Ubiquitination/ Proteasome
- Autophagy(1374)
- DUB(28)
- E1 Activating(1)
- E2 conjugating(1)
- E2 Ligases(0)
- E3 Ligase(8)
- Proteasome(123)
- p97(12)
- Mitophagy(104)
- ULK(10)
Products for Ubiquitination/ Proteasome
- Cat.No. Product Name Information
-
GC17865
Lomustine
Antineoplastic drug
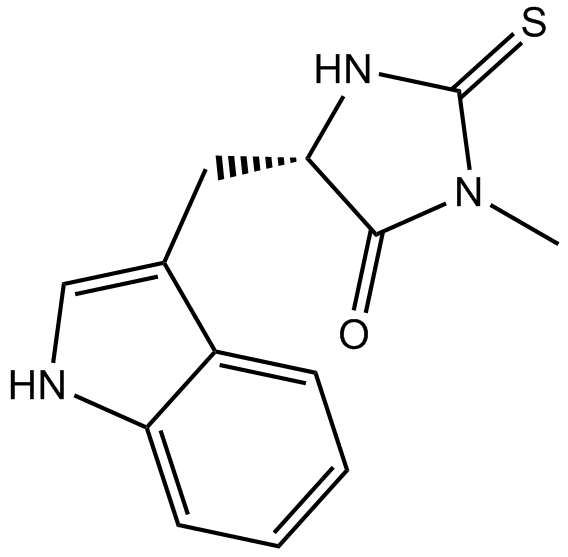
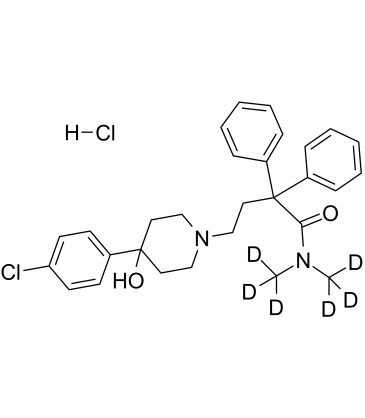
-
GC10330
Lonafarnib
A farnesyltransferase inhibitor with antitumor activity
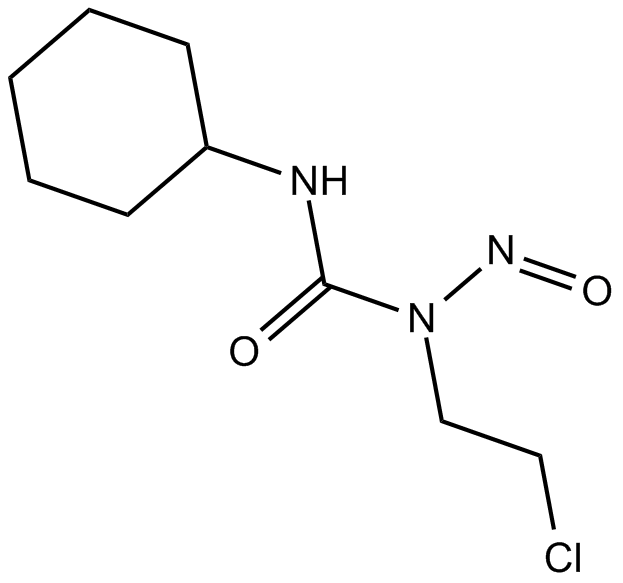
-
GC61003
Loperamide D6 hydrochloride
Loperamide D6 hydrochloride (R-18553 D6 hydrochloride) is a deuterium labeled Loperamide hydrochloride.
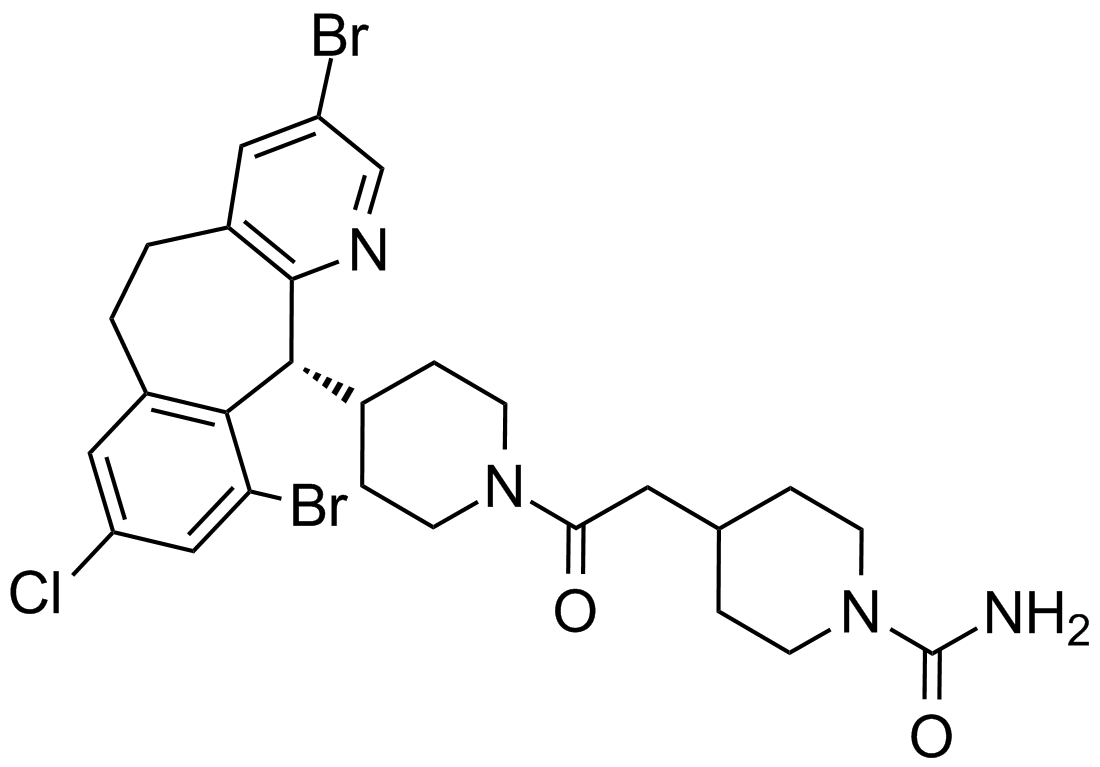
-
GC36479
Loperamide hydrochloride
Loperamide (hydrochloride) (R-18553 (hydrochloride)) is an opioid receptor agonist.
-
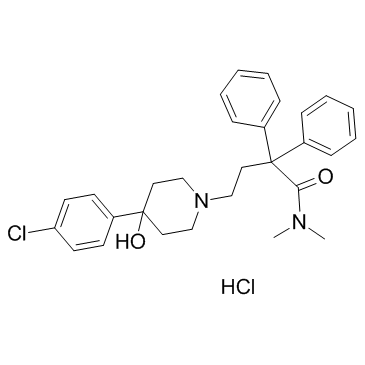
GC13717
Losmapimod
p38 MAPK inhibitor, orally active
-
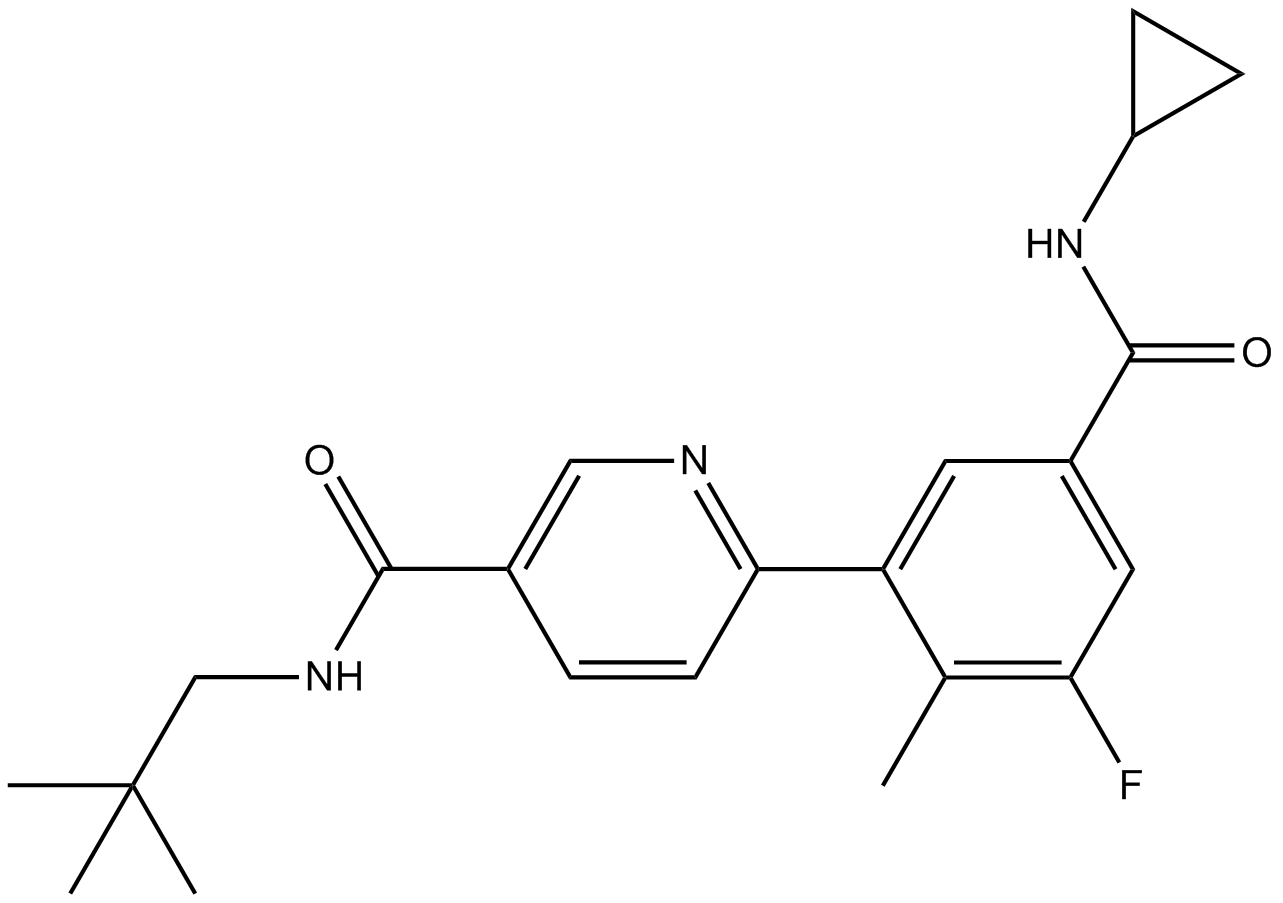
GC11633
Lovastatin
An inhibitor of HMG-CoA reductase
-
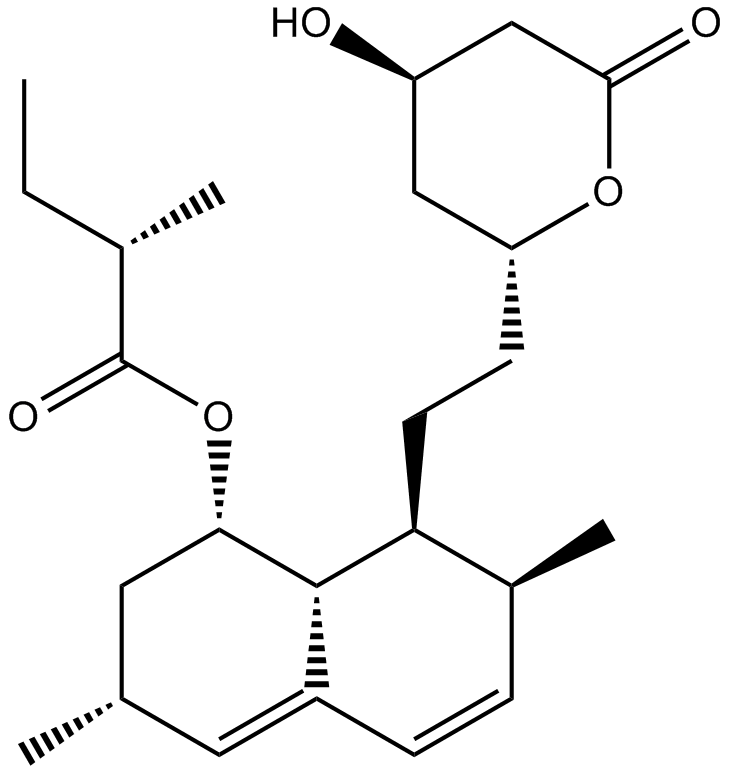
GC33126
Lucanthone
Lucanthone is an endonuclease inhibitor of Apurinic endonuclease-1 (APE-1).
-
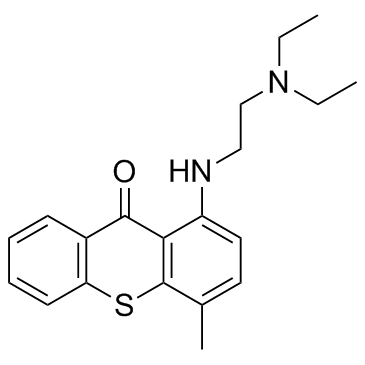
GC32152
Lumefantrine (Benflumetol)
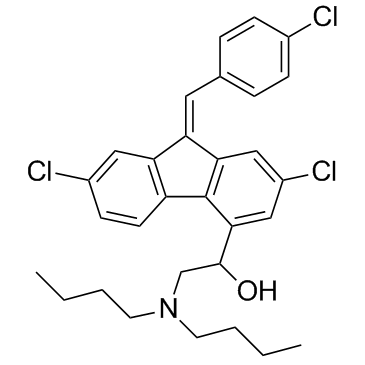
Lumefantrine (Benflumetol) is an antimalarial drug, used in combination with Artemether.
-
GC64363
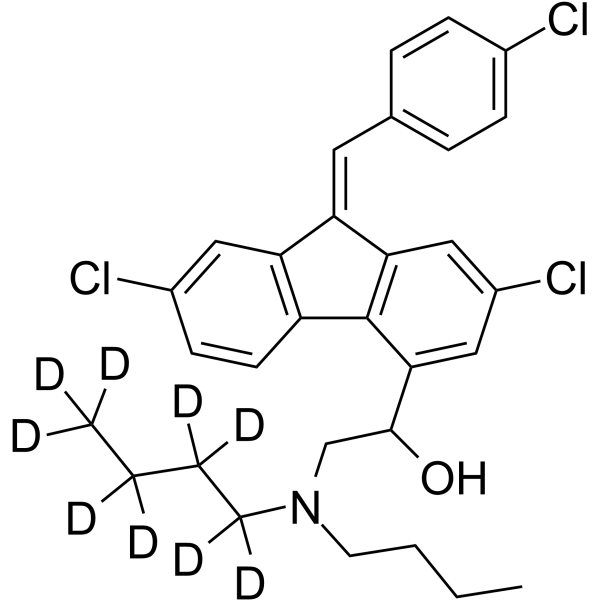
Lumefantrine-d9
-
GN10370
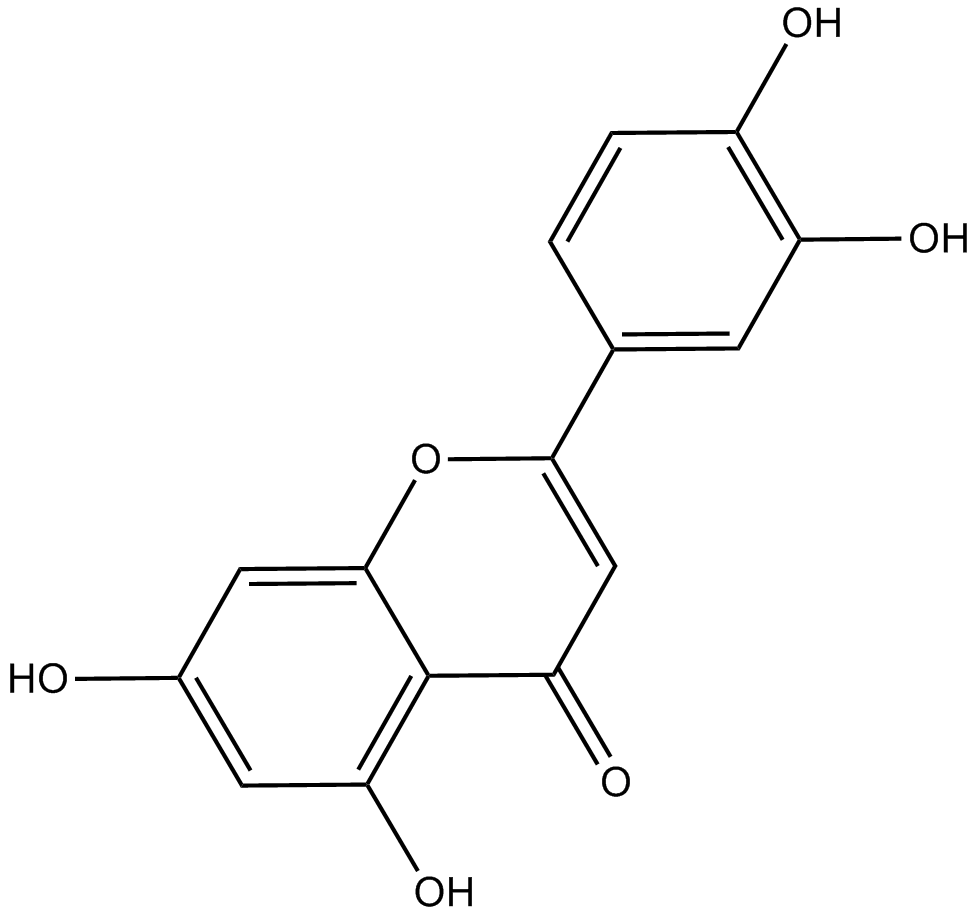
Luteolin
-
GC65432
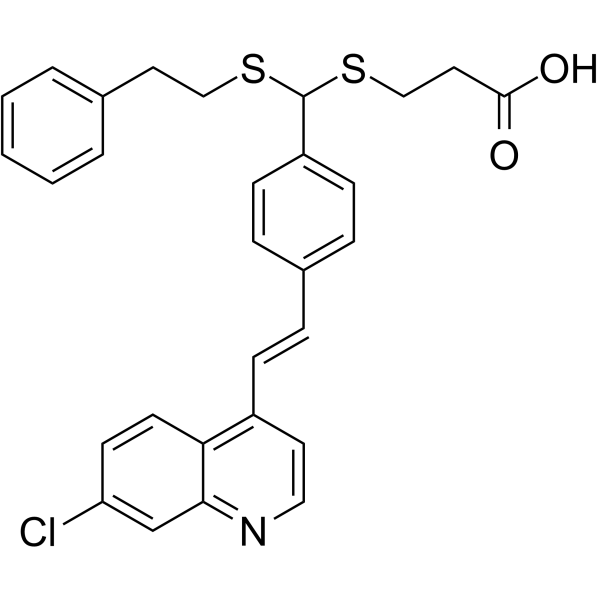
LV-320
LV-320 is a potent and uncompetitive ATG4B inhibitor with an IC50 of 24.5??M and a Kd of 16??M. LV-320 inhibits ATG4B enzymatic activity, blocks autophagic flux in cells, and is stable, non-toxic and active in vivo.
-
GC30884
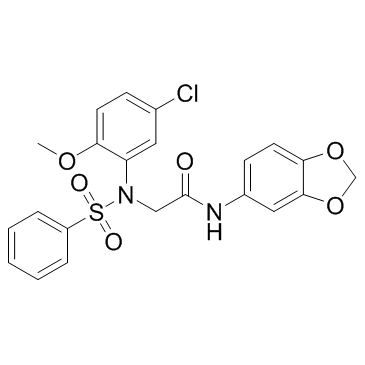
LX2343
An inhibitor of BACE1 and PI3K
-
GC64282
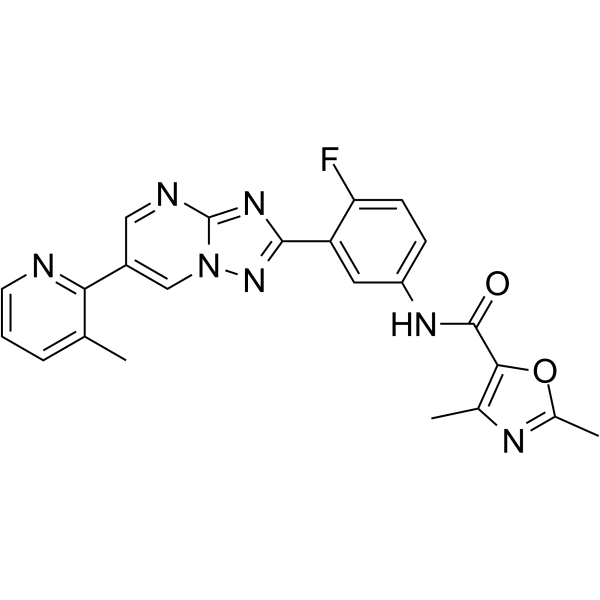
LXE408
LXE408 is an orally active, non-competitive and kinetoplastid-selective proteasome inhibitor.
-
GC12363
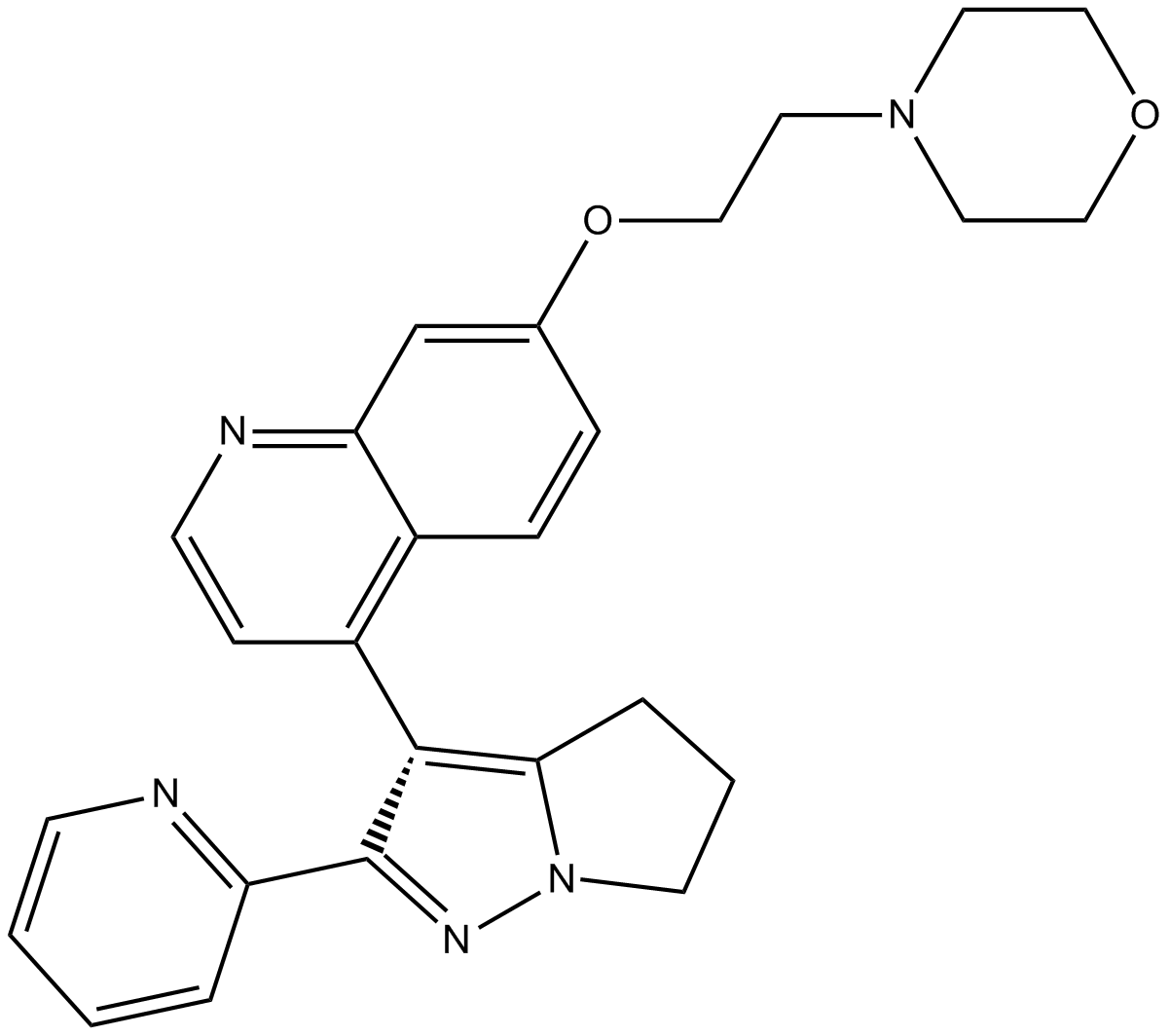
LY2109761
TβRI/II kinase inhibitor
-
GC15094
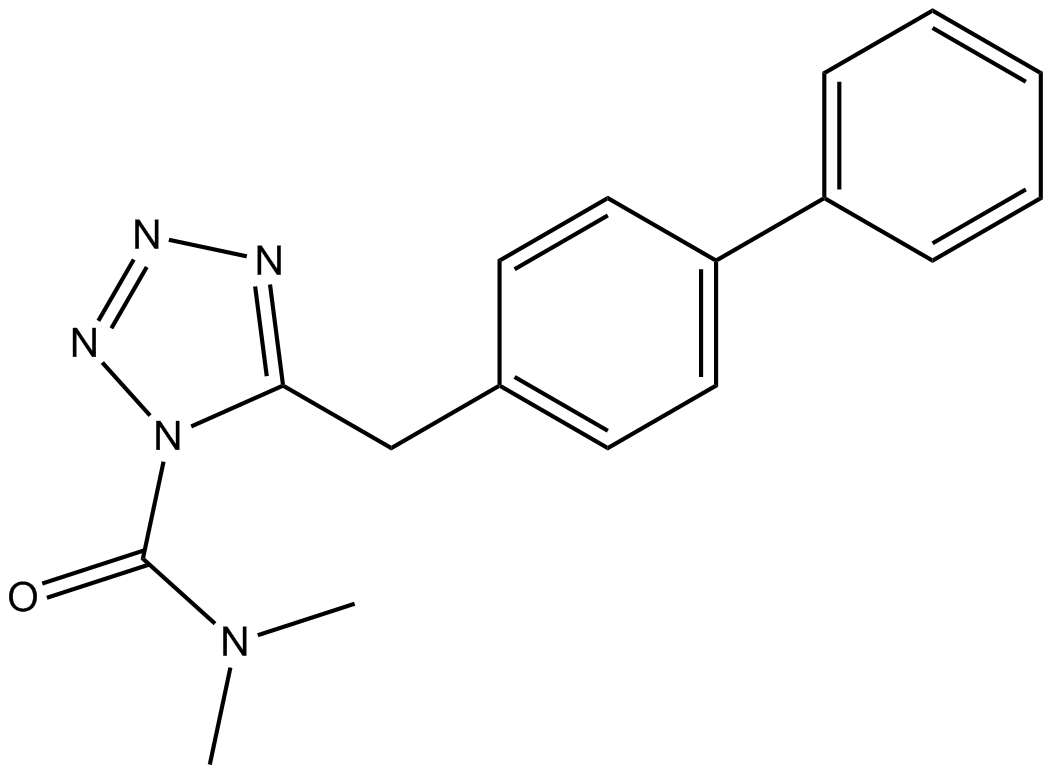
LY2183240
A potent, competitive inhibitor of anandamide uptake
-
GC16835
LY2228820
LY2228820 (LY2228820 dimesylate) is a selective, ATP-competitive inhibitor of p38 MAPK α/β with IC50s of 5.3 and 3.2 nM, respectively.
-
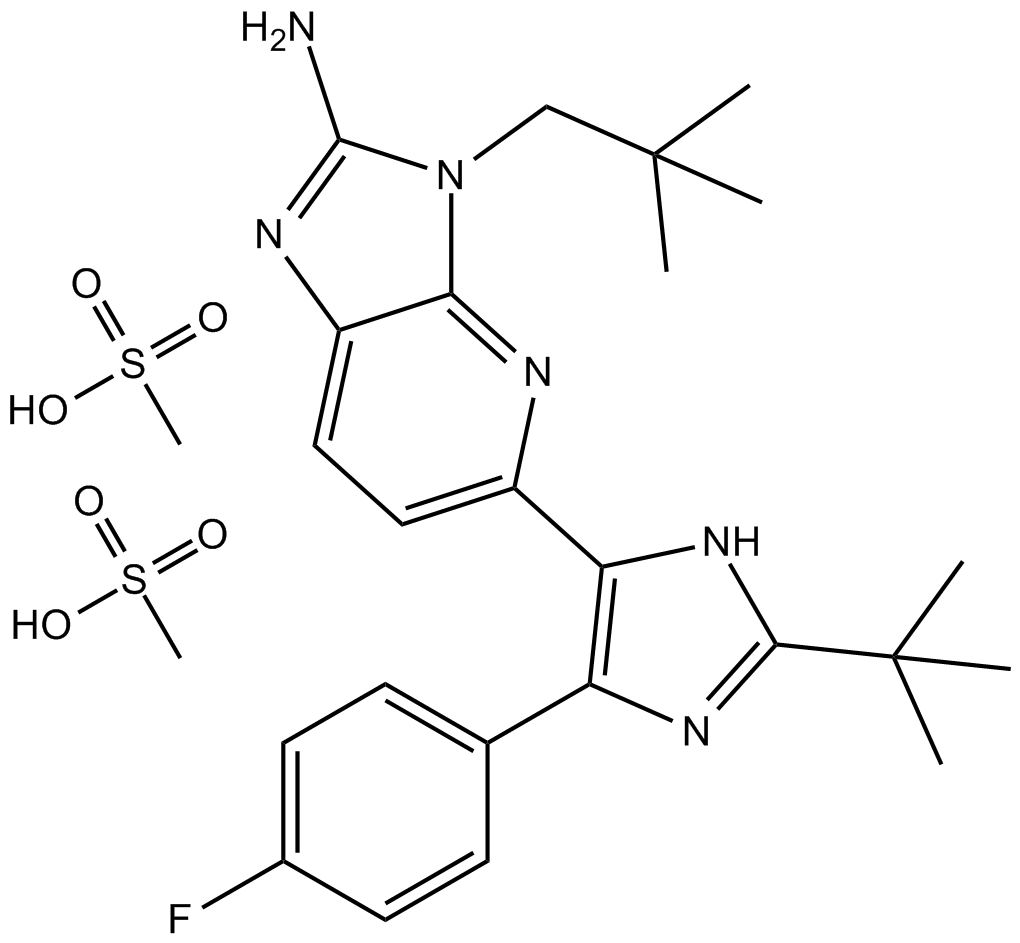
GC19421
LY2562175
LY2562175 is a potent and selective?FXR?agonist with an?EC50?of 193 nM
-
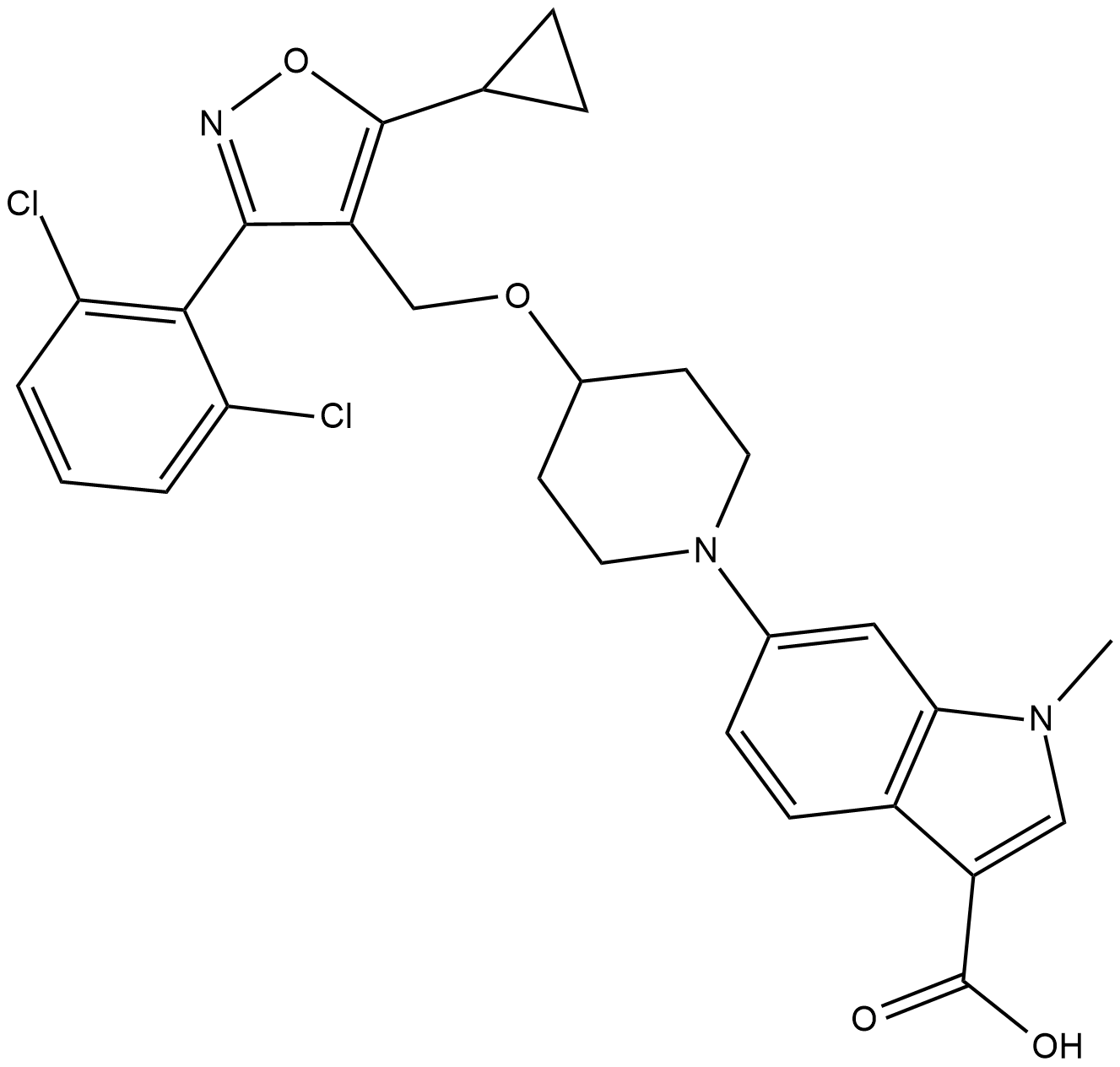
GC15741
LY2603618
LY2603618 (LY2603618) is a potent and selective inhibitor of Chk1 with an IC50 of 7 nM.
-
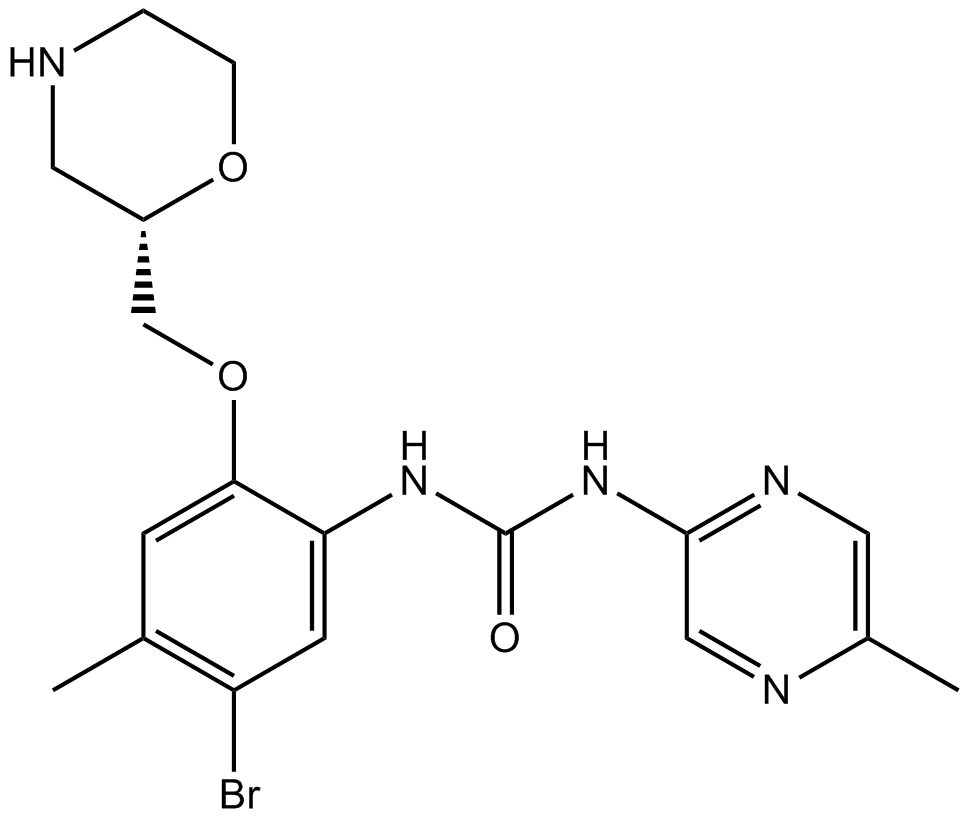
GC19232
LY2955303
LY2955303 is a potent and selective retinoic acid receptor gamma (RARγ) antagonist with a Ki of 1.09 nM.
-
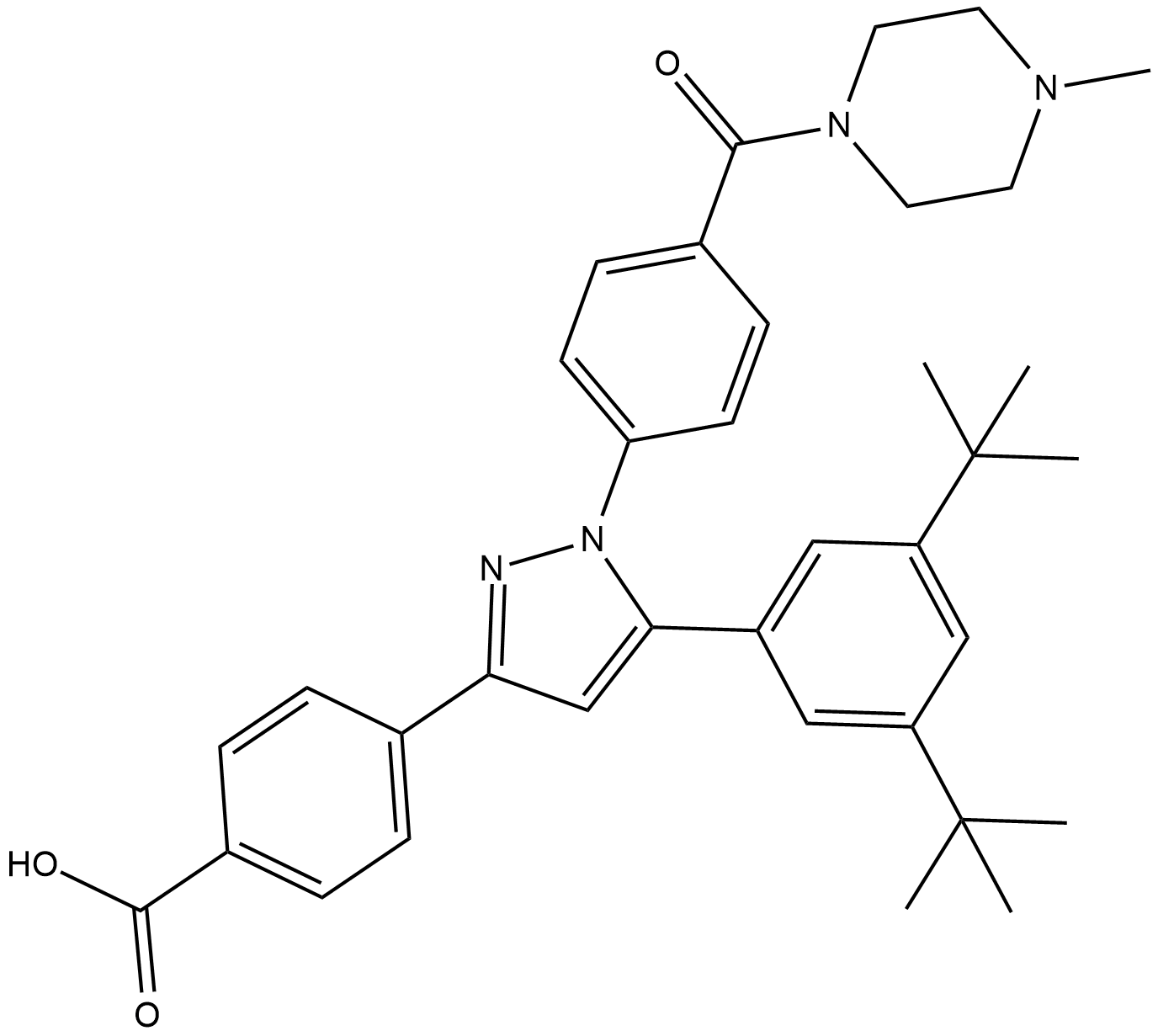
GC13980
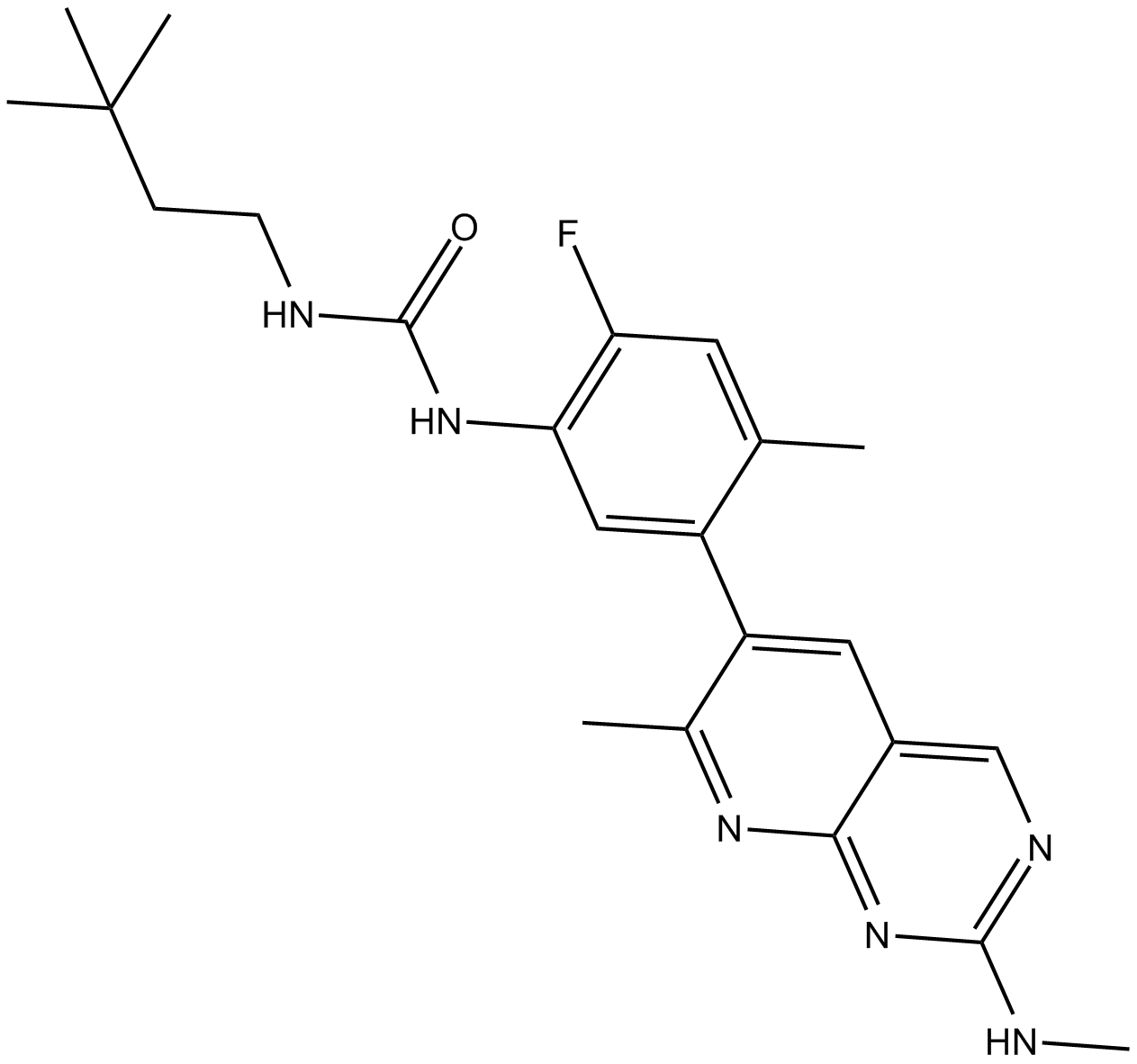
LY3009120
LY3009120 is a pan-RAF and RAF dimer inhibitor that inhibits all RAF isoforms and occupies both protomers in RAF dimers.
-
GC14371
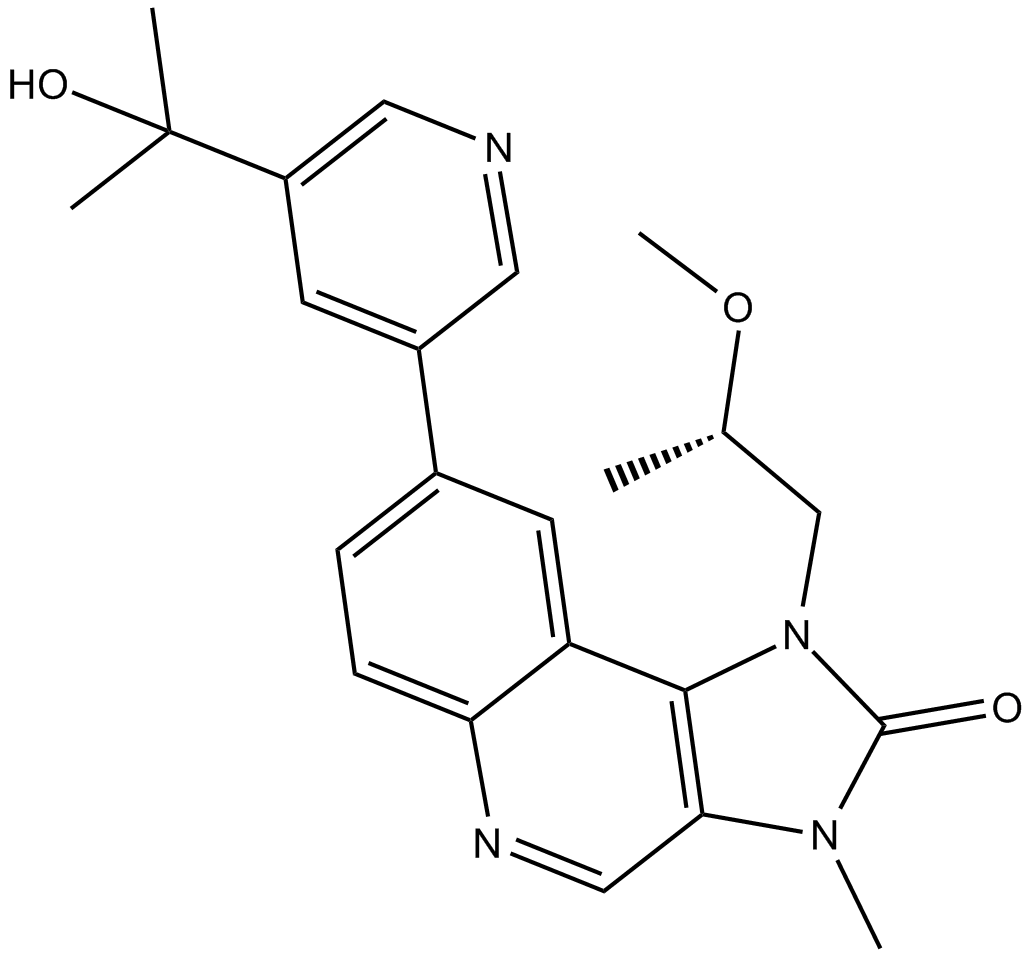
LY3023414
LY3023414 (LY3023414) potently and selectively inhibits class I PI3K isoforms, DNA-PK, and mTORC1/2 with IC50s of 6.07 nM, 77.6 nM, 38 nM, 23.8 nM, 4.24 nM and 165 nM for PI3Kα, PI3Kβ, PI3Kδ, PI3Kγ, DNA-PK and mTOR, respectively. LY3023414 potently inhibits mTORC1/2 at low nanomolar concentrations.
-
GN10396
Lycorine chloride

-
GC19236
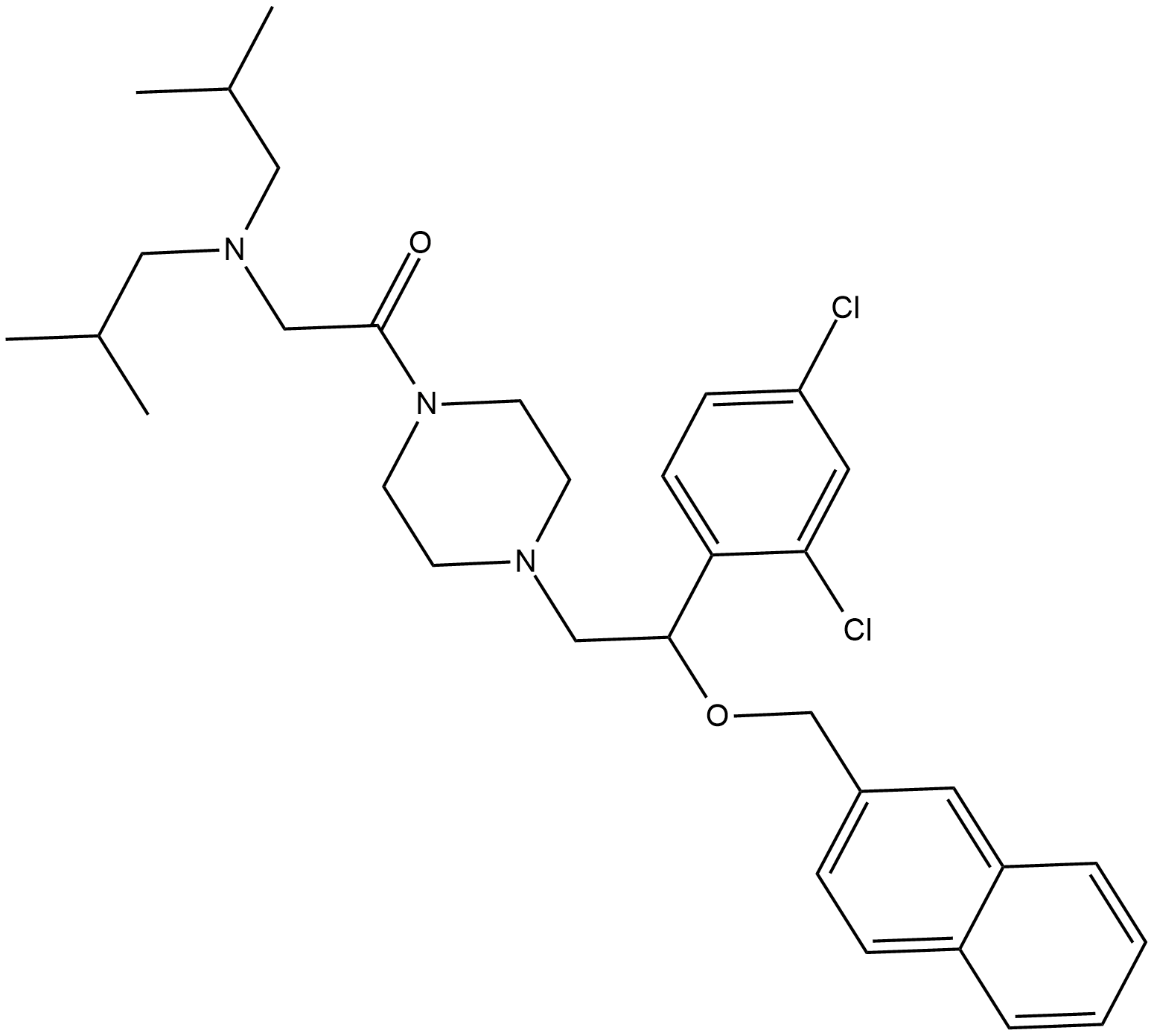
LYN-1604
LYN-1604 is a potent UNC-51-like kinase 1 (ULK1) agonist with an EC50 of 18.94 nM.
-
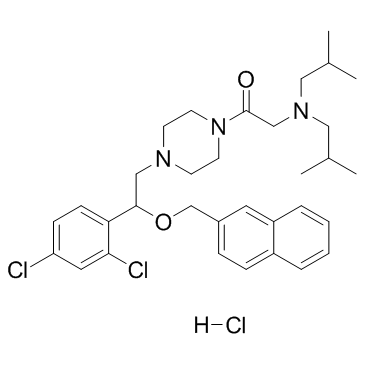
GC36517
LYN-1604 hydrochloride
LYN-1604 hydrochloride is a potent UNC-51-like kinase 1 (ULK1) activator (EC50=18.94 nM) for the research of triple negative breast cancer (TNBC).
-
GC19237
Lys01 trihydrochloride
Lys01 trihydrochloride (Lys01 trihydrochloride) is a novel lysosomal autophagy inhibitor with IC50 values of 3.6, 3.8, 6 and 7.9 μM for 1205Lu, c8161, LN229 and HT-29 cell line in the MTT assay.
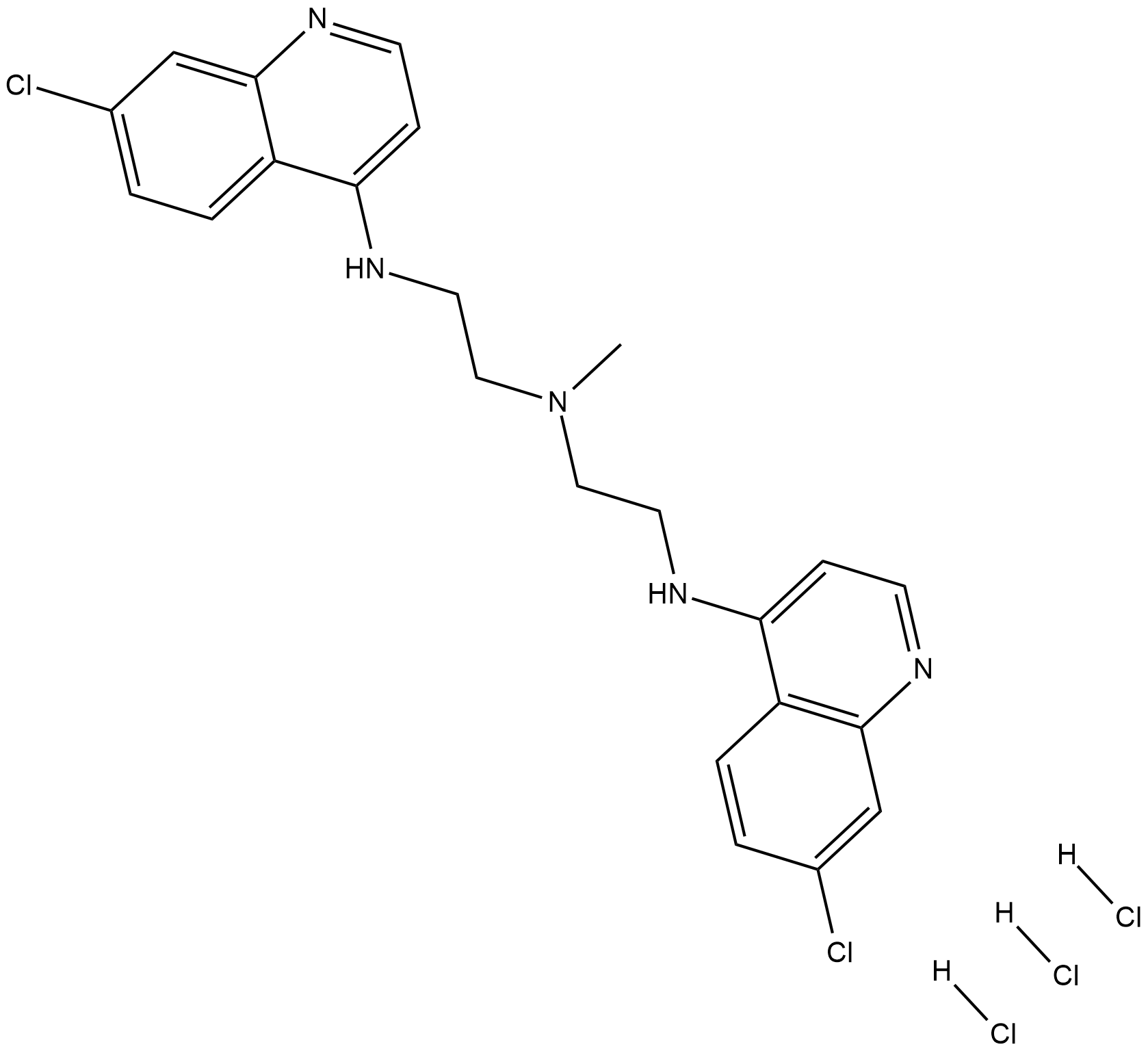
-
GN10301
Magnolol
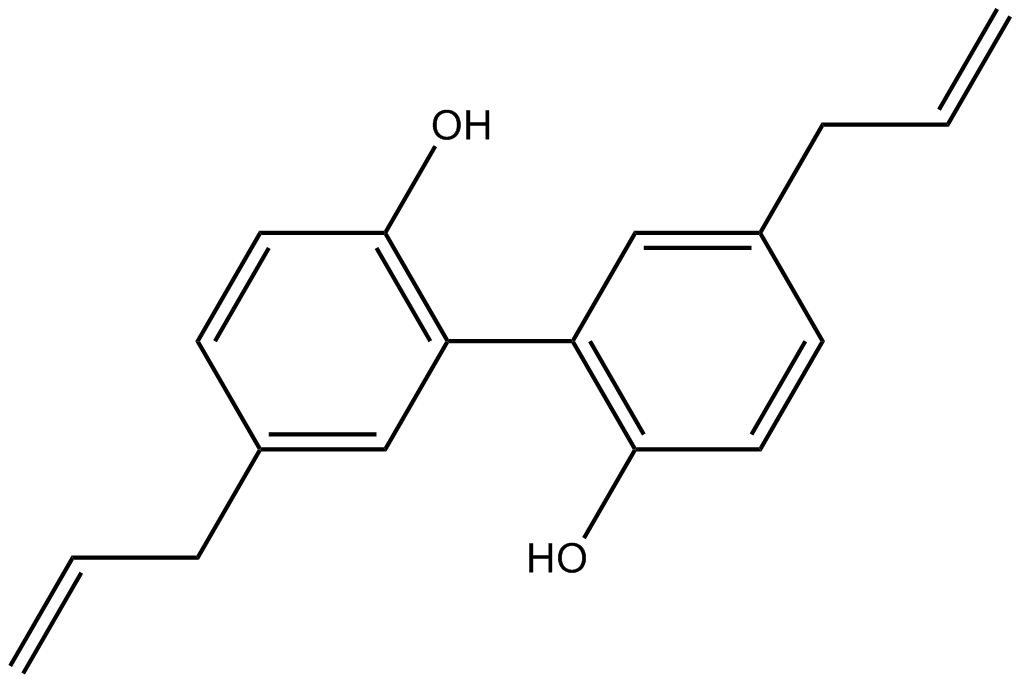
-
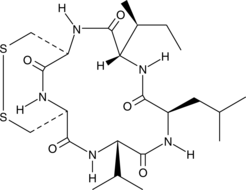
GC40007
Malformin A
Malformin A is a cyclopentapeptide fungal metabolite that has been found in A.
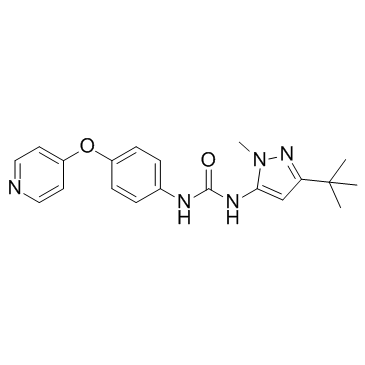
-
GC30676
MAPK13-IN-1
MPAK13-IN-1 is a MAPK13 (p38δ) inhibitor, with an IC50 of 620 nM.
-
GC12376
Maprotiline HCl
Maprotiline HCl is a selective noradrenalin re-uptake inhibitor and a tetracyclic antidepressant.
-
GC17874
Matrine
An alkaloid with diverse biological activities
-
GC10200
Mdivi 1
A mitochondrial division inhibitor
-
GC11640
MDL 28170
A cell permeable, selective calpain inhibitor
-
GC12385
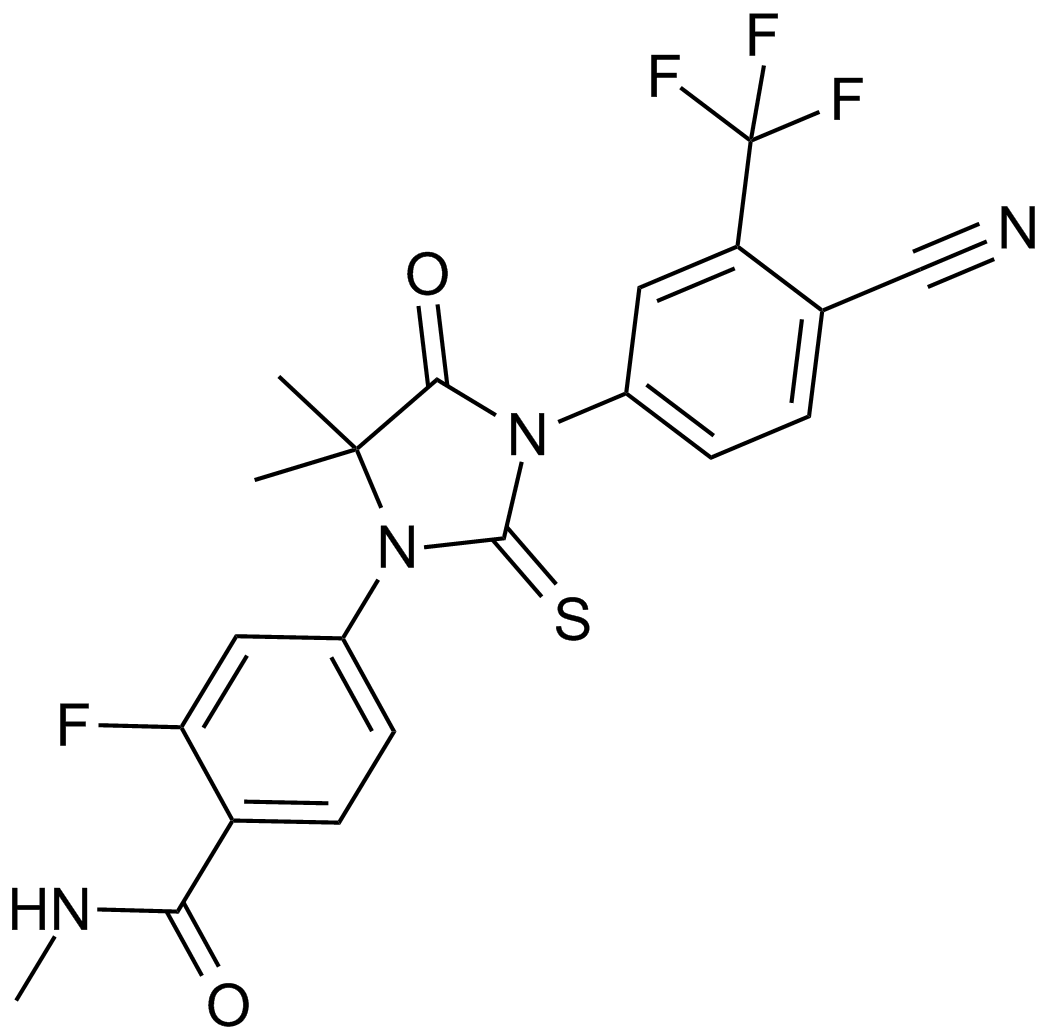
MDV3100 (Enzalutamide)
A non-steroidal androgen receptor antagonist
-
GC13252
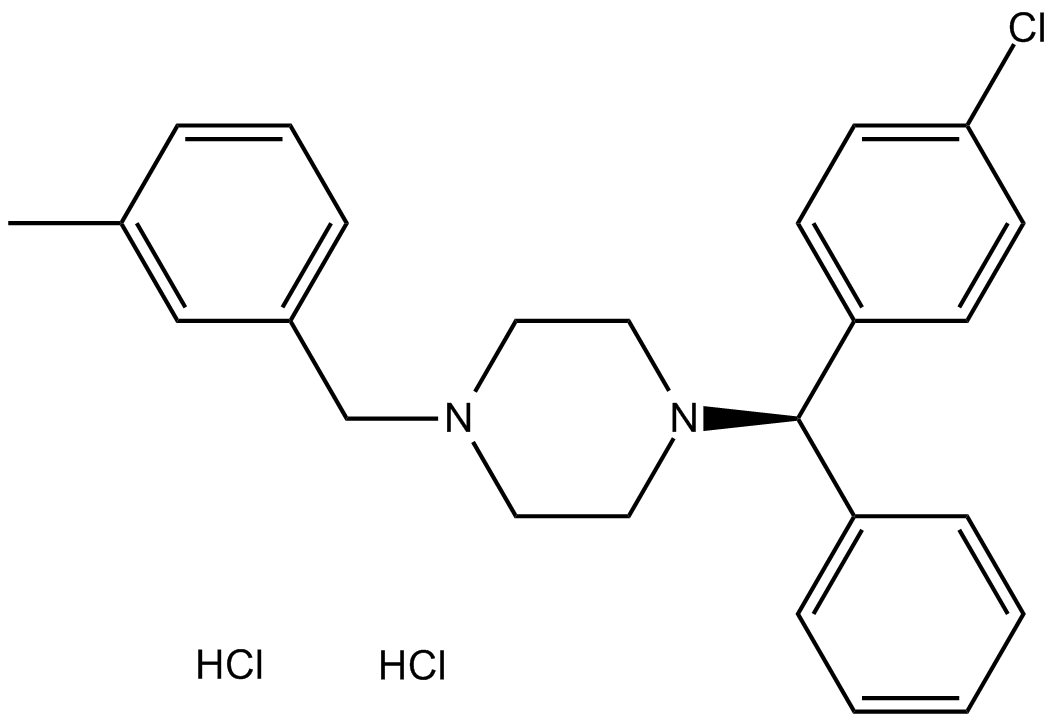
Meclizine 2HCl
Histamine H1 receptor antagonist
-
GC15854
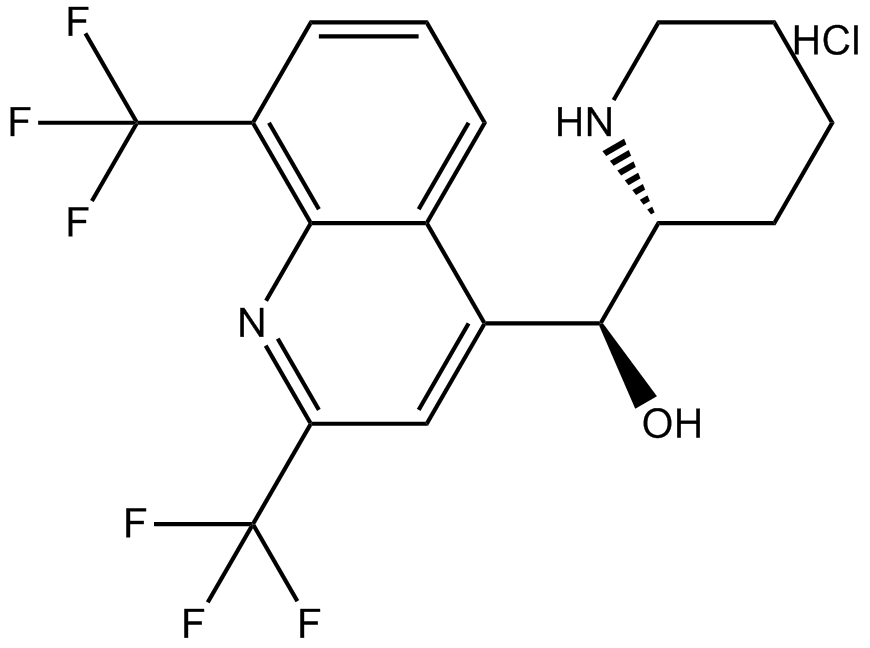
Mefloquine hydrochloride
An antimalarial compound
-
GC11688
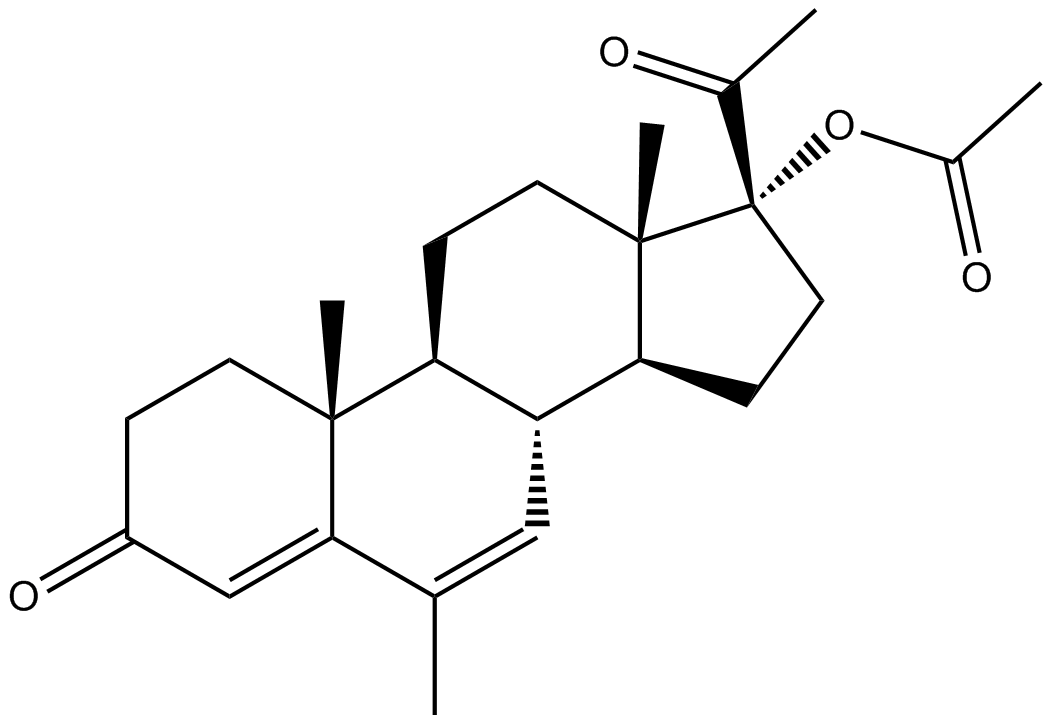
Megestrol Acetate
Anti-estrogen agent
-
GC34035
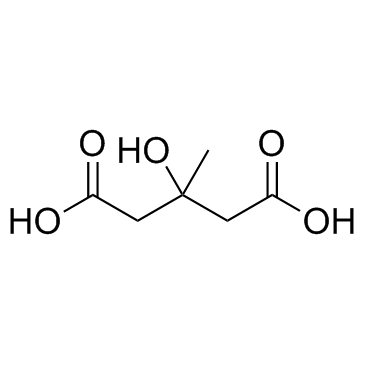
Meglutol (Dicrotalic acid)
Meglutol (Dicrotalic acid) is an antilipemic agent which lowers cholesterol, triglycerides, serum beta-lipoproteins and phospholipids, and inhibits the activity of hydroxymethylglutarryl CoA reductases, which is the rate limiting enzyme in the biosynthesis of cholesterol.
-
GC12843
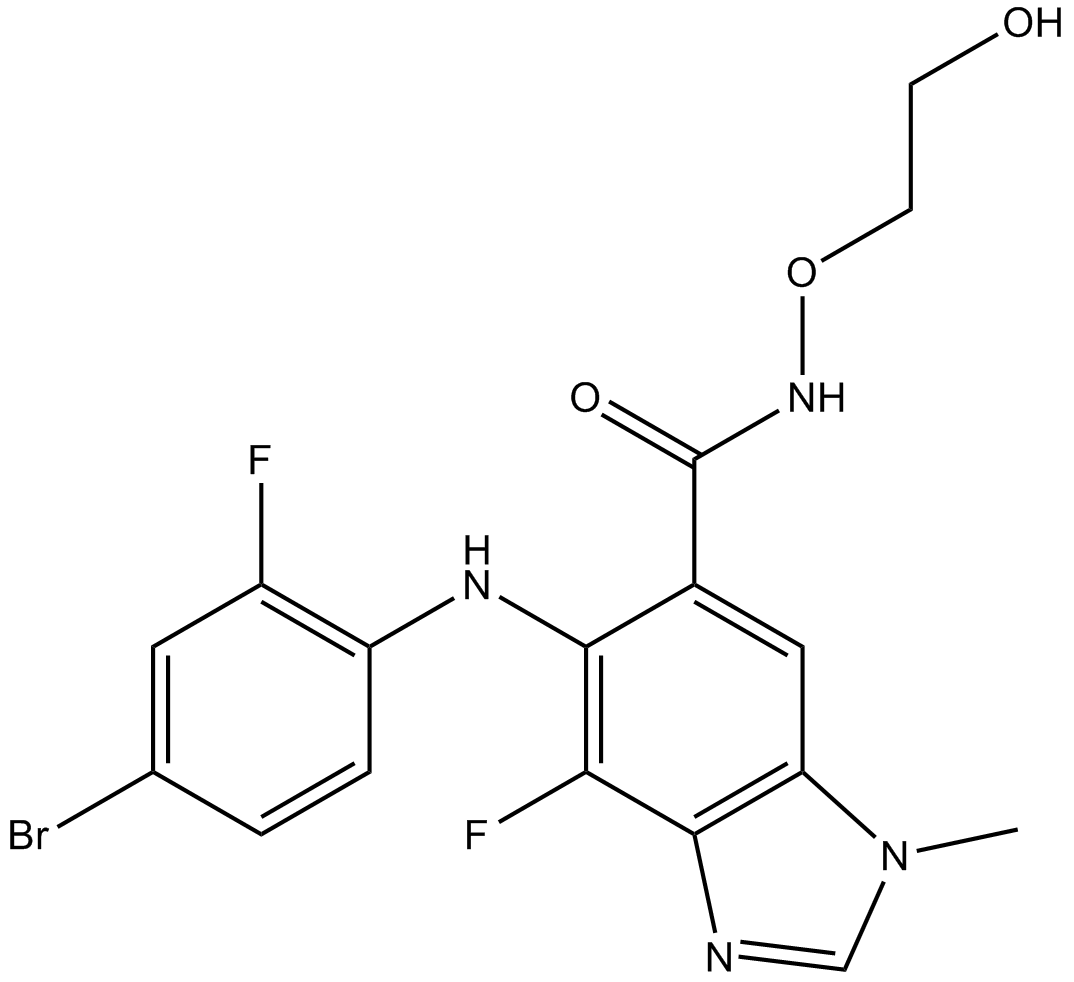
MEK162 (ARRY-162, ARRY-438162)
MEK162 (ARRY-162, ARRY-438162) (MEK162) is an oral and selective MEK1/2 inhibitor. MEK162 (ARRY-162, ARRY-438162) (MEK162) inhibits MEK with an IC50 of 12 nM.
-
GC15618
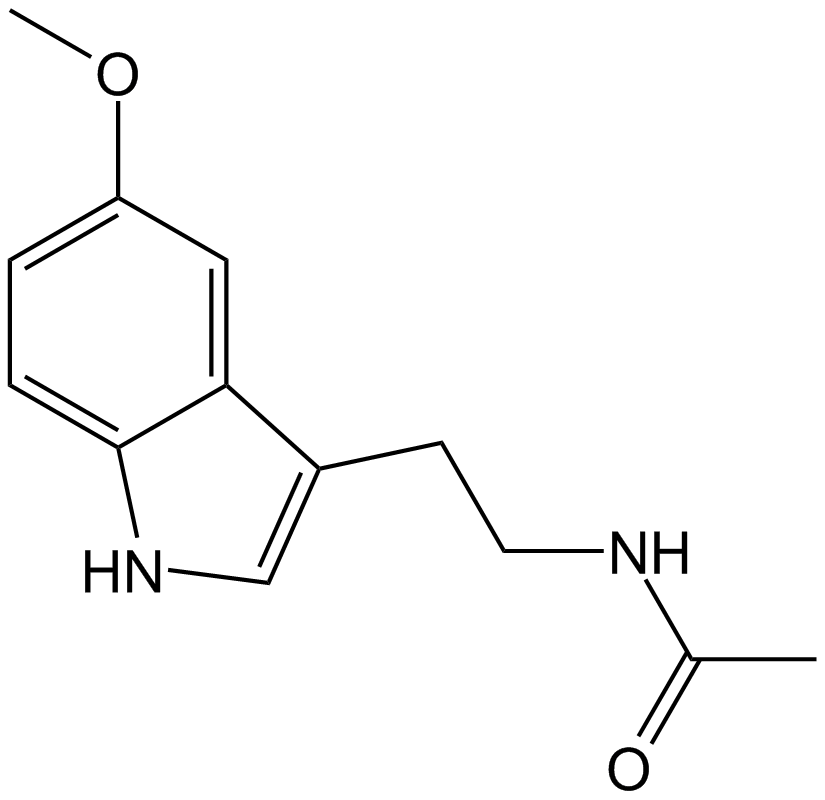
Melatonin
Melatonin receptors agonist
-
GC60241
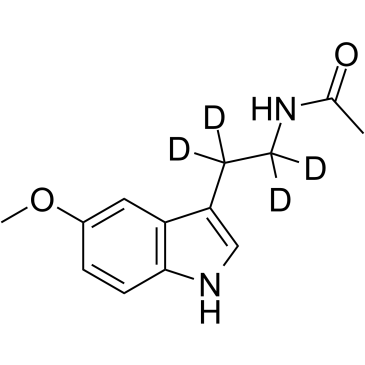
Melatonin D5
An internal standard for the quantification of melatonin
-
GC11762
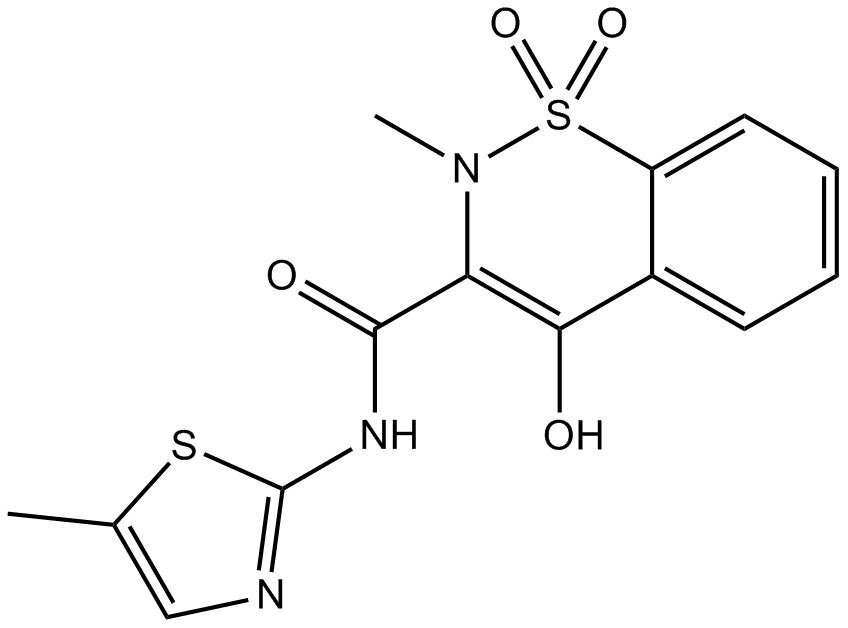
Meloxicam (Mobic)
Meloxicam (Mobic) is a non-steroidal antiinflammatory agent, inhibits COX activity, with IC50s of 0.49 μM and 36.6 μM for COX-2 and COX-1, respectively.
-
GC40065
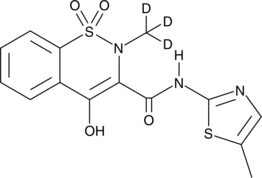
Meloxicam-d3
Meloxicam-d3 is intended for use as an internal standard for the quantification of meloxicam by GC- or LC-MS.
-
GC10198
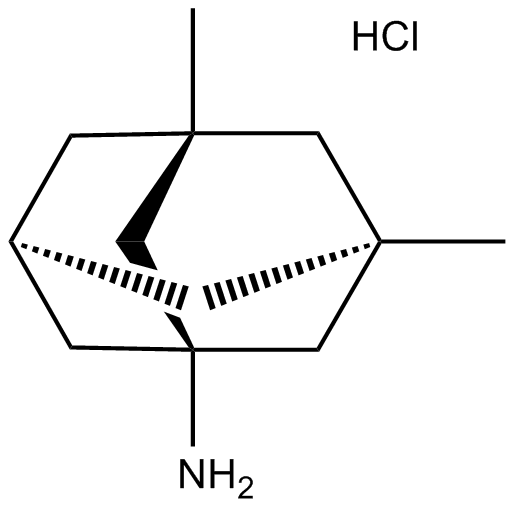
Memantine hydrochloride
NMDA receptor antagonist
-
GC15668
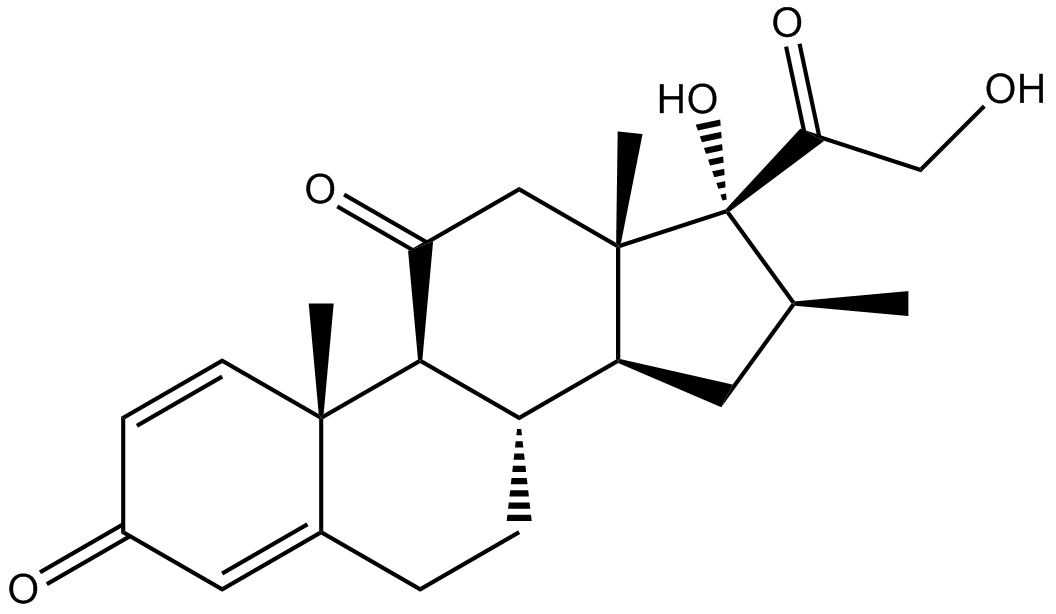
Meprednisone
Glucocorticoid receptor (GR) agonist
-
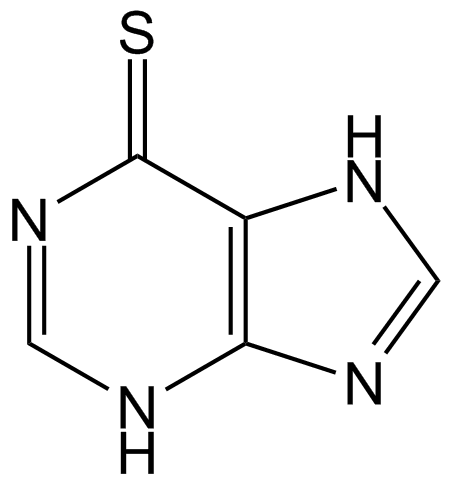
GC13704
Mercaptopurine (6-MP)
Mercaptopurine (6-MP) is a purine analogue which acts as an antagonist of the endogenous purines and has been widely used as antileukemic agent and immunosuppressive drug.
-
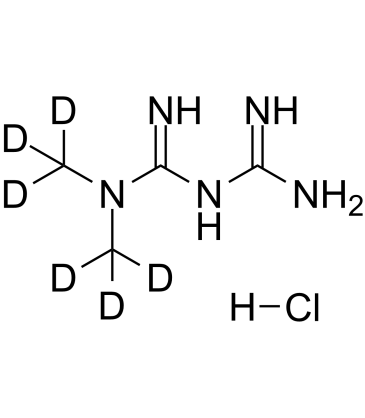
GC61045
Metformin D6 hydrochloride
An internal standard for the quantification of metformin
-
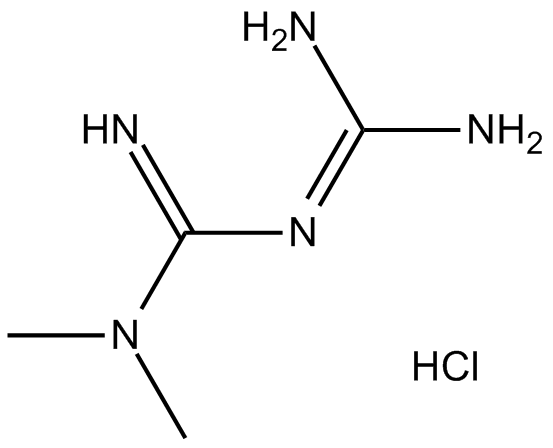
GC17443
Metformin HCl
Metformin HCl (1,1-Dimethylbiguanide hydrochloride) inhibits the mitochondrial respiratory chain in the liver, leading to activation of AMPK, enhancing insulin sensitivity for type 2 diabetes research.
-
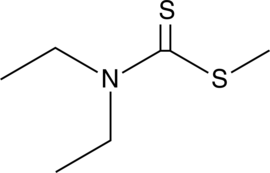
GC49241
Methyl Diethyldithiocarbamate
An active metabolite of disulfiram
-
GC15505
Methylprednisolone
A synthetic glucocorticoid
-
GC30356
Metofenazate (Methophenazine)
Metofenazate (Methophenazine) is a selective calmodulin inhibitor.
-
GC17411
Metyrapone
11-β hydroxylase (CYP11B1) inhibitor
-
GC11895
Mevastatin
HMG-CoA reductase inhibitor
-
GC10178
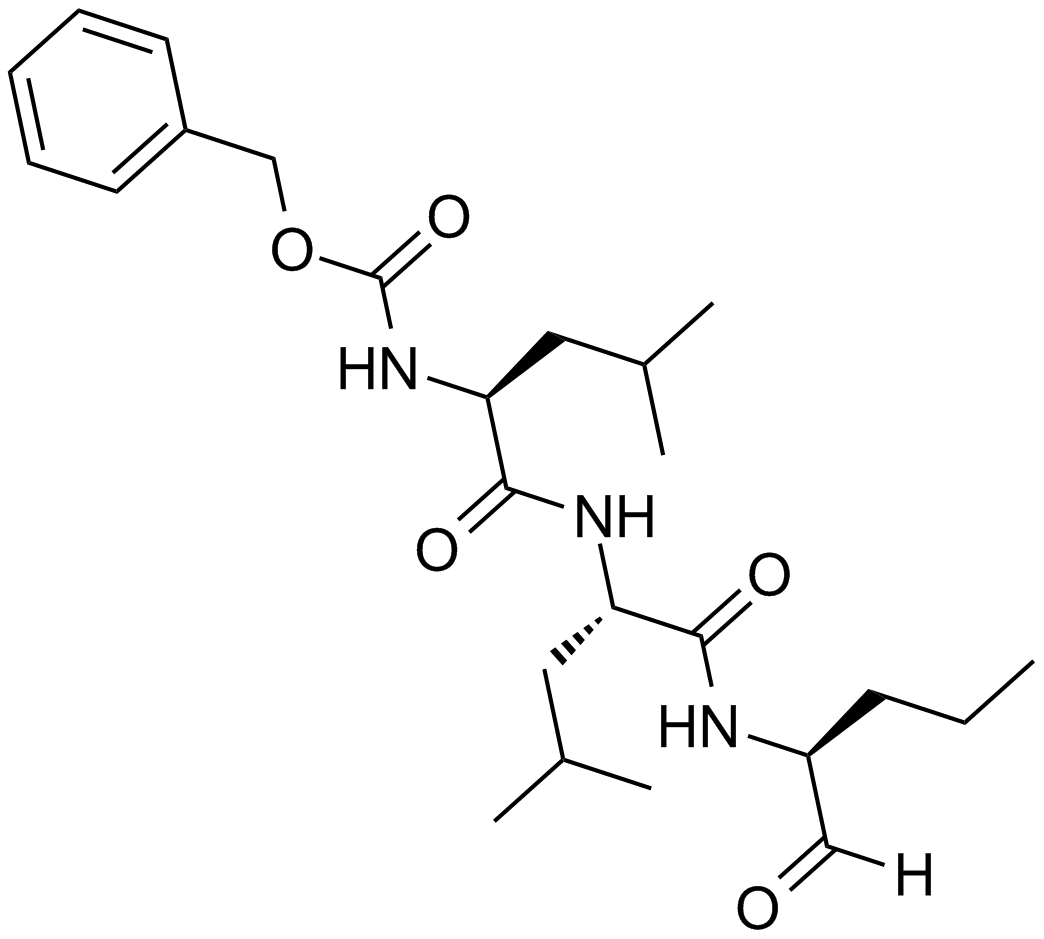
MG-115
A potent, reversible proteasome inhibitor
-
GC10383
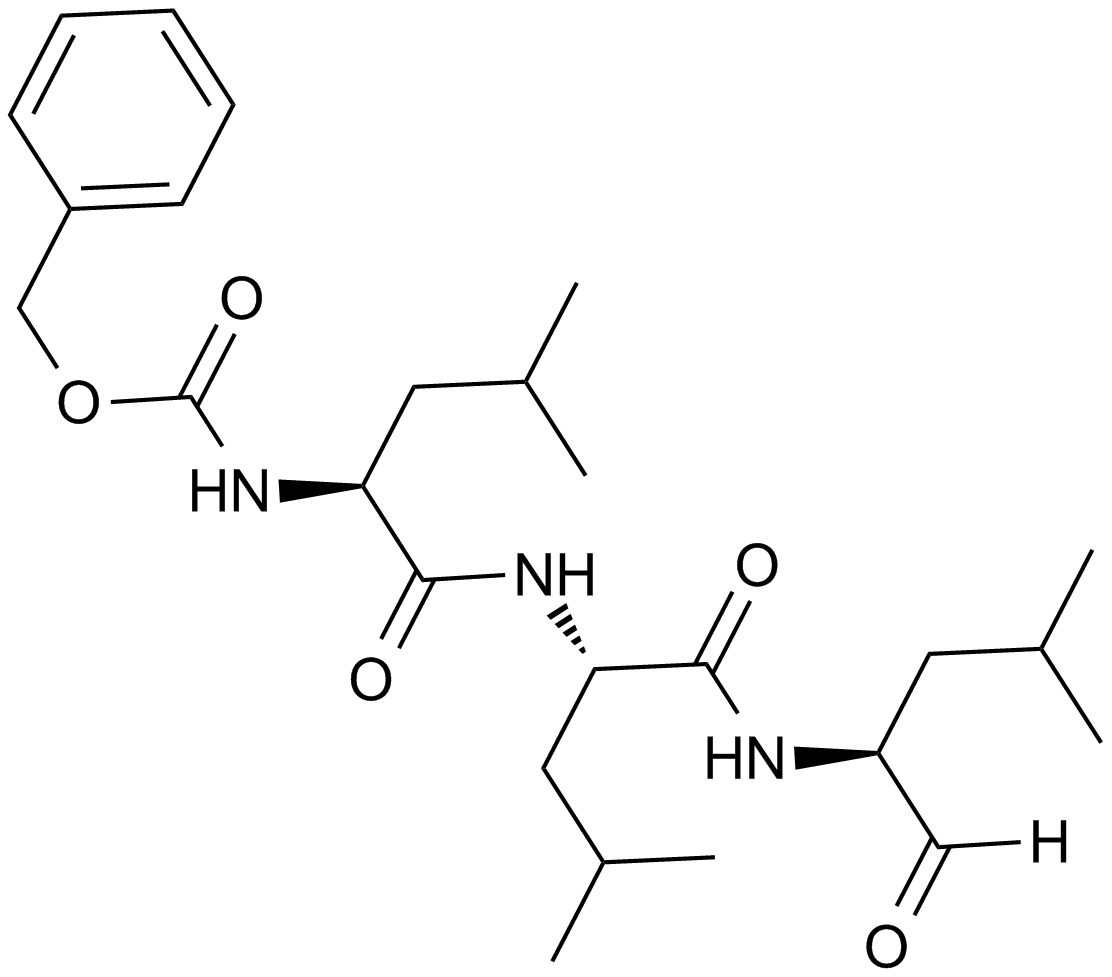
MG-132
A potent proteasome inhibitor
-
GC15517
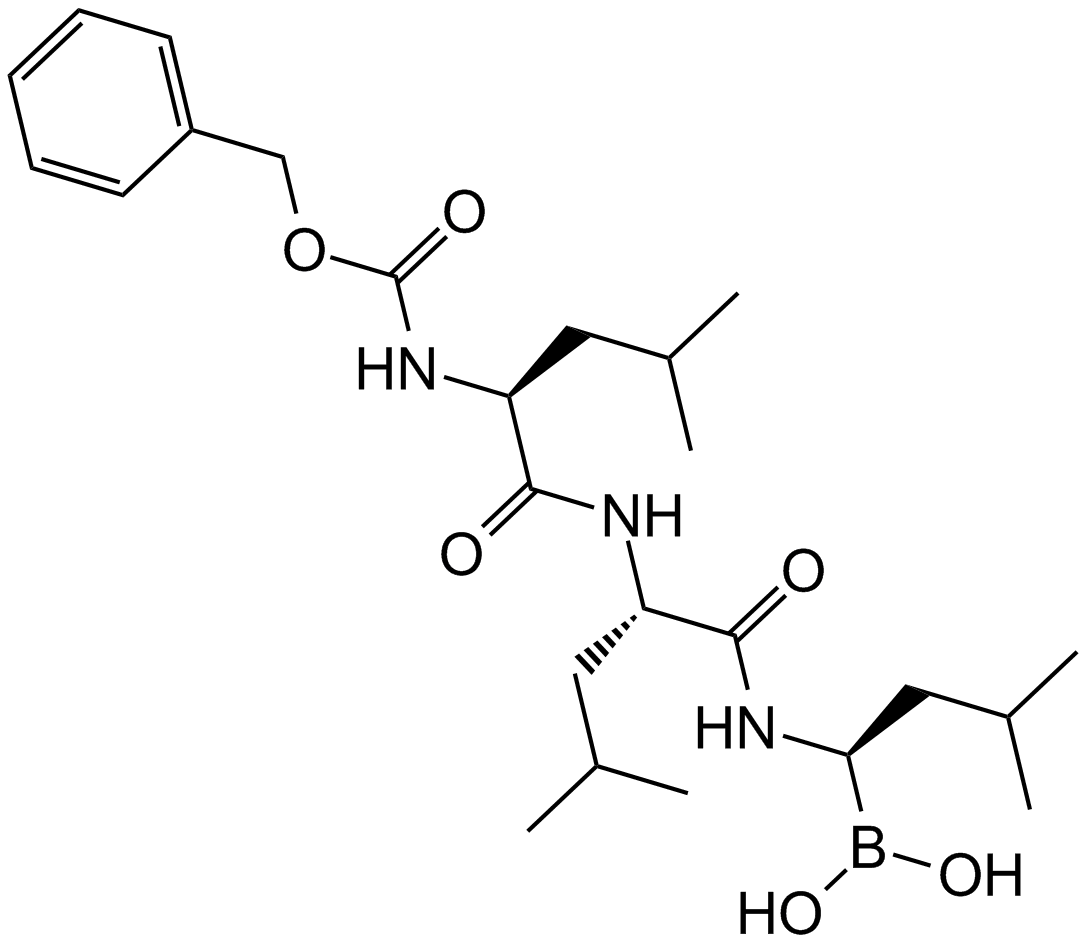
MG-262
A proteasome inhibitor with diverse biological activities
-
GC13790
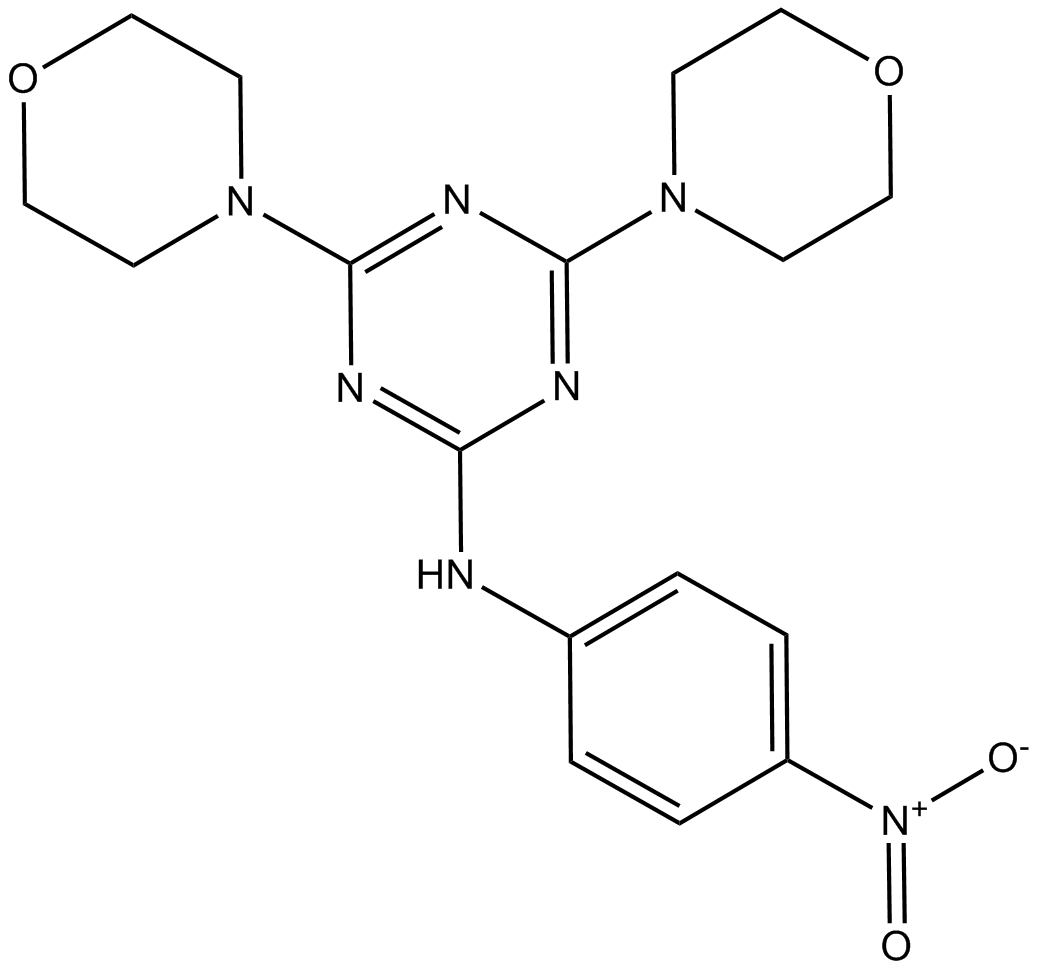
MHY1485
MHY1485 is a potent activator of mTOR, which inhibits autophagy and the fusion between autophagosomes and lysosomes.
-
GC11637
Mifepristone
Progesterone receptor antagonist
-
GC62564
Mito-LND
Mito-LND (Mito-Lonidamine) is an orally active and mitochondria-targeted inhibitor of oxidative phosphorylation (OXPHOS). Mito-LND inhibits mitochondrial bioenergetics, stimulates the formation of reactive oxygen species, and induces autophagic cell death in lung cancer cells.
-
GC16304
MK-2206 dihydrochloride
An allosteric Akt inhibitor
-
GC17162
MK-5108 (VX-689)
A potent inhibitor of Aurora kinases
-
GC14059
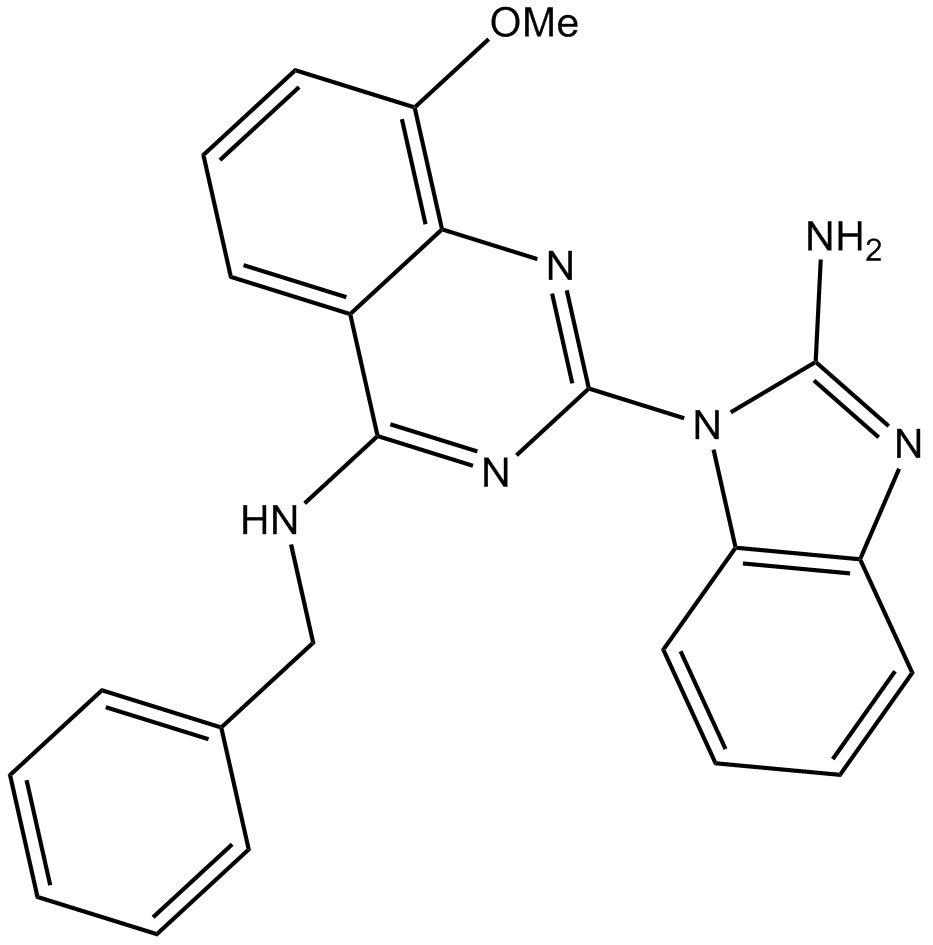
ML 240
ATP-competitive inhibitor of p97 ATPase
-
GC50067
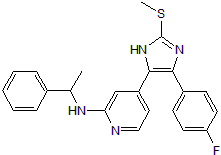
ML 3403
p38 inhibitor
-
GC17635
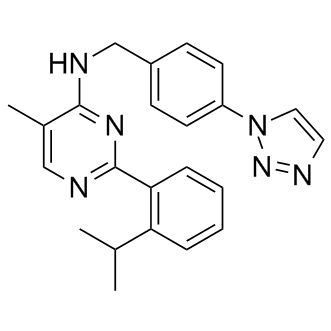
ML-323
USP1-UAF1 inhibitor
-
GC11749
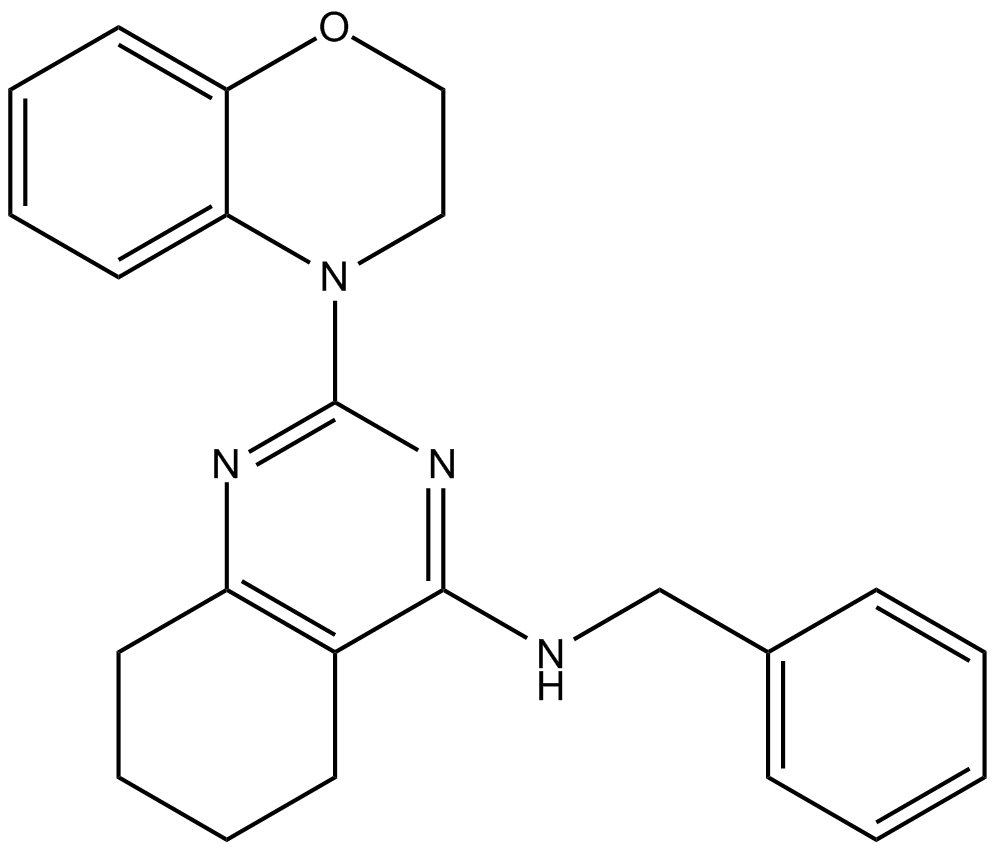
ML241
p97 ATPase inhibitor
-
GC13656
ML241 hydrochloride
p97 ATPase inhibitor
-
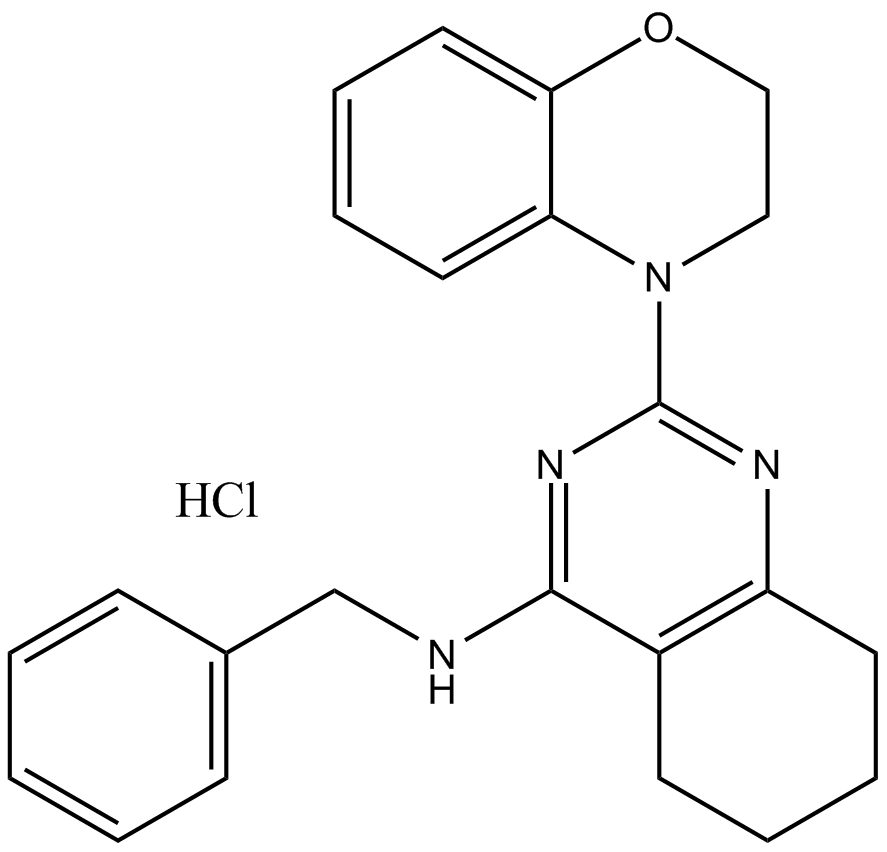
GC32785
ML327
ML327 is a blocker of MYC which can also de-repress E-cadherin transcription and reverse Epithelial-to-Mesenchymal Transition (EMT).
-
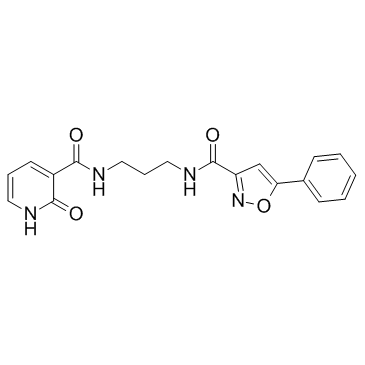
GC17329
MLCK inhibitor peptide 18
Selective competitive inhibitor of myosin light chain kinase
-
GC16146
MLN2238
A 20S proteasome inhibitor and an active metabolite of MLN9708
-
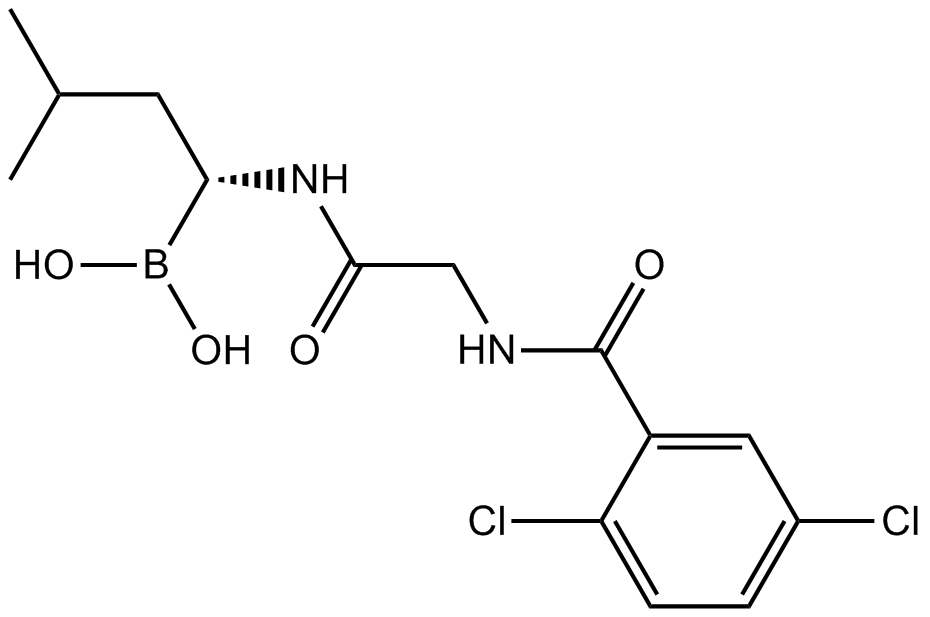
GC12690
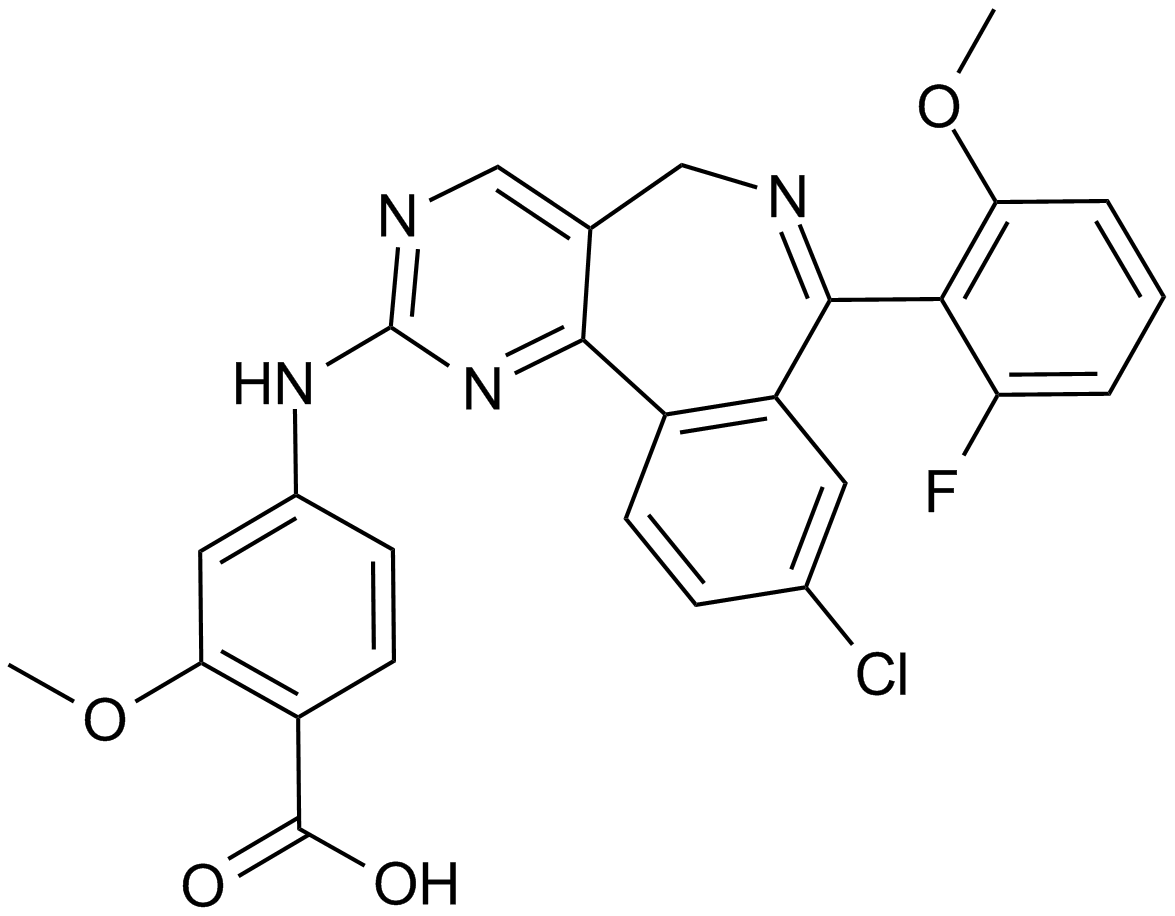
MLN8237 (Alisertib)
An Aurora A kinase inhibitor
-
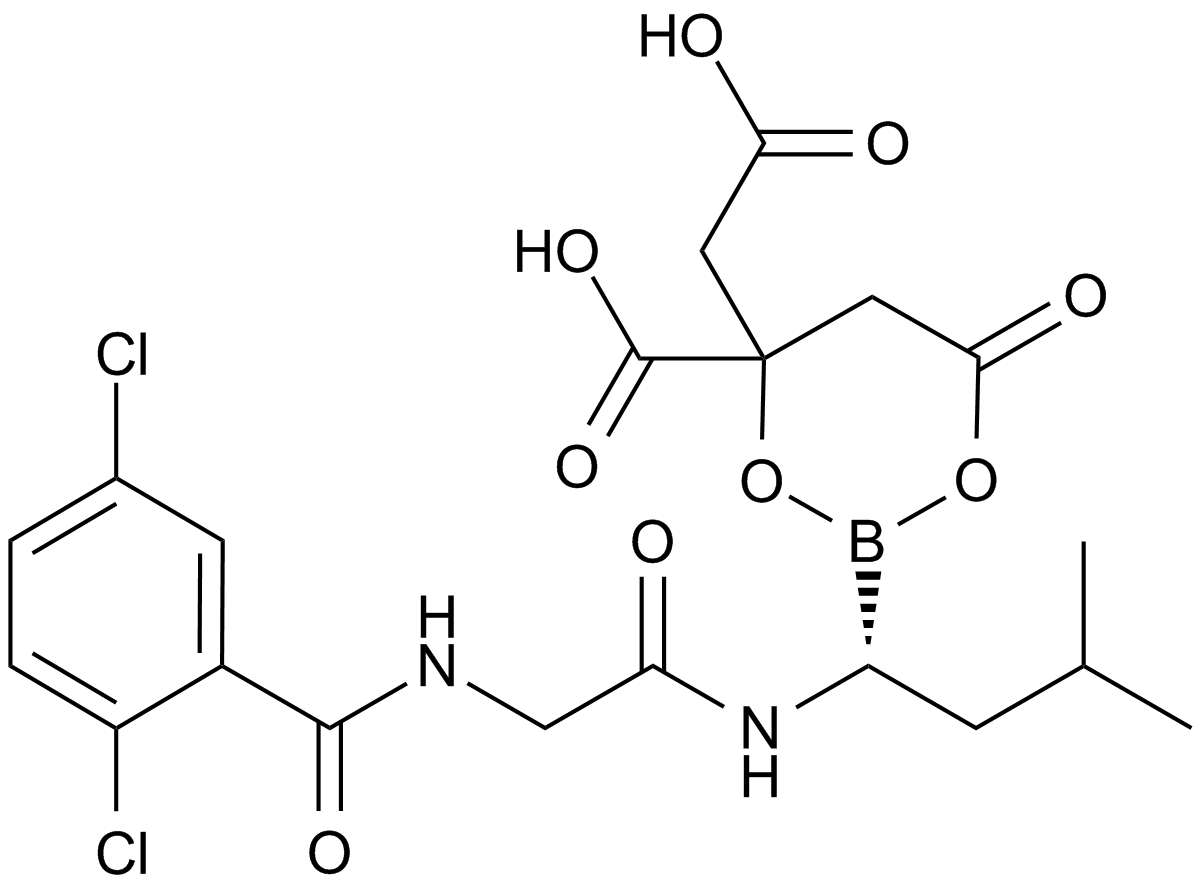
GC13718
MLN9708
A prodrug form of MLN2238
-
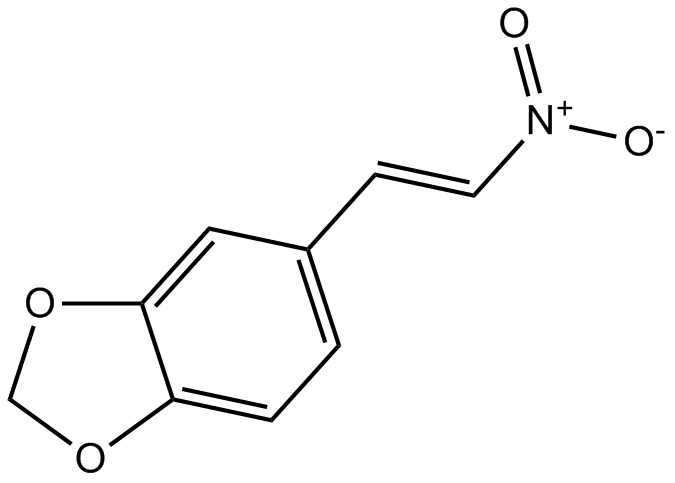
GC10048
MNS
Inhibitor of Src/Syk tyrosine kinases
-
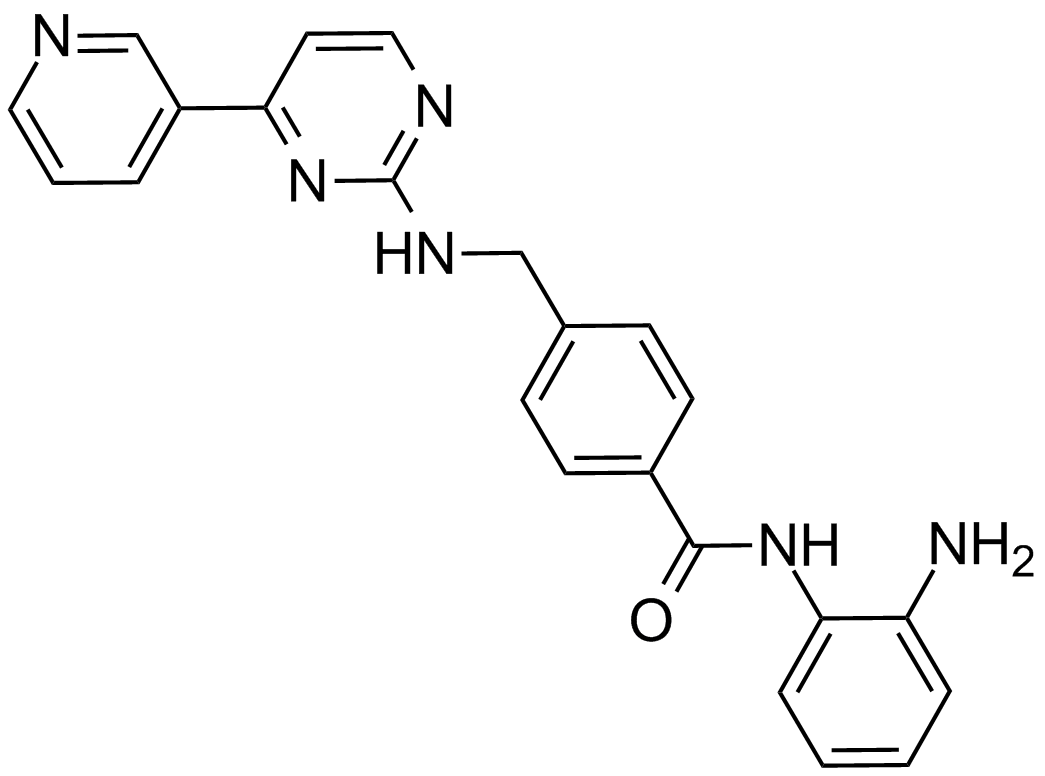
GC13055
Mocetinostat (MGCD0103, MG0103)
An orally available HDAC inhibitor
-
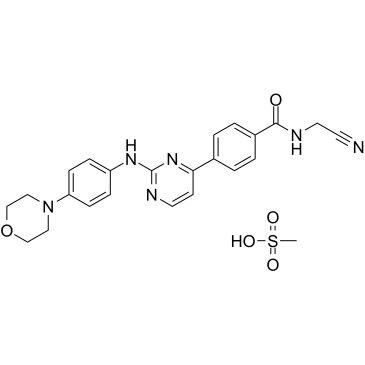
GC36646
Momelotinib Mesylate
Momelotinib Mesylate (CYT387 Mesylate) is an ATP-competitive inhibitor of JAK1/JAK2 with IC50 of 11 nM/18 nM, appr 10-fold selectivity versus JAK3.
-
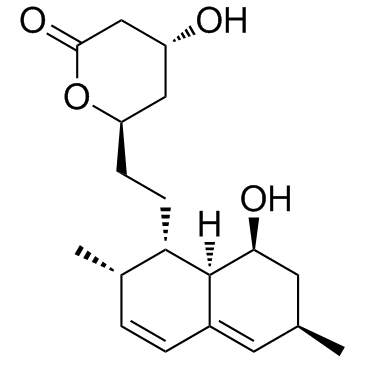
GC31586
Monacolin J (Antibiotic MB 530A)
Monacolin J (Antibiotic MB 530A) is an inhibitor of cholesterol biosynthesis, and inhibits the activity of HMG-CoA reductase.
-
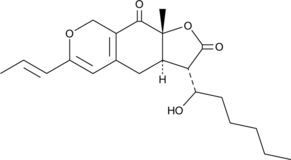
GC48676
Monascuspiloin
A fungal metabolite with anticancer activity
-
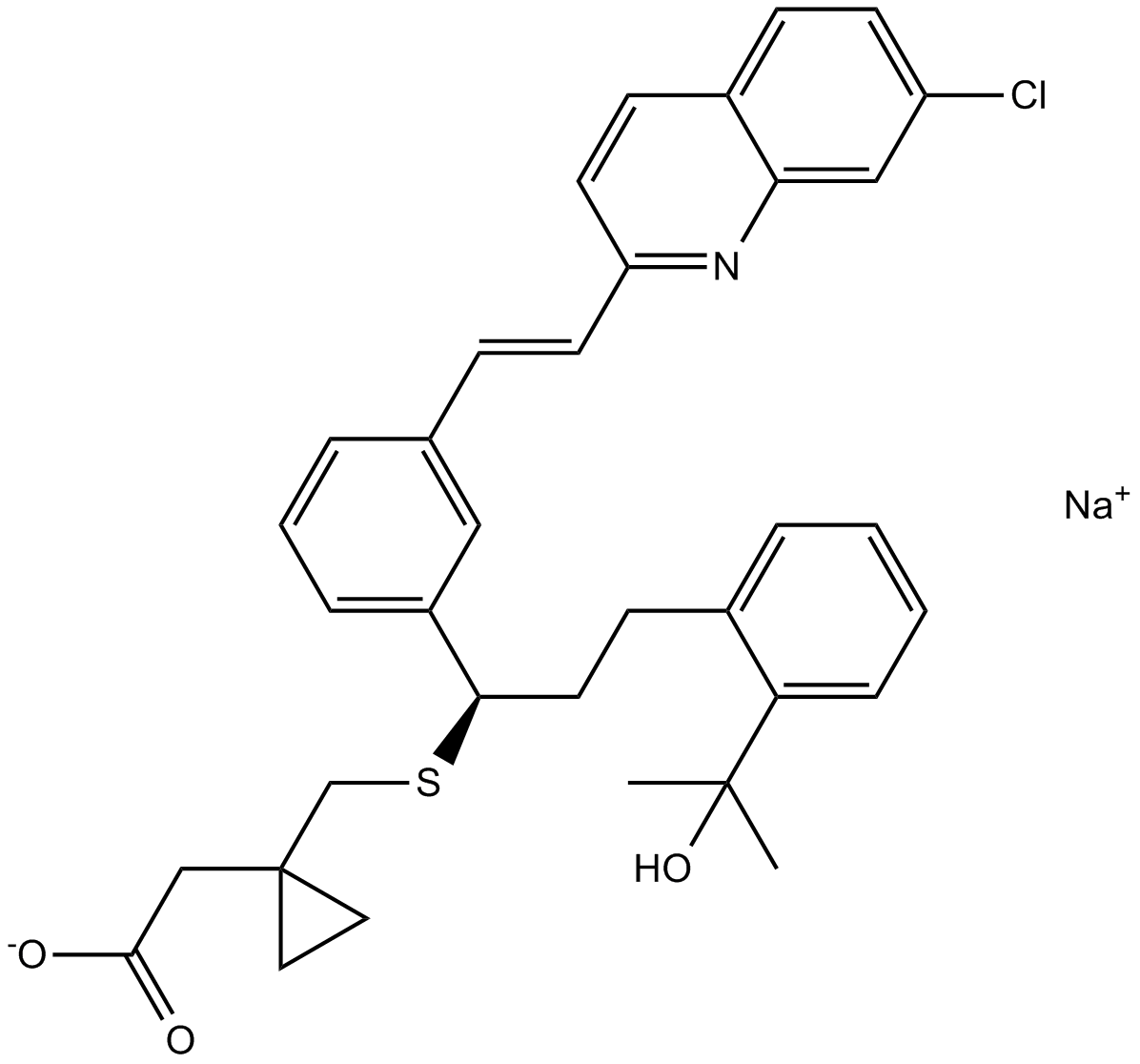
GC11006
Montelukast Sodium
Leukotriene receptor antagonist
-
GC17136
MRT67307
A kinase inhibitor
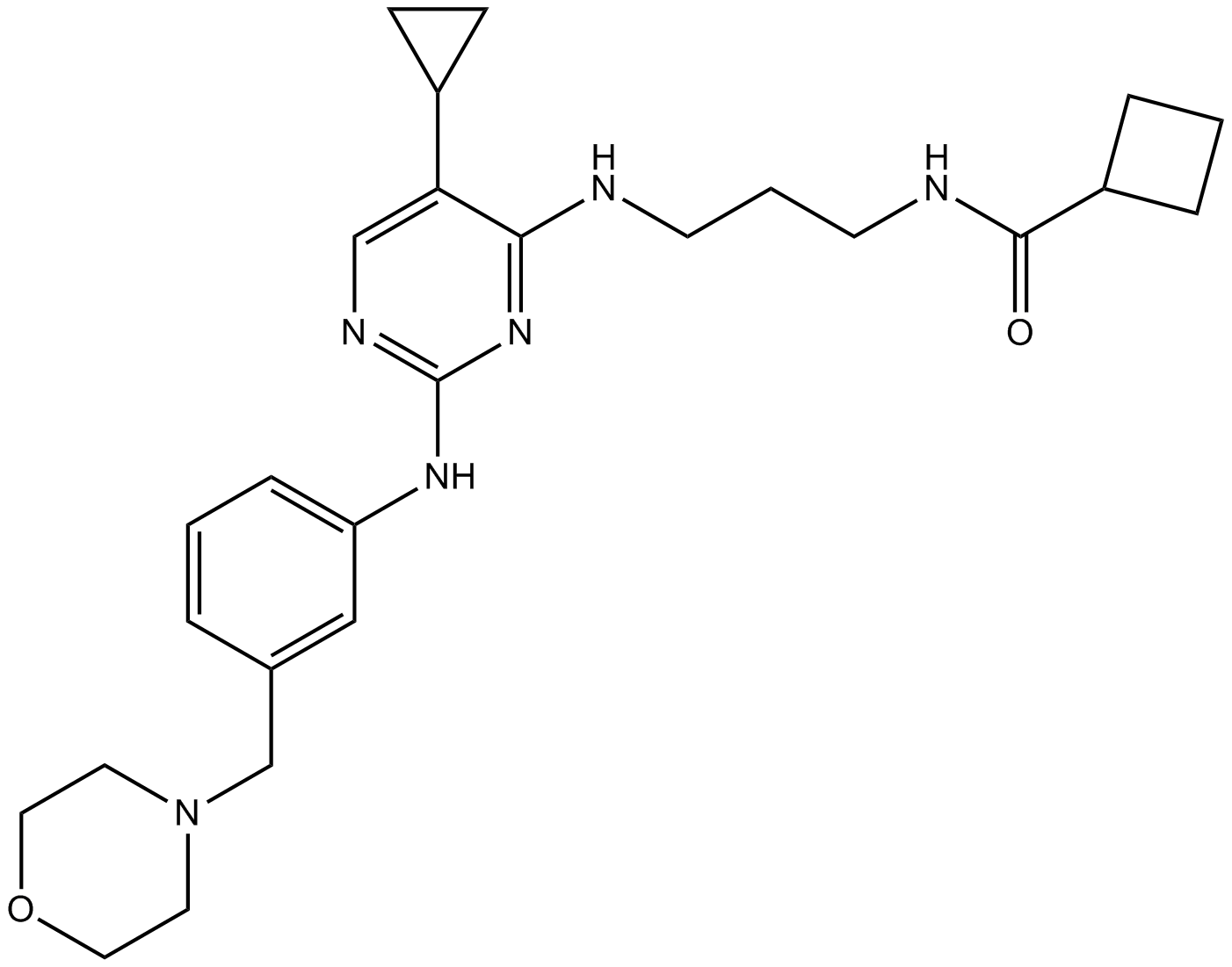
-
GC25650
MRT67307 HCl
MRT67307 is a potent and dual IKKε and TBK1 inhibitor with IC50 of 160 and 19 nM, respectively. MRT67307 potently inhibits ULK1 and ULK2 and blocks autophagy.
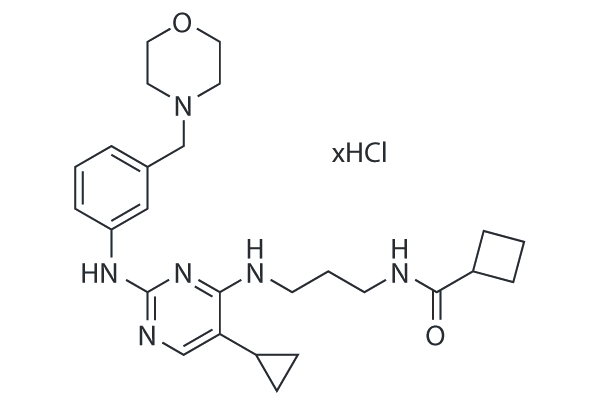
-
GC14086
MRT68921
A dual autophagy kinase ULK1/2 inhibitor
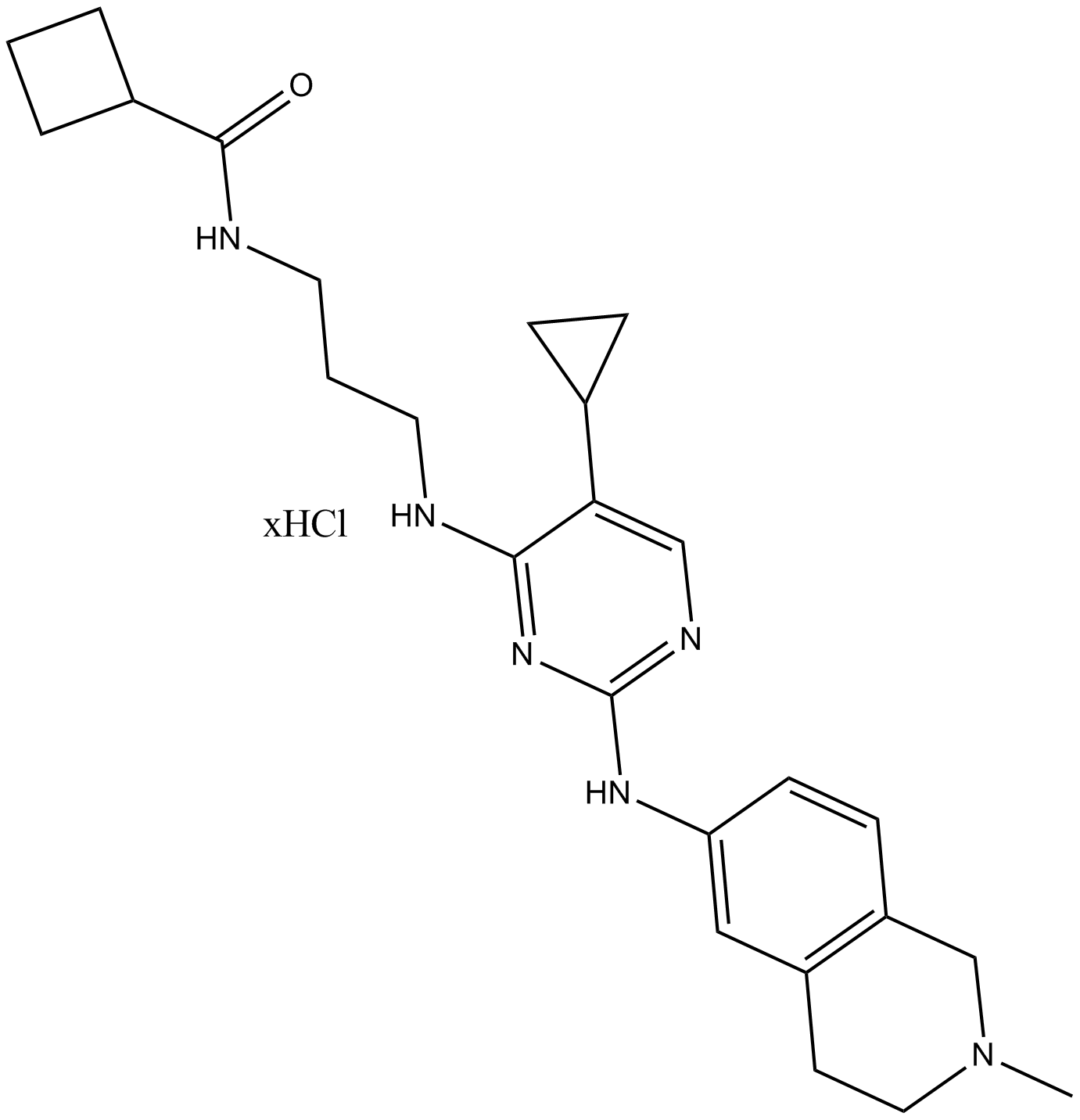
-
GC36654
MRT68921 dihydrochloride
MRT68921 dihydrochloride is a potent inhibitor of ULK1 and ULK2, with IC50 values of 2.9 nM and 1.1 nM, respectively.

-
GC25651
MRT68921 HCl
MRT68921 is a potent and dual autophagy kinase ULK1/2 inhibitor with IC50 of 2.9 nM and 1.1 nM, respectively.
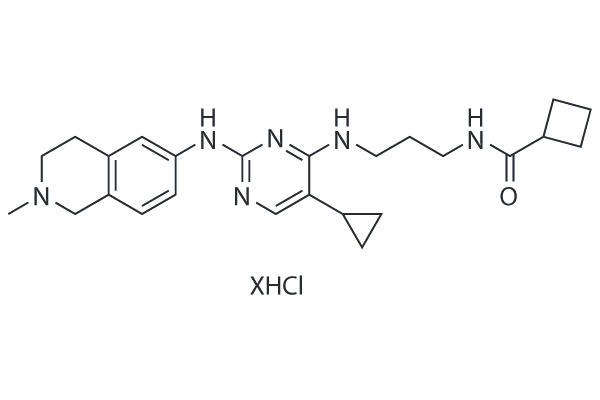
-
GC60257
MSC1094308
MSC1094308 is a non-competitive and reversible VPS4B/p97 (VCP) (I/II type AAA ATPase) allosteric inhibitor, with IC50 values of 0.71 μM and 7.2 μM for VPS4B and p97, respectively. MSC1094308 inhibits the D2 ATPase activity by binding to a drugable hotspot of p97. MSC1094308 can be used in study of cancer.
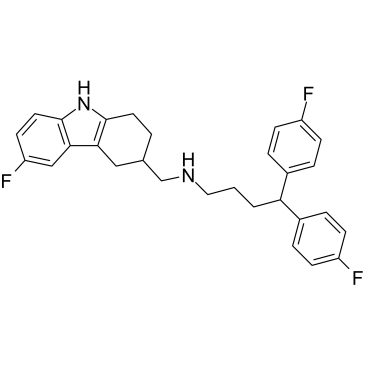
-
GC65115
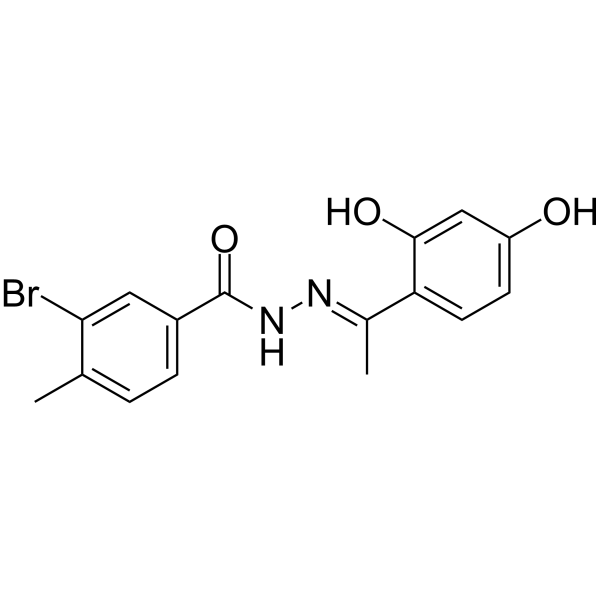
mTOR inhibitor-1
mTOR inhibitor-1 is a novel mTOR pathway inhibitor which can suppress cells proliferation and inducing autophagy.
-
GC62339
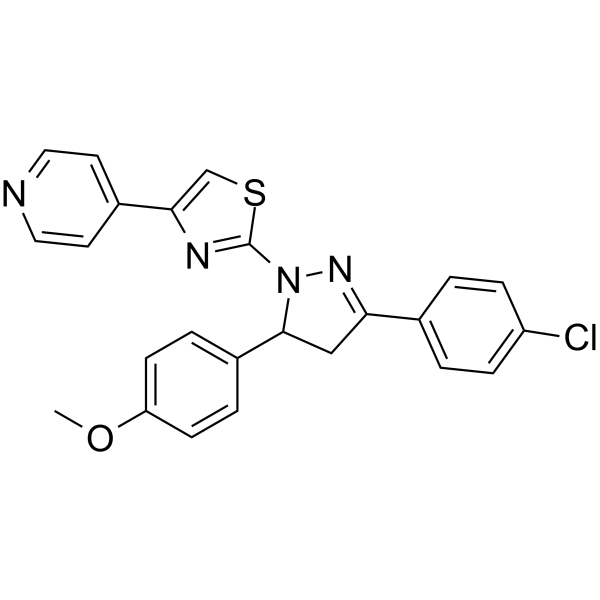
mTOR inhibitor-8
mTOR inhibitor-8 is an mTOR inhibitor and autophagy inducer. mTOR inhibitor-8 inhibits the activity of mTOR via FKBP12 and induces autophagy of A549 human lung cancer cells.
-
GC65297
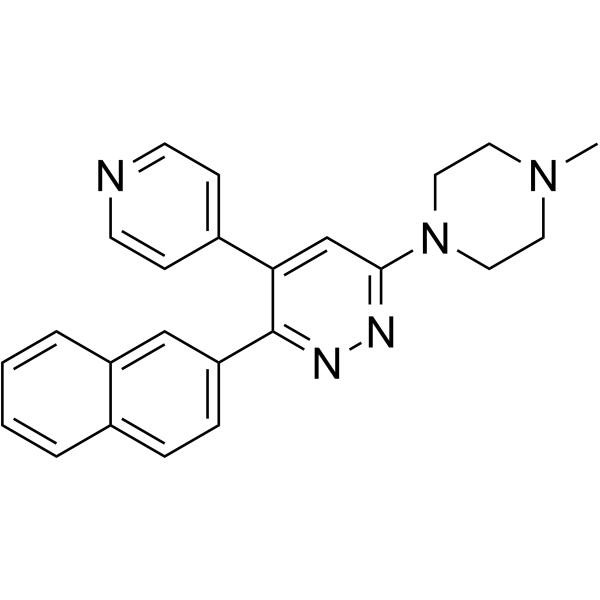
MW-150
MW150 (MW01-18-150SRM) is a selective, CNS penetrant, and orally active inhibitor of p38α MAPK with a Ki of 101 nM.
-
GC34918
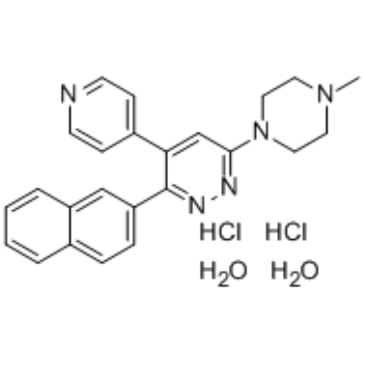
MW-150 dihydrochloride dihydrate
MW-150 dihydrochloride dihydrate (MW01-18-150SRM dihydrochloride dihydrate) is a selective, CNS penetrant, and orally active inhibitor of p38α MAPK with a Ki of 101 nM.
-
GC34094
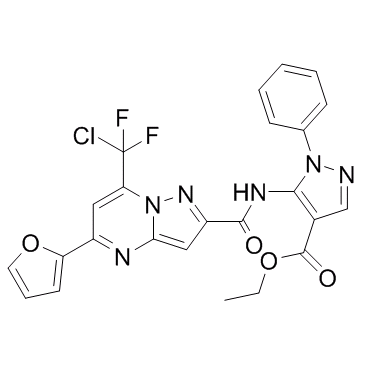
Mycro 3
Mycro 3 is a potent and selective inhibitor of Myc-associated factor X (MAX) dimerization. Mycro 3 also inhibit DNA binding of c-Myc. Mycro 3 could be used for the research of pancreatic cancer.
-
GN10634
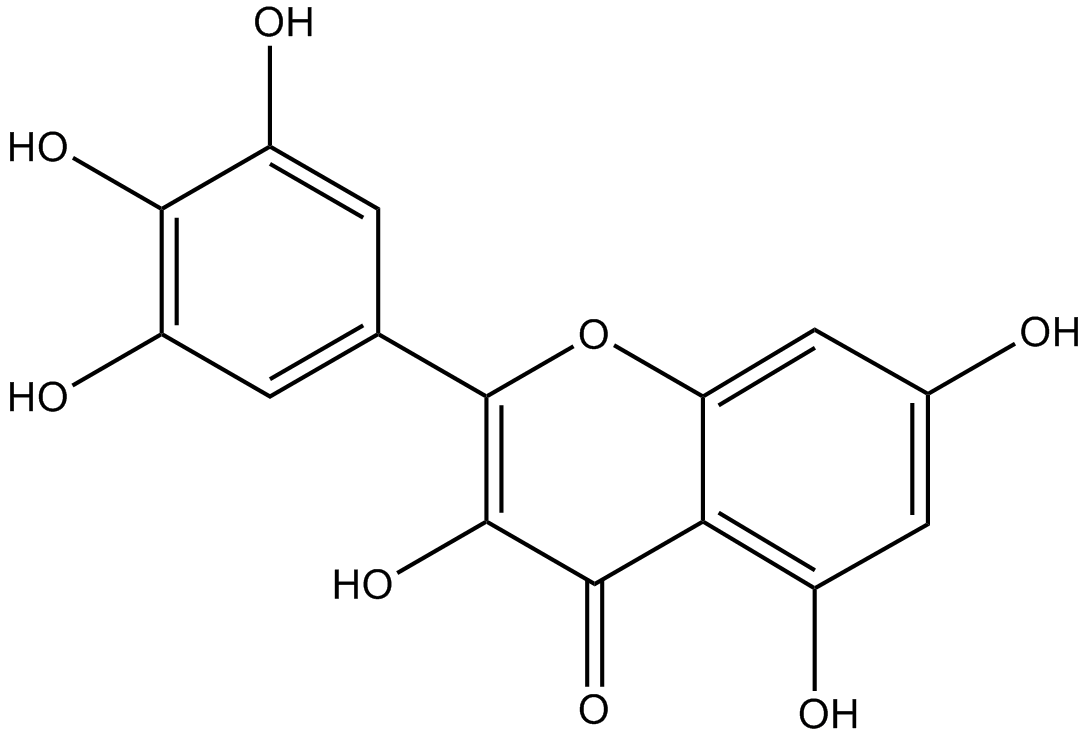
Myricetin
-
GC47735
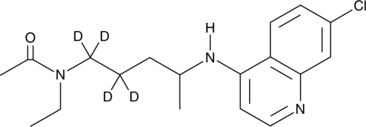
N-acetyl Desethylchloroquine-d4
An internal standard for the quantification of N-acetyl desethylchloroquine
-
GC36671
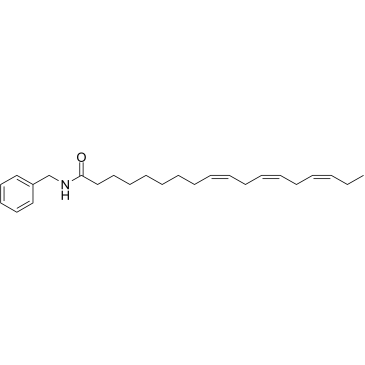
N-Benzyllinolenamide
N-Benzyllinolenamide is a natural macamide isolated from Lepidium meyenii, acts as an inhibitor of fatty acid amide hydrolase (FAAH) with an IC50 of 41.8 μM.
-
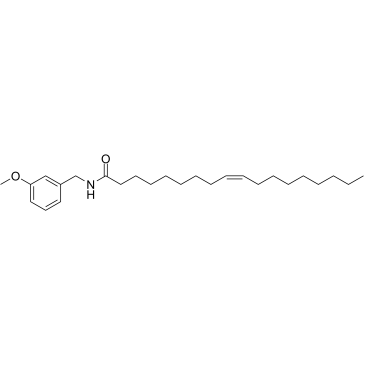
GC36703
N-Benzyloleamide
N-Benzyloleamide is a maccamide isolated from Lepidium meyenii (Maca).
-
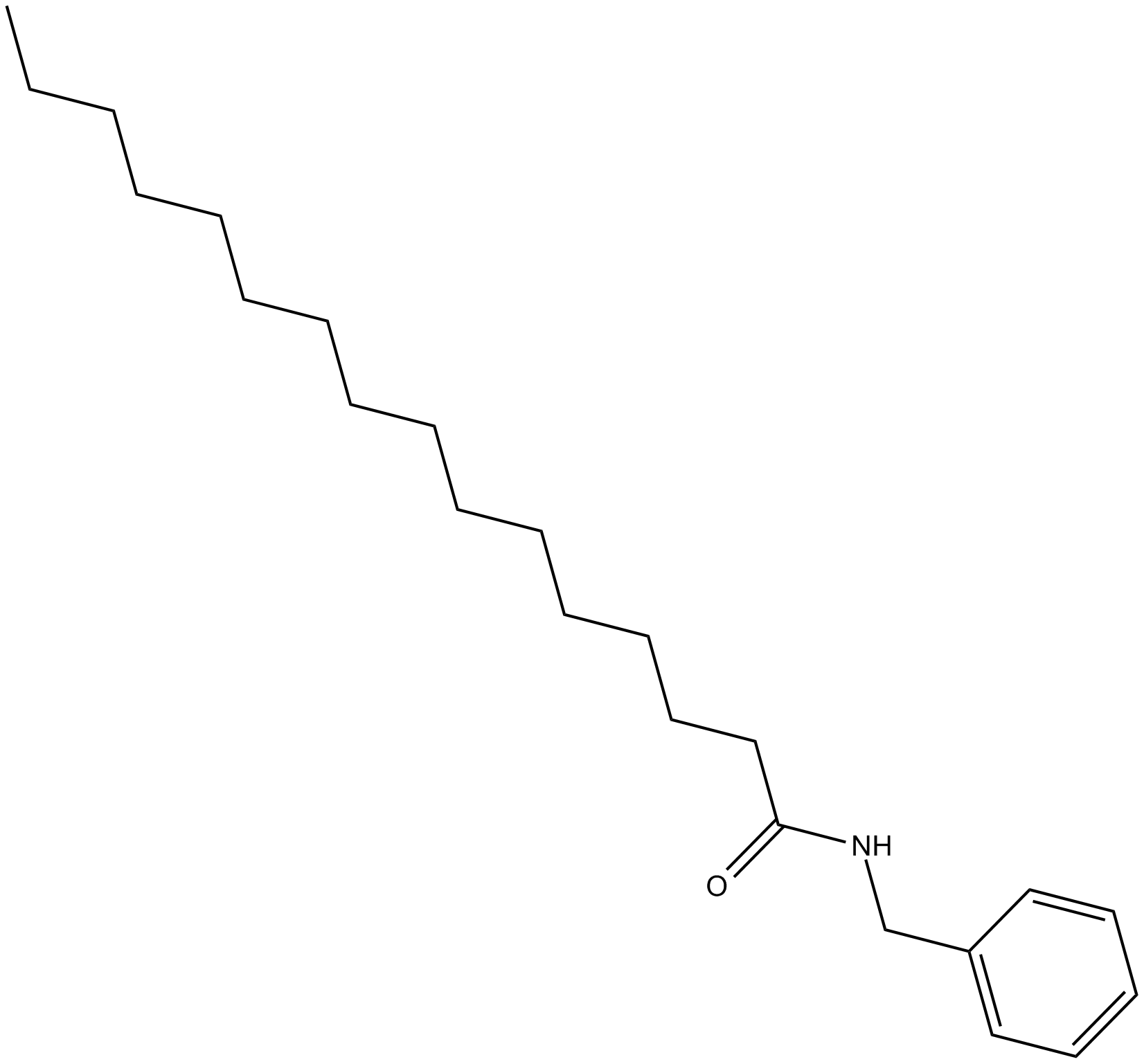
GC13063
N-Benzylpalmitamide
inhibitor of fatty acid amide hydrolase (FAAH)
-
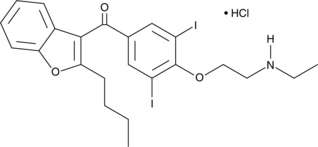
GC44346
N-Desethylamiodarone (hydrochloride)
N-Desethylamiodarone (hydrochloride) (N-desethylamiodarone hydrochloride) is a major active metabolite of Amiodarone.
-
GC50234
N106
Activator of SUMO-activating enzyme, E1 ligase
-
GC44290
NAADP (sodium salt)
Nicotinic acid adenine dinucleotide phosphate (NAADP) is a secondary messenger that induces calcium mobilization.
-
GC44291
NAB 2
NAB 2 is an inhibitor of α-synuclein-induced toxicity.
-
GC36690
Nampt-IN-3
Nampt-IN-3 (Compound 35) simultaneously inhibit nicotinamide phosphoribosyltransferase (NAMPT) and HDAC with IC50s of 31 nM and 55 nM, respectively. Nampt-IN-3 effectively induces cell apoptosis and autophagy and ultimately leads to cell death.
-
GC17975
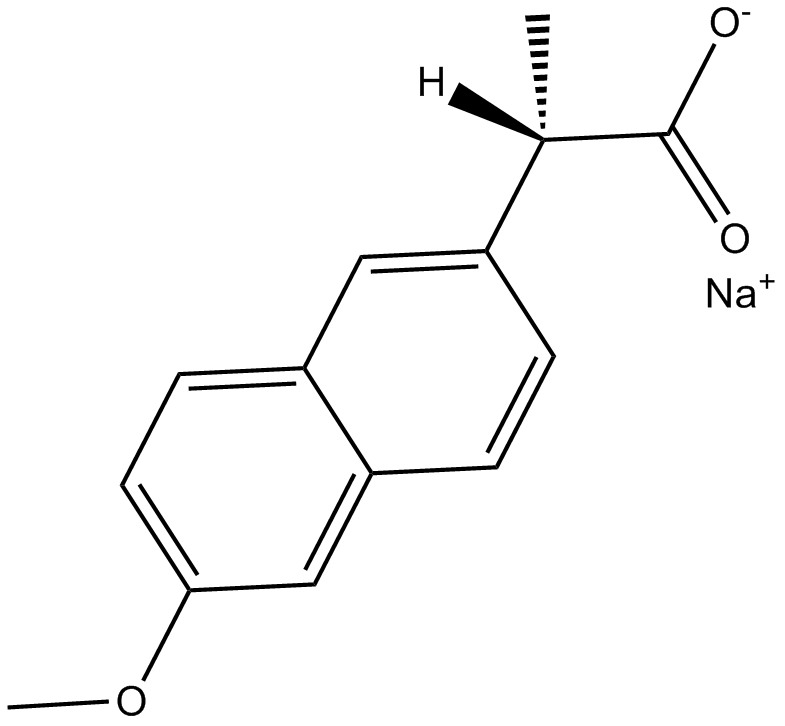
Naproxen Sodium
cyclooxygenase inhibitor
-
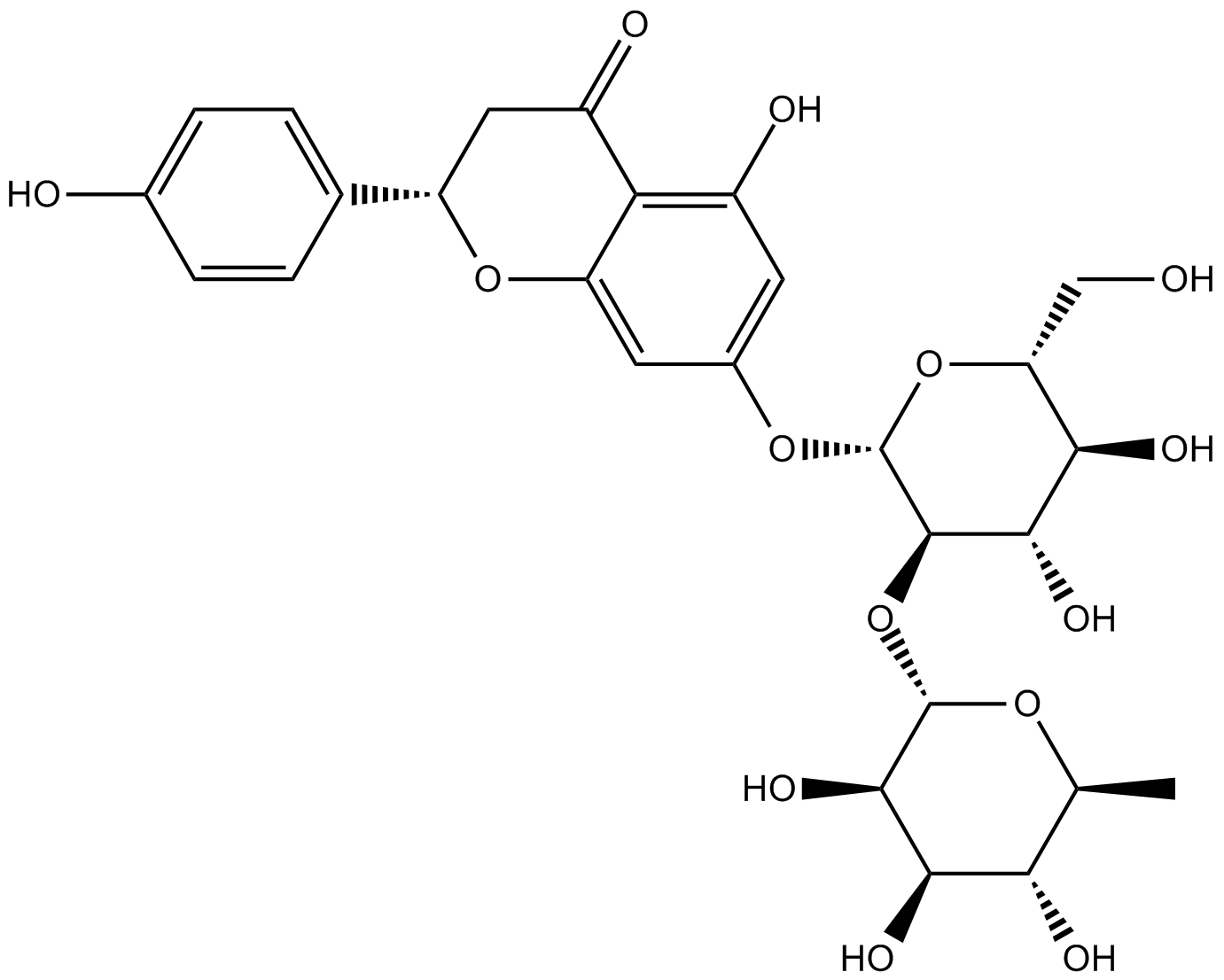
GN10257
Naringin
-
GC11008
Necrostatin-1
A RIP1 kinase inhibitor