Tyrosine Kinase
- IGFBR(1)
- Bcr-Abl(73)
- Ack1(4)
- Axl(6)
- ALK(61)
- BMX Kinase(4)
- Broad Spectrum Protein Kinase Inhibitor(11)
- c-FMS(35)
- c-Kit(65)
- c-MET(91)
- c-RET(7)
- CSF-1R(3)
- DDR1/DDR2 Receptor(1)
- EGFR(251)
- EphB4(2)
- FAK(34)
- FGFR(104)
- FLT3(102)
- HER2(19)
- IGF1R(33)
- Insulin Receptor(45)
- IRAK(32)
- ITK(12)
- Lck(2)
- LRRK2(23)
- PDGFR(117)
- PTKs/RTKs(5)
- Pyk2(6)
- ROR(41)
- Spleen Tyrosine Kinase (Syk)(35)
- Src(105)
- Tie-2 (3)
- Trk(37)
- VEGFR(203)
- Kinase(0)
- Discoidin Domain Receptor(15)
- DYRK(26)
- Ephrin Receptor(13)
- ROS(15)
- RET(34)
- TAM Receptor(28)
Products for Tyrosine Kinase
- Cat.No. Product Name Information
-
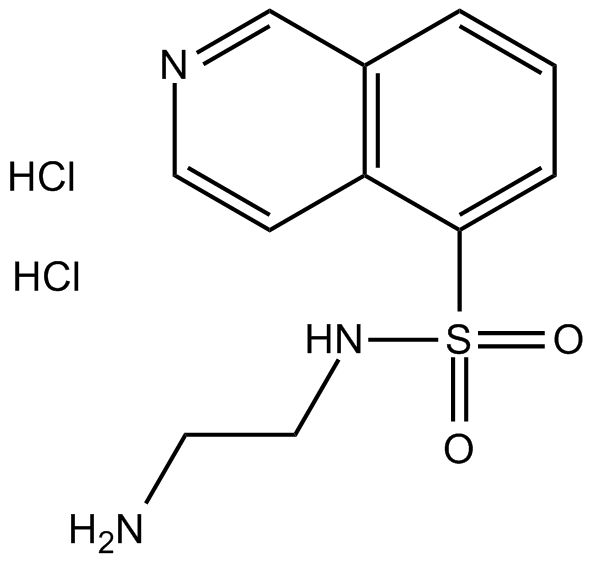
GC68030
FAK-IN-7
-
GC64836
Famitinib
Famitinib (SHR1020), an orally active multi-targeted kinase inhibitor, inhibits the activity of c-kit, VEGFR-2 and PDGFRβ with IC50 values of 2.3 nM, 4.7 nM and 6.6 nM, respectively. Famitinib exerts powerful antitumor activity in human gastric cancer cells and xenografts. Famitinib triggers apoptosis.
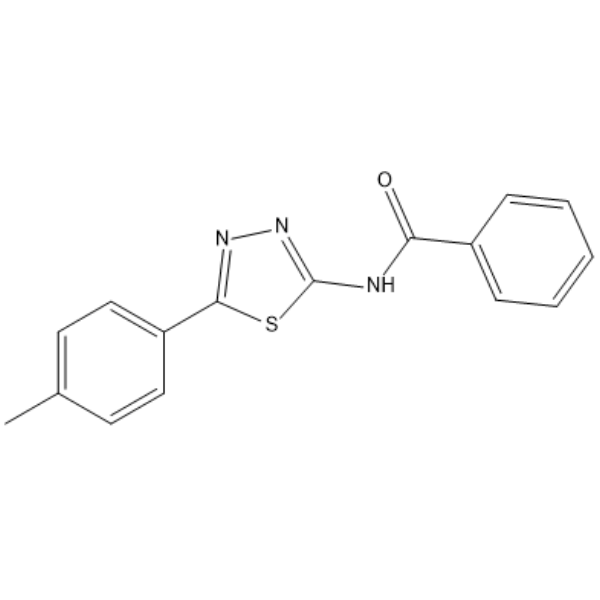
-
GC38437
Fangchinoline
An alkaloid with diverse biological activities
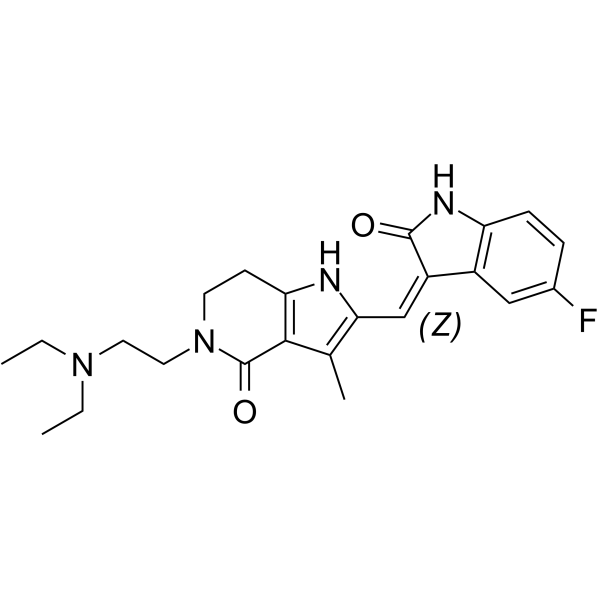
-
GC63395
FGFR-IN-1
FGFR-IN-1 is a potent FGFR inhibitor with an IC50 of <100 nM for FGFR1, FGFR2, and FGFR3, respectively (patent US20130338134A1, example 219).
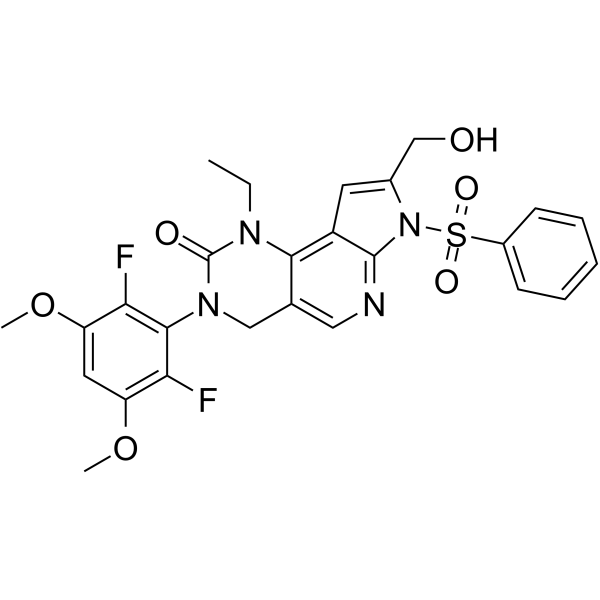
-
GC65212
FGFR1/DDR2 inhibitor 1
FGFR1/DDR2 inhibitor 1 is an orally active inhibitor of fibroblast growth factor receptor 1 (FGFR1) and discoindin domain receptor 2 (DDR2), with IC50 values of 31.1 nM and 3.2 nM, respectively. Antitumor activity.
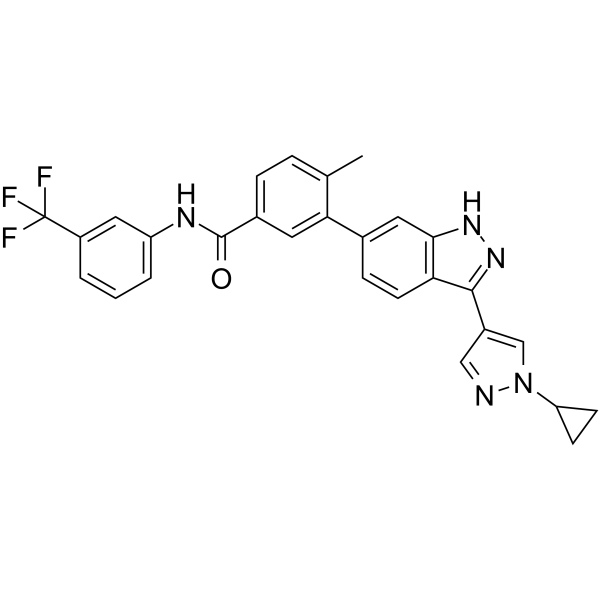
-
GC64475
FGFR2-IN-2
FGFR2-IN-2 (Compound 38) is a selective FGFR2 inhibitor with IC50s of 389, 29, and 758 nM for FGFR1, FGFR2, and FGFR3, respectively.
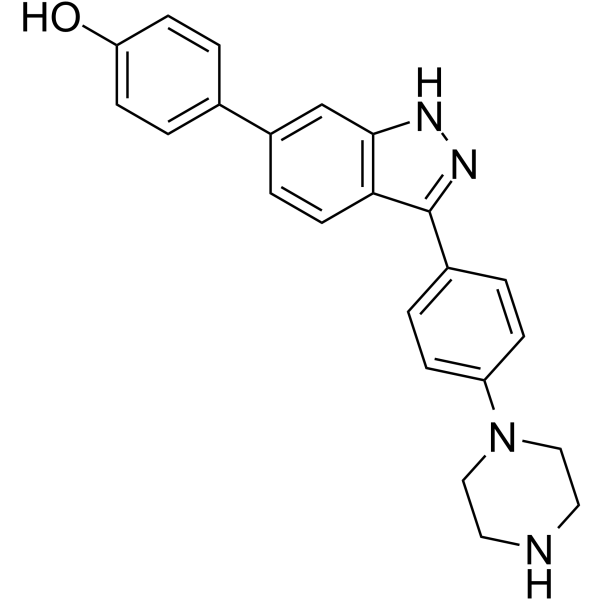
-
GC65992
FGFR2-IN-3
FGFR2-IN-3 is an orally active selective inhibitor of FGFR2.
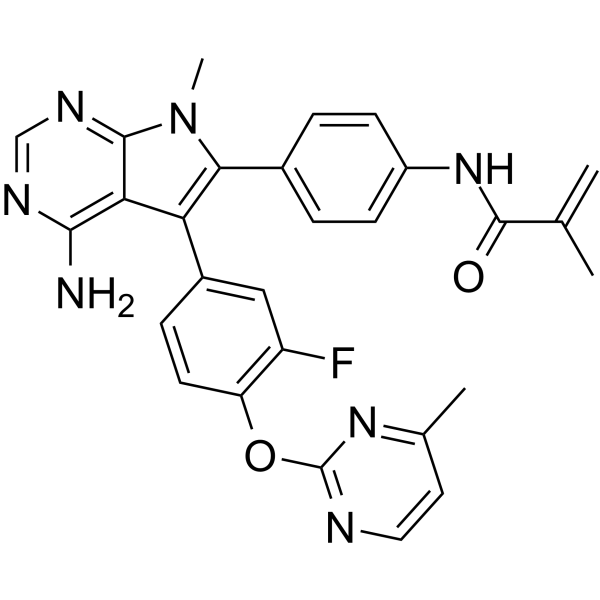
-
GC65991
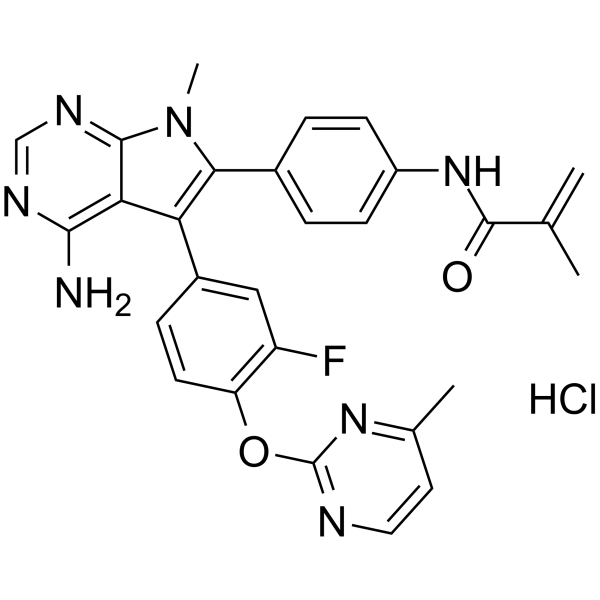
FGFR2-IN-3 hydrochloride
FGFR2-IN-3 hydrochloride is an orally active selective inhibitor of FGFR2.
-
GC19155
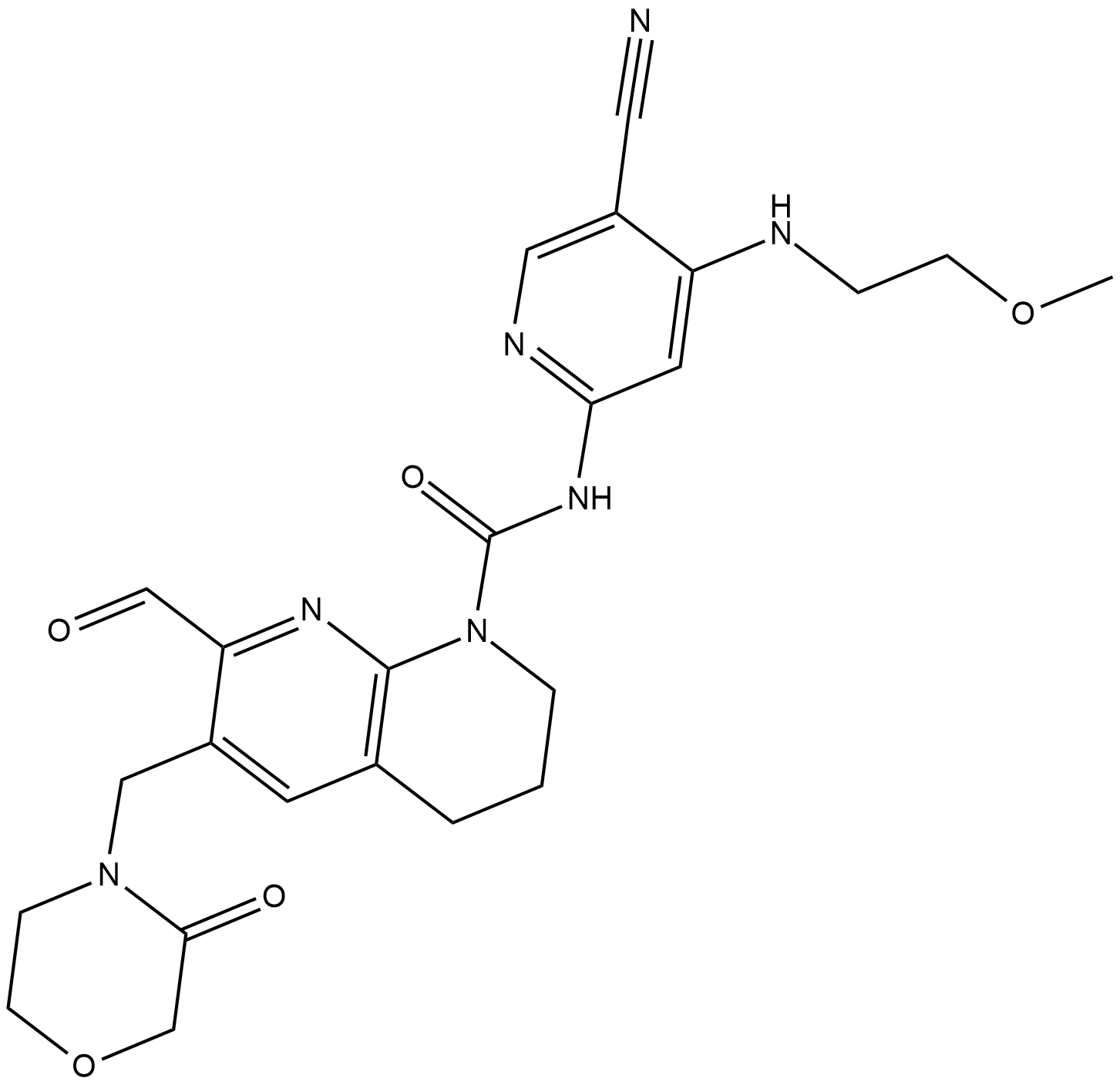
FGFR4-IN-1
FGFR4-IN-1 is a potent inhibiotr of FGFR4 with IC50 of 0.7 nM.
-
GC62602
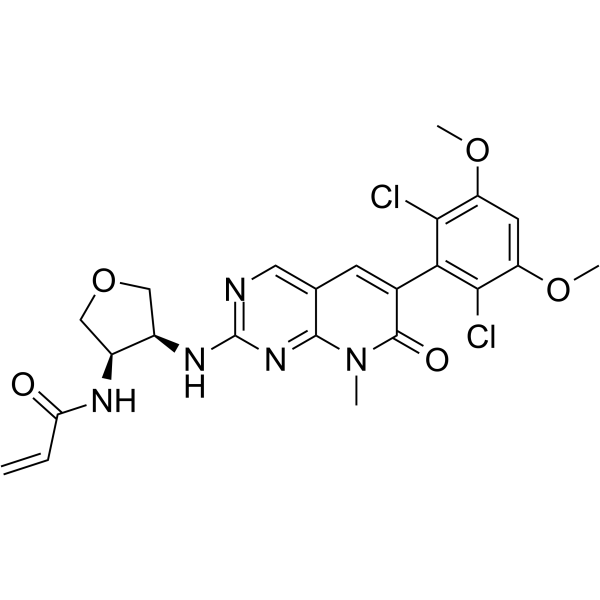
FGFR4-IN-5
FGFR4-IN-5 is a potent and selective covalent FGFR4 inhibitor with an IC50 of 6.5 nM. FGFR4-IN-5 exhibits strong anti-tumor activity in vivo and can be used for hepatocellular carcinoma research.
-
GC50051
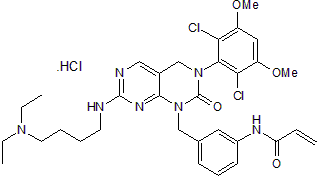
FIIN 1 hydrochloride
FIIN 1 hydrochloride is a potent, irreversible, selective FGFR inhibitor. FIIN 1 hydrochloride binds to FGFR1/2/3/4 and Flt1/4 with Kds of 2.8/6.9/5.4/120 nM and 32/120 nM respectively. The biochemical IC50s of FIIN 1 hydrochloride are 9.2, 6.2, 11.9, and 189 nM against FGFR1/2/3/4, respectively.
-
GC17323
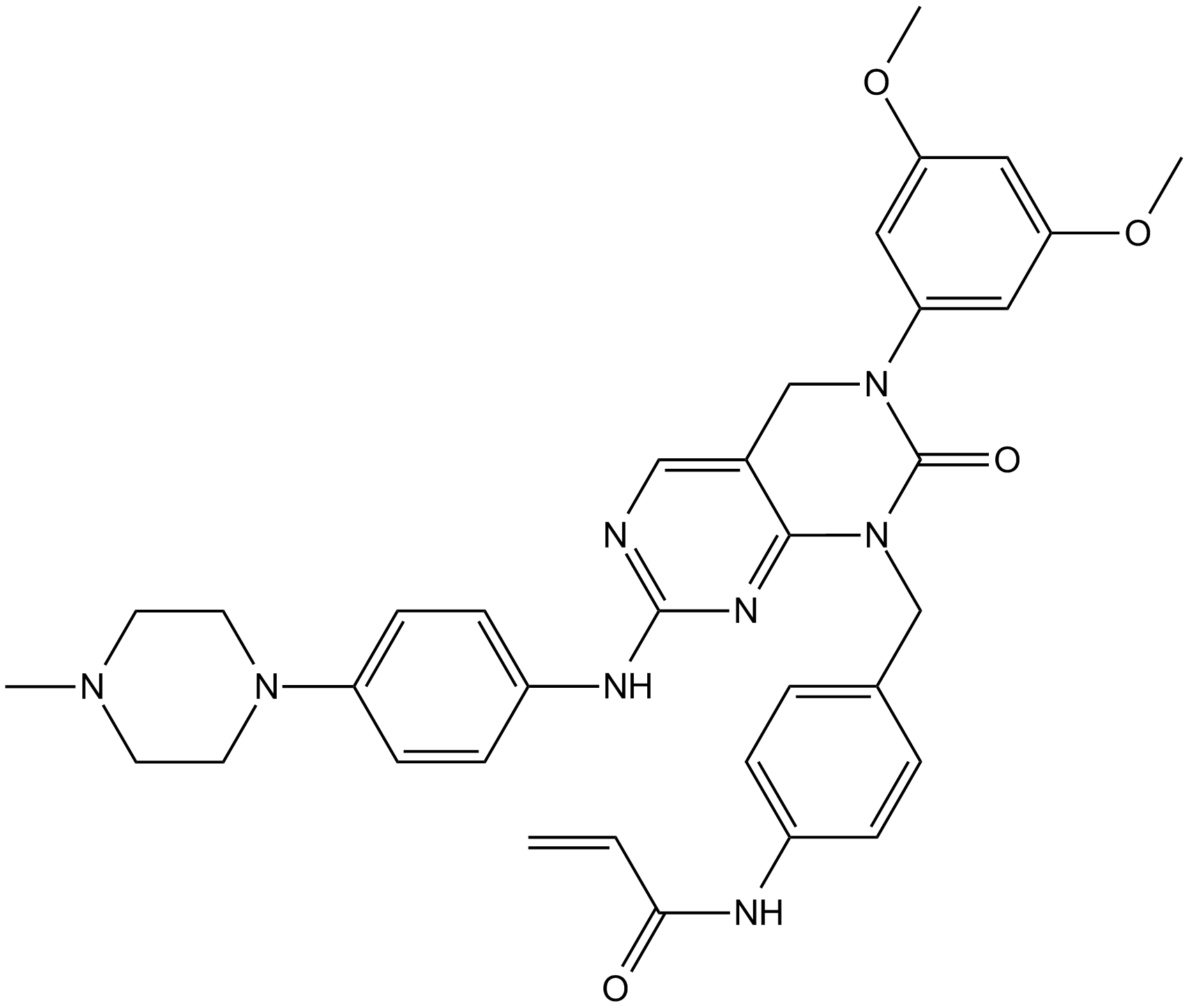
FIIN-2
Irreversible inhibitor of FGFR
-
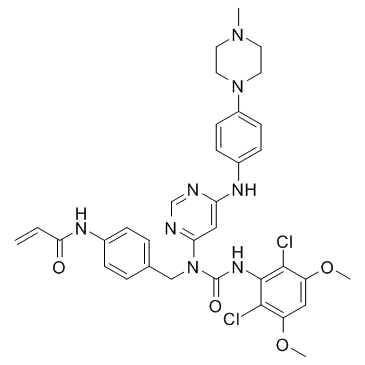
GC36044
FIIN-3
FIIN-3 is an irreversible inhibitor of FGFR with an IC50 of 13.1, 21, 31.4, and 35.3 nM for FGFR1, FGFR2, FGFR3 and FGFR4, respectively.
-
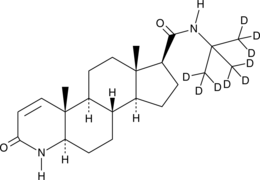
GC47350
Finasteride-d9
An internal standard for the quantification of finasteride
-
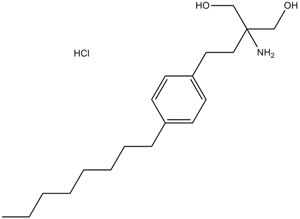
GC14807
Fingolimod(FTY720)
Fingolimod (FTY720) HCl (FTY720), an analog of sphingosine, is a potent sphingosine 1-phosphate (S1P) receptors modulator.
-
GC43676
FLT3 Inhibitor III
FLT3 Inhibitor III is a potent inhibitor of Fms-like tyrosine kinase 3 (FLT3; IC50 = 50 nM).
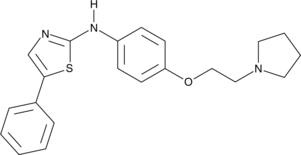
-
GC32968
FLT3-IN-1
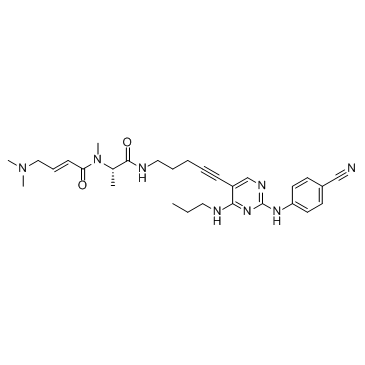
-
GC33242
FLT3-IN-1 Succinate
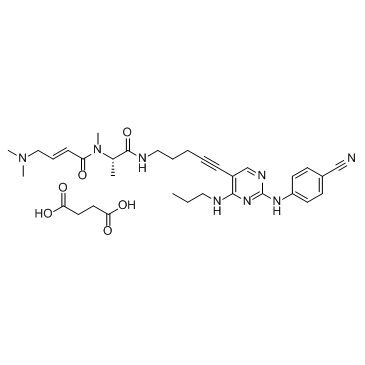
-
GC63936
FLT3-IN-10
FLT3-IN-10 (compound 7c) is a potent inhibitor of FMS-like tyrosine kinase 3 (FLT3). FLT3-IN-10 has the potential for the treatment of FLT3-mutated acute myeloid leukemia (AML).
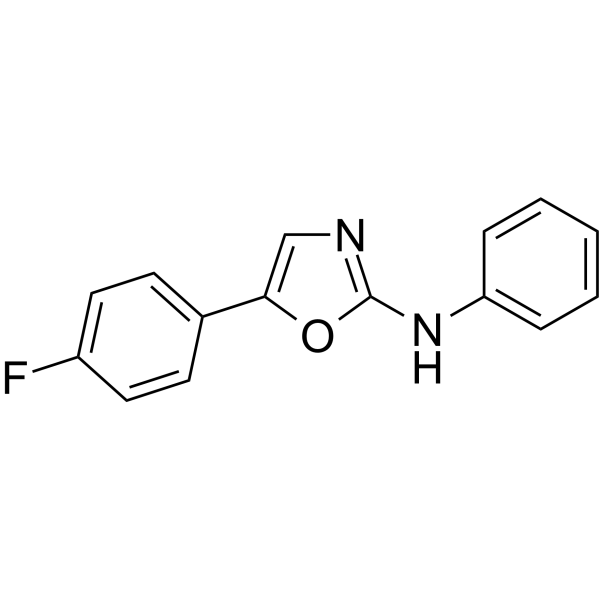
-
GC65984
FLT3-IN-16
FLT3-IN-16 is a potent FLT3 inhibitor with an IC50 of 1.1 μM. FLT3-IN-16 can be used for researching acute myeloid leukemia.
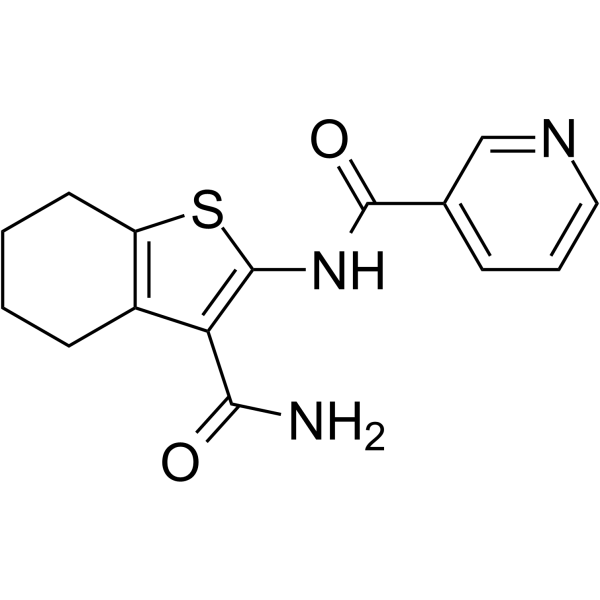
-
GC19158
FLT3-IN-2
FLT3-IN-2 is a FLT3 inhibitor with IC50 of < 1 uM, detailed information refer to WO 2012158957 A2 and WO 2007013896.
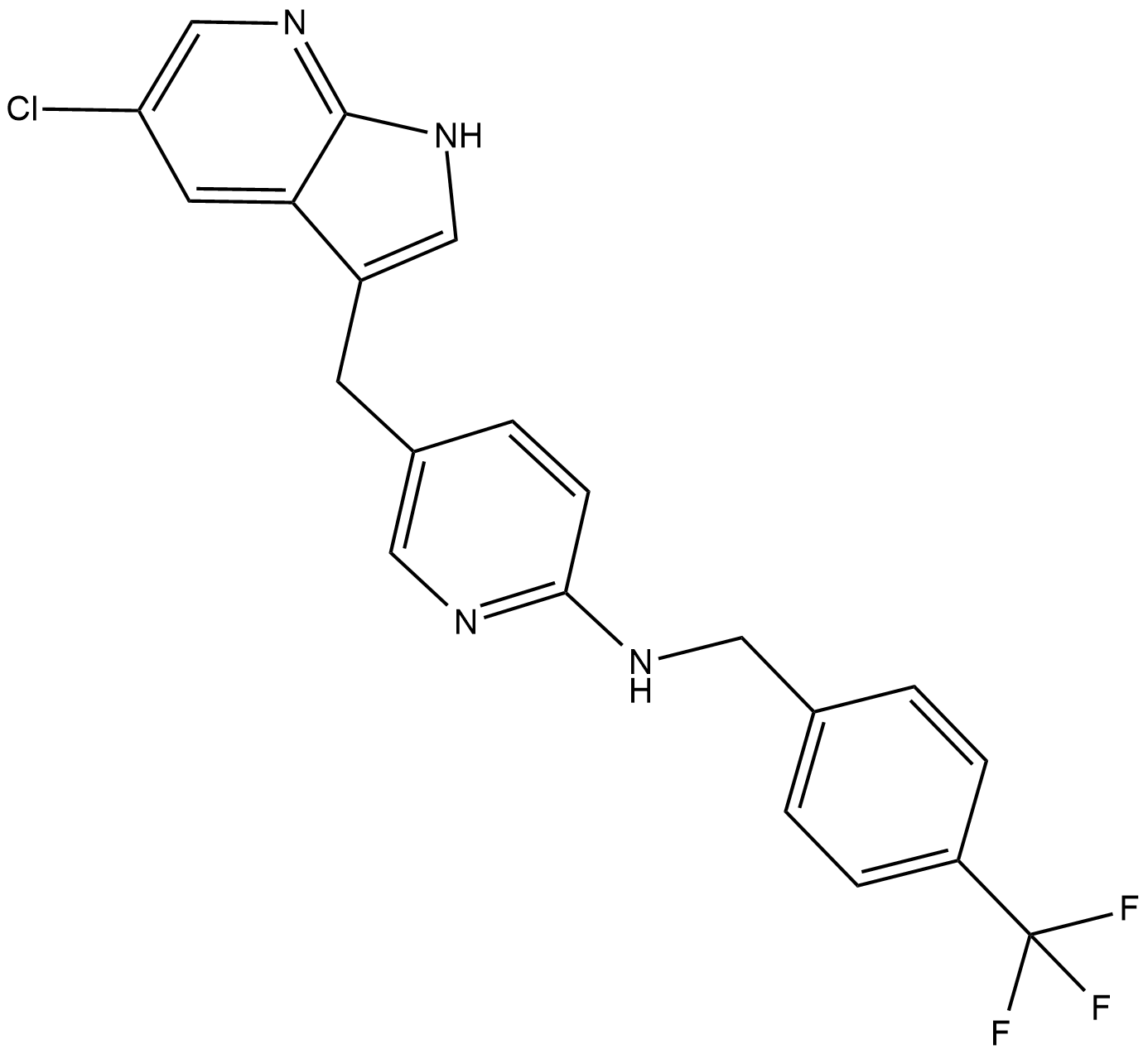
-
GC19772
FLT3-IN-3
FLT3-IN-3 is a potent FLT3 inhibitor with IC50s of 13 and 8 nM for FLT3 WT and FLT3 D835Y, respectively.
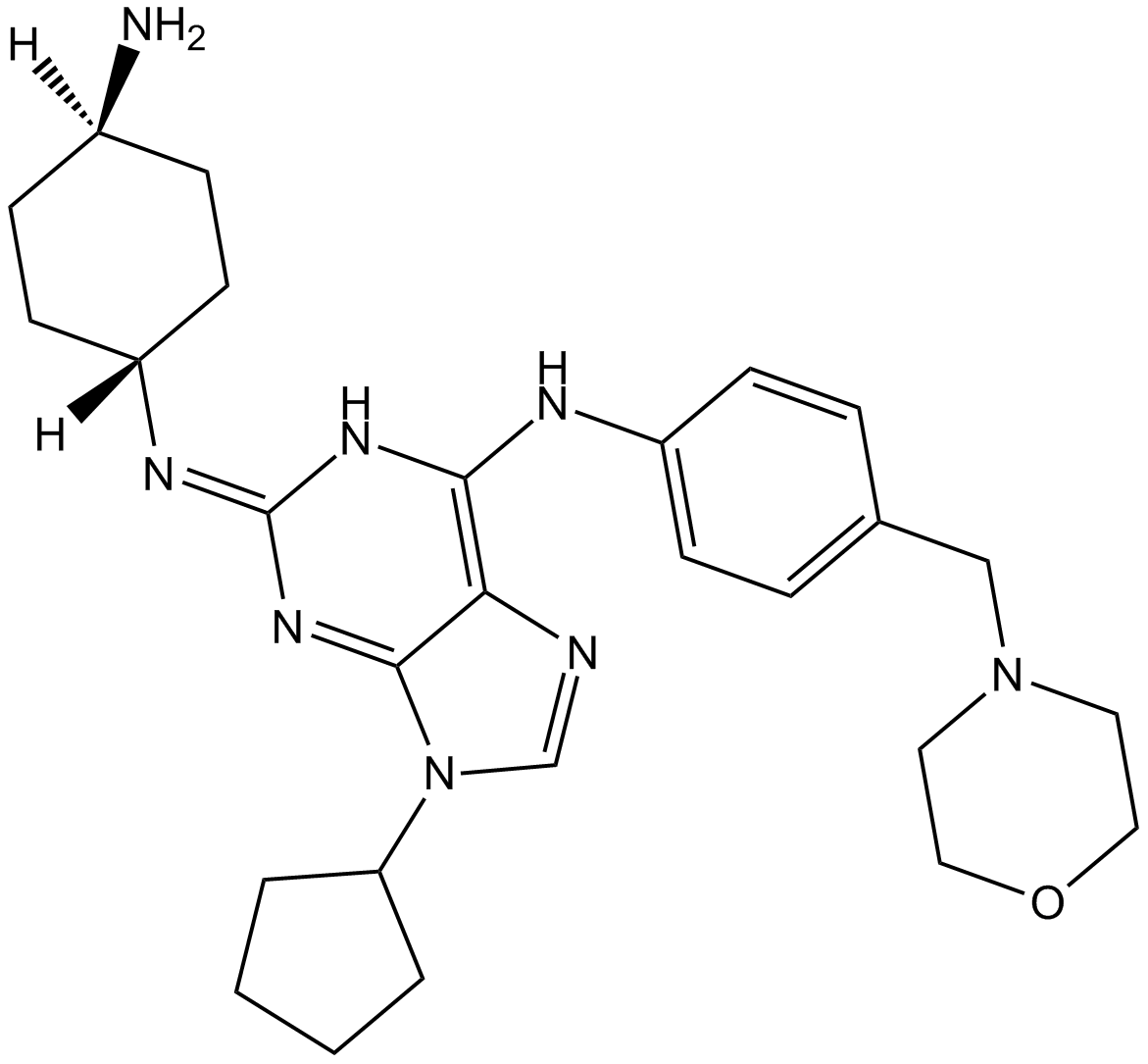
-
GC36055
FLT3-IN-4
FLT3-IN-4 is a potent and orally effective Fms-like tyrosine receptor kinase 3 (FLT3; IC50=7 nM) inhibitor for treating acute myelogenous leukemia.
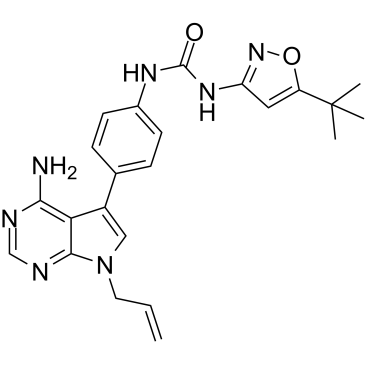
-
GC36056
FLT3-IN-6
FLT3-IN-6 is a potent and selective inhibitor of FLT3-ITD (FLT3 mutation) with an IC50 of 1.336 nM.
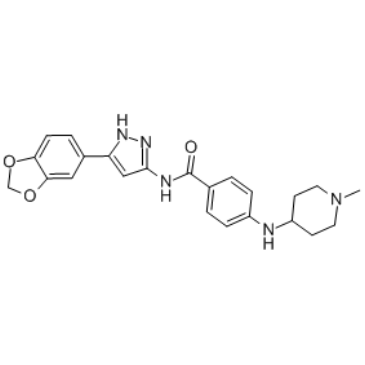
-
GC32867
Flumatinib (HHGV678)
Flumatinib (HHGV678) (HHGV678) is an orally available, selective inhibitor of Bcr-Abl. Flumatinib (HHGV678) inhibits c-Abl, PDGFRβ and c-Kit with IC50s of 1.2 nM, 307.6 nM and 665.5 nM, respectively.
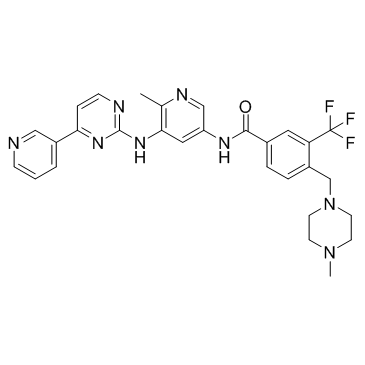
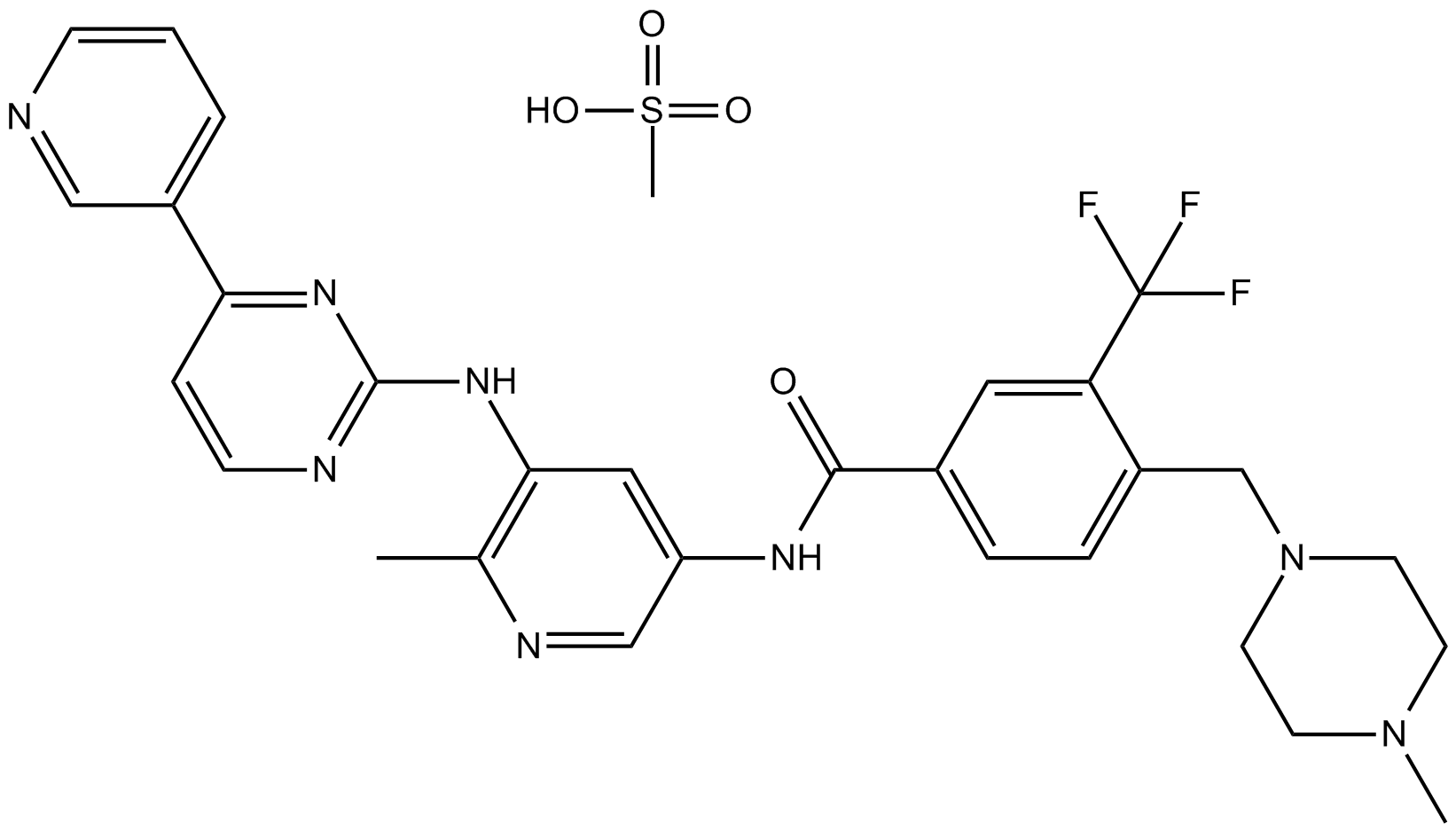
-
GC13914
Flumatinib mesylate
PDGRFβ inhibitor
-
GC33049
FN-1501
FN-1501 is a potent inhibitor of FLT3 and CDK, with IC50s of 2.47, 0.85, 1.96, and 0.28 nM for CDK2/cyclin A, CDK4/cyclin D1, CDK6/cyclin D1 and FLT3, respectively. FN-1501 has anticancer activity.
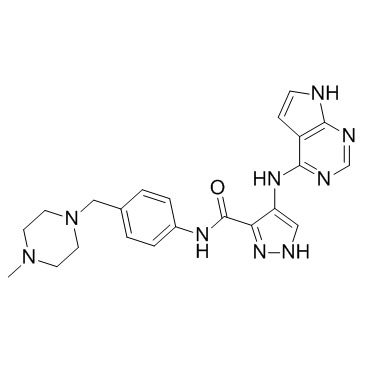
-
GC15735
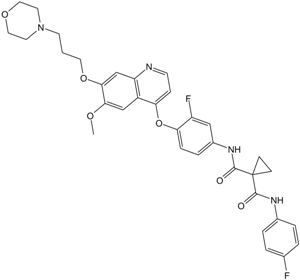
Foretinib (GSK1363089)
Foretinib (GSK1363089) is a multi-target tyrosine kinase inhibitor with IC50s of 0.4 nM and 0.9 nM for Met and KDR.
-
GN10527
Formononetin
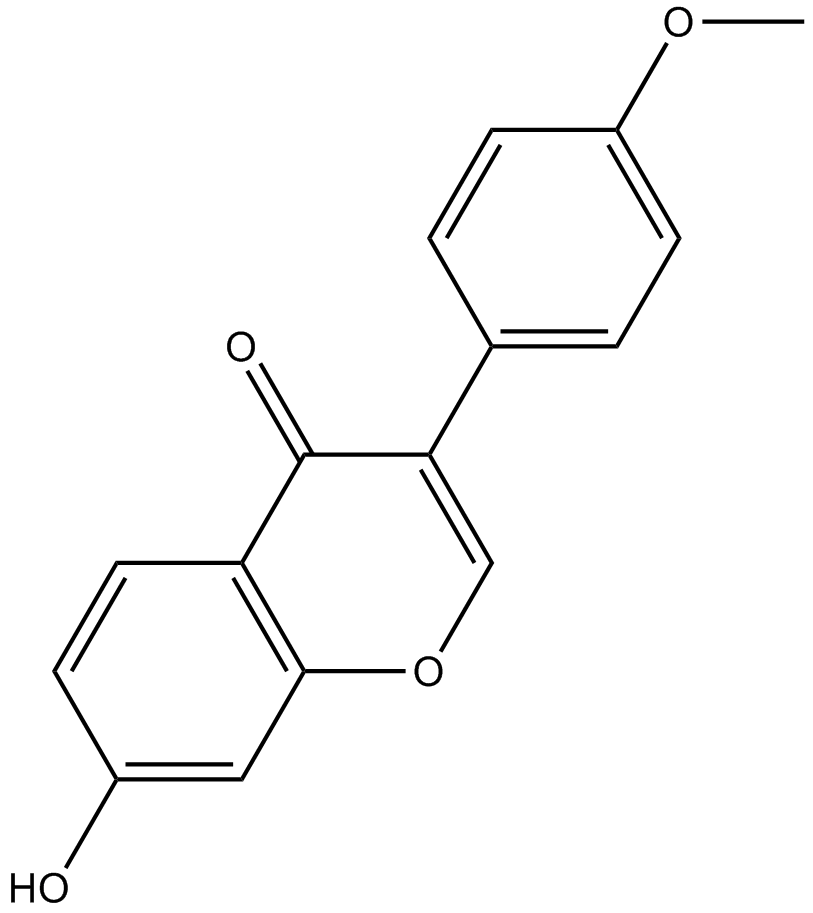
-
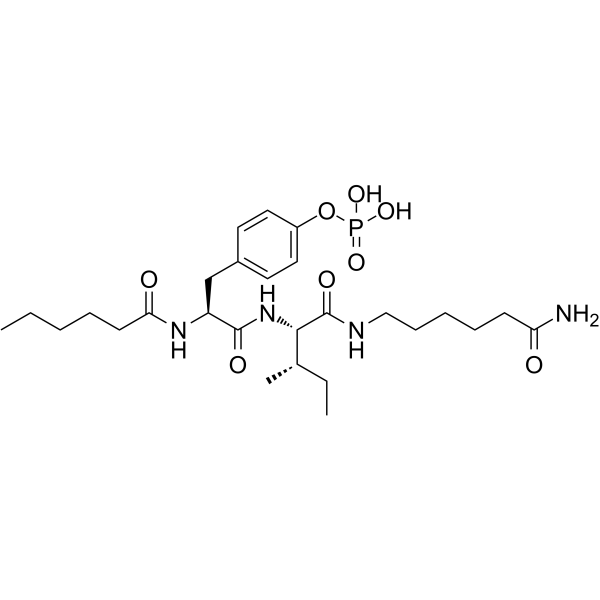
GC64708
Fosgonimeton
-
GC11044
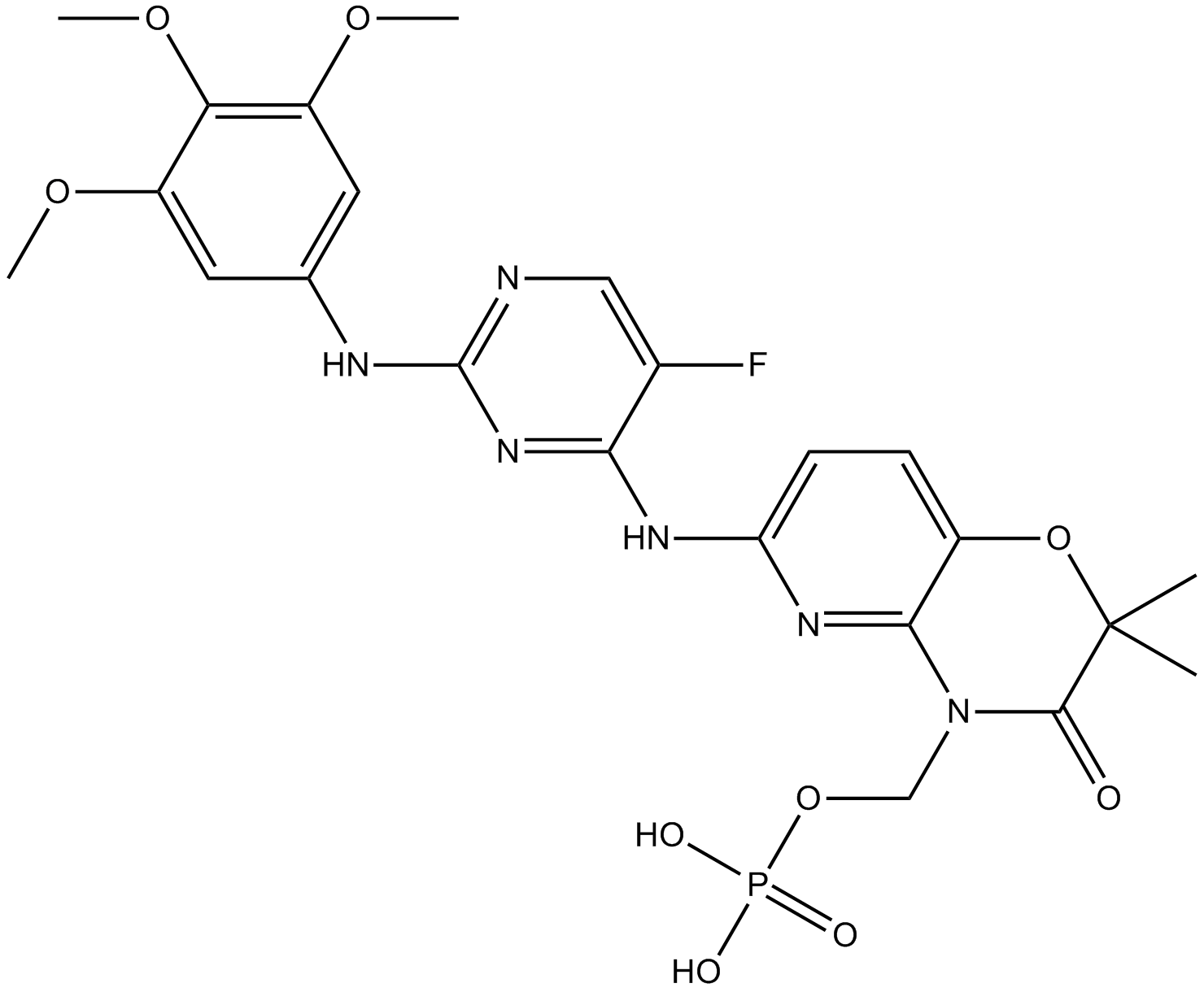
Fostamatinib (R788)
Fostamatinib (R788) (R788) is the oral prodrug of the active compound R406.
-
GC10355
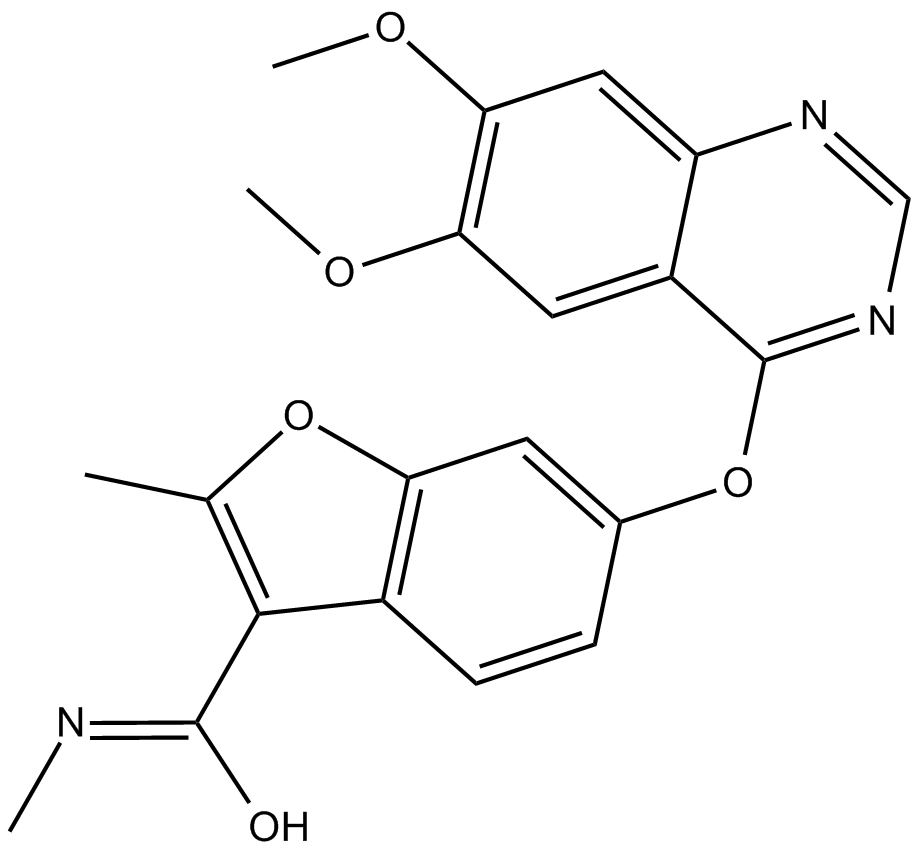
Fruquintinib(HMPL-013)
Fruquintinib(HMPL-013) (HMPL-013) is a highly potent and selective VEGFR 1/2/3 inhibitor with IC50s of 33, 0.35, and 35 nM, respectively.
-
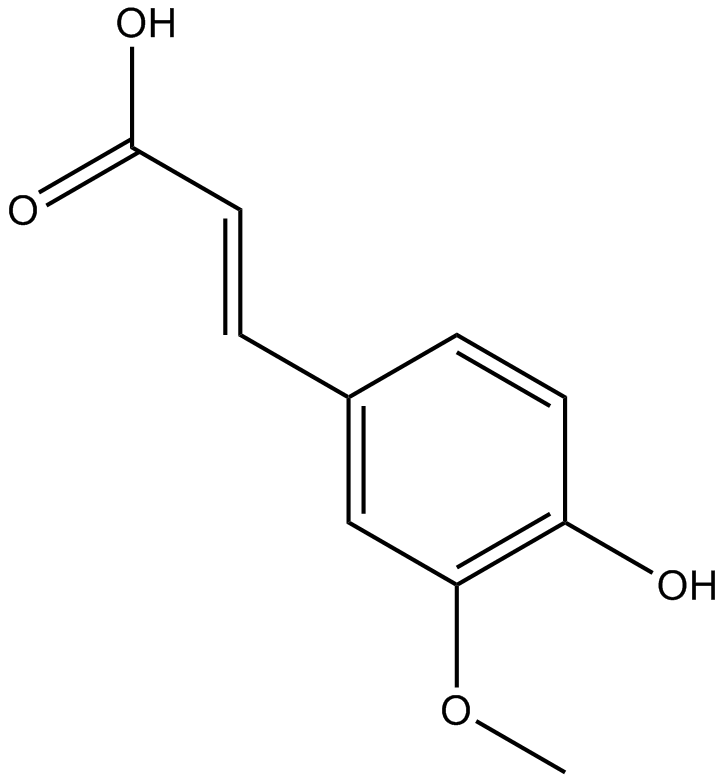
GN10540
Fumalic acid
-
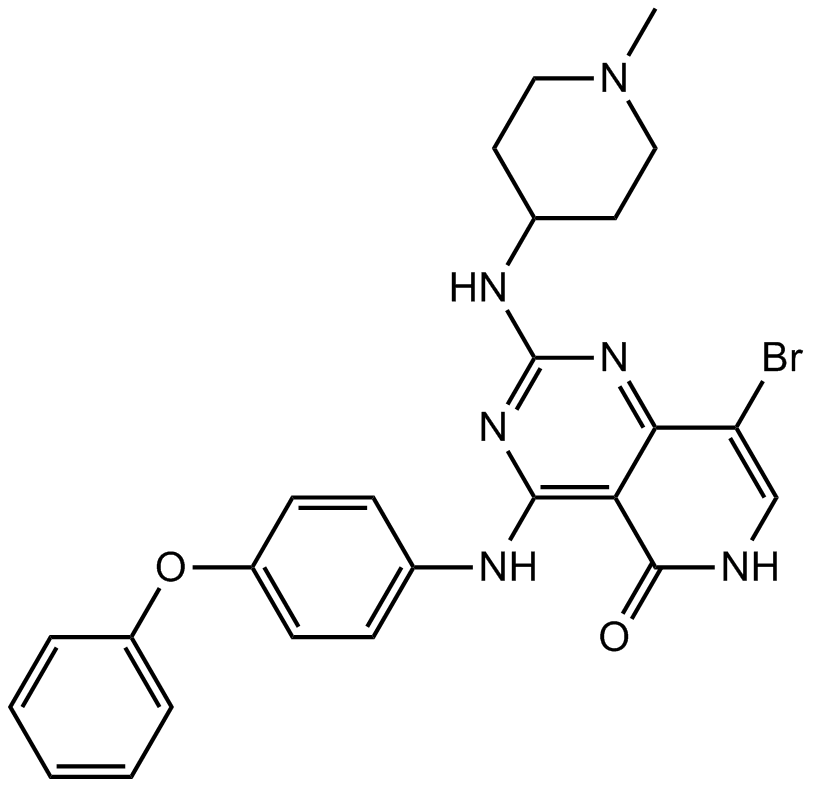
GC12178
G-749
FLT3 inhibitor
-
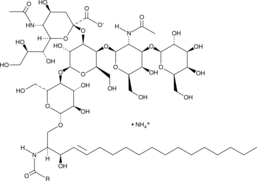
GC47392
Ganglioside GM1 Mixture (ovine) (ammonium salt)
A mixture of ganglioside GM1
-
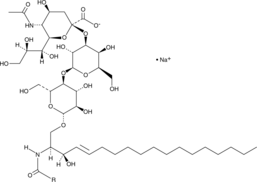
GC43732
Ganglioside GM3 Mixture (sodium salt)
Ganglioside GM3 is a monosialoganglioside that demonstrates antiproliferative and proapoptotic effects in tumor cells by modulating cell adhesion, proliferation, and differentiation.
-
GA21893
Gastric Inhibitory Polypeptide (1-30) amide (porcine)
Gastric Inhibitory Polypeptide (1-30) amide (porcine) is a full glucose-dependent insulinotropic polypeptide (GIP) receptor agonist with high affinity equal to native GIP(1-42).

-
GA21894
Gastric Inhibitory Polypeptide (3-42) (human)
Gastric Inhibitory Polypeptide (3-42) (human) acts as a glucose-dependent insulinotropic polypeptide (GIP) receptor antagonist, moderating the insulin secreting and metabolic actions of GIP in vivo.

-
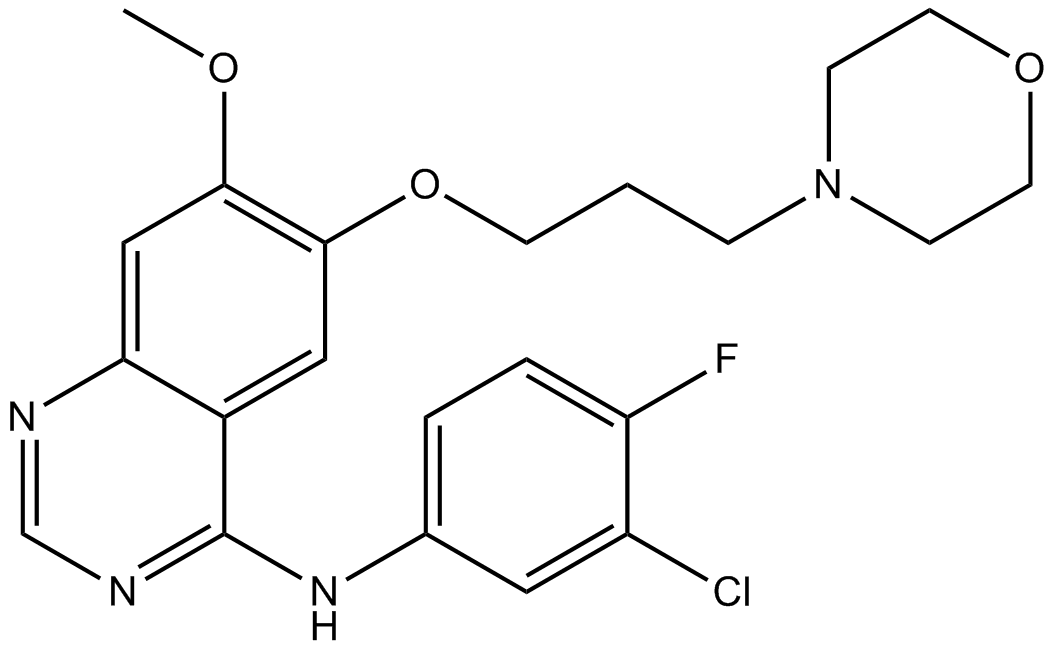
GC16737
Gefitinib (ZD1839)
Gefitinib (ZD1839), is a potent EGFR-TKI (EGFR tyrosine kinase inhibitor)
-
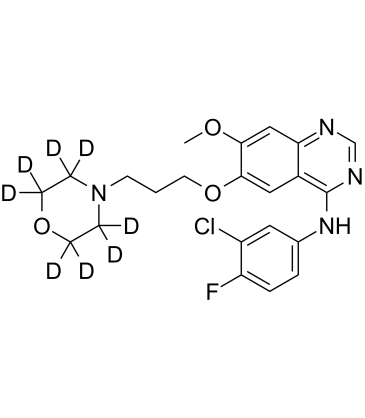
GC60868
Gefitinib D8
Gefitinib D8 (ZD1839 D8) is a deuterium labeled Gefitinib. Gefitinib is an EGFR tyrosine kinase inhibitor, with IC50 of 2-37 nM in NR6wtEGFR cells.
-
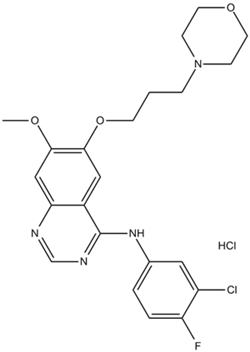
GC14295
Gefitinib hydrochloride
An EGFR inhibitor
-
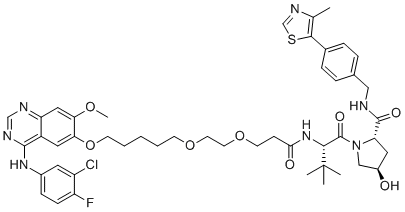
GC19509
Gefitinib-based PROTAC 3
A VHL-recruiting PROTAC
-
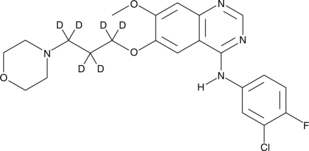
GC47395
Gefitinib-d6
An internal standard for the quantification of gefitinib
-
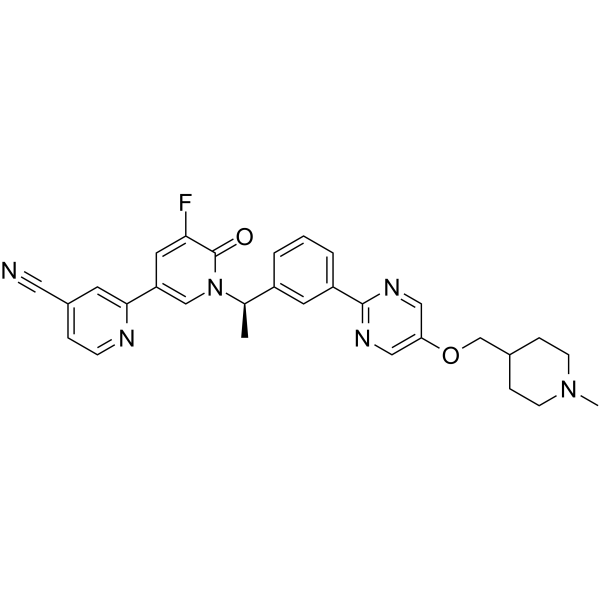
GC65478
Gemnelatinib
Gemnelatinib is a tyrosine kinase inhibitor (WO2018077227, implementation example 1). Gemnelatinib can be used for the research of cancer.
-
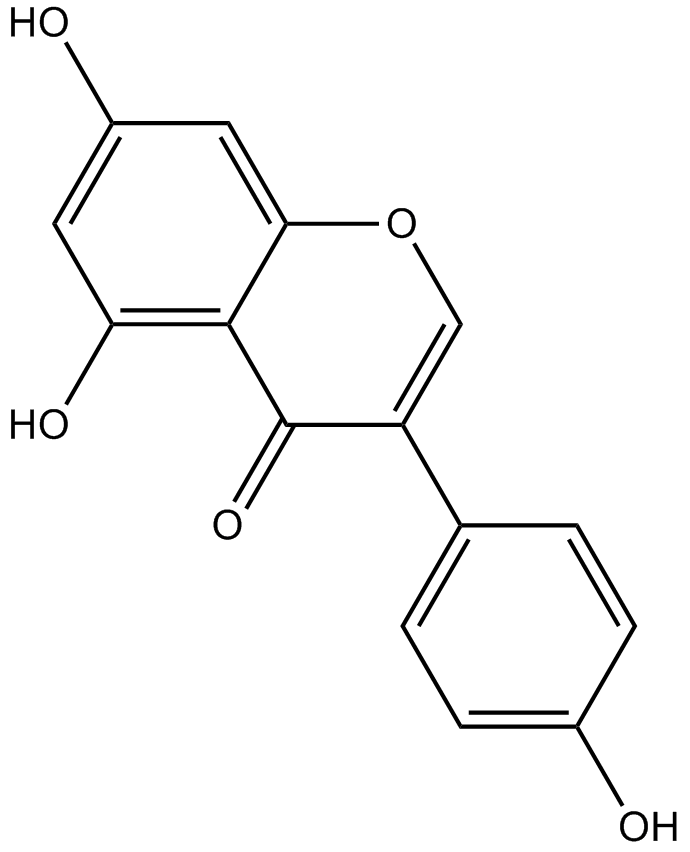
GC14102
Genistein
Genistein is an isoflavone belonging to the flavonoid group of compounds and is found in a number of plants.
-
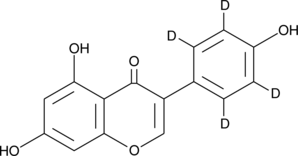
GC47398
Genistein-d4
An internal standard for the quantification of genistein
-
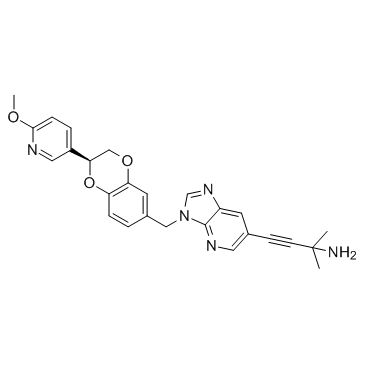
GC31842
GENZ-882706 (RA03546849)
GENZ-882706 (RA03546849) is a potent colony stimulating factor-1 receptor (CSF-1R) Inhibitor extracted from patent WO 2017015267A1.
-
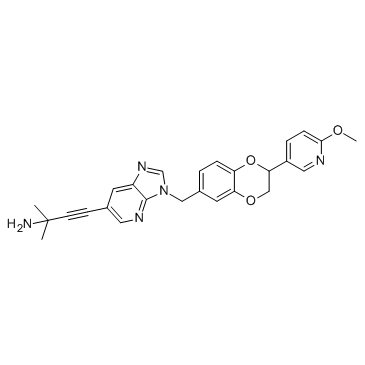
GC30295
GENZ-882706(Raceme) (GENZ-882706 racemate)
GENZ-882706(Raceme) (GENZ-882706 racemate) is the racemate of GENZ-882706.
-
GC62633
GGTI-2154
GGTI-2154 is a potent and selective inhibitor of geranylgeranyltransferase I (GGTase I), with an IC50 of 21 nM. GGTI-2154 shows more than 200-fold selectivity for GGTase I over FTase (IC50=5600 nM). GGTI-2154 can be used for the research of cancer.
-
GC19482
Gilteritinib
Gilteritinib (ASP2215, Xospata) for relapsed and /or refractory AML (R/R AML).
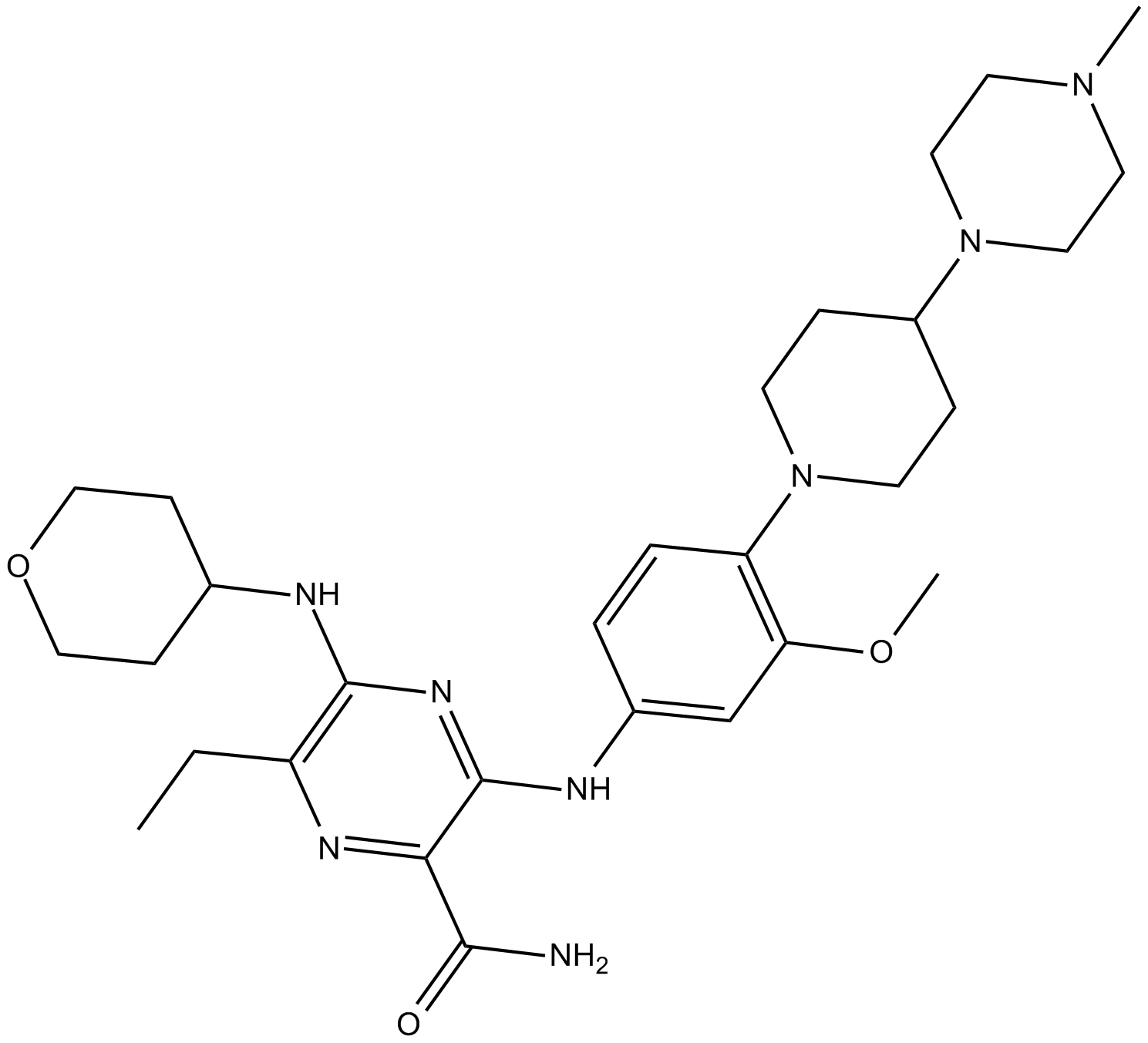
-
GC36135
Gilteritinib hemifumarate
Gilteritinib (ASP2215) hemifumarate is a potent and ATP-competitive FLT3/AXL inhibitor with IC50 of 0.29 nM/0.73 nM, respectively.
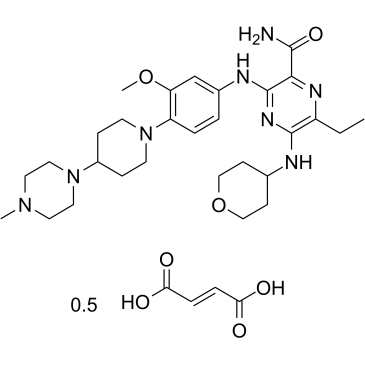
-
GN10156
Ginsenoside Rb1
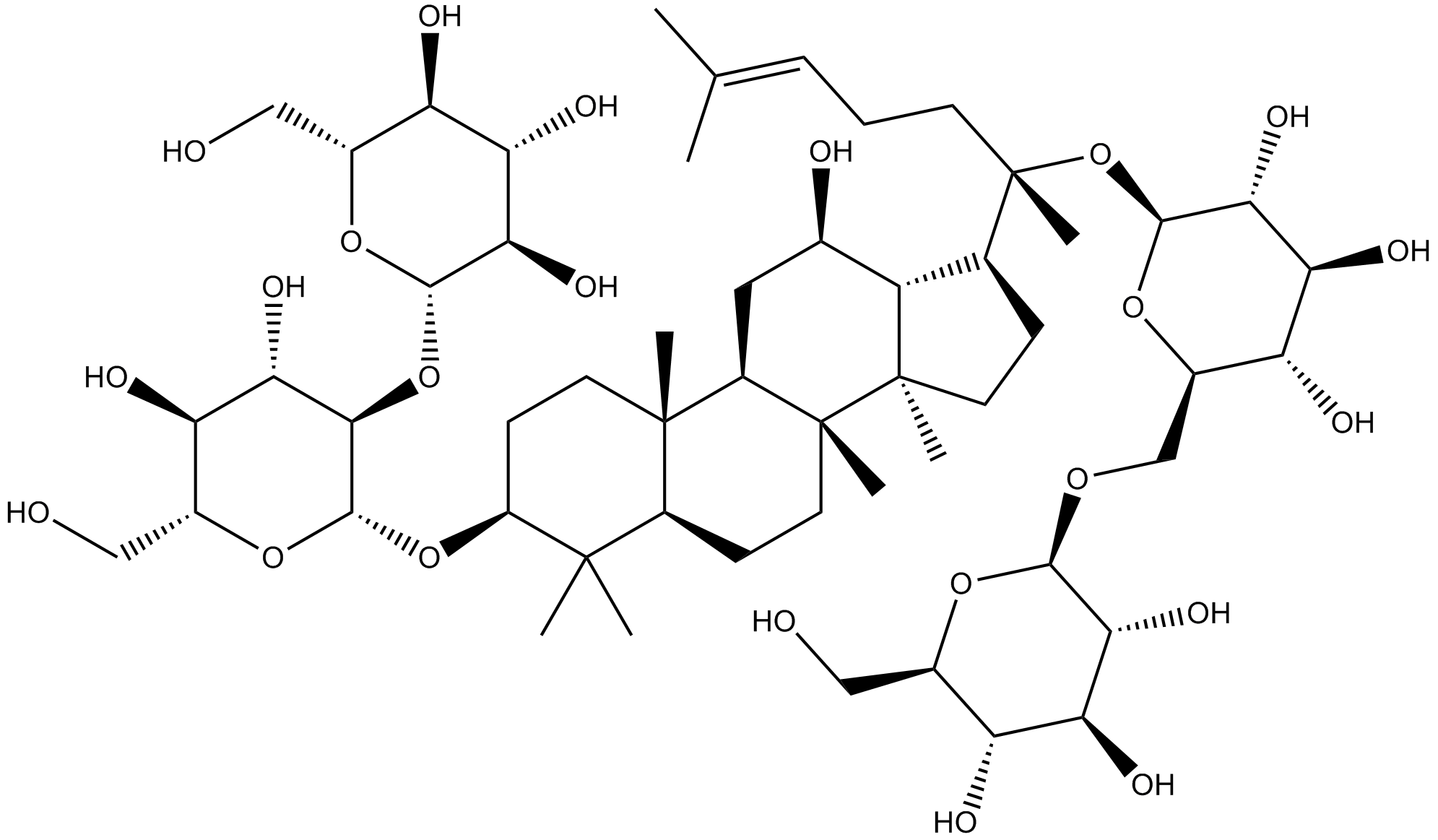
-
GC31706
Ginsenoside Rg5
A ginsenoside with diverse biological activities
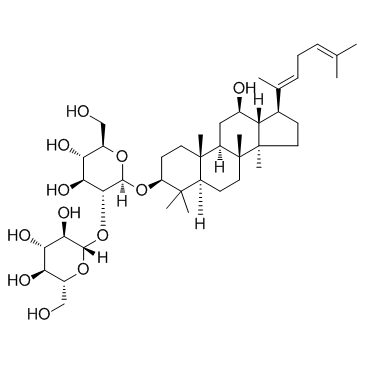
-
GC61569
GIP (1-30) amide, porcine TFA
GIP (1-30) amide, porcine TFA is a full glucose-dependent insulinotropic polypeptide (GIP) receptor agonist with high affinity equal to native GIP(1-42).

-
GC60175
GIP (1-30) amide,Human acetate
GIP (1-30) amide,human acetate is a glucose-dependent insulinotropic polypeptide (GIP) fragment.
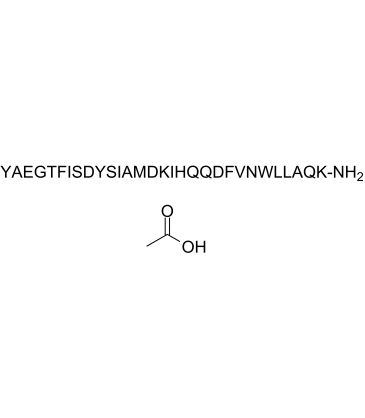
-
GC61741
GIP, human TFA
GIP, human TFA, a peptide hormone consisting of 42 amino acids, is a stimulator of glucose-dependent insulin secretion and a weak inhibitor of gastric acid secretion.

-
GC19166
Glesatinib hydrochloride
Glesatinib hydrochloride is an inhibitor of the MET and Axl receptor tyrosine kinase pathways, which drive tumour growth when altered.
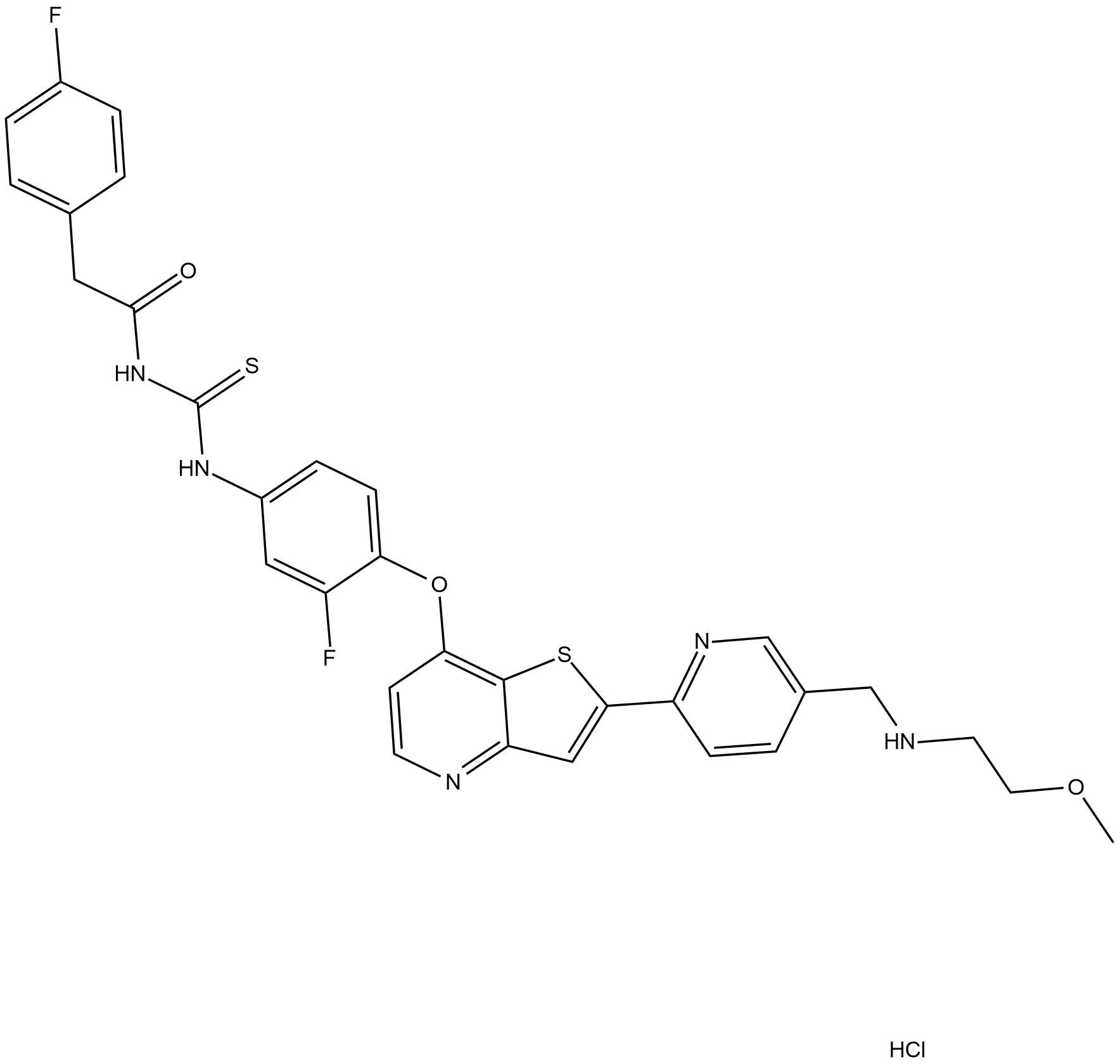
-
GC64947
Glumetinib
Glumetinib (SCC244) is a highly selective, orally bioavailable, ATP-competitive c-Met inhibitor with an IC50 of 0.42 nM. Glumetinib has greater than 2400-fold selectivity for c-Met over those 312 kinases evaluated, including the c-Met family member RON and highly homologous kinases Axl, Mer, TyrO3. Antitumor activity.
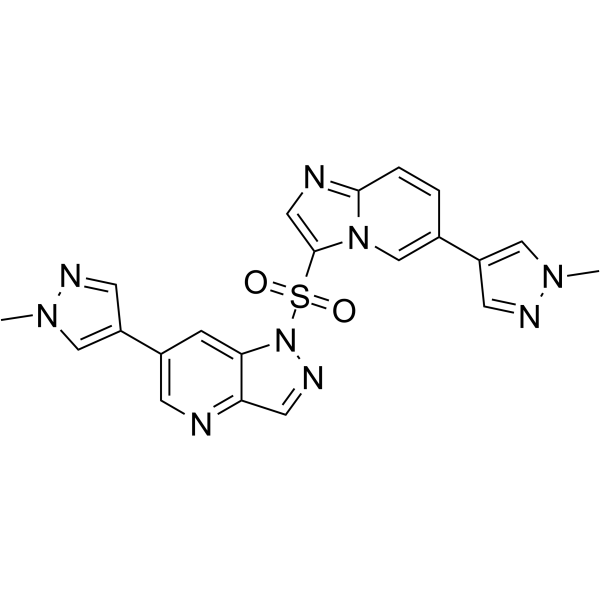
-
GC36167
GMB-475
GMB-475 is a degrader of BCR-ABL1 tyrosine kinase based on PROTAC, overcoming BCR-ABL1-dependent drug resistance. GMB-475 targets BCR-ABL1 protein and recruits the E3 ligase Von Hippel Lindau (VHL), resulting in ubiquitination and subsequent degradation of the oncogenic fusion protein.
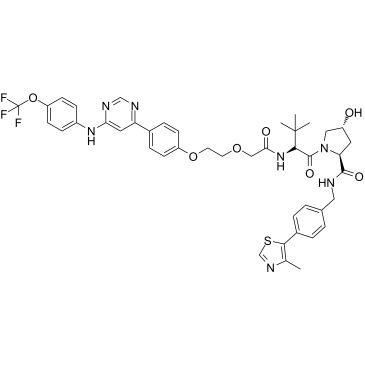
-
GC31871
GNE-0946
GNE-0946 is a potent and selective RORγ( RORc) agonist with an EC50 value of 4 nM for HEK-293 cell.
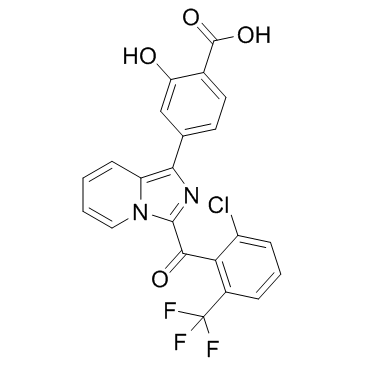
-
GC32053
GNE-4997
GNE-4997 is a potent and selective interleukin-2-inducible T-cell kinase (ITK) inhibitor with a Ki of 0.09 nM, and the correlation between the basicity of solubilizing elements in GNE-4997 and off-target antiproliferative effects reduces cytotoxicity.
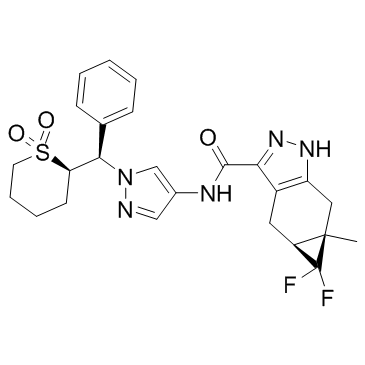
-
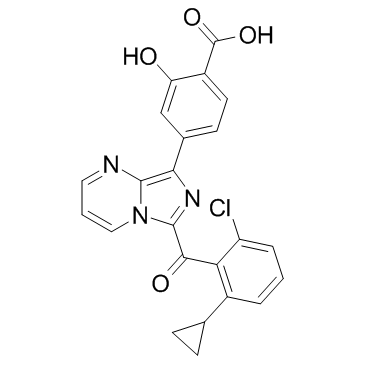
GC33882
GNE-6468
GNE-6468 is a highly potent and selective RORγ (RORc) inverse agonist with an EC50 value of 13 nM for HEK-293 cell.
-
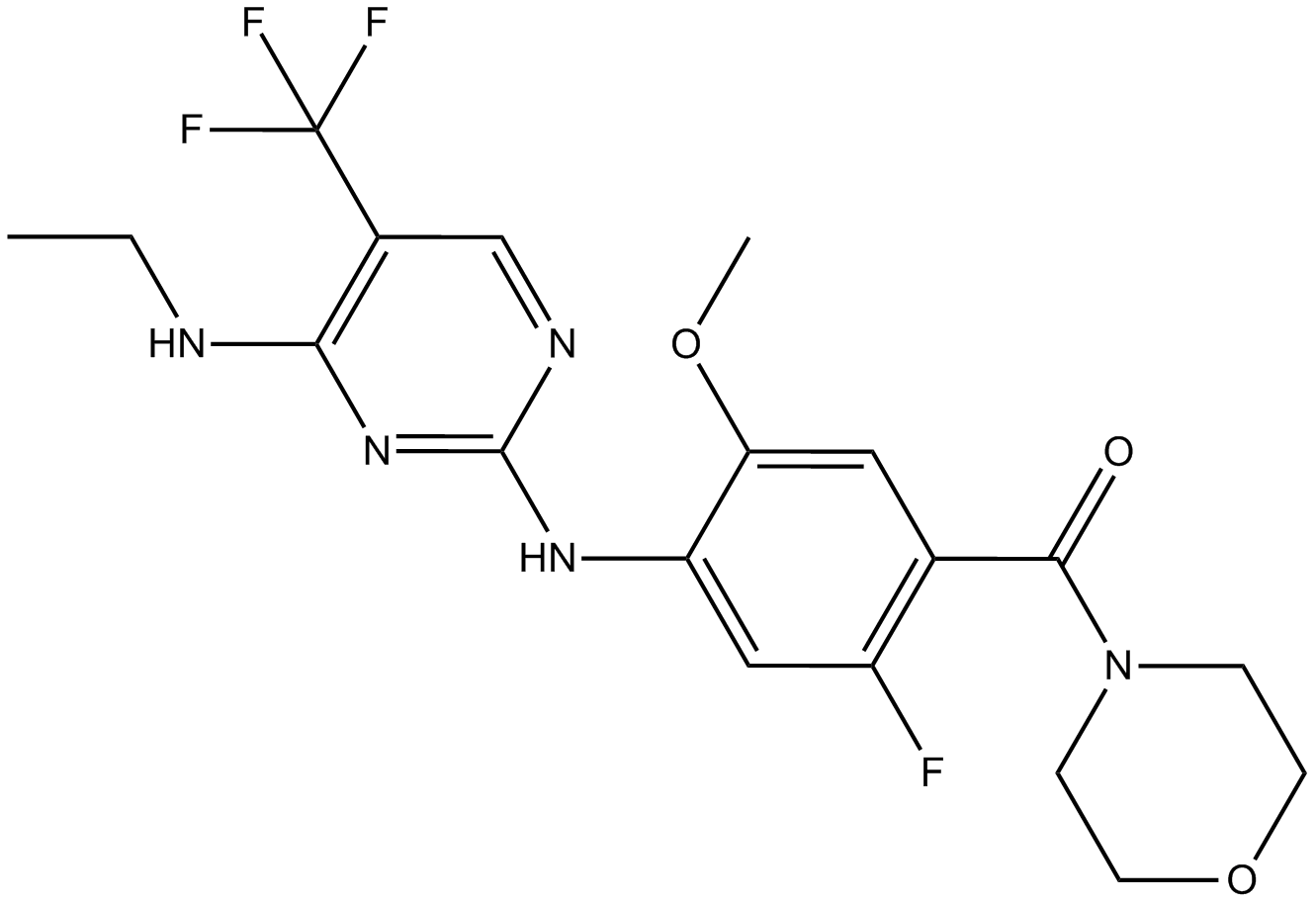
GC12121
GNE-7915
Potent and selective LRRK2 inhibitor
-
GC36171
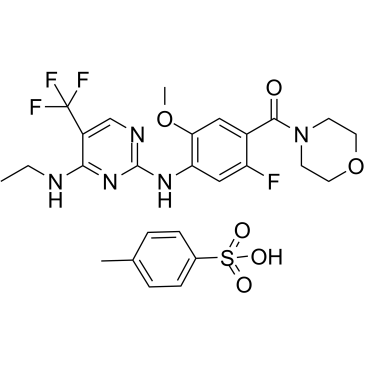
GNE-7915 tosylate
GNE-7915 tosylate is a potent, selective and brain-penetrant inhibitor of LRRK2 with an IC50 of 9 nM.
-
GC13868
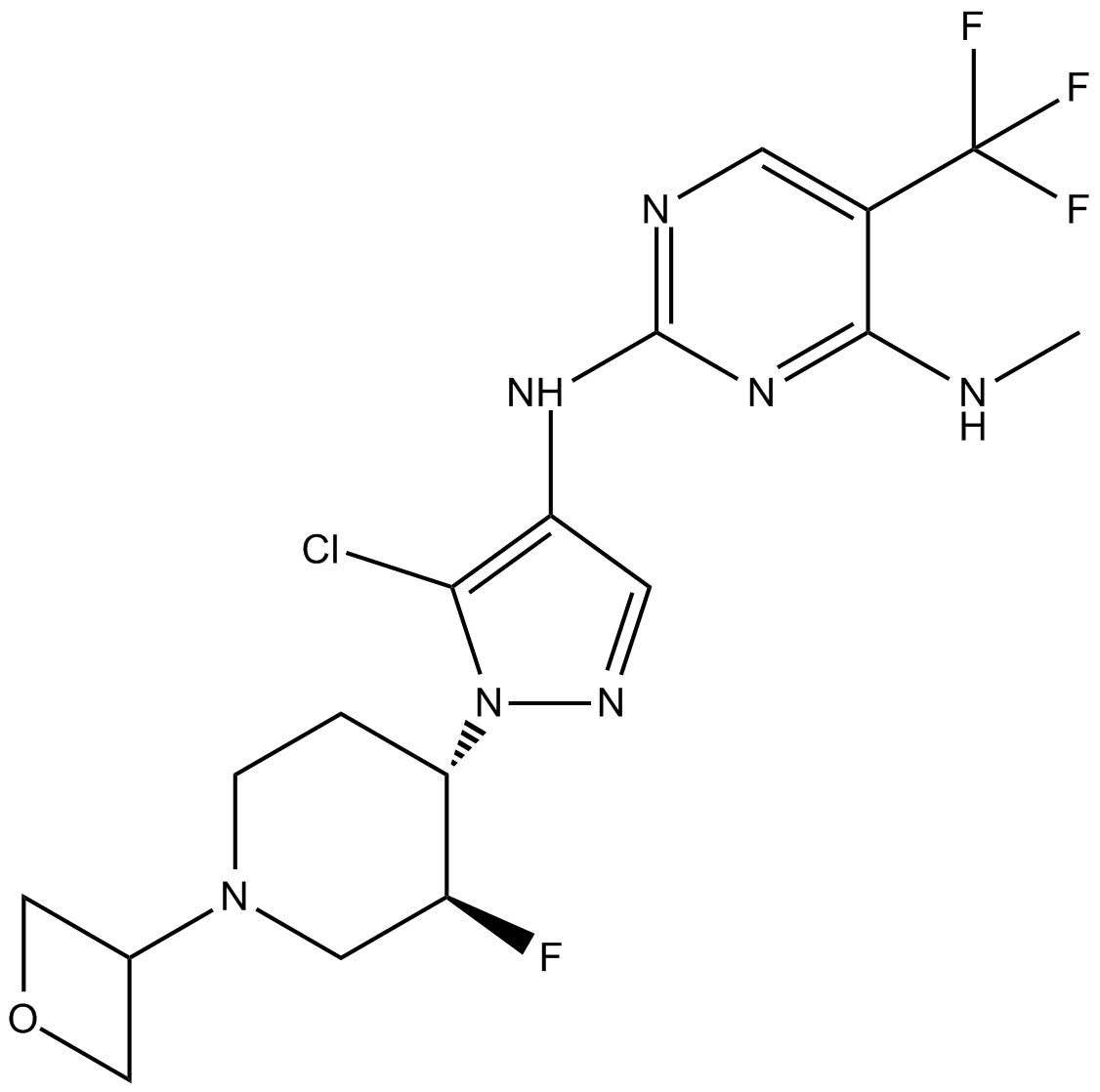
GNE-9605
LRRK2 inhibitor, brain-penetrant, potent and selective
-
GC15196
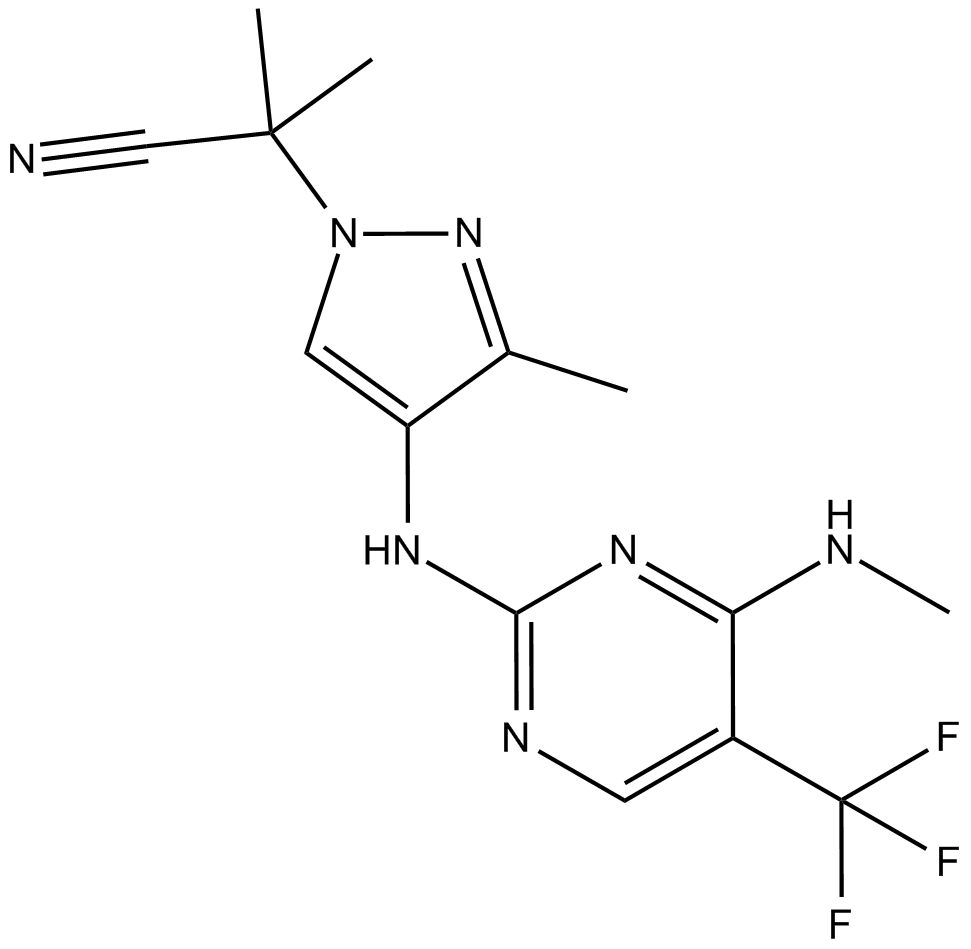
GNE0877
Potent and selective LRRK2 inhibitor
-
GC10858
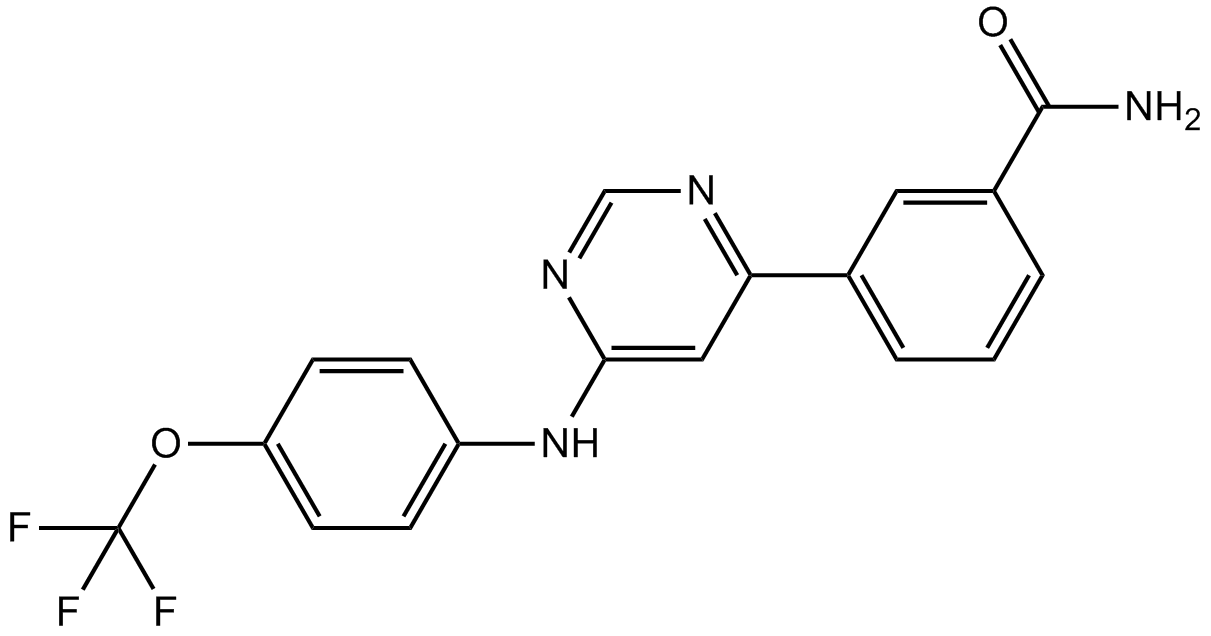
GNF 2
Bcr-Abl inhibitor
-
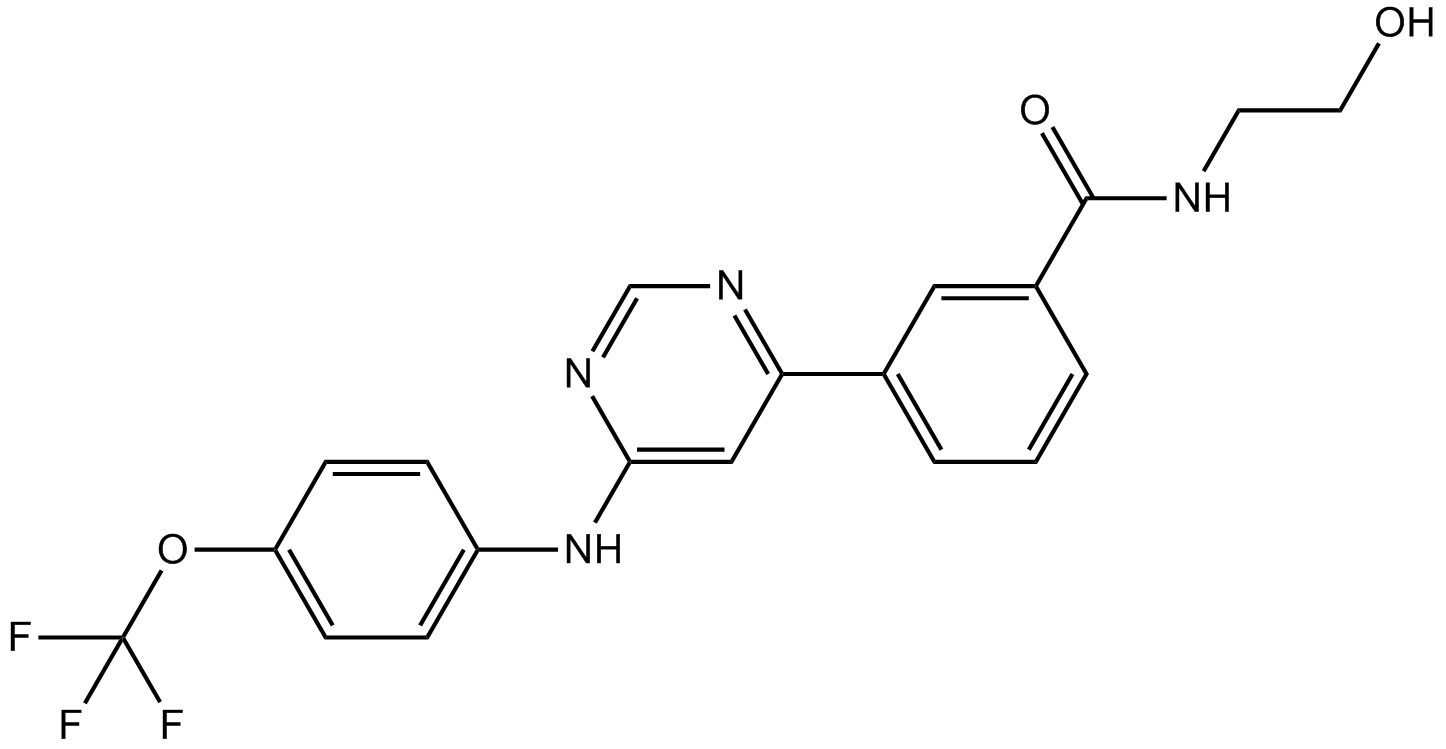
GC15079
GNF 5
Bcr-Abl inhibitor
-
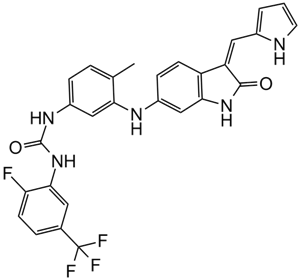
GC16343
GNF-5837
Pan-Trk inhibitor
-
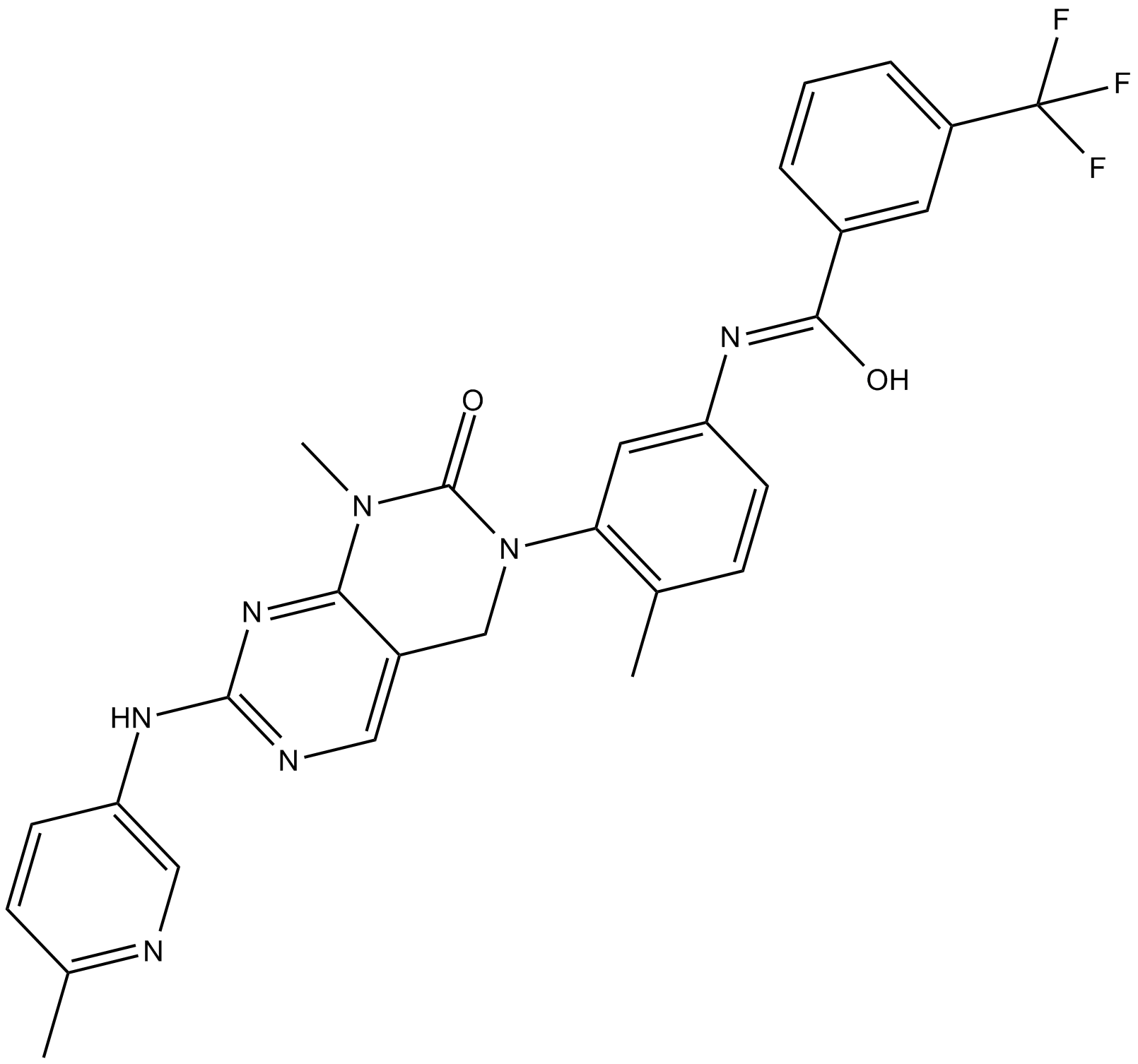
GC10607
GNF-7
Type II Bcr-Abl inhibitor
-
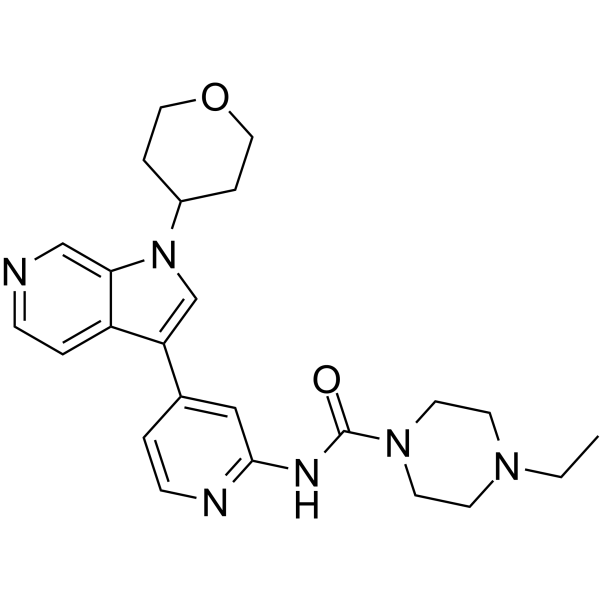
GC64724
GNF2133
GNF2133 is a potent, selective and orally active DYRK1A inhibitor with IC50s of 0.0062, >50 ?M for DYRK1A and GSK3β, respectively.
-
GC64725
GNF2133 hydrochloride
GNF2133 hydrochloride is a potent, selective and orally active DYRK1A inhibitor with IC50s of 0.0062, >50 ?M for DYRK1A and GSK3β, respectively.
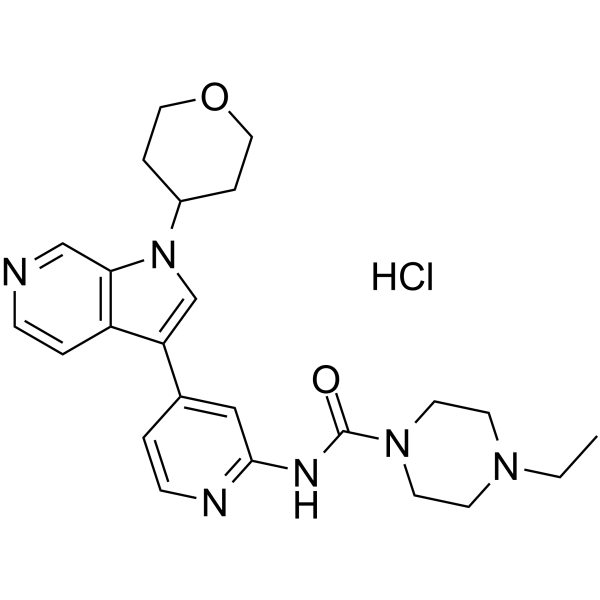
-
GC38787
GNF4877
GNF4877 is a potent DYRK1A and GSK3β inhibitor with IC50s of 6?nM and 16?nM, respectively, which leads to blockade of nuclear factor of activated T-cells (NFATc) nuclear export and increased β-cell proliferation (EC50 of 0.66?μM for mouse β (R7T1) cells).
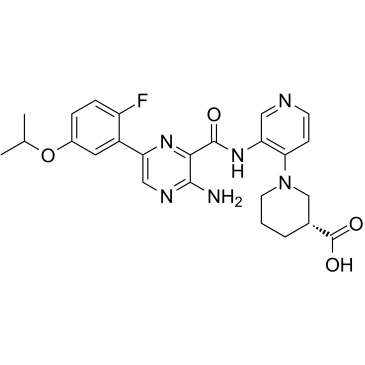
-
GC17715
Golvatinib (E7050)
Golvatinib (E7050) (E-7050) is a potent dual inhibitor of both c-Met and VEGFR2 kinases with IC50s of 14 and 16 nM, respectively.
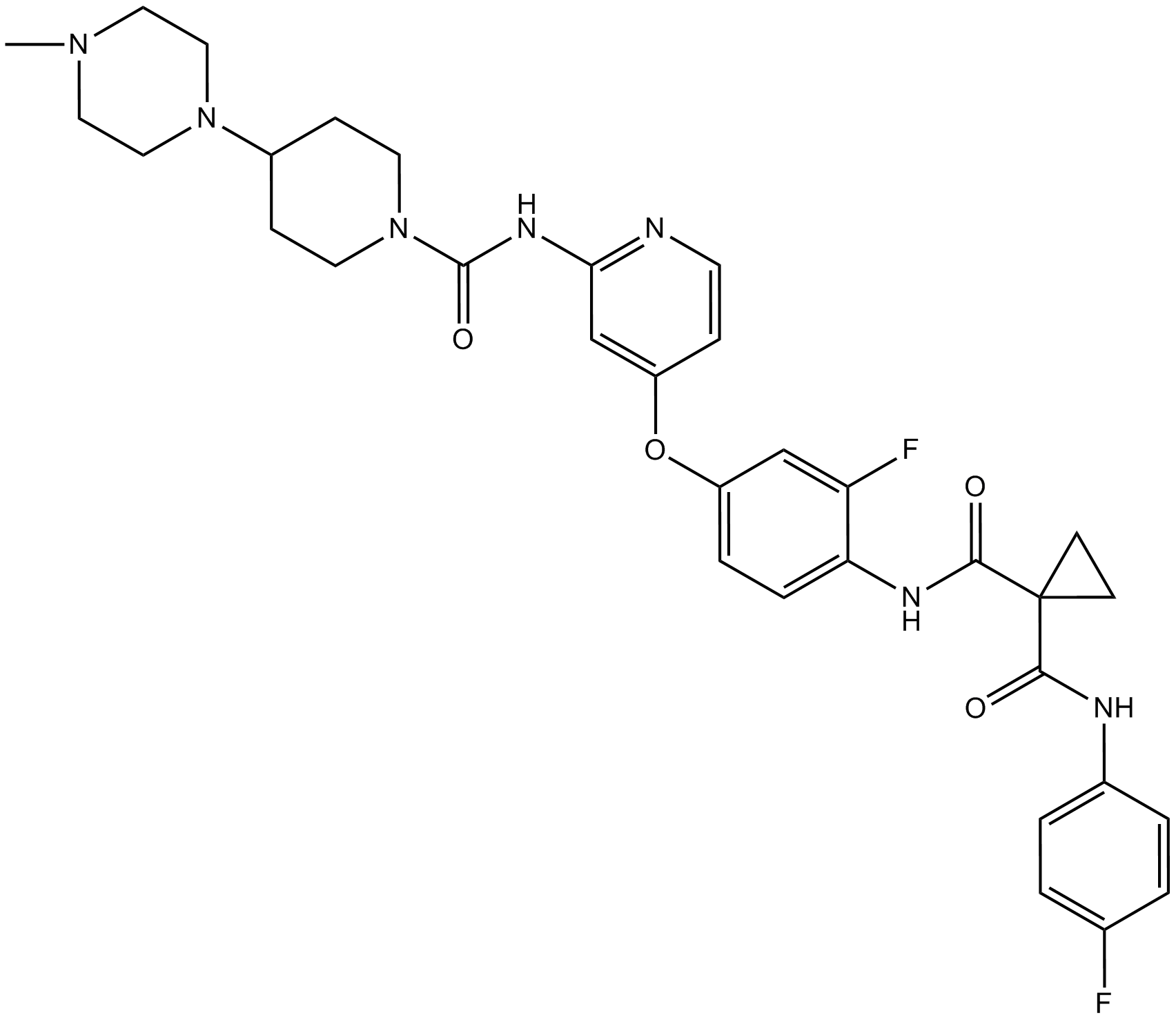
-
GC16430
GS-9973
GS-9973 (GS-9973) is an orally bioavailable, selective Syk inhibitor with an IC50 of 7.7 nM.
-
GC50429
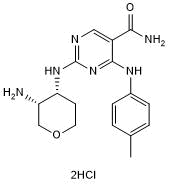
GSK 143
Syk inhibitor
-
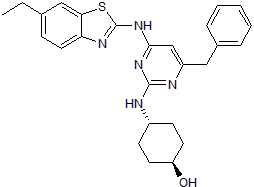
GC50156
GSK 2250665A
Itk inhibitor
-
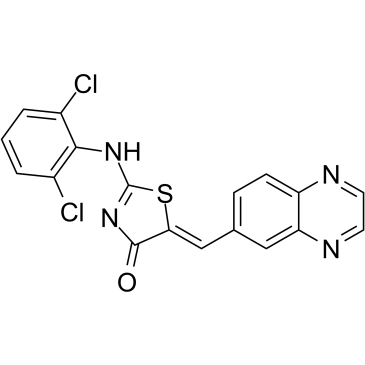
GC36194
GSK-626616
GSK-626616 is a potent, orally bioavailable inhibitor of DYRK3 (IC50=0.7 nM).
-
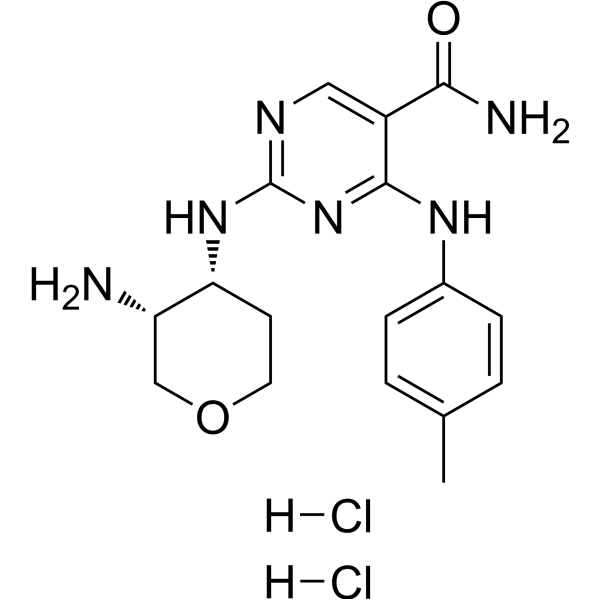
GC68447
GSK143 dihydrochloride
-
GC12273
GSK1838705A
IGF-IR/IR/ALK inhibitor, ATP-competitive
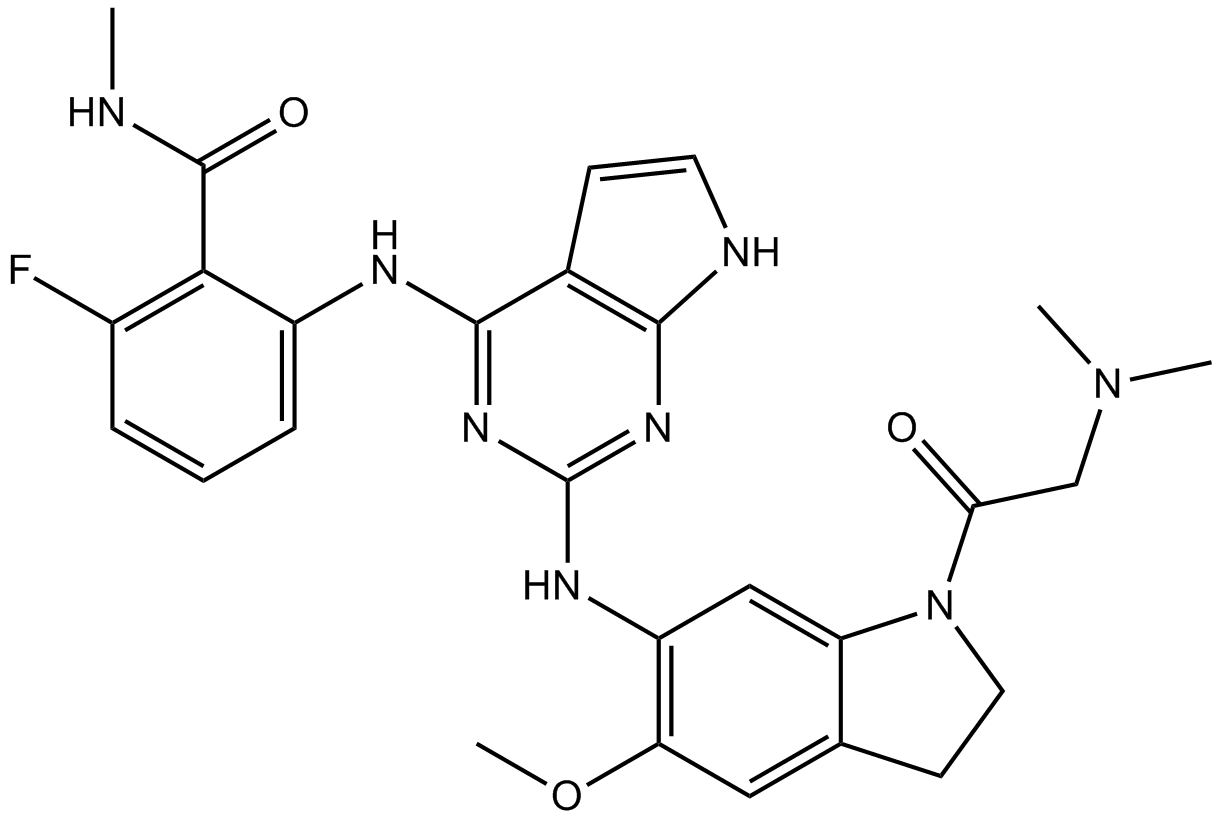
-
GC17612
GSK1904529A
Selective IGF-1R/IR inhibitor
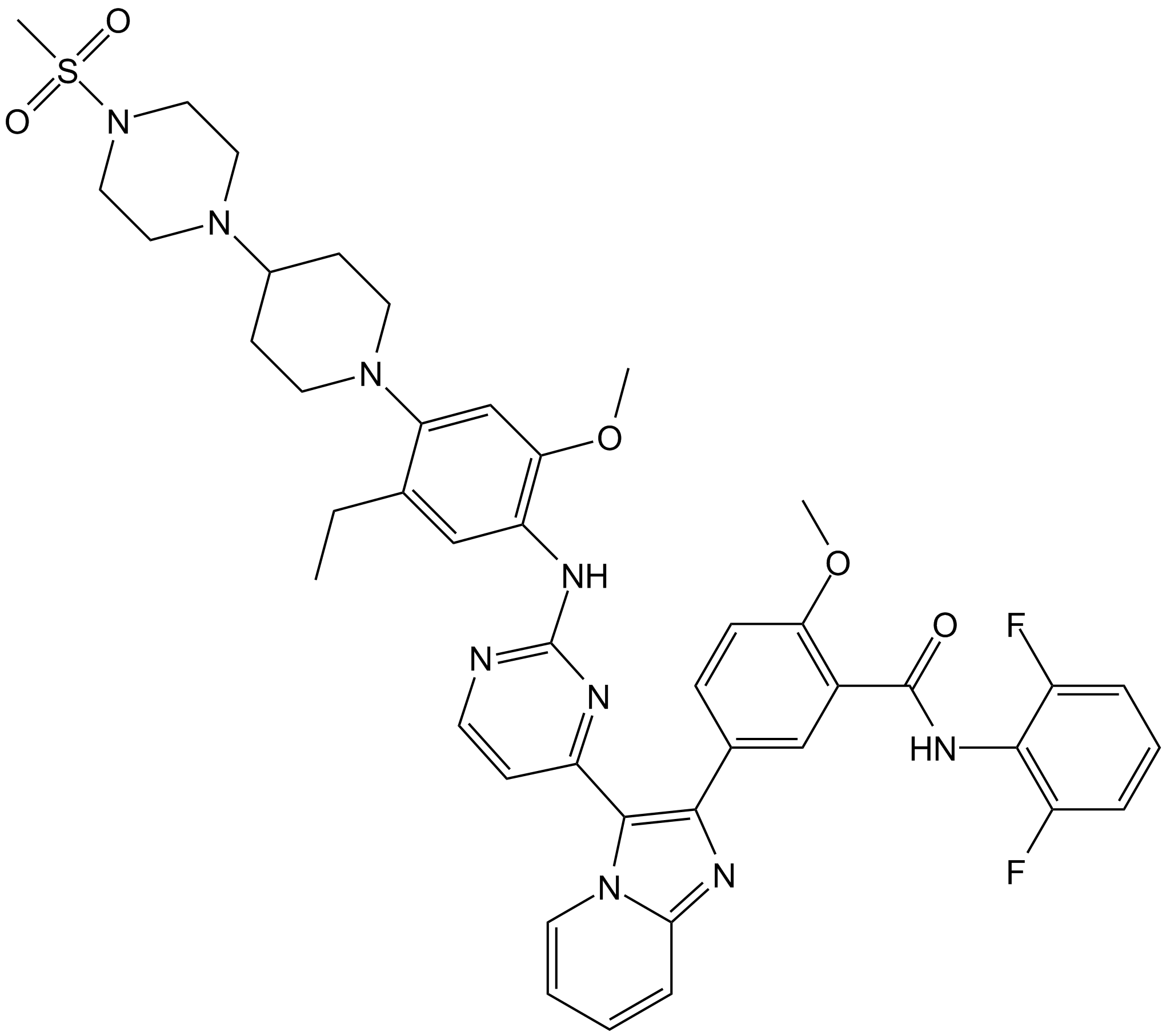
-
GC64888
GSK215
GSK215 is a potent and selective PROTAC focal adhesion kinase (FAK) degrader with a pDC50 of 8.4. GSK215 is designed by a binder for the VHL E3 ligase and the FAK inhibitor VS-4718. GSK215 induces rapid and prolonged FAK degradation, giving a long-lasting effect on FAK levels and a marked pharmacokinetic/pharmacodynamics (PK/PD) disconnect.
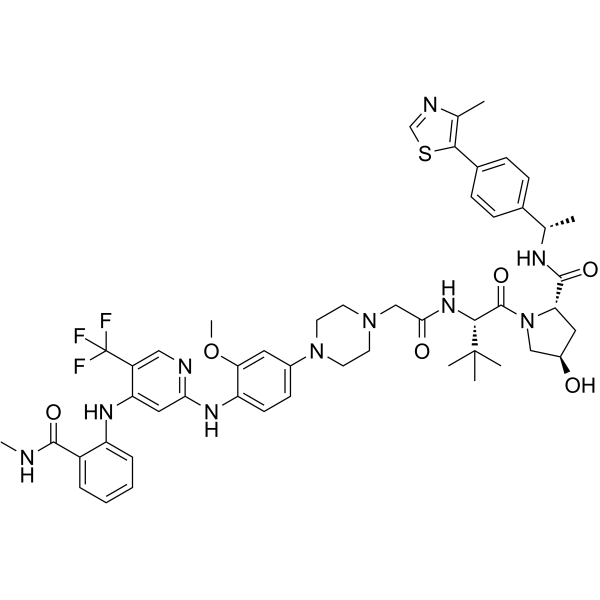
-
GC19179
GSK2256098
GSK2256098 is a selective FAK kinase inhibitor, which inhibits growth and survival of pancreatic ductal adenocarcinoma cells.
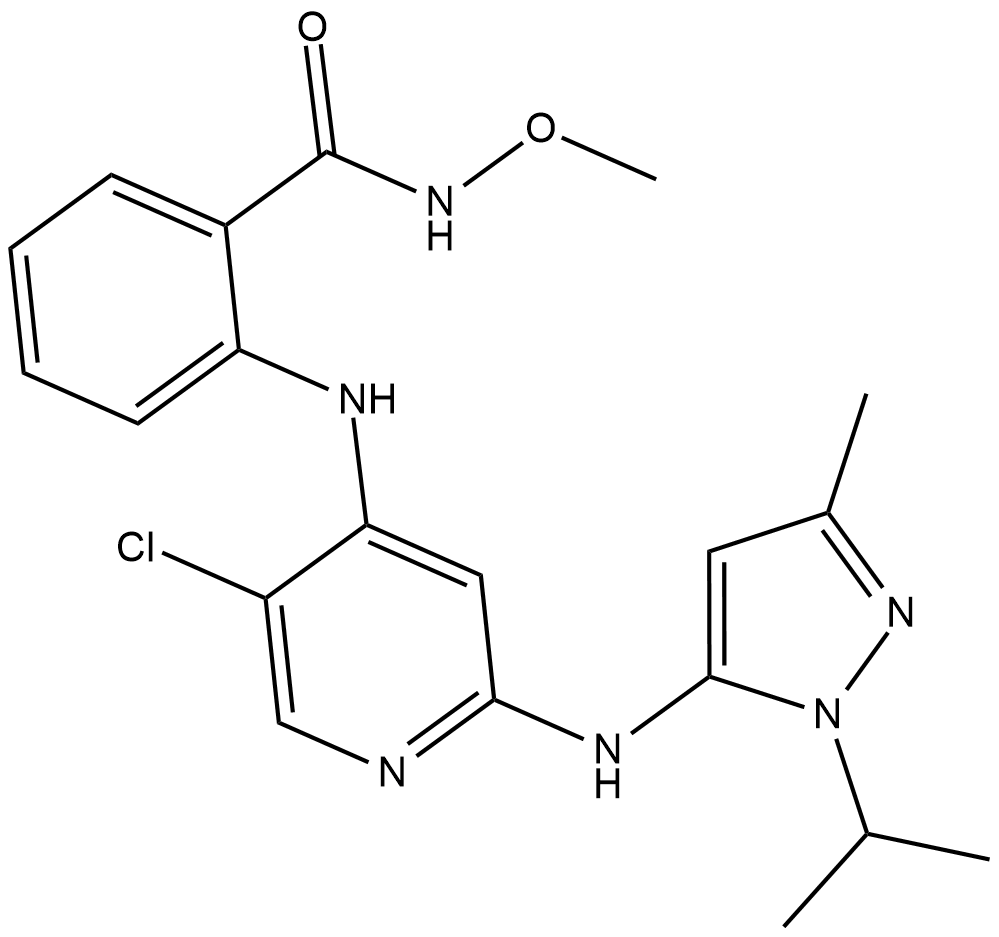
-
GC18049
GSK2578215A
LRRK2 inhibitor
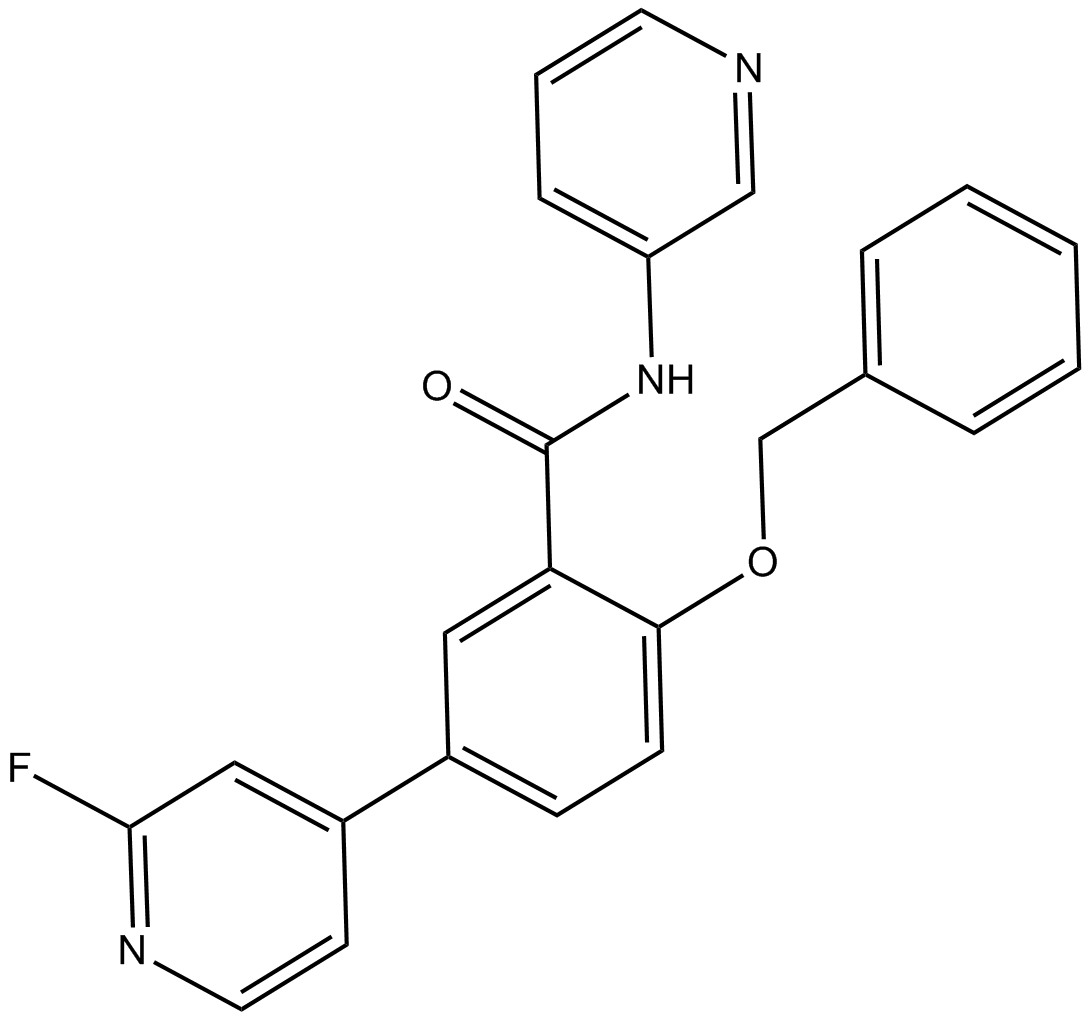
-
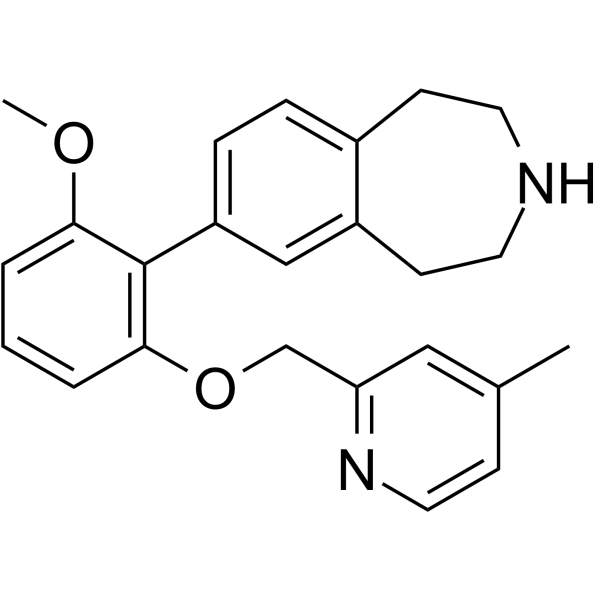
GC64889
GSK2646264
GSK2646264 (Compound 44) is a potent and selective spleen tyrosine kinase (SYK) inhibitor with a pIC50 of 7.1.
-
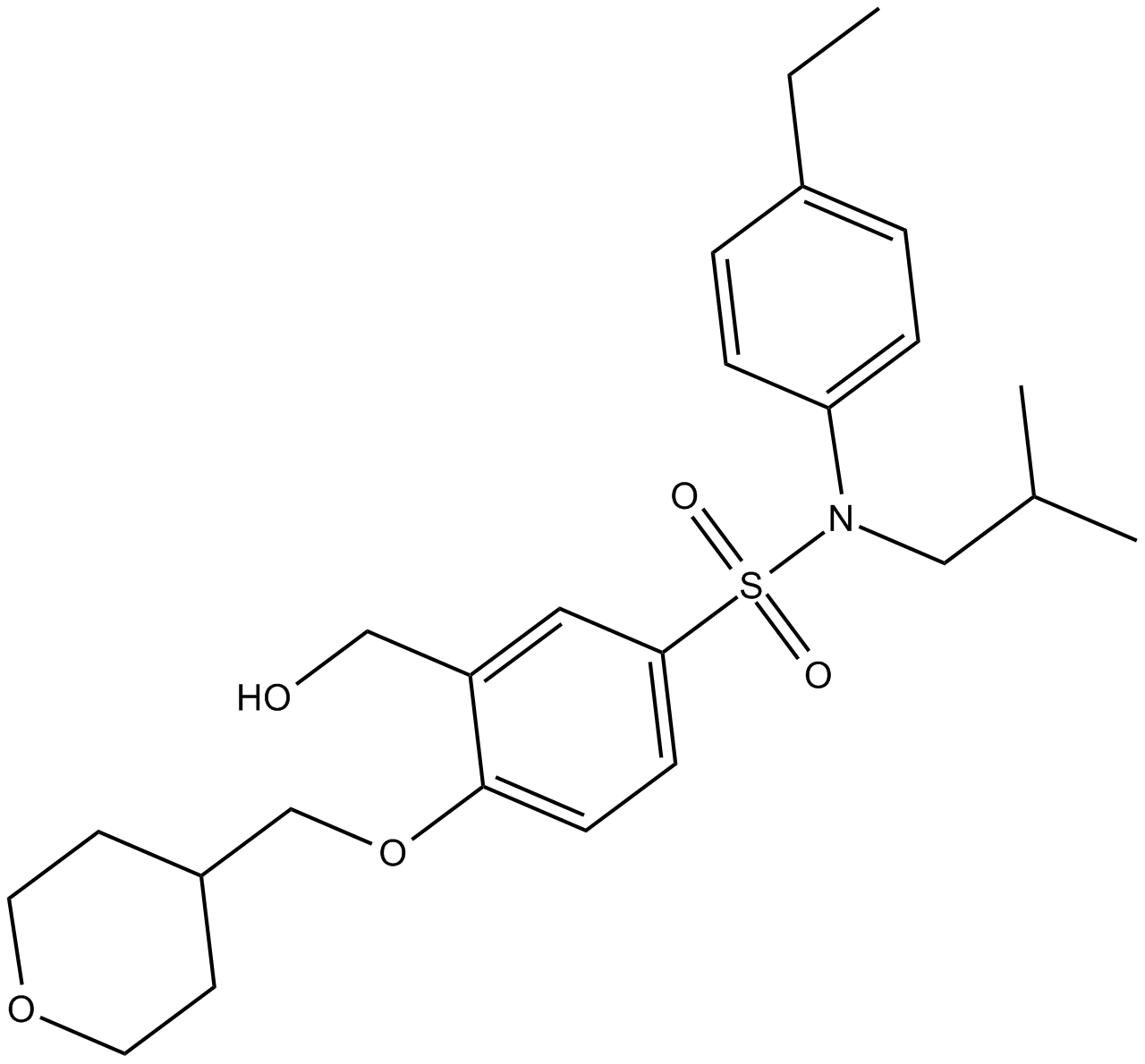
GC13905
GSK2981278
RORγ-selective inverse agonist
-
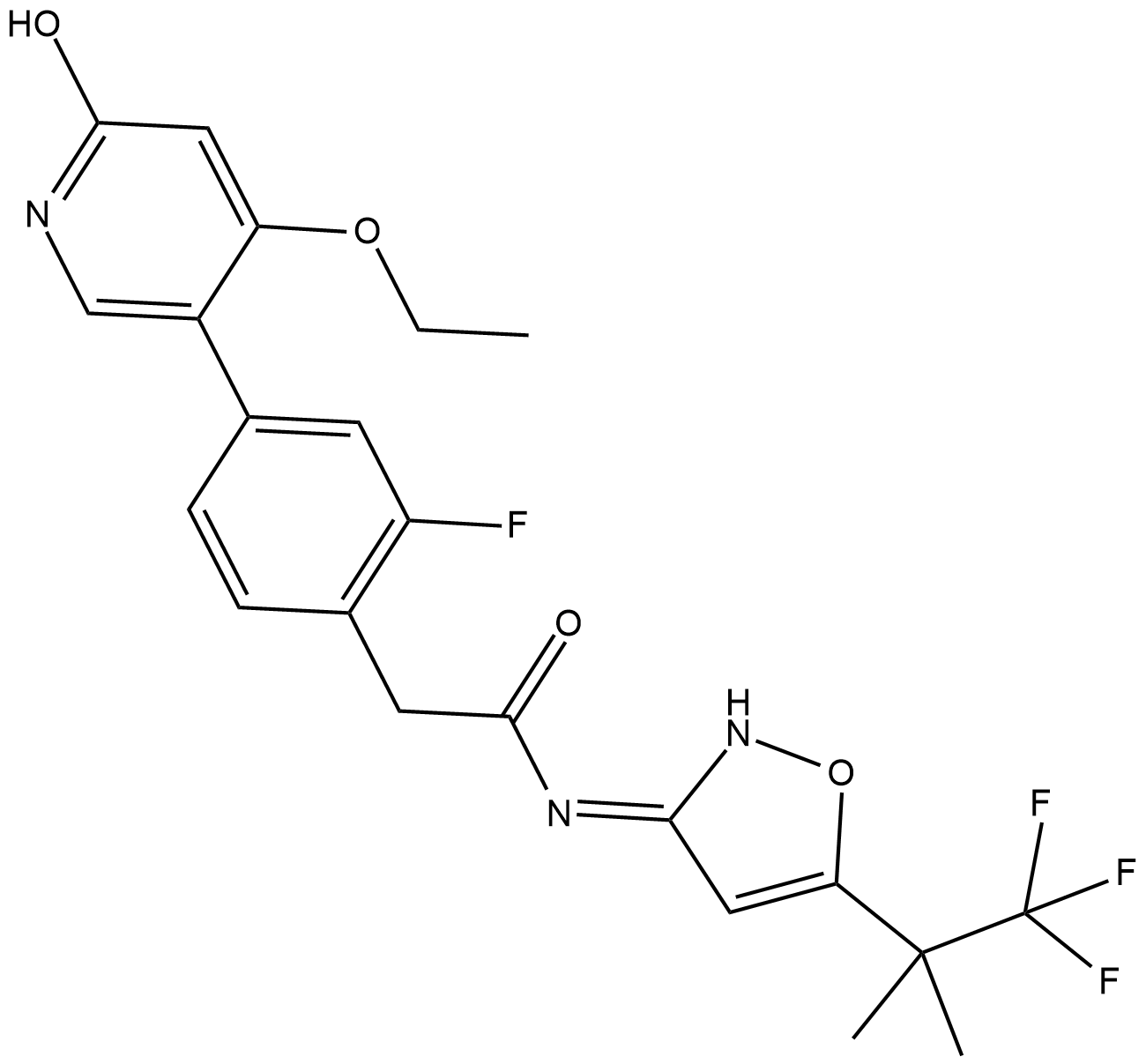
GC18492
GSK3179106
A RET kinase inhibitor
-
GC18591
GSK805
GSK805 is a potent, orally bioavailable retinoid-related orphan receptor gamma t (RORγt) inverse agonist that interacts with the receptor's putative ligand binding domain without exerting significant effects on DNA binding.
-
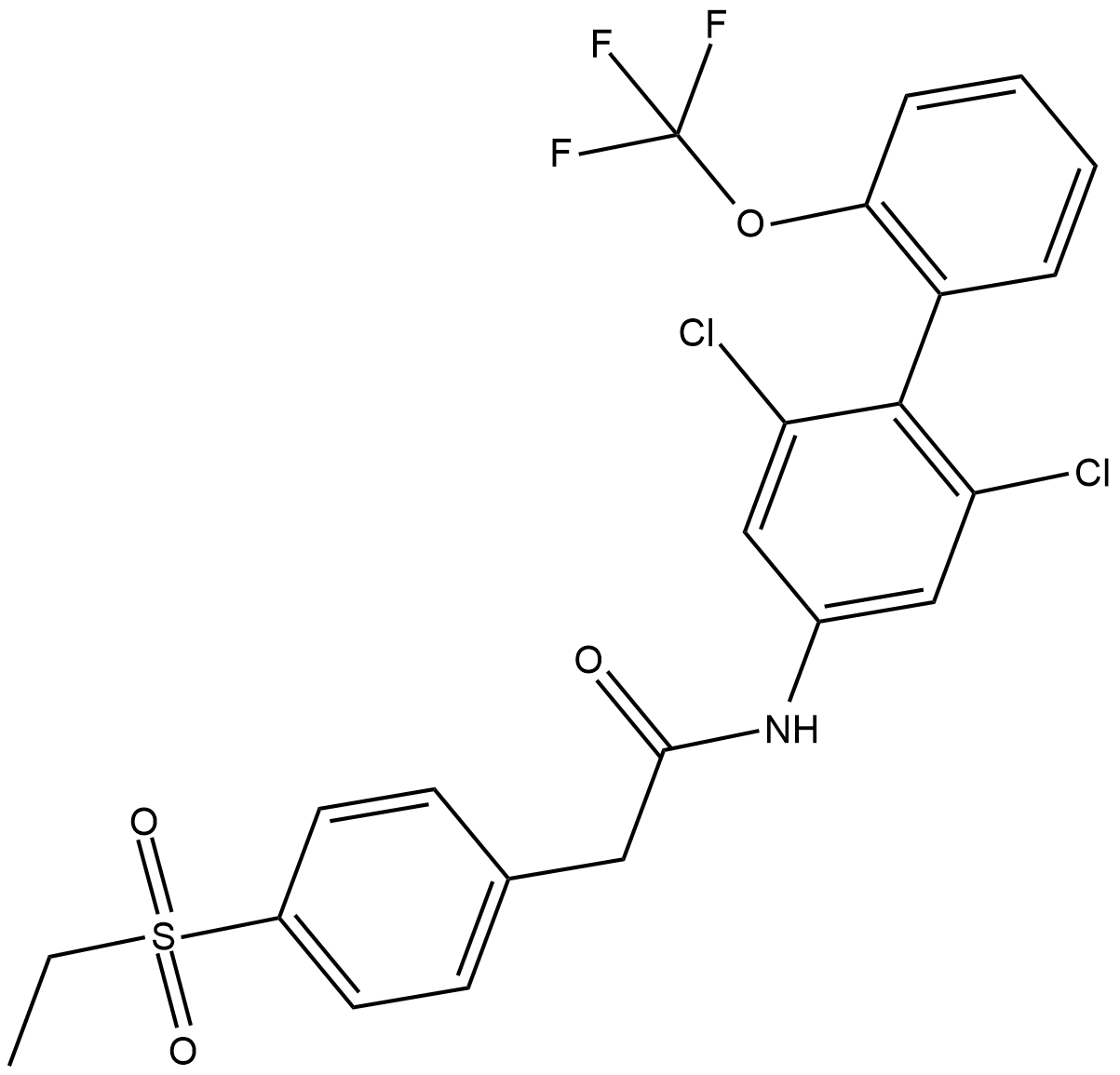
GC64875
Gunagratinib
Gunagratinib (ICP-192) is a low toxicity and orally active pan-FGFR (fibroblast growth factor receptors) inhibitor that potently and selectively inhibits FGFR activities irreversibly by covalent binding. Gunagratinib can be used for the research of cancer.
-
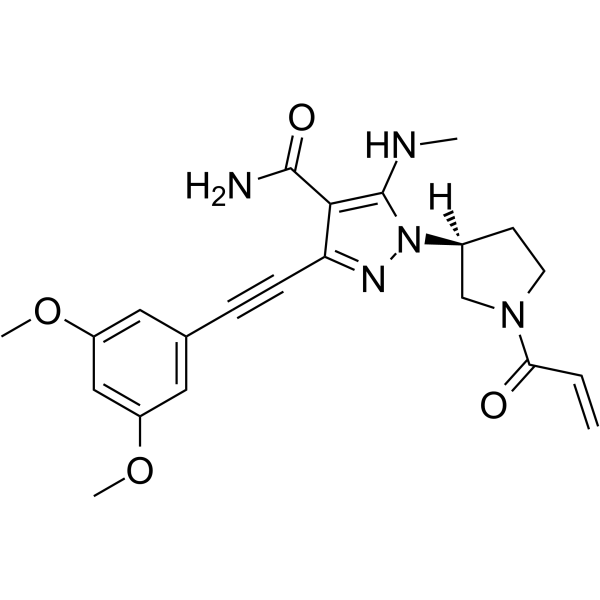
GC33041
Gusacitinib (ASN-002)
Gusacitinib (ASN-002) (ASN-002) is an orally active and potent dual inhibitor of spleen tyrosine kinase (SYK) and janus kinase (JAK) with IC50 values of 5-46 nM. Gusacitinib (ASN-002) has anti-cancer activity in both solid and hematological tumor types.
-
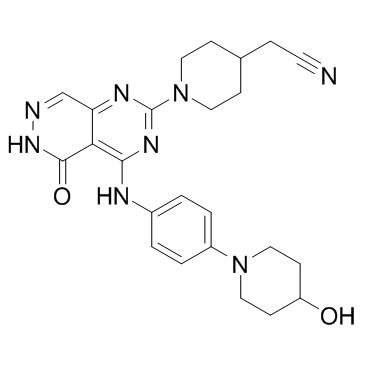
GC15927
GW 583340 dihydrochloride
dual EGFR/ErbB2 tyrosine kinase inhibitor
-
GC17286
GW2580
CFMS kinase/CSF-1R inhibitor,selective and ATP-competitive
-
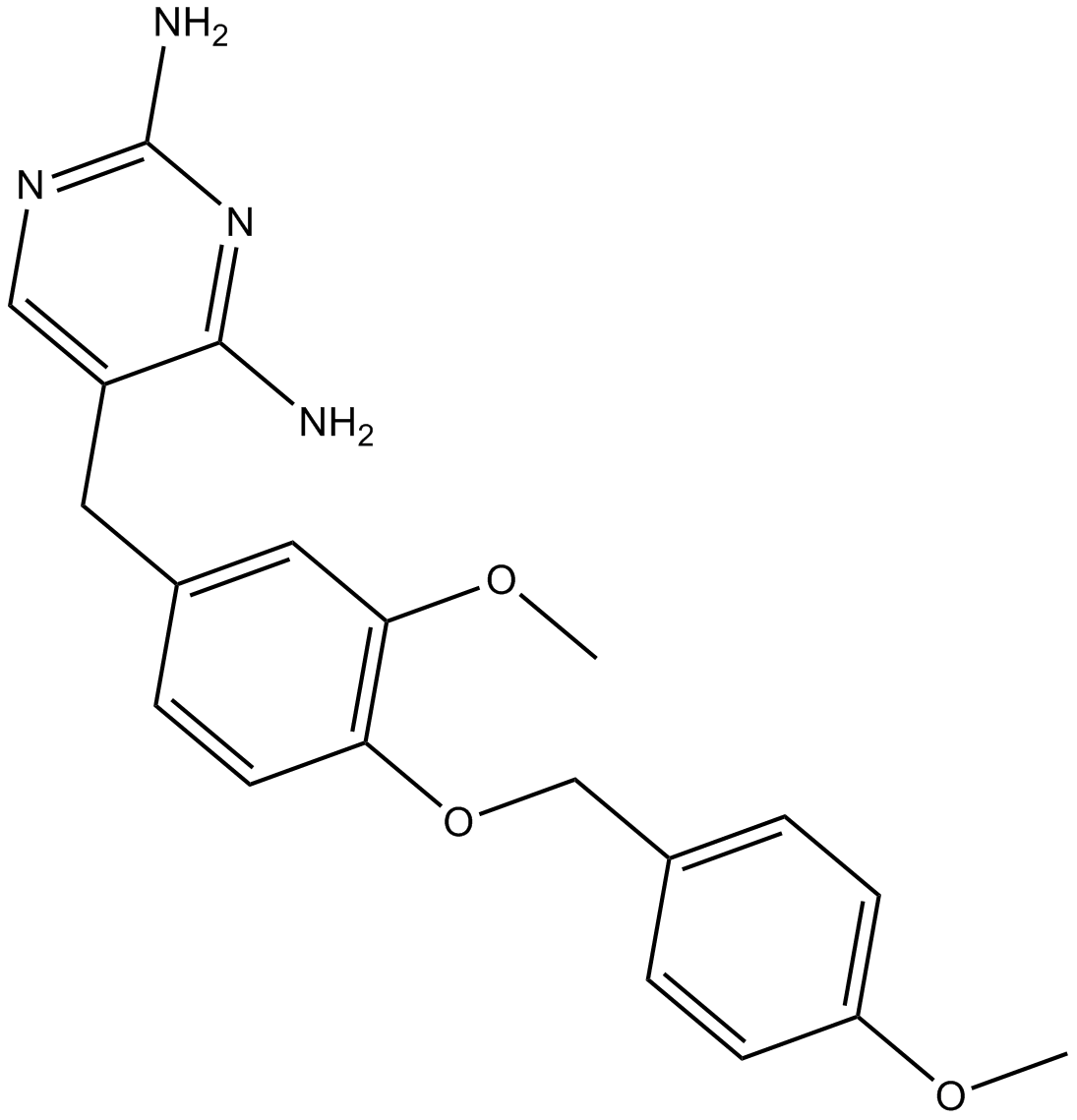
GC14123
GW441756
TrkA inhibitor,potent and selective
-
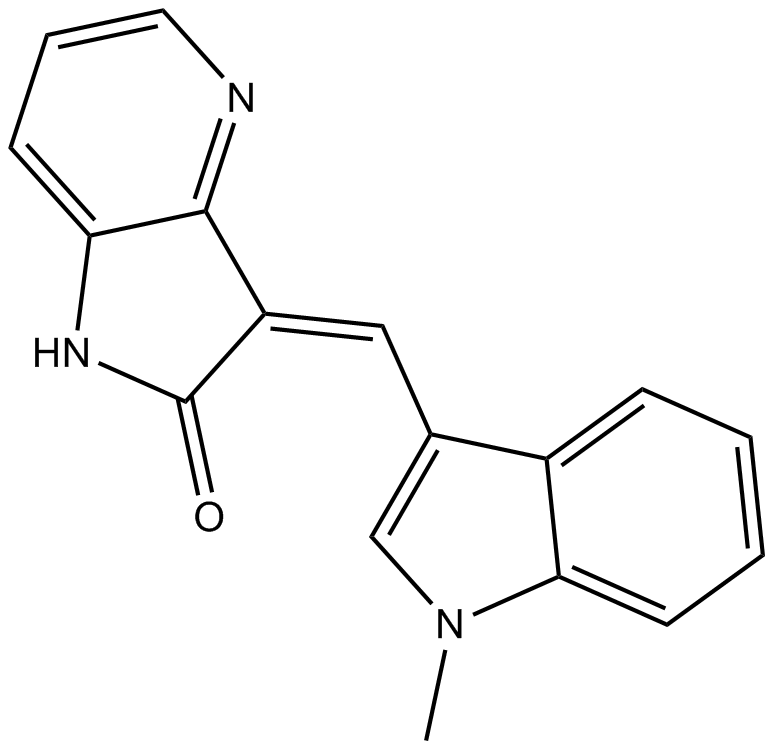
GC36203
GW806742X
GW806742X, an ATP mimetic and a potent MLKL (Mixed Lineage Kinase Domain-Like protein) inhibitor, binds the MLKL pseudokinase domain with a Kd of 9.3 μM.
-
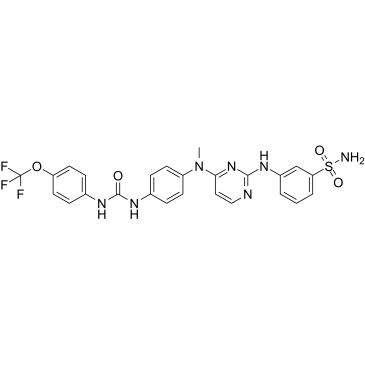
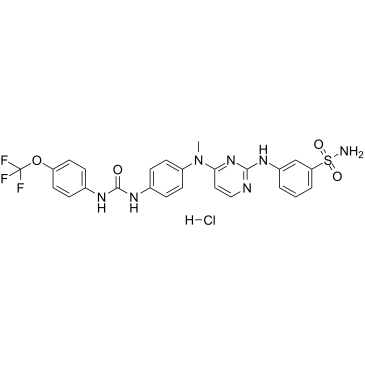
GC61454
GW806742X hydrochloride
GW806742X hydrochloride, an ATP mimetic and a potent MLKL (Mixed Lineage Kinase Domain-Like protein) inhibitor, binds the MLKL pseudokinase domain with a Kd of 9.3 μM. GW806742X hydrochloride has activity against VEGFR2 (IC50=2 nM). GW806742X hydrochloride retards MLKL membrane translocation and inhibits necroptosis.
-
GC10915
GZD824
Olverembatinib (GZD824) dimesylate is a potent and orally active pan-Bcr-Abl inhibitor. GZD824 potently inhibits a broad spectrum of Bcr-Abl mutants. GZD824 strongly inhibits native Bcr-Abl and Bcr-AblT315I with IC50s of 0.34 nM and 0.68 nM, respectively. GZD824 has antitumor activity.
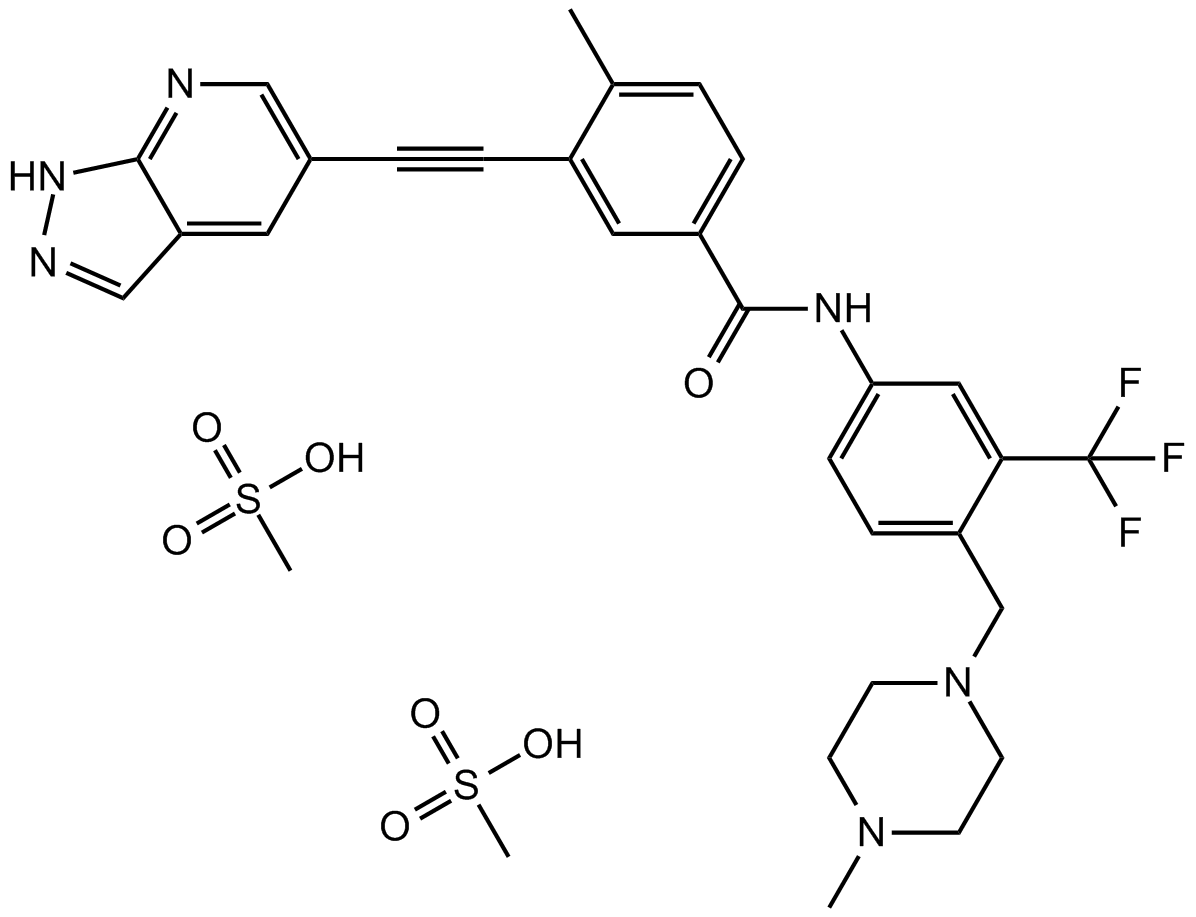
-
GC33201
GZD856
GZD856 formic is a potent and orally active PDGFRα/β inhibitor, with IC50s of 68.6 and 136.6 nM, respectively. GZD856 formic is also a Bcr-AblT315I inhibitor, with IC50s of 19.9 and 15.4?nM for native Bcr-Abl and the T315I mutant. GZD856 formic has antitumor activity.
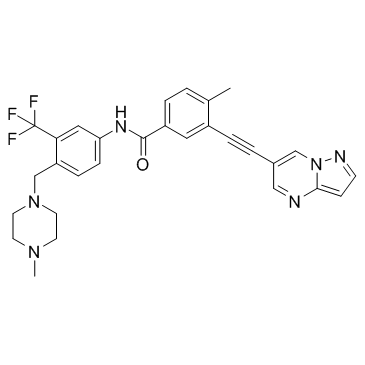
-
GC62453
GZD856 formic
GZD856 formic is a potent and orally active PDGFRα/β inhibitor, with IC50s of 68.6 and 136.6 nM, respectively. GZD856 formic is also a Bcr-AblT315I inhibitor, with IC50s of 19.9 and 15.4?nM for native Bcr-Abl and the T315I mutant. GZD856 formic has antitumor activity.
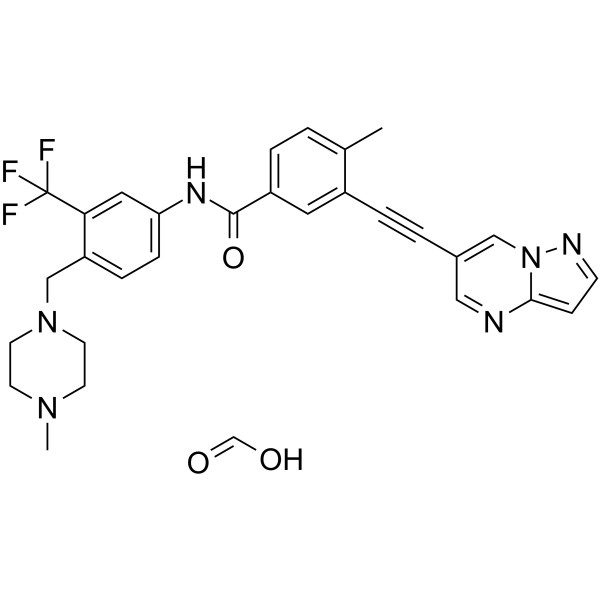
-
GC17923
H-7 dihydrochloride
H-7 dihydrochloride is a potent PKC (protein kinase C) inhibitor. At 100 μM, H-7 dihydrochloride completely inhibits both TPA (skin tumour promoter, 12-O-tetradecanoylphorbol-13-acetate) and phospholipase C-induced ODC (ornithine decarboxylase).
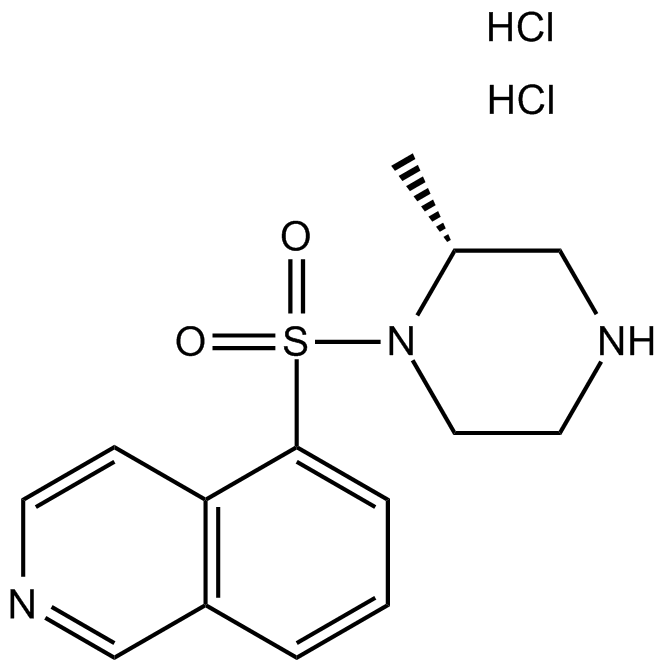
-
GC11420
H-9 dihydrochloride
Protein kinase inhibitor