PI3K/Akt/mTOR Signaling
- Akt(129)
- AMPK(75)
- CK2(11)
- DNA-PK(25)
- GSK-3(76)
- MELK(7)
- mTOR(118)
- PI3K(227)
- PI4K(15)
- PDK-1(18)
- PIKfyve(4)
- PKB(1)
- S6 Kinase(12)
Products for PI3K/Akt/mTOR Signaling
- Cat.No. Product Name Information
-
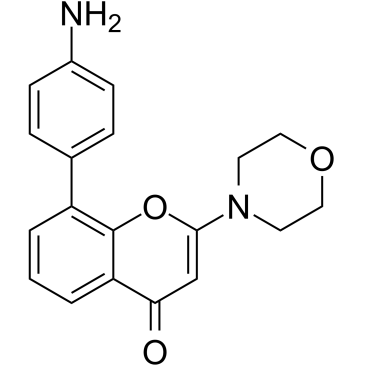
GC36426
LAS191954
LAS191954 is a potent, selective and orally active PI3Kδ inhibitor for inflammatory diseases treatment, with an IC50 of 2.6 nM.
-
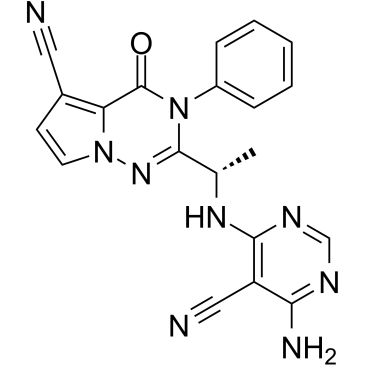
GC19471
Leniolisib
A potent and selective PI3Kδ inhibitor
-
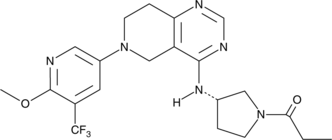
GC36447
Licochalcone E
Licochalcone E, a flavonoid compound isolated from Glycyrrhiza inflate, inhibits NF-κB and AP-1 transcriptional activity through the inhibition of AKT and MAPK activation.
-
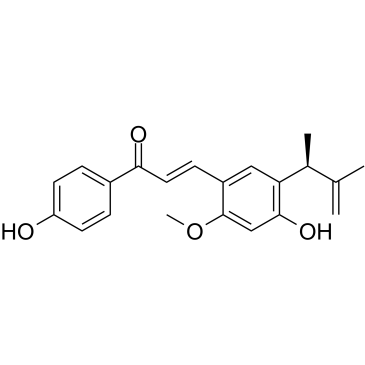
GC36456
Licoricidin
Licoricidin (LCD) is isolated from Glycyrrhiza uralensis Fisch, possesses anti-cancer activities.
-
GC16039
LJH685
RSK (p90 ribosomal S6 kinase) inhibitor
-
GC16601
LJI308
pan-RSK inhibitor
-
GC30770
LM22B-10
An activator of TrkB and TrkC
-
GC32510
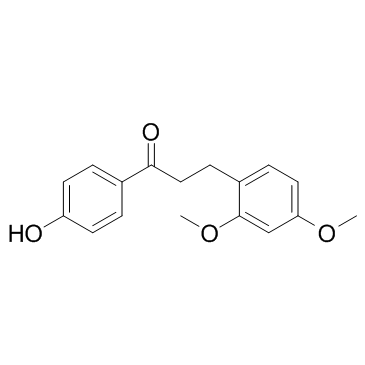
Loureirin A
Loureirin A is a flavonoid extracted from Dragon's Blood, can inhibit Akt phosphorylation, and has antiplatelet activity.
-
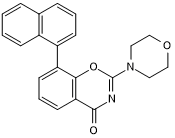
GC50276
LTURM 36
PI 3-kinase δ inhibitor
-
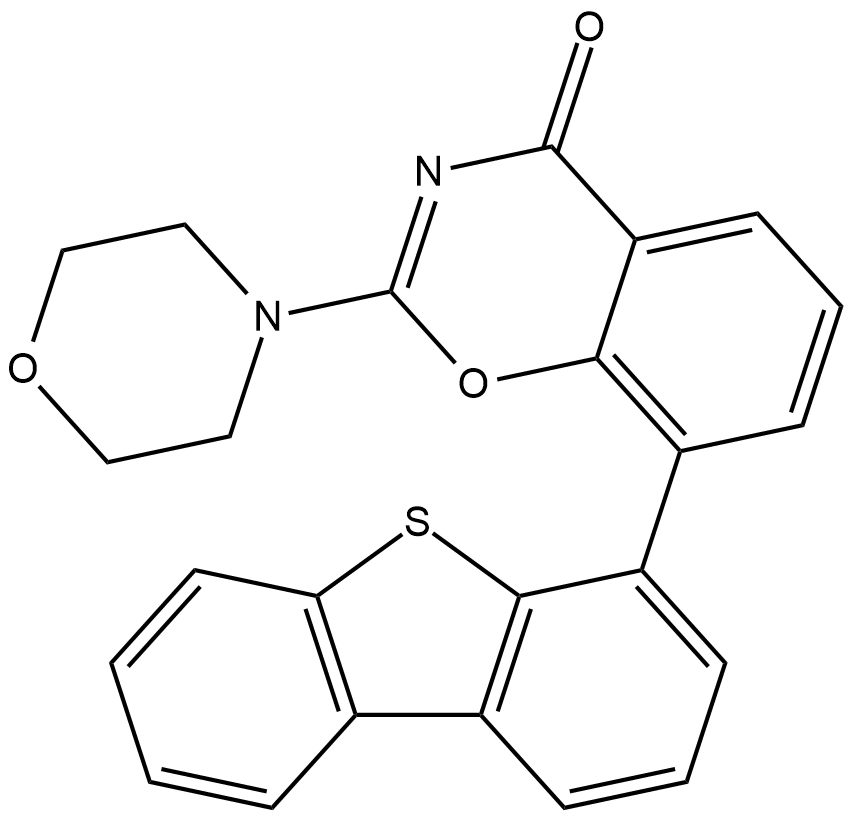
GC19227
LTURM34
LTURM34 is a specific DNA-PK inhibitor with an IC50 of 0.034 uM.
-
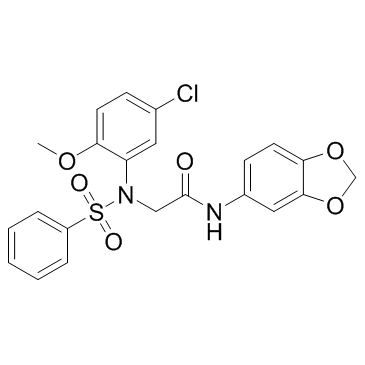
GC30884
LX2343
An inhibitor of BACE1 and PI3K
-
GC12045
LY 303511
An inhibitor of cell proliferation
-
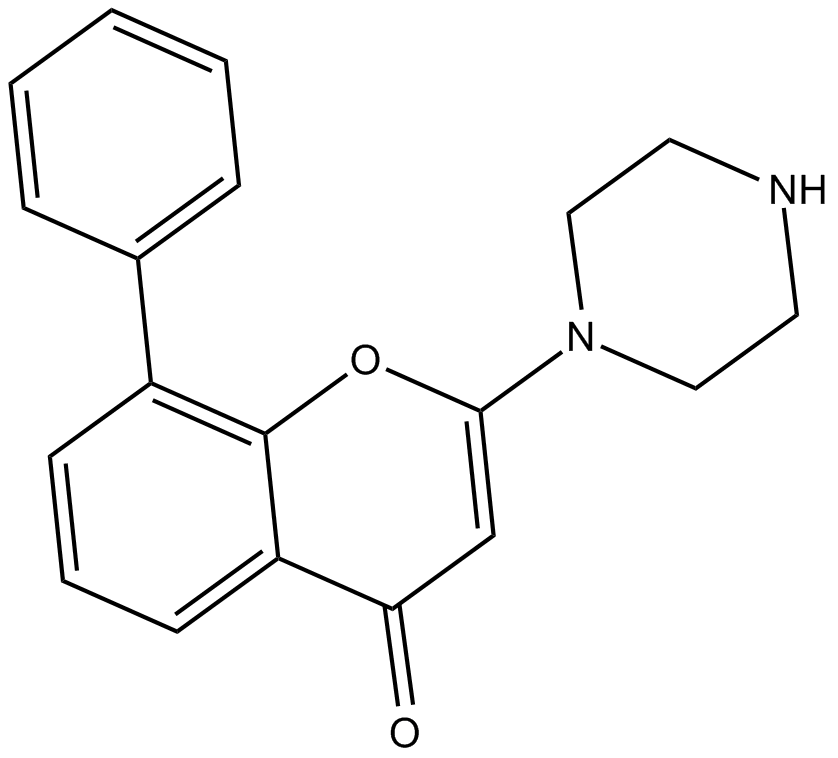
GC17187
LY2090314
A potent and selective inhibitor of GSK3
-
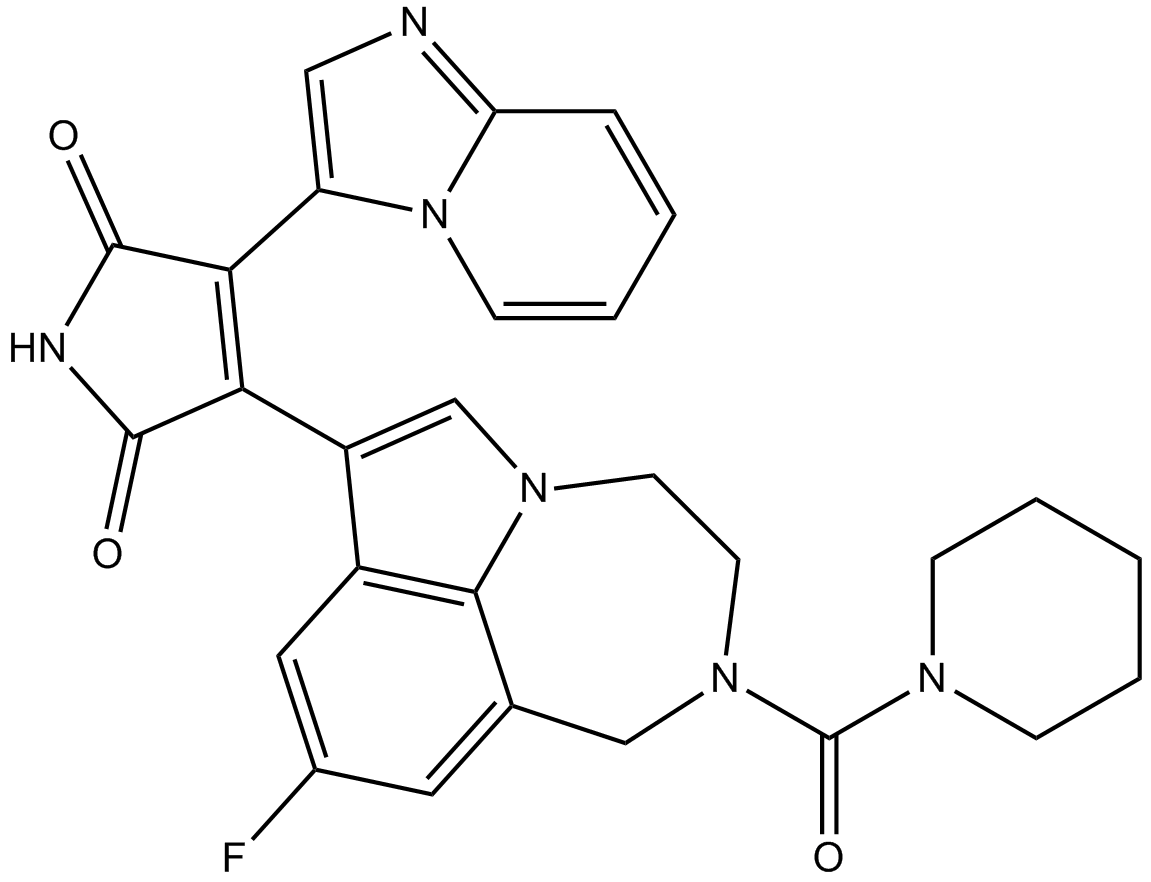
GC12089
LY2584702
LY2584702 is a selective ATP competitive inhibitor of p70S6K with an IC50 of 4 nM. In S6K1 enzyme assay, the IC50 of LY-2584702 is 2 nM.
-
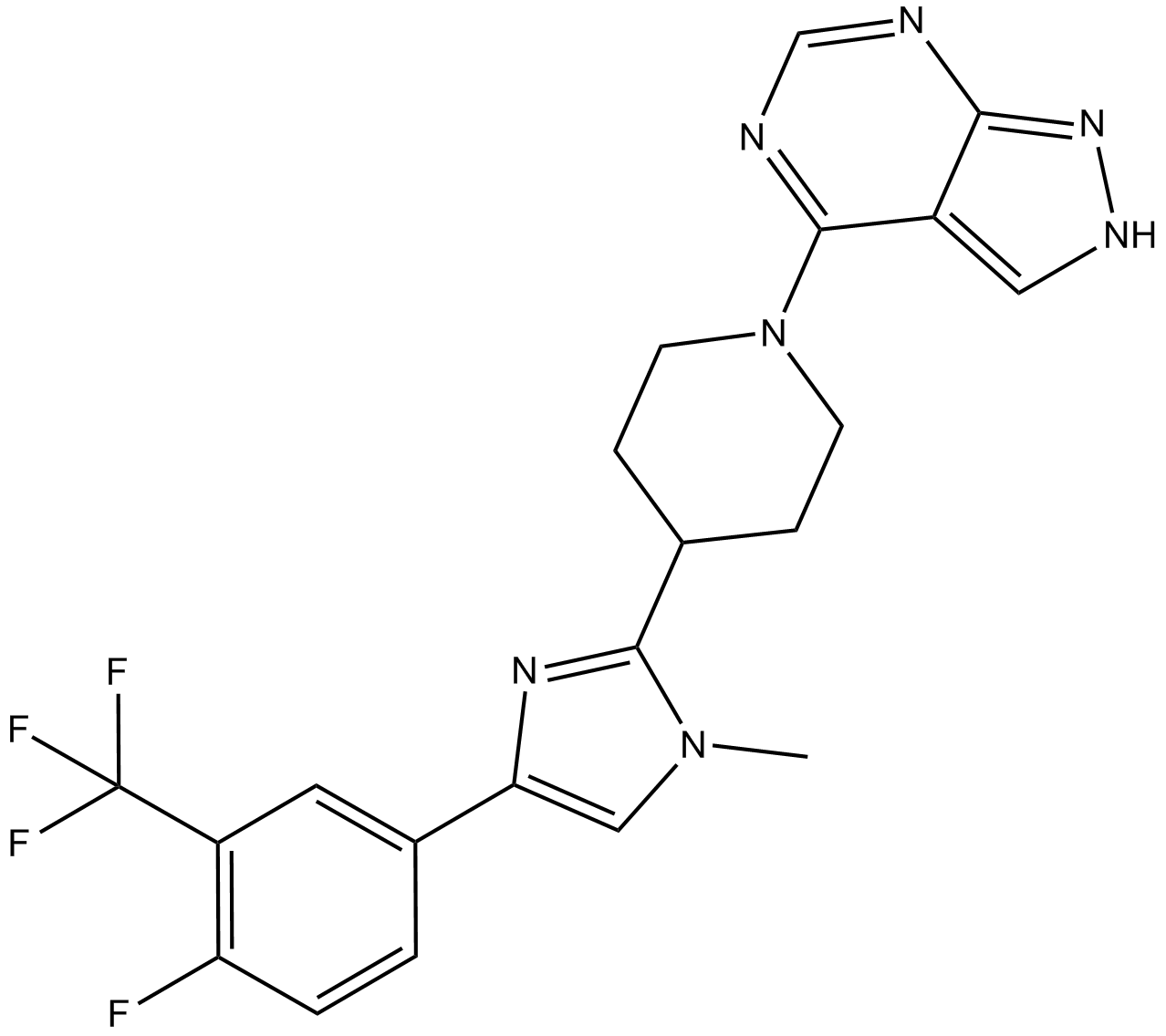
GC14371
LY3023414
LY3023414 (LY3023414) potently and selectively inhibits class I PI3K isoforms, DNA-PK, and mTORC1/2 with IC50s of 6.07 nM, 77.6 nM, 38 nM, 23.8 nM, 4.24 nM and 165 nM for PI3Kα, PI3Kβ, PI3Kδ, PI3Kγ, DNA-PK and mTOR, respectively. LY3023414 potently inhibits mTORC1/2 at low nanomolar concentrations.
-
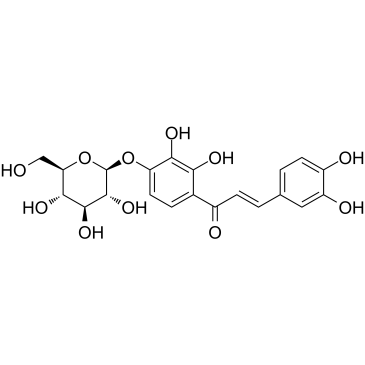
GC61029
Marein
A glucoside chalcone with diverse biological activities
-
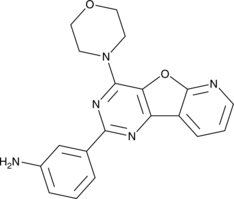
GC48707
MDK34597
MDK34597 (compound 35) is a potent PI3K p110α inhibitor with an pIC50 of 6.85.
-
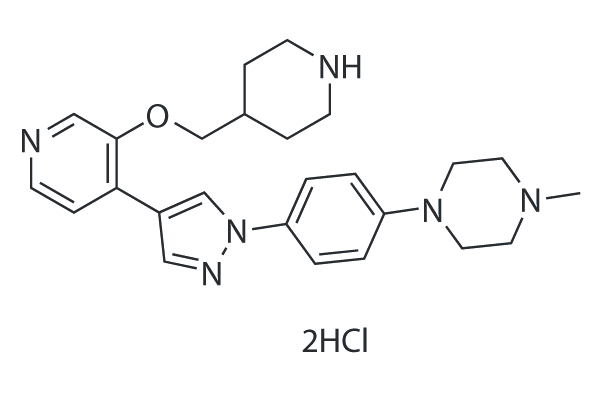
GC25626
MELK-8a Dihydrochloride
MELK-8a Dihydrochloride is a novel inhibitor of maternal embryonic leucine zipper kinase (MELK) with an IC50 of 2 nM.
-
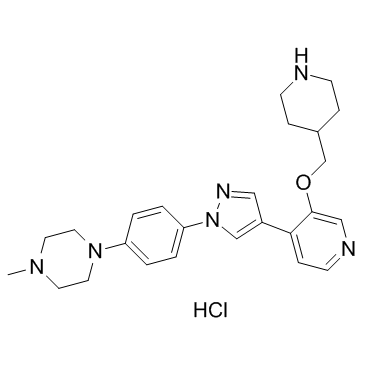
GC32736
MELK-8a hydrochloride
MELK-8a hydrochloride is a novel maternal embryonic leucine zipper kinase (MELK) inhibitor with an IC50 of 2 nM.
-
GC66065
MELK-IN-1
MELK-IN-1 is a potent inhibitor of maternal embryonic leucine zipper kinase (MELK) with an IC50 and a Ki of 3 nM and 0.39 nM, respectively.
-
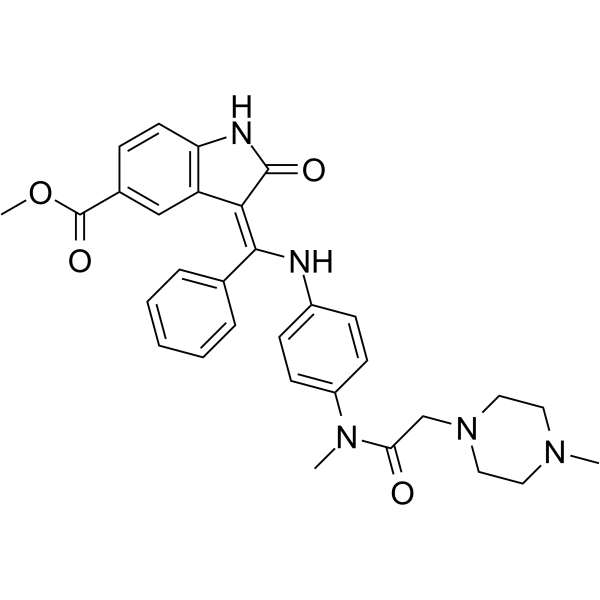
GC61045
Metformin D6 hydrochloride
An internal standard for the quantification of metformin
-
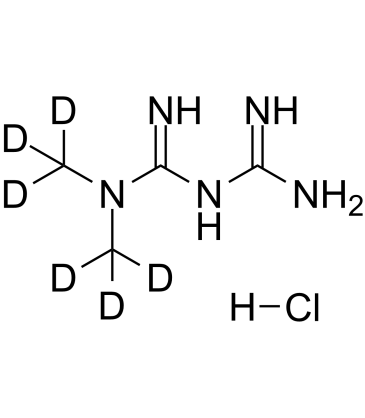
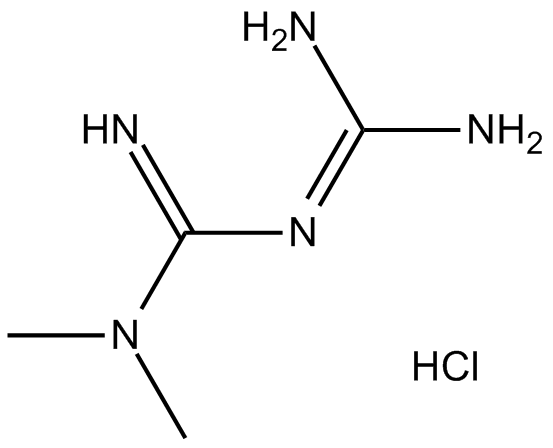
GC17443
Metformin HCl
Metformin HCl (1,1-Dimethylbiguanide hydrochloride) inhibits the mitochondrial respiratory chain in the liver, leading to activation of AMPK, enhancing insulin sensitivity for type 2 diabetes research.
-
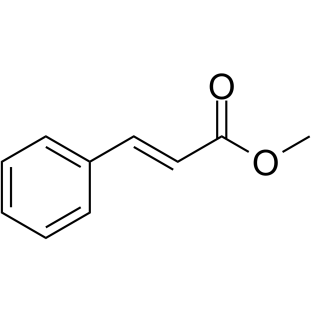
GC38815
Methyl cinnamate
Methyl cinnamate (Methyl 3-phenylpropenoate), an active component of Zanthoxylum armatum, is a widely used natural flavor compound.
-
GC64402
MHY-1685
MHY-1685, a novel mammalian target of rapamycin (mTOR) inhibitor, provides opportunities to improve hCSC-based myocardial regeneration.
-
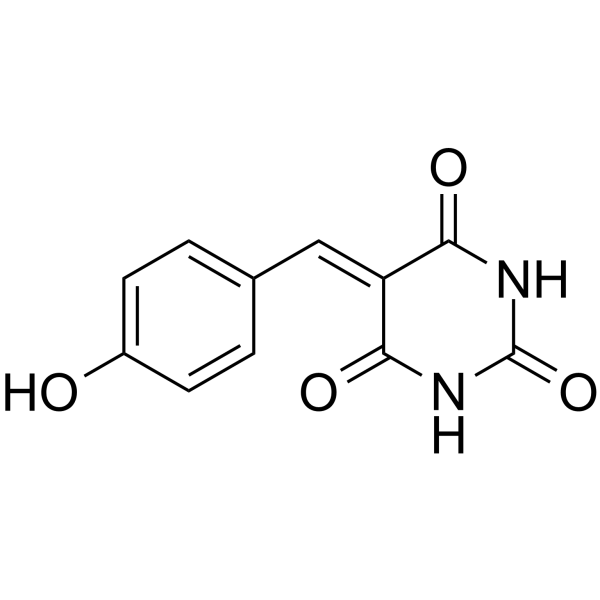
GC13790
MHY1485
MHY1485 is a potent activator of mTOR, which inhibits autophagy and the fusion between autophagosomes and lysosomes.
-
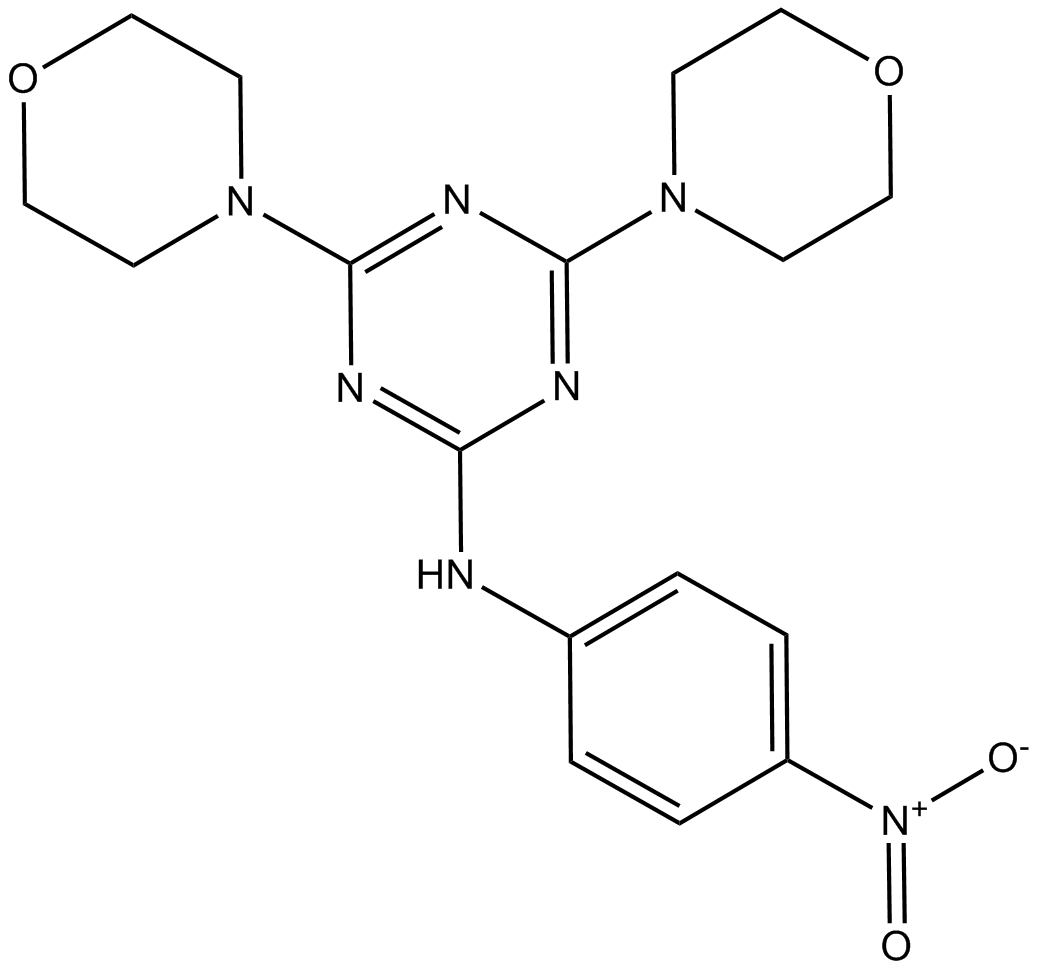
GC10811
Miltefosine
PI3K/Akt inhibitor
-
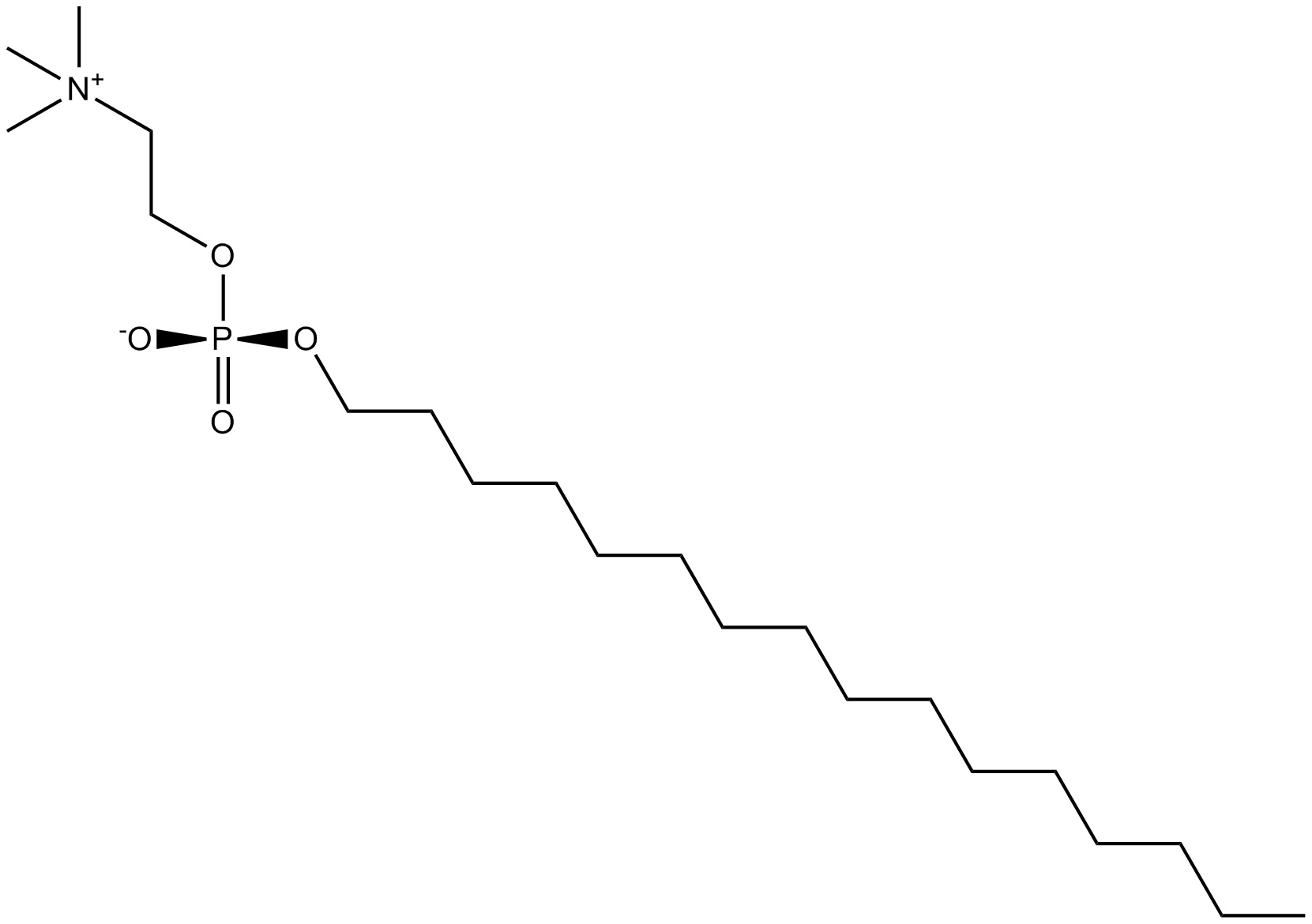
GC25638
Miransertib (ARQ 092) HCl
Miransertib (ARQ 092) HCl is a novel, orally bioavailable and selective AKT pathway inhibitor exhibiting a manageable safety profile among patients with advanced solid tumors.
-
GC16304
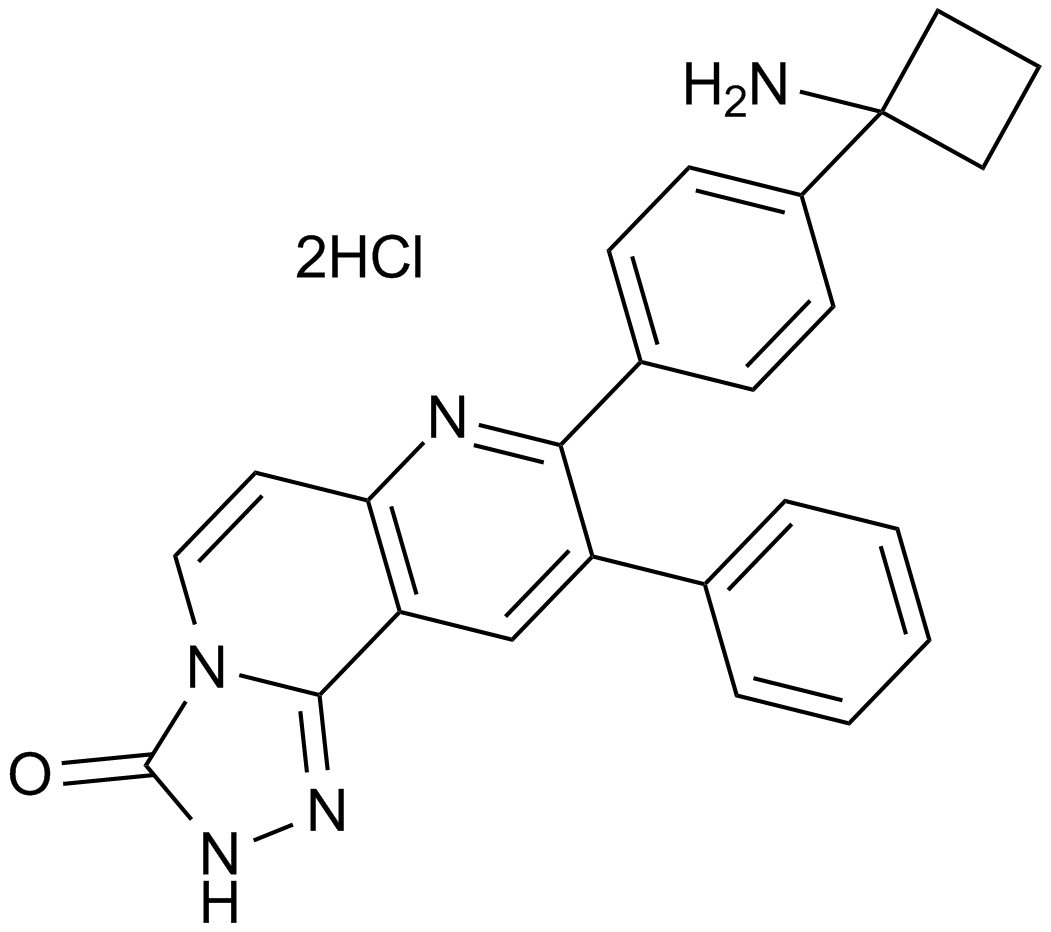
MK-2206 dihydrochloride
An allosteric Akt inhibitor
-
GC31361
MK-3903
MK-3903 is a potent and selective AMP-activated protein kinase (AMPK) activator with an EC50 of 8 nM.
-
GC31470
MK8722
MK8722 is a potent and systemic pan-AMPK activator.
-
GC65143
MKC-1
MKC-1 (Ro-31-7453) is an orally active and potent cell cycle inhibitor with broad antitumor activity. MKC-1 inhibits the Akt/mTOR pathway. MKC-1 arrests cellular mitosis and induces cell apoptosis by binding to a number of different cellular proteins including tubulin and members of the importin β family.
-
GC49440
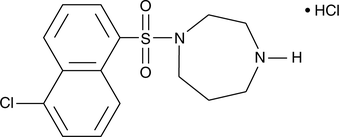
ML-9
A PKB/Akt inhibitor
-
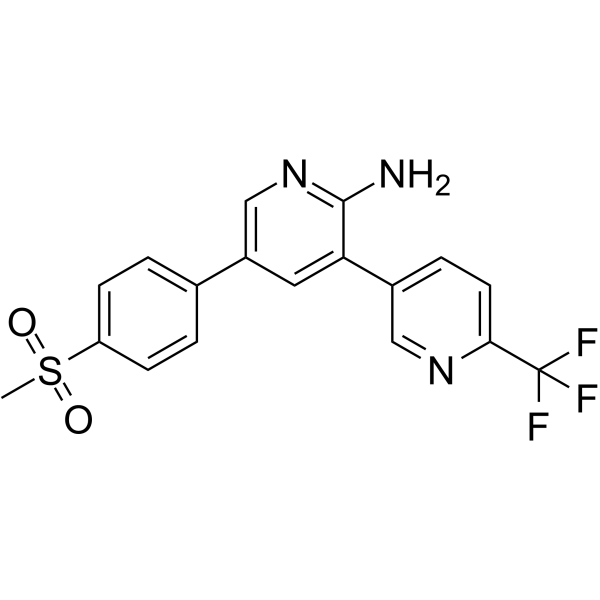
GC62109
MMV390048
MMV390048 is a representative of a new chemical class of Plasmodium PI4K inhibitor (Kdapp=0.3 ?M).
-
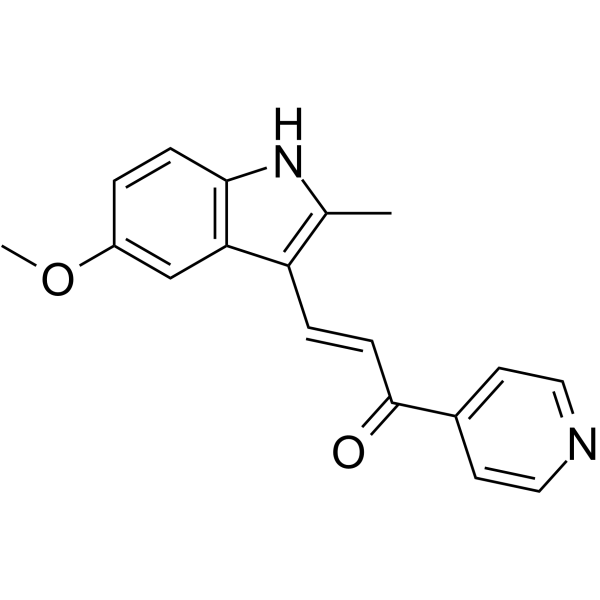
GC66013
MOMIPP
MOMIPP, a macropinocytosis inducer, is a PIKfyve inhibitor. MOMIPP penetrates the blood-brain barrier (BBB).
-
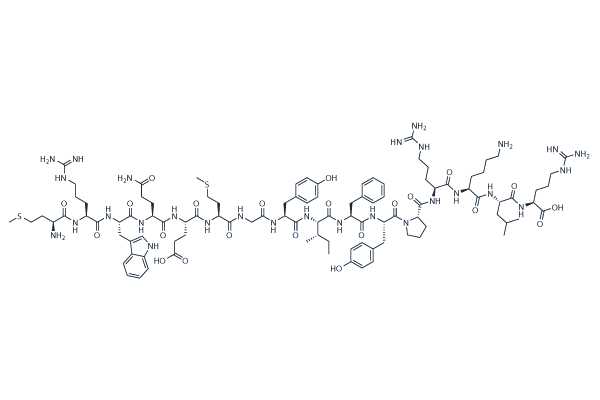
GC25648
MOTS-c
MOTS-c, a mitochondria-derived peptide (MDP), exerts antinociceptive and anti-inflammatory effects through activating AMPK pathway and inhibiting MAP kinases-c-fos signaling pathway.
-
GC65466
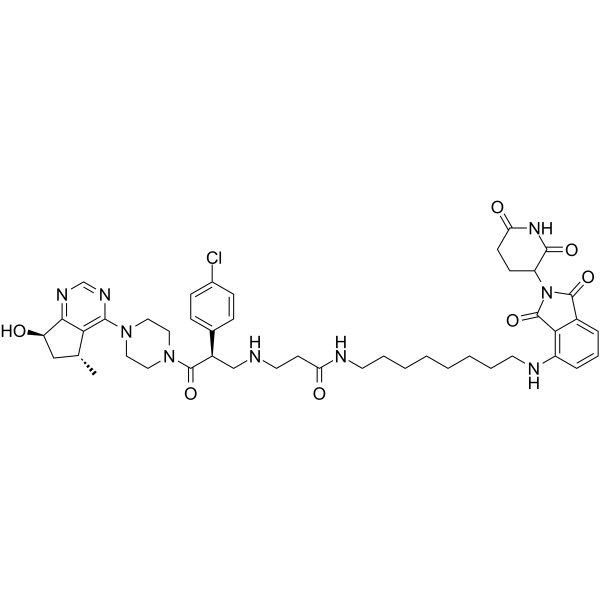
MS170
MS170 is a potent and selective PROTAC AKT degrader. MS170 depletes cellular total AKT (T-AKT) with the DC50 value of 32 nM. MS170 binds to AKT1, AKT2, and AKT3 with Kds of 1.3 nM, 77 nM, and 6.5 nM, respectively.
-
GC60258
MT 63-78
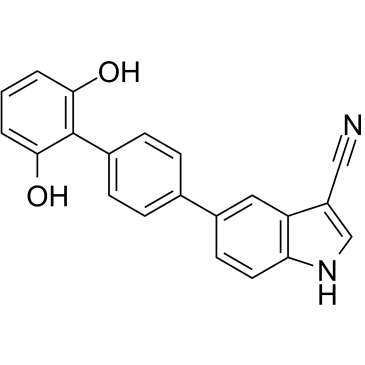
MT 63-78 is a specific and potent direct AMPK activator with an EC50 of 25 μM. MT 63–78 also induces cell mitotic arrest and apoptosis. MT 63-78 blocks prostate cancer growth by inhibiting the lipogenesis and mTORC1 pathways. MT 63-78 has antitumor effects.
-
GC64304
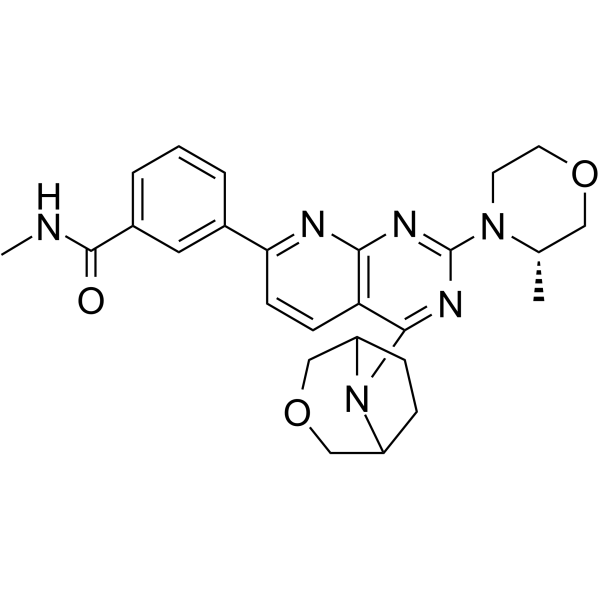
MTI-31
MTI-31 is a potent, orally active and highly selective inhibitor of mTORC1 and mTORC2.
-
GC65115
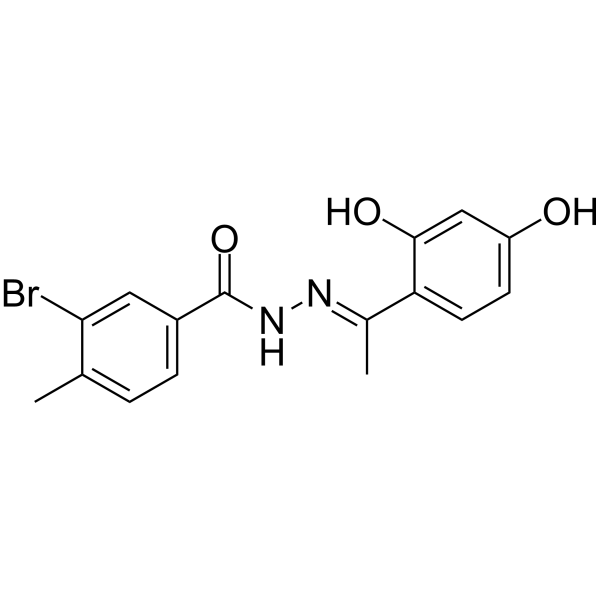
mTOR inhibitor-1
mTOR inhibitor-1 is a novel mTOR pathway inhibitor which can suppress cells proliferation and inducing autophagy.
-
GC62339
mTOR inhibitor-8
mTOR inhibitor-8 is an mTOR inhibitor and autophagy inducer. mTOR inhibitor-8 inhibits the activity of mTOR via FKBP12 and induces autophagy of A549 human lung cancer cells.
-
GC19256
MTX-211
MTX-211 is a dual inhibitor of EGFR and PI3K, used for the treatment of cancer and other diseases.
-
GC65442
Musk ketone
Musk ketone (MK) is a widely used artificial fragrance.
-
GC60262
N-Feruloyloctopamine
N-Feruloyloctopamine is an antioxidant constituent. N-Feruloyloctopamine significantly decreases thephosphorylationlevels of Akt and p38MAPK.
-
GC31536
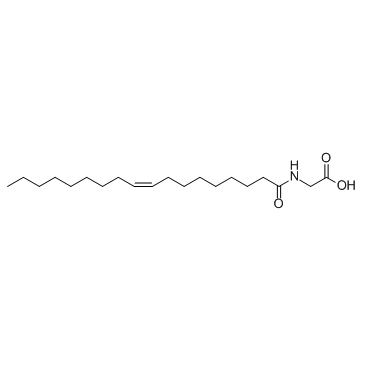
N-Oleoyl glycine
A putative substrate for peptidyl glycine αamidating enzyme
-
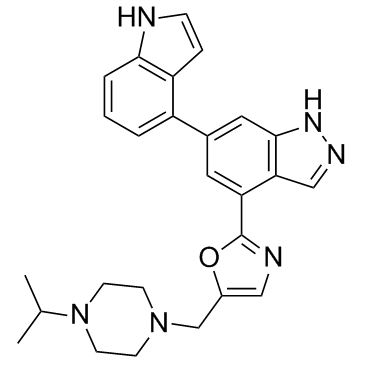
GC36714
Nemiralisib
Nemiralisib (GSK2269557 free base) is a potent and highly selective PI3Kδ inhibitor with a pKi of 9.9.
-
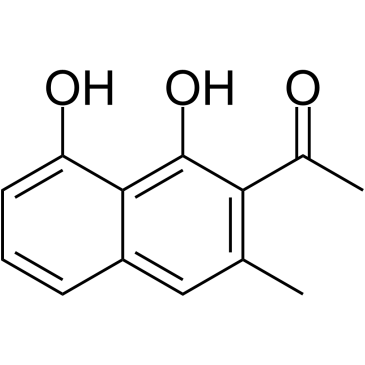
GC38567
Nepodin
A naphthol with diverse biological activities
-
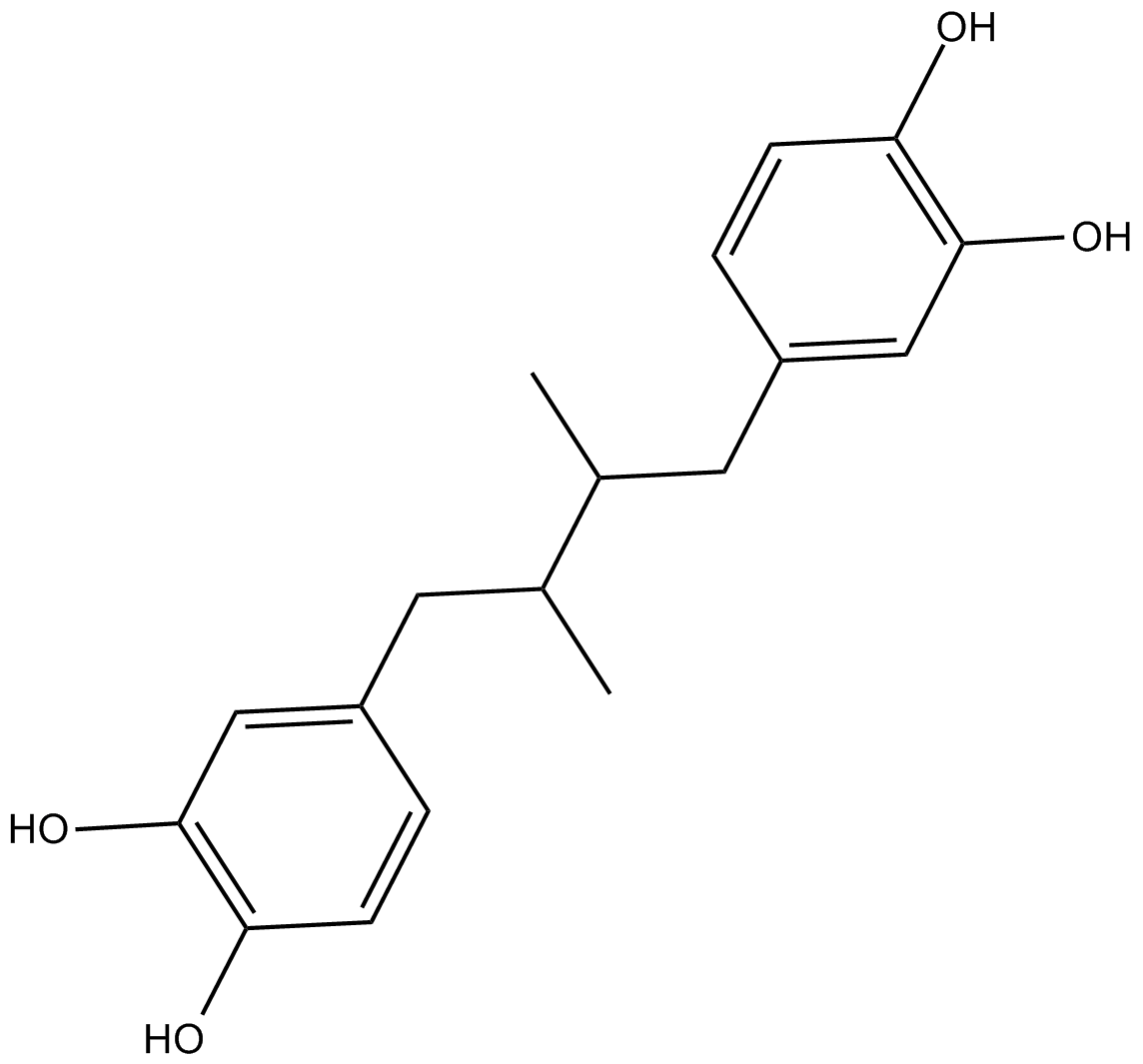
GC18089
Nordihydroguaiaretic acid
A non-selective LO inhibitor
-
GC11907
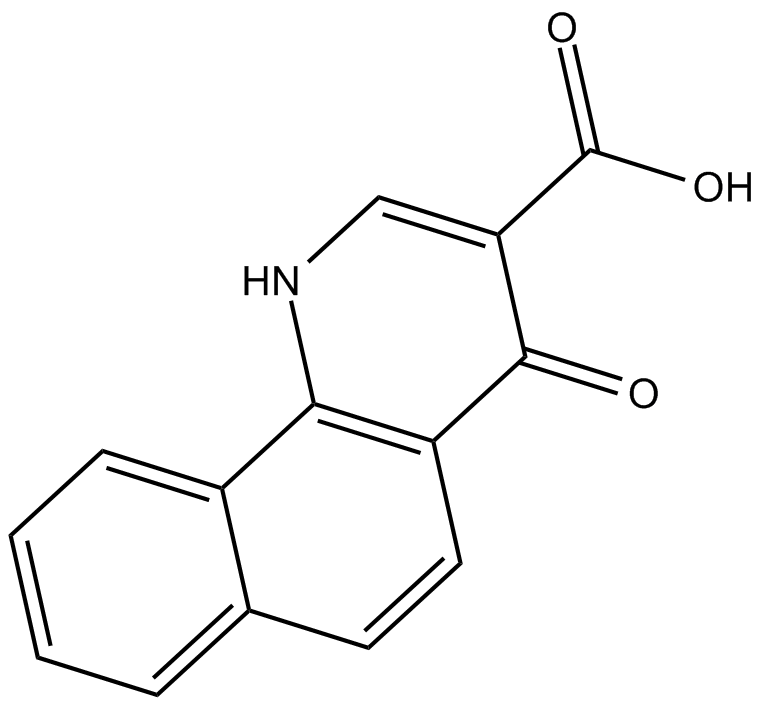
NSC 210902
NSC 210902 is a CK2 inhibitor, with an IC50 of 150 nM.
-
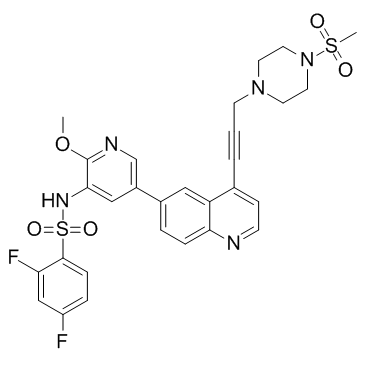
GC33014
NSC781406
A dual inhibitor of PI3K and mTOR
-
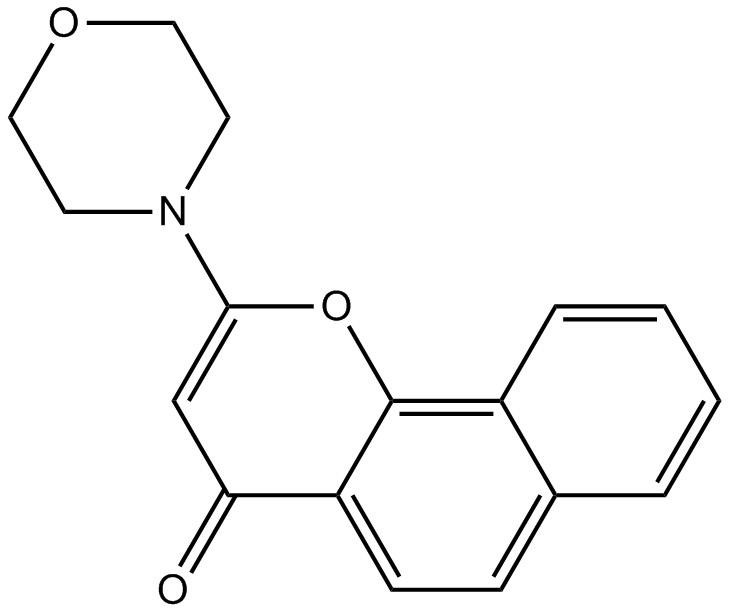
GC12332
NU 7026
DNPK inhibitor,ATP-competitive and potent
-
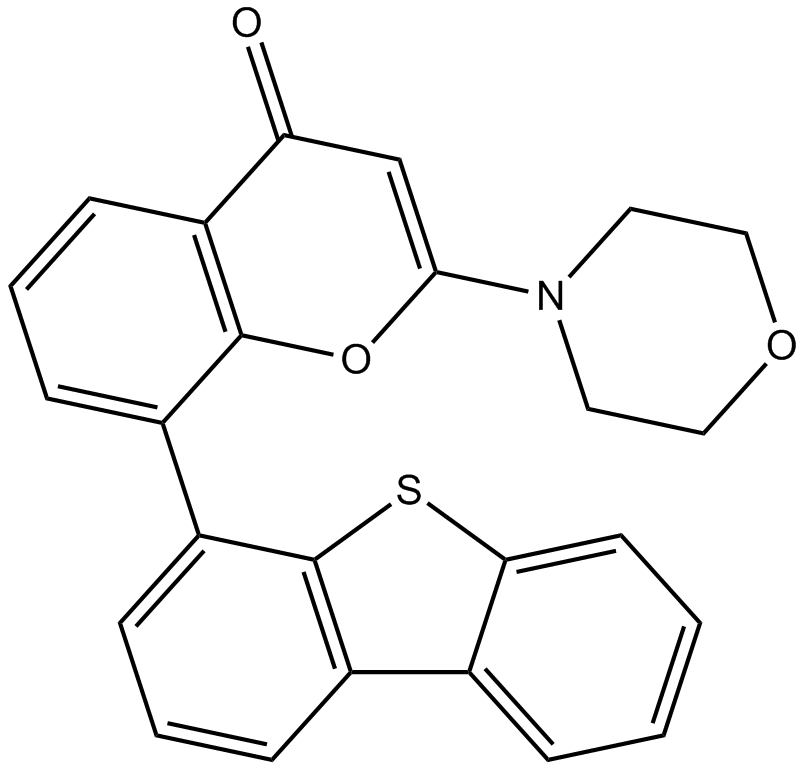
GC11251
NU7441 (KU-57788)
NU7441 (KU-57788) (NU7441) is a highly potent and selective DNA-PK inhibitor with an IC50 of 14 nM. NU7441 (KU-57788) is an NHEJ pathway inhibitor. NU7441 (KU-57788) also inhibits PI3K and mTOR with IC50s of 5.0 and 1.7 μM, respectively.
-
GC63121
NV-5138
NV-5138, a leucine analog, is the first selective and orally active brain mTORC1 activator, binding to Sestrin2.
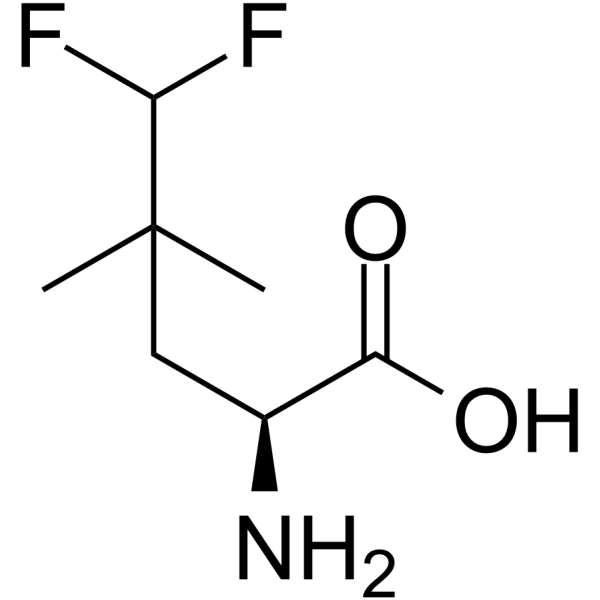
-
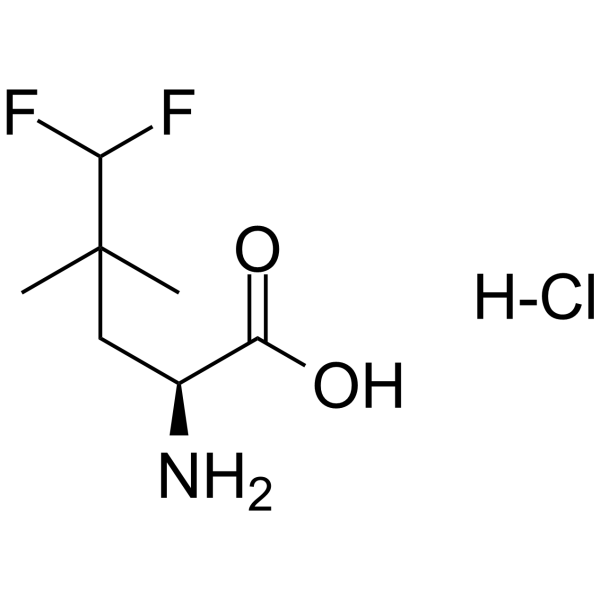
GC63122
NV-5138 hydrochloride
NV-5138 hydrochloride, a leucine analog, is the first selective and orally active brain mTORC1 activator, binding to Sestrin2.
-
GC13725
NVP-BAG956
A dual PDK1 and class I PI3K inhibitor

-
GC16145
NVP-BGT226
BGT226 (NVP-NVP-BGT226) is a PI3K (with IC50s of 4 nM, 63 nM and 38 nM for PI3Kα, PI3Kβ and PI3Kγ) /mTOR dual inhibitor which displays potent growth-inhibitory activity against human head and neck cancer cells.
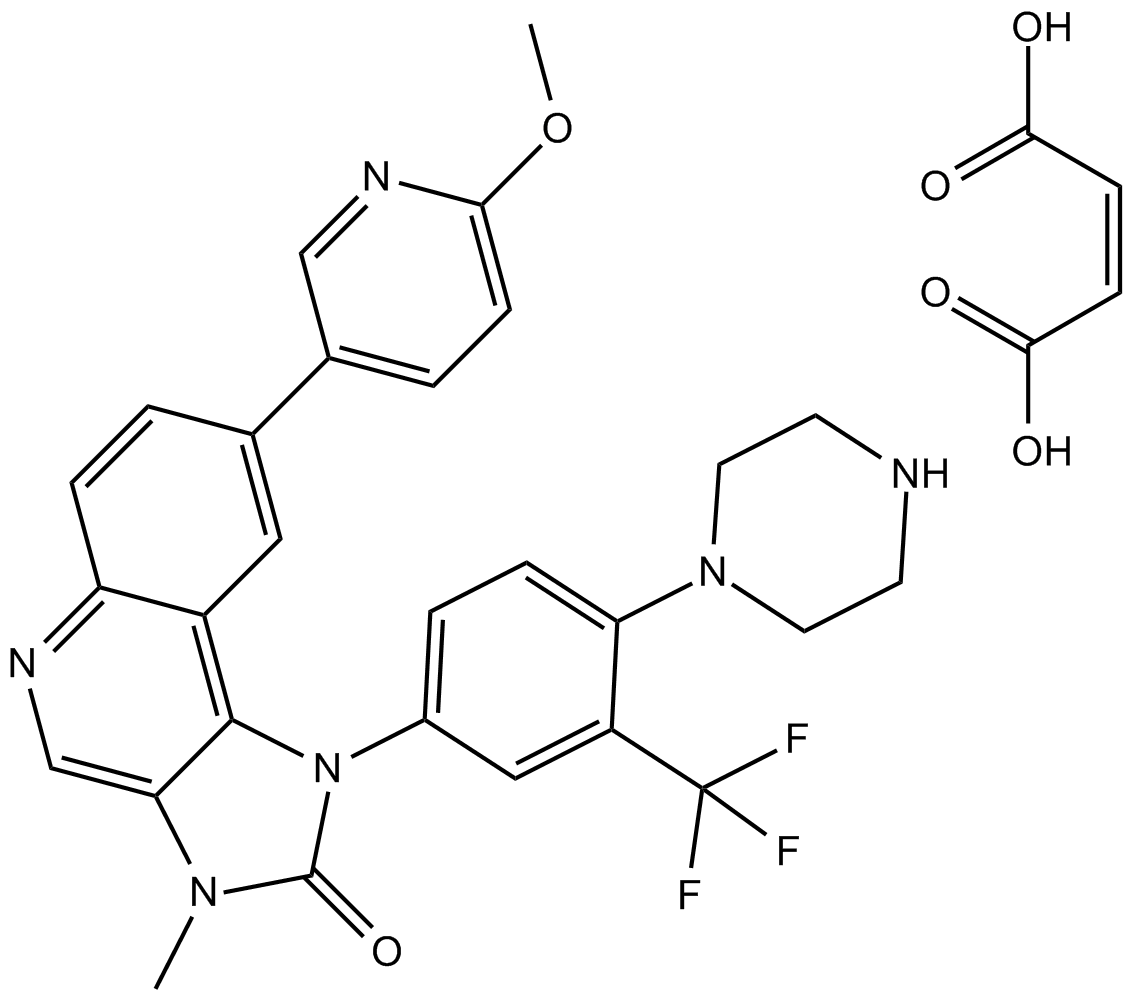
-
GC14504
NVP-BKM120 Hydrochloride
An inhibitor of class I PI3K isoforms
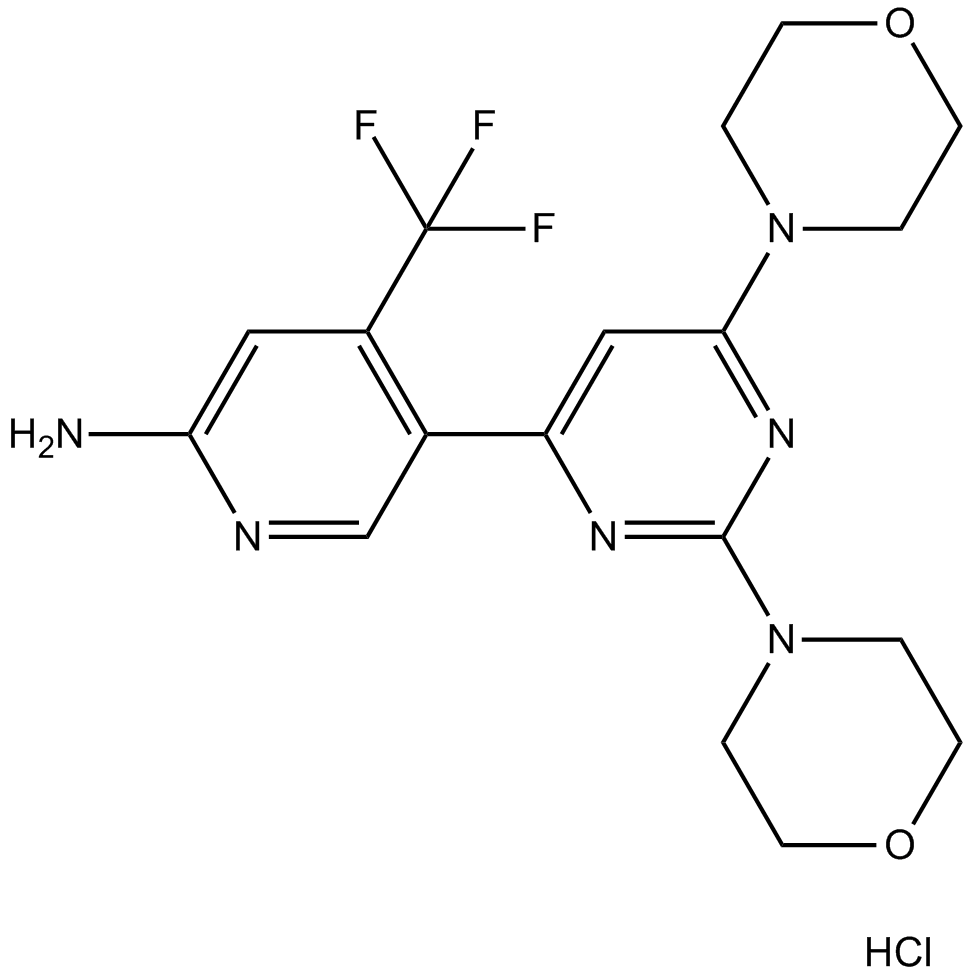
-
GC36786
NVP-QAV-572
NVP-QAV-572 is a PI3K inhibitor extracted from patent US7998990B2, Compound Example 8, has an IC50 of 10 nM.
-
GC62141
NVS-PI3-4
NVS-PI3-4 is a specific PI3Kγ inhibitor.
-
GC19269
O-304
O-304 is a small molecule AMPK activator.
-
GC36807
ON 146040
ON 146040 is a potent PI3Kα and PI3Kδ (IC50≈14 and 20 nM, respectively) inhibitor. ON 146040 also inhibits Abl1 (IC50<150 nM).
-
GN10692
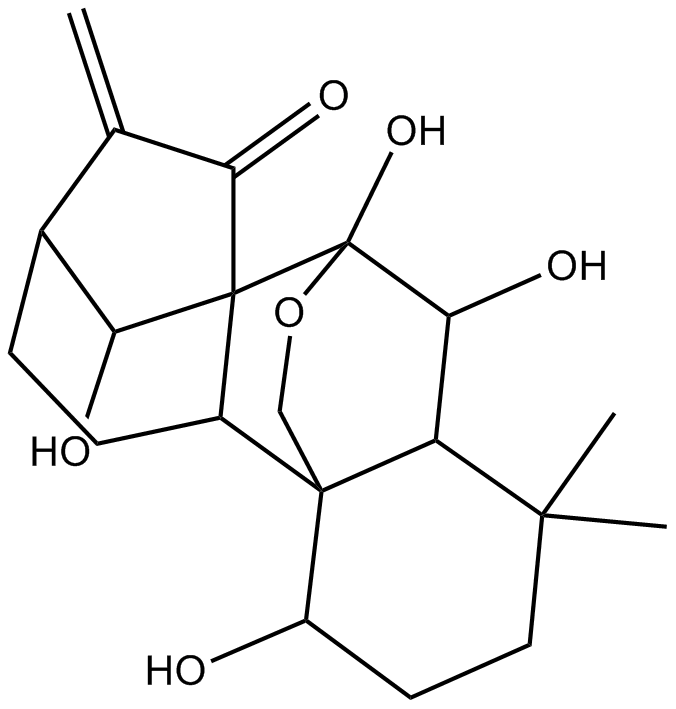
Oridonin
A diterpenoid with anti-inflammatory and anticancer properties
-
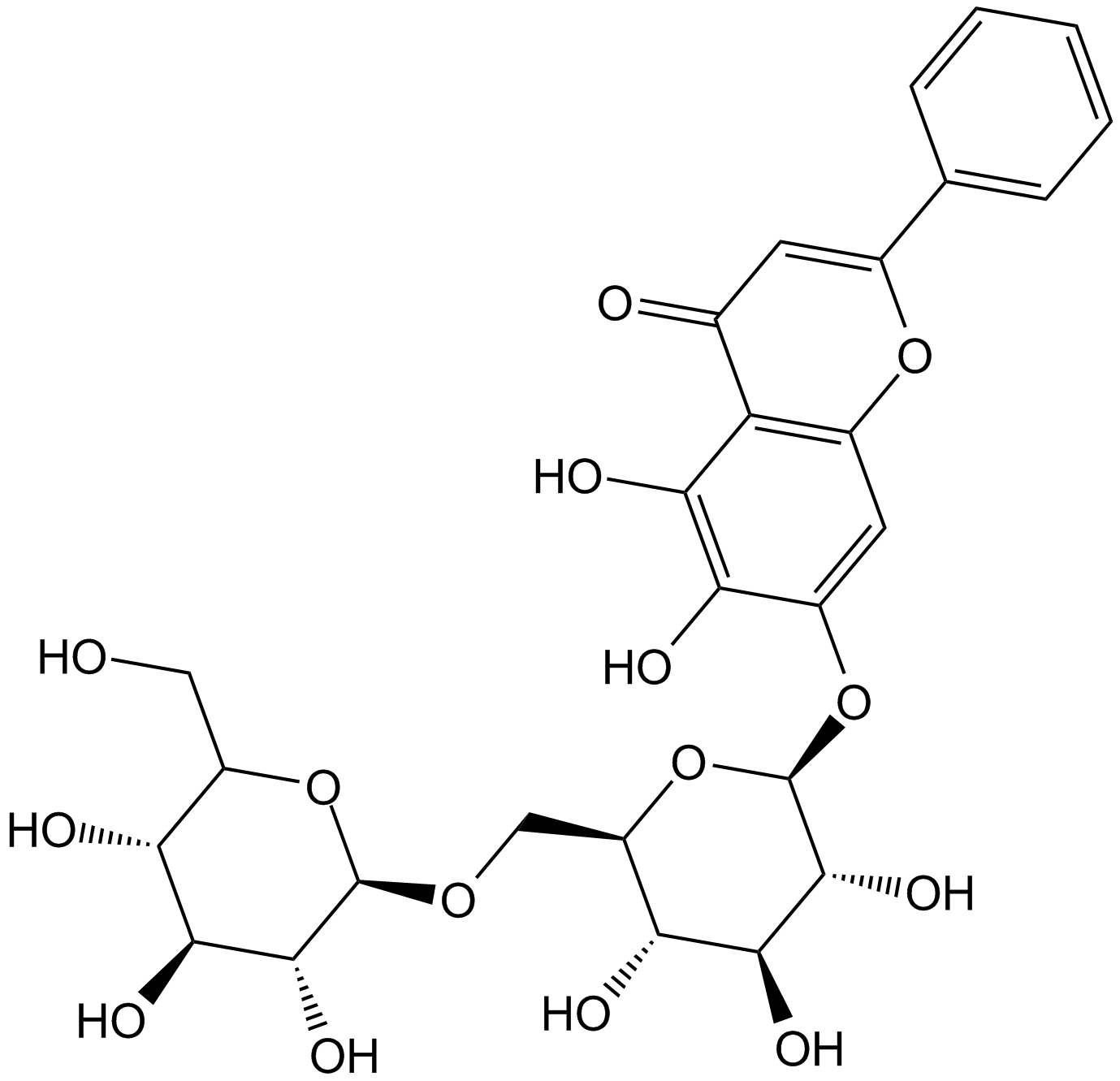
GN10732
Oroxin B
-
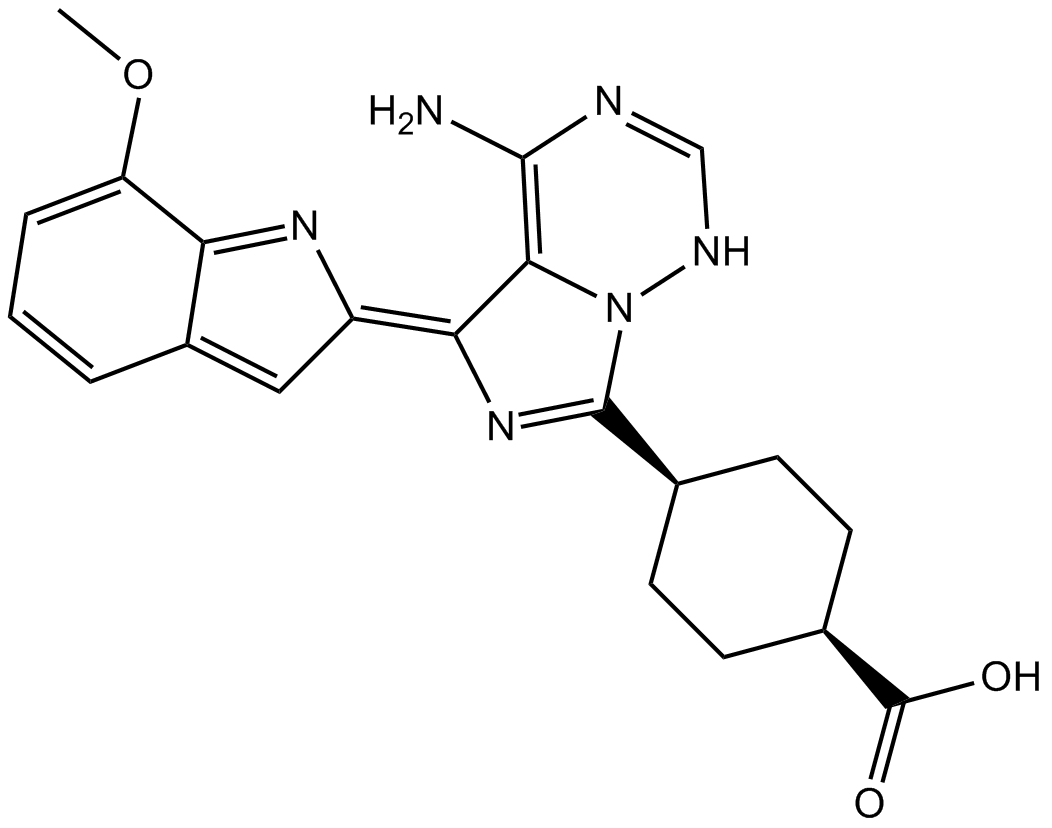
GC14071
OSI-027
MTORC1/ mTORC2 inhibitor
-
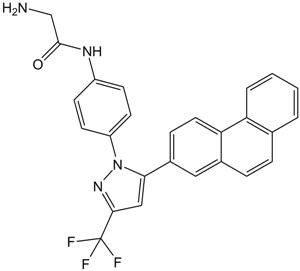
GC15742
OSU-03012 (AR-12)
Potent PDK1 inhibitor
-
GC10201
OTSSP167
MELK inhibitor
-
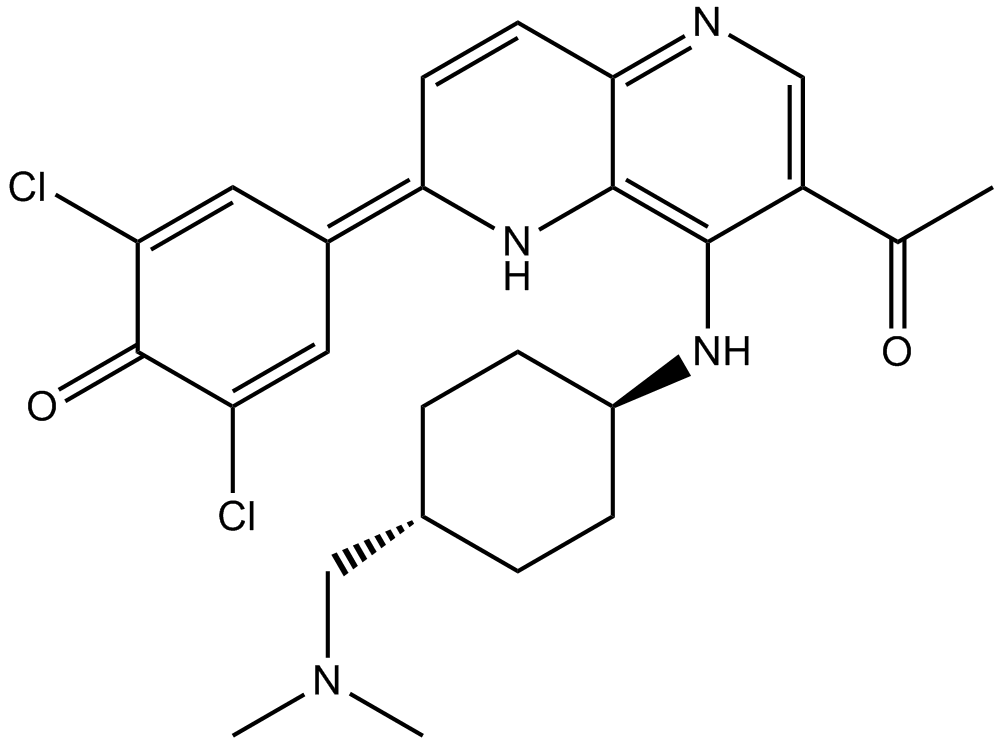
GC12155
OTSSP167 hydrochloride
MELK inhibitor,highly potent and selective
-
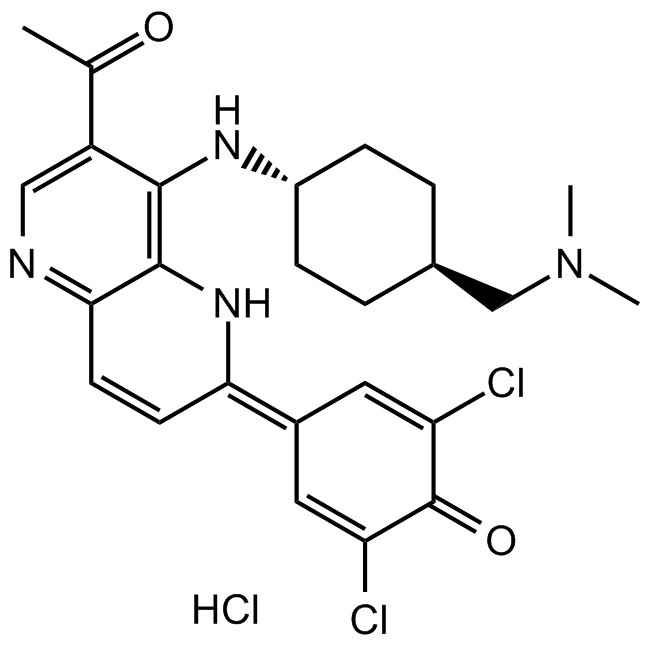
GC15740
OXA-01
mTORC1 and mTORC2 inhibitor
-
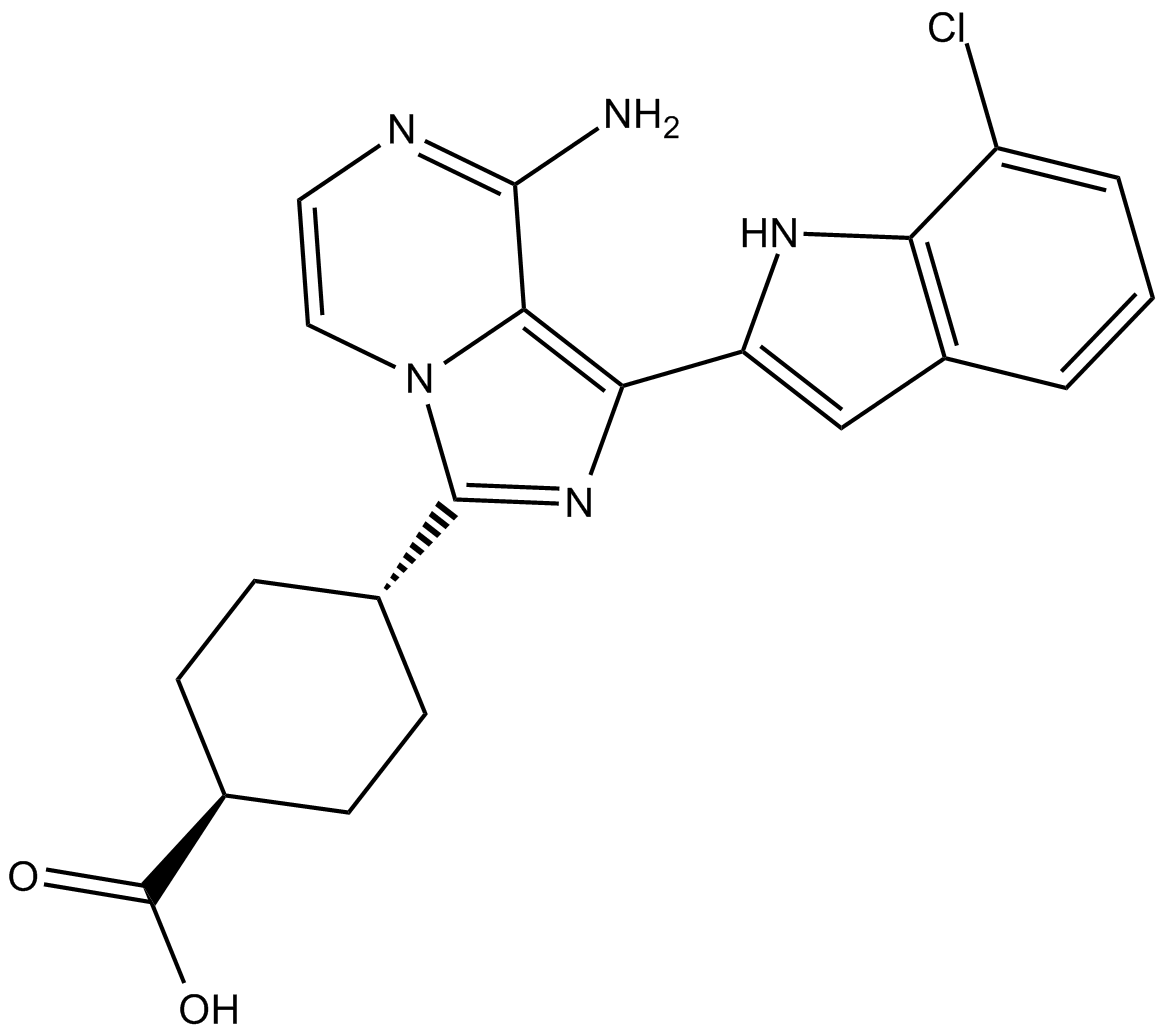
GC36832
P110δ-IN-1
P110δ-IN-1 is a potent and selective inhibitor of P110δ extracted from patent WO 2014055647 A1, with an IC50 of 8.4 nM.
-
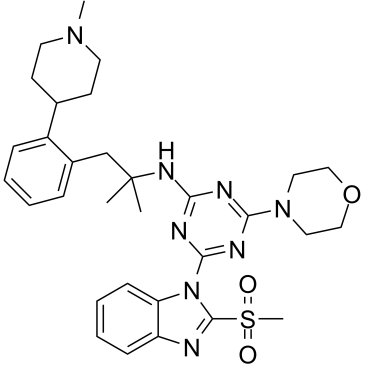
GN10752
Pachymic acid
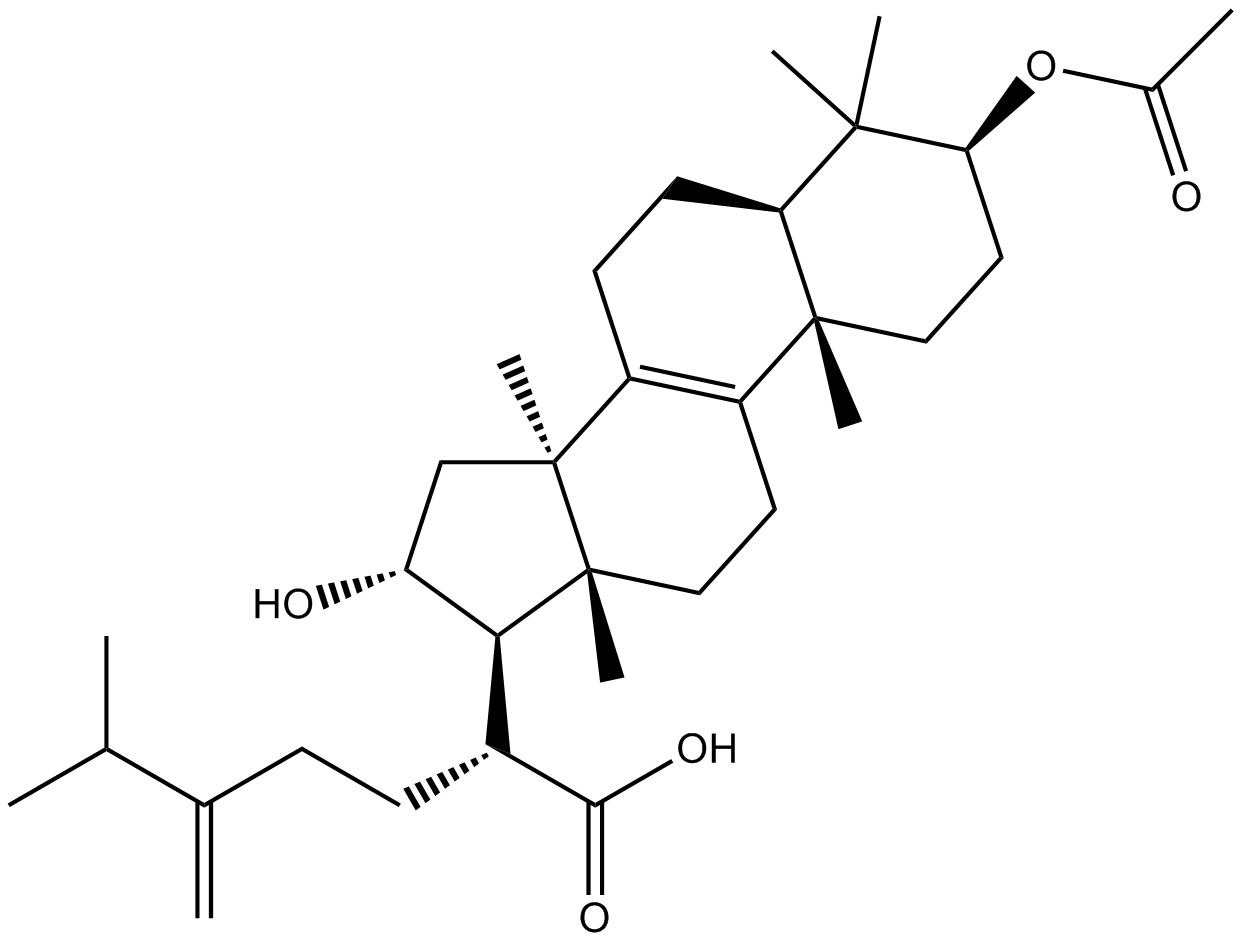
-
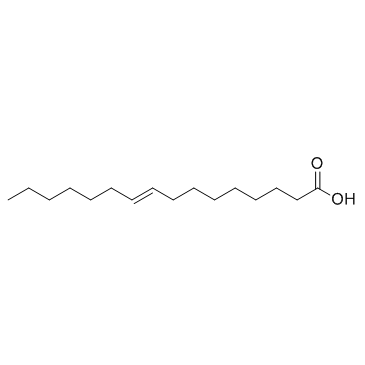
GC33765
Palmitelaidic Acid (9-trans-Hexadecenoic acid)
Palmitelaidic Acid (9-trans-Hexadecenoic acid) (9-trans-Hexadecenoic acid) is the trans isomer of palmitoleic acid.
-
GC12773
Palomid 529
PI3K/Akt/mTOR inhibitor
-
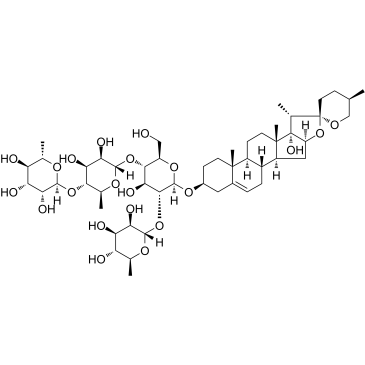
GC36855
Paris saponin VII
Paris saponin VII (Chonglou Saponin VII) is a steroidal saponin isolated from the roots and rhizomes of Trillium tschonoskii Maxim. Paris saponin VII-induced apoptosis in K562/ADR cells is associated with Akt/MAPK and the inhibition of P-gp. Paris saponin VII attenuates mitochondrial membrane potential, increases the expression of apoptosis-related proteins, such as Bax and cytochrome c, and decreases the protein expression levels of Bcl-2, caspase-9, caspase-3, PARP-1, and p-Akt. Paris saponin VII induces a robust autophagy in K562/ADR cells and provides a biochemical basis in the treatment of leukemia.
-
GC32916
Parsaclisib (INCB050465)
Parsaclisib (INCB050465) (INCB050465) is a potent, selective and orally active inhibitor of PI3Kδ, with an IC50 of 1 nM at 1 mM ATP. Parsaclisib (INCB050465) shows approximately 20000-fold selectivity over other PI3K class I isoforms. Parsaclisib (INCB050465) can be used for the research of relapsed or refractory B-cell malignancies.
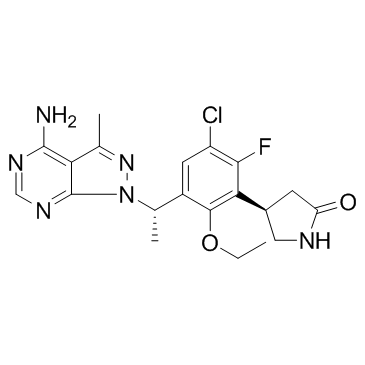
-
GC62593
Parsaclisib hydrochloride
Parsaclisib hydrochloride (INCB050465 hydrochloride) is a potent, selective and orally active inhibitor of PI3Kδ, with an IC50 of 1 nM at 1 mM ATP. Parsaclisib hydrochloride shows approximately 20000-fold selectivity over other PI3K class I isoforms. Parsaclisib hydrochloride can be used for the research of relapsed or refractory B-cell malignancies.
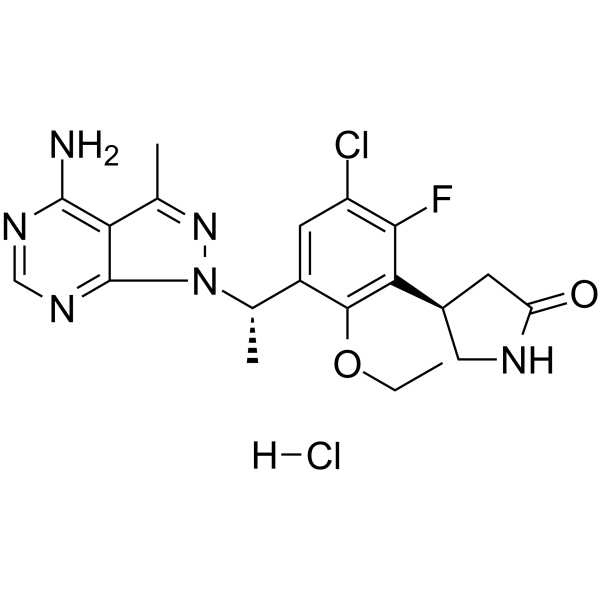
-
GC12866
PDK1 inhibitor
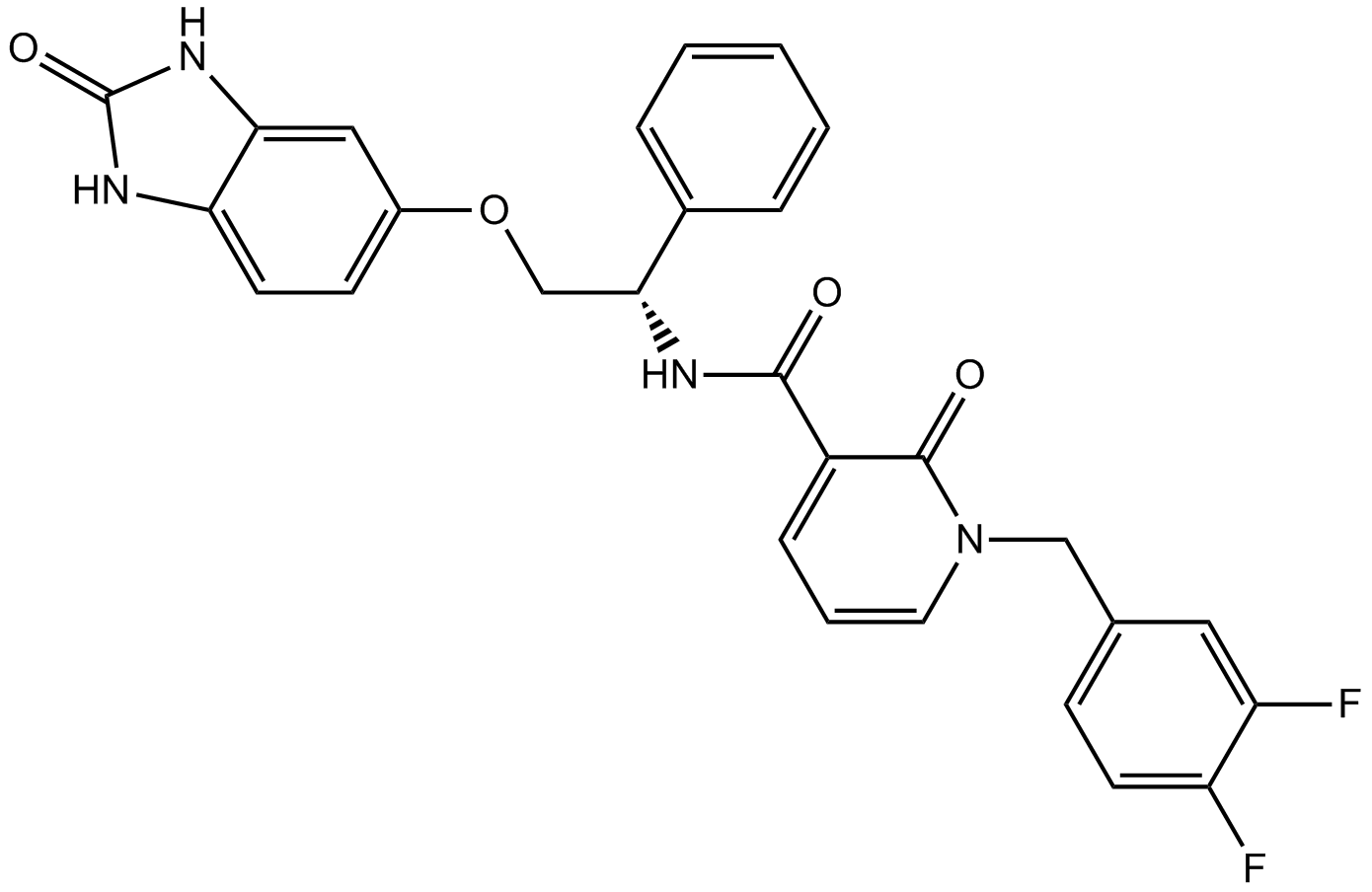
-
GC66474
PDK1-IN-RS2
PDK1-IN-RS2 is a mimic of peptide docking motif (PIFtide) and is a substrate-selective PDK1 inhibitor with a Kd of 9 μM. PDK1-IN-RS2 suppresses the activation of the downstream kinases S6K1 by PDK1.

-
GC44587
PDMP (hydrochloride)
PDMP is a ceramide analog first prepared in a search for inhibitors of glucosylceramide synthase.
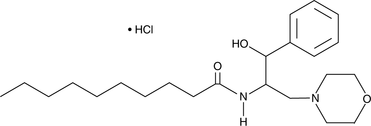
-
GC15680
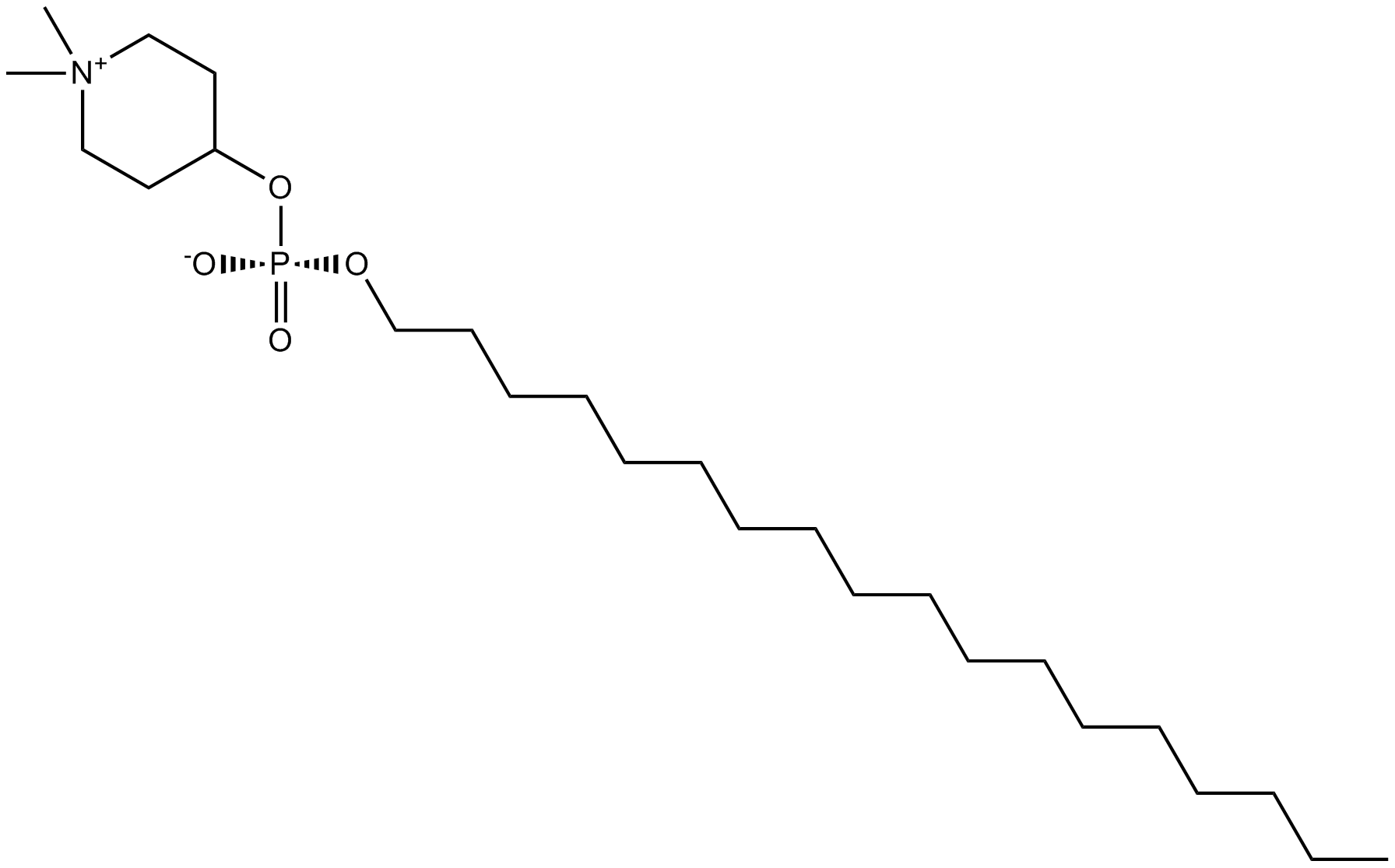
Perifosine
Akt inhibitor
-
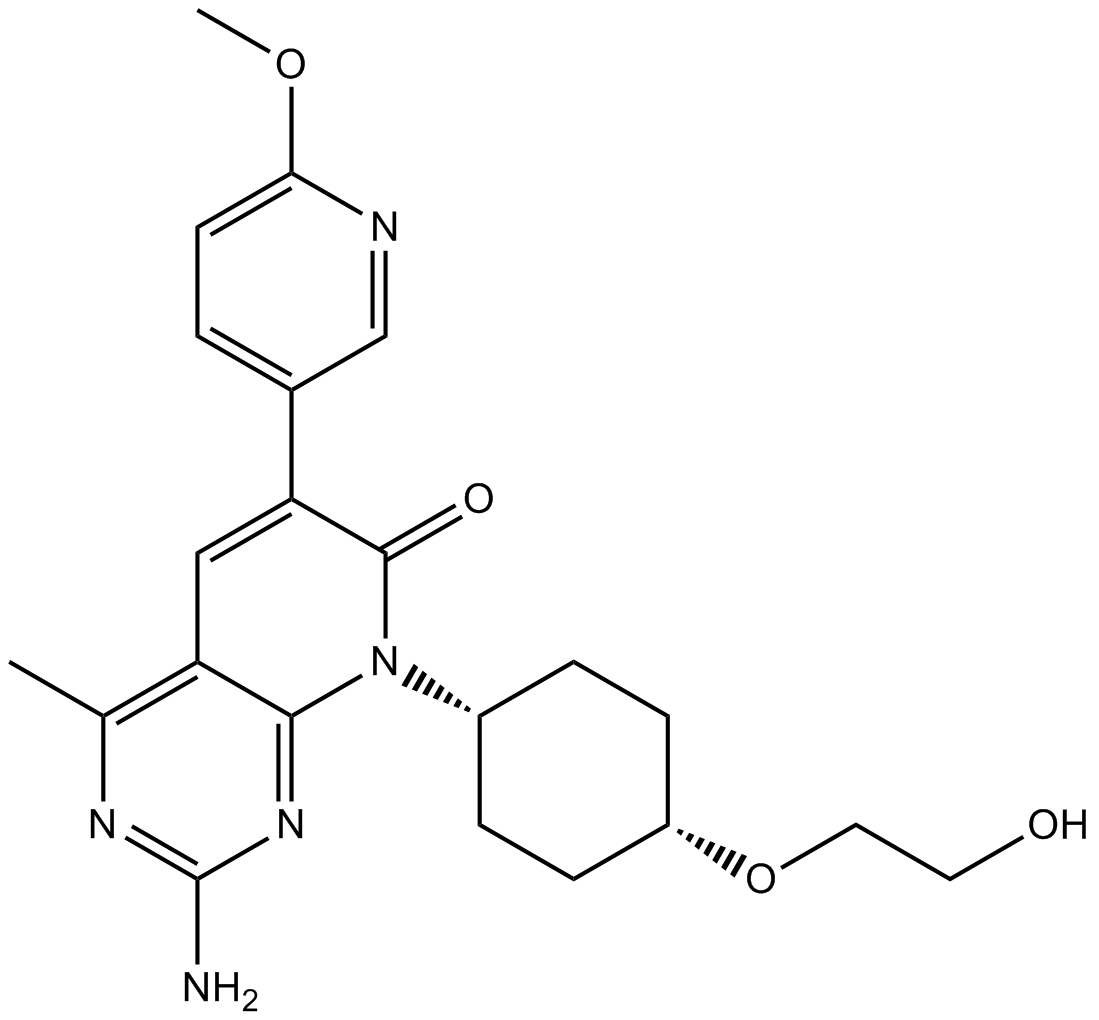
GC16417
PF-04691502
PI3K/mTOR (FRAP) inhibitor
-
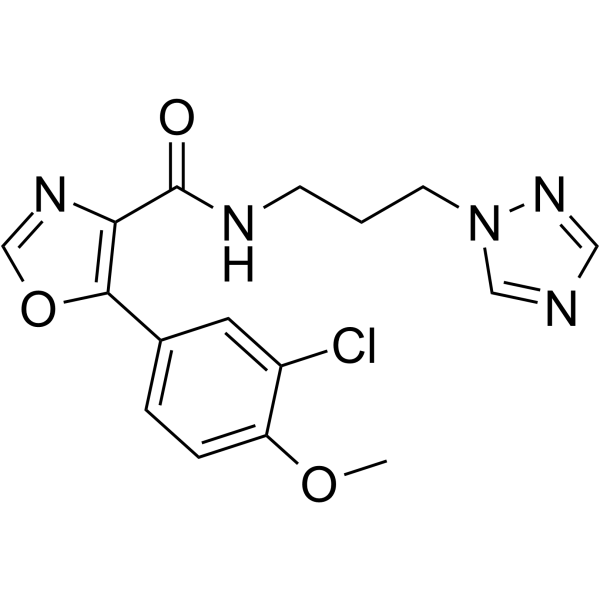
GC63446
PF-04802367
PF-04802367 (PF-367) is a highly selective GSK-3 inhibitor with an IC50 of 2.1 nM based on a recombinant human GSK-3β enzyme assay and 1.1 nM based on ADP-Glo assay.
-
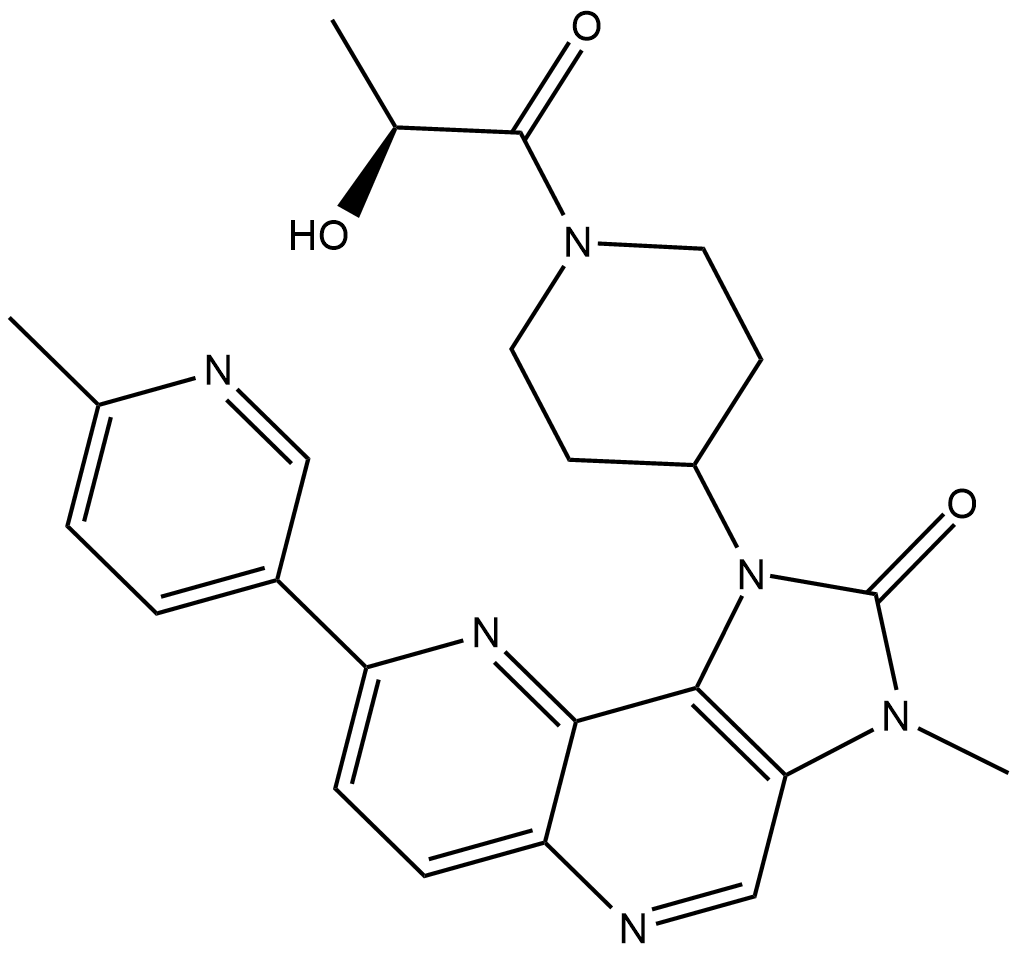
GC19283
PF-04979064
PF-04979064 is a potent and selective PI3K/mTOR dual kinase inhibitor with Kis of 0.13 nM and 1.42 nM for PI3Kα and mTOR, respectively.
-
GC15653
PF-05212384 (PKI-587)
PF-05212384 (PKI-587) (PKI-587) is a highly potent dual inhibitor of PI3Kα, PI3Kγ, and mTOR with IC50s of 0.4 nM, 5.4 nM and 1.6 nM, respectively. PF-05212384 (PKI-587) is equally effective in both complexes of mTOR, mTORC1 and mTORC2.
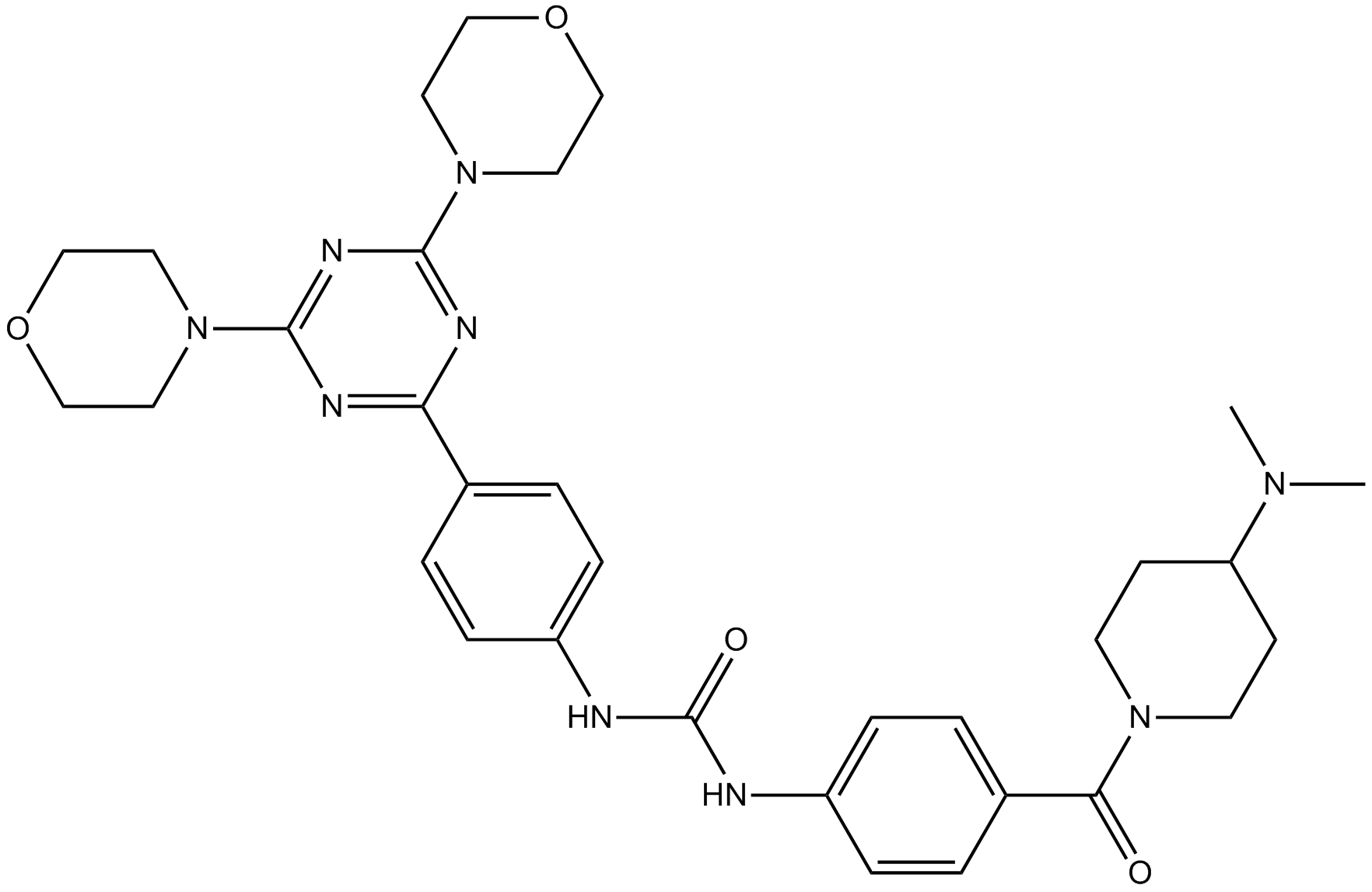
-
GC19286
PF-06409577
PF-06409577 is a potent and selective allosteric activator of AMPK α1β1γ1 isoform with an EC50 of 7 nM.
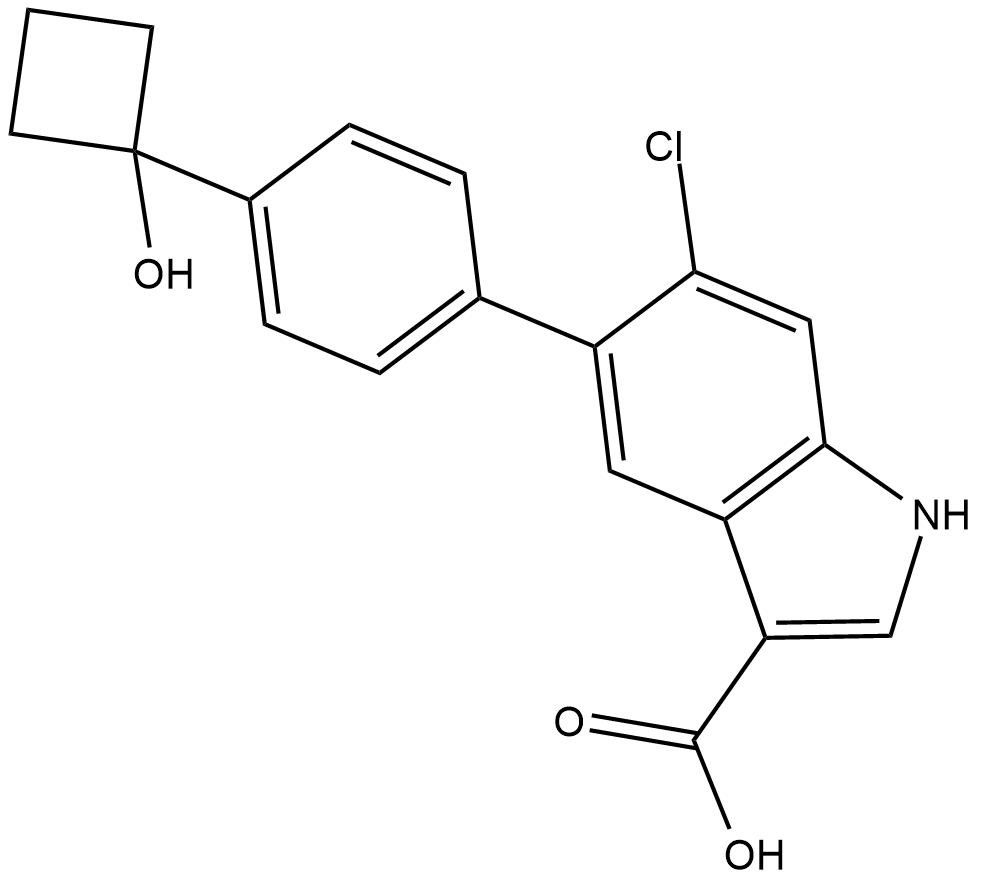
-
GC66002
PF-06685249
PF-06685249 (PF-249) is a potent and orally active allosteric AMPK activator with an EC50 of 12 nM for recombinant AMPK α1β1γ1. PF-06685249 can be used for diabetic nephropathy research.
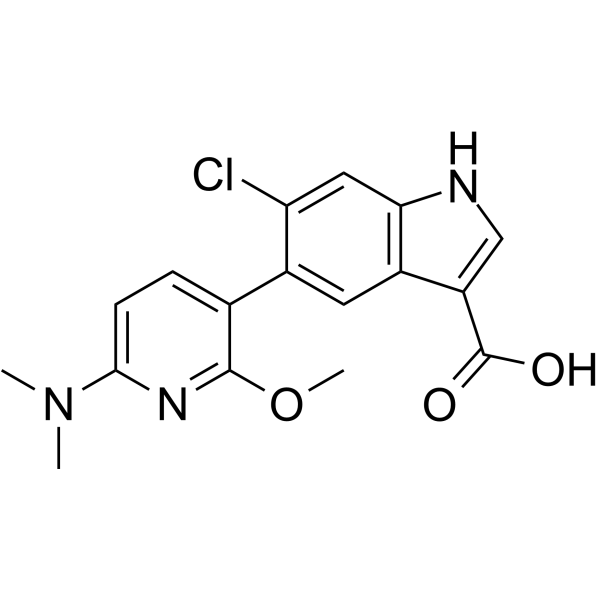
-
GC62556
PF-06843195
PF-06843195 is a highly selective PI3Kα inhibitor with an IC50 of 18 nM in Rat1 fibroblasts. The Kis of PF-06843195 for PI3Kα and PI3Kδ in biochemical kinase assay are less than 0.018 nM and 0.28 nM, respectively. PF-06843195 has great suppression of the PI3K/mTOR signaling pathway and durable antitumor efficacy.
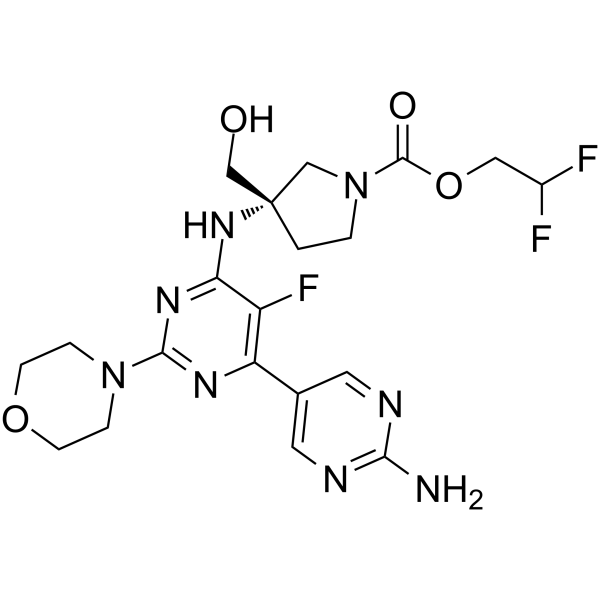
-
GC62660
PF-07104091
PF-07104091 is a CDK2/cyclin E1 inhibitor, a selective ATP-site inhibitor targeting the cyclin-bound activated state of the kinase [1].
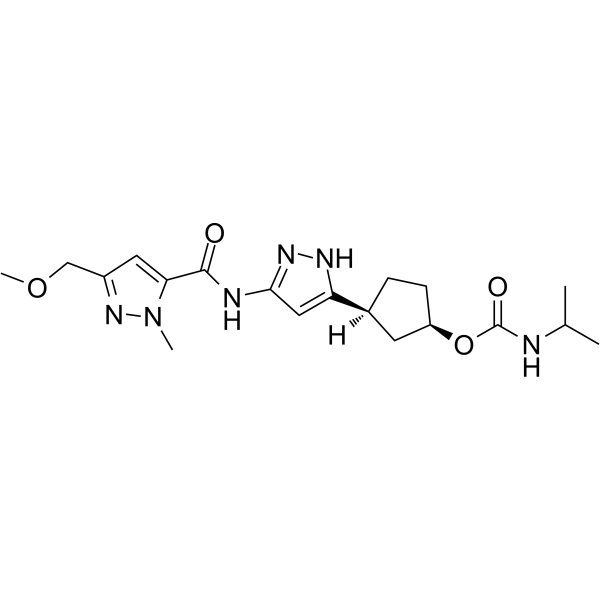
-
GC62661
PF-07104091 hydrate
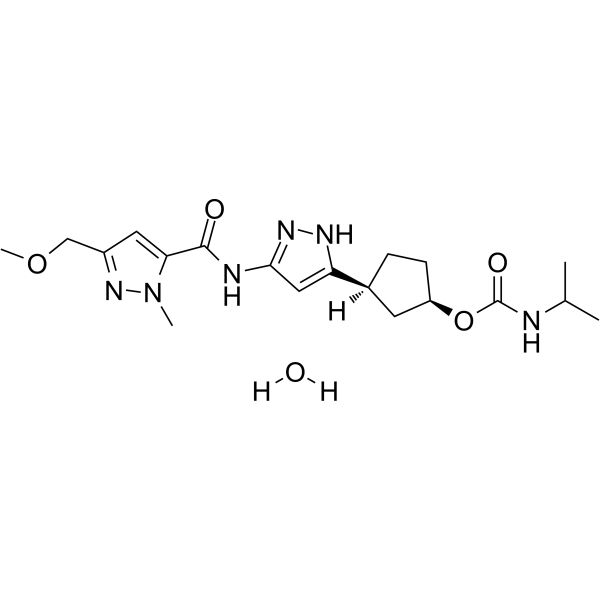
-
GC10689
PF-4708671
PF-4708671, is a novel cell-permeable inhibitor of S6K1, specifically inhibits the S6K1 isoform with a Ki of 20 nM and IC50 of 160 nM.
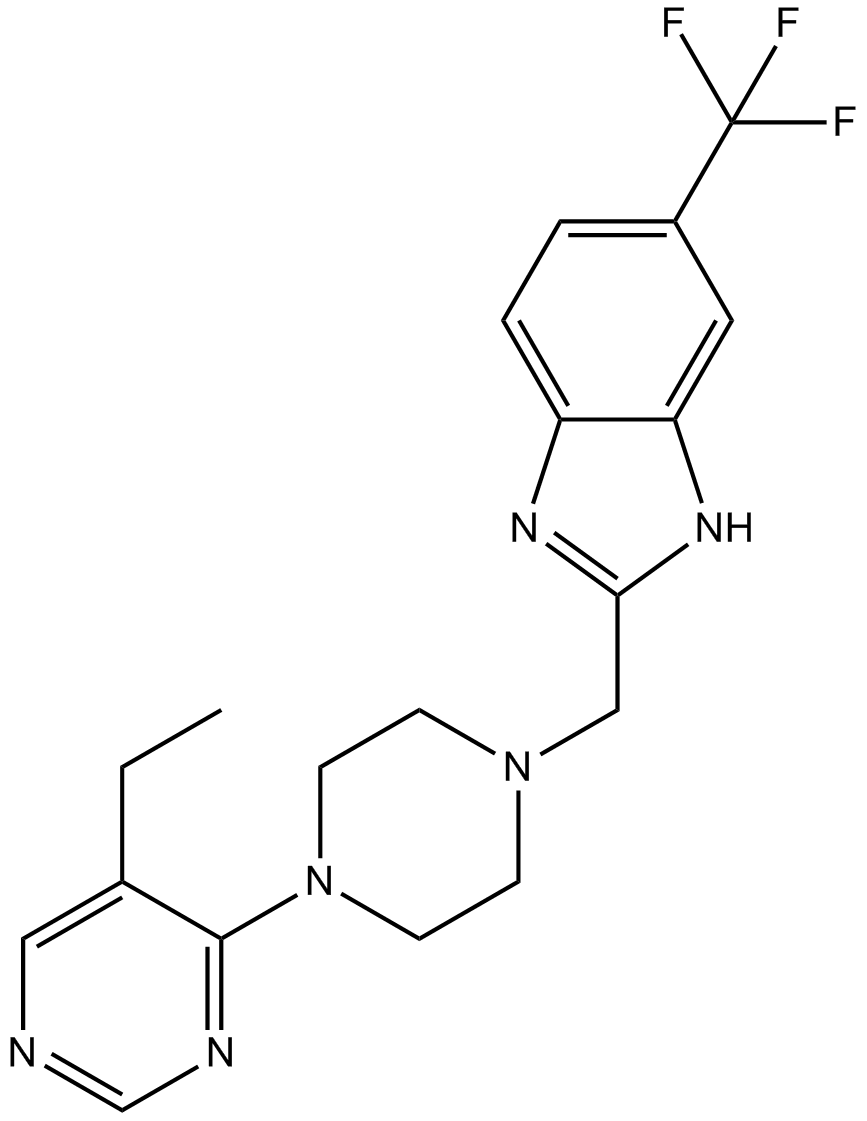
-
GC12375
PF-4989216
Selective oral PI3K inhibitor
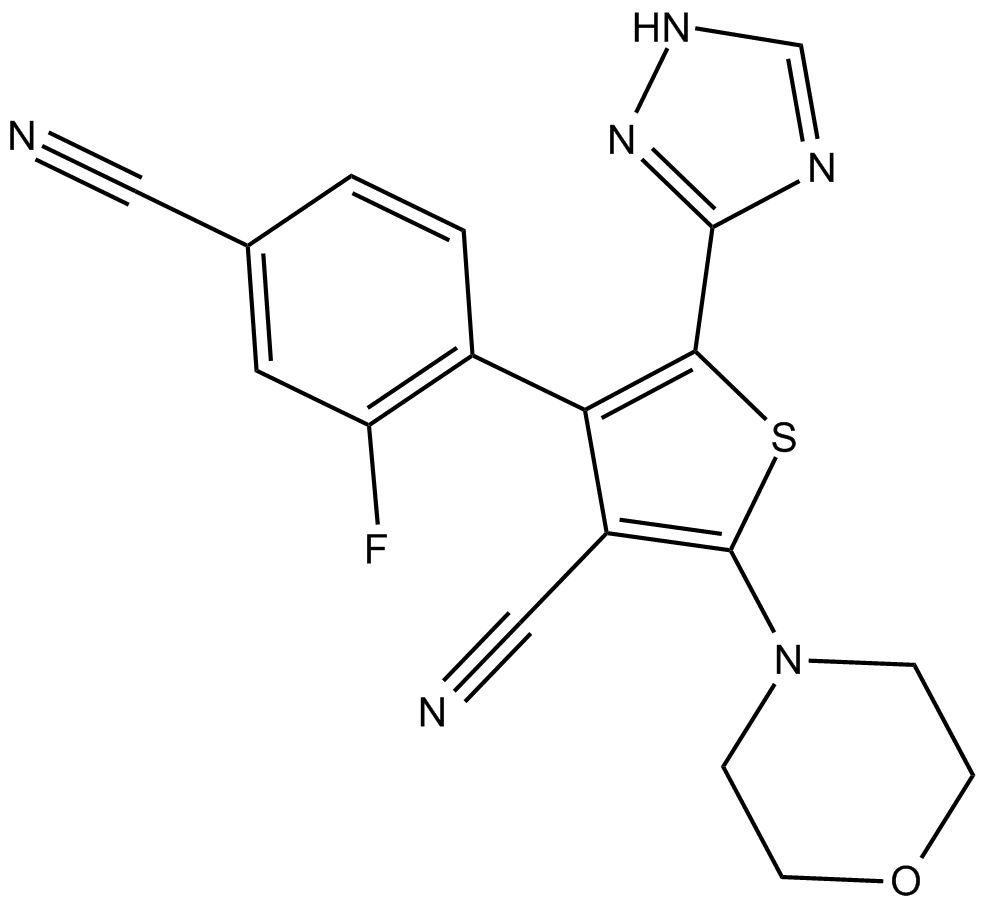
-
GC32892
PF-AKT400 (AKT protein kinase inhibitor)
PF-AKT400 (AKT protein kinase inhibitor) is a broadly selective, potent, ATP-competitive Akt inhibitor, displays 900-fold greater selectivity for PKBα (IC50=0.5 nM) than PKA (IC50=450 nM).
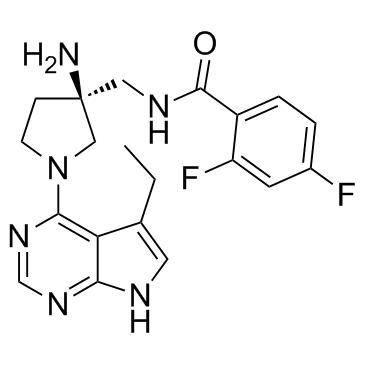
-
GC38170
Phellodendrine
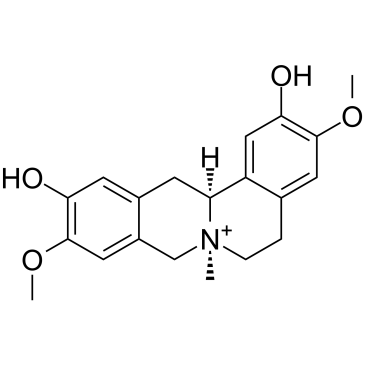
-
GC10461
Phenformin HCl
Phenformin HCl is an anti-diabetic drug from the biguanide class, can activate AMPK activity.
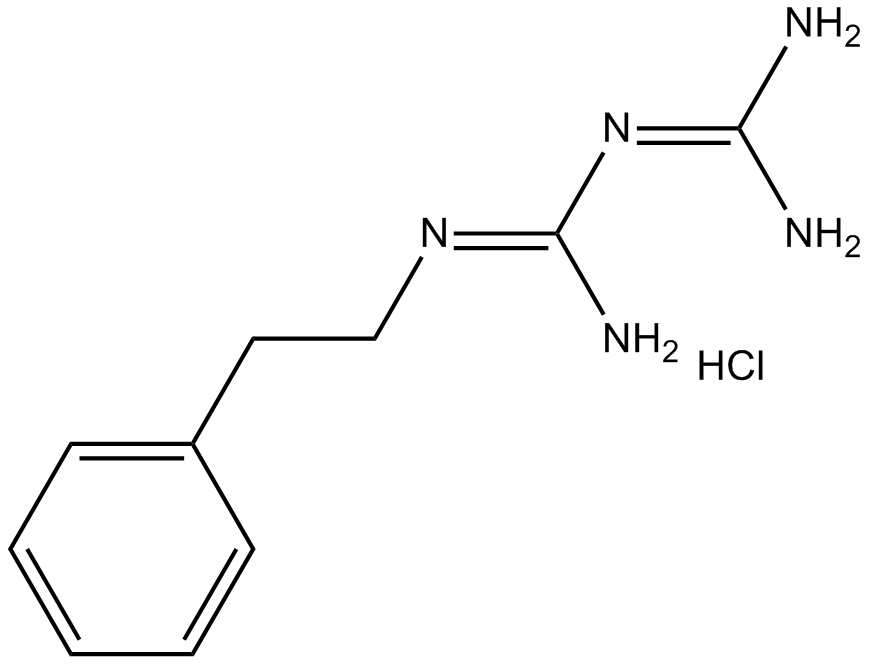
-
GC65375
Phospho-Glycogen Synthase Peptide-2(substrate) TFA
Phospho-Glycogen Synthase Peptide-2 (substrate) is peptide substrate?for glycogen synthase kinase-3 (GSK-3) and can be used for affinity purification of protein-serine kinases.
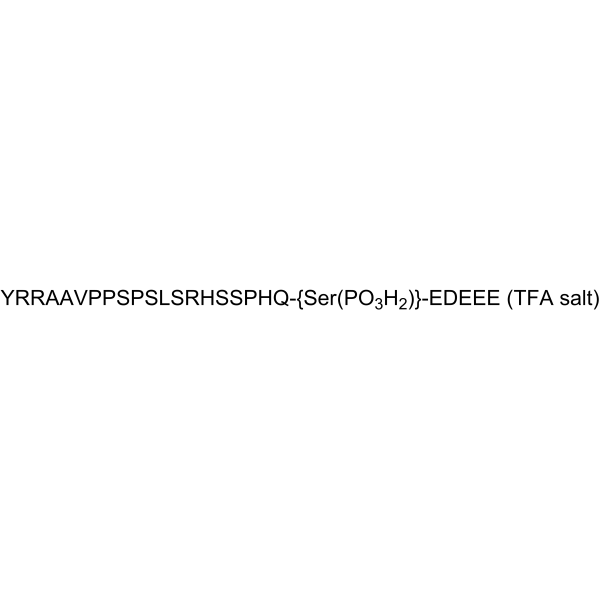
-
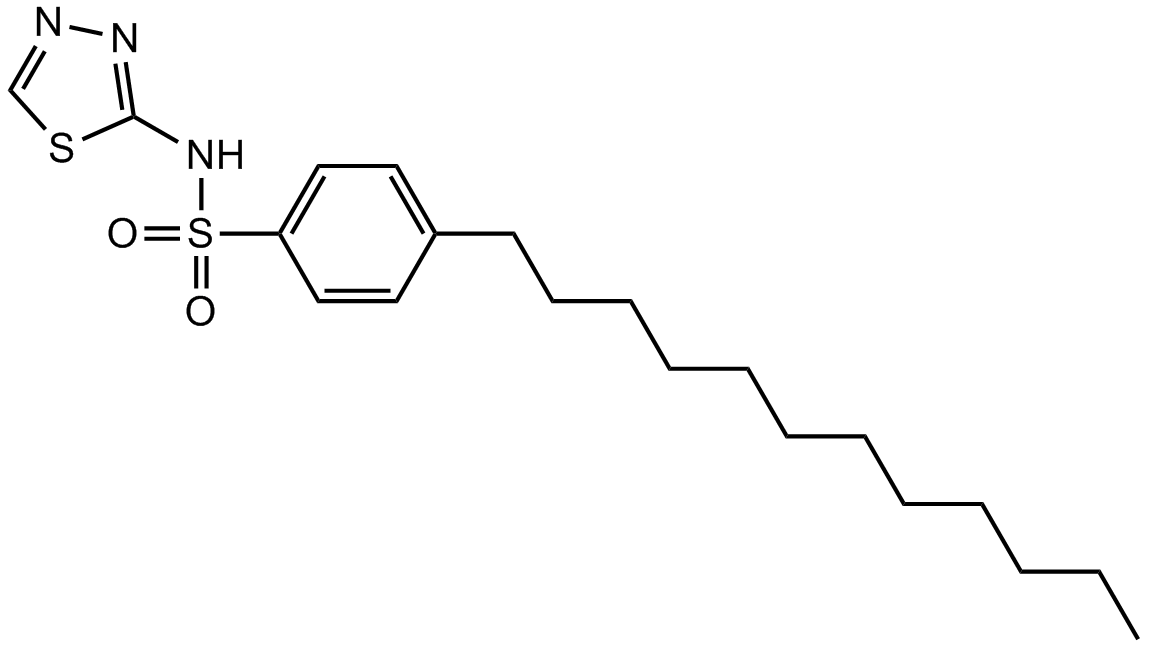
GC12094
PHT-427
Akt and PDPK1 inhibitor
-
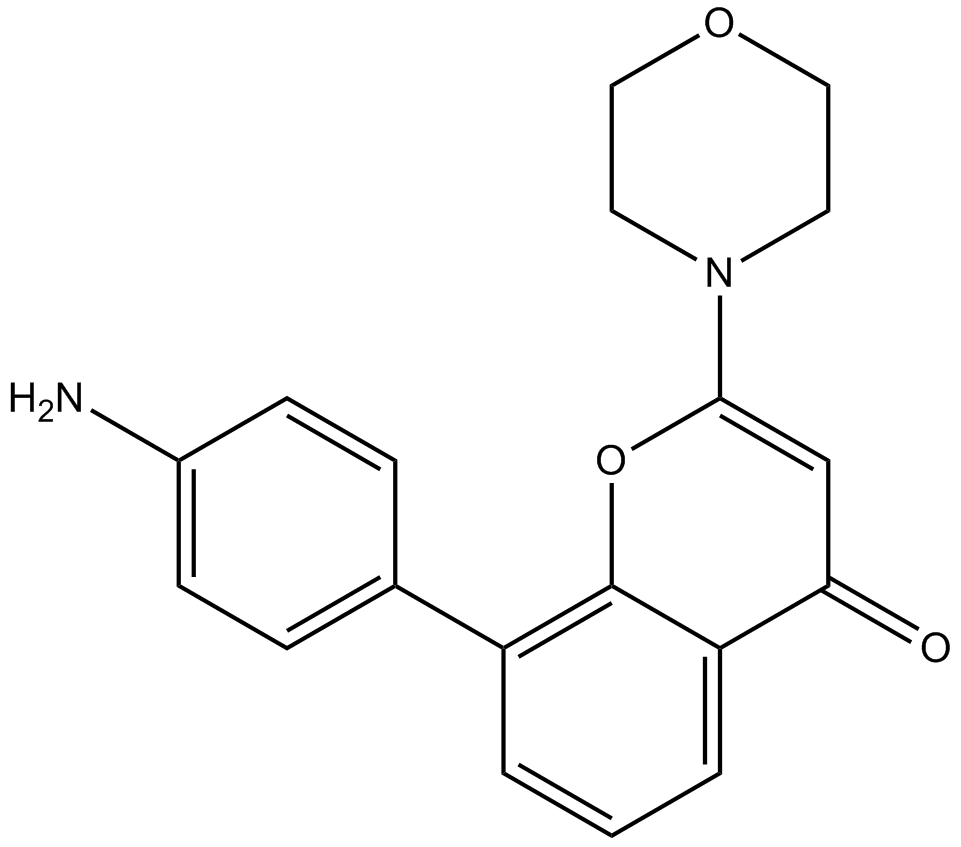
GC10386
PI 828
PI 3-Kinase inhibitor
-
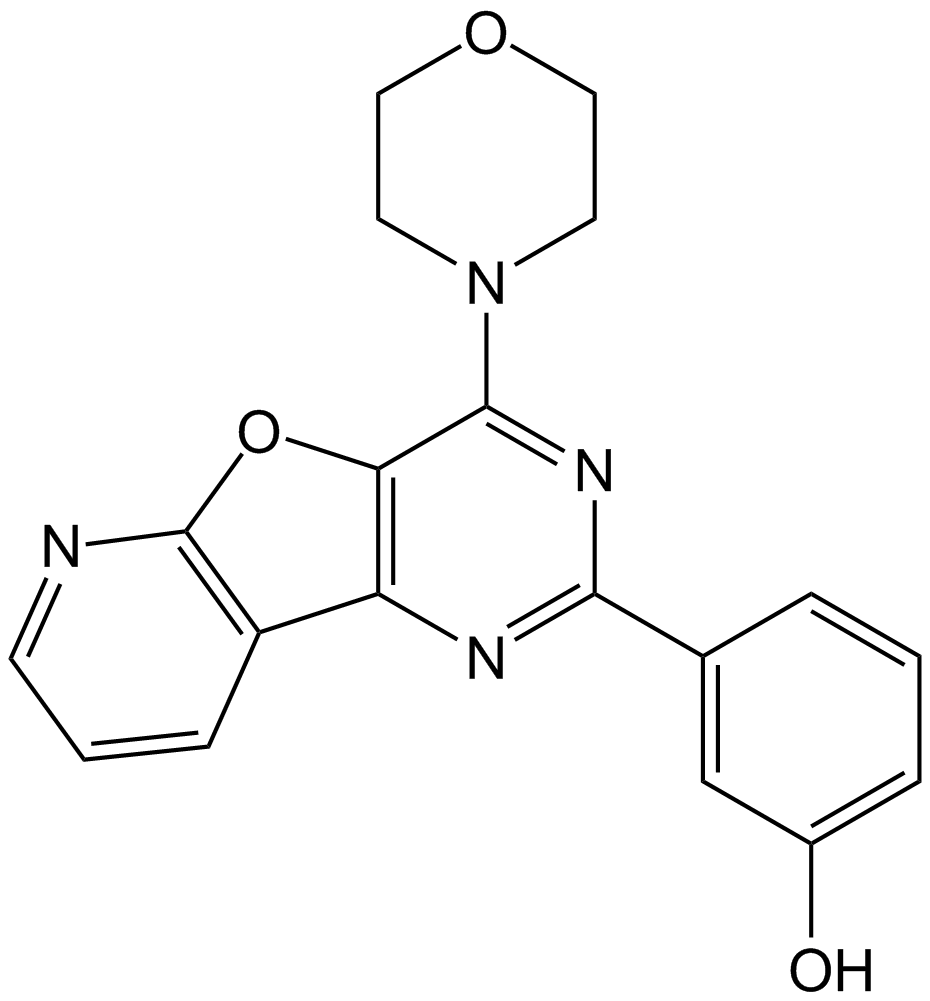
GC11165
PI-103
Class I PI3K, mTOR and DNA-PK inhibitor
-
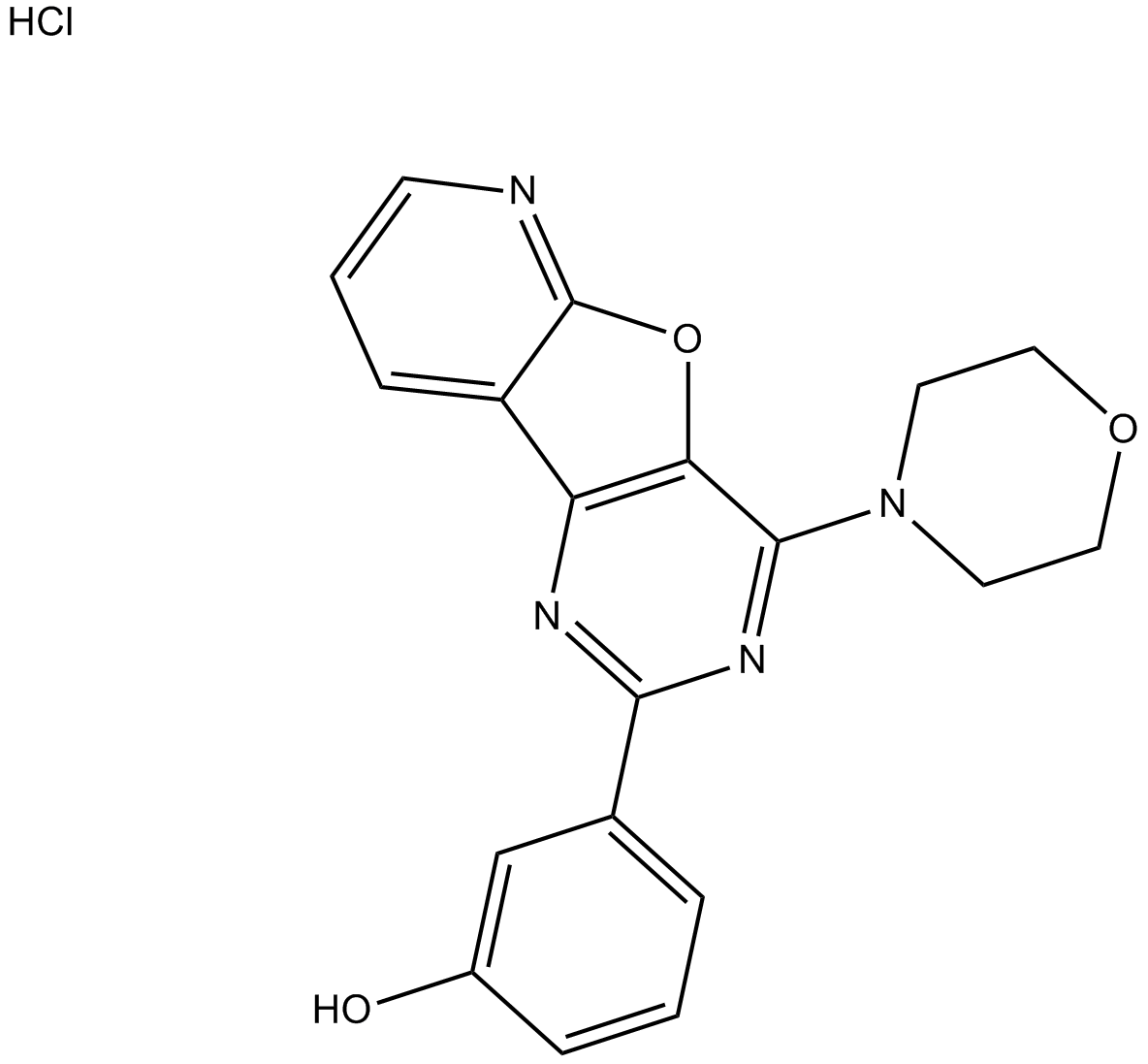
GC16379
PI-103 Hydrochloride
A potent, cell-permeable PI3K inhibitor
-
GC39155
PI-273
PI-273 is a first reversibly and specific phosphatidylinositol 4-kinase (PI4KIIα) inhibitor with an IC50 of 0.47 μM. PI-273 can inhibit breast cancer cell proliferation, block the cell cycle and induce cell apoptosis.
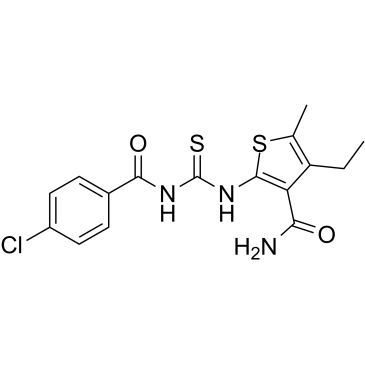
-
GC15310
PI-3065
P110δkinase inhibitor
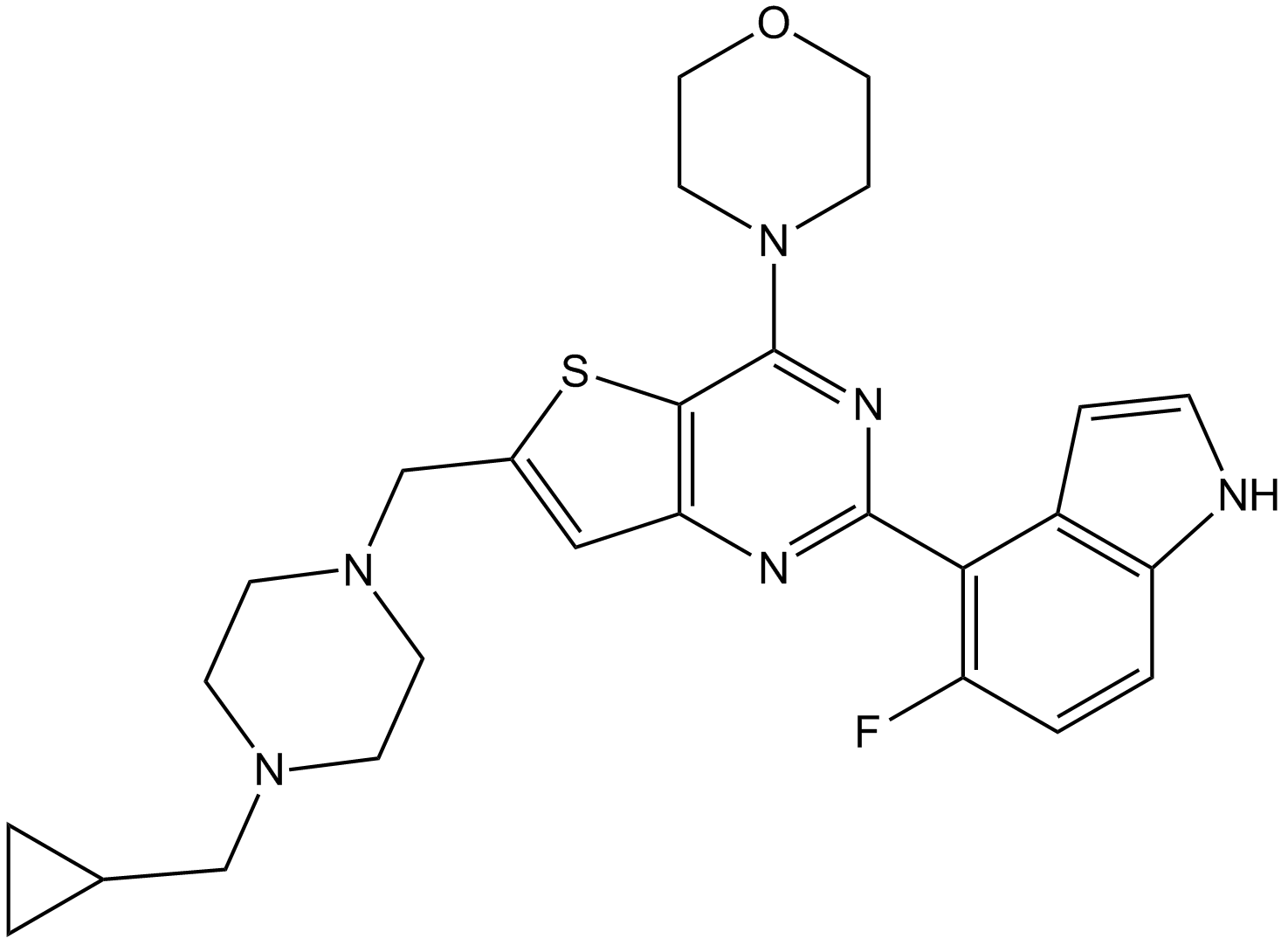
-
GC38578
PI-828
A PI3K inhibitor