HIV
HIV (human immunodeficiency virus) is a lentivirus that infect immune system. It cause immune system failure and lead to AIDS (acquired immnudeficiency syndrome).
Products for HIV
- Cat.No. Product Name Information
-
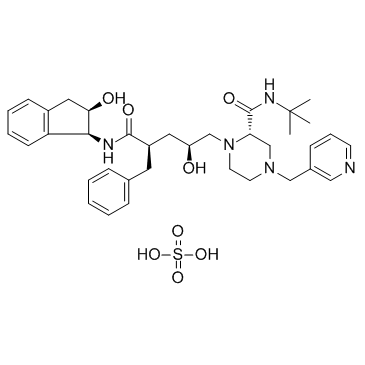
GC36310
Indinavir sulfate
Indinavir sulfate (MK-639) is an orally active and selective HIV-1 protease inhibitor with a Ki of 0.54 nM for PR.
-
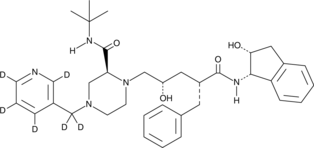
GC47455
Indinavir-d6
An internal standard for the quantification of indinavir
-
GC52395
Indolicidin (trifluoroacetate salt)
An antimicrobial peptide

-
GC18745
Integracin A
Integracin A is an inhibitor of HIV-1 integrase (IC50 = 3.2 μM in an HIV-1 integrase coupled assay).
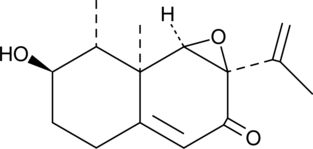
-
GC43905
Integracin B
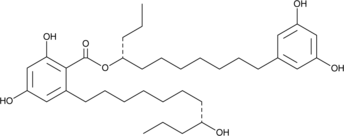
Integracin B is a fungal metabolite and an inhibitor of HIV-1 integrase (IC50 = 6.1 μM in an HIV-1 integrase coupled assay).
-
GC36320
InteriotherinA
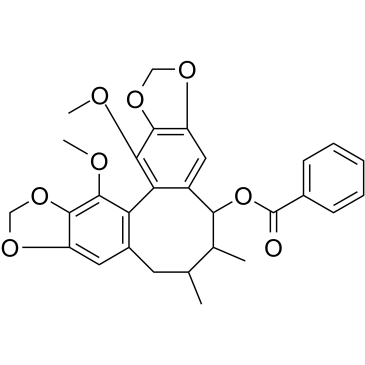
Interiotherin A is a lignan with a dibenzocyclooctadiene skeleton isolated from Kadsura interior.
-
GC32306
Islatravir (MK-8591)
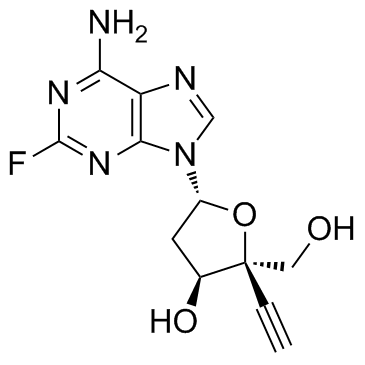
Islatravir (MK-8591) (MK-8591) is a potent anti-HIV-1 agent, acting as a nucleoside reverse transcriptase inhibitor, with EC50s of 0.068 nM, 3.1 nM and 0.15 nM for HIV-1 (WT), HIV-1 (M184V), HIV-1 (MDR), respectively.
-
GC18042
Isoprinosine
Isoprinosine is a potent, broad-spectrum antiviral compound for HIV infection.
-
GC36350
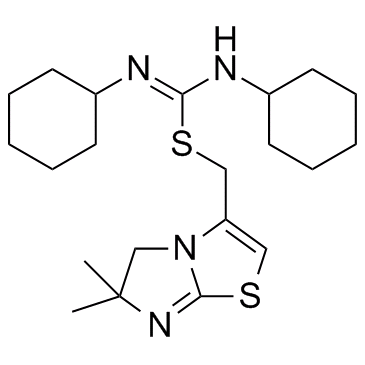
IT1t
IT1t is a potent CXCR4 antagonist; inhibits CXCL12/CXCR4 interaction with an IC50 of 2.1 nM.
-
GC10432
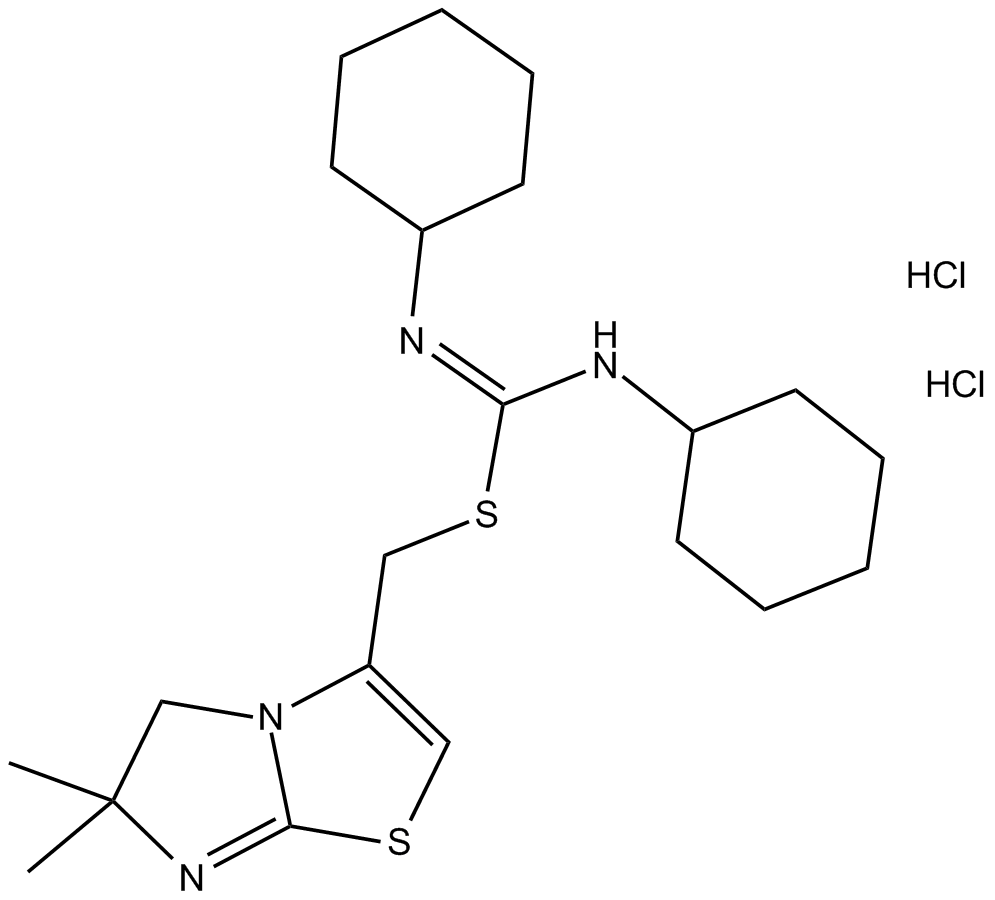
IT1t dihydrochloride
CXCR4 antagonist, potent
-
GC41236
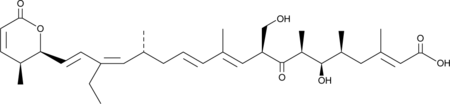
Kazusamycin A
Kazusamycin A is an antibiotic from Streptomyces and a hydroxy analog of Leptomycin B that demonstrates cytotoxic activity against various human and mouse tumor lines.
-
GC43995
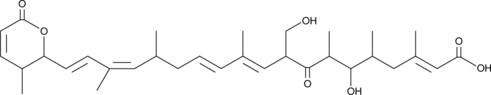
Kazusamycin B
Kazusamycin B is a bacterial metabolite originally isolated from Streptomyces.
-
GC60978
L-Chicoric Acid
A dicaffeoyl ester with diverse biological activities
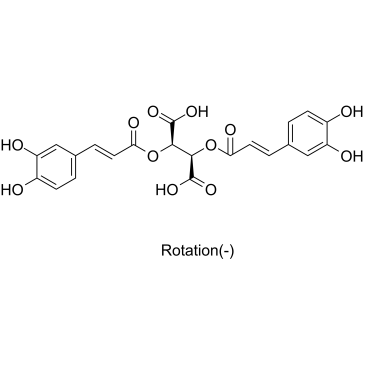
-
GC30862
L-Cycloserine ((S)-4-Amino-3-isoxazolidone)
L-Cycloserine ((S)-4-Amino-3-isoxazolidone) ((S)-4-Amino-3-isoxazolidone) irreversibly inhibits GABA pyridoxal 5′-phosphate-dependent aminitransferase in E.
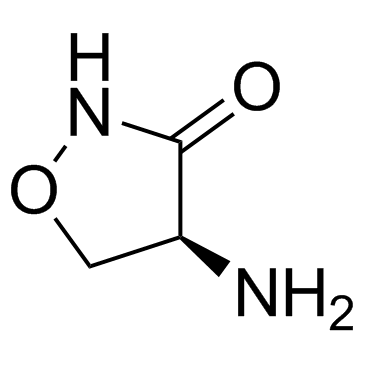
-
GC47572
L-Lysine lactam (hydrochloride)
A building block
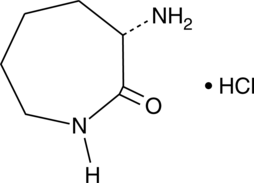
-
GC10317
Lamivudine
Nucleoside analog reverse transcriptase inhibitor
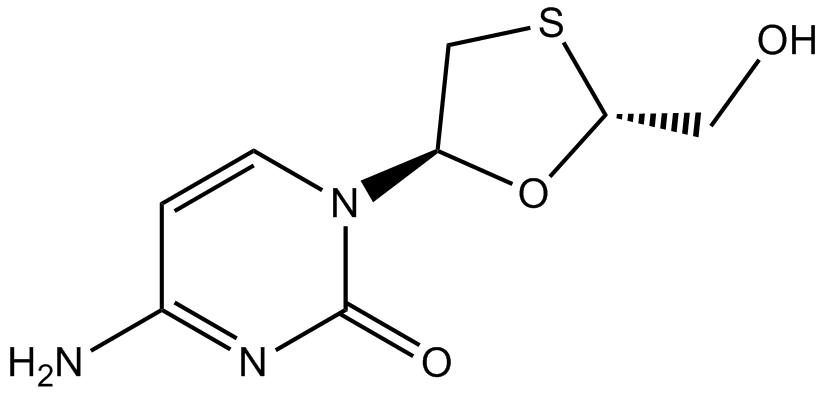
-
GC18345
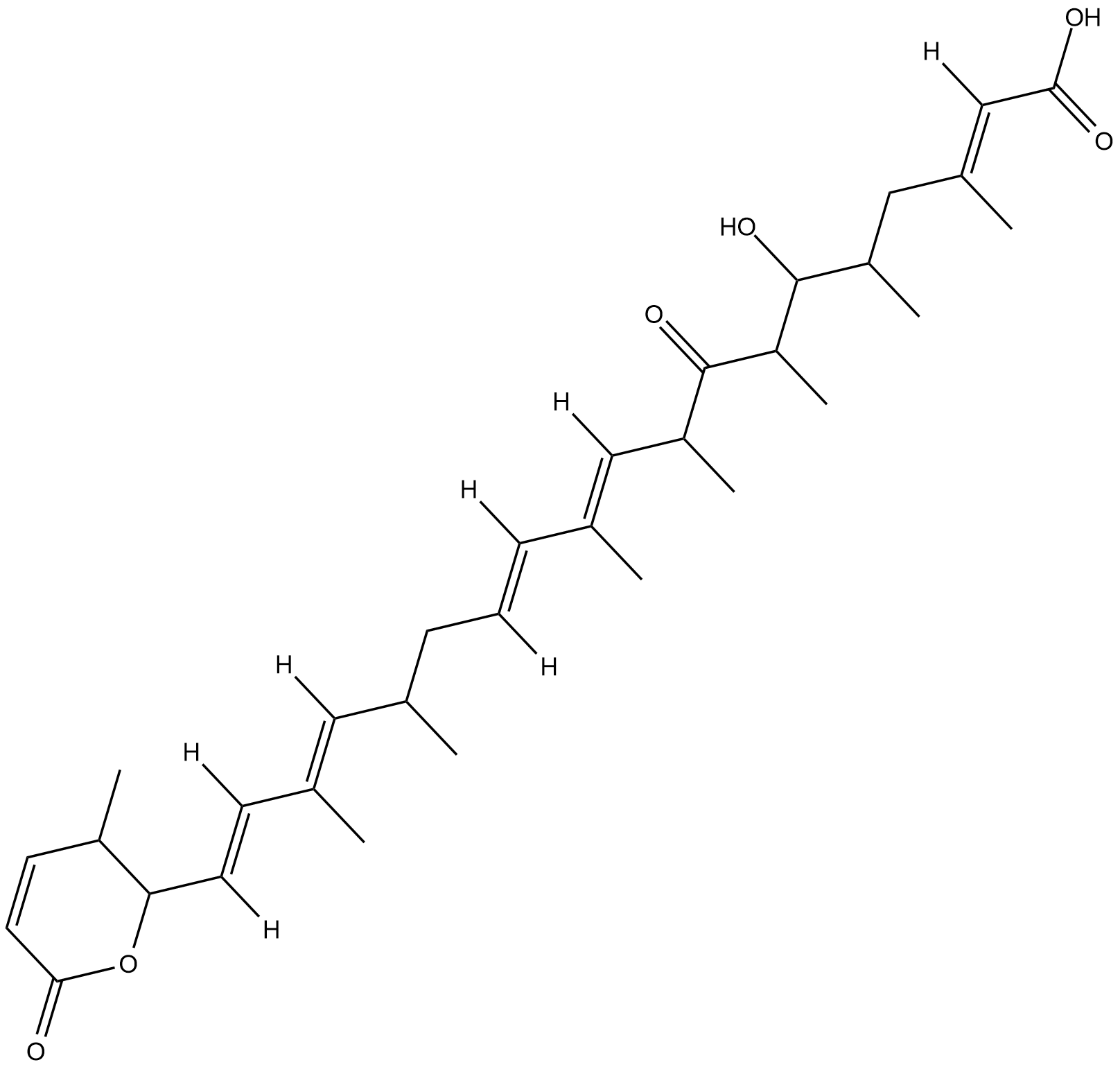
Leptomycin A
Leptomycin A is an inhibitor of CRM1 (exportin 1) that blocks CRM1 interaction with nuclear export signals, preventing the nuclear export of a broad range of proteins.
-
GC16596
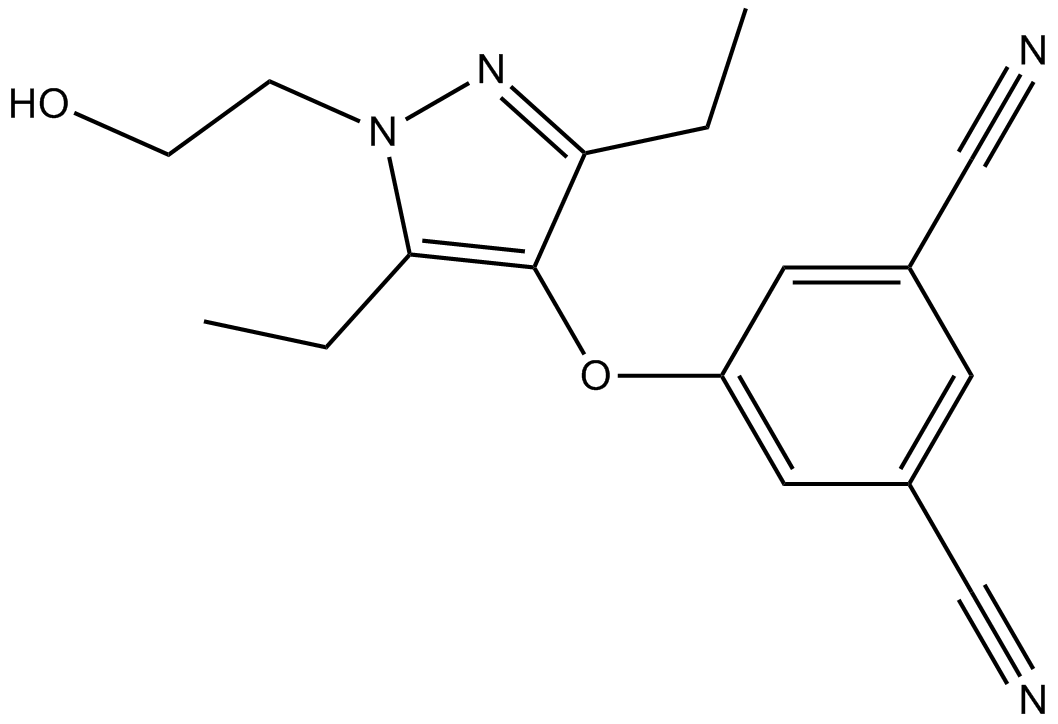
Lersivirine
An NNRTI
-
GC31715
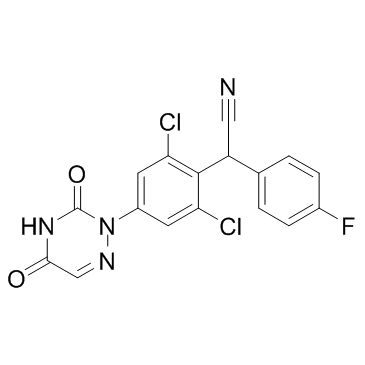
Letrazuril
Letrazuril is an anti-HIV agent.
-
GC16589
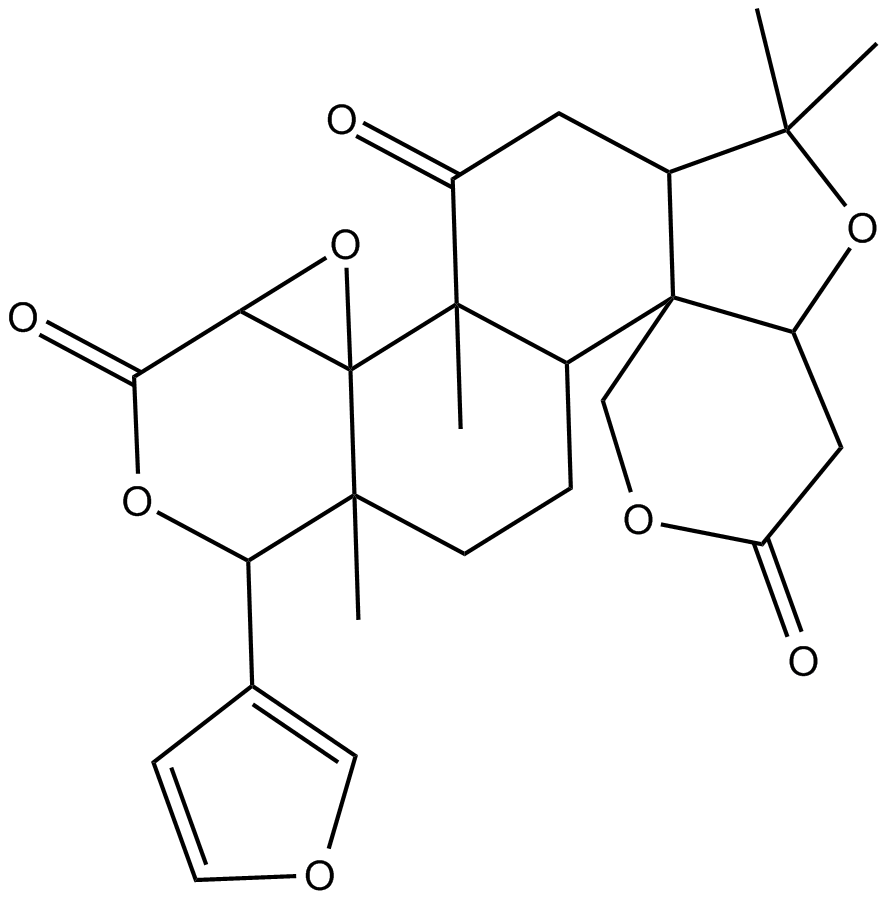
Limonin
A limonoid
-
GC13716
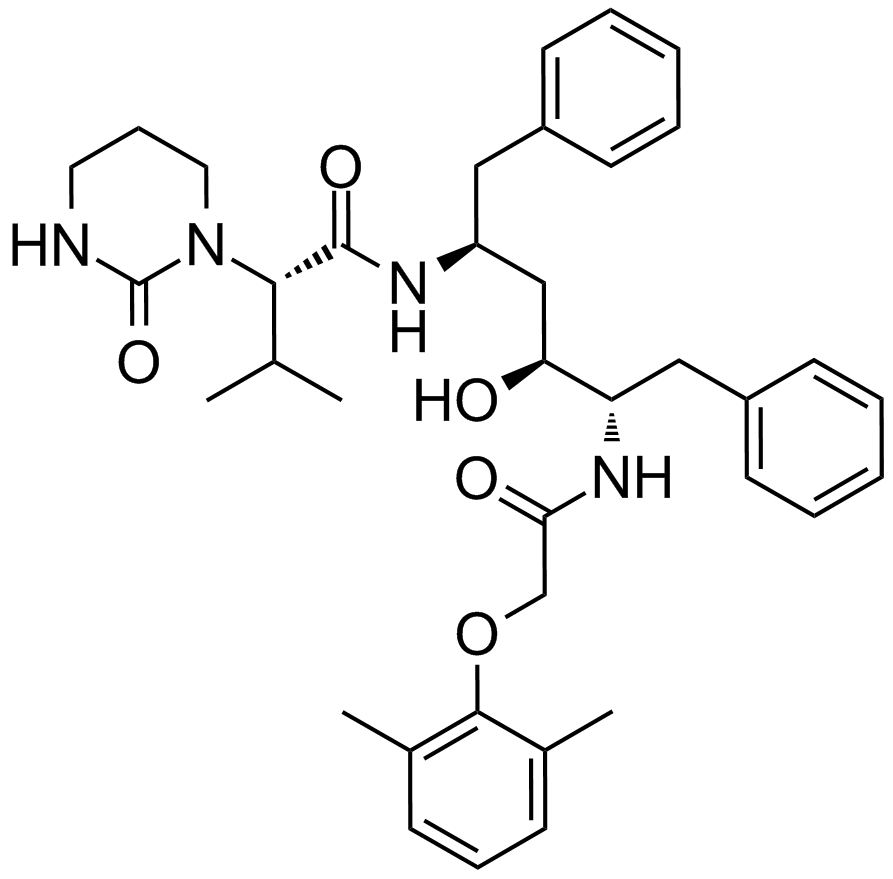
Lopinavir
A potent HIV-1 protease inhibitor
-
GC47573
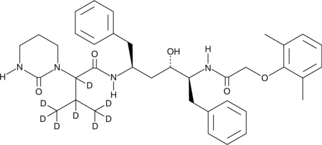
Lopinavir-d8
An internal standard for the quantification of lopinavir
-
GC32358
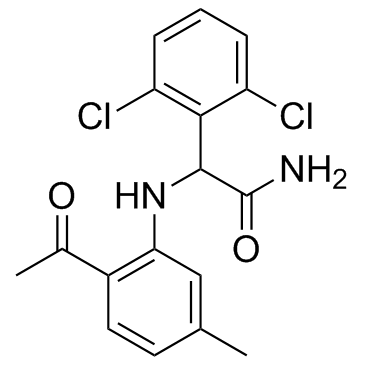
Loviride (R 89439)
Loviride (R 89439) (R 89439) is a non-nucleoside reverse transcriptase inhibitor (NNRTI), with an IC50 of 0.3 μM for reverse transcriptase from HIV-1.
-
GC44126
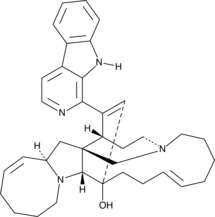
Manzamine A
Manzamine A is a β-carboline alkaloid with diverse activities that has been found in marine sponges, including X.
-
GC15090
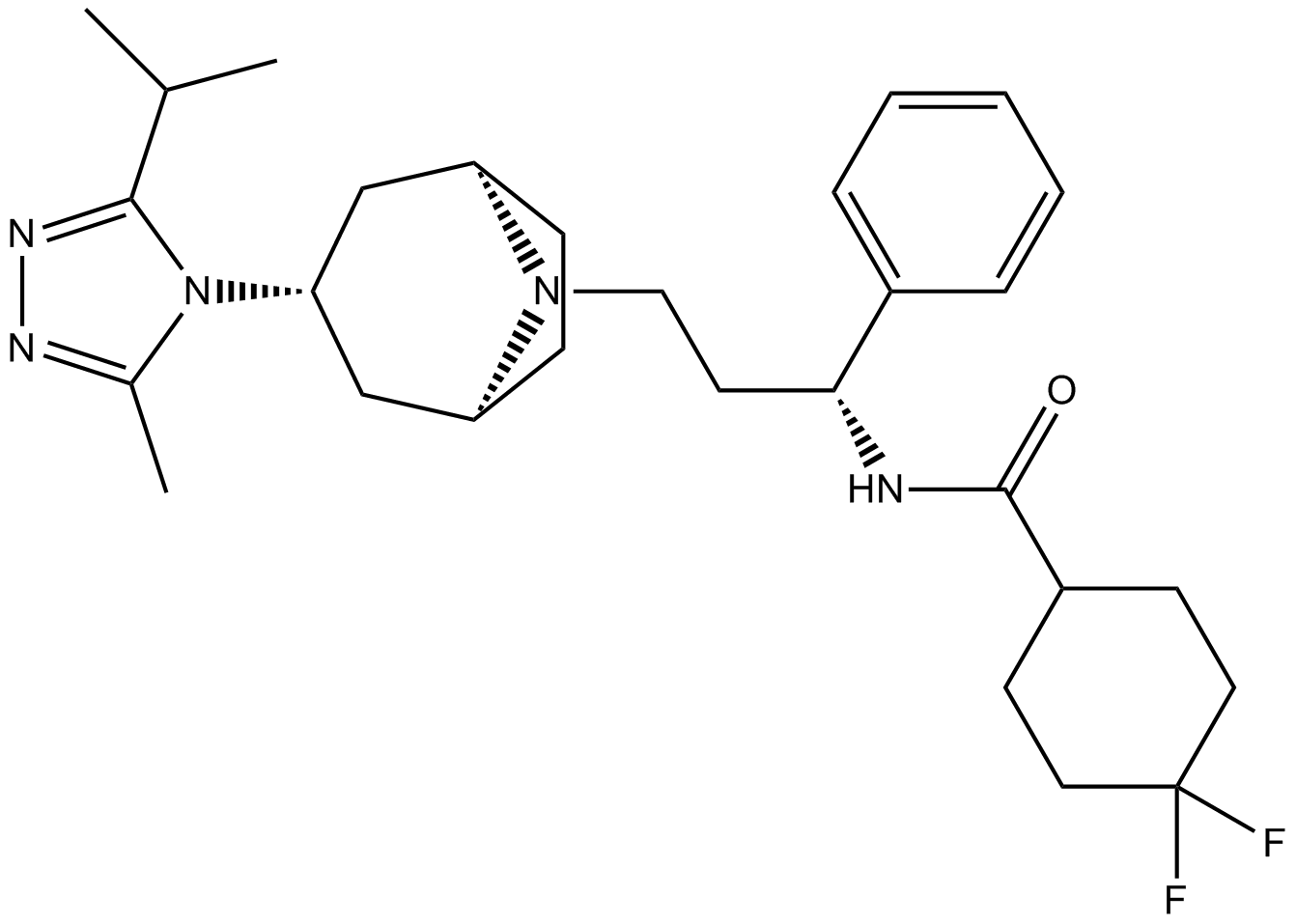
Maraviroc
Maraviroc, the first CCR5 antagonist, has been approved for the treatment of HIV infection, IC50 on hERG ion channel>10μM.
-
GC17632
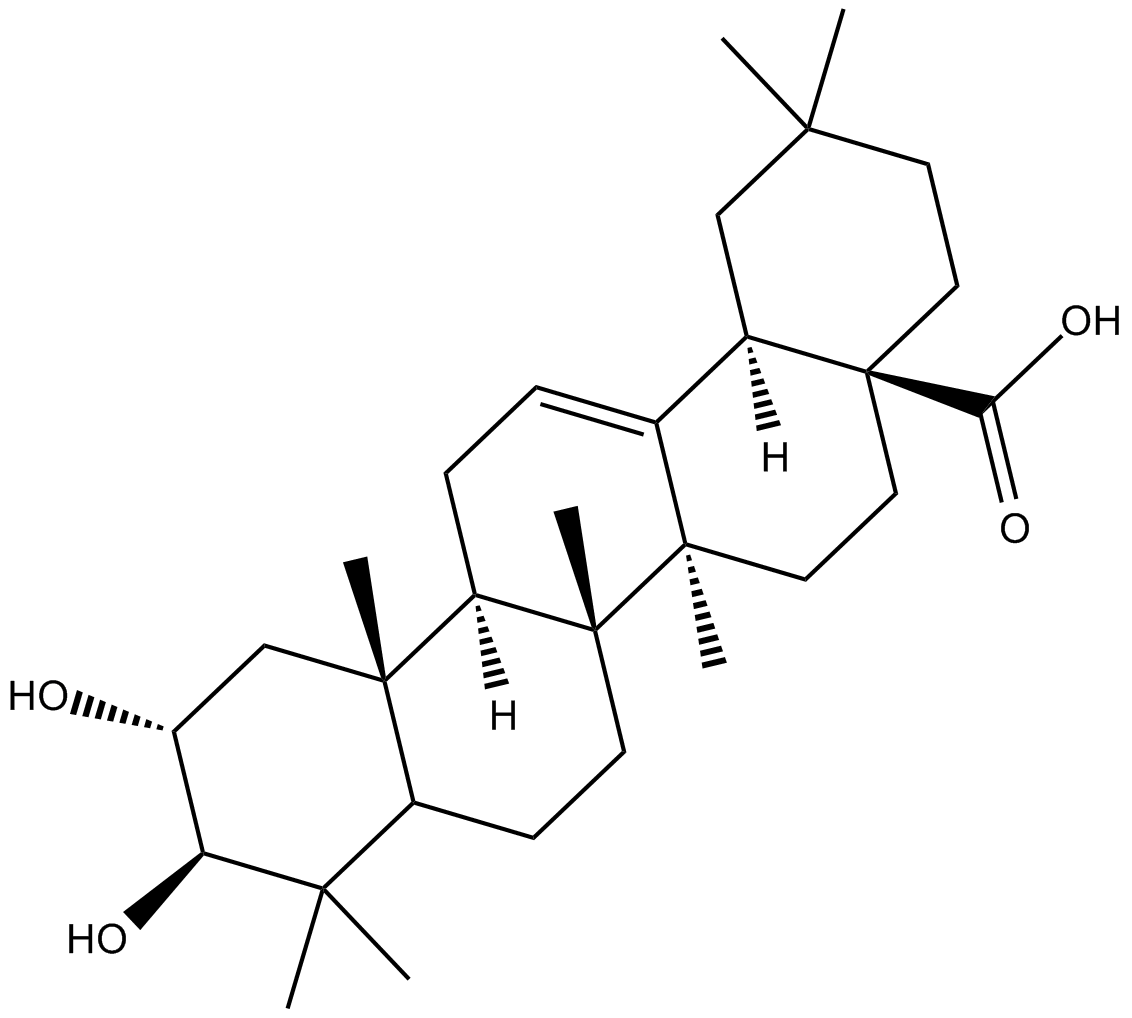
Maslinic Acid
inhibitor of NO, H2O2 , IL-6 and TNF-α
-
GC36550
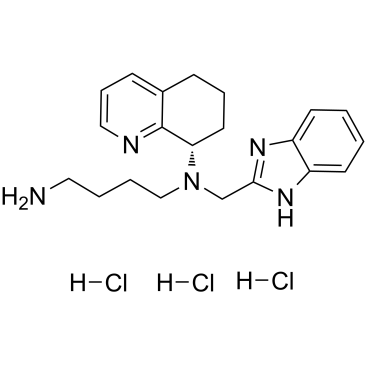
Mavorixafor trihydrochloride
Mavorixafor trihydrochloride (AMD-070 trihydrochloride) is a potent, selective and orally available CXCR4 antagonist, with an IC50 value of 13 nM against CXCR4 125I-SDF binding, and also inhibits the replication of T-tropic HIV-1 (NL4.3 strain) in MT-4 cells and PBMCs with an IC50 of 1 and 9 nM, respectively.
-
GC11688
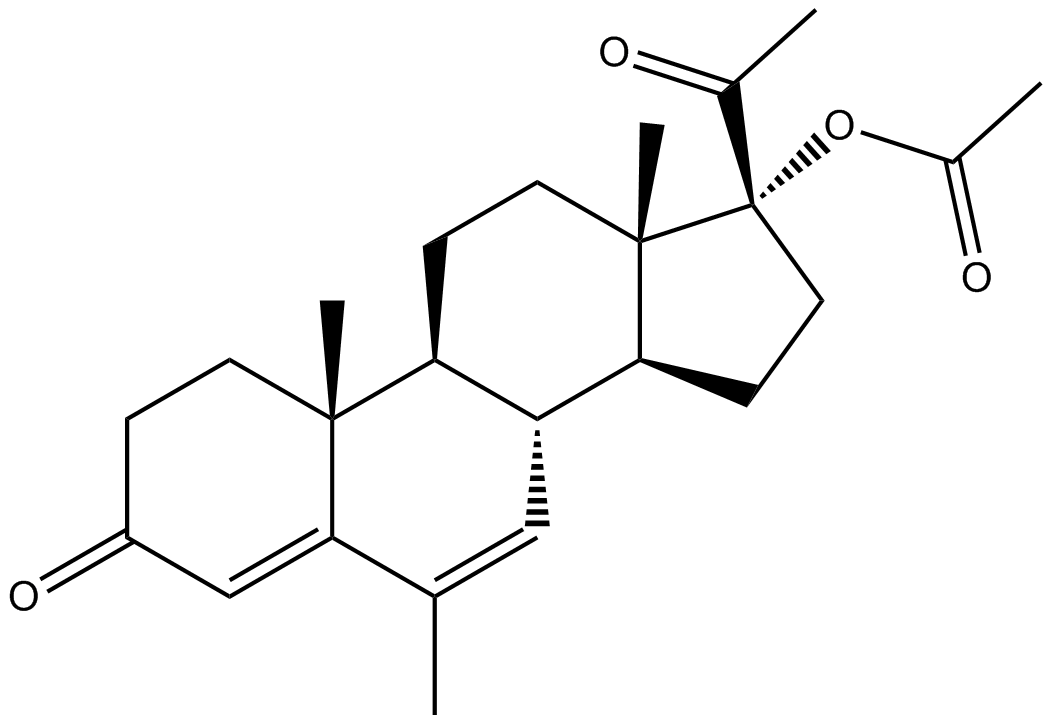
Megestrol Acetate
Anti-estrogen agent
-
GC10811
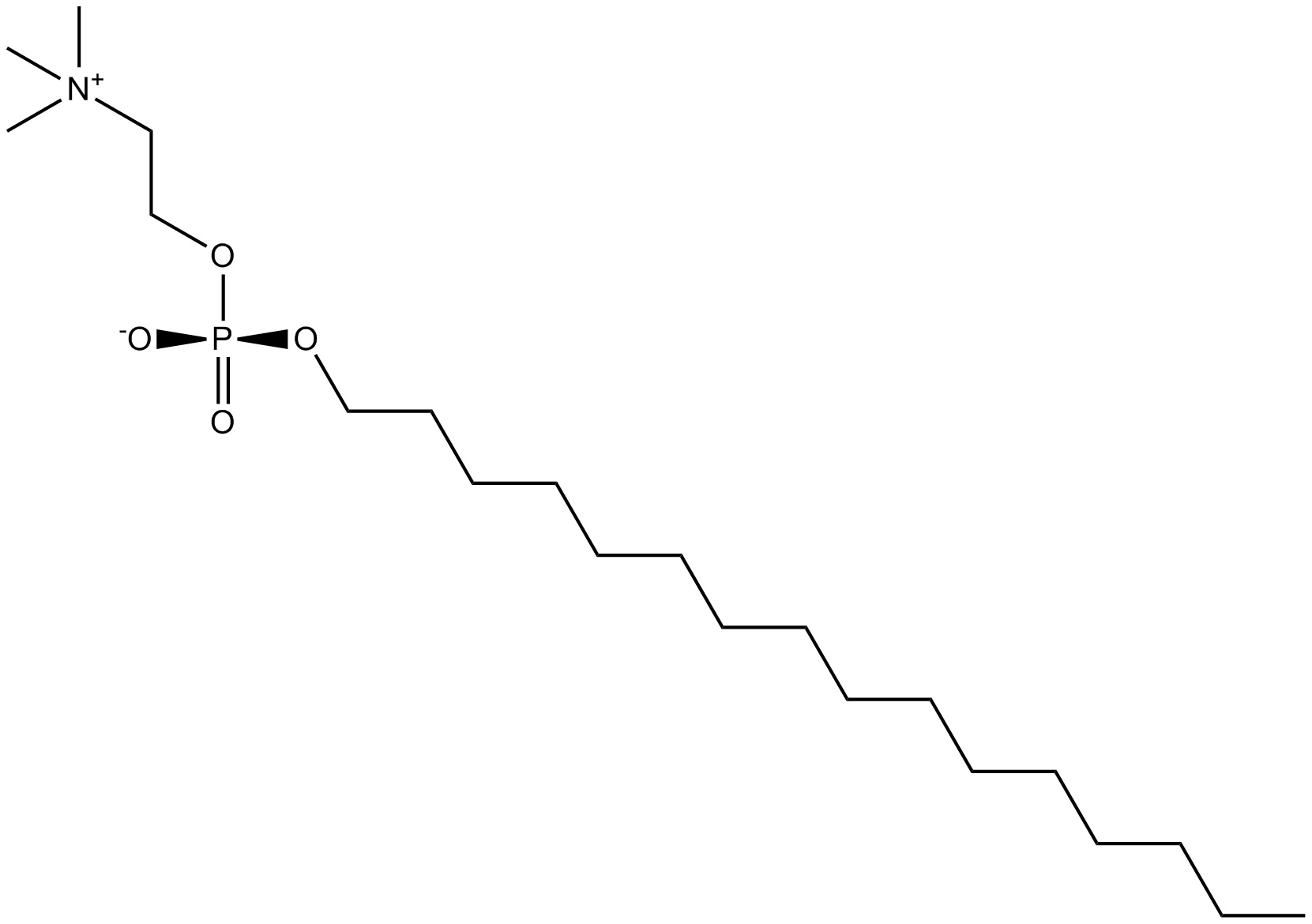
Miltefosine
PI3K/Akt inhibitor
-
GC39814
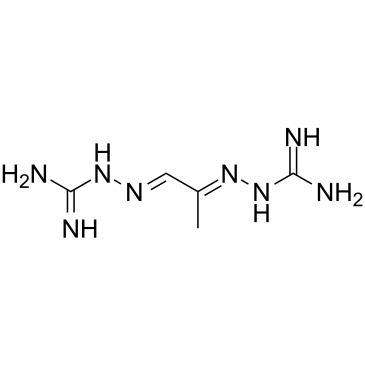
Mitoguazone
Mitoguazone (Methylglyoxal-bis(guanylhydrazone)) is a synthetic polycarbonyl derivative with potent antineoplastic activity.
-
GC45998
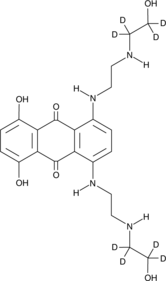
Mitoxantrone-d8
An internal standard for the quantification of mitoxantrone
-
GC32390
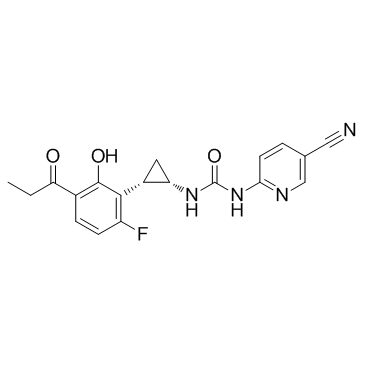
MIV-150 (PC 815)
MIV-150 (PC 815) is a nonnucleoside reverse transcriptase (NNRT) inhibitor, blocking HIV-1 and HIV-2 infections, with an EC50<1 nM against HIV-1/HIV-2MN.
-
GC15561
MK-2048
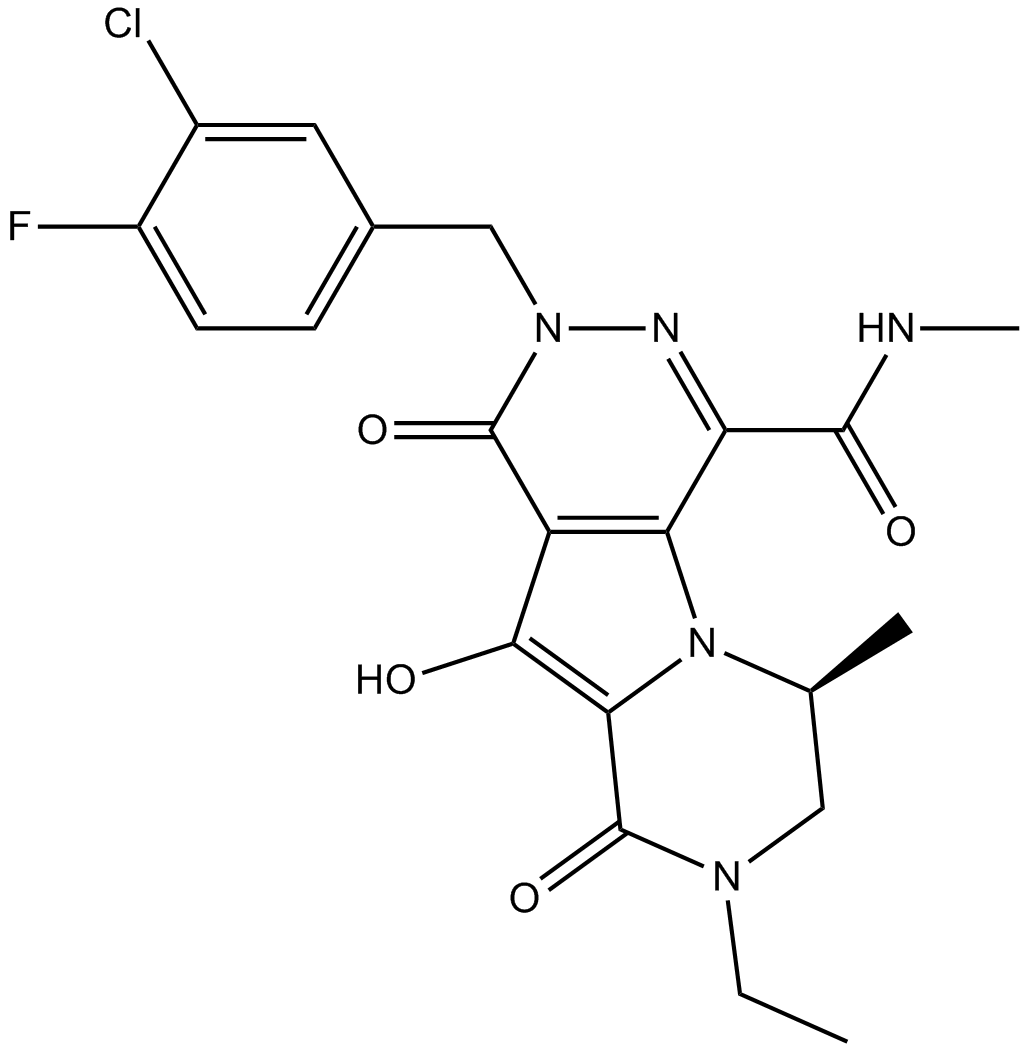
-
GC31881
MS417 (GTPL7512)
MS417 (GTPL7512) is a selective BET-specific BRD4 inhibitor, binds to BRD4-BD1 and BRD4-BD2 with IC50s of 30, 46 nM and Kds of 36.1, 25.4 nM, respectively, with weak selectivity at CBP BRD (IC50, 32.7 μM).
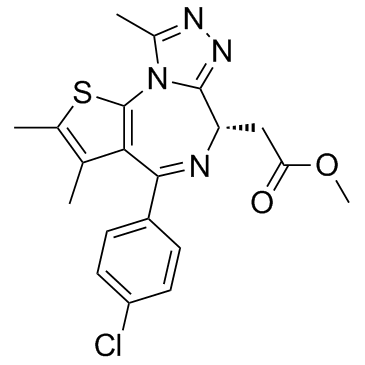
-
GC49388
N-Butyldeoxynojirimycin-d9 (hydrochloride)
An internal standard for the quantification of N-butyldeoxynojirimycin
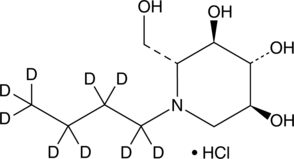
-
GC47741
Nalanthalide
A fungal metabolite with potassium channel inhibitory and antiviral activities
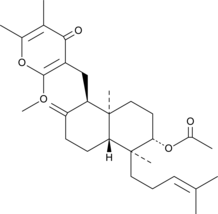
-
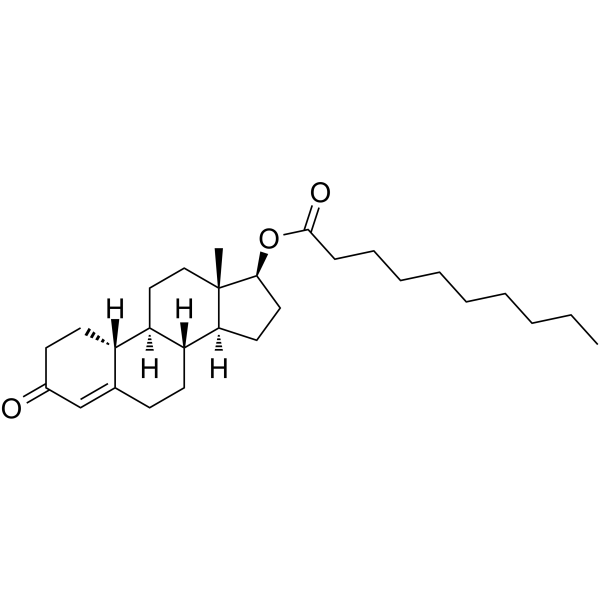
GC63095
Nandrolone decanoate
An Analytical Reference Standard
-
GC65905
NBD-14189
NBD-14189 is a potent HIV-1 entry antagonist with an IC50 of 89 nM against the HIV-1HXB2 pseudovirus. NBD-14189 binds to HIV-1 gp120 and shows potent antiviral activity (EC50<200 nM).

-
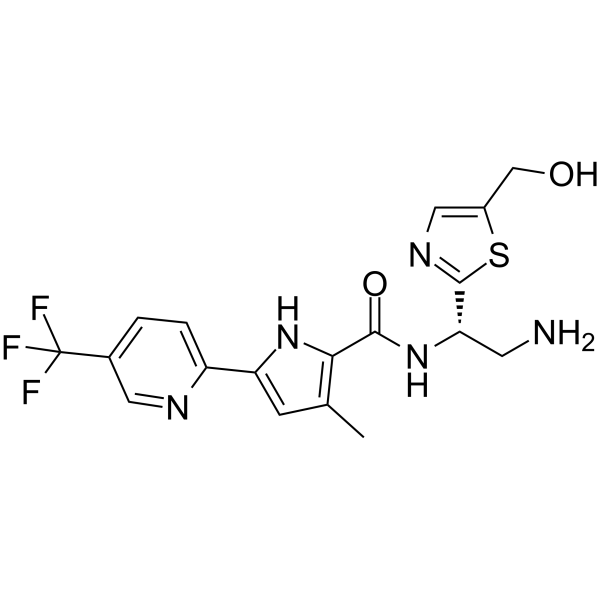
GC65903
NBD-14270
NBD-14270, a pyridine analogue, is a potent HIV-1 entry antagonist with an IC50 of 180 nM against 50 HIV-1 Env-pseudotyped viruses. NBD-14270 binds to HIV-1 gp120 and shows potent antiviral activity. NBD-14270 shows low cytotoxicity (CC50>100 μM).
-
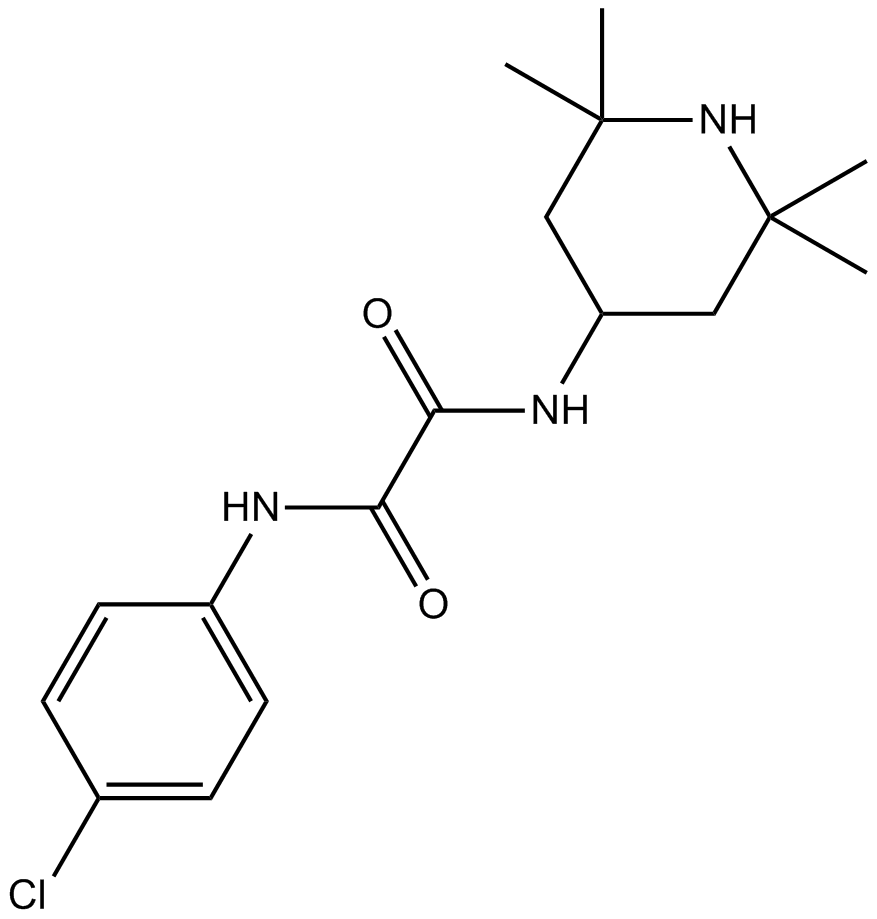
GC16035
NBD-556
A small molecule CD4 mimetic
-
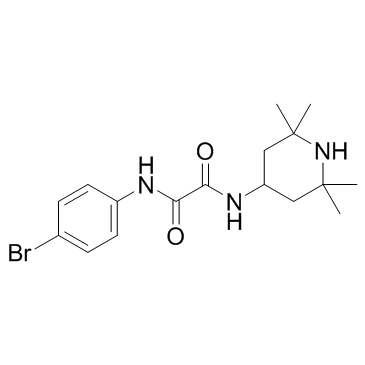
GC36702
NBD-557
NBD-557 is a potentially HIV-1 inhibitor.
-
GC17709
Nelfinavir
HIV-1 protease inhibitor
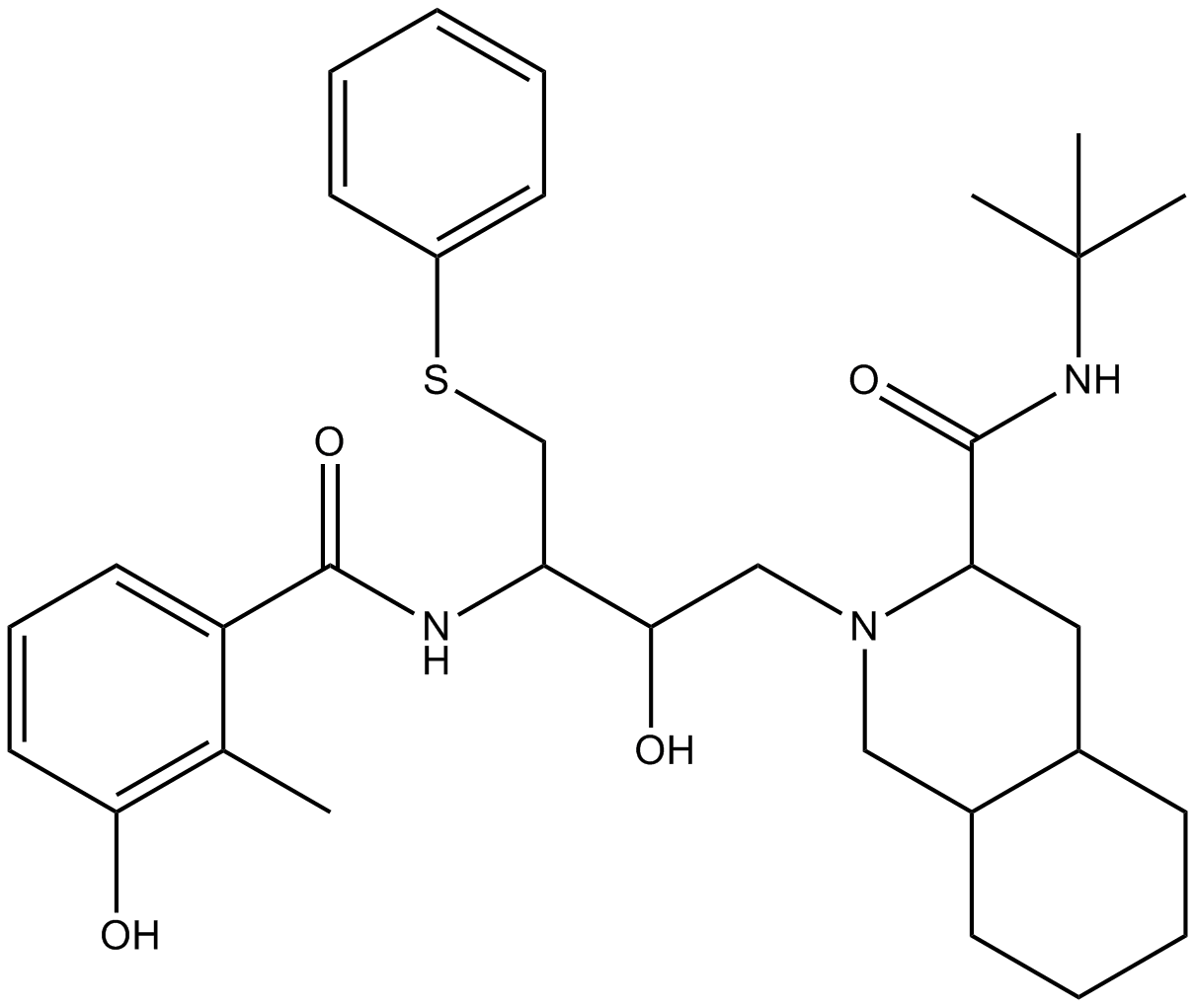
-
GC14084
Nelfinavir Mesylate
An HIV-1 protease inhibitor
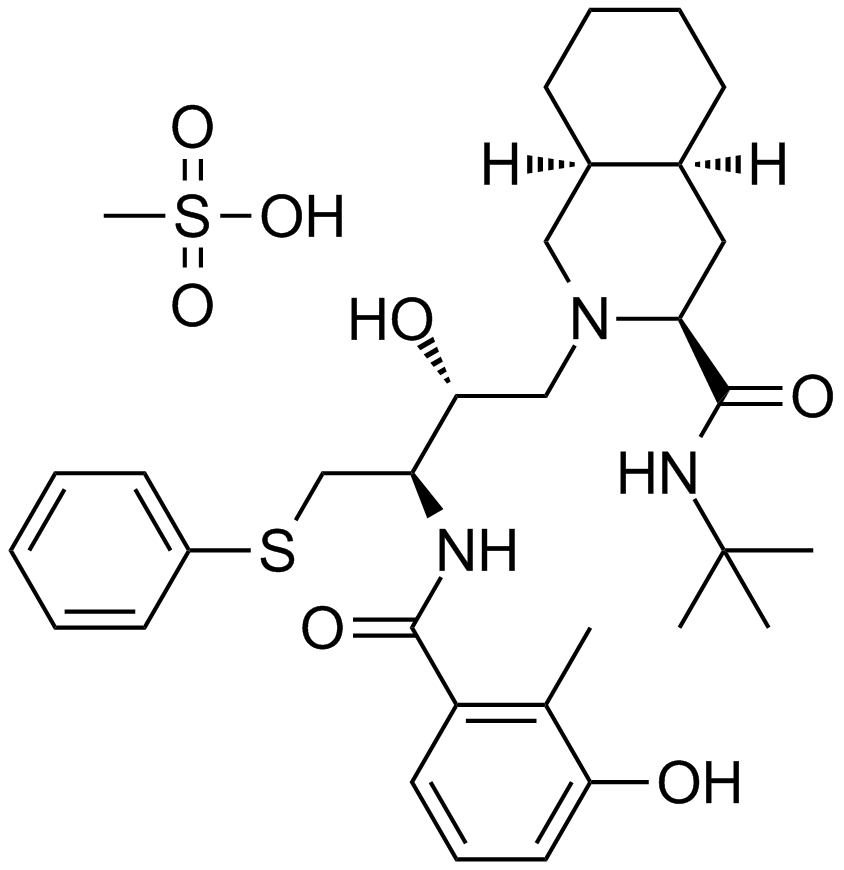
-
GC45926
Nelfinavir-d3
An internal standard for the quantification of nelfinavir
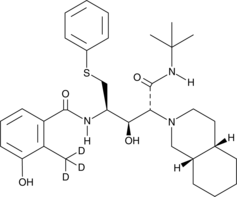
-
GC16387
Nevirapine
Non-nucleoside reverse transcriptase inhibitor
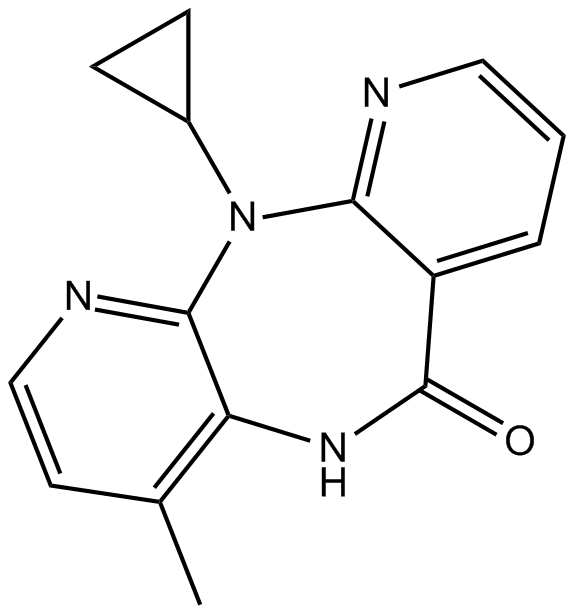
-
GC47770
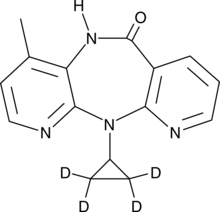
Nevirapine-d4
An internal standard for the quantification of nevirapine
-
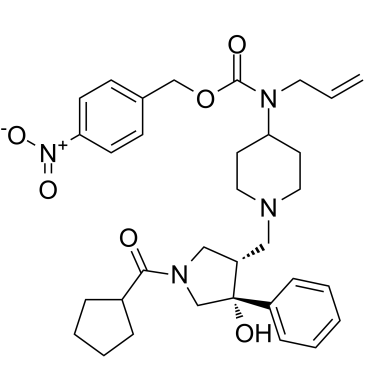
GC61879
Nifeviroc
Nifeviroc is an orally active CCR5 antagonist.
-
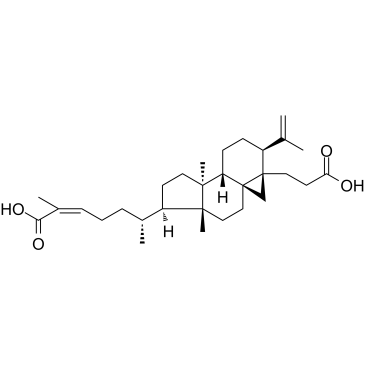
GC61134
Nigranoic acid
Nigranoic acid is a triterpenoid separated from Schisandra chinensis.
-
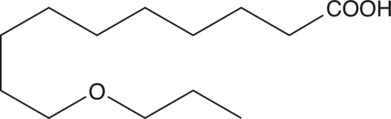
GC47810
O-11
A myristic acid analog with selective toxicity for trypanosomes
-
GC14784
O-Methylviridicatin
blocks activation of the HIV long terminal repeat by TNF-α
-
GN10184
Oleanolic acid
-
GC36798
Oleanonic acid
Oleanonic acid (3-Oxooleanolic acid) is a triterpenoid, inhibits infection by HIV-1 in in vitro infected PBMC, naturally infected PBMC and monocyte/macrophages with EC50 of 22.7 mM, 24.6 mM and 57.4 mM, respectively.
-
GC12821
Oltipraz
Nrf2 activator;An antischistosomal agent
-
GC62210
ONX-0914 TFA
ONX-0914 (PR-957) TFA is a selective inhibitor of low-molecular mass polypeptide-7 (LMP7), the chymotrypsin-like subunit of the immunoproteasome.
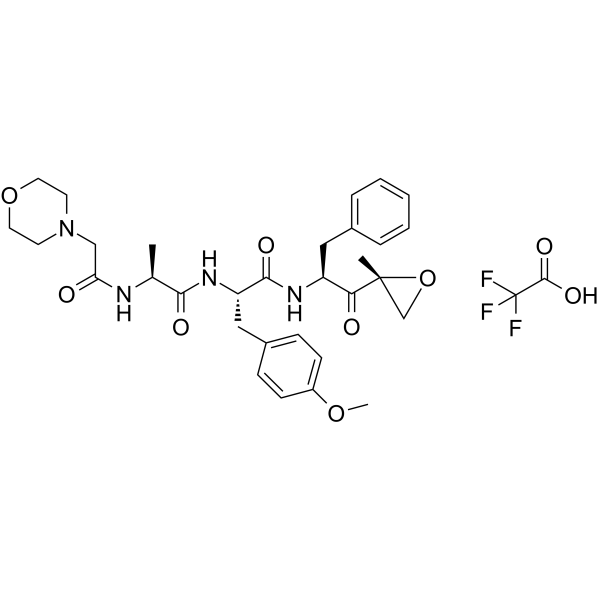
-
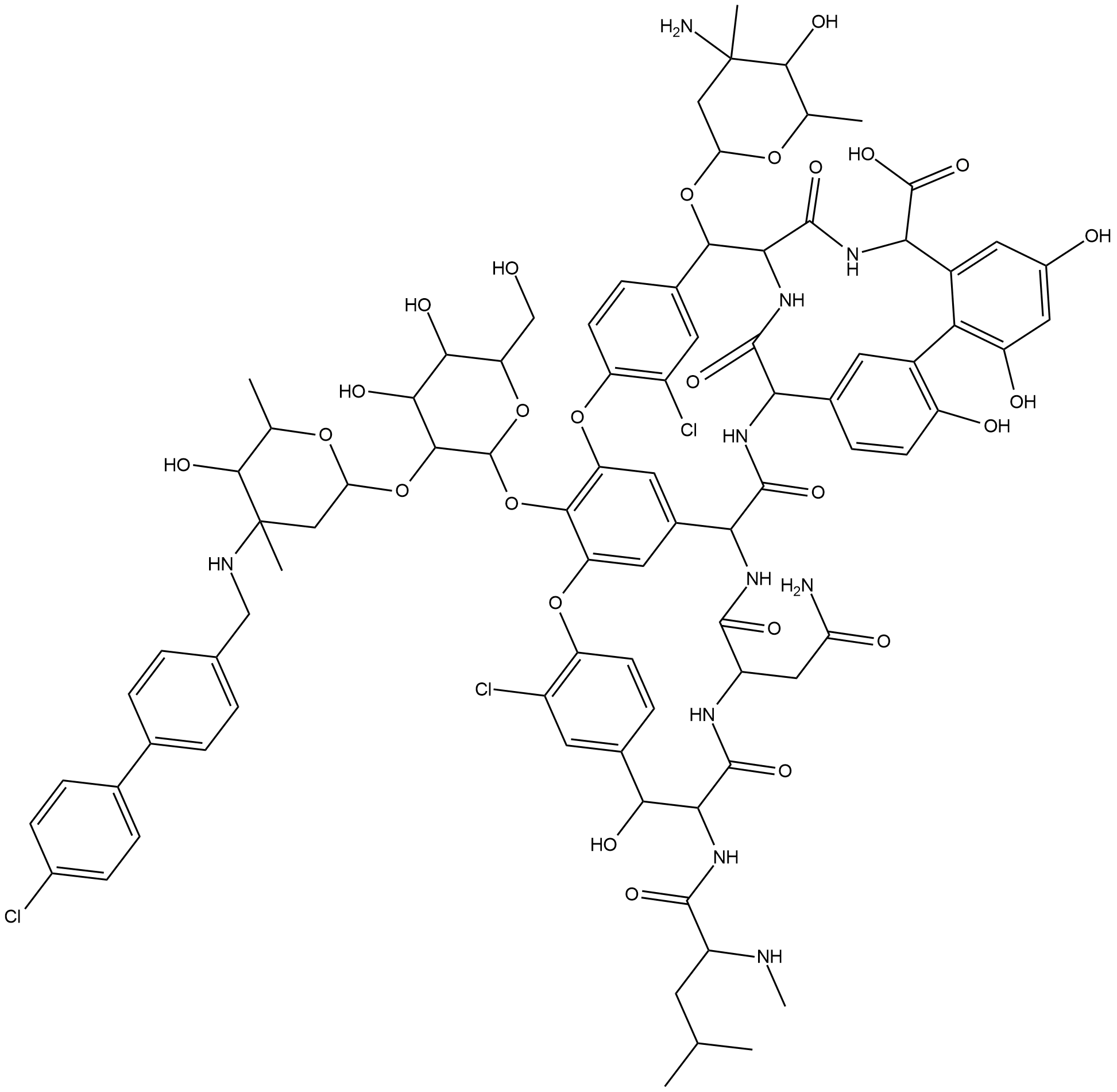
GP10160
Oritavancin(LY-333328)
Oritavancin(LY-333328) (LY 333328) is an orally active glycopeptide antibiotic with activity against gram-positive organisms.
-
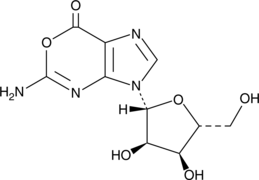
GC49073
Oxanosine
A guanosine analog with diverse biological activities
-
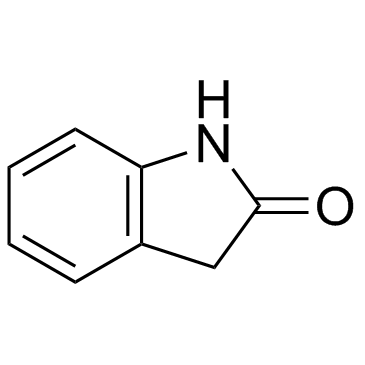
GC31140
Oxindole (Indolin-2-one)
Oxindole (Indolin-2-one) (Indolin-2-one) is an aromatic heterocyclic building block.
-
GC18964
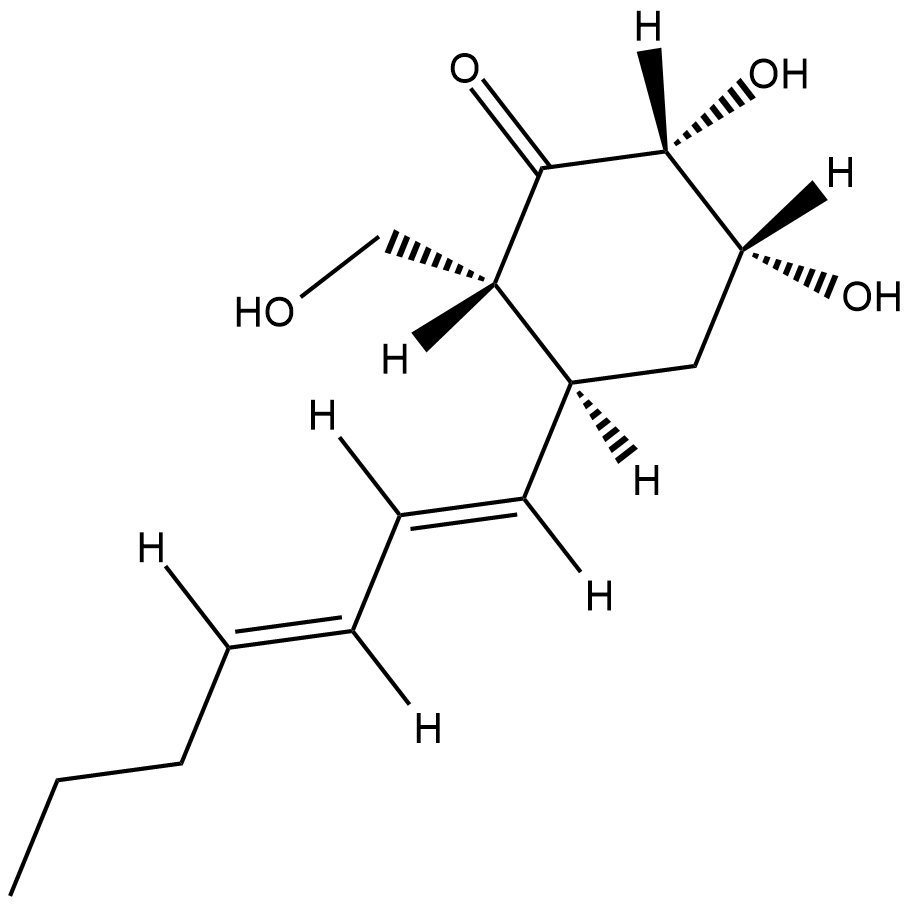
Palitantin
Palitantin is a fungal metabolite originally isolated from P.
-
GC12257
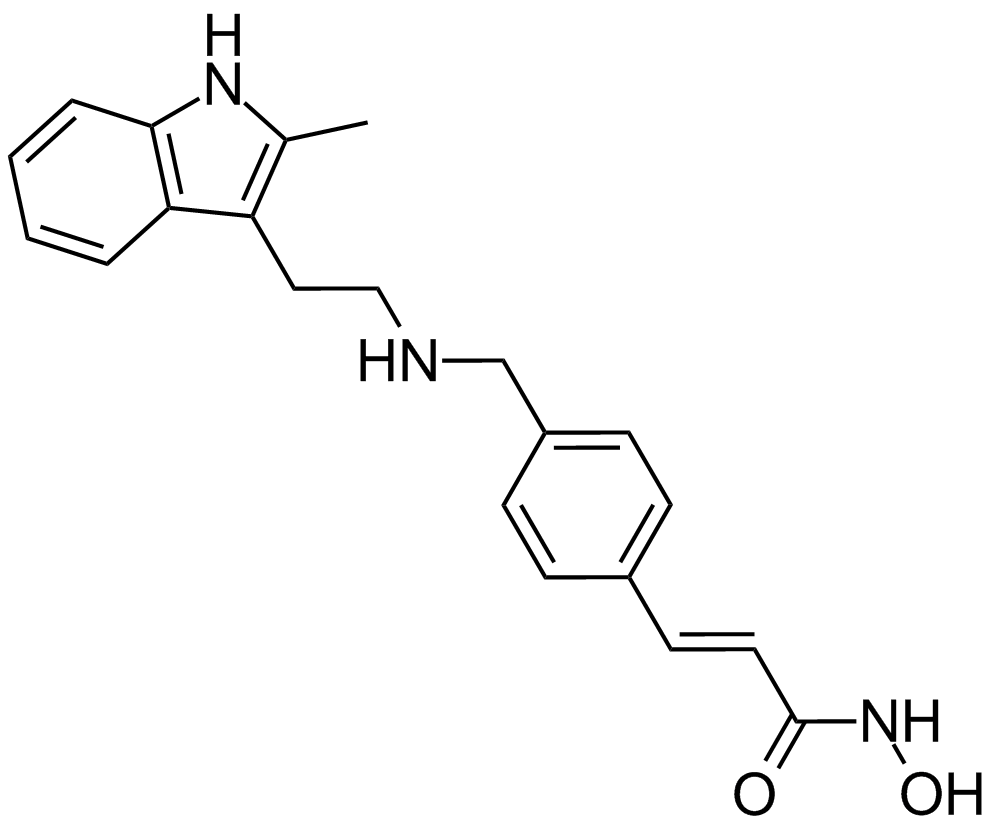
Panobinostat (LBH589)
A pan-HDAC inhibitor
-
GC61168
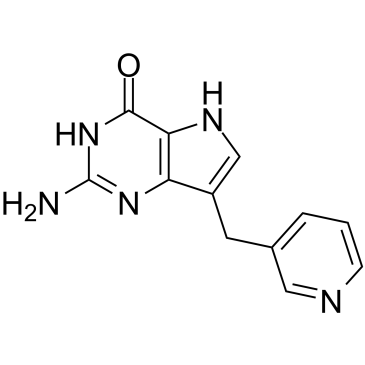
Peldesine
Peldesine (BCX 34) is a potent, competitive, reversible and orally active purine nucleoside phosphorylase (PNP) inhibitor with IC50s of 36 nM, 5 nM, and 32 nM for human, rat, and mouse red blood cell (RBC) PNP, respectively.
-
GC61633
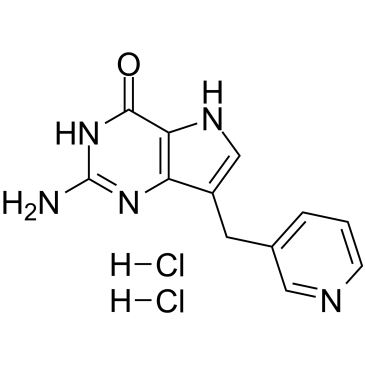
Peldesine dihydrochloride
Peldesine (BCX 34) dihydrochloride is a potent, competitive, reversible and orally active purine nucleoside phosphorylase (PNP) inhibitor with IC50s of 36 nM, 5 nM, and 32 nM for human, rat, and mouse red blood cell (RBC) PNP, respectively.
-
GC31685
Pentosan Polysulfate
Pentosan Polysulfate is an orally bioavailable medication with anti-inflammatory and pro-chondrogenic properties.

-
GC61171
Pentosan Polysulfate Sodium (W/W 43%)
Pentosan Polysulfate Sodium is an orally bioavailable, semi-synthetic medication with anti-inflammatory and pro-chondrogenic properties.

-
GC40685
Pentoxifylline
Pentoxifylline is a methylxanthine derivative.
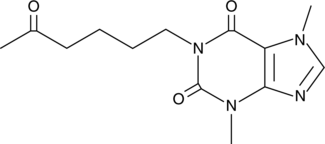
-
GC32309
Peptide T
Peptide T is an octapeptide from the V2 region of HIV-1 gp120.
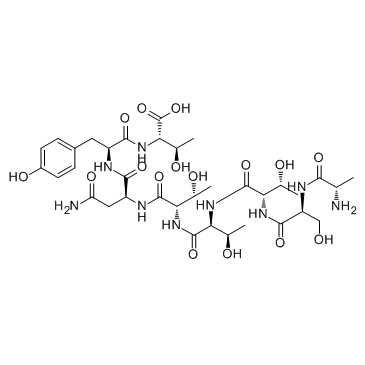
-
GC32335
Peptide T TFA
Peptide T (TFA) is an octapeptide from the V2 region of HIV-1 gp120.
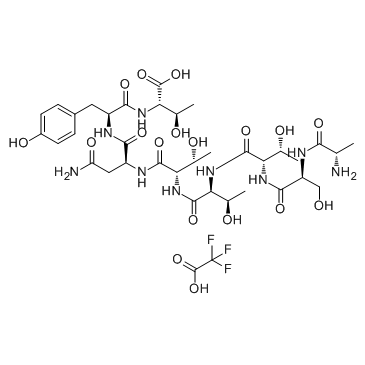
-
GC13875
Piperaquine
used in combination with artimesinin for inhibition of HIV-1 replication
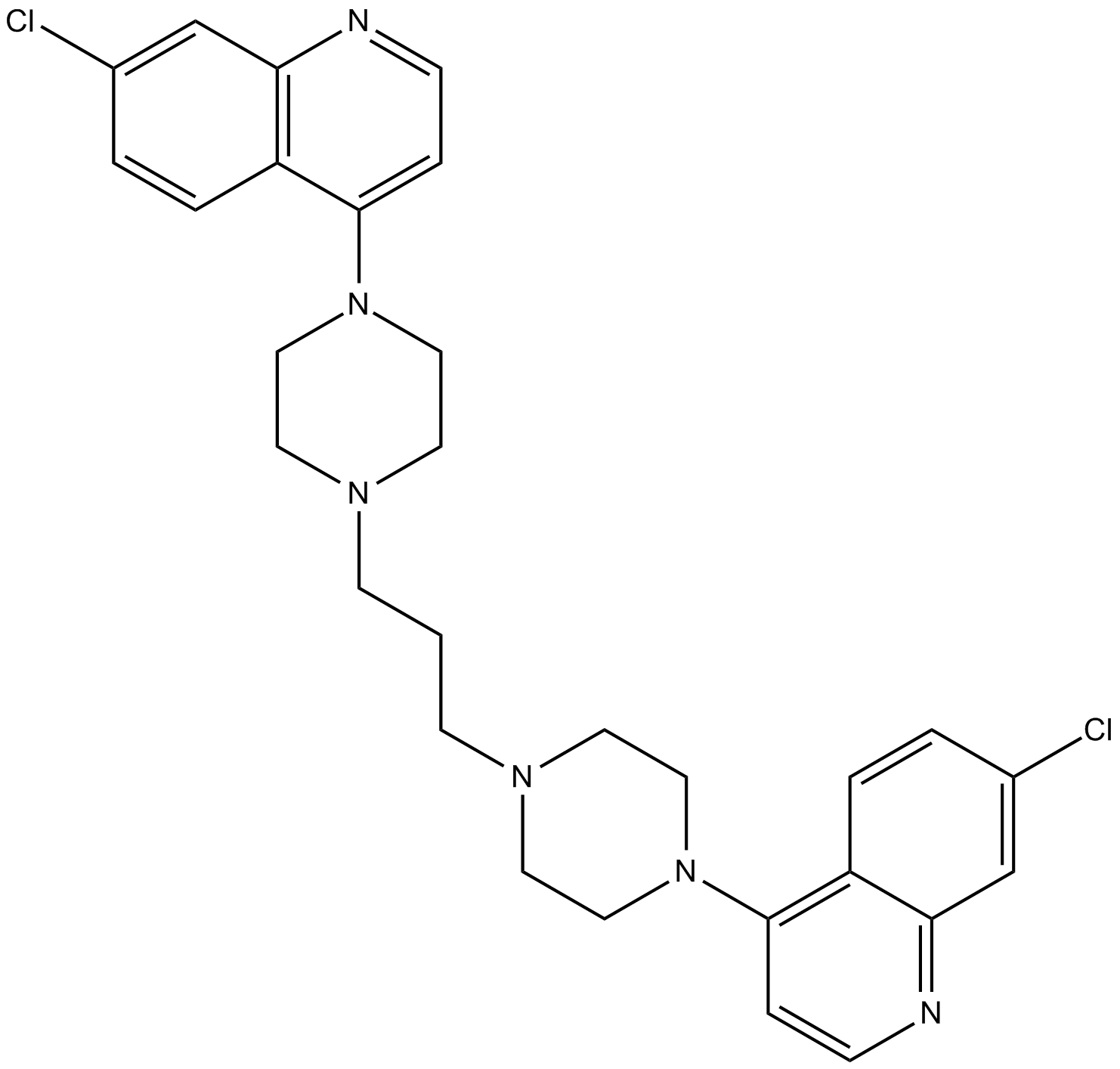
-
GC49441
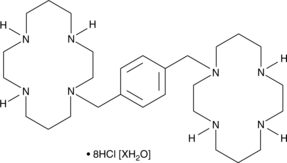
Plerixafor (hydrochloride hydrate)
An antagonist of CXCR4
-
GC47963
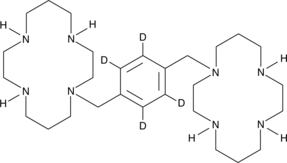
Plerixafor-d4
An internal standard for the quantification of plerixafor
-
GC63302
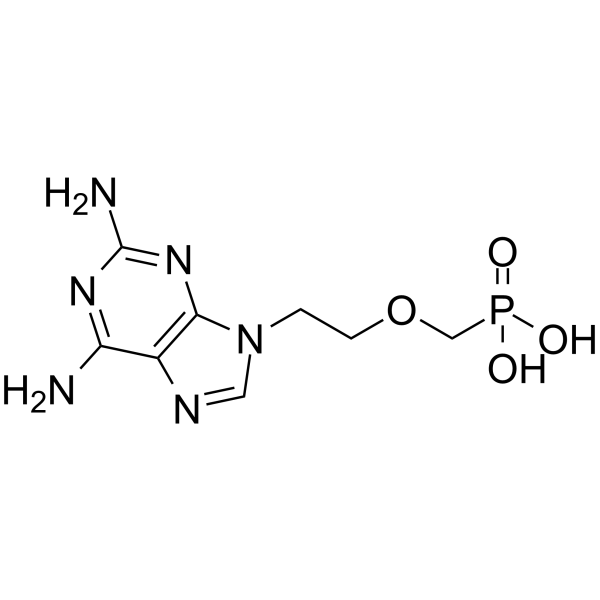
PMEDAP
PMEDAP is a potent inhibitor of human immunodeficiency virus (HIV) replication.
-
GC32327
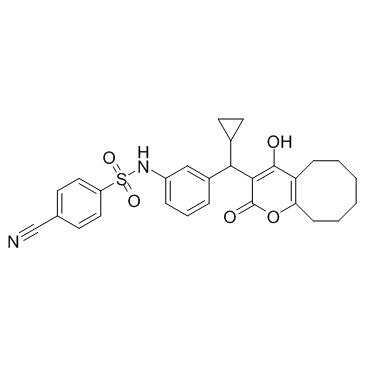
PNU-103017
PNU-103017 is an HIV protease inhibitor.
-
GC16825
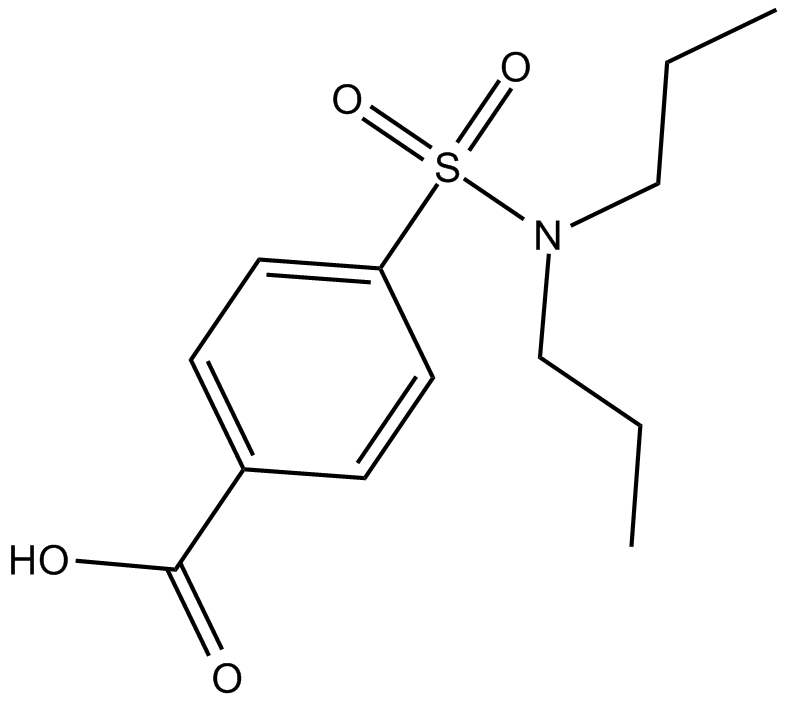
Probenecid
inhibitor of organic anion transport, MRP and pannexin-1 channel
-
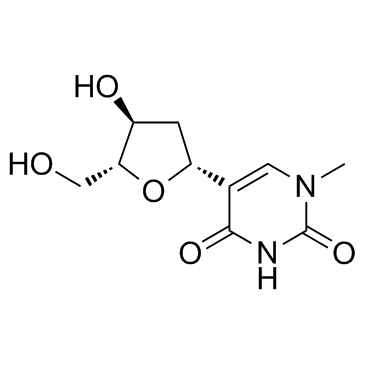
GC33944
Pseudothymidine (5-Methyl-2'-Deoxypseudouridin)
Pseudothymidine (5-Methyl-2'-Deoxypseudouridin) is a C-nucleoside analog of thymidine.
-
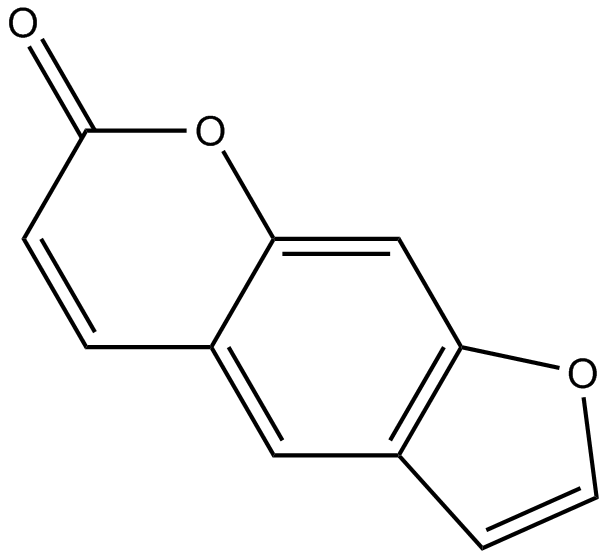
GN10451
Psoralen
-
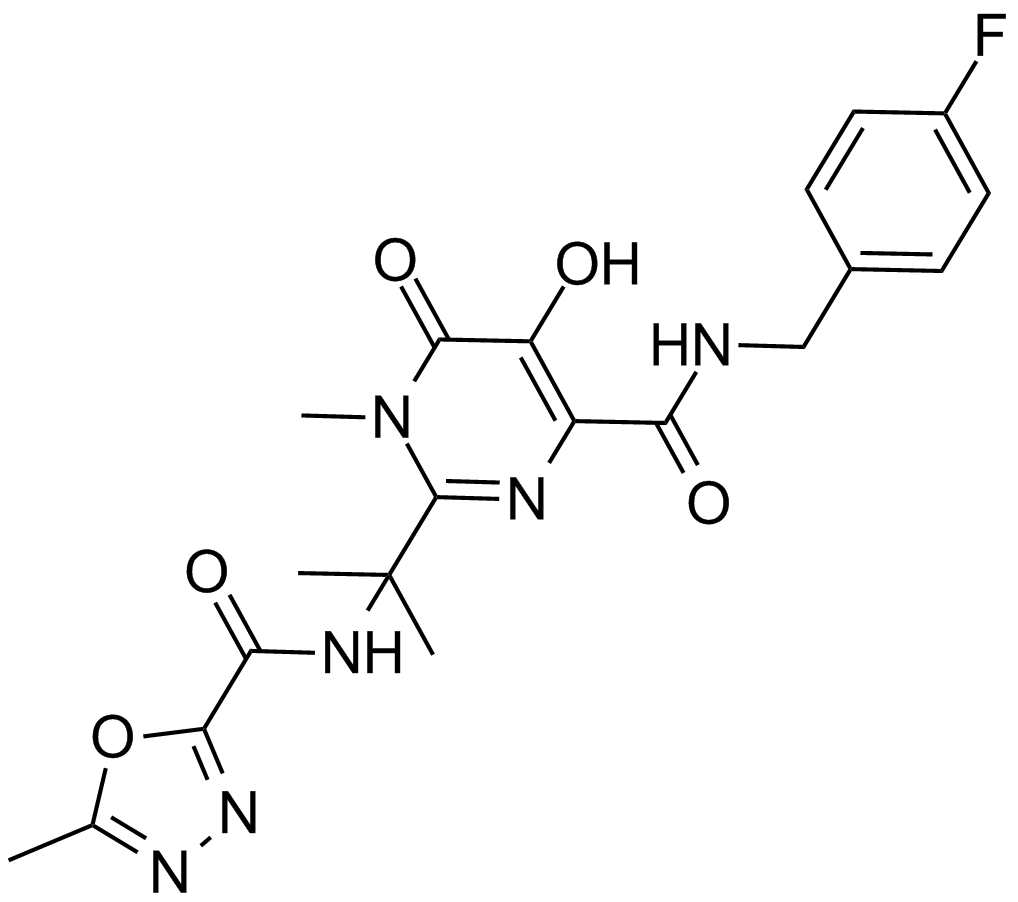
GC11956
Raltegravir (MK-0518)
-
GC15036
Raltegravir potassium salt
Raltegravir (MK 0518) potassium is a potent integrase (IN) inhibitor, used to treat HIV infection.
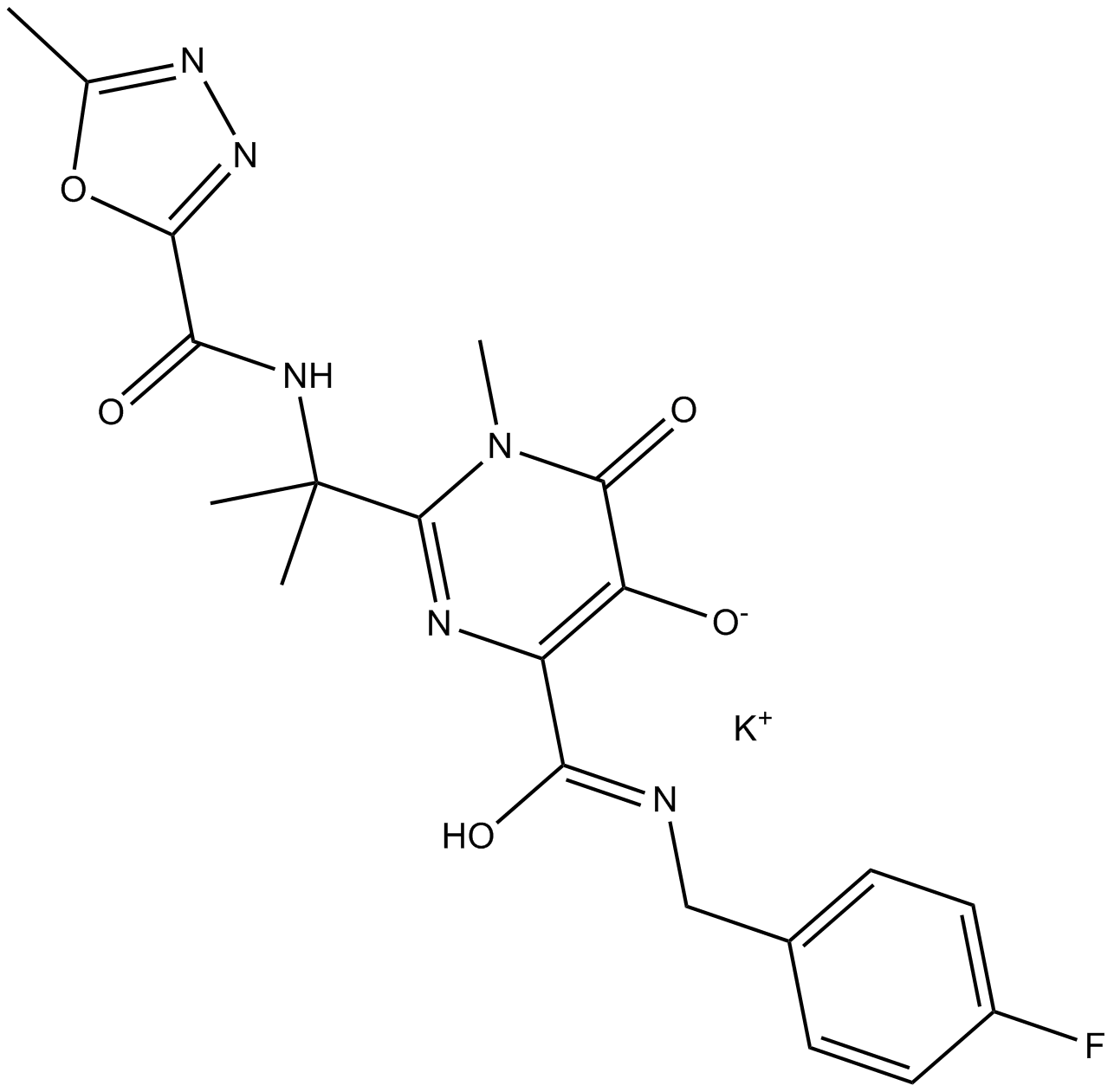
-
GC63523
Reverse transcriptase-IN-1
Reverse transcriptase-IN-1 (Compound 12z), a diarylbenzopyrimidine (DABP) analogue, is a potent, orally active HIV-1 nonnucleoside reverse transcriptase inhibitor.
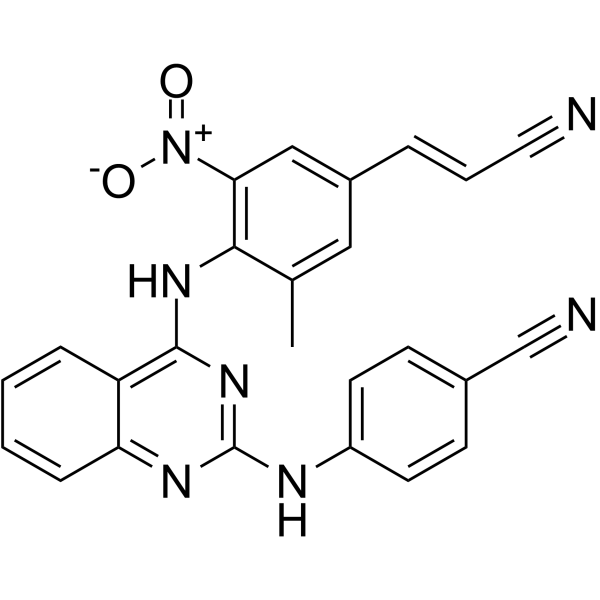
-
GC18659
Ribavirin 5'-monophosphate (lithium salt)
Ribavirin 5'-monophosphate inhibits viral DNA and RNA replication in vitro via the strong competitive inhibition of inosine monophosphate dehydrogenase (IMPDH), with a reported Ki value of 270 nM, and thus inhibits guanosine triphosphate synthesis.
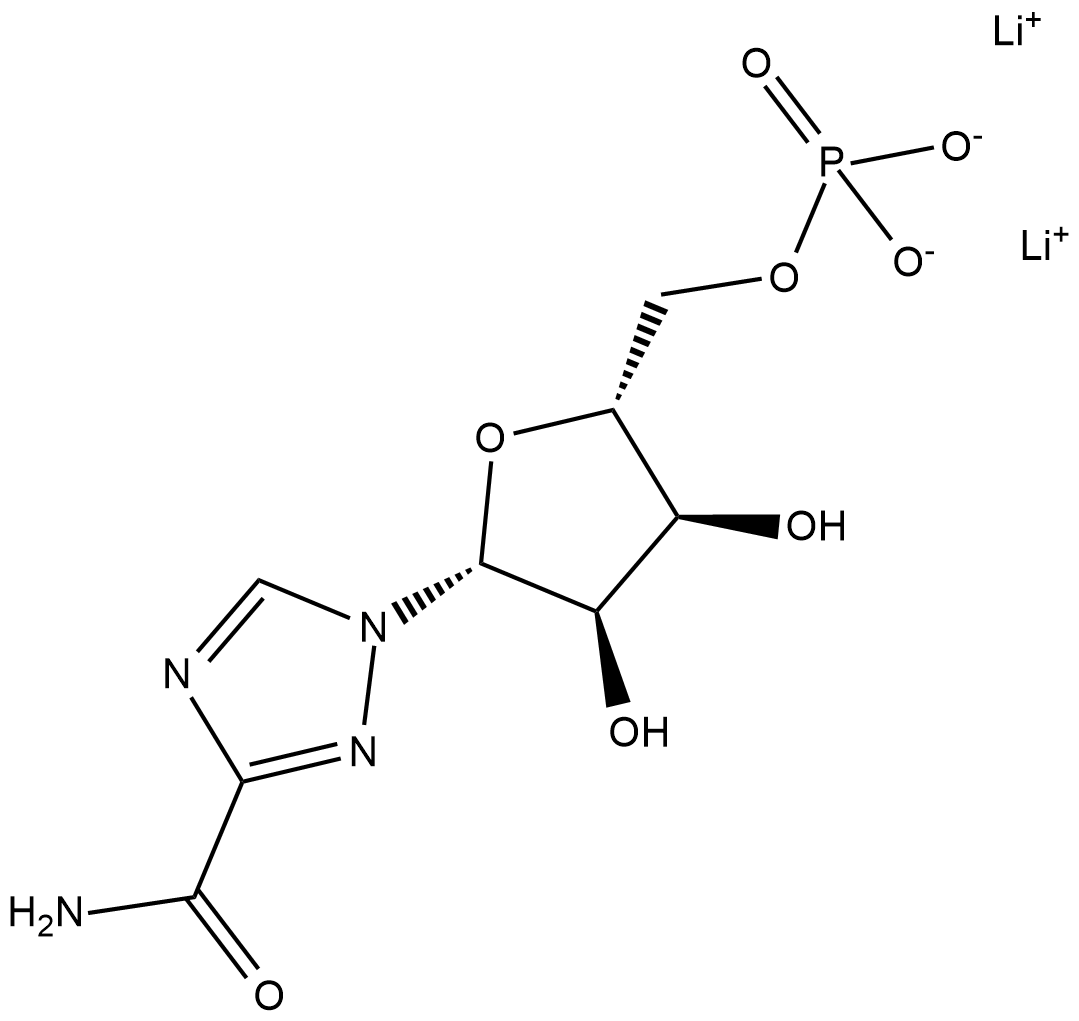
-
GC33932
RIG-1 modulator 1
RIG-1 modulator 1 is an anti-viral compound which can be useful for the treatment of viral infections including influenza virus, HBV, HCV and HIV extracted from patent WO 2015172099 A1.
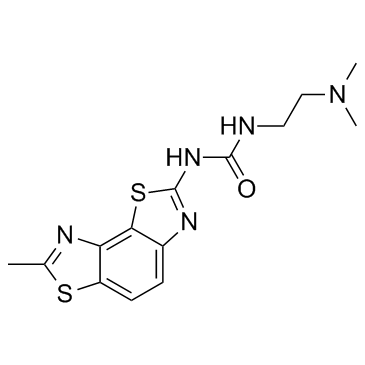
-
GC11059
Rilpivirine
A non-nucleoside reverse transcriptase inhibitor
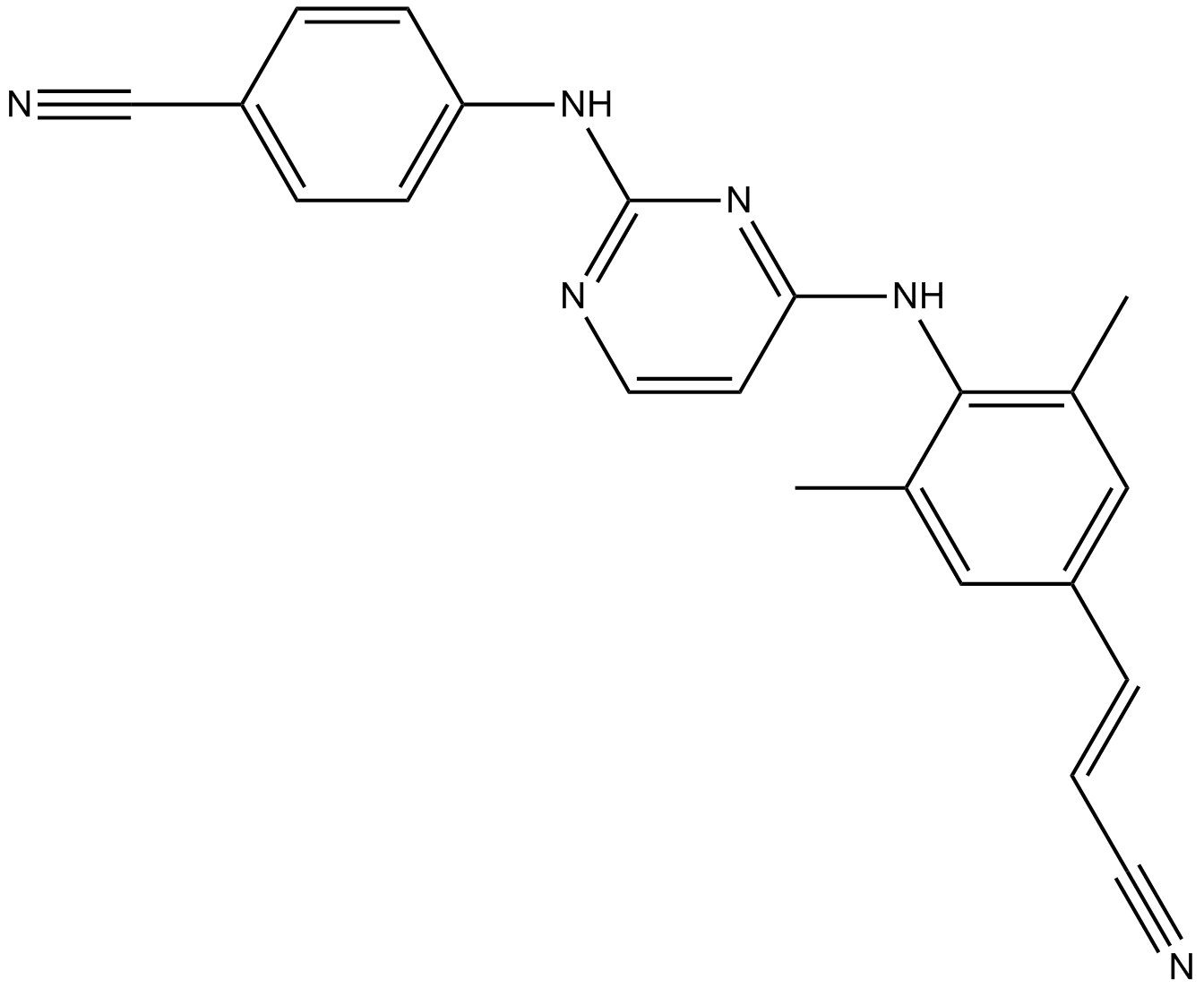
-
GC17303
Ritonavir
An HIV protease inhibitor
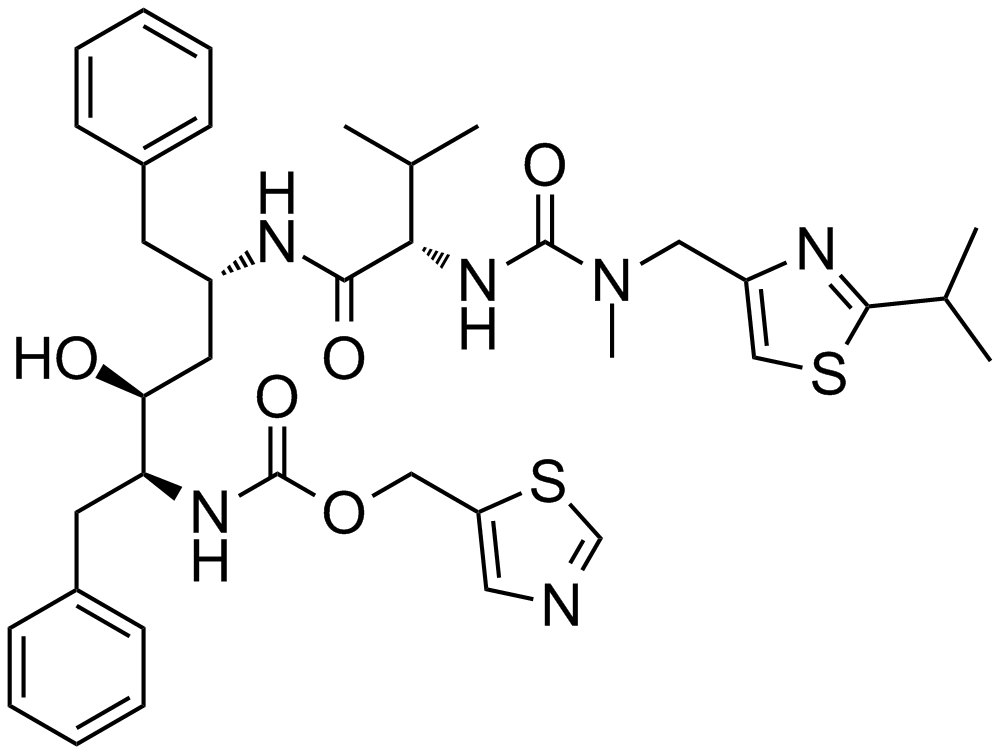
-
GC64331
Ritonavir-13C,d3
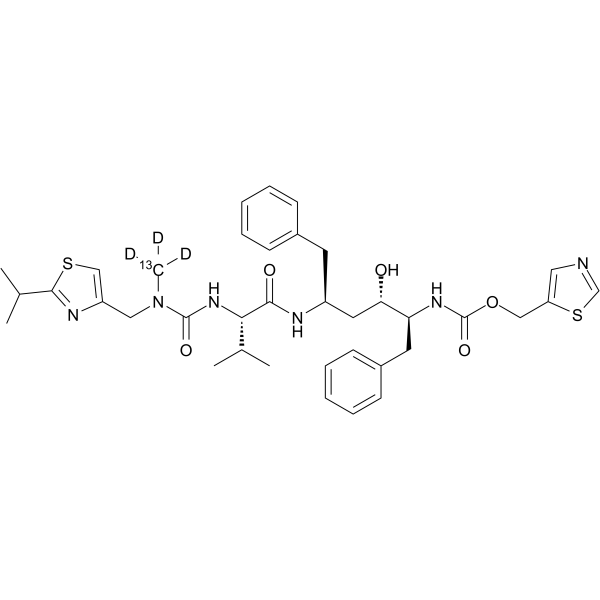
-
GC40221
Ritonavir-d6
Ritonavir-d6 is intended for use as an internal standard for the quantification of ritonavir by GC- or LC-MS.
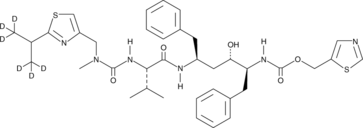
-
GC18382
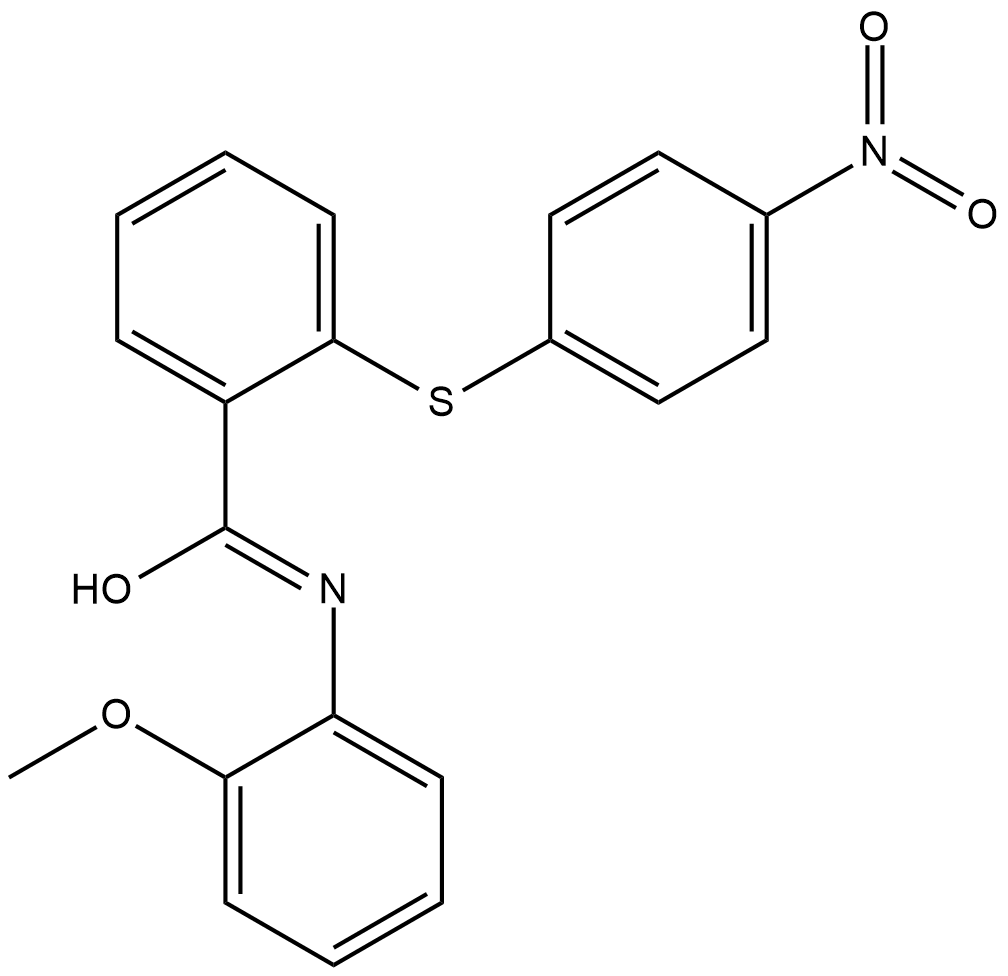
RN-18
RN-18 is an inhibitor of HIV-1 viral infectivity factor (Vif), an HIV-1 regulatory protein that does not have cellular homologs.
-
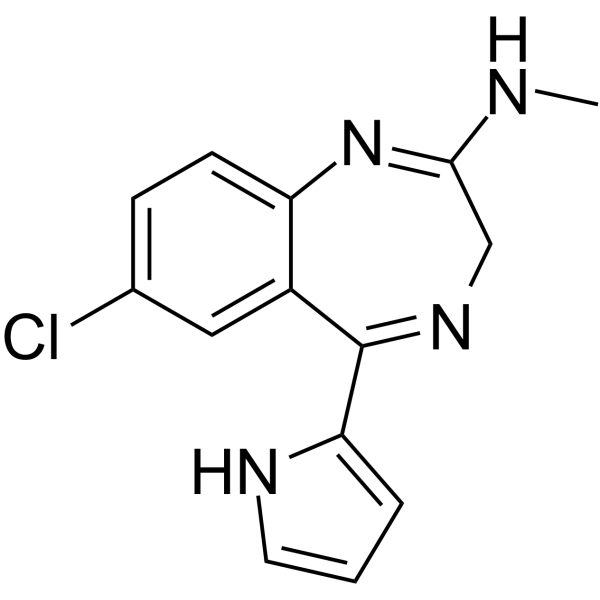
GC63979
Ro24-7429
Ro24-7429 is a potent and orally active HIV-1 transactivator protein Tat antagonist.
-
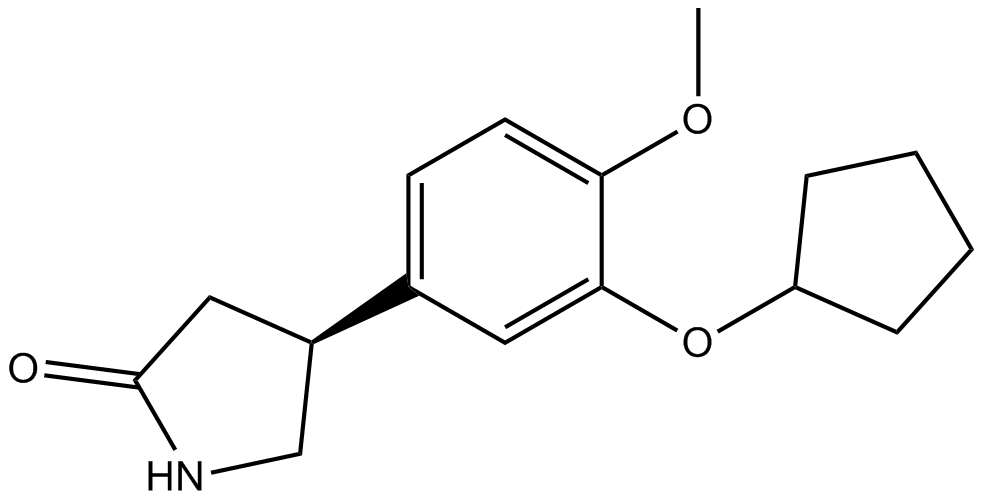
GC16629
Rolipram
A phosphodiesterase-4 inhibitor
-
GC14199
RVX-208
RVX-208 (RVX-208) is an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. The IC50s are 87 μM and 0.51 μM for BD1 and BD2, respectively.

-
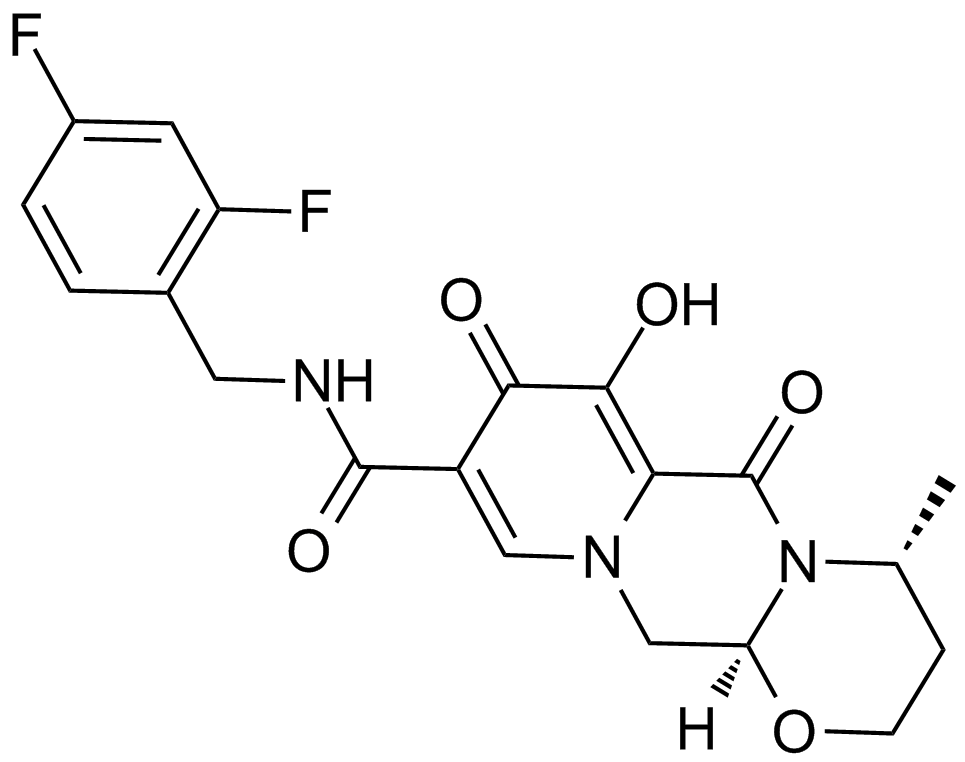
GC17906
S/GSK1349572
A potent inhibitor of HIV integrase
-
GC18016
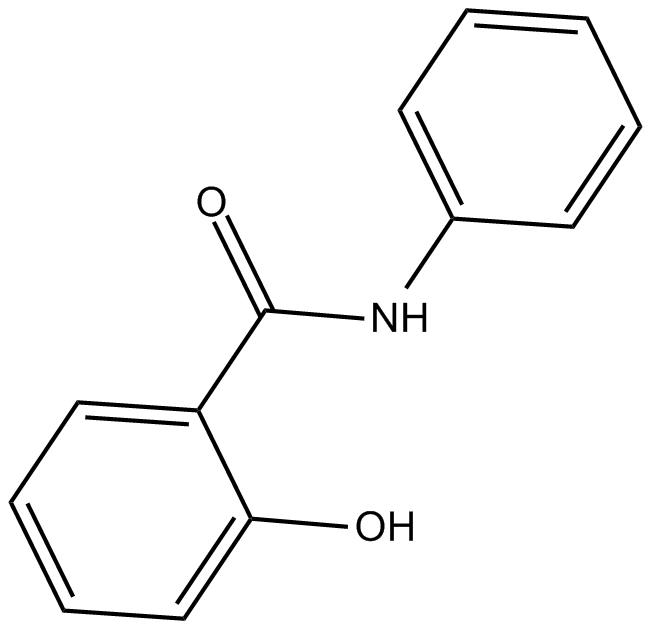
Salicylanilide
Antiviral,antibacterial,antifungal reagent
-
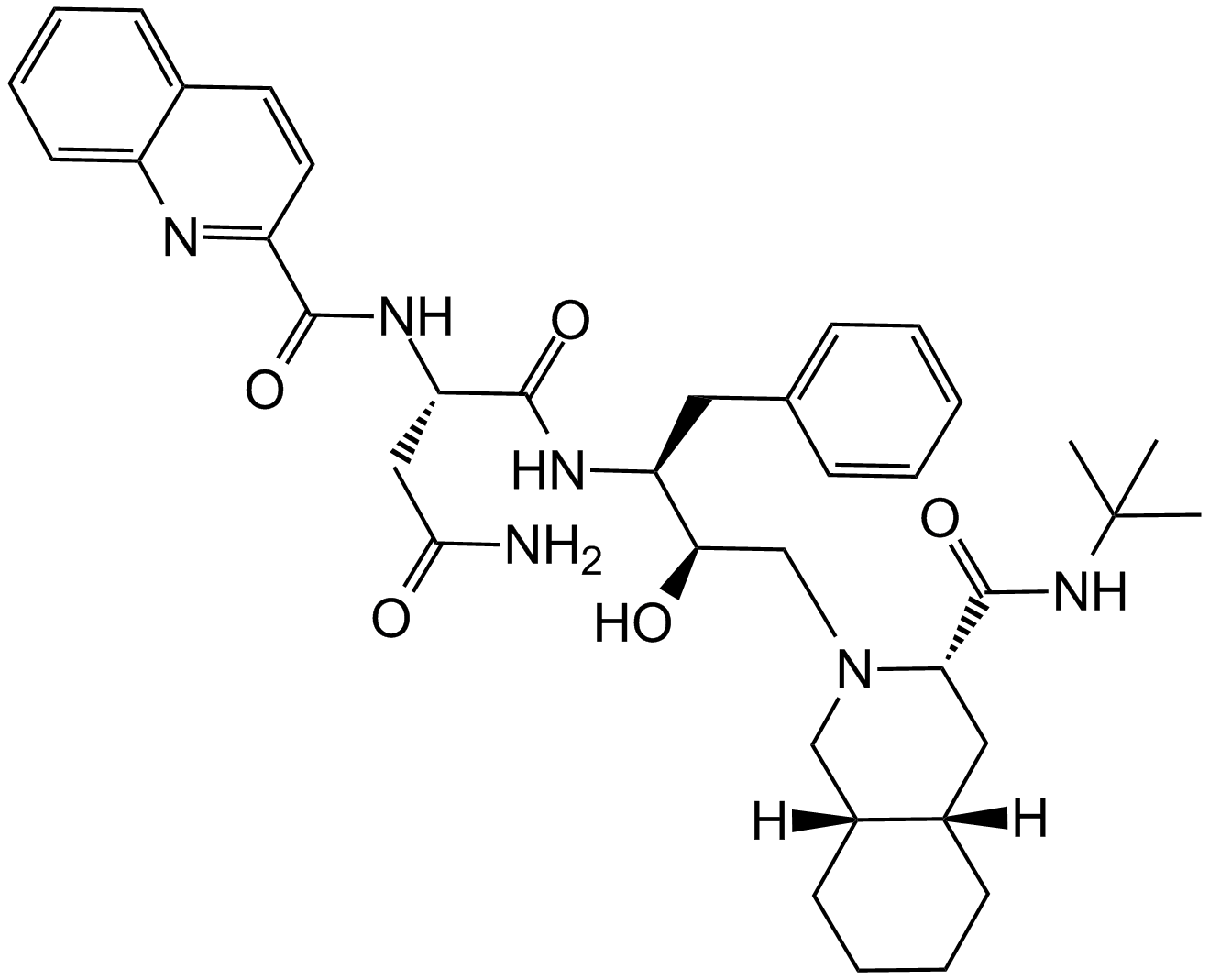
GC15335
Saquinavir
-
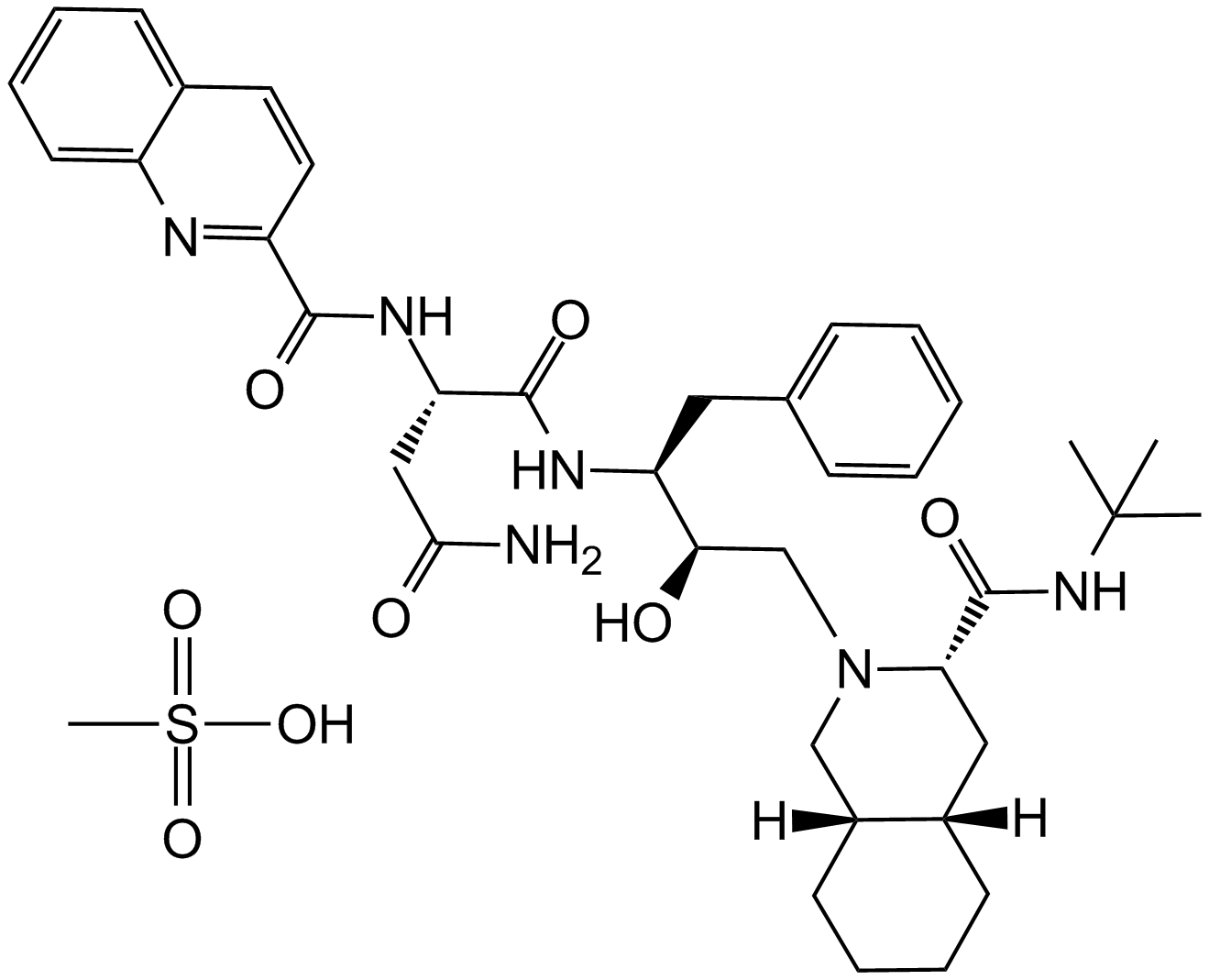
GC15785
Saquinavir mesylate
An HIV protease inhibitor
-
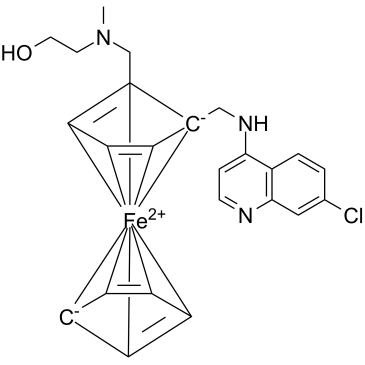
GC39642
SARS-CoV-IN-2
SARS-CoV-IN-2 is an effective inhibitor of SARS-CoV replication.
-
GC39563
SARS-CoV-IN-3
SARS-CoV-IN-3 is an effective inhibitor of SARS-CoV replication.
-
GN10001
Sennoside A
-
GN10216
Shikonin
-
GC44889
Siamycin I
Siamycin I is a tricyclic peptide originally isolated from Streptomyces and has antiviral and antibacterial activities.
-
GC32192
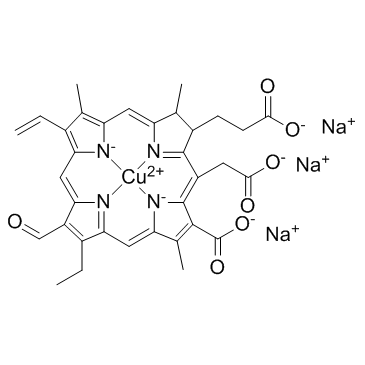
Sodium copper chlorophyllin
Sodium copper chlorophyllin exerts antiviral activities against Influenza virus and HIV with IC50s of 50 to 100 μM for both of them.
-
GC61284
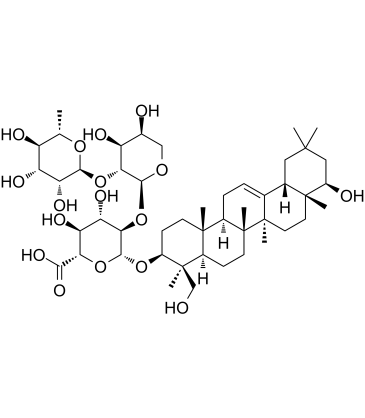
Soyasaponin II
Soyasaponin II is a saponin with antiviral activity.
-
GC60344
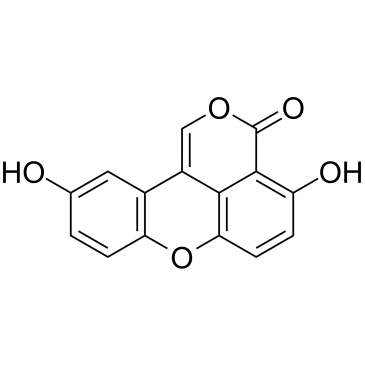
Sparstolonin B
Sparstolonin B acts as a selective TLR2 and TLR4 antagonist and selectively blocks TLR2- and TLR4-mediated inflammatory signaling.
-
GC48096
Sporogen-AO 1
A fungal metabolite with diverse biological activities