Ferroptosis
Ferroptosis is a unique iron-dependent form of nonapoptotic cell death. It is triggered by oncogenic RAS-selective lethal small molecule erastin. Acitvation of ferroptosis lead to nonapoptotic destruction of cancer cells.
Products for Ferroptosis
- Cat.No. Product Name Information
-
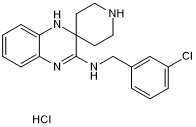
GC50345
Liproxstatin-1 hydrochloride
Potent ferroptosis inhibitor
-
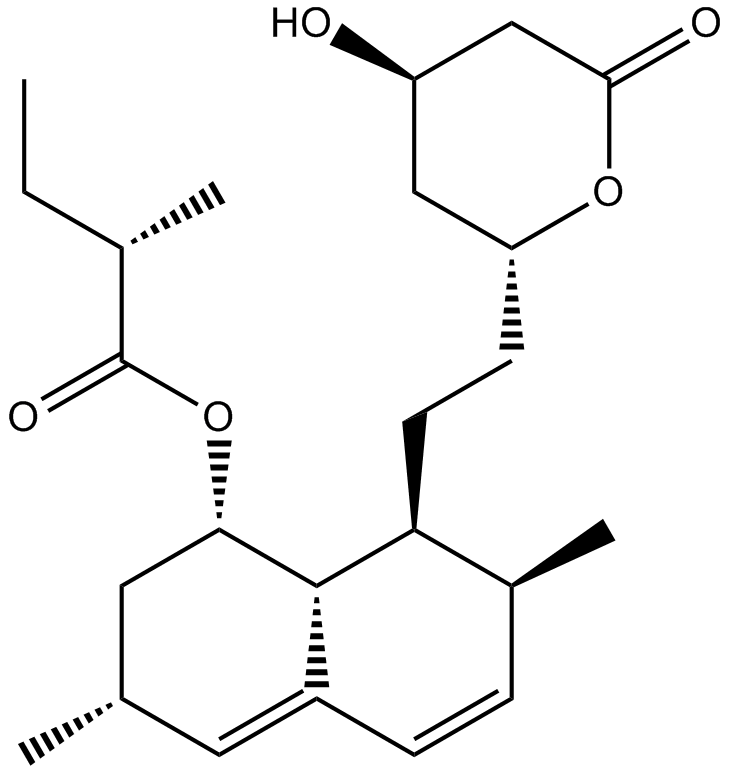
GC11633
Lovastatin
An inhibitor of HMG-CoA reductase
-
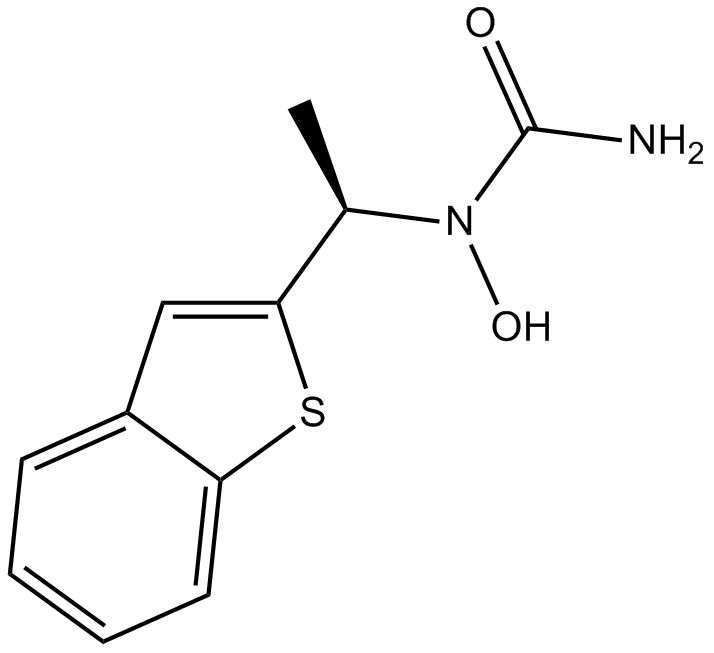
GC44214
ML-162
ML-162 is a GPX4 inhibitor that is selectively lethal to mutant RAS oncogene-expressing cell lines (IC50s = 25 and 578 nM for HRASG12V-expressing and wild-type BJ fibroblasts, respectively).
-
GC18705
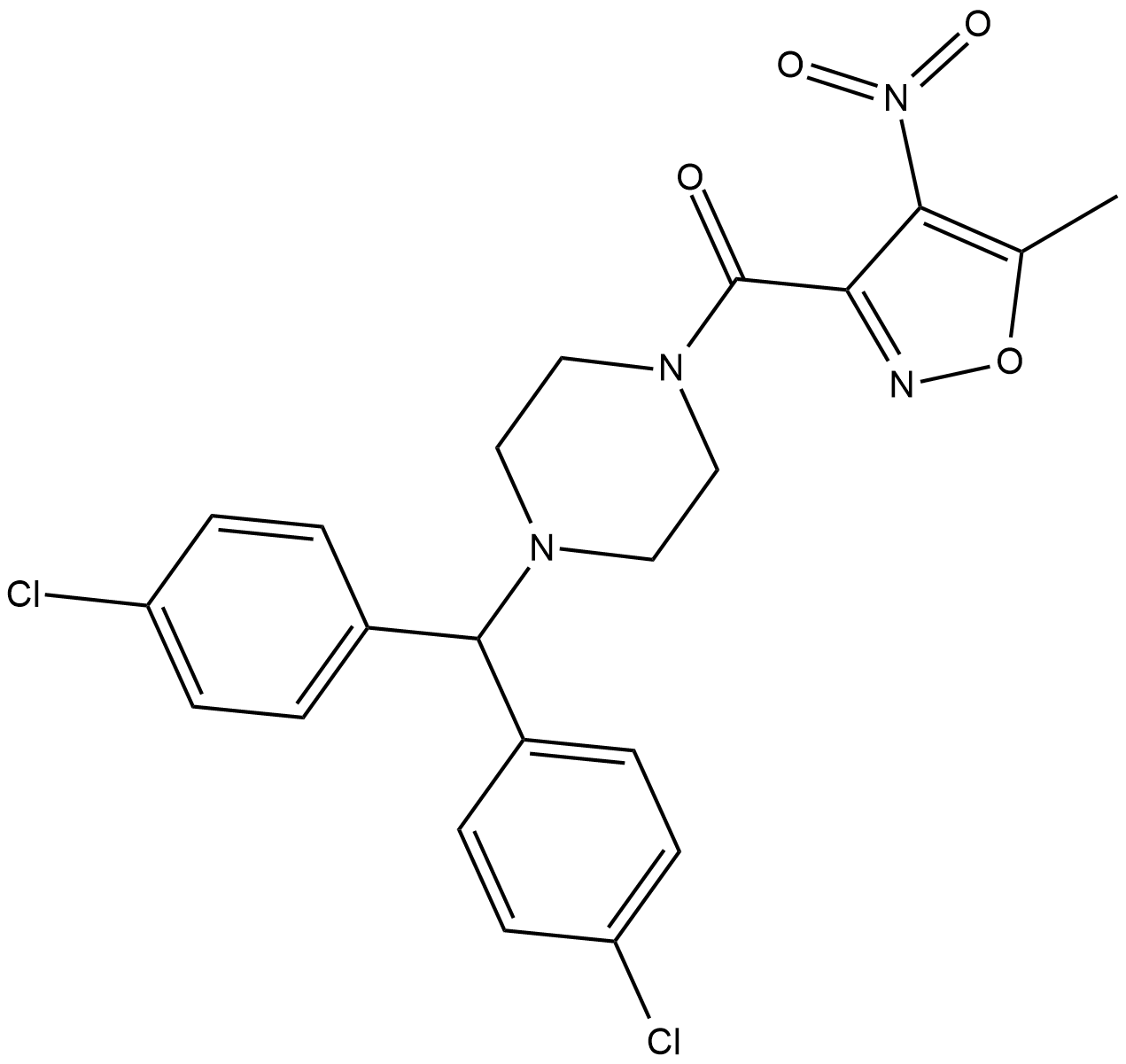
ML-210
A GPX4 inhibitor
-
GC19254
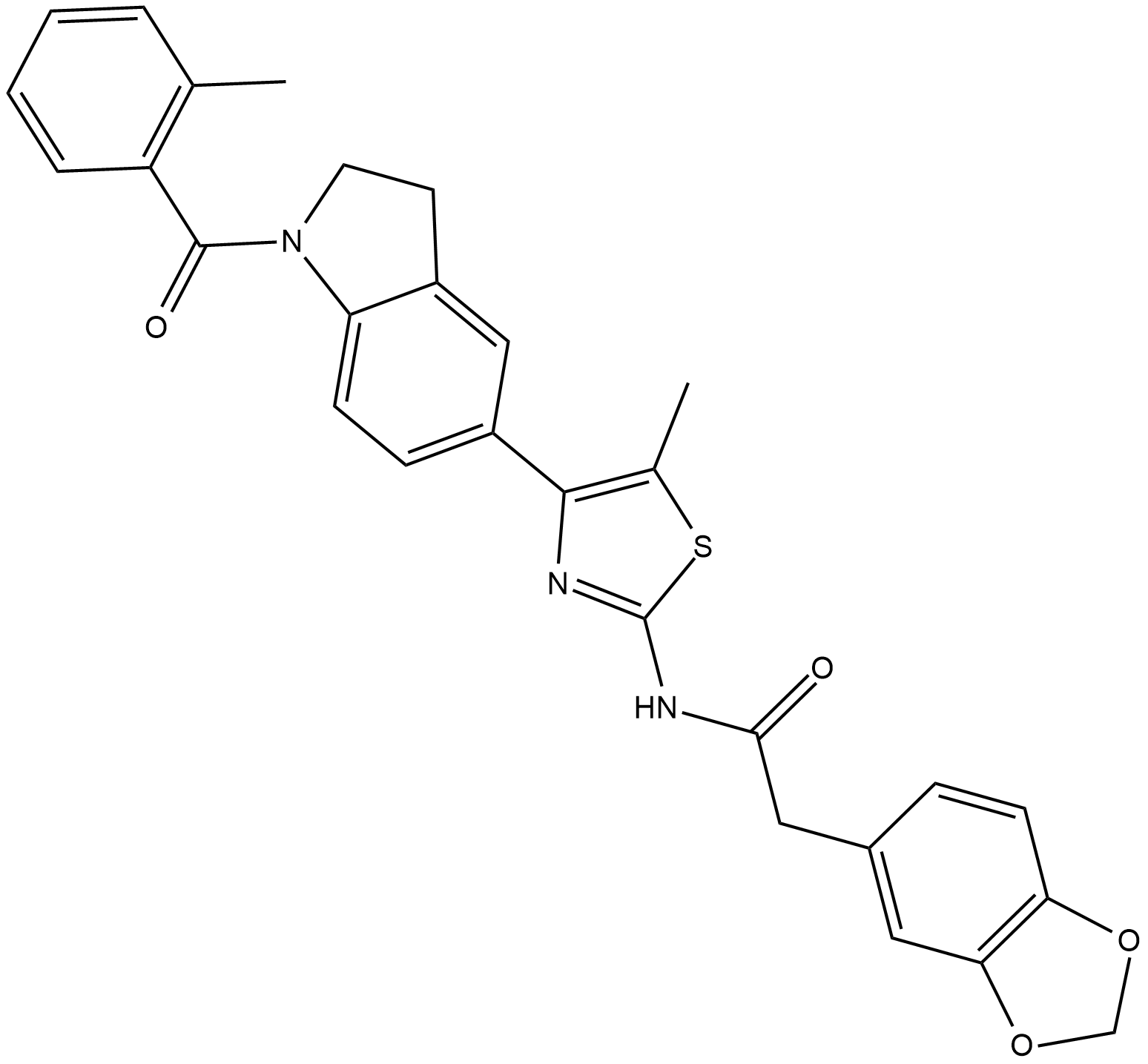
ML385
ML385 is a specific nuclear factor erythroid 2-related factor 2 (NRF2) inhibitor.
-
GC68479
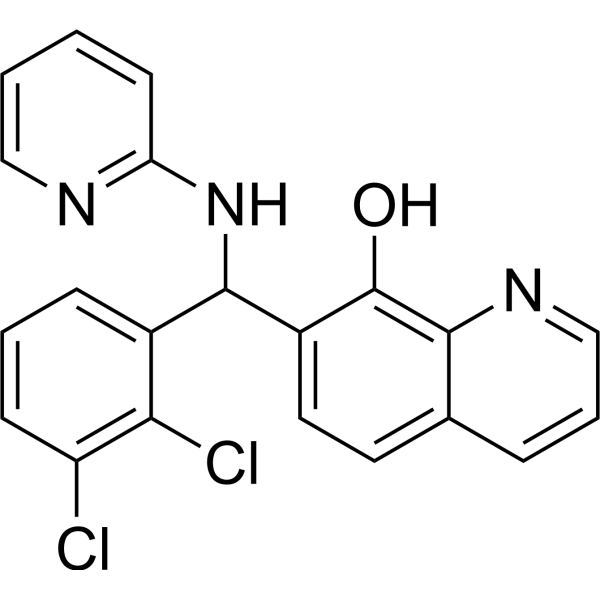
MMRi62
-
GC33469
NADPH tetracyclohexanamine
A reduced form of NADP+

-
GC34058
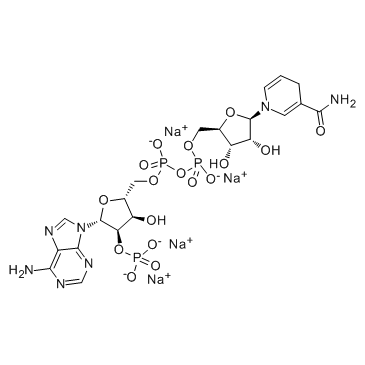
NADPH tetrasodium salt
A reduced form of NADP+
-
GC11008
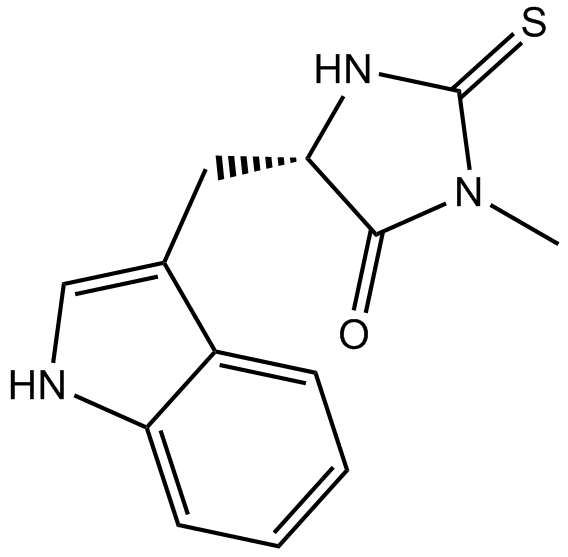
Necrostatin-1
A RIP1 kinase inhibitor
-
GC18089
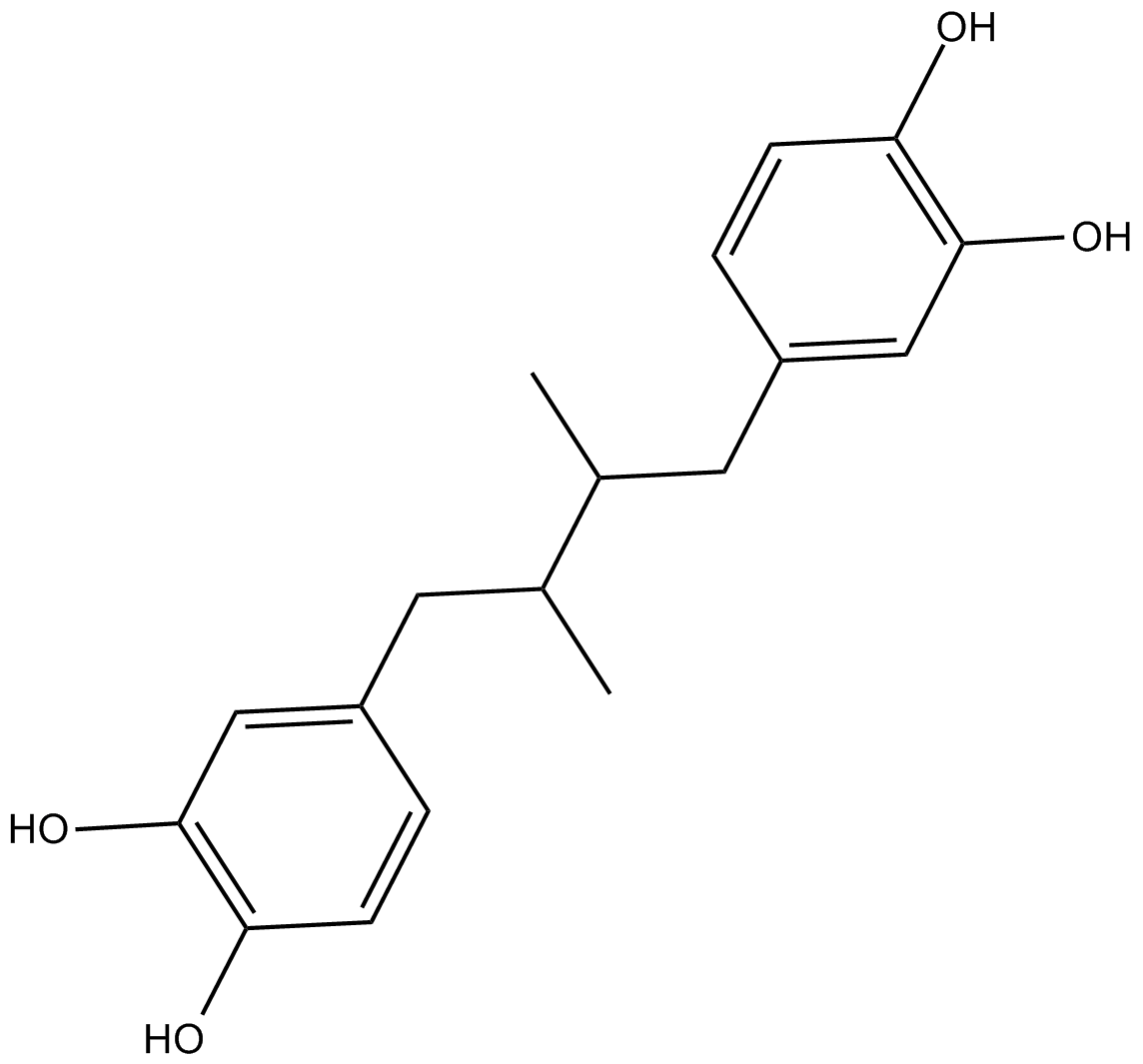
Nordihydroguaiaretic acid
A non-selective LO inhibitor
-
GC47870
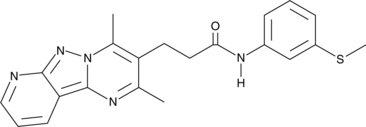
Pantothenate Kinase Inhibitor
A reversible inhibitor of pantothenate kinase
-
GC12205
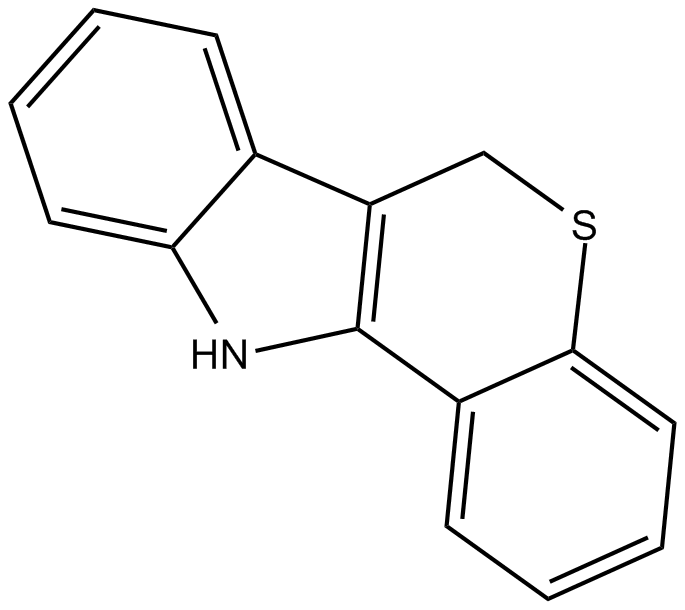
PD 146176
PD 146176 is one of the potent and selective inhibitors of reticulocyte 15-LOX-1.
-
GC10538
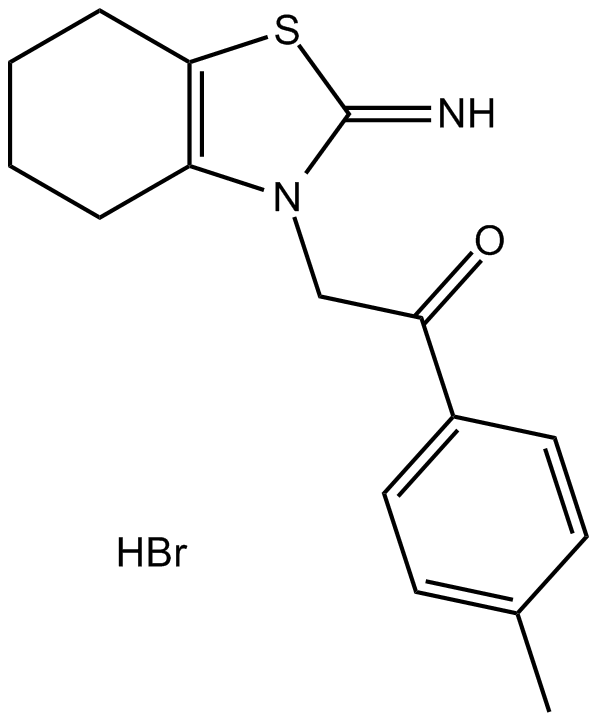
Pifithrin-α (PFTα)
Pifithrin-α(PFT-α) is a p53 inhibitor. Pifithrin-αwidely used in neuroscience to block neuronal apoptotic cell death. Pifithrin-αis also a potent stimulant of aryl hydrocarbon receptor (AhR)..
-
GC14948
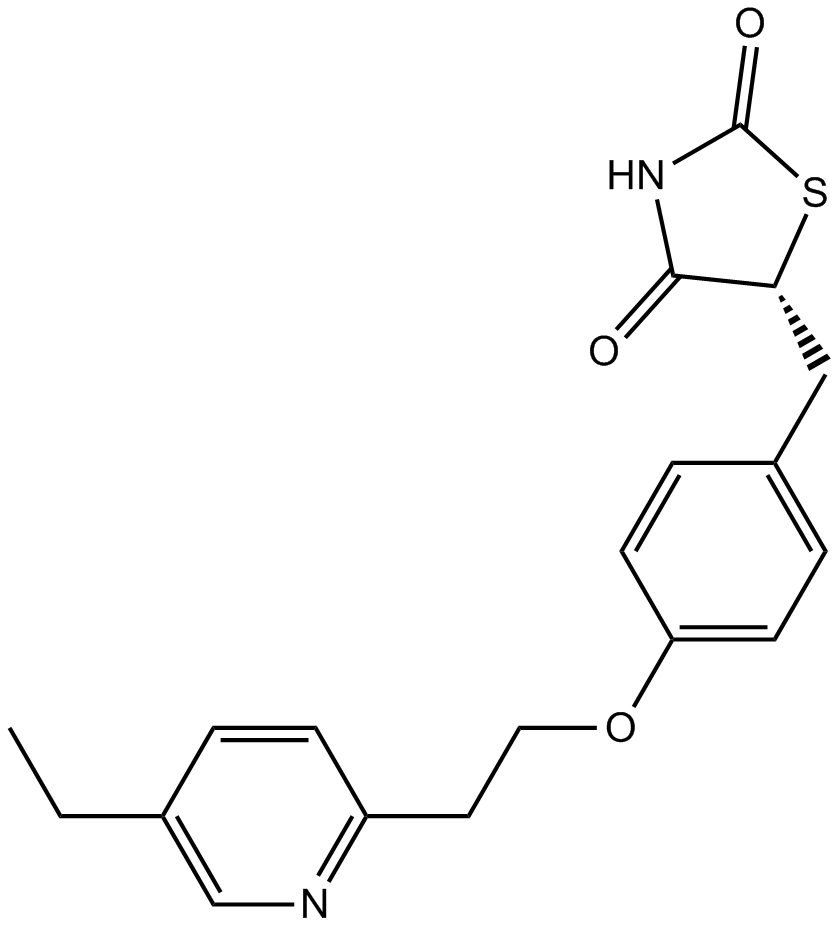
Pioglitazone
Pioglitazone (PIO) is a thiazolidinedione antidiabetic drug that acts as a PPARγ agonist. The EC50 of PPARγ in human and mouse was 0.93 and 0.99 µM, respectively..
-
GC61187
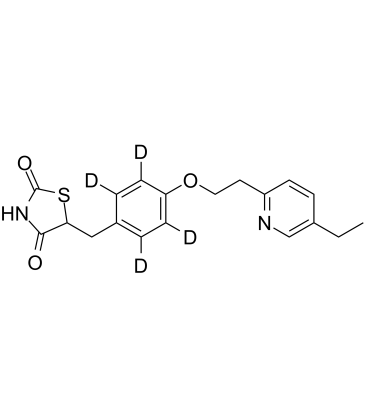
Pioglitazone D4
An internal standard for the quantification of pioglitazone
-
GC11152
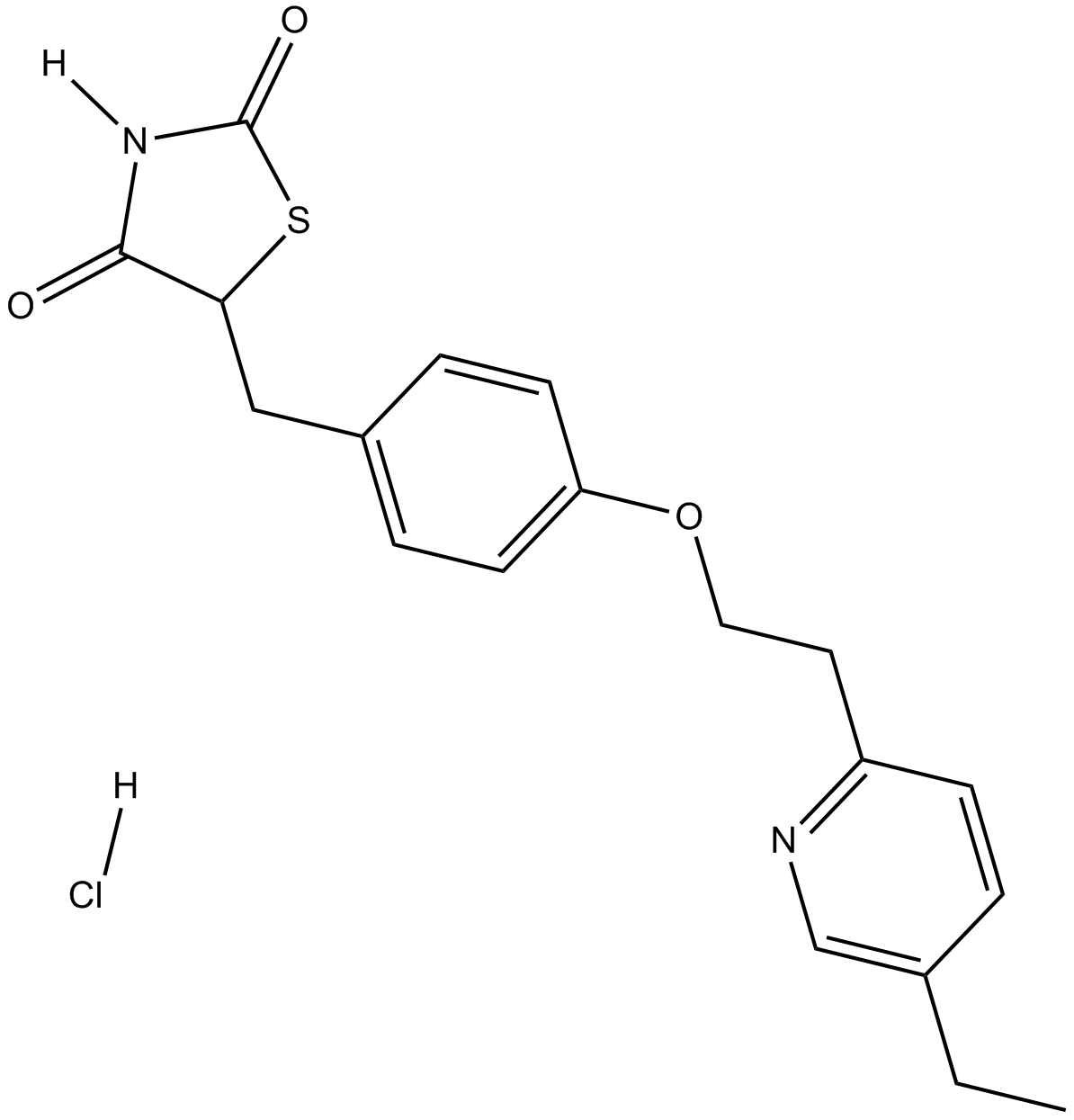
Pioglitazone HCl
A PPARγ agonist
-
GC36926
Piperazine Erastin
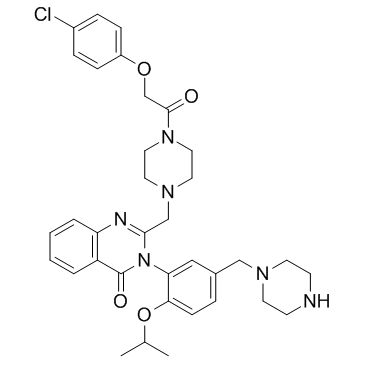
-
GC10282
Piperlongumine
An alkaloid with anticancer and antioxidant activities
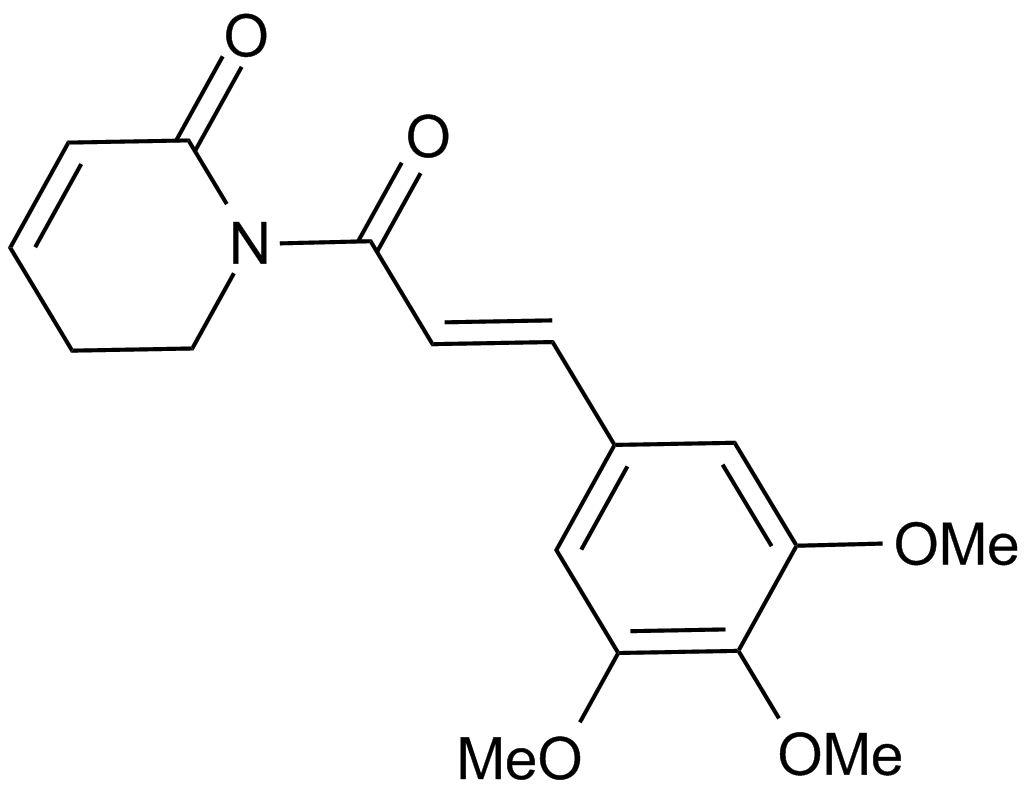
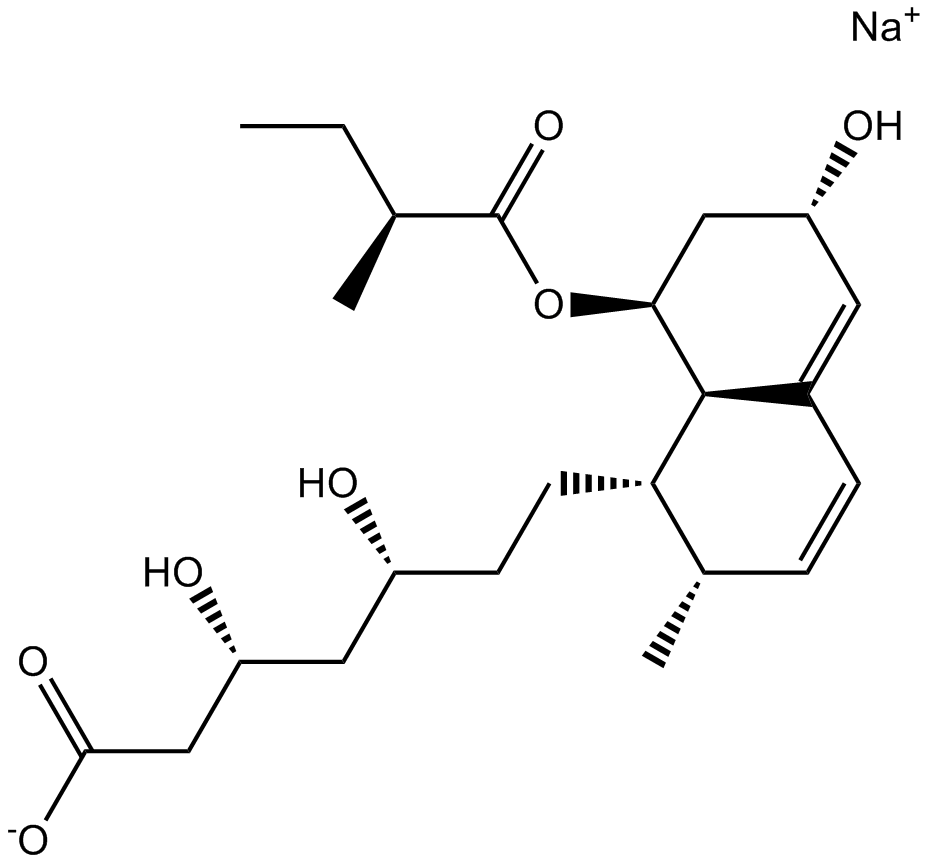
-
GC14538
Pravastatin sodium
A potent HMG-CoA reductase inhibitor
-
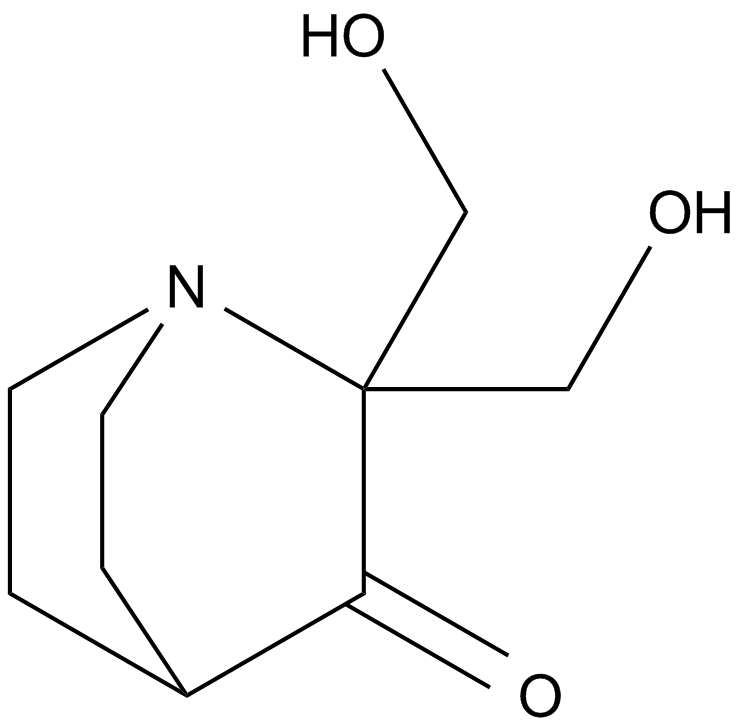
GC12086
PRIMA-1
A re-activator of the apoptotic function of mutant p53 proteins
-
GN10361
Pseudolaric Acid B
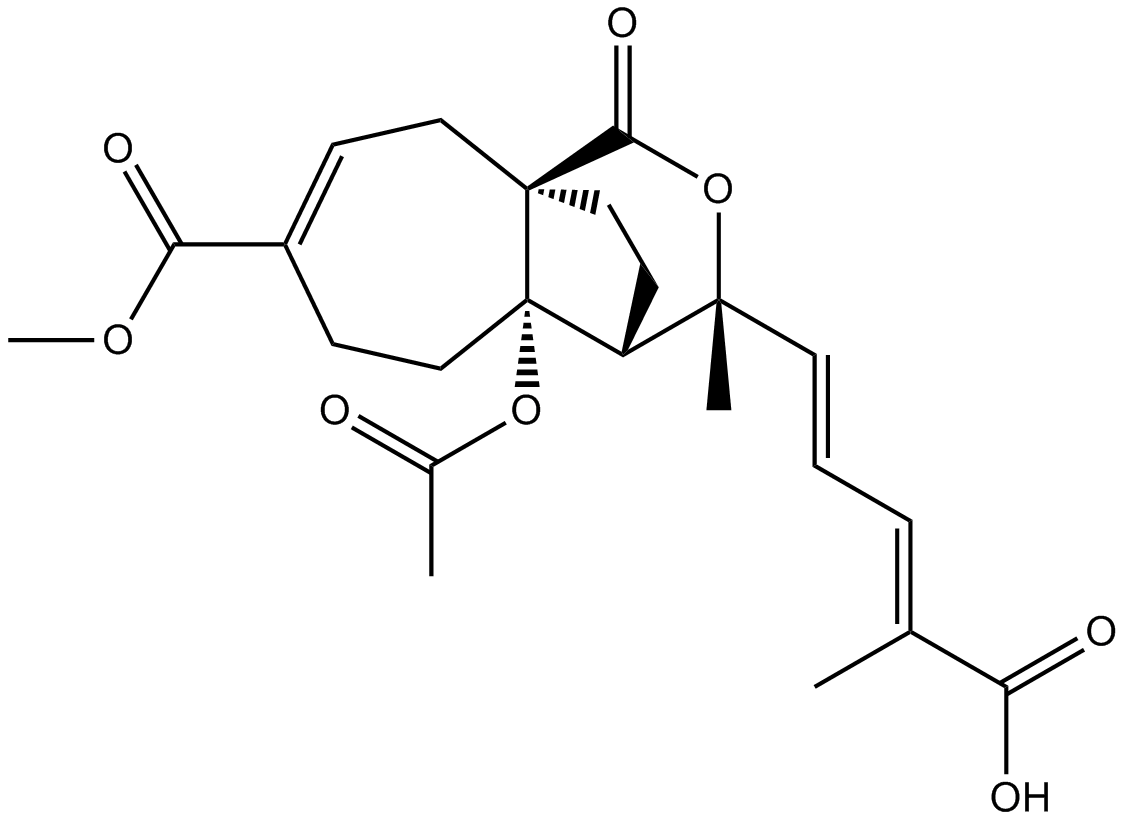
-
GC49691
PSP
A two-photon fluorescent probe for H2Sn
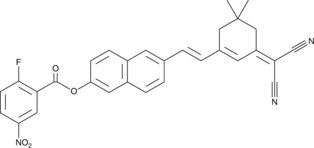
-
GC48029
RC363
An inhibitor of ferroptosis
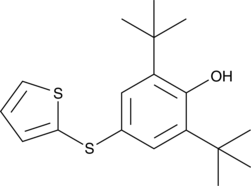
-
GC48030
RC574
An inhibitor of ferroptosis
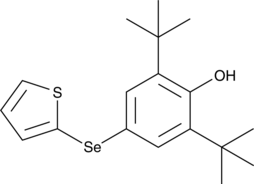
-
GC16444
Rosiglitazone
A PPARγ agonist
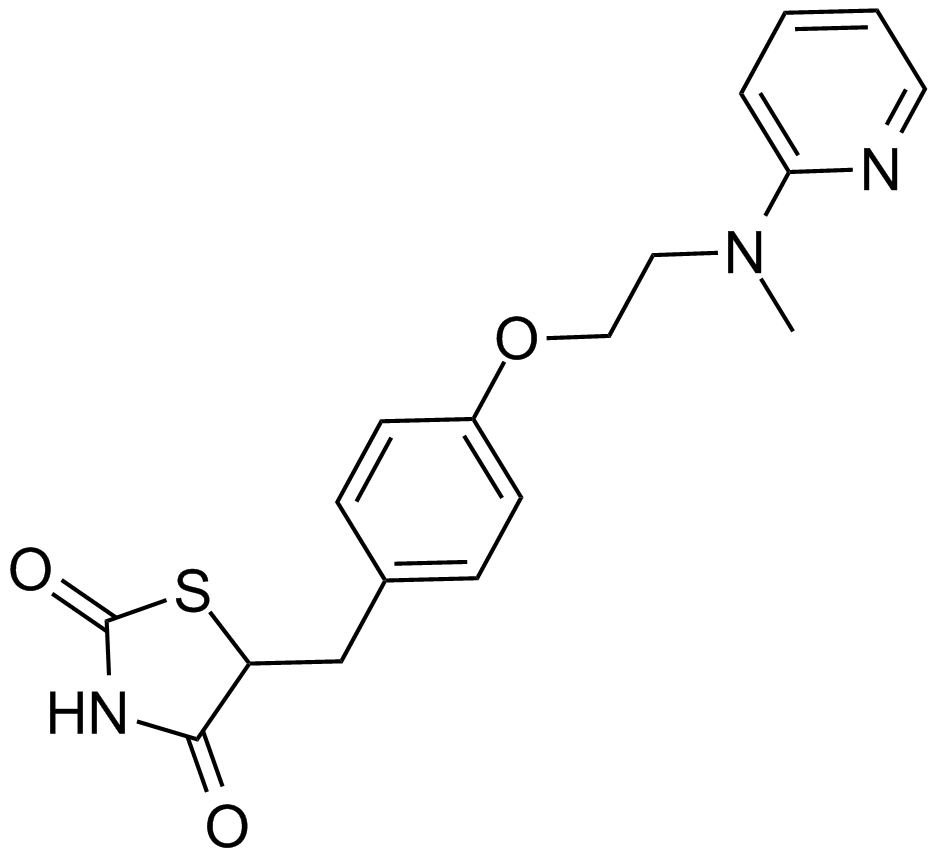
-
GC44851
Rosiglitazone (potassium salt)
Rosiglitazone (BRL 49653) potassium is an orally active selective PPARγ agonist (EC50: 60 nM, Kd: 40 nM).
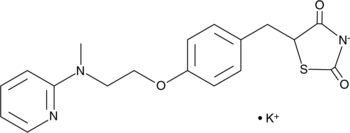
-
GC11318
Rosiglitazone maleate
A PPARγ agonist
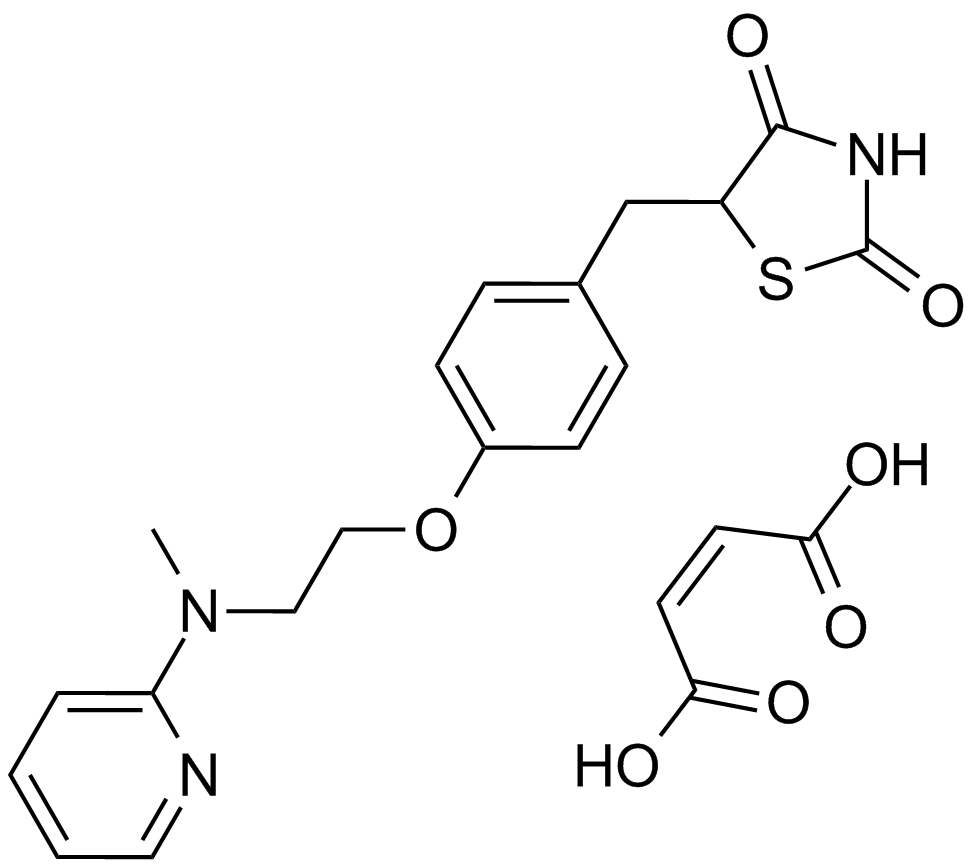
-
GC48059
Rosiglitazone-d3
An internal standard for the quantification of rosiglitazone
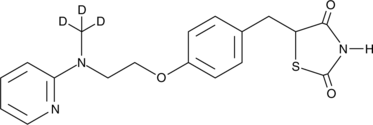
-
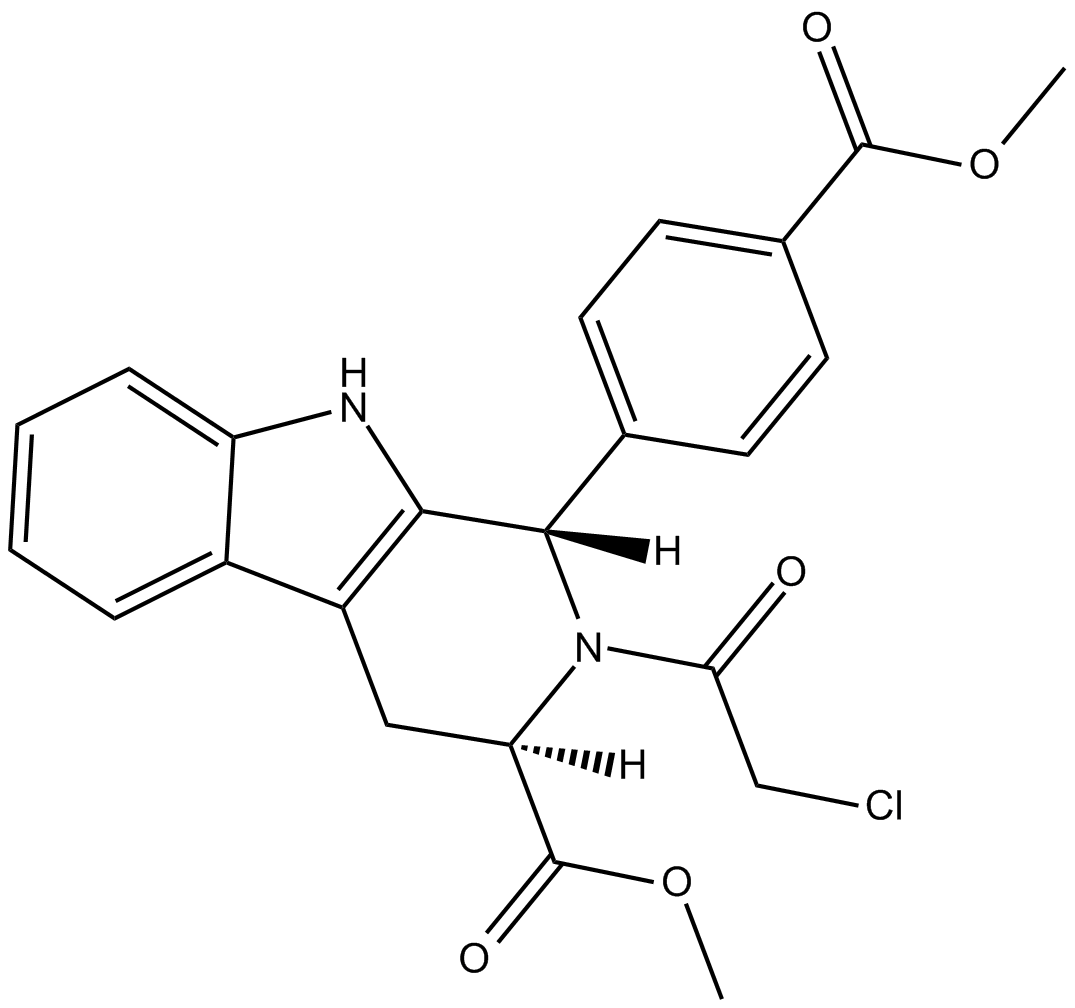
GC12431
RSL3
Glutathione peroxidase 4 inhibitor
-
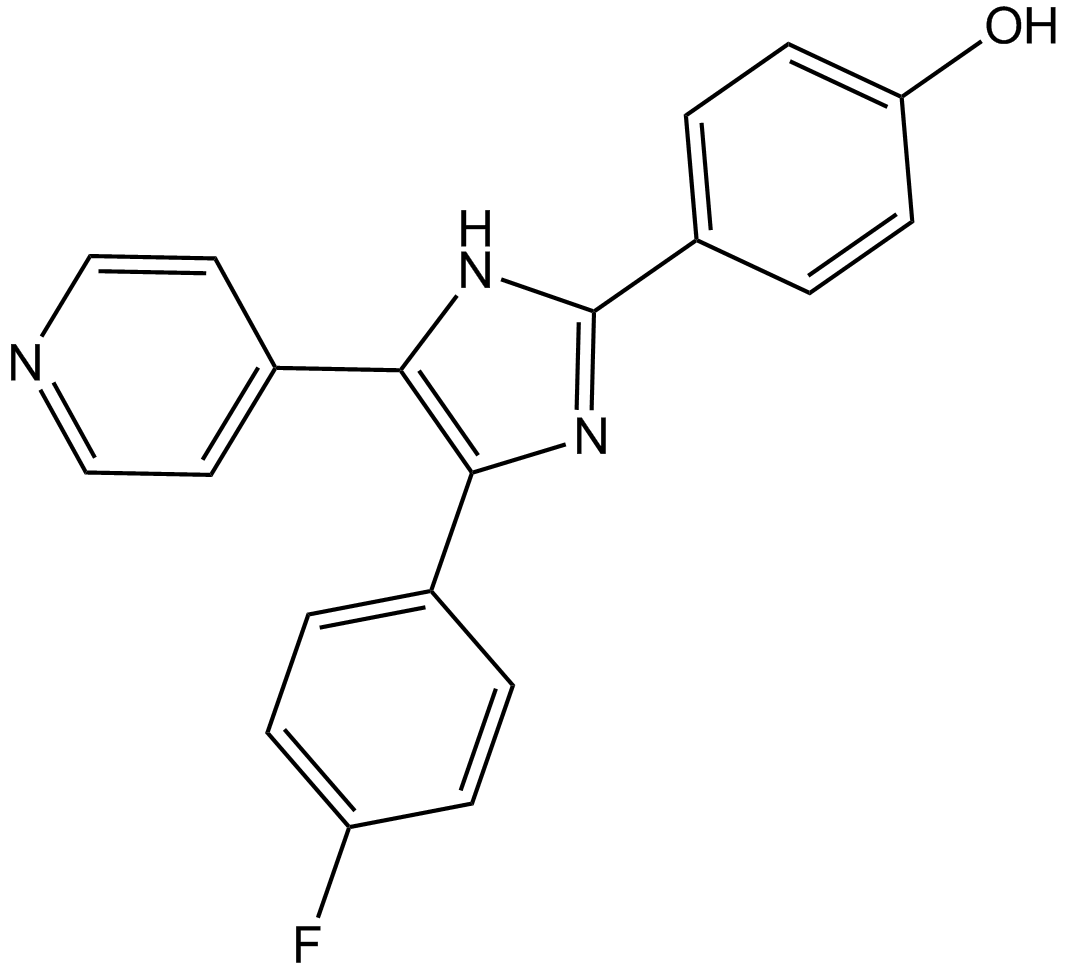
GC13968
SB202190 (FHPI)
SB202190 (FHPI) is a selective p38 MAP kinase inhibitor with IC50s of 50 nM and 100 nM for p38α and p38β2, respectively.
-
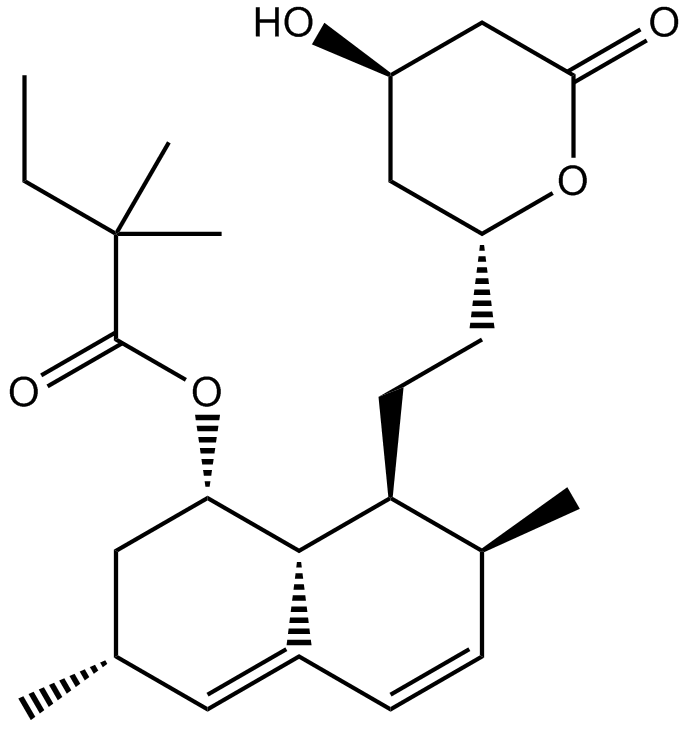
GC10585
Simvastatin (Zocor)
Simvastatin (Zocor) (MK 733) is a competitive inhibitor of HMG-CoA reductase with a Ki of 0.2 nM.
-
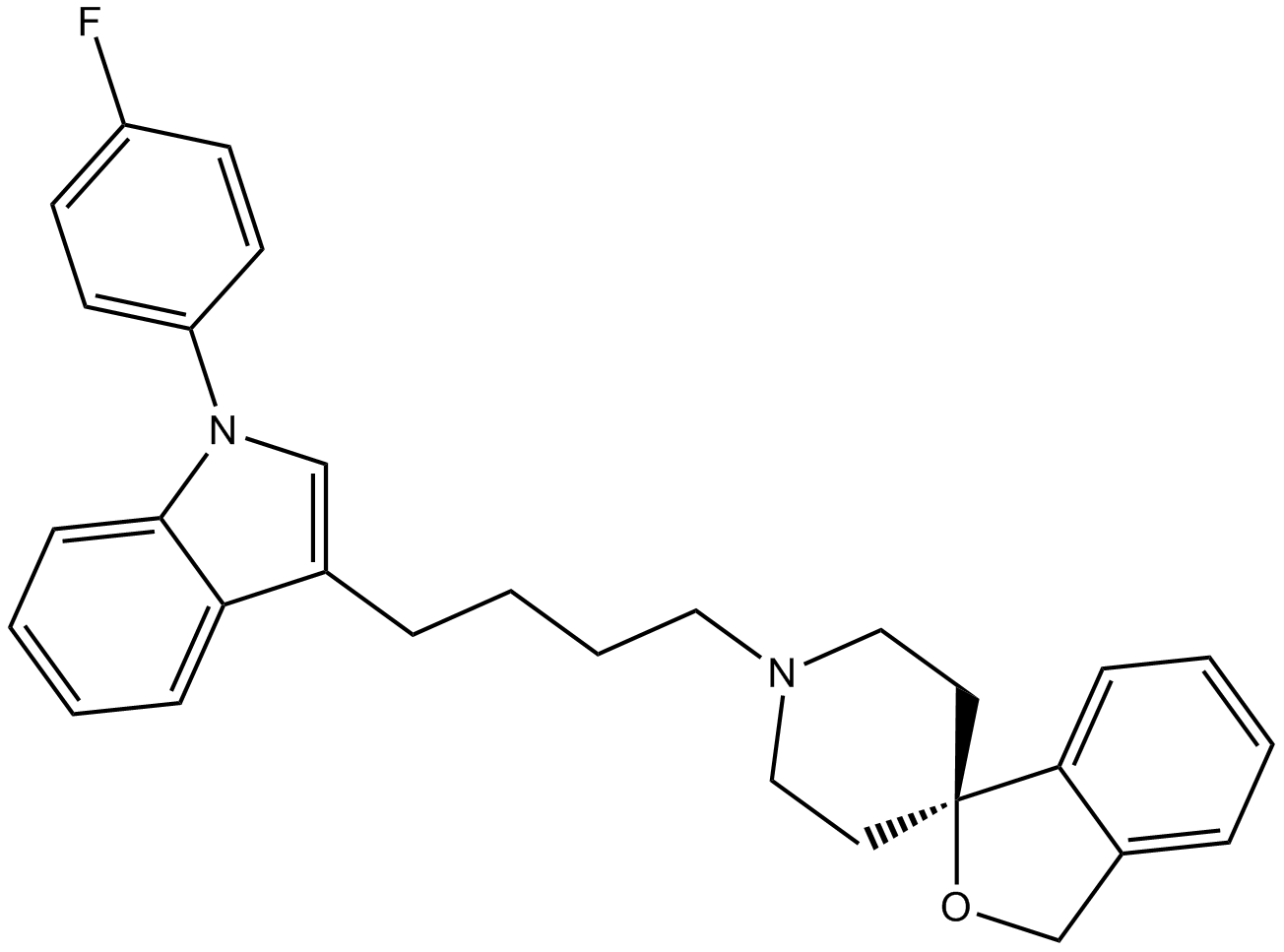
GC13083
Siramesine
Siramesine (Lu 28-179) is a potent sigma-2 receptor agonist. Siramesine has a subnanomolar affinity for sigma-2 receptors (IC50=0.12nM) and exhibits a 140-fold selectivity for sigma-2 receptors over sigma-1 receptors (IC50=17nM). Siramesine triggers cell death through destabilisation of mitochondria, but not lysosomes. Anti-cancer activity.
-
GC44893
Siramesine (fumarate)
Siramesine is an agonist of the sigma-2 (σ2) receptor (IC50 = 0.19 nM).
-
GC15222
Siramesine hydrochloride
Sigma-2 receptor agonist
-
GC17369
Sorafenib
Sorafenib acts as a multi-kinase inhibitor, targeting Raf-1 and B-Raf with IC50 values of 6 nM and 22 nM, respectively.
-
GC37664
Sorafenib (D3)
An internal standard for the quantification of sorafenib
-
GC37665
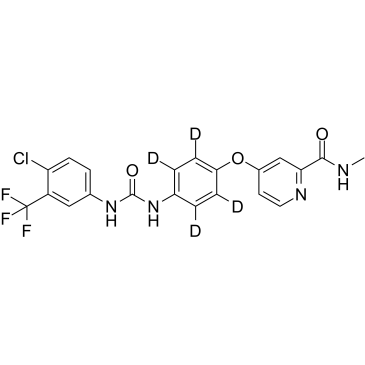
Sorafenib (D4)
Sorafenib (D4) (Bay 43-9006-d4) is the deuterium labeled Sorafenib. Sorafenib is a multikinase inhibitor IC50s of 6 nM, 20 nM, and 22 nM for Raf-1, B-Raf, and VEGFR-3, respectively.
-
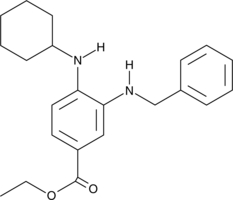
GC40922
SRS11-92
SRS11-92 is a ferroptosis inhibitor and a derivative of ferrostatin-1.
-
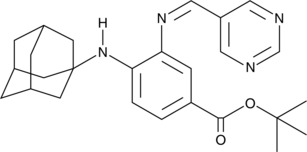
GC44947
SRS16-86
A ferroptosis inhibitor
-
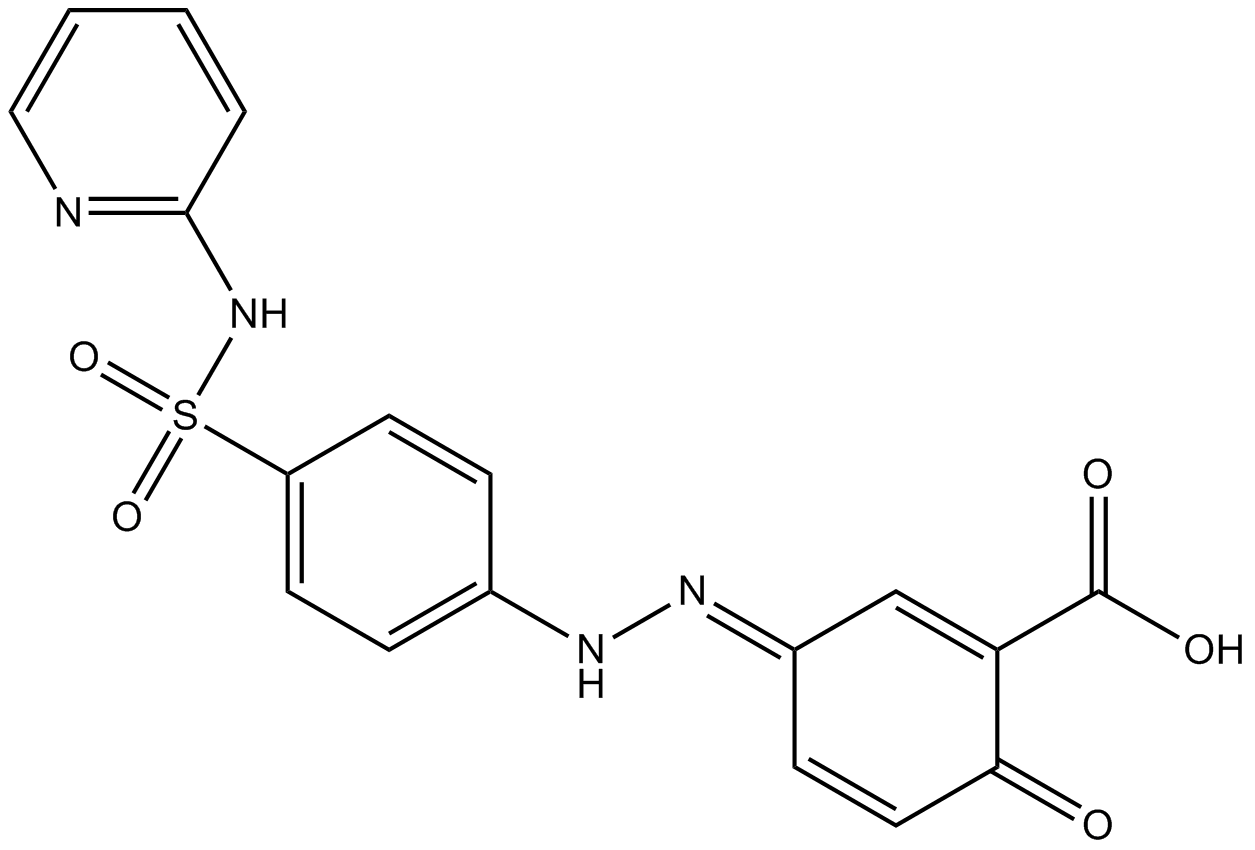
GC16868
Sulfasalazine
NF-κB activation inhibitor
-
GC34057
TBHQ (tert-Butylhydroquinone)
TBHQ (tert-Butylhydroquinone) (tert-Butylhydroquinone) is a widely used Nrf2 activator, protects against Doxorubicin (DOX)-induced cardiotoxicity through activation of Nrf2. TBHQ (tert-Butylhydroquinone) (tert-Butylhydroquinone) is also an ERK activator; rescues Dehydrocorydaline (DHC)-induced cell proliferation inhibitionin melanoma.
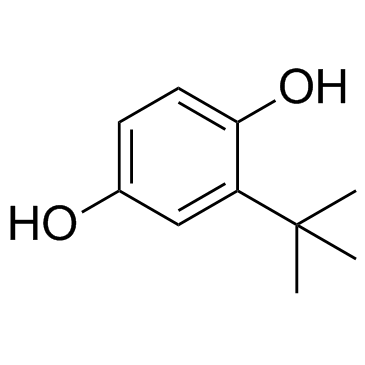
-
GN10289
Trigonelline
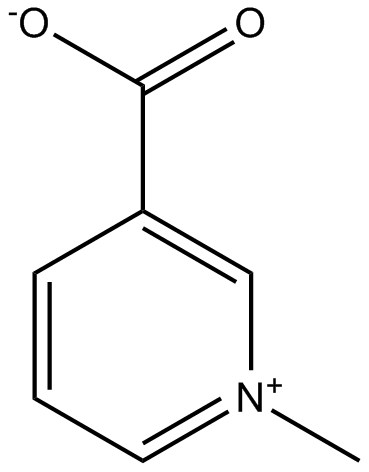
-
GC15272
Troglitazone
Selective PPARγ agonist
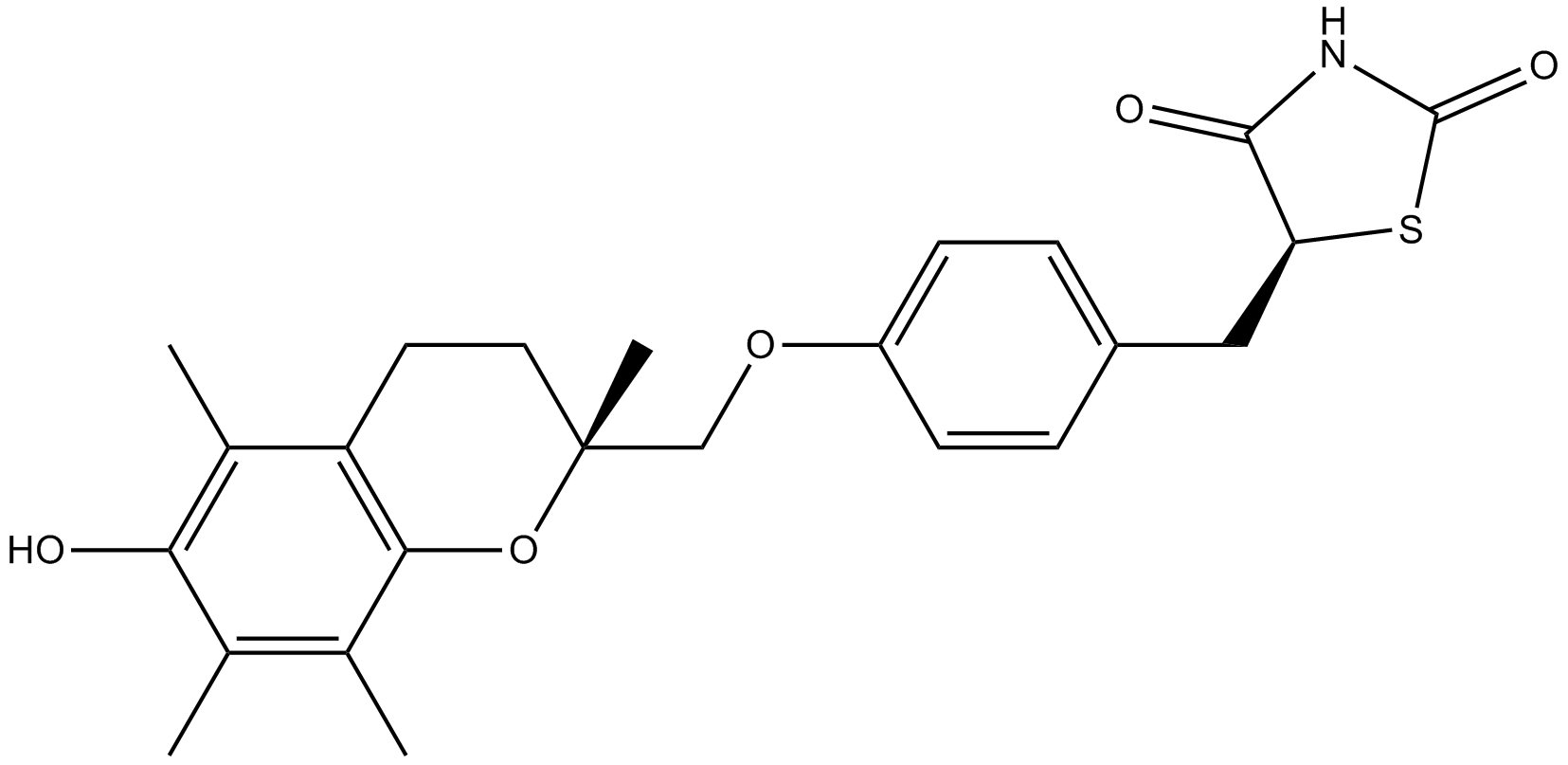
-
GC19457
Trolox
Trolox is a cell-permeable, water-soluble derivative of vitamin E with potent antioxidant properties.
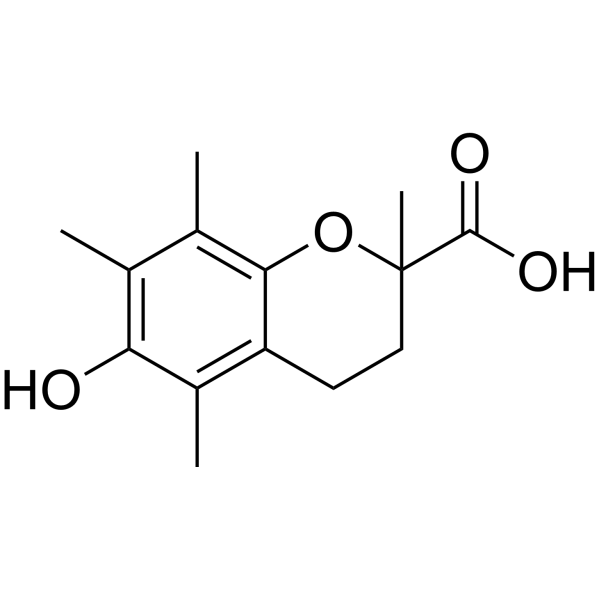
-
GC10451
U-73122
Inhibitor of phospholipase C, phospholipase A2, and 5-LO
-
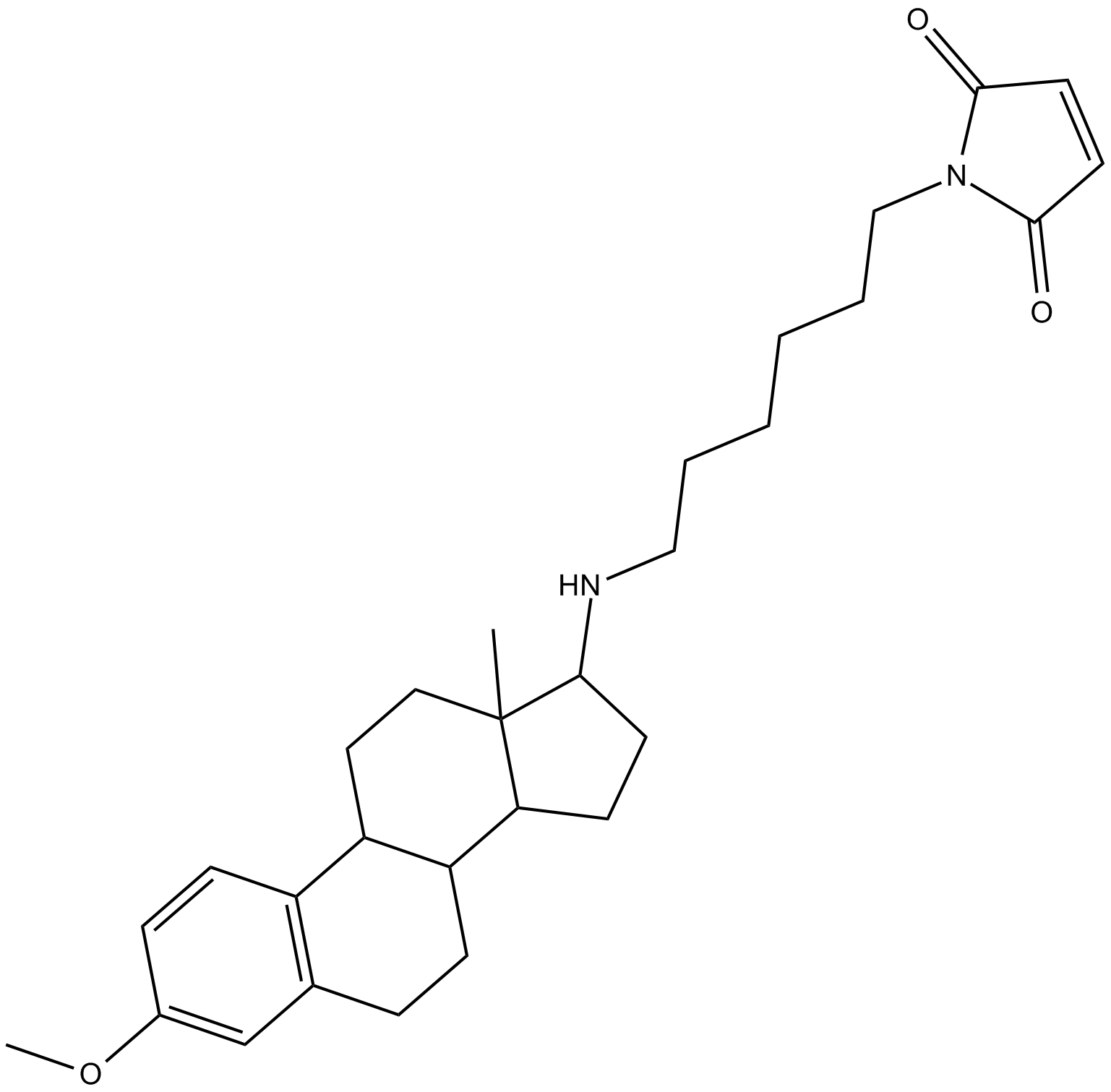
GC40193
UAMC-3203
UAMC-3203 is an inhibitor of ferroptosis that has an IC50 value of 10 nM for inhibition of erastin-induced ferroptosis in IMR-32 neuroblastoma cells.

-
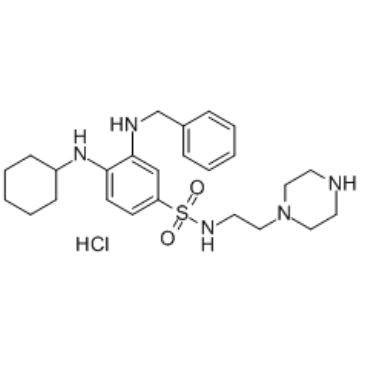
GC34939
UAMC-3203 hydrochloride
A ferroptosis inhibitor
-
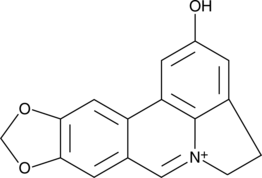
GC49308
Ungeremine
A betaine-type alkaloid with diverse biological activities
-
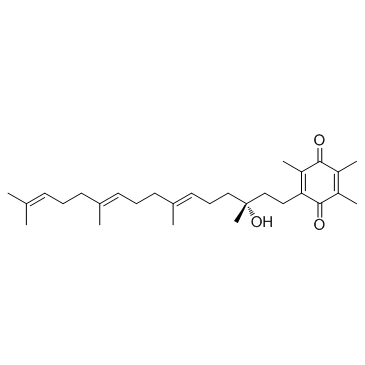
GC30512
Vatiquinone (EPI-743)
Vatiquinone (EPI-743) (Vatiquinone; α-Tocotrienol quinone; PTC-743; NCT04378075) is a potent cellular oxidative stress protectant, inhibits ferroptosis in cells, which could be used for the study for mitochondrial diseases.
-
GC16733
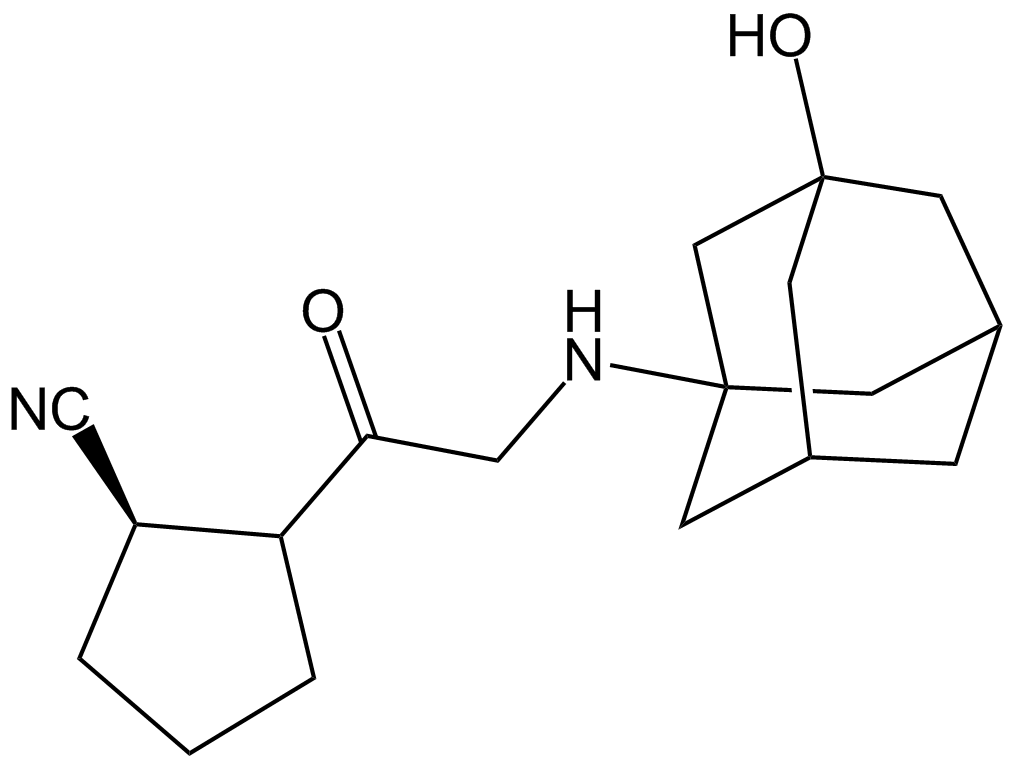
Vildagliptin (LAF-237)
A DPP-4 inhibitor
-
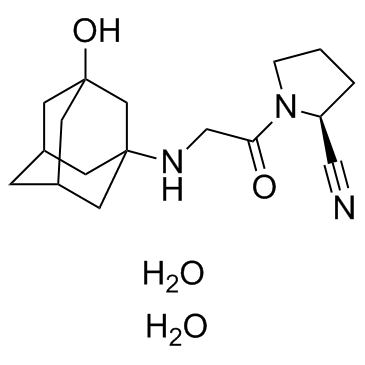
GC31492
Vildagliptin dihydrate (LAF237 dihydrate)
Vildagliptin dihydrate (LAF237 dihydrate) (LAF237 dihydrate) is a potent, stable, selective dipeptidyl peptidase IV (DPP-IV) inhibitor with an IC50 of 3.5 nM in human Caco-2 cells.
-
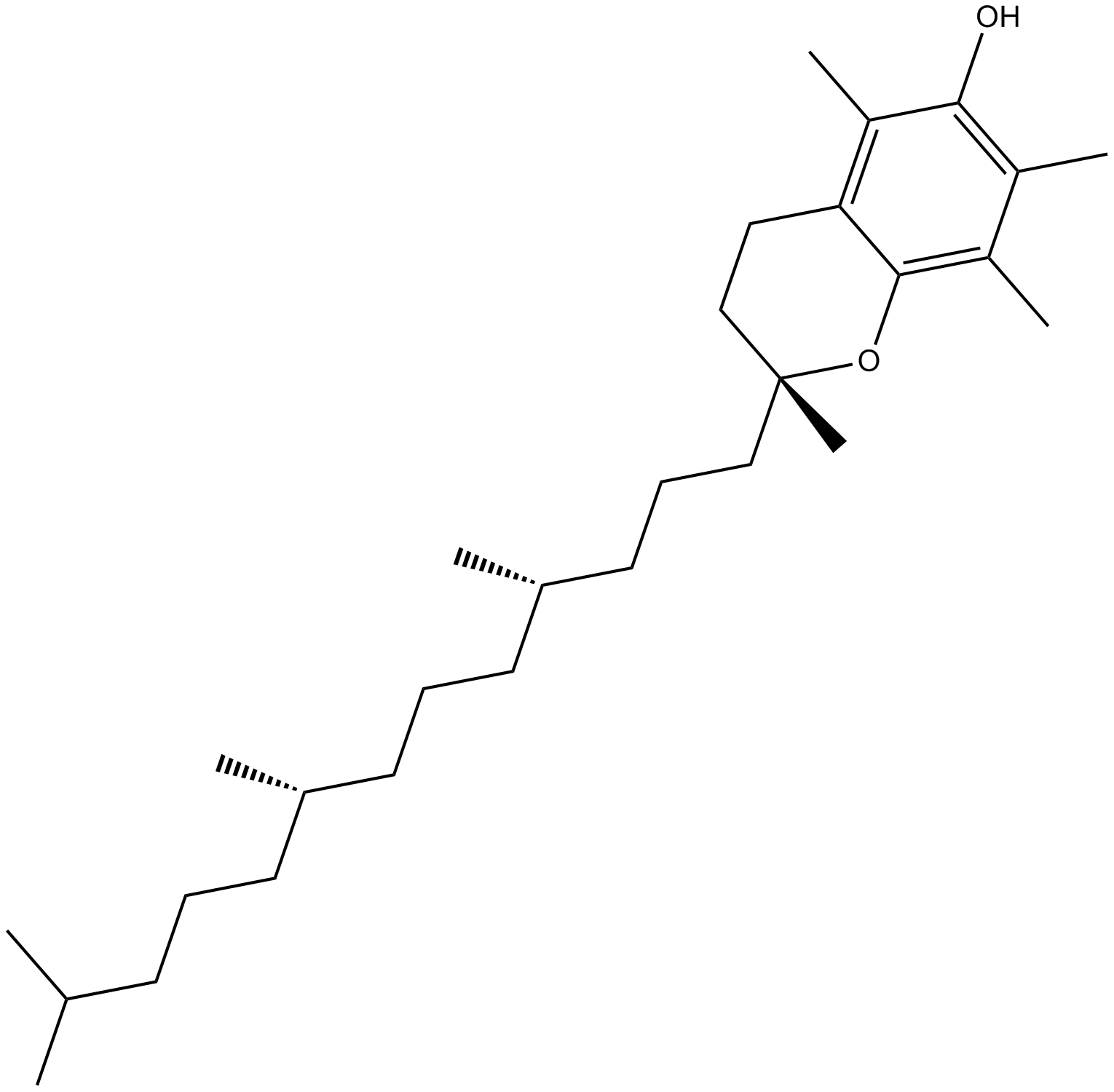
GC10042
Vitamin E
fat-soluble vitamin
-
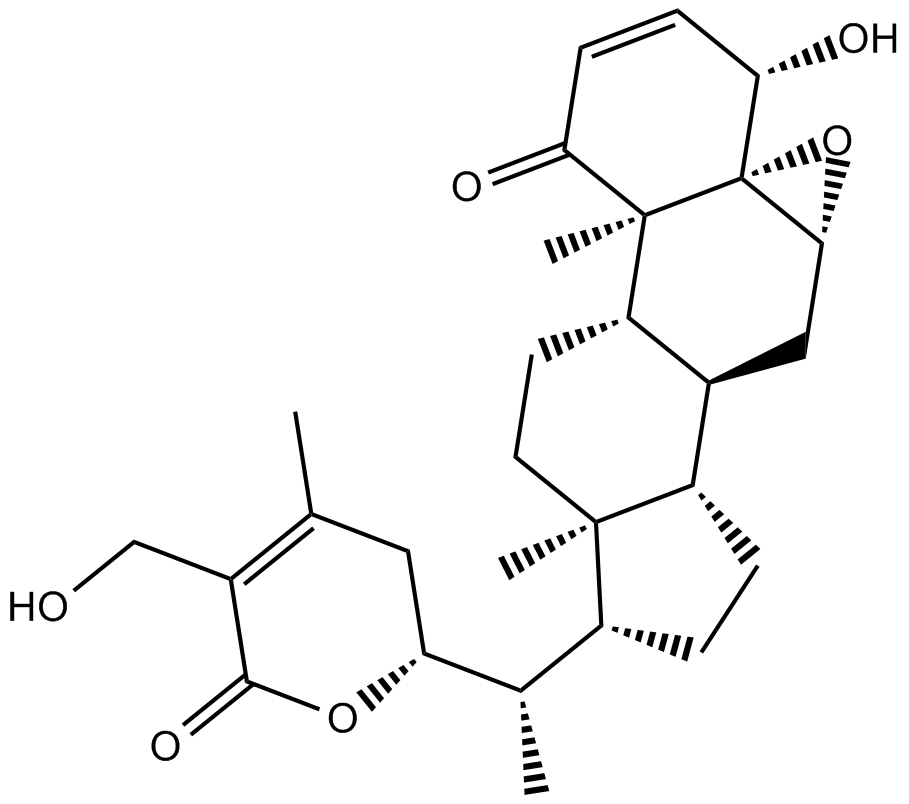
GC17309
Withaferin A
Prevents NF-κB activation by inhibiting activation of IKKβ
-
GC70157
YL-939
YL-939 is an effective inhibitor of ferroptosis. YL-939 inhibits ferroptosis by targeting PHB2/ferritin/iron axis.
-
GC16014
Zileuton
5-lipoxygenase (5-LOX) inhibitor, orally active