Small molecule inhibition of METTL3 as a strategy for the treatment of myeloid leukemia
Summary:
n6-methyladenosine (m6A) is an internal RNA-modifying enzyme primarily catalyzed by the METTL3-METTL14 methyltransferase complex. The m6A methyltransferase METTL3 has been implicated in the initiation and maintenance of acute myeloid leukemia (AML), but the potential application of targeting this enzyme remains unclear. Here, we present the identification and characterization of STM2457 and the crystal structure of STM2457 in complex with METTL3-METTL14. STM2457 is a potent and selective METTL3 catalytic inhibitor, and tumor treatment with STM2457 resulted in decreased AML growth and increased differentiation and apoptosis. Concomitant with selective reduction of m6A levels of leukemic mRNAs, expression consistent with translational defects was reduced. The investigators demonstrated that METTL3 inhibition in vivo resulted in impaired engraftment and prolonged survival in various mouse models of AMTL, particularly targeting key stem cell subsets of AML. Collectively, these results reveal METTL3 inhibition as a potential therapeutic strategy against AML, and targeting of RNA-modifying enzymes is a promising anticancer therapeutic avenue.
Research Content:
Divided into the following sections: STM2457 characterization + STM2457 cellular and molecular effects + STM2457 efficacy in vivo
1.Characterization of RNA Methyltransferase Inhibitor STM2457
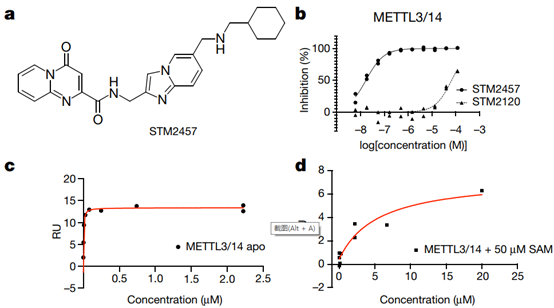
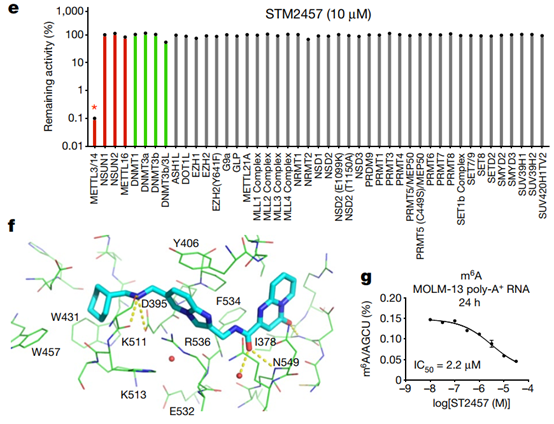
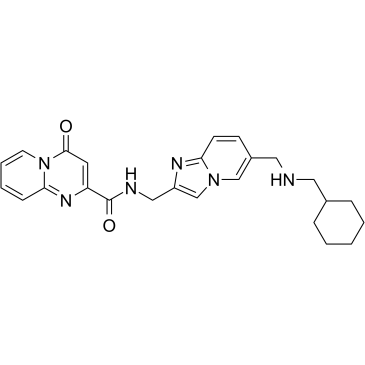
Fig. 1 | Characterization of the RNA methyltransferase inhibitor STM2457. a, Chemical structure of STM2457. b, Biochemical activity assay showing inhibition of the METTL3–METTL14 (METTL3/14) enzyme complex using a dose-range of STM2457 and STM2120. c, SPR assay showing binding affinity of STM2457 to the METTL3–METTL14 protein complex. RU, relative units. d, SPR assay showing binding affinity to the METTL3–METTL14 protein complex is reduced in the presence of SAM, indicative of SAM-competitive binding. e, STM2457 selectively inhibits METTL3–METTL14 in a methyltransferase profiling panel of 45 RNA (red bars), DNA (green bars) and protein methyltransferases (grey bars) (asterisk indicates less than 50% activity remaining) (n = 2). f, Crystal structure of METTL3–METTL14 (carbon atoms in green) in complex with STM2457 (carbon atoms in cyan). Hydrogen bonds (yellow dashed lines) and water molecules proximal to the inhibitor (red sphere) are shown (Protein Data Bank (PDB) ID 7O2I). g, Quantification of m6A levels on poly-A+-enriched RNA (expressed as m6A/AGCU ratio) after 24 h of treatment of MOLM-13 cells with the indicated STM2457 concentrations. Data are mean ± s.d., n = 3
To investigate the therapeutic potential of targeting the enzymatic activity of METTL3 as an anti-leukemia strategy, the researchers developed the small molecule STM2457. The researchers performed high-throughput screening of 250,000 different compounds. STM1760 (half inhibitory concentration = 51.7 μM) was one of only two non-s-adenosylmethionine (SAM) related results produced by the screen (Extended Data Fig. 1a,b). After optimizing potency, in vitro absorption-distribution metabolism-excretion (ADME) studies, and in vivo pharmacokinetic properties, we identified STM2457 (Figure 1a). The researchers also identified a structurally related molecule, STM2120 (half inhibitory concentration = 64.5 μM), which was 1000-fold less active against METTL3-METTL14 than STM2457. (Fig. 1b)
STM2457 is a potent METTL3-METTL14 catalytic inhibitor with a half inhibitory concentration of 16.9 nM (Fig. 1b). Direct binding to the METTL3-METTL14 heterodimer was confirmed by surface plasmon resonance (SPR) (Fig. 1c,d, Extended Data Fig. 1c–e). A cofactor-competitive binding mode was demonstrated using SAM in the SPR-run buffer (Fig. 1d). STM2457 is highly specific for METTL3 and has no inhibitory effect on other RNA methyltransferases (Extended Data Fig. 1f). Furthermore, STM2457 was more than 1000-fold selective for METTL3 when tested against 45 RNA, DNA and protein methyltransferases (Fig. 1e). The researchers further characterized the binding of STM2457 to METTL3 by X-ray crystallography, confirming the binding of STM2457 to the SAM binding site (Fig. 1f). The strong selectivity of STM2457 for SAM observed in the methyltransferase panel is structurally distinct from other known methyltransferase inhibitors, the avoidance of homocysteine binding used by SAM, the recombination of K513 on STM2457 binding, and Structural diversity of SAM-dependent methyltransferase cofactor binding sites.
2.Drug Inhibition of METTL3 Affects AML Cells
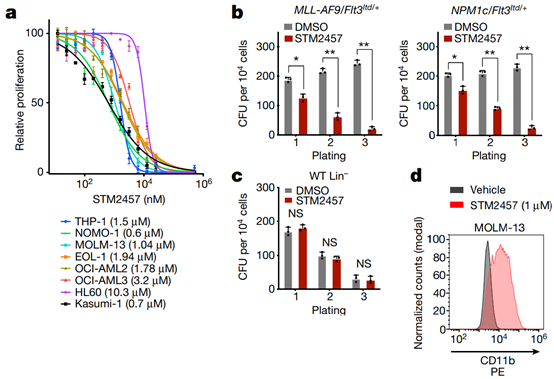
Fig. 2 | Pharmacological inhibition of METTL3 affects AML cells. a, STM2457 dose–response curves for a panel of AML cell lines. Data are mean ± s.d., n = 3. IC50 for each cell line is shown in brackets. b, Colony-forming efficiency of primary mouse MLL-AF9/Flt3Itd/+ and NPM1c/Flt3Itd/+ AML cells treated with vehicle or STM2457. Data are mean ± s.d., n = 3. CFU, colony-forming units. c, Colony-forming efficiency of wild-type (WT) Lin- cells treated with vehicle or STM2457. Data are mean ± s.d., n = 3. d, CD11b and MAC1 levels of MOLM-13 and MLL-AF9/Flt3Itd/+ primary mouse cells, respectively, treated with vehicle or STM2457. e, BrdU staining and cell cycle analysis in MOLM-13, HPC7 and MLL-AF9/Flt3Itd/+ primary mouse cells treated with vehicle or STM2457. f, Percentage of apoptotic cells in a panel of human AML cell lines following treatment with STM2457 at the indicated time points. Data are mean ± s.d., n = 3. g, Western blot analysis of BRD4, MYC, SP1 and GAPDH in MOLM-13 cells treated with the indicated doses of STM2457 or vehicle (n = 3). Two-tailed Student’s t-test; *P < 0.01, **P < 0.0001; NS, not significant. CFU, colony-forming units.
STM had an equivalent inhibitory effect on human and mouse METTL3 by 2457 (Extended Data Fig. 2b). The difference between the half inhibitory concentration of STM2457 and the cellular half inhibitory concentration is consistent with the competition of the Michalis constant (Km) (0.1 μM) of SAM9 of METTL3-METTL14 and the highly abundant intracellular SAM10. We further demonstrated on-target inhibition of STM2457 in cells by measuring a concentration-dependent decrease in poly-A+-enriched RNA (Fig. 1g). No changes were detected for other RNA modifications (Extended Data Fig. 2d). Pharmacokinetic analysis of STM2457 in mice following a single intraperitoneal dose of 50 mg/kg showed that the half-life of STM2457 was sufficient to ensure appropriate exposure levels within 24 hours in vivo (Extended Data Fig. 2e). Dose-dependent inhibition of poly-A+-enriched RNA in mouse spleen by m6A demonstrated a clear relationship between compound exposure and target inhibition in vivo (Extended Data Fig. 2f). These data demonstrate that STM2457 is a potent, specific and bioavailable METTL3 inhibitor suitable for in vivo studies.
3. STM2457 Reduces m6A Levels, Resulting in Defective mRNA Translation
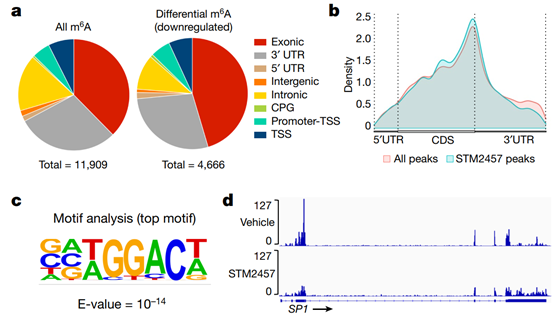
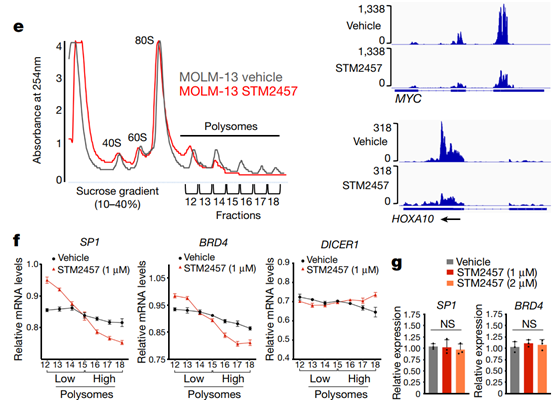
Fig. 3 | STM2457 reduces m6A levels and causes mRNA translation defects. a, Genomic distribution of all called m6A peaks (left) and downregulated m6A peaks (right) on poly-A+-enriched RNAs from STM2457-treated MOLM-13 cells. b, The distribution of all (red) or downregulated (light blue) m6A peaks of MOLM-13 cells upon treatment with STM2457. c, Motif analysis of the sequences under depleted peaks following treatment with STM2457 (hypergeometric test). d, Genomic visualization of the normalized m6A-meRIP signal for SP1, MYC and HOXA10 in MOLM-13 cells following treatment with vehicle or STM2457. e, Polysome fractionation analysis in MOLM-13 cells treated with vehicle or STM2457. Absorbance was continuously measured at 254 nm. f, Quantification by qPCR with reverse transcription (RT–qPCR) of SP1, BRD4 and DICER1 mRNAs in each polysome fraction (high and low molecular mass), presented as a percentage of total mRNA. Data are mean ± s.d., n = 3. g, RT–qPCR quantification of SP1 and BRD4 in total RNA samples isolated from MOLM-13 cells treated with vehicle or STM2457. Data are mean ± s.d., n = 3. Two-tailed Student’s t-test; *P < 0.05. UTR, untranslated region; CPG, CPG island; TSS, transcription start site; CDS, coding sequence.
To investigate its anti-leukemic potential, we examined the proliferation of a panel of human AML cell lines treated with STM2457 and detected a significant reduction in growth in a concentration-dependent manner (Fig. 2a), whereas STM2457 did not affect normal human cord blood CD34+ Colony-forming capacity of cells (Extended Data Fig. 3a). We also did not observe an effect of the control small molecule STM2120 on the proliferation of MOLM-13 cells, in contrast to our observations with STM2457 cells (Extended Data Fig. 3b). Furthermore, STM2457 treatment significantly reduced the clonal potential of primary mouse AML cells (Fig. 2b). But there is no effect on normal hematopoietic stem and progenitor cells. (Fig. 2c) Pharmacological inhibition of METTL3 also resulted in significant myeloid differentiation and cell cycle arrest in MOLM-13 and primary mouse AML cells (Fig. 2d,e). In contrast, we did not observe a similar effect to the non-leukemic hematopoietic cell line HPC7 (Figures 3e-3d). Furthermore, STM2457 treatment induced apoptosis in human and mouse AML models, but not in normal non-leukemic hematopoietic cells (Fig. 2f). To assess the effect of pharmacological inhibition of METTL3 on SP16, 12 and BRD4, two A known substrate for the METTL3 enzyme, the researchers treated MOLM-13 cells with STM2457 and observed a dose-dependent reduction in SP1 and BRD4 protein levels (Figure 2g). Notably, ectopic expression of SP1 significantly reduced the sensitivity of MOLM-13 cells to STM2457 (Extended Data Fig. 3f,g).
4.STM2457 Prevents the Expansion of AML and Reduces the Number of Key Leukemia Stem Cells in Vivo
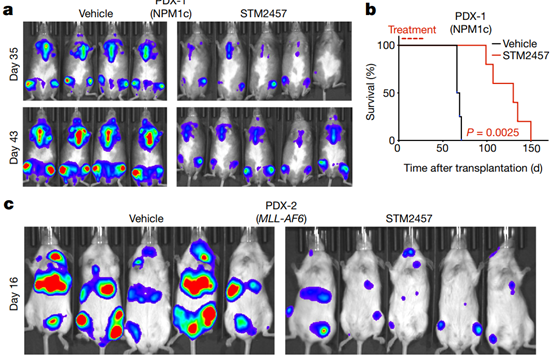
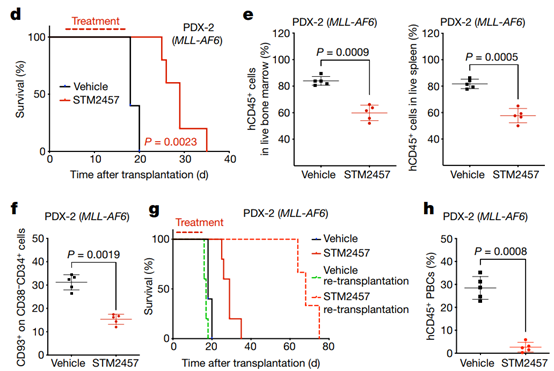
Fig. 4| STM2457 prevents AML expansion and reduces the number of key leukaemia stemcells in vivo. a, Bioluminescence imaging of AML PDX-1mice (NPM1c cells) treated with vehicle or 50mg kg-1 STM2457 (n = 5). b, Kaplan– Meier survival curves of AMLPDX-1 mice after treatment with vehicle or 50 mg kg-1 STM2457 at indicatedtimes (n = 5). c, Bioluminescence imaging ofAML PDX-2 mice (MLL-AF6 cells) treated with vehicle or 50mg kg-1 STM2457 (n = 5). d, Kaplan–Meier survival curves ofAML PDX-2 mice after treatment with vehicleor 50 mg kg-1 STM2457 at indicated times (n = 5). e, Percentage of cellsexpressing human CD45 (hCD45) inbone marrow and spleen of PDX-2 mice aftertreatment with vehicle or 50 mg kg-1 STM2457 (n = 5). f, Percentage of CD93+ cells among CD38-CD34+ cellsin bone marrow after treatment with vehicle or 50 mg kg-1 STM2457 (n = 5). g, Kaplan–Meier survival curves ofAML PDX-2 miceafter re-transplantation of MLL-AF6 cells. Mice were treated withvehicle or 50 mg kg-1 STM2457 atindicated times (n = 5). Treatment (in red) andsolid lines in red or black referto the primary transplantation shown in d. h, Percentage of cells expressinghuman CD45 in peripheral blood after re-transplantationof MLL-AF6 cells from AML PDX-2 mice (n = 5) as described in g. PBCs, peripheral blood cells.Two-tailed Student’s t-test; log-rank (Mantel–Cox) test was used forsurvival comparisons.
The data suggest that the catalytic function of METTL3 is important for leukemia growth, which is consistent with previous findings. Next, the researchers attempted to study the molecular mechanism by which STM2457 affects AML. RNA sequencing analysis of STM2457-treated MOLM-13 cells revealed that 1338 genes were up-regulated and 489 genes were down-regulated (Extended Data Fig. 4a). Gene Ontology (GO) analysis of differentially expressed genes revealed enrichment in pathways related to myeloid differentiation, cell cycle and leukemia progression (Extended Data Fig. 4b,c), closely matching our phenotypic observations Consistent. To examine the effect of METTL3 inhibition on m6A levels, we performed m6A-specific methylated RNA immunoprecipitation (m6A-meRIP-seq) in STM2457-treated MOLM-13 cells. 11909m6A peaks were found on poly-A+-enriched RNAs, of which 4666 were reduced after STM2457 treatment, indicating that they are dependent on METTL3 catalytic activity. We observed no major changes between the general m6A distribution and that after STM2457 treatment (Figures 3a-3b). After STM2457 treatment, through motif analysis, m6a-related DRACH motif 16 was identified as the highest candidate substrate sequence (Fig. 3c), verifying the specificity of STM2457 treatment.P-seq results with the meRIP-seq dataset from MOLM-13 cells in which METTL3 expression had been knocked out. We found substantial overlap in differentially m6a-methylated polya+RNAs as well as differential m6a peaks (Extended Data Fig. 5a-5b), including many known and previously unknown mettl3-specific m6a substrates (Fig. 3d). ). The observed differences in substrates and peaks may be due to more specific catalytic inhibition of METTL3 by METT457 compared to STMETTL3 knockdown, which may disrupt the entire m6A methyltransferase complex. m6A-meRIP and quantitative PCR (qPCR) validation revealed that STM2457 treatment resulted in decreased m6A levels in MeTTL3-dependent core leukemogenesis m6A substrates, including HOXA1018 and MYC19, but not in meTTL3-independent m6A substrates Differences were observed (Extended Data Fig. 5d), thus validating the specificity of our m6A profiles. GO analysis of differential m6A-meRIP candidate genes revealed chromatin mechanisms enriched for pathways involved in chromatin modification, DNA damage, and RNA splicing (Extended Data Fig. 5e), and the connection of m6A to nuclear biological processes. Previous studies have shown that genetic inhibition of METTL3 and METTL14 results in defective mRNA translation and ribosomal arrest. We therefore investigated whether isolated catalytic inhibition of METTL3 could lead to similar translational defects. Multimeric analysis showed that the high multimeric fraction was lost after treatment of MOLM-13 cells with STM2457 (Fig. 3e). Validation of the multimeric fraction using qPCR revealed a significant decrease in known mettl3-dependent substrates in the high molecular weight fraction and a significant increase in the low molecular weight fraction, indicating ribosome stalling (Fig. 3f). In contrast, DICER1, a mettl3-independent m6a substrate, was not affected by STM2457 treatment (Fig. 3f). Crucially, there was no change in overall RNA expression levels of the METTL3 biomarkers BRD4 and SP1. The protein levels were greatly reduced. Verify that the effect on mRNA is at the translational level, not at the transcriptional level. Furthermore, STM2457 treatment maintained the expression levels of METTL3 and DICER1 (Extended Data Fig. 5f) as well as the protein levels of METTL3, METTL14, DDX3X and DICER1 (Extended Data Fig. 5g), further suggesting that its effect on mRNA translation is not global, but rather a specific. Taken together, this mechanistic analysis suggests that catalytic inhibition of METTL3 results in defective gene expression, consistent with effects on mRNA translation efficiency of m6A substrates.
After finding positive evidence for a strong pharmacological inhibitory effect of METTL3 in vitro, we conducted in vivo studies using a clinically relevant AML model. Initially, the researchers used three different genotypes of human AML patient-derived xenografts (PDX). Daily treatment with STM2457 resulted in impaired engraftment and AML expansion in vivo and significantly prolonged lifespan in mice (Fig. 4a–d). There was no apparent toxicity or effect on mouse body weight (Extended Data Fig. 6f). The reduction of CD45+ cells in human bone marrow and spleen after treatment also confirmed the anti-leukemia effect (Fig. 4e). STM2457 potently inhibits METTL3 targets in vivo by selectively reducing key m6A substrates at the protein level, while METTL3 levels remain unchanged (Extended Data Fig. 6g). Furthermore, total m6A levels on poly-A+ enriched RNAs were significantly reduced after treatment with STM2457 (Extended Data Fig. 6h). In contrast to the PDX model, the investigators used a primary mouse MLL-AF9/Flt3Itd/+ in vivo model, with similar anti-leukemia observations with reduced engrafted AML cells, reduced spleen size, selective reduction of METTL3 biomarkers and reduced poly-A+ RNA was enriched (Extended Data Fig. 7a–d).
Having established the in vivo anti-leukemia effect of STM2457, we next investigated the effect of pharmacological inhibition of METTL3 on important leukemia stem cell subsets. CD93+ and L-GMP subsets are directly associated with the generation and maintenance of MLL rearrangement-driven AMLs in primary mouse and human patients. Notably, we observed that the CD93+ and L-GMP populations were significantly reduced after treatment with STM2457, suggesting a strong dependence on the catalytic function of METTL3 (Fig. 4f). In addition, the intensity of CD48+ increased after STM2457 treatment (Extended Data Fig. 7g), indicating a loss of self-renewal at the level of leukemia stem cells, which is consistent with our in vitro and in vitro observations. To demonstrate that METTL3 inhibition leads to functional impairment of leukemia stem cells, we performed retransplantation experiments using mouse or patient-derived AML cells in primary transplanted cells treated with vehicle or STM2457. The investigators observed that only after initial treatment with STM2457, patients' lifespan was significantly increased in peripheral blood, while the presence of AML cells in peripheral blood was significantly reduced (Figures 4g-4h). Taken together, pharmacological inhibition of METTL3 in vivo impairs AML expansion by affecting AML stem cells or leukemia dissemination.
In Conclusion:
The investigators assessed the potential toxicity of STM2457 in vivo at established anti-leukemic doses. No significant changes were observed in the numbers of bone marrow-derived hematopoietic stem and early progenitor cells (Lin−SCA1+KIT+), peripheral blood counts, or body weight in mice (Extended Data Fig. 8a–e). We also confirmed potent catalytic inhibition of METTL3, as m6A levels on poly-A+-enriched RNAs were significantly reduced after STM2457 treatment (Extended Data Fig. 8f). These data suggest that small molecule inhibition of METTL3 is detrimental to the maintenance of AML but has no significant or durable effect on normal hematopoiesis. In this article, the investigators have described STM2457, a bioavailable inhibitor of m6A action METTL3. The researchers have demonstrated that STM2457 catalytically inhibits METTL3 targeting of key stem cell populations in AML, reversing the AML phenotype and preventing or slowing AML progression. Our findings thus provide a promising strategy for targeting leukemic stem cell subsets responsible for AML persistence and relapse, and provide a rationale for future studies investigating METTL3 in combination with mainstream anti-AML therapies. Overall, the researchers' efforts further underscore the impact of m6A "hijacking" on AML status. To the researchers' knowledge, this is the first demonstration of the in vivo activity and therapeutic effect of an RNA methyltransferase inhibitor on cancer.










Comments