Ligand for E3 Ligase
The PROTAC molecule consists of a ligand, which binds an E3 ubiquitin ligase, connected by a linker to another ligand that binds the target protein.
The association between a protein and an E3 ligase, as induced by a PROTAC molecule, will lead to the transfer of ubiquitin and degradation of the targeted protein.
Targets for Ligand for E3 Ligase
Products for Ligand for E3 Ligase
- Cat.No. Product Name Information
-
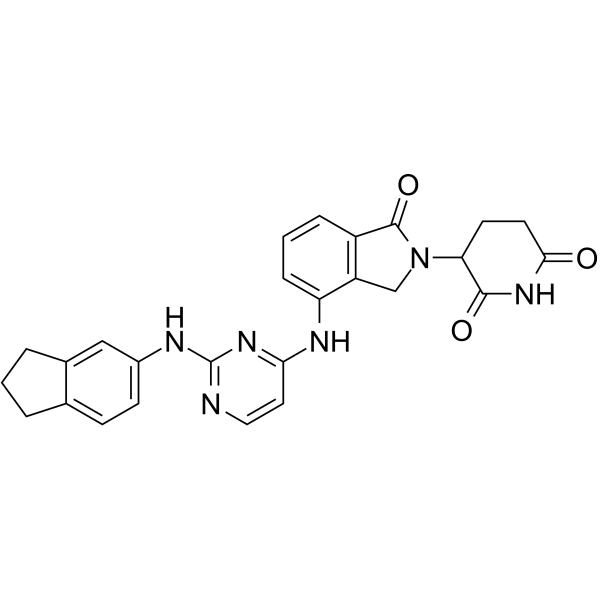
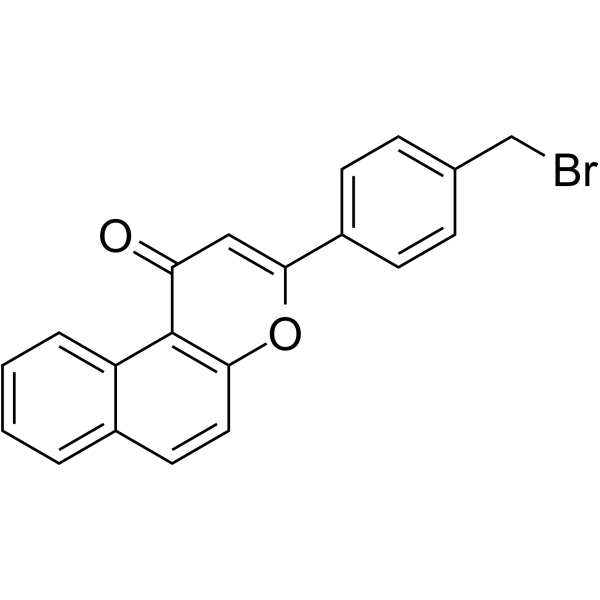
GC70184
β-Naphthoflavone-CH2-Br
Beta-naphthoflavone-CH2-Br is an aryl hydrocarbon receptor (AhR) ligand. It is used in the synthesis of PROTAC Beta-NF-JQ1.
-
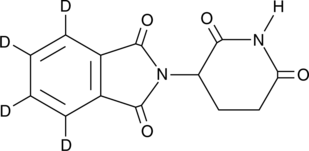
GC46008
(±)-Thalidomide-d4
An internal standard for the quantification of (±)-thalidomide
-
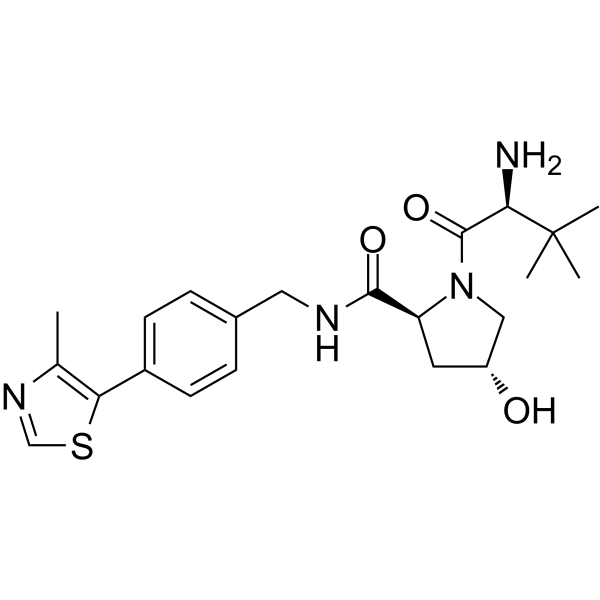
GC62283
(S,R,S)-AHPC
(S,R,S)-AHPC (VH032-NH2) is the VH032-based VHL ligand used in the recruitment of the von Hippel-Lindau (VHL) protein. (S,R,S)-AHPC can be connected to the ligand for protein (e.g., BCR-ABL1) by a linker to form PROTACs (e.g., GMB-475). GMB-475 induces the degradation of BCR-ABL1 with an IC50 of 1.11 μM in Ba/F3 cells.
-
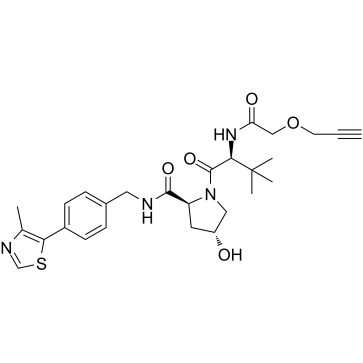
GC38724
(S,R,S)-AHPC-propargyl
(S,R,S)-AHPC-propargyl (VH032-propargyl) is a VHL ligand which is used in “click reaction” for PROTACs.
-
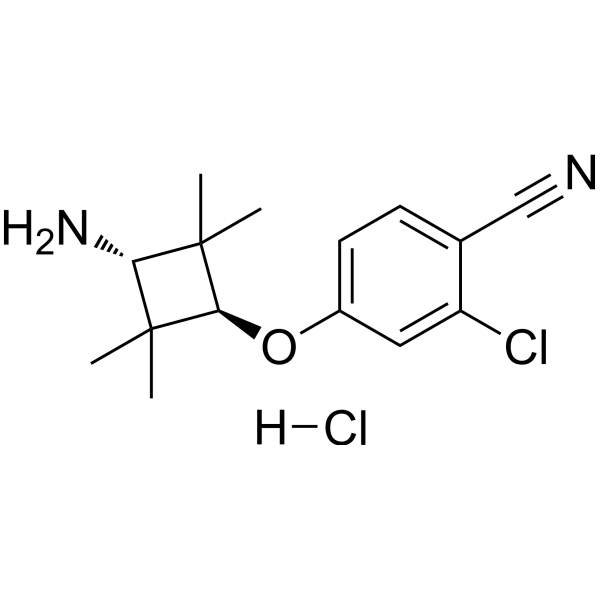
GC68681
AR antagonist 1 hydrochloride
AR antagonist 1 (compound 29) hydrochloride is an effective AR antagonist that binds to E3 ligand and has weak affinity for VHL protein. It is used in the synthesis of PROTAC ARD-266.
-
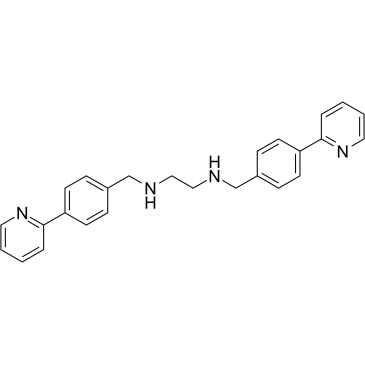
GC38055
BC-1215
An inhibitor of FBXO3
-
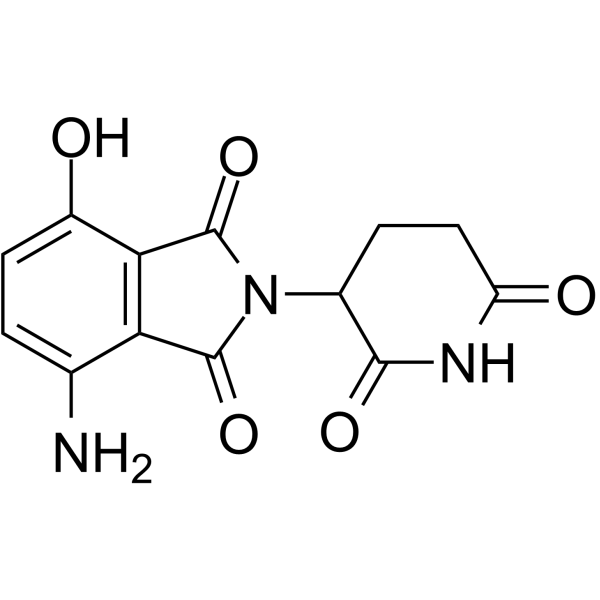
GC66442
CC-17369
CC-17369 (7-Hydroxy pomalidomide) is a metabolite of Pomalidomide. CC-17369 is the Pomalidomide -based cereblon (CRBN) ligand used in the recruitment of CRBN protein. CC-17369 can be connected to the ligand for protein by a linker to form PROTAC.
-
GC18862
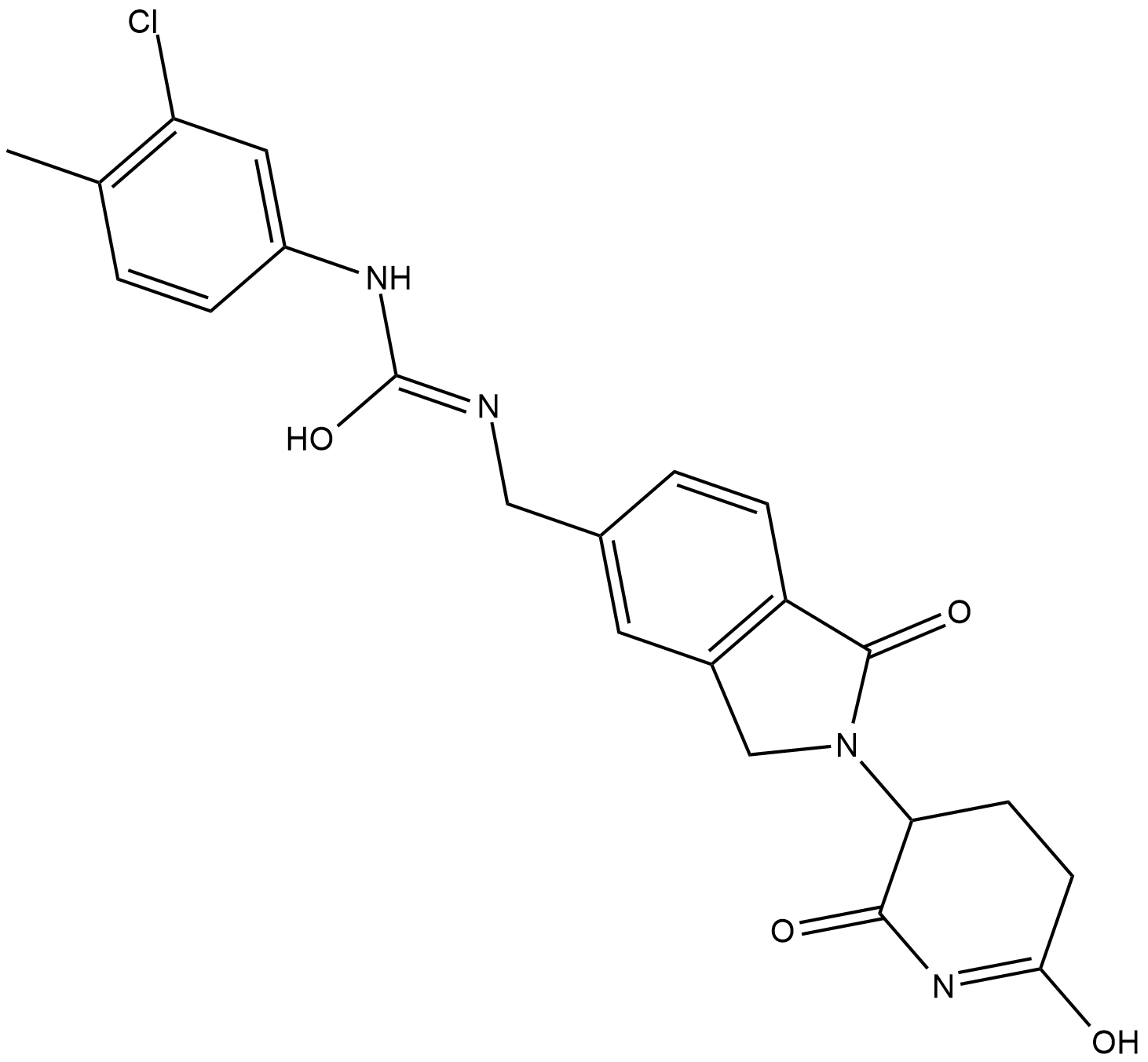
CC-885
CC-885 is a modulator of the E3 ligase protein cereblon with antiproliferative effects on human myeloid leukemia cell lines.
-
GC64851
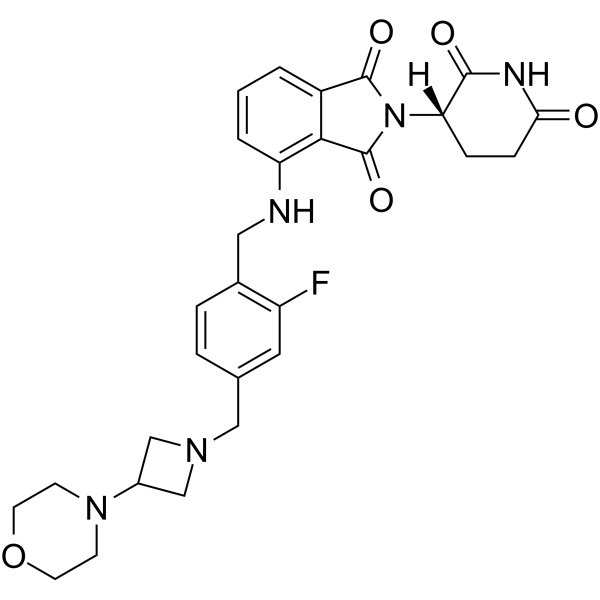
CC-99282
CC-99282 is a potent and orally active cereblon (CRBN) E3 ligase modulator (CELMoD). CC-99282 co-opts CRBN to induce potent and targeted degradation of Ikaros and Aiolos. CC-99282 can be used for researching non-Hodgkin lymphomas.
-
GC64835
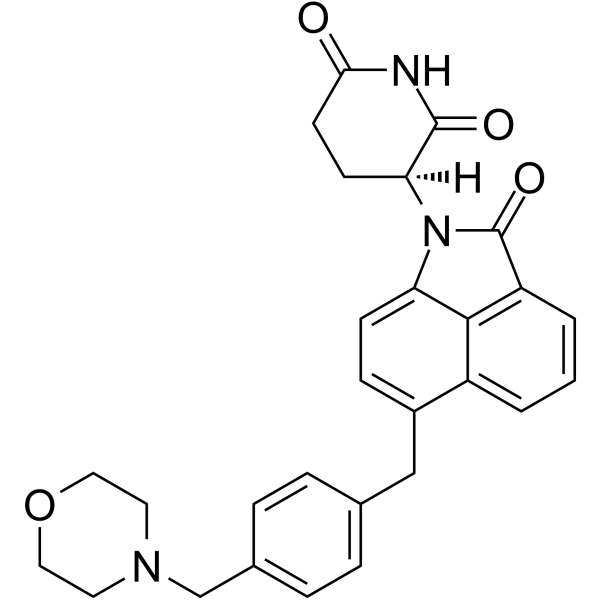
CFT7455
CFT7455 is an orally active zinc finger transcription factors Ikaros (IKZF1), Aiolos (IKZF3) degrader. CFT7455 is an anti-cancer agent that binds with high affinity to the cereblon E3 ligase (Kd of 0.9 nM) (WO2022032132A1; Compound 1).
-
GC50364
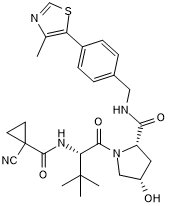
cis VH 298
Negative control for VH 298
-
GC62218
CRBN modulator-1
CRBN modulator-1, a Thalidomide analog and a CRBN modulator extracted from WO2020006262A1, compound 10, has an IC50 of 3.5 μM and a Ki of 0.98 μM.
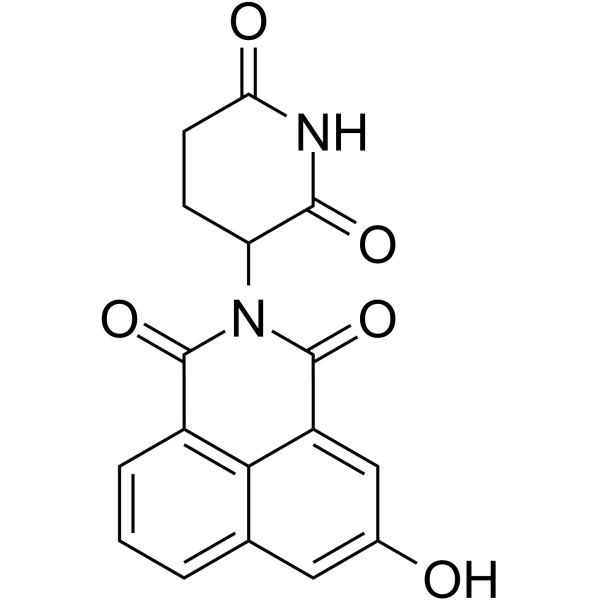
-
GC32711
E3 ligase Ligand 1
E3 ligase Ligand 1 (VHL ligand 2 hydrochloride) is the (S,R,S)-AHPC-based VHL ligand used in the recruitment of the von Hippel-Lindau (VHL) protein. E3 ligase Ligand 1 can be used to synthesize ARV-771, a von Hippel-Landau (VHL) E3 ligase-based BET PROTAC degrader. ARV-771 potently degrades BET protein in castration-resistant prostate cancer (CRPC) cells with a DC50 <1 nM.
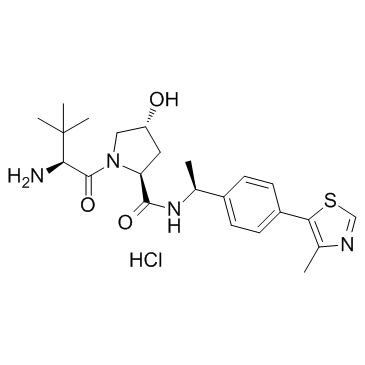
-
GC34291
E3 ligase Ligand 1 dihydrochloride
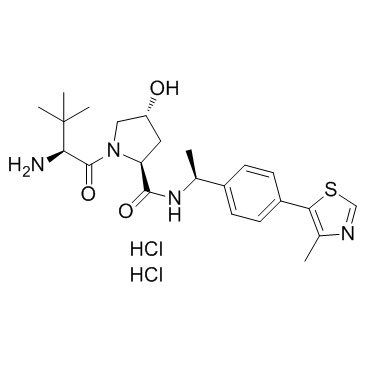
-
GC35924
E3 ligase Ligand 10
E3 ligase Ligand 10 is a ligand for E3 ubiquitin ligase. E3 ligase Ligand 10 can be connected to the ligand for protein by a linker to form PROTACs. PROTACs are inducers of ubiquitination-mediated degradation of cancer-promoting proteins.
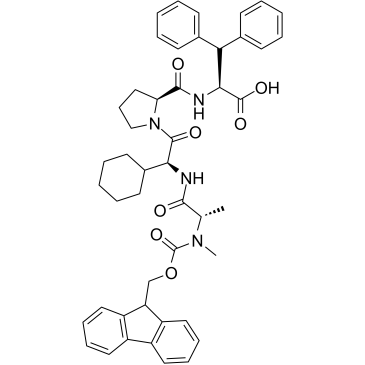
-
GC35925
E3 ligase Ligand 11
E3 ligase Ligand 11 is the LCL161 derivative based IAP ligand. E3 ligase Ligand 11 can be connected to the ABL ligand for protein by a linker to form SNIPER.
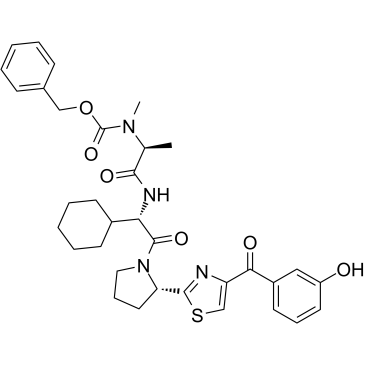
-
GC35926
E3 ligase Ligand 12
E3 ligase Ligand 12 is the LCL161 derivative based IAP ligand. E3 ligase Ligand 12 can be connected to the ABL ligand for protein by a linker to form SNIPER.
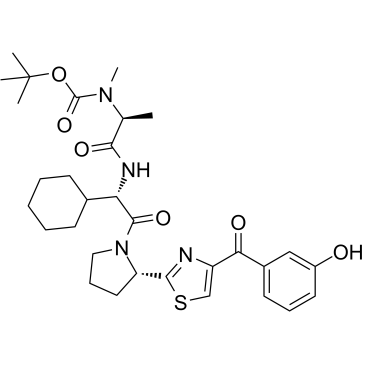
-
GC35927
E3 ligase Ligand 13
E3 ligase Ligand 13 is a ligand for E3 ubiquitin ligase. E3 ligase Ligand 13 can be connected to the ligand for protein by a linker to form PROTACs. PROTACs are inducers of ubiquitination-mediated degradation of cancer-promoting proteins.
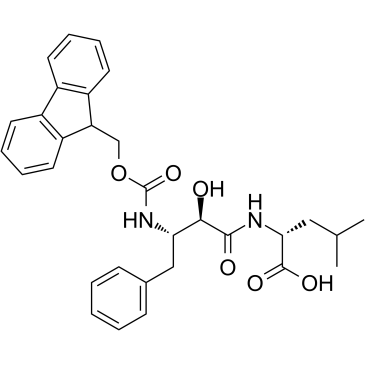
-
GC35928
E3 ligase Ligand 14
E3 ligase Ligand 14 is a ligand for E3 ubiquitin ligase. E3 ligase Ligand 14 can be connected to the ligand for protein by a linker to form PROTACs. PROTACs are inducers of ubiquitination-mediated degradation of cancer-promoting proteins.
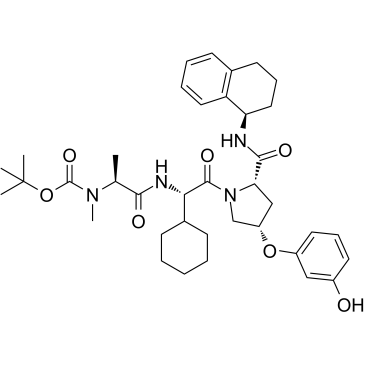
-
GC35929
E3 ligase Ligand 15
E3 ligase Ligand 15 (MDM2 ligand 2) is the Nutlin 3-based MDM2 ligand. E3 ligase Ligand 15 can be connected to the ligand for protein by a linker to form PROTACs.
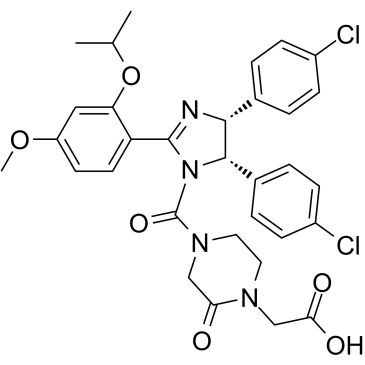
-
GC35930
E3 ligase Ligand 16
E3 ligase Ligand 16 (MDM2 ligand 1) is the Nutlin 3-based MDM2 ligand. E3 ligase Ligand 16 can be connected to the ligand for protein by a linker to form PROTAC.
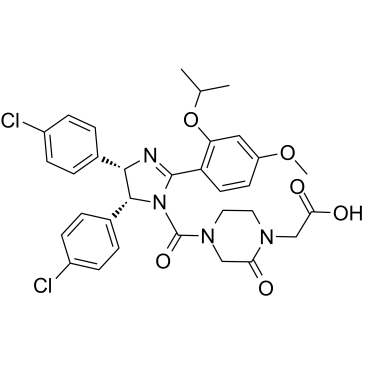
-
GC35932
E3 ligase Ligand 18
E3 ligase Ligand 18 is a ligand for E3 ubiquitin ligase. E3 ligase Ligand 18 can be connected to the ligand for protein by a linker to form PROTACs. PROTACs are inducers of ubiquitination-mediated degradation of cancer-promoting proteins.
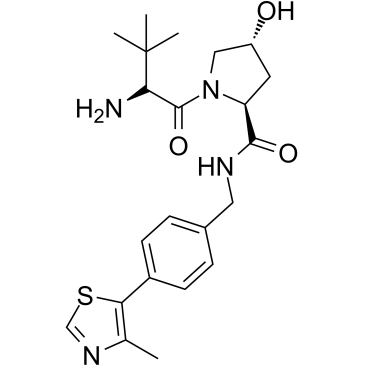
-
GC33101
E3 ligase Ligand 1A
E3 ligase Ligand 1A (VHL ligand 2) is the (S,R,S)-AHPC-based VHL ligand used in the recruitment of the von Hippel-Lindau (VHL) protein. E3 ligase Ligand 1A can be used to synthesize ARV-771, a von Hippel-Landau (VHL) E3 ligase-based BET PROTAC degrader. ARV-771 potently degrades BET protein in castration-resistant prostate cancer (CRPC) cells with a DC50 <1 nM.
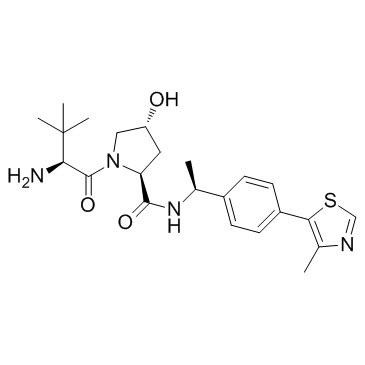
-
GC32848
E3 ligase Ligand 2
E3 ligase Ligand 2 (Cereblon ligand 2) is the Thalidomide-based Cereblon ligand used in the recruitment of CRBN protein. E3 ligase Ligand 2 (Cereblon ligand 2) can be connected to the ligand for protein by a linker to form PROTACs.
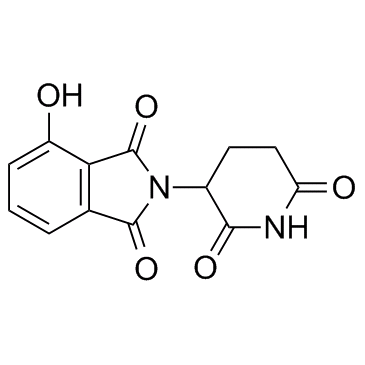
-
GC30029
E3 ligase Ligand 3
E3 ligase Ligand 3 (Cereblon ligand 3) is the Thalidomide-based Cereblon ligand used in the recruitment of CRBN protein.
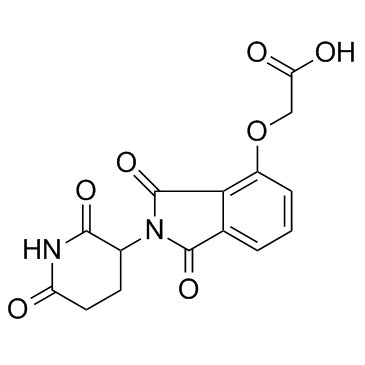
-
GC30164
E3 ligase Ligand 4
E3 ligase Ligand 4 (Cereblon ligand 4) is the Thalidomide-based Cereblon ligand used in the recruitment of CRBN protein. E3 ligase Ligand 4 (Cereblon ligand 4) can be connected to the ligand for IRAK4 protein by a linker to form PROTAC IRAK4 degrader-1.
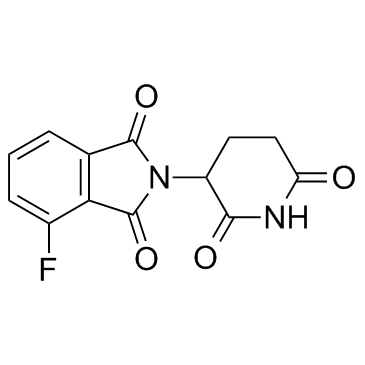
-
GC35933
E3 ligase Ligand 8
E3 ligase Ligand 8 is a ligand for E3 ubiquitin ligase. E3 ligase Ligand 8 can be connected to the ligand for protein by a linker to form PROTACs. PROTACs are inducers of ubiquitination-mediated degradation of cancer-promoting proteins.
-
GC34566
E3 ligase Ligand 9
E3 ligase Ligand 9 is a ligand for E3 ubiquitin ligase. E3 ligase Ligand 9 can be connected to the ligand for protein by a linker to form PROTACs or SNIPERs. PROTACs are inducers of ubiquitination-mediated degradation of cancer-promoting proteins.
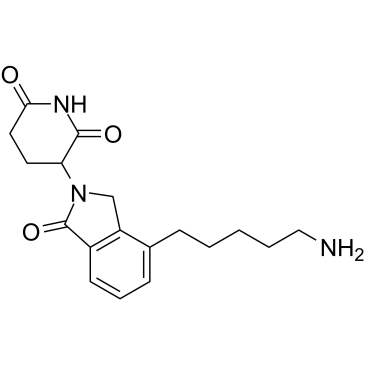
-
GC19195
Iberdomide
Iberdomide (CC-220) is a cereblon modulator with an IC50 of 60 nM.
-
GC60963
KB02-COOH
KB02-COOH is a fragment of synthesis of ubiquitin E3 ligase ligand KB02. KB02 can be used in the synthesis of PROTAC, such as KB02-JQ1 and KB02-SLF.
-
GC14976
Lenalidomide (CC-5013)
An analog of thalidomide
-
GC13473
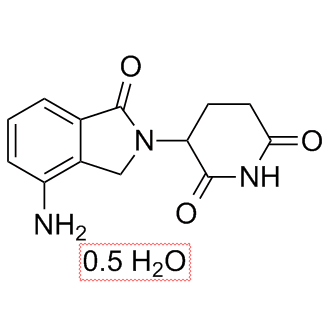
Lenalidomide hemihydrate
TNF-α secretion inhibitor
-
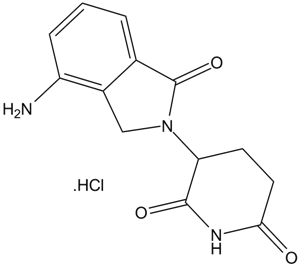
GC14015
Lenalidomide hydrochloride
An analog of thalidomide
-
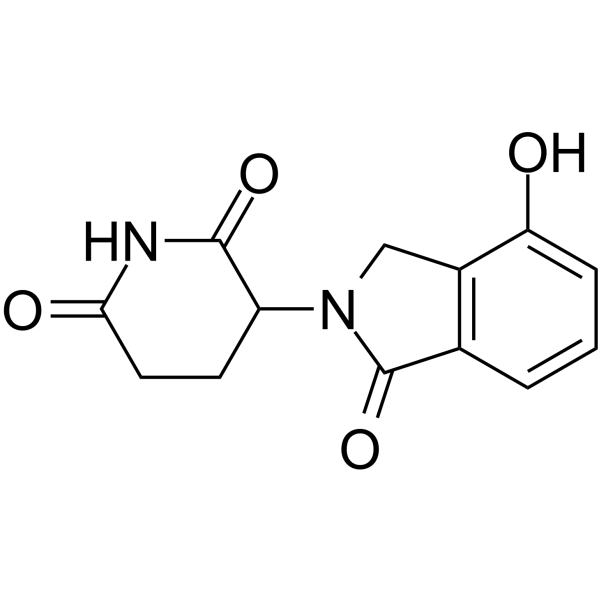
GC62624
Lenalidomide-4-OH
Lenalidomide-4-OH is the Lenalidomide-based cereblon (CRBN) ligand used in the recruitment of CRBN protein. Lenalidomide-4-OH can be connected to the ligand for protein by a linker to form PROTAC.
-
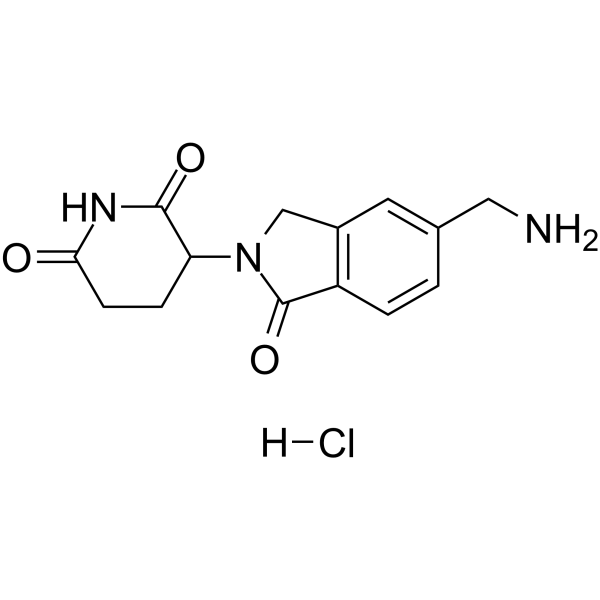
GC63806
Lenalidomide-5-aminomethyl hydrochloride
Lenalidomide-5-aminomethyl hydrochloride is the Lenalidomide-based cereblon (CRBN) ligand used in the recruitment of CRBN protein. Lenalidomide-5-aminomethyl hydrochloride can be connected to the ligand for protein by a linker to form PROTAC
-
GC62541
Lenalidomide-5-Br
Lenalidomide-5-Br is the Lenalidomide-based cereblon (CRBN) ligand used in the recruitment of CRBN protein. Lenalidomide-5-Br can be connected to the ligand for protein by a linker to form PROTAC.
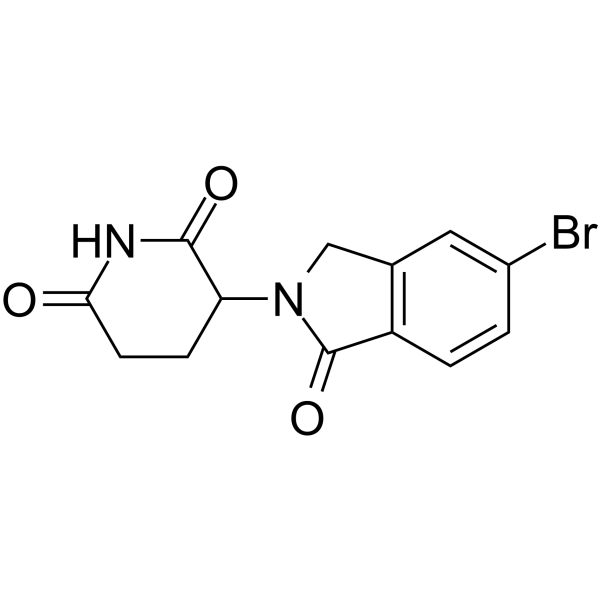
-
GC38579
Lenalidomide-Br
Lenalidomide-Br (Compound 41) is an analog of cereblon (CRBN) ligand Lenalidomide for E3 ubiquitin ligase, and is used in the recruitment of CRBN protein. Lenalidomide-Br can be connected to the ligand for protein by a linker to form PROTACs, such as the PROTAC STAT3 degrader SD-36.
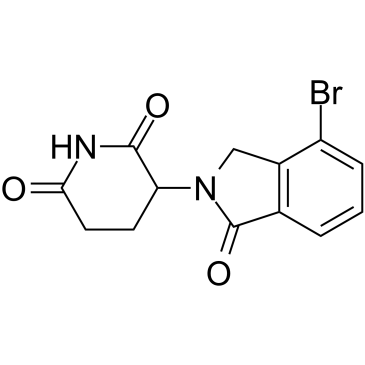
-
GC62147
Lenalidomide-I
Lenalidomide-I (Compound 72), an analog of cereblon (CRBN) ligand Lenalidomide for E3 ubiquitin ligase, is used in the recruitment of CRBN protein. Lenalidomide-I can be connected to the ligand for protein by a linker to form PROTACs, such as the PROTAC BET degrader QCA570.
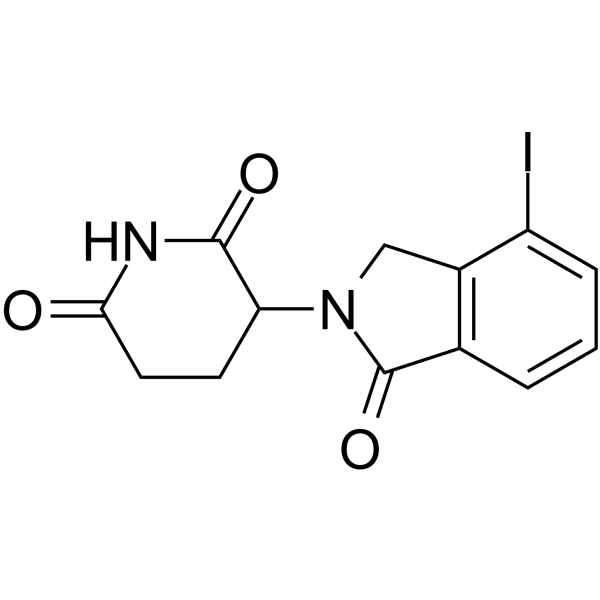
-
GC60223
Lenalidomide-OH
Lenalidomide-OH is an analog of cereblon (CRBN) ligand Lenalidomide for E3 ubiquitin ligase, and is used in the recruitment of CRBN protein. Lenalidomide-OH can be connected to the ligand for protein by a linker to form PROTACs, such as the PROTAC BTK degrader SJF620.
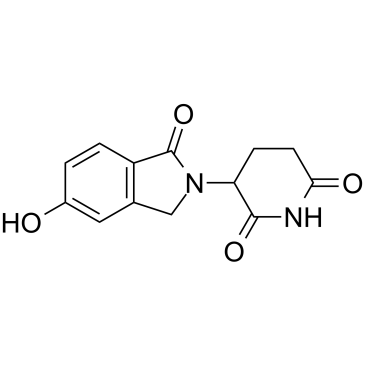
-
GC11697
Pomalidomide (CC-4047)
An inhibitor of cereblon
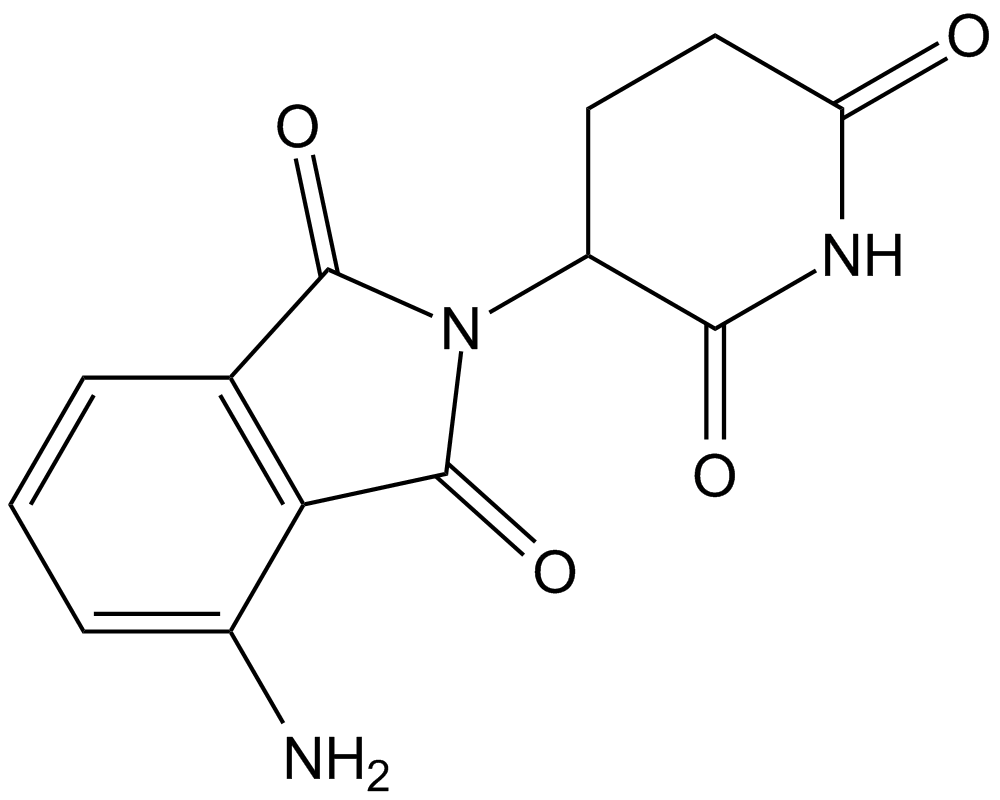
-
GC19302
Protein degrader 1 hydrochloride
(S,R,S)-AHPC (VH032-NH2) hydrochloride is the VH032-based VHL ligand used in the recruitment of the von Hippel-Lindau (VHL) protein. Protein degrader 1 hydrochloride can be connected to the ligand for protein (e.g., BCR-ABL1) by a linker to form PROTACs (e.g., GMB-475). GMB-475 induces the degradation of BCR-ABL1 with an IC50 of 1.11 μM in Ba/F3 cells.
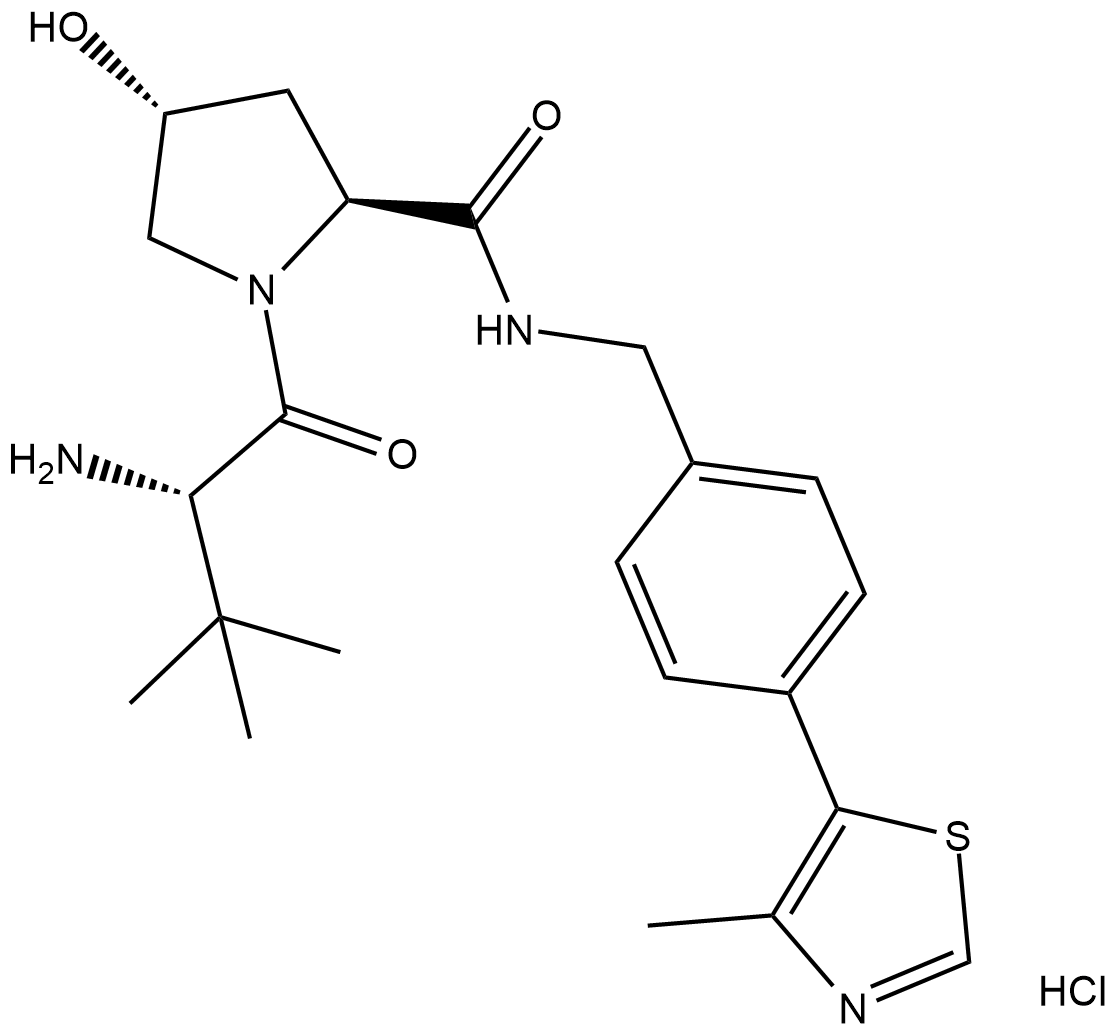
-
GC32433
Protein degrader 1 TFA
(S,R,S)-AHPC (VH032-NH2) TFA is the VH032-based VHL ligand used in the recruitment of the von Hippel-Lindau (VHL) protein. Protein degrader 1 TFA can be connected to the ligand for protein (e.g., BCR-ABL1) by a linker to form PROTACs (e.g., GMB-475). GMB-475 induces the degradation of BCR-ABL1 with an IC50 of 1.11 μM in Ba/F3 cells.
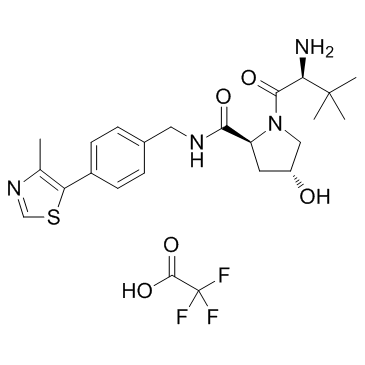
-
GC69906
SJPYT-195
SJPYT-195 is a cell-toxic degradation product of GSPT1 that can be used for synthesizing PROTAC.
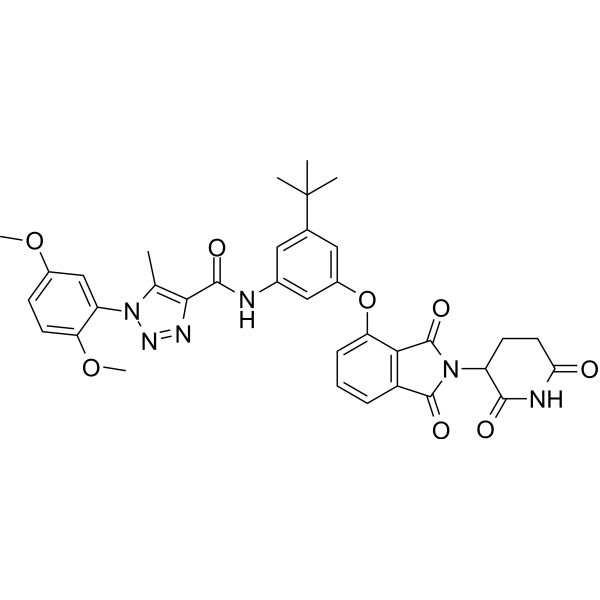
-
GC33068
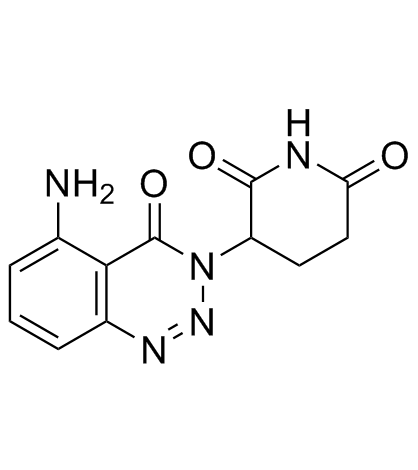
TD-106
TD-106 is a cereblon (CRBN) modulator, which can be used for targeted protein degradation. BRD4 PROTACs with TD-106 induce BRD4 degradation.
-
GC17054
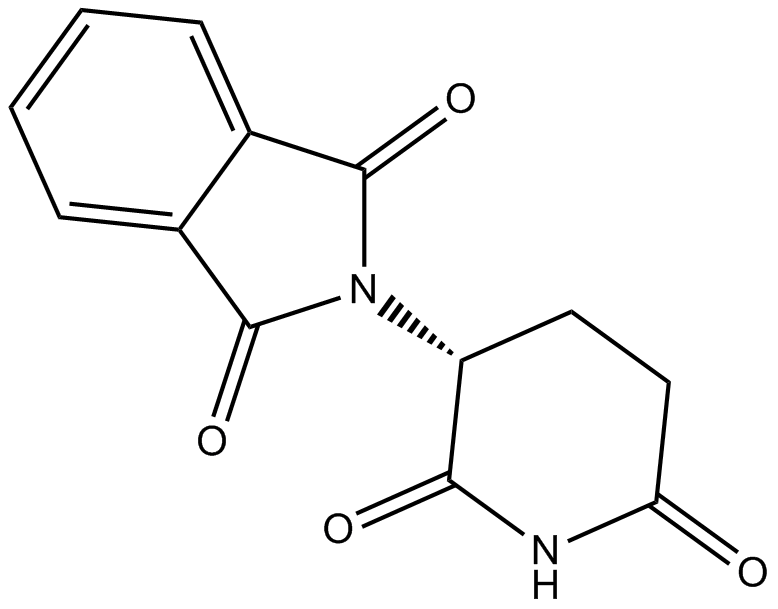
Thalidomide
An immunomodulatory compound with diverse biological activities
-
GC60360
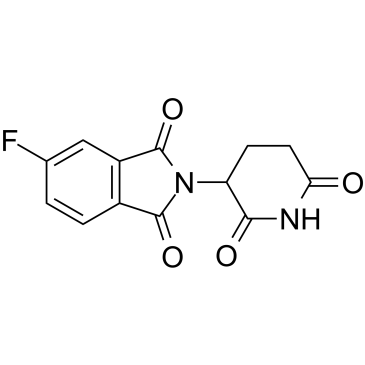
Thalidomide 5-fluoride
Thalidomide 5-fluoride is Thalidomide-based cereblon ligand?that incorporates to the ligand for IRAK4 protein by a linker to form PROTAC IRAK4 degrader-1.
-
GC63392
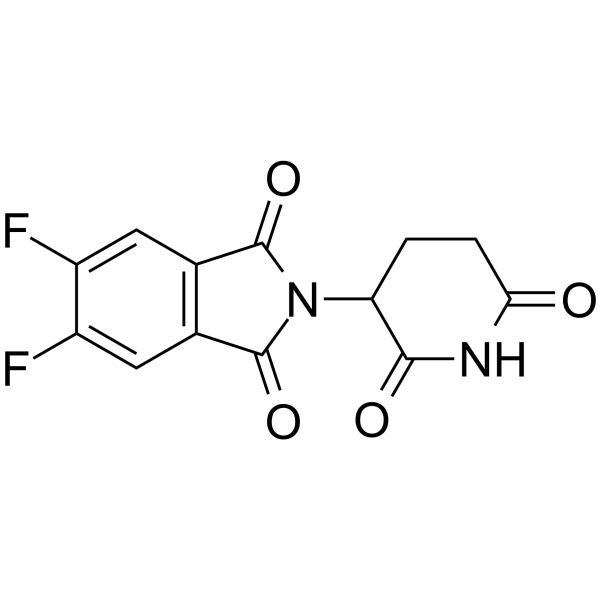
Thalidomide-5,6-F
Thalidomide-5,6-F is the Thalidomide-based cereblon ligand used in the recruitment of CRBN protein. Thalidomide-5,6-F can be connected to the ligand for protein by a linker to form PROTACs.
-
GC63810
Thalidomide-5-NH2-CH2-COOH
Thalidomide-5-NH2-CH2-COOH (compound 114) is a potent and selective inhibitor of tropomyosin receptor kinase (trk).
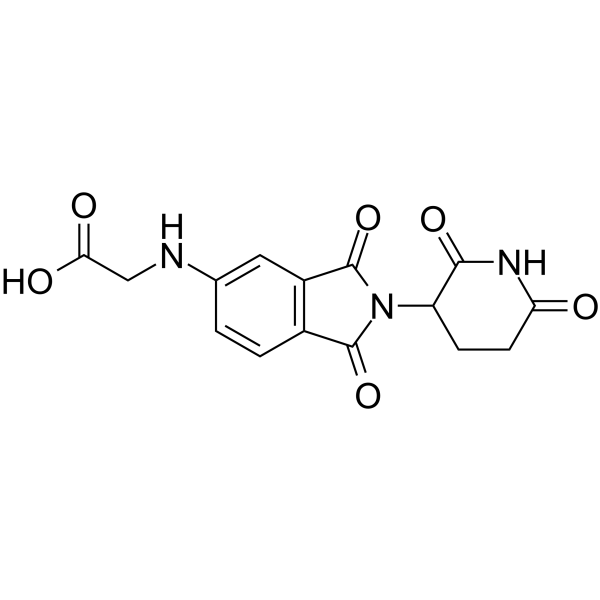
-
GC60361
Thalidomide-5-OH
Thalidomide-5-OH is the Thalidomide-based cereblon ligand used in the recruitment of CRBN protein. Thalidomide-5-OH can be connected to the ligand for protein by a linker to form PROTACs.
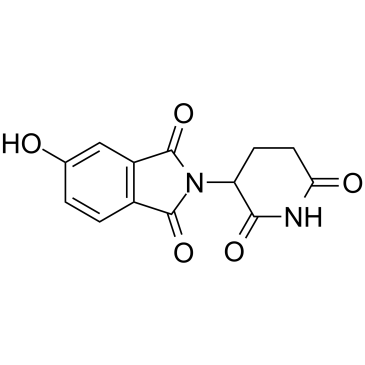
-
GC62378
Thalidomide-NH-CH2-COOH
Thalidomide-NH-CH2-COOH is the Thalidomide-based cereblon ligand used in the recruitment of CRBN protein. Thalidomide-NH-CH2-COOH can be connected to the ligand for protein by a linker to form PROTACs, such as THAL-SNS-032.
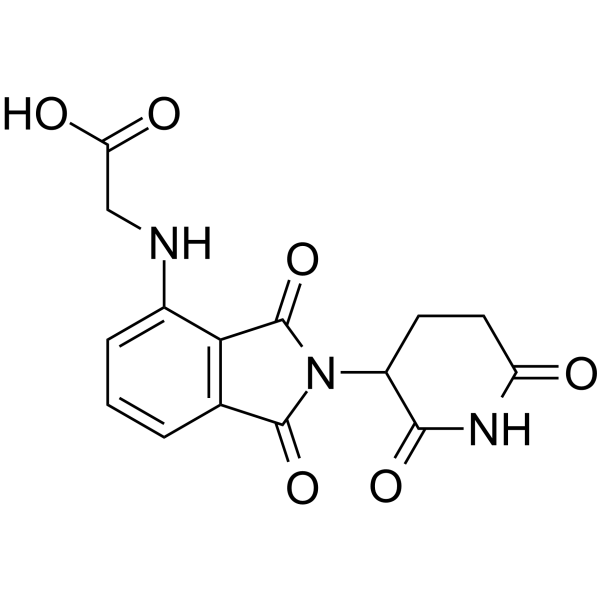
-
GC60362
Thalidomide-O-C8-Boc
Thalidomide-O-C8-Boc is the Thalidomide-based Cereblon ligand used in the recruitment of CRBN protein. Thalidomide-O-C8-Boc can be connected to the ligand for protein by a linker to form PROTACs.
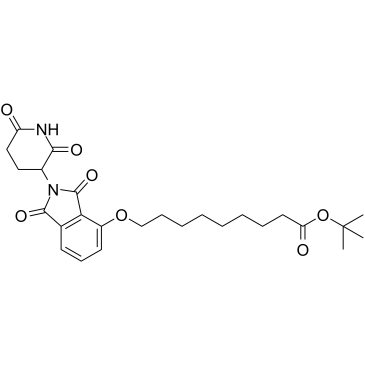
-
GC60363
Thalidomide-O-C8-COOH
Thalidomide-O-C8-COOH is the Thalidomide-based Cereblon ligand used in the recruitment of CRBN protein. Thalidomide-O-C8-COOH (Cereblon ligand 3) can be connected to the ligand for protein by a linker to form PROTACs.
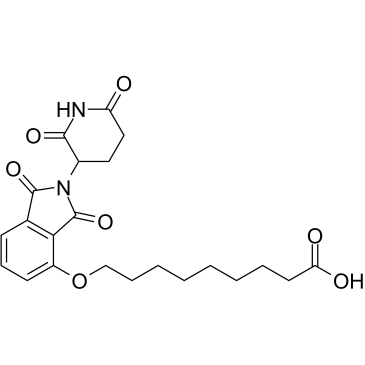
-
GC38862
Thalidomide-propargyl
Thalidomide-propargyl is the Thalidomide-based Cereblon ligand used in the recruitment of CRBN protein. Thalidomide-propargyl can be connected to the ligand for protein by a linker to form the IMiD containing PROTACs.
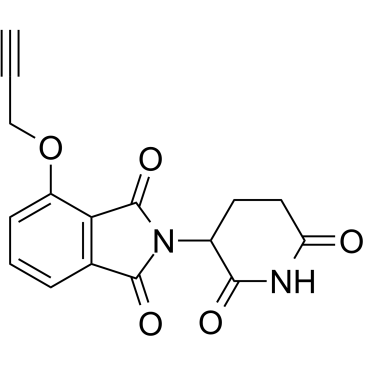
-
GC50623
VH 101, phenol
VH 101, phenol is the VH032-based VHL ligand. VH 101, phenol can be connected to the ligand for protein (e.g., SMARCA BD ligand) by a linker to form PROTACs (e.g., PROTAC 1). PROTAC 1 is a partial degrader of SMARCA2 and SMARCA4.
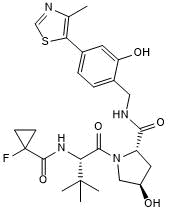
-
GC19375
VH-298
VH-298 is a highly potent inhibitor of the VHL:HIF-α interaction with a Kd value of 80 to 90 nM, used in PROTAC technology.
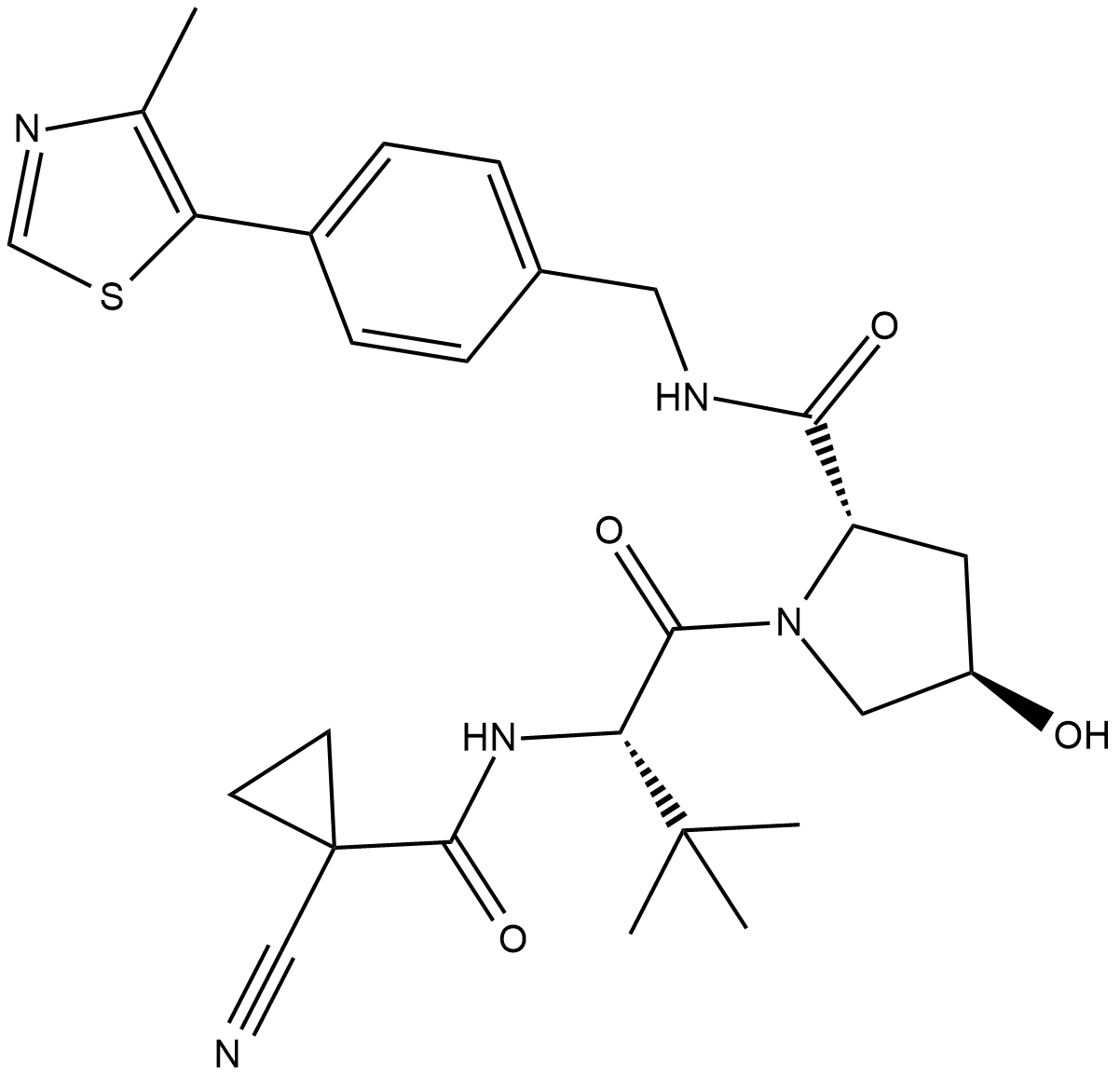
-
GC61452
VH032
VH032 is a VHL ligand used in the recruitment of the von Hippel-Lindau (VHL) protein. VH032 is a VHL/HIF-1α interaction inhibitor with a Kd
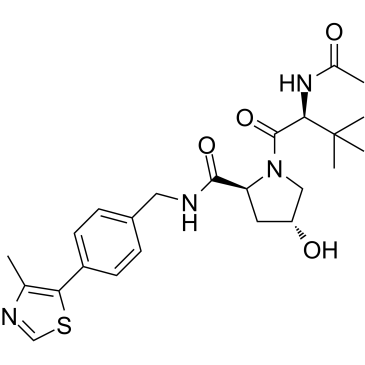
-
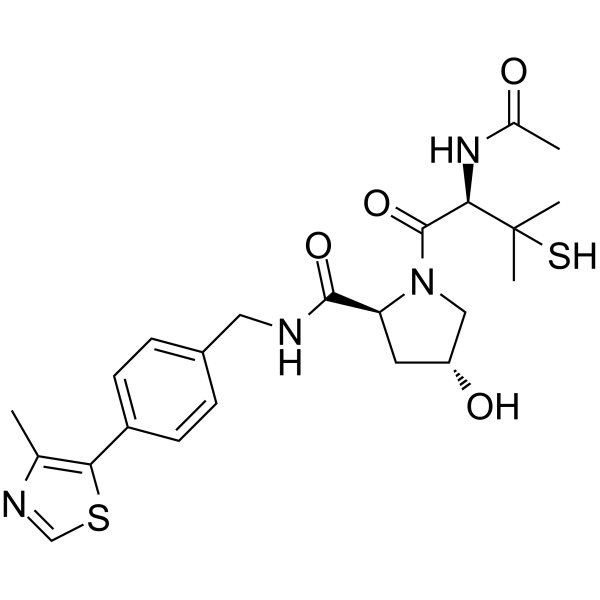
GC65392
VH032 thiol
VH032 thiol (VHL ligand 6) is a VHL ligand, which binds to pan-BET inhibitor JQ1 via a linker to form PROTAC.
-
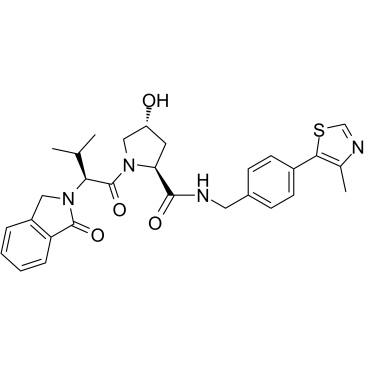
GC34855
VL285
VL285 is a potent VHL ligand with an IC50 of 0.34 μM.
-
GC64969
ZXH-1-161
ZXH-1-161 is a potent cereblon (CRBN) modulator with an IC50 value of 39 nM in MM1.S wild type cells. ZXH-1-161 has selective degradation activity towards GSPT1. ZXH-1-161 can be used for researching multiple myeloma.