Atherosclerosis
Products for Atherosclerosis
- Cat.No. Product Name Information
-
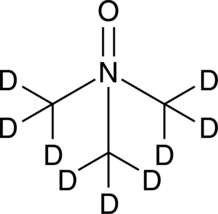
GC48283
α-Linolenic Acid-d14
An internal standard for the quantification of αLinolenic acid
-
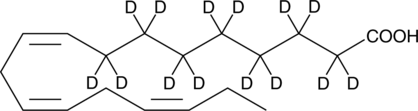
GC40467
(±)11-HETE
(±)11-HETE is one of the six monohydroxy fatty acids produced by the non-enzymatic oxidation of arachidonic acid.
-
GC41649
(±)13-HODE cholesteryl ester
(±)13-HODE cholesteryl ester was originally extracted from atherosclerotic lesions and shown to be produced by Cu2+-catalyzed oxidation of LDL.

-
GC41666
(±)9-HODE cholesteryl ester
(±)9-HODE cholesteryl ester was originally extracted from atherosclerotic lesions and shown to be produced by Cu2+-catalyzed oxidation of LDL.
-
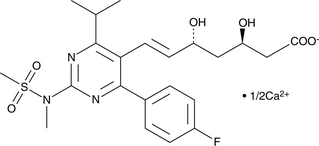
GC49690
(3R,5R)-Rosuvastatin (calcium salt)
A potential impurity found in bulk preparations of rosuvastatin
-
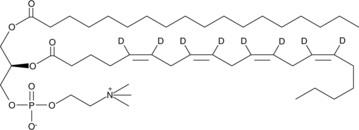
GC46496
1-Stearoyl-2-Arachidonoyl-d8-sn-glycero-3-PC
An internal standard for the quantification of 1-stearoyl-2-arachidonoyl-sn-glycero-3-PC
-
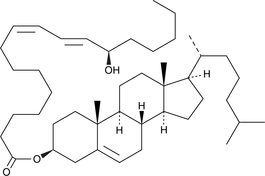
GC41893
13(R)-HODE cholesteryl ester
13(R)-HODE cholesteryl ester was originally extracted from atherosclerotic lesions.
-
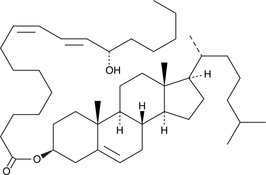
GC41895
13(S)-HODE cholesteryl ester
13(S)-HODE cholesteryl ester was originally extracted from atherosclerotic lesions.
-
GC41897
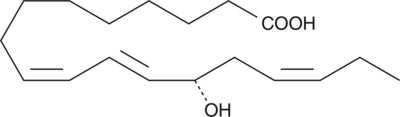
13(S)-HOTrE
13(S)-HOTrE is the 15-lipoxygenase (15-LO) product of linolenic acid.
-
GC49671
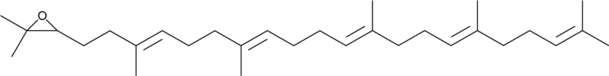
2,3-Oxidosqualene
An intermediate in the biosynthesis of sterols
-
GC41614
24-dehydro Cholesterol
24-dehydro Cholesterol is a molecule similar to cholesterol.

-
GC46243
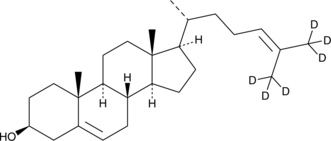
24-dehydro Cholesterol-d6
An internal standard for the quantification of 24-dehydro cholesterol
-
GC46528
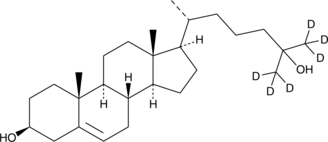
25-hydroxy Cholesterol-d6
An internal standard for the quantification of 25hydroxy cholesterol
-
GC42411
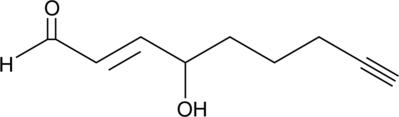
4-hydroxy Nonenal Alkyne
4-hydroxy Nonenal (4-HNE) is a major aldehyde produced during the lipid peroxidation of ω-6 polyunsaturated fatty acids, such as arachidonic acid and linoleic acid.
-
GC40053
5α,6α-epoxy Cholestanol
An oxysterol and a metabolite of cholesterol produced by oxidation

-
GC45357
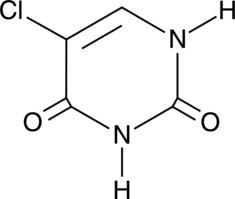
5-Chlorouracil
-
GC49629
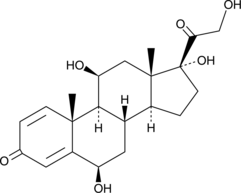
6β-hydroxy Prednisolone
A metabolite of prednisolone
-
GC40202
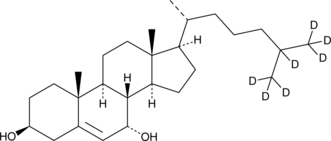
7α-hydroxy Cholesterol-d7
7α-hydroxy Cholesterol-d7 is intended for use as an internal standard for the quantification of 7α-hydroxy cholesterol by GC- or LC-MS.
-
GC46241
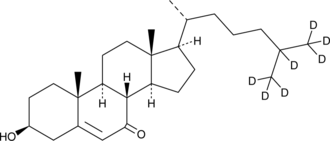
7-keto Cholesterol-d7
7-keto Cholesterol is a bioactive sterol and a major oxysterol component of oxidized LDL
-
GC42634
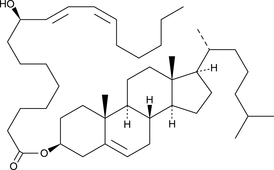
9(R)-HODE cholesteryl ester
9(R)-HODE cholesteryl ester was originally extracted from atherosclerotic lesions.
-
GC19460
9(S)-HODE
9(S)-HODE is produced by the lipoxygenation of linoleic acid in both plants and animals.

-
GC42635
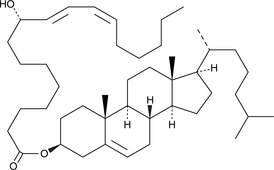
9(S)-HODE cholesteryl ester
9(S)-HODE cholesteryl ester was originally extracted from atherosclerotic lesions.
-
GC49784
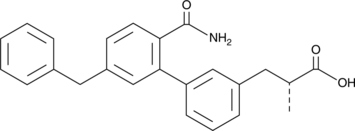
AZD 2716
An sPLA2 inhibitor
-
GC42890
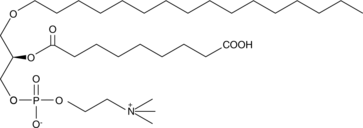
Azelaoyl PAF
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which promote the differentiation of monocytes via the nuclear receptor PPARγ.
-
GC46912
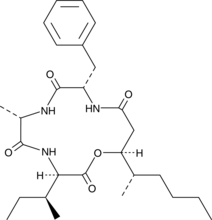
Beauveriolide III
A cyclodepsipeptide inhibitor of lipid droplet formation
-
GC42924
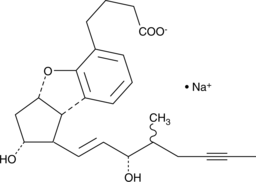
Beraprost (sodium salt)
Beraprost is an analog of prostacyclin in which the unstable enol-ether has been replaced by a benzofuran ether function.
-
GC42994
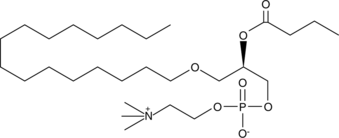
Butanoyl PAF
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which promote the differentiation of monocytes and activate polymorphonuclear leukocytes.
-
GC43136
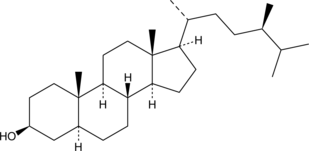
Campestanol
Campestanol is a phytosterol found in vegetables, fruits, nuts, and seeds.
-
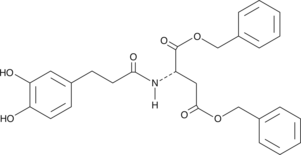
GC43160
CAY10485
Acyl-coenzyme A: cholesterol acyltransferase-1 and -2 (ACAT-1 and ACAT-2) catalyze the formation of cholesterol esters from cholesterol and long chain fatty acyl-coenzyme A, and may play a role in the development of atherosclerosis.
-
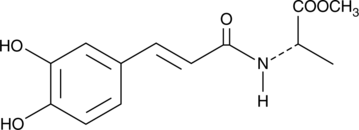
GC41636
CAY10487
The early stage of atherosclerosis is characterized by the aggregation of foam cells, so called a fatty streak, in the inner arterial wall.
-
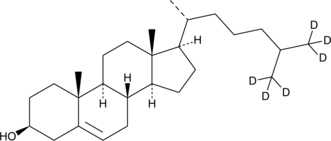
GC40203
Cholesterol-d6
Cholesterol-d6 is intended for use as an internal standard for the quantification of cholesterol by GC- or LC-MS.
-
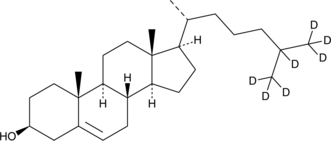
GC47085
Cholesterol-d7
An internal standard for the quantification of cholesterol
-
GC19433
Cholesteryl Heptadecanoate
Cholesteryl heptadecanoate is a cholesterol ester (CE) formed by the condensation of cholesterol with heptadecanoic acid, a C-17 saturated fatty acid that does not occur in any natural animal or vegetable fat at high concentrations.

-
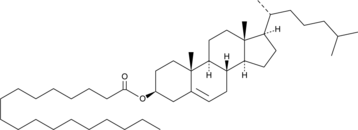
GC43263
Cholesteryl Stearate
Cholesteryl stearate is a cholesterol ester.
-
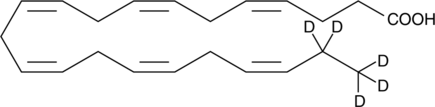
GC45439
Docosahexaenoic Acid-d5 MaxSpec• Standard
Docosahexaenoic Acid-d5 MaxSpec• Standard (DHA-d5) is the deuterium labeled Docosahexaenoic Acid.
-
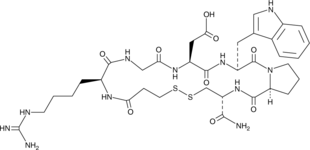
GC52193
Eptifibatide
Eptifibatide is a cyclic heptapeptide, acts as a competitive antagonist for the activated platelet glycoprotein IIb/IIIa receptor, with anti-platelet activity.
-
GC33760
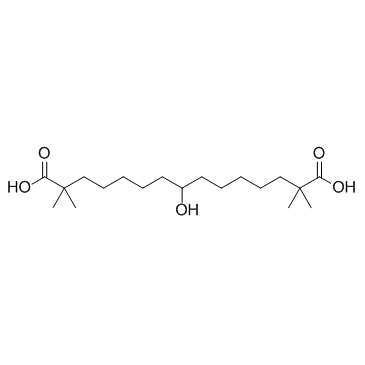
ETC-1002 (ESP-55016)
ETC-1002 (ESP-55016) (ETC-1002) is an ATP-citrate lyase (ACL) inhibitor.
-
GC47397
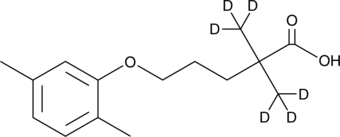
Gemfibrozil-d6
An internal standard for the quantification of gemfibrozil
-
GC48744
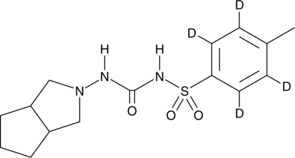
Gliclazide-d4
An internal standard for the quantification of gliclazide
-
GC18788
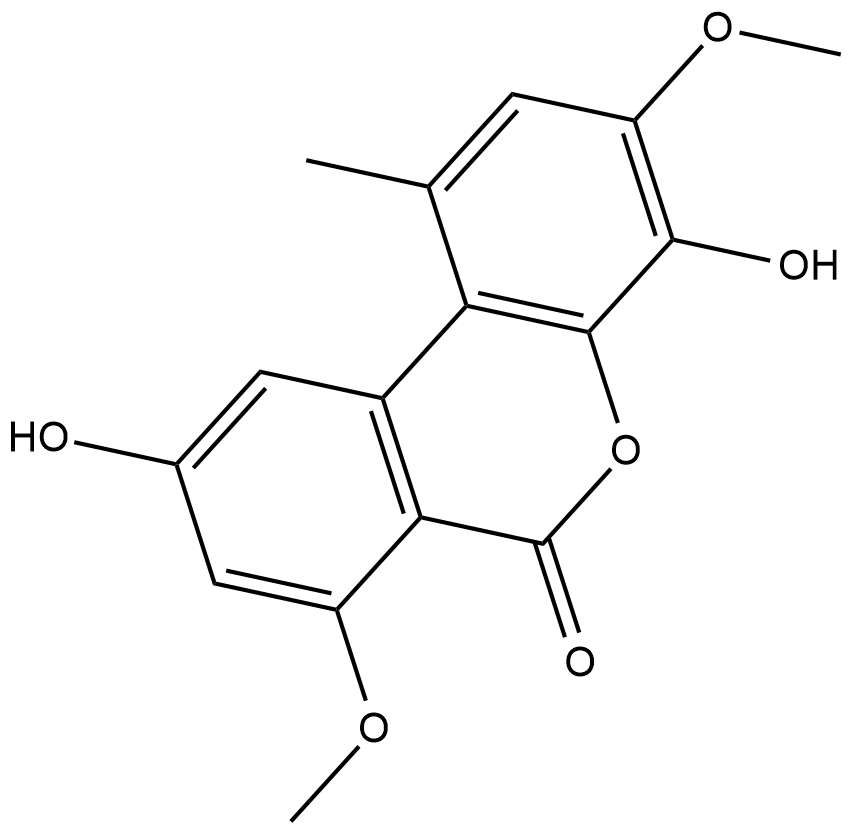
Graphislactone A
Graphislactone A is an antioxidant produced by Cephalosporium IFB-E001 and M.
-
GC43803
HA-1077 (hydrochloride)
Fasudil (HA-1077; AT877) dihydrochloride is a nonspecific RhoA/ROCK inhibitor and also has inhibitory effect on protein kinases, with an Ki of 0.33 μM for ROCK1, IC50s of 0.158 μM and 4.58 μM, 12.30 μM, 1.650 μM for ROCK2 and PKA, PKC, PKG, respectively. HA-1077 (hydrochloride) is also a potent Ca2+ channel antagonist and vasodilator.
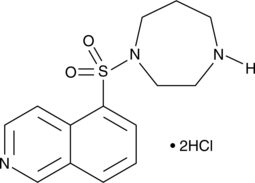
-
GC45469
Hexacosanoic Acid-d4
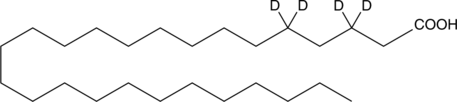
-
GC49496
Homocitrulline-d3
An internal standard for the quantification of homocitrulline
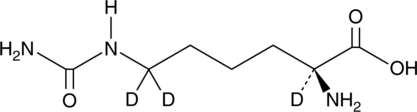
-
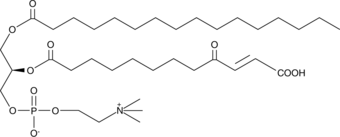
GC43997
KDdiA-PC
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which are cytotoxic and pro-atherogenic.
-
GC44011
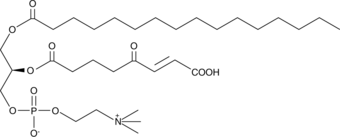
KOdiA-PC
Oxidized low-density lipoprotien (oxLDL) particles contain low molecular weight species which are cytotoxic and pro-atherogenic.
-
GC52497
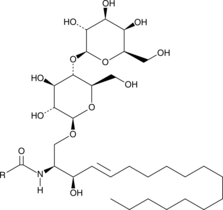
Lactosylceramide (porcine RBC)
A sphingolipid
-
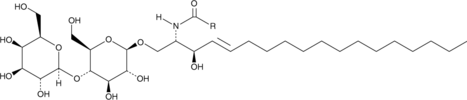
GC40142
Lactosylceramides (bovine buttermilk)
Lactosylceramide (LacCer) is an endogenous bioactive sphingolipid.
-
GC52281
Losartan Azide
A potential impurity in commercial preparations of losartan

-
GC44093
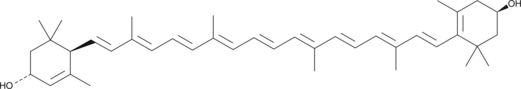
Lutein
Lutein is a natural yellow carotenoid, which can be found in plants, egg yolks, and in the human retina.
-
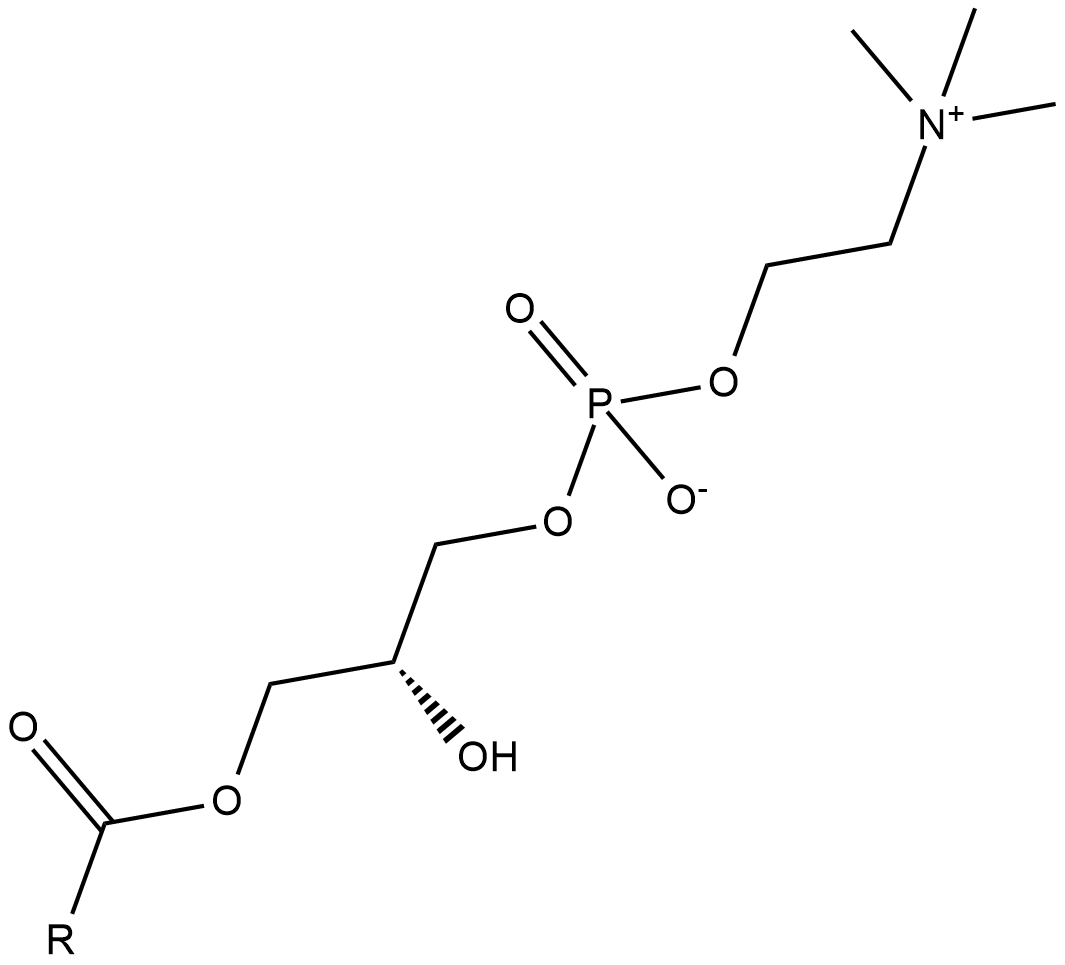
GC18241
Lysophosphatidylcholines
Lysophosphatidylcholines are produced by hydrolysis of the fatty acid of phosphatidylcholine at either the sn-1 or sn-2 position by phospholipase A2 (PLA2) or by lecithin-cholesterol acyltranferase (LCAT), which transfers the fatty acid to cholesterol.
-
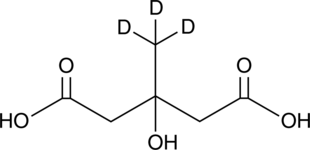
GC49486
Meglutol-d3
An internal standard for the quantification of meglutol
-
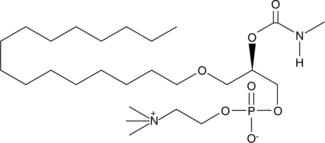
GC44180
Methylcarbamyl PAF C-16
Methylcarbamyl PAF C-16 is a stable analog of PAF C-16 with a half-life greater than 100 minutes in platelet poor plasma due to its resistance to degradation by PAF-AH.
-
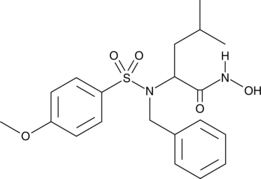
GC40624
MMP-3 Inhibitor VIII
Matrix metalloproteinases (MMPs) belong to a family of proteases that play a crucial role in tissue remodeling and repair by degrading extracellular matrix proteins to enable cell migration.
-
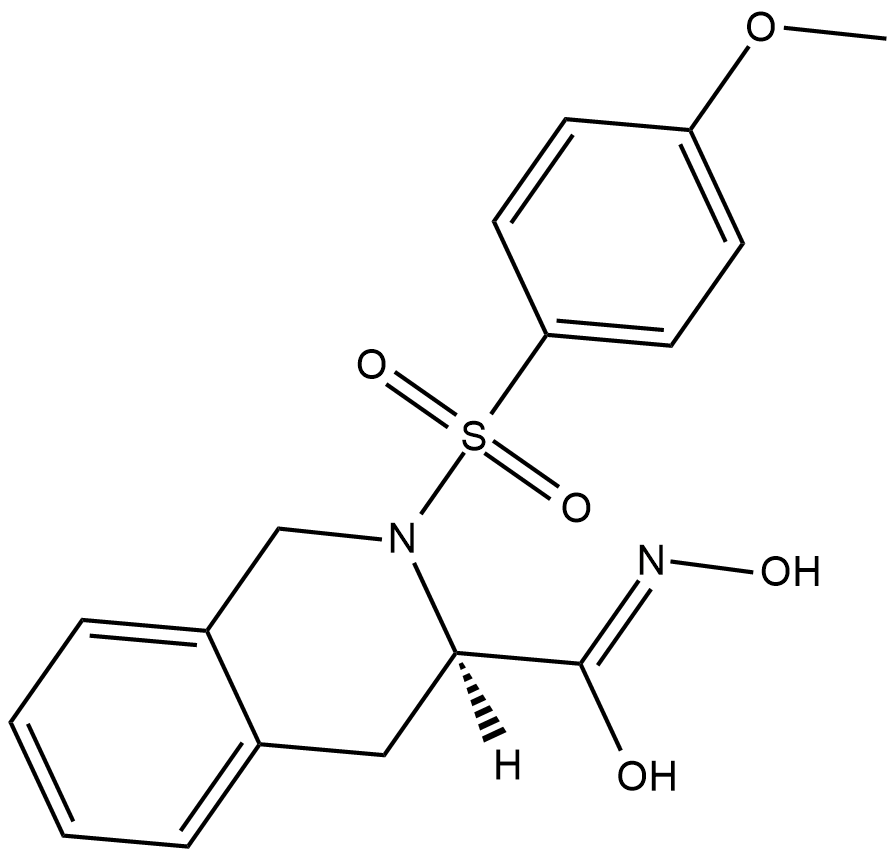
GC18616
MMP-8 Inhibitor I
MMP-8 Inhibitor I is a selective inhibitor of the neutrophil collagenase matrix metalloproteinase-8 (MMP-8) with an IC50 value of 4 nM.
-
GC20172
N-Isopropylphthalimide

-
GC44518
Osteocalcin (1-49) (human) (trifluoroacetate salt)
Osteocalcin (1-49) is a non-collagenous peptide that is secreted by osteoblasts and odontoblasts and comprises 1-2% of the total protein in bone.

-
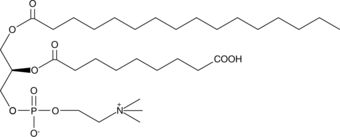
GC44573
PAz-PC
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which are cytotoxic and pro-atherogenic.
-
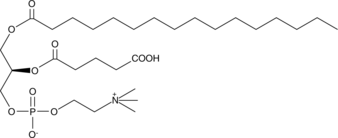
GC44616
PGPC
PGPC is an oxidized phospholipid that can be formed under conditions of oxidative stress.
-
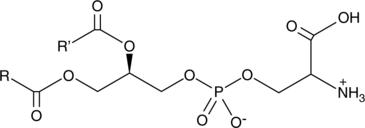
GC44633
Phosphatidylserines (bovine)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
-
GC18522
Phosphatidylserines (sodium salt)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
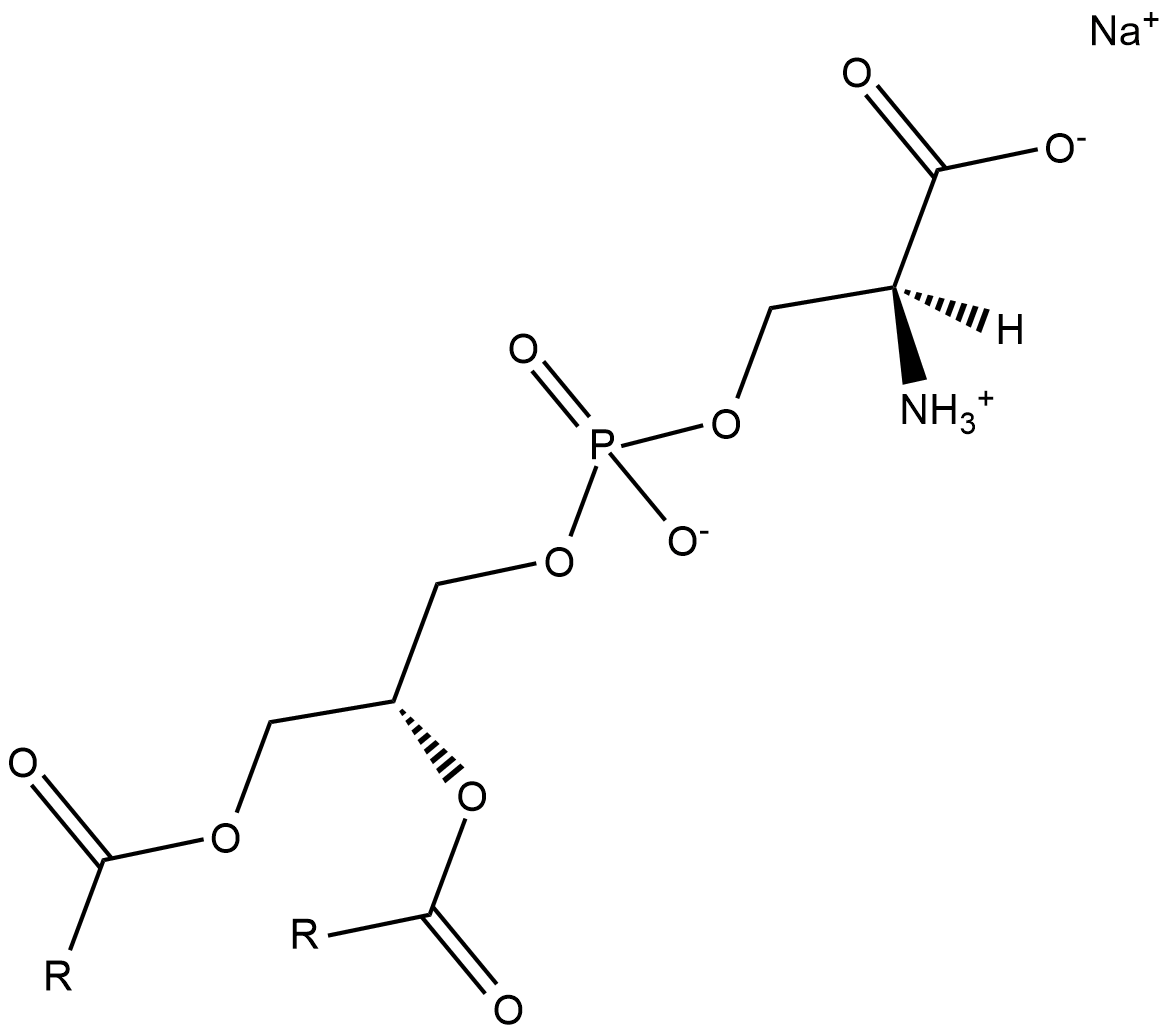
-
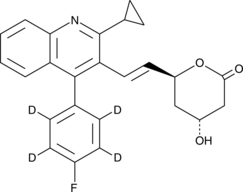
GC49340
Pitavastatin lactone-d4
An internal standard for the quantification of pitavastatin lactone
-
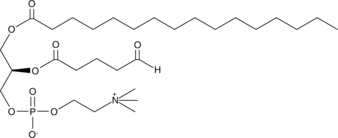
GC44669
POV-PC
Oxidized low-density lipoprotien (oxLDL) particles contain low molecular weight species which are cytotoxic and pro-atherogenic.
-
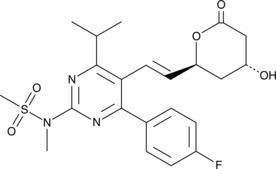
GC49107
Rosuvastatin lactone
The lactone form of rosuvastatin
-
GC48060
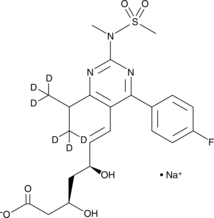
Rosuvastatin-d6 (sodium salt)
An internal standard for the quantification of rosuvastatin
-
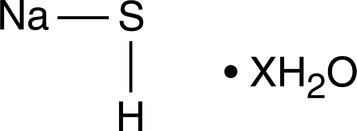
GC44910
Sodium Hydrogen Sulfide (hydrate)
Hydrogen sulfide (H2S) is, like nitric oxide, an important gaseous mediator that has significant effects on the immunological, neurological, cardiovascular and pulmonary systems of mammals.
-
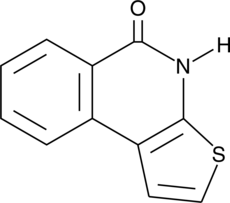
GC45057
TIQ-A
Poly(ADP-ribose) polymerase 1 (PARP1) is a critical DNA repair enzyme involved in DNA single-strand break repair via the base excision repair pathway.
-
GC48206
Trimethylamine-d9 N-oxide
An internal standard for the quantification of trimethylamine N-oxide