DNA Damage/DNA Repair
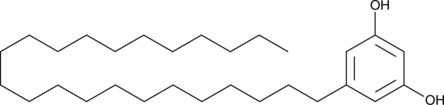
- MTH1(4)
- PARP(84)
- ATM/ATR(30)
- DNA Alkylating(20)
- DNA Ligases(3)
- DNA Methyltransferase(25)
- DNA-PK(30)
- HDAC(136)
- Nucleoside Antimetabolite/Analogue(140)
- Telomerase(17)
- Topoisomerase(151)
- tankyrase(5)
- Antifolate(38)
- CDK(250)
- Checkpoint Kinase (Chk)(31)
- CRISPR/Cas9(9)
- Deubiquitinase(72)
- DNA Alkylator/Crosslinker(71)
- DNA/RNA Synthesis(400)
- Eukaryotic Initiation Factor (eIF)(23)
- IRE1(23)
- LIM Kinase (LIMK)(9)
- TOPK(5)
- Casein Kinase(61)
- DNA Intercalating Agents(7)
- DNA/RNA Oxidative Damage(12)
Products for DNA Damage/DNA Repair
- Cat.No. Product Name Information
-
GC45353
4-oxo-27-TBDMS Withaferin A
4-oxo-27-TBDMS Withaferin A, a withaferin A derivative, exhibits potent antiproliferative effects on the tumor cells.4-oxo-27-TBDMS Withaferin A induces tumor cells apoptosis. 4-oxo-27-TBDMS Withaferin A is a anticancer agent.
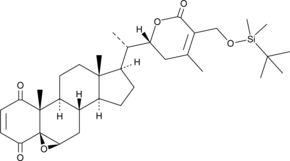
-
GC68557
4-Thio-2'-deoxyuridine
4-Thio-2'-deoxyuridine is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity, targeting inert lymphatic system malignancies. The anticancer mechanism in this process depends on inhibiting DNA synthesis and inducing cell apoptosis.
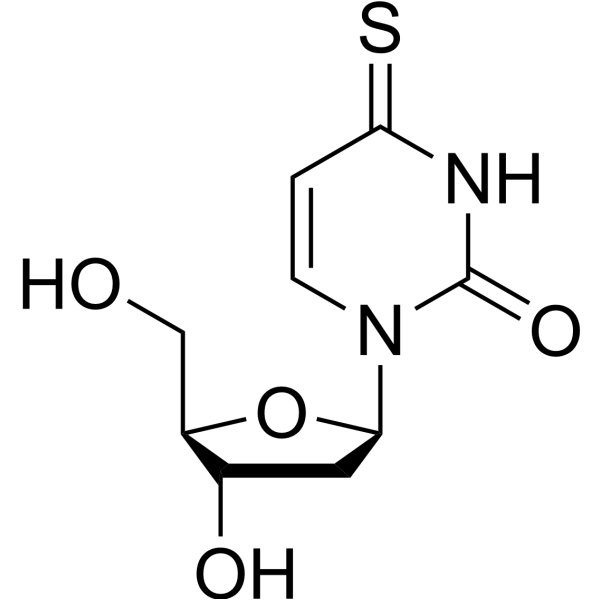
-
GC40477
4-Thiouracil
4-Thiouracil is a site-specific, photoactivatable probe used to detect RNA structures and nucleic acid-nucleic acid contacts.
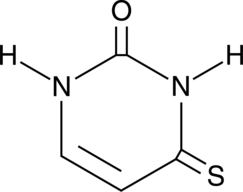
-
GC42474
4-Thiouridine
4-Thiouridine (4-SU) is a photoactivatable ribonucleoside analog that is widely used for RNA analysis, including short-range RNA-RNA crosslinking and nascent RNA labeling.
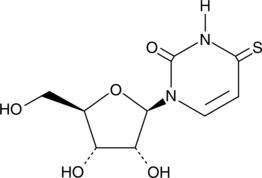
-
GC13104
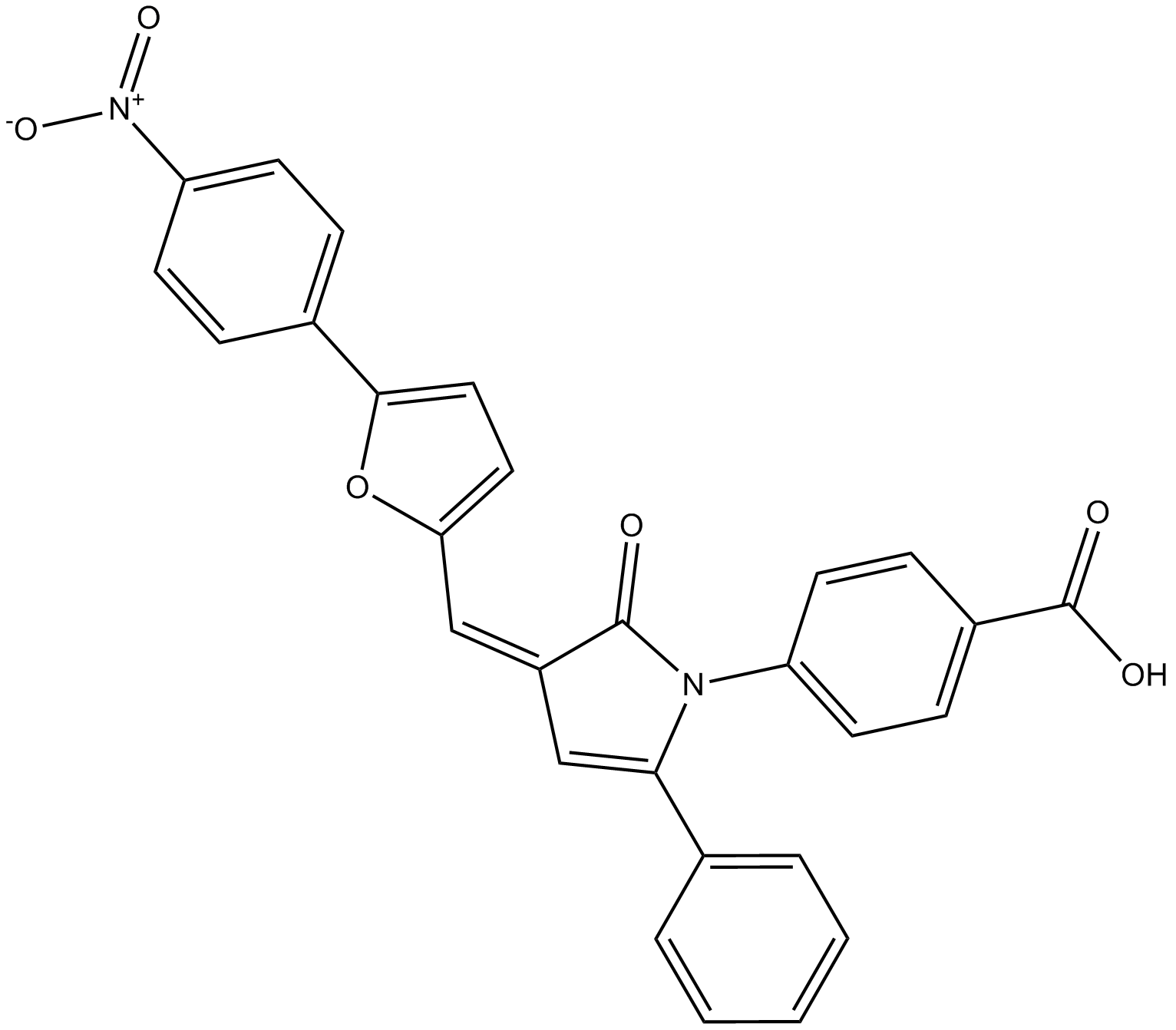
4E1RCat
Dual inhibitor of eIF4E:eIF4G and eIF4E:4E-BP1 interaction
-
GC30785
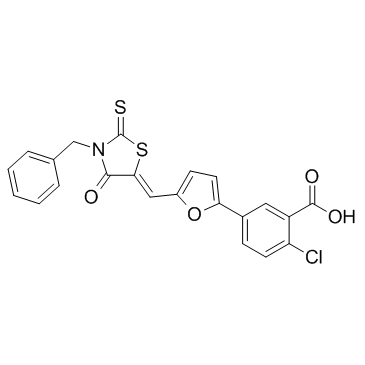
4E2RCat
4E2RCat is an inhibitor of eIF4E-eIF4G interaction with an IC50 of 13.5 μM.
-
GC10468
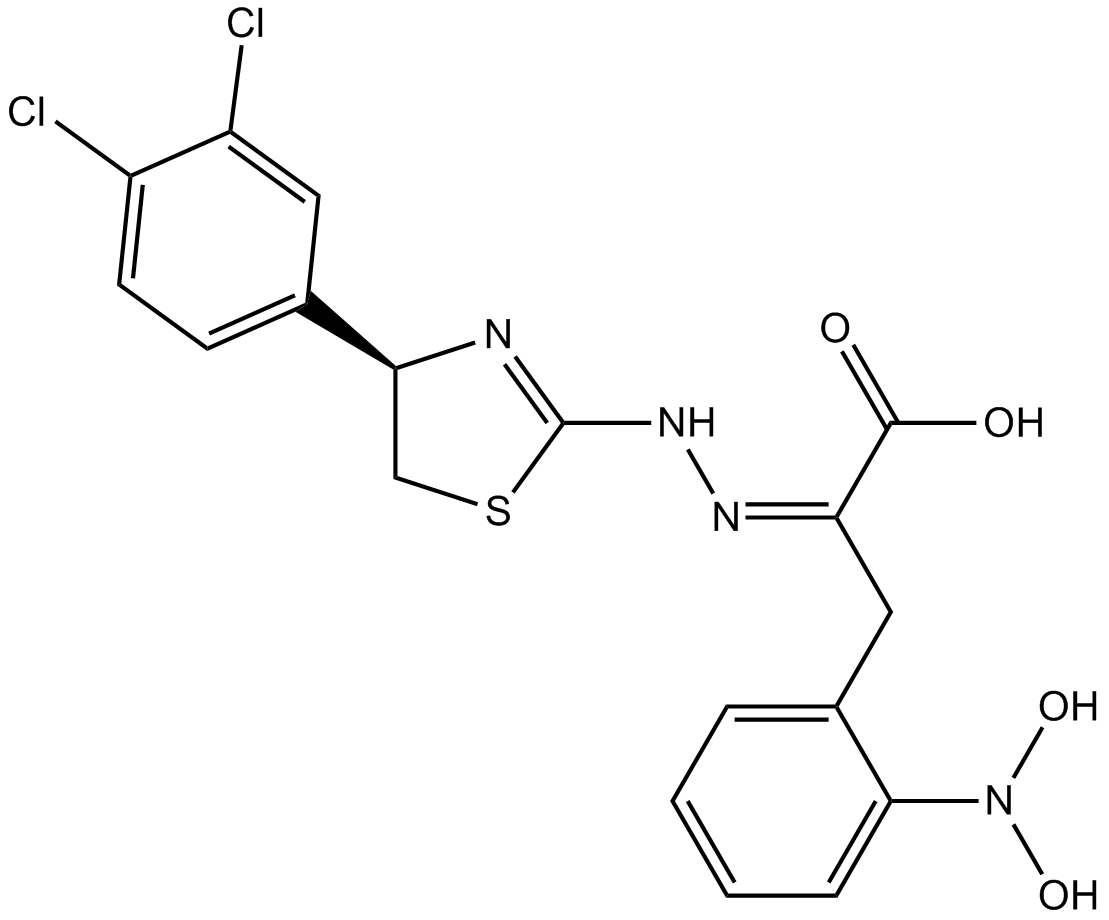
4EGI-1
Competitive eIF4E/eIF4G interaction inhibitor
-
GC12667
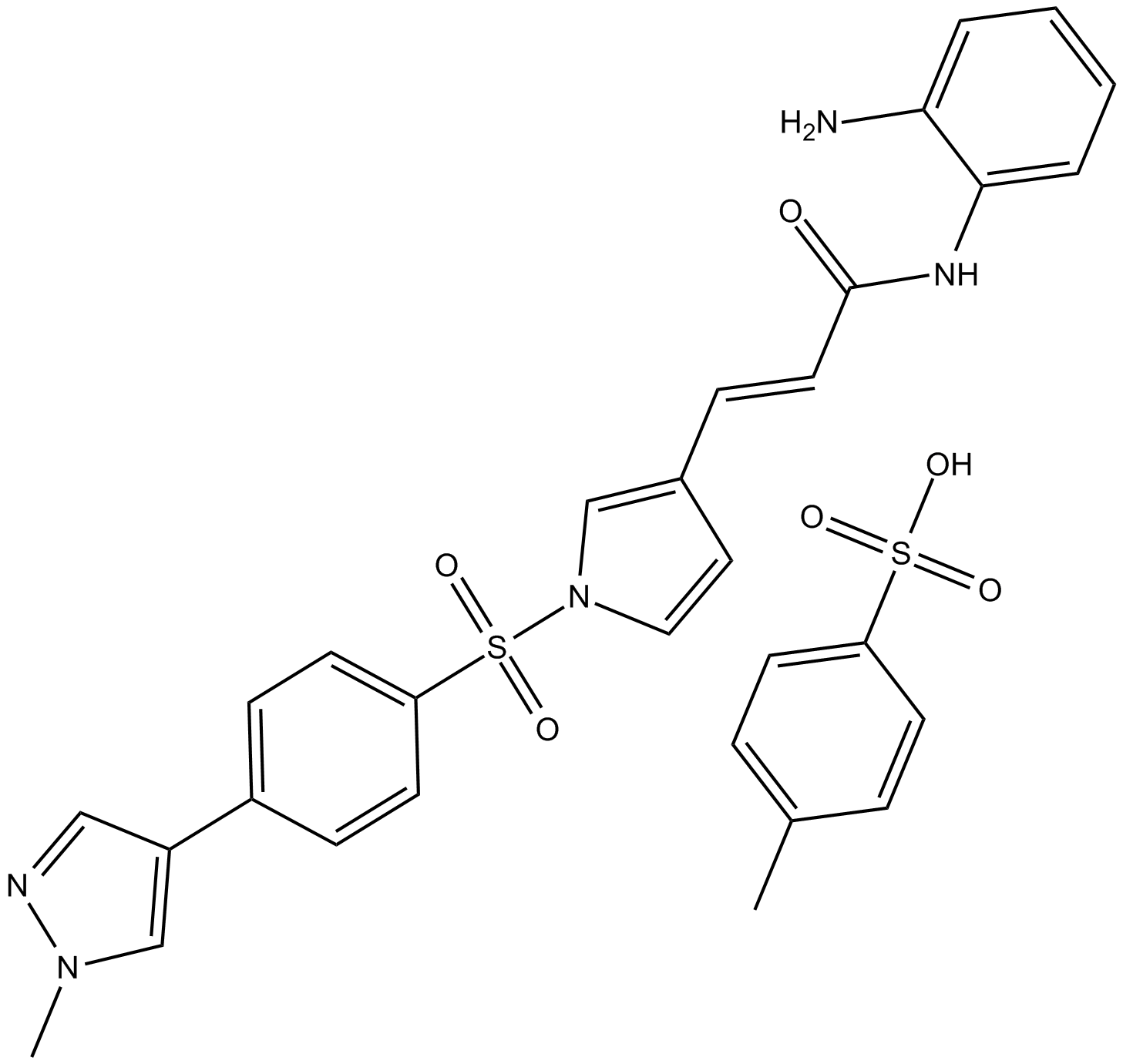
4SC-202
4SC-202 (4SC-202) is a selective class I HDAC inhibitor with IC50 of 1.20 μM, 1.12 μM, and 0.57 μM for HDAC1, HDAC2, and HDAC3, respectively. It also displays inhibitory activity against Lysine specific demethylase 1 (LSD1).
-
GC65520
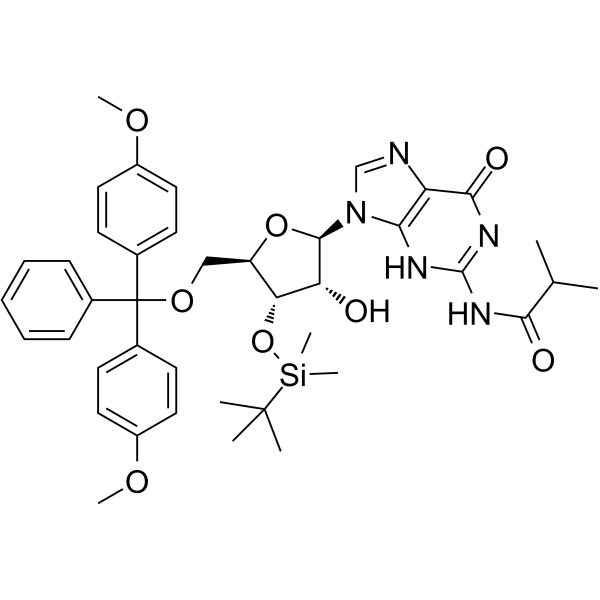
5'-DMT-3'-TBDMS-ibu-rG
5'-DMT-3'-TBDMS-ibu-rG is is a modified nucleoside.
-
GC65402
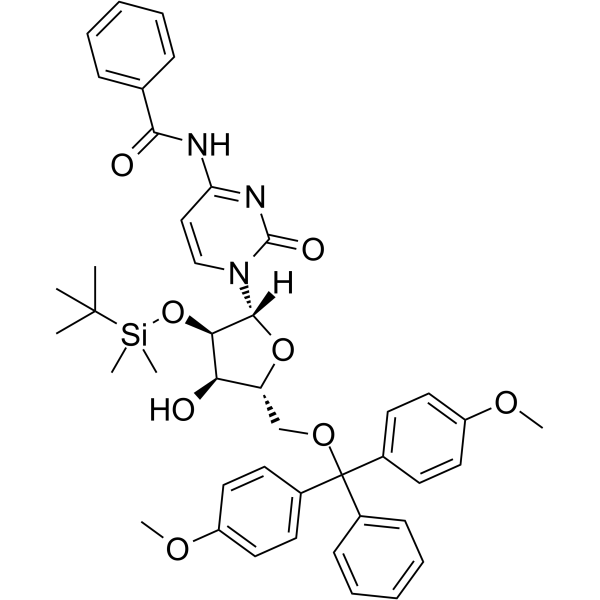
5'-O-DMT-2'-O-TBDMS-Bz-rC
5'-O-DMT-2'-O-TBDMS-Bz-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC65164
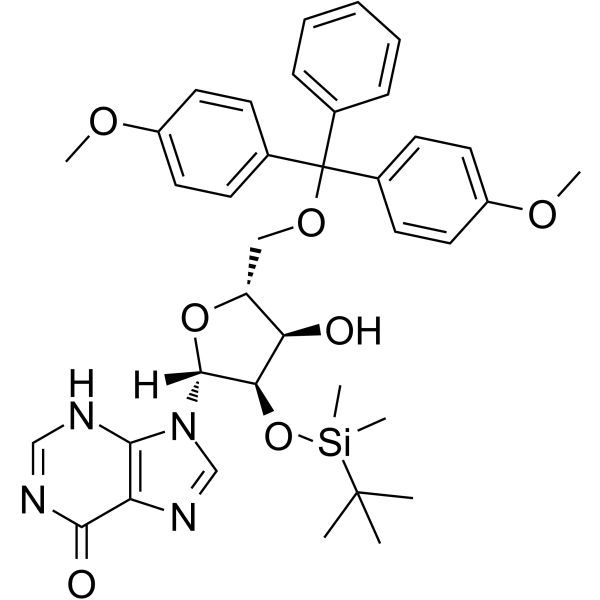
5'-O-DMT-2'-O-TBDMS-rI
5'-O-DMT-2'-O-TBDMS-rI is a modified nucleoside.
-
GC65396
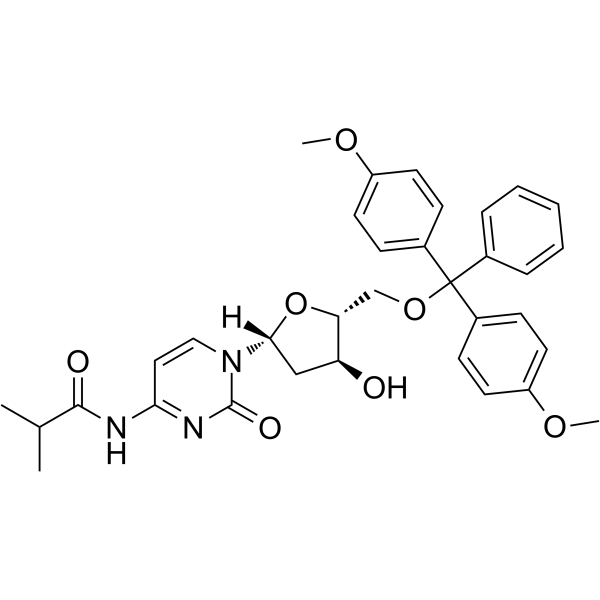
5'-O-DMT-ibu-dC
5'-O-DMT-ibu-dC can be used in the synthesis of oligodeoxyribonucleotides.
-
GC64982
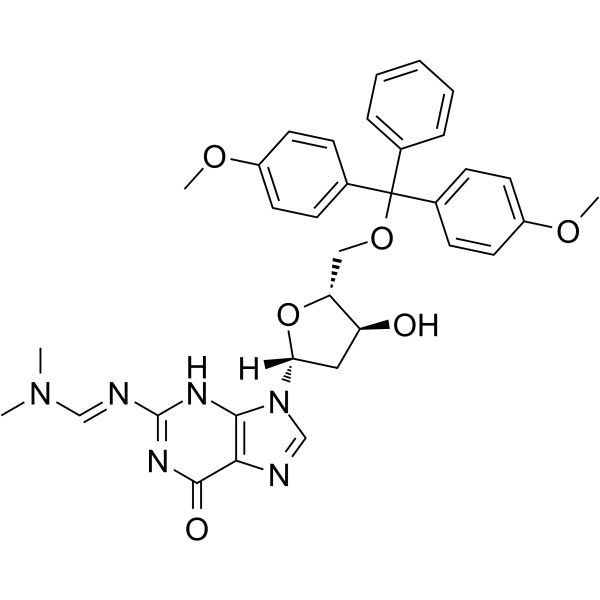
5'-O-DMT-N2-DMF-dG
5'-O-DMT-2'-O-TBDMS-rI is a modified nucleoside.
-
GC65482
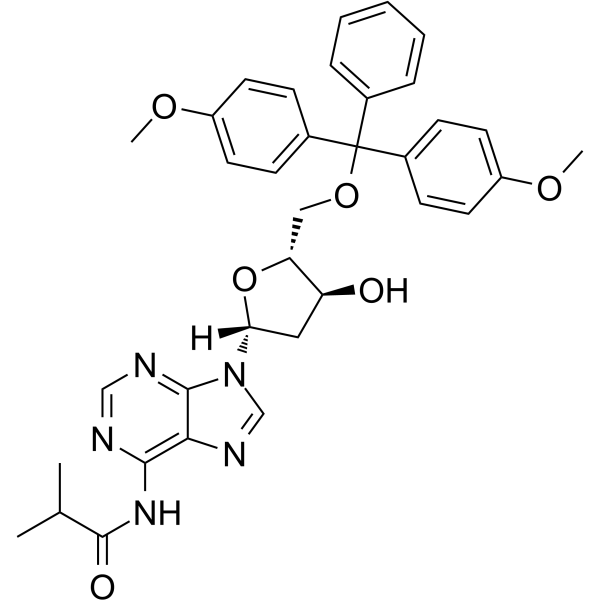
5'-O-DMT-N6-ibu-dA
5'-O-DMT-N6-ibu-dA can be used in the synthesis of oligodeoxyribonucleotides.
-
GC65562
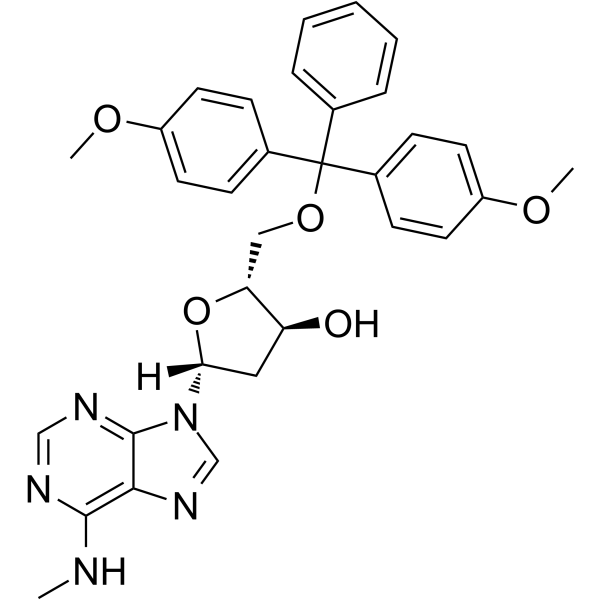
5'-O-DMT-N6-Me-2'-dA
5'-O-DMT-N6-Me-2'-dA is a nucleoside with protective and modification effects.
-
GC65401
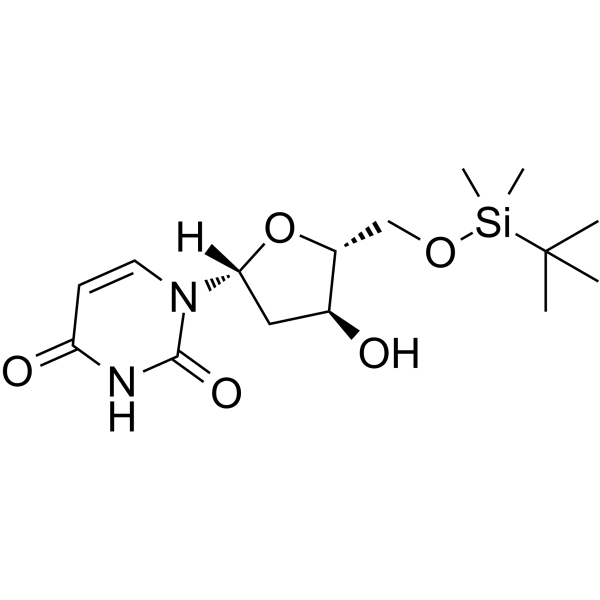
5'-O-TBDMS-dU
5'-O-TBDMS-dU can be used in the synthesis of oligoribonucleotides.
-
GC62157
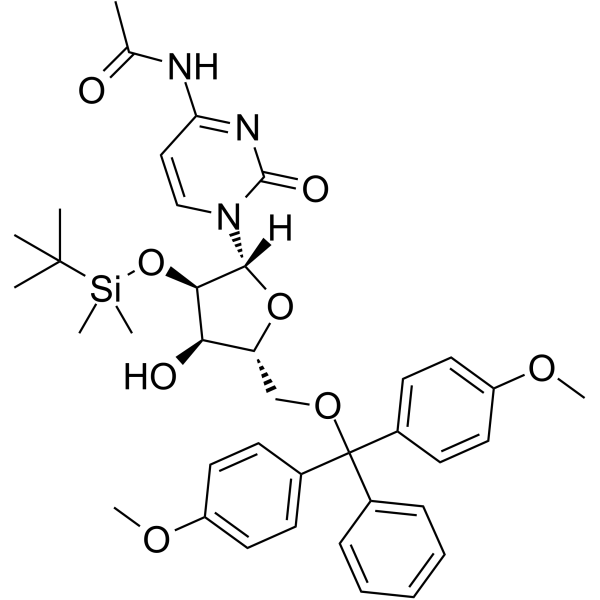
5’-O-DMT-2’-O-TBDMS-Ac-rC
5’-O-DMT-2’-O-TBDMS-Ac-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62149
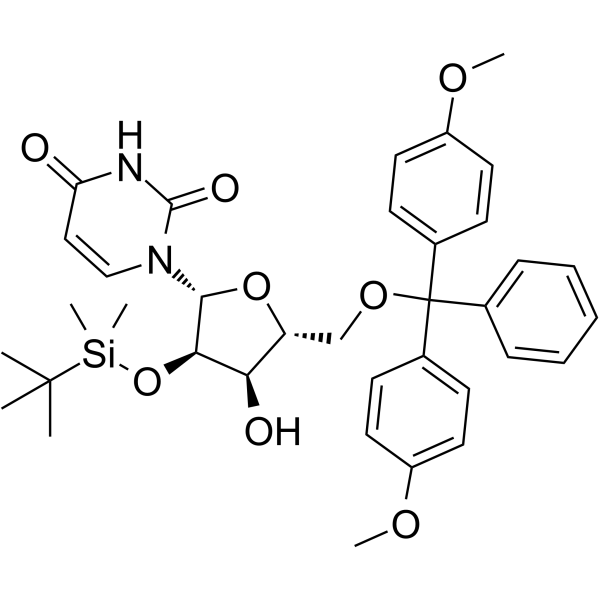
5’-O-DMT-2’-TBDMS-Uridine
5’-O-DMT-2’-TBDMS-Uridine is a deoxyribonucleoside used for the oligonucleotide synthesis.
-
GC62532
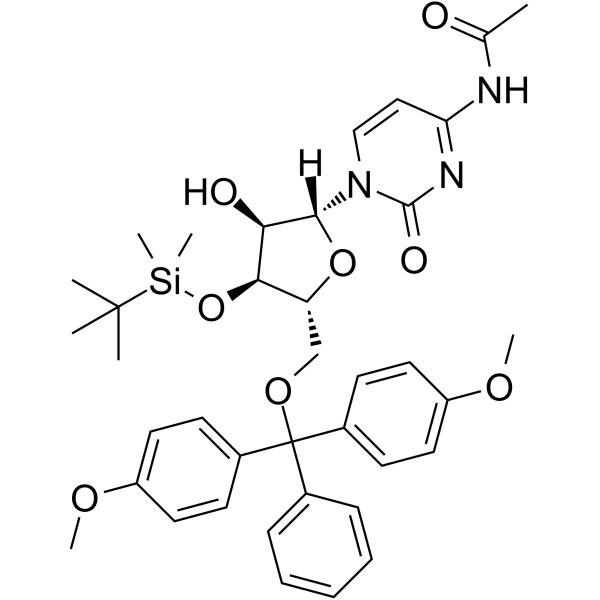
5’-O-DMT-3’-O-TBDMS-Ac-rC
5’-O-DMT-3’-O-TBDMS-Ac-rC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62573
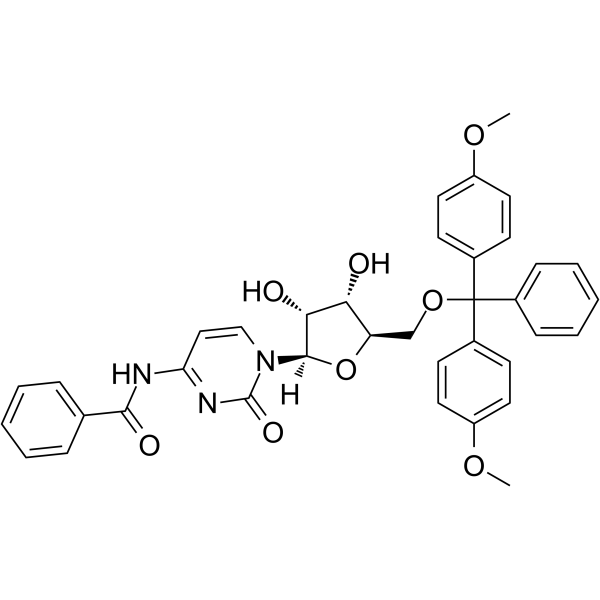
5’-O-DMT-Bz-rC
5'-O-DMT-Bz-Rc is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62574
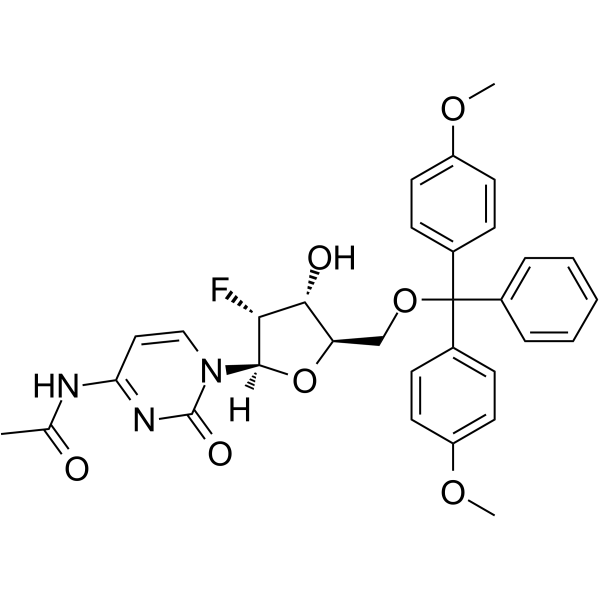
5’-O-DMT-N4-Ac-2’-F-dC
5’-O-DMT-N4-Ac-2’-F-dC is a modified nucleoside and can be used to synthesize DNA or RNA.
-
GC62575
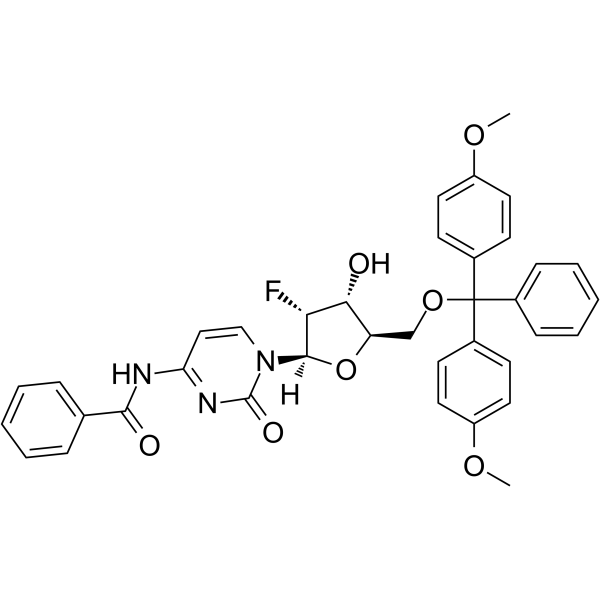
5’-O-DMT-N4-Bz-2’-F-dC
5’-O-DMT-N4-Bz-2’-F-dC is a nucleoside with protective and modification effects.
-
GC62576
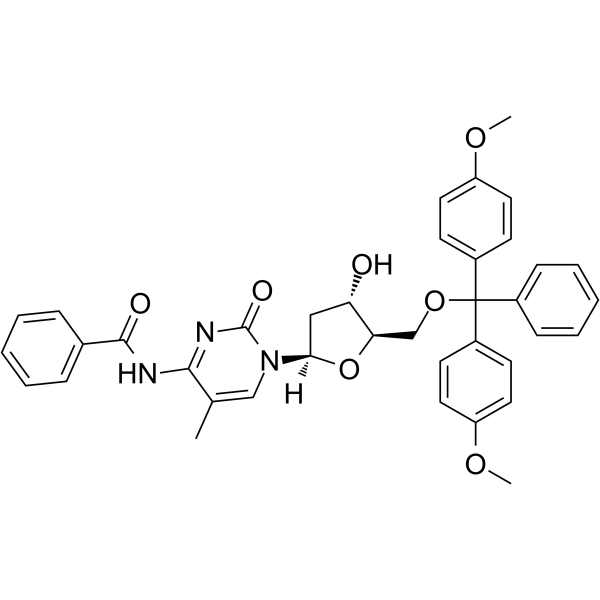
5’-O-DMT-N4-Bz-5-Me-dC
5’-O-DMT-N4-Bz-5-Me-dC is a modified nucleoside.
-
GC62812
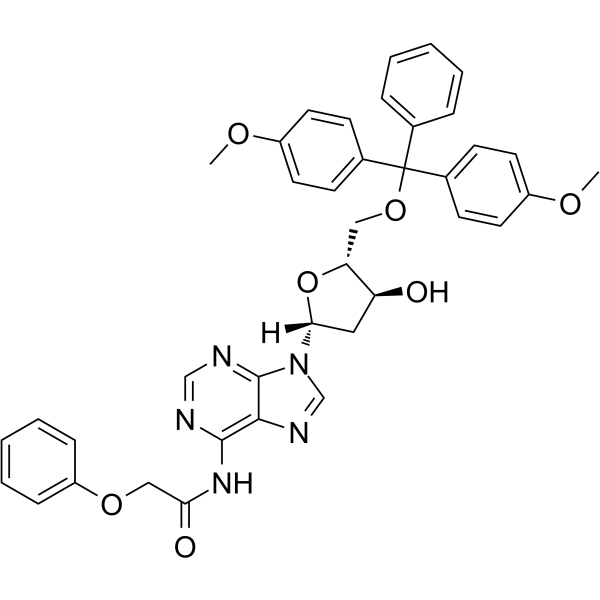
5’-O-DMT-PAC-dA
5’-O-DMT-PAC-dA can be used in the synthesis of oligoribonucleotides.
-
GC62813
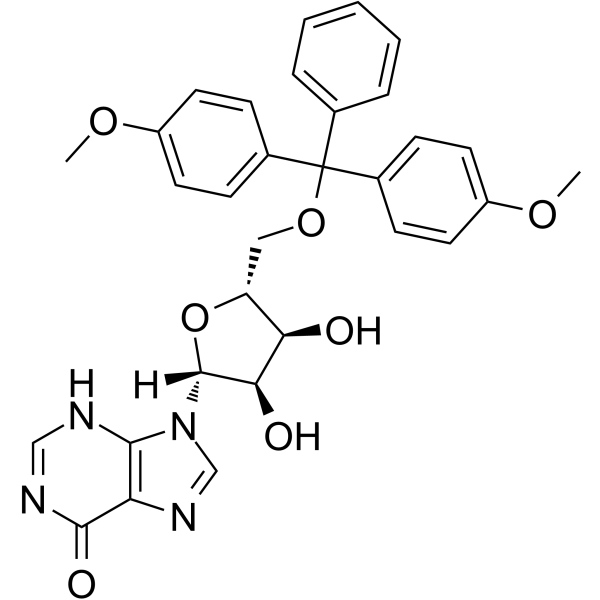
5’-O-DMT-rI
5'-O-DMT-Ri can be used in the synthesis of oligoribonucleotides.
-
GC62577
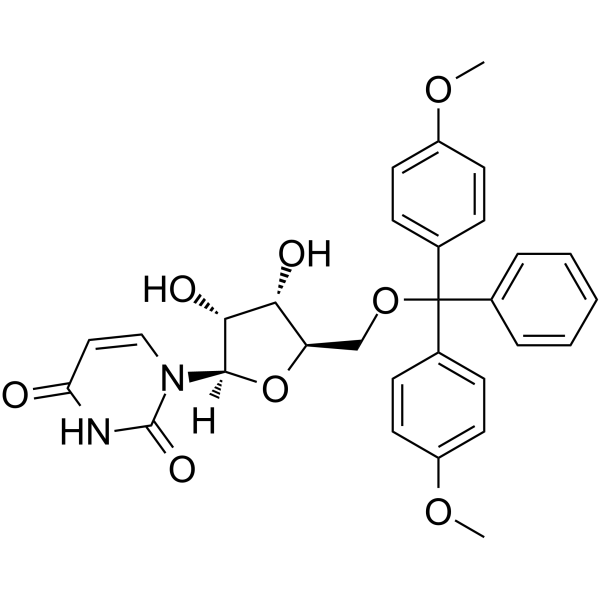
5’-O-DMT-rU
5’-O-DMT-rU is a modified nucleoside and can be used to synthesize RNA.
-
GC62578
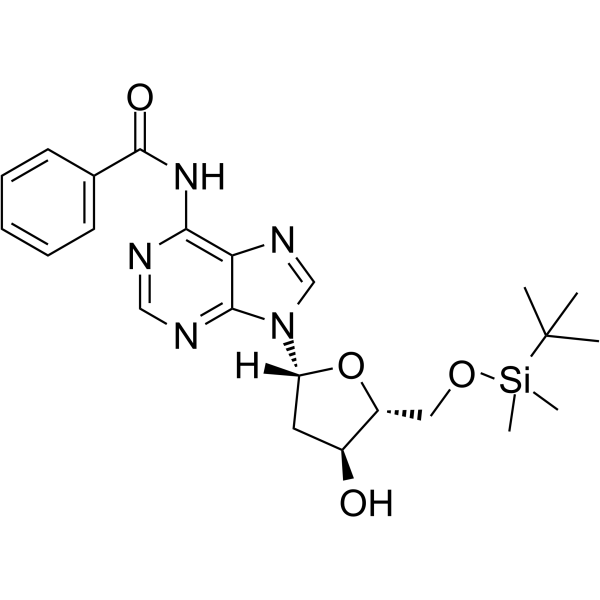
5’-O-TBDMS-Bz-dA
5’-O-TBDMS-Bz-dA is a nucleoside with protective and modification effects.
-
GC62579
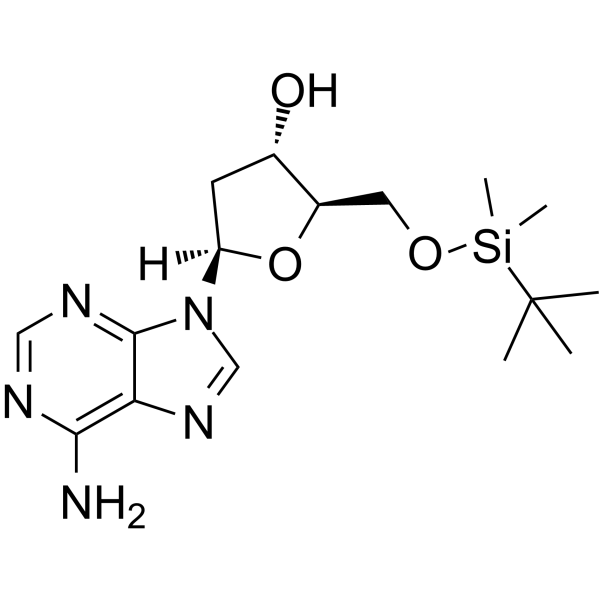
5’-O-TBDMS-dA
5’-O-TBDMS-dA is a modified nucleoside and can be used to synthesize DNA or RNA.
-
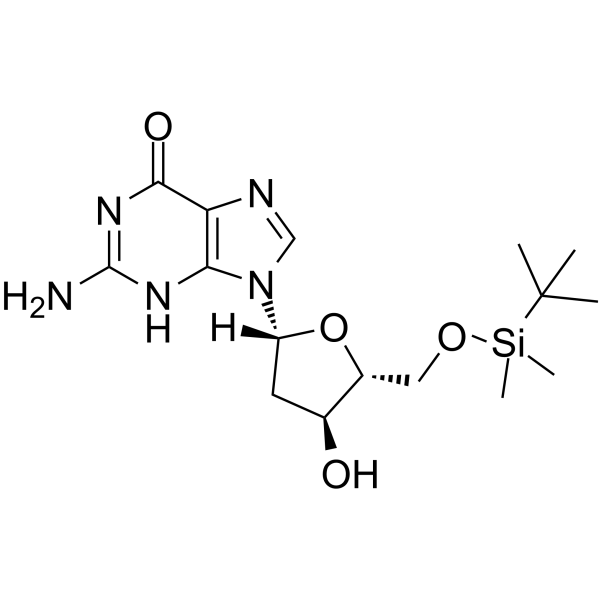
GC62580
5’-O-TBDMS-dG
5’-O-TBDMS-dG is a modified nucleoside.
-
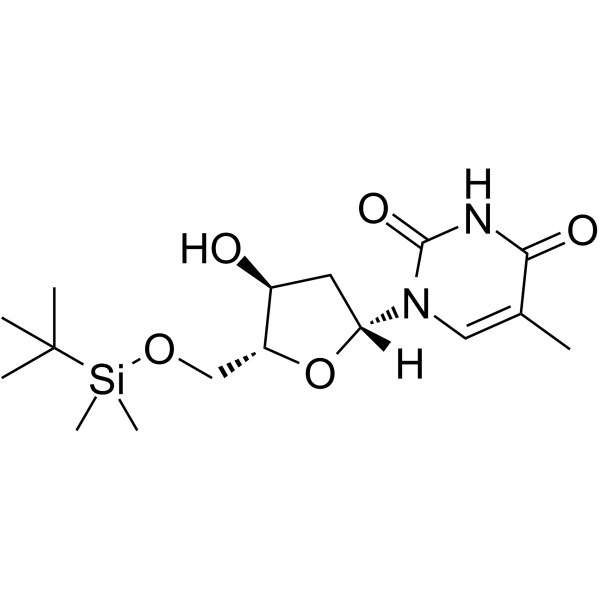
GC62581
5’-O-TBDMS-dT
5’-O-TBDMS-dT is a nucleoside with protective and modification effects.
-
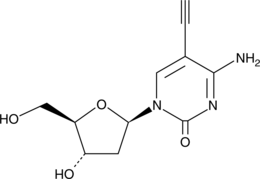
GC40641
5'-Ethynyl-2'-deoxycytidine
5'-Ethynyl-2'-deoxycytidine (EdC) is a nucleoside analog that inhibits replication of the herpes simplex virus-1 (HSV-1) KOS strain (ID50 = 0.2 μg/mL).
-
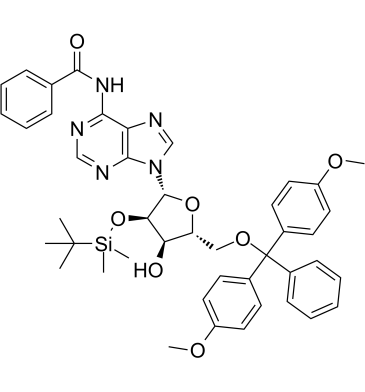
GC61432
5'-O-DMT-2'-O-TBDMS-N-Bz-Adenosine
5'-O-DMT-2'-O-TBDMS-N-Bz-Adenosine?is an adenosine derivative and can be used as an intermediate for oligonucleotides synthesis.
-
GC61726
5'-O-DMT-N4-Ac-dC
5'-O-DMT-N4-Ac-dC (N4-Acetyl-2'-deoxy-5'-O-DMT-cytidine, compound 7), a deoxynucleoside, can be used to synthesize of dodecyl phosphoramidite which is the raw material for dod‐DNA (amphiphilic DNA containing an internal hydrophobic region consisting of dodecyl phosphotriester linkages) synthesis.
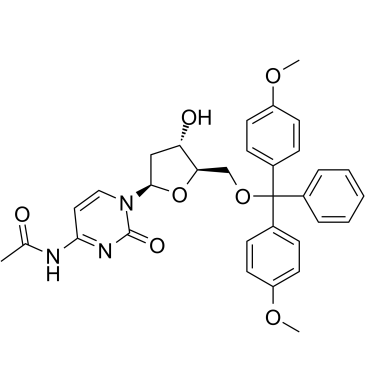
-
GC48662
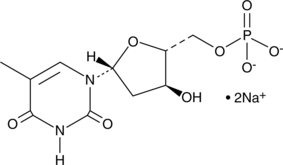
5'-Thymidylic Acid (sodium salt)
5'-Thymidylic Acid (sodium salt) is an endogenous metabolite.
-
GC35150
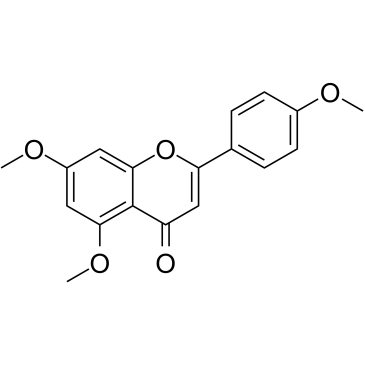
5,7,4'-Trimethoxyflavone
5,7,4'-Trimethoxyflavone is isolated from Kaempferia parviflora (KP) that is a famous medicinal plant from Thailand. 5,7,4'-Trimethoxyflavone induces apoptosis, as evidenced by increments of sub-G1 phase, DNA fragmentation, annexin-V/PI staining, the Bax/Bcl-xL ratio, proteolytic activation of caspase-3, and degradation of poly (ADP-ribose) polymerase (PARP) protein.5,7,4'-Trimethoxyflavone is significantly effective at inhibiting proliferation of SNU-16 human gastric cancer cells in a concentration dependent manner.
-
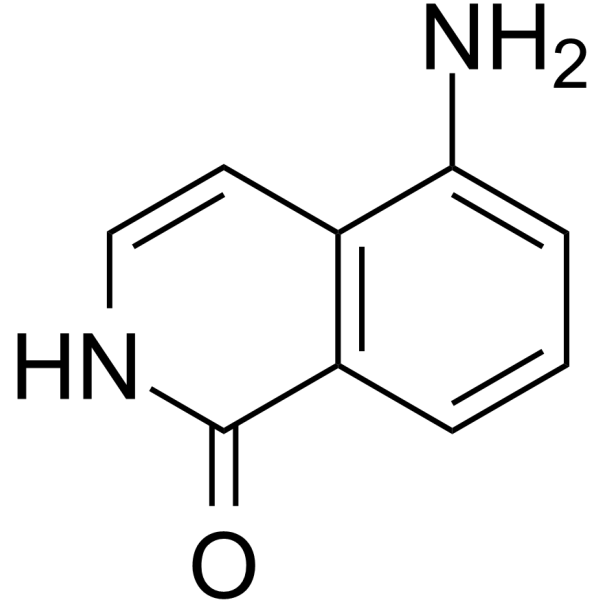
GC68161
5-AIQ
-
GC64952
5-Aza-7-deazaguanine
5-Aza-7-deazaguanine is a substrate for wild-type (WT) E. coli purine nucleoside phosphorylase and its Ser90Ala mutant in the synthesis of base-modified nucleosides.
-
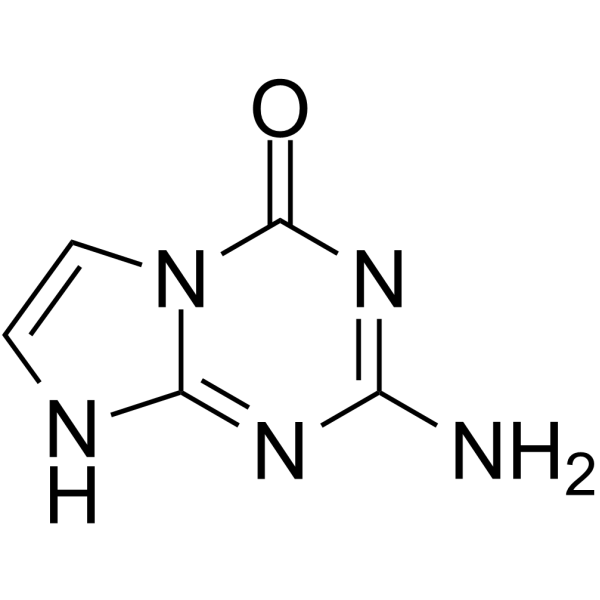
GC10946
5-Azacytidine
A DNA methyltransferase inhibitor
-
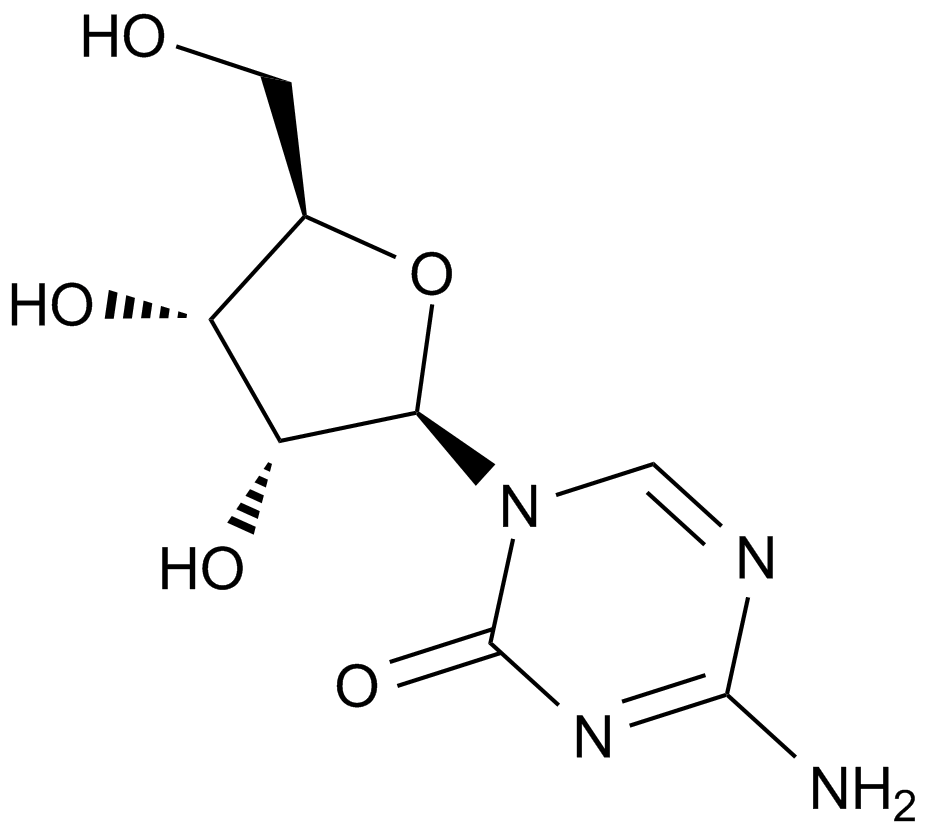
GC11940
5-BrdU
Synthetic thymidine analog
-
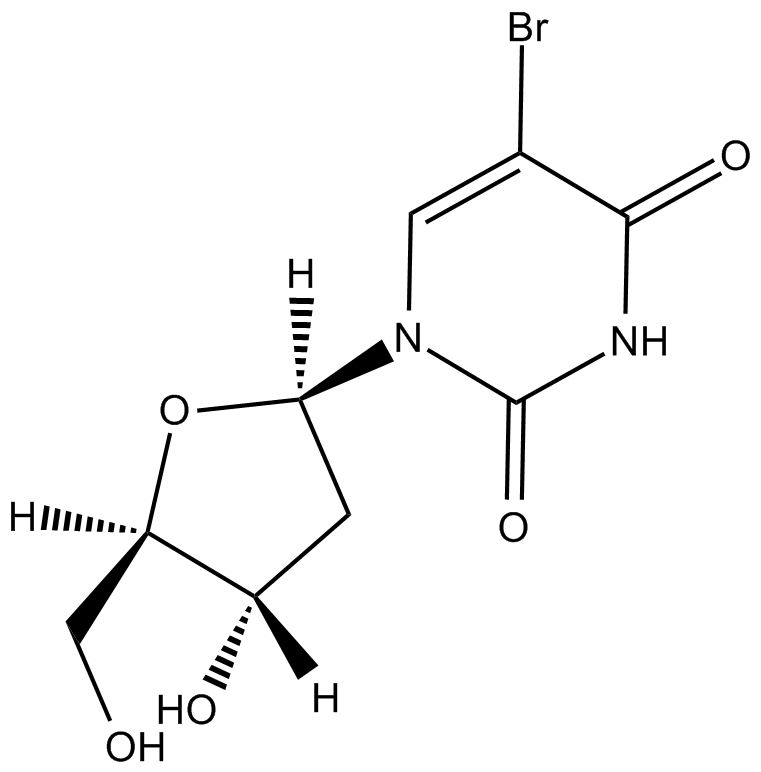
GC46681
5-Bromouridine
A brominated uridine analog
-
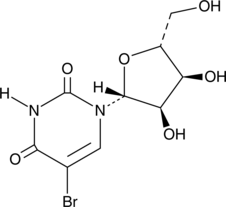
GC49245
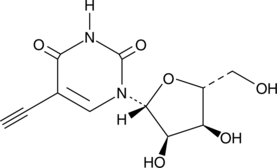
5-Ethynyluridine
5-Ethynyluridine (5-EU) is a potent cell-permeable nucleoside can be used to label newly synthesized RNA.
-
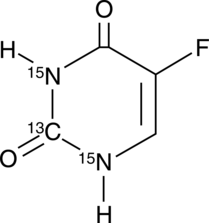
GC42545
5-Fluorouracil-13C,15N2
5-Fluorouracil-13C,15N2 is intended for use as an internal standard for the quantification of 5-flurouracil by GC- or LC-MS.
-
GC46033
5-Heneicosylresorcinol
An alkylresorcinol

-
GC62389
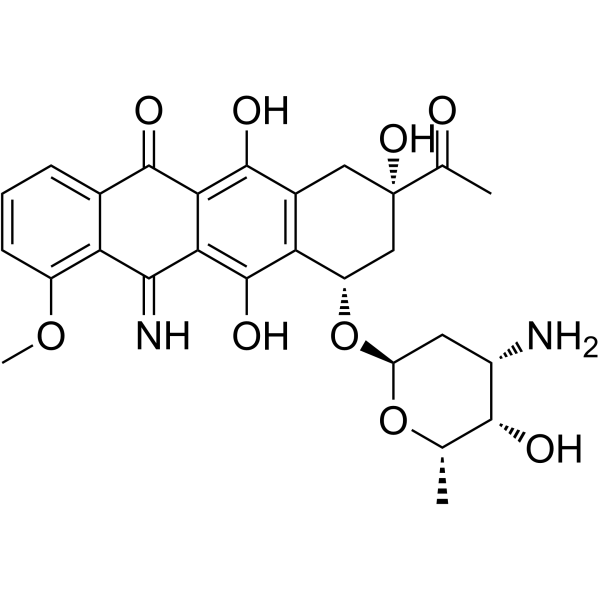
5-Iminodaunorubicin
5-Iminodaunorubicin is a quinone-modified anthracycline that retains antitumor activity. 5-Iminodaunorubicin produces protein-concealed DNA strand breaks in cancer cells.
-
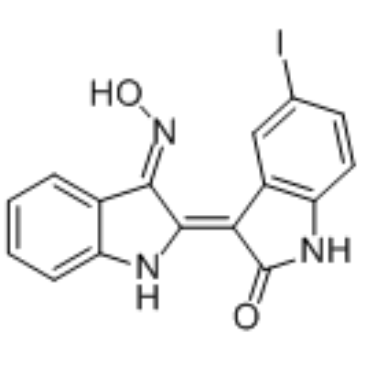
GC35162
5-Iodo-indirubin-3'-monoxime
5-Iodo-indirubin-3'-monoxime is a potent GSK-3β, CDK5/P25 and CDK1/cyclin B inhibitor, competing with ATP for binding to the catalytic site of the kinase, with IC50s of 9, 20 and 25 nM, respectively.
-
GC66658
5-Me-dC(Ac) amidite
5-Me-dC(Ac) amidite is used for synthesizing DNA or RNA.
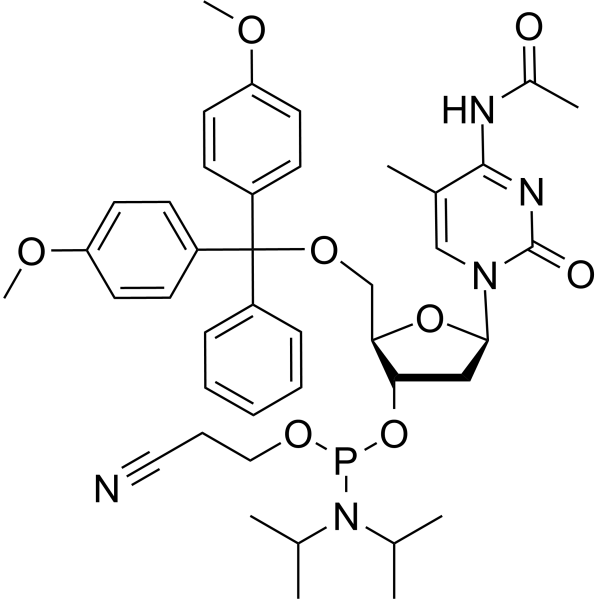
-
GC31187
5-Methoxyflavone
5-Methoxyflavone, belonged to Flavonoid family, is a DNA polymerase-beta inhibitor and neuroprotective agent against beta-amyloid toxicity.
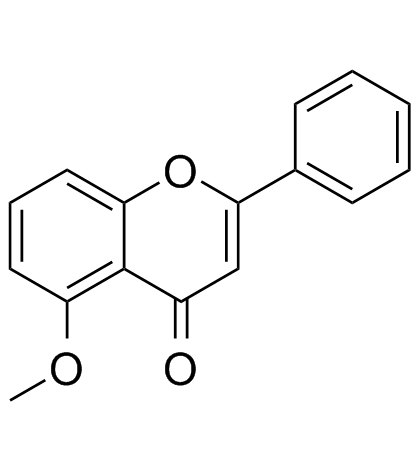
-
GC42562
5-Methyl-2'-deoxycytidine
5-Methyl-2'-deoxycytidine is a pyrimidine nucleoside that when incorporated into single-stranded DNA can act in cis to signal de novo DNA methylation.
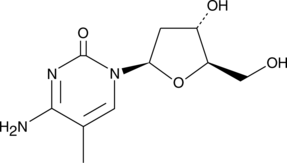
-
GC67671
5-Methylcytidine 5′-triphosphate trisodium
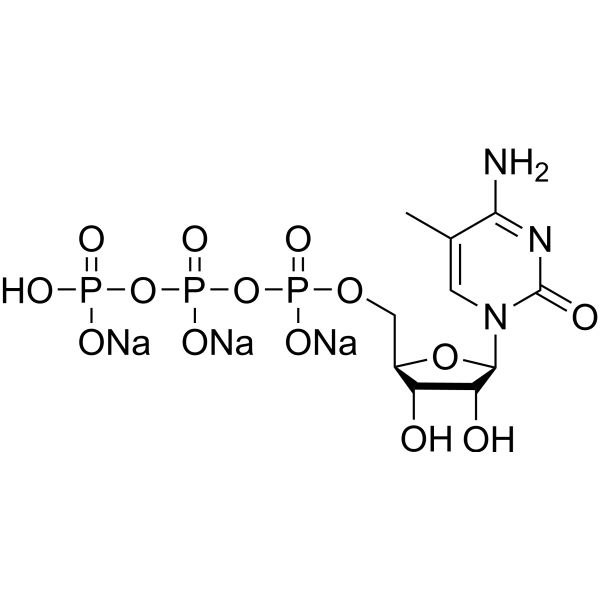
-
GC35166
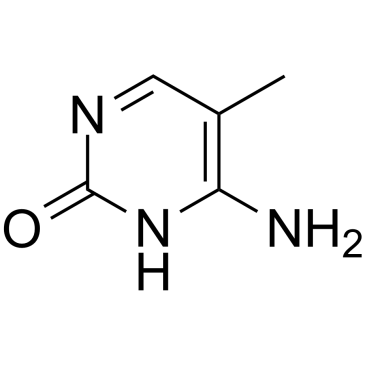
5-Methylcytosine
5-Methylcytosine is a well-characterized DNA modification, and is also predominantly in abundant non-coding RNAs in both prokaryotes and eukaryotes.
-
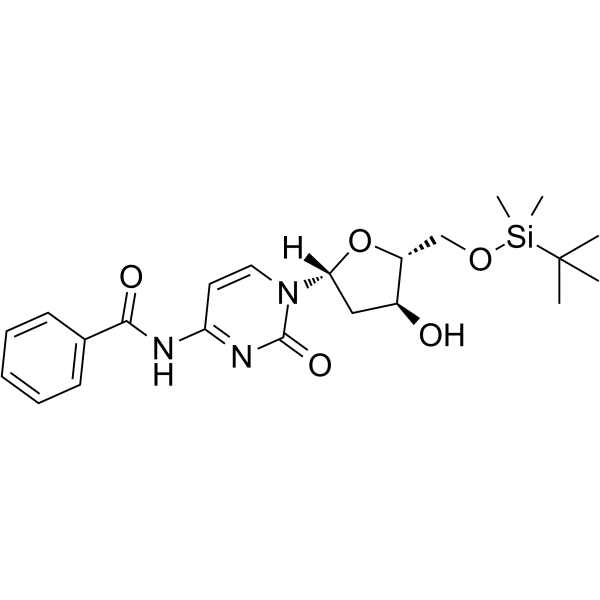
GC62547
5-O-TBDMS-N4-Benzoyl-2-deoxycytidine
5-O-TBDMS-N4-Benzoyl-2-deoxycytidine is a modified nucleoside.
-
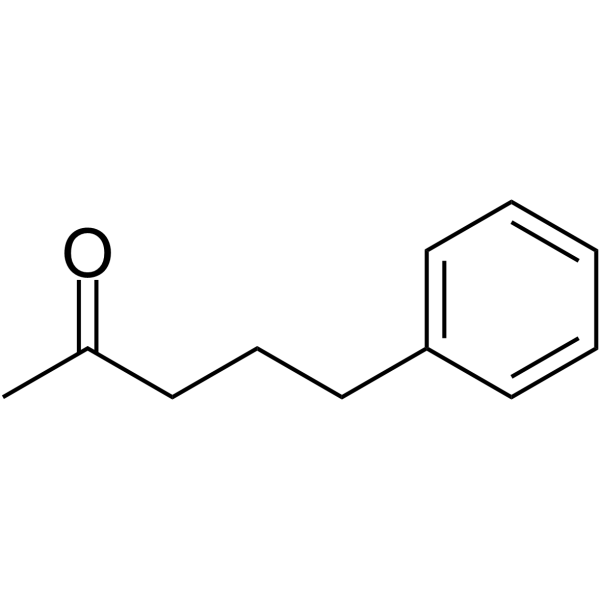
GC66055
5-Phenylpentan-2-one
5-Phenylpentan-2-one is a potent histone deacetylases (HDACs) inhibitor. 5-Phenylpentan-2-one can be used for urea cycle disorder research.
-
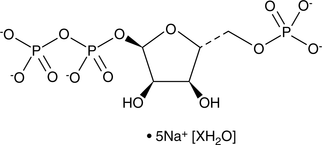
GC46713
5-Phospho-D-ribose 1-diphosphate (sodium salt hydrate)
An intermediate in several biochemical pathways
-
GC46079
5-Tricosylresorcinol
5-Tricosylresorcinolthe is the first cyst lipid.
-
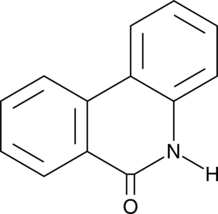
GC45772
6(5H)-Phenanthridinone
An inhibitor of PARP1 and 2
-
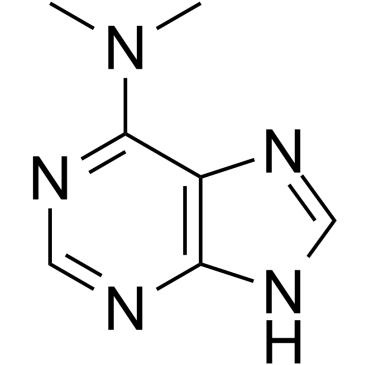
GC60532
6-(Dimethylamino)purine
6-(Dimethylamino)purine is a dual inhibitor of protein kinase and CDK.
-
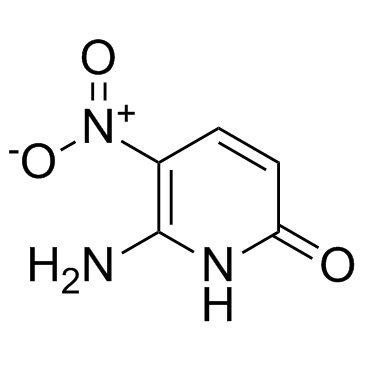
GC30548
6-Amino-5-nitropyridin-2-one
6-Amino-5-nitropyridin-2-one is a pyridine base and used as a nucleobase of hachimoji DNA, in which it pairs with 5-aza-7-deazaguanine.
-
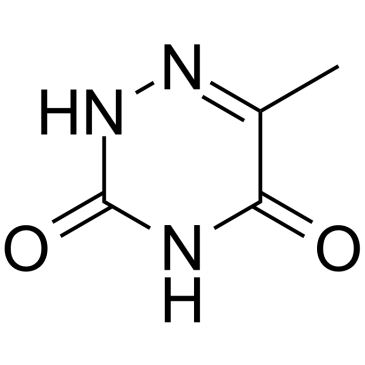
GC60031
6-Azathymine
6-Azathymine, a 6-nitrogen analog of thymine, is a potent D-3-aminoisobutyrate-pyruvate aminotransferase inhibitor.
-
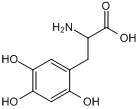
GC50249
6-Hydroxy-DL-DOPA
6-Hydroxy-DL-DOPA is a selective and effective allosteric inhibitor of the RAD52 ssDNA binding domain. 6-Hydroxy-DL-DOPA can be used for the research of cancer.
-
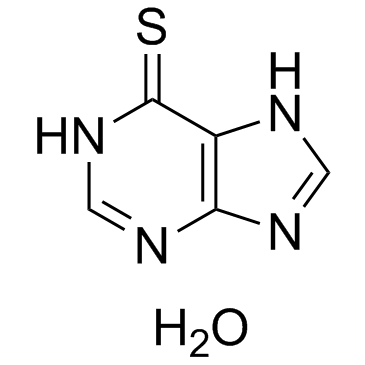
GC32975
6-Mercaptopurine hydrate
An inhibitor of purine synthesis and interconversion
-
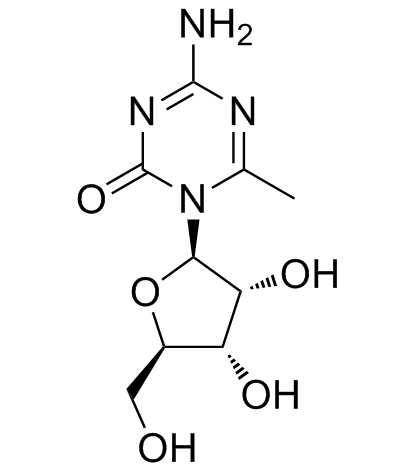
GC35180
6-Methyl-5-azacytidine
6-Methyl-5-azacytidine is a potent DNMT inhibitor.
-
GC49235
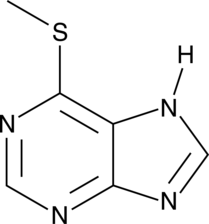
6-Methylmercaptopurine
A metabolite of 6-mercaptopurine
-
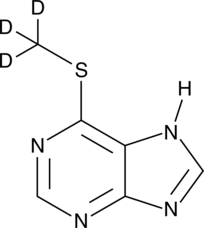
GC49488
6-Methylmercaptopurine-d3
An internal standard for the quantification of 6-MMP
-
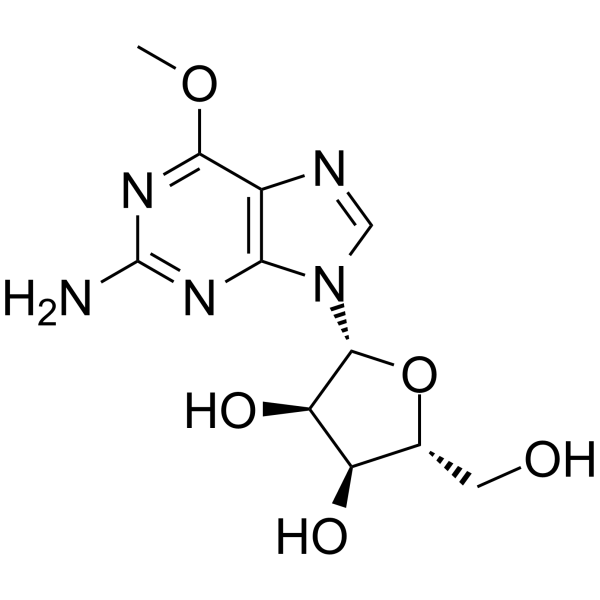
GC65082
6-O-Methyl Guanosine
6-O-Methyl Guanosine is a modified nucleoside.
-
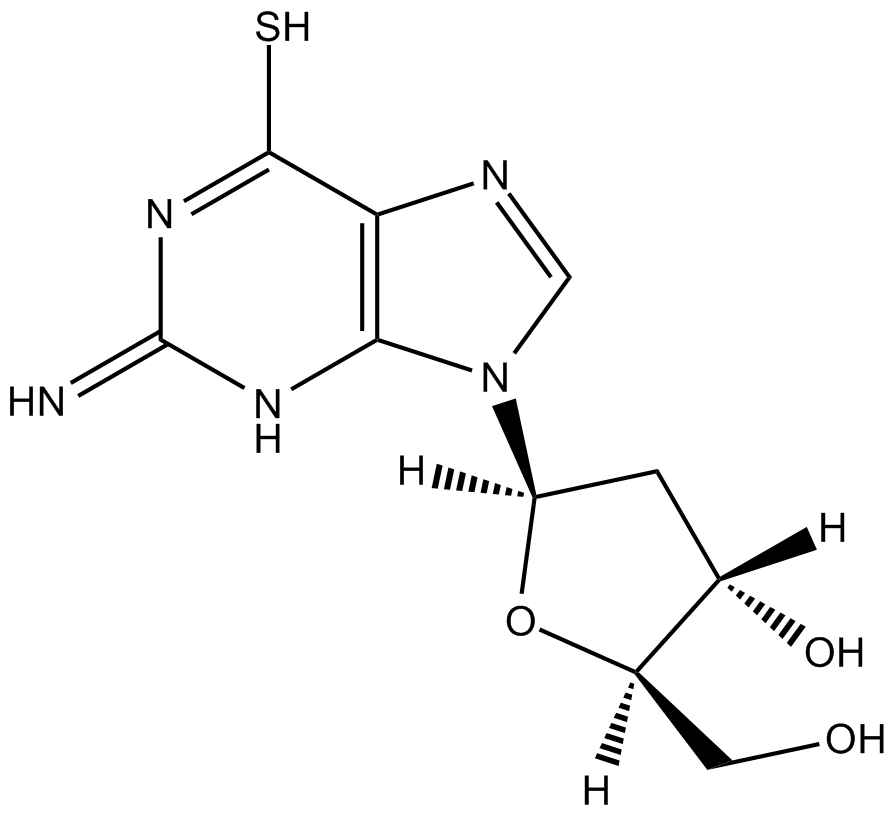
GC12193
6-Thio-dG
6-Thio-dG is a nucleoside analogue that can be incorporated into de novo-synthesized telomeres by telomerase.
-
GC35171
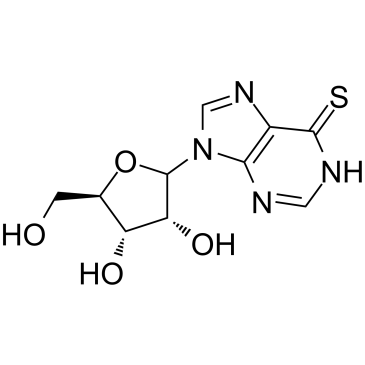
6-Thioinosine
6-Thioinosine (6TI) is a purine antimetabolite, acts as an anti-adipogenesis agent, downregulates mRNA levels of PPAR γ and C/EBPα, as well as PPAR γ target protein such as LPL, CD36, aP2, and LXRα.
-
GC39625
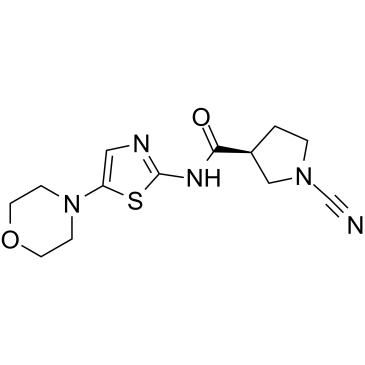
6RK73
6RK73 is a covalent irreversible and specific UCHL1 inhibitor with an IC50 of 0.23 ?M. 6RK73 shows almost no inhibition of UCHL3 (IC50=236 ?M). 6RK73 specifically inhibit UCHL1 activity in breast cancer.
-
GC46733
7,12-Dimethylbenz[a]anthracene
7,12-Dimethylbenz[a]anthracene has carcinogenic activity as a polycyclic aromatic hydrocarbon (PAH). 7,12-Dimethylbenz[a]anthracene is used to induce tumor formation in various rodent models.
![7,12-Dimethylbenz[a]anthracene Chemical Structure 7,12-Dimethylbenz[a]anthracene Chemical Structure](/media/struct/GC4/GC46733.png)
-
GC33577
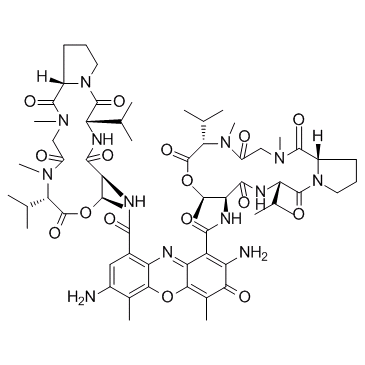
7-Aminoactinomycin D (7-AAD)
7-Aminoactinomycin D (7-AAD) (7-AAD) a fluorescent DNA stain, is a potent RNA polymerase inhibitor.
-
GC65408
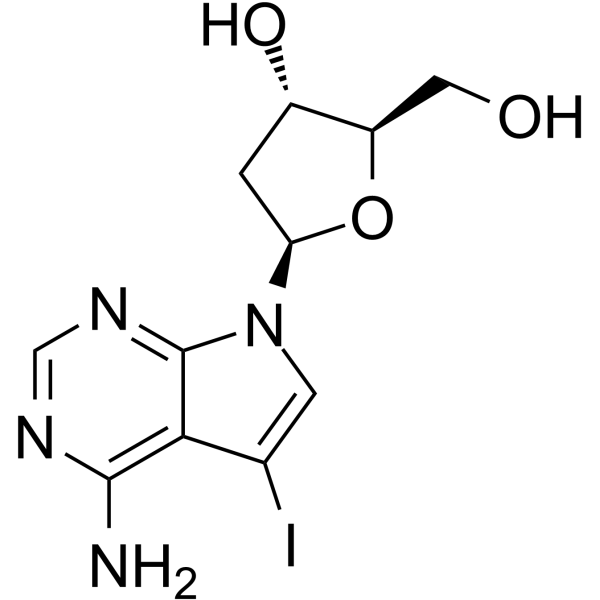
7-Deaza-2'-deoxy-7-iodoadenosine
7-Deaza-2'-deoxy-7-iodoadenosine is a modified oligonucleotide containing 7-Deazaadenine.
-
GC66060
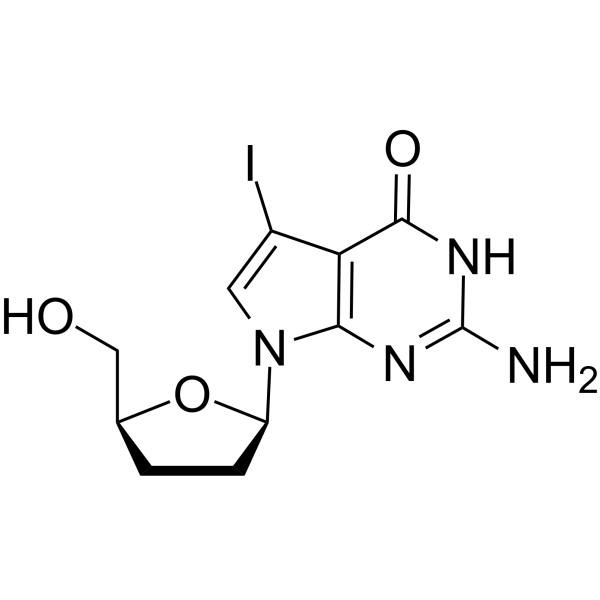
7-Iodo-2',3'-dideoxy-7-deaza-guanosine
7-Iodo-2',3'-dideoxy-7-deaza-guanosine is a dideoxynucleoside that can be used in DNA synthesis and sequencing reactions.
-
GC65675
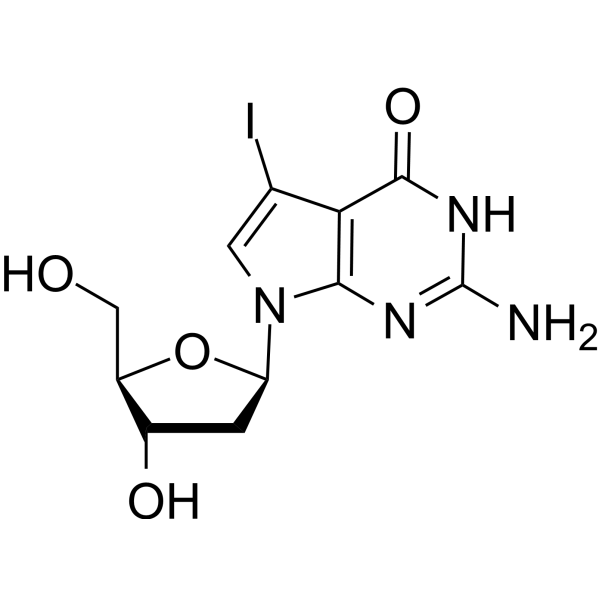
7-Iodo-7-deaza-2'-deoxyguanosine
7-Iodo-7-deaza-2'-deoxyguanosine (7-Deaza-7-Iodo-2'-deoxyguanosine) is a deoxyguanosine derivative that can be used in DNA synthesis and sequencing reactions.
-
GC30531
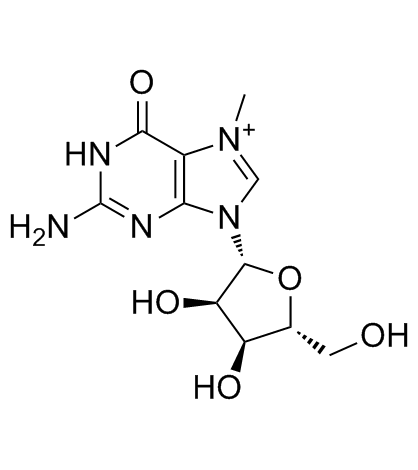
7-Methylguanosine
A nucleotide analog
-
GC49561
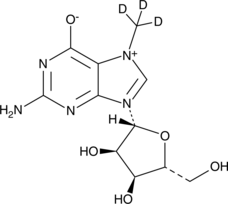
7-Methylguanosine-d3
An internal standard for the quantification of 7-methylguanosine
-
GC66058
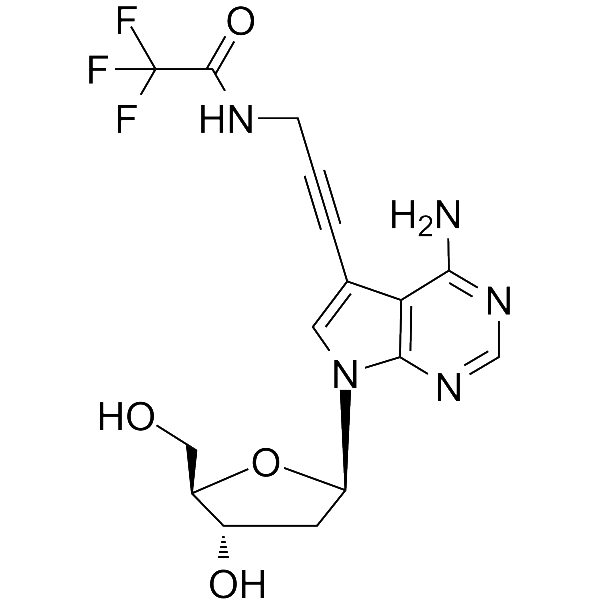
7-TFA-ap-7-Deaza-dA
7-TFA-ap-7-Deaza-dA is a modified nucleoside. 7-TFA-ap-7-Deaza-dA can be used in the synthesis of deoxyribonucleic acid or nucleic acid.
-
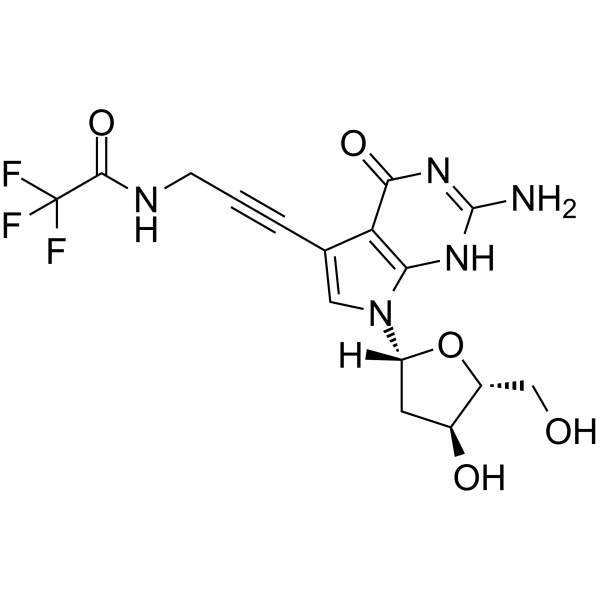
GC65525
7-TFA-ap-7-Deaza-dG
5'-O-TBDMS-dG is a modified nucleoside.
-
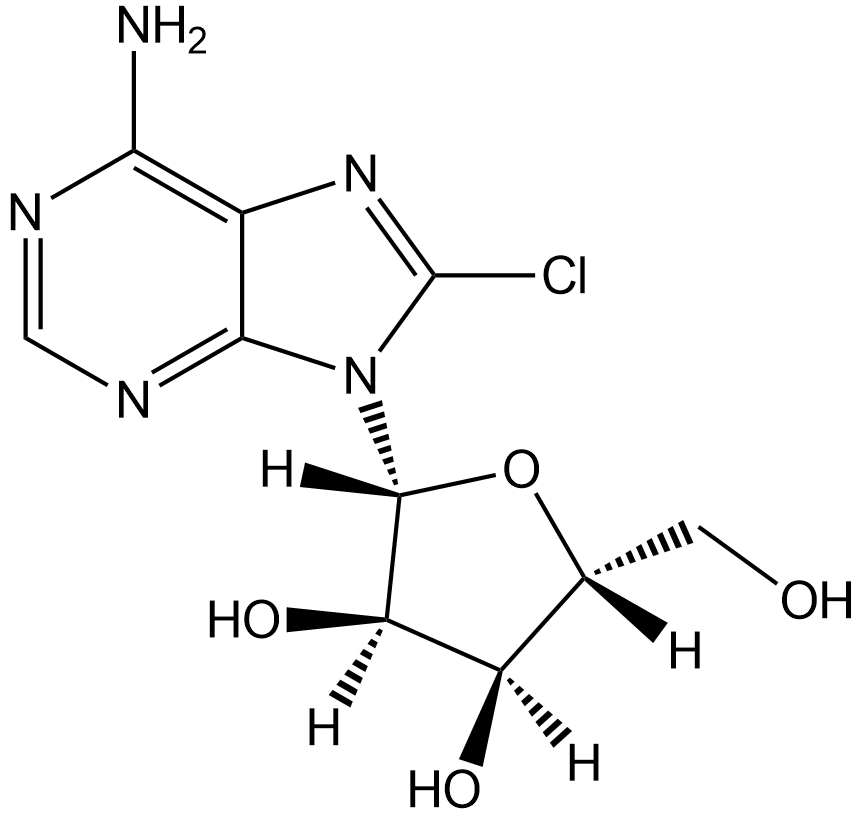
GC12777
8-Chloroadenosine
Nucleoside analog, inhibits RNA synthesis
-
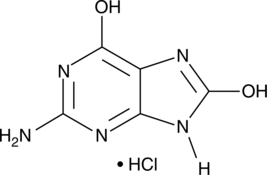
GC42627
8-Hydroxyguanine (hydrochloride)
8-Hydroxyguanine is produced by oxidative degradation of DNA by hydroxyl radical.
-
GC18426
8-Nitroguanine
8-Nitroguanine is a nitrative guanine derivative formed by oxidative damage to the guanine base in DNA by reactive nitrogen species (RNS) during inflammation and in vitro by reaction of DNA with peroxynitrite and other RNS reagents.
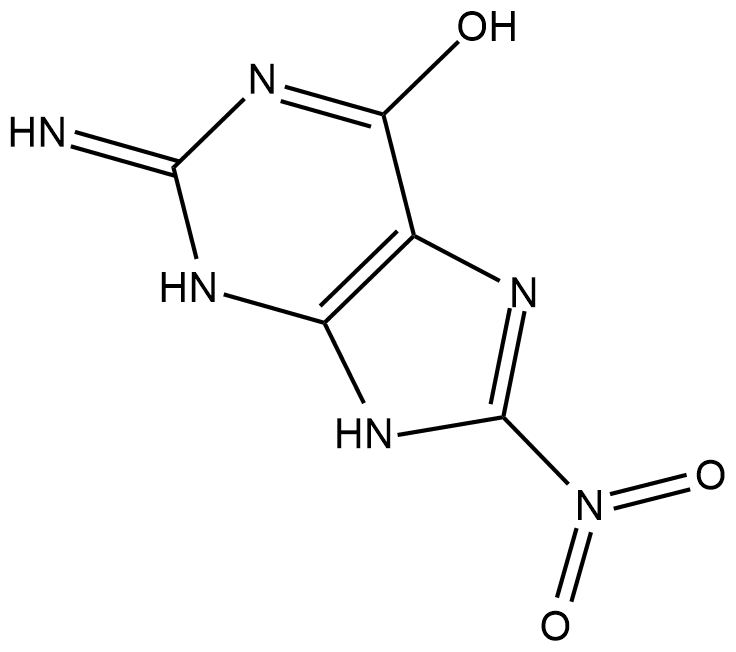
-
GC41643
9(Z),11(E),13(E)-Octadecatrienoic Acid
9(Z),11(E),13(E)-Octadecatrienoic Acid (α-ESA) is a conjugated polyunsaturated fatty acid commonly found in plant seed oil.

-
GC11903
9-amino Camptothecin
9-amino Camptothecin (9-amino-20(S)-camptothecin) is a topoisomerase I inhibitor with potent anticancer activity.
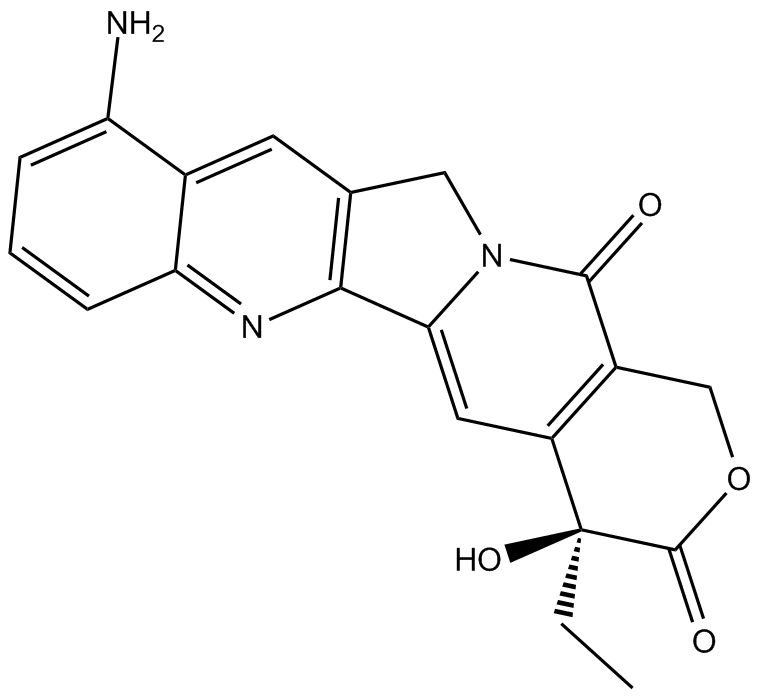
-
GC45367
9-Hydroxyellipticine (hydrochloride)
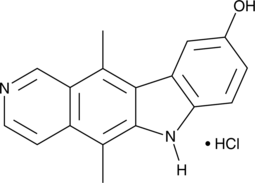
-
GN10035
9-Methoxycamptothecin
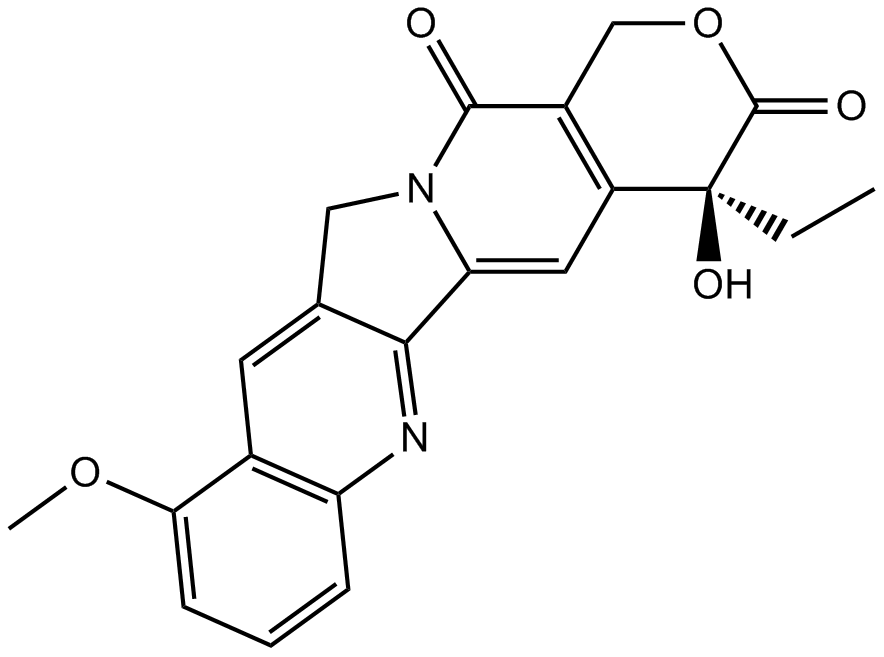
-
GC48840
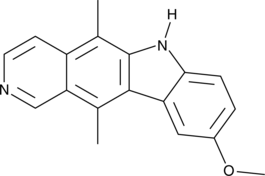
9-Methoxyellipticine
An alkaloid with anticancer activity
-
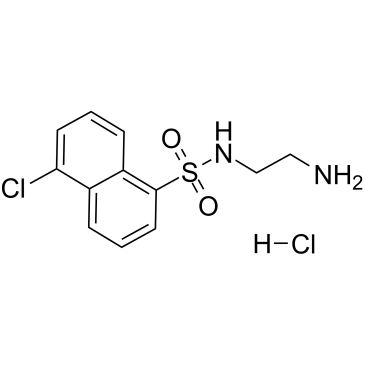
GC60037
A-3 hydrochloride
A-3 hydrochloride is a potent, cell-permeable, reversible, ATP-competitive non-selective antagonist of various kinases.
-
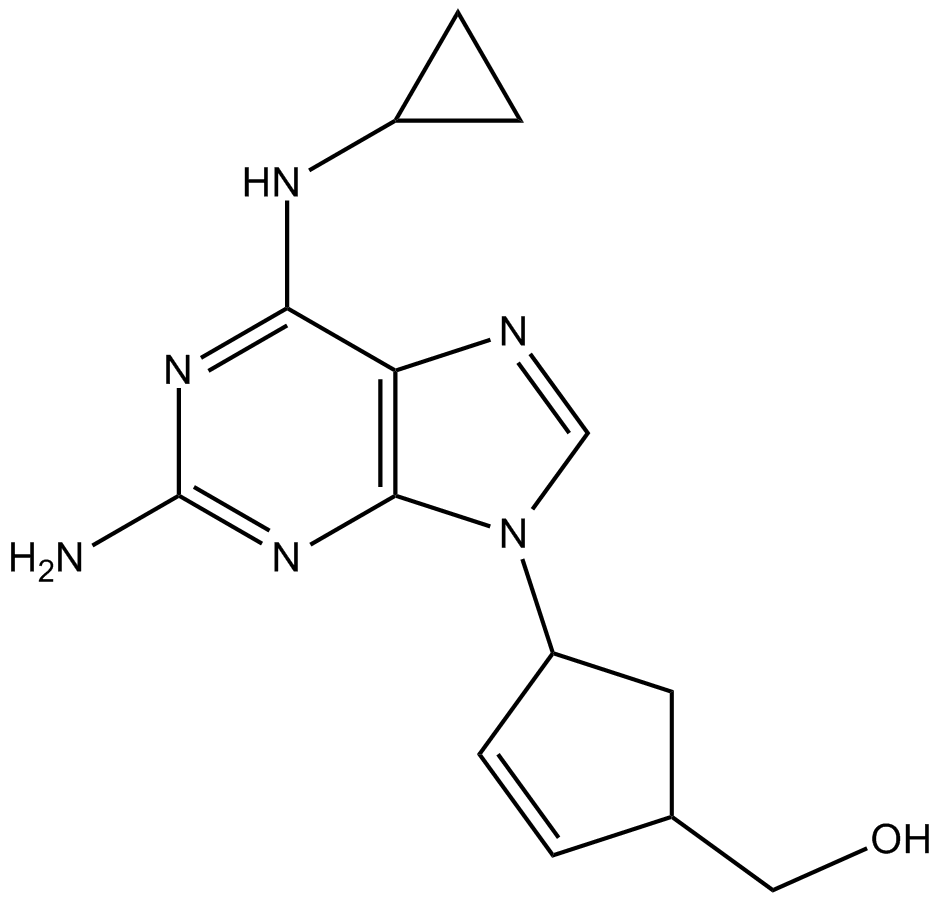
GC13805
Abacavir
-
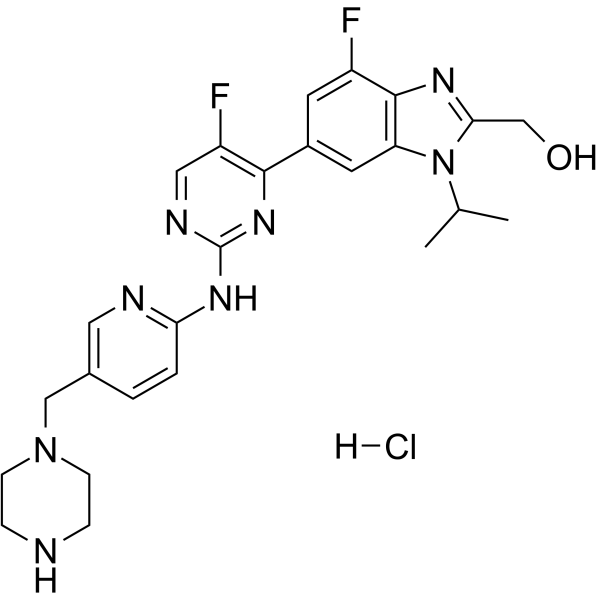
GC63728
Abemaciclib metabolite M18 hydrochloride
Abemaciclib metabolite M18 (LSN3106729) hydrochloride, the metabolite of Abemaciclib, is a CDK inhibitor with antitumor activity. Abemaciclib metabolite M18 hydrochloride and a CRBN ligand have been used to design PROTAC CDK4/6 degrader.
-
GC38749
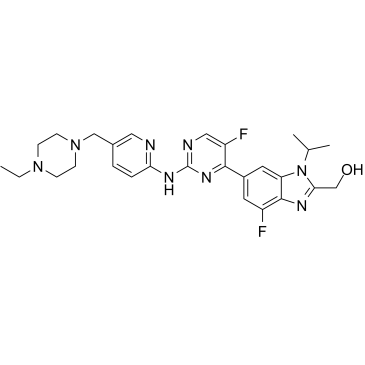
Abemaciclib metabolite M20
Abemaciclib metabolite M20 (LSN3106726), the active metabolite of Abemaciclib, is a selective CDK4/6 inhibitor for the treatment of cancer.
-
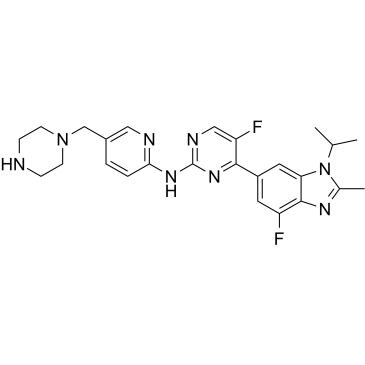
GC35218
Abemaciclib Metabolites M2
Abemaciclib Metabolites M2 (LSN2839567) is a metabolite of Abemaciclib, acts as a potent CDK4 and CDK6 inhibitor, with IC50s of 1.2 and 1.3 nM, respectively. Anti-cancer activity.
-
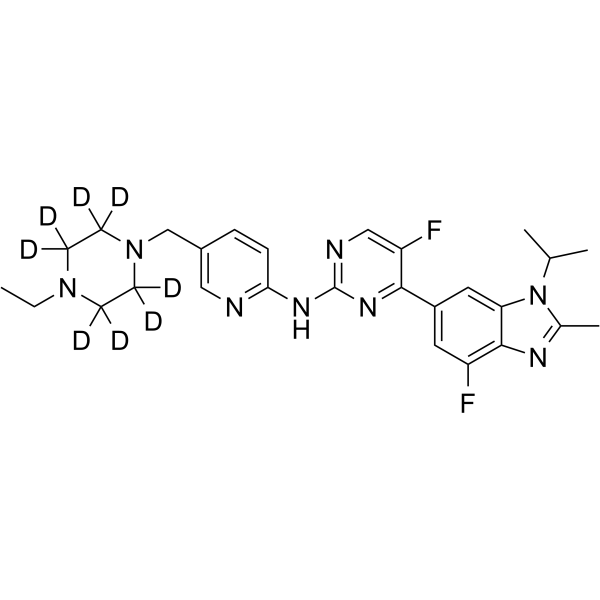
GC64280
Abemaciclib-d8
Abemaciclib-d8 (LY2835219-d8) is the deuterium labeled Abemaciclib. Abemaciclib (LY2835219) is a selective CDK4/6 inhibitor with IC50 values of 2 nM and 10 nM for CDK4 and CDK6, respectively.
-
GA20605
Ac-Lys-AMC
Ac-Lys-AMC is cleaved by trypsin.

-
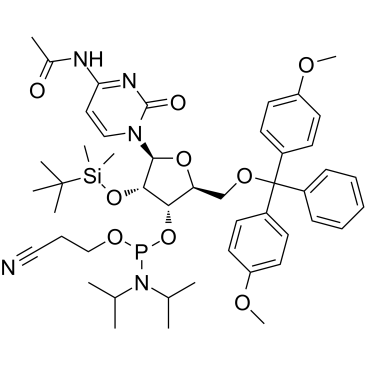
GC61763
Ac-rC Phosphoramidite
Ac-rC Phosphoramidite is used for the oligoribonucleotide phosphorodithioate modification (PS2-RNA).
-
GC34090
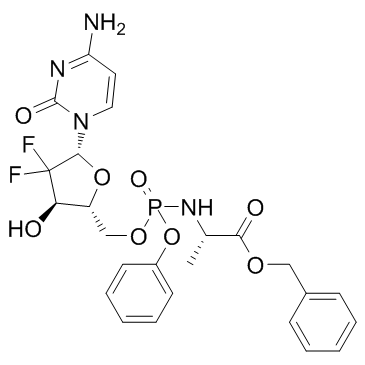
Acelarin (NUC-1031)
Acelarin (NUC-1031) (NUC-1031) is a ProTide transformation and enhancement of the widely-used nucleoside analogue, gemcitabine.
-
GC35238
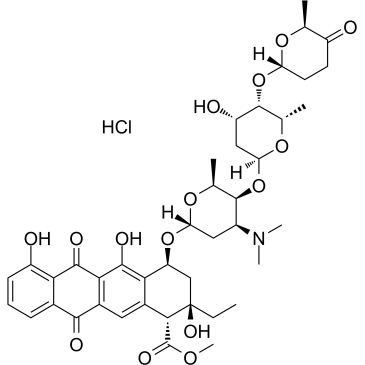
Aclacinomycin A hydrochloride
Aclacinomycin A hydrochloride (Aclarubicin hydrochloride), a fluorescent molecule and the first described non-peptidic inhibitor showing discrete specificity for the CTRL (chymotrypsin-like) activity of the 20S proteasome. Aclacinomycin A hydrochloride is also a dual inhibitor of topoisomerase I and II. An effective anthracycline chemotherapeutic agent for hematologic cancers and solid tumors.
-
GC49804
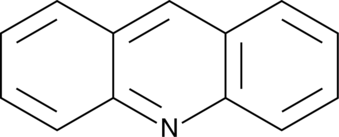
Acridine
An azaarene
-
GC16866
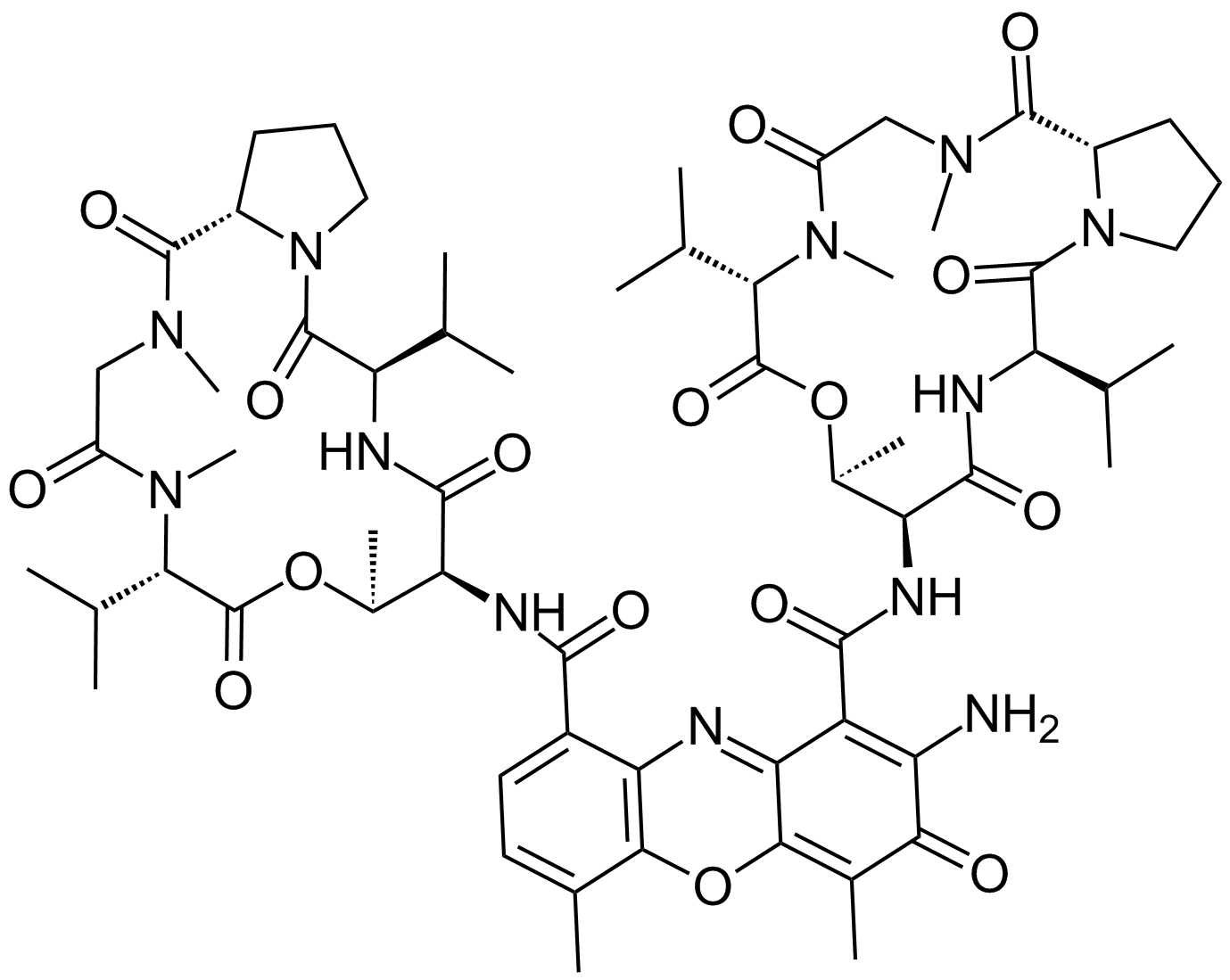
Actinomycin D
A DNA-interacting transcription blocker with anti-cancer activity
-
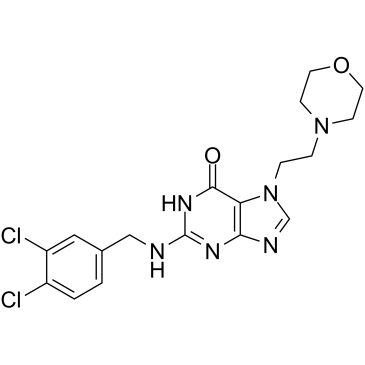
GC35247
ACX-362E
ACX-362E (ACX-362E) is a first-in-class, orally active DNA polymerase IIIC (pol IIIC) inhibitor, with a Ki of 0.325 μM for the DNA pol IIIC from C.
-
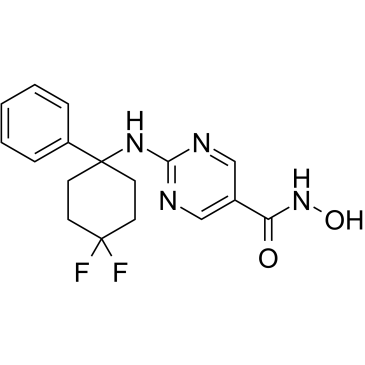
GC34458
ACY-1083
ACY-1083 is a selective and brain-penetrating HDAC6 inhibitor with an IC50 of 3 nM and is 260-fold more selective for HDAC6 than all other classes of HDAC isoforms. ACY-1083 effectively reverses chemotherapy-induced peripheral neuropathy.
-
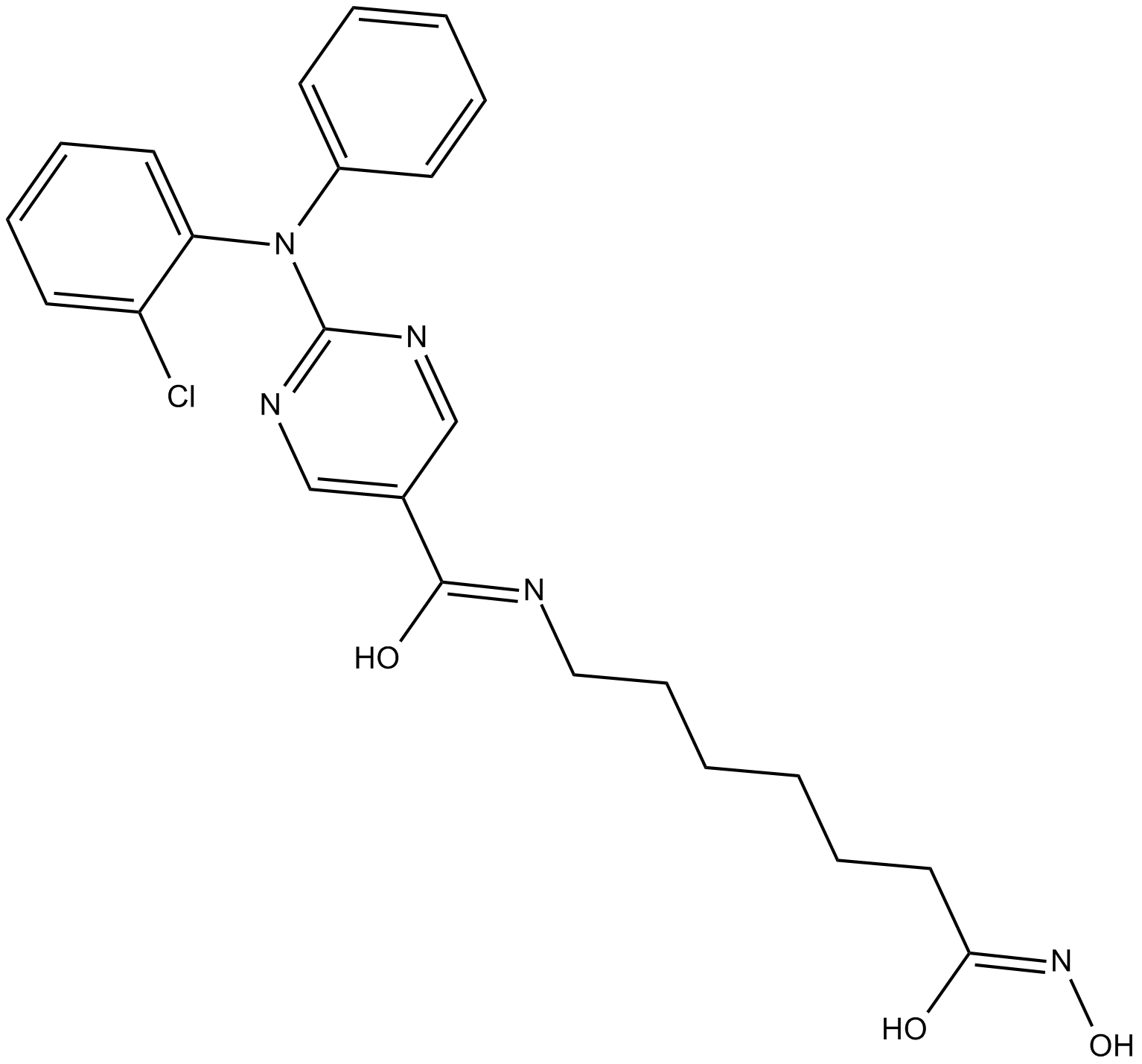
GC10417
ACY-241
ACY-241 (ACY241) is a second generation potent, orally active and high-selective HDAC6 inhibitor with an IC50 of 2.6 nM (IC50s of 35 nM, 45 nM, 46 nM and 137 nM for HDAC1, HDAC2, HDAC3 and HDAC8, respectively). ACY-241 has anticancer effects.
-
GC19020
ACY-738
ACY-738 is a potent, selective and orally-bioavailable HDAC6 inhibitor, with an IC50s of 1.7 nM; ACY-738 also inhibits HDAC1, HDAC2, and HDAC3, with IC50s of 94, 128, and 218 nM.



