KEAP1-Nrf2
The KEAP1-Nrf2 pathway is the major regulator of cytoprotective responses to chemical and/or oxidative stresses caused by reactive oxygen species (ROS) and electrophiles. Nrf2 (nuclear factor erythroid 2-related factor 2) is a transcription factor that binds together with small Maf proteins to the antioxidant response element (ARE) in the regulatory regions of cellular defense enzyme genes leading to the activation of a wide rang of cell defense processes; while KEAP1 (Kelch ECH associating protein 1), a negative repressor of Nrf2, is an adaptor protein for a Cul3-based ubiqutitin E3 ligase that binds to Nrf2 and promotes its degradation by the ubiquitin proteasome pathway.
Products for KEAP1-Nrf2
- Cat.No. Product Name Information
-
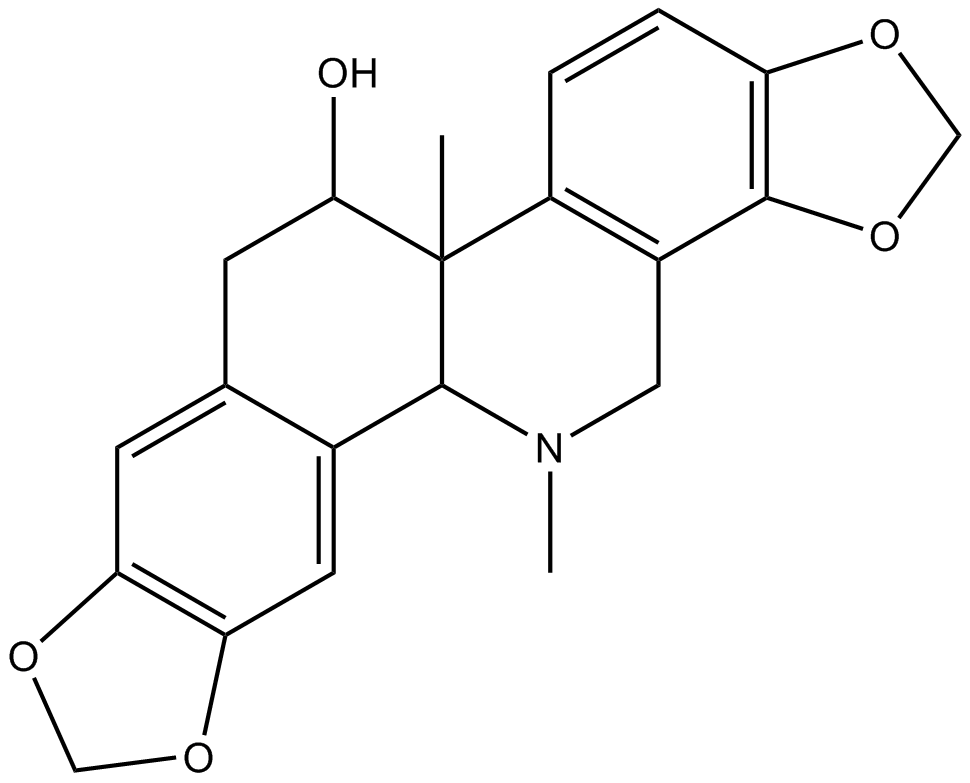
GN10654
(+)-Corynoline
-
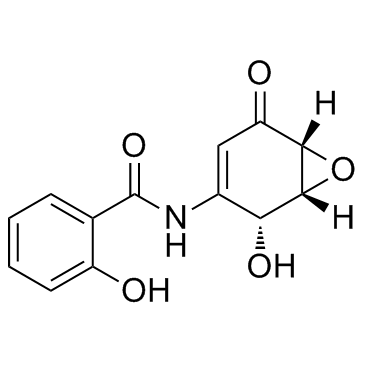
GC31691
(+)-DHMEQ
(+)-DHMEQ is an activator of antioxidant transcription factor Nrf2.
-
GC65610
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone
(R)-5-Hydroxy-1,7-diphenyl-3-heptanone is a diarylheptanoid that can be found in Alpinia officinarum.

-
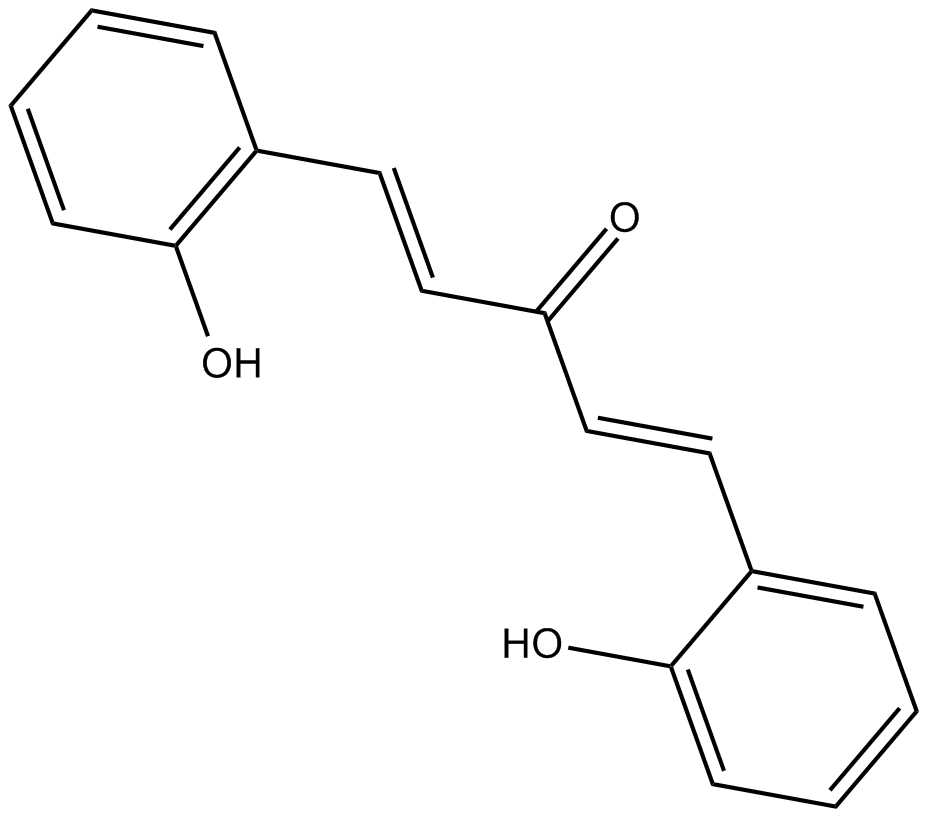
GC12545
2-HBA
indirect inducer of enzymes that catalyze detoxification reactions through the Keap1-Nrf2-ARE pathway.
-
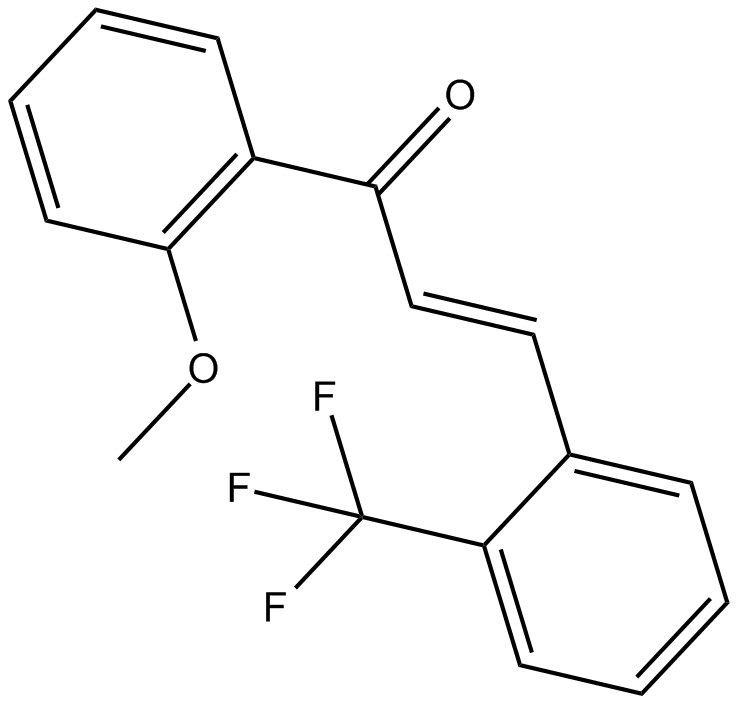
GC15355
2-Trifluoromethyl-2'-methoxychalcone
Nrf2 activator
-
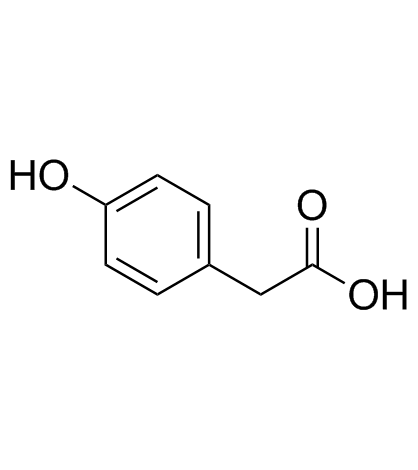
GC33815
4-Hydroxyphenylacetic acid
A phenolic acid with anti-inflammatory activity
-
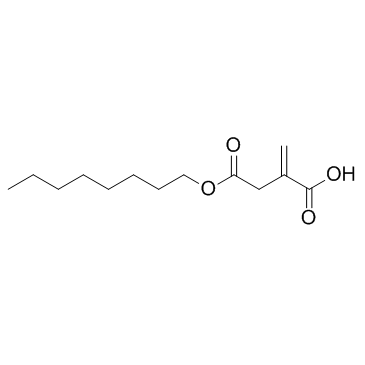
GC31648
4-Octyl Itaconate
4-Octyl Itaconate?(4-OI) is a cell-permeable itaconate derivative. Itaconate and 4-Octyl Itaconate?had similar thiol reactivity, making 4-Octyl Itaconate?a suitable itaconate surrogate to study its biological function.
-
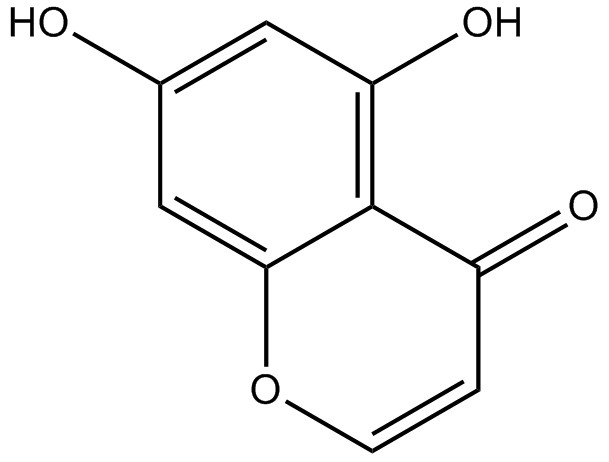
GN10629
5,7-dihydroxychromone
-
GC16120
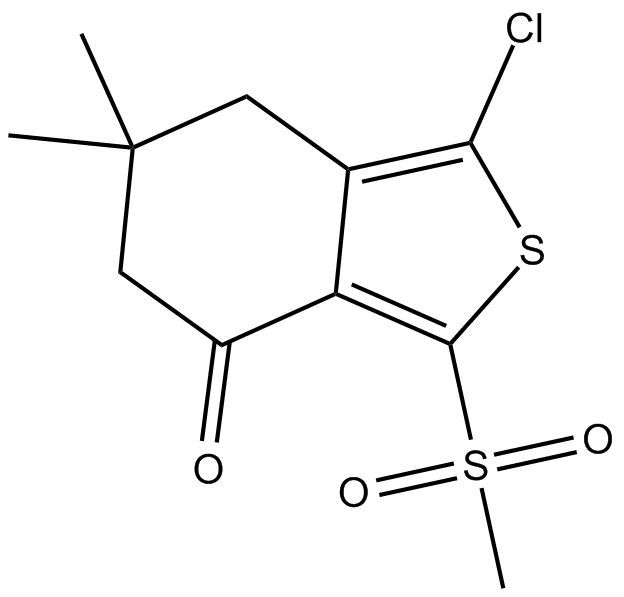
AI-3
Nrf2/Keap1 and Keap1/Cul3 interaction inhibitor
-
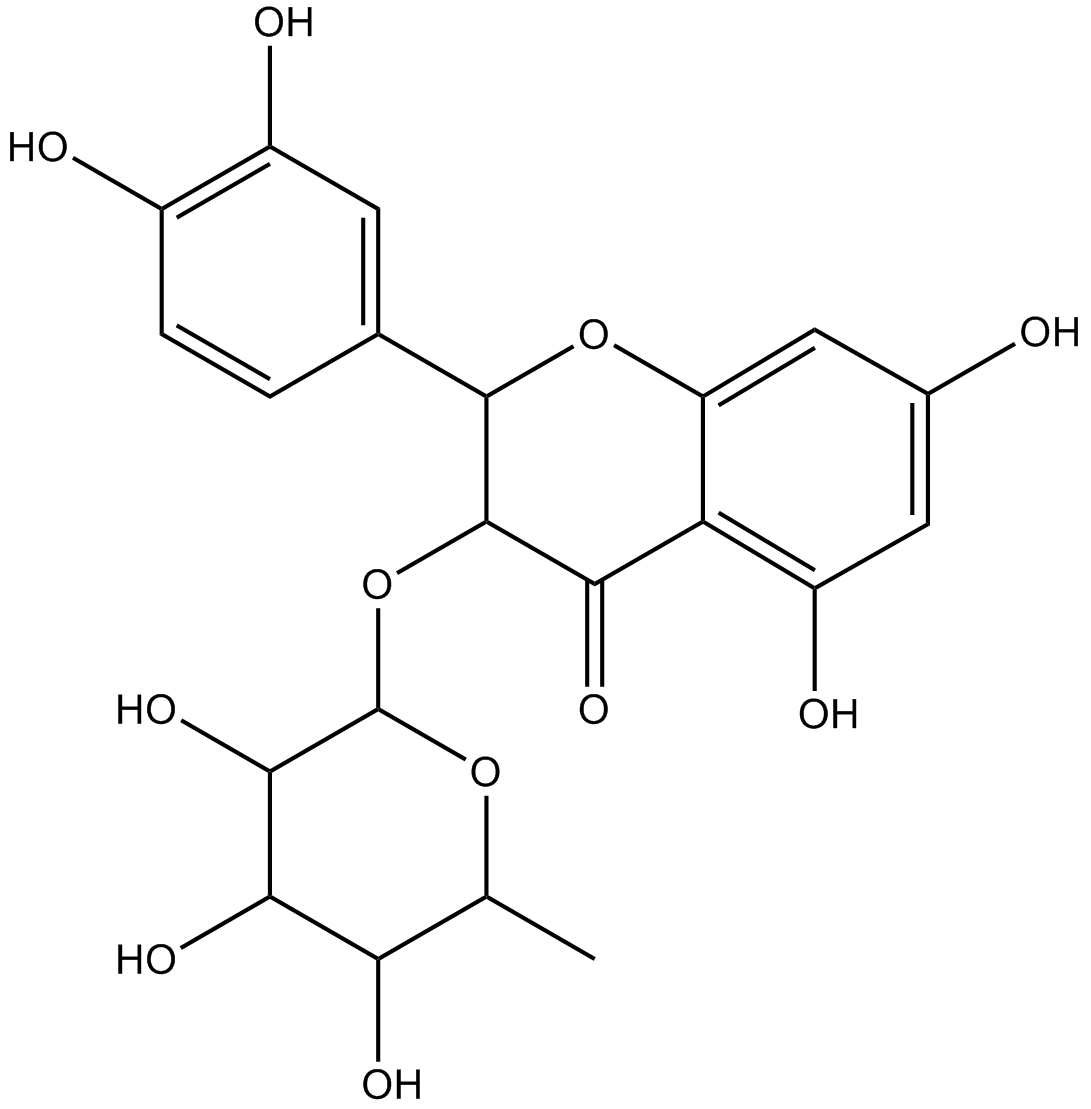
GN10415
Astilbin
-
GC15371
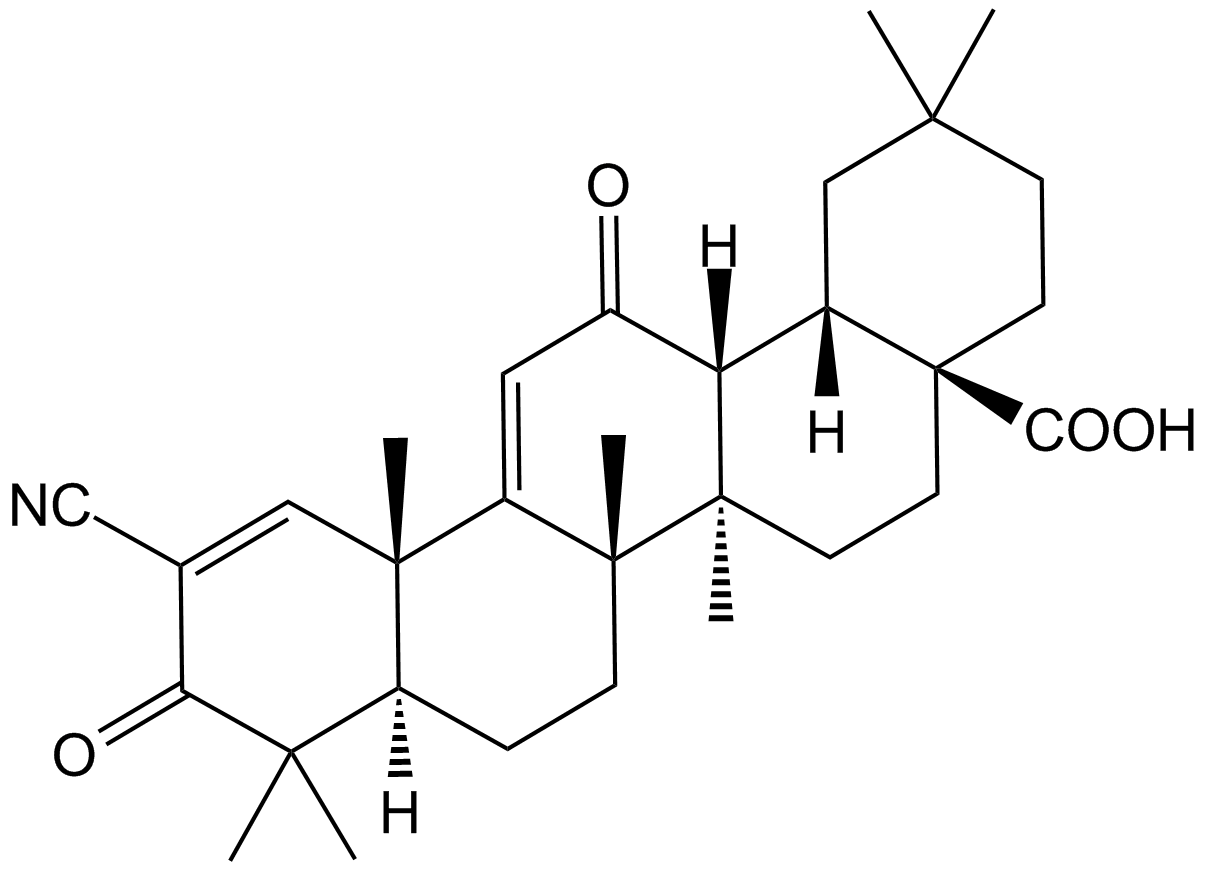
Bardoxolone
An anti-inflammatory compound that activates Nrf2/ARE signaling
-
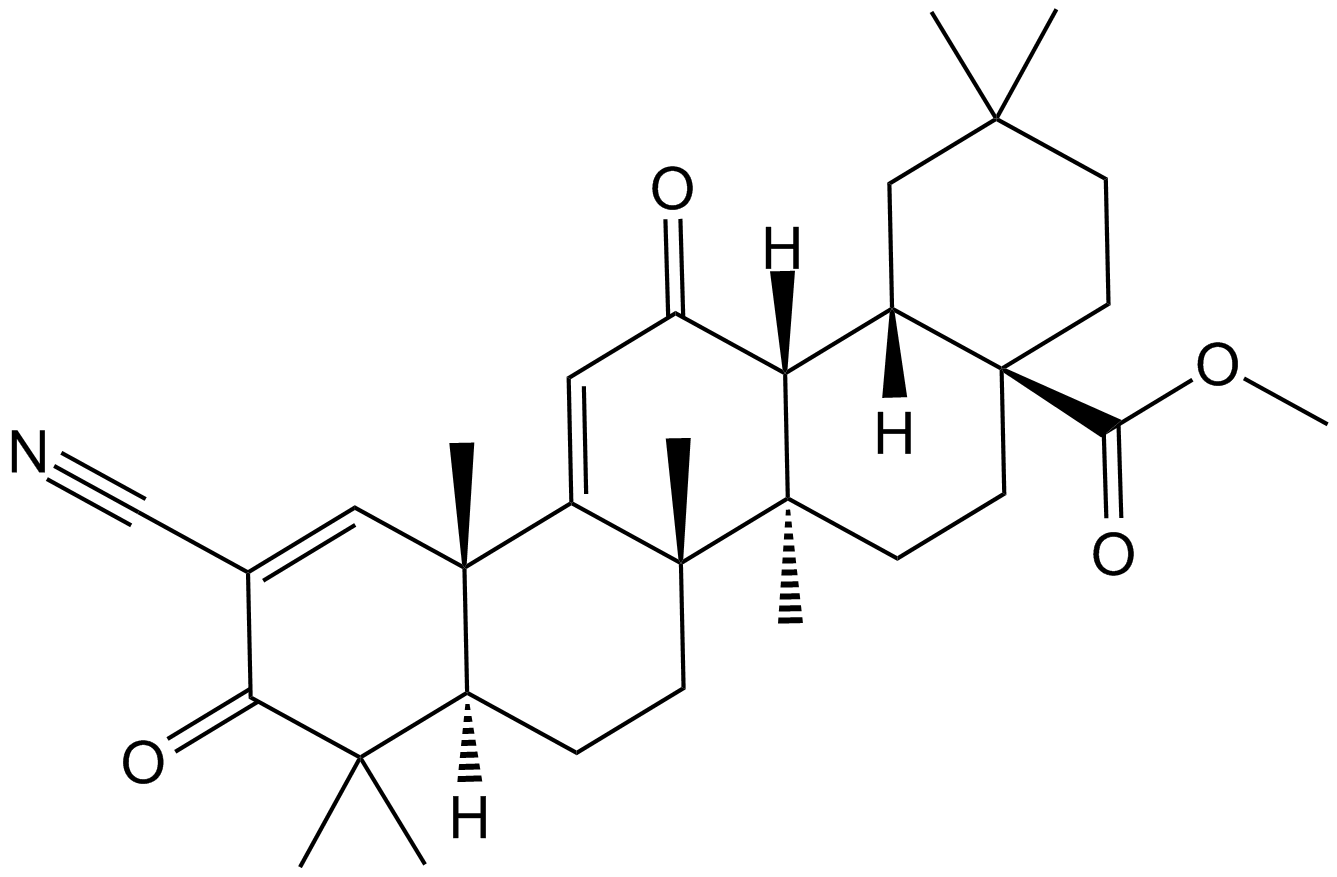
GC11572
Bardoxolone methyl
A synthetic triterpenoid with potent anticancer and antidiabetic activity
-
GC34070
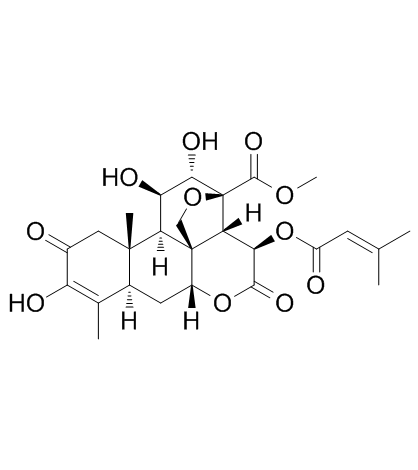
Brusatol (NSC 172924)
Brusatol (NSC 172924) (NSC172924) is a unique inhibitor of the Nrf2 pathway that sensitizes a broad spectrum of cancer cells to Cisplatin and other chemotherapeutic agents. Brusatol (NSC 172924) enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Brusatol (NSC 172924) can be developed into an adjuvant chemotherapeutic agent. Brusatol (NSC 172924) increases cellular apoptosis.
-
GC61636
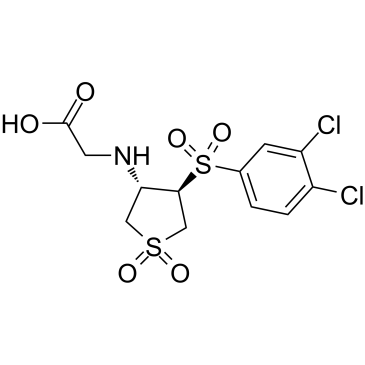
CBR-470-2
CBR-470-2, a glycine-substituted analog, can activate NRF2 signaling.
-
GC35629
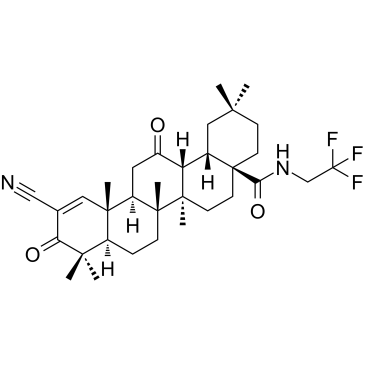
CDDO-dhTFEA
CDDO-dhTFEA (RTA dh404) is a synthetic oleanane triterpenoid compound which potently activates Nrf2 and inhibits the pro-inflammatory transcription factor NF-κB.
-
GC35630
CDDO-EA
CDDO-EA is an NF-E2 related factor 2/antioxidant response element (Nrf2/ARE) activator.

-
GC32723
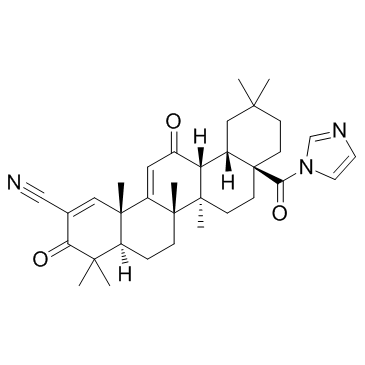
CDDO-Im (RTA-403)
CDDO-Im (RTA-403) (RTA-403) is an activator of Nrf2 and PPAR, with Kis of 232 and 344 nM for PPARα and PPARγ.
-
GC16625
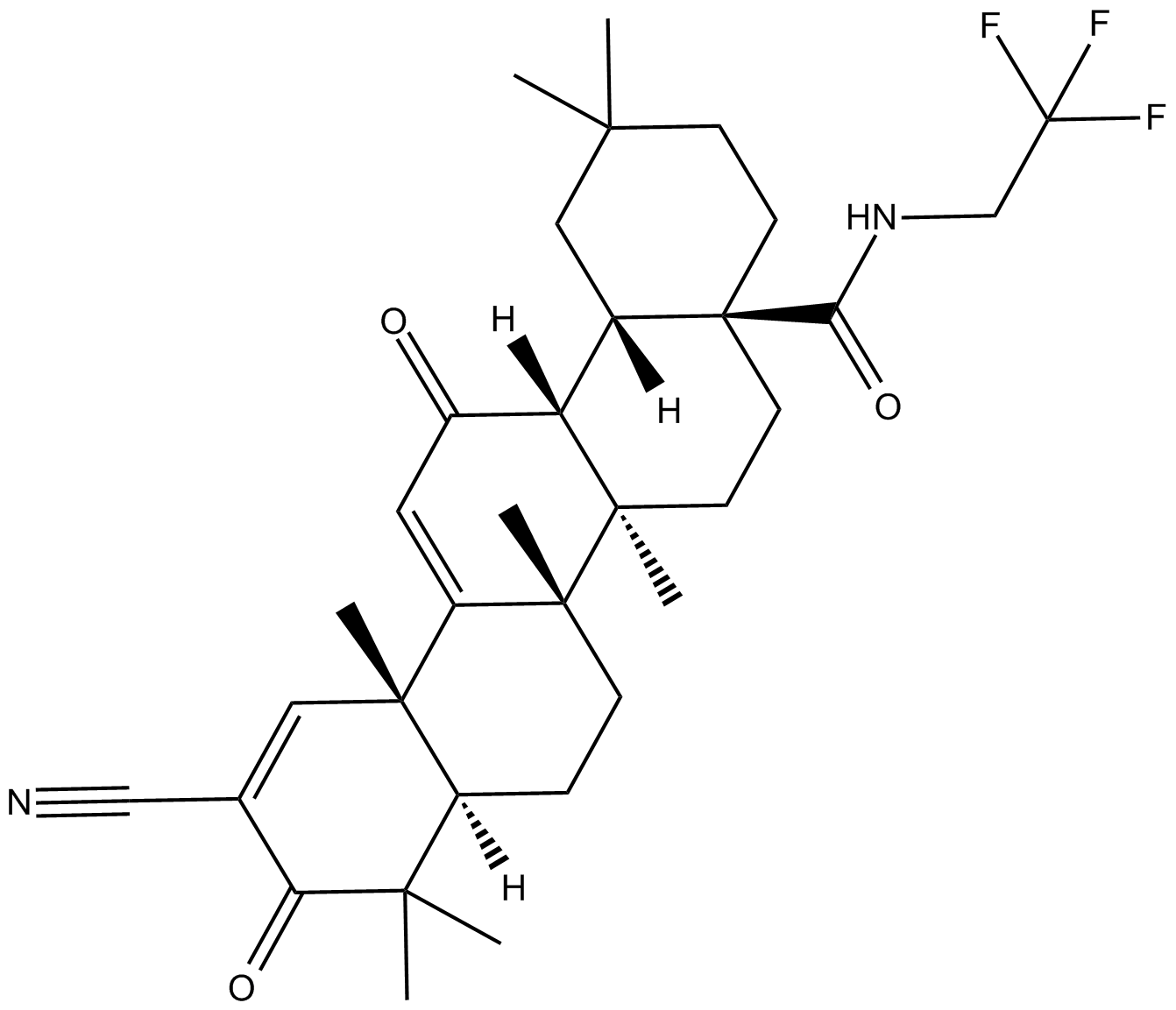
CDDO-TFEA
Nrf2 activator
-
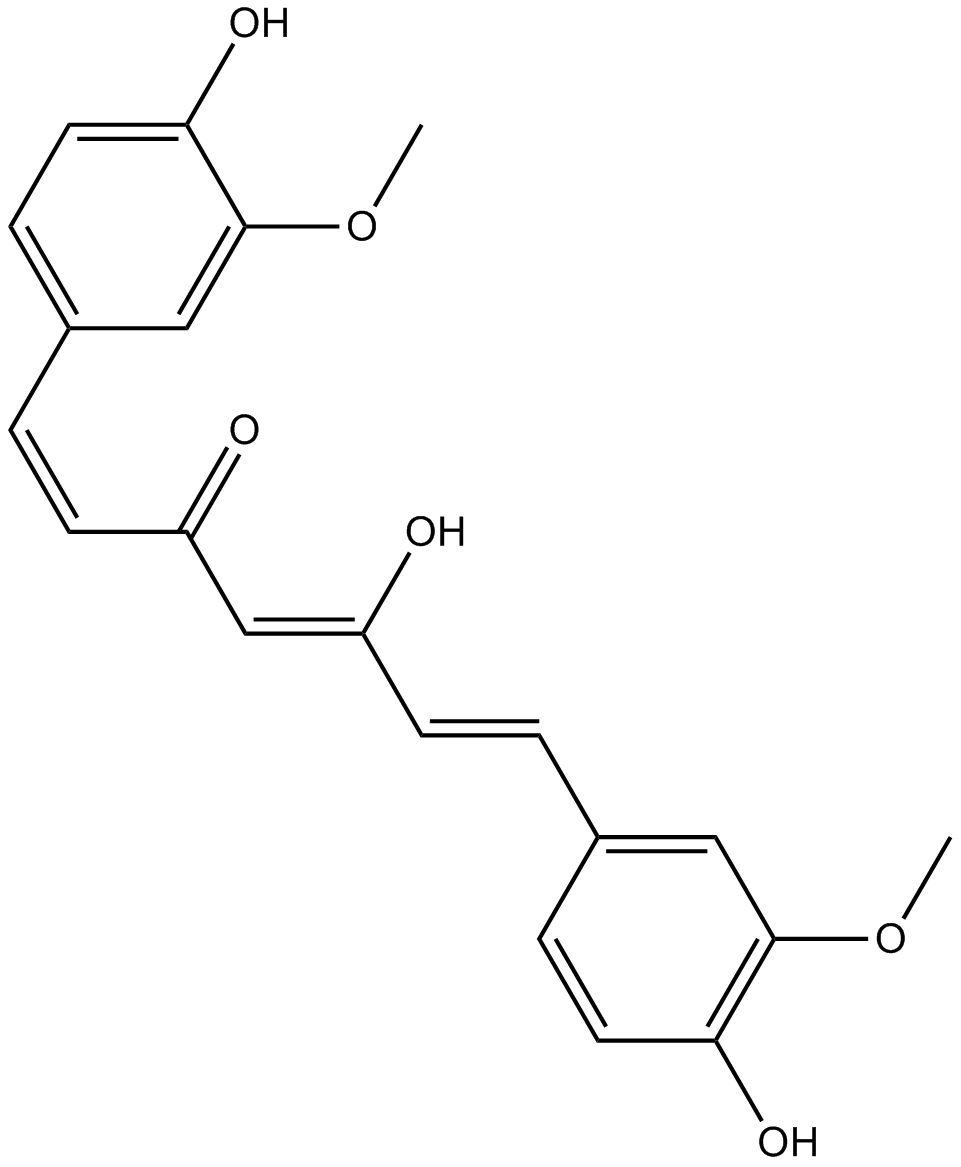
GC14787
Curcumin
A yellow pigment with diverse biological activities
-
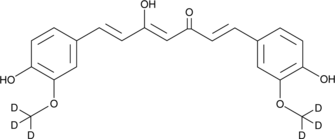
GC40226
Curcumin-d6
Curcumin-d6 is intended for use as an internal standard for the quantification of curcumin by GC- or LC-MS.
-
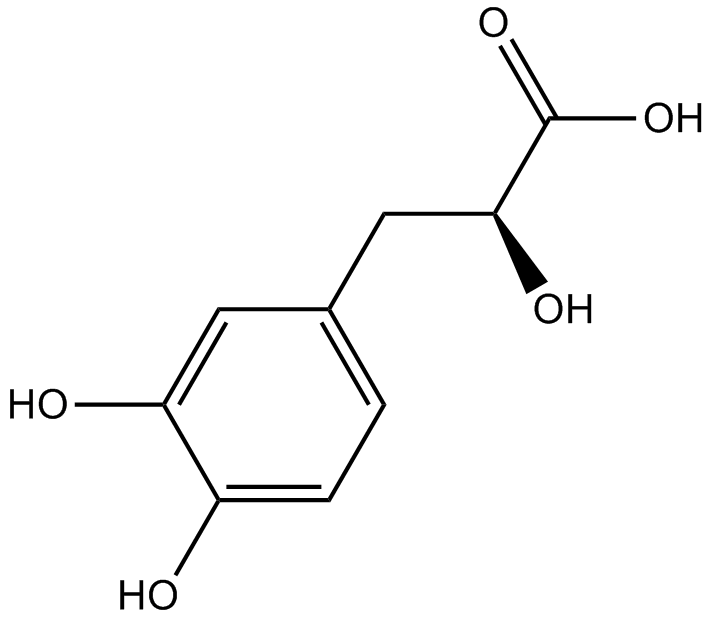
GN10318
Danshensu
-
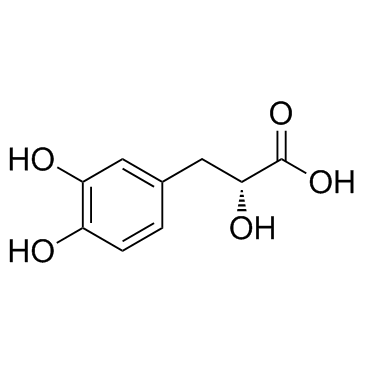
GC34010
Danshensu (Dan shen suan A)
Danshensu, an active ingredient of Salvia miltiorrhiza, shows wide cardiovascular benefit by activating Nrf2 signaling pathway.
-
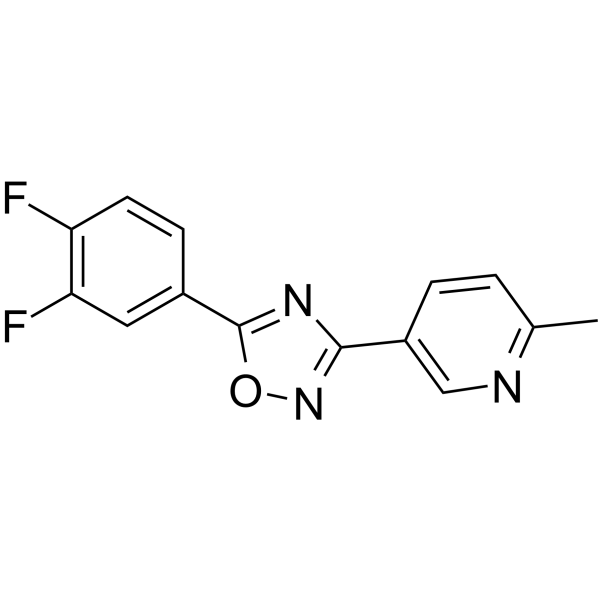
GC64971
DDO-7263
DDO-7263, a 1,2,4-Oxadiazole derivative, is a potent Nrf2-ARE activator.
-
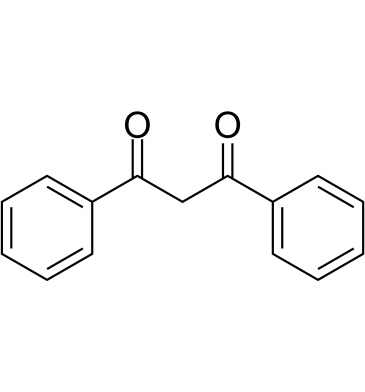
GC61419
Dibenzoylmethane
Dibenzoylmethane, a minor ingredient in licorice, activates Nrf2 and prevents various cancers and oxidative damage.
-
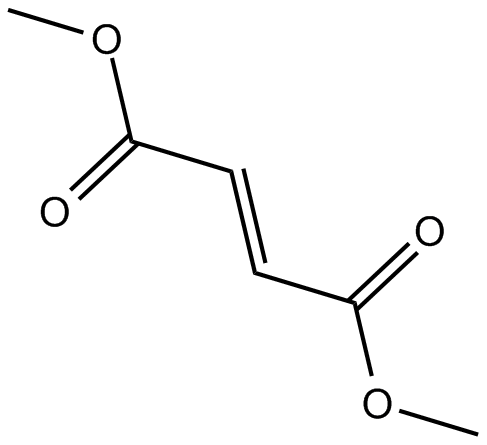
GC16590
Dimethyl Fumarate
nuclear factor (erythroid-derived)-like 2 (Nrf2) pathway activator
-
GC62936
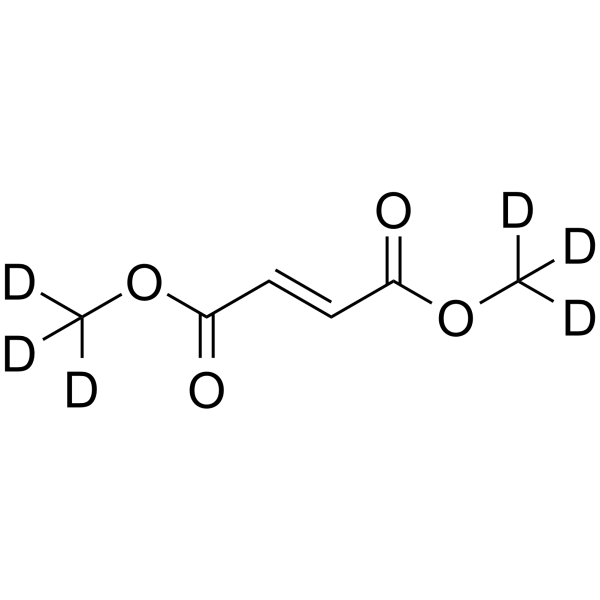
Dimethyl fumarate D6
Dimethyl fumarate D6 is a deuterium labeled Dimethyl fumarate.
-
GC25351
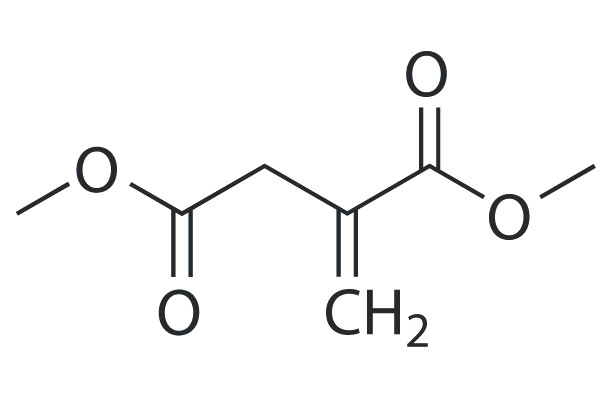
Dimethyl itaconate
Dimethyl itaconate can reprogram neurotoxic to neuroprotective primary astrocytes through the regulation of LPS-induced Nod-like receptor protein 3 (NLRP3) inflammasome and nuclear factor 2/heme oxygenase-1 (NRF2/HO-1) pathways.
-
GN10531
Dimethylfraxetin
-
GN10470
Eriodictyol
-
GC62957
Eriodictyol-7-O-glucoside
Eriodictyol-7-O-glucoside (Eriodictyol 7-O-β-D-glucoside), a flavonoid, is a potent free radical scavenger.
-
GC15605
Ezetimibe
Cholesterol transport inhibitor
-
GC60159
Ezetimibe ketone
Ezetimibe ketone (EZM-K) is a phase-I metabolite of Ezetimibe.
-
GC66354
Ezetimibe-d4-1
Ezetimibe-d4 is deuterium labeled Ezetimibe. Ezetimibe (SCH 58235) is a potent cholesterol absorption inhibitor. Ezetimibe is a Niemann-Pick C1-like1 (NPC1L1) inhibitor, and is a potent Nrf2 activator.
-
GN10741
Garcinone D
-
GN10038
Ginsenoside Rh3
-
GC33092
Hesperin
An isothiocyanate with diverse biological activities
-
GC11302
Hinokitiol
A tropolone with diverse biological activities

-
GC66004
K67
K67 specifically inhibits the interaction between Keap1 and S349-phosphorylated p62. K67 prevents p-p62 from blocking the binding of Keap1 and Nrf2. K67 effectively inhibits the proliferation of HCC cultures with high cellular S351-phosphorylated p62 by restoring the ubiquitination and degradation of Nrf2 driven by Keap1.
-
GC36390
Keap1-Nrf2-IN-1
Keap1-Nrf2-IN-1 is a Keap1 (Kelch-like ECH-associated protein 1)-Nrf2 (nuclear factor erythroid 2-related factor 2) protein-protein interaction inhibitor, and with an IC50 of 43 nM for Keap1 protein.
-
GC31321
KI696
KI696 is a high affinity probe that disrupts the Keap1/NRF2 interaction.
-
GC34642
KI696 isomer
KI696 isomer is the less active isomer of KI696.
-
GC44006
Kinsenoside
Kinsenoside is a glycoside originally isolated from A.
-
GN10491
Mangiferin
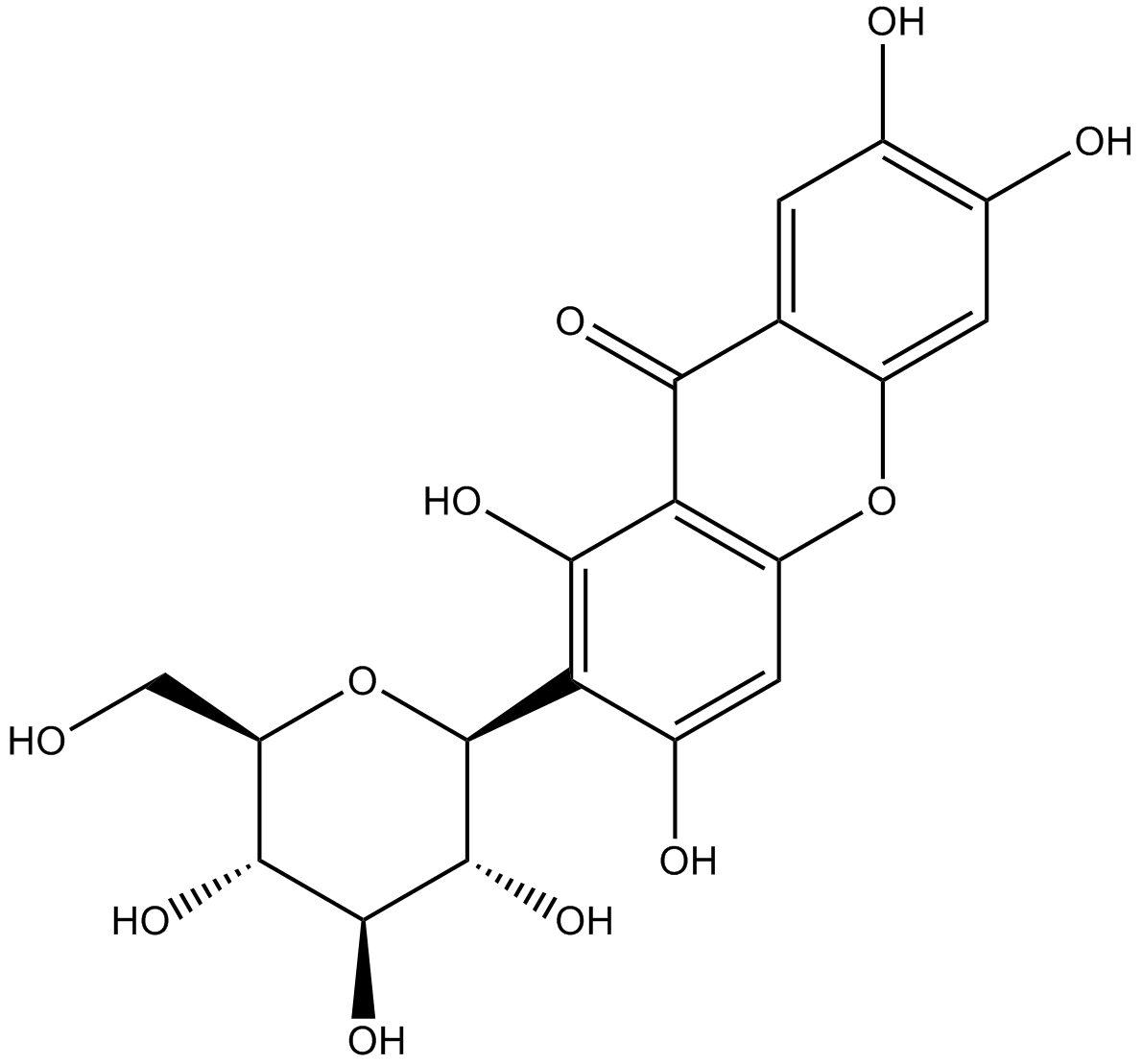
-
GC31783
Methyl 3,4-dihydroxybenzoate (Protocatechuic acid methyl ester)
Methyl 3,4-dihydroxybenzoate (Protocatechuic acid methyl ester) (Protocatechuic acid methyl ester; Methyl protocatechuate) is a major metabolite of antioxidant polyphenols found in green tea.

-
GC38819
ML334
An inhibitor of the Nrf2-Keap1 protein-protein interaction
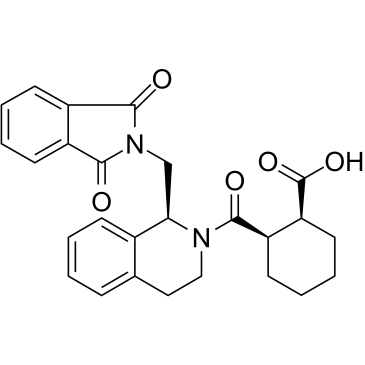
-
GC19254
ML385
ML385 is a specific nuclear factor erythroid 2-related factor 2 (NRF2) inhibitor.
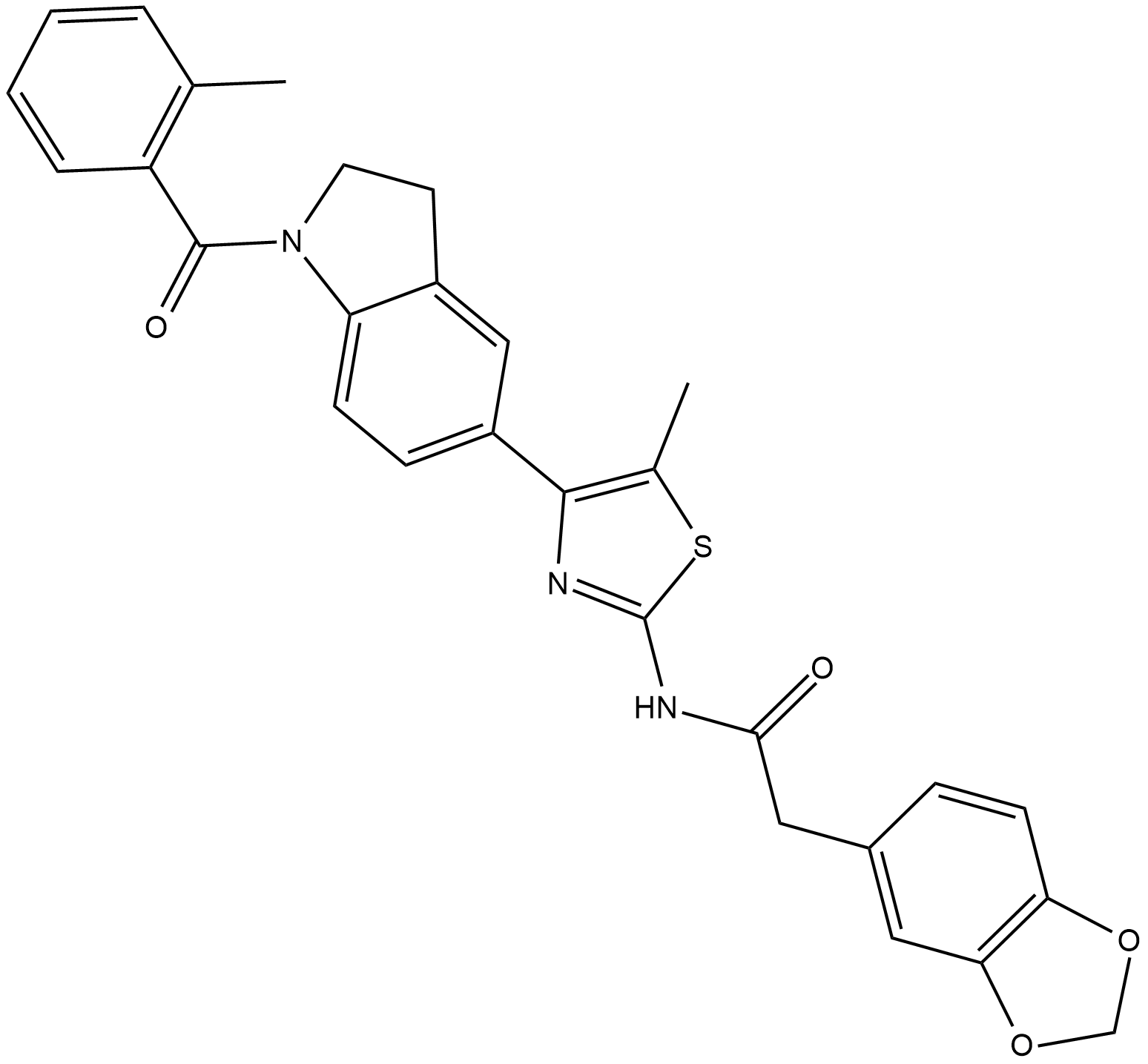
-
GC13058
NK 252
Nrf2 activator
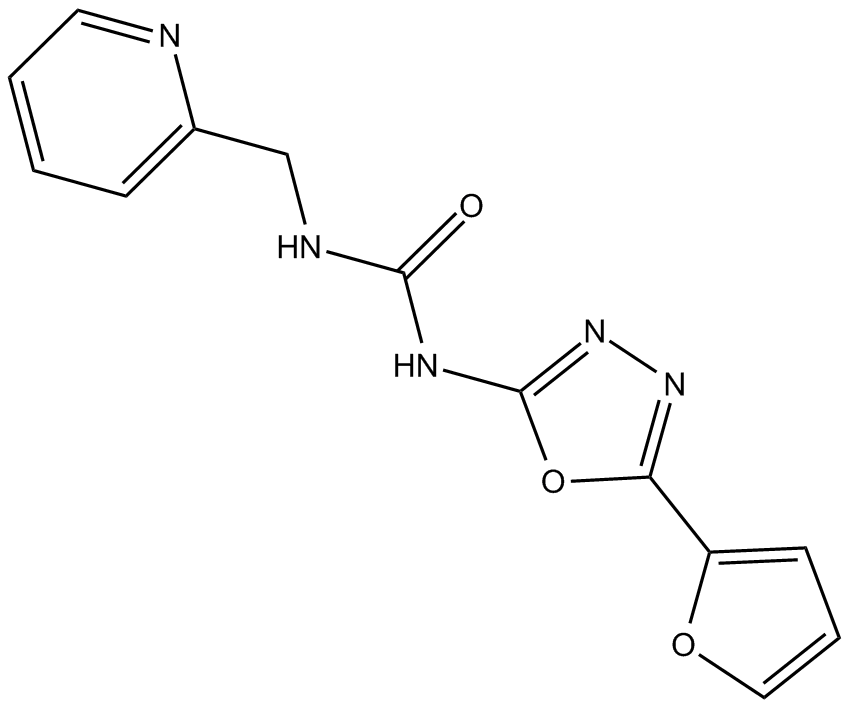
-
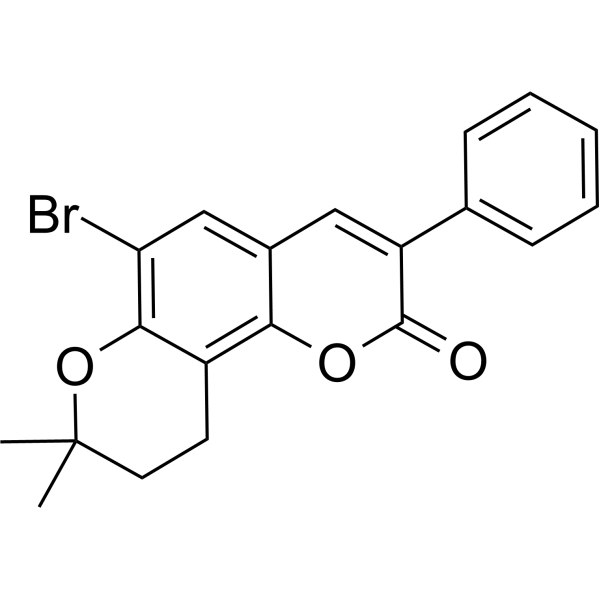
GC69590
Nrf2 activator-2
Nrf2 activator-2 (compound O15) is a derivative of sulforaphane, an effective Nrf2 activator with an EC50 of 2.9 μM in 293 T cells. Nrf2 activator-2 effectively inhibits the interaction between Keap1 and Nrf2, thereby showing an activating effect on Nrf2. Nrf2 activator-2 significantly reduces the level of ubiquitinated Nrf2 in cells.
-
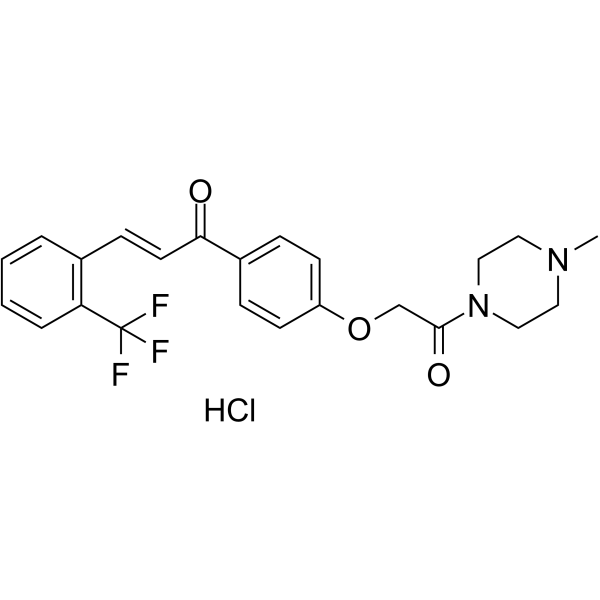
GC66054
Nrf2 activator-4
Nrf2 activator-4 (Compound 20a) is a highly potent, orally active Nrf2 activator with an EC50 of 0.63 μM. Nrf2 activator-4 suppresses reactive oxygen species against oxidative stress in microglia. Nrf2 activator-4 effectively recovers the learning and memory impairment in a scopolamine-induced mouse model.
-
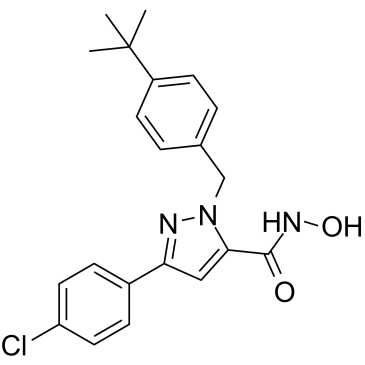
GC36773
Nrf2-IN-1
Nrf2-IN-1 is an inhibitor of nuclear factor-erythroid 2-related factor 2 (Nrf2). Nrf2-IN-1 is developed for the research of acute myeloid leukemia (AML).
-
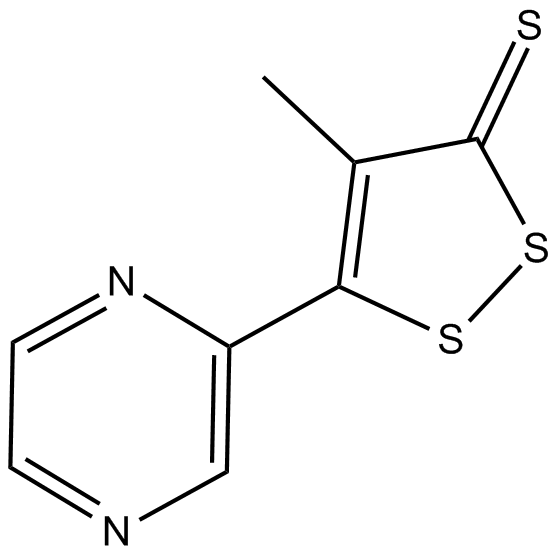
GC12821
Oltipraz
Nrf2 activator;An antischistosomal agent
-
GC13693
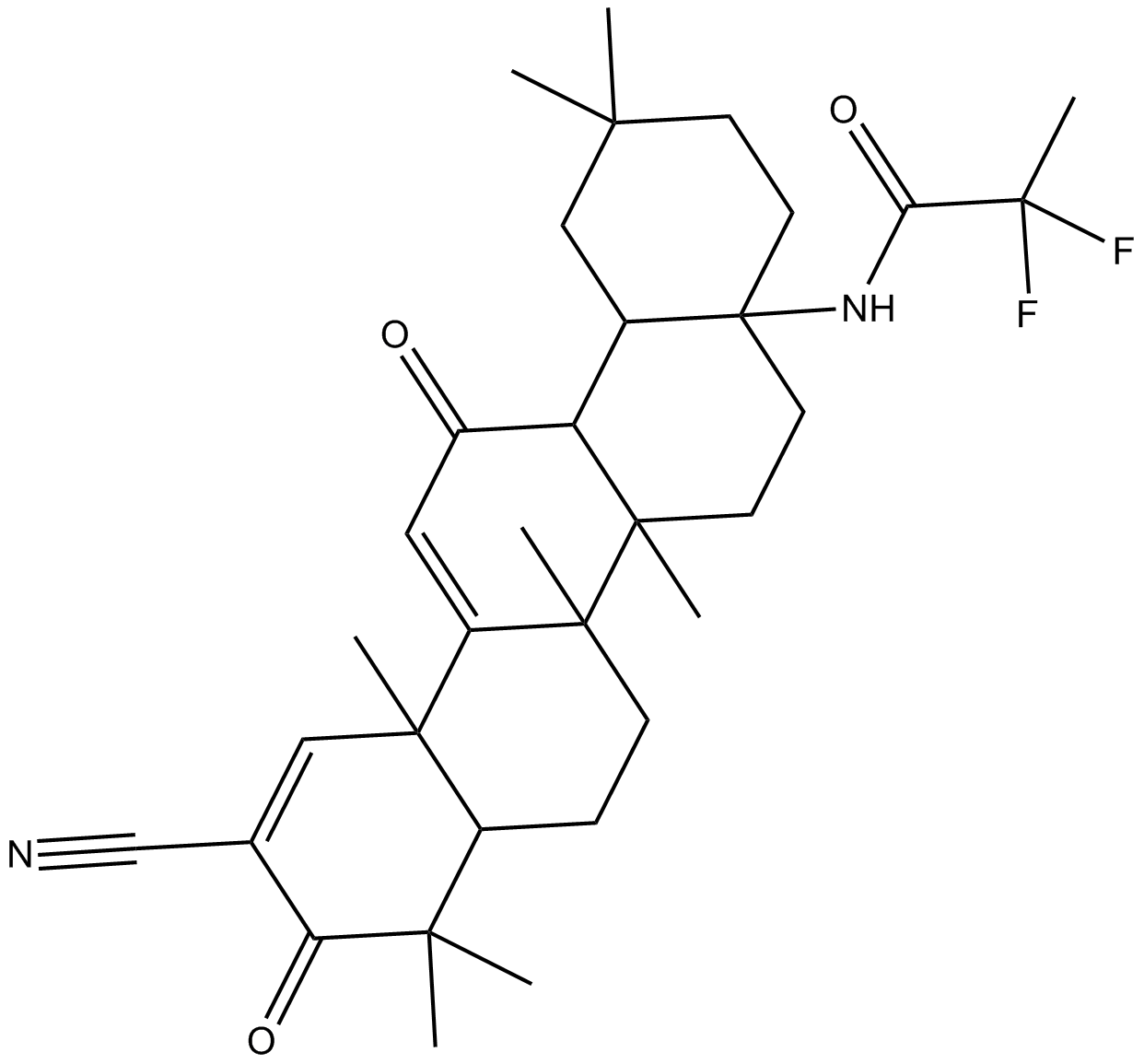
Omaveloxolone (RTA-408)
Omaveloxolone (RTA-408) (RTA 408) is an antioxidant inflammation modulator (AIM), which activates Nrf2 and suppresses nitric oxide (NO).
-
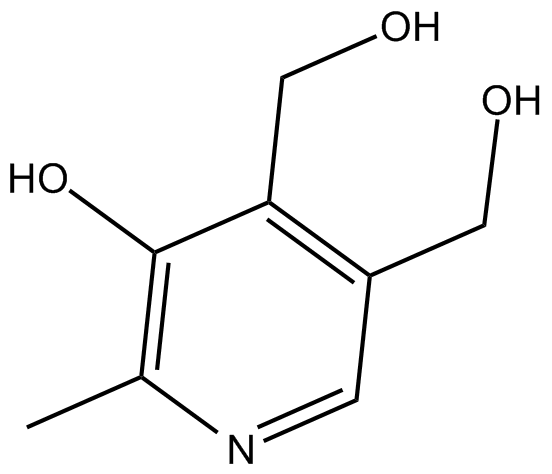
GC15457
Pyridoxine
A 4-methanol form of vitamin B6
-
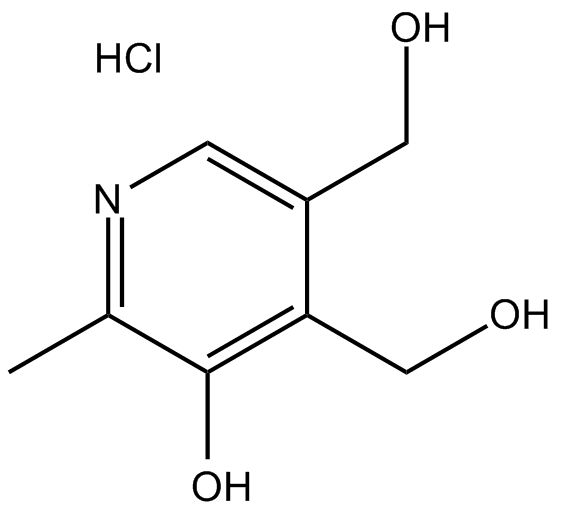
GC17393
Pyridoxine HCl
Pyridoxine HCl (Pyridoxol; Vitamin B6) is a pyridine derivative.
-
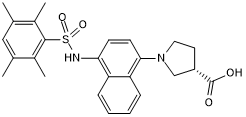
GC50242
RA 839
Nrf2 activator; inhibits Nrf2/Keap1 interaction
-
GC63933
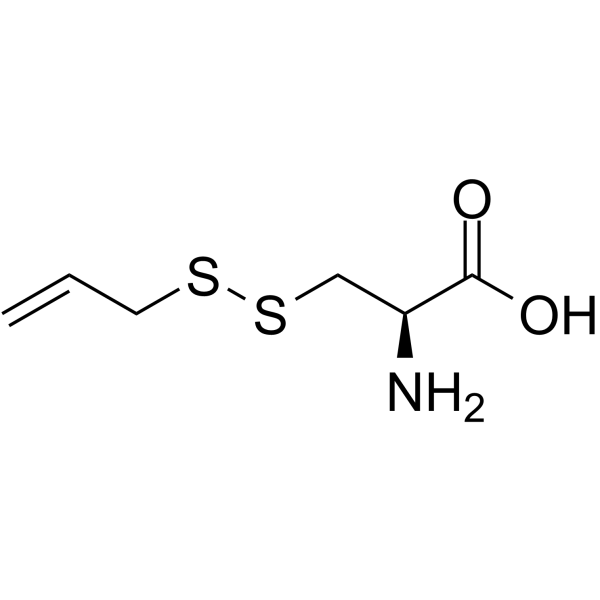
S-Allylmercaptocysteine
S-allylmercaptocysteine, an organic sulfur compound extracted from garlic, has anti-inflammatory and anti-oxidative effects for various pulmonary diseases.
-
GC14016
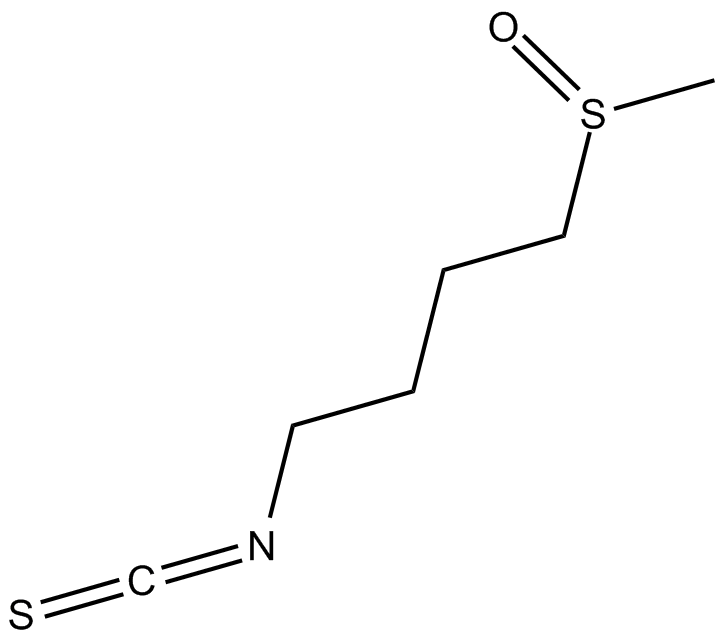
Sulforaphane
Sulforaphane (SFN) known as [1-isothiocyanato-4-(methylsulfinyl)butane].
-
GC14352
TAT 14
Nrf2 activator

-
GC34057
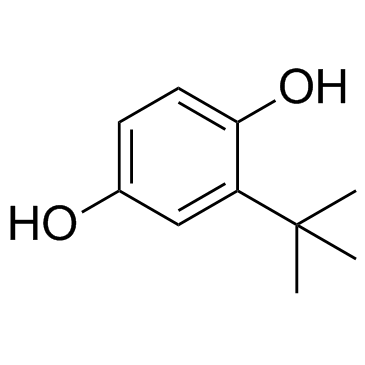
TBHQ (tert-Butylhydroquinone)
TBHQ (tert-Butylhydroquinone) (tert-Butylhydroquinone) is a widely used Nrf2 activator, protects against Doxorubicin (DOX)-induced cardiotoxicity through activation of Nrf2. TBHQ (tert-Butylhydroquinone) (tert-Butylhydroquinone) is also an ERK activator; rescues Dehydrocorydaline (DHC)-induced cell proliferation inhibitionin melanoma.
-
GC61338
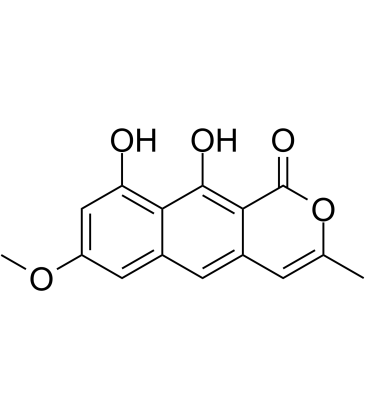
Toralactone
Toralactone, isolated from Cassia obtusifolia, mediates hepatoprotection via an Nrf2-dependent anti-oxidative mechanism.