G1
Products for G1
- Cat.No. Product Name Information
-
GC40675
2-deoxy-Artemisinin
2-deoxy-Artemisinin is an inactive metabolite of the antimalarial agent artemisinin.
-
GC45354
4β-Hydroxywithanolide E
A withanolide with anti-inflammatory and anticancer activities
-
GC42586
6α-hydroxy Paclitaxel
6α-hydroxy Paclitaxel is a primary metabolite of the anticancer compound paclitaxel, produced by the action of the cytochrome P450 isoform CYP2C8.
-
GC49393
all-trans-13,14-Dihydroretinol
A metabolite of all-trans retinoic acid

-
GC18539
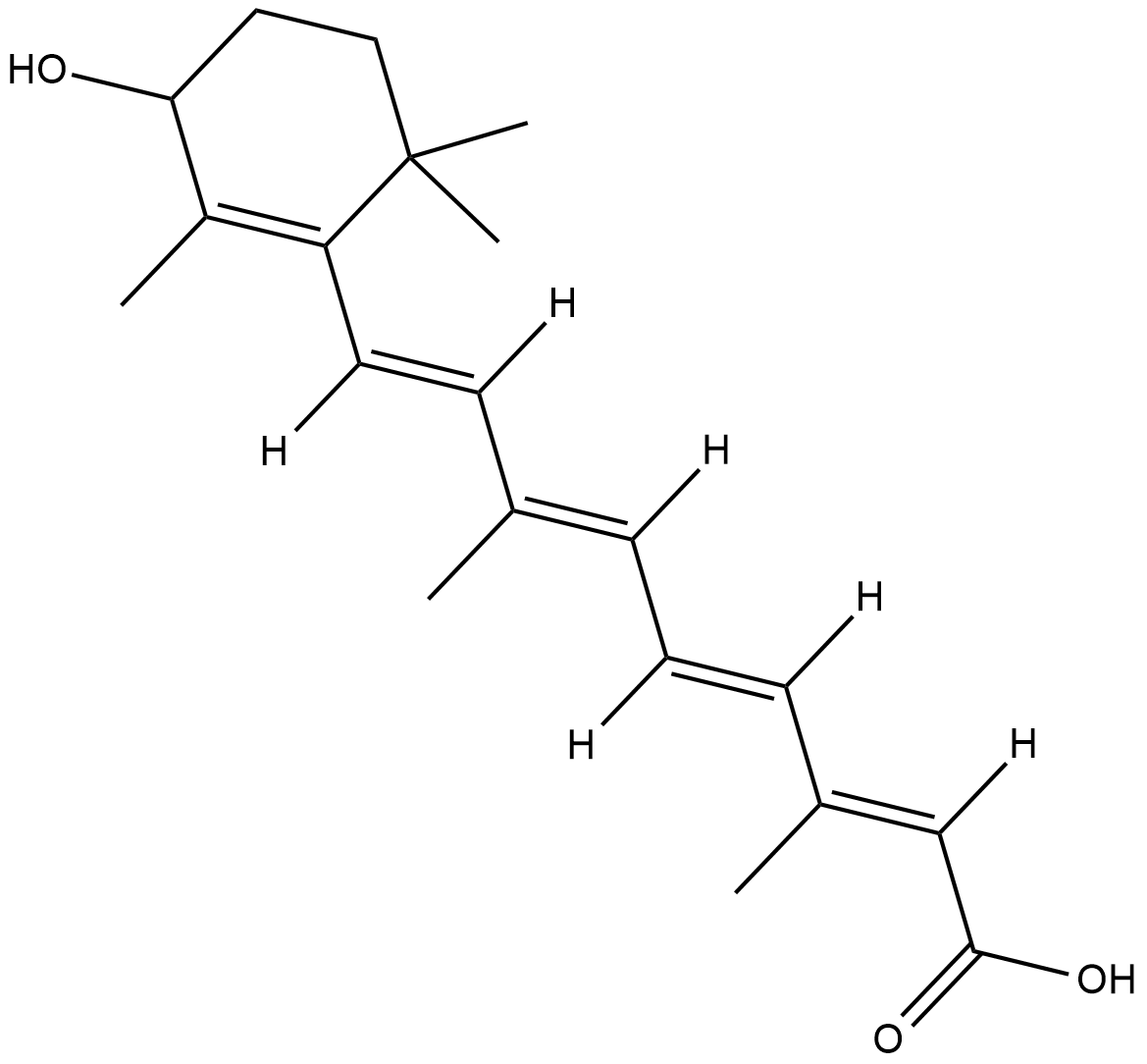
all-trans-4-hydroxy Retinoic Acid
all-trans-4-hydroxy Retinoic acid is a metabolite of all-trans retinoic acid formed by the cytochrome P450 (CYP) isoforms CYP26A1, B1, and C1.
-
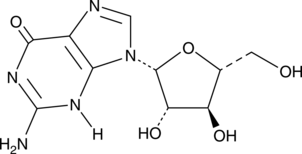
GC45385
Ara-G
-
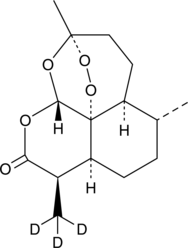
GC46882
Artemisinin-d3
An internal standard for the quantification of artemisinin
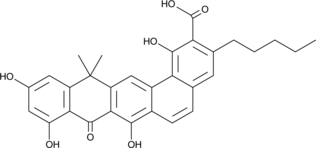
-
GC49042
Benastatin A
A bacterial metabolite with diverse biological activities
-
GC40009
Bostrycin
Bostrycin is an anthraquinone originally isolated from B.
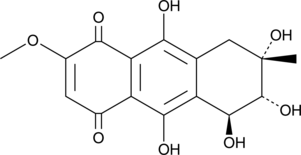
-
GC40708
C2 L-erythro Ceramide (d18:1/2:0)
C2 L-erythro Ceramide is a bioactive sphingolipid and a cell-permeable analog of naturally occurring ceramides.
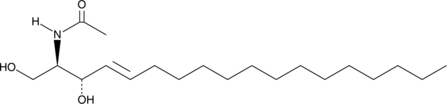
-
GC40709
C2 L-threo Ceramide (d18:1/2:0)
C2 L-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
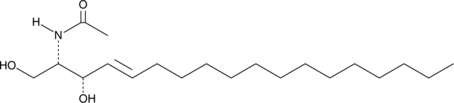
-
GC40795
CAY10503
CAY10503 is a proapoptotic, antiproliferative compound that is able to arrest cell cycle progression in the G0-G1 phase.
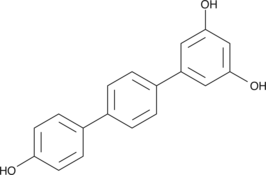
-
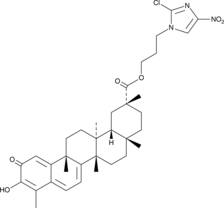
GC52245
CAY10792
An anticancer agent
-
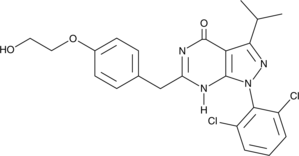
GC43217
CDK/CRK Inhibitor
CDK/CRK inhibitor is an inhibitor of cyclin-dependent kinases (CDK) and CDK-related kinases (CRK) with IC50 values ranging from 9-839 nM in vitro.
-
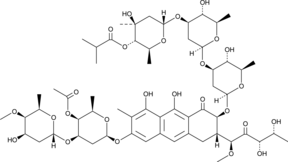
GC43265
Chromomycin A2
Chromomycin A2 is an aureolic acid that has been found in several marine actinomycetes and has antibacterial and anticancer activities.
-
GC43354
Cysmethynil
Post-translational protein prenylation is a 3-step process that occurs at the C-terminus of a number of proteins involved in cell growth control and oncogenesis.

-
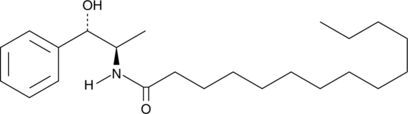
GC40296
D-erythro-MAPP
D-erythro-MAPP is a derivative of ceramide and an inhibitor of alkaline ceramidase (Ki = 2-13 μM; IC50 = 1-5 μM).
-
GC18693
Fascaplysin (chloride)
Fascaplysin (chloride) is an antimicrobial and cytotoxic red pigment, that can come from the marine sponge (Fascaplysin (chloride)opsis sp.

-
GC49344
Fisetin-d5
An internal standard for the quantification of fisetin
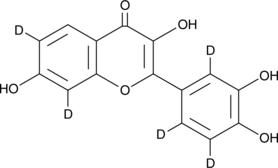
-
GC49089
FR900359
A cyclic depsipeptide and an inhibitor of Gαq, Gα11, and Gα14
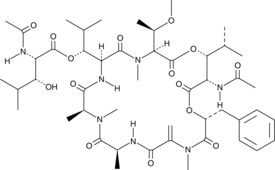
-
GC48387
Inostamycin A
A bacterial metabolite with anticancer activity
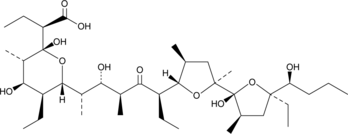
-
GC52472
Inostamycin A (sodium salt)
A bacterial metabolite with anticancer activity
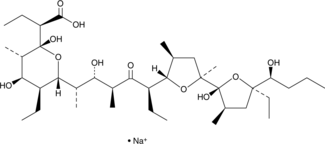
-
GC43995
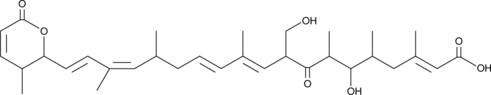
Kazusamycin B
Kazusamycin B is a bacterial metabolite originally isolated from Streptomyces.
-
GC47619
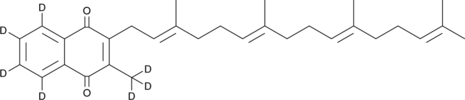
Menaquinone 4-d7
An internal standard for the quantification of menaquinone 4
-
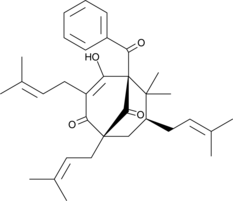
GC45523
Nemorosone
-
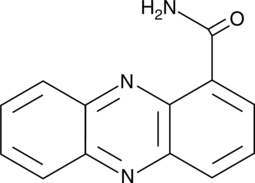
GC45538
Oxychlororaphine
-
GA23337
Oxythiamine . HCl
Thiamine antagonist and inhibitor of saccharomyces cerevisiae transketolase. RaÏs et al. indicated that oxythiamine inhibits the growth of Ehrlich's tumor cells.

-
GC45758
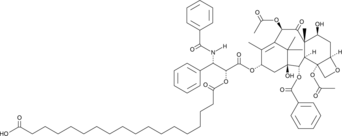
Paclitaxel octadecanedioate
A prodrug form of paclitaxel
-
GC47853
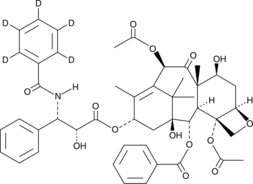
Paclitaxel-d5
An internal standard for the quantification of paclitaxel
-
GC18353
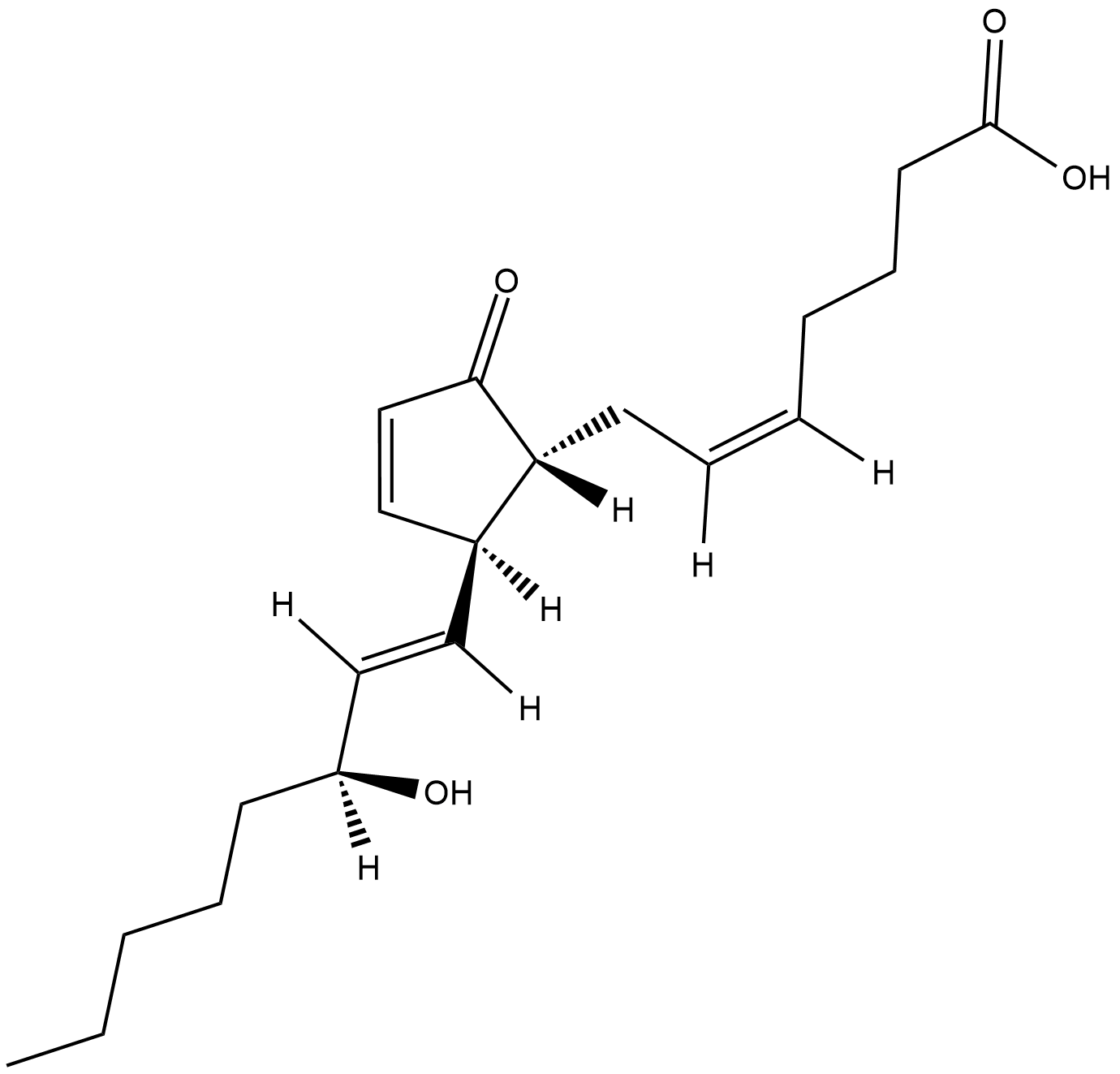
Prostaglandin A2
A naturally occurring prostaglandin with antiviral/antitumor activity
-
GC48019
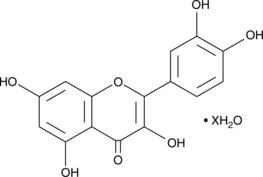
Quercetin (hydrate)
A flavonoid with diverse biological activities
-
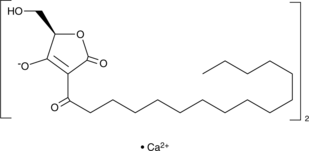
GC44846
RK-682 (calcium salt)
Protein tyrosine phosphatases (PTPs) remove phosphate from tyrosine residues of cellular proteins.
-
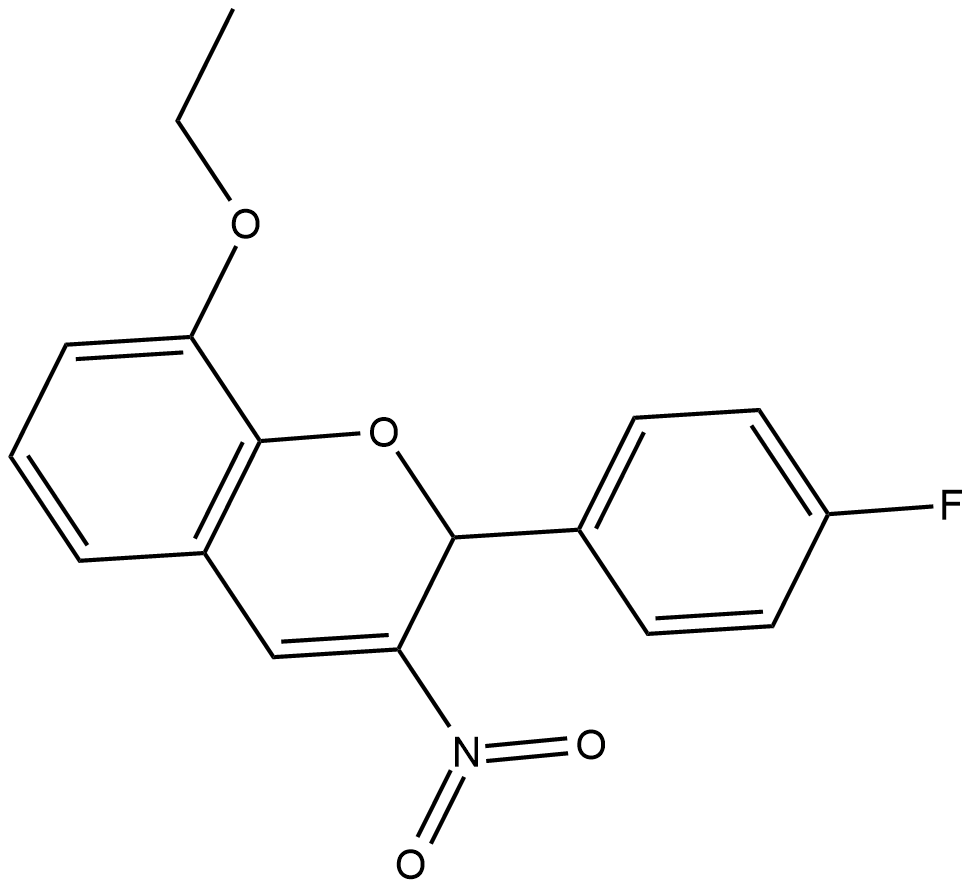
GC18852
S14161
D-Cyclins regulate the cell cycle by acting in a complex with cyclin dependent kinases (CDKs) to promote phosphorylation of the retinoblastoma protein and initiate cellular progression from the G1 to the S phase.
-
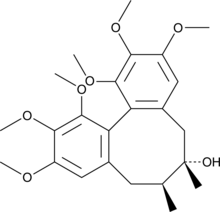
GC49674
Schizandrin
Schizandrin (Schizandrin), a dibenzocyclooctadiene lignan, is isolated from the fruit of Schisandra chinensis Baill.
-
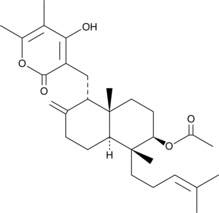
GC48076
Sesquicillin A
A fungal metabolite
-
GC49002
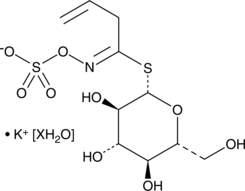
Sinigrin (hydrate)
A glucosinolate with diverse biological activities
-
GC48659
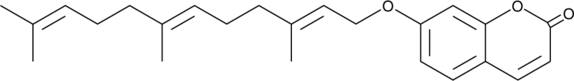
Umbelliprenin
A prenylated coumarin with diverse biological activities
-
GC40044
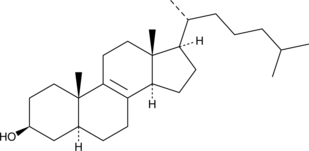
Zymostenol
Zymostenol is a late-stage precursor in the biosynthesis of cholesterol.