TGF-β / Smad Signaling
Transforming growth factor-beta (TGF-beta) is a multifunctional cytokine that regulates proliferation, migration, differentiation, and survival of many different cell types. Deletion or mutation of different members of the TGF-β family have been shown to cause vascular remodeling defect and absence of mural cell formation, leading to embryonic lethality or severe vascular disorders. TGF-β induces smooth muscle differentiation via Notch or SMAD2 and SMAD3 signaling in ES cells or in a neural crest stem cell line. TGF-β binds to TGF-βRI and to induce phosphorylation of SMAD2/3, thereby inhibiting proliferation, tube formation, and migration of endothelial cells (ECs).
TGF-β is a pluripotent cytokine with dual tumour-suppressive and tumour-promoting effects. TGF-β induces the epithelial-to-mesenchymal transition (EMT) leading to increased cell plasticity at the onset of cancer cell invasion and metastasis.
Targets for TGF-β / Smad Signaling
Products for TGF-β / Smad Signaling
- Cat.No. Product Name Information
-
GC44729
Prostratin
An HIV reactivator
-
GC37014
Protein Kinase C 19-31
Protein Kinase C 19-31, a peptide inhibitor of protein kinase C (PKC), derived from the pseudo-substrate regulatory domain of PKCa (residues 19-31) with a serine at position 25 replacing the wild-type alanine, is used as protein kinase C substrate peptide for testing the protein kinase C activity.

-
GC37015
Protein Kinase C 19-36
Protein Kinase C 19-36 is a pseudosubstrate peptide inhibitor of protein kinase C (PKC), with an IC50 of 0.18 μM.

-
GC66006
Protein kinase inhibitor H-7
Protein kinase inhibitor H-7 is a potent inhibitor of protein kinase C (PKC) and cyclic nucleotide dependent protein kinase, with a Ki of 6 μM for PKC.
-
GC11803
Pseudo RACK1
Activator of protein kinase C
-
GC69776
PT-262
PT-262 is an effective ROCK inhibitor with an IC50 value of approximately 5 μM. PT-262 induces loss of mitochondrial membrane potential and increases activation of caspase-3 and cell apoptosis. PT-262 inhibits phosphorylation of ERK and CDC2 through a p53-independent pathway. PT-262 blocks cellular cytoskeleton function and cell migration. PT-262 has anti-cancer activity.
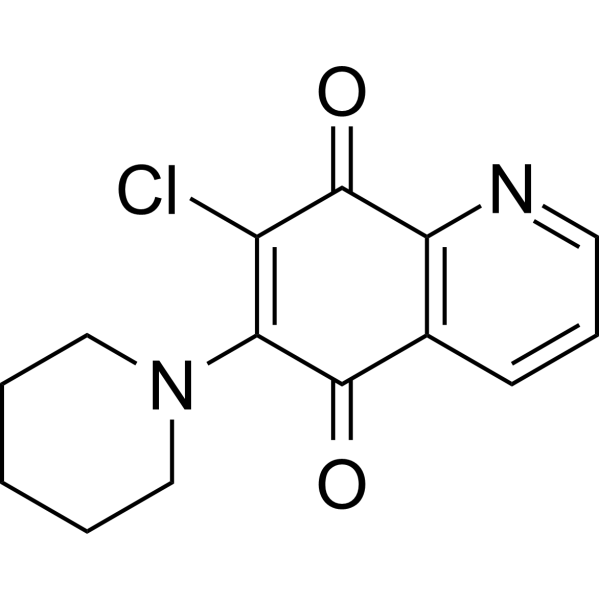
-
GC14583
R 59-022
Diacylglycerol (DAG) kinase inhibitor
-
GC69792
R 59-022 hydrochloride
R 59-022 (DKGI-I) hydrochloride is a DGK inhibitor (IC50: 2.8 μM). R 59-022 hydrochloride inhibits OAG phosphorylation to OAPA. It is also a 5-HT receptor antagonist that can activate protein kinase C (PKC). R 59-022 enhances the production of thromboxane in platelets induced by thrombin and inhibits phosphatidic acid production in neutrophils.
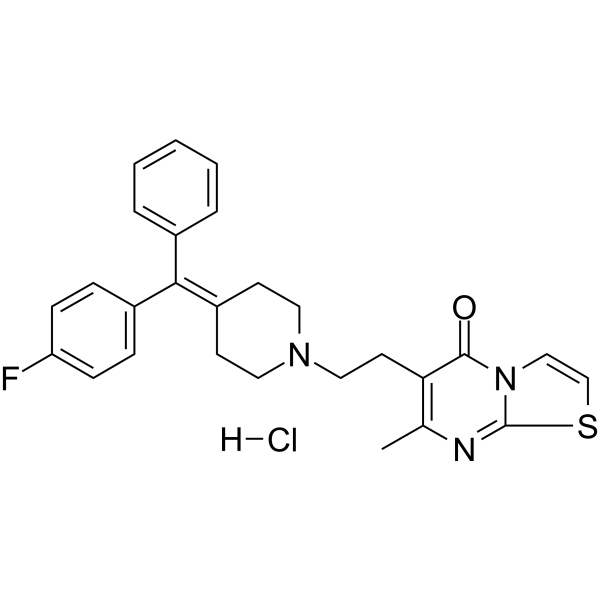
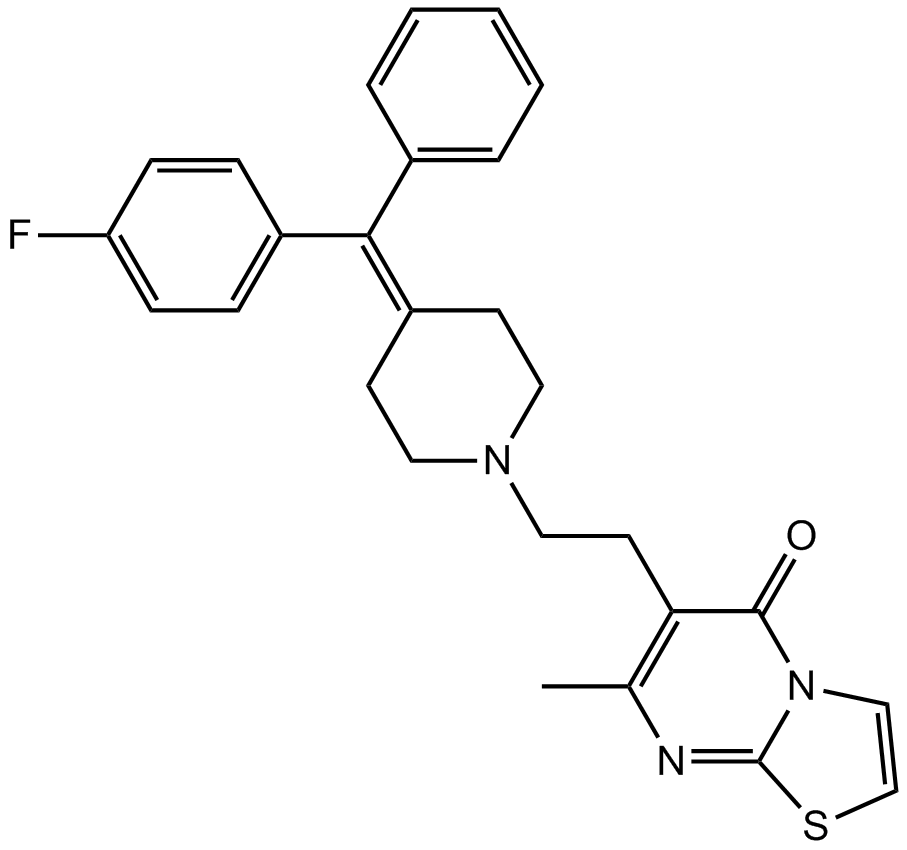
-
GC32840
R-268712
R-268712 is an orally active and selective ALK-5 inhibitor, with an IC50 of 2.5 nM.
-
GC18246
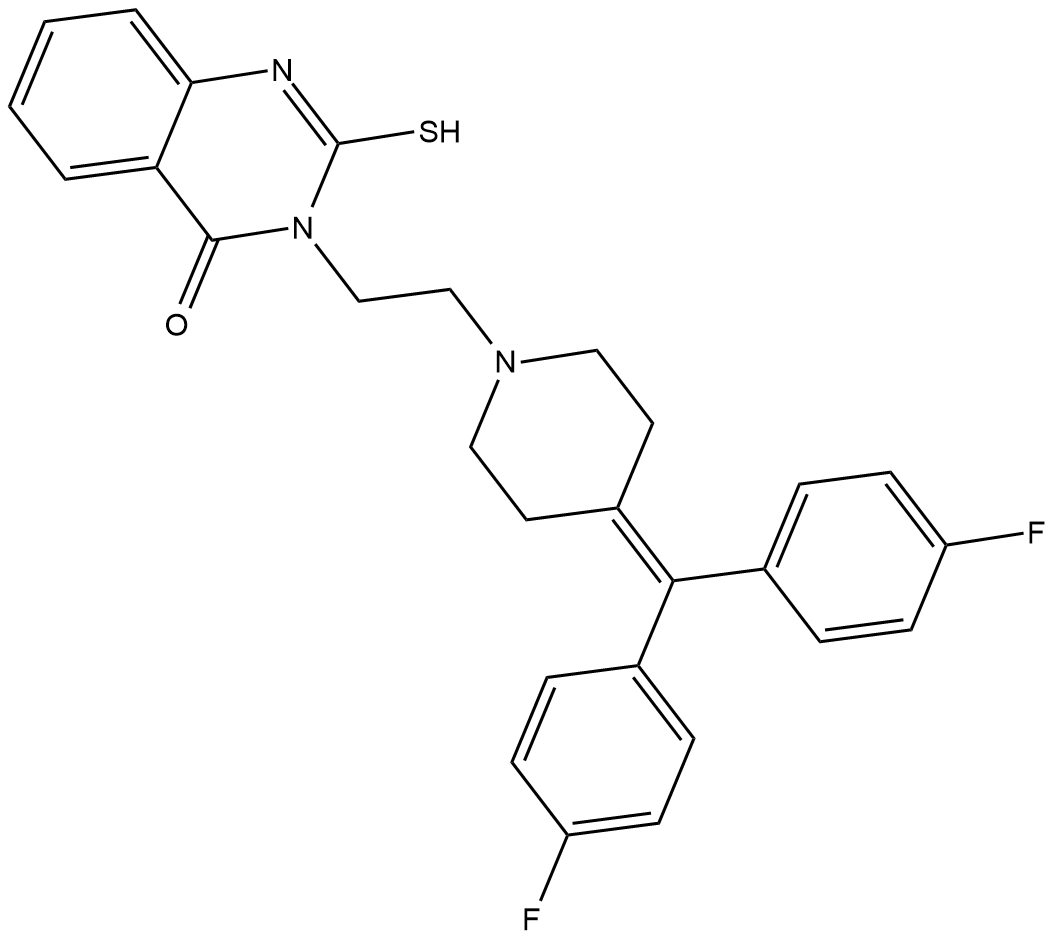
R-59-949
R-59-949 is an inhibitor of diacylglycerol kinase α (DGK-α) with an IC50 value of 300 nM in isolated platelet plasma membranes using exogenous diacylglycerol as a substrate.
-
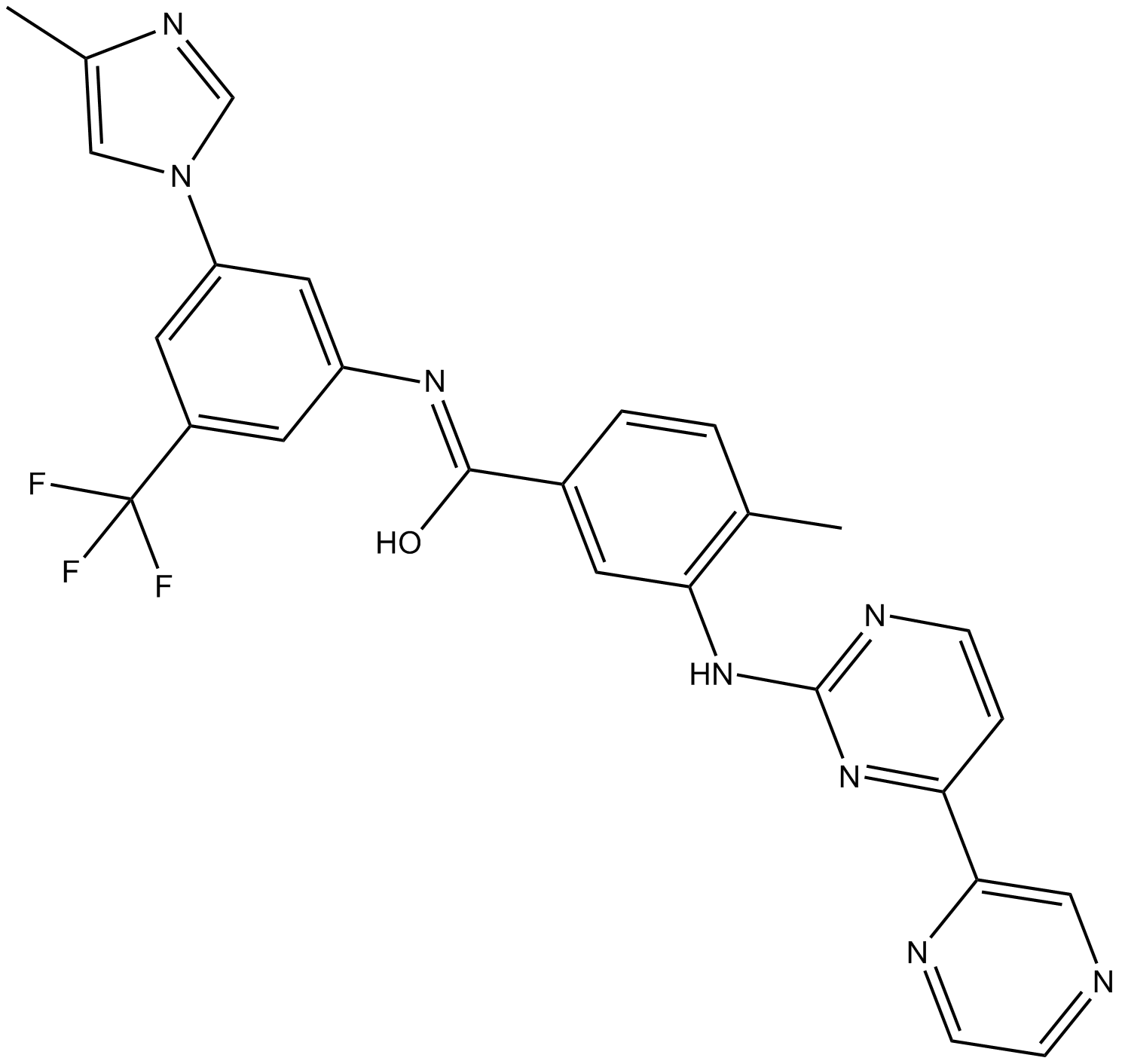
GC11140
Radotinib(IY-5511)
Bcr-Abl tyrosine kinase inhibitor
-
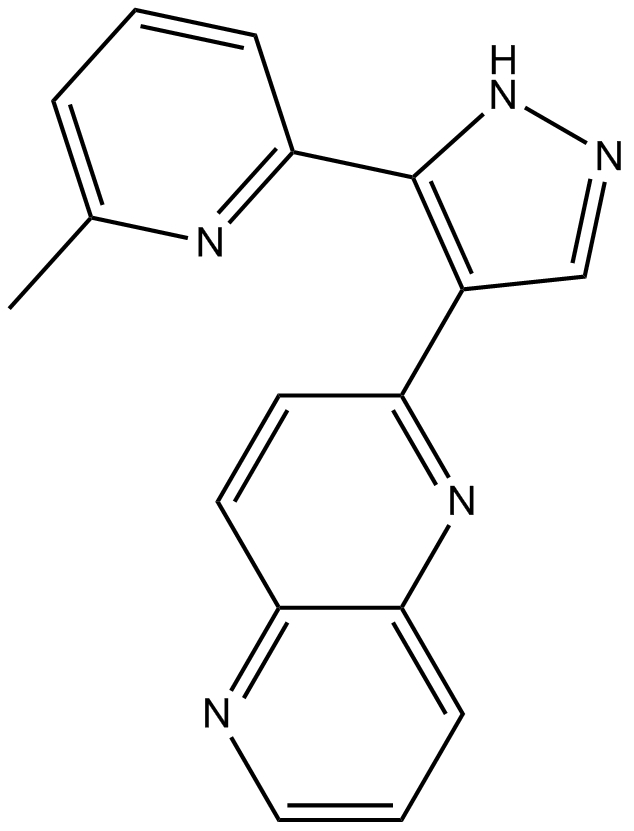
GC16793
RepSox
A selective inhibitor of ALK5
-
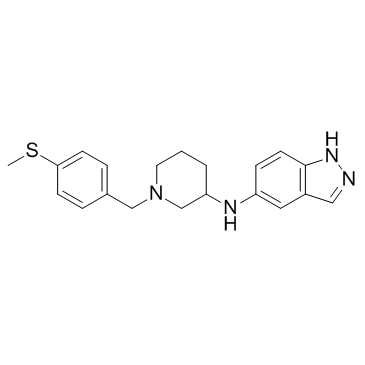
GC33310
Rho-Kinase-IN-1
Rho-Kinase-IN-1 is a Rho kinase (ROCK) inhibitor (Ki values of 30.5 and 3.9 nM for ROCK1 and ROCK2, respectively) extracted from US20090325960A1, compound 1.008.
-
GC66050
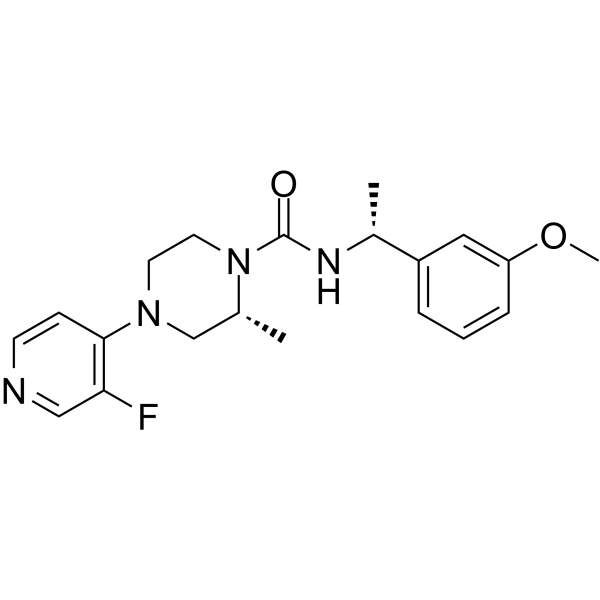
Rho-Kinase-IN-2
Rho-Kinase-IN-2 (Compound 23) is an orally active, selective, and central nervous system (CNS)-penetrant Rho Kinase (ROCK) inhibitor (ROCK2 IC50=3 nM). Rho-Kinase-IN-2 can be used in Huntington's research.
-
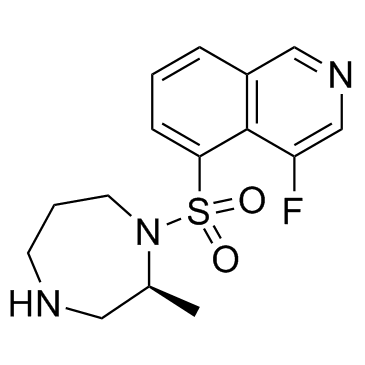
GC37534
Ripasudil free base
Ripasudil free base (K-115 free base) is a specific inhibitor of ROCK, with IC50s of 19 and 51 nM for ROCK2 and ROCK1, respectively.
-
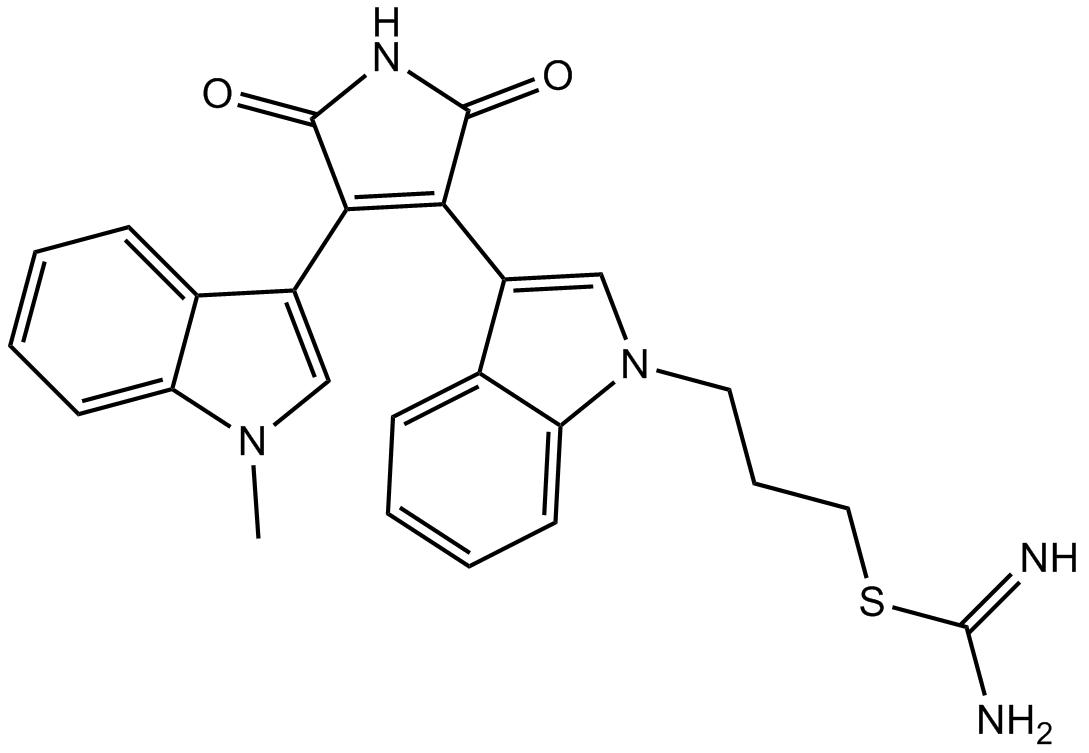
GC17322
Ro 31-8220
pan-PKC inhibitor
-
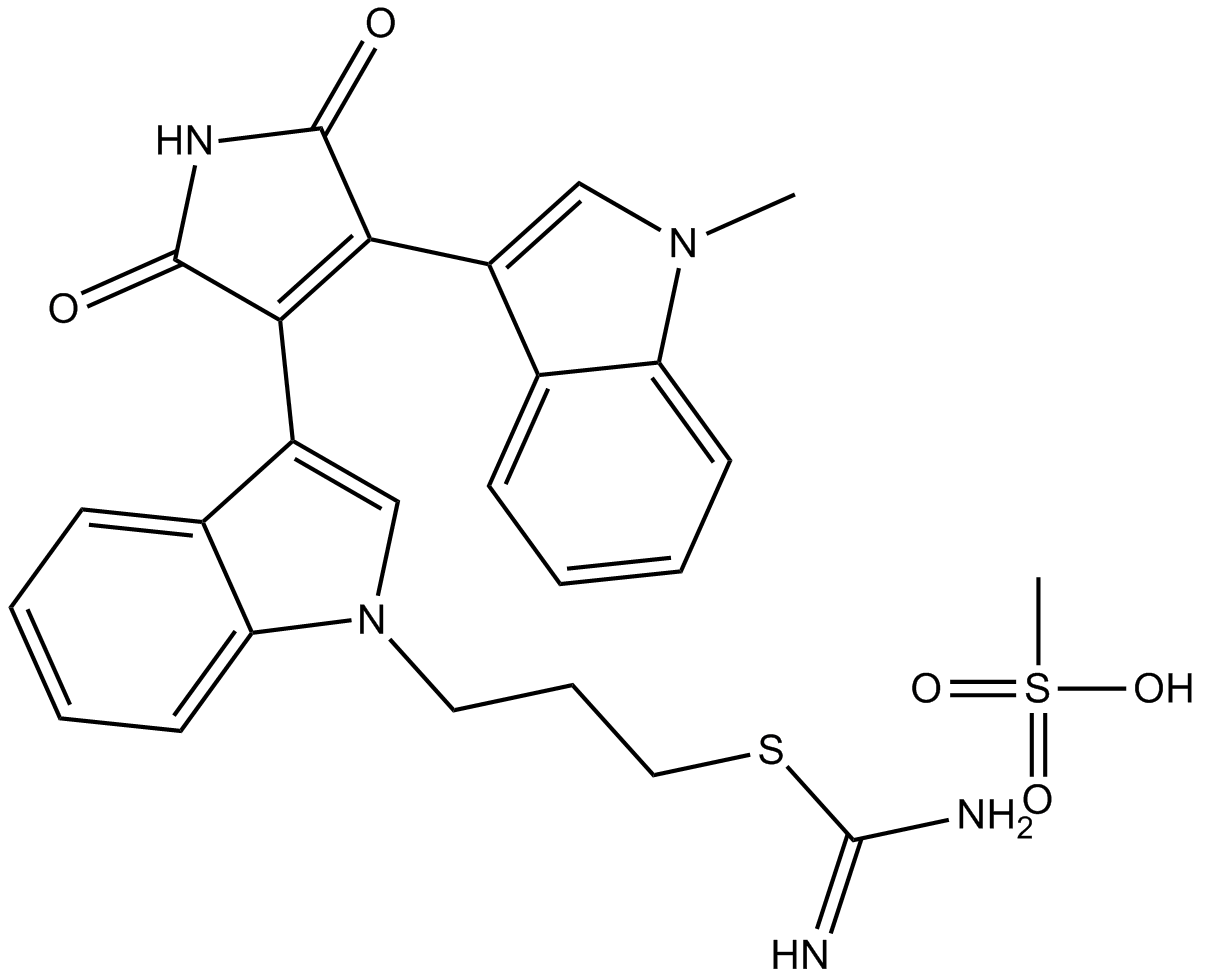
GC17014
Ro 31-8220 Mesylate
Pan-PKC inhibitor
-
GC10029
Ro 31-8220 methanesulfonate
Ro 31-8220 methanesulfonate is a potent PKC inhibitor, with IC50s of 5, 24, 14, 27, 24 and 23 nM for PKCα, PKCβI, PKCβII, PKCγ, PKCε and rat brain PKC, respectively.
-
GC13677
Ro 32-0432 hydrochloride
protein kinase C inhibitor
-
GC37551
ROCK inhibitor-2
ROCK inhibitor-2 is a selective dual ROCK1 and ROCK2 inhibitor with IC50s of 17 nM and 2 nM, respectively.
-
GC31952
ROCK-IN-1
ROCK-IN-1 is a potent inhibitor of ROCK, with an IC50 of 1.2 nM for ROCK2.
-
GC66338
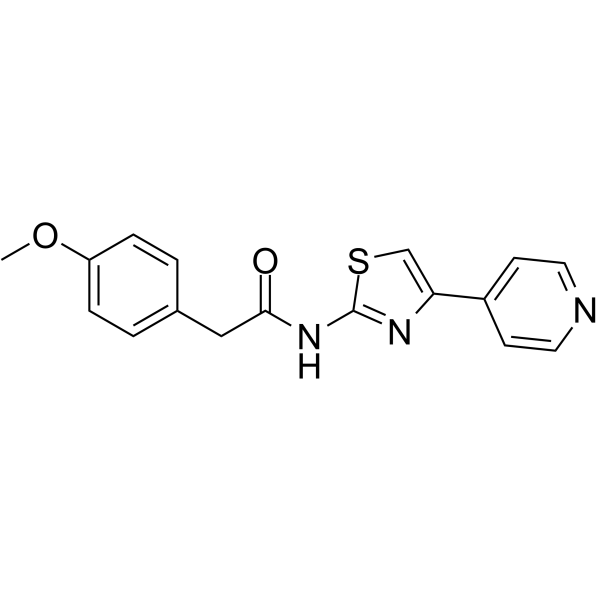
ROCK1-IN-1
ROCK1-IN-1 is a ROCK1 inhibitor with a Ki value of 540 nM. ROCK1-IN-1 can be used for the research of hypertension, glaucoma and erectile dysfunction.
-
GC31883
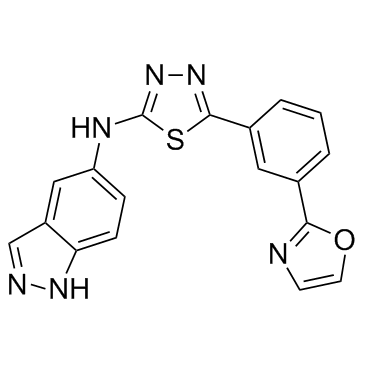
ROCK2-IN-2
ROCK2-IN-2 is a selective ROCK2 inhibitor extracted from patent US20180093978A1, Compound A-30, has an IC50 of <1 μM.
-
GC10820
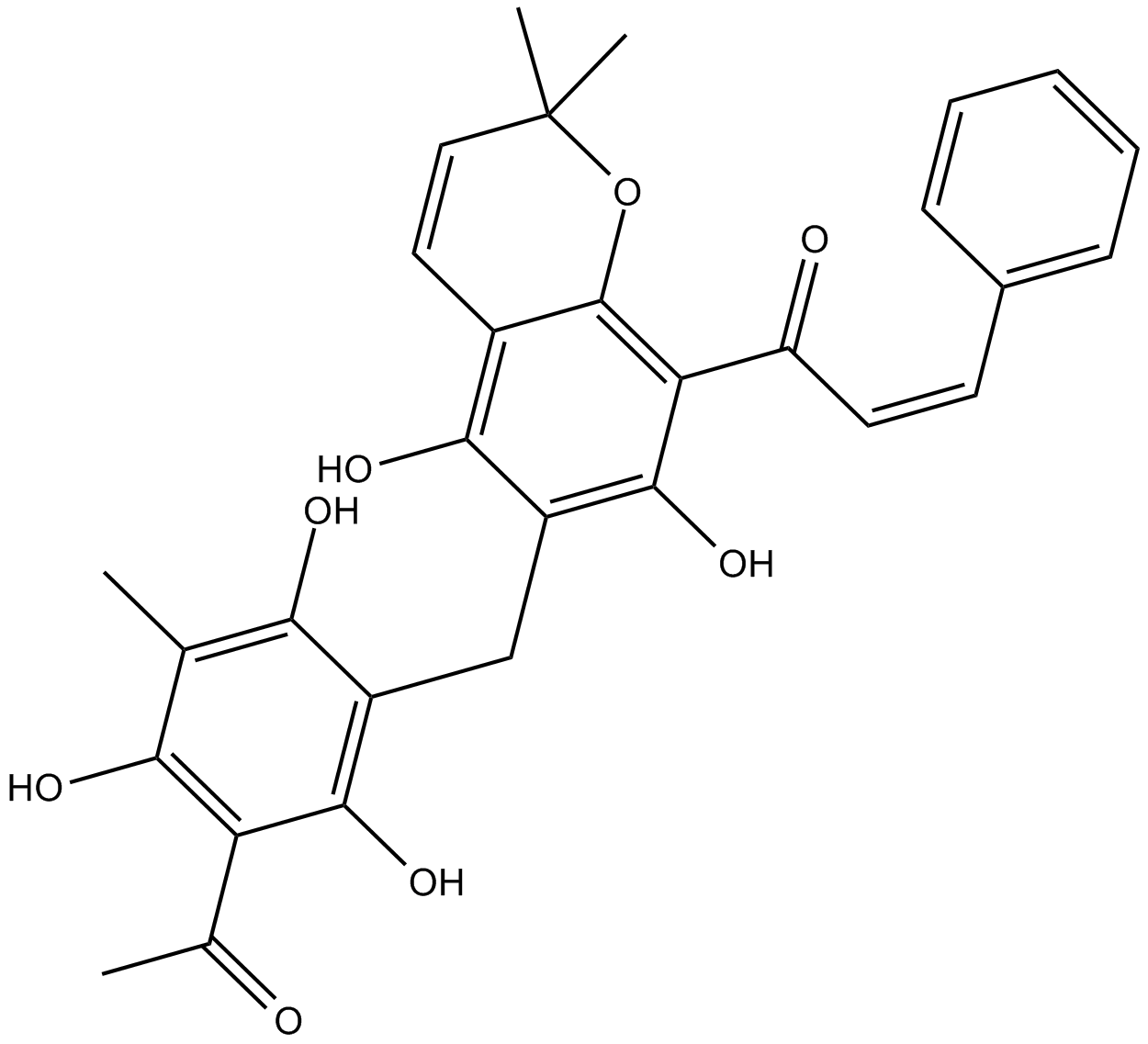
Rottlerin
PKC inhibitor
-
GC63177
Ruboxistaurin
Ruboxistaurin (LY333531) is an orally active, selective PKC beta inhibitor (Ki=2 nM).

-
GC16405
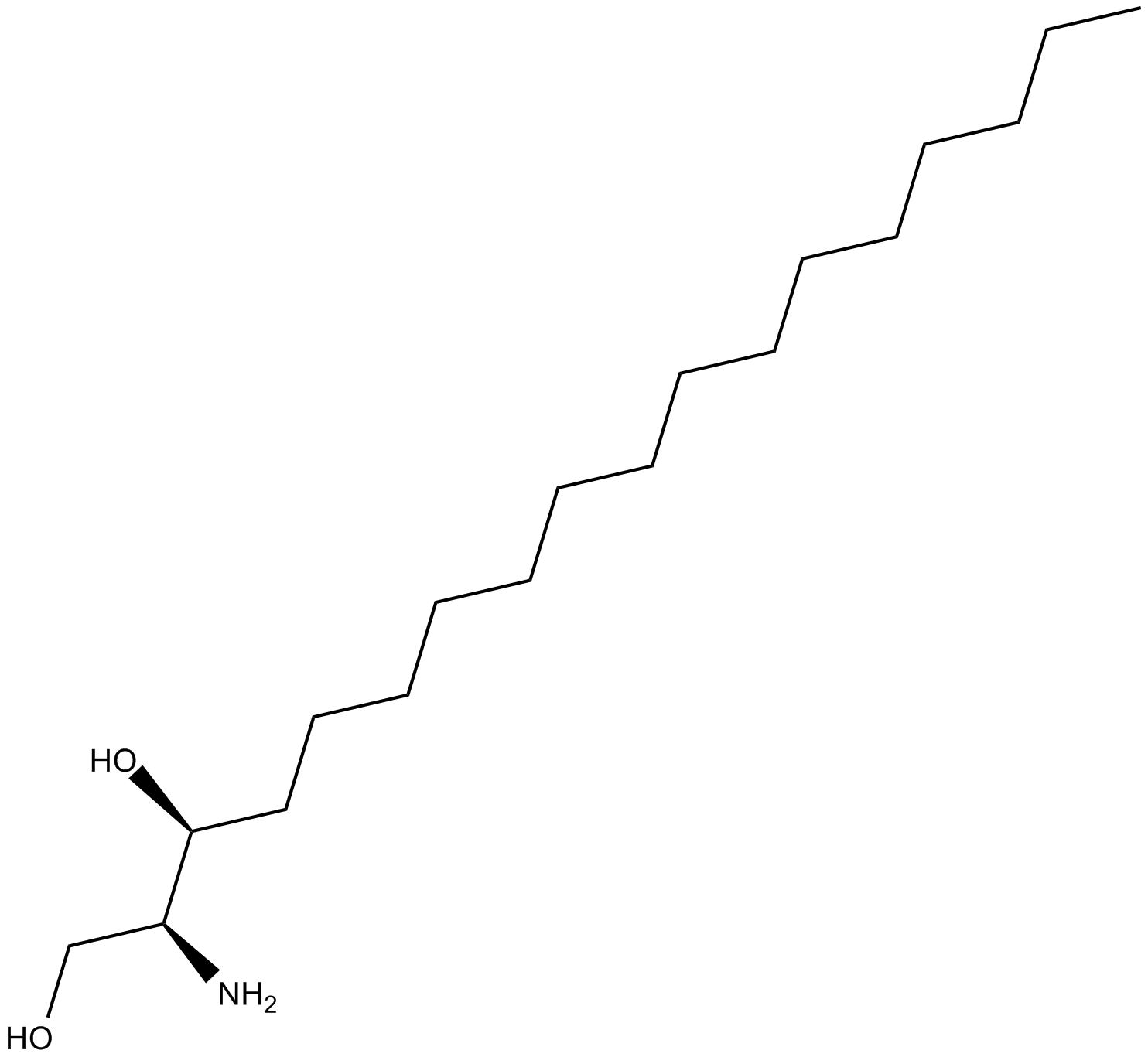
Safingol
PKC and sphingosine kinases inhibitor
-
GC63439
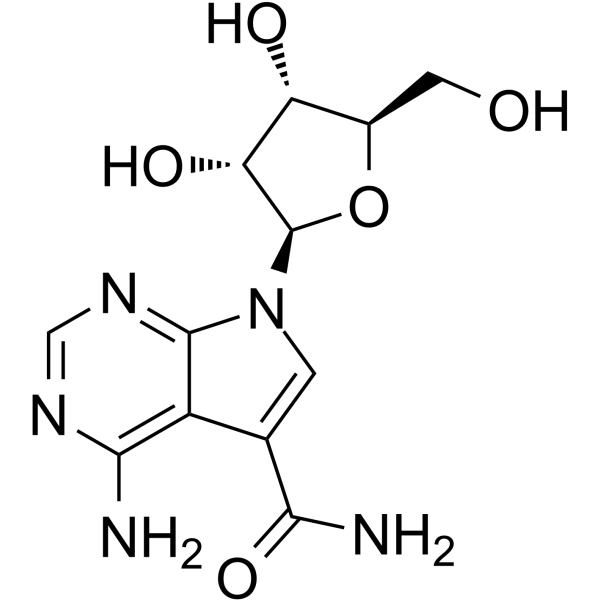
Sangivamycin
Sangivamycin (NSC 65346), a nucleoside analog, is a potent inhibitor of protein kinase C (PKC) with an Ki of 10 μM. Sangivamycin has potent antiproliferative activity against a variety of human cancers.
-
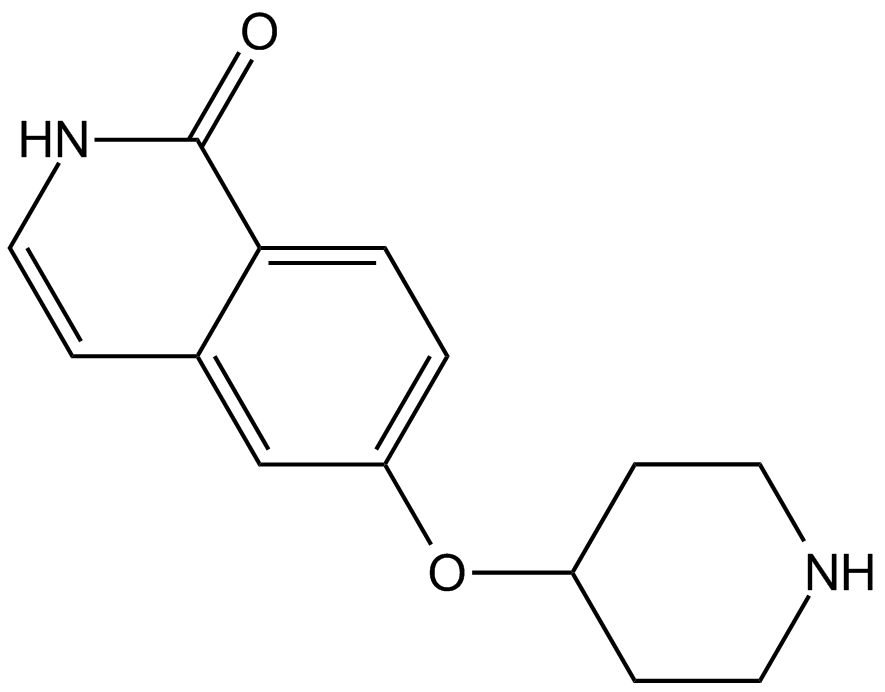
GC13759
SAR407899
ATP-competitive ROCK inhibitor
-
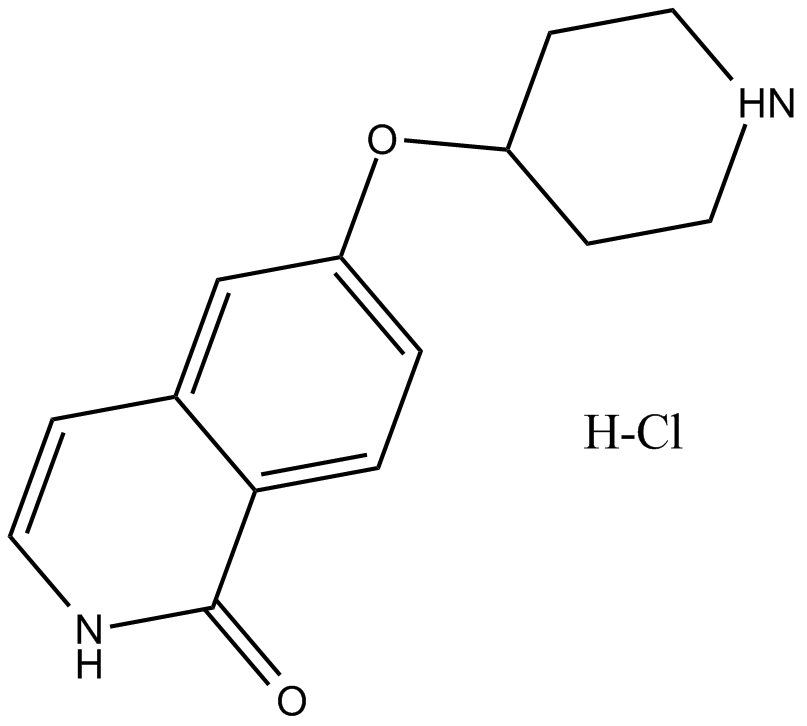
GC16289
SAR407899 hydrochloride
ATP-competitive ROCK inhibitor
-
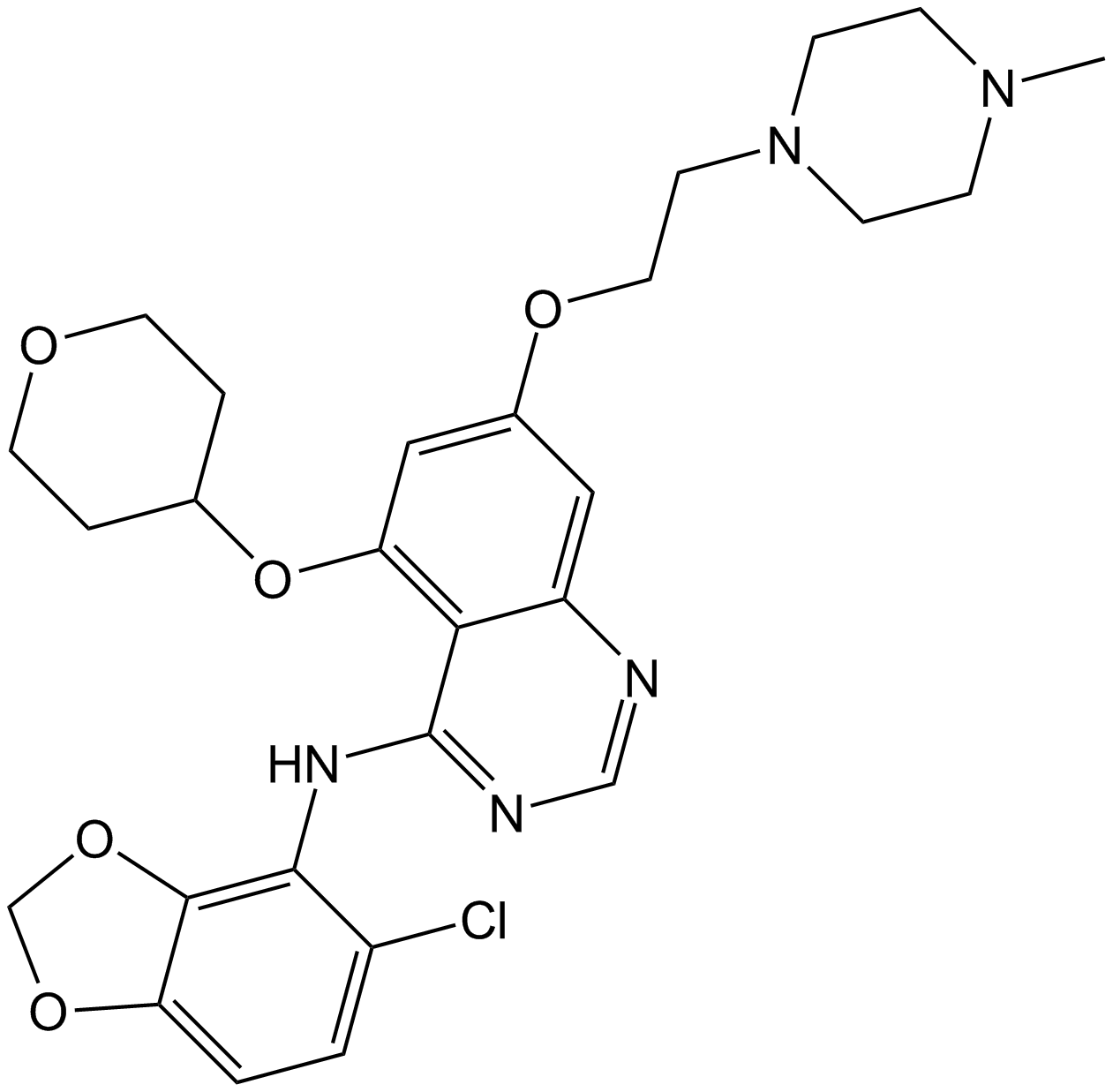
GC15197
Saracatinib (AZD0530)
Saracatinib (AZD0530) (AZD0530) is a potent Src family inhibitor with IC50s of 2.7 to 11 nM for c-Src, Lck, c-YES, Lyn, Fyn, Fgr, and Blk. Saracatinib (AZD0530) shows high selectivity over other tyrosine kinases.
-
GC11022
SB 218078
checkpoint kinase 1 (Chk1) inhibitor
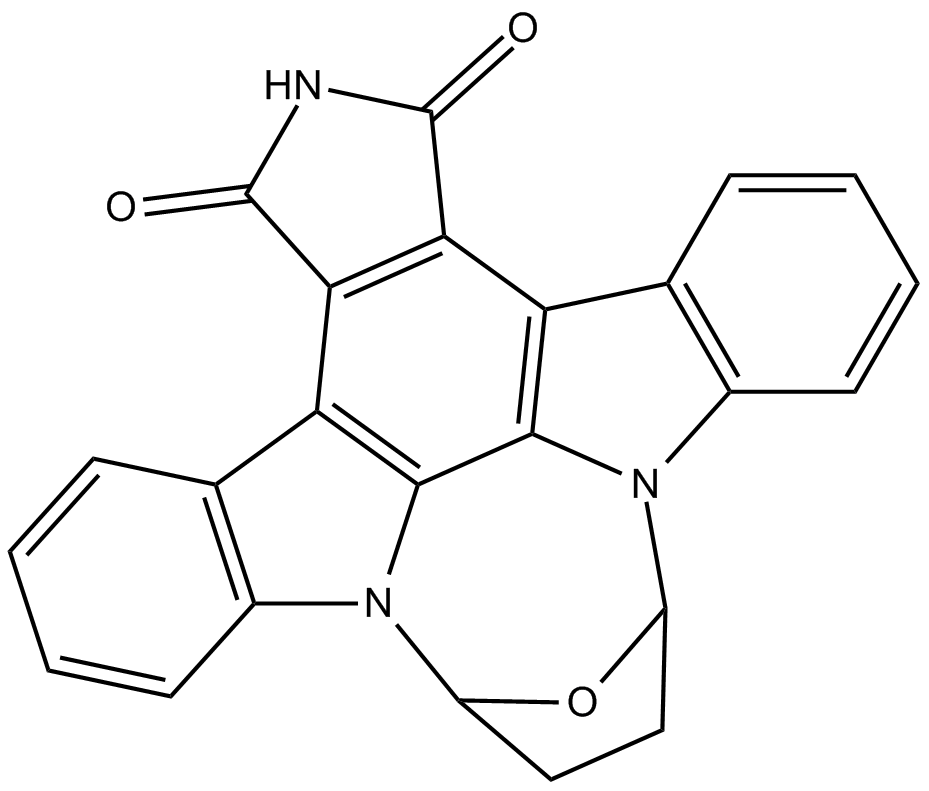
-
GC11545
SB 431542
SB-431542, a small molecule inhibitor
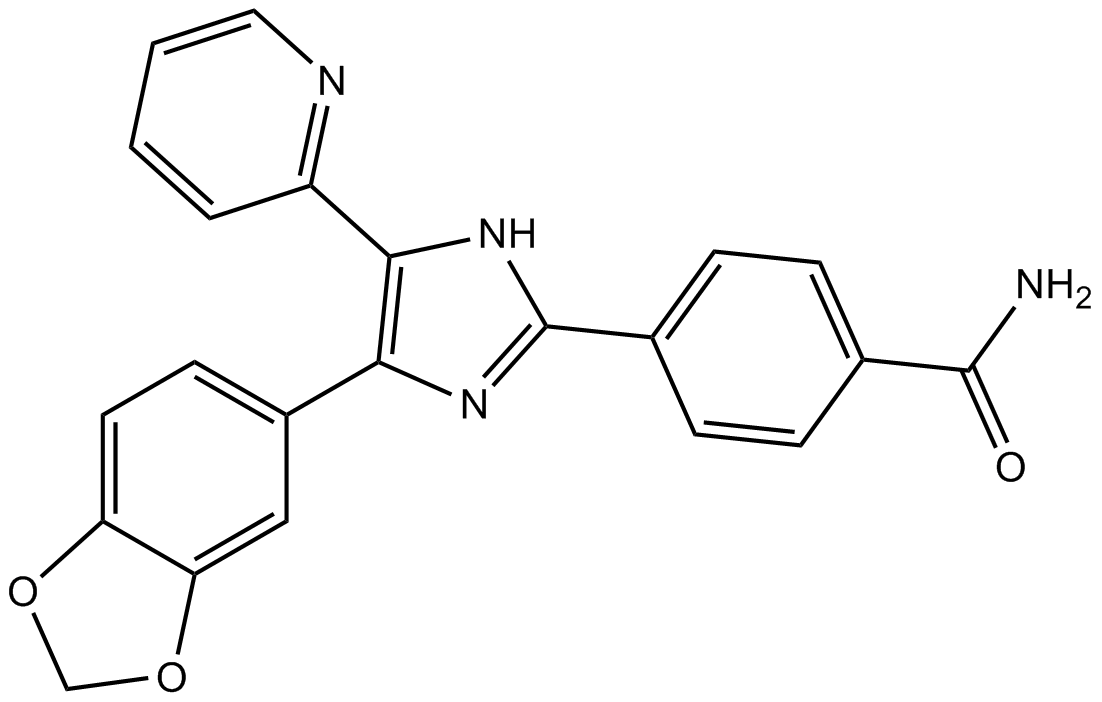
-
GC15170
SB 772077B dihydrochloride
Rho-kinase (ROCK) inhibitor
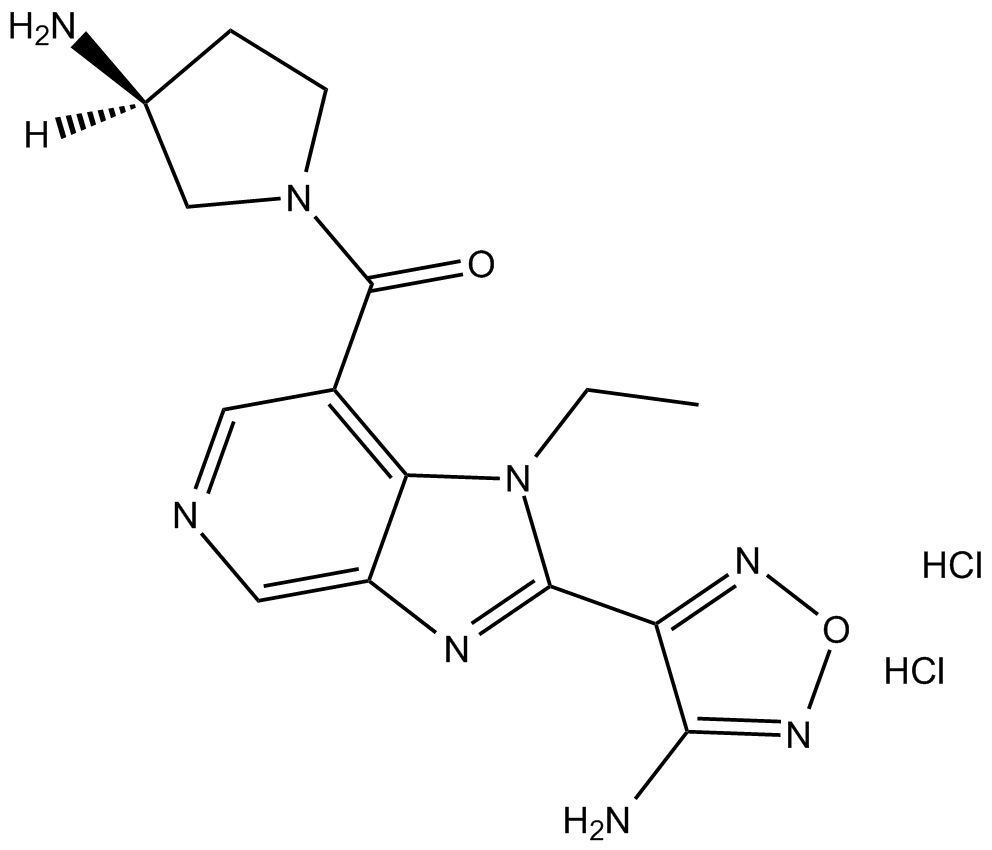
-
GC10421
SB-505124 hydrochloride
Inhibitor of receptors ALK4, ALK5, and ALK7
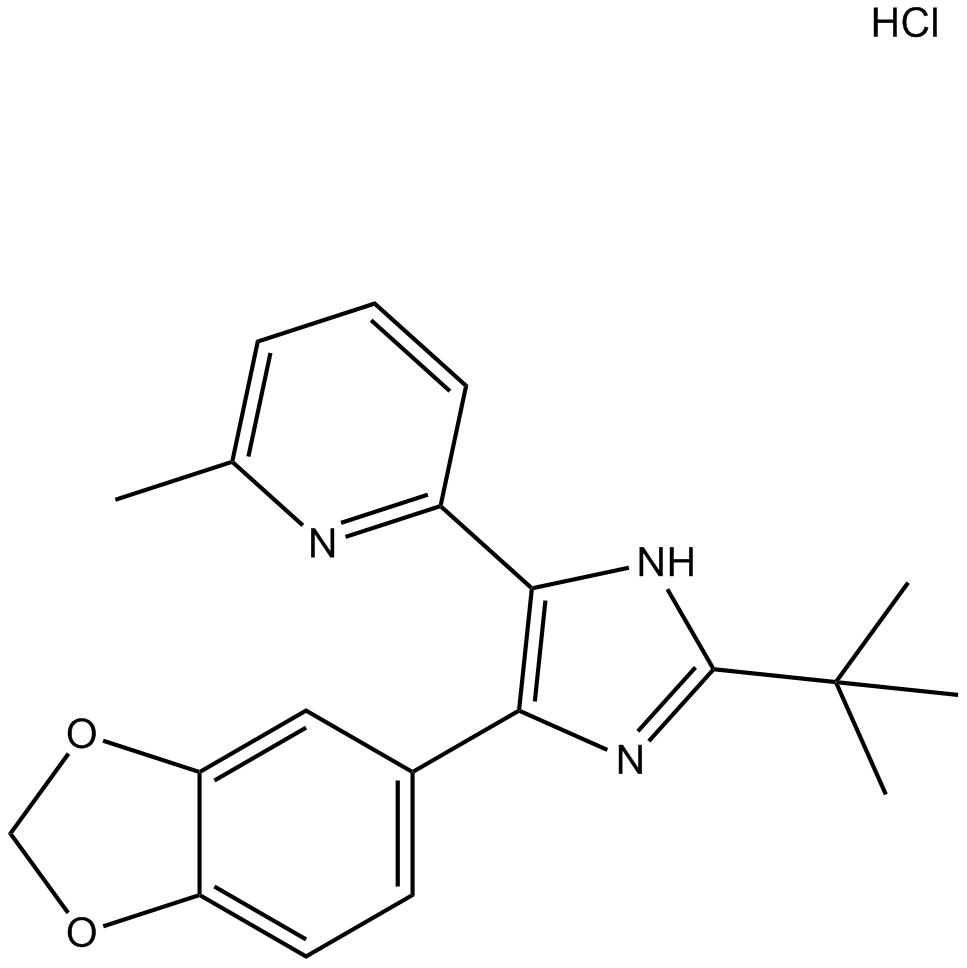
-
GC13769
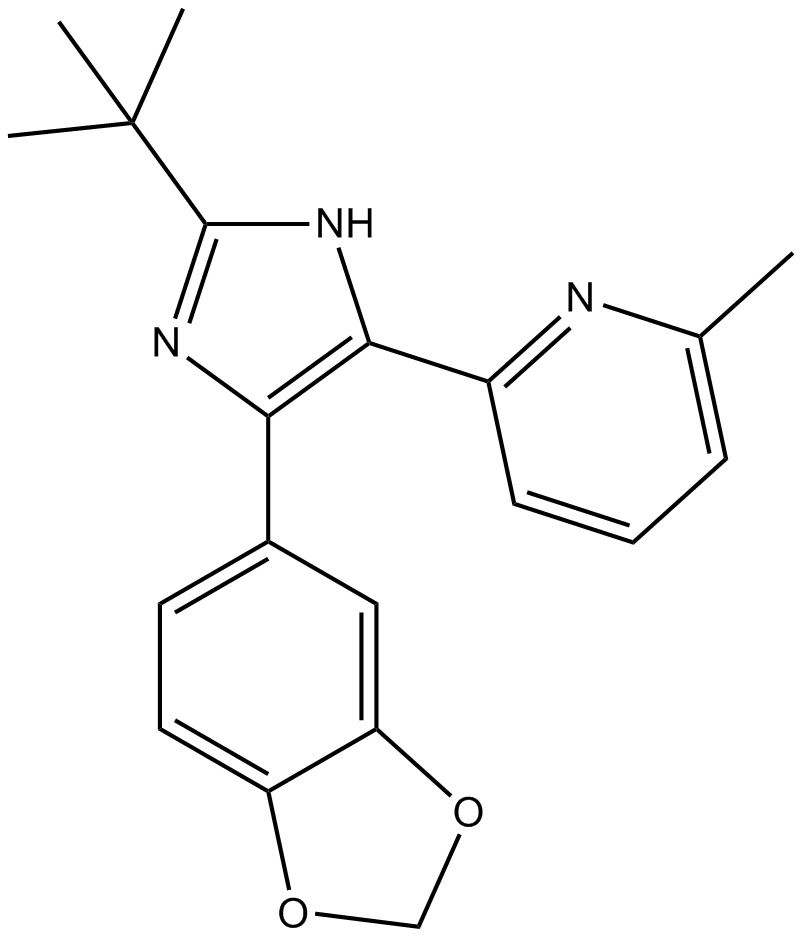
SB505124
ALK4/ALK5/ALK7 inhibitor
-
GC14349
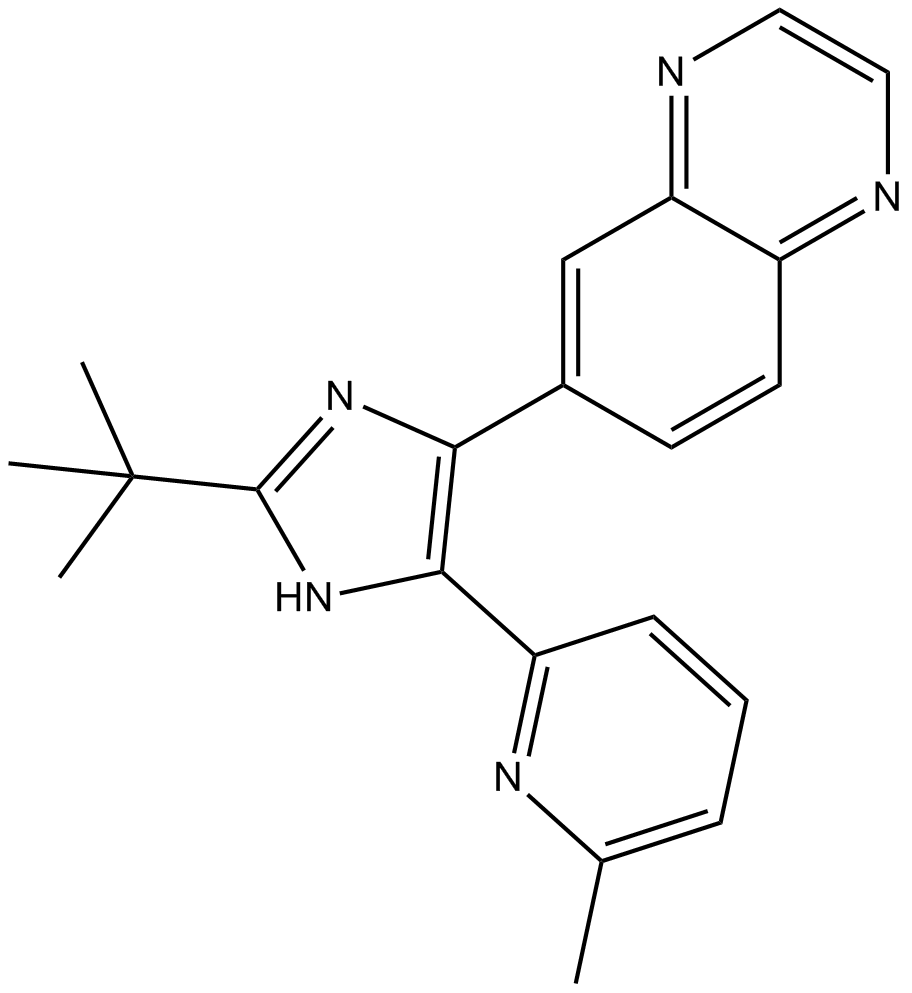
SB525334
(TGF-beta1) receptor inhibitor
-
GC13320
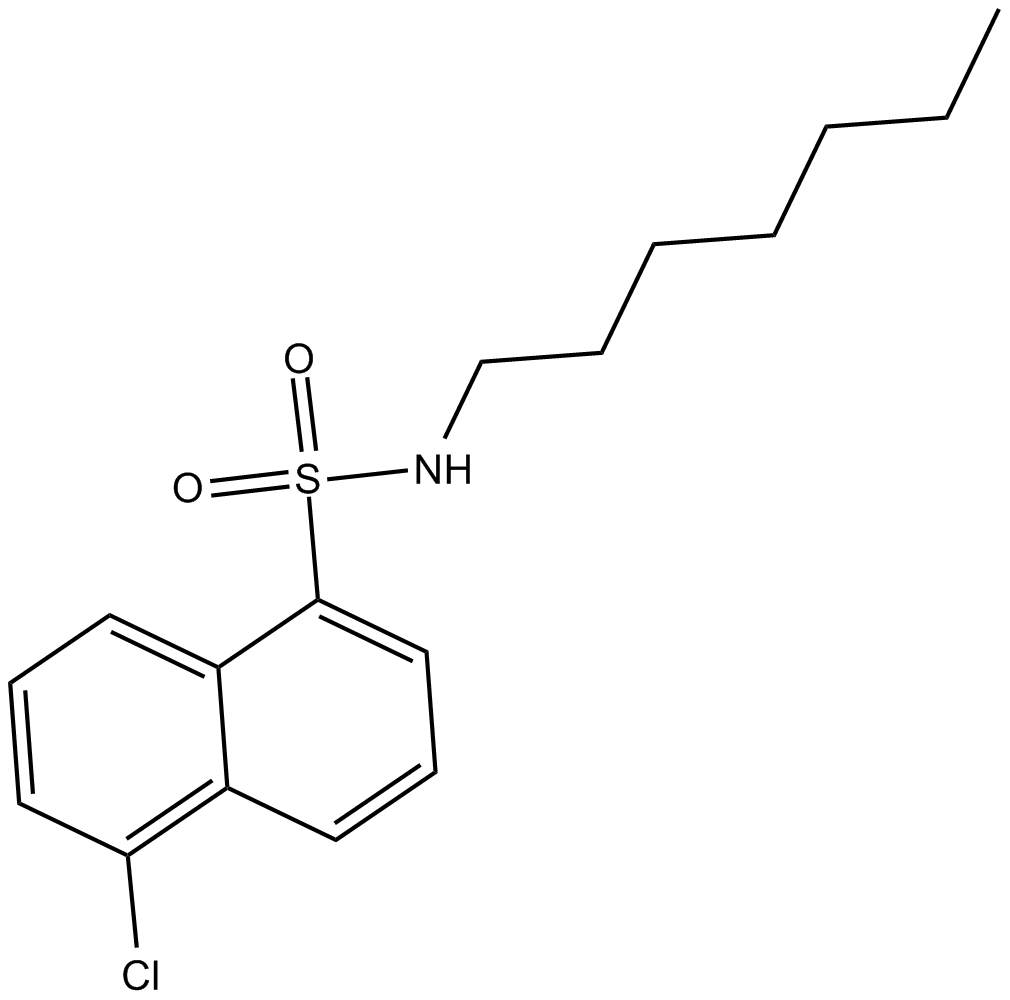
SC-10
Protein kinase C activator
-
GC18055
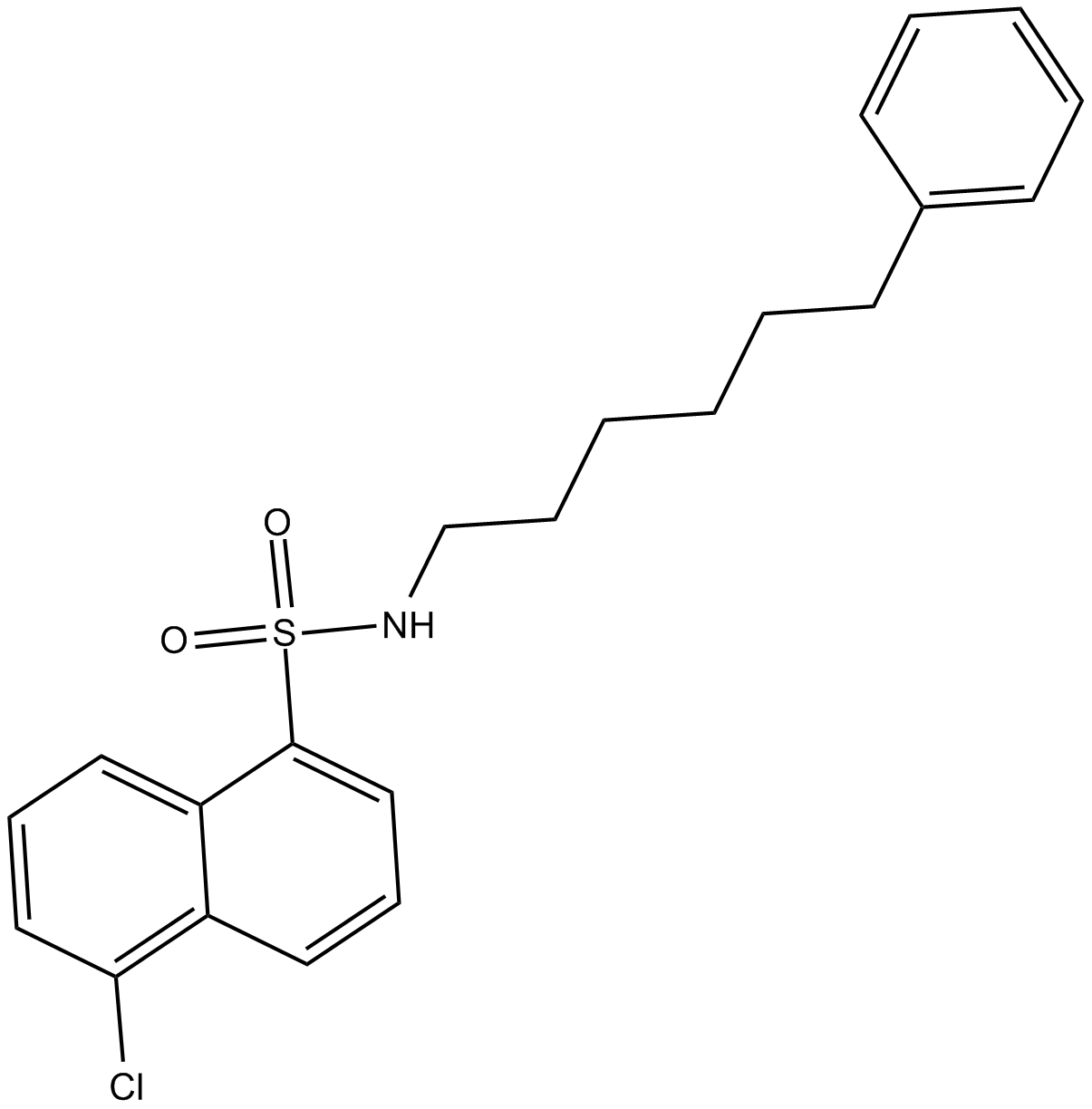
SC-9
protein kinase C activator
-
GC13904
SD-208
A potent, selective inhibitor of TGF-βRI
-
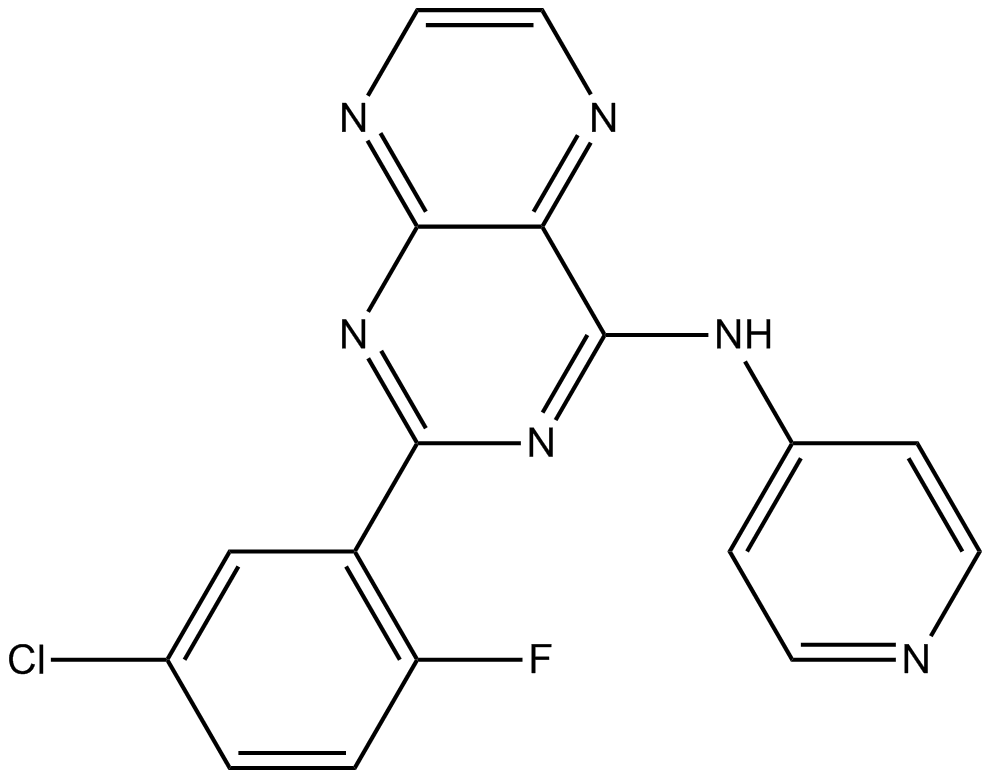
GC17191
SIS3 HCl
SIS3HCl is a potent and selective inhibitor of Smad3 with an IC50 of 3 μM for Smad3 phosphorylation.
-
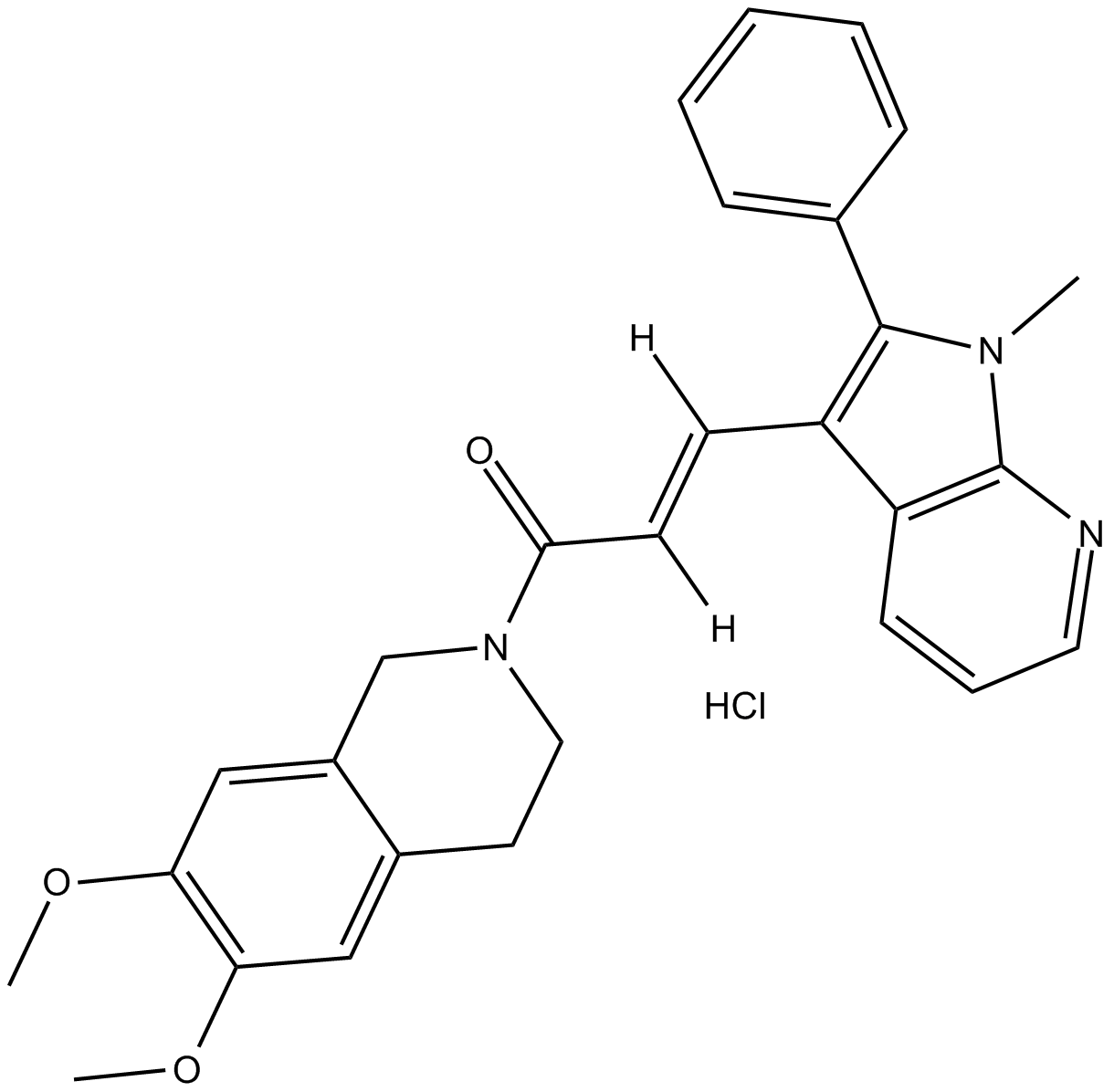
GC69903
SJ000063181
SJ000063181 is an effective BMP signal activator with an EC50 ≤1 µM. It can be used as a chemical probe to explore BMP signaling because it can penetrate zebrafish embryos.
-
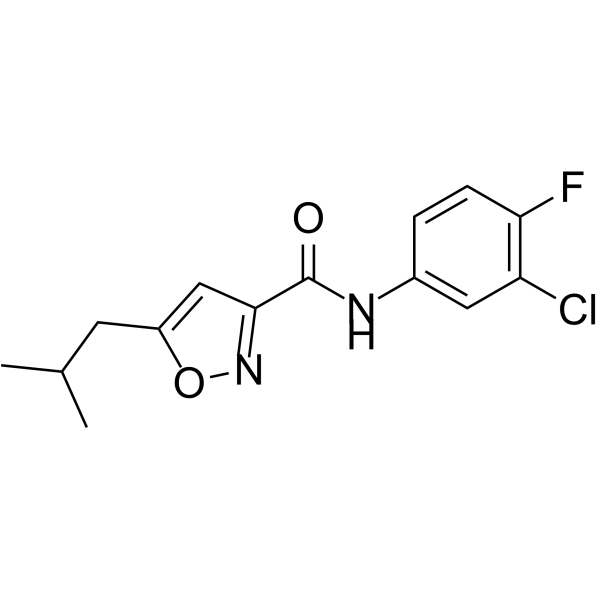
GC32788
SJ000291942
A BMP signaling activator
-
GC19447
SM 16
SM 16 is a ALK5/ALK4 kinase inhibitor

-
GC44903
SMAD3 Inhibitor, SIS3
SMAD3 Inhibitor, SIS3 is a potent and selective inhibitor of Smad3 phosphorylation.
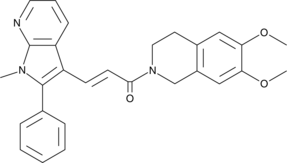
-
GC65592
SNIPER(ABL)-020
SNIPER(ABL)-020, conjugating Dasatinib (ABL inhibitor) to Bestatin (IAP ligand) with a linker, induces the reduction of BCR-ABL protein.
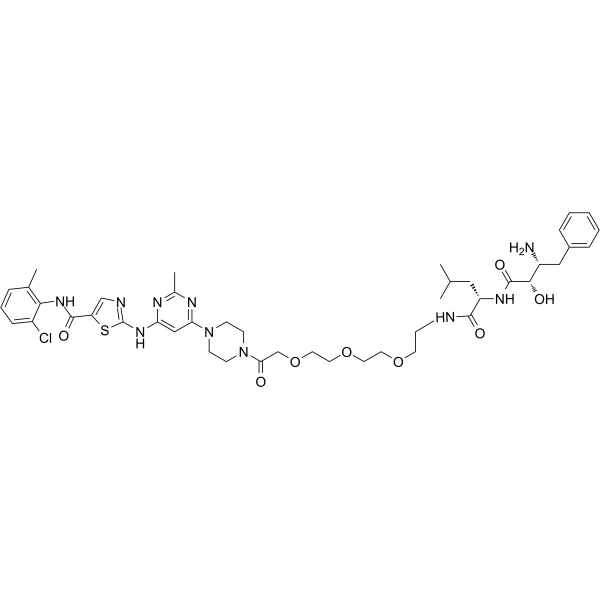
-
GC15520
Sotrastaurin (AEB071)
Sotrastaurin (AEB071) (AEB071) is a potent and orally-active pan-PKC inhibitor, with Kis of 0.22 nM, 0.64 nM, 0.95 nM, 1.8 nM, 2.1 nM and 3.2 nM for PKCθ, PKCβ, PKCα, PKCη, PKCδ and PKCε, respectively.
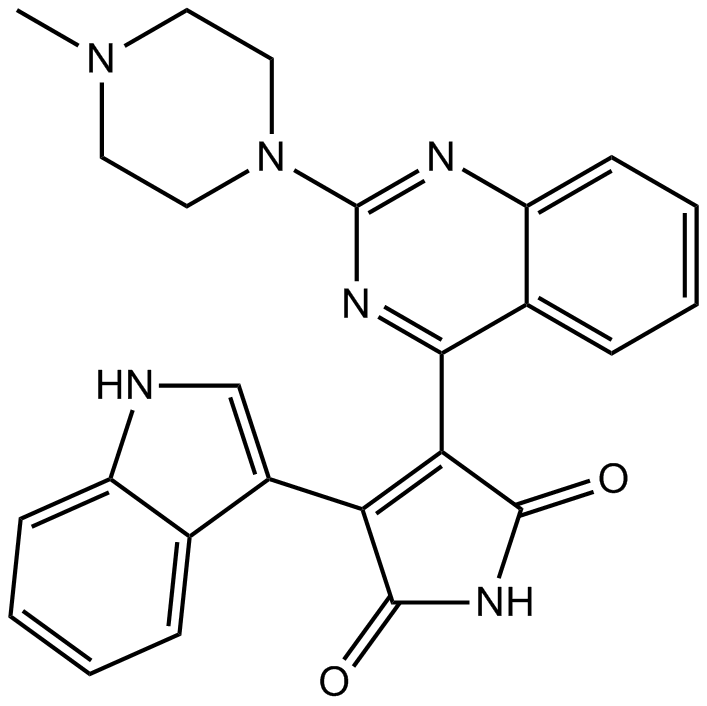
-
GC10722
SR-3677
Rho Kinase (ROCK-II) inhibitor
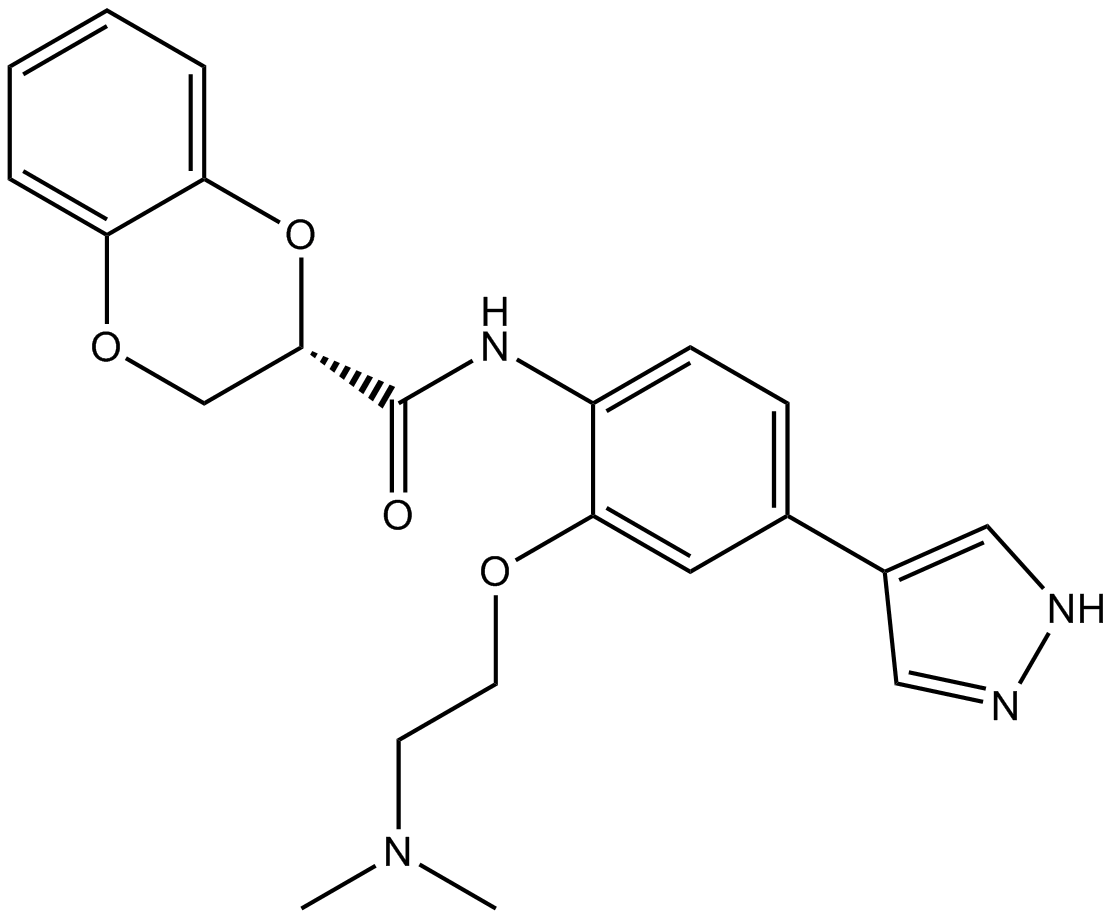
-
GC19338
SRI-011381 hydrochloride
An oral bioavailabile TGF-beta signaling agonist
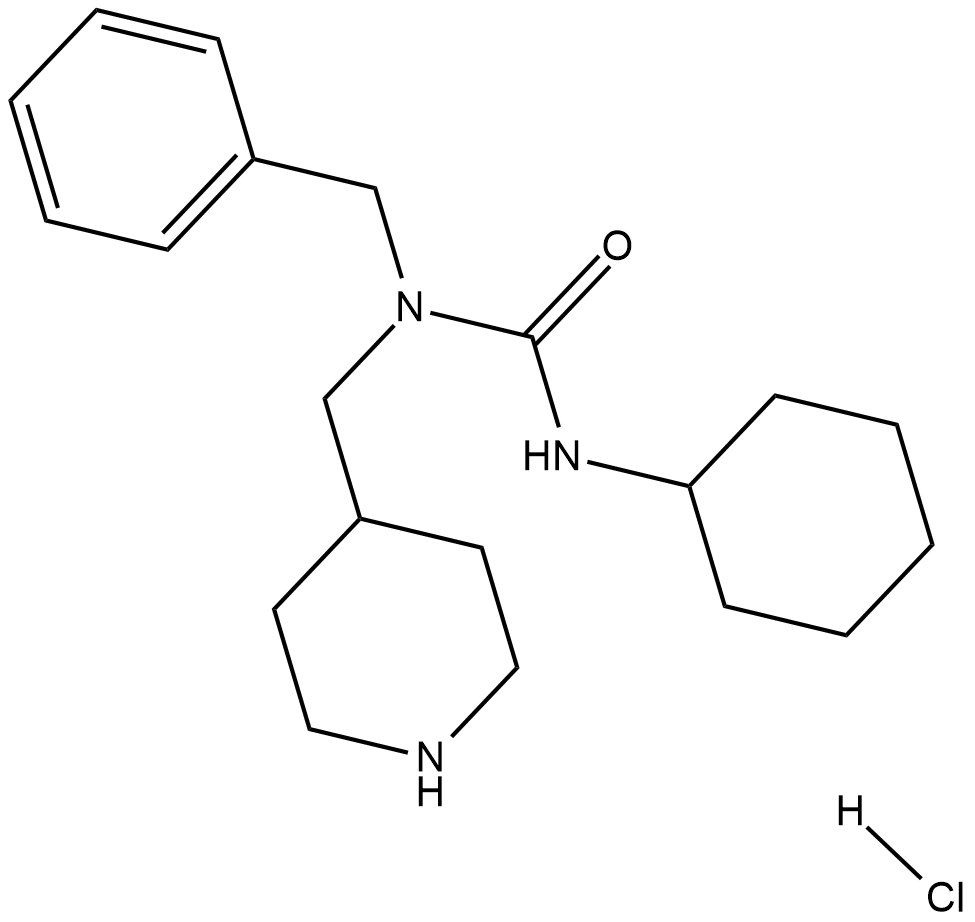
-
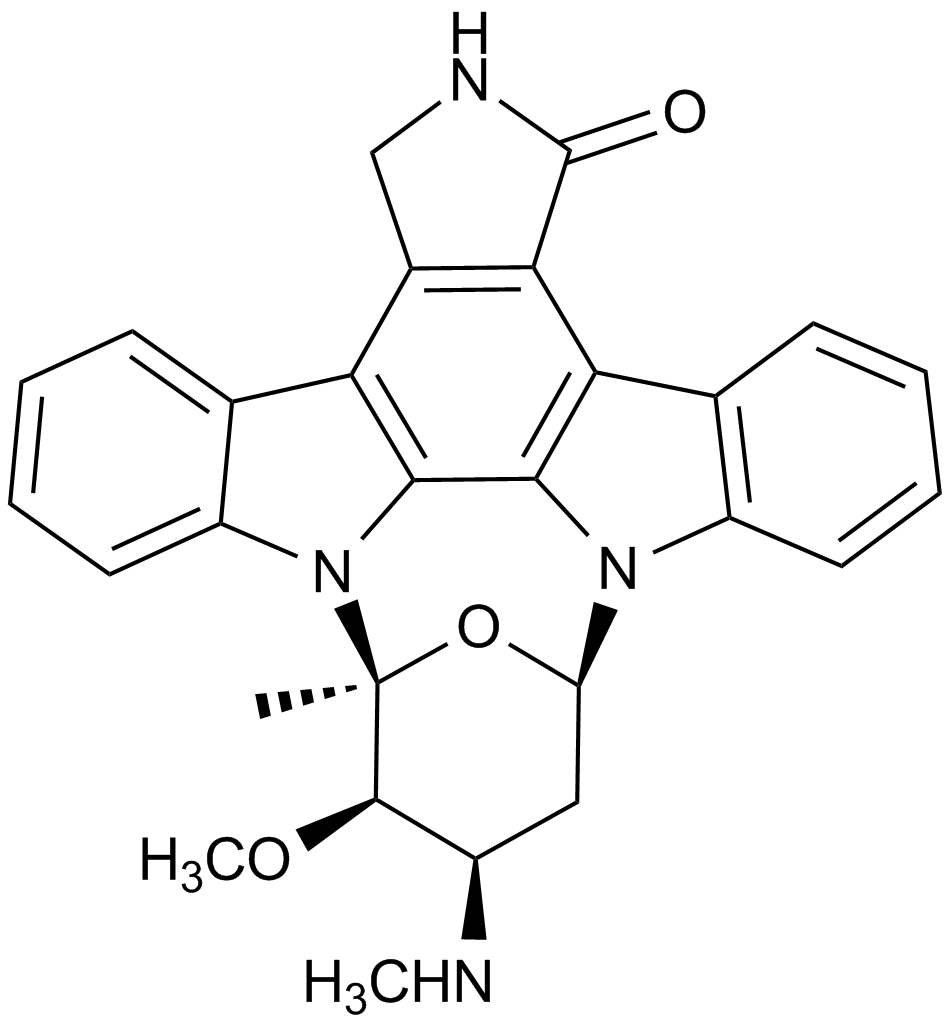
GC15299
Staurosporine(CGP 41251)
A potent inhibitor of protein kinase C
-
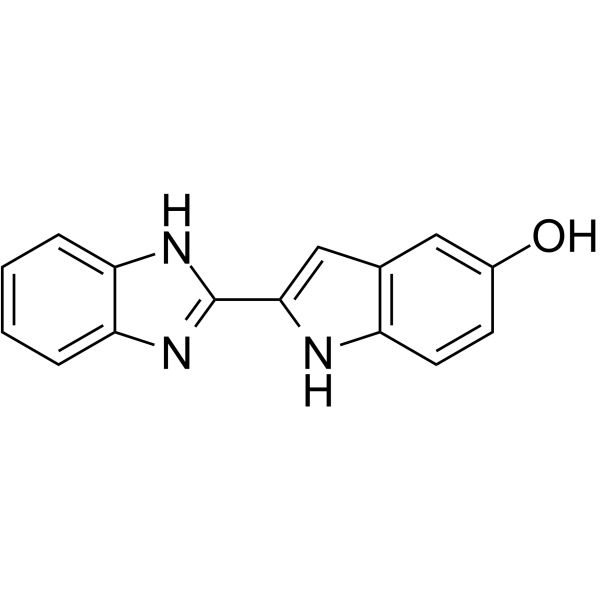
GC69981
SY-LB-35
SY-LB-35 is an effective bone morphogenetic protein (BMP) receptor agonist. It can stimulate C2C12 cells to significantly increase cell number and cell vitality, causing the cell cycle to transition into S phase and G2/M phase. SY-LB-35 stimulates typical Smad and atypical PI3K/Akt, ERK, p38, and JNK intracellular signaling pathways.
-
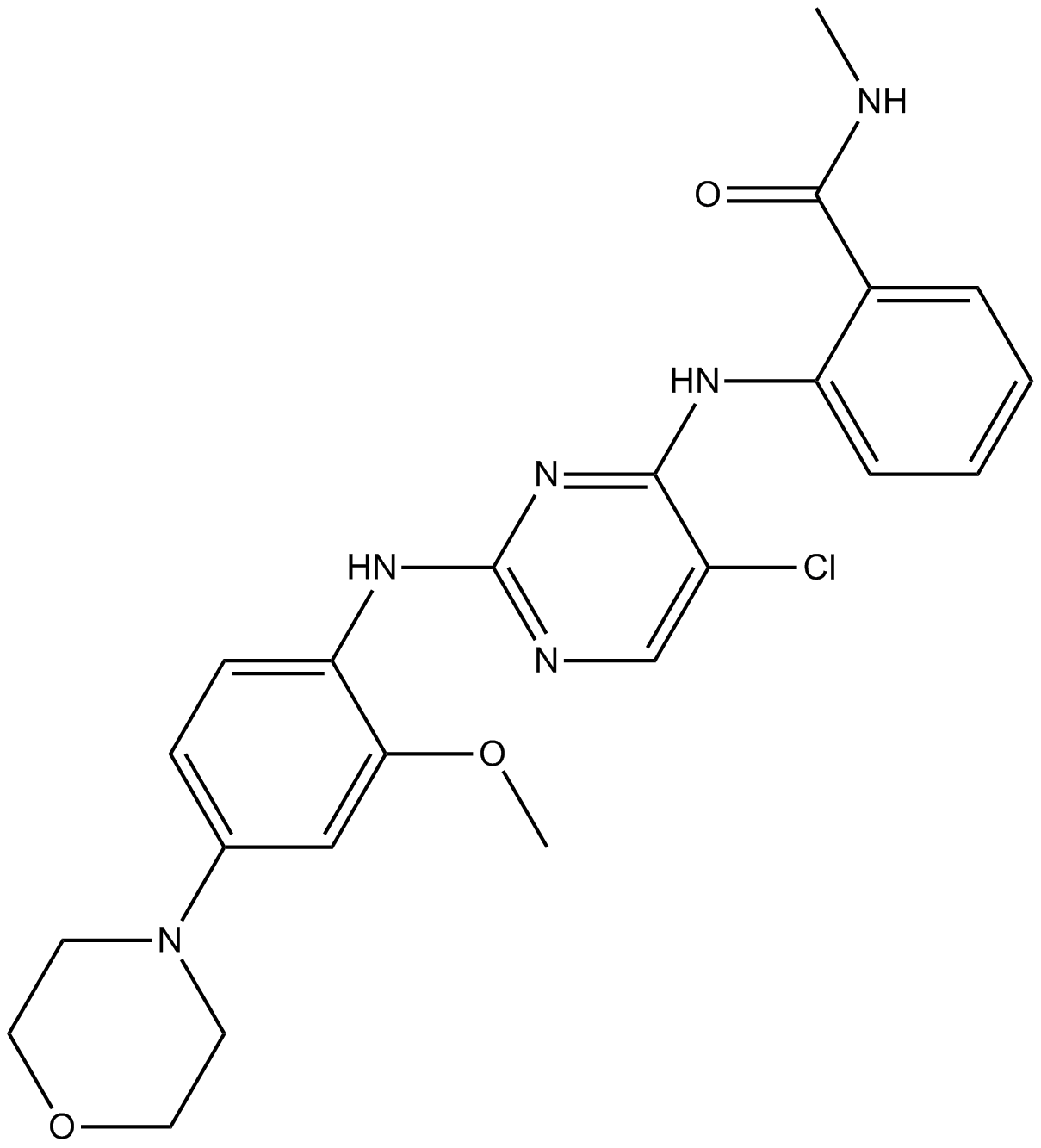
GC16821
TAE226(NVP-TAE226)
TAE226(NVP-TAE226) (TAE226) is a potent and ATP-competitive dual FAK and IGF-1R inhibitor with IC50s of 5.5 nM and 140 nM, respectively.
-
GC17901
Tamoxifen
Tamoxifen(TAM) serves as a selective estrogen receptor regulator (SERM), inhibiting estrogen's effects in breast cells while potentially stimulating estrogen activity in cells found in different tissues.
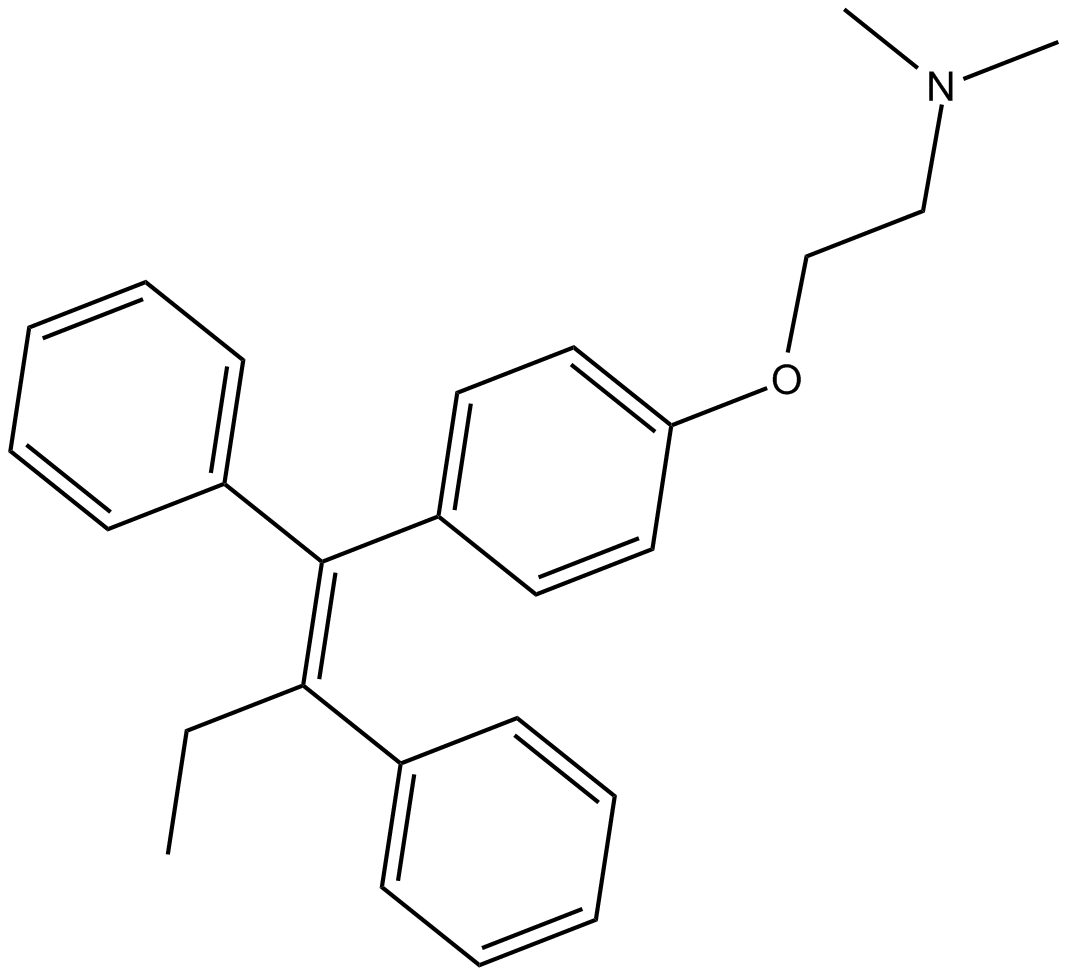
-
GC10682
TAS 301
inhibitor of smooth muscle cell migration and proliferation, inhibits PKC signaling
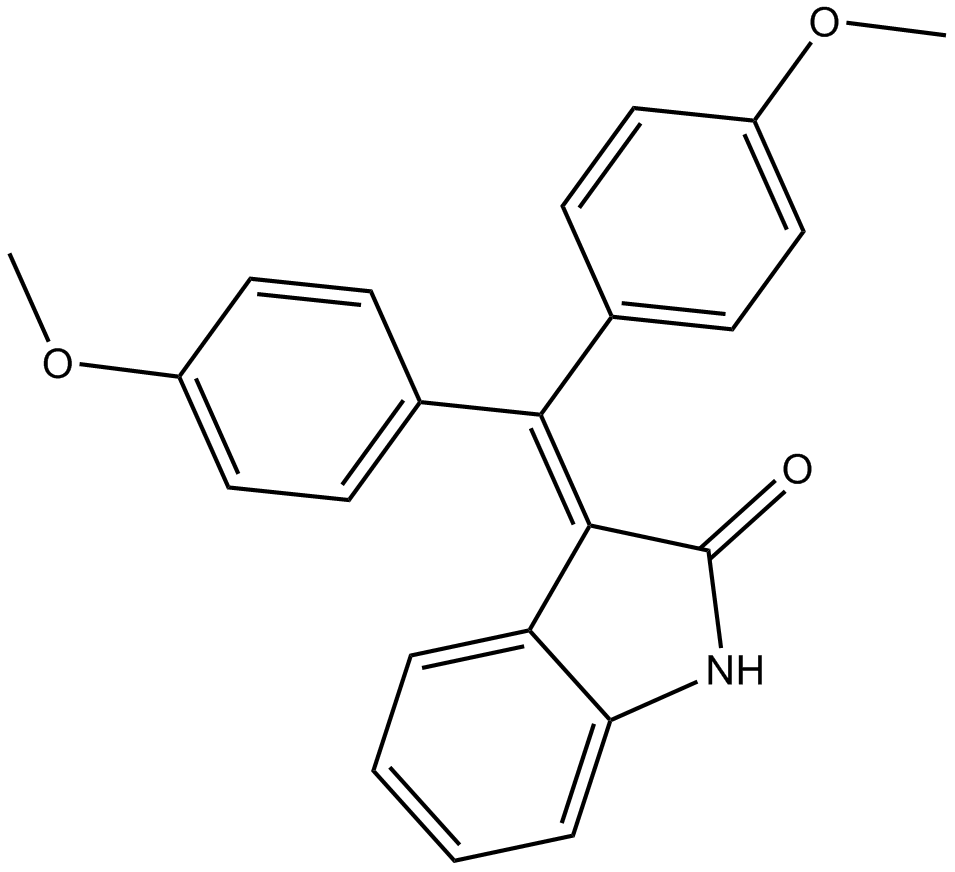
-
GC11181
Thiazovivin
ROCK inhibitor
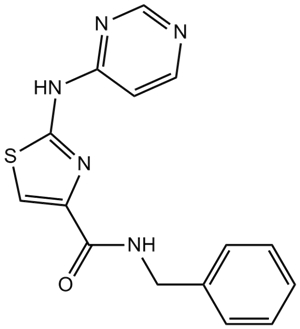
-
GC26000
TP0427736 HCl
TP0427736 is a potent inhibitor of ALK5 kinase activity with an IC50 of 2.72 nM and this effect is 300-fold higher than the inhibitory effect on ALK3 (IC50 = 836 nM for ALK3). It also inhibits Smad2/3 phosphorylation in A549 cells induced by TGF-β1 with an IC50 value of 8.68 nM.
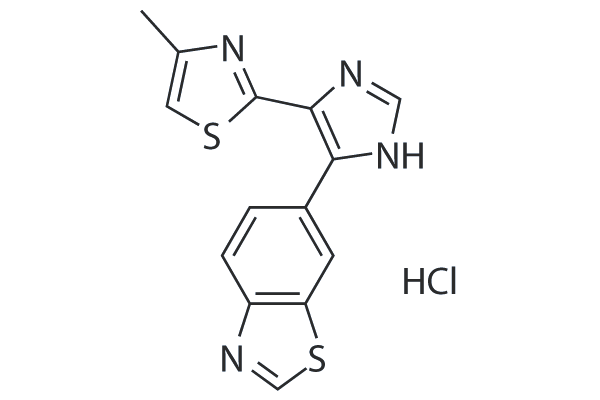
-
GC70050
Trabedersen
Trabedersen (AP 12009) is an antisense oligodeoxynucleotide that specifically inhibits TGF-β2 (TGF-beta/Smad). Trabedersen can be used to study malignant brain tumors and other solid tumors that overexpress TGF-β2, such as skin, pancreatic, and colon tumors.

-
GC40928
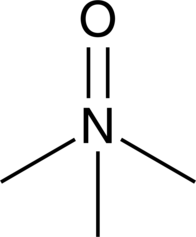
Trimethylamine N-oxide
Trimethylamine N-oxide (TMAO) is a product of the oxidation of TMA by flavin-containing mono-oxygenase 3 in the liver.
-
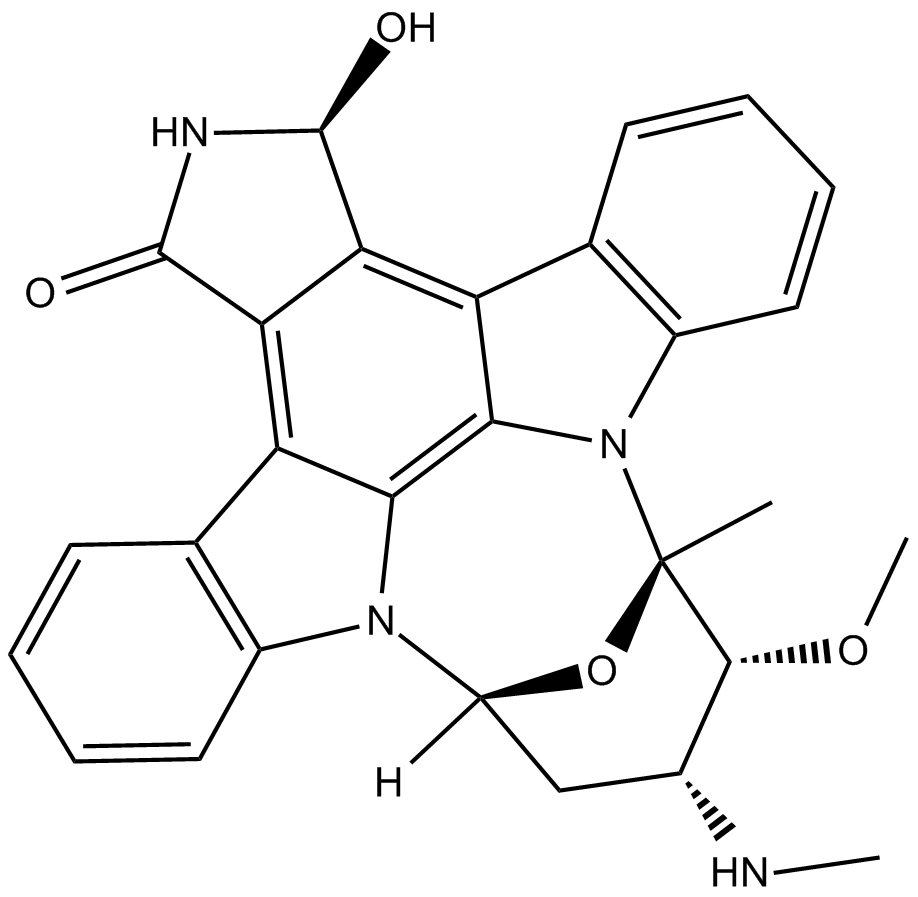
GC16678
UCN-02
protein kinase C inhibitor
-
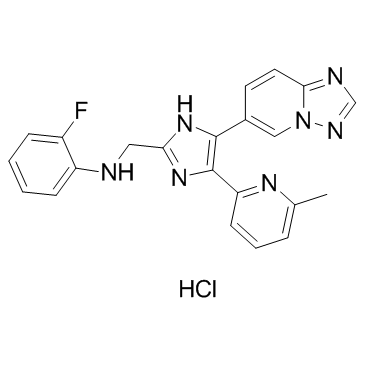
GC37880
Vactosertib Hydrochloride
Vactosertib Hydrochloride (EW-7197 Hydrochloride) is a potent, orally active and ATP-competitive activin receptor-like kinase 5 (ALK5) inhibitor with an IC50 of 12.9 nM. Vactosertib Hydrochloride also inhibits ALK2 and ALK4 (IC50 of 17.3 nM) at nanomolar concentrations. Vactosertib Hydrochloride has potently antimetastatic activity and anticancer effect.
-
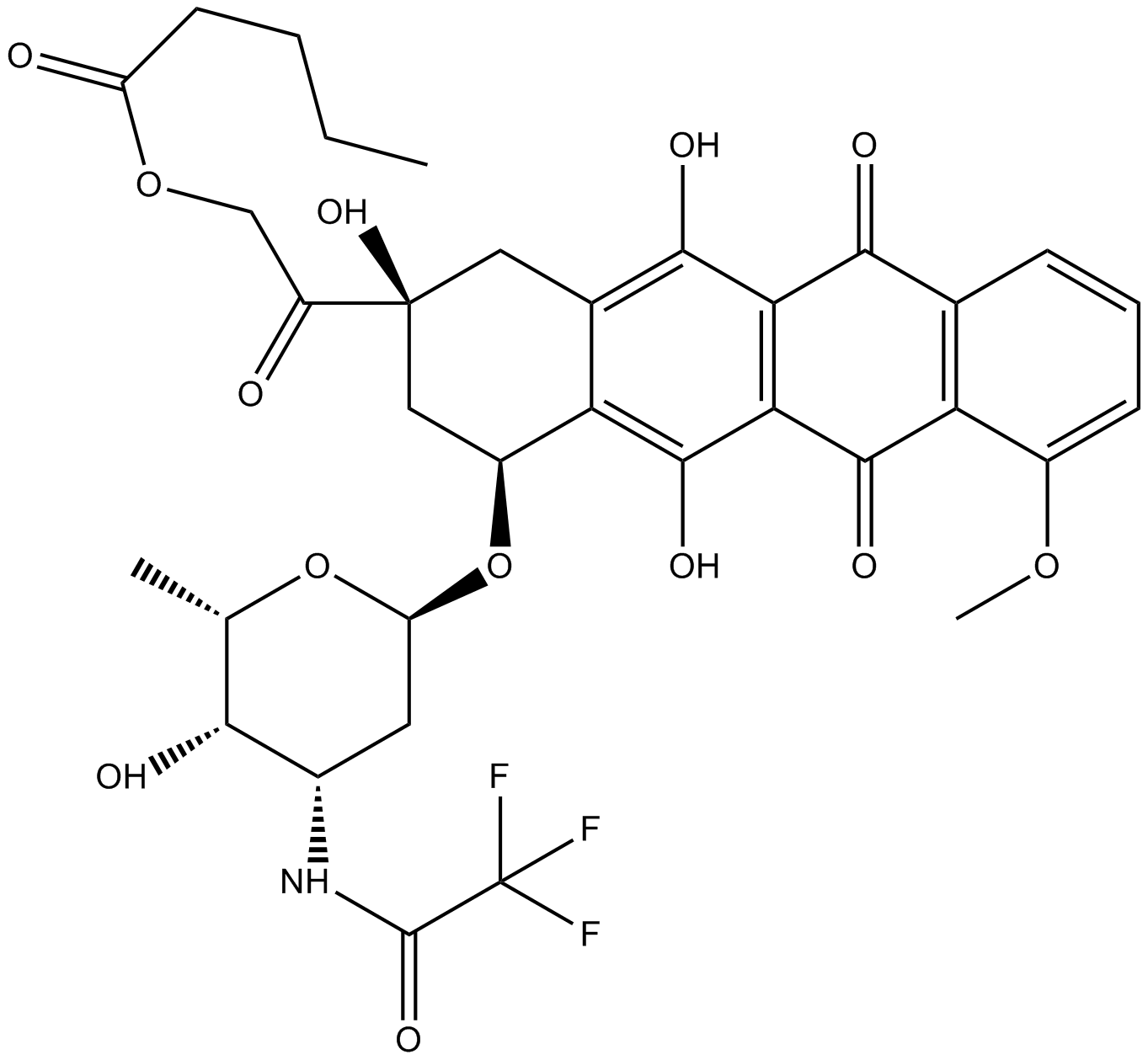
GC16934
Valrubicin
chemotherapy drug used to treat bladder cancer
-
GC70110
Vamotinib
Vamotinib (PF-114) is an effective, selective and orally active tyrosine kinase inhibitor. Vamotinib inhibits the self-phosphorylation of BCR/ABL and BCR/ABL-T315I. Vamotinib induces apoptosis. Vamotinib exhibits anti-proliferative and anti-tumor activity. Vamotinib has potential in the study of resistant Philadelphia chromosome-positive (Ph+) leukemia.
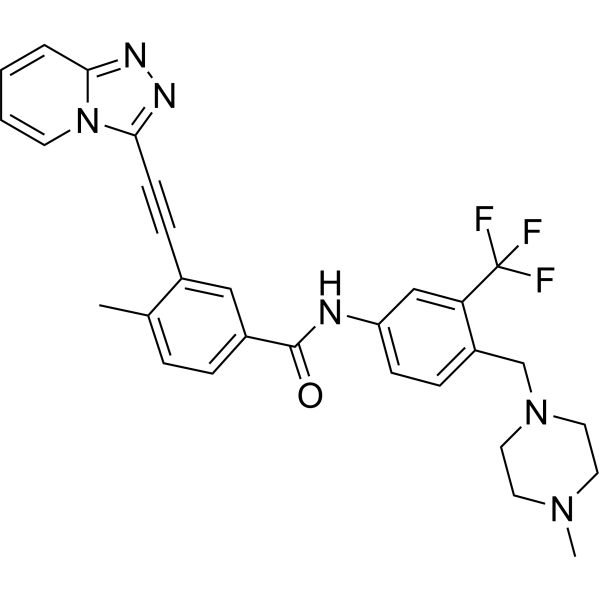
-
GC13232
Verbascoside
PKC/NF-κB inhibitor
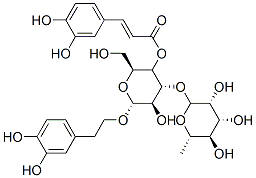
-
GC31912
VTX-27
VTX-27 is a selective protein kinase C θ (PKC θ) inhibitor, with Kis of 0.08 nM and 16 nM for PKC θ and PKC δ.
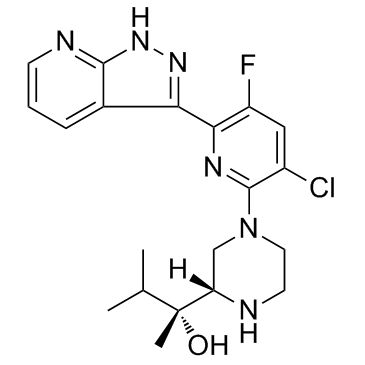
-
GC26055
WAY-301398
WAY-301398 (6-methoxynaphthalene) can bind to protein kinase C (PKC).
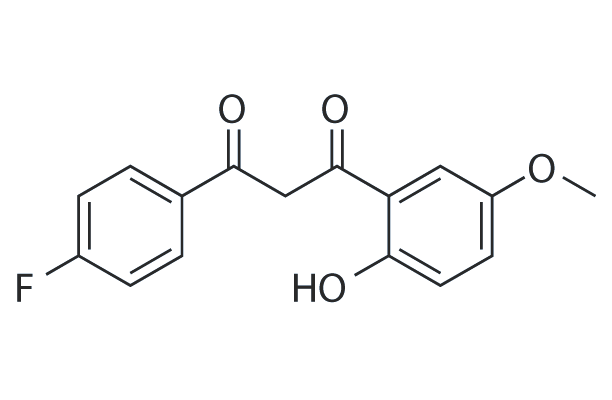
-
GC26065
WAY-354831
WAY-354831 exhibits antibacterial activity.
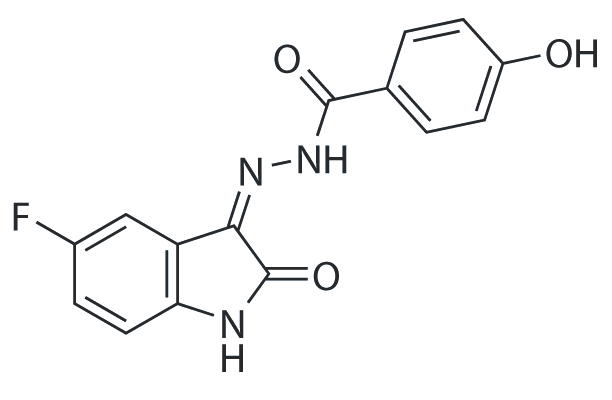
-
GC10970
WP1130
WP1130 (WP1130) is a cell-permeable deubiquitinase (DUB) inhibitor, directly inhibiting DUB activity of USP9x, USP5, USP14, and UCH37. WP1130 has been shown to downregulate the antiapoptotic proteins Bcr-Abl and JAK2.
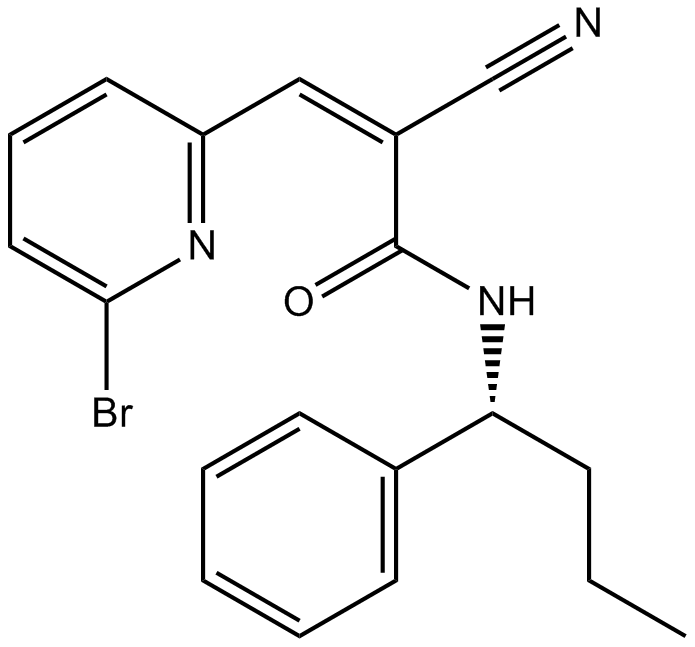
-
GC62146
XST-14
XST-14 is a potent, competitive and highly selective ULK1 inhibitor with an IC50 of 26.6 nM. XST-14 induces autophagy inhibition by reducing the phosphorylation of the ULK1 downstream substrate. XST-14 induces apoptosis in hepatocellular carcinoma (HCC) cells and has antitumor effects.
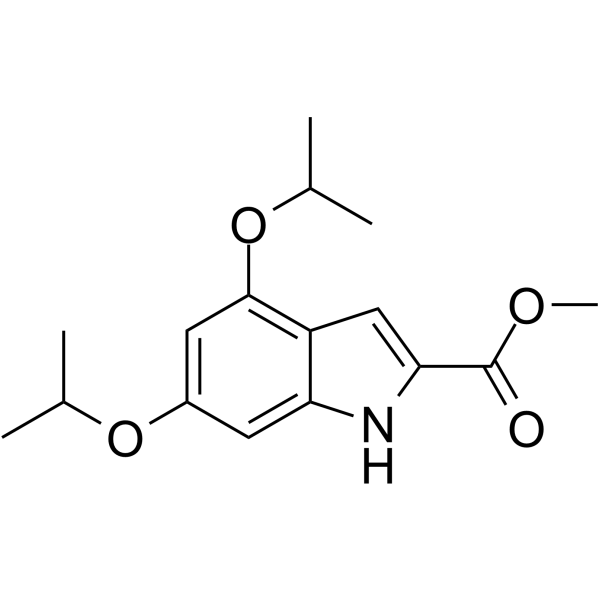
-
GC15712
Y-27632
A ROCK inhibitor,potent and selective
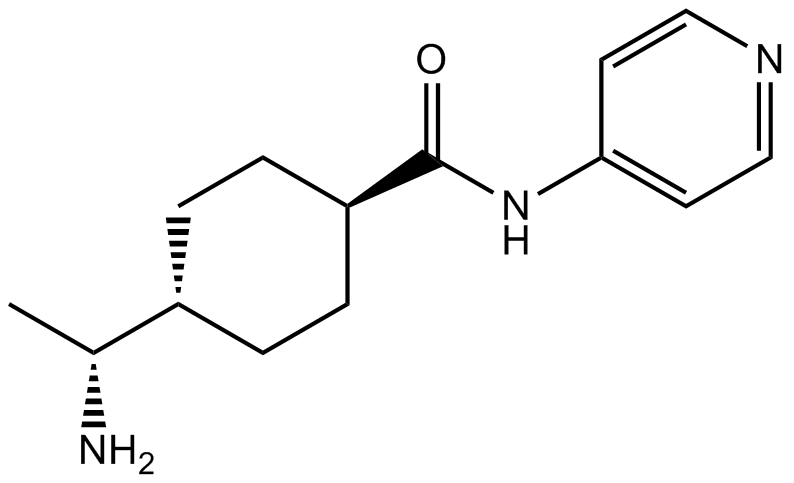
-
GC37947
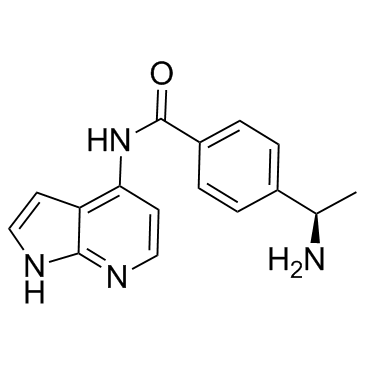
Y-33075
Y-33075 is a selective ROCK inhibitor derived from Y-27632, and is more potent than Y-27632, with an IC50 of 3.6 nM.
-
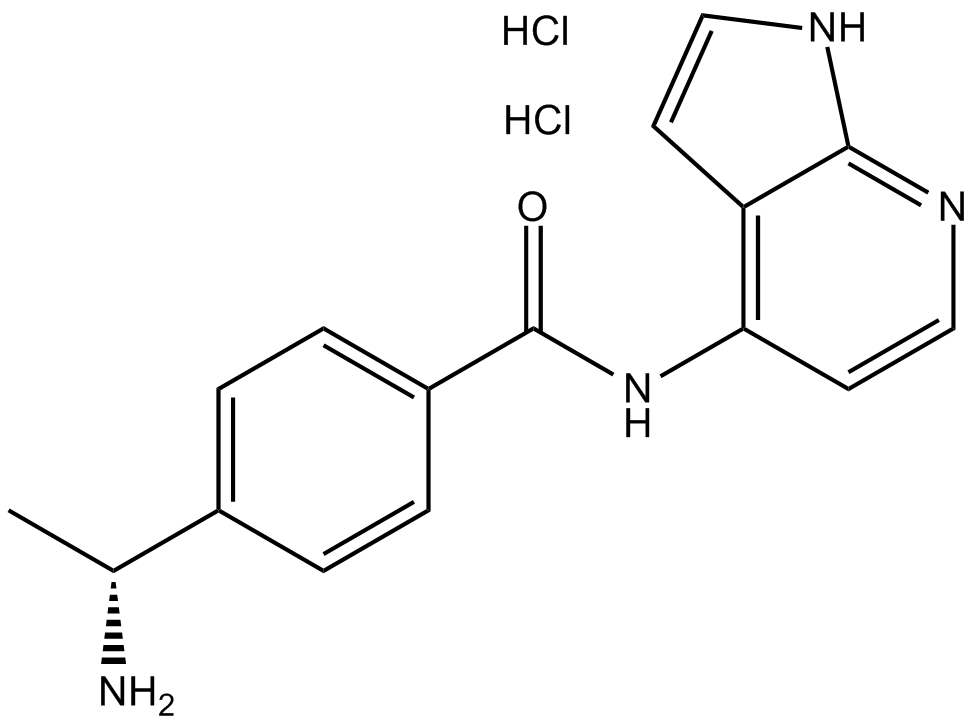
GC13423
Y-39983 dihydrochloride
ROCK family inhibitor
-
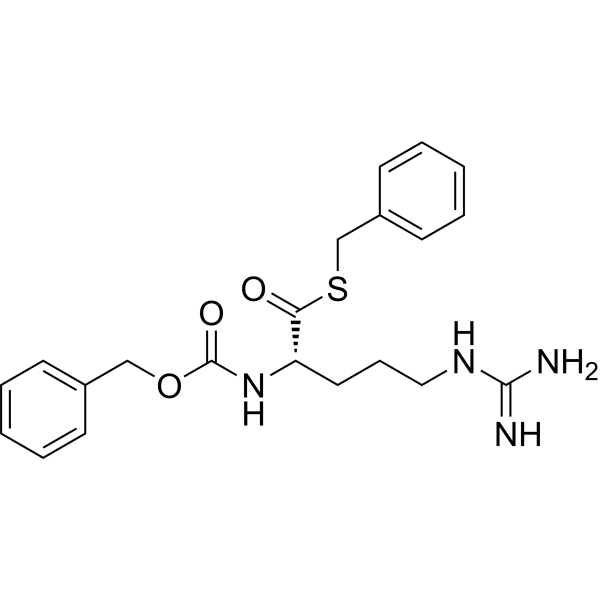
GC70166
Z-Arg-SBzl
Z-Arg-SBzl is a substrate for activating protein C in both cows and humans.
-
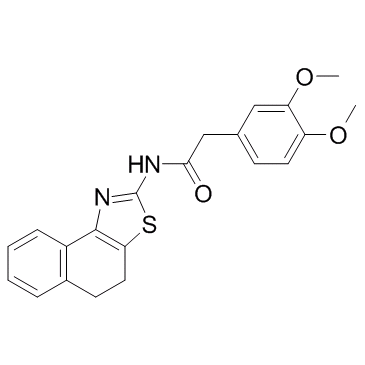
GC33120
ZINC00881524
A ROCK inhibitor
-
GC13273
ZIP
ZIP is a selective peptide inhibitor of PKMζ.
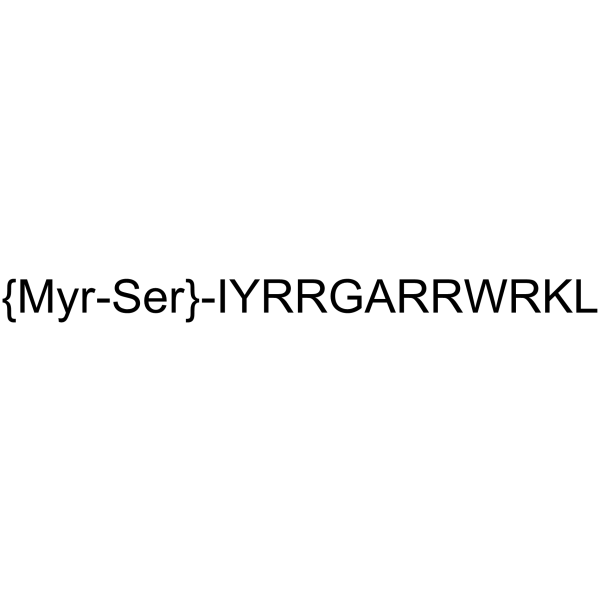
-
GC13461
ZIP (SCRAMBLED)
PKMζ inhibitor
-
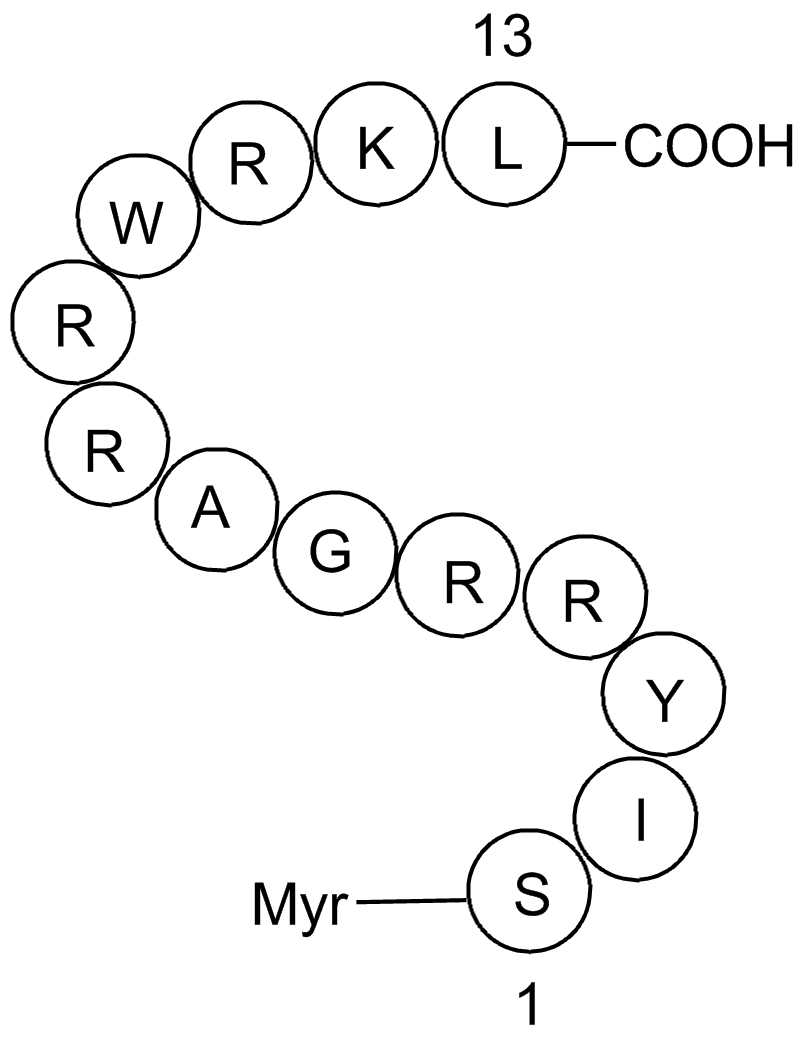
GC16510
ZIP, Biotinylated
ZIP with a biotin moiety covalently attached
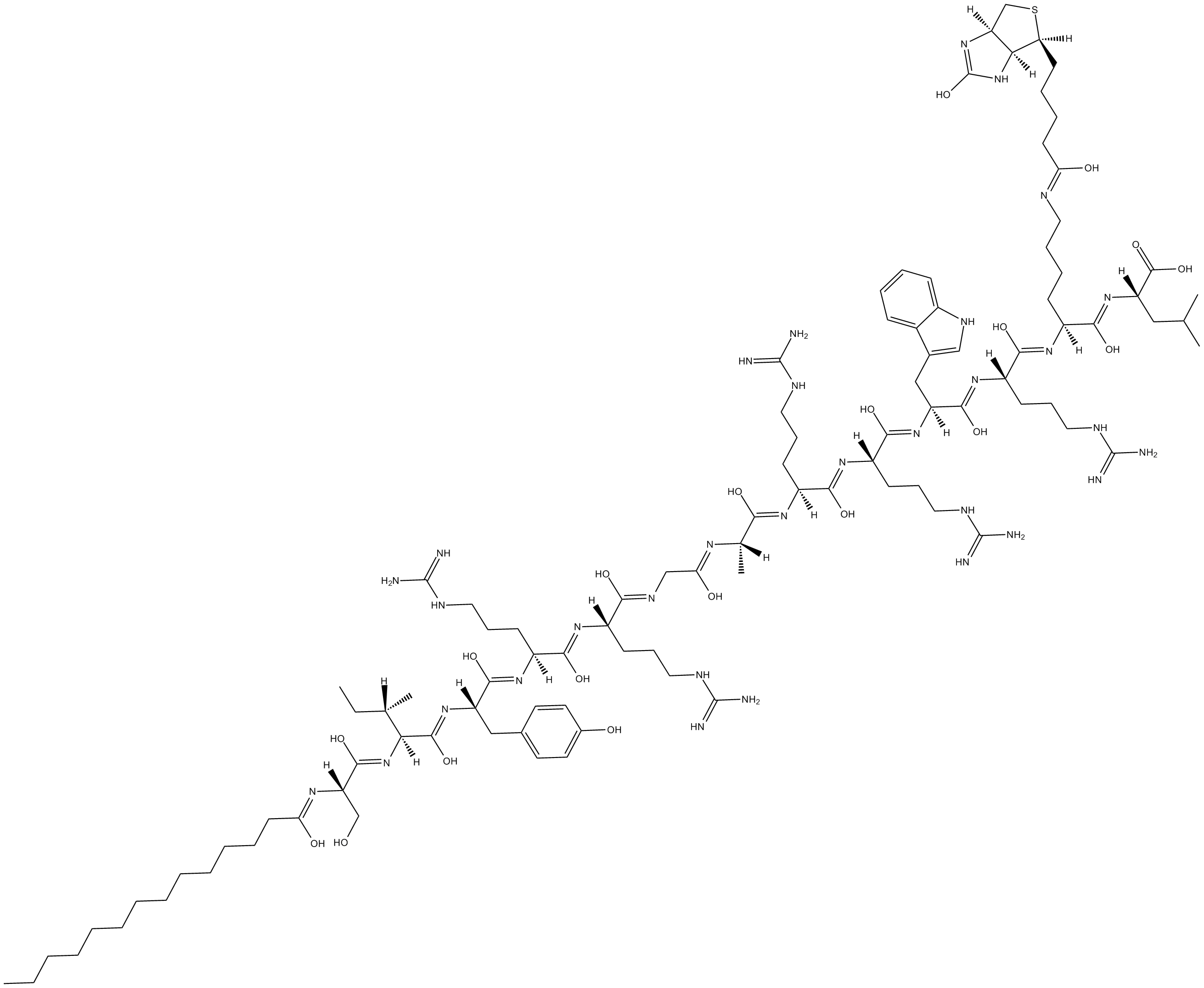
-
GC14701
Zoledronic Acid
Potent nitrogen-containing bisphosphonates
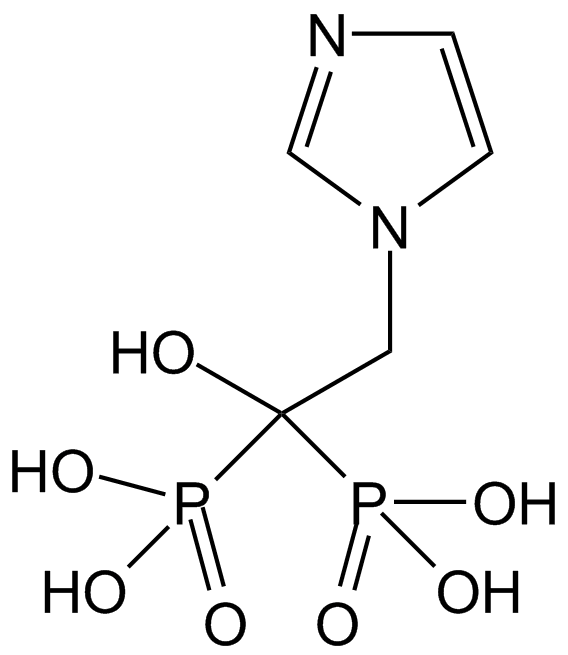
-
GC17727
[Ala107]-MBP (104-118)
Non-competitive inhibitor of PKC
![[Ala107]-MBP (104-118) Chemical Structure [Ala107]-MBP (104-118) Chemical Structure](/media/struct/GC1/GC17727.png)
-
GC15345
[Ala113]-MBP (104-118)
Non-competitive inhibitor of PKC
![[Ala113]-MBP (104-118) Chemical Structure [Ala113]-MBP (104-118) Chemical Structure](/media/struct/GC1/GC15345.png)
-
GC10343
α-Amyloid Precursor Protein Modulator
α-Amyloid Precursor Protein Modulator is a cell-permeable benzolactam-derived protein kinase C (PKC) activator with a Ki of 11.9 nM.
-
GC62478
ζ-Stat
ζ-Stat (NSC37044) is a specific and atypical PKC-ζ inhibitor, with an IC50 of 5 μM. ζ-Stat can reduce melanoma cell lines proliferation and induce apoptosis, and has antitumor activity in vitro.