Metabolism
- Sterol Biosynthesis(26)
- PPAR(188)
- 5-alpha Reductase(9)
- 5-Lipoxygenase(11)
- Adenosine Deaminase(6)
- Aminopeptidase(8)
- C14ɑ demethylase(2)
- Carbonic Anhydrase(58)
- CETP(9)
- Cholesterol absorption(1)
- CPT1(2)
- CYP3A/CYP450(22)
- Dehydrogenase(132)
- DHFR(6)
- DGAT(8)
- Dopamine β-hydroxylase(10)
- Enolase(11)
- FAAH(36)
- Factor Xa(50)
- Ferroptosis(164)
- Folate Analogue(2)
- Glucokinase(18)
- HLE(0)
- HMG-CoA Reductase(37)
- HSP(64)
- IDO(46)
- KRAS-PDEδ(3)
- MAO(6)
- Metabolic Enzymes(0)
- Neuronal Metabolism(8)
- Oxidative Phosphorylation(17)
- P450(127)
- PDE(237)
- Phospholipase(118)
- Procollagen C Proteinase(1)
- Saccharometabolism(1)
- SCD(18)
- SGLT(28)
- TPH(4)
- Transferase(177)
- Energy Metabolism(0)
- Inorganic Ions(0)
- ALP(1)
- Carbohydrates(18)
- Uric Acid(0)
- Muscle Metabolism(0)
- MDA(0)
- MPO(0)
- Transaminase(20)
- Glutathione Reductase(1)
- Thioredoxin Reductase(0)
- Catalase(8)
- monooxygenase(1)
- PKM2(1)
- aldehyde dehydrogenase(1)
- Squalene synthase(1)
- Hydrolase(47)
- ornithine decarboxylase(1)
- Amino acid metabolism(3)
- phosphatases(92)
- Pyruvate kinase(12)
- Others(4)
- MGL(1)
- Galactosidase(6)
- 12-Lipoxygenase(1)
- Fatty Acid Synthase (FASN)(9)
- Dihydroorotate Dehydrogenase(13)
- Bile Acids & Microbiome(94)
- Bone Growth & Remodeling(35)
- Carbohydrate Metabolism(126)
- Cofactors & Vitamins(64)
- Dyslipidemias(73)
- Inborn Errors of Metabolism(80)
- Metabolic Syndrome(16)
- Necroptosis(10)
- Necrosis(15)
- Nutrient Sensing(12)
- Phosphodiesterase(27)
- Reproductive Biology(126)
- Thermogenesis(6)
- Prolyl Hydroxylation Enzymes(1)
- Biliary System(4)
- Metabolic Disease(5)
- Fat Mass and Obesity-associated Protein (FTO)(4)
- SHIP(1)
Products for Metabolism
- Cat.No. Product Name Information
-
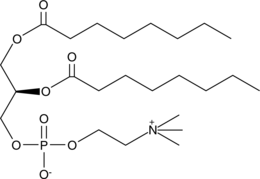
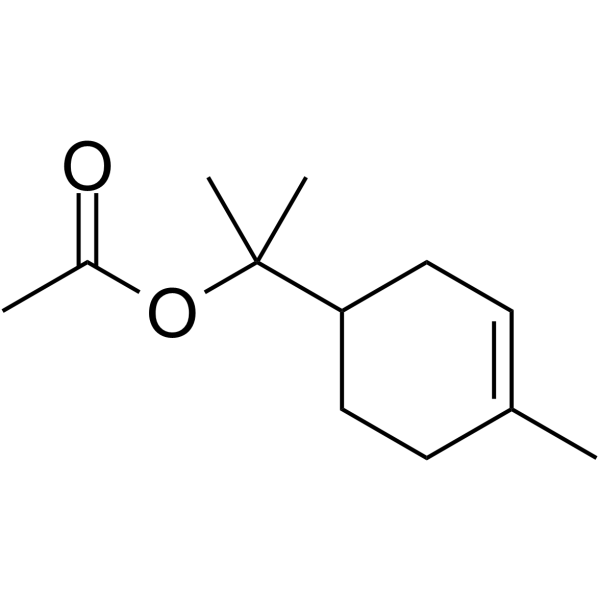
GC66197
α-?Terpinyl acetate
α-Terpinyl acetate is a monoterpene ester isolated from Laurus nobilis L. essential oil. α-Terpinyl acetate is a competitive P450 2B6 substrate which binding to the active site of P450 2B6 with a Kd value of 5.4 μM.
-
GC11123
α-CEHC
γTocopherol Metabolite, GTM, 2,7,8trimethyl2(βCarboxyEthyl)6Hydroxychroman
A major metabolite of δ-tocopherol
-
GC48276
α-Cyclodextrin (hydrate)
α-CD, NSC 269470, Schardinger α-Dextrin
A cyclic hexasaccharide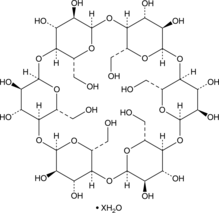
-
GC48278
α-D-Glucose-1,6-bisphosphate (potassium salt hydrate)
D-Glucose-1,6-diphosphate
A bis-phosphorylated derivative of α-D-glucose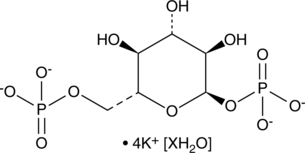
-
GC48279
α-D-Glucose-1-phosphate (sodium salt hydrate)
α-D-Glc 1-P
An intermediate in glycogen metabolism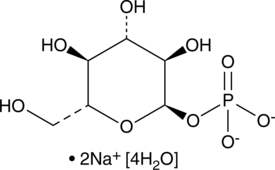
-
GC52253
α-Enolase (1-19)-biotin Peptide
Enolase-1 (1-19)-biotin
A biotinylated α-enolase peptide
-
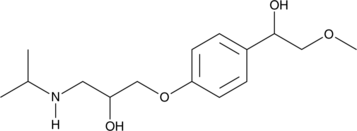
GC45208
α-hydroxy Metoprolol
α-hydroxy Metoprolol is an active metabolite of the β1-adrenergic receptor blocker metoprolol.
-
GC45210
α-Linolenic Acid (sodium salt)
ALA, C18:3 (9Z,12Z,15Z), C18:3 n-3
α-Linolenic acid (ALA) is an essential fatty acid found in leafy green vegetables.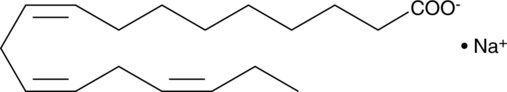
-
GC48283
α-Linolenic Acid-d14
ALAd14
An internal standard for the quantification of αLinolenic acid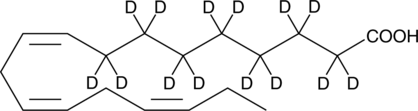
-
GC45602
α-Linolenic Acid-d5 MaxSpec• Standard
ALA-d5, C18:3 (9Z,12Z,15Z)-d5, C18:3 n-3-d5

-
GC49008
α-Muricholic Acid-d4
ω-MCA-d4
An internal standard for the quantification of α-muricholic acid
-
GC40480
α-Phenyl-α-(2-pyridyl)acetonitrile
NSC 16276, 2-Pyridylphenylacetonitrile
α-Phenyl-α-(2-pyridyl)thioacetamide, also known as antigastrin and SC-15396, is an inhibitor of gastric acid secretion.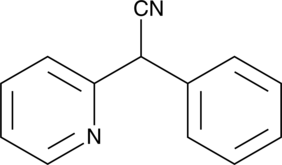
-
GC45232
β-D-Glucosamine Pentaacetate
N-Acetyl-β-D-Glucosamine tetraacetate, NSC 224432
β-D-Glucosamine pentaacetate is an N-acetylglucosamine derivative that has been shown to promote hyaluronic acid production.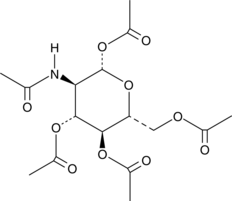
-
GC49769
β-Glucogallin
1-O-Galloyl-β-D-glucose
A plant metabolite and an aldose reductase 2 inhibitor
-
GC49890
β-Glycerophosphate-d5 (sodium salt hydrate)
Glycerol 2-Phosphate-d5
An internal standard for the quantification of β-glycerophosphate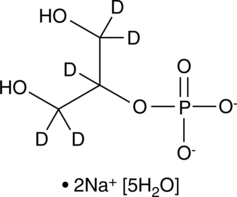
-
GC49647
β-Hyodeoxycholic Acid (hydrate)
3β,6α-dihydroxy-5β-Cholan-24-oic Acid, isoHDCA, Isohyodeoxycholic Acid
A 3β epimer of hyodeoxycholic acid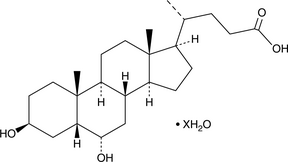
-
GC49007
β-Muricholic Acid-d4
ω-MCA-d4
An internal standard for the quantification of β-muricholic acid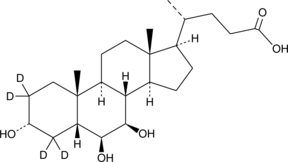
-
GC41502
β-Myrcene
NSC 406264
β-Myrcene is a terpene that has been found in Cannabis and has antioxidative properties.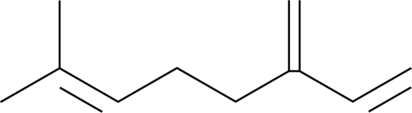
-
GC40551
γ-CEHC
γTocopherol Metabolite, GTM, 2,7,8trimethyl2(βCarboxyEthyl)6Hydroxychroman
The major tocopherol obtained from natural dietary sources is γ-tocopherol, whereas α-tocopherol is the form of Vitamin E typically obtained from synthetic supplements.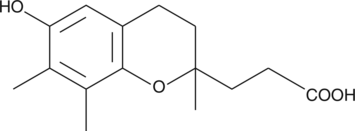
-
GC52400
γ-Glu-Ala (trifluoroacetate salt)
γ-Glutamylalanine, γ-L-Glutamyl-L-alanine
A dipeptide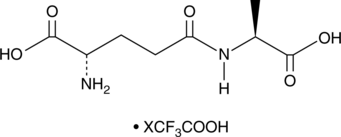
-
GC52404
γ-Glu-Phe (trifluoroacetate salt)
γ-Glutamylphenylalanine
A dipeptide with metabolism-altering activity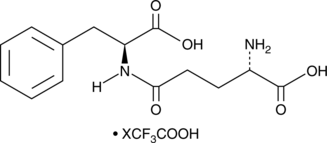
-
GC45239
δ4-Abiraterone
CB 7627, D4A
δ4-Abiraterone is a major metabolite of abiraterone. δ4-Abiraterone is an inhibitor of CYP17A1, 3b-hydroxysteroid dehydrogenase (3βHSD) and steroid-5a-reductase (SRD5A) and also an antagonist of androgen receptor.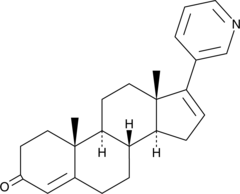
-
GC15975
α-Estradiol
Alfatradiol, α-Estradiol, 17-epi Estradiol, NSC 20293, 17α-Oestradiol
α-Estradiol is a weak estrogen and a 5α-reductase inhibitor which is used as a topical medication in the treatment of androgenic alopecia.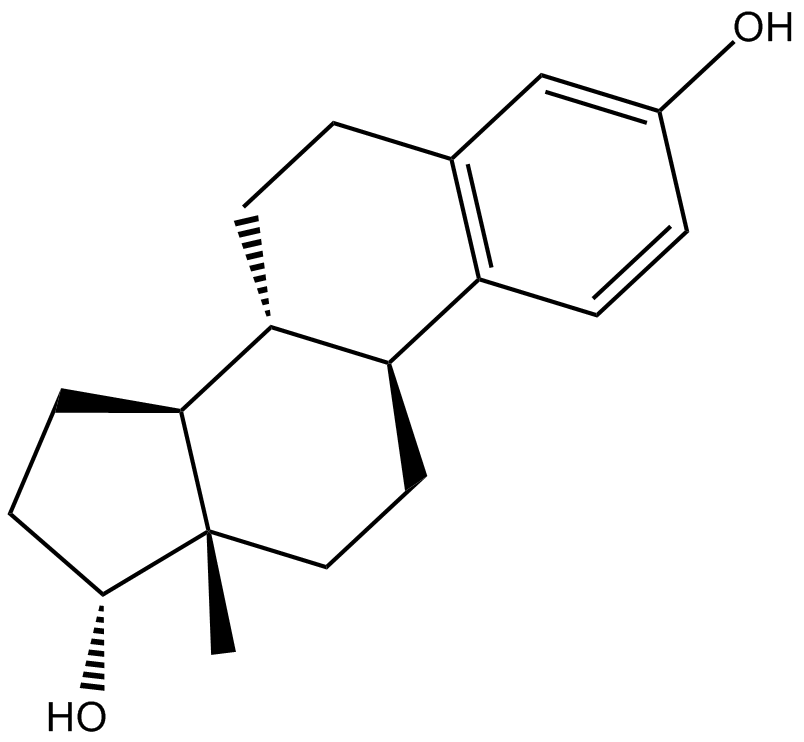
-
GC49009
ω-Muricholic Acid-d4
ω-MCA-d4
An internal standard for the quantification of ω-MCA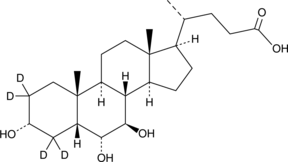
-
GC49296
τ-Fluvalinate
tau-Fluvalinate
A pyrethroid acaricide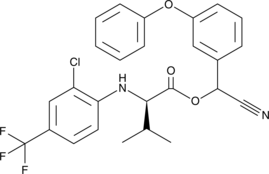
-
GC46314
(±)-β-Hydroxybutyrate-d4 (sodium salt)
βHydroxybutanoic Acidd4
An internal standard for the quantification of (±)βhydroxybutyrate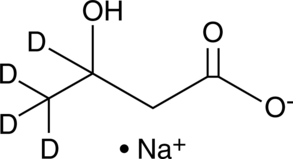
-
GC52010
(±)-10-hydroxy-12(Z),15(Z)-Octadecadienoic Acid
αHYA, (±)-10-hydroxy-12(Z),15(Z)-ODE
An oxylipin gut microbiota metabolite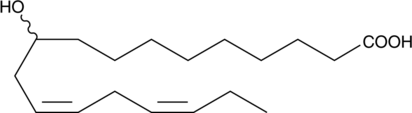
-
GC52013
(±)-10-hydroxy-12(Z)-Octadecenoic Acid
10-hydroxy-cis-12-Octadecenoic Acid
An oxylipin and metabolite of linoleic acid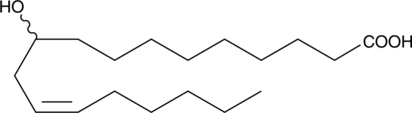
-
GC52421
(±)-10-hydroxy-12(Z)-Octadecenoic Acid-d5
10-hydroxy-cis-12-Octadecenoic Acid-d5
An internal standard for the quantification of (±)-10-hydroxy-12(Z)-octadecenoic acid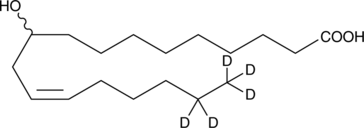
-
GC41661
(±)-4-hydroxy Propranolol β-D-Glucuronide
(±)-4-hydroxy Propranolol β-D-glucuronide is a metabolite of (±)-4-hydroxy propranolol, which is a metabolite of propranolol.
-
GC46284
(±)-Cotinine-d3
An internal standard for the quantification of cotinine
-
GC41671
(±)-Equol 4'-sulfate (sodium salt)
R,S-Equol 4'-Sulfate
(±)-Equol 4'-sulfate is a gut-mediated phase II metabolite of the isoflavonoid phytoestrogen (±)-equol.
-
GC41315
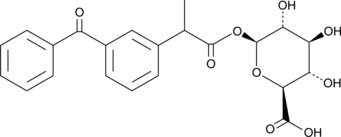
(±)-Ketoprofen Glucuronide
rac-Ketoprofen Acyl-β-D-glucuronide, (R,S)-Ketoprofen Glucuronide
(±)-Ketoprofen glucuronide is a phase II metabolite of the non-steroidal anti-inflammatory drug (NSAID) ketoprofen.
-
GC40954
(±)-N-3-Benzylnirvanol
(S)-N-3-benzyl Nirvanol
(±)-N-3-Benzylnirvanol is a racemic mixture of (+)-N-3-benzylnirvanol and (-)-N-3-benzylnirvanol.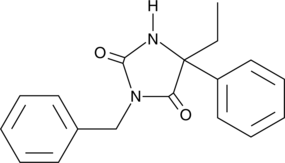
-
GC49875
(±)-N-desmethyl Venlafaxine (hydrochloride)
Wy 45494
A minor active metabolite of venlafaxine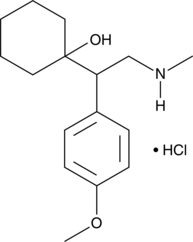
-
GC39271
(±)-Naringenin
SDihydrogenistein, NSC 11855, NSC 34875, Salipurol
A citrusderived flavonoid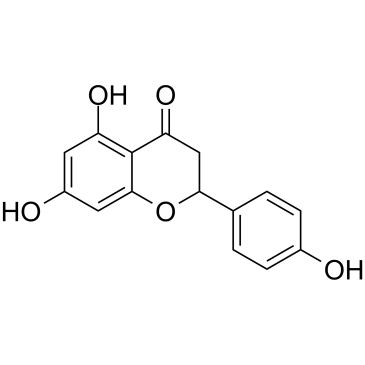
-
GC45618
(±)-trans-GK563
GK563
A GVIA iPLA2 inhibitor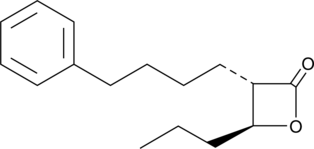
-
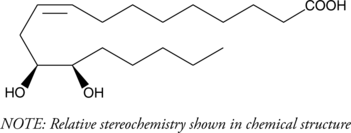
GC40802
(±)12(13)-DiHOME
Isoleukotoxin diol
(±)12(13)-DiHOME is the diol form of (±)12(13)-EpOME, a cytochrome P450-derived epoxide of linoleic acid also known as isoleukotoxin.
-
GC40270
(±)5(6)-DiHET
(±)5,6-DiHETrE
5(6)-DiHET is a fully racemic version of the enantiomeric forms biosynthesized from 5(6)-EET by epoxide hydrolases.
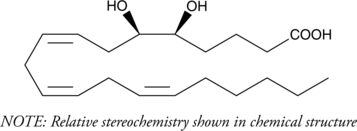
-
GC40282
(+)-5-trans Cloprostenol
DCloprostenol, (+)5,6trans Cloprostenol, (+)5trans16mchlorophenoxy tetranor PGF2α, (+)5trans16mchlorophenoxy tetranor Prostaglandin F2α
Cloprostenol is a synthetic derivative of prostaglandin F2α that is used in veterinary medicine as a luteolytic agent for the induction of estrus and in the treatment of reproductive disorders in cattle, swine, and horses.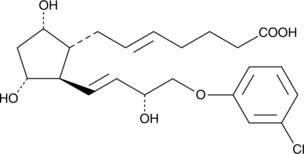
-
GC45260
(+)-Cloprostenol isopropyl ester
(+)-5-cis Cloprostenol isopropyl ester, (+)16mchlorophenoxy tetranor Prostaglandin F2α isopropyl ester
(+)-Cloprostenol is a synthetic analog of prostaglandin F2α (PGF2α).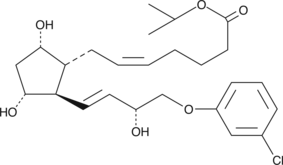
-
GC45262
(+)-Cloprostenol methyl ester
DCloprostenol methyl ester, (+)16mChlorophenoxy tetranor PGF2α methyl ester
(+)-Cloprostenol is a synthetic analog of prostaglandin F2α (PGF2α).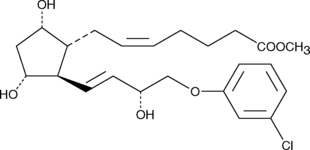
-
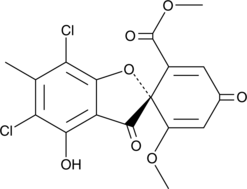
GC45264
(+)-Geodin
(+)-Geodin is a fungal metabolite.
-
GC12404
(+)-Ketoconazole
R 41400
Potent inhibitor of cytochrome P450c17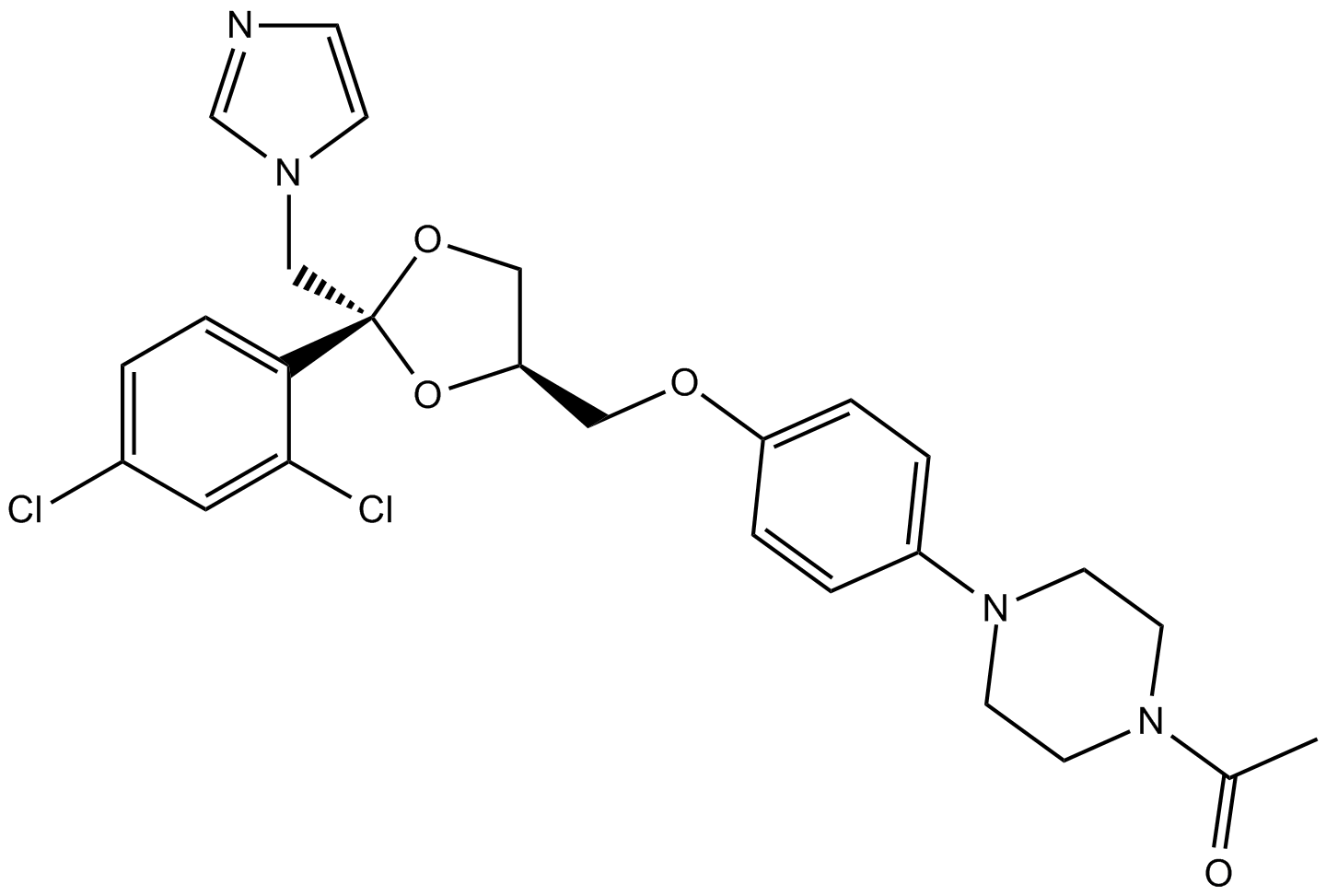
-
GC30424
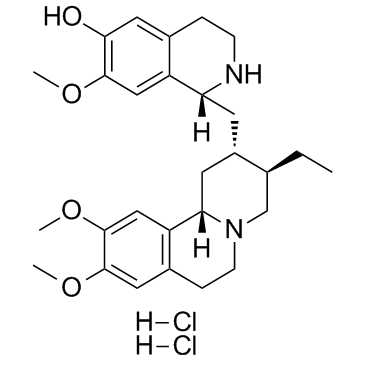
(-)-Cephaeline dihydrochloride (NSC 32944)
(-)-Cephaeline dihydrochloride (NSC 32944) is a selective CYP2D6 inhibtor with an IC50 of 121 μM.
-
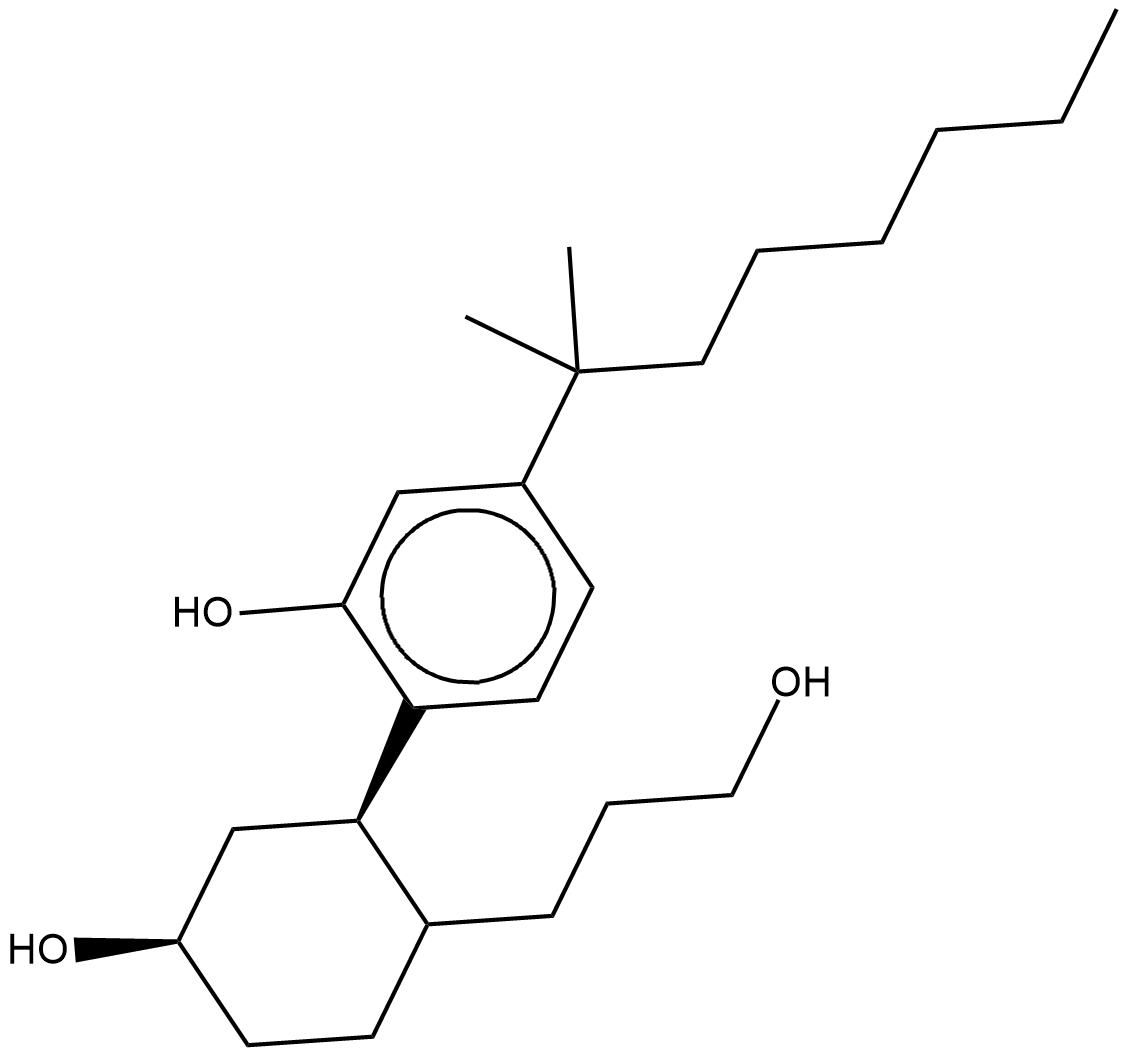
GC18343
(-)-CP 55,940
(-)-CP 55,940 is a potent and non-selective cannabinoid (CB) receptor agonist with Ki values of 0.5 to 5 and 0.69 to 2.8 nM for CB1 and CB2 receptors, respectively.
-
GC14049
(-)-Epigallocatechin gallate (EGCG)
EGCG
(-)-Epigallocatechin Gallate sulfate (EGCG) is a major polyphenol in green tea that inhibits cell proliferation and induces apoptosis.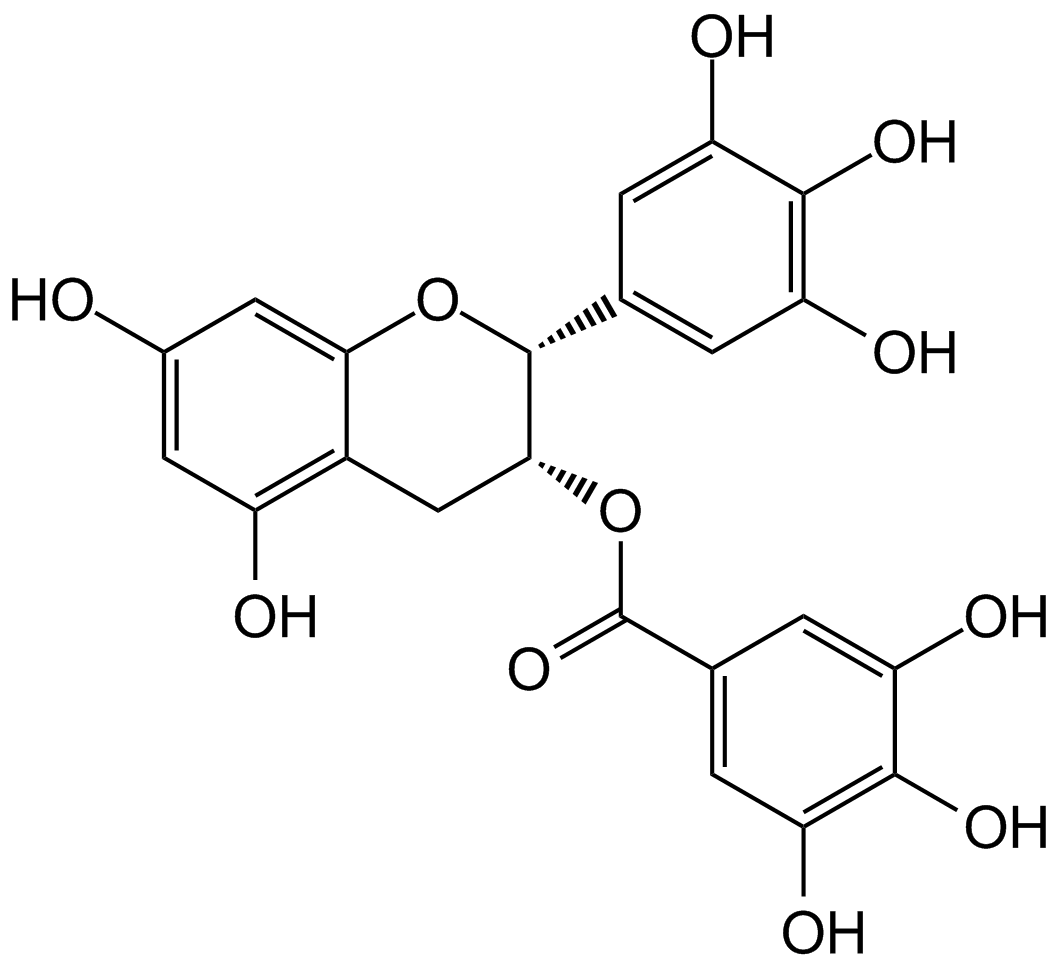
-
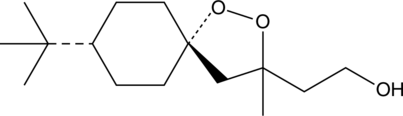
GC45248
(-)-FINO2
(-)-FINO2 is a ferroptosis-inducing peroxide compound that indirectly inhibits glutathione peroxidase 4 (GPX4) and oxidizes iron.
-
GC14520
(-)-p-Bromotetramisole Oxalate
(-)-p-Bromotetramisole, L-para-Bromotetramisole
(-)-p-Bromotetramisole Oxalate(L-p-bromotetramisole) is a potent and non-specific inhibitor of alkaline phosphatase and is also an inhibitor of protein tyrosine phosphatases.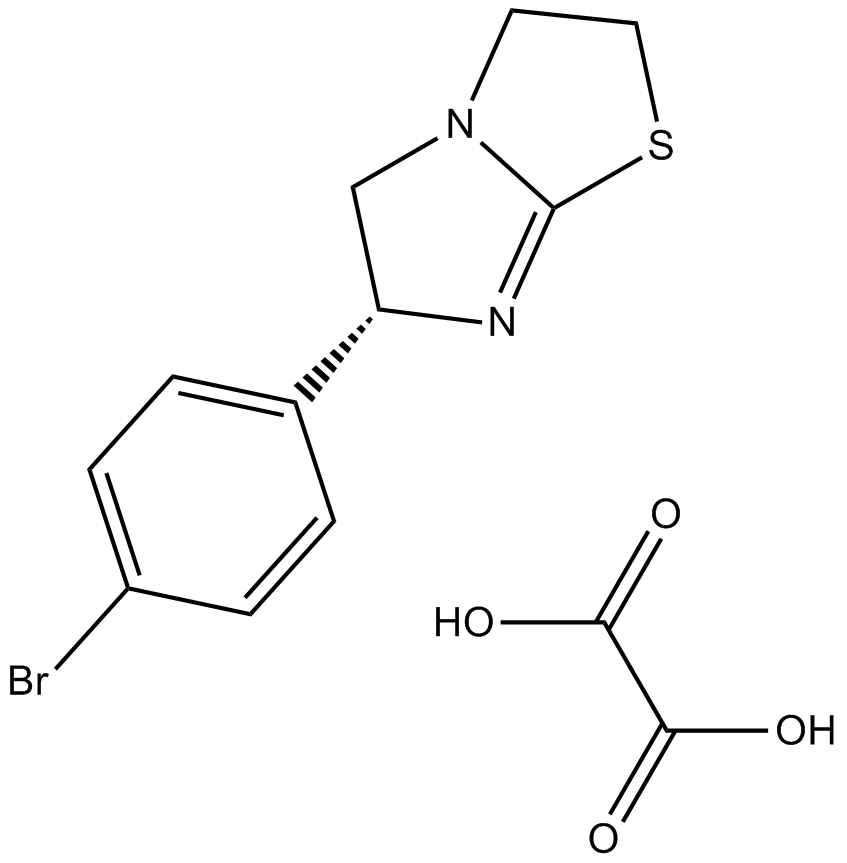
-
GC45252
(-)-Sitagliptin Carbamoyl Glucuronide
(R)-Sitagliptin Carbamoyl Glucuronide
(-)-Sitagliptin carbamoyl glucuronide is a minor phase II metabolite of the dipeptidyl peptidase 4 (DPP-4) inhibitor (-)-sitagliptin.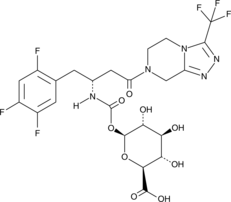
-
GC18622
(2'S)-Nicotine-1-oxide
Nicotine-N-oxide
(2'S)-Nicotine-1-oxide is a metabolite of nicotine .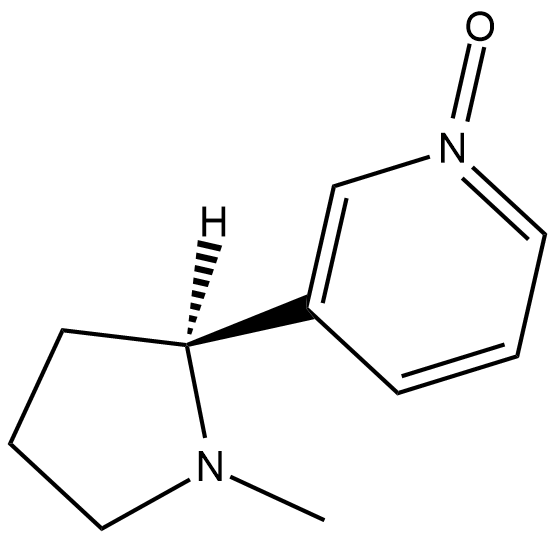
-
GC41691
(2R)-Glycerol-O-β-D-galactopyranoside
3-O-β-D-Galactopyranosyl-sn-glycerol
(2R)-Glycerol-O-β-D-galactopyranoside is a substrate for β-galactosidase, the lactose repressor, the galactose-binding protein, and the β-methylgalactoside transport system.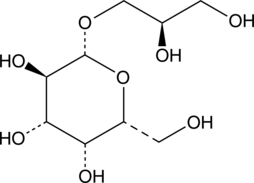
-
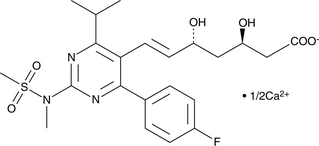
GC49690
(3R,5R)-Rosuvastatin (calcium salt)
A potential impurity found in bulk preparations of rosuvastatin
-
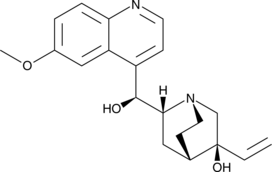
GC41694
(3S)-hydroxy Quinidine
(3S)-hydroxy Quinidine is an active quinidine metabolite.
-
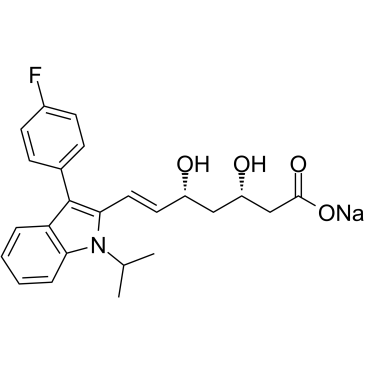
GC60396
(3S,5R)-Fluvastatin sodium
(3S,5R)-Fluvastatin sodium ((3S,5R)-XU 62-320) is the (3S,5R)-enantiomer?of Fluvastatin.
-
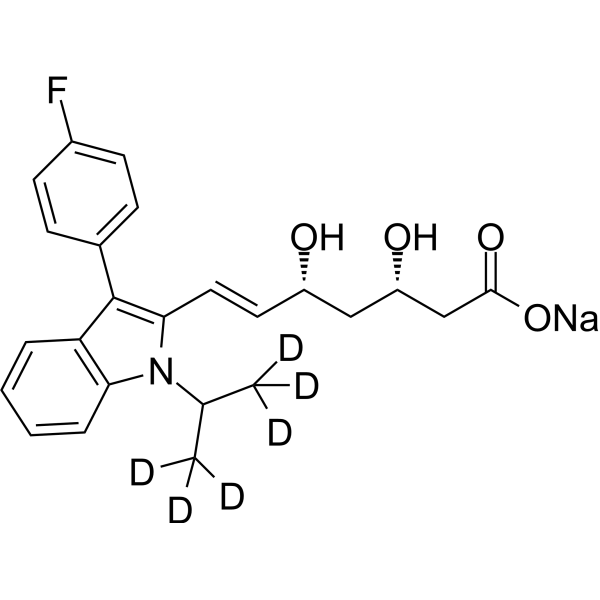
GC68317
(3S,5R)-Fluvastatin-d6 sodium
(3S,5R)-XU 62-320 D6
-
GC13751
(3S,5S)-Atorvastatin (sodium salt)
negetive control of Atorvastatin, HMG-CoA reductase inhibitor
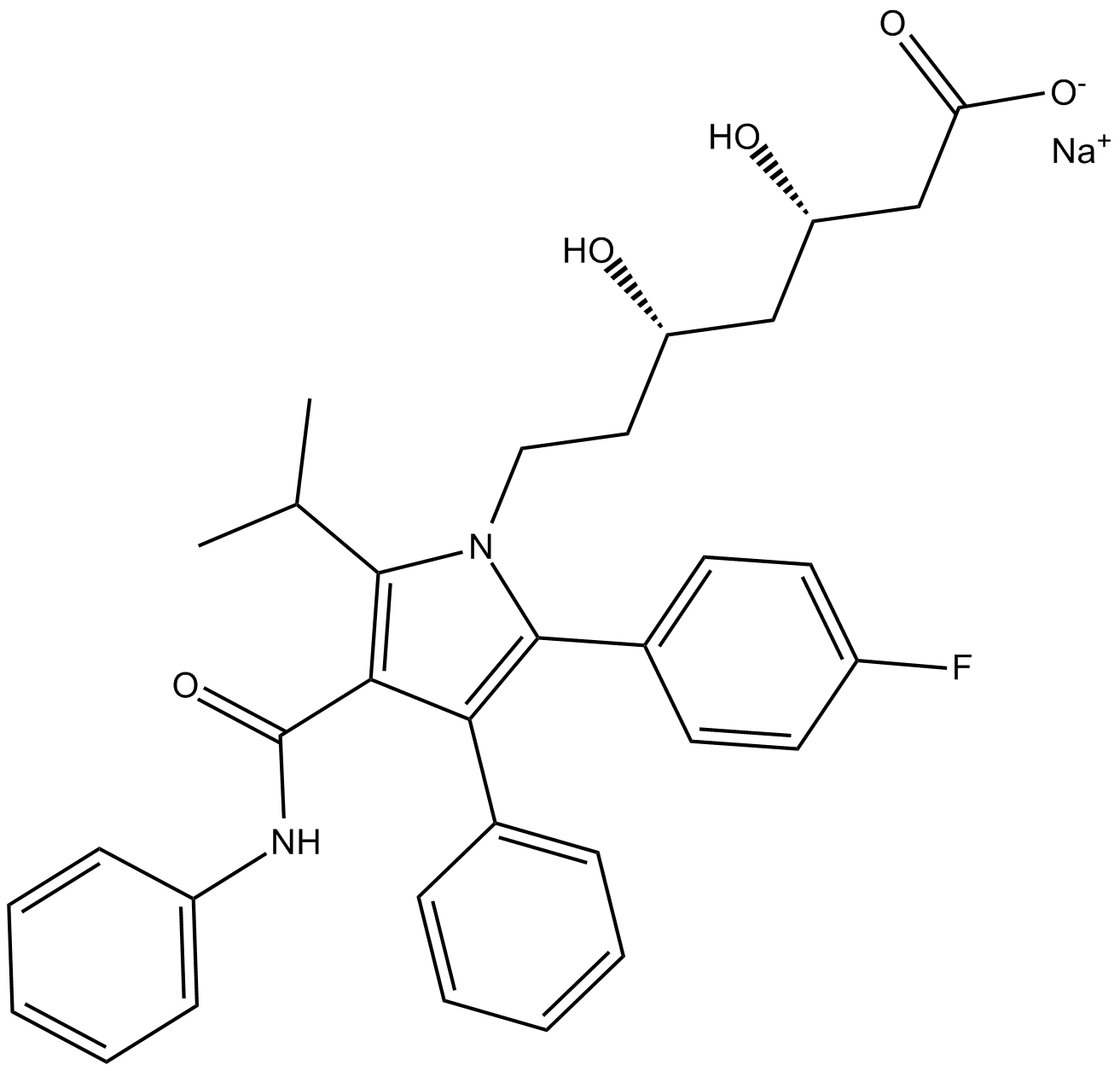
-
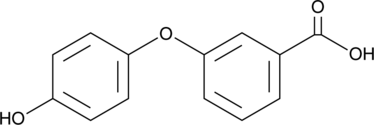
GC46333
(4′-Hydroxy)phenoxybenzoic Acid
A pyrethroid insecticide metabolite
-
GC48508
(4-Carboxybutyl-d4)triphenylphosphonium (bromide)
TPP-d4, 5-Triphenylphosphoniovaleric Acid-d4
An internal standard for the quantification of (4-carboxybutyl)triphenylphosphonium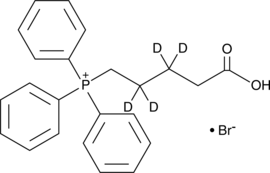
-
GC41695
(6R,S)-5,6,7,8-Tetrahydrofolic Acid (hydrochloride)
Tetrahydrofolate, THFA
(6R,S)-5,6,7,8-Tetrahydrofolic acid (THFA), the reduced form of folic acid, serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.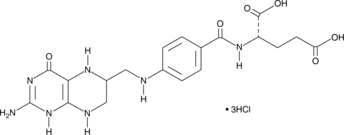
-
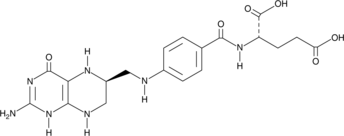
GC41088
(6S)-Tetrahydrofolic Acid
(6S)-Tetrahydrofolic acid is a diastereomer of tetrahydrofolic acid, a reduced form of folic acid that serves as a cofactor in methyltransferase reactions and is the major one-carbon carrier in one carbon metabolism.
-
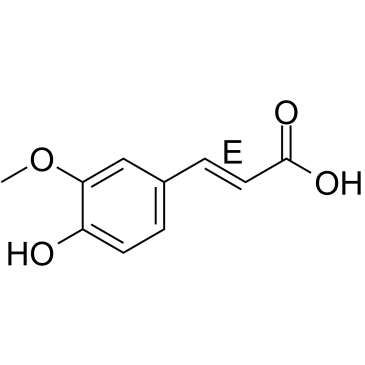
GC34980
(E)-Ferulic acid
(E)-Ferulic acid is a isomer of Ferulic acid which is an aromatic compound, abundant in plant cell walls. (E)-Ferulic acid causes the phosphorylation of β-catenin, resulting in proteasomal degradation of β-catenin and increases the expression of pro-apoptotic factor Bax and decreases the expression of pro-survival factor survivin. (E)-Ferulic acid shows a potent ability to remove reactive oxygen species (ROS) and inhibits lipid peroxidation. (E)-Ferulic acid exerts both anti-proliferation and anti-migration effects in the human lung cancer cell line H1299.
-
GC40286
(E,Z)-2-propyl-2-Pentenoic Acid
2-propyl-2-Pentenoate, 2-propylpenten-2-oic Acid, 2-ene-VPA
(E,Z)-2-propyl-2-Pentenoic acid is a bioactive metabolite of valproic acid that exhibits the same profile and potency of anticonvulsant activity in animal models as its parent compound without any observed teratogenicity and hepatotoxicity.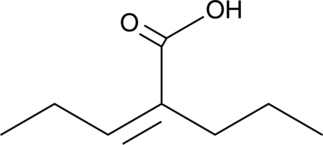
-
GC49189
(E/Z)-4-hydroxy Tamoxifen-d5
Afimoxifene-d5, 4-OHT-d5
An internal standard for the quantification of (E/Z)-4-hydroxy tamoxifen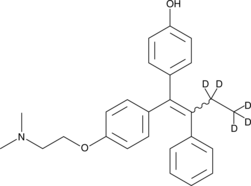
-
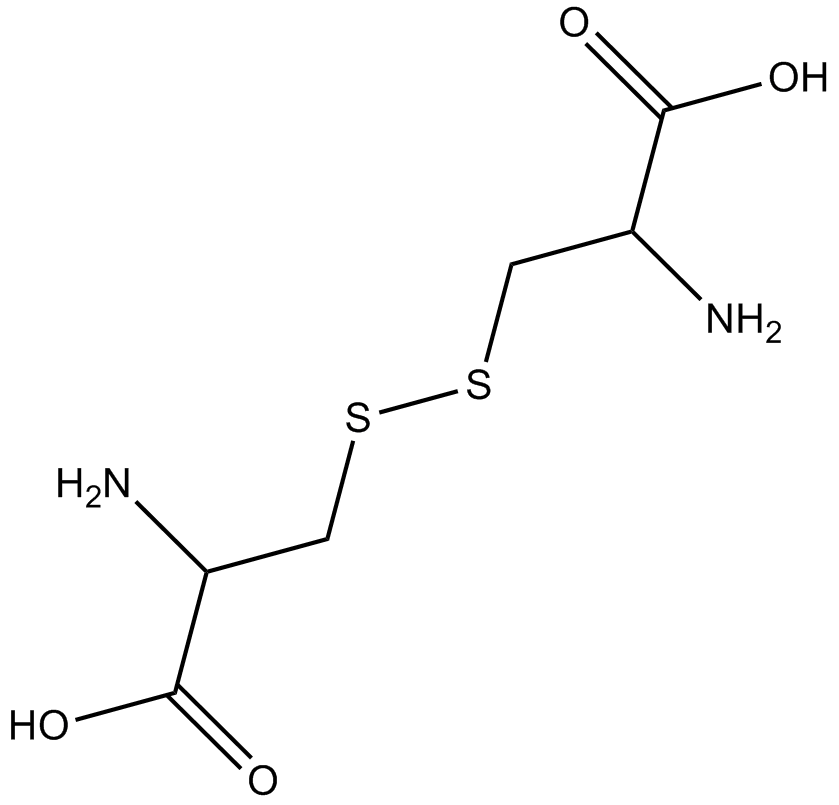
GA11210
(H-Cys-OH)2
(–)-Cystine, NSC 13203
-
GC34984
(R)-(-)-Rolipram
(R)-(–)-Rolipram, (R)-Rolipram
(R)-(-)-Rolipram is the R-enantiomer of Rolipram.
-
GC25001
(R)-Avanafil
R-Avanafil is a strong competitive inhibitor of phosphodiesterase 5 (PDE5) with a demonstrated in vitro IC 50 of 5.2 nM.
-
GC40678
(R)-Bromoenol lactone
(R)BEL
The phospholipases are an extensive family of lipid hydrolases that function in cell signaling, digestion, membrane remodeling, and as venom components.
-
GC52290
(R)-HTS-3
An inhibitor of LPCAT3
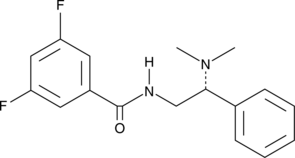
-
GC70609
(R)-IDO/TDO-IN-1
(R)-IDO/TDO-IN-1 (compound 25) is an indoleamine-2,3-dioxygenase (IDO) inhibitor, with good pharmacokinetic properties.

-
GC67864
(R)-Irsenontrine
(R)-E2027

-
GC41722
(R,S)-Carvedilol Glucuronide
(R,S)-Carvedilol glucuronide is a racemic mixture of the carvedilol metabolites (R)-carvedilol glucuronide and (S)-carvedilol glucuronide.
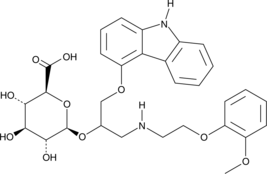
-
GC61548
(Rac)-5-Keto Fluvastatin
(Rac)-5-Keto Fluvastatin (3-Hydroxy-5-Keto Fluvastatin) is an impurity of Fluvastatin (XU 62320).
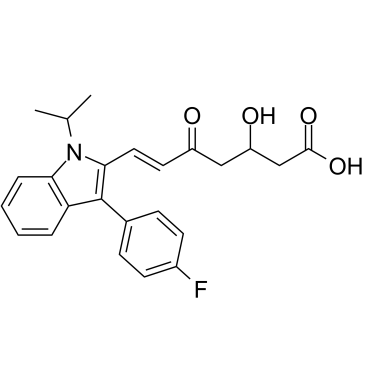
-
GC62743
(Rac)-EC5026
(Rac)-BPN-19186
(Rac)-EC5026 ((Rac)-BPN-19186) is a potent piperidine inhibitor of soluble epoxide hydrolase (sEH) extracted from patent WO2019156991A1, page 39, has a Ki of 0.06 nM.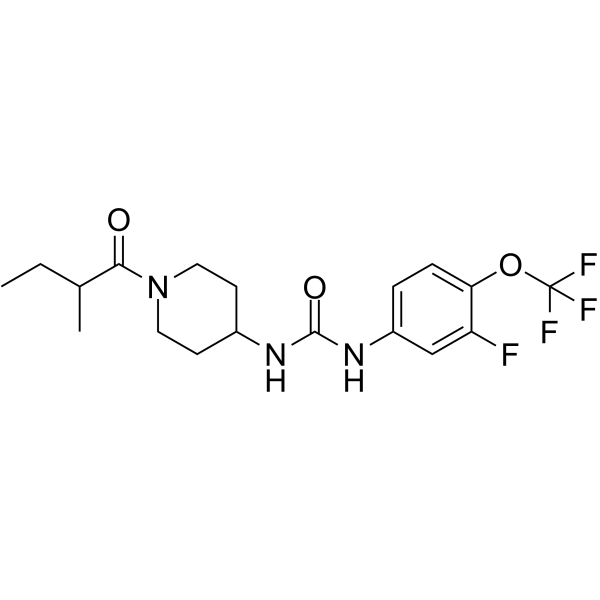
-
GC69796
(Rac)-Etavopivat
(Rac)-FT-4202
(Rac)-Etavopivat ((Rac)-FT-4202) is an isomer of Etavopivat. Etavopivat is an orally active activator of red blood cell pyruvate kinase-R (PKR), which can be used for research on sickle cell disease and other hemoglobin disorders.
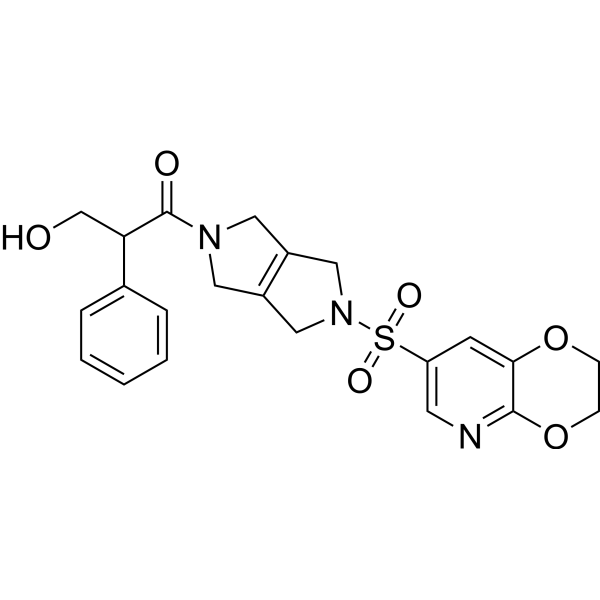
-
GC61750
(Rac)-Indoximod
(Rac)-Indoximod (1-Methyl-DL-tryptophan) is an indoleamine 2,3-dioxygenase (IDO) inhibitor.
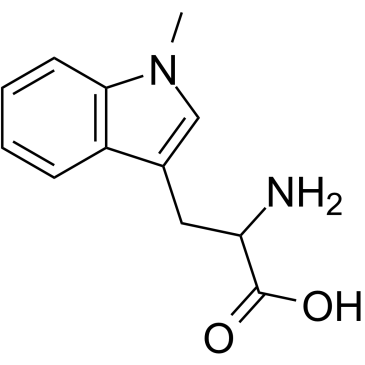
-
GC46347
(S)-(+)-Methoprene
Altosid, d-Methoprene, ZR 2458
An insect growth regulator and Met agonist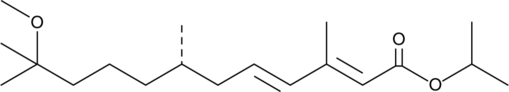
-
GC40679
(S)-Bromoenol lactone
(S)BEL
The phospholipases are an extensive family of lipid hydrolases that function in cell signaling, digestion, membrane remodeling, and as venom components.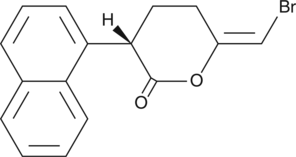
-
GC50321
(S)-C33
Potent PDE9 inhibitor
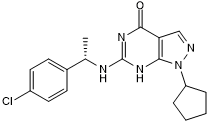
-
GC48719
(S)-Canadine
(–)-Canadine, (S)-Tetrahydroberberine
(S)-Canadine is an alkaloid and intermediate in the biosynthesis of berberine with insecticidal activity.
-
GC46352
(S)-DO271
An inactive control for DO264
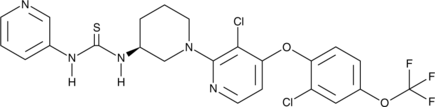
-
GC49520
(S)-Equol
4',7Dihydroxyisoflavan, (-)Equol, 4',7Isoflavandiol
An estrogen receptor β agonist
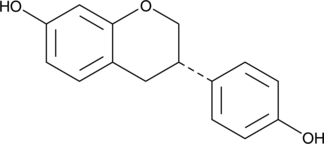
-
GC60417
(S)-Indoximod
NSC 77678, L-1-Methyltryptophan, 1-L-MT, L-1MT
(S)-Indoximod (1-Methyl-L-tryptophan) is an inhibitor of indoleamine 2,3-dioxygenase (IDO). (S)-Indoximod can be used for the research of cancer.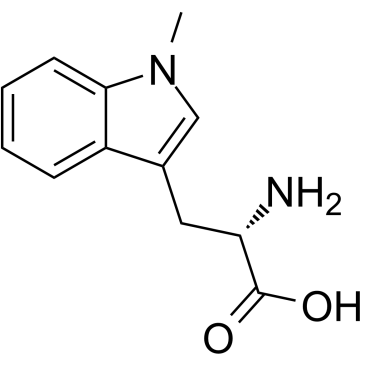
-
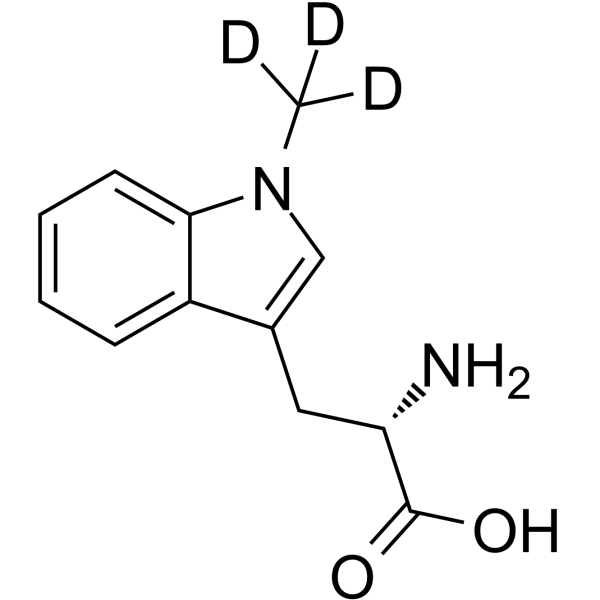
GC69891
(S)-Indoximod-d3
1-Methyl-L-tryptophan-d3; (S)-NLG-8189-d3
(S)-Indoximod-d3 is the deuterated form of (S)-Indoximod. (S)-Indoximod (1-Methyl-L-tryptophan) is an inhibitor of indoleamine 2,3-dioxygenase (IDO). (S)-Indoximod can be used for cancer research.
-
GC40145
(S)-Laudanosine
(+)-Laudanosine, L-Laudanosine, L-(+)-Laudanosine, NSC 35045
(S)-Laudanosine is the (S) enantiomer of laudanosine, a metabolite of the neuromuscular blocking agents atracurium and cisatracurium.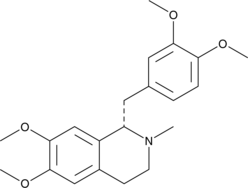
-
GC14486
(S)-Mephenytoin
(S)-5-Ethyl-3-methyl-5-phenylhydantoin
S-Mephenytoin (5-phenyl-5-ethyl-N-methylhydantoin) is an anticonvulsive drug which is metabolized by N-demethylation and 4-hydroxylation (of the aromatic ring).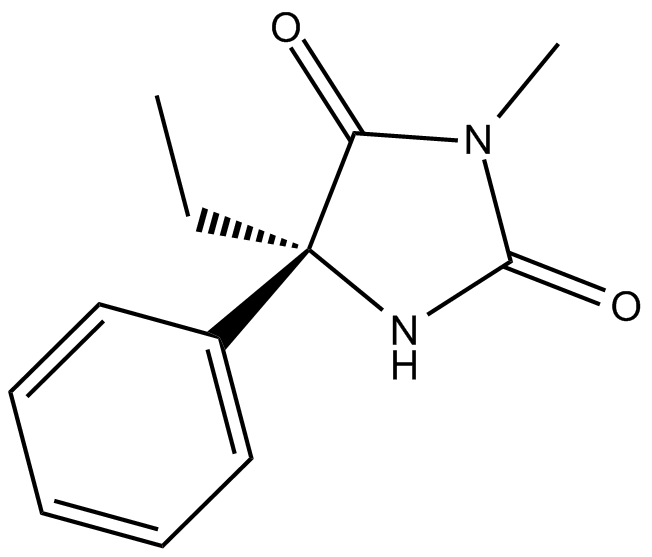
-
GC49179
(S)-O-Desmethyl Naproxen
(S)-6-Desmethyl Naproxen
A metabolite of (S)-naproxen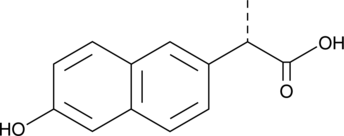
-
GC18596
(±)-2-propyl-4-Pentenoic Acid
2-Allylpentanoic Acid, 4-ene VPA
(±)-2-propyl-4-Pentenoic acid (4-ene VPA) is a major metabolite of valproic acid .

-
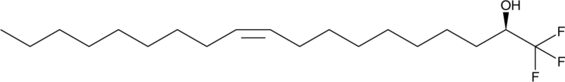
GC49034
1(R)-(Trifluoromethyl)oleyl alcohol
An oleic acid analog
-
GC46365
1,1'-(Azodicarbonyl)dipiperidine
ADDP, NSC 356027
A Mitsunobu reagent
-
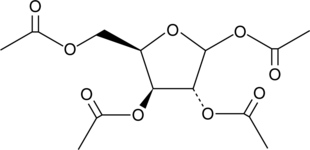
GC41762
1,2,3,5-Tetra-O-acetyl-D-xylofuranose
1,2,3,5-Tetra-O-acetyl-D-xylofuranose is the raw material for nucleotides synthesis.
-
GC46040
1,2,3-Trielaidoyl-rac-glycerol
Glycerol Trielaidate, TG(18:1(9E)/18:1(9E)/18:1(9E)), Trielaidin
A triacylglycerol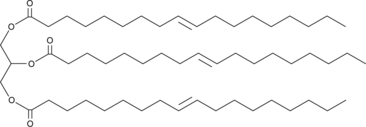
-
GC45698
1,2,3-Triheptanoyl-rac-glycerol
Glycerol Triheptanoate, TG(7:0/7:0/7:0), Triheptanoin
1,2,3-Triheptanoyl-rac-glycerol (Propane-1,2,3-triyl triheptanoate) is a synthetic medium-chain triglyceride (MCT) consisting of three odd-chain 7-carbon (heptanoate) fatty acids on a glycerol backbone.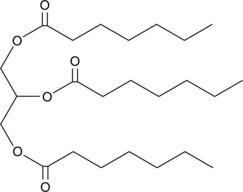
-
GC45285
1,2,3-Trihexanoyl-rac-glycerol
Glycerol trihexanoate, NSC 406885, Tricaproin, Trihexanoin
-
GC45956
1,2,3-Trinonanoyl-rac-glycerol
Glyceryl Trinonanoate, NSC 5647, Trinonanoin, Tripelargonin
A triacylglycerol
-
GC46009
1,2,3-Trioctanoyl-rac-glycerol-13C3
Glycerol Trioctanoate-13C3, TG(8:0/8:0/8:0)-13C3, Trioctanoin-13C3
An internal standard for the quantification of 1,2,3-trioctanoyl glycerol-13C3
-
GC46373
1,2,3-Triundecanoyl Glycerol
Glycerol Triundecanoate, TG(11:0/11:0/11:0), Triundecanoin, Triundecanoyl Glycerol
A triacylglycerol
-
GC41782
1,2-bis(heptanoylthio) Glycerophosphocholine
Diheptanoyl ThioPC, 1,2bis(Heptanoylthio)1,2dideoxysnglycero3phosphorylcholine
Diheptanoyl Thio-PC is a substrate for all phospholipase A2s (PLA2s) with the exception of cPLA2 and PAF-acetyl hydrolase (PAF-AH).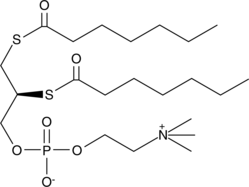
-
GC40236
1,2-Dilinoleoyl-sn-glycerol
1,2-Dioleoyl-sn-glycerol
1,2-Dilinoleoyl-sn-glycerol is a diacylglycerol (DAG) with linoleic acid (18:2) side chains attached at both the sn-1 and sn-2 positions.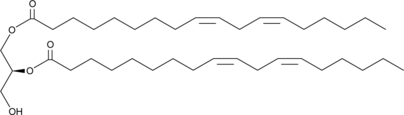
-
GC41808
1,2-Dioctanoyl PC
1,2-bis(O-octanoyl)-sn-glyceryl-Phophorylcholine, 1,2-DCPC, 1,2-Dicapryloyl-sn-glycero-3-PC, 1,2-Dioctanoyl Phosphatidylcholine
Phosphatidylcholine (PC) species are a common class of phospholipids that comprise the mammalian cell membrane.