Angiogenesis
Products for Angiogenesis
- Cat.No. Product Name Information
-
GC37975
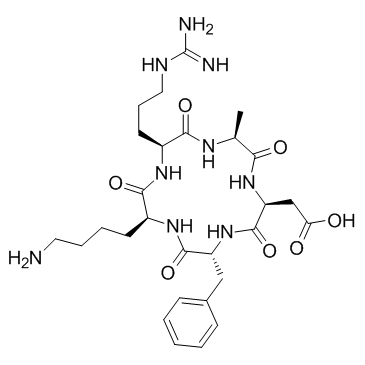
α2β1 Integrin Ligand Peptide
α2β1 Integrin Ligand Peptide interacts with the α2β1 integrin receptor on the cell membrane and mediates extracellular signals into cells.

-
GC38873
α2β1 Integrin Ligand Peptide TFA
-
GC68390
α5β1 integrin agonist-1
-
GC62380
αvβ1 integrin-IN-1
αvβ1 integrin-IN-1 (Compound C8) is a potent and selective αvβ1 integrin inhibitor with an IC50 of 0.63 nM.
-
GC62566
αvβ1 integrin-IN-1 TFA
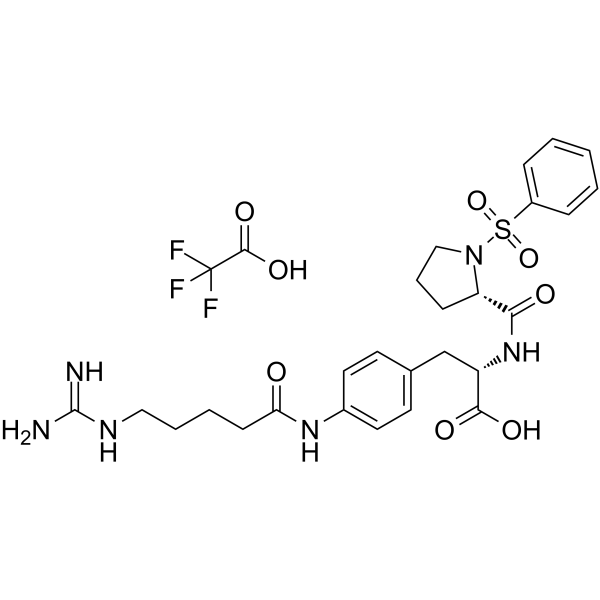
-
GC64932
αvβ5 integrin-IN-1
αvβ5 integrin-IN-1 is a first potent and selective αvβ5 integrin inhibitor (pIC50 = 8.2) .
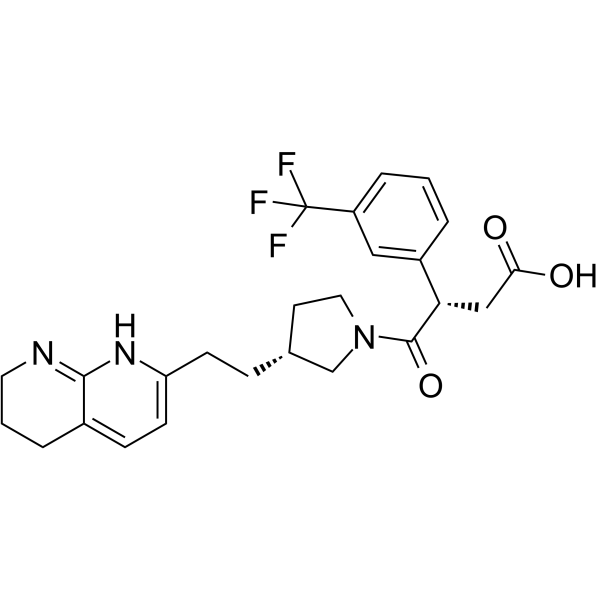
-
GC45269
(±)10(11)-DiHDPA
(±)10,11-DiHDPE
(±)10(11)-DiHDPA is produced from cytochrome P450 epoxygenase action on docosahexaenoic acid.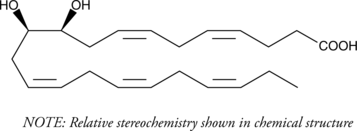
-
GC41212
(±)10(11)-EpDPA
(±)10,11-EDP, (±)10,11-EpDPE, (±)10,11-epoxy DPA, (±)10,11-epoxy Docosapentaenoic Acid
Cytochrome P450 metabolism of polyunsaturated fatty acids produces numerous bioactive epoxide regioisomers.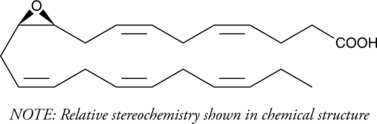
-
GC40466
(±)11(12)-EET
(±)11,12-EpETrE
(±)11(12)-EET is a fully racemic version of the R/S enantiomeric forms biosynthesized from arachidonic acid by cytochrome P450 enzymes.
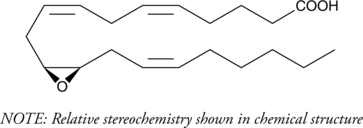
-
GC41648
(±)13(14)-DiHDPA
13(14)-DiHDPA, 13(14)-DiHDoPE, 13,14-DiHDPE
(±)13(14)-DiHDPA is a metabolite of docosahexaenoic acid that is produced via oxidation by cytochrome P450 epoxygenases.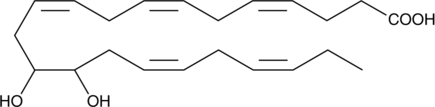
-
GC41191
(±)13(14)-EpDPA
(±)13,14-EDP, (±)13,14-EpDPE, (±)13,14-epoxy DPA, (±)13,14-epoxy Docosapentaenoic Acid
Cytochrome P450 metabolism of polyunsaturated fatty acids produces numerous bioactive epoxide regioisomers.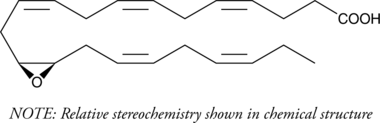
-
GC41653
(±)16(17)-DiHDPA
(±)16(17)-DiHDPA is produced from cytochrome P450 epoxygenase action on docosahexaenoic acid.
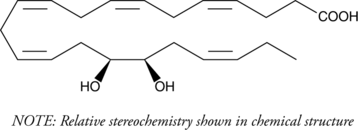
-
GC41655
(±)19(20)-EDP Ethanolamide
19,20-DHEA epoxide, 19,20-epoxy Docosapentaenoic Acid Ethanolamide, 19,20-EDP-EA, 19,20-EDP epoxide
(±)19(20)-EDP ethanolamide is an ω-3 endocannabinoid epoxide and cannabinoid (CB) receptor agonist (EC50s = 108 and 280 nM for CB1 and CB2, respectively).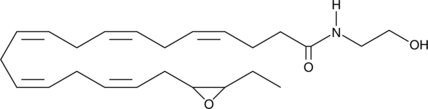
-
GC41203
(±)7(8)-EpDPA
(±)7,8-EDP, (±)7,8-EpDPE, (±)7,8-epoxy DPA, (±)7,8-epoxy Docosapentaenoic Acid
Docosahexaenoic acid is the most abundant ω-3 fatty acid in neural tissues, especially in the brain and retina.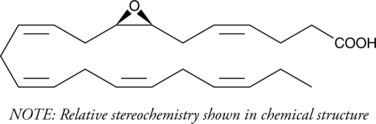
-
GC34069
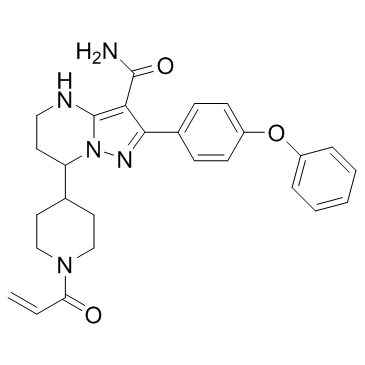
(±)-Zanubrutinib ((±)-BGB-3111)
(±)-Zanubrutinib ((±)-BGB-3111) ((±)-BGB-3111) is a potent, selective and orally available Bruton's tyrosine kinase (Btk) inhibitor.
-
GC67857
(R)-Elsubrutinib
(R)-ABBV-105

-
GC69837
(R/S)-Alicaforsen
(R/S)-ISIS-2302
(R/S)-Alicaforsen is the racemic form of Alicaforsen, which consists of both R and S configurations. Alicaforsen is a 20-base length antisense oligonucleotide that inhibits the production of ICAM-1, an important adhesion molecule involved in the migration and transport process of white blood cells to inflammatory sites.

-
GC63797
(S)-Sunvozertinib
(S)-DZD9008
(S)-Sunvozertinib ((S)-DZD9008), the S-enantiomer of Sunvozertinib, shows inhibitory activity against EGFR exon 20 NPH and ASV insertions, EGFR L858R/T790M mutation and Her2 exon20 YVMA insertion (IC50=51.2 nM, 51.9 nM, 1 nM, and 21.2 nM, respectively). (S)-Sunvozertinib also inhibits BTK.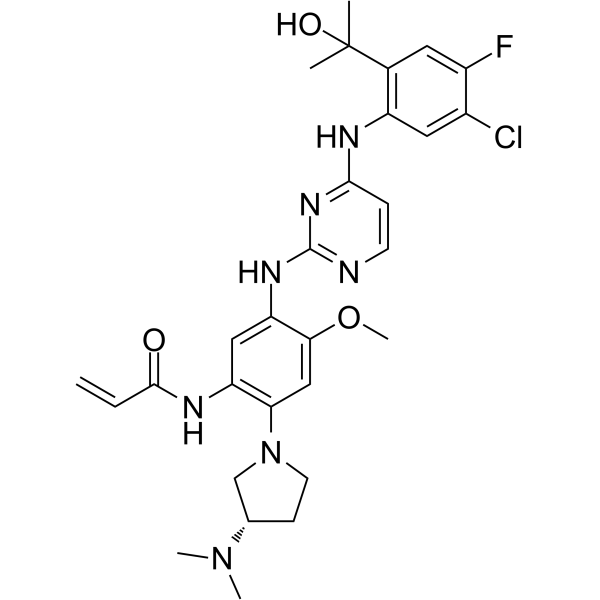
-
GC49808
12-methyl Tridecanoic Acid
iso-14:0, iso-C14:0, 12-MTA
A methylated fatty acid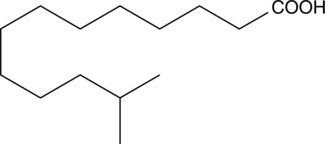
-
GC46474
18-Deoxyherboxidiene
RQN-18690A
A bacterial metabolite with antiangiogenic activity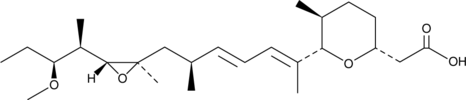
-
GC16195
2,4-DPD
2,4-Diethylpyridine dicarboxylate
Diethyl pyridine-2,4-dicarb is a potent prolyl 4-hydroxylase-directed proinhibitor.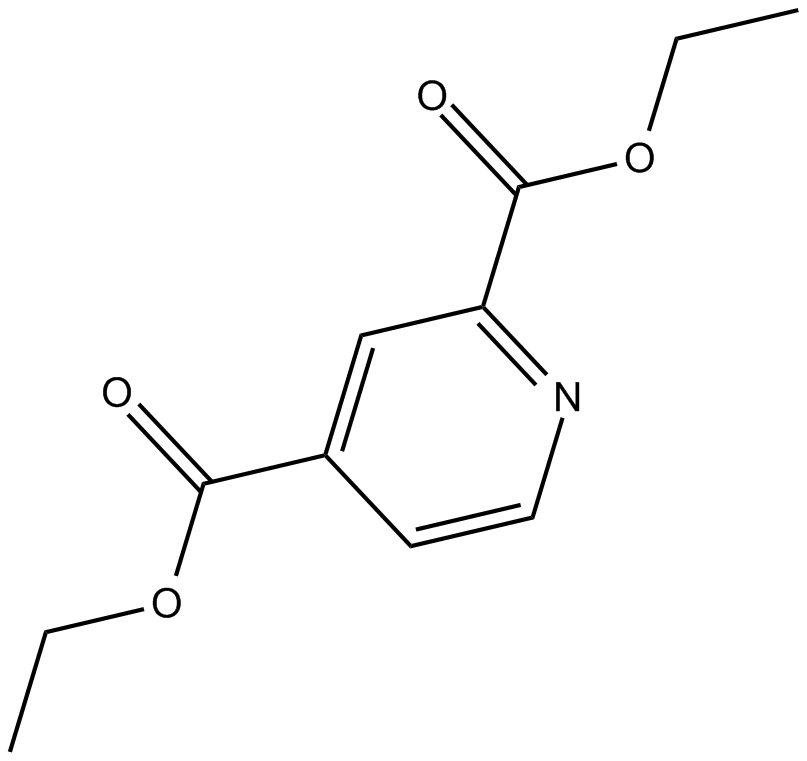
-
GC17368
2-Furoyl-LIGRLO-amide
Protease-activated receptor agonist
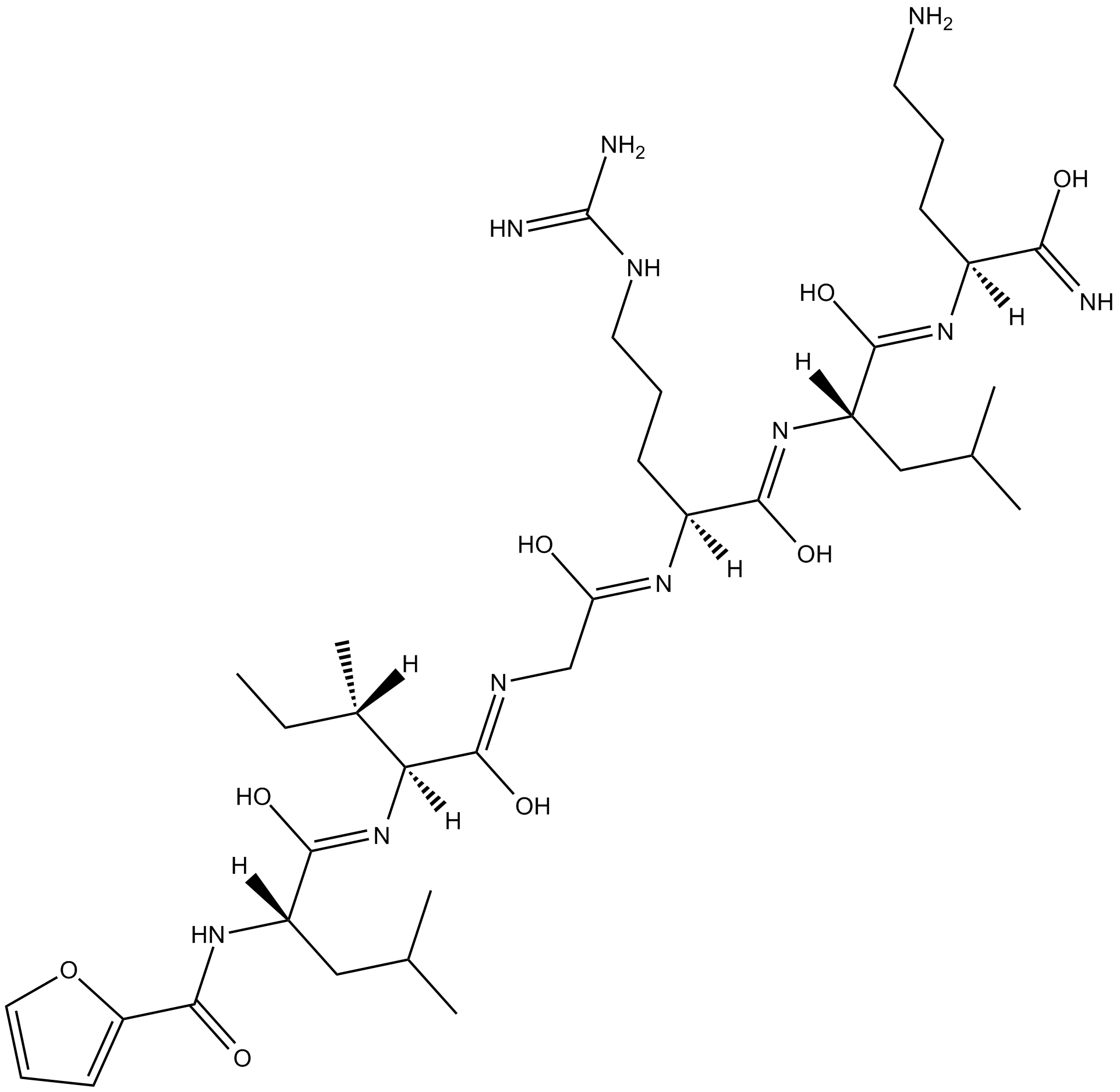
-
GC38731
2-Furoyl-LIGRLO-amide TFA
2-Furoyl-LIGRLO-amide TFA is a potent and selective proteinase-activated receptor 2 (PAR2) agonist with a pD2 value of 7.0.
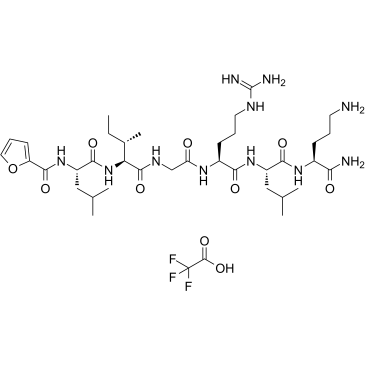
-
GC14282
3-acetyl-11-keto-β-Boswellic Acid
3-O-acetyl-11-keto-β-Boswellic acid,AKBA
3-acetyl-11-keto-β-Boswellic Acid (Acetyl-11-keto-β-boswellic acid) is an active triterpenoid compound from the extract of Boswellia serrate and a novel Nrf2 activator.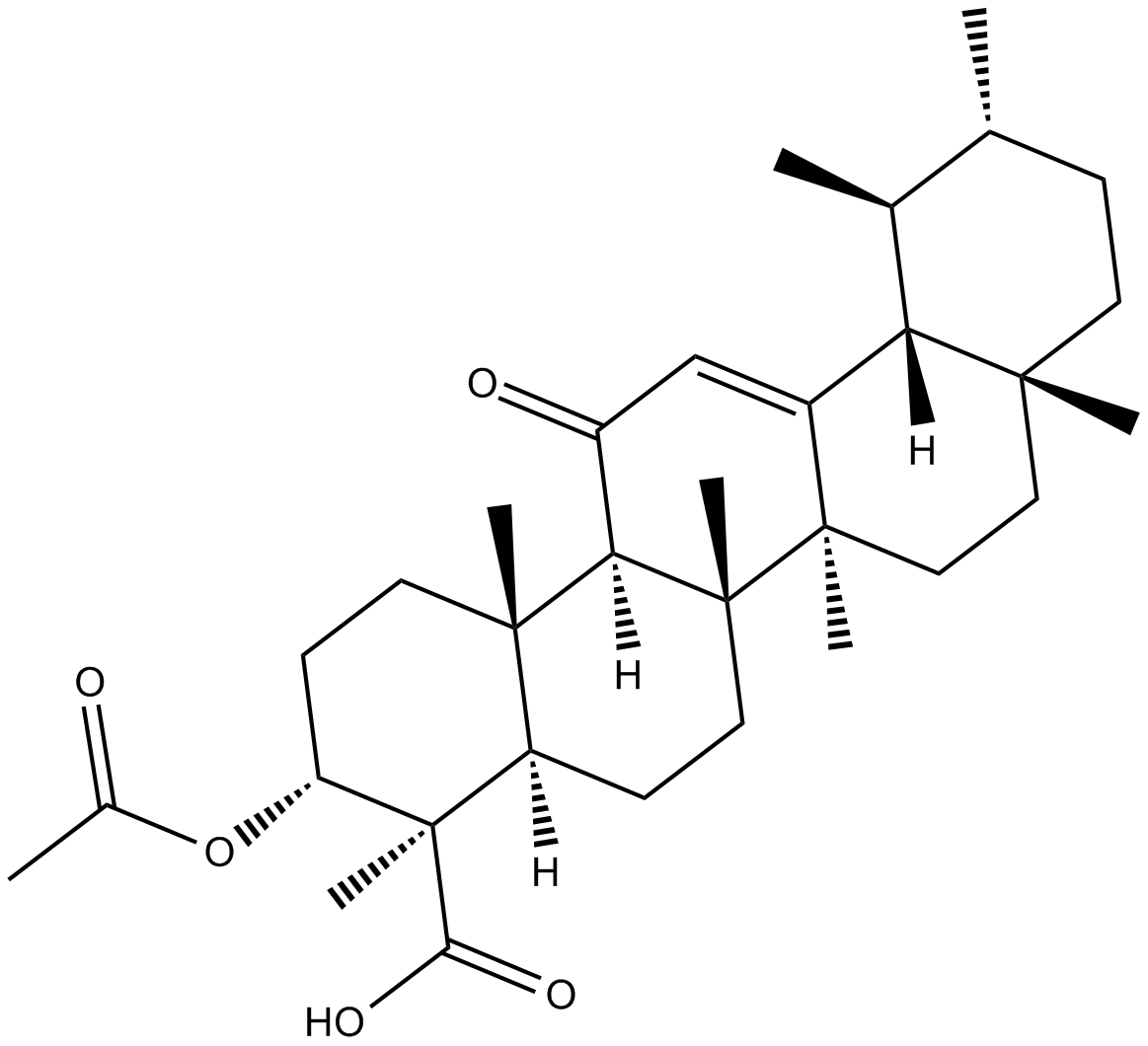
-
GC46583
3-Amino-2,6-Piperidinedione
α-Aminoglutarimide, 3-Aminoglutarimide, Glutamimide
An active metabolite of (±)-thalidomide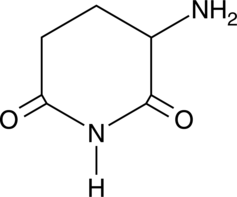
-
GC49562
4-methoxy Estrone
4-MeOE1, 4-methoxy E1
An active metabolite of estrone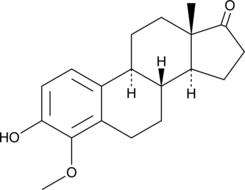
-
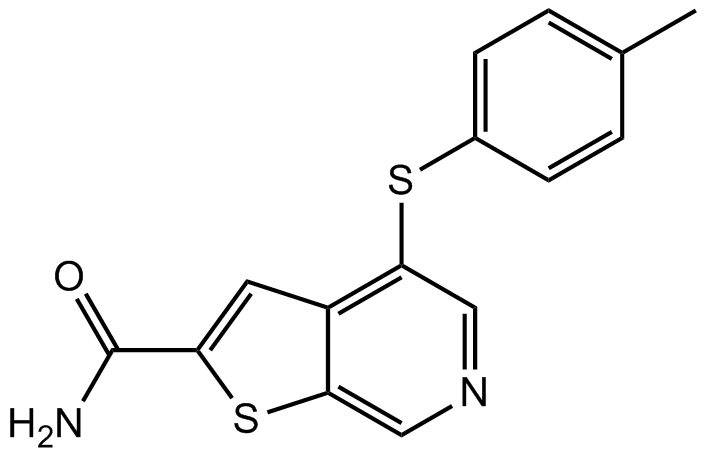
GC15557
A 205804
E-selectin/ICAM-1 expression inhibitor
-
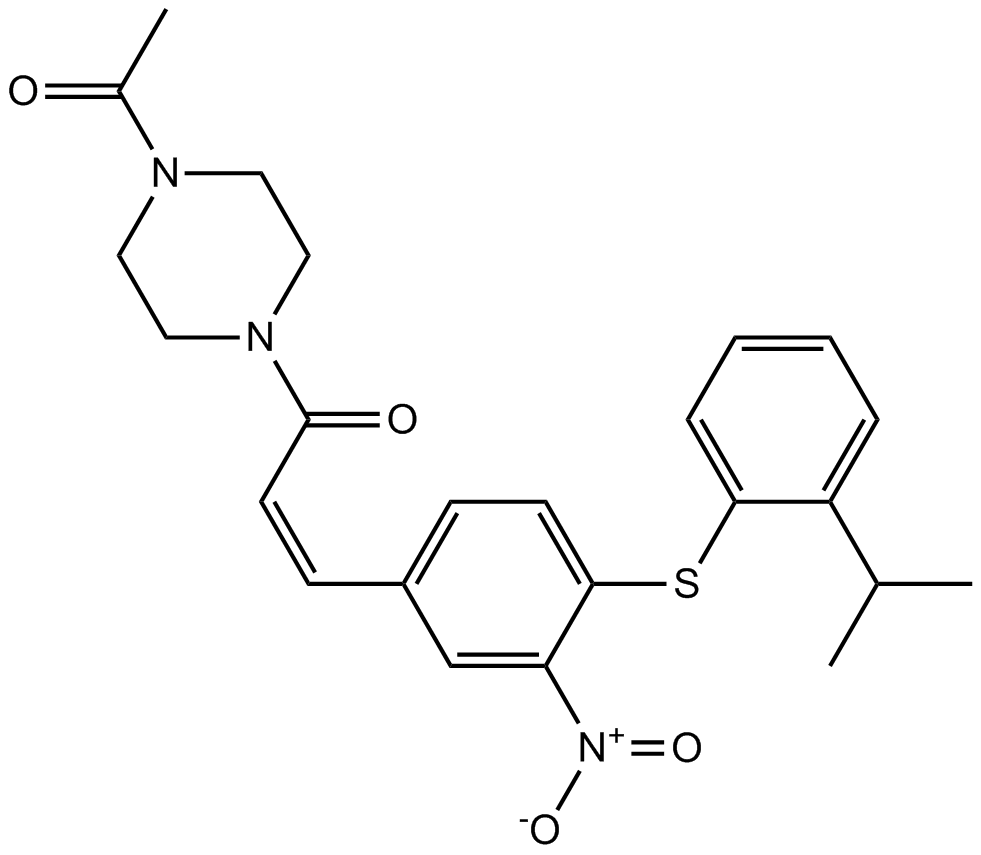
GC15192
A 286982
A LFA-1/ICAM-1 interaction inhibitor
-
GC65597
Abciximab
C7E3
Abciximab (C7E3), a chimeric mouse/human monoclonal antibody, is a glycoprotein (GP) IIb/IIIa inhibitor.
-
GC68594
Abituzumab
EMD 525797; DI17E6
Abituzumab (DI17E6) is a humanized monoclonal antibody (IgG2 type) against integrin αV. Abituzumab can effectively reduce the phosphorylation of FAK, Akt and ERK. Abituzumab can be used in cancer research, especially for prostate cancer.

-
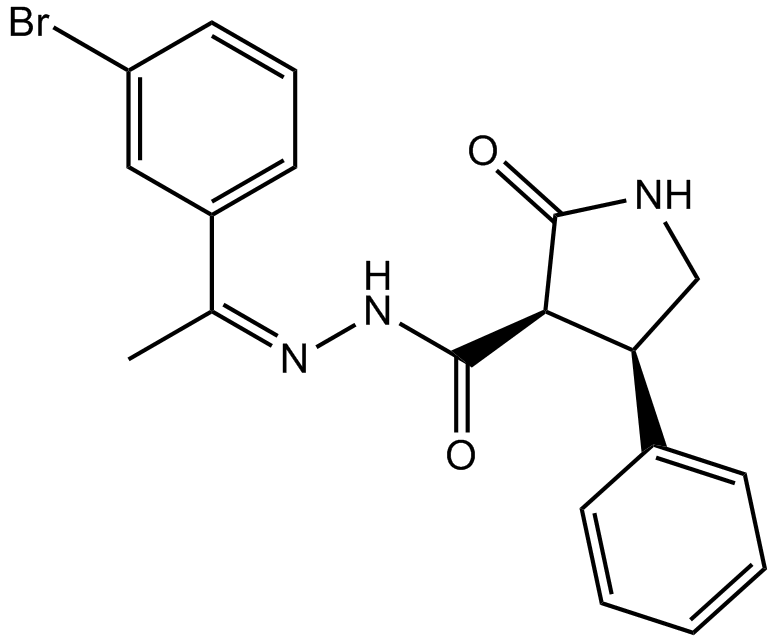
GC14831
AC 264613
PAR2 agonist,potent and selective
-
GC15290
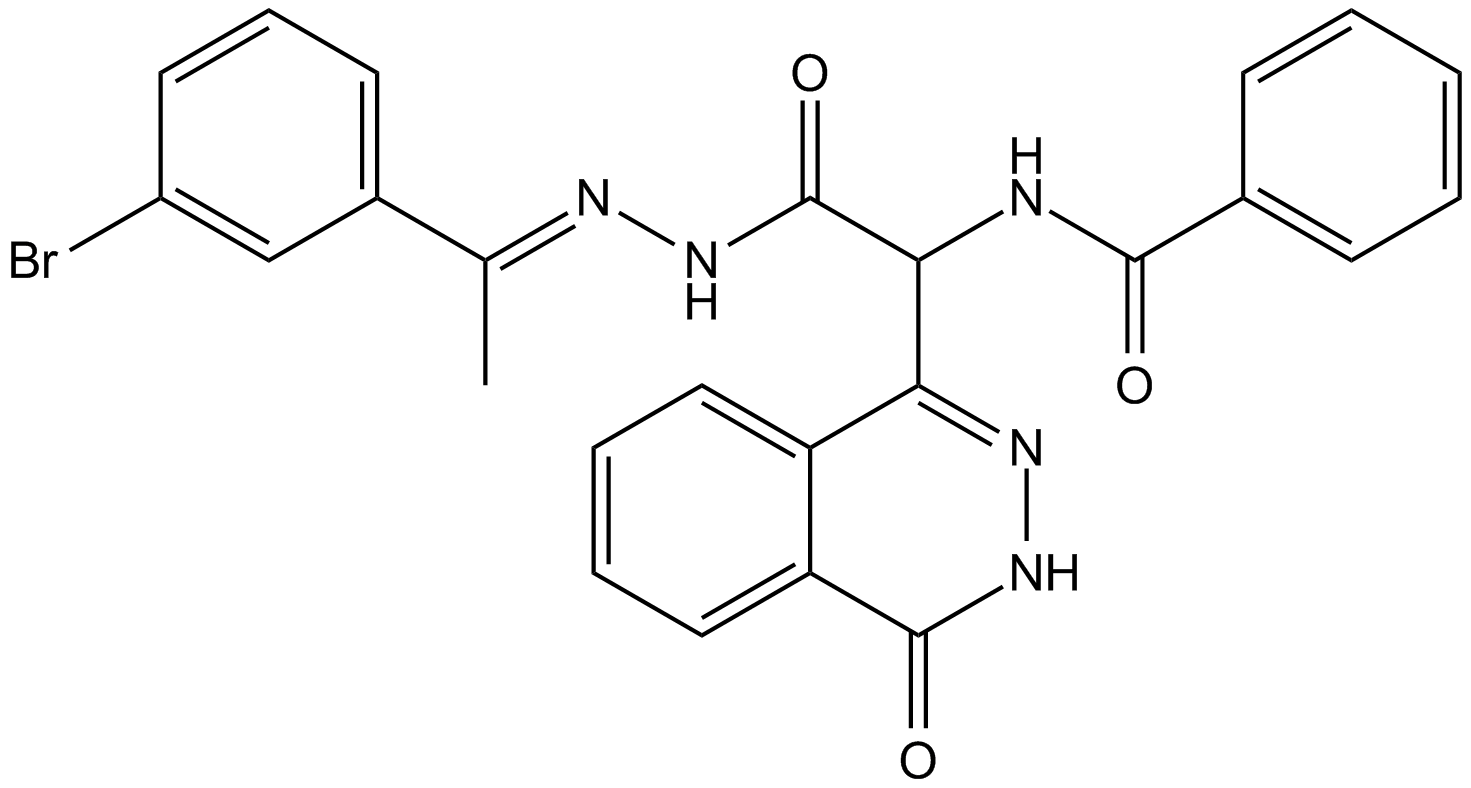
AC 55541
PAR2 agonist,potent and selective
-
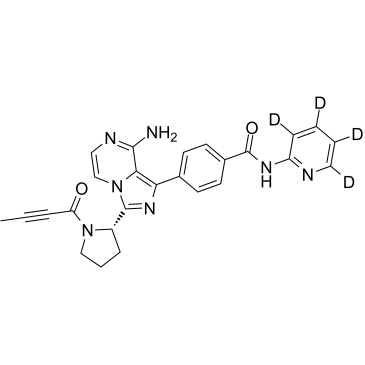
GC60551
Acalabrutinib D4
ACP-196-d4
-
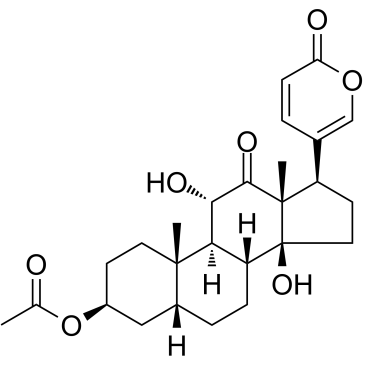
GC35230
Acetylarenobufagin
Acetylarenobufagin is a steroidal hypoxia inducible factor-1 (HIF-I) modulator.
-
GC15453
ACP-196
ACP-196
ACP-196 (ACP-196) is an orally active, irreversible, and highly selective second-generation BTK inhibitor. ACP-196 binds covalently to Cys481 in the ATP-binding pocket of BTK. ACP-196 demonstrates potent on-target effects and efficacy in mouse models of chronic lymphocytic leukemia (CLL).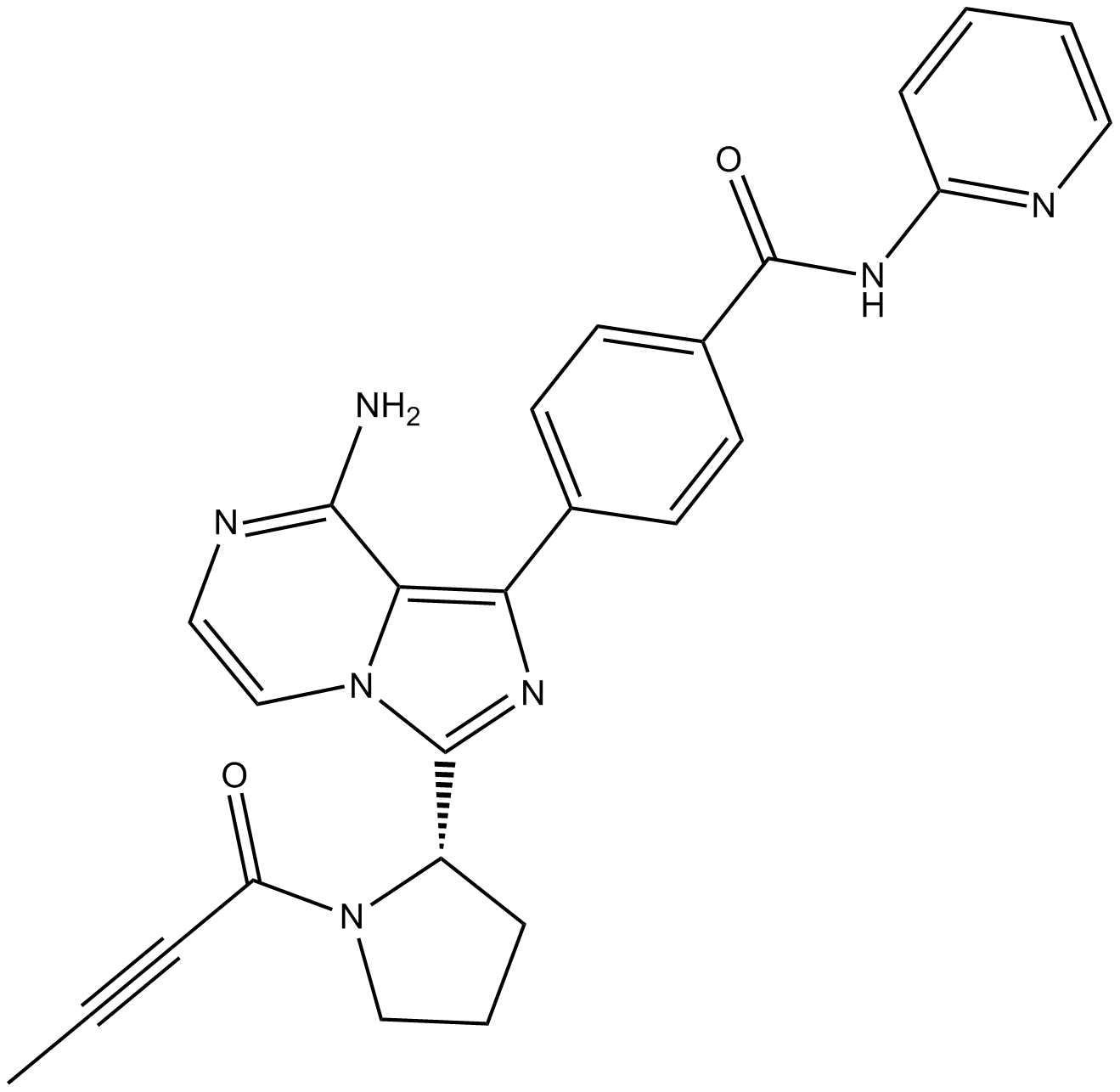
-
GC32083
Acriflavine
Acriflavine is a fluorescent dye for labeling high molecular weight RNA.

-
GC12487
Adaptaquin
HIF prolyl hydroxylase inhibitor
HIF-prolyl hydroxylase-2 (PHD2) inhibitor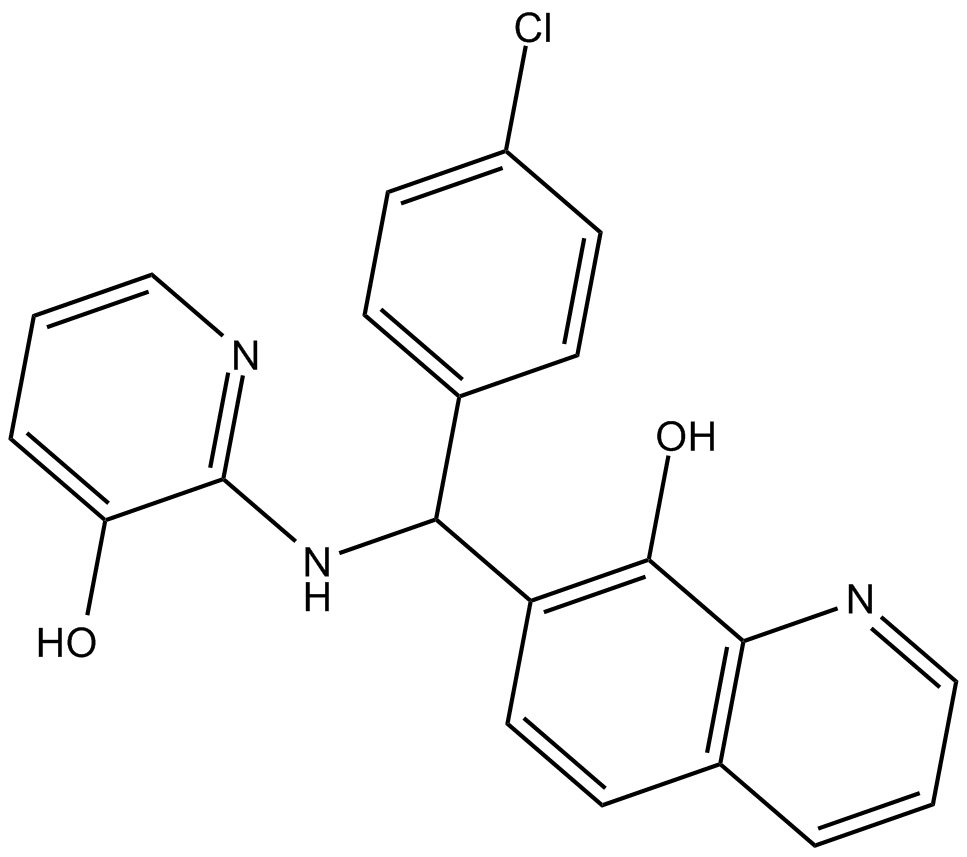
-
GC33416
AFP464
NSC710464 free base
AFP464 (NSC710464 free base), is an active HIF-1α inhibitor with an IC50 of 0.25 μM, also is a potent aryl hydrocarbon receptor (AhR) activator.
-
GC49799
Apatinib-d8
An internal standard for the quantification of apatinib
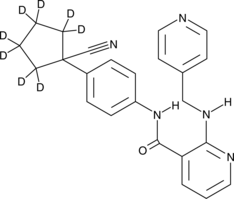
-
GC31679
ARQ 531
MK-1026
ARQ 531 (MK-1026) is a reversible non-covalent and orally active inhibitor of Bruton’s Tyrosine Kinase (BTK), with IC50s of 0.85 nM and 0.39 nM for WT-BTK and C481S-BTK, respectively.
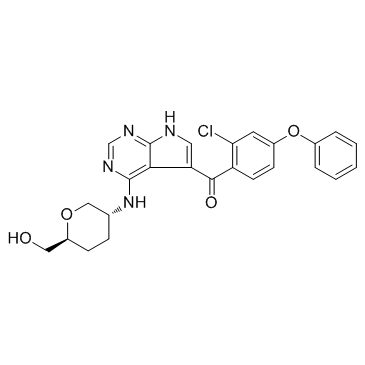
-
GC34133
ATN-161
ATN-161 is a novel integrin α5β1 antagonist, which inhibits angiogenesis and growth of liver metastases in a murine model.
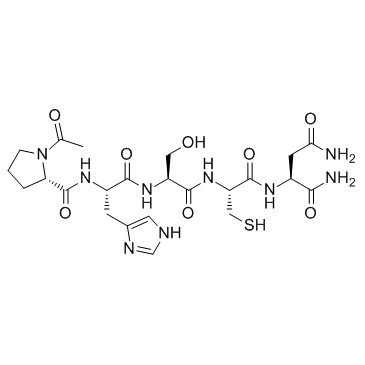
-
GC33837
ATN-161 trifluoroacetate salt (ATN-161 TFA salt)
Ac-PHSCN-NH2, PHSCN Peptide
ATN-161 trifluoroacetate salt (ATN-161 TFA salt) is a novel integrin α5β1 antagonist, which inhibits angiogenesis and growth of liver metastases in a murine model.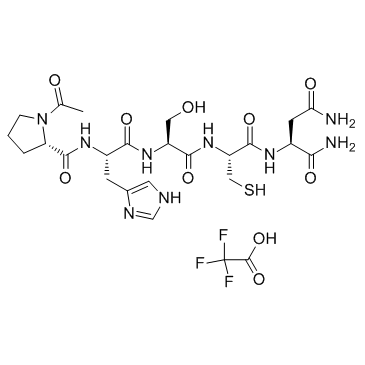
-
GC63658
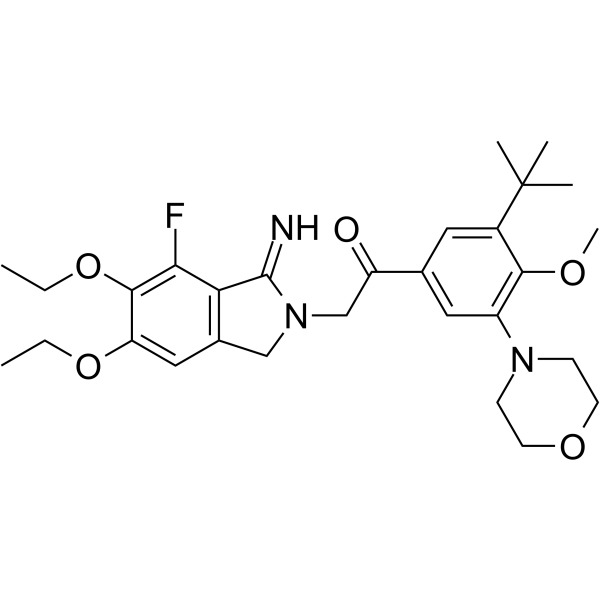
Atopaxar
Atopaxar (E5555) is a potent, orally active, selective and reversible thrombin receptor protease-activated receptor-1 (PAR-1) antagonist.
-
GC70685
Atuzabrutinib
Atuzabrutinib (SAR 444727) is a potent, selective reversible inhibitor of Btk (Bruton's tyrosine kinase) inhibitor.

-
GC13562
AVL-292
AVL292;AVL 292
A covalent BTK inhibitor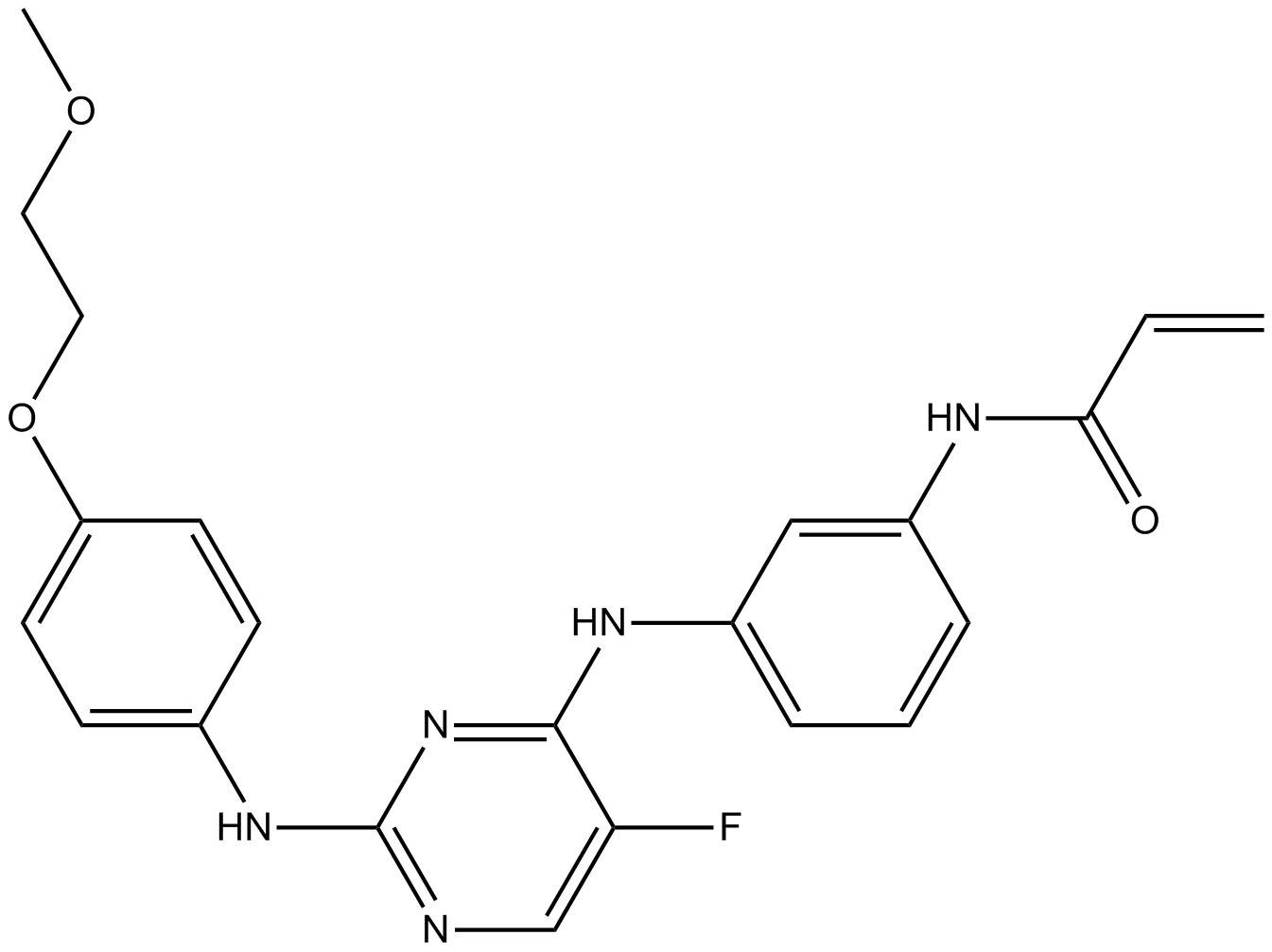
-
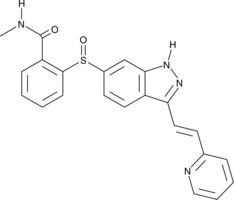
GC42887
Axitinib Sulfoxide
Axitinib sulfoxide is a major inactive metabolite of the tyrosine kinase inhibitor axitinib.
-
GC46899
Axitinib-13C-d3
AG-013736-13C-d3
An internal standard for the quantification of axitinib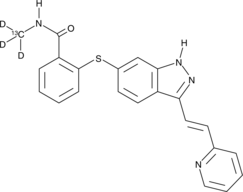
-
GC18152
AZ-3451
A potent, and selective PAR2 antagonist with Kd of 13.5 nM.
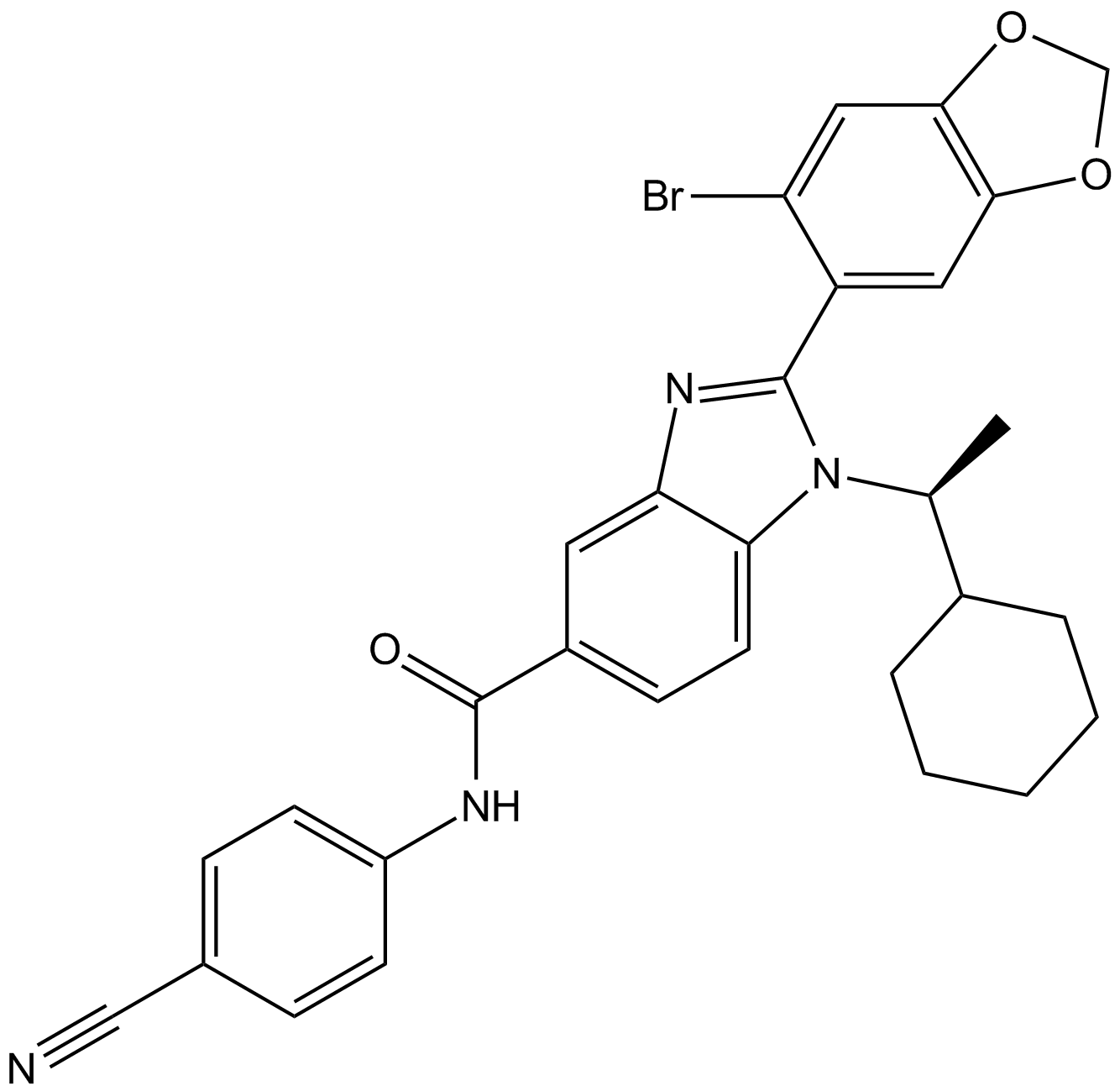
-
GC65312
AZ8838
AZ8838 is a potent, competitive, allosteric, orally active non-peptide small molecule antagonist of PAR2 with a pKi of 6.4 for hPAR2.
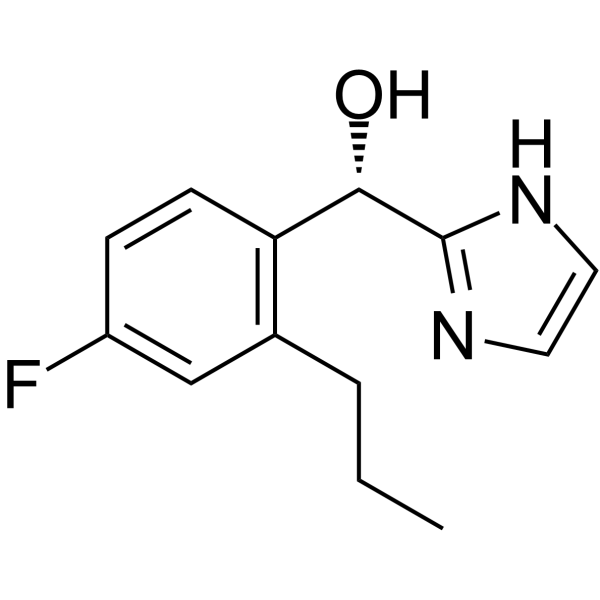
-
GC12698
BAY 87-2243
A HIF-1 inhibitor,potent and selective
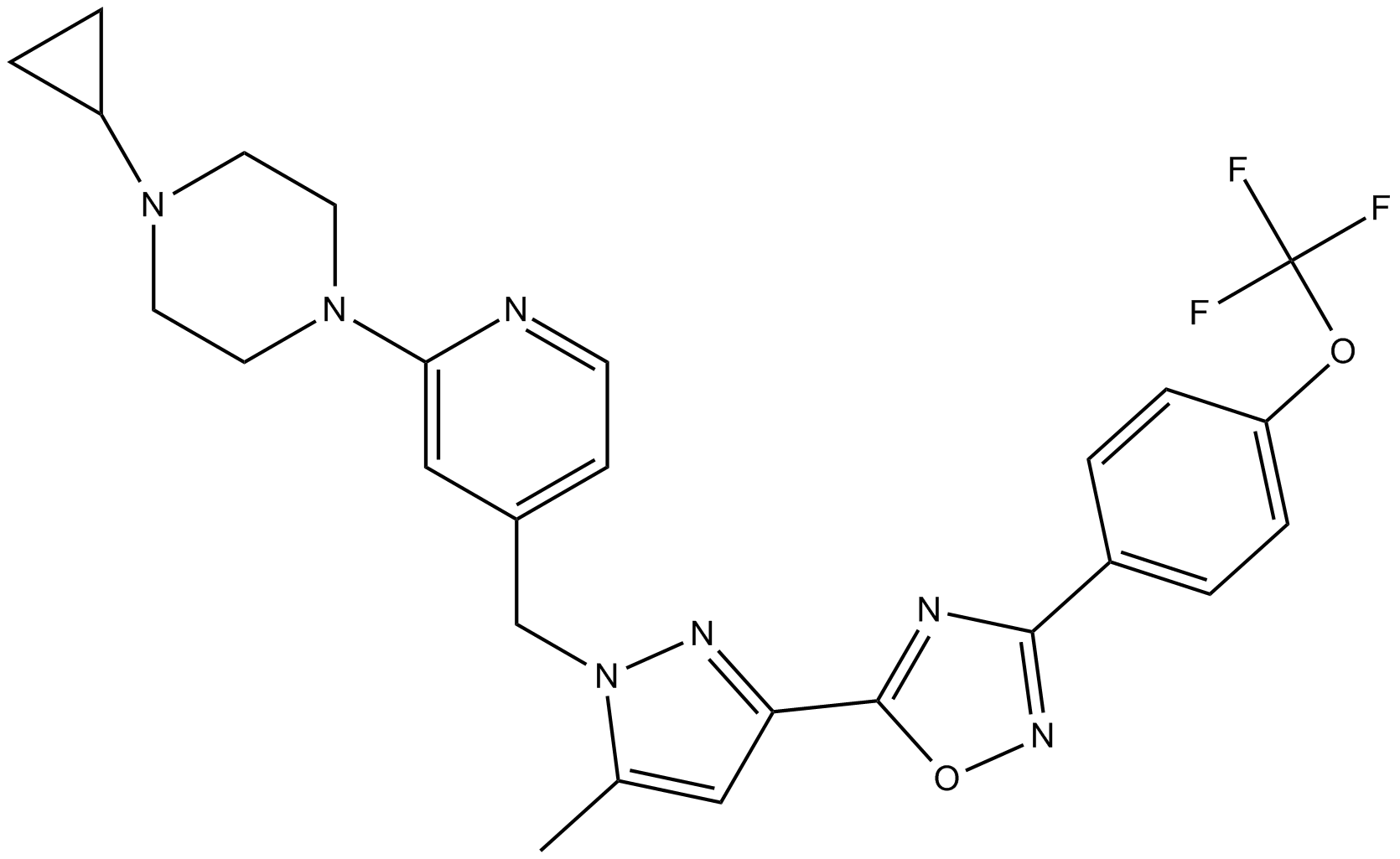
-
GC68754
Bersanlimab
BI-505
Bersanlimab (BI-505) is a fully human monoclonal antibody that targets intercellular adhesion molecule-1 (ICAM-1 or CD54). Bersanlimab has anti-cancer effects.

-
GC65909
Bexotegrast
PLN-74809
Bexotegrast is a potent inhibitor of αΝβ6 integrin. Bexotegrast can be used for researching fibrosis such as idiopathic pulmonary fibrosis (IPF) and nonspecific interstitial pneumonia (NSIP) (extracted from patent WO2020210404A1, compound 5).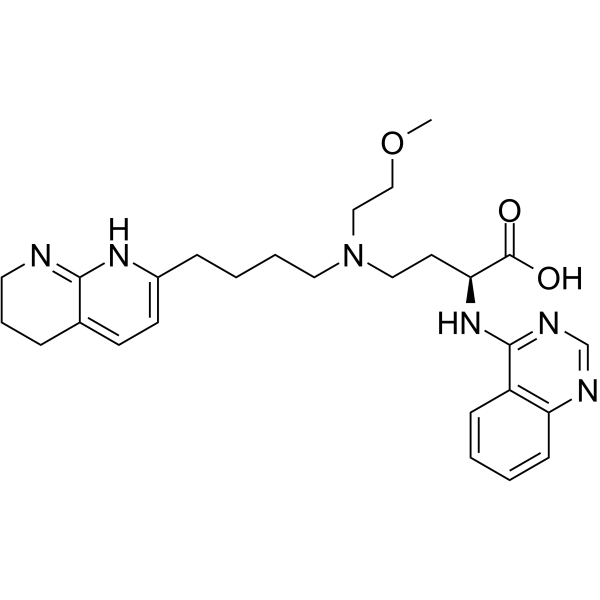
-
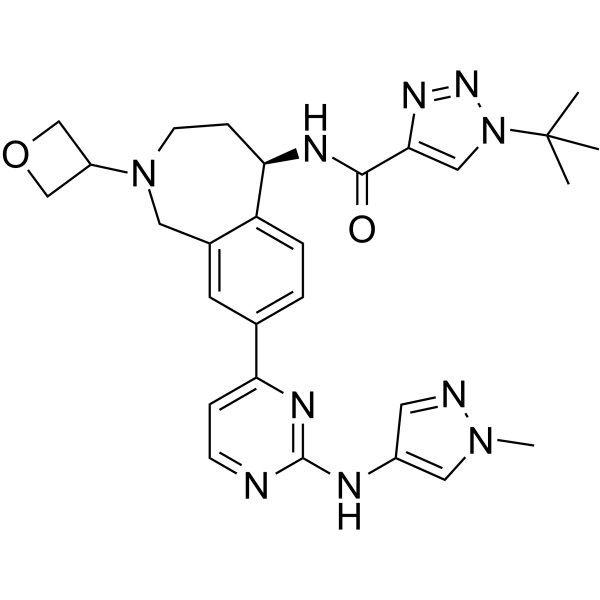
GC63846
BIIB091
BIIB091 is a highly selective, reversible and orally active BTK inhibitor for treating autoimmune diseases.
-
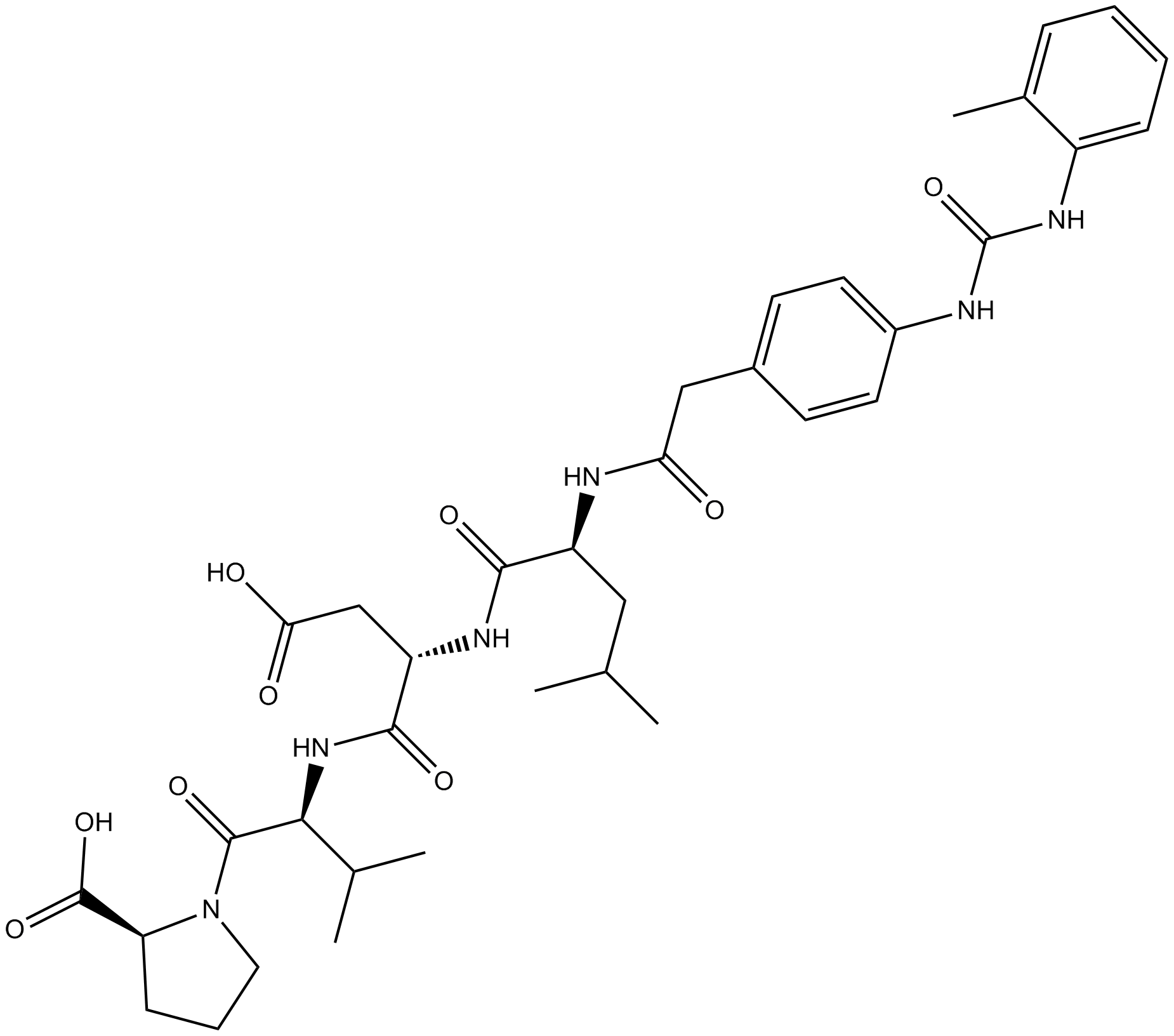
GC17560
BIO 1211
An α4β1 inhibitor,selective and high affinity
-
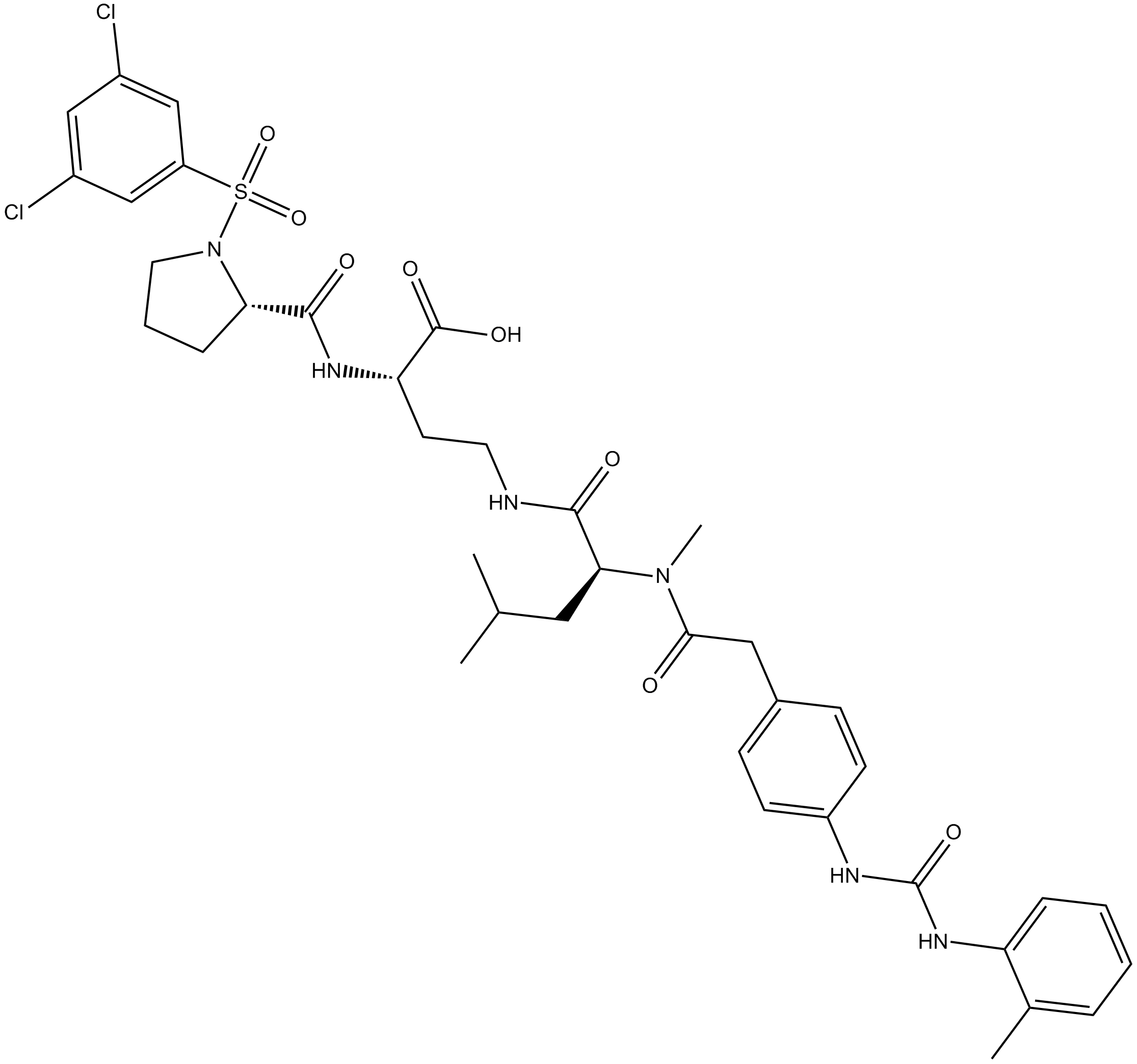
GC14208
BIO 5192
α4β1 inhibitor
-
GC60077
BIO5192 hydrate
BIO5192 hydrate is a selective and potent integrin α4β1 (VLA-4) inhibitor (Kd<10 pM).
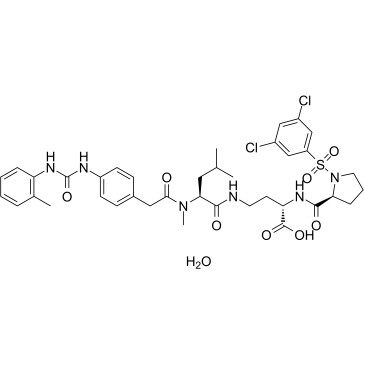
-
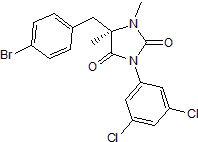
GC50078
BIRT 377
Potent negative allosteric modulator of LFA-1
-
GC68373
BLK-IN-2
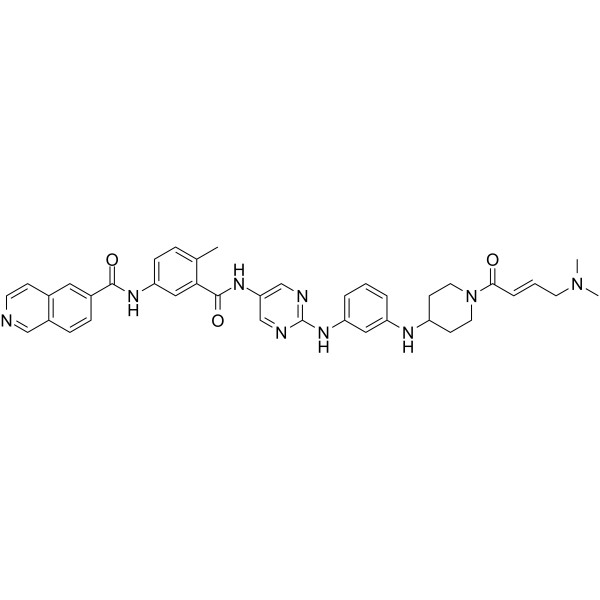
-
GC18717
BMS 986120
A selective PAR4 antagonist
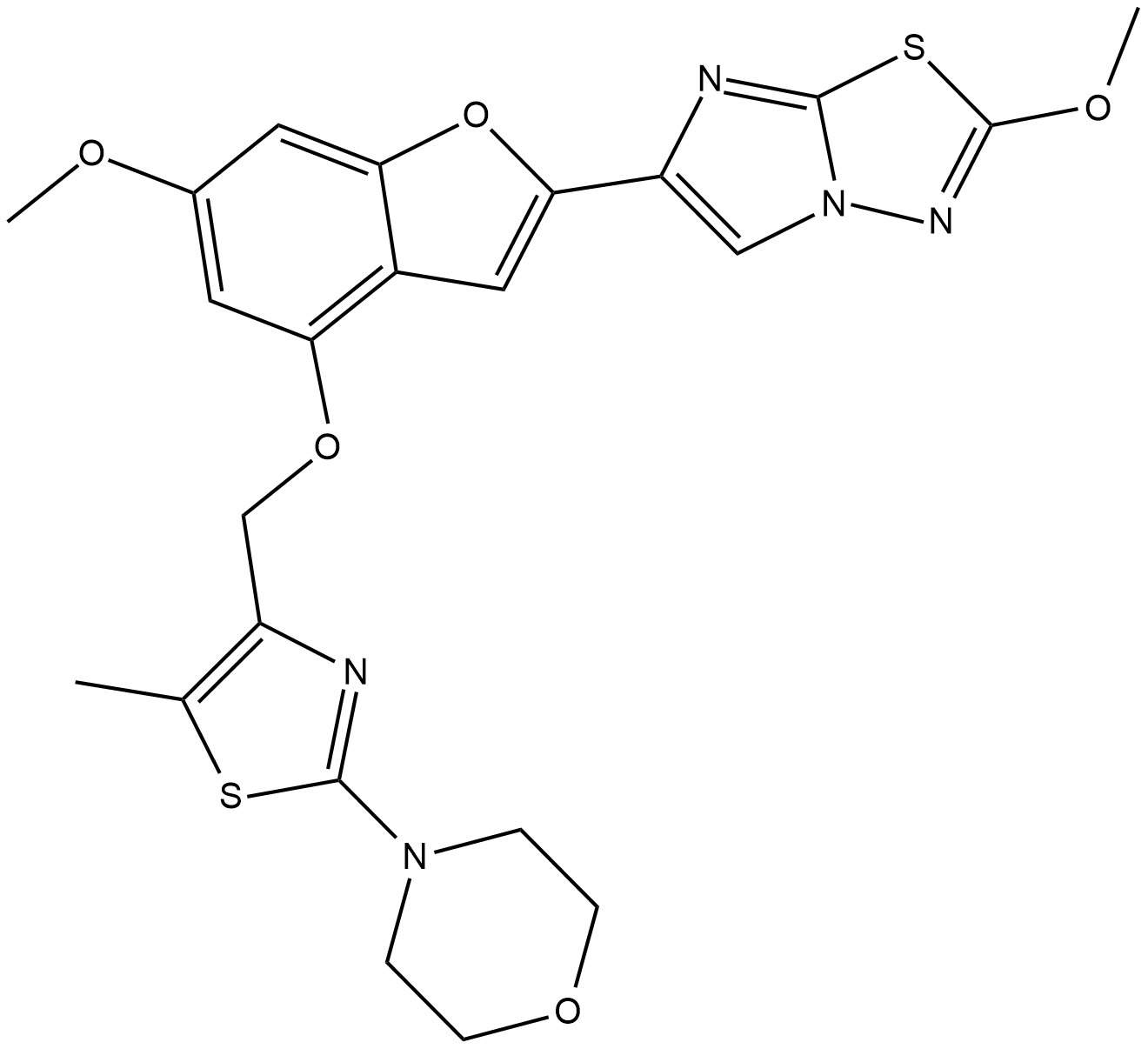
-
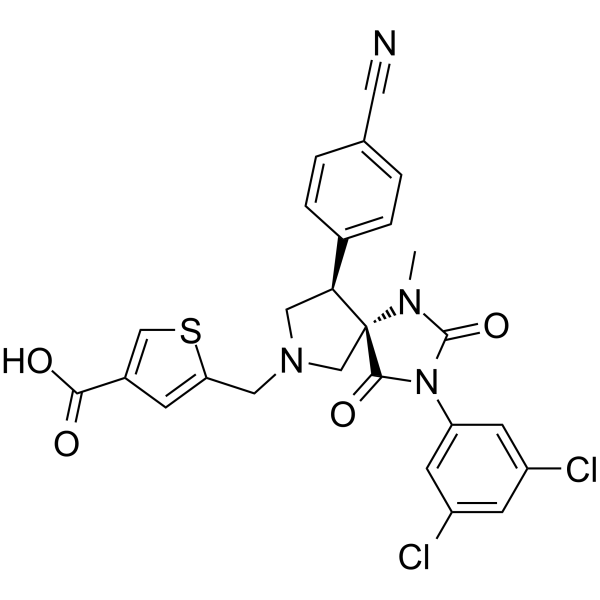
GC62872
BMS-587101
BMS-587101 is a potent and orally active antagonist of leukocyte function associated antigen-1 (LFA-1).
-
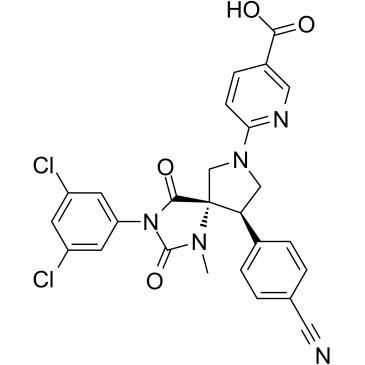
GC38893
BMS-688521
BMS-688521 is a highly potent, orally active inhibitor of the LFA-1/ICAM interaction, with an IC50 of 2.5 nM in the adhesion assay and an IC50 of 60 nM in the MLR assay.
-
GC31760
BMS-935177
BMS-935177 is a potent and selective reversible inhibitor of Bruton’s tyrosine kinase (Btk) with an IC50 of 3 nM.

-
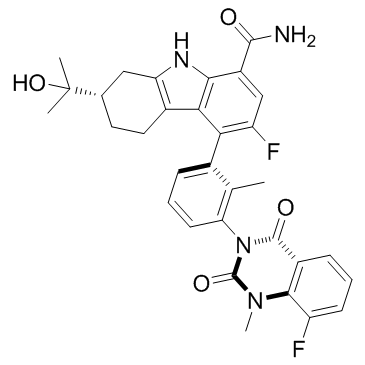
GC31713
BMS-986142
A BTK inhibitor
-
GC32844
BMS-986195
BMS-986195
BMS-986195 (BMS-986195) is a highly potent, selective covalent, irreversible inhibitor of Bruton's tyrosine kinase (BTK), with an IC50 of 0.1 nM.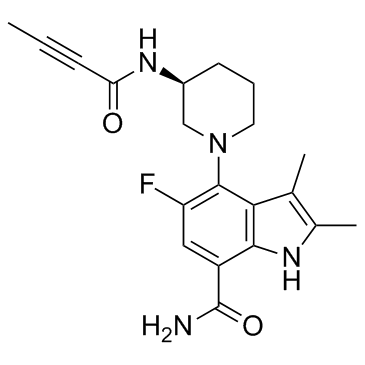
-
GC11063
BMX-IN-1
BMX Inhibitor 1
A selective BMX and BTK inhibitor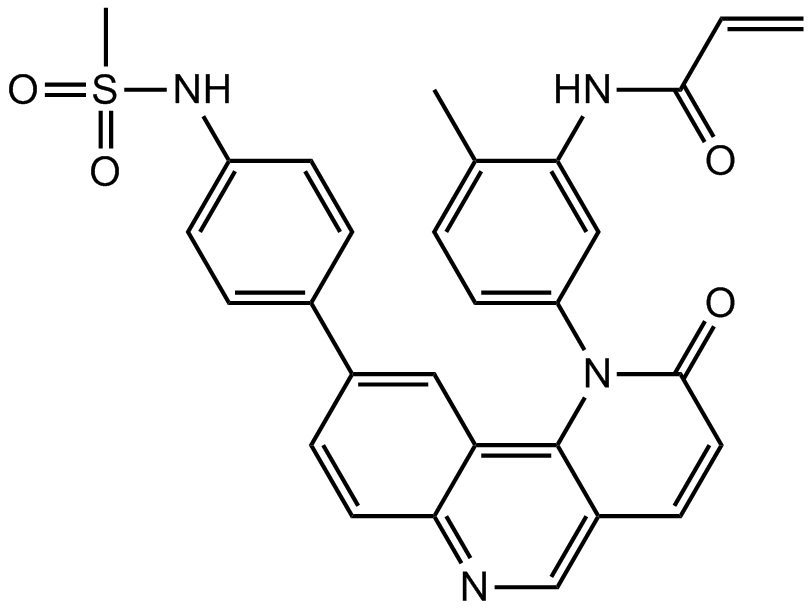
-
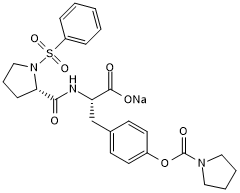
GC50325
BOP
BOP is a potent and selective dual α9β1/α4β1 integrin inhibitor with Kd values in the picomolar range.
-
GC19333
BTK IN-1
BTK-IN-1
BTK IN-1 is a potent BTK inhibitor, with an IC50 of <100 nM.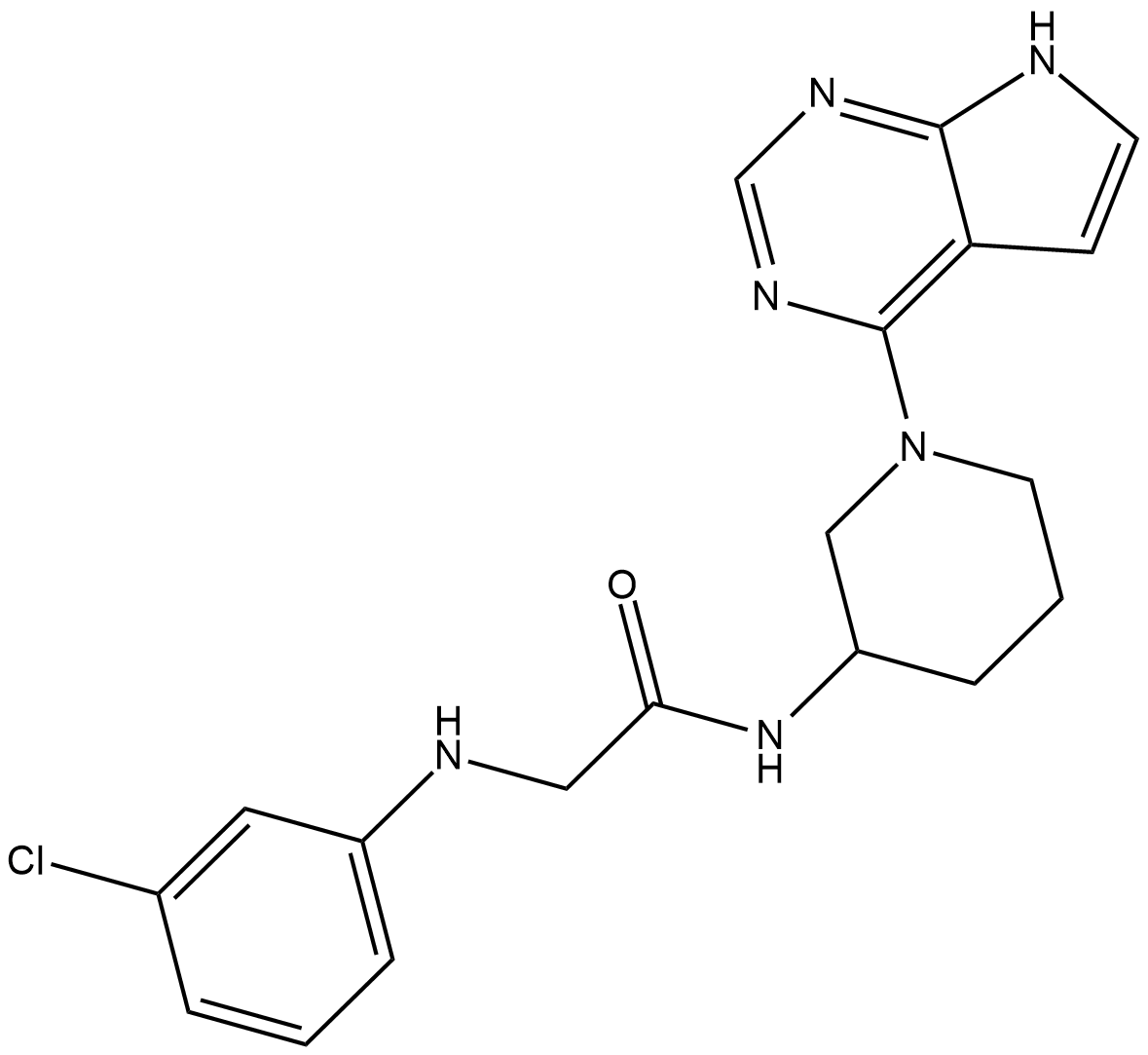
-
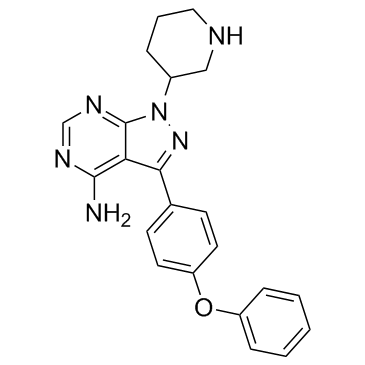
GC35561
Btk inhibitor 1
Btk inhibitor 1 is a racemate of IBT6A. IBT6A is an impurity of Ibrutinib. IBT6A can be used in synthesis of IBT6A Ibrutinib dimer and IBT6A adduct. Ibrutinib is a selective, irreversible Btk inhibitor with an IC50 of 0.5 nM.
-
GC35562
Btk inhibitor 1 hydrochloride
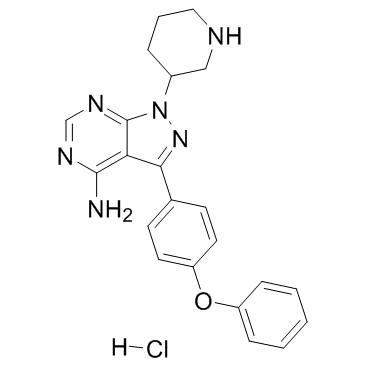
-
GC10924
Btk inhibitor 1 R enantiomer
Btk inhibitor 1 R enantiomer is an impurity of Ibrutinib. Btk inhibitor 1 R enantiomer can be used in synthesis of Btk inhibitor 1 R enantiomer Ibrutinib dimer and Btk inhibitor 1 R enantiomer adduct. Ibrutinib is a selective, irreversible Btk inhibitor with an IC50 of 0.5 nM.
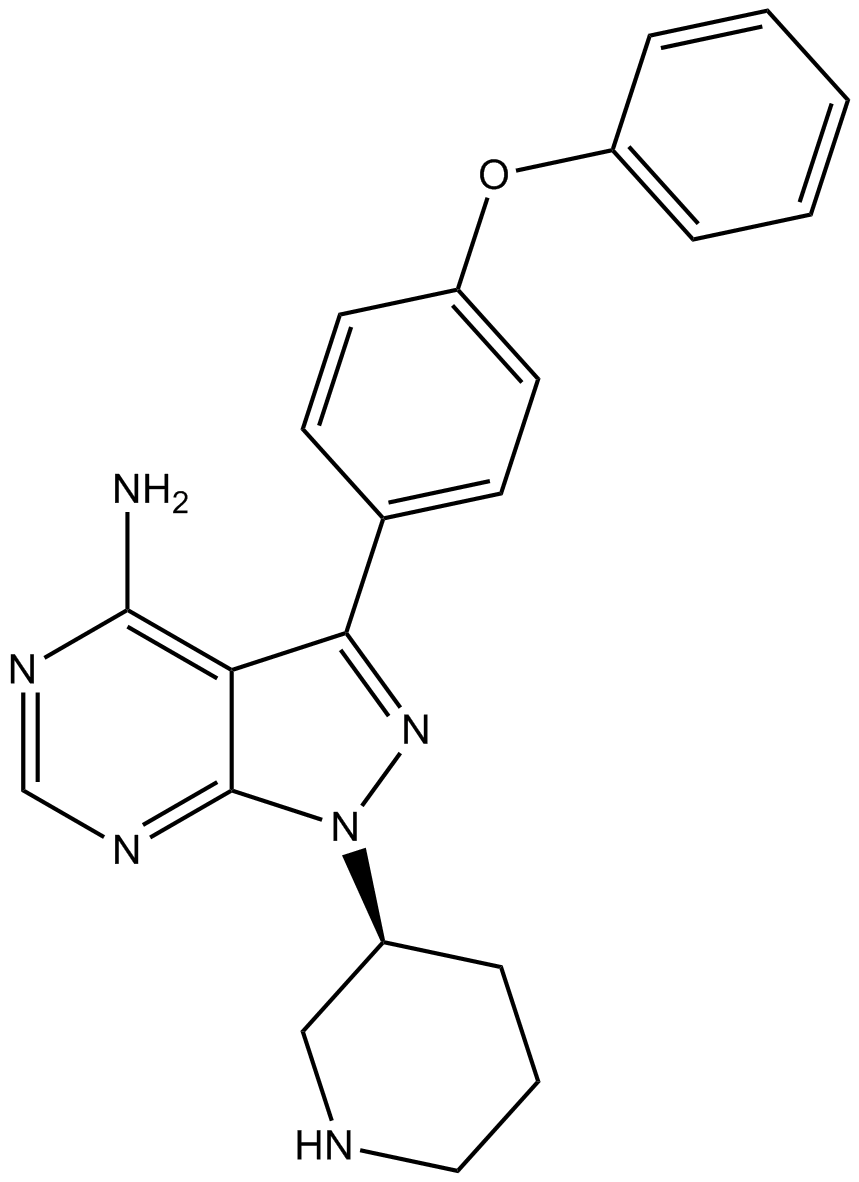
-
GC35563
Btk inhibitor 1 R enantiomer hydrochloride
Btk inhibitor 1 R enantiomer hydrochloride is an impurity of Ibrutinib. IBT6A can be used in synthesis of IBT6A Ibrutinib dimer and IBT6A adduct. Ibrutinib is a selective, irreversible Btk inhibitor with an IC50 of 0.5 nM.
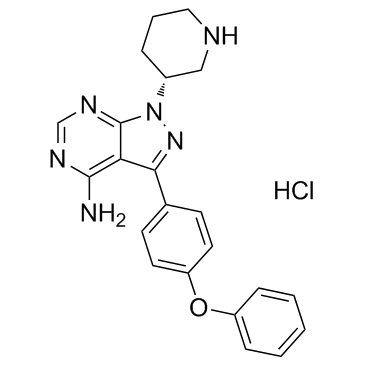
-
GC67940
BTK inhibitor 10

-
GC62498
BTK inhibitor 17
BTK inhibitor 17 is a potent and orally active irreversible BTK inhibitor with an IC50 of 2.1 nM.
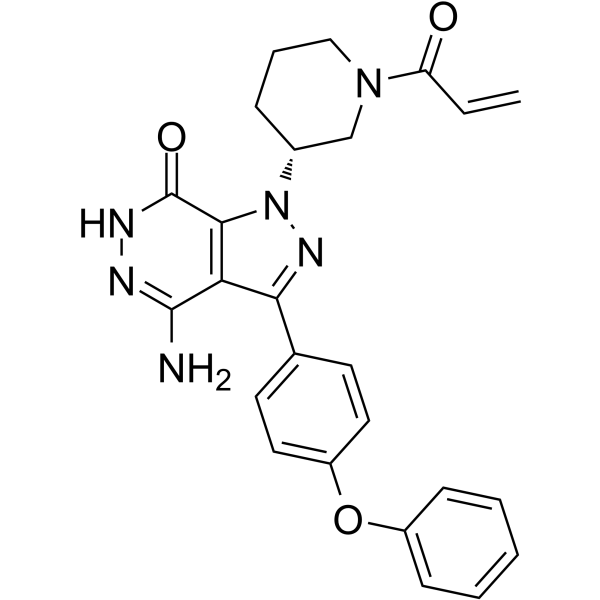
-
GC64360
BTK inhibitor 18
BTK inhibitor 18 is a potent, selective,orally active and covalent Btk inhibitor with a IC50 of 142 nM.
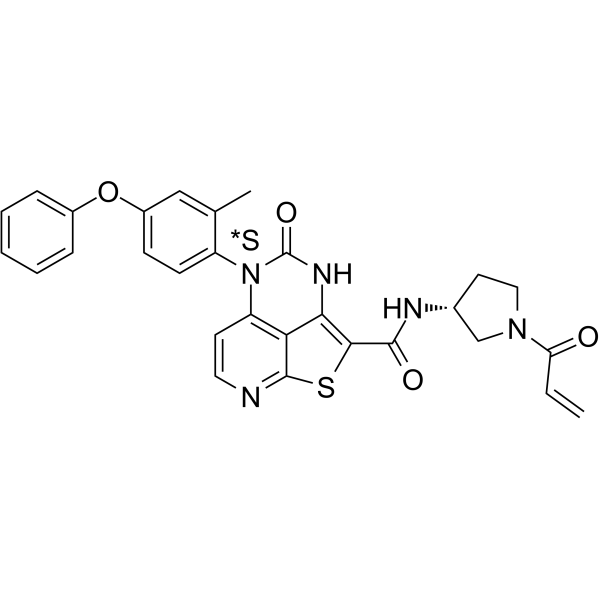
-
GC32007
Btk inhibitor 2
BGB-3111 analog
Btk inhibitor 2 (BGB-3111 analog) is a Bruton's tyrosine kinase (BTK) inhibitor extracted from patent US 20170224688 A1.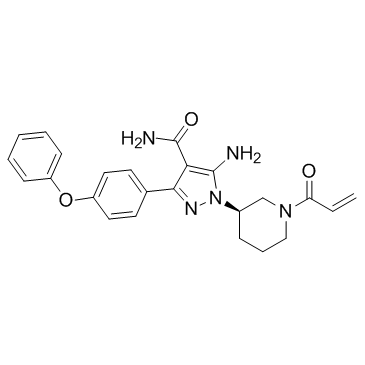
-
GC50075
BTT 3033
Selective inhibitor of integrin α2β1
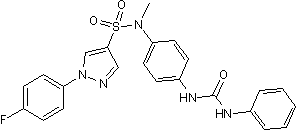
-
GC38128
c(phg-isoD-G-R-(NMe)k) TFA
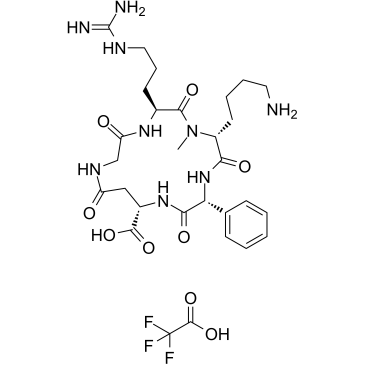
-
GC65455
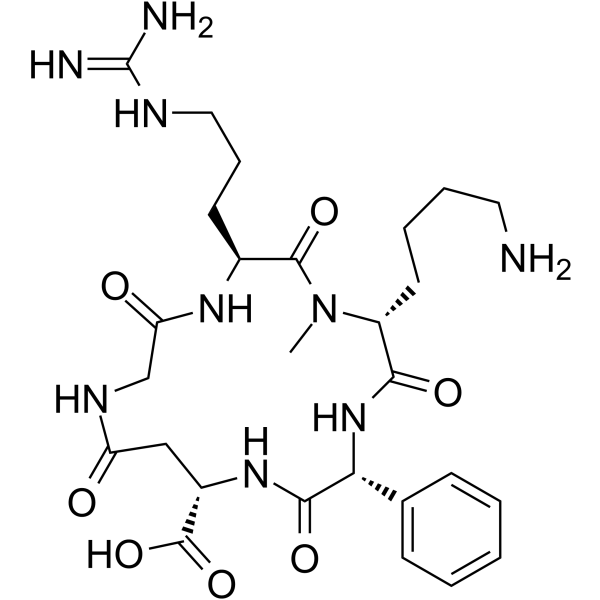
c(phg-isoDGR-(NMe)k)
c(phg-isoDGR-(NMe)k) is a selective and potent α5β1-integrin ligand with an IC50 of 2.9 nM.
-
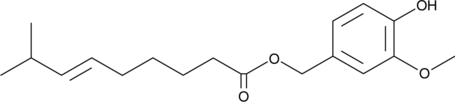
GC49433
Capsiate
A capsaicin analog with diverse biological activities
-
GC32042
Carotegrast
HCA2969
Carotegrast is an orally available α4 integrin receptor inhibitor with anti-inflammatories activities.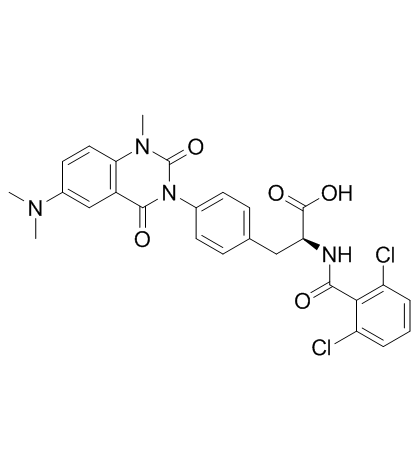
-
GC62143
Carotegrast methyl
AJM300
Carotegrast methyl (AJM300) is an orally active and selective α4 integrin antagonist.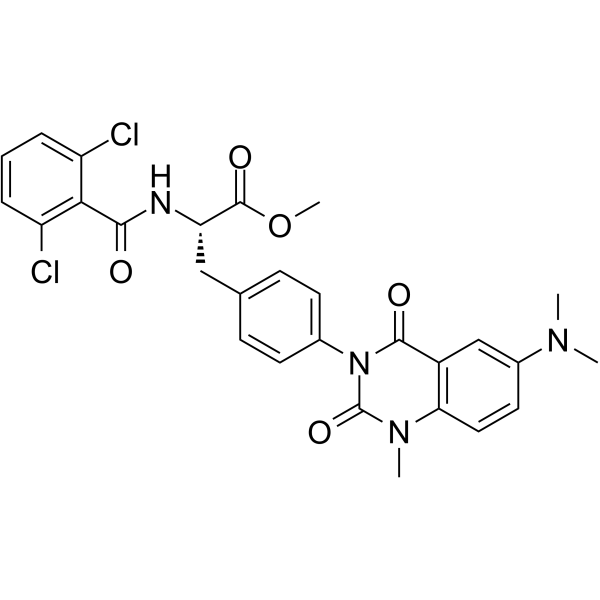
-
GC43198
CAY10717
CAY10717 is a multi-targeted kinase inhibitor that exhibits greater than 40% inhibition of 34 of 104 kinases in an enzymatic assay at a concentration of 100 nM.
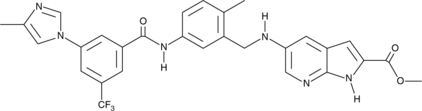
-
GC68856
Certepetide
CEND-1; iRGD
Certepetide (CEND-1) is a dual-function cyclic peptide (also known as iRGD). Certepetide is a penetrable tumor peptide that interacts with alpha-v integrins through its RGD motif and activates neuropilin-1 (NRP-1), thereby transforming the solid tumor microenvironment into temporary active molecular channels. Certepetide can accumulate in tumors and can be used for research on pancreatic cancer and other solid tumors.

-
GC33064
CG-806 (Luxeptinib)
CG-806
CG-806 (Luxeptinib) (CG-806) is an orally active, reversible, first-in-class, non-covalent and potent pan-FLT3/pan-BTK inhibitor. CG-806 (Luxeptinib) induces cell cycle arrest, apoptosis or autophagy in acute myeloid leukemia cells.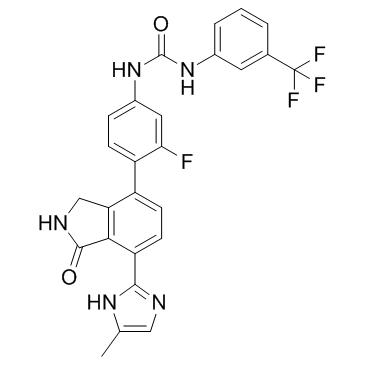
-
GC13365
CGI-1746
A potent, selective BTK inhibitor
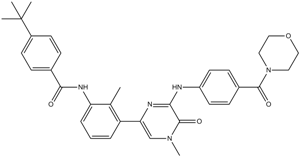
-
GC35683
CHMFL-BTK-01
CHMFL-BTK-01 (compound 9) is a highly selective irreversible BTK inhibitor, with an IC50 of 7 nM. CHMFL-BTK-01 (compound 9) potently inhibited BTK Y223 auto-phosphorylation.
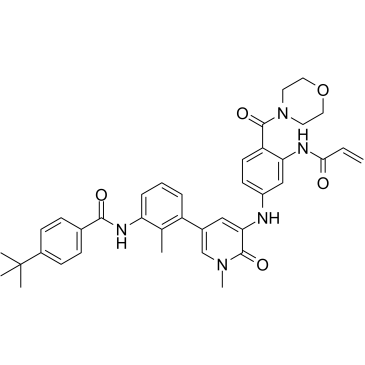
-
GC35684
CHMFL-EGFR-202
CHMFL-EGFR-202 is a potent, irreversible inhibitor of epidermal growth factor receptor (EGFR) mutant kinase, with IC50s of 5.3 nM and 8.3 nM for drug-resistant mutant EGFR T790M and WT EGFR kinases, respectively. CHMFL-EGFR-202 exhibits ?10-fold selectivity for EGFR L858R/T790M against the EGFR wild-type in cells. CHMFL-EGFR-202 adopts a covalent “DFG-in-C-helix-out” inactive binding conformation with EGFR, with strong antiproliferative effects against EGFR mutant-driven nonsmall-cell lung cancer (NSCLC) cell lines.
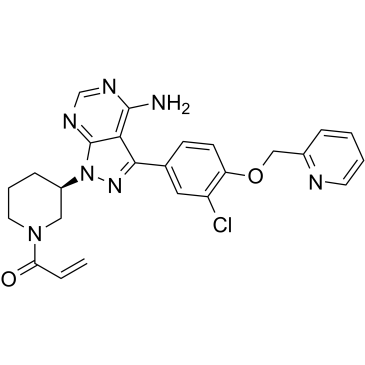
-
GC13559
Cilengitide
EMD 121974
Integrin inhibitor for αvβ3 and αvβ5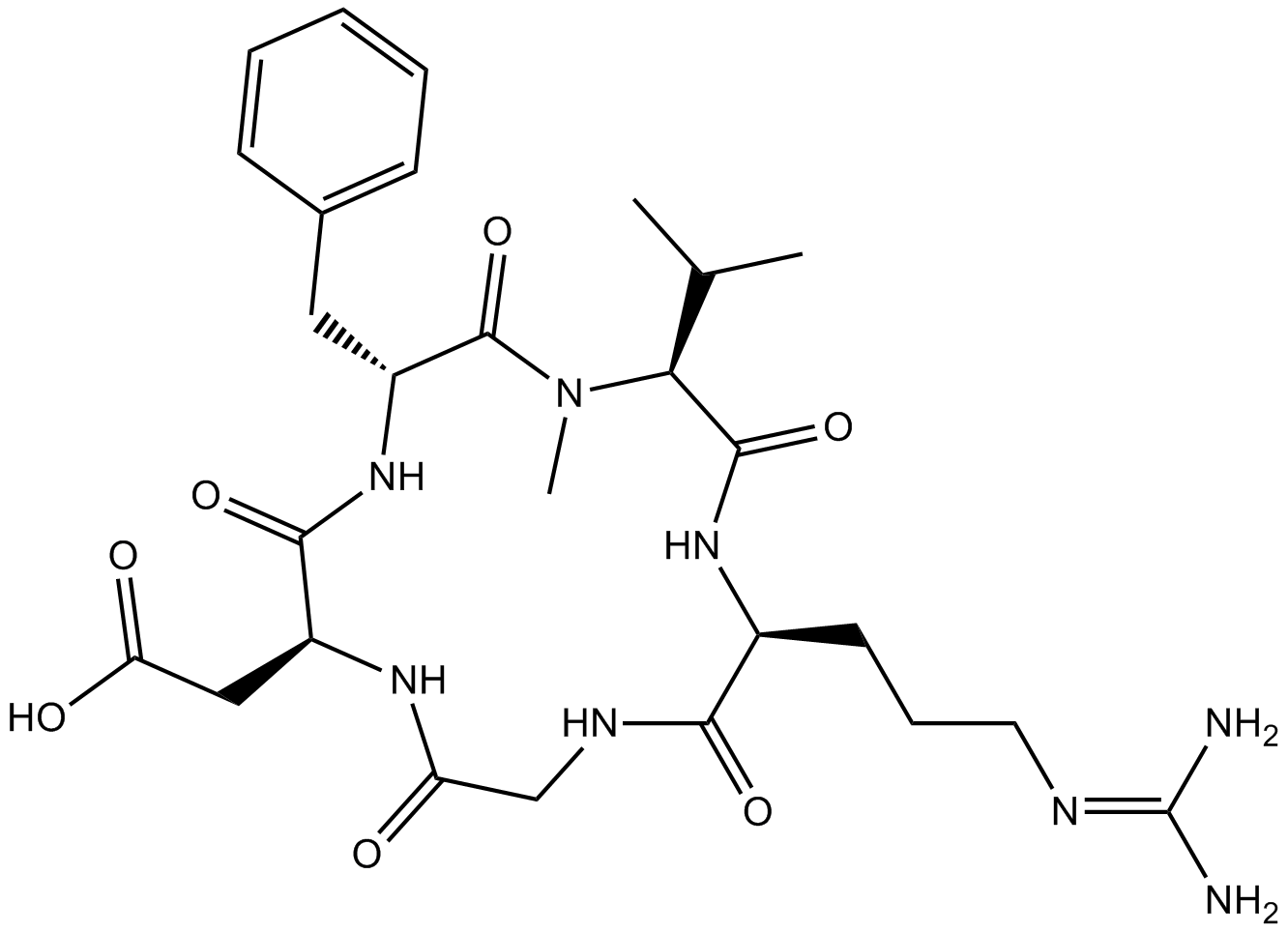
-
GC61520
Cilengitide TFA
EMD 121974 TFA
Cilengitide is a potent and selective integrin inhibitor for αvβ3 and αvβ5 receptor, with IC50 values of 4 nM and 79 nM, respectively.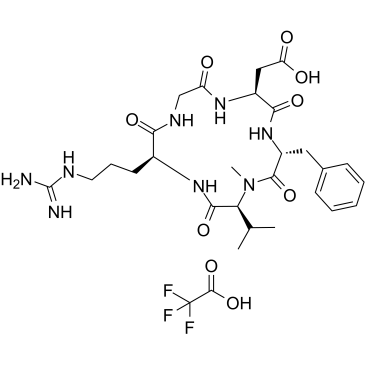
-
GC45880
Cimetidine-d3
An internal standard for the quantification of cimetidine
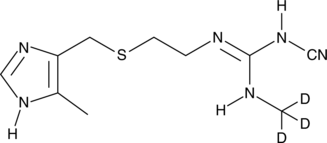
-
GC13439
CNX-774
BTK inhibitor, orally active, irreversible and selective
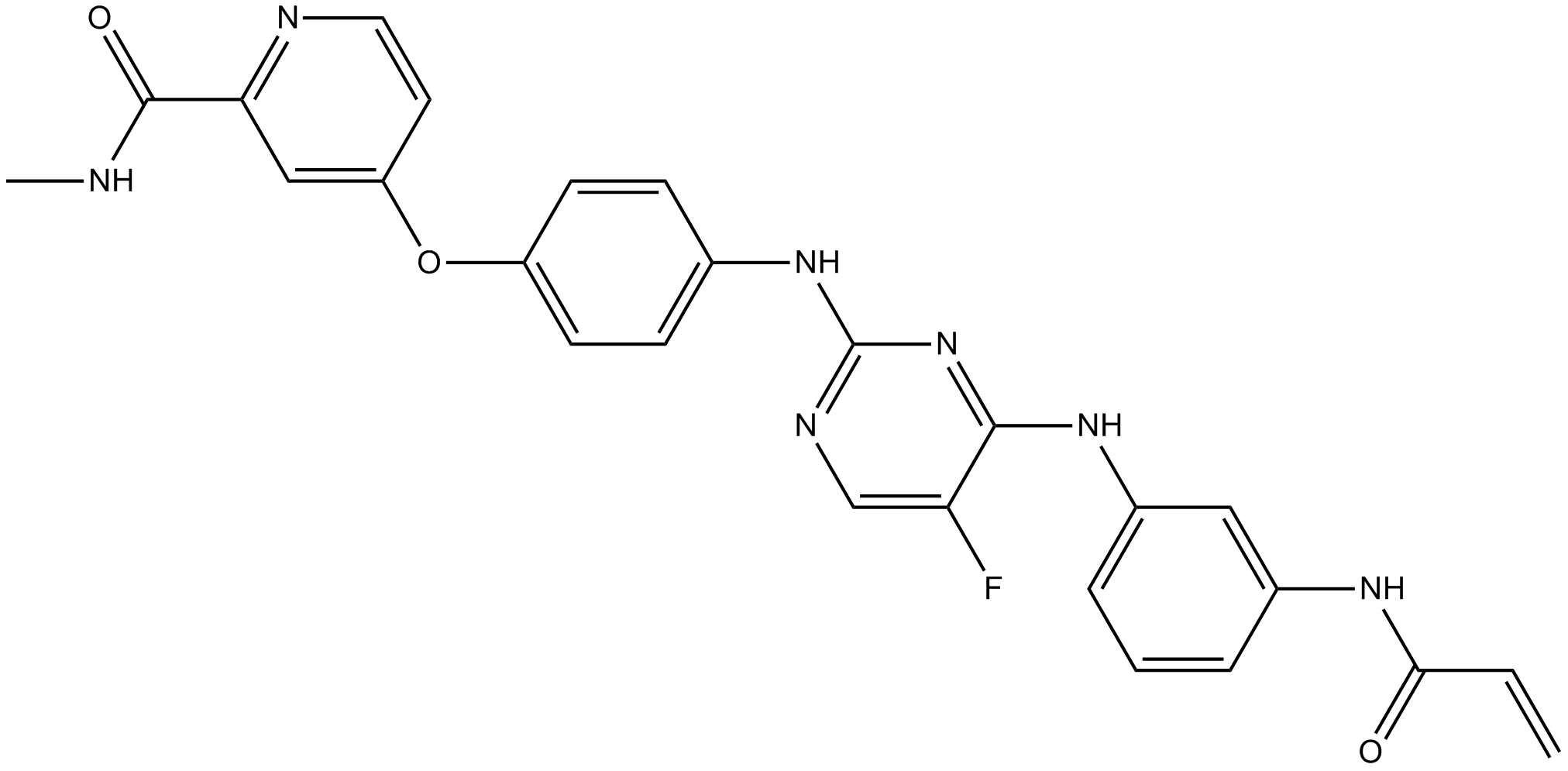
-
GC17631
Combretastatin A4
CA4, Combretastatin A4, CRC 8709
tubulin polymerization inhibitor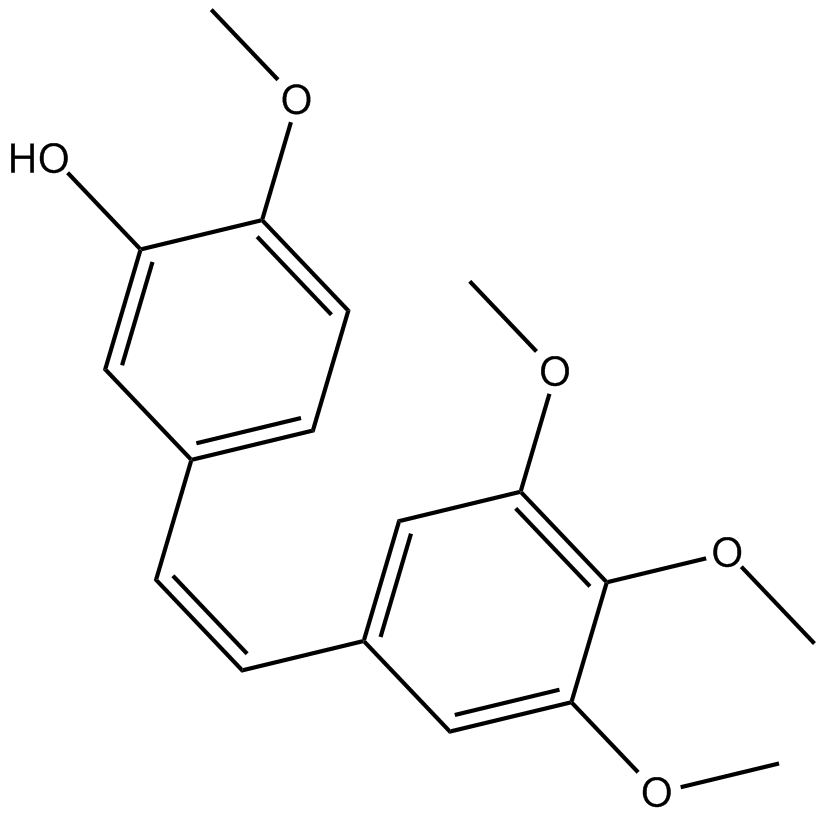
-
GN10535
Cucurbitacin B
Cuc B, NSC 49451, NSC 144154
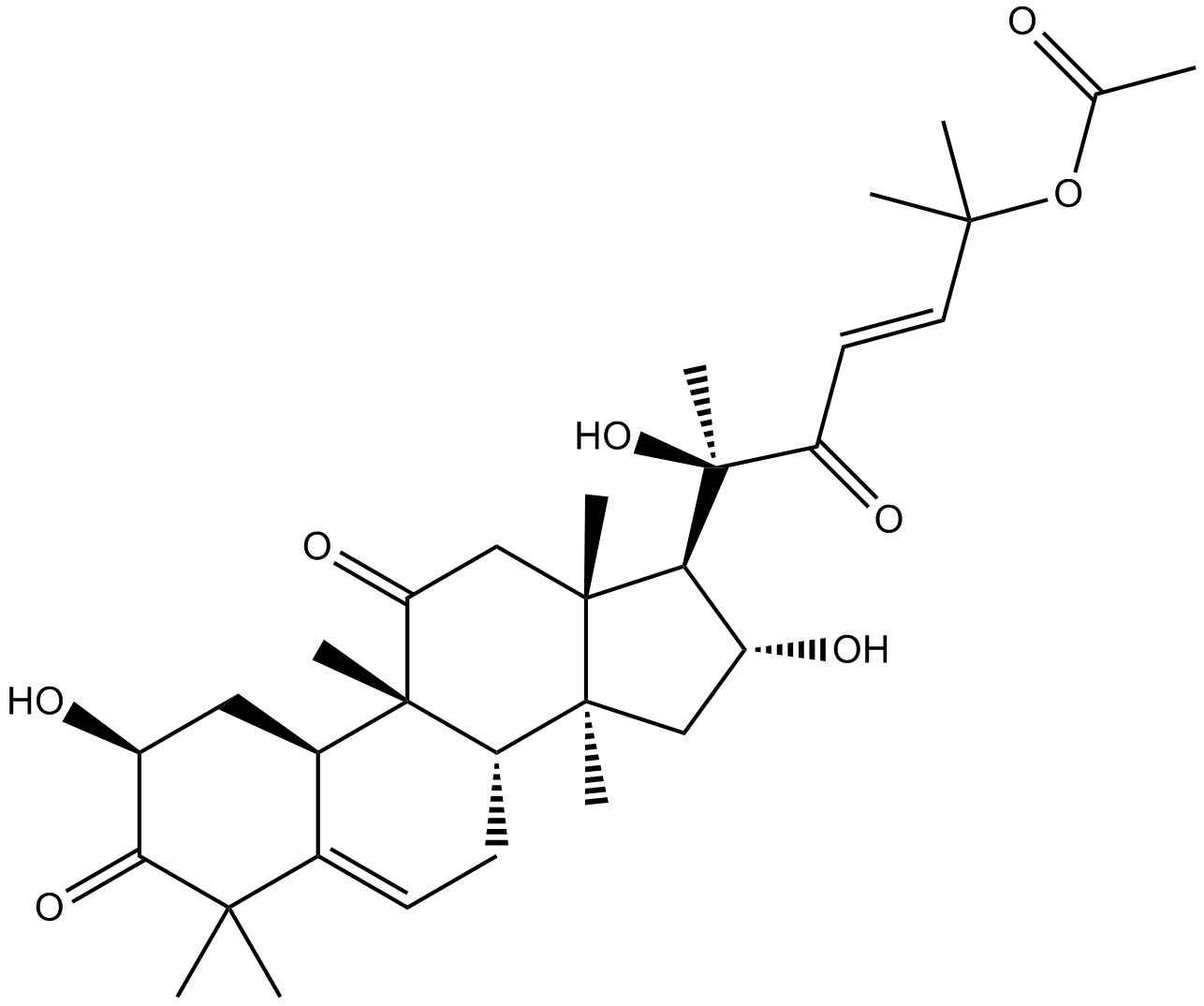
-
GC13050
CWHM-12
An inhibitor of αV integrins
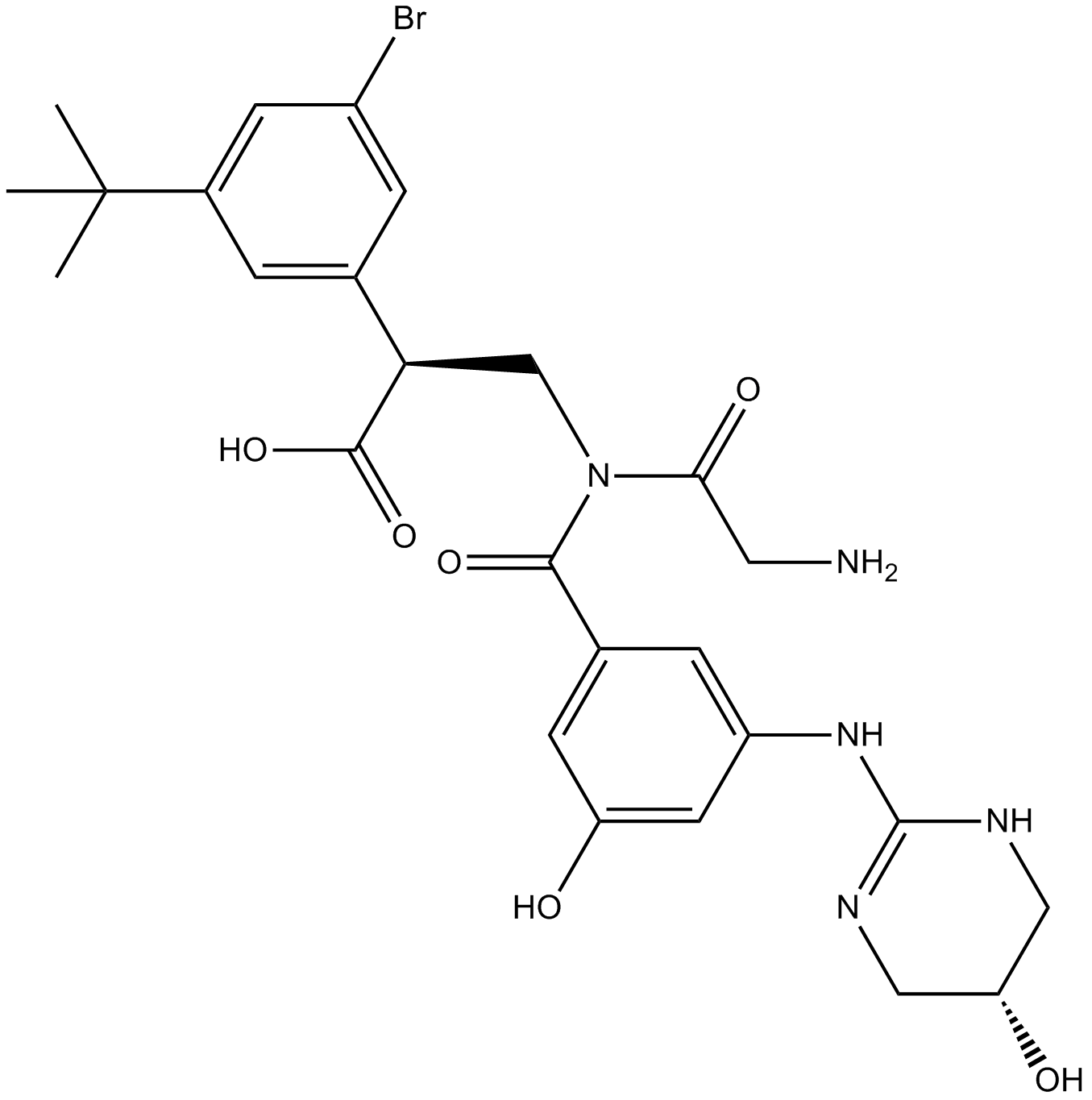
-
GC17610
Cyclo (-RGDfK)
Cyclo(-Arg-Gly-Asp-D-Phe-Lys)
An inhibitor of αvβ3 integrin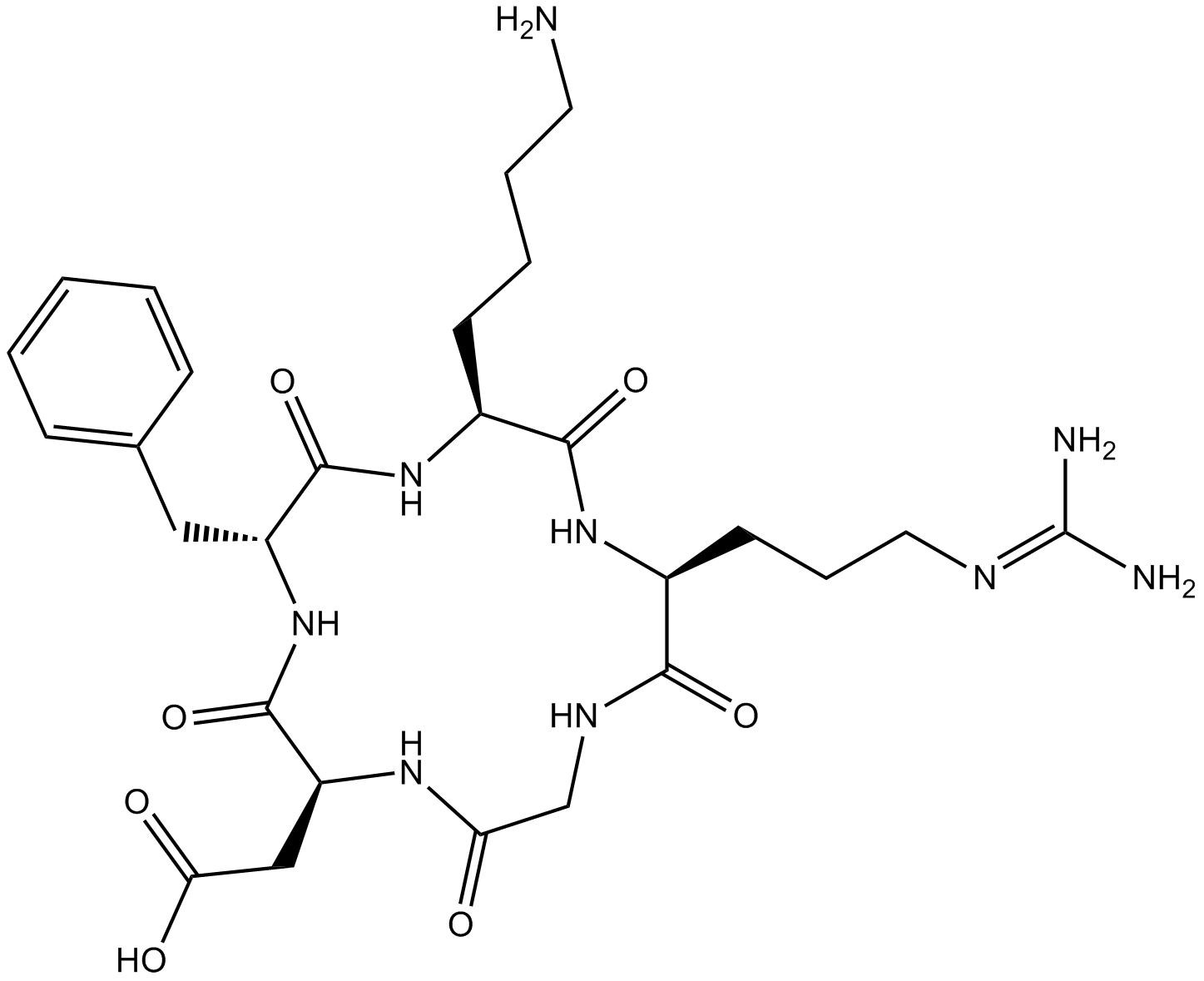
-
GA21306
Cyclo(-Arg-Gly-Asp-D-Tyr-Lys)
c(RGDyK) has been radioiodinated or modified with chelators for use as radiopharmaceutical.

-
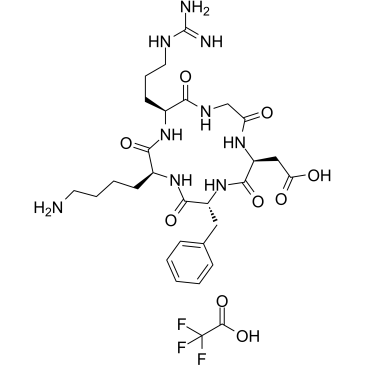
GC60117
Cyclo(-RGDfK) TFA
Cyclo(-RGDfK) TFA is a potent and selective inhibitor of the αvβ3 integrin, with an IC50 of 0.94 nM. Cyclo(-RGDfK) TFA potently targets tumor microvasculature and cancer cells through the specific binding to the αvβ3 integrin on the cell surface.
-
GC68921
Cyclo(Arg-Gly-Asp-D-Phe-Cys) TFA
Cyclo(RGDfC) TFA
Cyclo(Arg-Gly-Asp-D-Phe-Cys) (Cyclo RGDfC) TFA is a cyclic RGD peptide with high affinity for αvβ3 integrin, which can disrupt cell adhesion. Cyclo(Arg-Gly-Asp-D-Phe-Cys) TFA inhibits the expression of pluripotency genes in embryonic stem cells (ESC), and suppresses the tumorigenic potential of mESCs in vivo. Cyclo(Arg-Gly-Asp-D-Phe-Cys) TFA can be used for tumor-related research.

-
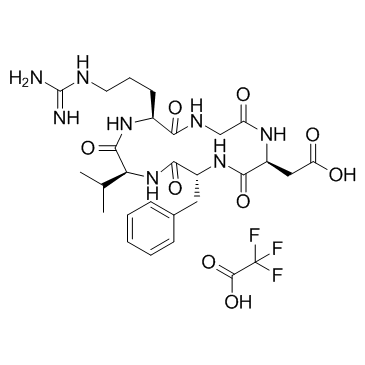
GC34141
Cyclo(Arg-Gly-Asp-D-Phe-Val) TFA
Cyclo(Arg-Gly-Asp-D-Phe-Val) (TFA) is an inhibitor of integrin αvβ3, with antitumor activity.
-
GC30111
Cyclo(RADfK)
Cyclo(RADfK) is a selective α(v)β(3) integrin ligand that has been extensively used for research, therapy, and diagnosis of neoangiogenesis.