Glycerophospholipids
Products for Glycerophospholipids
- Cat.No. Nombre del producto Información
-
GC42843
Arachidonoyl thio-PC
Arachidonoyl Thio-PC is a substrate for many phospholipase A2s (PLA2s) including sPLA2, cPLA2, and iPLA2.
-
GC42890
Azelaoyl PAF
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which promote the differentiation of monocytes via the nuclear receptor PPARγ.
-
GC42994
Butanoyl PAF
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which promote the differentiation of monocytes and activate polymorphonuclear leukocytes.
-
GC49856
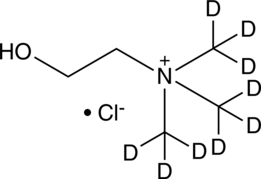
Choline-d9 (chloride)
An internal standard for the quantification of choline
-
GC43455
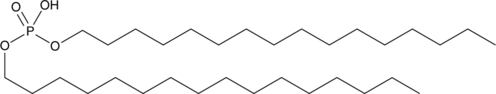
Dihexadecyl Phosphate
Dihexadecyl phosphate is a negatively-charged synthetic phospholipid that has been used to impart a negative charge to neutral liposomes.
-
GC49325
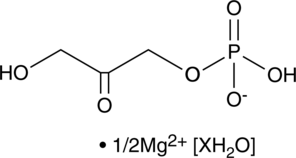
Dihydroxyacetone Phosphate (magnesium salt hydrate)
A glycerolipid precursor
-
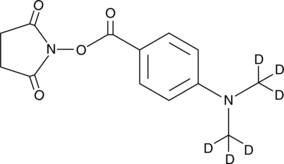
GC43499
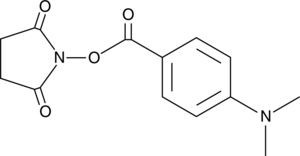
DMABA NHS ester
Phosphatidylethanolamine (PE) lipids are important components of cell membranes and biochemical pathways of fatty acid synthesis that contain abundant polyunsaturated fatty acyl (PUFA) groups.
-
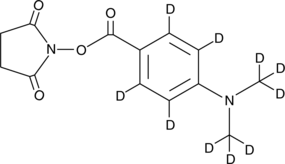
GC47247
DMABA-d10 NHS ester
An internal standard for the quantification of DMABA NHS ester
-
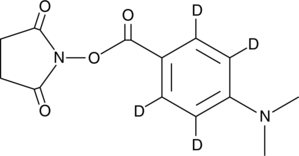
GC47248
DMABA-d4 NHS ester
An internal standard for the quantification of DMABA NHS ester
-
GC47249
DMABA-d6 NHS ester
An internal standard for the quantification of DMABA NHS ester
-
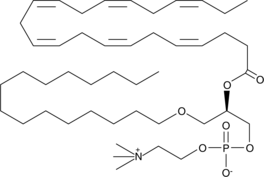
GC43557
Docosahexaenoyl PAF C-16
Docosahexaenoyl PAF C-16 is a PAF analog which contains docosahexaenoate at the sn-2 position rather than the acetate moiety found in PAF C-16.
-
GC49327
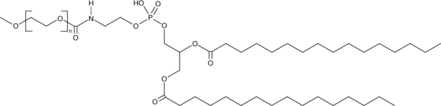
DPPE-MPEG(2000)
A PEGylated form of DPPE
-
GC52111
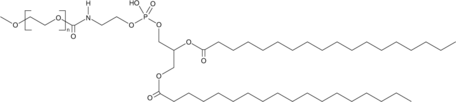
DSPE-MPEG(5000)
A PEGylated form of DSPE
-
GC52121
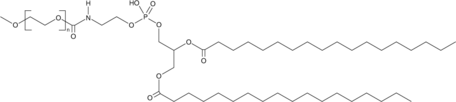
DSPE-MPEG(550)
A PEGylated form of DSPE
-
GC49001
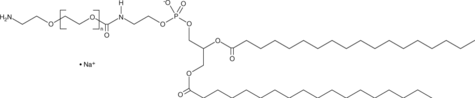
DSPE-PEG(2000)-amine (sodium salt)
A PEGylated derivative of DSPE
-
GC11000
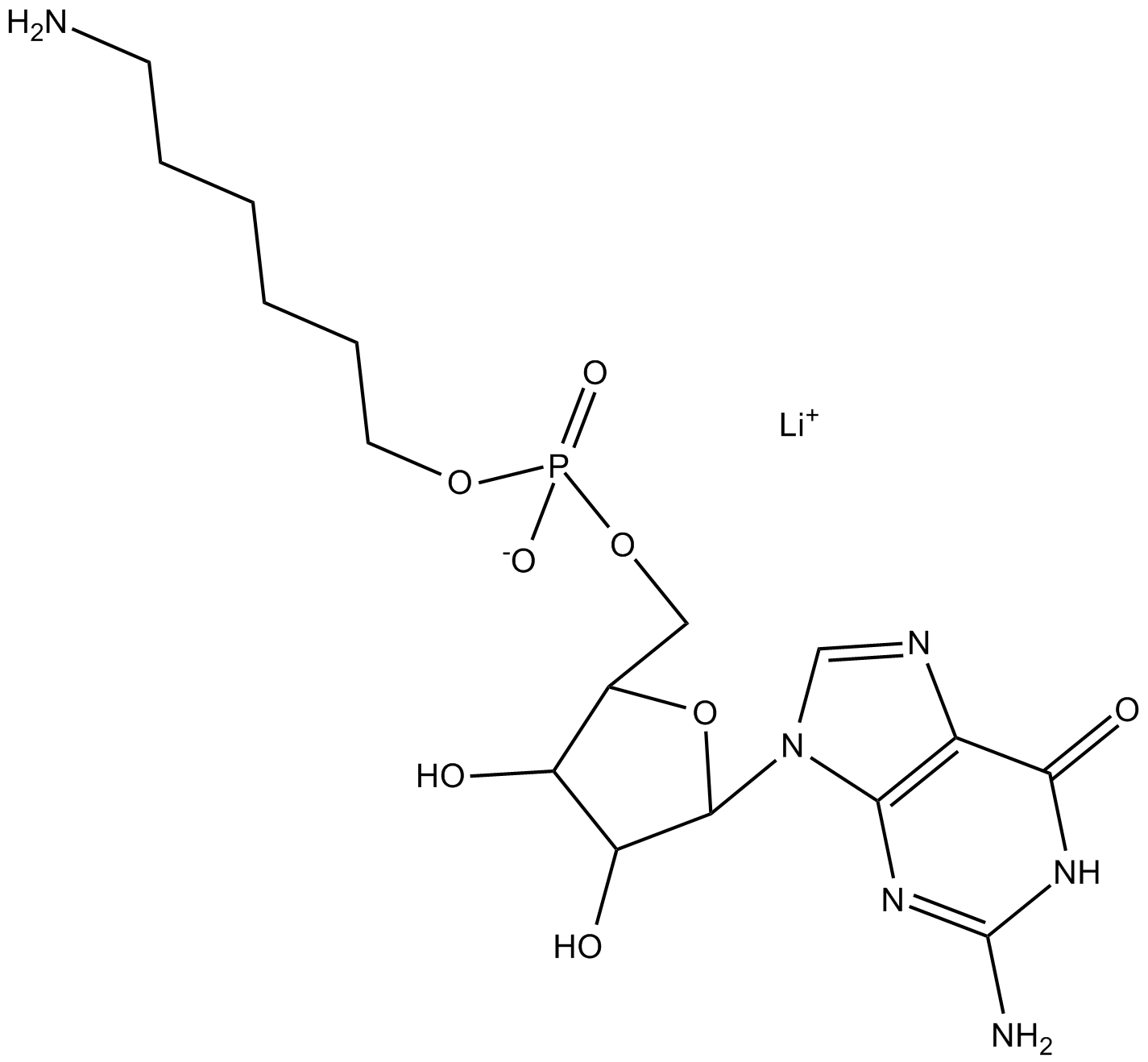
Eicosapentaenoyl PAF C-16
A PAF analog that contains EPA
-
GC43774
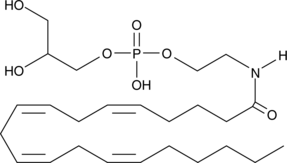
Glycerophospho-N-Arachidonoyl Ethanolamine
N-Acylated ethanolamines (NAE) are naturally-occurring lipids that have diverse bioactivities.
-
GC49273
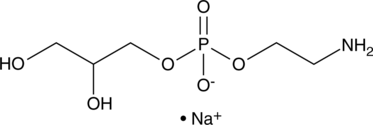
Glycerophosphorylethanolamine (sodium salt)
An active metabolite of phosphatidylethanolamine
-
GC43812
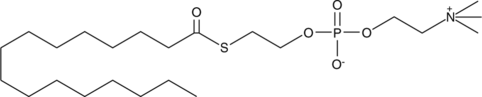
HEPC
Thioester analogs of glycerophospholipids, in combination with Ellman's reagent, are convenient colorimetric substrates for the measurement of phospholipase activity.
-
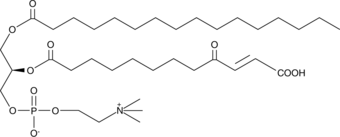
GC43997
KDdiA-PC
KDdiA-PC es un tipo de LDL oxidada (oxLDL) y es un ligando para el receptor eliminador de macrÓfagos CD36.
-
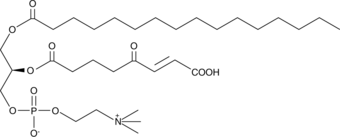
GC44011
KOdiA-PC
Oxidized low-density lipoprotien (oxLDL) particles contain low molecular weight species which are cytotoxic and pro-atherogenic.
-
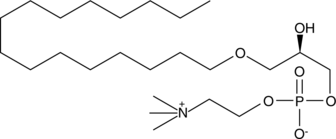
GC44103
Lyso-PAF C-16
Lyso-PAF C-16 can be formed by either the action of PAF-AH on PAF C-16, or by the action of a CoA-independent transacylase on 1-O-hexadecyl-2-acyl-glycerophosphocholine.
-
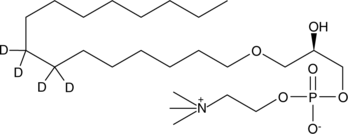
GC47587
Lyso-PAF C-16-d4
An internal standard for the quantification of LysoPAF C16
-
GC44104
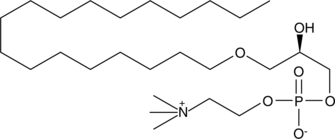
Lyso-PAF C-18
Lyso-PAF C-18 can be formed by either the action of PAF-AH on PAF C-18 or by the action of a CoA-independent transacylase on 1-O-octadecyl-2-acyl-glycerophosphocholine.
-
GC18241
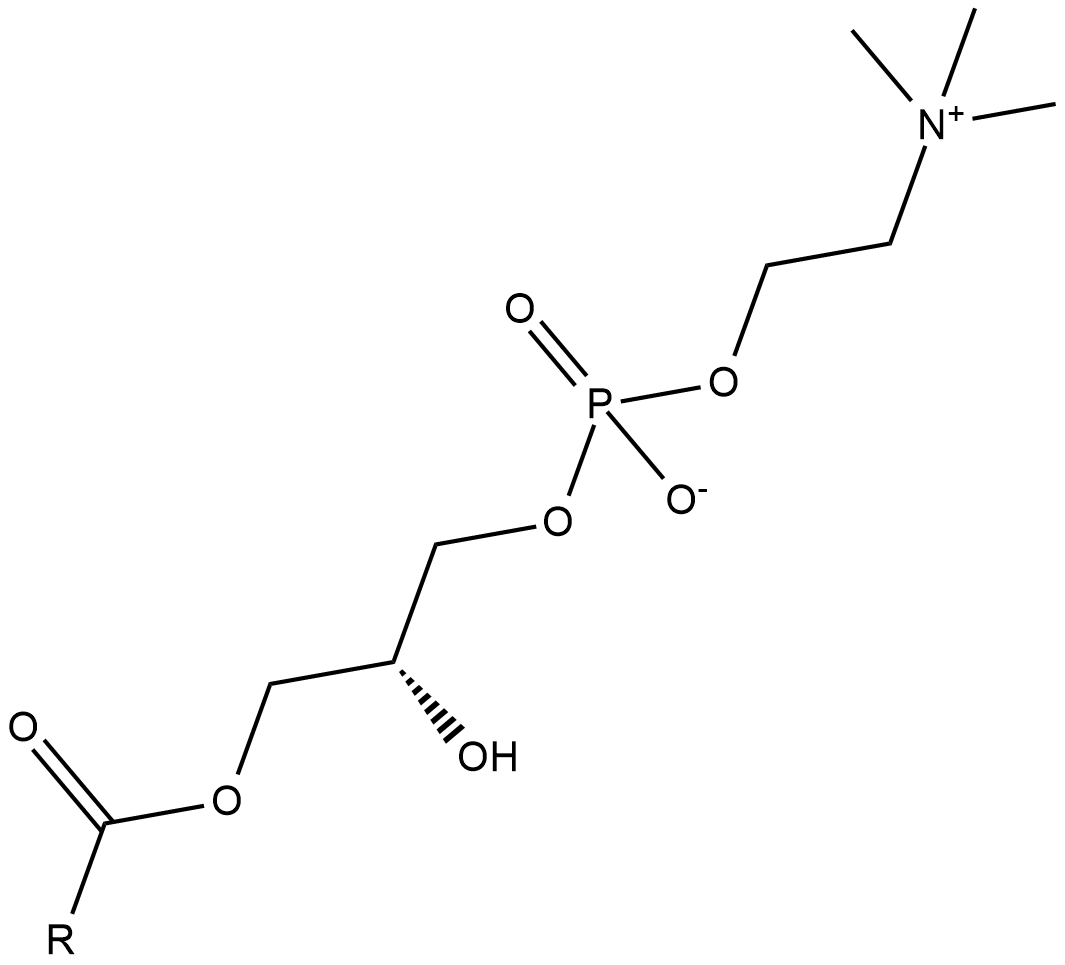
Lysophosphatidylcholines
Lysophosphatidylcholines are produced by hydrolysis of the fatty acid of phosphatidylcholine at either the sn-1 or sn-2 position by phospholipase A2 (PLA2) or by lecithin-cholesterol acyltranferase (LCAT), which transfers the fatty acid to cholesterol.
-
GC44180
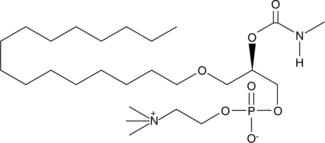
Methylcarbamyl PAF C-16
Methylcarbamyl PAF C-16 is a stable analog of PAF C-16 with a half-life greater than 100 minutes in platelet poor plasma due to its resistance to degradation by PAF-AH.
-
GC49661
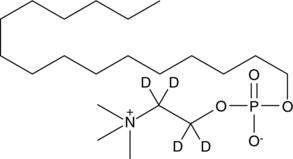
Miltefosine-d4
An internal standard for the quantification of miltefosine
-
GC44309
NAEPA
NAEPA, un derivado mimético del fosfato, es un agonista del receptor del Ácido lisofosfatÍdico (LPA).
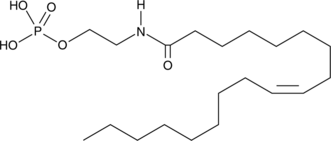
-
GC44499
Oleic Anhydride
Oleic anhydride is a fatty acid anhydride that inhibits sphingosine-induced phosphorylation of p32 in Jurkat T cells when used at concentrations ranging from 30 to 100 μM.
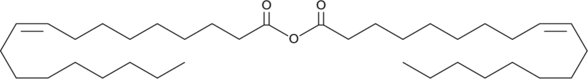
-
GC44500
Oleoyl 3-carbacyclic Phosphatidic Acid
Cyclic phosphatidic acids (cPAs) are naturally occurring analogs of lysophosphatidic acid (LPA) in which the sn-2 hydroxy group forms a 5-membered ring with the sn-3 phosphate.
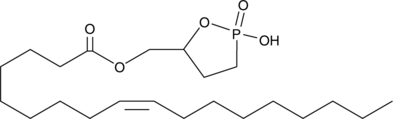
-
GC44540
PAF C-16 Carboxylic Acid
PAF C-16 is a naturally occurring phospholipid produced upon stimulation through two distinct pathways known as the "remodeling" and 'de novo' pathways.
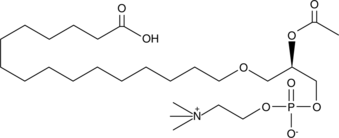
-
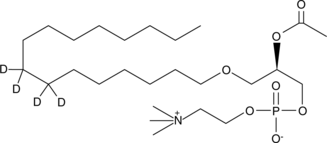
GC47855
PAF C-16-d4
An internal standard for the quantification of PAF C16
-
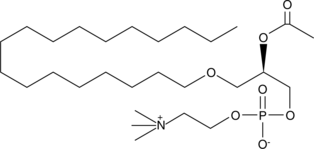
GC44541
PAF C-18
PAF C-18 is a naturally occurring phospholipid produced upon stimulation through two distinct pathways known as the "remodeling" and 'de novo' pathways.
-
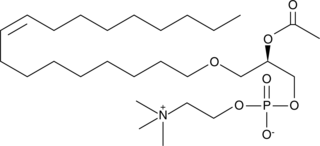
GC41450
PAF C-18:1
PAF C-18:1 is a naturally occurring phospholipid produced by cells upon stimulation and plays a role in the establishment and maintenance of the inflammatory response.
-
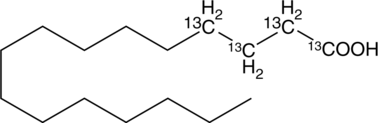
GC47859
Palmitic Acid-13C (C1, C2, C3, and C4 labeled)
El Ácido palmÍtico-13C (marcado con C1, C2, C3 y C4) es el Ácido palmÍtico marcado con 13C.
-
GC47862
Palmitic Acid-d5
An internal standard for the quantification of palmitic acid
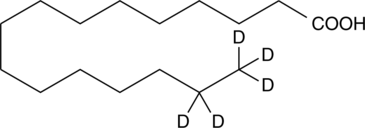
-
GC44545
Palmitoleoyl 3-carbacyclic Phosphatidic Acid
Cyclic phosphatidic acids (cPAs) are naturally occurring analogs of lysophosphatidic acid (LPA) in which the sn-2 hydroxy group forms a 5-membered ring with the sn-3 phosphate.
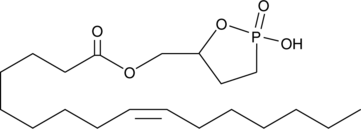
-
GC44546
Palmitoyl 3-carbacyclic Phosphatidic Acid
Cyclic phosphatidic acids (cPAs) are naturally occurring analogs of lysophosphatidic acid (LPA) in which the sn-2 hydroxy group forms a 5-membered ring with the sn-3 phosphate.
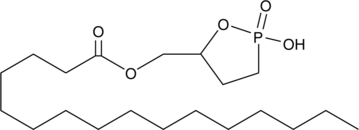
-
GC44549
Palmitoyl thio-PC
Thioester analogs of glycerophospholipids, in combination with Ellman's reagent, are convenient colorimetric substrates for the measurement of phospholipase (PL) activity.
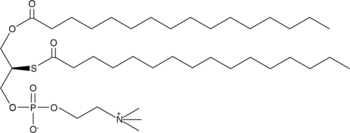
-
GC44573
PAz-PC
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which are cytotoxic and pro-atherogenic.
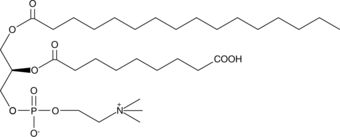
-
GC44616
PGPC
PGPC is an oxidized phospholipid that can be formed under conditions of oxidative stress.
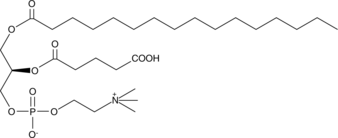
-
GC40252
Phosphatidic Acids (egg) (ammonium salt)
Phosphatidic acid is a phospholipid and an intermediate in glycerolipid biosynthesis.
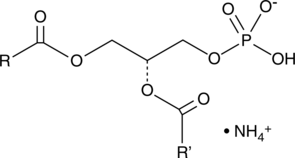
-
GC44631
Phosphatidylethanolamines (bovine)
Phosphatidylethanolamine is the most abundant phospholipid in prokaryotes and the second most abundant found in the membrane of mammalian, plant, and yeast cells, comprising approximately 25% of total mammalian phospholipids.
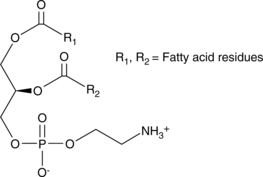
-
GC44632
Phosphatidylethanolamines (egg)
Phosphatidylethanolamine is the most abundant phospholipid in prokaryotes and the second most abundant found in the membrane of mammalian, plant, and yeast cells, comprising approximately 25% of total mammalian phospholipids.
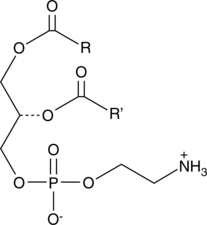
-
GC18560
Phosphatidylethanolamines (soy)
Phosphatidylethanolamine is the most abundant phospholipid in prokaryotes and the second most abundant found in the membrane of mammalian, plant, and yeast cells, comprising approximately 25% of total mammalian phospholipids.
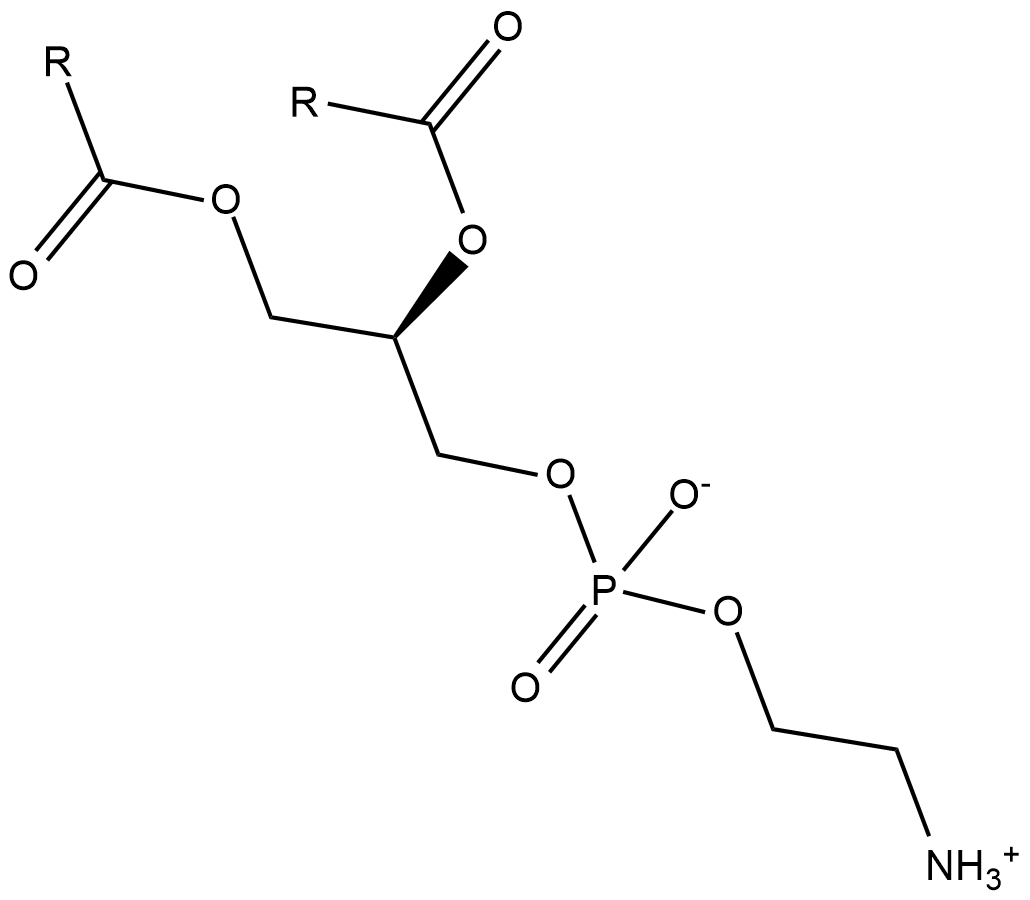
-
GC18377
Phosphatidylglycerols (sodium salt)
Phosphatidylglycerol is a naturally occurring anionic phospholipid and a constituent of plant, animal, and bacterial cell membranes.
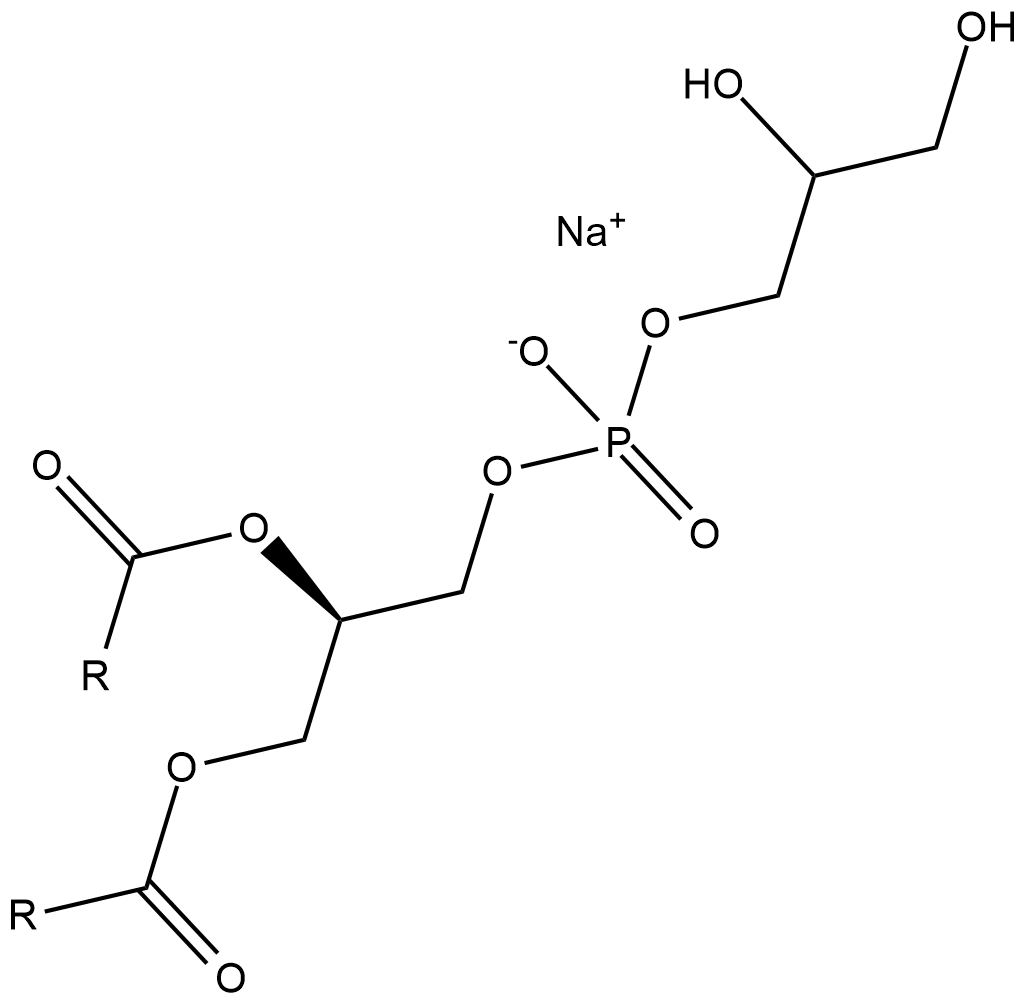
-
GC48355
Phosphatidylinositols (plant, wheat germ) (sodium salt)
A mixture of phosphatidylinositols
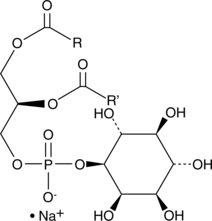
-
GC18636
Phosphatidylinositols (soy) (sodium salt)
Los fosfatidilinositoles (soja) (sal de sodio) son una mezcla de fosfatidilinositoles.
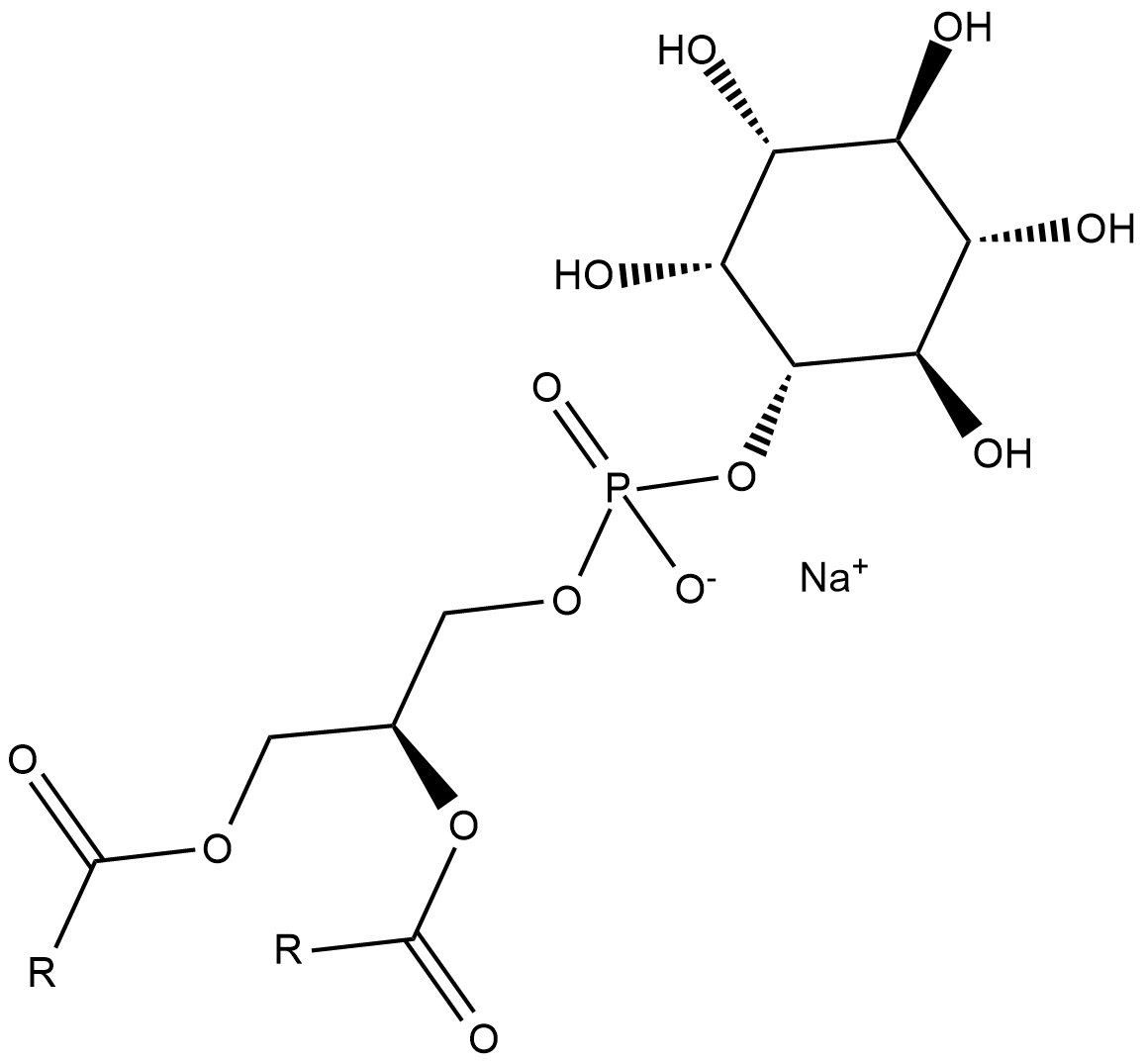
-
GC44633
Phosphatidylserines (bovine)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
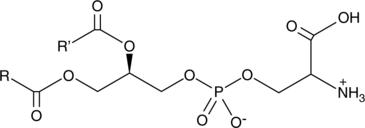
-
GC18522
Phosphatidylserines (sodium salt)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
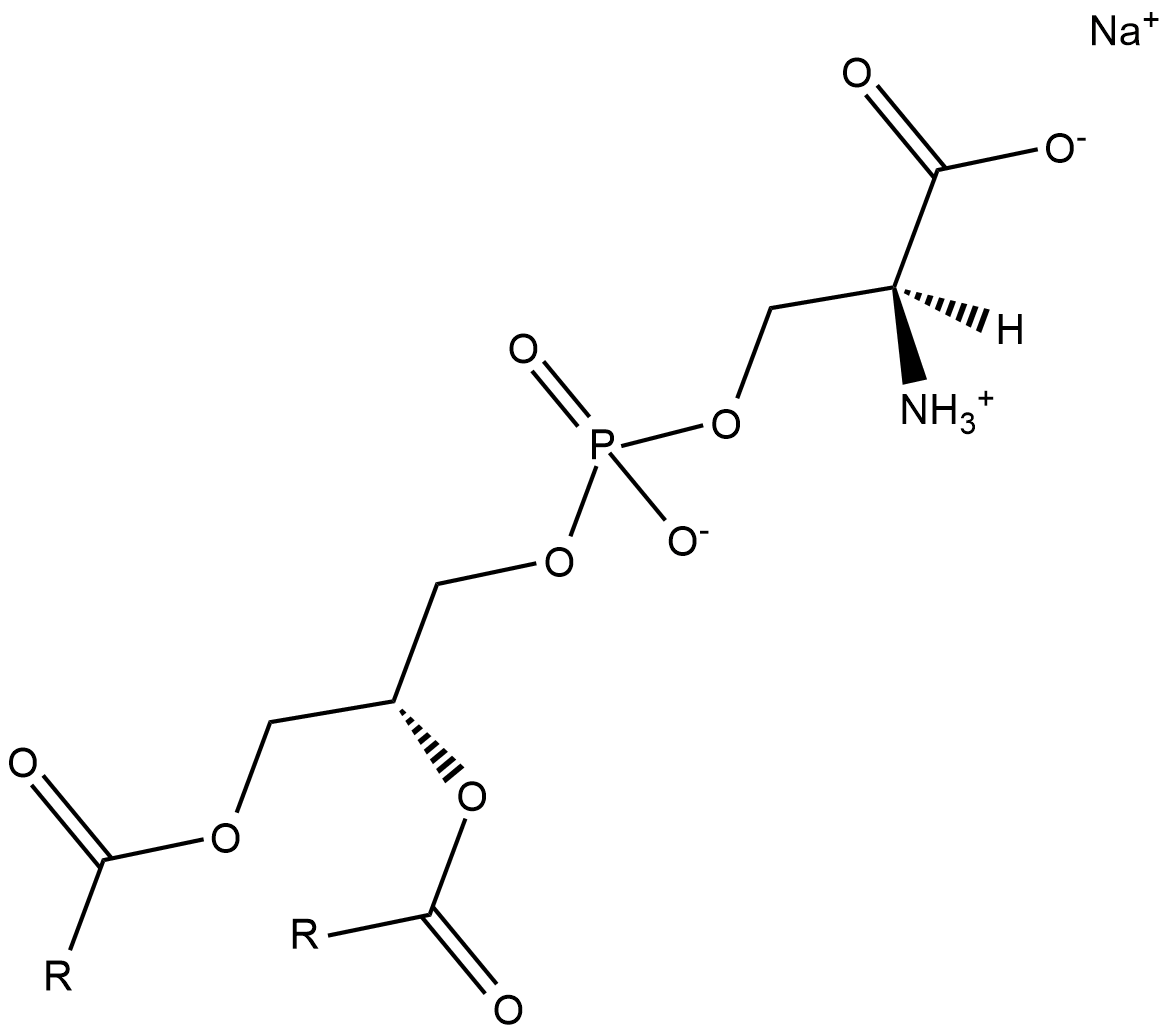
-
GC44669
POV-PC
Oxidized low-density lipoprotien (oxLDL) particles contain low molecular weight species which are cytotoxic and pro-atherogenic.
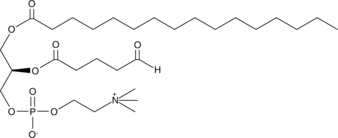
-
GC45680
Prodan
Prodan, un fluorÓforo solvatocrÓmico, se ha utilizado como indicador de membrana sensible al microambiente.
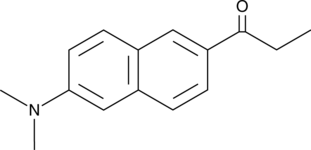
-
GC18872
Propargylcholine (bromide)
Propargylcholine is an alkynyl derivative of choline that has a propargyl group in place of one methyl group.
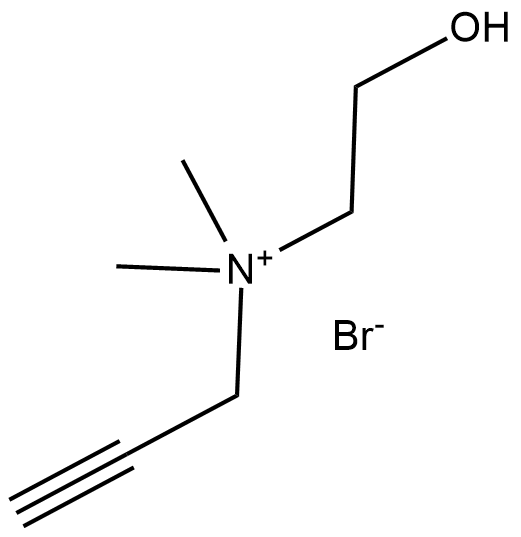
-
GC44740
PtdIns-(1,2-dioctanoyl) (sodium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
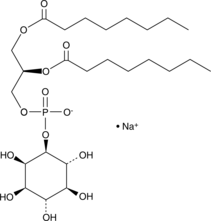
-
GC44741
PtdIns-(1,2-dipalmitoyl) (ammonium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
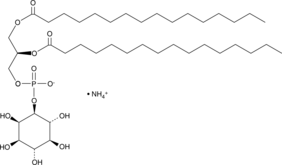
-
GC48370
PtdIns-(1,2-dipalmitoyl) MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
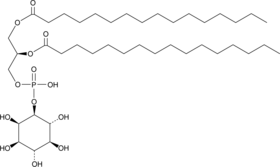
-
GC44743
PtdIns-(3)-P1 (1,2-dioctanoyl) (sodium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
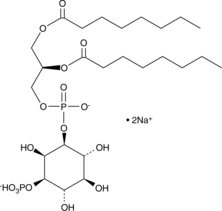
-
GC44748
PtdIns-(3,4,5)-P3 (1,2-dihexanoyl) (ammonium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
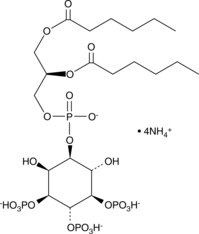
-
GC44750
PtdIns-(3,4,5)-P3 (1,2-dipalmitoyl) (sodium salt)
The phosphatidylinositol phosphates represent a small percentage of total membrane phospholipids.
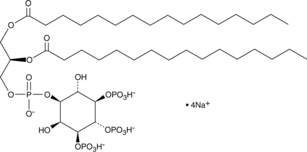
-
GC44753
PtdIns-(3,4,5)-P3-biotin (sodium salt)
The PtdIn phosphates play an important role in the generation and transduction of intracellular signals.
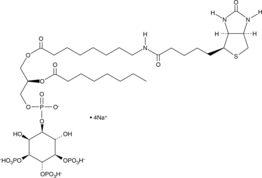
-
GC44759
PtdIns-(4)-P1 (1,2-dioctanoyl) (ammonium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
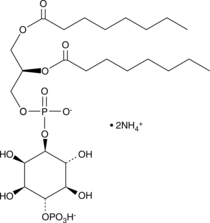
-
GC44905
SMI-481
SMI-481 es un inhibidor selectivo de la proteÍna de transferencia de fosfatidilinositol de levadura (PITP) Sec14.
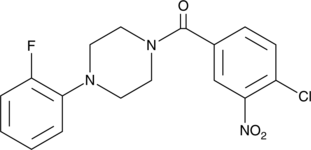
-
GC44969
Sulforhodamine 101 DHPE
Sulforhodamine 101 DHPE is a fluorescent probe made from the conjugation of the phospholipid 1,2-dipalmitoyl-sn-glycero-3-PE to sulforhodamine 101, a red fluorescent dye that displays excitation/emission spectra of 586/605 nm, respectively.