JAK/STAT Signaling

Targets for JAK/STAT Signaling
Products for JAK/STAT Signaling
- Cat.No. Product Name Information
-
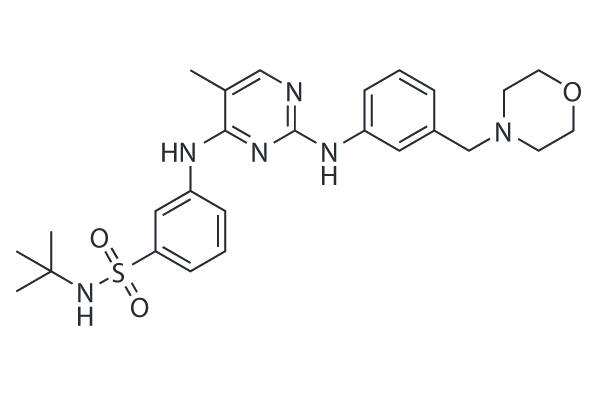
GC14170
Nifuroxazide
STAT inhibitor
-
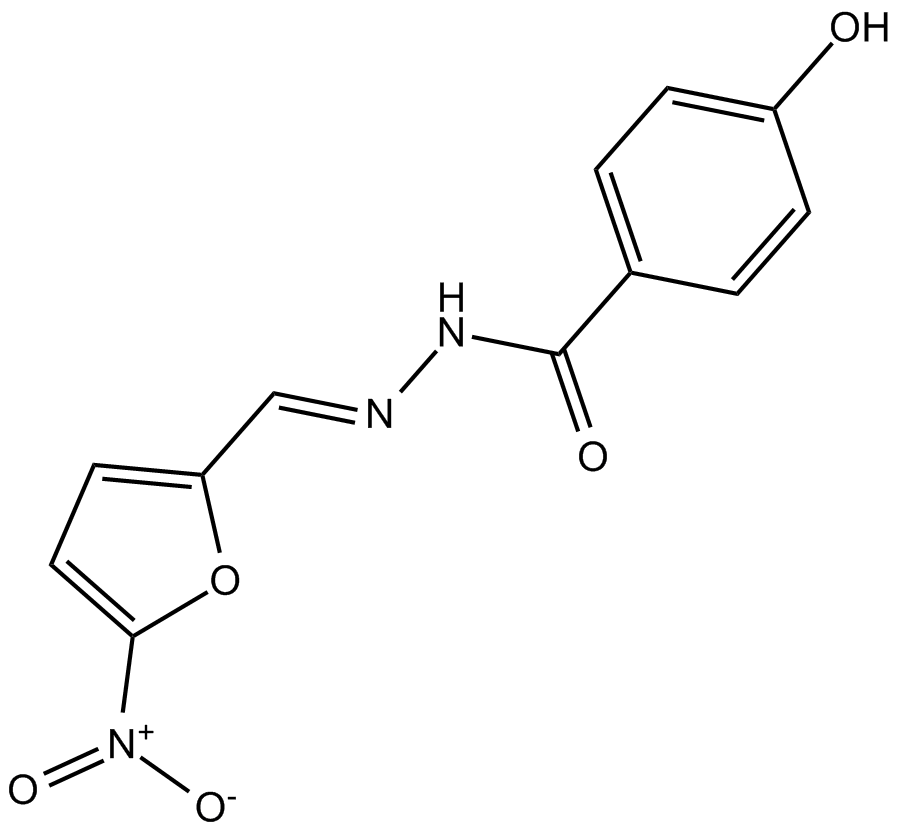
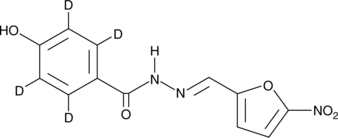
GC48839
Nifuroxazide-d4
An internal standard for the quantification of nifuroxazide
-
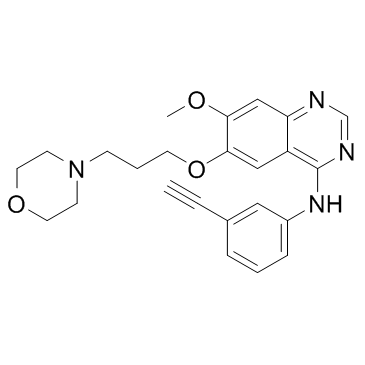
GC33131
NRC-2694
NRC-2694 is an epidermal growth factor receptor (EGFR) antagonist with anti-cancer and anti-proliferative properties.
-
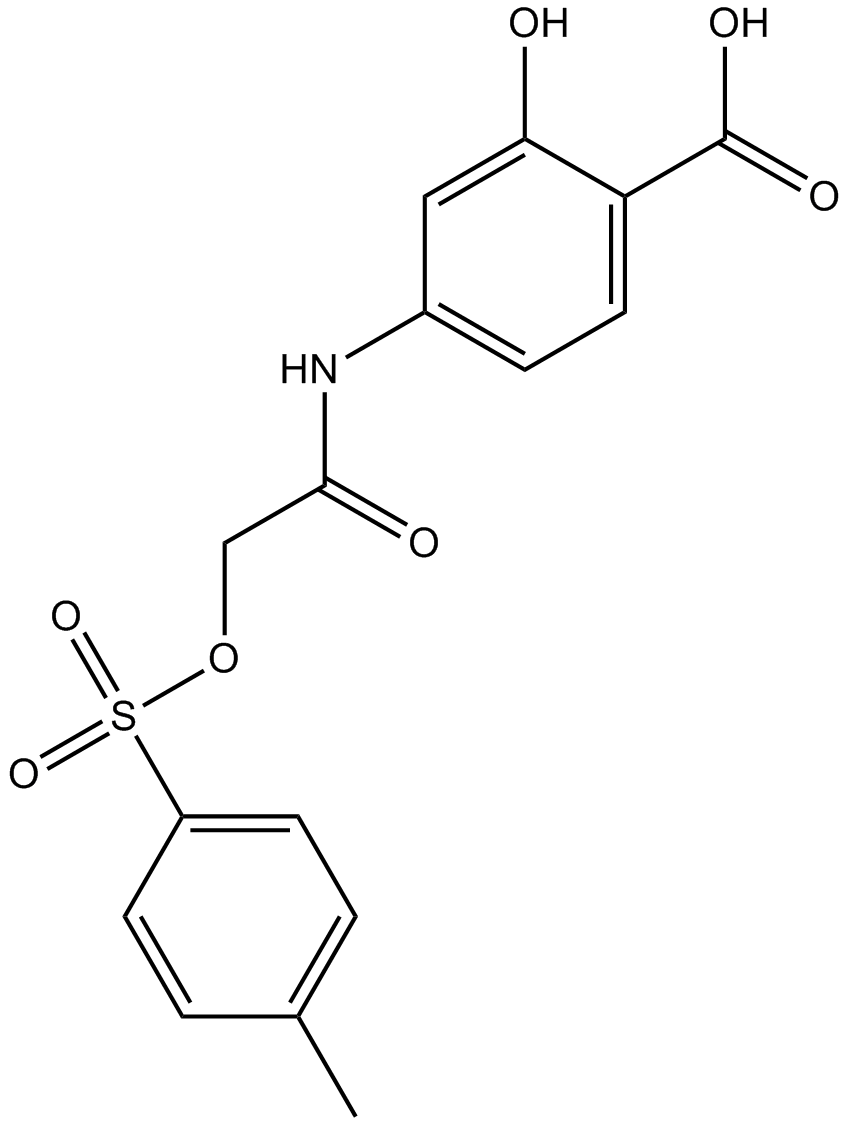
GC14653
NSC 74859
Stat3 inhibitor
-
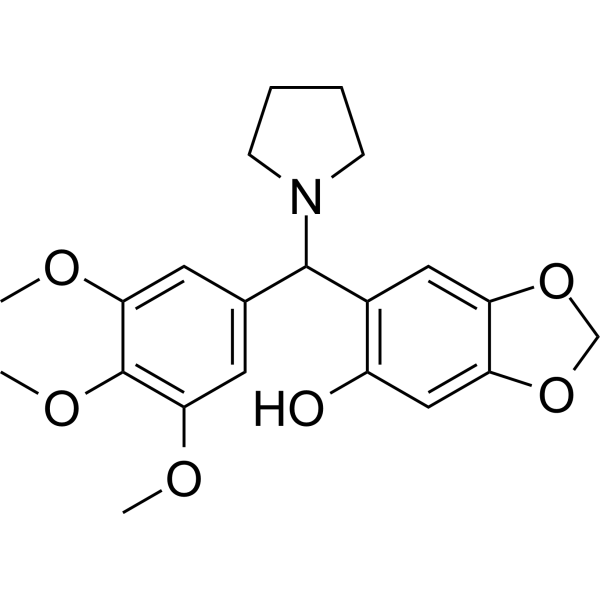
GC69594
NSC-370284
NSC-370284 is a selective inhibitor of both 10-11 translocation 1 (TET1) and 5-hydroxymethylcytosine (5hmC). It significantly inhibits TET1 expression levels by targeting STAT3/5.
-
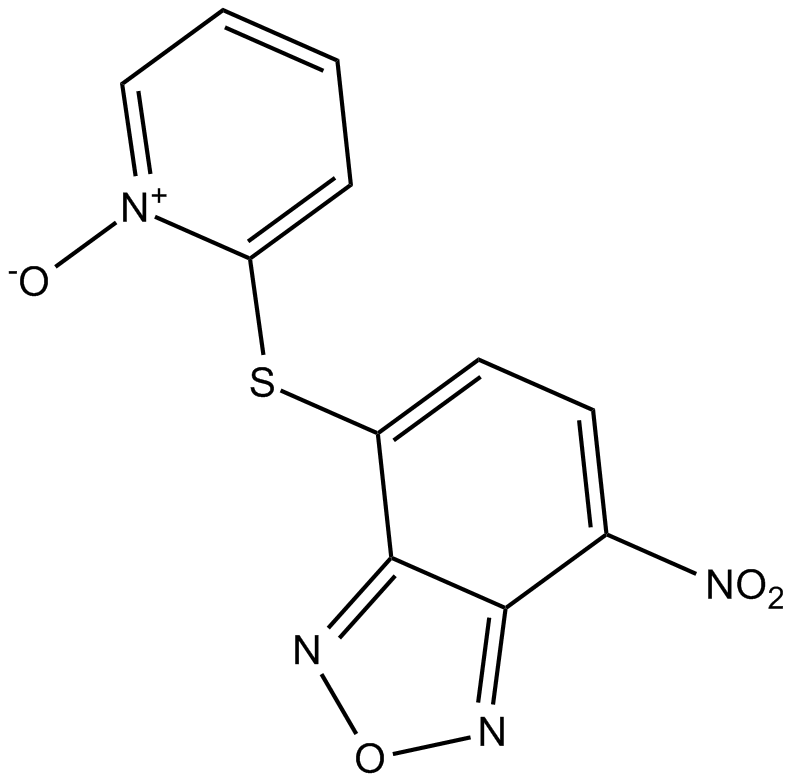
GC14103
NSC228155
EGFR activator
-
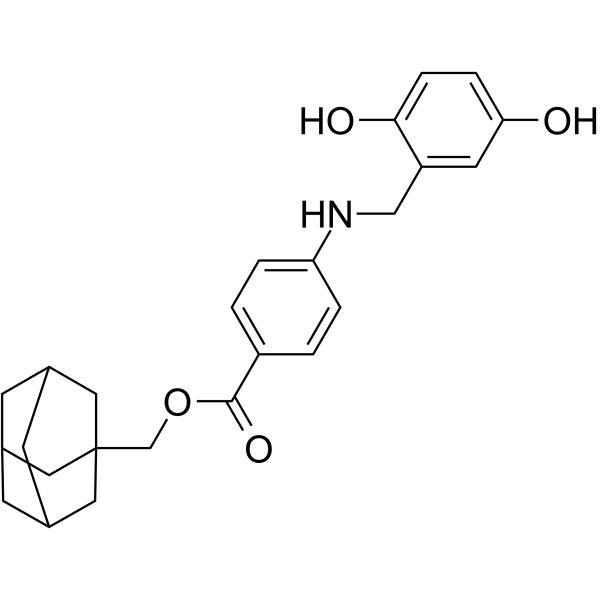
GC69596
NSC689857
NSC689857 is an effective inhibitor of EGFR and SCFSKP2, with an IC50 of 36 μM against Skp2-Cks1. NSC689857 can inhibit phosphorylation of p27 (IC50=30 μM). NSC689857 exhibits varying activity in different types of cancer, with higher resistance activity against leukemia cell lines compared to other cancer cells.
-
GC12712
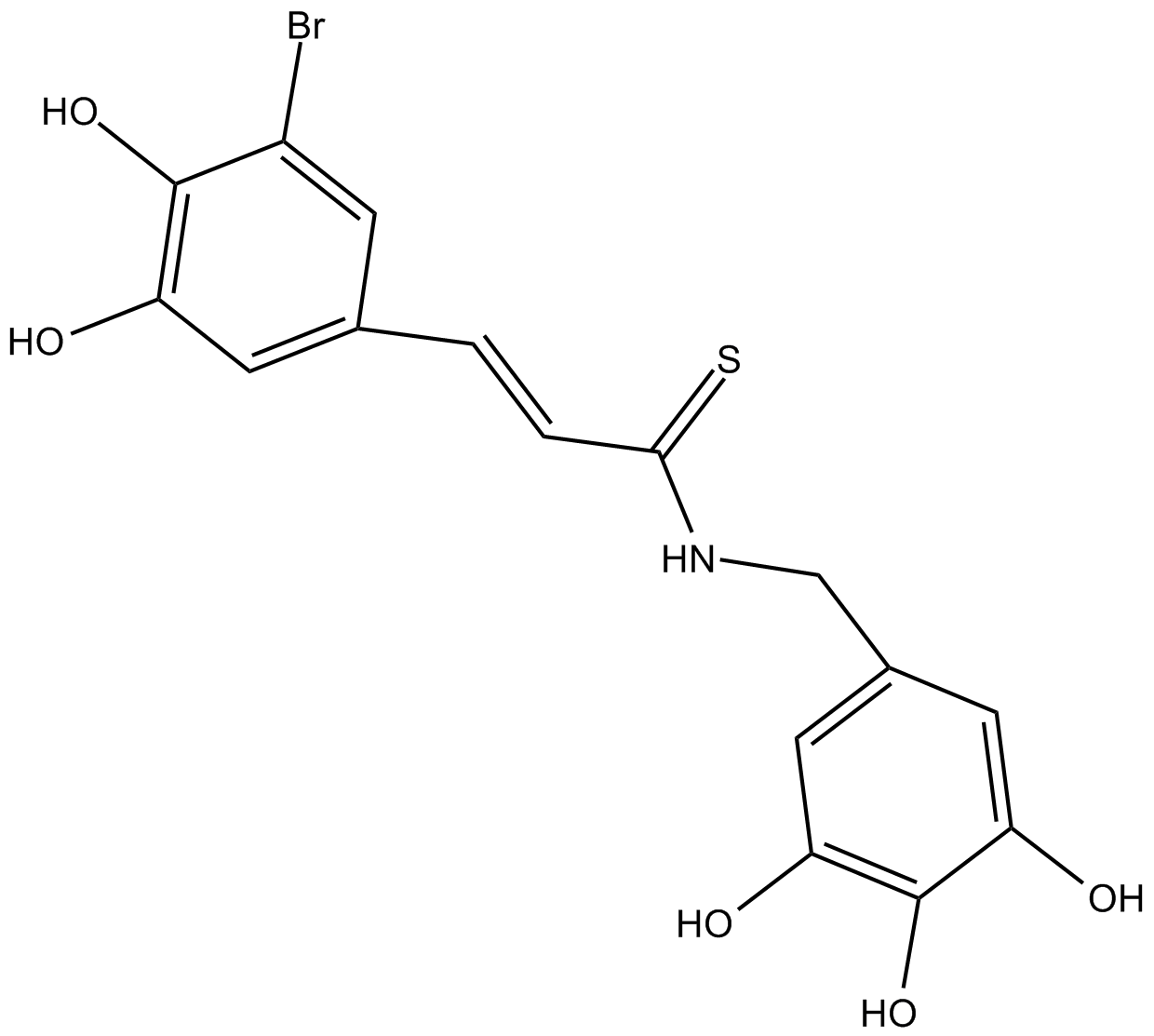
NT157
IRS-1/2 inhibitor, inhibits IGF-1R and STAT3 signaling pathway
-
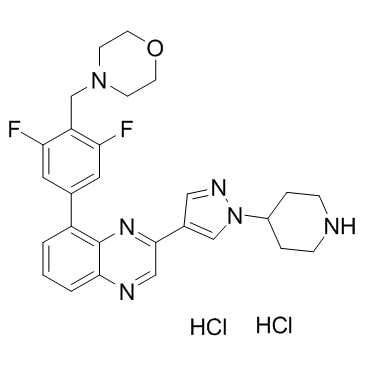
GC36783
NVP-BSK805 dihydrochloride
A potent, selective JAK2 inhibitor
-
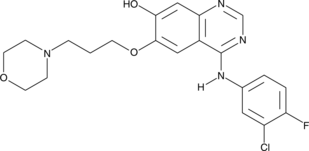
GC44491
O-Desmethyl Gefitinib
O-Desmethyl gefitinib is the major metabolite of gefitinib in human plasma, formed by the cytochrome P450 isoform CYP2D6.
-
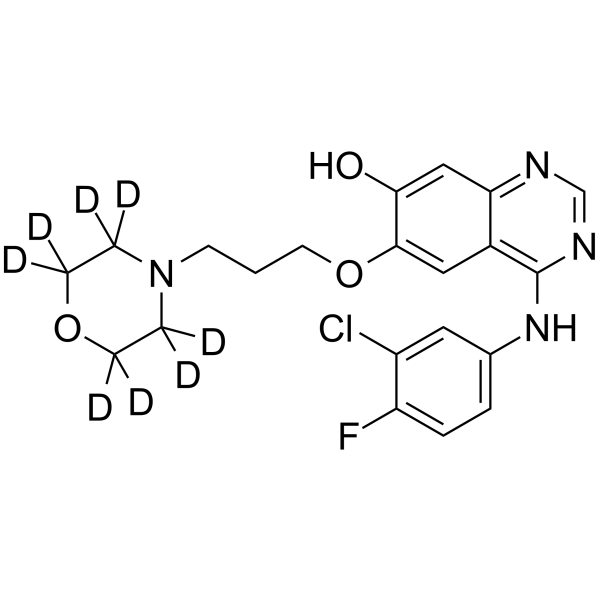
GC68321
O-Desmethyl gefitinib-d8
-
GC61627
Ochromycinone
Ochromycinone ((Rac)-STA-21) is a natural antibiotic and a STAT3 inhibitor.

-
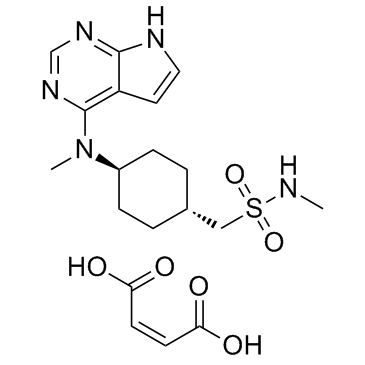
GC31677
Oclacitinib maleate (PF-03394197 maleate)
Oclacitinib maleate (PF-03394197 maleate) (PF-03394197 maleate) is a novel JAK inhibitor.
-
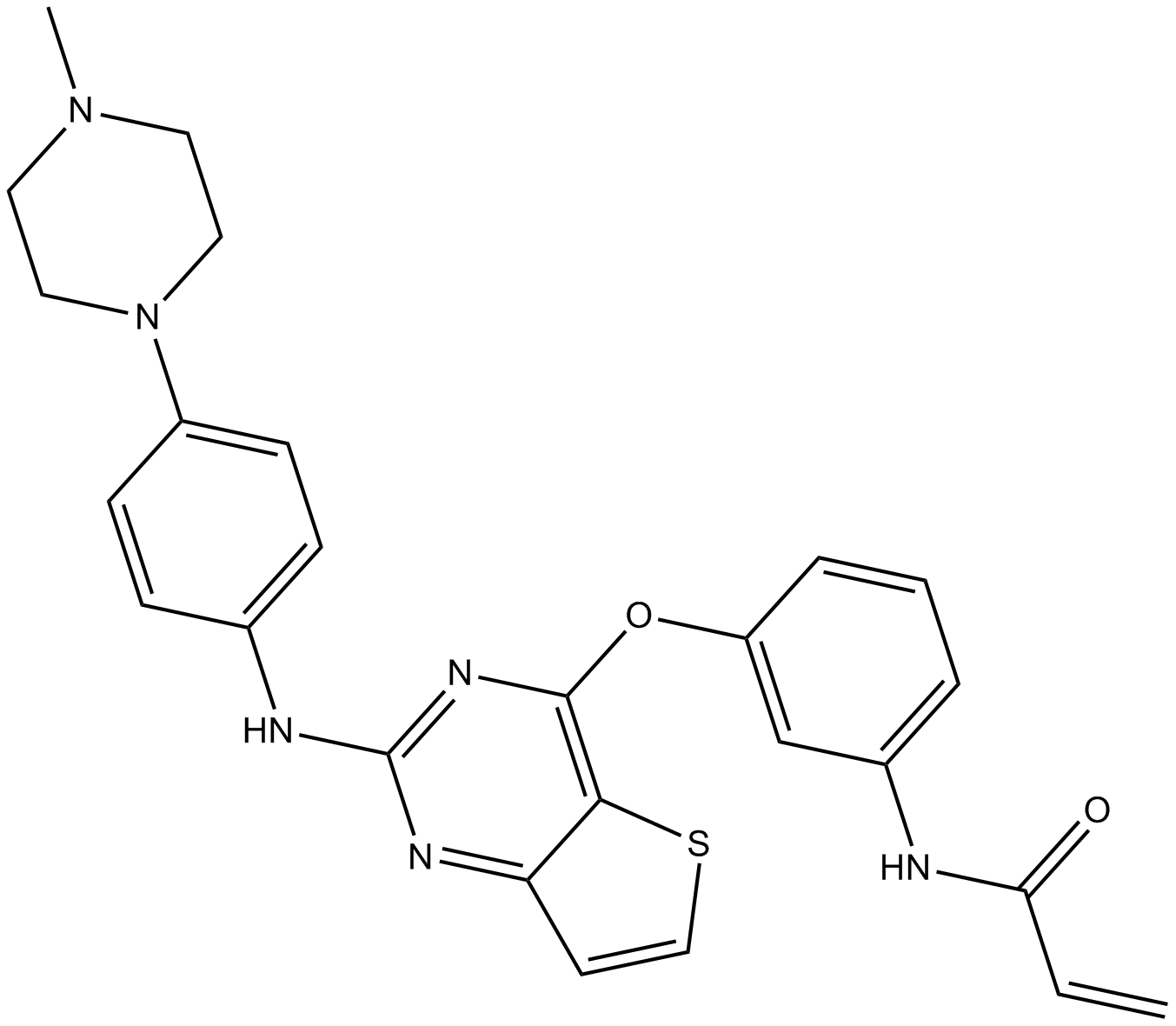
GC15370
Olmutinib (HM61713, BI 1482694)
Olmutinib (HM61713, BI 1482694) (HM61713; BI-1482694) is an orally active and irreversible third EGFR tyrosine kinase inhibitor that binds to a cysteine residue near the kinase domain. Olmutinib (HM61713, BI 1482694) is used for NSCLC.
-
GC64770
Oritinib
Oritinib (SH-1028), an irreversible third-generation EGFR TKI, overcomes T790M-mediated resistance in non-small cell lung cancer. Oritinib (SH-1028), a mutant-selective inhibitor of EGFR kinase activity, inhibits EGFRWT, EGFRL858R, EGFRL861Q, EGFRL858R/T790M, EGFRd746-750 and EGFRd746-750/T790M kinases, with IC50s of 18, 0.7, 4, 0.1, 1.4 and 0.89 nM, respectively.

-
GC17530
OSI-420
OSI-420 (OSI-420) is an active metabolite of Erlotinib. Erlotinib is a potent EGFR tyrosin kinase inhibitor.
-
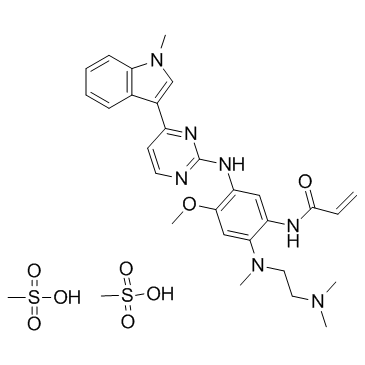
GC36819
Osimertinib dimesylate
Osimertinib dimesylate (AZD-9291 dimesylate) is an irreversible and mutant selective EGFR inhibitor with IC50s of 12 and 1 nM against EGFRL858R and EGFRL858R/T790M, respectively.
-
GC69635
OSM-SMI-10B
OSM-SMI-10B is a derivative of OSM-SMI-10. Co-incubation of OSM-SMI-10B with Oncostatin M (OSM) significantly reduces STAT3 phosphorylation induced by OSM in cancer cells.

-
GC41329
Pacritinib
FMS-like tyrosine kinase 3 (FLT3) and Janus kinase 2 (JAK2) are tyrosine kinases that mediate cytokine signaling and are frequently mutated in cancers, particularly acute myeloid leukemia.
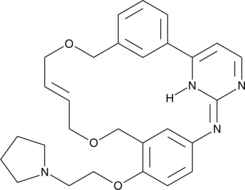
-
GC66405
Panitumumab
Panitumumab (ABX-EGF) is a fully human IgG2 anti-EGFR monoclonal antibody with anti-tumor activity. Panitumumab inhibits tumor cell proliferation, survival and angiogenesis. Panitumumab can be used in the research of cancers, such as colon cancer.

-
GC66333
Panitumumab (anti-EGFR)
Panitumumab (anti-EGFR) is a fully human IgG2 anti-EGFR monoclonal antibody with anti-tumor activity. Panitumumab (anti-EGFR) inhibits tumor cell proliferation, survival and angiogenesis. Panitumumab (anti-EGFR) can be used in the research of cancers, such as colon cancer.

-
GC69665
Patritumab
Patritumab (Human Anti-ERBB3 Recombinant Antibody) is a neutralizing monoclonal antibody targeting ERBB3. Patritumab has synergistic effects with Cetuximab and can effectively inhibit the phosphorylation of EGFR, HER2, HER3, ERK and AKT. Patritumab also induces apoptosis and inhibits the growth of pancreatic, non-small cell lung cancer and colorectal cancer xenograft tumors.

-
GC15925
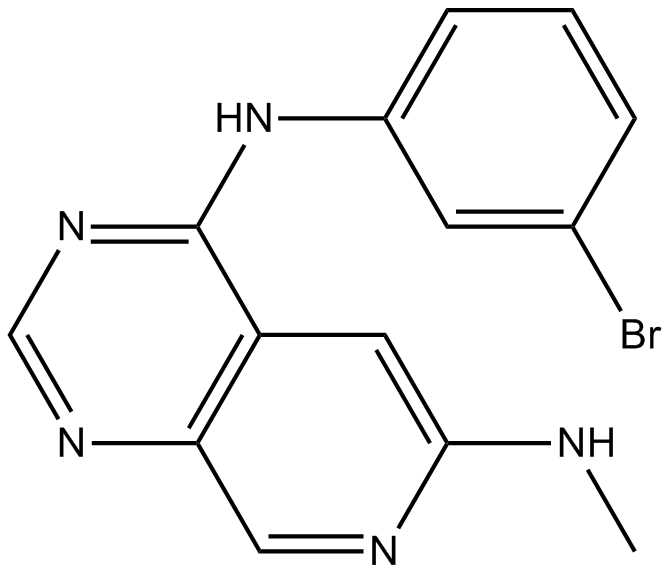
PD 158780
ErbB receptor family tyrosine kinase inhibitor
-
GC34105
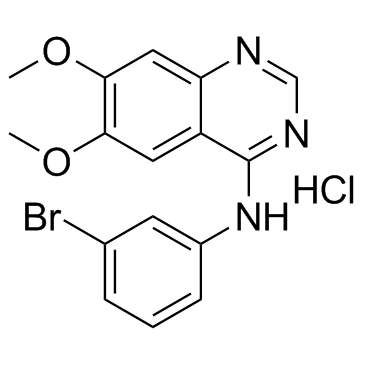
PD153035 Hydrochloride (ZM 252868)
A highly potent EGFR inhibitor
-
GC11015
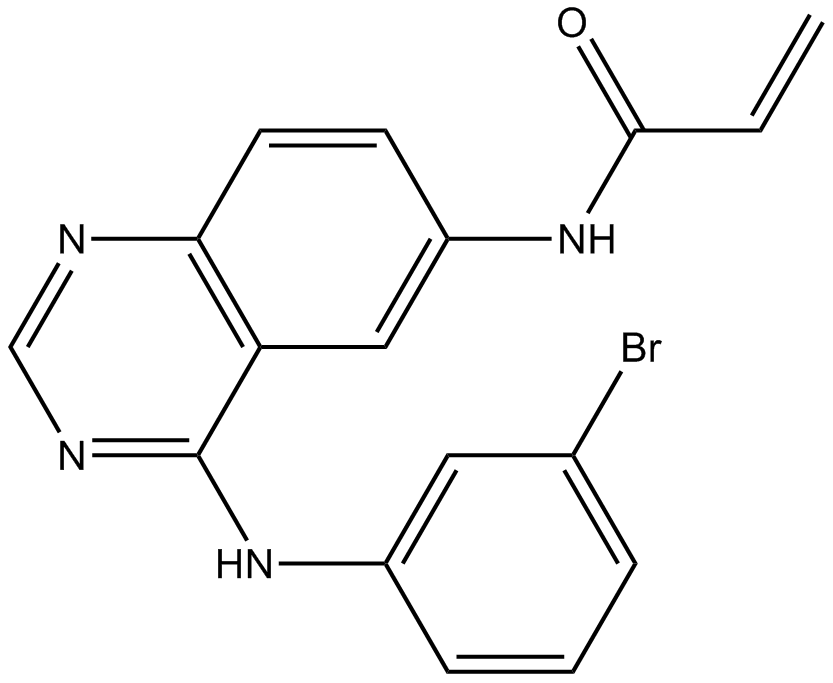
PD168393
EGFR inhibitor
-
GC10341
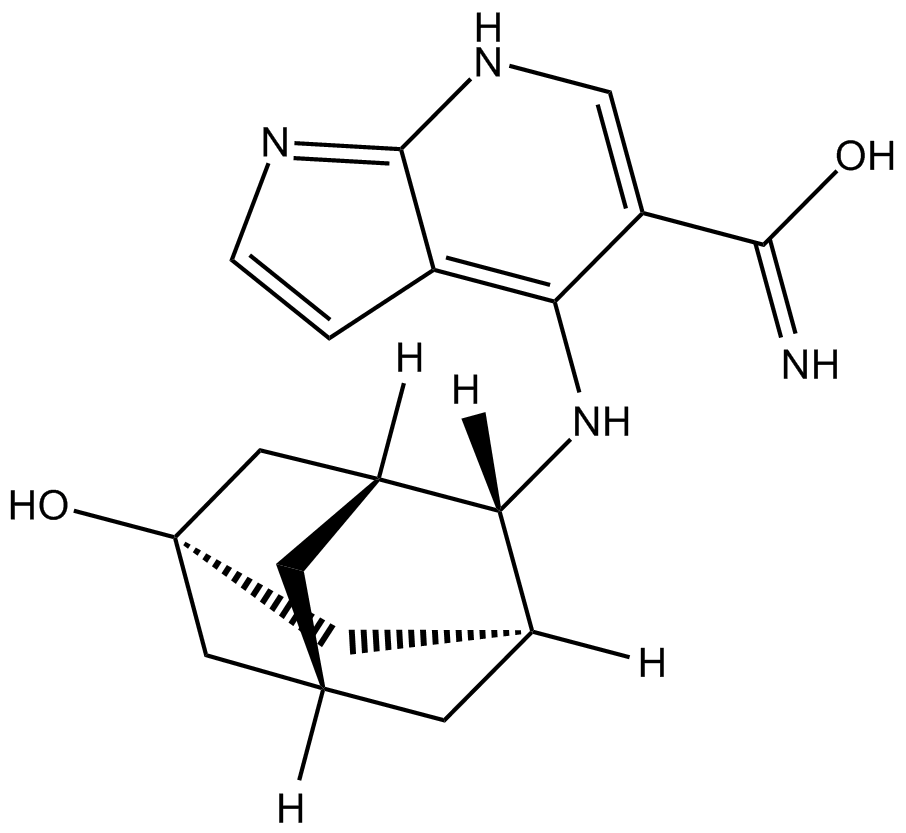
Peficitinb (ASP015K, JNJ-54781532)
Peficitinb (ASP015K, JNJ-54781532) (ASP015K) is an orally active JAK inhibitor, with IC50s of 3.9, 5.0, 0.7 and 4.8 nM for JAK1, JAK2, JAK3 and Tyk2, respectively.
-
GC17473
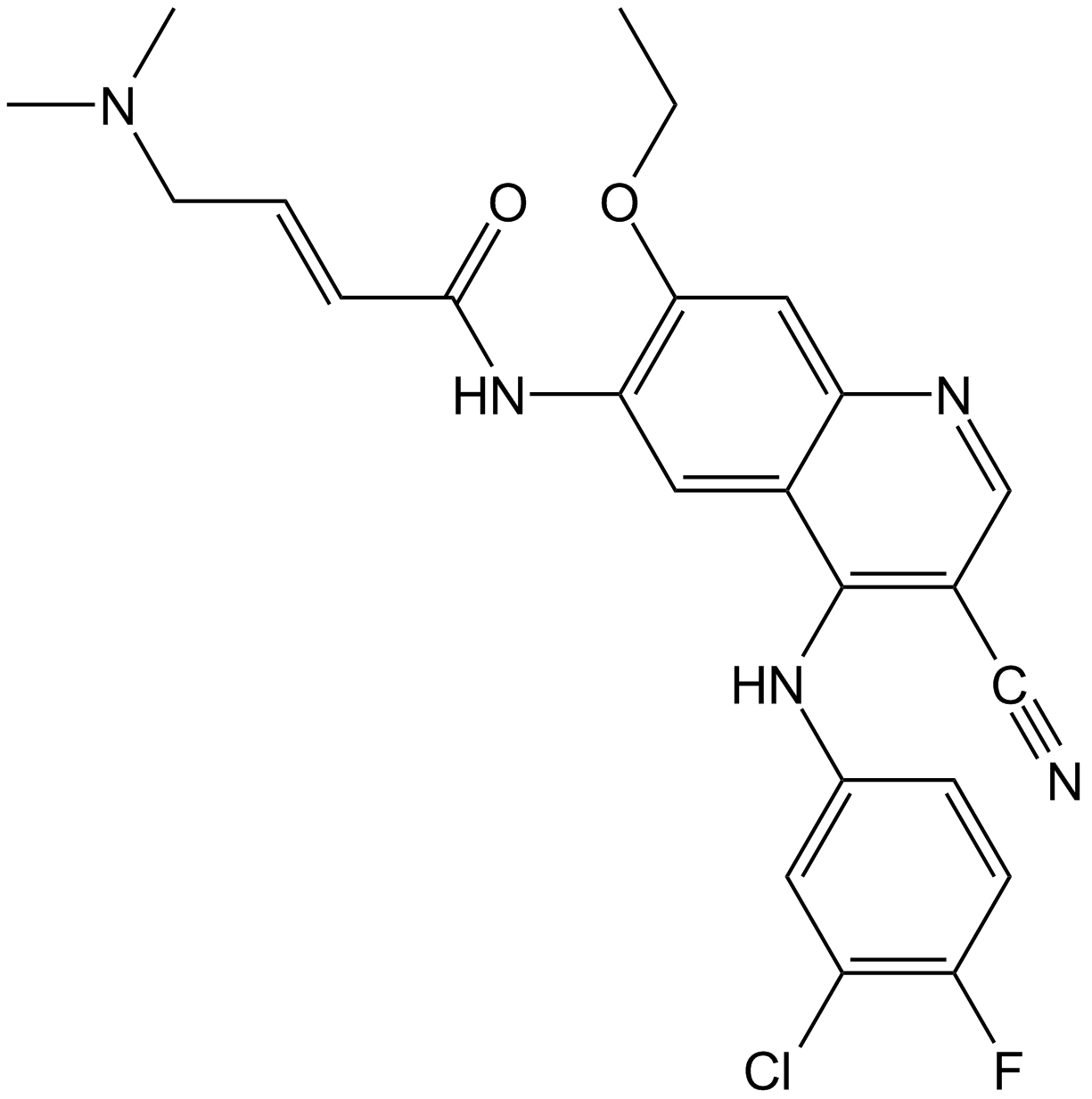
Pelitinib (EKB-569)
Pelitinib (EKB-569) (EKB-569;WAY-EKB 569) is an irreversible inhibitor of EGFR with an IC50 of 38.5 nM; also slightly inhibits Src, MEK/ERK and ErbB2 with IC50s of 282, 800, and 1255 nM, respectively.
-
GC34210
Pertuzumab (Anti-Human HER2, Humanized Antibody)
Pertuzumab (Anti-Human HER2, Humanized Antibody), the first of a new class of agents designated as HER dimerisation inhibitors, is a humanised IgG1 monoclonal antibody (mAb) that sterically binds domain II of the erbB2 receptor .

-
GC69690
Petosemtamab
Petosemtamab (MCLA 158) is a monoclonal antibody (mAb) that targets both EGFR (Kd: 0.22 nM) and LGR5 (Kd: 0.86 nM). Petosemtamab blocks EGFR signaling and receptor degradation in LGR5+ cancer cells. It can be used for research on solid tumors such as head and neck squamous cell carcinoma (HNSCC), metastatic colorectal cancer (CRC), etc.

-
GC14938
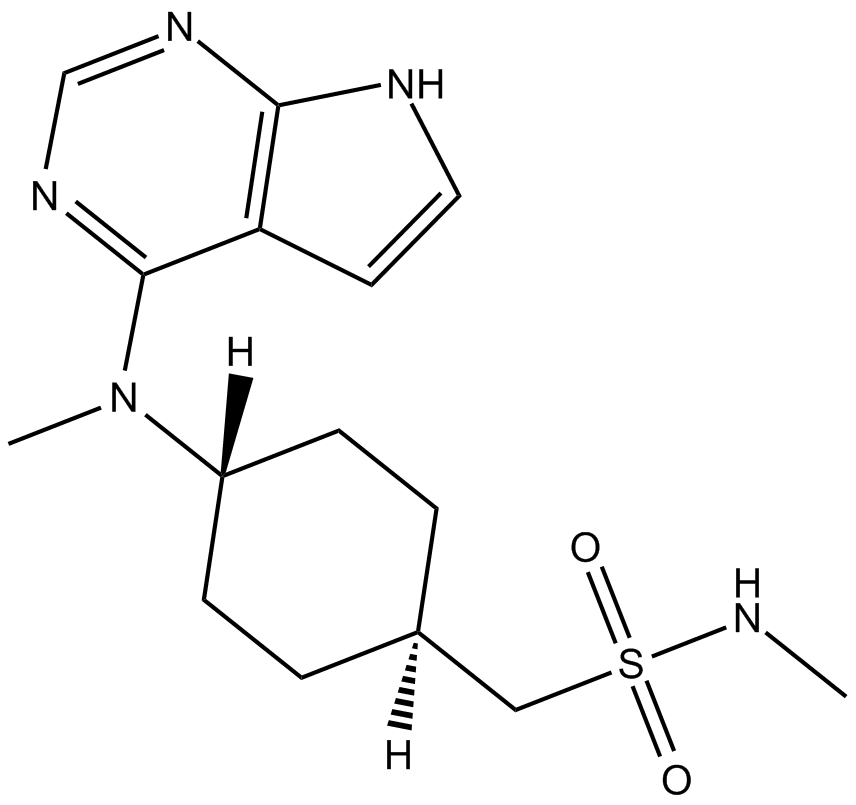
PF-03394197(Oclacitinib)
Novel Janus kinase inhibitor
-
GC36882
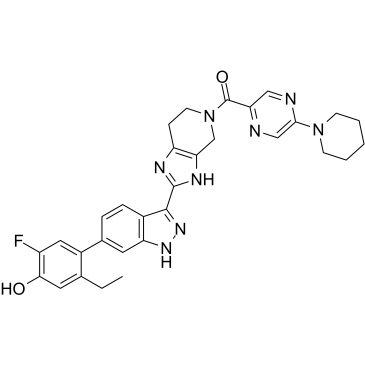
PF-06263276
PF-06263276 (PF 6263276) is a potent and selective pan-JAK inhibitor, with IC50s of 2.2 nM, 23.1 nM, 59.9 nM and 29.7 nM for JAK1, JAK2, JAK3 and TYK2, respectively.
-
GC32927
PF-06459988
PF-06459988 is an orally activity, irreversible and mutant-selective inhibitor of EGFR mutant forms. PF-06459988 demonstrates high potency and specificity to the T790M-containing double mutant EGFRs. PF-06459988 can be used for the research of cancer.
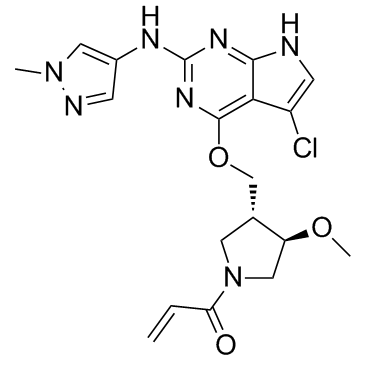
-
GC19288
PF-06651600
PF-06651600 (PF-06651600) is an orally active and selective JAK3 inhibitor with an IC50 of 33.1 nM.
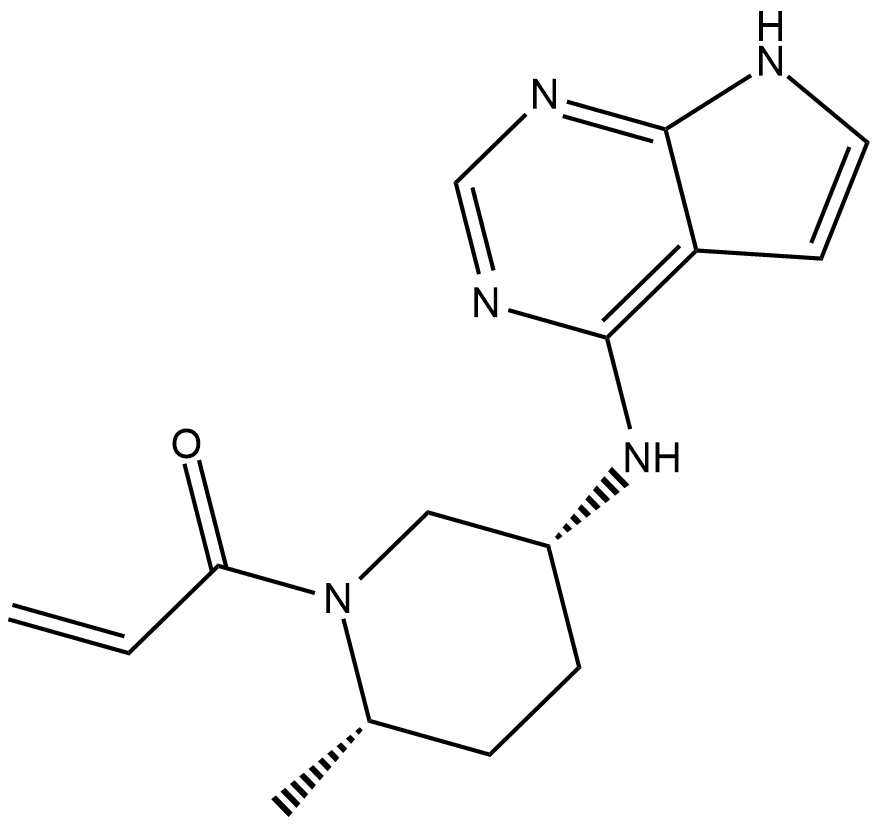
-
GC19405
PF-06700841 P-Tosylate
Brepocitinib (PF-06700841) P-Tosylate is a potent dual Janus kinase 1 (JAK1) and TYK2 inhibitor with IC50s of 17 nM and 23 nM, respectively.
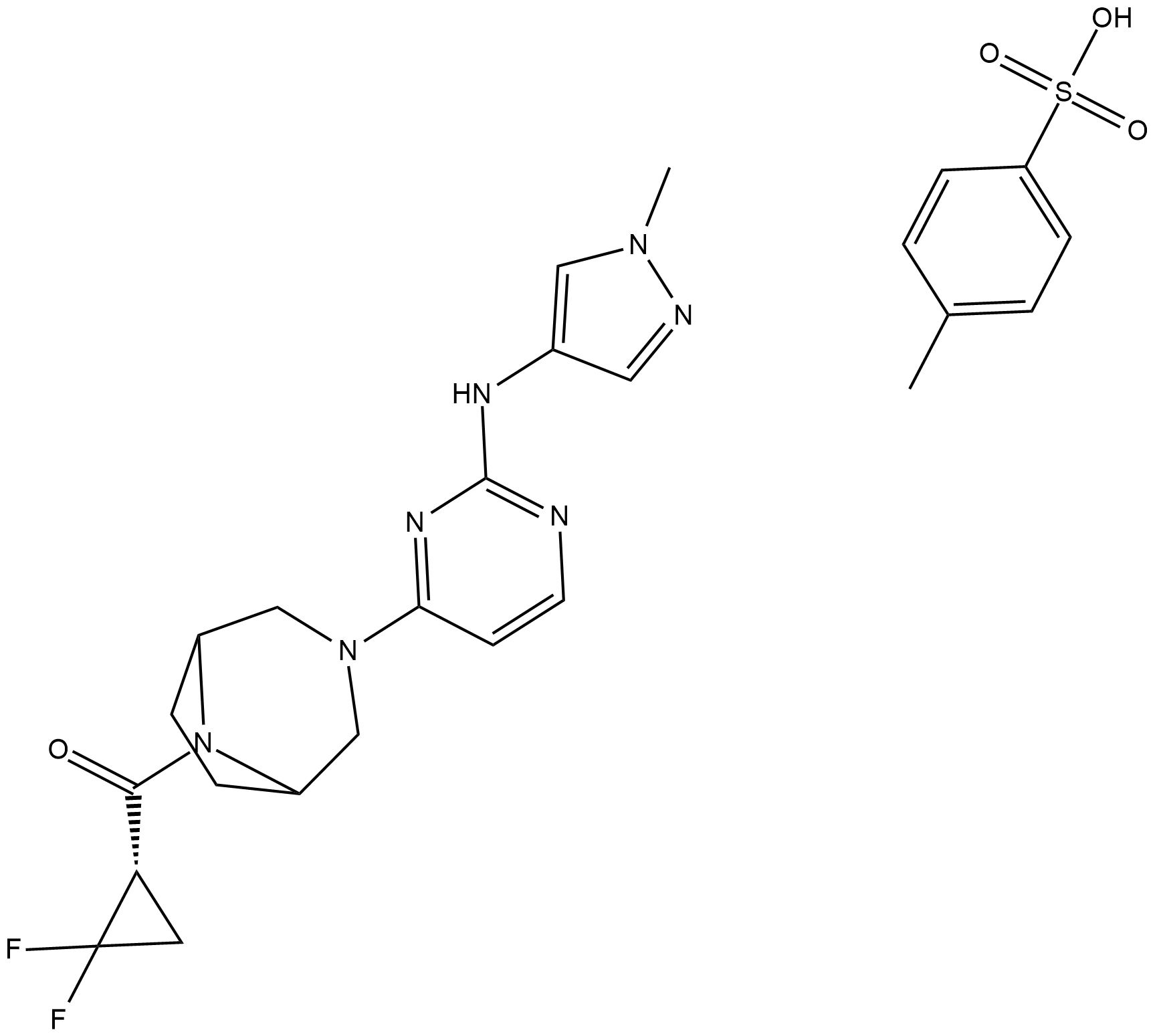
-
GN10314
Picroside I
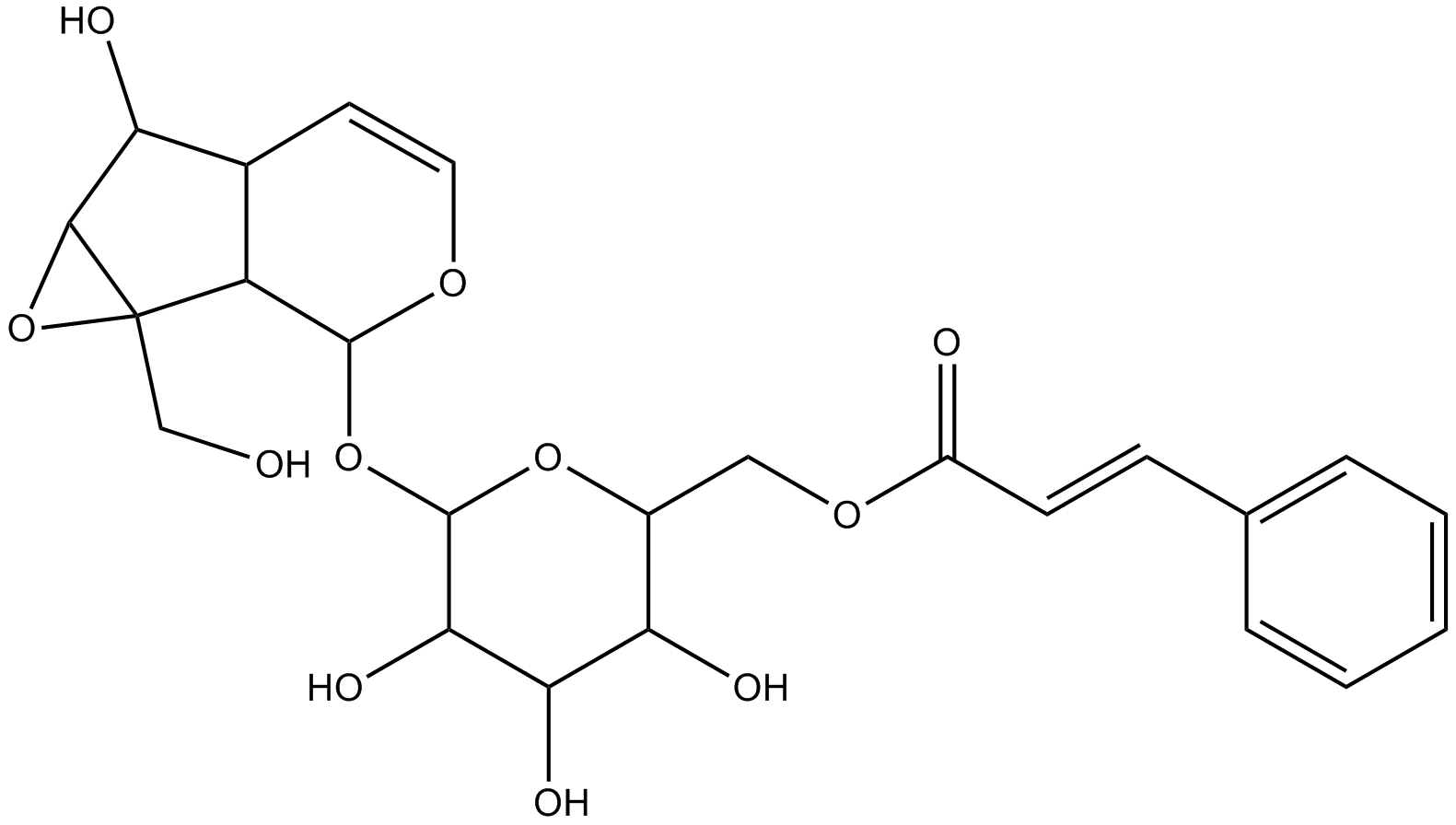
-
GC19419
PIM inhibitor 1 phosphate
Uzansertib (INCB053914) phosphate is an orally active, ATP-competitive pan-PIM kinase inhibitor with IC50s of 0.24 nM, 30 nM, 0.12 nM for PIM1, PIM2, PIM3, respectively. PIM inhibitor 1 phosphate has broad anti-proliferative activity against a variety of hematologic tumor cell lines.

-
GC36918
PIM-447 dihydrochloride
A pan-Pim kinase inhibitor

-
GC65278
PIM1-IN-1
PIM1-IN-1 is a potent and highly selective PIM1/3 inhibitor, with IC50s of 7, 5530 and 70 nM for PIM1, PIM2, and PIM3, respectively, inhibits the phosphorylation of BAD, a downstream target of PIM, with an EC50 of 262 nM. PIM1-IN-1 shows no obvious effect on FLT3 or hERG binding. Antiproliferative and anti-cancer activity.
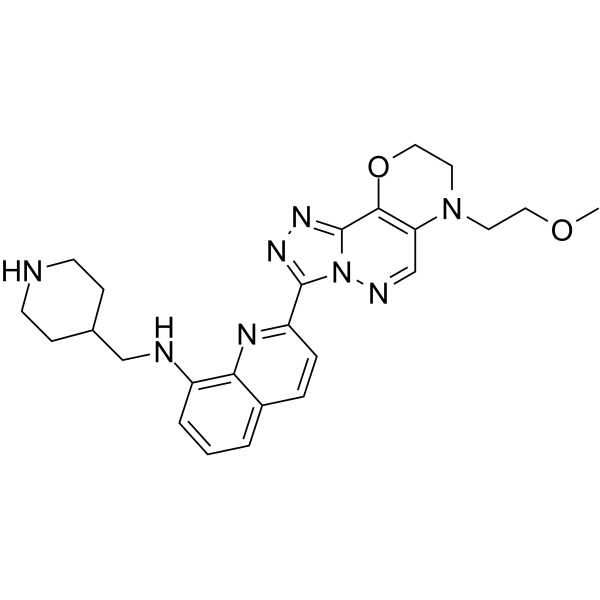
-
GC19398
PIM447
PIM447 is novel pan-PIM kinase inhibitor, including Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase.
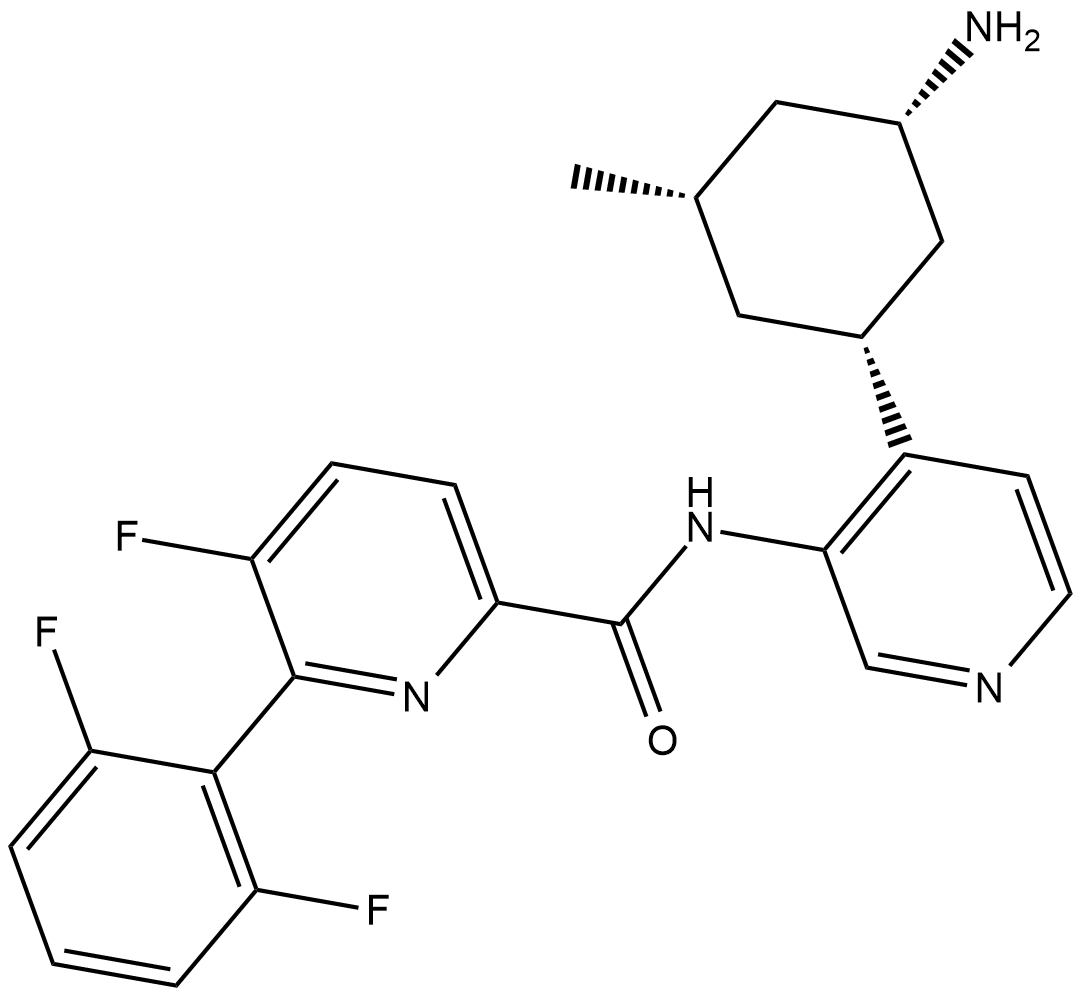
-
GC10643
Pimozide
dopamine receptors inhibitor

-
GC45756
Pimozide-d4
An internal standard for the quantification of pimozide
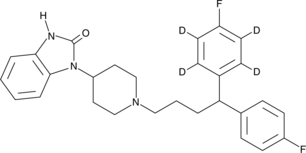
-
GC40915
PKI-166
An inhibitor of EGFR
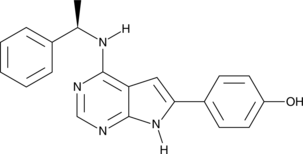
-
GC69728
Ponezumab
Ponezumab (PF-04360365) is a humanized monoclonal antibody against amyloid beta protein of the IgG2 class. Ponezumab can reduce Aβ levels in the central nervous system and improve performance in various learning and memory models in mice. Ponezumab can be used for research on Alzheimer's disease.

-
GC64701
Povorcitinib
Povorcitinib is a potent and selective inhibitor of JAK1.
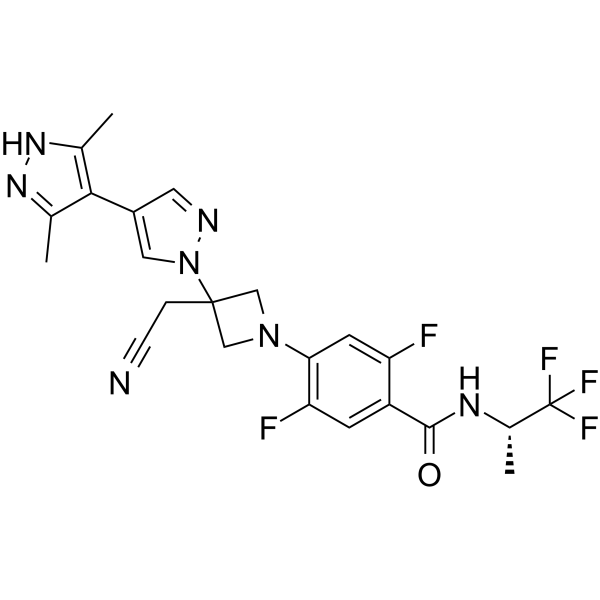
-
GC64700
Povorcitinib phosphate
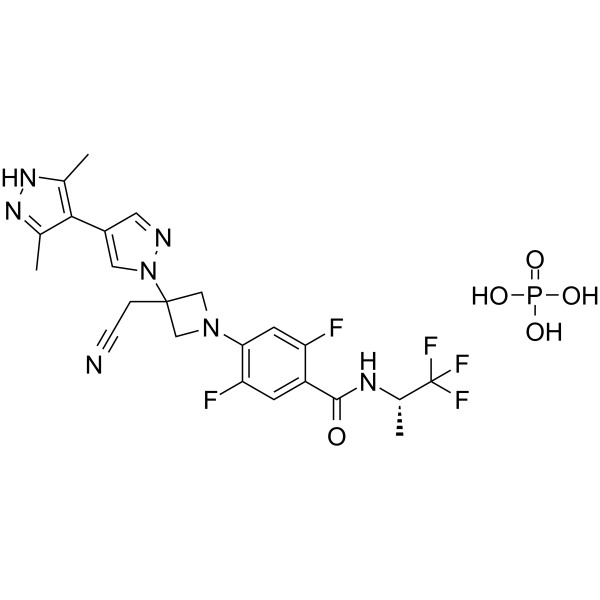
-
GC17916
Poziotinib
A irreversible pan-HER inhibitor
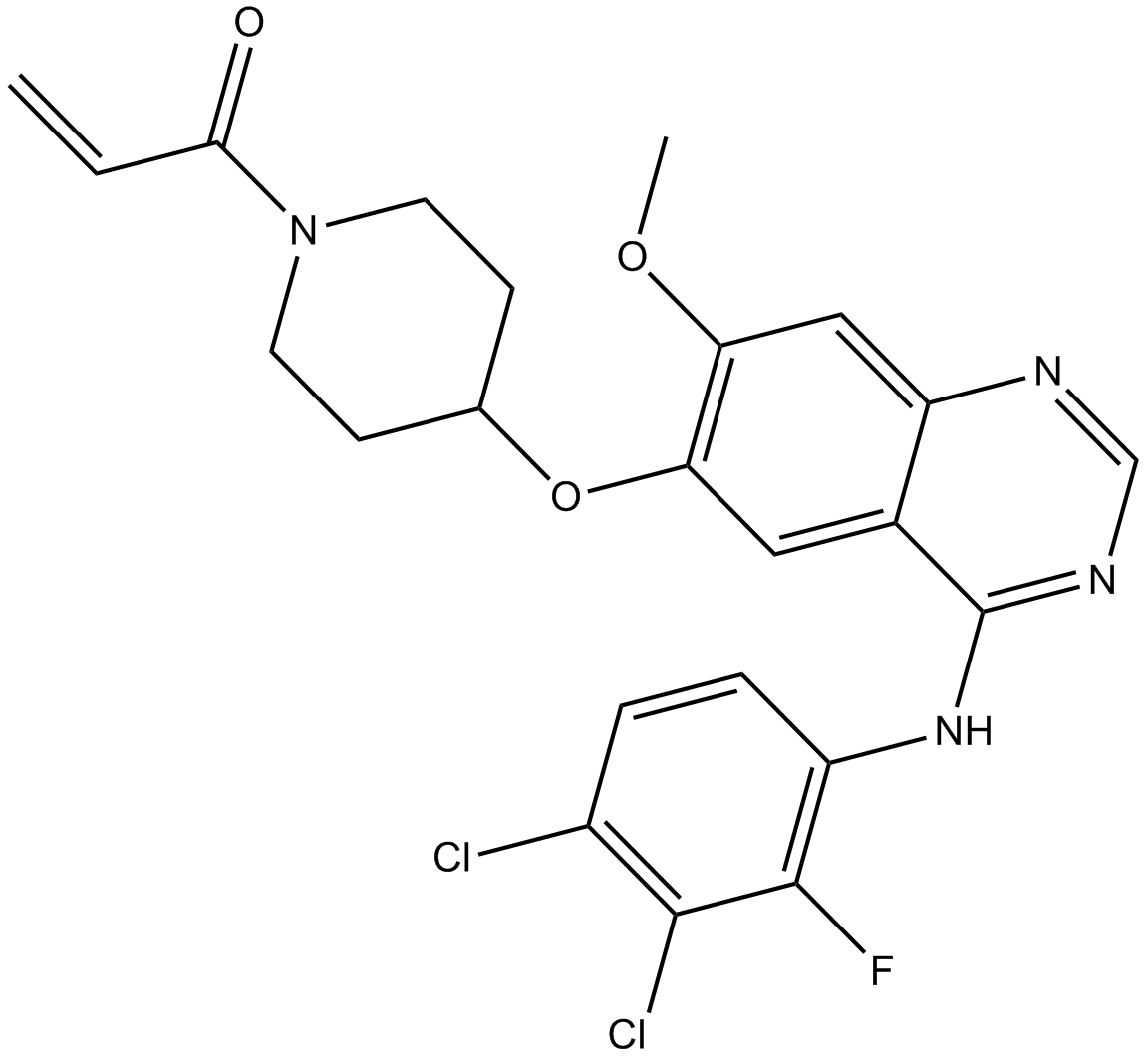
-
GC30601
Propanamide
Propanamide is a potent and selective JAK inhibitor extracted from patent WO2012122452A1, Compound II, has the potential for the skin disorders (such as cutaneous lupus) treatment.
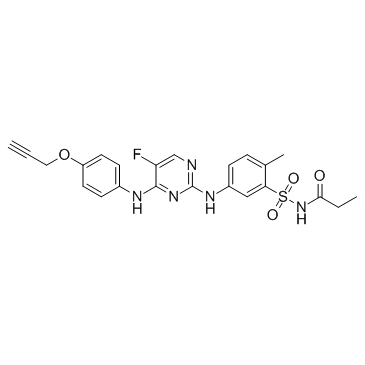
-
GC34742
Protosappanin A
Protosappanin A (PTA), an immunosuppressive ingredient and major biphenyl compound isolated from Caesalpinia sappan L, suppresses JAK2/STAT3-dependent inflammation pathway through down-regulating the phosphorylation of JAK2 and STAT3.
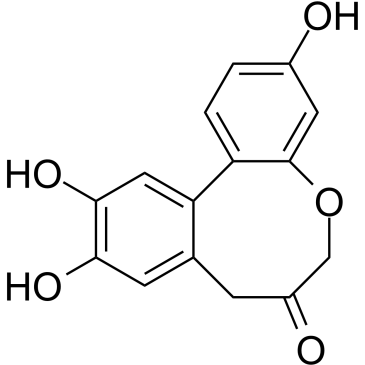
-
GC69780
Pumecitinib
Pumecitinib is a JAK inhibitor with anti-inflammatory activity.
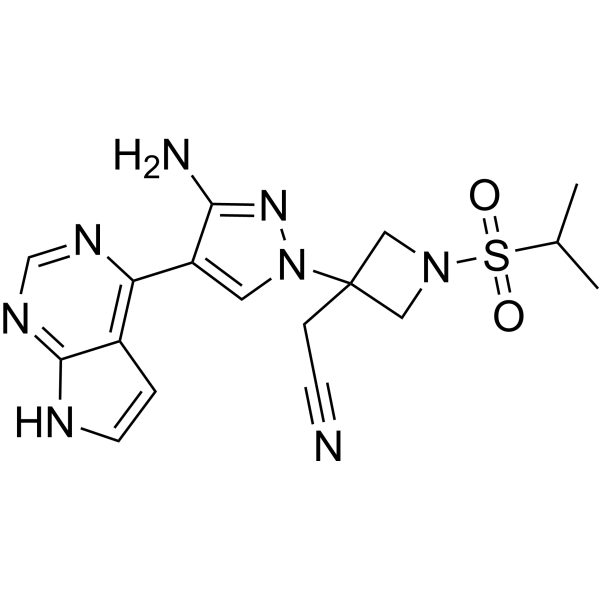
-
GC32733
Pyrotinib (SHR-1258)
Pyrotinib (SHR-1258) (SHR-1258) is a potent and selective EGFR/HER2 dual inhibitor with IC50s of 13 and 38 nM, respectively.
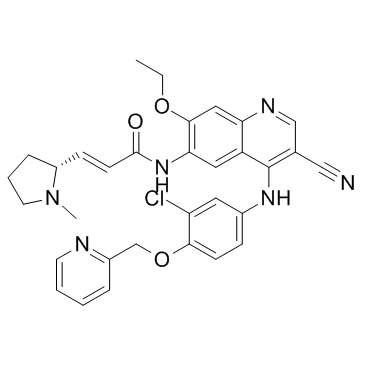
-
GC32989
Pyrotinib dimaleate (SHR-1258 dimaleate)
Pyrotinib dimaleate (SHR-1258 dimaleate) (SHR-1258 dimaleate) is a potent and selective EGFR/HER2 dual inhibitor with IC50s of 13 and 38 nM, respectively.
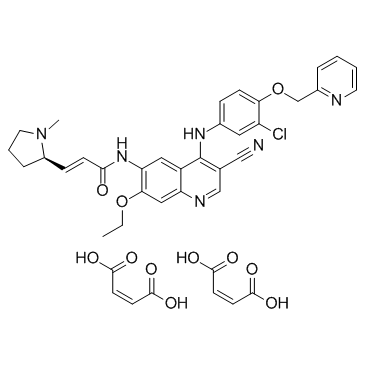
-
GC65201
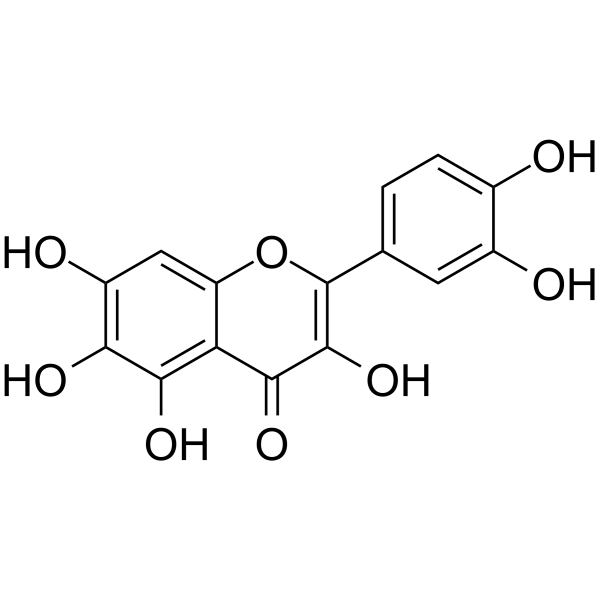
Quercetagetin
Quercetagetin (6-Hydroxyquercetin) is a flavonoid.
-
GC39081
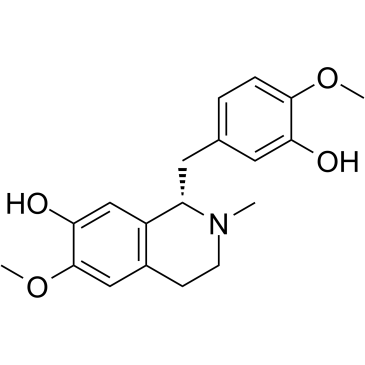
Reticuline
Reticuline shows anti-inflammatory effects through JAK2/STAT3 and NF-κB signaling pathways.
-
GC12038
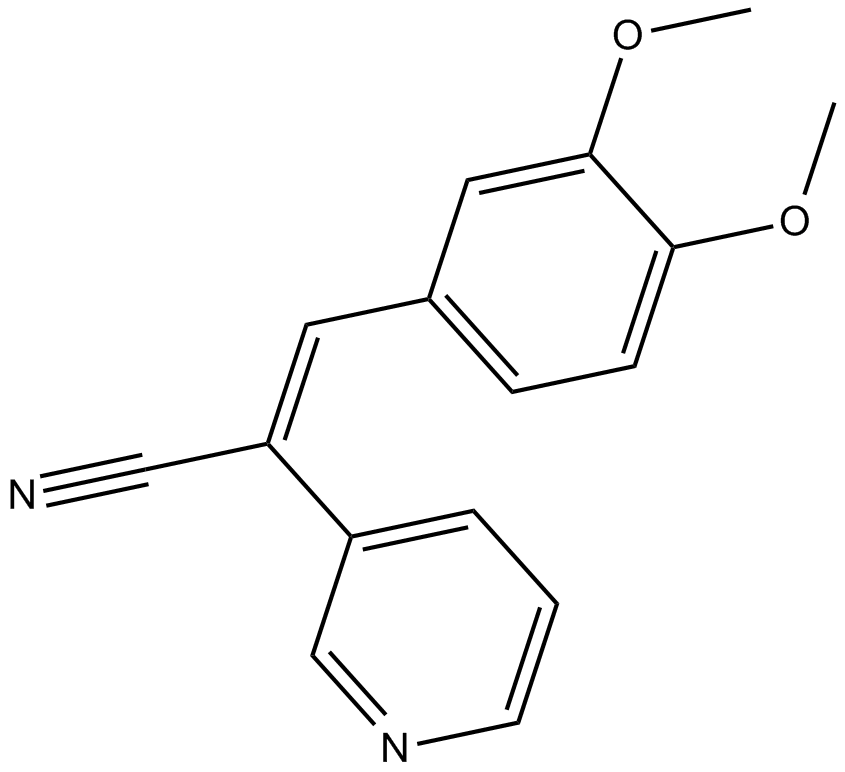
RG 13022
EGFR tyrosine kinase inhibitor
-
GC10217
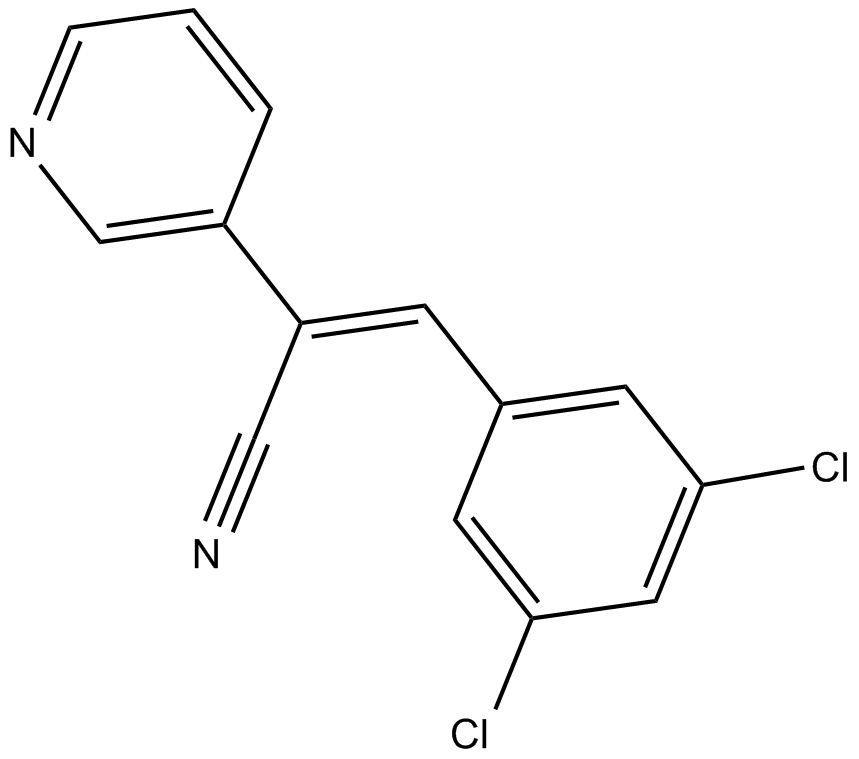
RG-14620
inhibitor of epidermal growth factor (EGF) receptor kinase
-
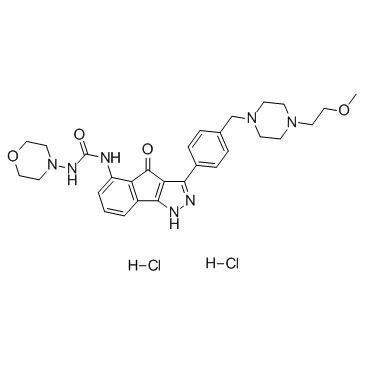
GC37522
RGB-286638
A multi-kinase inhibitor
-
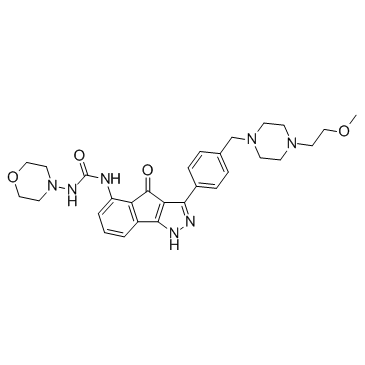
GC37523
RGB-286638 free base
A multi-kinase inhibitor
-
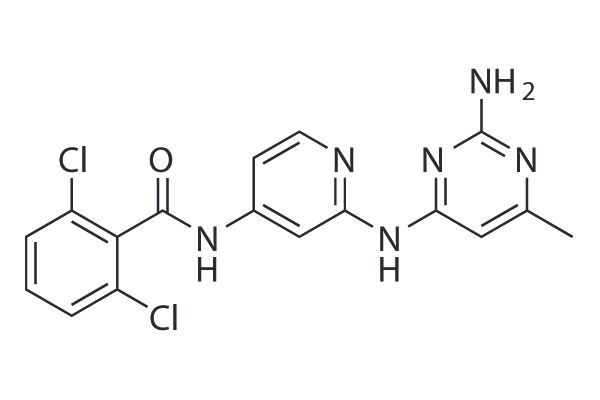
GC25869
RO495
RO495 (CS-2667) is a potent inhibitor of Non-receptor tyrosine-protein kinase 2 (TYK2).
-
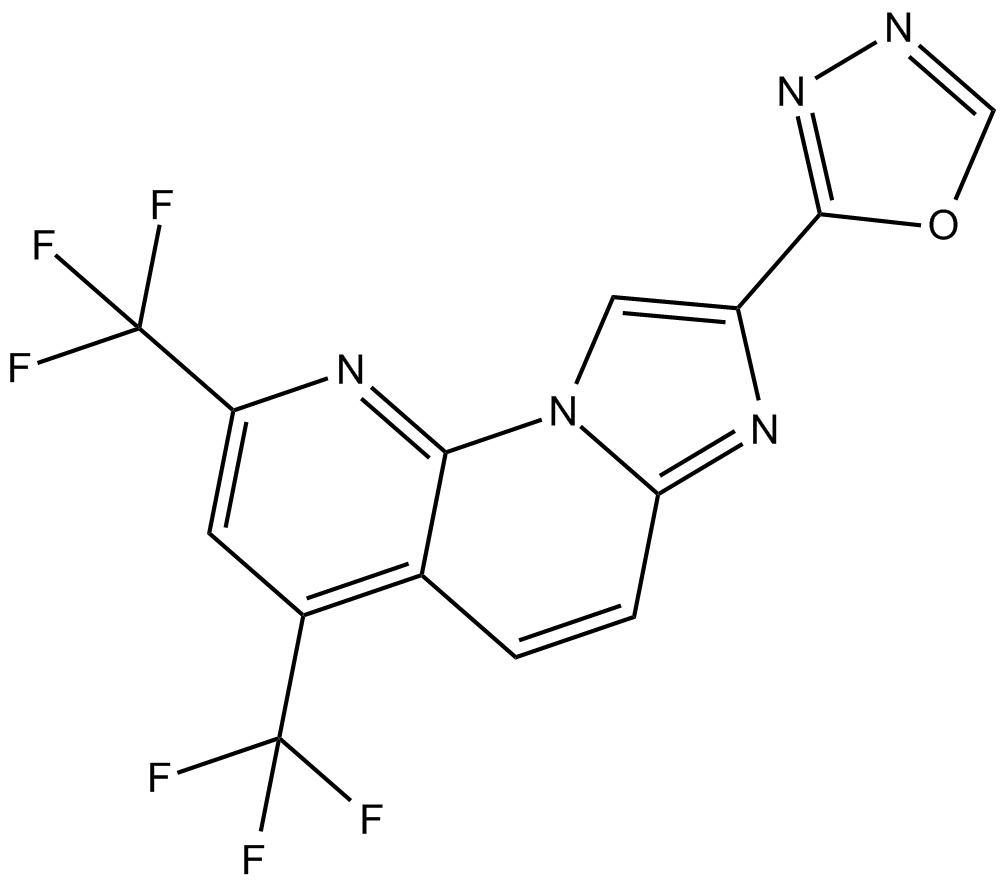
GC16956
RO8191
IFN-α receptor 2 agonist
-
GC33061
Rociletinib hydrobromide (CO-1686 (hydrobromide))
Rociletinib hydrobromide (CO-1686 (hydrobromide)) (CO-1686 hydrobromide) is an orally delivered kinase inhibitor that specifically targets the mutant forms of EGFR including T790M, and the Ki values for EGFRL858R/T790M and EGFRWT are 21.5 nM and 303.3 nM, respectively.
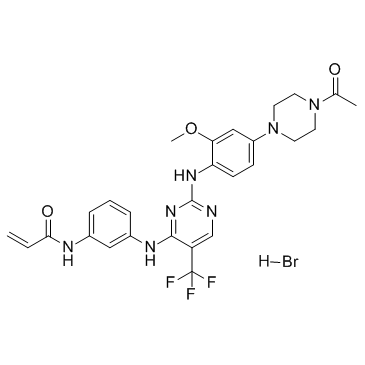
-
GC37568
RTC-5
RTC-5 (TRC-382) is an optimized phenothiazine with anti-cancer potency. RTC-5 demonstrates efficacy against a xenograft model of an EGFR driven cancer, its effects is attributed to concomitant negative regulation of PI3K-AKT and RAS-ERK signaling.

-
GC69843
Ruserontinib
Ruserontinib (SKLB1028) is an orally active inhibitor of EGFR, FLT3, and Abl kinases with an IC50 value of 55 nM against human FLT3. It has anti-tumor activity.
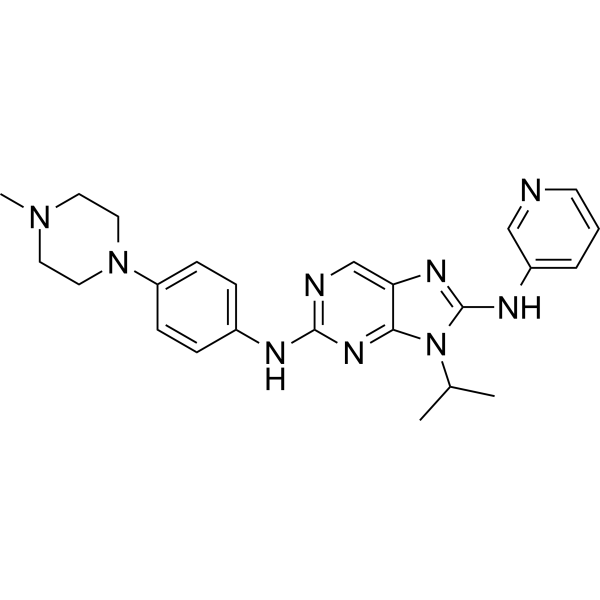
-
GC37575
Ruxolitinib sulfate
Ruxolitinib sulfate (INCB018424 sulfate) is the first potent, selective JAK1/2 inhibitor to enter the clinic with IC50s of 3.3 nM/2.8 nM, and has > 130-fold selectivity for JAK1/2 versus JAK3.
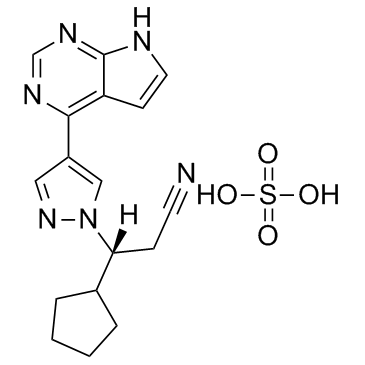
-
GN10164
Saikosaponin D
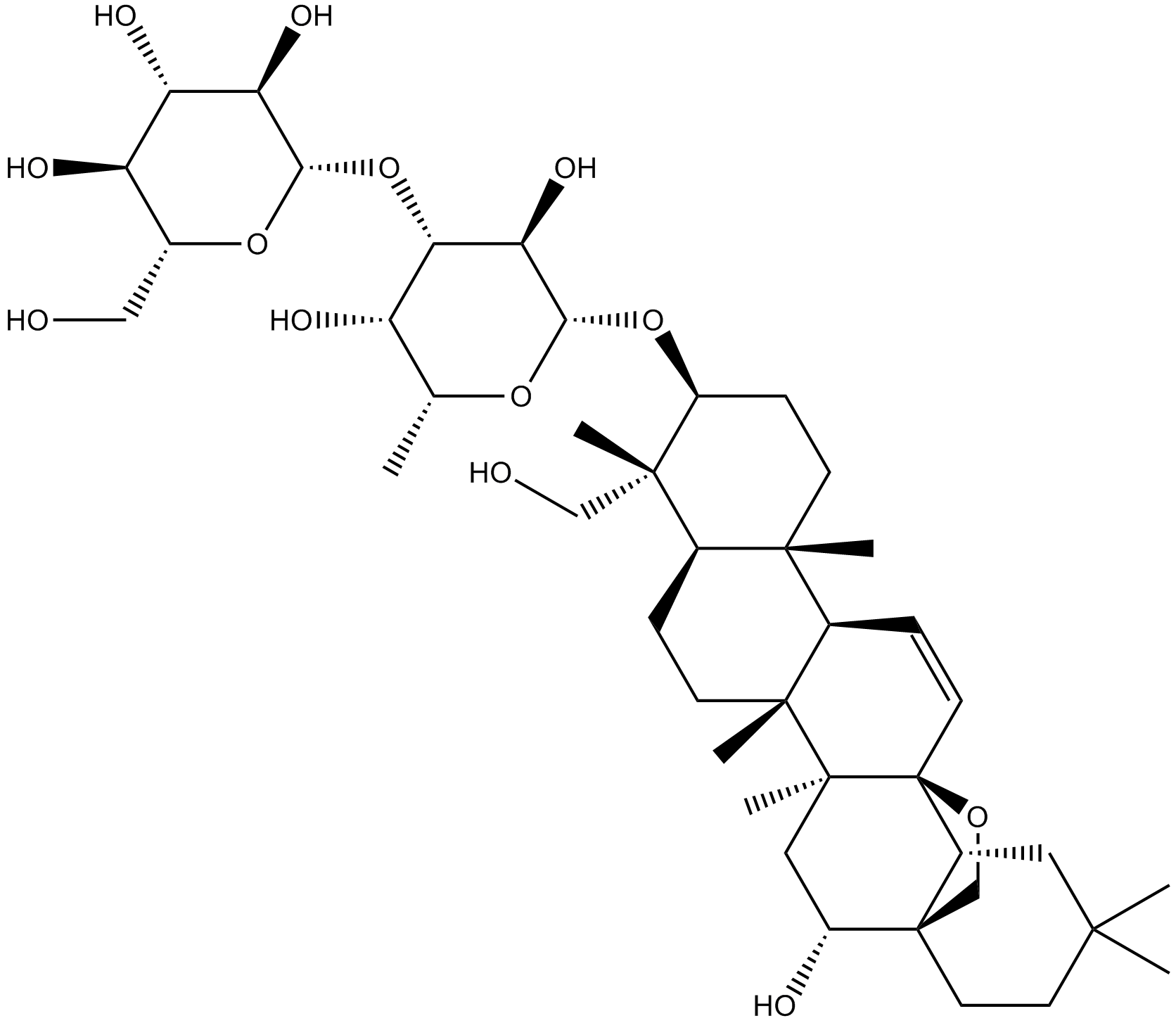
-
GC69854
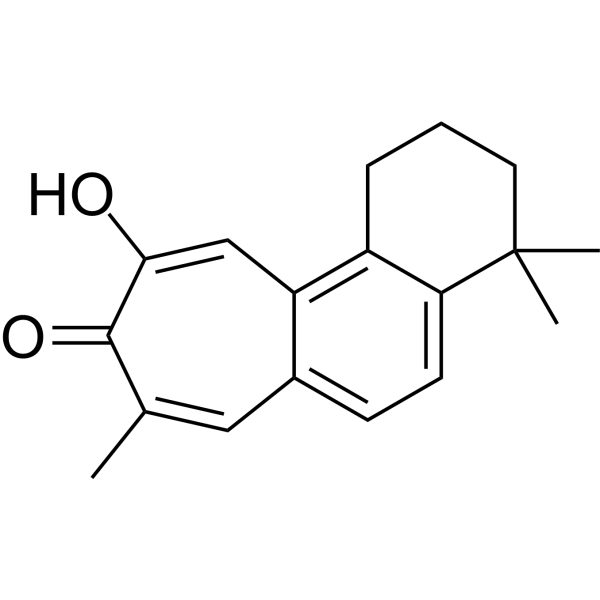
Salviolone
Salviolone is a natural diterpenoid derivative that can combat melanoma cells. Salviolone exhibits multifunctional effects on melanoma by blocking cell cycle progression, STAT3 signaling, and the malignant phenotype of A375 melanoma cells.
-
GC19320
SAR-20347
SAR-20347 is an inhibitor of TYK2, JAK1, JAK2 and JAK3 with IC50s of 0.6, 23, 26 and 41 nM, respectively.
-
GC25899
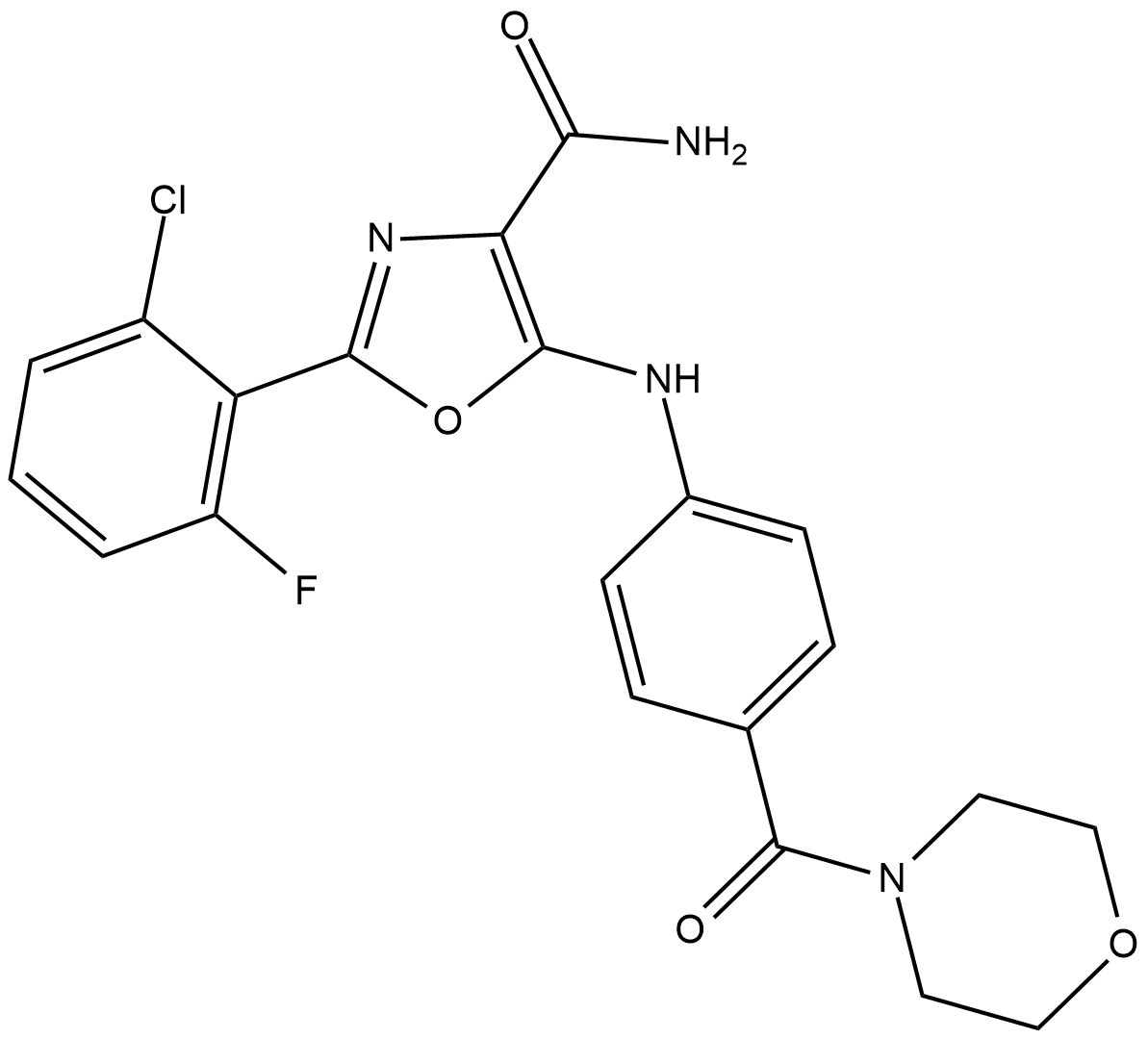
SC-1
SC-1 (1-(4-Chloro-3-(trifluoromethyl)phenyl)-3-(4-(4-cyanophenoxy)phenyl)urea, STAT3-IN-7), a Sorafenib analogue and potently inhibits the phosphorylation of STAT3, induces cell apoptosis through SHP-1 dependent STAT3 inactivation.
-
GC61524
SC-43
SC-43, a Sorafenib derivative, is a potent and orally active SHP-1 (PTPN6) agonist. SC-43 inhibits the phosphorylation of STAT3 and induces cell apoptosis. SC-43 has anti-fibrotic and anticancer effects.

-
GN10624
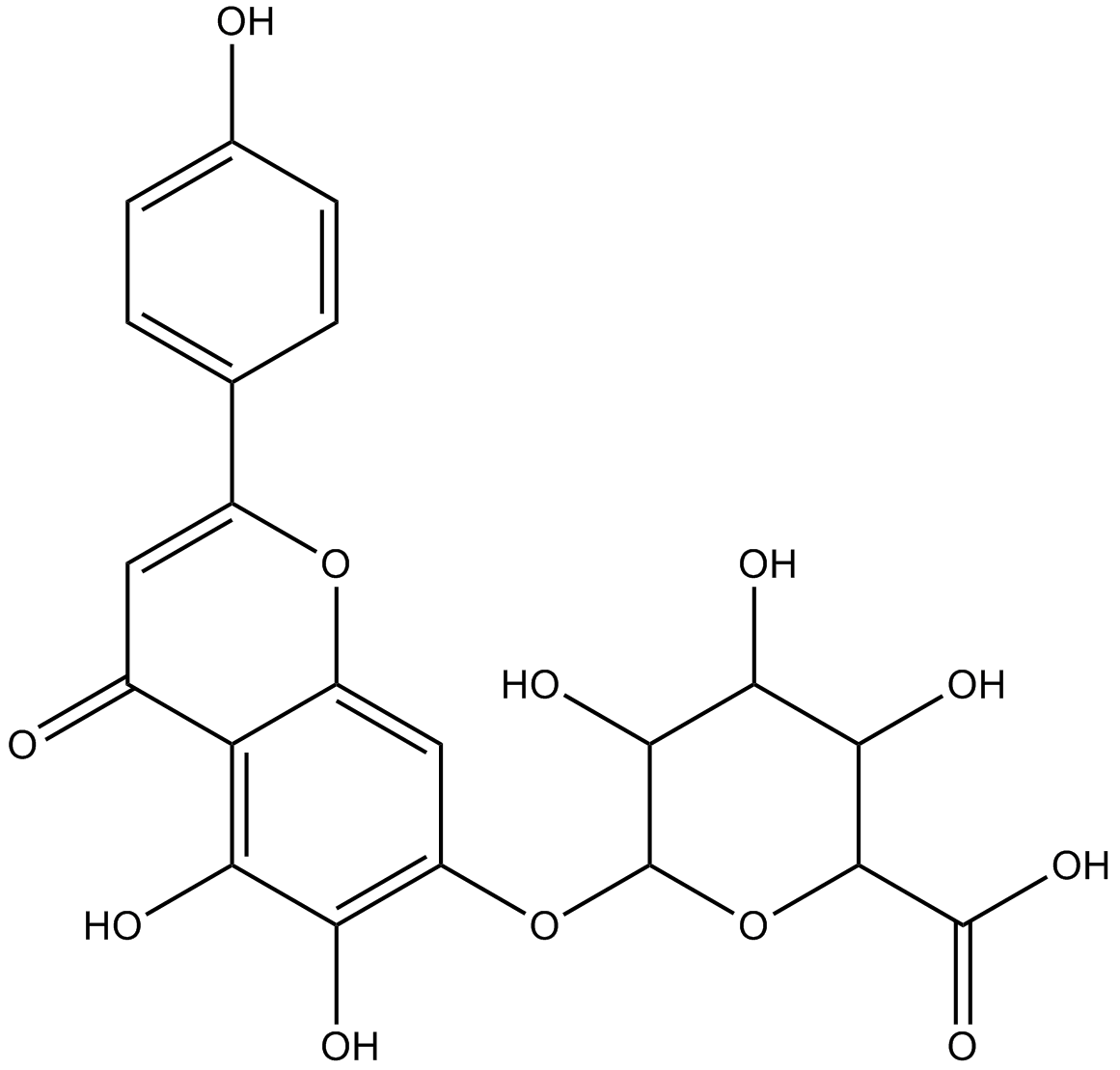
Scutellarin
-
GC44880
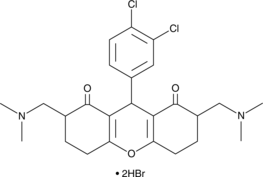
SD 1029
A JAK2 inhibitor
-
GC64774
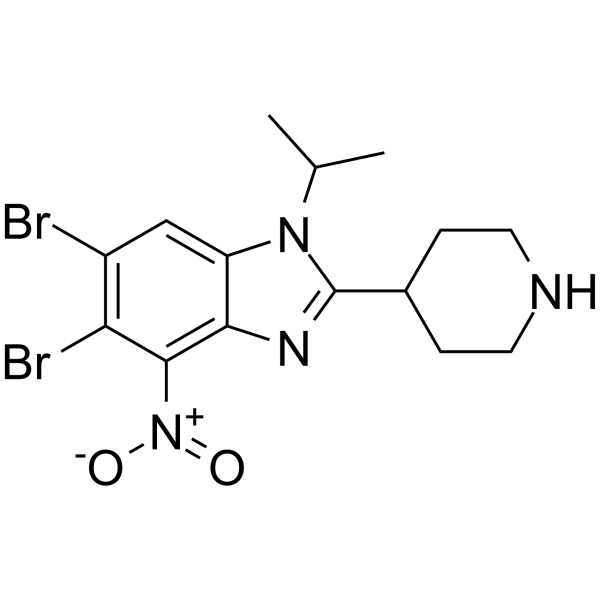
SEL24-B489
SEL24-B489 is a potent, type I, orally active, dual PIM and FLT3-ITD inhibitor, with Kd values of 2 nM for PIM1, 2 nM for PIM2 and 3 nM for PIM3, respectively.
-
GC68430
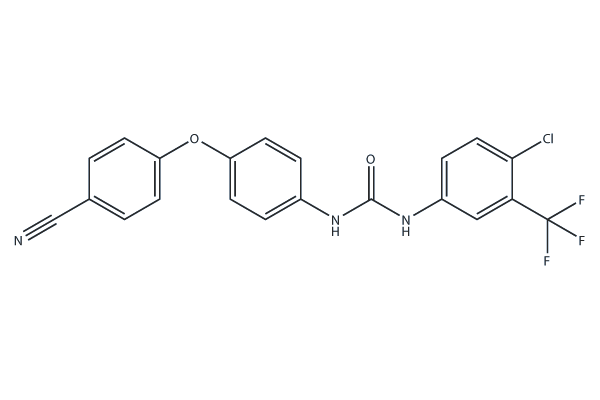
Selatinib
-
GC14658
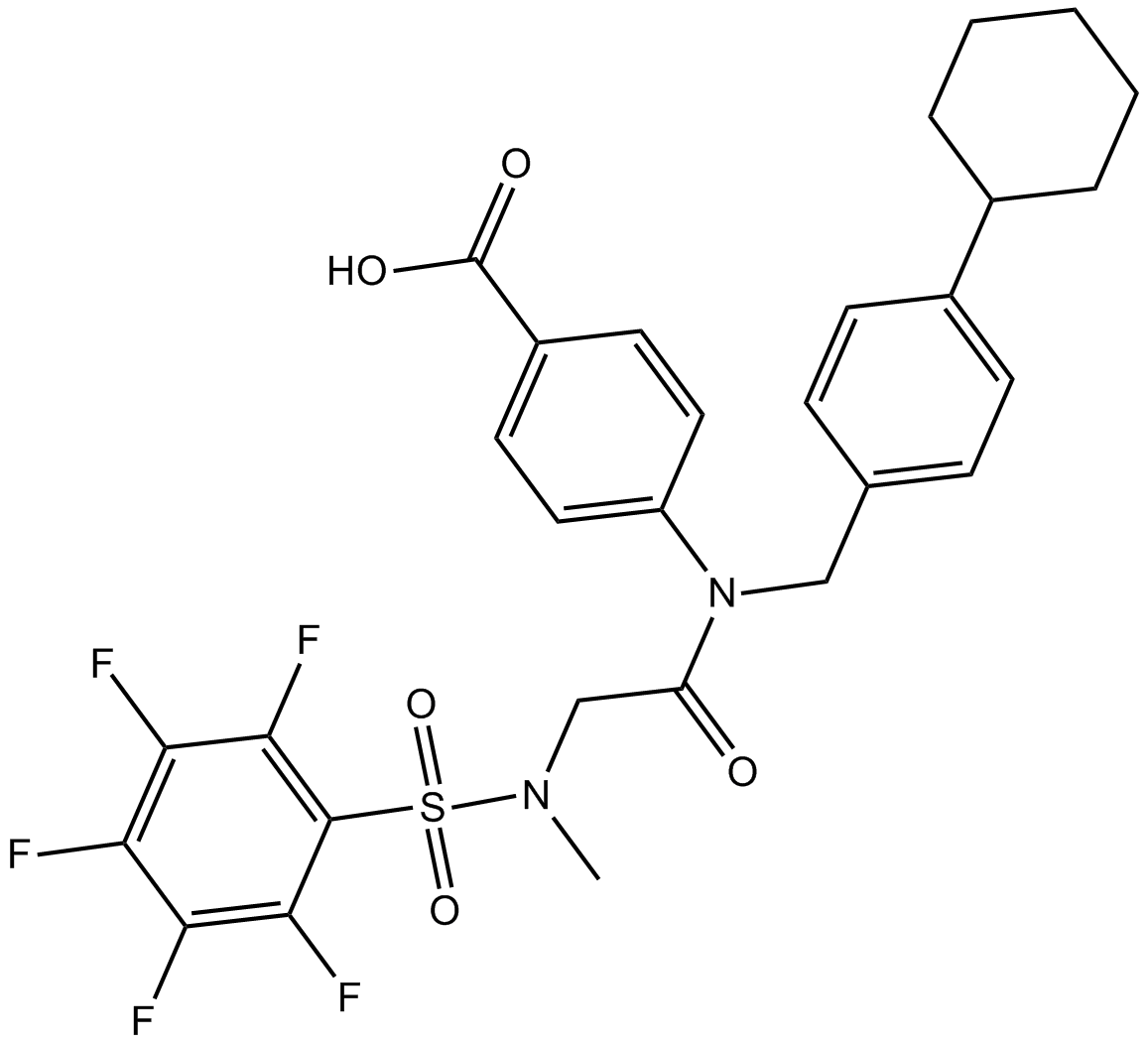
SH-4-54
STAT inhibitor, potent
-
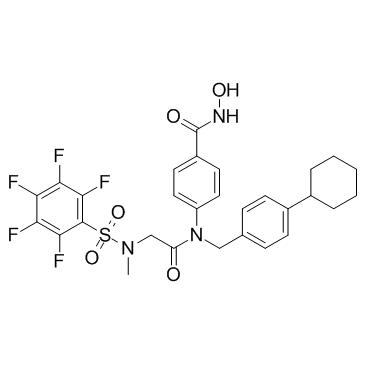
GC32928
SH5-07
SH5-07 is a hydroxamic acid based Stat3 inhibitor with an IC50 of 3.9 μM in in vitro assay.
-
GC37651
SMI-16a
A Pim-1 kinase inhibitor

-
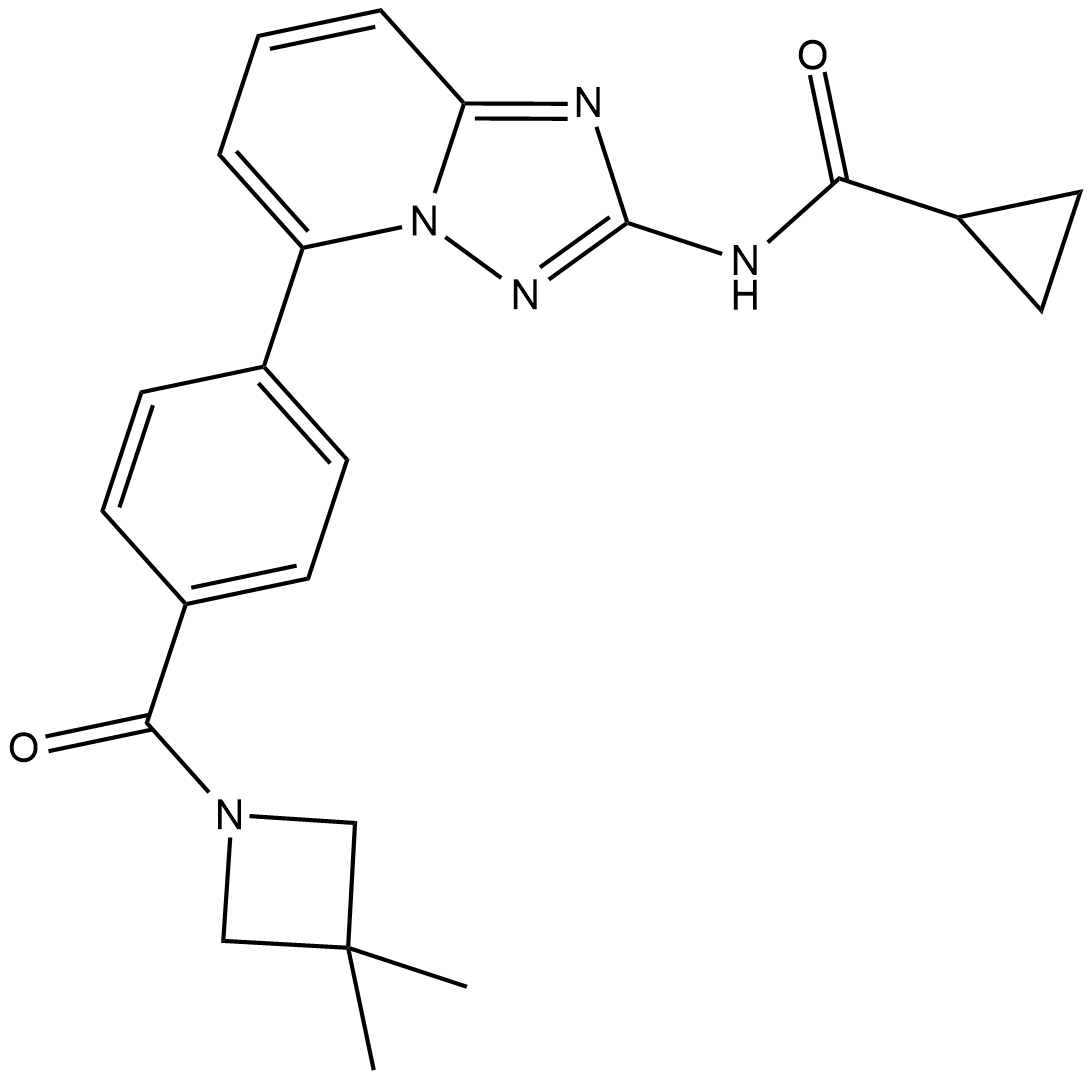
GC19334
Solcitinib
Solcitinib is an orally active, competitive, potent, selective JAK1 inhibitor, with an IC50 of 9.8 nM, and 11-, 55- and 23-fold selectivity over JAK2, JAK3 and TYK2, respectively; Solcitinib is used in the research of moderate-to-severe plaque-type psoriasis.
-
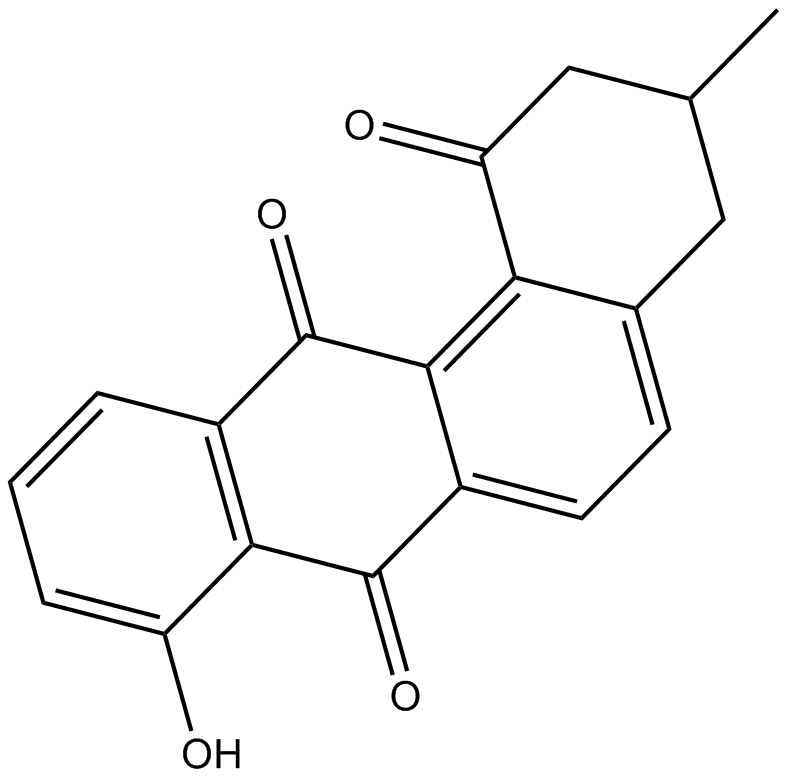
GC17761
STA-21
A STAT3 inhibitor
-
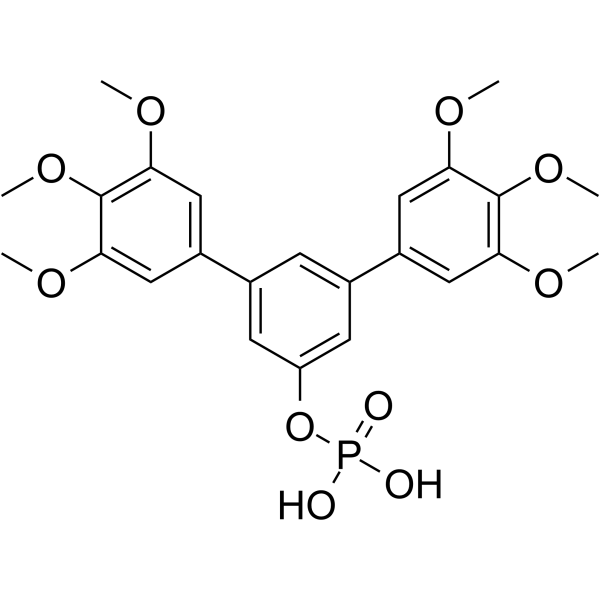
GC62559
Stafia-1
Stafia-1 is a potent STAT5a inhibitor (K i=10.9 μM, IC50=22.2 μM). Stafia-1 displays high selectivity over STAT5b and other STAT family members.
-
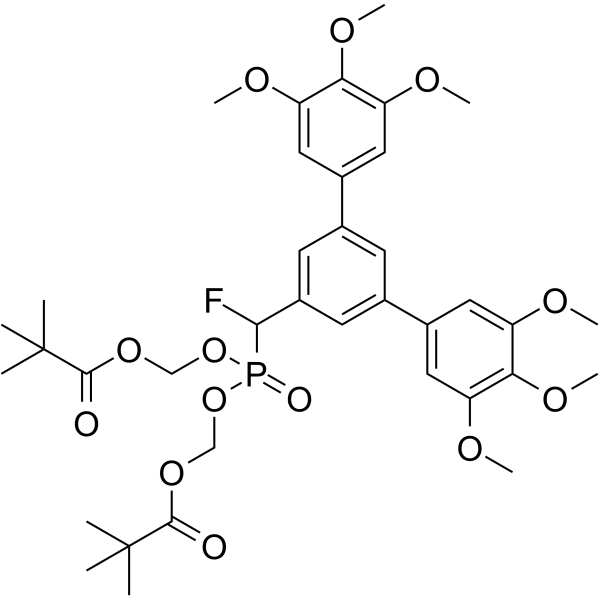
GC63203
Stafia-1-dipivaloyloxymethyl ester
Stafia-1-dipivaloyloxymethyl ester (compound 27, 0-200 μM) decreases pSTAT5a expression significantly, and has no obvious inhibition on pSTAT5b.
-
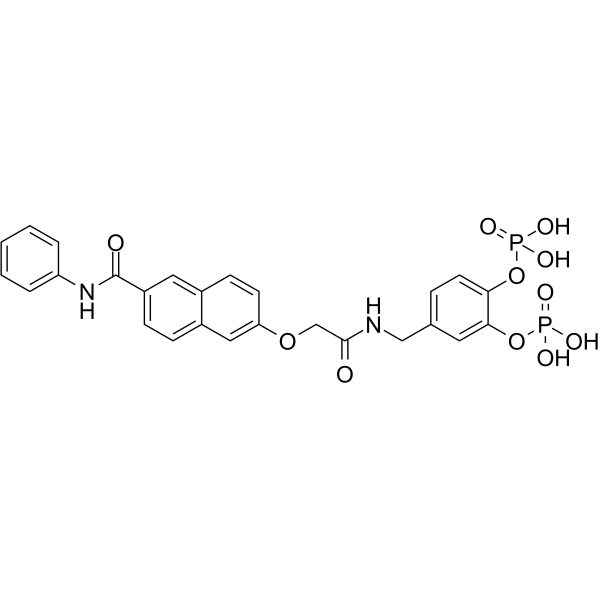
GC62254
Stafib-1
Stafib-1 is the first selective inhibitor of the STAT5b SH2 domain, with a Ki of 44 nM and an IC50 of 154 nM.
-
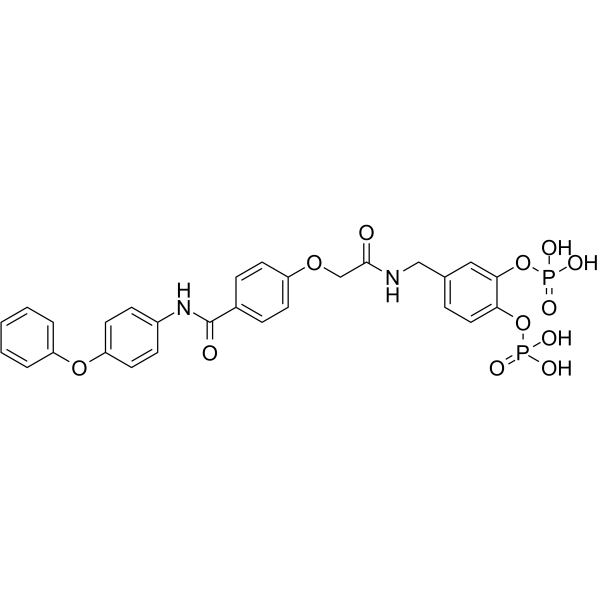
GC63204
Stafib-2
Stafib-2 is a potent and selctive inhibitor of the transcription factor STAT5b, with an IC50 of 82 nM and 1.7 μM for STAT5b and STAT5a, respectively.
-
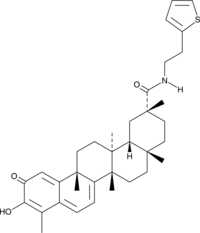
GC52293
STAT3 Inhibitor 4m
A STAT3 inhibitor
-
GC37688
STAT3-IN-1
STAT3-IN-1 (compound 7d) is an excellent, selective and orally active STAT3 inhibitor, with IC50 values of 1.82 μM and 2.14 μM in HT29 and MDA-MB 231 cells, respectively. STAT3-IN-1 (compound 7d) induces tumor apoptosis.
-
GC69952
STAT3-IN-11
STAT3-IN-11 (7a) is a selective inhibitor of STAT3 that can inhibit the phosphorylation of the pTyr705 site on STAT3. It can also inhibit downstream genes (Survivin and Mcl-1) without affecting upstream tyrosine kinases (Src and JAK2) and p-STAT1 expression. STAT3-IN-11 can induce apoptosis in cancer cells, making it a promising candidate for the discovery of STAT3 inhibitors and anti-tumor agents.
-
GC69953
STAT3-IN-12
STAT3-IN-12 is an effective STAT3 signaling inhibitor that can suppress the activation of the IL-6-induced JAK/STAT3 signaling pathway. It can inhibit cancer cell growth and migration, induce apoptosis (cell death), and block cell cycle progression. STAT3-IN-12 can be used in cancer-related research such as hepatocellular carcinoma (HCC) and esophageal cancer.
-
GC37689
STAT3-IN-3
STAT3-IN-3 is a potent and selective inhibitor of signal transducer and activator of transcription 3 (STAT3), with anti-proliferative activity. STAT3-IN-3 induces apoptosis in breast cancer cells. STAT3-IN-3 acts as a promising mitochondria-targeting STAT3 inhibitor for cancer research.
-
GC13647
STAT5 Inhibitor
STAT5 Inhibitor is a STAT5 inhibitor with an IC50 of 47 μM for STAT5β isoform.
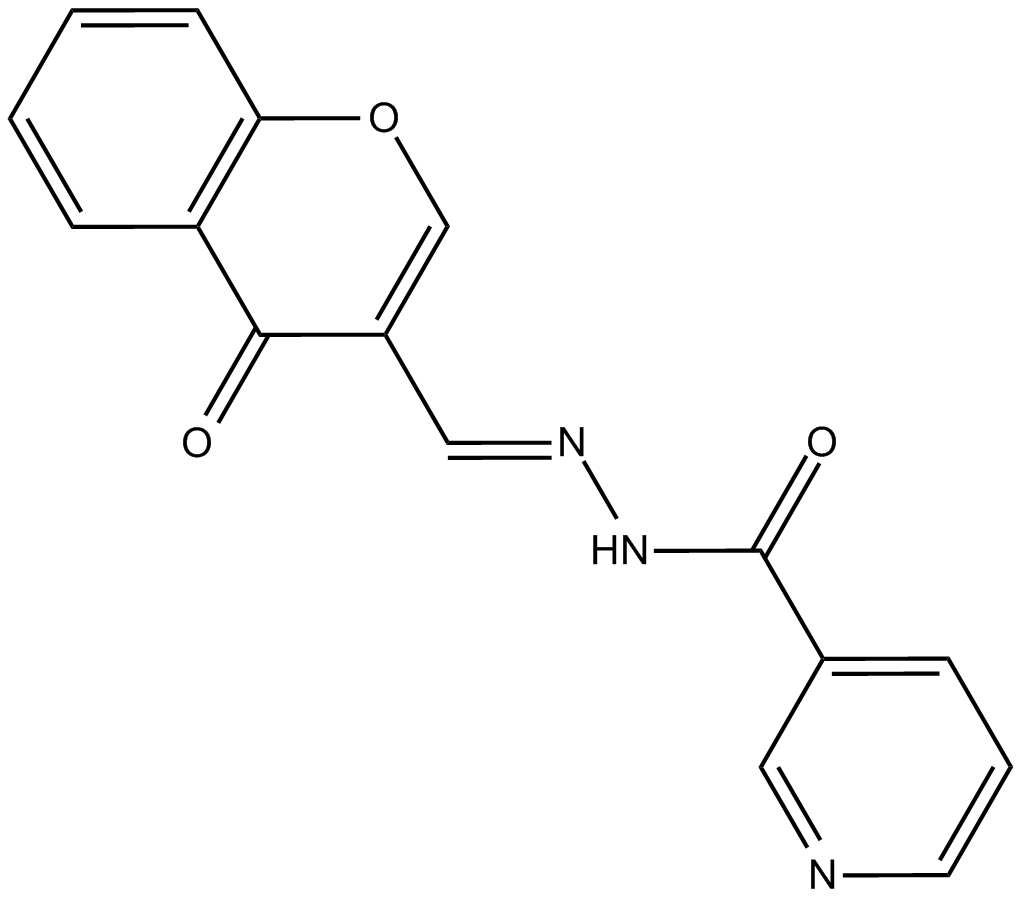
-
GC65344
STAT5-IN-2
STAT5-IN-2 is a STAT5 inhibitor, extracted from reference 1, example 17f. STAT5-IN-2 has potent antileukemic effect.
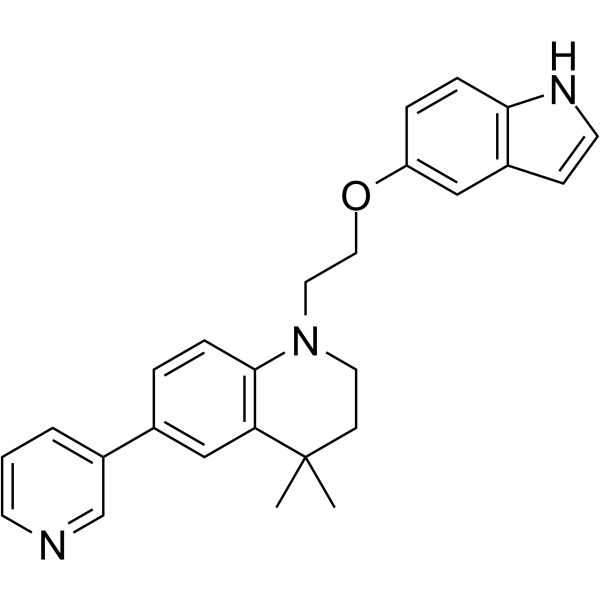
-
GC17886
Stattic
Stattic is the first non-peptide small molecule inhibitor of STAT3, which effectively inhibits STAT3 activation and nuclear translocation.
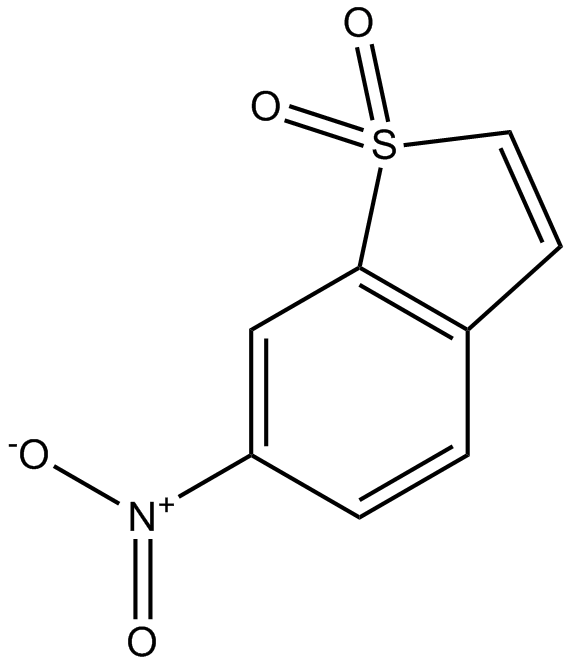
-
GC67779
STX-0119
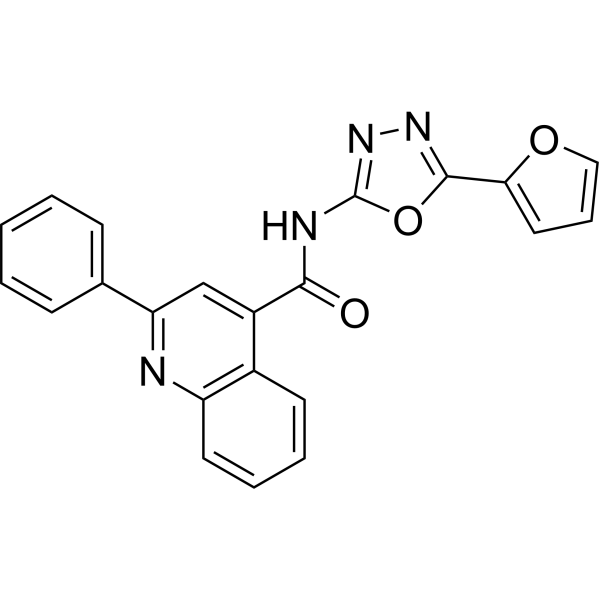
-
GC11172
TAK-285
HER2/EGFR(HER1) inhibitor
-
GC32100
Tarloxotinib bromide (TH-4000)
Tarloxotinib bromide (TH-4000) (TH-4000) is an irreversible EGFR/HER2 inhibitor.
-
GC65310
TAS0728
TAS0728 is a potent, selective, orally active, irreversible and covalent-binding HER2 inhibitor, with an IC50 of 13 nM. TAS0728 also shows IC50s of 4.9, 8.5, 31, 65, 33, 25 and 86 nM for BMX、HER4、BLK、EGFR、JAK3、SLK and LOK respectively. Furthermore, TAS0728 exhibits robust and sustained inhibition of the phosphorylation of HER2
-
GC32752
TAS6417
TAS6417 (CLN-081) is a highly effective, orally active and pan-mutation-selective EGFR tyrosine kinase inhibitor with a unique scaffold fitting into the ATP-binding site of the EGFR hinge region, with IC50 values ranging from 1.1-8.0 nM.
-
GC45004
TC 14012 (trifluoroacetate salt)
C-X-C chemokine receptor type 4 (CXCR4) is a receptor for the stromal cell-derived factor-1 which is designated as chemokine ligand 12 (CXCL12).
-
GC11990
TCS-PIM-1-4a
Pim inhibitor
-
GC41577
Tephrosin (synthetic)
Tephrosin (synthetic) is a natural rotenoid which has potent antitumor activities. Tephrosin (synthetic) induces degradation of of EGFR and ErbB2 by inducing internalization of the receptors.
-
GC31752
Tesevatinib (XL-647)
Tesevatinib (XL-647) (XL-647; EXEL-7647; KD-019) is an orally available, multi-target tyrosine kinase inhibitor; inhibits EGFR, ErbB2, KDR, Flt4 and EphB4 kinase with IC50s of 0.3, 16, 1.5, 8.7, and 1.4 nM.
-
GC39022
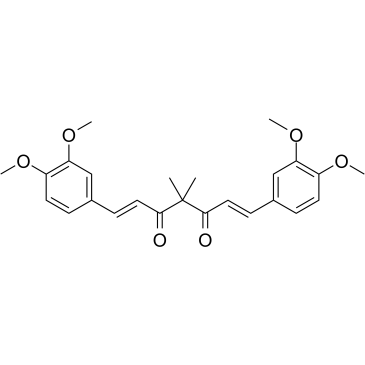
Tetramethylcurcumin
Tetramethylcurcumin (FLLL31), derived from curcumin, specifically suppresses the phosphorylation of STAT3 by binding selectively to Janus kinase 2 and the STAT3 Src homology-2 domain.
-
GC25992
TG-89
TG-89 is an inhibitor of JAK2 with IC50 of 11.2 μM.