DNA/RNA Synthesis
RNA synthesis, which is also called DNA transcription, is a highly selective process. Transcription by RNA polymerase II extends beyond RNA synthesis, towards a more active role in mRNA maturation, surveillance and export to the cytoplasm.
Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.DNA ligases discriminate against substrates containing RNA strands or mismatched base pairs at positions near the ends of the nickedDNA. Bleomycin (BLM) exerts its genotoxicity by generating free radicals, whichattack C-4′ in the deoxyribose backbone of DNA, leading to opening of the ribose ring and strand breakage; it is an S-independentradiomimetic agent that causes double-strand breaks in DNA.
First strand cDNA is synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H). Second strand cDNA synthesis is subsequently performed using DNA Polymerase I and RNase H. The remaining overhangs are converted into blunt ends using exonuclease/polymerase activity. After adenylation of the 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure is ligated to prepare the samples for hybridization. Cell cycle and DNA replication are the top two pathways regulated by BET bromodomain inhibition. Cycloheximide blocks the translation of mRNA to protein.
Targets for DNA/RNA Synthesis
Products for DNA/RNA Synthesis
- Cat.No. Product Name Information
-
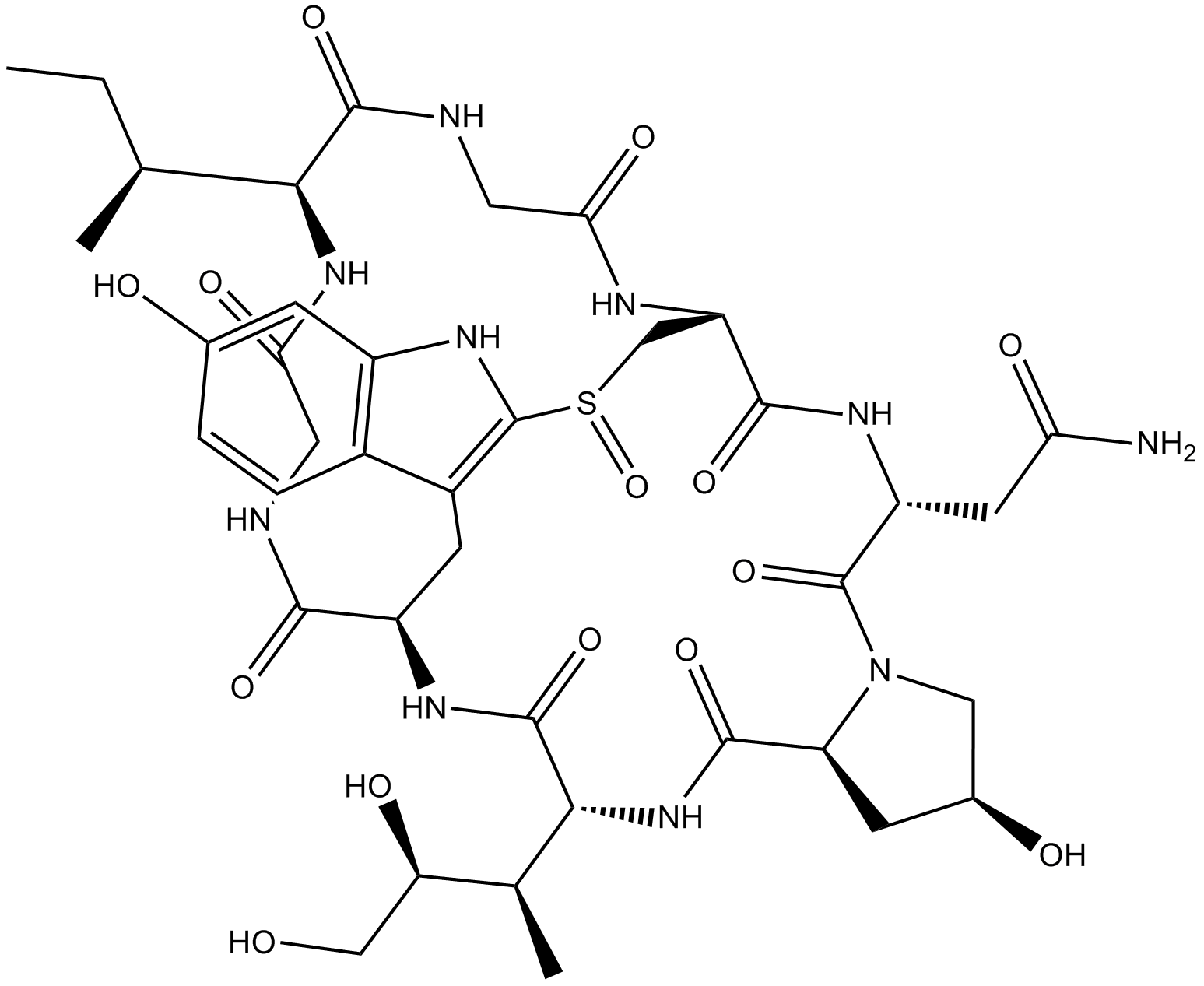
GC65372
Pseudouridine 5'-triphosphate trisodium
-
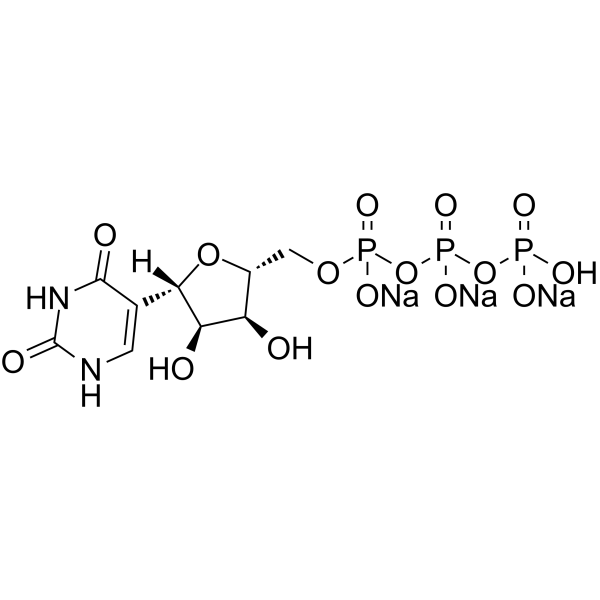
GC16367
PSI-7977
A prodrug form of PSI-7411
-
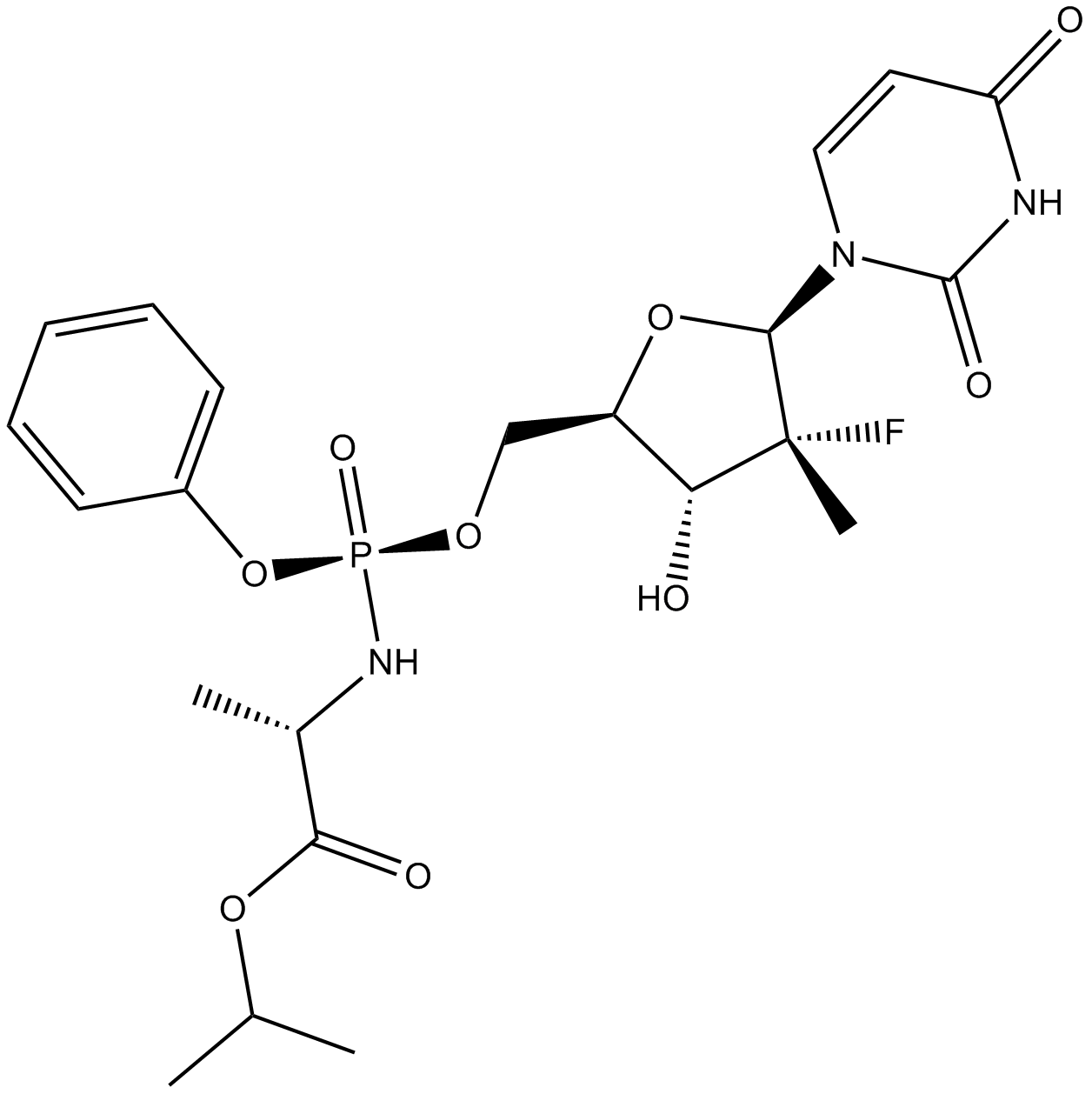
GC65335
PTC299
PTC299 is an orally active inhibitor of VEGFA mRNA translation that selectively inhibits VEGF protein synthesis at the post-transcriptional level. PTC299 is also a potent inhibitor of dihydroorotate dehydrogenase (DHODH). PTC299 shows good oral bioavailability and lack of off-target kinase inhibition and myelosuppression. PTC299 can be useful for the research of hematologic malignancies.
-
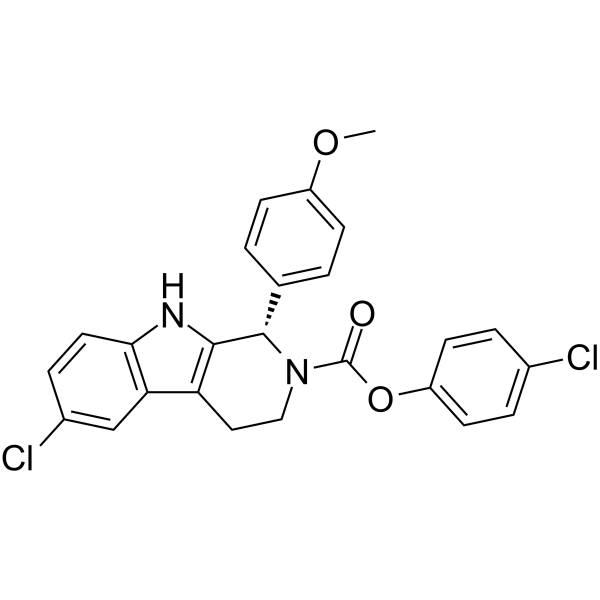
GC16384
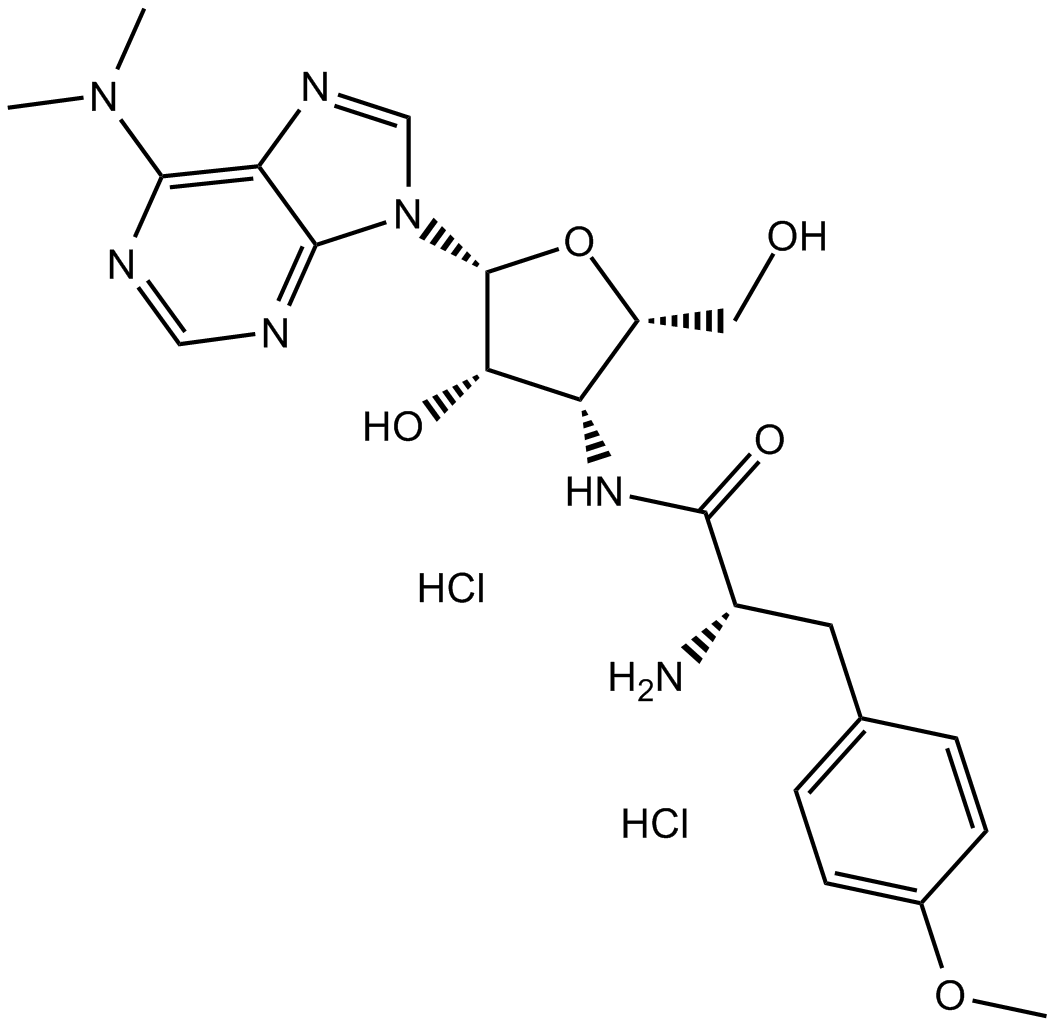
Puromycin dihydrochloride
Puromycin dihydrochloride is produced by Streptomyces alboniger, a grampositive actinomycete, through a series of enzymatic reactions
-
GC37042
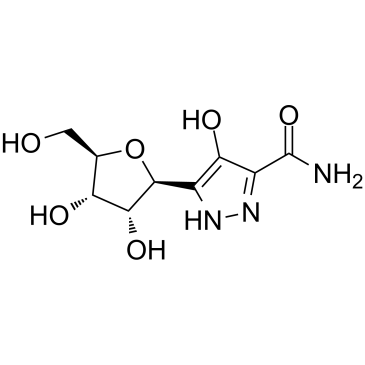
Pyrazofurin
Pyrazofurin, a pyrimidine nucleoside analogue with antineoplastic activity, inhibits cell proliferation and DNA synthesis in cells by inhibiting uridine 5'-phosphate (UMP) synthase. Pyrazofurin is an active, sensitive orotate-phosphoribosyltransferase inhibitor with IC50s between 0.06-0.37 ?M in the three squamous cell carcinoma (SCC) cell lines Hep-2, HNSCC-14B and HNSCC-14C.
-
GC45552
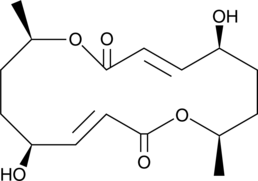
Pyrenophorol
-
GC45553
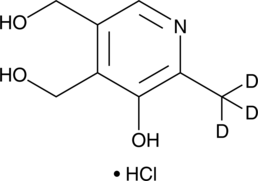
Pyridoxine-d3 (hydrochloride)
-
GC37044
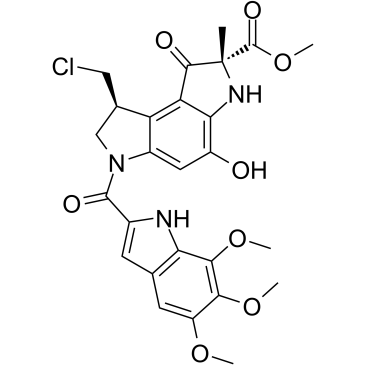
Pyrindamycin A
Pyrindamycin A is an antibiotic that inhibits DNA synthesis.
-
GC37051
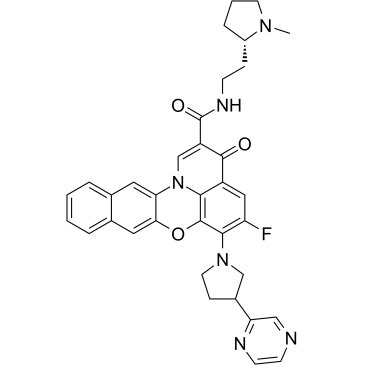
Quarfloxin
Quarfloxin (CX-3543), a fluoroquinolone derivative with antineoplastic activity, targets and inhibits RNA pol I activity, with IC50 values in the nanomolar range in neuroblastoma cells. Quarfloxin disrupts the interaction between the nucleolin protein and a G-quadruplex DNA structure in the ribosomal DNA (rDNA) template.
-
GC61666
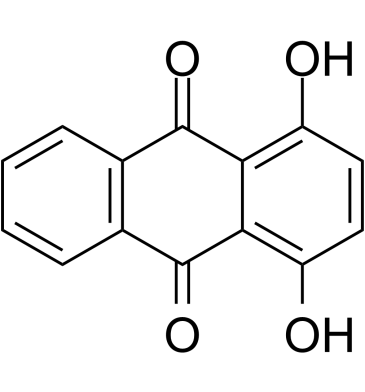
Quinizarin
Quinizarin (1,4-Dihydroxyanthraquinone), a part of the anticancer agents such as Doxorubicin, Daunorubicin, and Adriamycin, interacts with DNA by intercalating mode (Kd=86.1 μM).
-
GC17750
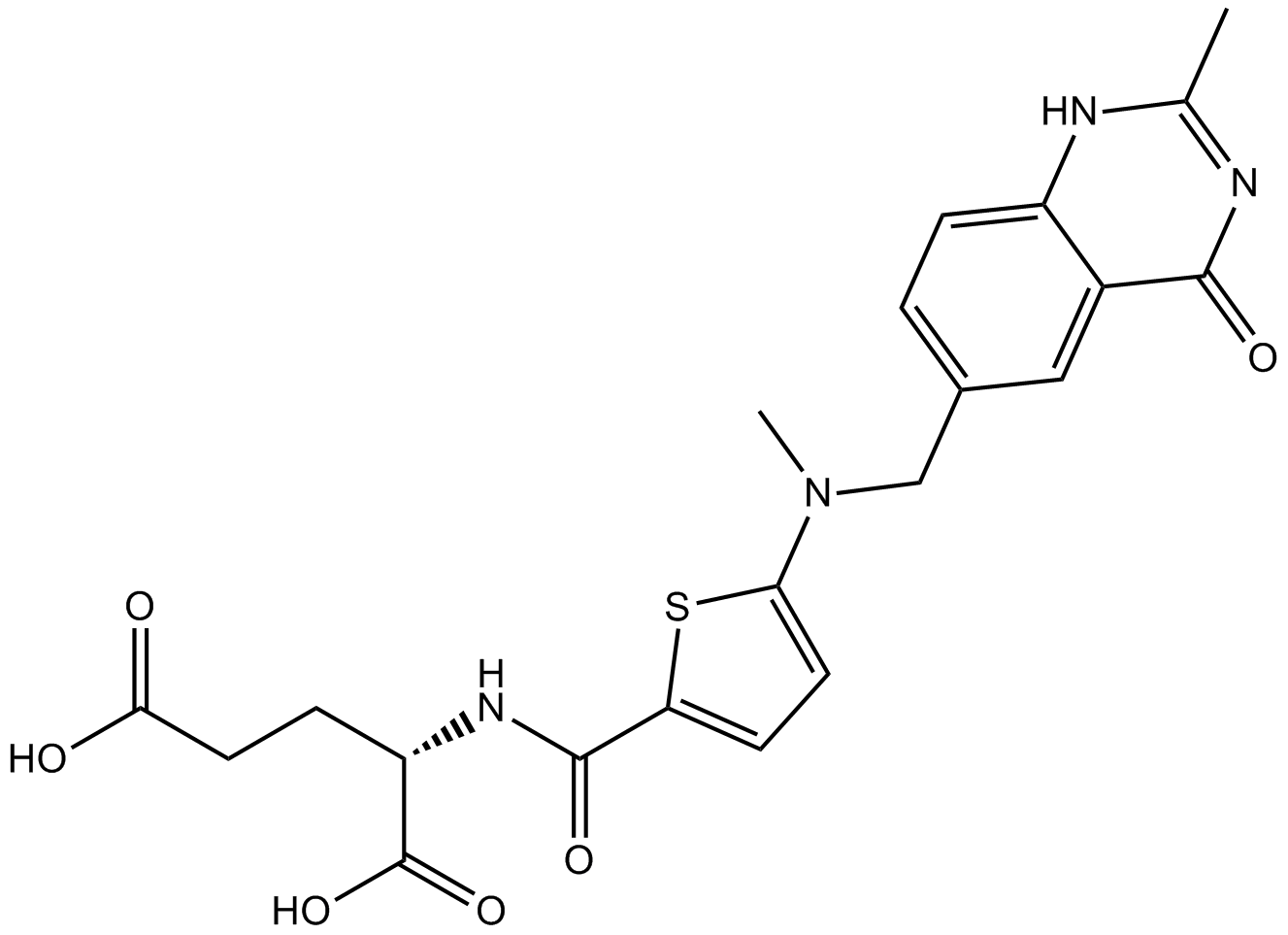
Raltitrexed
Thymidylate synthase inhibitor
-
GC18751
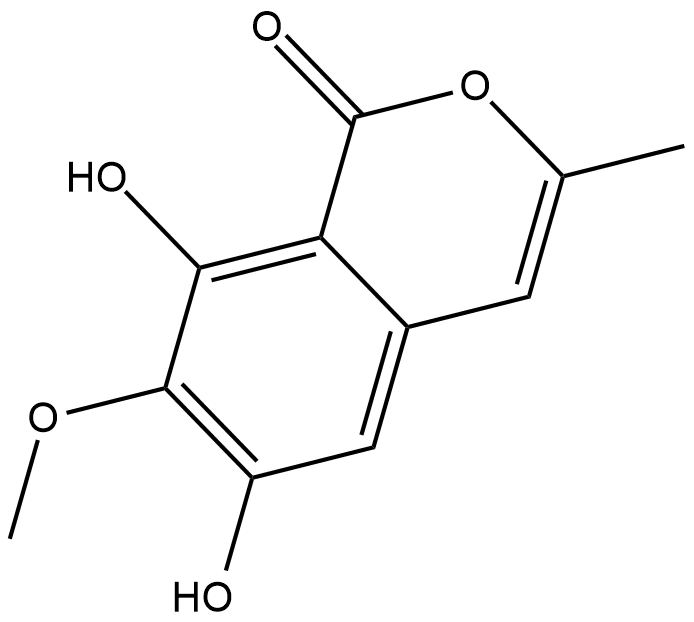
Reticulol
Reticulol is an isocoumarin derivative produced by certain species of Streptomyces that inhibits cAMP phosphodiesterase (IC50 = 41 uM).
-
GC19309
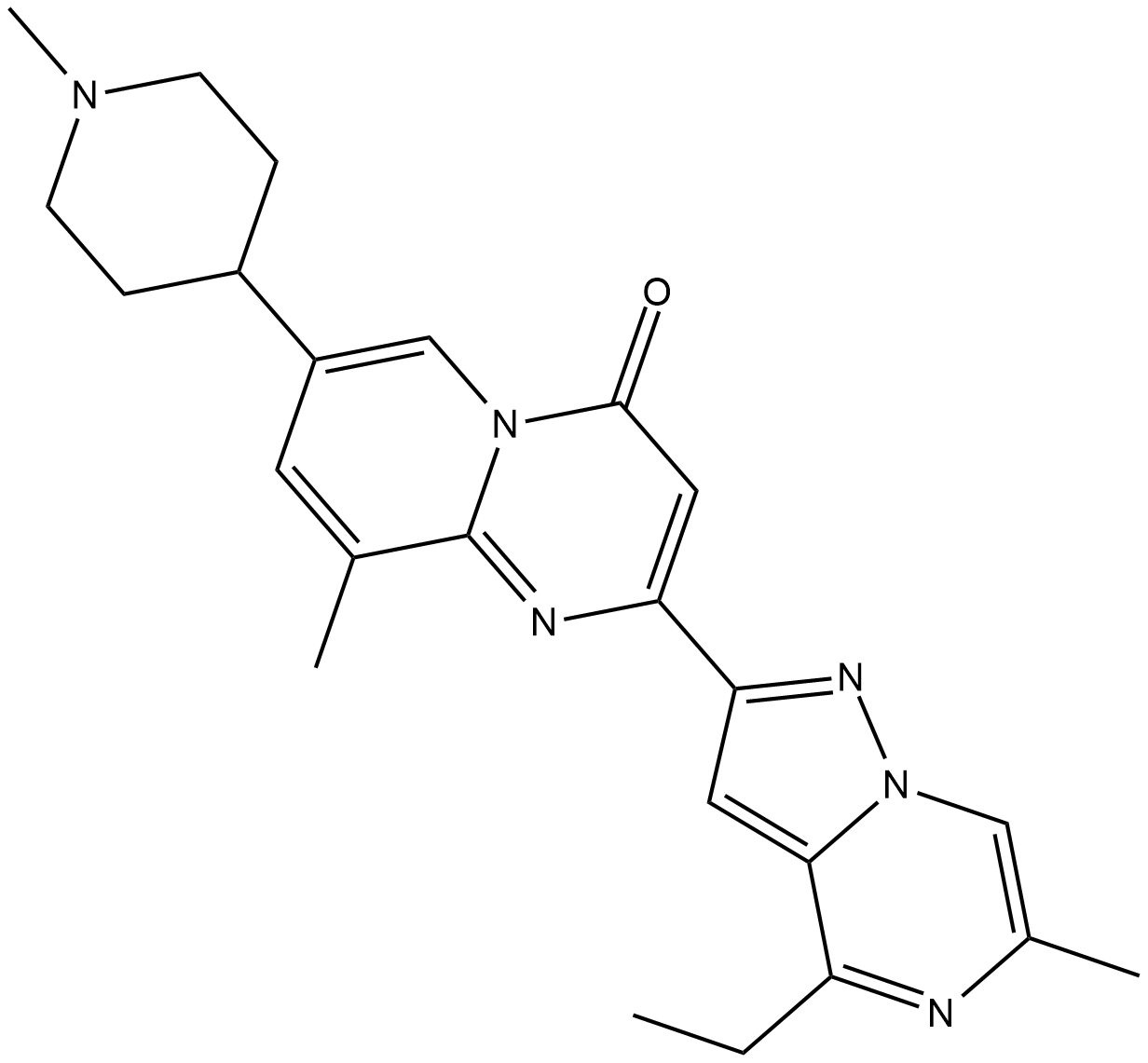
RG7800
RG7800 a small molecule SMN2 splicing modifier to enter human clinical trials to treat spinal muscular atrophy.
-
GC37521
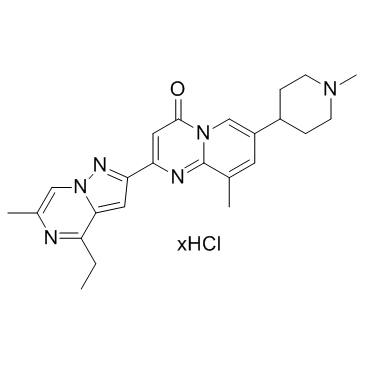
RG7800 hydrochloride
RG7800 hydrochloride is an orally active SMN2 splicing modulator, with EC1.5xs of 23 nM and 87 nM for SMN2 splicing and SMN protein; RG7800 hydrochloride has the potential to treat spinal muscular atrophy.
-
GC61246
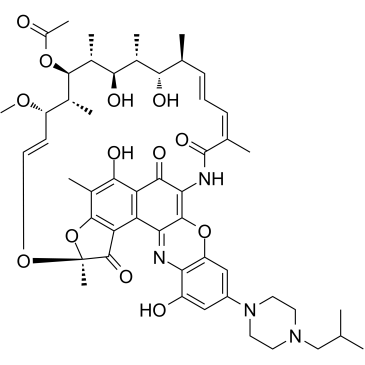
Rifalazil
Rifalazil (KRM-1648; ABI-1648), a rifamycin derivative, inhibits the bacterial DNA-dependent RNA polymerase and kills bacterial cells by blocking off the β-subunit in?RNA polymerase.
-
GC16970
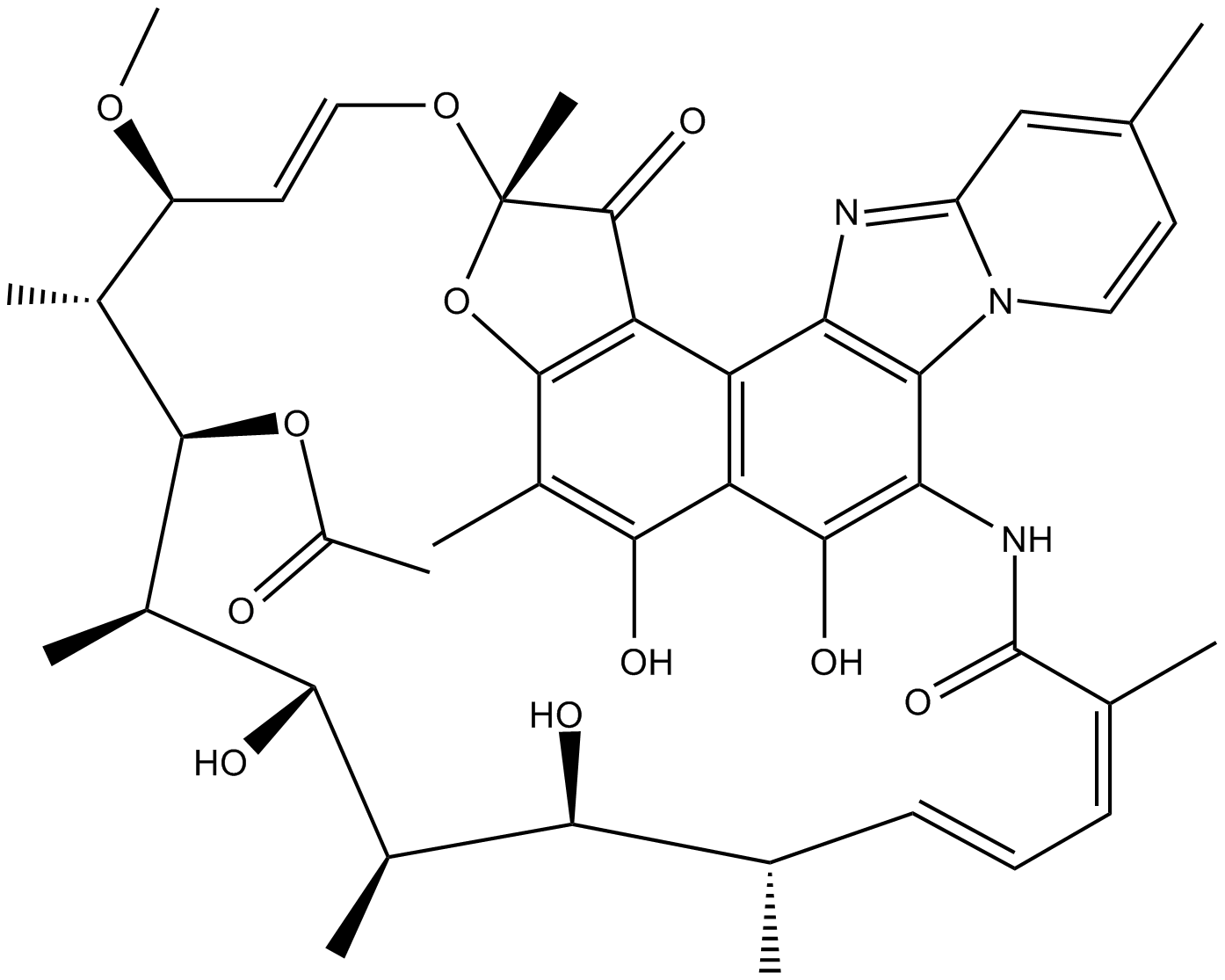
Rifaximin (Xifaxan)
Rifaximin (Xifaxan), a gastrointestinal-selective antibiotic, binds the β-subunit of bacterial DNA-dependent RNA polymerase, resulting in inhibition of bacterial RNA synthesis.
-
GC30845
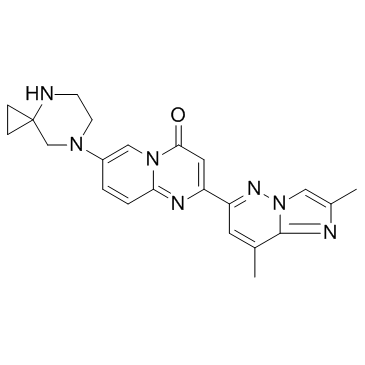
Risdiplam (RG7916)
Risdiplam (RG7916) (RG7916) is an orally administered, centrally and peripherally distributed SMN2 pre-mRNA splicing modifier that increases survival motor neuron (SMN) protein levels.
-
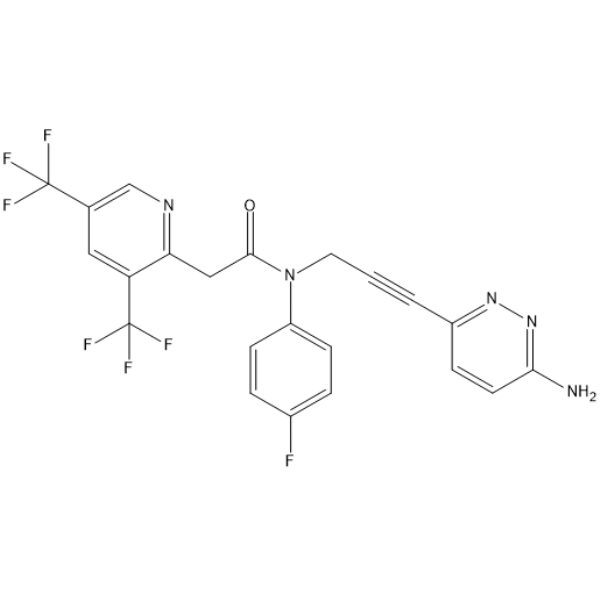
GC67865
RP-6685
-
GC14808
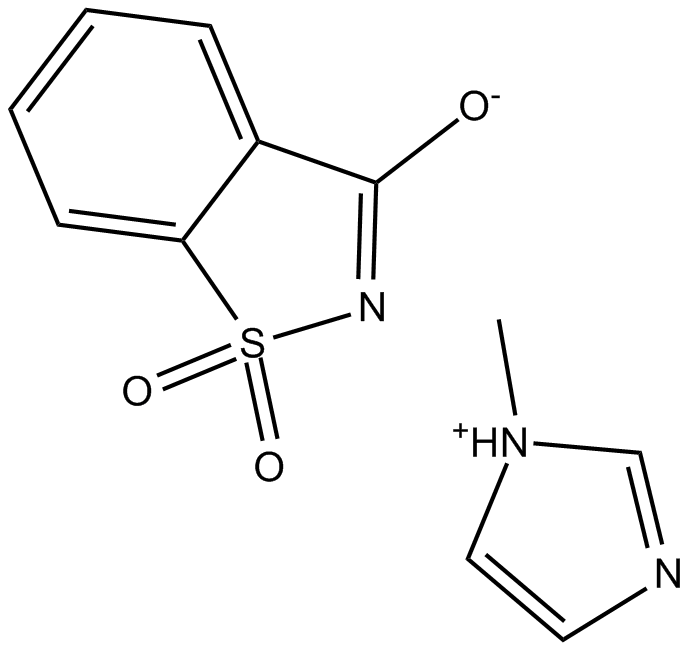
Saccharin 1-methylimidazole (SMI)
Saccharin 1-methylimidazole (SMI) is an activator for DNA/RNA Synthesis.
-
GC62693
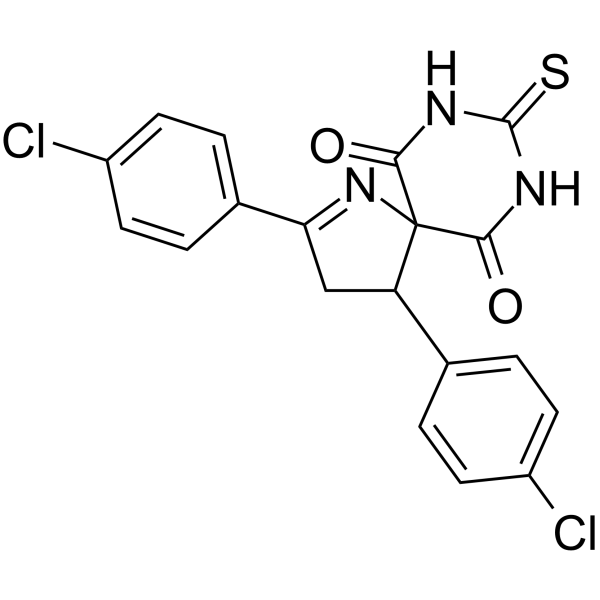
SCR130
SCR130 is a SCR7-based DNA nonhomologous end-joining (NHEJ) inhibitor. SCR130 inhibits the end-joining of DNA in a Ligase IV-dependent manner. SCR130 is specific to Ligase IV, and shows minimal or no effect on Ligase III and Ligase I mediated joining. SCR130 induces cell apoptosis and has anticancer activity.
-
GC12106
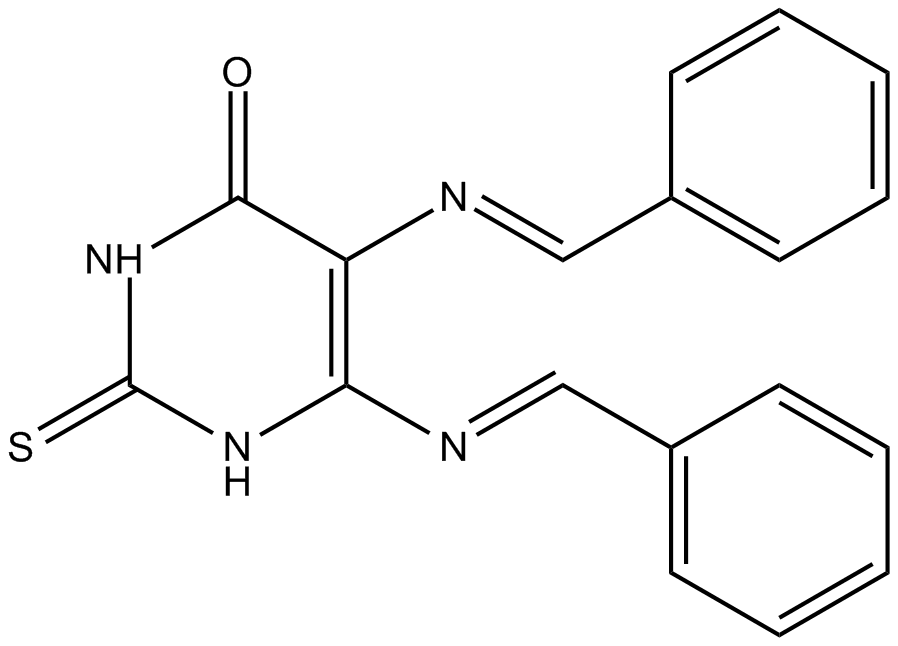
SCR7
DNA ligase IV inhibitor
-
GC61276
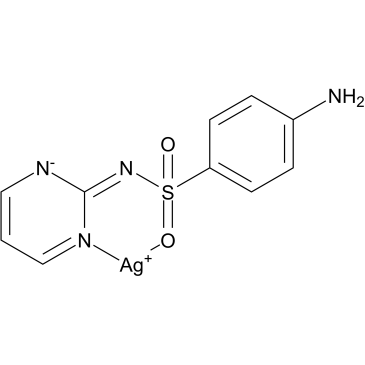
Silver sulfadiazine
Silver sulfadiazine (AgSD), a sulfonamide antibiotic, effects a dual inhibitory action on bacterial growth by its sulfa moiety (SD-SDZ) that prevents bacterial folate absorption and subsequent DNA synthesis.
-
GC30185
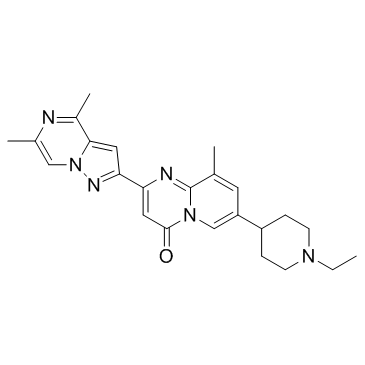
SMN-C3
SMN-C3 is an orally active SMN2 splicing modulator and has the potential to treat spinal muscular atrophy (SMA).
-
GC39552
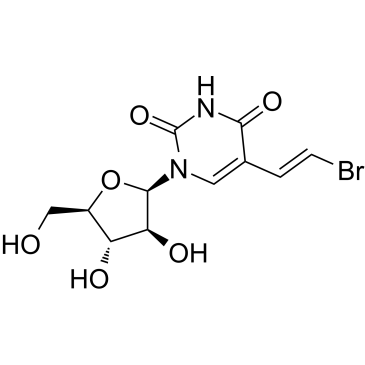
Sorivudine
Sorivudine (BV-araU) is an orally active synthetic pyrimidine nucleoside antimetabolite drug.
-
GC46223
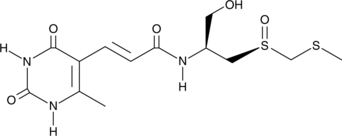
Sparsomycin
A bacterial metabolite with diverse biological activities
-
GC44953
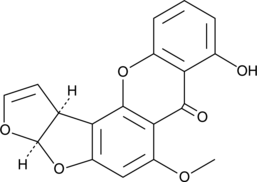
Sterigmatocystin
Sterigmatocystin is a mycotoxin produced by fungi of the genus Aspergillus[1].
-
GC46228
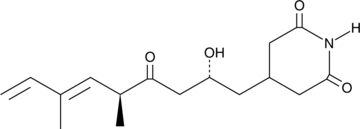
Streptimidone
A bacterial metabolite with diverse biological activities
-
GC17131
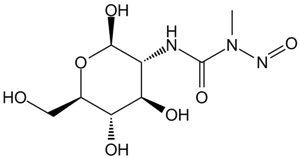
Streptozocin
A diabetogenic agent which targets beta cells
-
GC39510
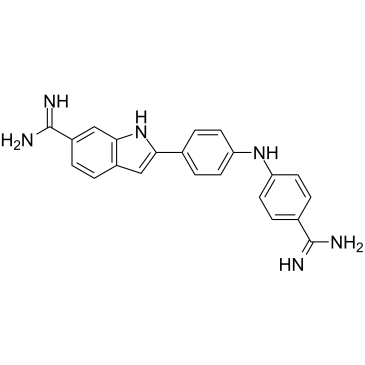
Synucleozid
Synucleozid (NSC 377363) is a potent inhibitor of the SNCA?mRNA that encodes α-synuclein protein (IC50=1.5 μM).
-
GC39511
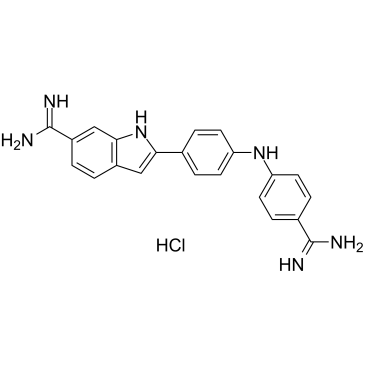
Synucleozid hydrochloride
Synucleozid hydrochloride (NSC 377363 hydrochloride) is a potent inhibitor of the?SNCA?mRNA?that encodes α-synuclein protein (IC50=1.5 μM).
-
GC12350
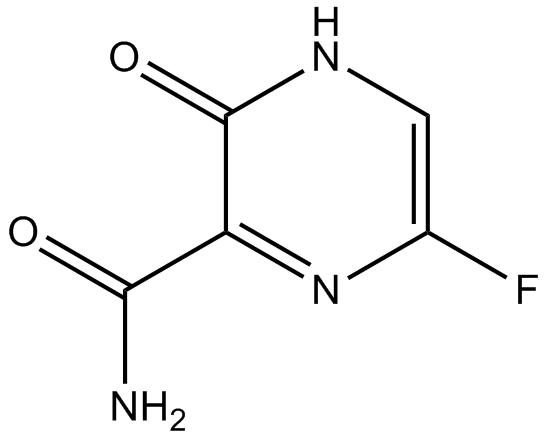
T 705
T 705 (T-705) is a potent viral RNA polymerase inhibitor, it is phosphoribosylated by cellular enzymes to its active form, T 705-ribofuranosyl-5′-triphosphate (RTP).
-
GC44981
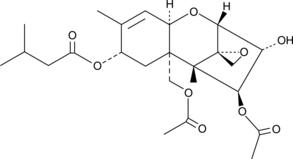
T-2 Toxin
T-2 toxin is a common trichothecene mycotoxin produced by Fusarium species, which can infect grain crops causing alimentary toxic aleukia in humans and animals.
-
GC61500
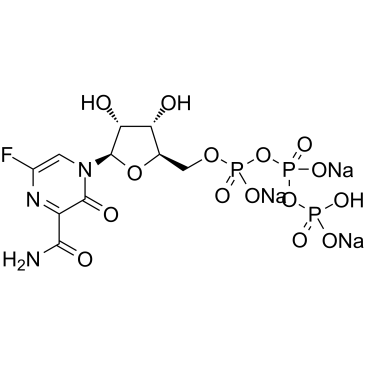
T-705RTP sodium
-
GC44984
TAF 10 Peptide
TAF10 is one of many protein factors or coactivators associated with RNA polymerase II activity.

-
GC39512
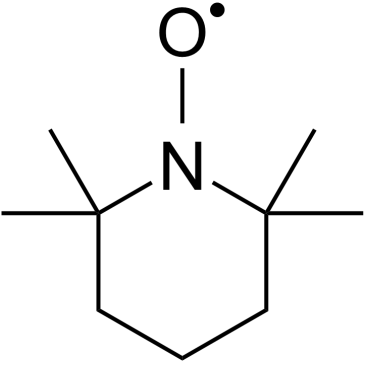
Tempo
Tempo is a classic nitroxide radical and is a selective scavenger of ROS that dismutases superoxide in the catalytic cycle. Tempo induces DNA-strand breakage. Tempo can be used as an organocatalyst for the oxidation of primary alcohols to aldehydes. Tempo has mutagenic and antioxidant effects.
-
GC64373
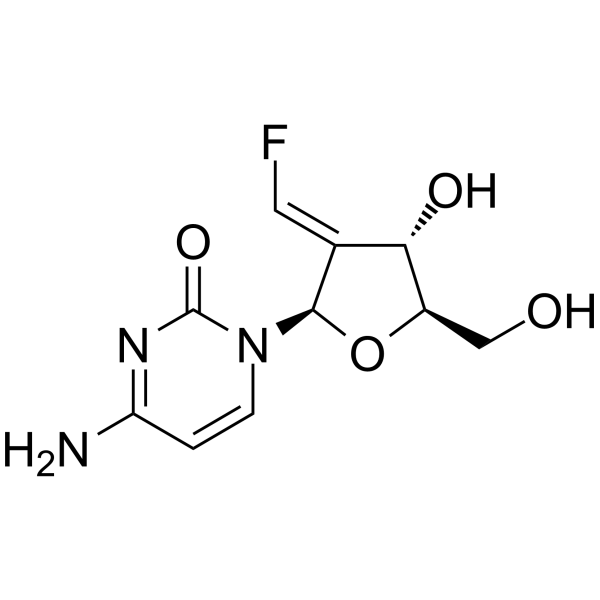
Tezacitabine
Tezacitabine is a cytostatic and cytotoxic antimetabolite and a nucleoside analogue. Tezacitabine irreversibly inhibits the ribonucleotide reductase and interferes with DNA replication and repair. Tezacitabine effectively induces cells apoptotic. Tezacitabine has the potential for leukemias and solid tumors (carcinomas) treatment.
-
GC17601
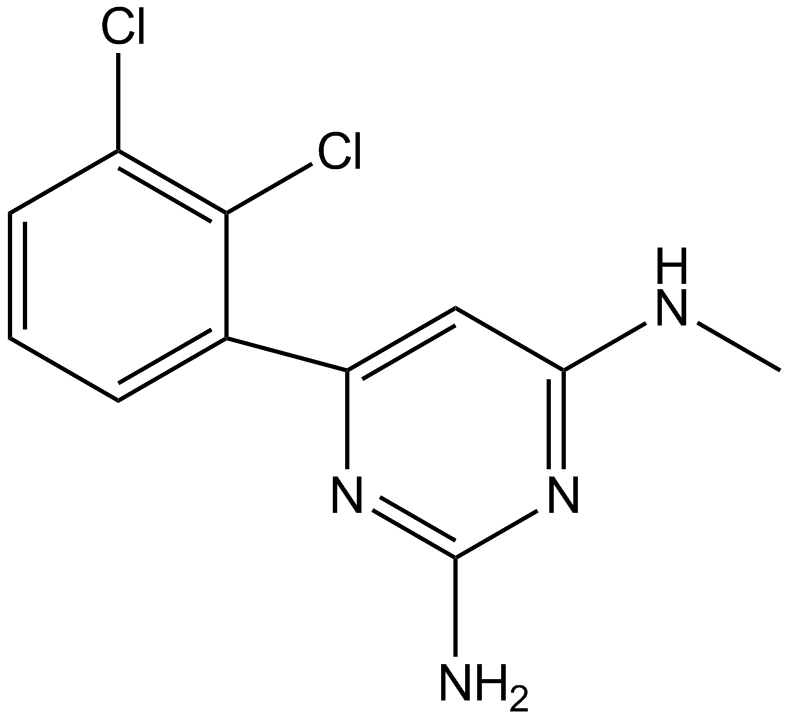
TH287
MTH1 inhibitor
-
GC37775
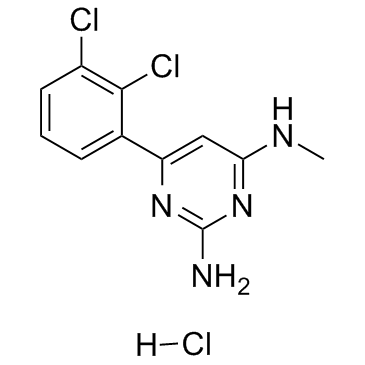
TH287 hydrochloride
TH287 hydrochloride is a potent and selective inhibitor of MTH1, with an IC50 of 0.8 nM. TH287 hydrochloride is highly selective towards MTH1, with no relevant inhibition of MTH2, NUDT5, NUDT12, NUDT14, NUDT16, dCTPase, dUTPase and ITPA at 100 μM. TH287 hydrochloride could act as a chemotherapeutic agent for cancer research.
-
GC12716
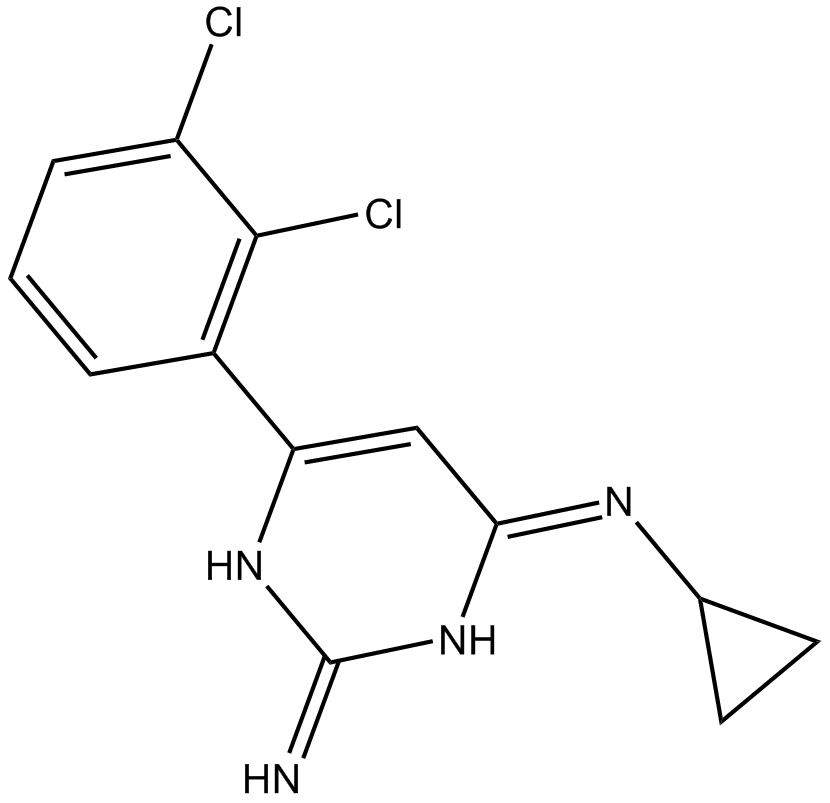
TH588
MTH1 inhibitor
-
GC37777
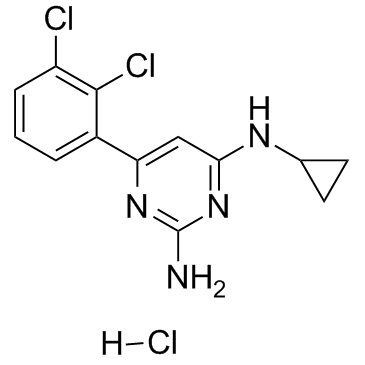
TH588 hydrochloride
TH588 hydrochloride is first-in-class nudix hydrolase family inhibitor that potently and selectively engage and inhibit the MTH1 (IC50= 5 nM).
-
GC31868
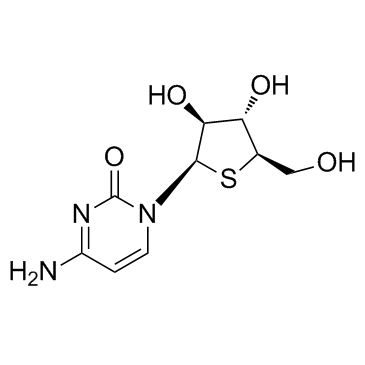
Thiarabine (OSI-7836)
Thiarabine (OSI-7836) (OSI-7836) shows potent anti-tumor activity and inhibition of DNA synthesis.
-
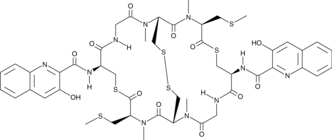
GC48165
Thiocoraline
A depsipeptide and DNA bis-intercalator with antibacterial and anticancer activities
-
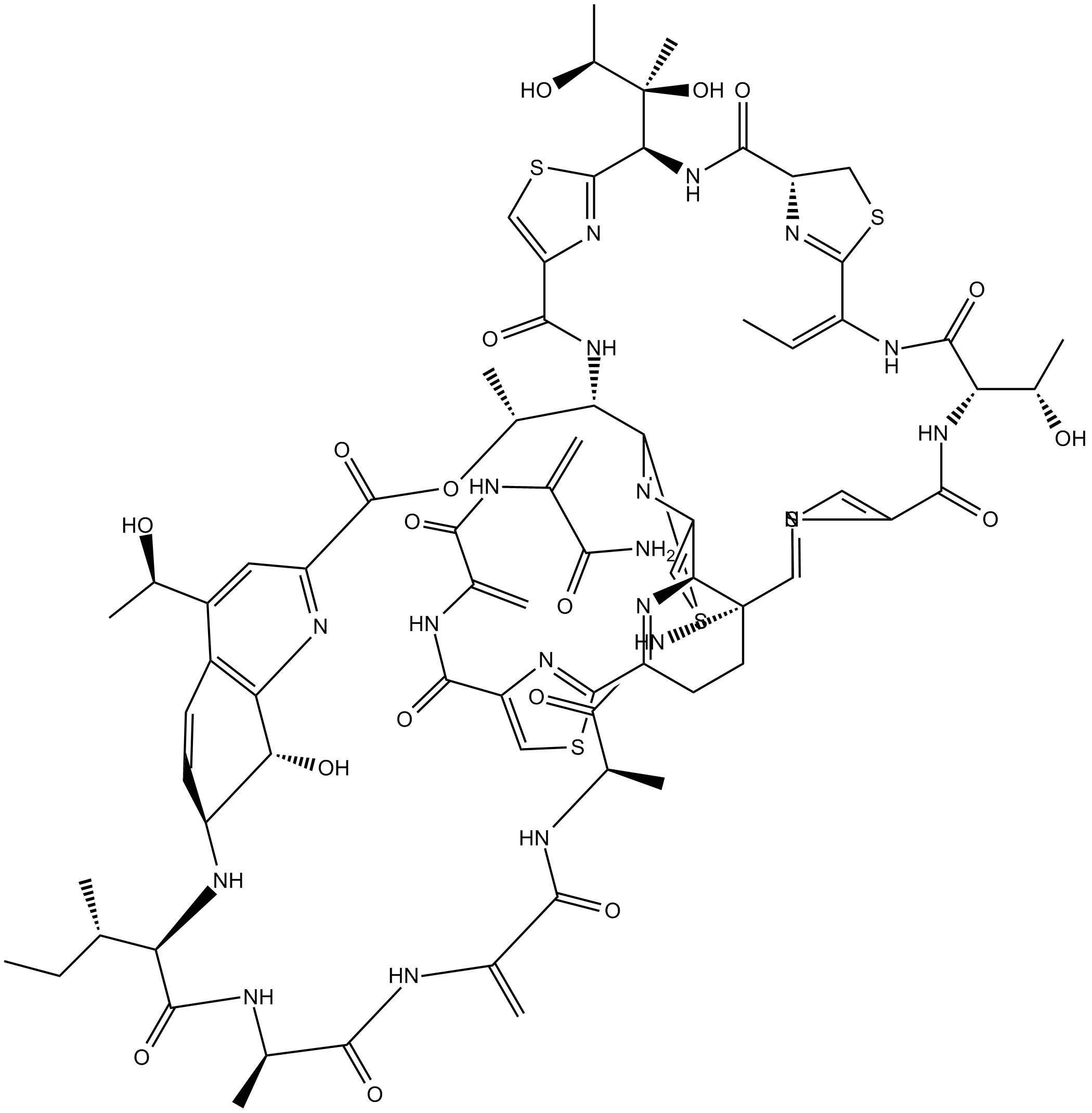
GC13002
Thiostrepton
Antibiotic that inhibits bacterial protein synthesis
-
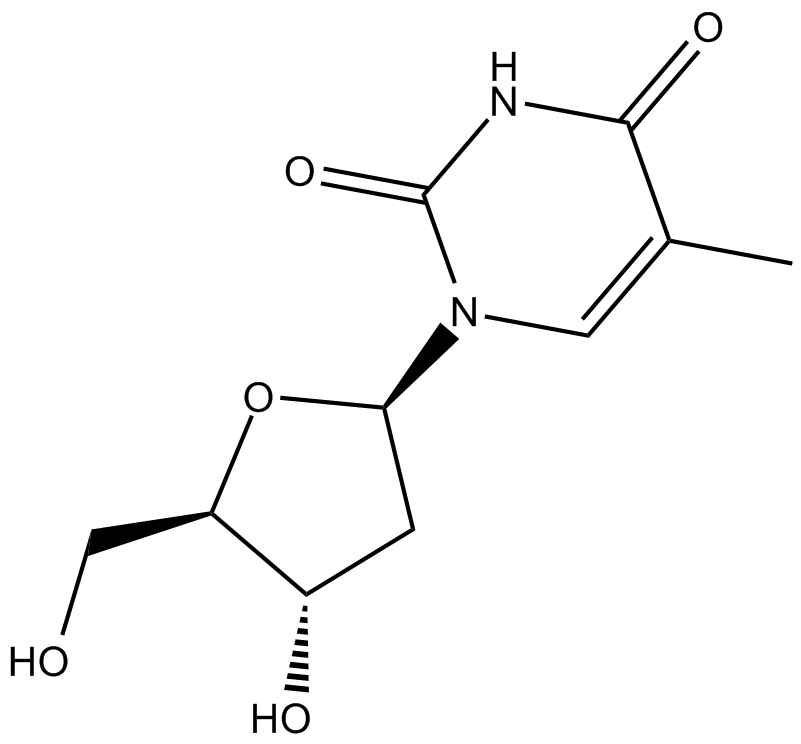
GC15815
Thymidine
pyrimidine nucleoside
-
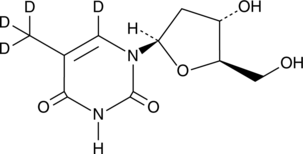
GC49529
Thymidine-d4
An internal standard for the quantification of thymidine
-
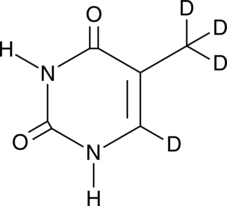
GC49474
Thymine-d4
An internal standard for the quantification of thymine
-
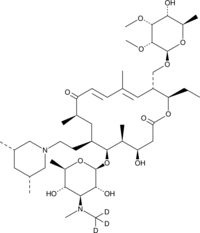
GC49076
Tilmicosin-d3
An internal standard for the quantification of tilmicosin
-
GC64897
Tofersen
Tofersen (BIIB067) is an antisense oligonucleotide that mediates RNase H-dependent degradation of superoxide dismutase 1 (SOD1) mRNA to reduce the synthesis of SOD1 protein.

-
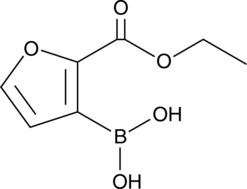
GC48190
TP-4748
A heterocyclic building block
-
GC17897
Triapine
Ribonucleotide reductase inhibitor
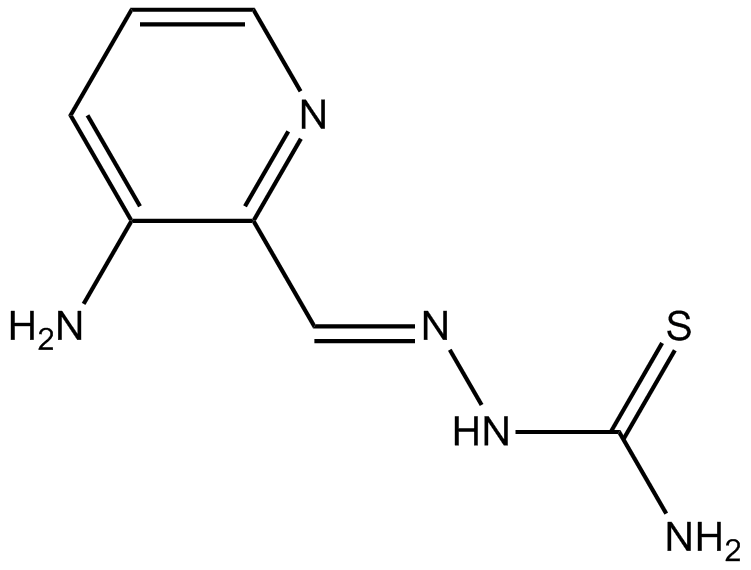
-
GC61348
Triazavirin
Triazavirin is a nucleoside analogue of nucleic acid and an antiviral agent.
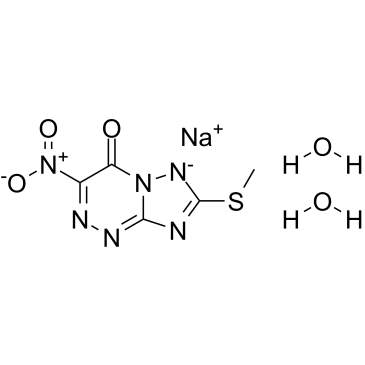
-
GC15392
Triciribine
Akt inhibitor,highly selective
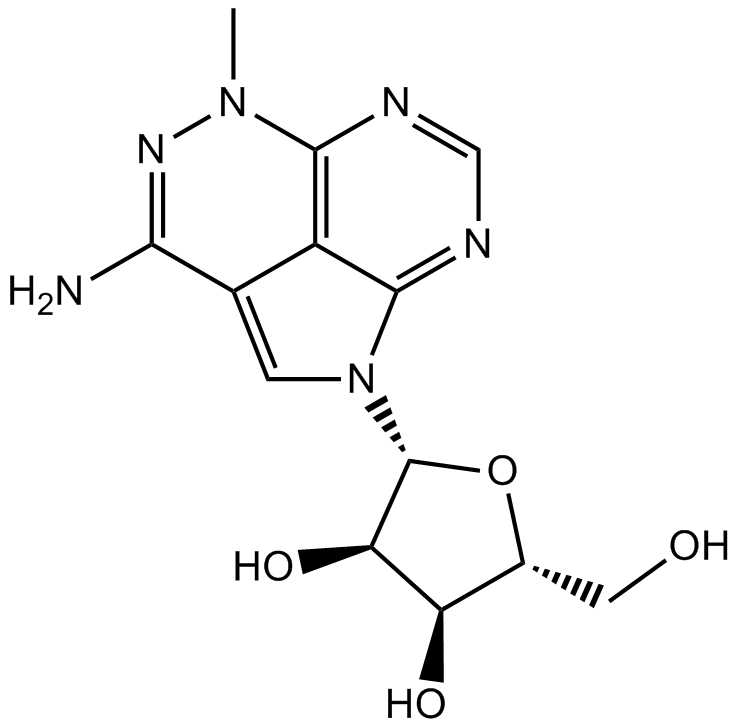
-
GC13399
Trimidox
specific ribonucleotide reductase inhibitor
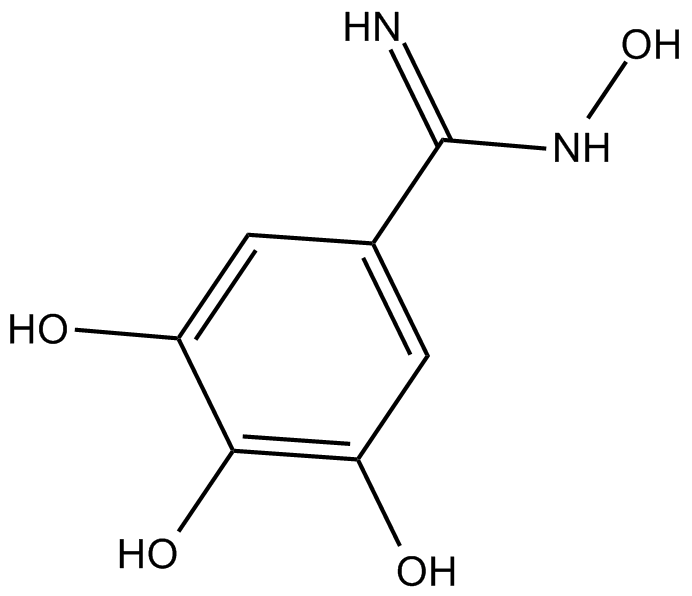
-
GC12044
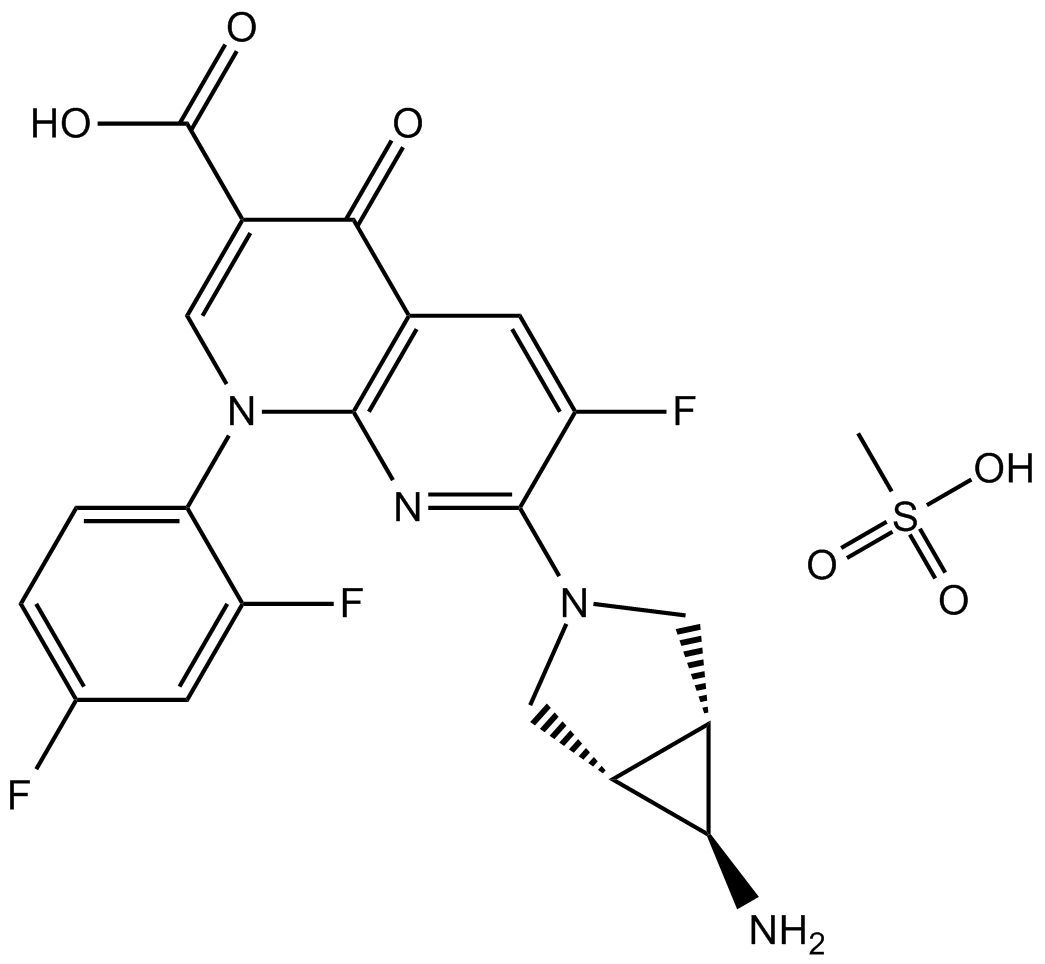
Trovafloxacin mesylate
Fluoroquinolone antibiotic
-
GC61359
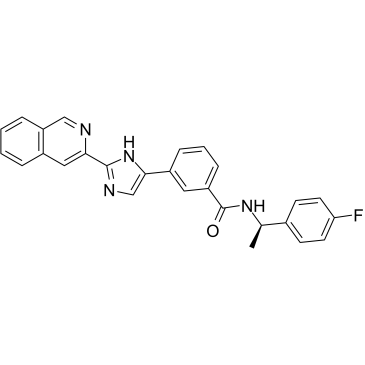
TTP-8307
TTP-8307 is a potent inhibitor of the replication of several rhino- and enteroviruses.
-
GC16879
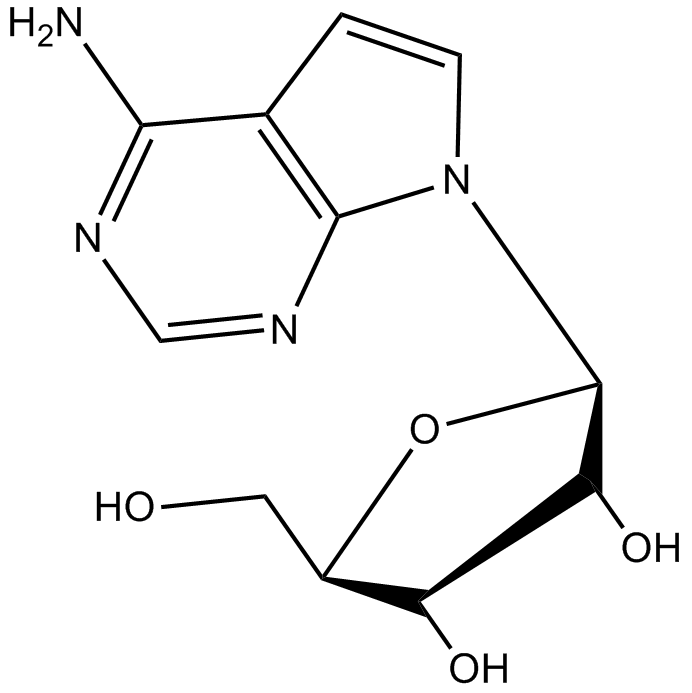
Tubercidin
Tubercidin is a pyrrolopyrimidine nucleoside analog with significant activity against schistosomal infections.
-
GC49835
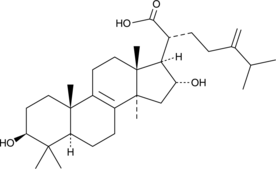
Tumulosic Acid
Tumulosic Acid, a triterpenoid, inhibits KLK5 protease activity (IC50= 14.84 μM).
-
GC41598
Tyrocidine Complex
Tyrocidine complex is a mixture of cyclic decapeptides originally isolated from B.
-
GC48987
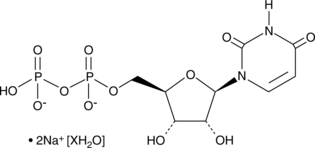
UDP (sodium salt hydrate)
An agonist of P2Y6
-
GC49132
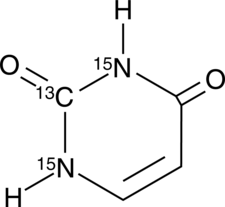
Uracil-13C,15N2
An internal standard for the quantification of uracil
-
GC49800
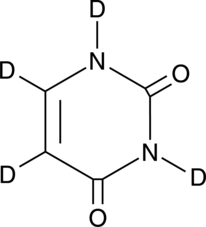
Uracil-d4
An internal standard for the quantification of uracil
-
GC13742
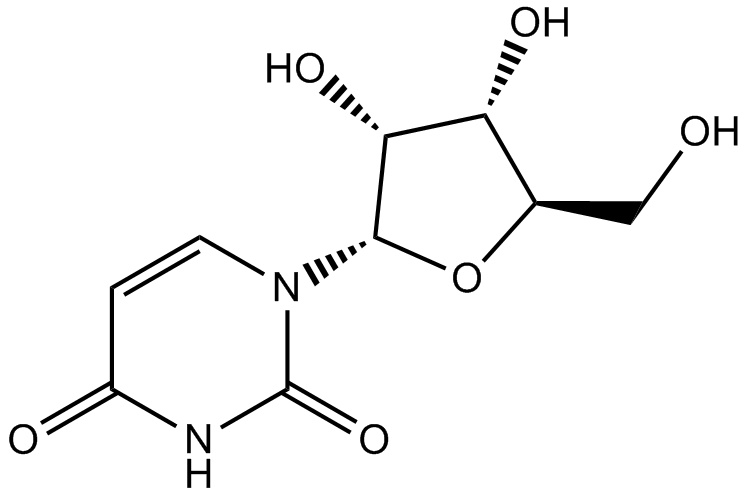
Uridine
DNA/RNA synthesis chemical
-
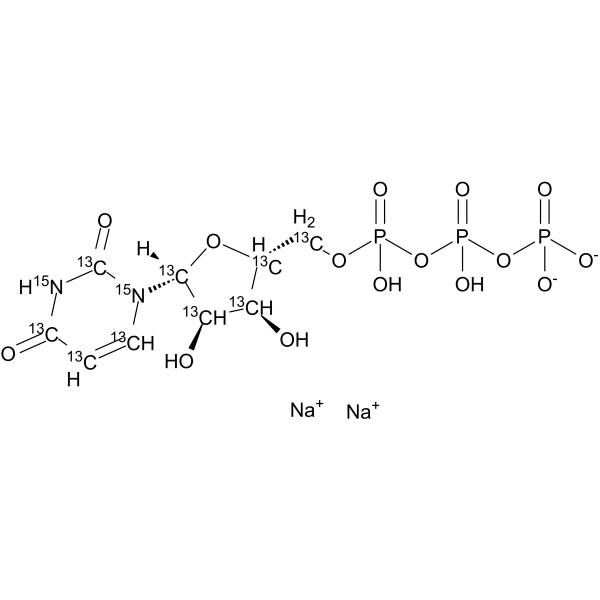
GC62546
Uridine triphosphate 13C9,15N2 sodium
Uridine triphosphate 13C9,15N2 (UTP 13C9,15N2) sodium is a labeled Uridine triphosphate sodium.
-
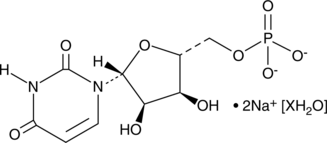
GC49297
Uridine-5’-monophosphate (sodium salt hydrate)
A ribonucleotide
-
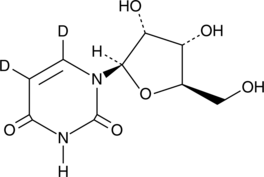
GC49528
Uridine-d2
Uridine-d2 is the deuterium labeled Uridine.
-
GC15168
Urolithin A
A gut-microbial metabolite of ellagic acid
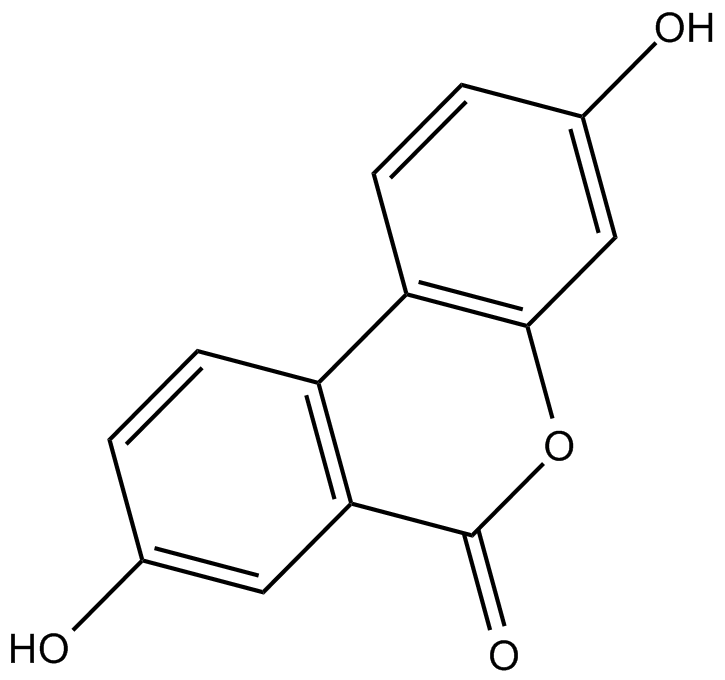
-
GC10339
VAL-083
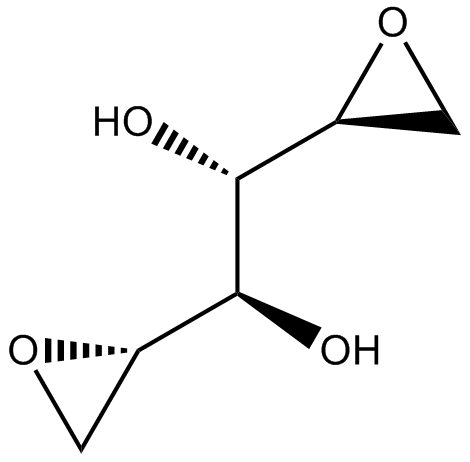
-
GC48533
Vanoxonin
A bacterial metabolite
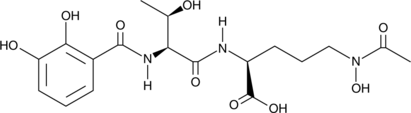
-
GC14851
Vidofludimus
DHODH inhibitor
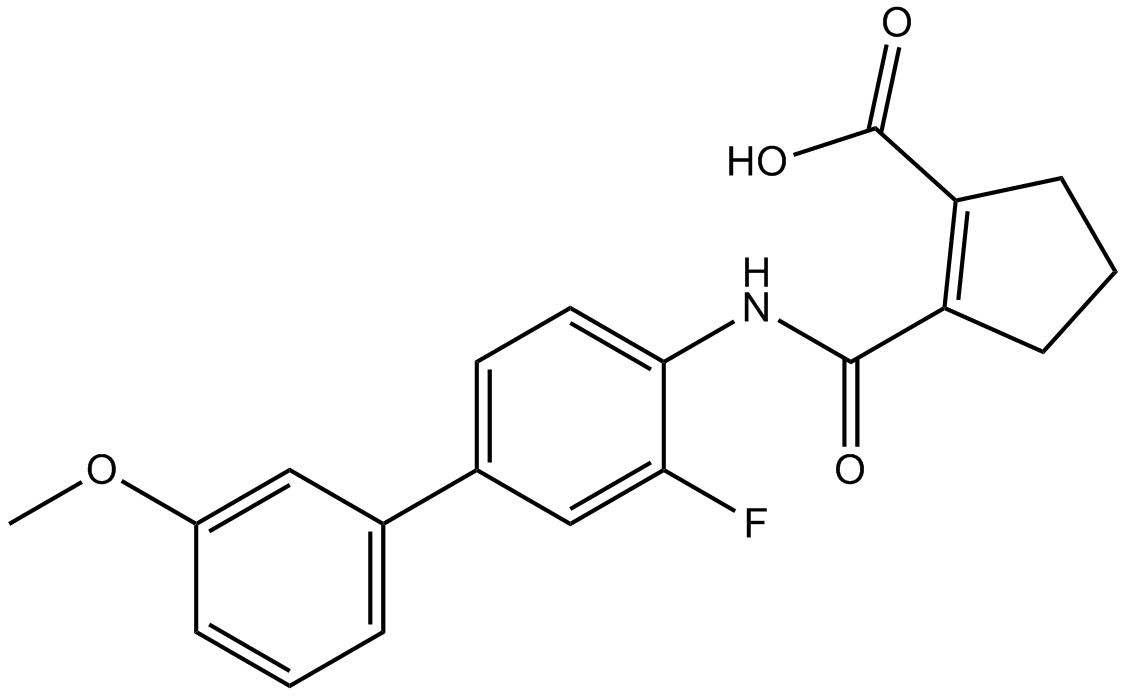
-
GC14663
Viomycin
Viomycin is a potent antibiotic against Mycobacteria. Viomycin rapidly inhibits polypeptide chain elongation when added to purified endogenous Escherichia coli polysomes actively engaged in polypeptide synthesis.
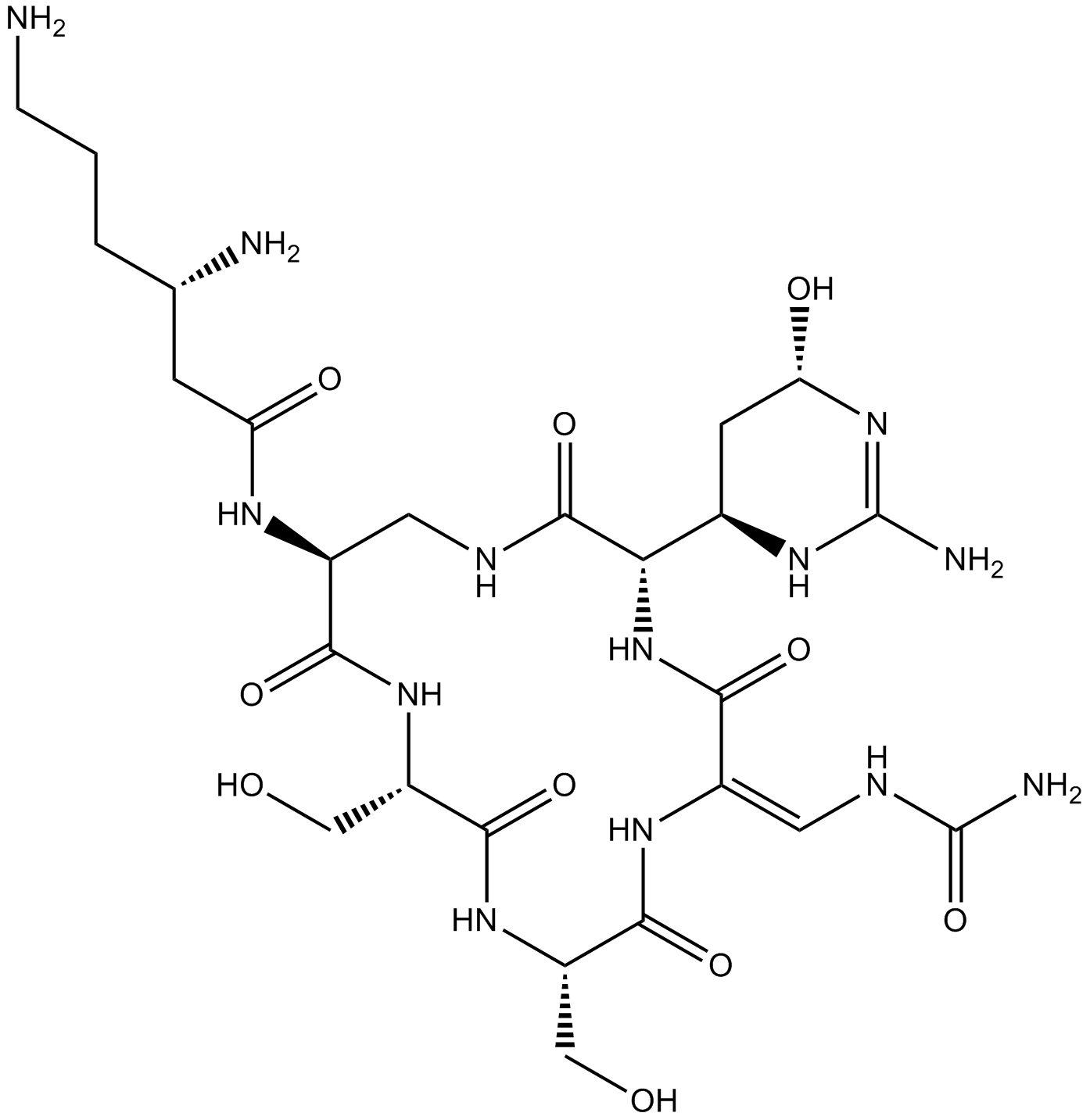
-
GC70142
Werner syndrome RecQ helicase-IN-3
Werner syndrome RecQ helicase-IN-3 is an effective orally active inhibitor of the Werner syndrome RecQ DNA helicase WRN, with an IC50 value of 0.06 µM. It exhibits anti-proliferative activity and has anti-cancer properties.
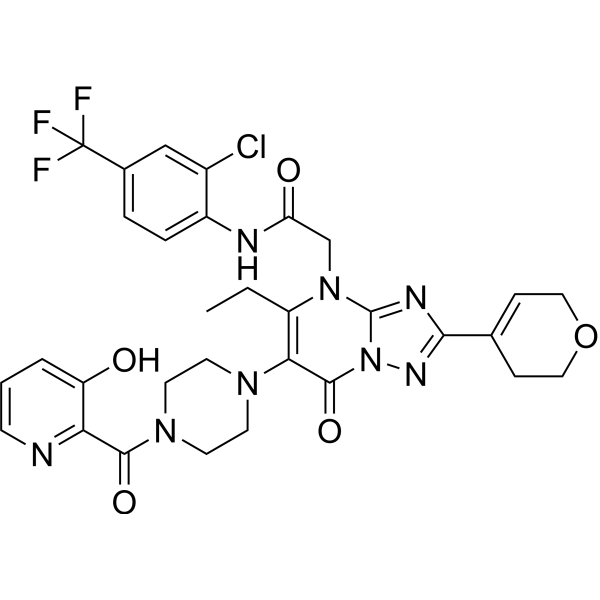
-
GC37940
Xanthopterin
A pteridine with antiproliferative and anticancer activities
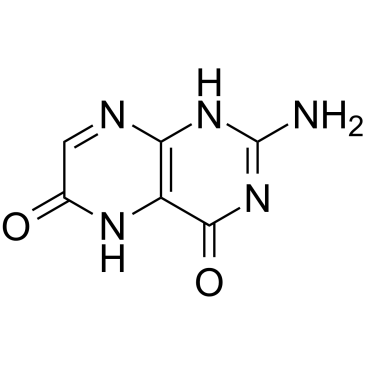
-
GC37941
Xanthopterin (hydrate)
Xanthopterin hydrate, an unconjugated pteridine compound, is the main component of the yellow granule in the Oriental hornet bear wings, produces a characteristic excitation/emission maximum at 386/456 nm.
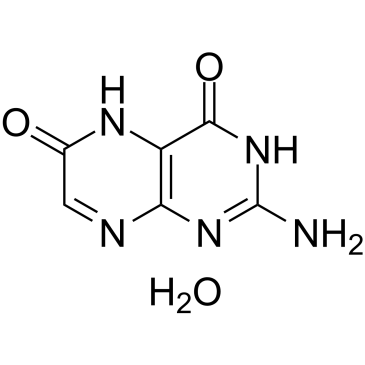
-
GC17386
YK-4-279
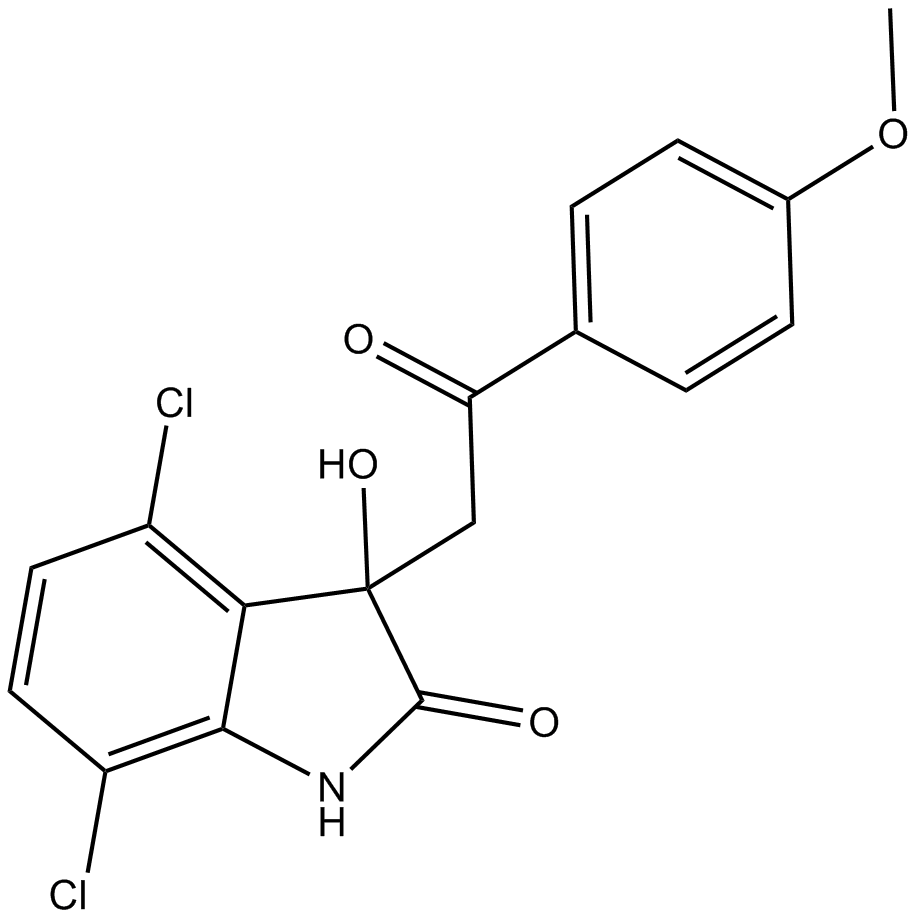
-
GC70172
ZIM
ZIM, a derivative of 4-Aminoantipyrine, is an effective DNA damage inducer that can cause genomic and chromosomal damage as well as induce cell death and activate phagocytosis. ZIM has potential for chemotherapy and can be used in cancer research.
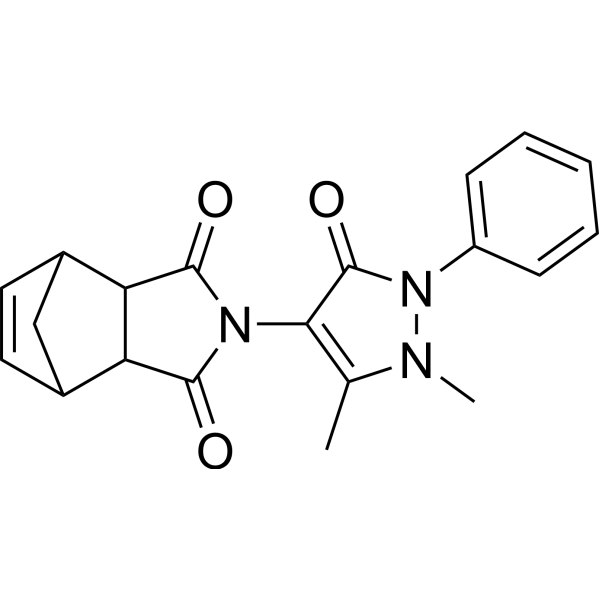
-
GC19391
Zoliflodacin
Zoliflodacin (ETX0914;AZD0914) is a novel spiropyrimidinetrione bacterial DNA gyrase/topoisomerase inhibitor.
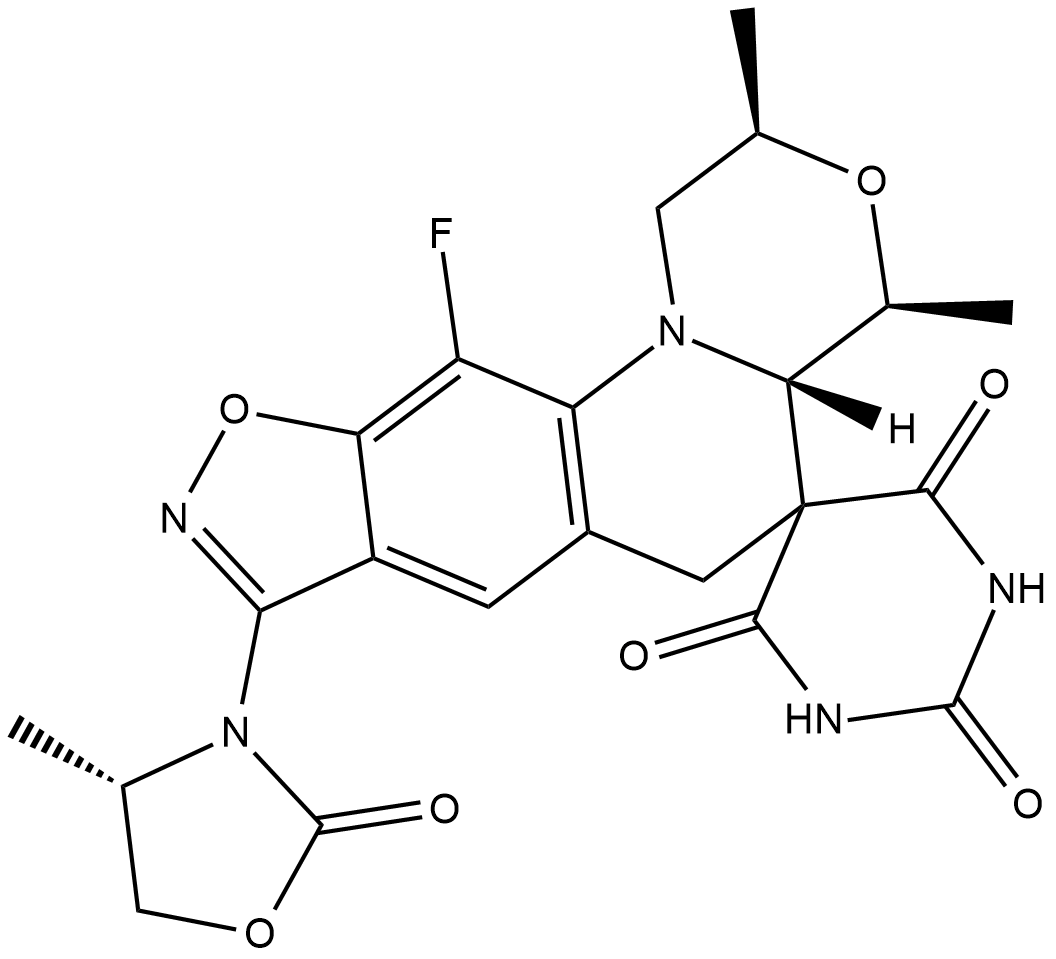
-
GC16277
α-Amanitin
A selective RNA polymerase II inhibitor