Cy5-UTP
A synthetic nucleotide called Cyanine 5- Uridine Triphosphate, commonly referred to as Cy5-UTP is utilised in biochemistry and molecular biology for a wide range of purposes, most notably in the tagging and identification of RNA. This UTP alternative with cyanine 5 tagging is used to label RNA probes by in vitro transcription using different RNA polymerases, for instance, Cy5-UTP functions like a substrate to T7 RNA polymerase to produce tagged probes. Additionally, following gel electrophoresis, Cyanine 5-labelled mRNA is easily detectable in ultraviolet light with no additional staining because the tagged product can be seen immediately by fluorescence . These techniques produce probes that can be used for a variety of molecular and cellular biological investigations, such as RNA localisation, measurement and detection. Also for in situ hybridization (FISH) and multicolour fluorescence analysis, particularly when dual-colour expression arrays are used with Cy5-UTP. This synthetic nucleotide has 650 and 665 nm of excitation and emission wavelengths, respectively.
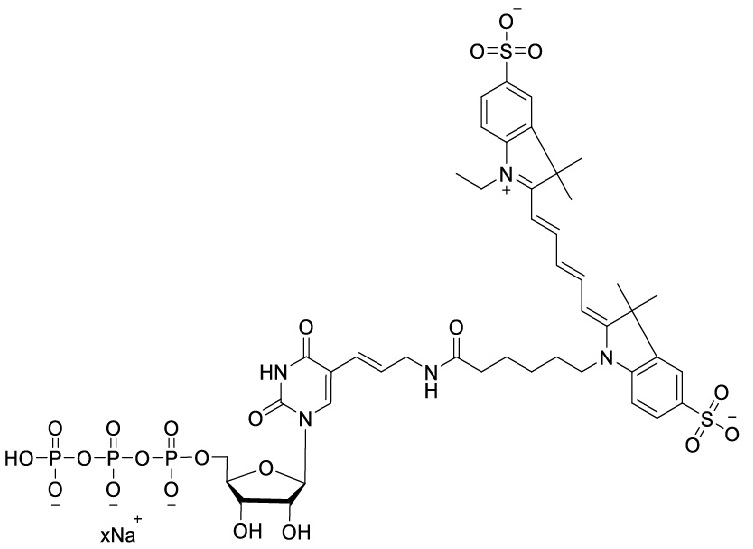
There are three primary parts to the molecular makeup of Cy5-UTP. The first is the uracil base, ribose sugar and a triphosphate group in uridine triphosphate (UTP), a ribonucleotide. Second, the sugar in UTP is linked to the cyanine dye (Cy5), a synthetic fluorophore, the luminous qualities of Cy5-UTP are due to this molecule. Lastly, Cy5 is attached to UTP via ribose sugar's 5' carbon, allowing for effective integration in RNA upon transcription . Some of the alternative names used for Cy5-UTP are Aminoallyl-UTP-Cy5; Triethylammonium salt; Cyanine 5-AA-UTP and Cyanine 5-Aminoallyluridine-5'-Triphosphate. It has a molecular formula; C45H58N5O22P3S2 (free acid) with 1178.01 g/mol (free acid) as its molecular weight and is soluble in water. The chemical structure of Cy5-UTP is given below.
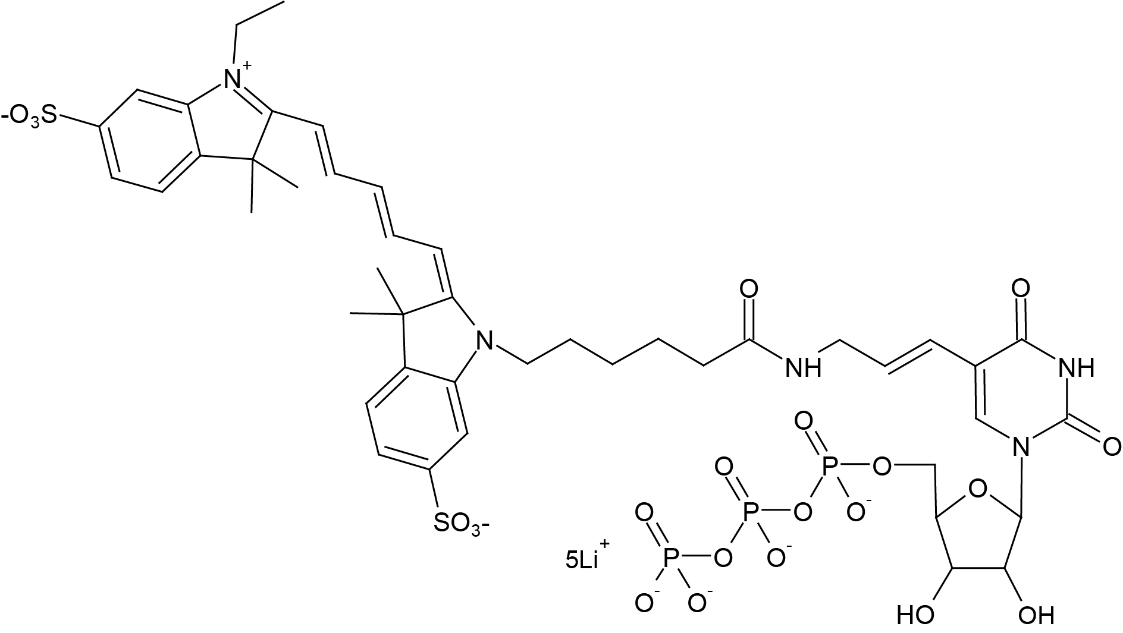
Figure 1: Chemical Structure of Cyanine 5-UTP;
Cyanine dyes are widely used in single-molecule research and high-resolution imaging, which are essential fluorophores for contemporary chemical and biological research. In single-molecule fluorescence resonance energy transfer (smFRET), the red-absorbing dye Cy5 is frequently utilised as an acceptor, with donors being Cy3 or similar yellow-emitting dyes . The process of integration of Cyanine 5-UTP in RNA molecules through in vitro transcription, where an RNA molecule is synthesised using a DNA template, involves the typical nucleotides employed in the reaction which are A, U, G and C. For labelling the RNA, scientists here use Cyanine 5-UTP instead of ordinary UTP. The transcription enzyme, i.e. RNA polymerase, then recognises Cy5-UTP as a substrate which then is incorporated in the chain of RNA and grows while adding it to points corresponding to the UTP residues of template DNA. Consequently, the newly created RNA has uridine residues that have been dye-labeled. When activated by a particular wavelength of light, usually between 650 and 670 nm, the Cy5 dye in the tagged RNA molecule generates fluorescence (Novakowski, 2019). Using the right tools, like a fluorescence microscope or fluorometer, this fluorescence is observed and measured.
Moreover, the use of oligonucleotide micro-arrays for mapping gene expression of amplified strands of RNA necessitates fast and reliable cRNA labelling techniques. For instance, oligonucleotide arrays are utilised to measure the levels of gene expression in tissues (i.e. control and diseased), where short sequences of 16–20 oligonucleotides are synthesised in situ in a higher density. RNA extracted from healthy and sick cells( samples) creates cDNA, which is then fluorescently dyed (either with Cy3 or Cy5 ). The microarray is co-hybridized with the labelled cDNA samples (Caudle et al., 2010). Using a confocal microscope and the right lasers for the best excitation of these dyes, fluorescent signals can be identified after hybridization, washing and drying creating an image file (Figure 2).
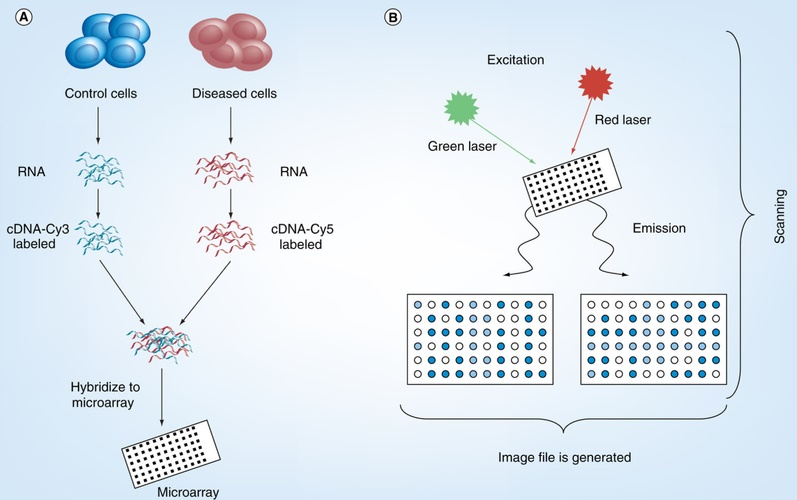
Figure 2: A general overview of the DNA microarray process; Source:
RNA is essential to cellular machinery like the large molecular complexes involved in the regulation of genetic information. Because of their complexity and size, RNA-related complexes have always been hard to research. Since these complexes are frequently difficult to purify using conventional biochemical techniques, therefore, tools like fluorescence and electron microscopy are crucial for studying them. Given problems like low permeability across cell membranes, random binding, cytotoxicity and significant molecular mass of RNA extensions, existing techniques for tagging RNA molecules are restricted. The investigation of low-abundance and brief RNA transcripts may be hampered by these issues. The use of Cy5-UTP-labeled probes for diagnostic applications, such as the identification of particular RNA sequences in samples from patients, has been investigated.
developed novel tagging techniques to insert modified nucleotides in various mRNA regions utilising yeast poly(A) and T7 RNA polymerase. These regions included the poly(A) tail, the body (with 5' UTR and a coding region along with 3' UTR) and the body and tail (BBT). Through the co-transcriptional integration of Cy5-UTP, the chemically altered UTP nucleotide has been randomly incorporated in the body of the RNA strand employing the enzymatic labelling approach, via T7 RNA polymerase (Figure 3).
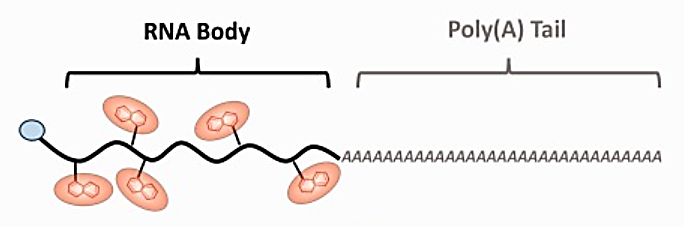
Figure 3: mRNA labelling with modified CNTP (Cy5-UTP) co-transcriptionally on RNA strand.
Using FLuc mRNA as a model, co-transcriptional body labelling shows a definite pattern of rising Cy5 intensity with percentage input of Cy5-UTP (0, 0.1, 0.5, 1.0, 5.0, and 10%). Additionally, measurement showed a linear relationship between the proportion of Cy5-UTP upon transcription and the number of fluorophores per extracted RNA .
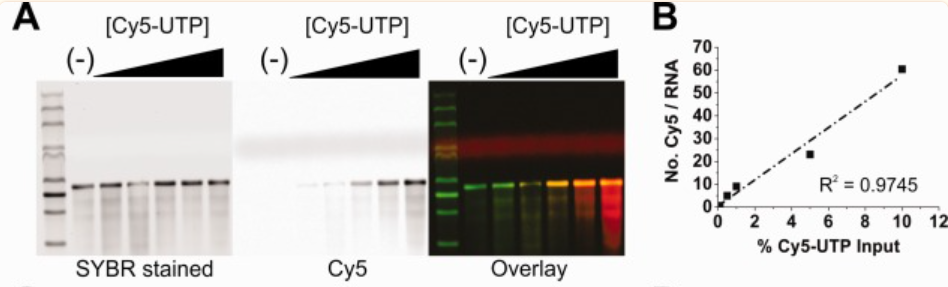
Figure 4: Stained gel images for total RNA (green in overlay) in the left panel and Cy5-labeled RNA (red in overlay) in the right panel show the level of RNA labelling.
The consistent distribution of the RNA output and the absence of an increase in side products above those seen for the 0% Cy5-UTP control transcription suggested that the CNTP is a well-tolerated substrate (Kim et al., 2017). This technique effectively marks the coding sequence-containing mRNA body. However, it was emphasised, that this method makes the mRNA non-coding, rendering it inappropriate for translation to useful protein. Recent research has however uncovered a potential problem with Cyanine dyes, particularly Cy5. As they are photoconvertible, they form derivatives that are blue-shifted when they are exposed to light. Concerns concerning the precision of multicolour imaging investigations have been raised in light of this phenomenon. Cho et al. looked into the process by which Cy5 undergoes photoconversion to Cy3, which happens when Cy5 is subjected to fluorescent imaging. The elimination of a C2H2 subunit from Cy5 was shown to be the crucial step; this mainly happens through a procedure which involves bond breakage and reconstitution.
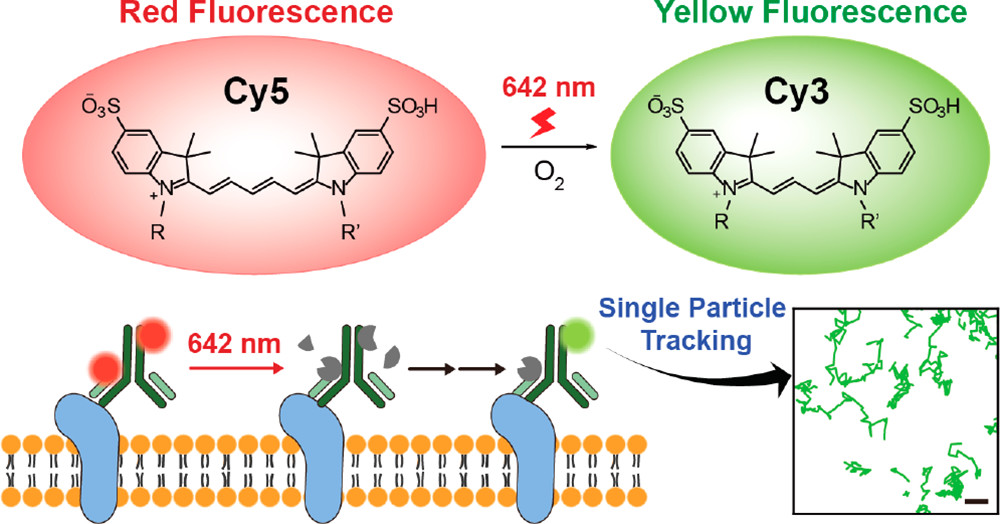
Figure 5: Cy5 to Cy3 photoconversion occurs in photoexcitation during fluorescence imaging;
Importantly, the scientists were able to verify through a variety of spectroscopic methods that the resulting luminous photoproduct is Cy3. Cy5 converts to Cy3 through a unique deletion of C2H2 from the polymethine chain during the photochemical process.
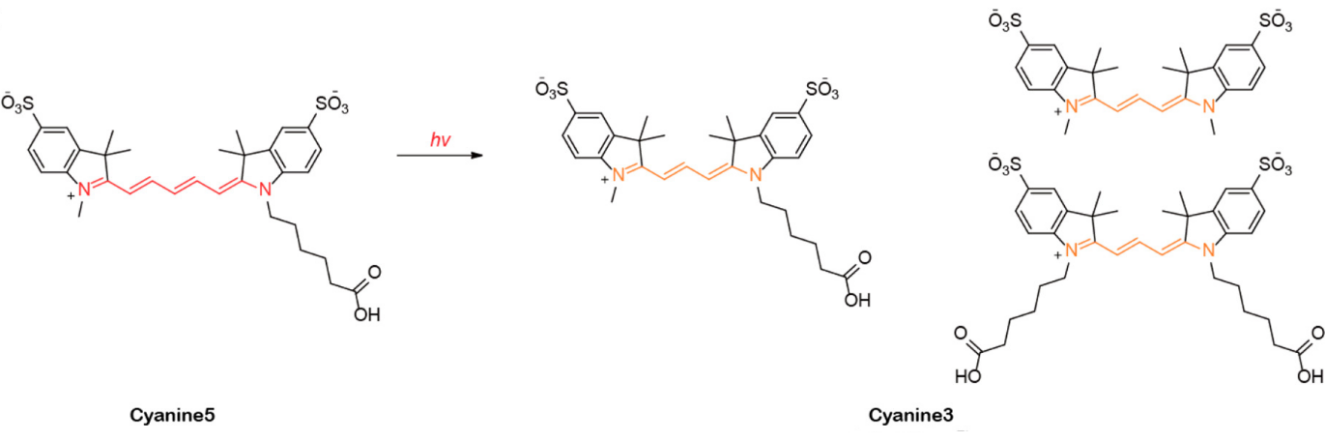
Figure 6: Photoconversion of Cy5 to Cy3 upon irradiation with a red laser.
The chemical process involved photooxidation causing the polymethine chain of Cy5 to be broken and the resulting carbonyl compounds and Fischer's base subsequently go through condensation to yield Cy3.





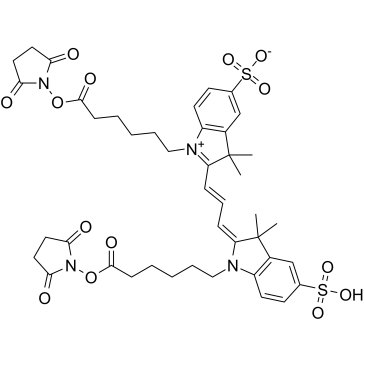
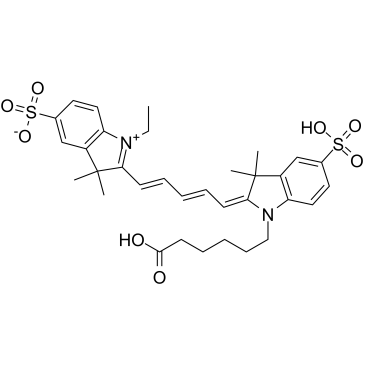





Commentaires