Lipoxygenase
Lipoxygenases (LOXs) are dioxygenases that catalyze the formation of corresponding hydroperoxides from polyunsaturated fatty acids such as linoleic acid and arachidonic acid. Thre are six LOX isoforms have been found in the humans and mice. 5-Lipoxygenase (5-LOX) is a distinct isoform playing an important role in asthma and inflammation. 5-LOX causes the constriction of bronchioles in response to cysteinyl leukotrienes such as LTC4, thus leading to asthma. 5-LOX also induces neutrophilic inflammation by its recruitment in response to LTB4. 12-Lipoxygenase (12-LOX) is an isoform expressed in epithelial cells and myeloid cells including platelets. 12-LOX can be found in the epithelial cells of the skin. 12-LOX is a potential target for novel anti-platelet therapeutics.15-Lipoxygenase (15-LOX) is expressed in epithelial cells and leukocytes, has different substrate specificity in the humans and mice.15-LOX-1 is a target of nonsteroidal anti-inflammatory drug-induced apoptosis in colorectal cancer cells.
Targets for Lipoxygenase
Products for Lipoxygenase
- Cat.No. Product Name Information
-
GC41136
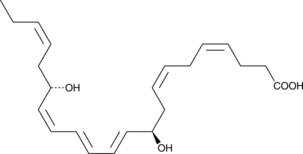
8(S),15(S)-DiHETE
8(S),15(S)-DiHETE is formed when 15(S)-HETE is subjected to further oxidation by 15-LO.
-
GC40382
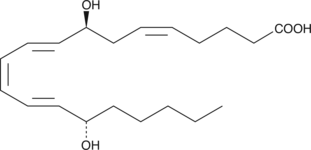
8(S)-HEPE
8(S)-HEPE is a monohydroxy fatty acid produced by lipoxygenase oxidation of EPA.
-
GC40463
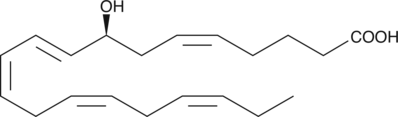
8(S)-HETE
8(S)-HETE is a major lipoxygenase product in PMA-treated murine epidermis.
-
GC42619
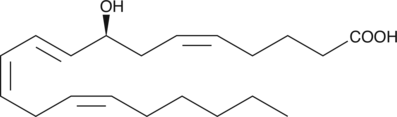
8(S)-HETrE
8(S)-HETrE is a monohydroxy polyunsaturated fatty acid produced by rabbit neutrophil lipoxygenase when dihomo-γ-linolenic acid is used as a substrate.

-
GC40542
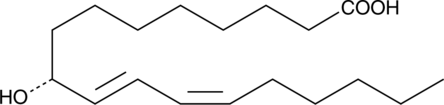
9(R)-HODE
9(R)-HODE is one of several monohydroxylated products of linoleic acid.
-
GC40029
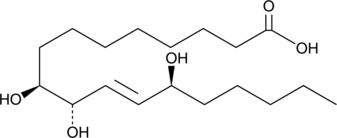
9(S),10(S),13(S)-TriHOME
9(S),10(S),13(S)-TriHOME is an oxylipin derived from linoleic acid.
-
GC46753
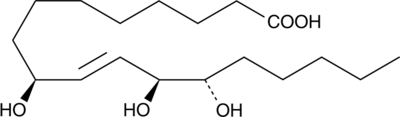
9(S),12(S),13(S)-TriHOME
An oxylipin
-
GC40383
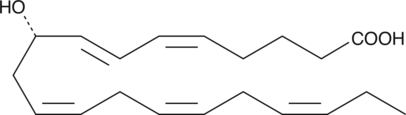
9(S)-HEPE
9(S)-HEPE is a monohydroxy fatty acid derived from EPA.
-
GC19460
9(S)-HODE
9(S)-HODE is produced by the lipoxygenation of linoleic acid in both plants and animals.

-
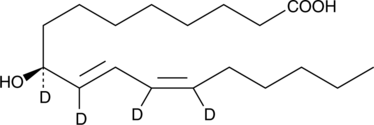
GC40250
9(S)-HODE-d4 MaxSpec® Standard
9(S)-HODE-d4 is intended for use as an internal standard for the quantification of 9(S)-HODE by GC- or LC-mass spectrometry.
-
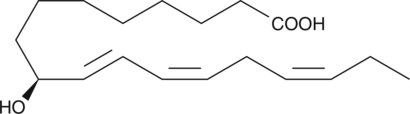
GC42636
9(S)-HOTrE
9(S)-HOTrE is a monohydroxy polyunsaturated fatty acid produced by the action of 5-lipoxygenase on α-linolenic acid.
-
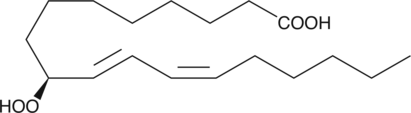
GC40357
9(S)-HpODE
9(S)-HpODE is produced by the action of arachidonate 5-LO on linoleic acid.
-
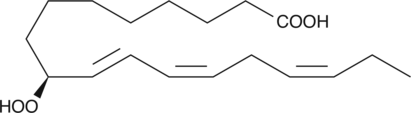
GC42637
9(S)-HpOTrE
9(S)-HpOTrE is a monohydroperoxy polyunsaturated fatty acid produced by the action of 5-lipoxygenase (5-LO) on α-linolenic acid.
-
GC42651
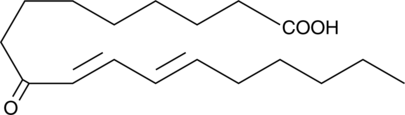
9-oxo-10(E),12(E)-Octadecadienoic Acid
9-oxo-10(E),12(E)-Octadecadienoic acid (9-oxoODA) is a natural agonist, abundant in tomatoes, that activates PPARα at 10-20 μM.
-
GC42653
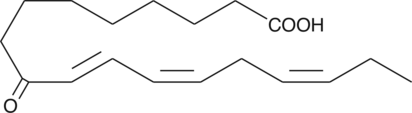
9-OxoOTrE
9-OxoOTrE is produced by the oxidation of 9-HpOTrE.
-
GC45960
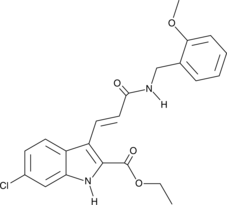
9c(i472)
9c(i472) is a potent inhibitor of 15-LOX-1 (15-lipoxygenase-1) with an IC50 value of 0.19 μM.
-
GC31938
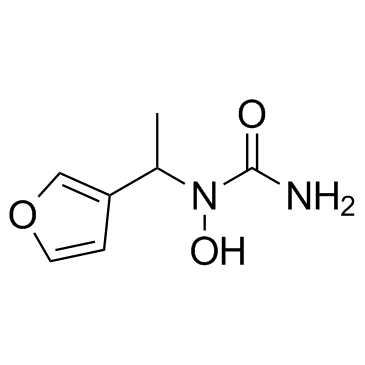
A-69412
A-69412 is a reversible, specific inhibitor of the 5-lipoxygenase (5-LO).
-
GC46798
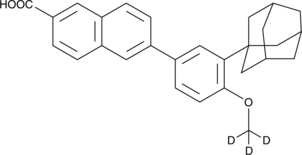
Adapalene-d3
An internal standard for the quantification of adapalene
-
GC41211
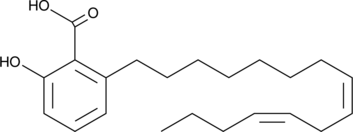
Anacardic Acid Diene
Anacardic acid diene is a polyunsaturated form of anacardic acid that has been found in cashew nut shell liquid.
-
GC41531
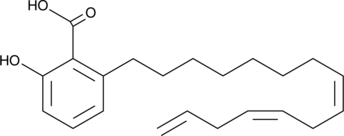
Anacardic Acid Triene
Anacardic acid triene is a polyunsaturated form of anacardic acid that has been found in cashew nut shell liquid.
-
GC35425
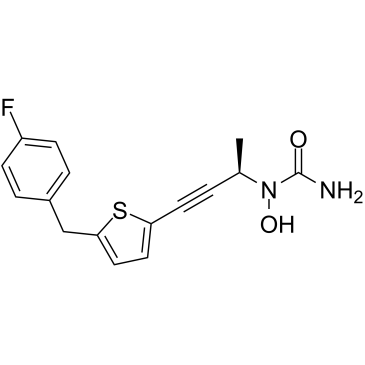
Atreleuton
A 5-LO inhibitor
-
GC32035
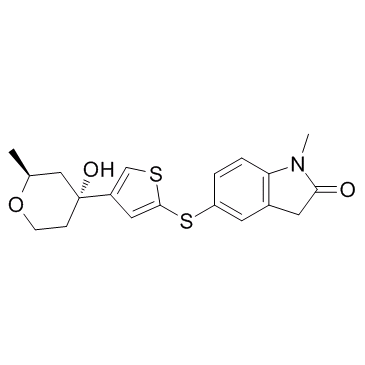
AZD 4407 (ZD 4407)
AZD 4407 (ZD 4407) is a potent 5-lipoxygenase inhibitor.
-
GC45951
BLX3887
A 15-LO-1 inhibitor
-
GC32002
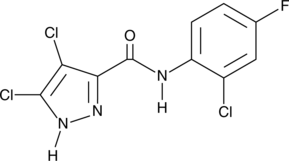
Bunaprolast (U66858)
Bunaprolast (U66858) (U66858) is a potent inhibitor of LTB4 production in human whole blood.
-
GC46105
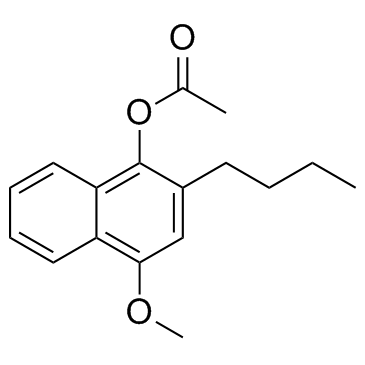
Butyrolactone II
A fungal metabolite
-
GN10792
Caffeic acid
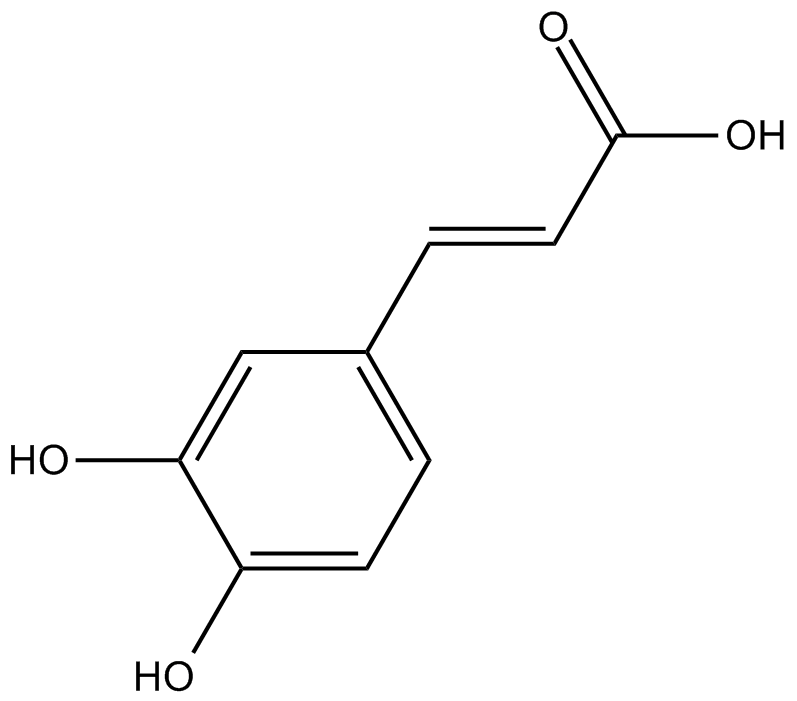
-
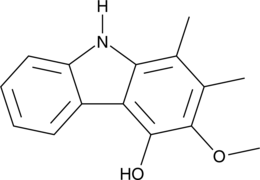
GC48893
Carbazomycin B
A bacterial metabolite with diverse biological activities
-
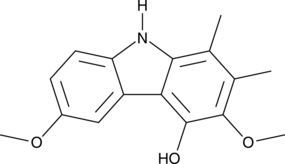
GC48850
Carbazomycin C
A bacterial metabolite with diverse biological activities
-
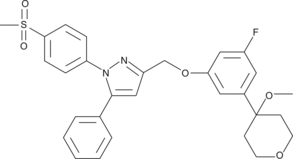
GC43153
CAY10416
Dual cyclooxygenase-2 (COX-2)/5-lipoxygenase (5-LO) inhibitors are potential therapeutic agents for inflammatory diseases and for prostate cancer.
-
GC41044
CAY10583
Leukotriene B4 (LTB4) promotes a number of leukocyte functions including aggregation, stimulation of ion fluxes, superoxide anion production, chemotaxis, and chemokinesis.
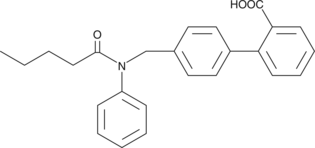
-
GC18688
CAY10589
Microsomal Prostaglandin E2 Synthase-1 (mPGES-1), with cyclooxygenase-2 (COX-2), synthesizes PGE2, which is directly involved in signaling during inflammation, fever and pain.
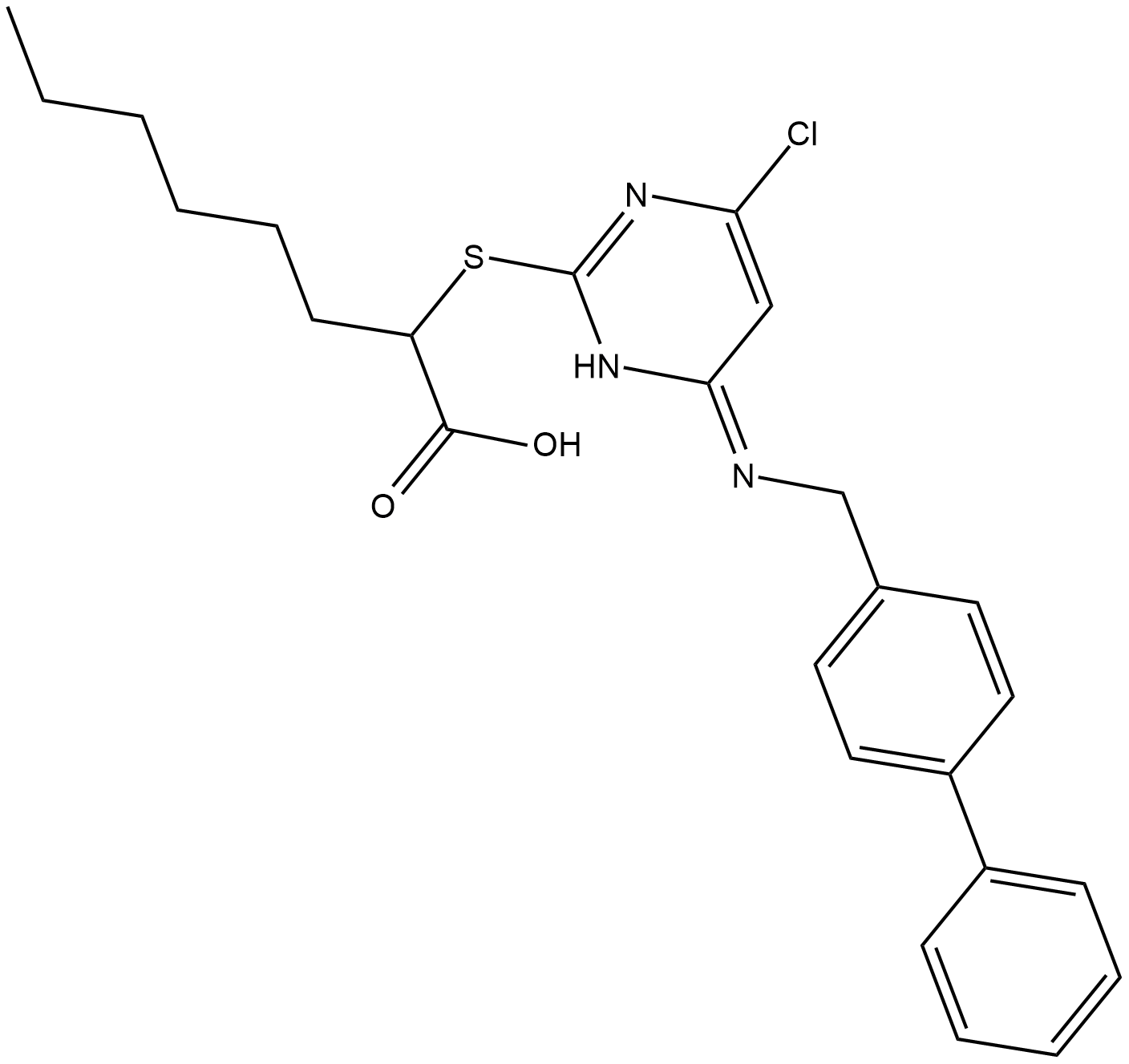
-
GC18877
CAY10606
5-Lipoxygenase (5-LO) initiates the synthesis of leukotrienes (LTs) from arachidonic acid, primarily in certain leukocyte populations.
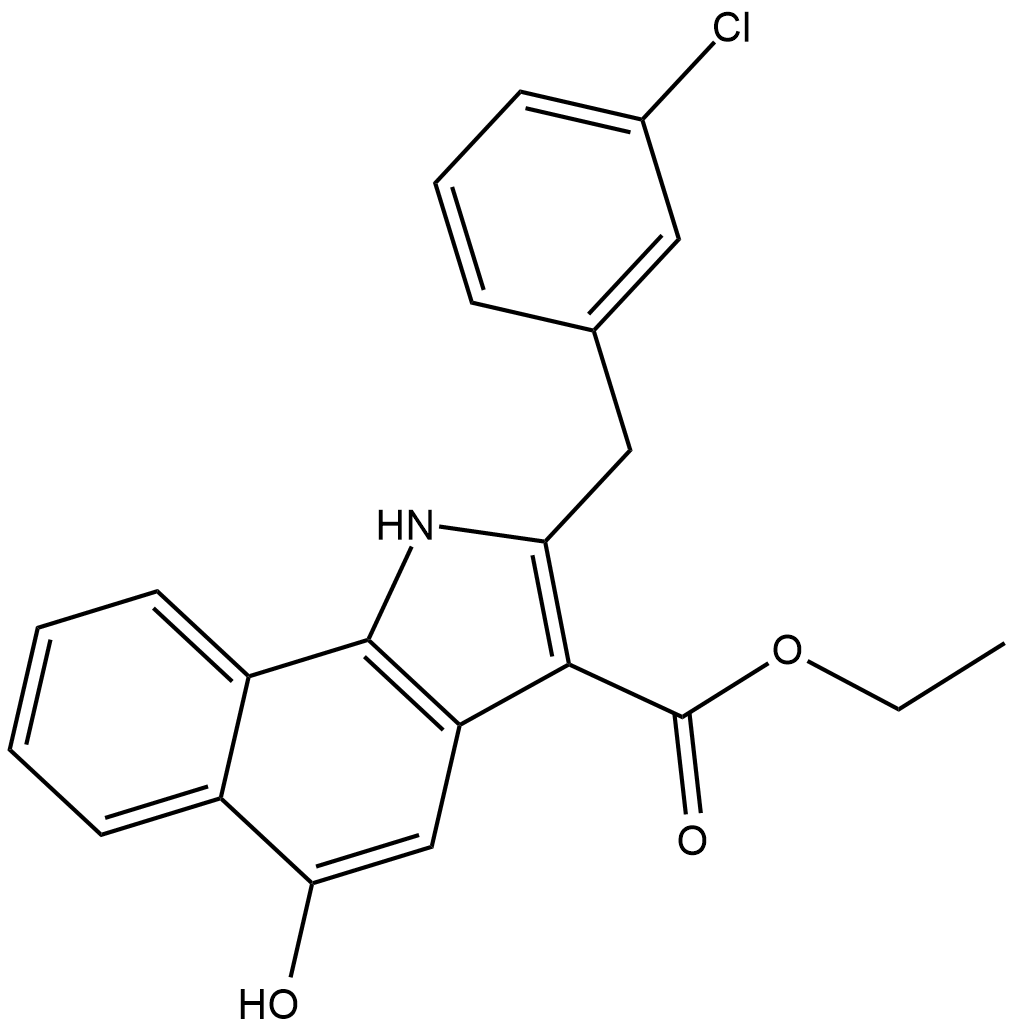
-
GC18480
CAY10649
5-Lipoxygenase (5-LO) catalyzes the biosynthesis of leukotrienes, which are involved in a variety of inflammatory responses, including neutrophil chemotaxis, vascular permeability, and smooth muscle contraction.
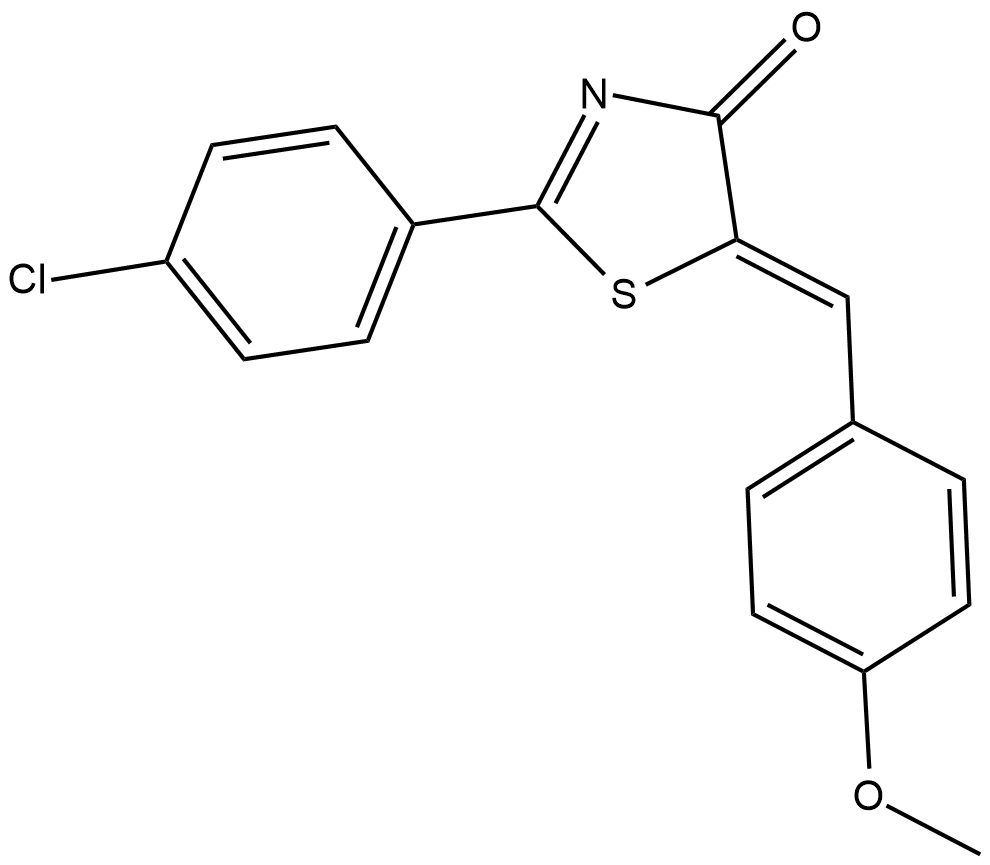
-
GC43194
CAY10698
Platelet-type 12-lipoxygenase catalyzes the formation of 12-HpETE from arachidonic acid.
-
GC49758
CAY10788
A CysLT1 receptor antagonist
-
GC31707
Chebulagic acid
Chebulagic acid is a COX-LOX dual inhibitor isolated from the fruits of Terminalia chebula Retz, on angiogenesis.
-
GC31235
Cirsiliol
A flavonoid with diverse biological activities
-
GC18728
CJ-13610
CJ-13610 is an inhibitor of 5-lipoxygenase (5-LO) that inhibits 5-LO product formation in human polymorphonuclear leukocytes (PMNLs) challenged with A23187 in vitro (IC50 = 70 nM).
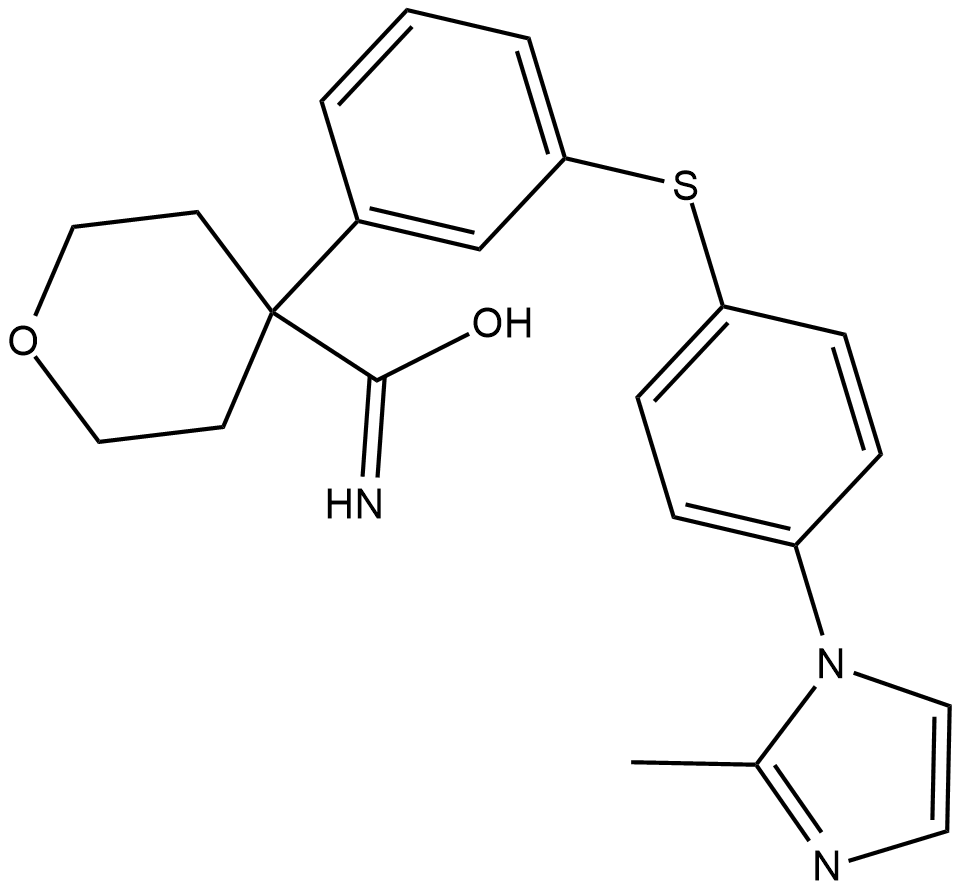
-
GC33905
CMI-392
CMI-392 is a dual 5-lipoxygenese inhibitor and platelet-activating factor (PAF) receptor antagonist with IC50s of 100 and 10 nM, respectively.
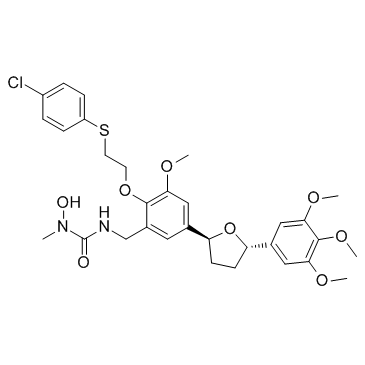
-
GC31961
CMI977 (LDP977)
CMI977 (LDP977) is a potent 5-Lipoxygenase (5-LO) inhibitor.
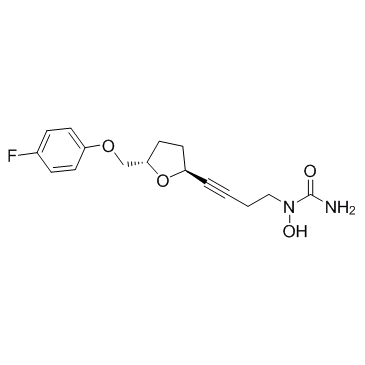
-
GC31967
COX/5-LO-IN-1
COX/5-LO-IN-1 (Atreleuton analog) is an inhibitor of cylooxygenase and 5-lipoxygenase (5-LO), used for the research of inflammatory and allergic disease states.
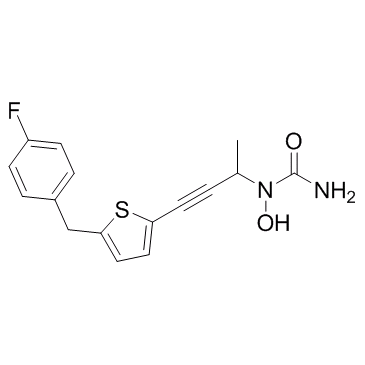
-
GC40697
DDA
DDA is a lipid peroxidation product of linolieic acid.
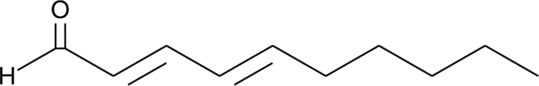
-
GC43453
Diflapolin
Diflapolin is a dual inhibitor of 5-lipoxygenase-activating protein (FLAP) and soluble epoxide hydrolase (sEH).
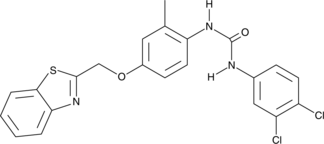
-
GC43459
Dihydro Montelukast
Dihydro montelukast is a potential impurity found in commercial montelukast preparations.
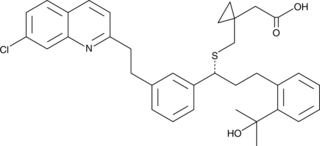
-
GC31940
Docebenone (AA 861)
Docebenone (AA 861) (AA 861) is a potent, selective and orally active 5-LO (5-lipoxygenase) inhibitor.
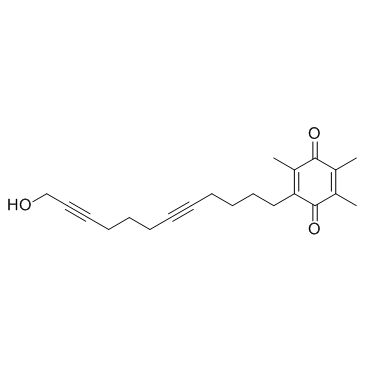
-
GC49590
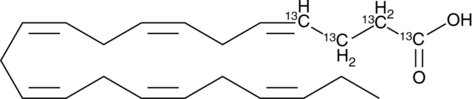
Docosahexaenoic Acid 1,2,3,4-13C
An internal standard for the quantification of DHA
-
GC31801
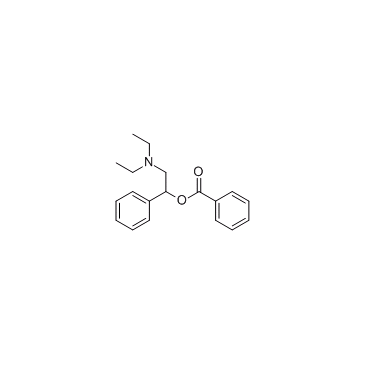
Enazadrem
Enazadrem is a 5-lipoxygenase inhibitor with antiinflammatory activities.
-
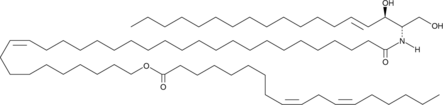
GC43616
EOS (d18:1/32:1/18:2)
EOS is a ceramide found in the outer layer of the epidermis in mammals.
-
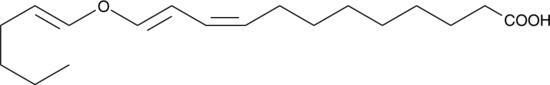
GC49778
Etheroleic Acid
A divinyl ether oxylipin
-
GC47312
Etherolenic Acid
A divinyl ether oxylipin

-
GC47323
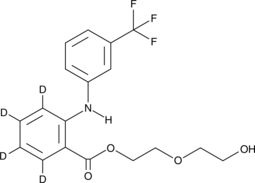
Etofenamate-d4
An internal standard for the quantification of etofenamate
-
GC49696
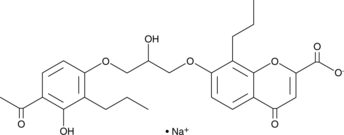
FPL 55712 (sodium salt)
A leukotriene receptor antagonist
-
GC39013
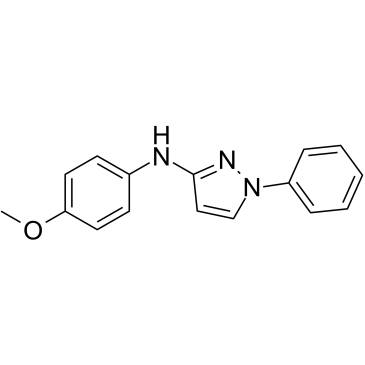
FPL 62064
FPL 62064 is a potent 5-lipoxygenase (5-LOX) and COX dual inhibitor, with IC50 values of 3.5 μM and 3.1 μM for RBL-1 cytosolic 5-lipoxygenase and prostaglandin synthetase (cyclooxygenase), respectively.
-
GC31947
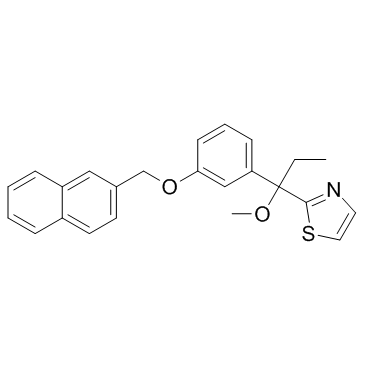
ICI 211965 (ZM-211965)
ICI 211965 (ZM-211965) (ZM-211965) is a selective and orally potent 5-Lipoxygenase (5-LPO) inhibitor.
-
GC12661
Indirubin-3'-oxime
Indirubin-3'-oxime is a potent GSK-3β inhibitor, and weakly inhibits 5-Lipoxygenase, with IC50s of 22 nM and 7.8-10 μM, respectively; Indirubin-3'-oxime also shows inhibitory activities against CDK5/p25 and CDK1/cyclin B, with IC50s of 100 and 180 nM.
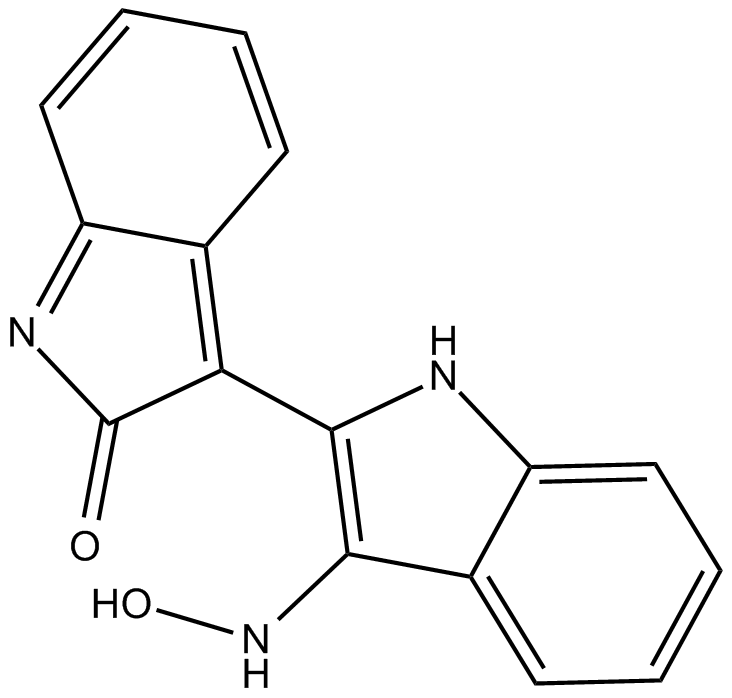
-
GC40840
Leukotriene A3 methyl ester
Biosynthesis of LTA3 occurs from 5,8,11-eicosatrienoic acid via the 5-LO pathway and it is the putative intermediate in the biosynthesis of 3-series leukotrienes.
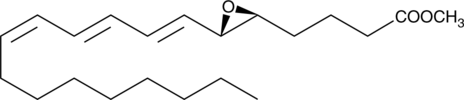
-
GC18835
Leukotriene A4 methyl ester
Leukotriene A4 (LTA4) is synthesized in mast cells, eosinophils, and neutrophils from arachidonic acid by 5-lipoxygenase (5-LO), which exhibits both lipoxygenase and LTA4 synthase activities.
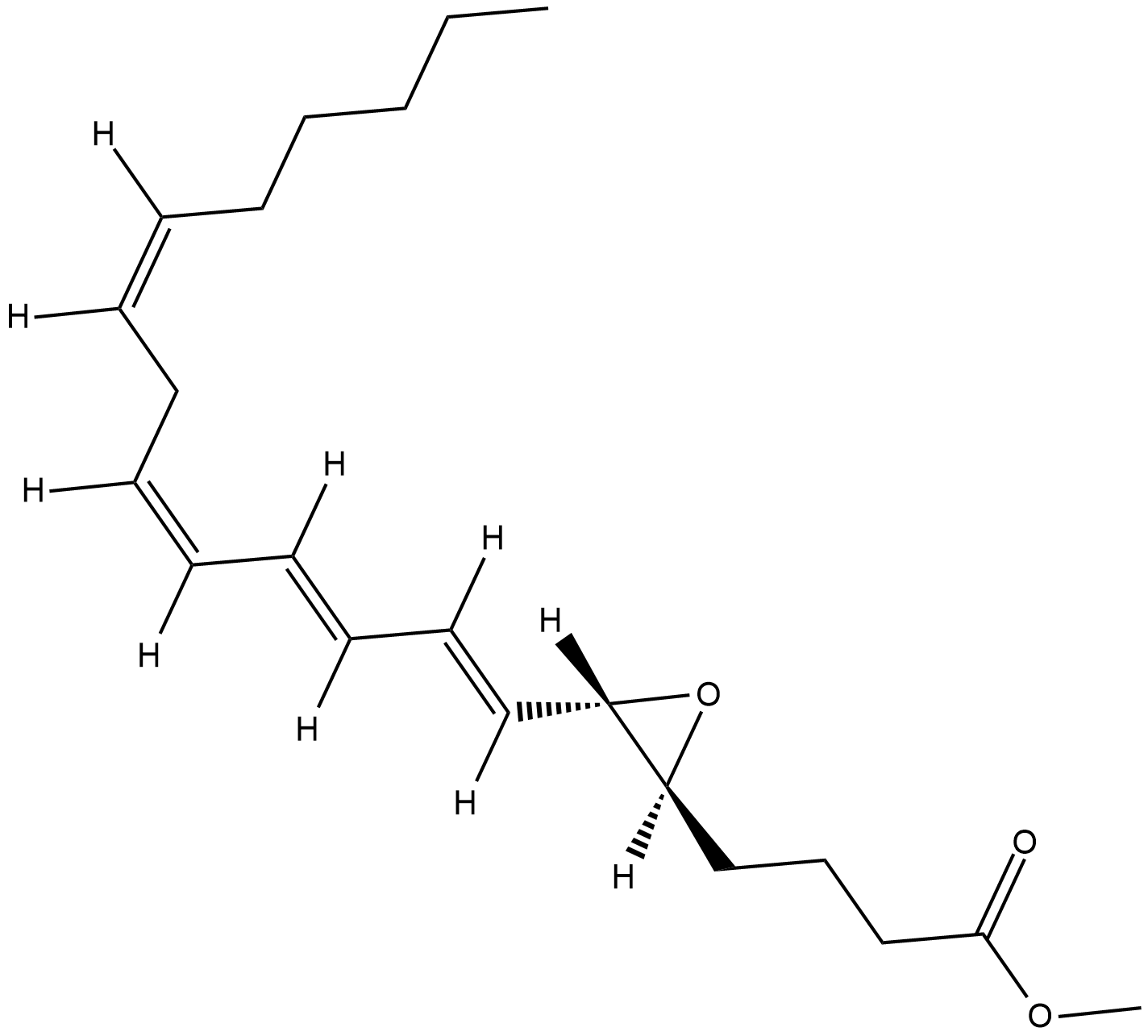
-
GC40278
Leukotriene B3
LTB3 is the LTA hydrolase metabolite of LTA3 in the leukotriene biosynthetic pathway.
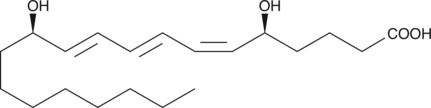
-
GC44052
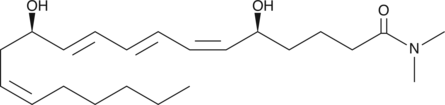
Leukotriene B4 dimethyl amide
LTB4 dimethyl amide is a moderate inhibitor of LTB4-induced degranulation of human neutrophils (Ki = 130 nM) and lysozyme release from rat PMNL.
-
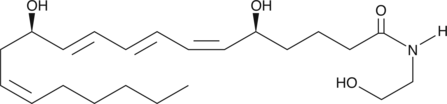
GC44053
Leukotriene B4 Ethanolamide
The effects of Leukotriene B4 (LTB4) are mediated by two known receptors, BLT1 and BLT2.
-
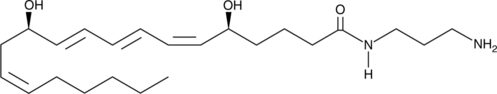
GC40631
Leukotriene B4-3-aminopropylamide
The effects of leukotriene B4 (LTB4) are mediated by two receptors, BLT1 and BLT2.
-
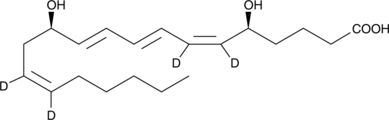
GC47556
Leukotriene B4-d4
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
GC49597
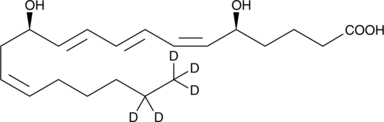
Leukotriene B4-d5
An internal standard for the quantification of LTB4
-
GC41107
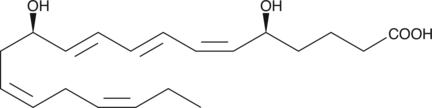
Leukotriene B5
Leukotriene B5 (LTB5) is a leukotriene with diverse biological activities.
-
GC44054
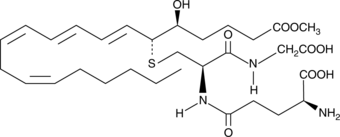
Leukotriene C4 methyl ester
Leukotriene C4 (LTC4) is the parent cysteinyl-leukotriene produced by the LTC4 synthase-catalyzed conjugation of glutathione to LTA4.
-
GC47559
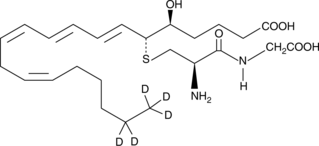
Leukotriene D4-d5
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
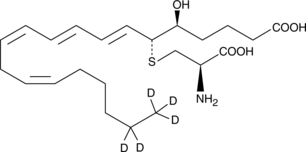
GC47560
Leukotriene E4-d5
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
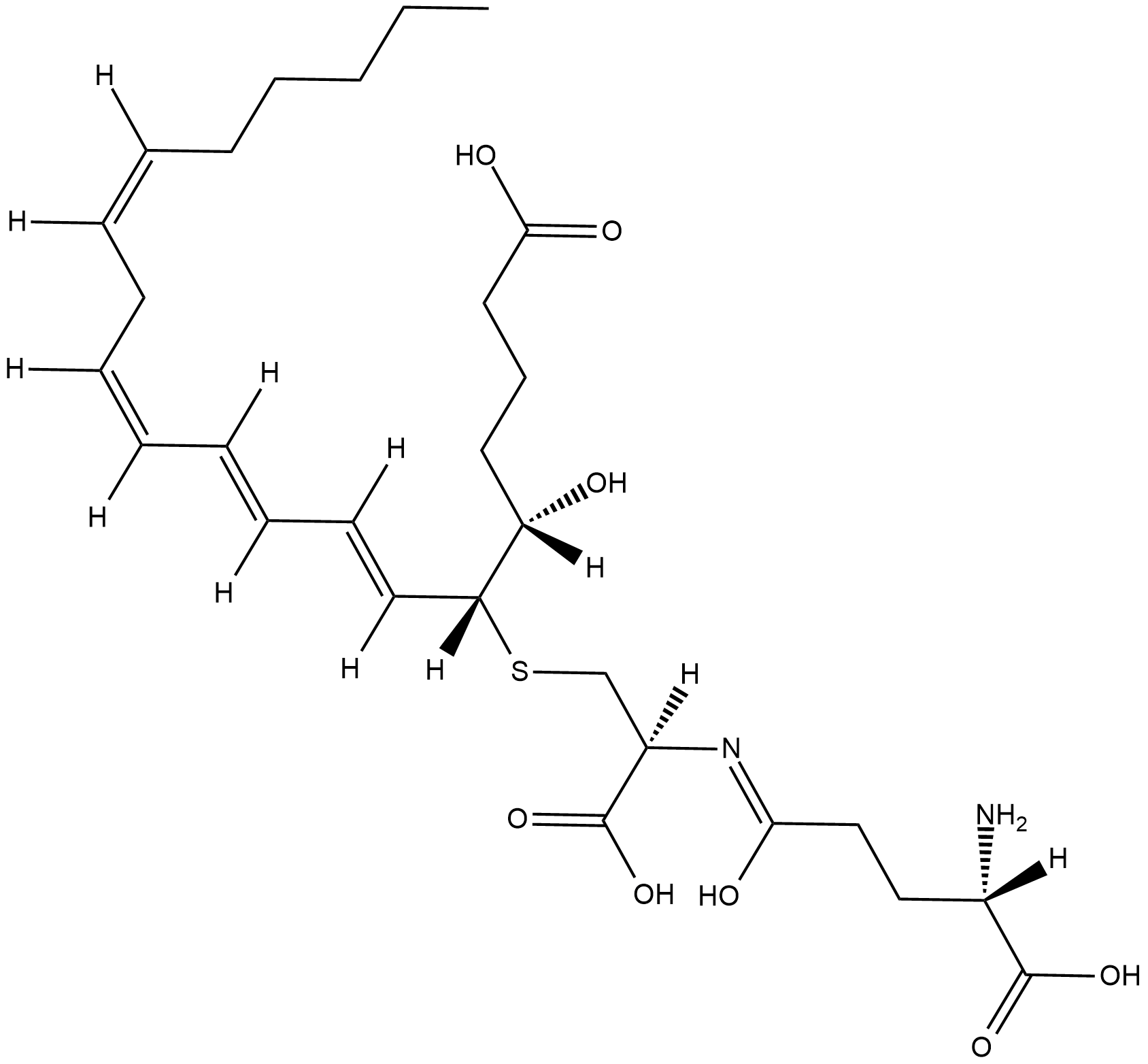
GC18845
Leukotriene F4
LTF4 is a cysteinyl-leukotriene produced in vitro, but not reported to date in vivo.
-
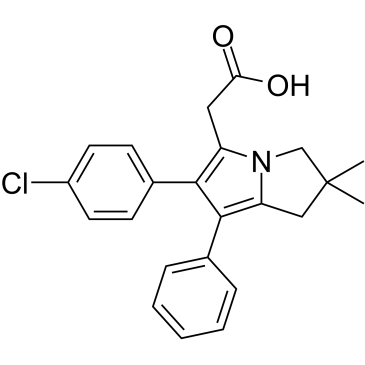
GC38196
Licofelone
A dual inhibitor of COX1/COX2 and 5LO
-
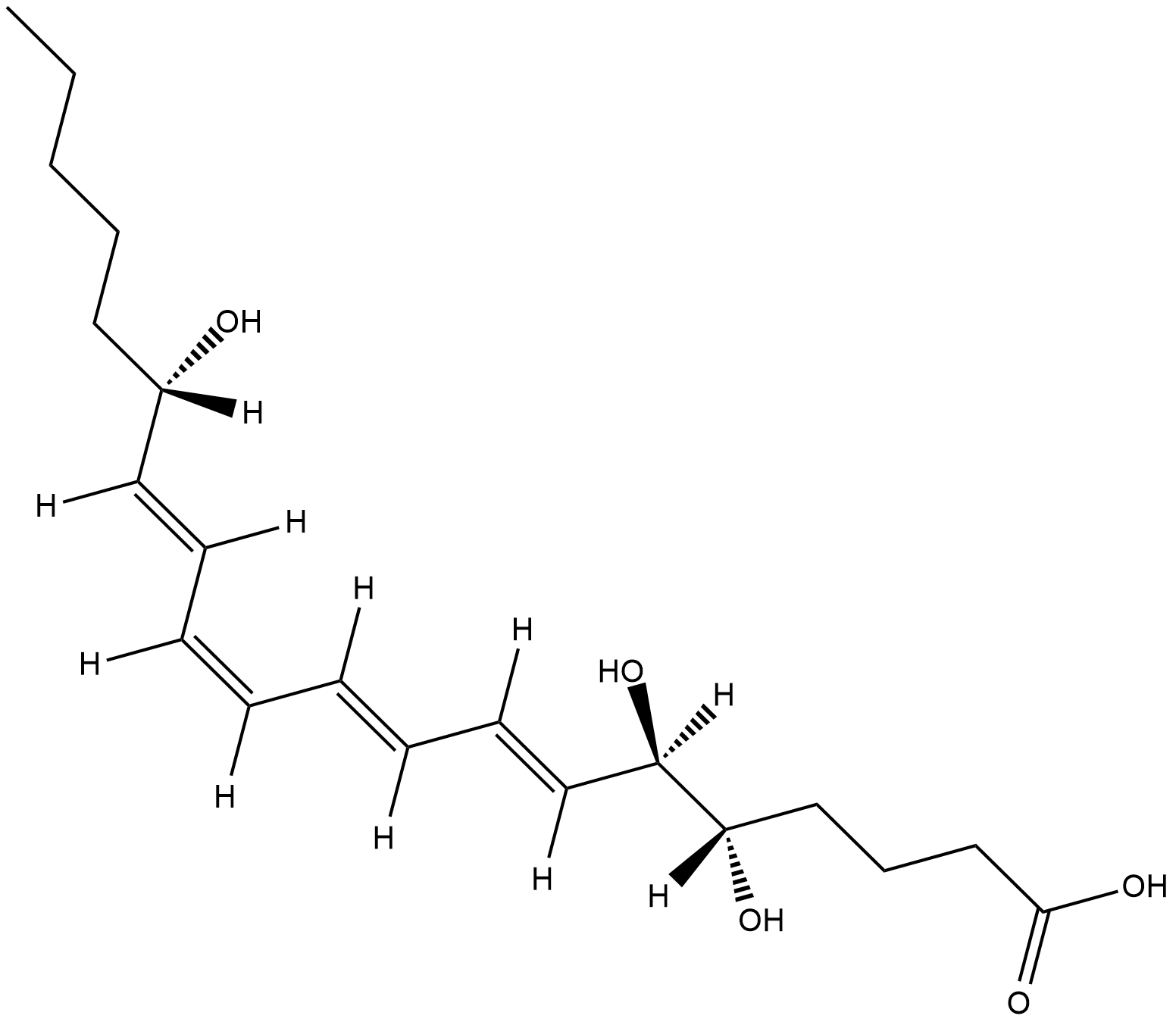
GC18552
Lipoxin A4
A trihydroxy fatty acid containing a conjugated tetraene
-
GC18680
Lipoxin A4 methyl ester
Lipoxin A4 methyl ester (LXA4 methyl ester) is a more lipid soluble, prodrug formulation of the transcellular metabolite LXA4.
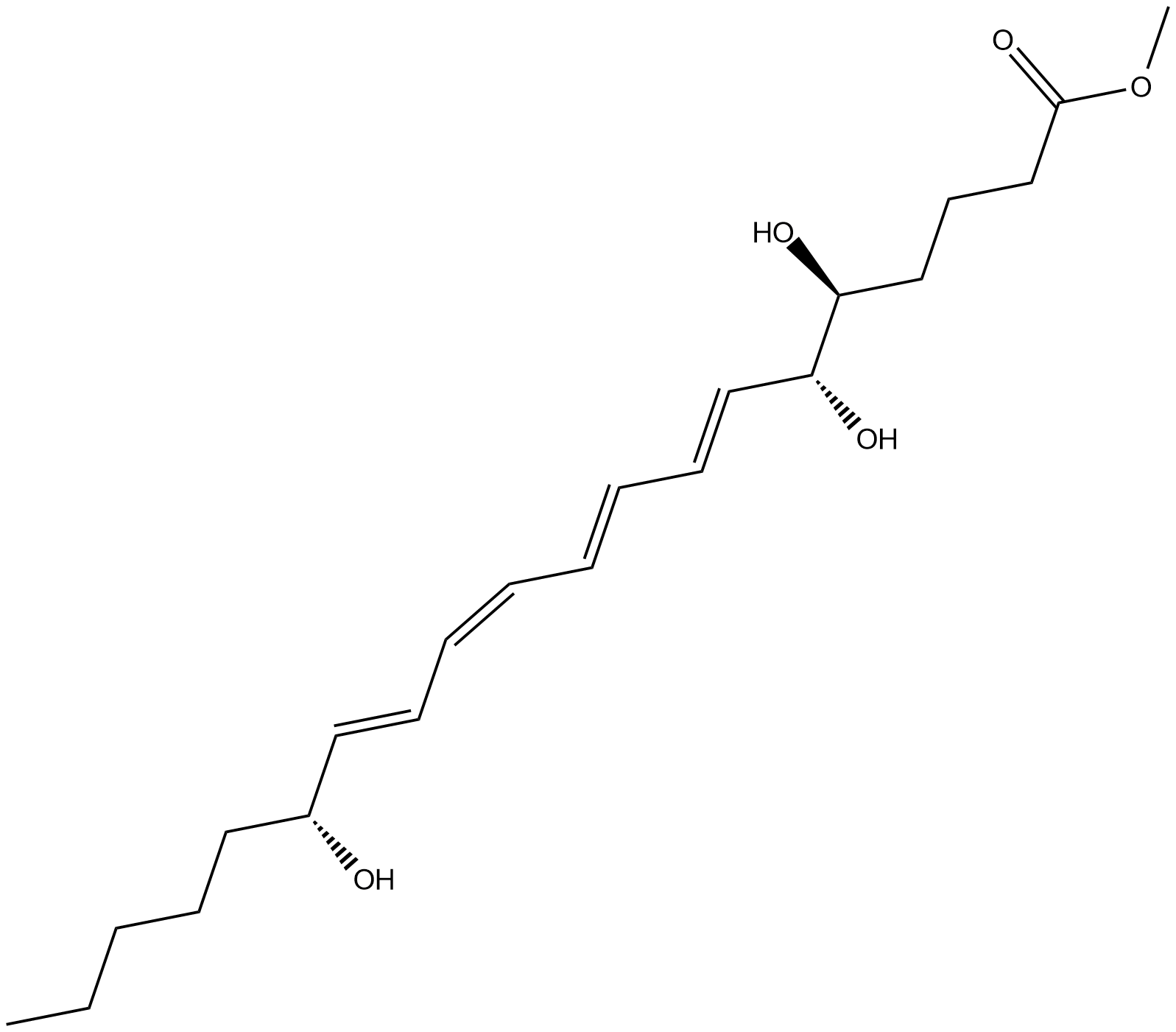
-
GC47569
Lipoxin A4-d5
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
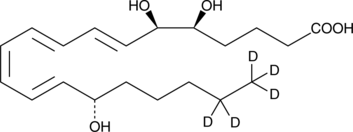
-
GC40609
Lipoxin A5
Lipoxin A5 (LXA5) is produced by enzymatic transformation of EPA by leukocytes.
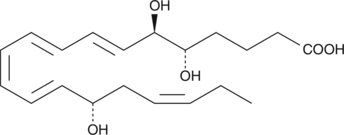
-
GC18681
Lipoxin B4
Lipoxin B4 (LXB4) is a positional isomer of LXA4 produced by the metabolism of 15-HETE or 15(S)-HpETE by human leukocytes.
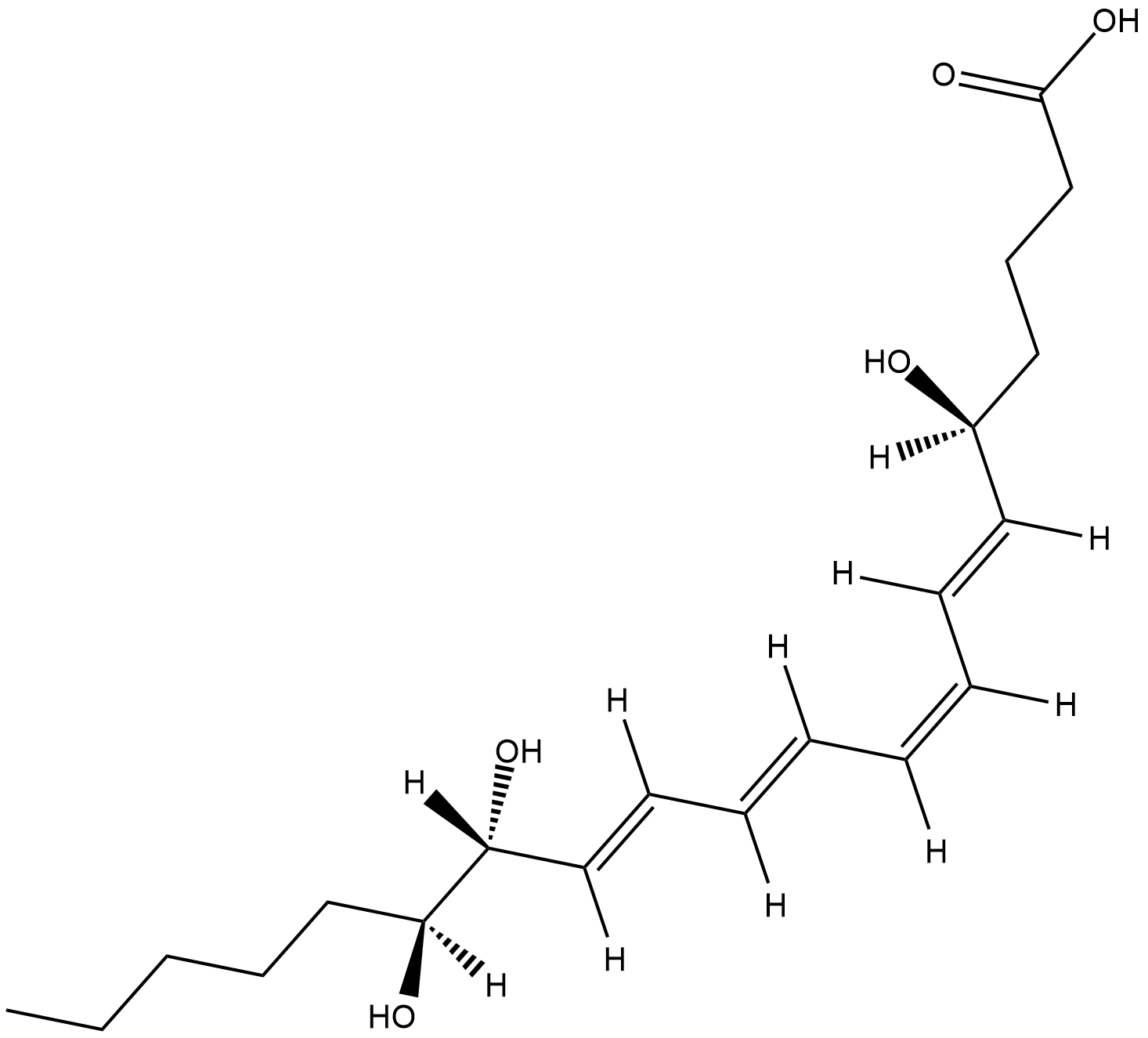
-
GC47570
Lipoxygenin
An inhibitor of 5-LO
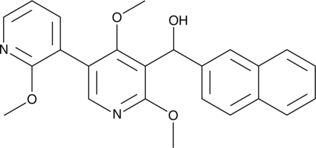
-
GC31978
Lonapalene (RS4317)
Lonapalene (RS4317) (RS4317) is a topically effective 5-lipoxygenase (5-LO) inhibitor.
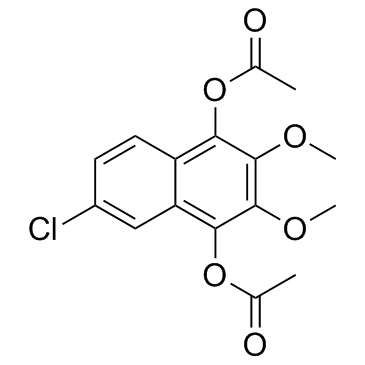
-
GC47575
Loratadine-d5
An internal standard for the quantification of loratadine
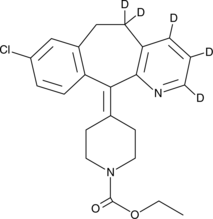
-
GC33907
LP117
LP117 is a novel and potent inhibitor of 5-Lipoxygenase (5-LO) product synthesis with an IC50 of 1.1 μM.
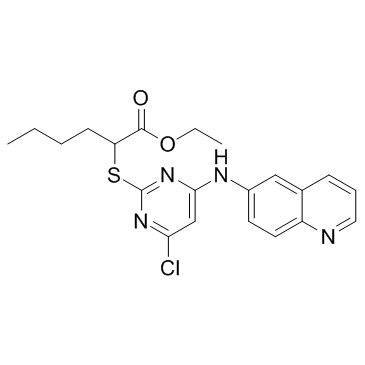
-
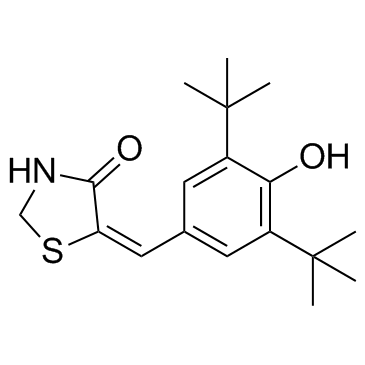
GC30610
LY 178002
LY 178002 is a potent inhibitor of 5-lipoxygenase (5-LPO), phospholipase A2, with IC50 of 0.6 μM for 5-lipoxygenase, inhibits cellular production of LTB4 by human polymorphonuclear leukocytes, and shows relatively weak inhibition on cyclooxygenase.
-
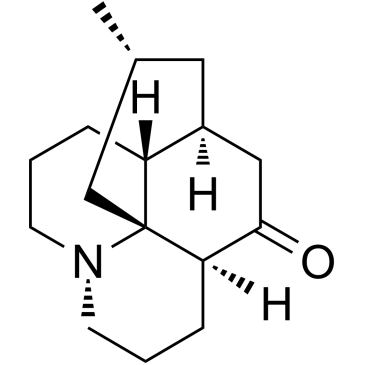
GC36516
Lycopodine
Lycopodine, a pharmacologically important bioactive component derived from Lycopodium clavatumspores, triggers apoptosis by modulating 5-lipoxygenase, and depolarizing mitochondrial membrane potential in refractory prostate cancer cells without modulating p53 activity. Lycopodine inhibits proliferation of HeLa cells through induction of apoptosis via caspase-3 activation.
-
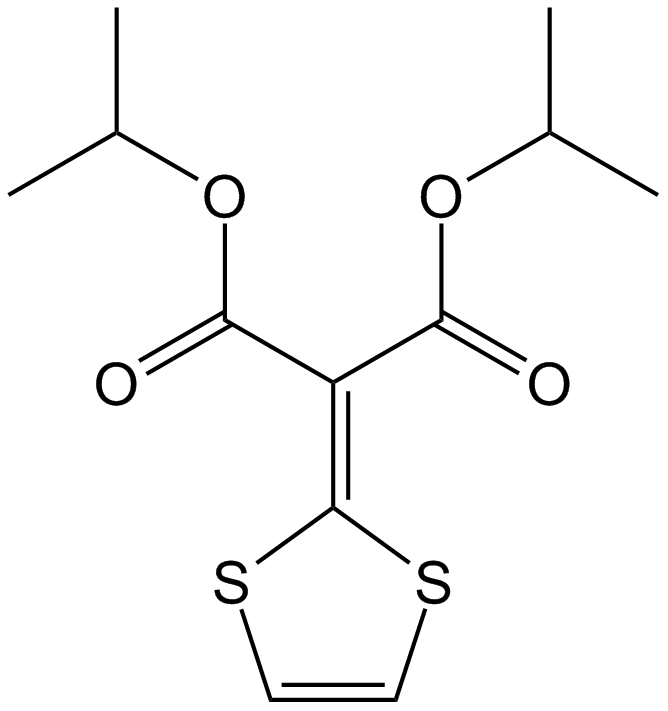
GC14751
Malotilate
Stimulates hepatocyte regeneration
-
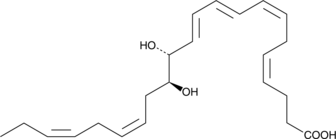
GC40980
Maresin 2
Docosahexaenoic acid is an ω-3 fatty acid that is abundant in the brain and the retina and is known to be important in early development.
-
GC44138
MCTR1
Maresin conjugates in tissue regeneration 1 (MCTR1) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid in macrophages at the site of inflammation.
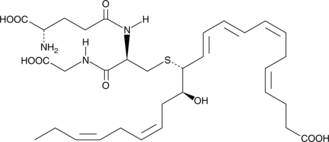
-
GC44139
MCTR2
Maresin conjugates in tissue regeneration 2 (MCTR2) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid in macrophages at the site of inflammation.
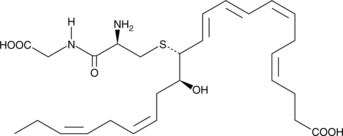
-
GC44140
MCTR3
Maresin conjugates in tissue regeneration 3 (MCTR3) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid in macrophages.
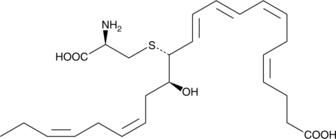
-
GC44212
MK-886 (sodium salt)
Arachidonic acid and selected other polyunsaturated fatty acids are stereoselectively oxygenated at carbon 5 by the non-heme iron containing enzyme 5-lipoxygenase (5-LO).
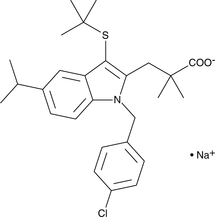
-
GC36628
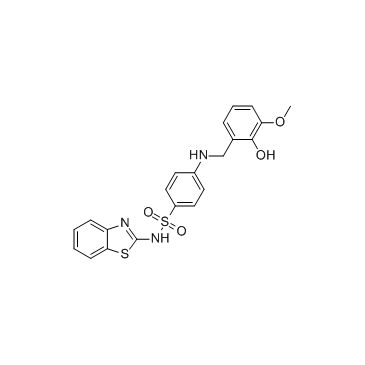
ML355
ML355 is a potent and selective inhibitor of 12-Lipoxygenase (12-LOX) with an IC50 of 0.34 μM, shows excellent selectivity over related lipoxygenases and cyclooxygenases, and possesses favorable ADME properties.
-
GC52105
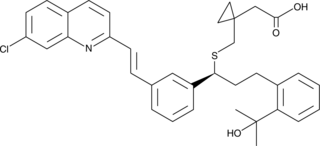
Montelukast
Montelukast (MK0476 free base) is a potent, selective and orally active antagonist of cysteinyl leukotriene receptor 1 (CysLT1).
-
GC47716
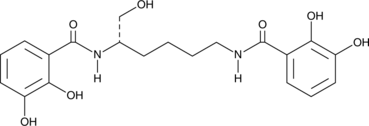
Myxochelin A
A microbial metabolite with diverse biological activities
-
GC44295
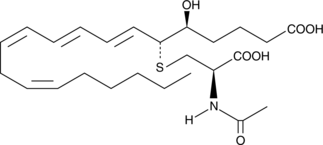
N-acetyl Leukotriene E4
N-acetyl LTE4 is the major inactive metabolite of LTE4 found in bile.
-
GC44415
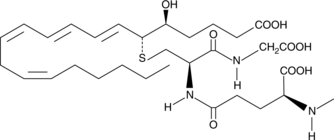
N-methyl Leukotriene C4
Produced by neutrophils, macrophages, mast cells, and by transcellular metabolism in platelets, leukotriene C4 (LTC4) is the parent cysteinyl leukotriene formed by the LTC4 synthase-catalyzed conjugation of glutathione to LTA4.
-
GC18089
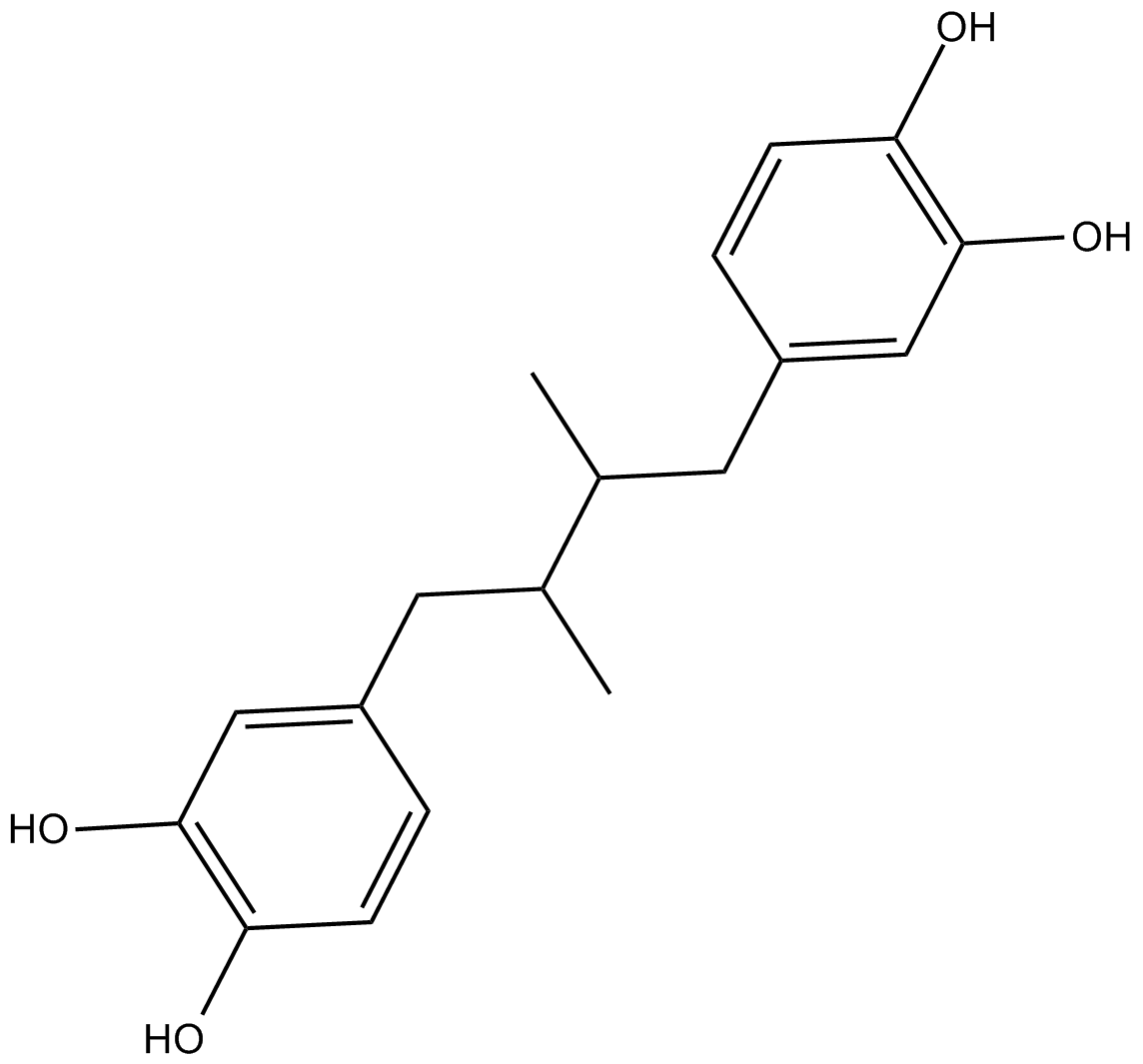
Nordihydroguaiaretic acid
A non-selective LO inhibitor
-
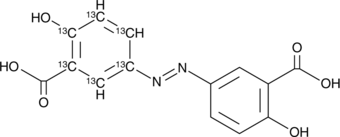
GC49098
Olsalazine-13C6
An internal standard for the quantification of olsalazine
-
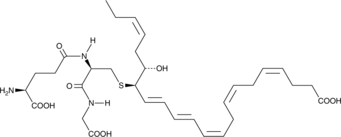
GC44579
PCTR1
Protectin conjugates in tissue regeneration 1 (PCTR1) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.
-
GC44580
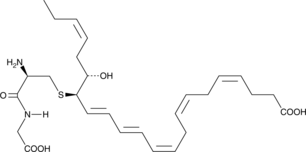
PCTR2
Protein conjugates in tissue regeneration 2 (PCTR2) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.
-
GC44581
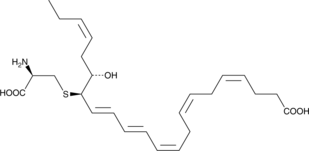
PCTR3
Protein conjugates in tissue regeneration 3 (PCTR3) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.
-
GC36883
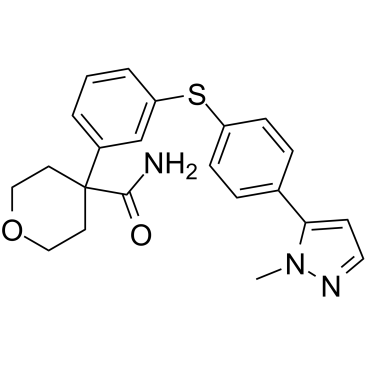
PF-4191834
PF-4191834 (PF-04191834) is an orally active, noniron chelating, and non-redox inhibitor of the 5-Lipoxygenase (5-LOX) (IC50=229 nM), displays ~300-fold selectivity for 5-LOX over 12-LOX and 15-LOX, shows no activity toward the cyclooxygenase enzymes, and is effective in inflammation and pain.
-
GC32038
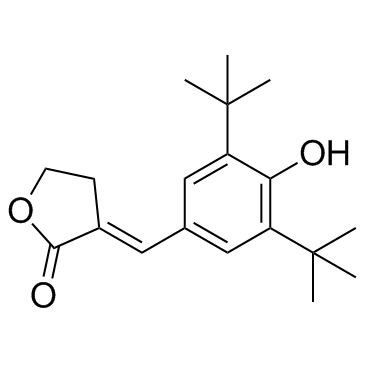
PGS-IN-1 (KME-4)
PGS-IN-1 (KME-4) is a potent inhibitor of prostaglandin synthetase (PGS) with an IC50 of 0.28 μM; also inhibits 5-lipoxygenase with an IC50 of 1.05 μM.
-
GC61180
Phenidone
Phenidone, an orally active dual inhibitor of cyclooxygenase (COX) and lipoxygenase (LOX), ameliorates rat paralysis in experimental autoimmune encephalomyelitis.
-
GC40981
Protectin D1
Protectin D1 is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.