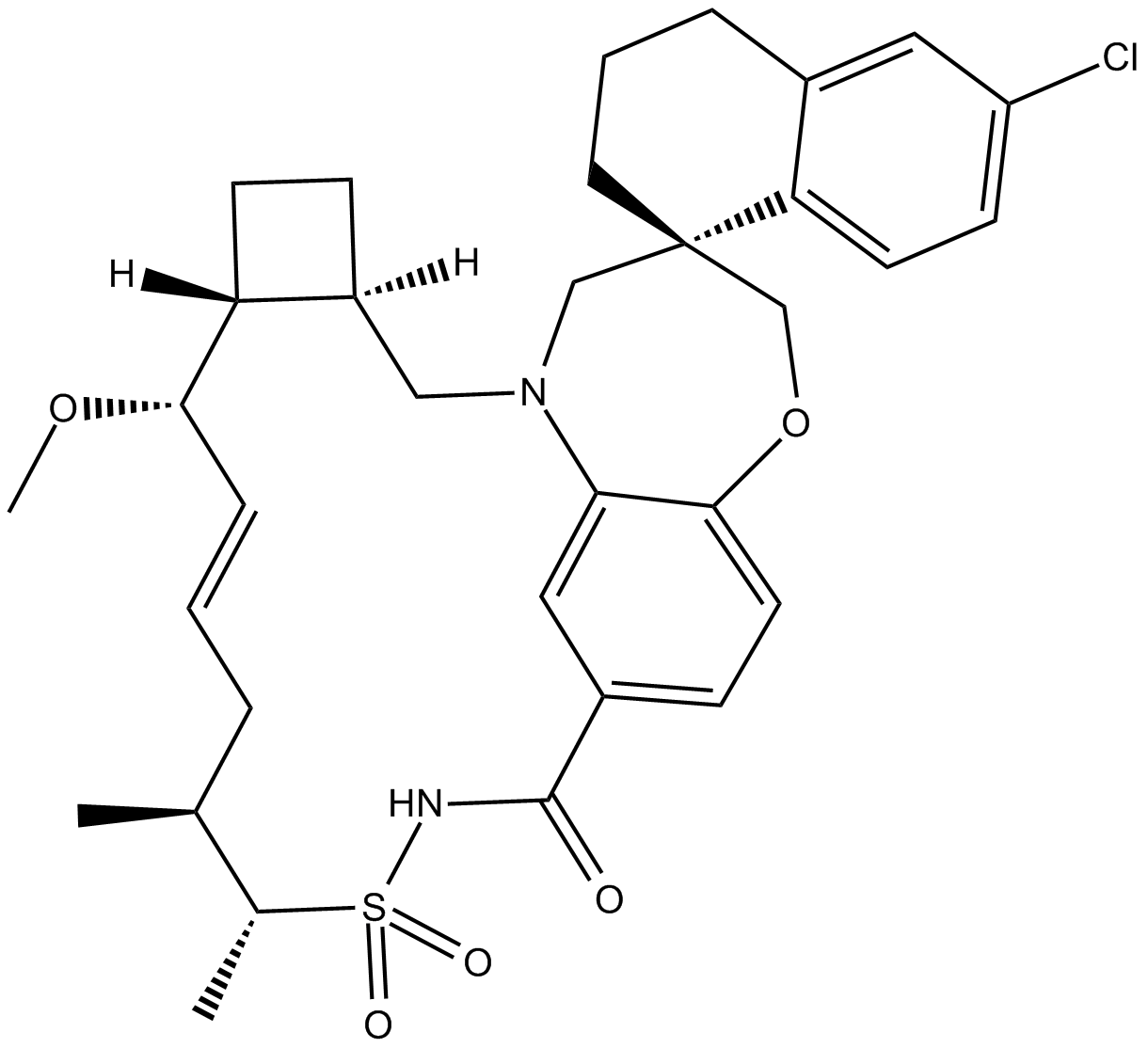
-
GC15828
AMG232
AMG232 (AMG 232) is a potent, selective and orally available inhibitor of p53-MDM2 interaction, with an IC50 of 0.6 nM. AMG232 binds to MDM2 with a Kd of 0.045 nM.
-
GC42785
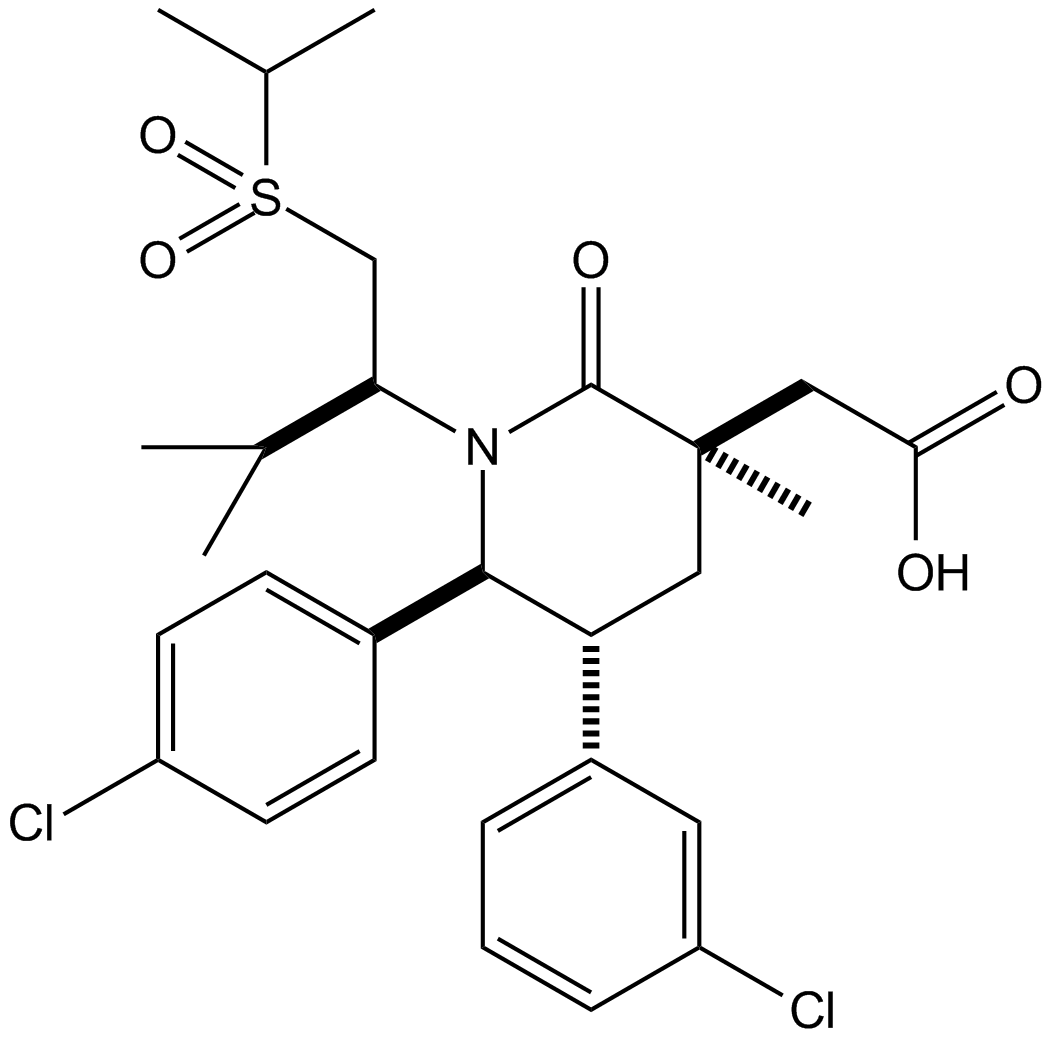
Amifostine (hydrate)
Amifostine (hydrate) (WR2721 trihydrate) is a broad-spectrum cytoprotective agent and a radioprotector. Amifostine (hydrate) selectively protects normal tissues from damage caused by radiation and chemotherapy. Amifostine (hydrate) is potent hypoxia-inducible factor-α1 (HIF-α1) and p53 inducer. Amifostine (hydrate) protects cells from damage by scavenging oxygen-derived free radicals. Amifostine (hydrate) reduces renal toxicity and has antiangiogenic action.
-
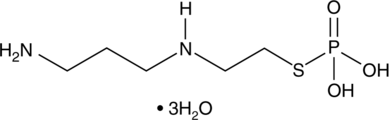
GC61804
Amifostine thiol
Amifostine thiol (WR-1065) is an active metabolite of the cytoprotector Amifostine. Amifostine thiol is a cytoprotective agent with radioprotective abilities. Amifostine thiol activates p53 through a JNK-dependent signaling pathway.
-
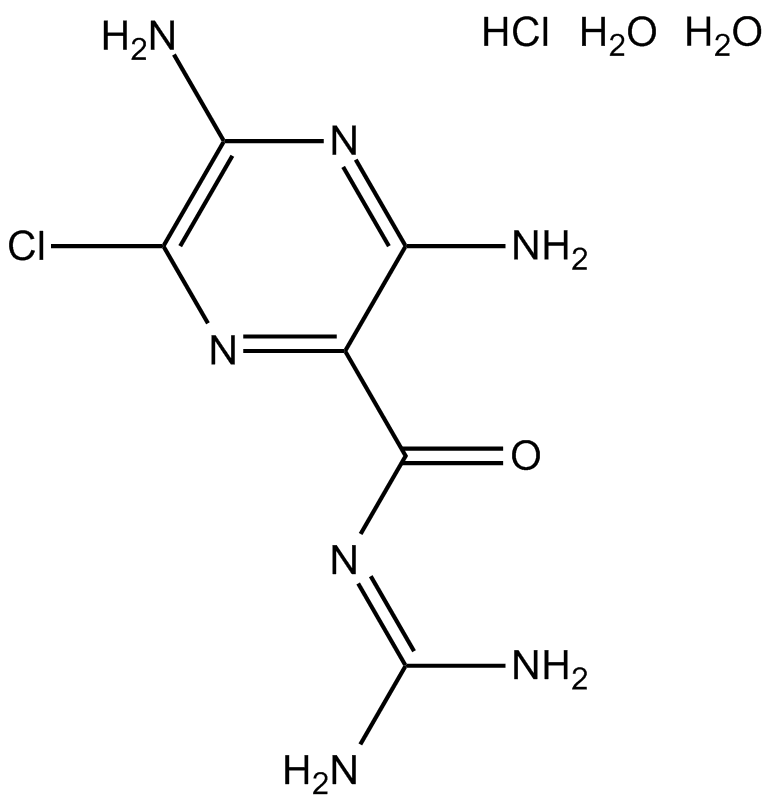
GC12051
Amiloride HCl dihydrate
Amiloride HCl dihydrate (MK-870 hydrochloride dihydrate) is an inhibitor of both epithelial sodium channel (ENaC[1]) and urokinase-type plasminogen activator receptor (uTPA[2]).
-
GC63932
Amsilarotene
Amsilarotene (TAC-101; Am 555S), an orally active synthetic retinoid, has selective affinity for retinoic acid receptor α (RAR-α) binding with Ki of 2.4, 400 nM for RAR-α and RAR-β. Amsilarotene induces the apoptotic of human gastric cancer, hepatocellular carcinoma and ovarian carcinoma cells. Amsilarotene can be used for the research of cancer.
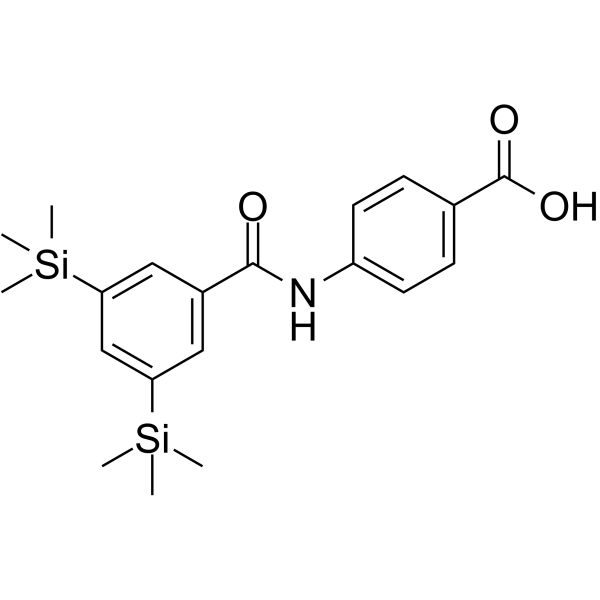
-
GC16391
Amuvatinib (MP-470, HPK 56)
A multi-targeted RTK inhibitor
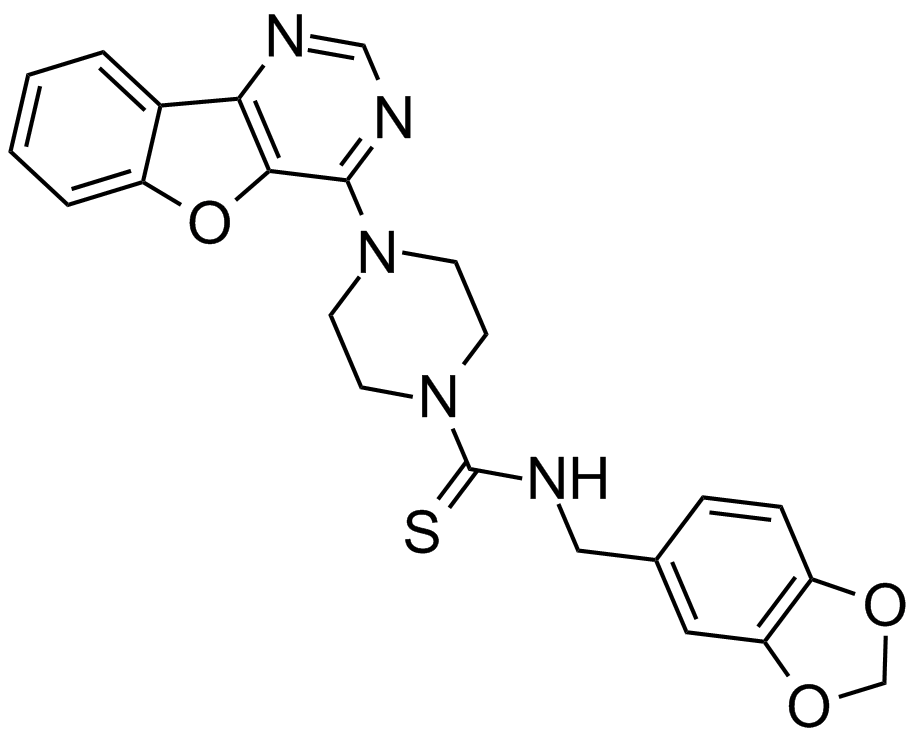
-
GC48339
Amycolatopsin A
A macrolide polyketide with antimycobacterial and anticancer activities
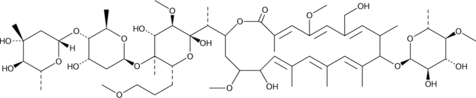
-
GC48341
Amycolatopsin B
A bacterial metabolite
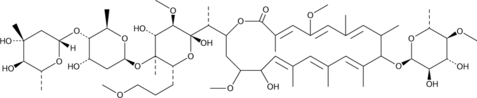
-
GC48350
Amycolatopsin C
A polyketide macrolide with antimycobacterial and anticancer activities
-
GC42806
Andrastin A
Andrastin A is a meroterpenoid farnesyltransferase inhibitor.
-
GN10045
Angelicin
-
GC60584
Angiotensin II human acetate
Angiotensin II human (Angiotensin II) acetate is a vasoconstrictor and a major bioactive peptide of the renin/angiotensin system.
-
GC42813
Anguinomycin A
Anguinomycin A is an antibiotic first isolated from a Streptomyces sp.
-
GC40614
Anhydroepiophiobolin A
Anhydroepiophiobolin A, an analog of Ophiobolin A, is a potent inhibitor of photosynthesis (I50s of 6.1 and 1 mM for photosynthesis in Chlorella and Spinach, respectively).
-
GC40214
Anhydroophiobolin A
Anhydroophiobolin A is an ophiobolin fungal metabolite that has been found in C.
-
GC11559
Anisomycin
JNK agonist, potent and specific
-
GC49259
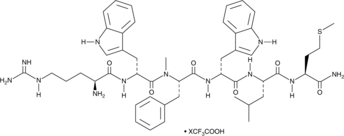
Antagonist G (trifluoroacetate salt)
A neuropeptide antagonist
-
GC66337
Anti-Mouse PD-L1 Antibody
Anti-Mouse PD-L1 Antibody is an anti-mouse PD-L1 IgG2b antibody inhibitor derived from host Rat.

-
GC35361
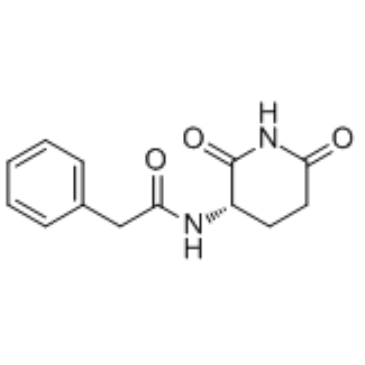
Antineoplaston A10
Antineoplaston A10, a naturally occurring substance in human body, is a Ras inhibitor potentially for the treatment of glioma, lymphoma, astrocytoma and breast cancer.
-
GC34172
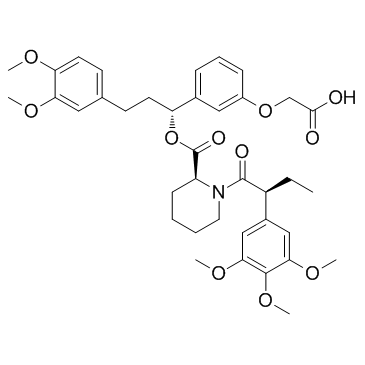
AP1867
AP1867 is a synthetic FKBP12F36V-directed ligand.
-
GC61745
AP1867-2-(carboxymethoxy)
AP1867-2-(carboxymethoxy), the AP1867 (a synthetic FKBP12F36V-directed ligand) based moiety, binds to CRBN ligand via a linker to form dTAG molecules.

-
GC15586
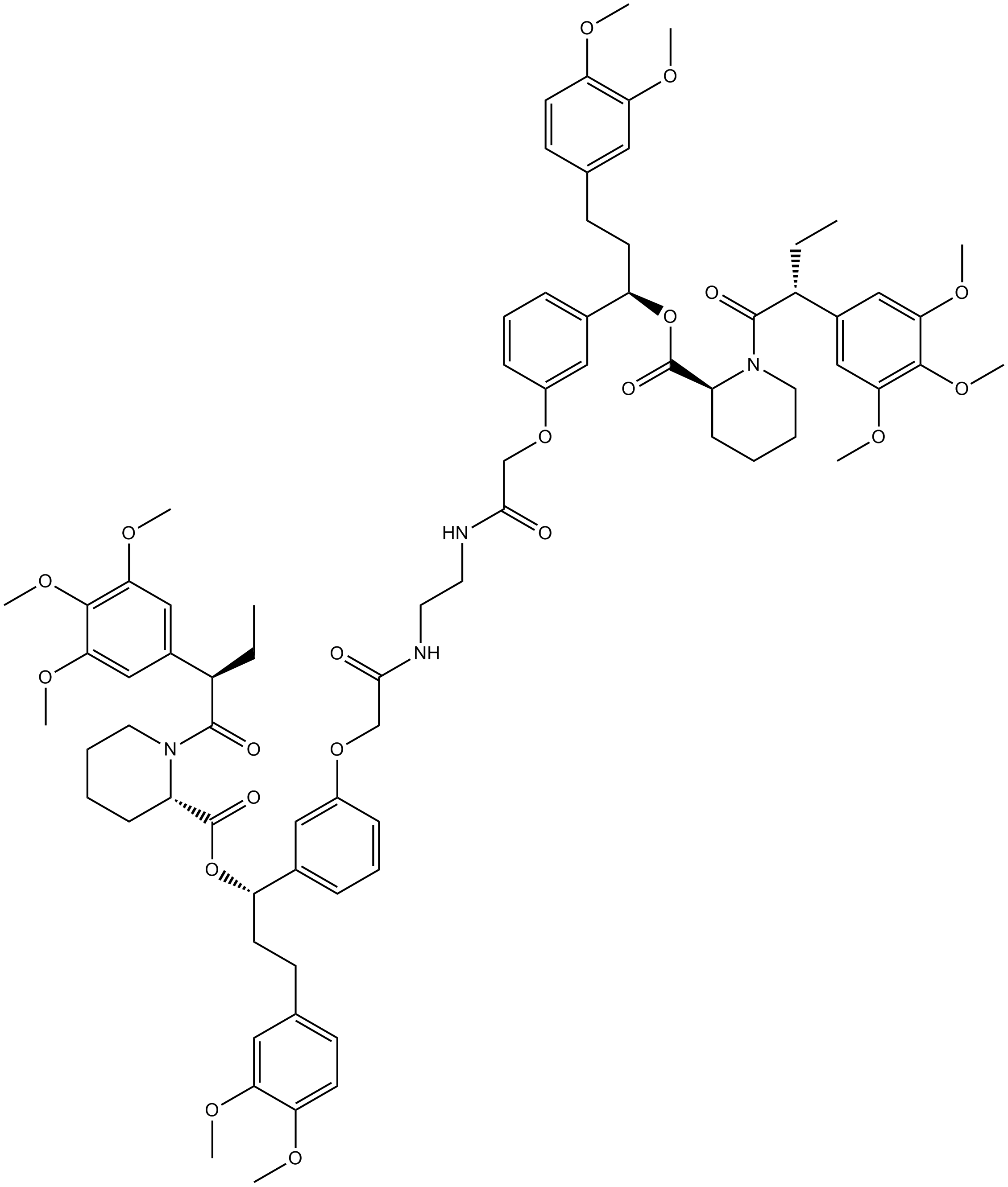
AP1903
AP1903 (AP1903) is a dimerizer agent that acts by cross-linking the FKBP domains. AP1903 (AP1903) dimerizes the Caspase 9 suicide switch and rapidly induces apoptosis.
-
GC14498
AP20187
Dimerizer,synthetic and cell-permeable
-
GC18518
Apcin
Apcin is an inhibitor of the E3 ligase activity of the mitotic anaphase-promoting complex/cyclosome (APC/C).
-
GC62419
Apcin-A
Apcin-A, an Apcin derivative, is an anaphase-promoting complex (APC) inhibitor. Apcin-A interacts strongly with Cdc20, and inhibits the ubiquitination of Cdc20 substrates. Apcin-A can be used to synthesize the PROTAC CP5V.
-
GC35367
APG-115
APG-115 (APG-115) is an orally active MDM2 protein inhibitor binding to MDM2 protein with IC50 and Ki values of 3.8 nM and 1 nM, respectively. APG-115 blocks the interaction of MDM2 and p53 and induces cell-cycle arrest and apoptosis in a p53-dependent manner.
-
GC62640
APG-1387
APG-1387, a bivalent SMAC mimetic and an IAP antagonist, blocks the activity of IAPs family proteins (XIAP, cIAP-1, cIAP-2, and ML-IAP). APG-1387 induces degradation of cIAP-1 and XIAP proteins, as well as caspase-3 activation and PARP cleavage, which leads to apoptosis. APG-1387 can be used for the research of hepatocellular carcinoma, ovarian cancer, and nasopharyngeal carcinoma.
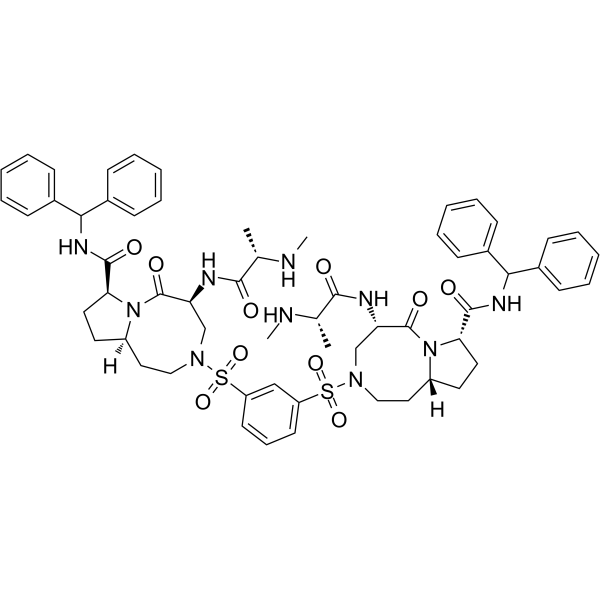
-
GC12961
Apicidin
A cell-permeable HDAC inhibitor
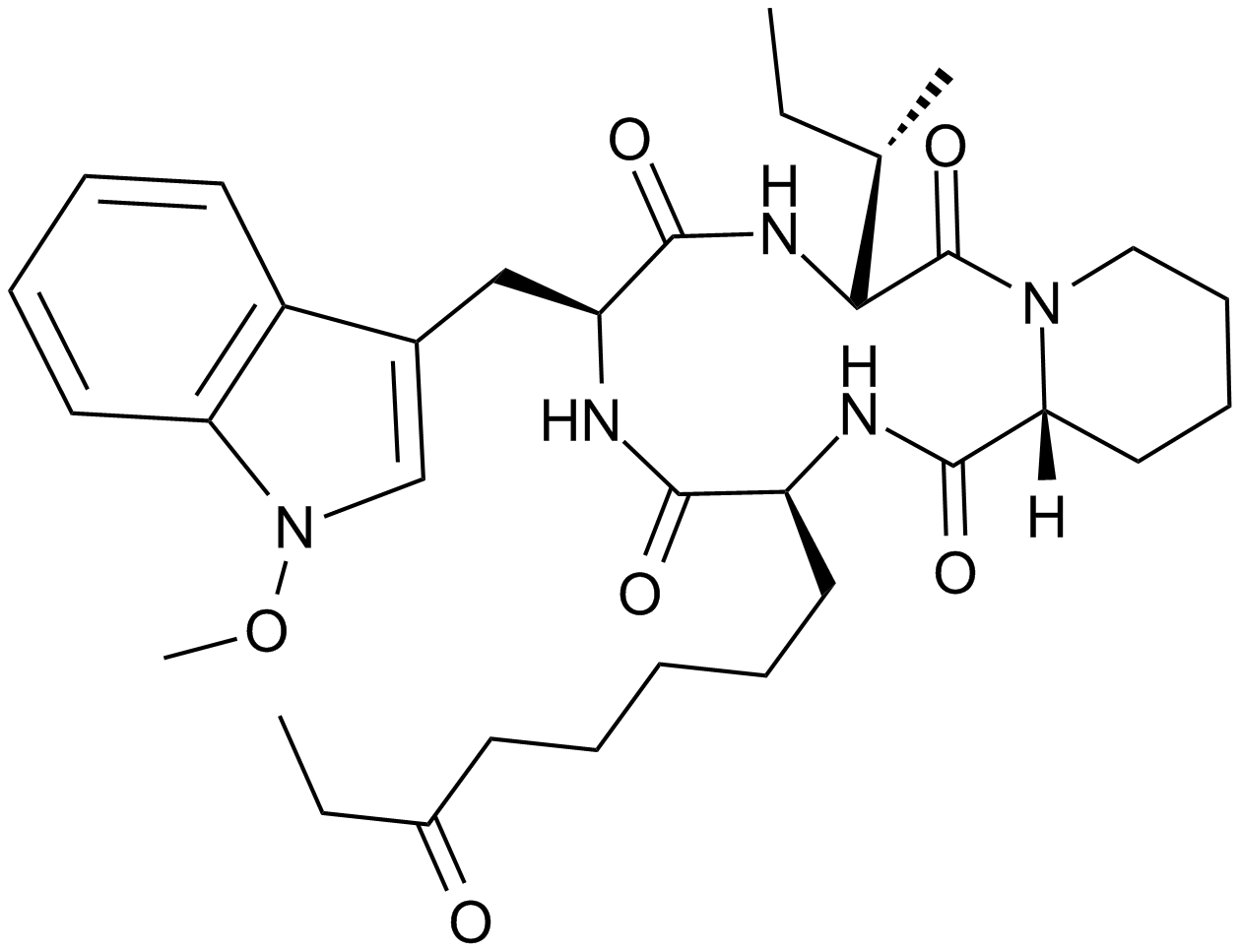
-
GC46862
Apigenin-d5
An internal standard for the quantification of apigenin
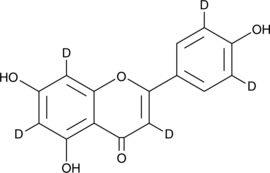
-
GC16237
Apocynin
Selective NADPH-oxidase inhibitor
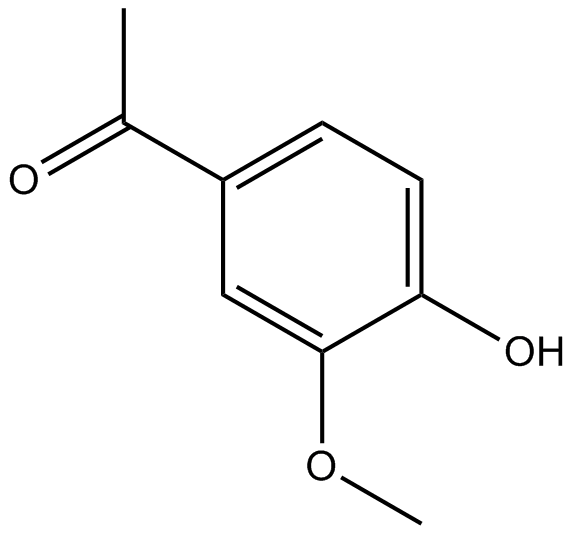
-
GC14080
Apogossypolone (ApoG2)
-
GC42827
Apoptolidin
Apoptolidin is an apoptosis inducer originally isolated from Nocardiopsis bacteria.
-
GC14209
Apoptosis Activator 2
An activator of caspases
-
GC14411
Apoptozole
inhibitor of heat shock protein 70 (Hsp70)
-
GC65004
Apostatin-1
Apostatin-1 (Apt-1) is a potent TRADD inhibitor.
-
GC35377
Apratastat
An inhibitor of ADAM17 and MMPs
-
GC10420
Apremilast (CC-10004)
An orally available PDE4 inhibitor
-
GC32692
APTO-253 (LOR-253)
APTO-253 (LOR-253) (LOR-253) is a small molecule that inhibits c-Myc expression, stabilizes G-quadruplex DNA, and induces cell cycle arrest and apoptosis in acute myeloid leukemia cells.
-
GC14590
AR-42 (OSU-HDAC42)
HDAC inhibitor,novel and potent
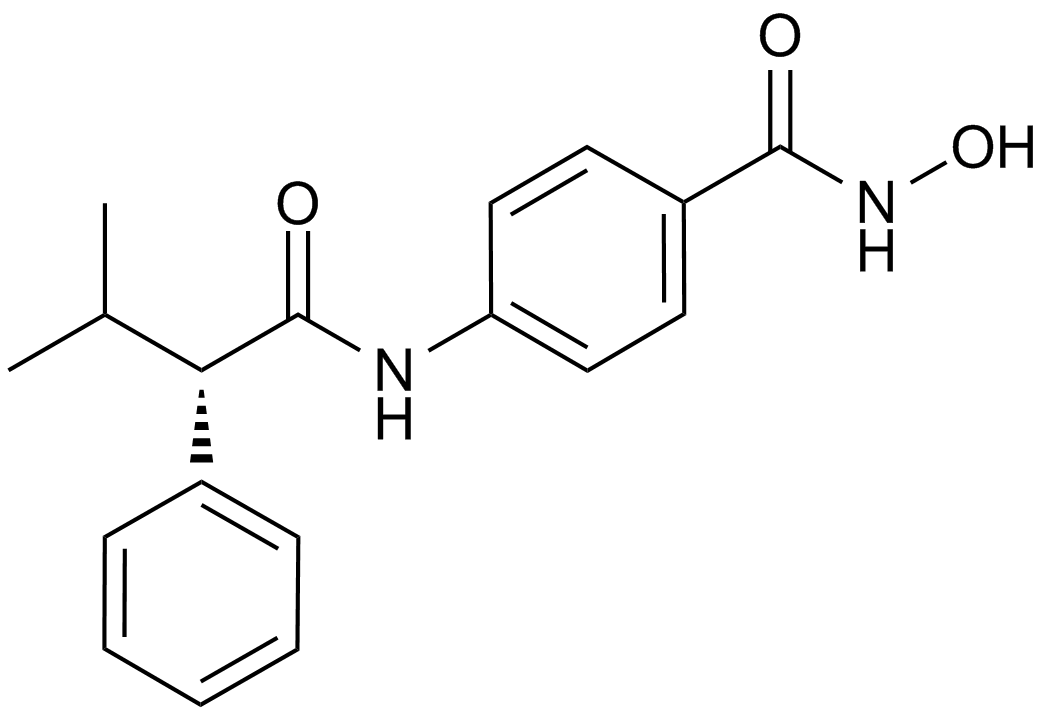
-
GC45385
Ara-G
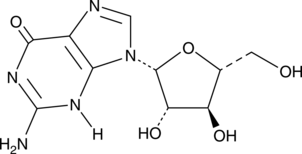
-
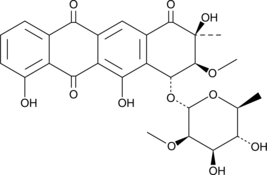
GC46878
Aranciamycin
A fungal metabolite with diverse biological activities
-
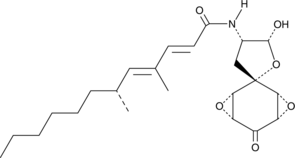
GC40116
Aranorosin
Aranorosin is a fungal metabolite originally isolated from P.
-
GC65163
Ardisiacrispin B
Ardisiacrispin B displays cytotoxic effects in multi-factorial drug resistant cancer cells via ferroptotic and apoptotic cell death.
-
GC49314
Arecaidine propargyl ester (hydrobromide)
A muscarinic M2 agonist
-
GC35388
Aristolactam I
Aristololactam I (AL-I), is the main metabolite of aristolochic acid I (AA-I), participates in the processes that lead to renal damage.
-
GC35395
Arnicolide D
Arnicolide D is a sesquiterpene lactone isolated from Centipeda minima. Arnicolide D modulates the cell cycle, activates the caspase signaling pathway and inhibits the PI3K/AKT/mTOR and STAT3 signaling pathways. Arnicolide D inhibits Nasopharyngeal carcinoma (NPC) cell viability in a concentration- and time-dependent manner.
-
GC19037
ARS-853
ARS-853 is a selective, covalent KRASG12C inhibitor with an IC50 of 2.5 uM.
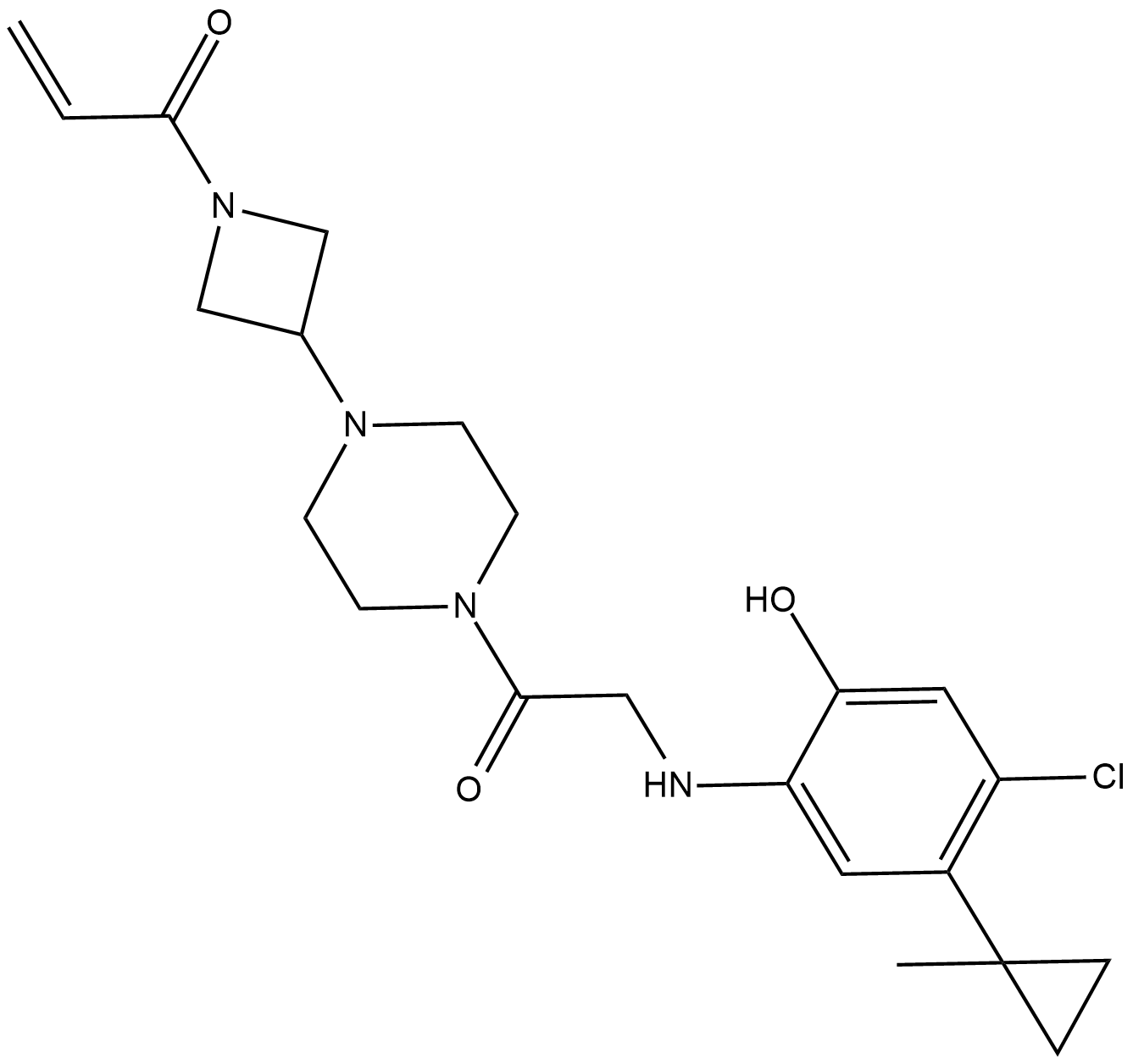
-
GC46882
Artemisinin-d3
An internal standard for the quantification of artemisinin
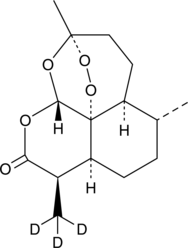
-
GC10040
Arylquin 1
potent secretagogue of the tumor suppressor protein prostate apoptosis response-4 (Par-4)
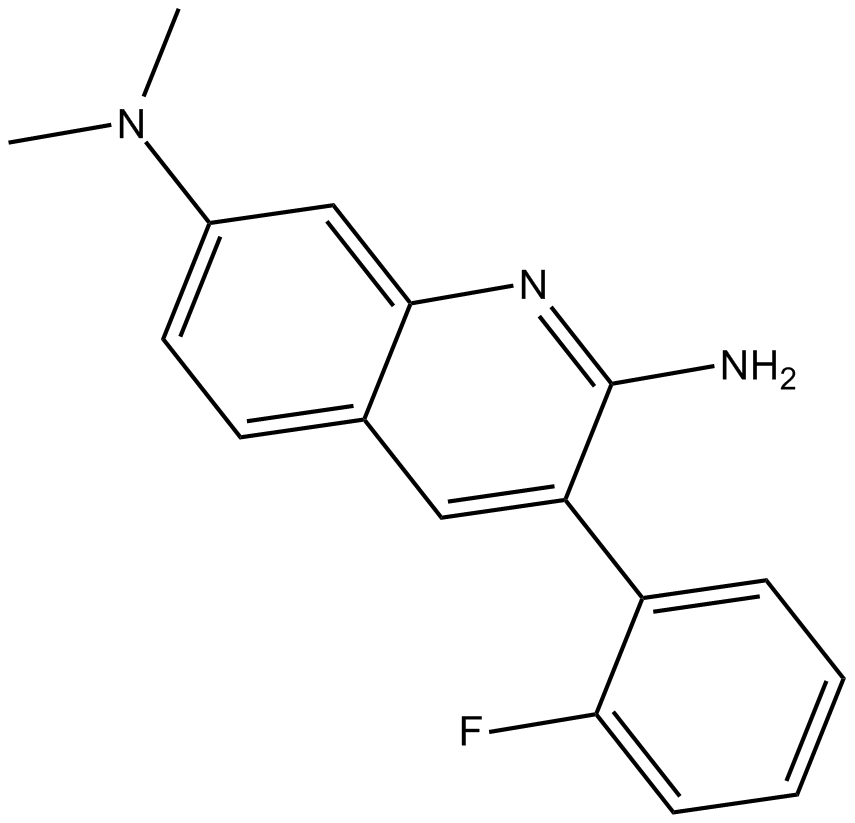
-
GC62615
AS-99
AS-99 is a first-in-class, potent and selective ASH1L histone methyltransferase inhibitor (IC50=0.79μM, Kd=0.89μM) with anti-leukemic activity. AS-99 blocks cell proliferation, induces apoptosis and differentiation, downregulates MLL fusion target genes, and reduces the leukemia burden in vivo.
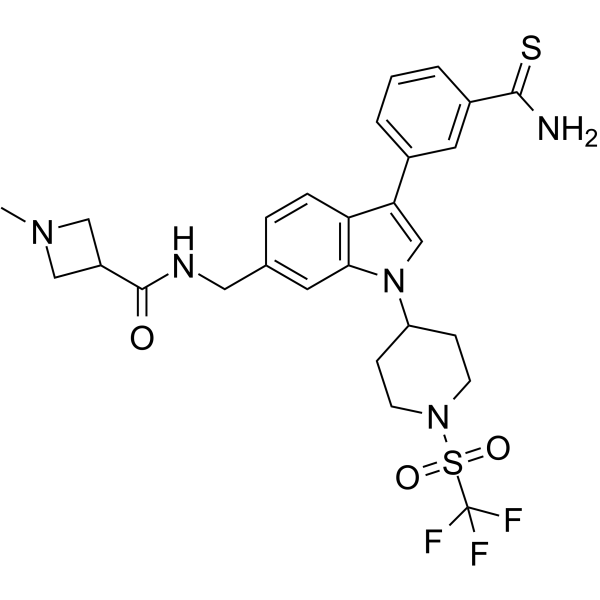
-
GC40715
Ascochlorin
Ascochlorin is an isoprenoid antibiotic and antiviral that has diverse effects on mammalian cells.
-
GC13215
Ascomycin(FK 520)
A potent macrolide immunosuppressant
-
GC12070
Ascorbic acid
An electron donor
-
GN10702
Asiatic acid
-
GN10534
Asiaticoside
-
GC19041
ASK1-IN-1
ASK1-IN-1 is a potent, orally available and selective ATP-competitive inhibitor of apoptosis signal-regulating kinase 1 (ASK1) with an IC50 of 2.87 nM.
-
GC62426
ASK1-IN-2
ASK1-IN-2 is a potent and orally active inhibitor of apoptosis signal-regulating kinase 1 (ASK1), with an IC50 of 32.8 nM.
-
GC42858
Aspergillin PZ
Aspergillin PZ is a fungal metabolite originally isolated from A.
-
GN10064
Asperosaponin VI
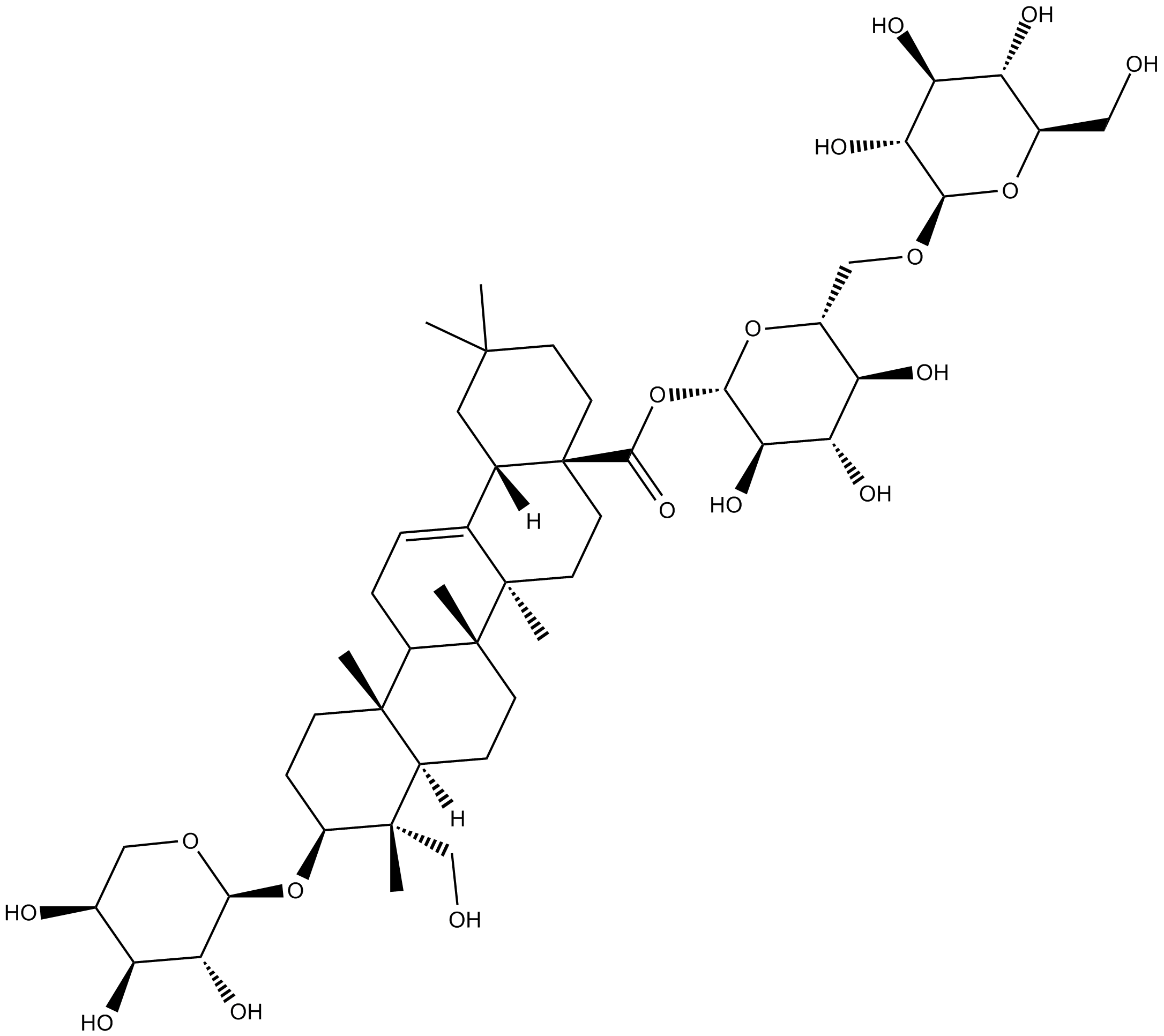
-
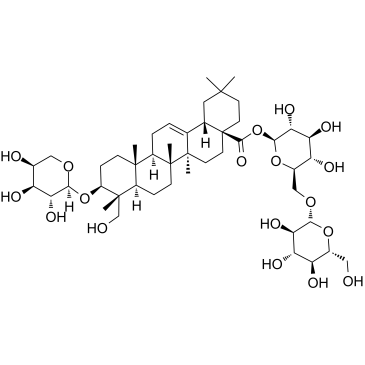
GC60603
Asperosaponin VI
A triterpenoid saponin with diverse biological activities
-
GC42860
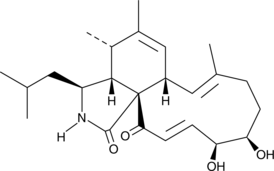
Aspochalasin D
Aspochalasin D is a co-metabolite originally isolated from A.
-
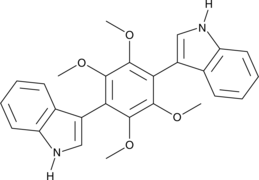
GC41640
Asterriquinol D dimethyl ether
Asterriquinol D dimethyl ether is a fungal metabolite that has been found in A.
-
GN10415
Astilbin
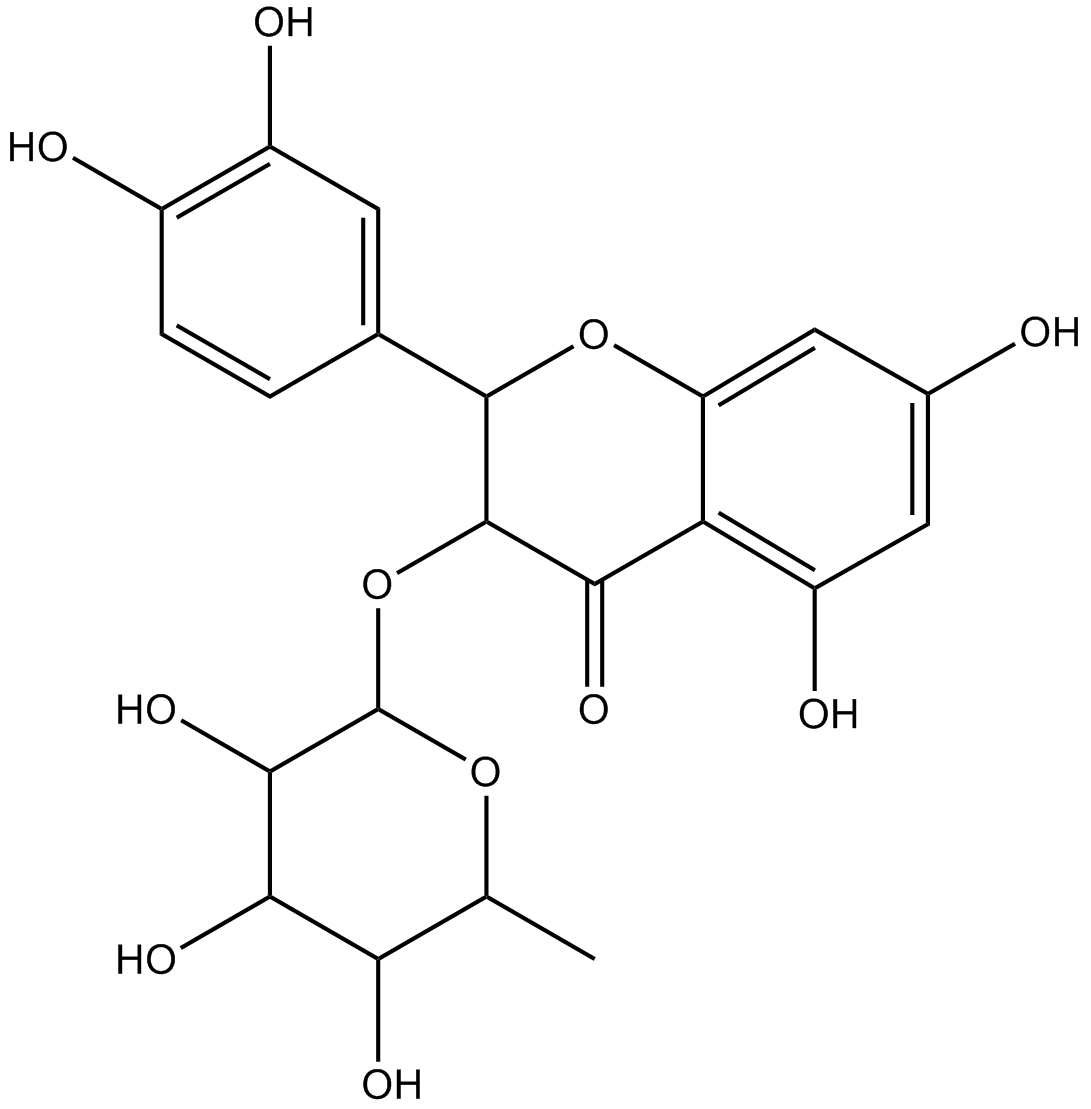
-
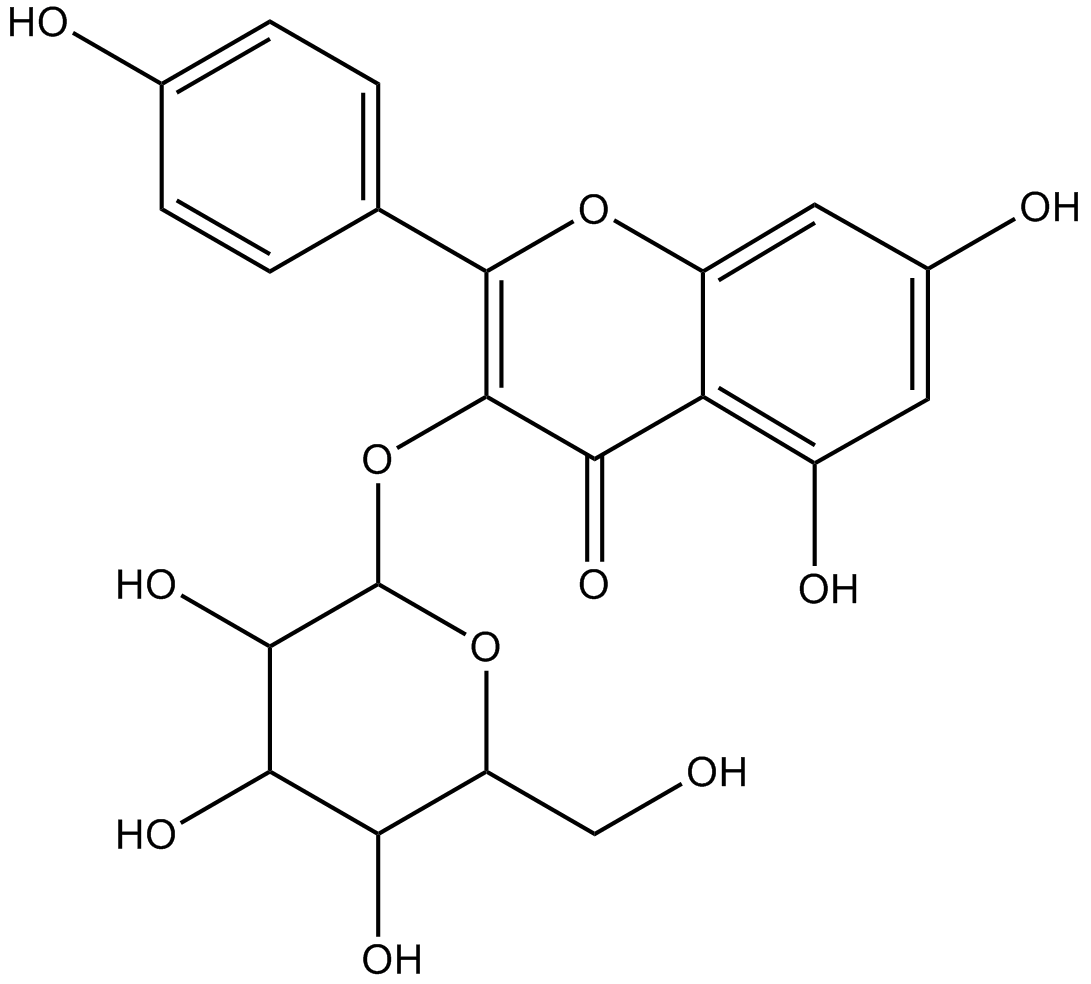
GN10561
astragalin
-
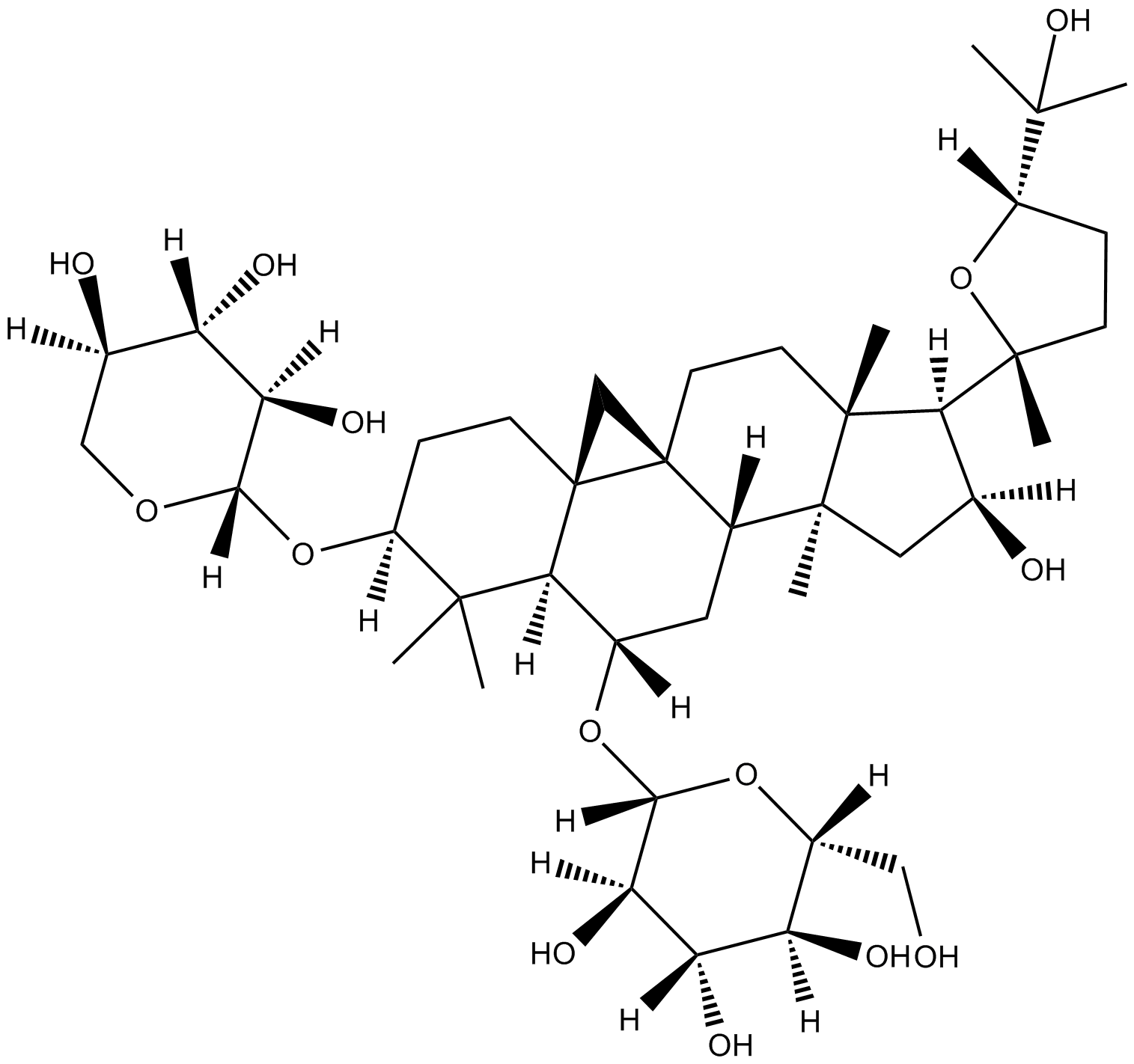
GC18109
Astragaloside A
anti-hypertension, positive inotropic action, anti-inflammation, and anti-myocardial injury
-
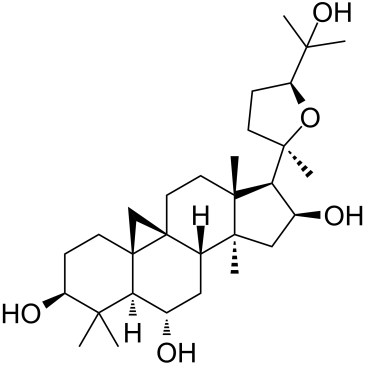
GC35415
Astramembrangenin
-
GC32803
ASTX660
ASTX660 is an orally bioavailable dual antagonist of cellular inhibitor of apoptosis protein (cIAP) and X-linked inhibitor of apoptosis protein (XIAP).
-
GC42863
Asukamycin
Asukamycin is polyketide isolated from the S.
-
GC11106
AT-101