Transcription Factors
Products for Transcription Factors
- Cat.No. Product Name Information
-
GC48292
α-MSH (human, mouse, rat, porcine, bovine, ovine) (trifluoroacetate salt)
α-MSH (α-Melanocyte-Stimulating Hormone) TFA, an endogenous neuropeptide, is an endogenous melanocortin receptor 4 (MC4R) agonist with anti-inflammatory and antipyretic activities.
-
GC52010
(±)-10-hydroxy-12(Z),15(Z)-Octadecadienoic Acid
An oxylipin gut microbiota metabolite
-
GC46347
(S)-(+)-Methoprene
An insect growth regulator and Met agonist
-
GC52026
10-oxo-12(Z),15(Z)-Octadecadienoic Acid
An oxylipin gut microbiota metabolite
-
GC42075
2,4-Pyridinedicarboxylic Acid (hydrate)
2,4-Pyridinedicarboxylic Acid (2,4-PDCA) is a compound that structurally mimics 2-oxoglutarate (2-OG, also known as α-ketoglutarate) and chelates zinc, thus affecting a range of enzymes.
-
GC46503
2-(1,8-Naphthyridin-2-yl)phenol
2-(1,8-Naphthyridin-2-yl)phenol is a selective enhancer of STAT1 transcription. 2-(1,8-Naphthyridin-2-yl)phenol can enhance the ability of IFN-γ to inhibit the proliferation of human breast cancer and fibrosarcoma cells.
-
GC46553
2-Nonylquinolin-4(1H)-one
A quinolone alkaloid with diverse biological activities
-
GC52446
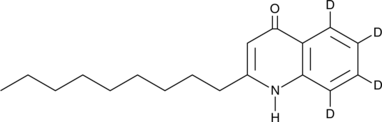
2-Nonylquinolin-4(1H)-one-d4
An internal standard for the quantification of 2-nonylquinolin-4(1H)-one
-
GC52129
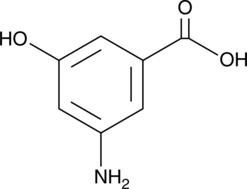
3-Amino-5-hydroxybenzoic Acid
-
GC18194
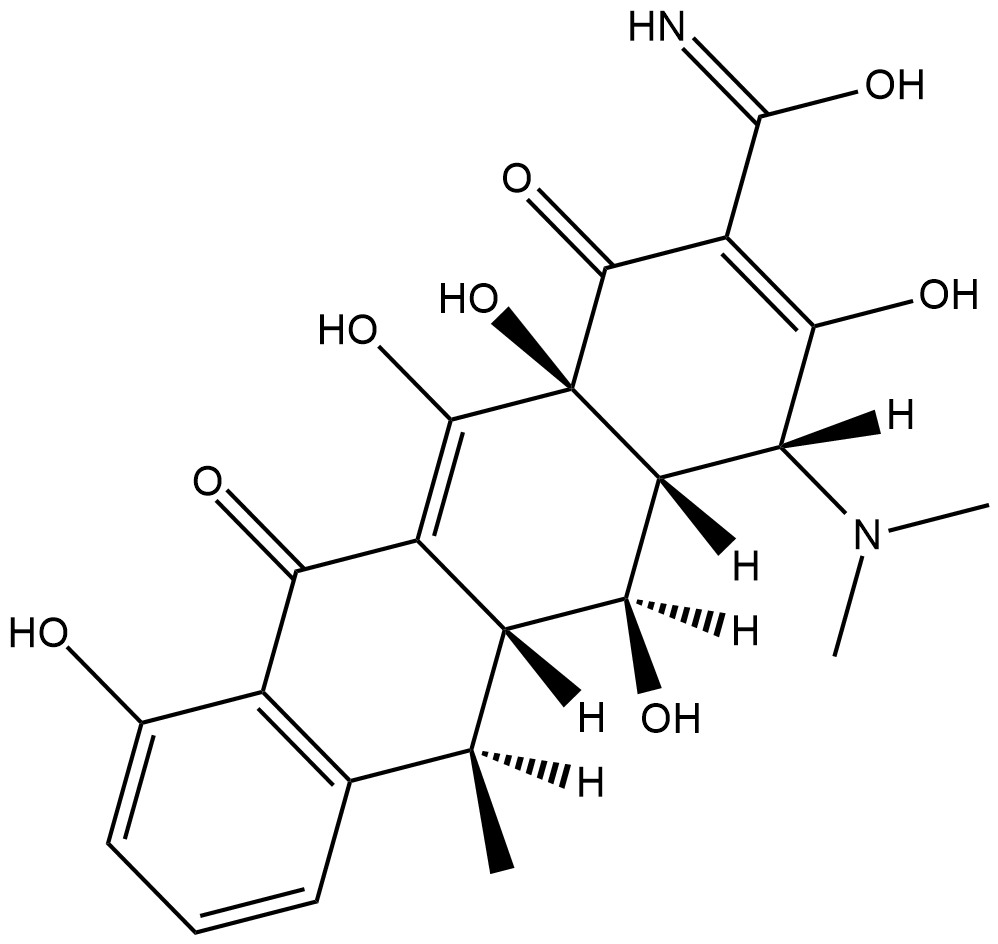
4-Epidoxycycline
4-Epidoxycycline is the 4-epimer hepatic metabolite of the antibiotic doxycycline .
-
GC48824
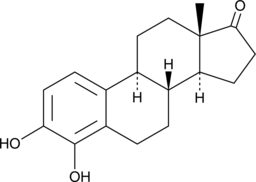
4-hydroxy Estrone
A metabolite of estrone
-
GC46673
4-octynyl Itaconate
4-octynyl Itaconate (ITalk) is a specific bioorthogonal probe for quantitative and site-specific chemoproteomic profiling of Itaconation in living cells.
-
GC52413
5-Aminosalicylic Acid-d7
An internal standard for the quantification of 5-aminosalicylic acid
-
GC45960
9c(i472)
9c(i472) is a potent inhibitor of 15-LOX-1 (15-lipoxygenase-1) with an IC50 value of 0.19 μM.
-
GC42743
AEM1
Cancer cell survival appears partly dependent on antioxidative enzymes, whose expression is regulated by the Keap1-Nrf2 pathway, to quench potentially toxic reactive oxygen species generated by their metastatic transformation.
-
GC16227
Anhydrotetracycline (hydrochloride)
powerful effector in both the tetracycline repressor (TetR) and reverse TetR (revTetR) systems
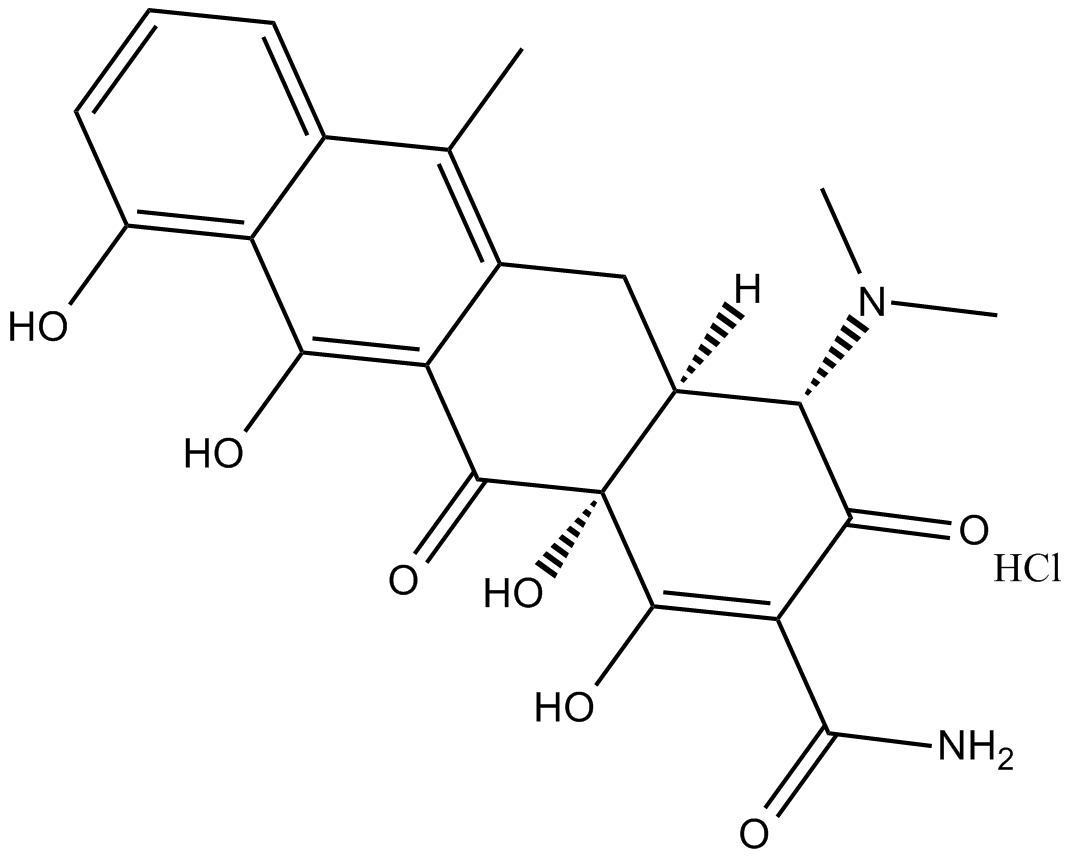
-
GC46862
Apigenin-d5
An internal standard for the quantification of apigenin
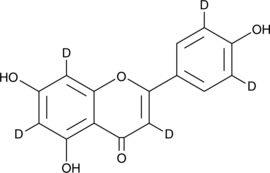
-
GC42880
Avenanthramide-C methyl ester
Avenanthramide-C methyl ester is an inhibitor of NF-κB activation that acts by blocking the phosphorylation of IKK and IκB (IC50 ~ 40 μM).
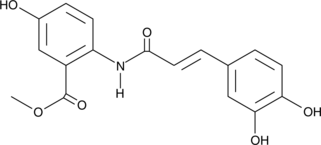
-
GC42925
Berteroin
Berteroin is a sulforaphane analog found in cruciferous vegetables including Chinese cabbage, rucola salad leaves, and mustard oil.
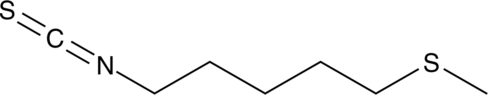
-
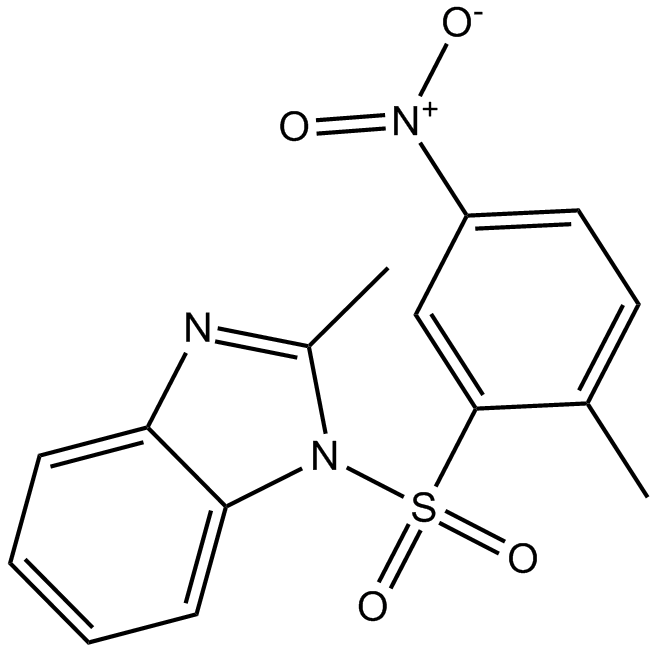
GC13231
BI 6015
HNF4α antagonist
-
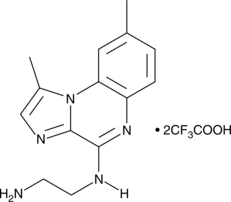
GC42953
BMS 345541 (trifluoroacetate salt)
BMS 345541 is a cell permeable inhibitor of the IκB kinases IKKα and IKKβ (IC50s = 4 and 0.3 μM).
-
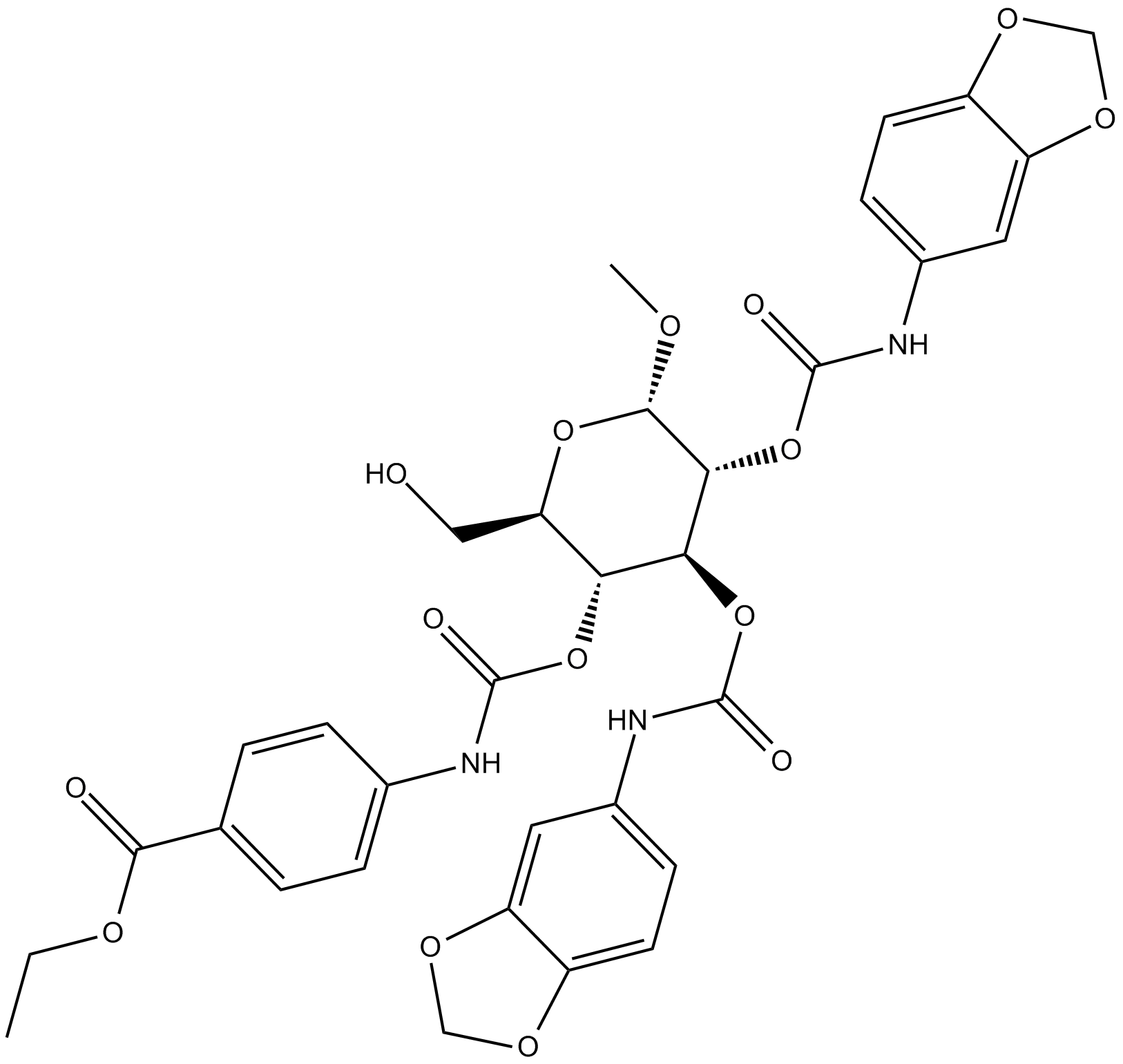
GC10233
BRD 7552
PDX1 inducer
-
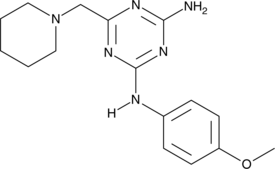
GC42975
BRD32048
ETS variant 1 (ETV1) is a transcription factor oncogene implicated in several cancers where it has been altered by chromosomal translocation, gene amplification, or lineage dysregulation.
-
GC42997
Butyrolactone I
A secondary metabolite from A
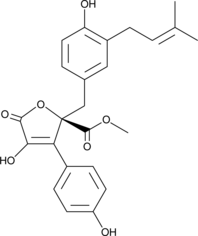
-
GC43110
C8 Galactosylceramide (d18:1/8:0)
C8 Galactosylceramide is a synthetic C8 short-chain derivative of known membrane microdomain-forming sphingolipids.

-
GC43176
CAY10575
CAY10575 (Compound 8) is an IKK2 inhibitor with an IC50 of 0.075 μM.
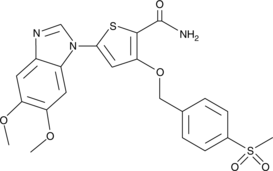
-
GC43189
CAY10681
Inactivation of the tumor suppressor p53 commonly coincides with increased signaling through NF-κB in cancer.
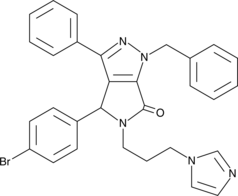
-
GC43190
CAY10682
(±)-Nutlin-3 blocks the interaction of p53 with its negative regulator Mdm2 (IC50 = 90 nM), inducing the expression of p53-regulated genes and blocking the growth of tumor xenografts in vivo.
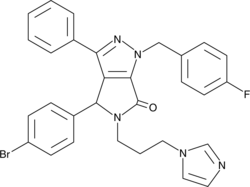
-
GC15474
CBFβ Inhibitor
CBFβ inhibitor
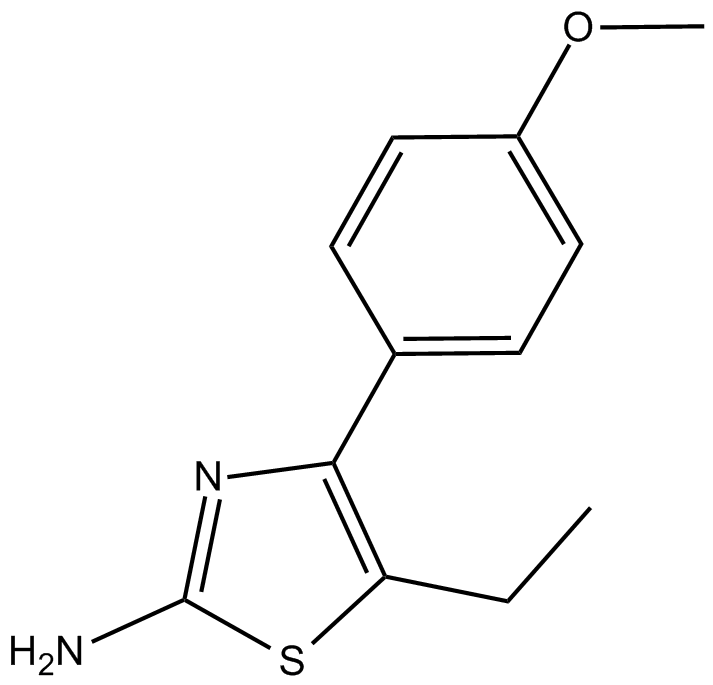
-
GC14266
CCG 203971
Antifibrotic agent
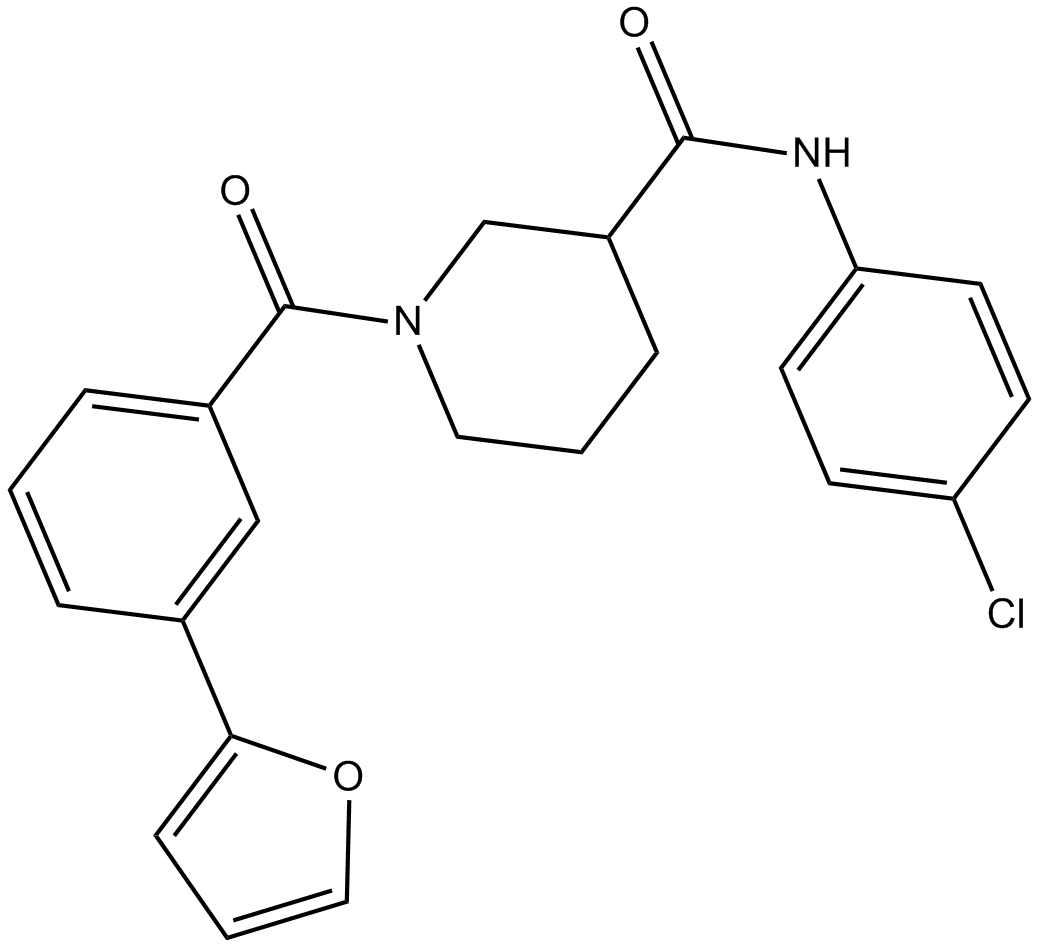
-
GC43212
CCG-232601
CCG-232601 is an inhibitor of the Rho/MRTF/SRF transcriptional pathway and a derivative of CCG-203971.
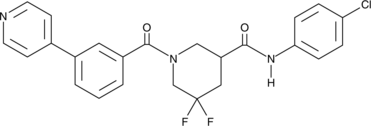
-
GC48982
CD532
An inhibitor of Aurora A kinase activity and the Aurora A-N-Myc protein-protein interaction
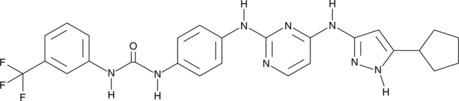
-
GC18392
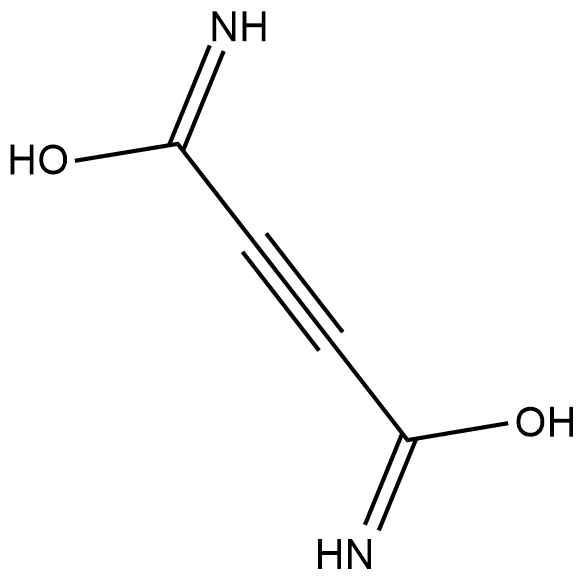
Cellocidin
Cellocidin is an antibiotic originally isolated from S.
-
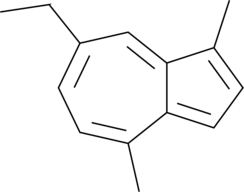
GC52081
Chamazulene
Chamazulene is a terpene that has been found in chamomile (M. recutita) flowers and has anti-inflammatory and antioxidant activities.
-
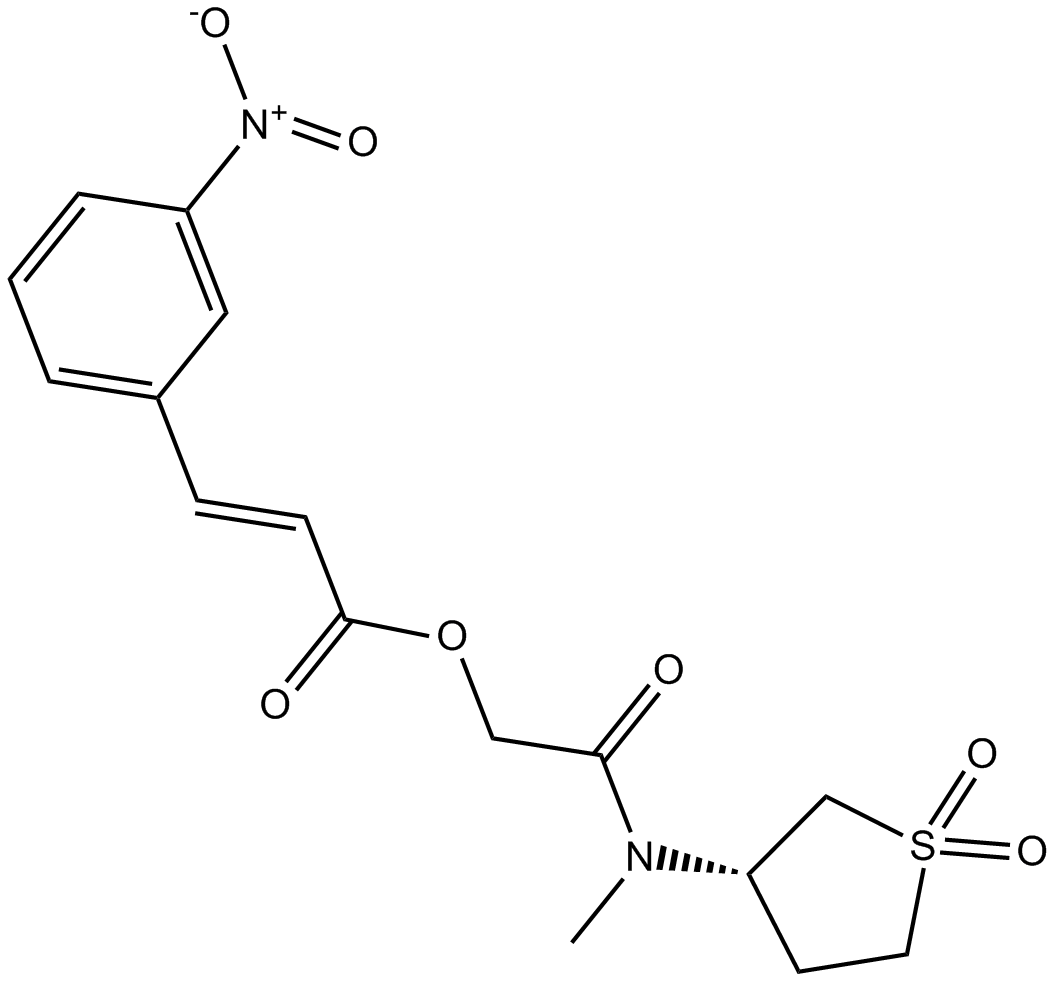
GC14685
CID 5951923
inhibitor of Krüppel-like factor 5 (KLF5) transcription factor
-
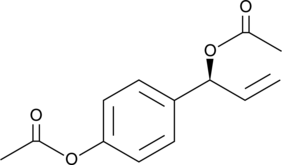
GC43368
D,L-1′-Acetoxychavicol Acetate
D,L-1′-Acetoxychavicol acetate is a natural compound first isolated from the rhizomes of ginger-like plants.
-
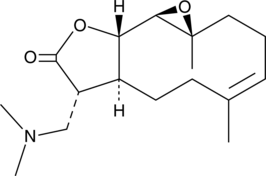
GC52194
Dimethylamino Parthenolide
-
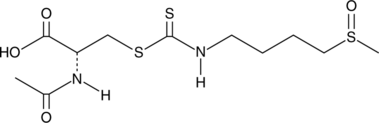
GC43493
DL-Sulforaphane N-acetyl-L-cysteine
Nrf2 activation of the antioxidant response element (ARE) is central to cytoprotective gene expression against oxidative and/or electrophilic stress.
-
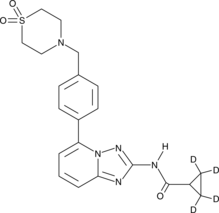
GC46148
Filgotinib-d4
An internal standard for the quantification of filgotinib
-
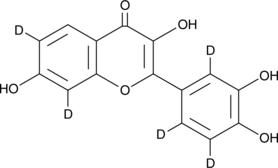
GC49344
Fisetin-d5
An internal standard for the quantification of fisetin
-
GC18652
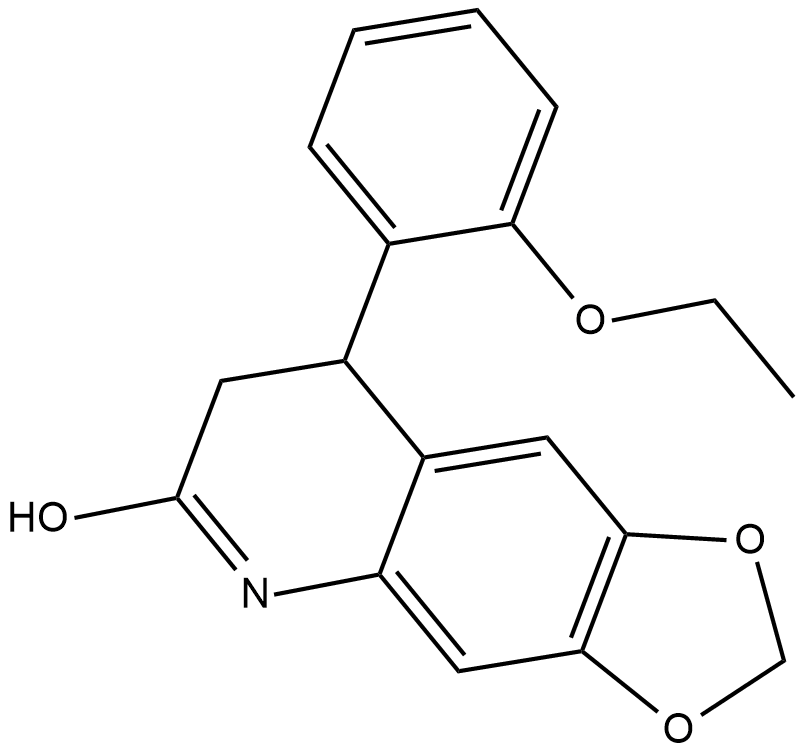
FQI 1
An inhibitor of Late SV40 Factor
-
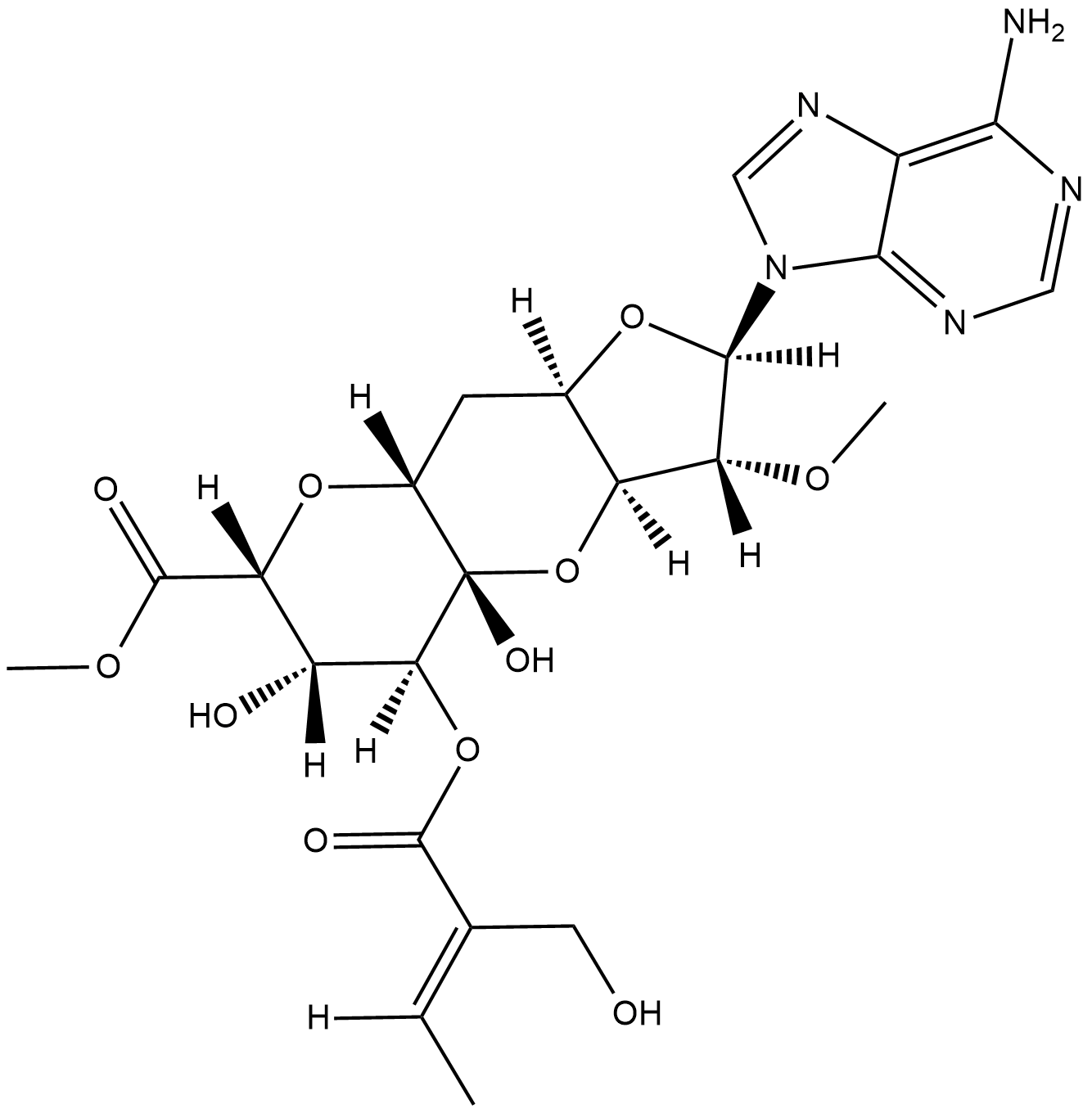
GC18796
Herbicidin A
Herbicidin A is an adenine nucleoside antibiotic originally isolated from S.
-
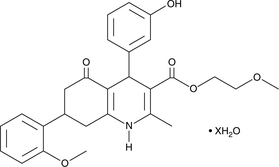
GC47435
HPI-1 (hydrate)
A Hedgehog pathway inhibitor
-
GC49742
Hsf1 Monoclonal Antibody (Clone 10H8)
For immunochemical analysis of Hsf1

-
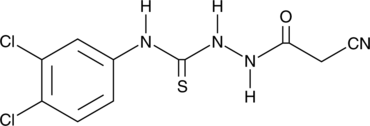
GC43893
iKIX1
An inhibitor of Mediator/Pdr1 interactions
-
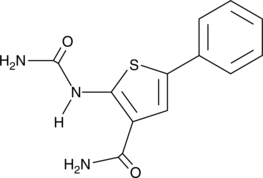
GC43894
IKK2 Inhibitor VI
Inhibitor of NF-κB kinase 2 (IKK2, also known as IKKβ) acts as part of an IKK complex in the canonical NF-κB pathway, phosphorylating inhibitors of NF-κB (IκBs) to initiate signaling.
-
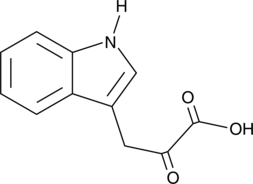
GC47456
Indole-3-pyruvic Acid
Indole-3-pyruvic Acid, a keto analogue of tryptophan, is an orally active AHR agonist.
-
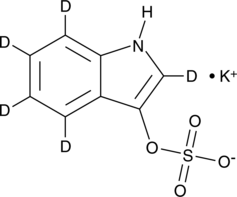
GC49290
Indoxyl Sulfate-d5 (potassium salt)
An internal standard for the quantification of indoxyl sulfate
-
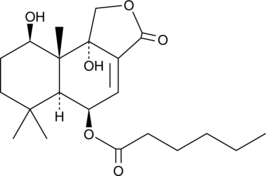
GC48618
Isonanangenine B
A drimane sesquiterpene lactone
-
GC11197
KL 001
cryptochrome protein (CRY) stabilizer
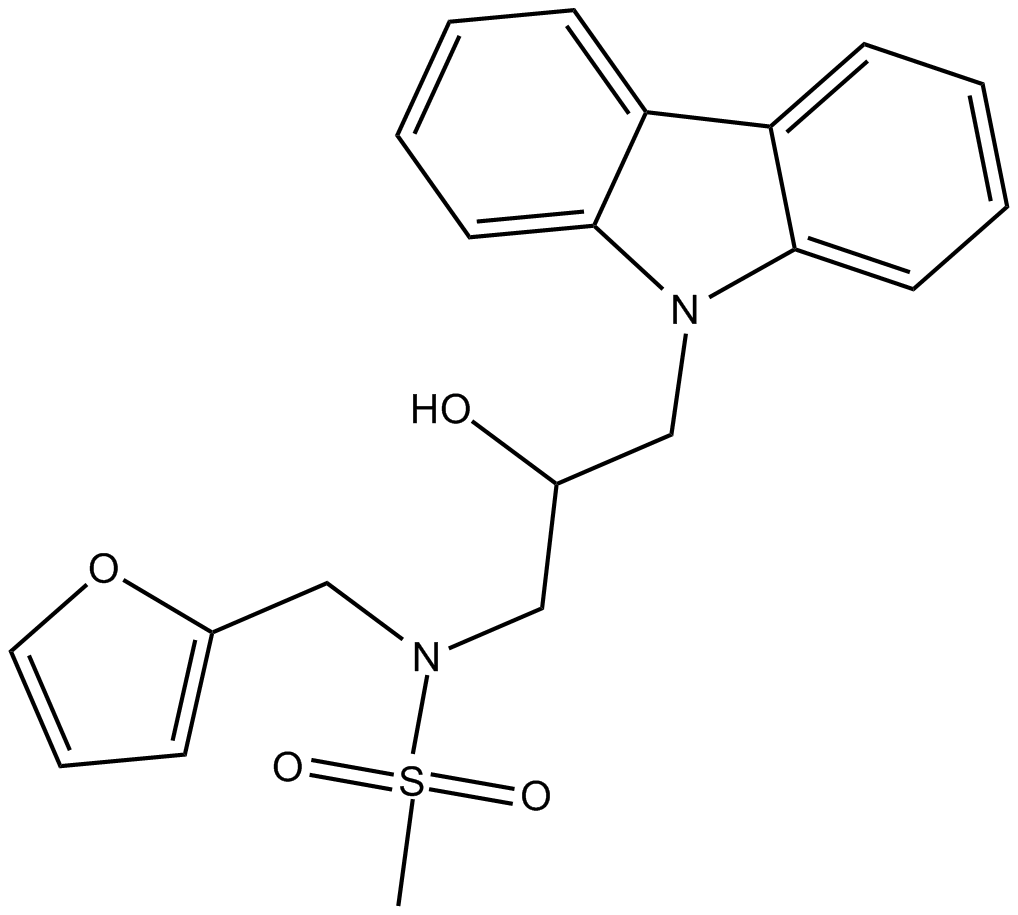
-
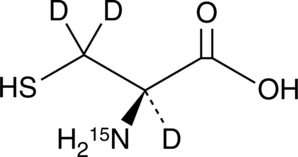
GC52283
L-Cysteine-15N-d3
An internal standard for the quantification of L-cysteine
-
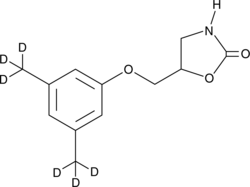
GC48907
Metaxalone-d6
An internal standard for the quantification of metaxalone
-
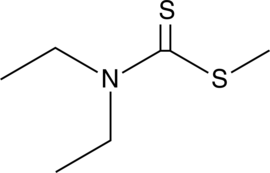
GC49241
Methyl Diethyldithiocarbamate
An active metabolite of disulfiram
-
GC44216
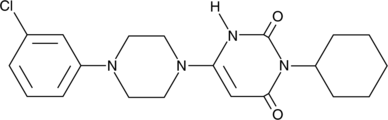
ML-180
ML-180 is an inverse agonist of liver receptor homolog-1 (LRH-1, IC50 = 3.7 μM) with maximum efficacy of 64% repression.
-
GC11307
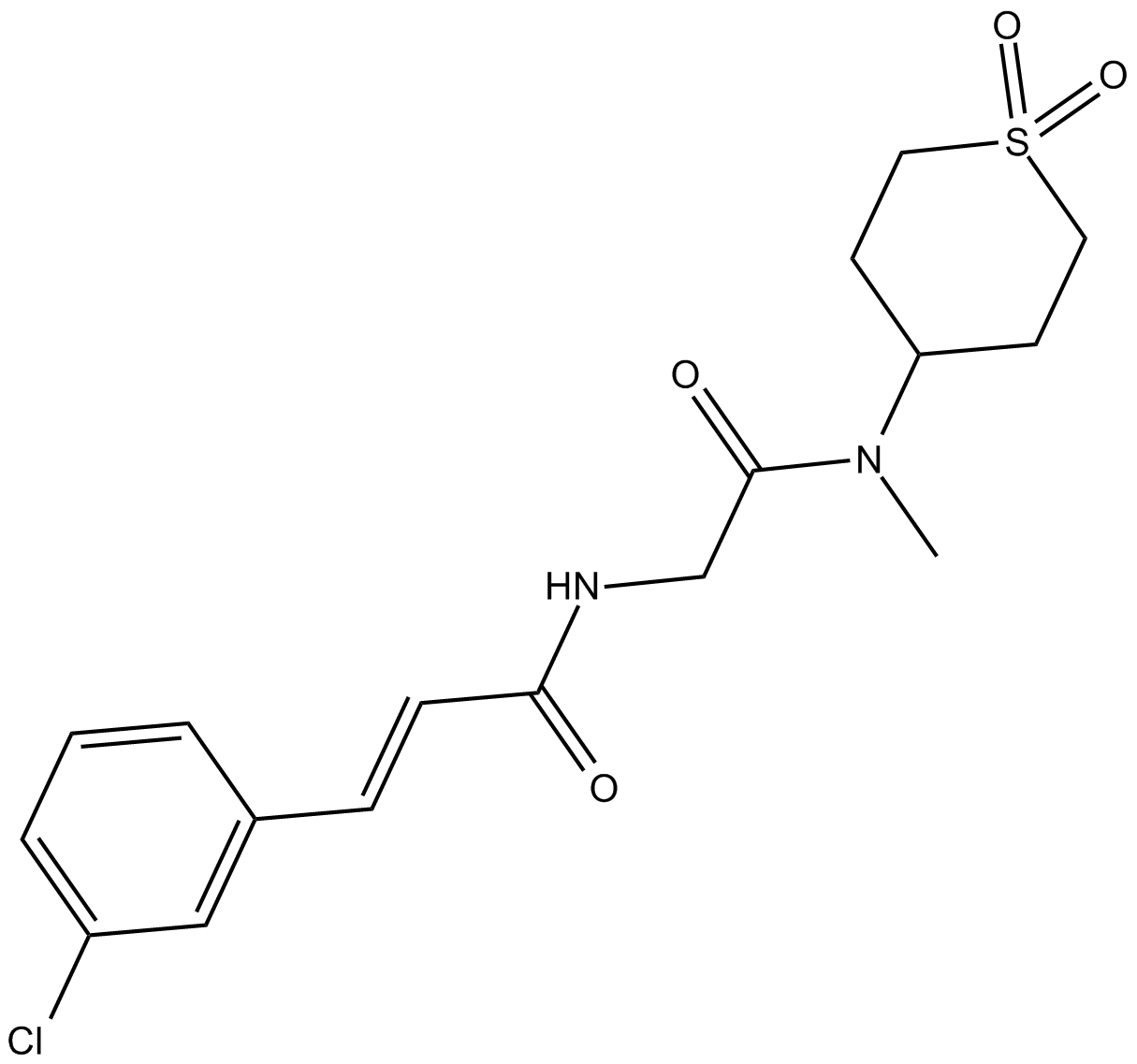
ML-264
Krüppel-like factor 5 (KLF5) inhibitor
-
GC14832
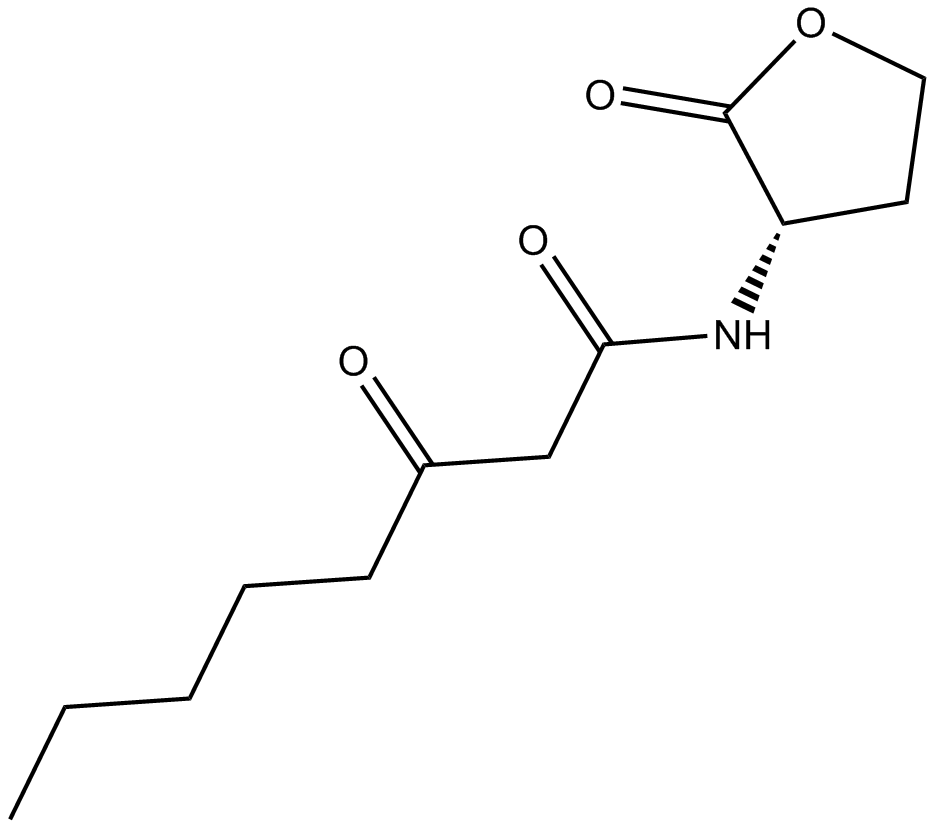
N-3-oxo-octanoyl-L-Homoserine lactone
A bacterial quorum sensing signal molecule
-
GC44459
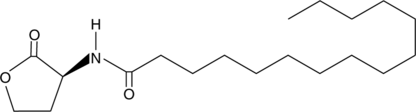
N-pentadecanoyl-L-Homoserine lactone
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.
-
GC44388
NF-κB Control
NF-κB inhibitor is a synthetic peptide corresponding to the nuclear localization sequence (NLS) of NF-κB p105 subunit (also known as p50) appended to a hydrophobic sequence to facilitate import into living cells.
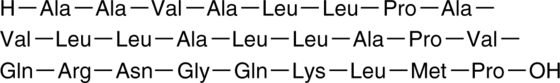
-
GC48985
NF-κB Inhibitor (trifluoroacetate salt)
A cell-permeable peptide that blocks nuclear import of NF-κB

-
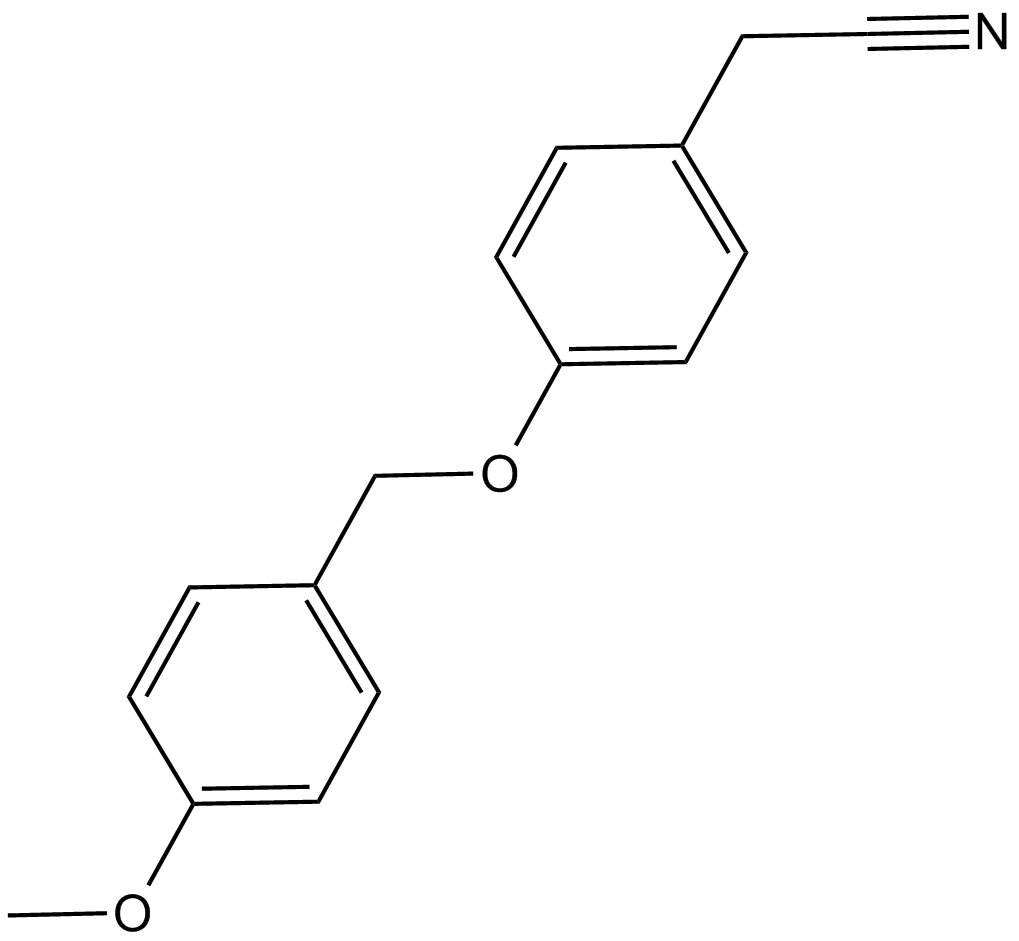
GC12499
O4I1
potent Oct3/4 inducer
-
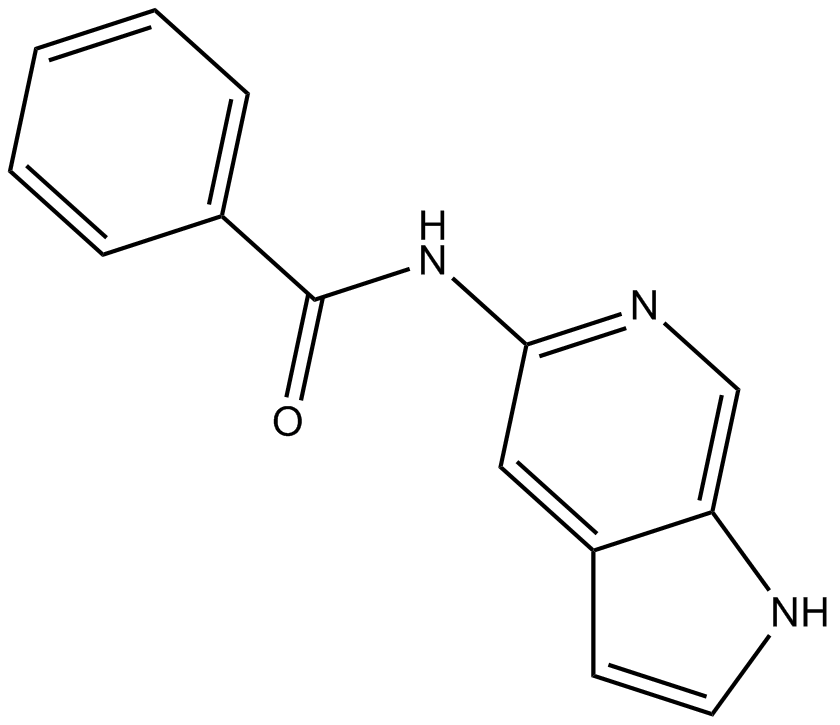
GC10151
OAC1
Oct4 activator
-
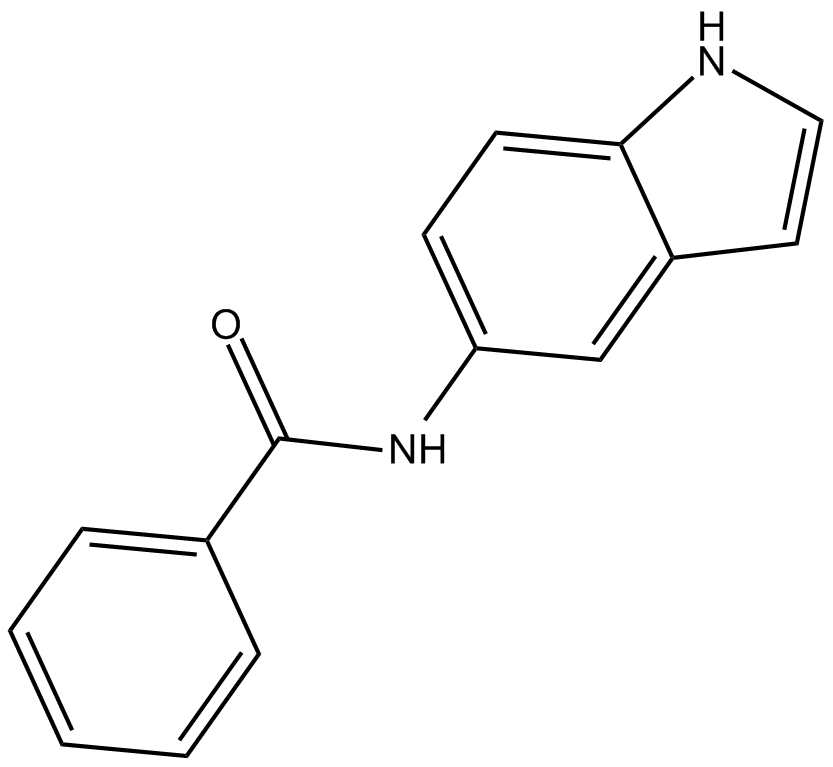
GC17327
OAC2
Oct4 activator
-
GC10724
OAC3
Oct4 activator
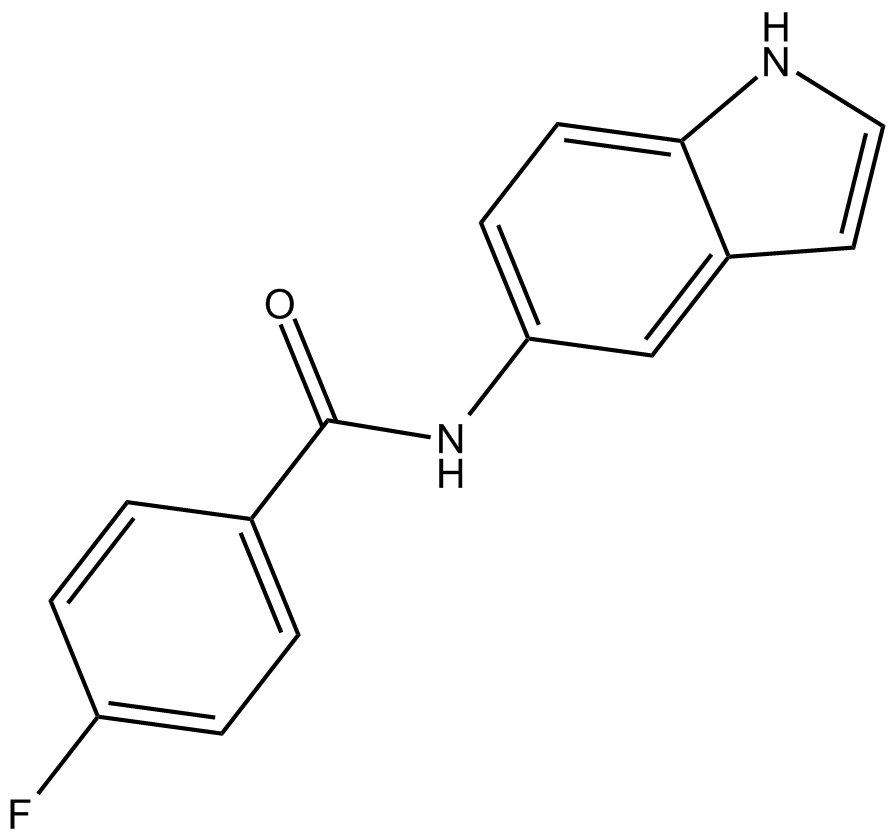
-
GC44652
PK7242 (maleate)
The protein p53, often called the 'guardian of the genome,' is a transcription factor that is activated in response to cellular stress (low oxygen levels, heat shock, DNA damage, etc.) and acts to prevent further proliferation of the stressed cell by promoting cell cycle arrest or apoptosis.
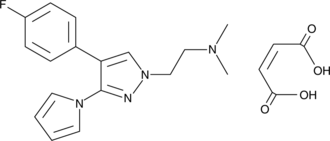
-
GC49860
Pyropheophorbide a methyl ester
Pyropheophorbide a methyl ester (Pyropheophorbide-a methyl ester), a chlorophyll-a derivative, is a potent photosensitizer that can be used in photodynamic therapy (PDT) of cancer. Pyropheophorbide a methyl ester has photodynamic activity and can induce apoptosis and inhibit tumor growth.

-
GC50256
RG 102240
Gene switch ligand for use in inducible gene expression systems
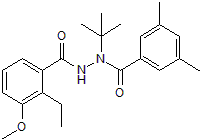
-
GC12727
Ro 5-3335
Core binding factor (CBF) inhibitor
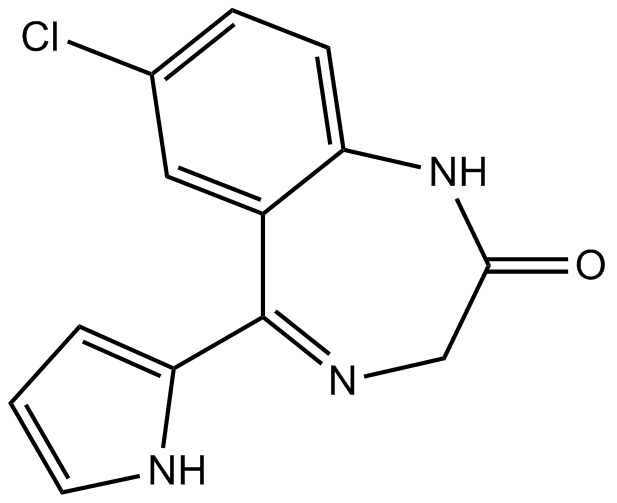
-
GC18624
Roslin-2
Roslin-2 (Benzylhexamethylenetetramine bromide) is a p53 reactivator with anticancer effects. Roslin-2 binds FAK, disrupts the binding of FAK and p53.
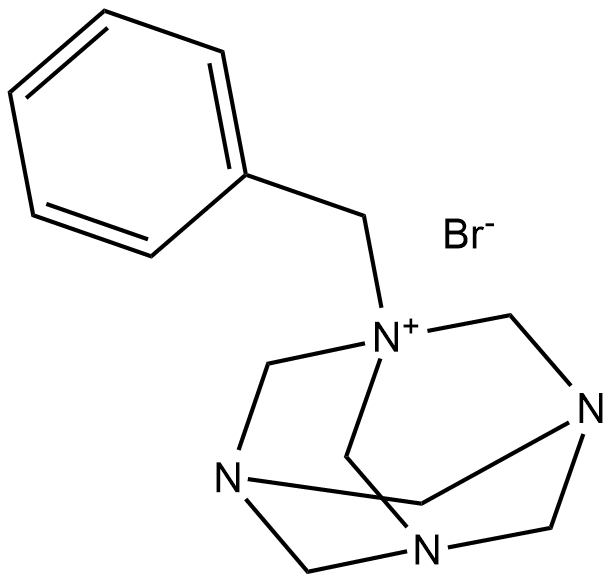
-
GC44888
SI-2
SI-2 is an inhibitor of steroid receptor coactivator 3 (SRC-3).
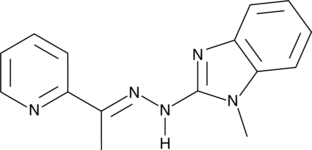
-
GC49002
Sinigrin (hydrate)
A glucosinolate with diverse biological activities
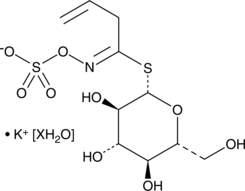
-
GC49049
SMU127
An agonist of TLR1/2
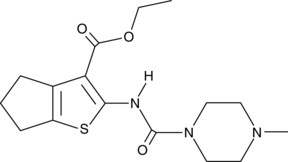
-
GC44943
sPLA2 Inhibitor
sPLA2 Inhibitor, a D-tyrosine derivative, is an orally active, potent secretory phospholipase A2 (sPLA2) inhibitor with an IC50 of 29 nM for human nonpancreatic secretory PLA2 isoform IIa (hnpsPLA2-IIa).
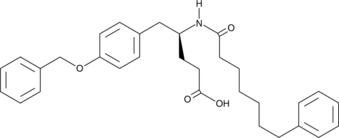
-
GC48098
SR 12343
An IKKβ NBD mimetic
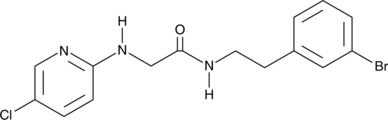
-
GC48099
SR 12460
An IKKβ NBD mimetic
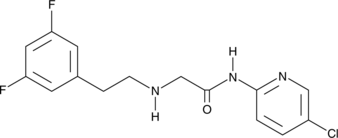
-
GC17839
Stauprimide
Small molecule that primes embryonic stem cells (ESCs) for differentiation
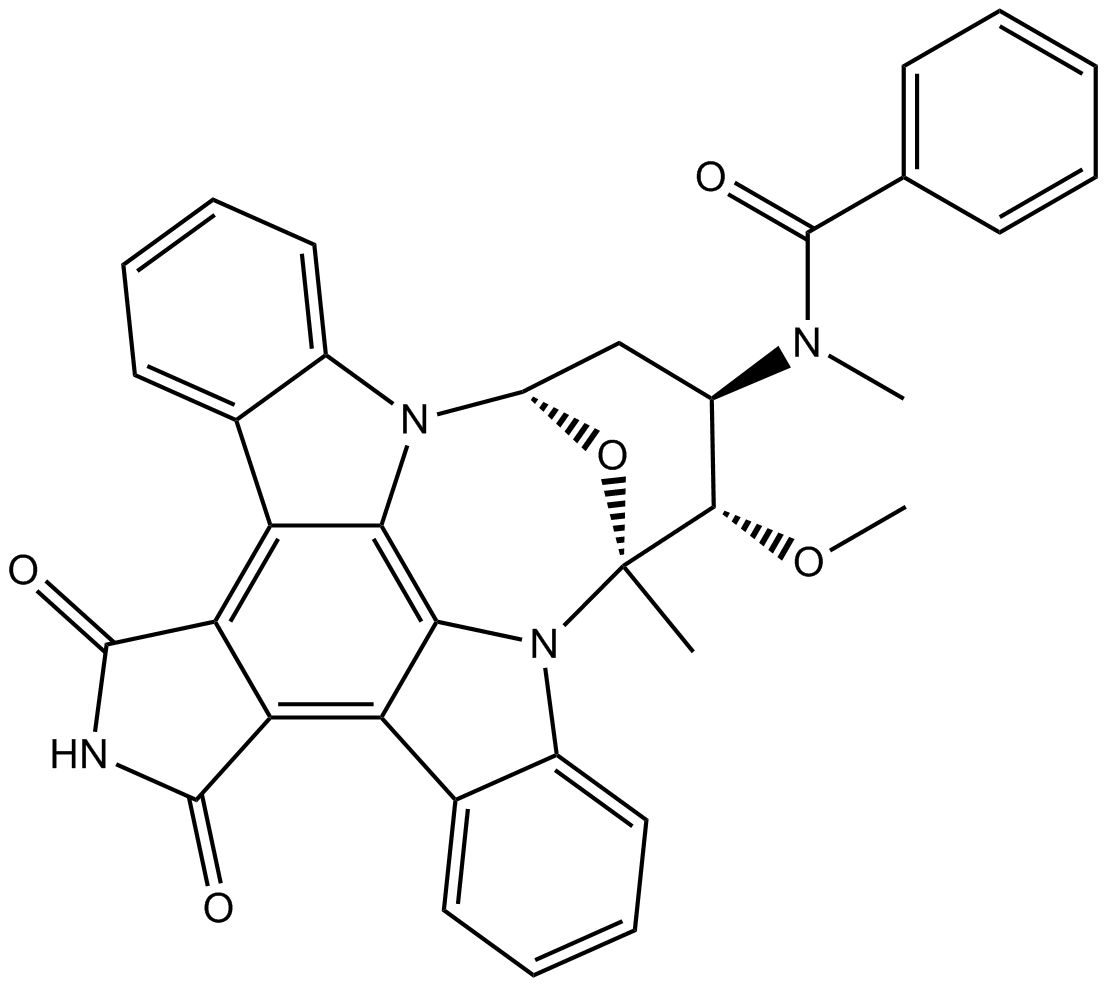
-
GC16165
T-5224
T-5224 is a non-peptidic small molecule AP-1 inhibitor

-
GC44984
TAF 10 Peptide
TAF10 is one of many protein factors or coactivators associated with RNA polymerase II activity.

-
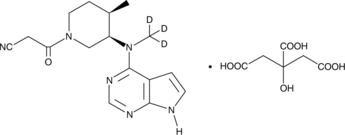
GC49692
Tofacitinib-d3 (citrate)
An internal standard for the quantification of tofacitinib
-
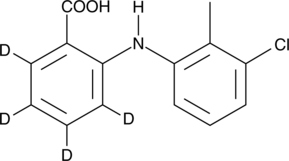
GC45965
Tolfenamic Acid-d4
An internal standard for the quantification of tolfenamic acid
-
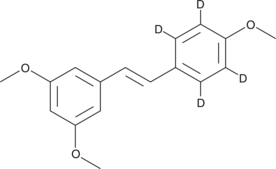
GC48195
trans-trismethoxy Resveratrol-d4
An internal standard for the quantification of trans-trismethoxy resveratrol
-
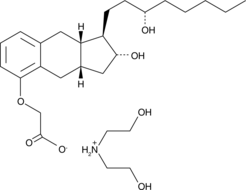
GC48202
Treprostinil (diethanolamine salt)
Treprostinil (UT-15C) diethanolamine is a potent EP2, DP1 and IP agonist with Ki values of 3.6, 4.4, 32.1, 212, 826, 2505 and 4680 nM for EP2, DP1, IP, EP1, EP4, EP3 and FP, respectively.
-
GC49835
Tumulosic Acid
Tumulosic Acid, a triterpenoid, inhibits KLK5 protease activity (IC50= 14.84 μM).
-
GC52180
WAY-169916
WAY-169916 is a pathway-selective ligand of ER (estrogen receptor) that acts by inhibiting NF-kB transcriptional activity.