Histone Methyltransferase
Histone methyltransferases are a group of enzymes that catalyze the methylation of histone lysine and arginine by adding methyl groups to specific histone arginine or lysine residues. Histone methyltransferaes can be classified into 3 classes, including SET domain lysine methyltransferases, non-SET domain lysine methyltransferases and arginine methyltransferases (PRMTs), all of which use S-adenosylmethionine as a cosubstrate for the transfer of the methyl group. Aberrant histone methylation has been associated with a wide range of human cancers (such as hematological malignancies), which leads to the development of novel cancer chemotherapies targeting cancer-associated histone methyltransferases (more than 20 lysine methyltransferases and 9 arginine methyltransferases in humans).
Products for Histone Methyltransferase
- Cat.No. Product Name Information
-
GC45908
(R)-BAY-598
An inhibitor of SMYD2
-
GC11340
(R)-PFI 2 hydrochloride
PFI-2 ((R)-(R)-PFI 2 hydrochloride) hydrochloride is a potent and selective SET domain containing lysine methyltransferase 7 (SETD7) inhibitor.
-
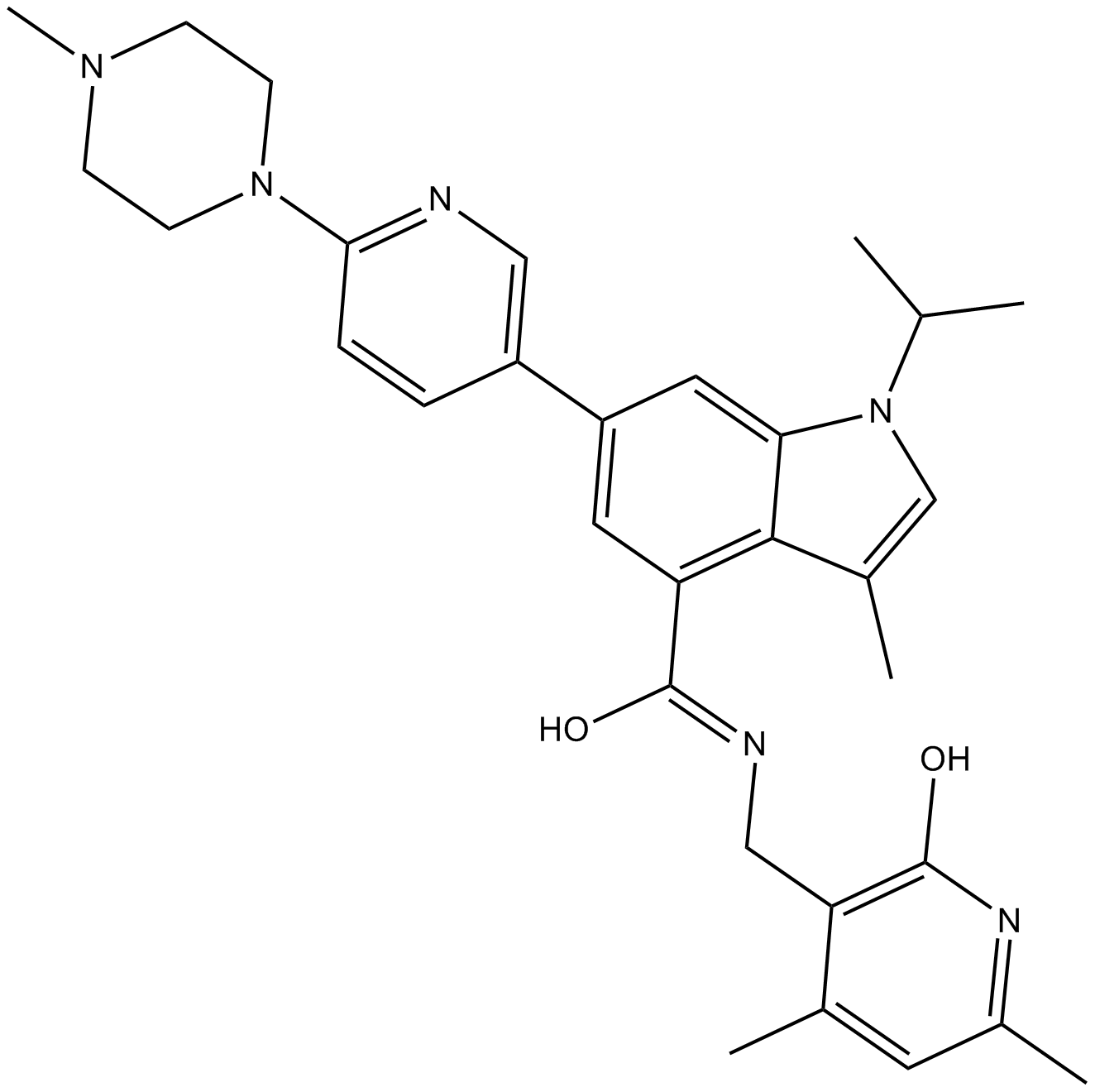
GC65045
(S)-MRTX-1719
(S)-MRTX-1719 (example 16-7) is the S-enantiomer of MRTX-1719. (S)-MRTX-1719 is a PRMT5/MTA complex inhibitor, with an IC50 of 7070 nM.
-
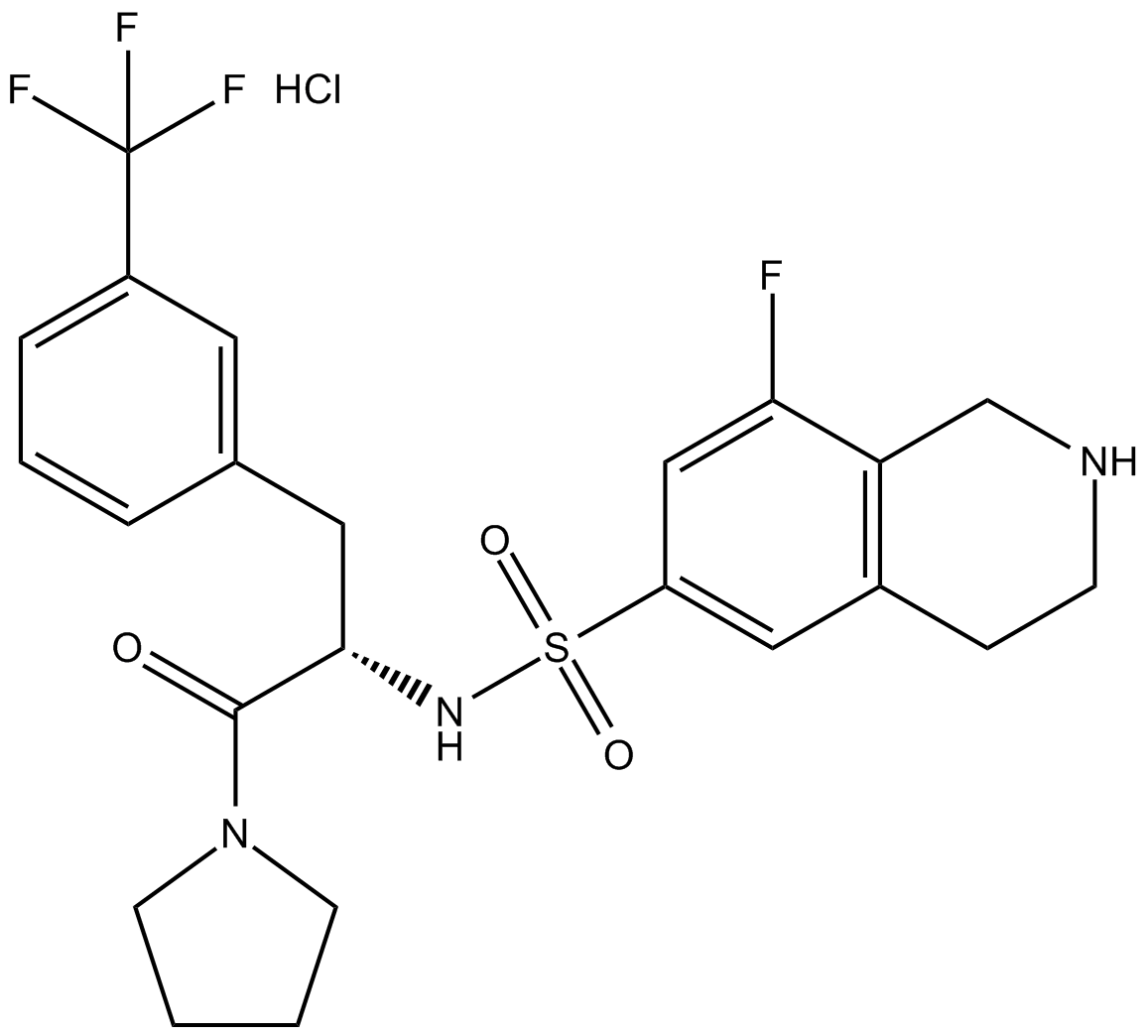
GC13634
(S)-PFI-2 (hydrochloride)
Negative control of (R)-PFI 2 hydrochloride
-
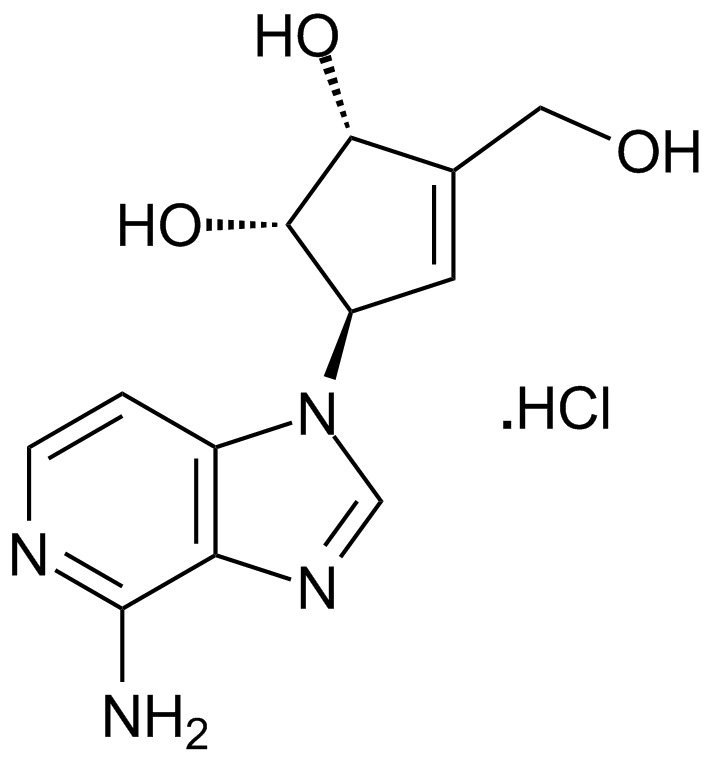
GC17907
3-Deazaneplanocin A (DZNep) hydrochloride
An inhibitor of the lysine methyltransferase EZH2
-
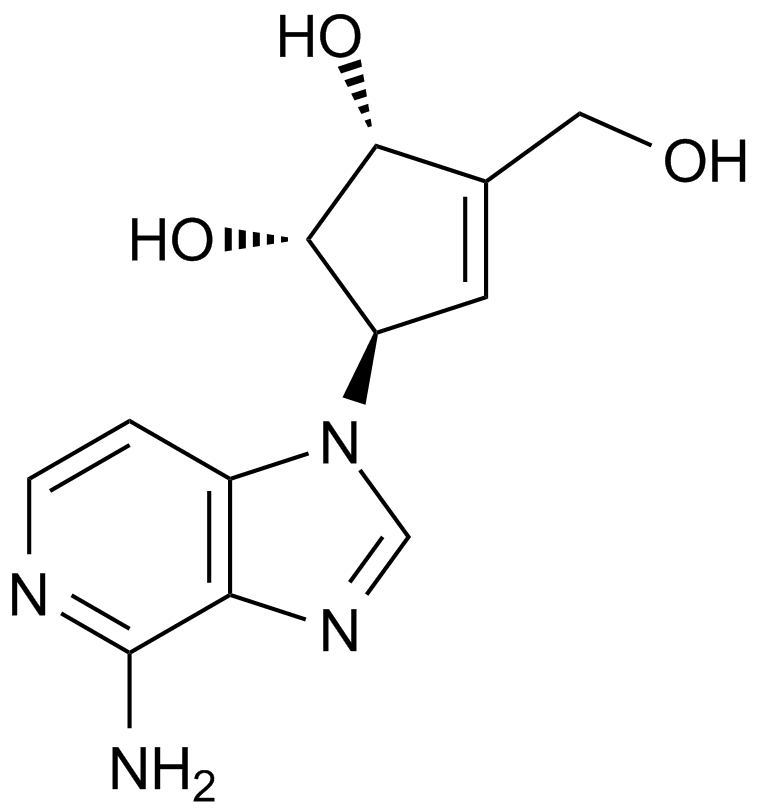
GC13145
3-Deazaneplanocin,DZNep
An inhibitor of lysine methyltransferase EZH2
-
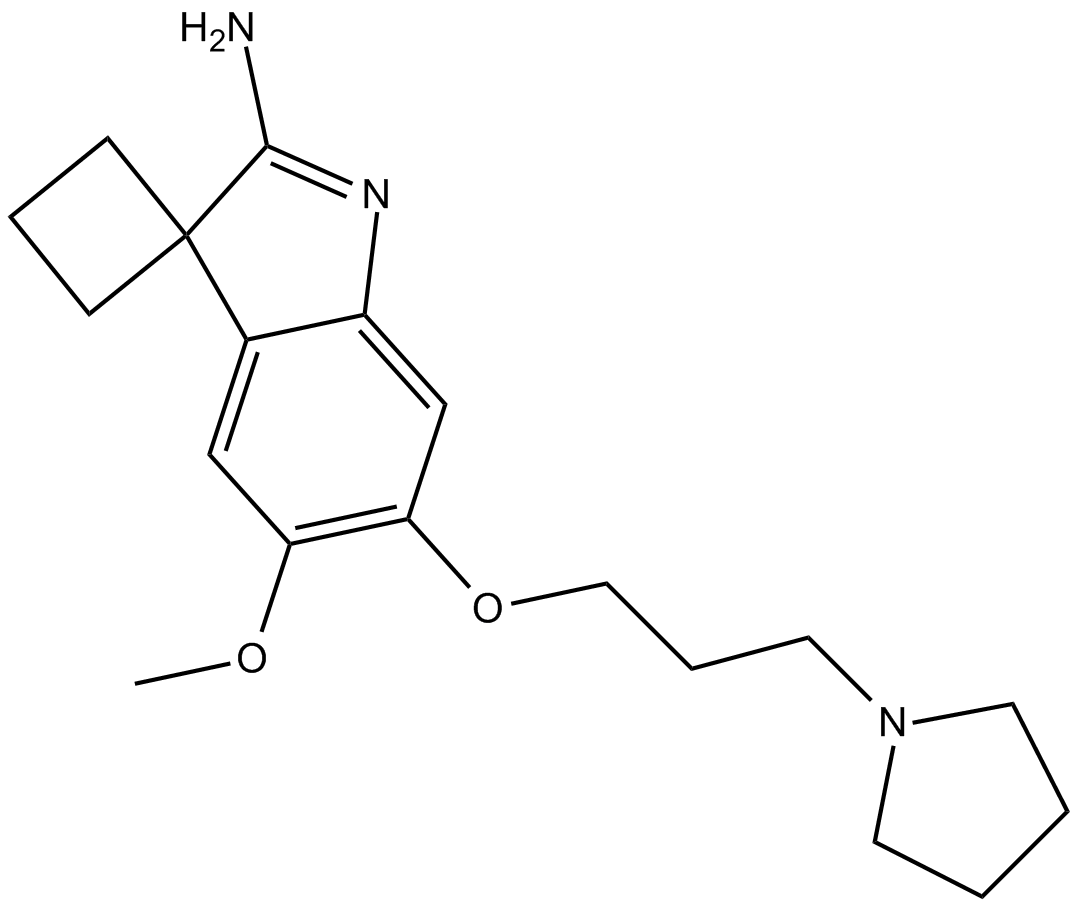
GC16015
A 366
G9a/GLP histone lysine methyltransferase inhibitor
-
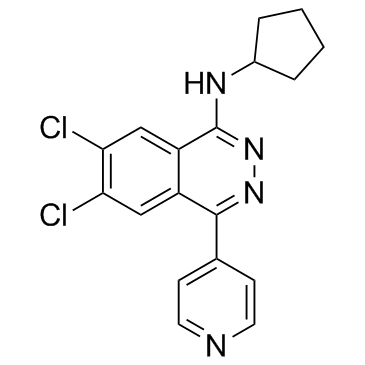
GC32861
A-196
A selective inhibitor of SUV420H1 and SUV420H2
-
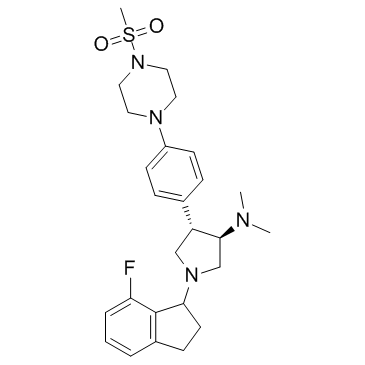
GC33187
A-395 (A395)
A-395 (A395) is an antagonist of polycomb repressive complex 2 (PRC2) protein-protein interactions that potently inhibits the trimeric PRC2 complex (EZH2-EED-SUZ12) with an IC50 of 18 nM.
-
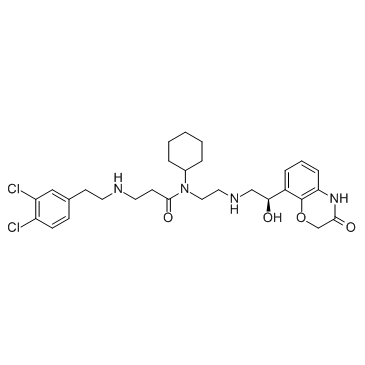
GC30503
A-893
A-893 is a cell-active inhibitor of Methyltransferase SMYD2, with an IC50 of 2.8 nM.
-
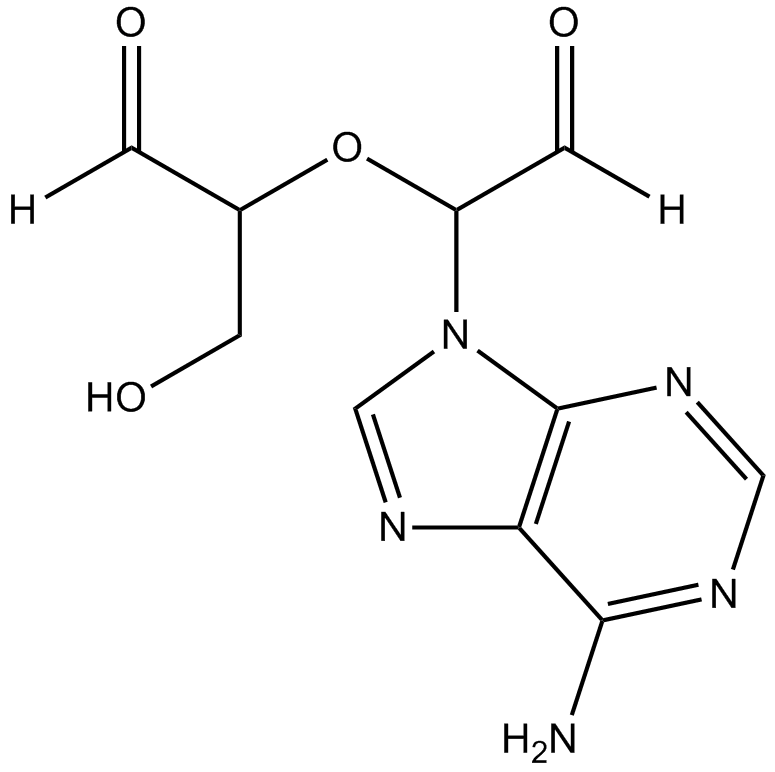
GC16509
Adox
Adox, a purine nucleoside analogue, is a potent inhibitor of S-Adenosylhomocysteine hydrolase (SAHH) (Ki=3.3 nM). Adenosine Dialdehyde exhibits potent anti-tumor activity in vivo and can be used for the cancer research.
-
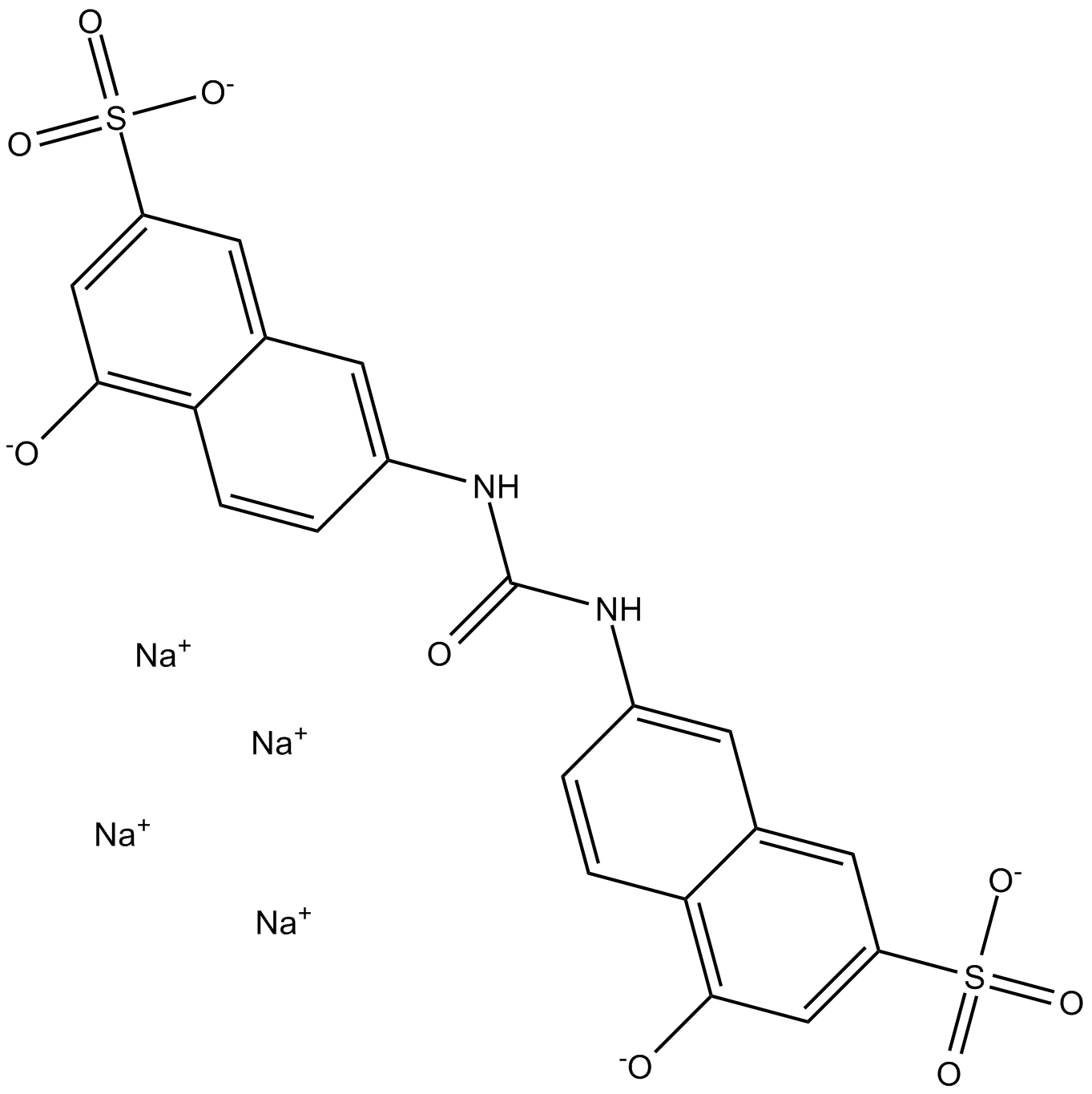
GC17275
AMI-1
A cell permeable inhibitor of PRMTs
-
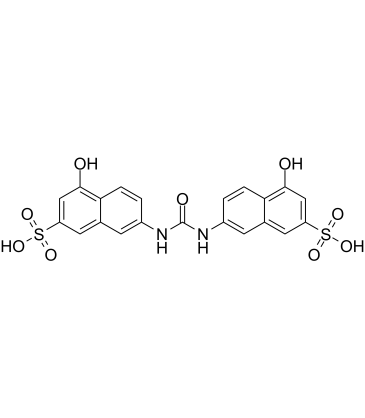
GC39840
AMI-1 free acid
AMI-1 free acid is a potent, cell-permeable and reversible inhibitor of protein arginine N-methyltransferases (PRMTs), with IC50s of 8.8 μM and 3.0 μM for human PRMT1 and yeast-Hmt1p, respectively. AMI-1 free acid exerts PRMTs inhibitory effects by blocking peptide-substrate binding.
-
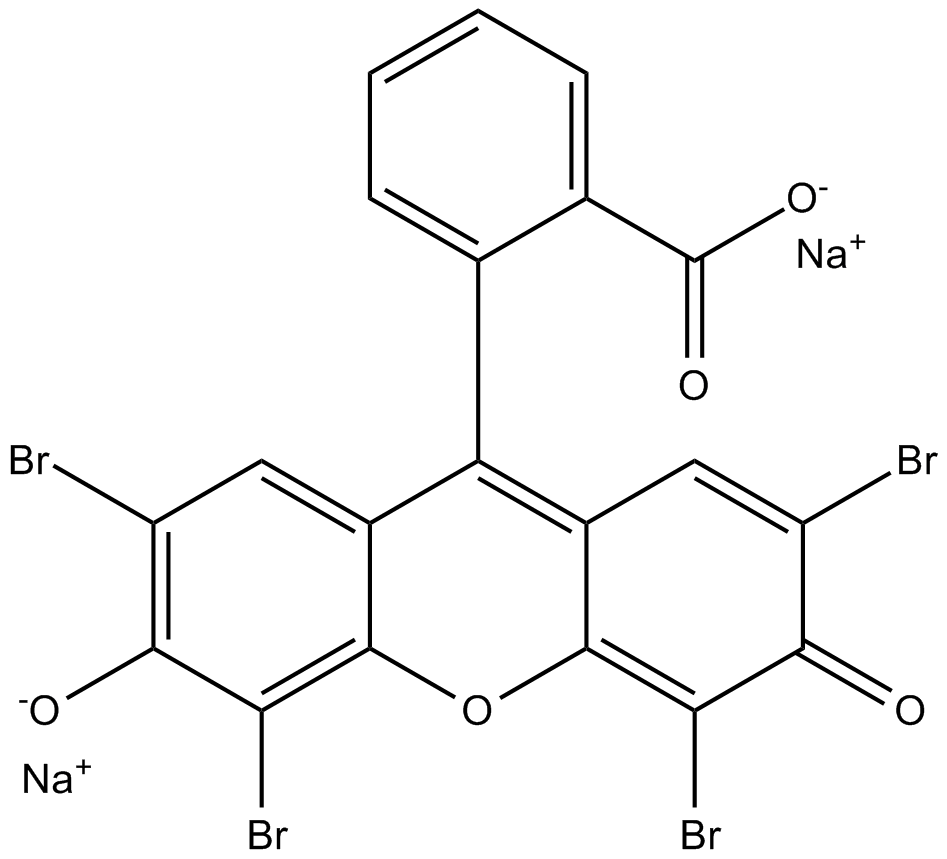
GC17546
AMI5
Eosin Y (disodium) is a soluble acid red dye molecule.
-
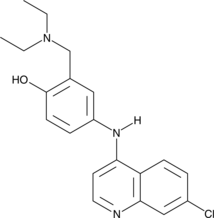
GC42790
Amodiaquine
Amodiaquine is an aminoquinoline antimalarial compound.
-
GC60579
Amodiaquine dihydrochloride
Amodiaquine dihydrochloride (Amodiaquin dihydrochloride), a 4-aminoquinoline class of antimalarial agent, is a potent and orally active histamine N-methyltransferase inhibitor with a Ki of 18.6 nM.
-
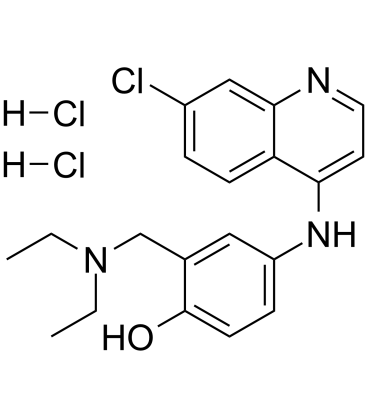
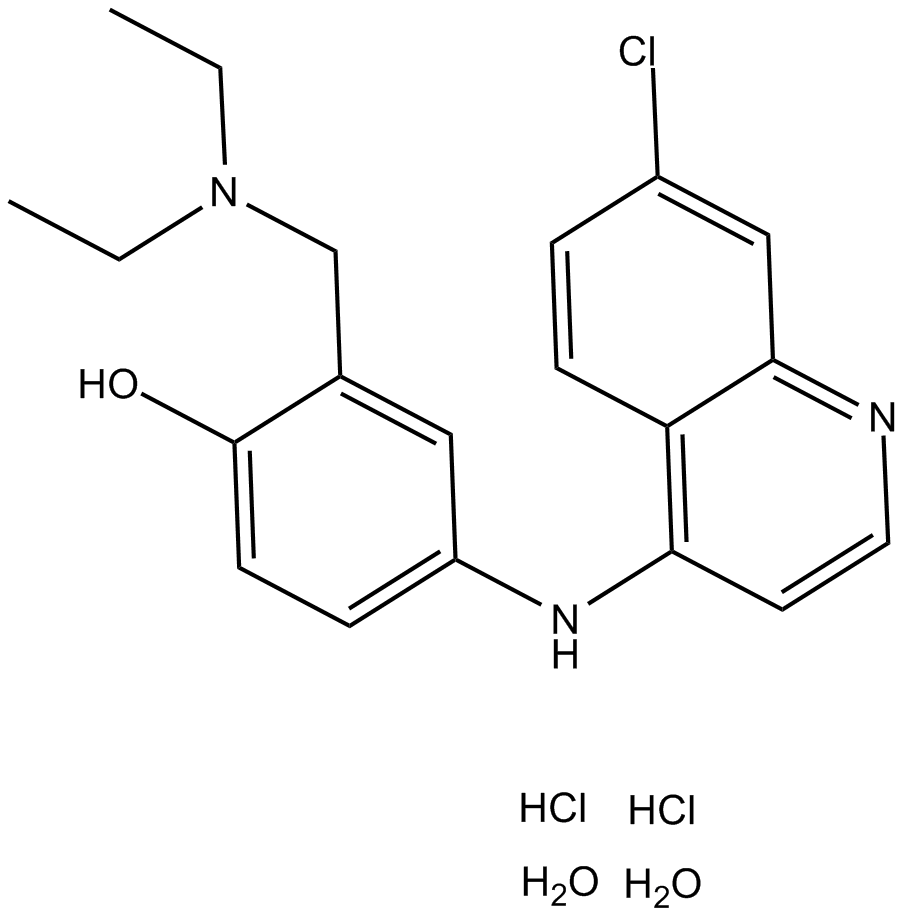
GC10905
Amodiaquine dihydrochloride dihydrate
histamine N-methyl transferase inhibitor
-
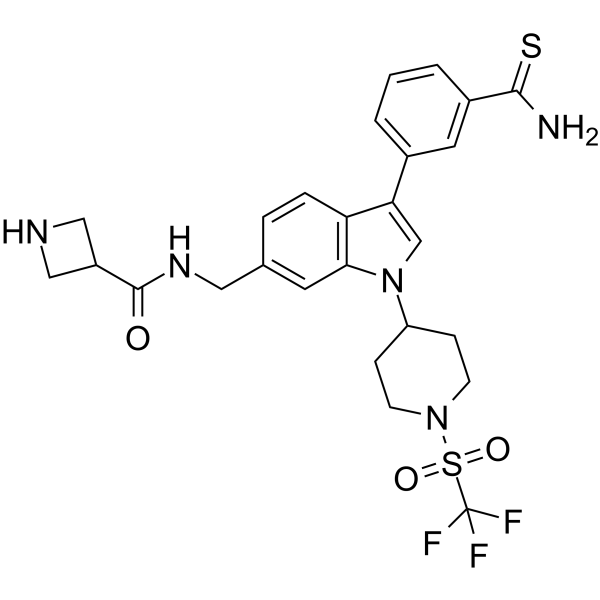
GC65918
AS-85
AS-85 is a potent ASH1L histone methyltransferase inhibitor (IC50=0.6 μM) with anti-leukemic activity. AS-85 strongly binds to the ASH1L SET domain, with the Kd value of 0.78μM.
-
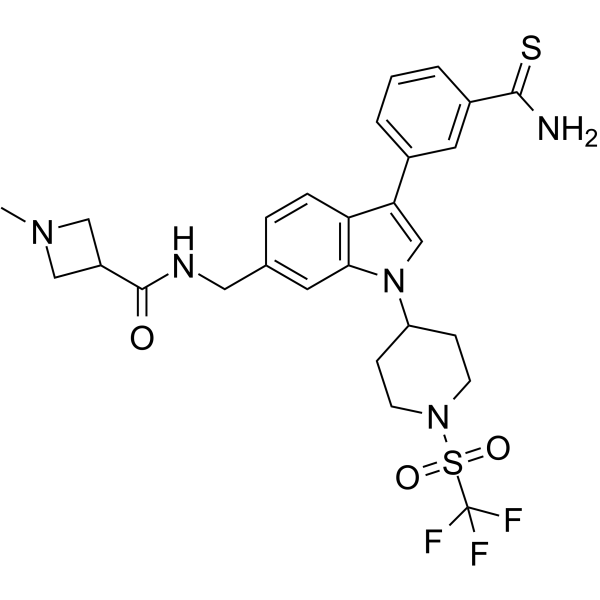
GC62615
AS-99
AS-99 is a first-in-class, potent and selective ASH1L histone methyltransferase inhibitor (IC50=0.79μM, Kd=0.89μM) with anti-leukemic activity. AS-99 blocks cell proliferation, induces apoptosis and differentiation, downregulates MLL fusion target genes, and reduces the leukemia burden in vivo.
-
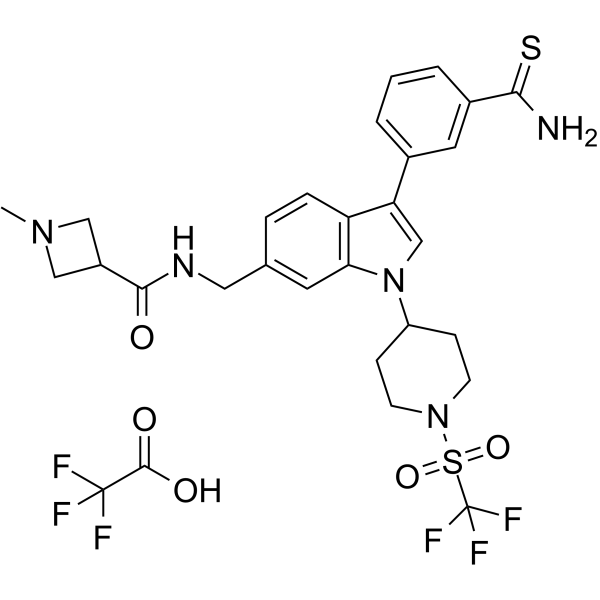
GC62849
AS-99 TFA
AS-99 TFA is a first-in-class, potent and selective ASH1L histone methyltransferase inhibitor (IC50=?0.79??M, Kd=?0.89??M) with anti-leukemic activity. AS-99 TFA blocks cell proliferation, induces apoptosis and differentiation, downregulates MLL fusion target genes, and reduces the leukemia burden in vivo.
-
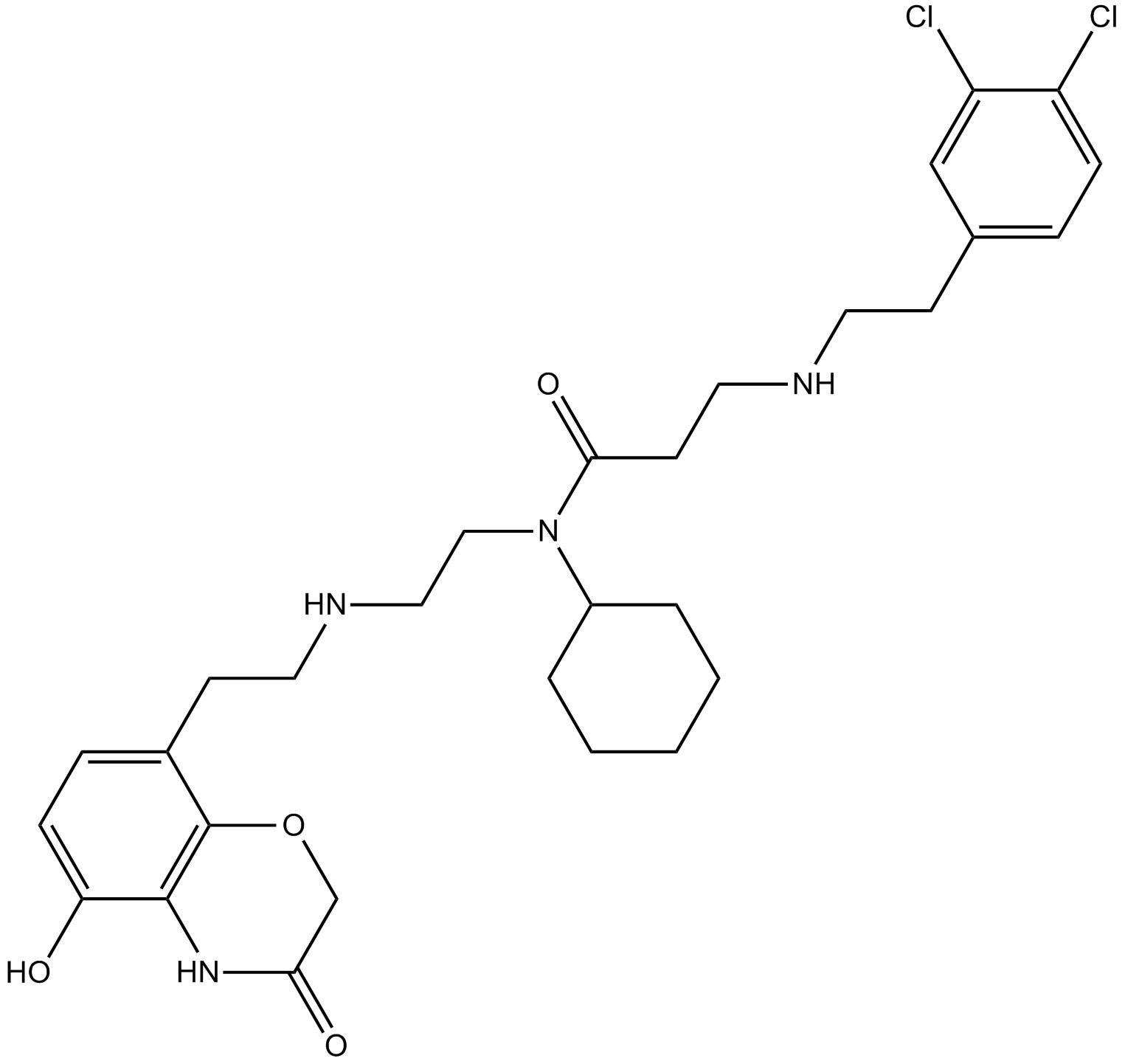
GC13744
AZ505
SMYD2 inhibitor,potent and selective
-
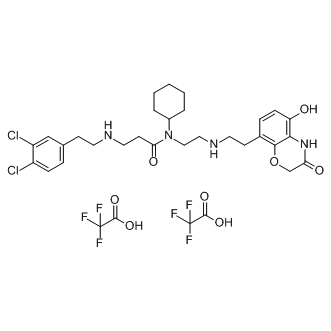
GC13103
AZ505 ditrifluoroacetate
SMYD2 inhibitor
-
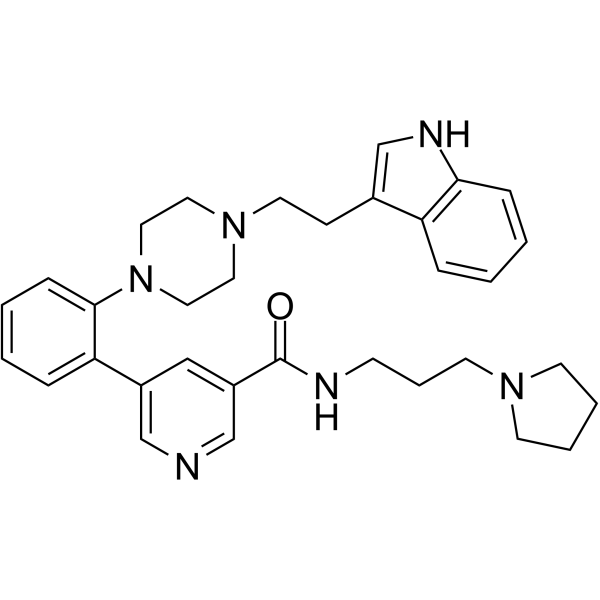
GC64124
AZ506
AZ506 is a potent SMYD2 inhibitor with an IC50 of 17 nM. AZ506 inhibits SMYD2 methyltransferase activity in cells, leading to a decrease in the SMYD2-mediated methylation signal.
-
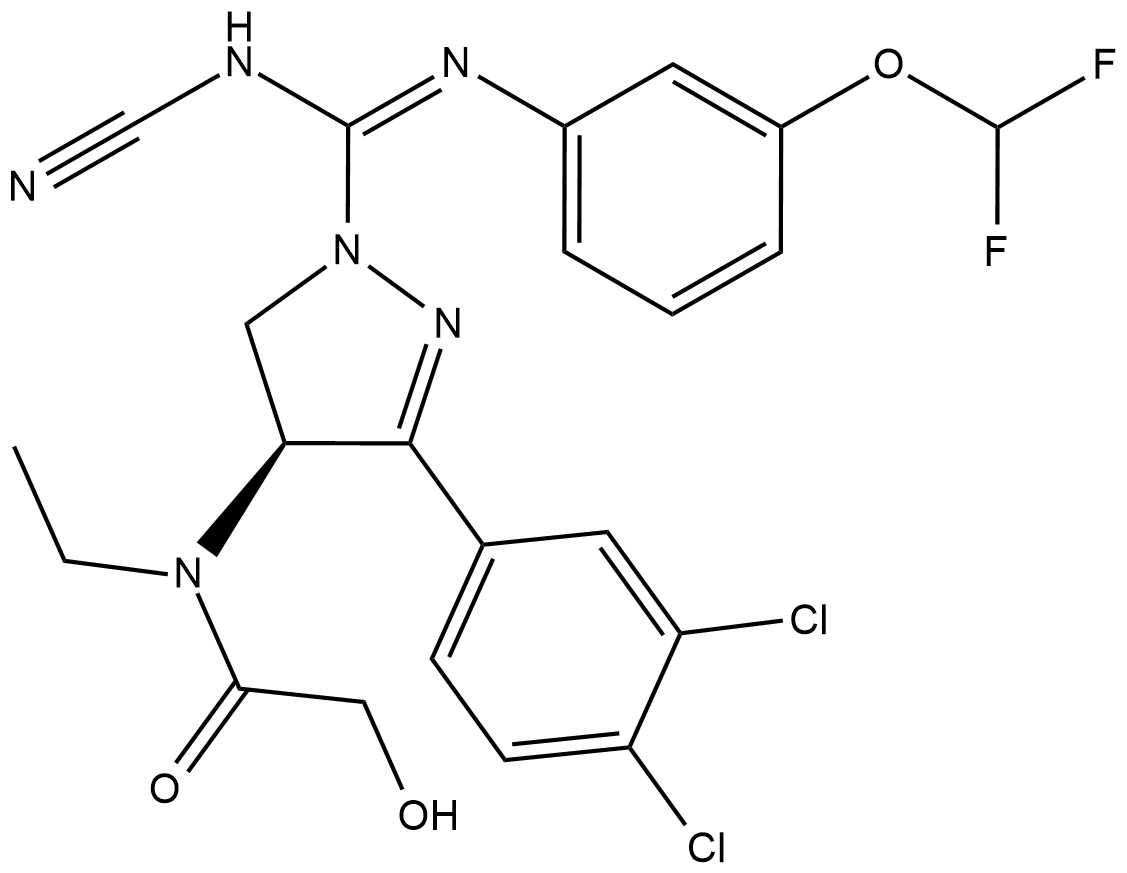
GC18159
BAY-598
A potent and selective SMYD2 inhibitor
-
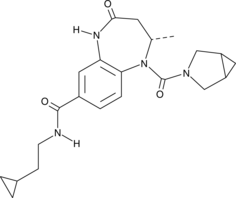
GC45389
BAY-6035
-
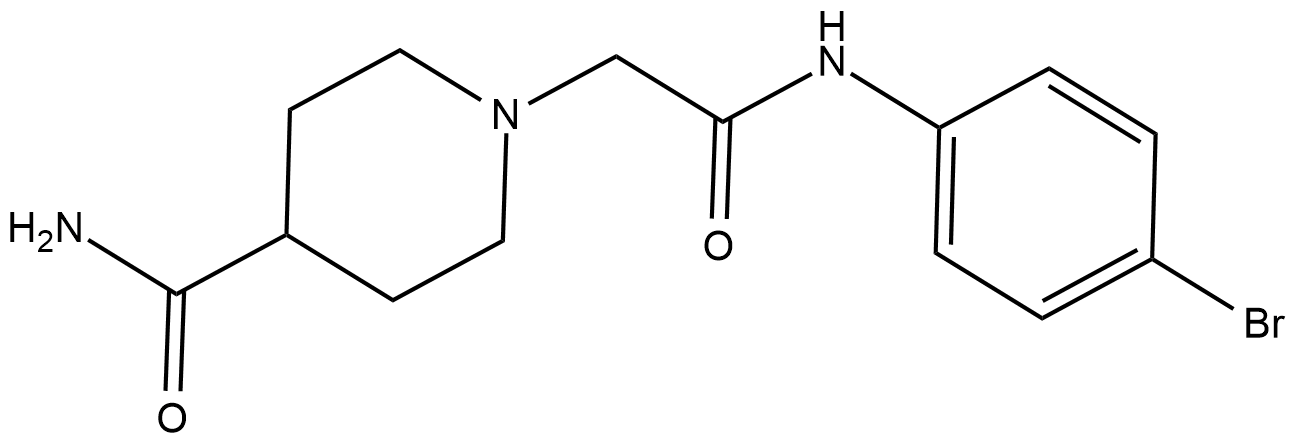
GC18161
BCI-121
A substrate-competitive SMYD3 inhibitor?
-
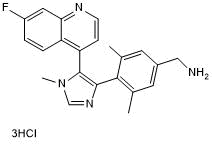
GC50540
BI 9321
Nuclear receptor-binding SET domain (NSD) 3 antagonist; selectively binds PWWP1 domain
-
GC39663
BI-9321 trihydrochloride
BI-9321 trihydrochloride is a potent, selective and cellular active nuclear receptor-binding SET domain 3 (NSD3)-PWWP1 domain antagonist with a Kd value of 166 nM. BI-9321 trihydrochloride is inactive against NSD2-PWWP1 and NSD3-PWWP2. BI-9321 trihydrochloride specifically disrupts histone interactions of the NSD3-PWWP1 domain with an IC50 of 1.2 μM in U2OS cells.
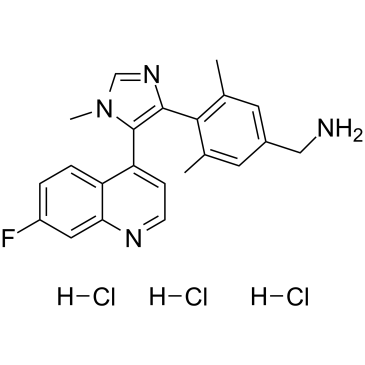
-
GC48463
Bisubstrate Inhibitor 78
An inhibitor of NNMT
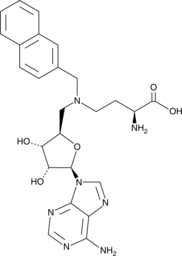
-
GC12171
BIX 01294
An inhibitor of G9a histone methyltransferase
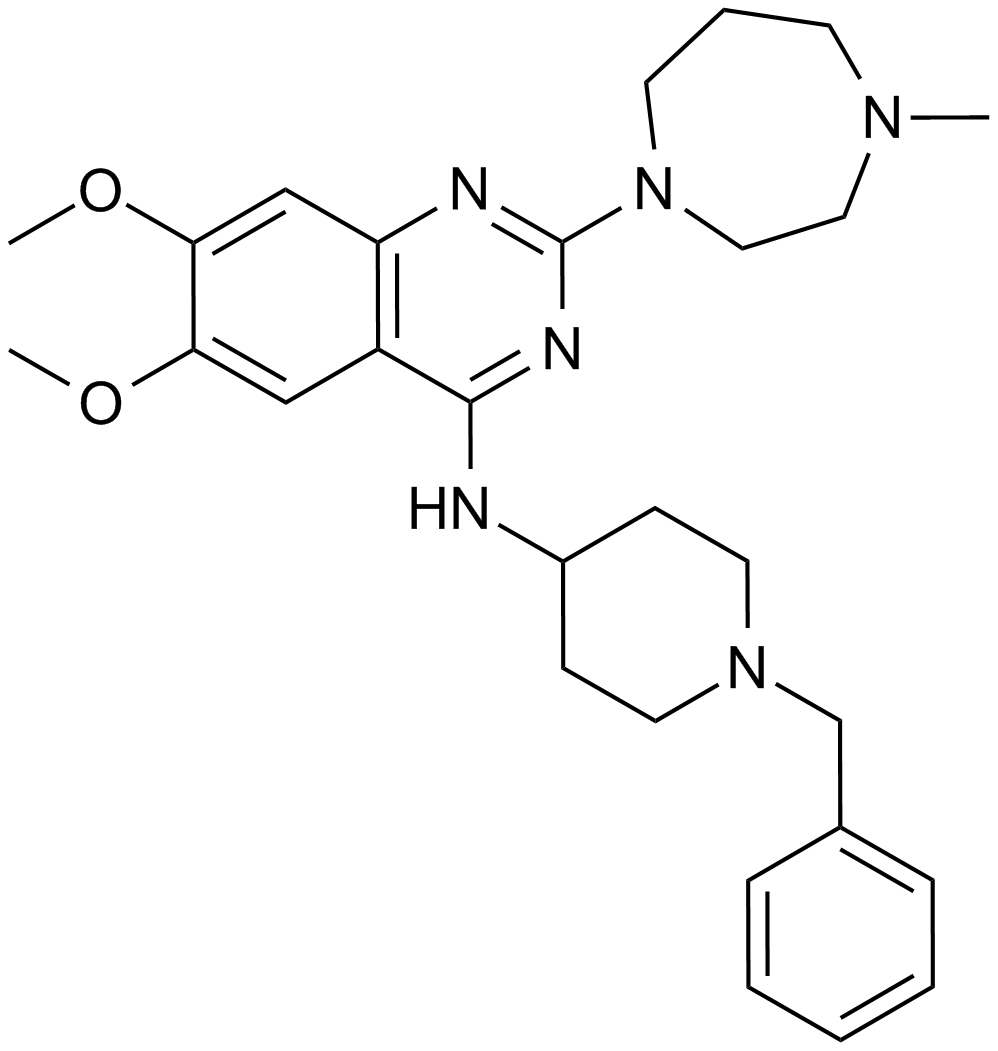
-
GC33301
BIX-01338 hydrate (BIX01338 hydrate)
BIX-01338 hydrate (BIX01338 hydrate) is a histone lysine methyltransferase inhibitor.
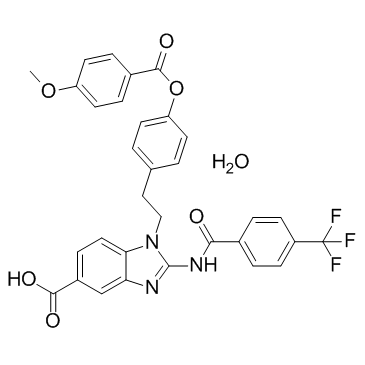
-
GC42944
BIX01294 (hydrochloride hydrate)
The methylation of lysine residues on histones plays a central role in determining euchromatin structure and gene expression.
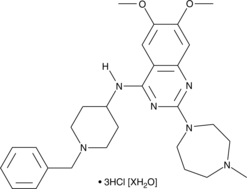
-
GC63731
BRD0639
BRD0639 is a first-in-class inhibitor of the PRMT5-substrate adaptor interaction. BRD0639 is a PRMT5 binding motif (PBM)-competitive agent that can support studies of PBM dependent PRMT5 activities.
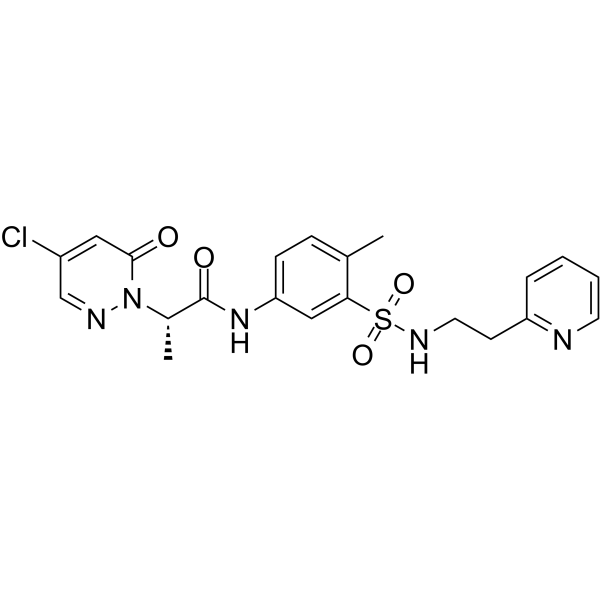
-
GC10259
BRD4770
novel G9a(EHMT2) inhibitor
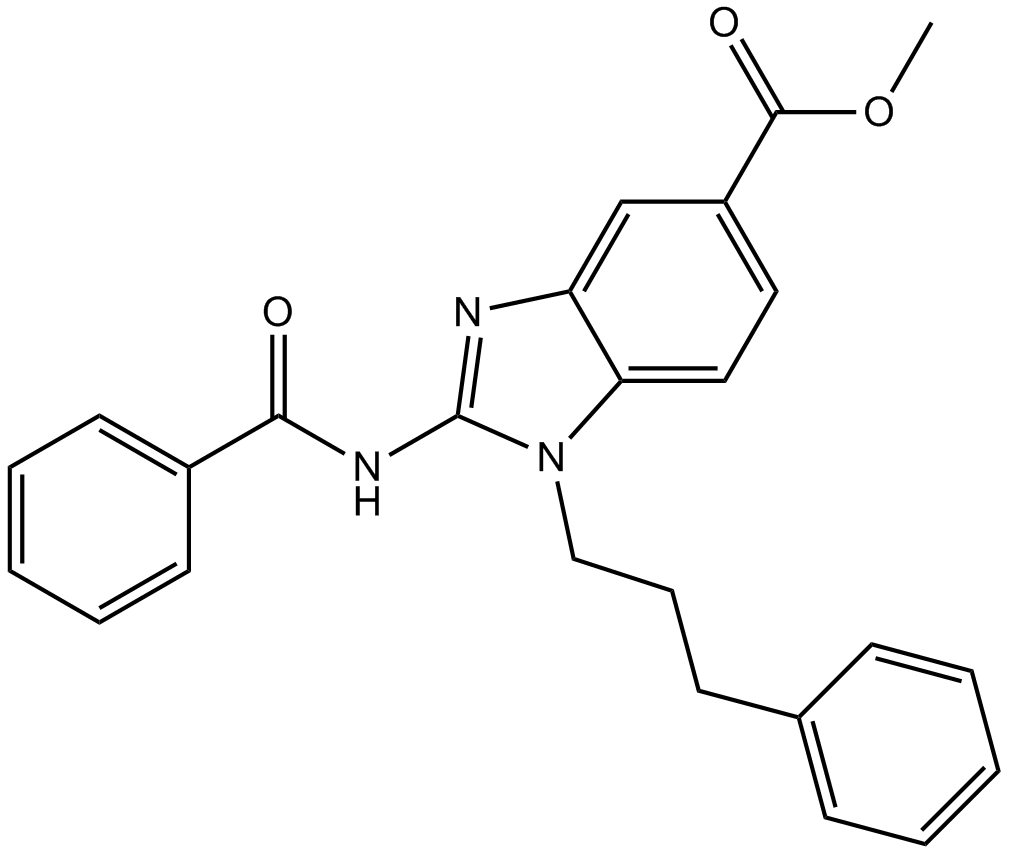
-
GC32988
BRD9539
An inhibitor of EHMT2/G9a and PRC2 in enzyme assays
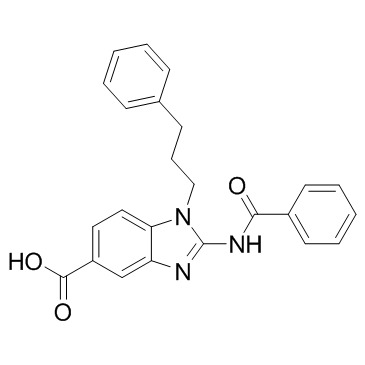
-
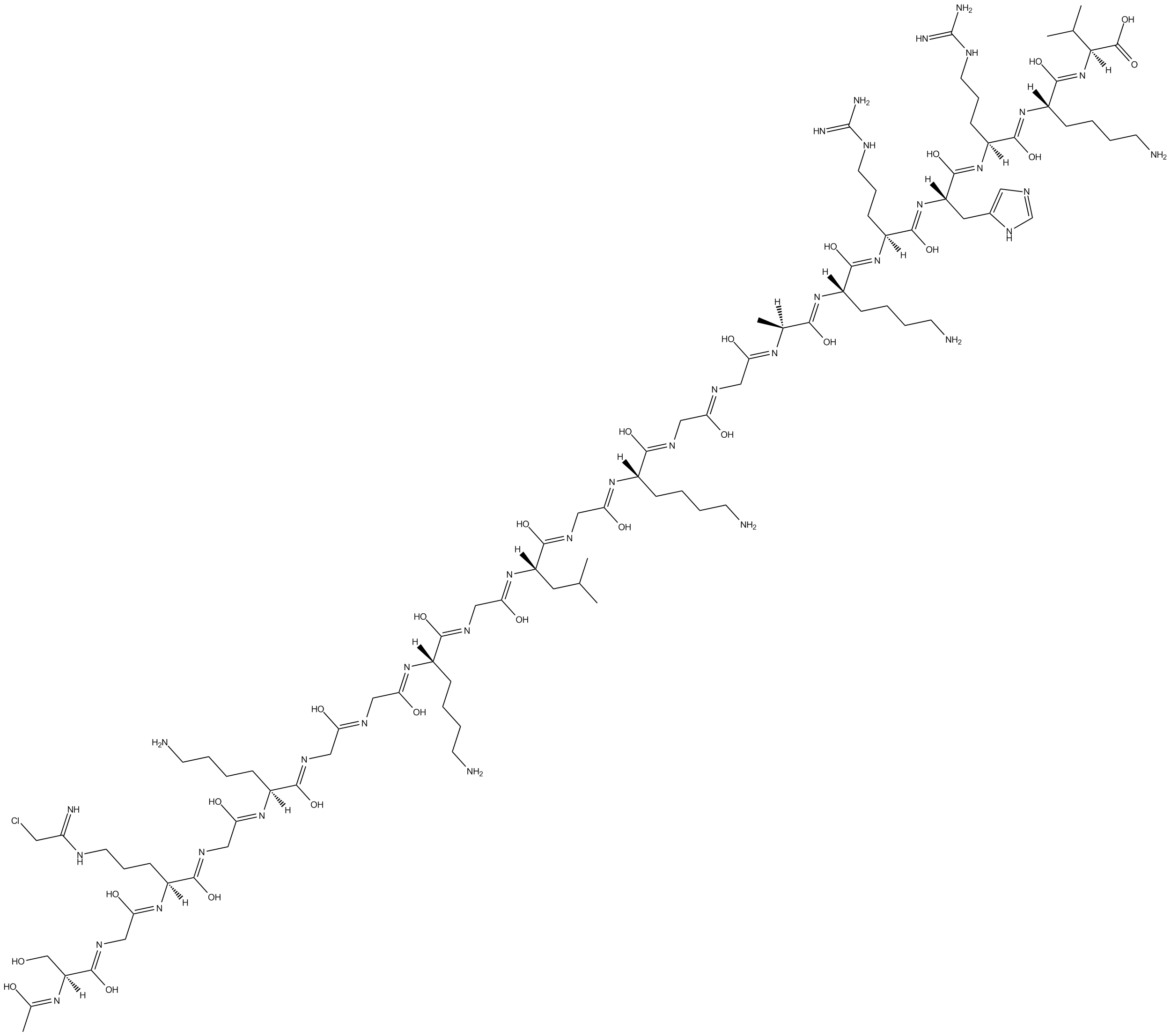
GC16130
C 21
Protein arginine methyltransferase 1 (PRMT1) inhibitor
-
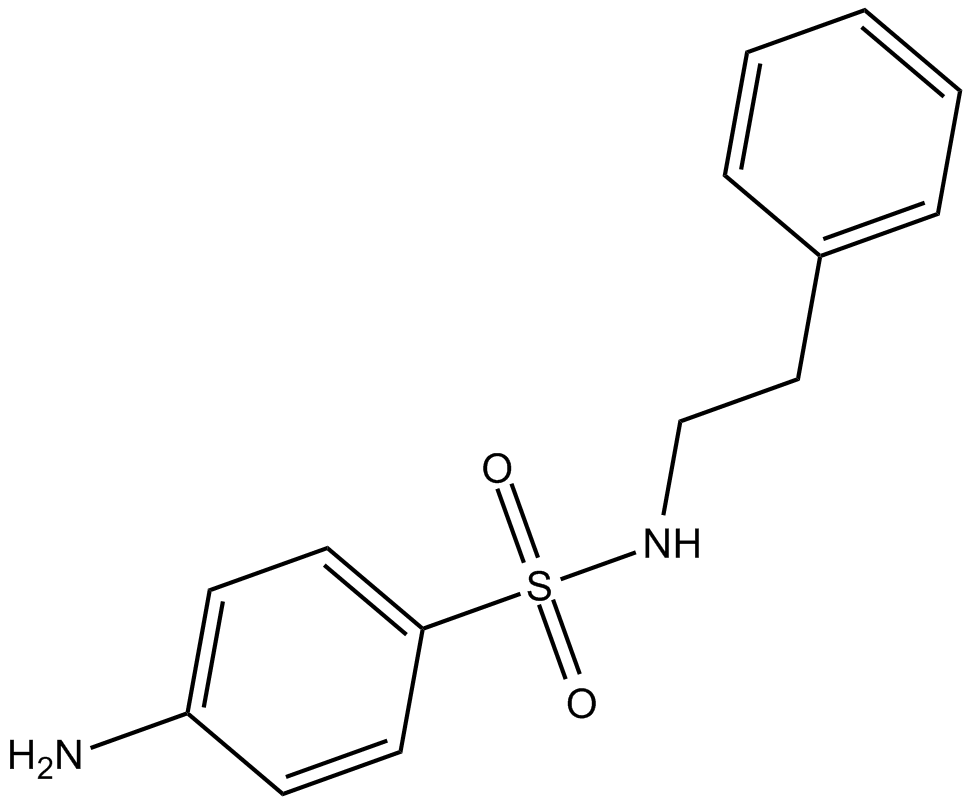
GC12005
C7280948
Novel PRMT1 inhibitor
-
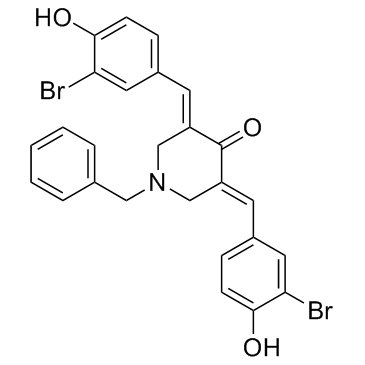
GC34108
CARM1-IN-1
A selective inhibitor of PRMT4/CARM1
-
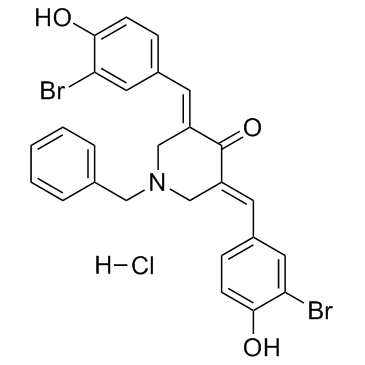
GC34424
CARM1-IN-1 hydrochloride
CARM1-IN-1 hydrochloride is a potent and specific CARM1(Coactivator-associated arginine methyltransferase 1) inhibitor with IC50 of 8.6 uM; shows very low activity against PRMT1 and SET7(IC50 > 600 uM).
-
GC18949
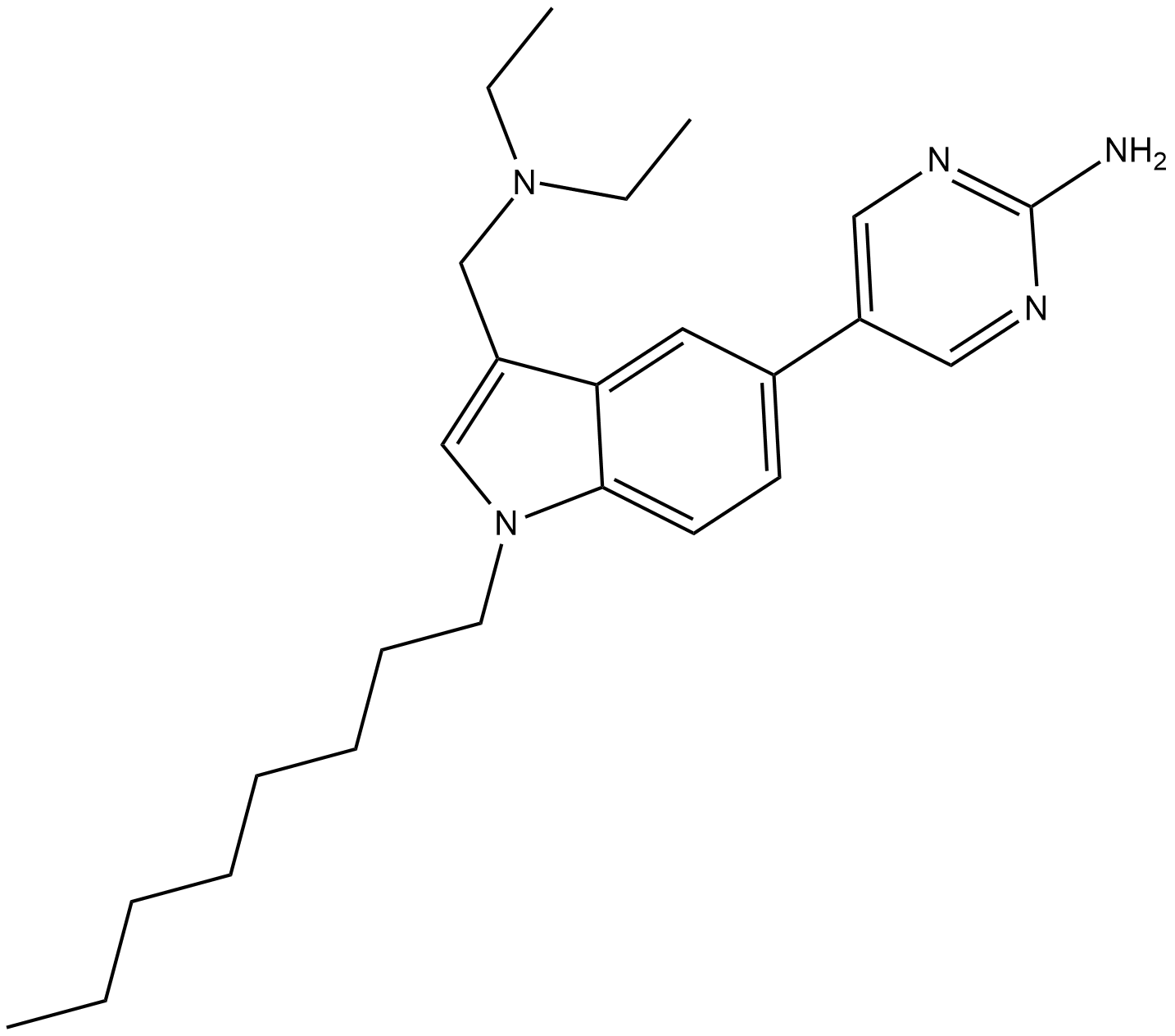
CAY10677
Post-translational protein prenylation is a 3-step process that occurs at the C-terminus of a number of proteins involved in cell growth control and oncogenesis.
-
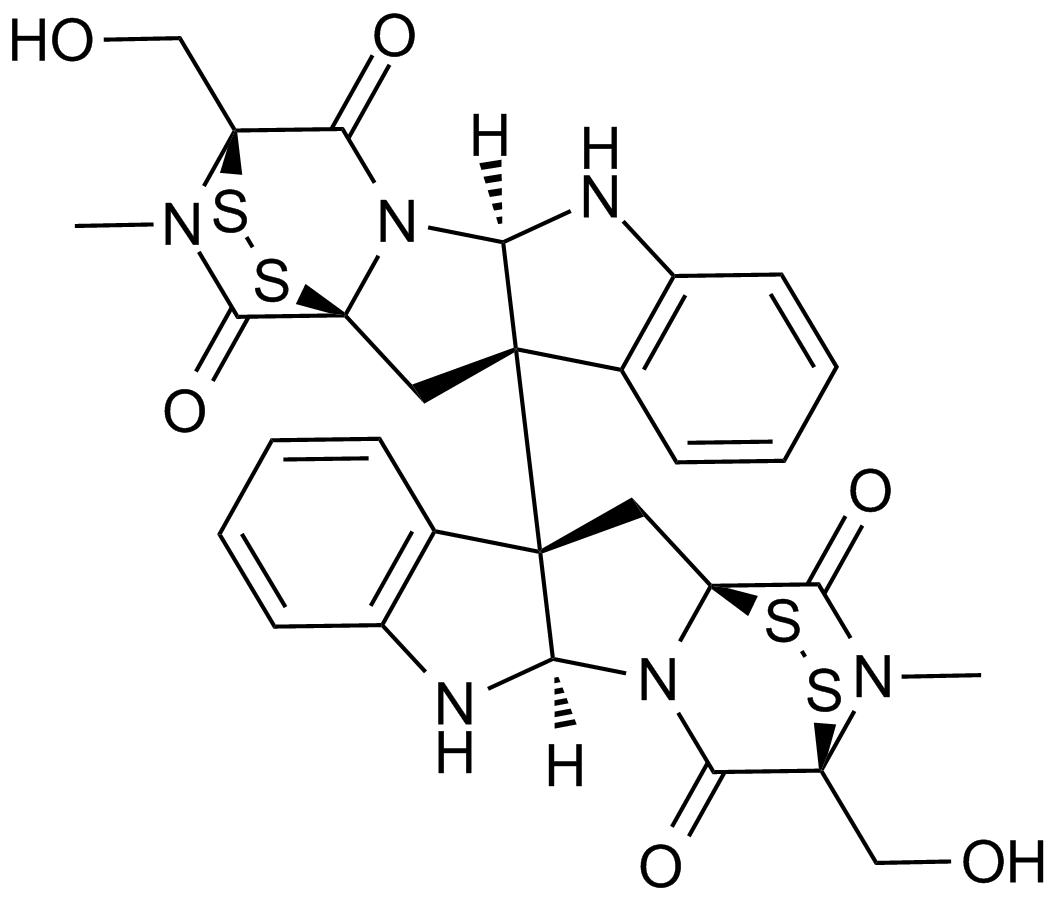
GC14936
Chaetocin
Inhibitor of lys9-specific HMTs
-
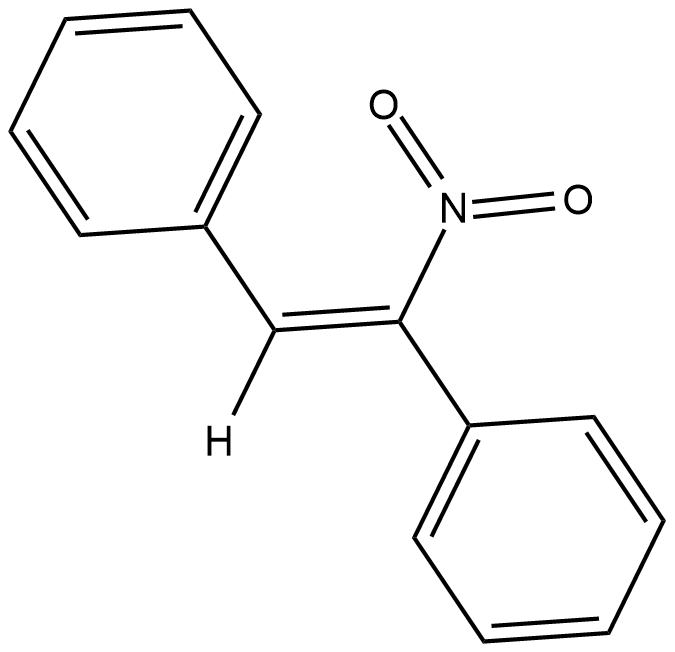
GC18577
CID-2818500
An inhibitor of PRMT1
-
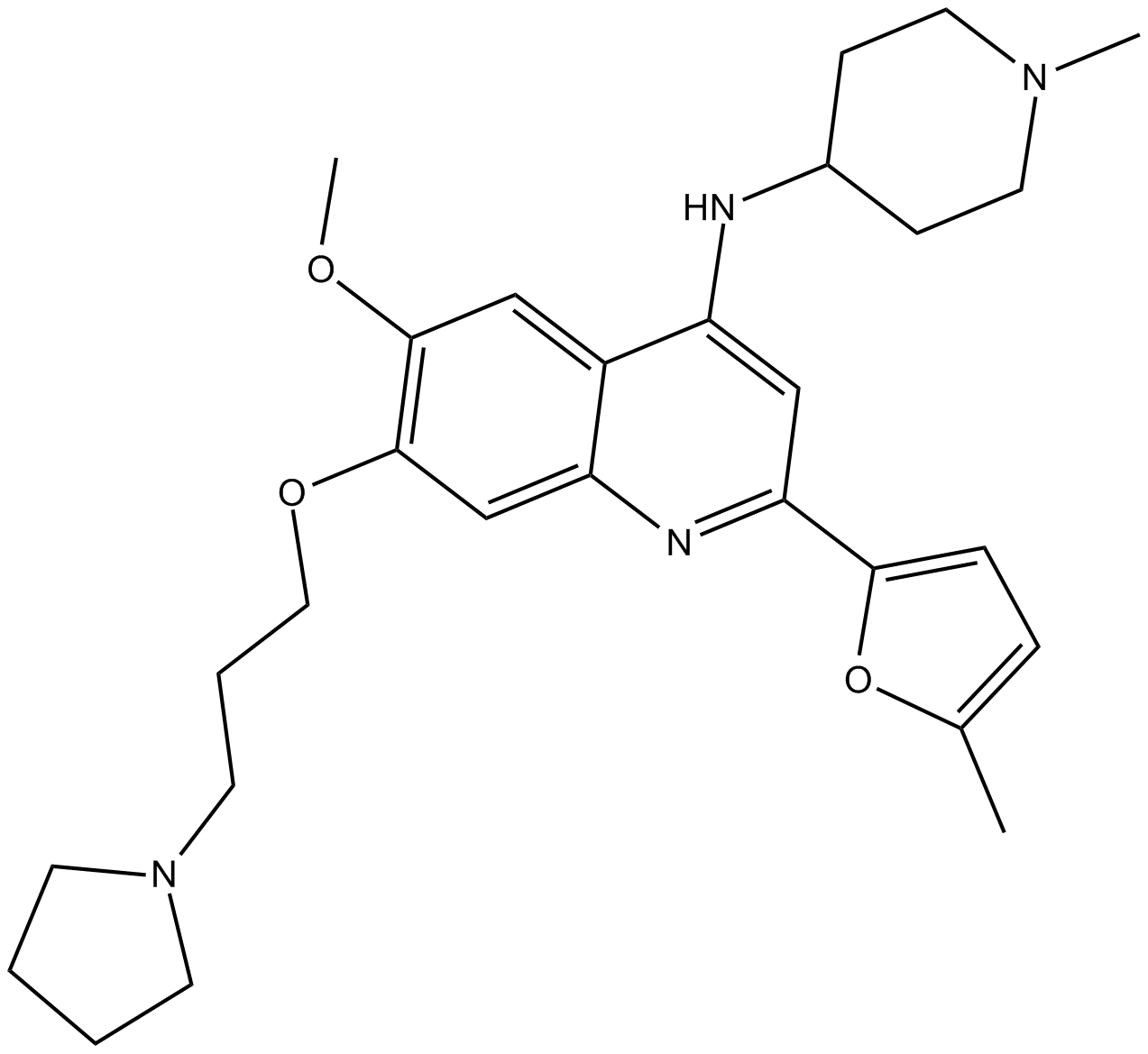
GC12367
CM-272
CM-272 is a first-in-class reversible dual inhibitor against G9a and DNMTs with IC50 values of 8 nM and 382 nM, respectively [1].
-
GC33320
CM-579
CM-579 is a first-in-class reversible, dual inhibitor of G9a and DNMT, with IC50 values of 16 nM, 32 nM for G9a and DNMT, respectively. Has potent in vitro cellular activity in a wide range of cancer cells.
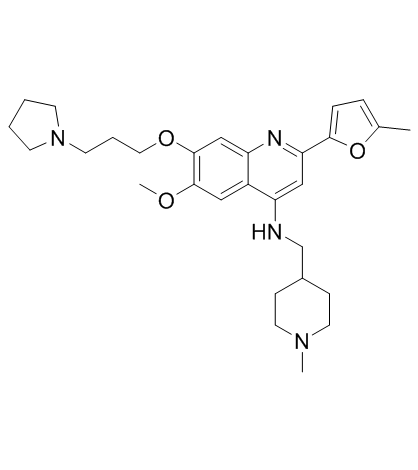
-
GC35714
CM-579 trihydrochloride
CM-579 trihydrochloride is a first-in-class reversible, dual inhibitor of G9a and DNMT, with IC50 values of 16 nM, 32 nM for G9a and DNMT, respectively. Has potent in vitro cellular activity in a wide range of cancer cells.
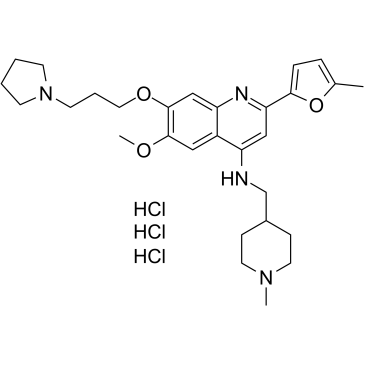
-
GC39665
CMP-5
CMP-5 is a potent, specific, and selective PRMT5 inhibitor, while displays no activity against PRMT1, PRMT4, and PRMT7 enzymes. CMP-5 selectively blocks S2Me-H4R3 by inhibiting PRMT5 methyltransferase activity on histone preparations. CMP-5 prevents Epstein-Barr virus (EBV)-driven B-lymphocyte transformation but leaving normal B cells unaffected.
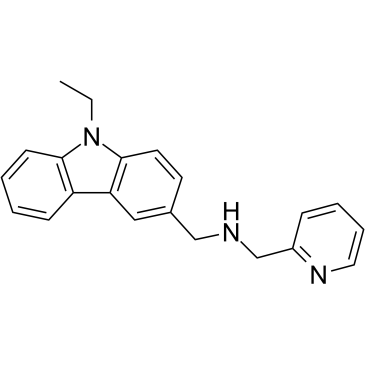
-
GC16298
CPI-1205
EZH2 inhibitor
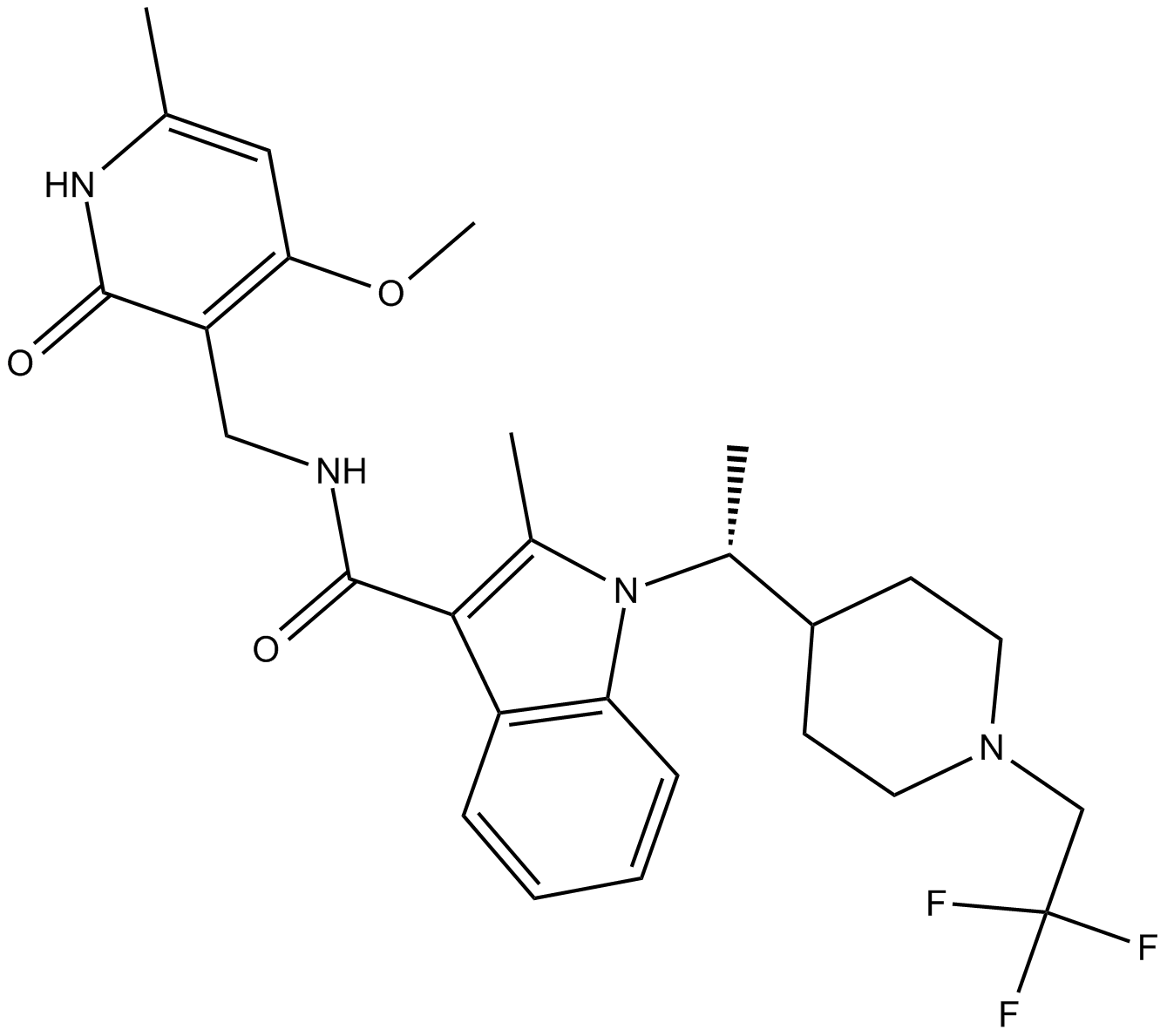
-
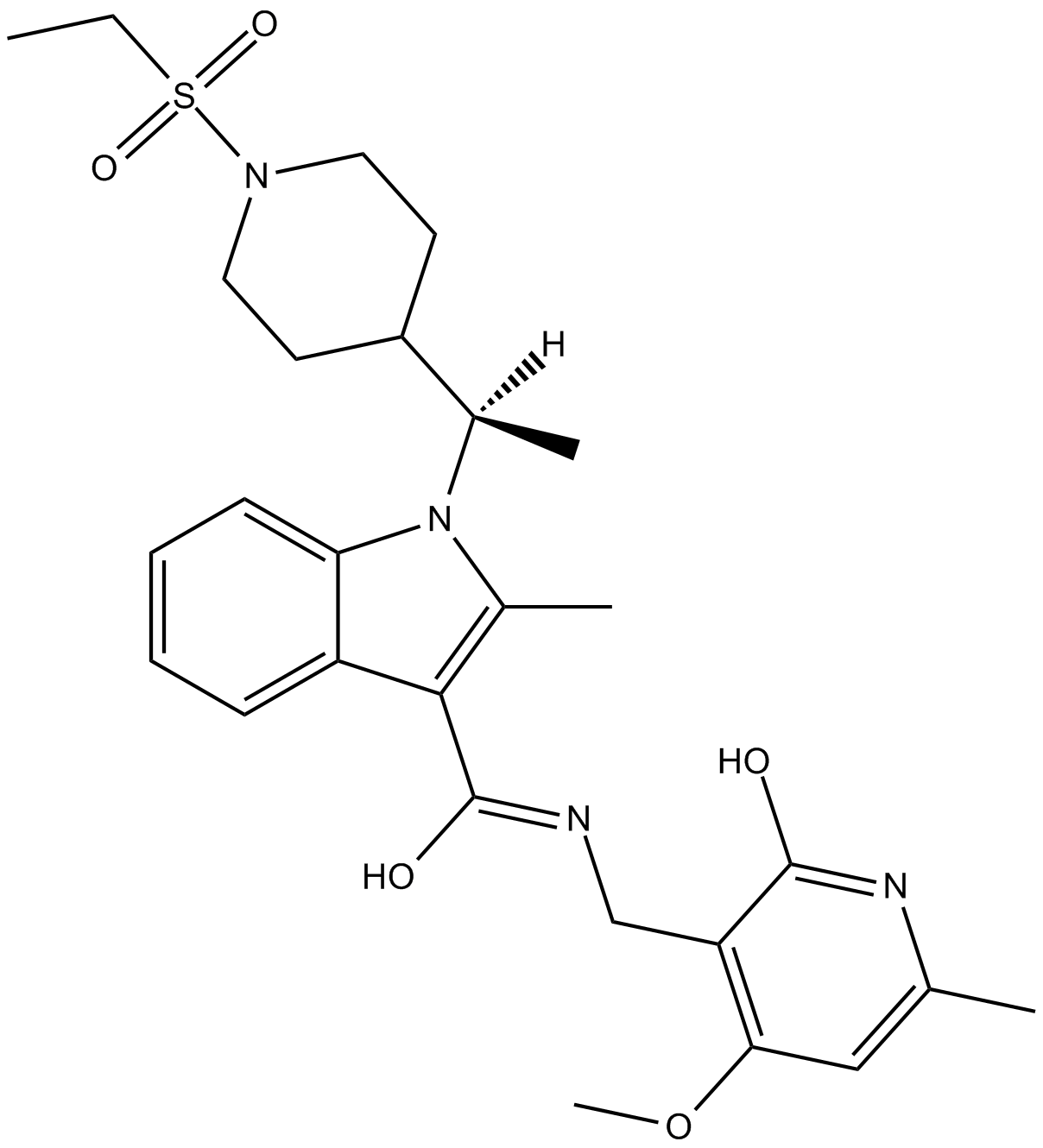
GC16599
CPI-169
EZH2 inhibitor
-
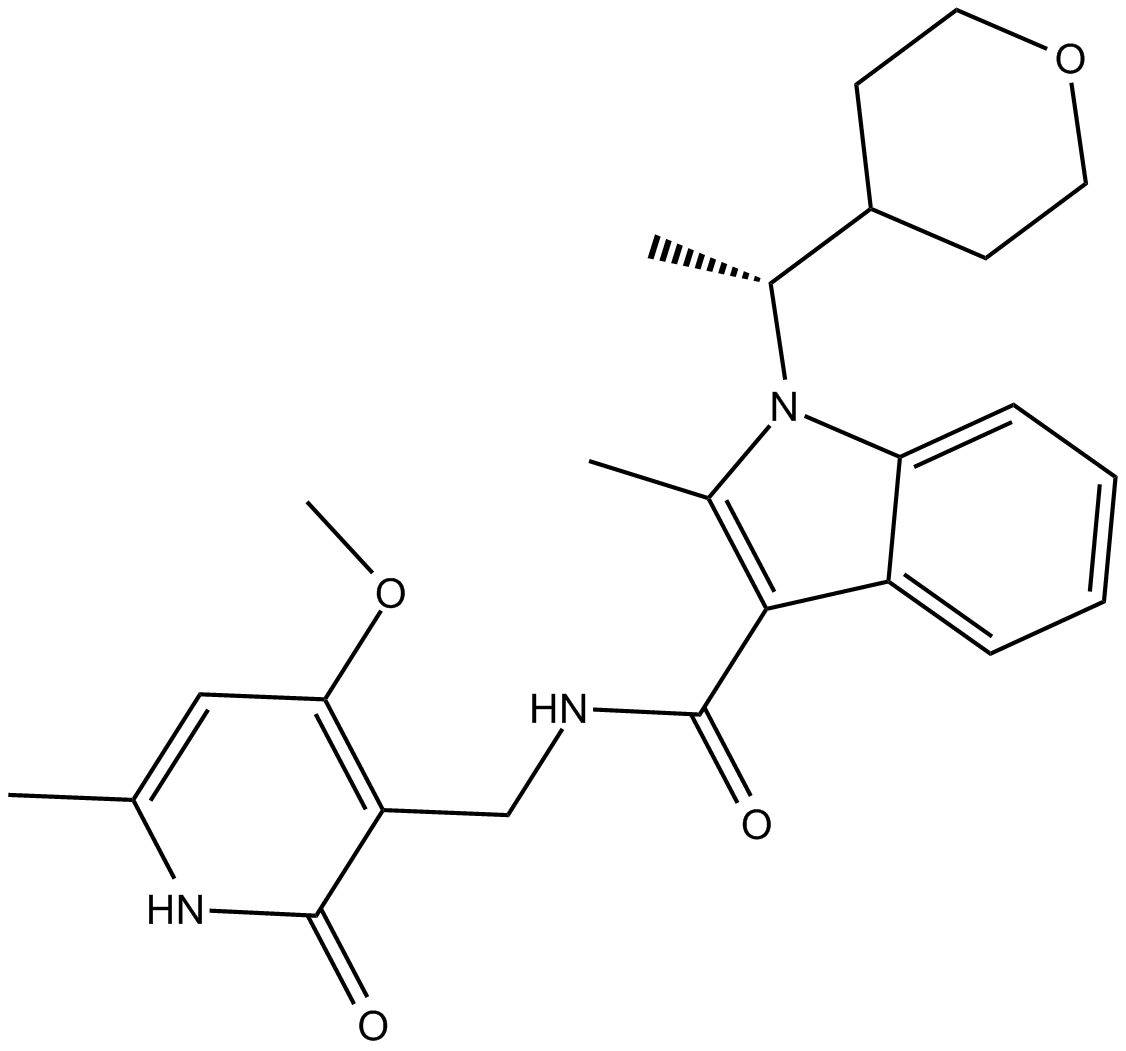
GC10021
CPI-360
EZH2 inhibitor
-
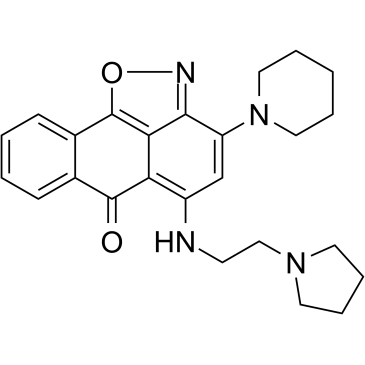
GC35742
CPUY074020
CPUY074020 is a potent and oral bioavailable inhibitor of histone methyltransferase G9a, with an IC50 of 2.18 μM. CPUY074020 possesses anti-proliferative activity.
-
GC43354
Cysmethynil
Post-translational protein prenylation is a 3-step process that occurs at the C-terminus of a number of proteins involved in cell growth control and oncogenesis.

-
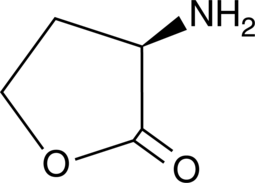
GC48967
D-Homoserine lactone
An enantiomer of L-homoserine lactone
-
GC65186
DC-S239
DC-S239 is a selective histone methyltransferase SET7 inhibitor with an IC50 value of 4.59 μM. DC-S239 also displays selectivity for DNMT1, DOT1L, EZH2, NSD1, SETD8 and G9a. DC-S239 has anticancer activity.
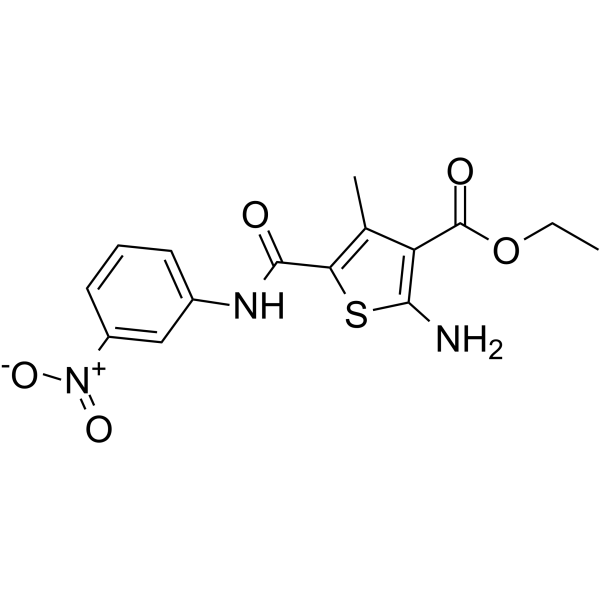
-
GC63662
DCLX069
DCLX069 is a selective protein arginine methyltransferase 1 (PRMT1) inhibitor with an IC50 value of 17.9 ?M. DCLX069 shows less active against PRMT4 and PRMT6. DCLX069 has anticancer effects.
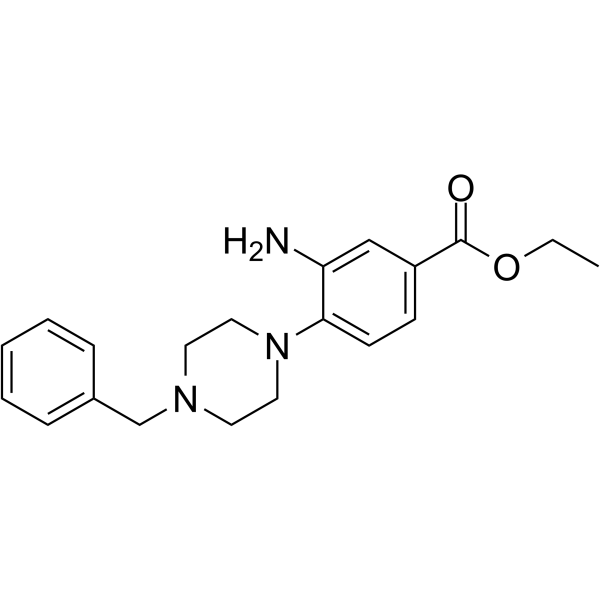
-
GC35816
DC_C66
DC_C66 is a cell-permeable, selective coactivator associated arginine methyltransferase 1 (CARM1) inhibitor with an IC50 of 1.8 μM. DC_C66 has a good selectivity for CARM1 against PRMT1 (IC50=21 μM), PRMT6 (IC50= 47μM), and PRMT5.
-
GC67863
DDO-2093 dihydrochloride

-
GC33208
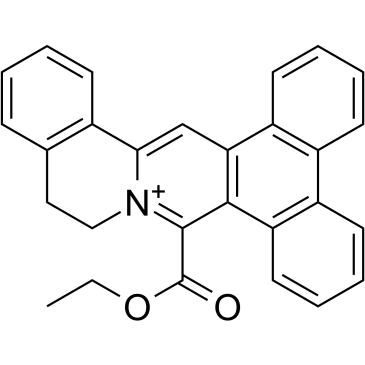
Dot1L-IN-1
Dot1L-IN-1 is a highly potent, selective and structurally novel Dot1L inhibitor with a Ki of 2 pM.
-
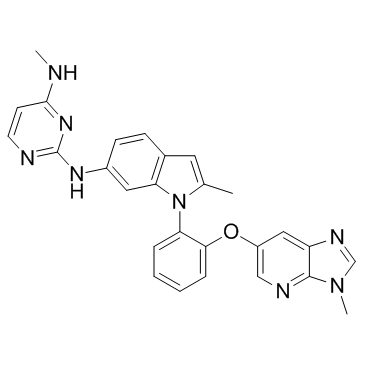
GC30530
Dot1L-IN-2
Dot1L-IN-2 is a potent, selective and orally bioavailable inhibitor of Dot1L (a histone methyltransferase), with an IC50 and Ki of 0.4 nM and 0.08 nM, respectively.
-
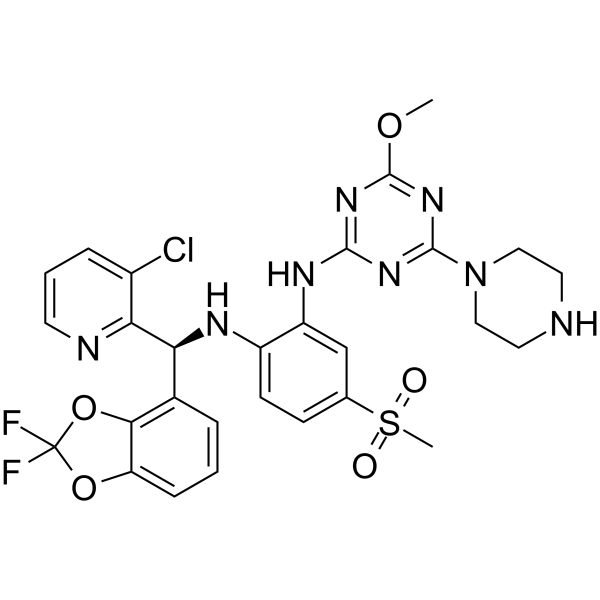
GC62613
Dot1L-IN-4
Dot1L-IN-4 is a potent disruptor of telomeric silencing 1-like protein (DOT1L) inhibitor with an IC50 SPA DOT1L of 0.11 nM.
-
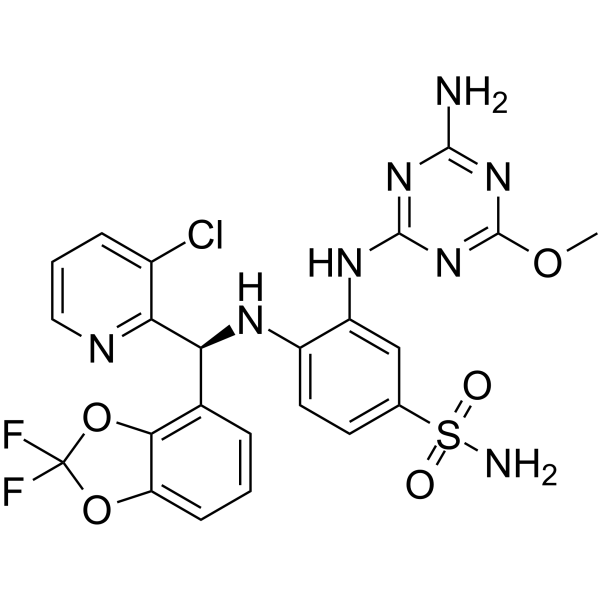
GC65962
Dot1L-IN-5
Dot1L-IN-5 is a potent disruptor of telomeric silencing 1-like protein (DOT1L) inhibitor with an IC50 SPA DOT1L of 0.17 nM.
-
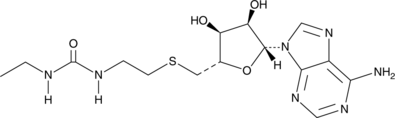
GC45927
DS-437
A dual inhibitor of PRMT5 and PRMT7
-
GC35914
DW14800
DW14800 is a protein arginine methyltransferase 5 (PRMT5) inhibitor, with an IC50 of 17 nM. DW14800 reduces H4R3me2s levels and enhances the transcription of HNF4α, but does not alter PRMT5 expression. Anti-cancer activity.
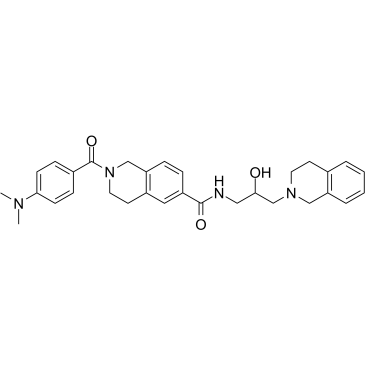
-
GC33220
EBI-2511
EBI-2511 is a highly potent and orally active EZH2 inhibitor, with an IC50 of 6 nM in Pfeffiera cell lines, respectively.
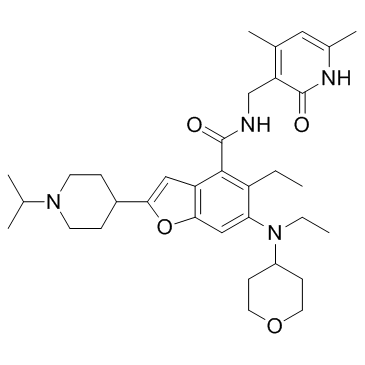
-
GC19130
EED226
EED226 is a potent, selective, and orally bioavailable embryonic ectoderm development (EED) inhibitor with an IC50 of 22 nM.
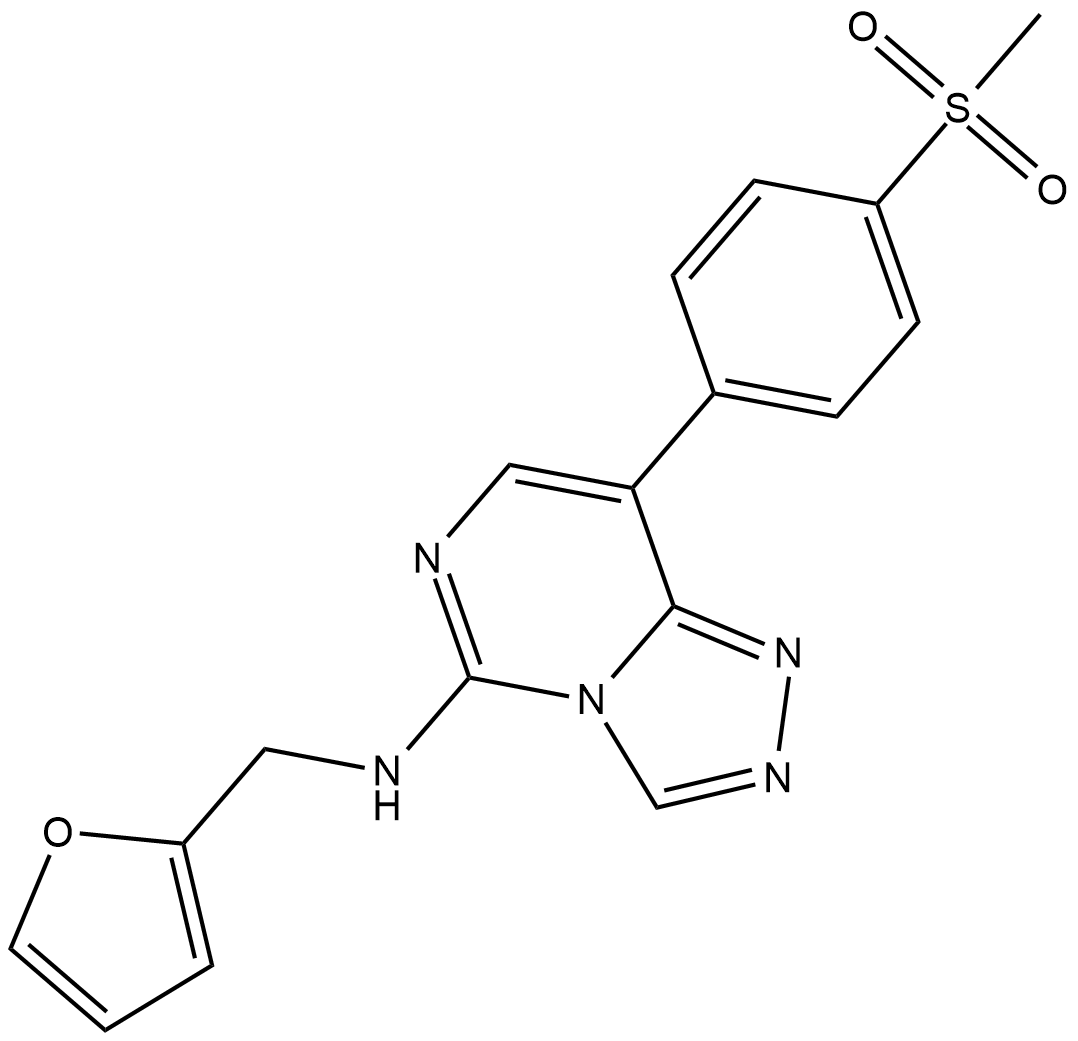
-
GC67934
EEDi-5285
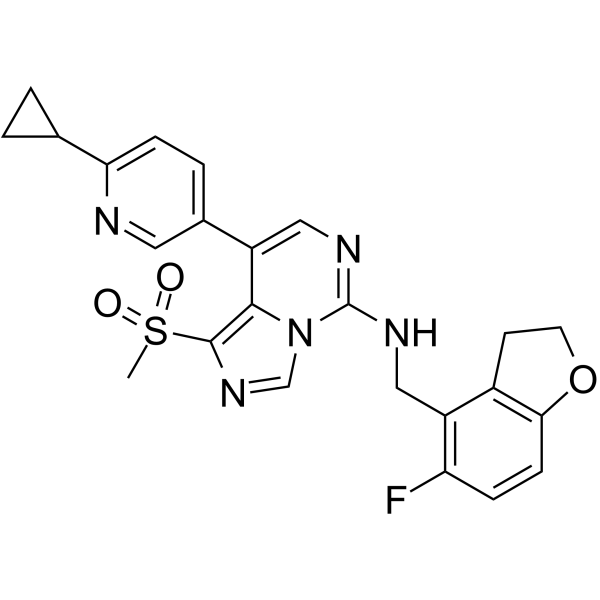
-
GC34567
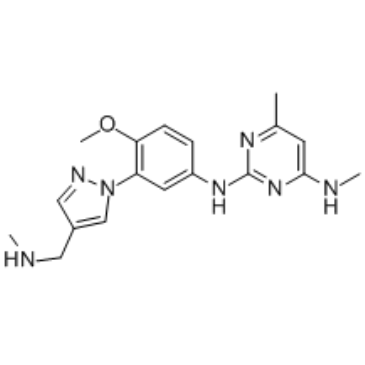
EHMT2-IN-1
EHMT2-IN-1 is a potent EHMT inhibitor, with IC50s of all <100 nM for EHMT1 peptide, EHMT2 peptide and cellular EHMT2. Used in the research of blood disorder or cancer.
-
GC34568
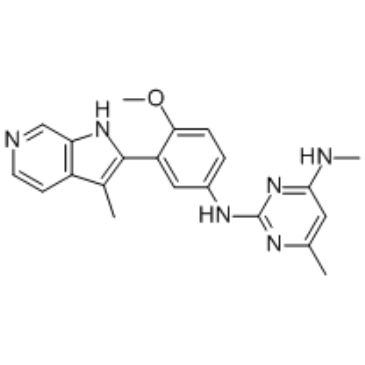
EHMT2-IN-2
EHMT2-IN-2 is a potent EHMT inhibitor, with IC50s of all <100 nM for EHMT1 peptide, EHMT2 peptide and cellular EHMT2. Used in the research of blood disease or cancer.
-
GC14756
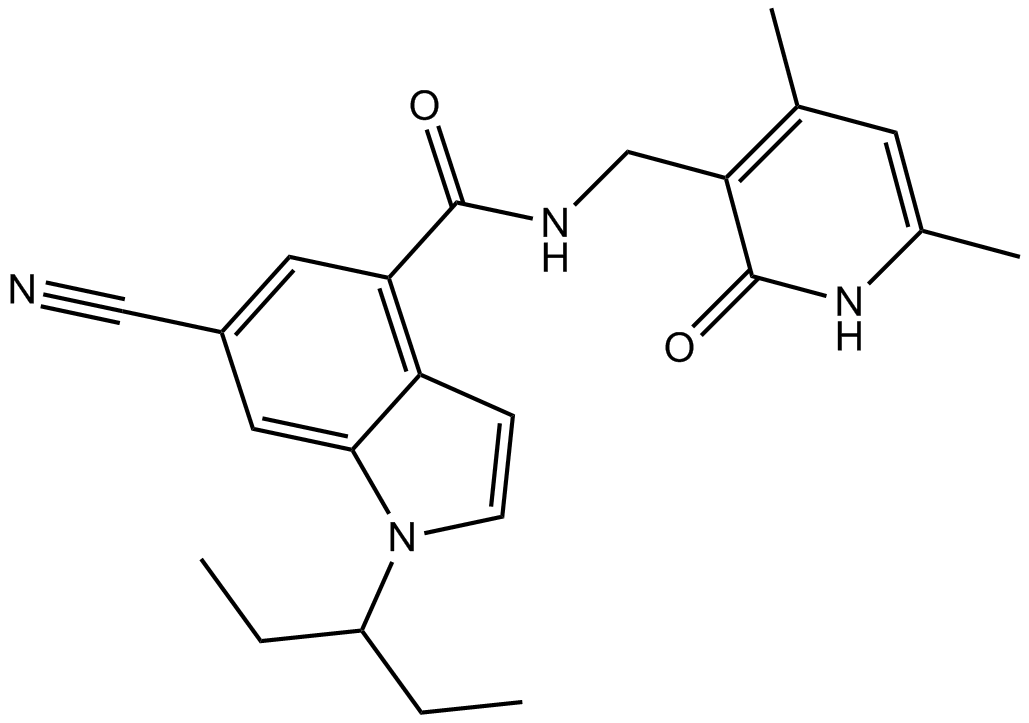
EI1
EZH2 inhibitor
-
GC17334
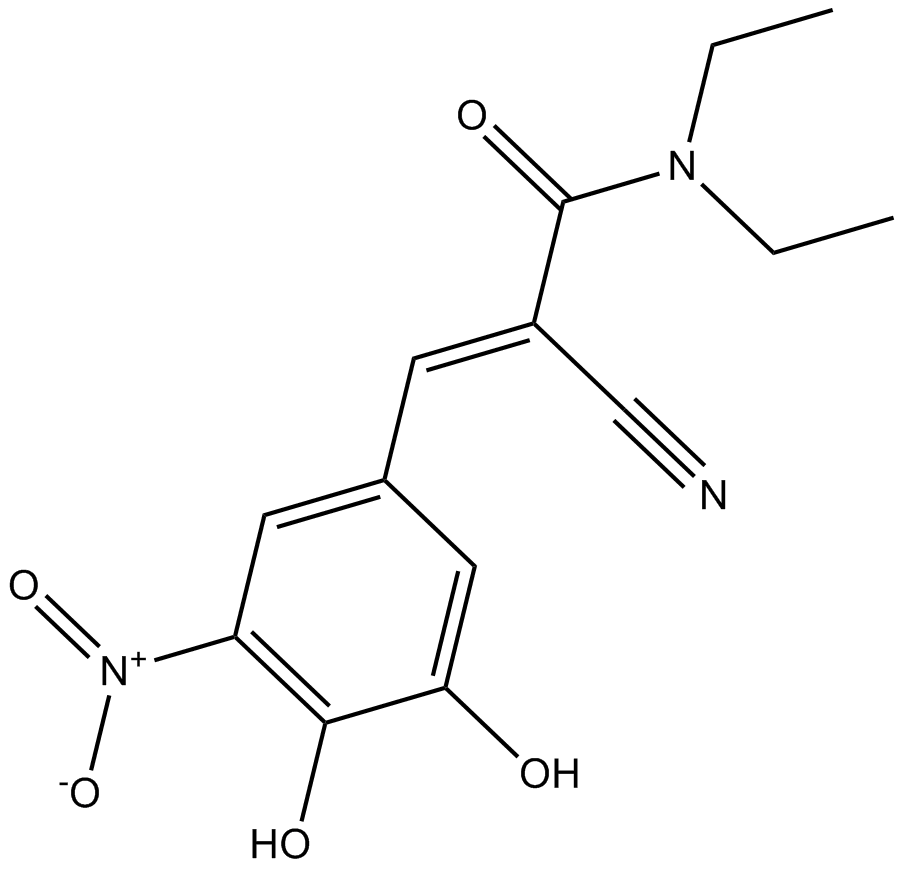
Entacapone
A reversible COMT inhibitor
-
GC47294
Entacapone-d10
An internal standard for the quantification of entacapone
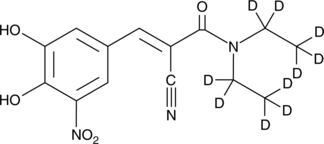
-
GC14062
EPZ-6438
A EZH2 inhibitor,potent and selective
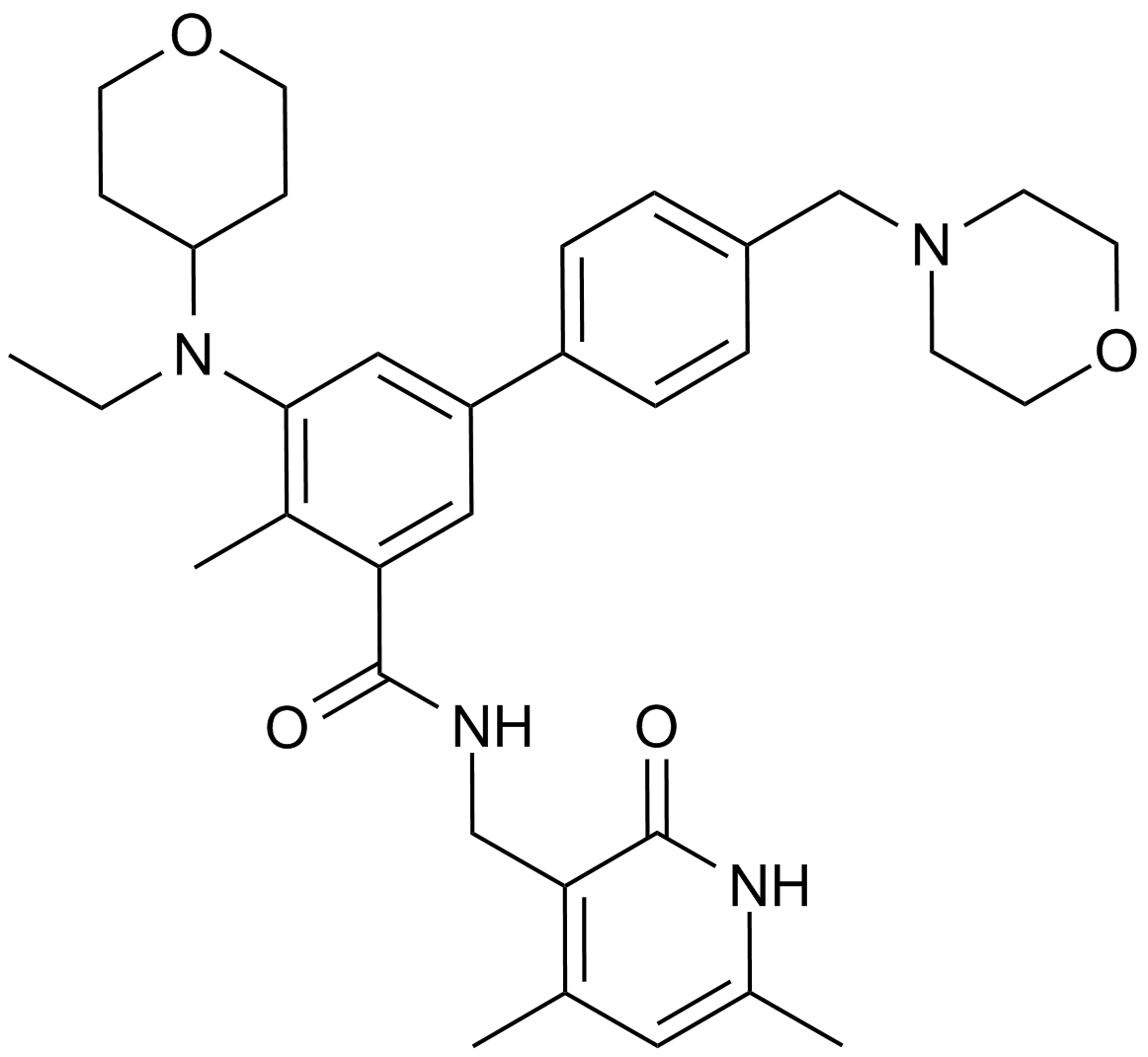
-
GC64343
EPZ-719
EPZ-719 is a novel and potent SETD2 inhibitor ( IC50 = 0.005 μM) with a high selectivity over other histone methyltransferases.
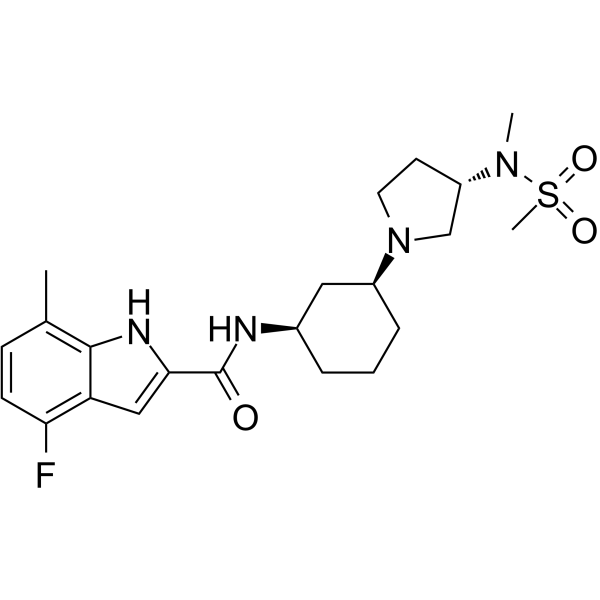
-
GC13383
EPZ004777
A potent inhibitor of DOT1L
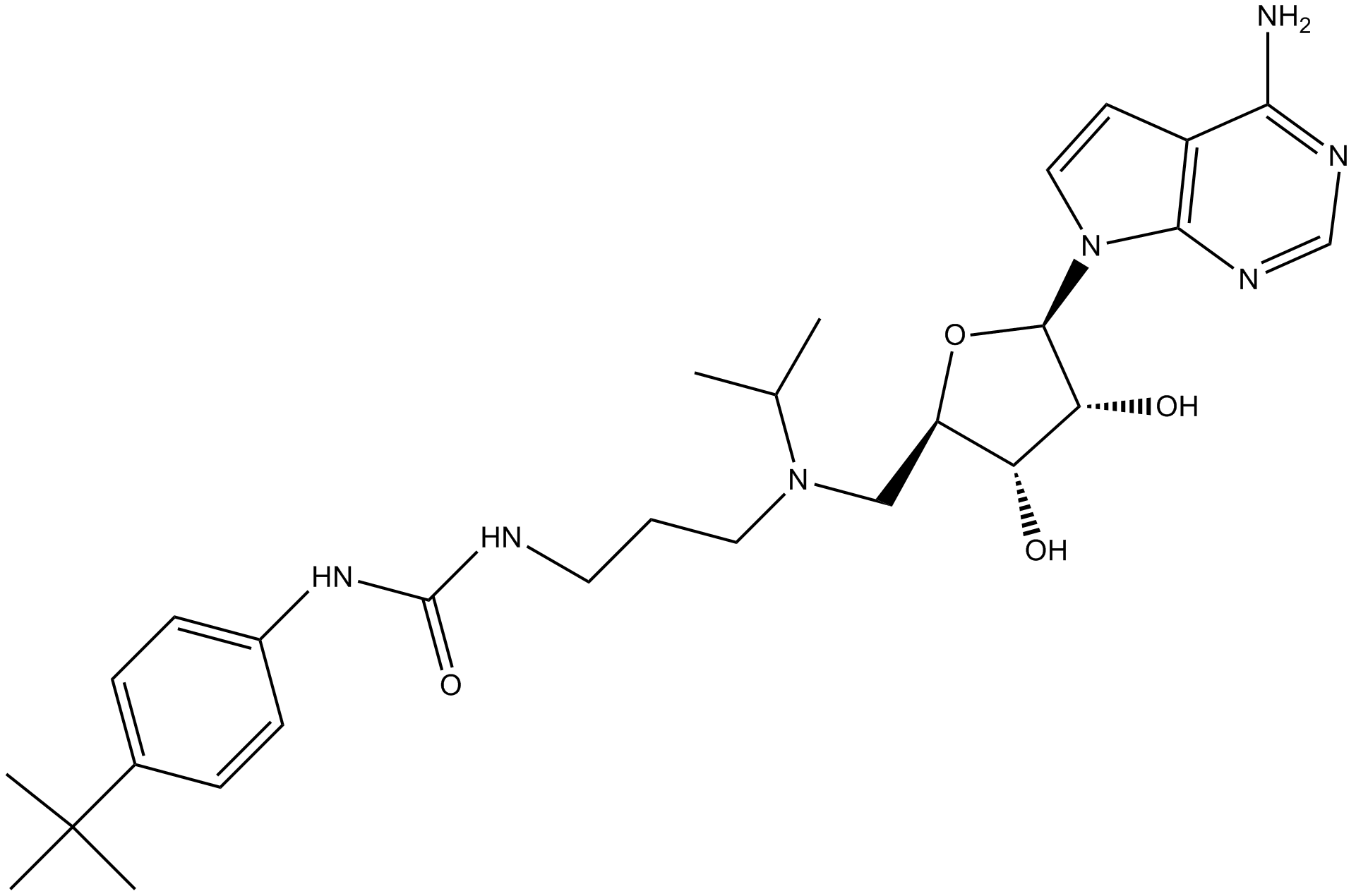
-
GC48980
EPZ004777 (formate)
A potent inhibitor of DOT1L

-
GC15259
EPZ004777 HCl
EPZ004777 HCl is a potent, selective DOT1L inhibitor with an IC50 of 0.4 nM.
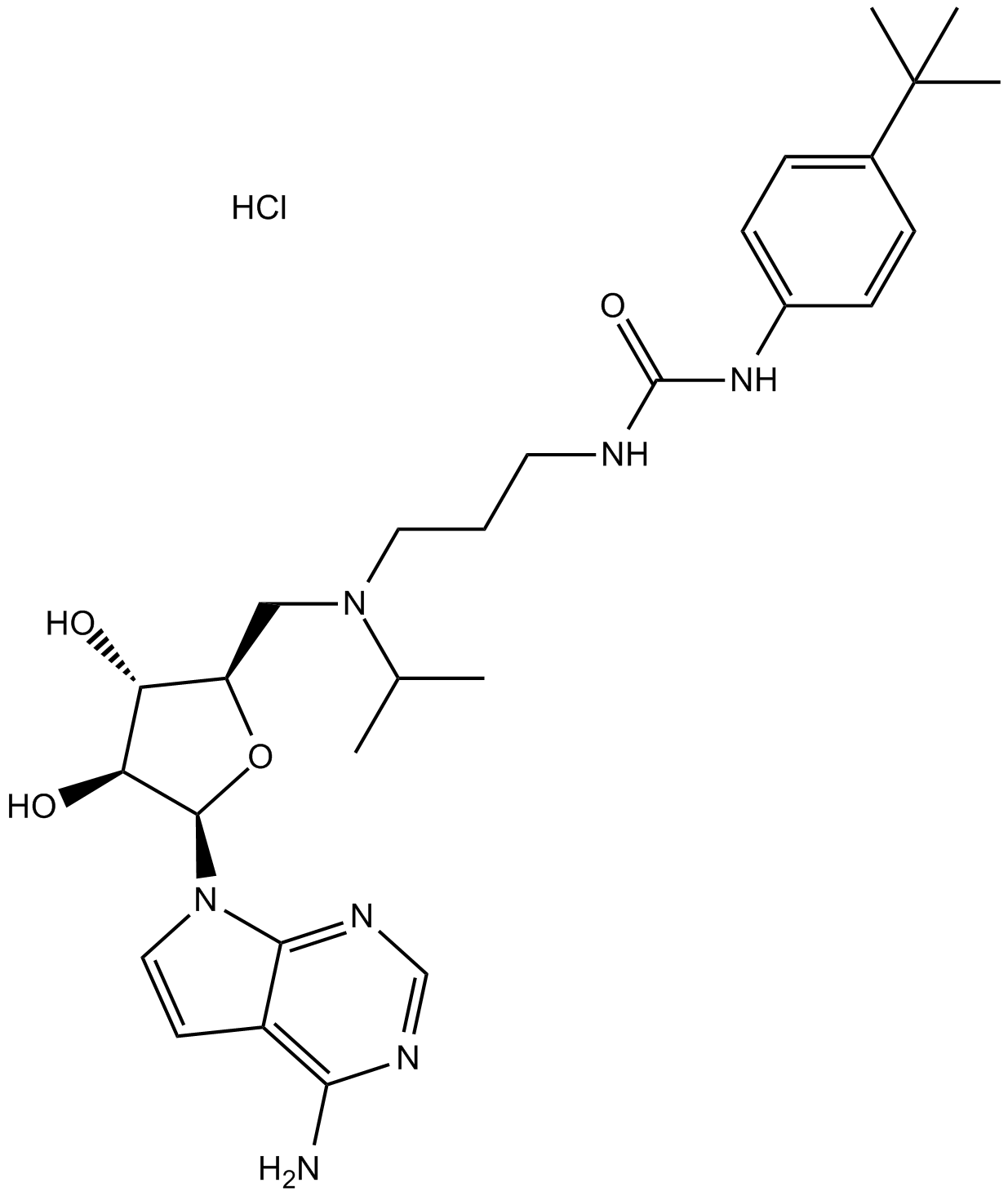
-
GC13878
EPZ005687
A potent, selective inhibitor of EZH2
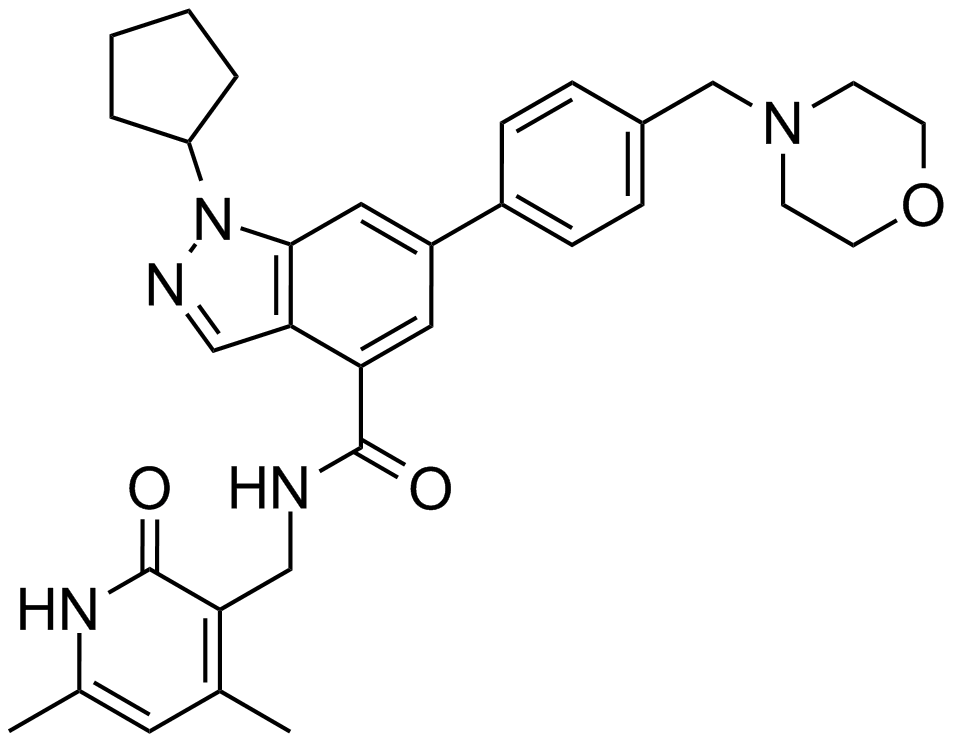
-
GC19141
EPZ011989
EPZ011989 is a potent, selective orally bioavailable EZH2 inhibitor with Ki 3000-fold selectivity over other HMTase.
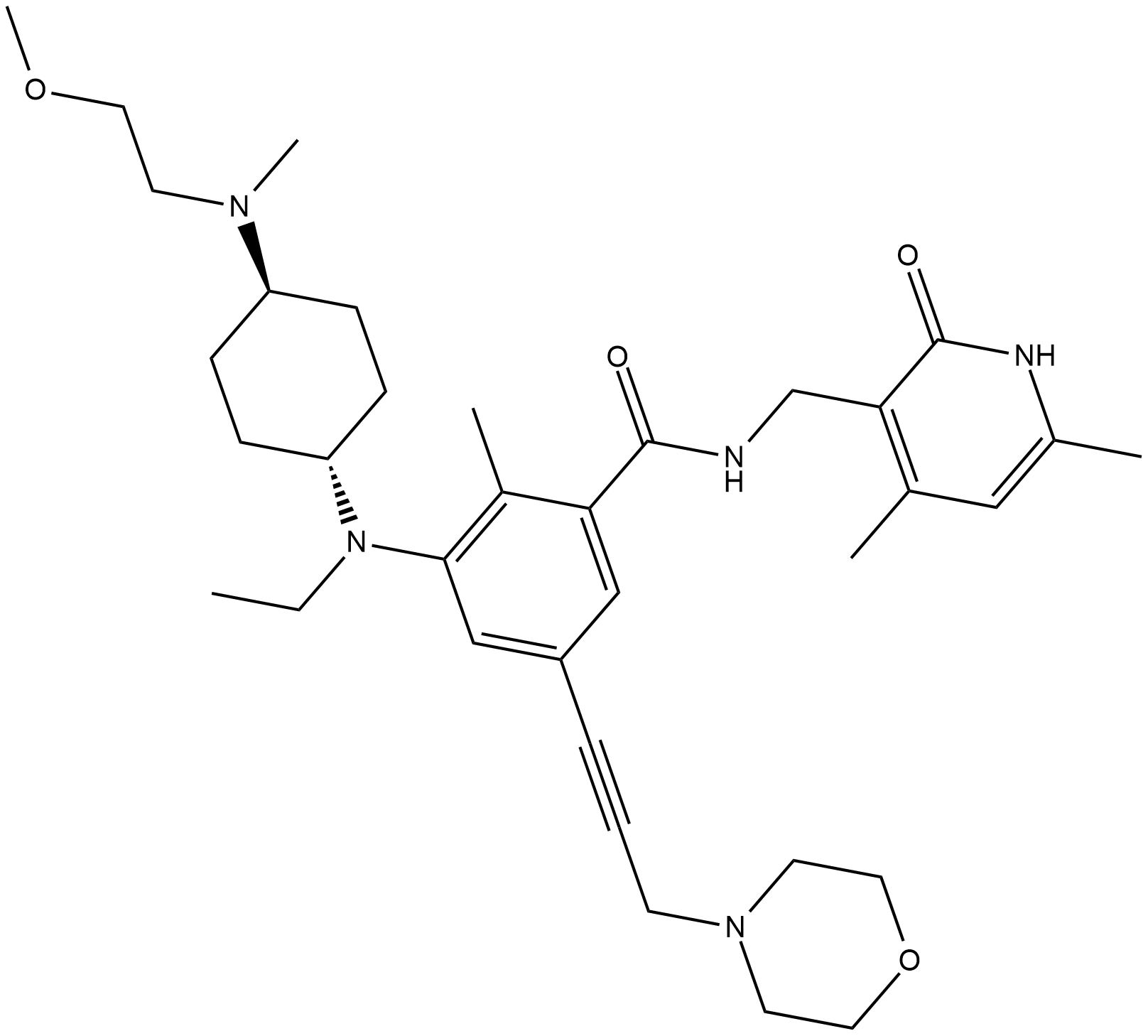
-
GC34136
EPZ011989 trifluoroacetate (EPZ-011989 trifluoroacetate)
EPZ-011989 trifluoroacetate is a potent and orally active Zeste Homolog 2 (EZH2) inhibitor with metabolic stability. EPZ-011989 trifluoroacetate has inhibitory inhibition for EZH2 with a Ki value of <3 nM. EPZ-011989 trifluoroacetate shows robust methyl mark inhibition and anti-tumor activity. EPZ-011989 trifluoroacetate can be used for the research of various cancers.
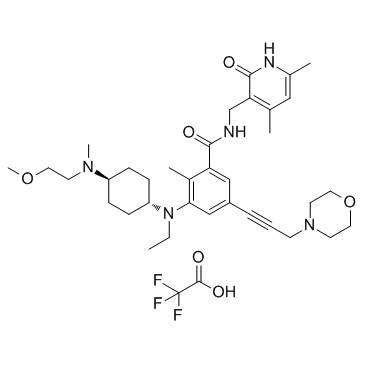
-
GC15302
EPZ015666
PRMT5 inhibitor
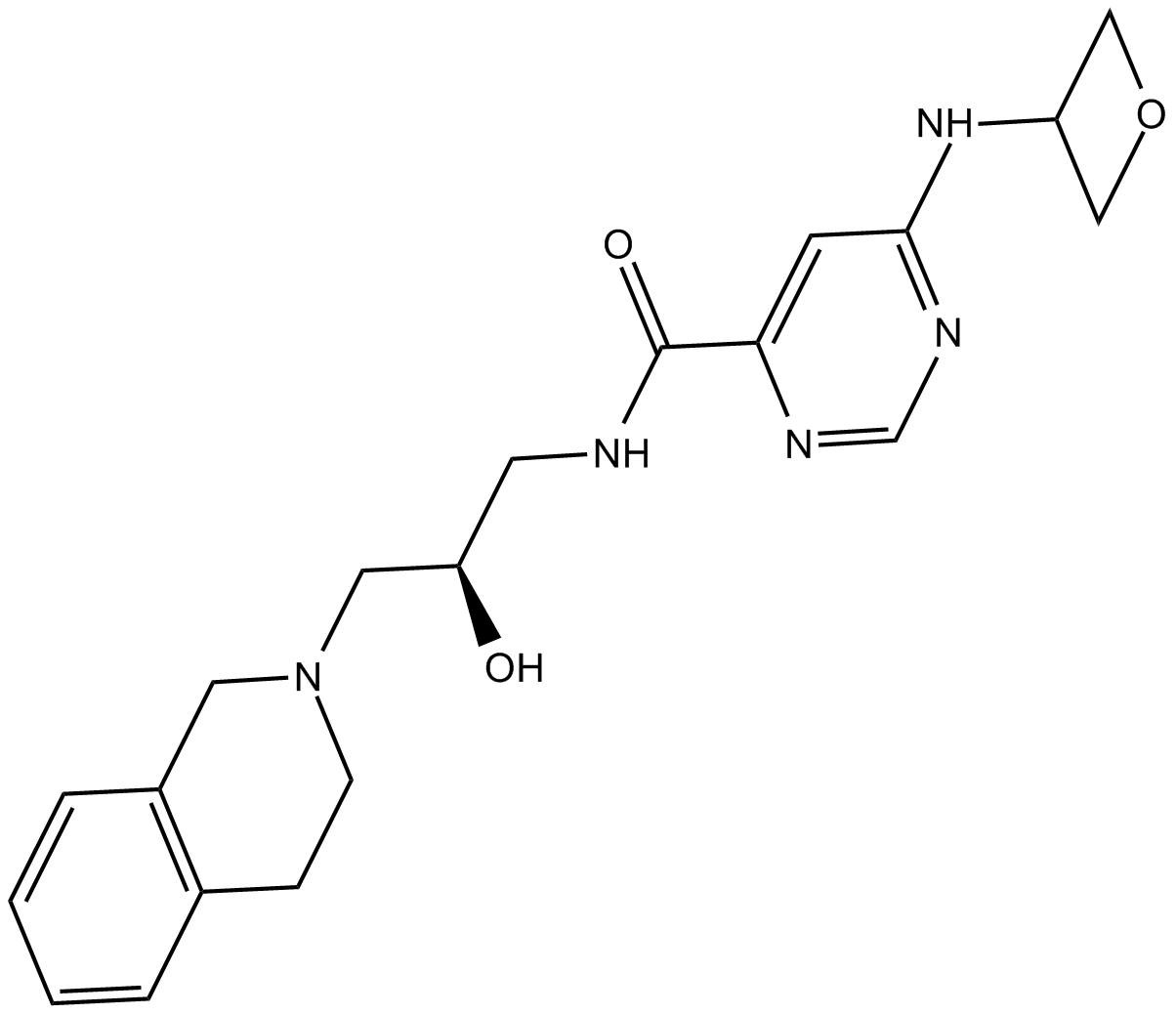
-
GC15102
EPZ020411
PRMT6 inhibitor
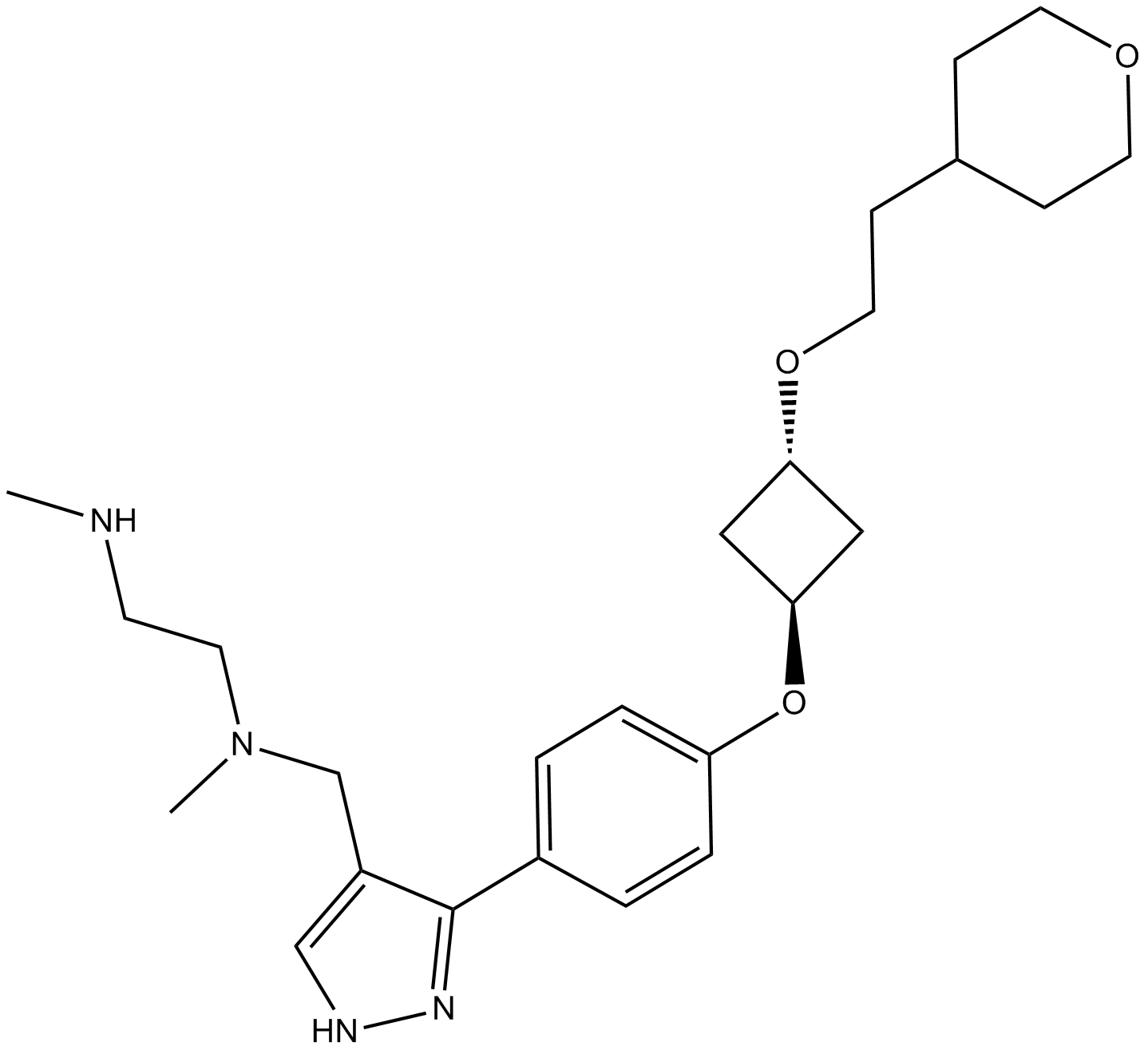
-
GC36000
EPZ020411 hydrochloride
EPZ020411 hydrochloride is a selective inhibitor of PRMT6 with an IC50 of 10 nM, it has >10 folds selectivity for PRMT6 over PRMT1 and PRMT8. EPZ020411 hydrochloride can be used for the research of cancer.
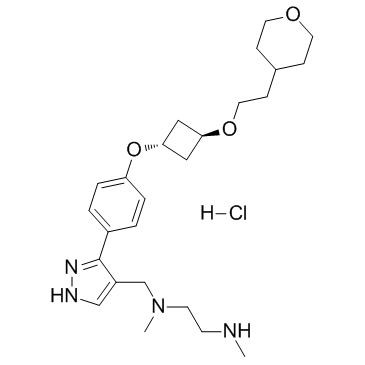
-
GC16224
EPZ031686
A SMYD3 inhibitor
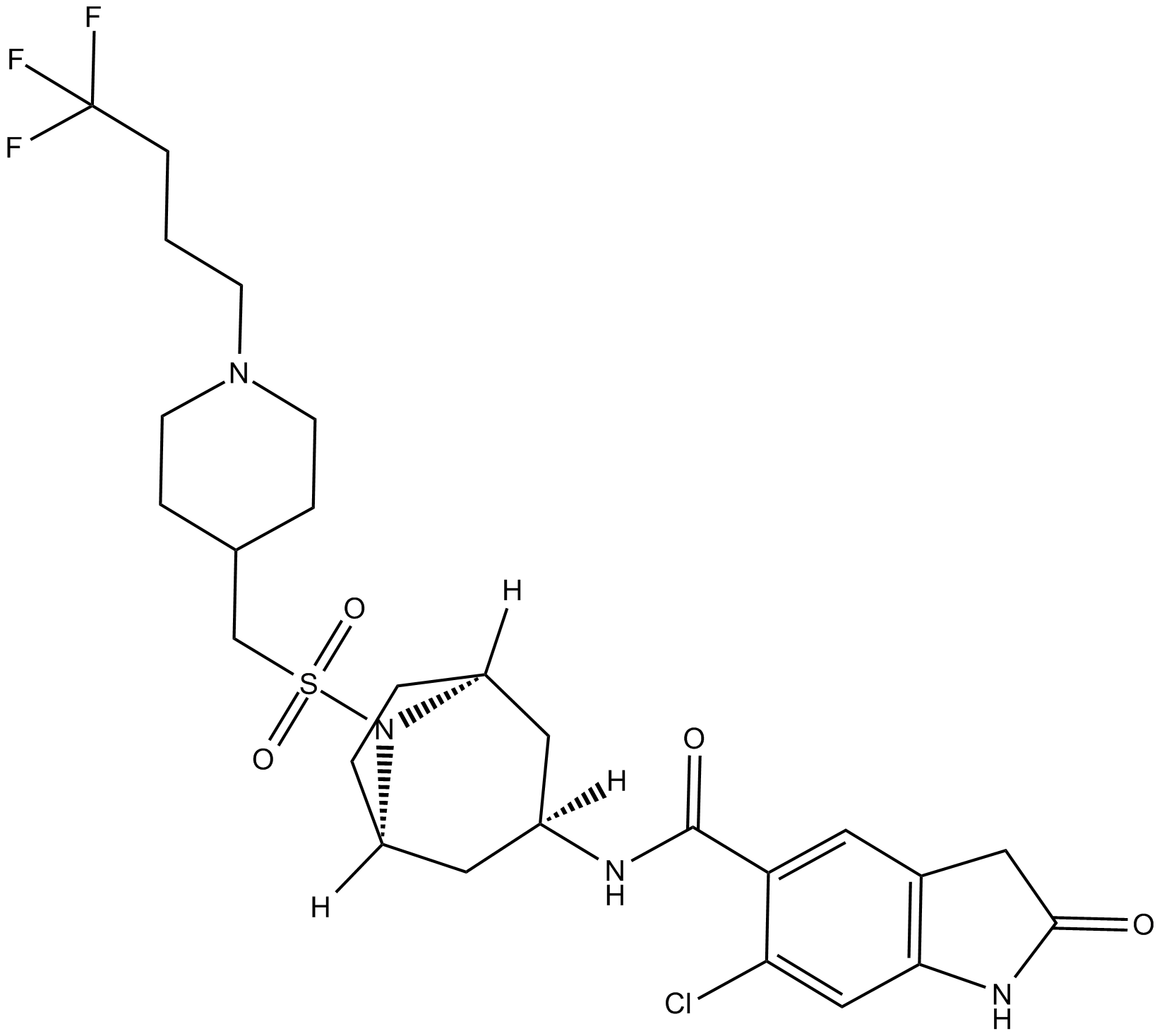
-
GC12932
EPZ5676
A highly potent DOT1L inhibitor
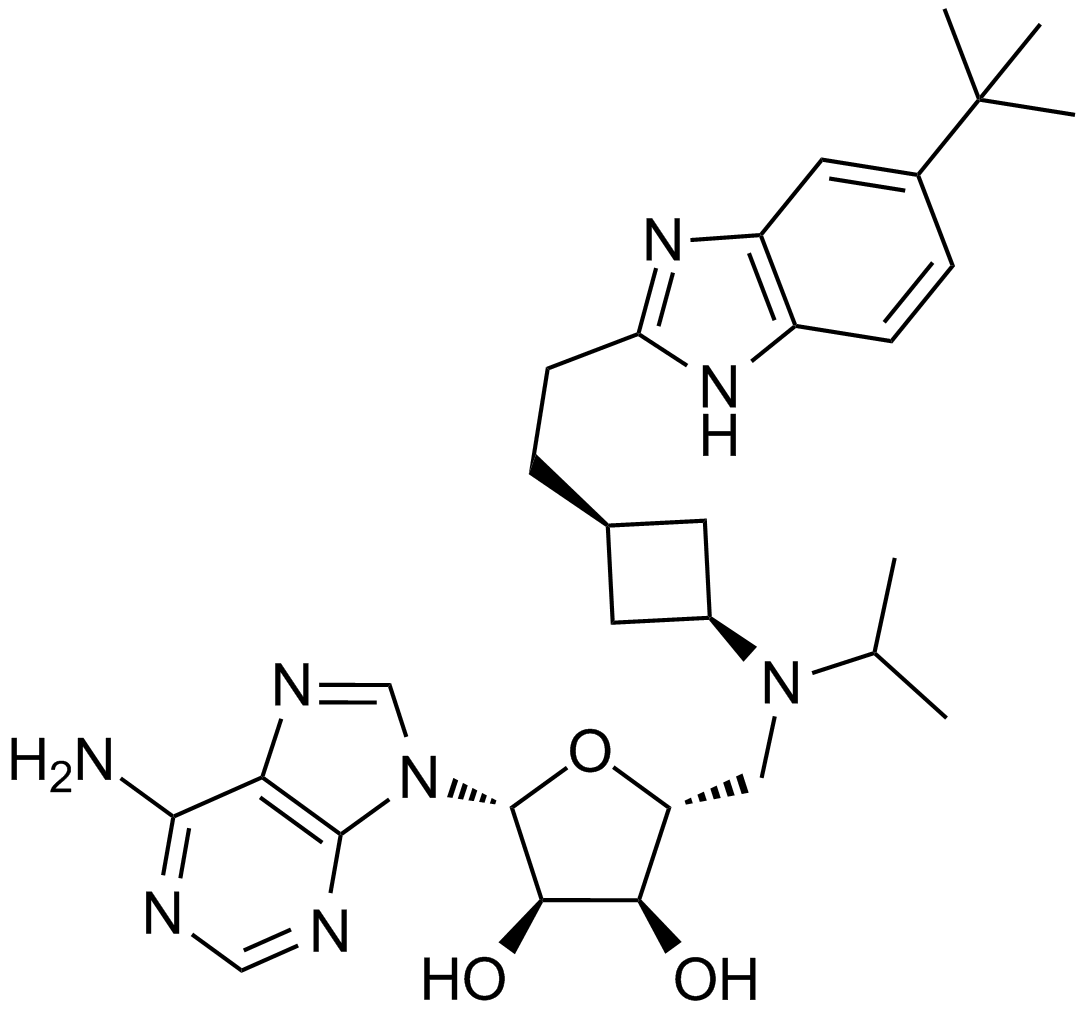
-
GC65980
EZH2-IN-13
EZH2-IN-13 is a potent EZH2 inhibitor, for details please refer to compound 73 in patent WO2017139404. EZH2-IN-13 can be used to study cancers or precancerous lesions associated with EZH2 activity.
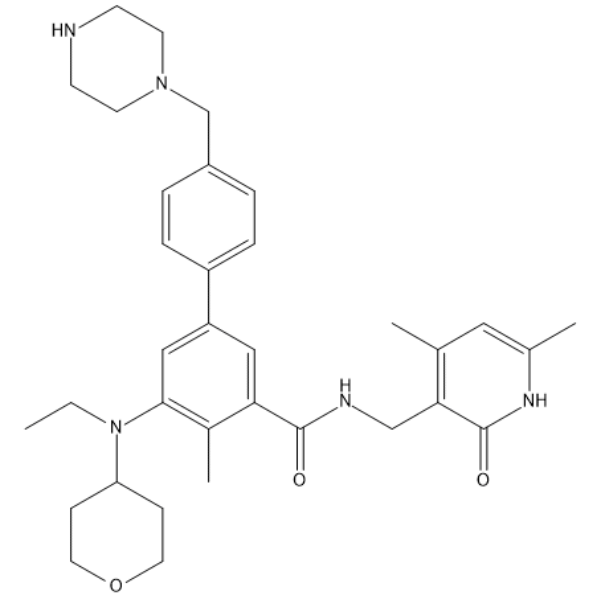
-
GC19149
EZM 2302
EZM 2302 is an inhibitor of coactivator-associated arginine methyltransferase 1 (CARM1) with an IC50 of 6 nM.
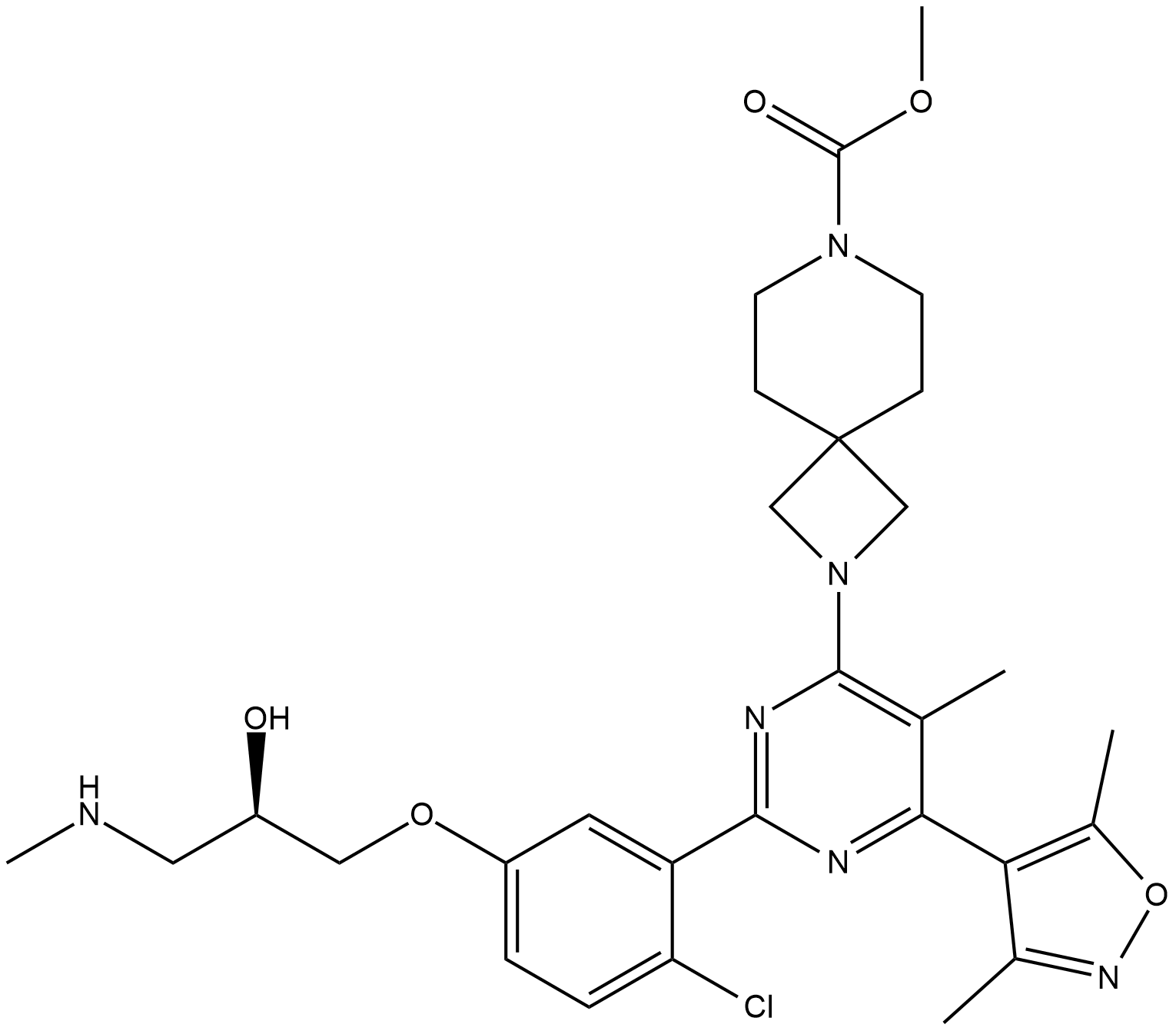
-
GC64900
EZM0414
EZM0414 is a potent, selective, orally bioavailable inhibitor of SETD2 (IC50=18 nM in SETD2 biochemical assay; IC50=34 nM in cellular assay). EZM0414 can be used for the research of relapsed or refractory multiple myeloma and diffuse large B-cell lymphoma.
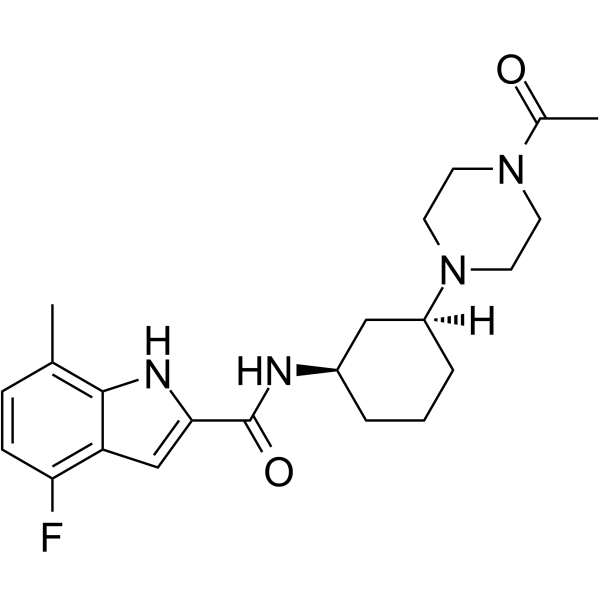
-
GC63545
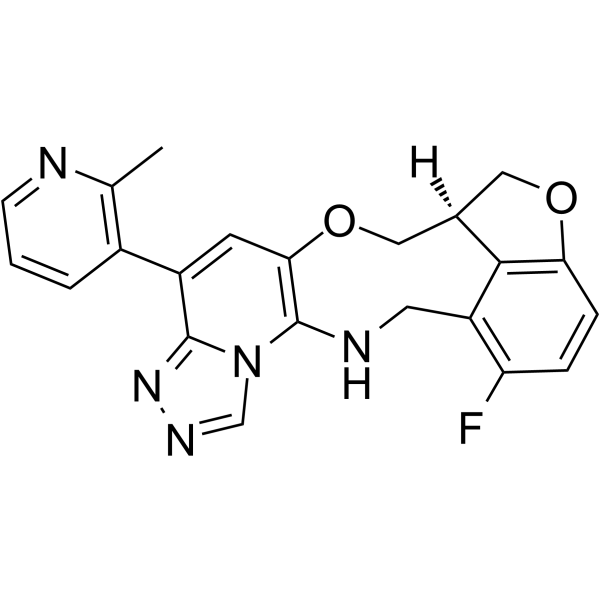
FTX-6058
FTX-6058 ((S)-FTX-6058) is a potent and orally active inhibitor of Embryonic Ectoderm Development (EED).
-
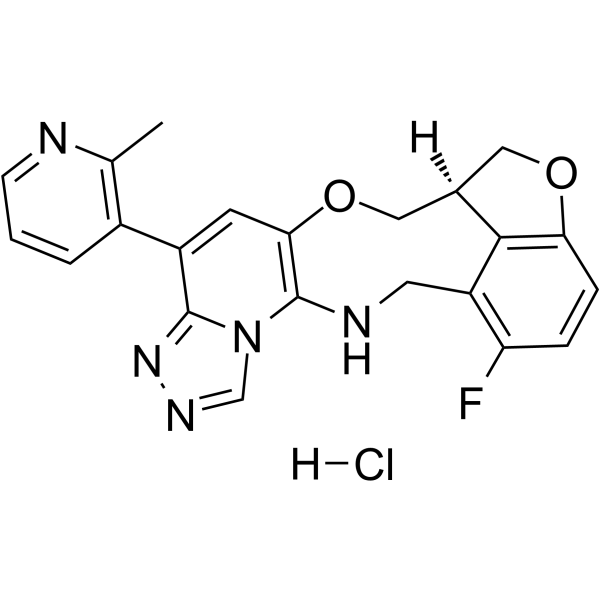
GC63546
FTX-6058 hydrochloride
(S)-Pociredir ((S)-FTX-6058) hydrochloride is a potent and orally active inhibitor of Embryonic Ectoderm Development (EED).
-
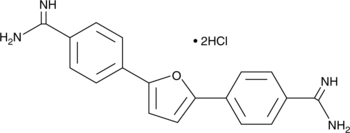
GC43715
Furamidine (hydrochloride)
Furamidine (hydrochloride) (DB75 dihydrochloride) is a selective protein arginine methyltransferase 1 (PRMT1) inhibitor with an IC50 of 9.4 μM.
-
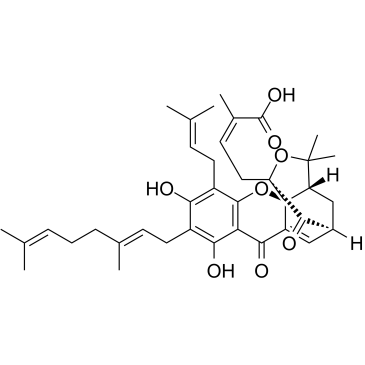
GC38062
Gambogenic acid
Gambogenic acid is an active ingredient in gamboge, with anticancer activity. Gambogenic acid acts as an effective inhibitor of EZH2, specifically and covalently binds to Cys668 within the EZH2-SET domain, and induces EZH2 ubiquitination.
-
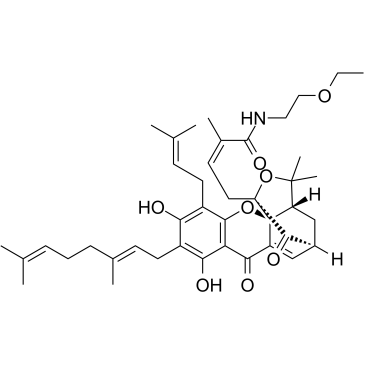
GC36168
GNA002
GNA002 is a highly potent, specific and covalent EZH2 (Enhancer of zeste homolog 2) inhibitor with an IC50 of 1.1 μM. GNA002 can specifically and covalently bind to Cys668 within the EZH2-SET domain, triggering EZH2 degradation through COOH terminus of Hsp70-interacting protein (CHIP)-mediated ubiquitination. GNA002 efficiently reduces EZH2-mediated H3K27 trimethylation, reactivates polycomb repressor complex 2 (PRC2)-silenced tumor suppressor genes.
-
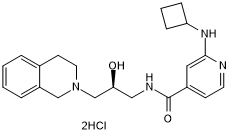
GC50697
GSK 591 dihydrochloride
-
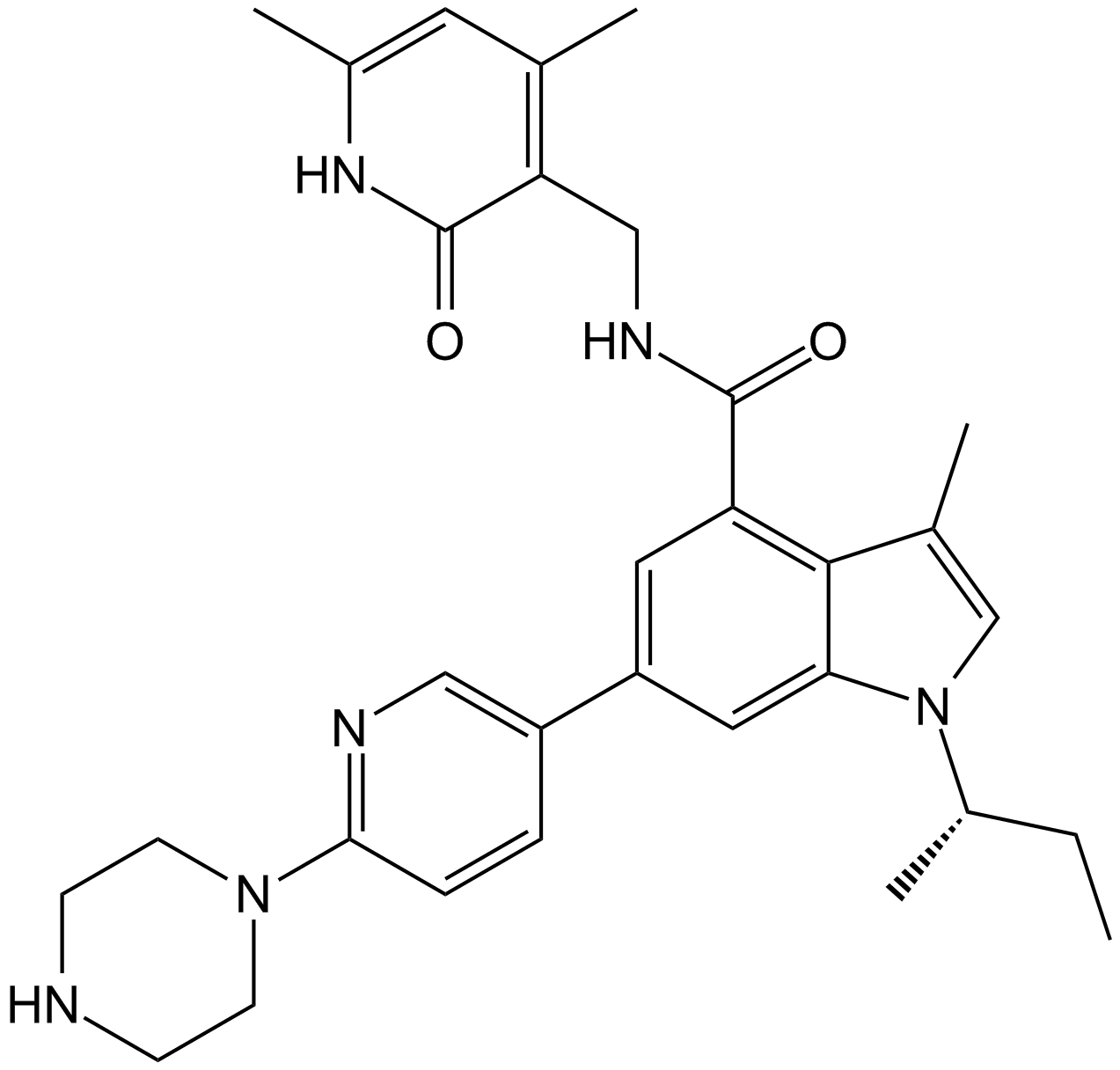
GC15783
GSK126
A selective EZH2 inhibitor
-
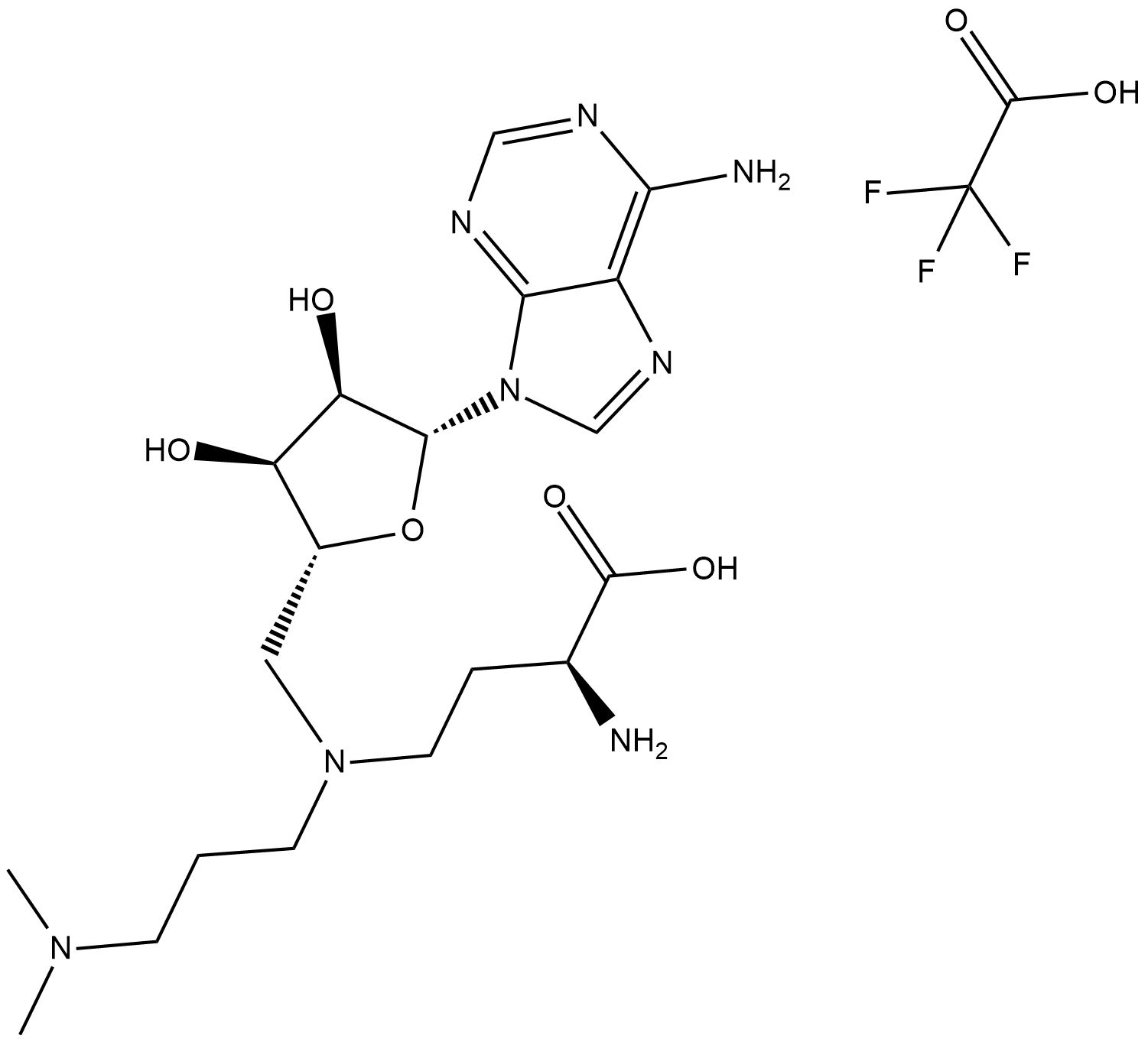
GC19181
GSK2807 Trifluoroacetate
GSK2807 Trifluoroacetate is a potent, selective and SAM-competitive inhibitor of SMYD3, with a Ki of 14 nM.
-
GC32693
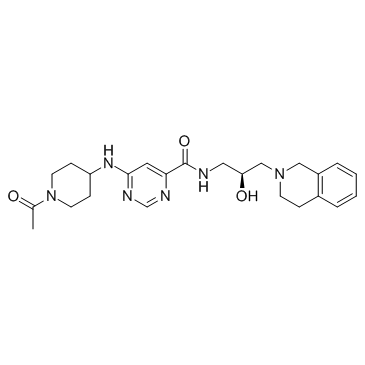
GSK3326595 (EPZ015938)
GSK3326595 (EPZ015938) (EPZ015938) is a potent, selective, reversible inhibitor of protein arginine methyltransferase 5 (PRMT5) with an IC50 of 6.2 nM.
-
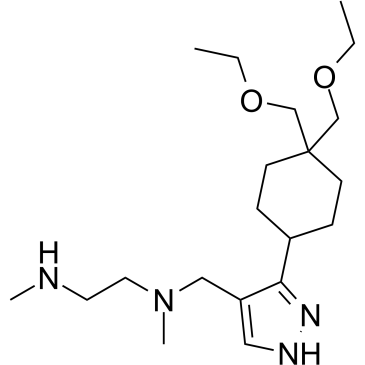
GC36191
GSK3368715
GSK3368715 (EPZ019997) is an orally active, reversible, and S-adenosyl-L-methionine (SAM) uncompetitive type I protein arginine methyltransferases (PRMTs) inhibitor (IC50=3.1 nM (PRMT1), 48 nM (PRMT3), 1148 nM (PRMT4), 5.7 nM (PRMT6), 1.7 nM (PRMT8)). GSK3368715 (EPZ019997) produces a shift in arginine methylation states, alters exon usage, and has strong anti-cancer activity.
-
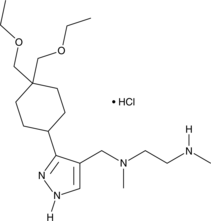
GC49398
GSK3368715 (hydrochloride)
An inhibitor of type I PRMTs
-
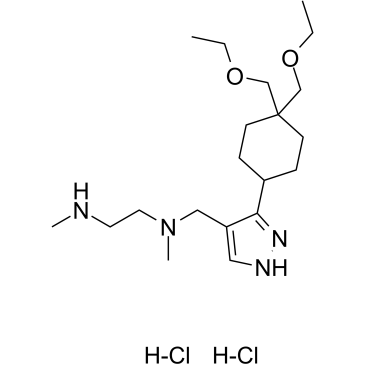
GC36192
GSK3368715 dihydrochloride
GSK3368715 dihydrochloride (EPZ019997 dihydrochloride) is an orally active, reversible, and S-adenosyl-L-methionine (SAM) uncompetitive type I protein arginine methyltransferases (PRMTs) inhibitor (IC50=3.1 nM (PRMT1), 48 nM (PRMT3), 1148 nM (PRMT4), 5.7 nM (PRMT6), 1.7 nM (PRMT8)). GSK3368715 dihydrochloride (EPZ019997 dihydrochloride) produces a shift in arginine methylation states, alters exon usage, and has strong anti-cancer activity.
-
GC17534
GSK343
A selective, cell-permeable EZH2 inhibitor
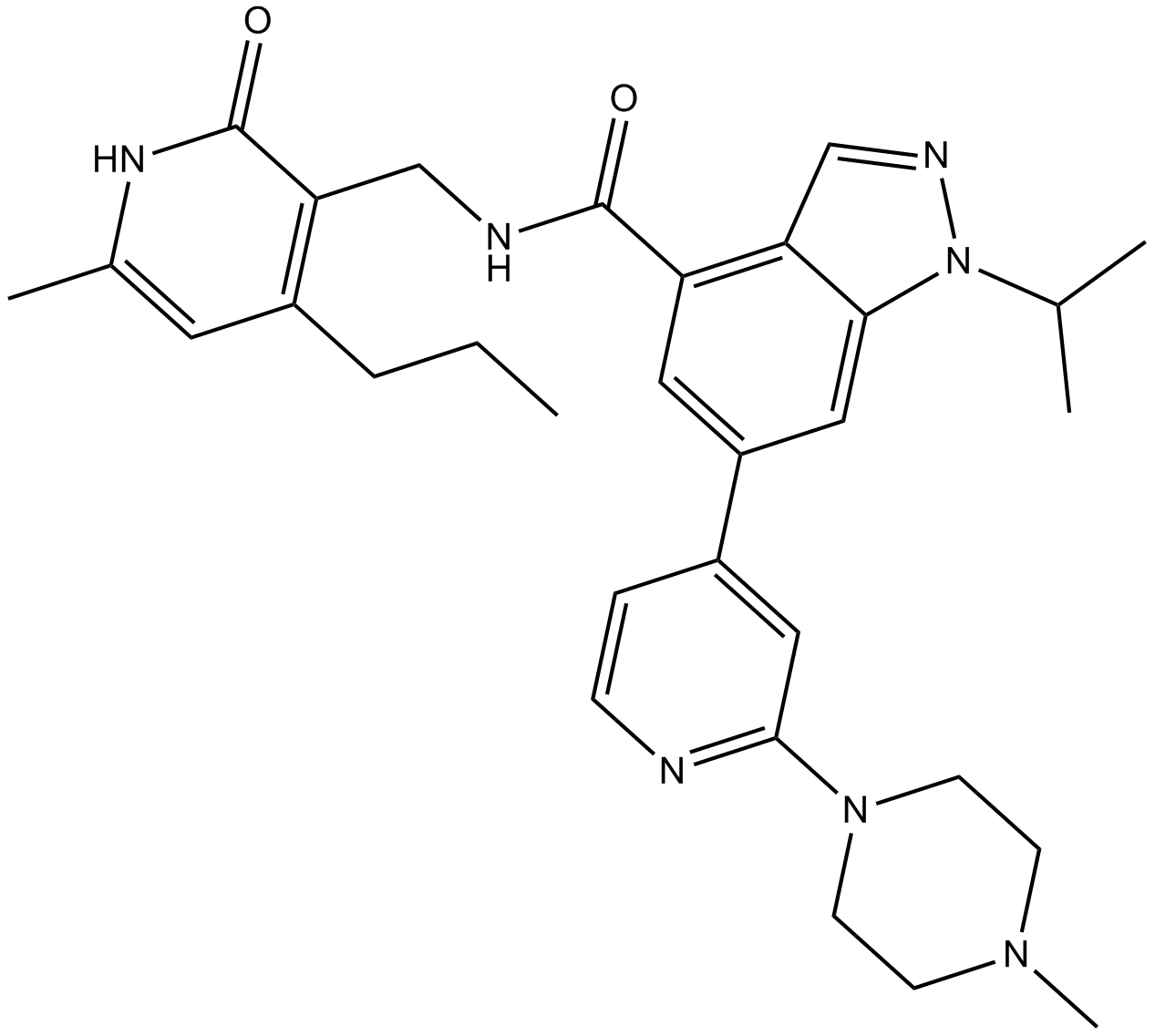
-
GC11414
GSK503
EZH2 inhibitor