Sirtuin
Silent information regulator 2 (Sir2) proteins, also known as sirtuins, are a family of nicotine adenine dinucleotide (NAD) dependent protein deacetylases in organisms ranging from bacteria to humans that are characterized by the presence of a unique and highly conserved catalytic domain of approximately 260 amino acids. Sirtuins are divided into 5 classes, including Class I (subclasses Ia, Ib and Ic), Class II, Class III, Class IV (subclasses IVa and IVb) and Class U (Gram-positive bacteria specific). The catalytic domain of sirtuins is comprised of two bilobed globular domains, the large domain containing the NAD-binding pocket and the small domain binding the acetyl-lysine substrate.
Products for Sirtuin
- Cat.No. Product Name Information
-
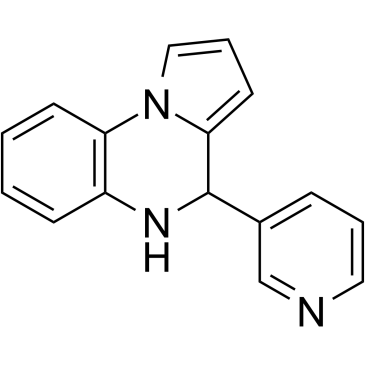
GC19013
3-TYP
3-TYP inhibit SIRT3 with an IC50 of 16 nM, and is more potent over SIRT1 and SIRT2 with IC50 of 88 nM and 92 nM, respectively.
-
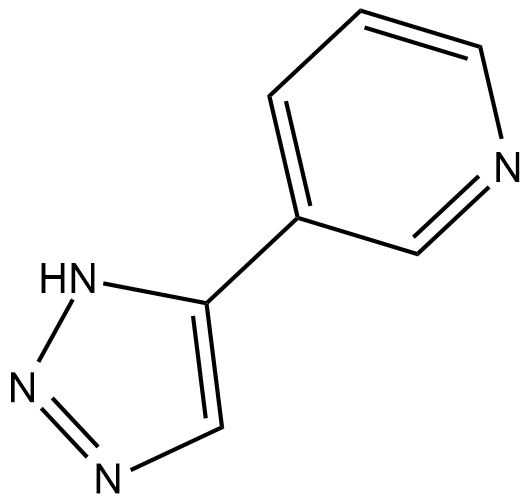
GC17922
4'-bromo-Resveratrol
Sirt1 and Sirt3 inhibitor
-
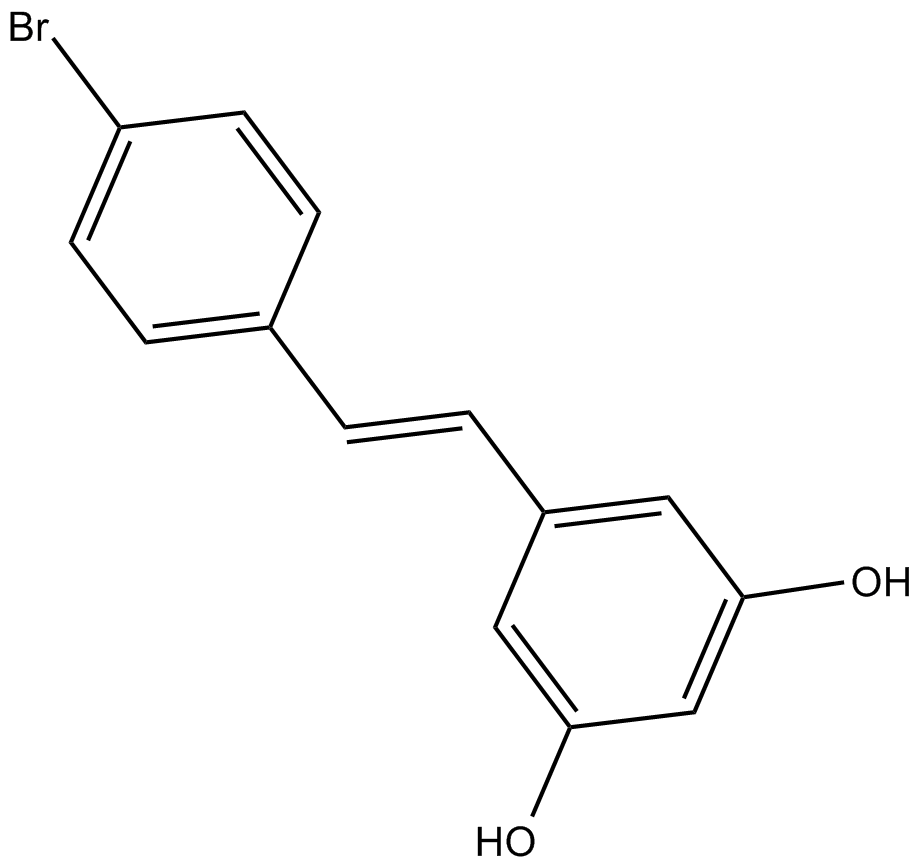
GC62651
7-Chloro-4-(piperazin-1-yl)quinoline
7-Chloro-4-(piperazin-1-yl)quinolone is an important scaffold in medicinal chemistry.
-
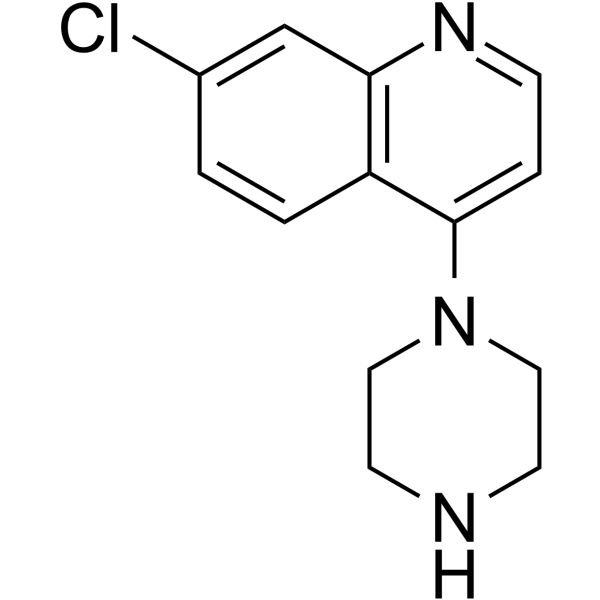
GC64527
ADTL-SA1215
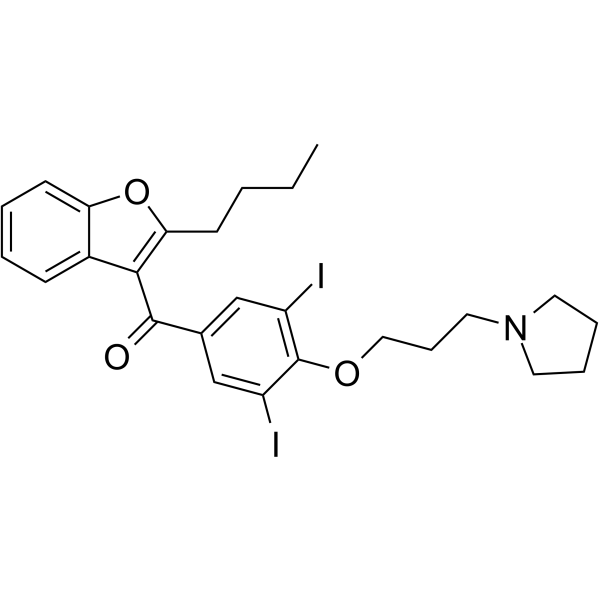
ADTL-SA1215 is a first-in-class specific small-molecule activator of SIRT3 that modulates autophagy in triple negative breast cancer.
-
GC17881
AGK 2
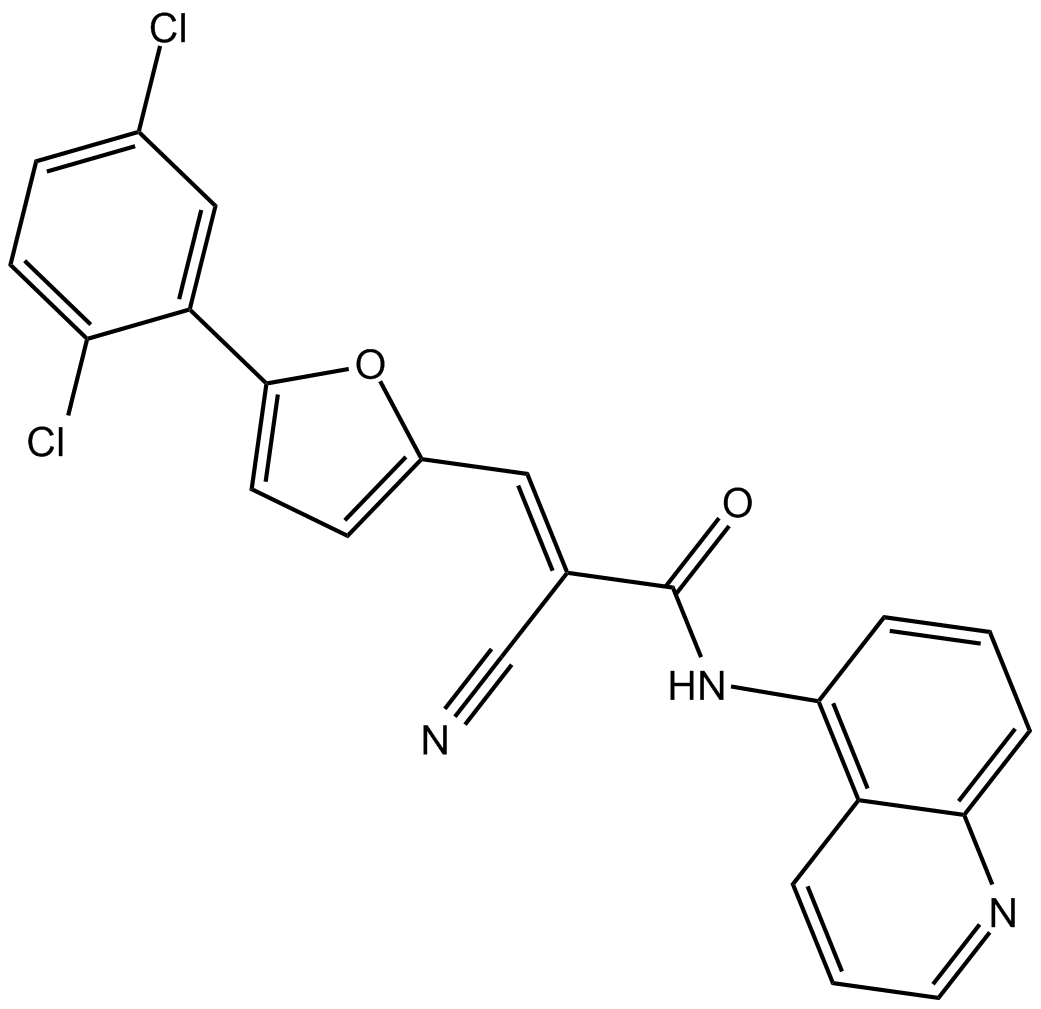
AGK2 is a selective SIRT2 inhibitor, with an IC50 of 3.
-
GC15931
AGK7
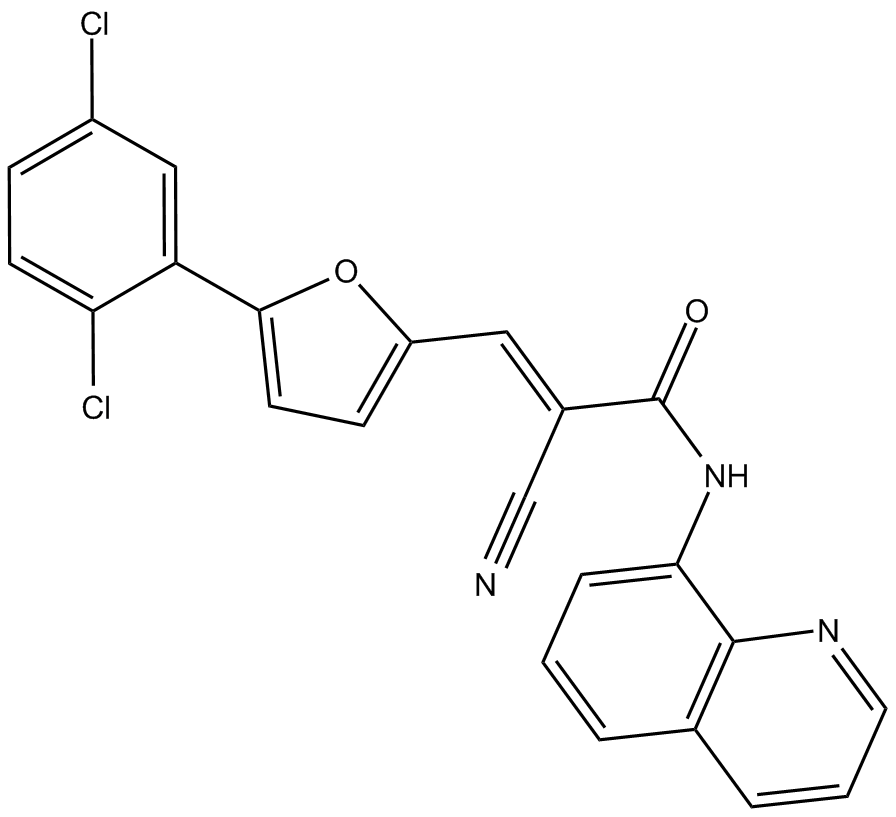
cell-permeable, selective inhibitor of SIRT2
-
GN10413
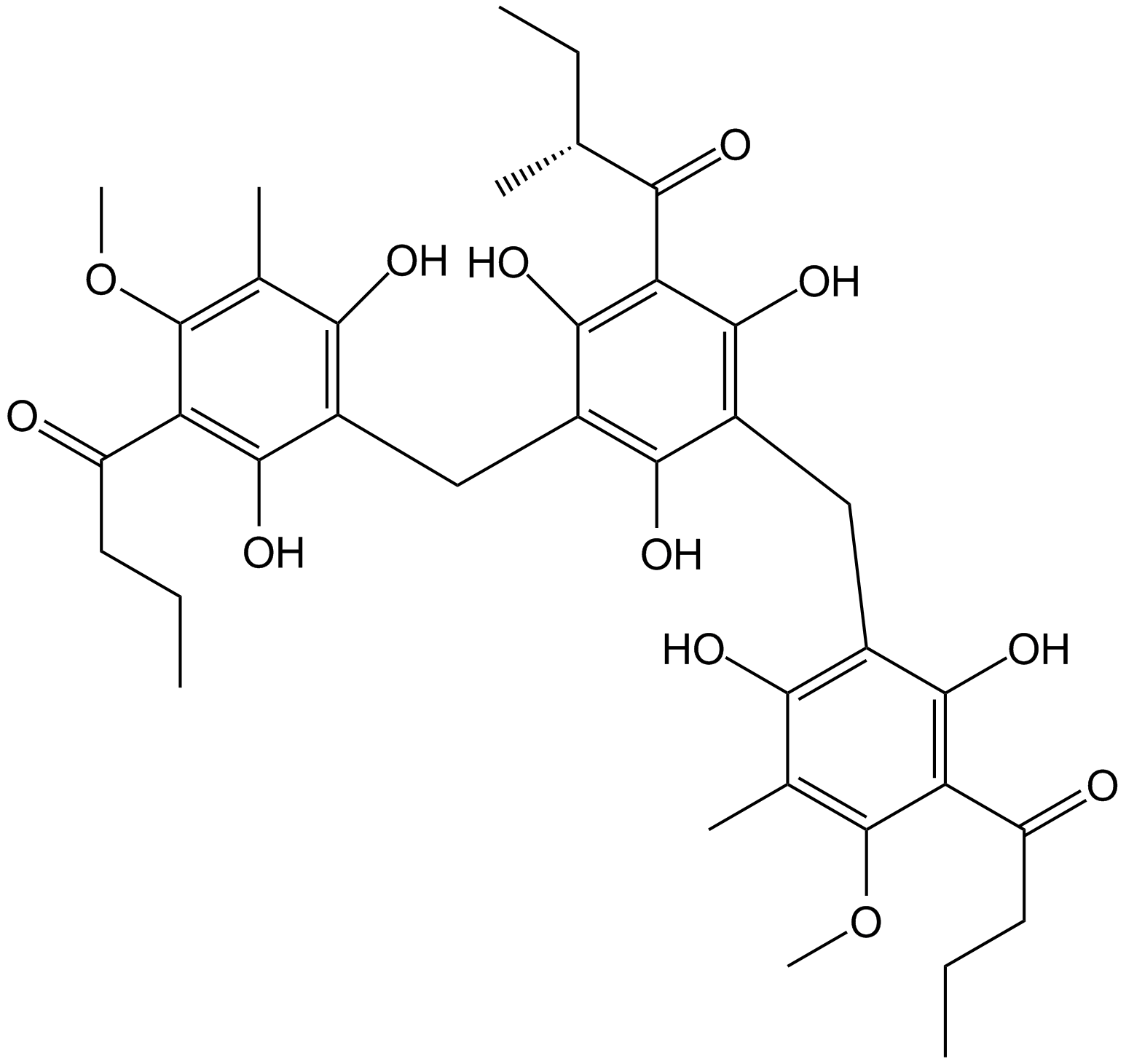
Agrimol B
-
GC10676
AK-7
selective and brain-permeable SIRT2 inhibitor
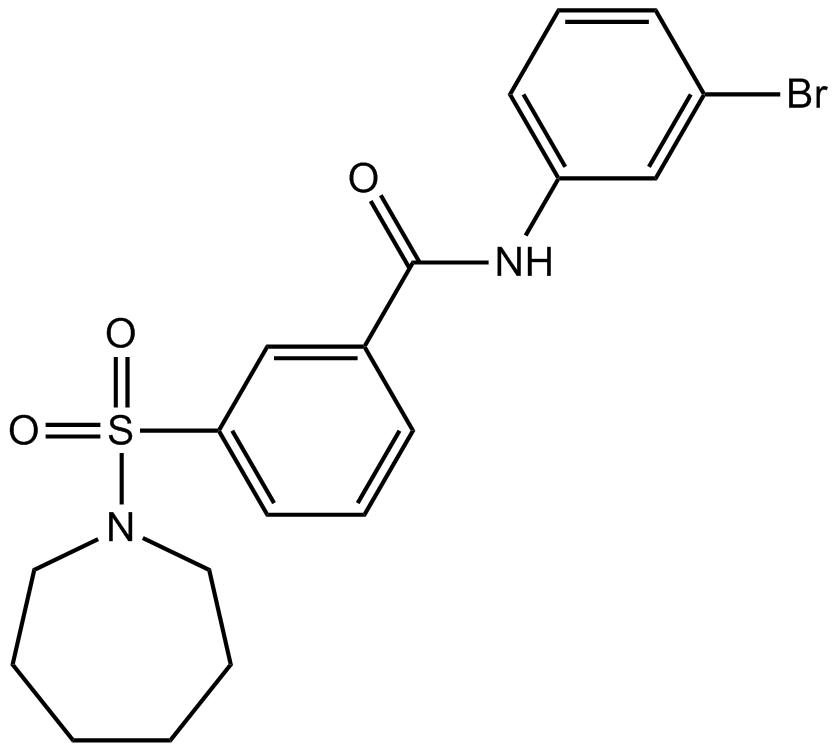
-
GC62886
Cannabisin F
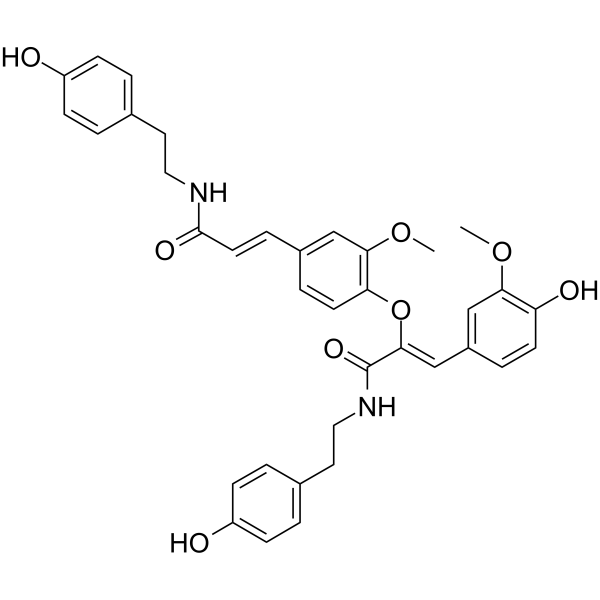
-
GC18228
CAY10602
Sirtuin 1 (SIRT1) is an evolutionarily conserved NAD-dependent protein deacetylase that has been implicated in cell metabolism, differentiation, stress and DNA damage response, and the control of multidrug resistance in cancer.
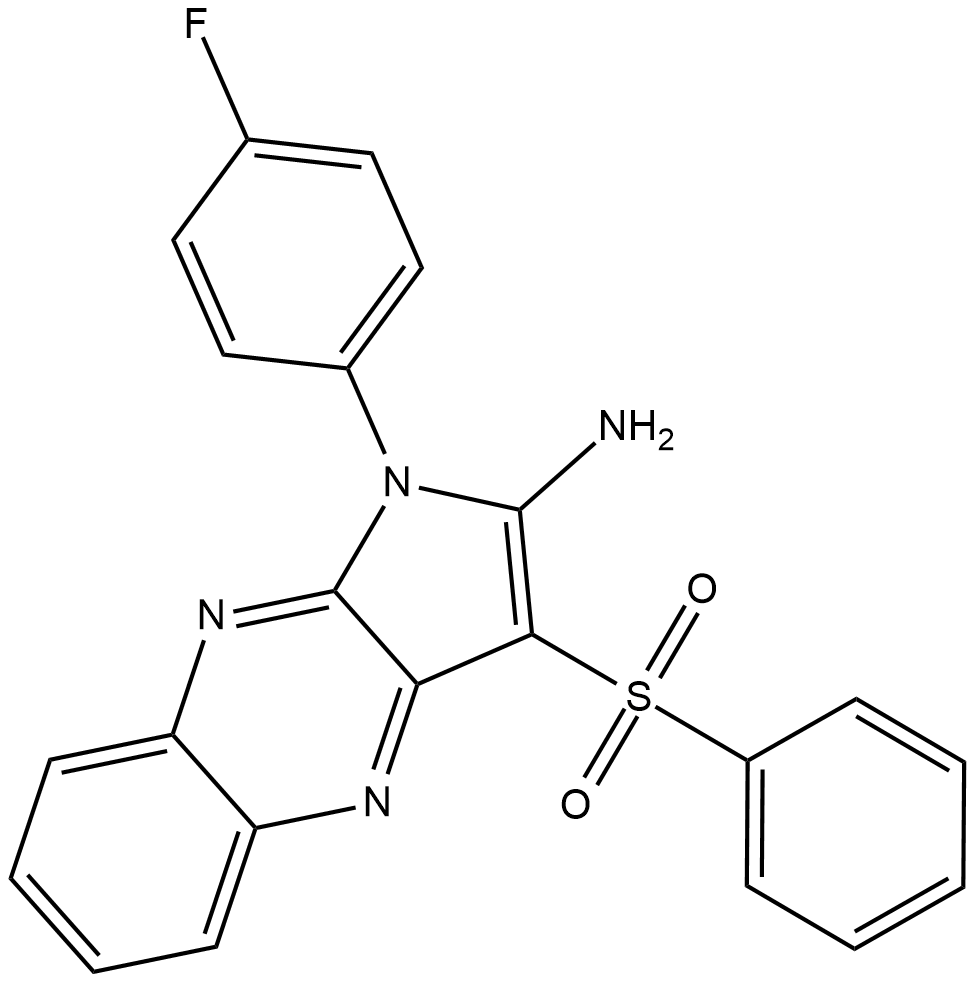
-
GC64255
CHIC35
CHIC35, an analog of EX-527, is a potent and selective inhibitor of SIRT1 (IC50=0.124 ?M).
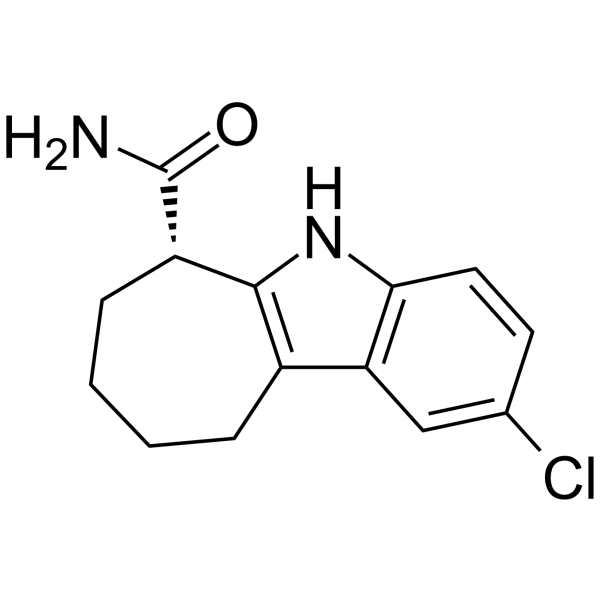
-
GC34187
Dihydrocoumarin (Hydrocoumarin)
Dihydrocoumarin (Hydrocoumarin) is a compound found in Melilotus officinalis. Dihydrocoumarin (Hydrocoumarin) is a yeast Sir2p inhibitor. Dihydrocoumarin (Hydrocoumarin) also inhibits human SIRT1 and SIRT2 with IC50s of 208 μM and 295 μM, respectively.
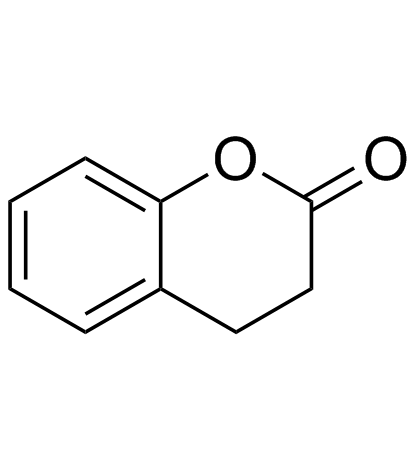
-
GC64575
Et-29
Et-29 is a potent and selective SIRT5 inhibitor (Ki=40 nM).
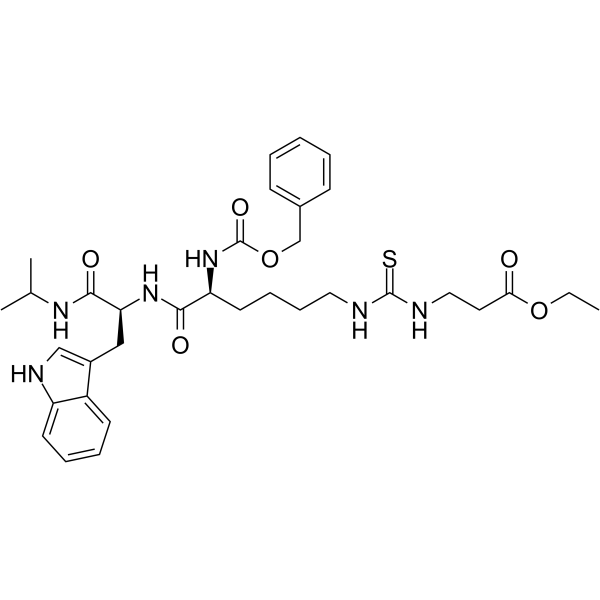
-
GC10635
EX 527 (SEN0014196)
A SIRT1 inhibitor
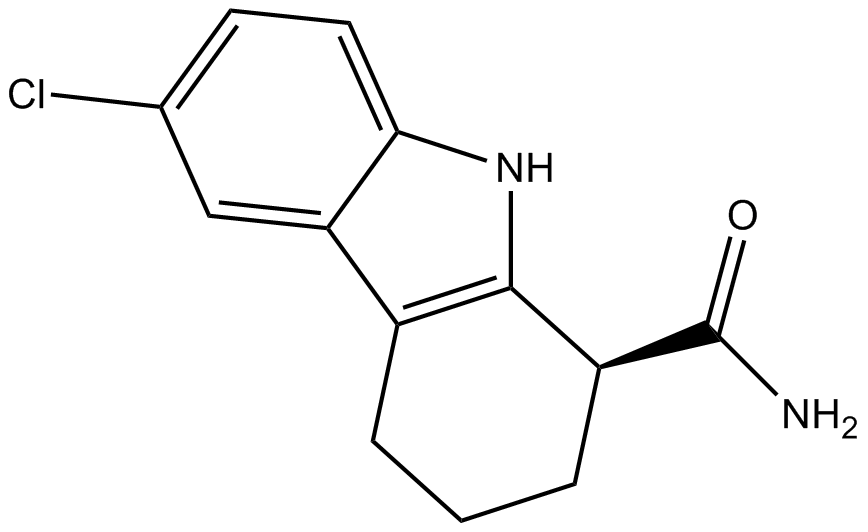
-
GC17126
EX-527 R-enantiomer
EX-527 R-enantiomer ((R)-EX-527) is a R-enantiomer of Selisistat. Selisistat (EX-527) is a potent and selective SIRT1 inhibitor with IC50 of 98 nM.
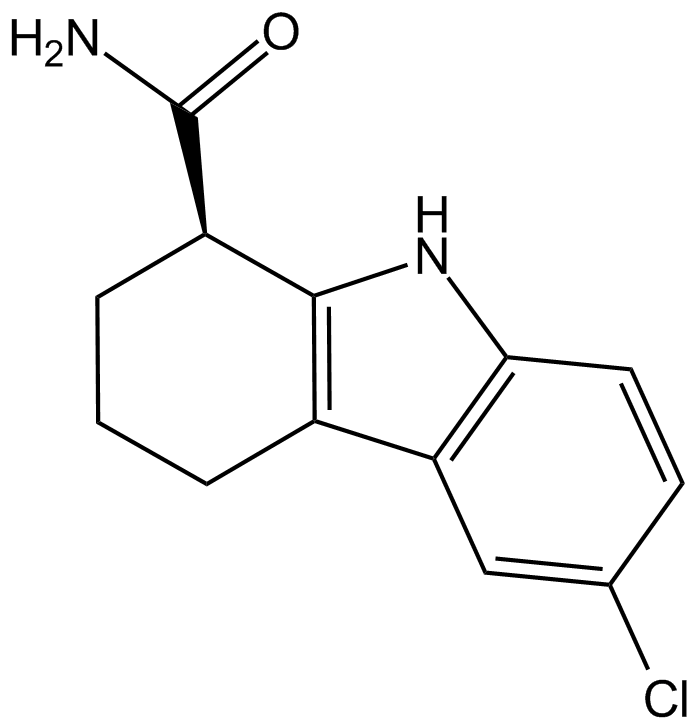
-
GC13417
EX-527 S-enantiomer
EX-527 S-enantiomer ((S)-EX-527) is a potent and selective SIRT1 inhibitor, with an IC50 of 98 nM.
-
GN10030
Fisetin
-
GC40900
Ganoderic Acid D
Ganoderic acid D is an oxygenated triterpenoid that has been found in G.
-
GC60172
Gardenia yellow
Gardenia yellow is an active member of crocin, increases mRNA expression of SIRT3, and acts as an orally active antidepressant agent.

-
GN10473
Ginkgolide C
-
GC14755
Inauhzin
SIRT1 inhibitor
-
GC14686
JFD00244
inhibitor of SIRT2
-
GC13062
JGB1741
SIRT1 inhibitor
-
GC34431
MC3482
MC3482 is a specific sirtuin5 (SIRT5) inhibitor.
-
GC65581
MIND4-19
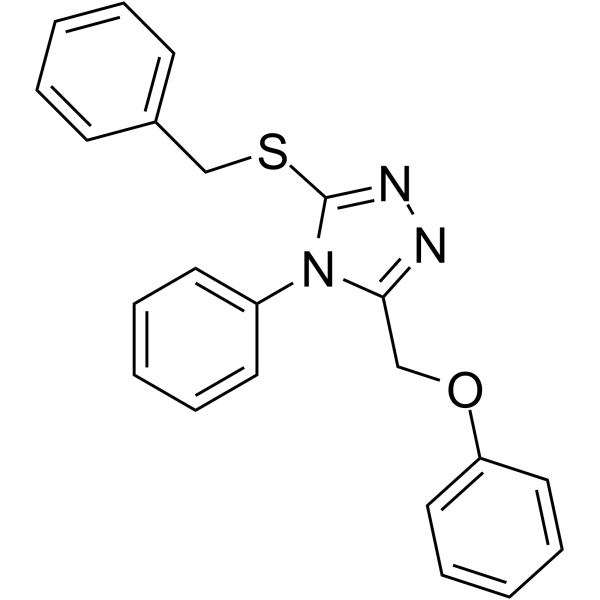
MIND4-19 is a potent SIRT2 inhibitor with an IC50 value of 7.0 μM.
-
GN10347
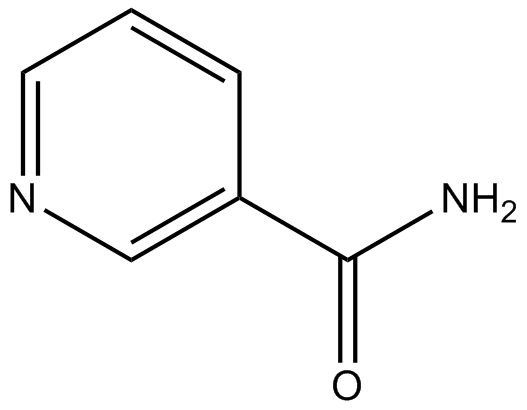
Nicotinamide (Vitamin B3)
Nicotinamide is an inhibitor of poly(ADP-ribose) polymerase (PARP-1) enzymes.
-
GC44401
Nicotinamide riboside
Nicotinamide riboside, a form of vitamin B3 and NAD+ precursor, is converted to bioavailable NAD+, via nicotinamide riboside kinase (NRK) and NMNAT, or by the action of nucleoside phosphorylase and NAM salvage.

-
GC36738
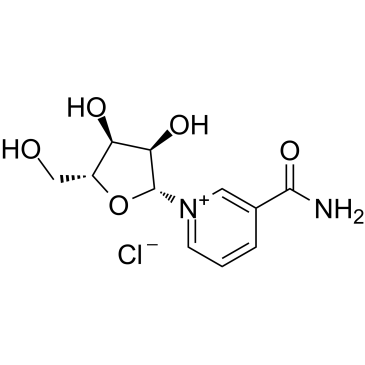
Nicotinamide riboside chloride
A nicotinamide adenine dinucleotide (NAD[+]) precursor vitamin
-
GN10420
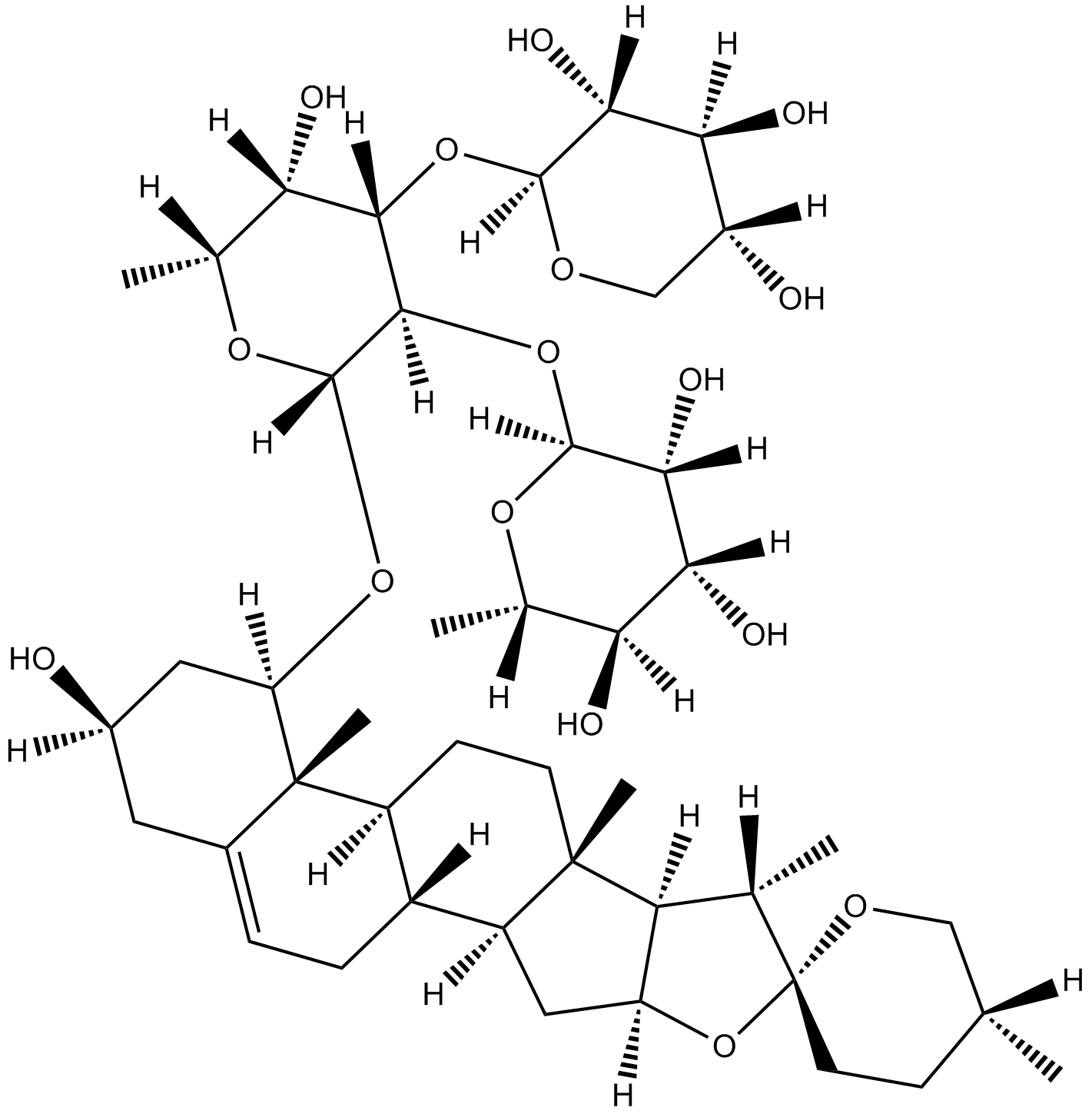
Ophiopogonin D'
-
GC32745
OSS_128167
OSS_128167 is a potent selective silencing regulatory protein 6 (SIRT6) inhibitor with an IC50 of 89 μM.
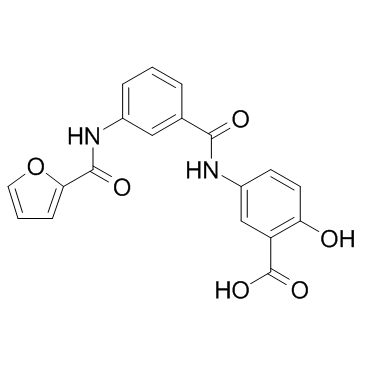
-
GC33418
PROTAC Sirt2 Degrader-1
PROTAC Sirt2 Degrader-1 is a SirReal-based PROTAC, acts as a Sirt2 degrader, composed of a highly potent and isotype-selective Sirt2 inhibitor, a linker, and a bona fide Cereblon ligand for E3 ubiquitin ligase. PROTAC Sirt2 Degrader-1 shows an IC50 of 0.25 μM for Sirt2, with no effect on Sirt1/Sirt3 (IC50s>100 μM).
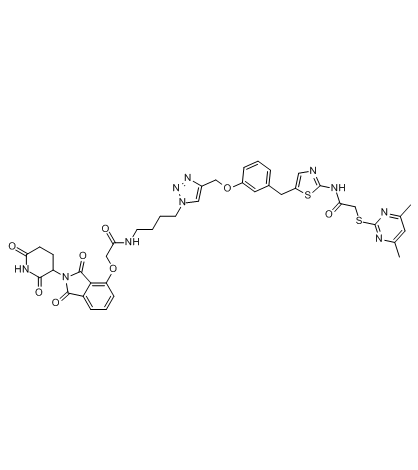
-
GC14553
Resveratrol
Resveratrol (trans-Resveratrol; SRT501) is a phytoalexin
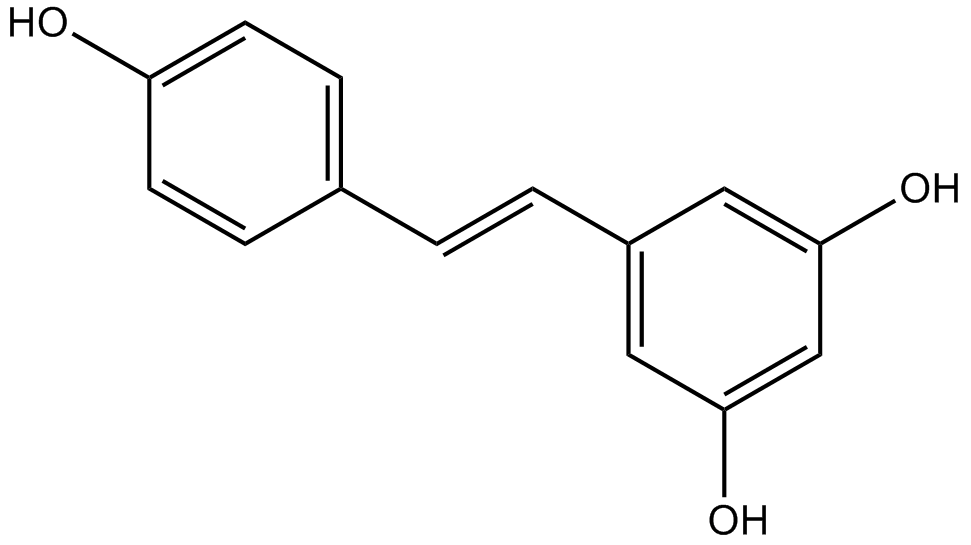
-
GC14817
Salermide
SIRT1 and SIRT2 inhibitor
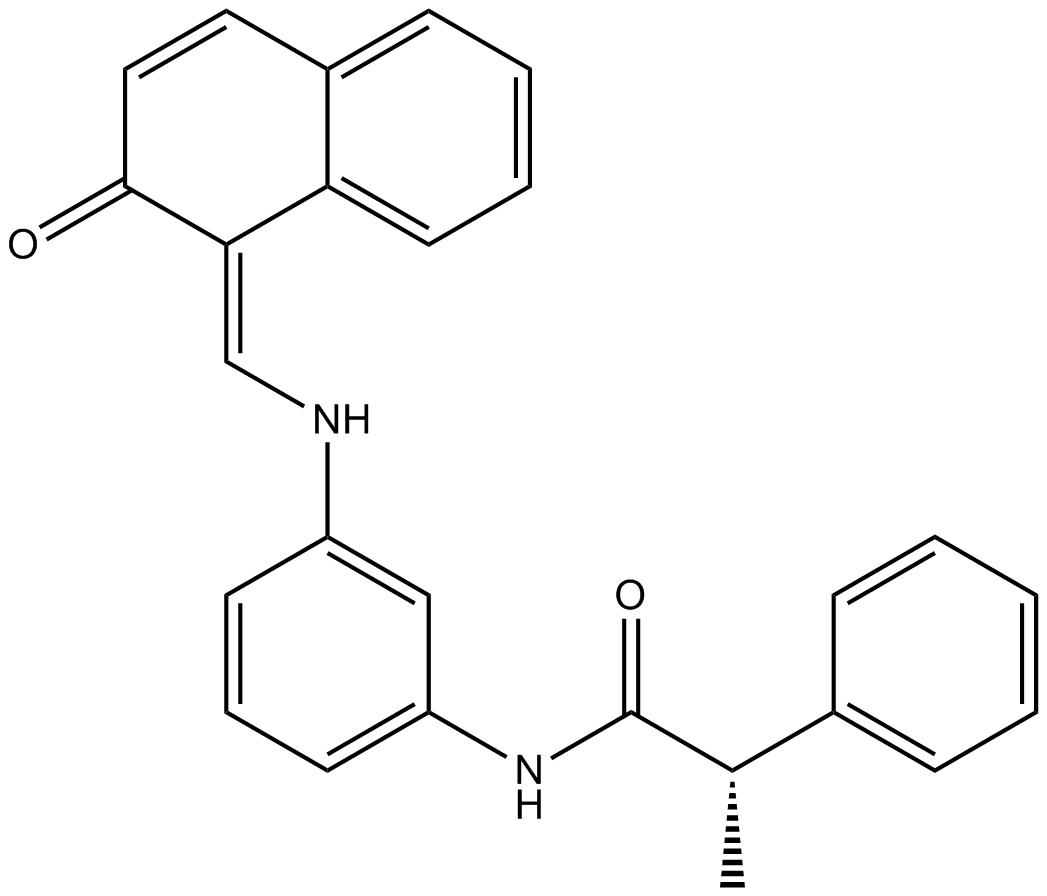
-
GC39435
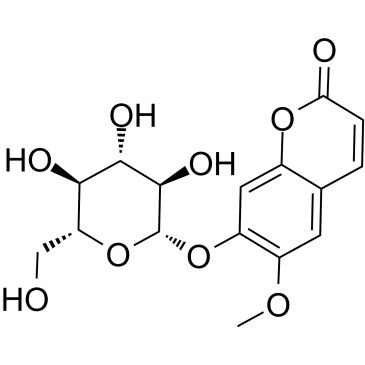
Scopolin
Scopolin is a coumarin isolated from Arabidopsis thaliana (Arabidopsis) roots.
-
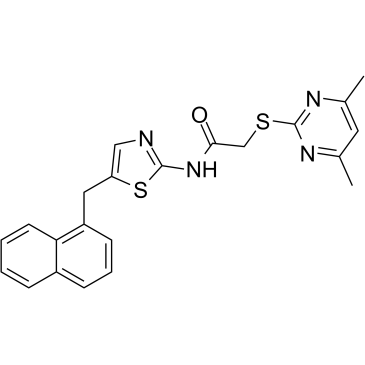
GC38491
SirReal2
A selective SIRT2 inhibitor
-
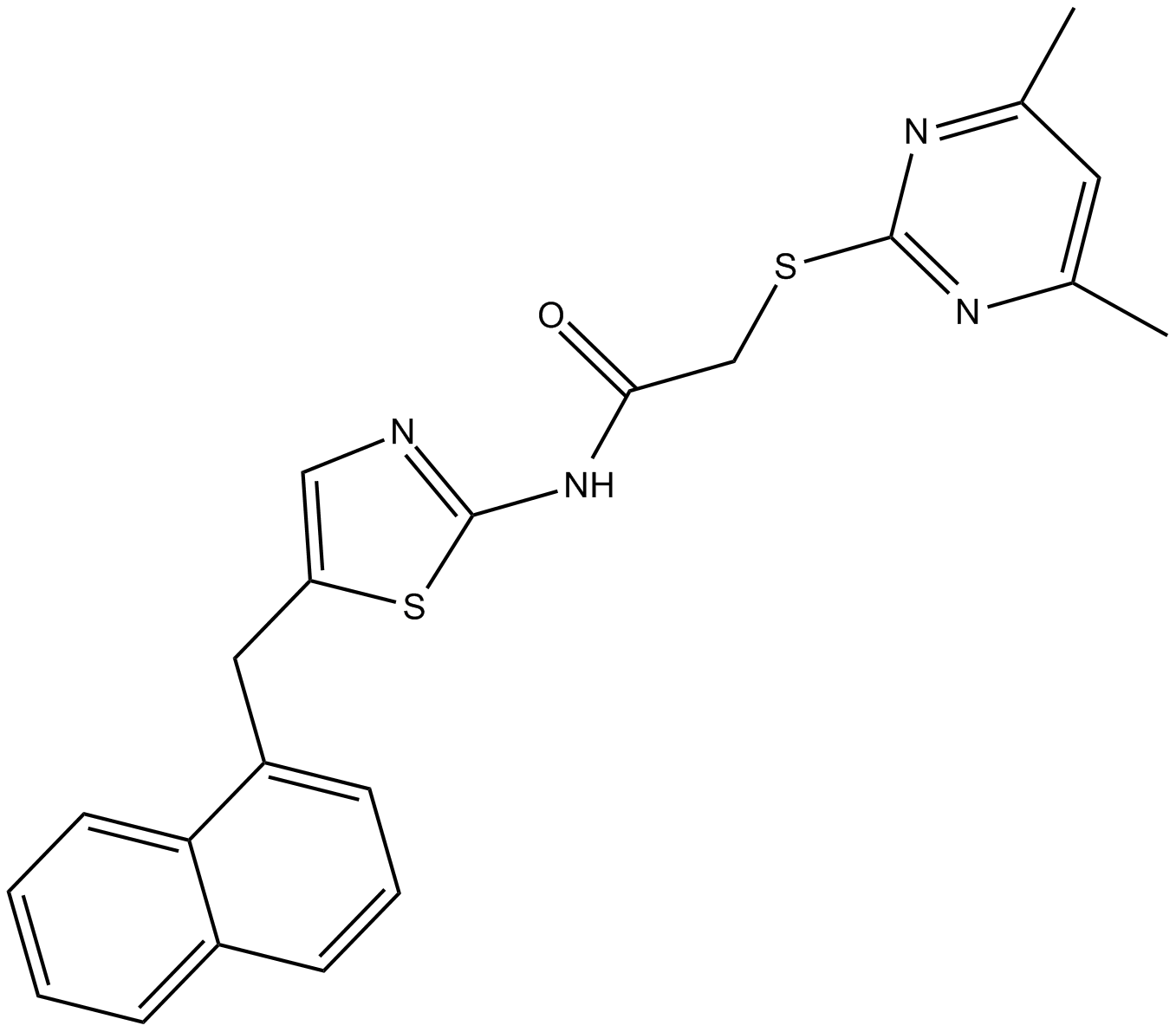
GC13942
SirReal2
Sirt2 inhibitor
-
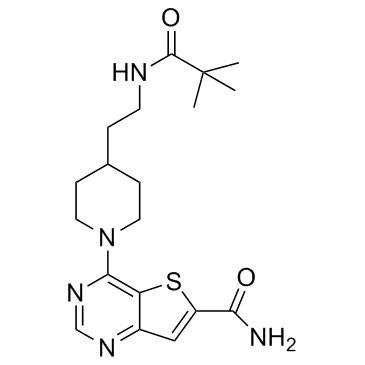
GC30733
SIRT-IN-1
SIRT-IN-1 is a potent inhibitor of SIRT1/2/3, with IC50s of 15, 10, 33 μM, respectively.
-
GC30693
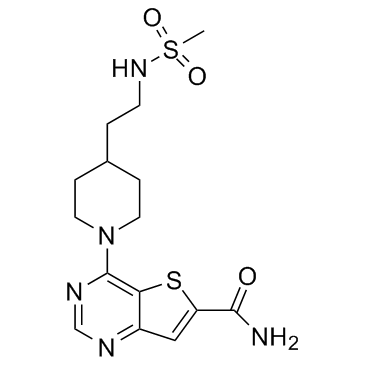
SIRT-IN-2
SIRT-IN-2 is a potent inhibitor of SIRT1/2/3, with IC50s of 4, 4, 7 μM, respectively.
-
GC61279
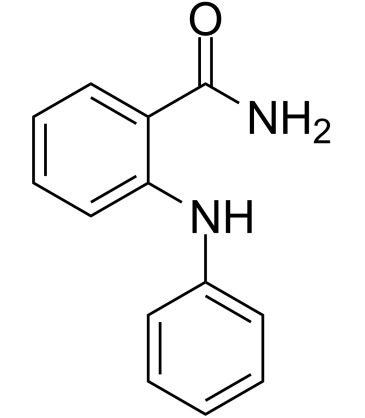
SIRT-IN-3
SIRT-IN-3 is a potent SIRT inhibitor, with an IC50 of 17 μM for SIRT1. SIRT-IN-3 shows about 4-fold and 14-fold selectivity for SIRT1 over SIRT2 and SIRT3, respectively (IC50 of 74 μM and 235 μM for SIRT2 and SIRT3, respectively).
-
GC69897
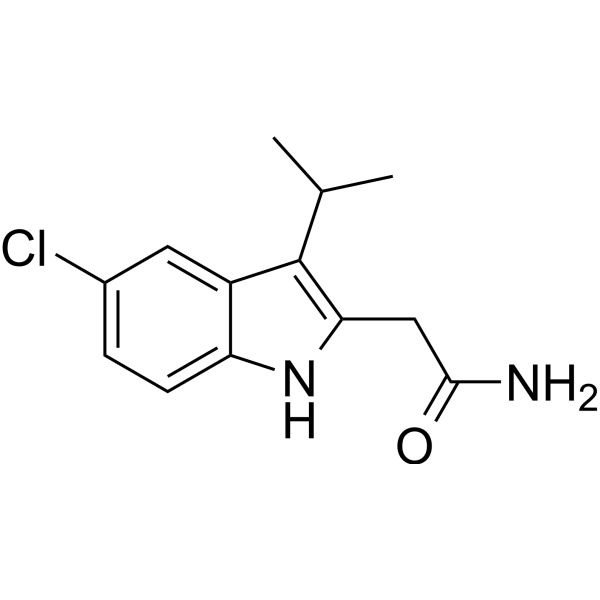
SIRT1-IN-2
SIRT1-IN-2 (compound 3h) is an effective and selective inhibitor of SIRT1 (Silent Information Regulator Factor 1), with an IC50 of 1.6 μM.
-
GC11716
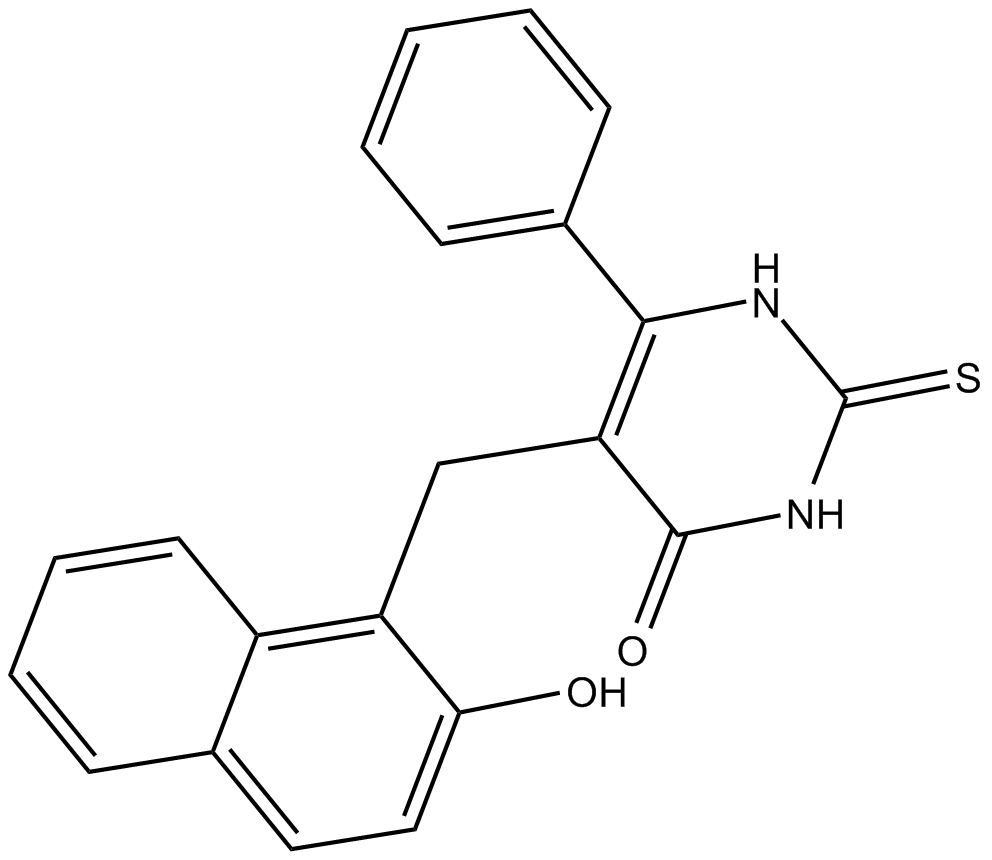
SIRT1/2 Inhibitor IV
SIRT1/2 Inhibitor IV is a SIRT1 and SIRT2 inhibitor with IC50 values of 56 μM and 59 μM, respectively. SIRT1/2 Inhibitor IV is a potent brain penetrant neutral sphingomyelinase (N-SMase) inhibitor (exosome inhibitor).
-
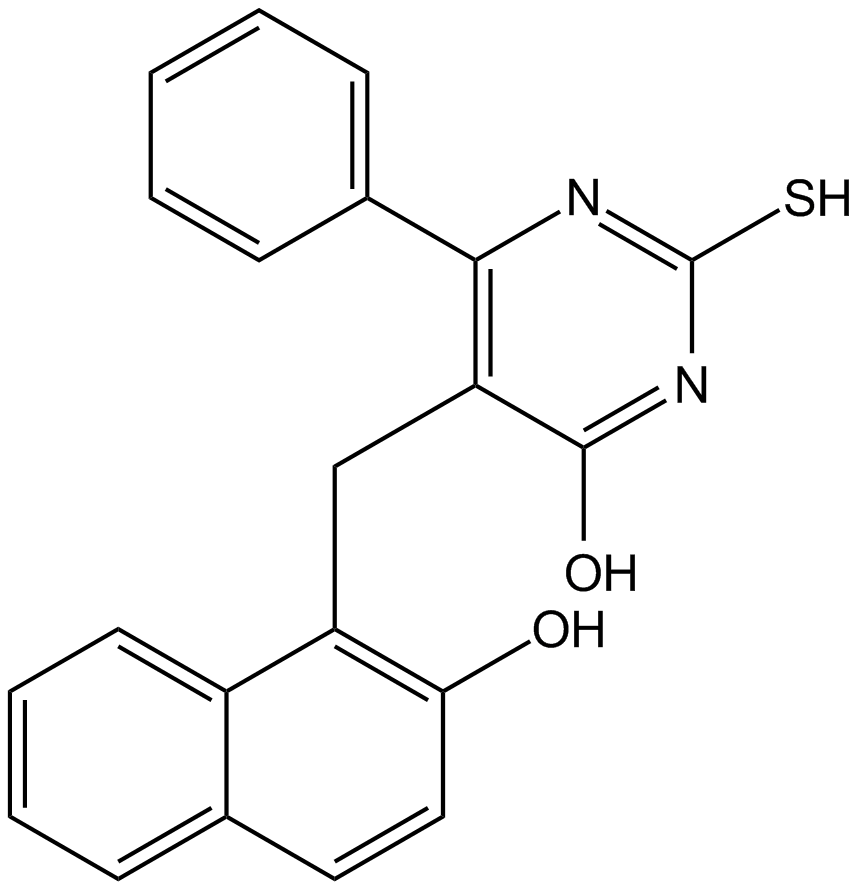
GC17145
SIRT1/2 Inhibitor IV
SIRT1 and SIRT2 inhibitor
-
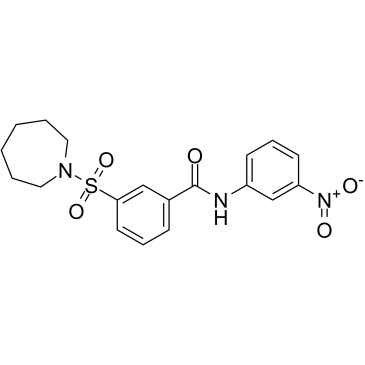
GC30922
SIRT2 Inhibitor II (AK-1)
SIRT2 Inhibitor II (AK-1) is a potent, specific and cell-permeable SIRT2 inhibitor, with an IC50 of 12.5 μM.
-
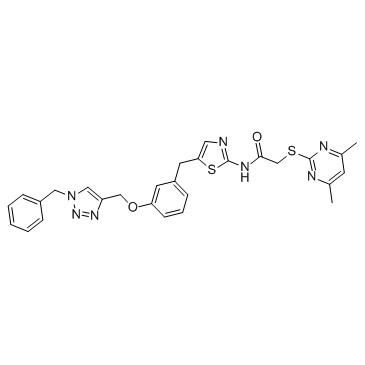
GC34785
Sirt2-IN-1
Sirt2-IN-1 (Compound 9) is a sirtuin 2 (Sirt2) inhibitor with an IC50 of 163 nM.
-
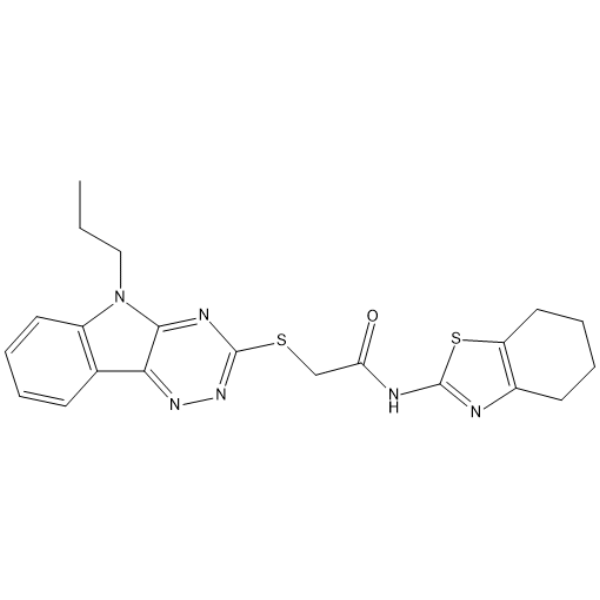
GC66026
SIRT2-IN-9
SIRT2-IN-9 (compound 12) is a selective inhibitor of SRIT2 with an IC50 value of 1.3 μM. SIRT2-IN-9 inhibits proliferative activity of MCF-7 breast cancer cells. SIRT2-IN-9 can be used for the research of cancer.
-
GC30251
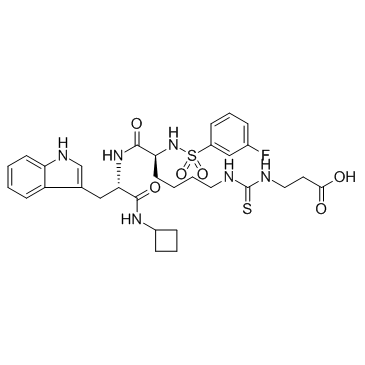
SIRT5 inhibitor
SIRT5 inhibitor is a potent Human Sirtuin 5 deacylase inhibitor, with an IC50 of 0.11 μM.
-
GC69898
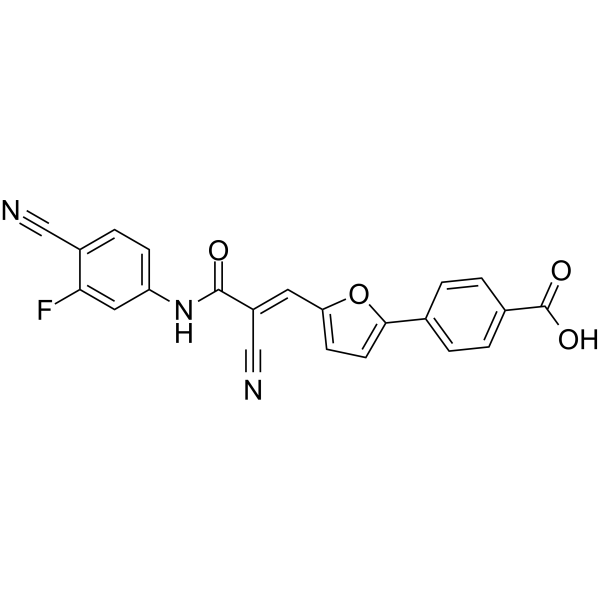
SIRT5 inhibitor 3
SIRT5 inhibitor 3 (compound 46) is an effective and competitive inhibitor of SIRT5, with an IC50 of 5.9 μM. It can inhibit the deacetylation activity of SIRT5. SIRT5 inhibitor 3 can be used for research on cancer and neurodegenerative diseases.
-
GC14945
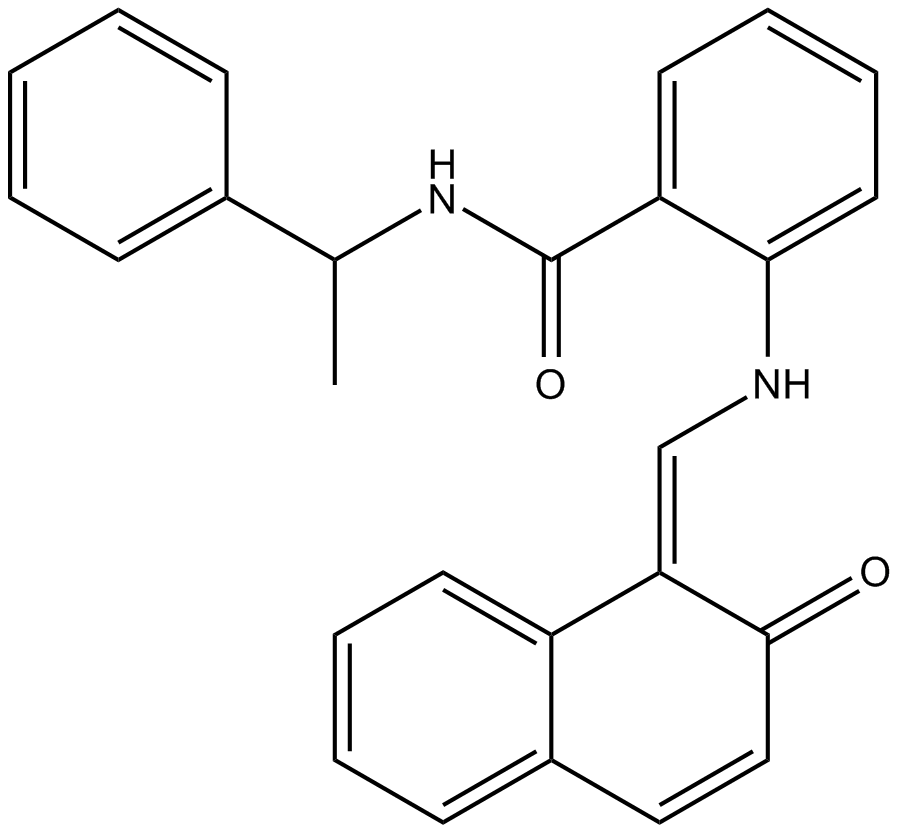
Sirtinol
Inhibitor of sirtuin deacetylases
-
GC30204
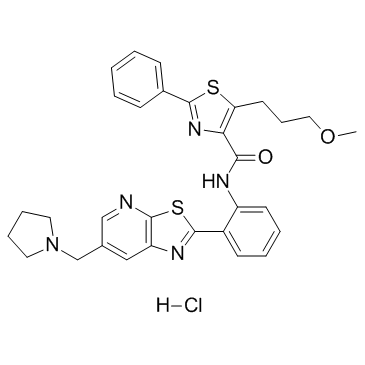
Sirtuin modulator 1
A SIRT1 activator
-
GC65030
Sirtuin modulator 2
Sirtuin modulator 2 (Compound 132) is a sirtuin modulator with an ED50 equal or less than 50 μM.
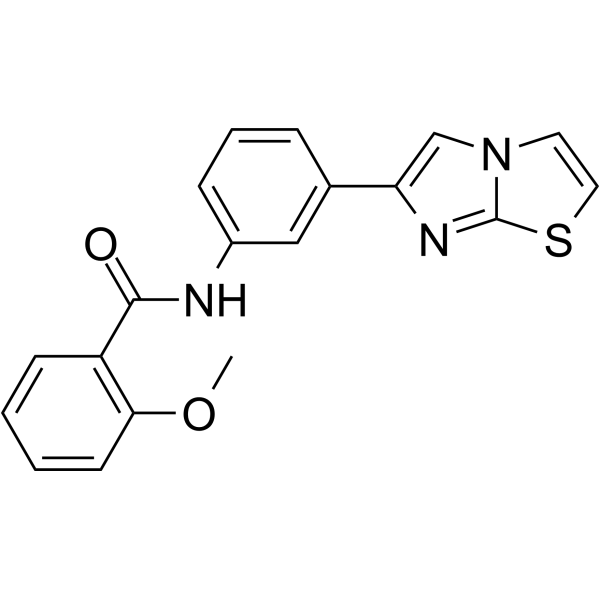
-
GC67778
Sirtuin modulator 3
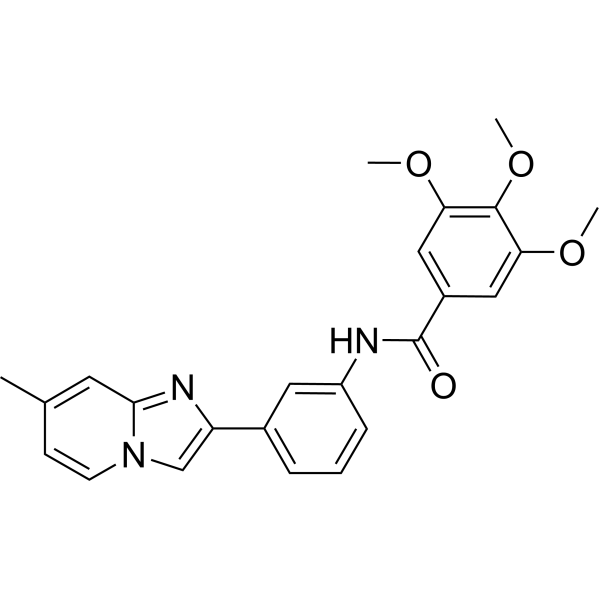
-
GC10555
Splitomicin
Inhibitor of yeast Sir2p
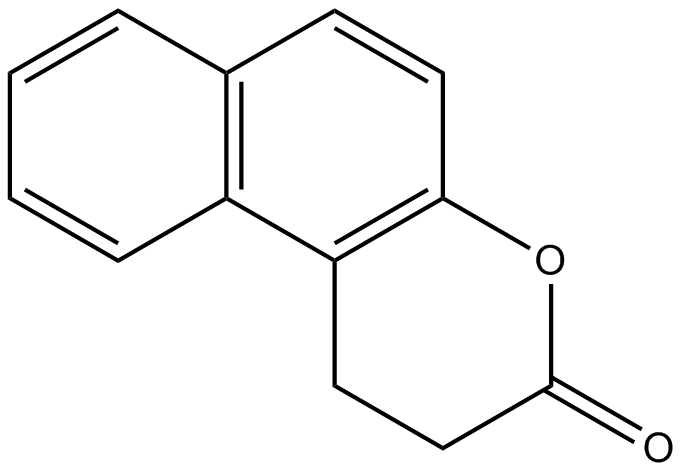
-
GC65019
SRT 1460
SRT 1460, a potent Sirtuin-1 (SIRT1) activator with an EC1.5 value of 2.9 μM, shows good selectivity for activation of SIRT1 versus SIRT2 and SIRT3 (EC1.5>300 μM), and is more potent than Resveratrol and the closest sirtuin homologues.
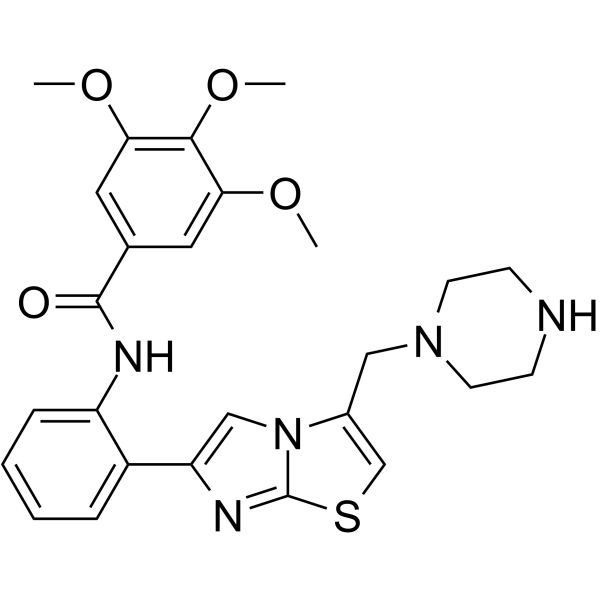
-
GC37677
SRT 1720
SRT 1720 is a selective activator of human SIRT1 with an EC1.5 of 0.16 μM, and shows less potent activities for SIRT2 and SIRT3 with EC1.5s of 37 μM and > 300 μM, respectively.
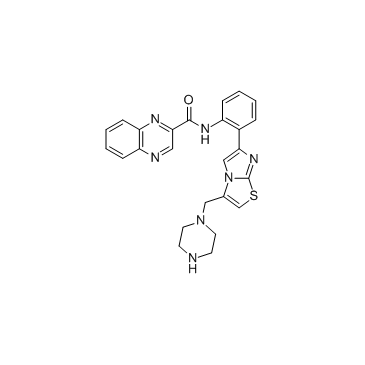
-
GC69950
SRT 1720 dihydrochloride
SRT 1720 dihydrochloride is a selective and orally active SIRT1 activator with an EC50 of 0.10 μM, showing weaker effects on SIRT2 and SIRT3.
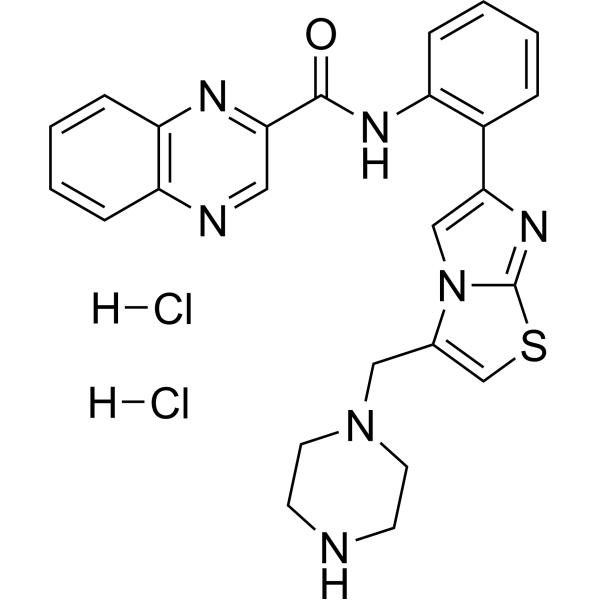
-
GC64946
SRT 2183
SRT 2183 is a selective Sirtuin-1 (SIRT1) activator with an EC1.5 value of 0.36 μM. SRT 2183 induces growth arrest and apoptosis, concomitant with deacetylation of STAT3 and NF-κB, and reduction of c-Myc protein levels.
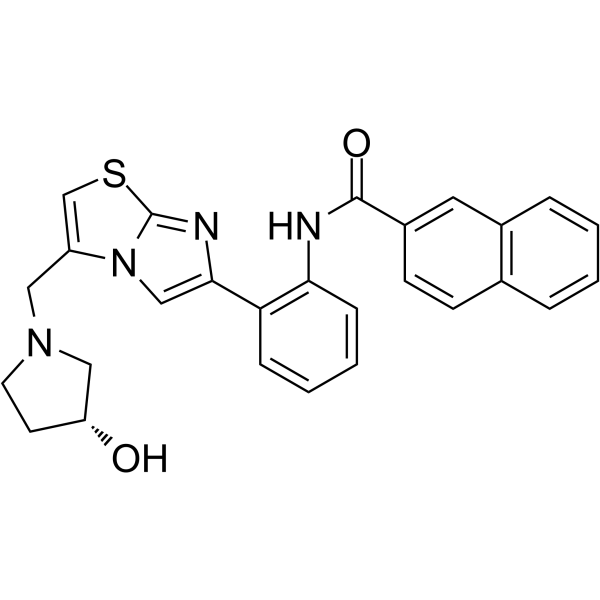
-
GC17101
SRT1720 HCl
SRT1720 HCl is an activator of SIRT1 (EC1.
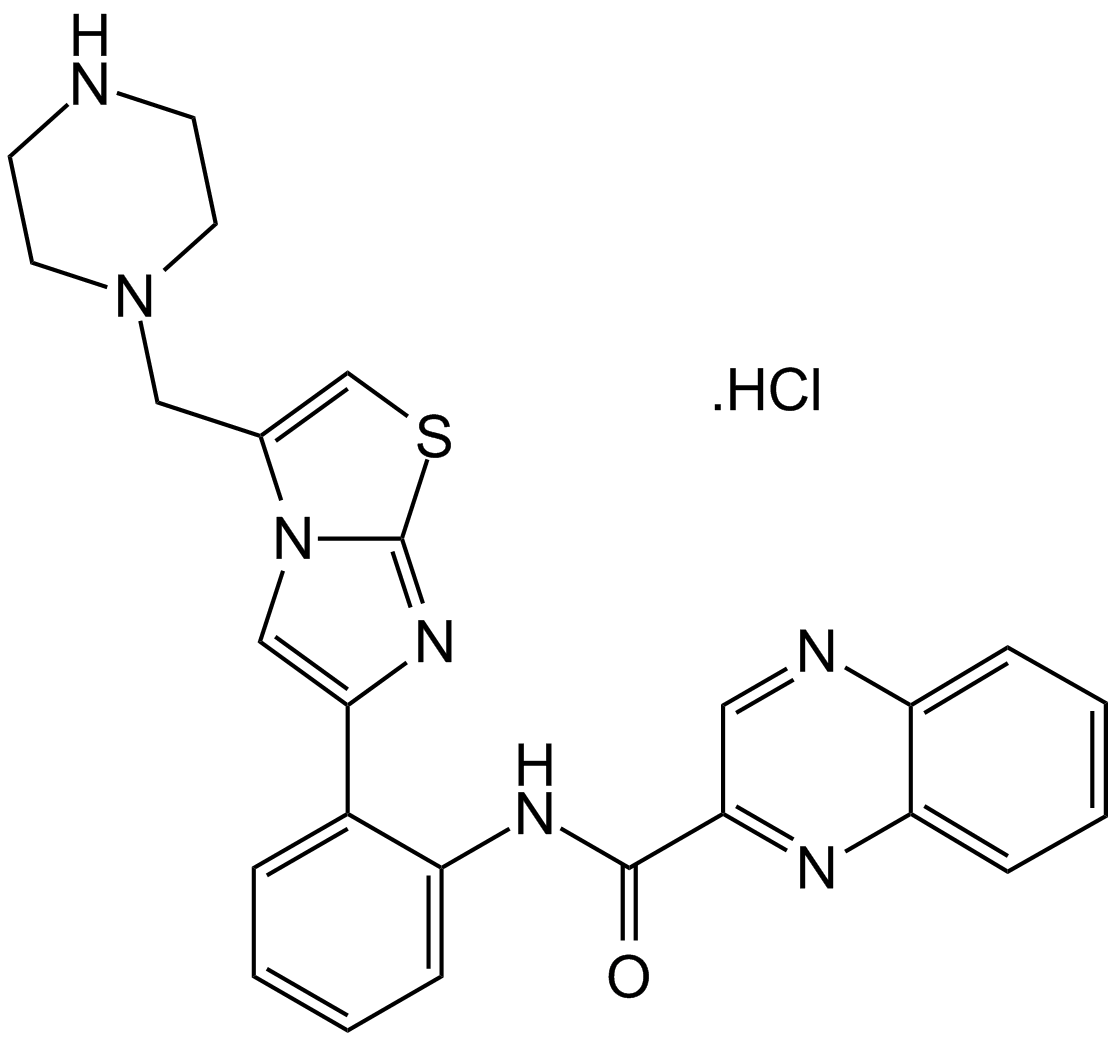
-
GC15109
SRT2104 (GSK2245840)
SRT2104 (GSK2245840) is a highly selective small-molecule activator of Sirtuin 1 (SIRT1).

-
GC14165
Tenovin-1
A small molecule activator of p53
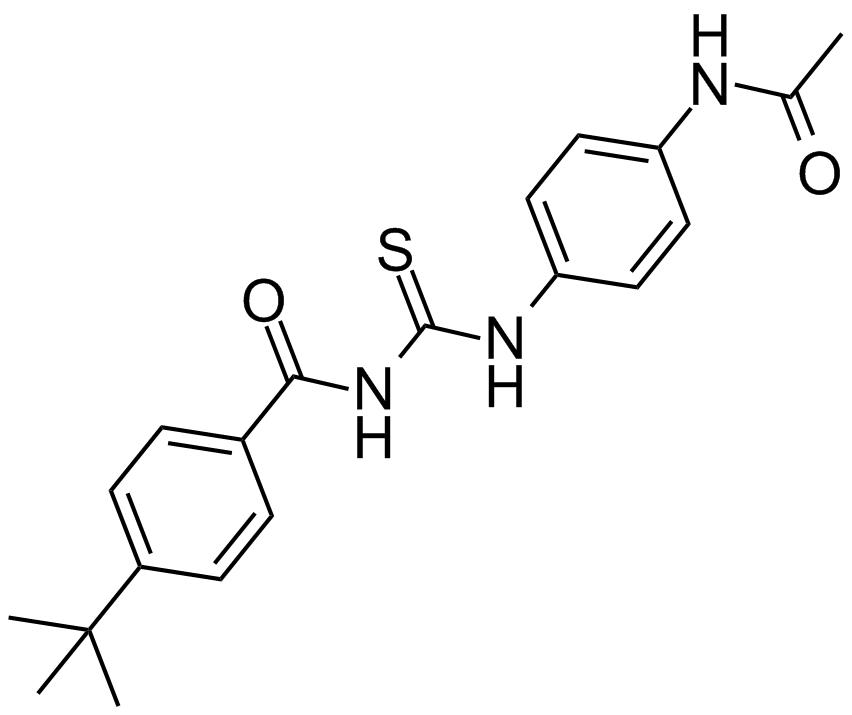
-
GC12337
Tenovin-3
p53 activator
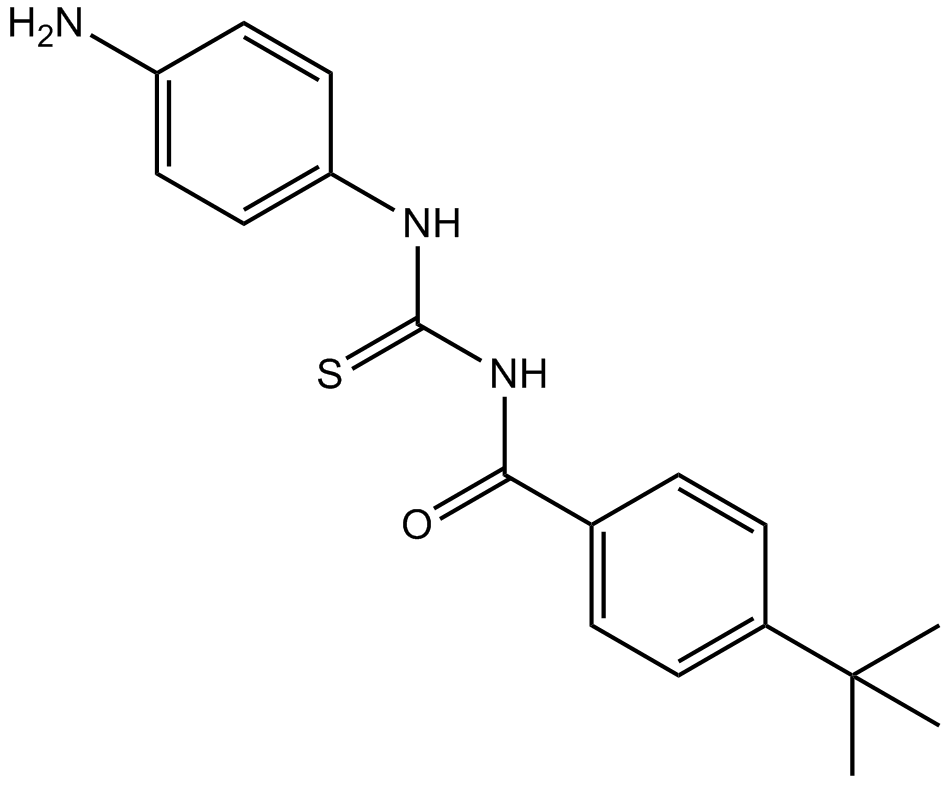
-
GC16436
Tenovin-6
SIRT inhibitor and p53 activator
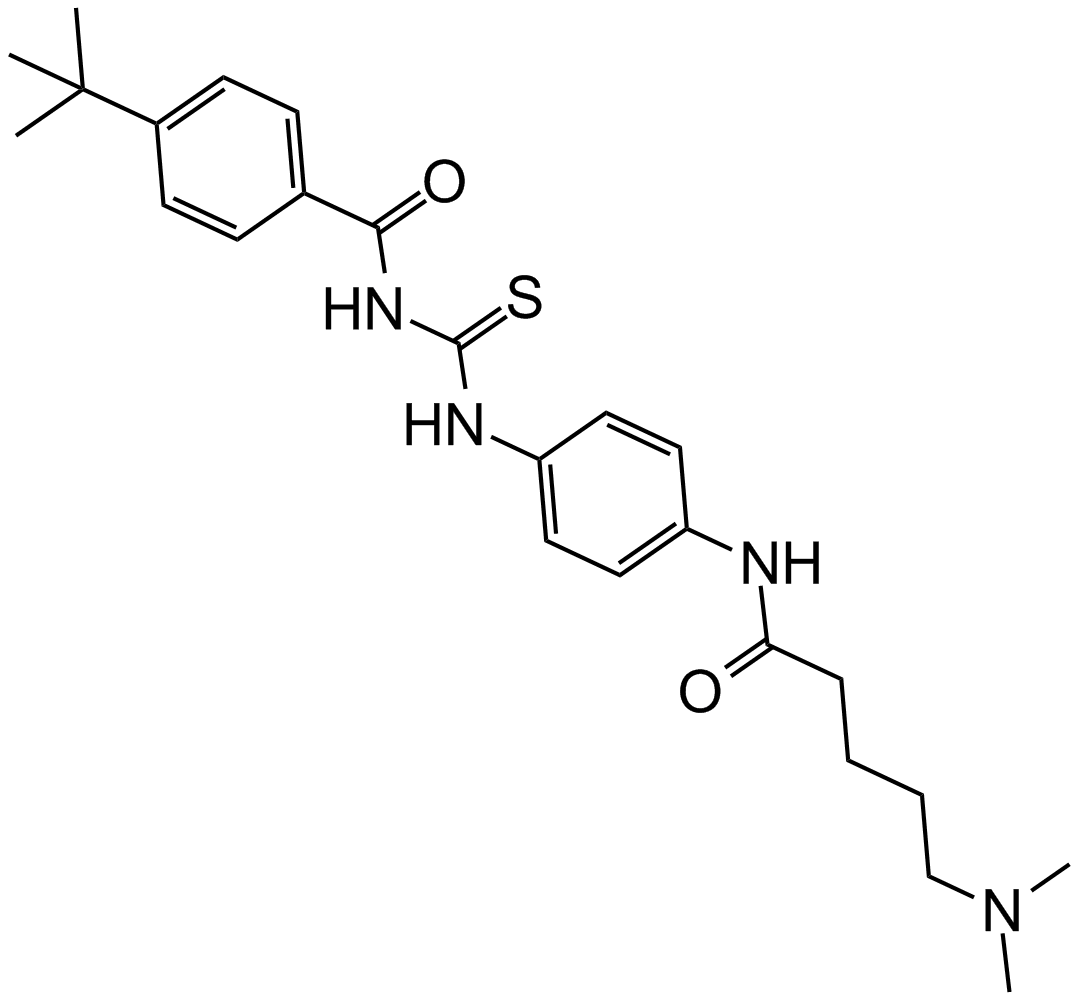
-
GC37761
Tenovin-6 Hydrochloride
Tenovin-6 Hydrochloride, an analog of Tenovin-1, is an activator of p53 transcriptional activity. Tenovin-6 Hydrochloride inhibits the protein deacetylase activities of purified human SIRT1, SIRT2, and SIRT3 with IC50s of 21 μM, 10 μM, and 67 μM, respectively. Tenovin-6 Hydrochloride also inhibits dihydroorotate dehydrogenase (DHODH).
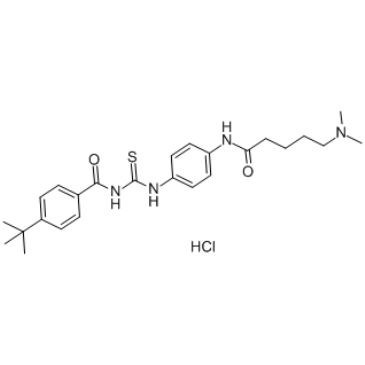
-
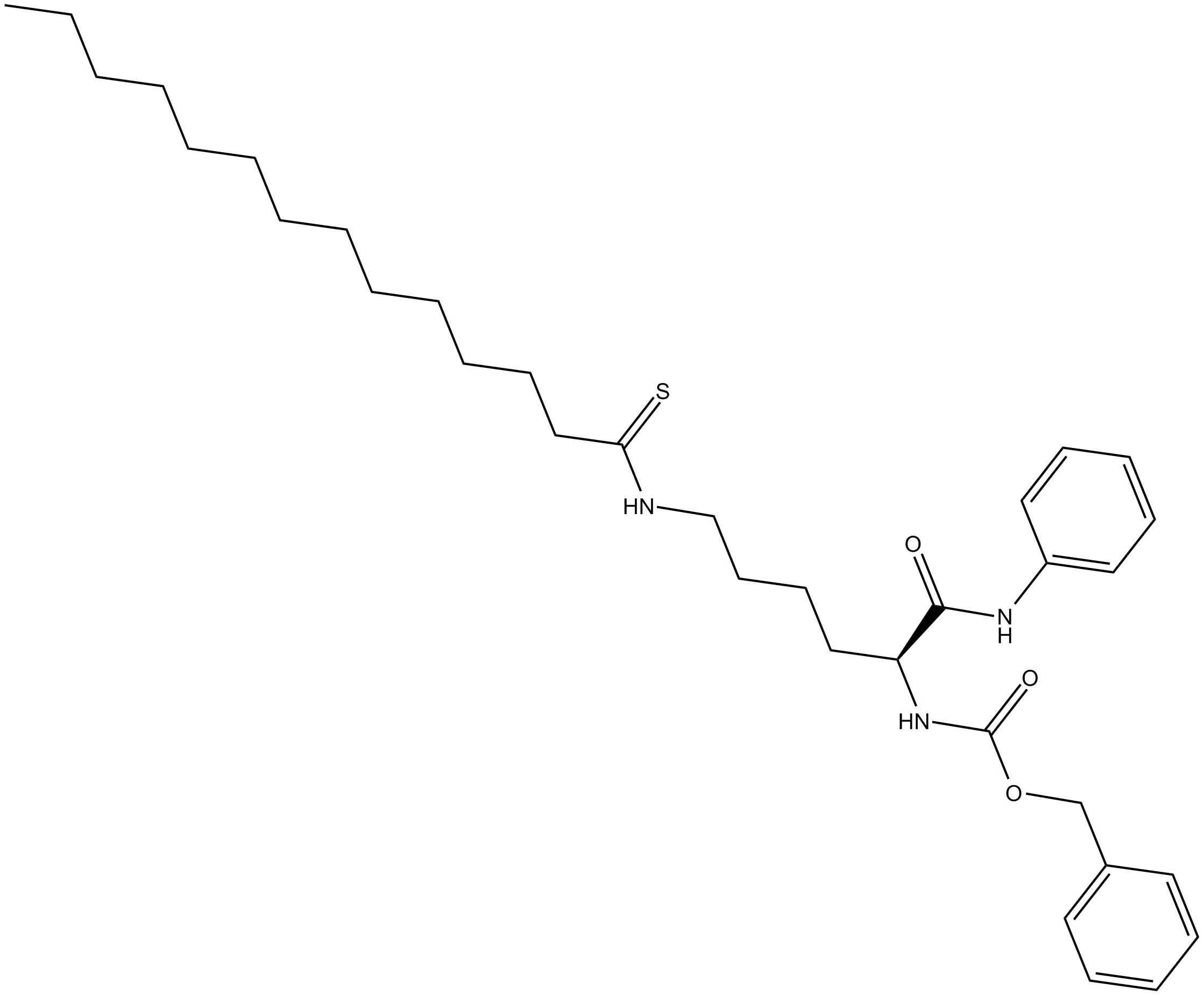
GC17724
Thiomyristoyl
A potent and selective SIRT2 inhibitor
-
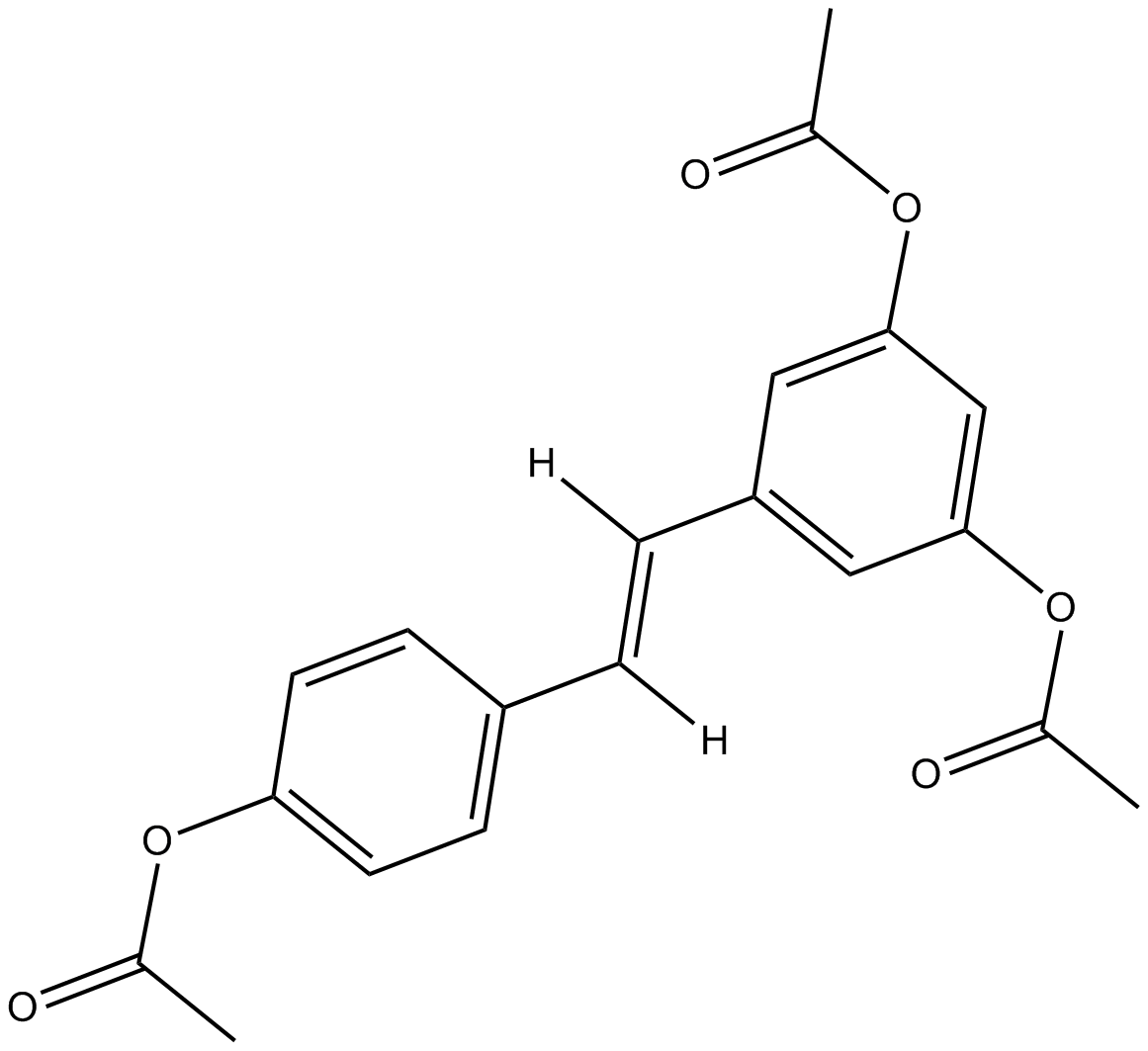
GC17704
Triacetyl Resveratrol
Cell-permeable resveratrol prodrug
-
GC34849
UBCS039
UBCS039 is the first synthetic, specific Sirtuin 6 (SIRT6) activator, inducing autophagy in human tumor cells, with an EC50 of 38 μM.