Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Targets for Chromatin/Epigenetics
- Bromodomain(52)
- Aurora Kinase(89)
- DNA Methyltransferase(40)
- HDAC(218)
- Histone Acetyltransferases(67)
- Histone Demethylases(98)
- Histone Methyltransferase(212)
- HIF(101)
- JAK(178)
- MBT Domains(1)
- PARP(128)
- Pim(35)
- Protein Ser/Thr Phosphatases(41)
- RNA Polymerase(8)
- Sirtuin(84)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(207)
- MicroRNA(13)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Products for Chromatin/Epigenetics
- Cat.No. Product Name Information
-
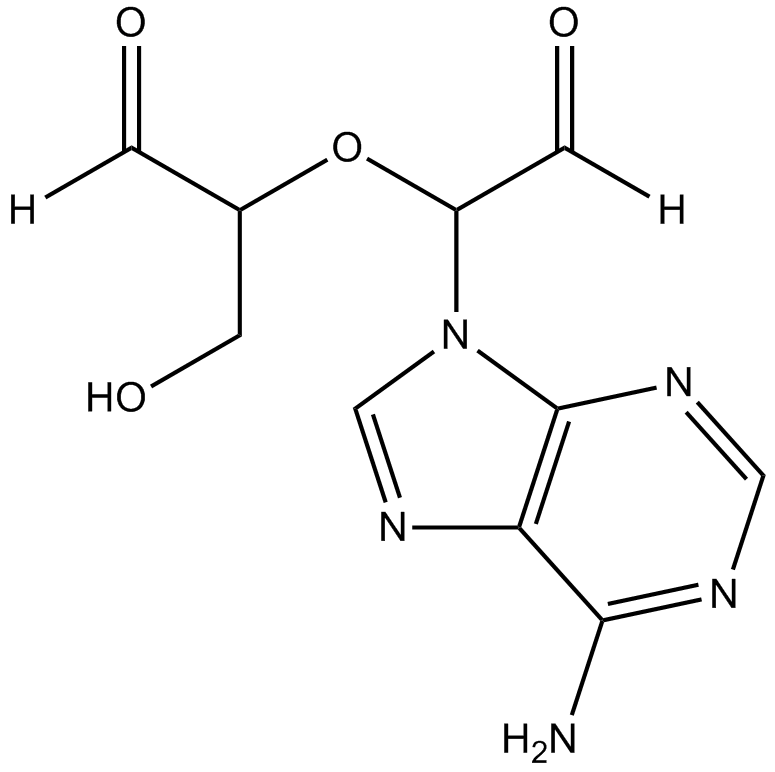
GC16509
Adox
Adox, a purine nucleoside analogue, is a potent inhibitor of S-Adenosylhomocysteine hydrolase (SAHH) (Ki=3.3 nM). Adenosine Dialdehyde exhibits potent anti-tumor activity in vivo and can be used for the cancer research.
-
GC64527
ADTL-SA1215
ADTL-SA1215 is a first-in-class specific small-molecule activator of SIRT3 that modulates autophagy in triple negative breast cancer.
-
GC65330
AES-135
AES-135, a hydroxamic acid-based pan-HDAC inhibitor, prolongs survival in an orthotopic mouse model of pancreatic cancer. AES-135 inhibits HDAC3, HDAC6, HDAC8, and HDAC11 with IC50s ranging from 190-1100 nM.
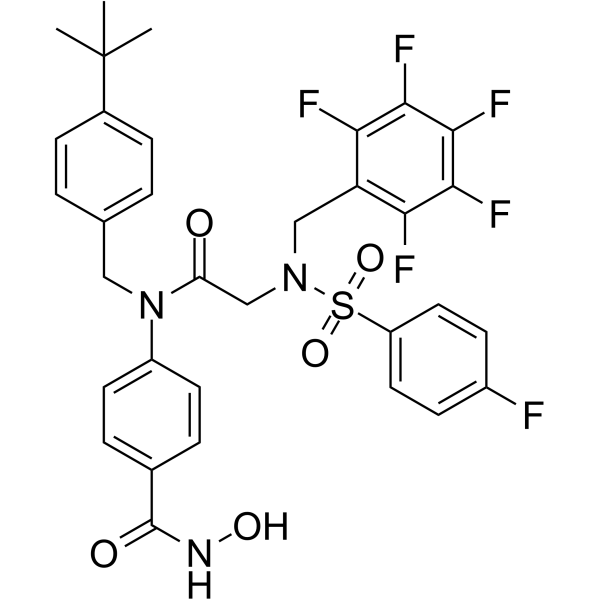
-
GC48775
AES-350
An inhibitor of class I and class IIb HDACs
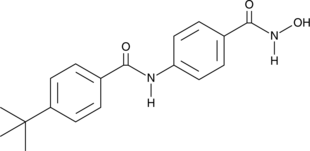
-
GC66379
AFM-30a hydrochloride
AFM-30a hydrochloride is a potent protein arginine deiminase 2 (PAD2) inhibitor and has excellent PAD2-selectivity. AFM-30a hydrochloride binds to PAD2 with an EC50 value of 9.5 μM. AFM-30a hydrochloride also inhibits H3 citrullination with an EC50 value of 0.4 μM. AFM-30a hydrochloride can be used for the research of certain cancers and a variety of autoimmune diseases including rheumatoid arthritis (RA), multiple sclerosis, lupus, and ulcerative colitis.
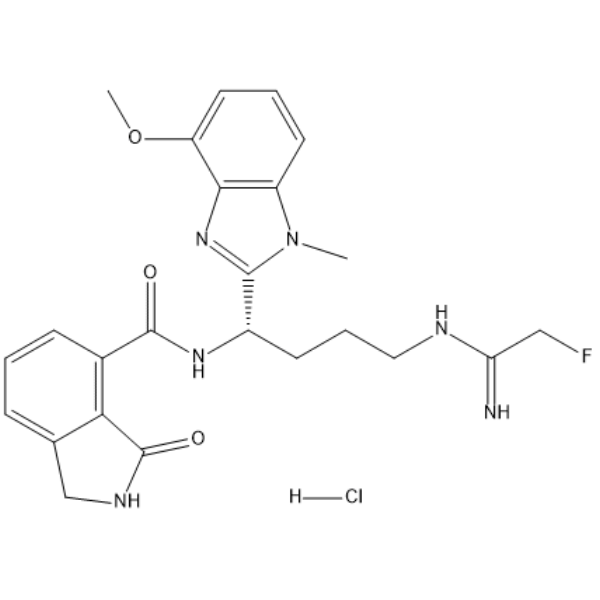
-
GC33416
AFP464
AFP464 (NSC710464 free base), is an active HIF-1α inhibitor with an IC50 of 0.25 μM, also is a potent aryl hydrocarbon receptor (AhR) activator.

-
GC16318
AG-14361
A PARP1 inhibitor
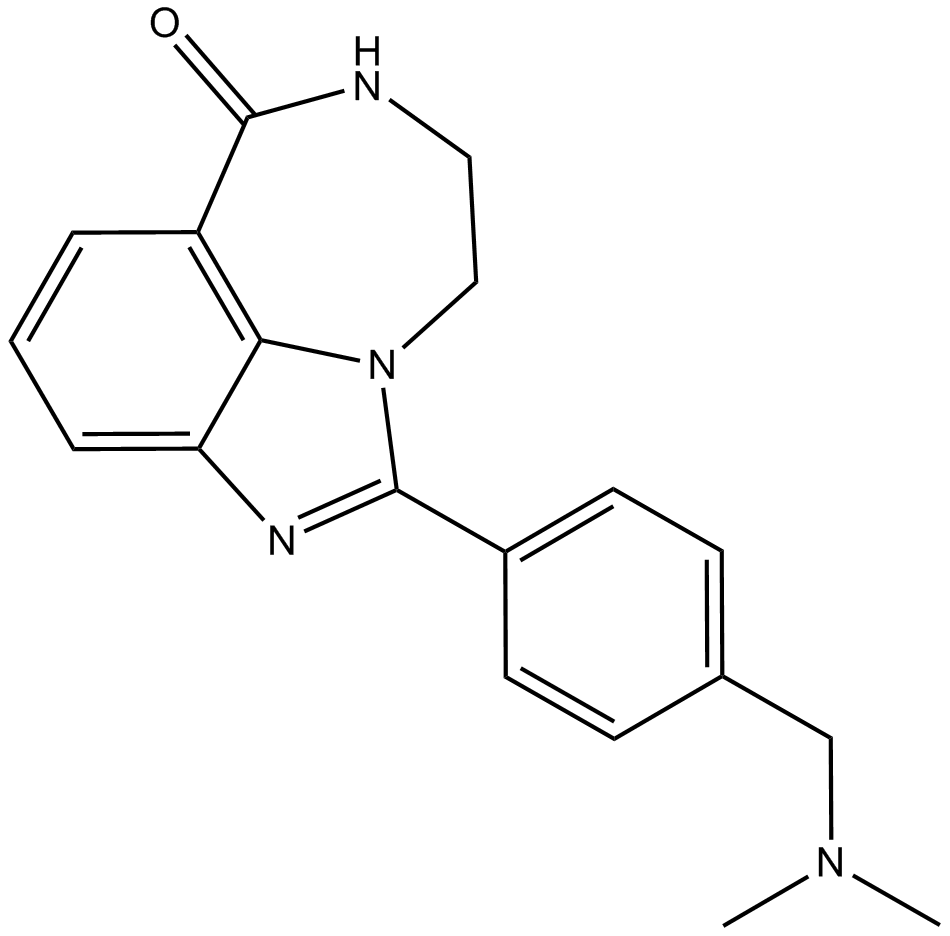
-
GC17881
AGK 2
AGK2 is a selective SIRT2 inhibitor, with an IC50 of 3.
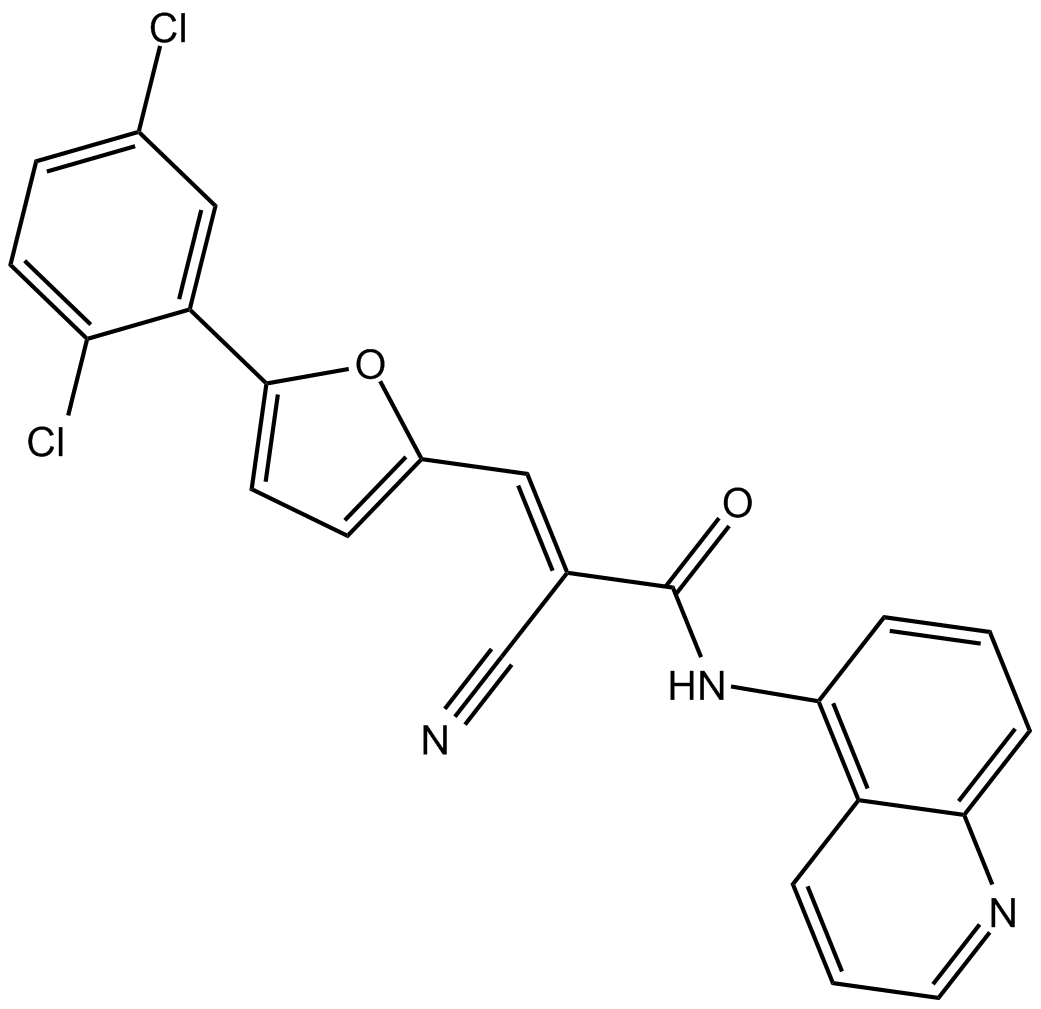
-
GC15931
AGK7
cell-permeable, selective inhibitor of SIRT2
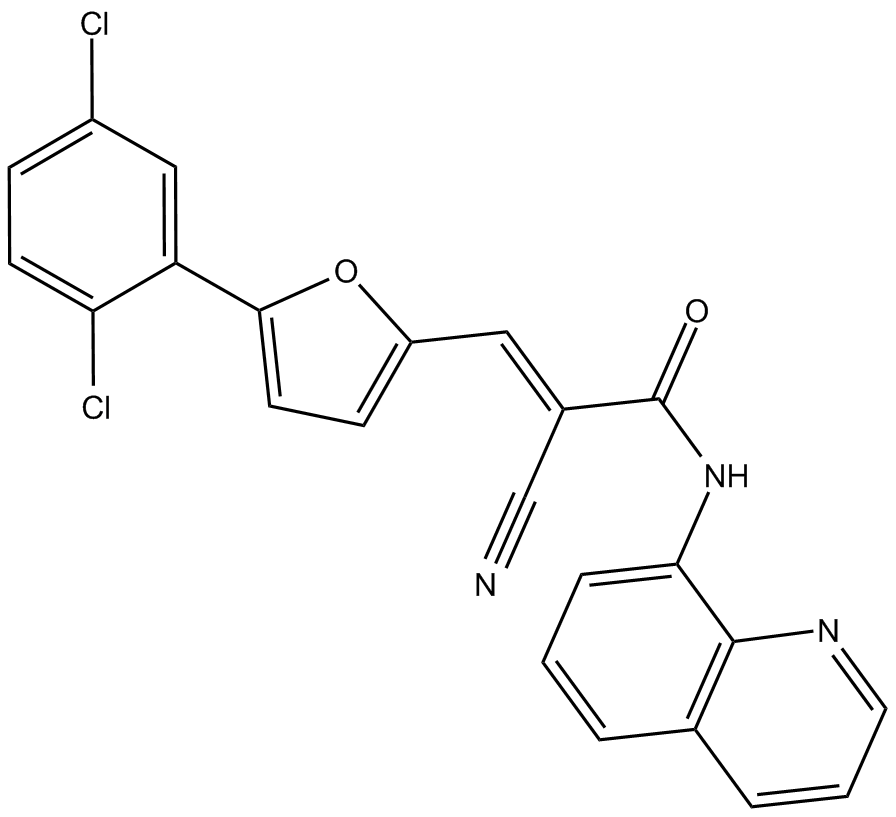
-
GN10413
Agrimol B
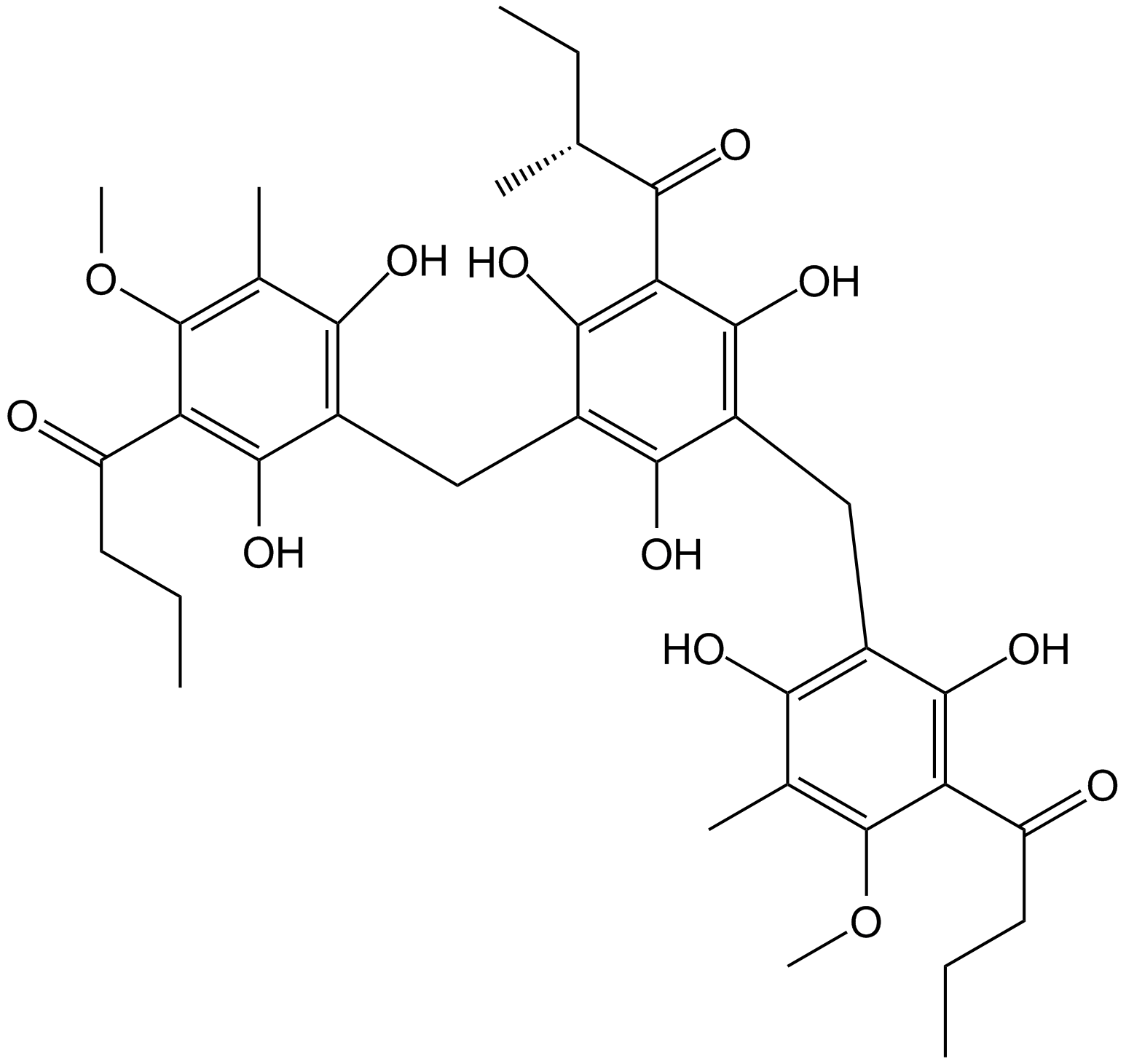
-
GC10676
AK-7
selective and brain-permeable SIRT2 inhibitor
-
GC62277
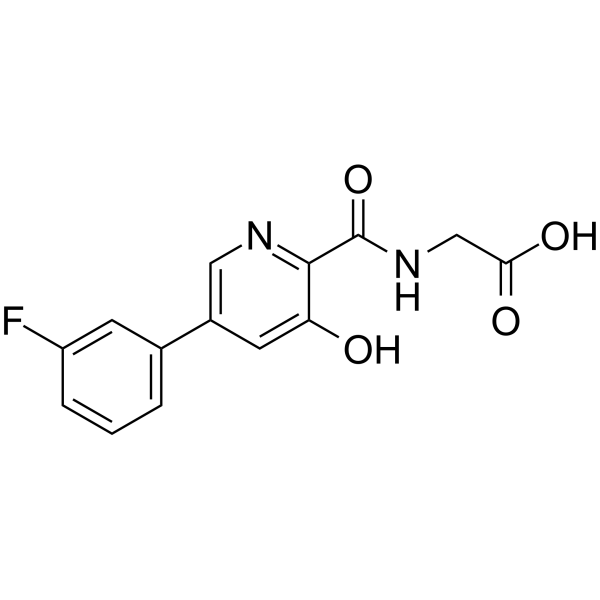
AKB-6899
Ro24-7429 is a potent and orally active HIV-1 transactivator protein Tat antagonist. Ro24-7429 is also a runt-related transcription factor 1 (RUNX1) inhibitor. Ro24-7429 has anti-HIV, antifibrotic and anti-inflammatory effects.
-
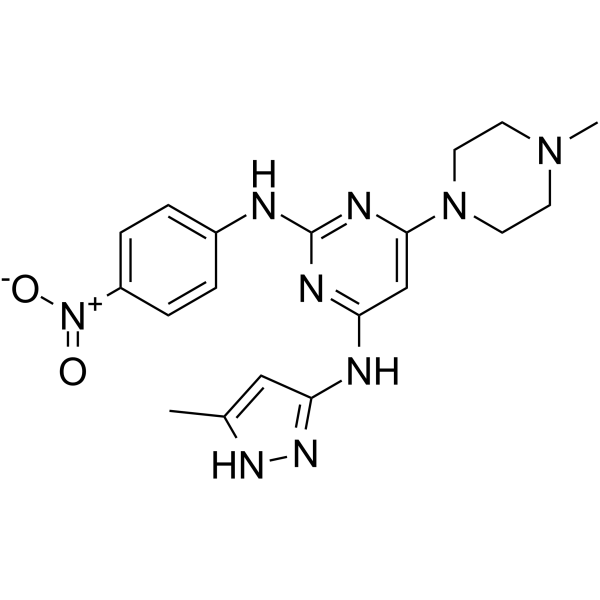
GC62433
AKI603
AKI603 is an inhibitor of Aurora kinase A (AurA), with an IC50 of 12.3 nM. AKI603 is developed to overcome resistance mediated by BCR-ABL-T315I mutation. AKI603 exhibits strong anti-proliferative activity in leukemic cells.
-
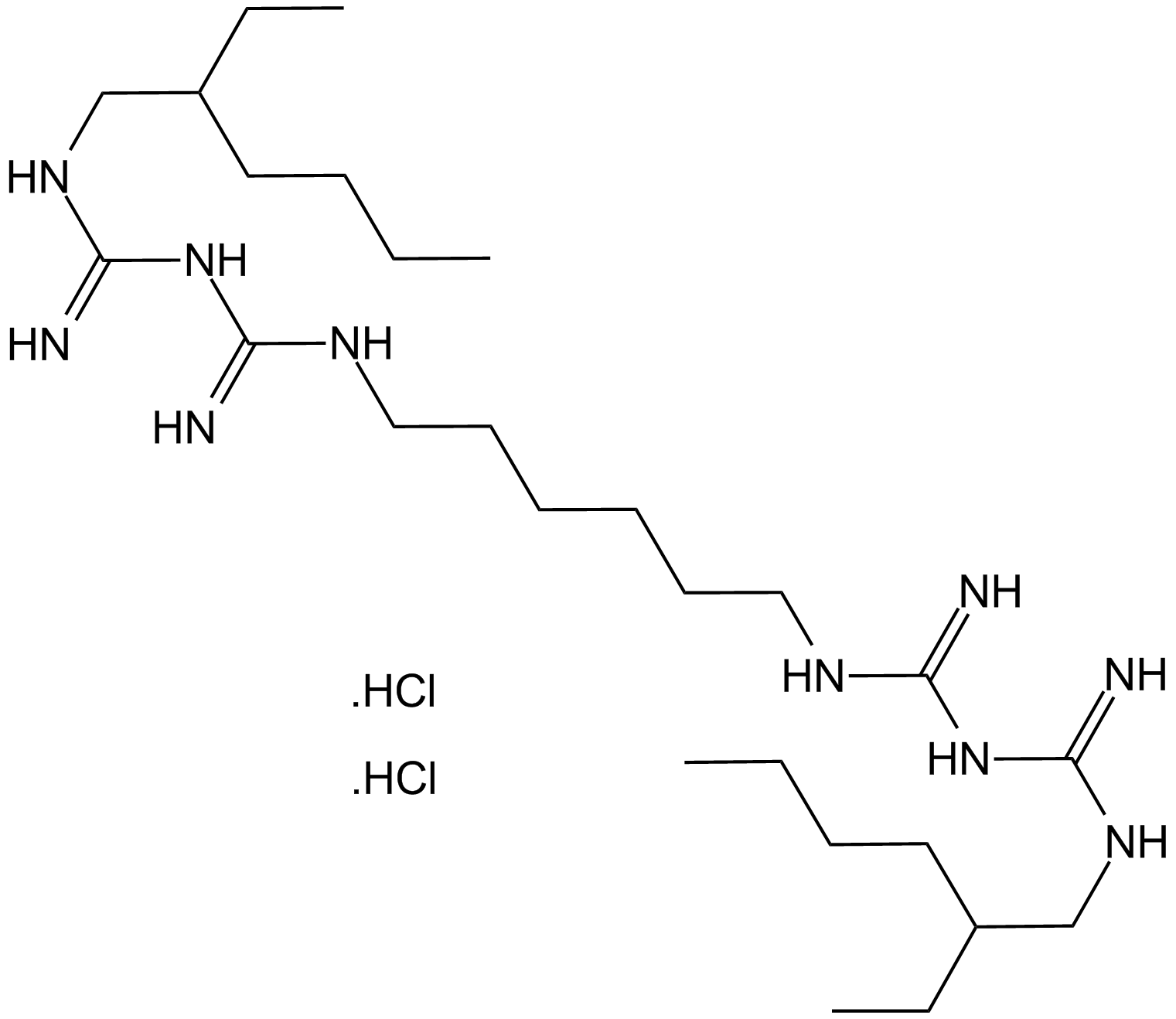
GC17344
Alexidine dihydrochloride
An alkyl bis(biguanide) antiseptic
-
GC49393
all-trans-13,14-Dihydroretinol
A metabolite of all-trans retinoic acid

-
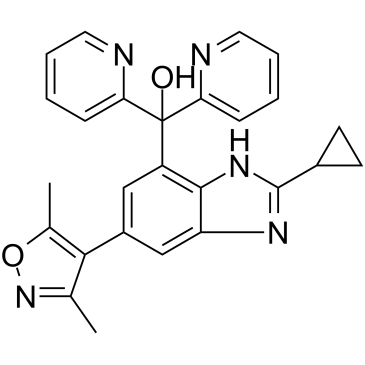
GC35297
Alobresib
A BET bromodomain inhibitor
-
GC67984
Alteminostat
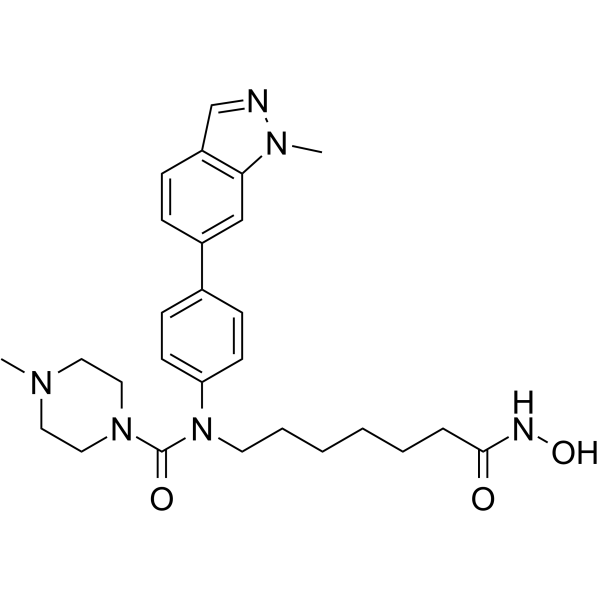
-
GC13198
AMG-900
A selective pan-Aurora kinase inhibitor
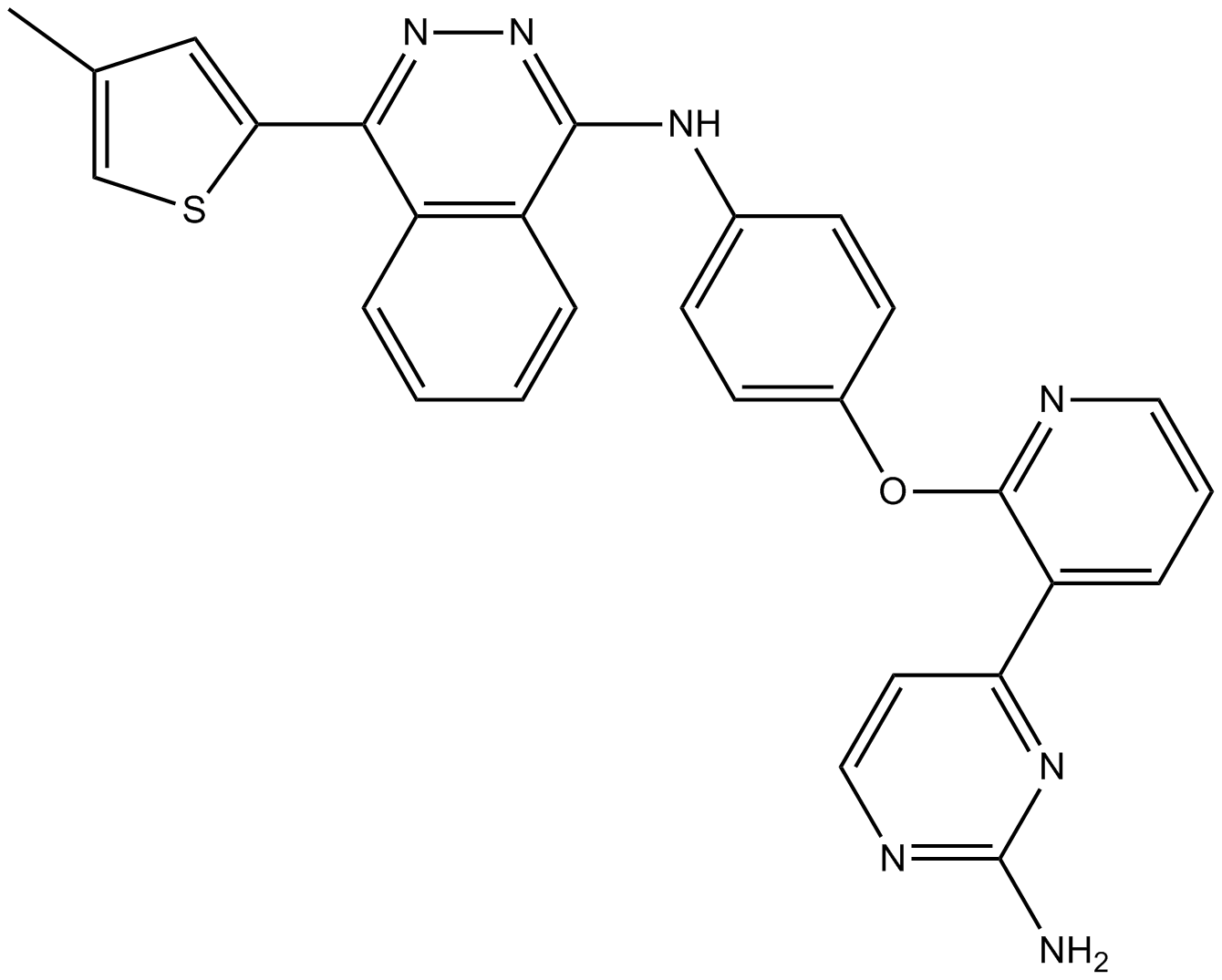
-
GC17275
AMI-1
A cell permeable inhibitor of PRMTs
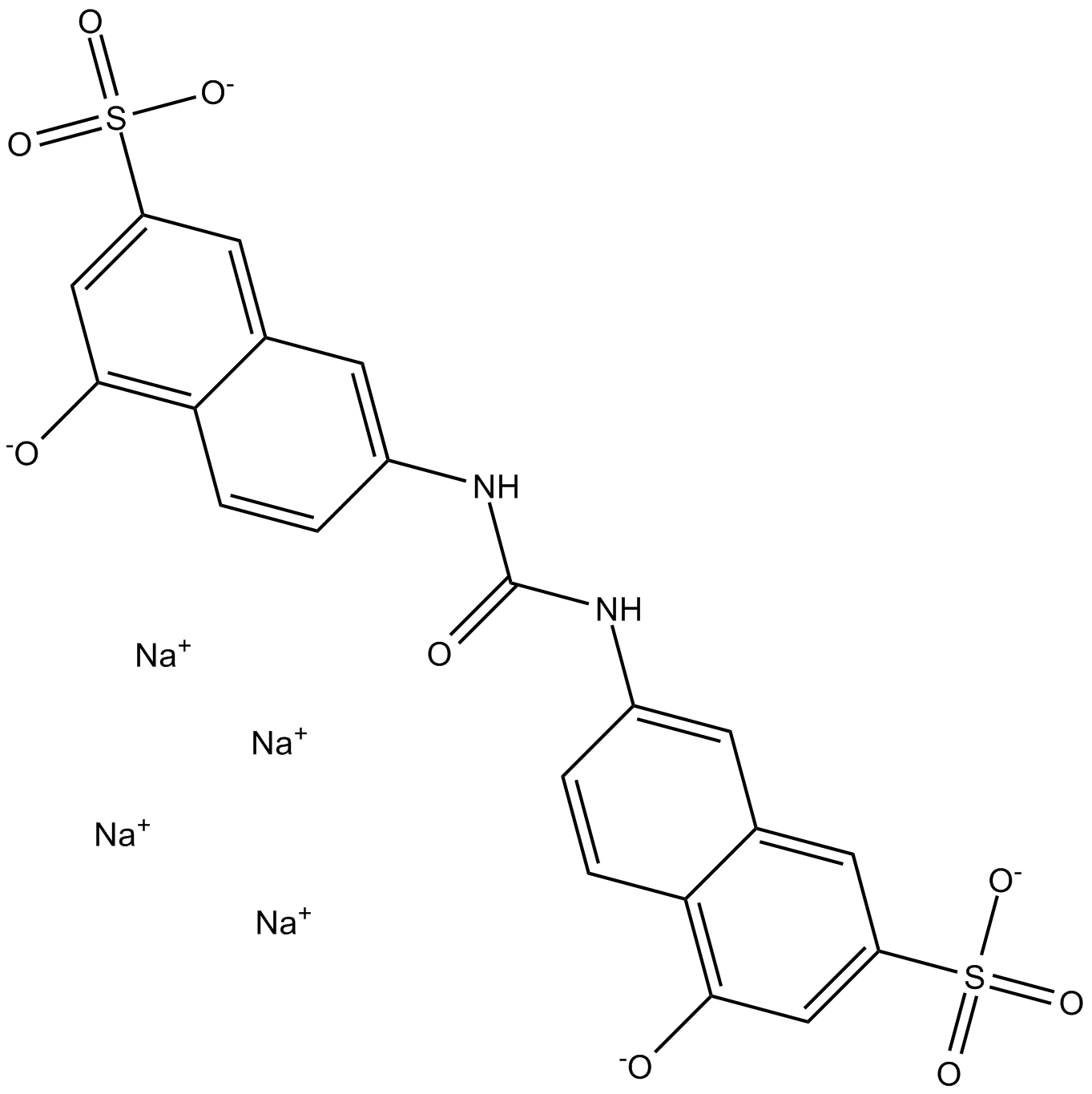
-
GC39840
AMI-1 free acid
AMI-1 free acid is a potent, cell-permeable and reversible inhibitor of protein arginine N-methyltransferases (PRMTs), with IC50s of 8.8 μM and 3.0 μM for human PRMT1 and yeast-Hmt1p, respectively. AMI-1 free acid exerts PRMTs inhibitory effects by blocking peptide-substrate binding.
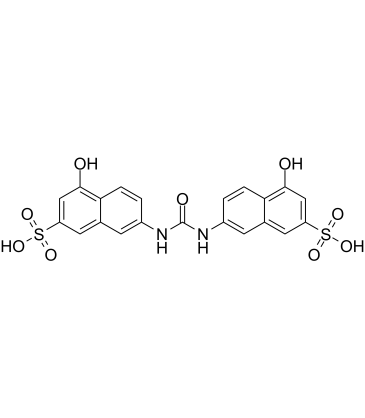
-
GC17546
AMI5
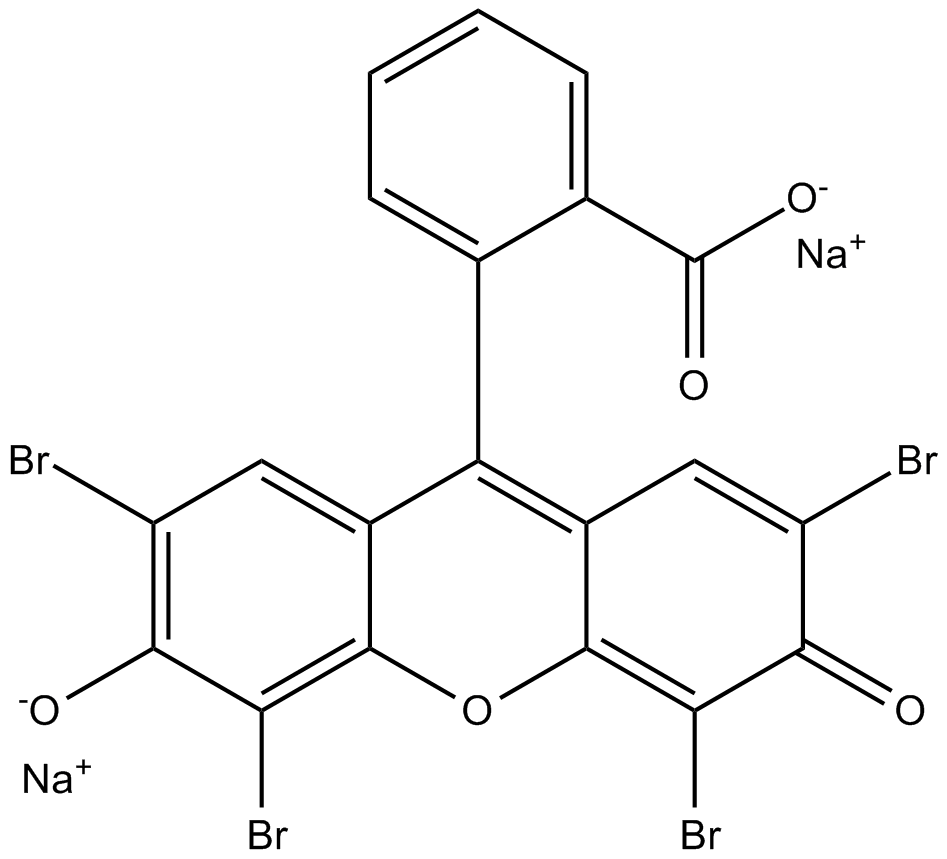
Eosin Y (disodium) is a soluble acid red dye molecule.
-
GC42785
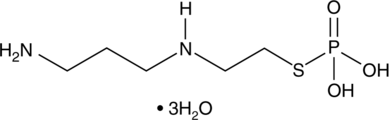
Amifostine (hydrate)
Amifostine (hydrate) (WR2721 trihydrate) is a broad-spectrum cytoprotective agent and a radioprotector. Amifostine (hydrate) selectively protects normal tissues from damage caused by radiation and chemotherapy. Amifostine (hydrate) is potent hypoxia-inducible factor-α1 (HIF-α1) and p53 inducer. Amifostine (hydrate) protects cells from damage by scavenging oxygen-derived free radicals. Amifostine (hydrate) reduces renal toxicity and has antiangiogenic action.
-
GC42790
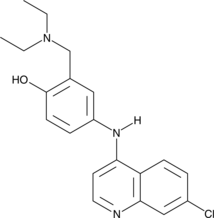
Amodiaquine
Amodiaquine is an aminoquinoline antimalarial compound.
-
GC60579
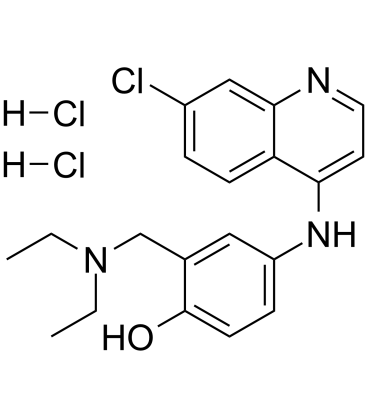
Amodiaquine dihydrochloride
Amodiaquine dihydrochloride (Amodiaquin dihydrochloride), a 4-aminoquinoline class of antimalarial agent, is a potent and orally active histamine N-methyltransferase inhibitor with a Ki of 18.6 nM.
-
GC10905
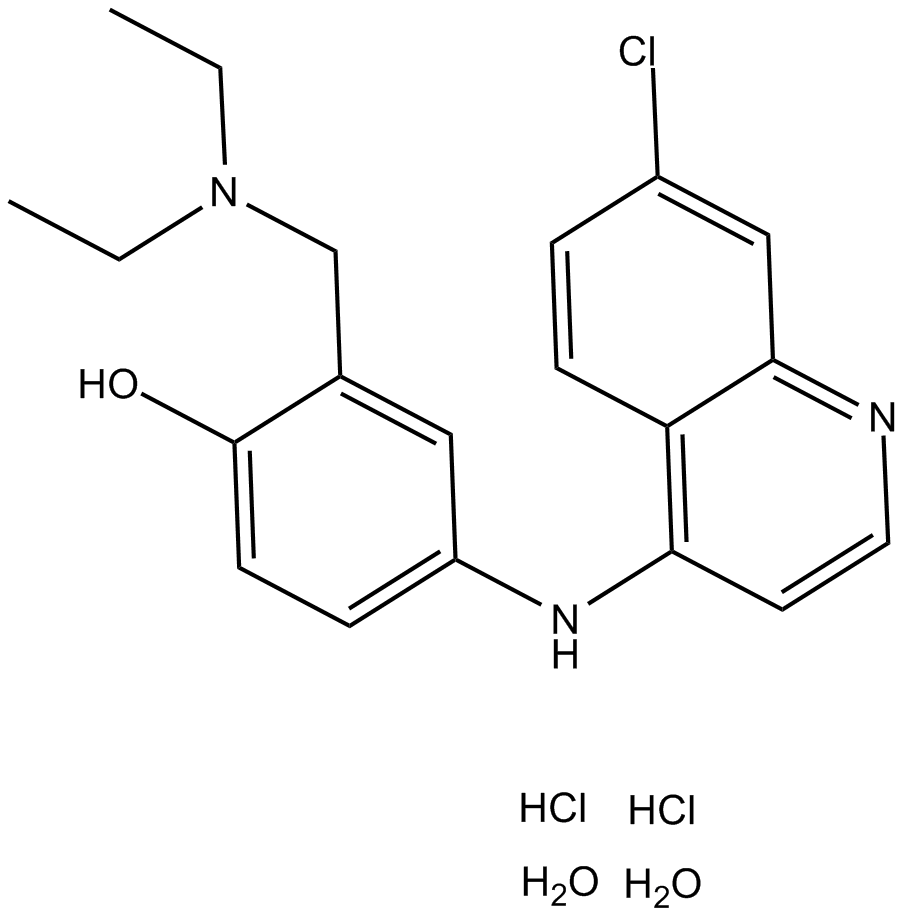
Amodiaquine dihydrochloride dihydrate
histamine N-methyl transferase inhibitor
-
GC67912
Amredobresib

-
GC17284
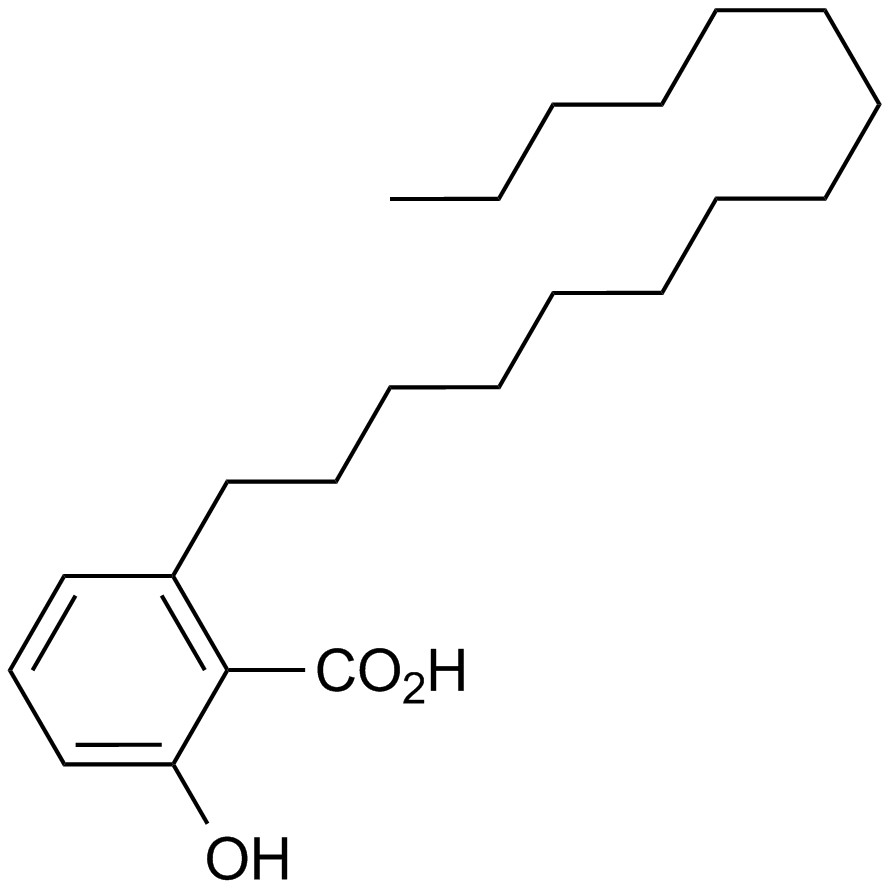
Anacardic acid
A histone acetyltransferase inhibitor
-
GC40674
APHA Compound 8
A class I and II HDAC inhibitor
-
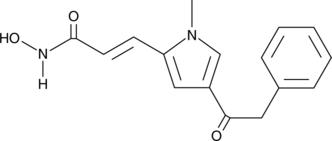
GC12961
Apicidin
A cell-permeable HDAC inhibitor
-
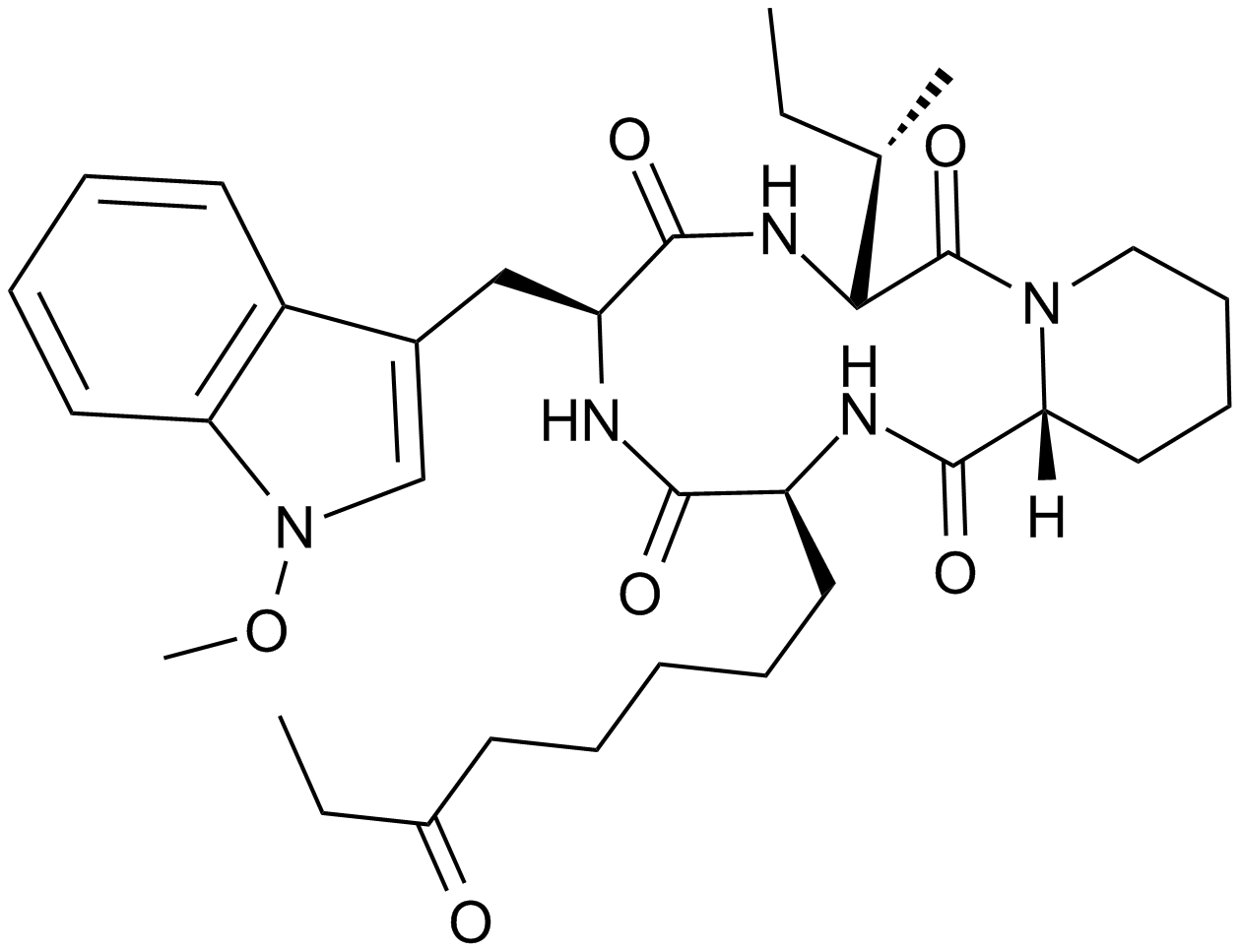
GC14590
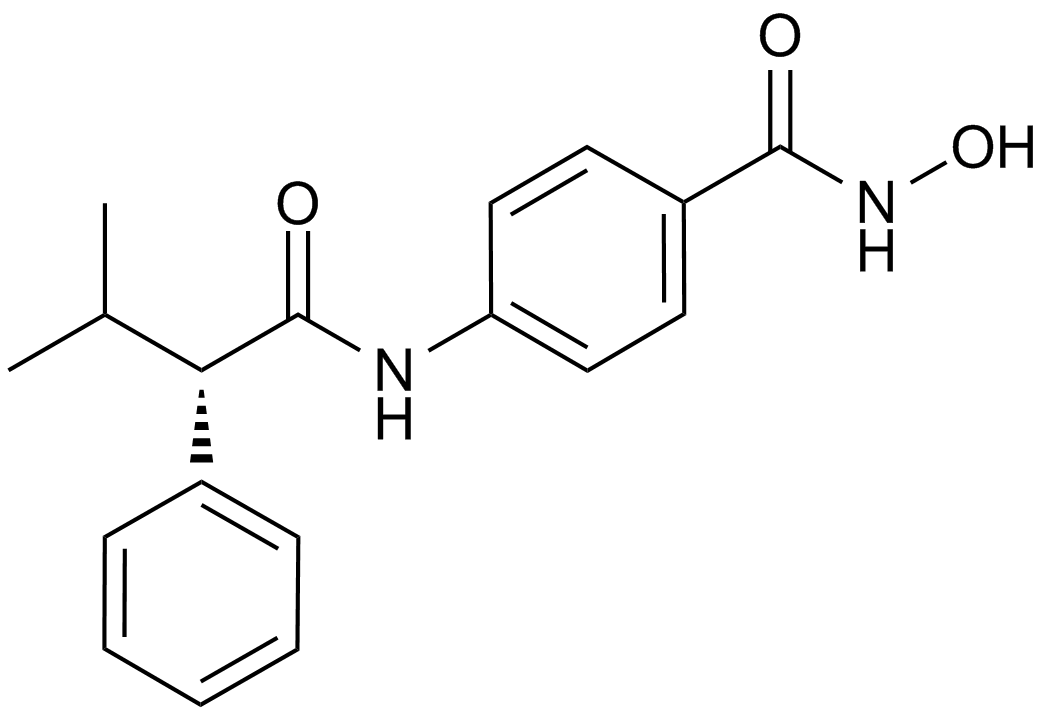
AR-42 (OSU-HDAC42)
HDAC inhibitor,novel and potent
-
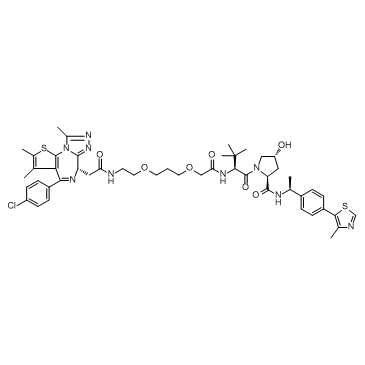
GC32685
ARV-771
ARV-771 is a potent BET PROTAC based on E3 ligase von Hippel-Lindau with Kds of 34 nM, 4.7 nM, 8.3 nM, 7.6 nM, 9.6 nM, and 7.6 nM for BRD2(1), BRD2(2), BRD3(1), BRD3(2), BRD4(1), and BRD4(2), respectively.
-
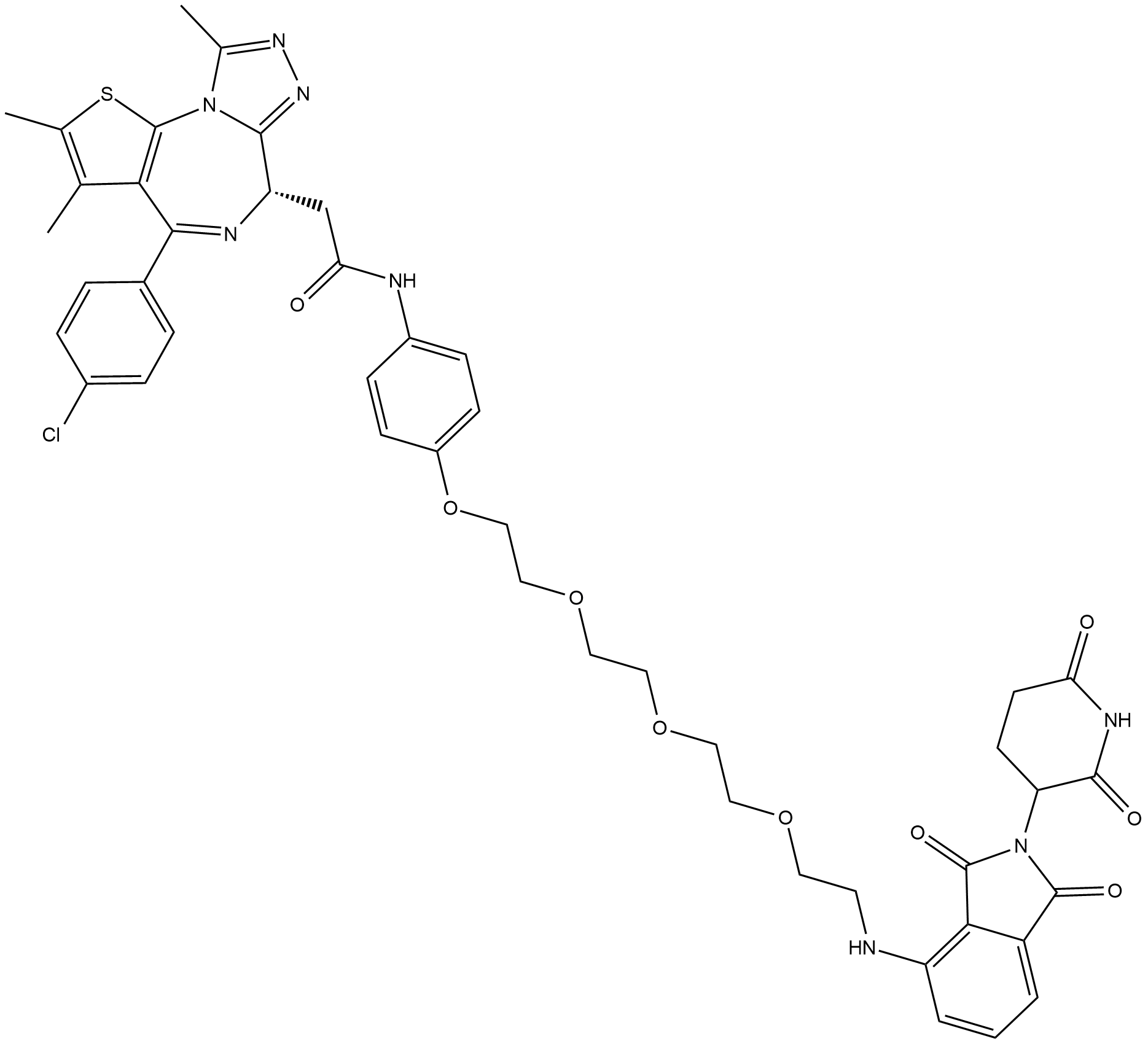
GC19038
ARV-825
ARV-825 is a BRD4 Inhibitor based on PROTAC technology.
-
GC65918
AS-85
AS-85 is a potent ASH1L histone methyltransferase inhibitor (IC50=0.6 μM) with anti-leukemic activity. AS-85 strongly binds to the ASH1L SET domain, with the Kd value of 0.78μM.
-
GC62615
AS-99
AS-99 is a first-in-class, potent and selective ASH1L histone methyltransferase inhibitor (IC50=0.79μM, Kd=0.89μM) with anti-leukemic activity. AS-99 blocks cell proliferation, induces apoptosis and differentiation, downregulates MLL fusion target genes, and reduces the leukemia burden in vivo.
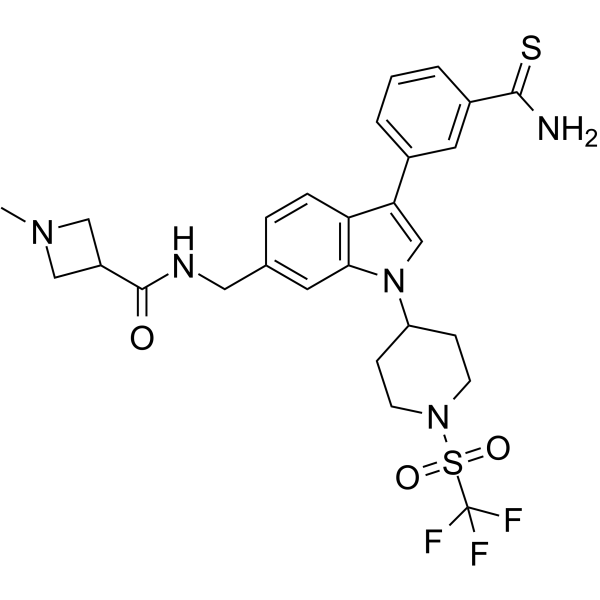
-
GC62849
AS-99 TFA
AS-99 TFA is a first-in-class, potent and selective ASH1L histone methyltransferase inhibitor (IC50=?0.79??M, Kd=?0.89??M) with anti-leukemic activity. AS-99 TFA blocks cell proliferation, induces apoptosis and differentiation, downregulates MLL fusion target genes, and reduces the leukemia burden in vivo.
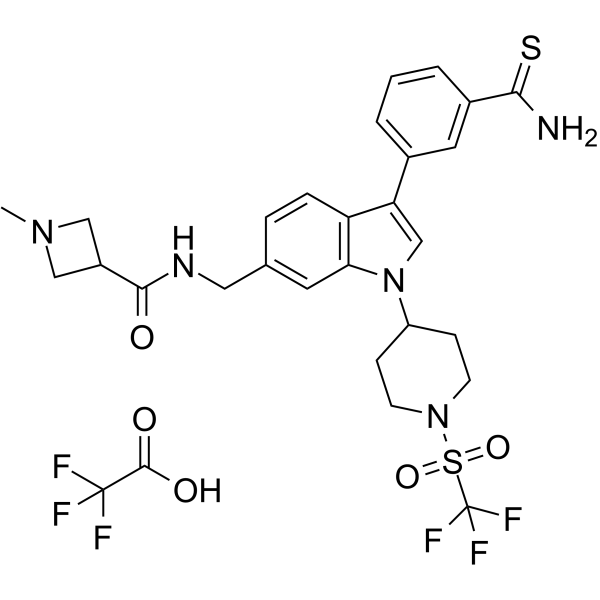
-
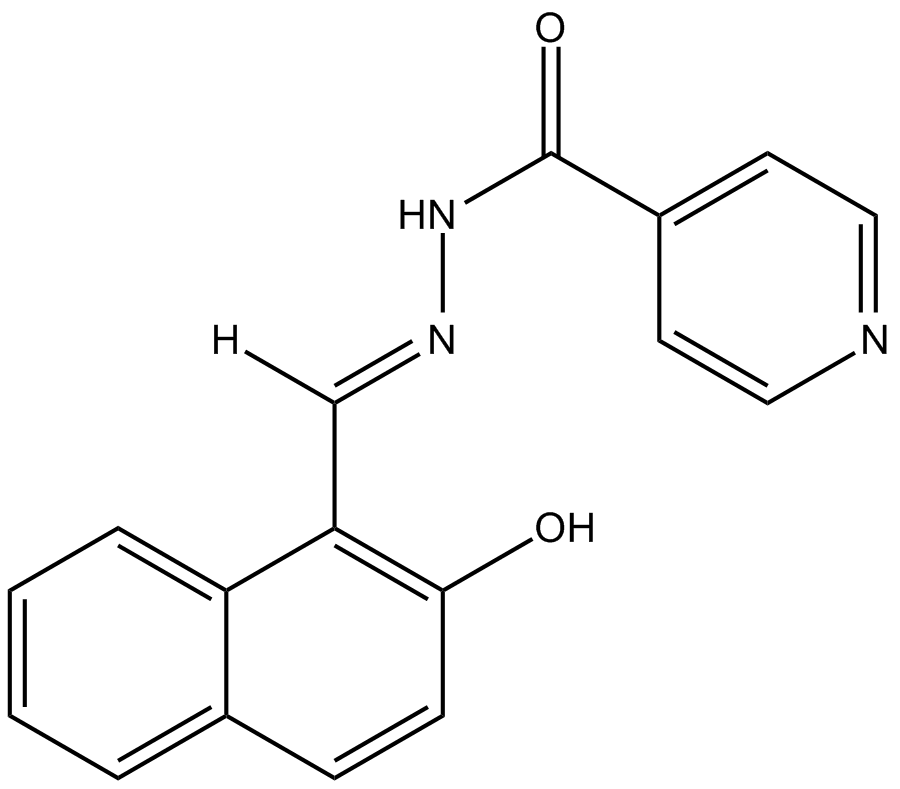
GC17820
AS8351
histone demethylase inhibitor
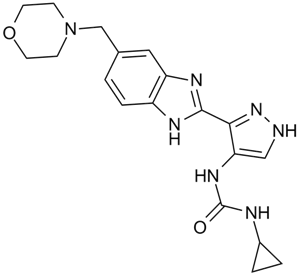
-
GC10638
AT9283
A broad spectrum kinase inhibitor
-
GC50066
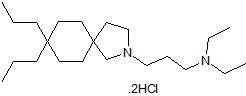
Atiprimod dihydrochloride
JAK2 inhibitor
-
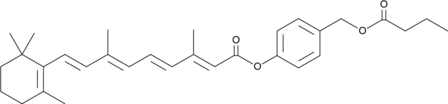
GC46892
ATRA-BA Hybrid
A prodrug form of all-trans retinoic acid and butyric acid
-
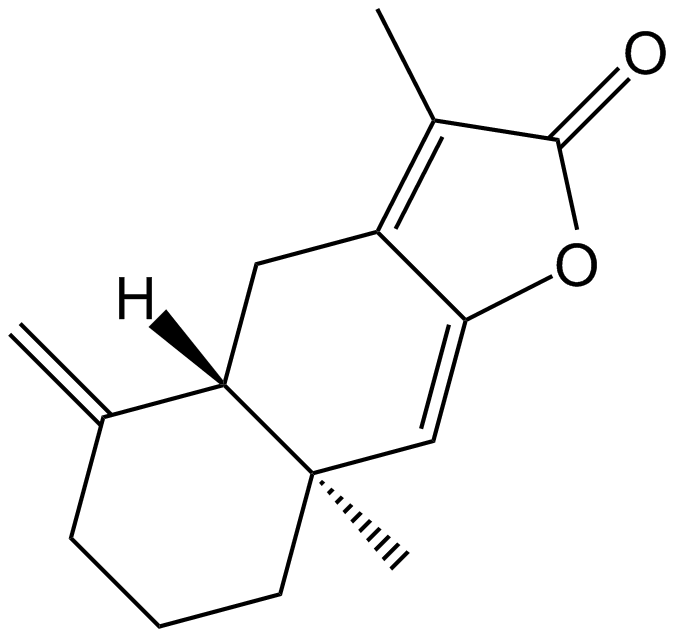
GN10627
Atractylenolide I
-
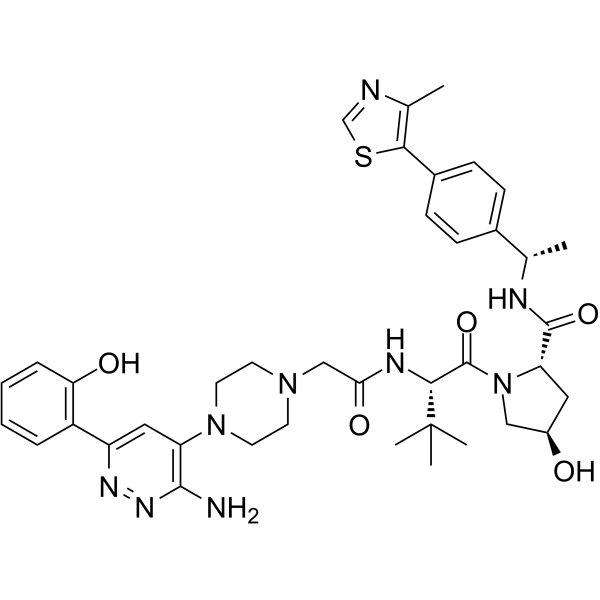
GC64271
AU-15330
AU-15330 is a proteolysis-targeting chimera (PROTAC) degrader of the SWI/SNF ATPase subunits, SMARCA2 and SMARCA4. AU-15330 induces potent inhibition of tumour growth in xenograft models of prostate cancer and synergizes with the AR antagonist enzalutamide. AU-15330 induces disease remission in castration-resistant prostate cancer (CRPC) models without toxicity.
-
GC39699
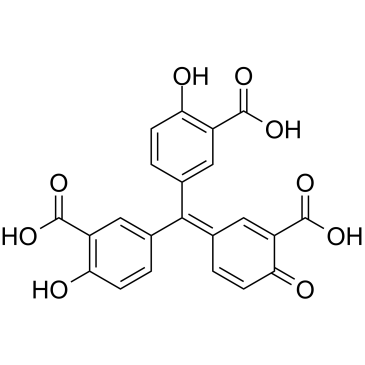
Aurintricarboxylic acid
Aurintricarboxylic acid is a nanomolar-potency, allosteric antagonist with selectivity towards αβ-methylene-ATP-sensitive P2X1Rs and P2X3Rs, with IC50s of 8.6 nM and 72.9 nM for rP2X1R and rP2X3R, respectively.
-
GC13332
Aurora A Inhibitor I
A potent and selective inhibitor of Aurora A kinase

-
GC33379
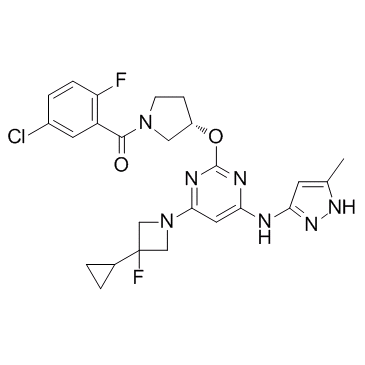
Aurora B inhibitor 1
Aurora B inhibitor 1 is an Aurora B (Aurora-1) inhibitor extracted from patent WO2007059299A1, compound 1-3, has a Ki value of <0.010 uM.
-
GC38441
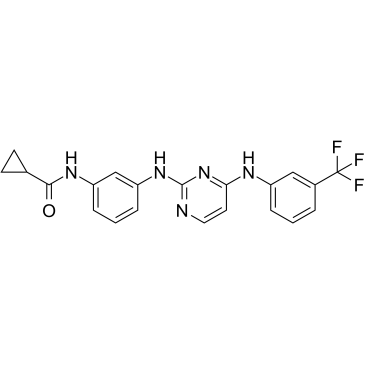
Aurora Kinase Inhibitor 3
A potent inhibitor of Aurora A kinase
-
GC40667
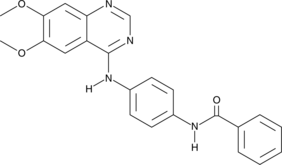
Aurora Kinase Inhibitor II
Aurora Kinase Inhibitor II is a selective and ATP-competitive Aurora kinase inhibitor with IC50s of 310 nM and 240 nM for Aurora A and Aurora B, respectively.
-
GC15711
Aurora Kinase Inhibitor III
Aurora Kinase Inhibitor III is a strong and selective Aurora A kinase inhibitor with an IC50 of 42 nM, and weakly inhibits EGFR with an IC50 of >10 μM.
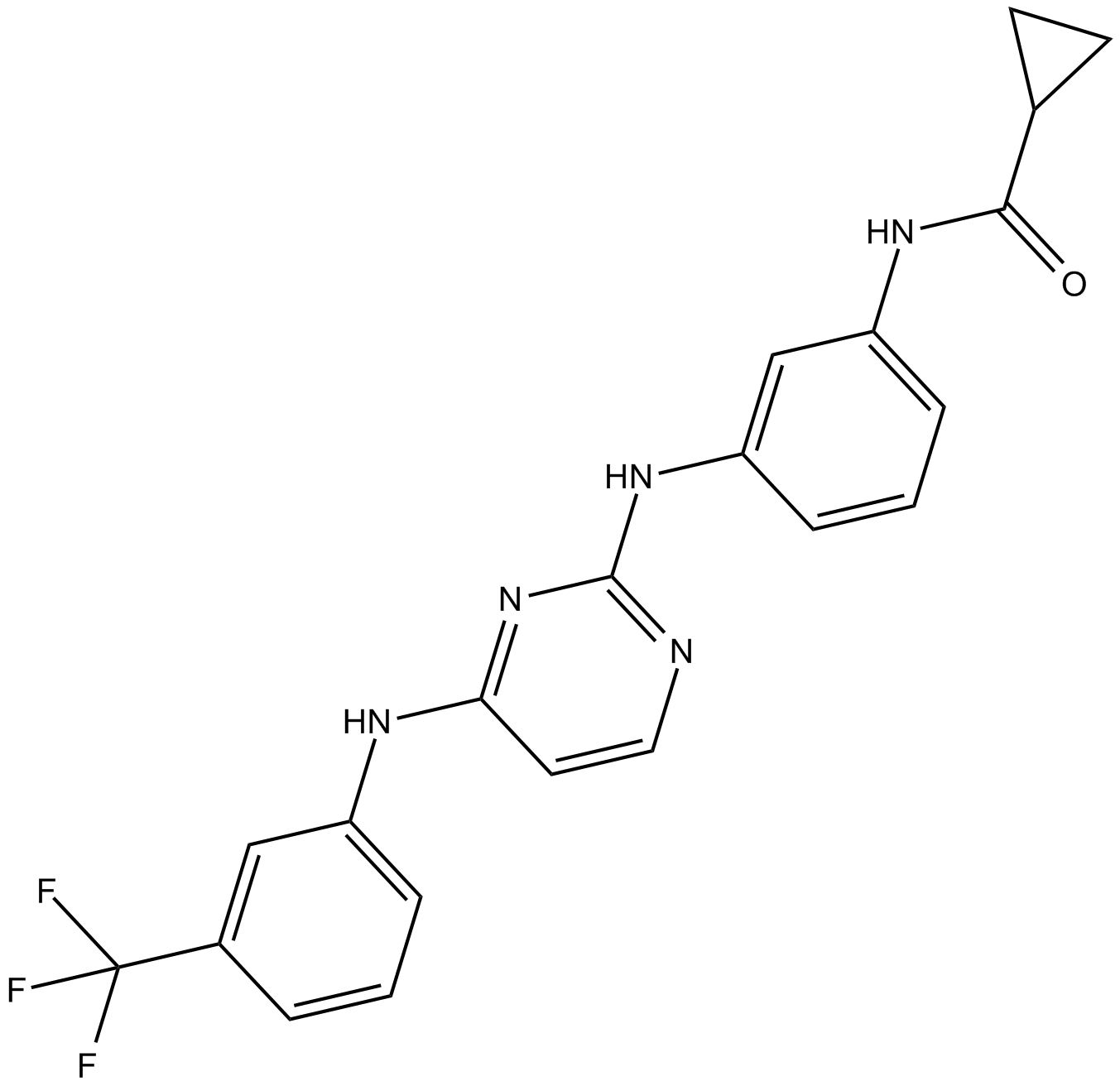
-
GC67899
Aurora kinase inhibitor-8

-
GC13433
AZ 960
A JAK2 inhibitor
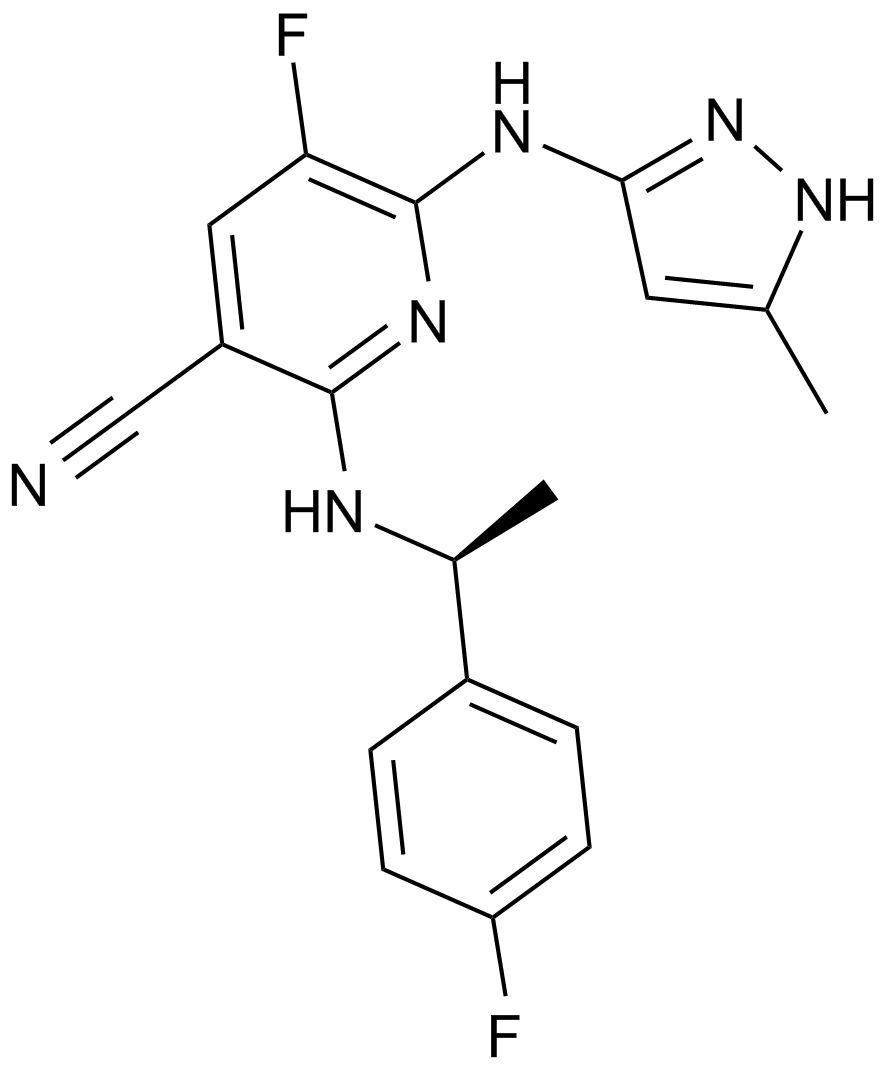
-
GC65899
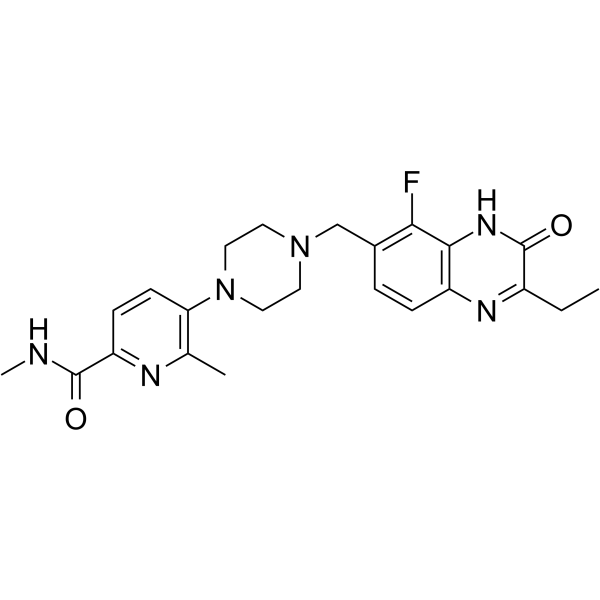
AZ3391
AZ3391 is a potent inhibitor of PARP. AZ3391 is a quinoxaline derivative. PARP family of enzymes play an important role in a number of cellular processes, such as replication, recombination, chromatin remodeling, and DNA damage repair. AZ3391 has the potential for the research of diseases and conditions occurring in tissues in the central nervous system, such as the brain and spinal cord (extracted from patent WO2021260092A1, compound 23).
-
GC13744
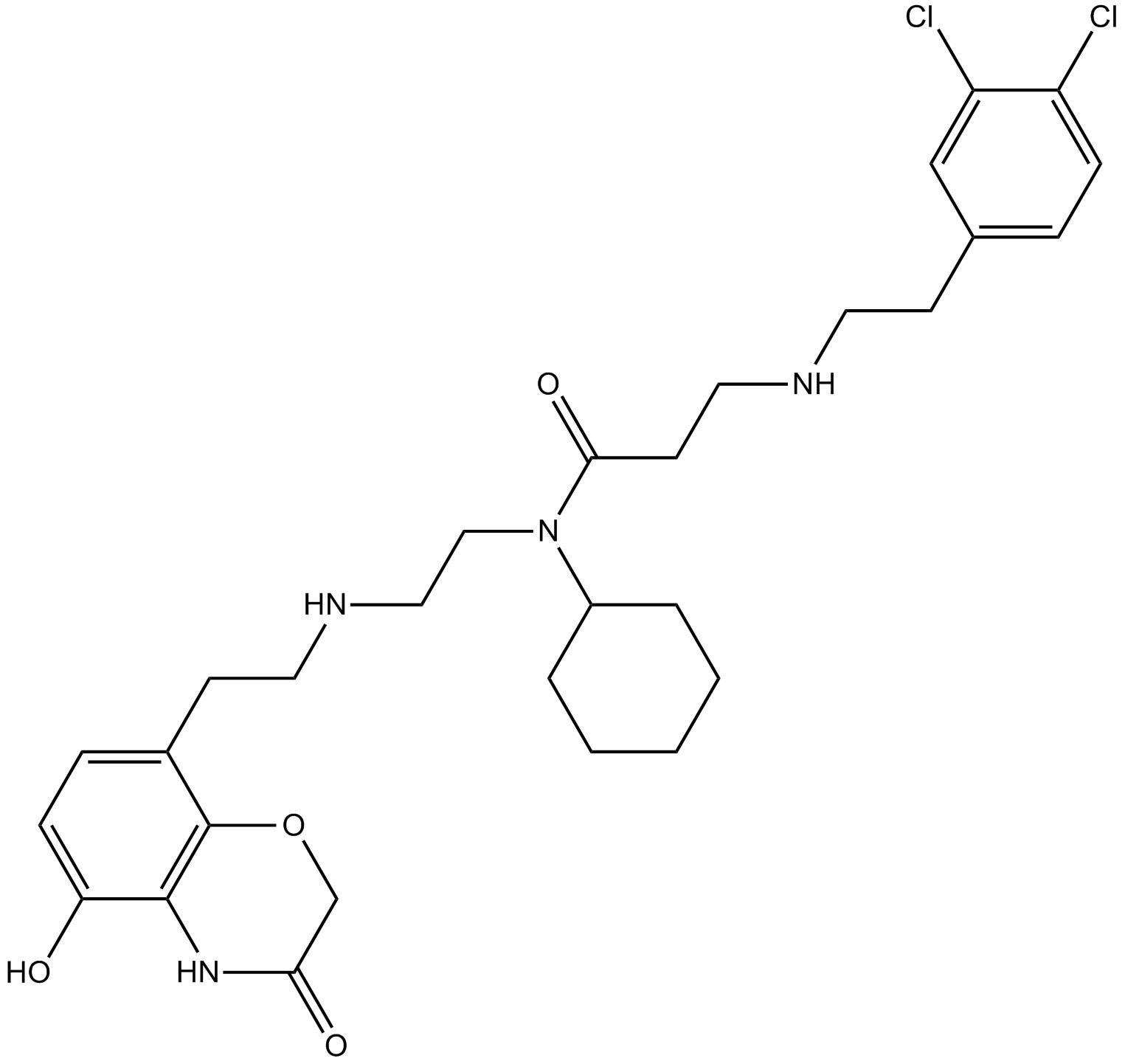
AZ505
SMYD2 inhibitor,potent and selective
-
GC13103
AZ505 ditrifluoroacetate
SMYD2 inhibitor
-
GC64124
AZ506
AZ506 is a potent SMYD2 inhibitor with an IC50 of 17 nM. AZ506 inhibits SMYD2 methyltransferase activity in cells, leading to a decrease in the SMYD2-mediated methylation signal.
-
GC16725
AZ6102
TNKS1/2 inhibitor
-
GC46900
AZ9482
A PARP inhibitor
-
GC48971
AZD 1152 (hydrochloride)
A prodrug for a potent Aurora B inhibitor
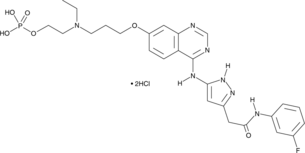
-
GC13014
AZD 5153
orally available, bivalent inhibitor of the bromodomain and extraterminal (BET) protein BRD4.
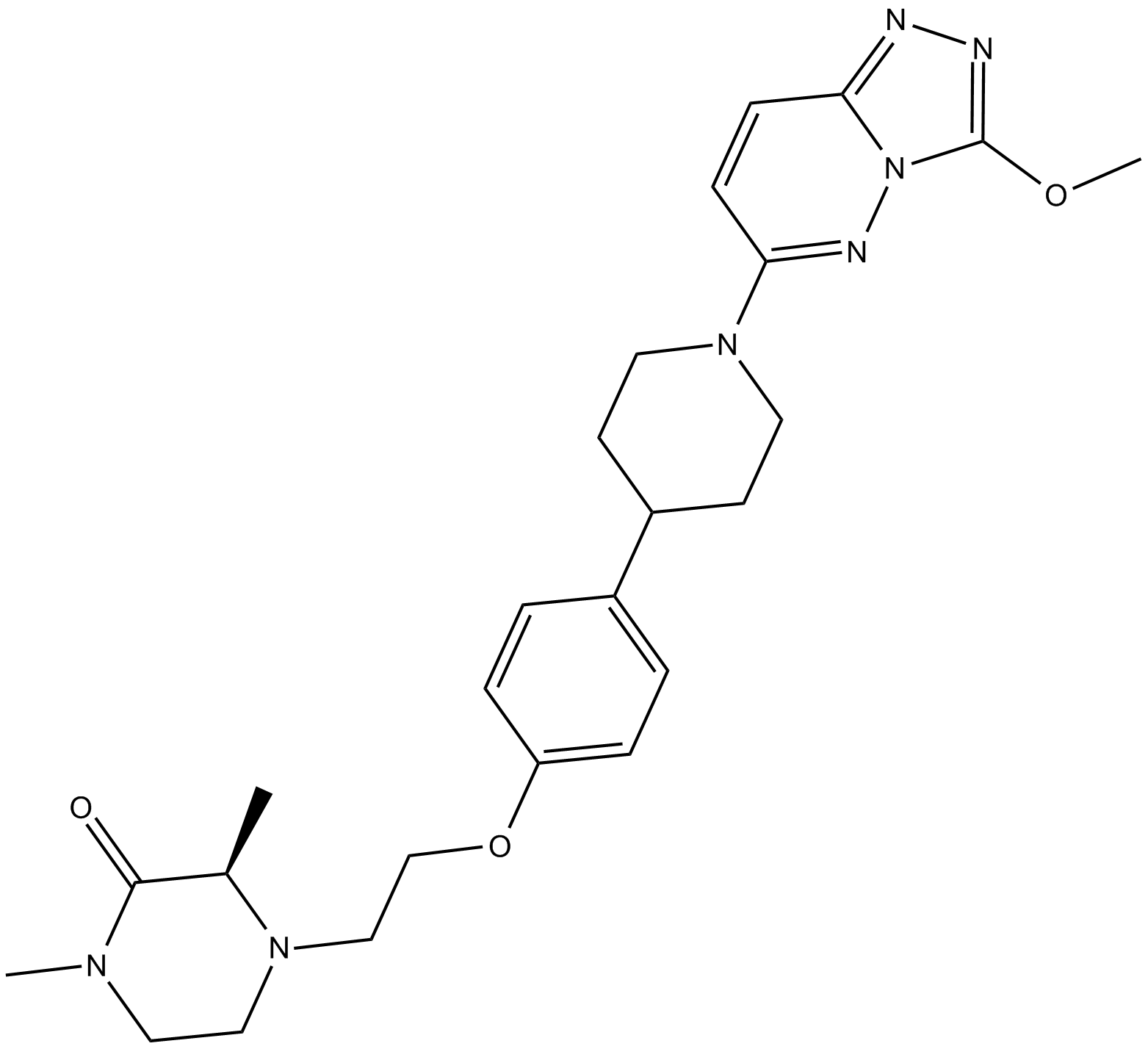
-
GC14709
AZD1152
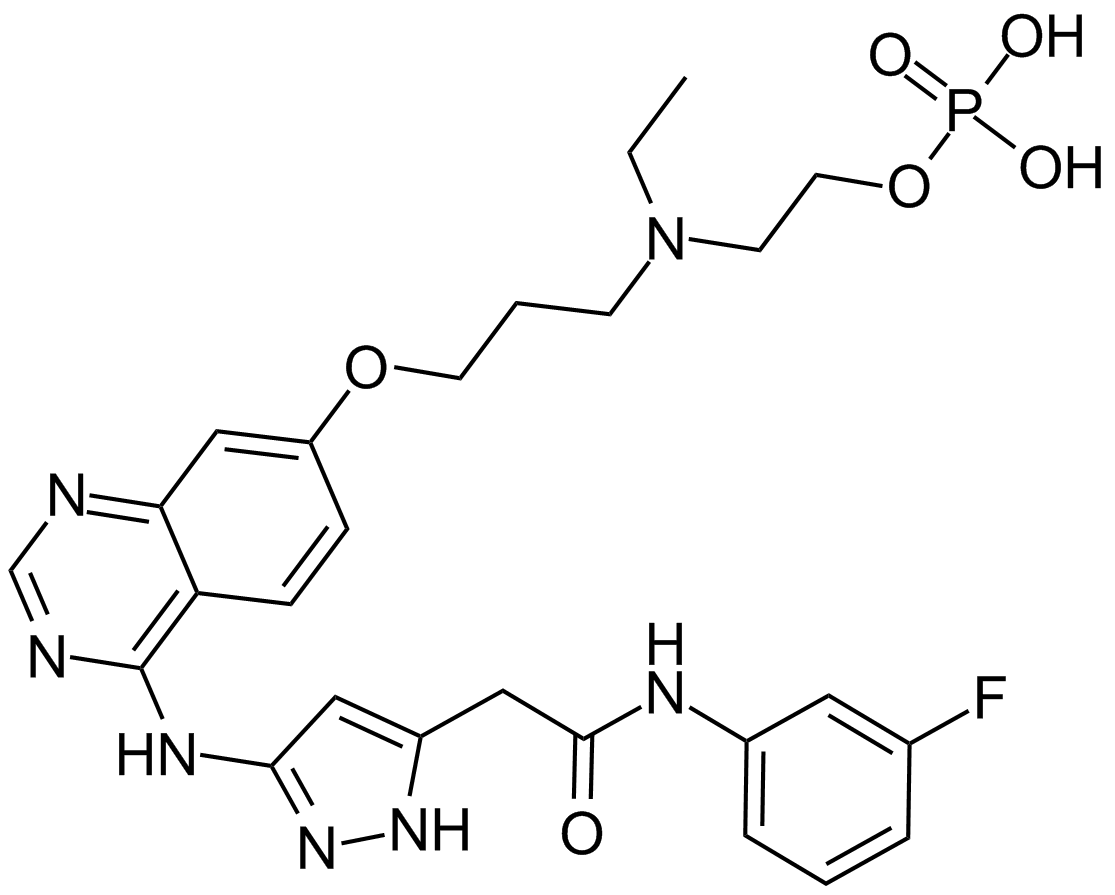
-
GC12660
AZD1208
A pan-Pim kinase inhibitor
-
GC35447
AZD1208 hydrochloride
AZD1208 hydrochloride is an orally bioavailable, highly selective PIM kinases inhibitor.
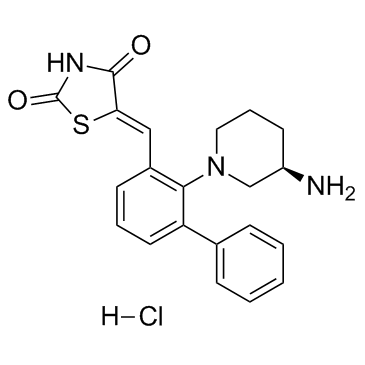
-
GC12504
AZD1480
A potent JAK2 inhibitor
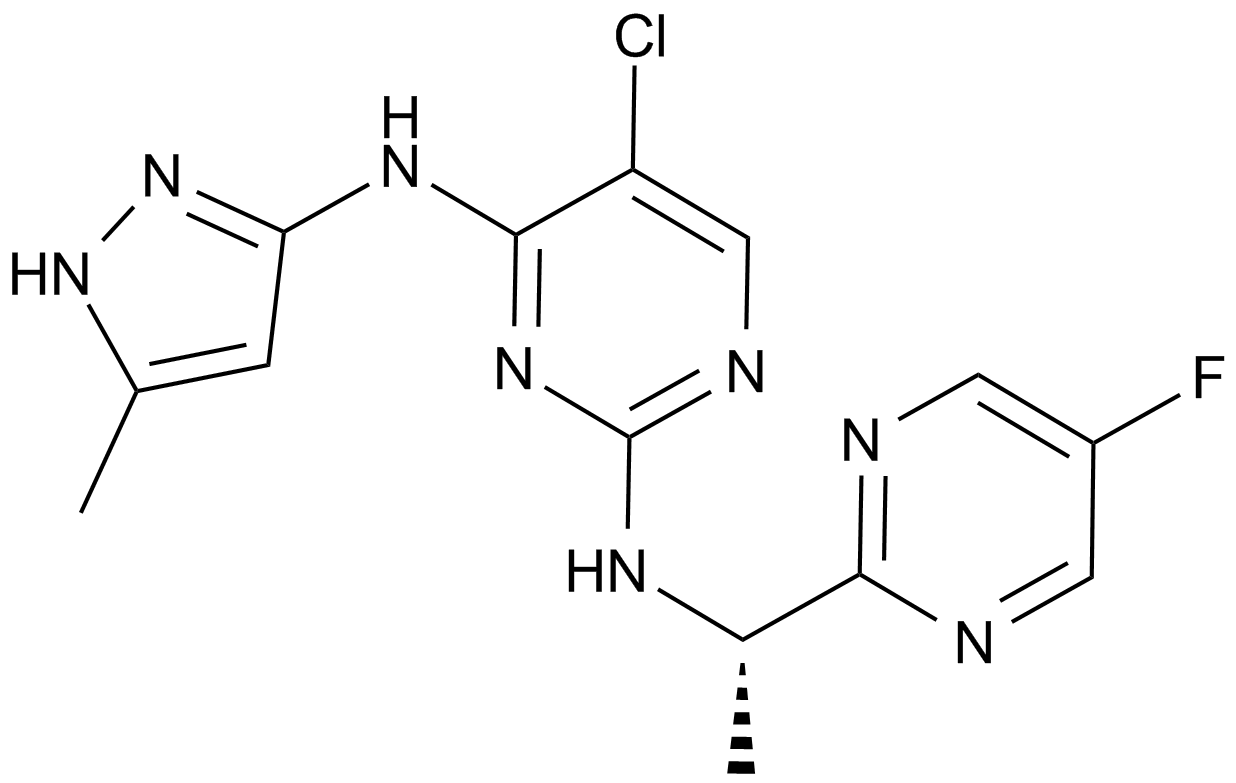
-
GC17965
AZD2461
A PARP inhibitor
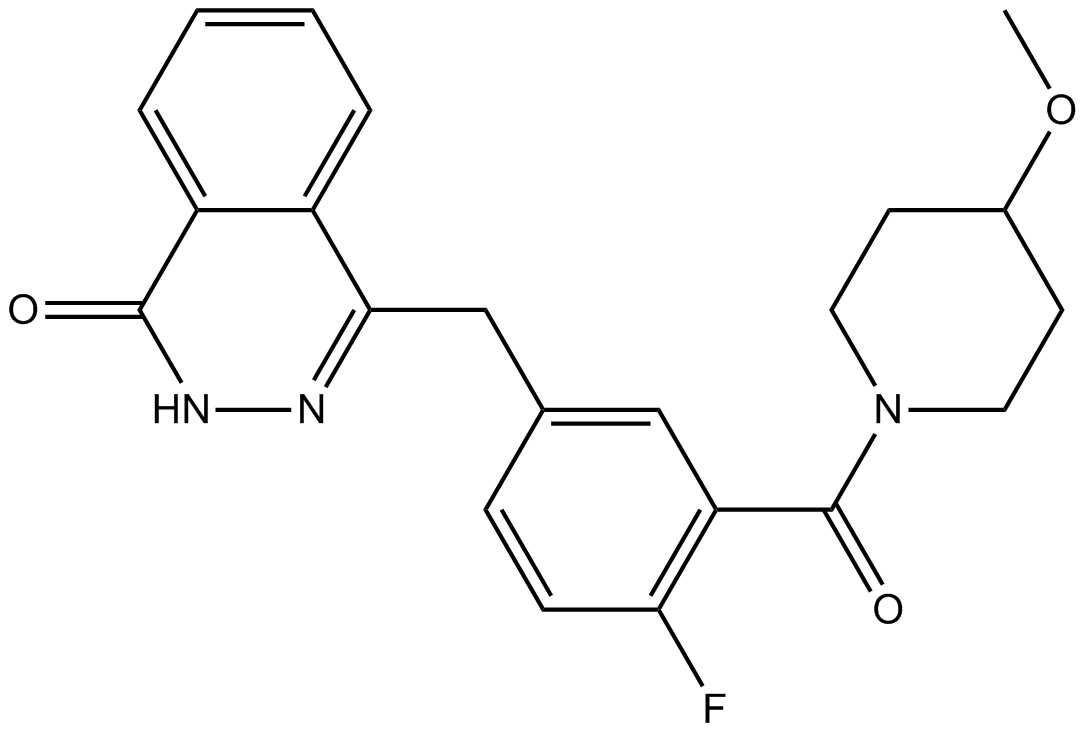
-
GC35448
AZD5153 6-Hydroxy-2-naphthoic acid
AZD5153 6-Hydroxy-2-naphthoic acid is the 6-Hydroxy-2-naphthoic acid of AZD5153. AZD5153 is a potent, selective, and orally available BET/BRD4 bromodomain inhibitor; disrupts BRD4 with an IC50 of 1.7 nM.
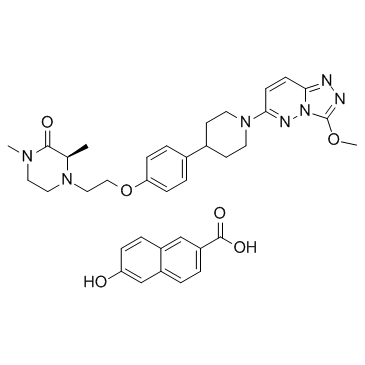
-
GC62310
AZD5305
AZD5305 is a potent, selective and oral active PARP inhibitor. AZD5305 is potent and efficacious in animal xenografts and PDX models.
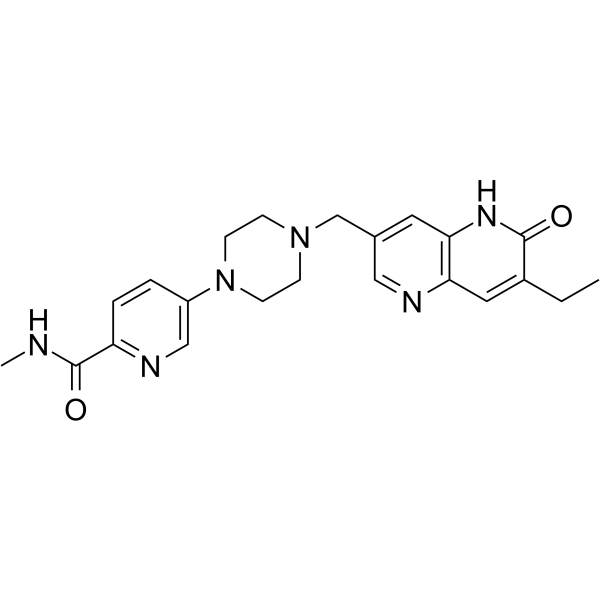
-
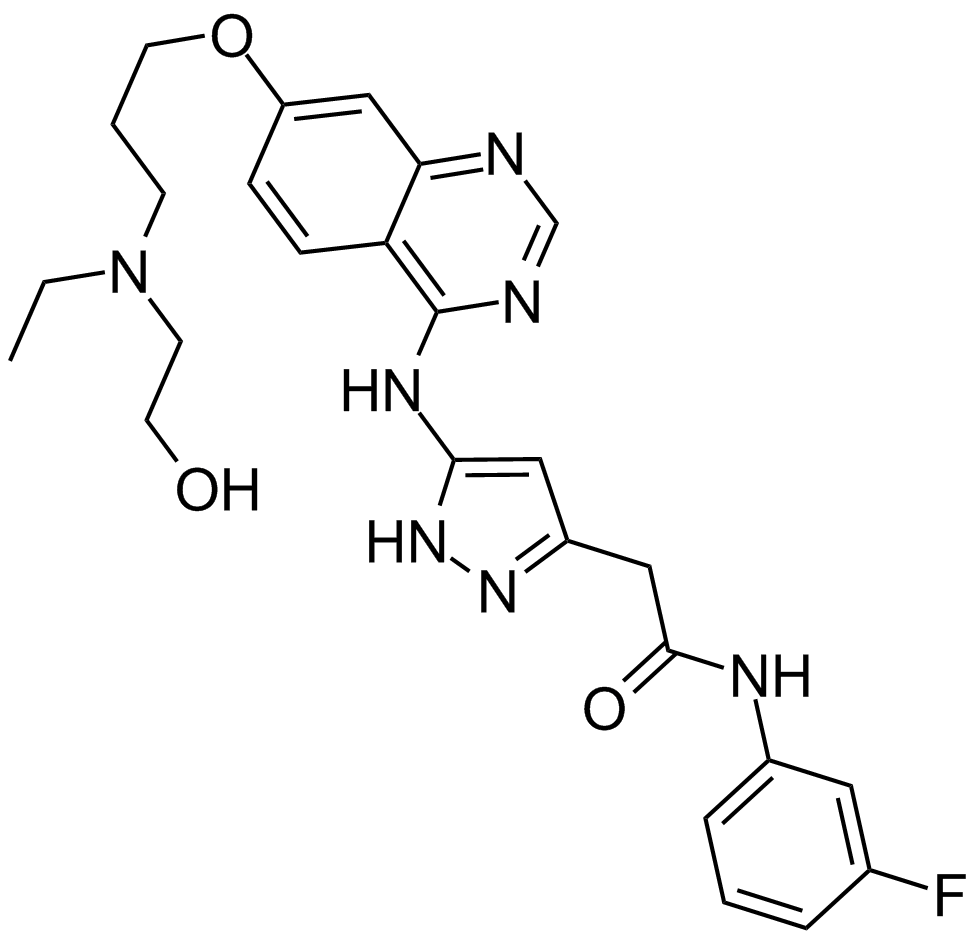
GC14955
Barasertib (AZD1152-HQPA)
A selective Aurora kinase B inhibitor
-
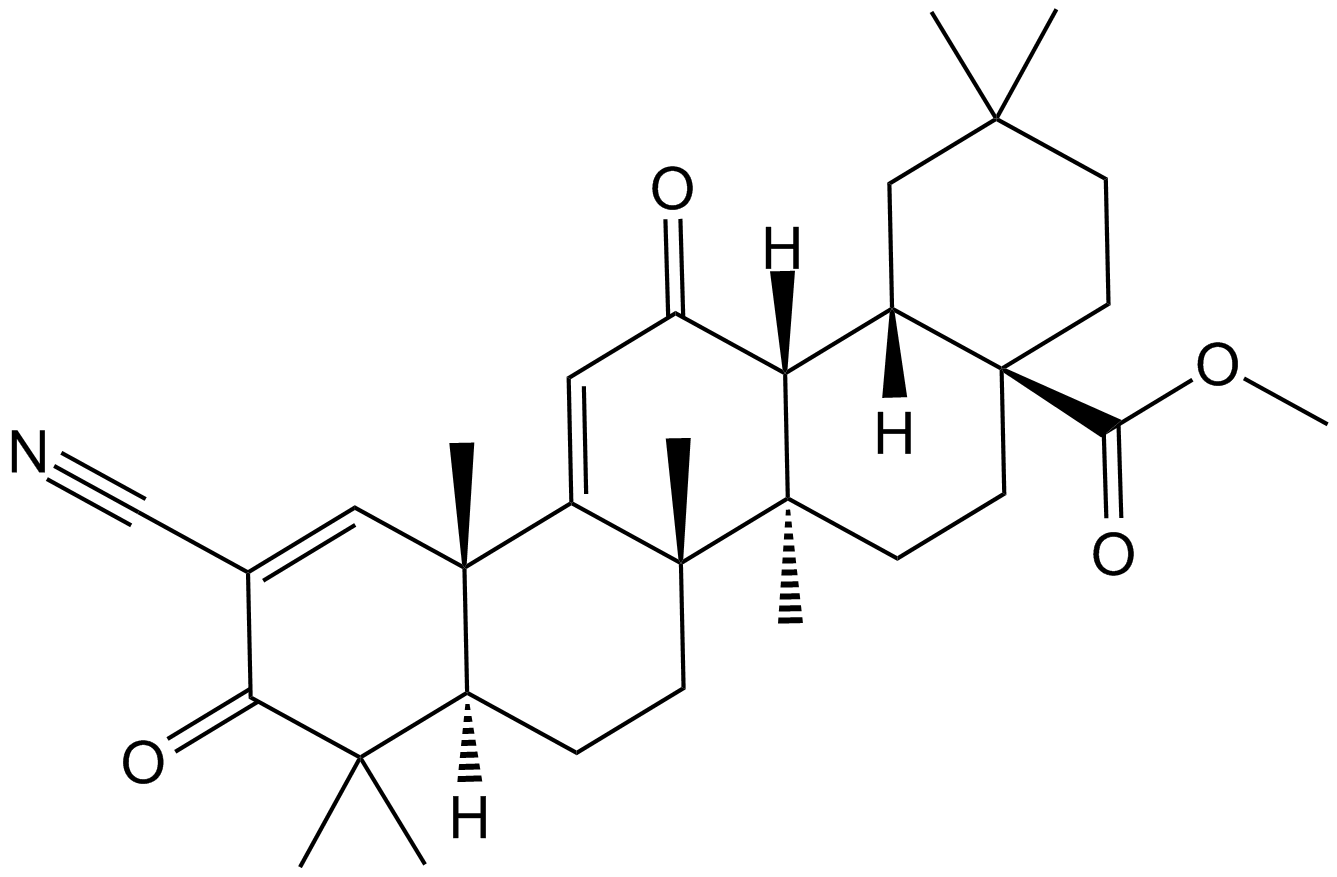
GC11572
Bardoxolone methyl
A synthetic triterpenoid with potent anticancer and antidiabetic activity
-
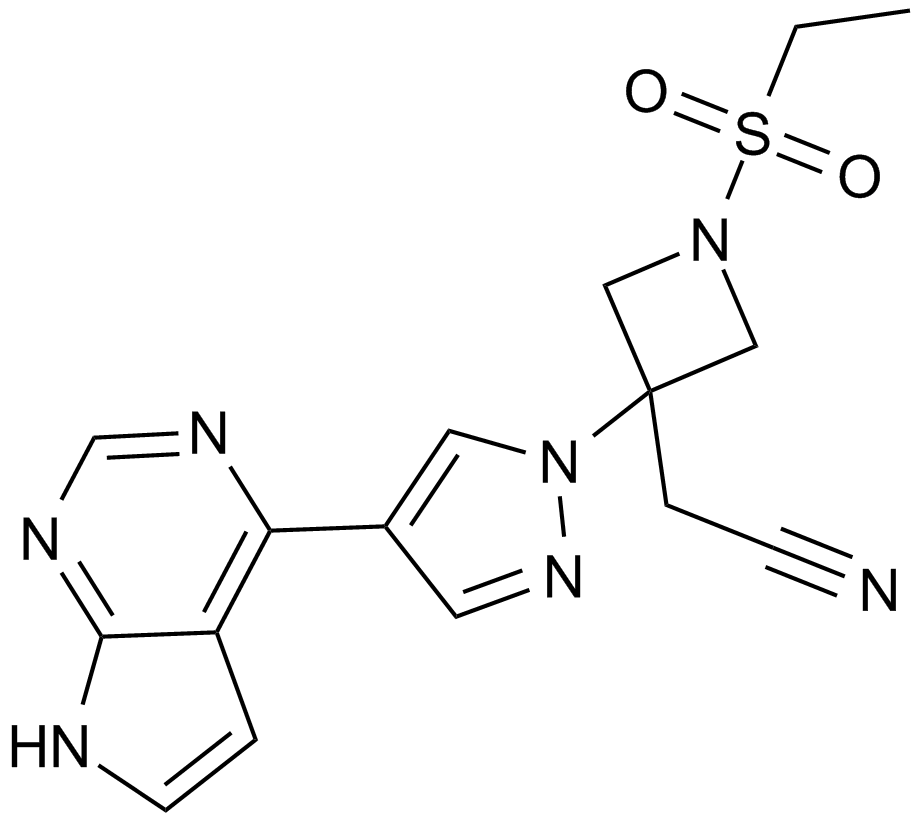
GC14844
Baricitinib (LY3009104, INCB028050)
A JAK1 and JAK2 inhibitor
-
GC13830
Baricitinib phosphate
A JAK1 and JAK2 inhibitor

-
GC12698
BAY 87-2243
A HIF-1 inhibitor,potent and selective
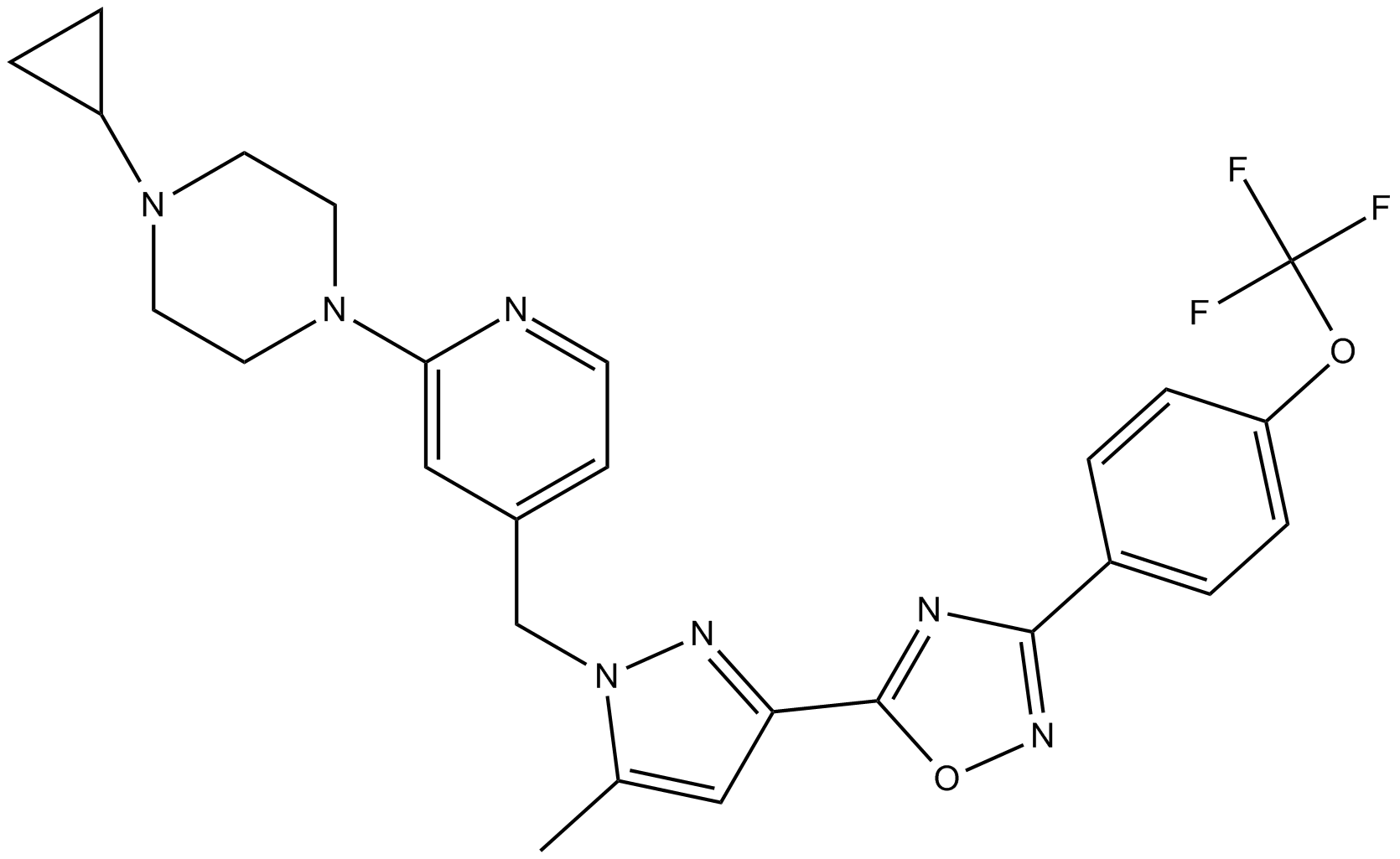
-
GC18508
BAY-299
BAY-299 is a potent and selective inhibitor of the bromodomain and PHD finger-containing (BRPF) family protein BRD1 (IC50 = 6 nM), also known as BRPF2, and the second bromodomain of transcription initiation factor TFIID subunits 1 (TAF1; IC50 = 13 nM).
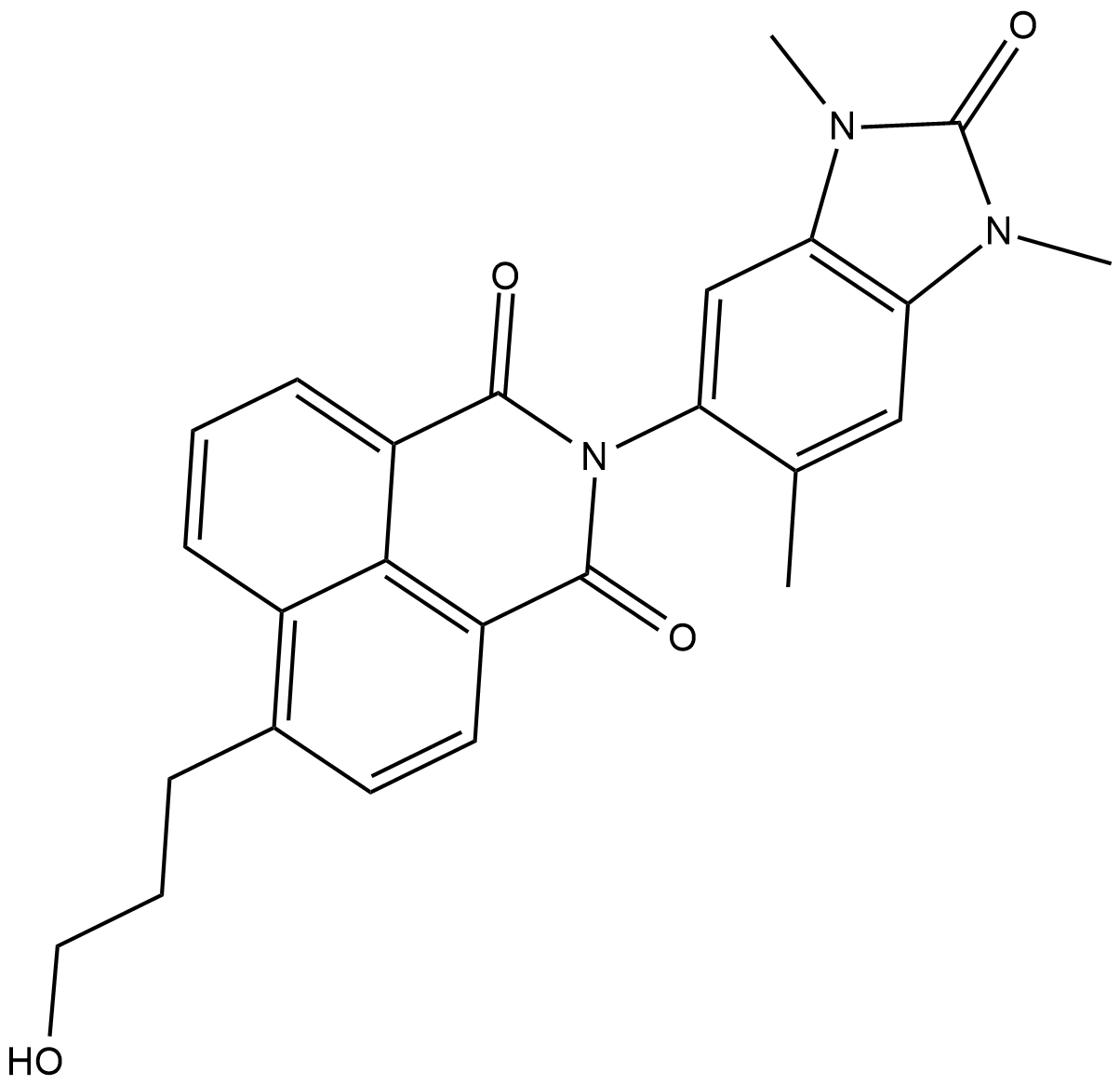
-
GC18159
BAY-598
A potent and selective SMYD2 inhibitor
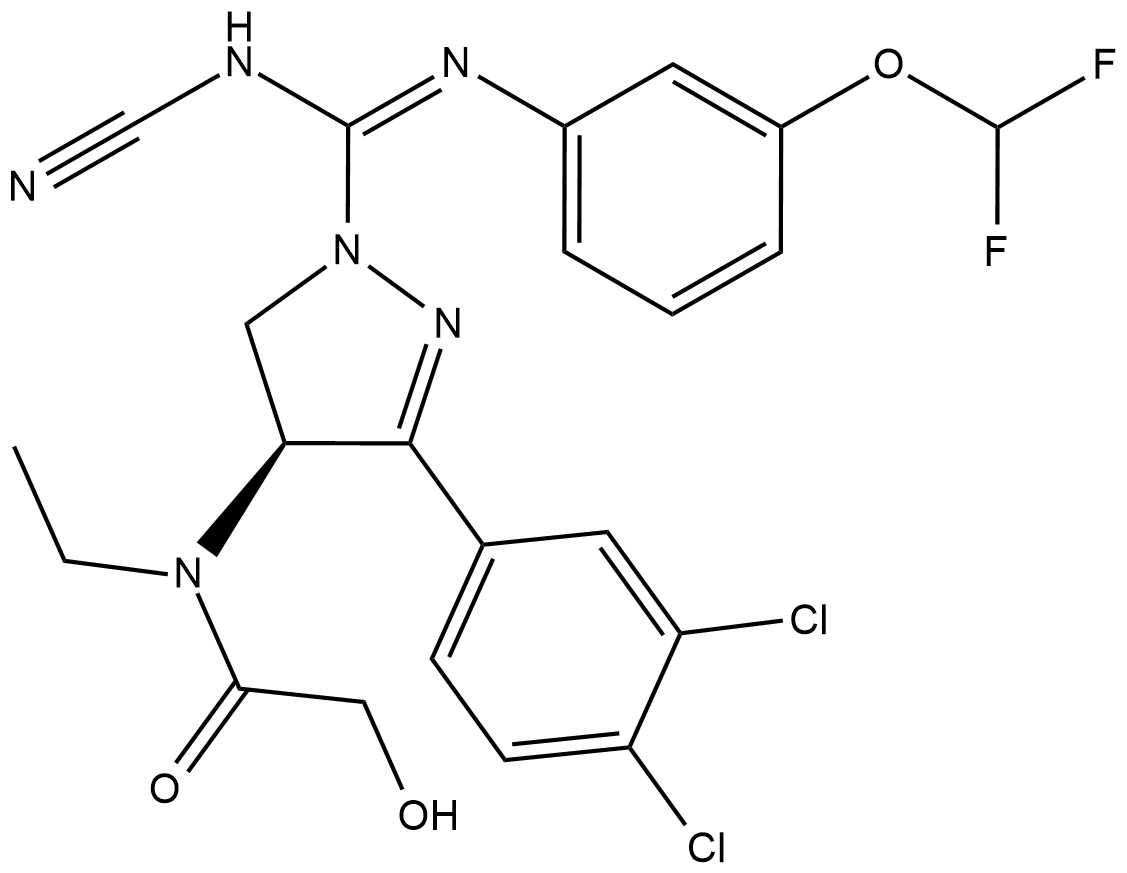
-
GC45389
BAY-6035
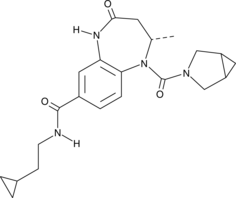
-
GC33104
BAY1238097
BAY1238097 is a potent and selective inhibitor of BET binding to histones and has strong anti-proliferative activity in different AML (acute myeloid leukemia) and MM (multiple myeloma) models through down-regulation of c-Myc levels and its downstream transcriptome (IC50 <100 nM).
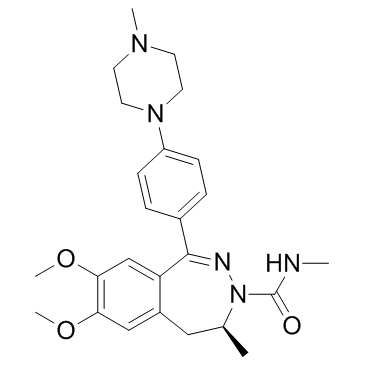
-
GC63694
BAZ1A-IN-1
BAZ1A-IN-1 is a potent inhibitor of BAZ1A (bromodomain-containing protein). BAZ1A-IN-1 shows a KD value of 0.52 μM against BAZ1A bromodomain. BAZ1A-IN-1 shows good anti-viability activity against cancer cell lines expressing a high level of BAZ1A, but weak or no activity against cancer cells with a low expression level of BAZ1A.
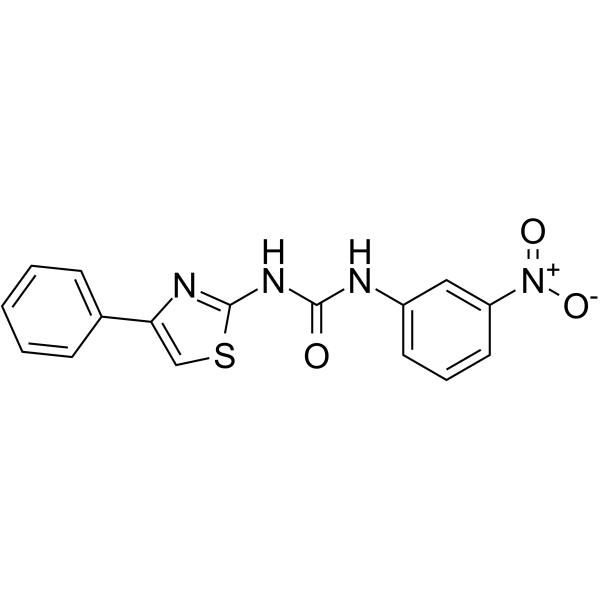
-
GC15175
BAZ2-ICR
Selective BAZ2 bromodomain inhibitor
-
GC35480
BB-Cl-Amidine hydrochloride
BB-Cl-Amidine hydrochloride is a peptidylarginine deminase (PAD) inhibitor.
-
GC42910
BB-F-Yne
BB-F-Yne is a cell-permeable derivative of the protein arginine diminase (PAD) inhibitor BB-Cl-amidine that contains an alkyne moiety for use in click chemistry reactions.
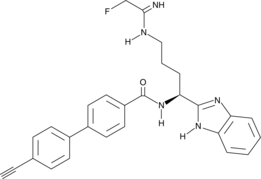
-
GC18161
BCI-121
A substrate-competitive SMYD3 inhibitor?
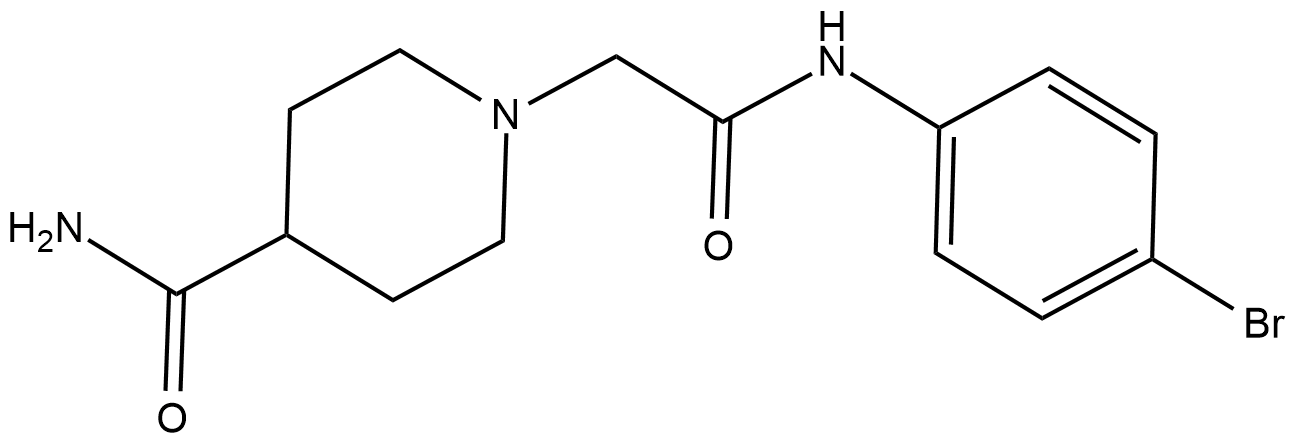
-
GC15962
Belinostat (PXD101)
Belinostat is an effective HDAC inhibitor with an IC50 of 27 nM in HeLa cell extracts.
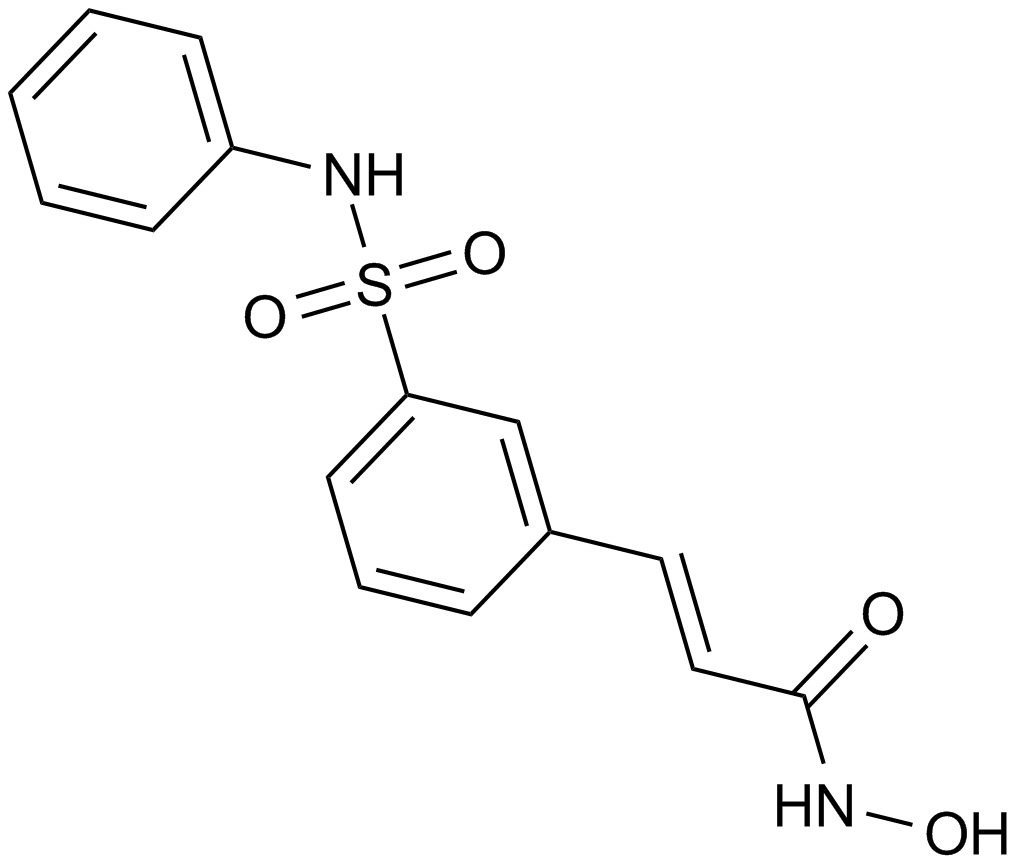
-
GC12844
Benzamide
poly (ADP-ribose) synthetase inhibitor
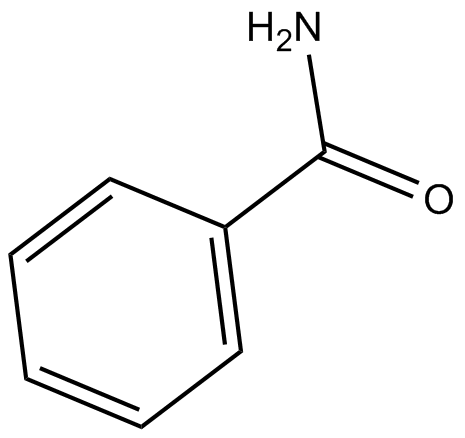
-
GC30763
Benzenebutyric acid (4-Phenylbutyric acid)
Benzenebutyric acid (4-Phenylbutyric acid) (4-PBA) is an inhibitor of HDAC and endoplasmic reticulum (ER) stress, used in cancer and infection research.
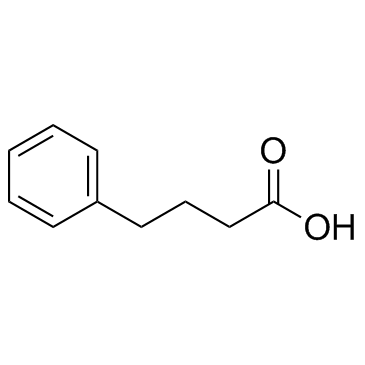
-
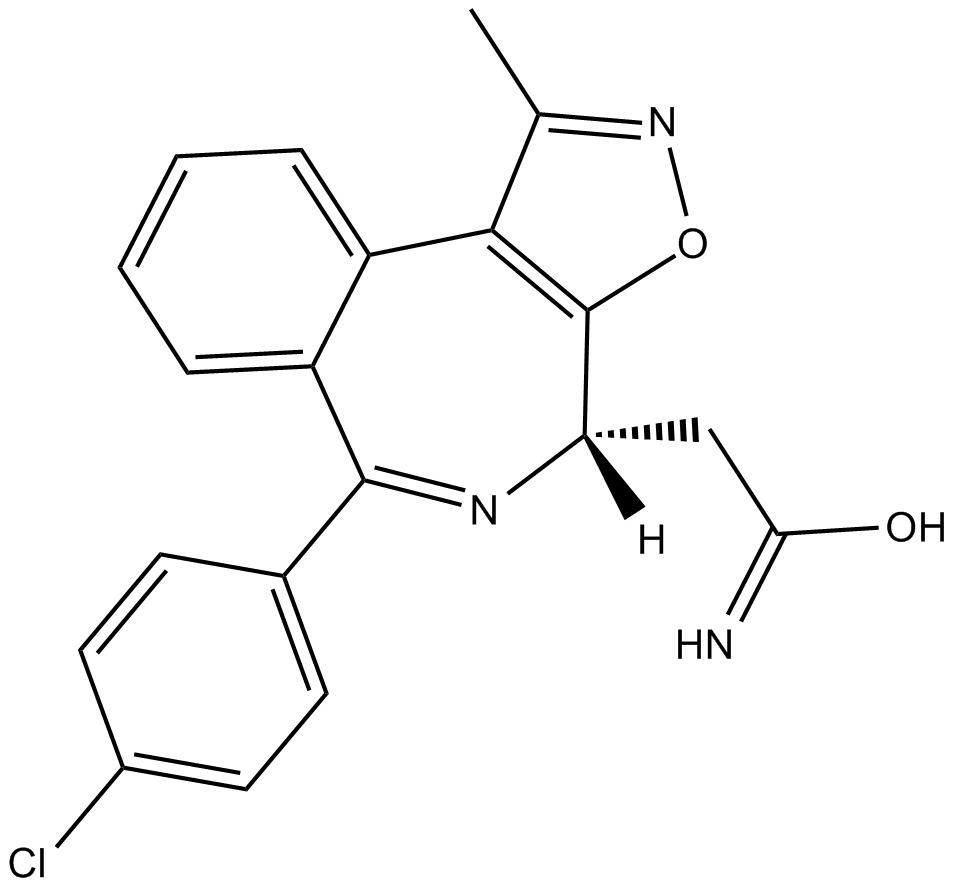
GC16299
BET bromodomain inhibitor
Potent and selective inhibitor for BRD4
-
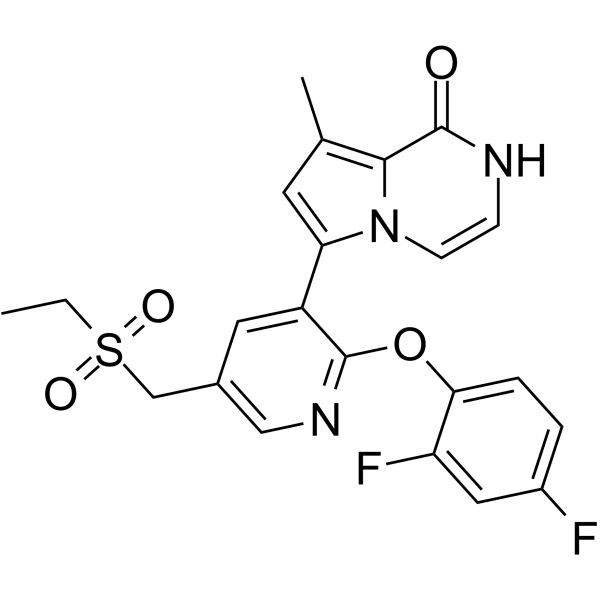
GC63528
BET bromodomain inhibitor 1
BET bromodomain inhibitor 1 is an orally active, selective bromodomain and extra-terminal (BET) bromodomain inhibitor with an IC50 of 2.6 nM for BRD4. BET bromodomain inhibitor 1 binds to BRD2(2), BRD3(2), BRD4(1), BRD4(2), and BRDT(2) with high affinities (Kd values of 1.3 nM, 1.0 nM, 3.0 nM, 1.6 nM, 2.1 nM, respectively). bromodomain inhibitor 1 has anti-cancer activity.
-
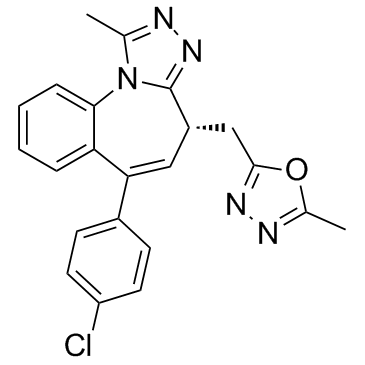
GC35505
BET-BAY 002
BET-BAY 002 is a potent BET inhibitor; shows efficacy in a multiple myeloma model.
-
GC35506
BET-BAY 002 S enantiomer
-
GC31831
BET-IN-1
BET-IN-1 is a pan-inhibitor of all eight BET bromodomains, and selectivity over other representative bromodomain-containing proteins.
-
GC65506
BETd-246
BETd-246 is a second-generation and PROTAC-based BET bromodomain (BRD) inhibitor connected by ligands for Cereblon and BET, exhibiting superior selectivity, potency and antitumor activity.
-
GC32791
BETd-260 (ZBC 260)
BETd-260 (ZBC 260) (ZBC 260) is a PROTAC connected by ligands for Cereblon and BET, with as low as 30 pM against BRD4 protein in RS4;11 leukemia cell line. BETd-260 (ZBC 260) potently suppresses cell viability and robustly induces apoptosis in hepatocellular carcinoma (HCC) cells.
-
GC12074
BG45
Novel HDAC3-selective inhibitor
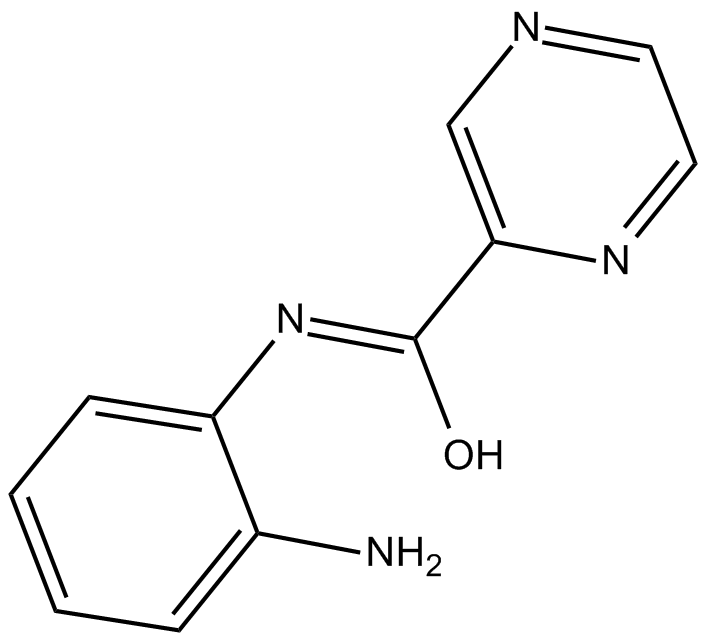
-
GC14380
BGP-15
PARP inhibitor
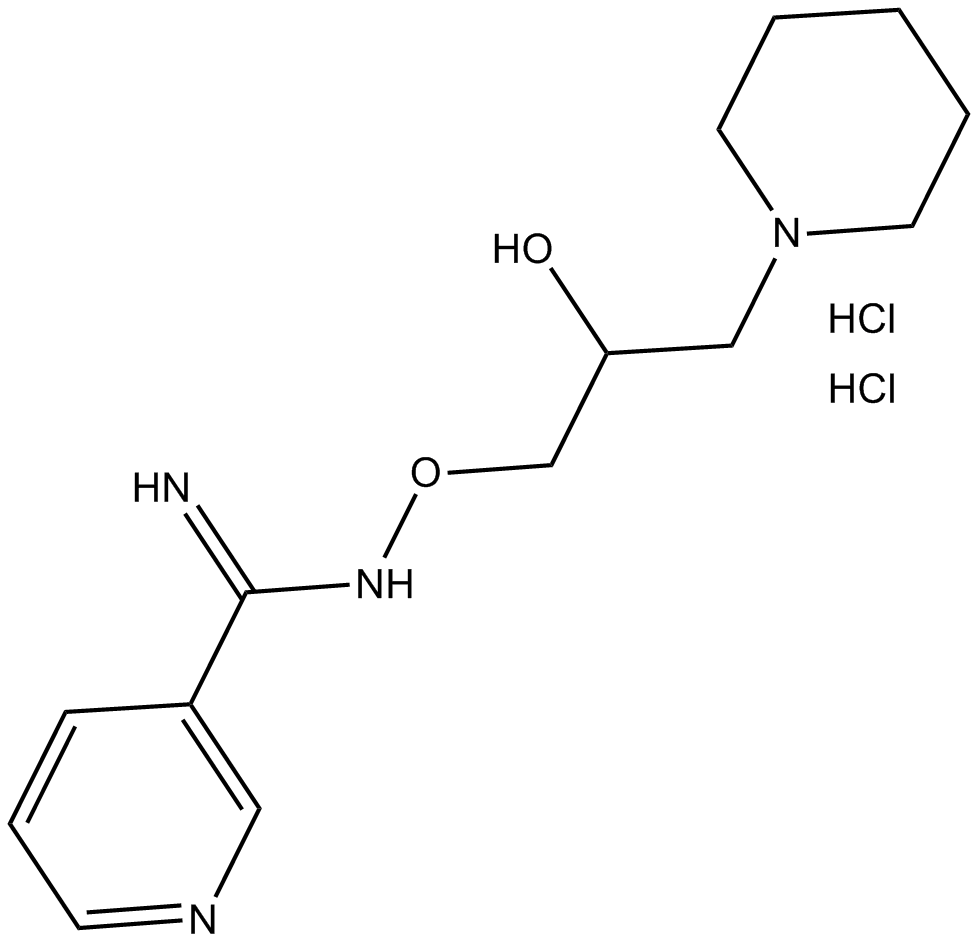
-
GC12450
BI 2536
A potent inhibitor of Plk1
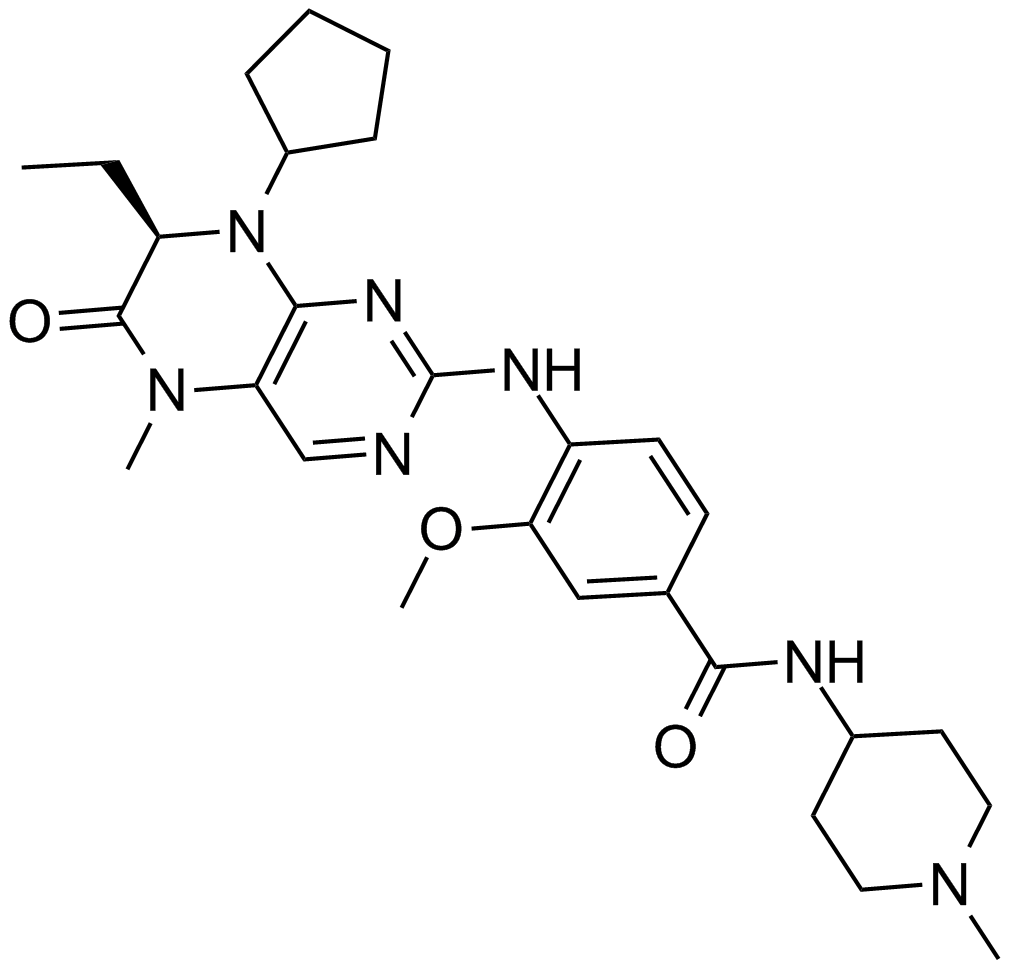
-
GC50540
BI 9321
Nuclear receptor-binding SET domain (NSD) 3 antagonist; selectively binds PWWP1 domain
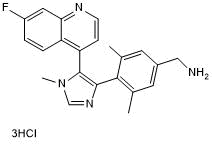
-
GC16726
BI-7273
BRD9 bromodomain inhibitor
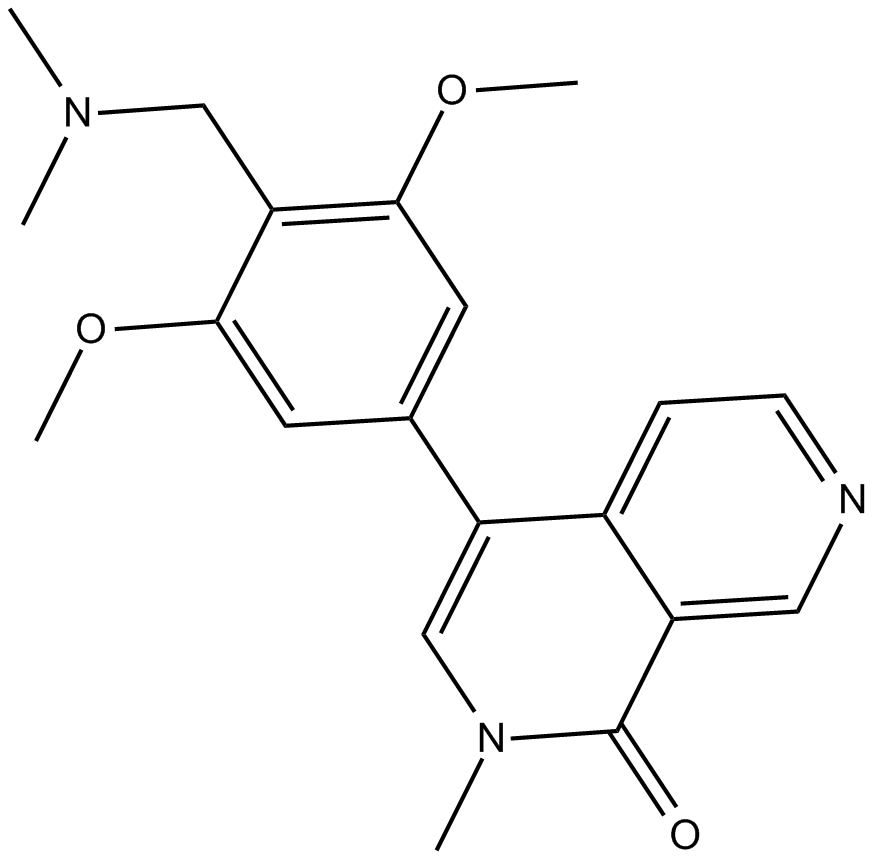
-
GC17828
BI-847325
dual inhibitor of MEK and Aurora kinases

-
GC39663
BI-9321 trihydrochloride
BI-9321 trihydrochloride is a potent, selective and cellular active nuclear receptor-binding SET domain 3 (NSD3)-PWWP1 domain antagonist with a Kd value of 166 nM. BI-9321 trihydrochloride is inactive against NSD2-PWWP1 and NSD3-PWWP2. BI-9321 trihydrochloride specifically disrupts histone interactions of the NSD3-PWWP1 domain with an IC50 of 1.2 μM in U2OS cells.
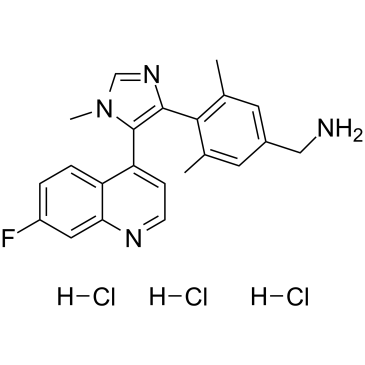
-
GC16817
BI-9564
BRD9/7 specific inhibitor
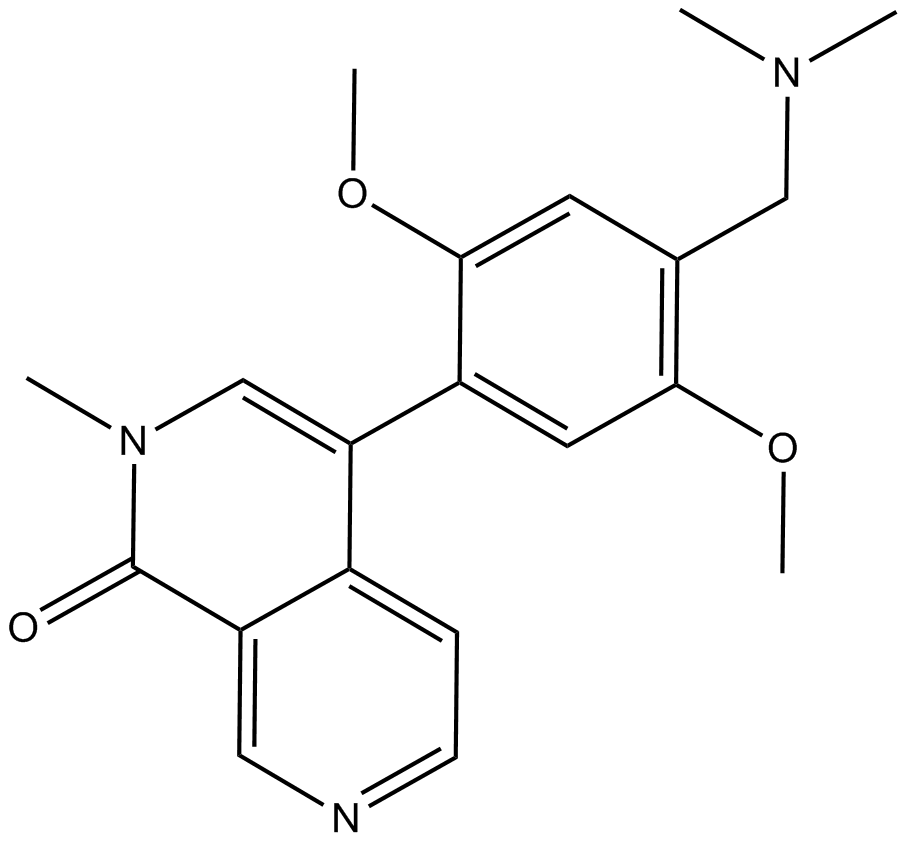
-
GC42933
Binucleine 2
Binucleine 2 is an isoform-specific and ATP-competitive inhibitor of Drosophila Aurora B kinase (Ki = 0.36 μM), a kinase involved in cell division.
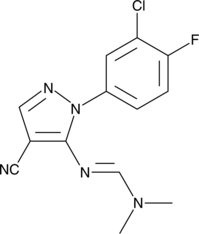
-
GC64789
Biotinylated-JQ1
Biotinylated-JQ1 (Biotin-JQ1) is a biotinylated derivative of JQ1 with high affinity for the bromodomain of BRD4. Biotinylated-JQ1 inhibits MM1.S multiple myeloma cells proliferation with the EC50?of 0.4 μM.
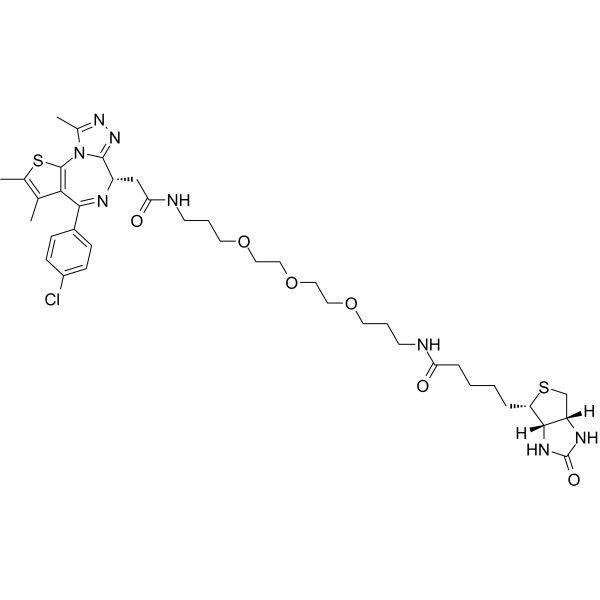
-
GC25139
Biphenyl-4-sulfonyl chloride
Biphenyl-4-sulfonyl chloride (p-Phenylbenzenesulfonyl, 4-Phenylbenzenesulfonyl, p-Biphenylsulfonyl) is a HDAC inhibitor with synthetic applications in palladium-catalyzed desulfitative C-arylation.
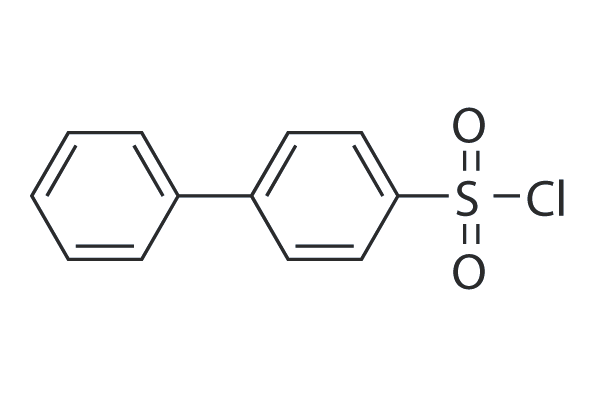
-
GC48463
Bisubstrate Inhibitor 78
An inhibitor of NNMT