Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Targets for Chromatin/Epigenetics
- Bromodomain(52)
- Aurora Kinase(89)
- DNA Methyltransferase(40)
- HDAC(218)
- Histone Acetyltransferases(67)
- Histone Demethylases(98)
- Histone Methyltransferase(212)
- HIF(101)
- JAK(178)
- MBT Domains(1)
- PARP(128)
- Pim(35)
- Protein Ser/Thr Phosphatases(41)
- RNA Polymerase(8)
- Sirtuin(84)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(207)
- MicroRNA(13)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Products for Chromatin/Epigenetics
- Cat.No. Product Name Information
-
GC33028
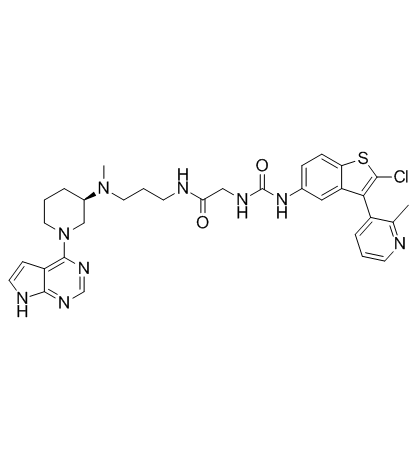
CF53
CF53 is a highly potent, selective and orally active inhibitor of BET protein, with a Ki of <1 nM, Kd of 2.2 nM and an IC50 of 2 nM for BRD4 BD1. CF53 binds to both the BD1 and BD2 domains of BRD2, BRD3, BRD4, and BRDT BET proteins with high affinities, very selective over non-BET bromodomain-containing proteins. CF53 shows potent anti-tumor activity both in vitro and in vivo.
-
GC65602
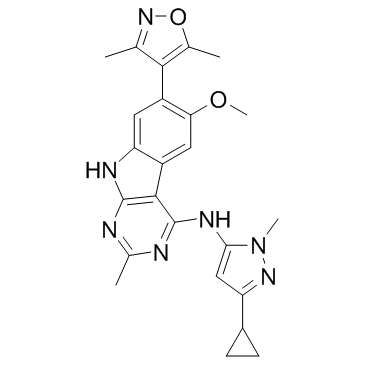
CFT8634
CFT8634 is a degrader targeting BRD9 extracted from patent WO2021178920A1 compound 174. CFT8634 can be used for the research of synovial sarcoma and SMARCB1-deleted solid tumors.
-
GC35668
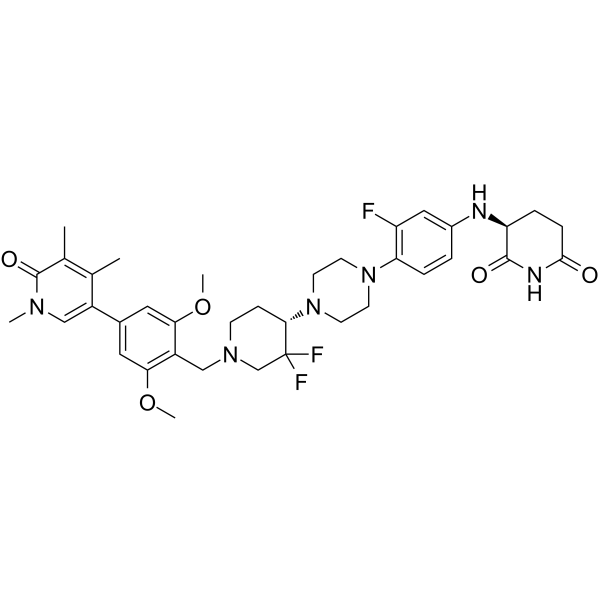
CG-200745
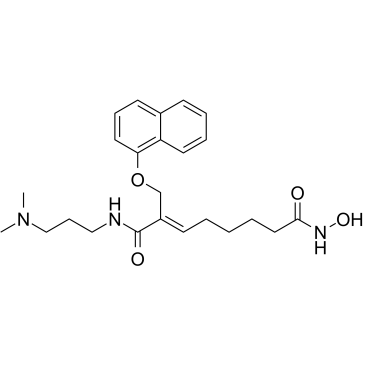
CG-200745 (CG-200745) is an orally active, potent pan-HDAC inhibitor which has the hydroxamic acid moiety to bind zinc at the bottom of catalytic pocket. CG-200745 inhibits deacetylation of histone H3 and tubulin. CG-200745 induces the accumulation of p53, promotes p53-dependent transactivation, and enhances the expression of MDM2 and p21 (Waf1/Cip1) proteins. CG-200745 enhances the sensitivity of Gemcitabine-resistant cells to Gemcitabine and 5-Fluorouracil (5-FU; ). CG-200745 induces apoptosis and has anti-tumour effects.
-
GC39679
CG347B
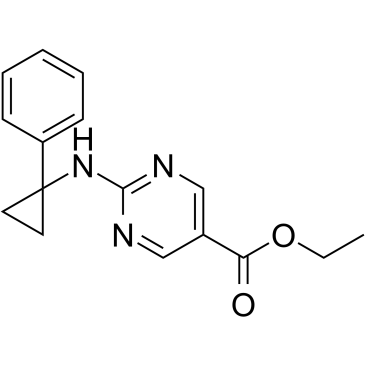
CG347B is a selective HDAC6 inhibitor, also involves in synthesis of other metalloenzyme inhibitors.
-
GC14936
Chaetocin
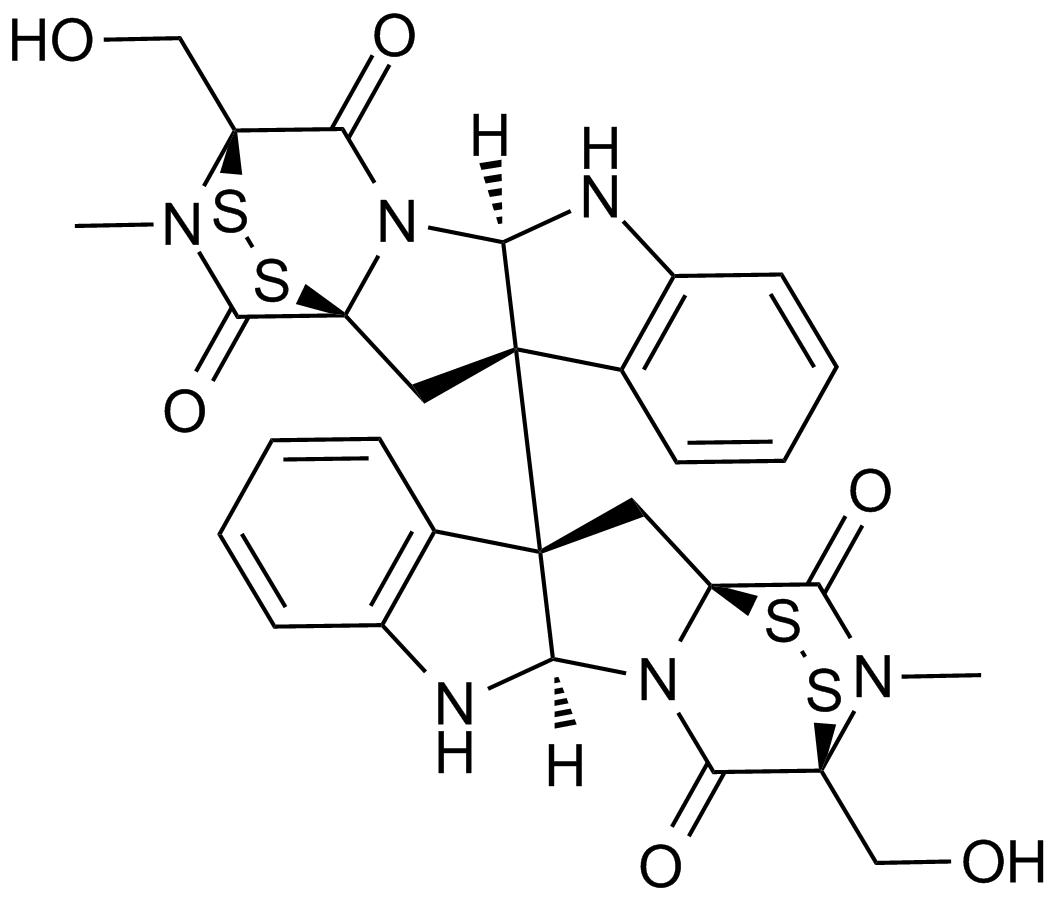
Inhibitor of lys9-specific HMTs
-
GC11975
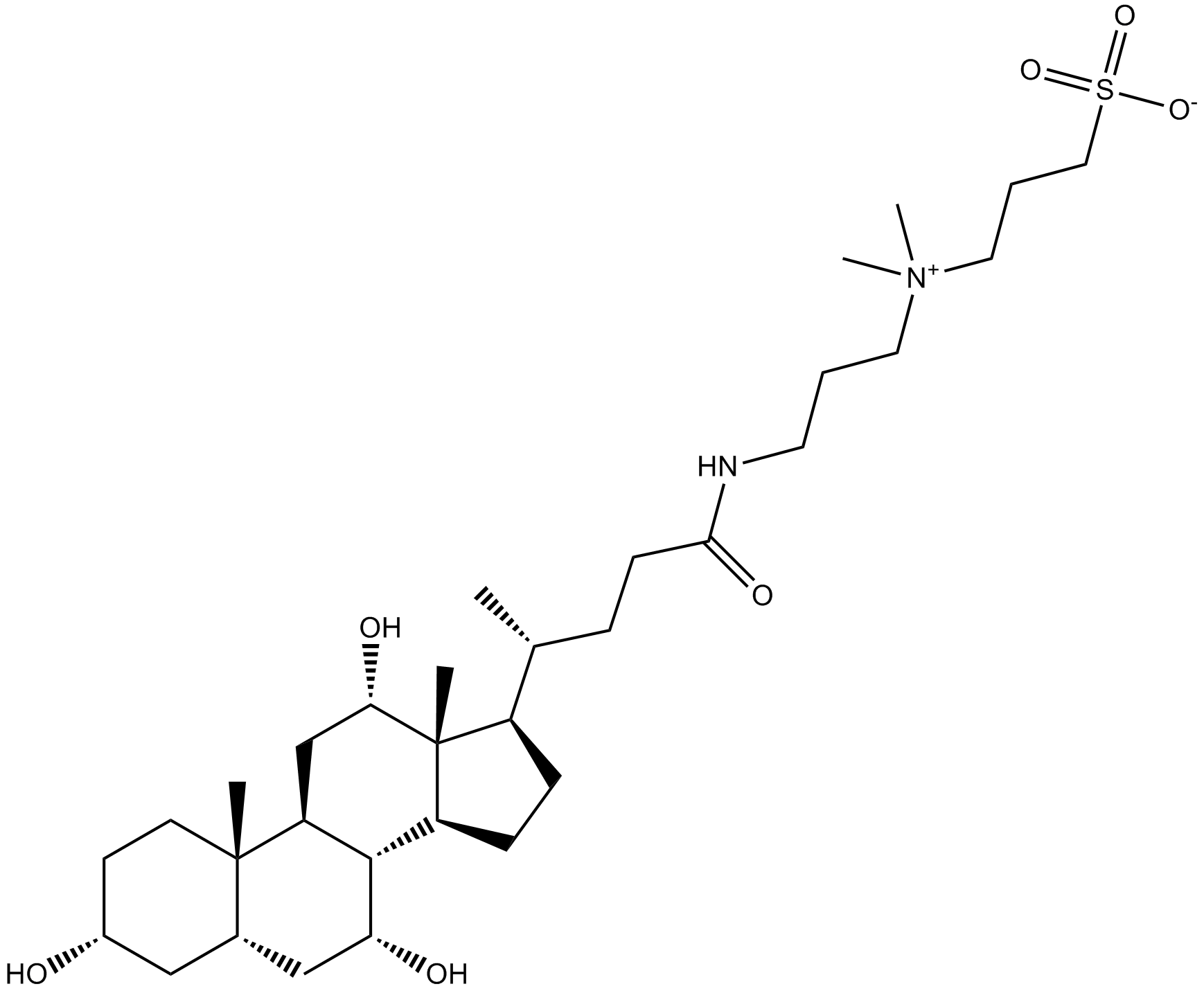
CHAPS
-
GC64942
CHDI-390576
CHDI-390576, a potent, cell permeable and CNS penetrant class IIa histone deacetylase (HDAC) inhibitor with IC50s of 54 nM, 60 nM, 31 nM, 50 nM for class IIa HDAC4, HDAC5, HDAC7, HDAC9, respectively, shows >500-fold selectivity over class I HDACs (1, 2, 3) and ~150-fold selectivity over HDAC8 and the class IIb HDAC6 isoform.
-
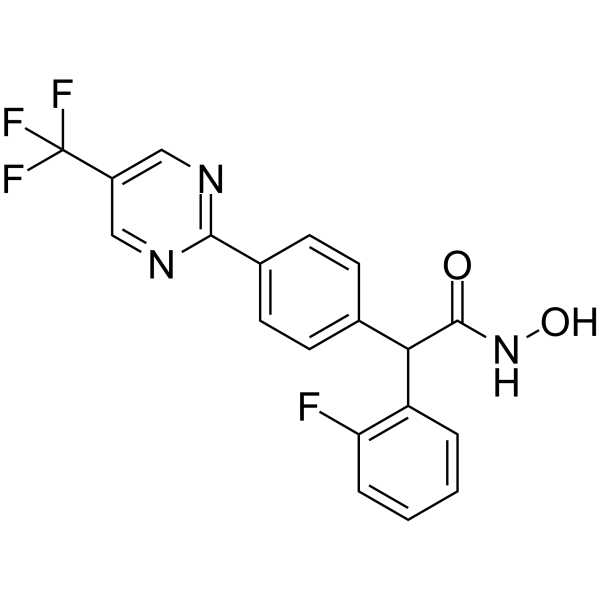
GC17405
Chetomin
An inhibitor of HIF signaling
-
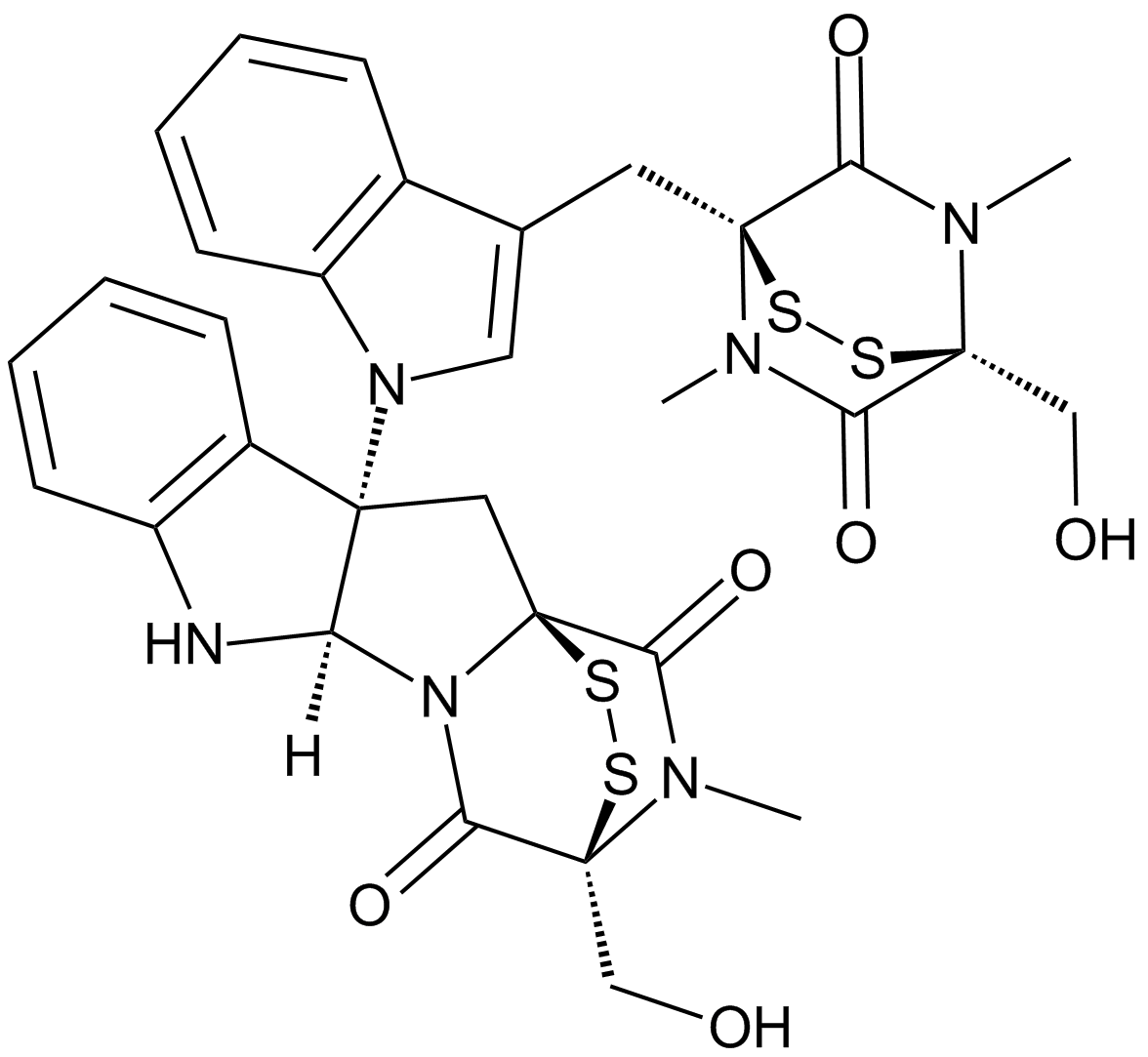
GC62145
Chiauranib
Chiauranib (CS2164) is an orally active multi-target inhibitor against tumor angiogenesis. Chiauranib potently inhibits the angiogenesis-related kinases (VEGFR1, VEGFR2, VEGFR3, PDGFRα and c-Kit), mitosis-related kinase Aurora B, and chronic inflammation-related kinase CSF-1R, with IC50 values ranging from 1-9 nM. Chiauranib has strongly anticancer effects.

-
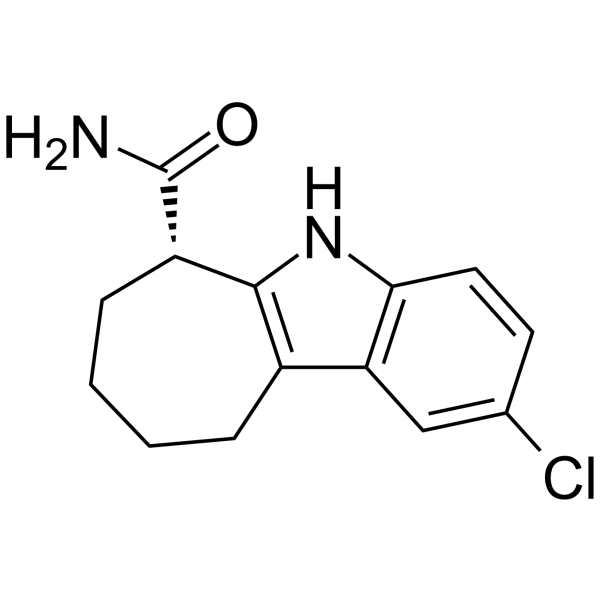
GC64255
CHIC35
CHIC35, an analog of EX-527, is a potent and selective inhibitor of SIRT1 (IC50=0.124 ?M).
-
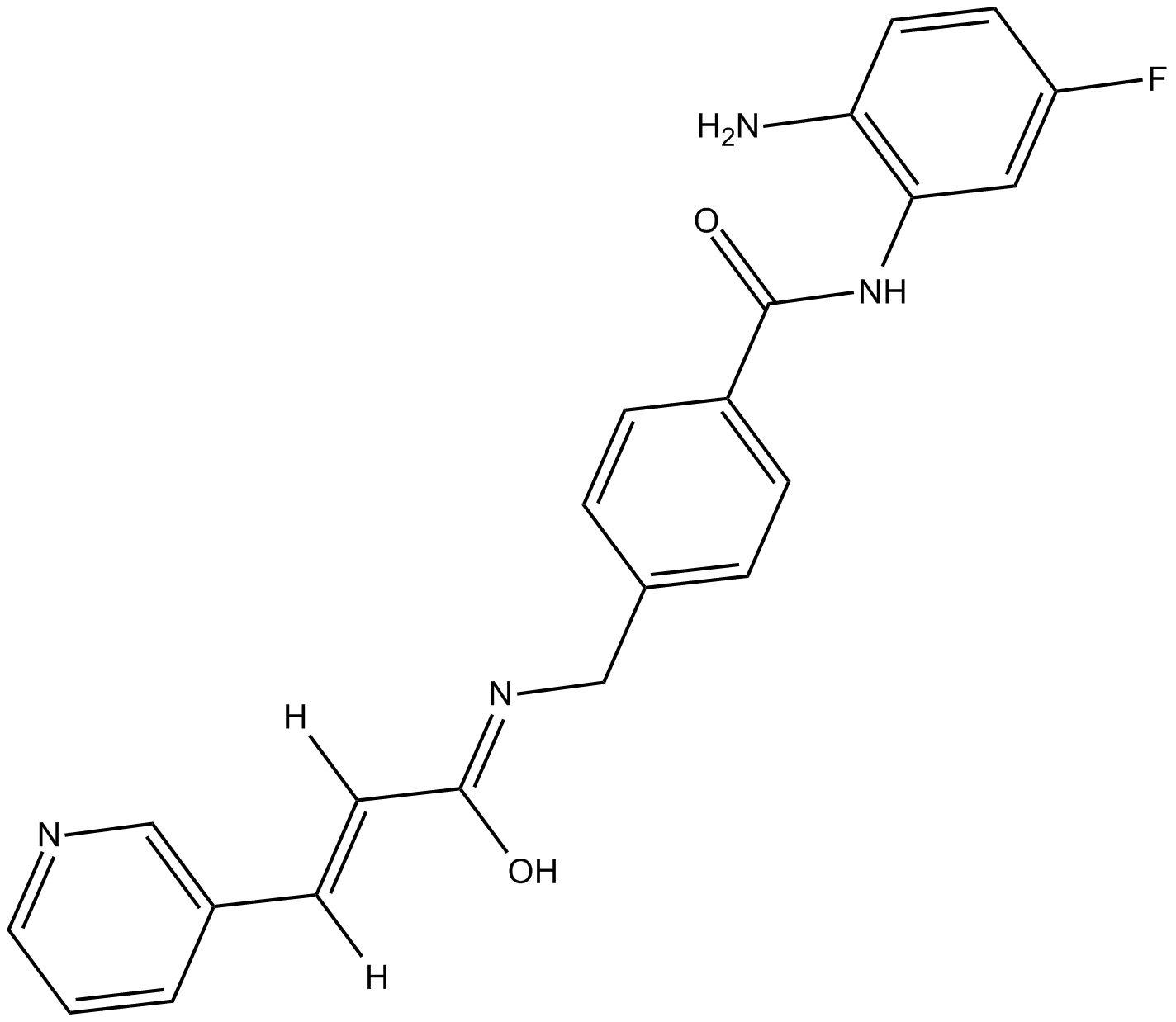
GC16042
Chidamide
Chidamide (Chidamide impurity) is an impurity of Chidamide. Chidamide is a potent and orally bioavailable HDAC enzymes class I (HDAC1/2/3) and class IIb (HDAC10) inhibitor.
-
GN10518
Chitosamine hydrochloride
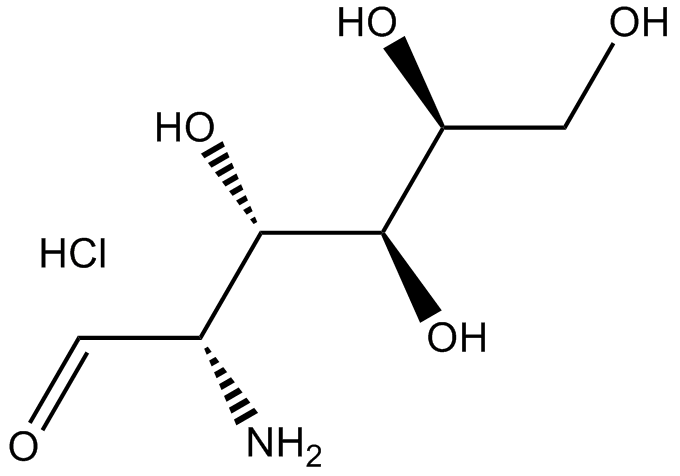
-
GC45717
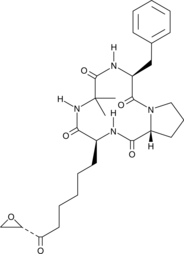
Chlamydocin
An HDAC inhibitor
-
GN10308
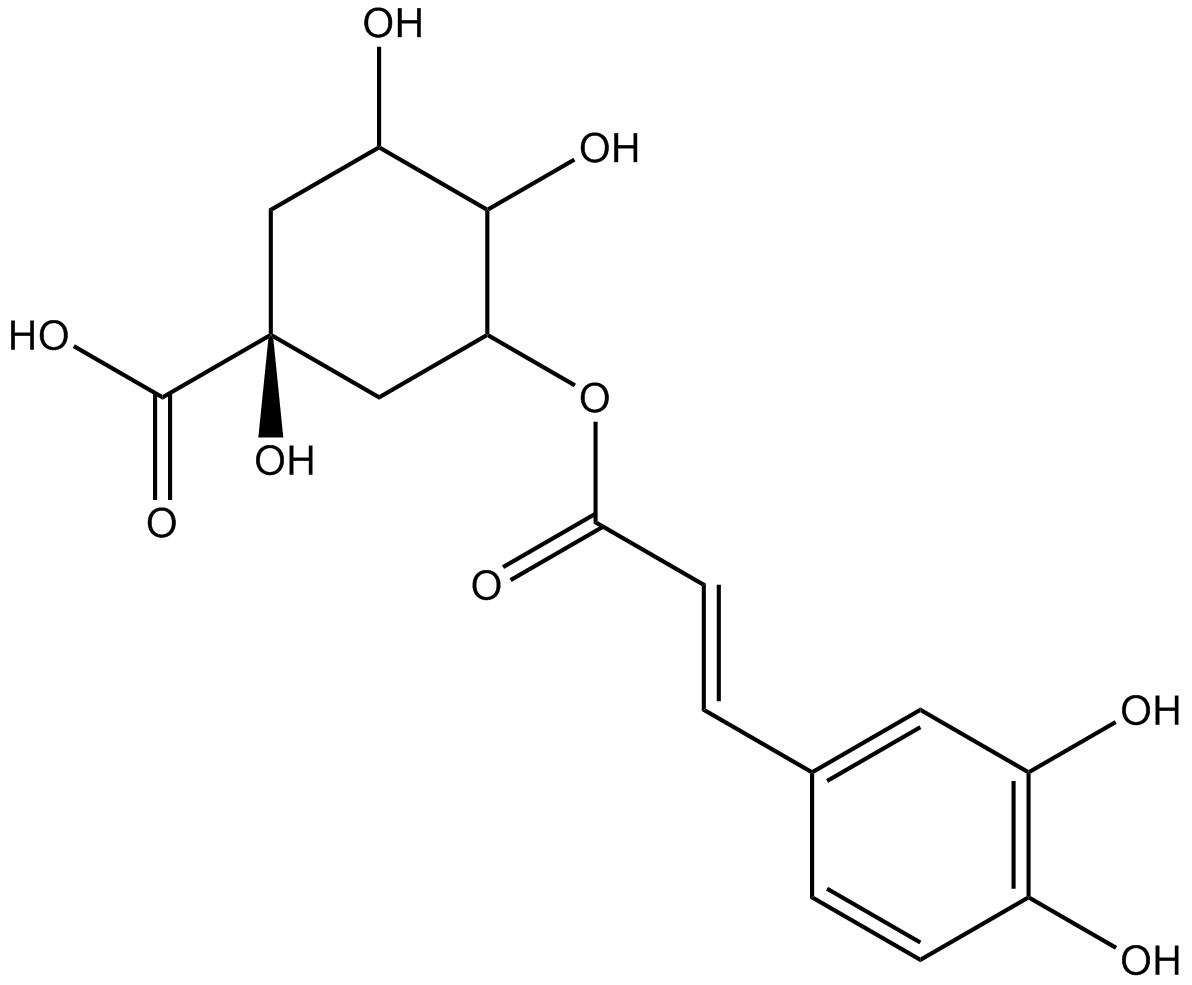
Chlorogenic acid
Chlorogenic acid is a major phenolic compound in coffee and tea.
-
GC11652
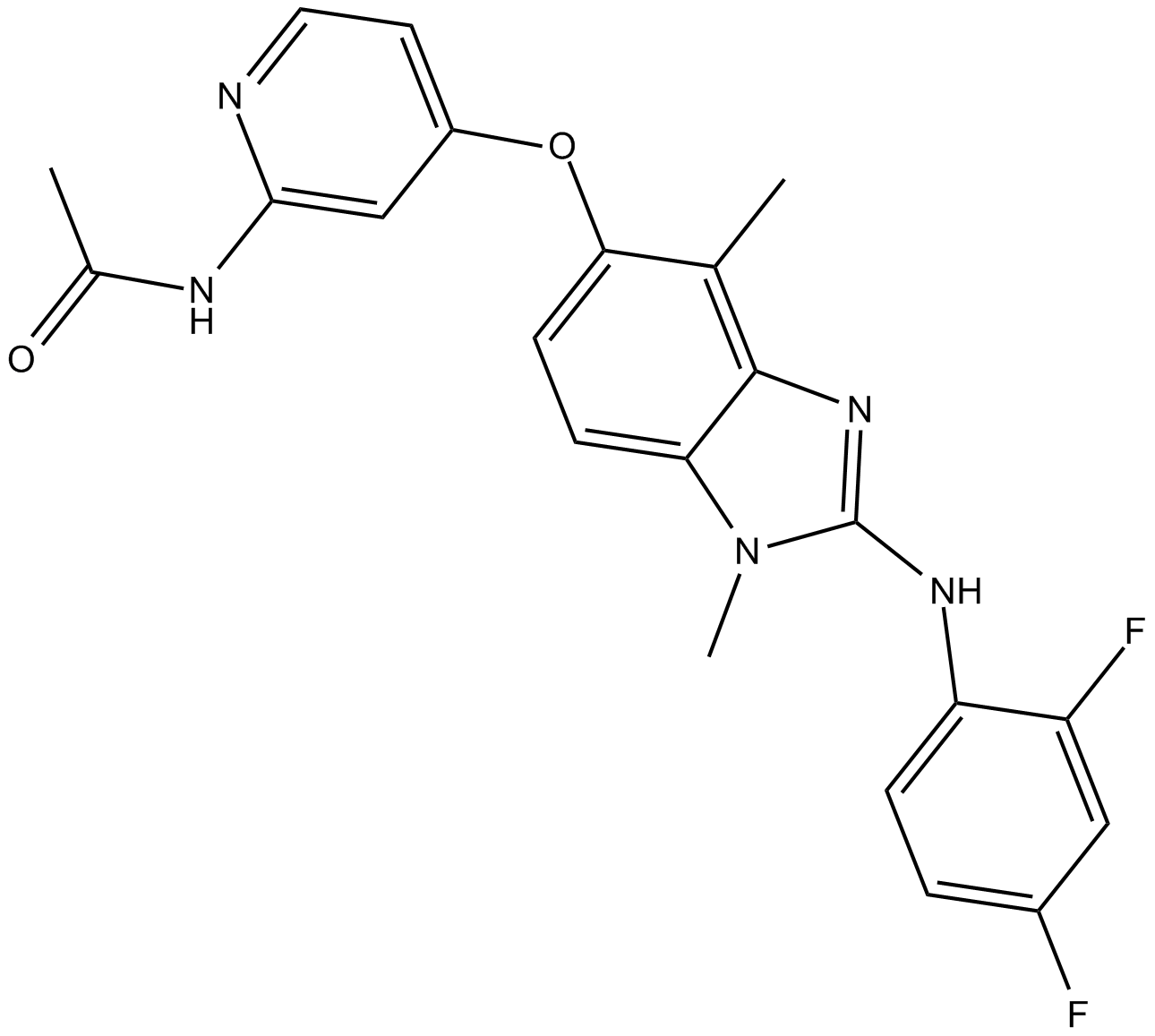
CHZ868
Type II JAK2 inhibitor
-
GC11539
CI 976
CI 976 (CI 976) is a potent, orally active, and selective inhibitor of ACAT (acyl coenzyme A:cholesterol acyltransferase) with an IC50s of 73 nM.
-
GC13408
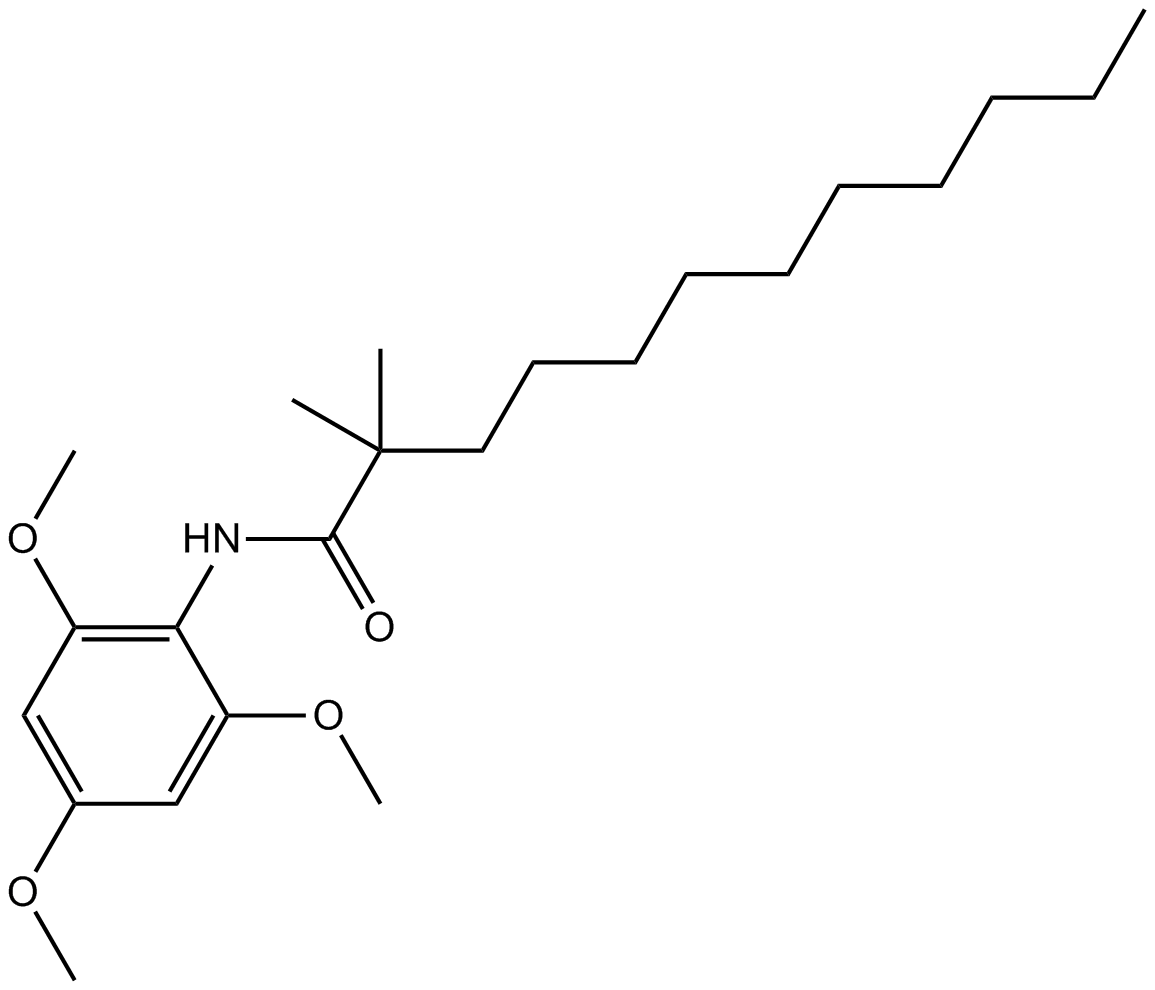
CI994 (Tacedinaline)
An inhibitor of HDAC1, -2, and -3
-
GC15718
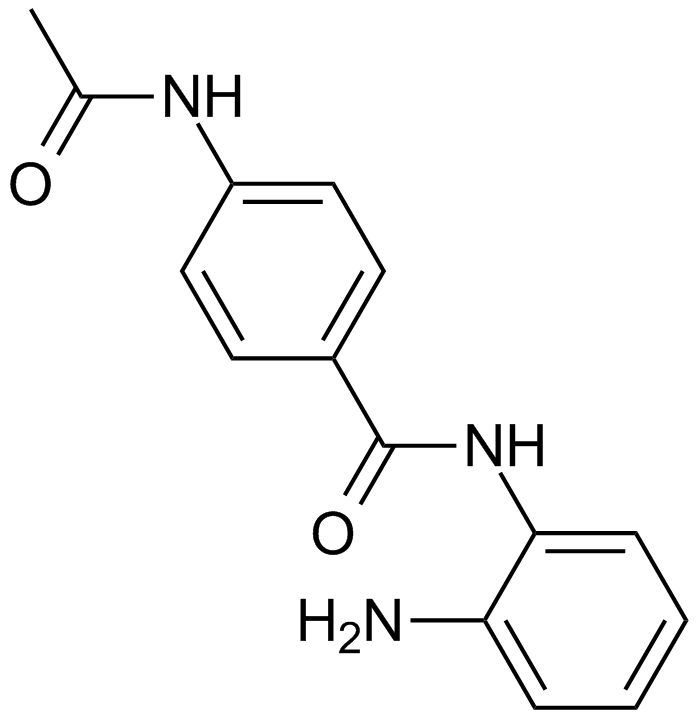
CID 2011756
An inhibitor of protein kinase D
-
GC18577
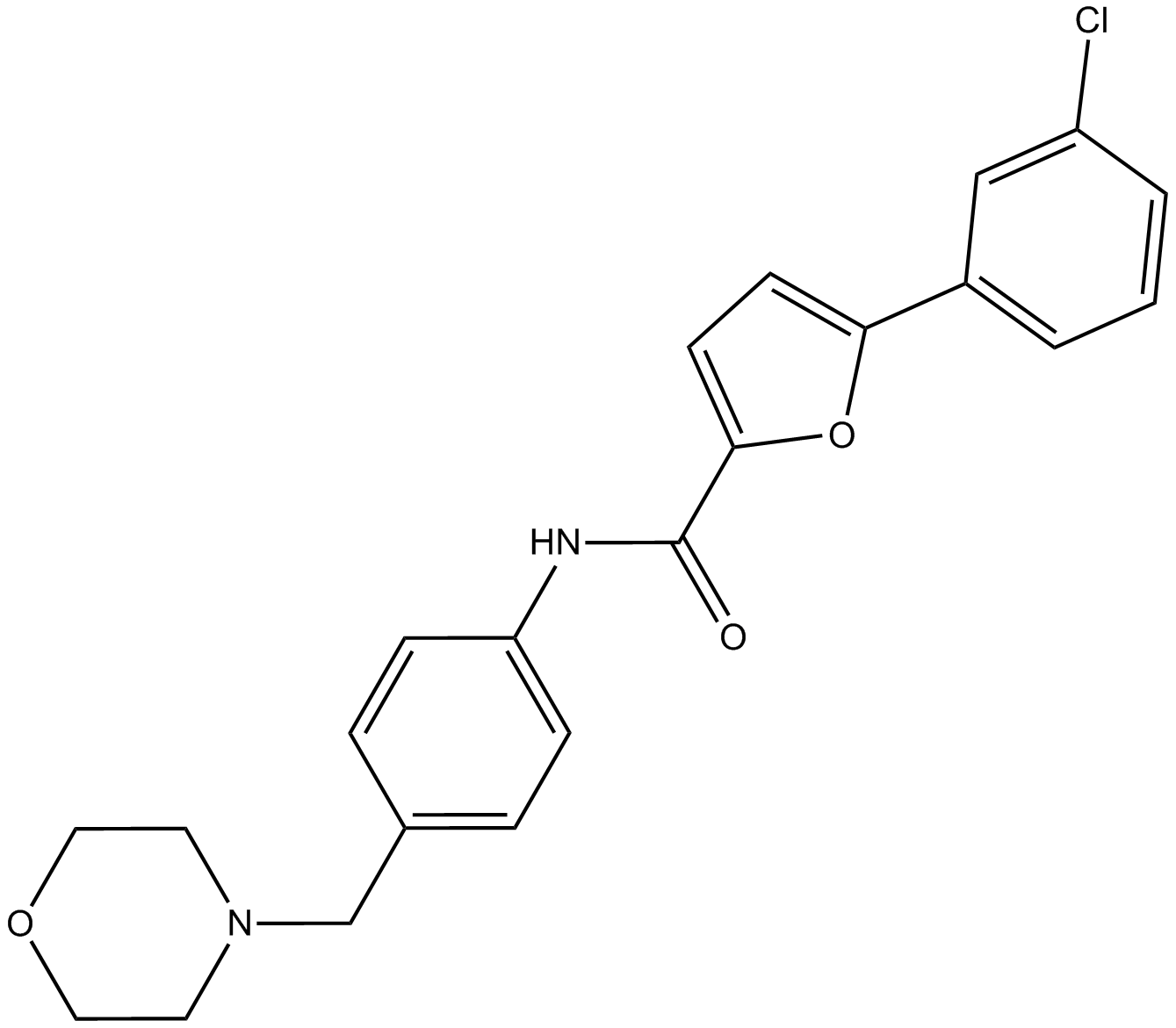
CID-2818500
An inhibitor of PRMT1
-
GC52351
Citrullinated α-Enolase (R8 + R14) (1-19)-biotin Peptide
A biotinylated and citrullinated α-enolase peptide

-
GC52363
Citrullinated Histone H3 (R2 + R8 + R17) (2-22)-biotin Peptide
A biotinylated and citrullinated histone H3 peptide

-
GC52367
Citrullinated Vimentin (G146R) (R144 + R146) (139-159)-biotin Peptide
A biotinylated and citrullinated mutant vimentin peptide

-
GC52370
Citrullinated Vimentin (R144) (139-159)-biotin Peptide
A biotinylated and citrullinated vimentin peptide

-
GC43275
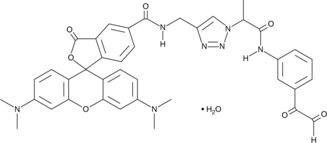
Citrulline-specific Probe-Rhodamine
Protein arginine deiminases (PADs) catalyze the posttranslational modification of arginine residues on proteins to form citrulline, which plays a large role in regulating gene expression.
-
GC39485
CK2/ERK8-IN-1
A dual inhibitor of CK2 and ERK8
-
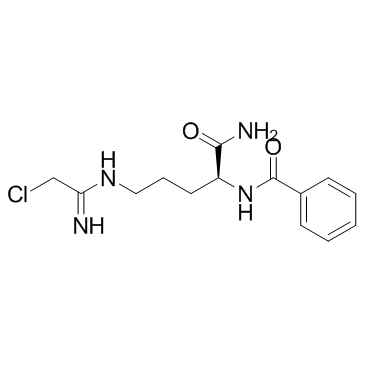
GC35706
Cl-amidine
Cl-amidine (N-α-benzoyl-N5-(2-chloro-1-iminoethyl)-l-ornithine amide), is a PAD4 inactivator with enhanced potency.
-
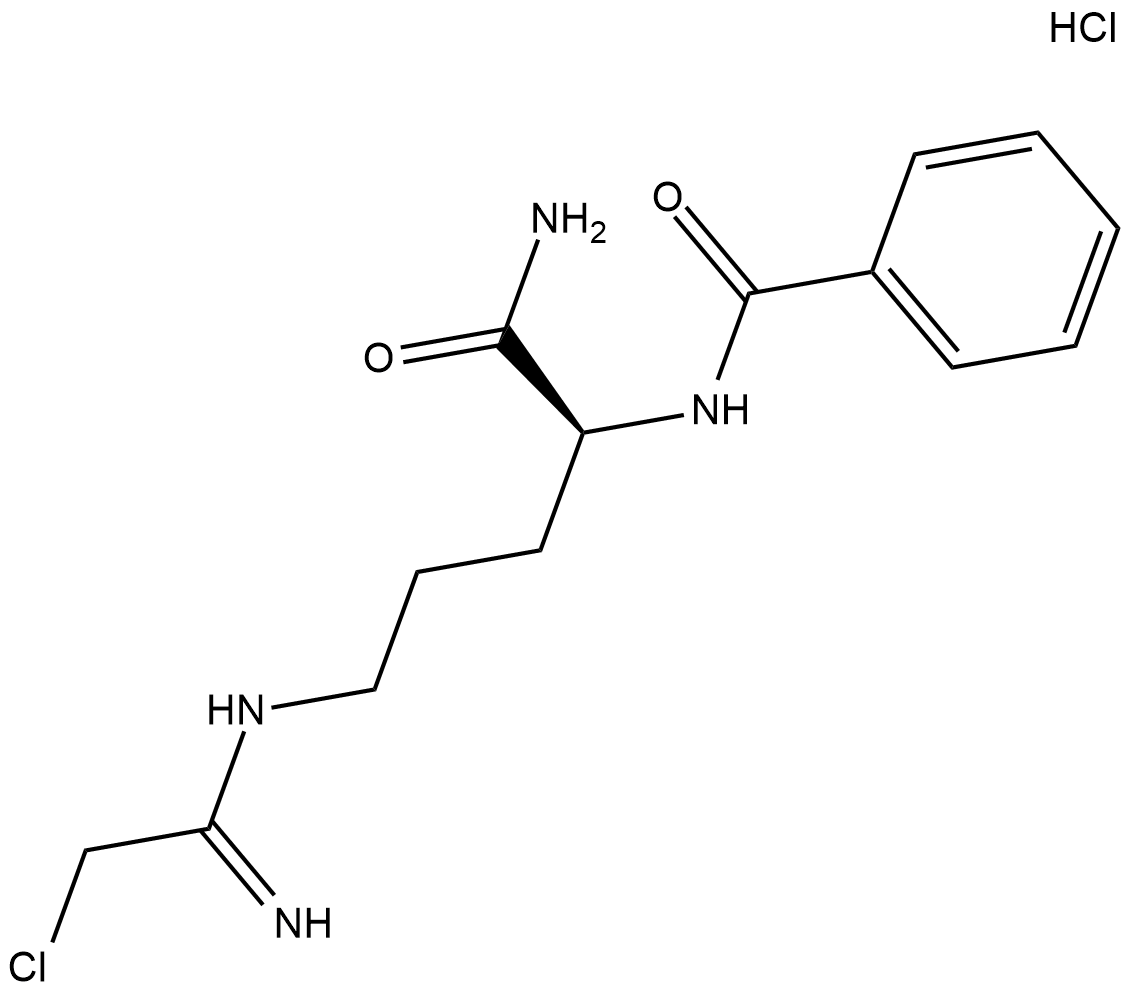
GC18925
Cl-Amidine (hydrochloride)
Cl-amidine is an inhibitor of protein arginine deiminases (PAD; IC50s = 0.8, 6.2, and 5.9 uM for PAD1, PAD3, and PAD4 in vitro, respectively).
-
GC11032
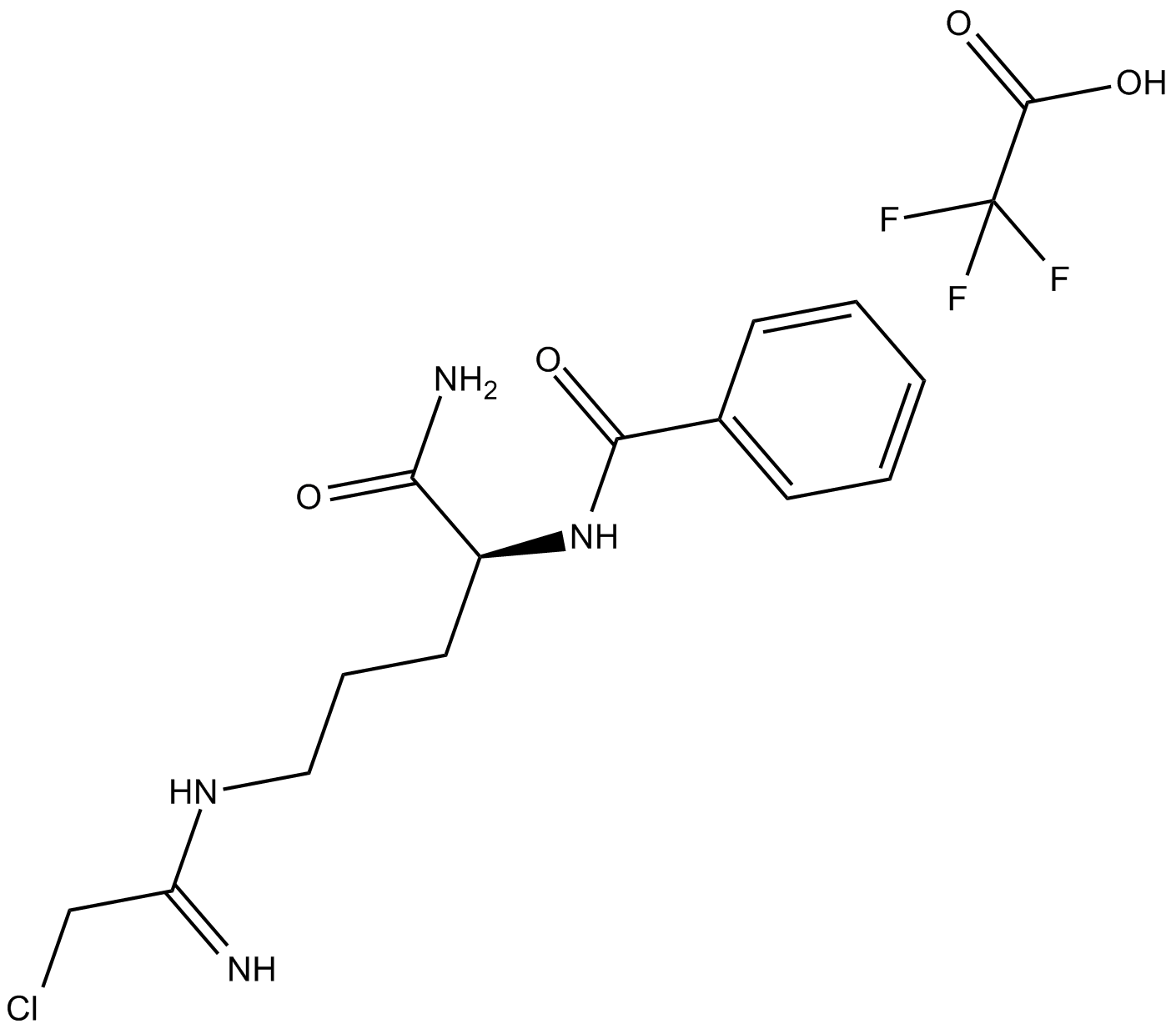
Cl-Amidine (trifluoroacetate salt)
Cl-Amidine (trifluoroacetate salt) is an orally active peptidylarginine deminase (PAD) inhibitor, with IC50 values of 0.8 μM, 6.2 μM and 5.9 μM for PAD1, PAD3, and PAD4, respectively.
-
GC12367
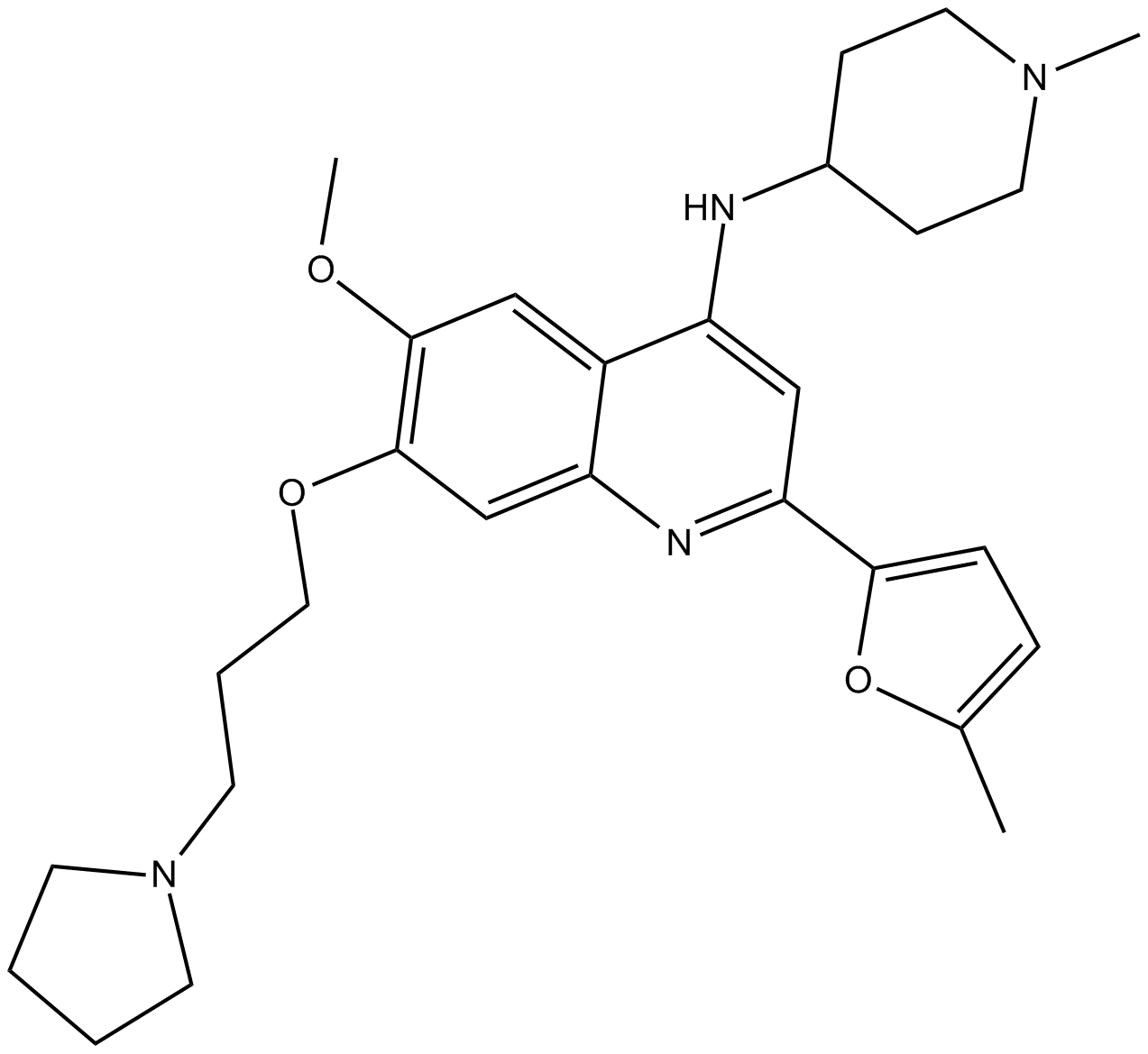
CM-272
CM-272 is a first-in-class reversible dual inhibitor against G9a and DNMTs with IC50 values of 8 nM and 382 nM, respectively [1].
-
GC33320
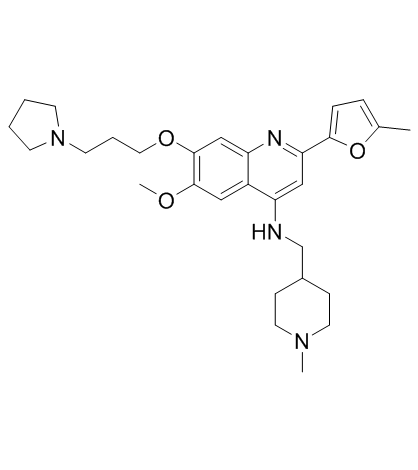
CM-579
CM-579 is a first-in-class reversible, dual inhibitor of G9a and DNMT, with IC50 values of 16 nM, 32 nM for G9a and DNMT, respectively. Has potent in vitro cellular activity in a wide range of cancer cells.
-
GC35714
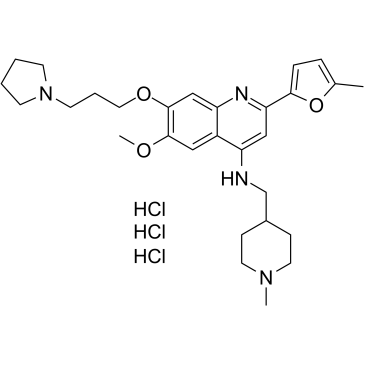
CM-579 trihydrochloride
CM-579 trihydrochloride is a first-in-class reversible, dual inhibitor of G9a and DNMT, with IC50 values of 16 nM, 32 nM for G9a and DNMT, respectively. Has potent in vitro cellular activity in a wide range of cancer cells.
-
GC65426
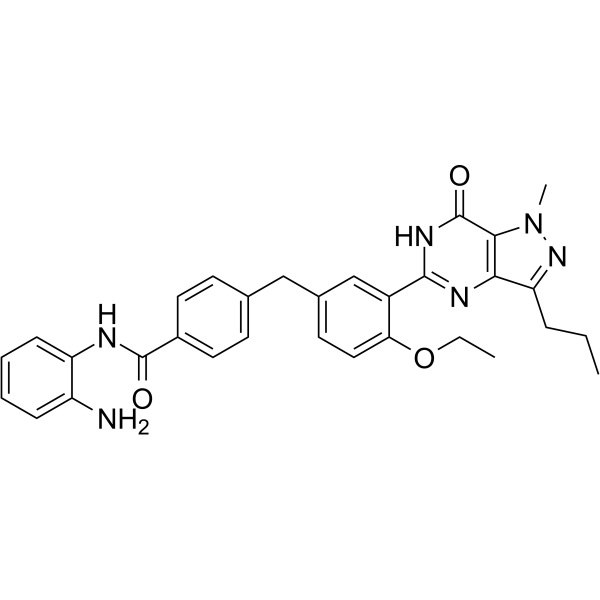
CM-675
CM-675 is a dual phosphodiesterase 5 (PDE5) and class I histone deacetylases-selective inhibitor, with IC50 values of 114 nM and 673 nM for PDE5 and HDAC1, respectively.
-
GC39665
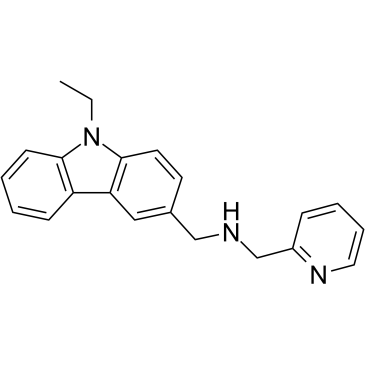
CMP-5
CMP-5 is a potent, specific, and selective PRMT5 inhibitor, while displays no activity against PRMT1, PRMT4, and PRMT7 enzymes. CMP-5 selectively blocks S2Me-H4R3 by inhibiting PRMT5 methyltransferase activity on histone preparations. CMP-5 prevents Epstein-Barr virus (EBV)-driven B-lymphocyte transformation but leaving normal B cells unaffected.
-
GC34165
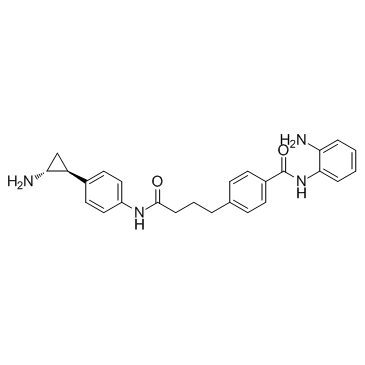
Corin
Corin is a dual inhibitor of histone lysine specific demethylase (LSD1) and histone deacetylase (HDAC), with a Ki(inact) of 110 nM for LSD1 and an IC50 of 147 nM for HDAC1.
-
GC43318
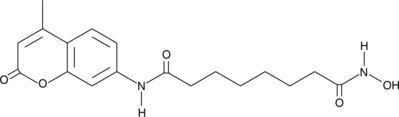
coumarin-SAHA
Suberoylanilide hydroxamic acid (SAHA) is a class I and class II histone deacetylase (HDAC) inhibitor that binds directly to the catalytic site of the enzyme thereby blocking substrate access.
-
GC62908
Coumermycin A1
Coumermycin A1 is a JAK2 signal activator.
-
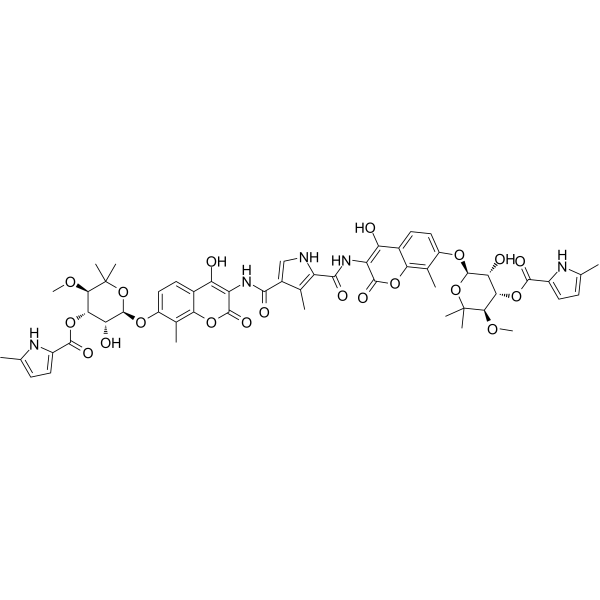
GC25304
CP2
CP2 is a cyclic peptide that inhibits the JmjC histone demethylases KDM4 with IC50 values of 42 nM and 29 nM for KDM4A and KDM4C, respectively.
-
GC16298
CPI-1205
EZH2 inhibitor
-
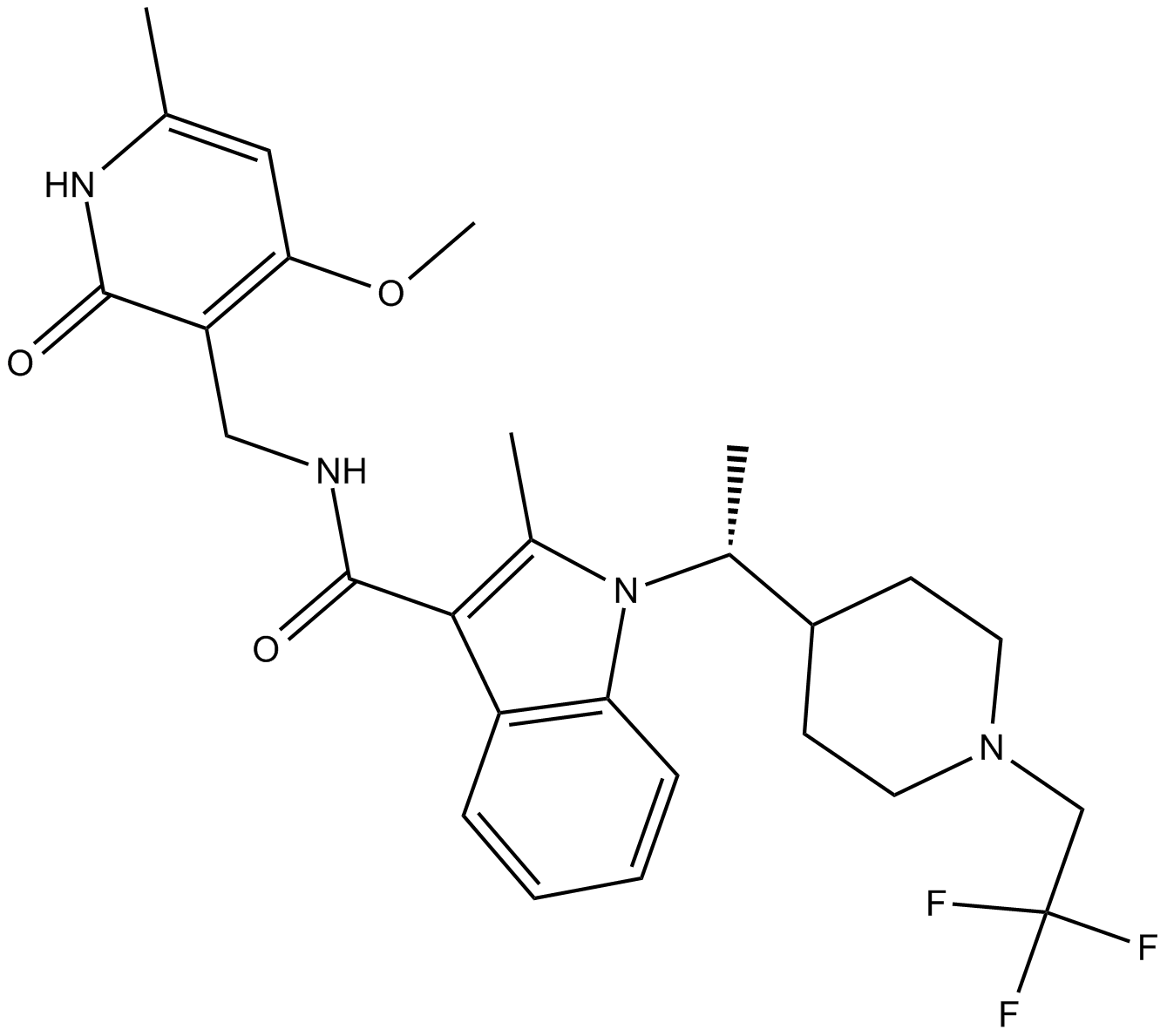
GC62669
CPI-1612
CPI-1612 is a highly potent, orally active EP300/CBP histone acetyltransferase (HAT) inhibitor with an IC50 of 8.1 nM for EP300 HAT. CPI-1612 has an anticancer activity.
-
GC16599
CPI-169
EZH2 inhibitor
-
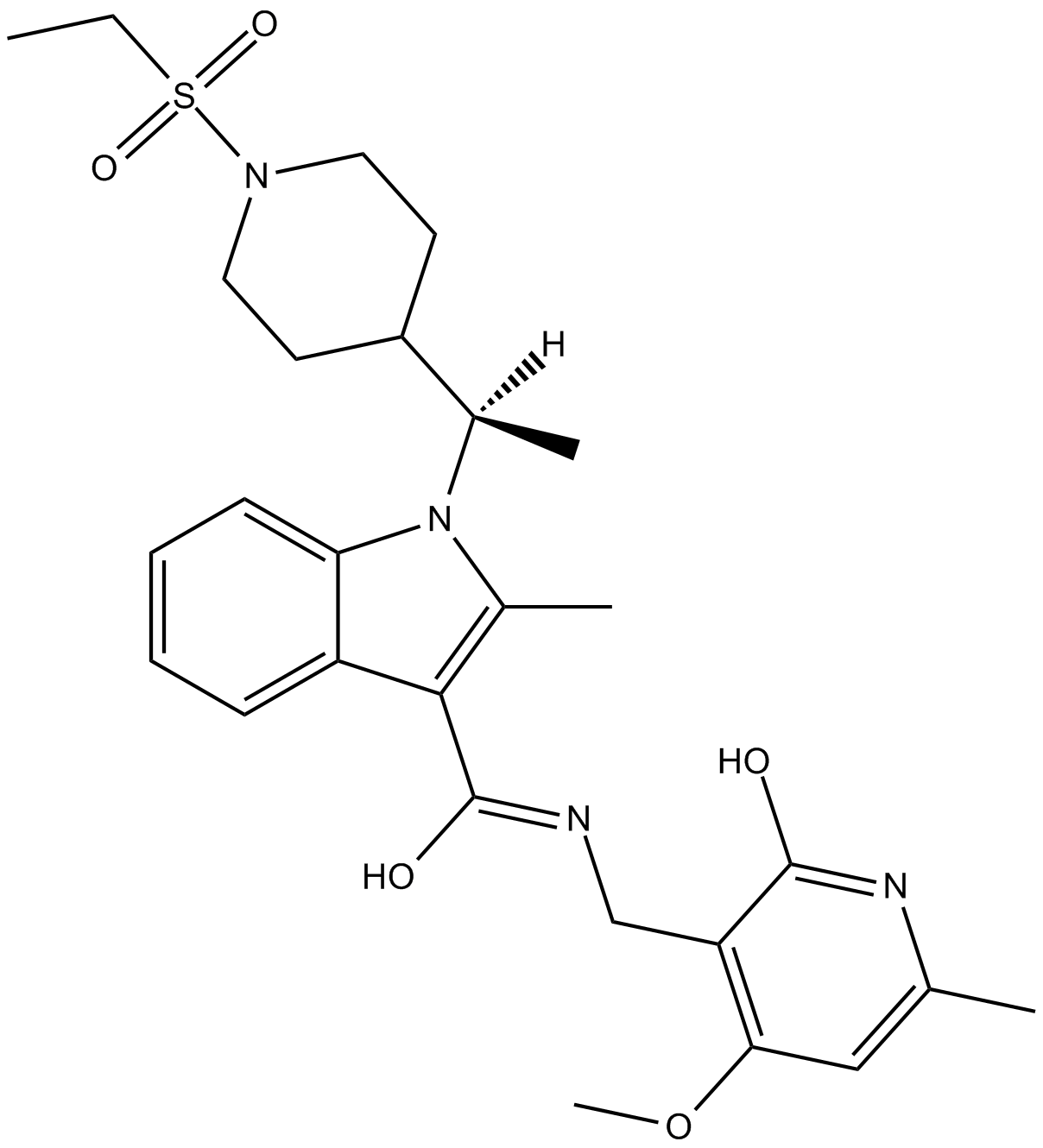
GC14699
CPI-203
BET bromodomain inhibitor
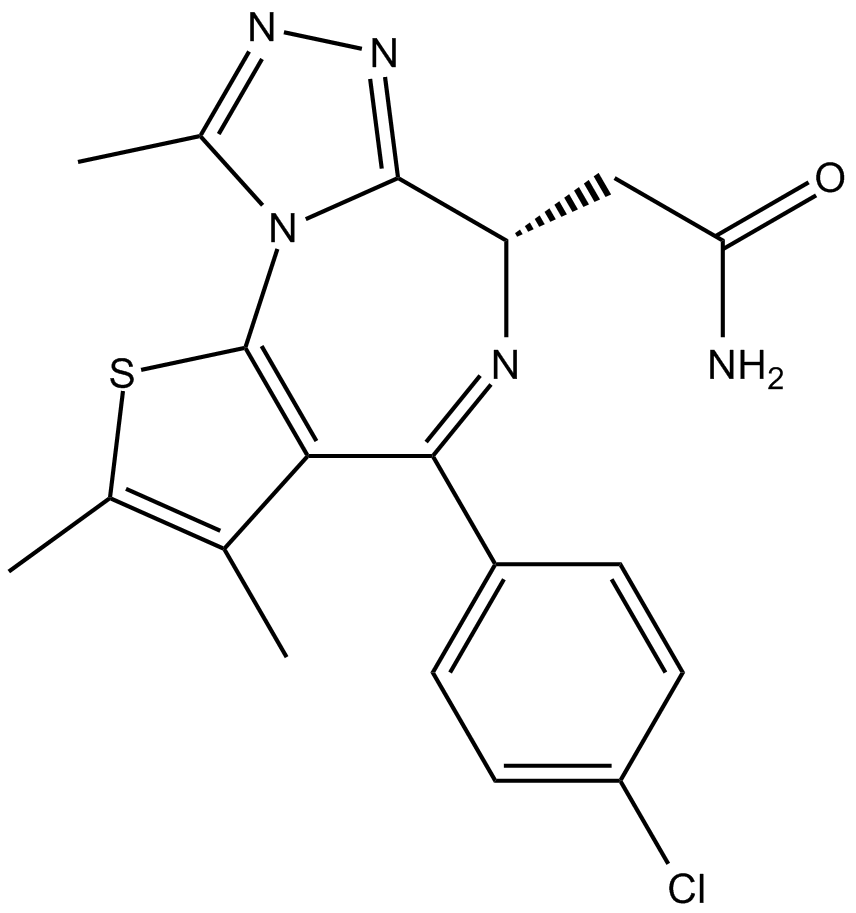
-
GC43320
CPI-268456
CPI-268456 (compound 141) is a potent BRD4 inhibitor with an IC50 of <0.5 μM.
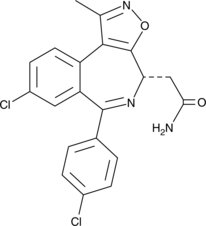
-
GC10021
CPI-360
EZH2 inhibitor
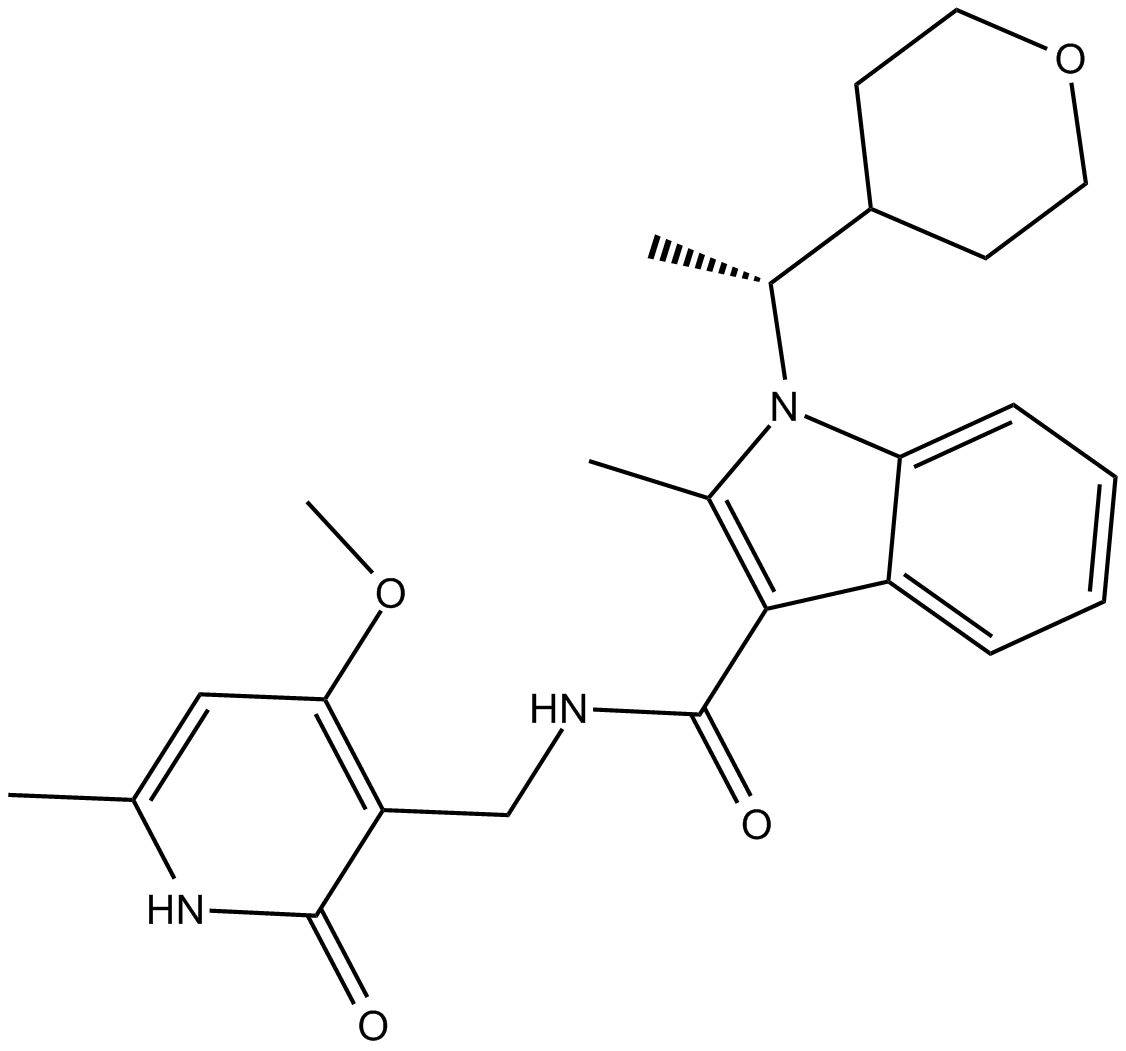
-
GC10774
CPI-455
KDM5 inhibitor
-
GC25305
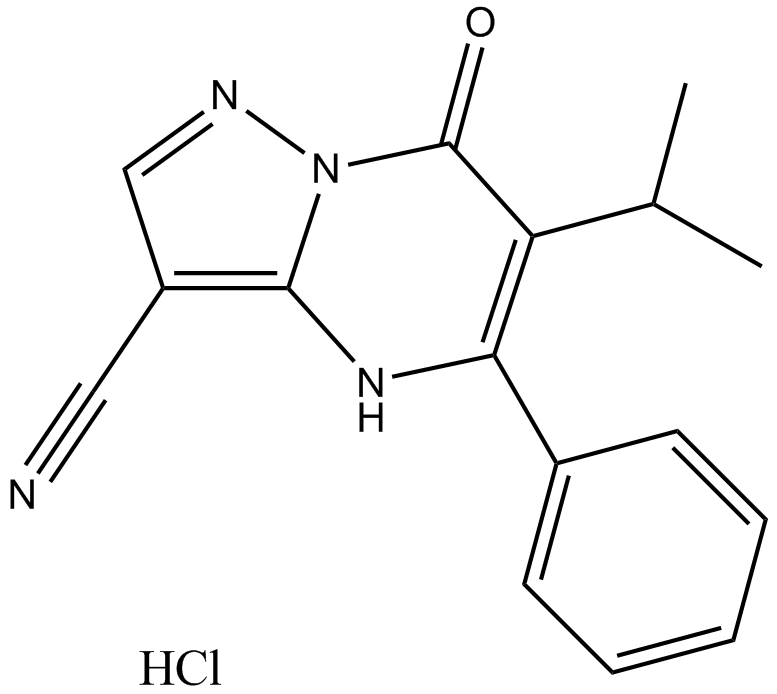
CPI-455 HCl
CPI-455 HCl is a specific KDM5 inhibitor with a half-maximal inhibitory concentration (IC50) of 10 ± 1 nM for full-length KDM5A in enzymatic assays, elevating global levels of H3K4 trimethylation (H3K4me3) and decreased the number of DTPs in multiple cancer cell line models treated with standard chemotherapy or targeted agents.This product has poor solubility, animal experiments are available, cell experiments please choose carefully!
-
GC10382
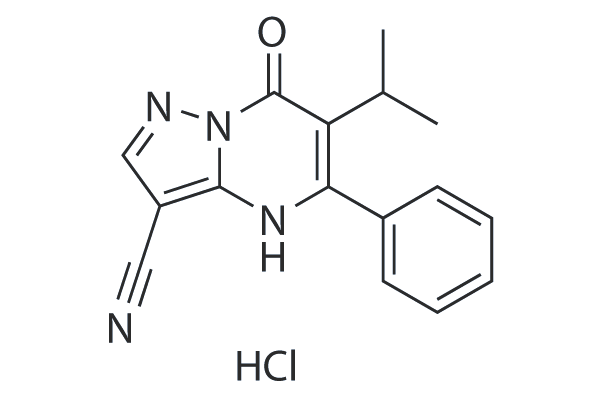
CPI-637
CBP/EP300 bromodomain inhibitor
-
GC39365
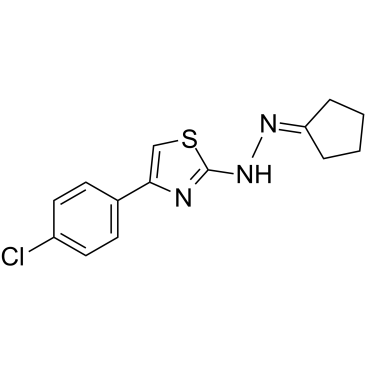
CPTH2
CPTH2 is a potent histone acetyltransferase (HAT) inhibitor. CPTH2 selectively inhibits the acetylation of histone H3 by Gcn5. CPTH2 induces apoptosis and decreases the invasiveness of a clear cell renal carcinoma (ccRCC) cell line through the inhibition of acetyltransferase p300 (KAT3B).
-
GC14428
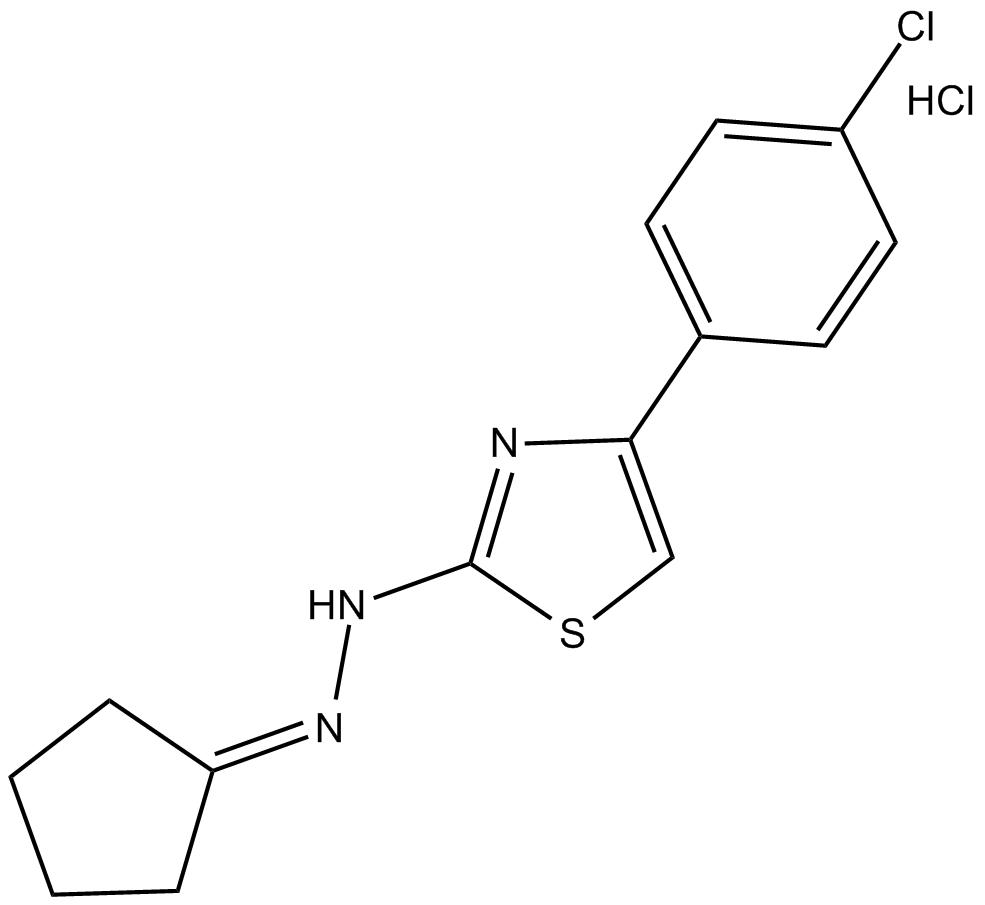
CPTH2 (hydrochloride)
inhibitor of HAT activity of Gcn5
-
GC35742
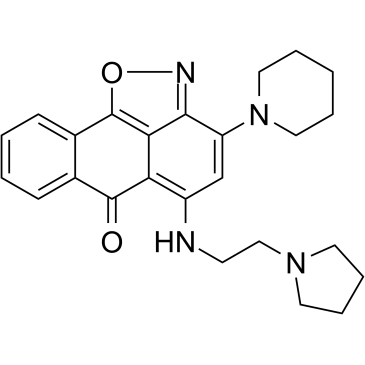
CPUY074020
CPUY074020 is a potent and oral bioavailable inhibitor of histone methyltransferase G9a, with an IC50 of 2.18 μM. CPUY074020 possesses anti-proliferative activity.
-
GC32565
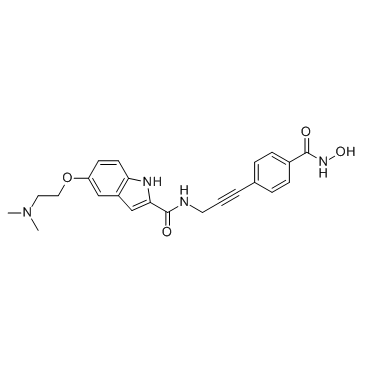
CRA-026440
CRA-026440 is a potent, broad-spectrum HDAC inhibitor.
-
GC67674
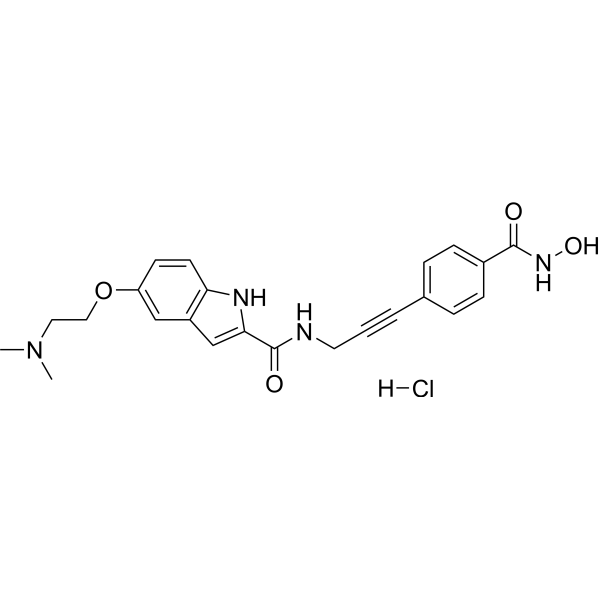
CRA-026440 hydrochloride
-
GC38412
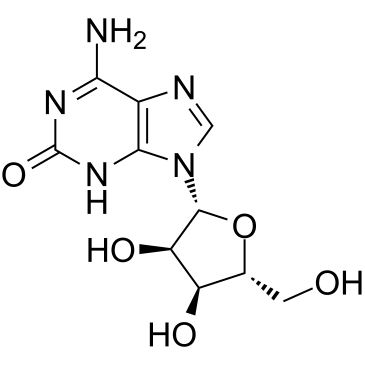
Crotonoside
A guanosine analog with diverse biological activities
-
GC14524
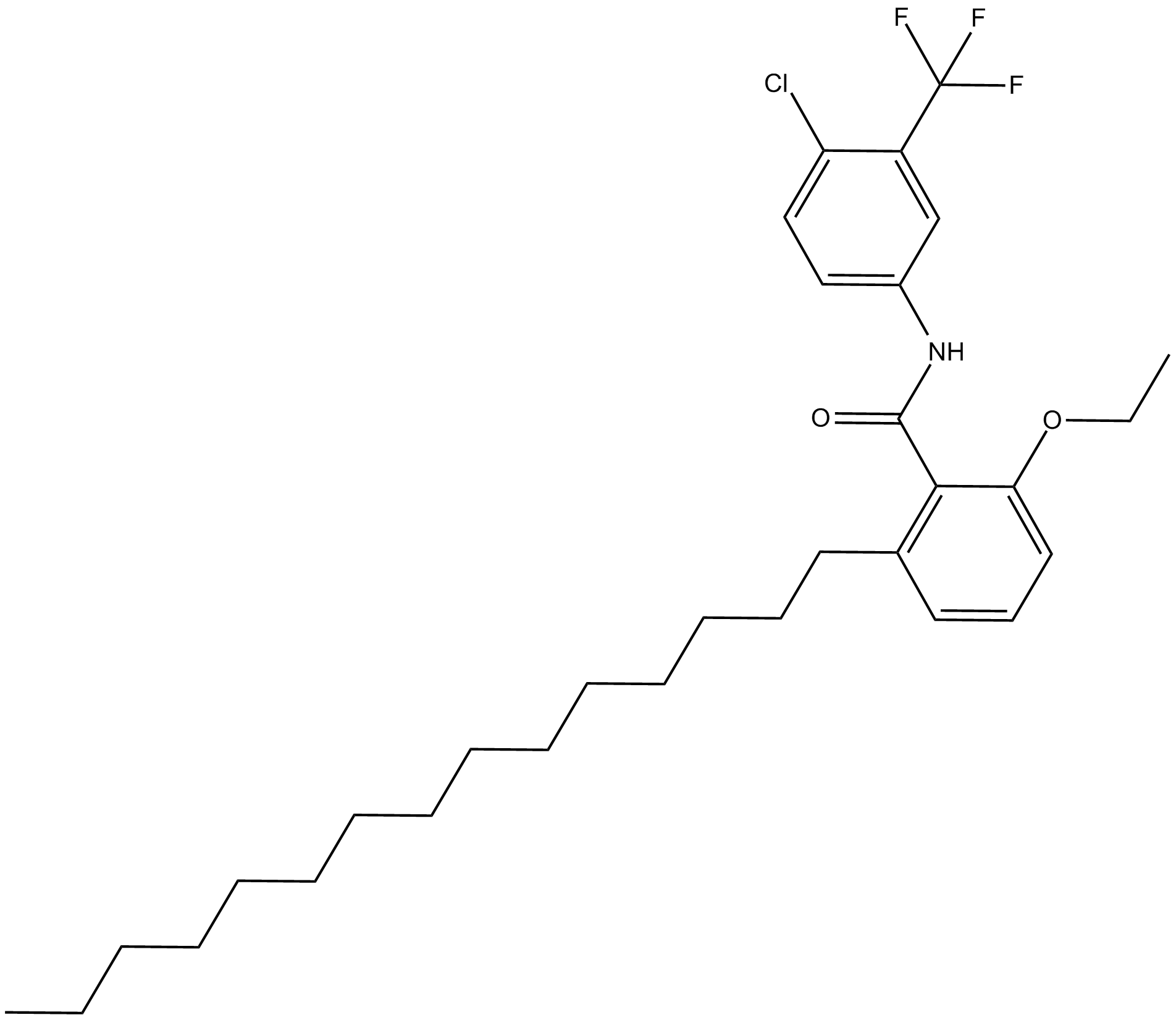
CTPB
selective activator of the histone acetyltransferase (HAT) p300
-
GC16008
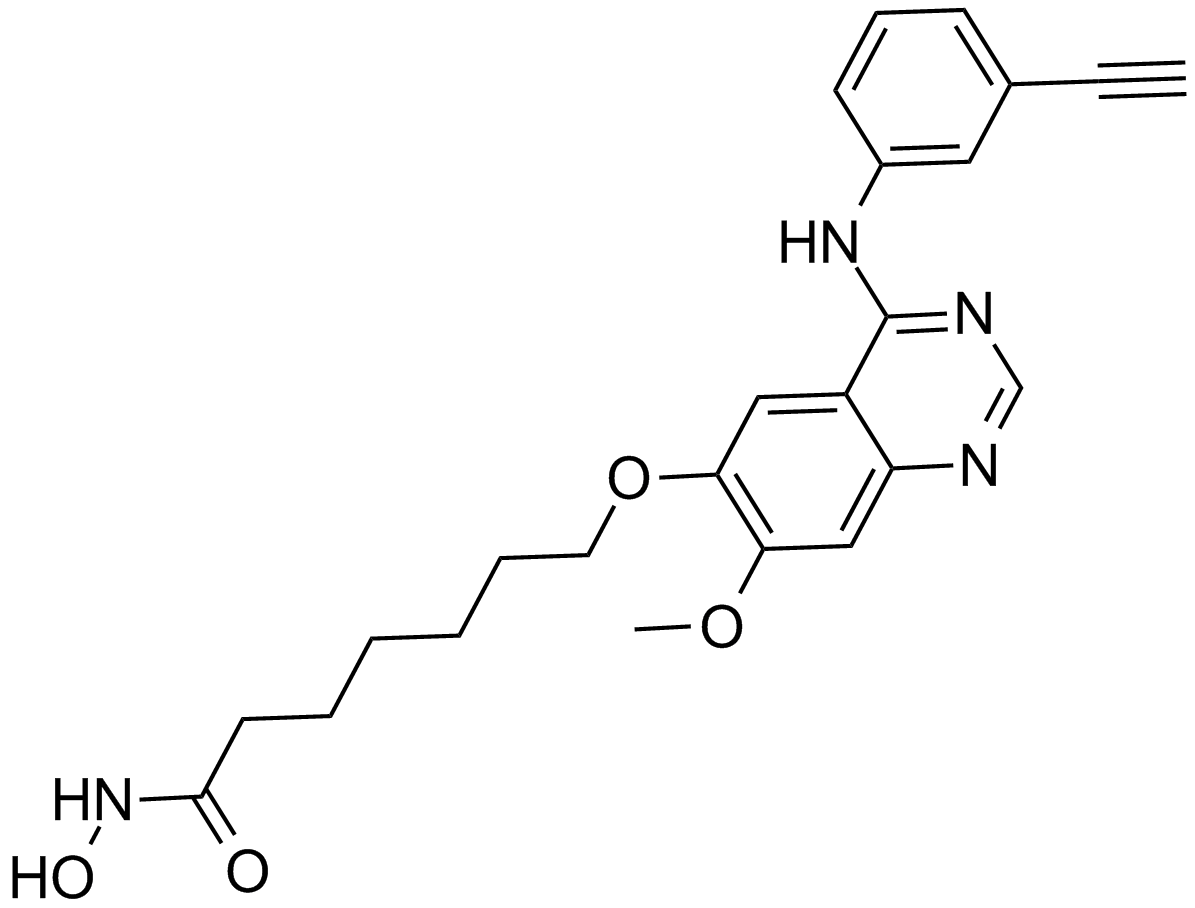
CUDC-101
A multi-target inhibitor of HDACs, EGFR, and HER2
-
GC12115
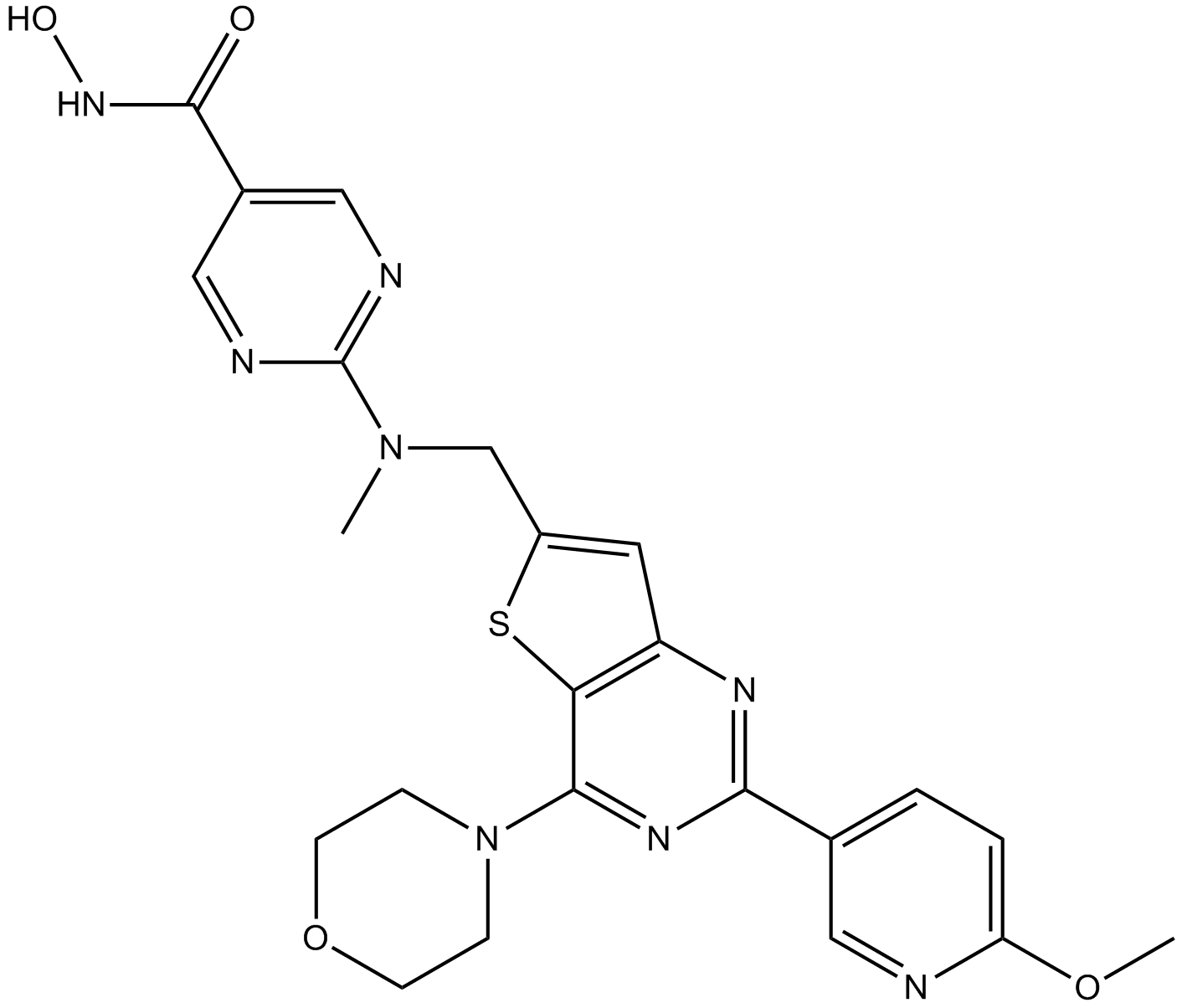
CUDC-907
A dual inhibitor of HDACs and PI3Ks
-
GN10442
Curculigoside
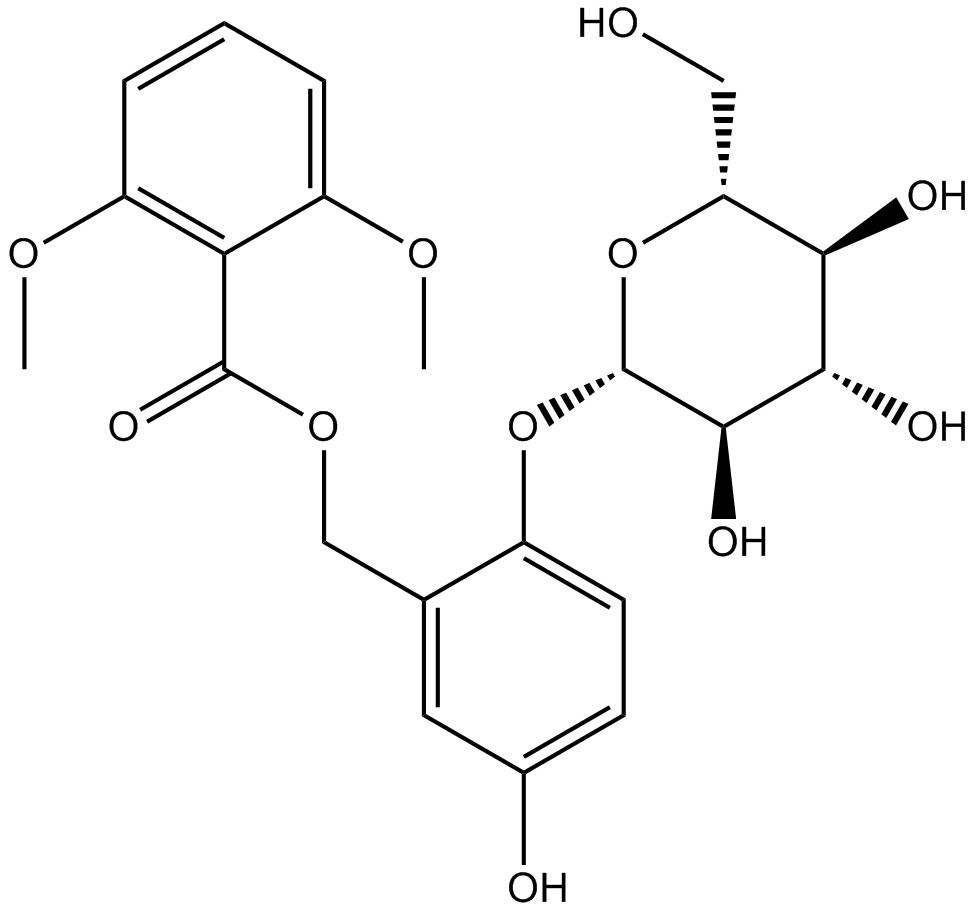
-
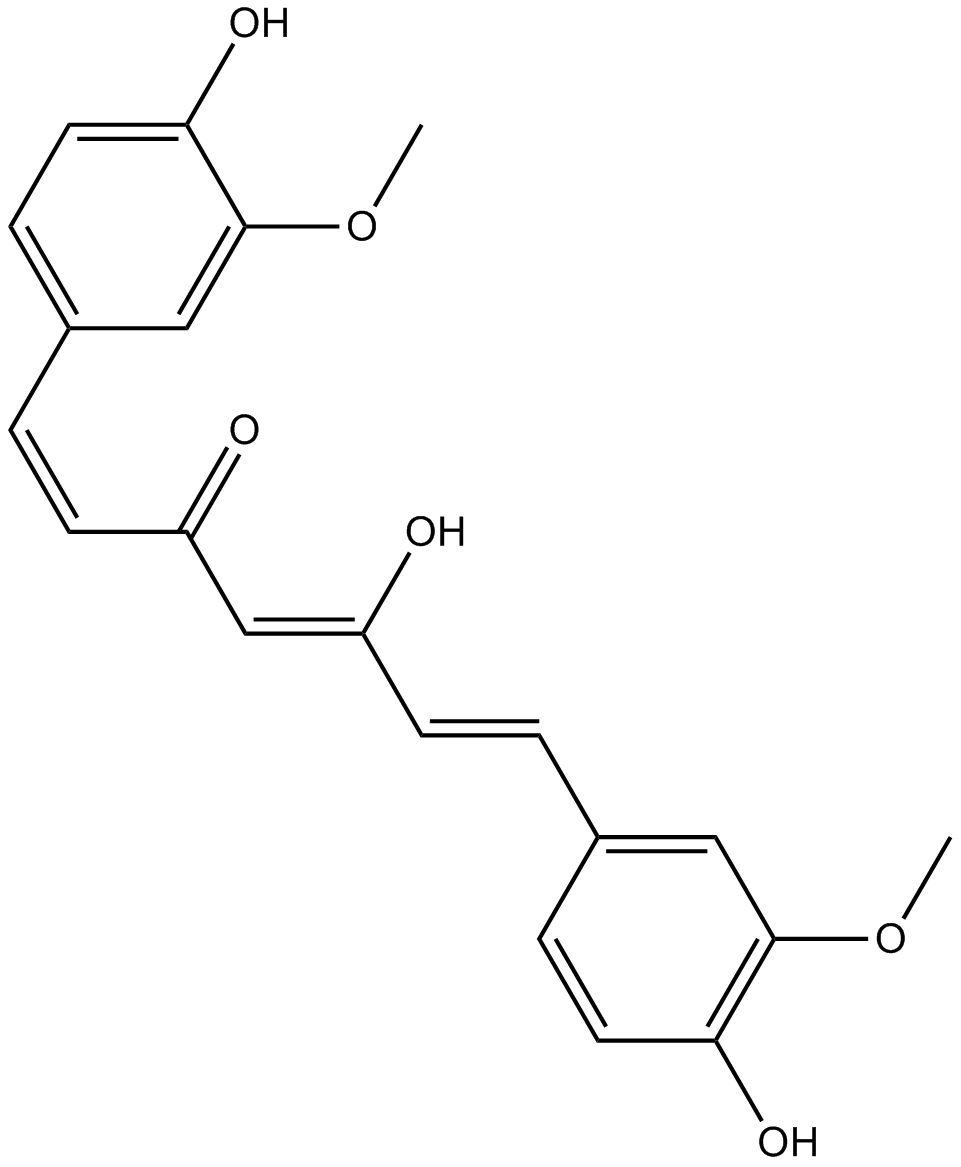
GC14787
Curcumin
A yellow pigment with diverse biological activities
-
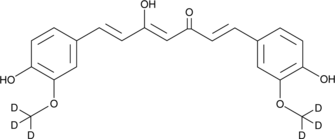
GC40226
Curcumin-d6
Curcumin-d6 is intended for use as an internal standard for the quantification of curcumin by GC- or LC-MS.
-
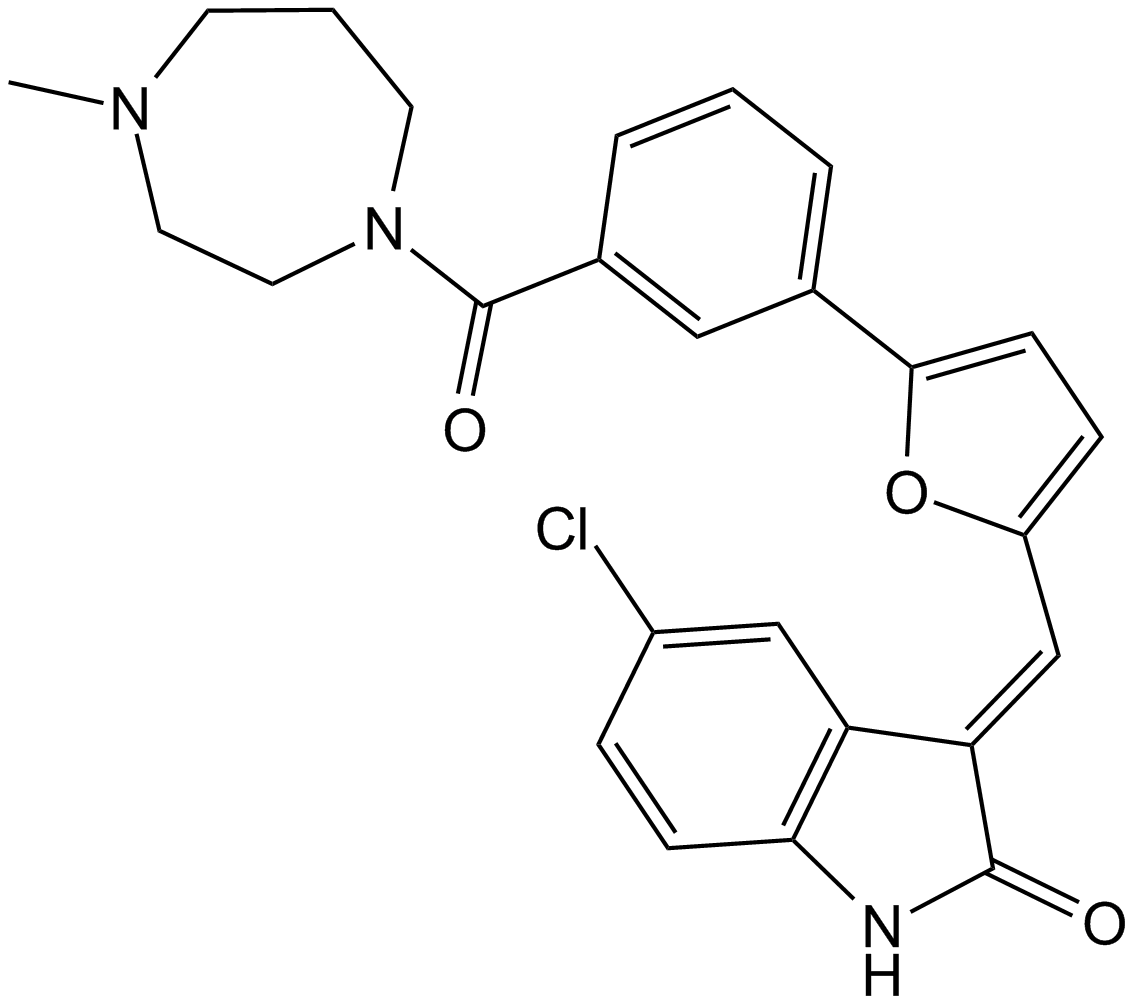
GC13056
CX-6258
A pan-Pim kinase inhibitor
-
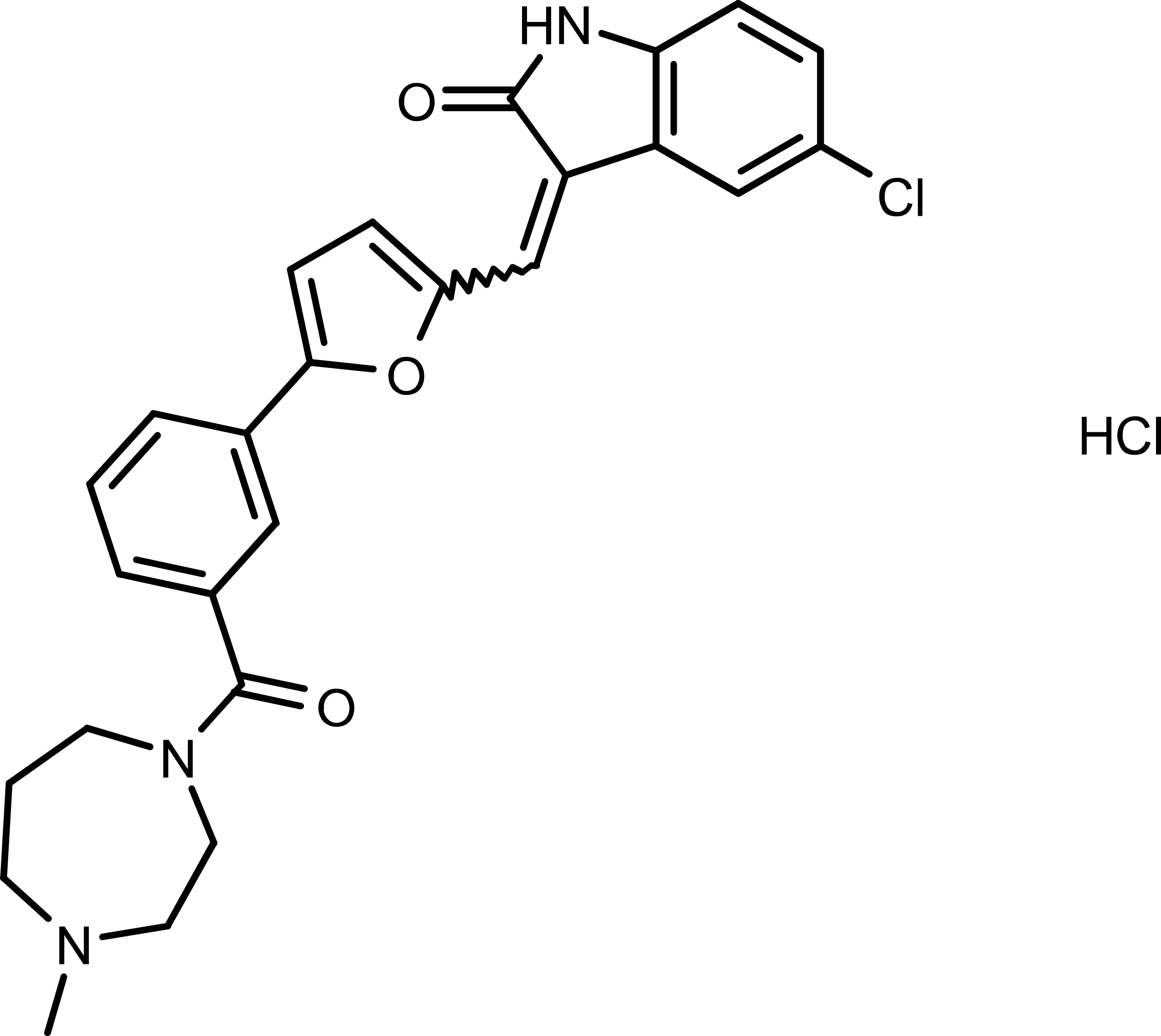
GC19747
CX-6258 HCl
CX-6258 HCl is a potent and kinase selective pan-Pim kinases inhibitor, with IC50s of 5 nM, 25 nM and 16 nM for Pim-1, Pim-2 and Pim-3, respectively.
-
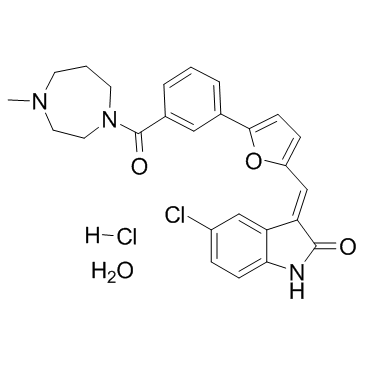
GC35762
CX-6258 hydrochloride hydrate
CX-6258 hydrochloride hydrate is a potent and kinase selective pan-Pim kinases inhibitor, with IC50s of 5 nM, 25 nM and 16 nM for Pim-1, Pim-2 and Pim-3, respectively.
-
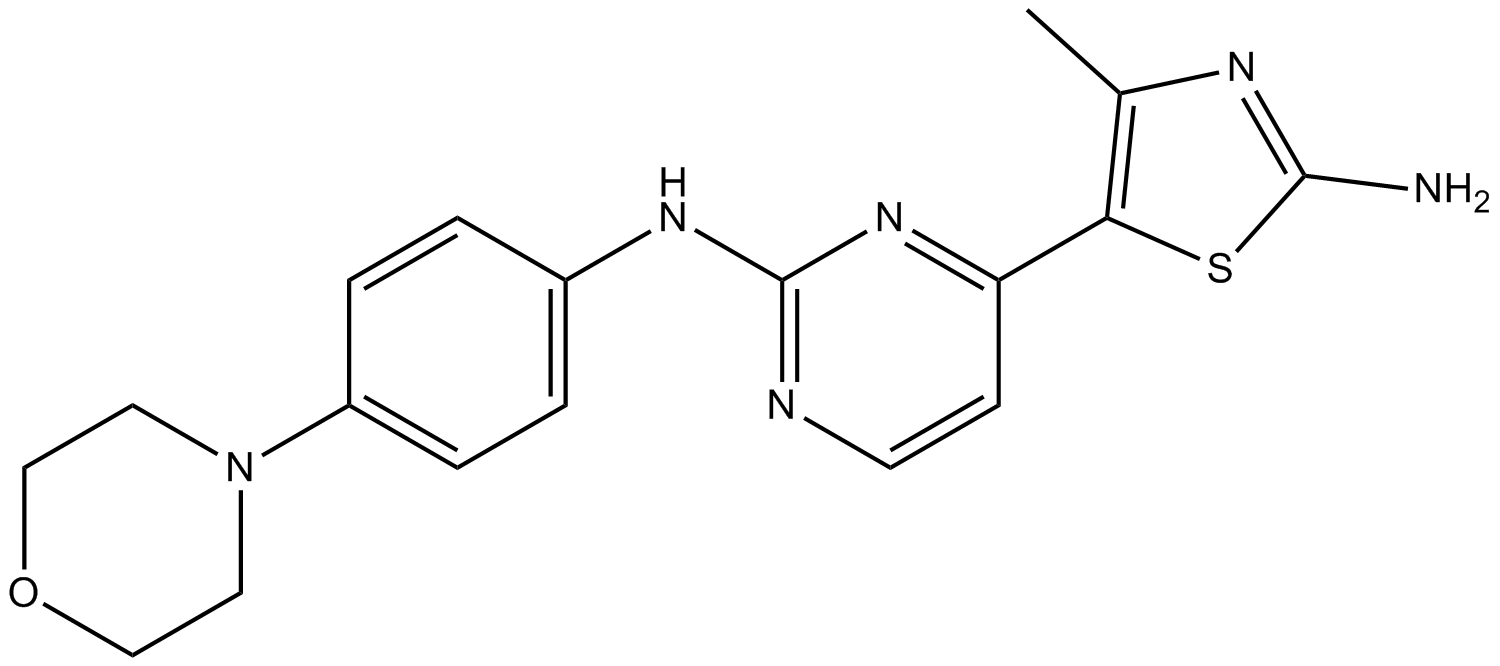
GC15106
CYC116
A potent Aurora kinase inhibitor
-
GC43354
Cysmethynil
Post-translational protein prenylation is a 3-step process that occurs at the C-terminus of a number of proteins involved in cell growth control and oncogenesis.

-
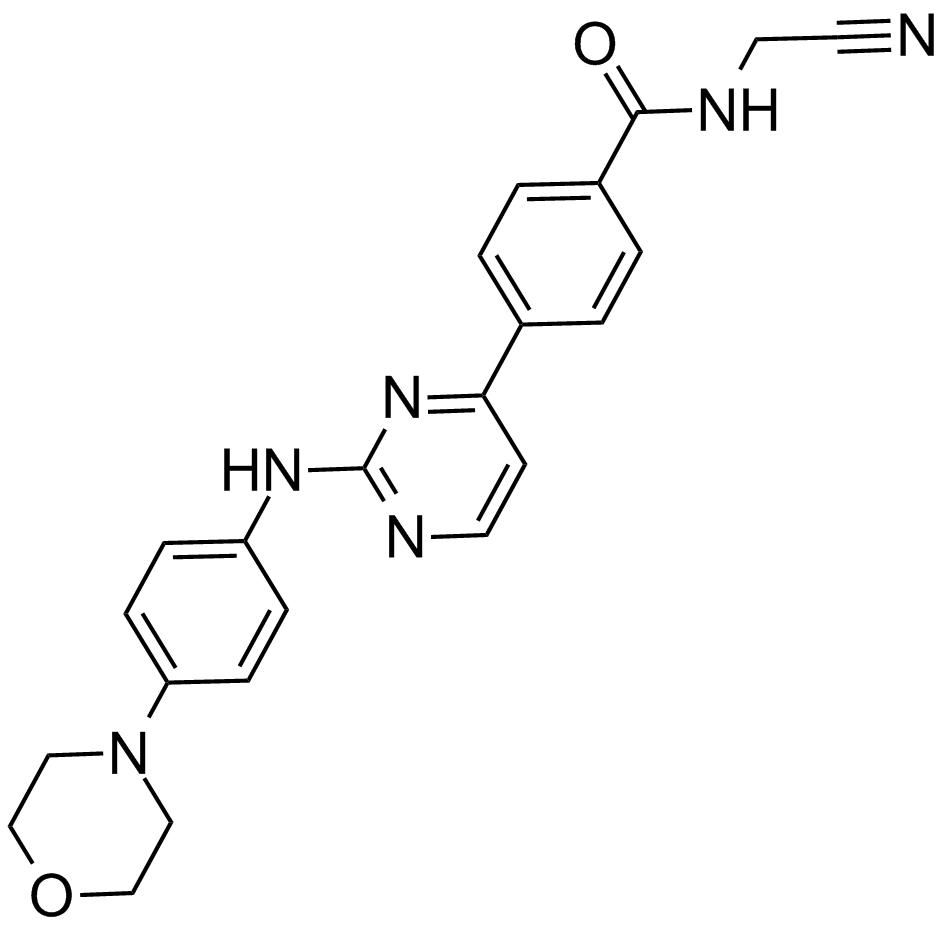
GC17050
CYT387
A potent inhibitor of JAK1 and JAK2
-
GC16468
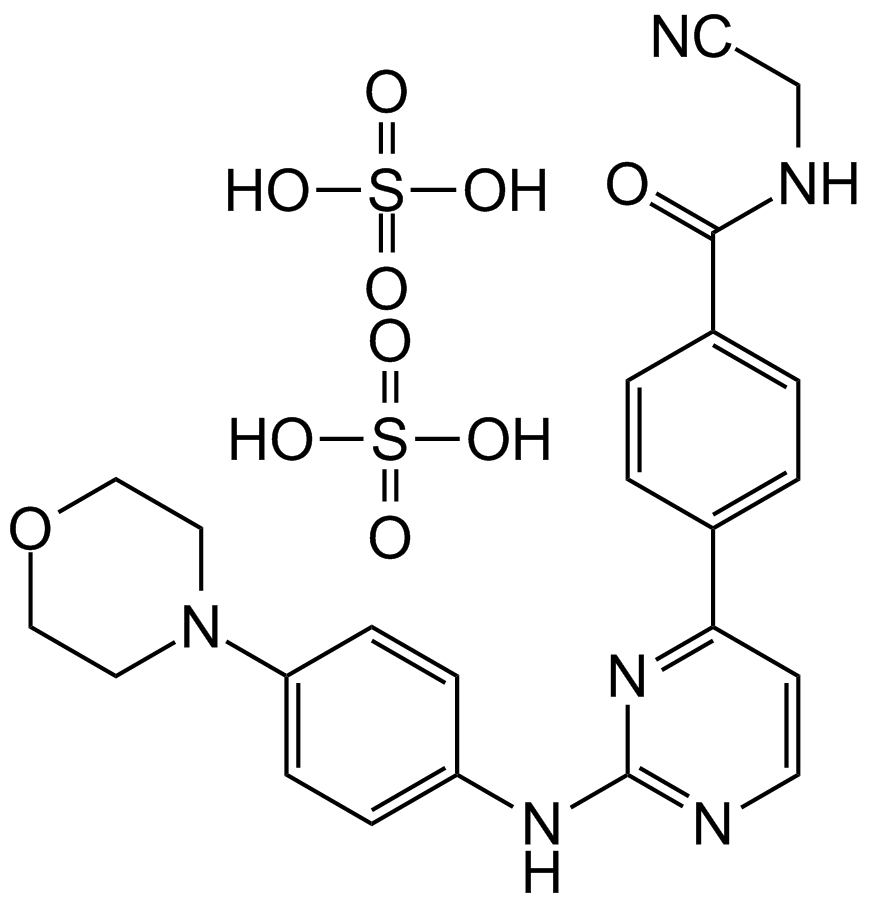
CYT387 sulfate salt
A potent inhibitor of JAK1 and JAK2
-
GC63780
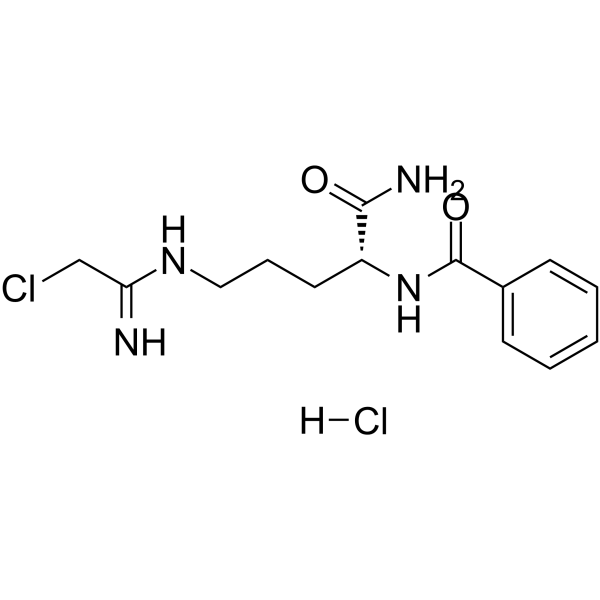
D-Cl-amidine hydrochloride
D-Cl-amidine hydrochloride is a potent and highly selective PAD1 inhibitor. D-Cl-amidine is well-torelated with no significant toxicity.
-
GC48967
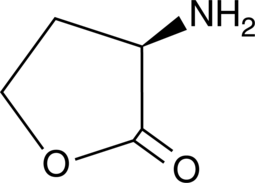
D-Homoserine lactone
An enantiomer of L-homoserine lactone
-
GC15830
Daminozide
Selective inhibitor of KDM2/7 histone demethylases
-
GC15217
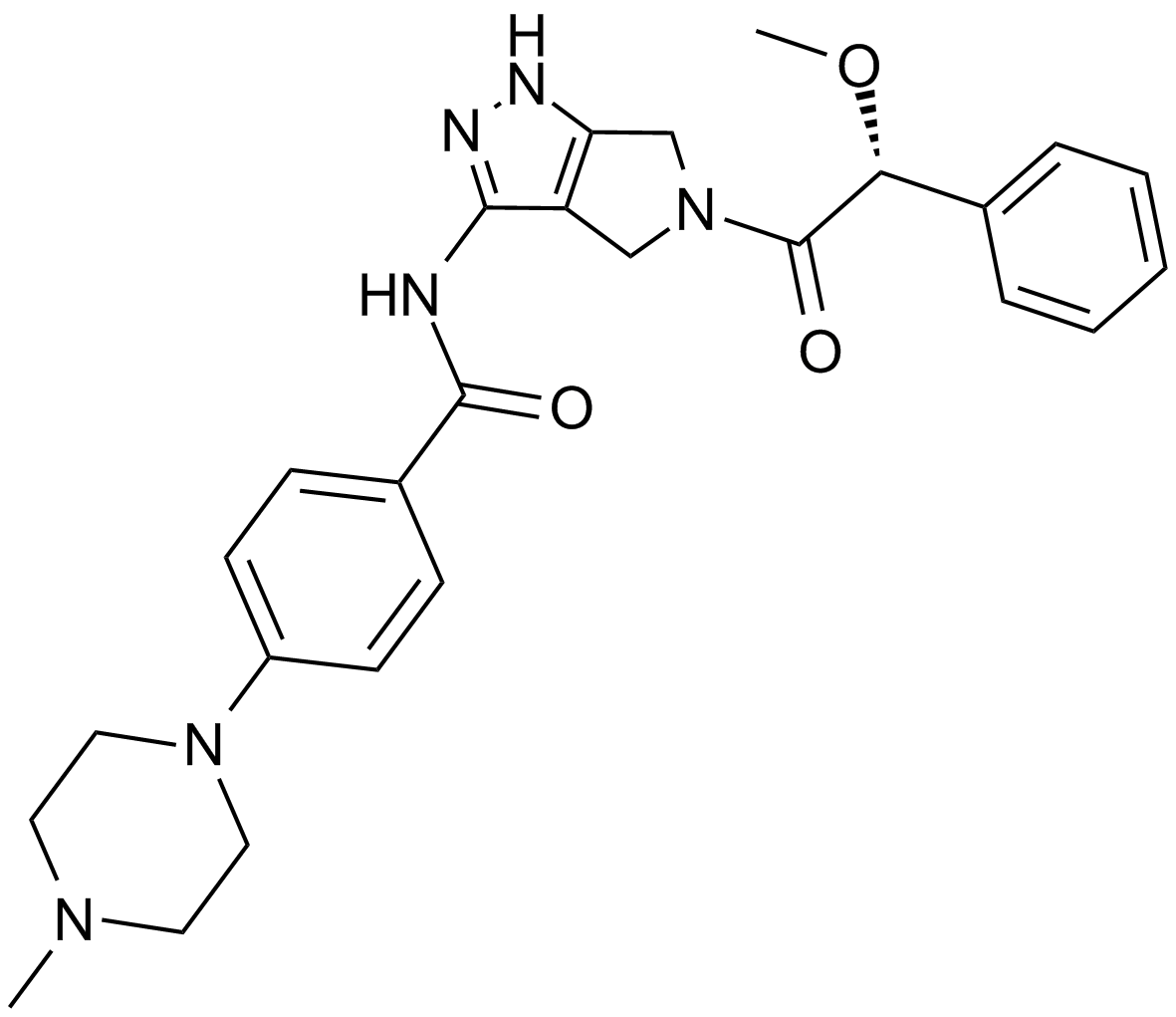
Danusertib (PHA-739358)
A pan-Aurora kinase and Abl inhibitor
-
GC16647
Daprodustat(GSK1278863)
Daprodustat(GSK1278863) (GSK1278863) is an orally active hypoxia-inducible factor prolyl hydroxylase (HIF-PH) inhibitor being developed for the treatment of anemia associated with chronic kidney disease.

-
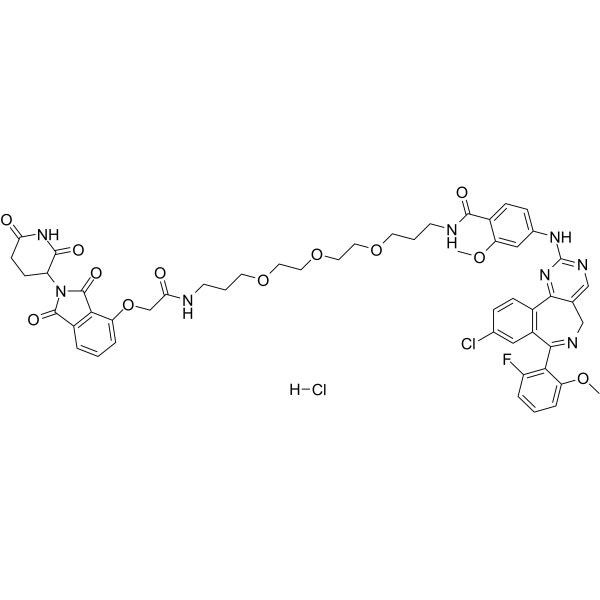
GC68147
dAURK-4 hydrochloride
-
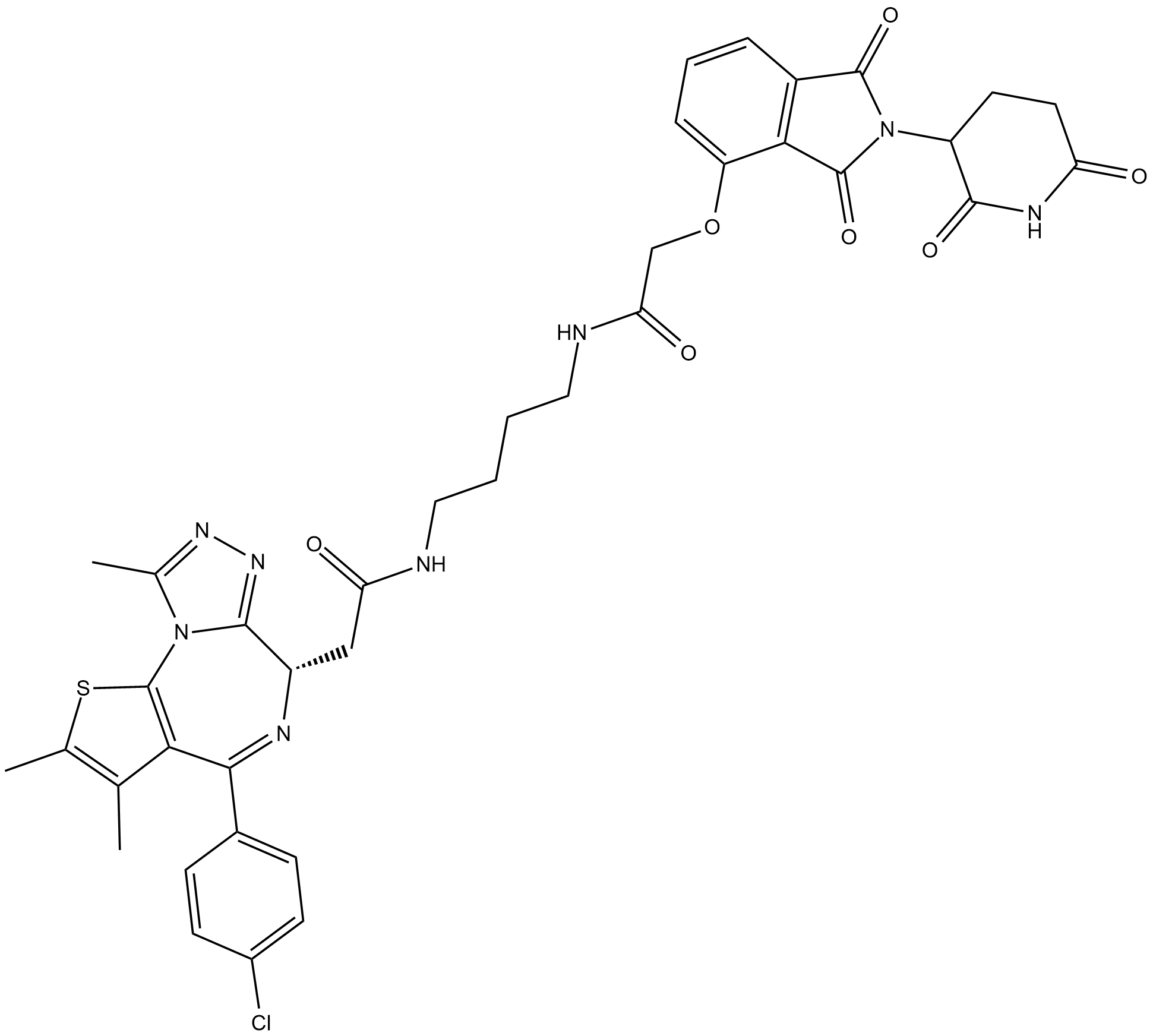
GC19119
dBET1
dBET1 is a potent BRD4 protein degrader based on PROTAC technology with an EC50 of 430 nM.
-
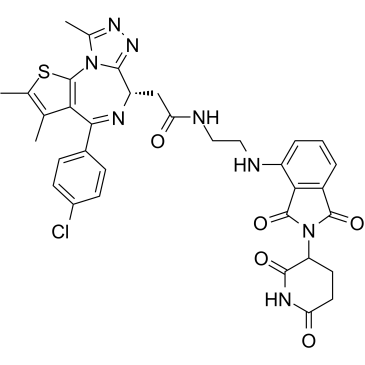
GC35815
dBET57
dBET57 is a potent and selective degrader of BRD4BD1 based on the PROTAC technology. dBET57 mediates recruitment to the CRL4Cereblon E3 ubiquitin ligase, with a DC50/5h of 500 nM for BRD4BD1, and is inactive on BRD4BD2.
-
GC32719
dBET6
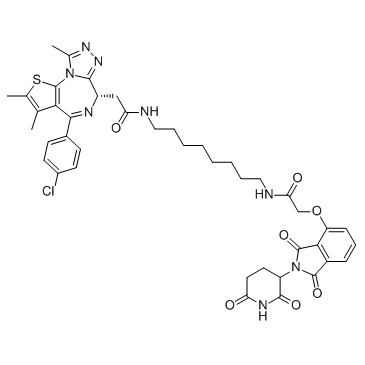
dBET6 is a highly potent, selective and cell-permeable PROTAC connected by ligands for Cereblon and BET, with an IC50 of 14 nM, and has antitumor activity.
-
GC33148
DC-05
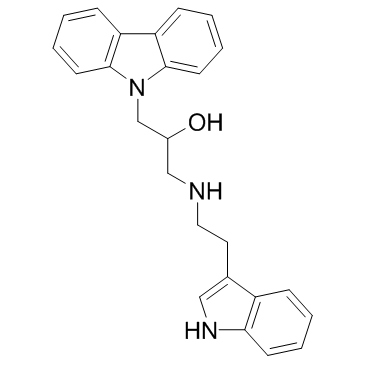
DC-05 is a DNA methyltransferase 1 (DNMT1) inhibitor, with an IC50 and a Kd of 10.3 μM and 1.09 μM, respectively.
-
GC65186
DC-S239
DC-S239 is a selective histone methyltransferase SET7 inhibitor with an IC50 value of 4.59 μM. DC-S239 also displays selectivity for DNMT1, DOT1L, EZH2, NSD1, SETD8 and G9a. DC-S239 has anticancer activity.
-
GC62211
dCBP-1
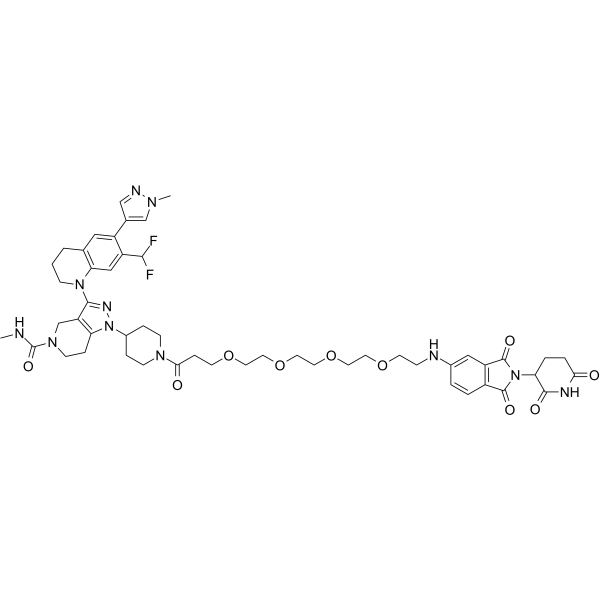
dCBP-1 is a potent and selective heterobifunctional degrader of p300/CBP based on Cereblon ligand. dCBP-1 is exceptionally potent at killing multiple myeloma cells and ablates oncogenic enhancer activity driving MYC expression.
-
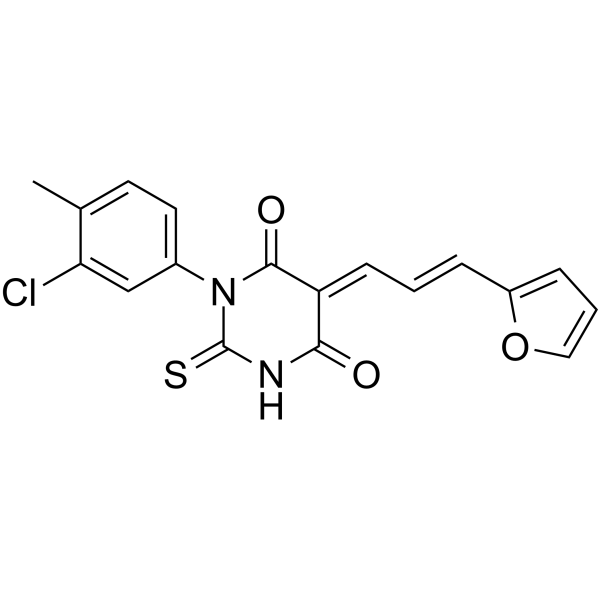
GC63627
DCH36_06
DCH36_06 is a potent and selective p300/CBP inhibitor with IC50s of 0.6 μM and 3.2 μM for p300 and CBP, respectively. DCH36_06 mediated p300/CBP inhibition leading to hypoacetylation on H3K18 in leukemic cells. Anti-tumor activity.
-
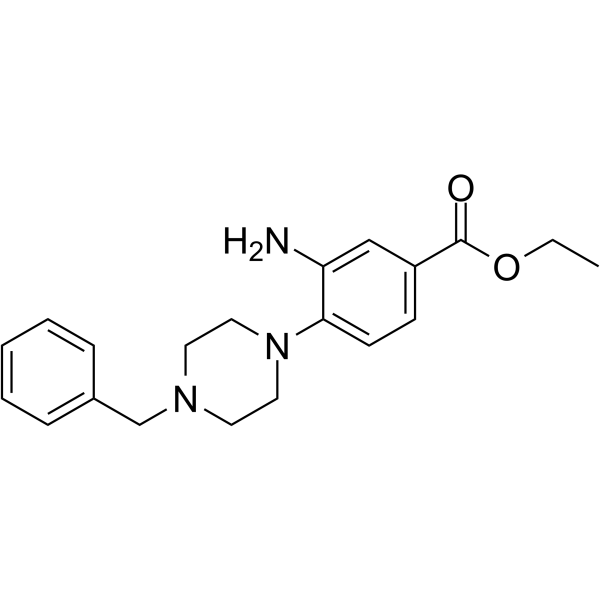
GC63662
DCLX069
DCLX069 is a selective protein arginine methyltransferase 1 (PRMT1) inhibitor with an IC50 value of 17.9 ?M. DCLX069 shows less active against PRMT4 and PRMT6. DCLX069 has anticancer effects.
-
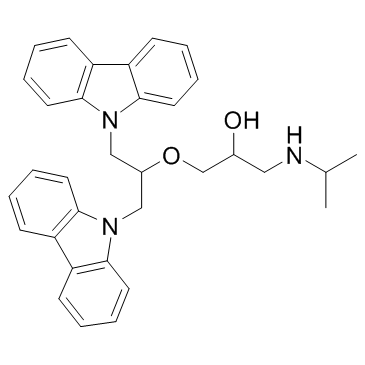
GC32872
DC_517
DC_517 is a DNA methyltransferase 1 (DNMT1) inhibitor, with an IC50 and a Kd of 1.7 μM and 0.91 μM, respectively.
-
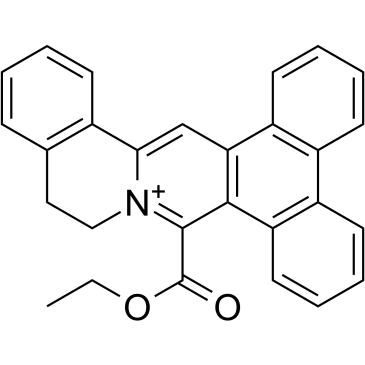
GC35816
DC_C66
DC_C66 is a cell-permeable, selective coactivator associated arginine methyltransferase 1 (CARM1) inhibitor with an IC50 of 1.8 μM. DC_C66 has a good selectivity for CARM1 against PRMT1 (IC50=21 μM), PRMT6 (IC50= 47μM), and PRMT5.
-
GC67863
DDO-2093 dihydrochloride

-
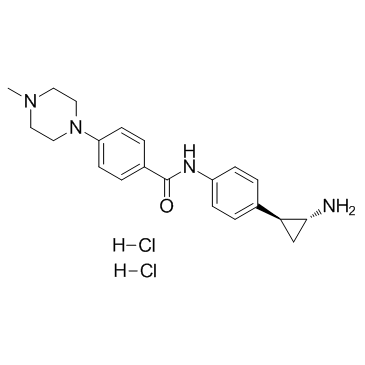
GC33013
DDP-38003 dihydrochloride
DDP-38003 dihydrochloride is an novel, orally available inhibitor of histone lysine-specific demethylase 1A (KDM1A/LSD1) with an IC50 of 84 nM.
-
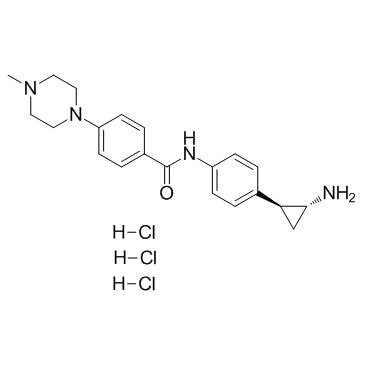
GC34296
DDP-38003 trihydrochloride
DDP-38003 trihydrochloride is an novel, orally available inhibitor of histone lysine-specific demethylase 1A (KDM1A/LSD1) with an IC50 of 84 nM.
-
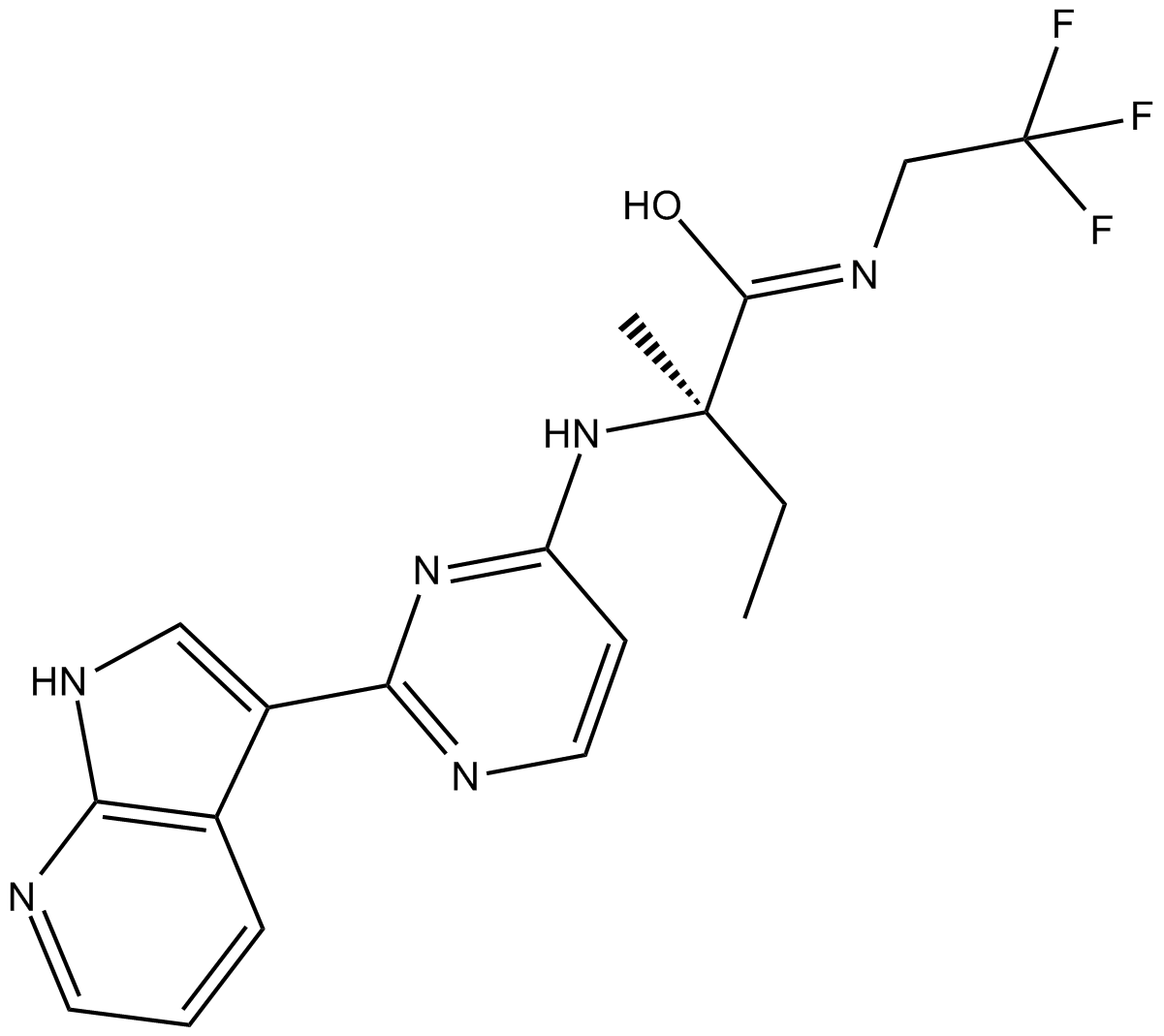
GC10770
Decernotinib(VX-509)
Decernotinib(VX-509) is a potent, orally active JAK3 inhibitor, with Kis of 2.5, 11, 13 and 11 nM for JAK3, JAK1, JAK2, and TYK2, respectively.
-
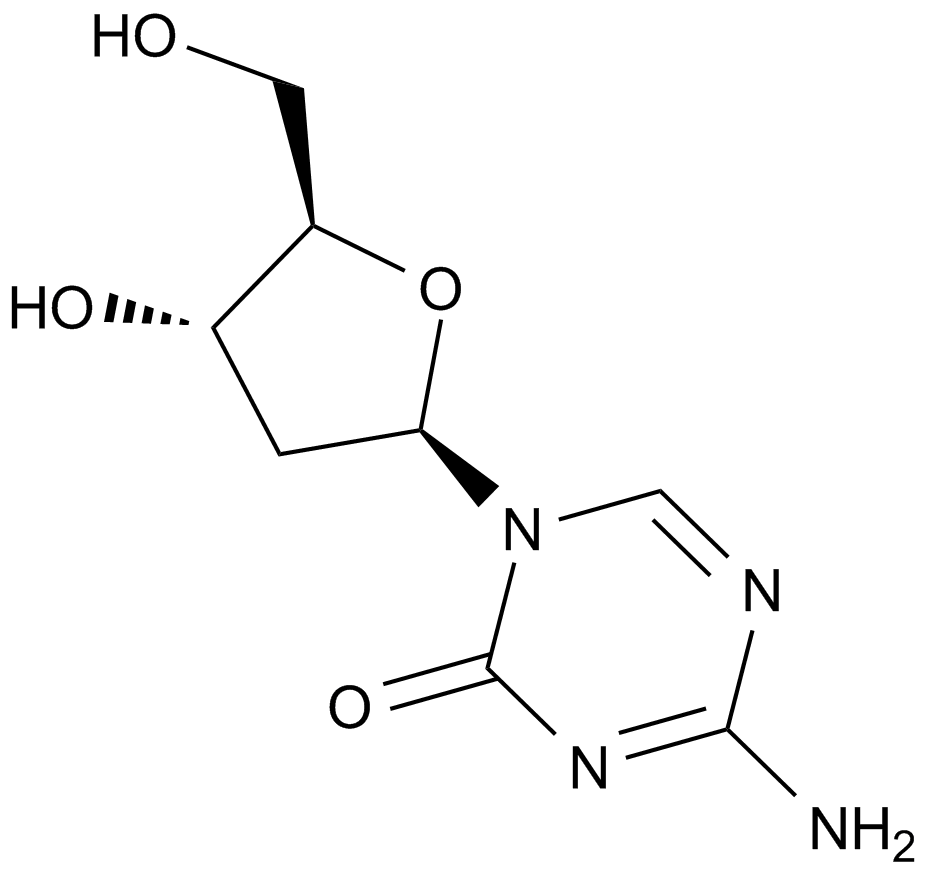
GC15255
Decitabine(NSC127716, 5AZA-CdR)
Decitabine(DAC) is a deoxycytidine analogue antimetabolite with oral bioactivity and functions as an inhibitor of DNA methyltransferase.
-
GC68955
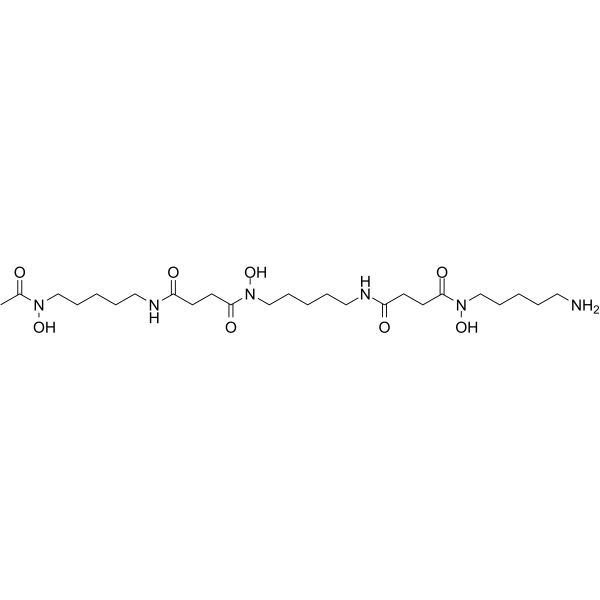
Deferoxamine
Deferoxamine (Deferoxamine B) is an iron chelator (binding Fe(III) and many other metal cations), widely used to reduce the accumulation and deposition of iron in tissues.
-
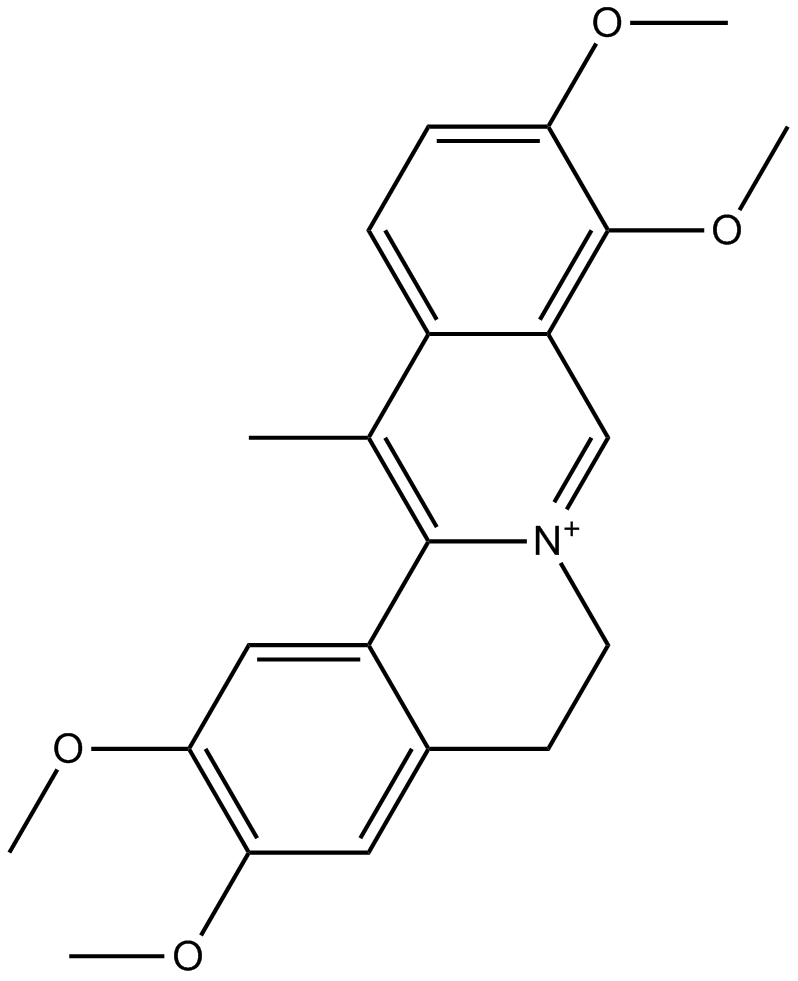
GN10040
Dehydrocorydaline
-
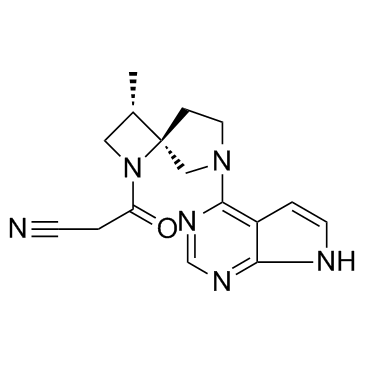
GC31660
Delgocitinib (JTE-052)
Delgocitinib (JTE-052) (JTE-052) is a specific JAK inhibitor with IC50s of 2.8, 2.6, 13 and 58 nM for JAK1, JAK2, JAK3 and Tyk2, respectively.
-
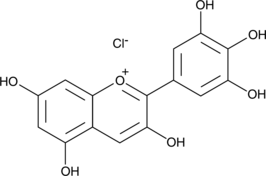
GC43406
Delphinidin (chloride)
Delphinidin (chloride) is an anthocyanidin, a natural plant pigment which serves as the precursor of certain anthocyanins that provide the blue-red colors of flowers, fruits, and red wine.
-
GC31230
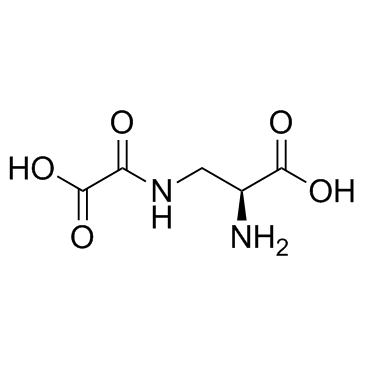
Dencichin (Dencichine)
Dencichin is a non-protein amino acid originally extracted from Panax notoginseng, and can inhibit HIF-prolyl hydroxylase-2 (PHD-2) activity.
-
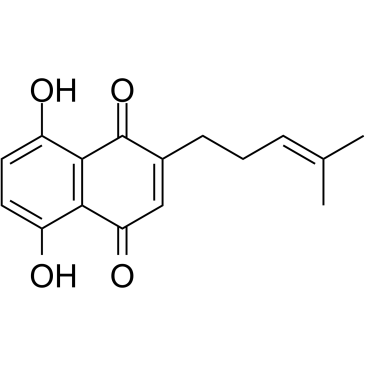
GC38767
Deoxyshikonin
A natural products with anti-tumor activity
-
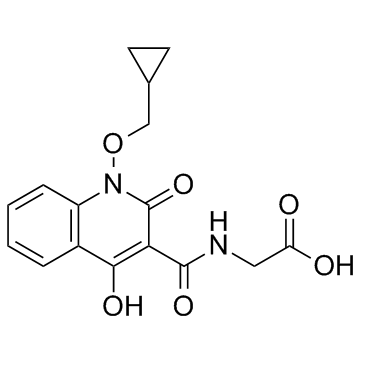
GC32449
Desidustat
An inhibitor of HIF-PH
-
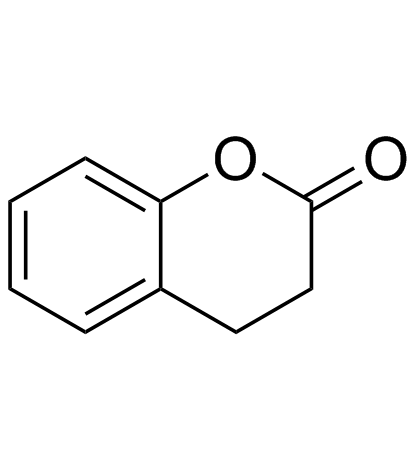
GC34187
Dihydrocoumarin (Hydrocoumarin)
Dihydrocoumarin (Hydrocoumarin) is a compound found in Melilotus officinalis. Dihydrocoumarin (Hydrocoumarin) is a yeast Sir2p inhibitor. Dihydrocoumarin (Hydrocoumarin) also inhibits human SIRT1 and SIRT2 with IC50s of 208 μM and 295 μM, respectively.
-
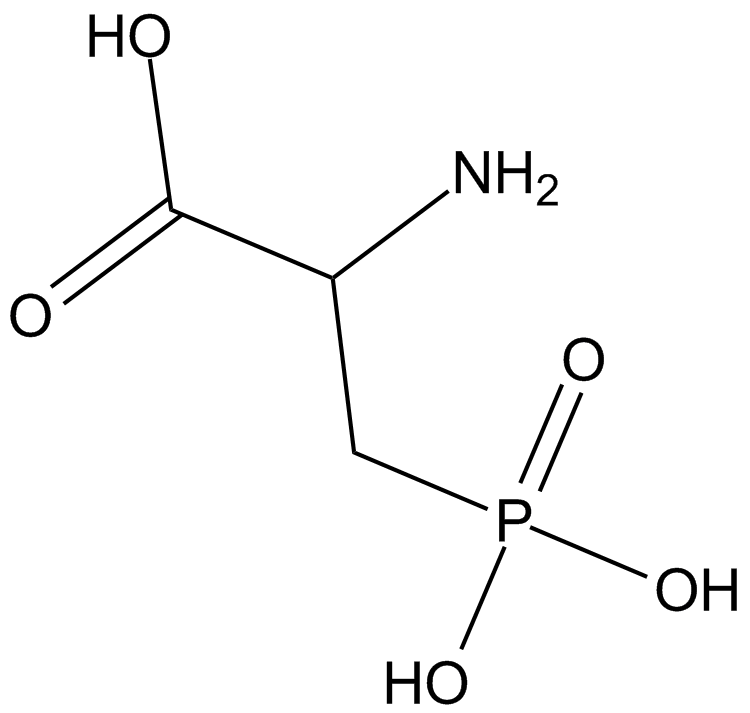
GC14024
DL-AP3
A metabotropic glutamate receptor antagonist
-
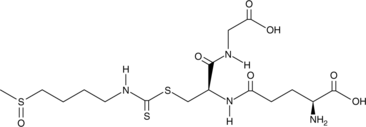
GC52368
DL-Sulforaphane Glutathione
A metabolite of sulforaphane
-
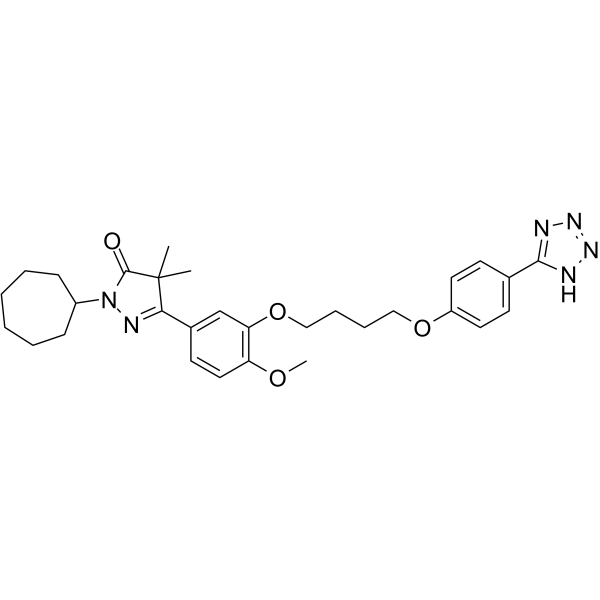
GC65546
DNMT3A-IN-1
DNMT3A-IN-1 is a potent and selective DNMT3A inhibitor.
-
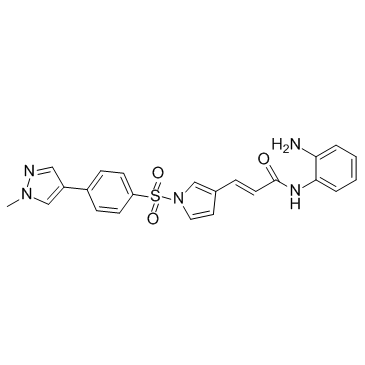
GC35893
Domatinostat
Domatinostat (4SC-202 free base) is a selective class I HDAC inhibitor with IC50 of 1.20 μM, 1.12 μM, and 0.57 μM for HDAC1, HDAC2, and HDAC3, respectively. It also displays inhibitory activity against Lysine specific demethylase 1 (LSD1).
-
GC14490
Donepezil HCl
Donepezil HCl (E2020) is a reversible, selective AChE inhibitor with an IC50 of 6.7 nM for AChE activity.
-
GC33208
Dot1L-IN-1
Dot1L-IN-1 is a highly potent, selective and structurally novel Dot1L inhibitor with a Ki of 2 pM.