Carbohydrate Metabolism
Products for Carbohydrate Metabolism
- Cat.No. Product Name Information
-
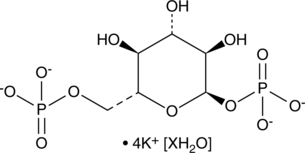
GC48278
α-D-Glucose-1,6-bisphosphate (potassium salt hydrate)
A bis-phosphorylated derivative of α-D-glucose
-
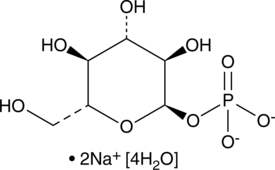
GC48279
α-D-Glucose-1-phosphate (sodium salt hydrate)
An intermediate in glycogen metabolism
-
GC52253
α-Enolase (1-19)-biotin Peptide
A biotinylated α-enolase peptide

-
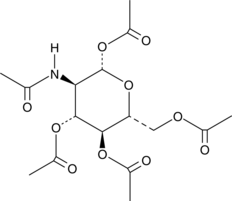
GC45232
β-D-Glucosamine Pentaacetate
β-D-Glucosamine pentaacetate is an N-acetylglucosamine derivative that has been shown to promote hyaluronic acid production.
-
GC49769
β-Glucogallin
A plant metabolite and an aldose reductase 2 inhibitor

-
GC46314
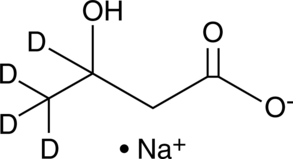
(±)-β-Hydroxybutyrate-d4 (sodium salt)
An internal standard for the quantification of (±)βhydroxybutyrate
-
GC45264
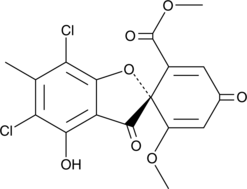
(+)-Geodin
(+)-Geodin is a fungal metabolite.
-
GC41691
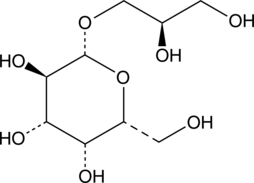
(2R)-Glycerol-O-β-D-galactopyranoside
(2R)-Glycerol-O-β-D-galactopyranoside is a substrate for β-galactosidase, the lactose repressor, the galactose-binding protein, and the β-methylgalactoside transport system.
-
GC41762
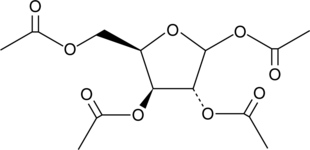
1,2,3,5-Tetra-O-acetyl-D-xylofuranose
1,2,3,5-Tetra-O-acetyl-D-xylofuranose is the raw material for nucleotides synthesis.
-
GC46009
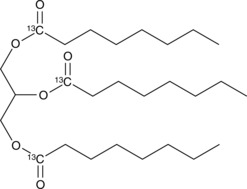
1,2,3-Trioctanoyl-rac-glycerol-13C3
An internal standard for the quantification of 1,2,3-trioctanoyl glycerol-13C3
-
GC41849
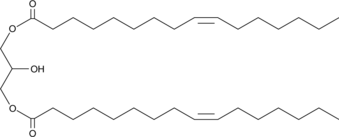
1,3-Dipalmitoleoyl-rac-glycerol
1,3-Dipalmitoleoyl-rac-glycerol is a diacylglycerol with palmitoleic acid at the sn-1 and sn-3 positions.
-
GC41858
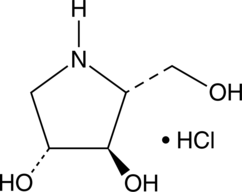
1,4-dideoxy-1,4-imino-D-Arabinitol (hydrochloride)
1,4-dideoxy-1,4-imino-D-Arabinitol (DAB) is an inhibitor of glycogen phosphorylase, a key enzyme in glycogenolysis.
-
GC18609
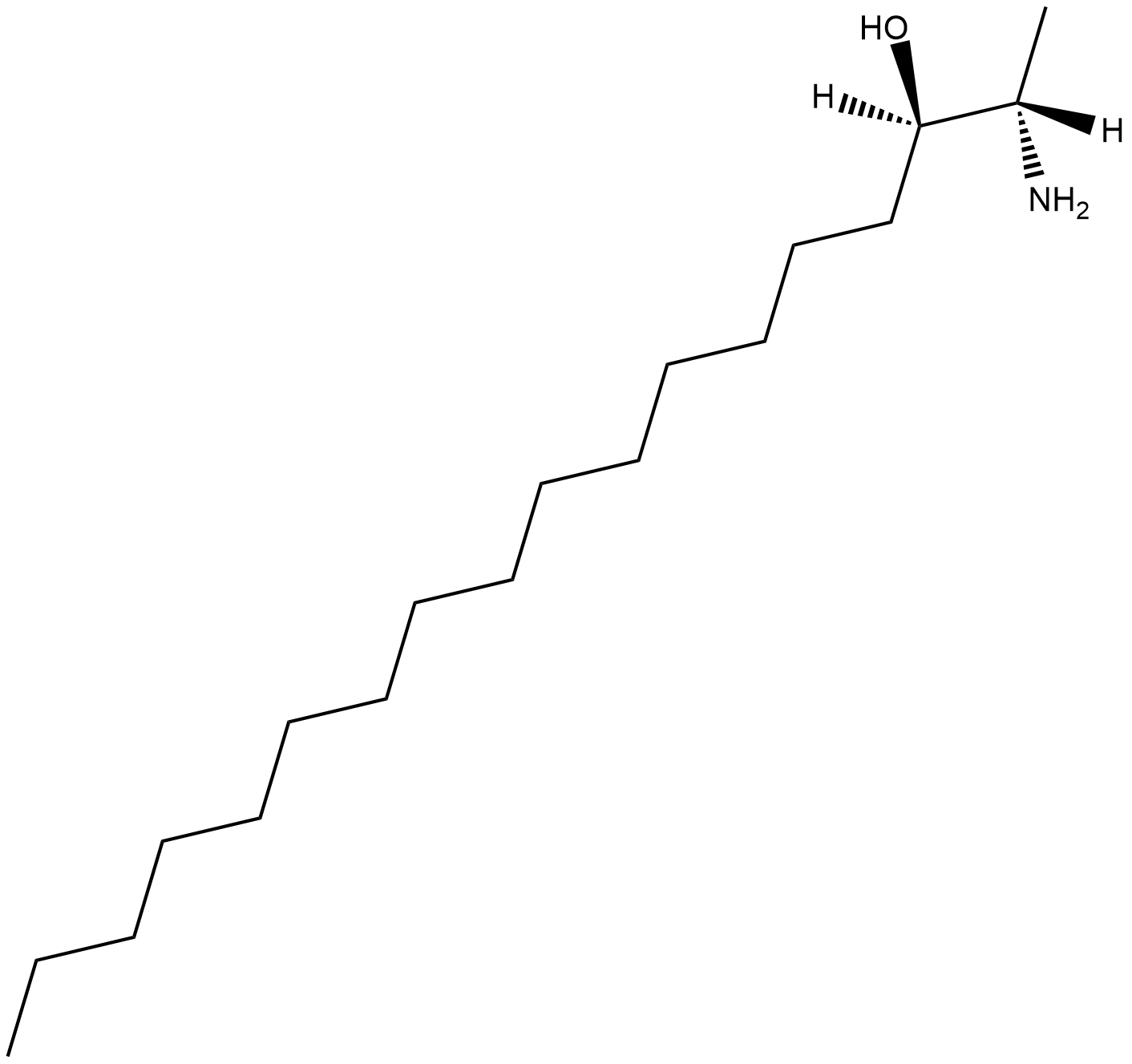
1-Deoxysphinganine (m18:0)
1-Deoxysphinganine (m18:0) (ES-285) is an antiproliferative (antitumoral) compound of marine origin. 1-Deoxysphinganine (m18:0) inhibits the growth of the prostate PC-3 and LNCaP cells through intracellular ceramide accumulation and PKCζ activation.
-
GC42049
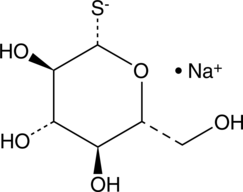
1-thio-β-D-Glucose (sodium salt)
1-thio-β-D-Glucose is an analog of β-D-glucose in which sulfur replaces the hydroxyl group at the one position.
-
GC42058
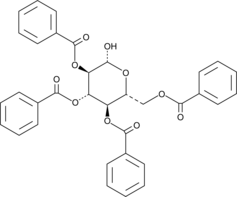
2,3,4,6-Tetra-O-benzoyl-β-D-glucopyranose
2,3,4,6-Tetra-O-benzoyl-β-D-glucopyranose is a D-glucopyranose derivative that has been used in glucosylation reactions.
-
GC48459
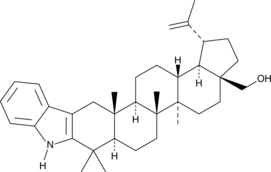
2,3-Indolobetulin
A derivative of betulin
-
GC48444
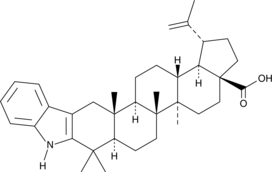
2,3-Indolobetulonic Acid
An inhibitor of α-glucosidase
-
GC41470
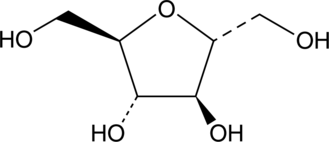
2,5-Anhydro-D-mannitol
2,5-Anhydro-D-mannitol is an antimetabolic fructose analogue that increases food intake in rats at doses of 50-800 mg/kg by inhibiting gluconeogenesis and glycogenolysis in the liver..
-
GC49272
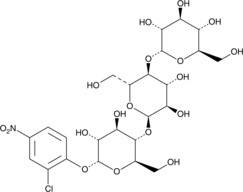
2-Chloro-4-nitrophenyl α-D-maltotrioside
A colorimetric α-amylase substrate
-
GC42137
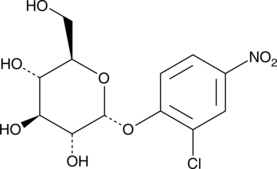
2-Chloro-4-nitrophenyl-α-D-glucopyranoside
Carbohydrates conjugated with 2-chloro-4-nitrophenyl (CNP) serve as chromogenic substrates in assays for enzymes that release CNP from the conjugated carbohydrate.
-
GC41528
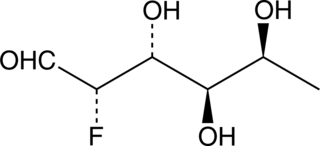
2-deoxy-2-fluoro L-Fucose
Fucose is a deoxyhexose that is present in many organisms.
-
GC49223
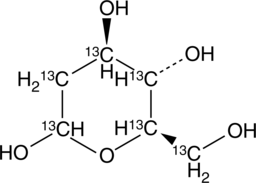
2-deoxy-D-Glucose-13C6
An internal standard for the quantification of 2-deoxy-D-glucose
-
GC19427
2-deoxy-D-Glucose-6-phosphate (sodium salt)
A synthetic analog of glucose used in cell metabolism and cancer research

-
GC42312
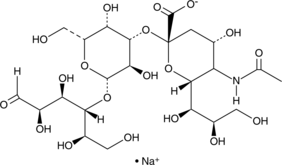
3'-Sialyllactose (sodium salt)
3'-Sialyllactose consists of the monosaccharide N-acetylneuraminic acid linked to the galactosyl subunit of lactose at the 3 position.
-
GC42204
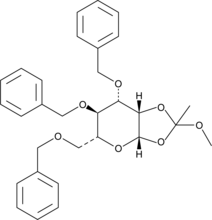
3,4,6-Tri-O-benzyl-β-D-Mannopyranose 1,2-(methyl orthoacetate)
3,4,6-Tri-O-benzyl-β-D-mannopyranose 1,2-(methyl orthoacetate) is a synthetic intermediate used in glycosylation reactions.
-
GC42261
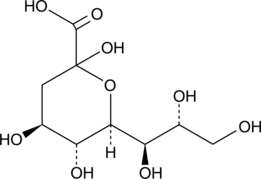
3-Deoxy-D-glycero-D-galacto-2-nonulosonic Acid
Sialic acids, commonly present as terminal carbohydrates on glycoconjugates, are essential for a variety of cellular functions including cell adhesion and signal recognition, as well as the formation and progression of tumors.
-
GC42308
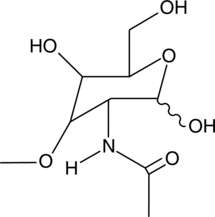
3-O-methyl-N-acetyl-D-Glucosamine
3-O-methyl-N-acetyl-D-Glucosamine (3-O-methyl-GlcNAc) is a competitive inhibitor of N-acetylglucosamine kinase and a non-competitive inhibitor of N-acetylmannosamine kinase (Kis = 17 and 80 μM in rat liver, respectively, in vitro).
-
GC42326
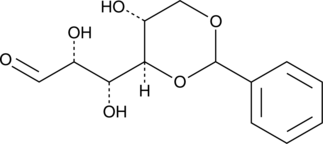
4,6-O-Benzylidene-D-glucose
4,6-O-Benzylidene-D-glucose is a useful intermediate for the synthesis of carbohydrates.
-
GC42434
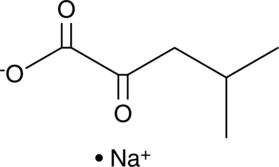
4-methyl-2-Oxovalerate (sodium salt)
4-methyl-2-Oxovalerate is an immediate precursor and metabolite of L-leucine.
-
GC42445
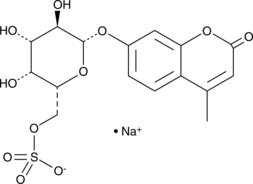
4-Methylumbelliferyl β-D-Galactopyranoside-6-sulfate (sodium salt)
4-Methylumbelliferyl β-D-Galactopyranoside-6-sulfate (sodium salt) is a fluorescent dye.
-
GC42446
4-Methylumbelliferyl β-D-N,N'-diacetylchitobioside
4-Methylumbelliferyl β-D-N,N'-diacetylchitobioside (4-μU-(GlcNAc)2) is a fluorogenic substrate for chitinases and chitobiosidases.
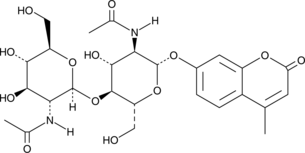
-
GC42442
4-Methylumbelliferyl 2-sulfamino-2-deoxy-α-D-Glucopyranoside (sodium salt)
4-Methylumbelliferyl 2-sulfamino-2-deoxy-α-D-Glucopyranoside (sodium salt) is a fluorogenic substrate of heparin sulphamidase, is desulfurized into 4-MU-α-GlcNH2.
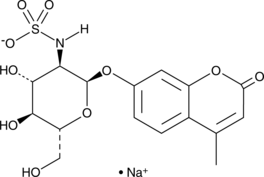
-
GC42449
4-Methylumbelliferyl-α-L-Iduronide (free acid)
4-Methylumbelliferyl-α-L-iduronide (free acid) is a fluorogenic substrate for α-L-iduronidase, an enzyme found in cell lysosomes that is involved in the degradation of glycosaminoglycans such as dermatan sulfate and heparin sulfate.
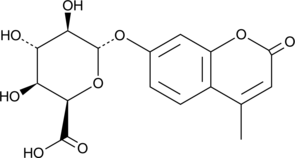
-
GC49114
4-Methylumbelliferyl-β-D-Galactoside
4-Methylumbelliferyl-β-D-Galactoside is a fluorescent substrate for β-galactosidase which, when cleaved, produces a water-soluble blue fluorescent coumarin fluorophore that can be detected using a fluoroenzymeter or fluorometer.
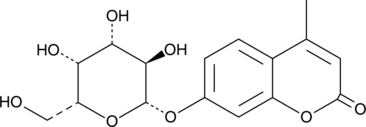
-
GC42452
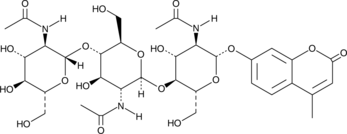
4-Methylumbelliferyl-β-D-N,N',N''-Triacetylchitotrioside
4-Methylumbelliferyl-β-D-N,N',N''-triacetylchitotrioside is a fluorogenic substrate for chitinases and chitotriosidases.
-
GC11870
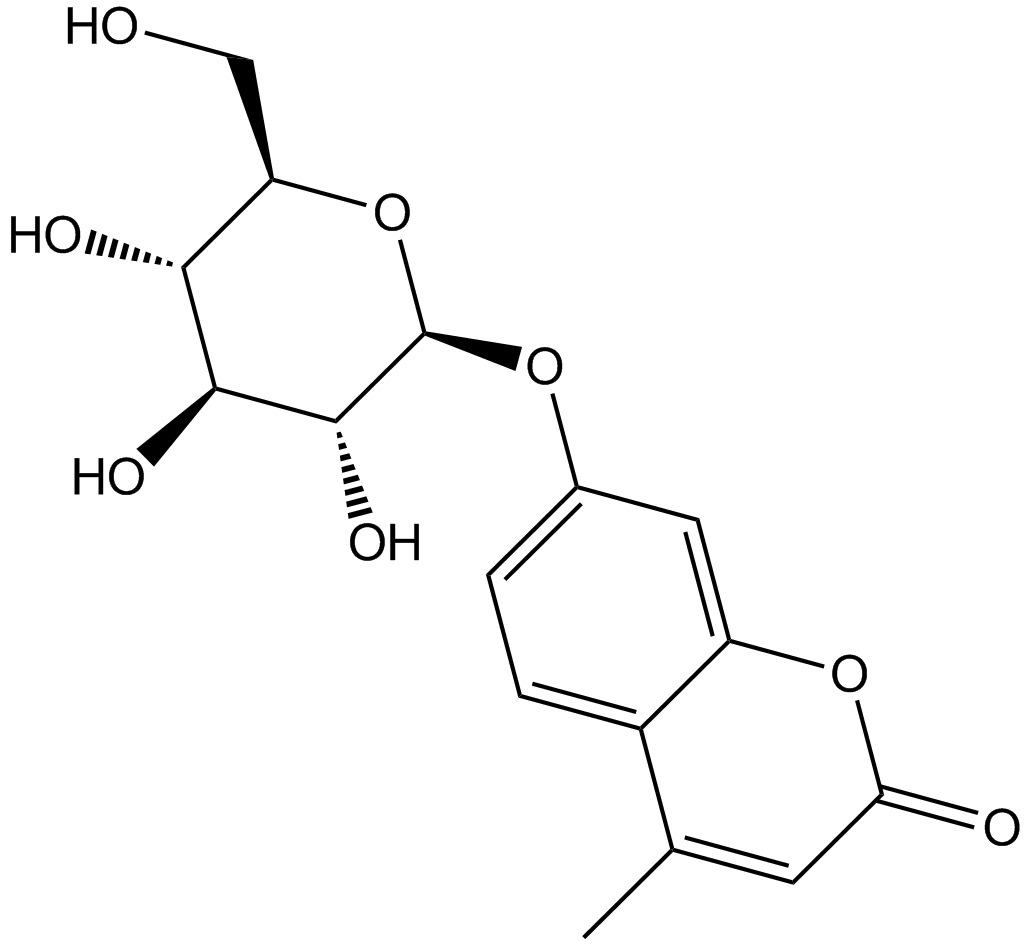
4-Methylumbelliferyl-β-D-Glucopyranoside
4-Methylumbelliferyl-β-D-Glucopyranoside (4-MUG) is a fluorogenic substrate of β-glucosidase and β-glucocerebrosidase (also known as glucosylceramidase).
-
GC52117
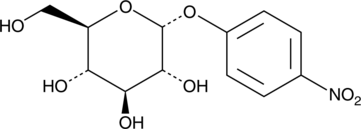
4-Nitrophenyl α-D-Glucopyranoside
A colorimetric α-glucosidase substrate
-
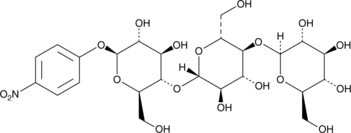
GC42462
4-Nitrophenyl β-D-Cellotrioside
4-Nitrophenyl β-D-cellotrioside is a chromogenic substrate for endoglucanases and cellobiohydrolases.
-
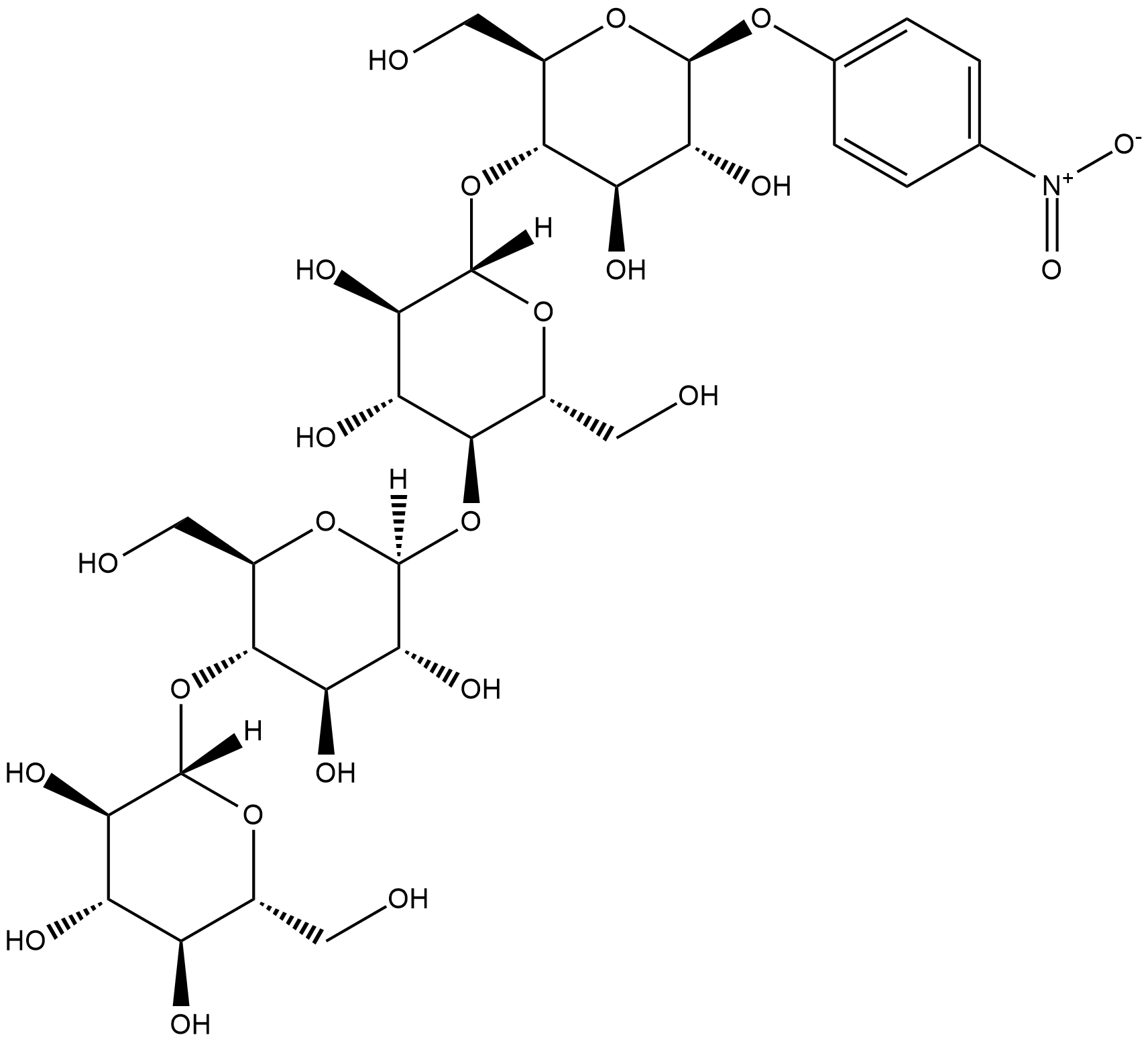
GC18209
4-Nitrophenyl β-D-Cellotetraoside
4-Nitrophenyl β-D-cellotetraoside is small molecule cellulose mimic that consists of a tetramer of D-glucose units joined by β-1-4-glycosidic bonds.
-
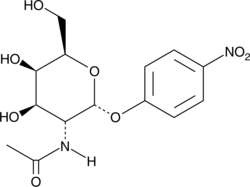
GC42463
4-Nitrophenyl-N-acetyl-α-D-galactosaminide
N-acetyl-D-galactosaminidase catalyzes the cleavage of N-acetylglucosamine (GlcNAc), the monomeric unit of the polymer chitin.
-
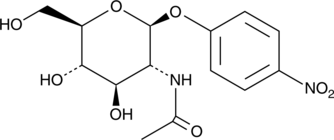
GC45841
4-Nitrophenyl-N-acetyl-β-D-glucosaminide
A chromogenic substrate for N-acetyl-β-glucosaminidase
-
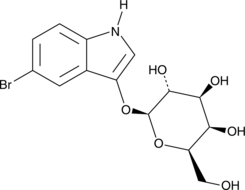
GC49877
5-bromo-3-indolyl-β-D-Galactopyranoside
5-bromo-3-indolyl-β-D-Galactopyranoside (Bluo-Gal) is a chromogenic substrate to detect bacterial β-D-galactosidase activity.
-
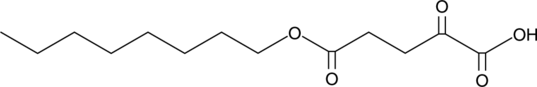
GC41310
5-Octyl-α-ketoglutarate
In addition to its role in the Krebs cycle, α-ketoglutarate (2-oxoglutarate) has roles as a substrate or modulator of enzymes.
-
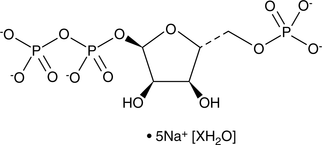
GC46713
5-Phospho-D-ribose 1-diphosphate (sodium salt hydrate)
An intermediate in several biochemical pathways
-
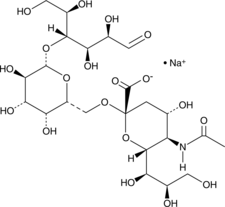
GC19536
6'-Sialyllactose Sodium Salt
6'-Sialyllactose (sodium), a predominant milk oligosaccharide, reduces the internalisation of Pseudomonas aeruginosa in human pneumocytes.
-
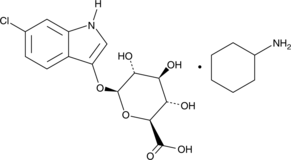
GC46722
6-Chloro-3-indolyl-β-D-Glucuronide (cyclohexylammonium salt)
A β-glucuronidase chromogenic substrate
-
GC18870
6-NBDG
6-NBDG is a non-hydrolyzable fluorescent glucose analog that is used to monitor glucose uptake and transport in living cells.
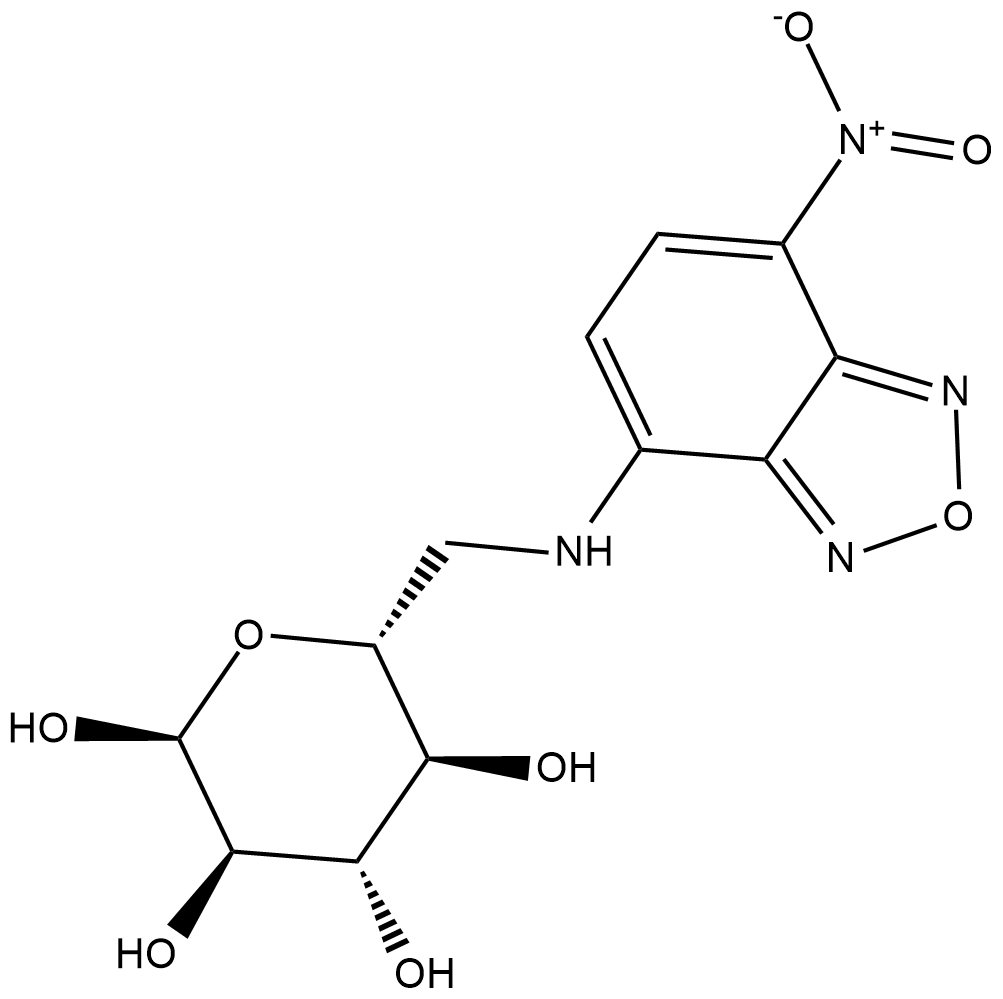
-
GC42620
8-Aminopyrene-1,3,6-trisulfonic Acid (sodium salt)
8-Aminopyrene-1,3,6-trisulfonic Acid (sodium salt) is a water-soluble anionic fluorescent dye.
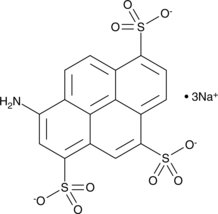
-
GC46780
Acesulfame-d4 (potassium salt)
An internal standard for the quantification of acesulfame
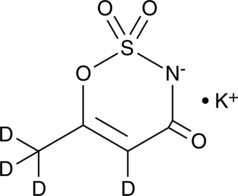
-
GC42736
ADP-Glucose (sodium salt)
An intermediate in polysaccharide synthesis
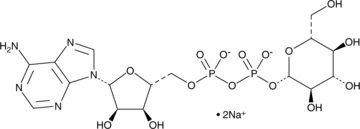
-
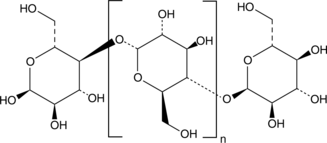
GC46852
Amylose
A polysaccharide
-
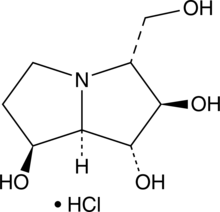
GC46896
Australine (hydrochloride)
A pyrrolizidine alkaloid
-
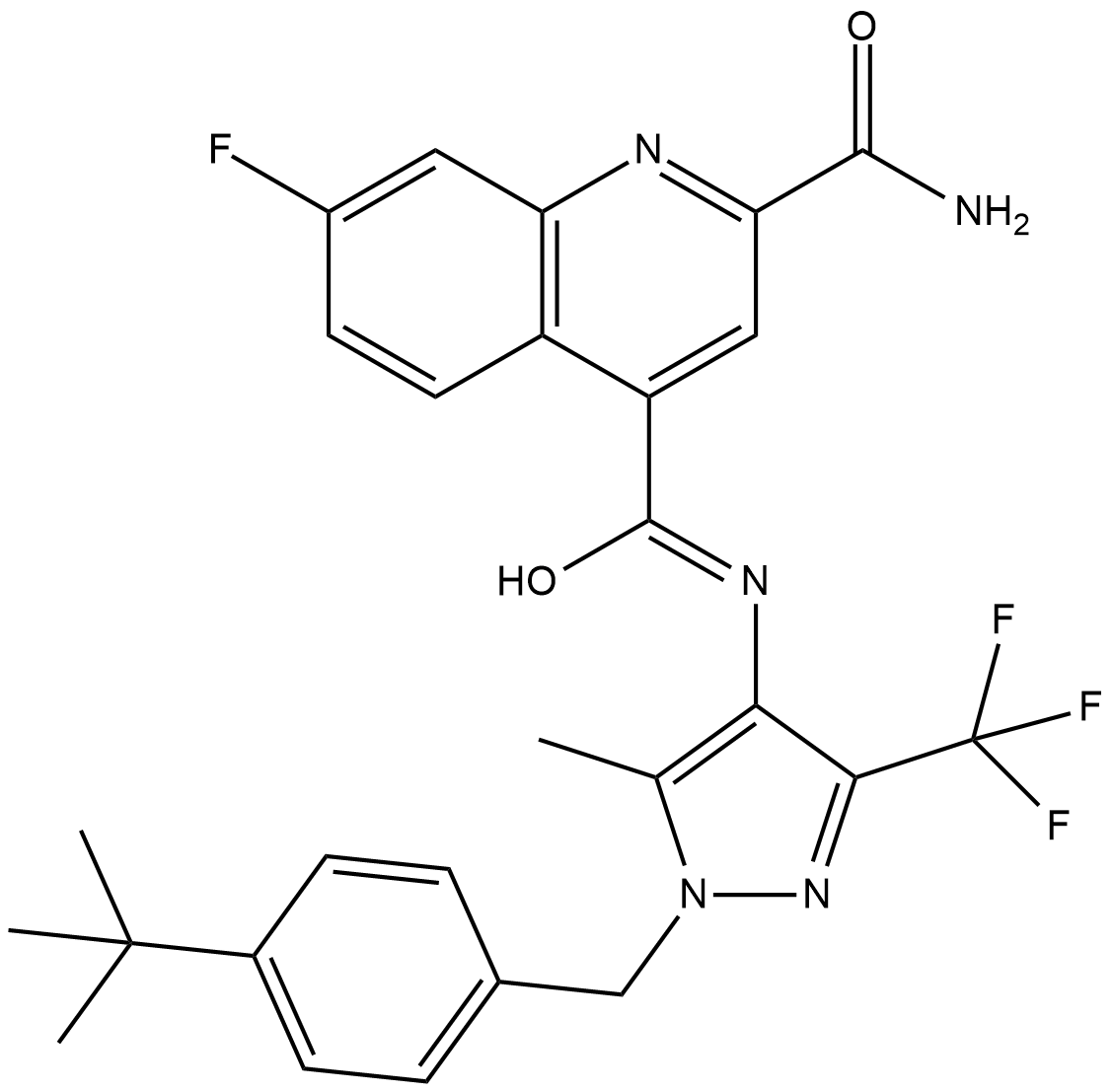
GC19000
BAY-588
BAY-588 is a glucose transporter inhibitor and derivative of BAY-876 .
-
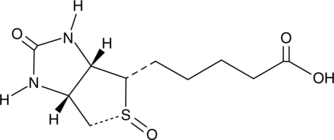
GC49200
Biotin (R)-Sulfoxide
A metabolite of biotin
-
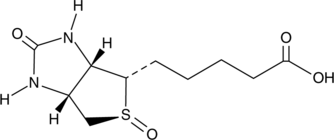
GC49170
Biotin (S)-sulfoxide
An inactive metabolite of biotin
-
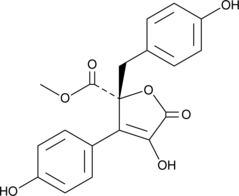
GC46105
Butyrolactone II
A fungal metabolite
-
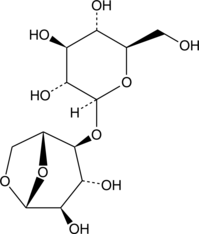
GC43226
Cellobiosan
Cellobiosan is an anhydro sugar formed during biofuel production from the fast pyrolysis of wood.
-
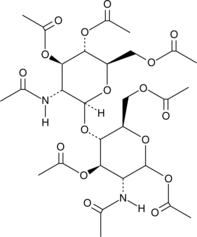
GC43238
Chitobiose Octaacetate
Chitobiose is a dimer of β-1,4 linked glucosamine units derived from chitin, which, in turn, is a long chain polymer of N-acetylglucosamine that is a primary component of fungal cell walls and arthropod exoskeletons.
-
GC47080
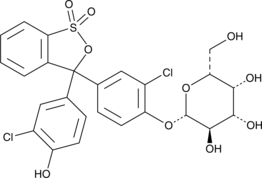
Chlorophenol Red β-D-Galactopyranoside
Chlorophenol Red β-D-Galactopyranoside is a long-wavelength dye, which has been widely used for colorimetric assays.
-
GC52351
Citrullinated α-Enolase (R8 + R14) (1-19)-biotin Peptide
A biotinylated and citrullinated α-enolase peptide

-
GC18572
Concanavalin A
Concanamycin A belongs to the concanamycins, a family of macrolide antibiotics isolated from Streptomyces diastatochromogenes that are highly active and selective inhibitors of the vacuolar proton-ATPase (v-[H+]ATPase).

-
GC49232
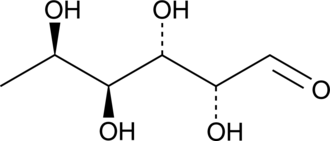
D-(+)-Fucose
A monosaccharide
-
GC41089
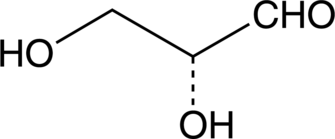
D-(+)-Glyceraldehyde
D-(+)-Glyceraldehyde is an intermediate in carbohydrate metabolism.
-
GC43365
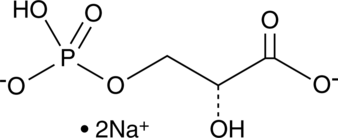
D-(-)-3-Phosphoglyceric Acid (sodium salt)
D-(-)-3-Phosphoglyceric acid (3-Phospho-D-glyceric acid) disodium is an important intermediate in the enzyme-catalysed process of glycolysis.
-
GC43366
D-(-)-Citramalic Acid (lithium salt)
D-(-)-Citramalic acid (lithium salt) is a biochemical intermediate involved in the anaerobic metabolism of glutamate through the methylaspartate pathway of C.
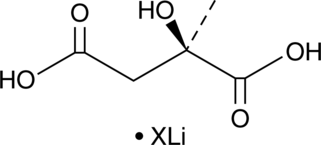
-
GC43430
D-Fructose-1,6-bisphosphate (sodium salt hydrate)
D-Fructose-1,6-bisphosphate is an intermediate in carbohydrate metabolism, including glycolysis and gluconeogenesis.
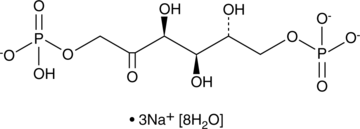
-
GC47204
D-Fructose-13C6
An internal standard for the quantification of D-fructose
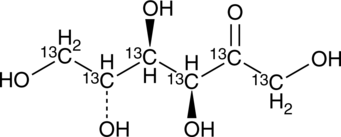
-
GC47205
D-Fructose-6-phosphate (sodium salt hydrate)
A sugar intermediate of the glycolytic pathway
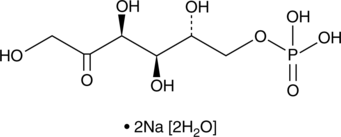
-
GC49492
D-Iditol
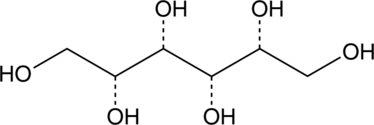
-
GC43501
D-Mannose-6-Phosphate (sodium salt hydrate)
D-Mannose-6-phosphate (M6P) is a phosphorylated aldohexose that has diverse physiological roles.
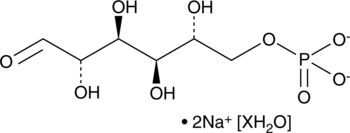
-
GC40754
D-Ribulose
Ribulose is a ketopentose, a monosaccharide containing five carbon atoms and a ketone functional group.
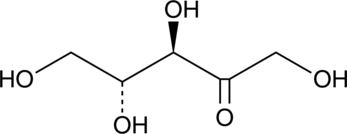
-
GC43571
D-Ribulose-5-phosphate (sodium salt)
D-Ribulose-5-phosphate is an intermediate in the pentose phosphate pathway.
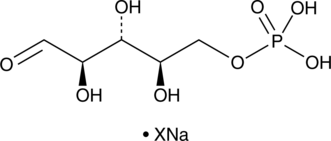
-
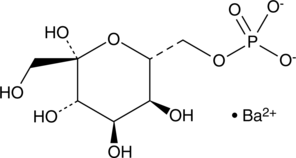
GC43577
D-Sedoheptulose-7-phosphate (barium salt)
D-Sedoheptulose-7-phosphate is an intermediate in the pentose phosphate pathway.
-
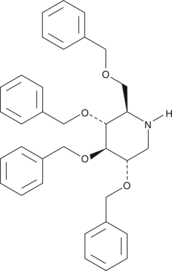
GC43409
Deoxynojirimycin Tetrabenzyl Ether
Deoxynojirimycin (dNM) tetrabenzyl ether is an intermediate for the synthesis of glucosylceramide synthase inhibitors such as 1-dNM, a glucose analog that potently inhibits α-glucosidase I and II.
-
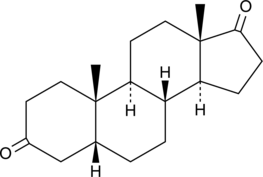
GC49762
Etiocholanedione
A neurosteroid
-
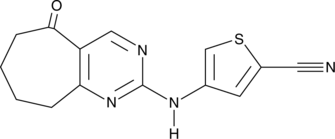
GC47387
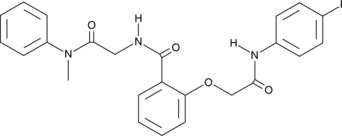
G6PDi-1
An inhibitor of G6PDH
-
GC43720
Galactinol (hydrate)
Galactinol (hydrate) is a marker for seed longevity.
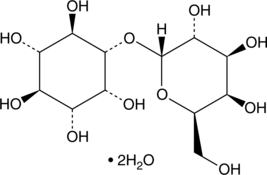
-
GC49228
Ganoderol B
A triterpenoid with diverse biological activities
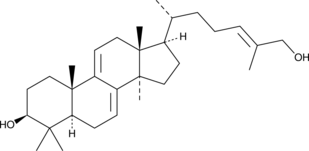
-
GC49106
Gentisein
A xanthone with diverse biological activities
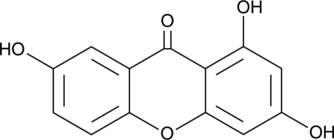
-
GC48994
Girard’s Reagent P-d5
An internal standard for the quantification of Girard’s reagent P
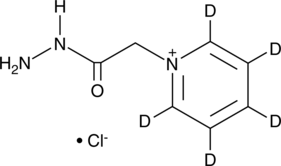
-
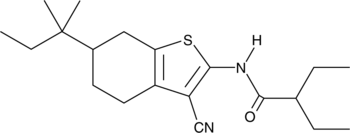
GC43765
Glucagon Receptor Antagonist I
Glucagon receptor antagonist I is a competitive antagonist of the glucagon receptor (GCGR; IC50 = 181 nM).
-
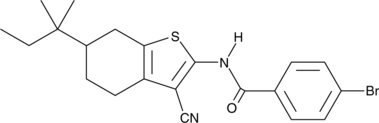
GC43766
Glucagon Receptor Antagonist Inactive Control
An inactive control for glucagon receptor antagonist I
-
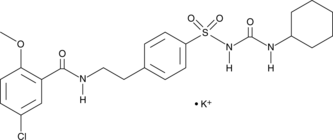
GC43773
Glyburide (potassium salt)
Glyburide is a sulfonylurea and an inhibitor of sulfonylurea receptor 1 (SUR1) linked to ATP-sensitive potassium channel Kir6.2 (IC50 = 4.3 nM).
-
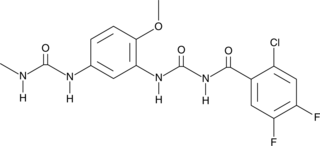
GC43777
Glycogen Phosphorylase Inhibitor
Glycogen phosphorylase in the liver, muscle, and brain initiate glycogenolysis by releasing glucose-1-phosphate from glycogen.
-
GC49744
GRP94 Monoclonal Antibody (Clone 9G10)
For immunochemical detection of GRP94

-
GC18875
GSK3β Inhibitor XVIII
An inhibitor of GSK3β
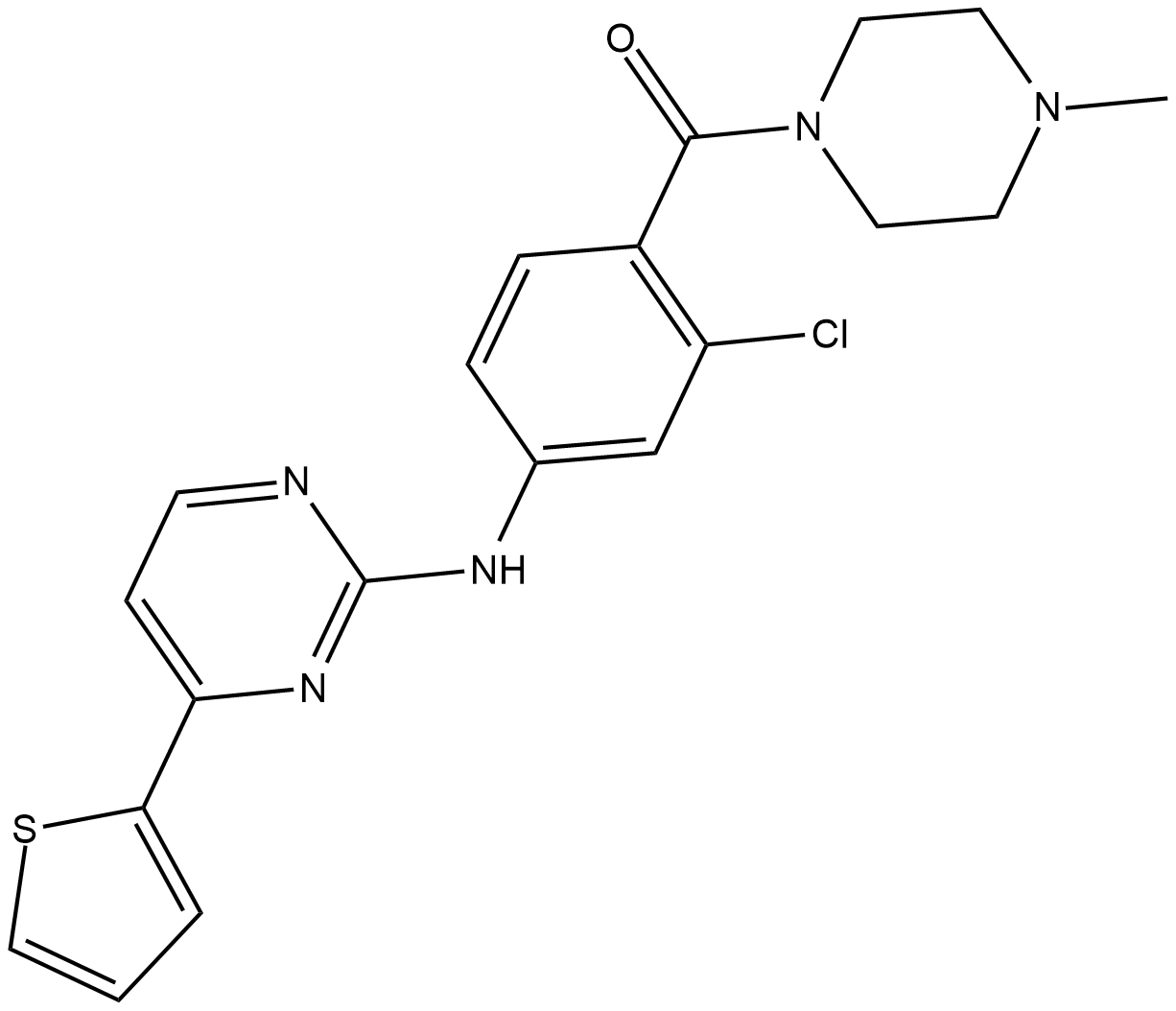
-
GC43816
Heptelidic Acid
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a key enzyme in carbohydrate metabolism, reversibly catalyzes the conversion of GAP to 1,3-bisphosphoglycerate and NAD+.
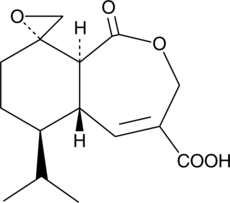
-
GC40299
L-(+)-Erythrose
L-(+)-Erythrose is an aldotetrose carbohydrate that has been used in glycation studies and to characterize erythrose reductase activity.
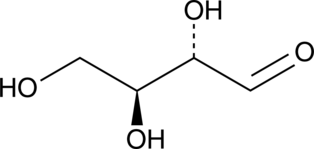
-
GC49373
L-Alanine-d3
An internal standard for the quantification of L-alanine
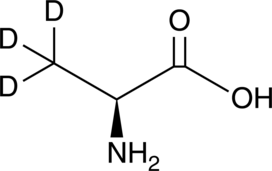
-
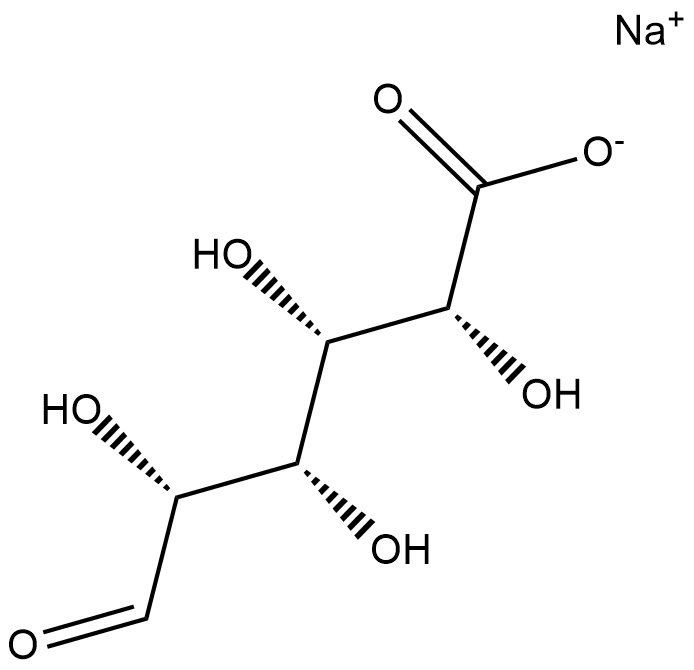
GC18807
L-Iduronic Acid (sodium salt)
L-Iduronic acid is an epimer of glucuronic acid and a monosaccharide component of glycosaminoglycans (GAGs), including heparin and chondroitin sulfate B, found on the outer cell membrane.
-
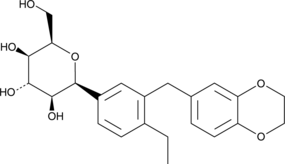
GC52079
Licogliflozin (mixed isomers)
A mixture of licogliflozin isomers
-
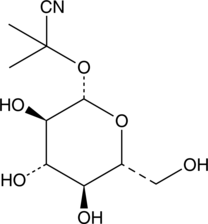
GC40659
Linamarin
Linamarin is a glucoside of acetone cyanohydrin found in the leaves and roots of cassava, lima beans, and flax.
-
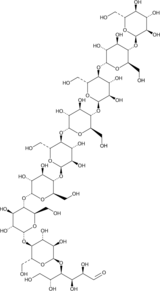
GC47592
Maltononaose
An oligosaccharide
-
GC47600
Massarigenin C
A fungal metabolite
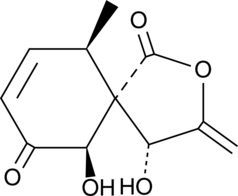
-
GC49472
Methylmalonic Acid-d3
Methylmalonic acid-d3 (Methylpropanedioic acid-d3) is the deuterium labeled Methylmalonic acid. Methylmalonic acid (Methylmalonate) is an indicator of Vitamin B-12 deficiency in cancer.
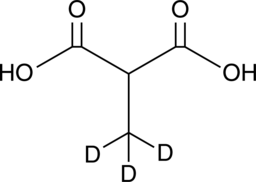
-
GC49521
MHY553
An agonist of PPARα
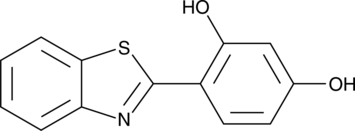
-
GC49083
N,N’-Diphenethylurea
A bacterial metabolite with diverse biological activities
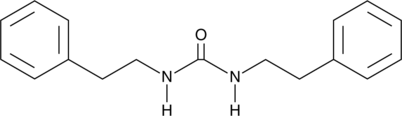
-
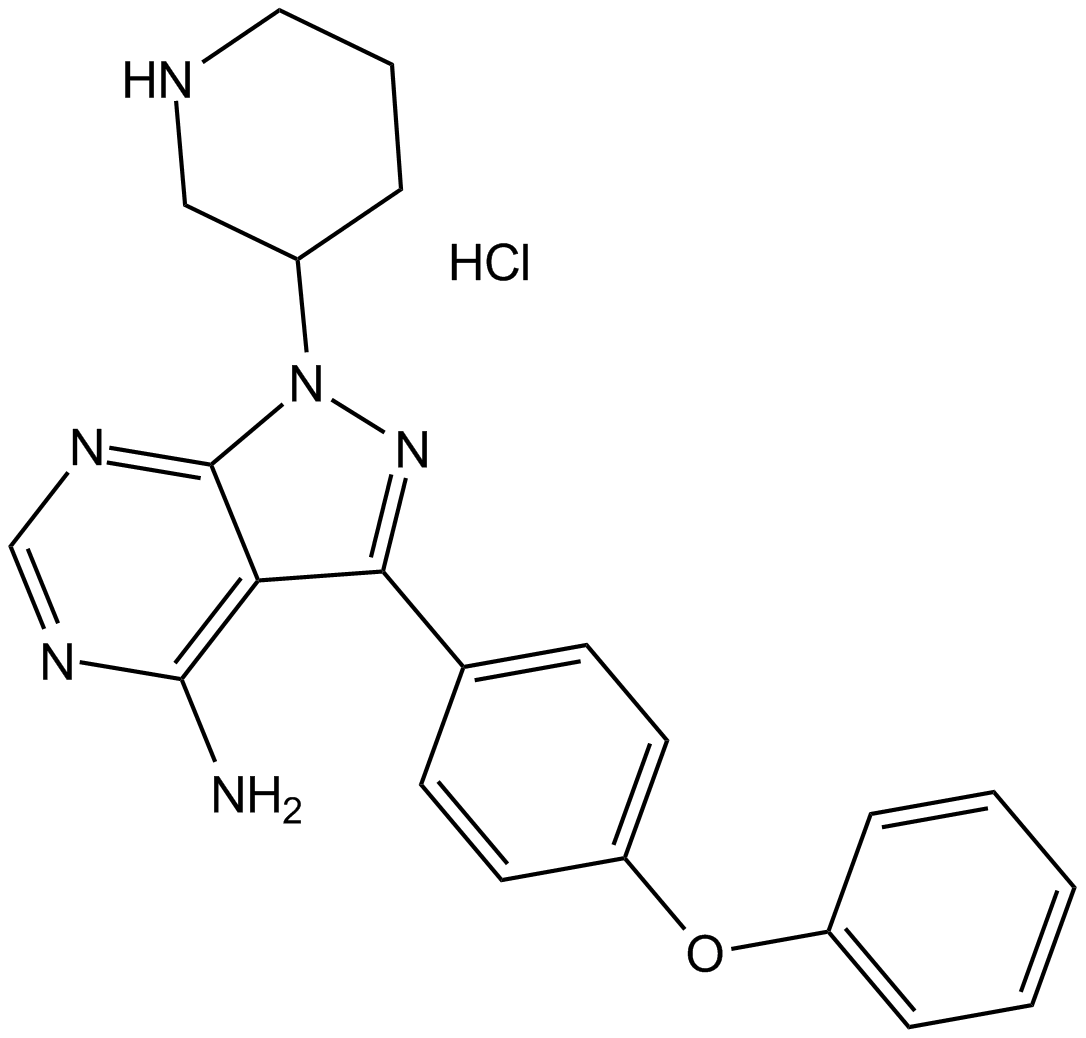
GC10879
N-acetyl-D-Lactosamine
N-acetyl-D-Lactosamine (LacNAc), a nitrogen-containing disaccharide, is an important component of various oligosaccharides such as glycoproteins and sialyl Lewis X.
-
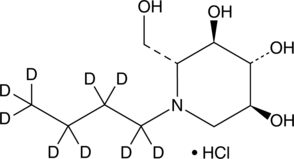
GC49388
N-Butyldeoxynojirimycin-d9 (hydrochloride)
An internal standard for the quantification of N-butyldeoxynojirimycin
-
GC44335
NCGC607
NCGC607 is a salicylic acid derivative and small molecule glucocerebrosidase (GCase) chaperone.