PKC
PKC (protein kinase C) is a serine/threonine protein kinase that mediates insulin signaling, actin cytoskeleton, cell mobility, apoptosis and tumorigenesis etc.
Products for PKC
- Cat.No. Product Name Information
-
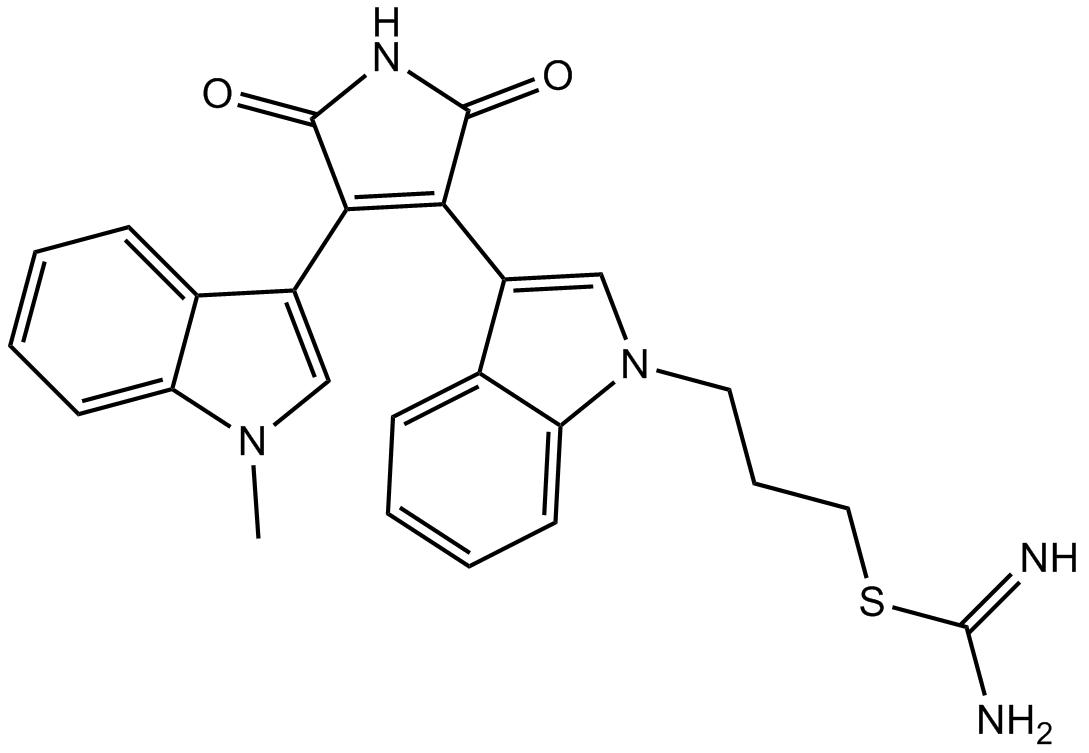
GC13890
(±)-Palmitoylcarnitine chloride
intermediate in mitochondrial fatty acid oxidation
-
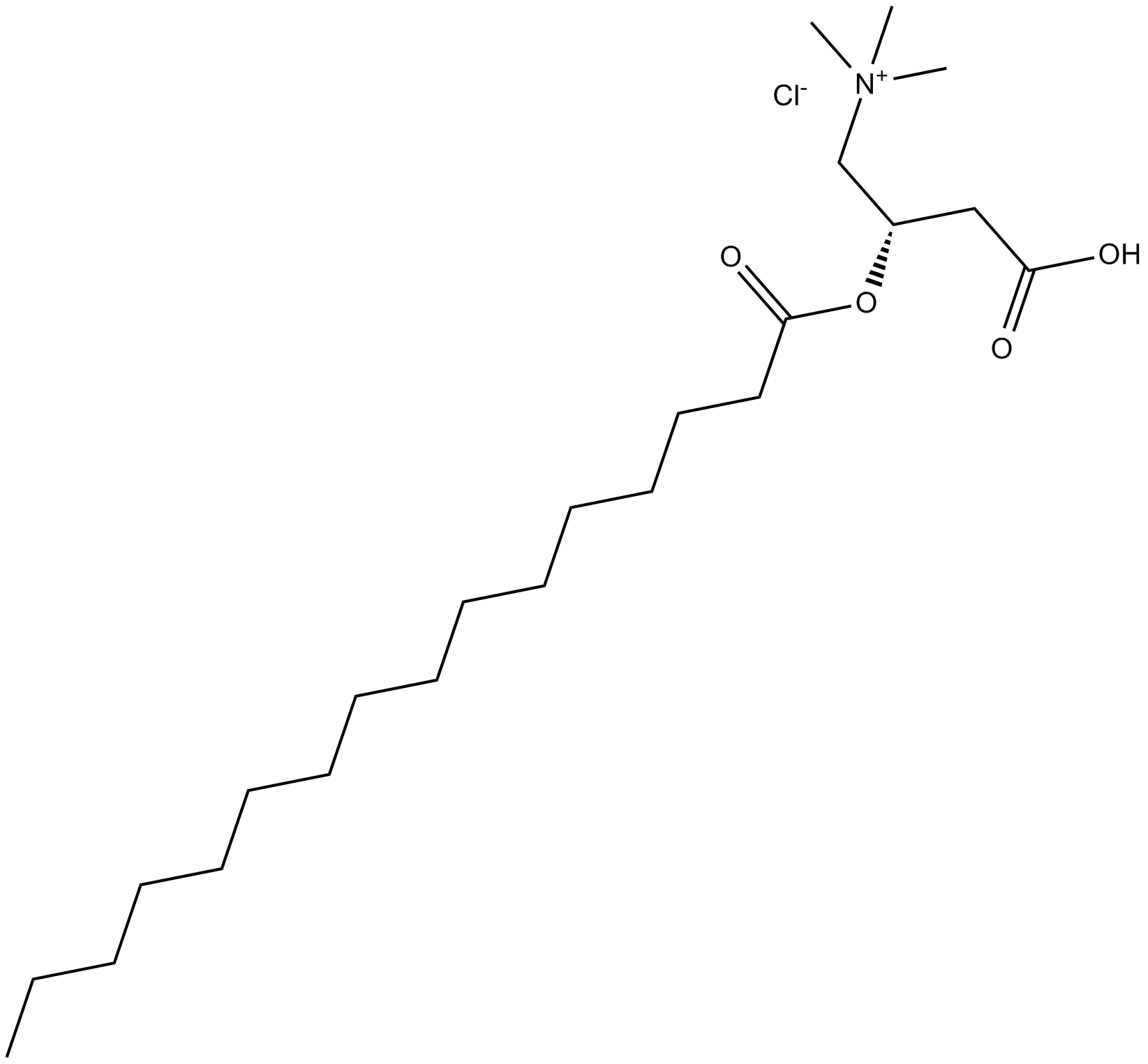
GC10603
(-)-epicatechin gallate
major catechin in green tea
-
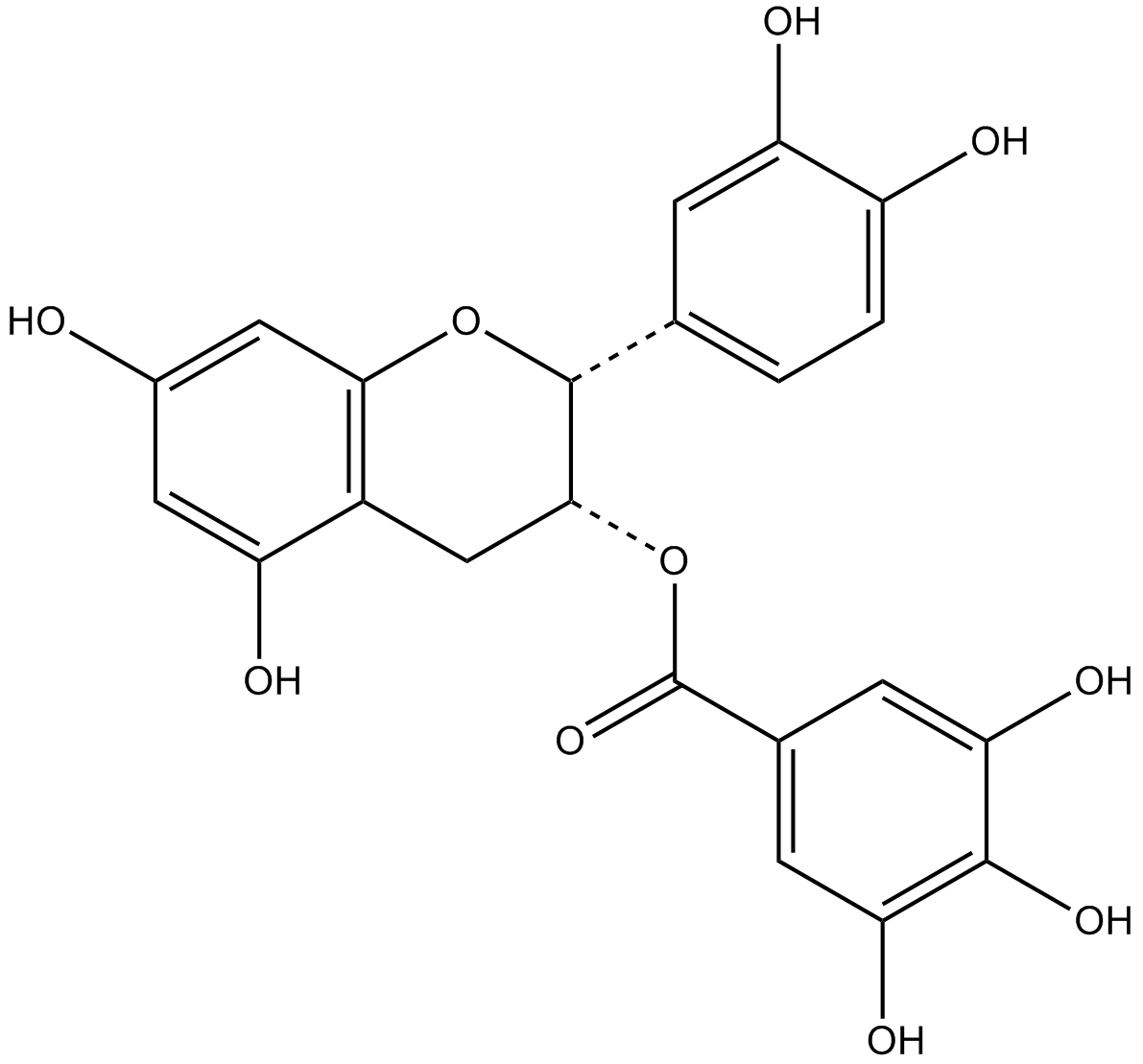
GC17242
(-)-epigallocatechin
green tea epicatechin
-
GC14049
(-)-Epigallocatechin gallate (EGCG)
(-)-Epigallocatechin Gallate sulfate (EGCG) is a major polyphenol in green tea that inhibits cell proliferation and induces apoptosis. In addition, it inhibits the activity of glutamate dehydrogenase 1/2 (GDH1/2, GLUD1/2) ..
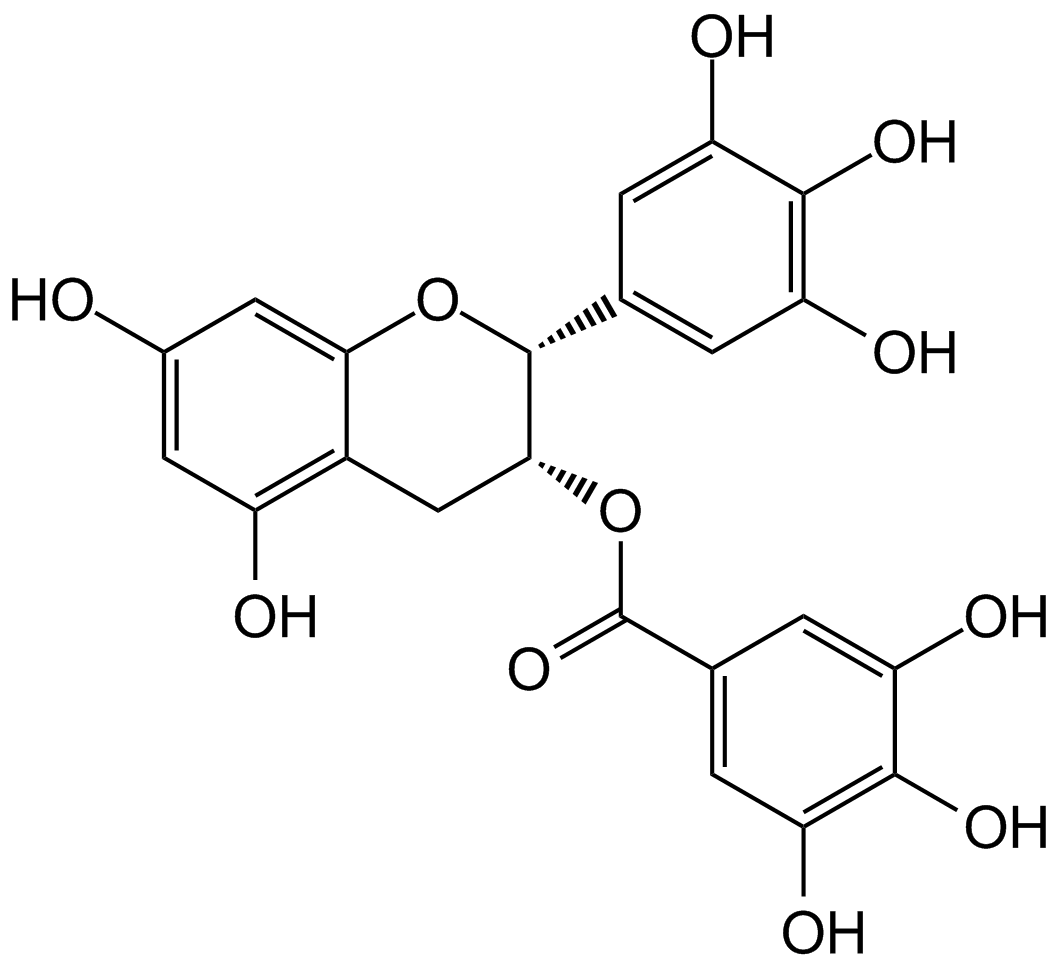
-
GC14012
(-)-Indolactam V
A protein kinase C activator
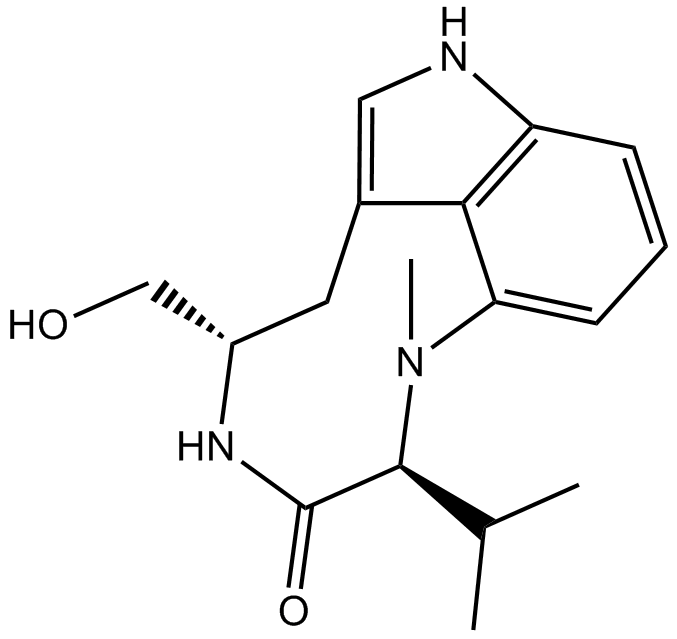
-
GC18062
1,2-Dilauroyl-sn-glycerol
A longchain DAG
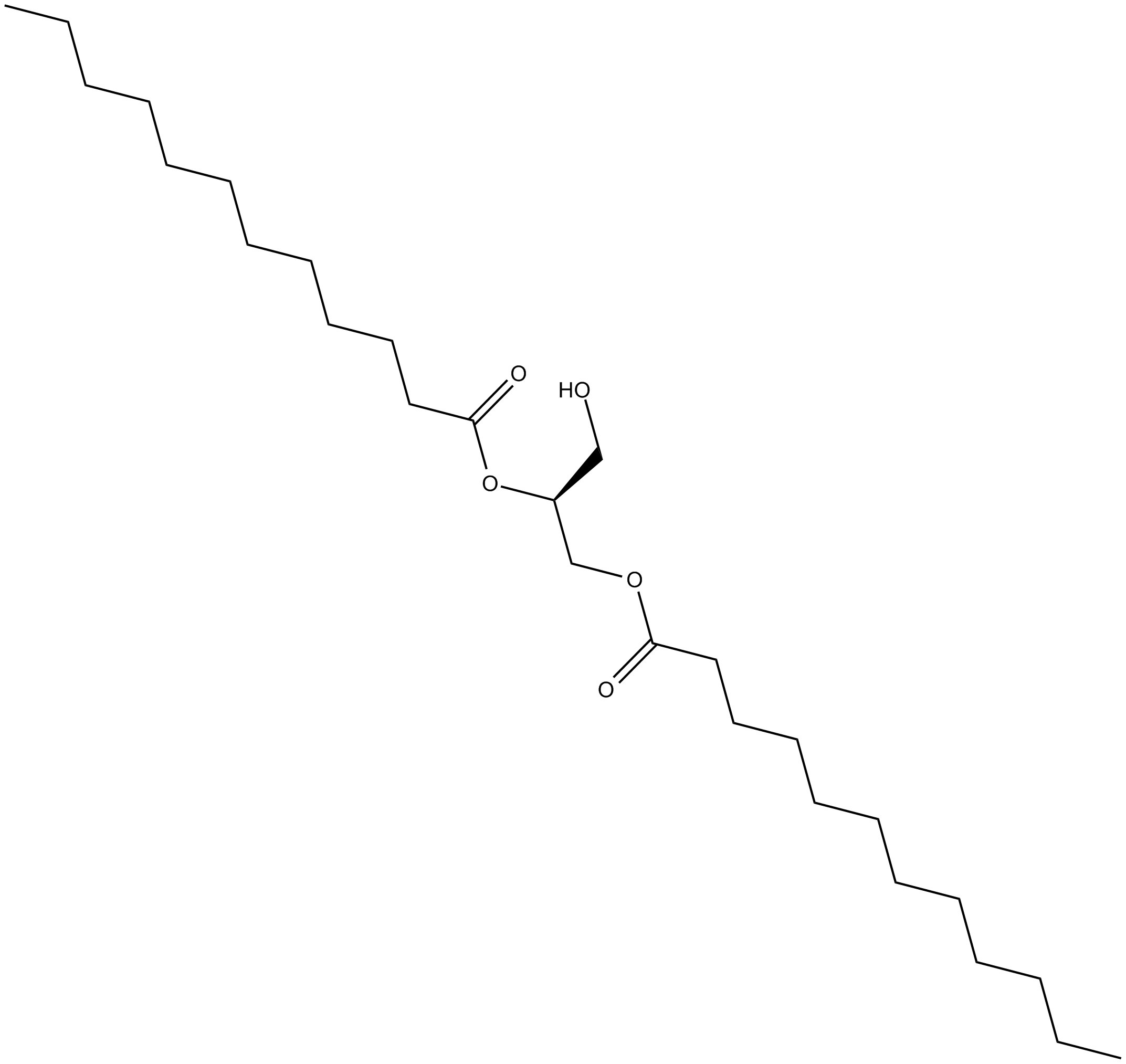
-
GC14134
1,2-Dimyristoyl-sn-glycerol
A longchain DAG
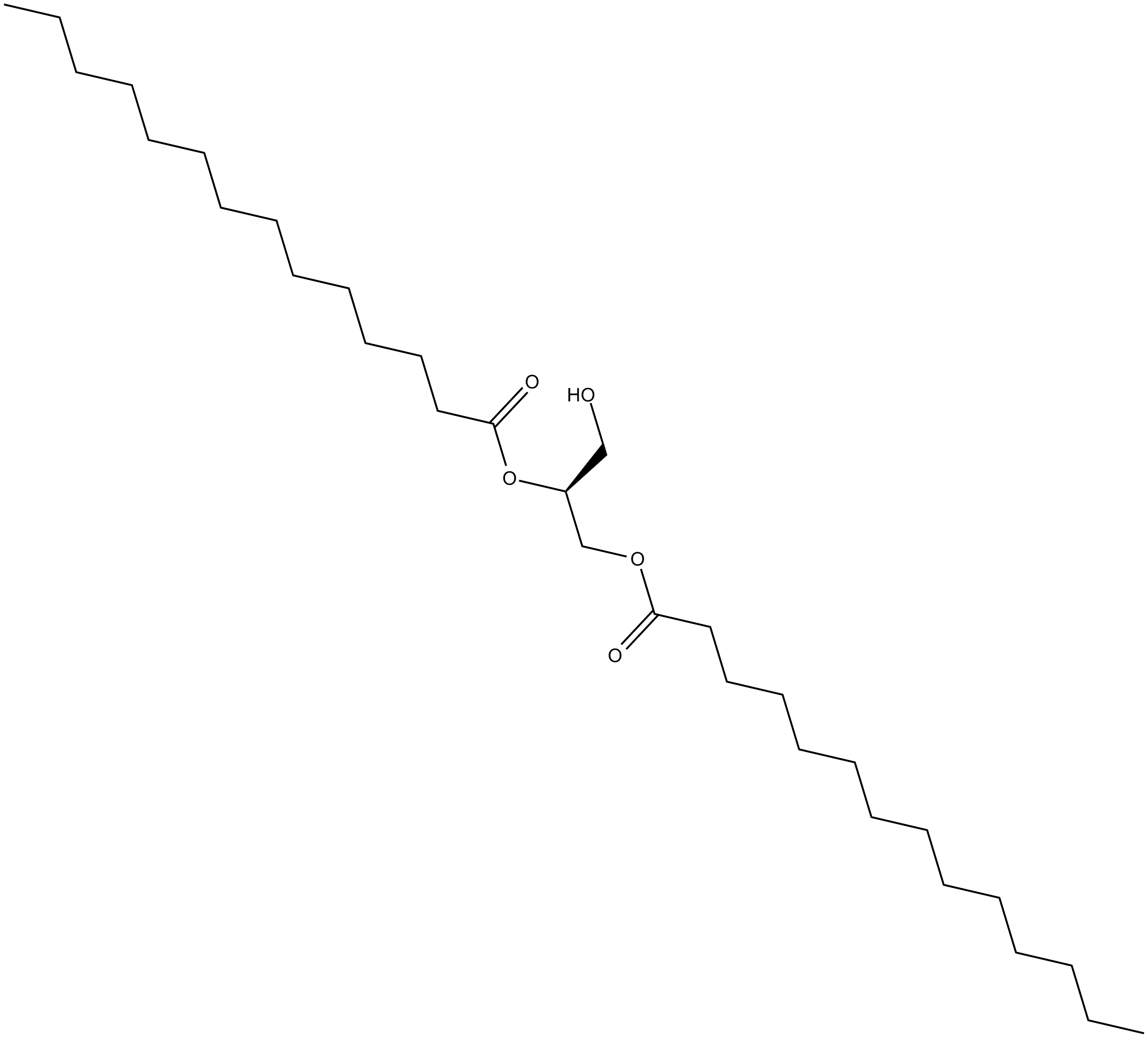
-
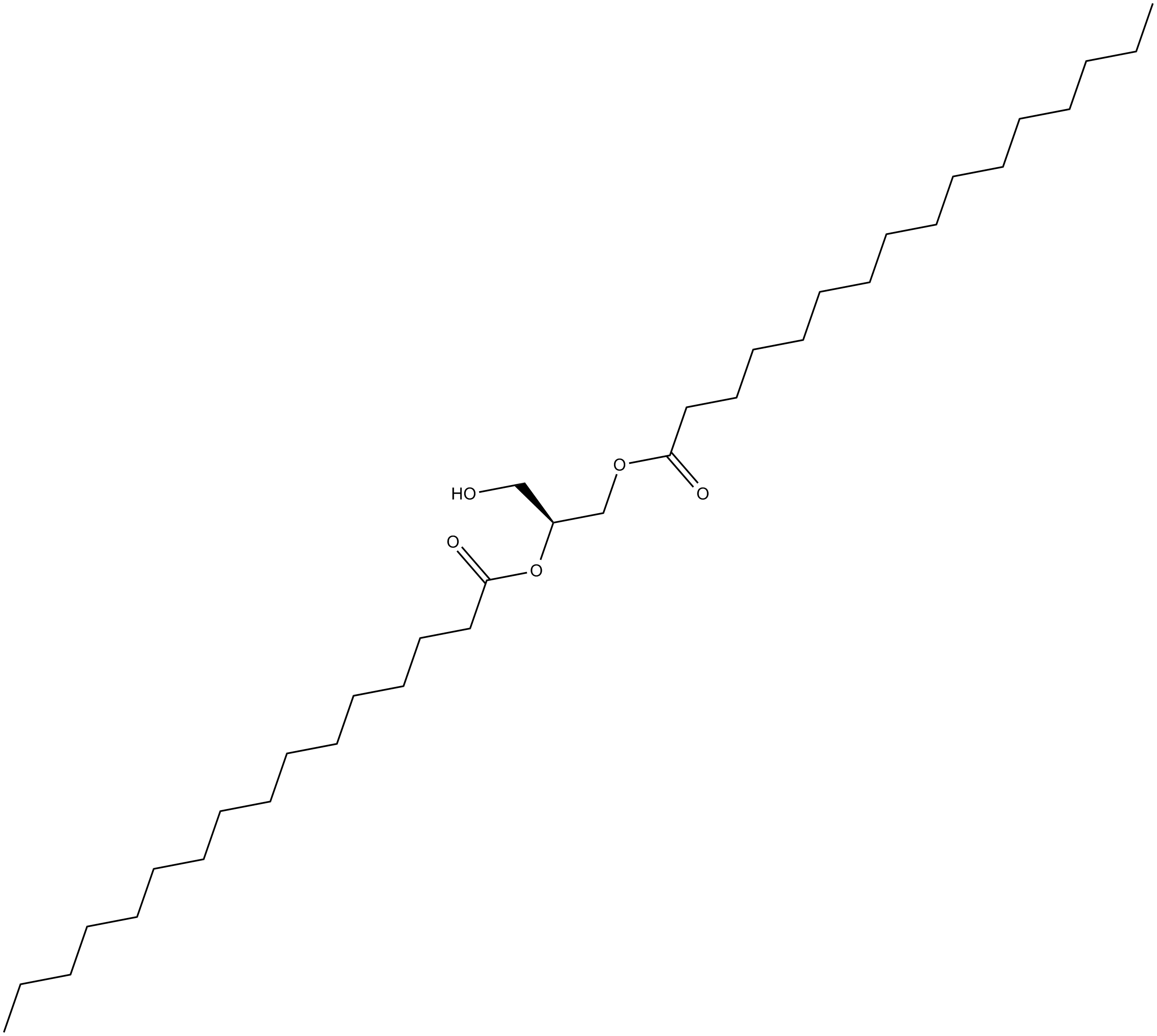
GC13877
1,2-Dipalmitoyl-sn-glycerol
weak activator of PKC
-
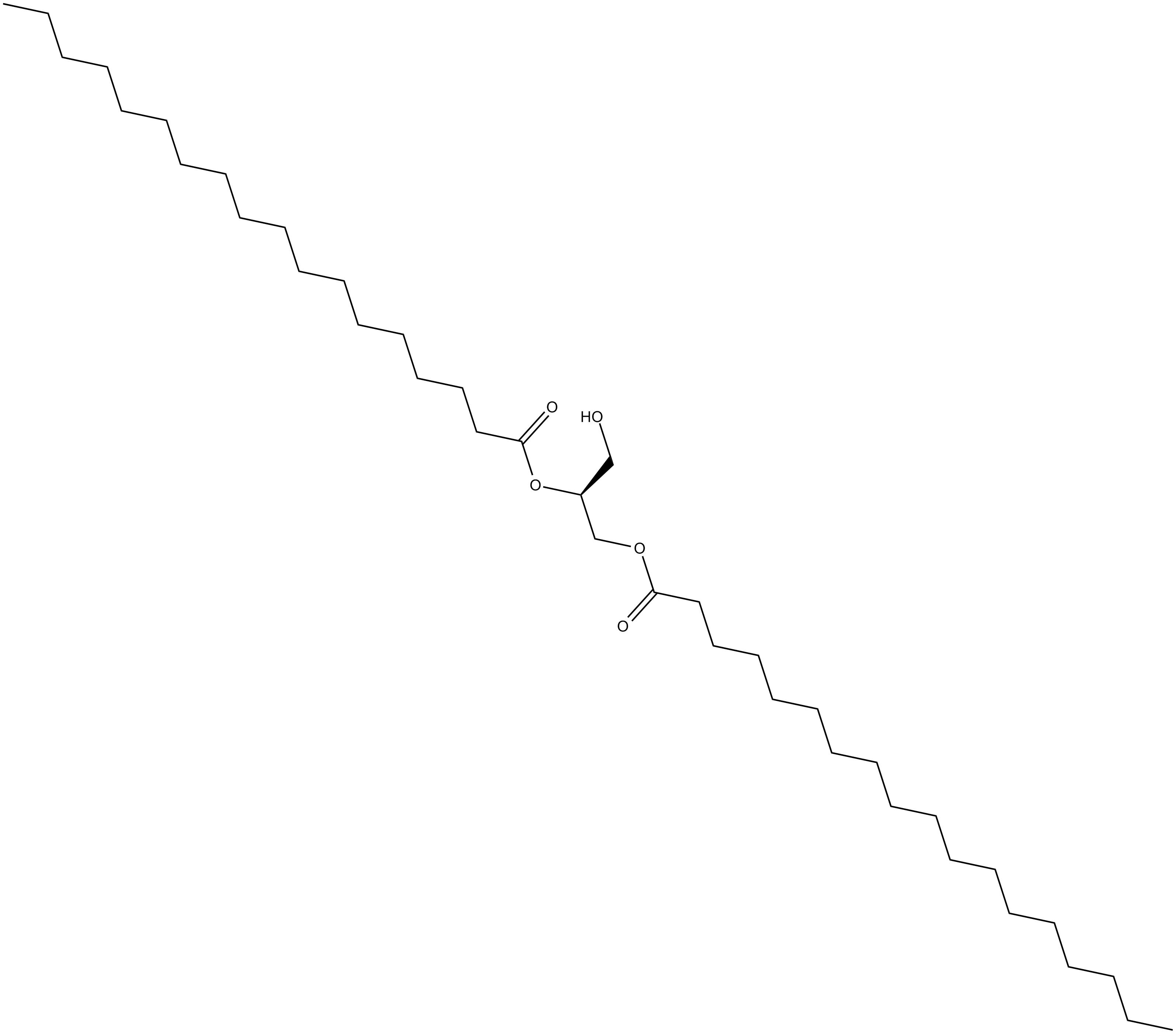
GC12662
1,2-Distearoyl-sn-glycerol
A diacylglycerol
-
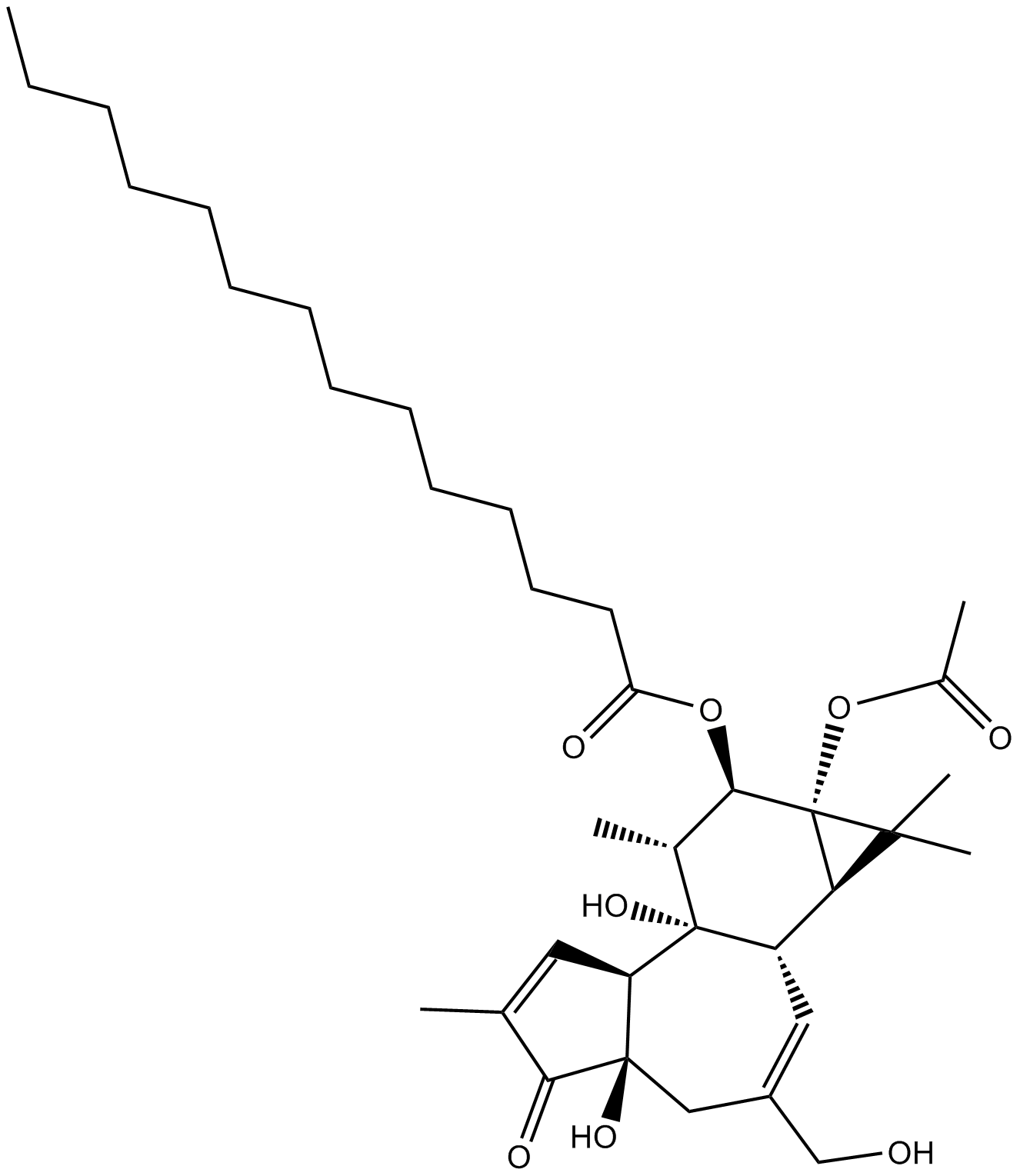
GN10444
12-O-tetradecanoyl phorbol-13-acetate
A PKC activator
-
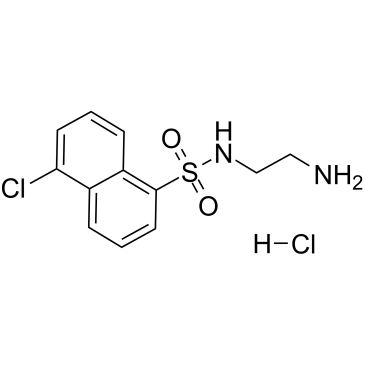
GC60037
A-3 hydrochloride
A-3 hydrochloride is a potent, cell-permeable, reversible, ATP-competitive non-selective antagonist of various kinases.
-
GC16475
Afuresertib
pan-AKT inhibitor
-
GC42747
Afuresertib (hydrochloride)
Afuresertib is a selective, orally bioavailable inhibitor of Akt1, 2, and 3 with Ki values of 0.08, 2, and 2.6 nM, respectively.
-
GC31959
AS2521780
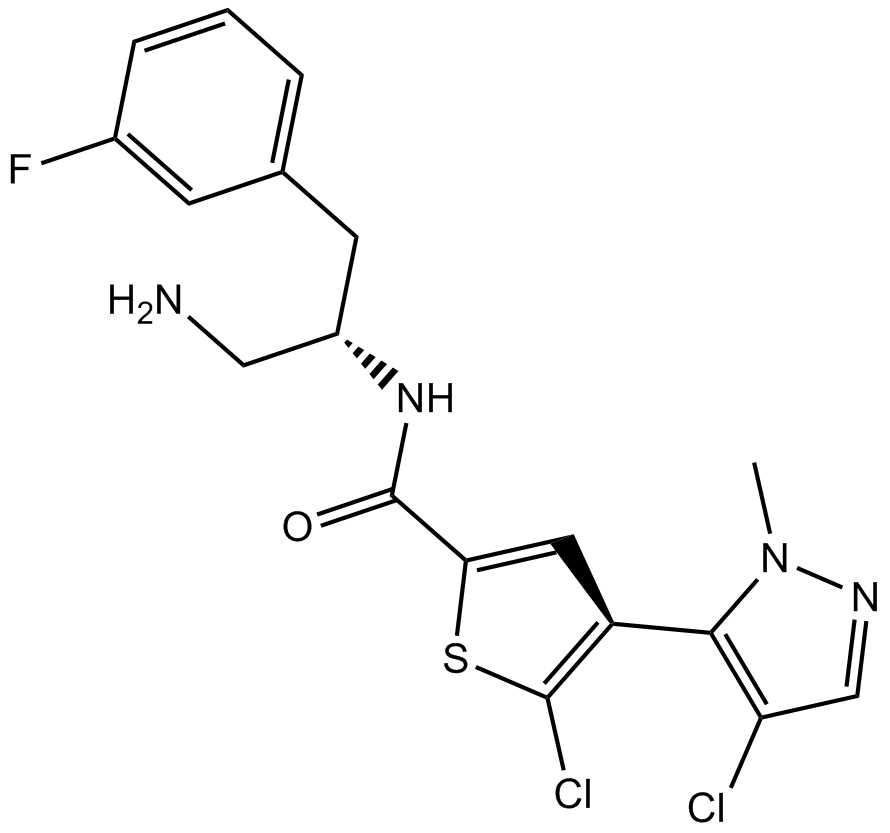
AS2521780 is a novel PKCθ selective inhibitor with an IC50 of 0.48 nM.
-
GC64815
Aurothiomalate sodium
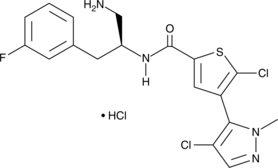
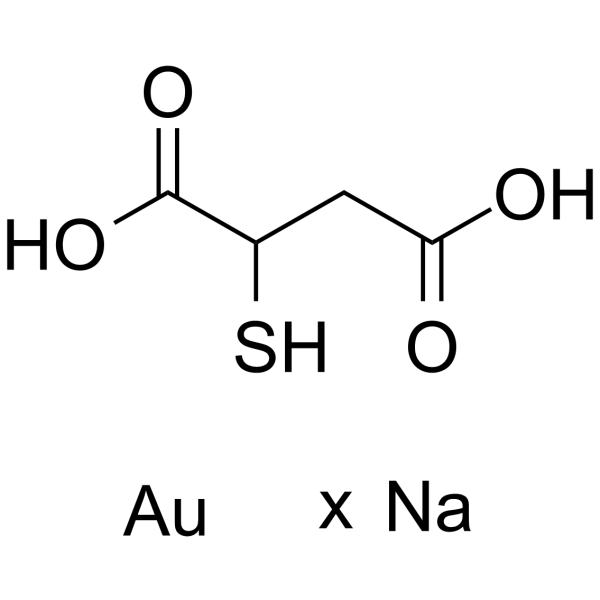
Aurothiomalate sodium is a potent and selective oncogenic PKCι signaling inhibitor.
-
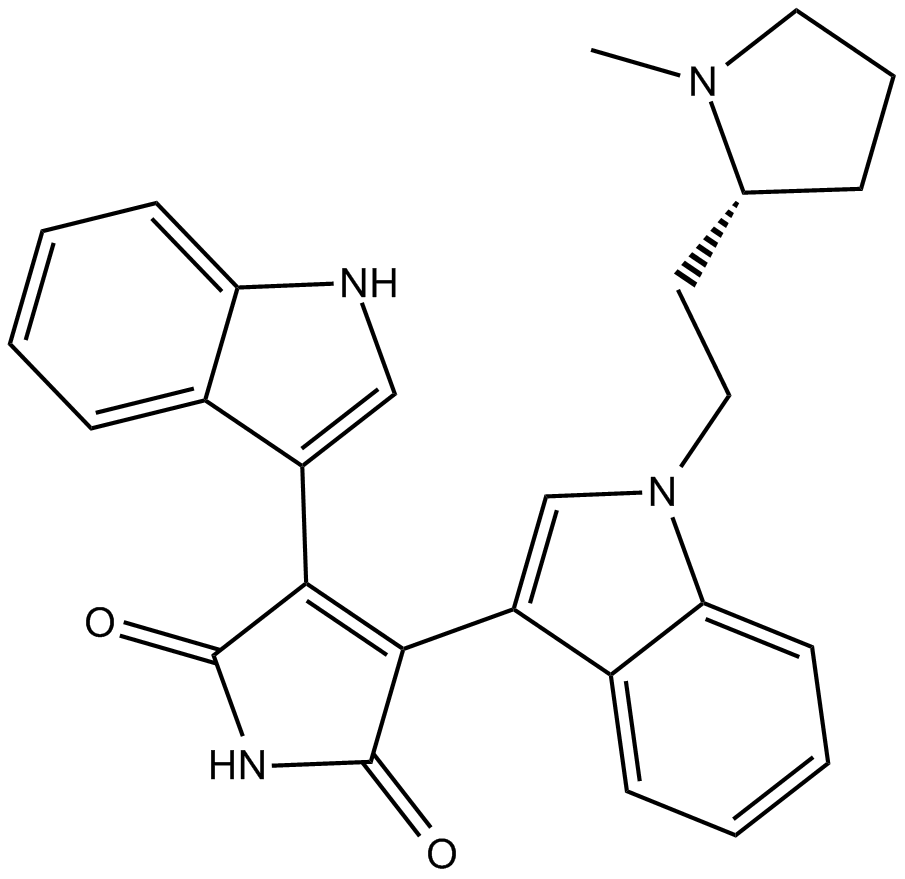
GC12135
Bisindolylmaleimide II
protein kinase C (PKC) inhibitor
-
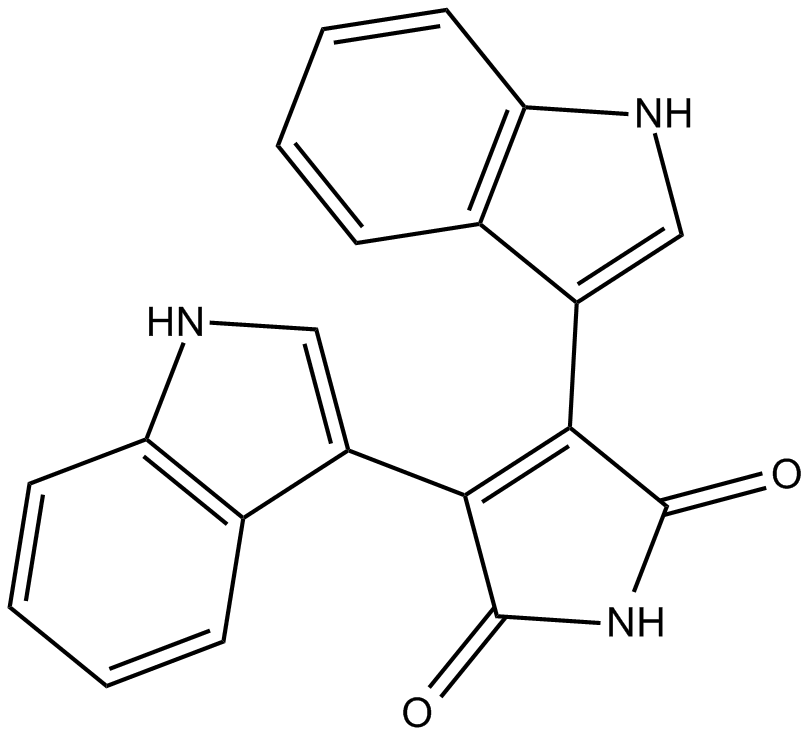
GC14716
Bisindolylmaleimide IV
protein kinase C (PKC) inhibitor
-
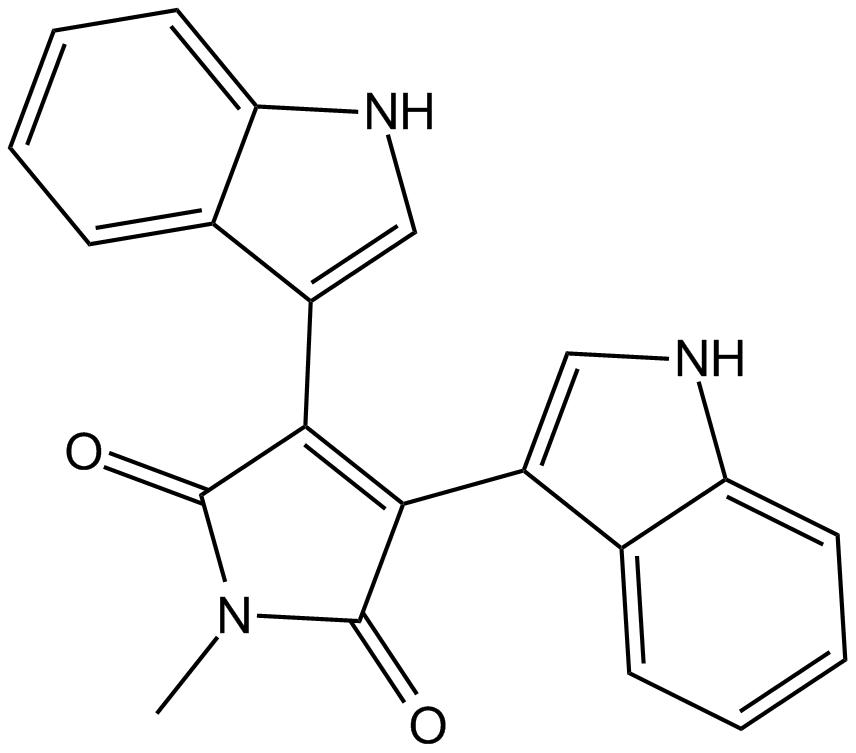
GC17239
Bisindolylmaleimide V
negative control for protein kinase C (PKC)-inhibitory activity
-
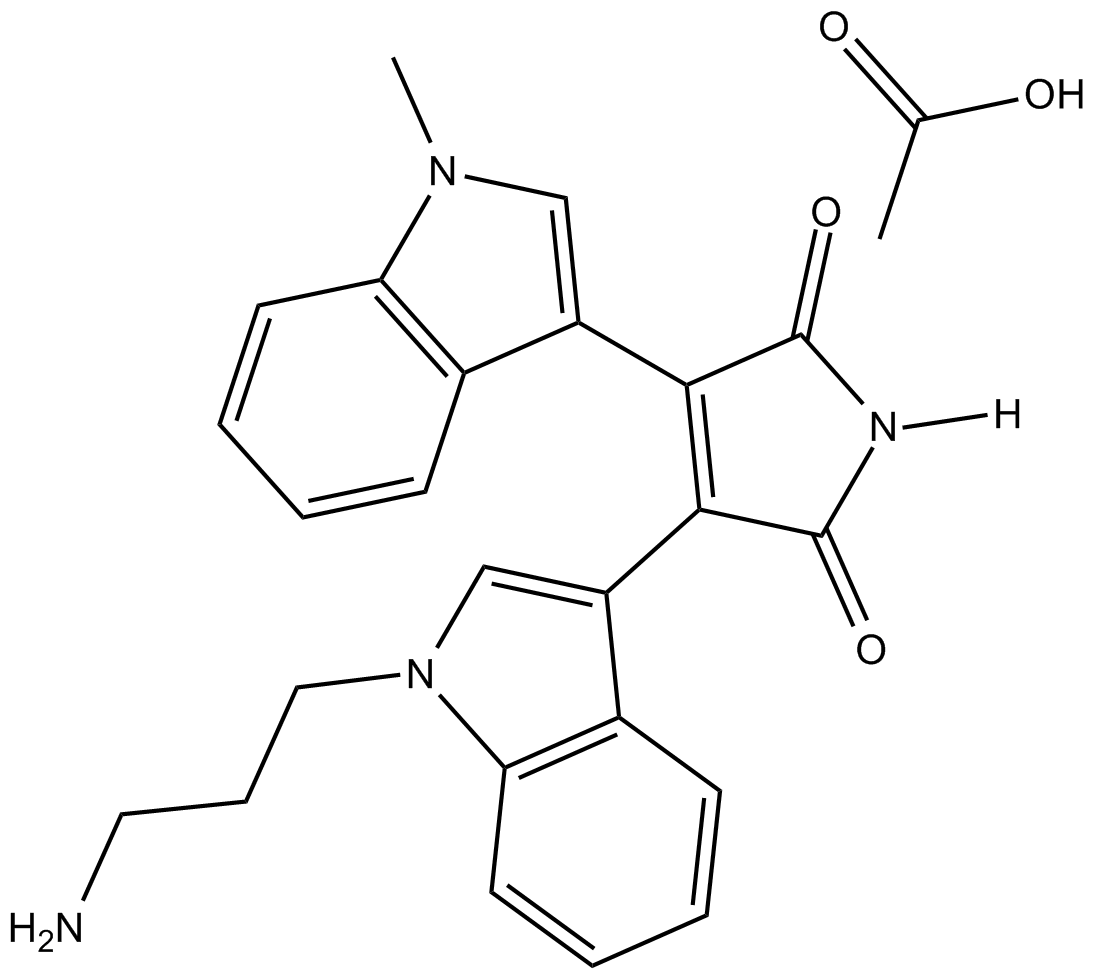
GC13226
Bisindolylmaleimide VIII (acetate)
A protein kinase C (PKC) inhibitor
-
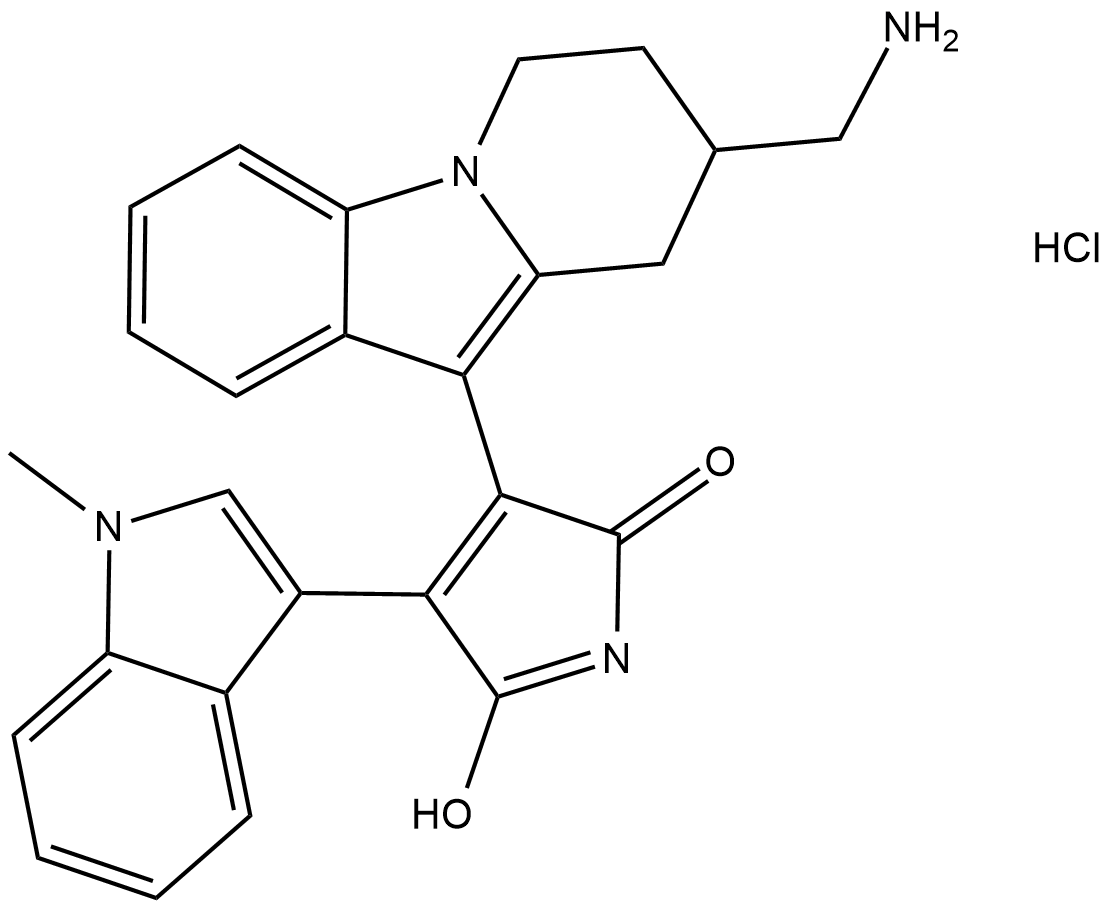
GC18354
Bisindolylmaleimide X (hydrochloride)
Bisindolylmaleimide X is a cell-permeable, reversible, ATP-competitive protein kinase C (PKC) inhibitor (IC50 = 15 nM, rat brain PKC).
-
GC35530
BJE6-106
BJE6-106 (B106) is a potent, selective 3rd generation PKCδ inhibitor with an IC50 of 0.05 μM and targets selectivity over classical PKC isozyme PKCα (IC50=50 μM).

-
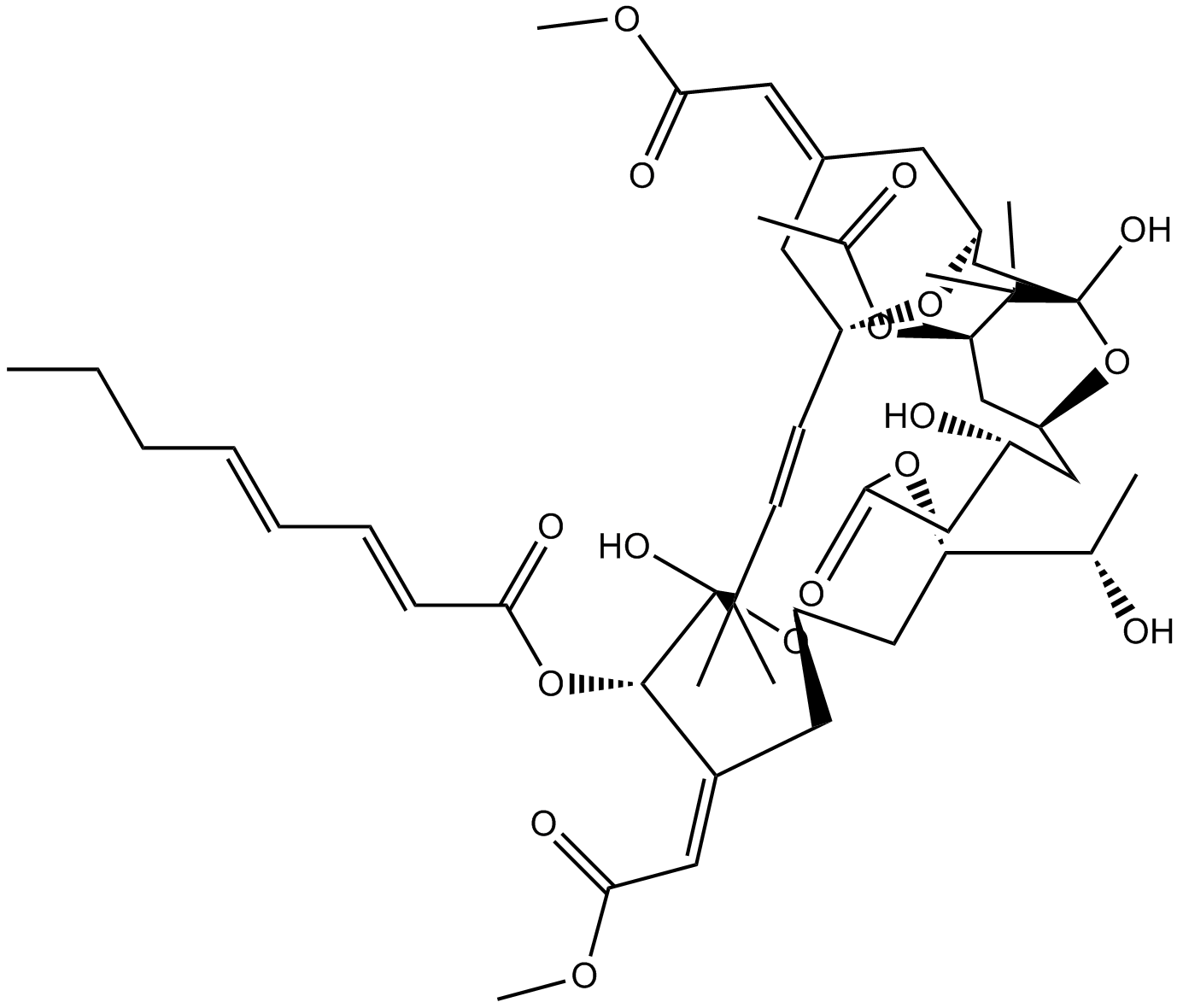
GC17670
Bryostatin 1
PKC activator
-
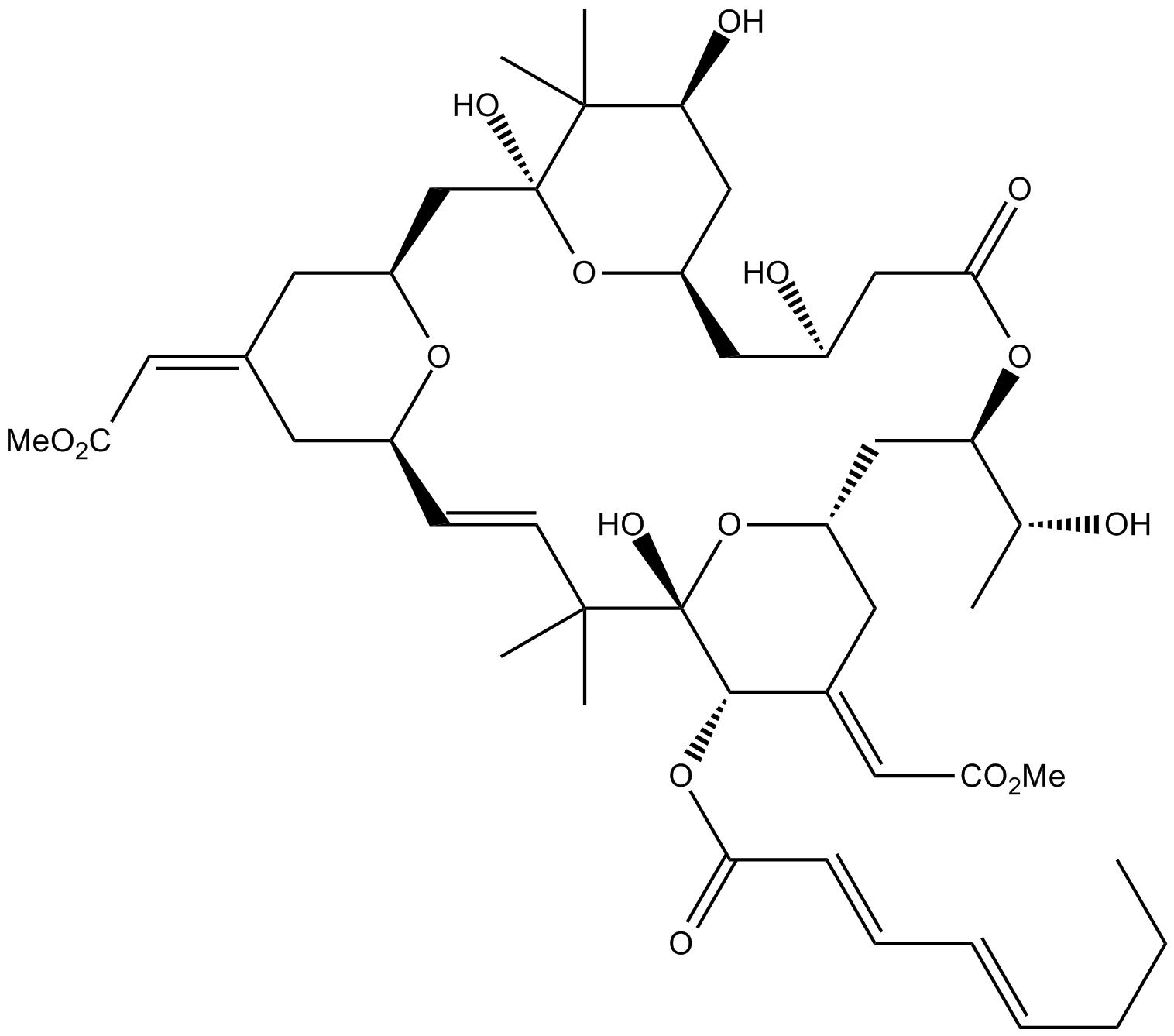
GC12842
Bryostatin 2
Protein kinase C (PKC) activator
-
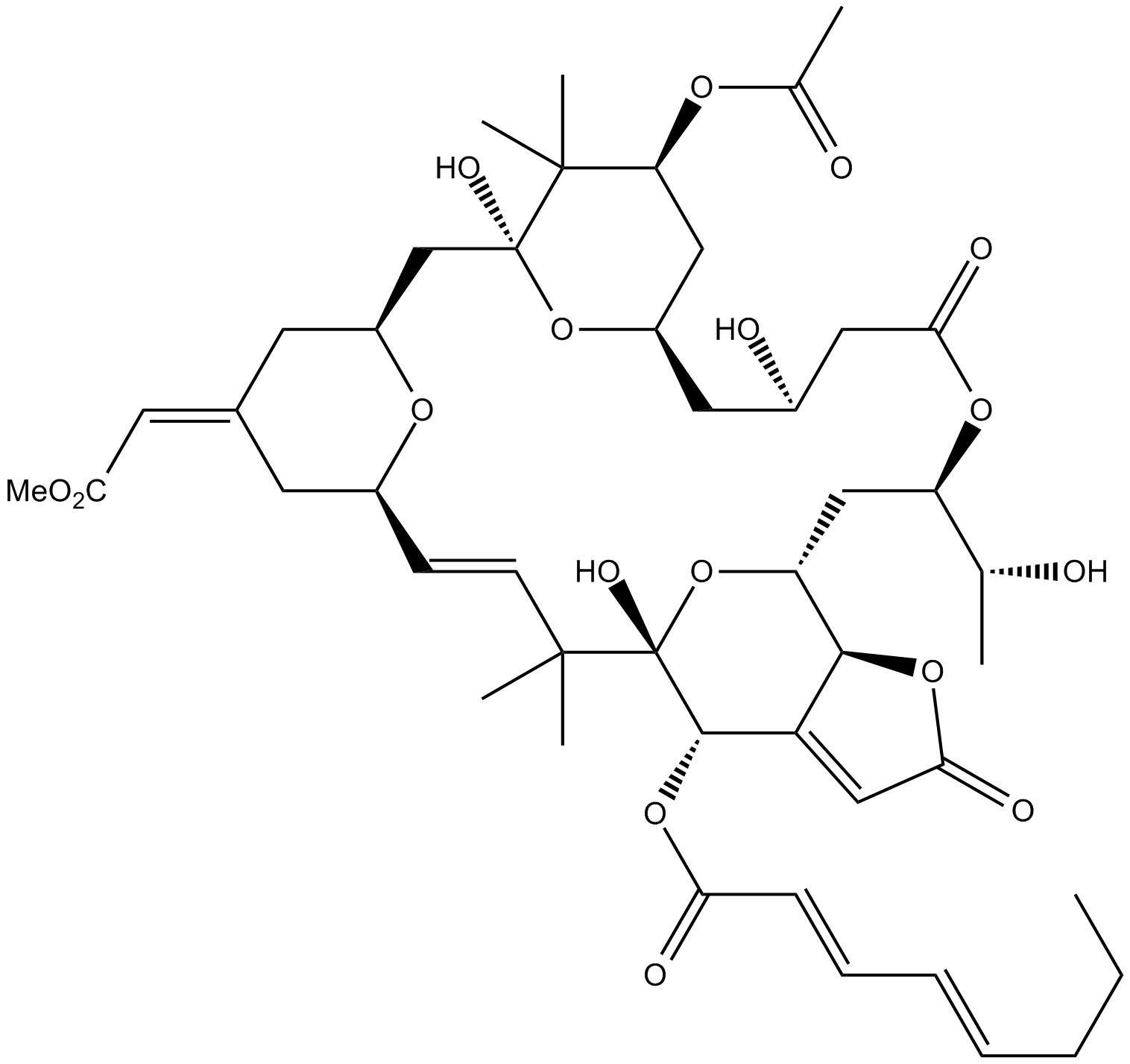
GC12695
Bryostatin 3
protein kinase C activator
-
GC11605
C-1
C-1 is a potent protein kinase inhibitor, with IC50s of 4 μM, 8 μM, 12 μM and 240 μM for cGMP-dependent protein kinase (PKG), cAMP-dependent protein kinase (PKA), protein kinase C (PKC) and MLC-kinase, respectively. C-1 also used as a ROCK inhibitor.

-
GC43105
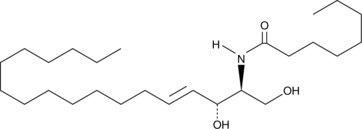
C8 Ceramide (d18:1.8:0)
C8 Ceramide (d18:1.8:0) (N-Octanoyl-D-erythro-sphingosine) is a cell-permeable analog of naturally occurring ceramides.
-
GC11159
Calphostin C
protein kinase C inhibitor
-
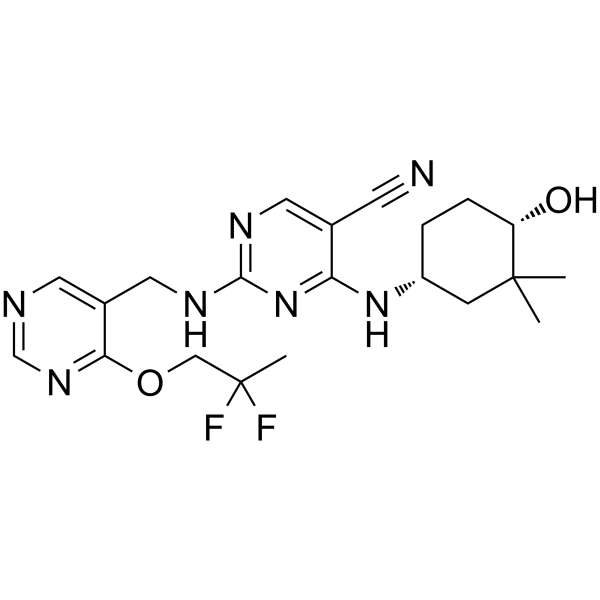
GC62612
CC-90005
CC-90005 is a potent, selective and orally active inhibitor of protein kinase C-θ (PKC-θ), with an IC50 of 8 nM.
-
GC18521
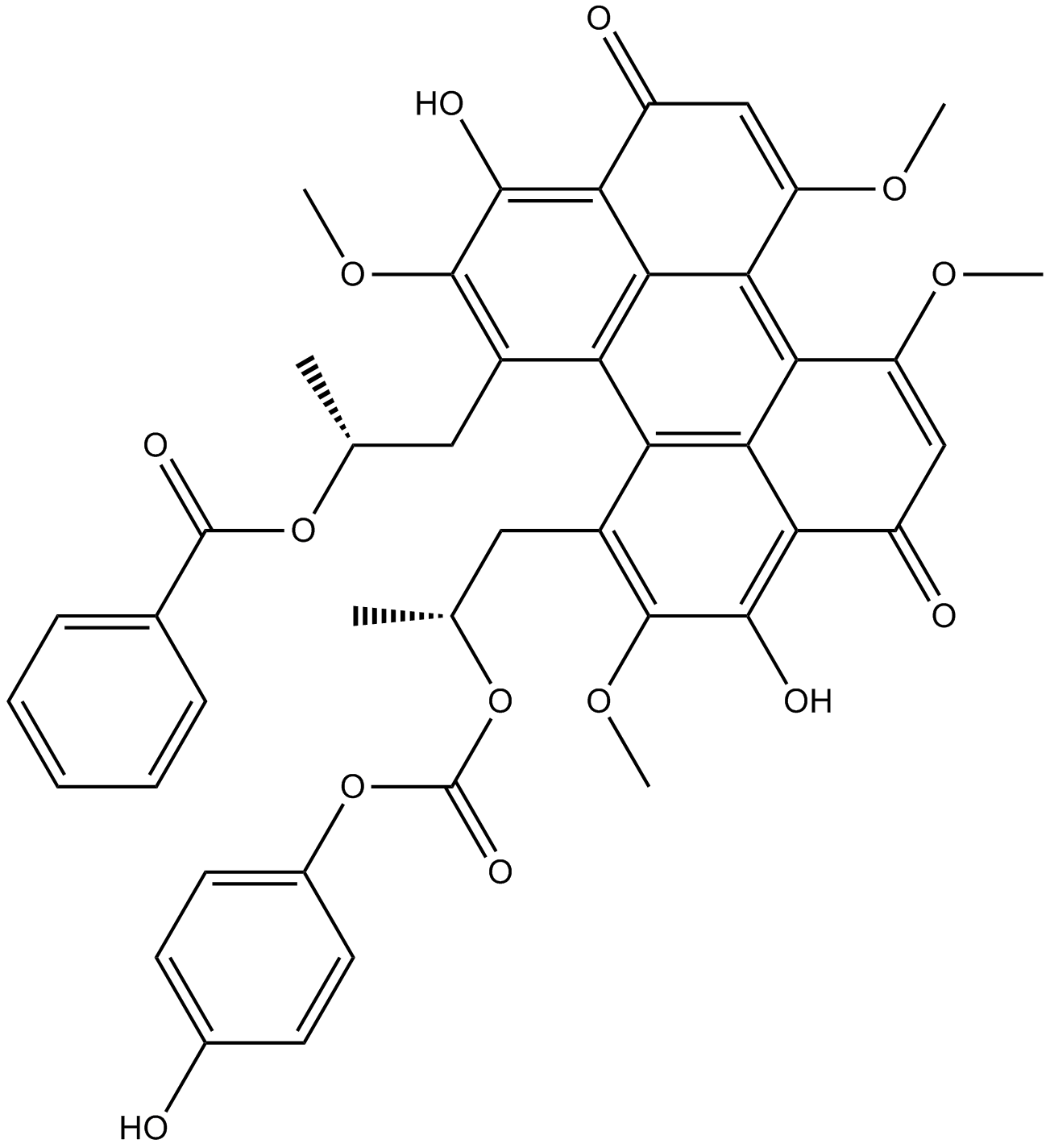
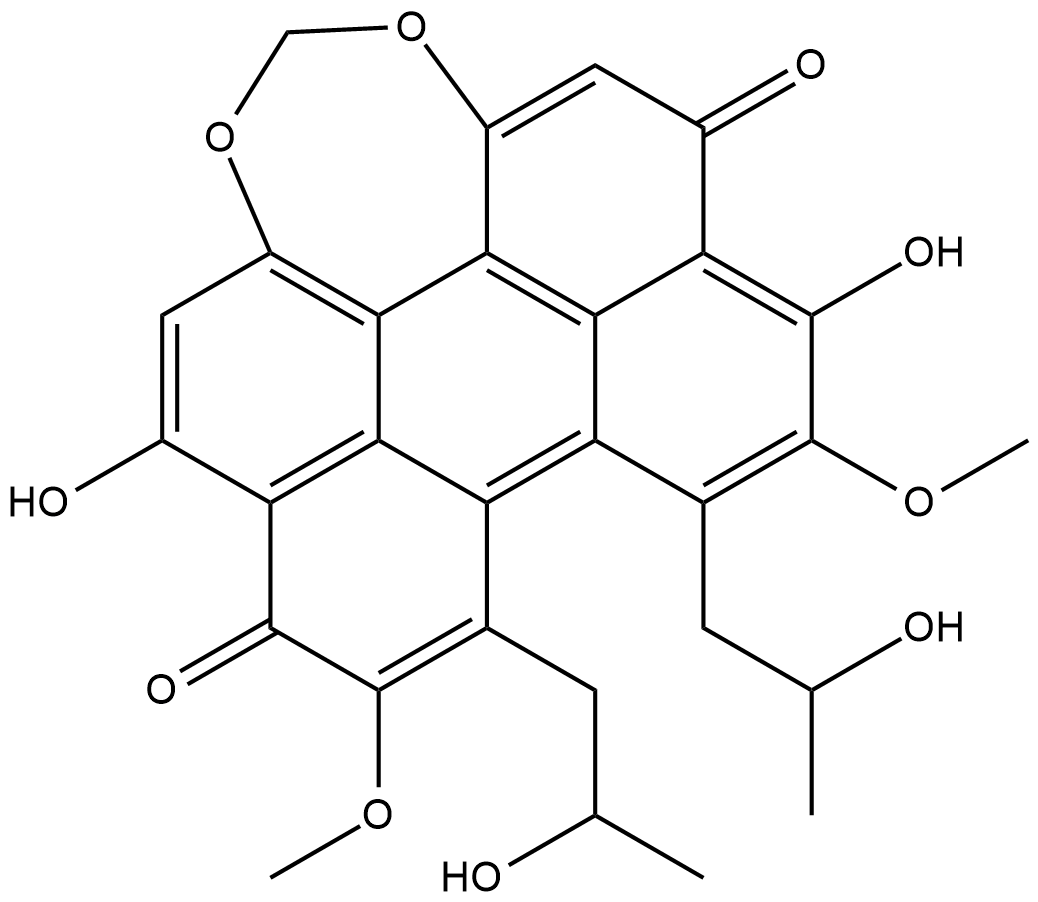
Cercosporin
Cercosporin is a pigmented phytotoxin isolated from the fungus C.
-
GC10687
CGP 53353
PKCβII inhibitor
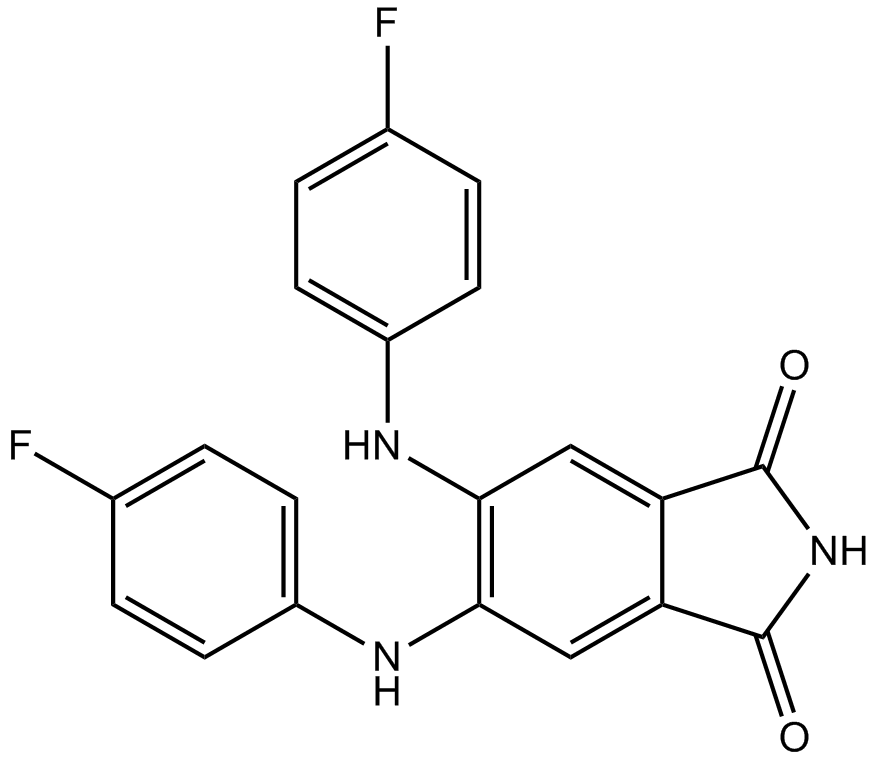
-
GC14650
CGP60474
A CDK inhibitor
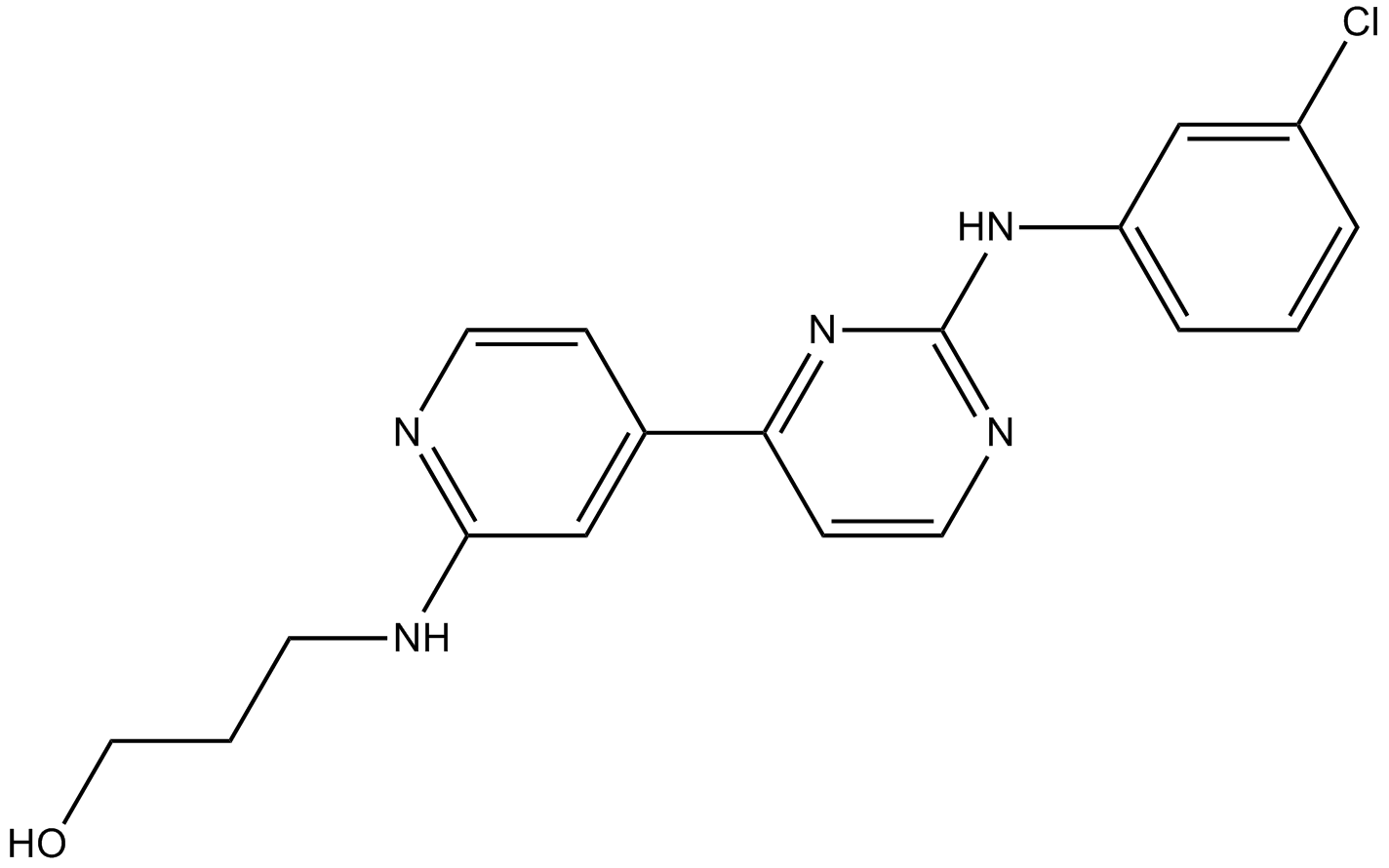
-
GN10463
Chelerythrine
-
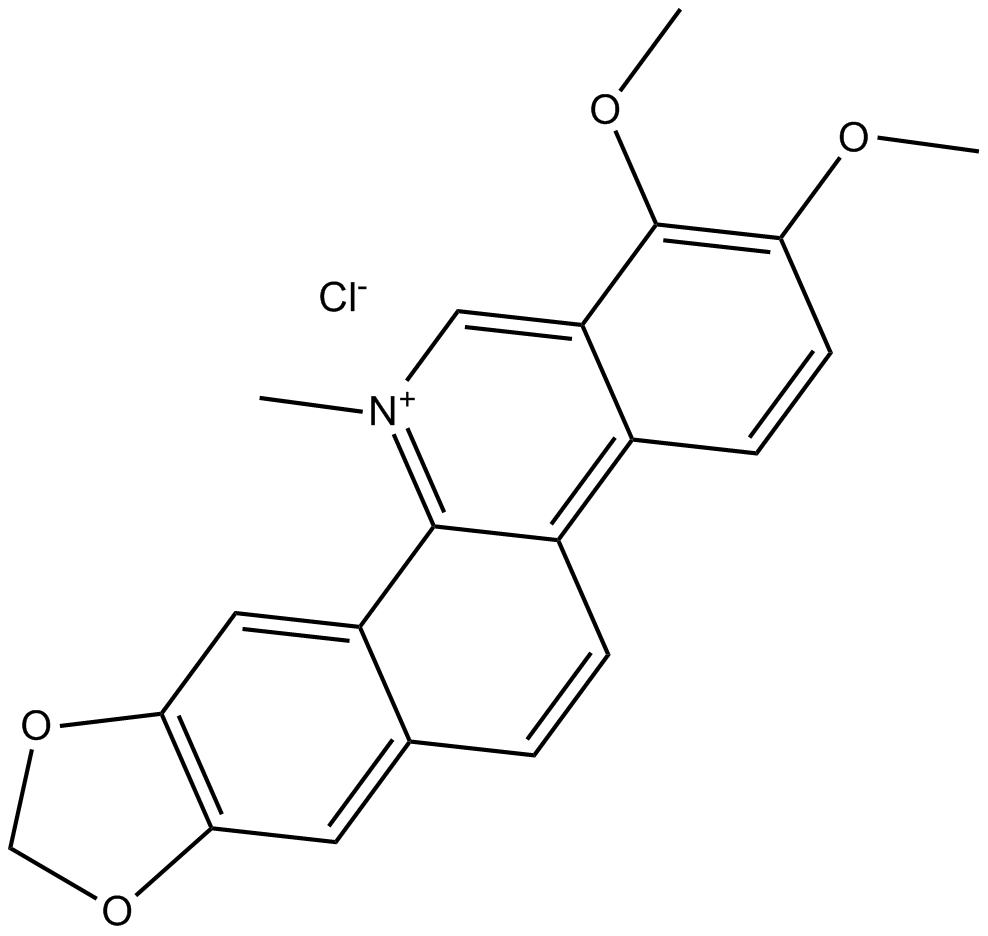
GC13065
Chelerythrine Chloride
Potent inhibitor of PKC and Bcl-xL
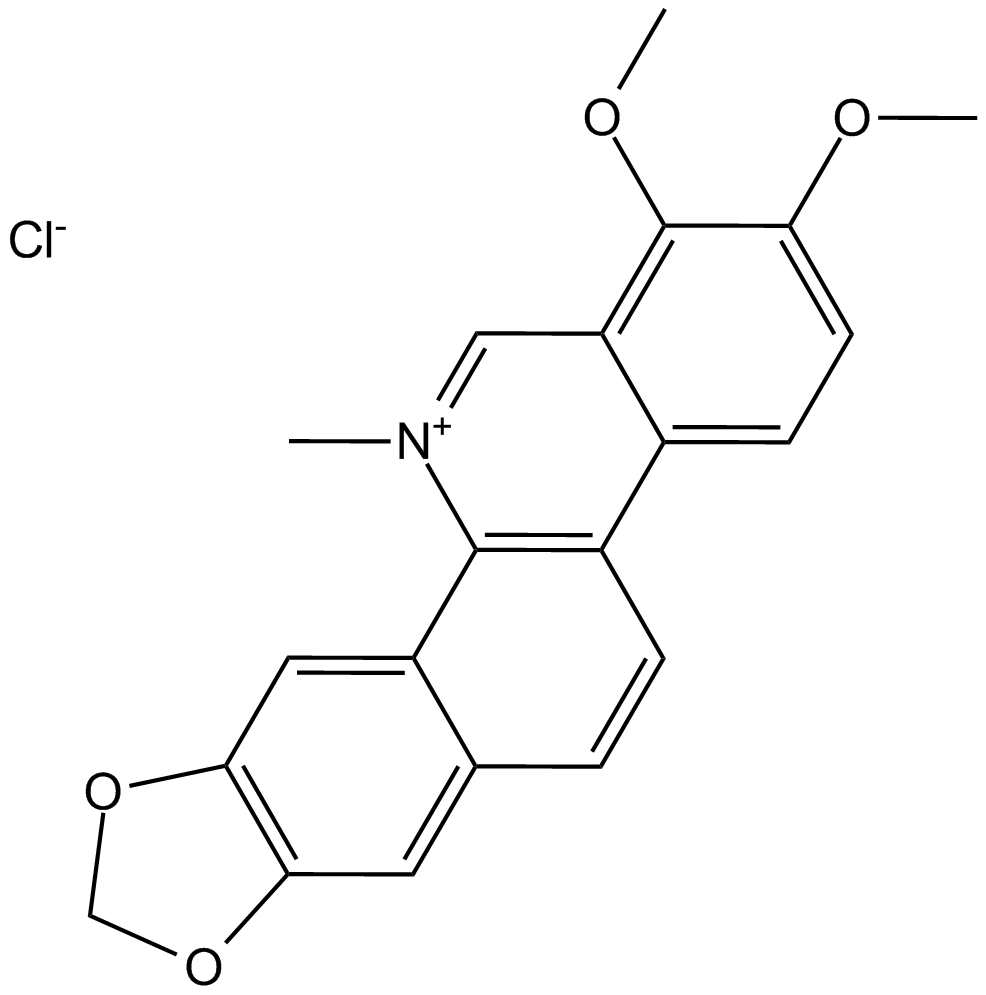
-
GC43286
CMPD101
A GRK2 and GRK3 inhibitor
-
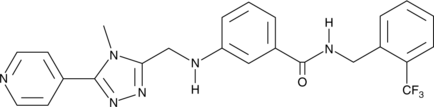
GC50704
CRT 0066854 hydrochloride
CRT 0066854 hydrochloride is a potent and selective atypical PKCs inhibitor.
-
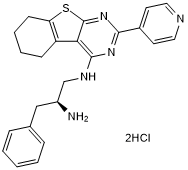
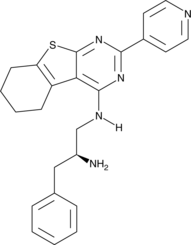
GC45414
CRT0066854
-
GC17591
D-erythro-Sphingosine (synthetic)
D-erythro-Sphingosine (synthetic) (Erythrosphingosine) is a very potent activator of p32-kinase with an EC50 of 8 μM, and inhibits protein kinase C (PKC). D-erythro-Sphingosine (synthetic) (Erythrosphingosine) is also a PP2A activator.

-
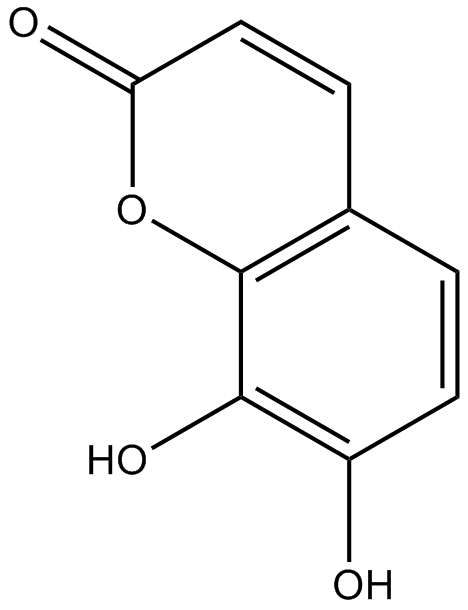
GN10336
Daphnetin
-
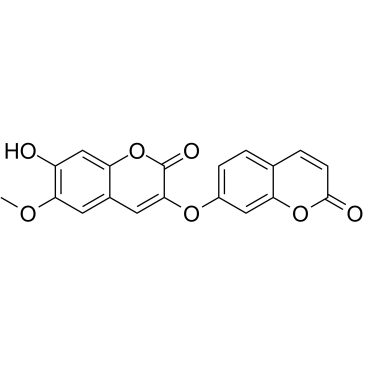
GC38186
Daphnoretin
A coumarin with diverse biological activities
-
GC38388
DCPLA-ME
DCPLA-ME, the methyl ester form of DCPLA, is a potent PKCε activator for use in the treatment of neurodegenerative diseases.

-
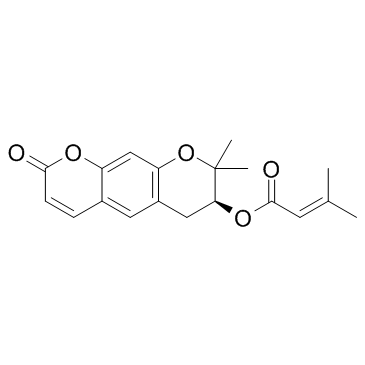
GC31892
Decursin ((+)-Decursin)
Decursin ((+)-Decursin) ((+)-Decursin ((+)-Decursin)) is a potent anti-tumor agent.
-
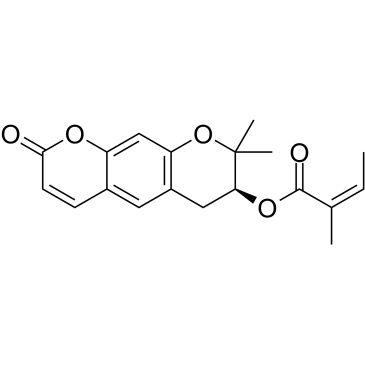
GC38085
Decursinol angelate
Decursinol angelate, a cytotoxic and protein kinase C (PKC) activating agent from the root of Angelica gigas, possesses anti-tumor and anti-inflammatory activities.
-
GC16354
Dequalinium Chloride
anti-tumor agent and PKC inhibitor

-
GC38482
Desmethylglycitein
Desmethylglycitein (4',6,7-Trihydroxyisoflavone), a metabolite of daidzein, sourced from Glycine max with antioxidant, and anti-cancer activities.Desmethylglycitein binds directly to CDK1 and CDK2 in vivo, resulting in the suppresses CDK1 and CDK2 activity. Desmethylglycitein is a direct inhibitor of protein kinase C (PKC)α, against solar UV (sUV)-induced matrix matrix metalloproteinase 1 (MMP1). Desmethylglycitein binds to PI3K in an ATP competitive manner in the cytosol, where it inhibits the activity of PI3K and downstream signaling cascades, leading to the suppression of adipogenesis in 3T3-L1 preadipocytes.

-
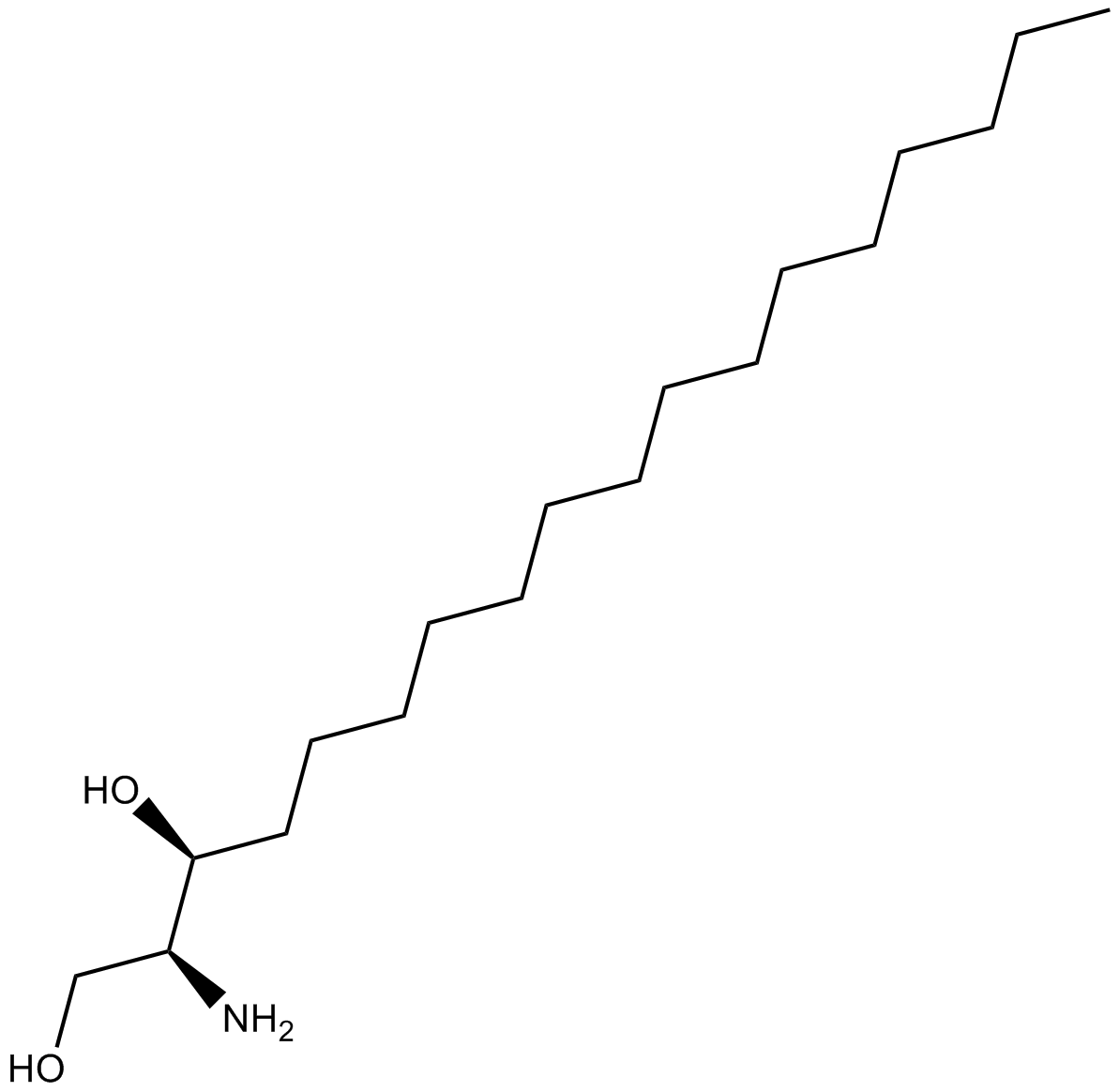
GC17767
Dihydrosphingosine
Dihydrosphingosine is a potent inhibitor of PKC and phospholipase A2 (PLA2).
-
GC11499
Enzastaurin (LY317615)
Enzastaurin (LY317615) (LY317615) is a potent and selective PKCβ inhibitor with an IC50 of 6 nM, showing 6- to 20-fold selectivity over PKCα, PKCγ and PKCε.
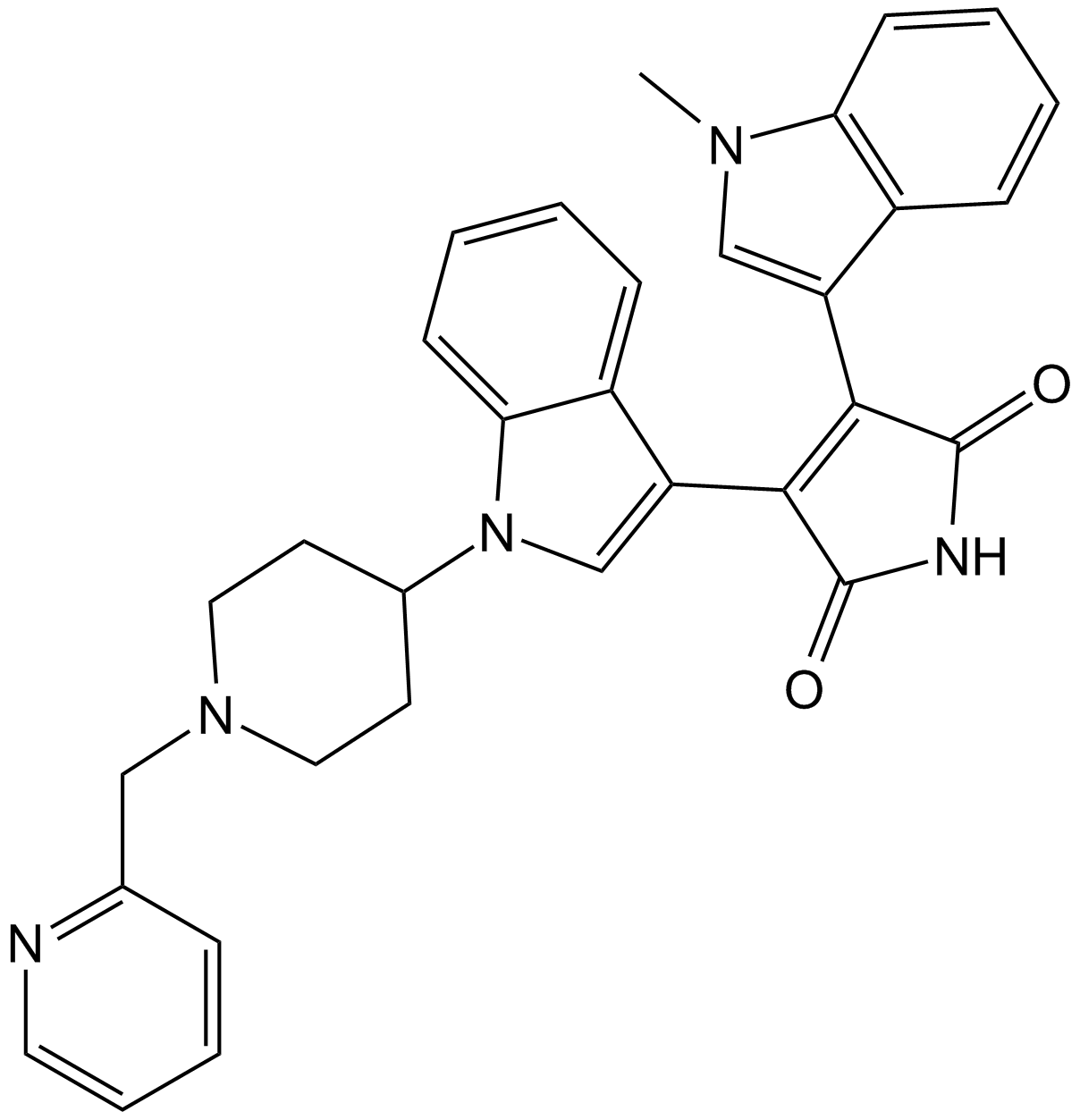
-
GC13869
Fasudil
Calcium antagonist

-
GC14289
Fasudil (HA-1077) HCl
Fasudil (HA-1077; AT877) Hydrochloride is a nonspecific RhoA/ROCK inhibitor and also has inhibitory effect on protein kinases, with an Ki of 0.33 μM for ROCK1, IC50s of 0.158 μM and 4.58 μM, 12.30 μM, 1.650 μM for ROCK2 and PKA, PKC, PKG, respectively. Fasudil (HA-1077) HCl is also a potent Ca2+ channel antagonist and vasodilator.
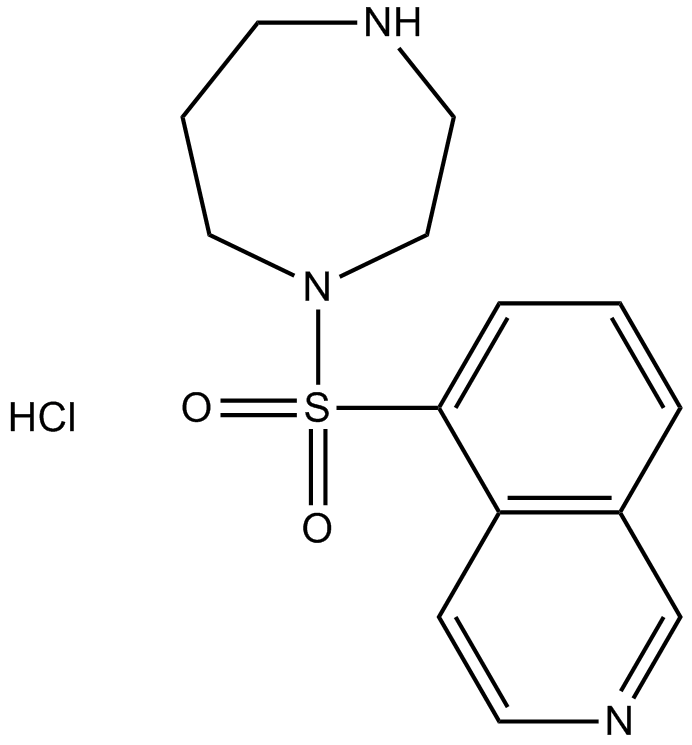
-
GC12027
FR 236924
FR 236924 (FR236924), a linoleic acid derivative, selectively and directly activates PKCε.
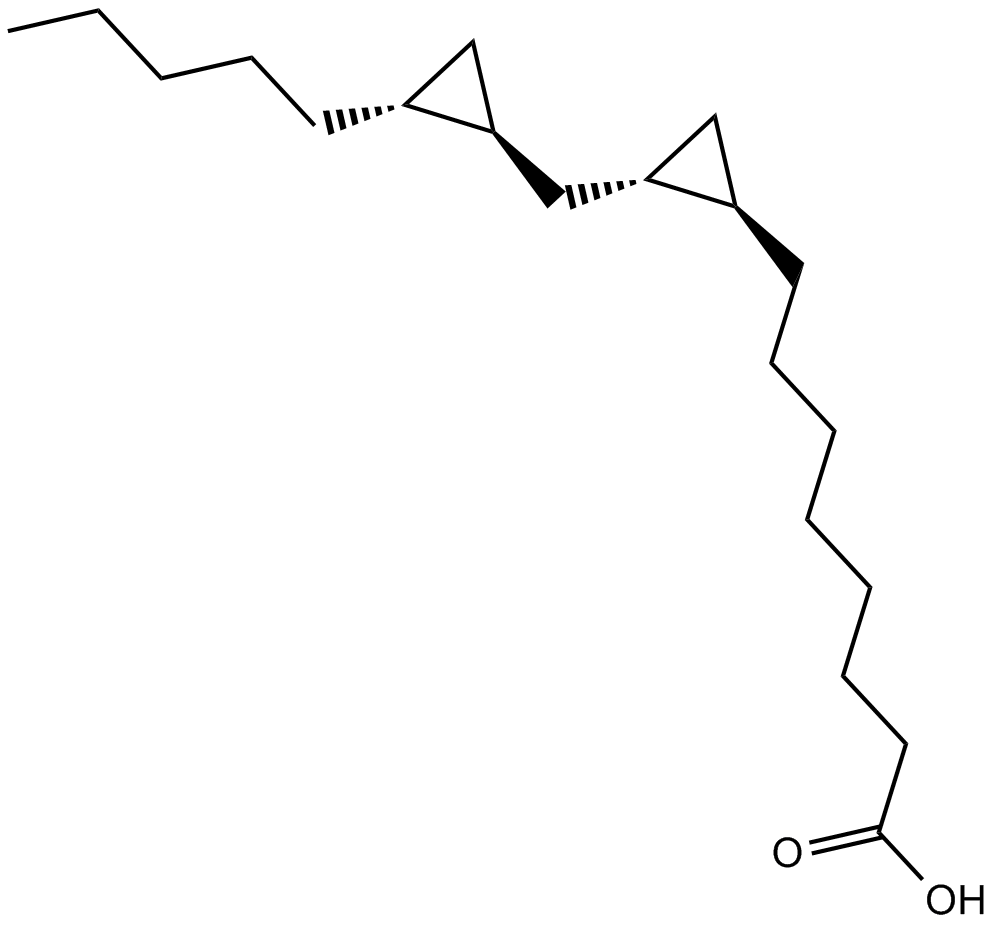
-
GC15431
GF 109203X (Bisindolylmaleimide I)
GF 109203X (GF109203X) is a highly selective, cell-permeable, and reversible protein kinase C (PKC) inhibitor with a Ki of 14 nM.

-
GC15564
Go 6976
PKCα/PKCβ1 inhibitor
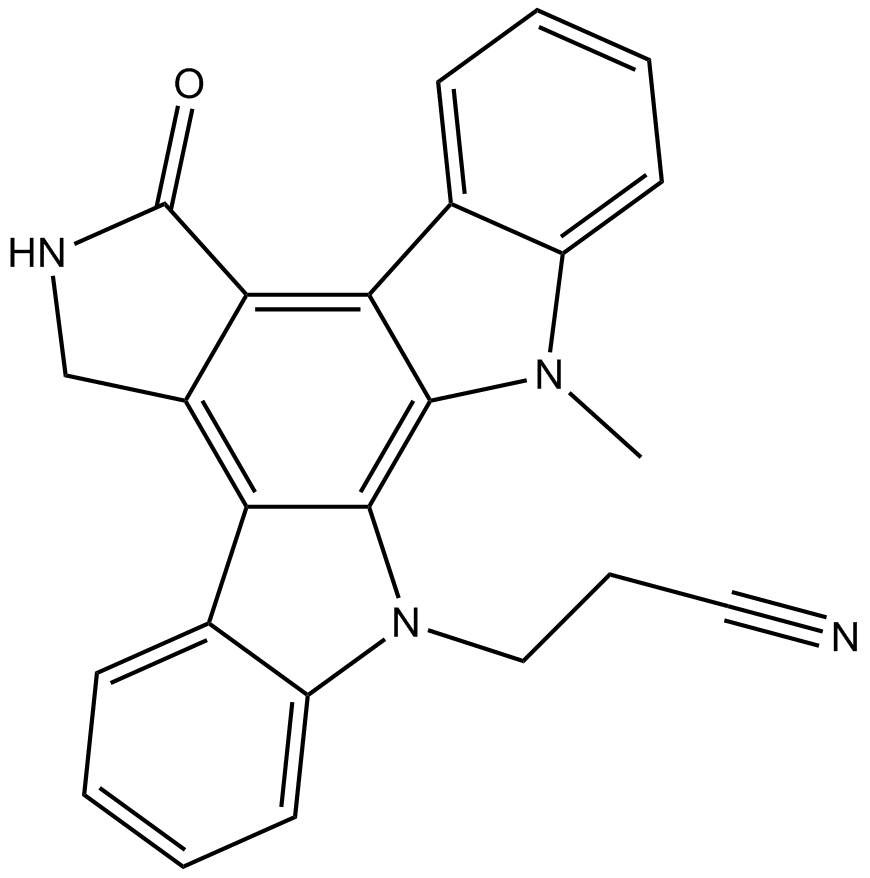
-
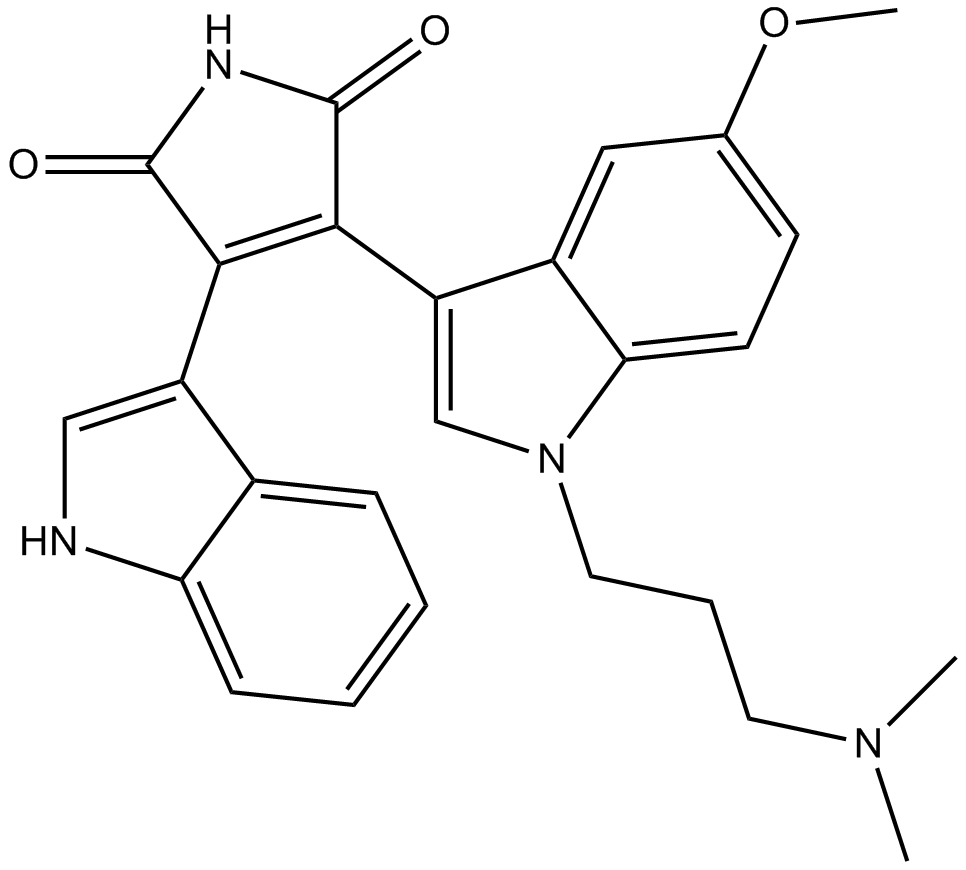
GC16907
Go 6983
Go 6983 (GÖ 6983) is one of the bisindolylmaleimide group of PKC inhibitor compounds, Go 6983 (GÖ 6983) was able to differentiate between PKC mu and other PKC isoenzymes.
-
GC10299
Hexadecyl Methyl Glycerol
protein kinase C activity inhibitor
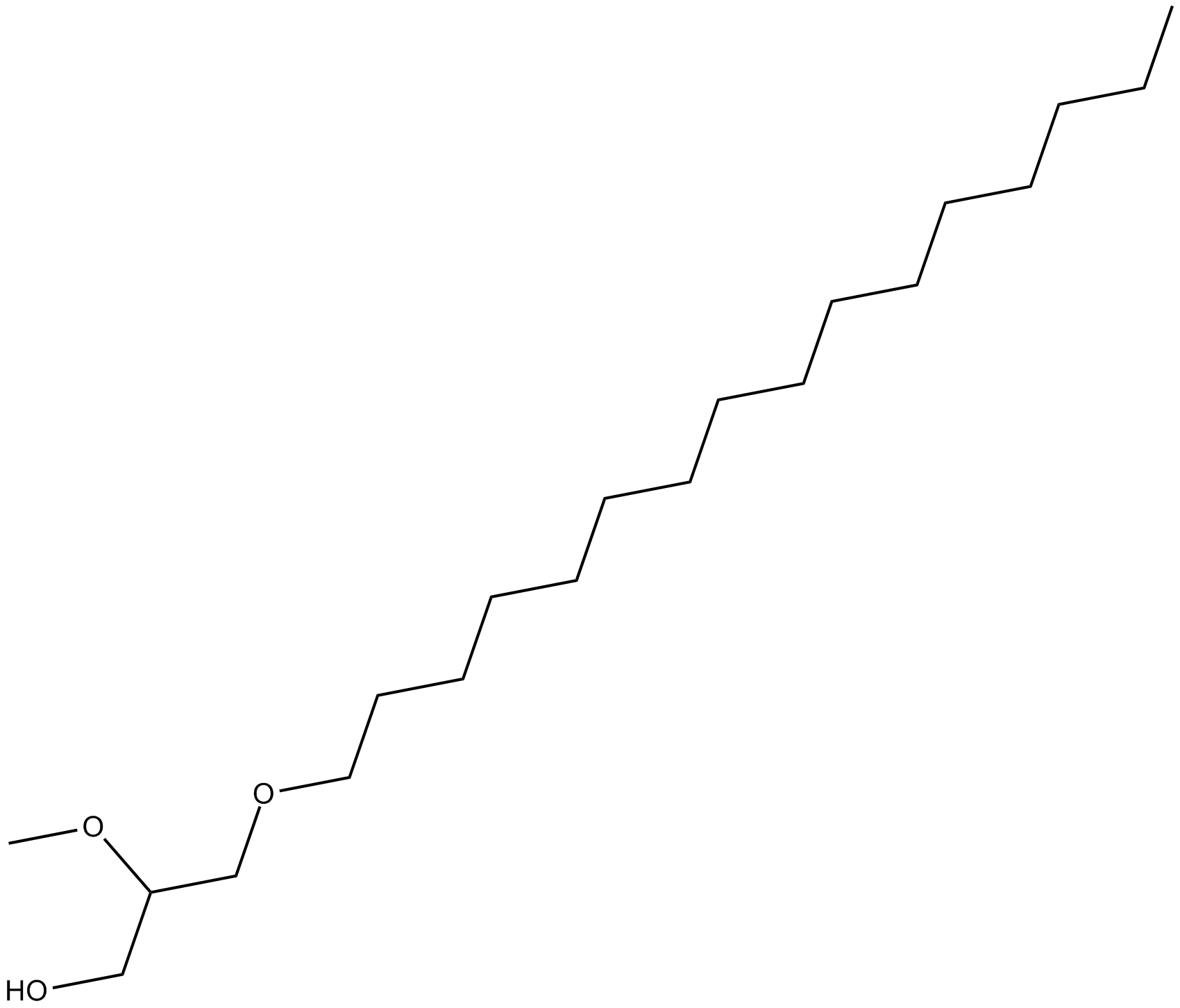
-
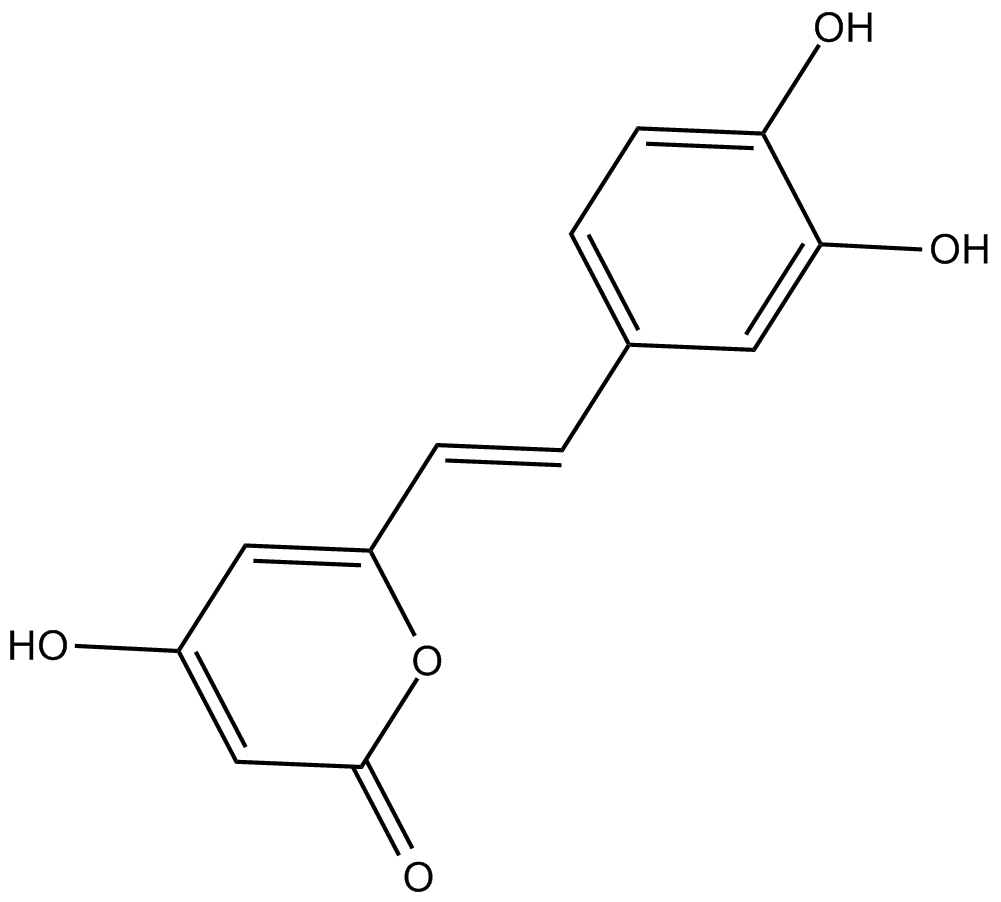
GC15018
Hispidin
protein kinase Cβ inhibitor
-
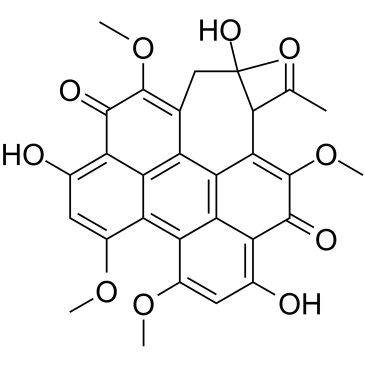
GC36282
Hypocrellin A
Hypocrellin A, a naturally occurring PKC inhibitor, has many biological and pharmacological properties, such as antitumour, antiviral, antibacterial, and antileishmanial activities.
-
GC15420
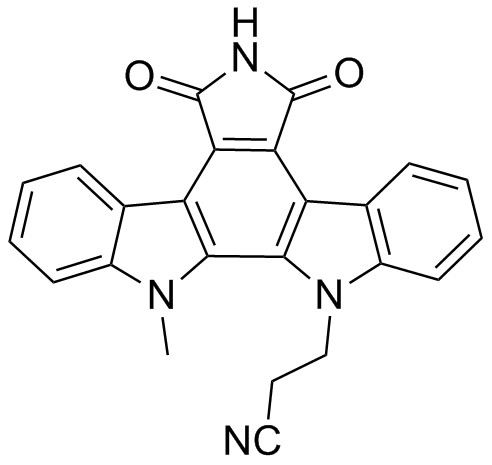
ICP 103
Protein kinase inhibitor
-
GC32870
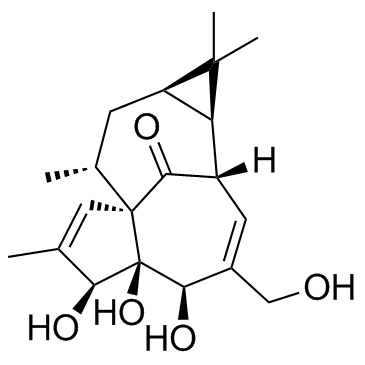
Ingenol ((-)-Ingenol)
Ingenol ((-)-Ingenol) is a PKC activator, with a Ki of 30 μM, with antitumor activity.
-
GC61776
Ingenol 3,20-dibenzoate
Ingenol 3,20-dibenzoate is a potent protein kinase C (PKC) isoform-selective agonist.

-
GC31656
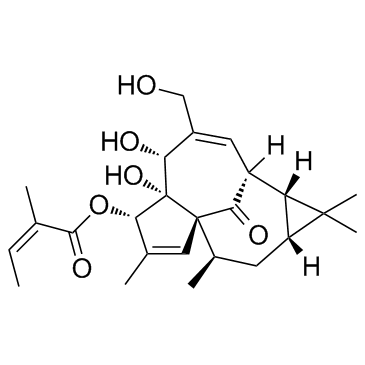
Ingenol Mebutate (Ingenol 3-angelate)
Ingenol Mebutate (Ingenol 3-angelate) is an active ingredient in Euphorbia peplus, acts as a potent PKC modulator, with Kis of 0.3, 0.105, 0.162, 0.376, and 0.171 nM for PKC-α, PKC-β, PKC-γ, PKC-δ, and PKC-ε, respectively, and has antiinflammatory and antitumor activity.
-
GC15148
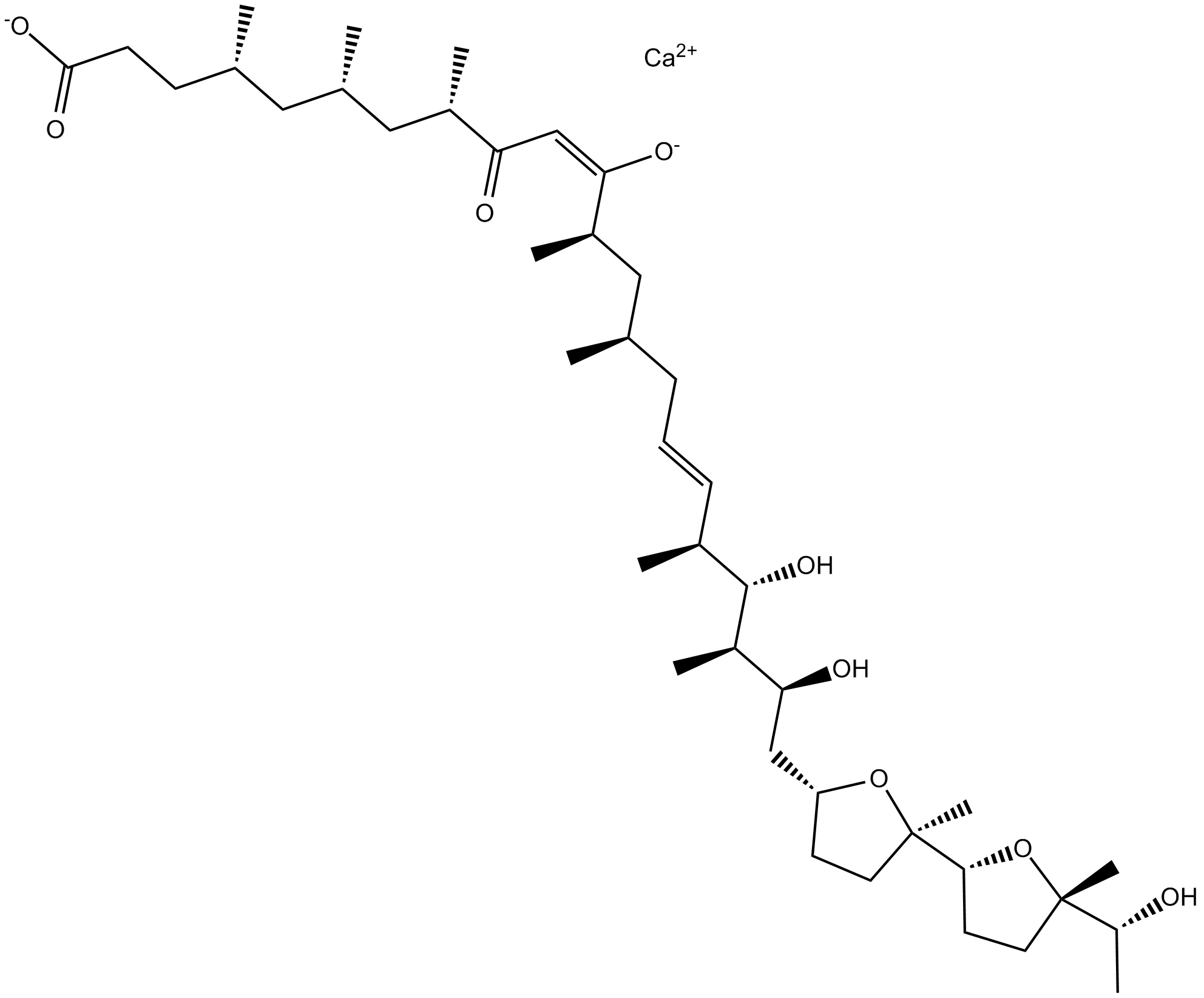
Ionomycin calcium salt
Lonomycin is a selective calcium ionophore derived from S. conglobatus that mobilizes intracellular calcium stores.
-
GC15446
Ionomycin free acid
Ionomycin free acid (SQ23377) is a potent, selective calcium ionophore and an antibiotic produced by Streptomyces conglobatus.
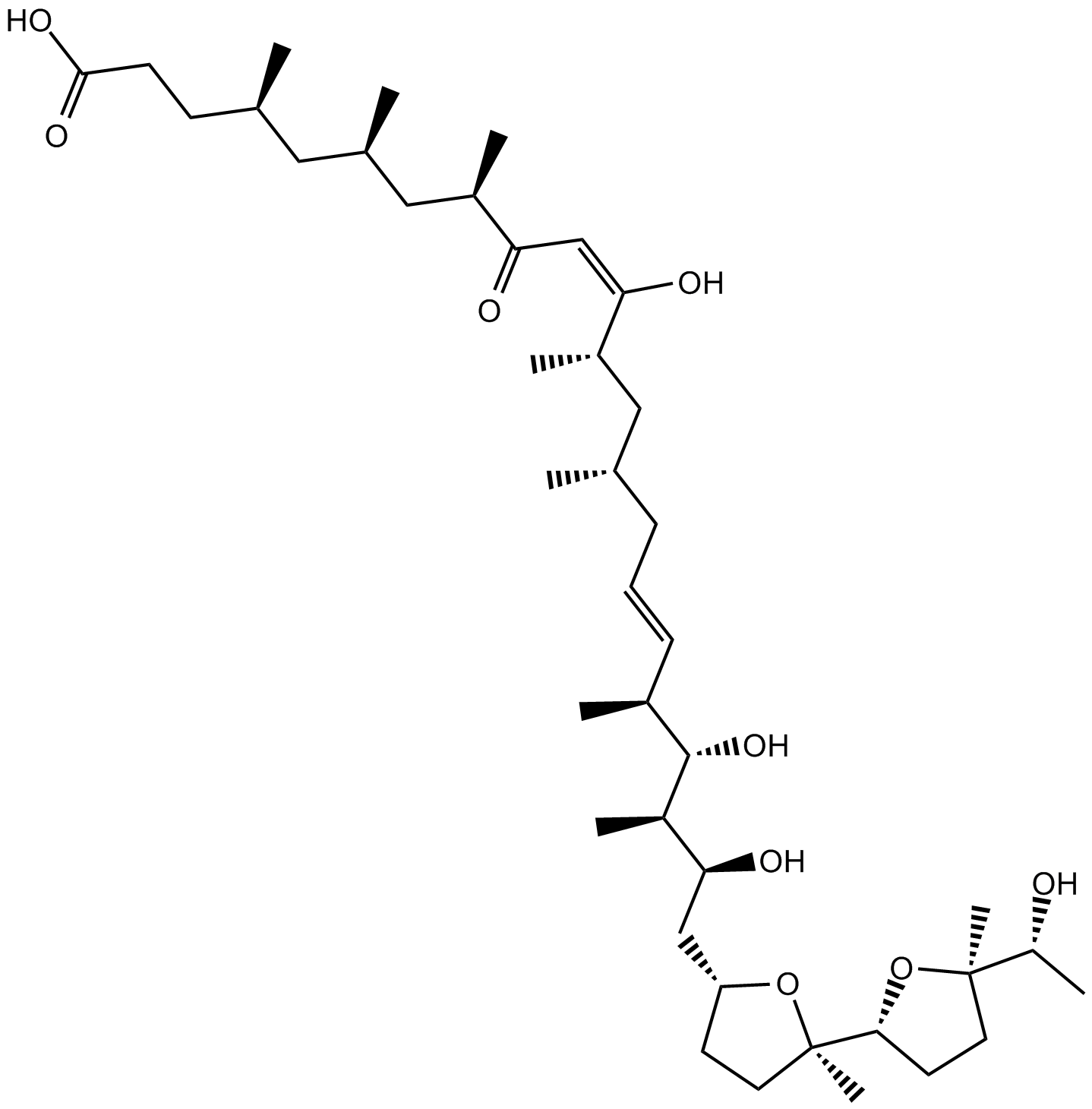
-
GC11362
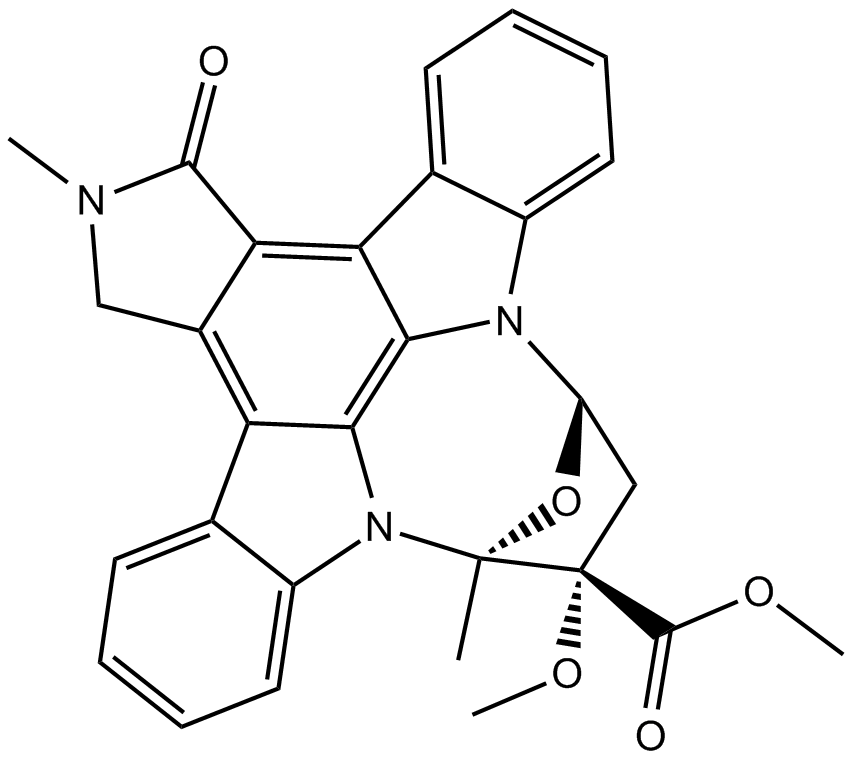
K 252a
A protein kinase inhibitor
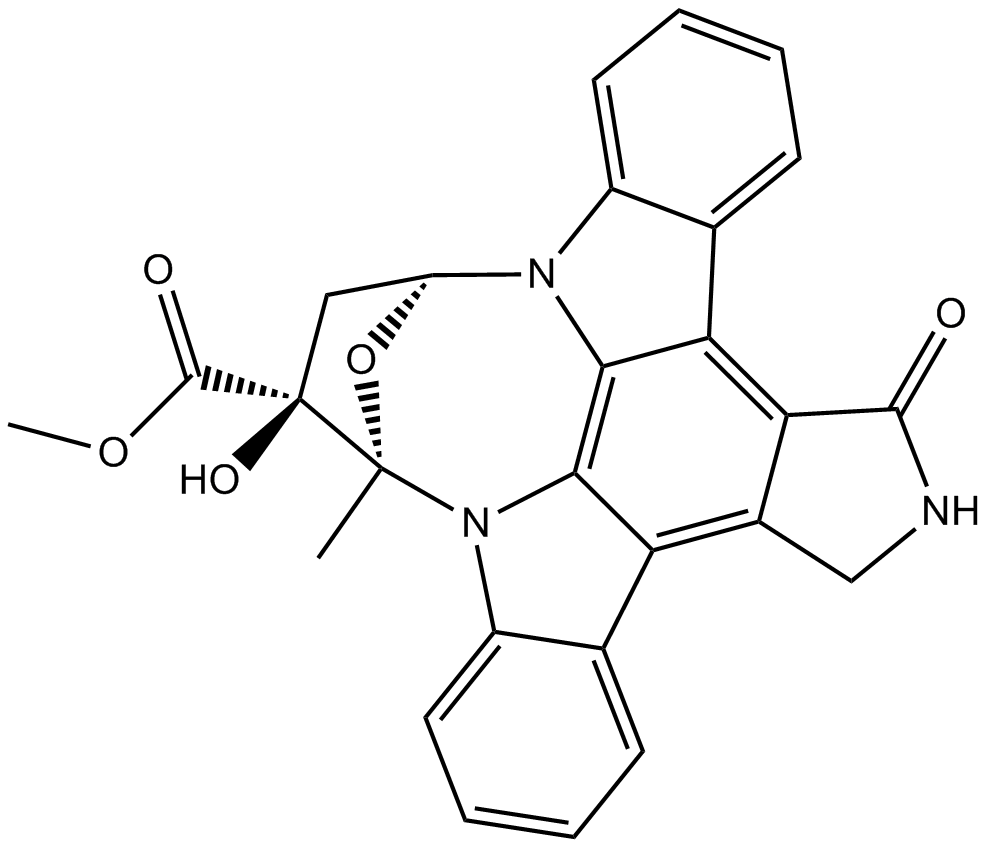
-
GC15281
K-252c
Protein kinase inhibitor
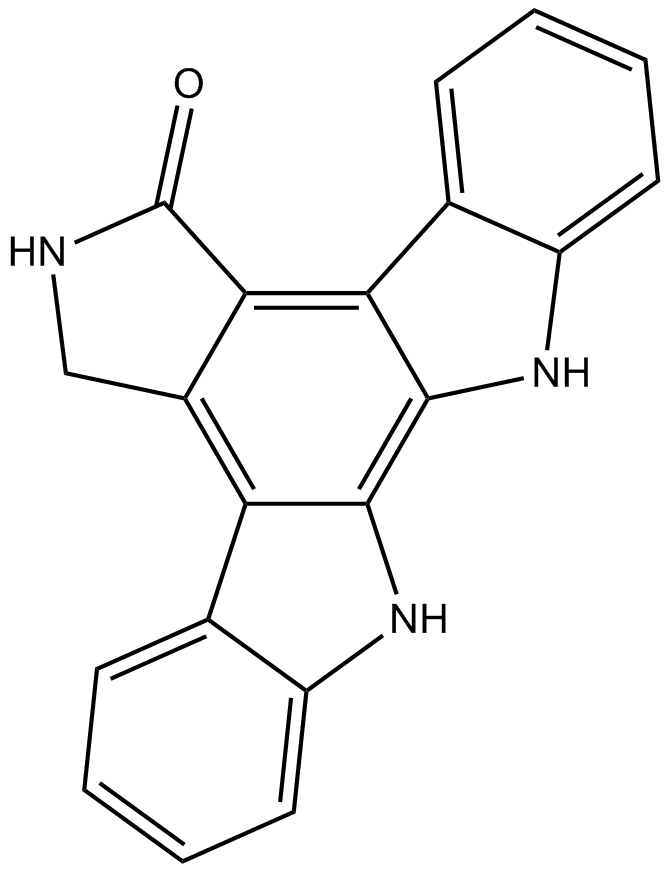
-
GC43993
K252b
K252b is an indolocarbazole isolated from the actinomycete Nocardiopsis, first described as an inhibitor of protein kinase C.
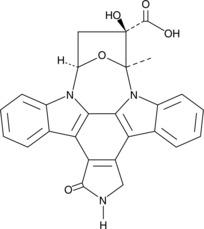
-
GC64263
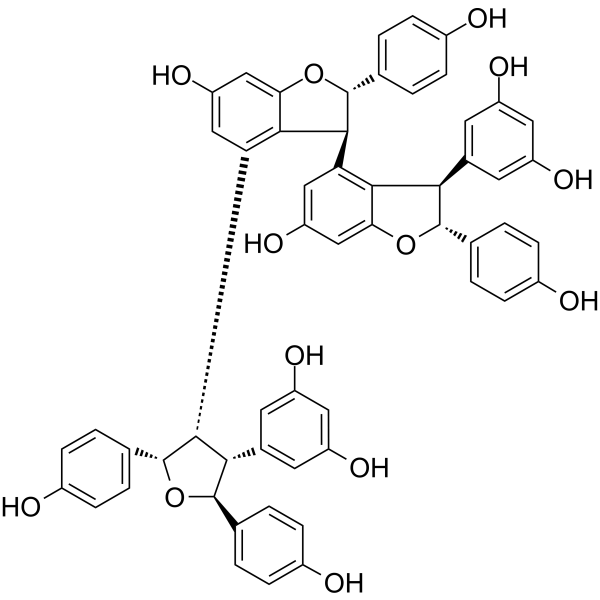
Kobophenol A
Kobophenol A, an oligomeric stilbene, blocks the interaction between the ACE2 receptor and S1-RBD with an IC50 of 1.81 μM and inhibits SARS-CoV-2 viral infection in cells with an EC50 of 71.6 μM.
-
GC17346
KT 5823
A protein kinase G inhibitor
-
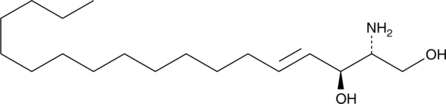
GC40770
L-erythro Sphingosine (d18:1)
L-erythro Sphingosine is a synthetic stereoisomer of sphingosine (d18:1).
-
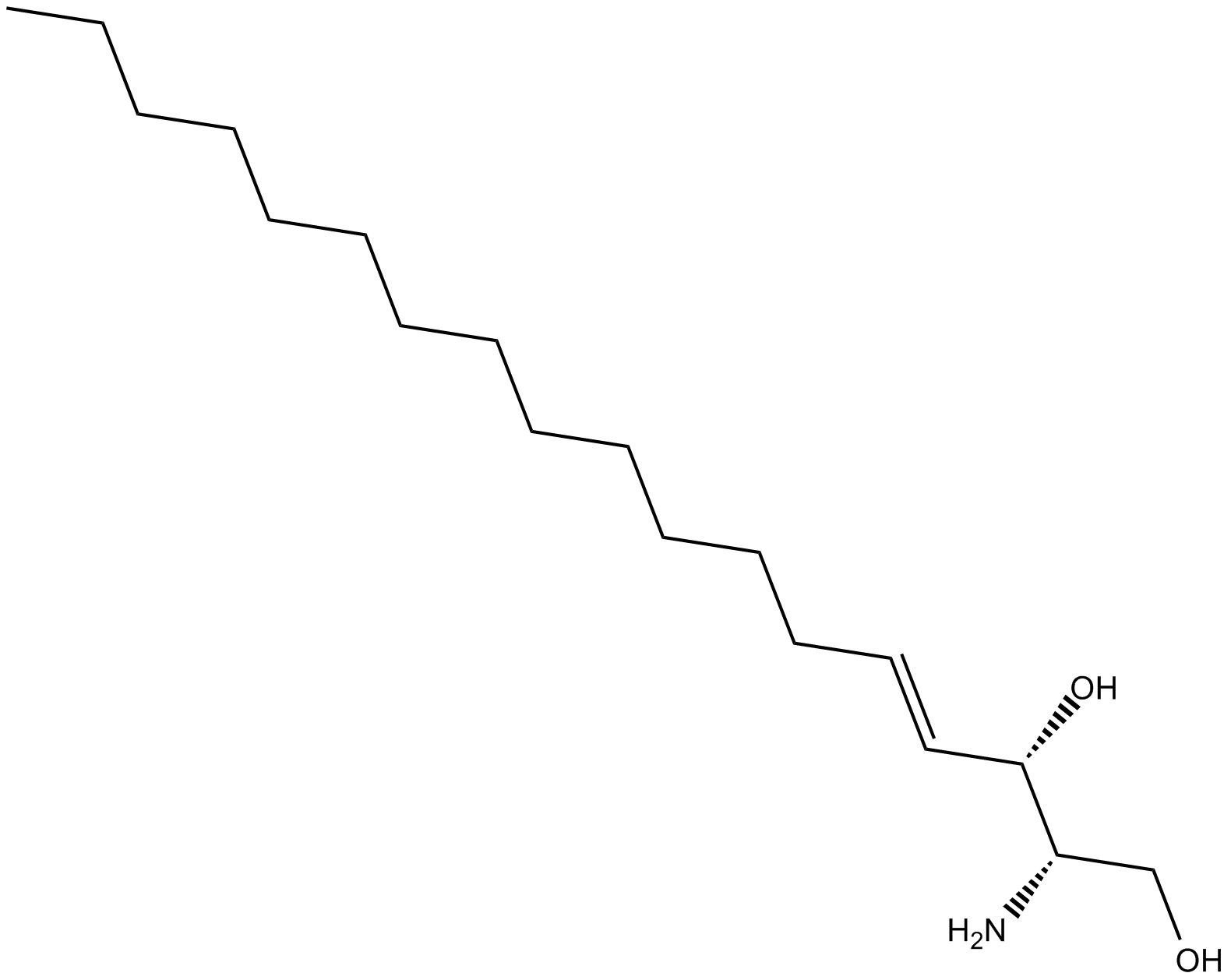
GC17815
L-threo-Sphingosine C-18
L-threo-Sphingosine C-18 is a potent MAPK inhibitor. L-threo-Sphingosine C-18 induces apoptosis and clear DNA fragmentation. L-threo-Sphingosine C-18 shows anticancer effect.
-
GC32811
LXS196
LXS196 (LXS196) is a potent, selective and orally active protein kinase C (PKC) inhibitor, with IC50 values of 1.9 nM, 0.4 nM and 3.1 μM for PKCα, PKCθ and GSK3β, respectively. LXS196 has the potential for uveal melanoma research.
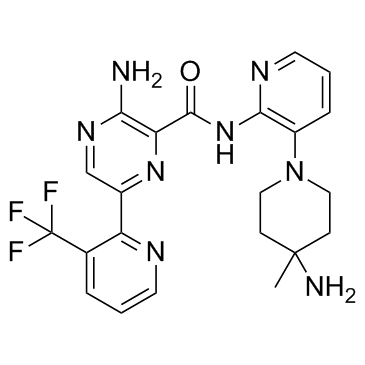
-
GC17563
LY 333531 hydrochloride
Ruboxistaurin (LY333531) hydrochloride is an orally active, selective PKC beta inhibitor (Ki=2 nM).
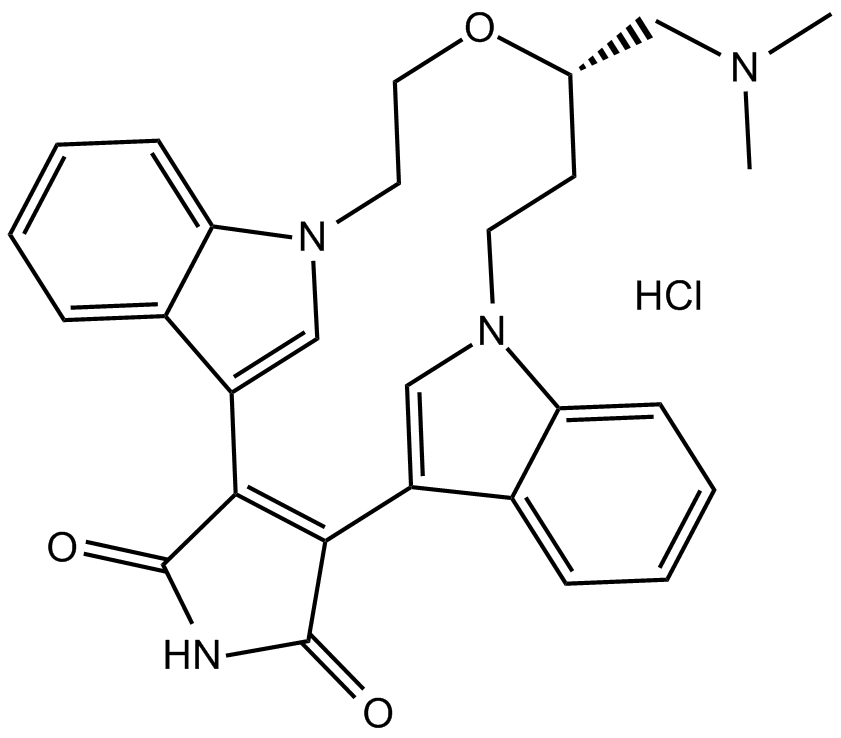
-
GC30545
Malantide
Malantide is a synthetic dodecapeptide derived from the site phosphorylated by cAMP-dependent protein kinase (PKA) on the β-subunit of phosphorylase kinase. Malantide is a highly specific substrate for PKA with a Km of 15 μM and shows protein inhibitor (PKI) inhibition >90% substrate phosphorylation in various rat tissue extracts. Malantide is also an efficient substrate for PKC with a Km of 16 μM.
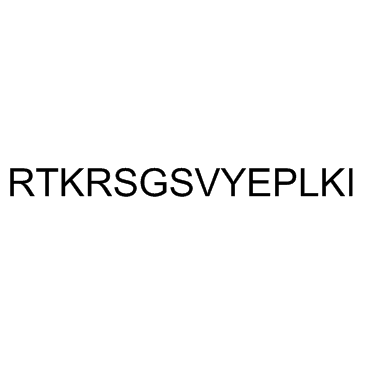
-
GC10496
Midostaurin (PKC412)
Midostaurin (PKC412) (PKC412; CGP 41251) is an orally active, reversible multi-targeted protein kinase inhibitor. Midostaurin (PKC412) inhibits PKCα/β/γ, Syk, Flk-1, Akt, PKA, c-Kit, c-Fgr, c-Src, FLT3, PDFRβ and VEGFR1/2 with IC50s ranging from 22-500 nM. Midostaurin (PKC412) also upregulates endothelial nitric oxide synthase (eNOS) gene expression. Midostaurin (PKC412) shows powerful anticancer effects.
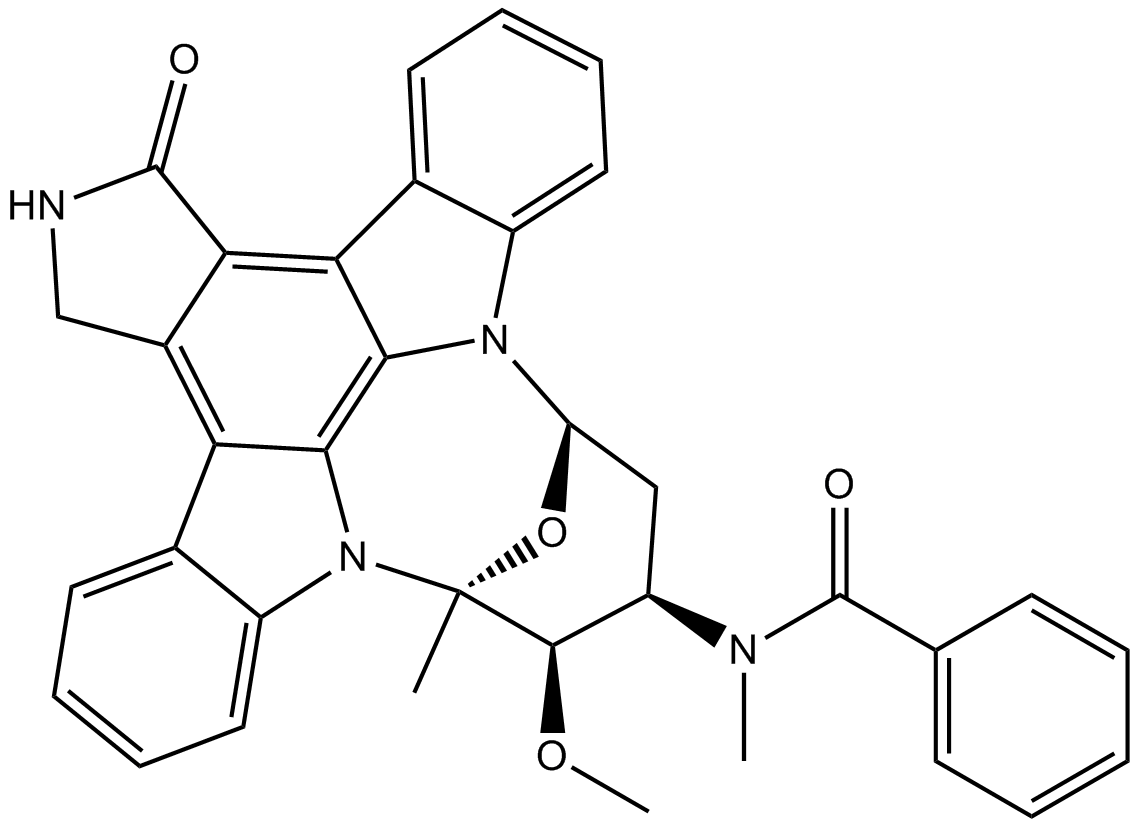
-
GC32714
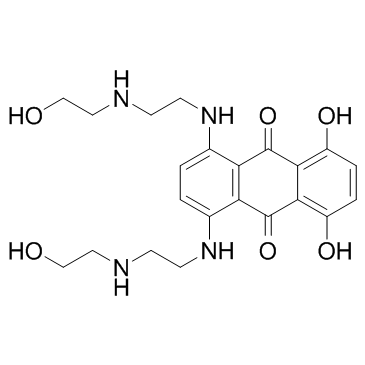
Mitoxantrone (mitozantrone)
Mitoxantrone (mitozantrone) is a potent topoisomerase II inhibitor.
-
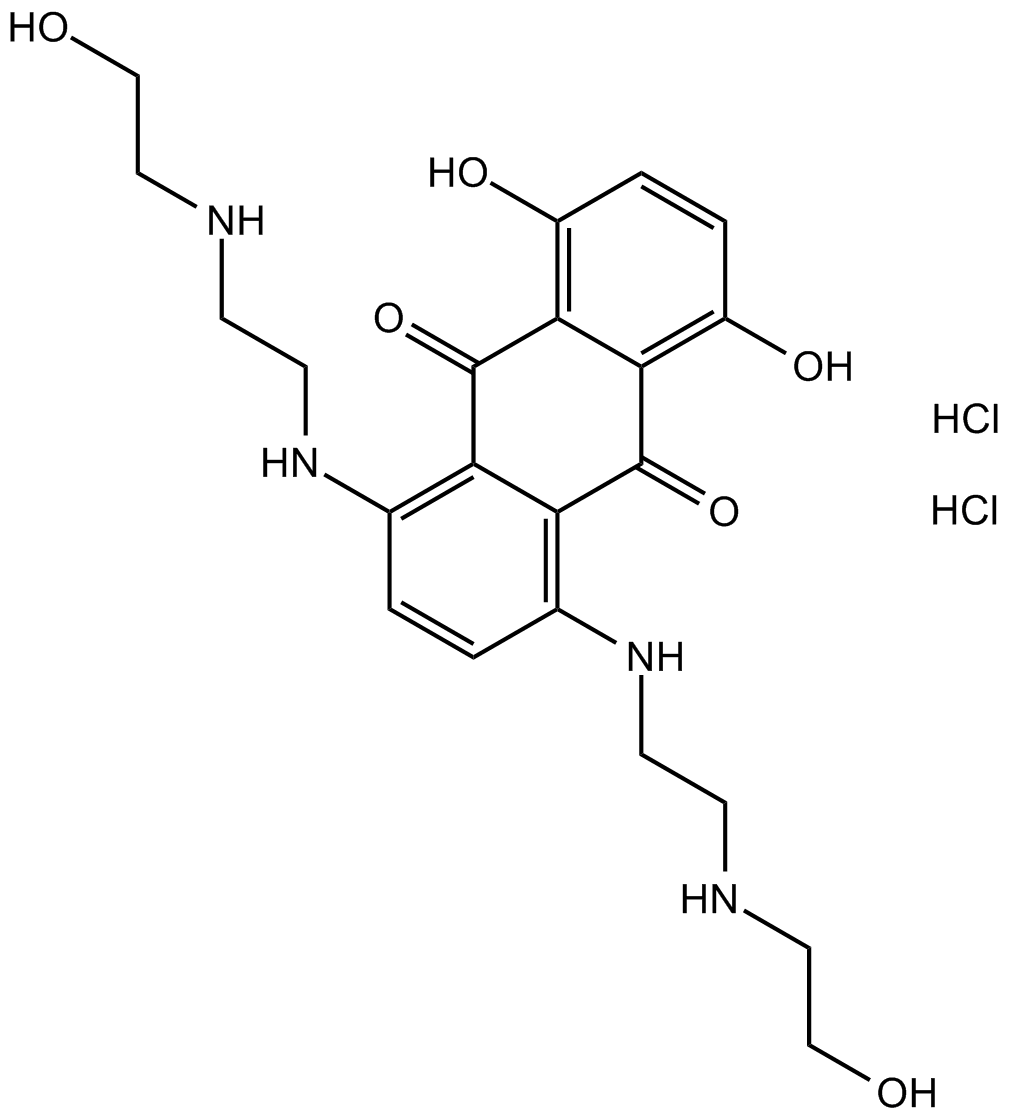
GC14363
Mitoxantrone HCl
Mitoxantrone HCl is a potent topoisomerase II inhibitor.
-
GC63778
Myelin Basic Protein TFA
Myelin Basic Protein (MHP4-14) TFA, a synthetic peptide comprising residues 4-14 of myelin basic protein, is a very selective PKC substrate (Km=7 μM).
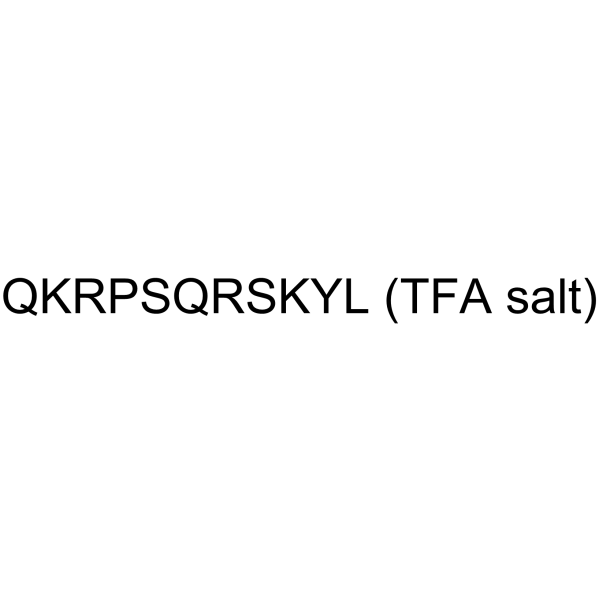
-
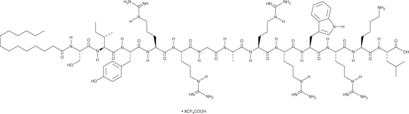
GC49269
Myr-ZIP
A PKMζ inhibitor
-
GC25662
N-Desmethyltamoxifen
N-Desmethyltamoxifen, the major metabolite of Tamoxifen in humans and a ten-fold more potent protein kinase C (PKC) inhibitor than Tamoxifen, also is a potent regulator of ceramide metabolism in human AML cells, limiting ceramide glycosylation, hydrolysis, and sphingosine phosphorylation.
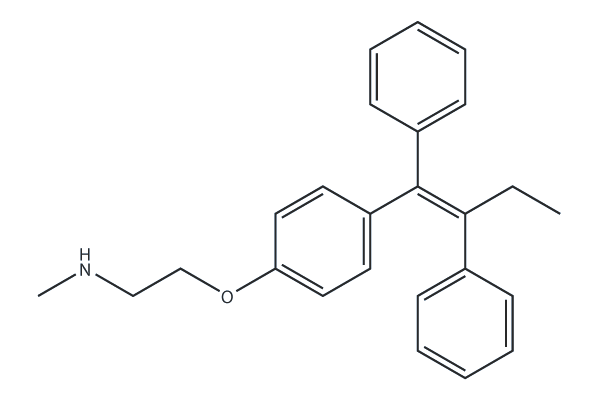
-
GC38931
N-Desmethyltamoxifen hydrochloride
N-Desmethyltamoxifen hydrochloride is the major metabolite of tamoxifen in humans. N-Desmethyltamoxifen, a poor antiestrogen, is a ten-fold more potent protein kinase C (PKC) inhibitor than Tamoxifen. N-Desmethyltamoxifen hydrochloride is also a potent regulator of ceramide metabolism in human AML cells, limiting ceramide glycosylation, hydrolysis, and sphingosine phosphorylation.

-
GC60274
O-Desmethyl Midostaurin
O-Desmethyl Midostaurin (CGP62221; O-Desmethyl PKC412) is the active metabolite of Midostaurin via cytochrome P450 liver enzyme metabolism. O-Desmethyl Midostaurin can be used as an indicator for Midostaurin metabolism in vivo. Midostaurin is a multi-targeted protein kinase inhibitor?with?IC50?ranging from 22-500 nM.

-
GC36833
p32 Inhibitor M36
p32 inhibitor M36 (M36) is a p32 mitochondrial protein inhibitor, which binds directly to p32 and inhibits p32 association with LyP-1.
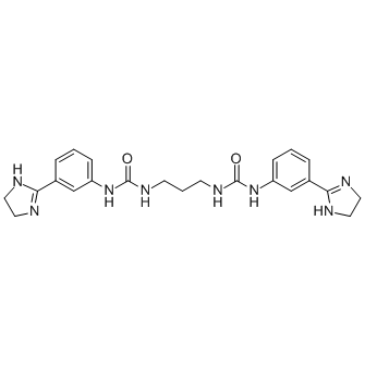
-
GC61585
Pep2m, myristoylated TFA
Pep2m, myristoylated TFA (Myr-Pep2m TFA) is a cell-permeable peptide.

-
GC17352
Phorbol 12,13-dibutyrate
Protein kinase C activator
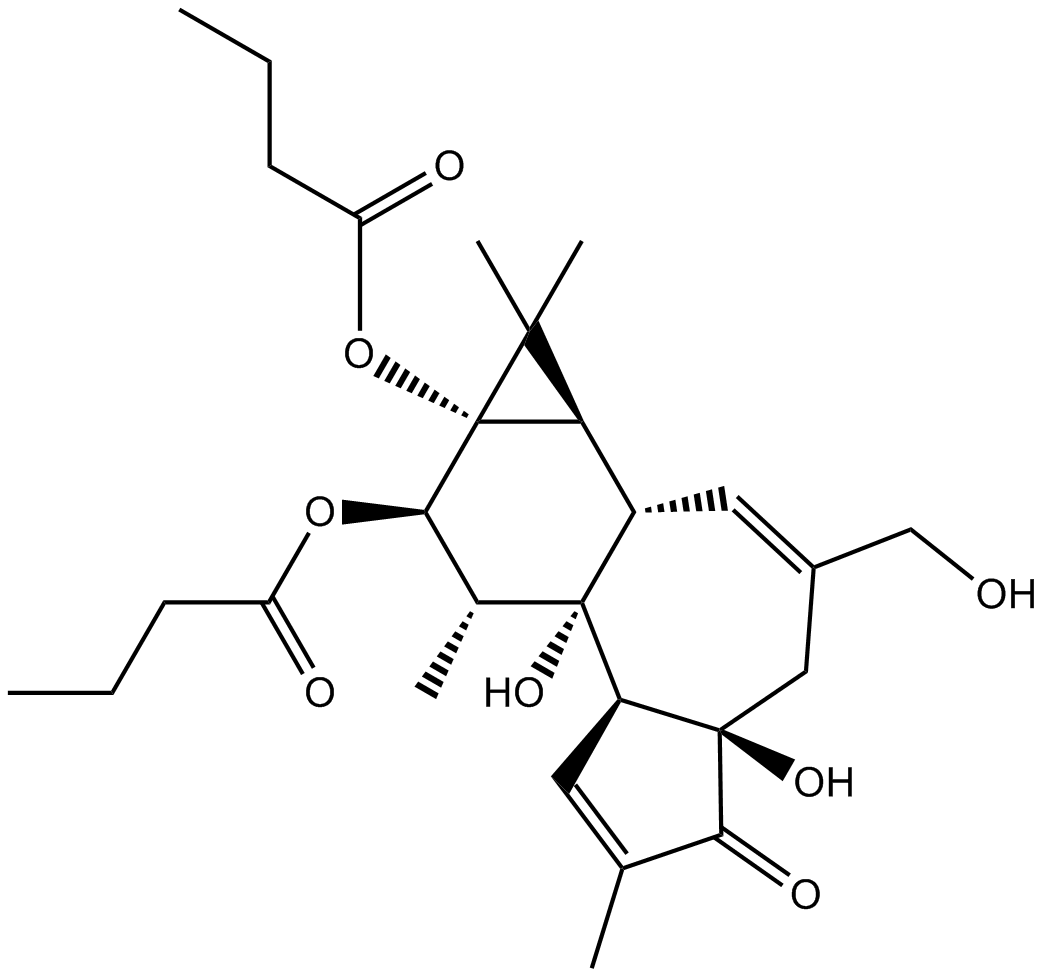
-
GC15471
PKC β pseudosubstrate
Selective cell-permeable peptide inhibitor of protein kinase C

-
GC10404
PKC ζ pseudosubstrate
PKC ζ pseudosubstrate is a selective cell-permeable inhibitor of PKC.
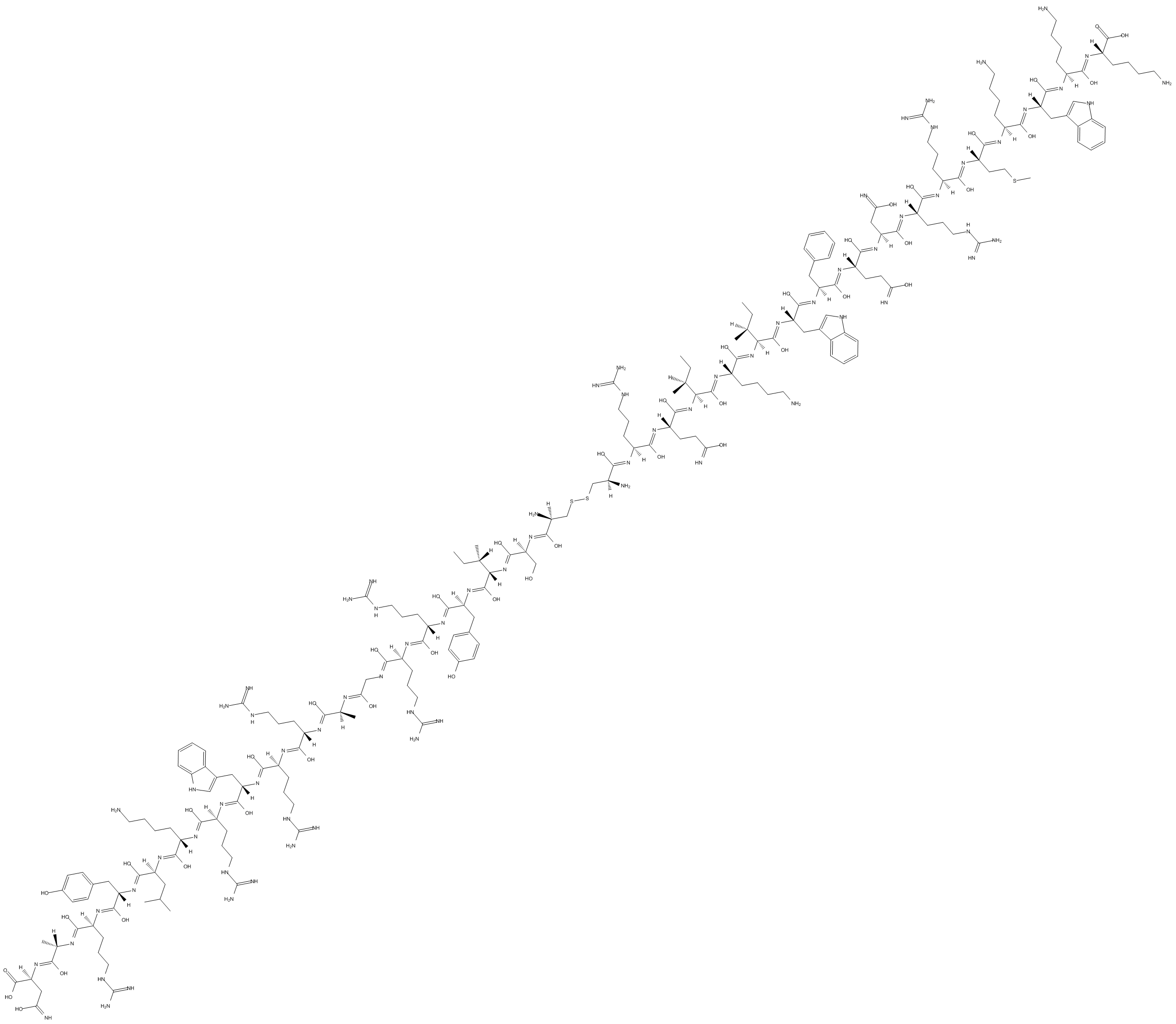
-
GC11671
PKC fragment (530-558)
Potent activator of protein kinase C

-
GC44655
PKCε Inhibitor Peptide
PKCε Inhibitor Peptide (ε-V1-2), a PKCε-derived peptide, is a selective PKCε inhibitor.
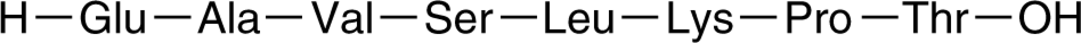
-
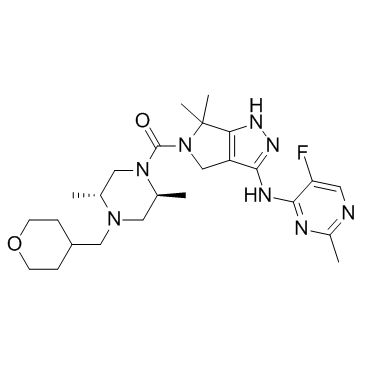
GC31711
PKC-IN-1
PKC-IN-1 is a potent, ATP-competitive and reversible inhibitor of conventional PKC enzymes with Kis of 5.3 and 10.4 nM for human PKCβ and PKCα, and IC50s of 2.3, 8.1, 7.6, 25.6, 57.5, 314, 808 nM for PKCα, PKCβI, PKCβII, PKCθ, PKCγ, PKC mu and PKCε, respectively.
-
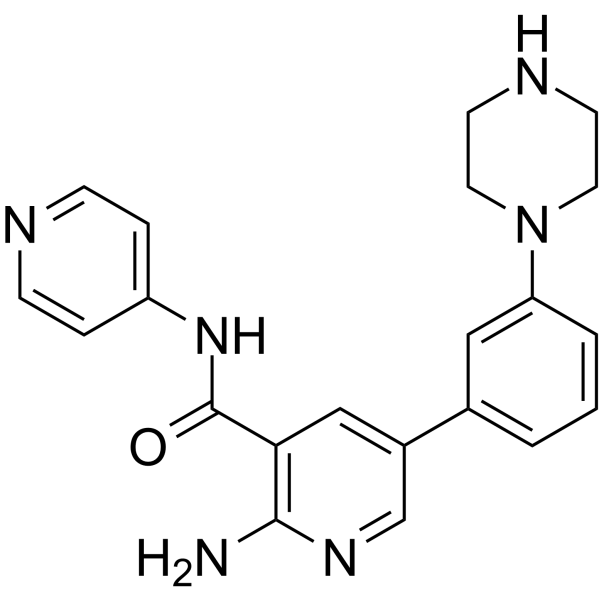
GC65126
PKC-iota inhibitor 1
PKC-iota inhibitor 1 (compound 19) is a protein kinase C-iota (PKC-ι ?) inhibitor with an IC50 value of 0.34 μM.
-
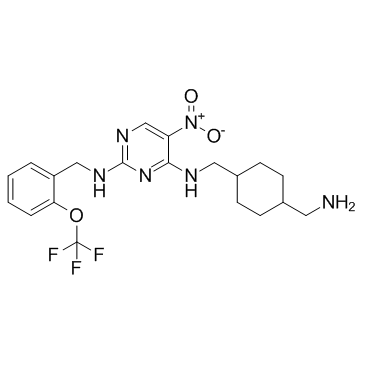
GC30200
PKC-theta inhibitor
PKC-theta inhibitor is a selective PKC-θinhibitor, with an IC50 of 12 nM.
-
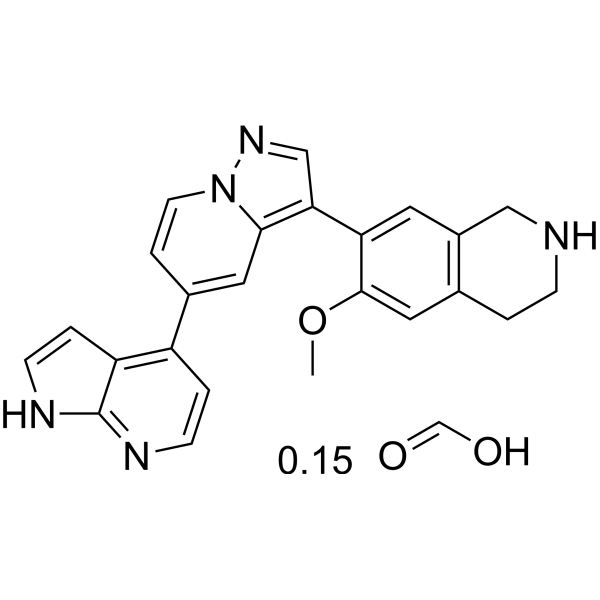
GC67686
PKCiota-IN-2 formic
-
GC36974
Procyanidin A1
Procyanidin A1 (Proanthocyanidin A1) is a procyanidin dimer, which inhibits degranulation downstream of protein kinase C activation or Ca2+ influx from an internal store in RBL-213 cells.

-
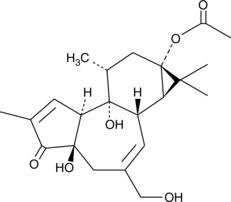
GC44729
Prostratin
An HIV reactivator
-
GC37014
Protein Kinase C 19-31
Protein Kinase C 19-31, a peptide inhibitor of protein kinase C (PKC), derived from the pseudo-substrate regulatory domain of PKCa (residues 19-31) with a serine at position 25 replacing the wild-type alanine, is used as protein kinase C substrate peptide for testing the protein kinase C activity.

-
GC37015
Protein Kinase C 19-36
Protein Kinase C 19-36 is a pseudosubstrate peptide inhibitor of protein kinase C (PKC), with an IC50 of 0.18 μM.

-
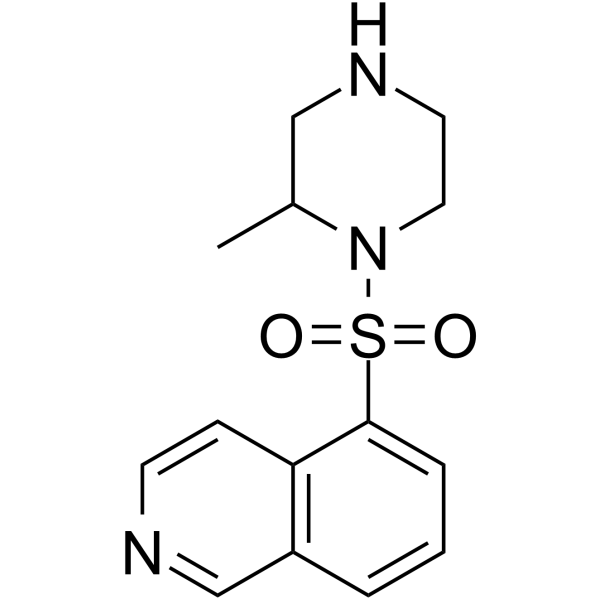
GC66006
Protein kinase inhibitor H-7
Protein kinase inhibitor H-7 is a potent inhibitor of protein kinase C (PKC) and cyclic nucleotide dependent protein kinase, with a Ki of 6 μM for PKC.
-
GC11803
Pseudo RACK1
Activator of protein kinase C
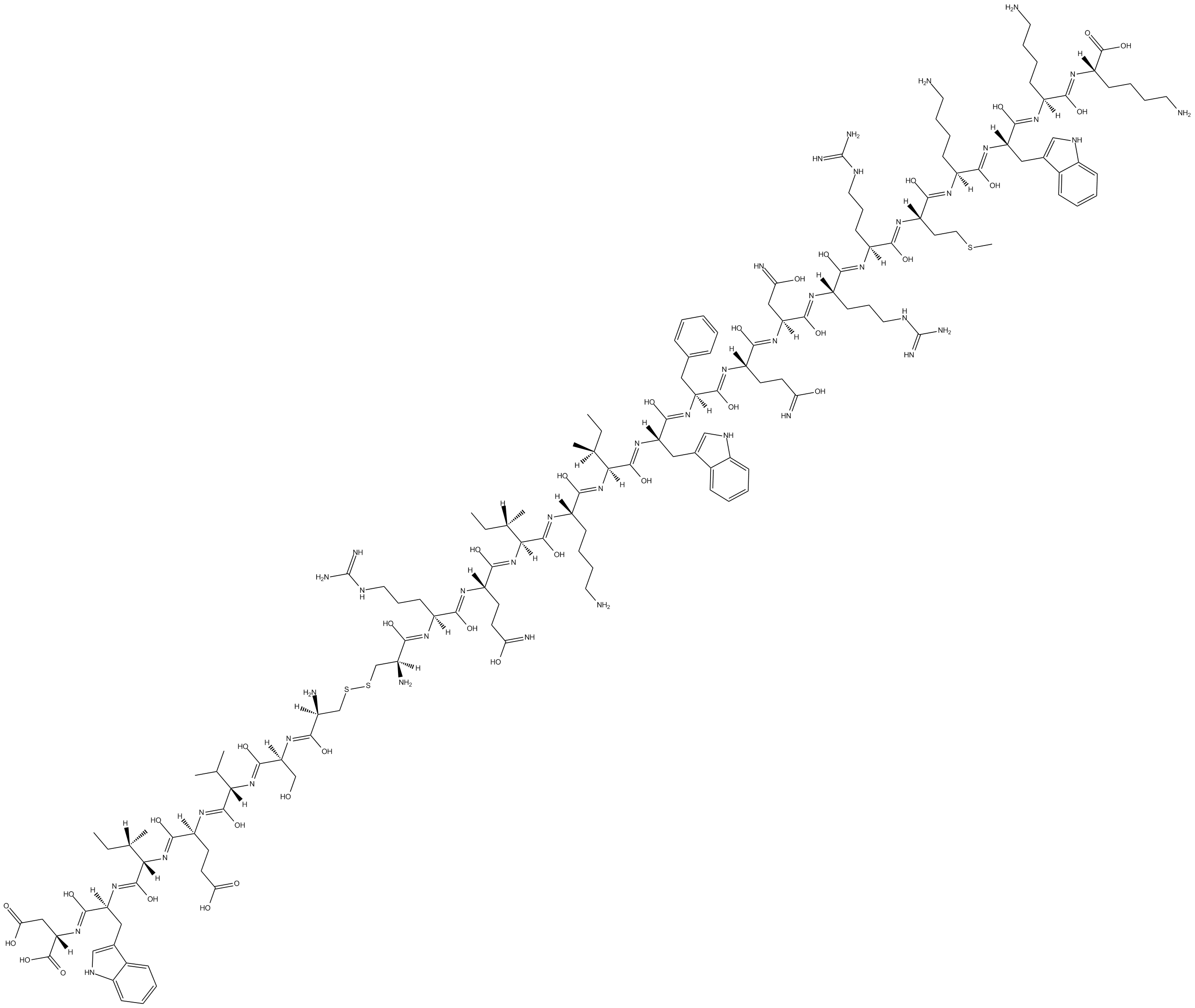
-
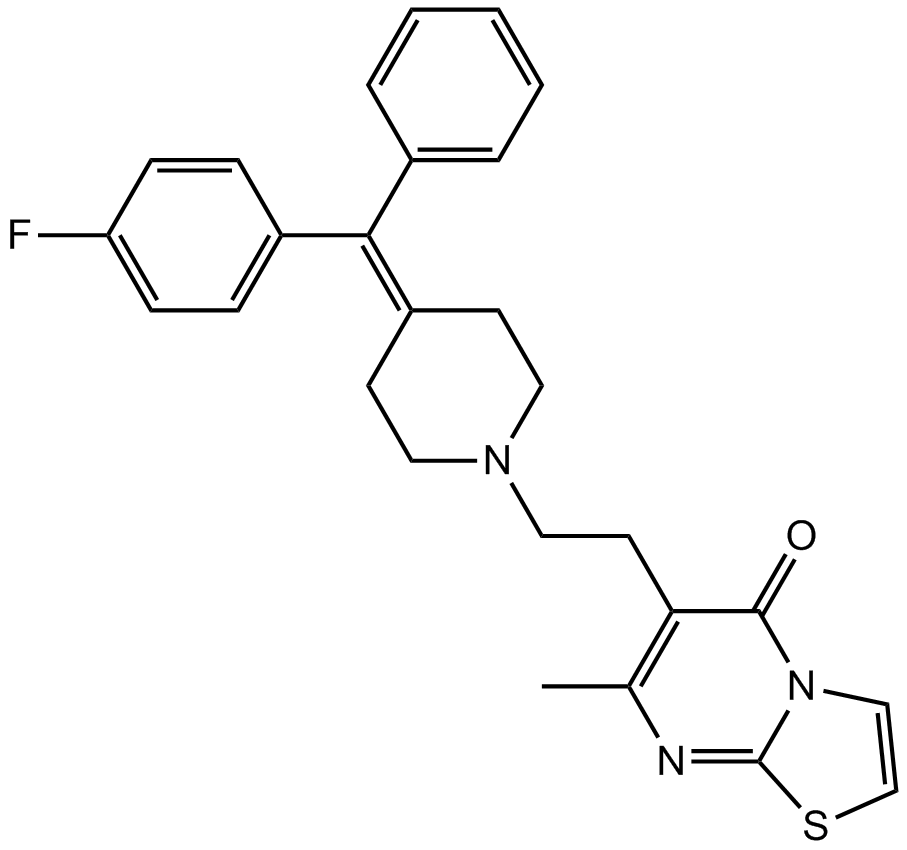
GC14583
R 59-022
Diacylglycerol (DAG) kinase inhibitor
-
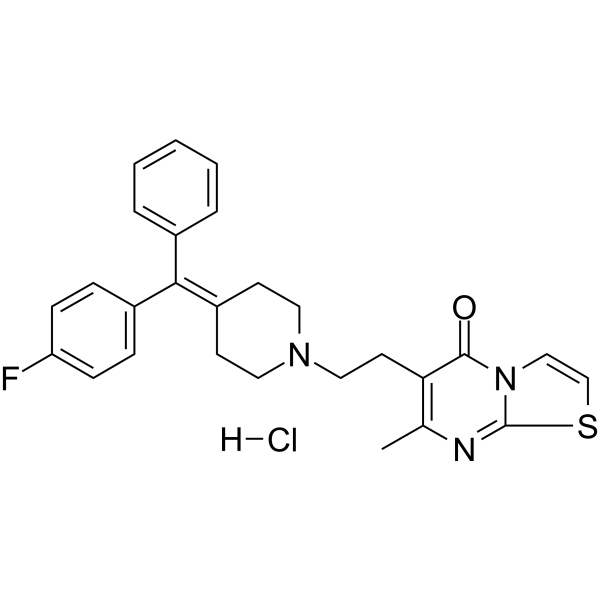
GC69792
R 59-022 hydrochloride
R 59-022 (DKGI-I) hydrochloride is a DGK inhibitor (IC50: 2.8 μM). R 59-022 hydrochloride inhibits OAG phosphorylation to OAPA. It is also a 5-HT receptor antagonist that can activate protein kinase C (PKC). R 59-022 enhances the production of thromboxane in platelets induced by thrombin and inhibits phosphatidic acid production in neutrophils.
-
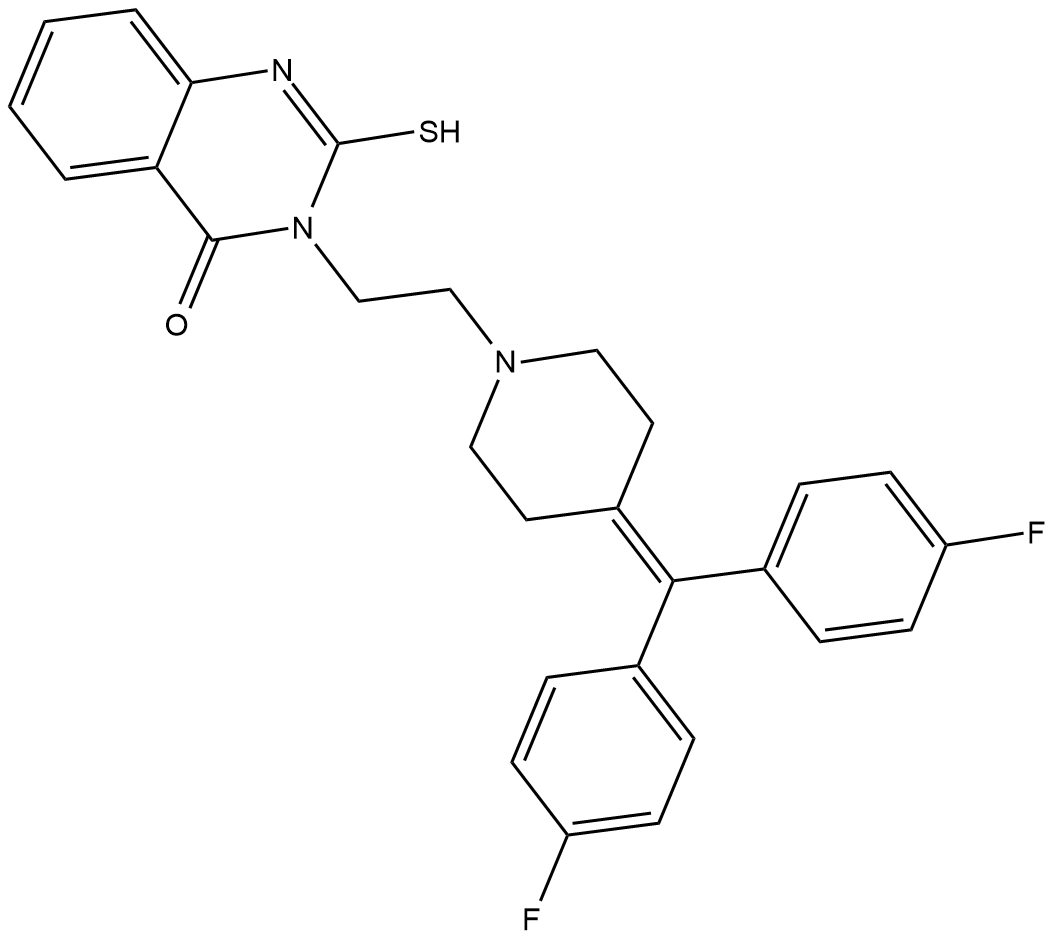
GC18246
R-59-949
R-59-949 is an inhibitor of diacylglycerol kinase α (DGK-α) with an IC50 value of 300 nM in isolated platelet plasma membranes using exogenous diacylglycerol as a substrate.
-
GC17322
Ro 31-8220
pan-PKC inhibitor