Histone Methyltransferase
Histone methyltransferases are a group of enzymes that catalyze the methylation of histone lysine and arginine by adding methyl groups to specific histone arginine or lysine residues. Histone methyltransferaes can be classified into 3 classes, including SET domain lysine methyltransferases, non-SET domain lysine methyltransferases and arginine methyltransferases (PRMTs), all of which use S-adenosylmethionine as a cosubstrate for the transfer of the methyl group. Aberrant histone methylation has been associated with a wide range of human cancers (such as hematological malignancies), which leads to the development of novel cancer chemotherapies targeting cancer-associated histone methyltransferases (more than 20 lysine methyltransferases and 9 arginine methyltransferases in humans).
Products for Histone Methyltransferase
- Cat.No. Product Name Information
-
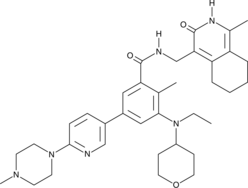
GC14585
GSK591
PRMT5 inhibitor
-
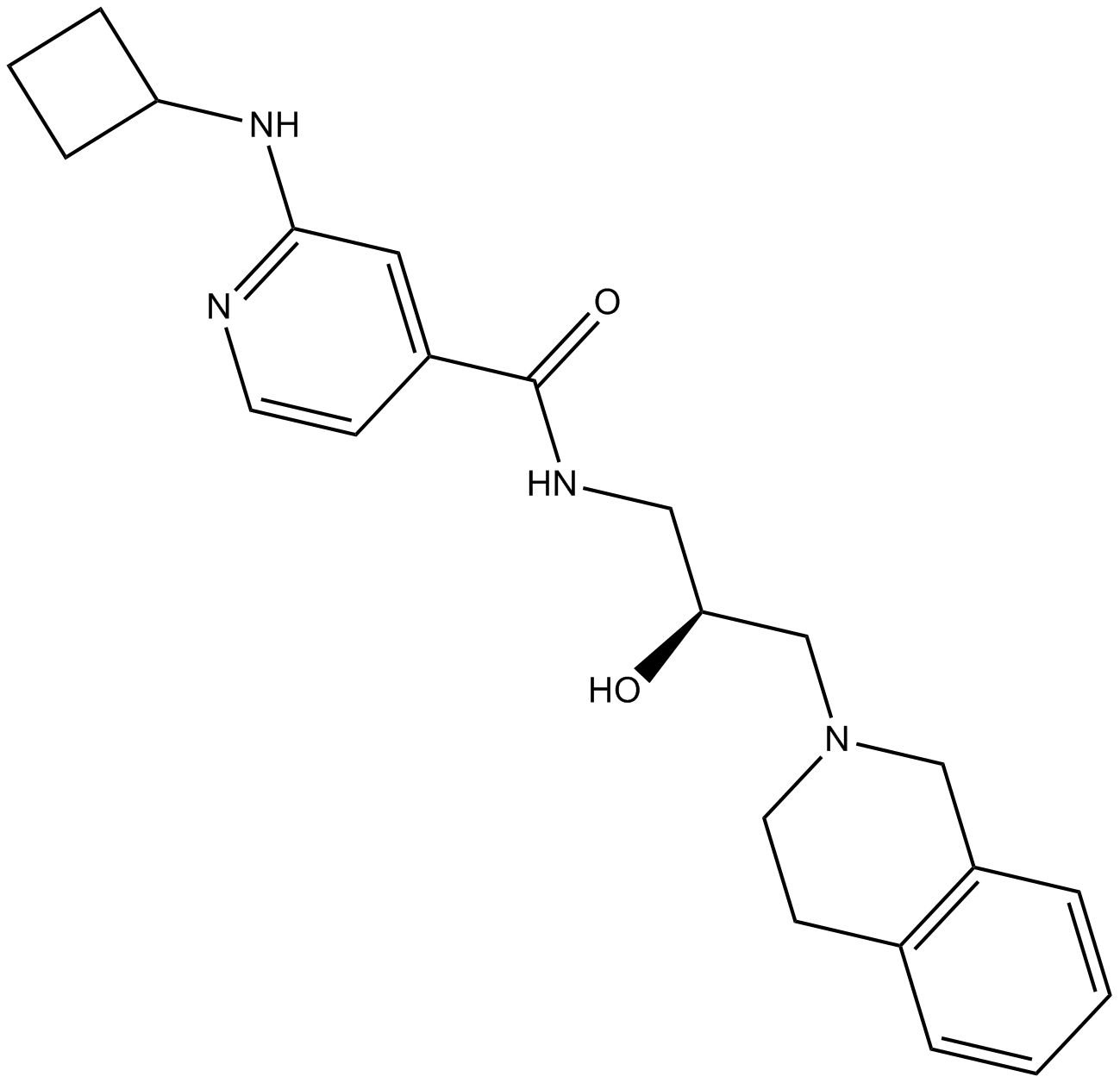
GP10020
Histone-H2A-(107-122)-Ac-OH
Histone-H2A peptide
-
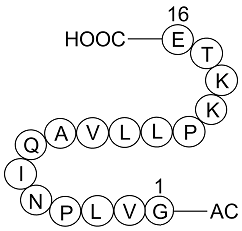
GC17023
HLCL-61
HLCL-61 is a first-in-class inhibitor of protein arginine methyltransferase 5 (PRMT5).
-
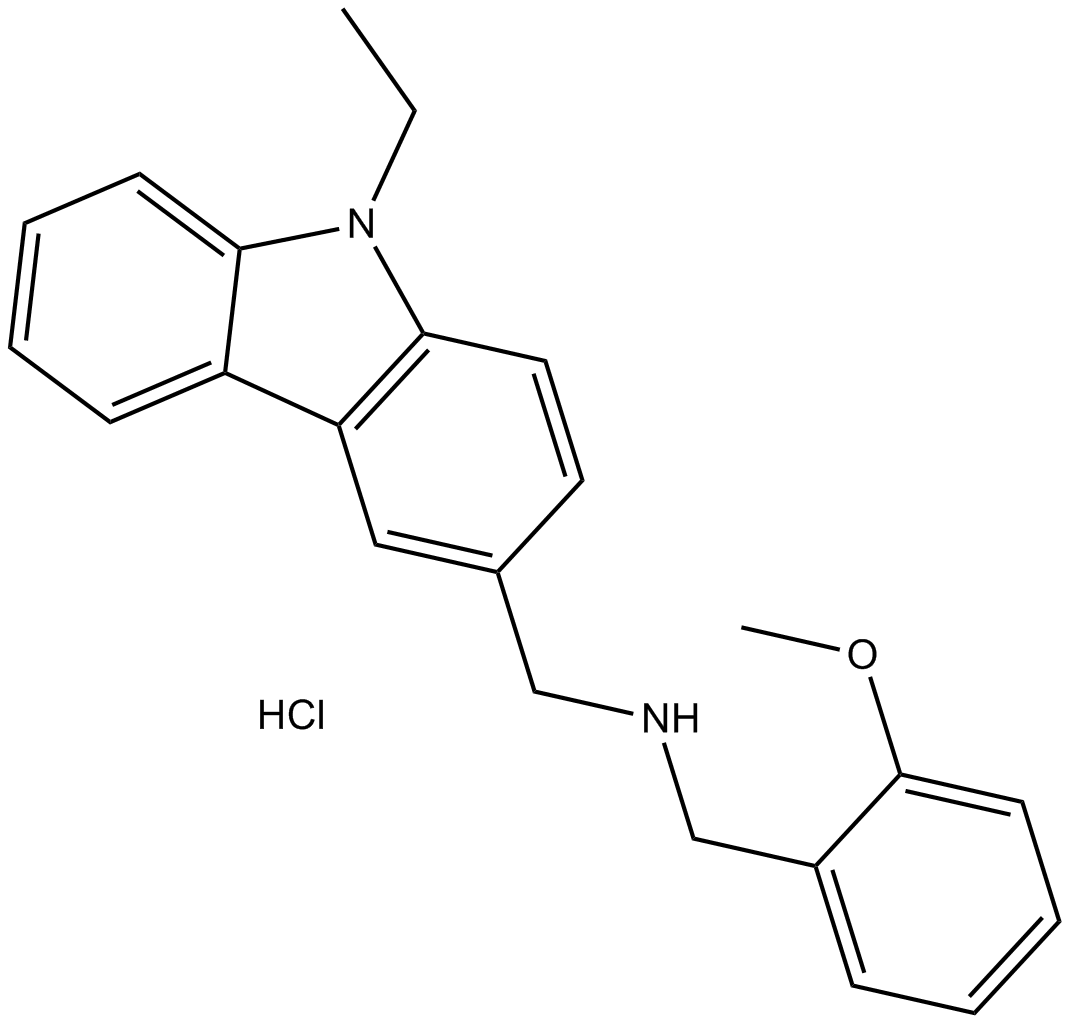
GC19465
JNJ-64619178
JNJ-64619178 is a PRMT5 inhibitor

-
GC19211
JQEZ5
JQEZ5 is a novel and potent EZH2 inhibitor.
-
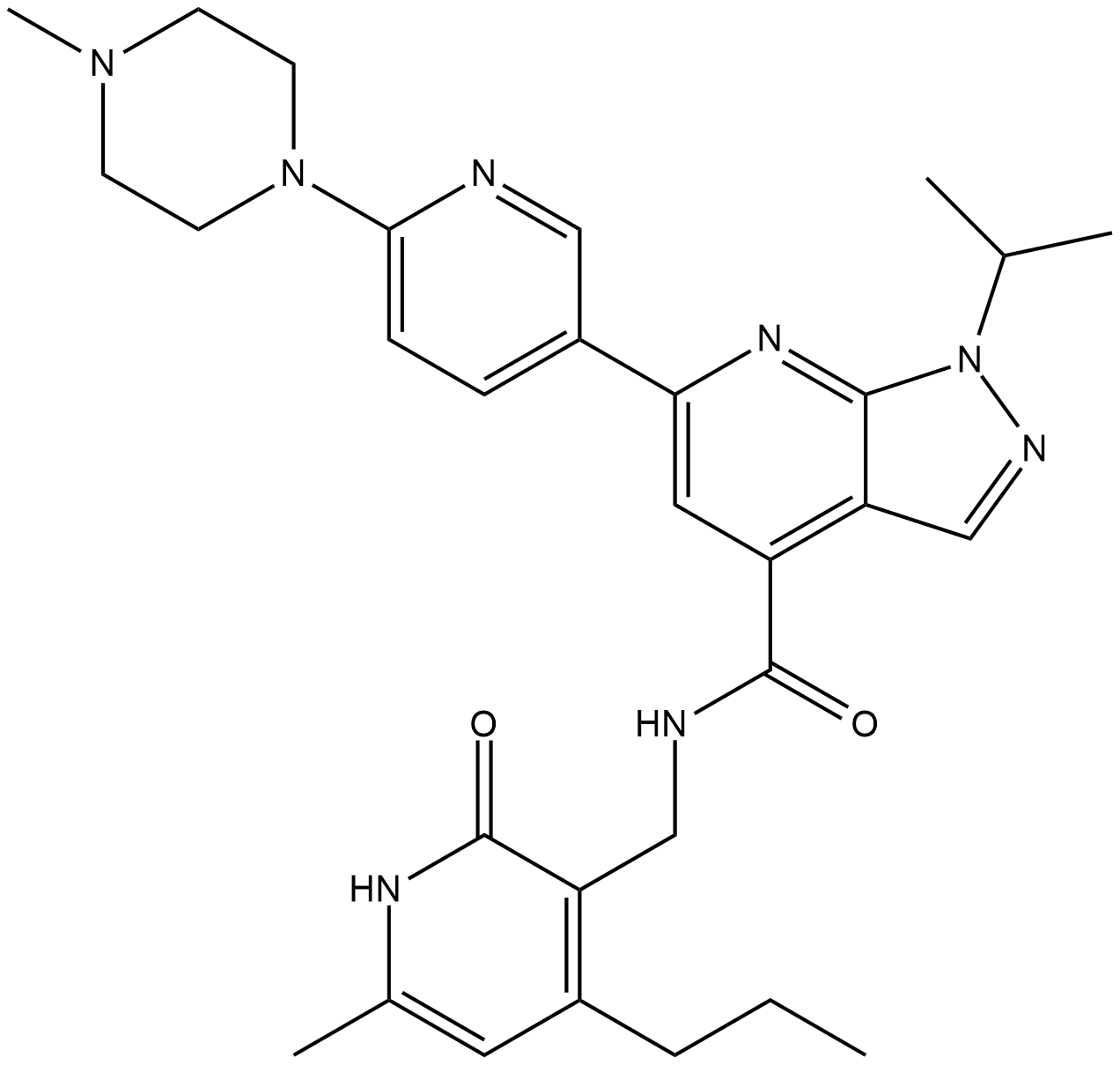
GC33184
LLY-283
A PRMT5 inhibitor
-
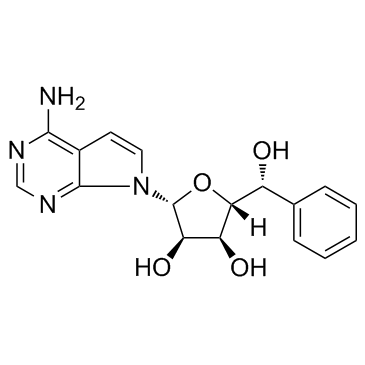
GC16261
LLY507
SMYD2 inhibitor
-
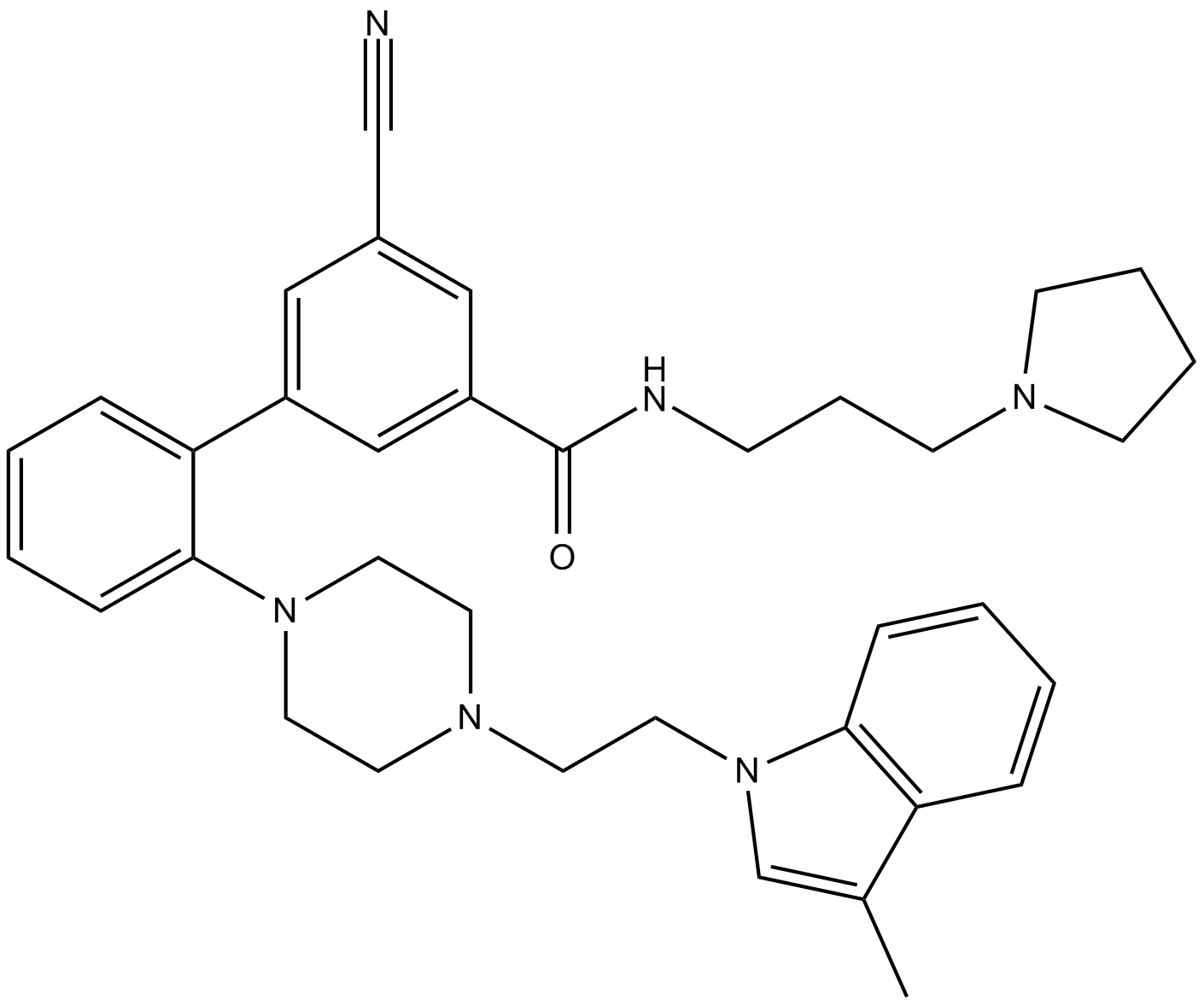
GC63056
MAK683-CH2CH2COOH
MAK683-CH2CH2COOH binds to EED (embryonic ectoderm development protein). MAK683-CH2CH2COOH and a VHL ligand for the E3 ubiquitin ligase have been used to design PROTAC EED degrader-1 and PROTAC EED degrader-2.
-
GC65254
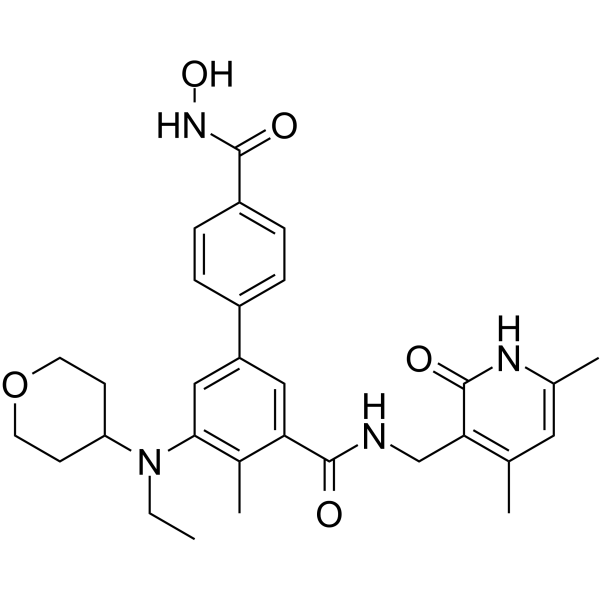
MC4355
MC4355 is a dual inhibitor of EZH2 and histone deacetylase (HDAC).
-
GC61058
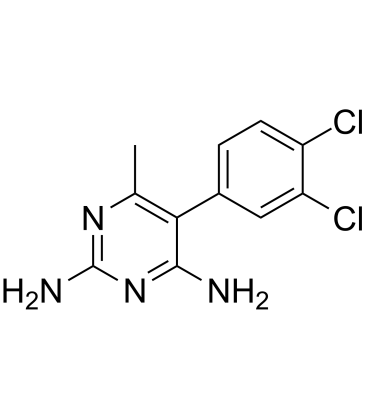
Metoprine
Metoprine (BW 197U) is a potent histamine N-methyltransferase (HMT) inhibitor.
-
GC14652
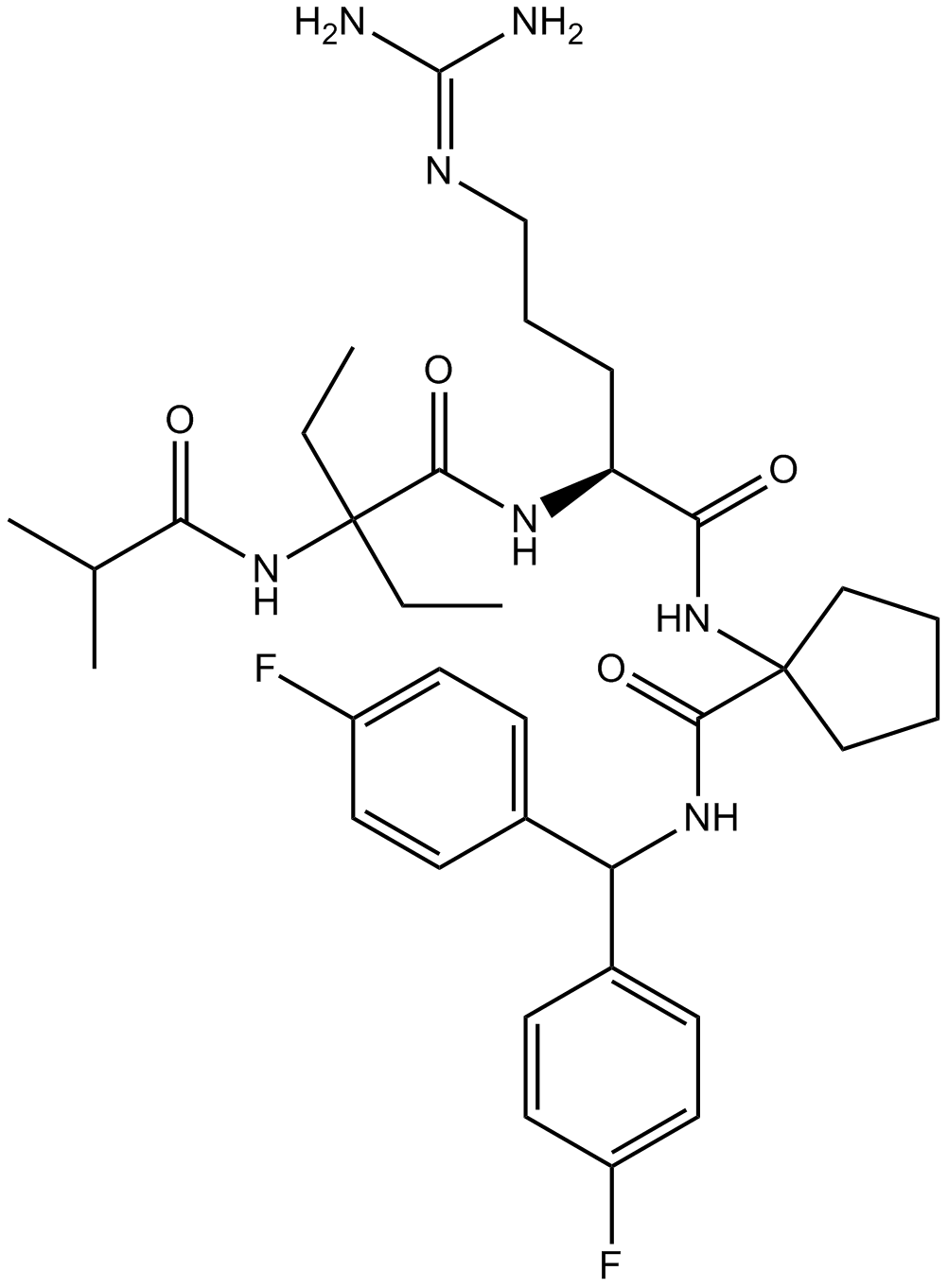
MM-102
MLL1 inhibitor,high-affinity peptidomimetic
-
GC36632
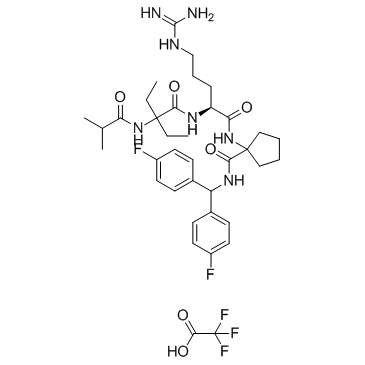
MM-102 TFA
MM-102 TFA (HMTase Inhibitor IX TFA) is a potent WDR5/MLL interaction inhibitor, achieves IC50 = 2.4 nM with an estimated Ki 200 times more potent than the ARA peptide.
-
GC67758
MM-401 TFA
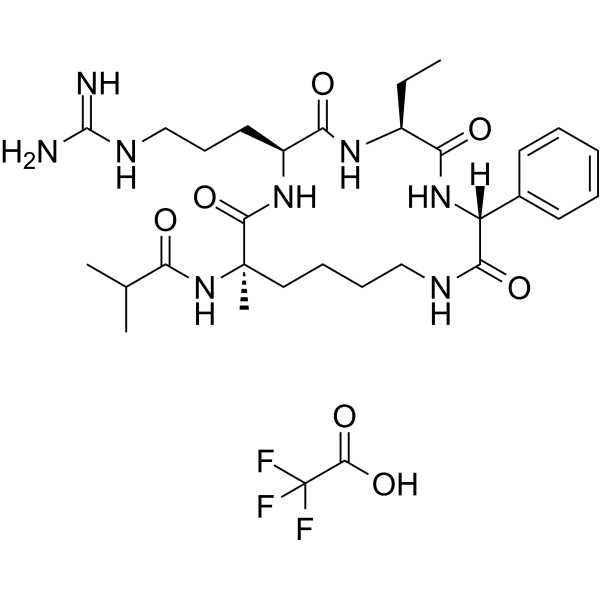
-
GC33278
MM-589
MM-589 is a potent inhibitor of WD repeat domain 5 (WDR5) and mixed lineage leukemia (MLL) protein-protein interaction. MM-589 binds to WDR5 with an IC50 of 0.90 nM and inhibits the MLL H3K4 methyltransferase activity with an IC50 of 12.7 nM.
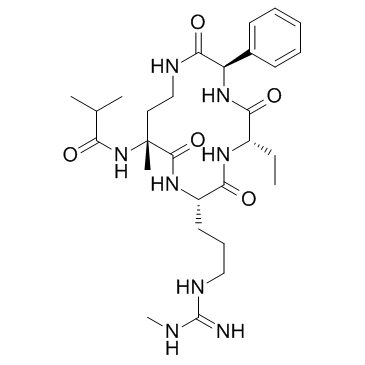
-
GC65037
MM-589 TFA
MM-589 TFA is a potent inhibitor of WD repeat domain 5 (WDR5) and mixed lineage leukemia (MLL) protein-protein interaction. MM-589 binds to WDR5 with an IC50 of 0.90 nM and inhibits the MLL H3K4 methyltransferase activity with an IC50 of 12.7 nM.
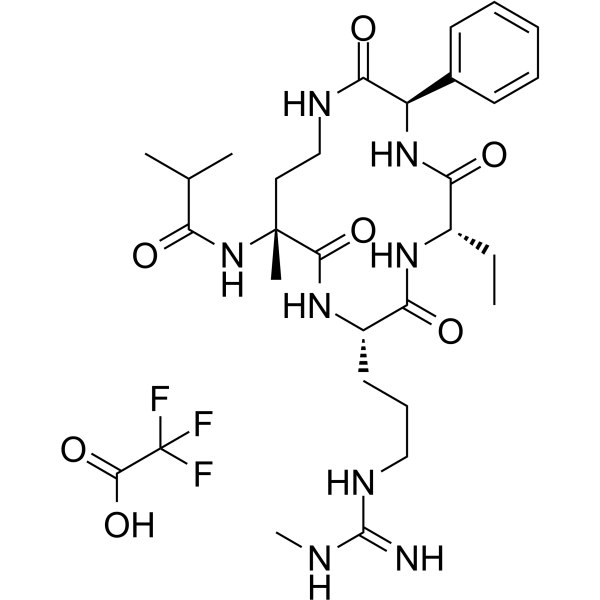
-
GC61468
MR837
MR837 is an inhibitor of NSD2-PWWP1. MR837 can bind with human nuclear receptor binding SET domain protein 2 (PWWP domain).
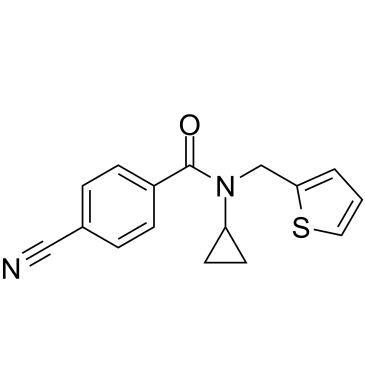
-
GC50576
MRK 740
Potent PRDM9 inhibitor
-
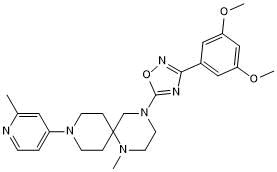
GC63558
MRTX-1719
MRTX-1719 is a potent first-in-class selective inhibitor of the PRMT5/MTA complex, with an IC50 of less than 10 nM in PRMT5/MTA MTAPDEL SDMA cells.
-
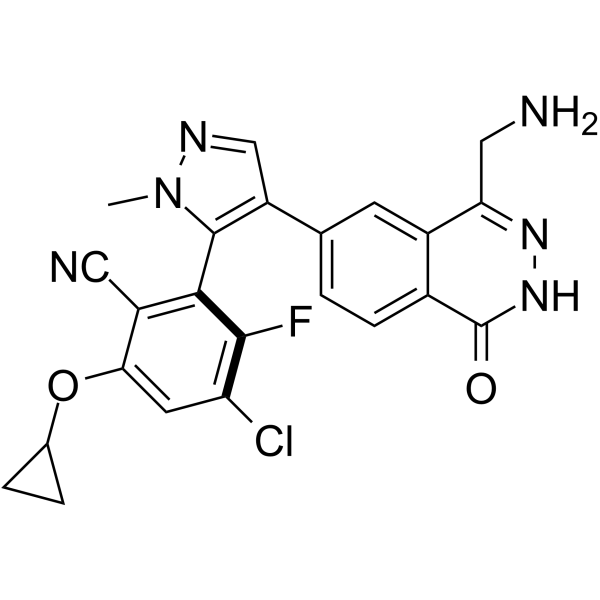
GC69499
MRTX-1719 hydrochloride
MRTX-1719 hydrochloride is an effective, novel, and selective inhibitor of the PRMT5/MTA complex. Its IC50 value for PRMT5/MTA MTAPDEL SDMA cell lines is <10 nM.
-
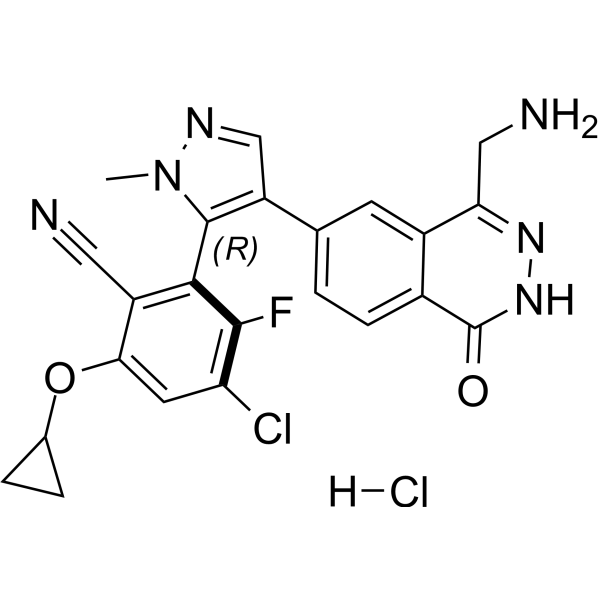
GC62715
MRTX9768
MRTX9768 is a potent, selective, orally active, first-in-class PRMT5-MTA complex inhibitor.
-
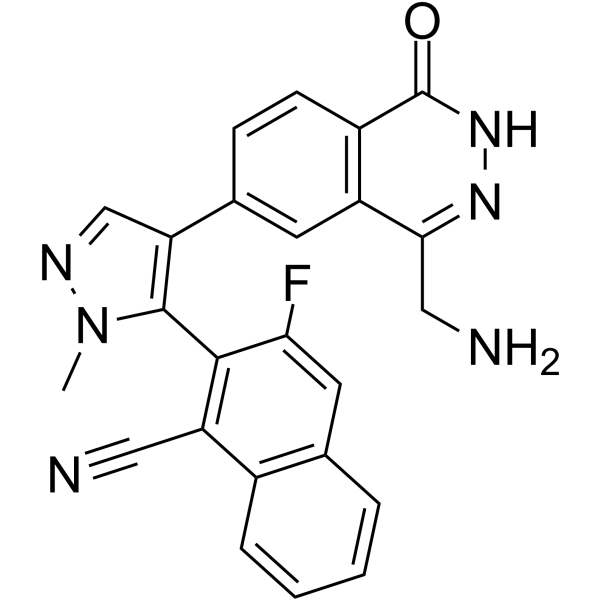
GC63080
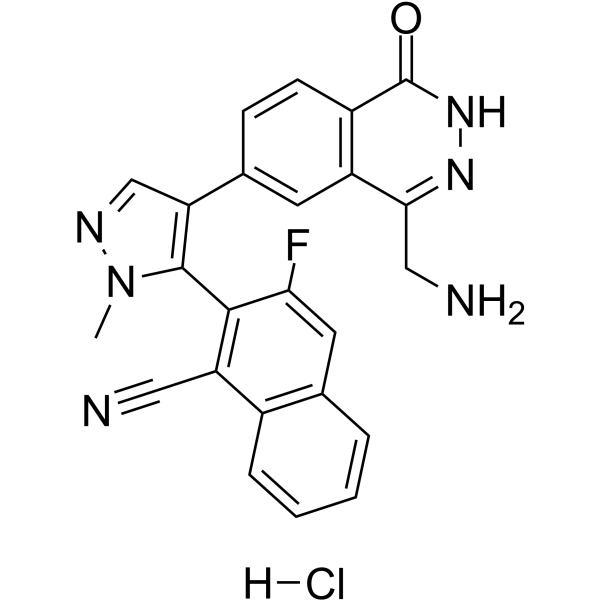
MRTX9768 hydrochloride
MRTX9768 hydrochloride is a potent, selective, orally active, first-in-class PRMT5-MTA complex inhibitor.
-
GC10099
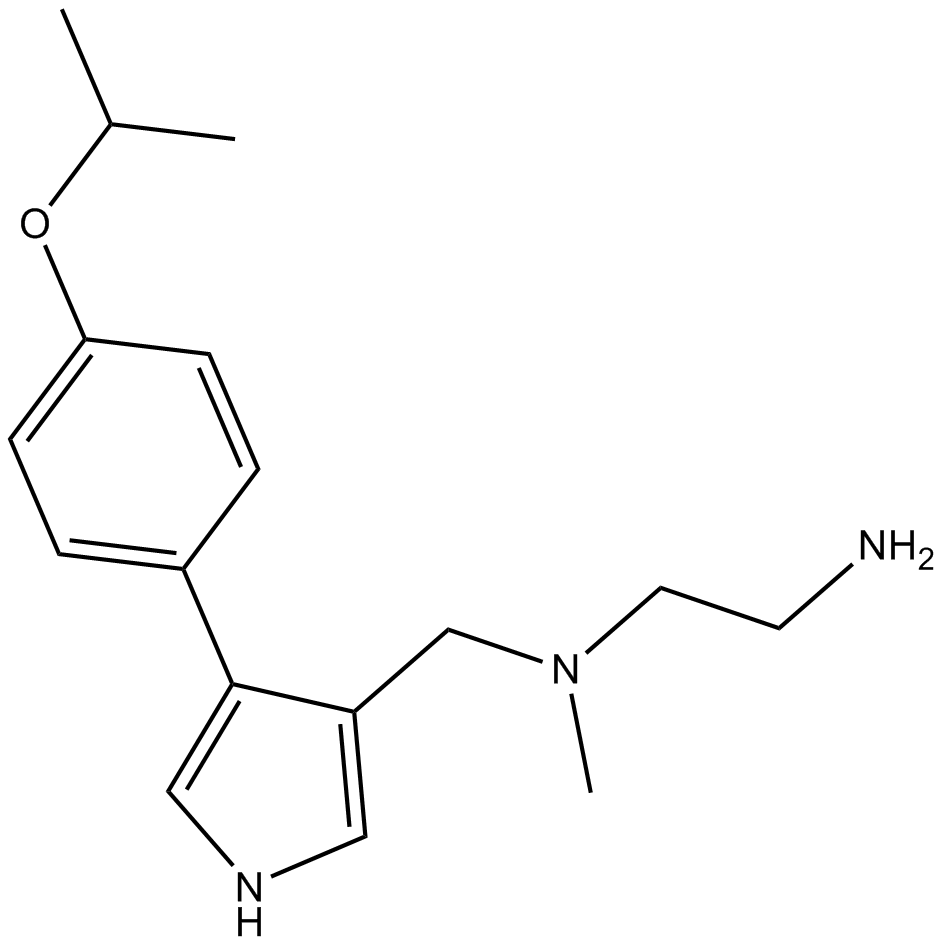
MS023
MS023 is a potent, selective, and cell-active inhibitor of type I protein arginine methyltransferases (PRMTs).
-
GC16432
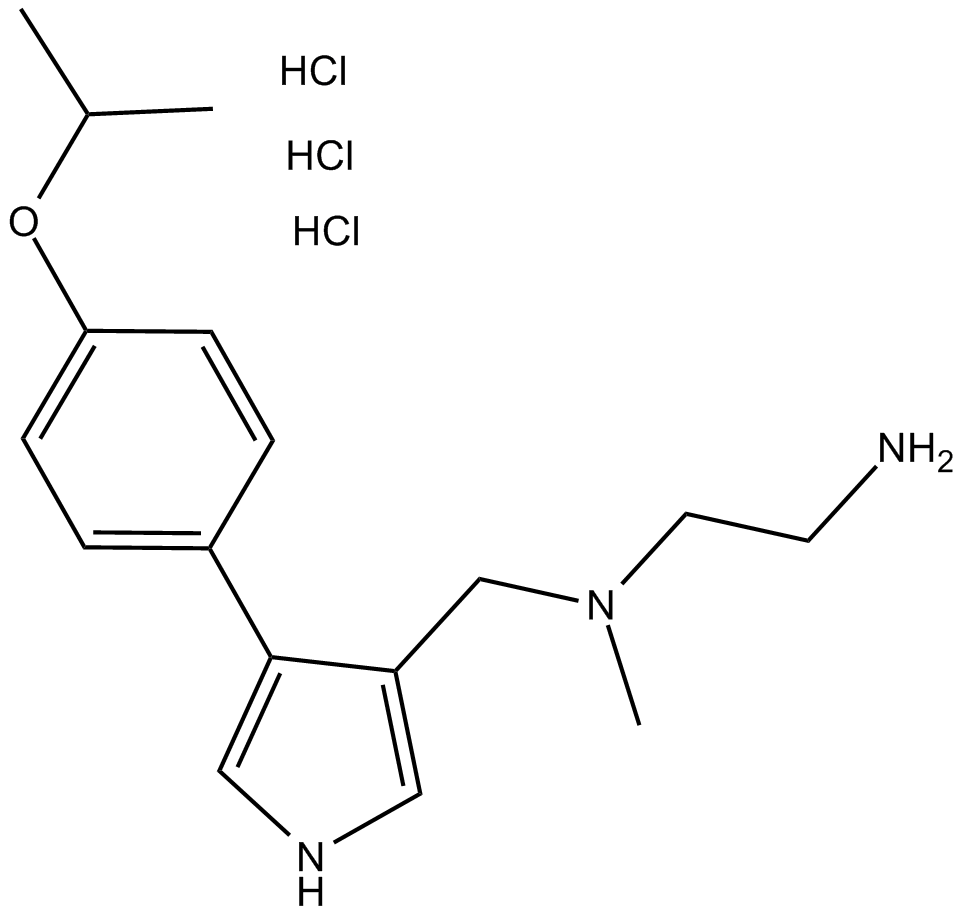
MS023 (hydrochloride)
type I PRMTs inhibitor
-
GC36655
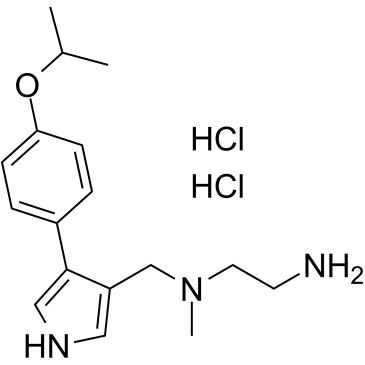
MS023 dihydrochloride
MS023 dihydrochloride is a potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases (PRMTs) inhibitor, with IC50s of 30, 119, 83, 4 and 5 nM for PRMT1, PRMT3, PRMT4, PRMT6, and PRMT8, respectively.
-
GC36656
MS049
MS049 is a potent, selective, and cell-active dual inhibitor of PRMT4 and PRMT6 with IC50s of 34 nM and 43 nM, respectively. MS049 reduces levels of Med12me2a and H3R2me2a in HEK293 cells. MS049 is not toxic and does not affect the growth of HEK293 cells.
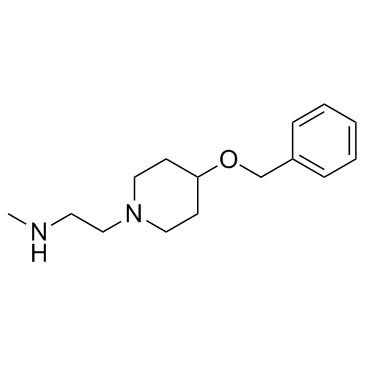
-
GC14240
MS049 (hydrochloride)
MS049 (hydrochloride) is a potent, selective, and cell-active dual inhibitor of PRMT4 and PRMT6 with IC50s of 34 nM and 43 nM, respectively. MS049 (hydrochloride) reduces levels of Med12me2a and H3R2me2a in HEK293 cells. MS049 (hydrochloride) is not toxic and does not affect the growth of HEK293 cells.
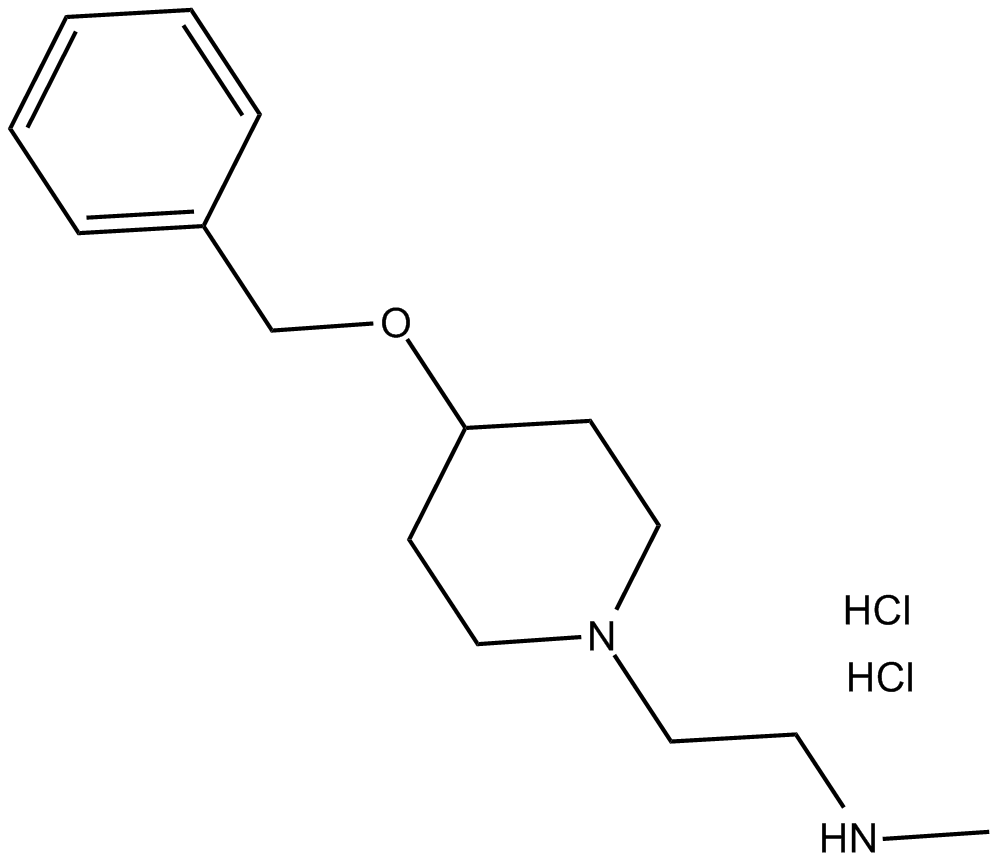
-
GC64295
MS67
MS67 is a potent and selective WD40 repeat domain protein 5 (WDR5) degrader with a Kd of 63 nM. MS67 is inactive against other protein methyltransferases, kinases, GPCRs, ion channels, and transporters. MS67 shows potent acticancer effects.

-
GC69504
MS8815
MS8815 is a selective PROTAC degrader of zeste homolog 2 (EZH2). It has inhibitory activity against EZH2 with an IC50 value of 8.6 nM. MS8815 can be used for research on triple-negative breast cancer (TNBC).
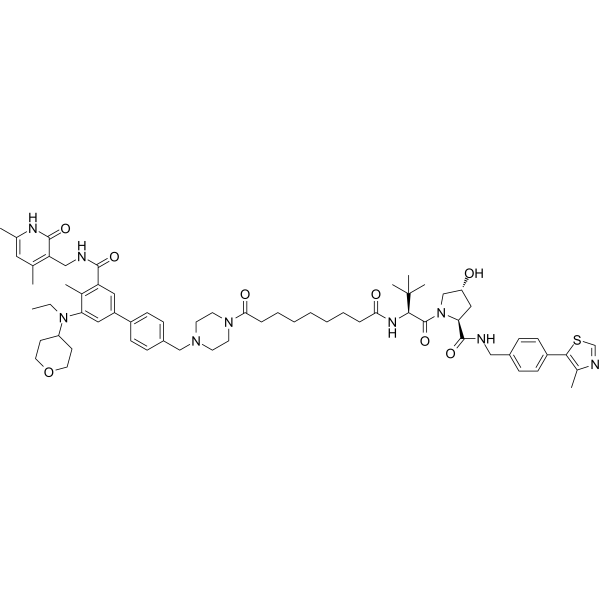
-
GC61142
NSC745885
NSC745885 an effective anti-tumor agent, shows selective toxicity against multiple?cancer?cell lines but not normal cells.?NSC745885 is an effective?down-regulator of EZH2 via proteasome-mediated degradation. NSC745885 provides possibilities for the study of advanced?bladder?and oral squamous cell carcinoma (OSCC) cancers.
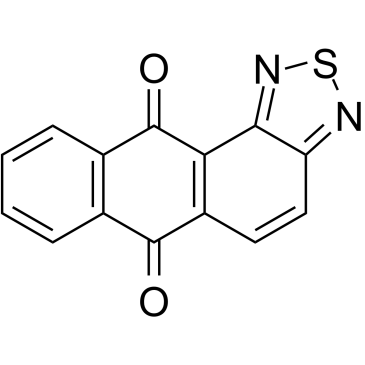
-
GC64772
NV03
NV03 is a potent and selective antagonist of UHRF1 (Ubiquitin-like with PHD and RING finger domains 1)- H3K9me3 interaction by binding to UHRF1 tandem tudor domain, with a Kd of 2.4 μM. NV03 has anticancer activity.
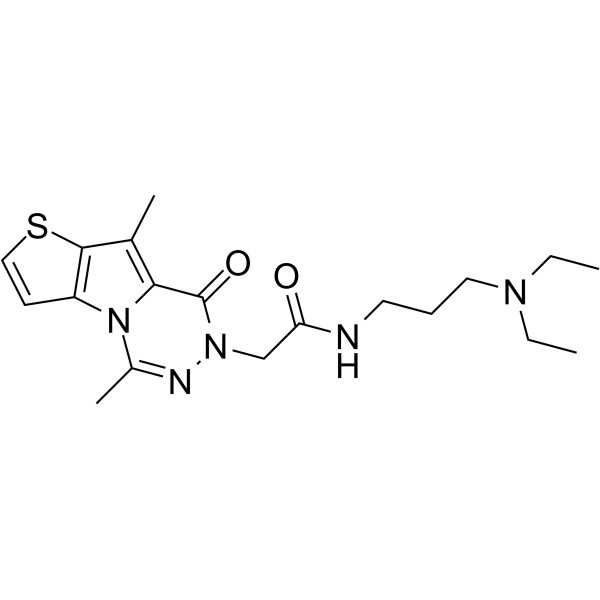
-
GC16397
OICR-9429
Wdr5-MLL interaction antagonist
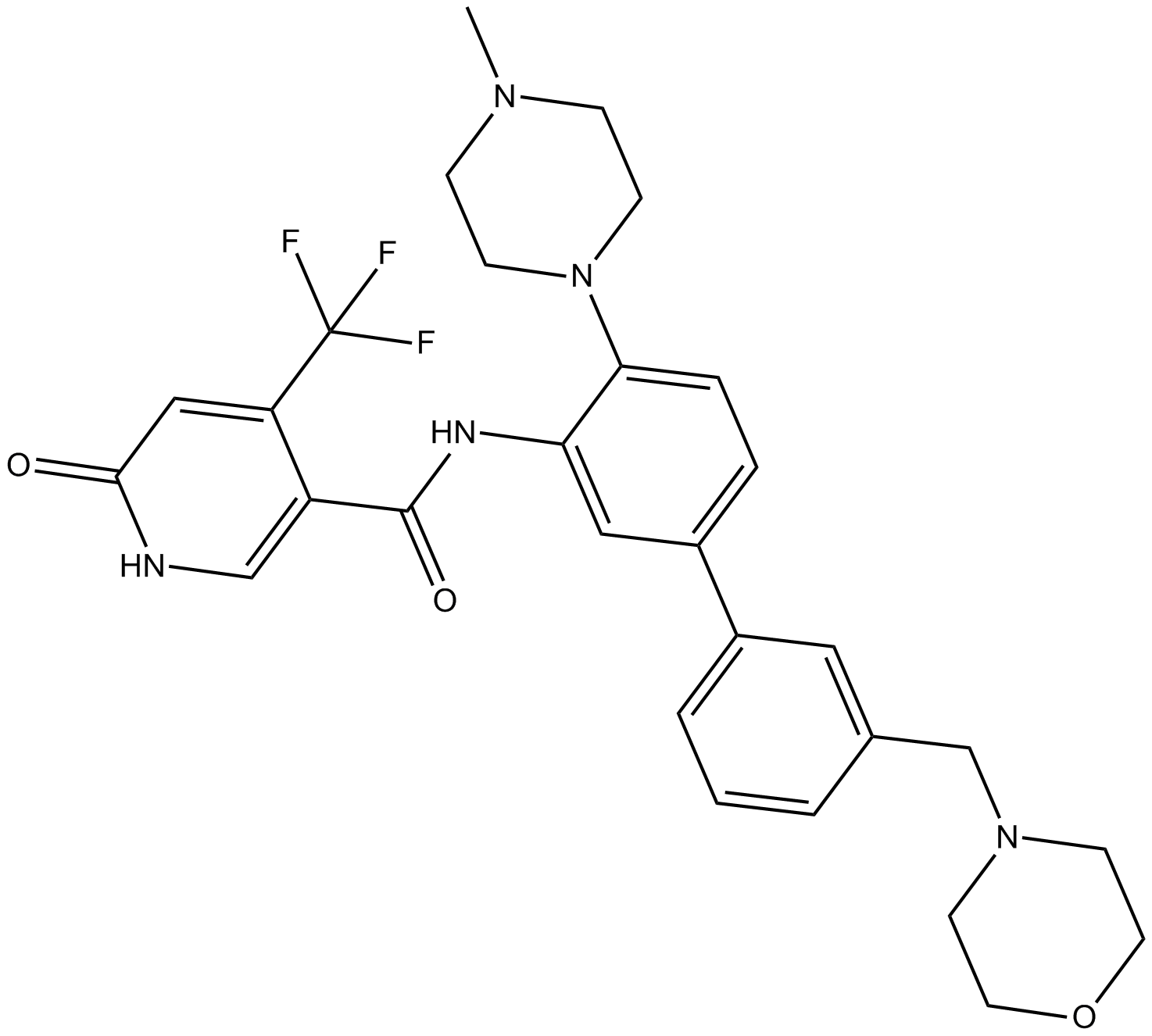
-
GC69636
OTS193320
OTS193320 is an imidazopyridine compound, a SUV39H2 methyltransferase activity inhibitor. OTS193320 reduces the global histone H3 lysine 9 trimethylation level in breast cancer cells and induces apoptosis cell death. Compared to single-agent OTS193320 or DOX, the combination of OTS193320 with Doxorubicin (DOX; A) can lead to a decrease in γ-H2AX levels and cancer cell viability.
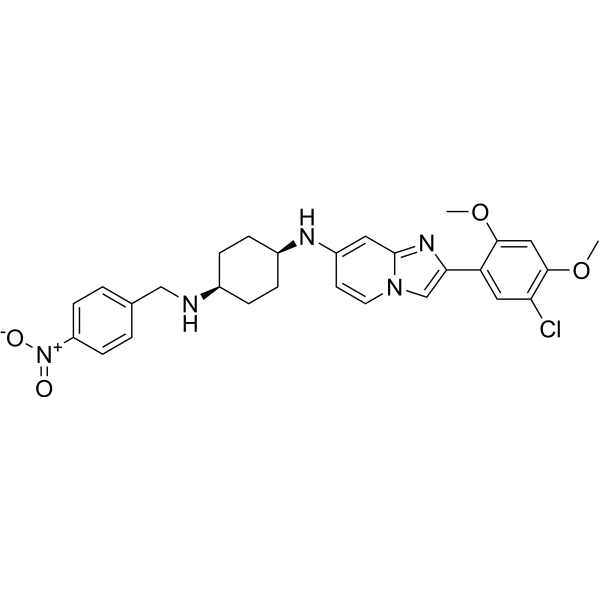
-
GC50367
PF 06726304 acetate
Highly potent and SAM-competitive EZH2 inhibitor
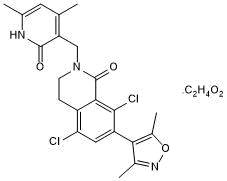
-
GC32977
PF-06726304
PF-06726304 is a potent and selective EZH2 inhibitor. PF-06726304 inhibits wild-type and Y641N mutant EZH2 with Kis of 0.7 and 3.0 nM, respectively. PF-06726304 displays robust antitumor growth activity.
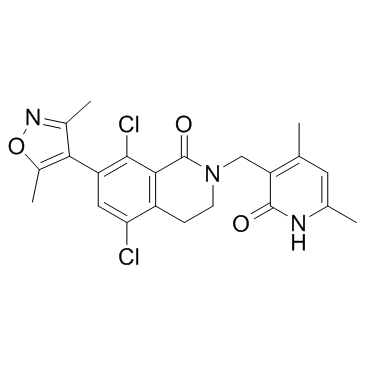
-
GC17956
PFI-2
SETD7 methyltransferase inhibitor
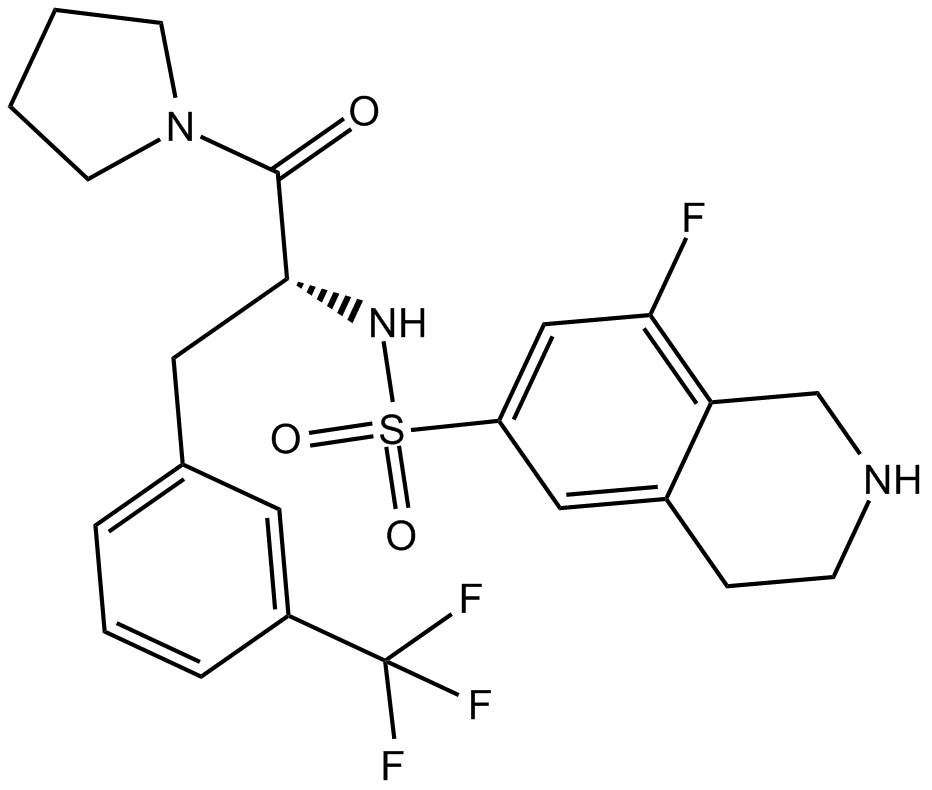
-
GC12530
PFI-2 (hydrochloride)

-
GC64941
PR5-LL-CM01
PR5-LL-CM01 is a potent protein arginine methyltransferase 5 (PRMT5) inhibitor (IC50= 7.5 μM). Anti-tumor activies.
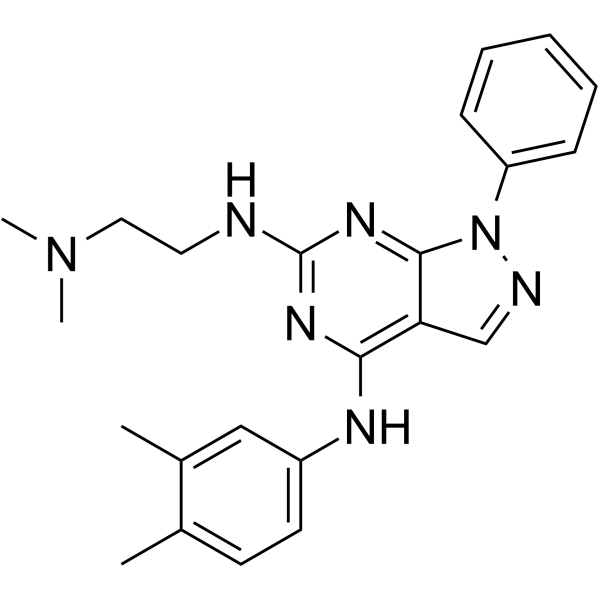
-
GC36969
PRMT5-IN-1
PRMT5 IN-1, a hemiaminal, is a potent, selective protein arginine methyltransferase 5 (PRMT5) inhibitor with an IC50 of 11 nM for PRMT5/MEP50. PRMT5 IN-1 can be converted to aldehydes and react with C449 to form covalent adducts under physiological conditions.
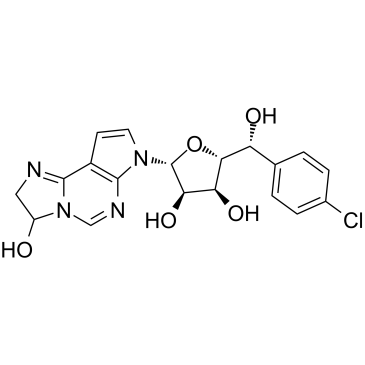
-
GC65027
PRMT5-IN-20
PRMT5-IN-20 is a selective protein arginine methyltransferase 5 (PRMT5) inhibitor with anti-tumor activity.
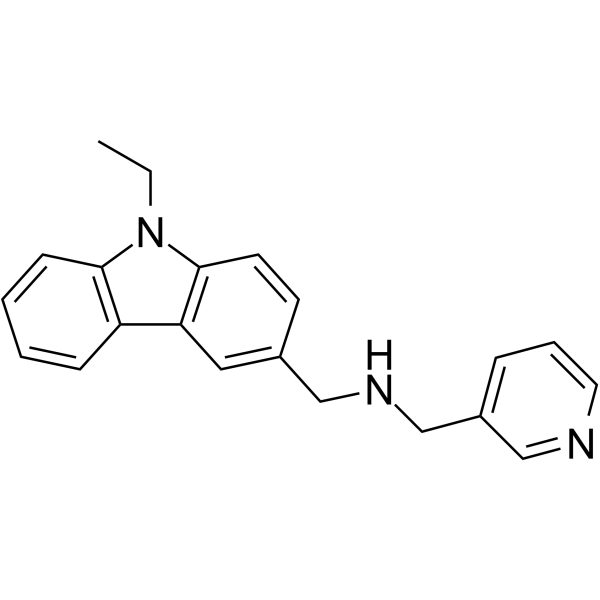
-
GC62187
PROTAC EED degrader-1
PROTAC EED degrader-1 is a von Hippel-Lindau-based PROTAC targeting EED with a pKD of 9.02. PROTAC EED degrader-1 is a polycomb repressive complex 2 (PRC2) inhibitor (pIC50=8.17) targeting the EED subunit.
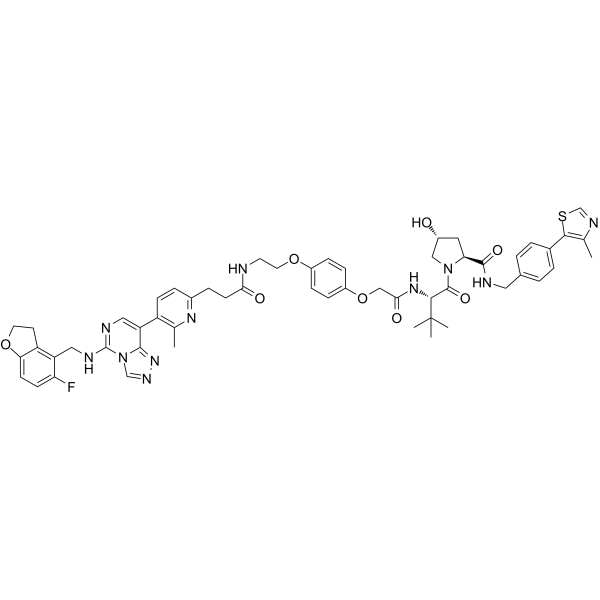
-
GC62717
PROTAC EED degrader-2
PROTAC EED degrader-2 is a von Hippel-Lindau-based PROTAC targeting EED with a pKD of 9.27. PROTAC EED degrader-2 is a polycomb repressive complex 2 (PRC2) inhibitor (pIC50=8.11) targeting the EED subunit.
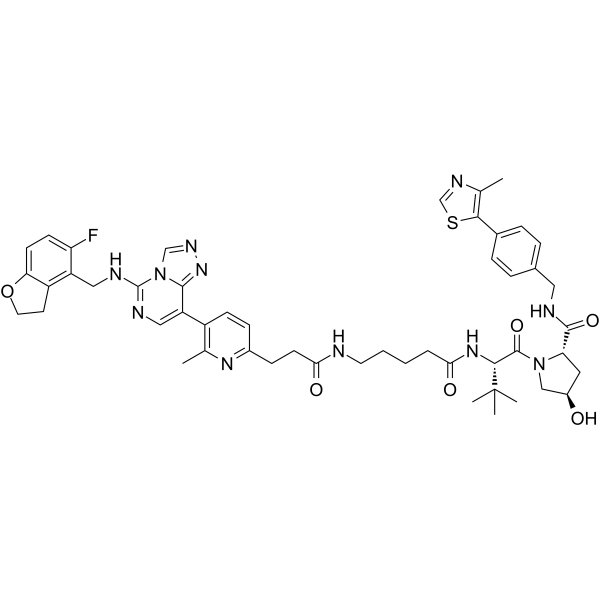
-
GC65509
PROTAC EZH2 Degrader-1
PROTAC EZH2 Degrader-1 (Compound 150d), a potent PROTAC EZH2 Degrader, exerts inhibitory effect on EZH2 methyltransferase activity with the IC50 of 2.7 nM. EZH2 plays an important role in many tumorigenesis and development processes.
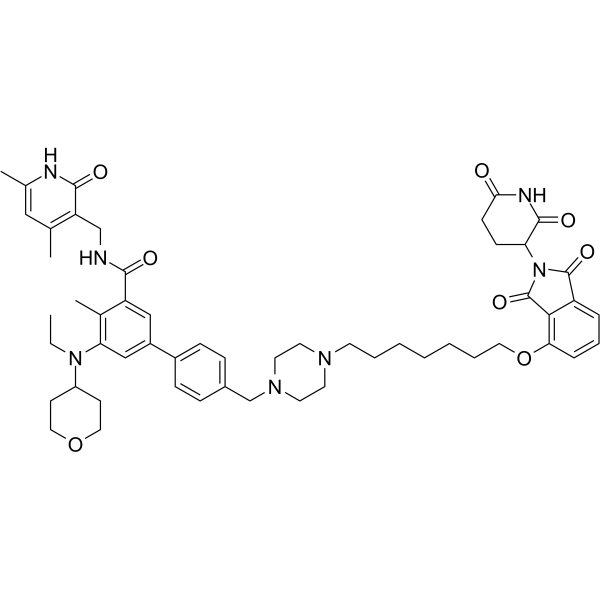
-
GC46213
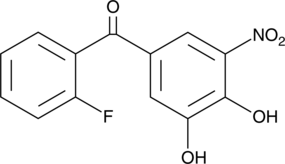
Ro 41-0960
A COMT inhibitor
-
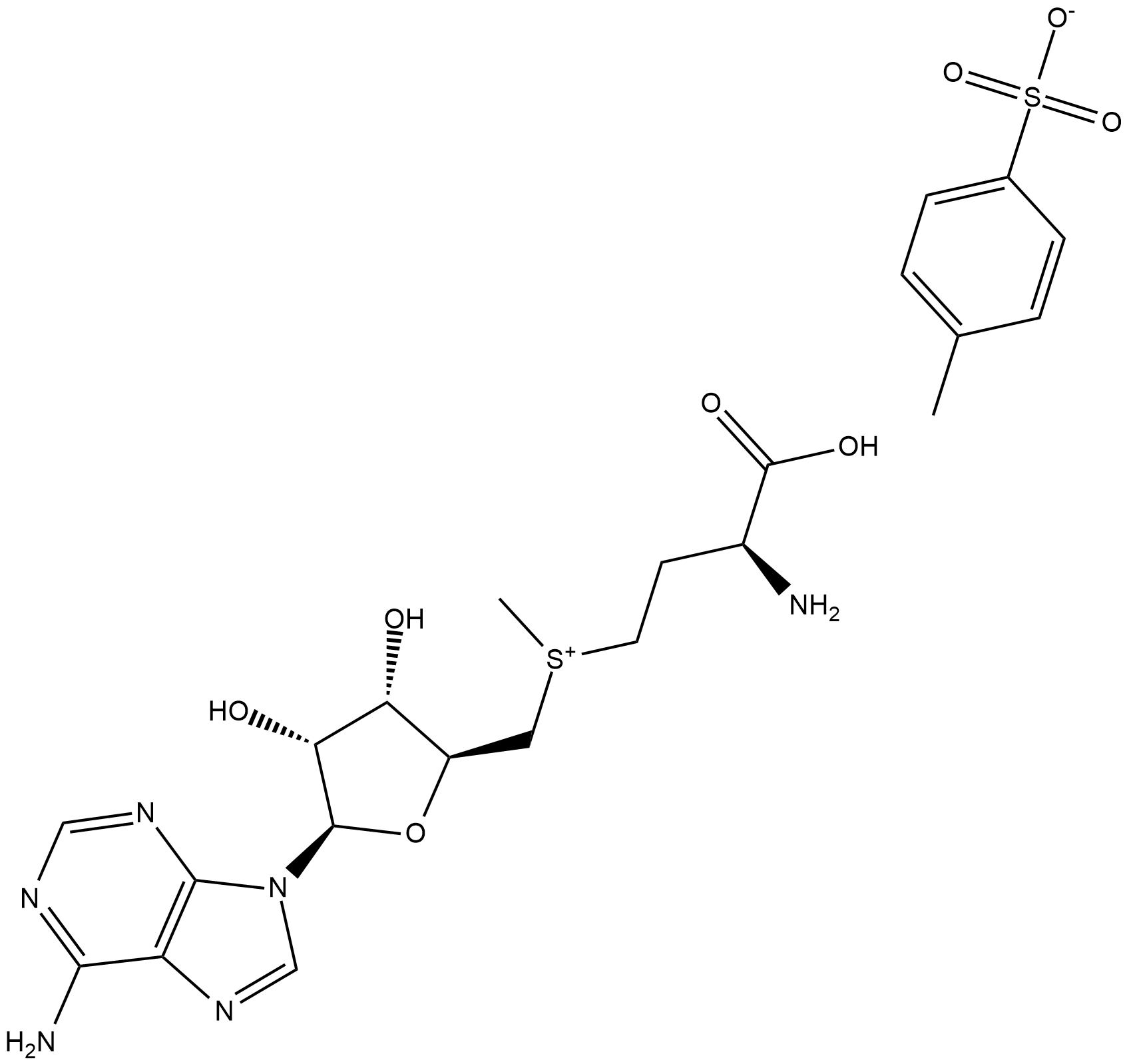
GC18650
S-(5'-Adenosyl)-L-methionine (tosylate)
S-(5'-Adenosyl)-L-methionine (SAM) is a ubiquitous methyl donor involved in a wide variety of biological reactions, including those mediated by DNA and protein methyltransferases.
-
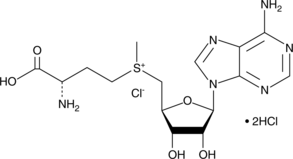
GC44859
S-(5'-Adenosyl)-L-methionine chloride (hydrochloride)
S-(5'-Adenosyl)-L-methionine chloride (SAM) is a ubiquitous methyl donor involved in a wide variety of biological reactions, including those mediated by DNA and protein methyltransferases.
-
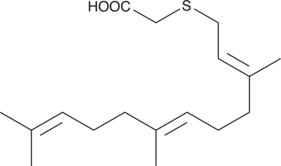
GC44886
S-Farnesyl Thioacetic Acid
S-Farnesyl thioacetic acid is an analog of S-farnesyl cysteine which behaves as a competitive inhibitor of isoprenylated protein methyltransferase (also known as S-adenosylmethionine-dependent methyltransferase).
-
GC62555
SETD2-IN-1 TFA
SETD2-IN-1 TFA is a potent, selective and orally active inhibitor of SETD2 which is a human histone methyltransferase. SETD2-IN-1 TFA has anti-proliferative effects.
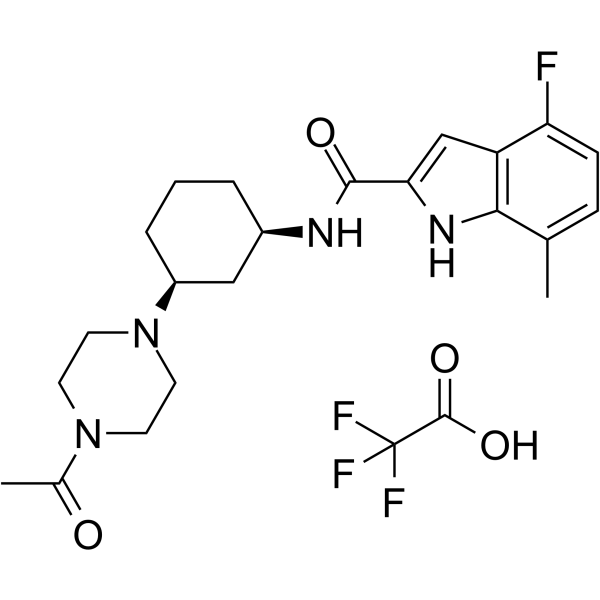
-
GC64893
SETDB1-TTD-IN-1
SETDB1-TTD-IN-1 is a potent, selective and endogenous binder competitive inhibitor of SET domain bifurcated protein 1 tandem tudor domain (SETDB1-TTD), with a Kd of 88 nM. SETDB1-TTD-IN-1 can be used for the research of biological functions and disease associations of SETDB1-TTD.
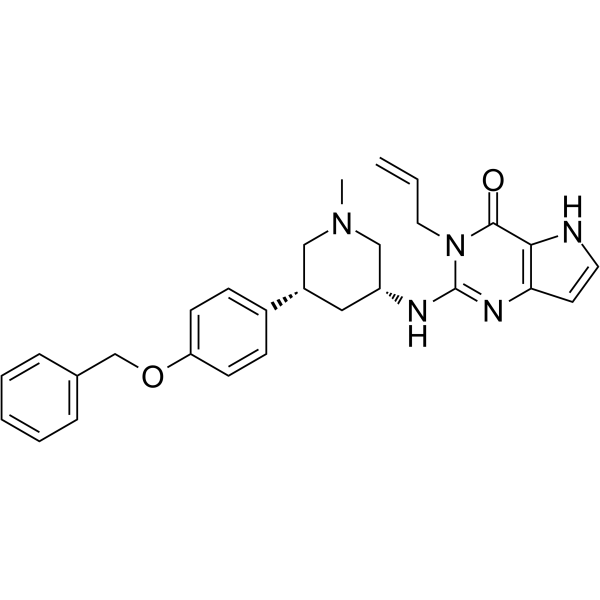
-
GC64894
SETDB1-TTD-IN-1 TFA
SETDB1-TTD-IN-1 TFA is a potent, selective and endogenous binder competitive inhibitor of SET domain bifurcated protein 1 tandem tudor domain (SETDB1-TTD), with a Kd of 88 nM. SETDB1-TTD-IN-1 TFA can be used for the research of biological functions and disease associations of SETDB1-TTD.
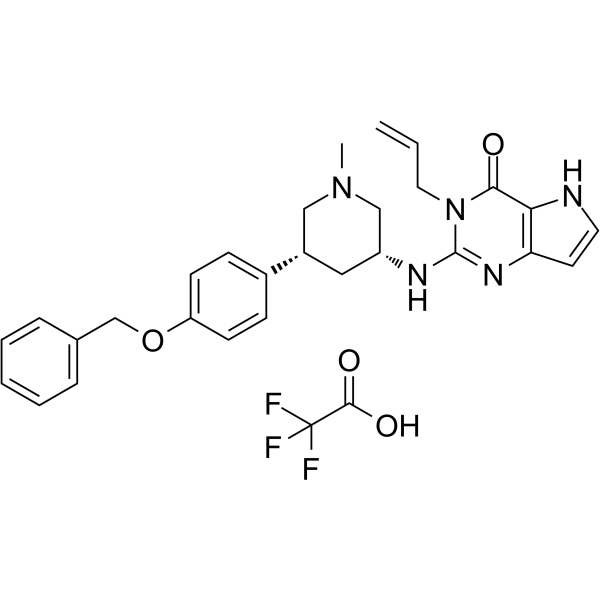
-
GC11491
SGC 0946
A potent inhibitor of DOT1L
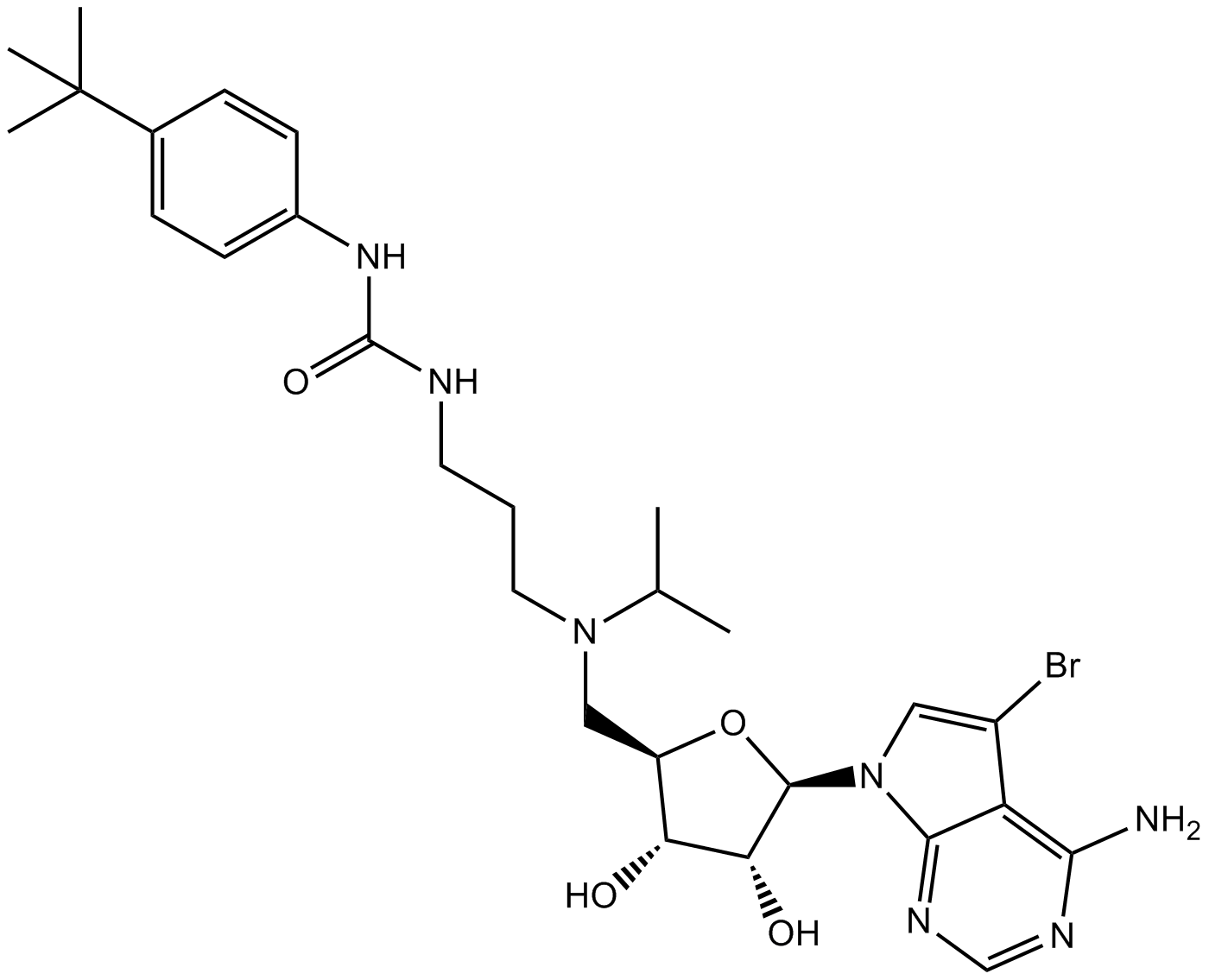
-
GC50699
SGC 6870
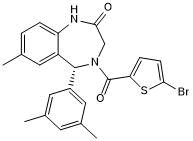
-
GC19329
SGC2085
SGC2085 is a potent and selective coactivator associated arginine methyltransferase 1 (CARM1) inhibitor with an IC50 of 50 nM.
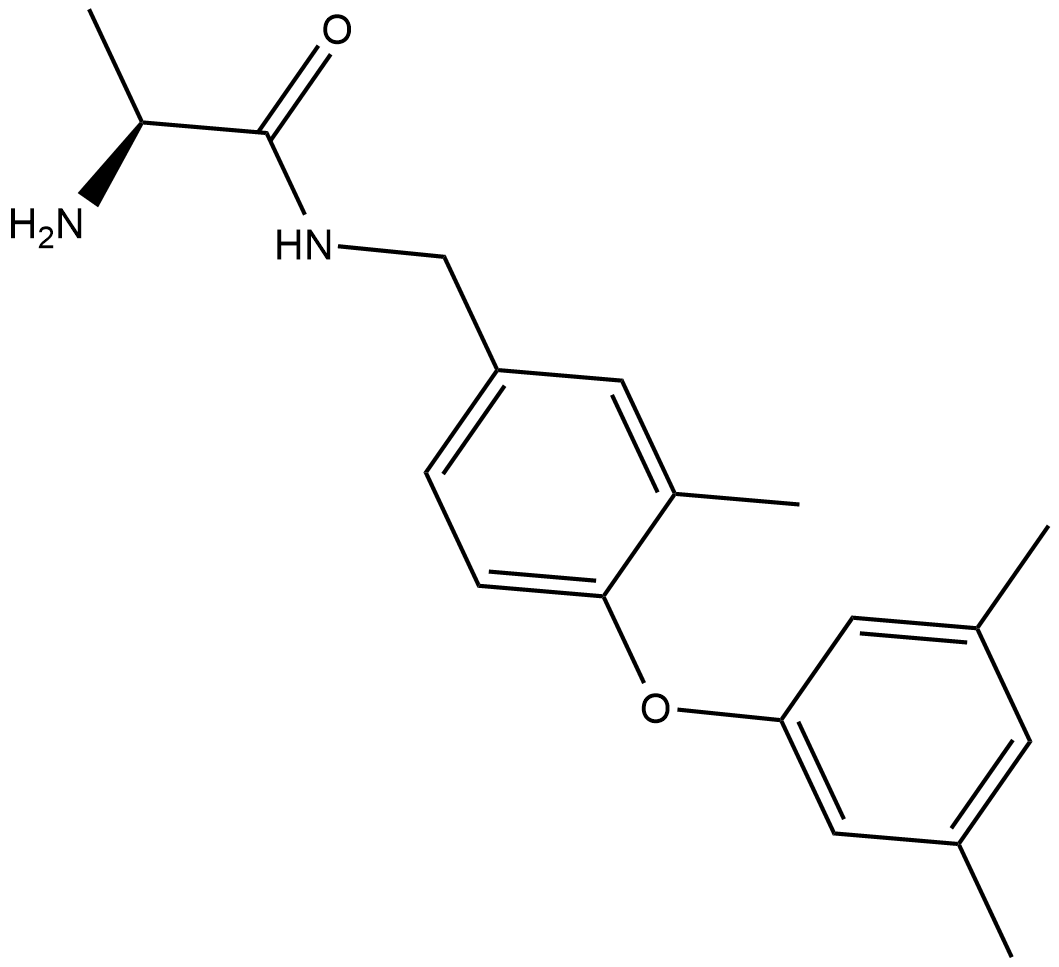
-
GC18428
SGC2085 (hydrochloride)
SGC2085 is an inhibitor of protein arginine methyltransferase 4 (PRMT4/CARM1; IC50 = 50 nM).
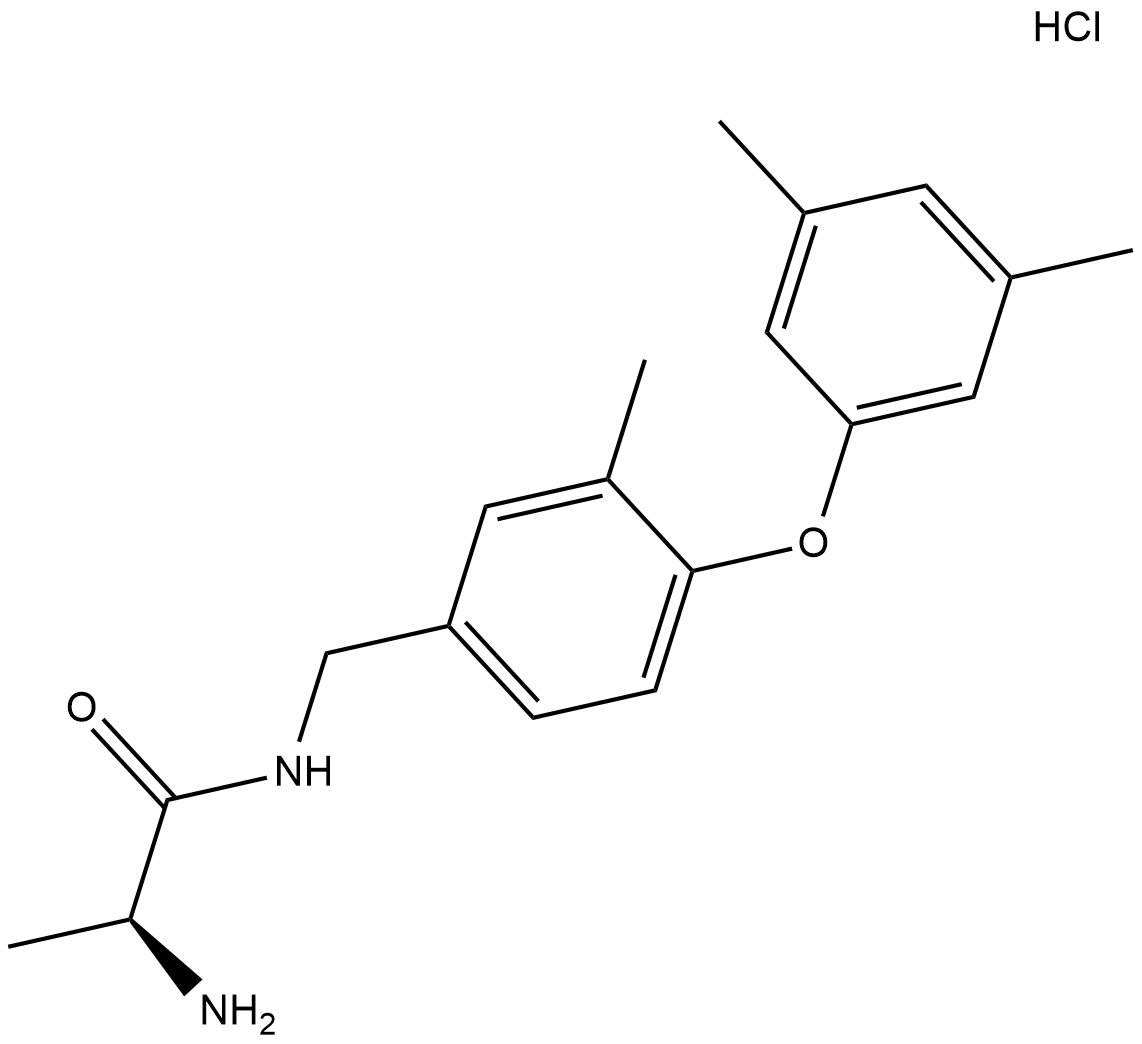
-
GC12874
SGC707
PRMT3 inhibitor
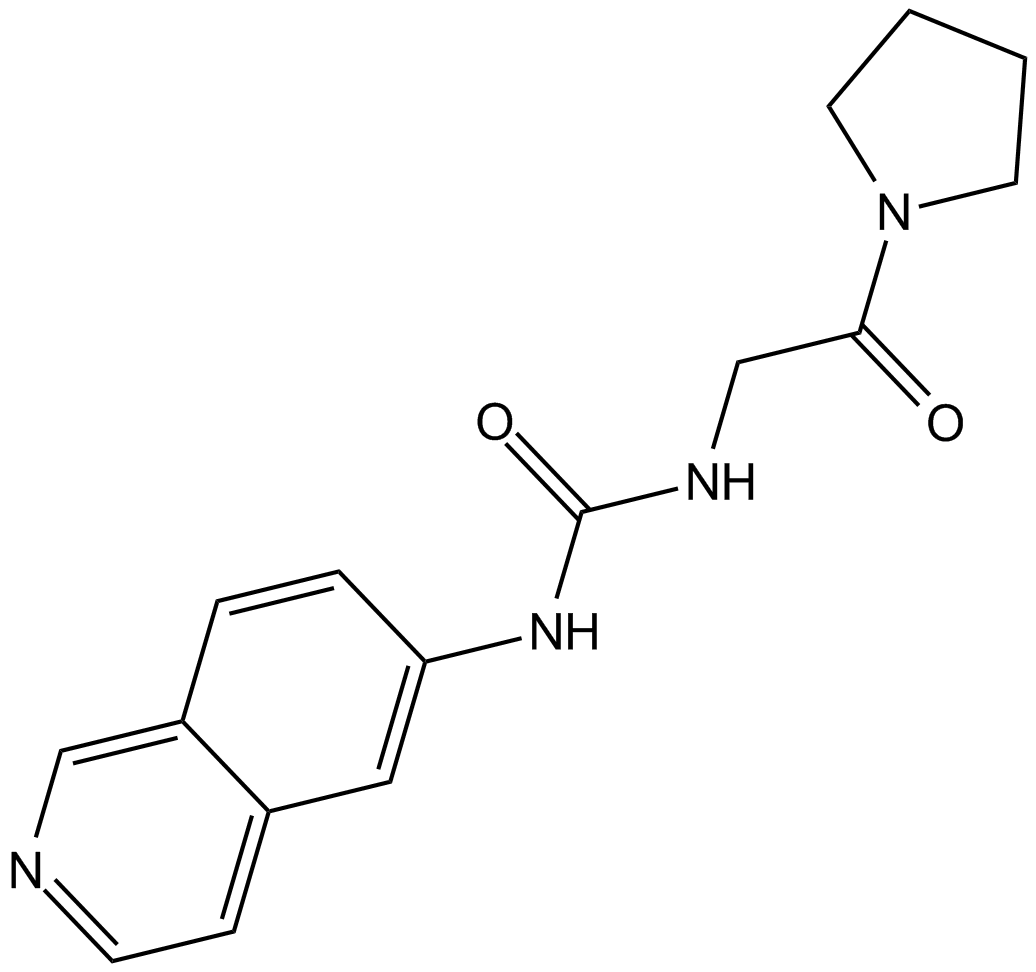
-
GC14364
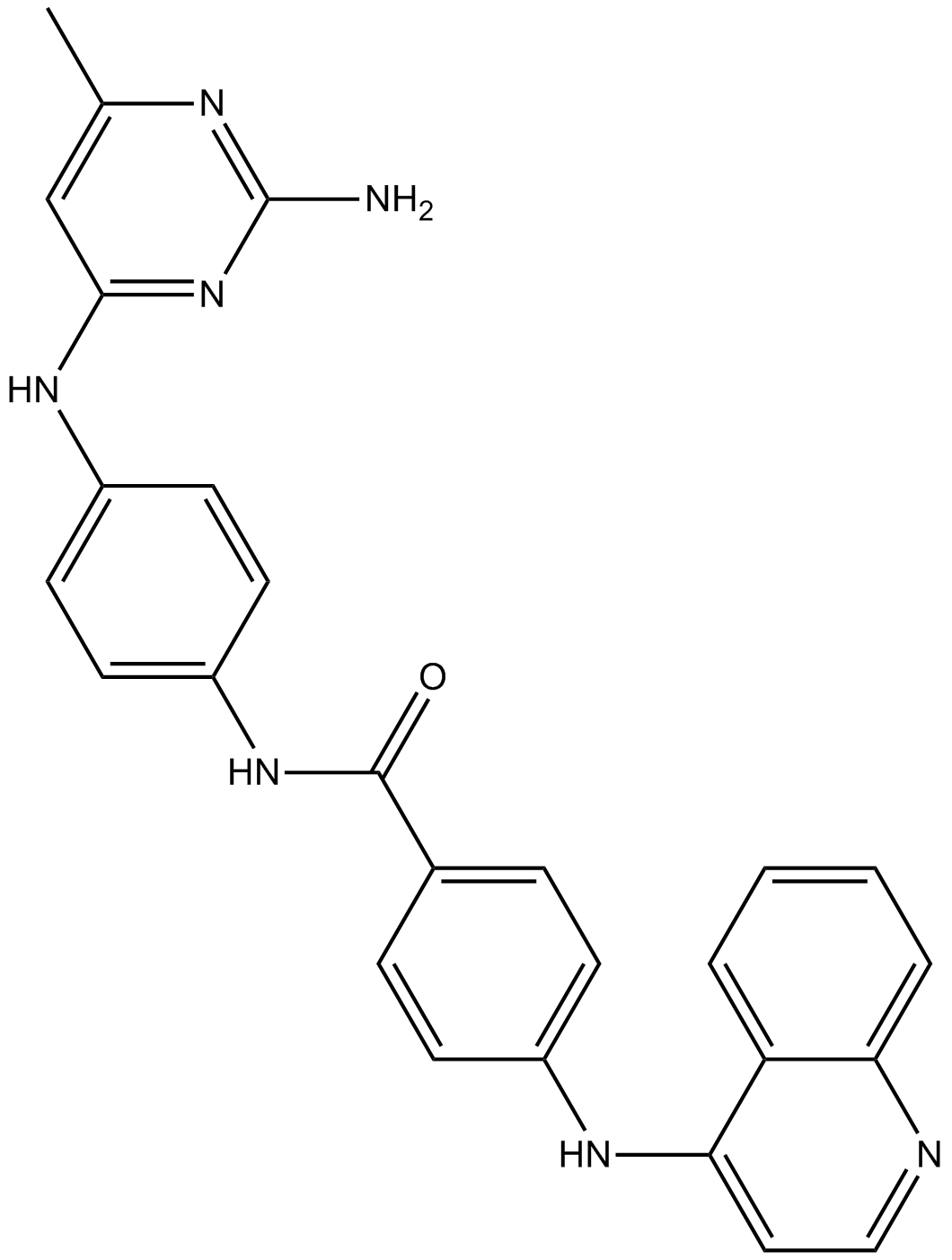
SGI-1027
DNMT inhibitor
-
GC34784
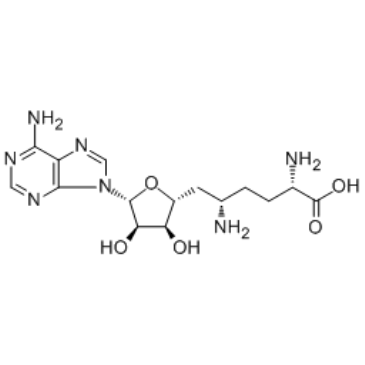
Sinefungin
A methyltransferase inhibitor
-
GC37654
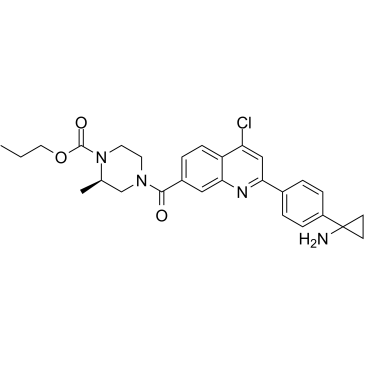
SMYD3-IN-1
SMYD3-IN-1 (compound 29) is an irreversible and selective inhibitor of SMYD3 (SET and MYND domain containing 3), with an IC50 of 11.7 nM.
-
GC18642
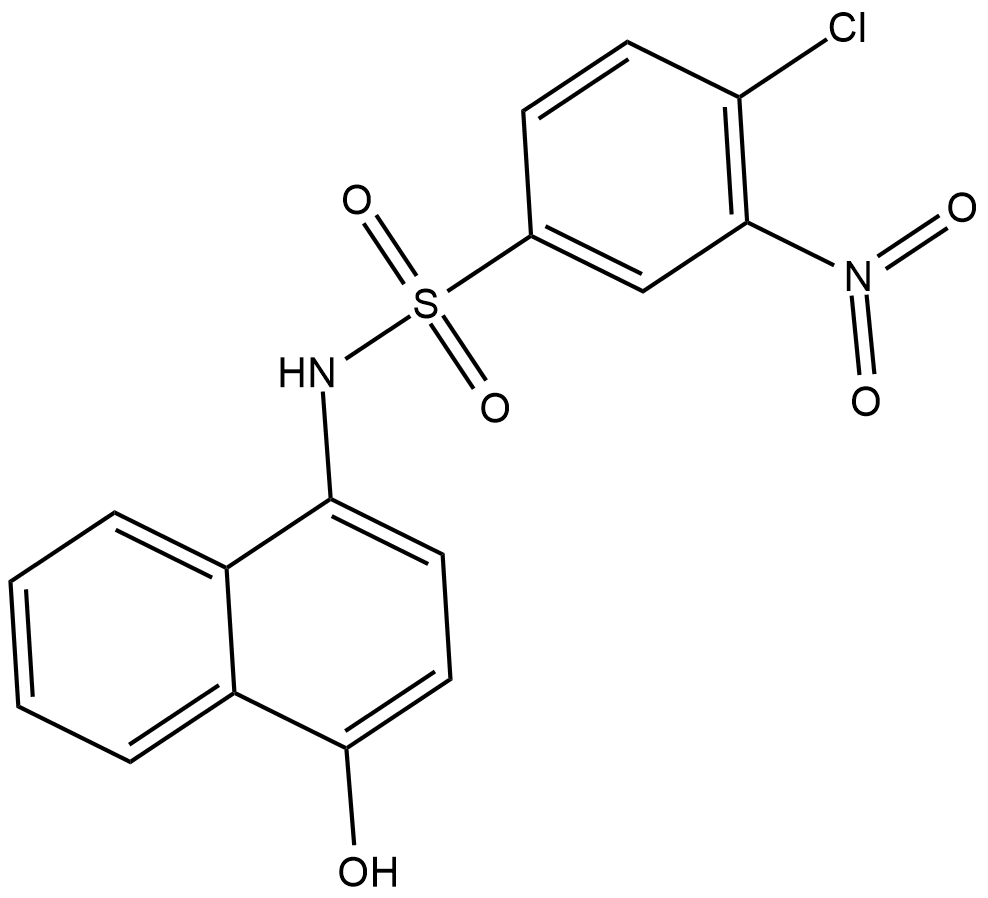
SW155246
SW155246 is an inhibitor of DNA methyltransferase 1 (DNMT1; IC50 = 1.2 μM).
-
GC62390
SW2_110A
SW2_110A is a selective chromobox 8 chromodomain (CBX8 ChD) inhibitor with a Kd of 800 nM. SW2_110A shows minimal 5-fold selectivity for CBX8 ChD over all other CBX paralogs in vitro.
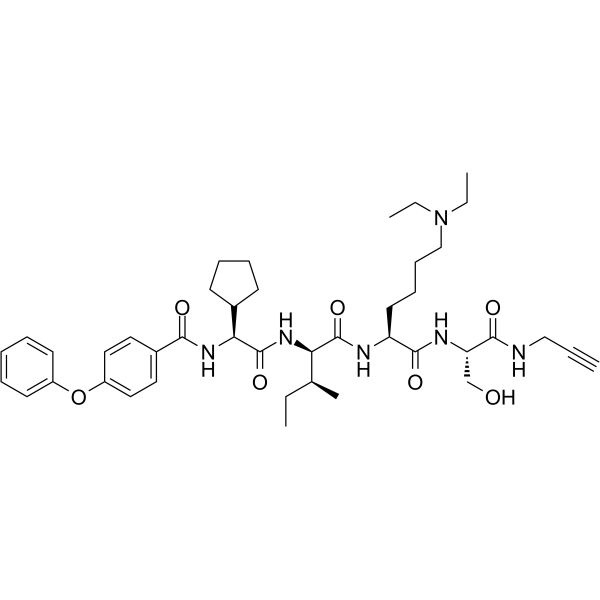
-
GC69980
SW2_152F
SW2_152F is an effective selective inhibitor of chromobox 2 chromodomain (CBX2 ChD), with a Kd of 80 nM. In vitro, SW2_152F exhibits selectivity for CBX2 ChD that is 24-1000 times higher than other CBX paralogs.
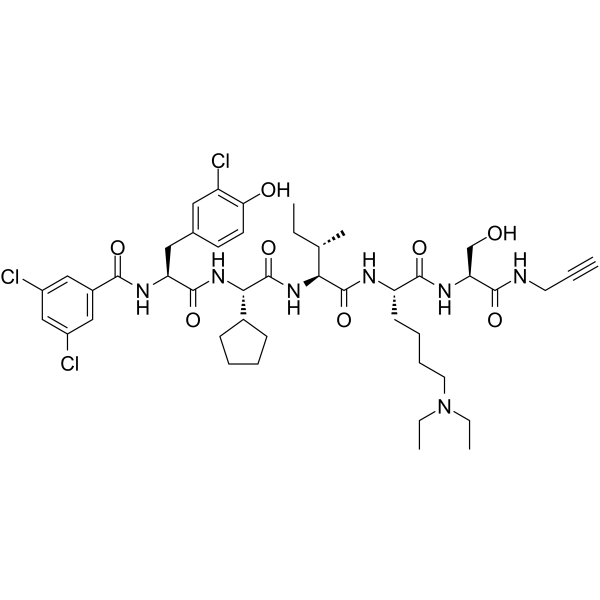
-
GC67676
Tazemetostat de(methylene morpholine)-O-C3-O-C-COOH
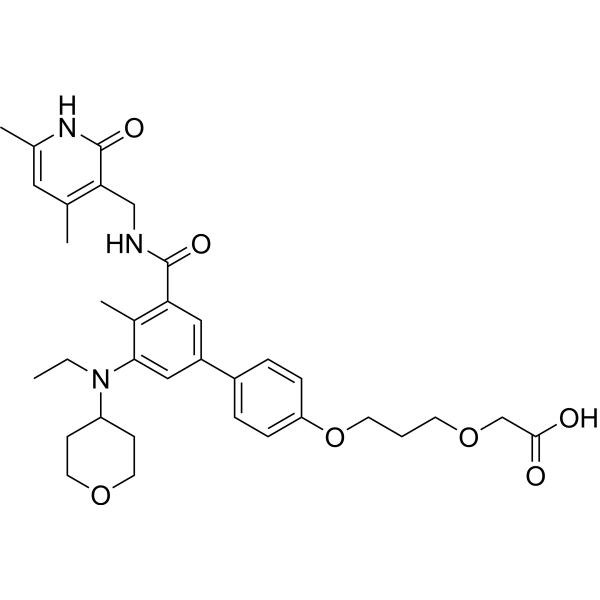
-
GC38163
Tazemetostat hydrobromide
Tazemetostat hydrobromide (EPZ-6438 hydrobromide) is a potent, selective and orally available EZH2 inhibitor. Tazemetostat hydrobromide inhibits the activity of human polycomb repressive complex 2 (PRC2)-containing wild-type EZH2 with a Ki value of 2.5 nM. Tazemetostat hydrobromide inhibits EZH2 with IC50s of 11 and 16 nM in peptide assay and nucleosome assay, respectively. Tazemetostat hydrobromide inhibits Rat EZH2 with an IC50 of 4 nM. Tazemetostat hydrobromide also inhibits EZH1 with an IC50 of 392 nM.
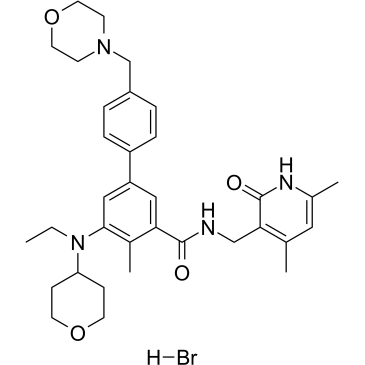
-
GC17672
TC-E 5003
PRMT1 inhibitor, potent
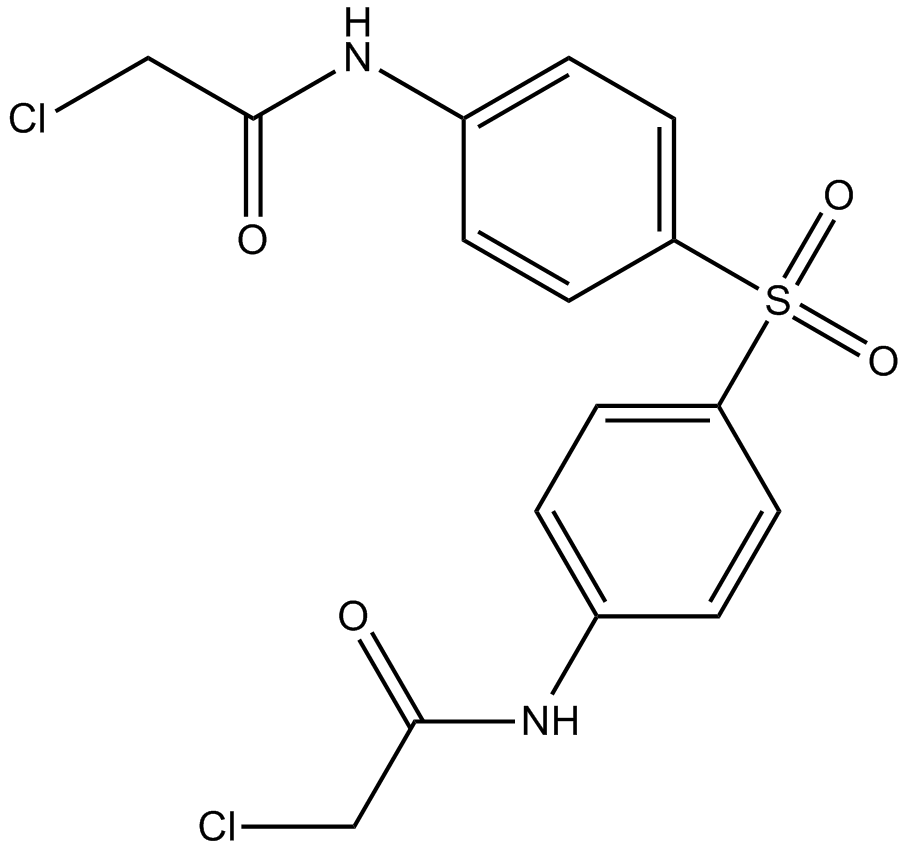
-
GC70041
TNG908
TNG908 is a blood-brain barrier penetrable MTAP-cooperative PRMT5 inhibitor. TNG908 has 15 times higher selectivity for MTAPnull cell lines than for MTAPWT cell lines and can be used in cancer research.
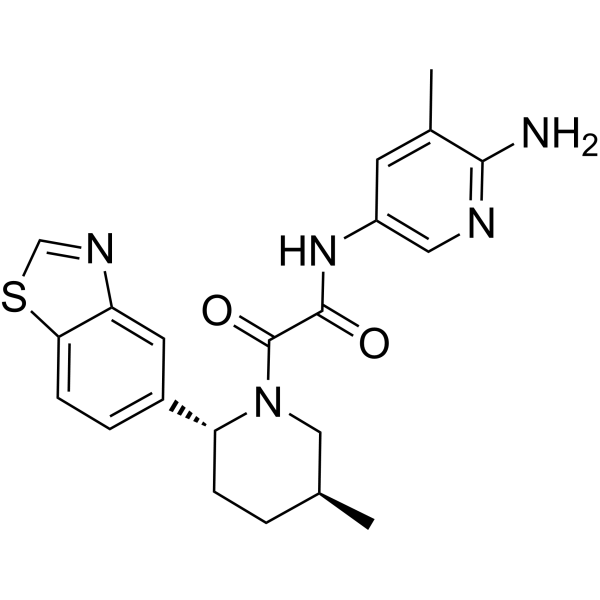
-
GC45763
Tolcapone-d4
An internal standard for the quantification of tolcapone
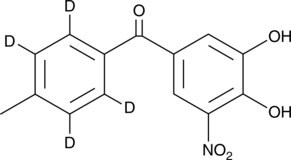
-
GC41263
TP-064
TP-064 is a potent inhibitor of protein arginine methyltransferase 4 (PRMT4; IC50 in vitro.
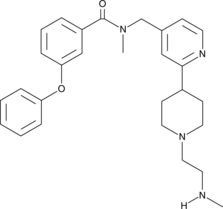
-
GC15688
UNC 0224
A potent inhibitor of G9a histone methyltransferase
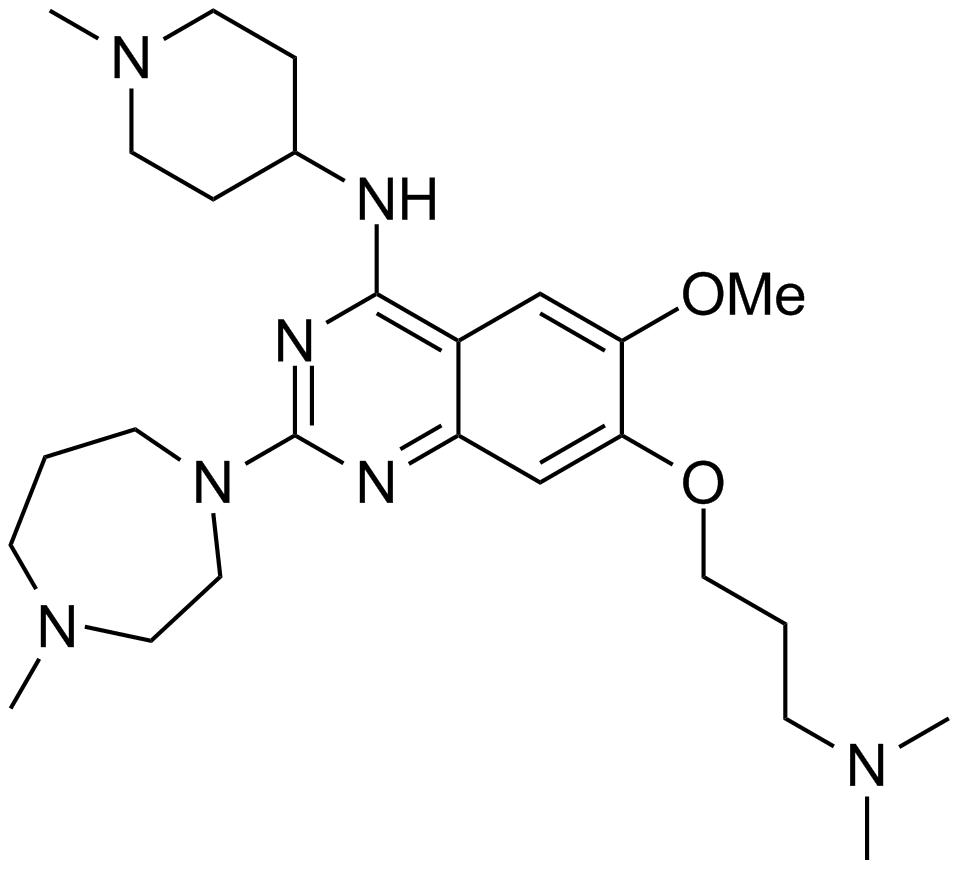
-
GC12548
UNC 0631
G9a inhibitor
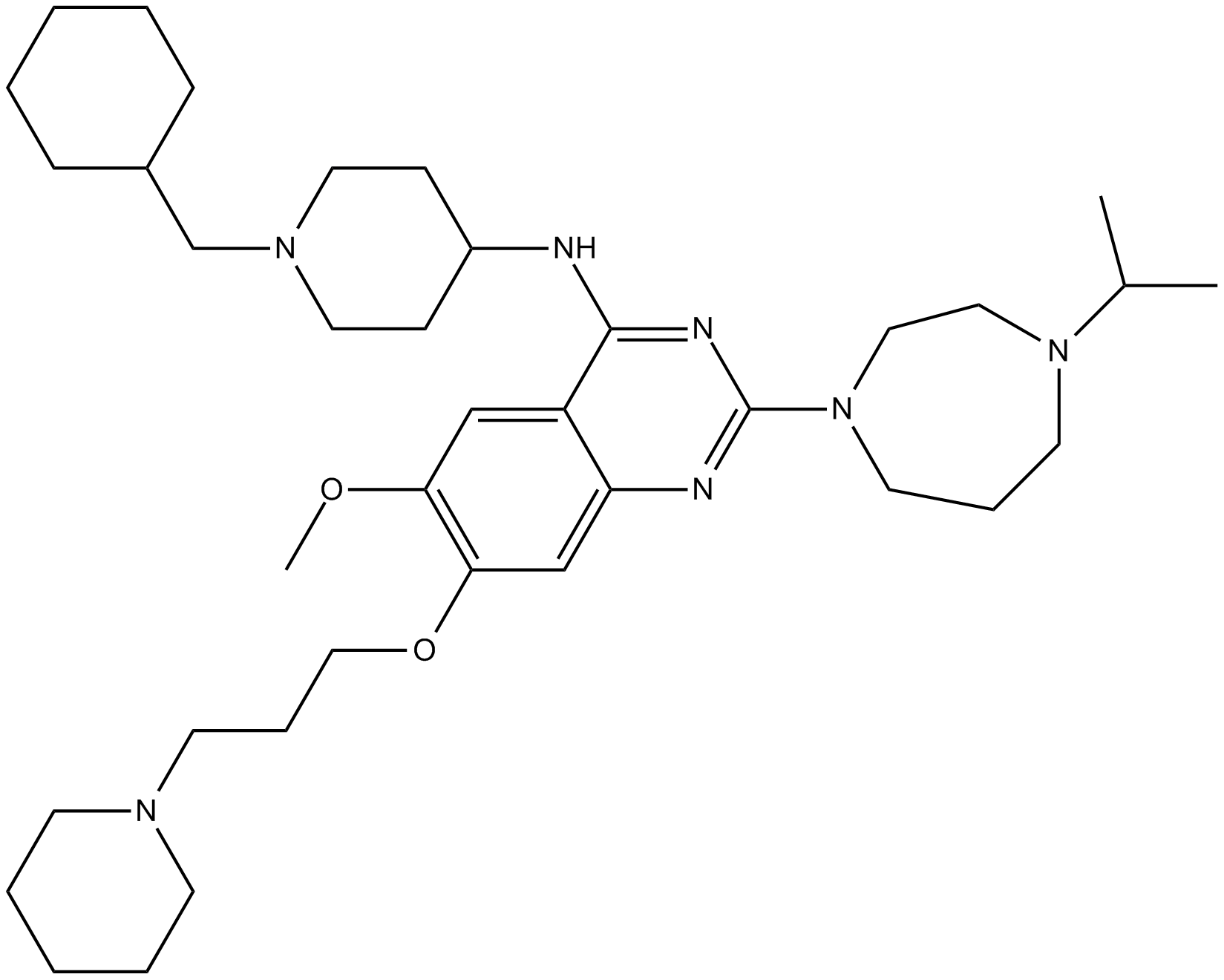
-
GC14249
UNC 0642
G9a and GLP histone lysine methyltransferase inhibitor
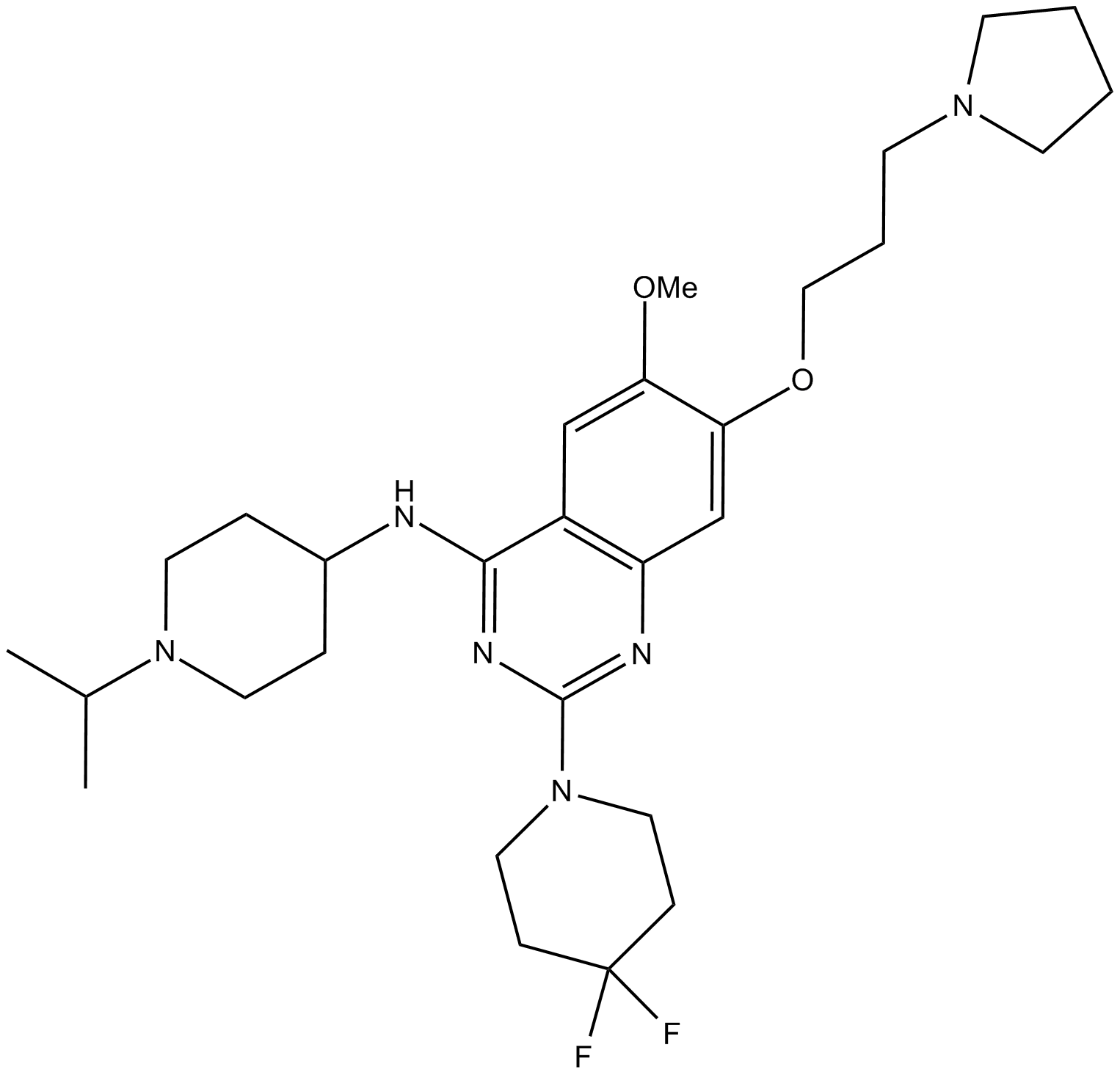
-
GC16518
UNC 0646
An inhibitor of G9a/GLP methyltransferases
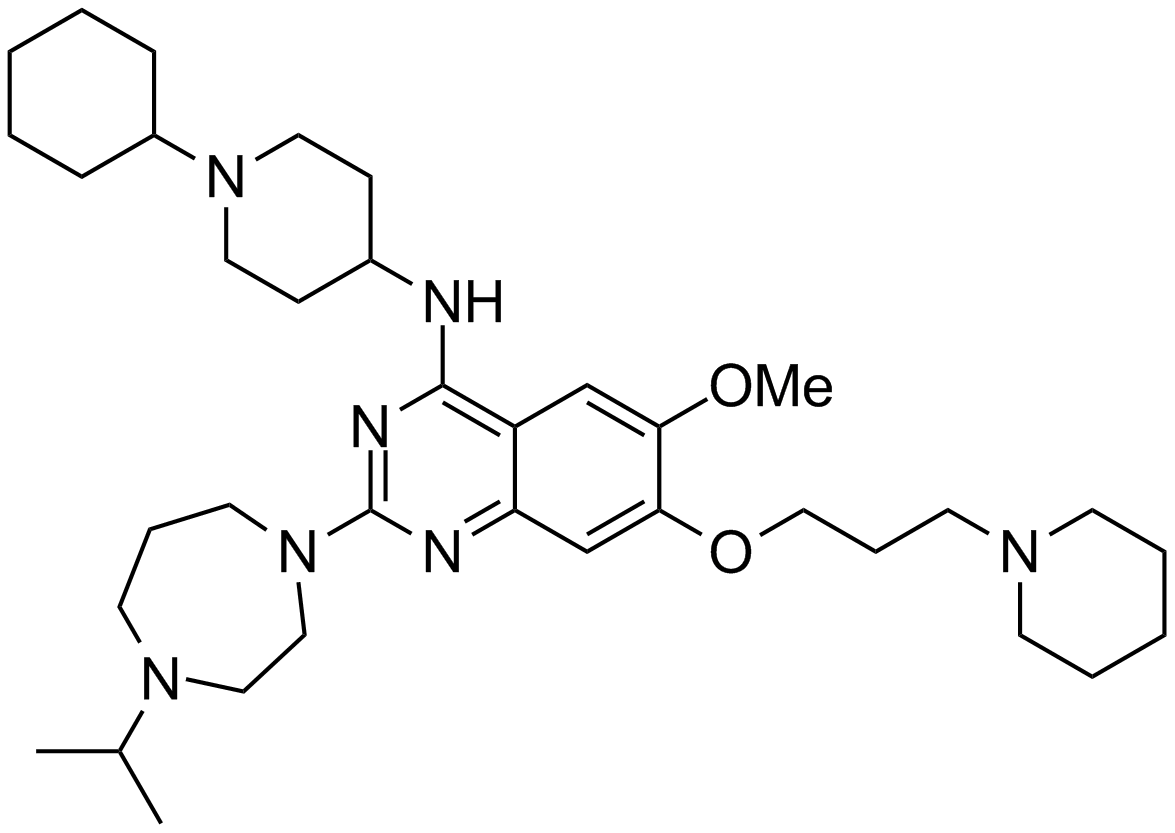
-
GC50082
UNC 2399
UNC 2399, a biotinylated UNC1999, is a selective EZH2 degrader, maintaining high in vitro potency for EZH2, with an IC50 of 17 nM.
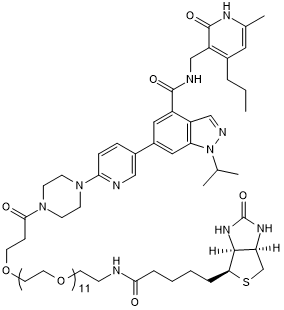
-
GC15652
UNC 2400
negative control of UNC1999
-
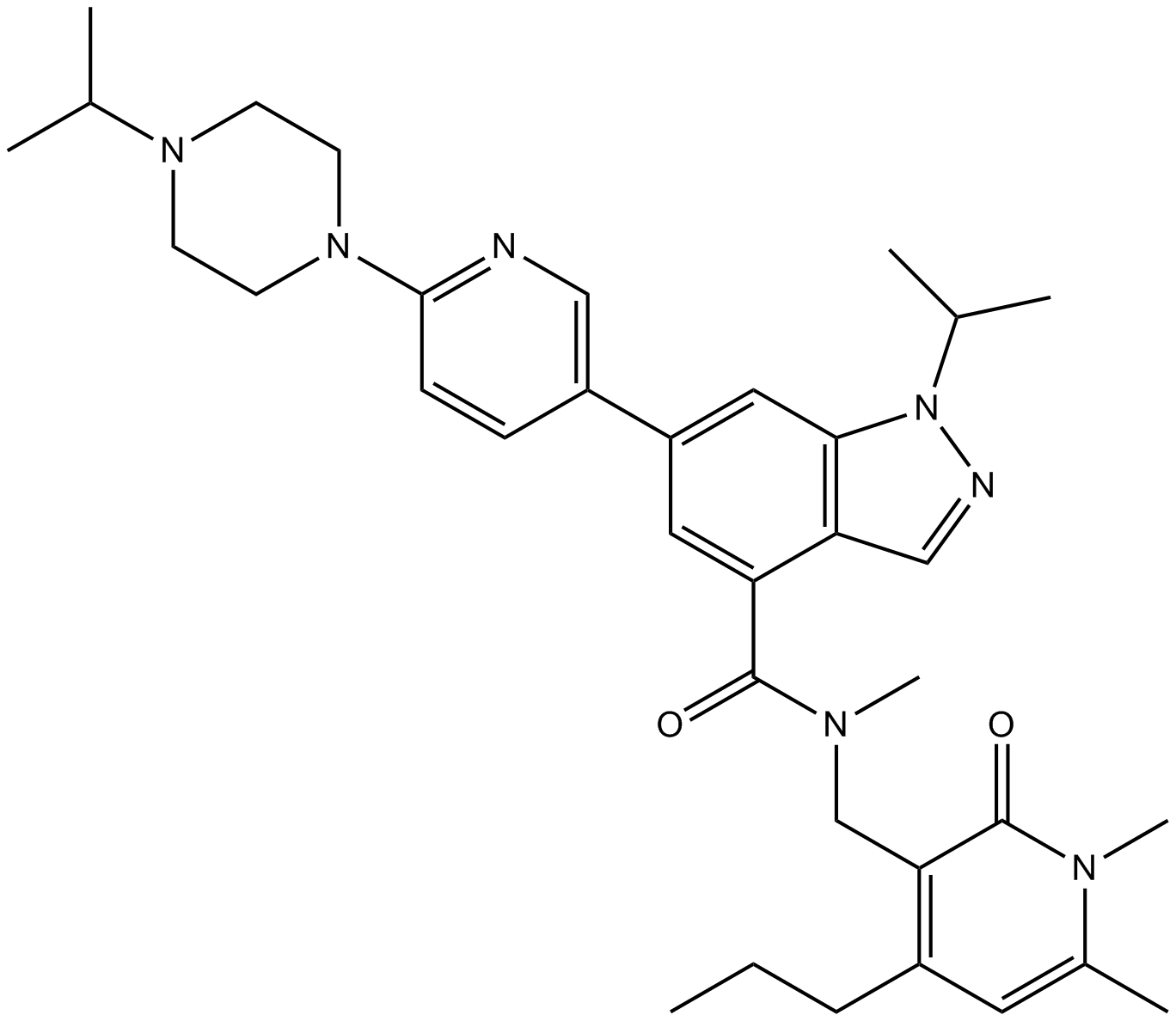
GC37856
UNC0321
UNC0321 is a potent and selective histone methyltransferase G9a inhibitor with a Ki of 63 pM and with assay-dependent IC50 values of 6-9 nM. UNC0321 also inhibits GLP with assay-dependent IC50 values of 15-23 nM. UNC0321 is inactive against SET7/9, SET8/PreSET7, PRMT3 and JMJD2E.
-
GC16741
UNC0379
N-lysine methyltransferase SETD8 inhibitor
-
GC34160
UNC0379 trifluoroacetate (UNC-0379 trifluoroacetate)
UNC0379 trifluoroacetate (UNC-0379 trifluoroacetate) is a selective, substrate-competitive inhibitor of lysine methyltransferase SETD8 (KMT5A) with an IC50 of 7.3 μM, KD value of 18.3 μM.
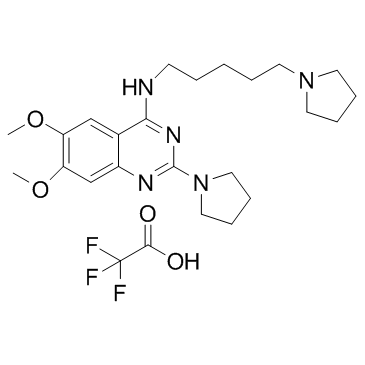
-
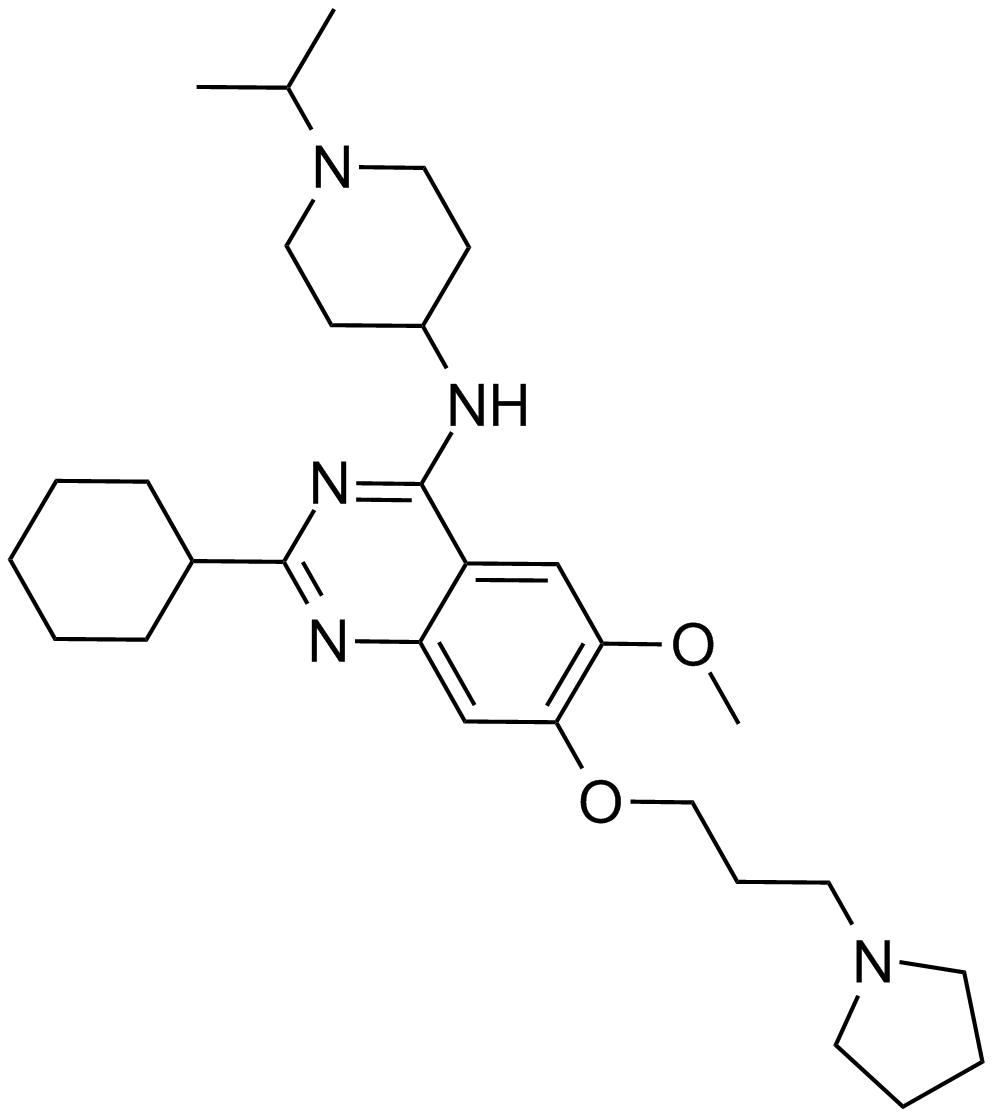
GC15610
UNC0638
A G9a and GLP histone methyltransferase inhibitor
-
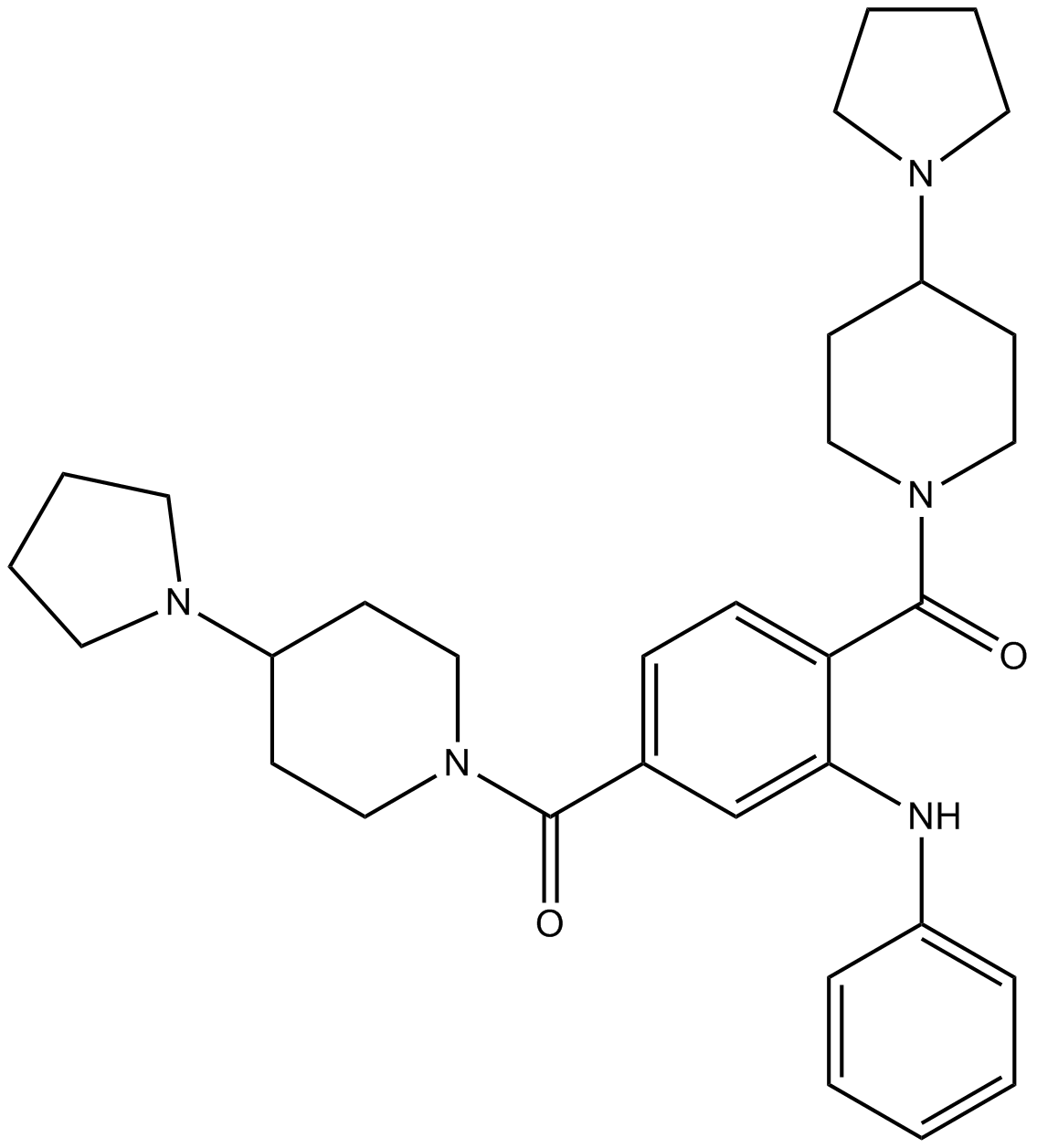
GC16794
UNC1215
Potent L3MBTL3 domain inhibitor
-
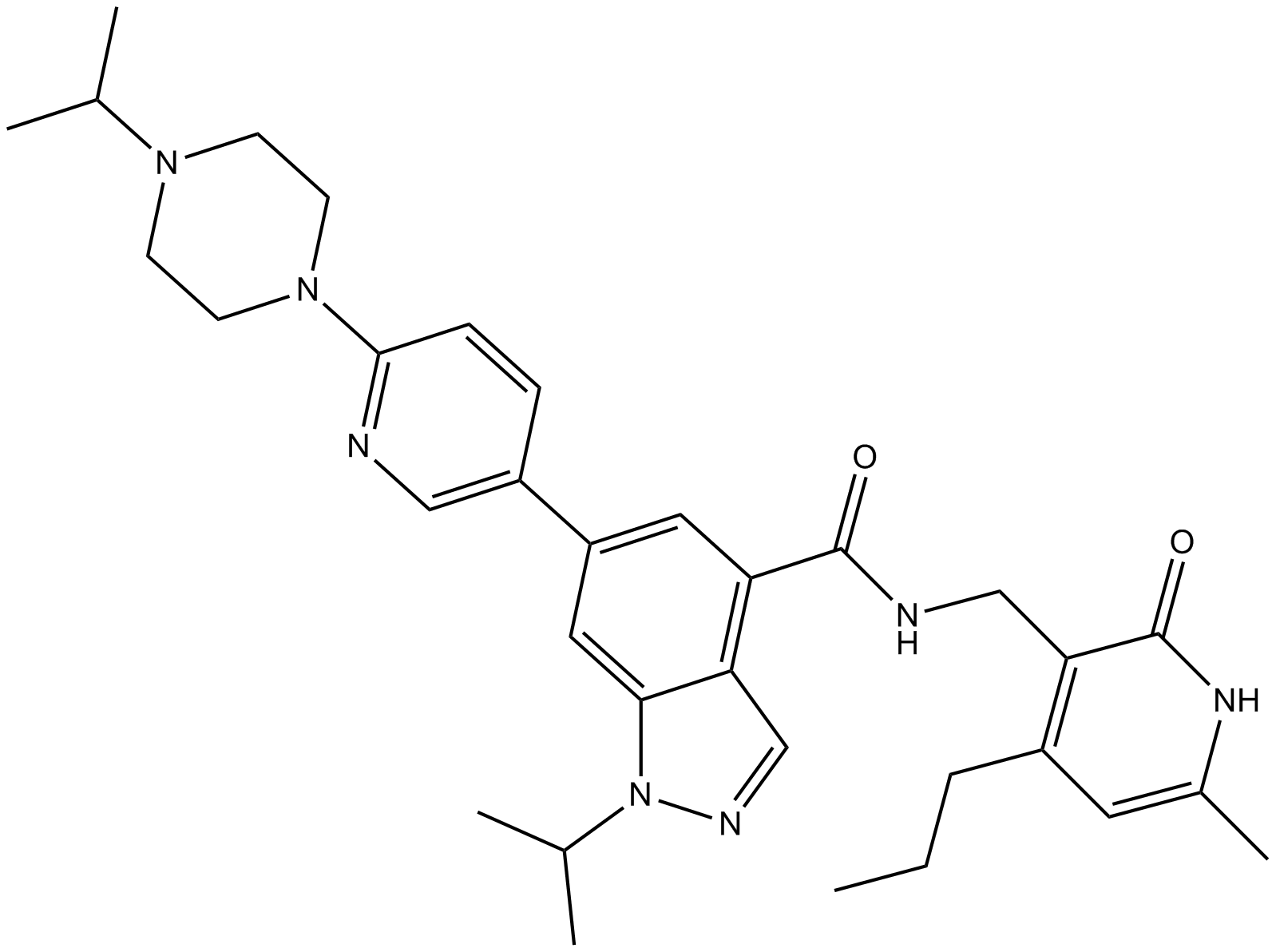
GC11375
UNC1999
EZH2 inhibitor
-
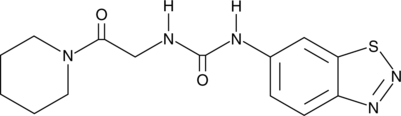
GC45117
UNC2327
An allosteric inhibitor of PRMT3
-
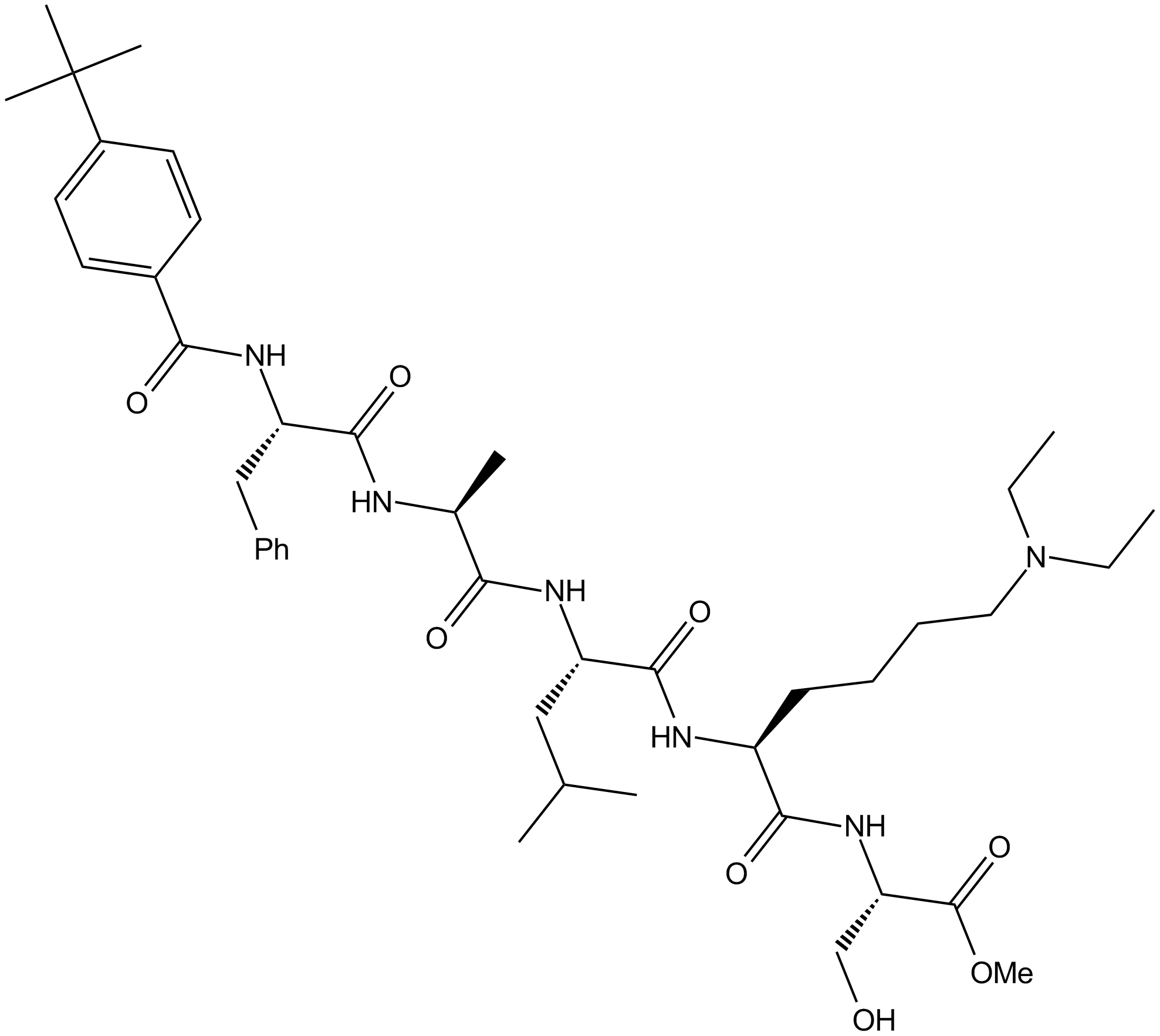
GC17207
UNC3866
a potent antagonist of CBX4 and CBX7 chromodomains
-
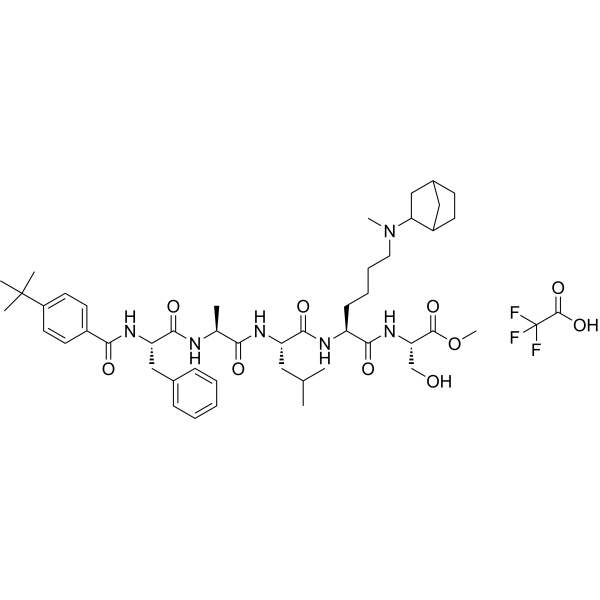
GC70091
UNC4976 TFA
UNC4976 TFA is a positive allosteric modulator (PAM) peptide that binds to the chromodomain and nucleic acid binding domain of CBX7. UNC4976 TFA antagonizes the specific recruitment of CBX7 to target genes by H3K27me3, while increasing non-specific binding to DNA and RNA.
-
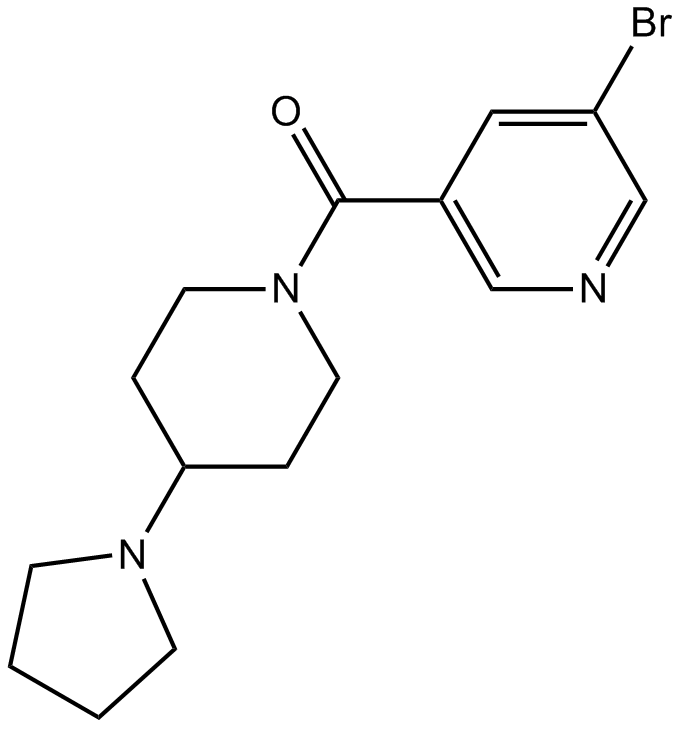
GC18096
UNC669
L3MBTL antagonist,potent and selective
-
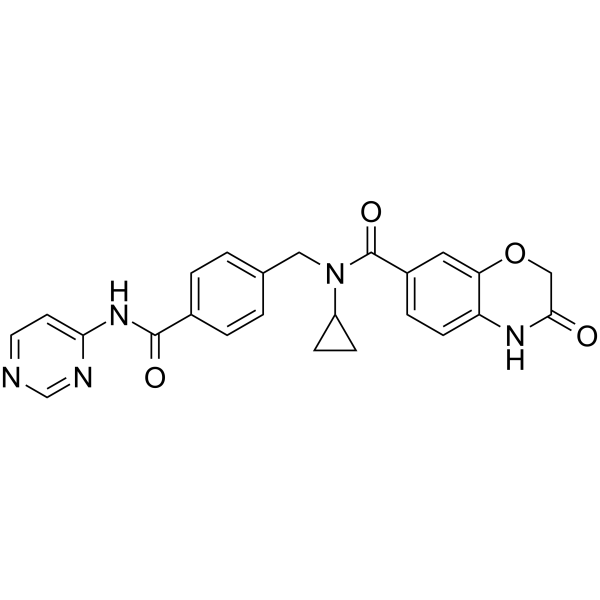
GC64083
UNC6934
UNC6934, a chemical probe targeting the PWWP domain, alters NSD2 nucleolar localization.
-
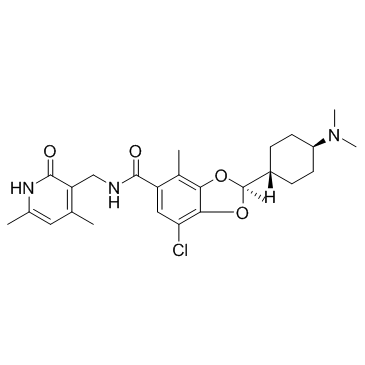
GC33397
Valemetostat
A dual EZH1 and EZH2 inhibitor
-
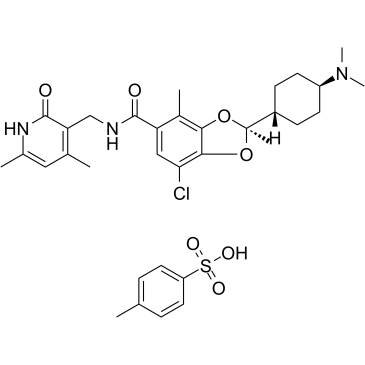
GC37881
Valemetostat tosylate
Valemetostat tosylate (DS-3201 tosylate), a first-in-class EZH1/2 dual inhibitor. Valemetostat tosylate can be used for the research of relapsed/refractory peripheral T-cell lymphoma.
-
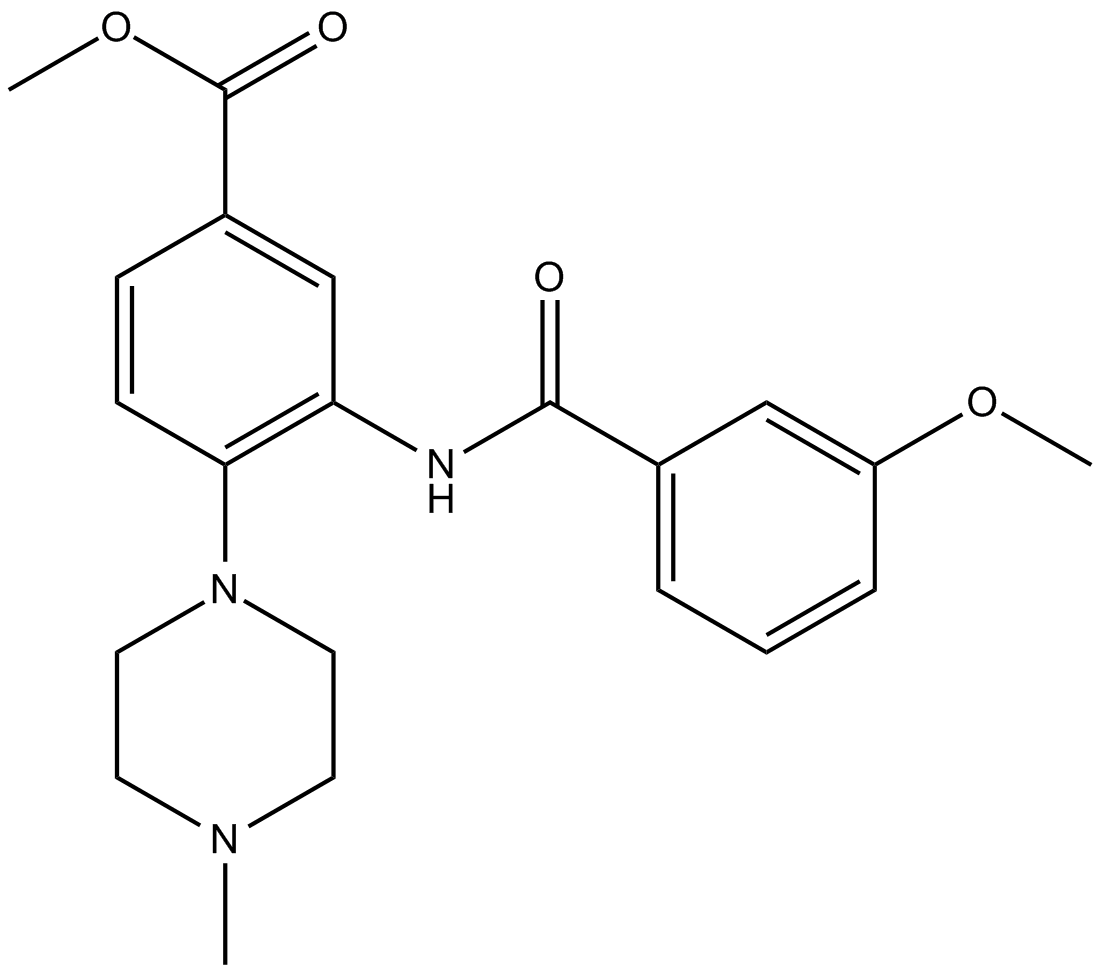
GC14763
WDR5 0103
WD repeat-containing protein 5 (WDR5) antagonist
-
GC62558
WDR5-IN-1
WDR5-IN-1 is a potent and selective WD repeat domain 5 (WDR5) inhibitor, with a Kd of <0.02 nM. WDR5-IN-1 inhibits MLL1 histone methyltransferase (HMT) activity with an IC50 of 2.2 nM. WDR5-IN-1 diminishes MYC recruitment at WDR5-displaced genes and exhibits potent anti-proliferative effects in CHP-134 (neuroblastoma) and Ramos (Burkitt’s lymphoma) lines.
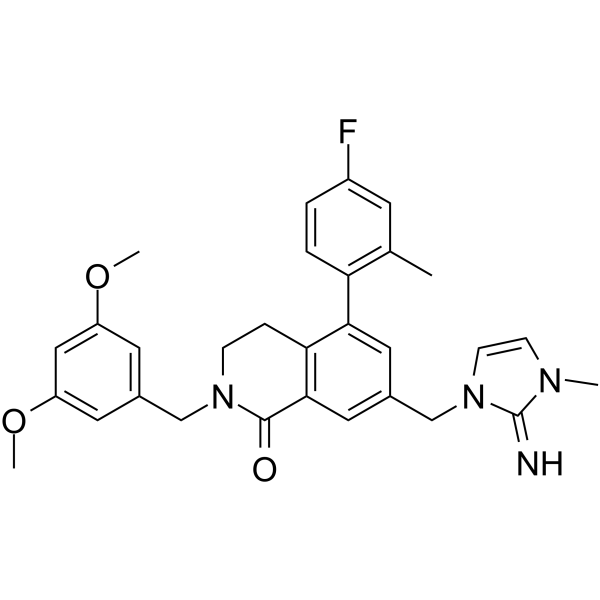
-
GC19388
XY1
XY1 is a very close analogue of SGC707 (a potent, selective, and non-competitive inhibitor of PRMT3 with IC50 of 31 nM), but XY1 is completely inactive.

-
GC45609
ZLD1039
An inhibitor of EZH2