Bacterial Diseases
Products for Bacterial Diseases
- Cat.No. Nom du produit Informations
-
GC47464
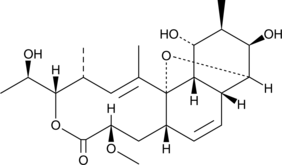
Isodeoxycholic Acid
isoDCA
A bile acid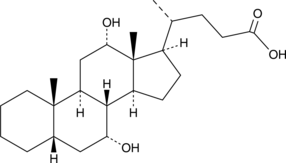
-
GC47466
Isolithocholic Acid
Isolithocholate, iso-LCA
L'acide isolithocholique (acide β-lithocholique) est un isomère de l'acide lithocholique.
-
GC49306
Isopimaric Acid
7,15-Isopimaradien-18-oic Acid, (+)-Isopimaric Acid
L'acide isopimarique est un puissant ouvreur de canaux K+ (BK) activés par le calcium À grande conductance.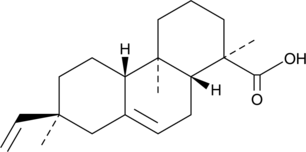
-
GC48554
Isoreserpiline
3-Isoreserpilinic Acid, methyl ester, Neoreserpiline
An indole alkaloid with diverse biological activities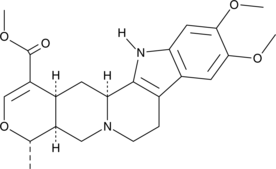
-
GC49142
Isorhoifolin
Apigenin-7-O-rutinoside
L'isorhoifoline est un glycoside flavonoÏde des feuilles de periploca nigrescens.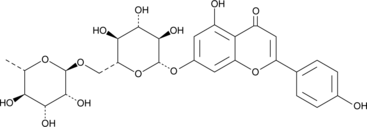
-
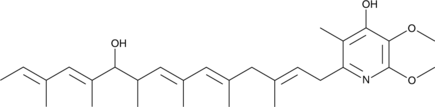
GC47473
IT-143A
A bacterial metabolite
-
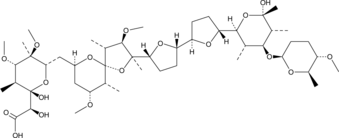
GC47523
K-41
A fungal metabolite with antibiotic and antiparasitic activities
-
GC44004
Kijanimicin
NSC 329515
Kijanimicin is an antibiotic first isolated from the fermentation broth of A.
-
GC47530
Kocurin
Baringolin, PM181104
A peptide antibiotic
-
GC41627
Kumbicin C
La kumbicine C est un composé bis-indolyl benzénoÏde d'un champignon du sol australien, Aspergillus kumbius.
-
GC18900
L-156,602
PD 124966
L-156,602 is an antagonist of the complement component 5A (C5a) receptor (IC50 = 1.7 ug/L in a radioligand binding assay using neutrophil membranes).
-
GC44020
L-681,217
L-681,217 is an antibiotic originally isolated from S.
-
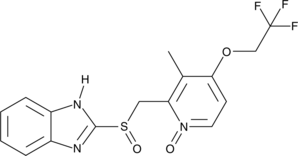
GC49640
Lansoprazole N-oxide
A potential impurity found in bulk preparations of lansoprazole
-
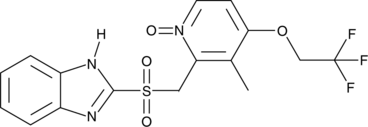
GC52119
Lansoprazole sulfone N-oxide
A potential impurity found in commercial preparations of lansoprazole
-
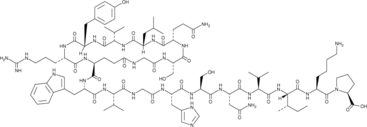
GC44034
Lariatin A
Lariatin A is an antimycobacterial lasso peptide originally isolated from R.
-
GC49753
LCS3
LCS3 est un inhibiteur réversible et non compétitif de la glutathion disulfure réductase (GSR) et de la thiorédoxine réductase 1 (TXNRD1) (IC50 = 3,3 μ M et 3,8 μ M, respectivement). LCS3 présente une activité anti-tumorale et induit l'apoptose. LCS3 peut être utilisé dans la recherche sur l'adénocarcinome pulmonaire (LUAD).
-
GC18190
Lecanoric Acid
NSC 249981
L'acide lécanorique est un inhibiteur de l'histidine-décarboxylase isolé du champignon.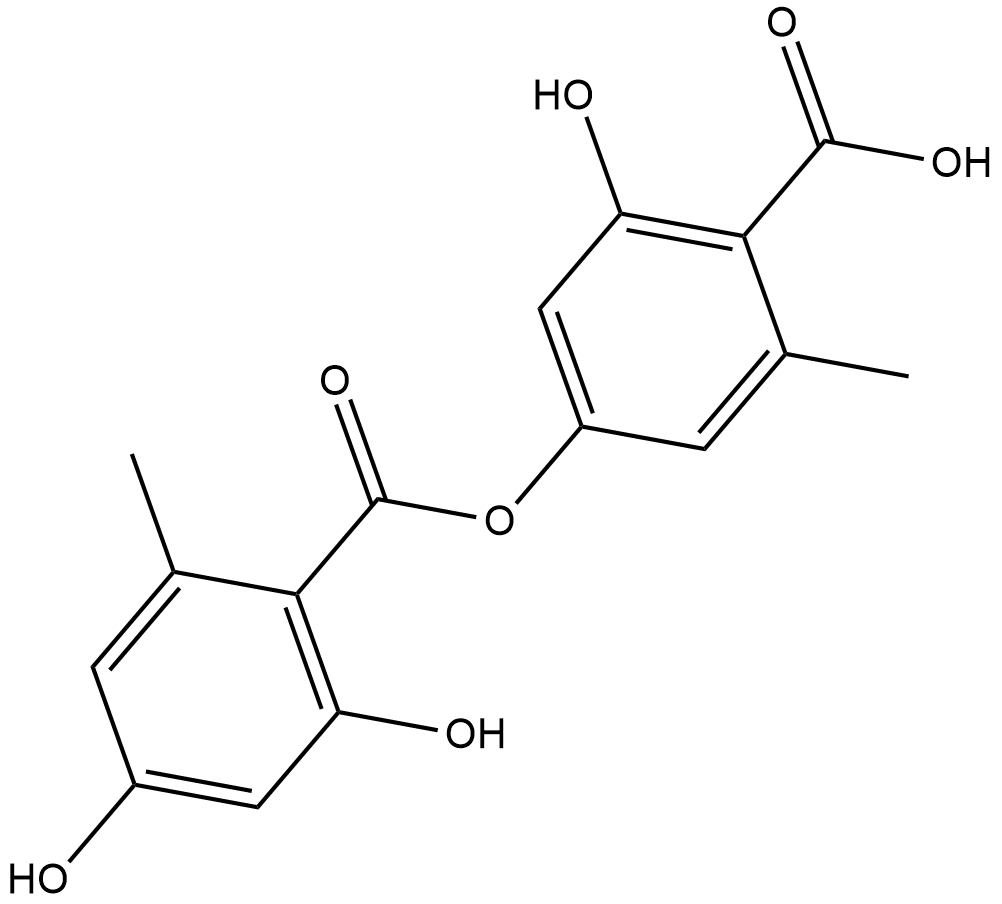
-
GC49751
Lefamulin
BC-3781
La léfamuline (BC-3781) est un antibiotique actif par voie orale.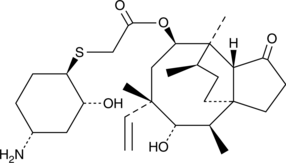
-
GC46166
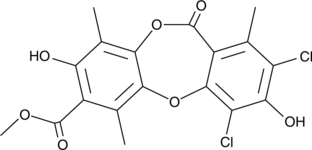
Leoidin
A depsidone
-
GC44050
Leucinostatin A
P168
La leucinostatine A (antibiotique P168) est un nonapeptide exerÇant une activité remarquable notamment contre Candida albicans et Cryptococcus neoformans.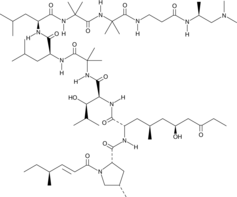
-
GC44051
Leucomycin A13
Kitasamycin A13
Leucomycin A13 is a macrolide antibiotic and a component of the leucomycin complex originally isolated from S.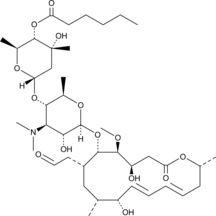
-
GC41234
Leucomycin A4
Kitasamycin A4
Leucomycin A4 is a macrolide antibiotic that has been found in S.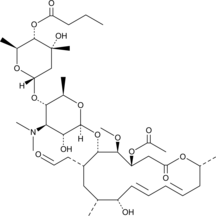
-
GC19008
Leucomycin A5
Kitasamycin A5, Leukomycin A5, Turimycin H4
Leucomycin A5 is a metabolite from the leucomycin complex, which was originally isolated from S.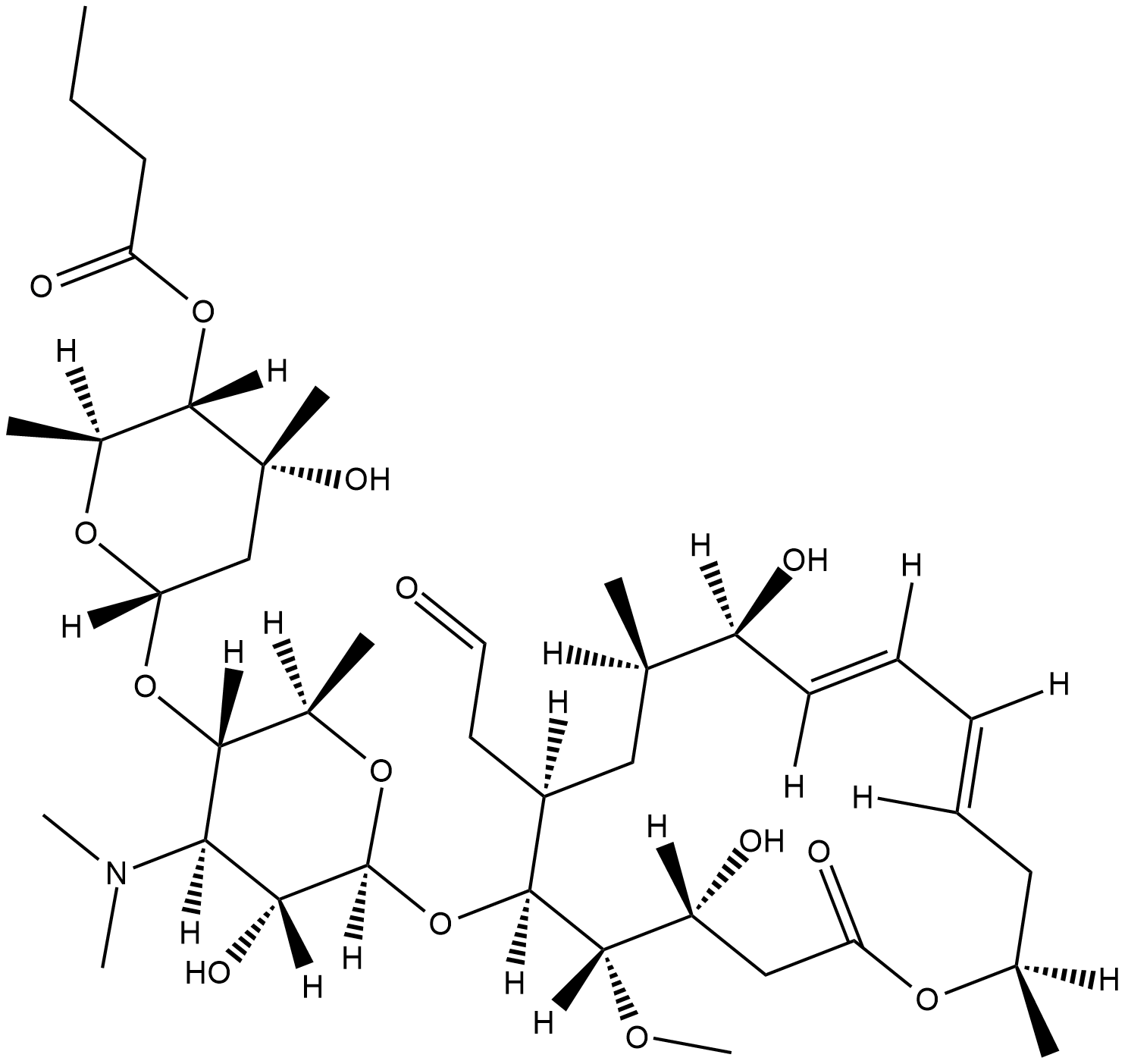
-
GC40144
Levofloxacin N-oxide
Le N-oxyde de lévofloxacine est un métabolite mineur de la lévofloxacine.
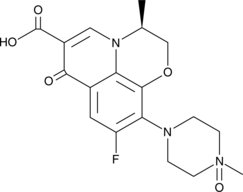
-
GC44066
Lincomycin
U-10149
La lincomycine, un antibiotique lincosamide, est un agent antimicrobien utilisé pour la recherche d'infections bactériennes À Gram positif.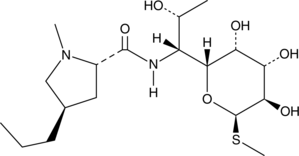
-
GC48344
Linearmycin A
A polyene antibiotic
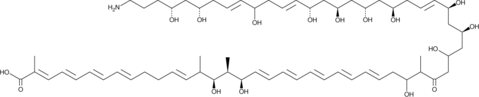
-
GC48342
Linearmycin B
A polyene antibiotic
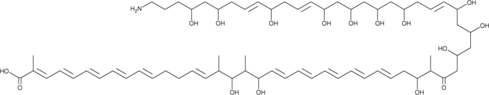
-
GC41517
Linoleic Acid ethyl ester
C18:2 (cis-9,12) ethyl ester, Ethyl cis-9,12-Octadecadienoate, Ethyl Linoleate, Telfairic Acid ethyl ester
L'ester éthylique de l'acide linoléique inhibe le développement des lésions athérosclérotiques et l'expression des médiateurs inflammatoires.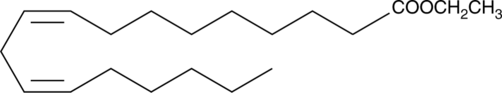
-
GC44078
LL-37 amide (trifluoroacetate salt)
CAP-18, hCAP-18, Cathelicidin, FALL-39
LL-37 is a cationic and α-helical antimicrobial peptide expressed in human bone marrow, testis, granulocytes, and gingival epithelium and is upregulated in psoriatic lesions.
-
GC44098
Lydicamycin
Antibiotic CC 12
La lydicamycine est un antibiotique isolé du bouillon de fermentation d'une souche d'actinomycète identifiée comme Streptomyces lydicus.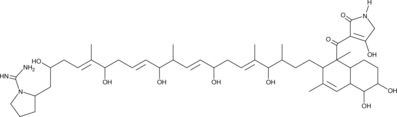
-
GC44101
Lyso-Globotriaosylceramide (d18:1)
Globotriaosylsphingosine (d18:1), Lyso-Ceramide Trihexoside, Lyso-Gb3
Lyso-globotriaosylceramide is a form of globotriaosylceramide that is lacking the fatty acyl group.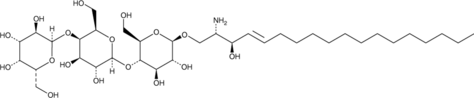
-
GC18331
Maculosin
Cyclo(L-Pro-L-Tyr)
La maculosine est une phytotoxine spécifique À l'hÔte de la centaurée maculée d'Alternaria alternata. La maculosine est une molécule de détection de quorum impliquée dans la communication cellule-cellule par Pseudomonas aeruginosa. La maculosine agit également comme une molécule de signalisation régulant l'expression des gènes de virulence chez Lactobacillus reuteri. La maculosine présente des propriétés antioxydantes, anticancéreuses et non toxiques. La maculosine présente une activité cytotoxique contre les lignées cellulaires du cancer du foie humain, avec une CI50 de 48,90 μg/mL.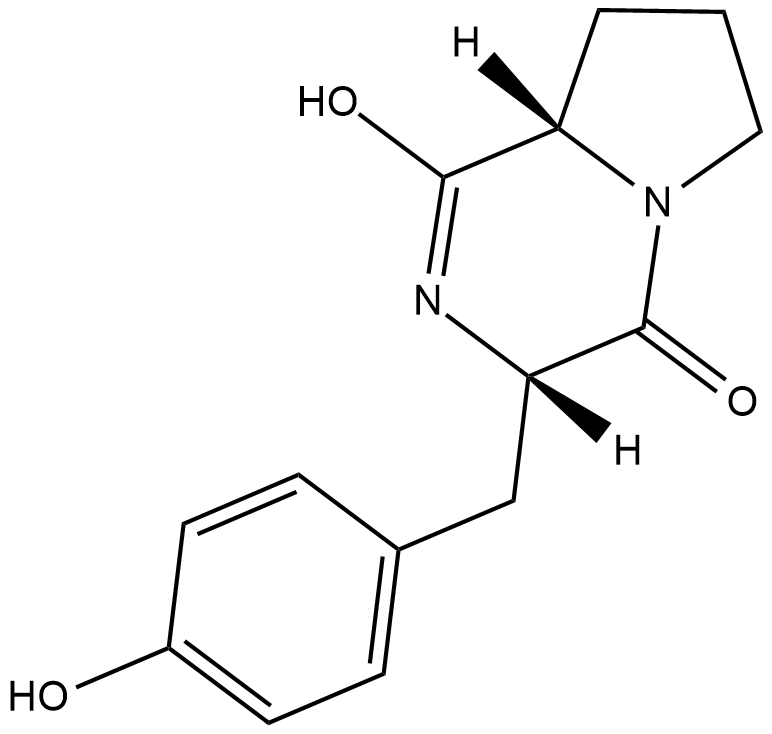
-
GC49208
Maduramicin (ammonium salt)
Antibiotic X-14868A, CL 259,971, Maduramycin, Prinicin
La maduramicine (sel d'ammonium) (Maduramycin ammonium) est isolée de l'actinomycète Actinomadura rubra.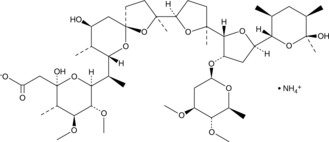
-
GC46031
Malabaricone B
NSC 287967, NSC 630196
Malabaricone B, un phénolique végétal naturel, est un inhibiteur de l'α-glucosidase actif par voie orale avec une IC50 de 63,7 μM.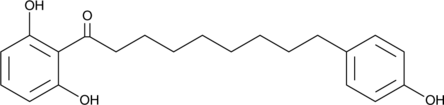
-
GC46028
Malabaricone C
NSC 287968
Malabaricone C est un inhibiteur naturel de la sphingomyéline synthase (SMS) avec des IC50 de 3 et 1,5 μM pour SMS 1 et 2, respectivement .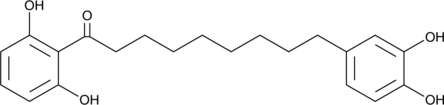
-
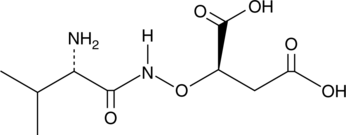
GC41566
Malioxamycin
Malioxamycin is an antibiotic produced by S.
-
GC44126
Manzamine A
Keramamine A
Manzamine A is a β-carboline alkaloid with diverse activities that has been found in marine sponges, including X.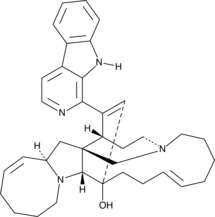
-
GC52434
Marbofloxacin-d8 (hydrochloride)
An internal standard for the quantification of marbofloxacin
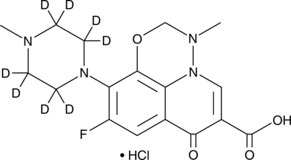
-
GC45505
Martinomycin
LL-D 37187α
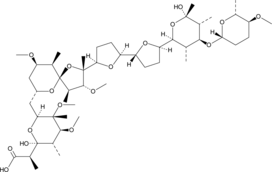
-
GC44138
MCTR1
13-glutathionyl-14-hydroxy Docosahexaenoic Acid, Maresin Conjugates in Tissue Regeneration 1, Maresin Sulfido Conjugate 1
Maresin conjugates in tissue regeneration 1 (MCTR1) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid in macrophages at the site of inflammation.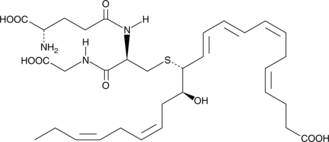
-
GC44139
MCTR2
13-cysteinylglycinyl-14-hydroxy Docosahexaenoic Acid, Maresin Conjugates in Tissue Regeneration 2, Maresin Sulfido Conjugate 2
Maresin conjugates in tissue regeneration 2 (MCTR2) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid in macrophages at the site of inflammation.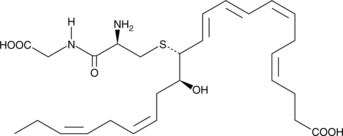
-
GC44140
MCTR3
13-cysteinyl14-hydroxyDocosahexaenoic Acid, Maresin Conjugates in Tissue Regeneration 3, Maresin Sulfido Conjugate 3
Maresin conjugates in tissue regeneration 3 (MCTR3) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid in macrophages.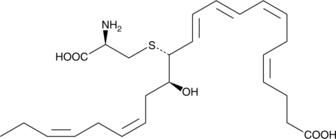
-
GC44150
Meclocycline
GS-2989, NSC 78502
Meclocycline is a tetracycline antibiotic.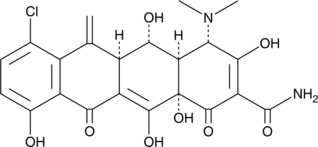
-
GC45755
Meropenem-d6
An internal standard for the quantification of meropenem
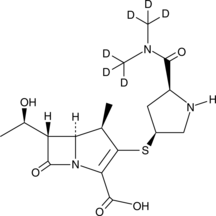
-
GC52088
Methylchloroisothiazolinone (hydrochloride)
CMIT, MCI, MCIT
A biocide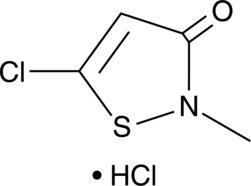
-
GC48857
Methylchloroisothiazolinone/Methylisothiazolinone Mixture
CMIT/MIT, MCI/MI, MCIT/MIT
A mixture of isothiazolinone biocides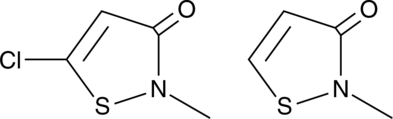
-
GC47670
Metronidazole-d4
An internal standard for the quantification of metronidazole
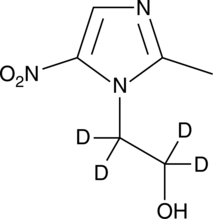
-
GC47671
Mezlocillin (sodium salt)
A β-lactam antibiotic
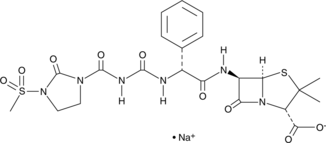
-
GC52165
Minosaminomycin
MSM
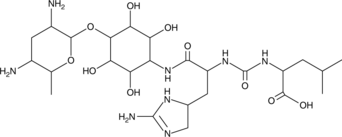
-
GC44241
Moenomycin Complex
Bambermycin, Flavomycin, Flavophospholipol, Gainpro
Moenomycin complex is a mixture of moenomycins A, A12, C1, C3 and C4, which are antibiotics isolated from several strains of Streptomyces that directly inhibit bacterial peptidoglycan glycosyltransferases.
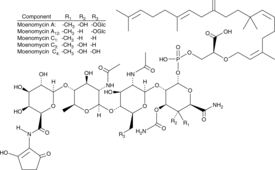
-
GC47700
Monocerin
(+)-Monocerin
La monocérine est un dérivé de l'isocoumarine.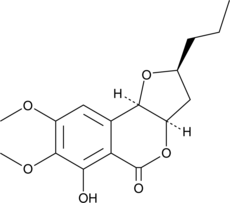
-
GC46174
Monomethylsulochrin
1,2-Secotrypacidin, Methylsulochrin
La monométhylsulochrine est un puissant métabolite antibactérien du champignon endophyte Aspergillus fumigatus, isolé des feuilles d'Albizia lucidior (fabaceae).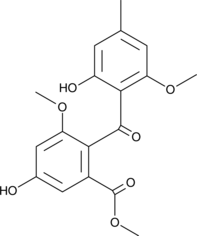
-
GC44246
Moxifloxacin N-Sulfate (sodium salt)
Moxifloxacin N-sulfate is a metabolite of moxifloxacin.
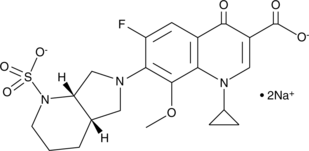
-
GC47711
Myceliothermophin E
A fungal metabolite with anticancer and antimicrobial activities
-
GC47716
Myxochelin A
(S)-Myxochelin A
A microbial metabolite with diverse biological activities
-
GC47728
N,O-Diacetyltyramine
4-(2-acetamidoethyl) Phenyl Acetate
A fungal metabolite with diverse biological activities
-
GC44266
N-(3-hydroxy-7-cis tetradecenoyl)-L-Homoserine lactone
N-(3-hydroxy-7-cis tetradecenoyl)-L-HSL
N-(3-hydroxy-7-cis tetradecenoyl)-L-Homoserine lactone is a long-chain N-acyl-homoserine lactone (AHL) produced by some Gram-negative bacteria and is involved in quorum sensing.
-
GC49187
N-3-hydroxydecanoyl-DL-Homoserine lactone
3OH-C10-DL-HSL, 3OH-C10-HSL, OH-C10-HSL
A bacterial quorum-sensing signaling molecule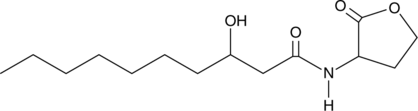
-
GC41575
N-3-hydroxyoctanoyl-L-Homoserine lactone
OHC8HSL
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.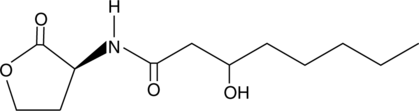
-
GC45939
N-3-oxo-decanoyl-L-Homoserine lactone
N-3-oxo-decanoyl-L-HSL
A bacterial quorum-sensing signaling molecule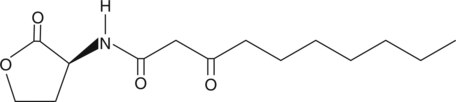
-
GC44286
N-3-oxo-hexadec-11(Z)-enoyl-L-Homoserine lactone
3oxoC16:1Δ11cis(L)HSL
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.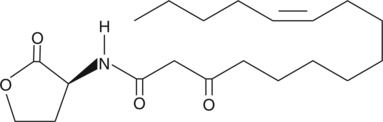
-
GC44287
N-3-oxo-hexadecanoyl-L-Homoserine lactone
3oxo-C16-HSL, N3oxopalmitoylLHomoserine lactone
La N-3-oxo-hexadécanoyl-L-homoserine lactone est une molécule de signalisation pour coordonner les comportements de groupe À haute densité chez de nombreuses bactéries.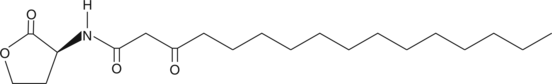
-
GC14832
N-3-oxo-octanoyl-L-Homoserine lactone
3-oxo-C8-HSL,N-β-oxo-octanoyl-L-Homoserine lactone,N-(3-Oxooctanoyl)-L-homoserine; N-(3-OXOOCTANOYL)-L-HOMOSERINE LACTONE
La N-3-oxo-octanoyl-L-homosérine lactone, un signal de détection de quorum, est un autoinducteur d'Agrobacterium.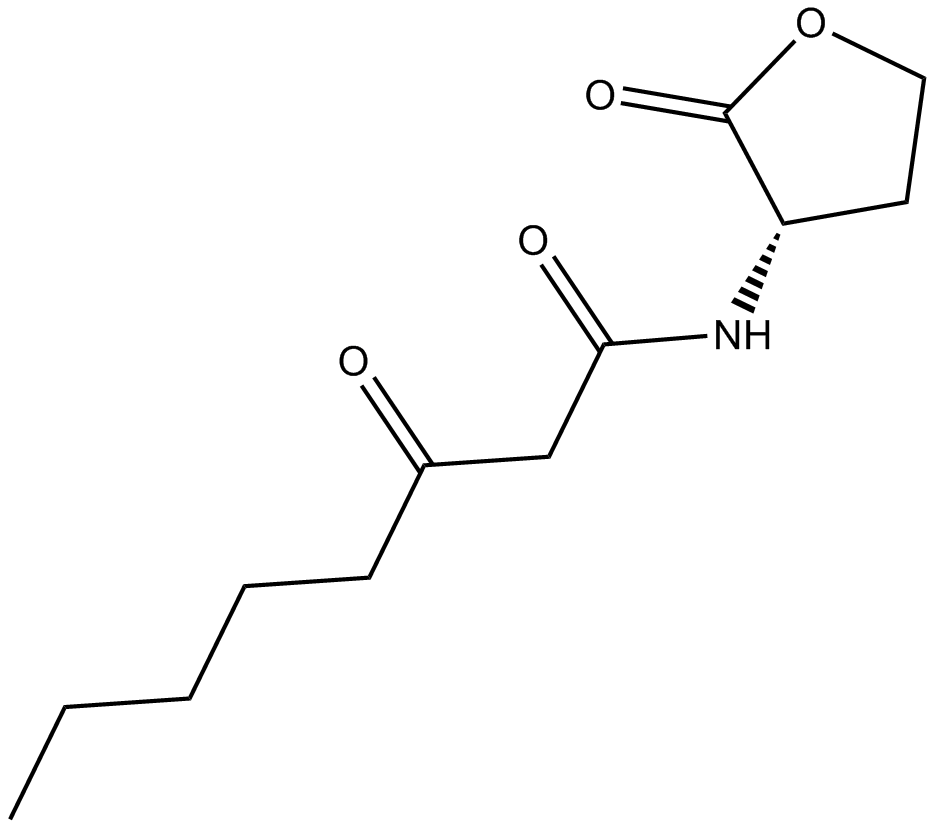
-
GC47732
N-3-oxo-pentanoyl-L-Homoserine lactone
3-oxo-C5-HSL
A bacterial quorum-sensing signaling molecule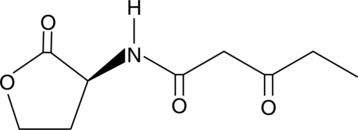
-
GC40925
N-3-oxo-tetradec-7(Z)-enoyl-L-Homoserine lactone
3oxoC14:1Δ7cis(L)HSL, N-3-oxo-tetradec-7(Z)-enoyl-L-HSL
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.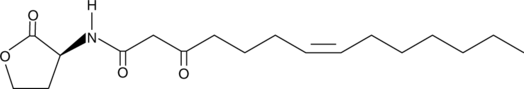
-
GC44288
N-3-oxo-tetradecanoyl-L-Homoserine lactone
3-oxo-C14-HSL; N-3-oxo-myristoyl-L-Homoserine lactone
La N-3-oxo-tétradécanoyl-L-homosérine lactone (oxo-C14-HSL) est un inducteur rhizobactérien et peut améliorer la défense de base contre les nématodes.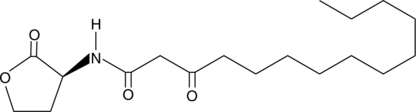
-
GC44294
N-acetyl Dapsone
NSC 27184
N-acetyl Dapsone is a metabolite of dapsone, an anti-inflammatory and antibacterial compound that is widely used in the treatment of leprosy, malaria, acne, and various immune disorders.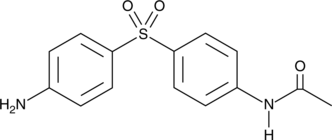
-
GC44334
N-butyryl-L-Homocysteine thiolactone
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.
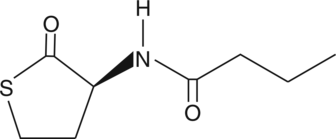
-
GC44336
N-cis-hexadec-9Z-enoyl-L-Homoserine lactone
N-(2-oxotetrahydrofuran-3S-yl) Palmitoleyl Amide
Quorum sensing is a regulatory process used by bacteria for controlling gene expression in response to increasing cell density.
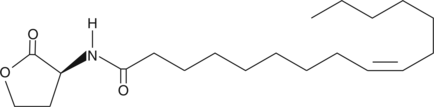
-
GC41081
N-cis-octadec-9Z-enoyl-L-Homoserine lactone
C18:1Δ9 cis(L)HSL, N(2oxotetrahydrofuran3Syl) Oleyl Amide
Quorum sensing is a regulatory process used by bacteria for controlling gene expression in response to increasing cell density.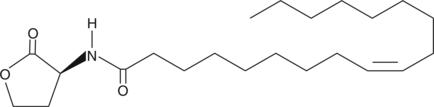
-
GC44337
N-cis-tetradec-9Z-enoyl-L-Homoserine lactone
C14:1Δ9cis(L)HSL, N(2oxotetrahydrofuran3Syl) Myristoleyl Amide
Quorum sensing is a regulatory process used by bacteria for controlling gene expression in response to increasing cell density.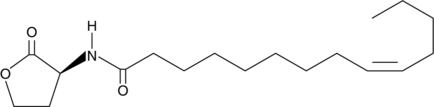
-
GC40852
N-Demethylerythromycin A
N-Demethylerythromycin A is a metabolite of erythromycin.
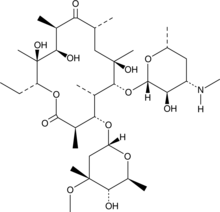
-
GC49162
N-Demethylvancomycin (hydrochloride)
A51568A, Norvancomycin
A glycopeptide antibiotic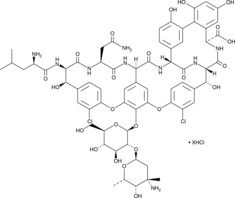
-
GC52073
N-formyl-2-Aminophenol
2’-Hydroxyformanilide, NSC 93900
A bacterial secondary metabolite with antioxidant activity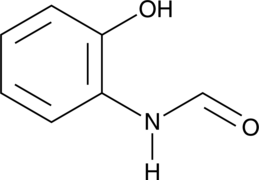
-
GC18770
N-hexadecanoyl-L-Homoserine lactone
C16-HSL N-palmitoyl-L-Homoserine
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density
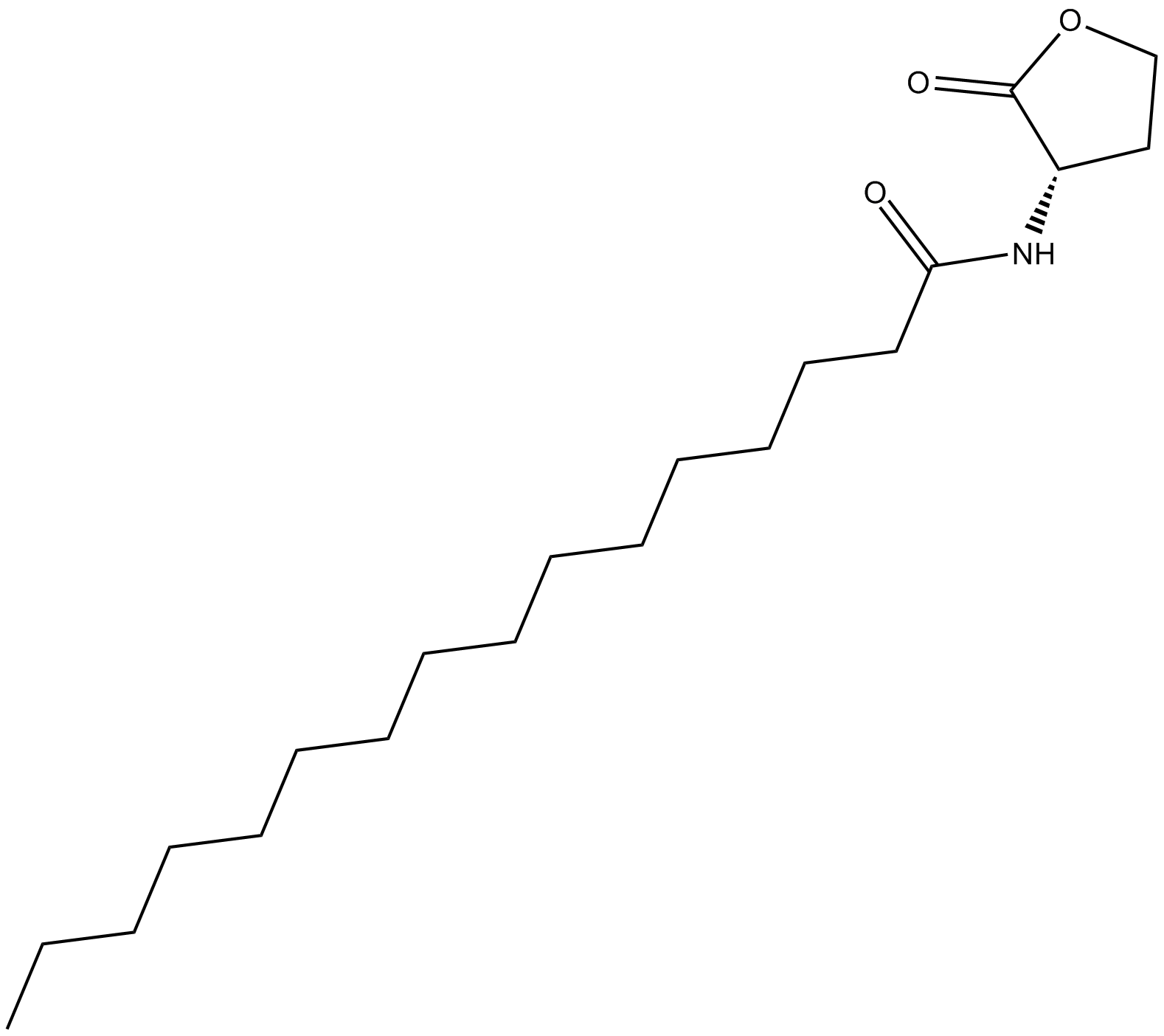
-
GC47772
N-hexanoyl-DL-Homoserine lactone
C6-HSL, N-hexanoyl Homoserine lactone
La N-hexanoyl-DL-homosérine lactone est une molécule de détection de quorum bactérienne produite dans la rhizosphère.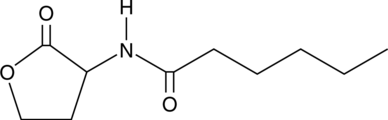
-
GC52007
N-hydroxylamine Dapsone
Dapsone hydroxylamine, DDS-NHOH
An active metabolite of dapsone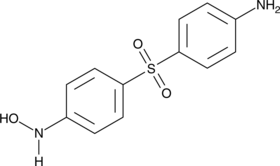
-
GC49394
N-Nitroso Fluoxetine
A derivative of fluoxetine
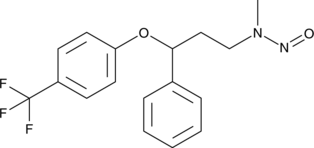
-
GC41082
N-octadecanoyl-L-Homoserine lactone
C18-HSL, N-octadecanoyl-L-HSL
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.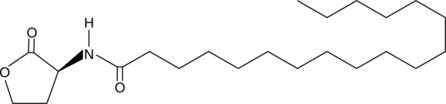
-
GC44459
N-pentadecanoyl-L-Homoserine lactone
C15HSL
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.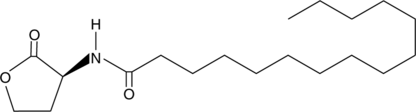
-
GC40615
N-phenylacetyl-L-Homoserine lactone
C8HSL, N-phenylacetyl-L-HSL
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.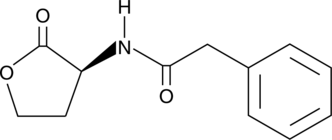
-
GC47804
N-Phenylpiperidin-4-amine
4-Anilinopiperidine, NSC 80678
A building block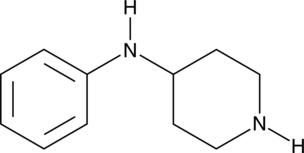
-
GC47805
N-Phenylthiourea
1-Phenyl-2-thiourea, NSC 5779, Phenylthiocarbamide, Phenylthiourea, PTC, PTU
A tyrosinase inhibitor and building block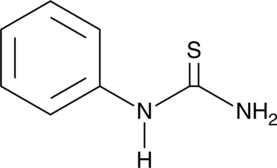
-
GC44472
N-tridecanoyl-L-Homoserine lactone
C13HSL
Quorum sensing is a regulatory system used by bacteria for controlling gene expression in response to increasing cell density.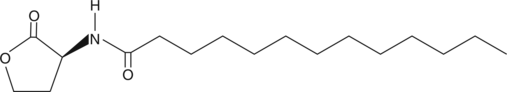
-
GC48516
Nafcillin-d5 (sodium salt)
An internal standard for the quantification of nafcillin
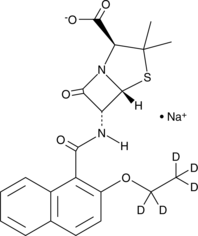
-
GC49144
NAG-thiazoline
GlcNAc-thiazoline
An OGA inhibitor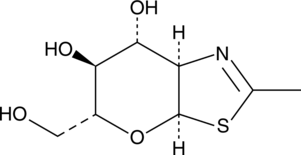
-
GC48619
Nanangenine B
A drimane sesquiterpene
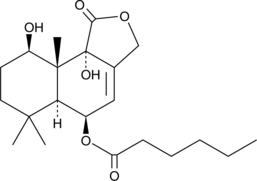
-
GC48667
Nanangenine C
A drimane sesquiterpene
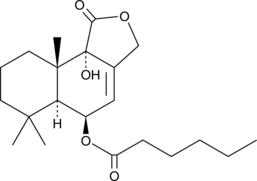
-
GC48660
Nanangenine F
A drimane sesquiterpene
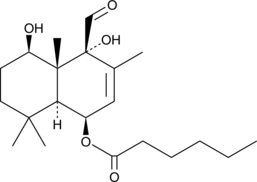
-
GC47747
Napyradiomycin A1
La napyradiomycine A1 est un composé énantiosélectif des napyradiomycines.
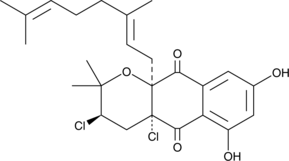
-
GC40081
Nargenicin
Antibiotic 47444
Nargenicin is a macrolide antibiotic that selectively inhibits the growth of S.
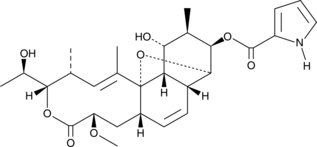
-
GC18489
Neamine (hydrochloride)
Neomycin A
La néamine (chlorhydrate), un produit de dégradation de la néomycine, est un antibiotique aminoglycoside À large spectre.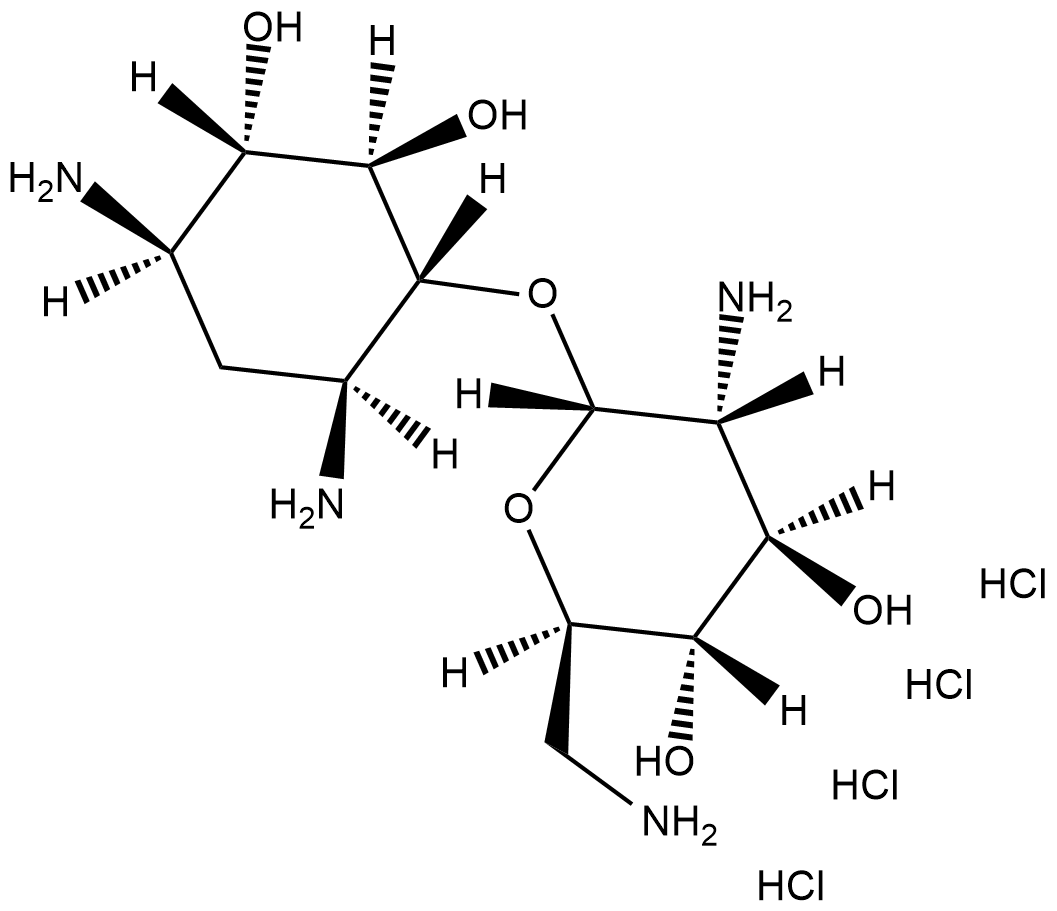
-
GC47760
Nebularine
Desaminoadenosine, NSC 65423, Purine riboside, Purinosine, Ribosylpurine
A purine ribonucleoside with diverse biological activities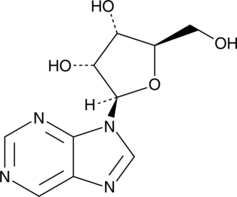
-
GC40970
Neohydroxyaspergillic Acid
NSC 613947
L'acide néohydroxyaspergillique, un antibiotique, est actif contre certains bactériophages.
-
GC49151
Neospiramycin I
A macrolide antibiotic
-
GC48839
Nifuroxazide-d4
An internal standard for the quantification of nifuroxazide
-
GC14823
Nitrocefin
Nitrocephin
La nitrocéfine est un substrat chromogénique de la β-lactamase qui subit un changement de couleur distinctif du jaune au rouge lorsque la liaison amide dans le cycle β-lactame est hydrolysée par la β-lactamase.
-
GC48896
Nitrofurantoin-13C3
An internal standard for the quantification of nitrofurantoin
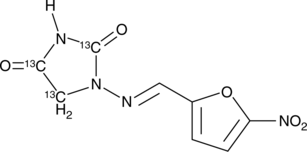
-
GC47787
Nocardamine
Desferrioxamine E, Proferrioxamine E
A ferrioxamine siderophore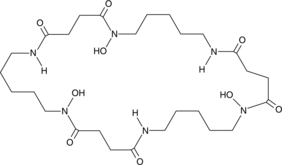
-
GC44437
Nodusmicin
Nodusmicin is a macrolide antibiotic.