MDR multidrug resistance
MDR (multidrug resistance) is a property that first identified in tumor cell, hat the resistance of tumor cell is to a variety of chemotherapy agent due to active efflux via transporter proteins.
Products for MDR multidrug resistance
- Cat.No. Product Name Information
-
GC45246
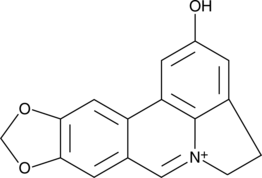
(-)-Chaetominine
(-)-Chaetominine is a cytotoxic alkaloid originally isolated from Chaetomium sp.
-
GC41865
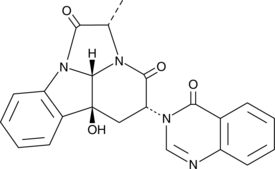
10'-Desmethoxystreptonigrin
10'-Desmethoxystreptonigrin is an antibiotic originally isolated from Streptomyces and a derivative of the antibiotic streptonigrin.
-
GC45324
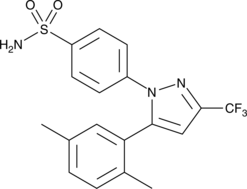
2,5-dimethyl Celecoxib
-
GC42616
7-oxo Staurosporine
7-oxo Staurosporine is an antibiotic originally isolated from S.
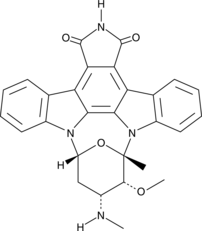
-
GC49275
8-Oxycoptisine
8-Oxycoptisine is a natural protoberberine alkaloid with anti-cancer activity.
-
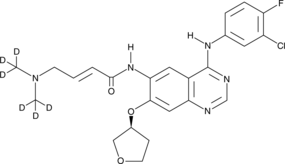
GC46809
Afatinib-d6
An internal standard for the quantification of afatinib
-
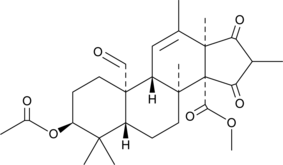
GC42806
Andrastin A
Andrastin A is a meroterpenoid farnesyltransferase inhibitor.
-
GC43004
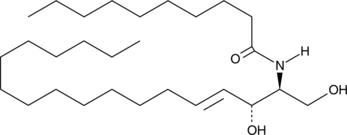
C10 Ceramide (d18:1/10:0)
C10 ceramide is an endogenous, bioactive sphingolipid produced by ceramide synthase.
-
GC43060
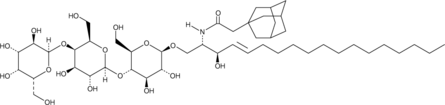
C2 Adamantanyl Globotriaosylceramide (d18:1/2:0)
C2 Adamantanyl globotriaosylceramide (AdaGb3) is a bioactive sphingolipid and water-soluble form of globotriaosylceramide that contains an adamantanyl group in place of the fatty acyl chain.
-
GC45395
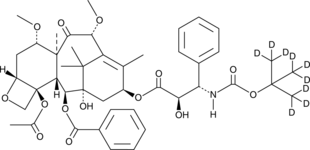
Cabazitaxel-d9
Cabazitaxel-d9 is deuterium labeled Cabazitaxel. Cabazitaxel is a semi-synthetic derivative of the natural taxoid 10-deacetylbaccatin III with potential antineoplastic activity.
-
GC45679
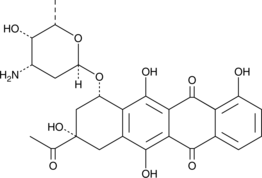
Carubicin
An anthracycline with anticancer activity
-
GC40795
CAY10503
CAY10503 is a proapoptotic, antiproliferative compound that is able to arrest cell cycle progression in the G0-G1 phase.
-
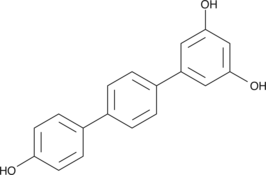
GC18901
CAY10647
CAY10647 is an aryl-benzoyl-imidazole which prevents proliferation of multidrug resistant cancer cells in vitro (IC50 = 28-63 nM) and inhibits melanoma xenograph tumor growth in vivo (10-30 mg/kg).
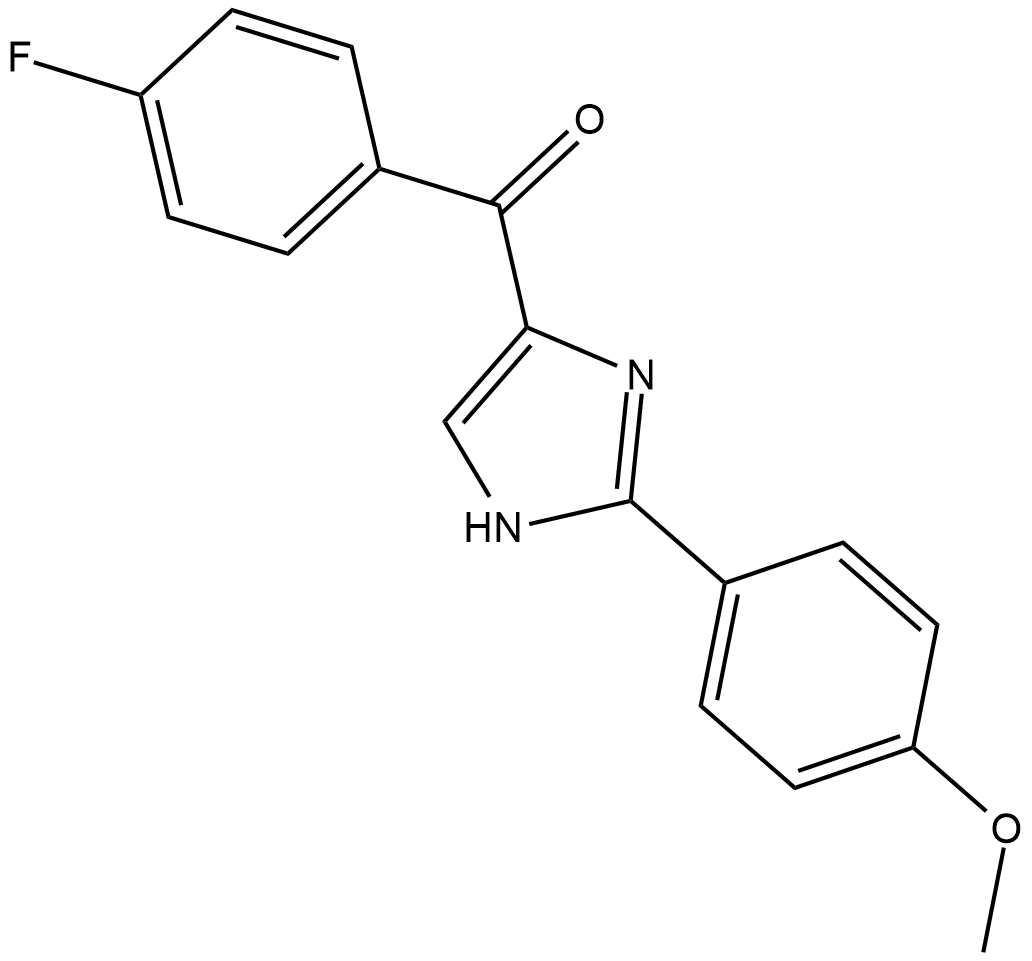
-
GC45792
Chlorido[N,N'-disalicylidene-1,2-phenylenediamine]iron(III)
An inducer of ferroptosis
![Chlorido[N,N'-disalicylidene-1,2-phenylenediamine]iron(III) Chemical Structure Chlorido[N,N'-disalicylidene-1,2-phenylenediamine]iron(III) Chemical Structure](/media/struct/GC4/GC45792.png)
-
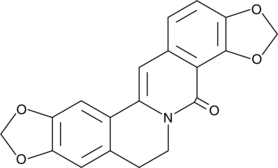
GC46004
Evoxanthine
An alkaloid
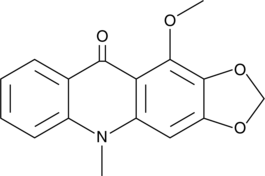
-
GC49106
Gentisein
A xanthone with diverse biological activities
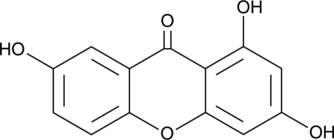
-
GC47409
Glycocholic Acid-d4
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
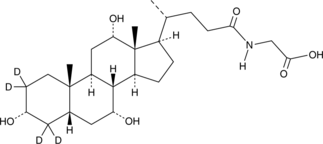
-
GC49670
Indium (III) thiosemicarbazone 5b
An anticancer agent
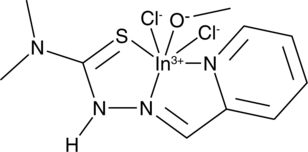
-
GC44129
MC 70
A P-glycoprotein inhibitor
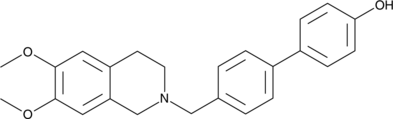
-
GC48479
Migrastatin
A fungal metabolite with antimuscarinic and anticancer activities
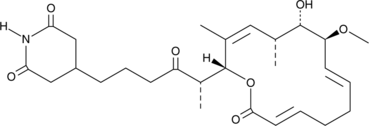
-
GC44195
Milbemycin A4 oxime
Milbemycin A4 oxime is a derivative of milbemycin A4 and a component of milbemycin oxime, compounds that both have insecticidal and nematocidal activity.
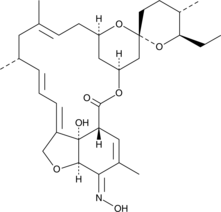
-
GC41564
MPT0B014
An inhibitor of tubulin polymerization
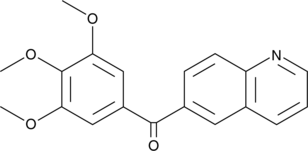
-
GC45784
ONT-093
A P-glycoprotein inhibitor
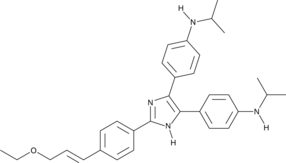
-
GC49251
Oxaliplatin-d10
An internal standard for the quantification of oxaliplatin
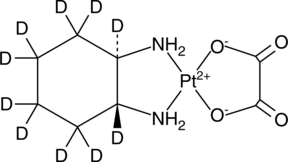
-
GC44613
PF-6274484
PF-6274484 is an inhibitor of the EGF receptor (EGFR; IC50s = 0.18 and 0.14 nM for wild-type EGFR and inhibitor-resistant EGFRL858R/T790M, respectively).
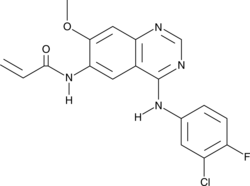
-
GC44615
PGP-4008
PGP-4008 is a selective inhibitor of P-glycoprotein (P-gp) that does not affect the activity of multidrug resistance-related protein 1 (MRP1).
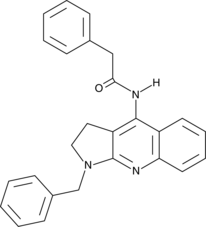
-
GC41629
Piperafizine A
Piperafizine A is a natural methylated diketopiperazine first isolated from an actinomycete, Streptoverticillium.
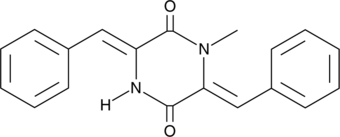
-
GC52455
Pixantrone-d8 (maleate)
An internal standard for the quantification of pixantrone
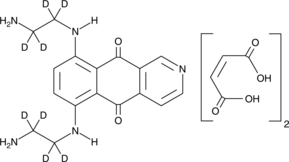
-
GC52104
Ponatinib (hydrochloride)
An inhibitor of native and mutant Bcr-Abl
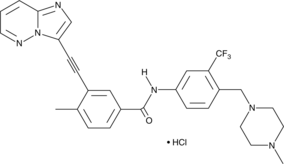
-
GC17403
Pyronaridine Tetraphosphate
An antimalarial agent
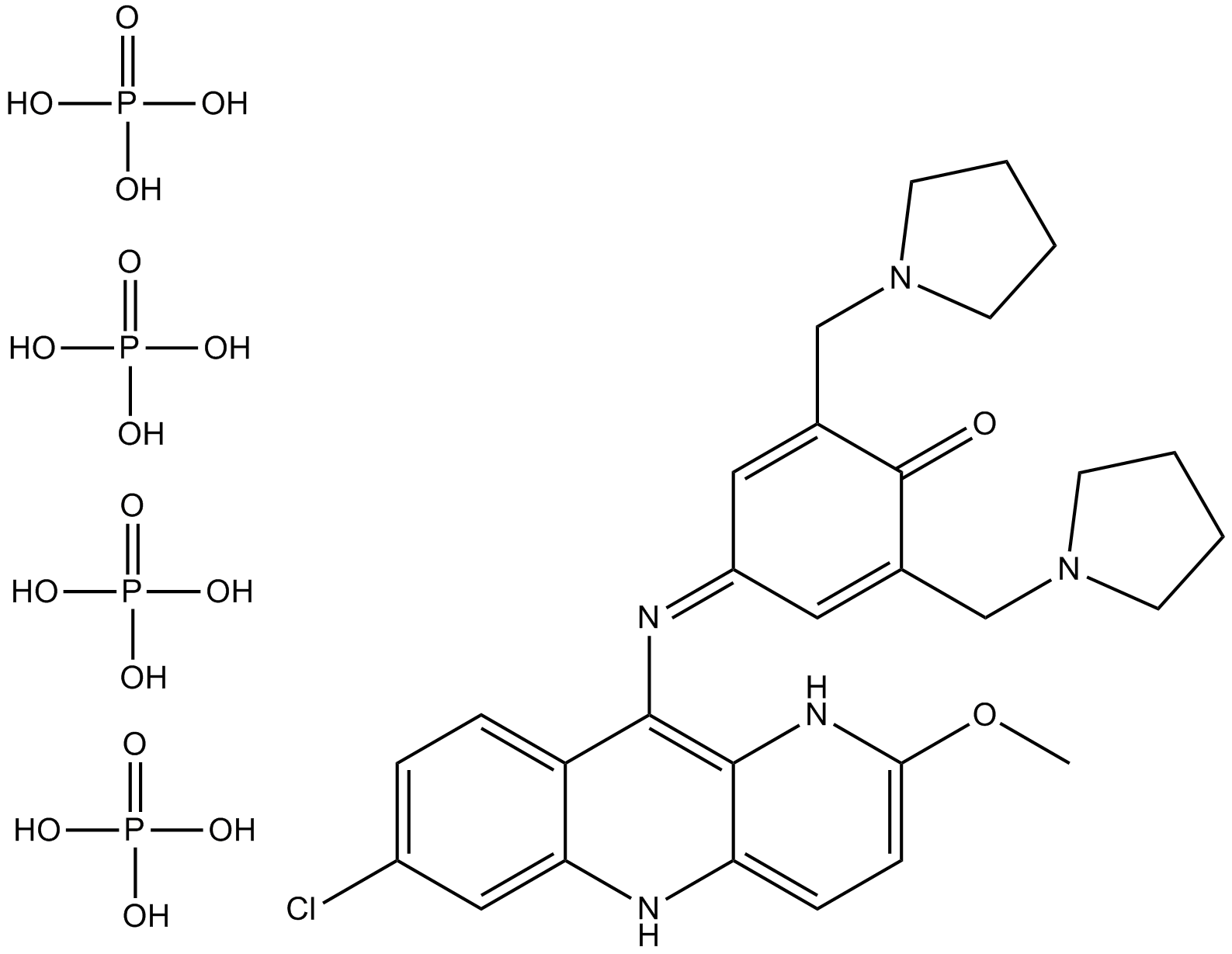
-
GC44821
Reversin 121
Reversin 121 is a hydrophobic peptide chemosensitizer that can reverse P-glycoprotein-mediated multidrug resistance.
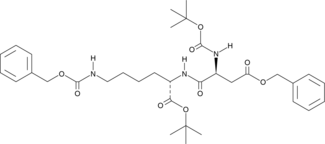
-
GC45688
STX140
An estrogen sulfamate
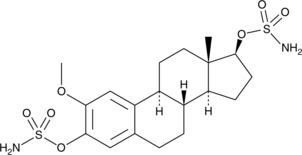
-
GC48119
Suprafenacine
An inhibitor of tubulin polymerization with anticancer activity
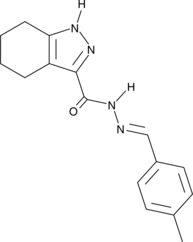
-
GC48219
Ulipristal Acetate-d6
An internal standard for the quantification of ulipristal acetate
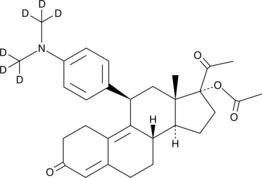
-
GC49308
Ungeremine
A betaine-type alkaloid with diverse biological activities