DNA Damage/DNA Repair
- MTH1(4)
- PARP(84)
- ATM/ATR(30)
- DNA Alkylating(20)
- DNA Ligases(3)
- DNA Methyltransferase(25)
- DNA-PK(30)
- HDAC(136)
- Nucleoside Antimetabolite/Analogue(140)
- Telomerase(17)
- Topoisomerase(151)
- tankyrase(5)
- Antifolate(38)
- CDK(250)
- Checkpoint Kinase (Chk)(31)
- CRISPR/Cas9(9)
- Deubiquitinase(72)
- DNA Alkylator/Crosslinker(71)
- DNA/RNA Synthesis(400)
- Eukaryotic Initiation Factor (eIF)(23)
- IRE1(23)
- LIM Kinase (LIMK)(9)
- TOPK(5)
- Casein Kinase(61)
- DNA Intercalating Agents(7)
- DNA/RNA Oxidative Damage(12)
Products for DNA Damage/DNA Repair
- Cat.No. Product Name Information
-
GC30782
ACY-775
An HDAC6 inhibitor
-
GC30526
ACY-957
ACY-957 is an orally active and selective inhibitor of HDAC1 and HDAC2, with IC50s of 7 nM, 18 nM, and 1300 nM against HDAC1/2/3, respectively, and shows no inhibition on HDAC4/5/6/7/8/9.
-
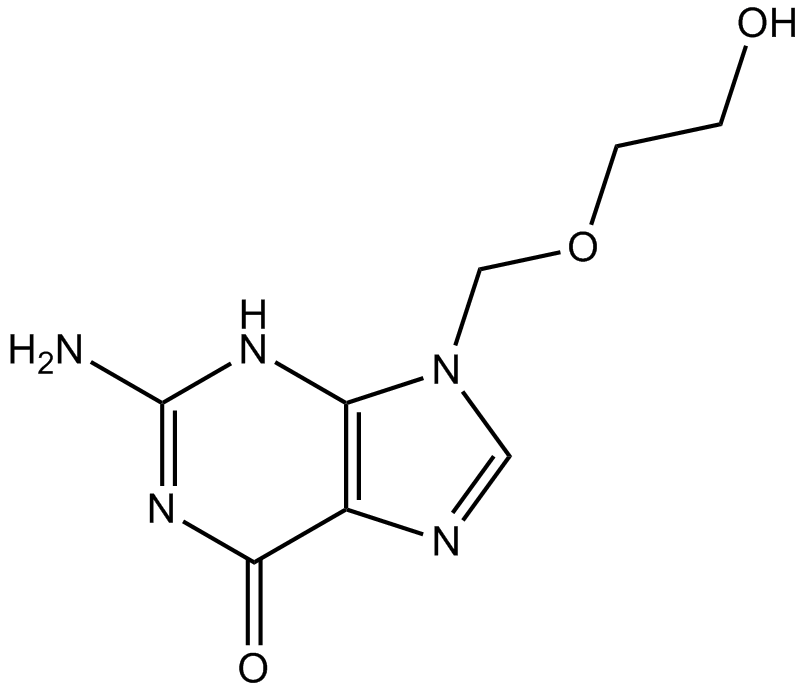
GC17186
Acyclovir
Antiviral agent
-
GC13432
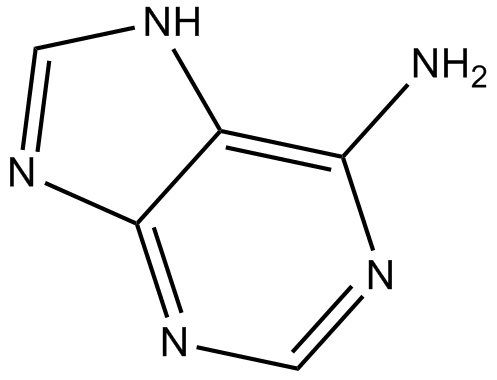
Adenine
High affinity adenine receptor agonist
-
GC11825
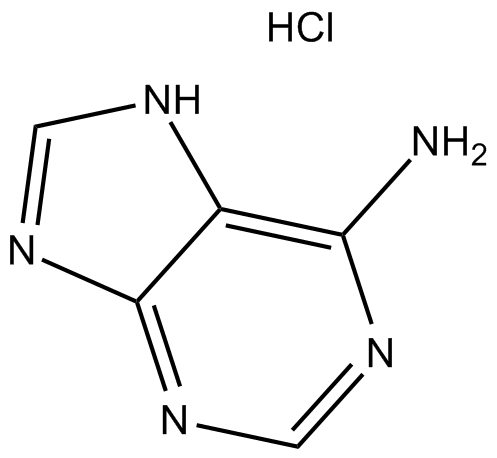
Adenine HCl
Adenine HCl (6-Aminopurine hydrochloride), a purine, is one of the four nucleobases in the nucleic acid of DNA. Adenine HCl acts as a chemical component of DNA and RNA. Adenine HCl also plays an important role in biochemistry involved in cellular respiration, the form of both ATP and the cofactors (NAD and FAD), and protein synthesis.
-
GC17278
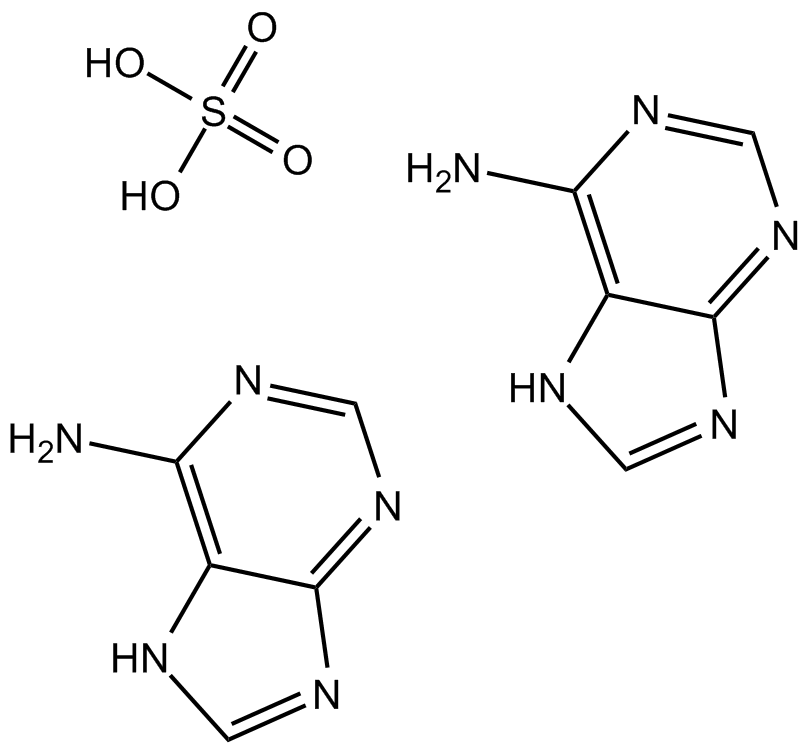
Adenine sulfate
Adenine sulfate (6-Aminopurine hemisulfate), a purine, is one of the four nucleobases in the nucleic acid of DNA. Adenine sulfate acts as a chemical component of DNA and RNA. Adenine sulfate also plays an important role in biochemistry involved in cellular respiration, the form of both ATP and the cofactors (NAD and FAD), and protein synthesis.
-
GC67946
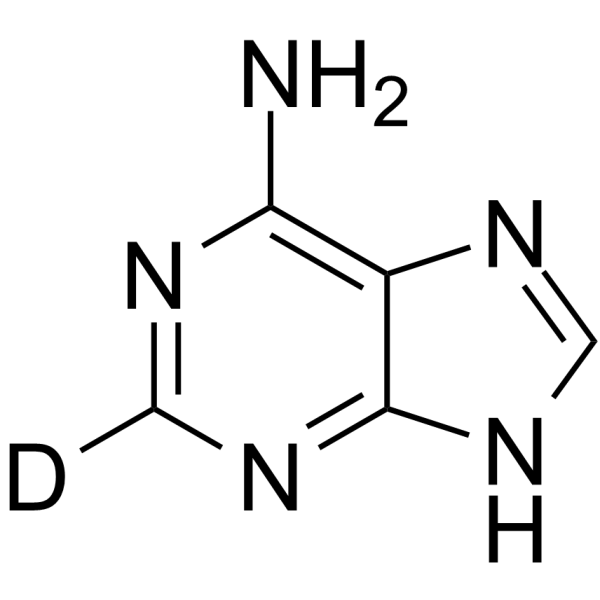
Adenine-d1
-
GC14106
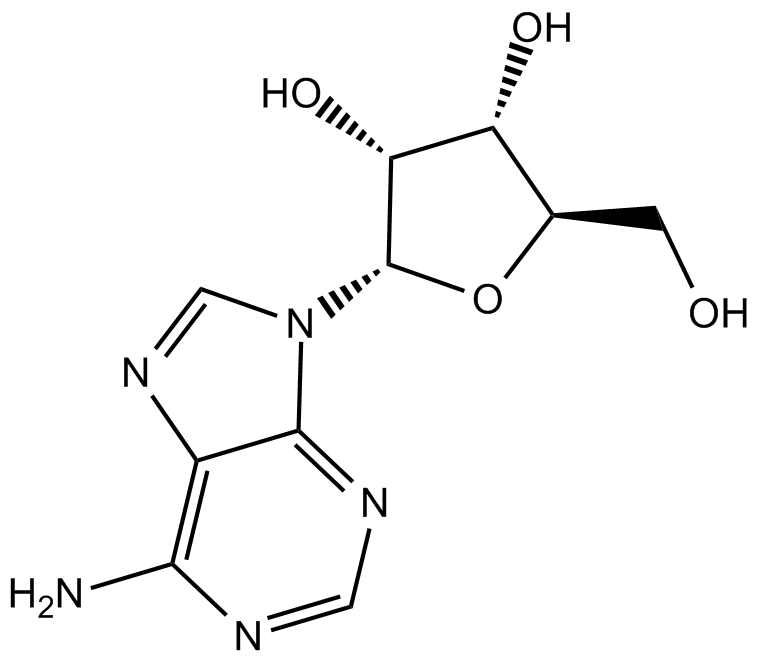
Adenosine
nucleoside
-
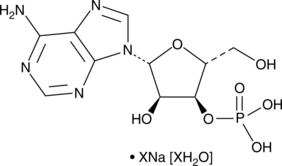
GC49004
Adenosine 3’-monophosphate (sodium salt hydrate)
A nucleotide
-
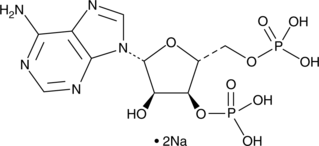
GC42732
Adenosine 3',5'-diphosphate (sodium salt)
Adenosine 3',5'-diphosphate (sodium salt) is an hydroxysteroid sulfotransferases inhibitor.
-
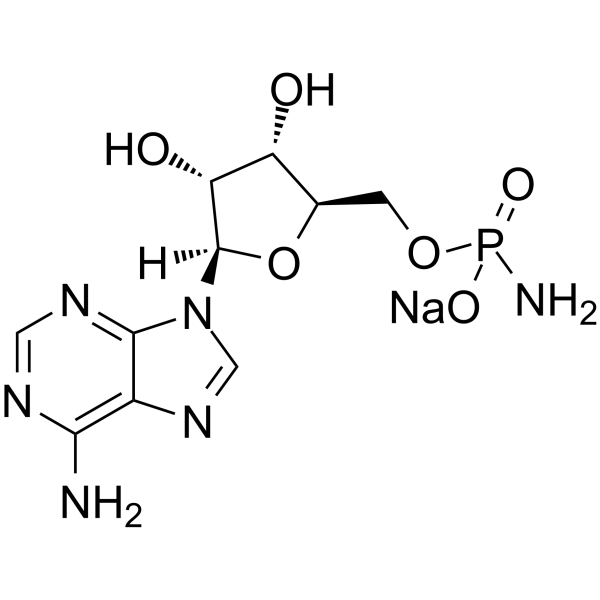
GC65462
Adenosine 5′-monophosphoramidate sodium
Adenosine 5′-monophosphoramidate sodium is an adenosine derivative and can be used as an intermediate for nucleotide synthesis.
-
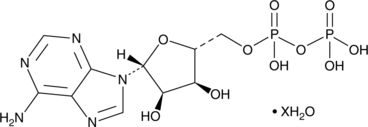
GC49285
Adenosine 5’-methylenediphosphate (hydrate)
An inhibitor of ecto-5’-nucleotidase
-
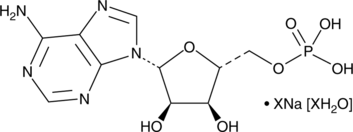
GC42733
Adenosine 5'-monophosphate (sodium salt hydrate)
Adenosine 5'-monophosphate (AMP) is a central nucleotide with functions in metabolism and cell signaling.
-
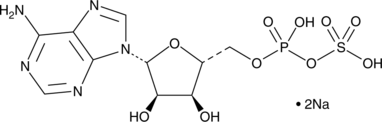
GC42734
Adenosine 5'-phosphosulfate (sodium salt)
Adenosine 5'-phosphosulfate is an ATP and sulfate competitive inhibitor of ATP sulfurylase in humans, S.
-
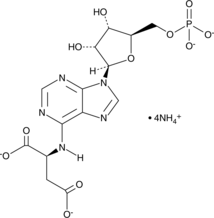
GC49231
Adenylosuccinic Acid (ammonium salt)
A purine nucleotide and an intermediate in the purine nucleotide cycle
-
GC65330
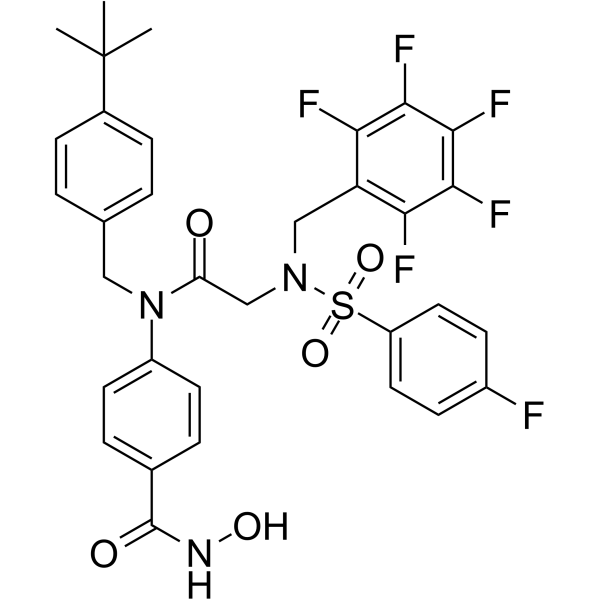
AES-135
AES-135, a hydroxamic acid-based pan-HDAC inhibitor, prolongs survival in an orthotopic mouse model of pancreatic cancer. AES-135 inhibits HDAC3, HDAC6, HDAC8, and HDAC11 with IC50s ranging from 190-1100 nM.
-
GC46810
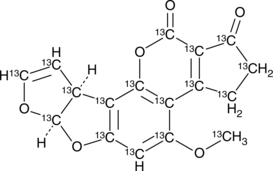
Aflatoxin B1-13C17
An internal standard for the quantification of aflatoxin B1
-
GC62470
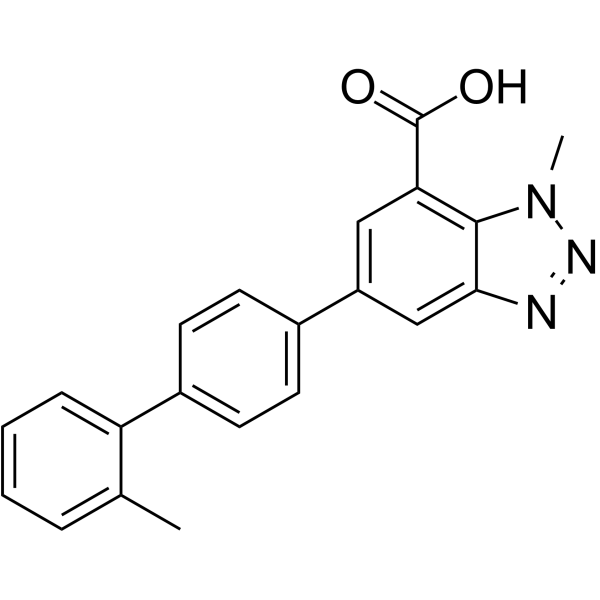
AG-636
AG-636 is a potent, reversible, selective and orally active dihydroorotate dehydrogenase (DHODH) inhibitor with an IC50 of 17 nM. AG-636 has strong anticancer effects.
-
GC42765
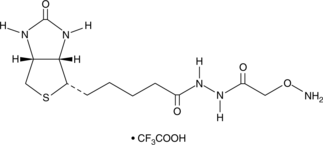
Aldehyde Reactive Probe (trifluoroacetate salt)
DNA is continually damaged by endogenous and environmental agents leading to the formation of abasic (apurinic/apyrimidinic, AP) sites that are disruptive to DNA synthesis.
-
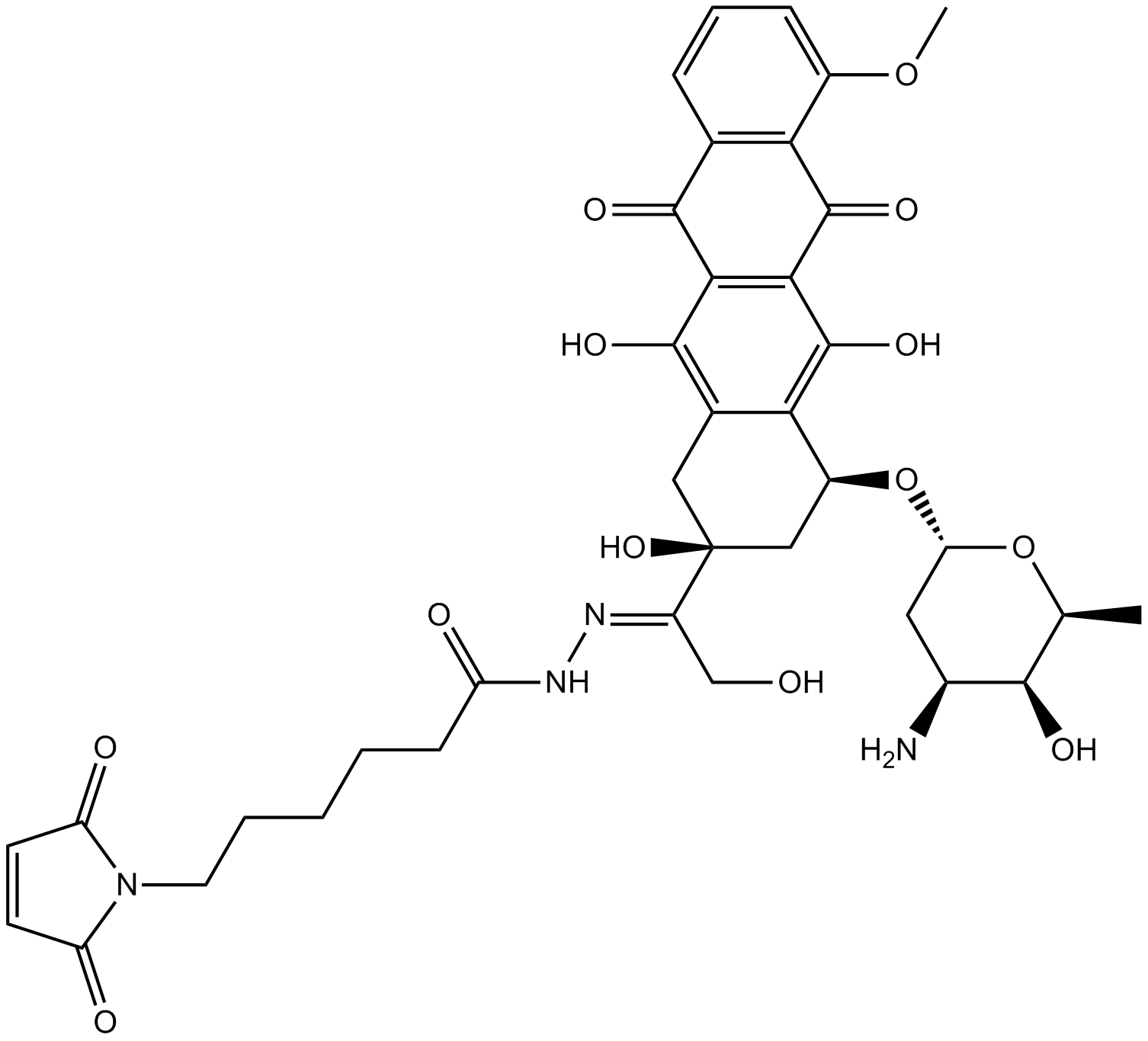
GC14858
Aldoxorubicin
An albumin-binding prodrug of doxorubicin
-
GC15841
Alsterpaullone
CDKs and GSK3β inhibitor
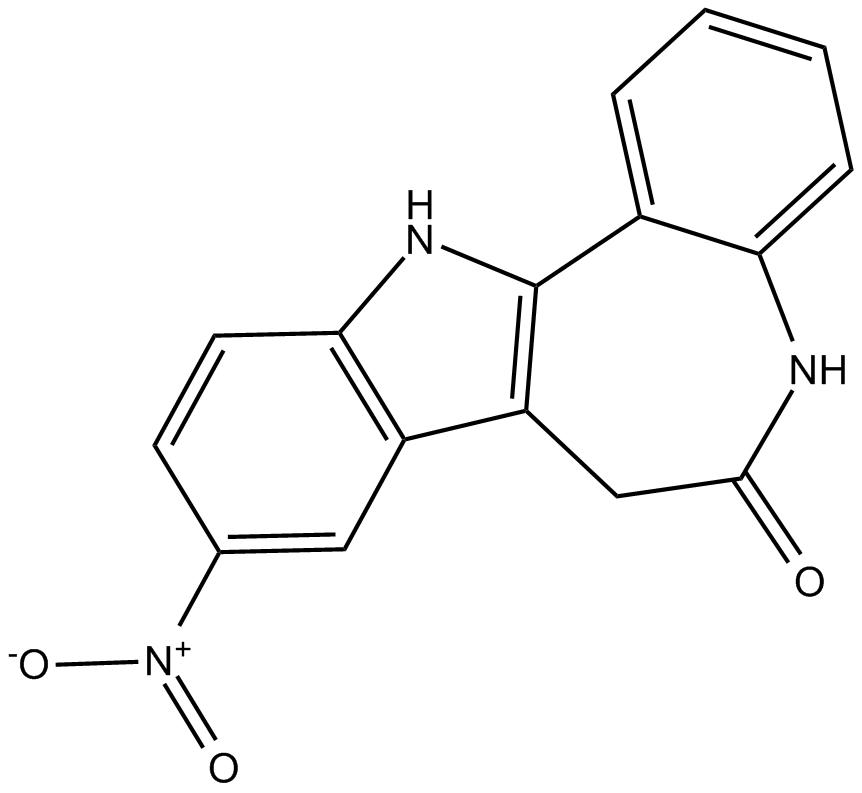
-
GC67984
Alteminostat
-
GC10779
Alternariol
A mycotoxin with antifungal and phytotoxic activity
-
GC18437
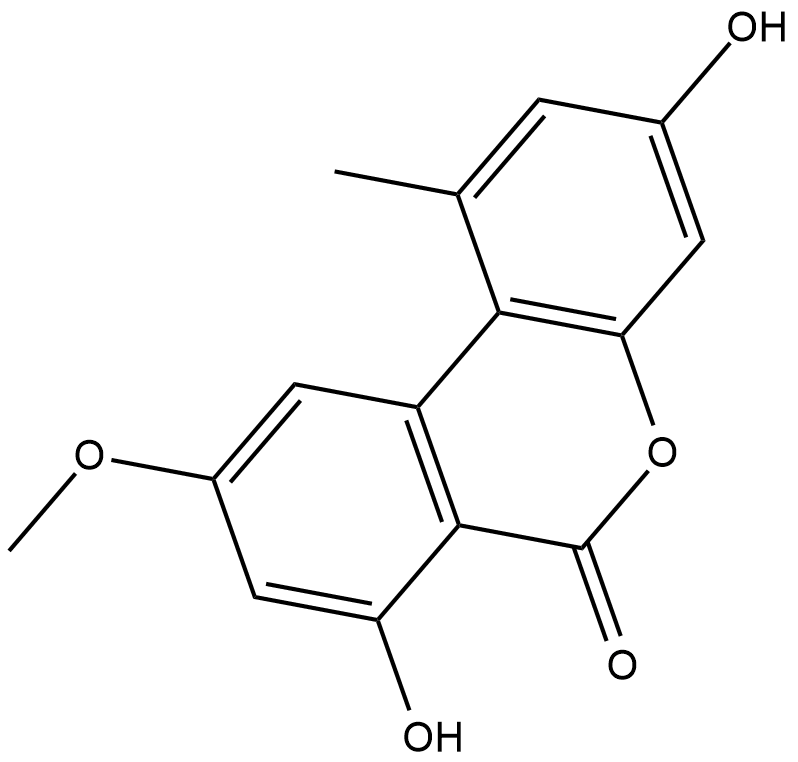
Alternariol monomethyl ether
Alternariol monomethyl ether, isolated from the roots of Anthocleista djalonensis (Loganiaceae), is an important taxonomic marker of the plant species.
-
GC49039
Althiomycin
A thiazole antibiotic
-
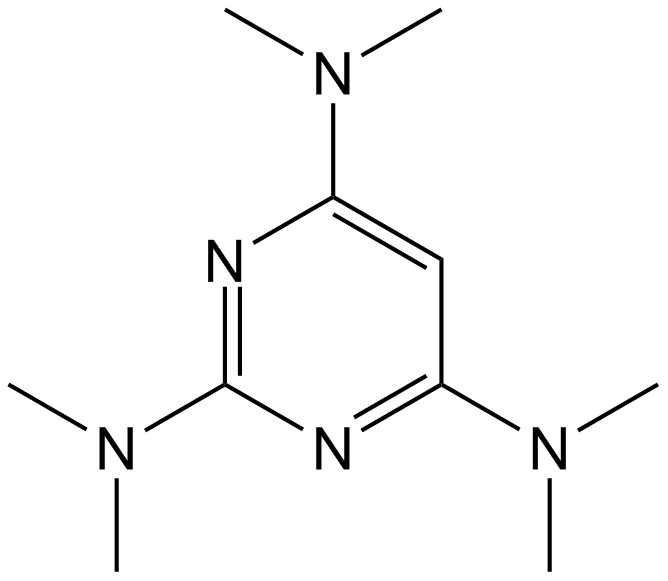
GC14564
Altretamine
Antineoplastic agent
-
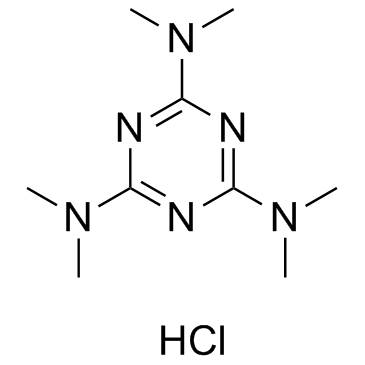
GC35310
Altretamine hydrochloride
Altretamine hydrochloride is an alkylating antineoplastic agent.
-
GC64638
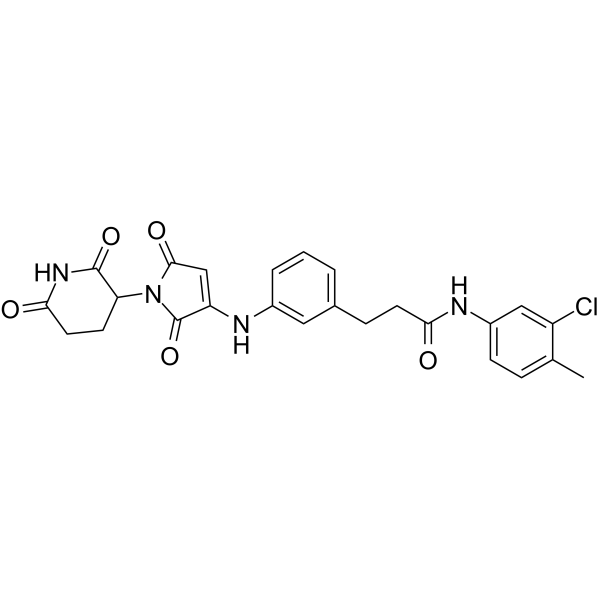
ALV1
ALV1 is a potent Ikaros and Helios degrader.
-
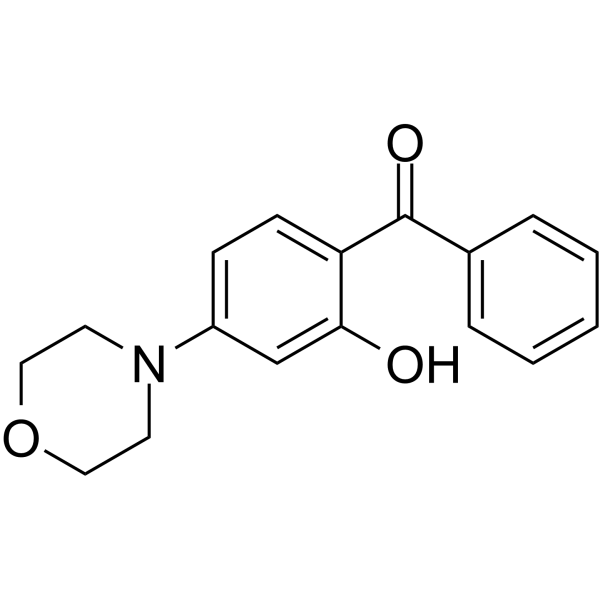
GC64818
AMA-37
AMA-37, an Arylmorpholine analog, is ATP-competitive DNA-PK inhibitor, with IC50 values of 0.27 μM (DNA-PK), 32 μM (p110α), 3.7 μM (p110β), and 22 μM (p110γ), respectively.
-
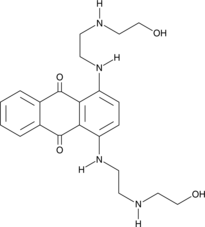
GC42783
Ametantrone
Ametantrone (NSC 196473) is an antitumor agent that intercalates into DNA and induces topoisomerase II (TOP2)-mediated DNA break.
-
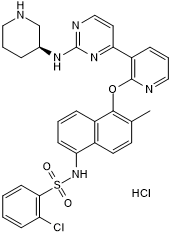
GC50366
AMG 18 hydrochloride
AMG 18 hydrochloride (AMG-18 Hydrochloride) is a mono-selective IRE1α inhibitor that allosterically attenuates IRE1α RNase activity with an IC50 of 5.9 nM.
-
GC14899
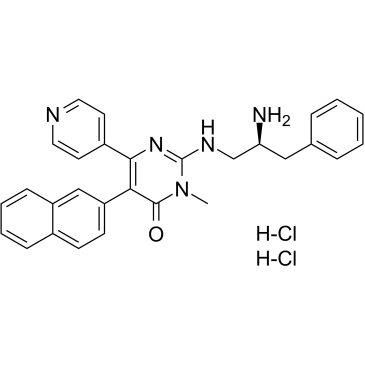
AMG 548
P38α inhibitor,potent and selective
-
GC14974
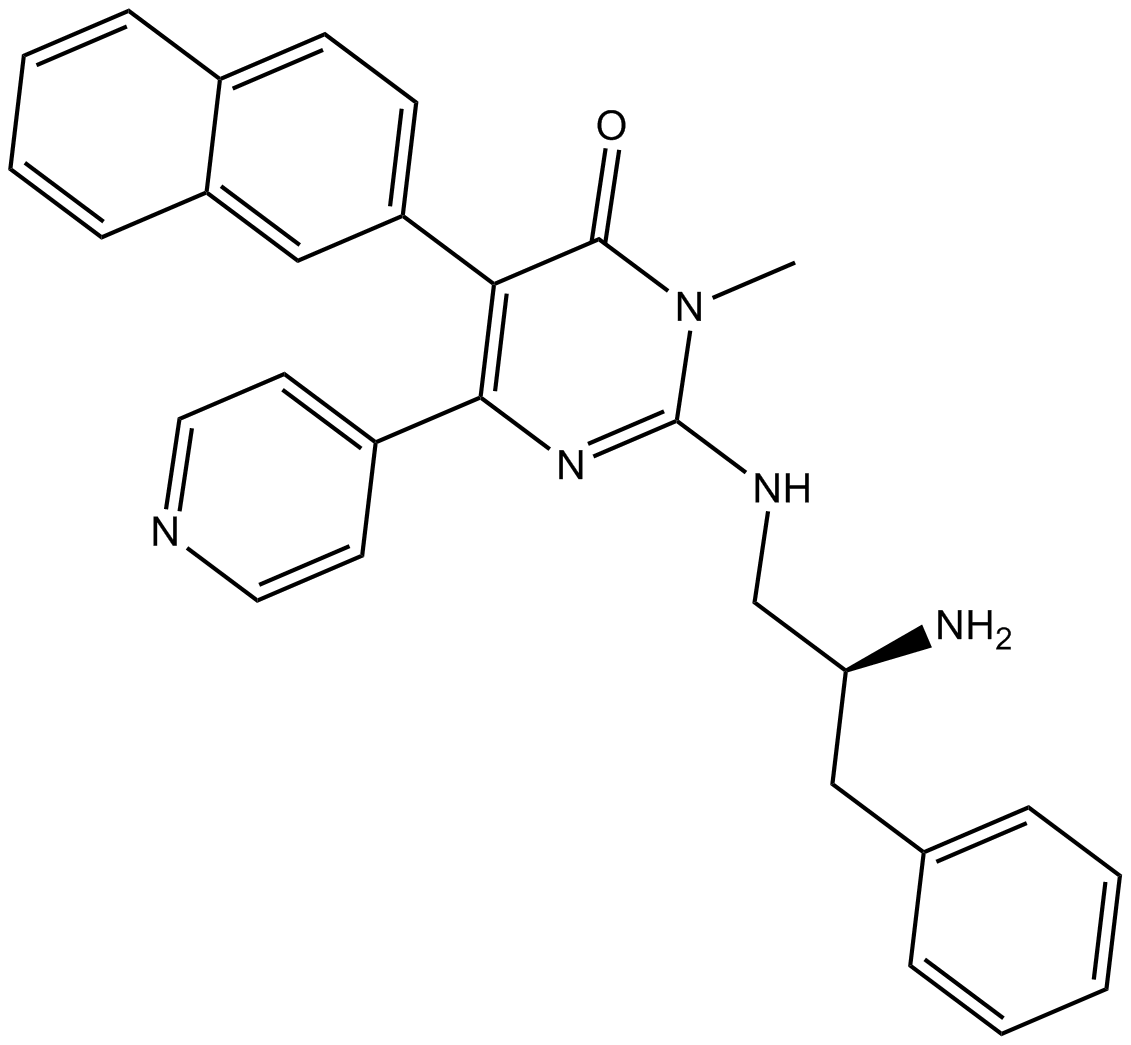
AMG 925
FLT3/CDK4 inhibitor,potent and selective
-
GC35315
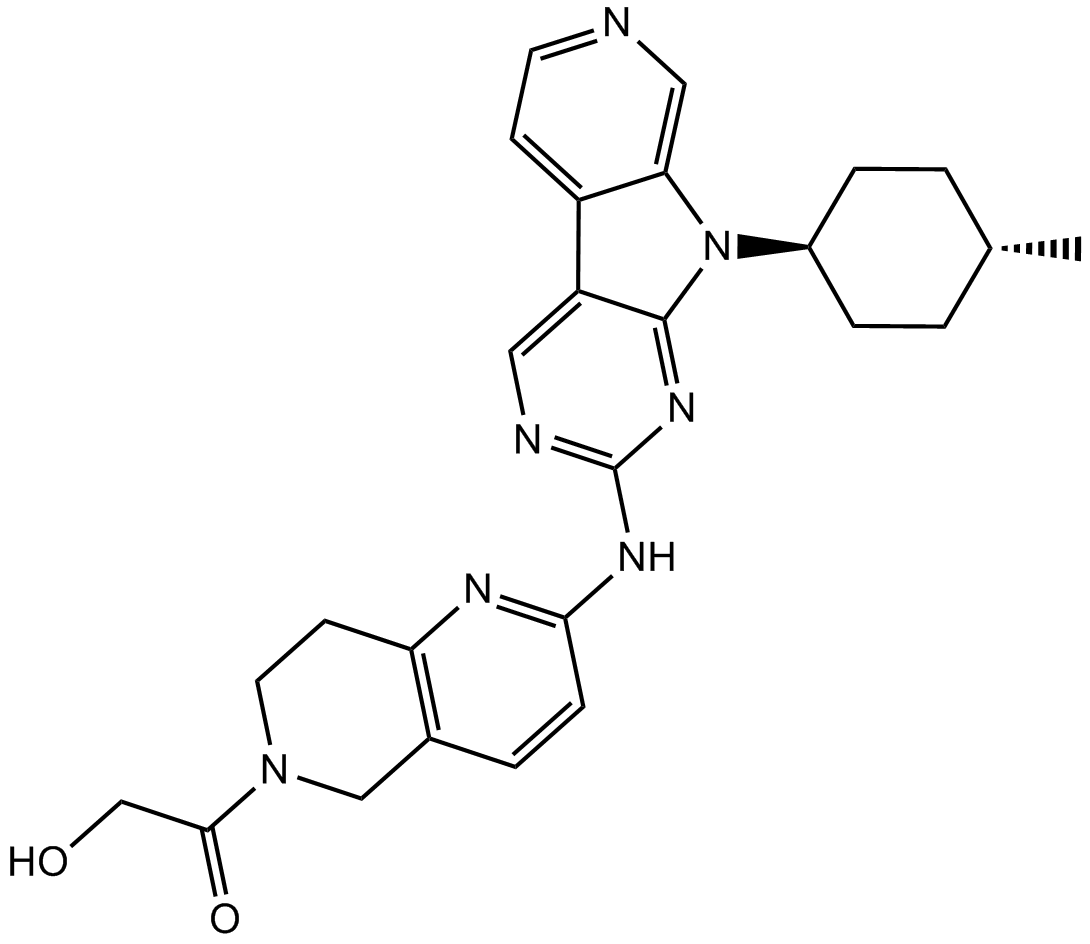
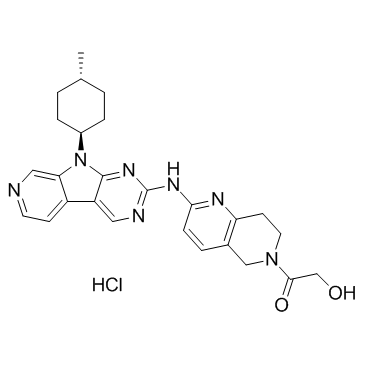
AMG 925 HCl
AMG 925 HCl is a potent, selective, and orally available FLT3/CDK4 dual inhibitor with IC50s of 2±1 nM and 3±1 nM, respectively.
-
GC38518
AMG-548 dihydrochloride
AMG-548 dihydrochloride, an orally active and selective p38α inhibitor (Ki=0.5 nM), shows slightly selective over p38β (Ki=36 nM) and >1000 fold selective against p38γ and p38δ.
-
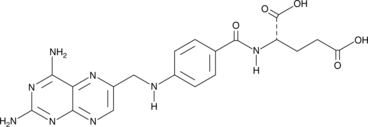
GC42789
Aminopterin
Aminopterin is a synthetic folic acid derivative whose metabolite is a competitive inhibitor of dihydrofolate reductase, which is a cofactor for nucleic acid synthesis.
-
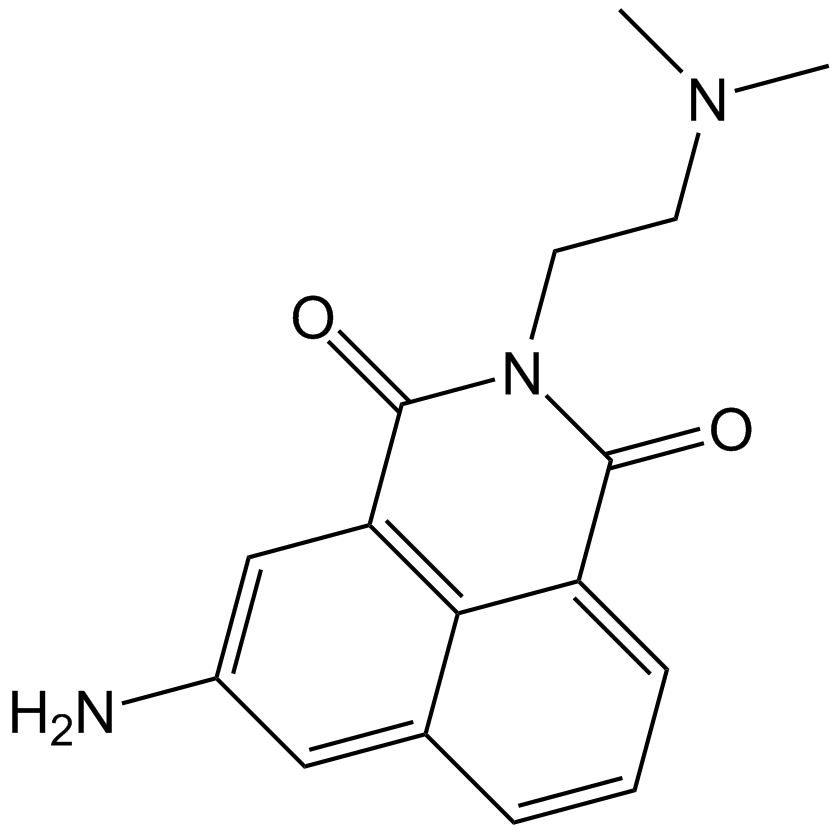
GC11019
Amonafide
DNA intercalator,Topo II inhibitor
-
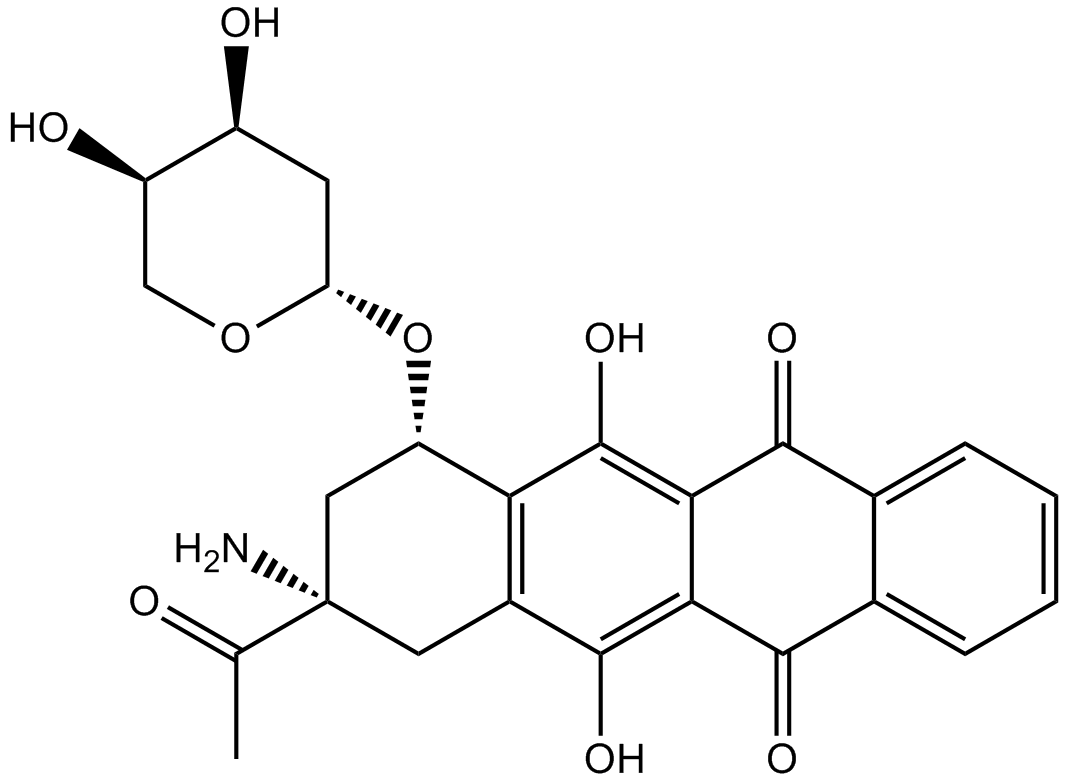
GC15644
Amrubicin
A synthetic anthracycline antibiotic. It inhibits DNA topoisomerase II. Antineoplastic.
-
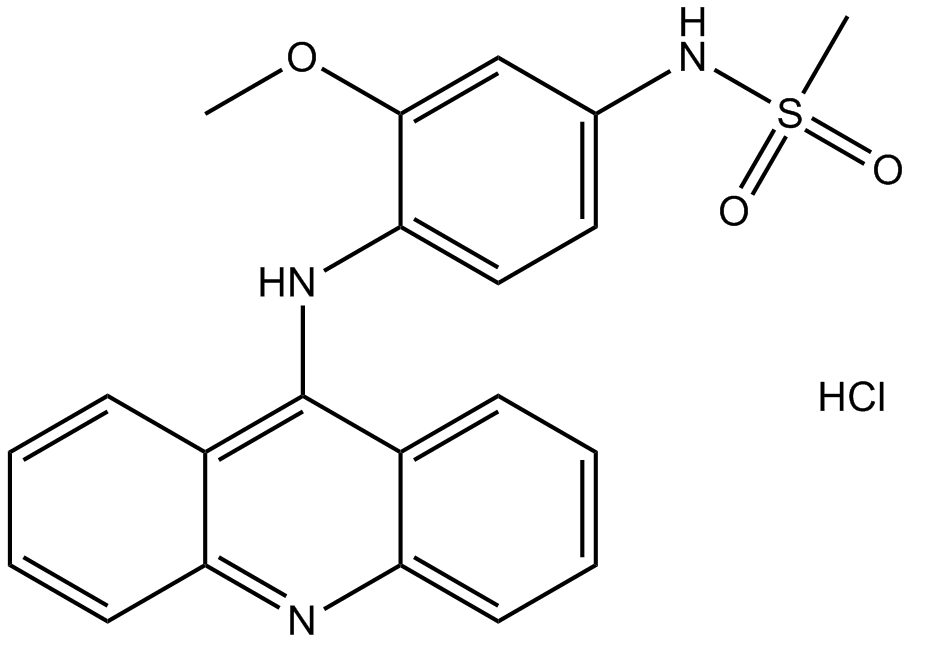
GC12326
Amsacrine
Topoisomerase 2 inhibitor

-
GC11747
Amsacrine hydrochloride
Topoisomerase 2 inhibitor
-
GC39284
ANI-7
ANI-7 is an activator of aryl hydrocarbon receptor (AhR) pathway. ANI-7 inhibits the growth of multiple cancer cells, and potently and selectively inhibits the growth of MCF-7 breast cancer cells with a GI50 of 0.56 μM. ANI-7 induces CYP1-metabolizing mono-oxygenases by activating AhR pathway, and also induces DNA damage, checkpoint Kinase 2 (Chk2) activation, S-phase cell cycle arrest, and cell death in sensitive breast cancer cell lines.
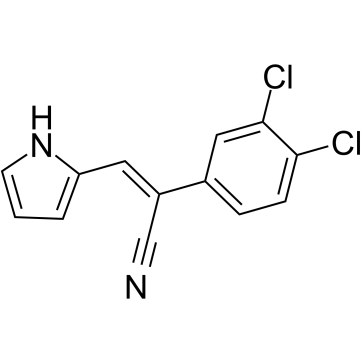
-
GC11559
Anisomycin
JNK agonist, potent and specific
-
GC68667
Anticancer agent 73
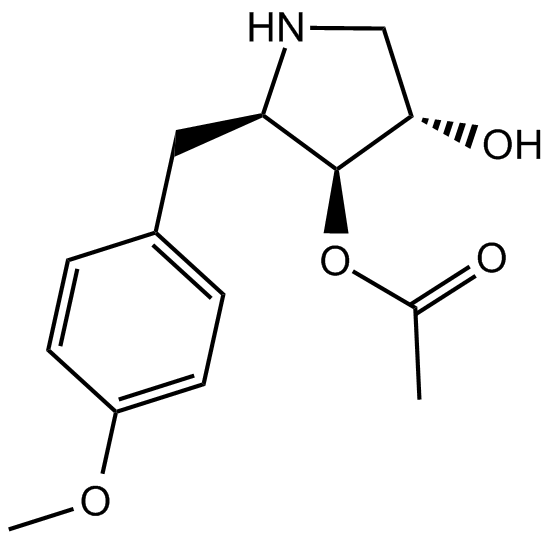
Anticancer agent 73 (compound CIB-3b) is an anticancer drug that targets TAR RNA binding protein 2 (TRBP) and disrupts its interaction with Dicer. Anticancer agent 73 can rebalance the expression profile of oncogenic or tumor-suppressive miRNAs. Anticancer agent 73 has inhibitory effects on the proliferation and metastasis of hepatocellular carcinoma (HCC) cells both in vitro and in vivo.
-
GC40032
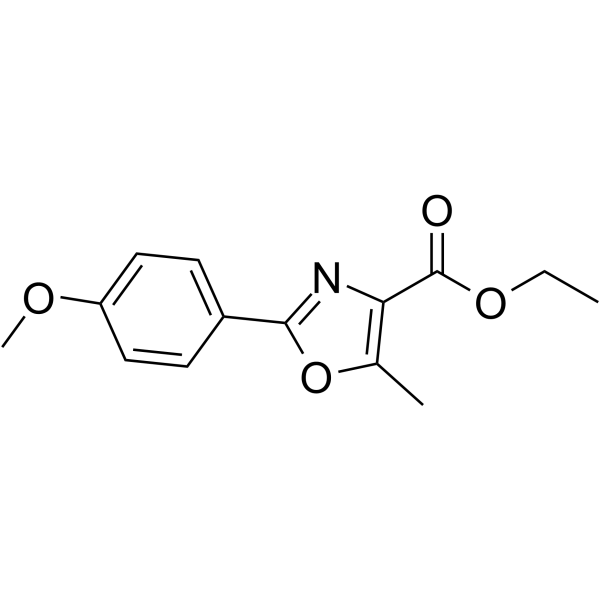
Antipain (hydrochloride)
Antipain (hydrochloride) is a protease inhibitor isolated from Actinomycetes.
-
GC68448
AOH1160
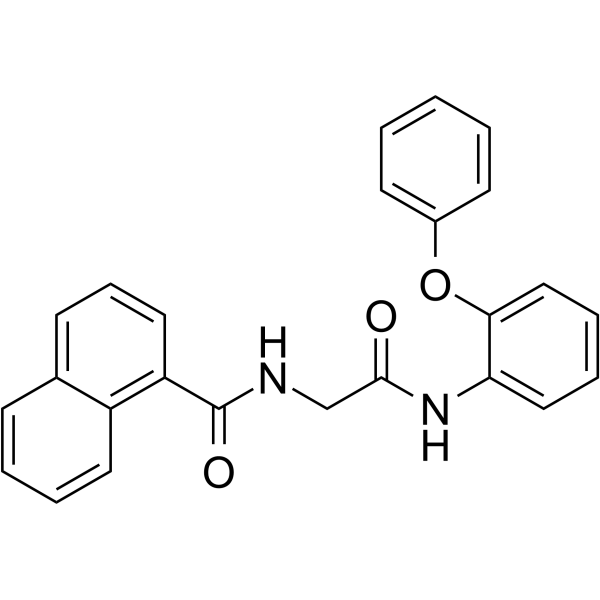
-
GC67876
APE1-IN-1

-
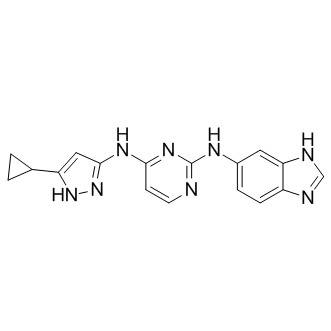
GC17139
APY29
IRE1α inhibitor
-
GC45385
Ara-G
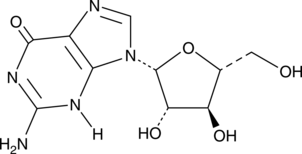
-
GC62425
ARN-21934
ARN-21934 is a potent, highly selective, blood-brain barrier (BBB) penetrant inhibitor for human topoisomerase II α over β. ARN-21934 inhibits DNA relaxation with an IC50 of 2 μM as compared to the anticancer agent Etoposide (IC50=120 μM). ARN-21934 exhibits a favorable in vivo pharmacokinetic profile and is a promising lead compound for anticancer research.
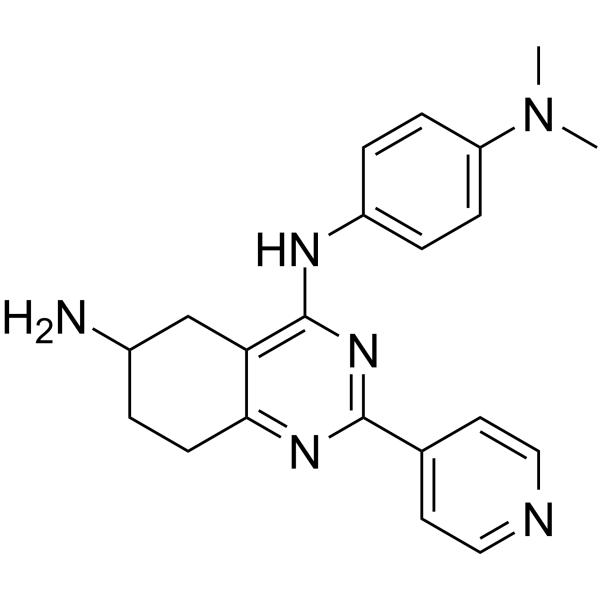
-
GC46880
ARN24139
A topoisomerase II poison
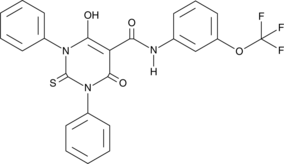
-
GC38736
AS2863619
A dual inhibitor of Cdk8 and Cdk19
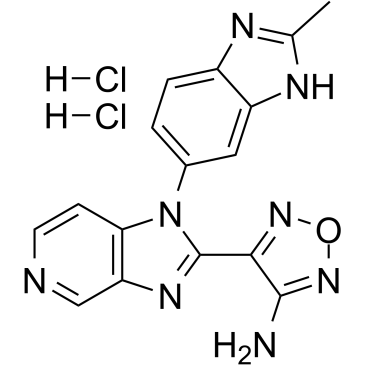
-
GC38737
AS2863619 free base
AS2863619 free base enables conversion of antigen-specific effector/memory T cells into Foxp3+ regulatory T (Treg) cells for the treatment of various immunological diseases.
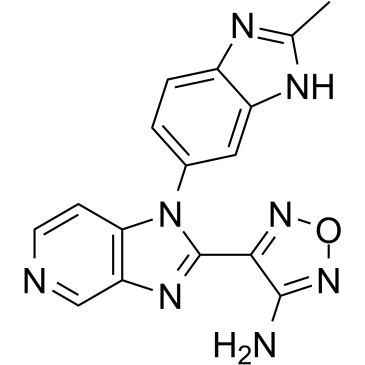
-
GC38463
ASLAN003
ASLAN003 (ASLAN003) is an orally active and potent Dihydroorotate Dehydrogenase (DHODH) inhibitor with an IC50 of 35 nM for human DHODH enzyme. ASLAN003 inhibits protein synthesis via activation of AP-1 transcription factors. ASLAN003 induces apoptosis and substantially prolongs survival in acute myeloid leukemia (AML) xenograft mice.
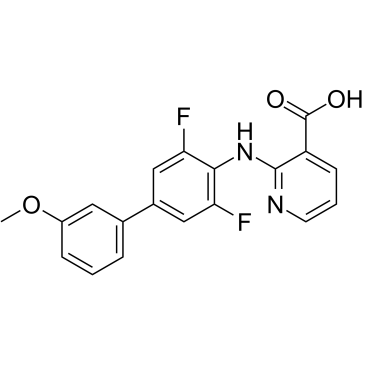
-
GC61821
AT-130
AT-130, a phenylpropenamide derivative, is a potent hepatitis B virus (HBV) replication non-nucleoside inhibitor.
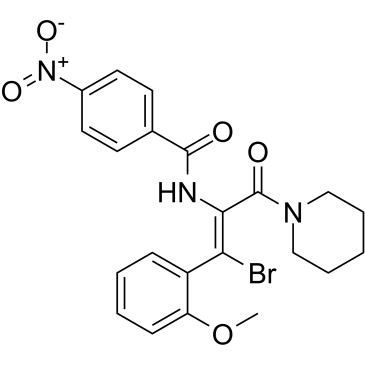
-
GC15870
AT7519
Multi-CDK inhibitor
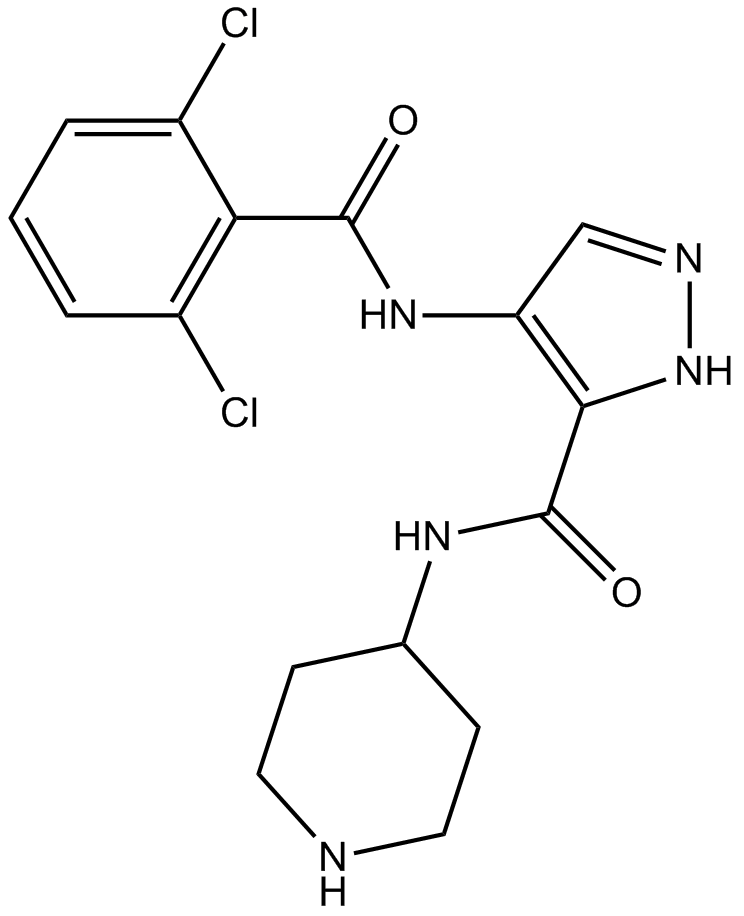
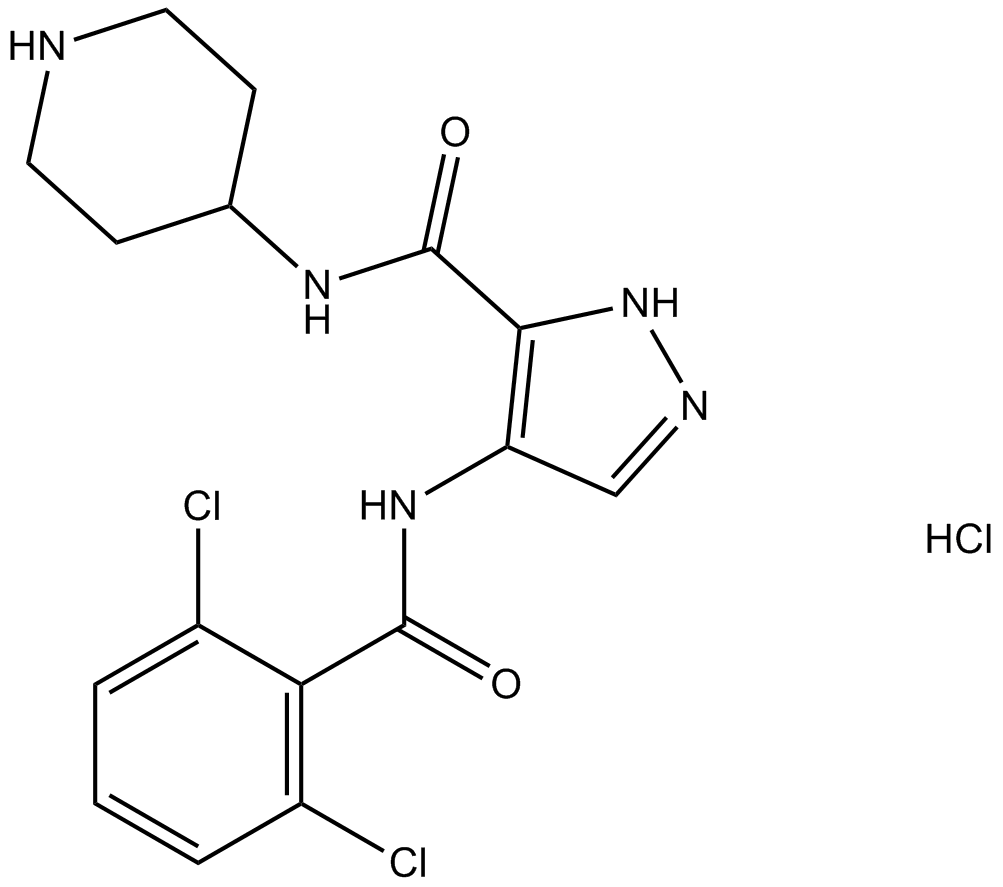
-
GC13998
AT7519 Hydrochloride
A Cdk inhibitor
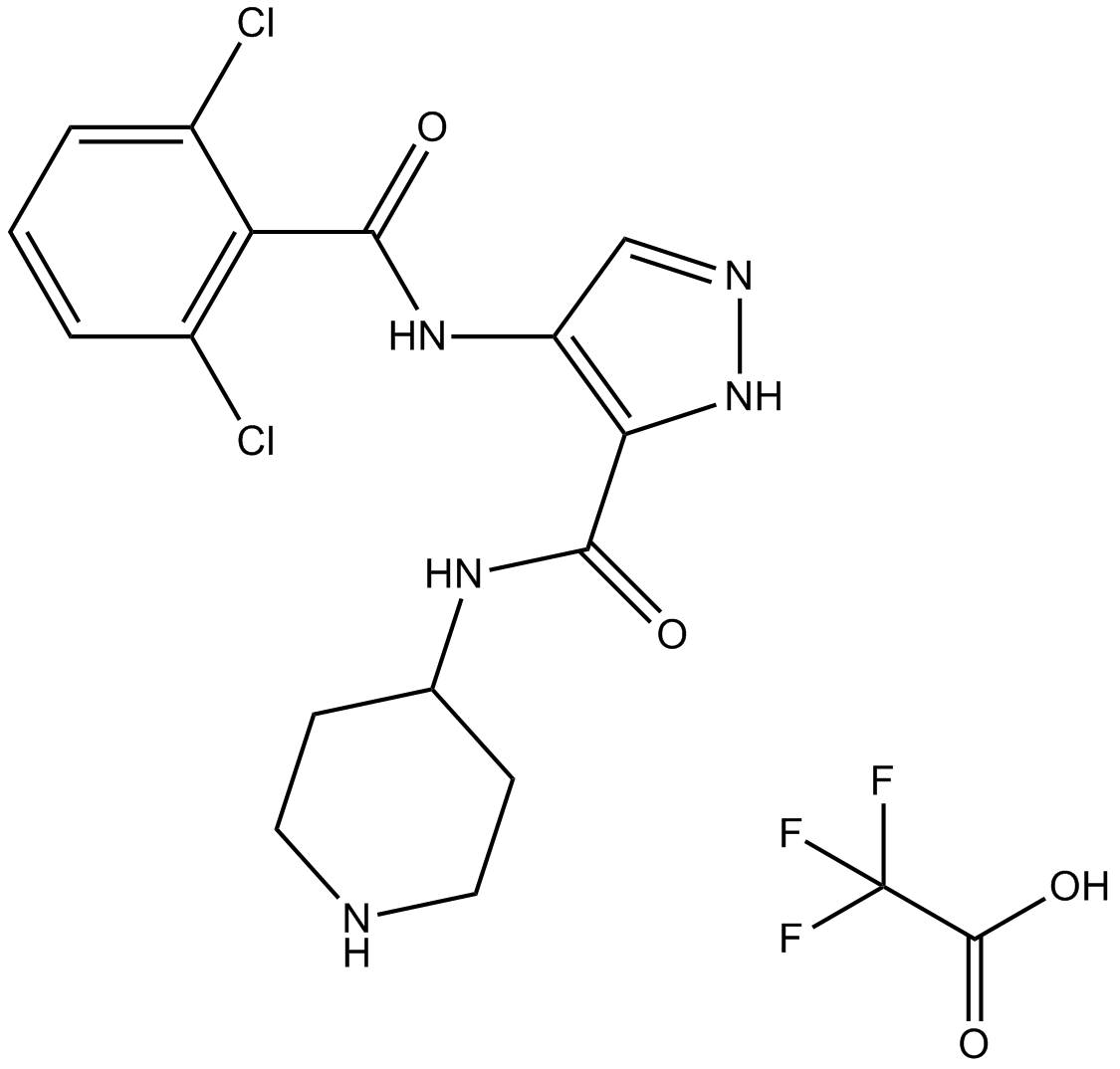
-
GC15597
AT7519 trifluoroacetate
-
GC65327
ATM Inhibitor-5
ATM Inhibitor-5 [formula (1)] is a potent inhibitor of serine/threonine protein kinase ATM (extracted from patent WO2022058351A1).
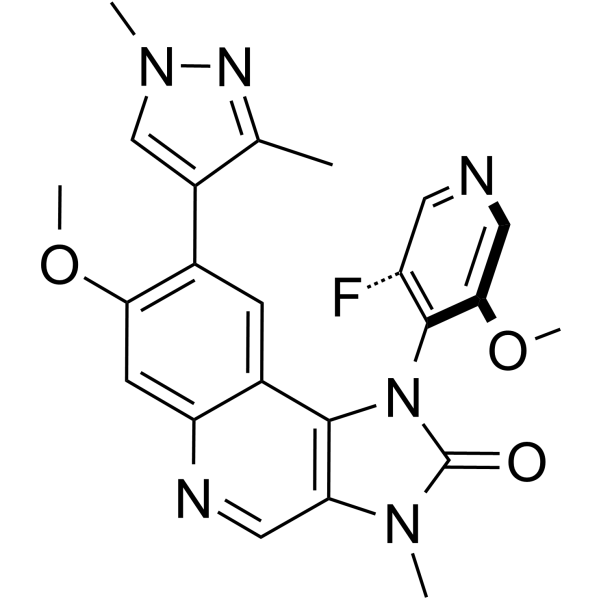
-
GC65514
ATR inhibitor 1
ATR inhibitor 1 is a ATR inhibitor extracted from patent WO2015187451A1, compound I-l, has a Ki value below 1 ?Μ.
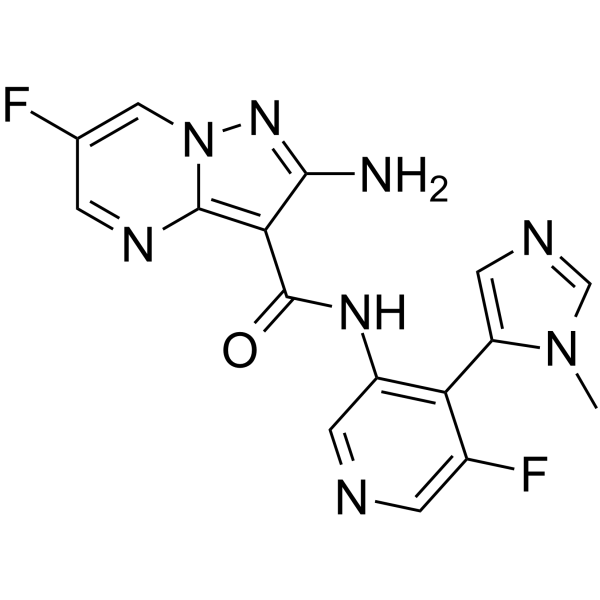
-
GC68707
ATR-IN-4
ATR-IN-4 is an effective ATR inhibitor. ATR-IN-4 inhibits the growth of human prostate cancer cells DU145 and human lung cancer cells NCI-H460, with IC50 values of 130.9 nM and 41.33 nM, respectively (excerpted from patent CN112142744A, compound 13).
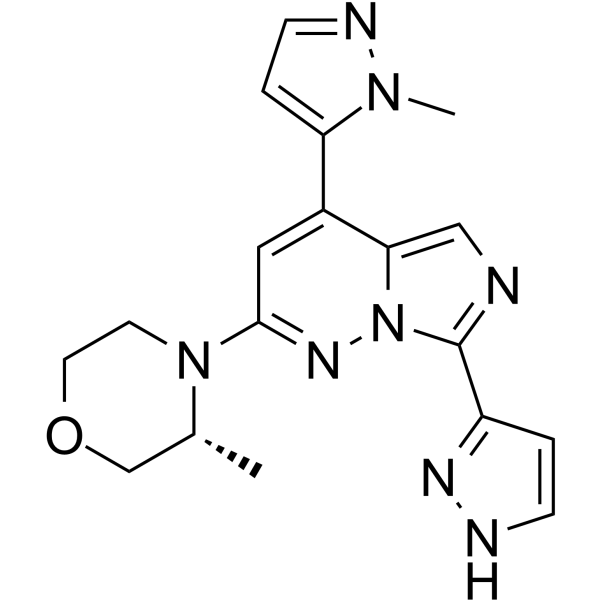
-
GC34422
Atuveciclib (BAY-1143572)
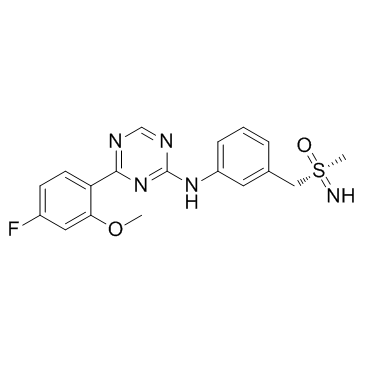
-
GC34059
Atuveciclib Racemate (BAY-1143572 Racemate)
Atuveciclib Racemate (BAY-1143572 Racemate) (BAY-1143572 Racemate) is the racemate mixture of Atuveciclib. Atuveciclib is a potent and highly selective, oral P-TEFb/CDK9 inhibitor which supresses CDK9/CycT1 with an IC50 of 13 nM.
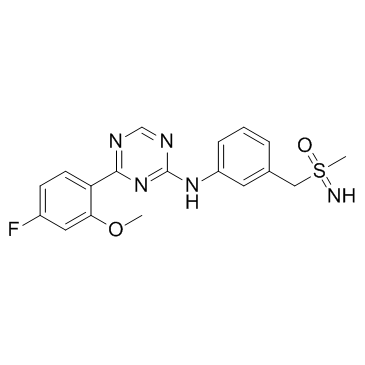
-
GC34421
Atuveciclib S-Enantiomer (BAY-1143572 S-Enantiomer)
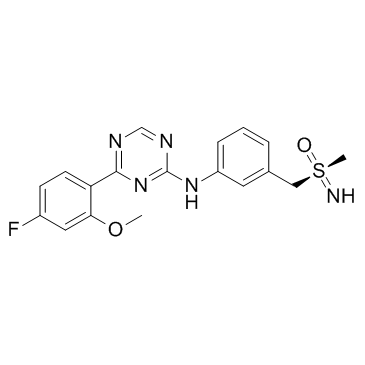
-
GC39699
Aurintricarboxylic acid
Aurintricarboxylic acid is a nanomolar-potency, allosteric antagonist with selectivity towards αβ-methylene-ATP-sensitive P2X1Rs and P2X3Rs, with IC50s of 8.6 nM and 72.9 nM for rP2X1R and rP2X3R, respectively.
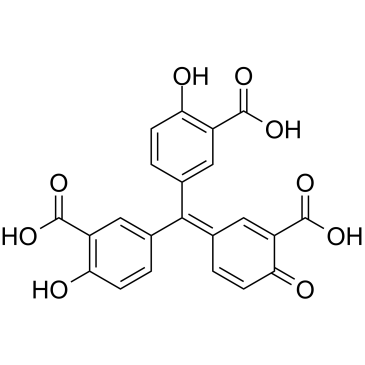
-
GC46895
Aurintricarboxylic Acid (ammonium salt)
A protein synthesis inhibitor with diverse biological activities
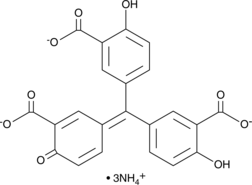
-
GC40005
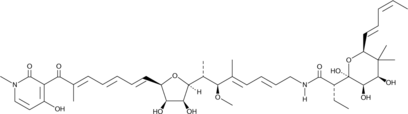
Aurodox
Aurodox is a polyketide antibiotic originally isolated from S.
-
GC60062
AV-153 free base
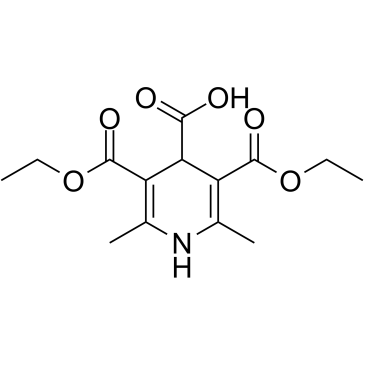
AV-153 free base, a 1,4-dihydropyridine (1,4-DHP) derivative, is an antimutagenic. AV-153 free base intercalates to DNA in a single strand break and reduces DNA damage, stimulates DNA repair in human cells in vitro. AV-153 free base interacts with thymine and cytosine and has an influence on poly(ADP)ribosylation. AV-153 free base has anti-cancer activity.
-
GC67960
AVG-233
-
GC63449
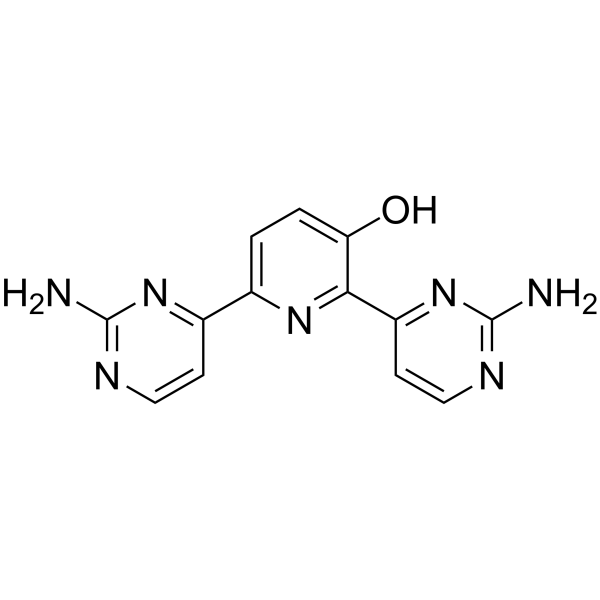
Avotaciclib
Avotaciclib (BEY1107) is a potent and orally active inhibitor of cyclin dependent kinase 1 (CDK1). Avotaciclib can be used for the research of locally advanced or metastatic pancreatic cancer.
-
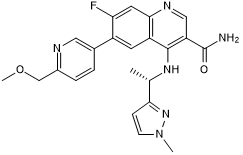
GC50424
AZ 5704
Potent and selective ATM kinase inhibitor; orally bioavailable
-
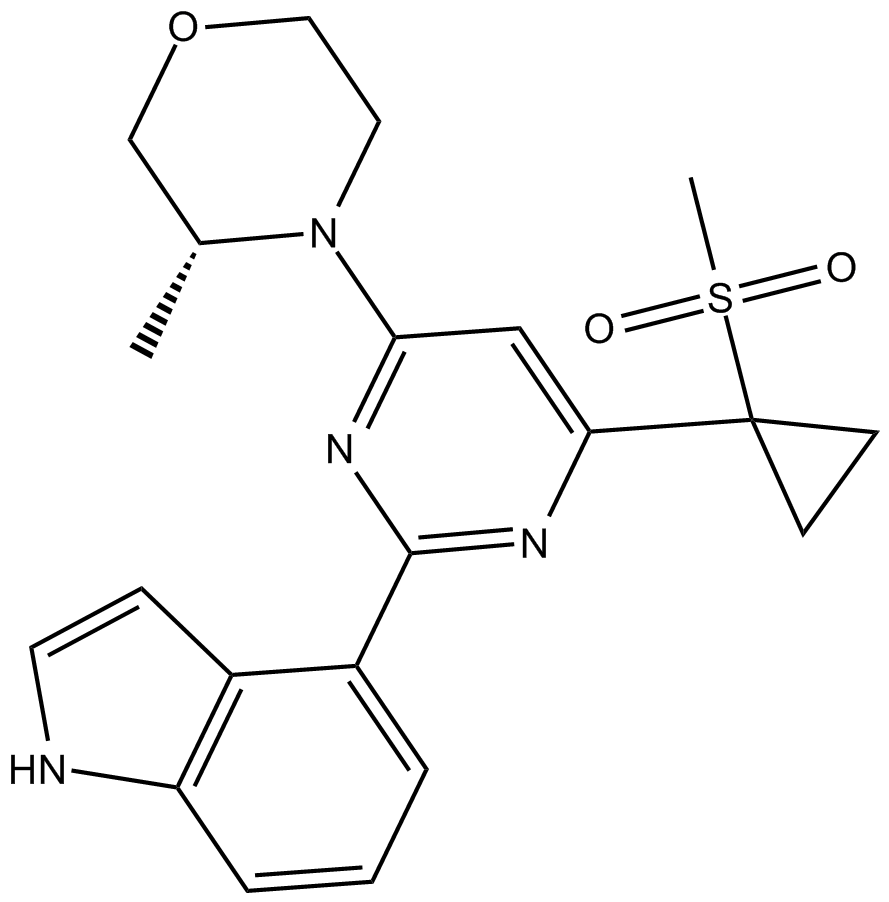
GC14949
AZ20
A potent, selective ATR inhibitor
-
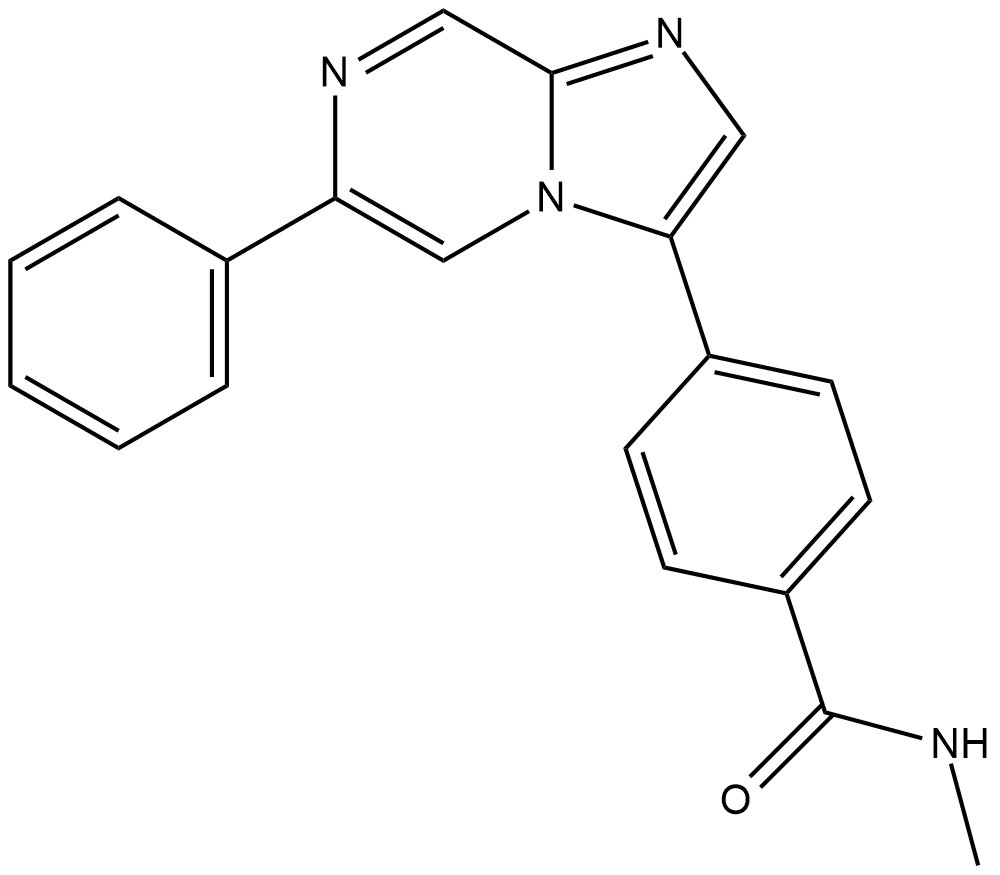
GC19047
AZ32
AZ32 is an orally bioavailable and blood-brain barrier-penetrating ATM inhibitor with an IC50 of <6.2 nM for ATM enzyme, and an IC50 of 0.31 uM for ATM in cell.
-
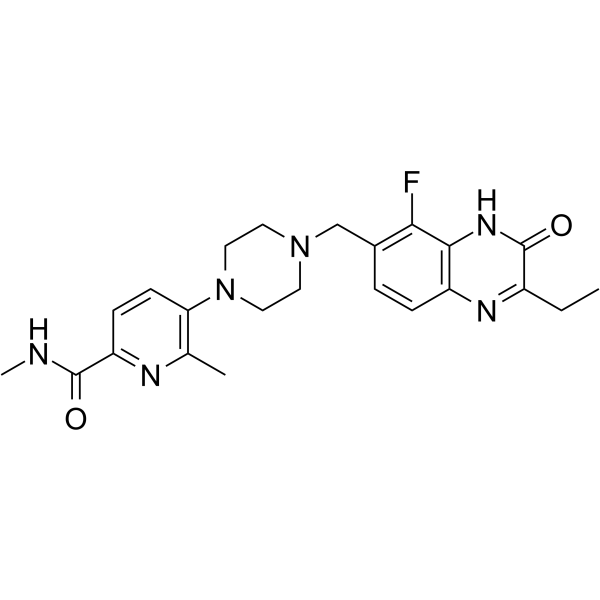
GC65899
AZ3391
AZ3391 is a potent inhibitor of PARP. AZ3391 is a quinoxaline derivative. PARP family of enzymes play an important role in a number of cellular processes, such as replication, recombination, chromatin remodeling, and DNA damage repair. AZ3391 has the potential for the research of diseases and conditions occurring in tissues in the central nervous system, such as the brain and spinal cord (extracted from patent WO2021260092A1, compound 23).
-
GC16725
AZ6102
TNKS1/2 inhibitor
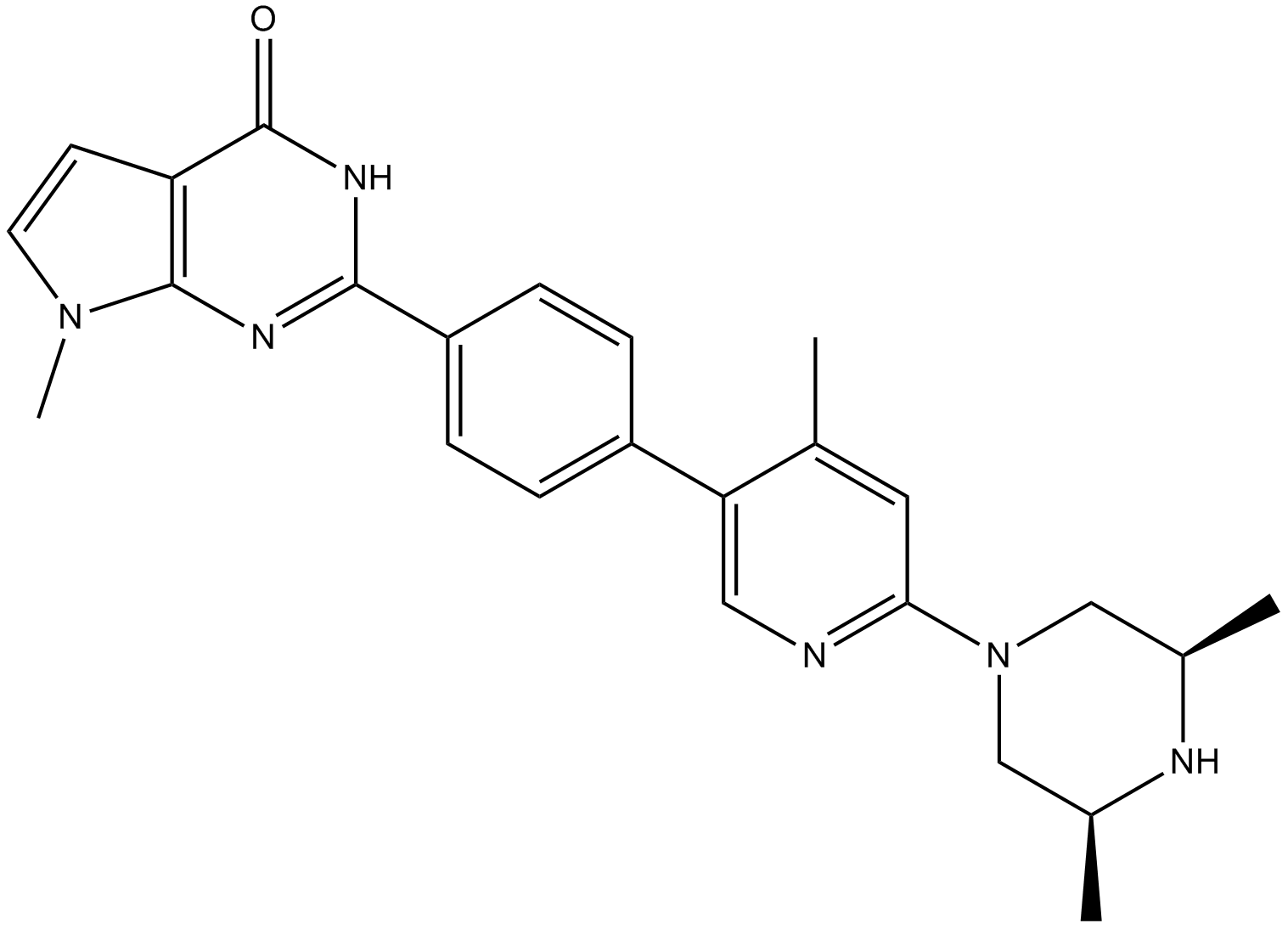
-
GC46900
AZ9482
A PARP inhibitor
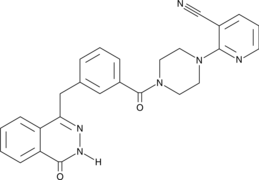
-
GC11843
Azaguanine-8
Azaguanine-8 is a purine analogue that shows antineoplastic activity. Azaguanine-8 functions as an antimetabolite and easily incorporates into ribonucleic acids, interfering with normal biosynthetic pathways, thus inhibiting cellular growth.
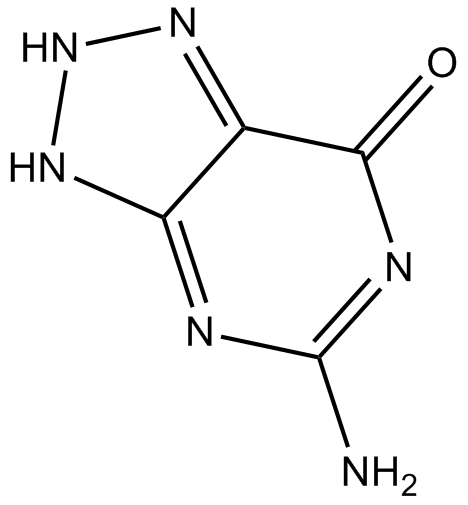
-
GC15033
Azathioprine
purine synthesis and GTP-binding protein Rac1 activation inhibitor
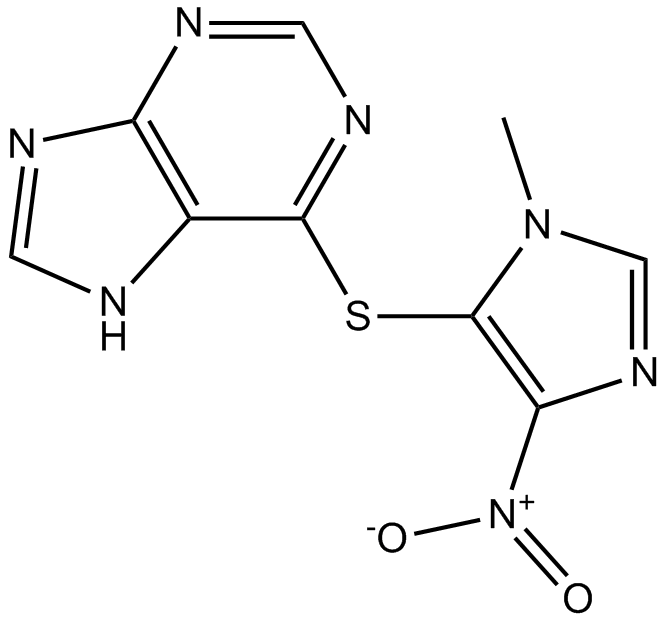
-
GC50119
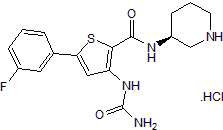
AZD 7762 hydrochloride
Potent and selective ATP-competitive inhibitor of Chk1 and Chk2; also enhances CRISPR-Cpf1-mediated genome editing
-
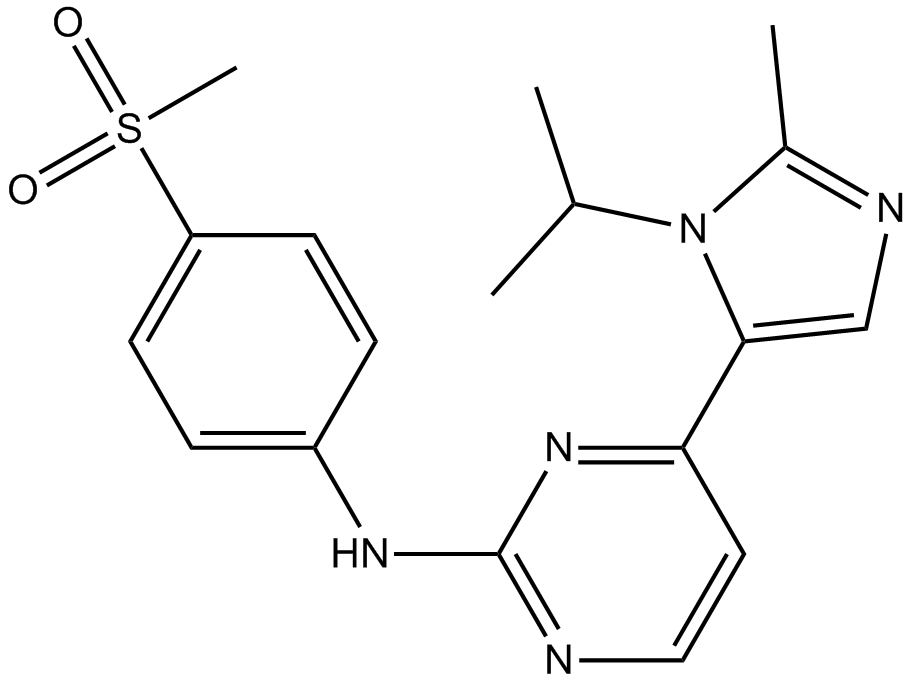
GC12438
AZD-5438
Potent CDK1/2/9 inhibitor
-
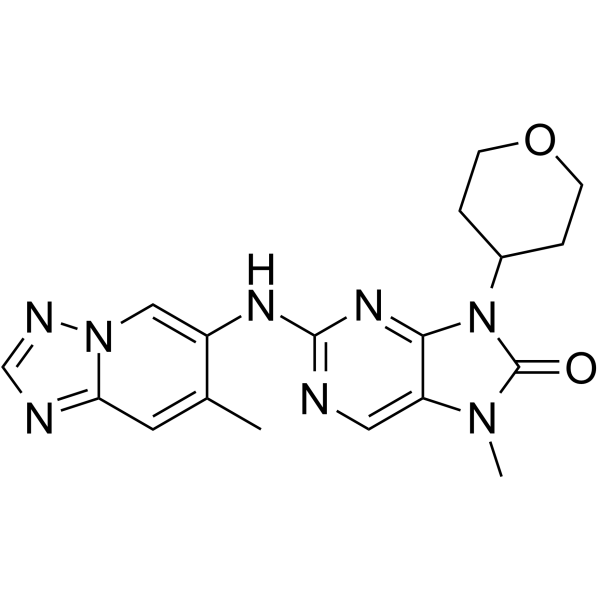
GC64938
AZD-7648
AZD-7648 is a potent, orally active, selective DNA-PK inhibitor with an IC50 of 0.6 nM. AZD-7648 induces apoptosis and shows antitumor activity.
-
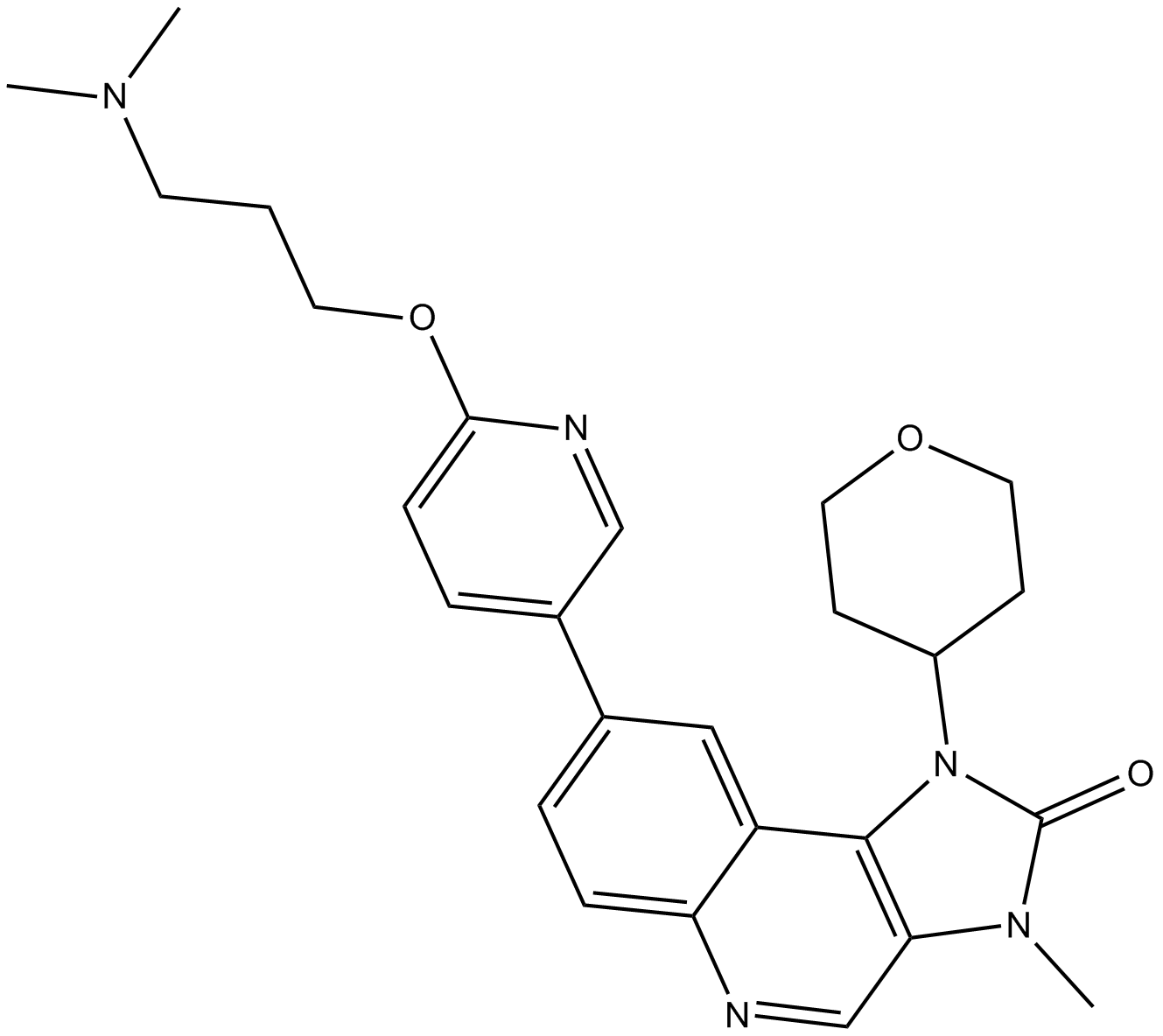
GC11593
AZD0156
ATM kinase inhibitor
-
GC19468
AZD1390
AZD1390 is a potent and selective ATM inhibitor

-
GC32717
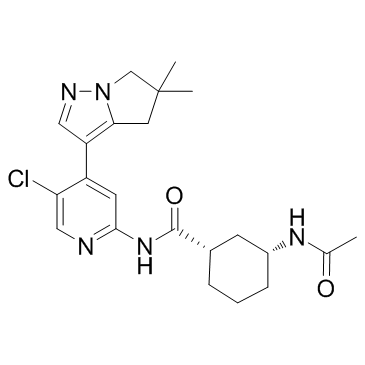
AZD4573
AZD4573 is a potent and highly selective CDK9 inhibitor (IC50 of <4 nM) that enables transient target engagement for the treatment of hematologic malignancies.
-
GC16941
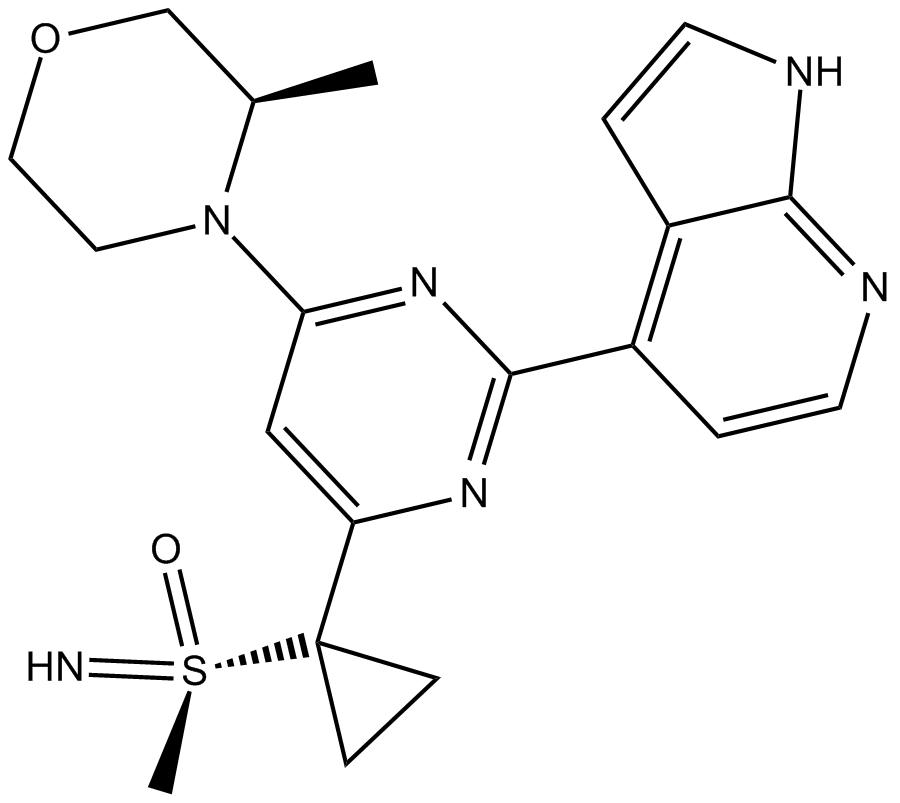
AZD6738
AZD6738 (AZD6738) is an orally active and bioavailable inhibitor of ATR kinase with an IC50 of 1 nM.
-
GC10546
AZD7762
Checkpoint kinase inhibitor,ATP competitive
-
GC60616
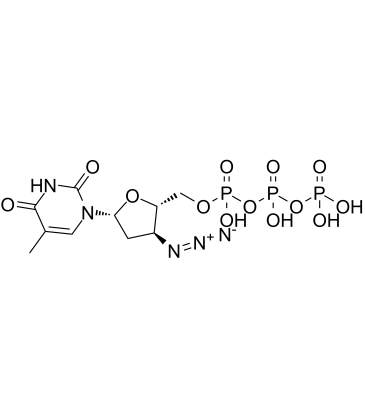
AZT triphosphate
AZT triphosphate (3'-Azido-3'-deoxythymidine-5'-triphosphate) is a active triphosphate metabolite of Zidovudine (AZT).
-
GC60617
AZT triphosphate TEA
AZT triphosphate TEA (3'-Azido-3'-deoxythymidine-5'-triphosphate TEA) is a active triphosphate metabolite of Zidovudine (AZT).
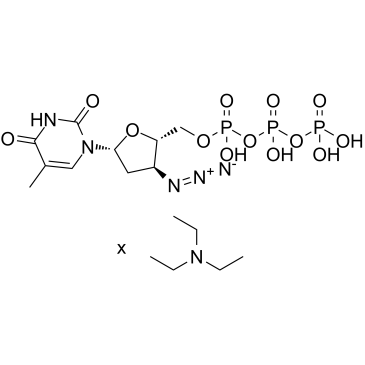
-
GC33406
B I09
B I09 is an IRE-1 RNase inhibitor, with an IC50 of 1230 nM.
-
GC33240
Banoxantrone D12 (AQ4N D12)
Banoxantrone D12 (AQ4N D12) is the deuterium labeled banoxantrone. Banoxantrone is a novel bioreductive agent that can be reduced to a stable, DNA-affinic compound AQ4, which is a potent topoisomerase II inhibitor.
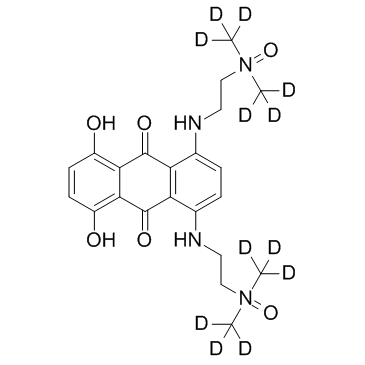
-
GC34169
Banoxantrone D12 dihydrochloride (AQ4N D12 dihydrochloride)
Banoxantrone-d12 (AQ4N-d12) dihydrochloride is the deuterium labeled Banoxantrone dihydrochloride. Banoxantrone is a novel bioreductive agent that can be reduced to a stable, DNA-affinic compound AQ4, which is a potent topoisomerase II inhibitor.
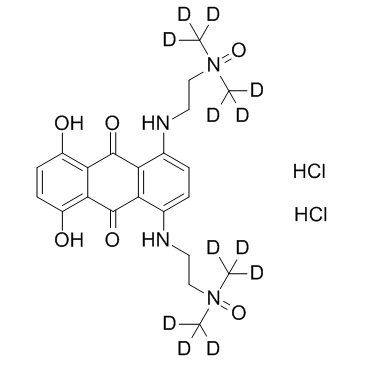
-
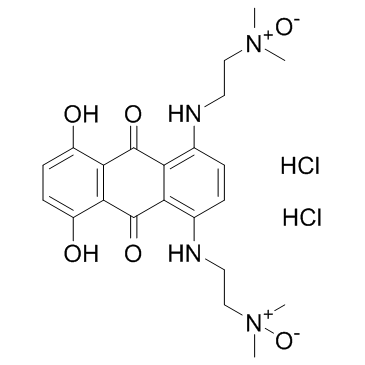
GC34085
Banoxantrone dihydrochloride (AQ4N dihydrochloride)
Banoxantrone dihydrochloride (AQ4N dihydrochloride) is a novel bioreductive agent that can be reduced to a stable, DNA-affinic compound AQ4, which is a potent topoisomerase II inhibitor.
-
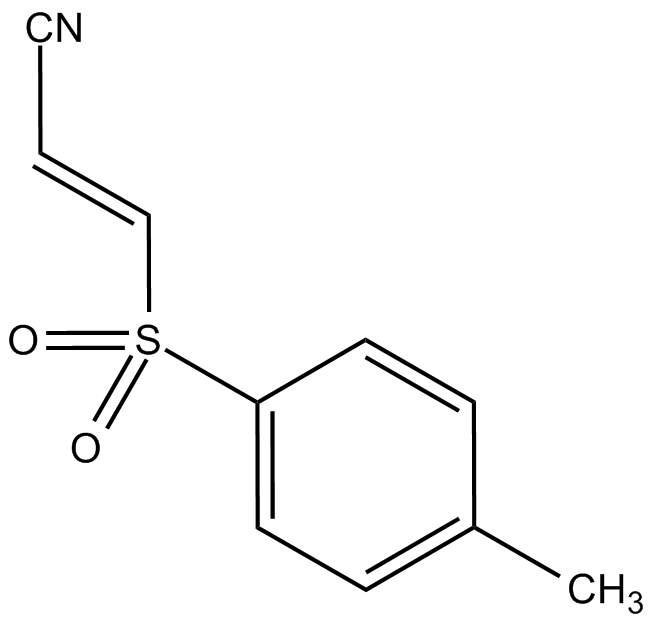
GC13035
Bay 11-7821
A selective and irreversible NF-κB inhibitor
-
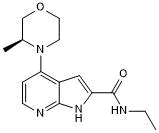
GC50656
BAY 707
BAY 707 is a substrate-competitive, highly potent and selective inhibitor of MTH1(NUDT1) with an IC50 of 2.3 nM. BAY 707 has a good pharmacokinetic (PK) profile to other MTH1 compounds and is well-tolerated in mice, but shows a clear lack of in vitro or in vivo anticancer efficacy.
-
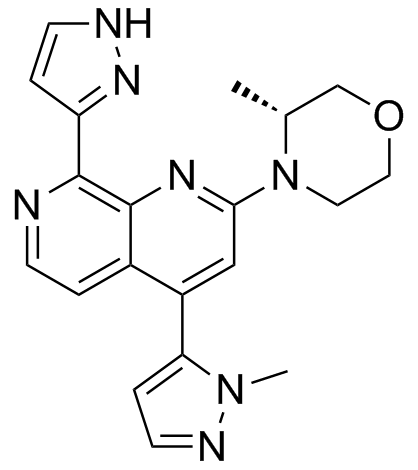
GC33420
BAY-1895344
BAY-1895344 (BAY-1895344) is a potent, orally active and selective ATR inhibitor with an IC50 of 7 nM. BAY-1895344 has anti-tumor activity. BAY-1895344 can be used for the research of solid tumors and lymphomas.
-
GC34374
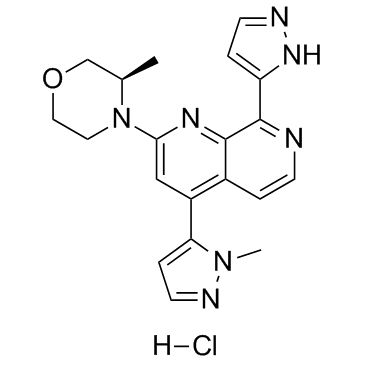
BAY-1895344 hydrochloride
An ATR inhibitor
-
GC32838
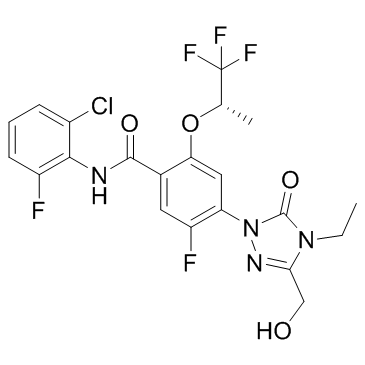
BAY-2402234
A DHODH inhibitor
-
GC63763
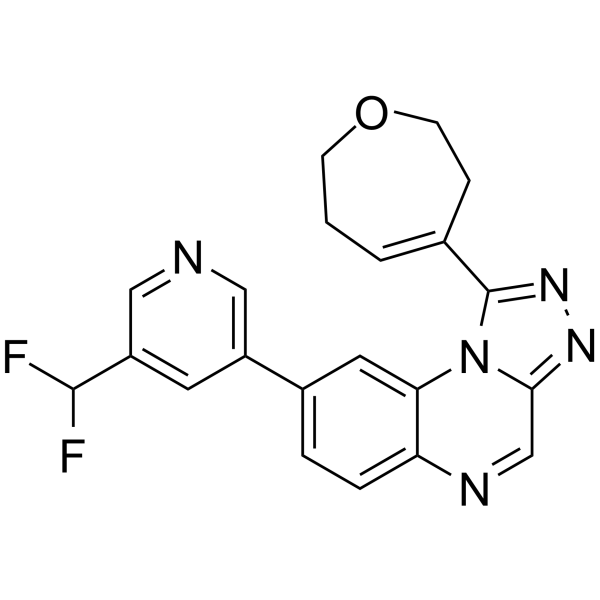
BAY-8400
BAY-8400 is an orally active, potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (IC50=81 nM). BAY-8400 can be used for the research of cancer.
-
GC65259
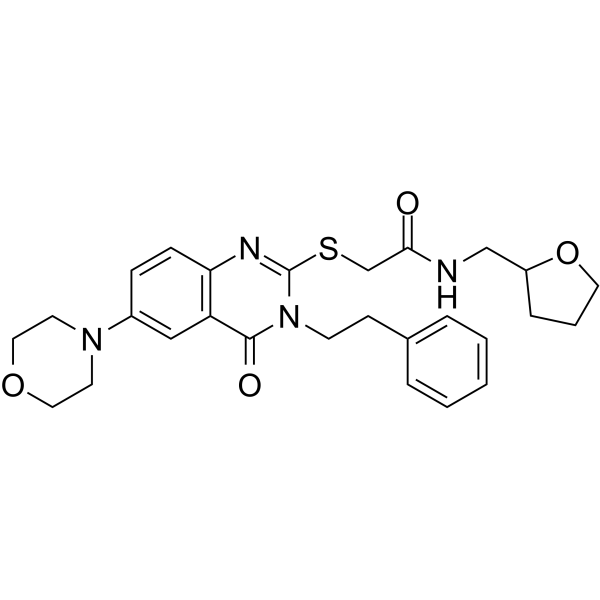
BC-1471
BC-1471 is a STAM-binding protein (STAMBP) deubiquitinase inhibitor.
-
GC50572
BC-LI-0186
Leucyl-tRNA synthase (TRS)/Ras-related GTP-binding protein D (RagD) interaction inhibitor
-
GC62863
BCH001
BCH001, a quinoline derivative, is a specific PAPD5 inhibitor.