NOD-like Receptor (NLR)
Nucleotide oligomerization domain (NOD)-like receptors (NLRs) are a specialized group of intracellular proteins that play a critical role in the regulation of the host innate immune response. NLRs act as scaffolding proteins that assemble signaling platforms that trigger nuclear factor-κB and mitogen-activated protein kinase signaling pathways and control the activation of inflammatory caspases. Importantly, mutations in several members of the NLR family have been linked to a variety of inflammatory diseases consistent with these molecules playing an important role in host-pathogen interactions and the inflammatory response. NLRs including Nod1 and Nod2 are thought to be kept in an inactive state by intra-molecular interactions.
The central role of the Nod-like receptor (NLR) protein family became increasingly appreciated in innate immune responses. NLRs are classified as part of the signal transduction ATPases with numerous domains (STAND) clade within the AAA+ ATPase family.
Targets for NOD-like Receptor (NLR)
Products for NOD-like Receptor (NLR)
- Cat.No. Product Name Information
-
GC50569
NLRP3-IN-2
NLRP3 inflammasome inhibitor

-
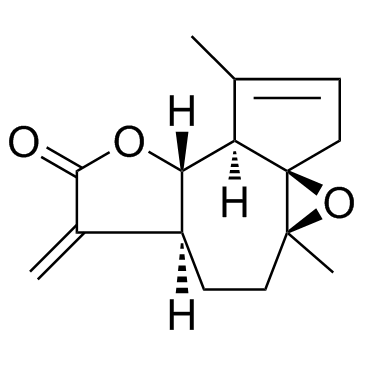
GC35385
Arglabin
A sesquiterpene lactone
-
GC62507
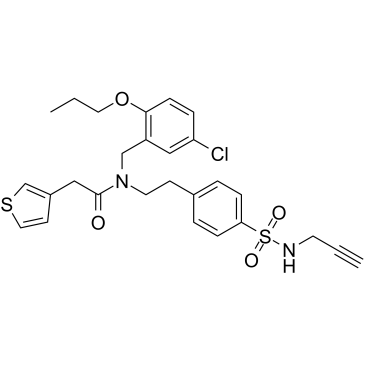
BMS-986299
BMS-986299 (compound 112) is a first-in-class NLRP3 inflammasome agonist with an EC50 of 1.28 μM.
-
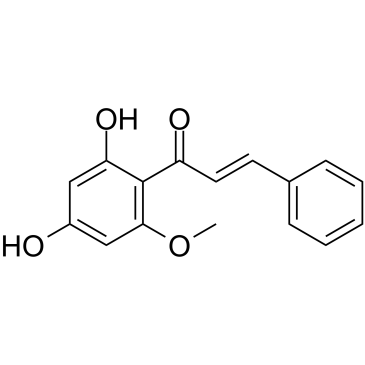
GC38059
Cardamonin
A chalconoid with anti-inflammatory and anti-tumor activity
-
GC50342
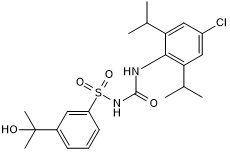
CP 424174
Inhibitor of IL-1β post-translational processing; indirectly inhibits NLRP3
-
GC19113
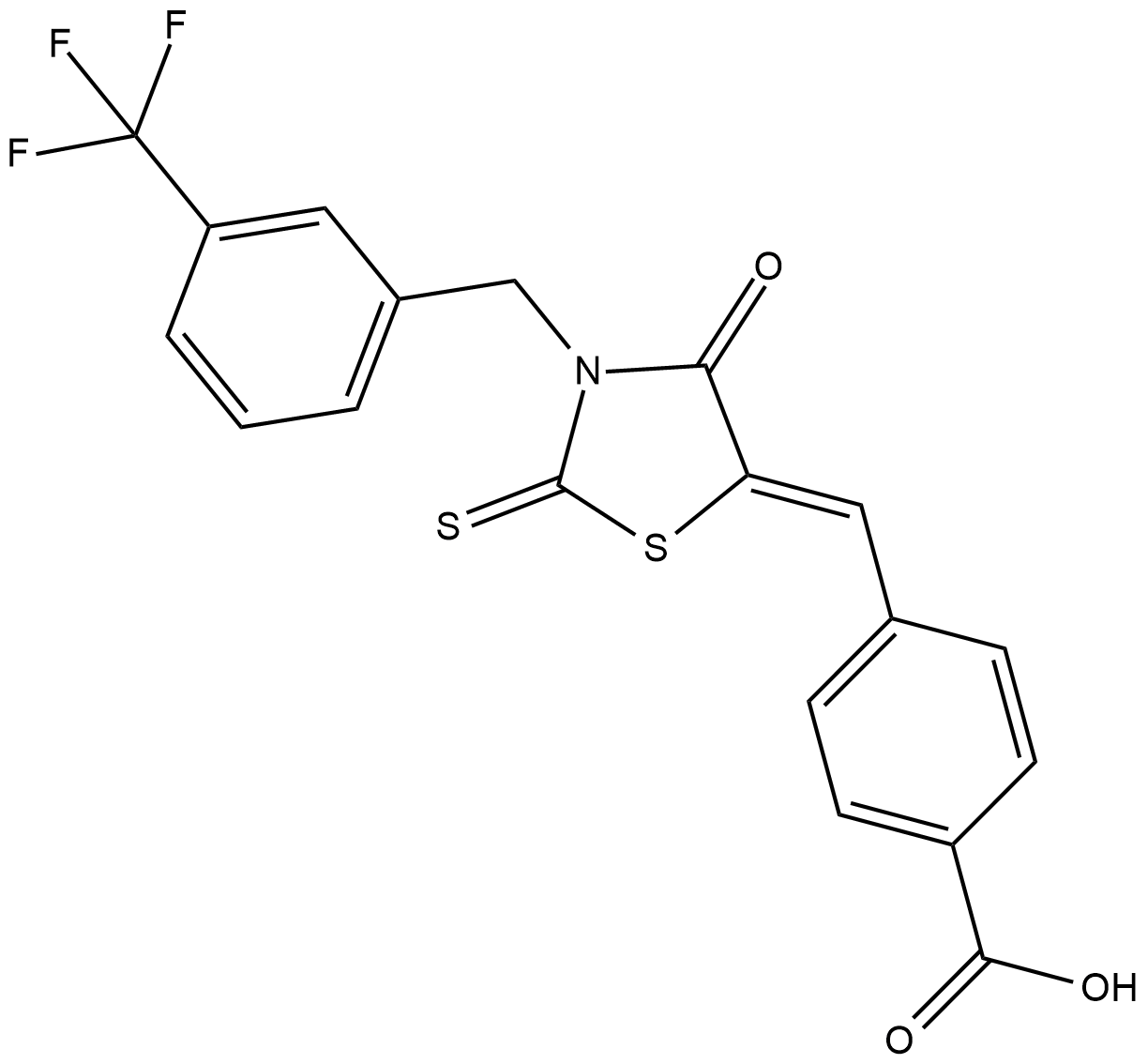
CY-09
CY-09, a specific inhibitor of the NLRP3 inflammasome, directly targets NLRP3 and has an IC50 value of 18.
-
GC34555
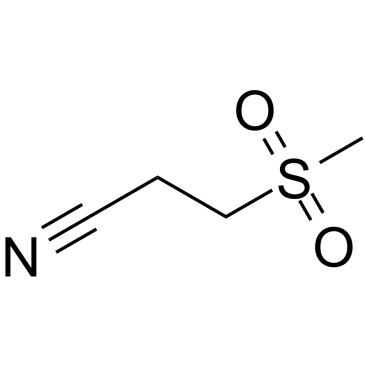
Dapansutrile
An NLRP3 inflammasome inhibitor
-
GC60887
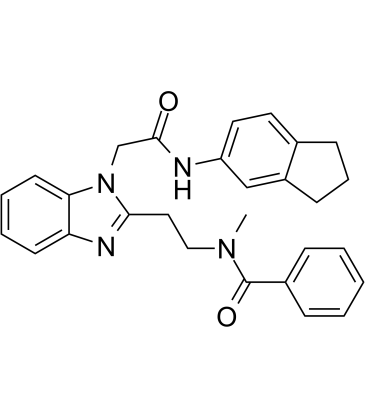
GSK717
GSK717 is a potent, selective NOD2 (nucleotide-binding oligomerization domain 2) inhibitor.
-
GC19201
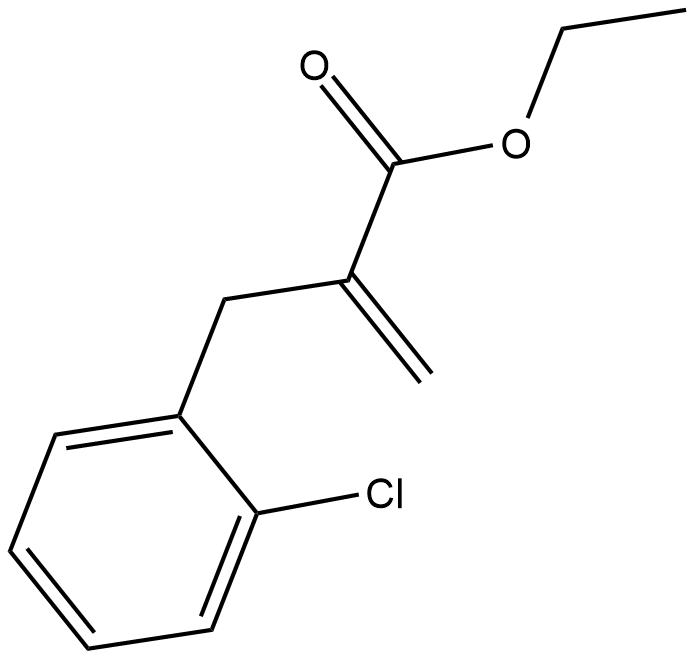
INF39
INF39 is an irreversible and noncytotoxic NLRP3 inhibitor.
-
GC60958
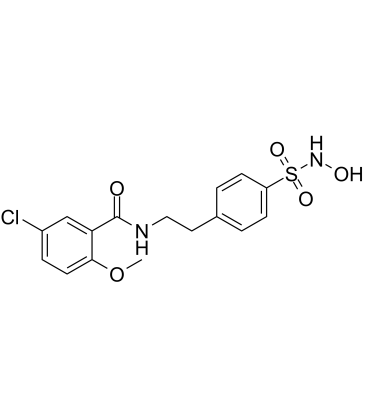
JC-171
JC-171 is a selective NLRP3 inflammasome inhibitor, with an IC50 of 8.45 μM for inhibiting LPS/ATP-induced interleukin-1β (IL-1β) release from J774A.1 macrophages.
-
GC62300
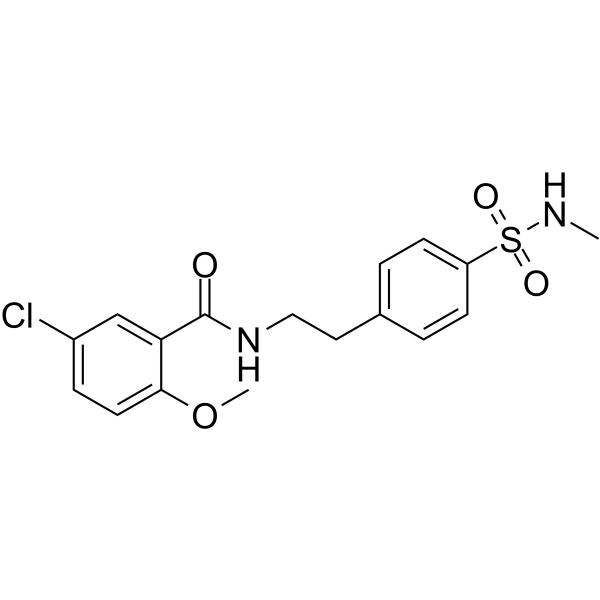
JC124
JC124 is a specific NLRP3 inflammasome inhibitor.
-
GC62454
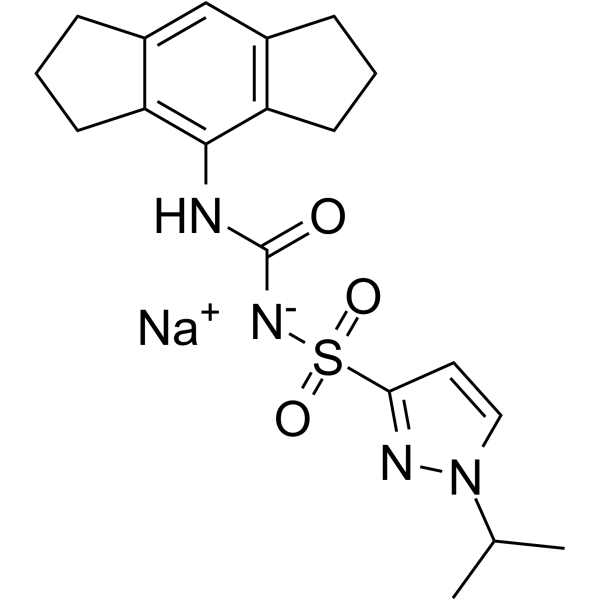
MCC7840 sodium
Emlenoflast (MCC7840) sodium, a sulfonylurea, is a potent and selective inhibitor of NLRP3 inflammasome, with an IC50 of <100 nM.
-
GC31644
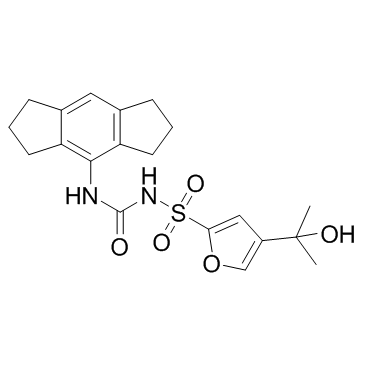
MCC950 (CP-456773)
MCC950 (CP-456773) (CP-456773; CRID3) is a potent and selective NLRP3 inhibitor with IC50s of 7.5 and 8.1 nM in BMDMs and HMDMs, respectively.
-
GC10634
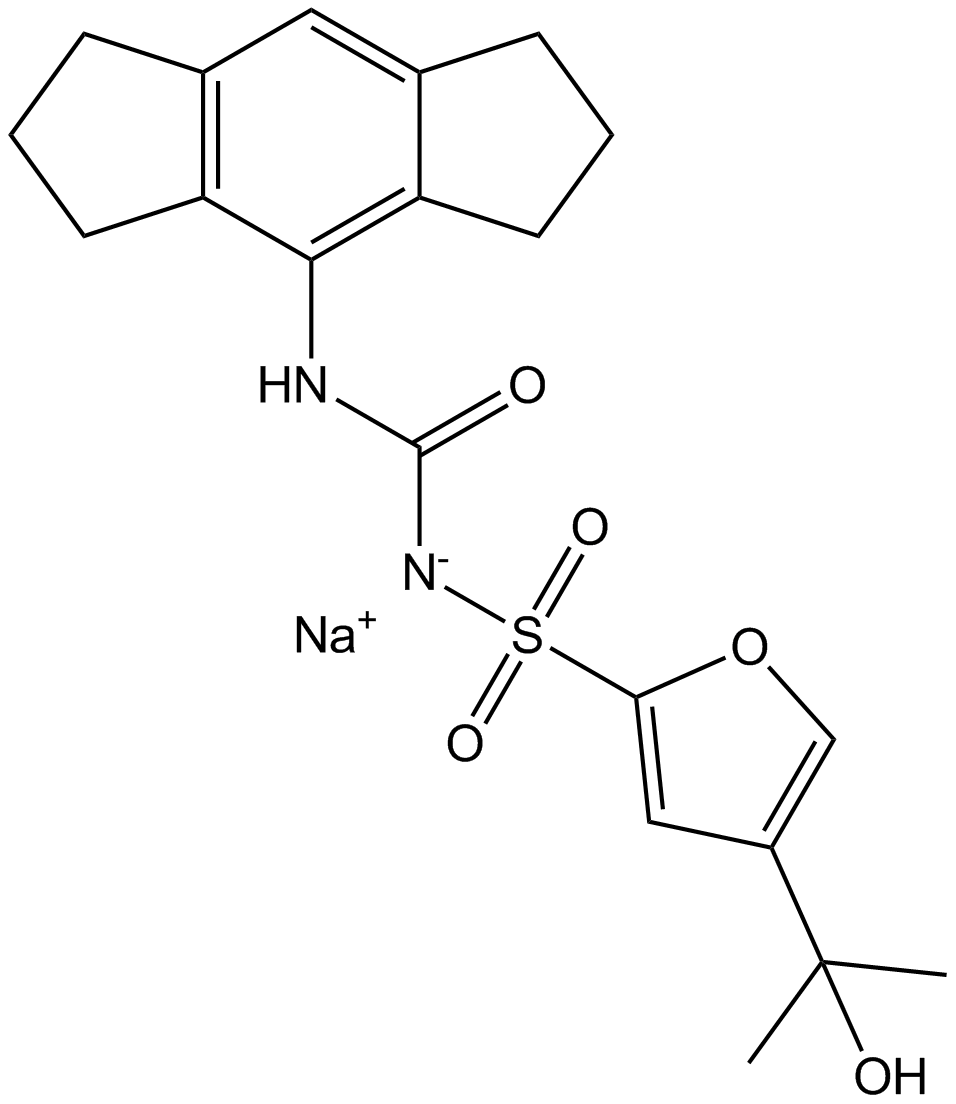
MCC950 sodium
MCC950 is a potent, selective, small molecule inhibitor of NLRP3.
-
GC16641
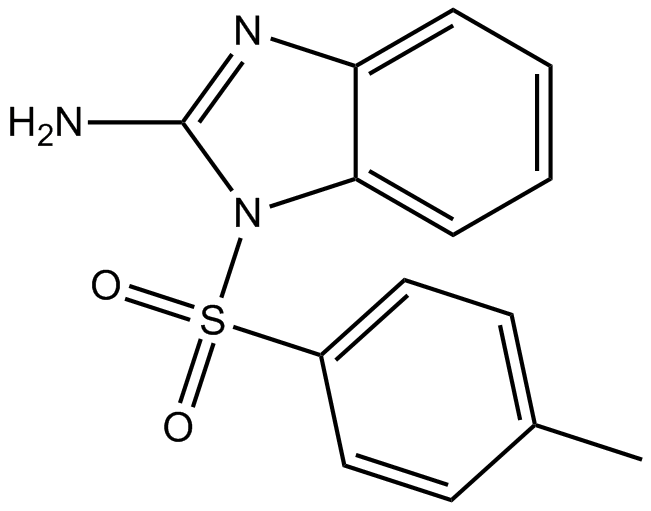
ML130 (Nodinitib-1)
ML130 (Nodinitib-1) (ML130;CID-1088438) is a NOD1 inhibitor with an IC50 of 0.56 μM.
-
GC50556
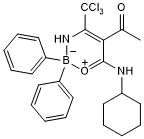
NBC 19
Potent NLRP3 inflammasome inhibitor
-
GC50611
NBC 6
NBC 6 is a potent, selective NLRP3 inflammasome inhibitor (IC50= 574 nM) that acts independently of Ca2+.
-
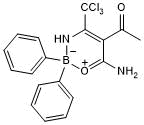
GC39719
Nigericin
K+/H+ ionophore,promoting K+/H+ exchange across mitochondrial membrane

-
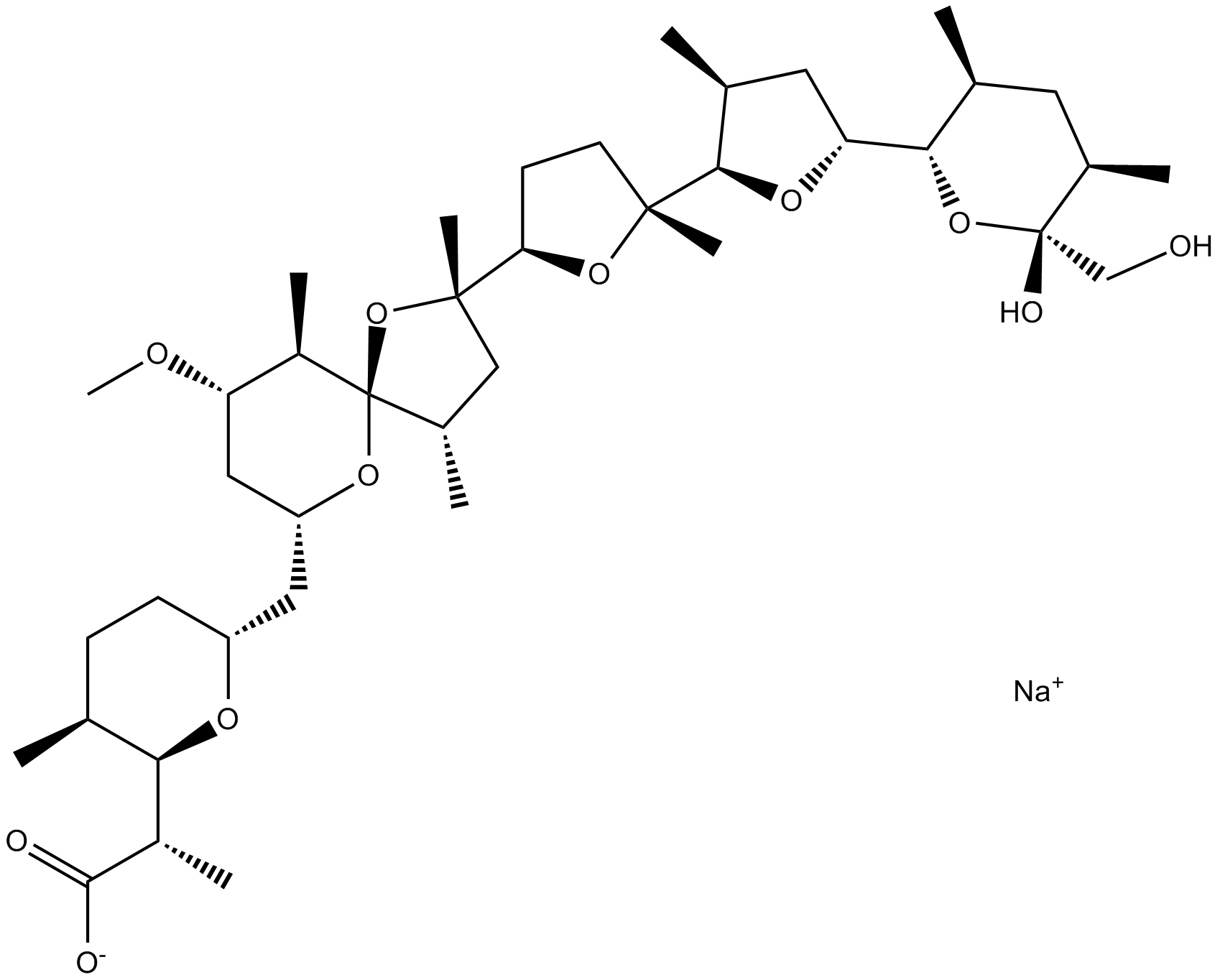
GC15811
Nigericin sodium salt
Nigericin sodium salt is an electrically neutral K +/H +?ionophore from Streptomyctus hygroscopicus an antibiotic that is lipophilic and selectively causes potassium to drain from the mitochondrial membrane.
-
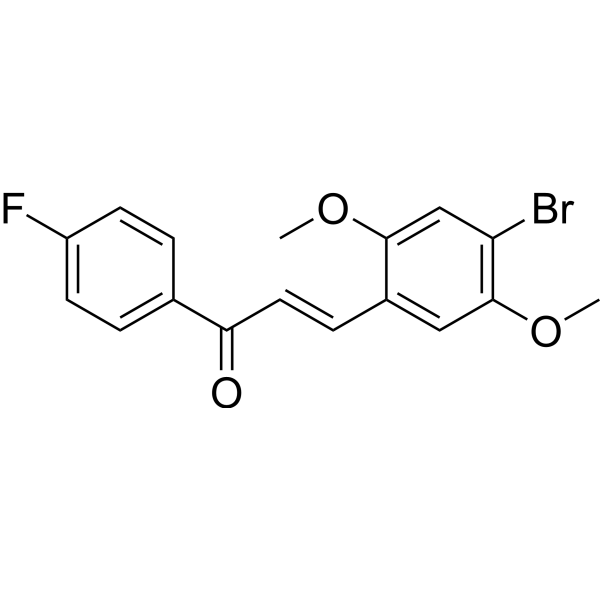
GC69570
NLRP3-IN-10
NLRP3-IN-10 is a potent inhibitor of NLRP3 that inhibits IL-1β with an IC50 of 251.1 nM. NLRP3-IN-10 can reduce ASC speck formation and inhibit NLRP3 inflammasome activation.
-
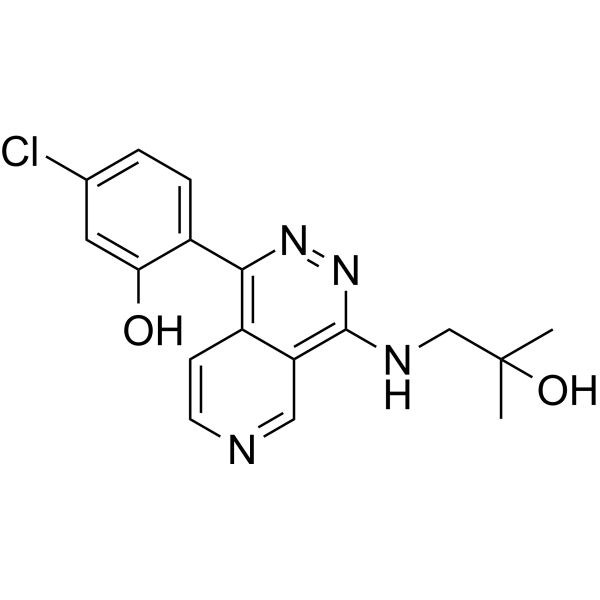
GC69571
NLRP3-IN-11
NLRP3-IN-11 is an inhibitor of the NLRP3 protein. It has biological activity with an IC50 value of <0.3 μM. NLRP3-IN-11 can be used for research on inflammatory and degenerative diseases, including NASH, atherosclerosis and other cardiovascular diseases, Alzheimer's disease, Parkinson's disease, diabetes, gout and many other autoimmune inflammatory diseases.
-
GC69572
NLRP3-IN-13
NLRP3-IN-13 (compound C77 in reference patent) is an effective selective NLRP3 inhibitor (IC50: 2.1 μM). It inhibits NLRP3 and NLRC4 inflammasomes, and suppresses IL-1β production mediated by NLRP3. NLRP3-IN-13 also inhibits NLRP3 ATPase activity. It can be used for research on neuroinflammatory diseases.
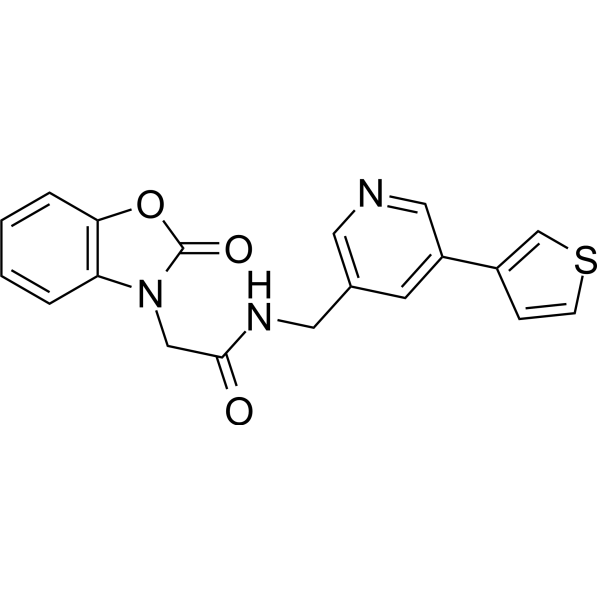
-
GC69573
NLRP3-IN-18
NLRP3-IN-18 (compound 13) is an effective NLRP3 inhibitor with an IC50 value of ≤ 1.0 µM[1].
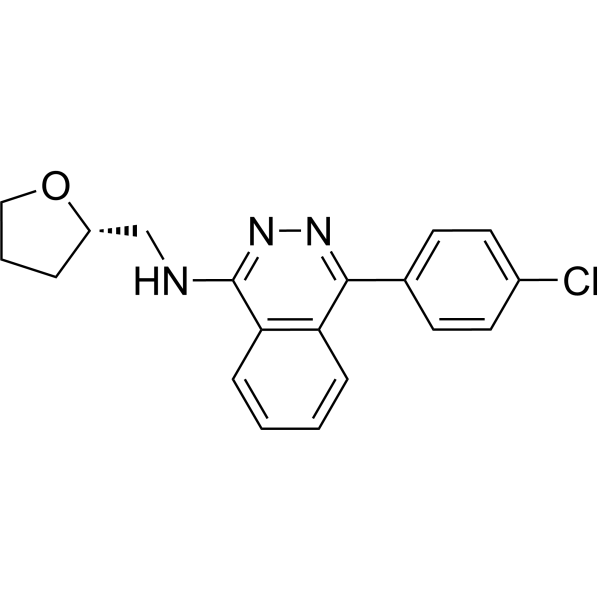
-
GC69569
NLRP3/AIM2-IN-2
NLRP3/AIM2-IN-2 is a novel potent inhibitor with species-specific effects on NLRP3 and AIM2 inflammasome-dependent cell pyroptosis, with an IC50 value of 0.2392 ±0.0233μM.
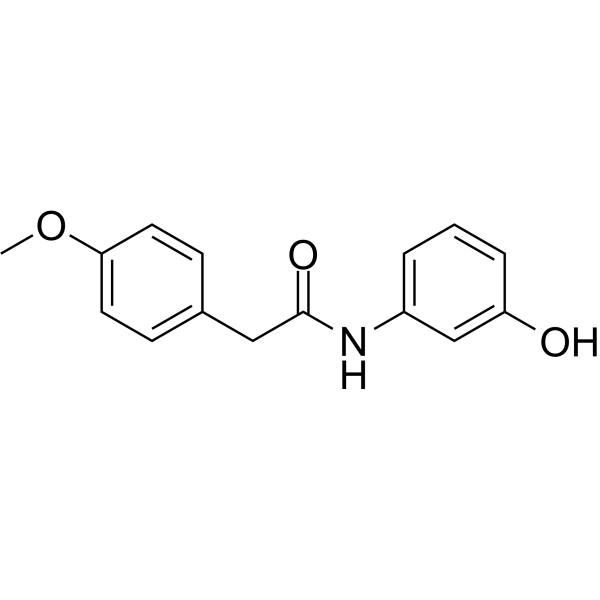
-
GC31657
NOD-IN-1
An inhibitor of NOD1 and NOD2
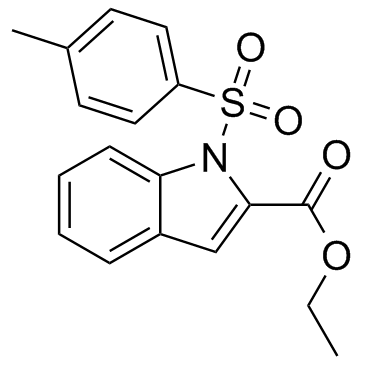
-
GC65995
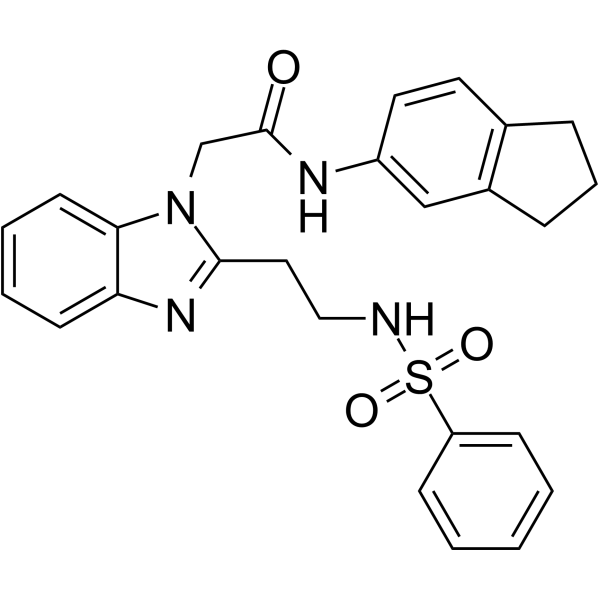
NOD2 antagonist 1
NOD2 antagonist 1 (compound 32) is a potent and selective NOD2 antagonist with an IC50 of 5.23 μM. NOD2 antagonist 1 inhibits Muramyl dipeptide (MDP)-induced IL-8 secretion in THP-1 cells and inhibits MDP-induced IL-6, IL-10, TNF-α release in PBMCs.
-
GC63377
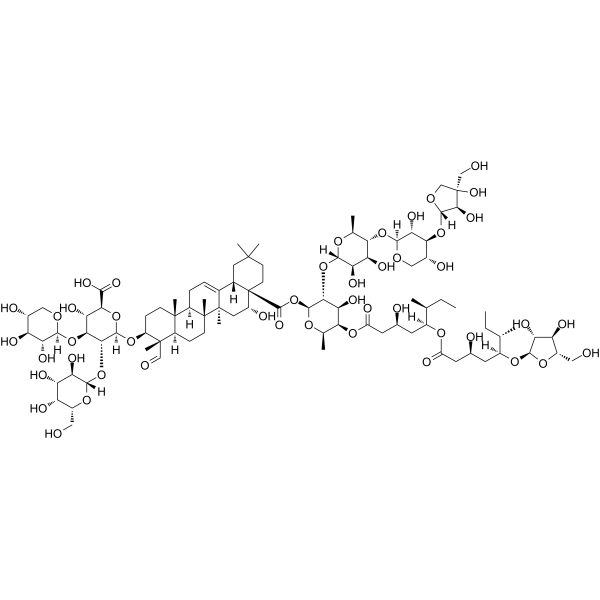
QS-21
QS-21, an immunostimulatory saponin, could be used as a potent vaccine adjuvant.
-
GN10172
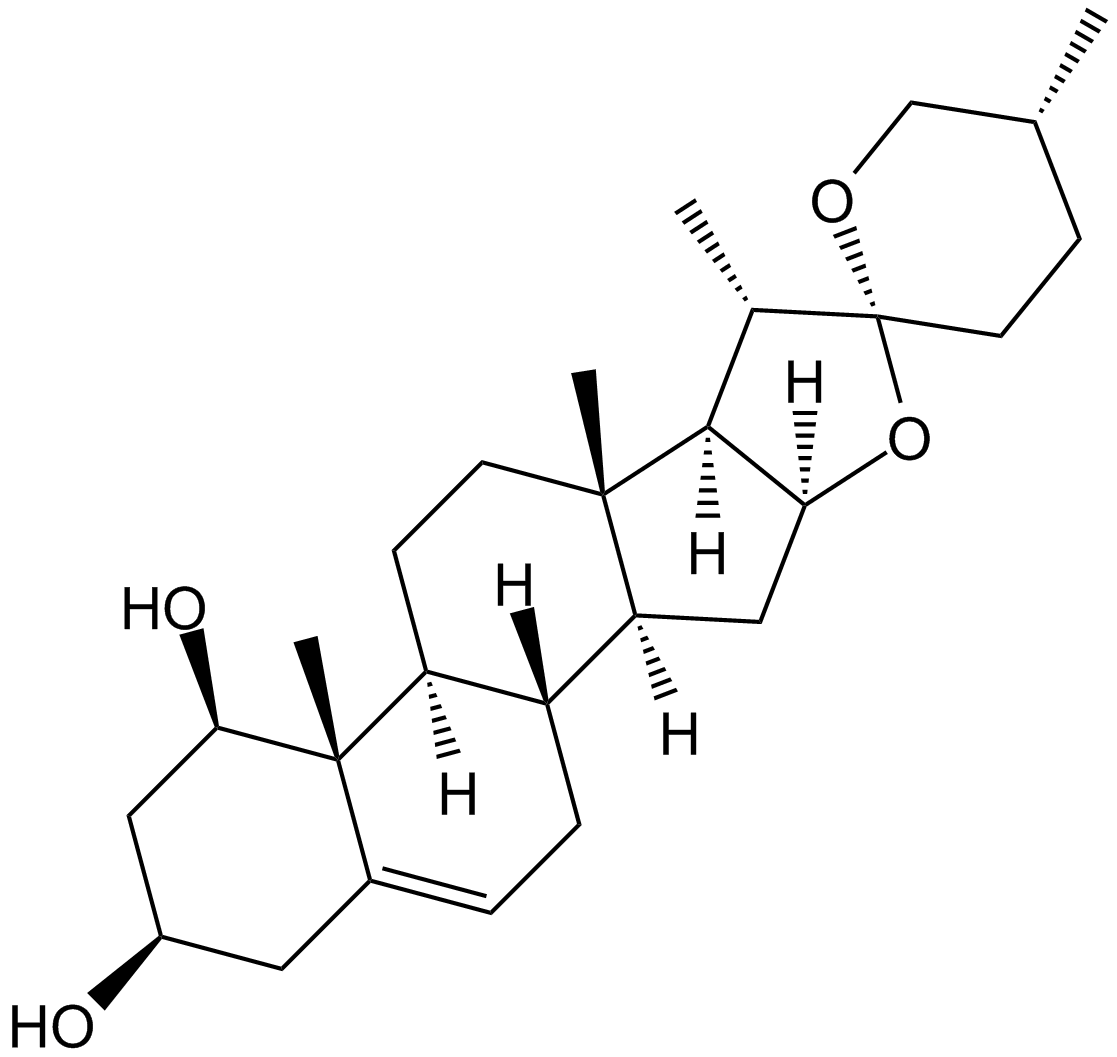
Ruscogenin
-
GC64089
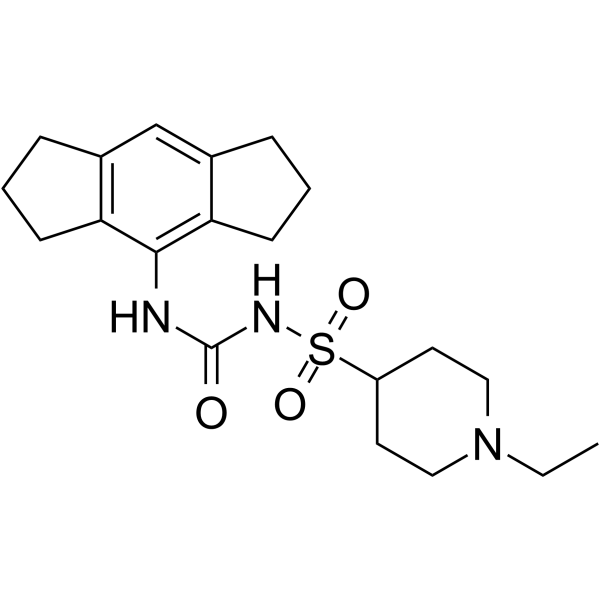
Selnoflast
Selnoflast (example 6) is a NLRP3 inhibitor (extracted from patent WO2019008025).
-
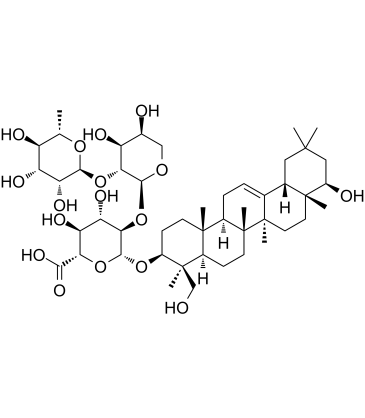
GC61284
Soyasaponin II
Soyasaponin II is a saponin with antiviral activity.
-
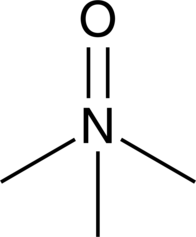
GC40928
Trimethylamine N-oxide
Trimethylamine N-oxide (TMAO) is a product of the oxidation of TMA by flavin-containing mono-oxygenase 3 in the liver.
-
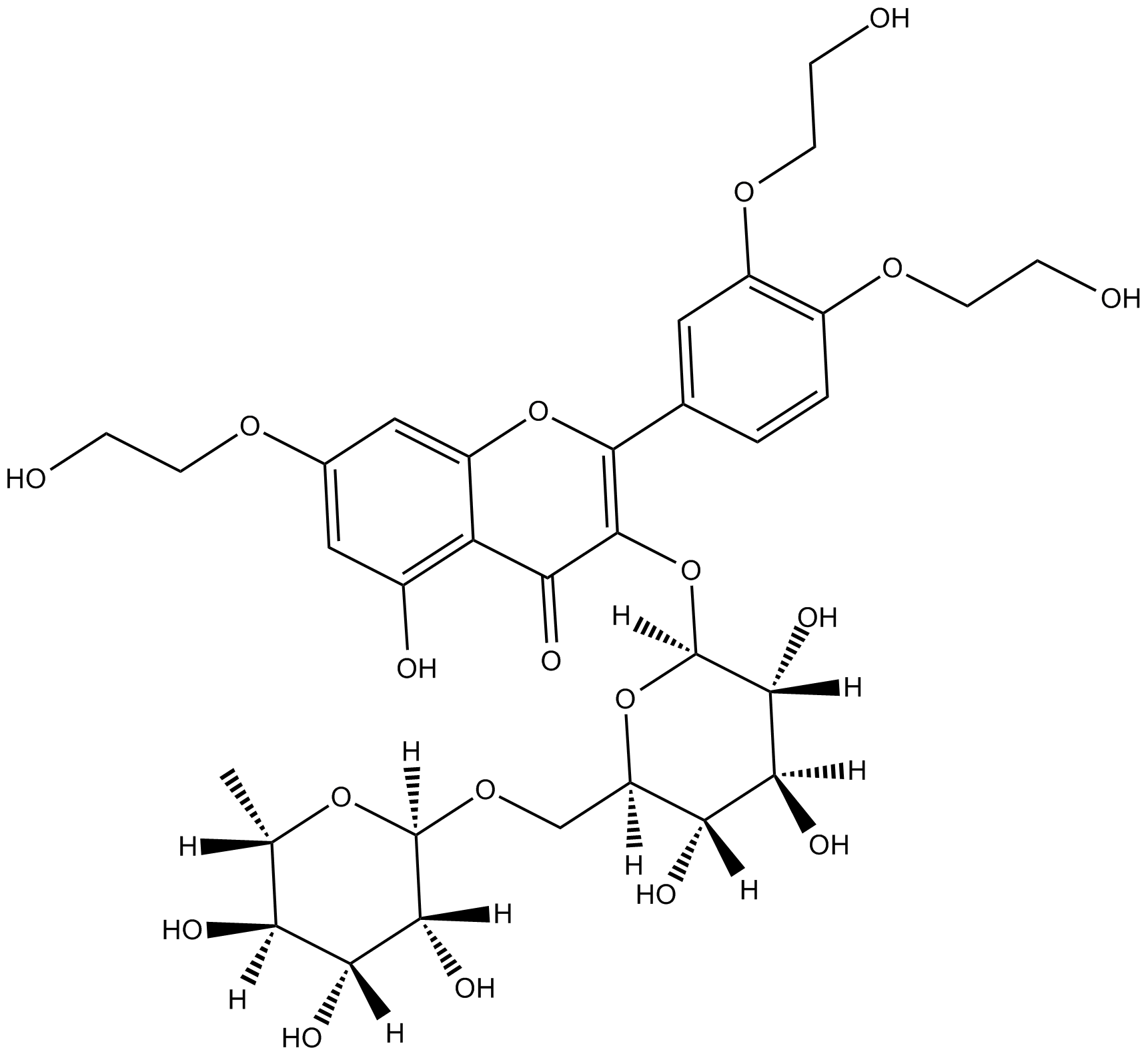
GC15933
Troxerutin
flavonoid
-
GC39311
YQ128
YQ128 is a potent and selective second-generation NLRP3 (NOD-like receptor P3) inflammasome inhibitor with an IC50 of 0.30 ?M.