Immunology/Inflammation
The immune and inflammation-related pathway including the Toll-like receptors pathway, the B cell receptor signaling pathway, the T cell receptor signaling pathway, etc.
Toll-like receptors (TLRs) play a central role in host cell recognition and responses to microbial pathogens. TLR4 initially recruits TIRAP and MyD88. MyD88 then recruits IRAKs, TRAF6, and the TAK1 complex, leading to early-stage activation of NF-κB and MAP kinases [1]. TLR4 is endocytosed and delivered to intracellular vesicles and forms a complex with TRAM and TRIF, which then recruits TRAF3 and the protein kinases TBK1 and IKKi. TBK1 and IKKi catalyze the phosphorylation of IRF3, leading to the expression of type I IFN [2].
BCR signaling is initiated through ligation of mIg under conditions that induce phosphorylation of the ITAMs in CD79, leading to the activation of Syk. Once Syk is activated, the BCR signal is transmitted via a series of proteins associated with the adaptor protein B-cell linker (Blnk, SLP-65). Blnk binds CD79a via non-ITAM tyrosines and is phosphorylated by Syk. Phospho-Blnk acts as a scaffold for the assembly of the other components, including Bruton’s tyrosine kinase (Btk), Vav 1, and phospholipase C-gamma 2 (PLCγ2) [3]. Following the assembly of the BCR-signalosome, GRB2 binds and activates the Ras-guanine exchange factor SOS, which in turn activates the small GTPase RAS. The original RAS signal is transmitted and amplified through the mitogen-activated protein kinase (MAPK) pathway, which including the serine/threonine-specific protein kinase RAF followed by MEK and extracellular signal related kinases ERK 1 and 2 [4]. After stimulation of BCR, CD19 is phosphorylated by Lyn. Phosphorylated CD19 activates PI3K by binding to the p85 subunit of PI3K and produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) from PIP2, and PIP3 transmits signals downstream [5].
Central process of T cells responding to specific antigens is the binding of the T-cell receptor (TCR) to specific peptides bound to the major histocompatibility complex which expressed on antigen-presenting cells (APCs). Once TCR connected with its ligand, the ζ-chain–associated protein kinase 70 molecules (Zap-70) are recruited to the TCR-CD3 site and activated, resulting in an initiation of several signaling cascades. Once stimulation, Zap-70 forms complexes with several molecules including SLP-76; and a sequential protein kinase cascade is initiated, consisting of MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAPKK), and MAP kinase (MAPK) [6]. Two MAPK kinases, MKK4 and MKK7, have been reported to be the primary activators of JNK. MKK3, MKK4, and MKK6 are activators of P38 MAP kinase [7]. MAP kinase pathways are major pathways induced by TCR stimulation, and they play a key role in T-cell responses.
Phosphoinositide 3-kinase (PI3K) binds to the cytosolic domain of CD28, leading to conversion of PIP2 to PIP3, activation of PKB (Akt) and phosphoinositide-dependent kinase 1 (PDK1), and subsequent signaling transduction [8].
References
[1] Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature immunology, 2010, 11(5): 373-384.
[2] Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650.
[3] Packard T A, Cambier J C. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation[J]. F1000Prime Rep, 2013, 5(40.10): 12703.
[4] Zhong Y, Byrd J C, Dubovsky J A. The B-cell receptor pathway: a critical component of healthy and malignant immune biology[C]//Seminars in hematology. WB Saunders, 2014, 51(3): 206-218.
[5] Baba Y, Matsumoto M, Kurosaki T. Calcium signaling in B cells: regulation of cytosolic Ca 2+ increase and its sensor molecules, STIM1 and STIM2[J]. Molecular immunology, 2014, 62(2): 339-343.
[6] Adachi K, Davis M M. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells[J]. Proceedings of the National Academy of Sciences, 2011, 108(4): 1549-1554.
[7] Rincón M, Flavell R A, Davis R A. The Jnk and P38 MAP kinase signaling pathways in T cell–mediated immune responses[J]. Free Radical Biology and Medicine, 2000, 28(9): 1328-1337.
[8] Bashour K T, Gondarenko A, Chen H, et al. CD28 and CD3 have complementary roles in T-cell traction forces[J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2241-2246.
Targets for Immunology/Inflammation
- Cyclic GMP-AMP Synthase(1)
- Apoptosis(137)
- 5-Lipoxygenase(18)
- TLR(106)
- Papain(2)
- PGDS(1)
- PGE synthase(26)
- SIKs(10)
- IκB/IKK(83)
- AP-1(2)
- KEAP1-Nrf2(47)
- NOD1(1)
- NF-κB(265)
- Interleukin Related(129)
- 15-lipoxygenase(2)
- Others(10)
- Aryl Hydrocarbon Receptor(35)
- CD73(16)
- Complement System(46)
- Galectin(30)
- IFNAR(19)
- NO Synthase(78)
- NOD-like Receptor (NLR)(37)
- STING(84)
- Reactive Oxygen Species(434)
- FKBP(14)
- eNOS(4)
- iNOS(24)
- nNOS(21)
- Glutathione(37)
- Adaptive Immunity(144)
- Allergy(129)
- Arthritis(25)
- Autoimmunity(134)
- Gastric Disease(64)
- Immunosuppressants(27)
- Immunotherapeutics(3)
- Innate Immunity(411)
- Pulmonary Diseases(76)
- Reactive Nitrogen Species(43)
- Specialized Pro-Resolving Mediators(42)
- Reactive Sulfur Species(24)
Products for Immunology/Inflammation
- Cat.No. Product Name Information
-
GC41207
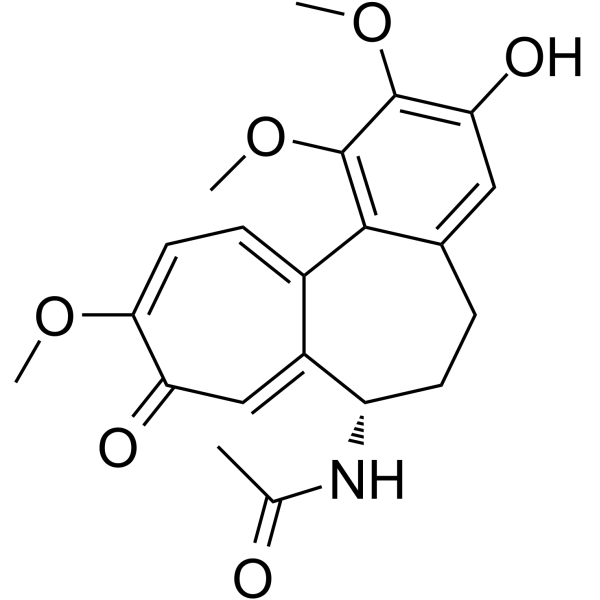
17(R)-HDHA
Resolvins are a group of polyhydroxylated metabolites of docosahexaenoic acid (DHA) found in the inflammatory exudates of aspirin-treated experimental animals.
-
GC49178
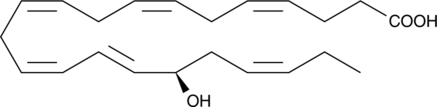
17(R)-Protectin D1
An aspirin-triggered epimer of protectin D1
-
GC41951
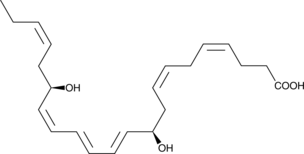
17(R)-Resolvin D1
Resolvins are a family of potent lipid mediators derived from both eicosapentaenoic acid and docosahexaenoic acid.
-
GC41227
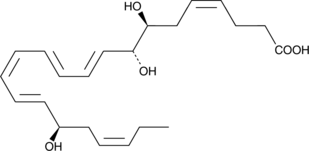
17(R)-Resolvin D1 methyl ester
17(R)-Resolvin D1 (17(R)-RvD1) is an aspirin-triggered epimer of RvD1 that reduces human polymorphonuclear leukocyte transendothelial migration, the earliest event in acute inflammation, with equipotency to RvD1 (EC50 = ~30 nM).
-
GC45307
17(R)-Resolvin D3
-
GC41952
17(R)-Resolvin D4
17(R)-Resolvin D4 (17(R)-RvD4) is an aspirin-triggered epimer of RvD4 .
-
GC41208
17(S)-HDHA
17(S)-HDHA is a primary mono-oxygenation product of docosahexaenoic acid in human whole blood, human leukocytes, and mouse brain.
-
GC40975
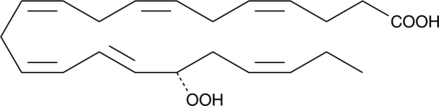
17(S)-HpDHA
17(S)-HpDHA is a mono-oxygenation product of docosahexaenoic acid in human whole blood, human leukocytes, human glial cells, and mouse brain.
-
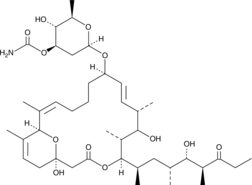
GC41958
17-hydroxy Venturicidin A
17-hydroxy Venturicidin A is a macrolide fungal metabolite originally isolated from Streptomyces.
-
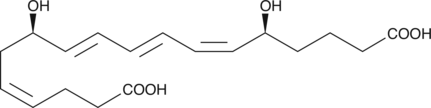
GC41980
18-carboxy dinor Leukotriene B4
18-carboxy dinor Leukotriene B4 (18-carboxy dinor LTB4) is a β-oxidation metabolite of LTB4.
-
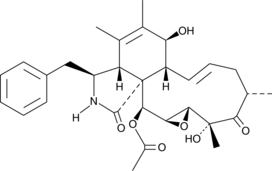
GC41982
19,20-Epoxycytochalasin C
19,20-Epoxycytochalasin C is a fungal metabolite originally isolated from Nemania sp.
-
GC41983
19,20-Epoxycytochalasin D
19,20-Epoxycytochalasin D is a fungal metabolite that has been found in the endophytic fungus Nemania sp.
-
GC41160
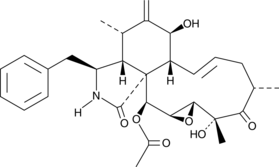
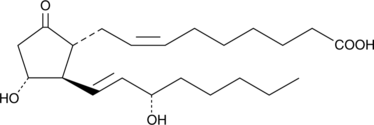
1a,1b-dihomo Prostaglandin E2
1a,1b-dihomo Prostaglandin E2 (PGE2) is a rare polyunsaturated fatty acid first identified in extracts of sheep vesicular gland microsomes, known to contain COX, incubated with adrenic acid.
-
GC38728
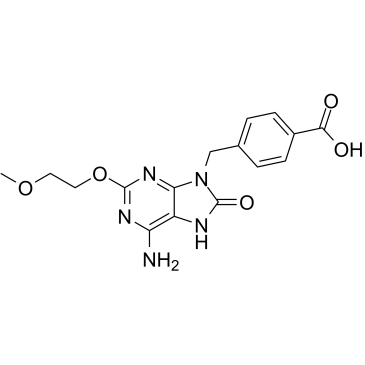
1V209
1V209 (TLR7 agonist T7) is a Toll-like receptor 7 (TLR7) agonist and has anti-tumor effects. 1V209 can be conjugated with various polysaccharides to improve its water solubility, and enhance its efficacy, and maintain low toxicity.
-
GC52501
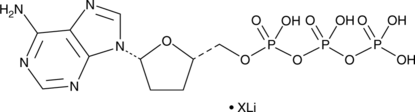
2',3'-Dideoxyadenosine 5'-triphosphate (lithium salt)
An inhibitor of reverse transcriptases and DNA polymerases
-
GC67628
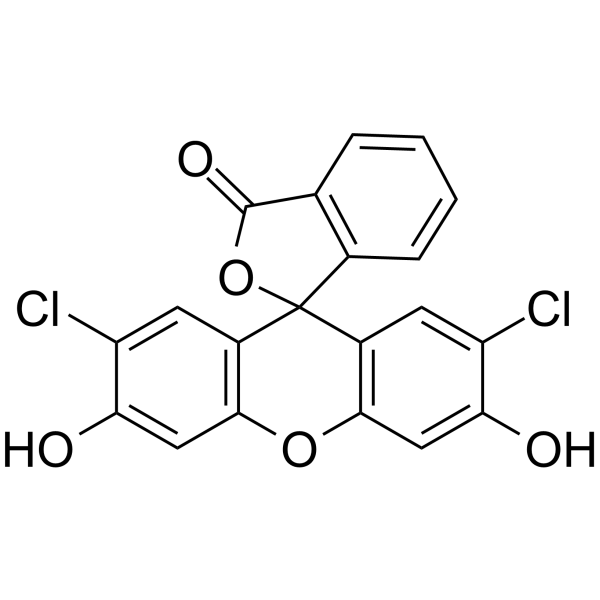
2',7'-Dichlorofluorescein
2',7'-Dichlorofluorescein acts as a fluorescent probe (Ex=496 nm and Em=525 nm) for reactive oxygen species (ROS) measurement.
-
GC49823
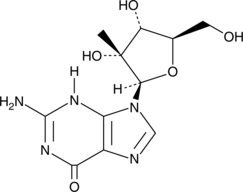
2′-C-β-Methylguanosine
An active nucleoside metabolite of BMS-986094
-
GC49514
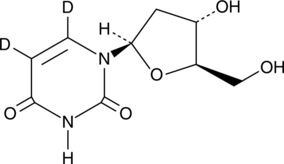
2′-Deoxyuridine-d2
An internal standard for the quantification of 2’-deoxyuridine
-
GC52122
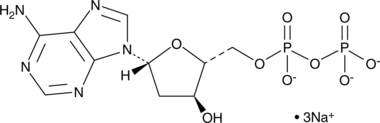
2’-Deoxyadenosine-5’-diphosphate (sodium salt)
A nucleotide diphosphate
-
GC46508
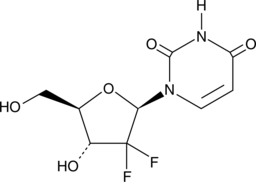
2',2'-Difluoro-2'-deoxyuridine
An active metabolite of gemcitabine
-
GC42079
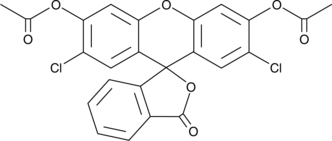
2',7'-Dichlorofluorescein diacetate
2',7'-Dichlorofluorescein diacetate is as a cell-permeable fluorogenic probe to quantify reactive oxygen species (ROS) and nitric oxide (NO).
-
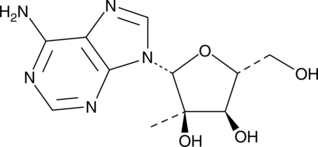
GC41281
2'-C-Methyladenosine
2'-C-Methyladenosine is an inhibitor of hepatitis C virus (HCV) replication (IC50 = 0.3 μM in Huh-7 human hepatoma cells) that is not cytotoxic at concentrations up to 100 μM.
-
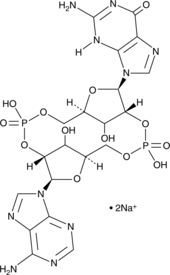
GC42080
2'2'-cGAMP (sodium salt)
2'2'-cGAMP is a synthetic dinucleotide (CDN) that contains non-canonical 2'5'-phosphodiester bonds.
-
GC42090
2'3'-cGAMP (sodium salt)
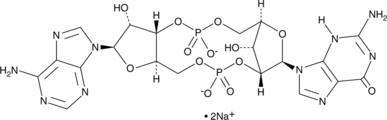
2'3'-cGAMP is a second messenger produced from ATP and GTP by cGMP-AMP synthase (cGAS) in the cytoplasm of mammalian cells in response to the presence of DNA.
-
GC40415
2,3-dinor-11β-Prostaglandin F2α
2,3-dinor-11β-Prostaglandin F2α (2,3-dinor-11β-PGF2α) was recovered from the urine of both normal monkeys and humans when infused with radiolabeled PGD2, where it represented approximately 1% and 4% of the infused radiolabeled dose, respectively.
-
GC49671
2,3-Oxidosqualene
An intermediate in the biosynthesis of sterols
-
GC68044
2,4,6-Trihydroxybenzaldehyde
-
GC68452
2,4,6-Triiodophenol

-
GC46522
2,4-Dichlorobenzenesulfonyl chloride
A heterocyclic building block
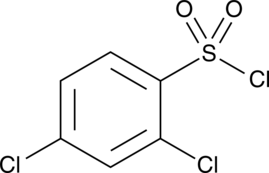
-
GC42076
2,5-Deoxyfructosazine (hydrochloride)
2,5-Deoxyfructosazine is a pyrazine derivative that can be found in cured tobacco and is used as a flavoring agent in the food and tobacco industry.
-
GC39325
2,5-Dihydroxyacetophenone
2,5-Dihydroxyacetophenone, isolated from Rehmanniae Radix Preparata, inhibits the production of inflammatory mediators in activated macrophages by blocking the ERK1/2 and NF-κB signaling pathways.
-
GC46057
2,5-Dihydroxycinnamic Acid phenethyl ester
An inhibitor of 5-LO
-
GC46502
2-(1-(Thiophen-2-yl)ethylidene)hydrazinecarbothioamide
An antimicrobial agent
-
GC42112
2-Acetyl-5-tetrahydroxybutyl Imidazole
Sphingosine-1-phosphate (S1P) lyase catalyzes the irreversible decomposition of S1P to hexadecanaldehyde and phosphoethanolamine.
-
GC46533
2-Amino-6-chloropurine
A precursor in the synthesis of nucleoside analogs
-
GC52029
2-Aminoflubendazole
-
GC42123
2-Aminopurine (hydrochloride)
2-Aminopurine (hydrochloride) is a fluorescent analog of guanosine.
-
GC64739
2-Aminoquinoline
2-Aminoquinoline (2-Quinolinamine) is a promising compound as bioavailable nNOS inhibitor but suffers from low human nNOS inhibition, low selectivity versus human eNOS, and significant binding to other CNS targets.
-
GC42135
2-chloro Palmitic Acid
2-chloro Palmitic Acid, an inflammatory lipid mediator, interferes with protein palmitoylation,induces ER-stress markers, reduced the ER ATP content, and activates transcription and secretion of IL-6 as well as IL-8.2-chloro Palmitic Acid disrupts the mitochondrial membrane potential and induces procaspase-3 and PARP cleavage.2-chloro Palmitic Acid can across blood-brain barrier (BBB) and compromises ER- and mitochondrial functions in the human brain endothelial cell line hCMEC/D3.
-
GC42136
2-chloro Stearic Acid
2-chloro Stearic acid is a bioactive fatty acid that accumulates in primary human monocytes and neutrophils as well as murine neutrophils stimulated with phorbol 12-myristate 13-acetate.
-
GC40675
2-deoxy-Artemisinin
2-deoxy-Artemisinin is an inactive metabolite of the antimalarial agent artemisinin.
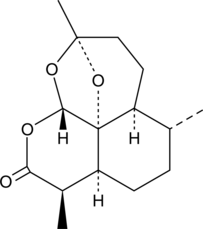
-
GC40634
2-epi-Abamectin
2-epi-Abamectin is a degradation product of abamectin.
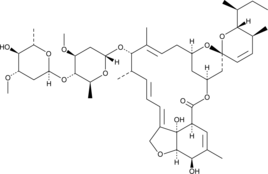
-
GC46544
2-Fluoro-4-iodo benzonitrile
A building block
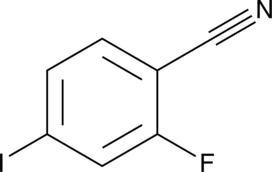
-
GC45912
2-heptyl-3-hydroxy-4(1H)-Quinolone
A bacterial quorum-sensing signaling molecule
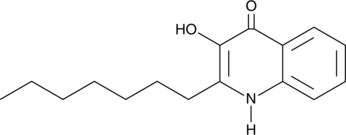
-
GC18391
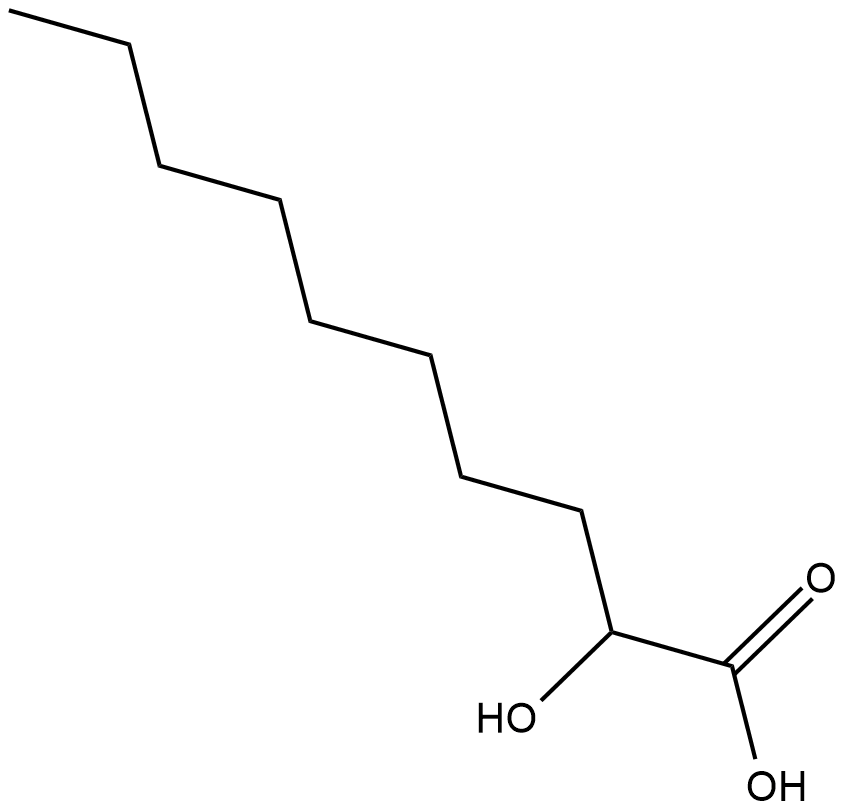
2-hydroxy Decanoic Acid
2-hydroxy Decanoic acid is a fatty acid found in the lipophilic portion of the lipopolysaccharide fraction of P.
-
GC42166
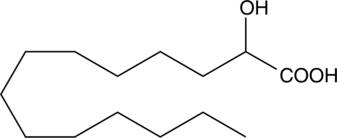
2-hydroxy Myristic Acid
2-hydroxy Myristic acid is a hydroxy fatty acid that has been found in bovine, human, and horse milk, cow and buffalo cheeses, sea bass filet, seal oil, human vernix caseosa, and wool wax.
-
GC42171
2-hydroxy Stearic Acid methyl ester
2-hydroxy Stearic acid is a hydroxylated fatty acid methyl ester that broadens phase transition in dimyristoylphosphatidylcholine (DMPC) lipid membranes.

-
GC40944
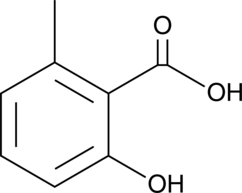
2-hydroxy-6-Methylbenzoic Acid
2-hydroxy-6-Methylbenzoic acid is a constituent of G.
-
GC48910
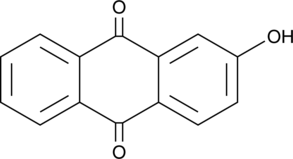
2-Hydroxyanthraquinone
An anthraquinone with antibacterial and estrogenic activities
-
GC17084
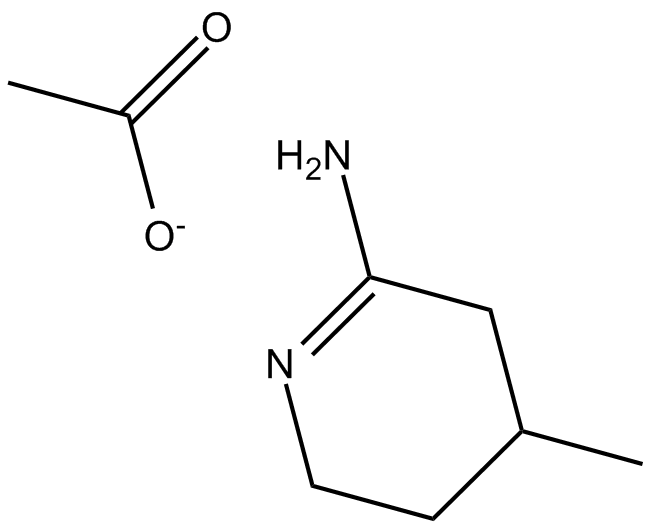
2-Imino-4-methylpiperidine (acetate)
2-Imino-4-methylpiperidine (acetate) is a potent and orally active NO synthase (NOS) isoforms inhibitor with IC50s of 0.1 μM, 1.1 μM, and 0.2 μM for human iNOS (hiNOS), heNOS and hnNOS, respectively.
-
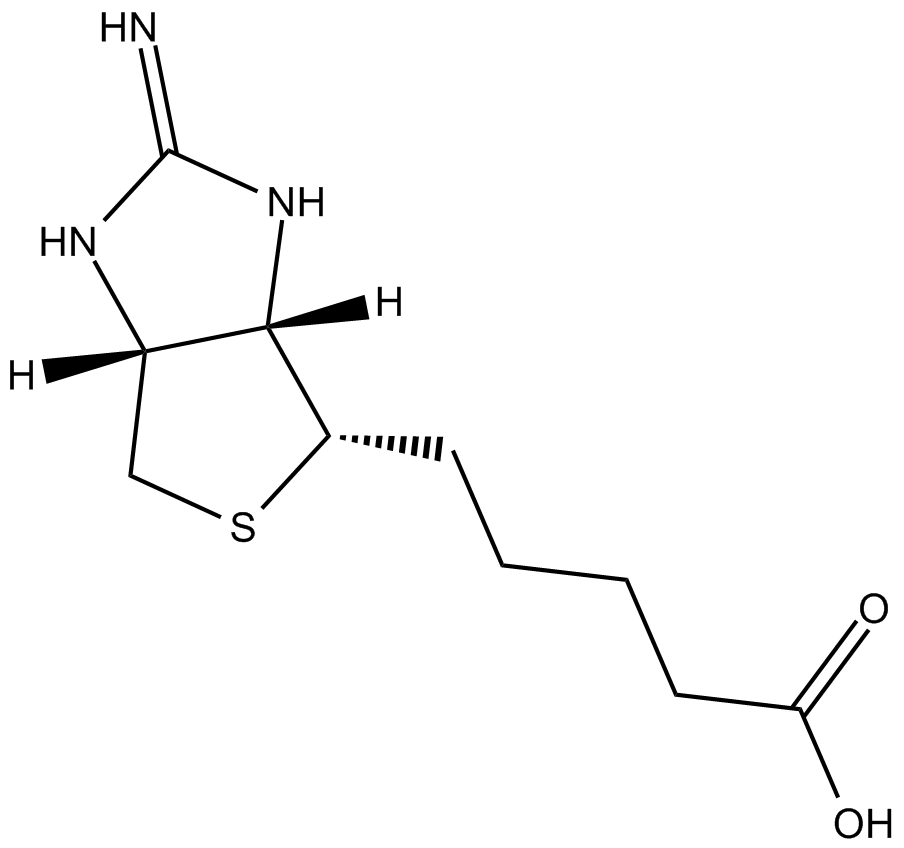
GC16664
2-Iminobiotin
iNOS and nNOS inhibitor
-
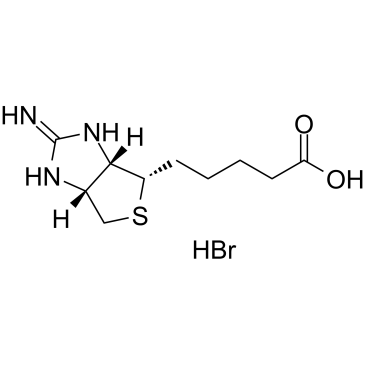
GC39326
2-Iminobiotin hydrobromide
2-Iminobiotin hydrobromide (Guanidinobiotin hydrobromide) is a biotin (vitamin H or B7) analog.
-
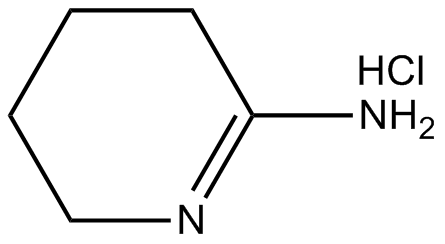
GC12166
2-Iminopiperidine hydrochloride
iNOS inhibitor
-
GC52140
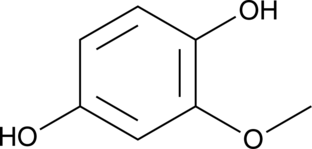
2-Methoxyhydroquinone
-
GC42186
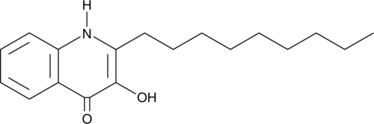
2-nonyl-3-hydroxy-4-Quinolone
2-nonyl-3-hydroxy-4-Quinolone is a quinolone compound produced by P.
-
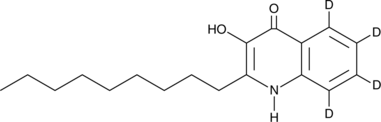
GC52445
2-nonyl-3-hydroxy-4-Quinolone-d4
An internal standard for the quantification of 2-nonyl-3-hydroxy-4-quinolone
-
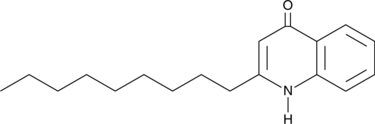
GC46553
2-Nonylquinolin-4(1H)-one
A quinolone alkaloid with diverse biological activities
-
GC52446
2-Nonylquinolin-4(1H)-one-d4
An internal standard for the quantification of 2-nonylquinolin-4(1H)-one
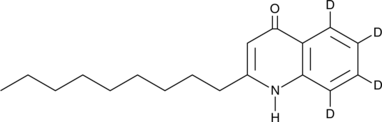
-
GC49786
2-NP-AOZ
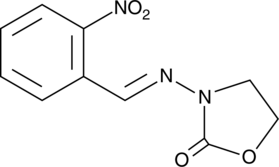
-
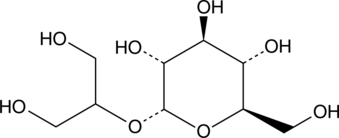
GC49283
2-O-(α-D-Glucopyranosyl)glycerol
A compatible solute
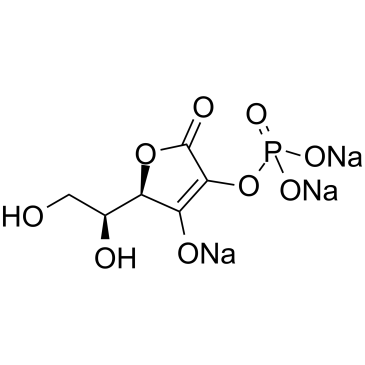
-
GC35095
2-Phospho-L-ascorbic acid trisodium salt
2-Phospho-L-ascorbic acid trisodium salt (2-Phospho-L-ascorbic acid trisodium) is a long-acting vitamin C derivative that can stimulate collagen formation and expression.
-
GC17951
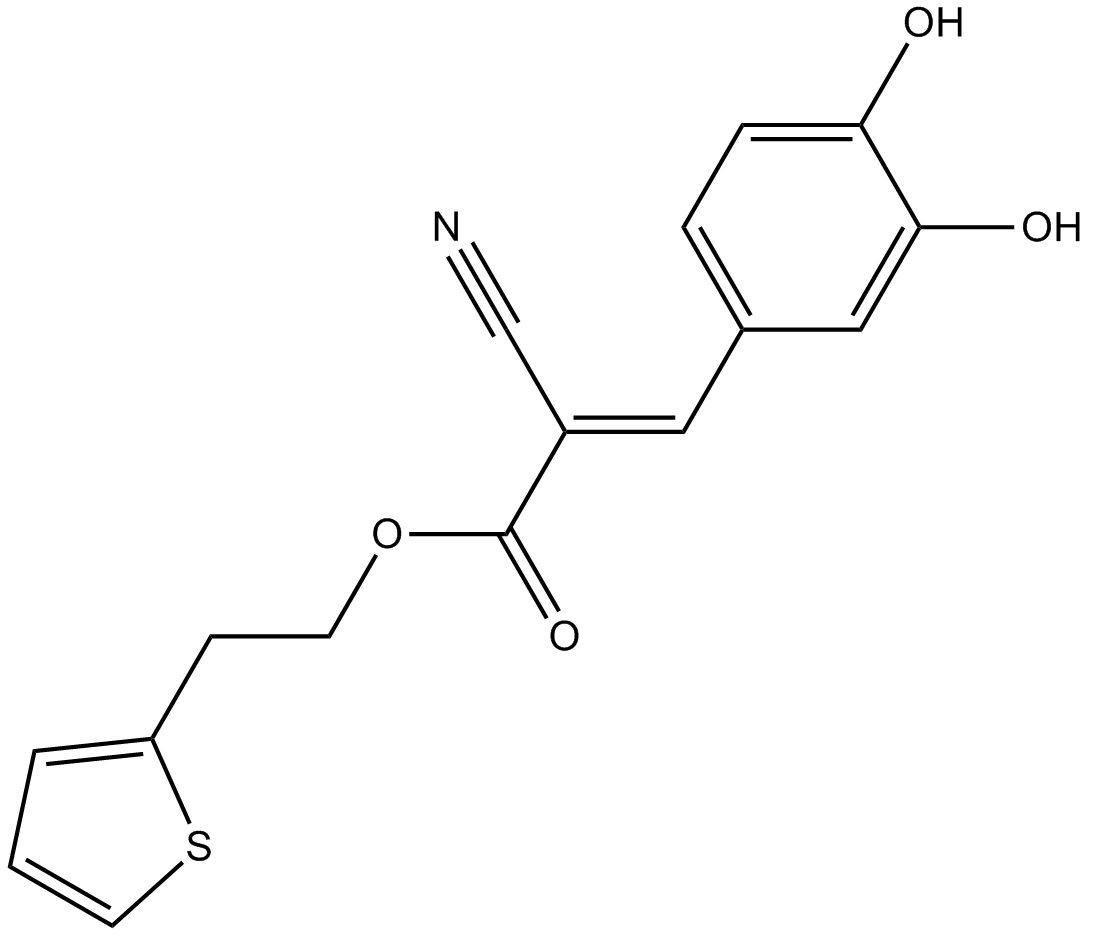
2-TEDC
5-, 12-, and 15-lipoxygenase inhibitor
-
GC68043
2-tert-Butyl-1,4-benzoquinone

-
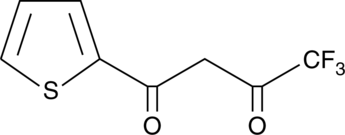
GC42193
2-Thenoyltrifluoroacetone
2-Thenoyltrifluoroacetone (TTFA) is an inhibitor of respiration in animals and bacteria.
-
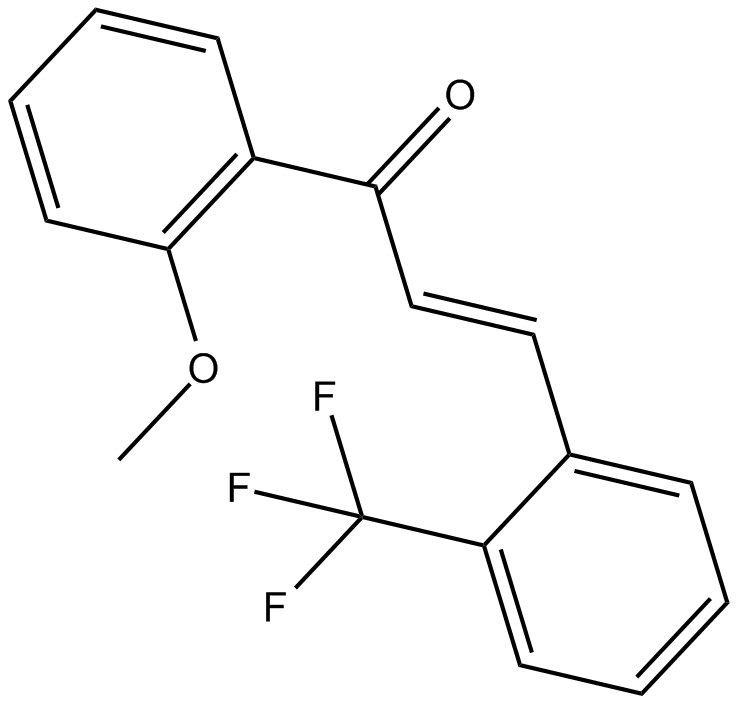
GC15355
2-Trifluoromethyl-2'-methoxychalcone
Nrf2 activator
-
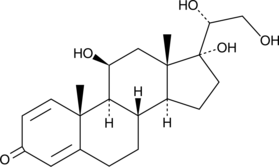
GC40960
20α-dihydro Prednisolone
20α-dihydro Prednisolone is a metabolite of prednisolone.
-
GC42082
20-carboxy Leukotriene B4
20-carboxy LTB4 is a metabolite of LTB4 in human neutrophils.
-
GC41387
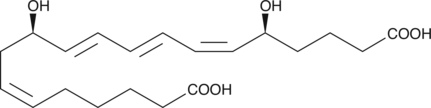
20-hydroxy Arachidic Acid
20-hydroxy Arachidic acid is a hydroxylated fatty acid that has been found in the suberin component of silver birch (B.
-
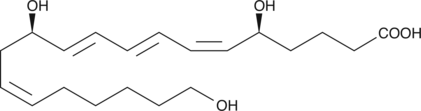
GC41421
20-hydroxy Leukotriene B4
20-hydroxy LTB4 is a metabolite of LTB4 in human neutrophils.
-
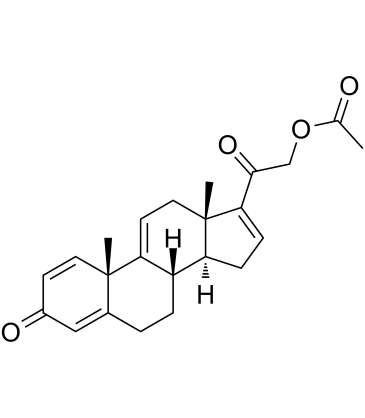
GC60467
21-Acetoxypregna-1,4,9(11),16-tetraene-3,20-dione
21-Acetoxypregna-1,4,9(11),16-tetraene-3,20-dione is an intermediate of delta 9,11 steroids synthesis, for example, Vamorolone?
-
GC42087
21-desacetyl Deflazacort
21-desacetyl Deflazacort is the active glucocorticoid derived from the prodrug deflazacort.
-
GC40571
24(S),25-epoxy Cholesterol
24(S),25-epoxy Cholesterol is an oxysterol and the most abundant oxysterol in mouse ventral midbrain.
-
GC49365
25-Desacetyl Rifampicin
A major active metabolite of rifampicin

-
GC48482
28-Acetylbetulin
A lupane triterpenoid with anti-inflammatory and anticancer activities
-
GC48503
28-Deoxybetulin methyleneamine
A derivative of betulin
-
GC46549
2F-Peracetyl-Fucose
An inhibitor of protein fucosylation
-
GC49005
2S-Eriodictyol
A flavanone with antioxidant activity
-
GC25014
3',3'-cGAMP
3',3'-cGAMP (3',3'-cyclic GMP-AMP, Cyclic GMP-AMP, cGAMP) activates the endoplasmic reticulum (ER)-resident receptor stimulator of interferon genes (STING), thereby inducing an antiviral state and the secretion of type I IFNs.
-
GC49122
3′-deoxy Thymidine
An antiviral nucleoside analog
-
GC49871
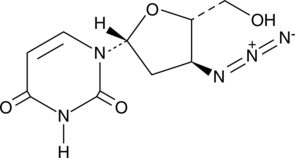
3’-Azido-2’,3’-dideoxyuridine
An antiviral nucleoside analog
-
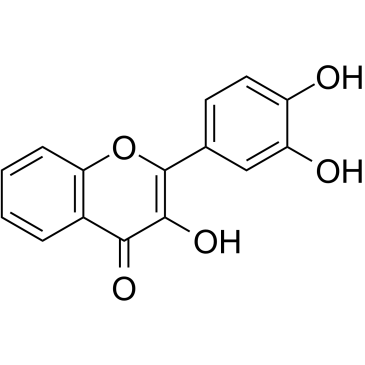
GC34451
3',4'-Dihydroxyflavonol
3',4'-Dihydroxyflavonol (DiOHF) is an effective antioxidant, which reduces superoxide and improves nitric oxide (NO) function in diabetic rat mesenteric arteries.
-
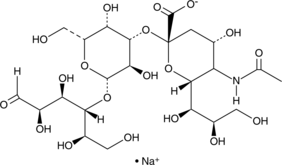
GC42312
3'-Sialyllactose (sodium salt)
3'-Sialyllactose consists of the monosaccharide N-acetylneuraminic acid linked to the galactosyl subunit of lactose at the 3 position.
-
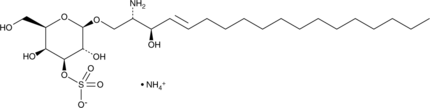
GC42242
3'-sulfo Galactosylsphingosine (ammonium salt)
3'-sulfo Galactosylsphingosine is a form of sulfatide that is lacking the fatty acyl group.
-
GC42245
3'3'-cGAMP (sodium salt)
3'3'-cGAMP is a second messenger produced in bacteria by specific dinucleotide cyclases.
-
GN10006
3,4-Dihydroxybenzaldehyde
-
GC33992
3,4-Dimethoxycinnamic acid (O-Methylferulic acid)
3,4-Dimethoxycinnamic acid (O-Methylferulic acid) (O-Methylferulic acid) is a monomer extracted and purified from Securidaca inappendiculata Hassk.
-
GC61673
3,5-Di-tert-butylphenol
3,5-Di-tert-butylphenol is an volatile organic compound with anti-biofilm and antifungal activities.

-
GC46577
3,5-Dihydroxybenzaldehyde
A building block
-
GC64762
3,6-Dihydroxyflavone
3,6-Dihydroxyflavone is an anti-cancer agent. 3,6-Dihydroxyflavone dose- and time-dependently decreases cell viability and induces apoptosis by activating caspase cascade, cleaving poly (ADP-ribose) polymerase (PARP). 3,6-Dihydroxyflavone increases intracellular oxidative stress and lipid peroxidation.
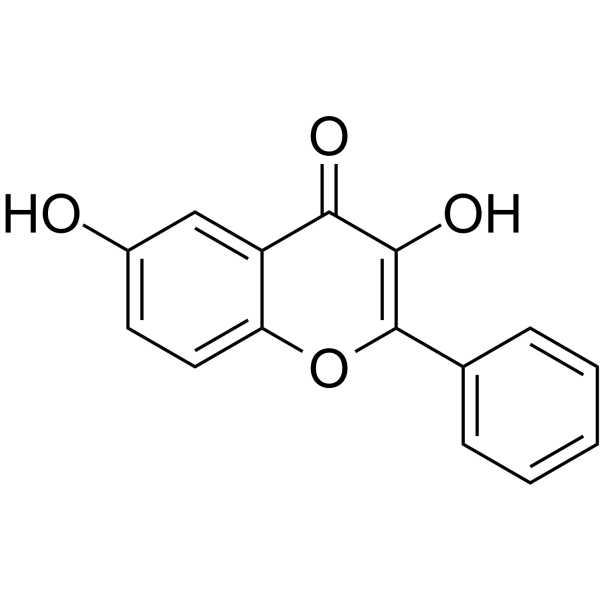
-
GC49169
3,8’-Biapigenin
A biflavonoid with diverse biological activities
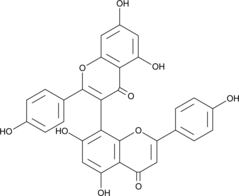
-
GC52324
3-(3-Hydroxyphenyl)propionic Acid sulfate
A metabolite of certain phenols and glycosides
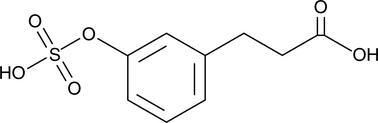
-
GC14282
3-acetyl-11-keto-β-Boswellic Acid
3-acetyl-11-keto-β-Boswellic Acid (Acetyl-11-keto-β-boswellic acid) is an active triterpenoid compound from the extract of Boswellia serrate and a novel Nrf2 activator.
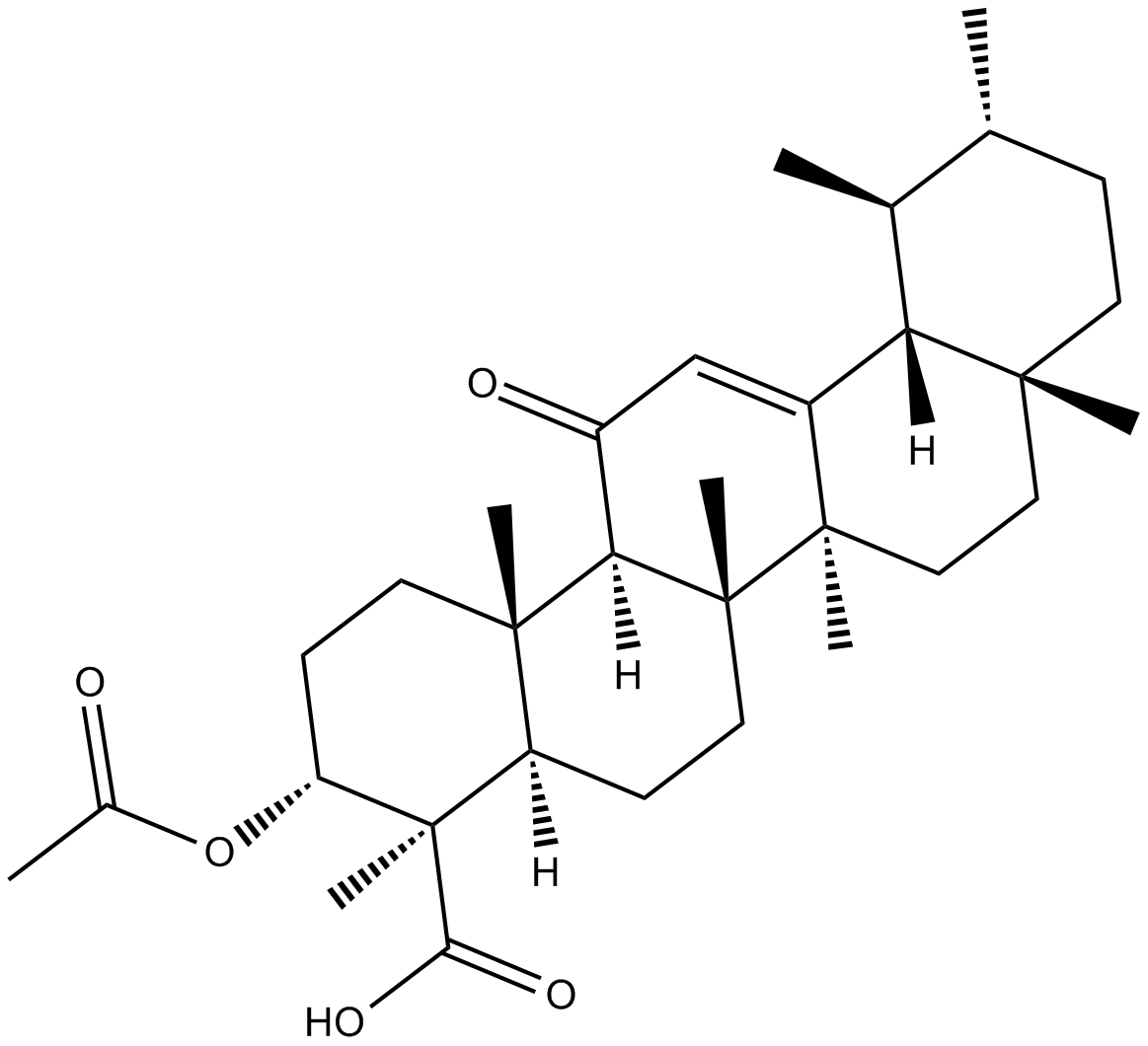
-
GC68081
3-Amino-1,2,4-triazine

-
GC46583
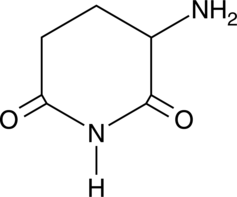
3-Amino-2,6-Piperidinedione
An active metabolite of (±)-thalidomide
-
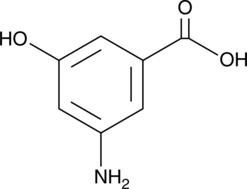
GC52129
3-Amino-5-hydroxybenzoic Acid
-
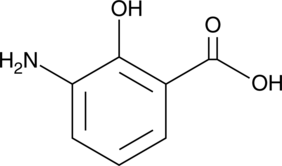
GC49849
3-Aminosalicylic Acid
A salicylic acid derivative
-
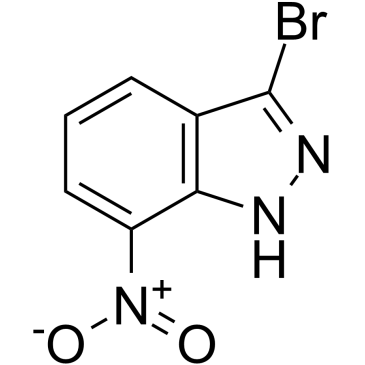
GC38208
3-Bromo-7-nitroindazole
A potent inhibitor of nNOS
-
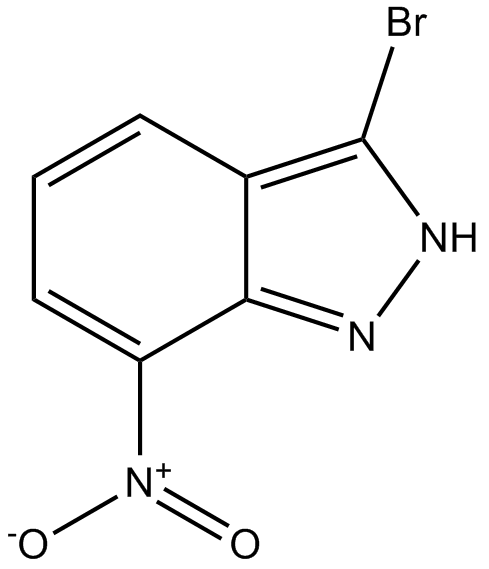
GC10385
3-Bromo-7-nitroindazole
nNOS inhibitor
-
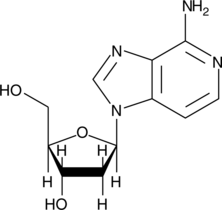
GC42259
3-Deaza-2'-deoxyadenosine
3-Deaza-2'-deoxyadenosine strongly inhibits lymphocyte-mediated cytolysis with low cytotoxicity when applied at 100 μM.
-
GC62794
3-Demethylcolchicine
3-Demethylcolchicine, a colchicine metabolite, possesses a hydroxy-group on its carbon ring that could participate in radical scavenging and markedly inhibits the carrageenin edema.