Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(15)
- Caspase(77)
- 14.3.3 Proteins(3)
- Apoptosis Inducers(71)
- Bax(15)
- Bcl-2 Family(136)
- Bcl-xL(13)
- c-RET(15)
- IAP(32)
- KEAP1-Nrf2(73)
- MDM2(21)
- p53(137)
- PC-PLC(6)
- PKD(8)
- RasGAP (Ras- P21)(2)
- Survivin(8)
- Thymidylate Synthase(12)
- TNF-α(141)
- Other Apoptosis(1145)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(6)
- PD-1/PD-L1 interaction(60)
- ASK1(4)
- PAR4(2)
- RIP kinase(47)
- FKBP(22)
Products for Apoptosis
- Cat.No. Product Name Information
-
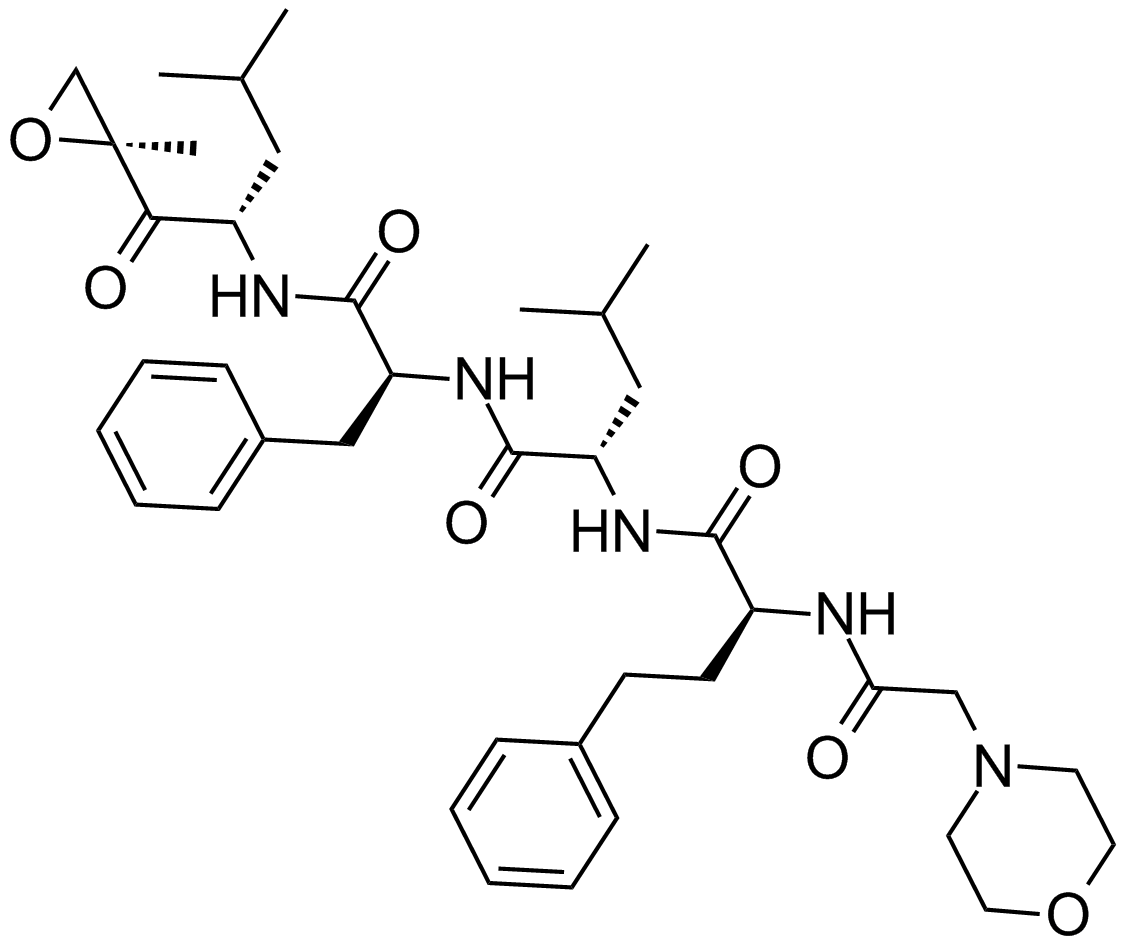
GC12426
Birinapant (TL32711)
An antagonist of cIAP1, cIAP2, and XIAP
-
GN10037
Bisdemethoxycurcumin
-
GC68308
Bisdemethoxycurcumin-d8
-
GC35530
BJE6-106
BJE6-106 (B106) is a potent, selective 3rd generation PKCδ inhibitor with an IC50 of 0.05 μM and targets selectivity over classical PKC isozyme PKCα (IC50=50 μM).

-
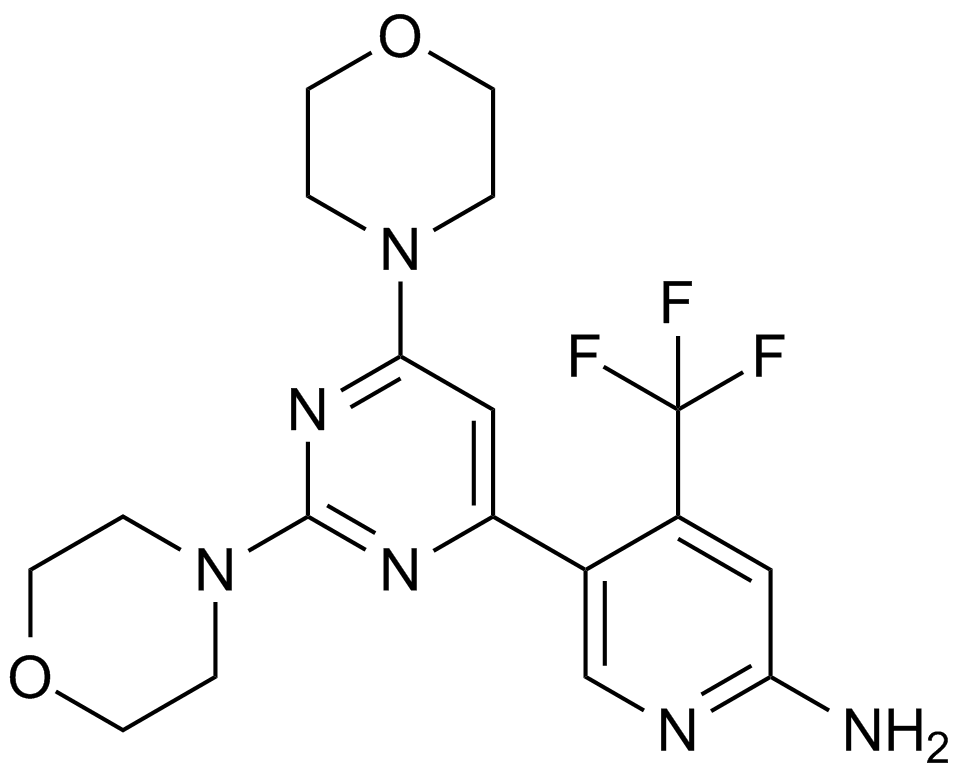
GC11931
BKM120
An inhibitor of class I PI3K isoforms
-
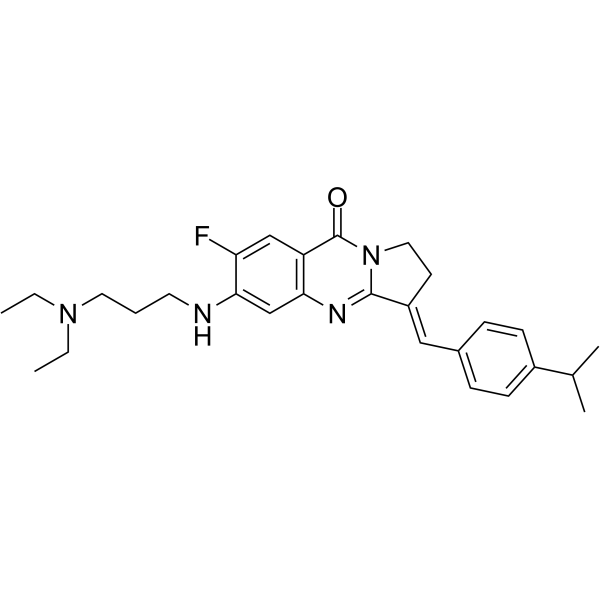
GC65428
BLM-IN-1
BLM-IN-1 (compound 29) is an effective Bloom syndrome protein (BLM) inhibitor, with a strong BLM binding KD of 1.81 μM and an IC50 of 0.95 μM for BLM. Induces DNA damage response, as well as apoptosis and proliferation arrest in cancer cells.
-
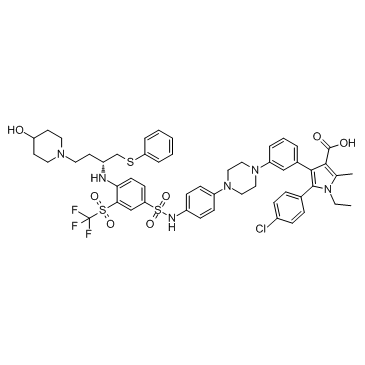
GC33407
BM 957
BM 957 is a potent Bcl-2 and Bcl-xL inhibitor, with Kis of 1.2, <1 nM and IC50s of 5.4, 6.0 nM respectively.
-
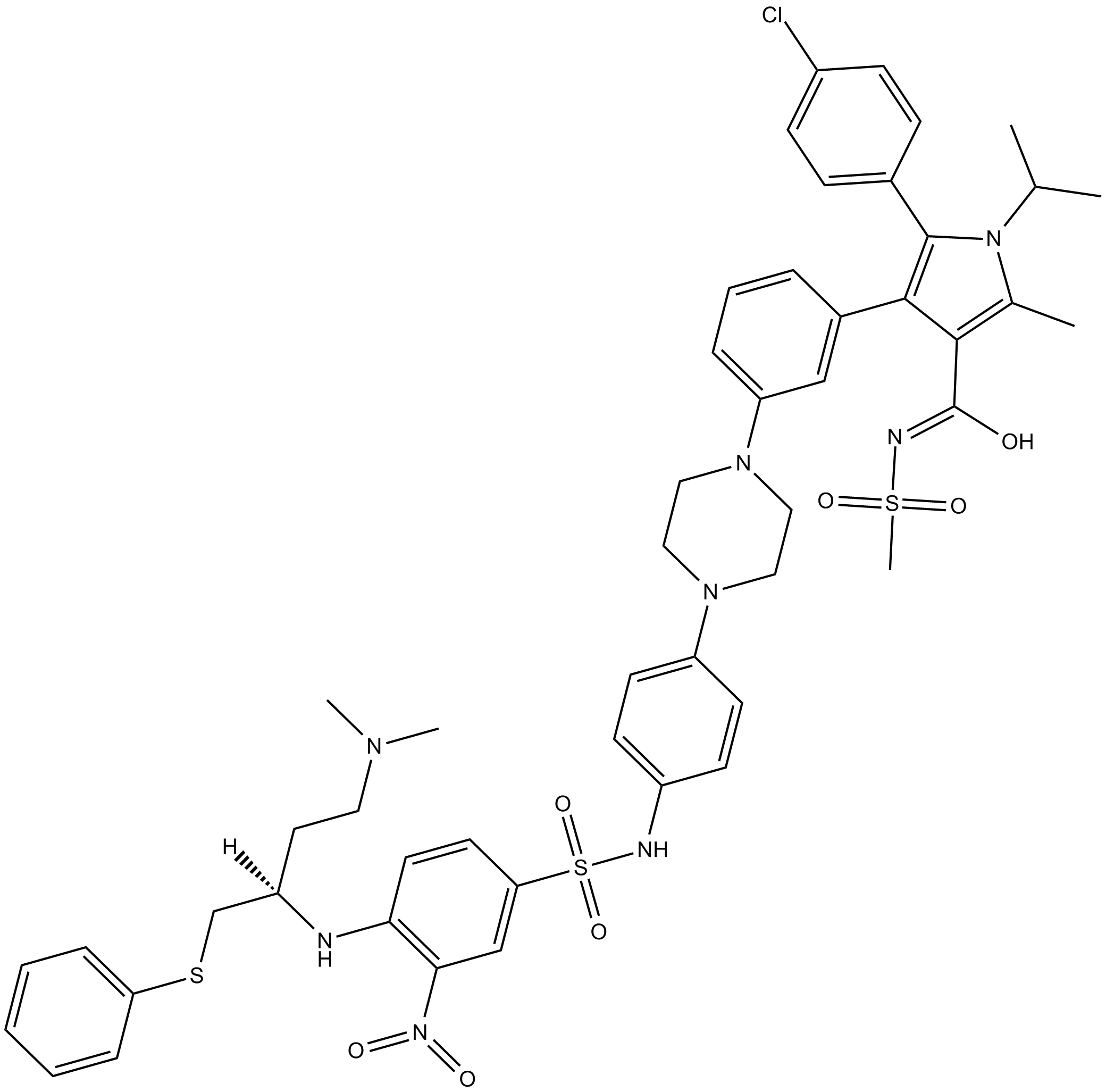
GC13498
BM-1074
-
GC62871
BM-1244
BM-1244 (APG-1252-M1) is a potent Bcl-xL/Bcl-2 inhibitor with Kis of 134 and 450 nM for Bcl- xL and Bcl-2, respectively. BM-1244 inhibits senescent fibroblasts (SnCs) with an EC50 of 5 nM. (From patent WO2019033119A1).
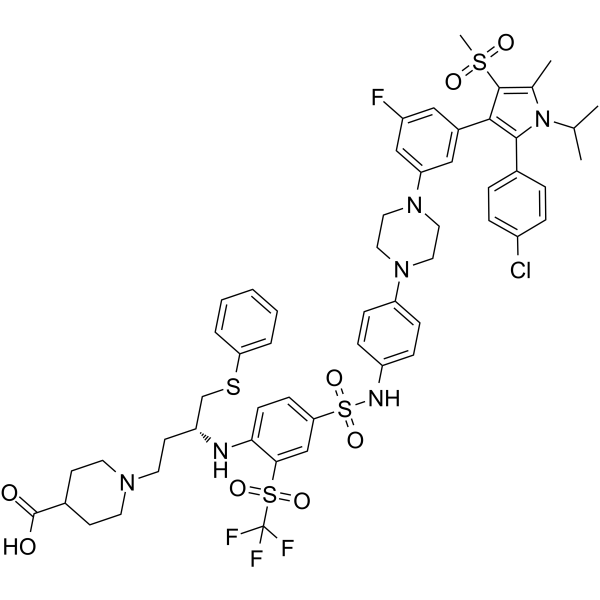
-
GC12822
BML-210(CAY10433)
BML-210(CAY10433) is a novel HDAC inhibitor, and its mechanism of action has not been characterized.
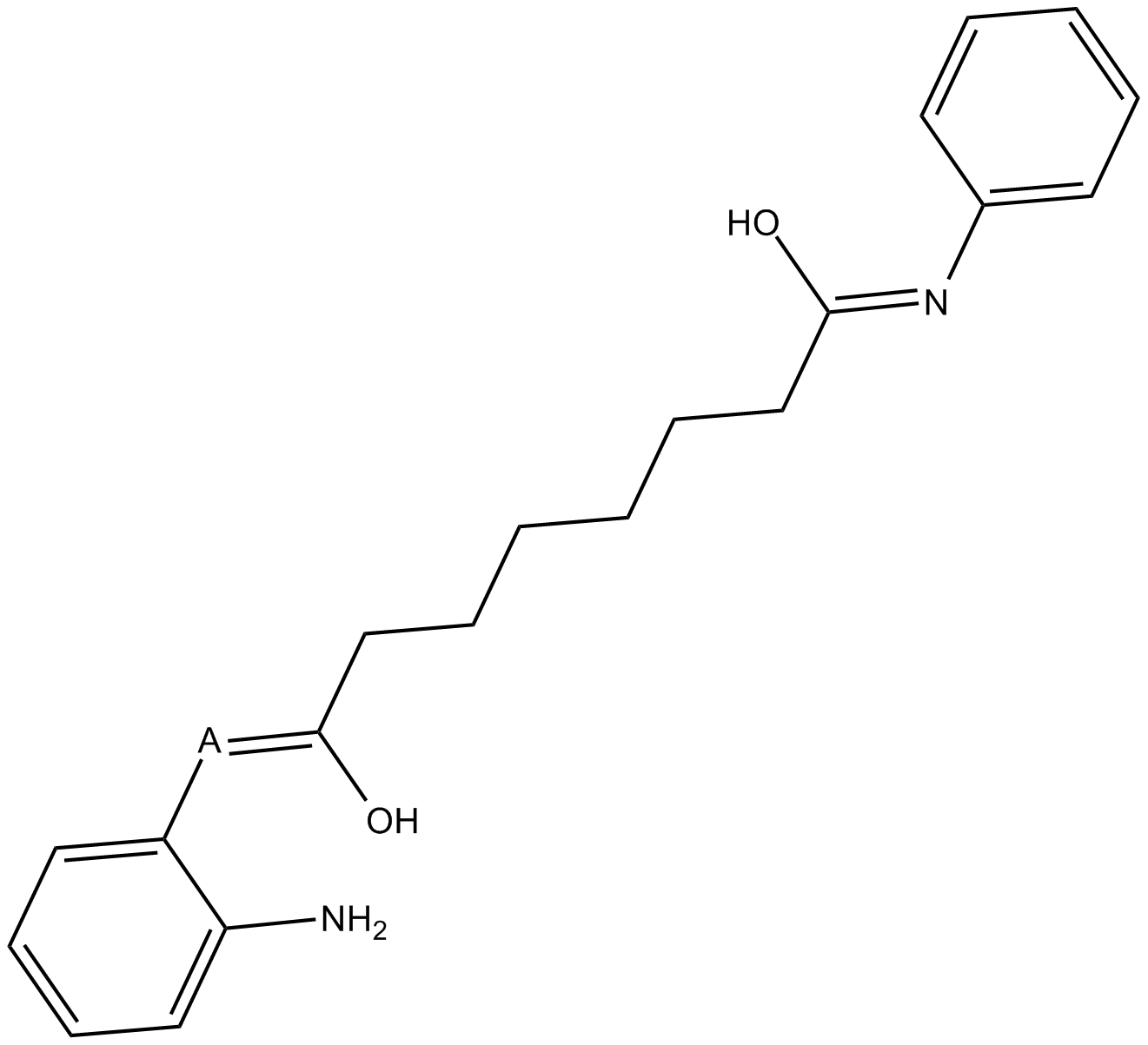
-
GC11648
BML-277
Chk2 inhibitor,potent and highly selective
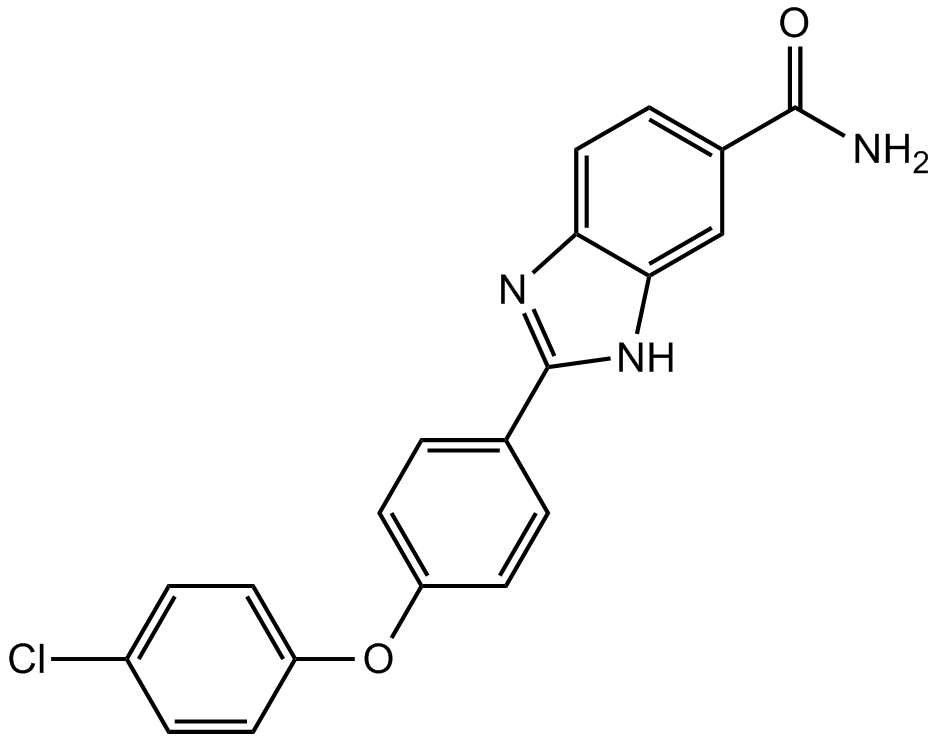
-
GC42953
BMS 345541 (trifluoroacetate salt)
BMS 345541 is a cell permeable inhibitor of the IκB kinases IKKα and IKKβ (IC50s = 4 and 0.3 μM).
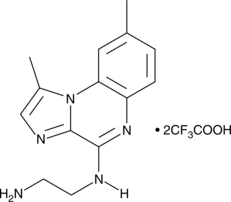
-
GC25160
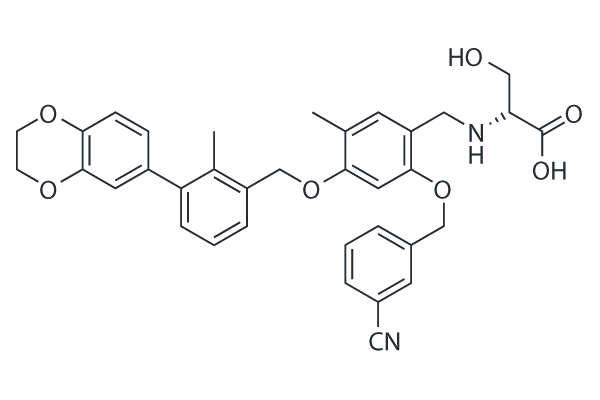
BMS-1001
BMS-1001 is a potent inhibitor of PD-1/PD-L1 interaction with EC50 of 253 nM. BMS-1001 alleviates the inhibitory effect of the soluble PD-L1 on the T-cell receptor-mediated activation of T-lymphocytes.
-
GC38740
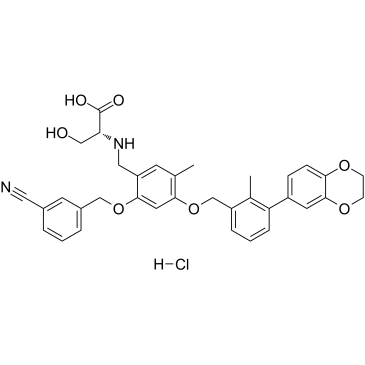
BMS-1001 hydrochloride
BMS-1001 hydrochloride is an orally active human PD-L1/PD-1 immune checkpoint inhibitor.
-
GC31753
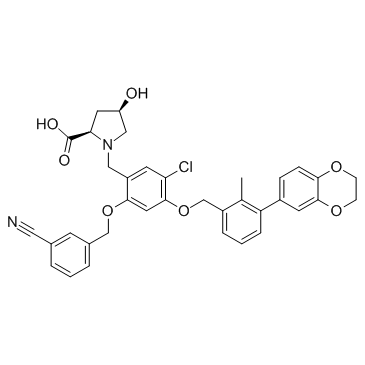
BMS-1166 (PD-1/PD-L1-IN1)
BMS-1166 (PD-1/PD-L1-IN1) is a potent PD-1/PD-L1 immune checkpoint inhibitor.
-
GC38131
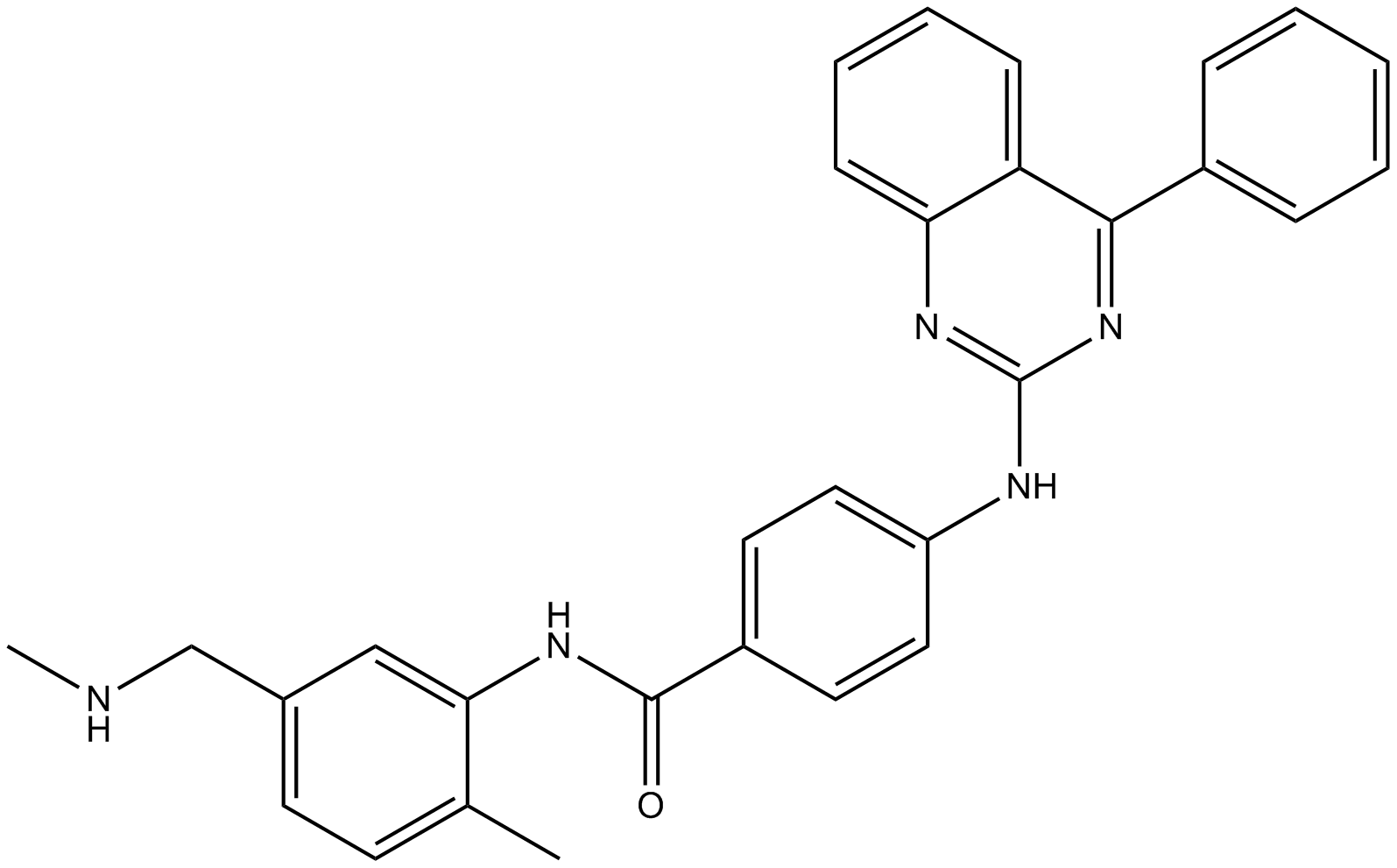
BMS-1166 hydrochloride
BMS-1166 hydrochloride is a potent PD-1/PD-L1 immune checkpoint inhibitor.

-
GC13628
BMS-833923
An orally bioavailable Smo inhibitor
-
GC62682
BMSpep-57 hydrochloride
BMSpep-57 hydrochloride is a potent and competitive macrocyclic peptide inhibitor of PD-1/PD-L1 interaction with an IC50 of 7.68?nM. BMSpep-57 hydrochloride binds to PD-L1 with Kds of 19 nM and 19.88 nM in MST and SPR assays, respectively. BMSpep-57 hydrochloride facilitates T cell function by in creasing IL-2 production in PBMCs.
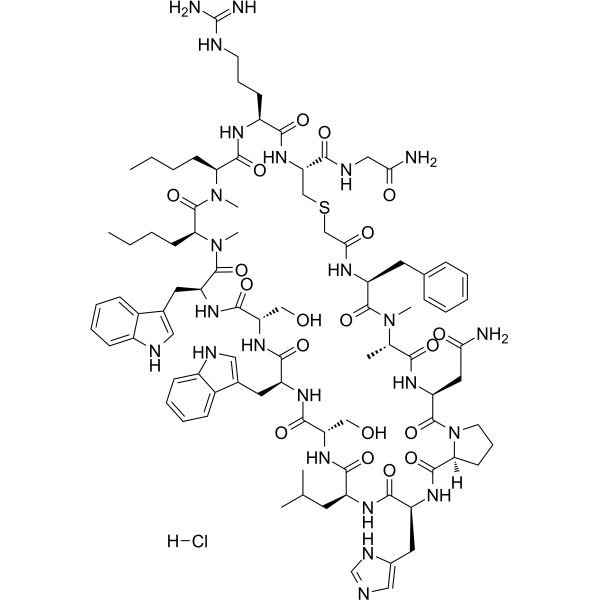
-
GA20897
Boc-Arg(Boc)₂-OH
An amino acid building block

-
GC16774
Boc-D-FMK
An irreversible pan-caspase inhibitor
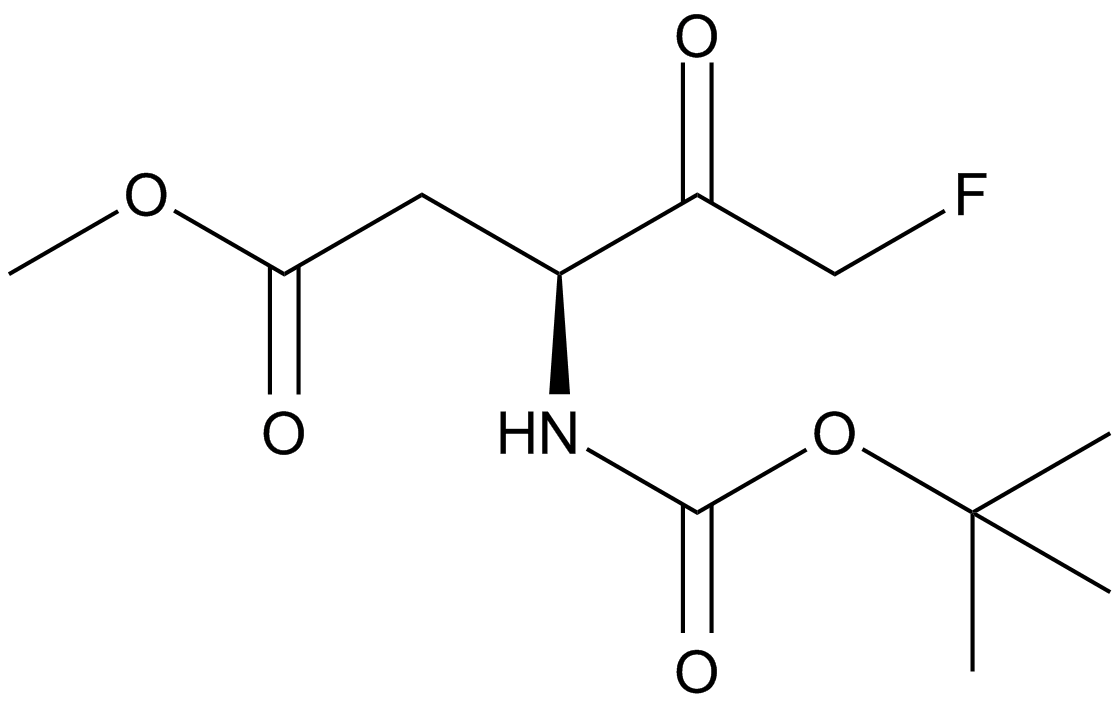
-
GC33501
Bornyl acetate
An acetate form of borneol
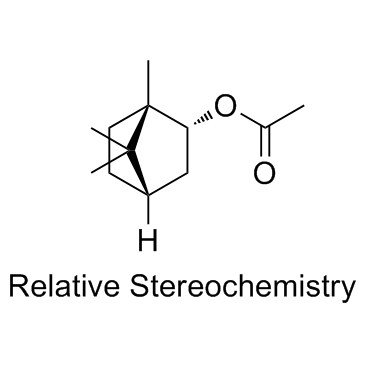
-
GC11040
Borrelidin
threonyl-tRNA synthetase (ThrRS) inhibitor
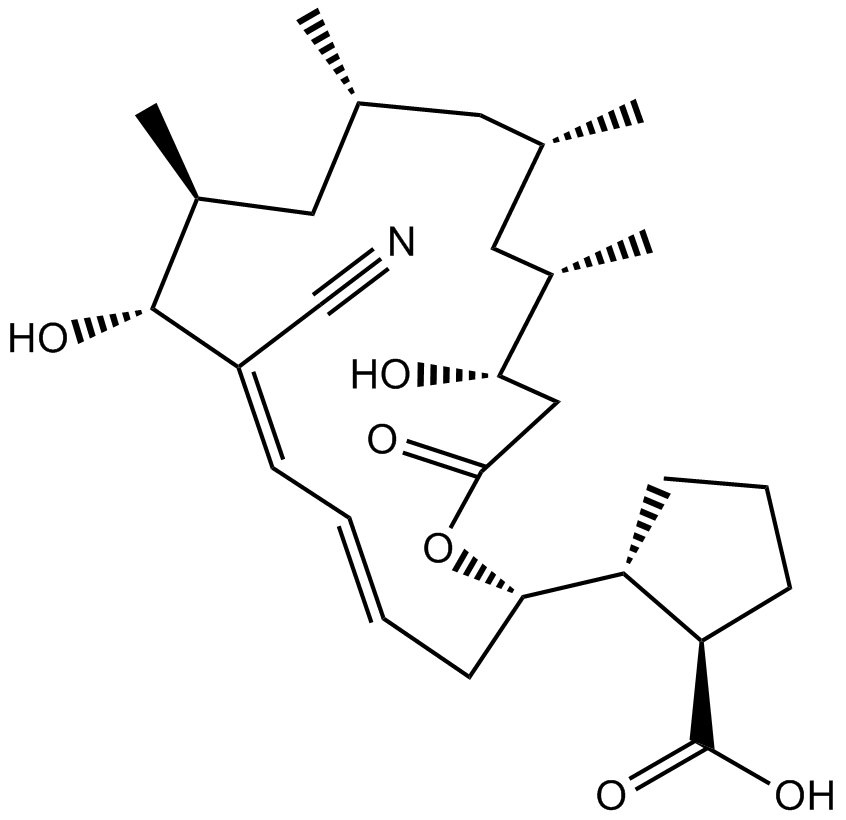
-
GC17644
Bortezomib (PS-341)
Bortezomib (PS-341), as a dipeptide boronic acid proteasome inhibitor with antitumor activity, can potently inhibiit 20S proteasome with Ki of 0.6 nM by targeting a threonine residue..
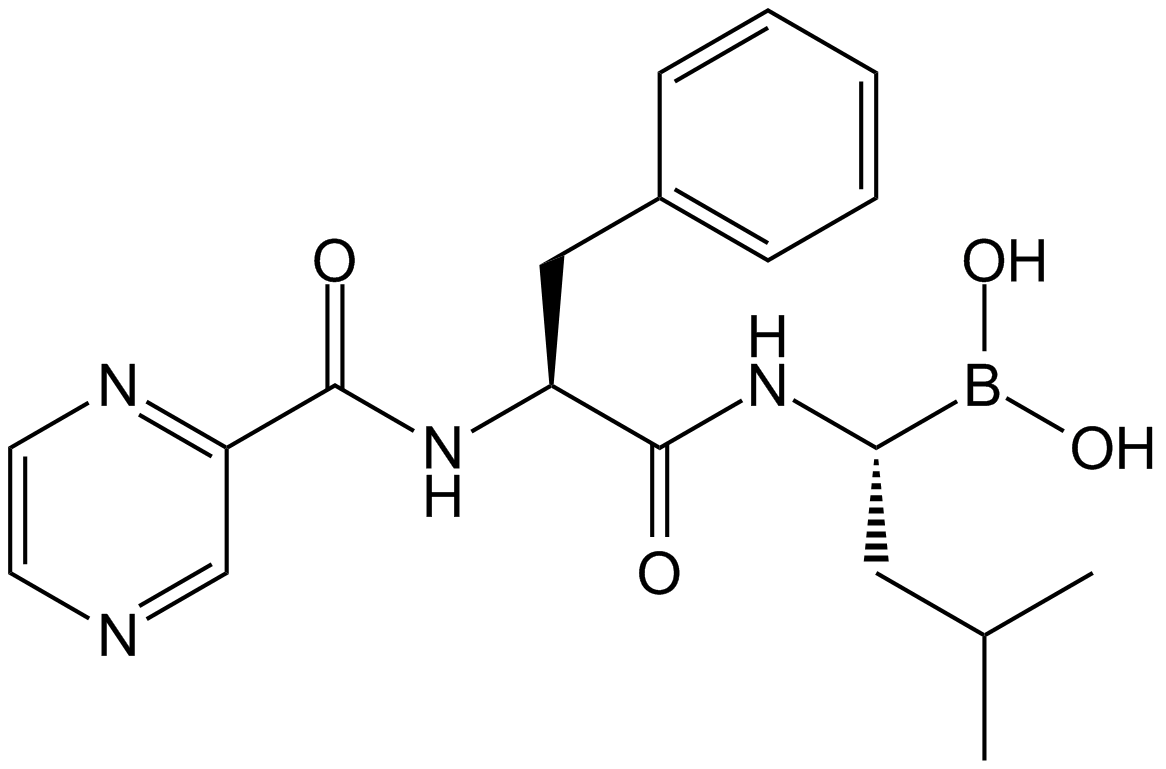
-
GC65010
Bortezomib-d8
Bortezomib-d8 (PS-341-d8) is the deuterium labeled Bortezomib. Bortezomib (PS-341) is a reversible and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki=0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is the first proteasome inhibitor anticancer agent. Anti-cancer activity.
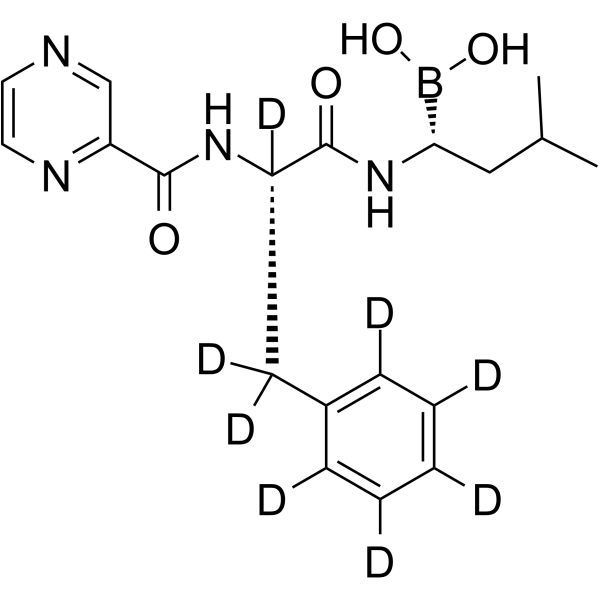
-
GC40009
Bostrycin
Bostrycin is an anthraquinone originally isolated from B.
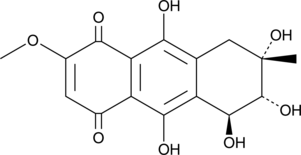
-
GC42969
bpV(phen) (potassium hydrate)
bpV(phen) (potassium hydrate), a insulin-mimetic agent, is a potent protein tyrosine phosphatase (PTP) and PTEN inhibitor with IC50s of 38 nM, 343 nM and 920 nM for PTEN, PTP-β and PTP-1B, respectively.
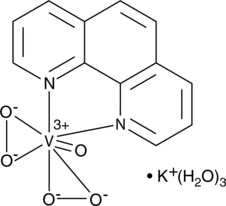
-
GC42974
Brassinin
Brassinin (BSN) is a phytoalexin isolated from B.
-
GC11632
Brassinolide
A plant growth regulator
-
GC52101
Brazilein
Brazilein is an important immunosuppressive component isolated from Caesalpinia sappan L.
-
GN10802
Brazilin
-
GC68288
Brentuximab

-
GC35554
Brevilin A
A sesquiterpene lactone with anticancer activity

-
GC35555
Britannin
Britannin, isolated from Inula aucheriana, is a sesquiterpene lactone.
-
GC62536
Bromelain
Bromelain is an anti-inflammatory drug derived from pineapple stem that acts through down-regulation of plasma kininogen, inhibition of Prostaglandin E2 expression, degradation of advanced glycation end product receptors and regulation of angiogenic biomarkers as well as antioxidant action upstream in the COX-pathway.

-
GC35559
Bruceine D
Bruceine D is a Notch inhibitor with anti-cancer activity and induces apoptosis in several human cancer cells.
-
GC34070
Brusatol (NSC 172924)
Brusatol (NSC 172924) (NSC172924) is a unique inhibitor of the Nrf2 pathway that sensitizes a broad spectrum of cancer cells to Cisplatin and other chemotherapeutic agents. Brusatol (NSC 172924) enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Brusatol (NSC 172924) can be developed into an adjuvant chemotherapeutic agent. Brusatol (NSC 172924) increases cellular apoptosis.
-
GC38014
BT2
An Mcl-1 inhibitor
-
GC38467
BTdCPU
BTdCPU is a potent heme-regulated eIF2α kinase (HRI) activator. BTdCPU promotes eIF2α phosphorylation and induced apoptosis in resistant cell.
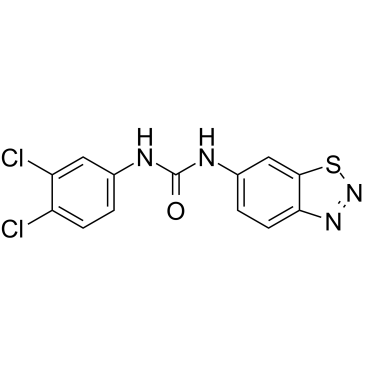
-
GC34511
BTR-1
BTR-1 is an active anti-cancer agent, causes S phase arrest, and affects DNA replication in leukemic cells. BTR-1 activates apoptosis and induces cell death.
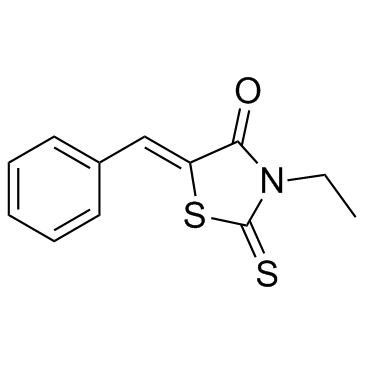
-
GC11141
BTZO 1
migration inhibitory factor (MIF) binder
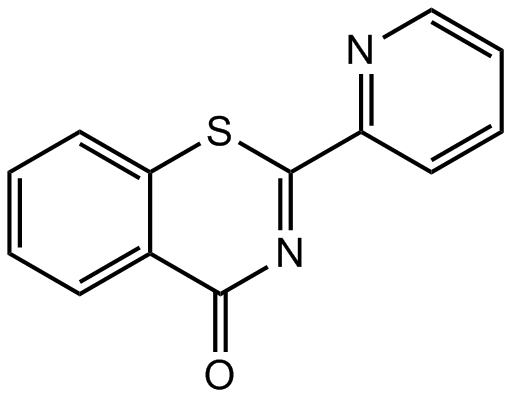
-
GC48376
Burnettramic Acid A
A fungal metabolite with diverse biological activities
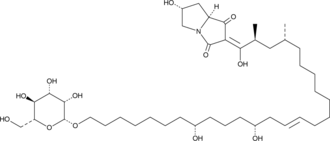
-
GC48409
Burnettramic Acid A aglycone
A fungal metabolite with anticancer activity
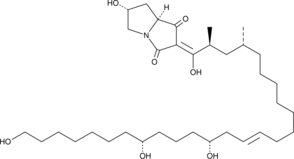
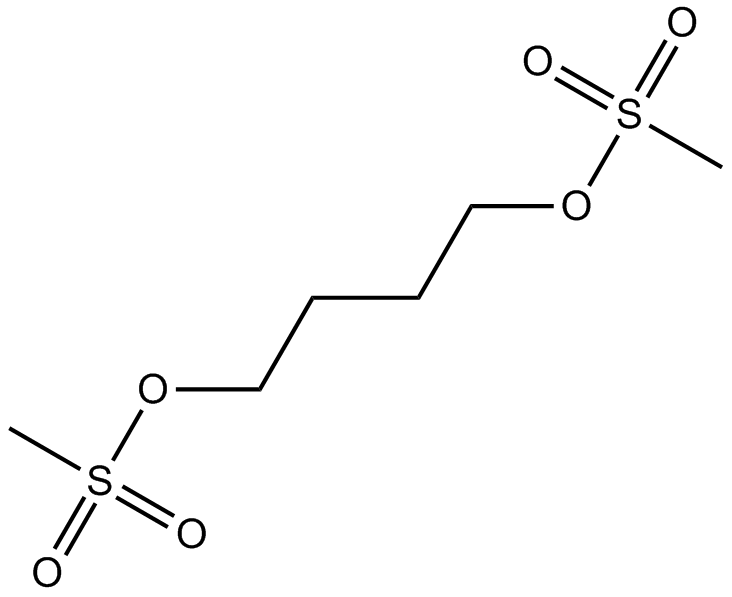
-
GC13671
Busulfan
DNA alkylating agent
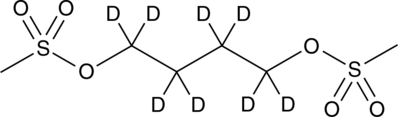
-
GC46962
Busulfan-d8
An internal standard for the quantification of busulfan
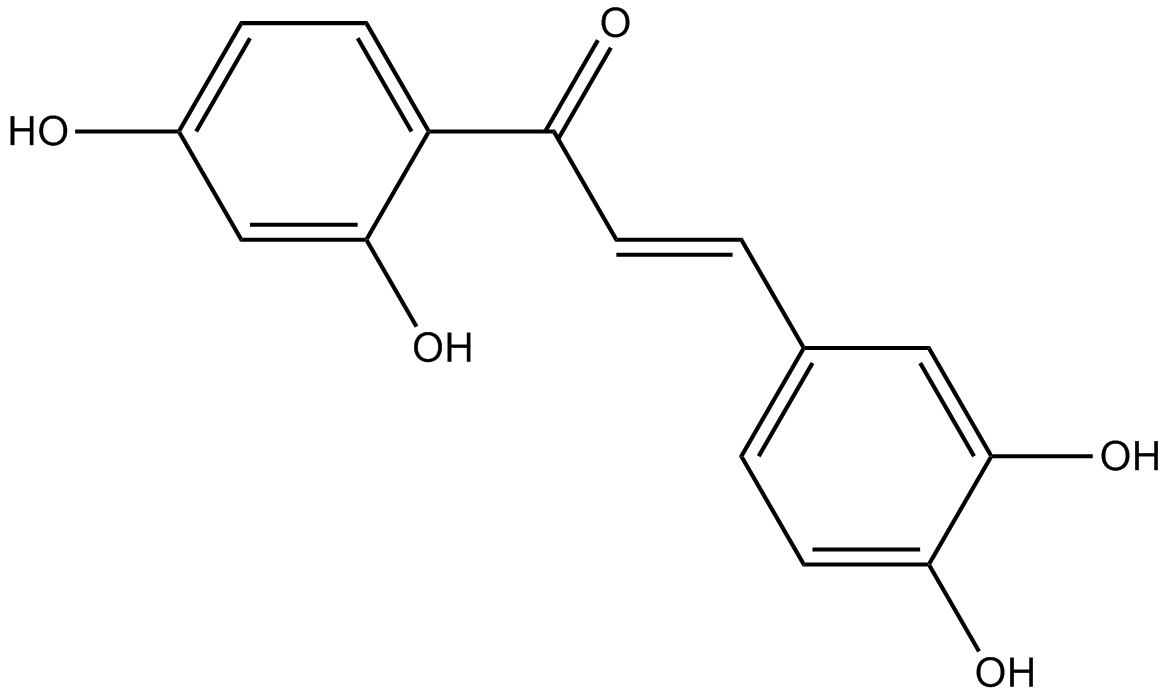
-
GC10944
Butein
Protein kinase inhibitor
-
GC46104
Butyric Acid-d7
An internal standard for the quantification of sodium butyrate
-
GC12333
BV6
Selective inhibitor of IAP proteins
-
GC48433
BX-320
An inhibitor of PDK1
-
GC16818
BX-912
PDK1 inhibitor,potent and ATP-competitive
-
GC31806
Bz 423 (BZ48)
Bz 423 (BZ48) is a pro-apoptotic 1,4-benzodiazepine with therapeutic properties in murine models of lupus demonstrating selectivity for autoreactive lymphocytes, and activates Bax and Bak.
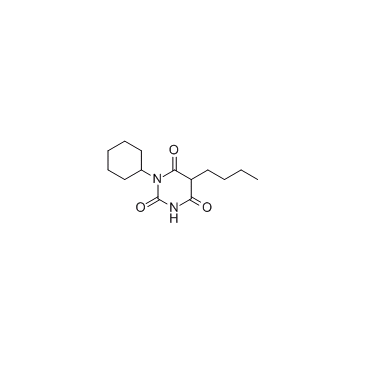
-
GC33826
C 87
C 87 is a novel small-molecule TNFα inhibitor; potently inhibits TNFα-induced cytotoxicity with an IC50 of 8.73 μM.
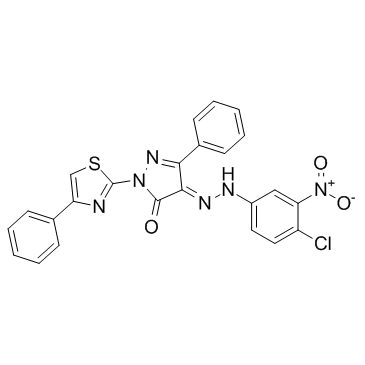
-
GC19095
C-DIM12
C-DIM12 is a synthetic Nurr1 activaor induces Nurr1 and DA gene expression in cell lines and primary neurons.
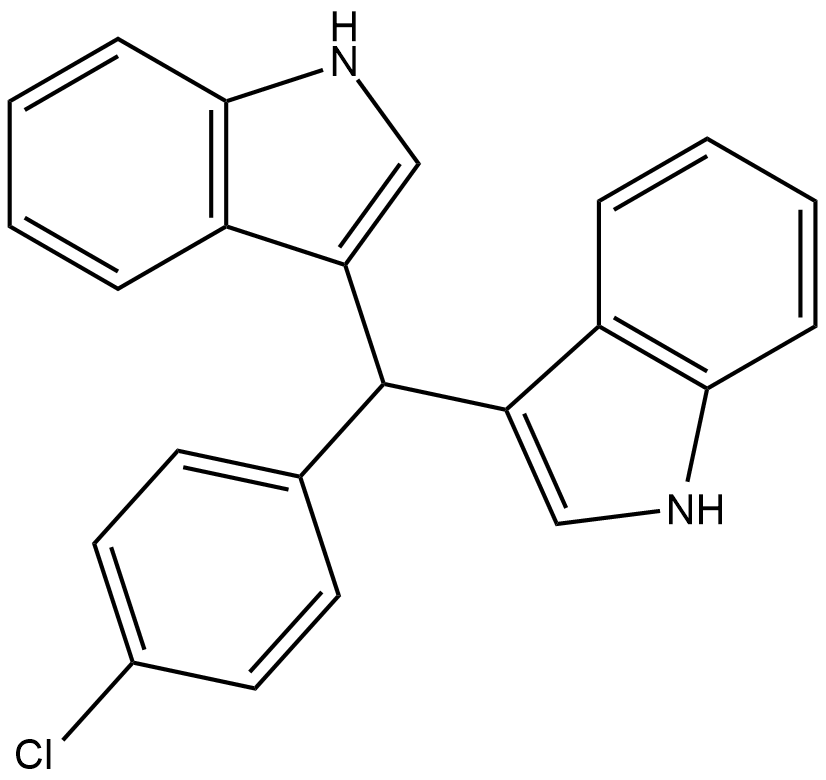
-
GC43028
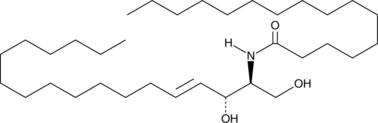
C16 Ceramide (d18:1/16:0)
C16 ceramide (d18:1/16:0), as an endogenous ceramide, generated by ceramide synthase 6 (CerS6), that acts as a lipid second messenger to regulate apoptosis and stress signaling. C16-ceramide plays a pivotal role in inducing insulin resistance.
-
GC46976
C16 Ceramide-d7 (d18:1-d7/16:0)
An internal standard for the quantification of C-16 ceramide
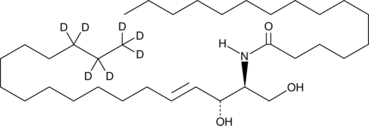
-
GC43052
C18 Phytoceramide (t18:0/18:0)
C18 Phytoceramide (t18:0/18:0) (Cer(t18:0/18:0)) is a bioactive sphingolipid found in S.
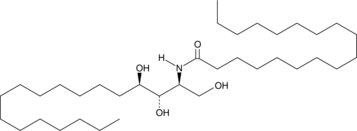
-
GC40141
C18 Phytoceramide-d3 (t18:0/18:0-d3)
C18 Phytoceramide-d3 (t18:0/18:0-d3) is intended for use as an internal standard for the quantification of C18 phytoceramide (t18:0/18:0) by GC- or LC-MS.

-
GC43065
C2 Phytoceramide (t18:0/2:0)
C2 Phytoceramide is a bioactive semisynthetic sphingolipid that inhibits formyl peptide-induced oxidant release (IC50 = 0.38 μM) in suspended polymorphonuclear cells.
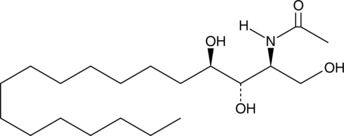
-
GC43069
C22 Ceramide (d18:1/22:0)
C-22 ceramide is an endogenous bioactive sphingolipid.
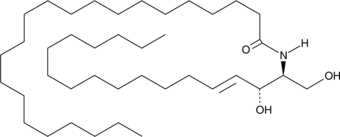
-
GC43075
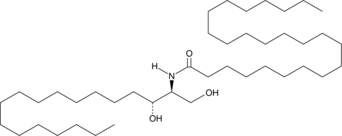
C24 dihydro Ceramide (d18:0/24:0)
C24 dihydro Ceramide is a sphingolipid that has been found in the stratum corneum of human skin.
-
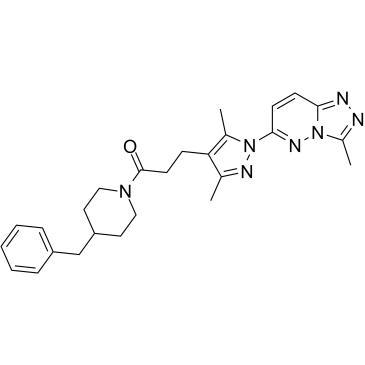
GC34513
C25-140
C25-140, a first-in-class, orally active, and fairly selective TRAF6-Ubc13 inhibitor, directly binds to TRAF6, and blocks the interaction of TRAF6 with Ubc13.
-
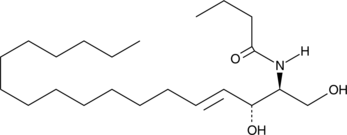
GC43084
C4 Ceramide (d18:1/4:0)
C4 Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
-
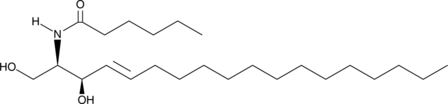
GC40688
C6 D-threo Ceramide (d18:1/6:0)
C6 D-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides., C6 D-threo Ceramide is cytotoxic to U937 cells in vitro (IC50 = 18 μM).
-
GC40689
C6 L-erythro Ceramide (d18:1/6:0)
C6 L-erythro Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
-
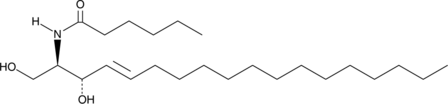
GC40690
C6 L-threo Ceramide (d18:1/6:0)
C6 L-threo Ceramide (d18:1/6:0) is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
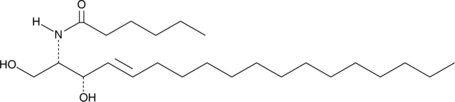
-
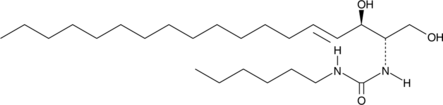
GC45616
C6 Urea Ceramide
An inhibitor of neutral ceramidase
-
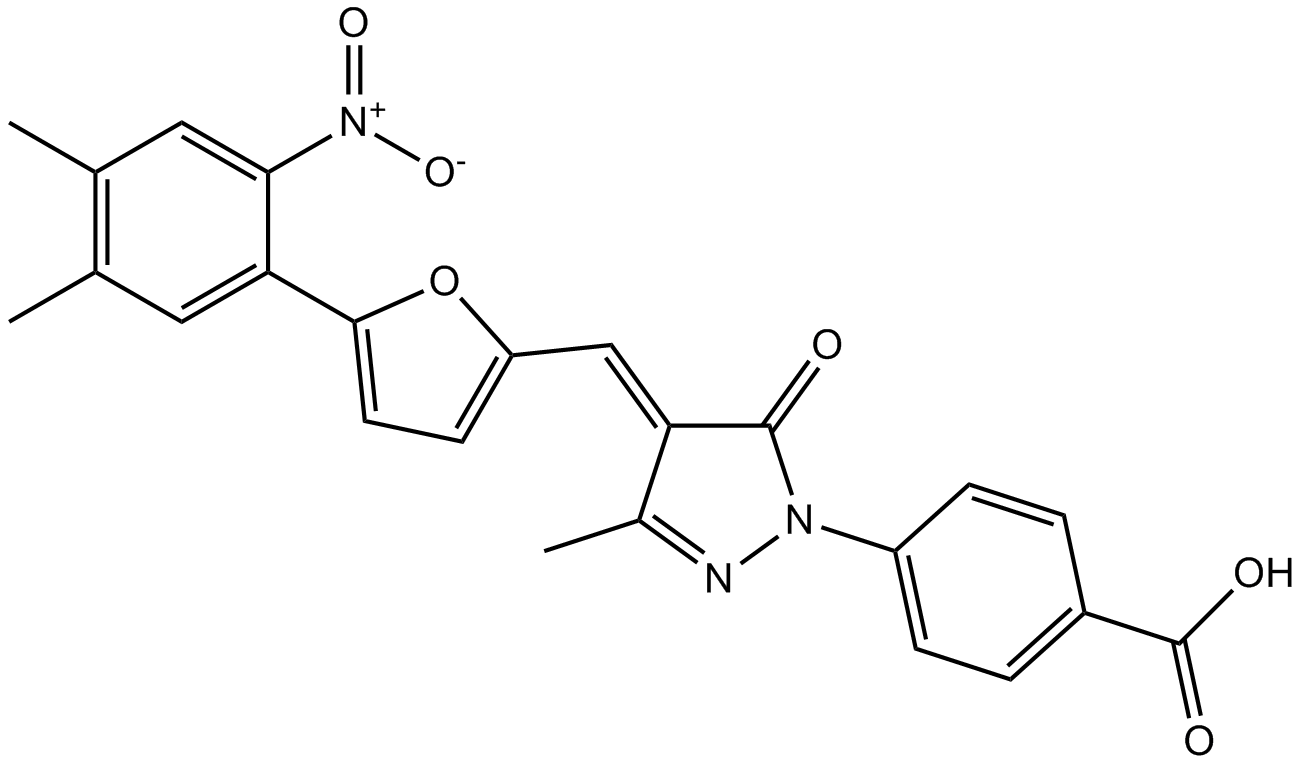
GC12733
C646
C646, a potent and selective p300/CBP histone acetyltransferase inhibitor (Ki 400 nM), has been shown to have pleiotropic activity, including neuroprotective, anti-cancer and anti-epithelial-mesenchymal transition (anti-EMT) effects..
-
GC43105
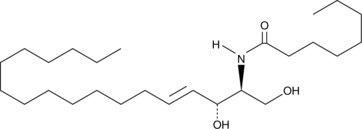
C8 Ceramide (d18:1.8:0)
C8 Ceramide (d18:1.8:0) (N-Octanoyl-D-erythro-sphingosine) is a cell-permeable analog of naturally occurring ceramides.
-
GC43109
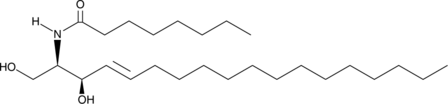
C8 D-threo Ceramide (d18:1/8:0)
C8 D-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
-
GC43110
C8 Galactosylceramide (d18:1/8:0)
C8 Galactosylceramide is a synthetic C8 short-chain derivative of known membrane microdomain-forming sphingolipids.

-
GC43111
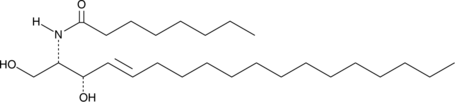
C8 L-threo Ceramide (d18:1/8:0)
C8 L-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
-
GC33218
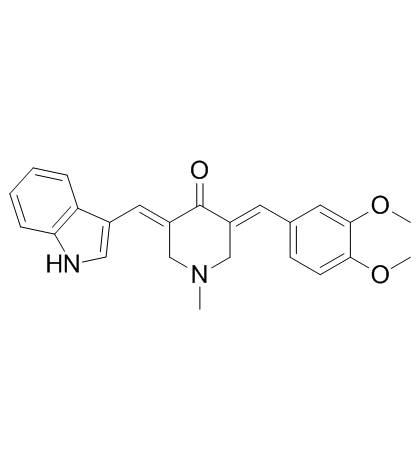
CA-5f
A potent late-stage macroautophagy/autophagy inhibitor
-
GC15779
Cabozantinib (XL184, BMS-907351)
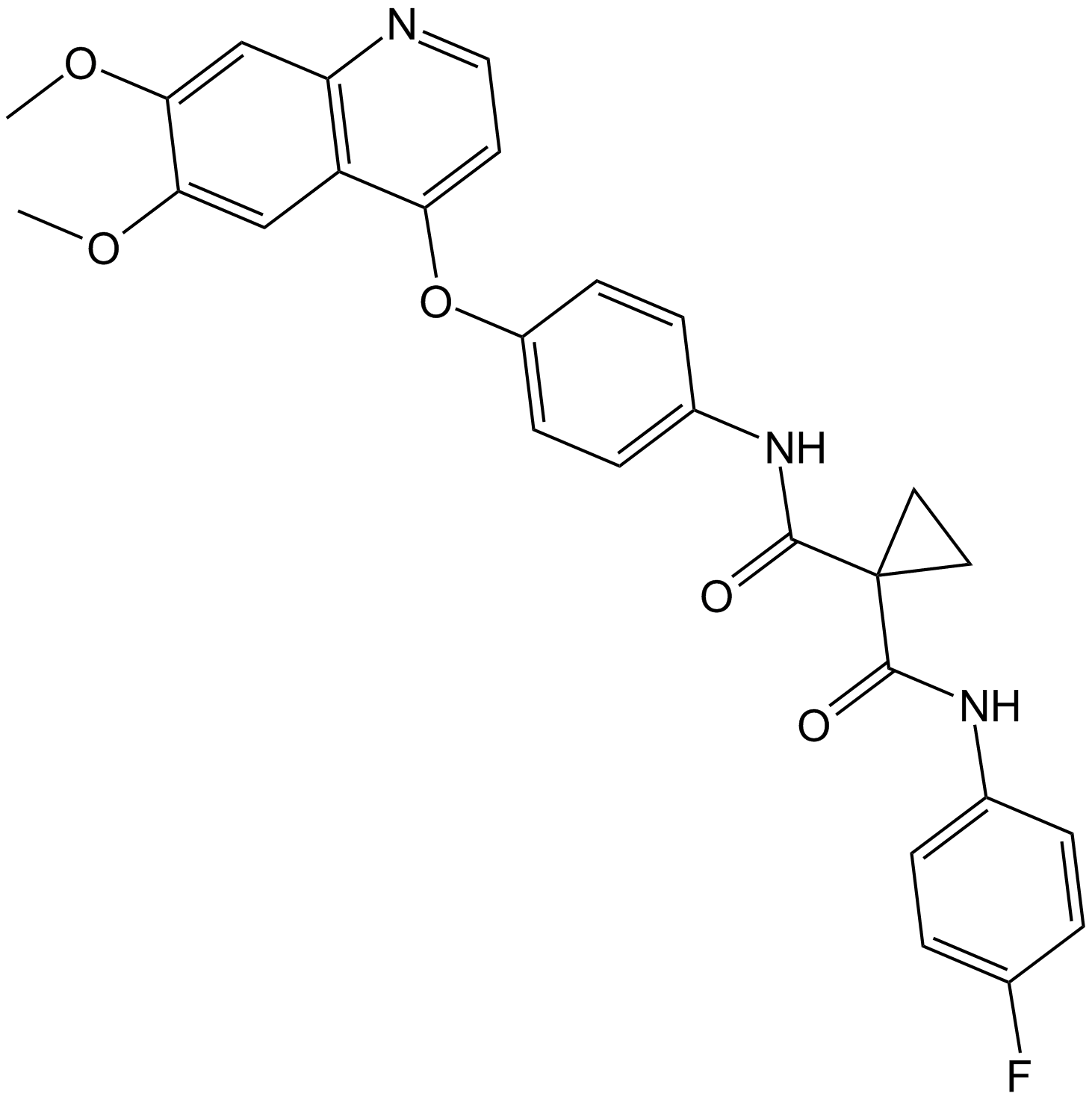
Cabozantinib (XL184, BMS-907351) is a potent and orally active inhibitor of VEGFR2 and MET, with IC50 values of 0.035, and 1.3 nM, respectively. Cabozantinib (XL184, BMS-907351) displays strong inhibition of KIT, RET, AXL, TIE2, and FLT3 (IC50=4.6, 5.2, 7, 14.3, and 11.3 nM, respectively). Cabozantinib (XL184, BMS-907351) shows antiangiogenic activity. Cabozantinib (XL184, BMS-907351) disrupts tumor vasculature and promotes tumor and endothelial cell apoptosis.
-
GC12531
Cabozantinib malate (XL184)
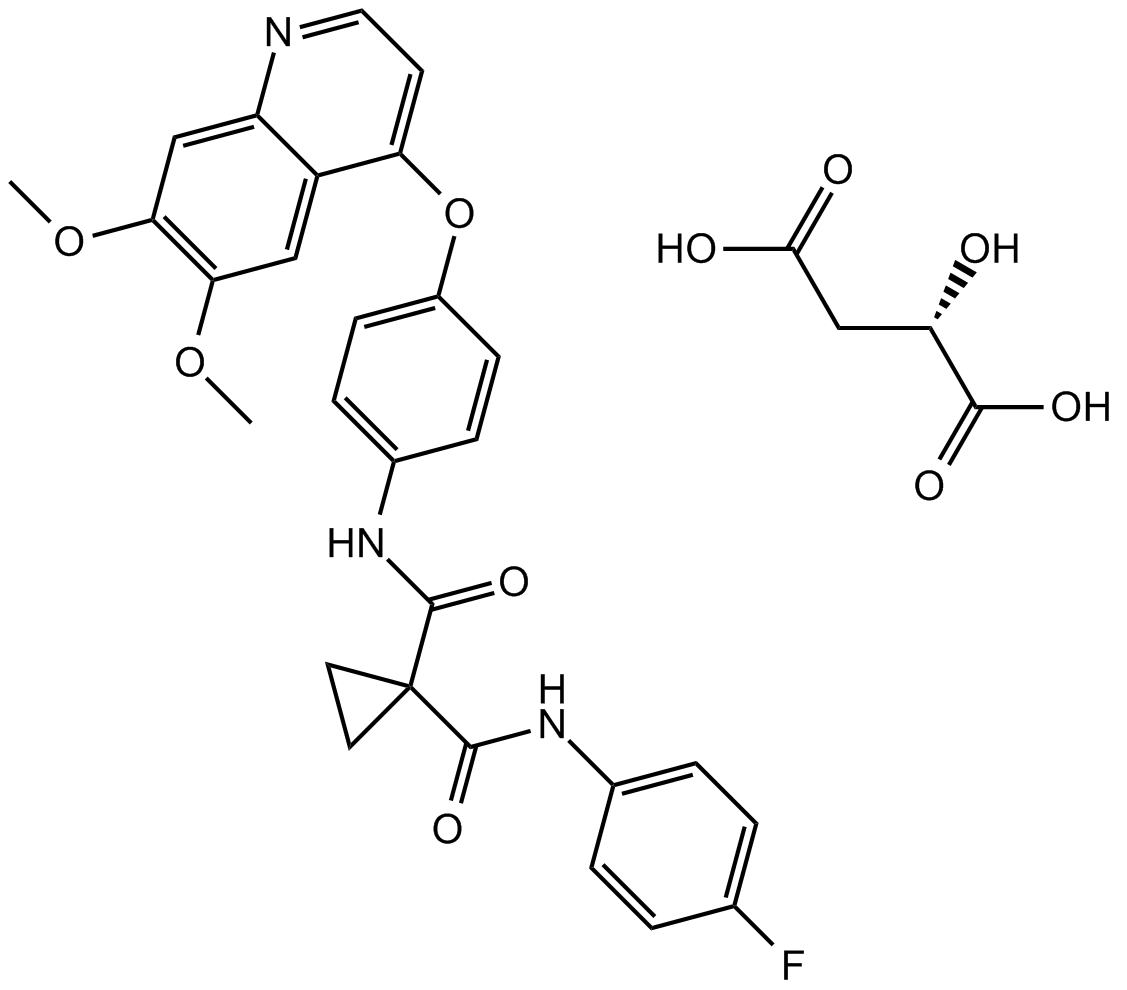
Cabozantinib malate (XL184) (XL184 S-malate) is a potent multiple receptor tyrosine kinases inhibitor that inhibits VEGFR2, c-Met, Kit, Axl and Flt3 with IC50s of 0.035, 1.3, 4.6, 7 and 11.3 nM, respectively.
-
GC10692
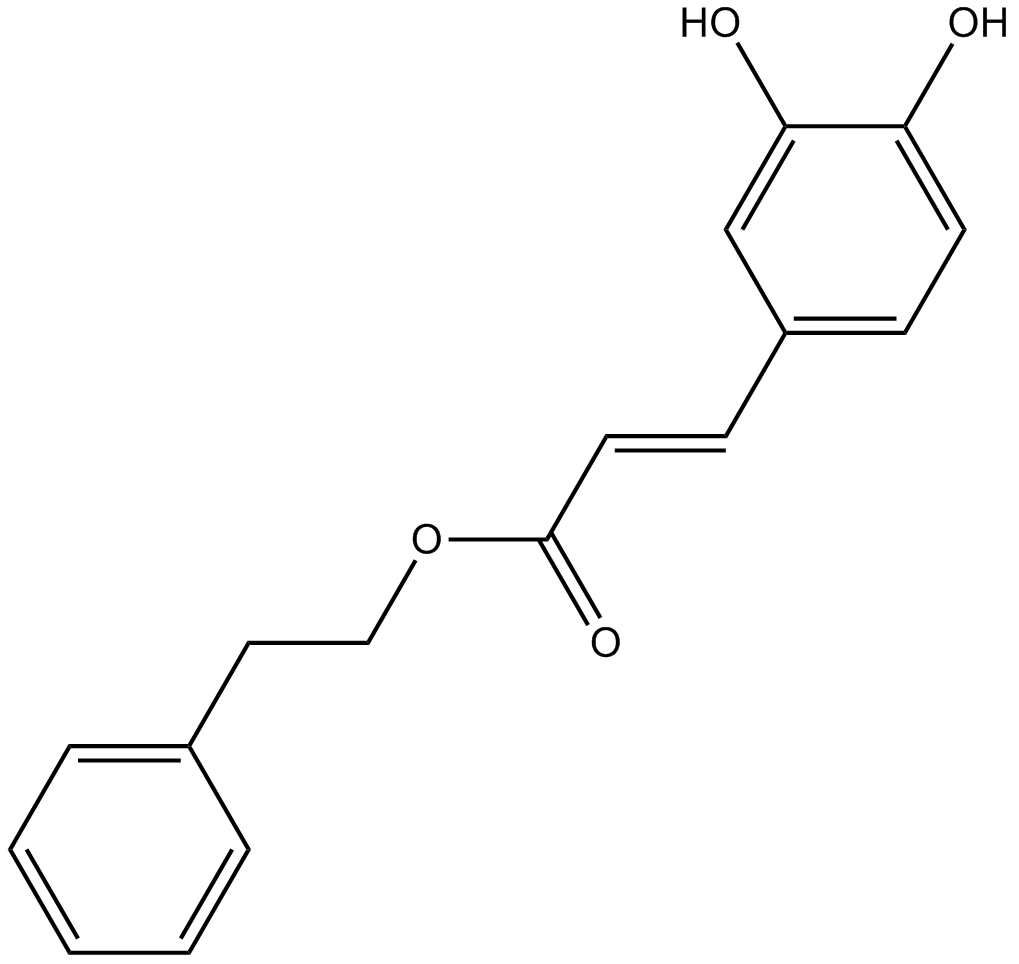
Caffeic Acid Phenethyl Ester
NF-κB activation inhibitor
-
GC18604
Calcein Blue AM
Calcein Blue AM is a fluorogenic dye that is used to assess cell viability.
-
GC43121
Calcein Orange™ Diacetate
Calcein Orange Diacetate is a fluorogenic dye that is used to assess cell viability.

-
GC43123
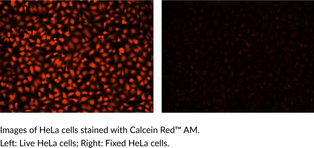
Calcein Red™ AM
Calcein Red? AM is a fluorogenic dye that is used to assess cell viability.
-
GC60668
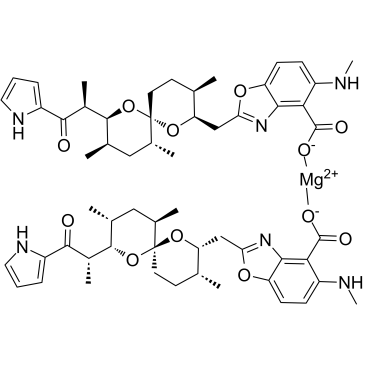
Calcimycin hemimagnesium
Calcimycin (A-23187) hemimagnesium is an antibiotic and a unique divalent cation ionophore (like calcium and magnesium).
-
GC15161
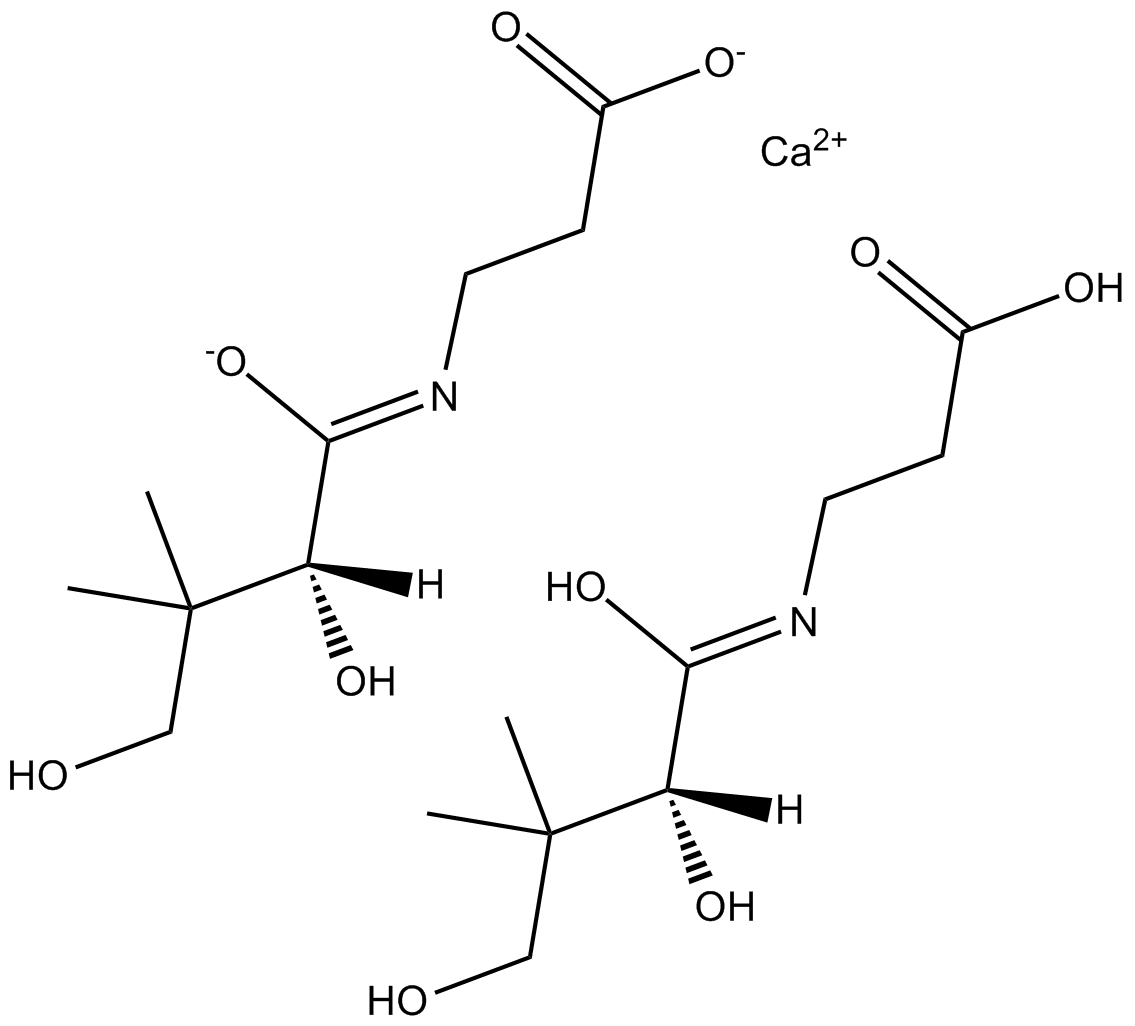
Calcium D-Panthotenate
Calcium D-Panthotenate (Vitamin B5 calcium salt), a vitamin, can reduce the patulin content of the apple juice.
-
GC30240
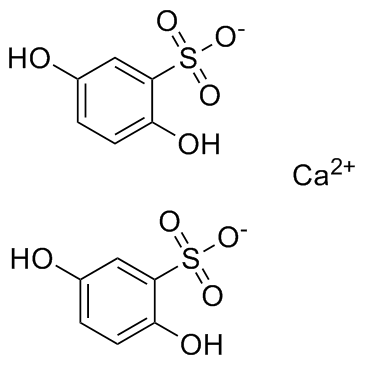
Calcium dobesilate
Calcium dobesilate, a vasoprotective, is widely used in chronic venous disease, diabetic retinopathy and the symptoms of haemorrhoidal attack in many countries.
-
GC19086
Calicheamicin
Calicheamicin is a cytotoxic agent that causes double-strand DNA breaks.
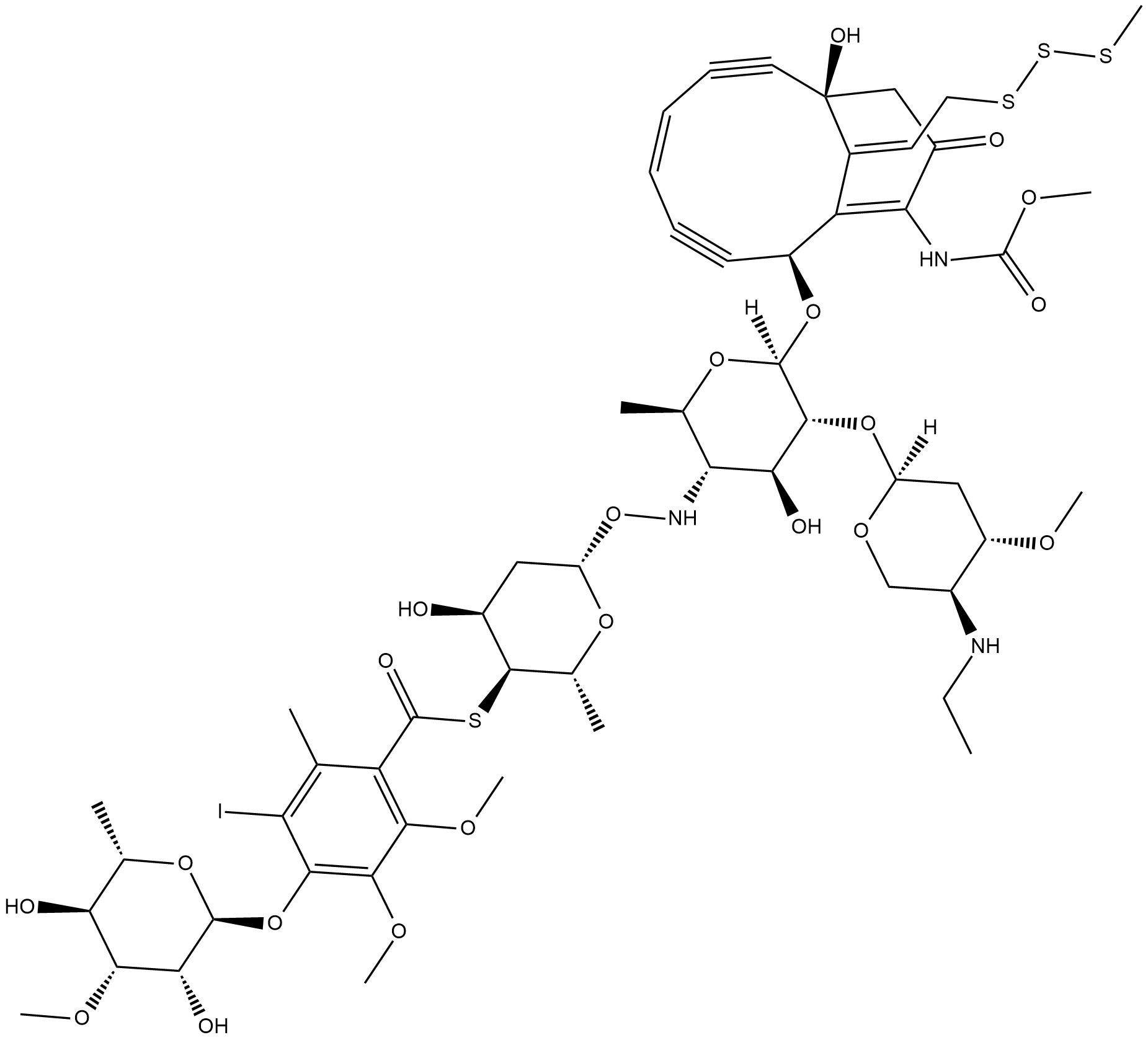
-
GC65081
CALP1 TFA
CALP1 TFA is a calmodulin (CaM) agonist (Kd of 88 ?M) with binding to the CaM EF-hand/Ca2+-binding site.
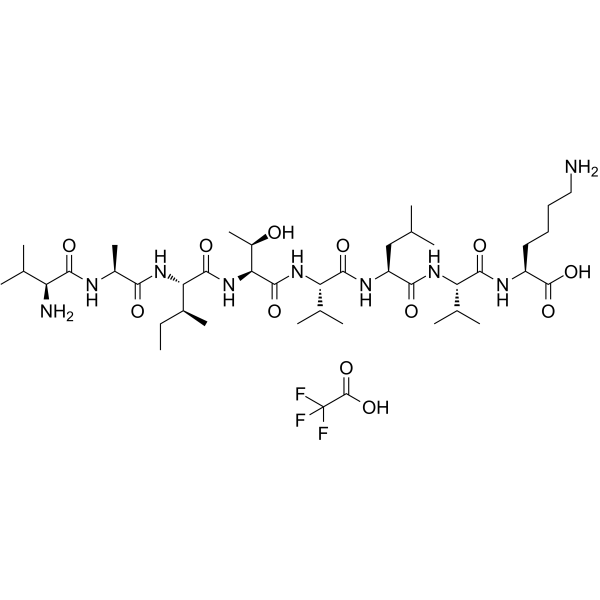
-
GC18315
Calpain Inhibitor VI
Calpain inhibitor VI is an inhibitor of the calcium-dependent cysteine proteases u-calpain (calpain-1; IC50 = 7.5 nM) and m-calpain (calpain-2; IC50 = 78 nM).
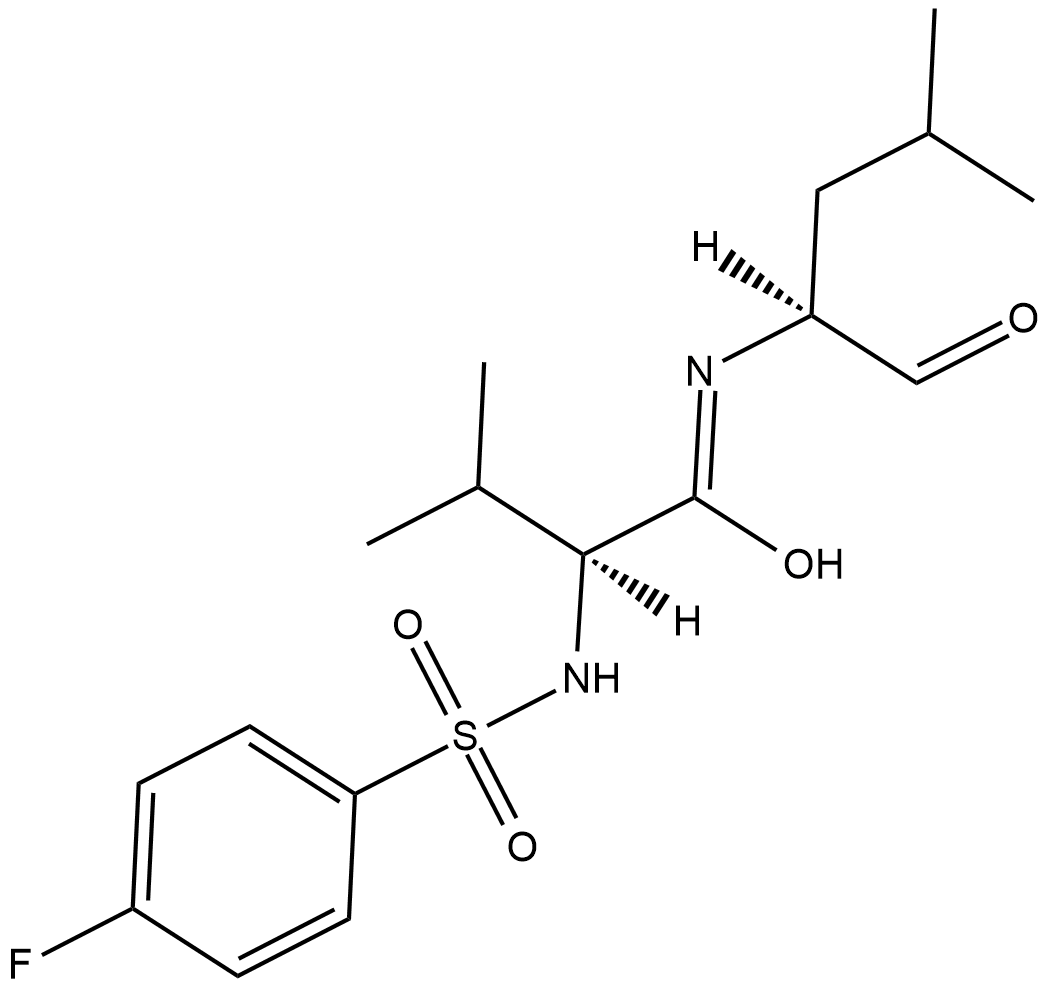
-
GC10342
Calpeptin
A calpain inhibitor
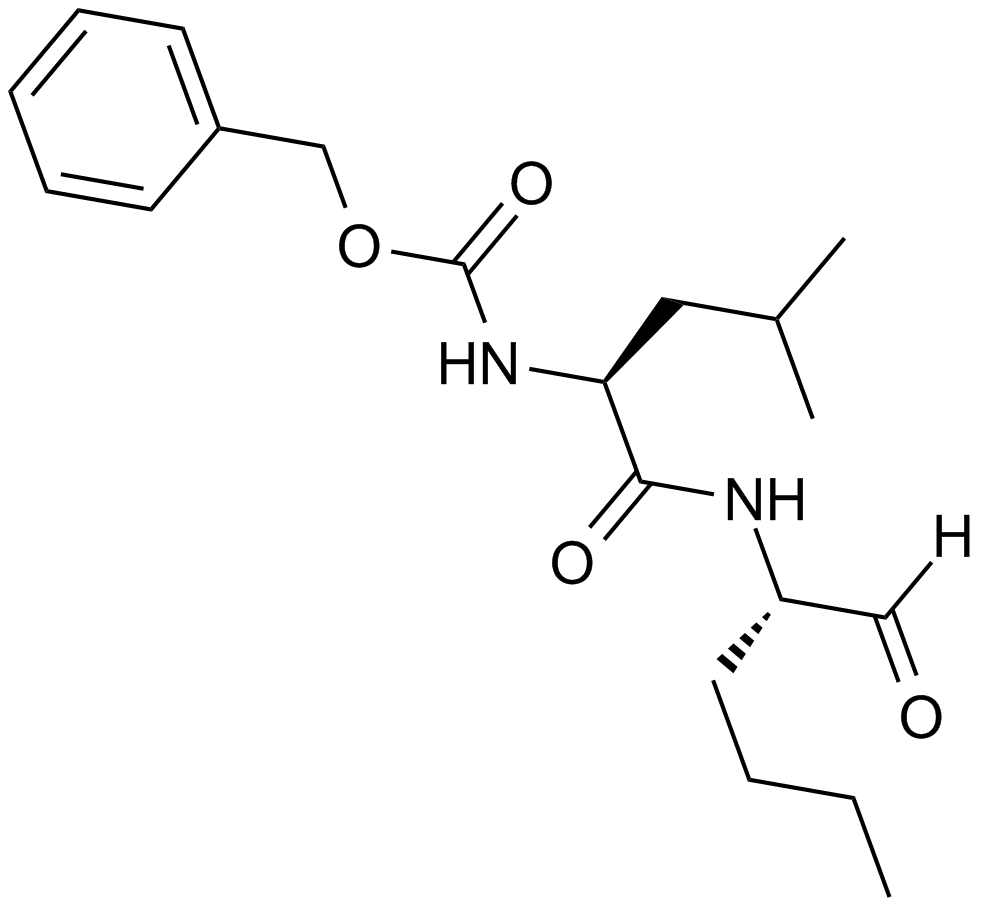
-
GN10667
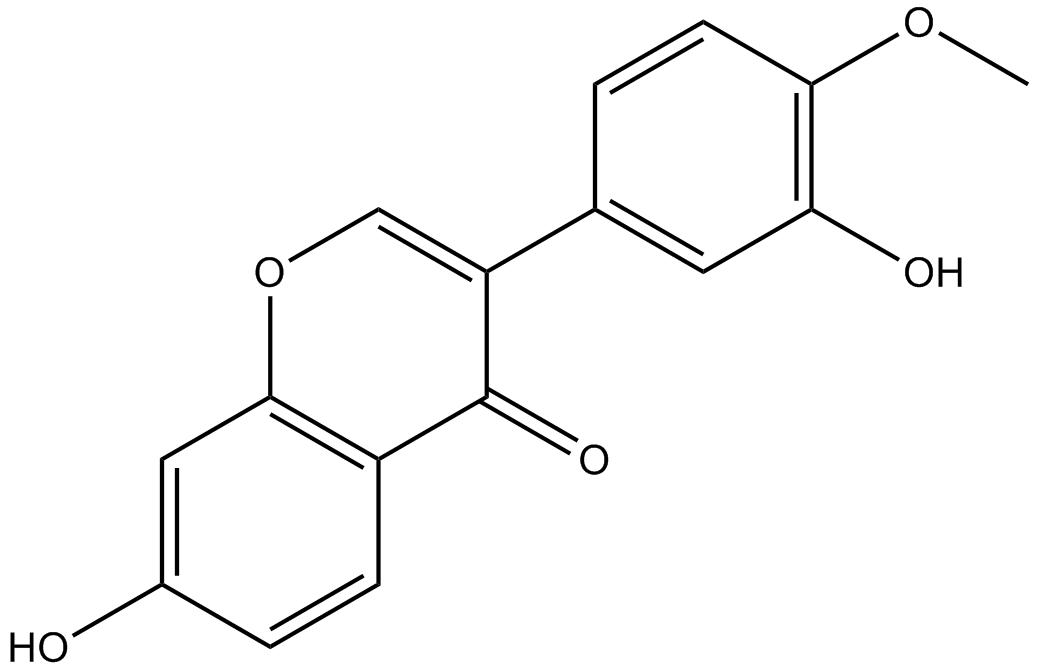
Calycosin
Calycosin (CA, 7, 3-dihydroxy-4-methoxy isoflavone, C16H12O5) is one of the flavonoids extracted from astragalus root, also known as the typical phytoestrogens.
-
GC35598
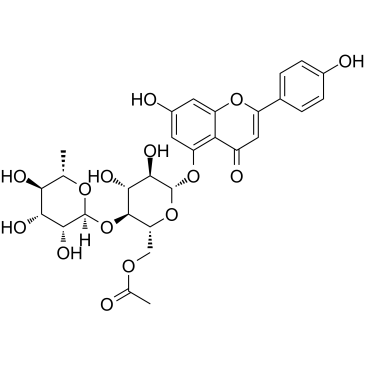
Camellianin A
Camellianin A, the main flavonoid in A. nitida leaves, displays anticancer activity and angiotensin converting enzyme (ACE)-inhibitory activity. Camellianin A inhibits the proliferation of the human Hep G2 and MCF-7 cell lines and induces the significant increase of the G0/G1 cell population.
-
GC62253
Camrelizumab
Camrelizumab (SHR-1210) is a potent humanied high-affinity IgG4-κ monoclonal antibody (mAb) to PD-1. Camrelizumab?binds PD-1 at a high affinity of 3 nM and inhibits the binding interaction of PD-1 and PD-L1 with an IC50 of 0.70 nM. Camrelizumab acts as?anti-PD-1/PD-L1 agent and can be used for cancer research, including NSCLC, ESCC, Hodgkin lymphoma, and advanced HCC et,al.

-
GC12318
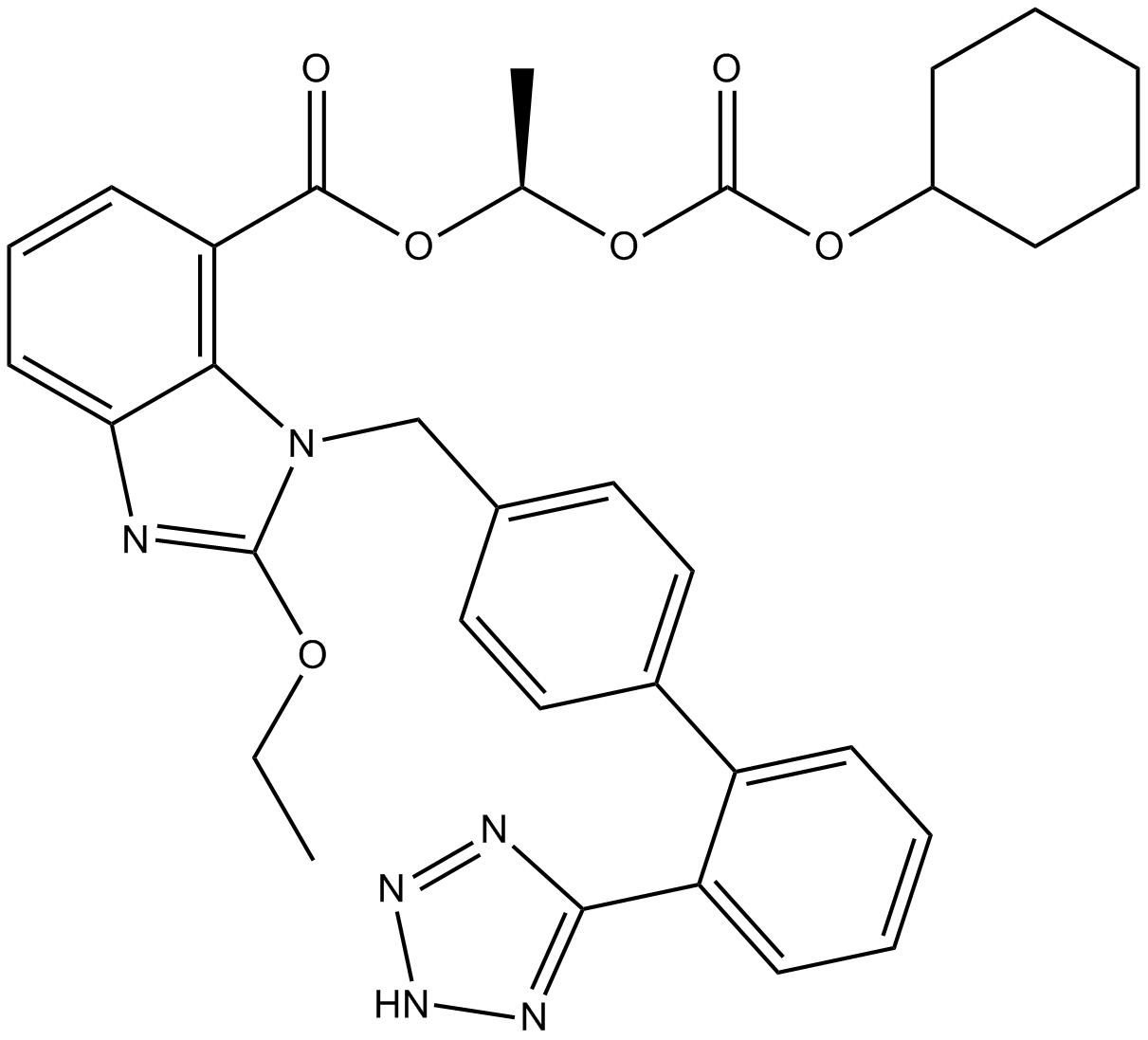
Candesartan Cilexetil
Prodrug of the angiotensin II receptor 1 (AT1) antagonist candesartan
-
GC15866
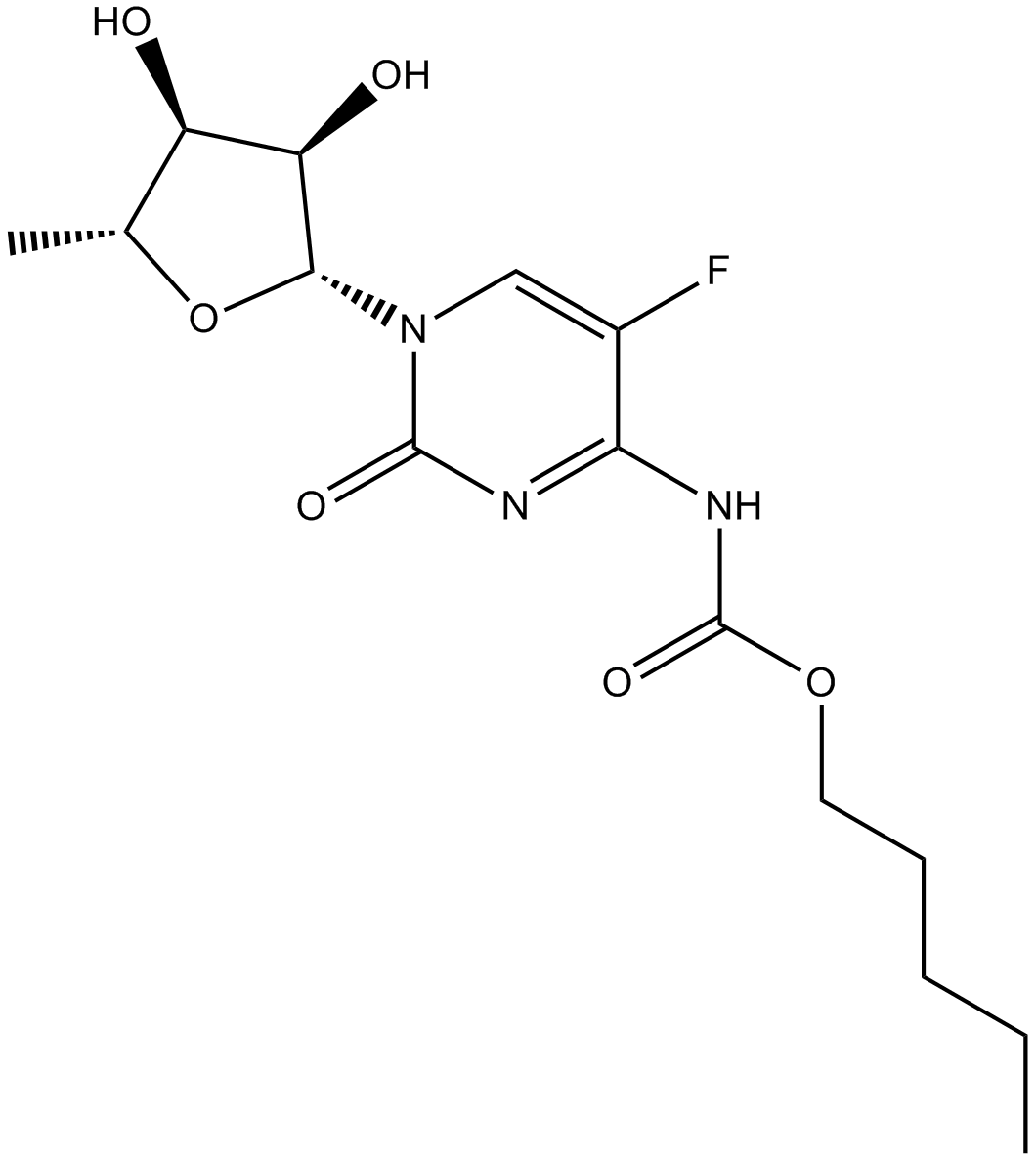
Capecitabine
DNA, RNA and protein synthesis inhibitor
-
GC14065
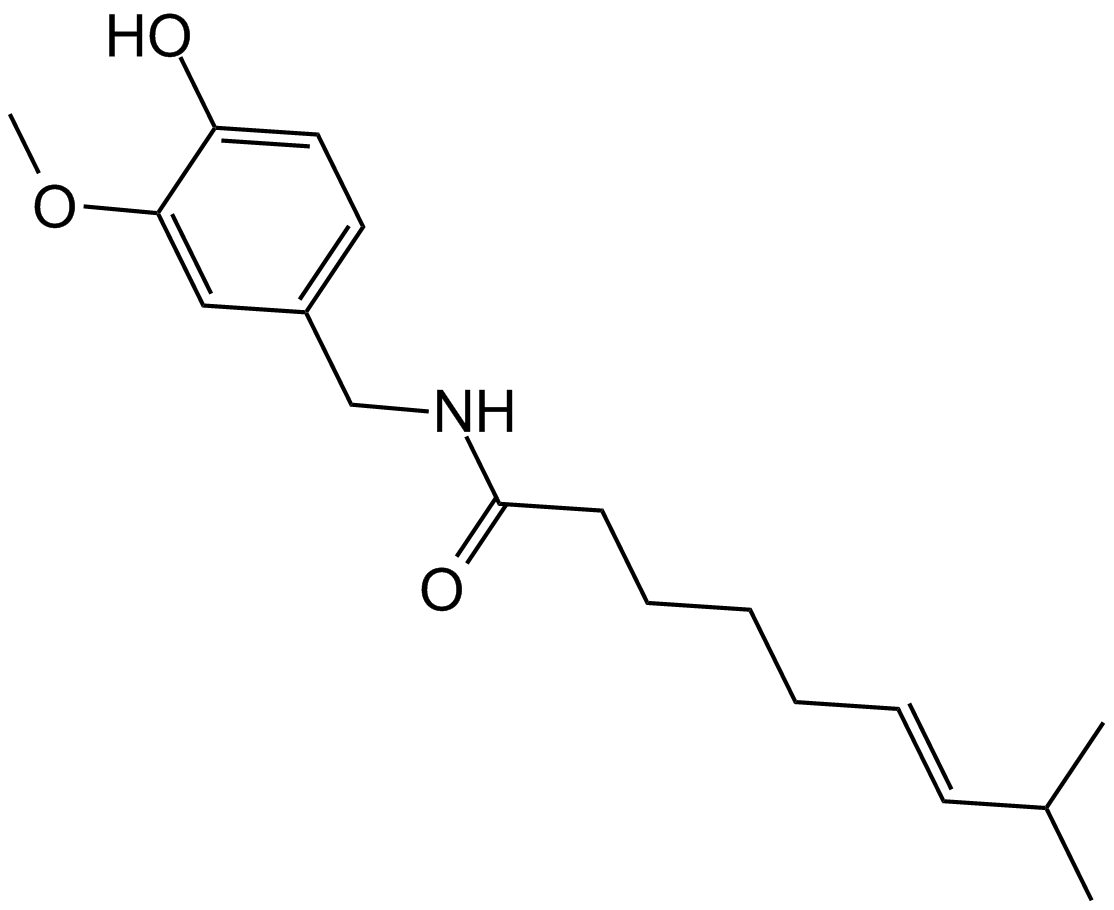
Capsaicin
A terpene alkaloid with diverse biological activities
-
GC17918
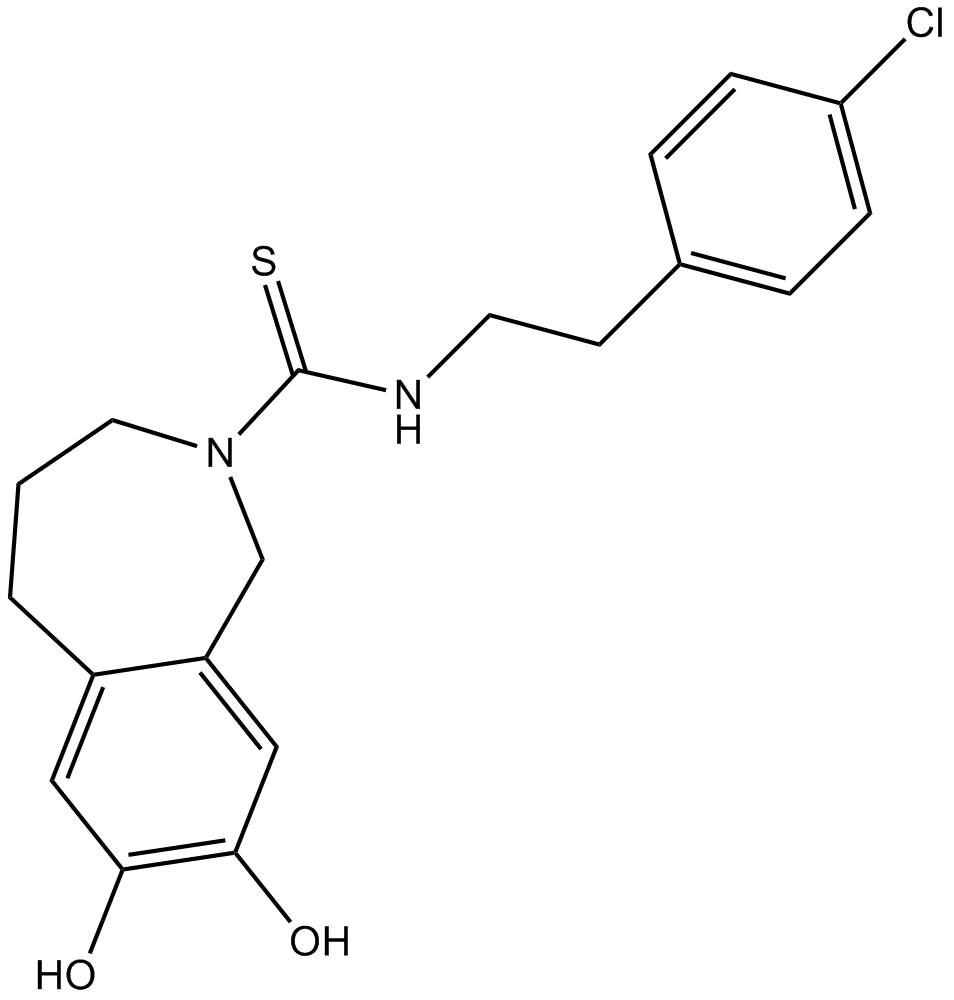
Capsazepine
A TRPV1 antagonist
-
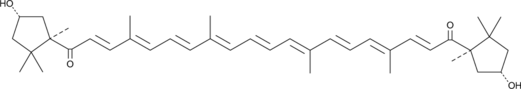
GC49415
Capsorubin
A carotenoid with diverse biological activities
-
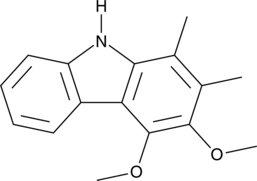
GC48878
Carbazomycin A
A bacterial metabolite with diverse biological activities
-
GC48893
Carbazomycin B
A bacterial metabolite with diverse biological activities
-
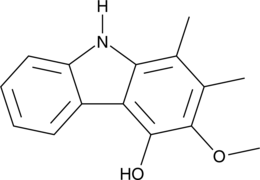
GC48850
Carbazomycin C
A bacterial metabolite with diverse biological activities
-
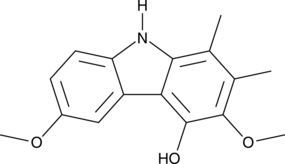
GC48826
Carbazomycin D
A bacterial metabolite with diverse biological activities
-
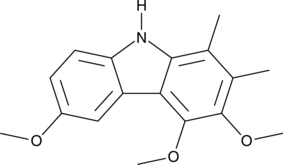
GC49147
Carboxyphosphamide
An inactive metabolite of cyclophosphamide
-
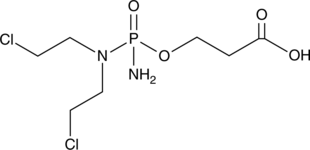
GC18069
Cardamonin
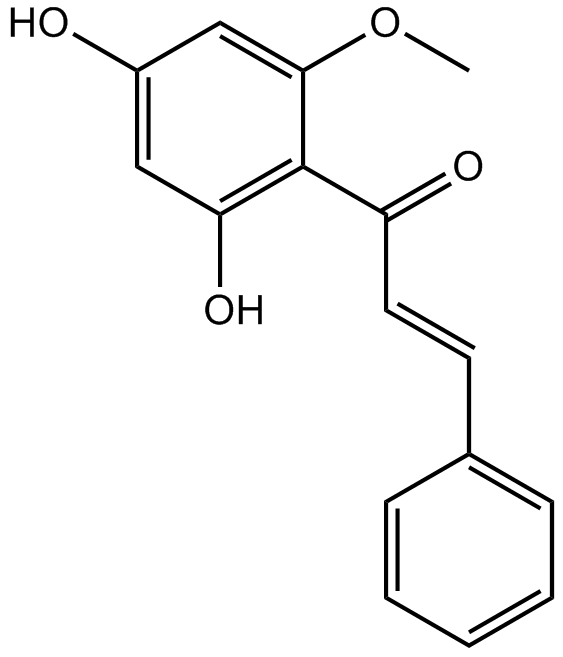
Cardamonin ((E)-Cardamomin) is a novel antagonist of hTRPA1 cation channel with an IC50 of 454 nM.
-
GC18449
Cardanol monoene
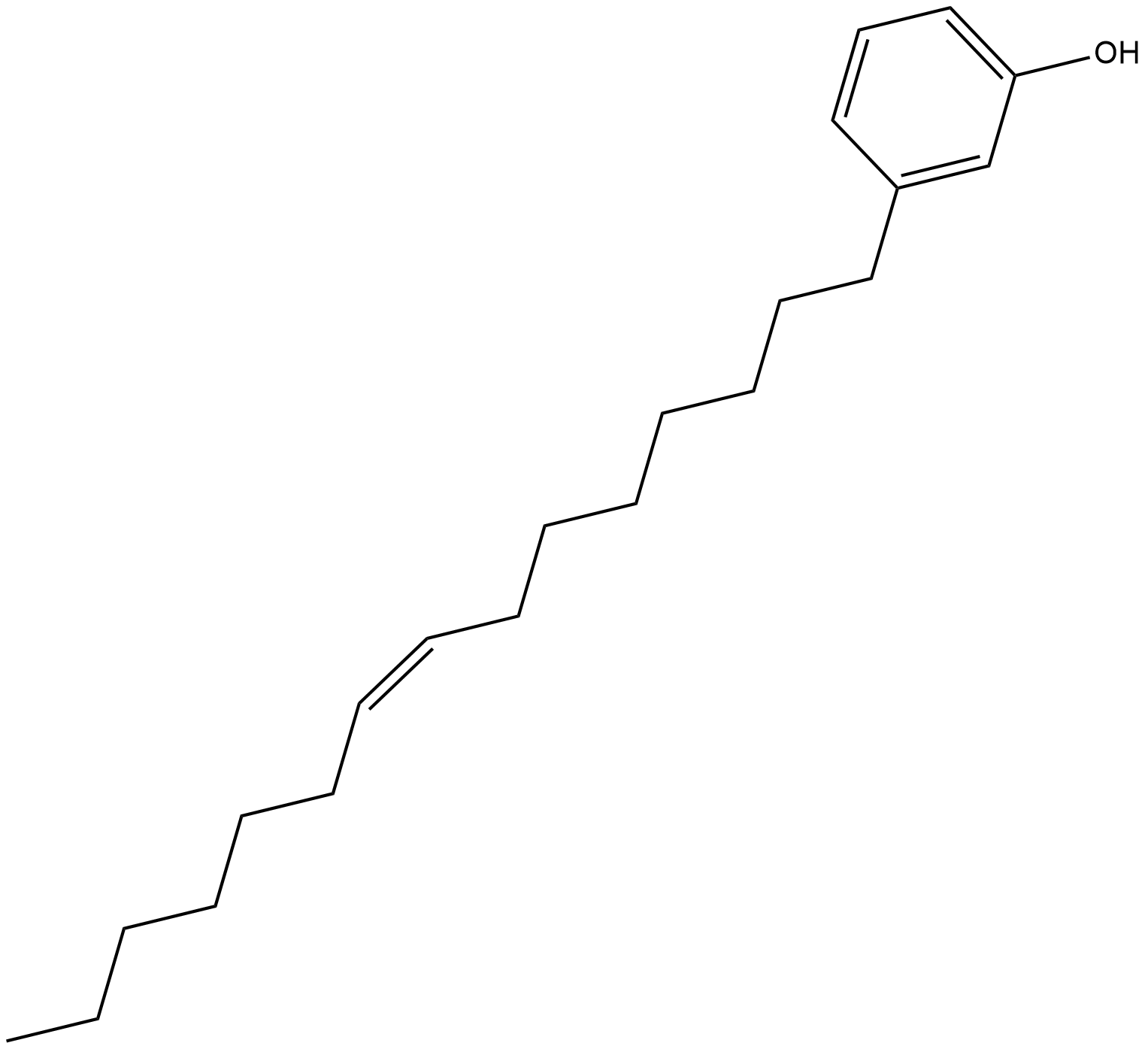
Cardanol monoene is a phenol found in cashew nut shell liquid that reversibly inhibits tyrosinase with an IC50 value of 56 uM in vitro.
-
GC15089
Carfilzomib (PR-171)
A proteasome inhibitor