ADC Cytotoxin
ADC Cytotoxin are a large collection of toxins for antibody-drug conjugate (ADC) development projects. The toxins that can be used as payloads should be soluble, amenable to conjugation, and stable. There are myriads of cellular toxins in natural or synthesized, but only a few have been found adaptable for an ADC concept.
The toxins targeting tubulin filaments include Maytansinoids, Auristatins, Taxol derivatives, etc. Amatoxins are a class of transcription-inhibiting agents. They bind to RNA polymerase II, leading to cell apoptosis. Protein toxins function in the similar MOA (mechanism of action) with cellular toxins, they could inhibit protein synthesis and induce cell death. While enzyme-based ADCs alter the microenvironment of disease tissues to disturb their functions.
Targets for ADC Cytotoxin
Products for ADC Cytotoxin
- Cat.No. Product Name Information
-
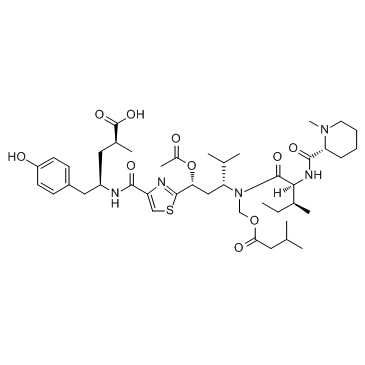
GC34955
(+)-CBI-CDPI1
(+)-CBI-CDPI1 is an enhanced functional analog of CC-1065. (+)-CBI-CDPI1 is a DNA alkylating agent. (+)-CBI-CDPI1 is an antibody drug conjugates (ADCs) toxin.
-
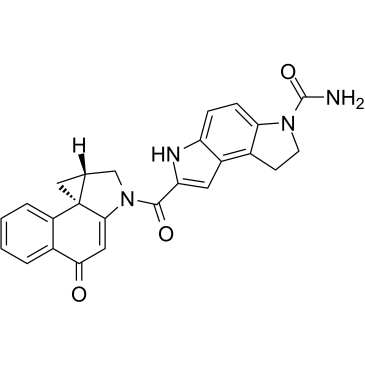
GC34956
(+)-CBI-CDPI2
(+)-CBI-CDPI2 is an enhanced functional analog of CC-1065. (+)-CBI-CDPI1 is a DNA alkylating agent. (+)-CBI-CDPI2 is an antibody drug conjugates (ADCs) toxin.
-
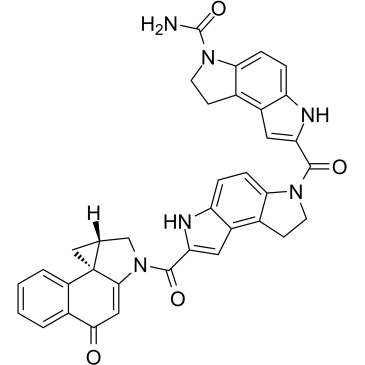
GC60421
(S)-Seco-Duocarmycin SA
(S)-Seco-Duocarmycin SA is a DNA alkylator, cytotoxic to cancer cells, and acts as a ADC cytotoxin for antibody-drug conjugates.
-
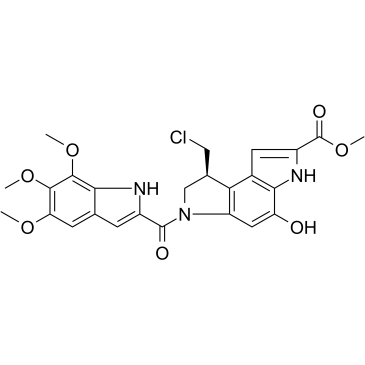
GC35044
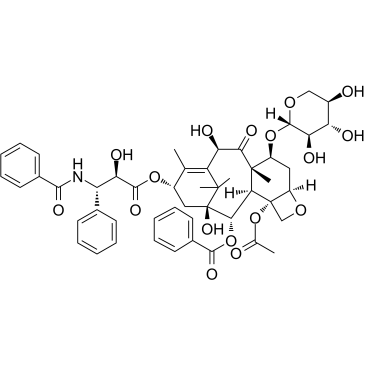
10-Deacetyl-7-xylosyl paclitaxel
10-Deacetyl-7-xylosyl paclitaxel is a Paclitaxel (a microtubule stabilizing agent; enhances tubulin polymerization) derivative with improved pharmacological features.
-
GC14858
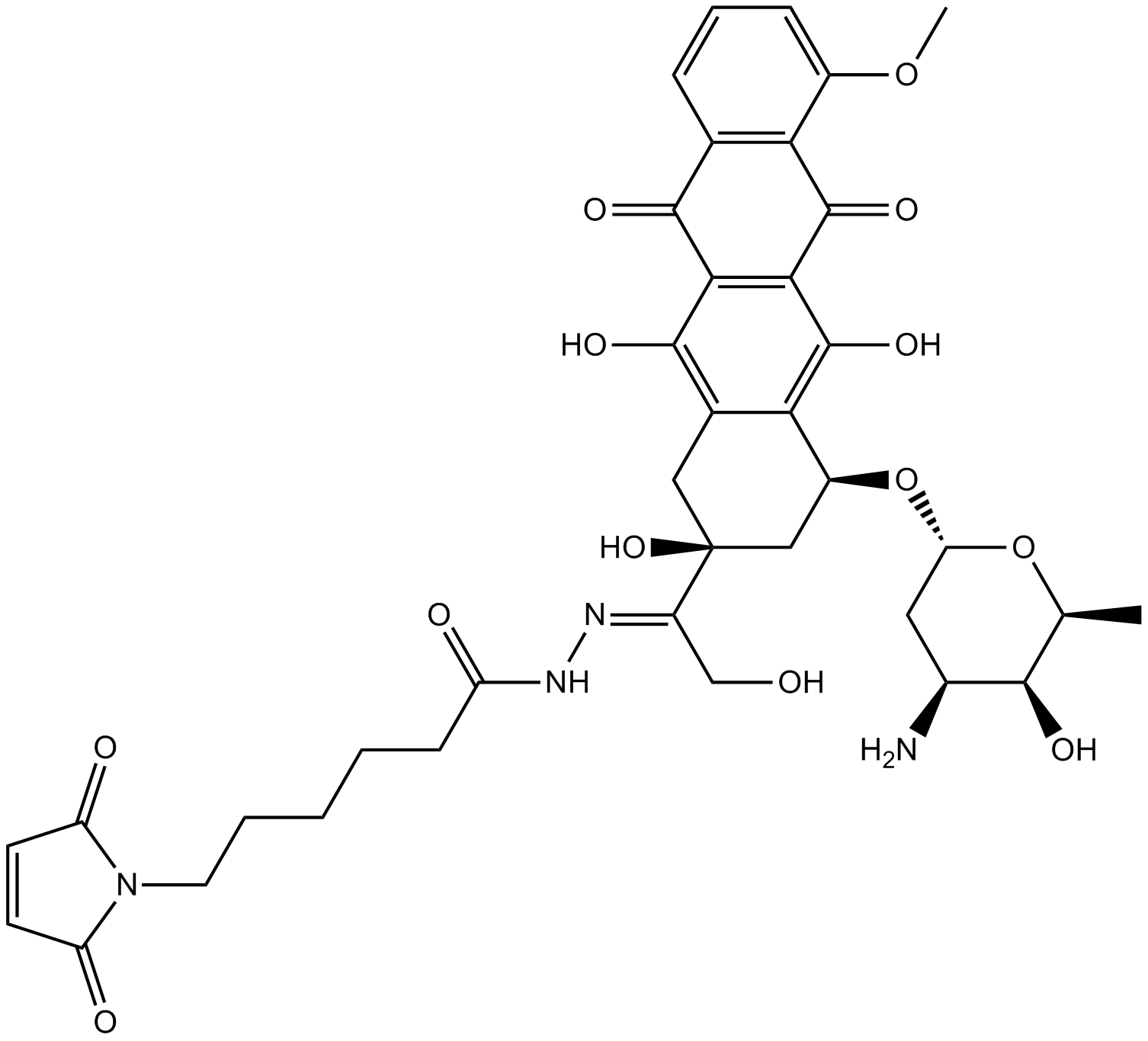
Aldoxorubicin
An albumin-binding prodrug of doxorubicin
-
GC61563
Aminohexylgeldanamycin
Aminohexylgeldanamycin (AHGDM), a Geldanamycin derivative, is a potent HSP90 inhibitor. Aminohexylgeldanamycin shows antiangiogenic and antitumor activities.
-
GC11947
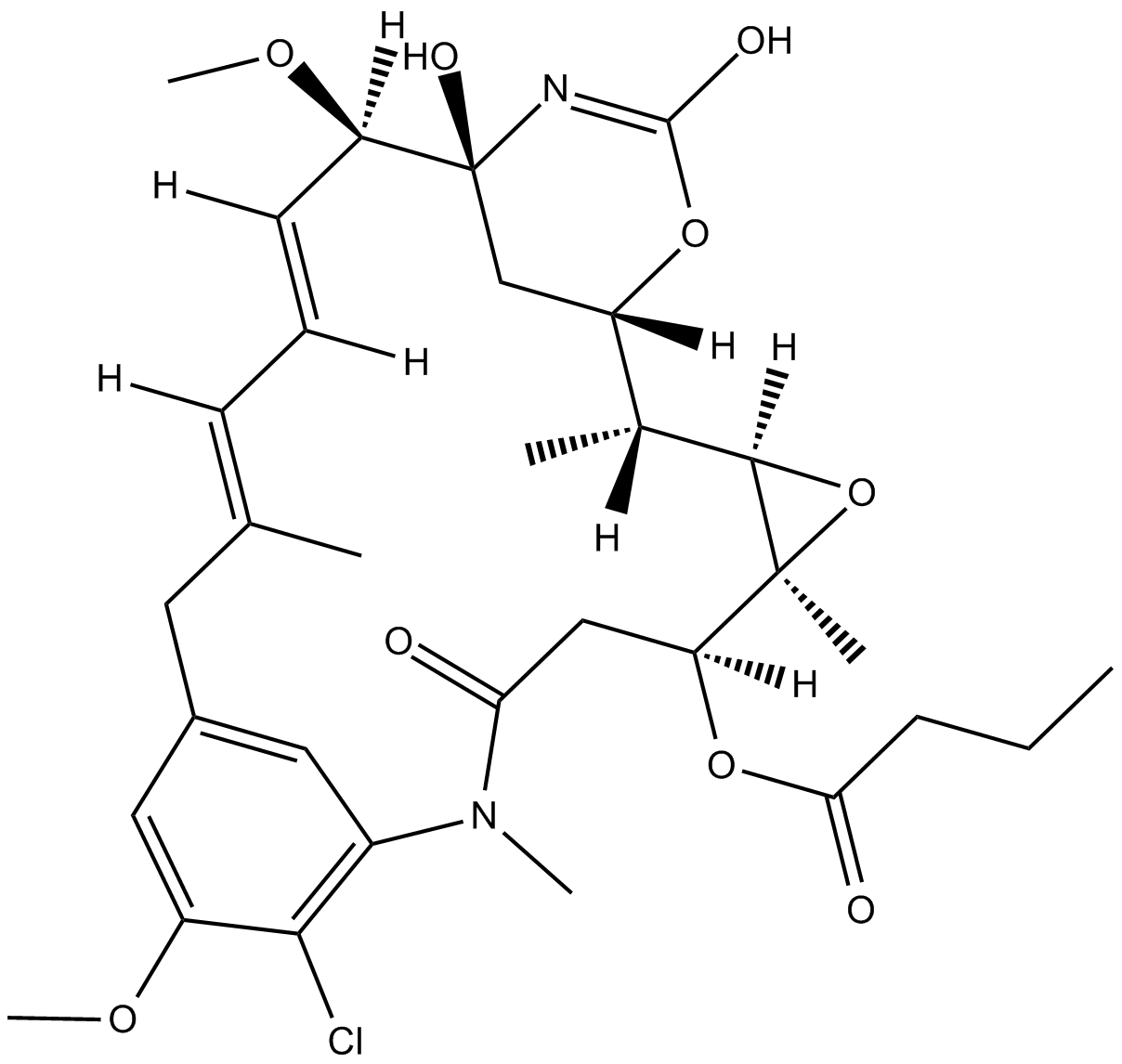
Ansamitocin P-3
A microtubule depolymerizing agent
-
GC35429
Auristatin E
Auristatin E is a cytotoxic tubulin modifier with potent and selective antitumor activity; MMAE analog and cytotoxin in Antibody-drug conjugates. Auristatin E inhibits cell division by blocking the polymerisation of tubulin.
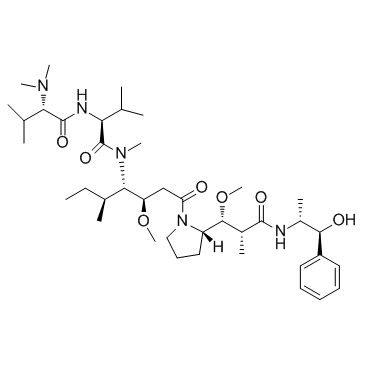
-
GC35430
Auristatin F
Auristatin F is a potent cytotoxin. Auristatin F, a potent microtubule inhibitor and vascular damaging agent (VDA), can be used in antibody-drug conjugates (ADC).
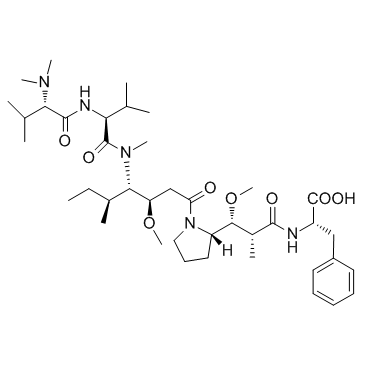
-
GC35574
C-11
C-11 is a tubulin inhibitor and acts as an ADC cytotoxin, displays cytotoxicity for carcinoma cell lines.
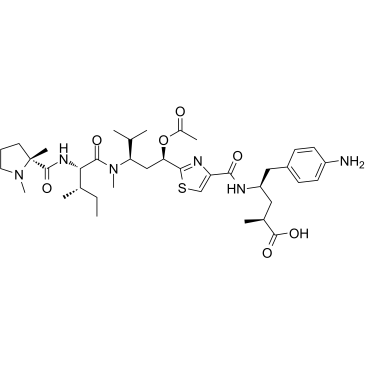
-
GC19086
Calicheamicin
Calicheamicin is a cytotoxic agent that causes double-strand DNA breaks.
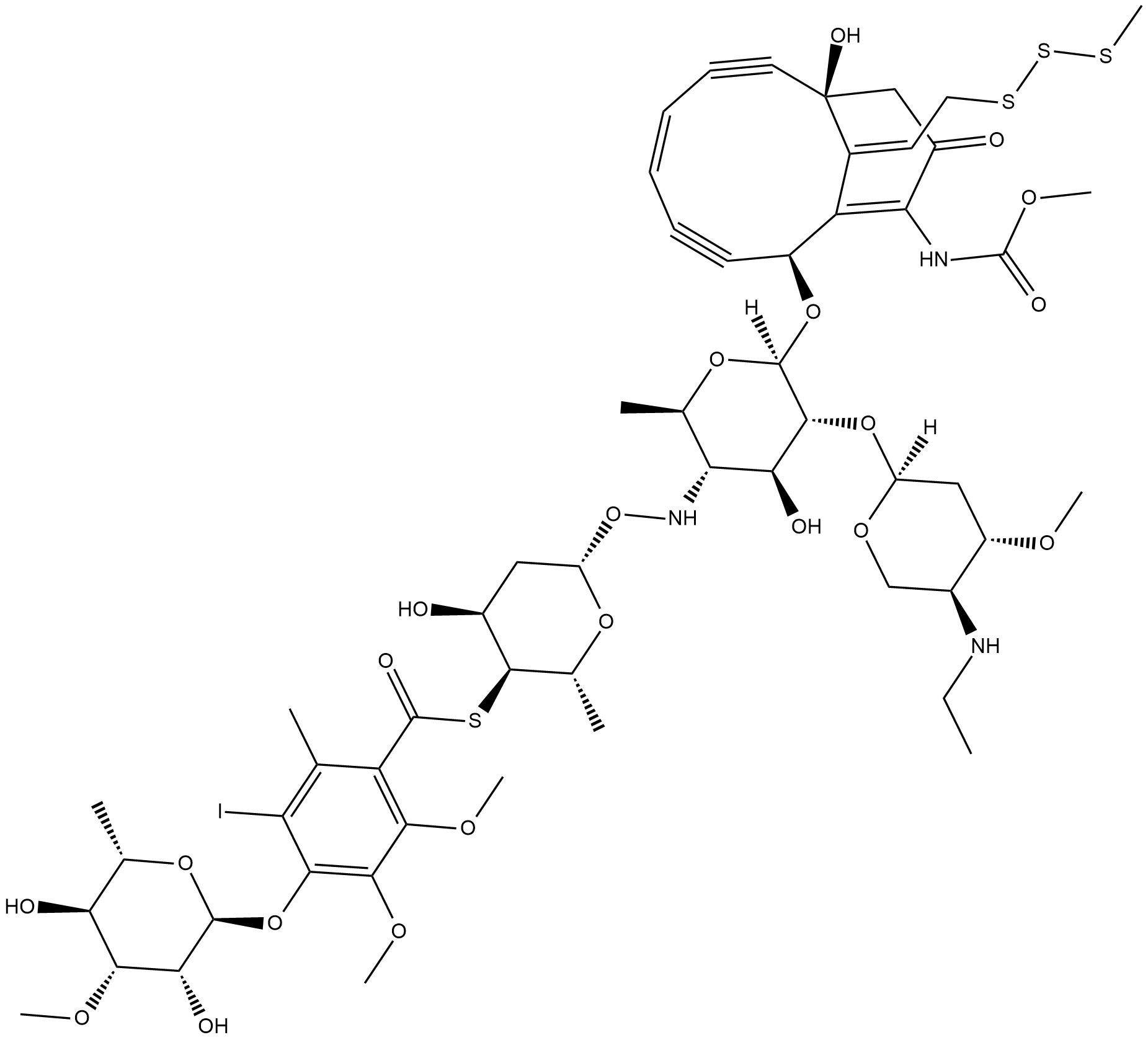
-
GC38016
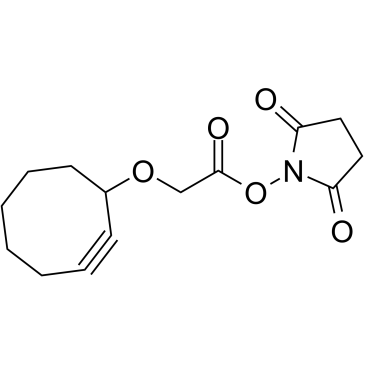
Cyclooctyne-O-NHS ester
Cyclooctyne-O-NHS ester is a cleavable ADC linker used in the synthesis of antibody-drug conjugates (ADCs).
-
GC38631
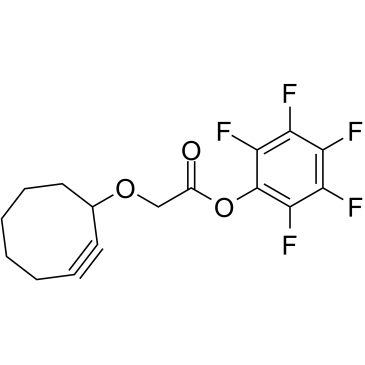
Cyclooctyne-O-PFP ester
Cyclooctyne-O-PFP ester is a cleavable ADC linker used in the synthesis of antibody-drug conjugates (ADCs).
-
GC35797
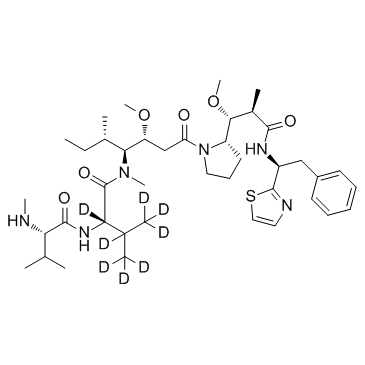
D8-MMAD
-
GC35798
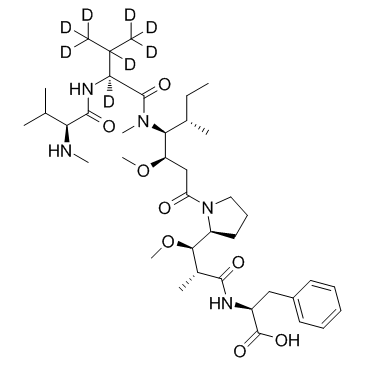
D8-MMAF
D8-MMAF hydrochloride is a deuterated form of MMAF hydrochloride. MMAF Hydrochloride, a potent tubulin polymerization inhibitor, is used as a antitumor agent and a cytotoxic component of antibody-drug conjugates (ADCs).
-
GC35799
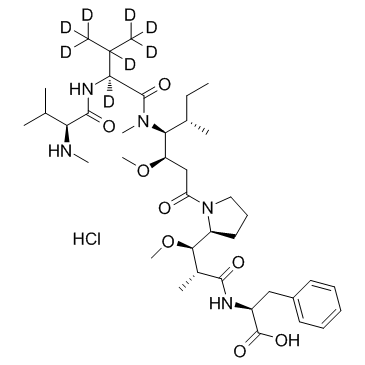
D8-MMAF hydrochloride
-
GC11175
Daun02
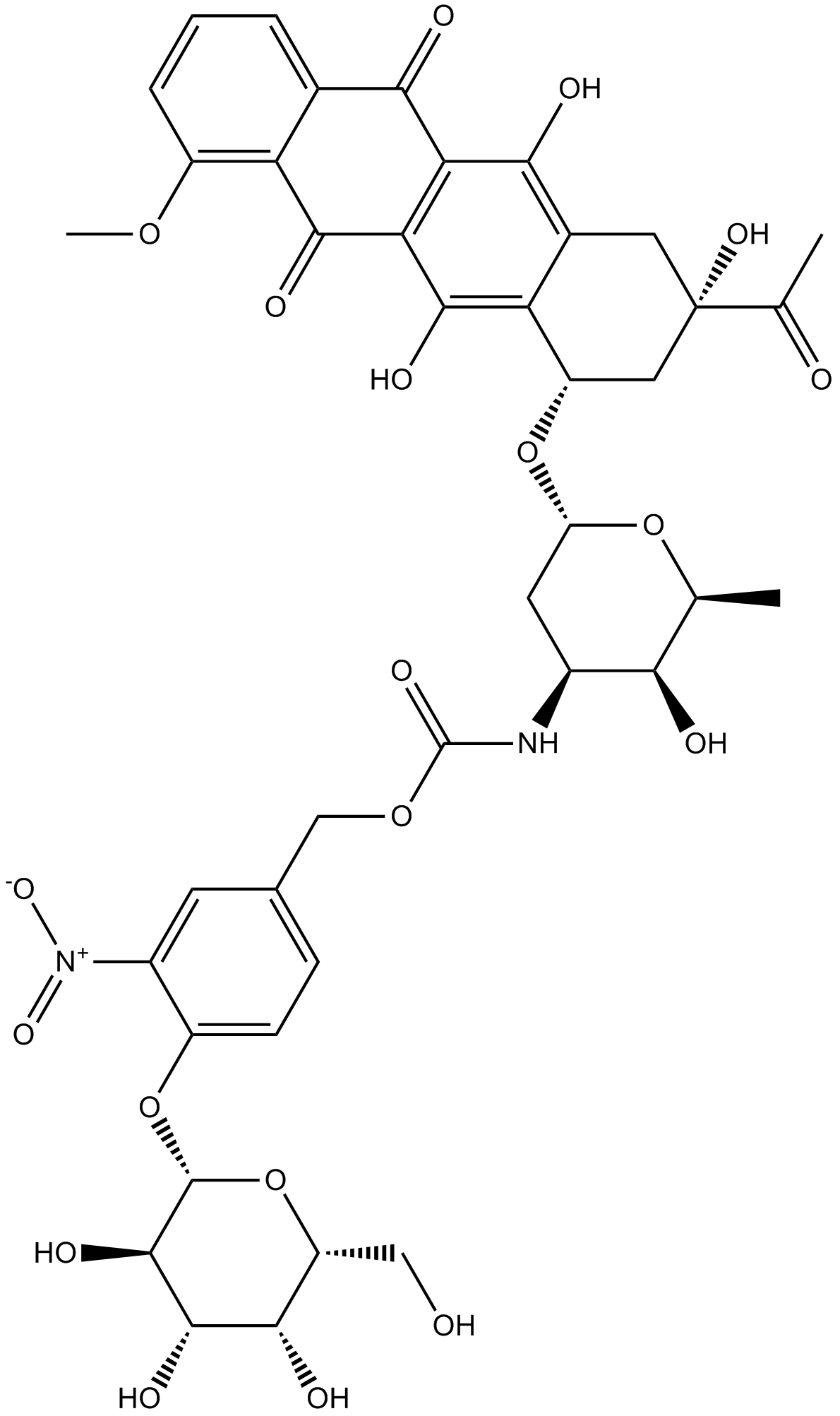
-
GC12828
Daunorubicin
DNA topoisomerase II inhibitor
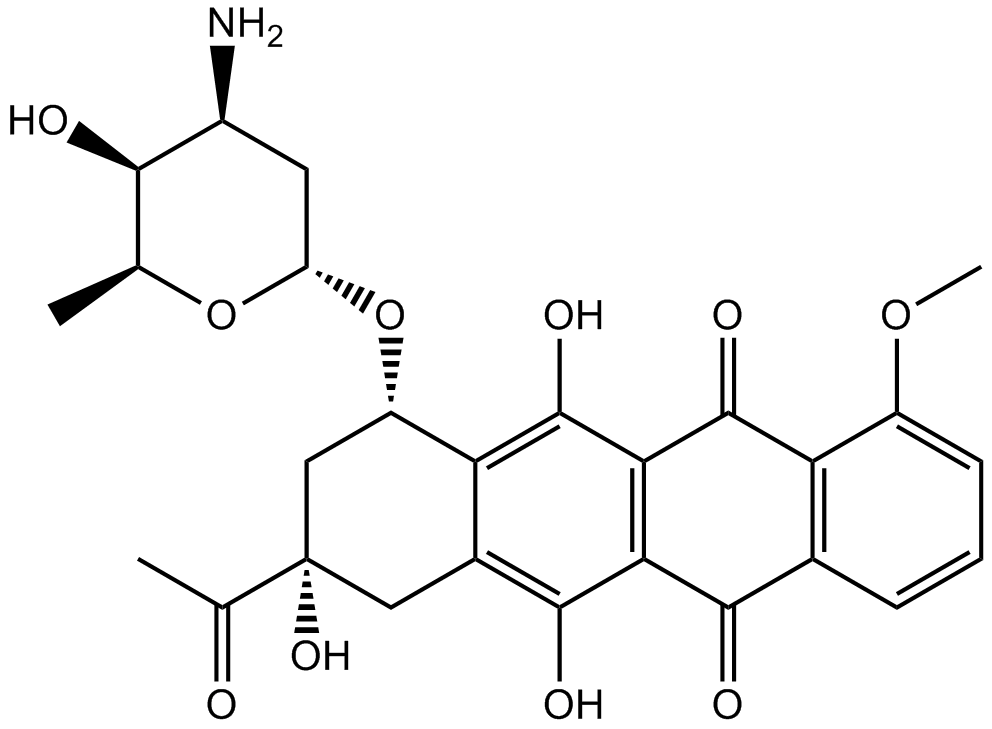
-
GC10354
Daunorubicin HCl
Daunorubicin (Daunomycin) hydrochloride is a topoisomerase II inhibitor with potent anti-tumor activity.
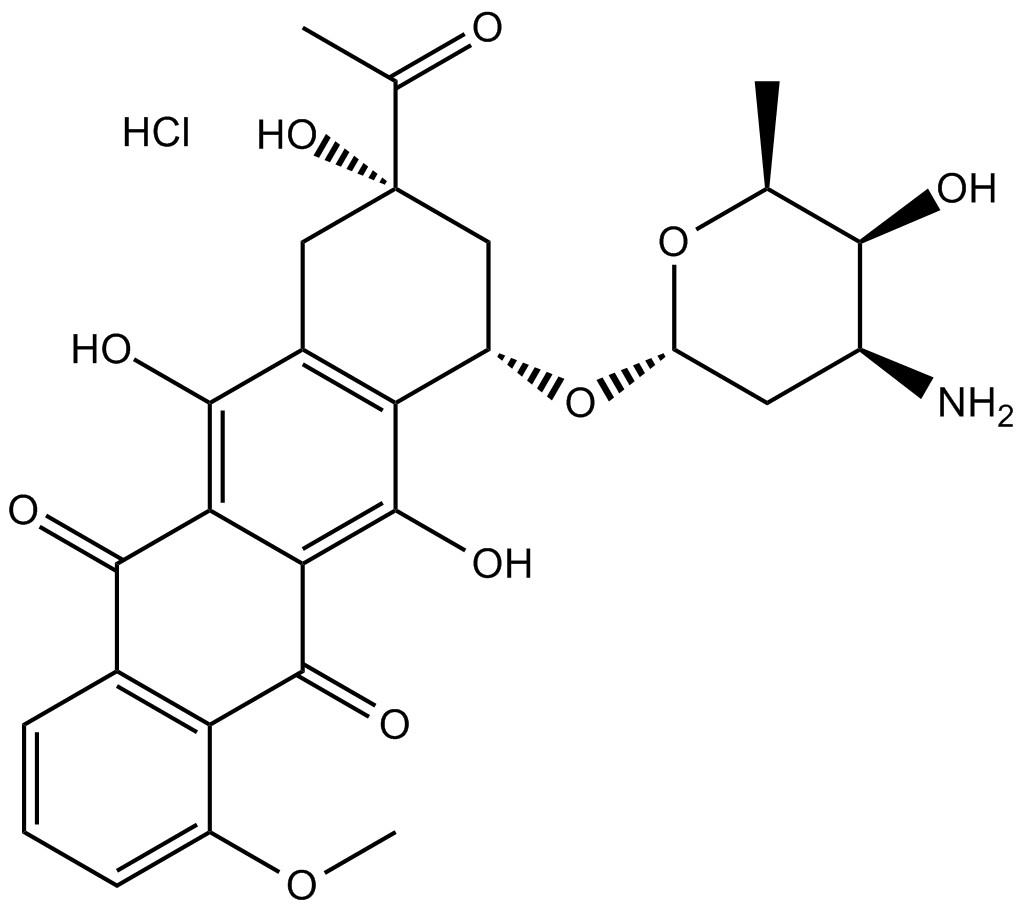
-
GC38538
DC0-NH2
DC0-NH2 is an effector moiety for ADC and a simplified analog of DC1 with better stability. DC0-NH2 is about 1000-fold more cytotoxic than commonly used anticancer drugs (ex. Doxorubicin). DC0-NH2 can bind to the minor groove of DNA, followed by alkylation of adenine residues by its propabenzindole (CBI) component.
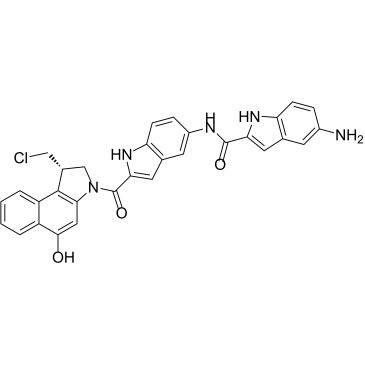
-
GC34185
DC1
DC1, an analogue of the minor groove-binding DNA alkylator CC-1065, is a ADC Cytotoxin. DC1 can be used in synthesis of antibody-drug conjugates for the targeted treatment of cancer.
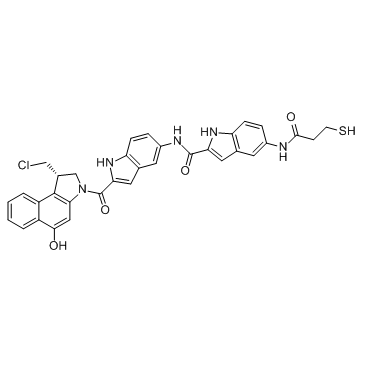
-
GC34190
DC1-SMe
DC1Sme, a DC1 derivative, exhibits IC50 values of 22 pM, 10 pM, 32 pM and 250 pM for Ramos, Namalwa, HL60/s and COLO 205 cancer cells, respectively. DC1, an analogue of the minor groove-binding DNA alkylator CC-1065, is a ADC Cytotoxin. DC1 can be used in synthesis of antibody-drug conjugates for the targeted treatment of cancer.
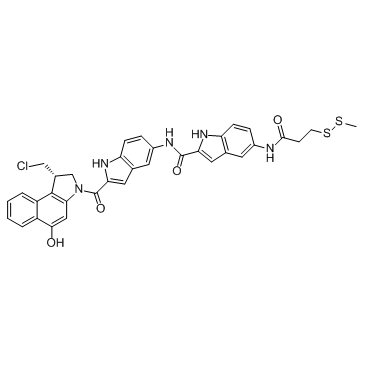
-
GC35817
DC41
DC41 is a DC1 derivative. DC1, a simplified analogue of CC-1065, is an antibody conjugate of cytotoxic DNA alkylators for the targeted treatment of cancer.
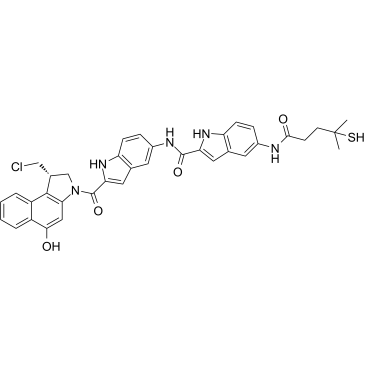
-
GC35818
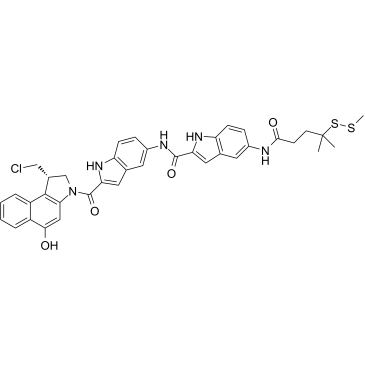
DC41-SMe
DC41-SMe, a DC1 derivative, shows cytotoxicity in Ramos, Namalwa, and HL60/s cells with IC50s ranging from 18-25 pM. DC1, a simplified analogue of CC-1065, is an antibody conjugate of cytotoxic DNA alkylators for the targeted treatment of cancer.
-
GC60143
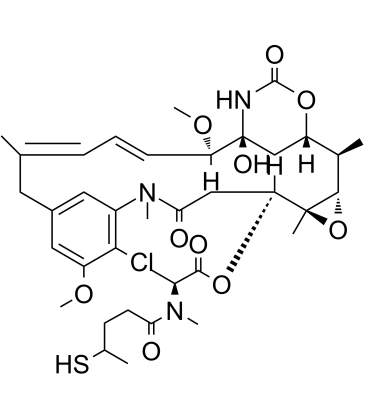
DM3
DM3 (Maytansinoid DM3) is a maytansine analog bearing disulfide or thiol groups and a tubulin inhibitor, and is a cytotoxic moiety of antibody-drug conjugates (ADCs).
-
GC38355
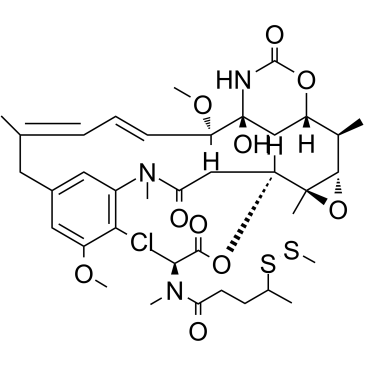
DM3-SMe
DM3-SMe is a maytansine derivative and a tubulin inhibitor, and is a cytotoxic moiety of antibody-drug conjugates (ADCs), which can be linked to antibody through disulfide bond or stable thioether bond. DM3-SMe shows highly cytotoxic activity in vitro with an IC50 of 0.0011 nM.
-
GC38468
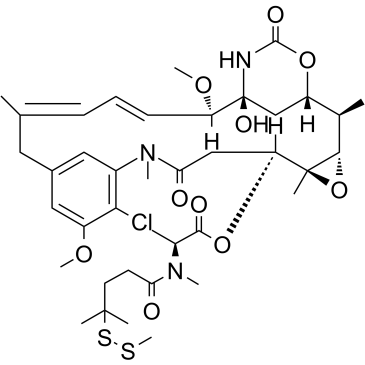
DM4-SMe
DM4-SMe is a metabolite of antibody-maytansin conjugates (AMCs) and a tubulin inhibitor, and also a cytotoxic moiety of antibody-drug conjugates (ADCs), which can be linked to antibody through disulfide bond or stable thioether bond. DM4-SMe inhibits KB cells with an IC50 of 0.026 nM.
-
GC16273
Dolastatin 10
Antitumor agent
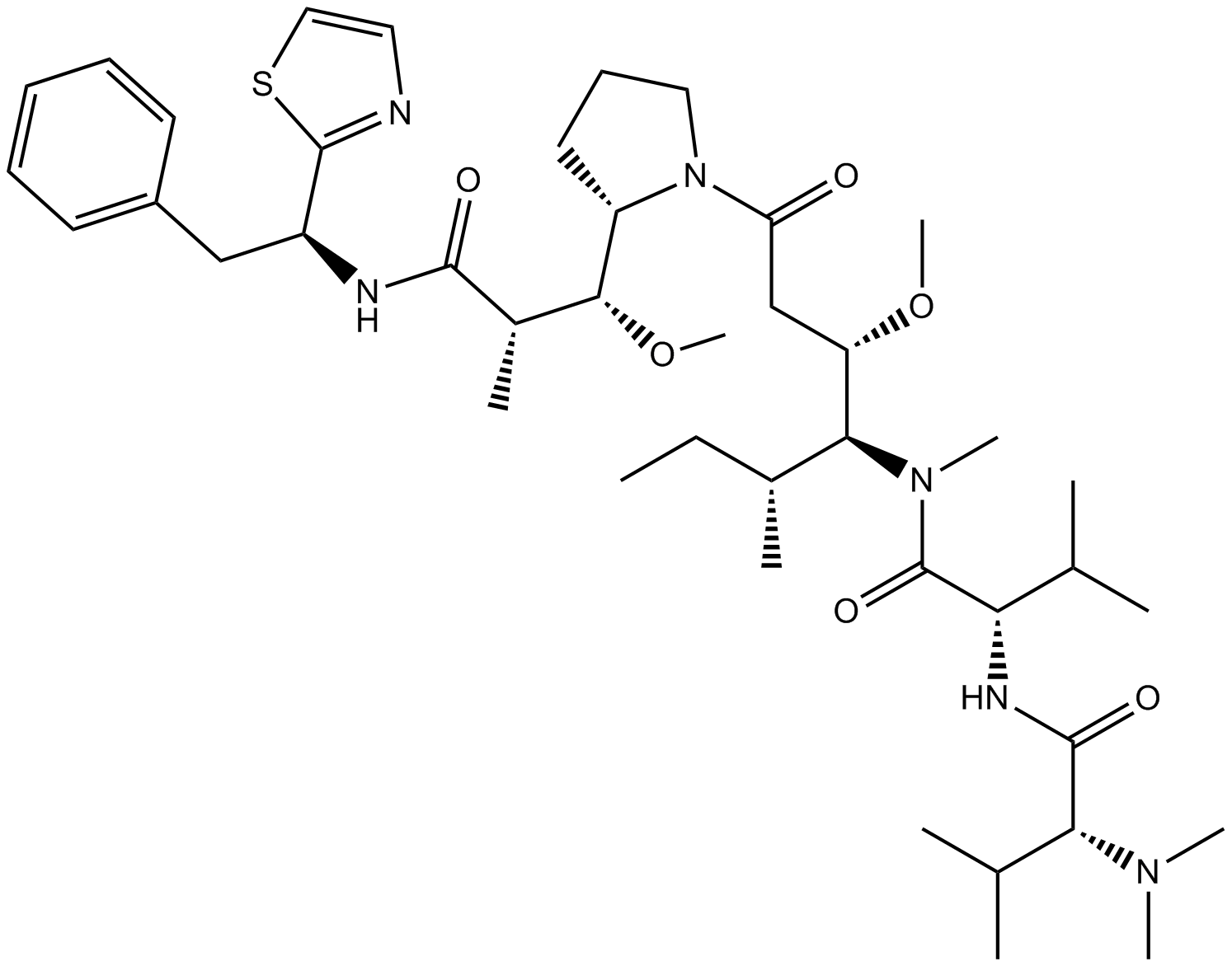
-
GA21397
Dolastatin 15
Dolastatin 15 originally isolated from the sea hare Dolabella auricularia belongs to a family of antimitotic and antineoplastic depsipeptides inducing apoptosis in various malignant cell types. It was shown to inhibit the growth of the P388 lymphocytic leukemia cell line efficiently, with an ED?? value of 2.4 ng/mL.

-
GC17567
Doxorubicin (Adriamycin) HCl
Doxorubicin (Hydroxydaunorubicin) hydrochloride, a cytotoxic anthracycline antibiotic, is an anti-cancer chemotherapy agent.
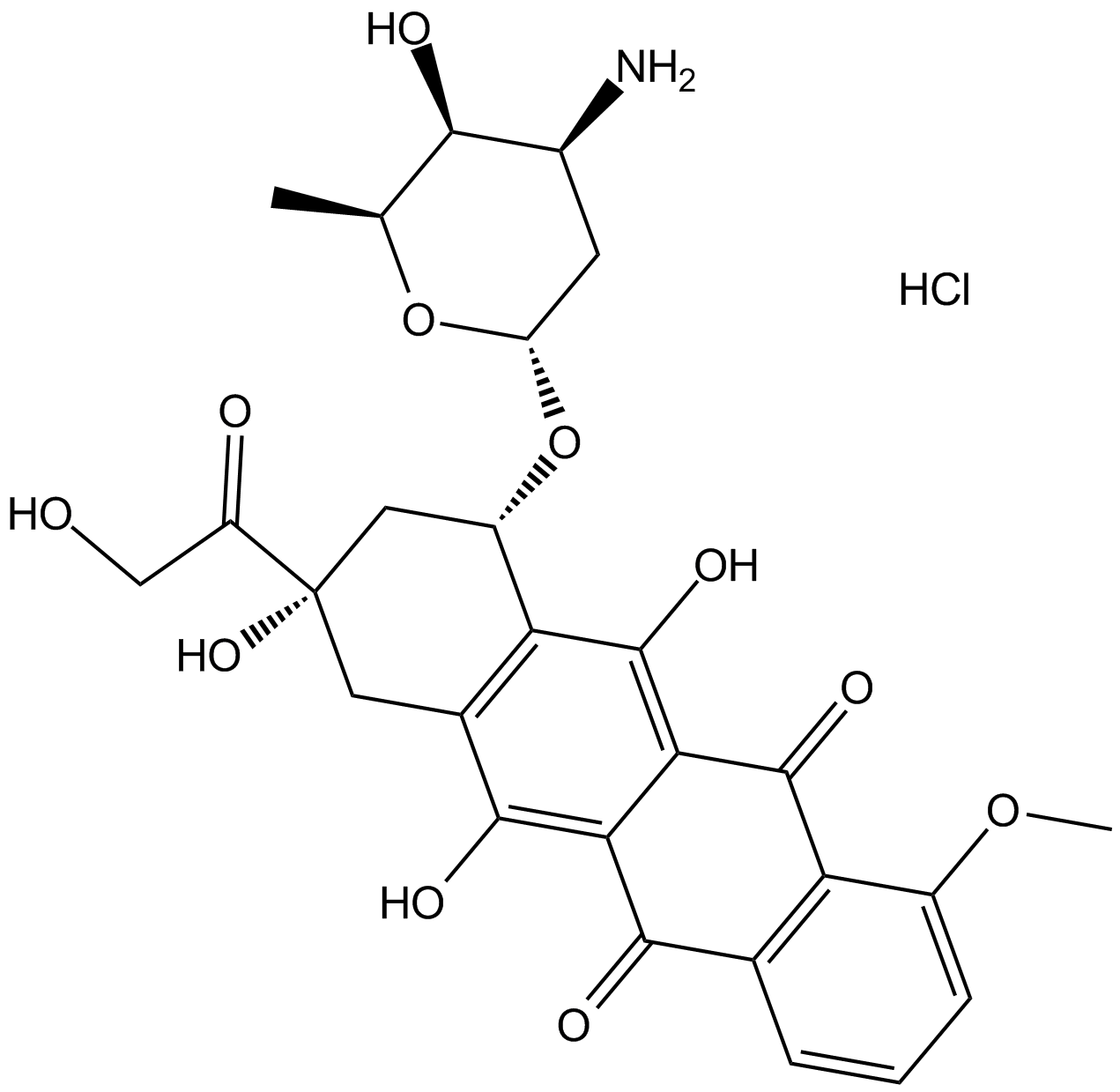
-
GC38774
DRF-1042
DRF-1042 is an orally active derivative of Camptothecin. DRF-1042 acts to inhibit DNA topoisomerase I. DRF-1042 shows good anticancer activity against a panel of human cancer cell lines including multi-drug resistance (MDR) phenotype.

-
GC35908
Duocarmycin A
Duocarmycin A, which is one of well-known antitumor antibiotics, is a DNA alkylator and efficiently alkylates adenine N3 at the 3′ end of AT-rich sequences in the DNA. Duocarmycin A, as a chemotherapeutic agent, results HLC-2 cells typically apoptotic changes, including chromatin condensation, sub-G1 accumulation in DNA histogram pattern, and decrease in procaspase-3 and 9 levels.
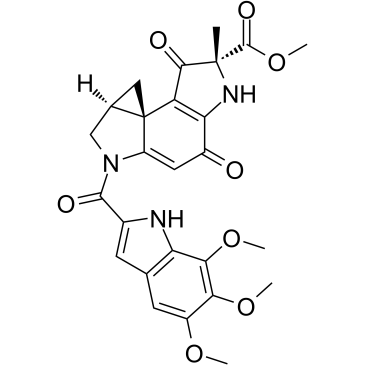
-
GC38080
Duocarmycin Analog
Duocarmycin Analog is an analog of Duocarmycin, and used as an DNA alkylator and ADC cytotoxin.
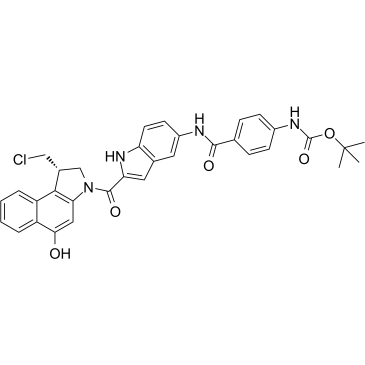
-
GC35909
Duocarmycin GA
Duocarmycin GA is an antibody drug conjugates (ADCs) toxin. Duocarmycin is a DNA alkylating agent that binds in the minor groove. Duocarmycin GA can be used against multi-drug resistant cell lines.
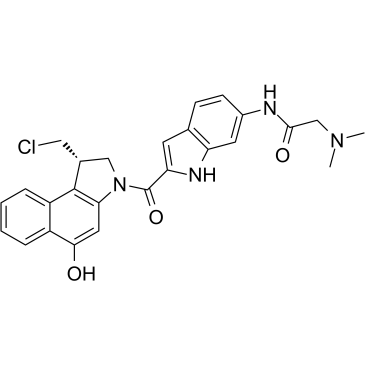
-
GC35910
Duocarmycin MA
Duocarmycin MA is an antibody drug conjugates (ADCs) toxin. Duocarmycin is a DNA alkylating agent that binds in the minor groove. Duocarmycin MA can be used against multi-drug resistant cell lines.
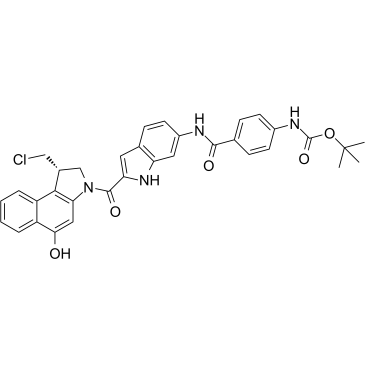
-
GC35911
Duocarmycin MB
Duocarmycin MB is an antibody drug conjugates (ADCs) toxin. Duocarmycin is a DNA alkylating agent that binds in the minor groove. Duocarmycin MB can be used against multi-drug resistant cell lines.
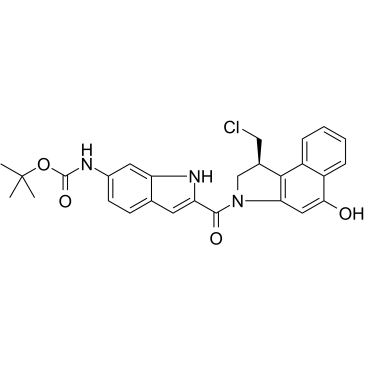
-
GC35912
Duocarmycin SA
Duocarmycin SA is a potent antitumor antibiotic with an IC50 of 10 pM. Duocarmycin SA is an extremely potent cytotoxic agent capable of inducing a sequence-selective alkylation of duplex DNA. Duocarmycin SA demonstrates synergistic cytotoxicity against glioblastoma multiforme (GBM) cells treated with proton radiation in vitro.
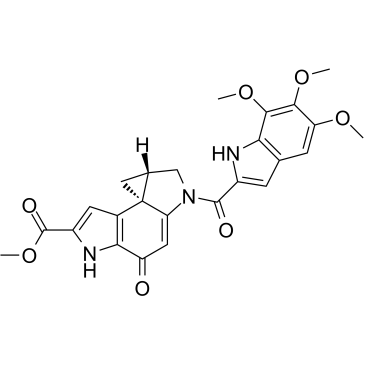
-
GC32187
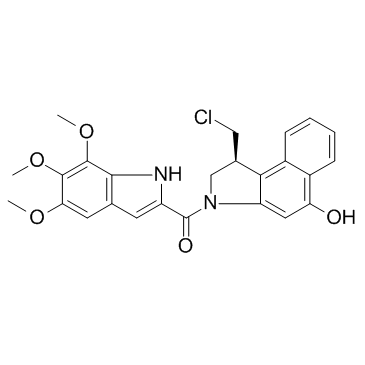
Duocarmycin TM
Duocarmycin TM (CBI-TMI) is a potent antitumor antibiotic. Duocarmycin TM induces a sequence-selective alkylation of duplex DNA.
-
GC32926
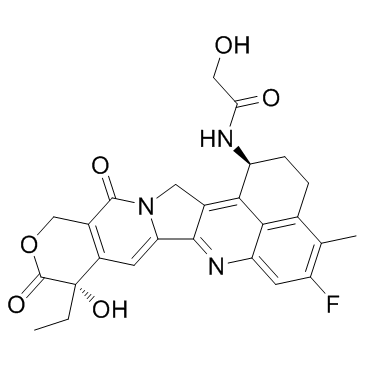
Dxd (Exatecan derivative)
Dxd (Exatecan derivative) (Exatecan derivative for ADC) is a potent DNA topoisomerase I inhibitor, with an IC50 of 0.31 μM, used as a conjugated drug of HER2-targeting ADC (DS-8201a).
-
GC62212
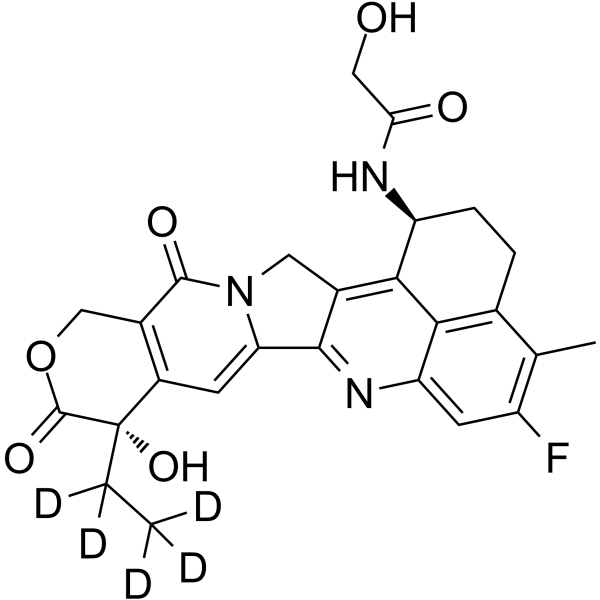
Dxd-D5
Dxd-d5 (Exatecan-d5 derivative for ADC) is a deuterium labeled Dxd. Dxd is a potent DNA topoisomerase I inhibitor, with an IC50 of 0.31 μM, used as a conjugated drug of HER2-targeting ADC (DS-8201a) .
-
GC43882
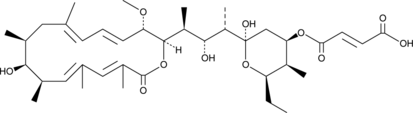
Hygrolidin
Hygrolidin is a macrocyclic lactone originally isolated from S.
-
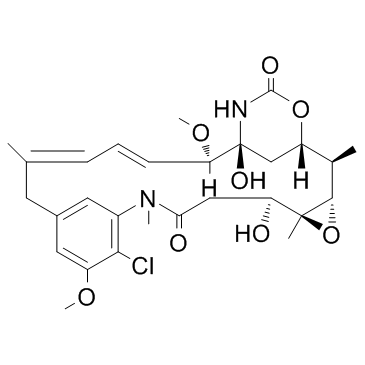
GC32895
Maytansinol (Ansamitocin P-0)
Maytansinol (Ansamitocin P-0) inhibits microtubule assembly and induces microtubule disassembly in vitro.
-
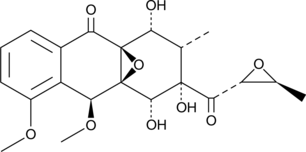
GC47620
Mensacarcin
A bacterial metabolite with anticancer activity
-
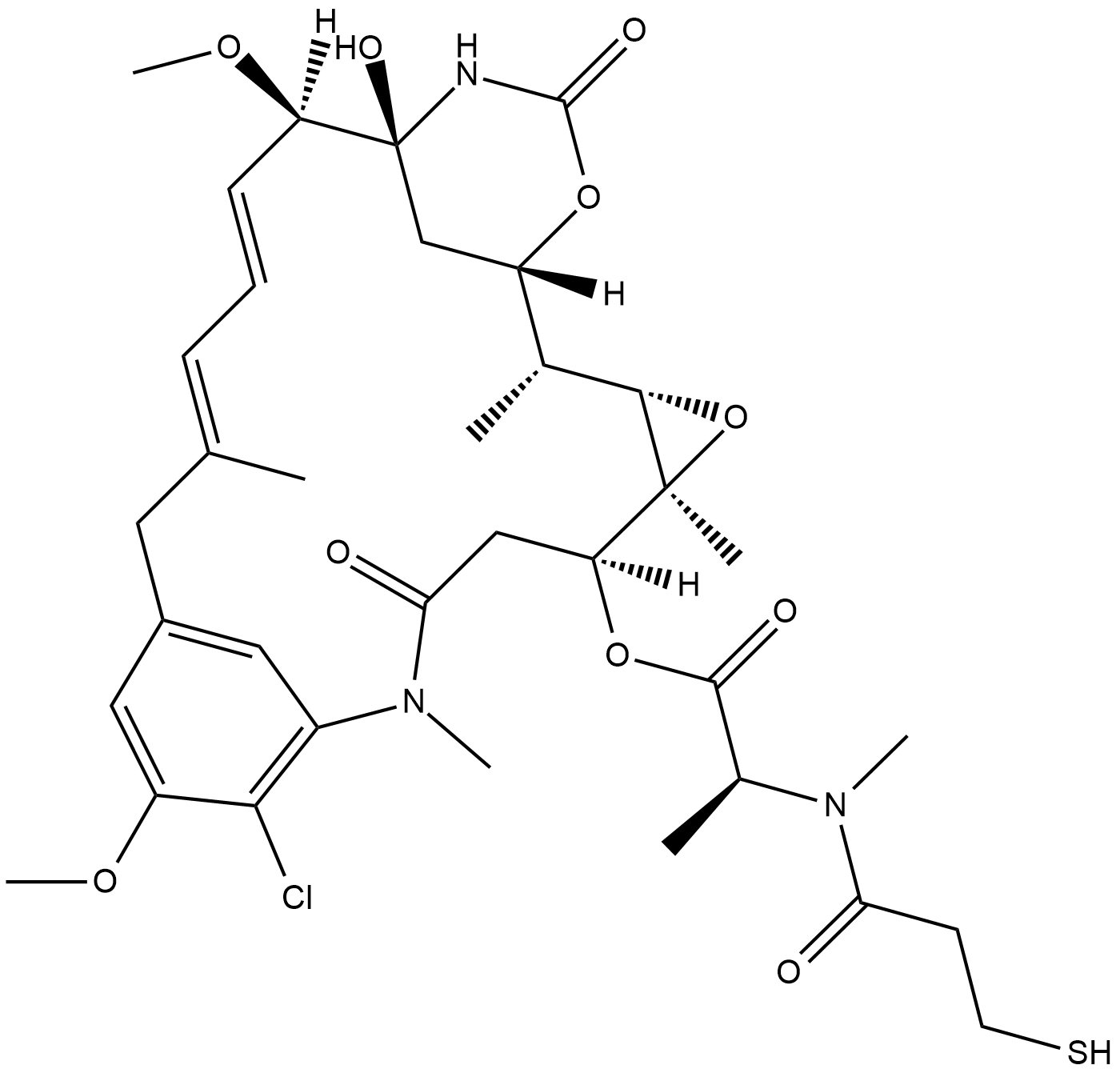
GC19244
Mertansine
Mertansine (DM1) is a microtubulin inhibitor which binds at the tips of microtubules and suppresses the dynamicity of microtubules..
-
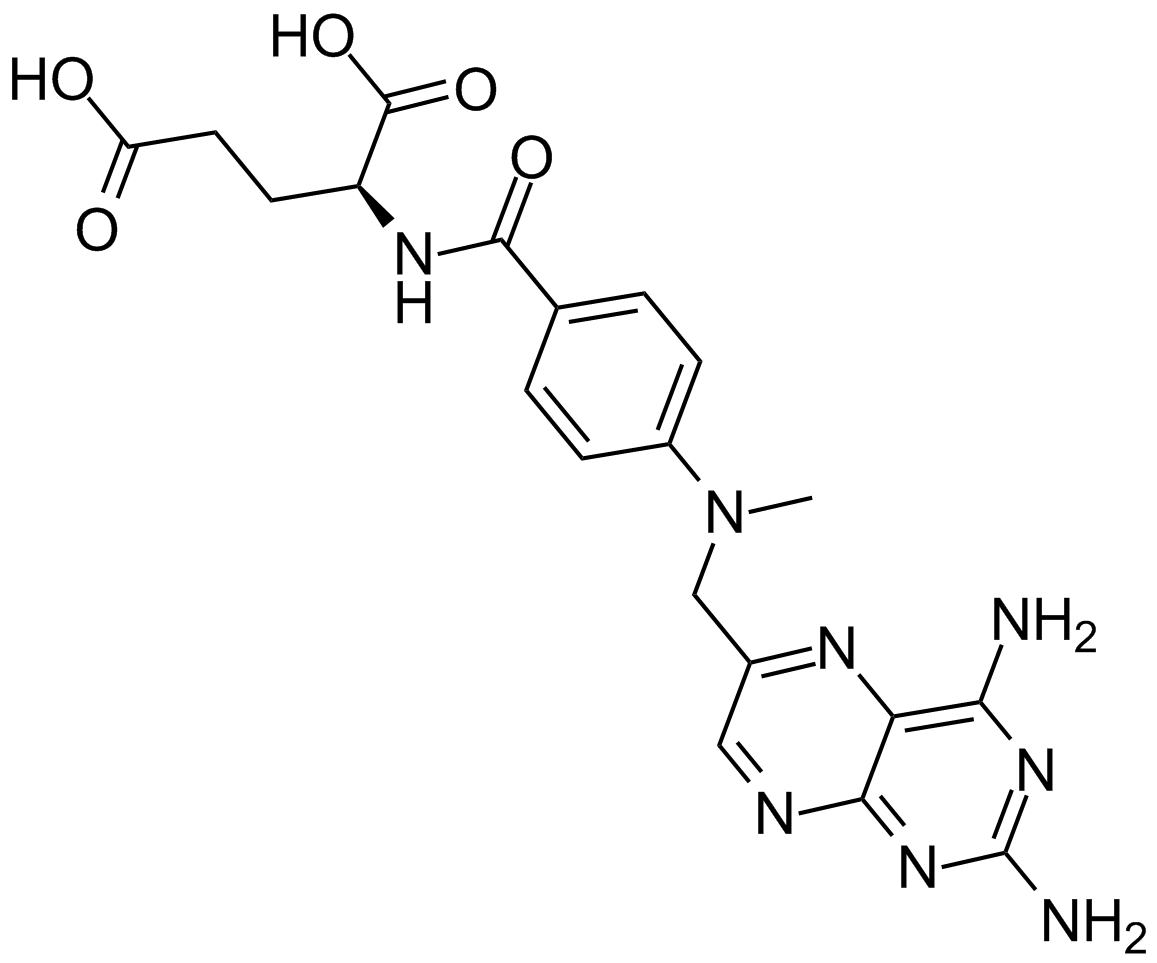
GC10405
Methotrexate
Methotrexate, a folate antagonist, is a potent anti-inflammatory agent when used weekly in low concentrations, the anti-phlogistic action of which is due to increased adenosine release at inflamed sites.
-
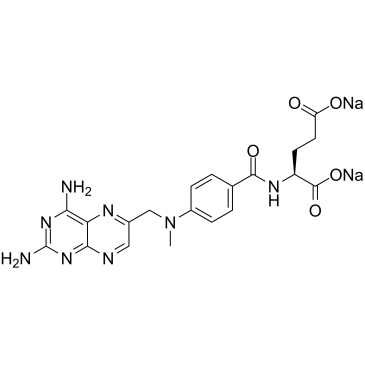
GC61047
Methotrexate disodium
Methotrexate (Amethopterin) disodium, an antimetabolite and antifolate agent, inhibits the enzyme dihydrofolate reductase, thereby preventing the conversion of folic acid into tetrahydrofolate, and inhibiting DNA synthesis.
-
GC15798
MMAD
-
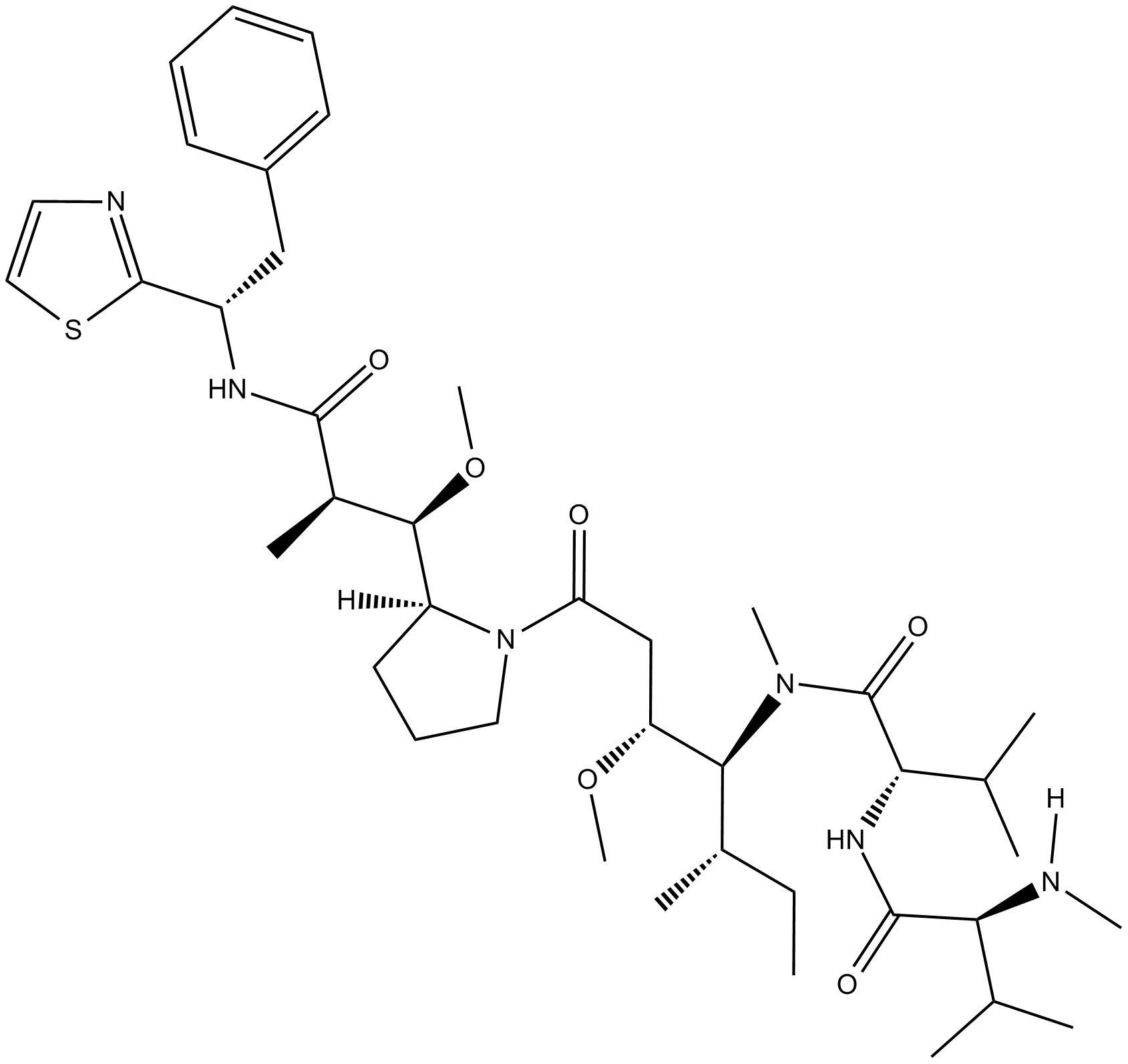
GC63775
MMAD-d8
D8-MMAD is a deuterated form of MMAD, which is a microtubule disrupting agent.
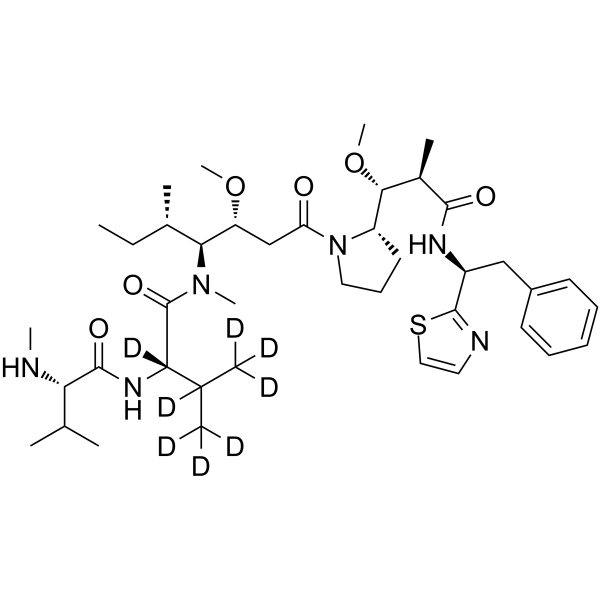
-
GC10034
MMAF
Anti-mitotic/anti-tubulin/antineoplastic agent
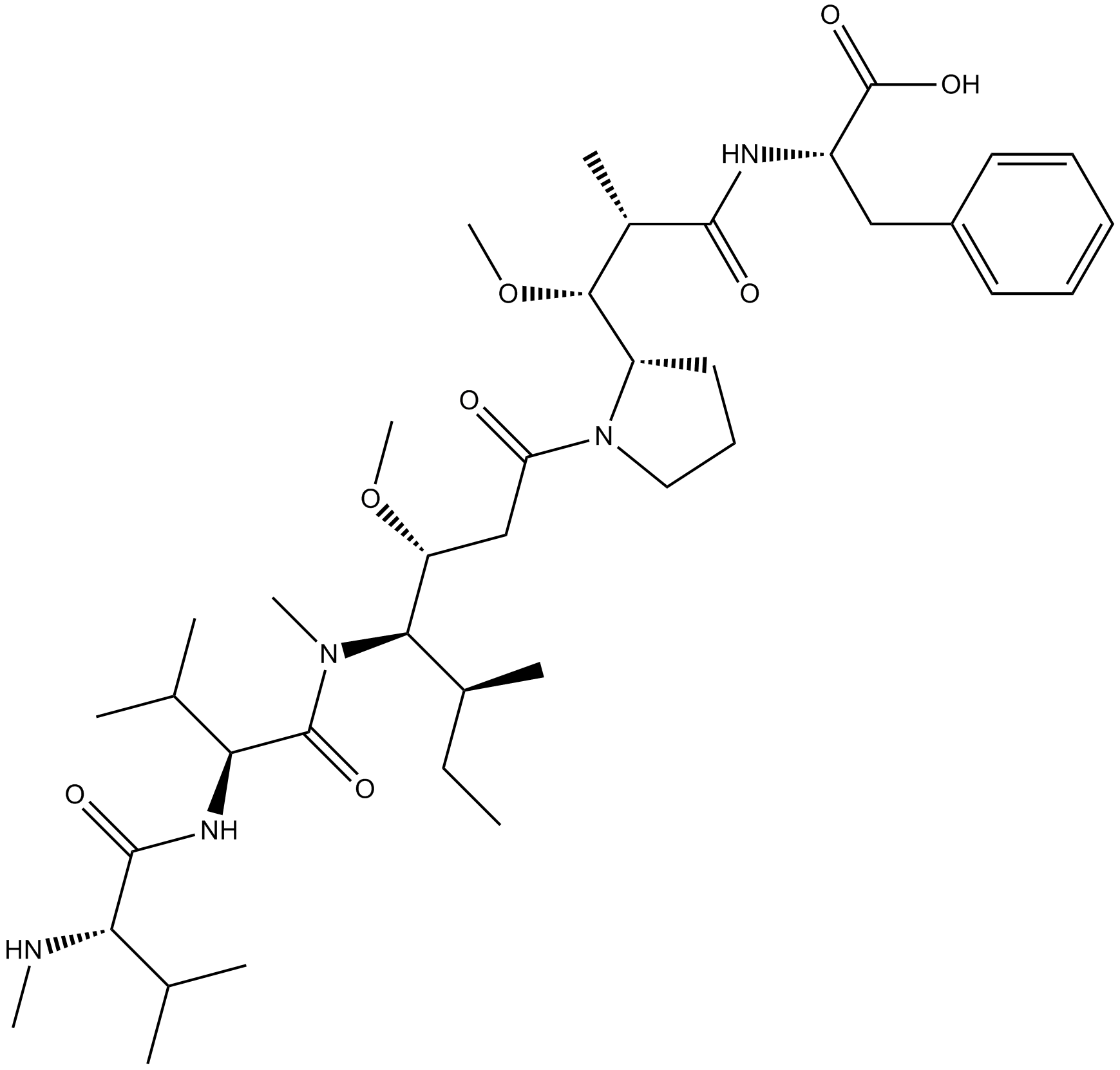
-
GC36633
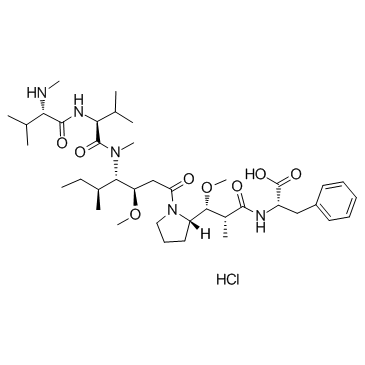
MMAF Hydrochloride
MMAF (Monomethylauristatin F) hydrochloride is a potent tubulin polymerization inhibitor and is used as a antitumor agent. MMAF hydrochloride is widely used as a cytotoxic component of antibody-drug conjugates (ADCs) such as Vorsetuzumab mafodotin and SGN-CD19A.
-
GC38397
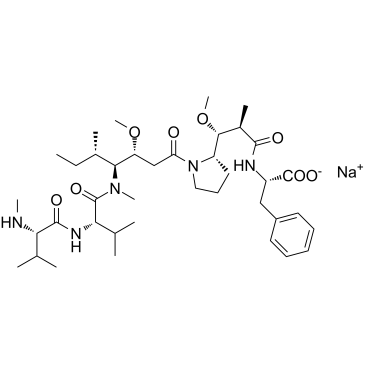
MMAF sodium
MMAF sodium (Monomethylauristatin F sodium) is a potent tubulin polymerization inhibitor and is used as a antitumor agent. MMAF sodium (Monomethylauristatin F sodium) is widely used as a cytotoxic component of antibody-drug conjugates (ADCs) such as Vorsetuzumab mafodotin and SGN-CD19A.
-
GC36634
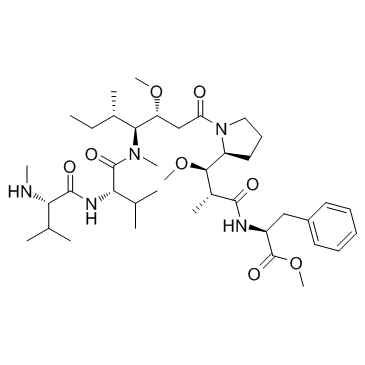
MMAF-OMe
MMAF-Ome, an antitubulin agent, is also an ADC cytotoxin. MMAF-Ome inhibits several tumor cell lines with IC50s of 0.056 nM, 0.166 nM, 0.183 nM, and 0.449 nM for MDAMB435/5T4, MDAMB361DYT2, MDAMB468, and Raji (5T4-) cell lines, respectively.
-
GC17276
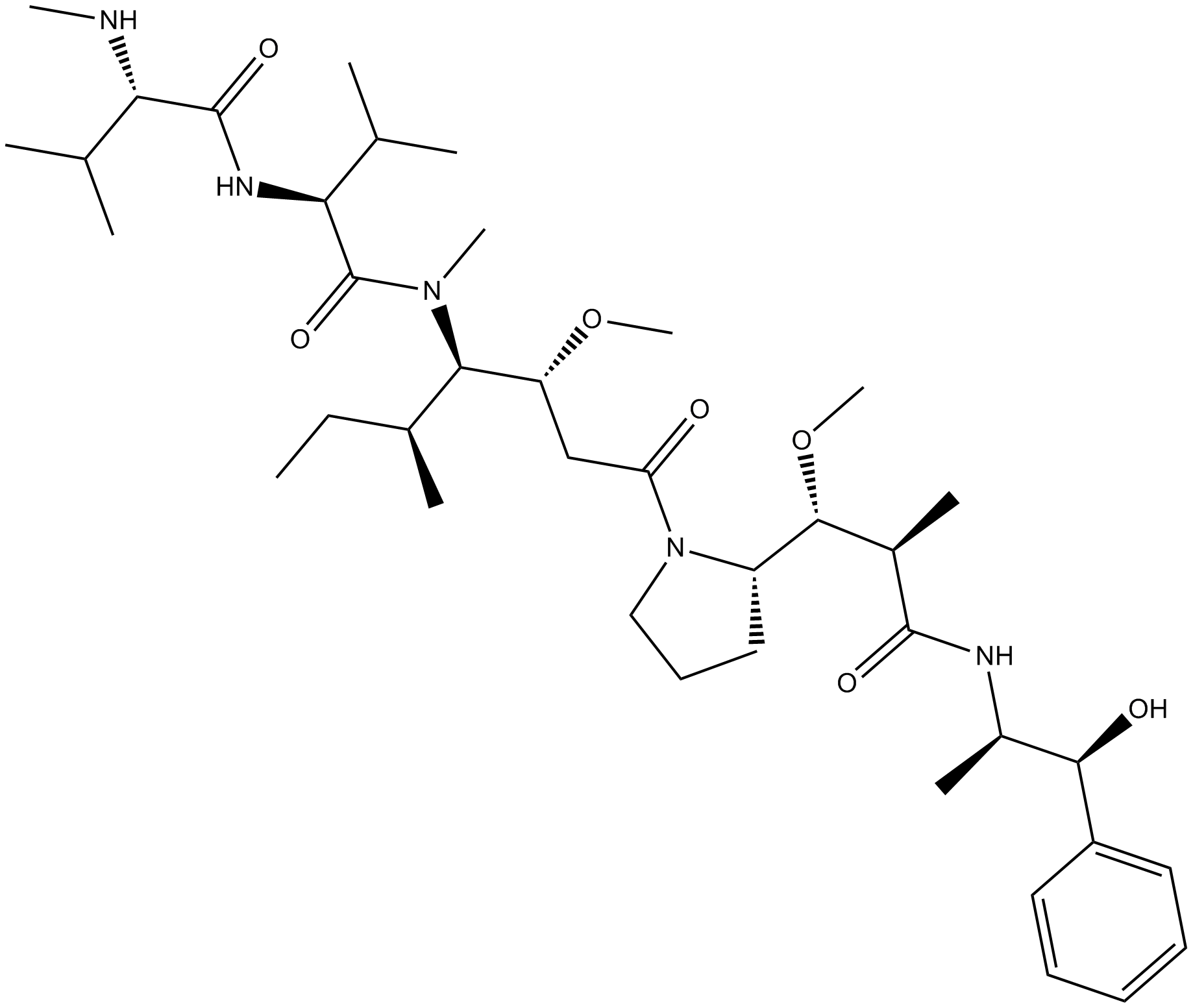
Monomethyl auristatin E
A potent antimitotic compound
-
GC12511
Paclitaxel (Taxol)
A potent mitotic inhibitor
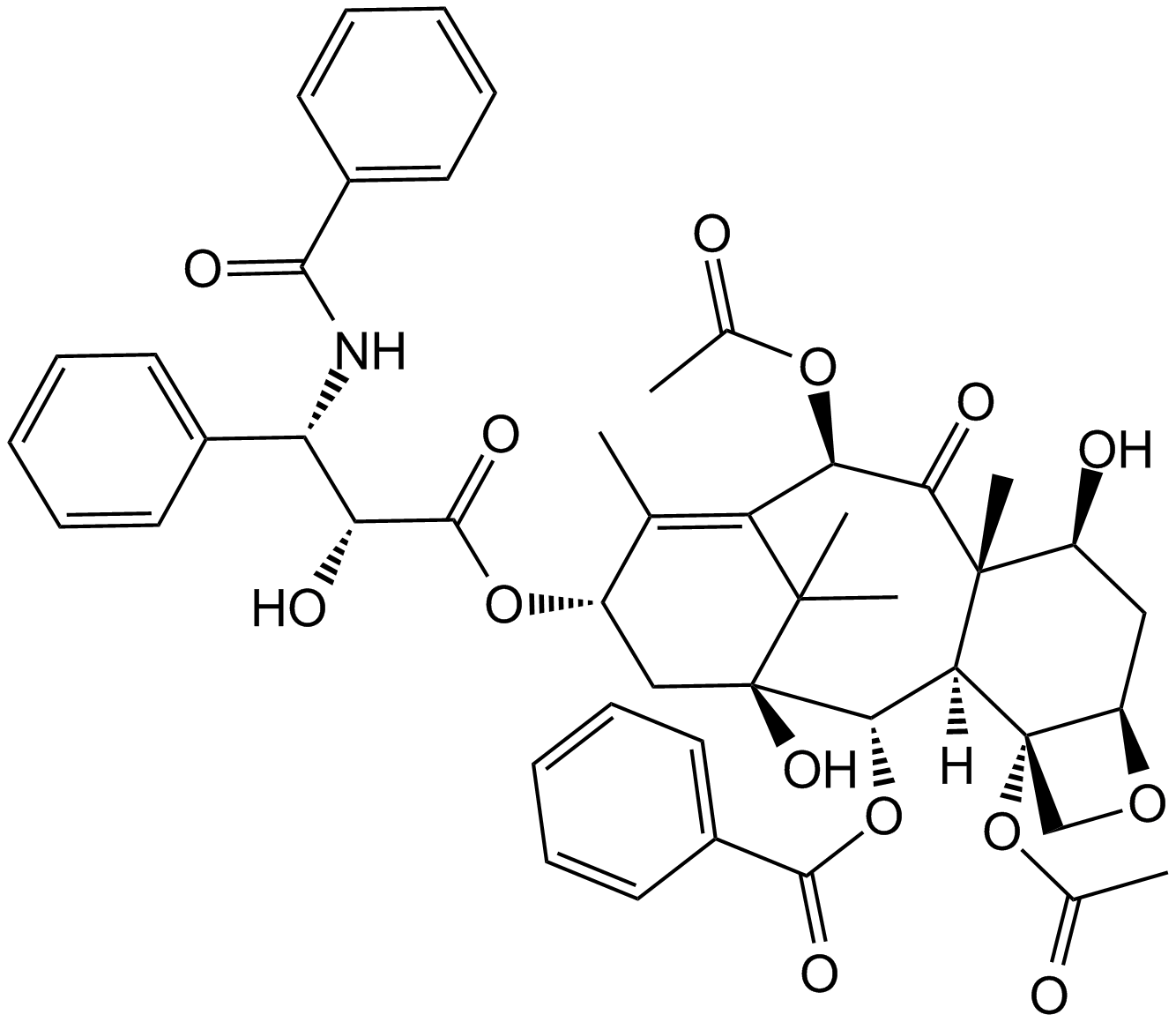
-
GC33136
PF-06380101
PF-06380101 (Aur0101), an auristatin microtubule inhibitor, is a cytotoxic Dolastatin 10 analogue. PF-06380101 (Aur0101) shows excellent potencies in tumor cell proliferation assays and differential ADME properties when compared to other synthetic auristatin analogues that are used in the preparation of ADCs.
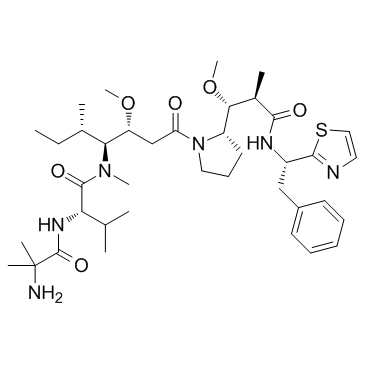
-
GC36941
PNU-159682
PNU-159682, a metabolite of the anthracycline Nemorubicin, is a highly potent DNA topoisomerase II inhibitor with excellent cytotoxicity. PNU-159682 acts as a more potent and tolerated?ADC cytotoxin than Doxorubicin for ADC synthesis. PNU-159682 can be used in EDV-nanocell technology to overcome drug resistance.
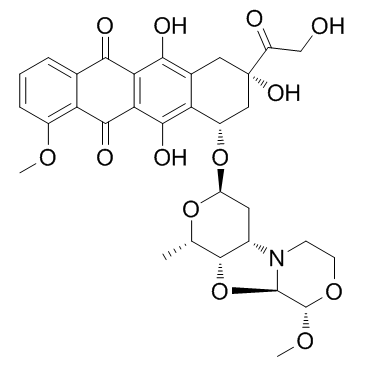
-
GC66071
PNU-159682 carboxylic acid
PNU-159682 carboxylic acid (compound 53) is a potent ADCs cytotoxin and encodes a member of the C-type lectin/C-type lectin-like domain (CTL/CTLD) superfamily. PNU-159682 carboxylic acid has protein fold and diverse functions, such as cell adhesion, cell-cell signalling, glycoprotein turnover, and roles in inflammation and immune response.
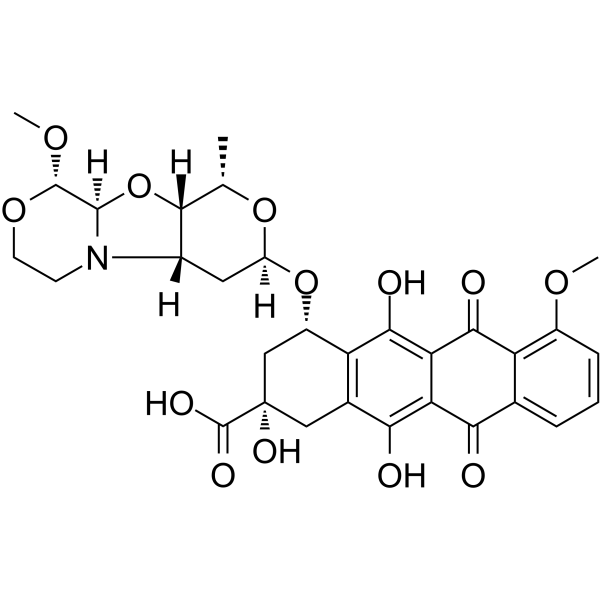
-
GC61213
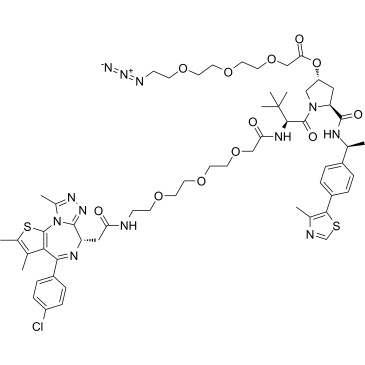
PROTAC BRD4 Degrader-5-CO-PEG3-N3
PROTAC BRD4 Degrader-5-CO-PEG3-N3 (Compound 2) is a PROTAC-linker Conjugate for PAC, comprises the BRD4 degrader GNE-987 and PEG-based linker.
-
GC44809
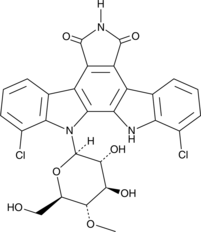
Rebeccamycin
Rebeccamycin is an indolocarbazole antibiotic produced by S.
-
GC69863
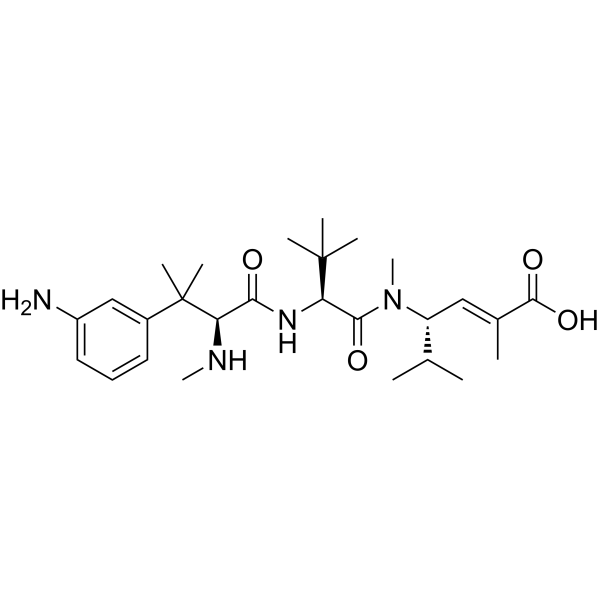
SC209
SC209 is an ADC cytotoxin derived from patent WO2021247798, used for synthesizing anti-EGFR antibody-active molecule conjugated ADCs. SC209 is a metabolite of STRO-002.
-
GC63184
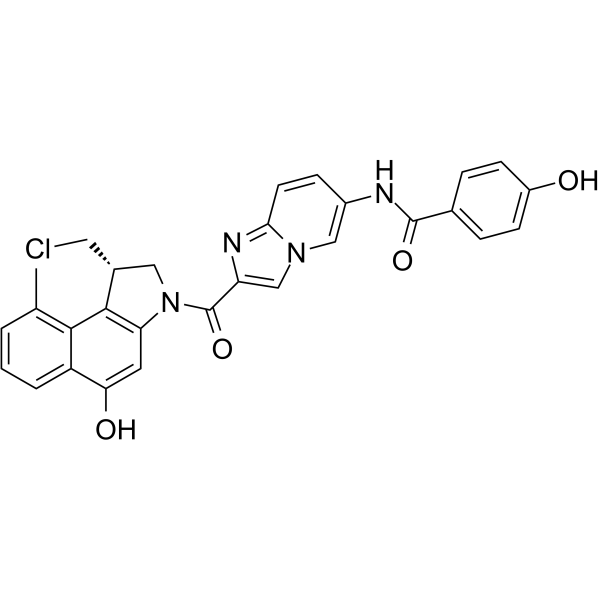
Seco-DUBA
Seco-DUBA is a duocarmycin (DUBA) prodrug containing two hydroxyl groups, which can each be used for coupling to an antibody via a linker.
-
GC38349
Seco-Duocarmycin SA
Seco-Duocarmycin SA is a DNA alkylator, and is used as an ADC cytotoxin.
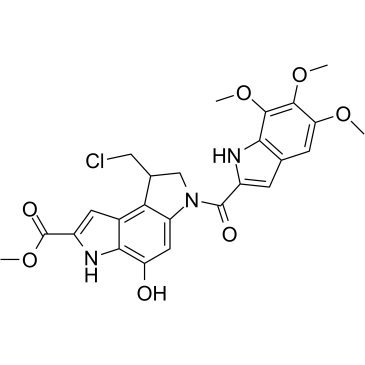
-
GC38354
Seco-Duocarmycin TM
Seco-Duocarmycin TM is a DNA alkylator agent belonging to Duocarmycins family that inhibits DNA synthesis. Seco-Duocarmycin TM is a cytotoxic agent, used as the cytotoxic component in antibody-drug conjugates (ADC).
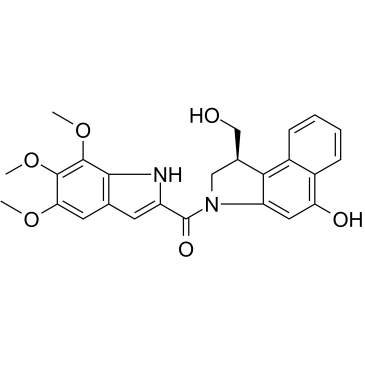
-
GC62691
SG3199
SG3199 is a cytotoxic DNA minor groove interstrand crosslinking pyrrolobenzodiazepine (PBD) dimer. SG3199 is the released warhead component of the ADC payload Tesirine (SG3249).
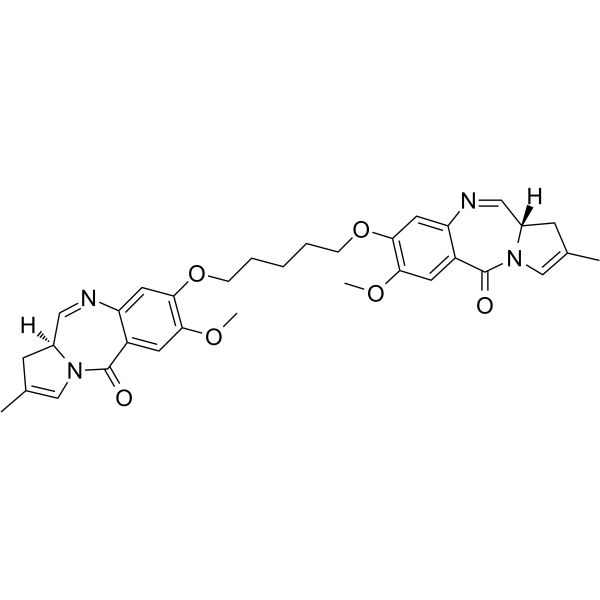
-
GC63189
SGD-1882
SGD-1882 is a cytotoxic, DNA minor-groove crosslinking agent pyrrolobenzodiazepine (PBD) dimer, acting as the payload for ADCs.
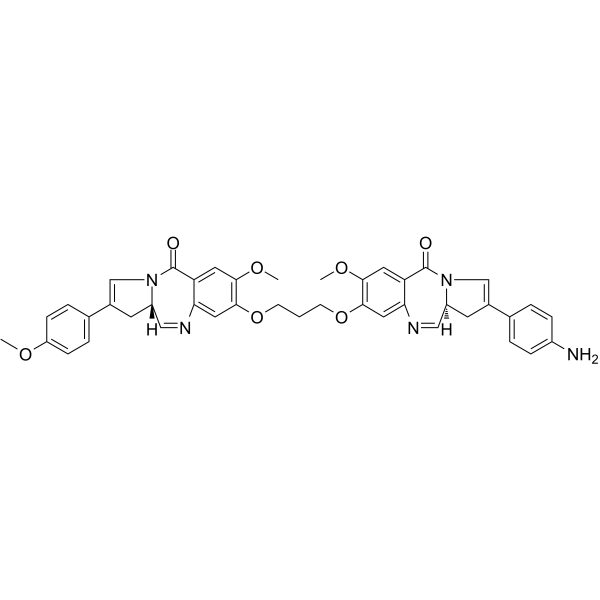
-
GC34086
SJG-136 (NSC-694501)
SJG-136 (NSC-694501) is a DNA cross-linking agent, with an XL50 of 45 nM for pBR322 DNA. SJG-136 (NSC-694501) has potent antitumor activity.
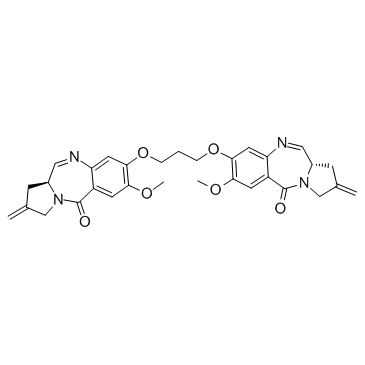
-
GN10449
SN-38 (NK012)
-
GC32753
Taltobulin (HTI-286)
Taltobulin (HTI-286) (HTI-286), a synthetic analogue of the tripeptide hemiasterlin, is a potent antimicrotubule agent that circumvents P-glycoprotein-mediated resistance in vitro and in vivo. Taltobulin (HTI-286) inhibits the polymerization of purified tubulin, disrupts microtubule organization in cells, and induces mitotic arrest, as well as apoptosis.
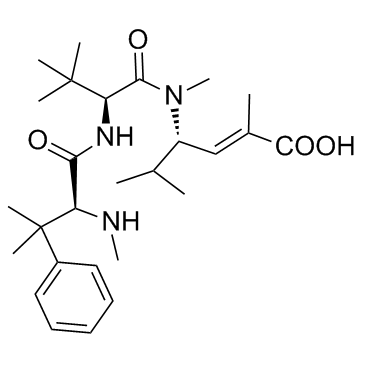
-
GC61314
Taltobulin hydrochloride
Taltobulin hydrochloride (HTI-286 hydrochloride), a synthetic analogue of the tripeptide hemiasterlin, is a potent antimicrotubule agent that circumvents P-glycoprotein-mediated resistance in vitro and in vivo. Taltobulin hydrochloride inhibits the polymerization of purified tubulin, disrupts microtubule organization in cells, and induces mitotic arrest, as well as apoptosis.
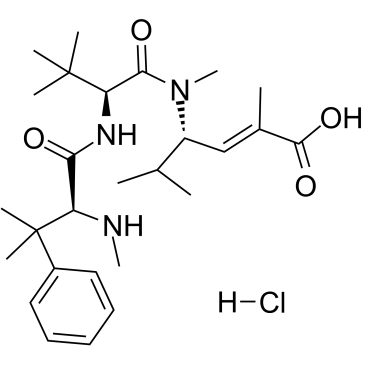
-
GC34303
Taltobulin hydrochloride (HTI-286 hydrochloride)
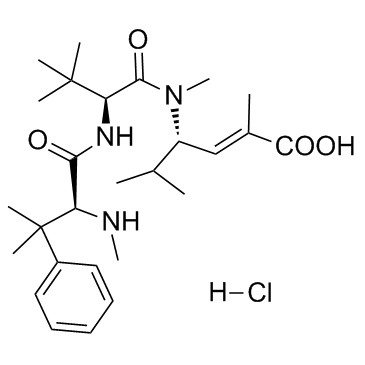
-
GC34135
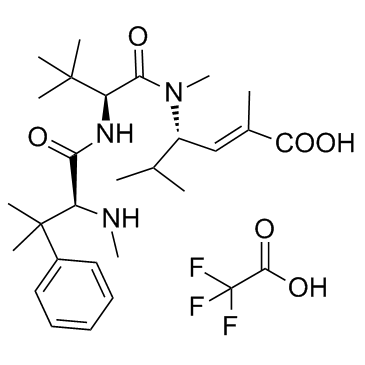
Taltobulin trifluoroacetate (HTI-286 trifluoroacetate)
Taltobulin trifluoroacetate (HTI-286 trifluoroacetate) (HTI-286 trifluoroacetate), a synthetic analogue of the tripeptide hemiasterlin, is a potent antimicrotubule agent that circumvents P-glycoprotein-mediated resistance in vitro and in vivo. Taltobulin trifluoroacetate (HTI-286 trifluoroacetate) inhibits the polymerization of purified tubulin, disrupts microtubule organization in cells, and induces mitotic arrest, as well as apoptosis.
-
GC62288
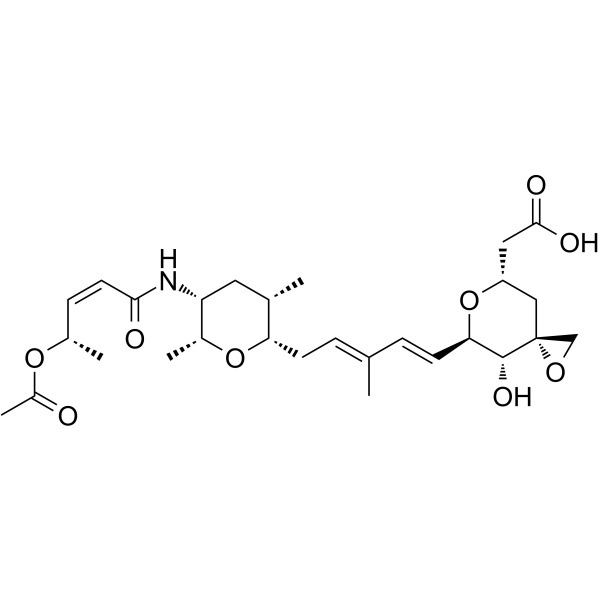
Thailanstatin A
Thailanstatin A is an ultra-potent inhibitor of eukaryotic RNA splicing (IC50=650 nM). Thailanstatin A exerts effects via non-covalent binding to the SF3b subunit of the U2 snRNA subcomplex of the spliceosome and shows low-nM to sub-nM IC50s against multiple cancer cell lines. Thailanstatin A, a payload for ADCs, is conjugated to the lysines on trastuzumab yielding “linker-less” ADC.
-
GC70043
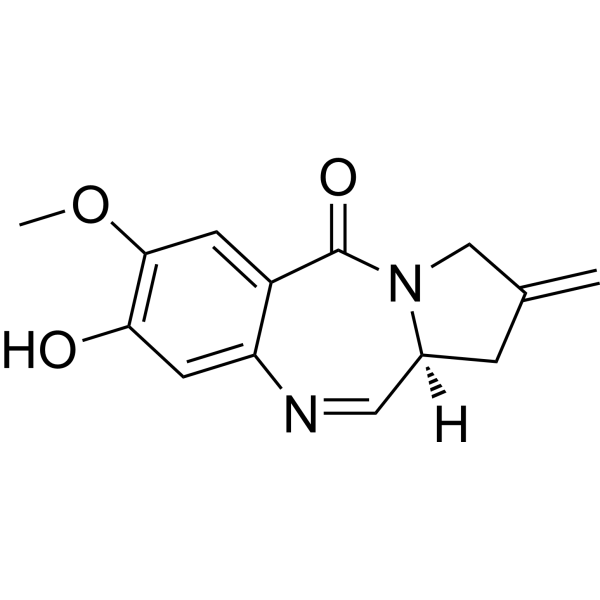
Tomaymycin DM
Tomaymycin DM is a DNA alkylating agent and a derivative of Tomaymycin. It is a monomer of PBD and can serve as an effective payload for tumor-targeting antibody-drug conjugates (ADCs).
-
GC33155
Tubulysin A (TubA)
Tubulysin A (TubA)(TubA) is a myxobacterial product that can function as an antiangiogenic agent in many in vitro assays; anti-microtubule, anti-mitotic, an apoptosis inducer, anticancer, anti-angiogenic, and antiproliferative.