HIF
Hypoxia inducible factors (HIFs) are heterodimeric proteins belonging to the basic helix-loop-helix-PAS (bHLH/PAS) family of transcription factors that mediate the primary transcriptional response to stress caused by hypoxia (accessible O2 level < 2%). HIFs are characterized by containing two subunits, including O2-labile α subunit (HIFα) and constitutively expressed β subnit (HIFβ). Mammalian HIFα consists of three isoforms, including HIF1α, HIF2α and HIF3α, which play an essential role in the regulation of HIF activity by O2 availability. Under normal O2 tension, HIFα is hydroxylated at the two conserved proline residues within the O2-dependent degradation (ODD) domain by prolylhydroxylase domain proteins (PHDs) and undergoes proteosomal degradation catalyzed by a complex formed by E3 ubiquitin ligase and the von Hippel-Lindau protein (pVHL); while, under hypoxia, HIFα stabilizes with PHDs being deactivated and hence induce transcription of genes with adaptive functions.
Products for HIF
- Cat.No. Product Name Information
-
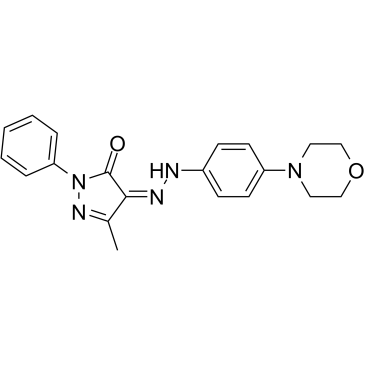
GC16195
2,4-DPD
Diethyl pyridine-2,4-dicarb is a potent prolyl 4-hydroxylase-directed proinhibitor.
-
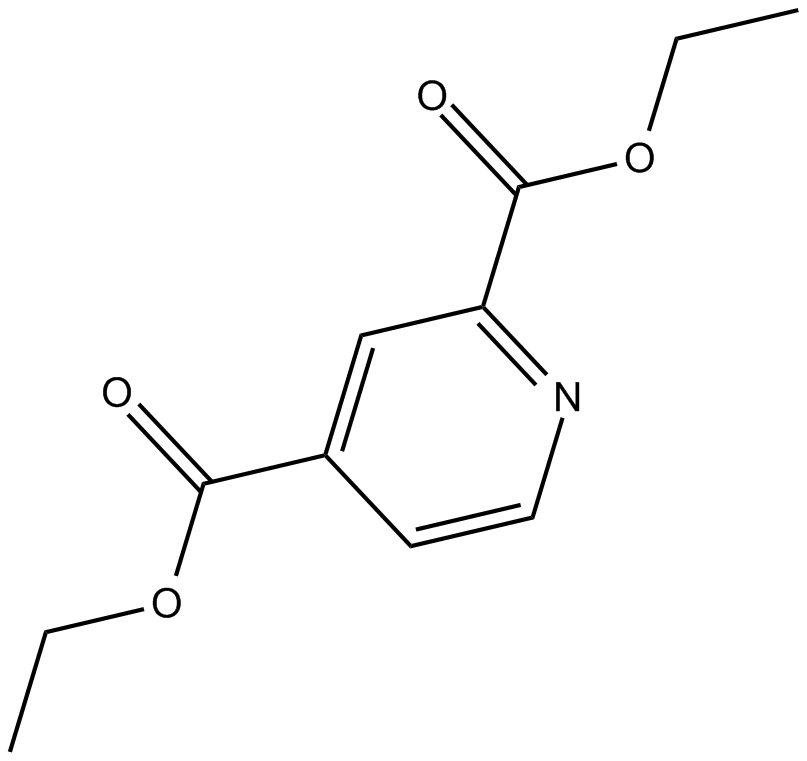
GC14282
3-acetyl-11-keto-β-Boswellic Acid
3-acetyl-11-keto-β-Boswellic Acid (Acetyl-11-keto-β-boswellic acid) is an active triterpenoid compound from the extract of Boswellia serrate and a novel Nrf2 activator.
-
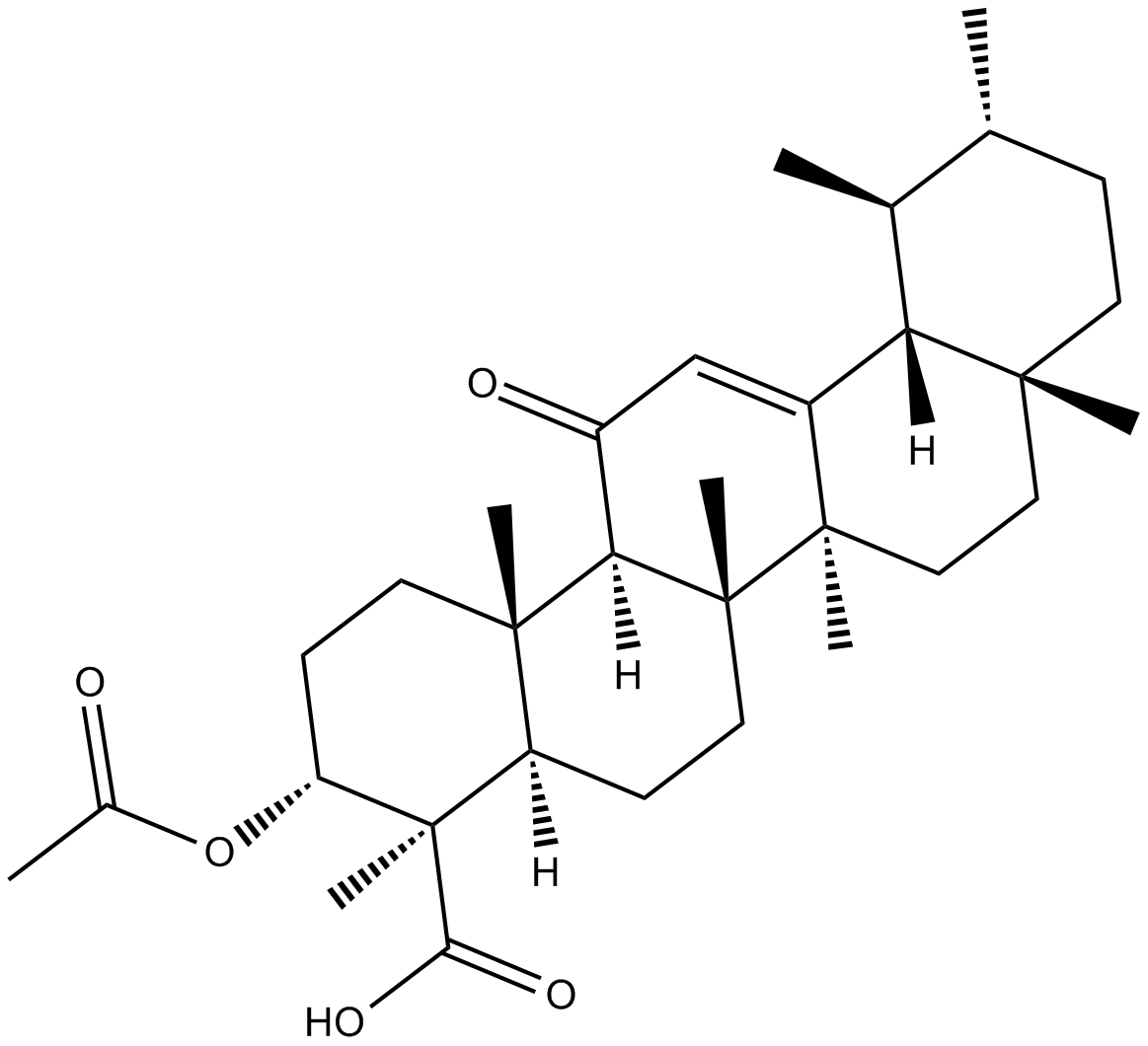
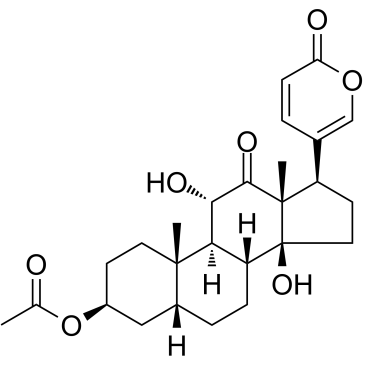
GC35230
Acetylarenobufagin
Acetylarenobufagin is a steroidal hypoxia inducible factor-1 (HIF-I) modulator.
-
GC32083
Acriflavine
Acriflavine is a fluorescent dye for labeling high molecular weight RNA.

-
GC12487
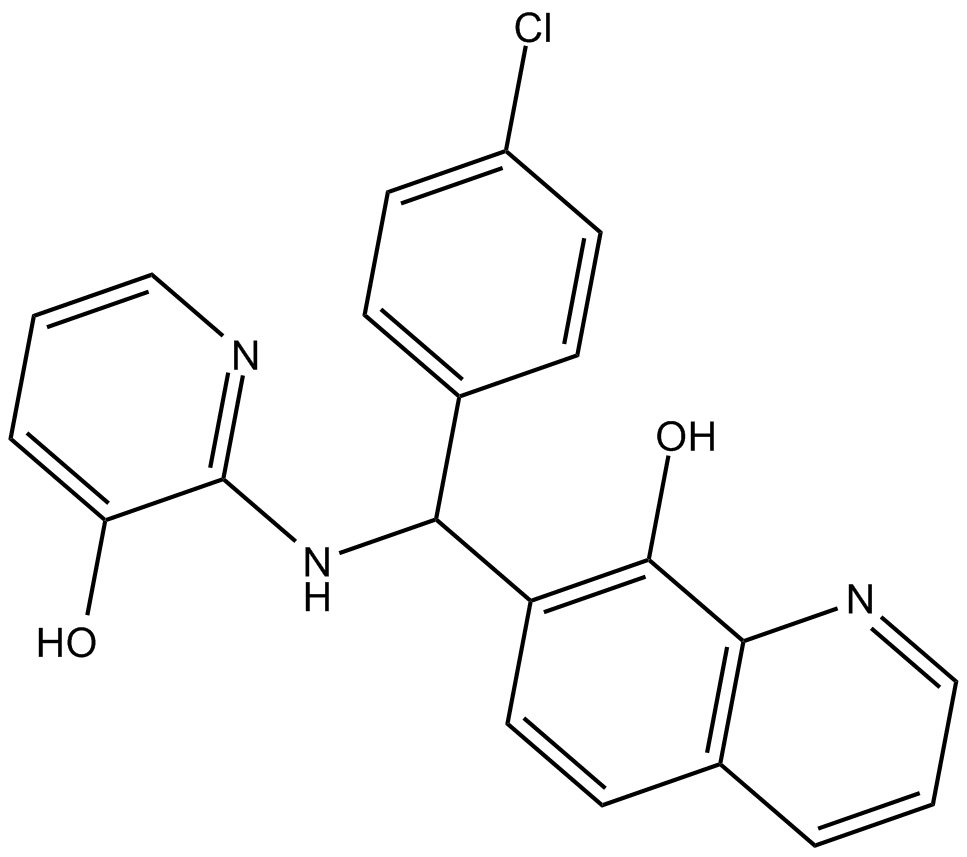
Adaptaquin
HIF-prolyl hydroxylase-2 (PHD2) inhibitor
-
GC33416
AFP464
AFP464 (NSC710464 free base), is an active HIF-1α inhibitor with an IC50 of 0.25 μM, also is a potent aryl hydrocarbon receptor (AhR) activator.

-
GC12698
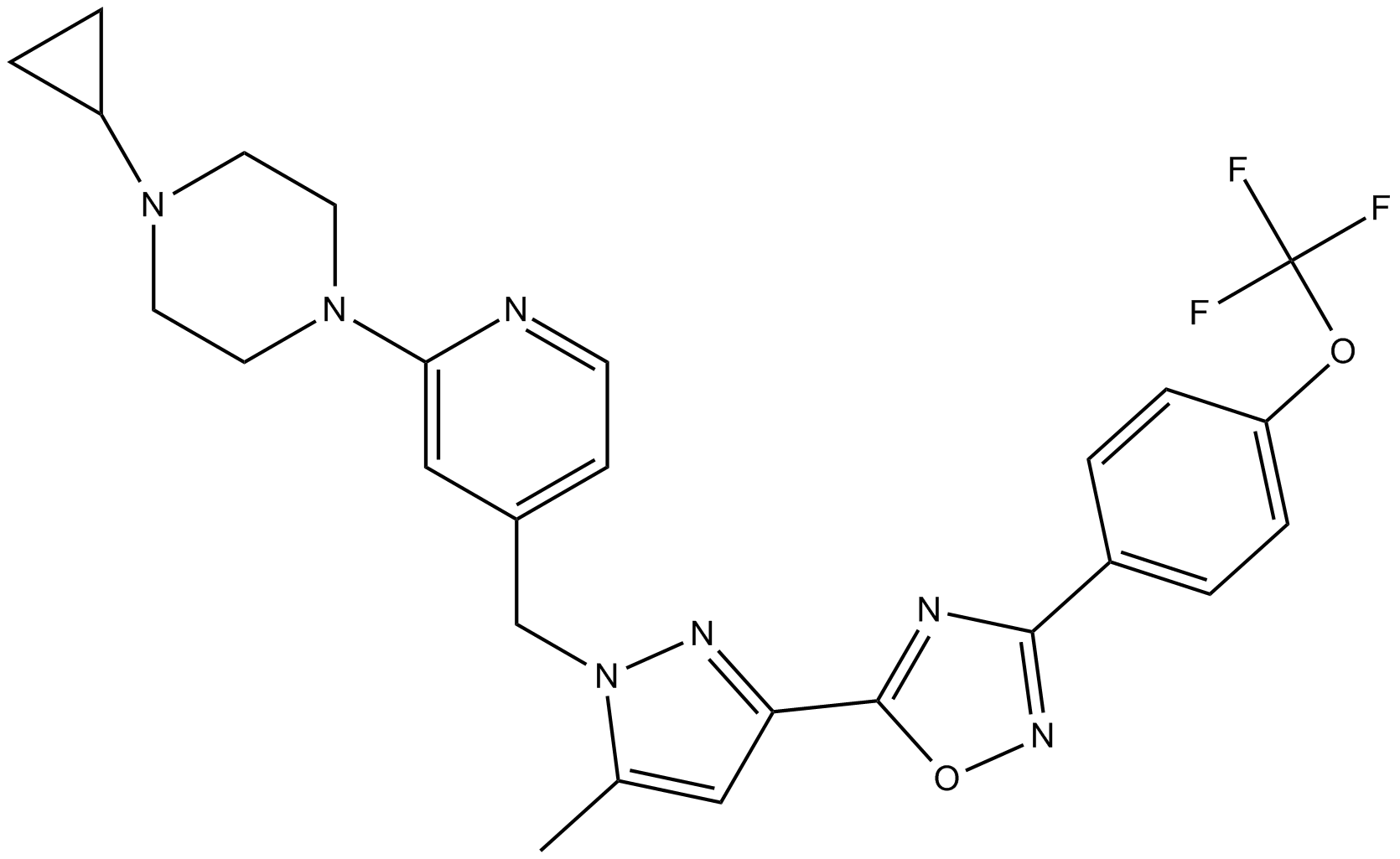
BAY 87-2243
A HIF-1 inhibitor,potent and selective
-
GC16647
Daprodustat(GSK1278863)
Daprodustat(GSK1278863) (GSK1278863) is an orally active hypoxia-inducible factor prolyl hydroxylase (HIF-PH) inhibitor being developed for the treatment of anemia associated with chronic kidney disease.

-
GC31230
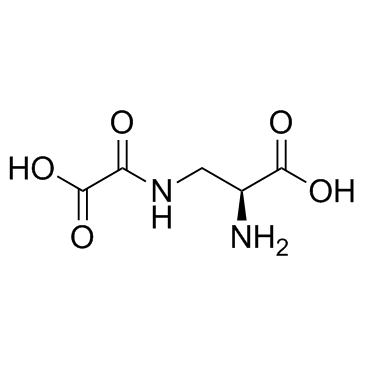
Dencichin (Dencichine)
Dencichin is a non-protein amino acid originally extracted from Panax notoginseng, and can inhibit HIF-prolyl hydroxylase-2 (PHD-2) activity.
-
GC38767
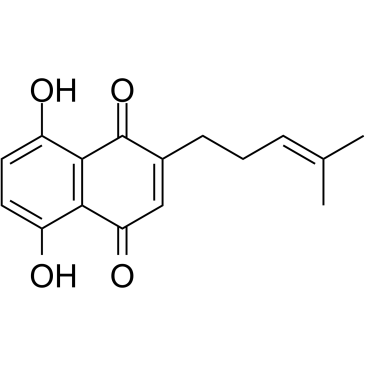
Deoxyshikonin
A natural products with anti-tumor activity
-
GC32449
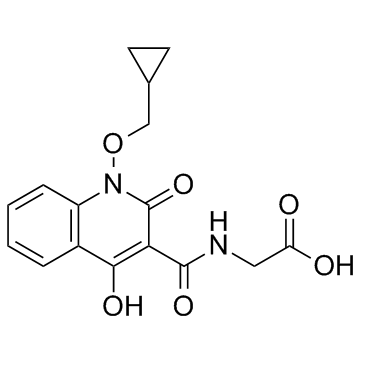
Desidustat
An inhibitor of HIF-PH
-
GC18236
Echinomycin
An inhibitor of HIF1mediated gene transcription

-
GC33043
EL-102
EL-102 is a hypoxia-induced factor 1 (Hif1α) inhibitor. EL-102 induces apoptosis, inhibits tubulin polymerisation and shows activities against prostate cancer. EL-102 can be used for the research of cancer.
-
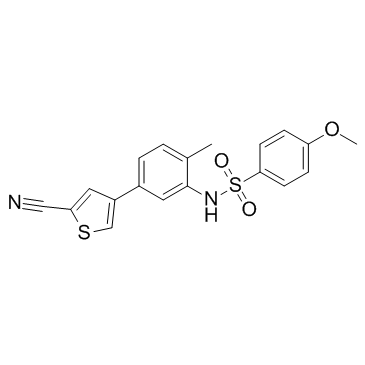
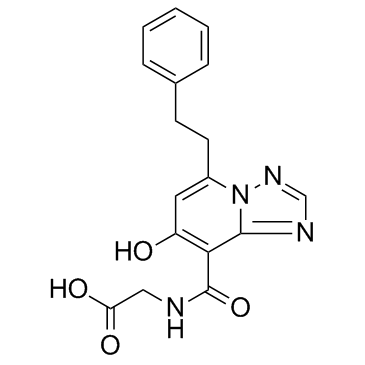
GC33634
Enarodustat (JTZ-951)
Enarodustat (JTZ-951) is a potent and orally active hypoxia-inducible factor prolyl hydroxylase inhibitor, with an EC50 of 0.22 μM.
-
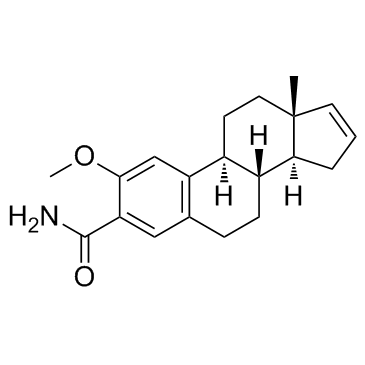
GC33170
ENMD-119 (ENMD 1198)
ENMD-119 (ENMD 1198) (IRC-110160), an orally active microtubule destabilizing agent, is a 2-methoxyestradiol analogue with antiproliferative and antiangiogenic activity. ENMD-119 (ENMD 1198) is suitable for inhibiting HIF-1alpha and STAT3 in human HCC cells and leads to reduced tumor growth and vascularization.
-
GC16638
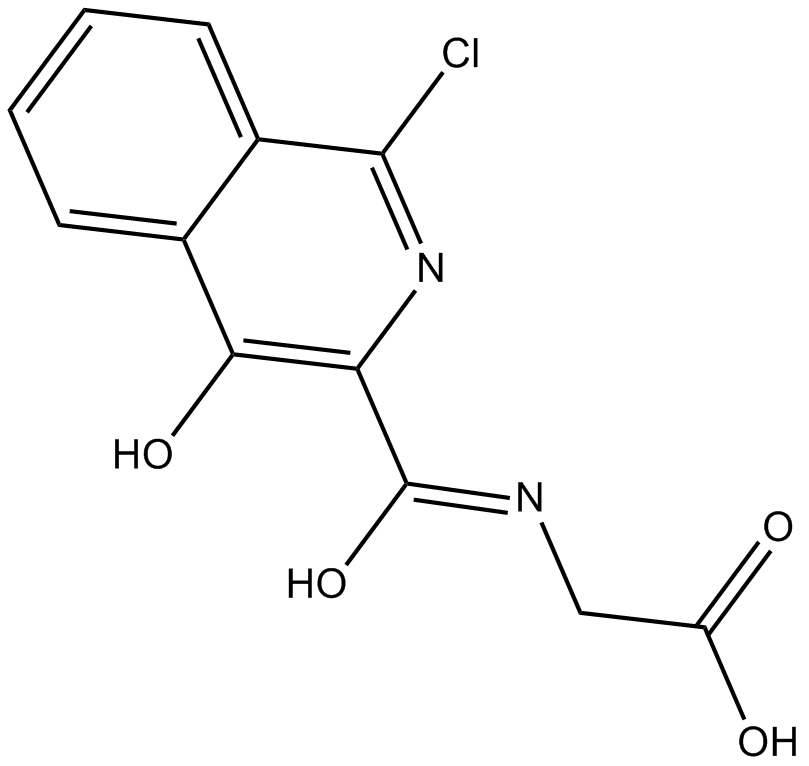
FG2216
HIF-prolyl hydroxylase inhibitor
-
GC38044
Fraxinellone
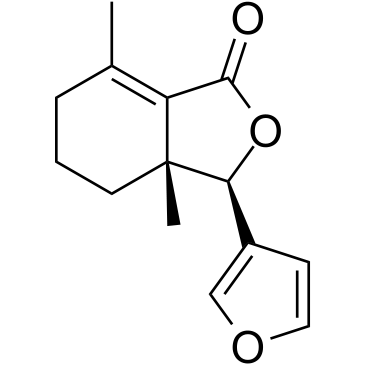
-
GC30263
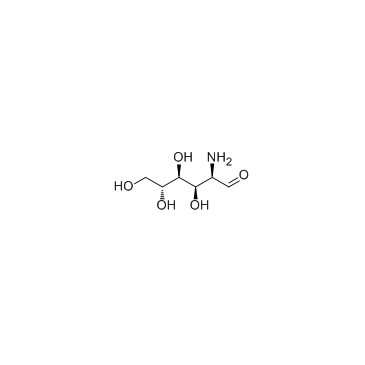
Glucosamine (D-Glucosamine)
Glucosamine (D-Glucosamine) (D-Glucosamine (D-Glucosamine)) is an amino sugar and a prominent precursor in the biochemical synthesis of glycosylated proteins and lipids, is used as a dietary supplement.
-
GC34595
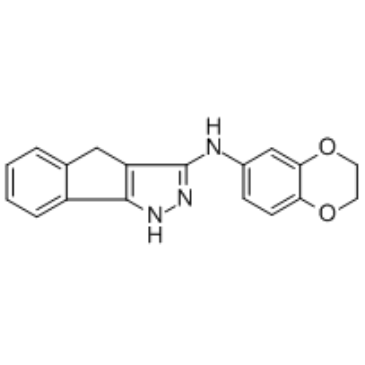
GN44028
A HIF-1 inhibitor
-
GC39146
HIF-1 inhibitor-1
An inhibitor of HIF-1 signaling

-
GC31358
HIF-2α-IN-1
HIF-2α-IN-1 is a HIF-2α inhibitor has an IC50 of less than 500 nM in HIF-2α scintillation proximity assay.
-
GC11767
Hydralazine HCl
Hydralazine HCl is a orally active antihypertensive agent, reduces peripheral resistance directly by relaxing the smooth muscle cell layer in arterial vessel.
-
GC32948
IDF-11774
A novel HIF-1 inhibitor
-
GC12255
IOX4
PHD2 inhibitor
-
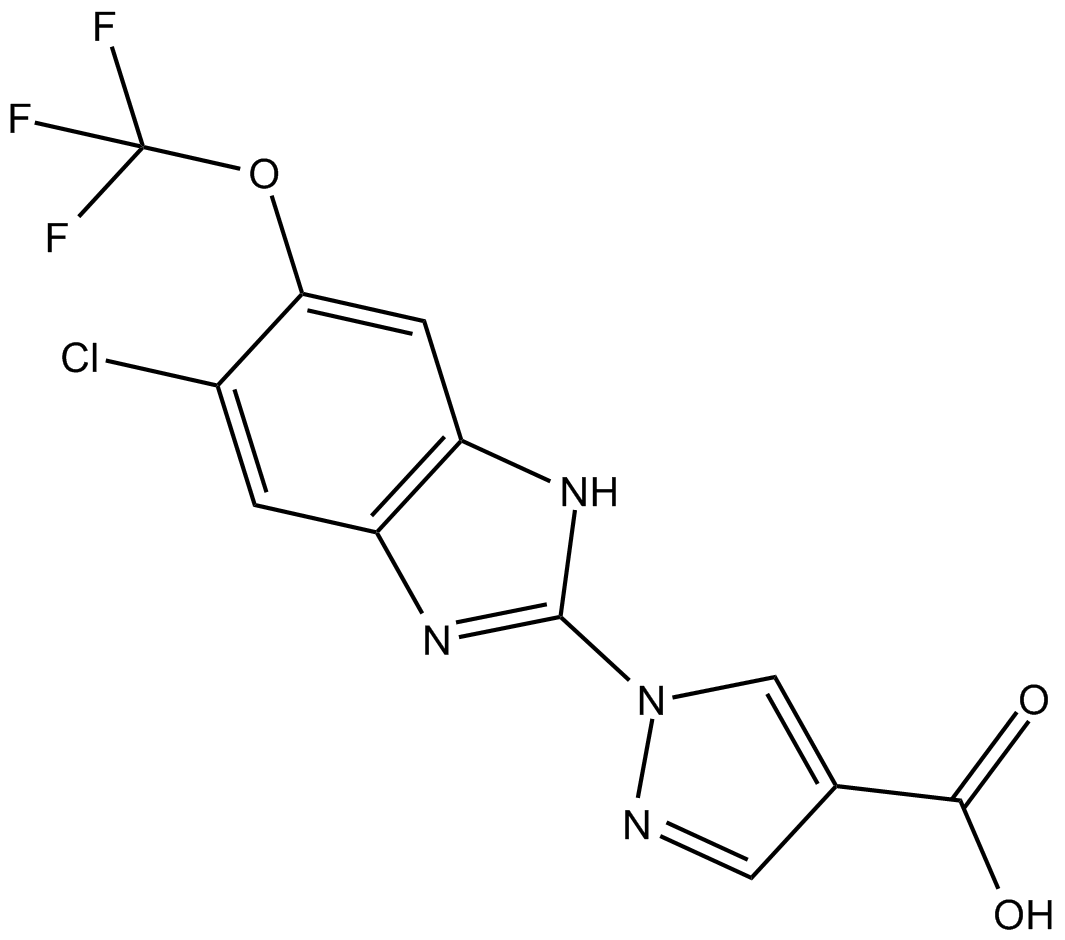
GC15379
JNJ-42041935
Hypoxia-inducible factor (HIF) prolyl hydroxylase (PHD) inhibitor
-
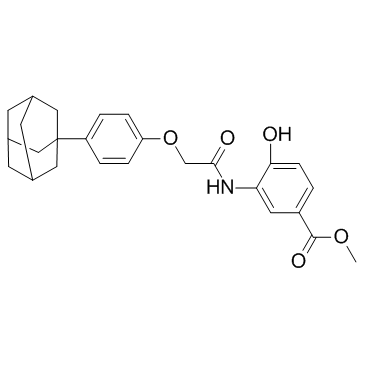
GC32724
LW6 (HIF-1α inhibitor)
LW6 (HIF-1α inhibitor) (HIF-1α inhibitor) is a novel HIF-1 inhibitor with an IC50 of 4.4 μM. LW6 (HIF-1α inhibitor) decreases HIF-1α protein expression without affecting HIF-1β expression.
-
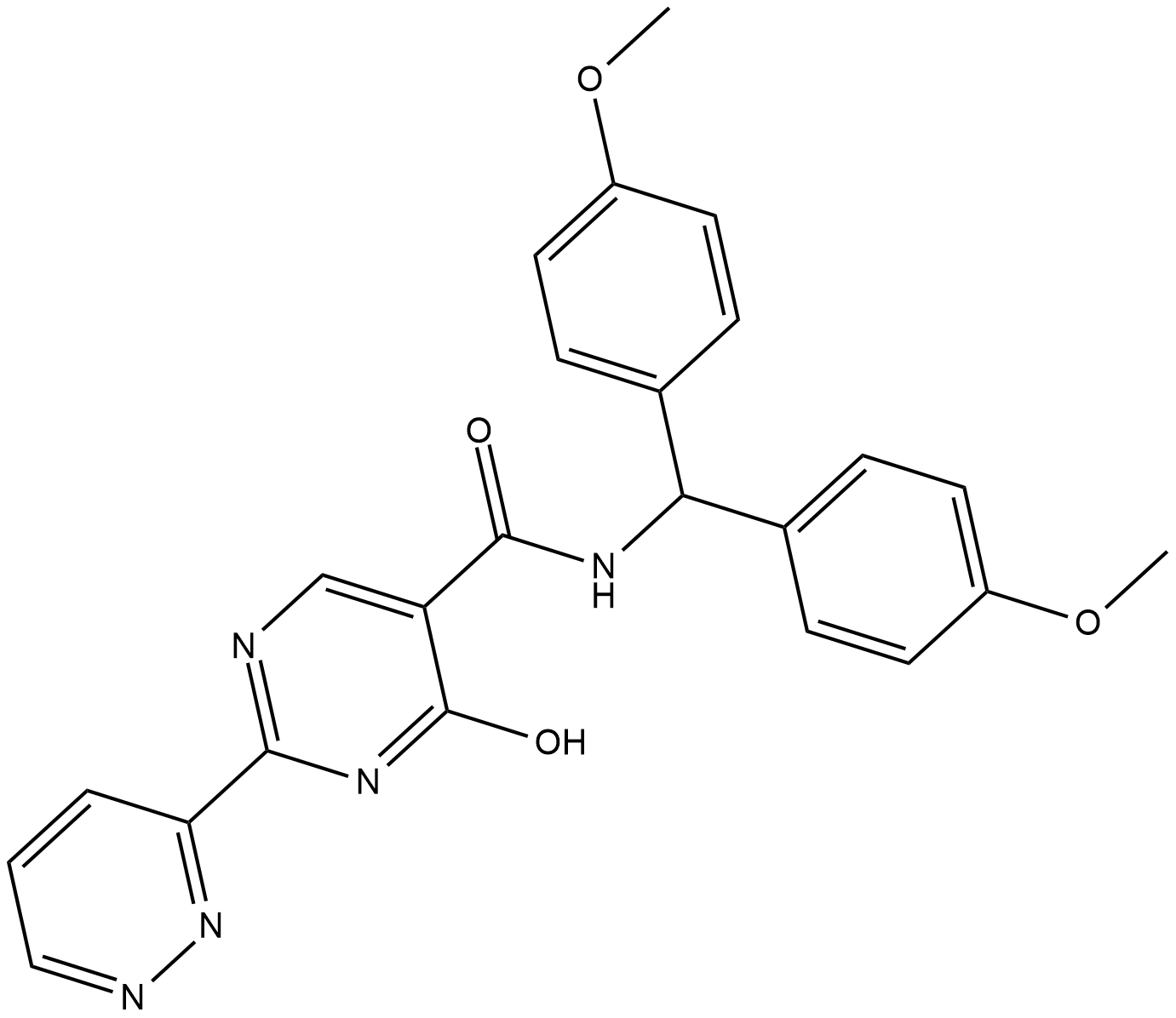
GC19251
MK-8617
MK-8617 is an orally active pan-inhibitor of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) with an IC50 of 1 nM for PHD2.
-
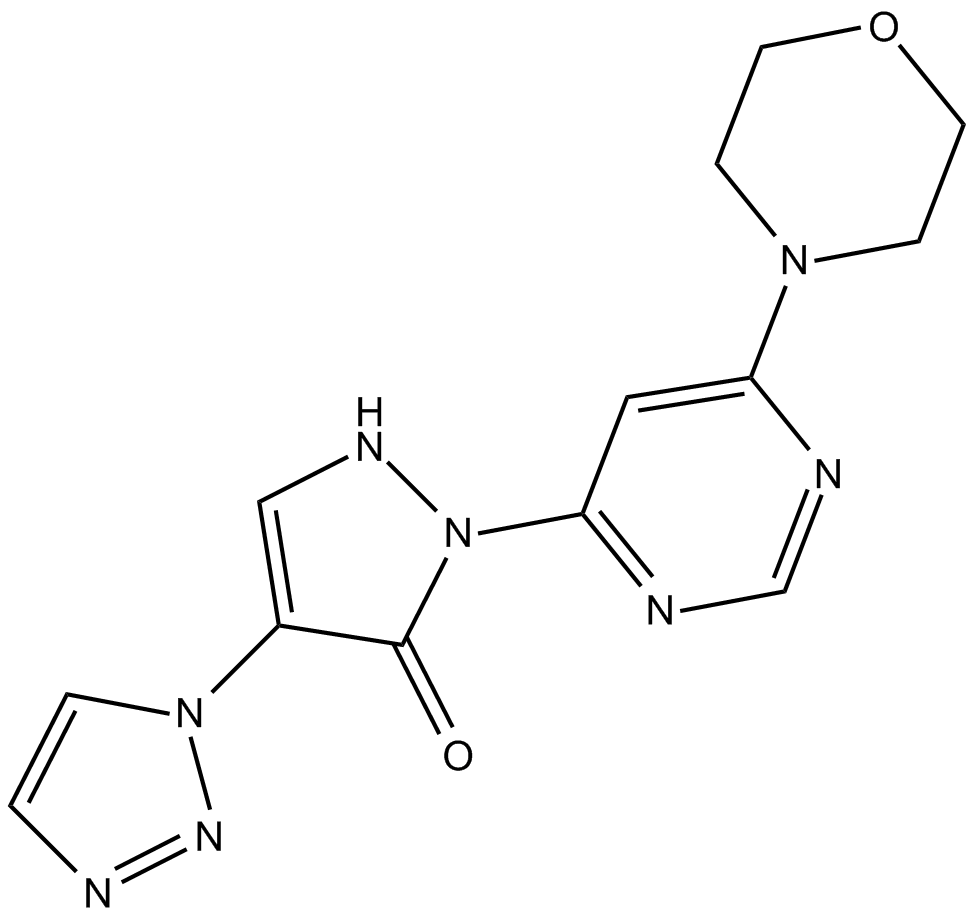
GC10046
Molidustat (BAY85-3934)
Molidustat (BAY85-3934) (BAY 85-3934) is a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase (HIF-PH) with mean IC50 values of 480 nM for PHD1, 280 nM for PHD2, and 450 nM for PHD3.
-
GC17358
Octyl-α-ketoglutarate
prolyl hydroxylases (PHD) activator
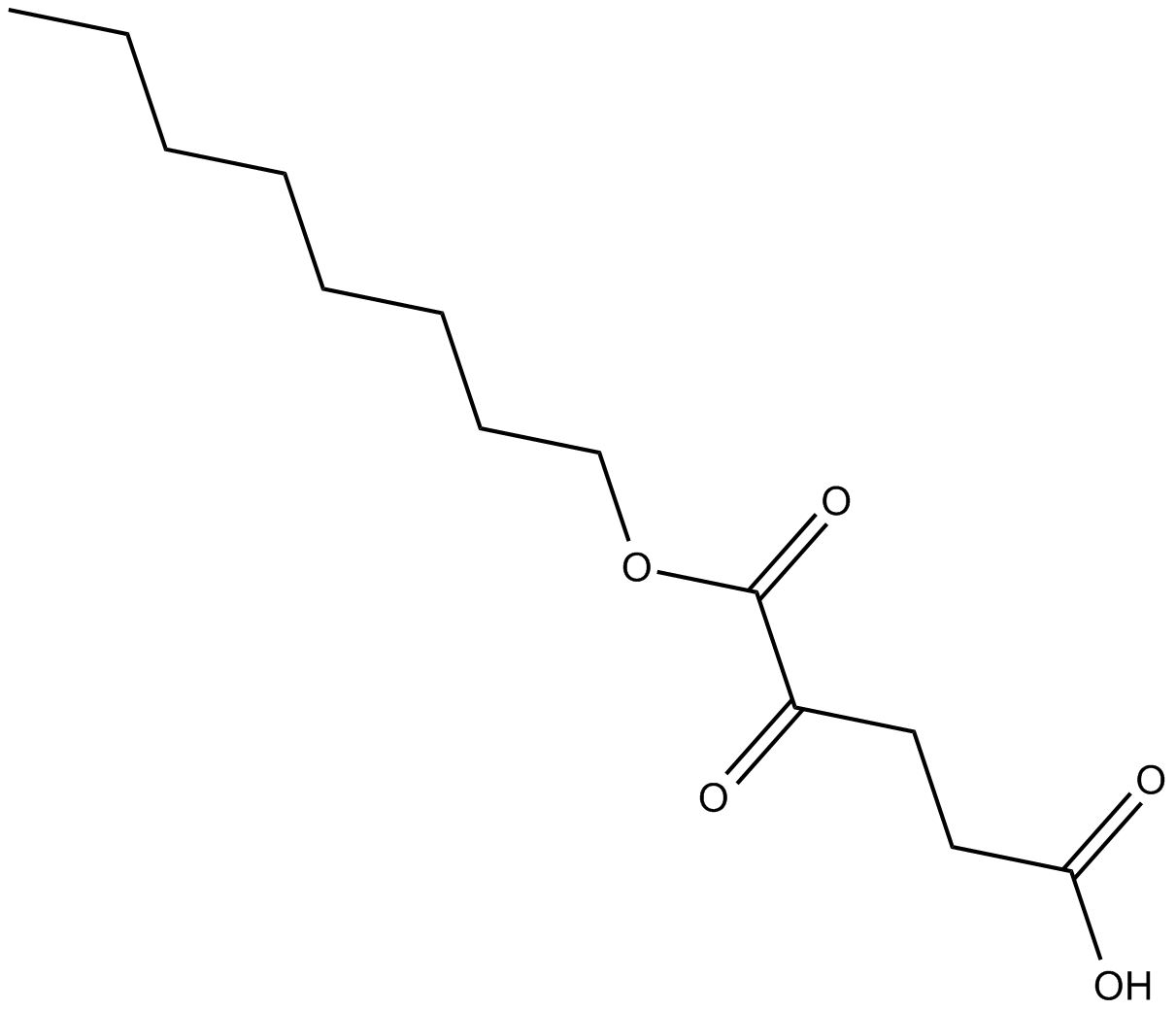
-
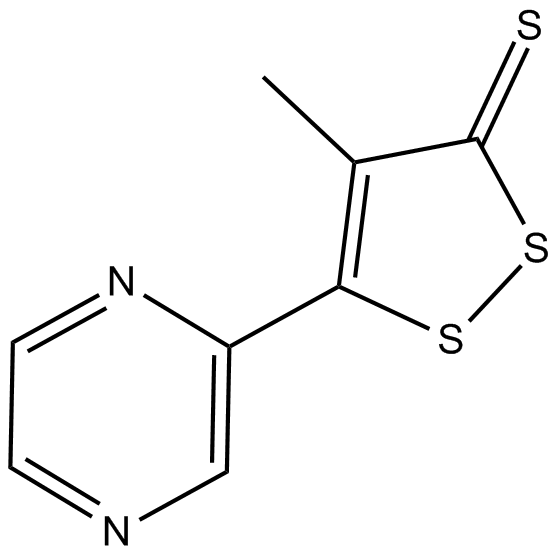
GC12821
Oltipraz
Nrf2 activator;An antischistosomal agent
-
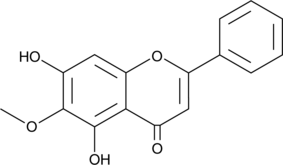
GC41625
Oroxylin A
Oroxylin A is a flavonoid that has been found in S.
-
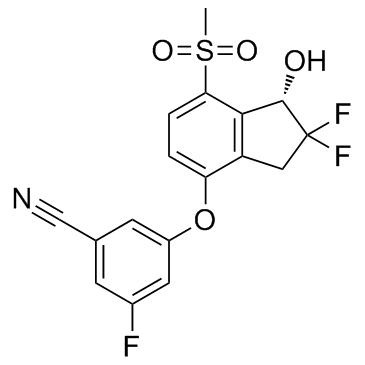
GC32680
PT-2385
PT-2385 is a selective HIF-2α inhibitor with a Ki of less than 50 nM.
-
GC37034
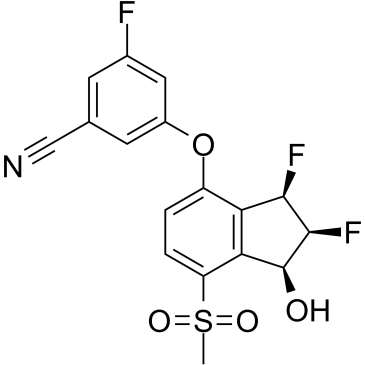
PT2977
PT2977 (PT2977) is an orally active and selective HIF-2α inhibitor with an IC50 of 9 nM. PT2977, as a second-generation HIF-2α inhibitor, increases potency and improves pharmacokinetic profile. PT2977 is a potential treatment for clear cell renal cell carcinoma (ccRCC).
-
GC11031
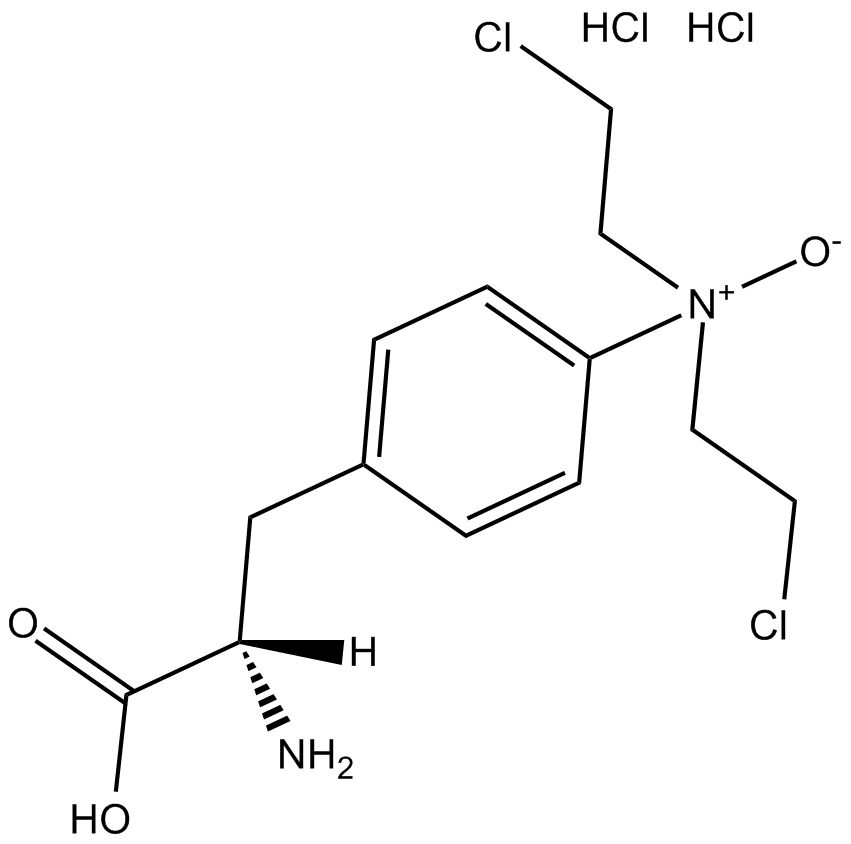
PX-478 2HCl
PX-478 is a selective inhibitor that suppresses constitutive and hypoxia-induced HIF-1α levels.
-
GC37066
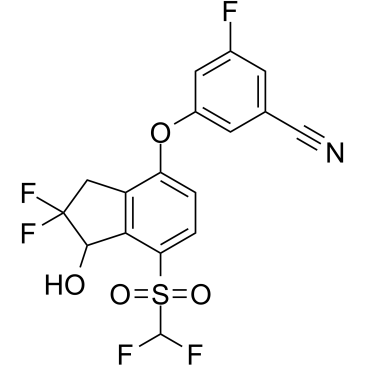
Rac-PT2399
A potent and specific HIF-2α inhibitor
-
GC32876
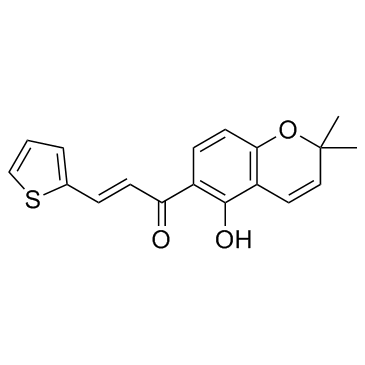
SYP-5
A HIF-1 inhibitor
-
GC32945
THS-044
THS-044 binding stabilizes the HIF2α PAS-B folded state, for regulating HIF2 activity in endogenous and clinical settings.
-
GC32228
Tilorone dihydrochloride
An interferon-inducing antiviral agent
-
GC37801
TM6089
TM6089 is a unique Prolyl Hydroxylase (PHD) inhibitor which stimulates HIF activity without iron chelation and induces angiogenesis and exerts organ protection against ischemia.
-
GC31498
TP0463518
TP0463518 is a potent hypoxia-inducible factor prolyl hydroxylases (PHDs) inhibitor
-
GC16811
Vadadustat
HIF-PH inhibitor
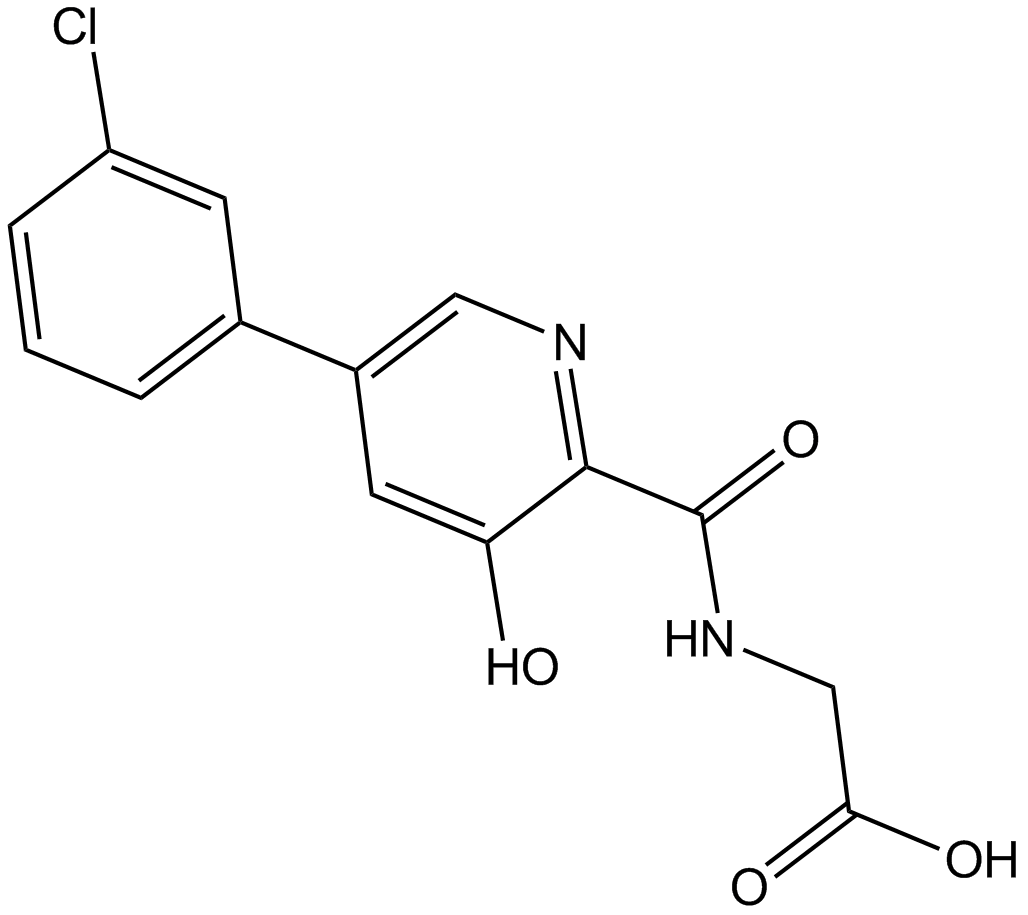
-
GC37889
VCE-004.8
VCE-004.8 (VCE-004.8) is an orally active, specific PPARγ and CB2 receptor dual agonist.
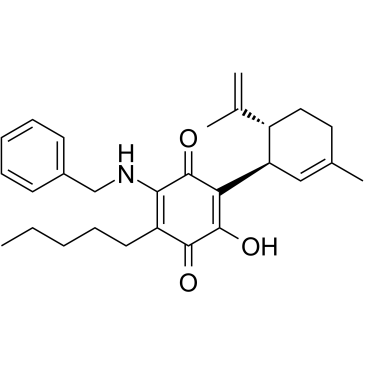
-
GC37966
ZINC13466751
ZINC13466751 is a potent inhibitor of HIF-1α/von Hippel-Lindau interaction with an IC50 of 2.0 ?M.