TGF-β / Smad Signaling
Transforming growth factor-beta (TGF-beta) is a multifunctional cytokine that regulates proliferation, migration, differentiation, and survival of many different cell types. Deletion or mutation of different members of the TGF-β family have been shown to cause vascular remodeling defect and absence of mural cell formation, leading to embryonic lethality or severe vascular disorders. TGF-β induces smooth muscle differentiation via Notch or SMAD2 and SMAD3 signaling in ES cells or in a neural crest stem cell line. TGF-β binds to TGF-βRI and to induce phosphorylation of SMAD2/3, thereby inhibiting proliferation, tube formation, and migration of endothelial cells (ECs).
TGF-β is a pluripotent cytokine with dual tumour-suppressive and tumour-promoting effects. TGF-β induces the epithelial-to-mesenchymal transition (EMT) leading to increased cell plasticity at the onset of cancer cell invasion and metastasis.
Targets for TGF-β / Smad Signaling
Products for TGF-β / Smad Signaling
- Cat.No. Product Name Information
-
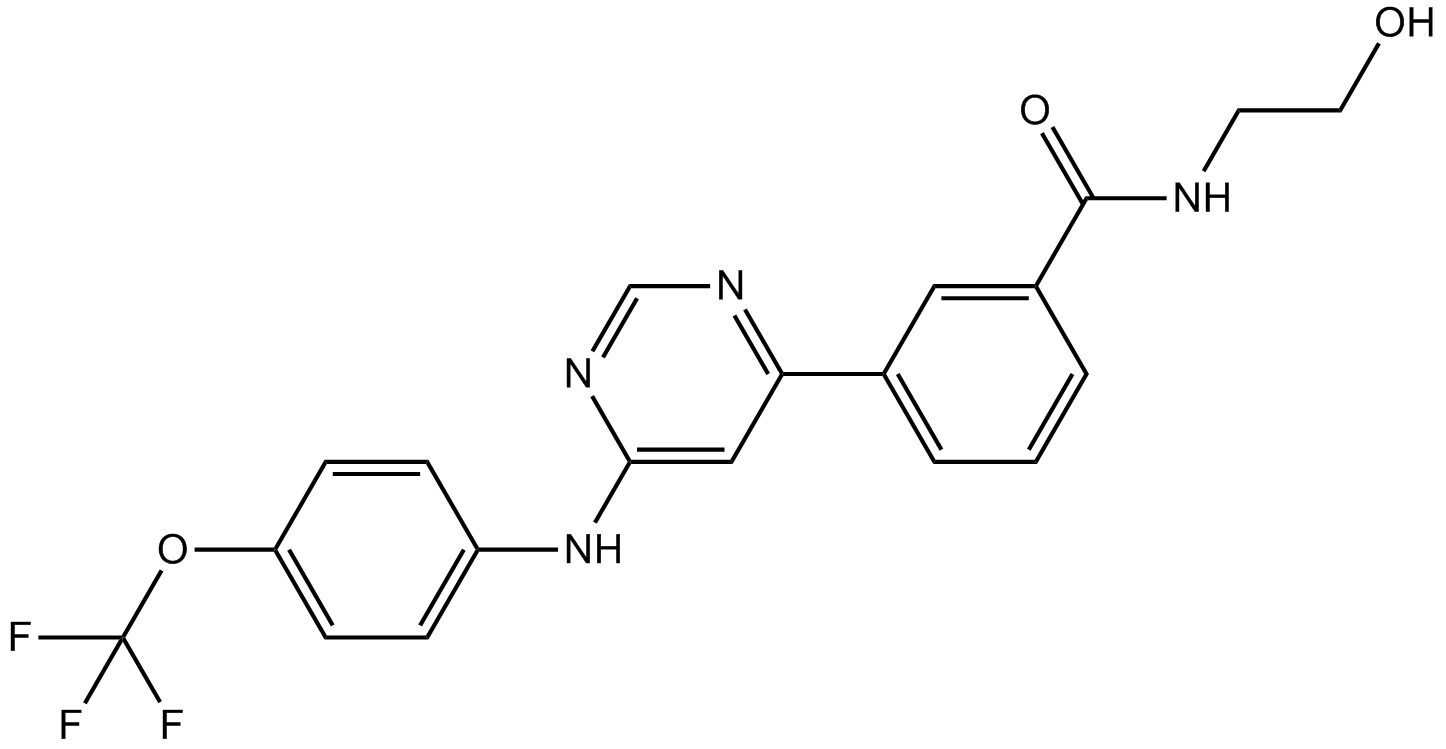
GC15079
GNF 5
Bcr-Abl inhibitor
-
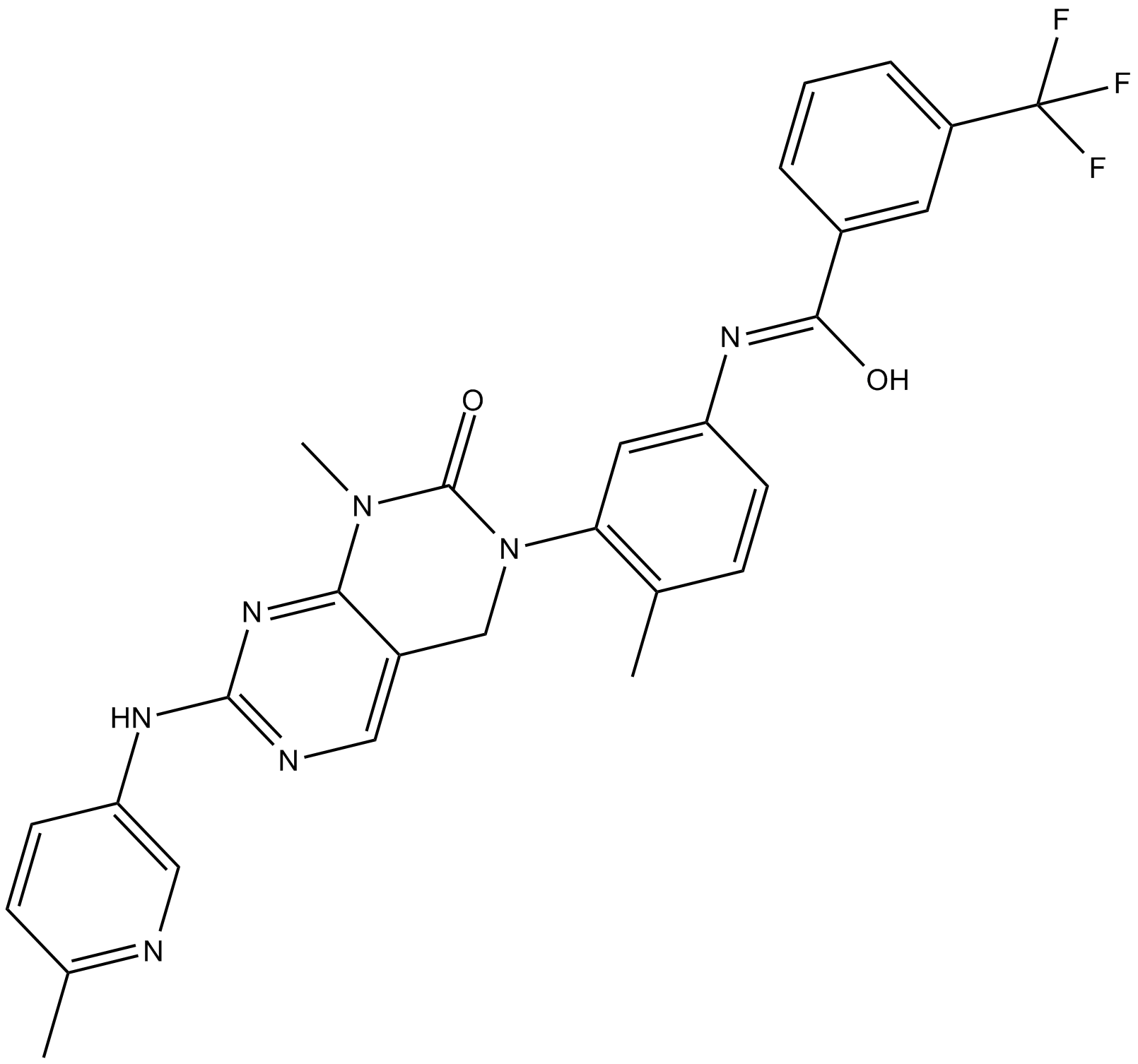
GC10607
GNF-7
Type II Bcr-Abl inhibitor
-
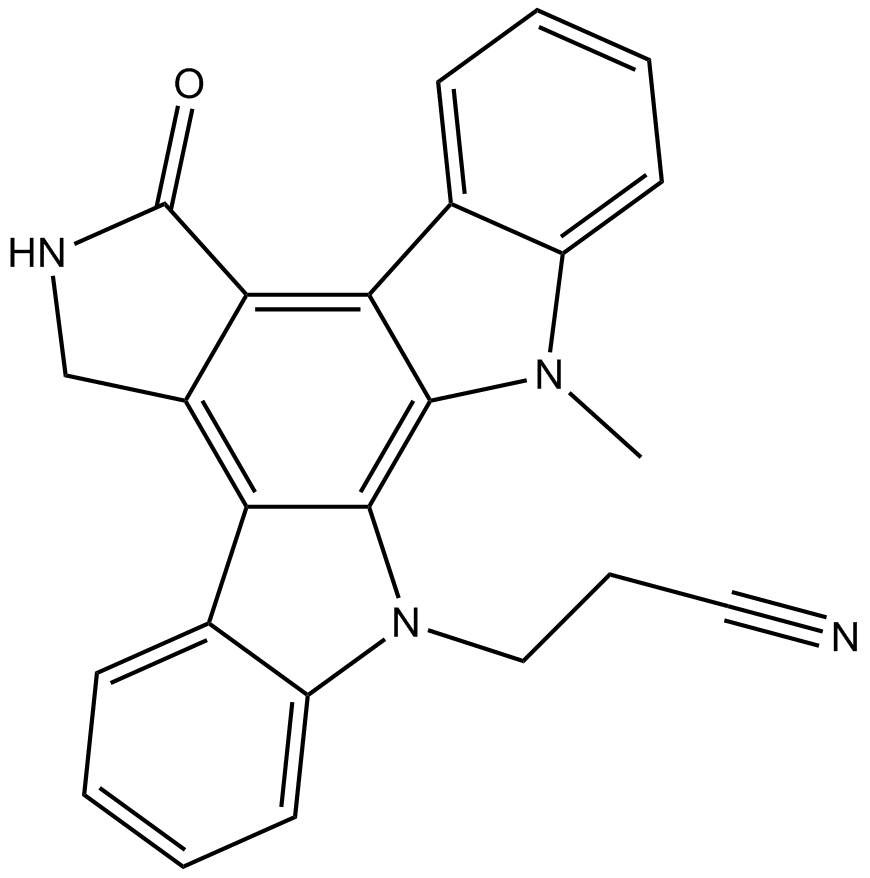
GC15564
Go 6976
PKCα/PKCβ1 inhibitor
-
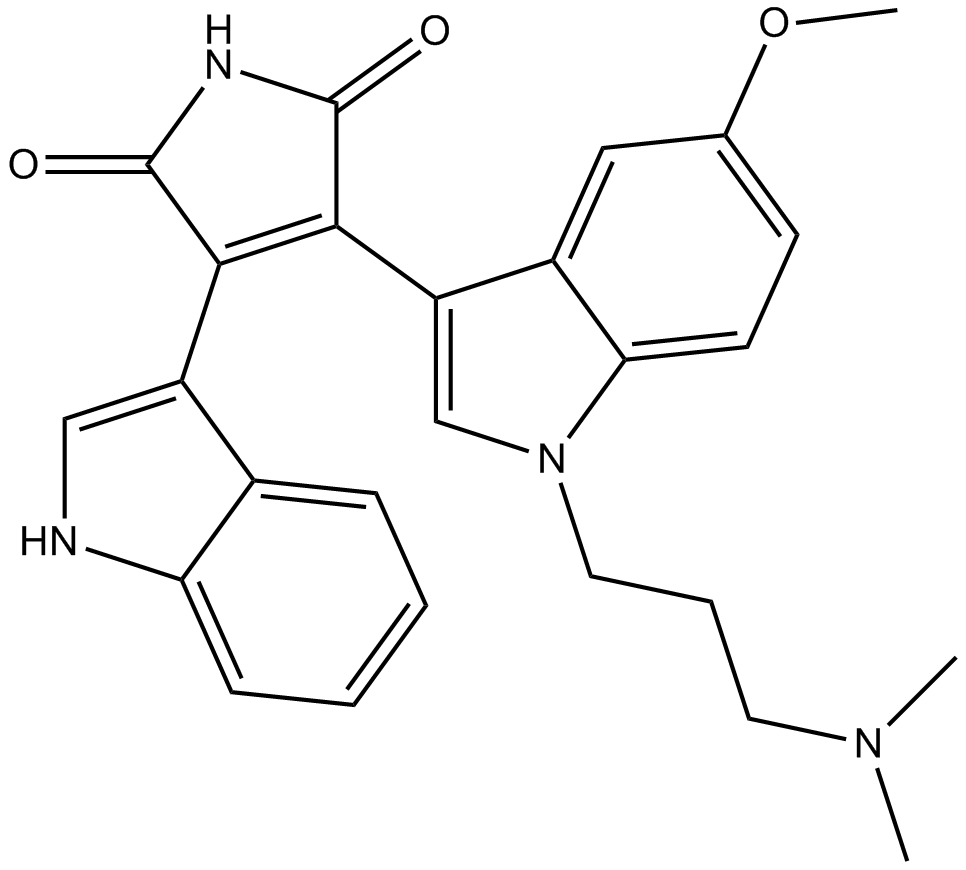
GC16907
Go 6983
Go 6983 (GÖ 6983) is one of the bisindolylmaleimide group of PKC inhibitor compounds, Go 6983 (GÖ 6983) was able to differentiate between PKC mu and other PKC isoenzymes.
-
GC38791
GSK-25
GSK-25 is a potent, selective and orally bioavailable ROCK1 inhibitor (IC50=7 nM).
-
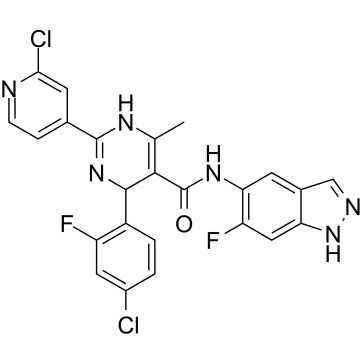
GC19177
GSK180736A
GSK180736A is a G protein-coupled receptor kinase 2 (GRK2) inhibitor with an IC50 of 0.77 uM.
-
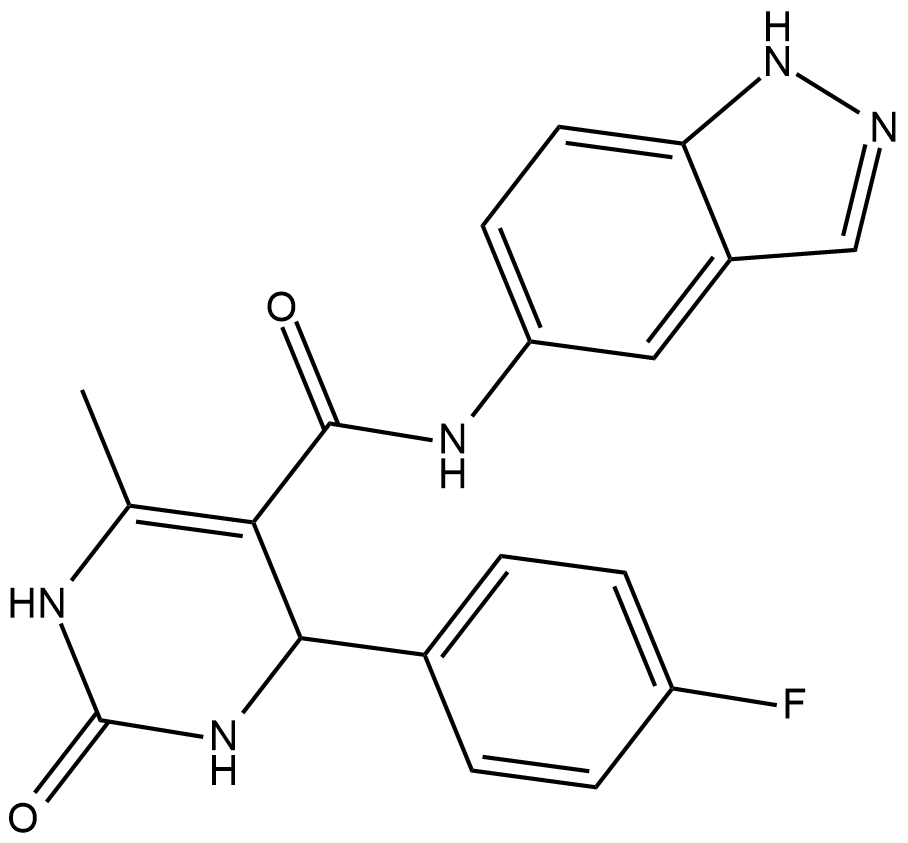
GC17936
GSK269962A
ROCK inhibitor
-
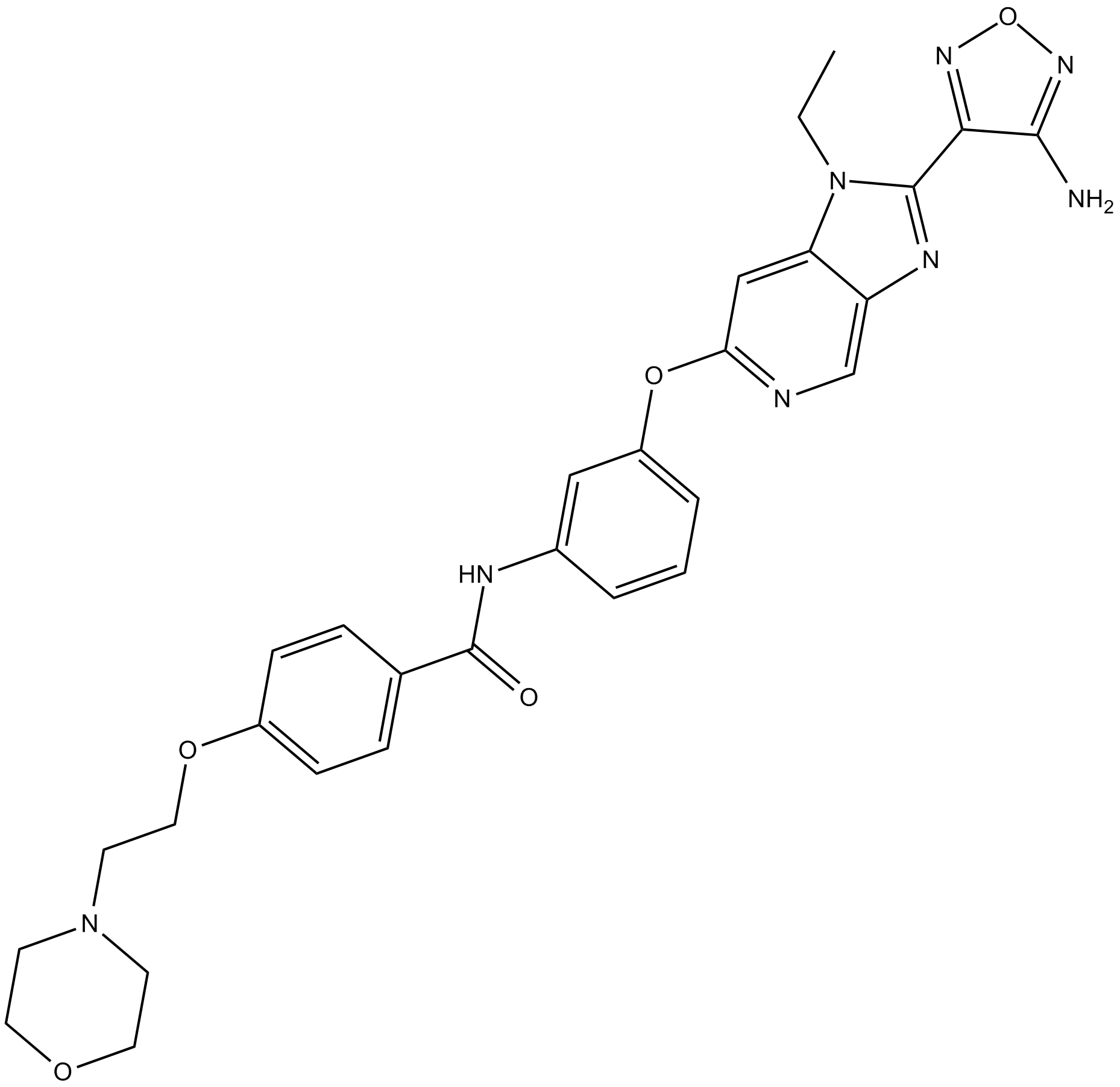
GC25482
GSK269962A HCl
GSK269962A HCl (GSK269962B, GSK269962) is a selective ROCK(Rho-associated protein kinase) inhibitor with IC50 values of 1.6 and 4 nM for ROCK1 and ROCK2, respectively.
-
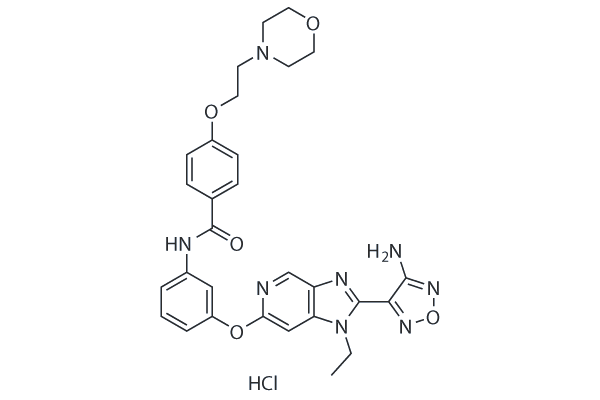
GC18119
GSK429286A
Selective ROCK1/ROCK2 inhibitor
-
GC11878
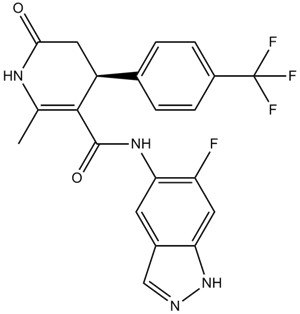
GW788388
ALK5 inhibitor,potent and selective
-
GC10915
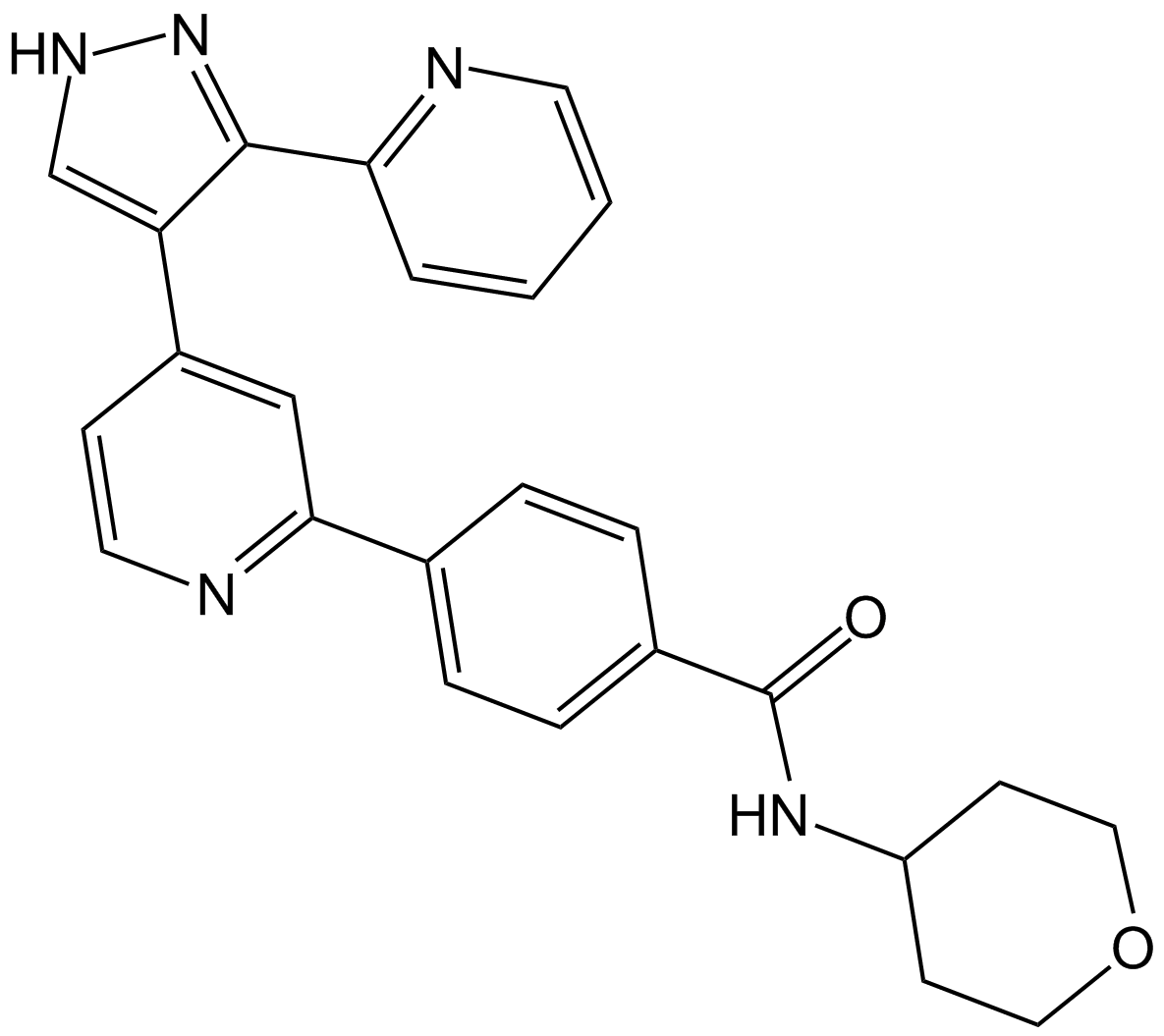
GZD824
Olverembatinib (GZD824) dimesylate is a potent and orally active pan-Bcr-Abl inhibitor. GZD824 potently inhibits a broad spectrum of Bcr-Abl mutants. GZD824 strongly inhibits native Bcr-Abl and Bcr-AblT315I with IC50s of 0.34 nM and 0.68 nM, respectively. GZD824 has antitumor activity.
-
GC33201
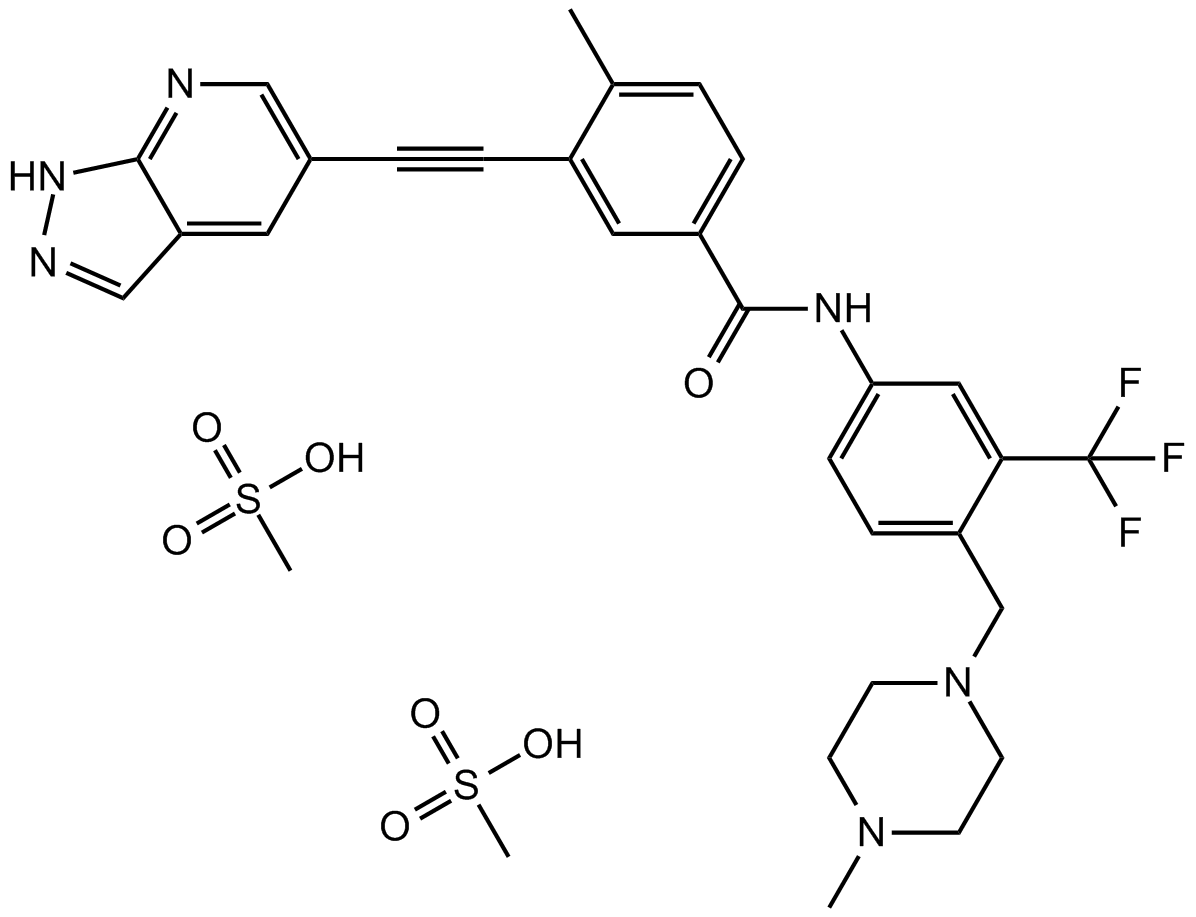
GZD856
GZD856 formic is a potent and orally active PDGFRα/β inhibitor, with IC50s of 68.6 and 136.6 nM, respectively. GZD856 formic is also a Bcr-AblT315I inhibitor, with IC50s of 19.9 and 15.4?nM for native Bcr-Abl and the T315I mutant. GZD856 formic has antitumor activity.
-
GC36207
H-1152
H-1152 is a membrane-permeable and selective ROCK inhibitor, with a Ki value of 1.6 nM, and an IC50 value of 12 nM for ROCK2.
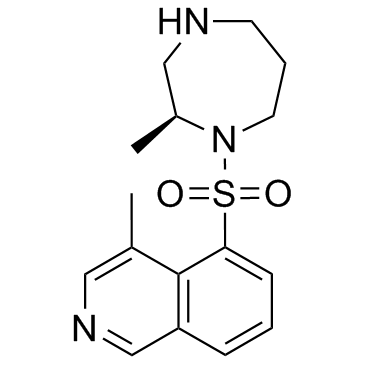
-
GC36208
H-1152 dihydrochloride
H-1152 dihydrochloride is a membrane-permeable and selective ROCK inhibitor, with a Ki value of 1.6 nM, and an IC50 value of 12 nM for ROCK2.
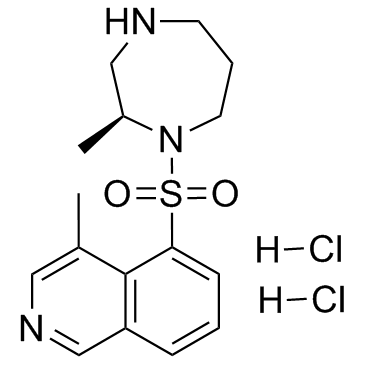
-
GC31650
Halofuginone (RU-19110)
Halofuginone (RU-19110) (RU-19110), a Febrifugine derivative, is a competitive prolyl-tRNA synthetase inhibitor with a Ki of 18.3 nM.
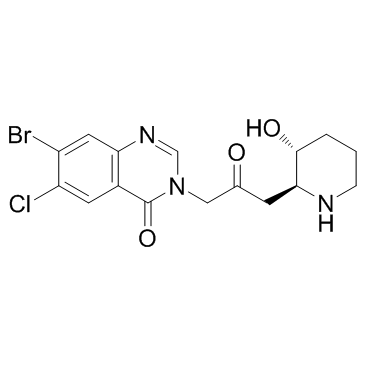
-
GC31950
Halofuginone hydrobromide (RU-19110 (hydrobromide))
Halofuginone (RU-19110) hydrobromid, a Febrifugine derivative, is a competitive prolyl-tRNA synthetase inhibitor with a Ki of 18.3 nM.
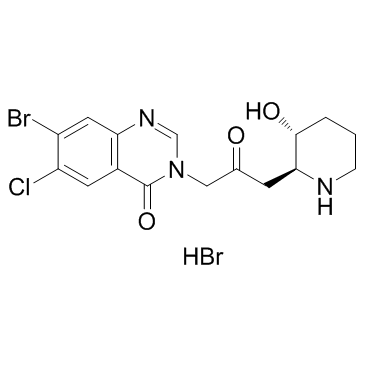
-
GC10299
Hexadecyl Methyl Glycerol
protein kinase C activity inhibitor
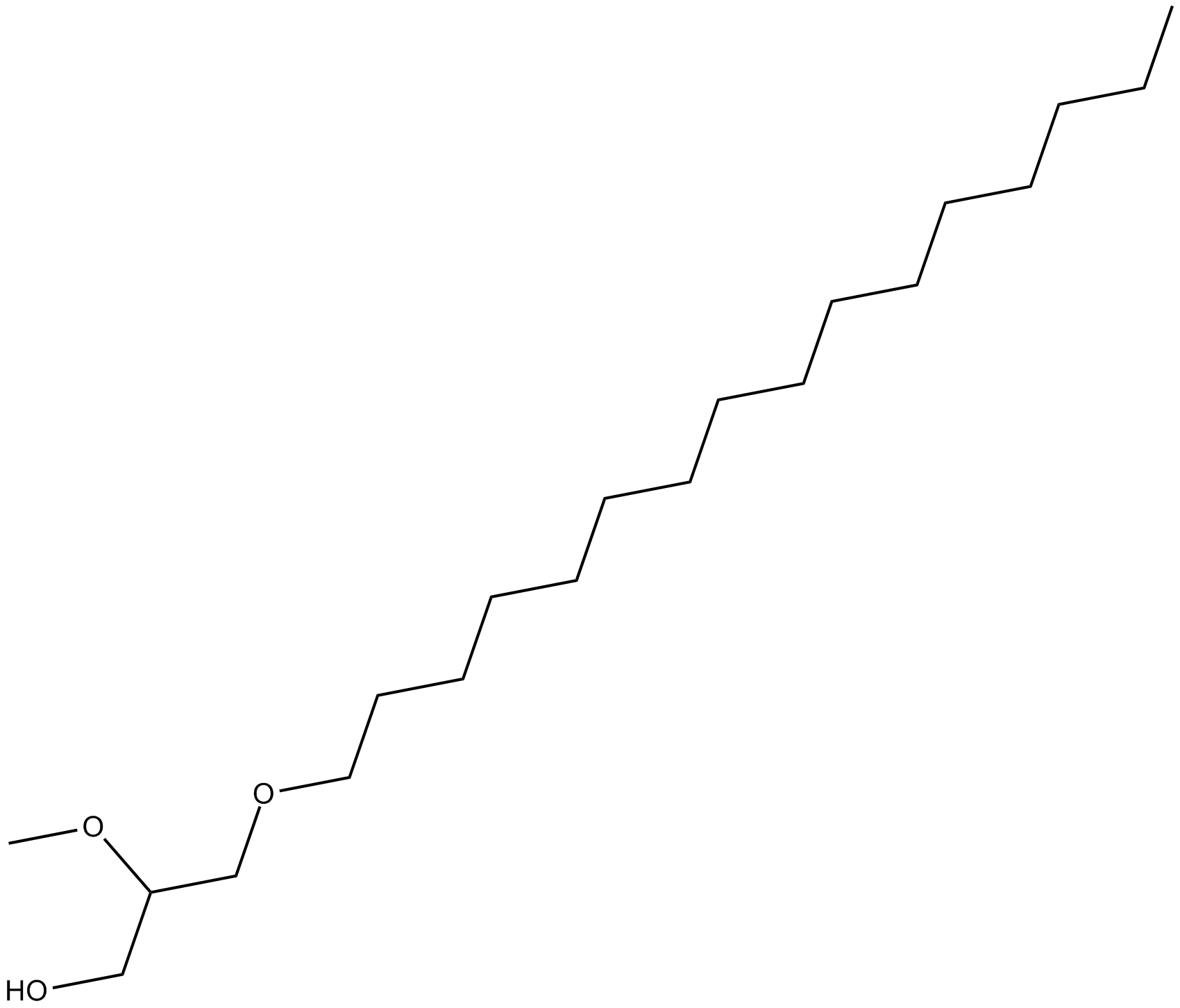
-
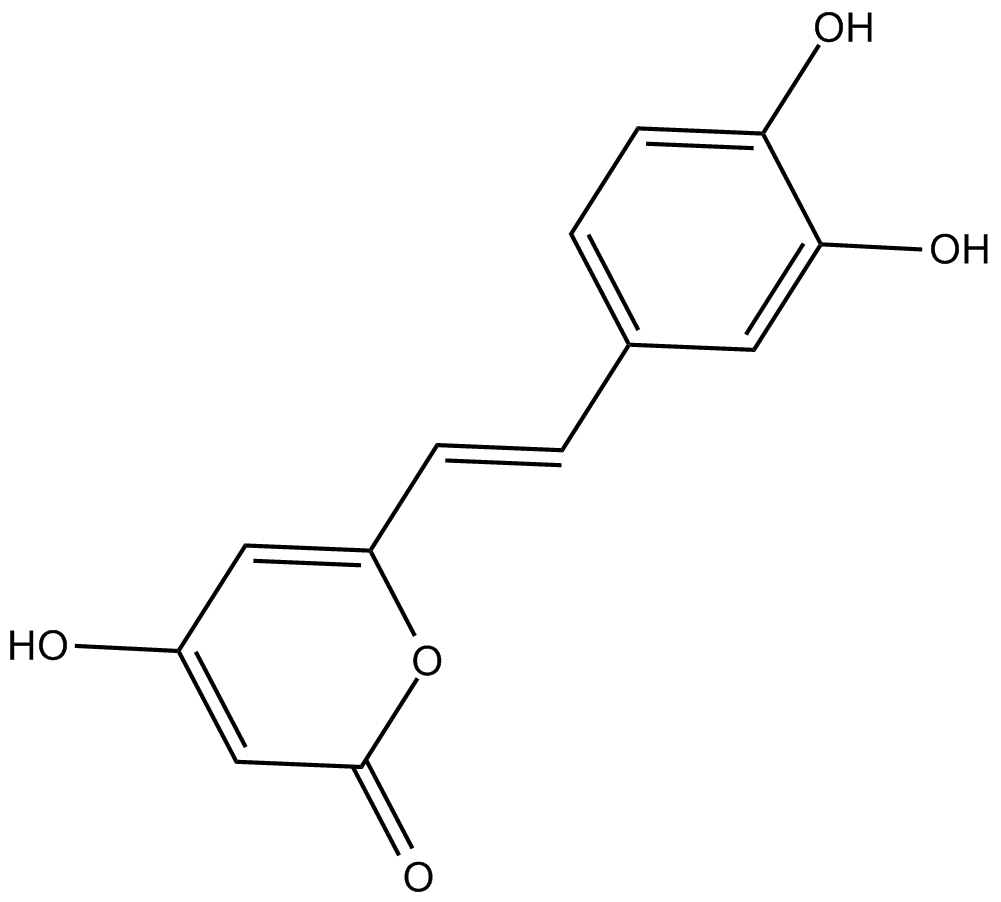
GC15018
Hispidin
protein kinase Cβ inhibitor
-
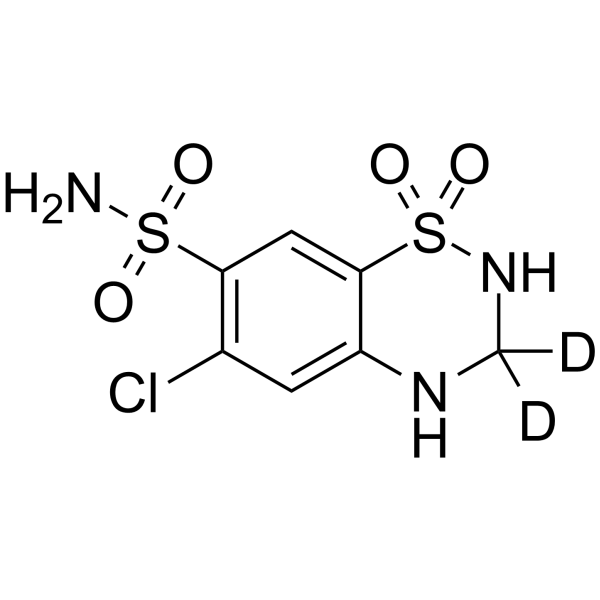
GC64326
Hydrochlorothiazid-d2
-
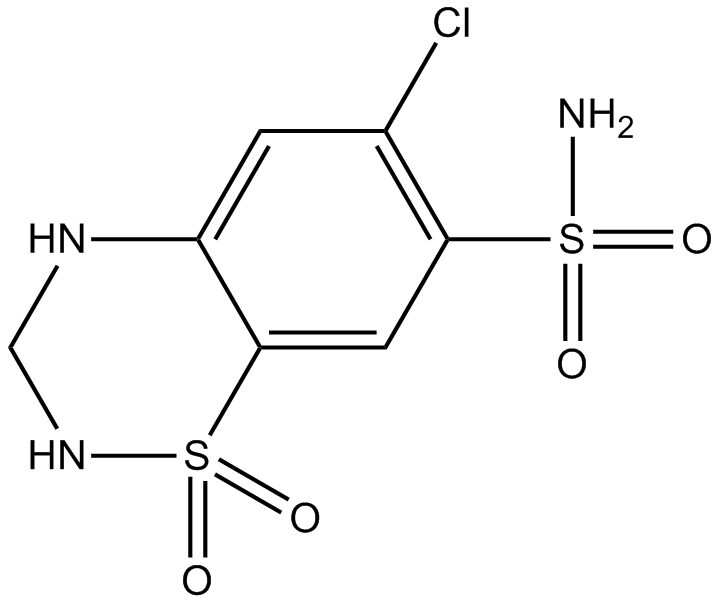
GC17523
Hydrochlorothiazide
diuretic drug of the thiazide class
-
GC36282
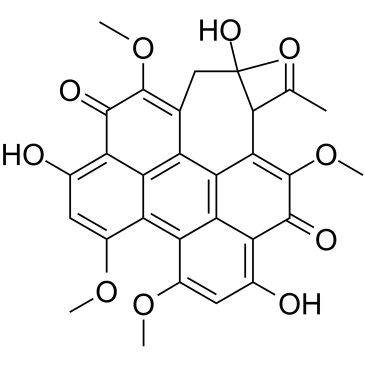
Hypocrellin A
Hypocrellin A, a naturally occurring PKC inhibitor, has many biological and pharmacological properties, such as antitumour, antiviral, antibacterial, and antileishmanial activities.
-
GC15420
ICP 103
Protein kinase inhibitor
-
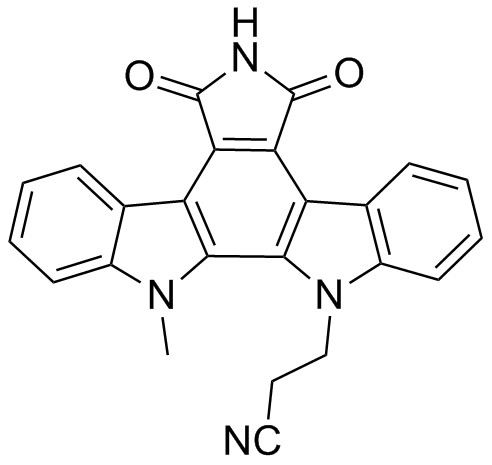
GC10314
Imatinib (STI571)
Imatinib (STI571) (STI571) is an orally bioavailable tyrosine kinases inhibitor that selectively inhibits BCR/ABL, v-Abl, PDGFR and c-kit kinase activity. Imatinib (STI571) (STI571) works by binding close to the ATP binding site, locking it in a closed or self-inhibited conformation, therefore inhibiting the enzyme activity of the protein semicompetitively. Imatinib (STI571) also is an inhibitor of SARS-CoV and MERS-CoV.
-
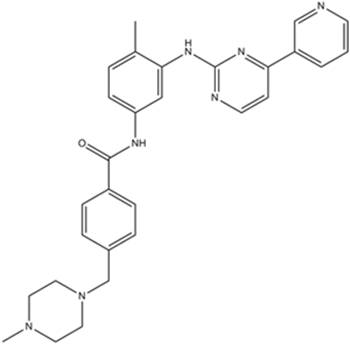
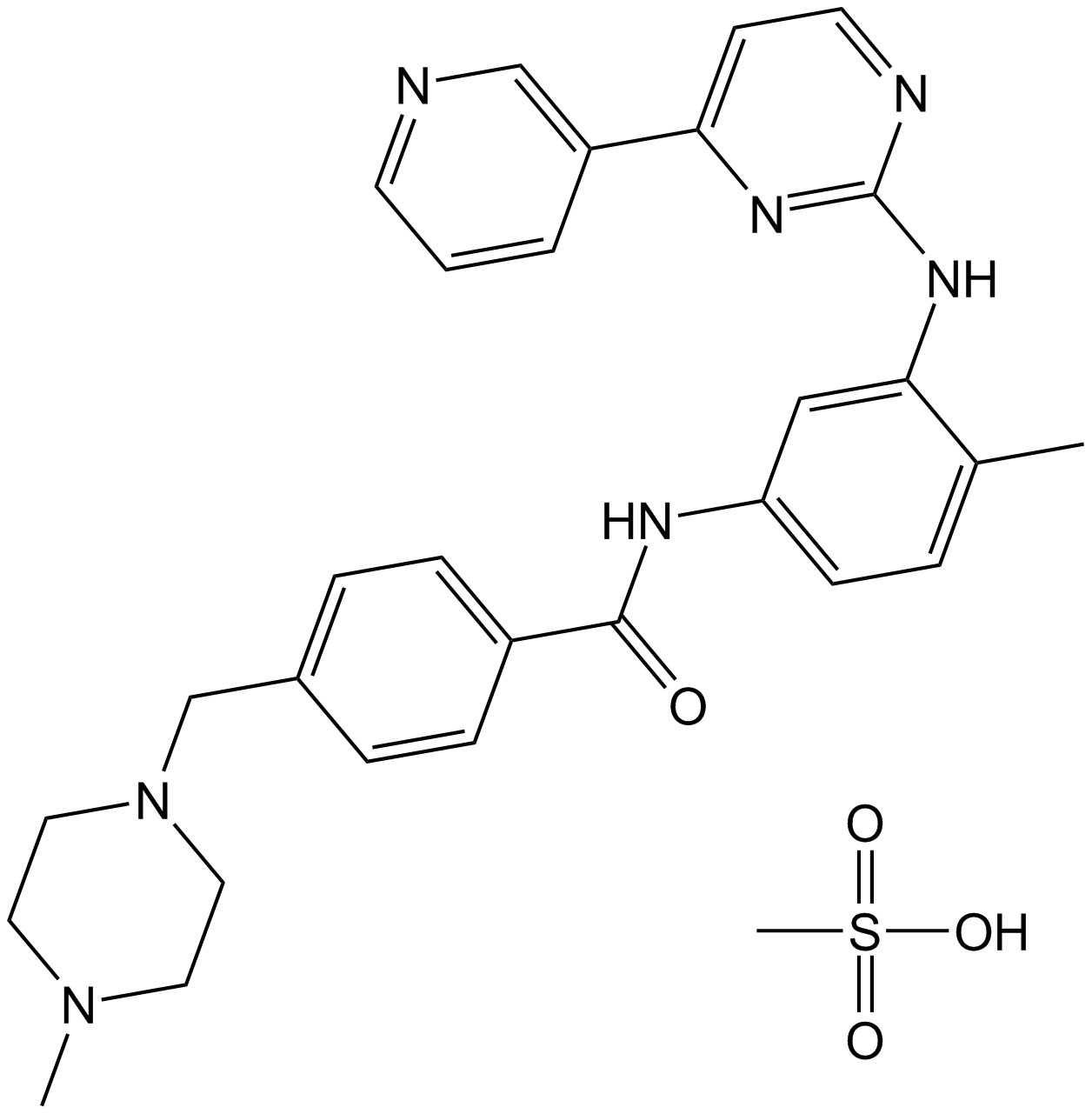
GC11759
Imatinib Mesylate (STI571)
Imatinib Mesylate (STI571) (STI571 Mesylate) is a tyrosine kinases inhibitor that inhibits c-Kit, Bcr-Abl, and PDGFR (IC50=100 nM) tyrosine kinases.
-
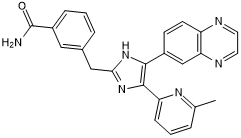
GC50317
IN 1130
Potent and selective inhibitor of TGF-βRI
-
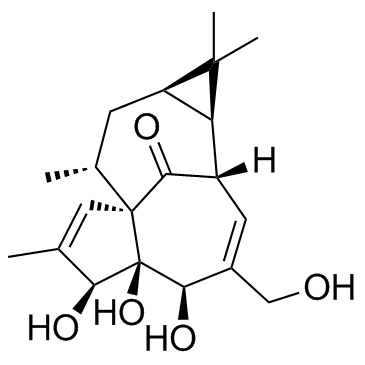
GC32870
Ingenol ((-)-Ingenol)
Ingenol ((-)-Ingenol) is a PKC activator, with a Ki of 30 μM, with antitumor activity.
-
GC61776
Ingenol 3,20-dibenzoate
Ingenol 3,20-dibenzoate is a potent protein kinase C (PKC) isoform-selective agonist.

-
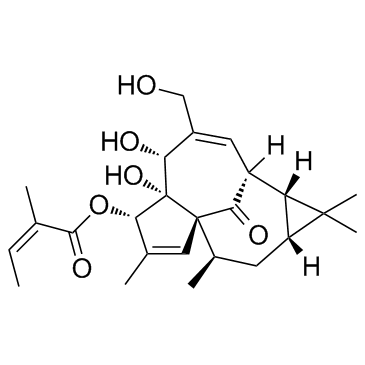
GC31656
Ingenol Mebutate (Ingenol 3-angelate)
Ingenol Mebutate (Ingenol 3-angelate) is an active ingredient in Euphorbia peplus, acts as a potent PKC modulator, with Kis of 0.3, 0.105, 0.162, 0.376, and 0.171 nM for PKC-α, PKC-β, PKC-γ, PKC-δ, and PKC-ε, respectively, and has antiinflammatory and antitumor activity.
-
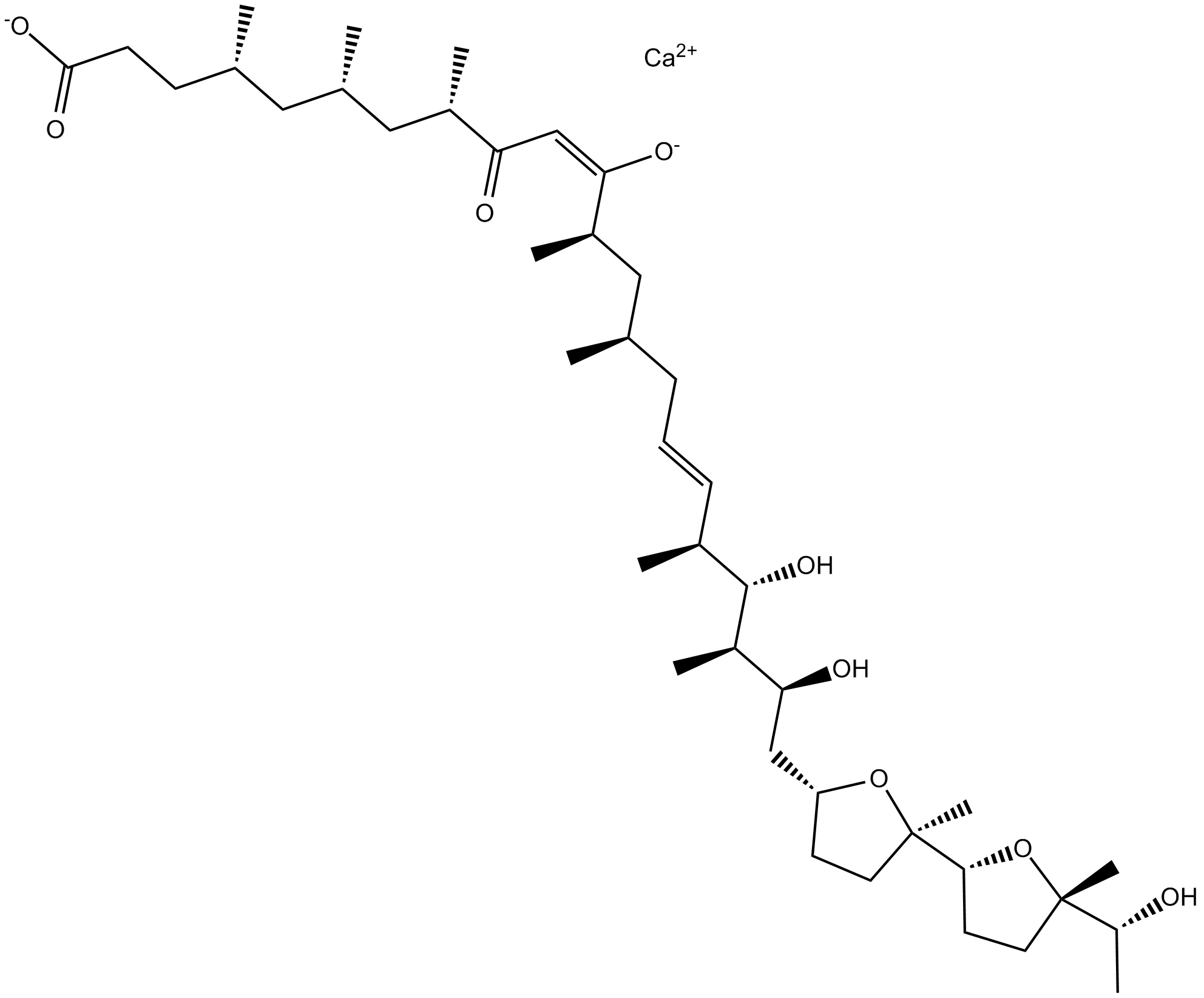
GC15148
Ionomycin calcium salt
Lonomycin is a selective calcium ionophore derived from S. conglobatus that mobilizes intracellular calcium stores.
-
GC15446
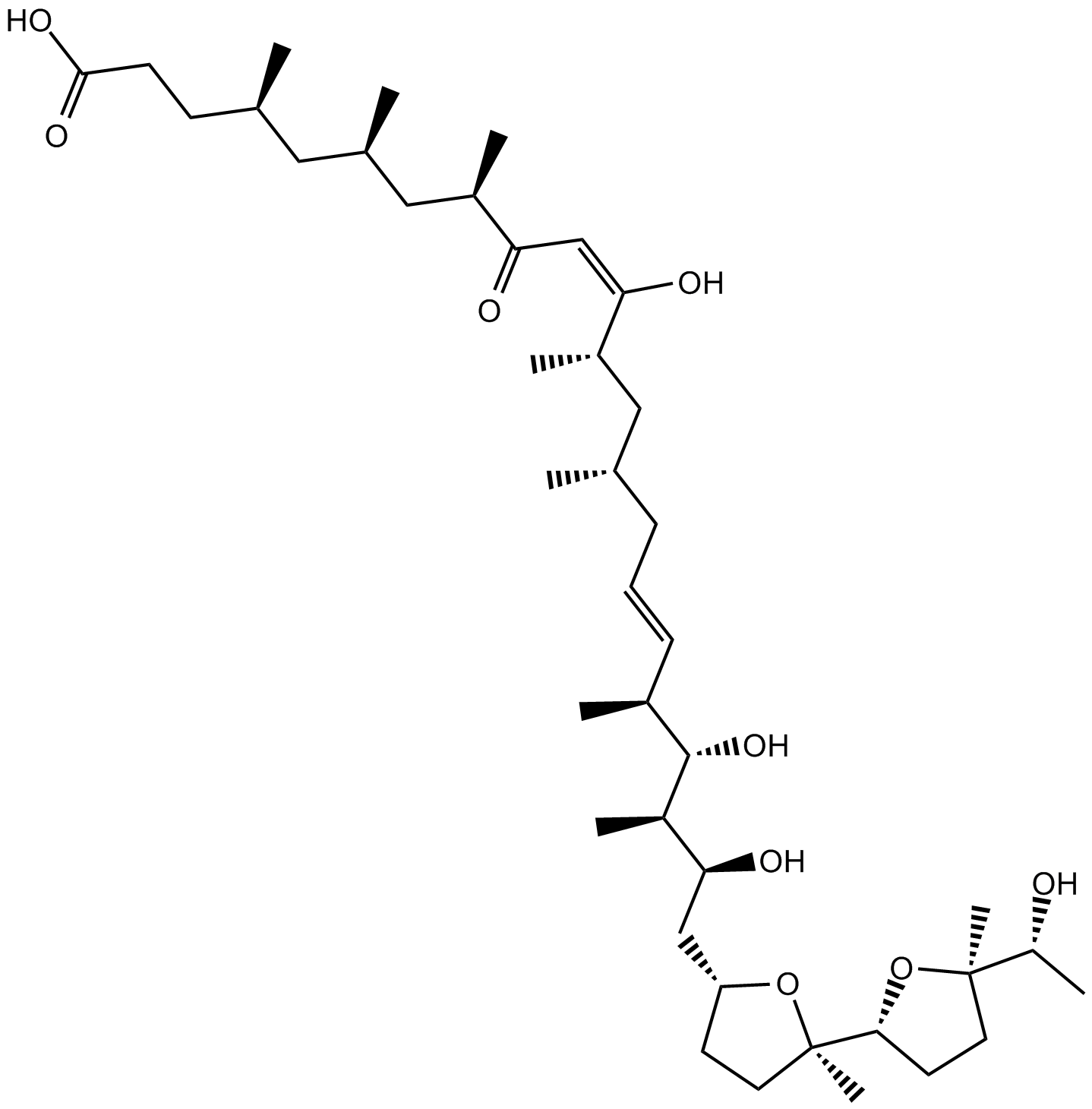
Ionomycin free acid
Ionomycin free acid (SQ23377) is a potent, selective calcium ionophore and an antibiotic produced by Streptomyces conglobatus.
-
GC60210
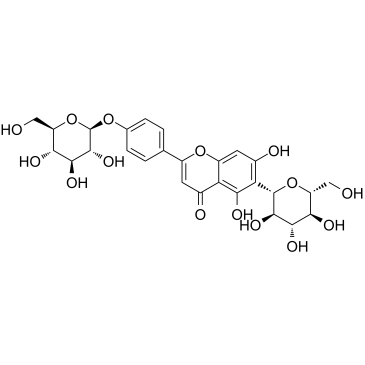
Isosaponarin
Isosaponarin is a flavone glycoside isolated from wasabi leaves.
-
GC12108
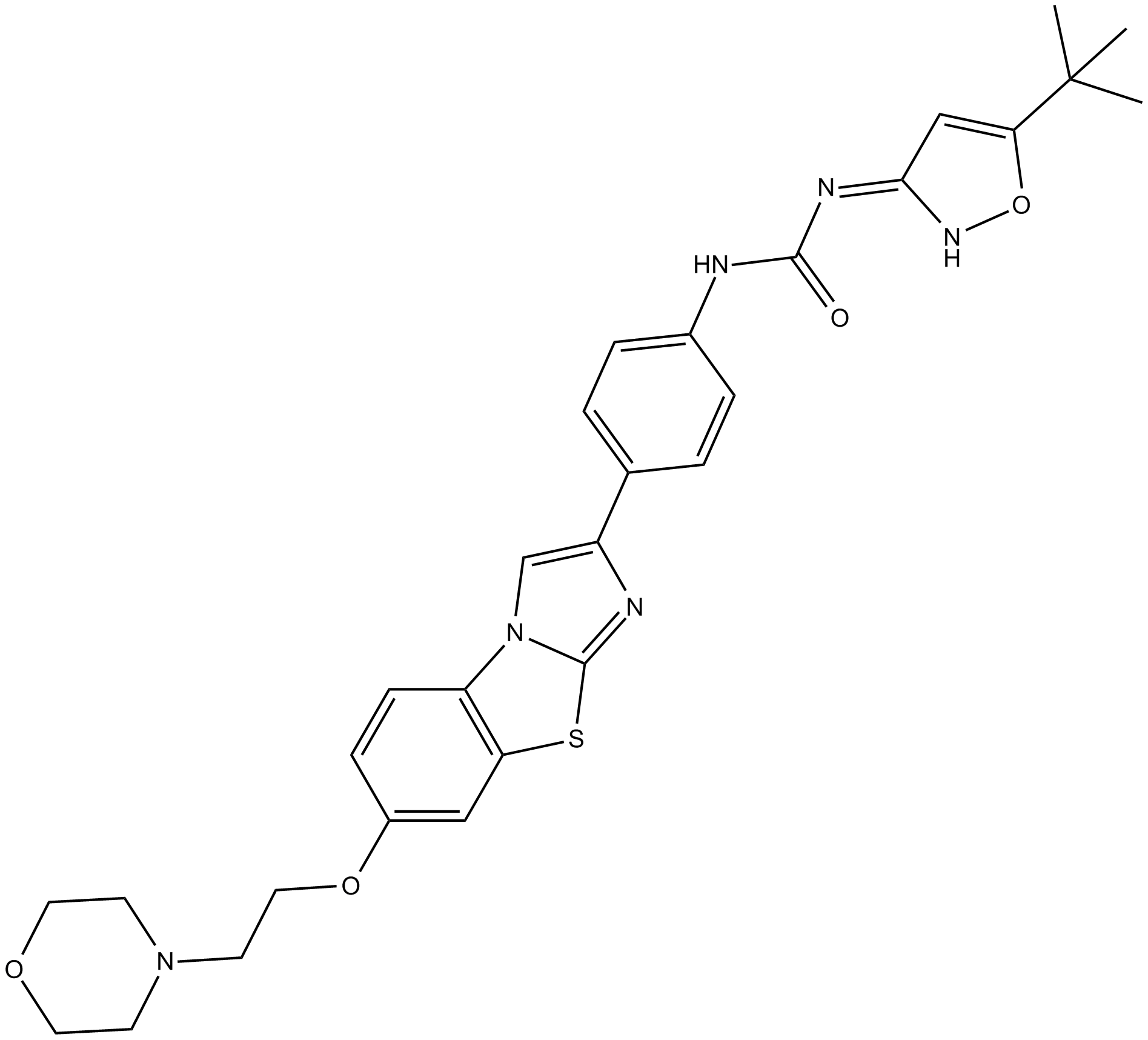
ITD 1
First selective TGFβ inhibitor
-
GC11362
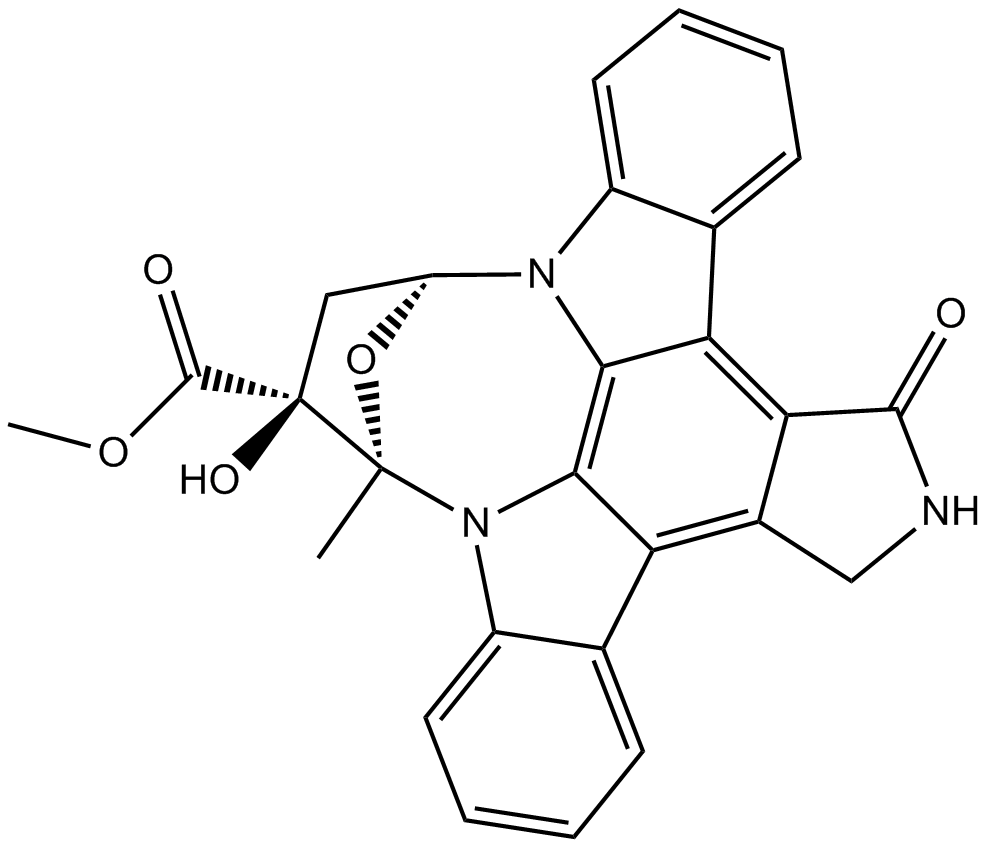
K 252a
A protein kinase inhibitor
-
GC15281
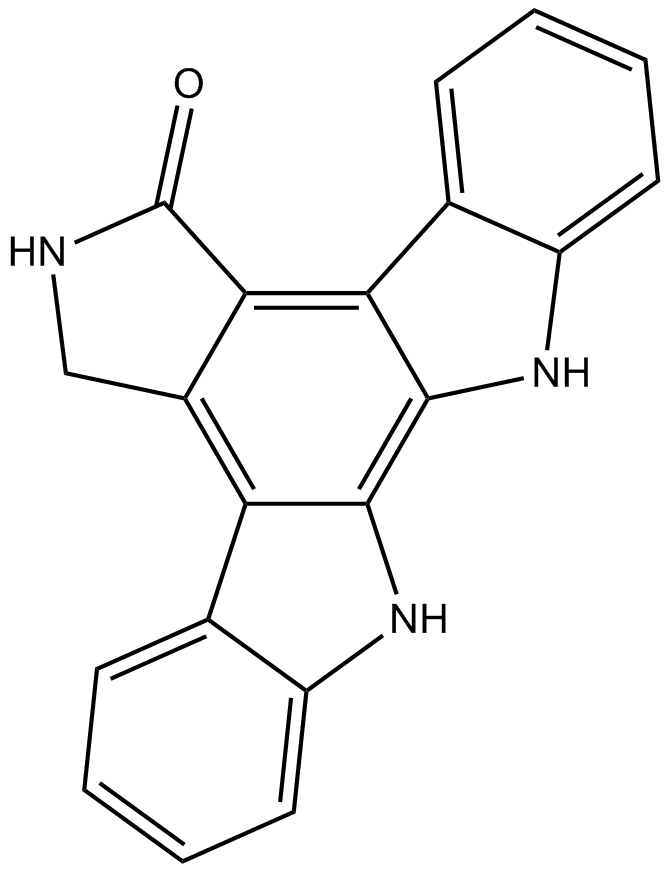
K-252c
Protein kinase inhibitor
-
GC15567
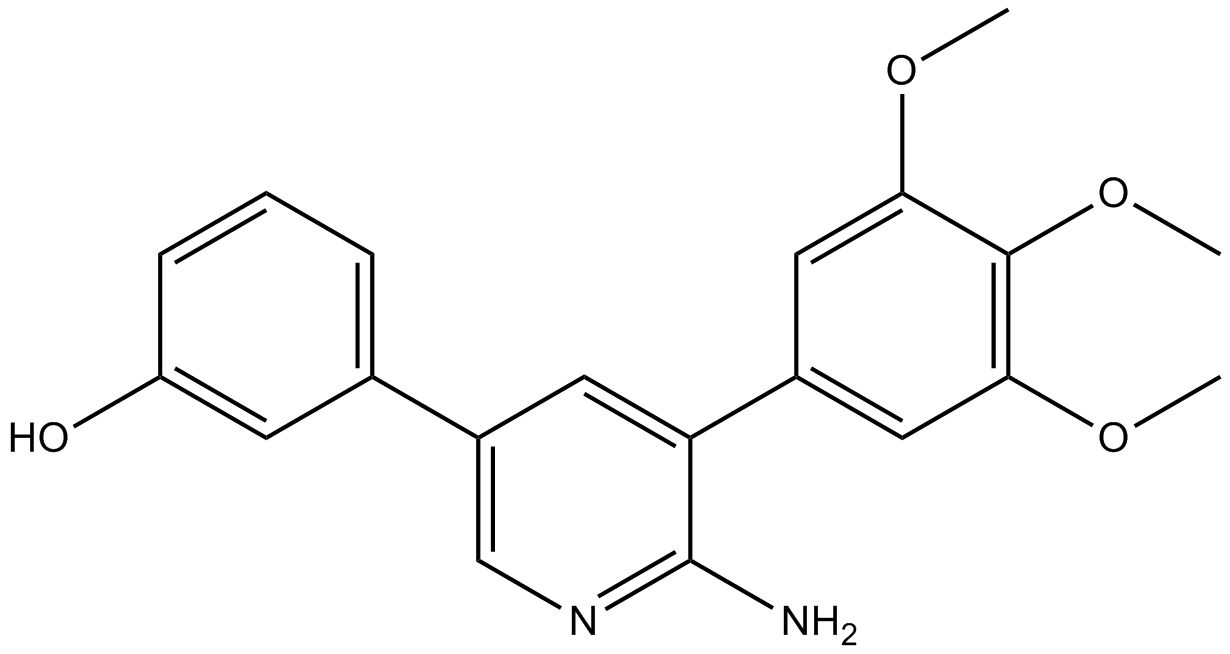
K02288
ALK inhibitor
-
GC43993
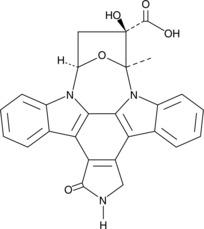
K252b
K252b is an indolocarbazole isolated from the actinomycete Nocardiopsis, first described as an inhibitor of protein kinase C.
-
GC15057
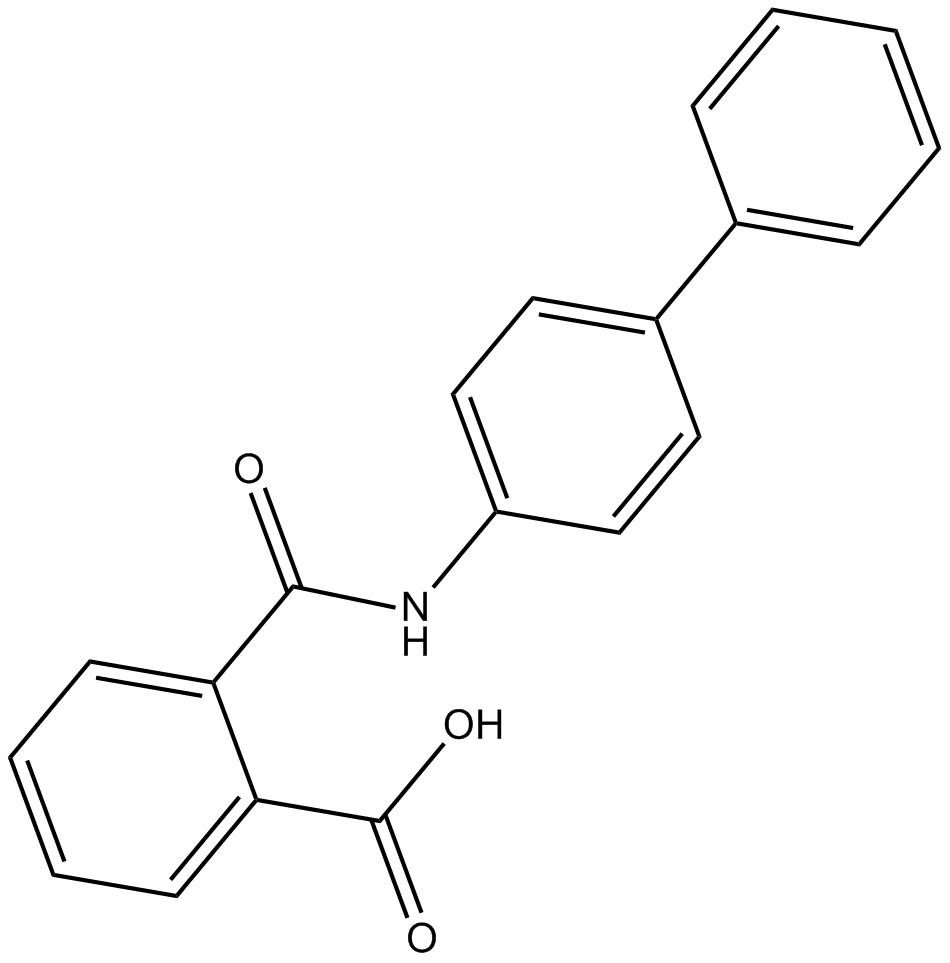
Kartogenin
Promote differentiation of multipotent MSCs into chondrocytes
-
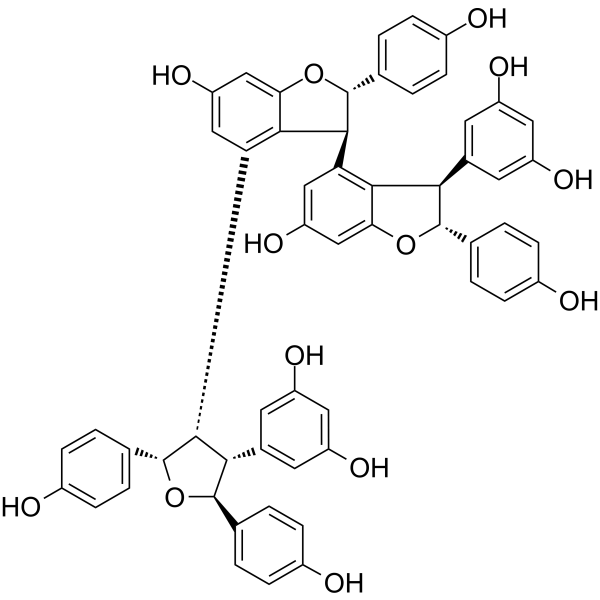
GC64263
Kobophenol A
Kobophenol A, an oligomeric stilbene, blocks the interaction between the ACE2 receptor and S1-RBD with an IC50 of 1.81 μM and inhibits SARS-CoV-2 viral infection in cells with an EC50 of 71.6 μM.
-
GC17346
KT 5823
A protein kinase G inhibitor
-
GC12817
K–115 hydrochloride dihydrate
K-115 hydrochloride dihydrate (K-115) is a specific inhibitor of ROCK, with IC50s of 19 and 51 nM for ROCK2 and ROCK1, respectively.
-
GC40770
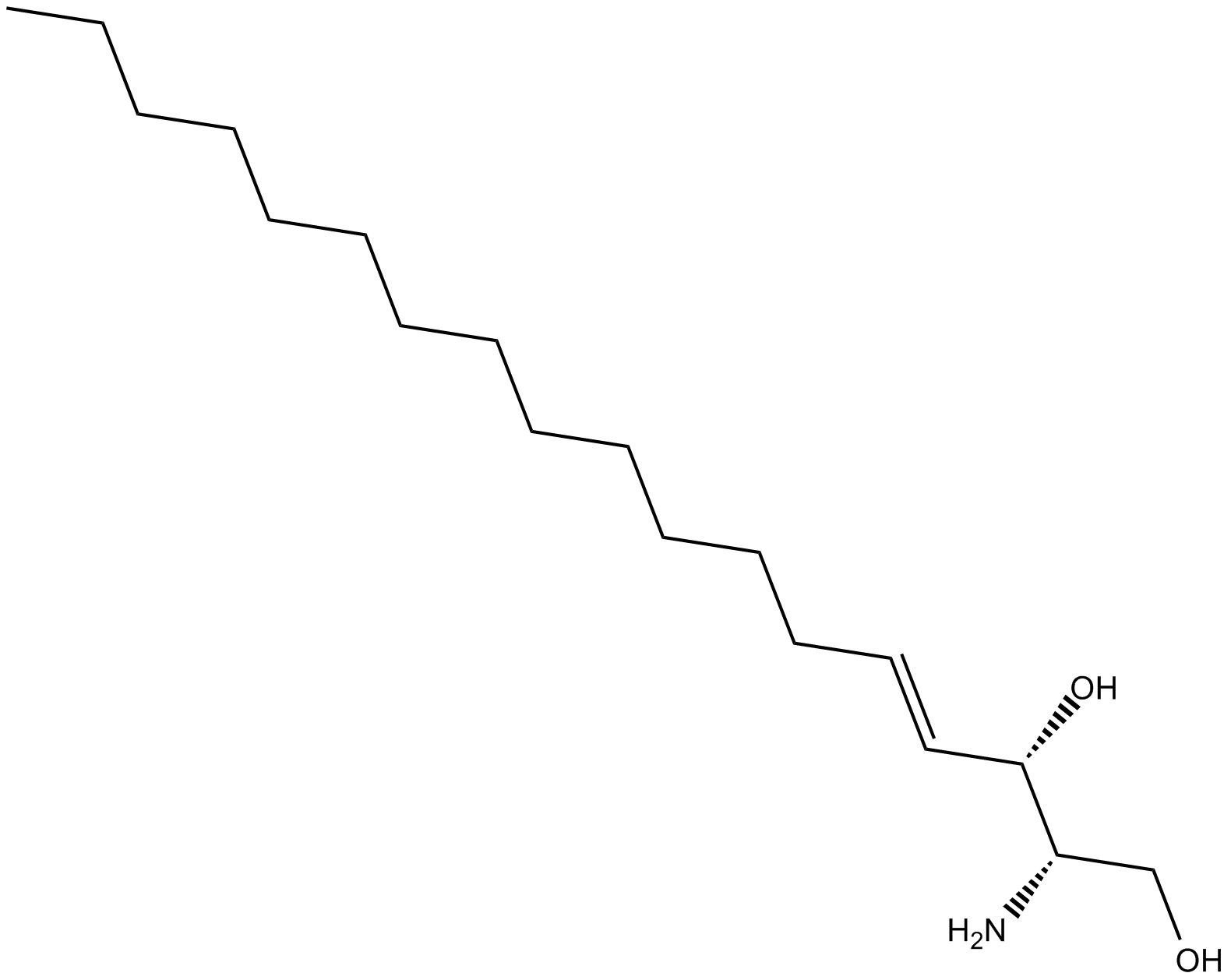
L-erythro Sphingosine (d18:1)
L-erythro Sphingosine is a synthetic stereoisomer of sphingosine (d18:1).
-
GC17815
L-threo-Sphingosine C-18
L-threo-Sphingosine C-18 is a potent MAPK inhibitor. L-threo-Sphingosine C-18 induces apoptosis and clear DNA fragmentation. L-threo-Sphingosine C-18 shows anticancer effect.
-
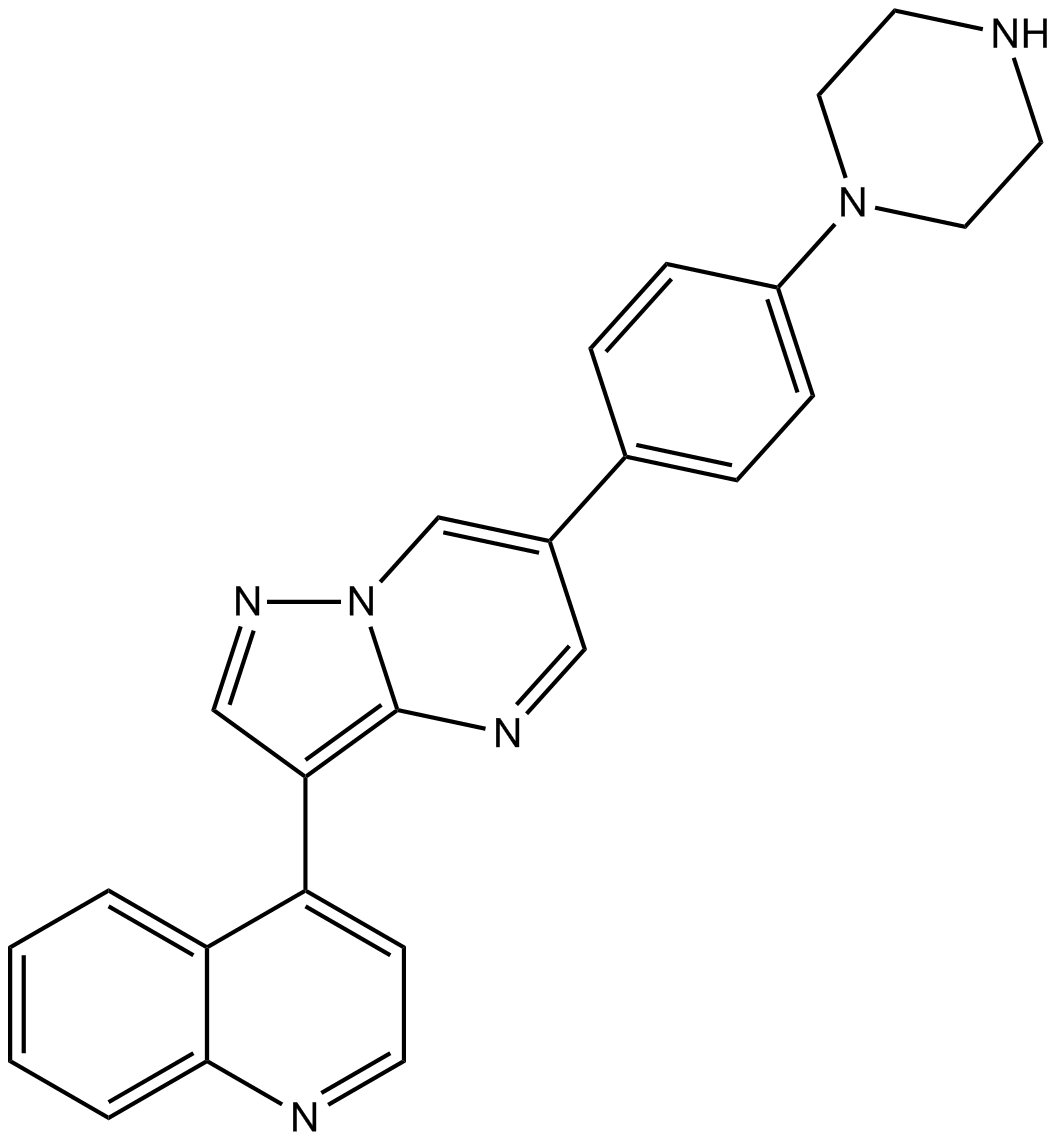
GC16580
LDN-193189
ALK inhibitor,potent and selective
-
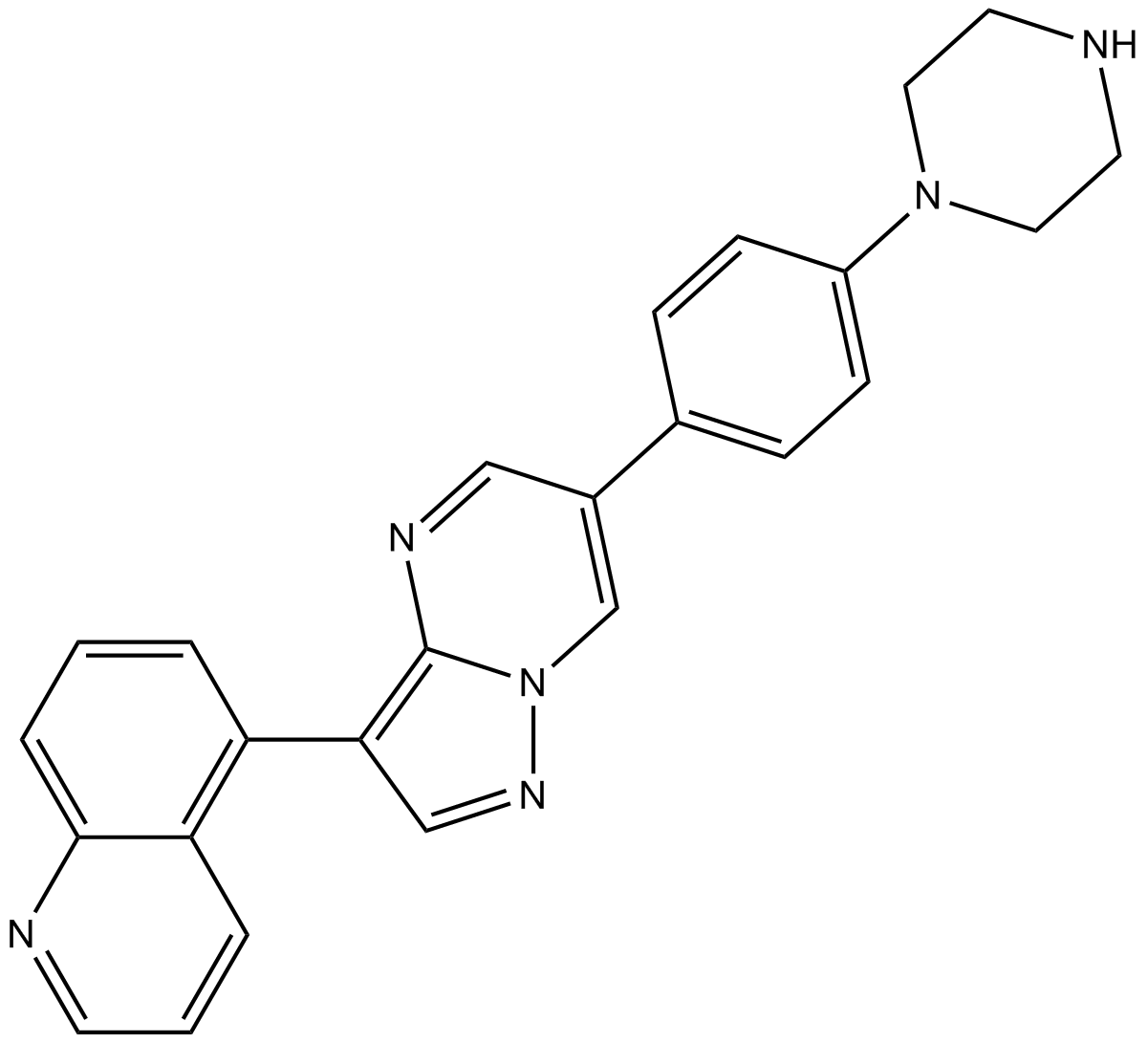
GC17035
LDN-212854
BMP receptor inhibitor,potent and selective
-
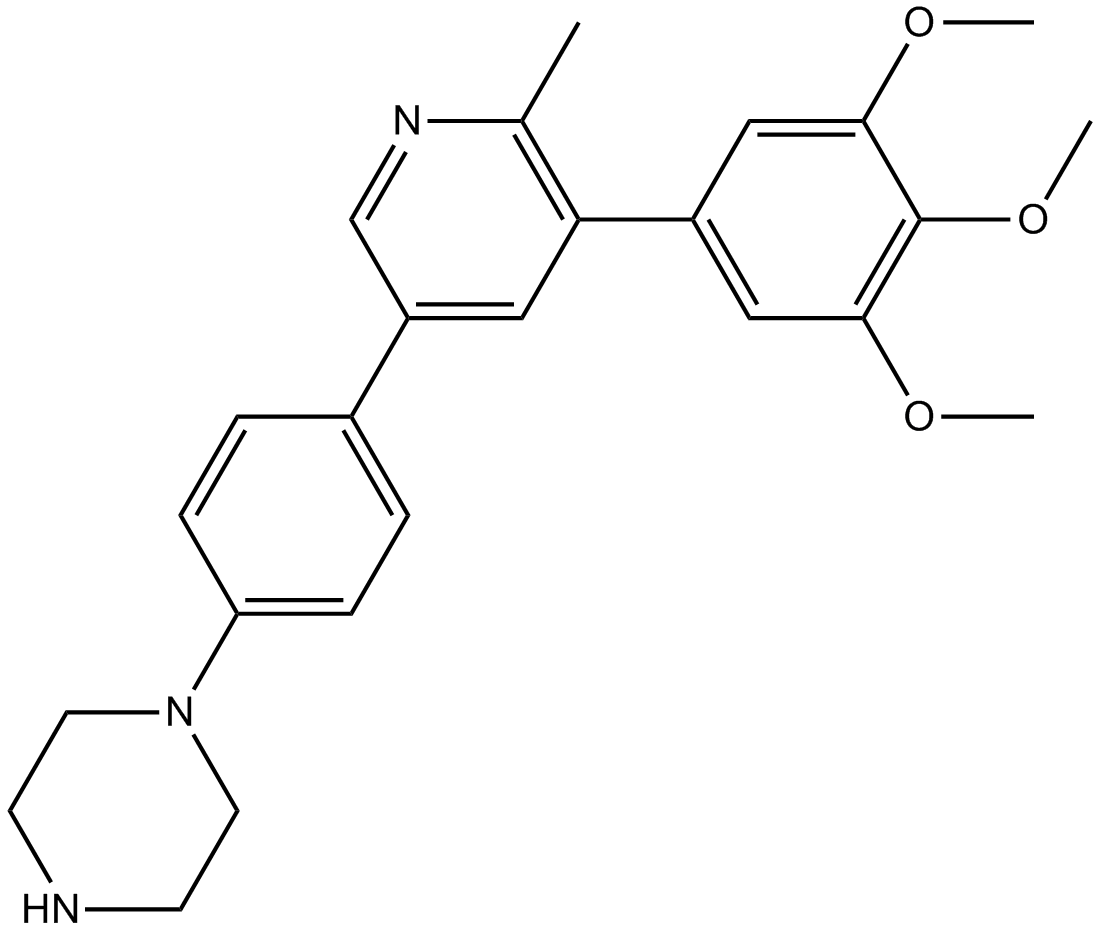
GC13225
LDN-214117
potent and selective ALK2 inhibitor
-
GC14931
LDN193189 Hydrochloride
An inhibitor of BMP receptors ALK1, ALK2, ALK3, and ALK6

-
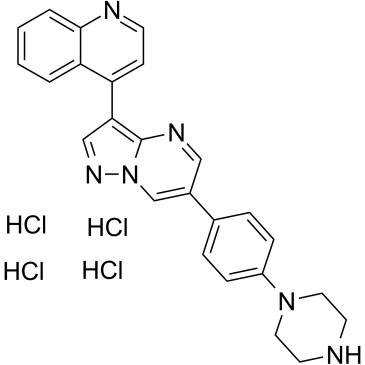
GC36433
LDN193189 Tetrahydrochloride
A selective BMP type I receptor inhibitor
-
GC39398
LSKL, Inhibitor of Thrombospondin (TSP-1) (TFA)
LSKL, Inhibitor of Thrombospondin (TSP-1) TFA is a latency-associated protein (LAP)-TGFβ derived tetrapeptide and a competitive TGF-β1 antagonist. LSKL, Inhibitor of Thrombospondin (TSP-1) TFA inhibits the binding of TSP-1 to LAP and alleviates renal interstitial fibrosis and hepatic fibrosis. LSKL, Inhibitor of Thrombospondin (TSP-1) TFA suppresses subarachnoid fibrosis via inhibition of TSP-1-mediated TGF-β1 activity, prevents the development of chronic hydrocephalus and improves long-term neurocognitive defects following subarachnoid hemorrhage (SAH). LSKL, Inhibitor of Thrombospondin (TSP-1) TFA can readily crosse the blood-brain barrier.

-
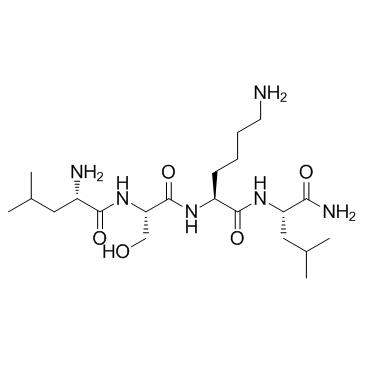
GC32728
LSKL, Inhibitor of Thrombospondin TSP-1
LSKL, Inhibitor of Thrombospondin TSP-1 is a latency-associated protein (LAP)-TGFβ derived tetrapeptide and a competitive TGF-β1 antagonist. LSKL, Inhibitor of Thrombospondin TSP-1 inhibits the binding of TSP-1 to LAP and alleviates renal interstitial fibrosis and hepatic fibrosis. LSKL, Inhibitor of Thrombospondin TSP-1 suppresses subarachnoid fibrosis via inhibition of TSP-1-mediated TGF-β1 activity, prevents the development of chronic hydrocephalus and improves long-term neurocognitive defects following subarachnoid hemorrhage (SAH). LSKL, Inhibitor of Thrombospondin TSP-1 can readily crosse the blood-brain barrier.
-
GC69412
Luspatercept
Luspatercept (ACE-536) is a recombinant modified ActRIIB fusion protein that can bind to the ligands of the transforming growth factor β superfamily. Luspatercept increases red blood cell count and promotes maturation of red blood cell precursors. Luspatercept binds with GDF11, inhibiting the Smad2/3 signaling pathway. Luspatercept can be used for research on anemia.

-
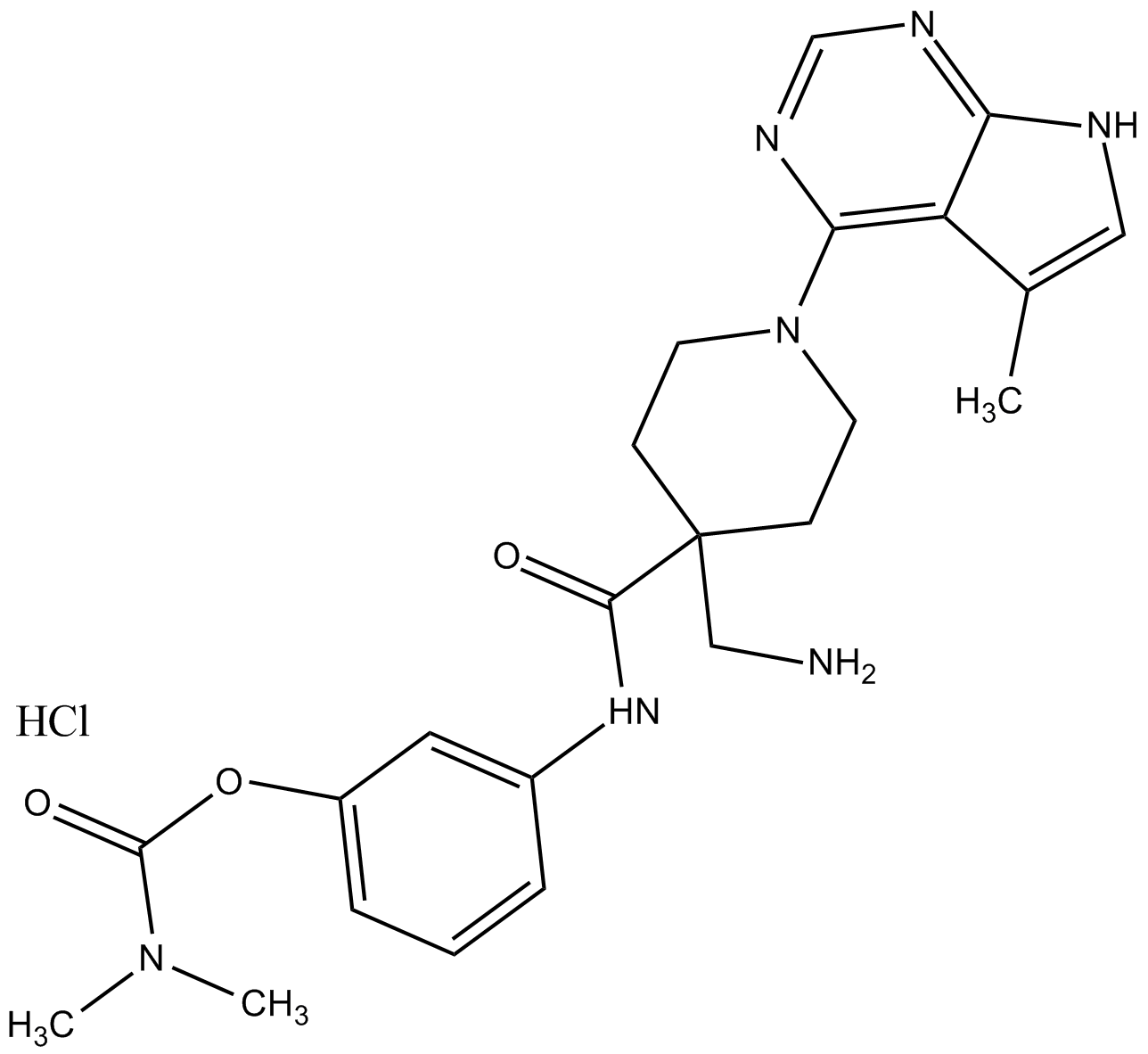
GC12402
LX7101 HCL
LX7101 HCL is a potent inhibitor of LIMK and ROCK2 with IC50 values of 24, 1.6 and 10 nM for LIMK1, LIMK2 and ROCK2, respectively; also inhibits PKA with an IC50 less than 1 nM.
-
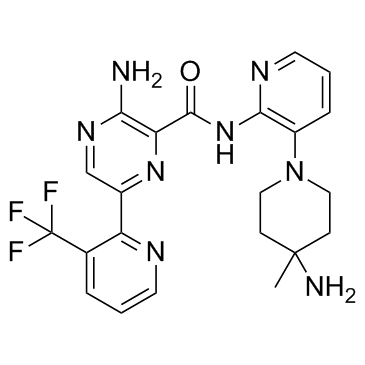
GC32811
LXS196
LXS196 (LXS196) is a potent, selective and orally active protein kinase C (PKC) inhibitor, with IC50 values of 1.9 nM, 0.4 nM and 3.1 μM for PKCα, PKCθ and GSK3β, respectively. LXS196 has the potential for uveal melanoma research.
-
GC17563
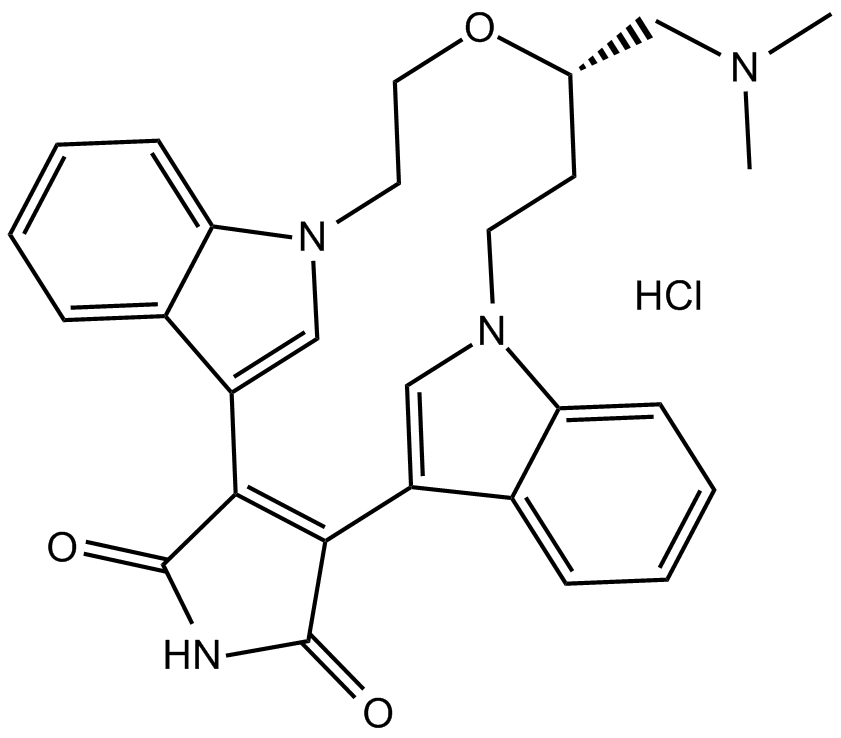
LY 333531 hydrochloride
Ruboxistaurin (LY333531) hydrochloride is an orally active, selective PKC beta inhibitor (Ki=2 nM).
-
GC12363
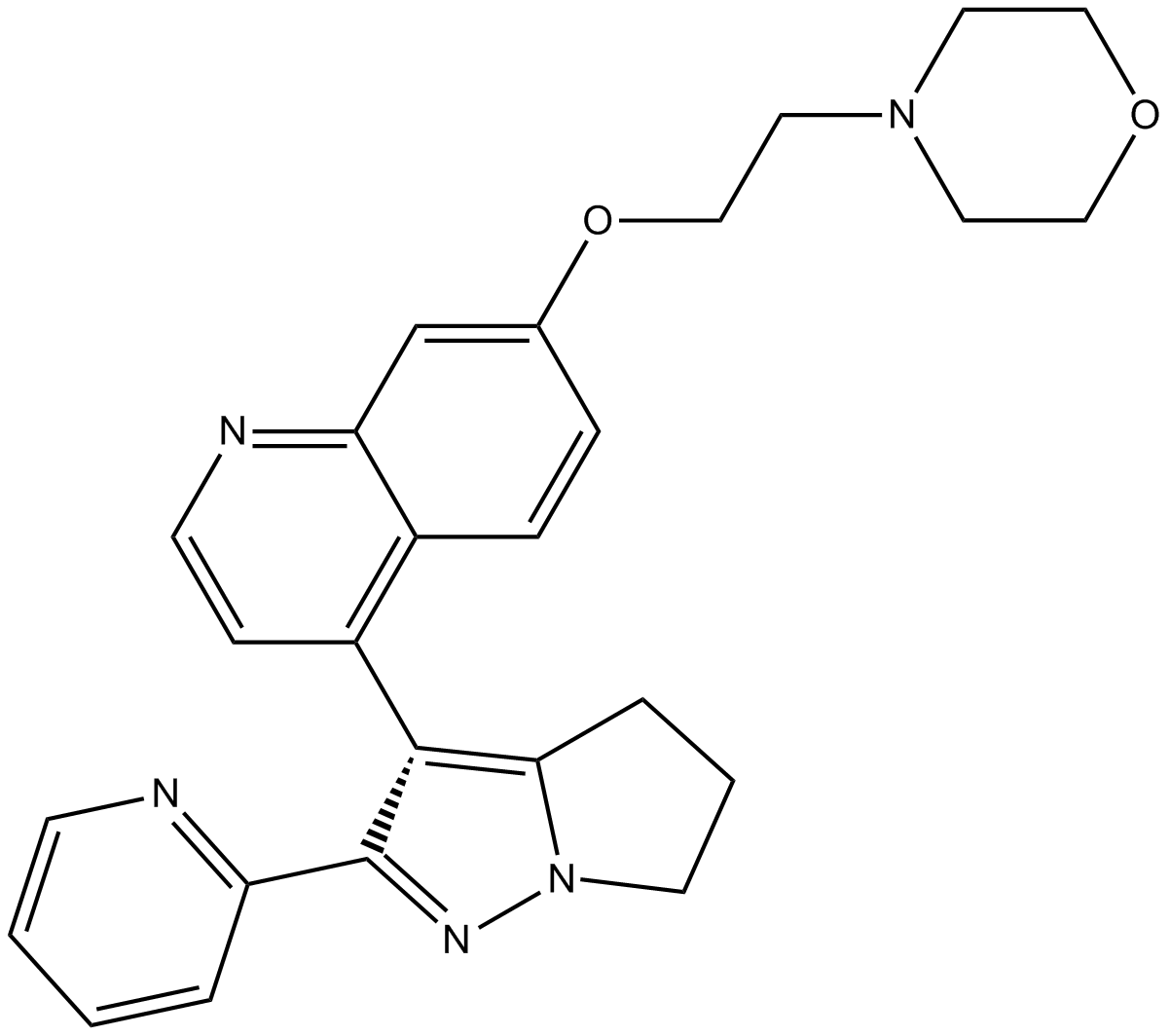
LY2109761
TβRI/II kinase inhibitor
-
GC18015
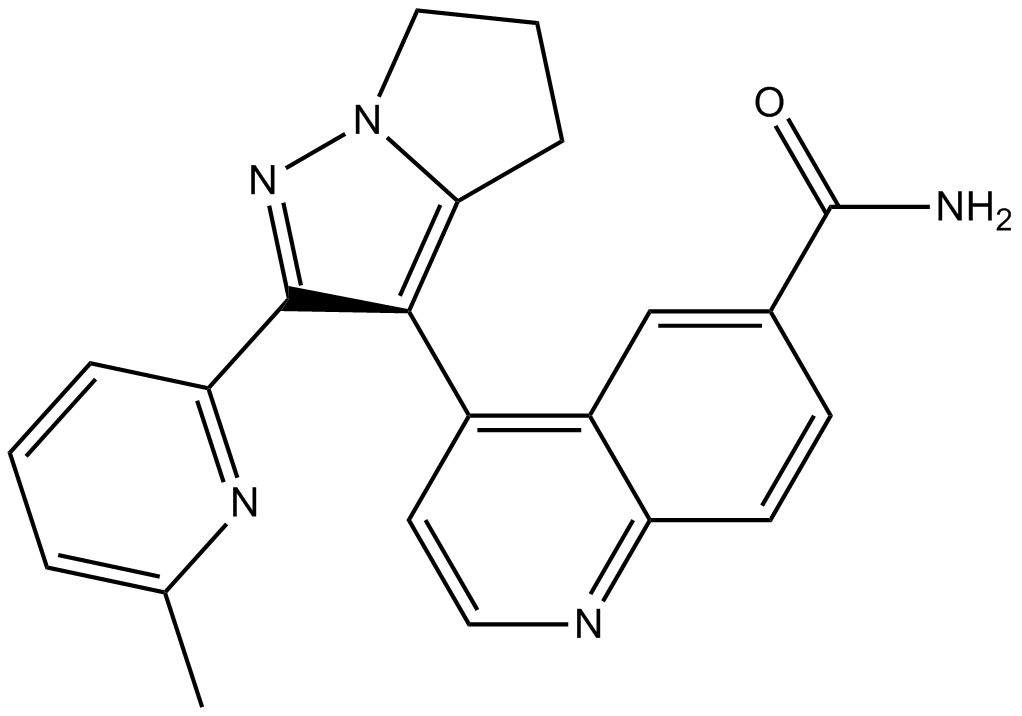
LY2157299
LY2157299 (LY2157299) is an oral and selective TGF-β receptor type I (TGF-βRI) kinase inhibitor with an IC50 of 56 nM.
-
GC19234
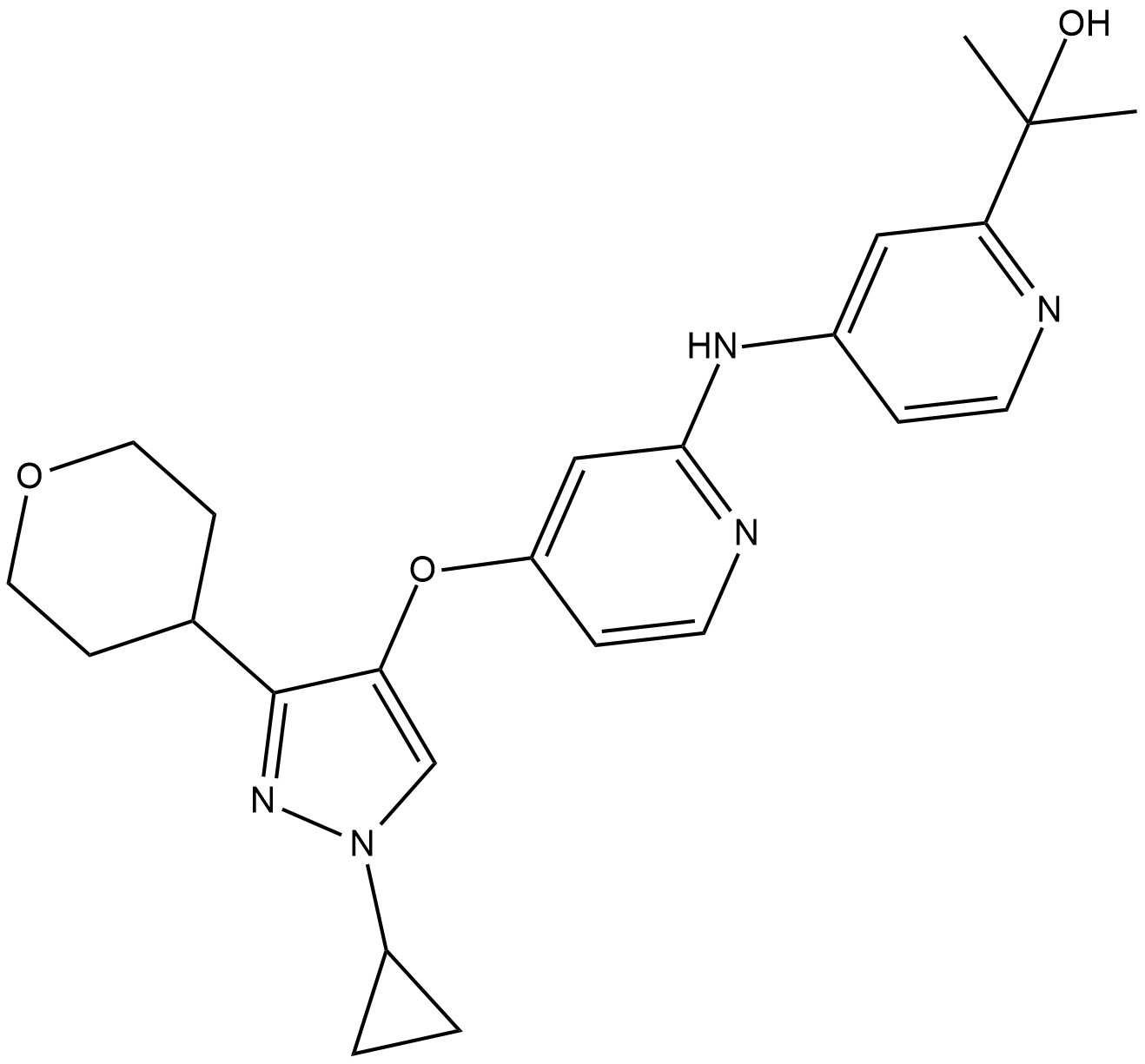
LY3200882
LY3200882 is a novel and highly selective inhibitor of TGF-β receptor type 1 (TGFβRI).
-
GC11604
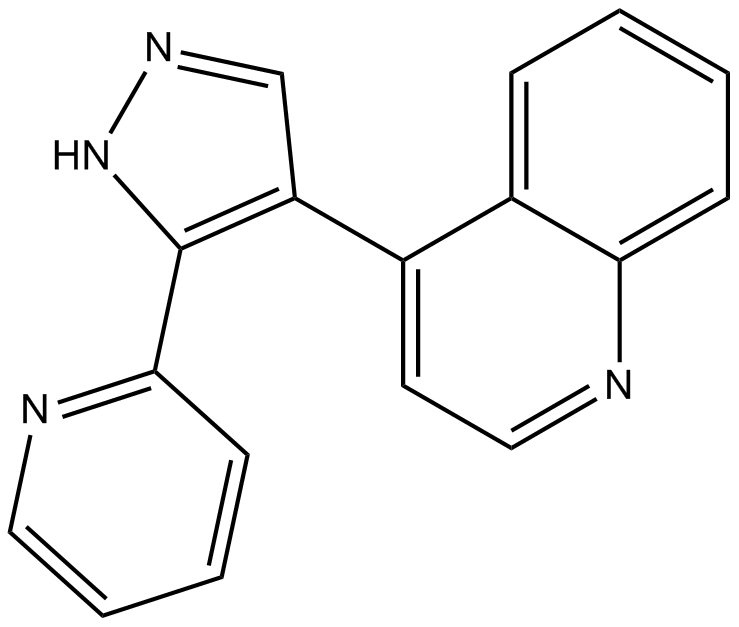
LY364947
Inhibitor of TGF-β type I receptor kinase domain
-
GC30545
Malantide
Malantide is a synthetic dodecapeptide derived from the site phosphorylated by cAMP-dependent protein kinase (PKA) on the β-subunit of phosphorylase kinase. Malantide is a highly specific substrate for PKA with a Km of 15 μM and shows protein inhibitor (PKI) inhibition >90% substrate phosphorylation in various rat tissue extracts. Malantide is also an efficient substrate for PKC with a Km of 16 μM.
-
GC10496
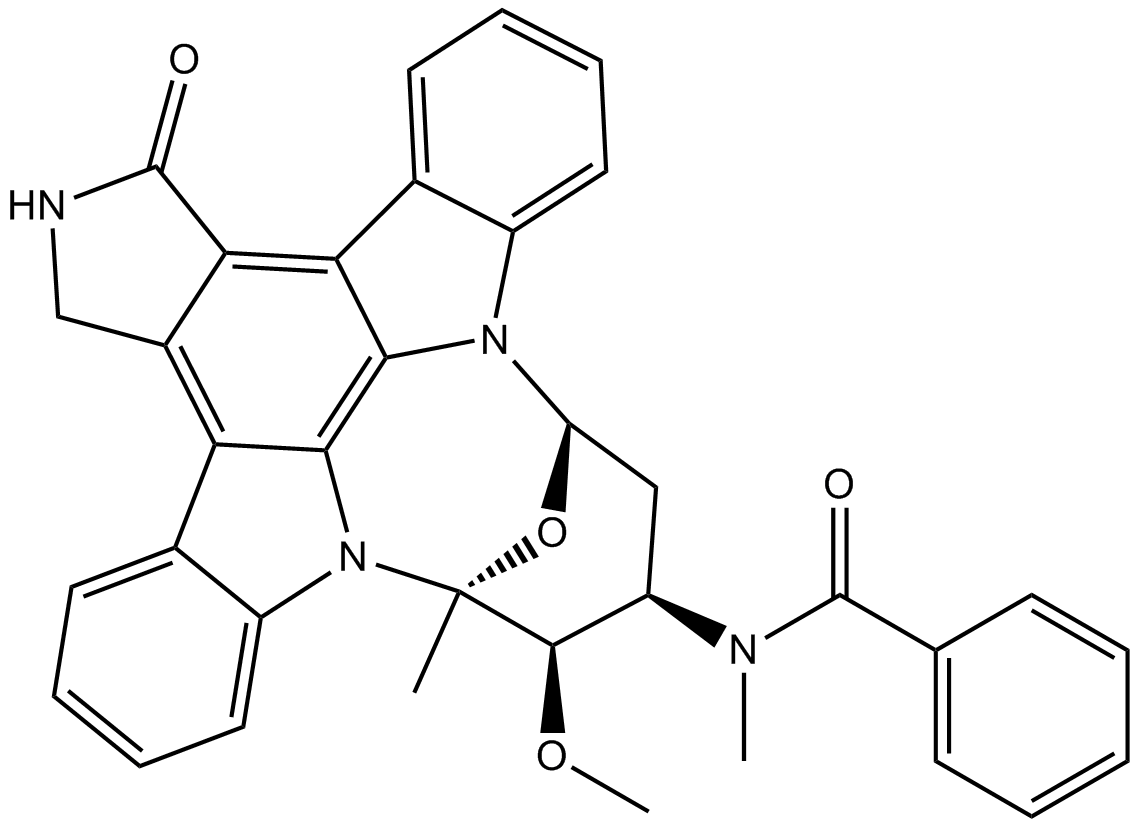
Midostaurin (PKC412)
Midostaurin (PKC412) (PKC412; CGP 41251) is an orally active, reversible multi-targeted protein kinase inhibitor. Midostaurin (PKC412) inhibits PKCα/β/γ, Syk, Flk-1, Akt, PKA, c-Kit, c-Fgr, c-Src, FLT3, PDFRβ and VEGFR1/2 with IC50s ranging from 22-500 nM. Midostaurin (PKC412) also upregulates endothelial nitric oxide synthase (eNOS) gene expression. Midostaurin (PKC412) shows powerful anticancer effects.
-
GC32714
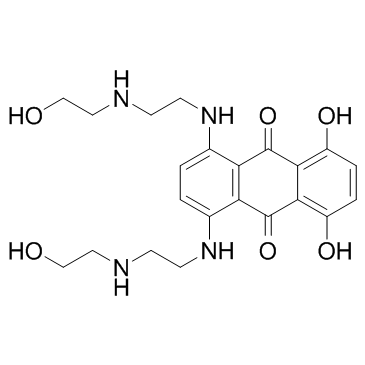
Mitoxantrone (mitozantrone)
Mitoxantrone (mitozantrone) is a potent topoisomerase II inhibitor.
-
GC14363
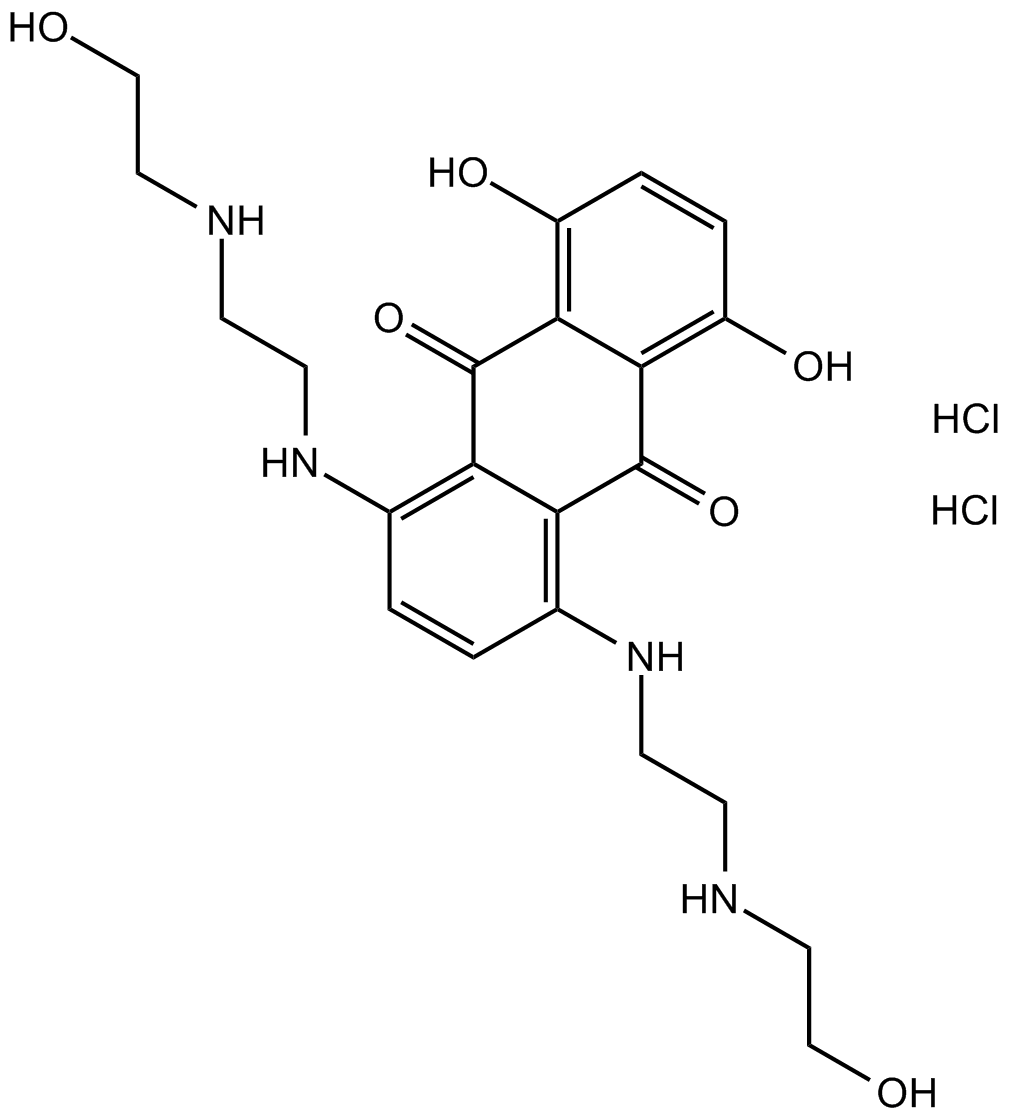
Mitoxantrone HCl
Mitoxantrone HCl is a potent topoisomerase II inhibitor.
-
GC17582
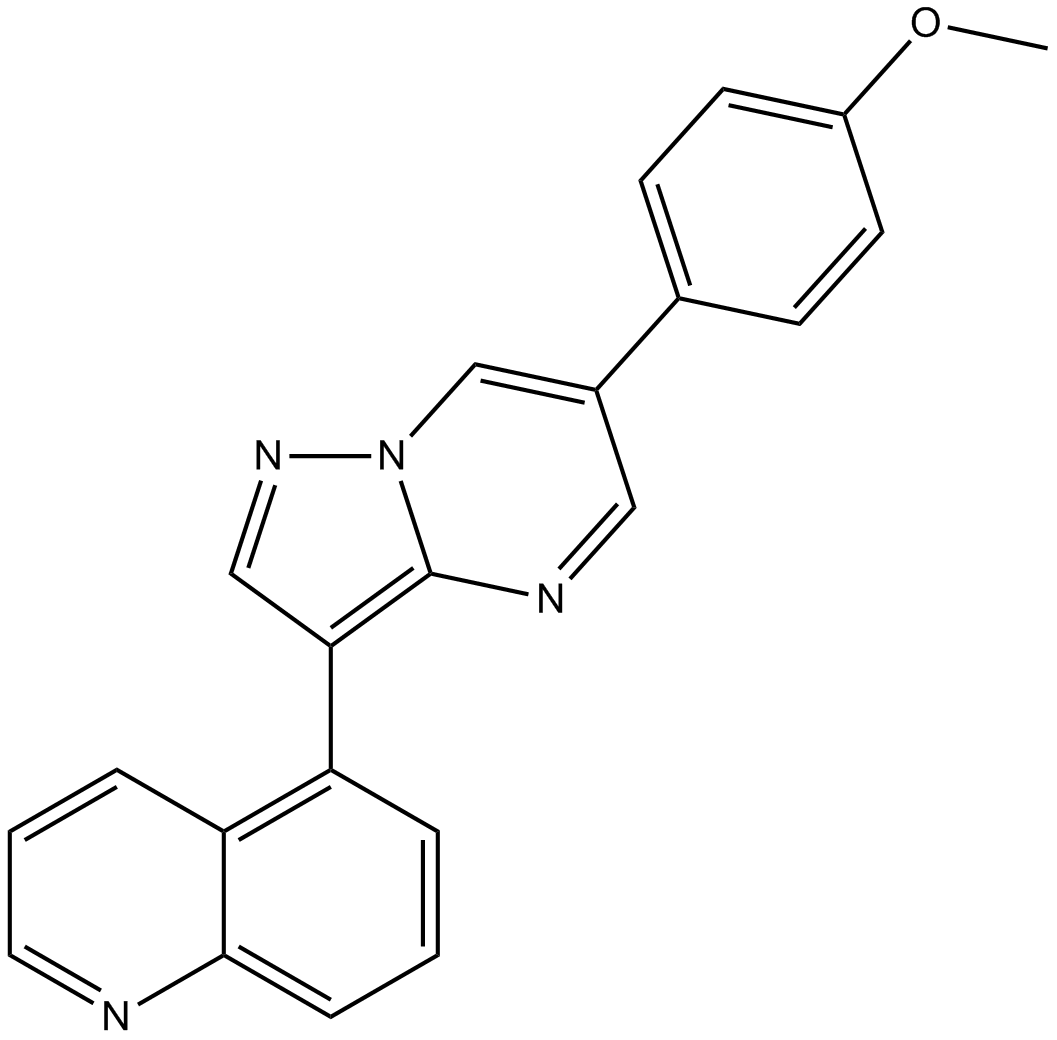
ML347
BMP receptor inhibitor,potent and selective
-
GC65585
Mongersen
Mongersen (GED-0301) is a specific and orally active SMAD7 antisense oligonucleotide.

-
GC63778
Myelin Basic Protein TFA
Myelin Basic Protein (MHP4-14) TFA, a synthetic peptide comprising residues 4-14 of myelin basic protein, is a very selective PKC substrate (Km=7 μM).
-
GC49269
Myr-ZIP
A PKMζ inhibitor
-
GC44303
N-Acetylpuromycin
N-Acetylpuromycin is a non-ribotoxic form of the antibiotic puromycin that is formed in puromycin-resistant S.
-
GC25662
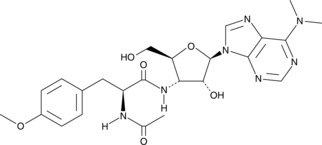
N-Desmethyltamoxifen
N-Desmethyltamoxifen, the major metabolite of Tamoxifen in humans and a ten-fold more potent protein kinase C (PKC) inhibitor than Tamoxifen, also is a potent regulator of ceramide metabolism in human AML cells, limiting ceramide glycosylation, hydrolysis, and sphingosine phosphorylation.
-
GC38931
N-Desmethyltamoxifen hydrochloride
N-Desmethyltamoxifen hydrochloride is the major metabolite of tamoxifen in humans. N-Desmethyltamoxifen, a poor antiestrogen, is a ten-fold more potent protein kinase C (PKC) inhibitor than Tamoxifen. N-Desmethyltamoxifen hydrochloride is also a potent regulator of ceramide metabolism in human AML cells, limiting ceramide glycosylation, hydrolysis, and sphingosine phosphorylation.

-
GC10716
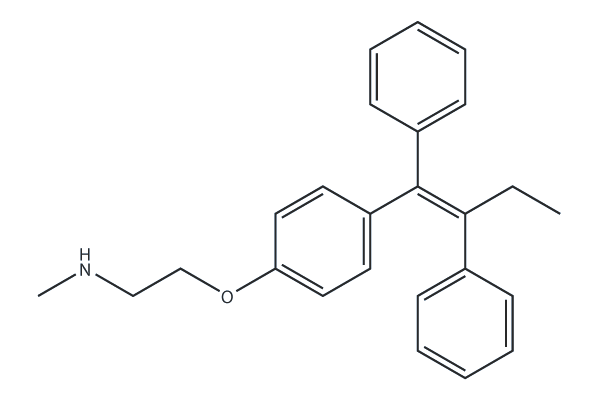
Netarsudil (AR-13324)
ROCK inhibitor
-
GC13514
NG25
An inhibitor of MAP4K2 and TAK1

-
GC25669
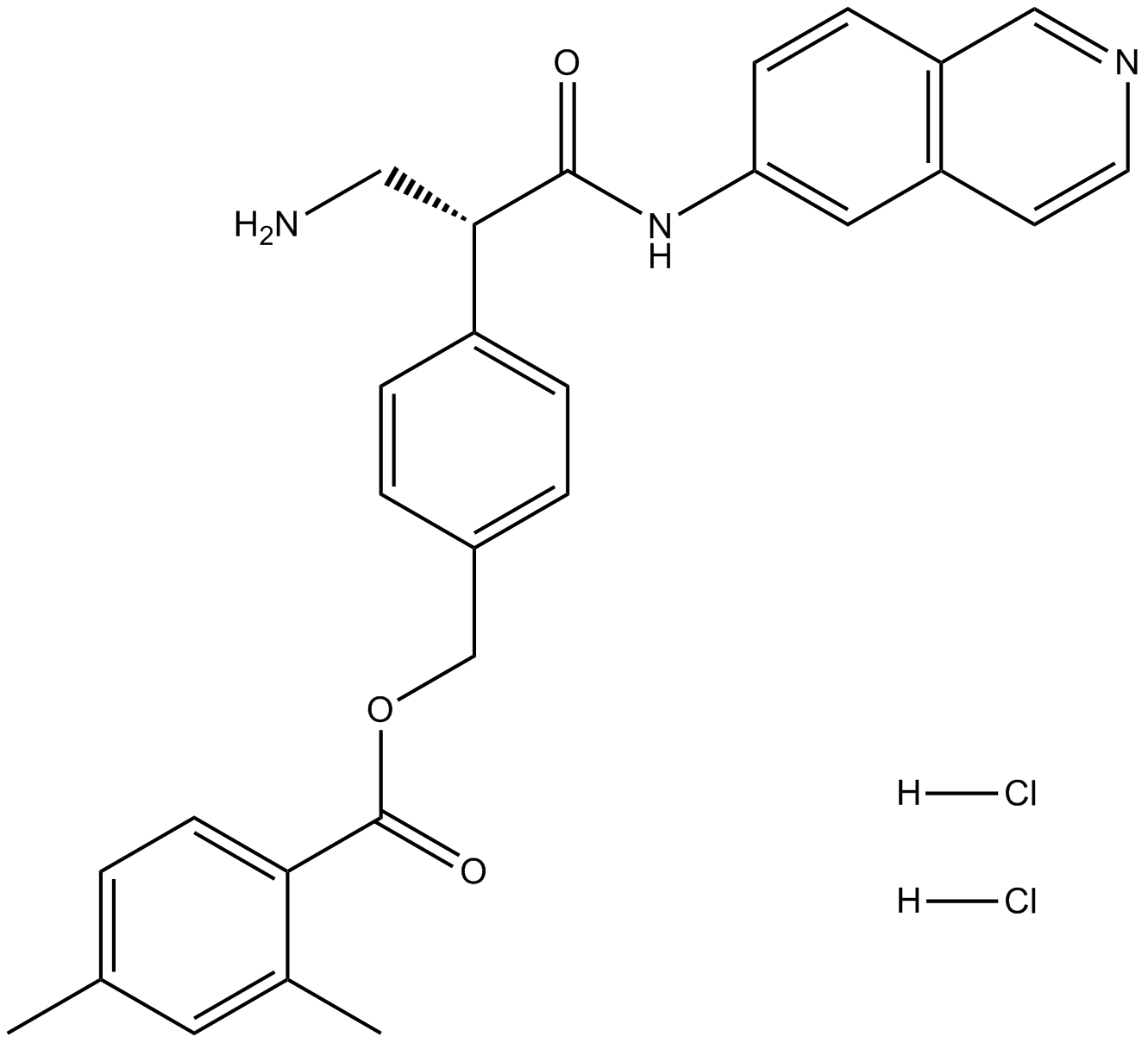
Nilotinib hydrochloride
Nilotinib hydrochloride (AMN-107) is the hydrochloride salt form of nilotinib, an orally bioavailable Bcr-Abl tyrosine kinase inhibitor with antineoplastic activity.
-
GC14075
Nocodazole
A tubulin production inhibitor,anti-neoplastic agent
-
GC60274
O-Desmethyl Midostaurin
O-Desmethyl Midostaurin (CGP62221; O-Desmethyl PKC412) is the active metabolite of Midostaurin via cytochrome P450 liver enzyme metabolism. O-Desmethyl Midostaurin can be used as an indicator for Midostaurin metabolism in vivo. Midostaurin is a multi-targeted protein kinase inhibitor?with?IC50?ranging from 22-500 nM.

-
GC36807
ON 146040
ON 146040 is a potent PI3Kα and PI3Kδ (IC50≈14 and 20 nM, respectively) inhibitor. ON 146040 also inhibits Abl1 (IC50<150 nM).
-
GN10378
Oxymatrine
-
GC36833
p32 Inhibitor M36
p32 inhibitor M36 (M36) is a p32 mitochondrial protein inhibitor, which binds directly to p32 and inhibits p32 association with LyP-1.
-
GC12637
PD 180970
P210bcr/abl tyrosine kinase inhibitor
-
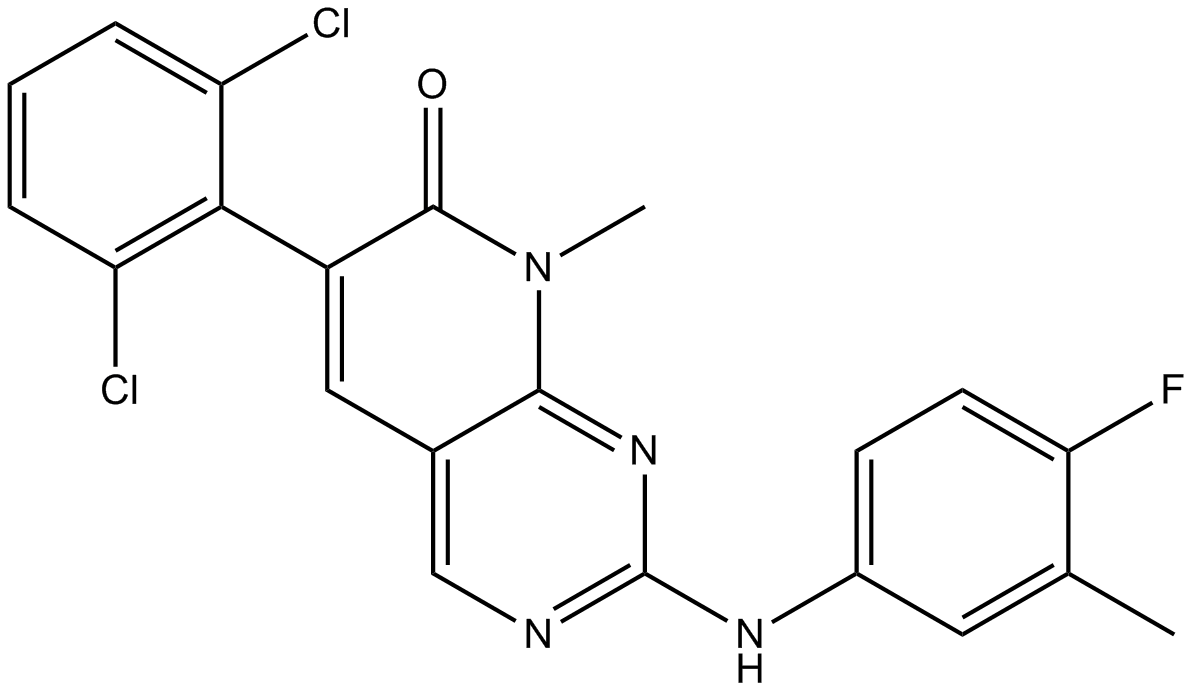
GC13592
PD173955
Dual Src/Abl kinase inhibitor, ATP-competitive,
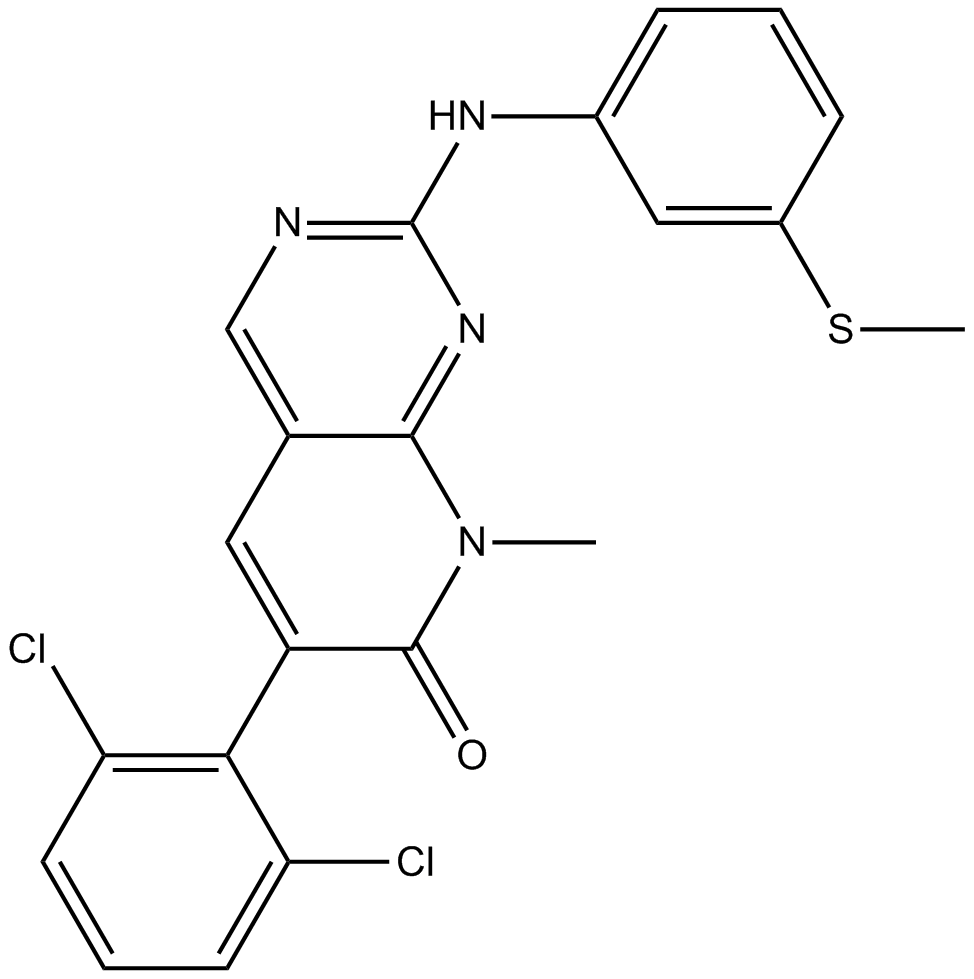
-
GC33901
Pentanoic acid
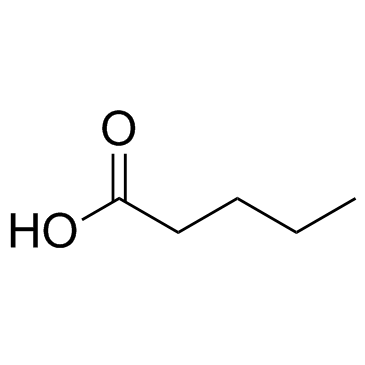
-
GC61585
Pep2m, myristoylated TFA
Pep2m, myristoylated TFA (Myr-Pep2m TFA) is a cell-permeable peptide.

-
GC14767
PF-00562271
PF-562271 (VS-6062) besylate is a potent ATP-competitive, reversible inhibitor of FAK and Pyk2 kinase, with an IC50 of 1.5 nM and 13 nM, respectively.
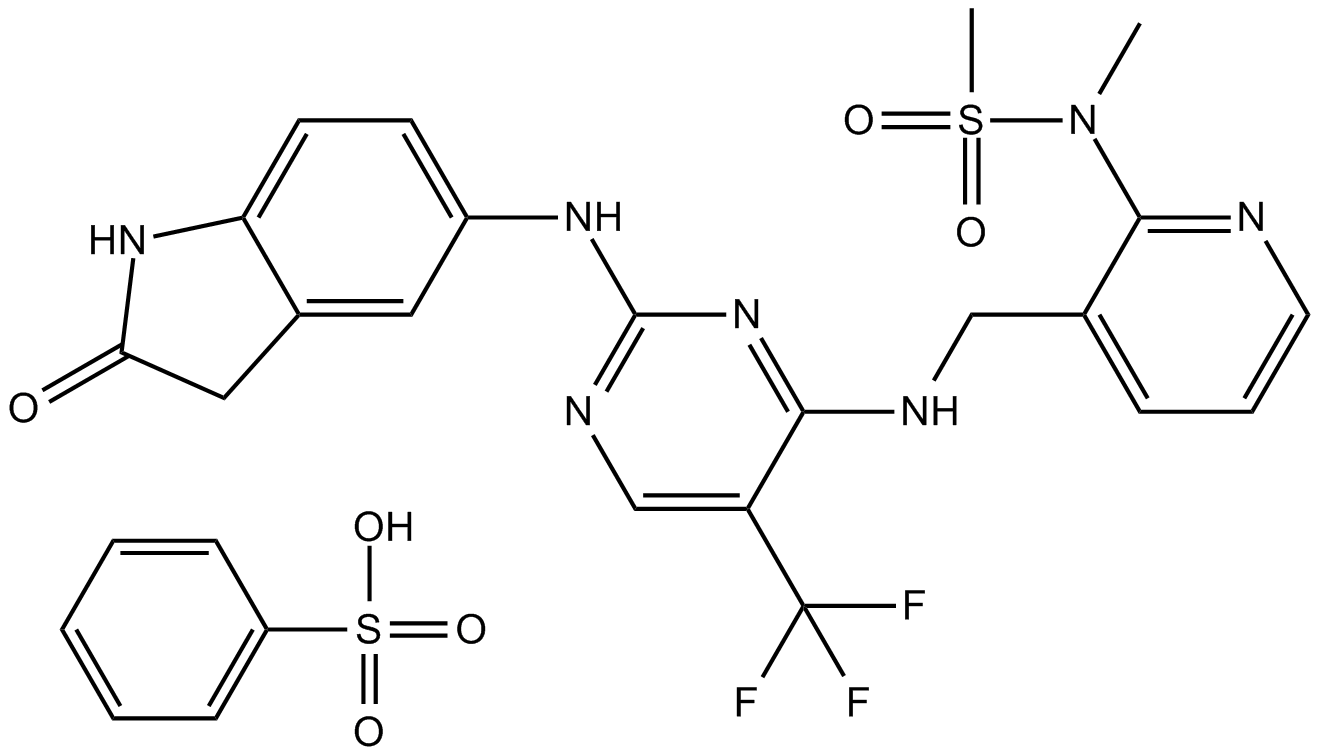
-
GC14407
PF-431396
Pyk2 and FAK inhibitor
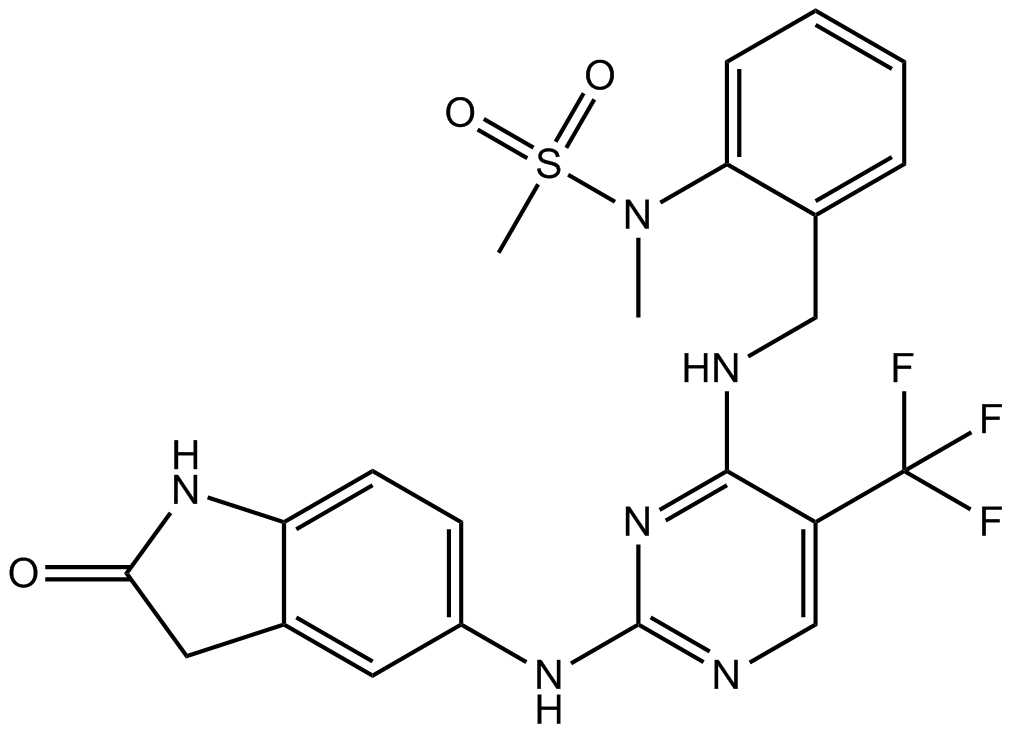
-
GC15380
PF-562271
ATP-competitive FAK inhibitor, reversible
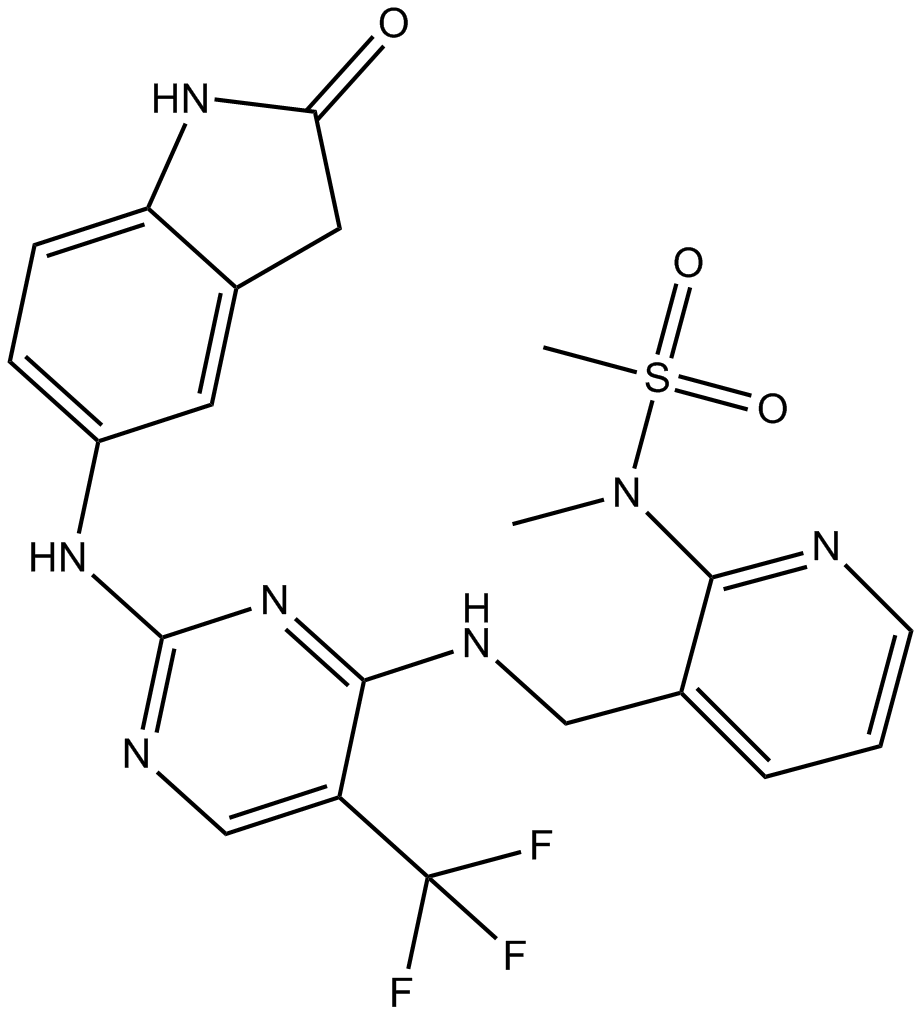
-
GC10810
PF-562271 HCl
PF-562271 (VS-6062) hydrochloride is a potent, ATP-competitive and reversible FAK and Pyk2 kinase inhibitor with IC50s of 1.5 nM and 13 nM, respectively.
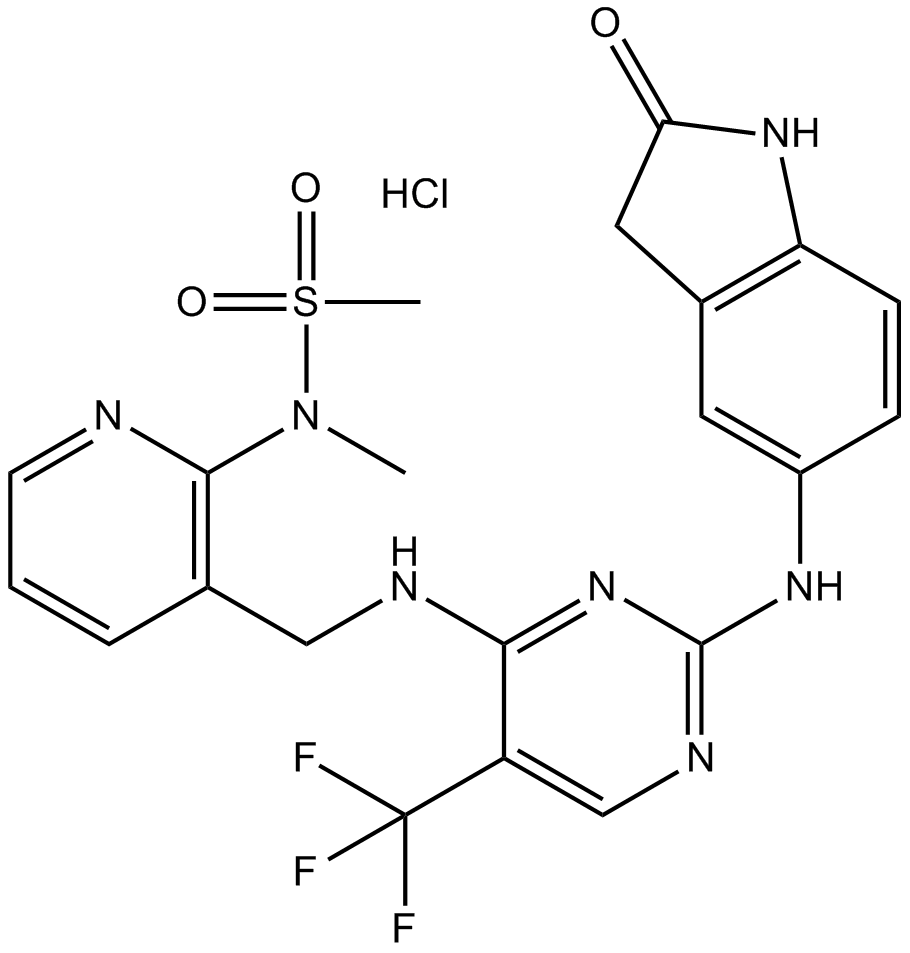
-
GC17352
Phorbol 12,13-dibutyrate
Protein kinase C activator
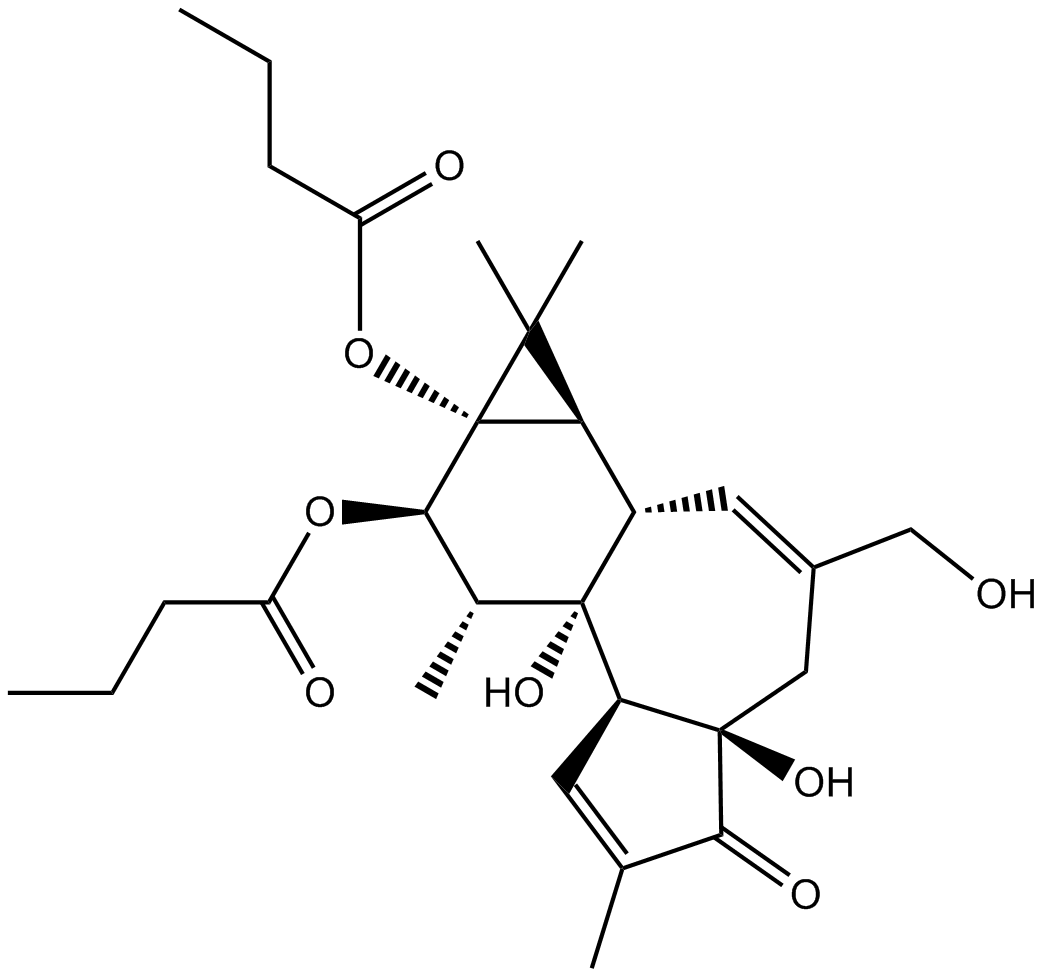
-
GC12790
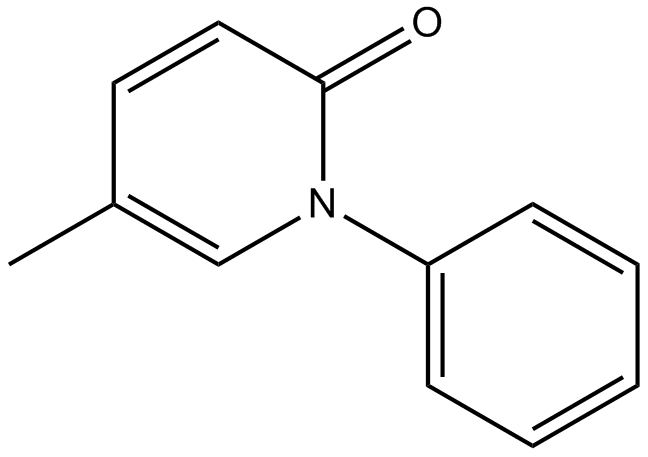
Pirfenidone
TGF-β production inhibitor
-
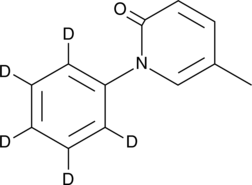
GC40197
Pirfenidone-d5
Pirfenidone-d5 is intended for use as an internal standard for the quantification of pirfenidone by GC- or LC-MS.
-
GC15471
PKC β pseudosubstrate
Selective cell-permeable peptide inhibitor of protein kinase C

-
GC10404
PKC ζ pseudosubstrate
PKC ζ pseudosubstrate is a selective cell-permeable inhibitor of PKC.
-
GC11671
PKC fragment (530-558)
Potent activator of protein kinase C

-
GC44655
PKCε Inhibitor Peptide
PKCε Inhibitor Peptide (ε-V1-2), a PKCε-derived peptide, is a selective PKCε inhibitor.
-
GC31711
PKC-IN-1
PKC-IN-1 is a potent, ATP-competitive and reversible inhibitor of conventional PKC enzymes with Kis of 5.3 and 10.4 nM for human PKCβ and PKCα, and IC50s of 2.3, 8.1, 7.6, 25.6, 57.5, 314, 808 nM for PKCα, PKCβI, PKCβII, PKCθ, PKCγ, PKC mu and PKCε, respectively.
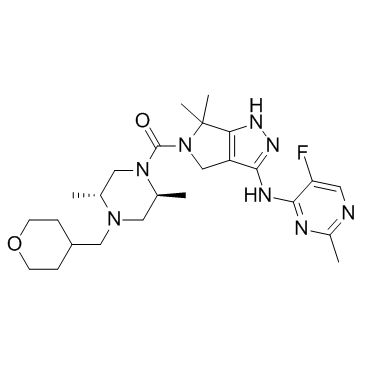
-
GC65126
PKC-iota inhibitor 1
PKC-iota inhibitor 1 (compound 19) is a protein kinase C-iota (PKC-ι ?) inhibitor with an IC50 value of 0.34 μM.
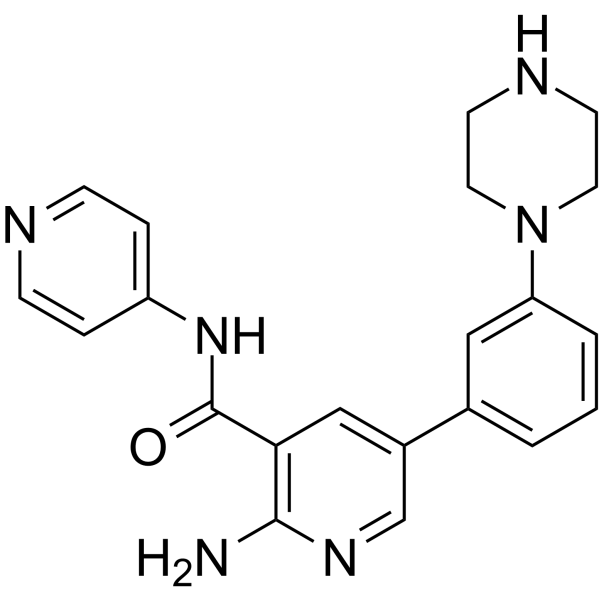
-
GC30200
PKC-theta inhibitor
PKC-theta inhibitor is a selective PKC-θinhibitor, with an IC50 of 12 nM.
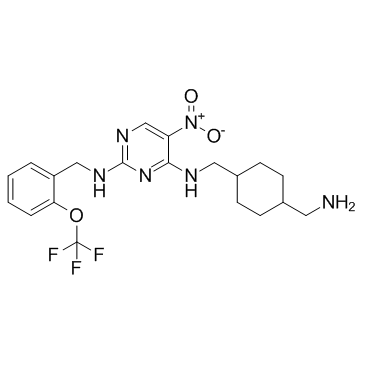
-
GC67686
PKCiota-IN-2 formic
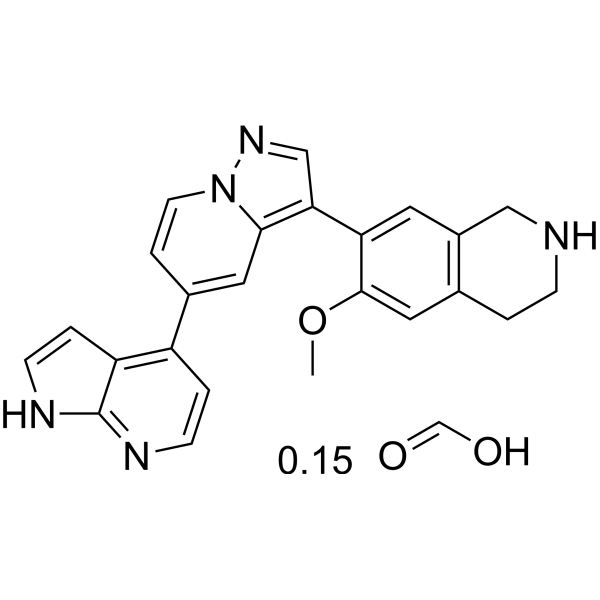
-
GC14396
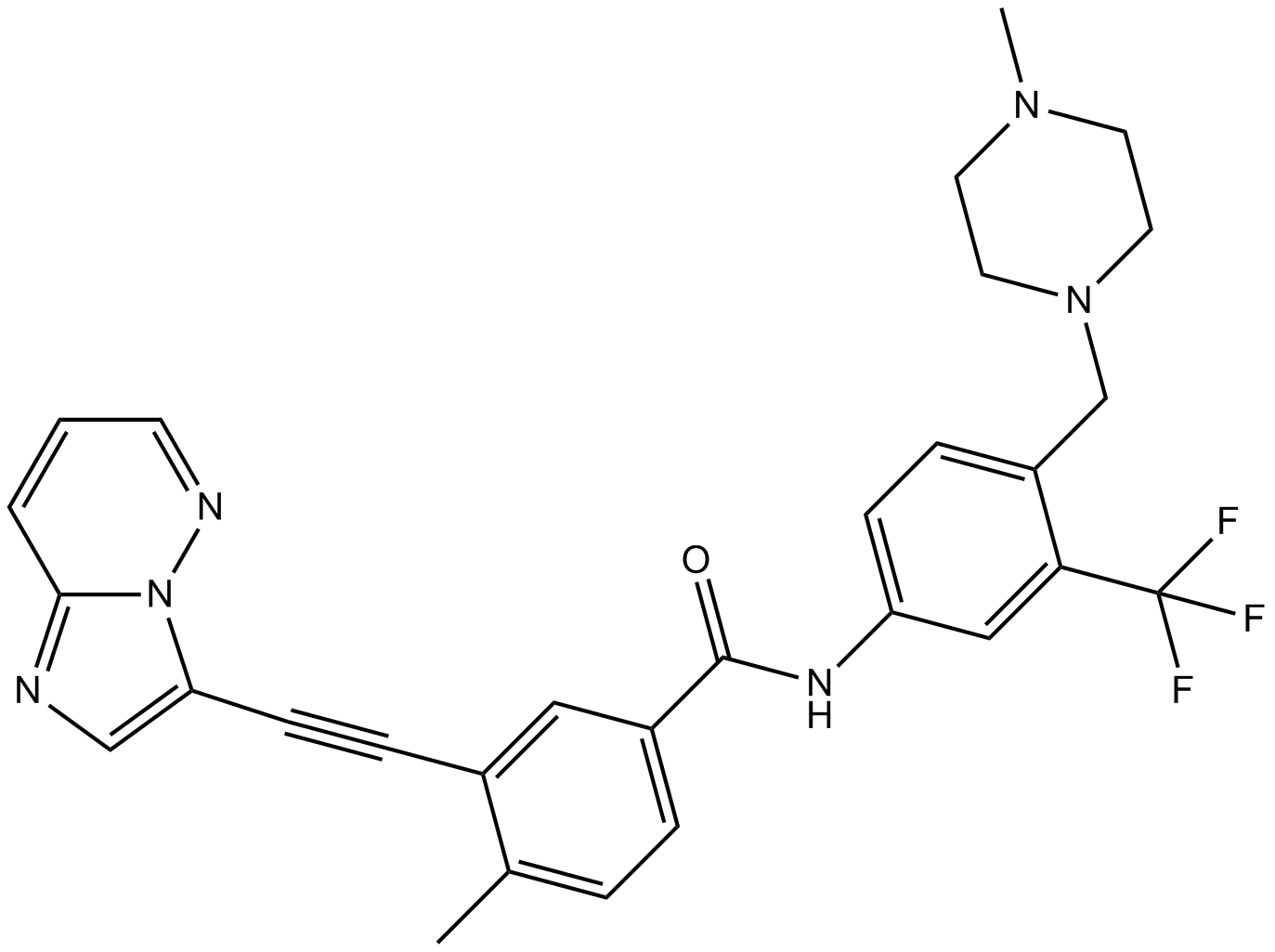
Ponatinib (AP24534)
Ponatinib (AP24534) (AP24534) is an orally active multi-targeted kinase inhibitor with IC50s of 0.37 nM, 1.1 nM, 1.5 nM, 2.2 nM, and 5.4 nM for Abl, PDGFRα, VEGFR2, FGFR1, and Src, respectively.
-
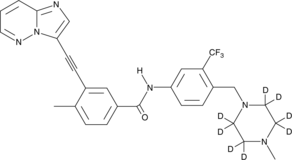
GC45828
Ponatinib-d8
An internal standard for the quantification of ponatinib
-
GC68339
Ponsegromab

-
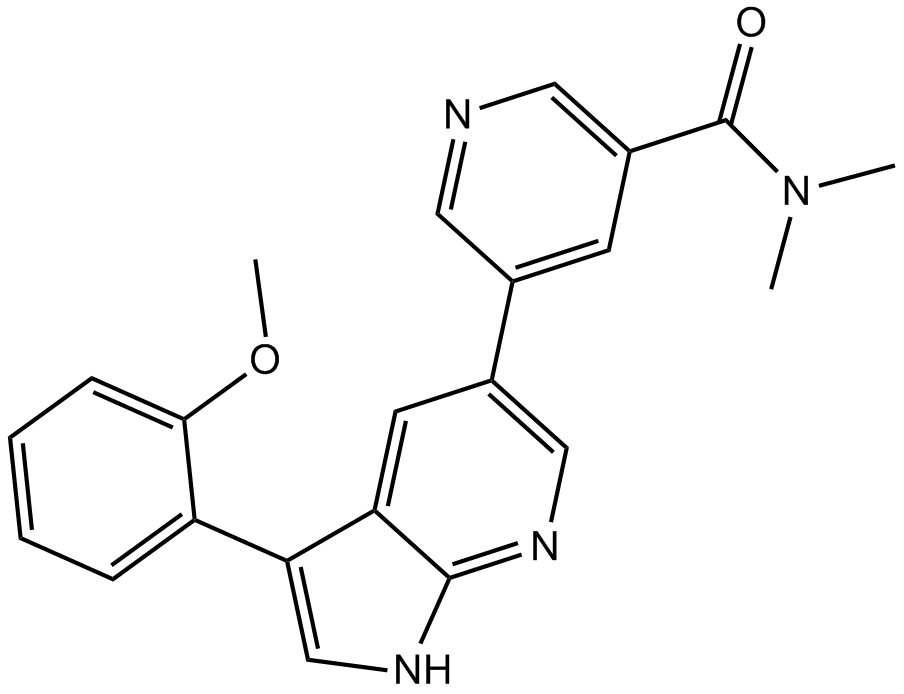
GC12779
PPY A
Abl kinases inhibitor
-
GC36974
Procyanidin A1
Procyanidin A1 (Proanthocyanidin A1) is a procyanidin dimer, which inhibits degranulation downstream of protein kinase C activation or Ca2+ influx from an internal store in RBL-213 cells.



